Abstract
Peritoneal metastasis (PM) is most frequent in gastric cancer (GC) and cancer-associated fibroblasts (CAFs) play a critical role in this process. However, the concrete mechanism of crosstalk between CAFs and cancer cells in PM of GC remains unclear. Microarray sequencing of GC focus and PM lesions was performed, and biglycan (BGN) was screened for further study. Clinically, BGN expression was higher in GC tissues than adjacent normal tissues, and high expression correlated with poor prognosis. In vitro experiments demonstrated that BGN promoted tumor progression and the transformation of mesothelial cells (MCs) into cancer-associated fibroblasts like cells (CAFLCs). In turn, CAFLCs-derived fibroblast activation protein (FAP) facilitated the proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) of GC cells. GC-derived BGN combined with toll like receptor 2 (TLR2)/TLR4 on MCs to activate the NF-κB pathway and promote the transformation of MCs into CAFLCs by the recovery experiment, coimmunoprecipitation assay, nuclear and cytoplasmic protein extraction assay. CAFLCs-derived FAP could activate the JAK2/STAT3 signaling pathway in GC. Finally, activated STAT3 promoted BGN transcription in GC, resulting in a BGN/FAP-STAT3 positive feedback loop. Taken together, mutual interaction between tumor cells and activated MCs mediated by a BGN/FAP-STAT3 positive feedback loop facilitates PM of GC and provides a potential biomarker and therapeutic target for GC metastasis.
Keywords: biglycan, cancer-associated fibroblasts, fibroblast activation protein, gastric cancer, peritoneal metastasis
Introduction
Gastric cancer (GC) is the fifth most frequently diagnosed cancer and the third leading cause of cancer-related death worldwide 1. In advanced GC cases, 14% of patients have peritoneal metastasis (PM) during primary surgery, and the prognosis of patients with GC PM is extremely poor with a median survival time less than six months 2. Thus, there is an urgent need to explore the mechanism by which GC PM occurs to identify a poor prognostic factor which could contribute to PM of GC.
The metastatic niche in PM is primarily composed of abdominopelvic cavity surface, which is lined with a single layer of mesothelial cells (MCs). An intact peritoneum acts as the first line of defense against bacterial and tumor invasion 3. The interaction between tumor cells and human peritoneal MCs (HPMCs) promotes peritoneum secondary tumor implantation 4. Growing evidence displays that HPMCs are converted into CAFs through the mesothelial-to-mesenchymal transition (MMT), damaging the intact peritoneum and allowing tumor cells to colonize the peritoneum 5-8. CAF markers, which mainly consist of fibroblast activation protein (FAP), α-smooth muscle actin (α-SMA), and Vimentin, promote tumor progression 9-11. Studies also show that tumor cells exhibit more malignant behavior when co-cultured with CAFs 12-14.
Microarray sequencing is used to analyze differential expression of mRNAs between PM lesion and GC primary foci. Remarkably, according to microarray sequencing between PM foci and primary lesion, biglycan (BGN) ranked the fourth highest depending on fold change among 71 differential expression genes to screen for further studies. BGN, which is an extracellular protein (ECM) 15, is highly expressed in many human cancers, and predicts poor prognosis 16. In microarray and bioinformatic analysis data, BGN is associated with the epithelial-mesenchymal transition (EMT) process by integrating the TGFβ/Snail and TNFα/NF-κB pathway in the tumor microenvironment (TME) 17. In advanced bladder cancer, the expression of BGN correlates with the EMT process 18. EMT increases the ability of tumor epithelial cells to migrate and invade, enabling primary tumors to spread and form secondary tumor metastases 19, 20.
In the TME, Toll-like receptor 2 (TLR2) and Toll-like receptor 4 (TLR4) are considered the natural receptors of BGN, promoting tubular formation in endothelial cells by activating NF-κB signaling pathway 21. In colon cancer, BGN combines with TLR4 to activate NF-κB 22. Bioinformatics analysis of co-expression genes in BGN networks shows that BGN interacts with 42 neighboring genes, including FAP 16. While FAP is rarely detected in normal human tissue or benign tumors, this gene is up-regulated in stroma fibroblasts from more than 90% of malignant epithelial tumors 11. Studies demonstrate that FAP is one of the biomarkers for CAFs 9-11. However, it remains unknown whether BGN can combine with TLR2/TLR4 to activate NF-κB and promote the transformation of HPMCs into cancer-associated fibroblasts like cells (CAFLCs). In addition, the underlying mechanism by which CAFLCs impacts the PM of GC requires further research.
In this study, BGN expression in GC tissues was higher than paired adjacent normal tissues (PANT), and high expression correlated with poor prognosis of GC patients. In vitro experiments demonstrated that BGN promoted the GC progression and the transformation of MCs into CAFLCs. In turn, CAFLCs-derived FAP facilitated the proliferation, migration, invasion, and EMT of GC cells. FAP also rescued the effect of BGN on tumor progression. GC-derived BGN was shown to combine with TLR2/TLR4 on MCs to activate NF-κB and promote the transformation of MCs into CAFLCs. Moreover, CAFLCs-derived FAP activated the JAK2/STAT3 pathway in GC. Finally, STAT3 combined with the BGN promoter to induce BGN transcription, resulting in a BGN/FAP-STAT3 positive feedback loop in GC. This study shows that mutual interaction between tumor cells and activated MCs mediated by a BGN/FAP-STAT3 positive feedback loop facilitates PM of GC, providing a poor prognostic factor which could contribute to peritoneal metastasis of gastric cancer.
Materials and Methods
Microarray sequencing of GC focus and PM focus
Three pairs of tissue samples containing GC focus and PM focus were frozen and crushed using Biopulverizer, and the tissue samples were homogenized using Mini-Bead-Beater-16. Then Quick Amp Labeling, One-Color (Agilent p/n 5190-0442), RNeasy Mini Kit (Qiagen p/n 74104), and NanoDrop ND-1000 were utilized for labeling reaction, RNA purifying, and labeled cDNA QC respectively. After hybridization and microarray cleaning, scanning was performed using Agilent Microarray Scanner (Agilent p/n G2565BA). Finally, Agilent feature extraction software was used to extract all data.
Patient tissue samples and follow-up
There were 65 sets of GC tissue and their PANT (more than 5 cm distance to cancer margin) samples in this study, all of which underwent radical surgery at Zhongnan Hospital of Wuhan University (Wuhan, China) from January 2016 to May 2018. Moreover, all of inclusion patients were diagnosed with primary GC by histopathology, and no patient received neo-adjuvant chemotherapy or local radiotherapy before radical operation. All of the severed tissue specimens were preserved in liquid nitrogen as soon as possible, and then stored at -80 °C. The last follow-up visit was June 10, 2021. TNM stages of GC were determined depending on the 8th edition of the AJCC Cancer Staging Manual. All of tissue samples were obtained with written informed from the patients, and the study was executed with the ethical approval for scientific research projects under medical ethics committee of Zhongnan Hospital of Wuhan University (Scientific Ethical Approval NO. 2019079).
Cell lines, cell culture, and reagents
The human gastric cancer cell lines AGS, BGC823, HGC27, MGC803, MKN45, SGC7901, the human gastric epithelial cell line GES1, HPMCs line HMrSV5 were gained from the Cell Bank of Wuhan University in Wuhan. In the incubator, all cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA) under a humidified atmosphere of 5% CO2 at 37 °C. To obtain CAFLCs, in the co-cultured model, the non-contact co-culture transwell system of six-well plate (Corning, USA) enabled the exchange of cell-secreted factors in the culture medium, while the penetration of cells themselves not allowed. 3×105 HMrSV5 cells were seeded in the upper chamber, and 3×105 cells GC cells or GES1 cells were seeded in the lower chamber of the co-cultured model in the DMEM containing 10% FBS for 12 hours (h) respectively. Then the cells were washed with phosphate-buffered saline (PBS, Gibco, USA) twice in the co-cultured model, and the culture medium was replaced with free-FBS DMEM until the morphological changes of HMrSV5.
Recombinant human FAP protein (rFAP) (ABclonal, China) was diluted with PBS and utilized at an ultimate concentration of 200 ng/ml. Stattic (STAT3 inhibitors), BAY 11-7082 (NF-κB inhibitors), C29 (TLR2 inhibitors), TAK242 (TLR4 inhibitors), PT100 (FAP inhibitors) was dissolved in Dimethyl sulfoxide (DMSO, Sinopharm, China), the above final concentrations were 10 μM, 5 μM, 50 μM, 50 μM, 100 μM separately. And our team purchased the above inhibitors from MedChemExpress, USA.
Immunohistochemistry (IHC) staining
The expression of BGN and FAP in gastric tissue specimens including primary lesion, and PM lesion were detected by IHC. FAP, and Ki67 expression were measured in vivo xenograft assay by IHC. Specimens were treated with antibodies against human BGN (1:100, Novus Biologicals, USA), FAP (1:200, Bioswamp, China), Ki67 (1:200, Cell Signaling Technology, USA). IHC staining was conducted on paraffin-embedded formalin-fixed samples according to the instructions recommended by the manufacturer.
Cell transfection
Lentiviral-mediated BGN overexpression and knockdown (Genechem, China) were constructed in GC cell lines. For knockdown of BGN, a total of three siRNAs and scrambled siRNA (JTS scientific, China) were transfected into GC cells with lipofectamine 2000 reagent (Invitrogen, USA) in Opti-MEM (Gibco, USA) for the sake of screening out the optimal knockdown effects. Then, according to the selected siRNA, lentivirus mediated knockdown of BGN vectors (anti-BGN) and their negative control lentivirus vectors (anti-NC-BGN) were constructed and transfected into GC cells. Moreover, lentivirus mediated overexpression of BGN vectors (Oe-BGN) and their negative control lentivirus vectors (Oe-NC-BGN) were also contracted and transfected into GC cells, the stable transfection cell lines expressed green fluorescent protein (GFP) and anti-puromycin. Following the manufacturer's protocol.
Two human short hairpin knockdown RNA of FAP (sh-FAP) plasmids and control plasmids (sh-control) were purchased from the Genechem, China.
Quantification of secreting protein through enzyme-linked immunosorbent assay (ELISA)
The BGN and FAP concentrations in different culture supernatants were estimated by ELISA kits (Bioswamp, China) following the manufacturer's instructions.
RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNA was isolated from GC cell lines, human tissue samples, primary GC and PM xenograft tumor cells by Trizol reagent (Invitrogen, USA) according to the instructions recommended by the manufacturer. The concentration of RNA was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific, USA). Then, 1μg of total RNA from every specimen was reverse transcribed into cDNA by the PrimeScriptTM RT reagent kit (Vazyme, China). The SYBR-Green PCR Master Mix (Vazyme, China) and the cDNA were utilized for subsequent qRT-PCR by the BioRad IQ5 Real time PCR machine (BioRad, USA) to analysis mRNA expression. And Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was considered as an internal control. The 2-ΔΔCt method was used to calculate relative RNA expression. The sequences of primers involved in the assay were designed in Table S1 (supplementary Table 1).
Western blot analysis
The total proteins in cells were washed with cold PBS twice and lysed in RIPA buffer containing protease inhibitor cocktail (Thermo Scientific, USA). Protein concentrations were quantified using the BCA Protein Assay Kit (Beyotime, China) after protein extraction. The whole proteins were separated by 10% SDS-PAGE and then electrotransferred onto PVDF membranes (Millipore, USA). After being blocked for 2h with 5% non-fat milk in tris-buffered saline including 0.1% tween-20 (TBST), the PVDF membranes were incubated with diluted primary antibodies in antibody dilatation buffer at 4 °C overnight. The following primary antibodies used were anti-BGN (1:1000, Novus Biologicals, USA), anti-TLR2 (1:1000, Proteintech, USA), anti-TLR4 (1:1000, Proteintech, USA), anti-FAP (1:1000, Bioswamp, China), anti-NF-κB (1:1000, Proteintech, USA), anti-p-NF-κB (1:1000, phosphor Ser536, Cell Signaling Technology, USA), anti-α-SMA (1:1000, Abcam, USA), anti-JAK2 (1:1000, Abcam, USA), anti-STAT3 (1:1000, Cell Signaling Technology, USA), anti-p-JAK2 (1:1000, phosphor Y1007+Y1008, Abcam, USA), anti-p-STAT3 (1:1000, phosphor Tyr705, Cell Signaling Technology, USA), anti-E-cadherin (1:1000, Proteintech, USA), Vimentin (1:1000, Proteintech, USA), anti-GAPDH (1:5000, Proteintech, USA), anti-PCNA (1:1000, Cell Signaling Technology, USA), anti-β-Actin (1:1000, Cell Signaling Technology, USA). The second day the PVDF membranes were washed fifth in TBST solution for 10 minutes each time, and subsequently incubated with diluted HRP-conjugated secondary antibodies for 1h at room temperature. Finally, the protein bands were scanned by the ChemiDocTM XRS+System (Bio-Rad, USA), and quantification was conducted using the Bio-Rad Image Lab software.
Nuclear and cytoplasmic protein extraction assay
Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) was used to detect subcellular distribution of p65. Firstly, after co-culture with Oe-BGN AGS or Oe-NC-BGN AGS for 96h, HMrSV5 were used for subsequent experiments. The cells were fully resuspended in cytoplasmic protein extraction reagent A on ice for 15 minutes. Then cytoplasmic protein extraction reagent B was added and resuspended on ice for 1 minute. Next, the above cell suspension solution was centrifuged at 16000g for 5 minutes at 4 °C. The extracted supernatant was cytoplasmic protein. The sediment was fully resuspended using nuclear protein extraction reagent on ice for 30 minutes. Then, the above cell suspension solution was centrifuged at 16000g for 10 minutes at 4 °C. Finally, the extracted supernatant was nuclear protein.
Coimmunoprecipitation (Co-IP) assay
Co-IP assay was used to analyze the binding relationship between the BGN and TLR2/TLR4 proteins. Firstly, HMrSV5 after being co-cultured with Oe-BGN AGS for 96h were used for subsequent experiments. We conducted Co-IP assay using the Co-IP Kit (Abison, Guangzhou, China) following the instructions recommended by the manufacturer. The cells were washed three times with cold PBS, then lysed in ice-cold lysis buffer on ice for at least 5 minutes. Next, the above cell suspension solution was centrifuged at 12000rpm for 10 minutes, and the supernatant was incubated with anti-BGN antibody, IgG, or TLR2 antibody, TLR4 antibody in shaking table for overnight at 4 °C. Next, the Protein A/G agarose beads were used to capture the antigen-antibody complexes for 12h at 4 °C, and then the beads were washed with PBS, and the complexes were boiled and subjected to western blot analysis.
Dual-luciferase report assay
To assess whether rFAP regulate BGN expression via STAT3, A 2000-bp DNA fragment of the human BGN gene promoter containing STAT3 binding sites was amplified via PCR and cloned into pPRO-RB-Report-Basic plasmid (double report gene vector, RiboBio, China), including wild type (WT) of BGN promoter region plasmid, namely BGN-WT, mutation (MUT) of STAT3 potential binding BGN promoter region plasmid, namely BGN-MUT1 (-539 to -549), BGN-MUT2 (-1405 to -1415), BGN-MUT3 (-1468 to -1478), and BGN-negative control (BGN-NC) plasmid. Firstly, GC cells were seeded in six-well plates were transfected with BGN-NC or BGN-WT separately in culture solution containing stattic or DMSO for 24h, then the transfectants were suspended and replaced with fresh culture solution containing rFAP for 24h. Secondly, to further evaluate whether STAT3 regulate BGN expression directly, the human plasmid expression enhanced GFP STAT3 (STAT3-Ex) and corresponding negative control (STAT3-NC) were purchased from RiboBio, China. GC cells were seeded in six-well plates, and co-transfected with different plasmids containing BGN-WT, BGN-MUT1, BGN-MUT2, BGN-MUT3, BGN-NC, STAT3-Ex, STAT3-NC, then the transfectants were suspended and replaced with fresh culture solution. Finally, the total cells were lysed 48h after transfection, and relative luciferase activity were calculated by dual-luciferase reporter system (Progema, USA) following the instructions recommended by the manufacturer.
Chromatin immunoprecipitation (CHIP) assay
To assess the interaction between STAT3 and BGN promoter region, we conducted CHIP assays using a SimpleCHIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, #9003, USA) following the instructions recommended by the manufacturer. Firstly, the cells were fixed for protein/DNA crosslinking and lysed. Then the DNA was sheared using sonication and immunoprecipitated by anti-STAT3 antibody, and the normal rabbit immunoglobulin G (IgG) in the kit was used as a negative control. Subsequently, the unbound material was washed to purify bound antibody-protein/DNA material, then STAT3-bound DNA was released using proteinase K digestion and further purified by filtration. The purified DNA was analyzed by qRT-PCR via using the designed primers, and the primers contained the BGN promoter region spanning the sites of their corresponding binding with STAT3 (Table s1). Finally, the PCR products were analyzed on 2% agarose gel, and the gel containing ethidium bromide staining was visualized using Tanon-1600 Gel Image System (Tanon, China).
Immunofluorescence staining (IF)
The cells were seeded on 15 mm round coverslip in 24-well plates and cultured for 24h. Then the cells were washed with PBS, fixed with 4% paraformaldehyde for 30 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes and blocked in 3% bovine serum albumin for 20 minutes at room temperature. Sequentially, the cells were treated with primary antibodies including anti-FAP (1:100, Bioswamp, China), anti-Vimentin (1:100, Proteintech, USA), anti-WT1 (1:200, Cell Signaling Technology, USA), anti-mesothelin (1:200, Abcam, USA) at 4 °C overnight. The cells were rewashed with PBS and incubated with fluorescent secondary antibody for 50 minutes in the absence of light at room temperature. After being washed with PBS, the cell nuclei were stained with DAPI (10 μg/ml) for 10 minutes. Finally, the Olympus BX5 fluorescence microscope (Olympus Optical, Japan) was used to take the images of cells.
Cell proliferation assay
Cell Counting Kit-8 (CCK8) and plate colony formation assay were performed to measure the cell proliferation. For CCK8 assay, the cells were seeded in 96-well plates with the cell density of 3.0×103 cells/well in 100 μL culture solution. After incubating for 0h, 24h, 48h, 72h, 96h, each well was put in 10 μL CCK8 (Biosharp, China) and incubated for 2h at 37°C. Finally, the Multiskan FC microplate absorbance reader (Thermo Fisher, USA) was used to determine the OD values at a wavelength of 450 nm. For plate colony formation assay, 1.0×103 viable cells were planted in 6-well plates and incubated for two weeks. Then the colonies were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.5% crystal violet for 20 minutes. Finally, the colonies were photographed and recorded the number in 6-well plates.
Wound healing assay
The wound healing assay was operated to detect the migratory ability of GC cells. The cells were planted in 6-well plates and incubated with the DMEM containing 10% FBS until they reached 90% confluence. Several vertical lines were scratched on the cell surface using 10 μL pipette tips, then the plates were washed with PBS to remove non-adherent cells and cells were cultured with the serum-free DMEM. Finally, the migrating cells photographs were captured after incubation for 0h, 24h. The area between both edges of cell free was detected using Image J software (NIH, USA).
Migration assay and invasion assay
The migrated and invasive ability of tumor cells were estimated by Transwell migration and invasion assay 23-25. The migration assay and invasion assay were operated using a 24-well Transwells (8 μm pore size, Coring, USA). Cell migration assay was performed using 24-well Transwells uncoated with Matrigel, and Cell invasion assay was performed using 24-well Transwells pre-coated with Matrigel. Firstly, the upper chamber was precoated with 1:8 dilution of Matrigel (Corning, USA) in the Transwell invasion assay following the instructions recommended by the manufacturer, while the upper chamber was not treated with Matrigel in Transwell migration assay. For both assays, 1.0×105 viable cells were seeded in upper chamber and cultured with serum-free DMEM, while 700 μL DMEM containing 10% FBS was added to the lower chambers. After 48h of incubation, the cells and Matrigel above the membrane in the upper chamber were carefully removed with cotton swabs. Then the migrated or invaded cells on the bottom surface of membrane were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Finally, the stained cells in 5 random fields were counted and photographed under the inverted microscope at ×200 magnification to operate a quantitative analysis.
In vivo xenograft assay
The 6-weeks old female BALB/c-nu mice were purchased from Gempharmatech (Jiangsu, China). All animal experiments were approved by laboratory animal welfare and ethics review of Wuhan University center for animal experiment (WP20210360). Firstly, the tumor growth assay was performed and randomly divided into five groups (n = 5 per group), namely anti-NC-BGN HGC27 (5.0×106 cells) group, anti-BGN HGC27 (5.0×106 cells) group, anti-BGN HGC27 (5.0×106 cells) and CAFLCs/sh-control (5.0×106 cells) group, anti-BGN HGC27 (5.0×106 cells) and CAFLCs/sh-FAP (5.0×106 cells) group, CAFLCs (5.0×106 cells) group, which were resuspended in 200 μl of serum-free DMEM and Matrigel (Corning, USA) (1:1) and inoculated into the flank of each mice. For the tumor peritoneal dissemination assay, the number of cells in above groups was 1.0×107 and also resuspended in 200 μl of serum-free DMEM and Matrigel (1:1), then injected into peritoneal cavity of each mouse. The mice in tumor growth assay and tumor peritoneal dissemination assay were sacrificed and necropsied to evaluate tumor burden after thirty days and forty days later respectively. Finally, the subcutaneous tumors and the intraperitoneal disseminated tumors were photographed and weighted to perform statistical analysis. Furthermore, the intraperitoneal disseminated tumors were detected by IHC staining, and qRT-PCR assay.
Statistics analysis
All experiments were conducted at least in triplicate and results were presented as standard error of the mean (SEM). All statistical data were analyzed using Statistical Product and Service Solutions (SPSS) software version 22.0 (IBM SPSS, USA) and GraphPad prism software version 8.0 (GraphPad Software, USA) for windows. Pearson's correlation analysis was used to evaluate the correlation between mRNA expression of BGN and FAP in GC tissues. The relationship between the expression of BGN, FAP, and their pathological characteristics were analyzed using Chi-square test. Differences between the two groups were evaluated using the Student's t test. The Kaplan-Meier survival curve was drawn for survival analysis, and difference between curves was determined by the log rank test. Univariate and multivariate Cox regression analysis were performed to discriminate the independent factors of prognosis. A two-sided P < 0.05 was considered as statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
BGN expression is associated with poor overall survival (OS) in GC
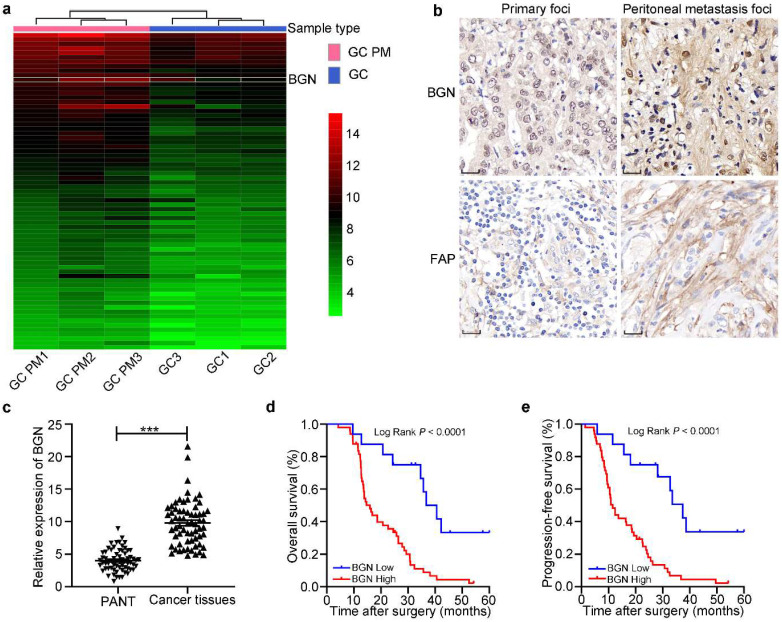
A heatmap of mRNA microarray sequencing between PM lesion and primary foci showed that the expression of BGN in PM of GC lesion was higher than that in primary GC lesion (Fig. 1a). BGN ranked the fourth highest depending on fold change among 71 differential expression genes to screen for further studies. IHC staining displayed that BGN expression was obviously higher in PM foci than GC primary foci, and was distributed in cytoplasm of tumor cells and stroma (Fig. 1b). In addition, qRT-PCR showed that BGN expression was higher in 65 GC tissues than matched PANT (Fig. 1c). To explore whether BGN expression was associated with GC progression, the correlation between BGN expression and clinicopathological parameters was assessed (Table 1). Importantly, high expression of BGN correlated significantly with T stage, LNM (lymph node metastasis), poor tumor differentiation degrees, advanced tumor-node-metastasis (TNM) stage, and postoperative PM (P < 0.05, respectively). Survival analysis demonstrated that patients with higher BGN expression had lower overall survival (OS) (P < 0.0001) (Fig. 1d) and progression-free survival (PFS) (P < 0.0001) (Fig. 1e). Univariate and multivariate analyses indicated that BGN expression was an independent prognostic factor correlating with poor PFS (HR = 4.733, 95%CI = 2.153-10.408, P = 0.000) and OS (HR = 4.851, 95%CI = 2.156-10.914, P = 0.000) (Table 2). These data showed that BGN facilitated tumor progression and influenced the prognosis of GC patients.
Figure 1.
The expression and prognosis of BGN in gastric cancer. *P < 0.05, **P < 0.01, ***P < 0.001. a The heatmap of mRNA microarray sequencing between PM foci and GC primary lesion was analyzed in three pairs. Thus, BGN was screened for more study. b The representative IHC staining images for BGN and FAP expression were shown in GC primary foci and PM foci. Original magnification 400×, scale bar 20 μm. c The relative expression of BGN was measured by qRT-PCR in 65 GC tissues and paired adjacent normal tissues. d-e The association of BGN expression between five-year overall survival (d) and five-year progression-free survival (e) was analyzed by Kaplan-Meier survival analysis.
Table 1.
Correlation between BGN and FAP expression levels and clinicopathologic characteristics of GC patients.
| Parameters | n (%) | BGN expression | P | FAP expression | P | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Gender | |||||||
| Male | 39 (60.0) | 8 | 31 | 0.347 | 13 | 26 | 0.583 |
| Female | 26 (40.0) | 8 | 18 | 7 | 19 | ||
| Age (years) | |||||||
| <60 | 22 (33.8) | 6 | 16 | 0.722 | 5 | 17 | 0.315 |
| ≥60 | 43 (66.2) | 10 | 33 | 15 | 28 | ||
| Tumor size (cm) | |||||||
| <5 | 42 (64.6) | 8 | 34 | 0.159 | 14 | 28 | 0.545 |
| ≥5 | 23 (35.4) | 8 | 15 | 6 | 17 | ||
| Tumor differentiation | |||||||
| Moderate/Well | 23 (35.4) | 9 | 14 | 0.044 | 12 | 11 | 0.006 |
| Poor | 42 (64.6) | 7 | 35 | 8 | 34 | ||
| T stage | |||||||
| T1-2 | 25 (38.5) | 10 | 15 | 0.023 | 12 | 13 | 0.017 |
| T3-4 | 40 (61.5) | 6 | 34 | 8 | 32 | ||
| LNM | |||||||
| N0-1 | 28 (43.1) | 11 | 17 | 0.017 | 14 | 14 | 0.003 |
| N2-3 | 37 (56.9) | 5 | 32 | 6 | 31 | ||
| TNM stagea | |||||||
| I/II | 26 (40.0) | 11 | 15 | 0.007 | 14 | 12 | 0.001 |
| III/IV | 39 (60.0) | 5 | 34 | 6 | 33 | ||
| Intraoperative DM | |||||||
| Absence | 58 (89.2) | 15 | 43 | 0.836 | 19 | 39 | 0.571 |
| Presence | 7 (10.8) | 1 | 6 | 1 | 6 | ||
| CEA (ng/ml) | |||||||
| <5 | 44 (67.7) | 9 | 35 | 0.260 | 14 | 30 | 0.791 |
| ≥5 | 21 (32.3) | 7 | 14 | 6 | 15 | ||
| CA19-9 (U/ml) | |||||||
| <37 | 41 (63.1) | 9 | 32 | 0.515 | 11 | 30 | 0.368 |
| ≥37 | 24 (36.9) | 7 | 17 | 9 | 15 | ||
| Postoperative PM | |||||||
| Absence | 28 (43.1) | 11 | 17 | 0.017 | 14 | 14 | 0.003 |
| Presence | 37 (56.9) | 5 | 32 | 6 | 31 | ||
| Overall | 65 (100) | 16 | 49 | 20 | 45 | ||
Notes: boldface indicates P < 0.05; a, the 8th edition of the AJCC Cancer Staging Manual. Abbreviations: BGN, biglycan; FAP, fibroblast activation protein; T stage, tumor invasion stage; LNM, lymph node metastasis; TNM, tumor-node-metastasis; DM, distant metastasis; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; PM, peritoneal metastasis.
Table 2.
Univariate and multivariate analyses of clinicopathologic parameters associated with progression-free survival and overall survival.
| Parameters | Progression-free survival | Overall survival | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Gender (Female vs Male) | 1.029 | 0.599-1.768 | 0.918 | 0.958 | 0.558-1.646 | 0.877 | ||||||
| Age (<60 vs ≥60) | 1.005 | 0.568-1.780 | 0.985 | 0.965 | 0.573-1.791 | 0.965 | ||||||
| Tumor size (≥5 cm vs <5 cm) | 1.011 | 0.582-1.758 | 0.969 | 1.001 | 0.576-1.740 | 0.997 | ||||||
| Tumor differentiation (Poor vs Moderate/Well) | 1.030 | 0.585-1.813 | 0.919 | 0.948 | 0.579-1.793 | 0.948 | ||||||
| T stage (T3-4 vs T1-2) | 1.869 | 1.048-3.335 | 0.034 | 0.668 | 0.289-1.542 | 0.344 | 1.834 | 1.033-3.256 | 0.038 | 0.675 | 0.287-1.589 | 0.368 |
| LNM (N2-3 vs N0-1) | 1.609 | 0.936-2.766 | 0.085 | 1.580 | 0.920-2.714 | 0.097 | ||||||
| TNM stagea (III/IV vs I/II) | 2.226 | 1.262-3.925 | 0.006 | 1.129 | 0.480-2.654 | 0.781 | 2.201 | 1.250-3.877 | 0.006 | 1.061 | 0.441-2.554 | 0.894 |
| Intraoperative DM (Presence vs Absence) | 6.452 | 2.588-16.083 | 0.000 | 6.400 | 2.251-18.198 | 0.000 | 6.269 | 2.525-15.562 | 0.000 | 7.583 | 2.524-22.782 | 0.000 |
| CEA (High vs Low) | 0.800 | 0.442-1.446 | 0.460 | 0.814 | 0.450-1.472 | 0.496 | ||||||
| CA19-9 (High vs Low) | 1.228 | 0.707-2.131 | 0.466 | 1.249 | 0.719-2.170 | 0.429 | ||||||
| Postoperative PM (Presence vs Absence) | 6.873 | 3.530-13.384 | 0.000 | 6.248 | 2.998-13.020 | 0.000 | 6.494 | 3.391-12.434 | 0.000 | 5.850 | 2.817-12.146 | 0.000 |
| BGN (High vs Low) | 4.095 | 1.958-8.565 | 0.000 | 4.733 | 2.153-10.408 | 0.000 | 3.910 | 1.877-8.145 | 0.000 | 4.851 | 2.156-10.914 | 0.000 |
| FAP (High vs Low) | 2.459 | 1.318-4.591 | 0.005 | 1.809 | 0.899-3.640 | 0.096 | 2.497 | 1.328-4.696 | 0.005 | 2.018 | 0.988-4.120 | 0.054 |
Notes: boldface indicates P < 0.05; a, the 8th edition of the AJCC Cancer Staging Manual. Abbreviations: BGN, biglycan; FAP, fibroblast activation protein; T stage, tumor invasion stage; LNM, lymph node metastasis; TNM, tumor-node-metastasis; DM, distant metastasis; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; PM, peritoneal metastasis.
BGN promotes the proliferation, migration, invasion, and EMT of GC cells
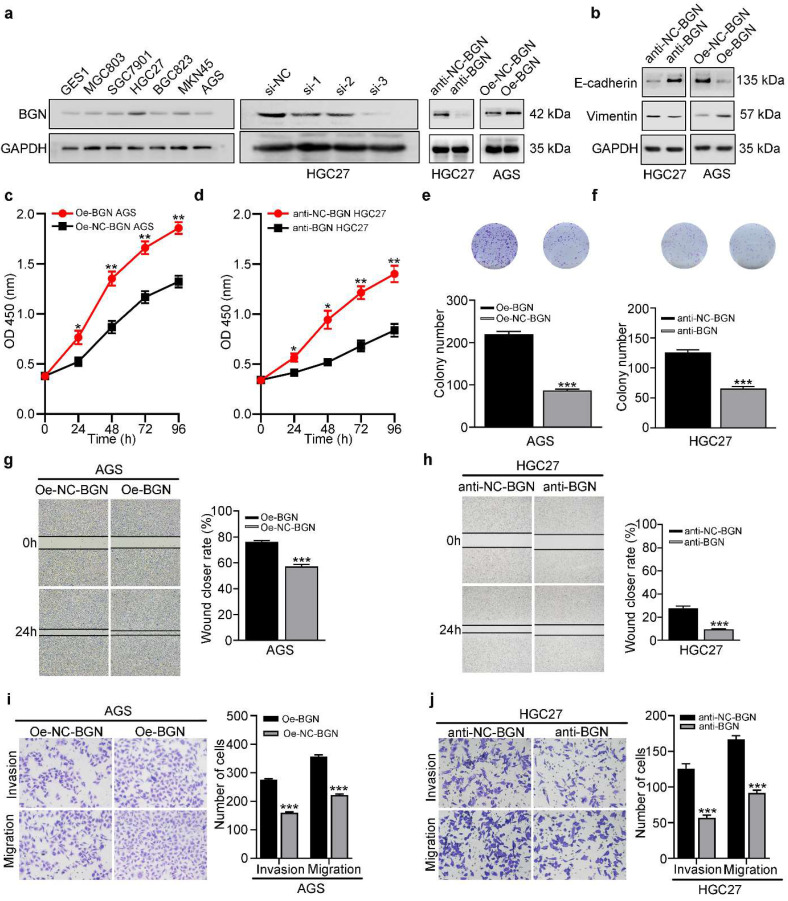
To further explore the function of BGN, we firstly investigated the expression of BGN in GC cell lines. In contrast to GES-1, BGN was up-regulated in GC cell lines (Fig. 2a). While the maximum expression of BGN was detected in HGC27, the minimum expression was detected in AGS (Fig. 2a). Thus, these two cell lines were selected for additional experiments. Three different siRNAs were designed and transfected into HGC27, and si-3 had the optimal knockdown effect (Fig. 2a). Therefore, the si-3 sequence and corresponding negative control were selected to construct lentivirus-based vectors. Lentivirus-mediated BGN overexpression was constructed in AGS. Then the down-regulation and up-regulation expression of BGN effects were detected by western blot (Fig. 2a). Gain and loss of function experiments demonstrated that overexpression of BGN in AGS promoted the proliferation (Fig. 2c, 2e), migration (Fig. 2g, 2i), and invasion (Fig. 2i) of GC cells, while knockdown of BGN in HGC27 inhibited the proliferation (Fig. 2d, 2f), migration (Fig. 2h, 2j), and invasion (Fig. 2j) of GC cells.
Figure 2.
BGN promotes the proliferation, migration, invasion, and EMT of GC cells. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± SEM (n = 3). a BGN expression was validated in six GC cell lines, the human gastric epithelial cell line (GES1). Three different siRNAs of BGN were designed and transfected into HGC27, and si-3 had the optimal knockdown effect. Lentiviral-based BGN knockdown stable HGC27 cell line (anti-BGN HGC27), lentiviral-based BGN overexpression stable AGS cell line (Oe-BGN AGS), and negative control stable cell lines were verified by western blot. b-j The effect of BGN on the proliferation, migration, invasion, and EMT of GC cells was detected with lentiviral-based BGN overexpression and knockdown stable cell lines by the CCK8 assay (c-d), colony formation assay (e-f), wound healing assay (g-h), Transwell migration and invasion assay (i-j), and western blot analysis (b), respectively. Representative photographs of wound healing (Original magnification 40×), migratory or invaded cells (Original magnification 200×).
In the context of neoplasia, EMT is closely linked with migration and invasion of cancer cells and plays an important role in cancer metastasis. To assess whether EMT is implicated in BGN-induced migration and invasion, western blot was used to measure expression of the epithelia marker (E-cadherin), and the mesenchymal marker (Vimentin). Overexpression of BGN in AGS significantly facilitated Vimentin expression and decreased E-cadherin expression (Fig. 2b), while knockdown of BGN in HGC27 dramatically inhibited Vimentin expression but improved E-cadherin expression (Fig. 2b). Taken together, these results indicate that BGN can promote the proliferation, migration, invasion, and EMT of GC cells.
BGN promotes the transformation of HMrSV5 into CAFLCs through TLR2/TLR4/NF-κB signaling pathway
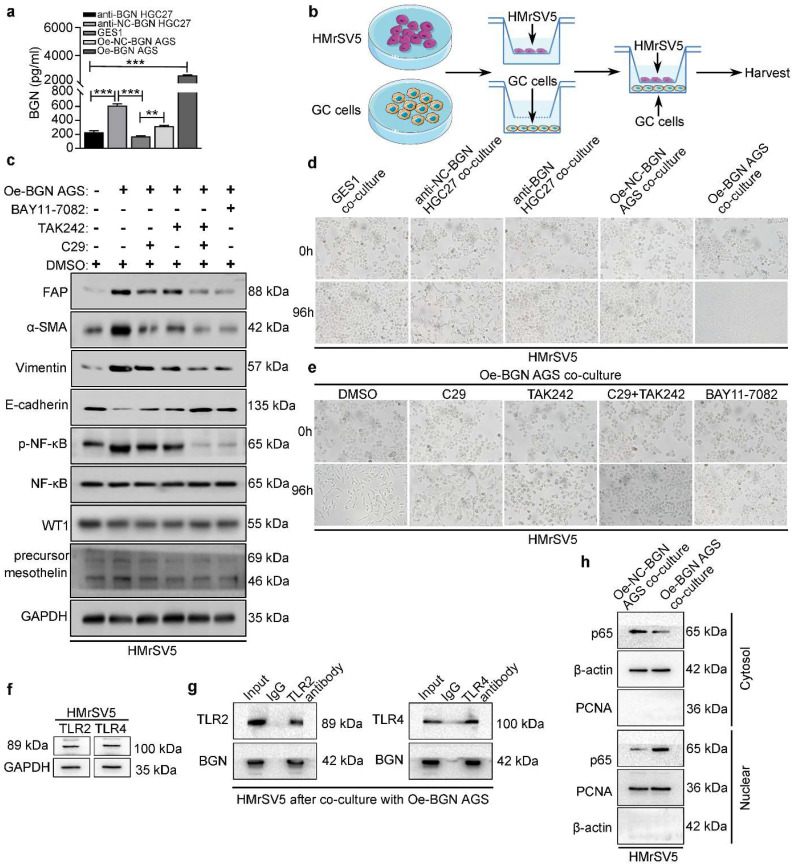
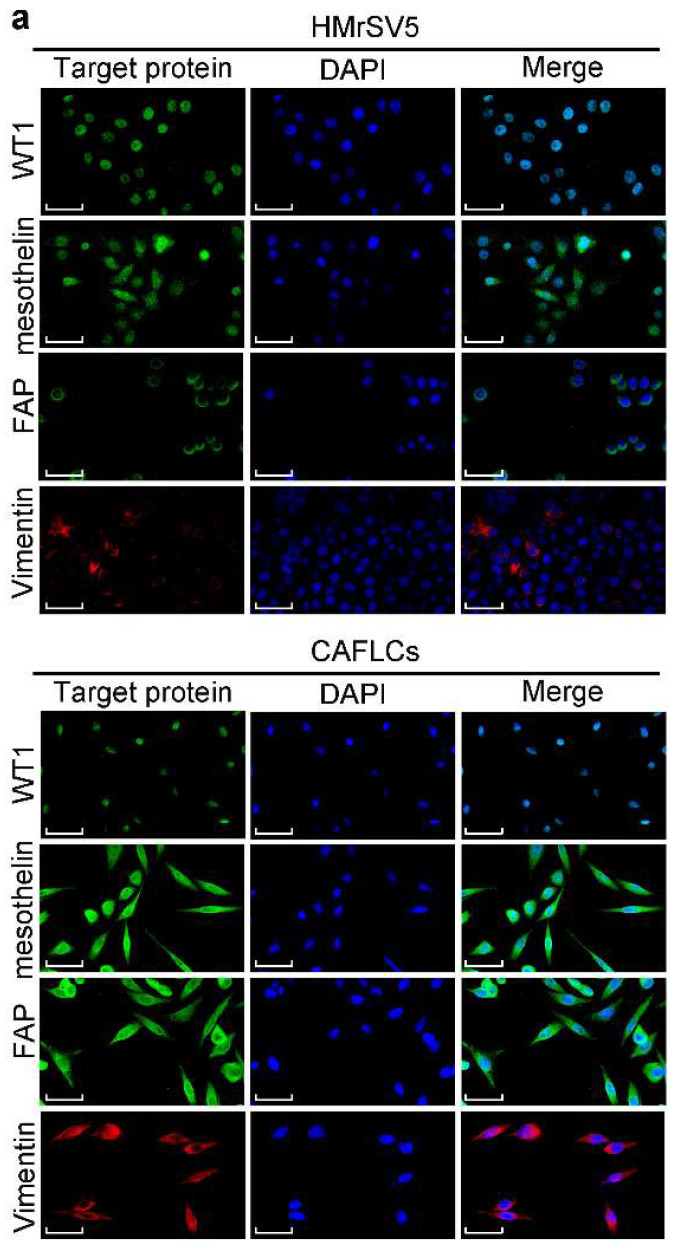
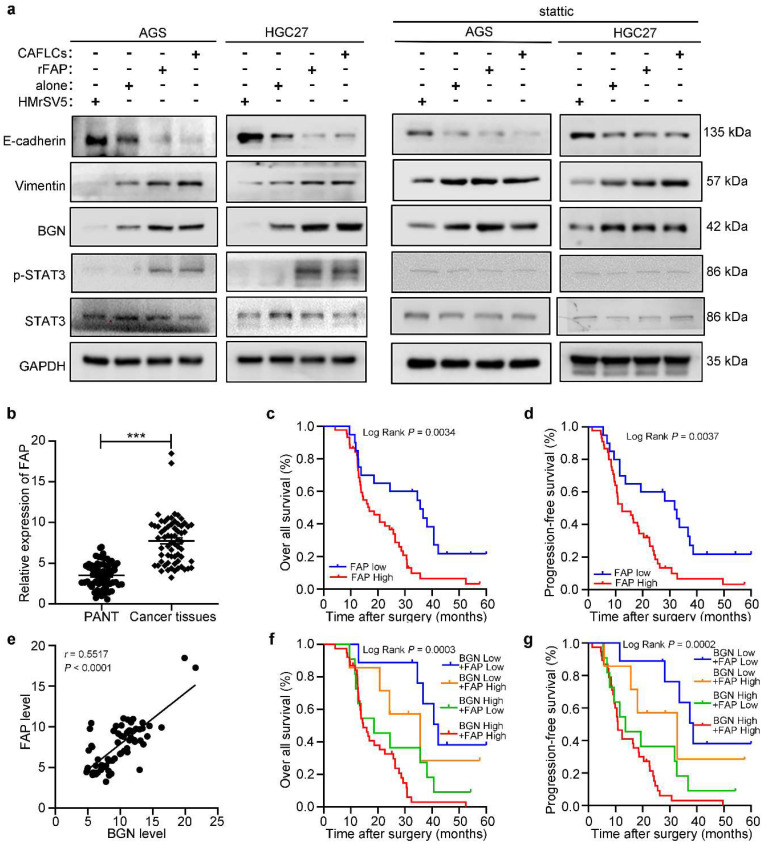
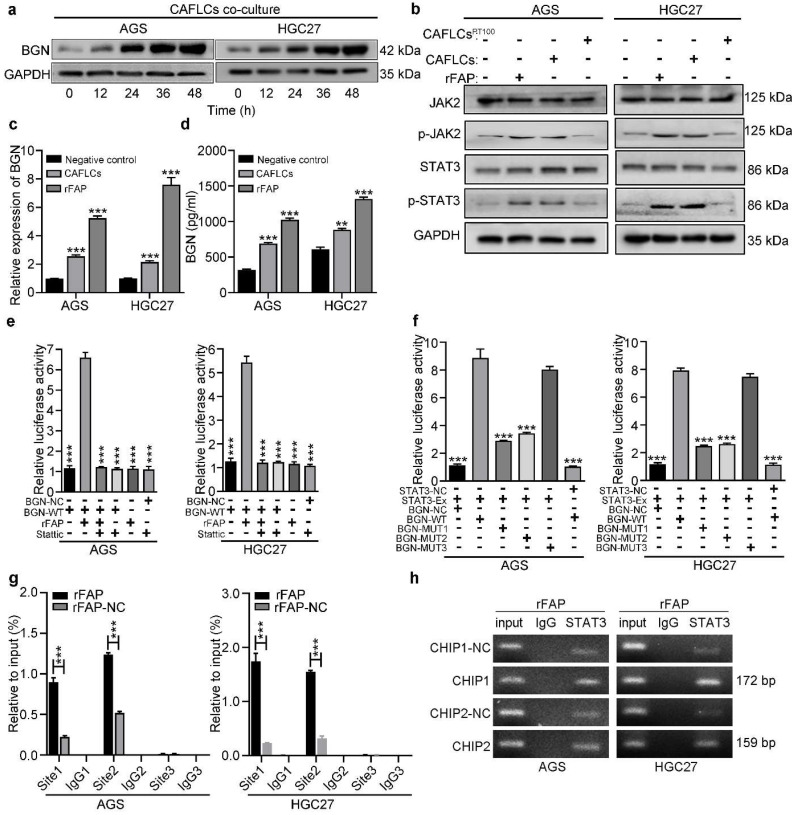
To further assess the function of BGN in TME, the concentration of BGN in culture medium of GES1, anti-BGN HGC27, anti-NC-BGN HGC27, Oe-BGN AGS, and Oe-NC-BGN AGS, was measured by ELISA. BGN levels were highest in supernatant of Oe-BGN AGS (Fig. 3a). Then we took advantage of co-culture experiment (Fig. 3b). Each of the cell lines listed above was co-cultured with HMrSV5 respectively. After co-culture with HMrSV5 for 96h, we observed that the normal cobble-stone morphology of HMrSV5 transmuted into spindle-like morphology in the co-culture group of Oe-BGN AGS, and partial changes occurred in the anti-NC-BGN HGC27 group (Fig. 3d). Vimentin, α-SMA, and FAP expression were significantly up-regulated in HMrSV5 after co-culture with Oe-BGN AGS, while E-cadherin expression was down-regulated compared with HMrSV5 after incubation with DMSO (Fig. 3c). Similar findings were observed in IF assay (Fig. 4a). Moreover, WT1 and mesothelin were two main markers of mesothelial cells, Western blot and IF assays showed that both HMrSV5 and HMrSV5 after co-culture with Oe-BGN AGS expressed WT1 and mesothelin (Fig. 3c, Fig. 4a). In summary, these data show that GC cells-derived BGN can convert HMrSV5 into CAFLCs by MMT in TME.
Figure 3.
In co-cultured model, BGN promotes the transformation of HMrSV5 into CAFLCs through TLR2/TLR4/NF-κB signaling pathway. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± SEM (n = 3). a ELISA assay of BGN protein secretion of GES1, anti-BGN HGC27, anti-NC-BGN HGC27, Oe-BGN AGS, and Oe-NC-BGN AGS. b Schema for representing the experiment procedures. c and e The effect of Oe-BGN AGS on the transformation of HMrSV5 was analyzed by western blot (c) and bright-field images (e). HMrSV5 were incubated with DMSO, C29, TAK242, C29+TAK242, or BAY11-7082 for 24h prior to co-culture with Oe-BGN AGS for 96h. Representative bright-field photographs (Original magnification 200×). d The effect of GES1, anti-BGN HGC27, anti-NC-BGN HGC27, Oe-BGN AGS, or Oe-NC-BGN AGS on the transformation of HMrSV5 for 96h was analyzed by bright-field images. Representative bright-field photographs (Original magnification 200×). f TLR2 and TLR4 expression were validated in HMrSV5 by western blot. g Coimmunoprecipitation (Co-IP) assays were used to identify interaction between BGN and TLR2/TLR4 in HMrSV5 after co-culture with Oe-BGN AGS for 96h. h Nuclear and cytoplasmic protein extraction assays were evaluated by western blot in HMrSV5 after co-culture with Oe-BGN AGS or Oe-NC-BGN AGS for 96h.
Figure 4.
The expression of related proteins between CAFLCs and HMrSV5 are detected by Immunofluorescence assays. a Immunofluorescence assays of WT1, mesothelin, FAP, and Vimentin in CAFLCs and HMrSV5 were detected respectively. Original magnification 400×, scale bar 50 μm.
BGN is the ligand for TLR2/TLR4, and NF-κB is an extremely important downstream signaling pathway of TLR2/TLR4. However, whether GC cells-derived BGN can combine with TLR2/TLR4 receptors to transform HMrSV5 into CAFLCs remains to be explored. Expression of TLR2/TLR4 in HMrSV5 was detected at the protein level (Fig. 3f). Next, HMrSV5 were incubated with DMSO, C29 (TLR2 inhibitor), TAK242 (TLR4 inhibitor), C29+TAK242, or BAY11-7082 (NF-κB inhibitor) for 24h prior to co-culture with Oe-BGN AGS. Compared with DMSO, C29+TAK242 or BAY11-7082 fully blocked the transformation of HMrSV5 into CAFLCs, while C29 or TAK242 partially blocked the transformation by the recovery experiment (Fig. 3c, 3e). Moreover, the Co-IP results indicated that BGN interacted with TLR2 and TLR4 in HMrSV5 after co-culture with Oe-BGN AGS (Fig. 3g). Meanwhile, Nuclear and cytoplasmic protein extraction assays were used to detect cytosolic and nuclear p65 distribution in HMrSV5 after co-culture with Oe-BGN AGS or Oe-NC-BGN AGS. The results indicated that p65 nuclear translocation markedly increased in HMrSV5 after being co-cultured with Oe-BGN AGS compared with HMrSV5 after being co-cultured with Oe-NC-BGN AGS (Fig. 3h). Therefore, Oe-BGN AGS-derived BGN promoted p65 nuclear translocation of HMrSV5 in co-culture model. Taken together, these results show that GC cells-derived BGN can convert HMrSV5 into CAFLCs through TLR2/TLR4/NF-κB signaling pathway in TME.
CAFLCs-derived FAP promotes tumor progression in TME
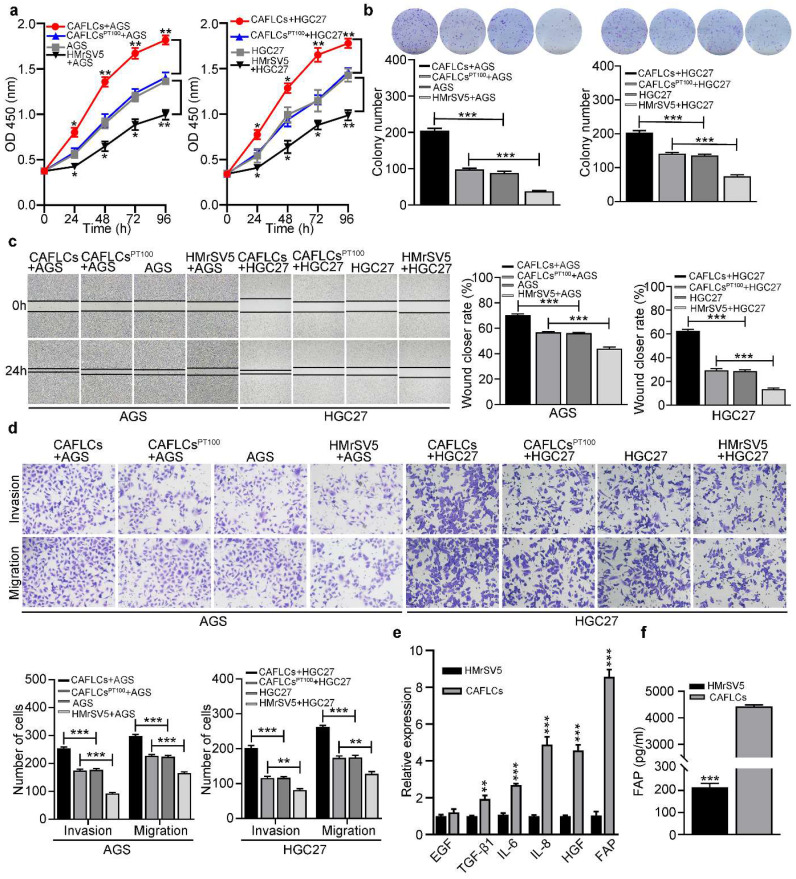
To further investigate the role of HMrSV5-derived CAFLCs in TME, HGC27 or AGS were separately co-cultured with CAFLCs or HMrSV5 for 24h. While CAFLCs increased the proliferation (Fig. 5a, 5b), migration (Fig. 5c, 5d), invasion (Fig. 5d), and EMT (Fig. 6a) of GC cells, HMrSV5 inhibited these effects. Next, the mechanism by which CAFLCs promote tumor progression was assessed. Given the important role of CAFs derived-cytokines in tumorigenesis and development, changes in the expression of six key cytokines in CAFs were assessed 9, 10, 26, 27. In contrast to HMrSV5, FAP was most up-regulated in CAFLCs (Fig. 5e). ELISA results also showed that FAP was more abundant in supernatant of CAFLCs than HMrSV5 supernatants (Fig. 5f). To evaluate whether CAFLCs-derived FAP had pro-tumorigenic activities in vitro, HGC27 or AGS were co-cultured with CAFLCs or CAFLCsPT100 (CAFs pretreated with PT100) for 24h. The results showed that CAFLCs derived-FAP evidently promoted proliferation (Fig. 5a, 5b), migration (Fig. 5c, 5d), invasion (Figure 5d), and EMT (Fig. 6a) of GC cells.
Figure 5.
In co-cultured model, CAFLCs-derived FAP facilitates the proliferation, migration, and invasion of GC cells. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± SEM (n = 3). a-d The effect of HMrSV5 or CAFLCs-derived FAP on the proliferation, migration and invasion of GC cells (AGS or HGC27) was detected by the CCK8 assay (a), colony formation assay (b), wound healing assay (c), Transwell migration and invasion assay (d), respectively. HGC27 or AGS were separately co-cultured with CAFLCs, HMrSV5, or CALCFsPT100 for 24h. Representative photographs of wound healing (Original magnification 40×), migratory or invaded cells (Original magnification 200×). e Relative expression levels of representative CAFLCs-secreted key cytokines were measured by qRT-PCR. f ELISA assay of FAP protein secretion of CAFLCs and HMrSV5.
Figure 6.
The expression and prognosis of FAP in gastric cancer. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± SEM (n = 3). a Western blot analysis of GC cells (AGS or HGC27) alone, rFAP-supplemented GC cells, HMrSV5-co-cultured GC cells, CAFLCs-co-cultured GC cells in the presence or absence of stattic. b The relative expression of FAP was detected by qRT-PCR in 65 GC tissues and paired adjacent normal tissues. c-d The association of FAP expression between five-year overall survival (c) and five-year progression-free survival (d) was analyzed by Kaplan-Meier survival analysis. e The correlation of BGN expression with FAP expression in GC tissues. f-g The association of both BGN and FAP expression between five-year overall survival (f) and five-year progression-free survival (g) were analyzed by Kaplan-Meier survival analysis.
Clinically, IHC staining was performed to measure FAP expression in primary GC foci and PM foci from patient, and found to be markedly higher in PM foci than GC primary foci (Fig. 1b). Then FAP expression in 65 GC tissues was higher than matched PANT via qRT-PCR (Fig. 6b). FAP expression had similar results with BGN in GC, including clinicopathological parameters (Table 1), OS (P = 0.0034) (Fig. 6c), PFS (P = 0.0037) (Fig. 6d), and univariate analyses (Table 2). Moreover, we showed that BGN was positively correlated with FAP expression by qRT-PCR in GC tissues (Fig. 6e), and patients with high expression of both BGN and FAP had the worst survival outcome (Fig. 6f, 6g).
FAP promotes BGN expression in GC cells by activating STAT3, and creating a positive feedback loop
Findings indicated that BGN and FAP expression were positively correlated, and bioinformatics analysis of co-expression genes showed that BGN interacted directly with FAP 16. Interestingly, CAFLCs could promote BGN expression in GC cells in a time-dependent manner (Fig. 7a). These findings inspired us to explore whether CAFLCs derived-FAP could regulate BGN expression. HGC27 or AGS were separately incubated with rFAP or co-cultured with CAFLCs for 48h. The results displayed that rFAP or CAFLCs could promote the expression of BGN in GC cells (Fig. 6a, 7c, 7d). As previously reported, JAK2/STAT3 signaling pathway could be activated by CAFs or FAP stimulation 9, 28-30. Thus, it was speculated that CAFLCs-derived FAP could promote BGN expression in GC cells through JAK2/STAT3 signaling pathway. HGC27 or AGS were separately co-cultured with CAFLCs, CAFLCsPT100, or incubated with rFAP for 48h. The results showed that p-JAK2 and p-STAT3 expression in GC cells significantly increased after co-culture with CAFLCs or incubation with rFAP but markedly decreased after co-culture with CAFLCsPT100 (Fig. 7b). Thus, CAFLCs-derived FAP played a vital role in GC cells by activating JAK2/STAT3 signaling pathway. Next, HGC27 or AGS were separately treated with stattic (a STAT3 inhibitor) for 24h prior to co-culture with CAFLCs, HMrSV5, or rFAP. The results displayed that rFAP or CAFLCs could promote BGN and p-STAT3 expression, and EMT of GC cells, while stattic significantly inhibited these effects (Fig. 6a). Taken together, these results suggest that CAFLCs-derived FAP boosted BGN expression and EMT of GC cells by activating JAK2/STAT3 signaling pathway.
Figure 7.
FAP promotes BGN expression in GC cells by activating STAT3. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± SEM (n = 3). a Western blot of BGN from GC cells (AGS or HGC27) co-cultured with CAFLCs in a time-dependent manner. b Western blot analysis of GC cells (AGS or HGC27) alone, rFAP-supplemented GC cells, CAFLCs-co-cultured GC cells, and CAFLCsPT100-co-cultured GC cells for 48h. c The relative expression of BGN was detected in GC cells (AGS or HGC27) alone, rFAP-supplemented GC cells, CAFLCs-co-cultured GC cells for 48h. d The expression of BGN was detected in GC cells (AGS or HGC27) alone, rFAP-supplemented GC cells, CAFLCs-co-cultured GC cells for 48h by ELISA assay. e GC cells (AGS or HGC27) were transfected with a BGN promoter reporter plasmid (pPRO-RB-Report-Basic plasmid) for 24h. GC cells were incubated with or without stattic (10 μM) for 24h before rFAP (200 ng/ml) stimulation, relative luciferase activity of BGN promoter was detected after rFAP stimulation for 24h. f A reporter plasmid for BGN was constructed by cloning BGN promoter region (WT or NC) or identified STAT3 binding site mutants (MUT-1, MUT-2, MUT-3), and the human plasmid expression STAT3 (STAT3-Ex, STAT3-NC) were co-transfected with GC cells (AGS or HGC27), relative luciferase activity of BGN promoter was detected 48h after transfection. g-h GC cells (AGS and HGC27) were stimulated with rFAP. The qRT-PCR of CHIP products demonstrated the direct binding ability of STAT3 to BGN promoter region in GC cells, the input (2%) (g). The qRT-PCR of CHIP products was analyzed including negative control (NC), CHIP1, CHIP2, and CHIP3. The values were normalized to input (2%), and agarose gel (2%) (h).
A dual-luciferase assay was performed to further explore whether FAP could directly activate the BGN promoter region by STAT3 signaling pathway. HGC27 or AGS were transfected with dual-luciferase vector of BGN (WT or NC), and the transfected cells were incubated with or without stattic before rFAP simulation. FAP was shown to promote BGN transcription in GC cells by activating STAT3 (Fig. 7e). Bioinformatics prediction was conducted using Jaspar to uncover potential transcription binding sites on the BGN promoter region and find three putative STAT3-binding sites. A dual-luciferase vector of BGN-MUT that included BGN-MUT1 (-539 to -549), BGN-MUT2 (-1405 to -1415), and BGN-MUT3 (-1468 to -1478), and a plasmid of STAT3-Ex and STAT3-NC were constructed. To detect involvement of STAT3 in BGN regulation, every BGN report, including BGN-NC, BGN-WT, BGN-MUT1, BGN-MUT2, and BGN-MUT3, were co-transfected with STAT3-Ex or STAT3-NC in HGC27 or AGS, and promoter activity was assessed 48h after transfection. The result showed that BGN-MUT1 and BGN-MUT2 obviously decreased STAT3-Ex activation of the BGN promoter in contrast to the activation observed in response to BGN-WT or BGN-MUT3 (Fig. 7f). Findings from the dual-luciferase assay implied that BGN-MUT1 (-539 to -549) or BGN-MUT2 (-1405 to -1415) might contain a STAT3-binding site. Three primer sets were designed for potential STAT3 binding sites, site1, site2, and site3 (table S1) using a CHIP assay. Findings indicated that STAT3 bound directly to two STAT3 binding sites, site1 and site2 (Fig. 7g, 7h). These results demonstrated that CAFLCs-derived FAP could promote BGN transcription by activating STAT3 in GC cells to form a positive feedback loop.
GC cells-derived BGN and CAFLCs-derived FAP promote GC PM in vivo
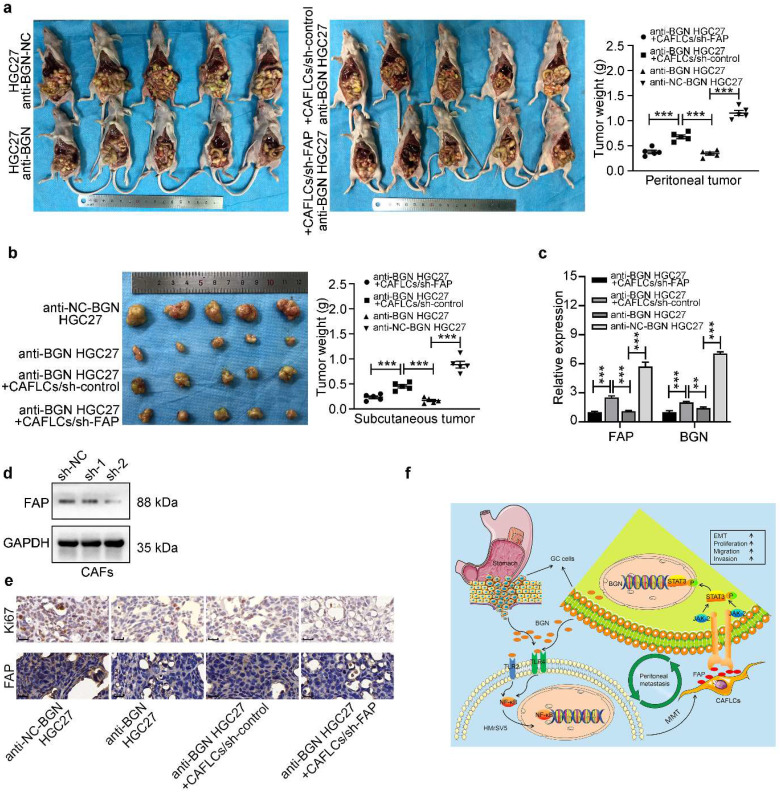
To confirm the in vitro results, an in vivo xenograft gastric cancer nude mouse model was performed. Anti-NC-BGN HGC27, anti-BGN HGC27, anti-BGN HGC27+CAFLCs/sh-control, anti-BGN HGC27+CAFLCs/sh-FAP, and CAFLCs groups were injected subcutaneously or intraperitoneally into nude mice. Plasmid was transfected to knockdown expression of FAP into CAFLCs. As shown Fig. 8d, FAP (sh-2) obtained a better knockdown effect. Thus, the second plasmid (sh2-FAP) was selected for the experiment. No tumors were formed after subcutaneous or intraperitoneal injection of CAFLCs alone (data not shown).
Figure 8.
Co-injection of GC cells-derived BGN and CAFLCs-derived FAP promote GC PM in vivo. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± SEM (n = 3). a-b The morphological characteristics of peritoneal and subcutaneous tumor xenograft, tumor weight in anti-NC-BGN HGC27 group, anti-BGN HGC27 group, anti-BGN HGC27+CAFLCs/sh-control group, anti-BGN HGC27+CAFLCs/sh-FAP group. Each group had five mice. c The relative expression of BGN and FAP mRNA of peritoneal tumor from anti-NC-BGN HGC27 group, anti-BGN HGC27 group, anti-BGN HGC27+CAFLCs/sh-control group, anti-BGN HGC27+CAFLCs/sh-FAP group. d Two different shRNAs of FAP were designed and transfected into CAFLCs, and FAP (sh-2) obtained a better knockdown effect by western blot. e IHC analyzed Ki67 and FAP expression in peritoneal tumor from anti-NC-BGN HGC27 group, anti-BGN HGC27 group, anti-BGN HGC27+CAFLCs/sh-control group, anti-BGN HGC27+CAFLCs/sh-FAP group. Original magnification 400×, scale bar 20 μm. f Schematic illustration of mutual interaction between tumor cells and activated MCs in the tumor microenvironment. Our study proposed the mechanism model that a BGN/FAP-STAT3 positive feedback loop facilitated PM of GC.
For the subcutaneous xenograft model (Fig. 8b), the weight of tumors in anti-NC-BGN HGC27 group were noticeably heavier than those in anti-BGN HGC27 group, anti-BGN HGC27+CAFLCs/sh-control group, anti-BGN HGC27+CAFLCs/sh-FAP group, further analysis showed the weight of tumors in anti-BGN HGC27+CAFLCs/sh-control group were obviously heavier than those in anti-BGN HGC27 group, anti-BGN HGC27+CAFLCs/sh-FAP group, while two groups of anti-BGN HGC27, anti-BGN HGC27+CAFLCs/sh-FAP were not statistically significant in tumor weights.
For the tumor peritoneal dissemination assay (Fig. 8a), the tumors produced by anti-NC-BGN HGC27 were dramatically heavier and wider than those anti-BGN HGC27, anti-BGN HGC27+CAFLCs/sh-control, anti-BGN HGC27+CAFLCs/sh-FAP, further analysis showed the tumors produced by anti-BGN HGC27+CAFLCs/sh-control were evidently heavier and wider than those anti-BGN HGC27, anti-BGN HGC27+CAFLCs/sh-FAP, while two groups of anti-BGN HGC27, anti-BGN HGC27+CAFLCs/sh-FAP were not statistically significant in tumor weights and peritoneal dissemination.
Taken together, the results indicated that co-injection of anti-BGN HGC27+CAFLCs/sh-control rescued the inhibitory effect of knockdown BGN expression on subcutaneous tumor growth or intraperitoneal tumor dissemination, while co-injection of anti-BGN HGC27+CAFLCs/sh-FAP eliminated the rescued effect. Thus, GC cells-derived BGN and CAFLCs-derived FAP played a critical role in regulating tumor growth and intraperitoneal dissemination of GC in co-injection. IHC staining analysis and qRT-PCR were performed in different groups of intraperitoneal disseminated tumors. The result showed that anti-BGN HGC27 group inhibited FAP, BGN, and Ki67 expression of PM foci compared with anti-NC-BGN HGC27 group; while anti-BGN HGC27+CAFLCs/sh-control group promoted FAP, BGN, and Ki67 expression of PM foci compared with anti-BGN HGC27 group and anti-BGN HGC27+CAFLCs/sh-FAP group (Fig. 8c, 8e). Taken together, these results verified that GC cells-derived BGN promoted Ki67, FAP expression of intraperitoneal disseminated tumors in TME. At the same time, CAFLCs-derived FAP boosted the proliferation, BGN expression of GC cells to form a positive feedback loop, resulting in tumorigenesis and progression of GC PM in TME.
In this study, BGN was screened for further research using microarray sequencing. Both in vitro and in vivo experiments demonstrated that BGN promoted tumor progression and transformation of MCs into CAFLCs through the TLR2/TLR4/NF-κB signaling pathway. In turn, CAFLCs-derived FAP facilitated the proliferation, migration, invasion, and EMT of GC cells by activating JAK2/STAT3 signaling pathway. Activated STAT3 promoted the transcription of BGN in GC, thus forming a BGN/FAP-STAT3 positive feedback loop facilitating PM of GC (Fig. 8f).
Discussion
According to Paget's “seed and soil” theory, metastasis occurs not only relies on competent cancer cells (the “seed”) but also requires a predetermined invitation by certain organs (the “soil”) 31. Thus, metastases are influenced by various factors and mysterious cellular crosstalk between cancer "seed" and "soil", now the "soil" has been known as the TME 32. The TME is rather complex, and extremely different from microenvironment of normal tissue, and HPMCs are a key component of the peritoneal TME for PM. The complex and dynamic bidirectional interactions between tumor cells and HMPCs-derived CAFs play an important role in PM. This study defined the molecular and cellular mechanisms that allow BGN to induce GC cells proliferation, migration, invasion, and EMT capacity, and convert HMrSV5 into CAFLCs. CAFLCs and CAFLCs-derived FAP were found, in turn, to facilitate the proliferation, migration, invasion, EMT, and BGN expression of GC cells through the JAK2/STAT3 signaling pathway. These results revealed that a positive feedback loop of cancer-TME-cancer promoted PM of GC through BGN/FAP-STAT3 interaction in TME.
Clinically, expression of BGN and FAP were evidently up-regulated in GC tissues compared with corresponding PANT, and BGN was positively correlated with FAP expression in GC tissues. IHC revealed that BGN and FAP in PM foci tissues were significantly higher than in primary GC lesion. BGN and FAP were also positively correlated with advanced TNM, poor tumor differentiation, LNM, and postoperative PM according to the 8th edition of the AJCC Cancer Staging Manual. In addition, BGN was defined as independent prognostic factors associated with poor clinical outcomes. Studies have found that high expression of BGN in colorectal cancer 33, GC 34, prostate cancer 35, lung cancer 36, endometrial cancer 37, and melanoma 38, and linked expression to poor prognosis. Moreover, high FAP expression is shown in GC 39, esophageal squamous cell carcinoma 40, colorectal cancer 41, pancreatic cancer 42, and non-small cell lung adenocarcinoma 43, and linked with poor prognosis. In accordance with Paget's “seed and soil” theory 31, high expression of BGN in GC tissues allows tumor cells to infiltrate and break through the stomach's serosa to colonize the peritoneum by transforming of HMPCs into CAFLCs through the interaction between cancer cells and stroma in the peritoneal TME.
PM derived from the gastrointestinal tract and ovary malignant tumors, various factors can transform HPMCs into CAFs, including malignant ascites 44, cancer-derived exosomes 4, 45, transforming growth factor-β 5, 46, peritoneal injury 13, tumor necrosis factor-α 47. In this study, BGN promoted the proliferation, migration, invasion, and EMT of GC cells. Recently studies showed that BGN enhanced migration of GC cells by the regulation of EMT 34. EMT enhances the tumor-initiating and potential of cancer cells metastasis 20. In vitro recovery experiment results from our study revealed that GC cells-derived BGN acted on the TLR2/TLR4 receptor of HMrSV5 and activated the downstream signaling molecule, NF-κB, which promoted the transformation of HMrSV5 into CAFLCs. Moreover, from the molecular mechanistic perspective, the Co-IP results demonstrated that BGN interacted with TLR2 and TLR4. Meanwhile, HMrSV5 after being co-cultured with Oe-BGN AGS increased p65 phosphorylation, facilitated p65 nuclear translocation to exert its biological functions. Hu et al. demonstrated that GC cells-secreted BGN could activate endothelial cells, stimulate tubular formation, and induce VEGF via TLR2/TLR4 and NF-κB signaling pathway 21. CAFs are a major constituent of TME with various functions, including ECM deposition and remodeling, crosstalk with tumor cells and TME, and progression of malignancies 9, 10, 28, 48. It was also demonstrated that HMrSV5-derived CAFLCs could boost the proliferation, migration, invasion, and EMT of GC cells, while HMrSV5 suppressed these effects.
Given the importance role of cytokines in cell-cell reciprocal interaction, FAP was identified as the most notably up-regulated cytokine in this study. Prior studies have found various concentration of secretory FAP in circulation 49. However, the plasma concentration of FAP in non-tumor patients was not significantly different in patients with a variety of cancers. This may be because FAP expression is transient in response to diverse stimuli, even though it is highly expressed in cancer tissues 30. In the current study, FAP concentrations in the supernatant of CAFLCs were significantly higher than HMrSV5. Our study demonstrated that CAFLCs and CAFLCs-derived FAP could boost the proliferation, migration, invasion, and EMT of GC cells. Liu et al. showed that exogenous FAP and HELF-derived FAP had a similar effect 39. Bioinformatics analysis of co-expression genes showed that interaction networks of BGN could interact with FAP 16. As expected, CAFLCs-derived FAP promoted BGN expression of GC cells in co-culture model. CAFs or CAFs-derived FAP are involved in the development of tumor-initiation and metastasis by complicated mechanisms in the TME, and JAK2/STAT3 signaling pathway plays an important role in this process 27, 29, 48, 50. In this study, CAFLCs-derived FAP promoted BGN expression and EMT of GC cells through JAK2/STAT3 signaling pathway. It is well known that STAT3 is a key transcription factor that regulates tumor progression and metastasis 51, 52. The findings shown here demonstrate an underlying association between FAP and BGN that is mediated by STAT3. CHIP and dual-luciferase reporter assays confirmed that STAT3 could directly bind two sites in the BGN promoter regions. In brief, this study provides strong evidence that FAP/STAT3-induced BGN activation is critical for GC cells proliferation, migration, invasion, and EMT capacity in this feedback loop, suggesting that the BGN/FAP-STAT3 signal axis plays a vital role in CAFLCs-medicated progression of GC PM.
In this study, BGN was screened for further research by microarray sequencing. In vitro experiments demonstrated that BGN promoted tumor progression and transformation of MCs into CAFLCs by TLR2/TLR4/NF-κB signaling pathway. In turn, CAFLCs-derived FAP facilitated the proliferation, migration, invasion, and EMT of GC cells by activating JAK2/STAT3 signaling pathway. Activated STAT3 could promote the transcription of BGN in GC, forming a BGN/FAP-STAT3 positive feedback loop that facilitated PM of GC. In conclusion, the mutual interaction between tumor cells and activated MCs mediated by a BGN/FAP-STAT3 positive feedback loop facilitates PM of GC, providing a poor prognostic factor which could contribute to peritoneal metastasis of gastric cancer.
Supplementary Material
Supplementary table.
Acknowledgments
Funding
This work was supported by grants from the Health Commission of Hubei Province Scientific Research Project (No. WJ2019H012), Improvement Project for Theranostic ability on Difficulty miscellaneous disease (Tumor) (No. ZLYNXM202018), National Key Clinical Specialty Construction Project and National Natural Science Fund Youth Fund of China (No. 81702411).
Availability of data and materials
The data and materials can be found in the supplementary documents.
Ethics approval and consent to participate
The study was executed with the ethical approval for scientific research projects under medical ethics committee of Zhongnan Hospital of Wuhan University (Scientific Ethical Approval NO. 2019079) and obtained written informed consent from all patients. All animal experiments were approved by laboratory animal welfare and ethics review of Wuhan University center for animal experiment (WP20210360).
Author contributions
Conception and design: Haitao Wu, Zhenxian Xiang, Shuyi Wang, Chaogang Yang, Bin Xiong.
Data analysis: Haitao Wu, Zhenxian Xiang, Guoquan Huang, Bin Xiong.
Experimental operation: Haitao Wu, Guoquan Huang, Zhenxian Xiang, Qiuming He, Jialin Song, Rongzhang Dou.
Manuscript writing: Haitao Wu.
Manuscript revision: Haitao Wu, Guoquan Huang, Zhenxian Xiang, Qiuming He.
Abbreviations
- CAFs
cancer-associated fibroblasts
- CAFLCs
cancer-associated fibroblasts like cells
- GC
gastric cancer
- PM
peritoneal metastasis
- EMT
epithelial-mesenchymal transition
- MCs
mesothelial cells
- BGN
biglycan
- FAP
fibroblast activation protein
- HPMCs
human peritoneal mesothelial cells
- MMT
mesothelial-to-mesenchymal transition
- α-SMA
α-smooth muscle actin
- ECM
extracellular protein
- TME
tumor microenvironment
- TLR
Toll-like receptor
- PANT
paired adjacent normal tissues
- rFAP
recombinant human FAP protein
- IHC
Immunohistochemistry
- anti-BGN
lentivirus mediated knockdown of BGN vectors
- anti-NC-BGN
lentivirus mediated negative control knockdown of BGN vectors
- Oe-BGN
lentivirus mediated overexpression of BGN vectors
- Oe-NC-BGN
lentivirus mediated negative control overexpression of BGN vectors
- GFP
green fluorescent protein
- sh
short hairpin knockdown RNA
- qRT-PCR
quantitative real-time polymerase chain reaction
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- WT
wild type
- MUT
mutation
- CHIP
chromatin immunoprecipitation
- IF
immunofluorescence staining
- CCK8
Cell Counting Kit-8
- OS
overall survival
- PFS
progression-free survival
- T stage
tumor invasion stage
- LNM
lymph node metastasis
- TNM
tumor-node-metastasis
- DM
distant metastasis
- CEA
carcinoembryonic antigen
- CA19-9
carbohydrate antigen 19-9
- h
hours
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- DMSO
Dimethyl sulfoxide
- ELISA
enzyme-linked immunosorbent assay
- PBS
phosphate-buffered saline
- TBST
tris-buffered saline tween-20
- SEM
standard error of the mean
- SPSS
Statistical Product and Service Solutions
- NC
negative control
- Co-IP
Coimmunoprecipitation
- STAT3-Ex
human plasmid expression STAT3
- STAT3-NC
human plasmid negative control expression STAT3
- IgG
Immunoglobulin G
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Thomassen I, van Gestel YR, van Ramshorst B, Luyer MD, Bosscha K, Nienhuijs SW. et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int J Cancer. 2014;134:622–8. doi: 10.1002/ijc.28373. [DOI] [PubMed] [Google Scholar]
- 3.Capobianco A, Cottone L, Monno A, Manfredi AA, Rovere-Querini P. The peritoneum: healing, immunity, and diseases. J Pathol. 2017;243:137–47. doi: 10.1002/path.4942. [DOI] [PubMed] [Google Scholar]
- 4.Deng G, Qu J, Zhang Y, Che X, Cheng Y, Fan Y. et al. Gastric cancer-derived exosomes promote peritoneal metastasis by destroying the mesothelial barrier. FEBS Lett. 2017;59:2167–79. doi: 10.1002/1873-3468.12722. [DOI] [PubMed] [Google Scholar]
- 5.Rynne-Vidal A, Au-Yeung CL, Jiménez-Heffernan JA, Pérez-Lozano ML, Cremades-Jimeno L, Bárcena C, Mesothelial-to-mesenchymal transition as a possible therapeutic target in peritoneal metastasis of ovarian cancer. 2017; 242: 140-51. [DOI] [PMC free article] [PubMed]
- 6.Sandoval P, Jiménez-Heffernan JA, Rynne-Vidal Á, Pérez-Lozano ML, Gilsanz Á, Ruiz-Carpio V. et al. Carcinoma-associated fibroblasts derive from mesothelial cells via mesothelial-to-mesenchymal transition in peritoneal metastasis. J Pathol. 2013;231:517–31. doi: 10.1002/path.4281. [DOI] [PubMed] [Google Scholar]
- 7.Gordillo CH, Sandoval P. Mesothelial-to-Mesenchymal Transition Contributes to the Generation of Carcinoma-Associated Fibroblasts in Locally Advanced Primary Colorectal Carcinomas. Cancers (Basel) 2020;12:499. doi: 10.3390/cancers12020499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rynne-Vidal A, Jiménez-Heffernan JA, Fernández-Chacón C, López-Cabrera M, Sandoval P. The Mesothelial Origin of Carcinoma Associated-Fibroblasts in Peritoneal Metastasis. Cancers (Basel) 2015;7:1994–2011. doi: 10.3390/cancers7040872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahai E, Astsaturov I. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–86. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park D, Sahai E, Rullan A. SnapShot: Cancer-Associated Fibroblasts. Cell. 2020;181:486. doi: 10.1016/j.cell.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: Cancer-associated fibroblasts and their markers. 2020; 146: 895-905. [DOI] [PMC free article] [PubMed]
- 12.Wang L, Xu Z, Hu C, Chen S, Du Y, Huang L. et al. Peritoneal metastatic gastric carcinoma cells exhibit more malignant behavior when co-cultured with HMrSV5 cells. Aging (Albany NY) 2020;12:3238–48. doi: 10.18632/aging.102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki M, Fushida S, Harada S, Tsukada T, Kinoshita J, Oyama K. et al. Establishing a xenograft mouse model of peritoneal dissemination of gastric cancer with organ invasion and fibrosis. BMC Cancer. 2017;17:23. doi: 10.1186/s12885-016-2991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang TC, Li X, He YN, Wang YH, Shen JJ, Wang SL. et al. Cancer-associated fibroblasts-derived HAPLN1 promotes tumour invasion through extracellular matrix remodeling in gastric cancer. Gastric Cancer. 2022;25:346–59. doi: 10.1007/s10120-021-01259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher LW, Heegaard AM, Vetter U, Vogel W, Just W, Termine JD. et al. Human biglycan gene. Putative promoter, intron-exon junctions, and chromosomal localization. J Biol Chem. 1991;266:14371–7. [PubMed] [Google Scholar]
- 16.Zhao SF, Yin XJ, Zhao WJ, Liu LC, Wang ZP. Biglycan as a potential diagnostic and prognostic biomarker in multiple human cancers. Oncol Lett. 2020;19:1673–82. doi: 10.3892/ol.2020.11266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Zhong A, Li S, Meng X, Wang X, Xu F. et al. The integrated pathway of TGFβ/Snail with TNFα/NFκB may facilitate the tumor-stroma interaction in the EMT process and colorectal cancer prognosis. Sci Rep. 2017;7:4915. doi: 10.1038/s41598-017-05280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz GB, Grimm T, Sers C, Riemer P, Elmasry M, Kirchner T. et al. Prognostic value and association with epithelial-mesenchymal transition and molecular subtypes of the proteoglycan biglycan in advanced bladder cancer. Urol Oncol. 2019;37:530. doi: 10.1016/j.urolonc.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Zang MD, Wang HX, Li JF, Su LP, Yan M. et al. Biglycan stimulates VEGF expression in endothelial cells by activating the TLR signaling pathway. Mol Oncol. 2016;10:1473–84. doi: 10.1016/j.molonc.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang HC, Cai BH, Suen CS, Lee HY, Hwang MJ, Liu FT. et al. BGN/TLR4/NF-B Mediates Epigenetic Silencing of Immunosuppressive Siglec Ligands in Colon Cancer Cells. Cells. 2020;9:397. doi: 10.3390/cells9020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Zhang L, Li M, Zhang Y, Sun M, Wang L. et al. Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nat Commun. 2022;13:1248. doi: 10.1038/s41467-022-28913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh JM, Venters C, Di C, Pinto AM, Wan L, Younis I, Cai Z. et al. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat Commun. 2020;11:1. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G. et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. doi: 10.1038/ncomms9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahai E, Astsaturov I. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–86. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida GJ. Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathways. J Exp Clin Cancer Res. 2020;39:112. doi: 10.1186/s13046-020-01611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev. 2021;101:147–76. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Lin Y, Shi Y, Li B, Liu W, Yin W. et al. FAP Promotes Immunosuppression by Cancer-Associated Fibroblasts in the Tumor Microenvironment via STAT3-CCL2 Signaling. Cancer Res. 2016;76:4124–35. doi: 10.1158/0008-5472.CAN-15-2973. [DOI] [PubMed] [Google Scholar]
- 30.Puré E, Blomberg R. Pro-tumorigenic roles of fibroblast activation protein in cancer: back to the basics. Oncogene. 2018;37:4343–57. doi: 10.1038/s41388-018-0275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 32.Long Y, Lu Z, Xu S, Li M, Wang X, Zhang Z. Self-Delivery Micellar Nanoparticles Prevent Premetastatic Niche Formation by Interfering with the Early Recruitment and Vascular Destruction of Granulocytic Myeloid-Derived Suppressor Cells. Nano Lett. 2020;20:2219–29. doi: 10.1021/acs.nanolett.9b03883. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y, Bian S, Zhou X, Cui Y, Wang W, Wen L. et al. Single-Cell Multiomics Sequencing Reveals Prevalent Genomic Alterations in Tumor Stromal Cells of Human Colorectal Cancer. Cancer Cell. 2020;38:818–28. doi: 10.1016/j.ccell.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Pinto F, Santos-Ferreira L. The Extracellular Small Leucine-Rich Proteoglycan Biglycan Is a Key Player in Gastric Cancer Aggressiveness. Cancers (Basel) 2021;13:1330. doi: 10.3390/cancers13061330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobsen F, Kraft J, Schroeder C, Hube-Magg C, Kluth M, Lang DS. et al. Up-regulation of Biglycan is Associated with Poor Prognosis and PTEN Deletion in Patients with Prostate Cancer. Neoplasia. 2017;19:707–15. doi: 10.1016/j.neo.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morimoto H, Hida Y. Biglycan, tumor endothelial cell secreting proteoglycan, as possible biomarker for lung cancer. Thorac Cancer. 2021;12:1347–57. doi: 10.1111/1759-7714.13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, Wang X, Zhang Y, Che X, Liu Z, Zhang L. et al. Biglycan enhances the ability of migration and invasion in endometrial cancer. Arch Gynecol Obstet. 2016;293:429–38. doi: 10.1007/s00404-015-3844-5. [DOI] [PubMed] [Google Scholar]
- 38.Andrlová H, Mastroianni J, Madl J, Kern JS, Melchinger W, Dierbach H. et al. Biglycan expression in the melanoma microenvironment promotes invasiveness via increased tissue stiffness inducing integrin-β1 expression. Oncotarget. 2017;8:42901–16. doi: 10.18632/oncotarget.17160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Huang C, Peng C, Xu F, Li Y, Yutaka Y. et al. Stromal fibroblast activation protein alpha promotes gastric cancer progression via epithelial-mesenchymal transition through Wnt/ β-catenin pathway. BMC Cancer. 2018;18:1099. doi: 10.1186/s12885-018-5035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higashino N, Koma YI, Hosono M, Takase N, Okamoto M, Kodaira H. et al. Fibroblast activation protein-positive fibroblasts promote tumor progression through secretion of CCL2 and interleukin-6 in esophageal squamous cell carcinoma. Lab Invest. 2019;99:777–92. doi: 10.1038/s41374-018-0185-6. [DOI] [PubMed] [Google Scholar]
- 41.Ma TH, Gao CC, Xie R, Yang XZ, Dai WJ, Zhang JL. et al. Predictive values of FAP and HGF for tumor angiogenesis and metastasis in colorectal cancer. Neoplasma. 2017;64:880–6. doi: 10.4149/neo_2017_609. [DOI] [PubMed] [Google Scholar]
- 42.Wen Z, Liu Q, Wu J, Xu B, Wang J, Liang L. et al. Fibroblast activation protein α-positive pancreatic stellate cells promote the migration and invasion of pancreatic cancer by CXCL1-mediated Akt phosphorylation. Ann Transl Med. 2019;7:532. doi: 10.21037/atm.2019.09.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreno-Ruiz P, Corvigno S, Te Grootenhuis NC, La Fleur L, Backman M, Strell C. et al. Stromal FAP is an independent poor prognosis marker in non-small cell lung adenocarcinoma and associated with p53 mutation. Lung Cancer. 2021;155:10–9. doi: 10.1016/j.lungcan.2021.02.028. [DOI] [PubMed] [Google Scholar]
- 44.Wei M, Yang T, Chen X, Wu Y, Deng X, He W. et al. Malignant ascites-derived exosomes promote proliferation and induce carcinoma-associated fibroblasts transition in peritoneal mesothelial cells. Oncotarget. 2017;8:42262–71. doi: 10.18632/oncotarget.15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Li B, Li Q, Wei S, He Z, Huang X. et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018;9:854. doi: 10.1038/s41419-018-0928-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Liu C, Chang X, Qi Y, Zhu Z, Yang X. Fibrosis of mesothelial cell-induced peritoneal implantation of ovarian cancer cells. Cancer Manag Res. 2018;10:6641–7. doi: 10.2147/CMAR.S183043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Z, Yao J, Wang F, Xu Q. TNF-alpha and the phenotypic transformation of human peritoneal mesothelial cell. Chin Med J (Engl) 2002;115:513–7. [PubMed] [Google Scholar]
- 48.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18:99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 49.Aertgeerts K, Levin I, Shi L, Snell GP, Jennings A, Prasad GS. et al. Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J Biol Chem. 2005;280:19441–4. doi: 10.1074/jbc.C500092200. [DOI] [PubMed] [Google Scholar]
- 50.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–96. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garg M, Shanmugam MK, Bhardwaj V, Goel A, Gupta R, Sharma A. et al. The pleiotropic role of transcription factor STAT3 in oncogenesis and its targeting through natural products for cancer prevention and therapy. Med Res Rev. 2020 doi: 10.1002/med.21761. doi: 10.1002/med.21761. [DOI] [PubMed] [Google Scholar]
- 52.Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in Cancer Immunotherapy. Mol Cancer. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table.
Data Availability Statement
The data and materials can be found in the supplementary documents.