Abstract
The relationship between ingestion of diets amended with a Pb-contaminated soil and the composition of the fecal microbiome was examined in a mouse model. Mice consumed diets amended with a Pb-contaminated soil in its native (untreated) state or after treatment for remediation with phosphoric acid or triple superphosphate alone or in combination with iron-waste material or biosolids compost. Subacute dietary exposure of mice receiving treated soil resulted in modulation of the fecal intestinal flora, which coincided with reduced relative Pb bioavailability in the bone, blood and kidney and differences in Pb speciation compared to untreated soil. Shifts in the relative abundance of several phyla including Verrucomicrobia, Tenericutes, Firmicutes, Proteobacteria, and TM7 (Candidatus Saccharibacteria) were observed. Because the phyla persist in the presence of Pb, it is probable that they are resistant to Pb. This may enable members of the phyla to bind and limit Pb uptake in the intestine. Families Ruminococcaceae, Lachnospiraceae, Erysipelotrichaceae, Verrucomicrobiaceae, Prevotellaceae, Lactobacilaceae, and Bacteroidaceae, which have been linked to health or disease, also were modulated. This study is the first to explore the relationship between the murine fecal microbiome and ingested Pb contaminated soils treated with different remediation options designed to reduce bioavailability. Identifying commonalities in the microbiome that are correlated with more positive health outcomes may serve as biomarkers to assist in the selection of remediation approaches that are more effective and pose less risk.
Keywords: Fecal microbiome, smelting soil, lead, triple superphosphate, phosphoric acid, biosolids, iron-rich waste, bioavailability
1. Introduction
For centuries, mining and smelting of lead (Pb) has contaminated soils, sediments and ground and surface waters worldwide (Moore & Luoma, 1990; Smith et al., 2021; Kozlov & Zvereva, 2007; Rowan et al., 1995; Eichler et al., 2014; Gamiño-Gutiérrez et al., 2013; Gillanet al., 2015; Jones et al., 1991). In children, soil and dust continue to be major sources of Pb exposure (Dong, Taylor, & Gulson, 2020; Fry, Wheeler, Gillings, Flegal, & Taylor, 2020). As an example, air emissions near the Port Pirie smelter site contained up to 16 μg/m3 Pb which has been linked to elevated Pb in children (Taylor, 2019). In 2021, ~76% of two-year old Port Pirie children tested had blood lead levels > 5 μg/dL (Government of South Australia, 2022). The predominant role of these routes for Pb exposure is reflected in the integrated exposure uptake biokinetic model (IEUBK; USEPA, 1994; Hogan et al., 1998; Laidlaw et al., 2017) and in more recently developed models such as the Stochastic Human Exposure and Dose Simulation (SHEDS) Multimedia IEUBK model (Zartarian et al., 2017). Mouse (Kang et al., 2016; Juhasz et al., 2014; Bradham et al., 2018; Bradham et al., 2019; Breton et al., 2013), rat (Freeman et al., 1994; Brown et al., 2004), juvenile swine (Casteel, Weis, Henningsen, & Brattin, 2006), minipig (Marschner, Welge, Hack, Wittsiepe, & Wilhelm, 2006) and goat (Cretacci & Parsons, 2010) models have been used to assess Pb bioavailability, however the role of the gut microbiome in soil Pb bioavailability has not been explored. Both abiotic and biotic processes can alter the physical and chemical properties of Pb present in soil. Ultimately these transformations can affect the bioavailability of Pb that is ingested or inhaled. For example, phosphate solubilizing microbes in soil can promote the formation of poorly soluble Pb-phosphate compounds (Park et al., 2011; Teng et al., 2019; Yuan et al., 2017; Zhang et al., 2019). In addition, the presence of Pb in soil can eliminate sensitive species and select for resistant or tolerant organisms (Gillan et al., 2015; Hynninen et al., 2009; Jie et al., 2016; Van Houdt et al., 2009). These Pb-induced changes in the microbial landscape may affect the biogeochemical cycling of Pb in soil. Through resistance mechanisms, Pb is sequestered in less soluble forms (phosphates, sulfides, or carbonates) or can be adsorbed to exopolysaccharide or on the cell surface making it less available for biological uptake and transformation (George & Wan, 2020). Similarly, ingestion of Pb can alter the intestinal microbiome, producing both structural and functional anomalies that impact gut metabolism (Breton et al., 2013; Gao et al., 2017). For example, soil Pb exposure of individuals residing near a mining and smelting area correlated with changes in the intestinal microbiota (Shao & Zhu, 2020). Furthermore, increased Pb exposure has been shown to concurrently select for microbial Pb resistance genes and antibiotic resistance genes (El-Sayed, 2016; Kang & So, 2016; Koc et al., 2013; Matyar, 2012; Fatih Matyar et al., 2014). This Pb-induced change in intestinal microbiota may increase the risk of antibiotic failure in disease treatment (Bengtsson-Palme et al., 2018; Wright, 2010).
As part of a larger effort to understand the role of the gastrointestinal tract microbiota in the bioavailability of Pb in soil, various remediation strategies were examined for their role in modulating the composition of the fecal microbiomes of mice that ingested diets amended with remediated soils. Because soil Pb is an important source of Pb exposure in children, reducing the likelihood of soil ingestion has long been a central element in strategies to prevent childhood Pb poisoning (Gailey et al., 2020). This goal has often been attained by removal and replacement of Pb-contaminated soil (Henry et al., 2015). However, this is a complex and costly strategy that can be difficult to implement on a large scale. Alternatively, in situ solidification and stabilization of Pb can be used to remediate contaminated soils (National Research Council, 1997). Typically, chemical remediation of a Pb-contaminated soil involves in situ conversion of Pb into a poorly soluble form that, if ingested, will have decreased bioavailability (Bolan et al., 2014). Lead-contaminated soils are often remediated by treatment with phosphorus-containing compounds (e.g., rock phosphate, triple superphosphate [TSP], phosphoric acid [PA]) to promote formation of recalcitrant Pb phosphates, such as pyromorphite (Basta et al., 2001; Bradham et al., 2018; Bradham et al., 2016; Brown et al., 2004; Hettiarachchi et al., 2000; Scheckel et al., 2013).
Interest in remediation to reduce the bioavailability of Pb in treated soils has prompted the use of animal models to assess the efficacy of soil treatment. For example, the mouse model has been used to compare the relative bioavailability (RBA) of Pb in soil from a Superfund site in Joplin, Missouri, that was amended with phosphate alone on in combination with iron-rich waste material or biosolids compost (Bradham et al., 2018). This study found that treatment of soil with phosphate produced changes in the speciation of Pb in soil. When measured in the mouse model, the RBA estimates for Pb in treated soils were significantly lower than that obtained for Pb in the untreated soil. Treating soil with phosphoric acid reduced Pb RBA in bone by 32%. Triple superphosphate treated soil resulted in a 50% reduction in Pb RBA in bone; addition of iron waste or biosolids reduced RBA further to 64% and 74%, respectively. These findings suggested that remediation with phosphate could be an effective approach for the long-term reduction of Pb bioavailability. However, these studies do not provide any insights into other possible effects of soil treatment on potential interactions between Pb and organisms that ingest untreated or treated soil. Although there exists some information on the effects of ingested Pb on the intestinal microbiome (Breton et al., 2013; Gao et al., 2017; Xia et al., 2018), it is unclear how remediation-induced changes in Pb speciation might affect microorganisms resident in the gastrointestinal tract. This study is the first to explore the relationship between the fecal microbiome and ingested Pb contaminated soils with and without remediation treatments in a mouse model. While treating the soil with PA, or TSP with and without biosolids and/or iron-rich waste has been linked to a reduction in Pb bioavailability in the mouse, better understanding of the impact of remediated soil compared to the native soil on the mouse fecal microbiome has not been explored. Understanding this relationship between microbiome composition and the speciation of Pb in treated soils may provide new insights into the utility of various remediation methods. This information may help risk assessors and risk managers in selection of an optimal treatment for a specific contaminated site.
2. Materials and Methods
2.1. Soil and remediation treatment
Soils were obtained from a field trial designed to determine remediation alternatives for Pb smelter emission contaminated soil (Joplin, MO, USA). Site information, soil treatments, sample collection and processing are provided in Bradham et al. (2018) and summarized in Table 1. Briefly, three amendments were applied to field trial soil: (1) soil amended with 1% P as phosphoric acid (PA) , (2) 3.2% P as TSP (TSP), (3) 2.5% Fe as iron-rich waste material and 1% P as TSP (Fe/TSP), and (4) 10% biosolids compost and 1% P as TSP (C/TSP). Soil collected from the field trial site before treatment was designated as untreated soil (control). PA treated soil was collected 3 years after application and samples of soils treated with TSP, Fe/TSP, or C/TSP were collected 16 y after treatment. Metal concentrations in untreated and treated smelter impacted soil are provided in Table 1 (Bradham et al., 2018).
Table 1.
Summary of soil treatments, elemental analysis (mean±95% confidence limits) and relative lead bioavailability (means ± standard error). Reduction in lead bioavailability related to each soil treatment is calculated relative to untreated soil [(slopetreated/slopeuntreated) x 100] and shown as a percent (Bradham et al., 2018).
| Soil Treatments |
Control | 3.2% P as TSP Triple Superphosphate |
2.5% Fe as Fe rich waste + 1% P as Triple Superphosphate |
10% C as Biosolids + 1% P as Triple Superphosphate |
1% P as Phosphoric Acid |
|---|---|---|---|---|---|
| Abbreviation | Control | TSP | Fe/TSP | C/TSP | PA |
| Duration (years) | 0 | 16 | 16 | 16 | 3 |
| Elemental Analysis of Soils (ppm) | |||||
| Pb | 3055.2 ± 1321.1 | 2136.8 ± 1291.3 | 1992.5 ± 1743.8 | 2684.7 ± 1763.9 | 3088.9 ± 1079.5 |
| Zn | 3813.3 ± 1316.8 | 2950.6 ± 1363.2 | 2587.9 ± 2060.2 | 3354.5 ± 1787.6 | 3579.7 ± 1094.1 |
| Cd | 17.1 ± 6.0 | 12.9 ± 5.7 | 10.5 ± 6.8 | 14.5 ± 7.2 | 15.7 ± 4.1 |
| Cu | 34. 8 ± 16.5 | 21.1 ± 6.9 | 19.6 ± 6.4 | 38.0 ± 9.8 | 28.2 ± 10.0 |
| Ni | 13.6 ± 1.8 | 13.0 ± 2.3 | 19.2 ± 4.9 | 11.5 ± 2.3 | 11.5 ± 0.6 |
| As | 7.8 ± 2.4 | 6.2 ± 0.8 | 5.2 ± 1.0 | 5.9 ± 0.6 | 4.1 ± 0.4 |
| P | 683.2 ± 190.8 | 15024.0 ± 9177.1 | 3785.6 ± 2503.4 | 8085.4 ± 2796.3 | 6687.2 ± 1979.4 |
| Soil Treatment Effect on Relative Lead Bioavailability | |||||
| Bone | 100 | 54 ± 0.09 | 45 ± 0.11 | 28 ± 0.09 | 69 ± 0.09 |
| Blood | 100 | 57 ± 0.07 | 45 ± 0.08 | 47 ± 0.07 | 52 ± 0.08 |
| Kidney | 100 | 68 ± 0.09 | 53 ± 0.10 | 55 ± 0.08 | 67 ± 0.07 |
2.2. Assessment of Pb bioavailability in the mouse
A mouse-based assay for determination of soil Pb bioavailability has been described (Bradham et al., 2016). Briefly, twenty-eight day old female C57BL/6 mice (Charles River Laboratories, Raleigh, NC, USA) were acclimated for 12-13 days and then housed in groups of three in metabolic cages (3 cages per soil treatment [control, TSP, Fe/TSP, C/TSP, or PA] for a total of nine mice for each treatment; Lab Products, Seaford, DE). During the 9-day assay, mice had free access to drinking water and powdered AIN-93G rodent diet (Dyets, Bethlehem, PA) that was amended with 0.6% (w/w) of soil treated with PA, TSP with and with biosolids or iron rich waste or untreated soil (control). Test diets contained 21.6 (control), 22.2 (TSP), 14.3 (Fe/TSP), 24.2 (C/TSP), and 24.2 (PA) ppm Pb. For each metabolic cage, intakes of diet and water were monitored daily and each day’s feces from each cage were combined to produce a cumulative cage sample (3 cumulative fecal samples for each soil treatment). Relative bioavailability of Pb was determined by calculating linear regression slopes (m) for cumulative Pb intake (mg) and tissue Pb level (mg kg−1 or mg l−1) for treated and untreated soils and the following equation, mtreated/muntreated (Bradham et al., 2018). Homogenized fecal samples were stored at −20° C until processed for DNA extraction. Compared to mice that received control AIN-93G rodent diet, ingestion of soil-amended diets did not have significant adverse effects on absolute or relative changes in body weight or on diet or water consumption.
2.3. 16S rRNA gene sequencing
Homogenized fecal samples were shipped frozen (−20° C) to the DNA Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory (Lemont, Illinois). There, DNA was extracted and the 16S rRNA gene V4 region of Bacteria and Archaea was amplified by polymerase chain reaction (PCR) with universal primers using extracted DNA as template. The PCR and amplicon sequencing were performed on a MiSeq DNA sequencer platform (Illumina, Inc., San Diego, CA) according to procedures used by the Earth Microbiome Project (Thompson et al., 2017).
2.4. Data analysis
DNA sequences were processed with mothur software using a standard set of commands (Schloss et al., 2009). The split.cluster command was used to construct the distance matrix and sequences having >97% nucleotide identities were clustered within an Operational Taxonomic Unit (OTU). Taxonomic identifications used the SILVA v135 non-redundant 16S rRNA sequence data base (Quast et al., 2012). Sequences were randomly subsampled to obtain a constant 5,000 sequences sample−1 for microbiome comparisons.
Statistical analyses by permutation of ranks, and calculations of diversity and evenness, were done with Primer-e 7 (Clarke & Warwick, 2001). 16S rRNA gene sequences OTU−1 sample−1 were transformed (fourth root) and a Bray-Curtis resemblance matrix and non-metric multidimensional scaling (nMDS) plots generated (Clarke etal., 2014). OTU, family, and phylum level dendrograms based on Bray-Curtis resemblance and similarity profile (SIMPROF) permutations were applied to highlight statistically significant clusters. PERMANOVA, a permutational multivariate analysis of variance, was used to test the significance of Bray-Curtis dissimilarities between treatments (Anderson, 2008). The community diversity (Shannon, H), evenness (Pielou, J), and richness (Margalef, S) indices were obtained from Primer for each replicate of the treatment. Treatment effects and difference among the means were determined with the one-way analysis of variance (ANOVA) followed by a Tukey test in R (R Core Team, 2014). A significance level of p<0.05 was selected for detection of treatment effects.
3. Results
3.1. Fecal microbiome similarities
Within replicate treatments, the fecal microbiomes were 82% to 93% similar to each other when aggregated at the genus, family and phylum levels for all treatments (Table S1). When considering all OTUs, similarity was reduced to 34-36%; when the 50 most prevalent OTUs were analyzed, similarity increased to 88-90%. Diversity (Shannon, H), richness (Margalef, S) and evenness (Pielou, J) indices, calculated at the genus and family levels, revealed only limited significant differences with C/TSP (Table 2). Diversity of the fecal microbiome differed in mice on an amended diet of C/TSP treated soil compared to control at the genus (p=0.0529) and family (p=0.0063) levels. At the family level, diversity in the microflora of mice that received diets amended with TSP (p=0.0895), Fe/TSP (p=0.0334), and PA (p=0.0633) were less similar to the C/TSP treated group; at the genus level, microflora from Fe/TSP (p=0.0198) and PA (p=0.0771) treated soil fed mice were different from those that received C/TSP treated soil. C/TSP treatment also contributed to changes in fecal community evenness compared to control (p=0.0471) and Fe/TSP (p=0.0739) treatments.
Table 2.
Fecal microbiome diversity indices. Richness (Margalef, S), evenness (Pielou, J), and diversity (Shannon, H) and were determined for the fecal microbiome of mice that received treated or untreated (control) soils in their diet. ANOVA, p<0.05, significant**; 0.05<p<0.08 marginally significant*; Tukey multiple comparison of means, a significantly different than b within index p<.
| Taxon | Diversity Index |
Control | TSP | Fe/TSP | C/TSP | PA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | sem1 | Value | sem | Value | sem | Value | sem | Value | sem | ||
| Genus | S | 81.5 | 0.7 | 80.7 | 4.7 | 89.5 | 5.4 | 83.7 | 1.0 | 75.2 | 3.4 |
| J* | 0.5 | 0.0 | 0.5 | 0.0 | 0.5 | 0.0 | 0.6 | 0.0 | 0.5 | 0.0 | |
| H** | 2.3b* | 0.0 | 2.4 | 0.0 | 2.3b** | 0.1 | 2.5a | 0.0 | 2.3b* | 0.0 | |
| Family | S | 56.50 | 0.77 | 56.33 | 3.41 | 62.00 | 4.84 | 56.17 | 1.34 | 52.17 | 1.85 |
| J** | 0.54b** | 0.01 | 0.56 | 0.01 | 0.54b* | 0.02 | 0.59a | 0.01 | 0.56 | 0.01 | |
| H** | 2.16b** | 0.02 | 2.23b* | 0.03 | 2.21b* | 0.04 | 2.37a | 0.04 | 2.22b* | 0.03 | |
Standard error of mean, sem
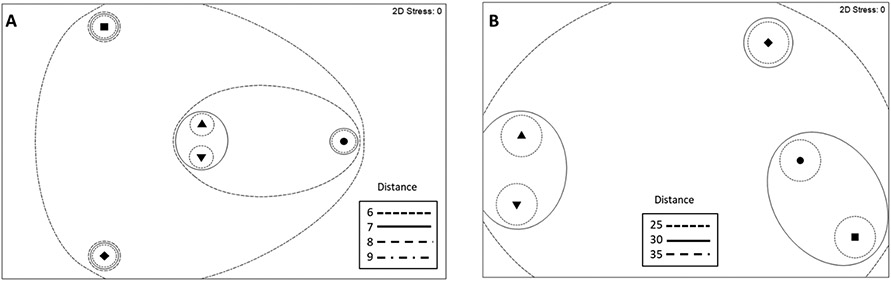
Bray-Curtis dissimilarity analysis, using non-metric multidimensional scaling, examined the relatedness of fecal microbiomes from mice that consumed diets amended with untreated or treated soils at the OTU and the family levels (Fig. 1). OTUs from feces of mice that consumed Fe/TSP and PA amended diets were more closely related as were TSP and untreated soil groups. C/TSP separated from the other treatment groups. Similar results were observed when the OTUs were aggregated at the family level. However, fecal microbiomes from mice that consumed PA amended diets clustered more closely with those from mice that consumed diets amended with untreated and TSP treated soil. PERMANOVA analysis revealed an overall treatment effect at the OTU level and when aggregated at the genus, family, and phylum levels (Table 3). In pairwise comparisons at the OTU, genus, and family levels, fecal microbiomes of mice that consumed diets amended with TSP, Fe/TSP, C/TSP and PA treated soil differed significantly from those with untreated soil. All other pairwise comparisons among treatments revealed significant differences at all taxon levels except when aggregated at the phylum level, whereby only the ingestion of diet amended with PA-treated soil resulted in a significant change to the fecal microbiome compared to the fecal microbiome with untreated soil. Microflora from mice that ingested diet amended with PA treated soil also were significantly different from microflora in feces of mice that ingested diet amended with Fe/TSP or C/TSP treated soil.
Fig. 1.
Bray-Curtis resemblance matrix and non-metric multidimensional scaling (nMDS) of fecal microbiome among remediated and control treatments for all OTUs (A) and when aggregated at the family level (B). Dietary amendment of soil treatments are as follows: untreated, triangle; TSP, inverted triangle; Fe/TSP, square; C/TSP, diamond; PA, circle.
Table 3.
Treatment effects on the murine fecal microbiome. PERMANOVA and t-tests were performed to determine differences in the fecal microbiome at each taxon level among treatments with dietary treated (TSP, Fe/TSP, C/TSP, PA) or untreated (control) soil; p<0.05, significant; 0.05<p<0.07 marginally significant (italics).
| Group Comparison |
df | t | P(perm) | Unique permutations |
|---|---|---|---|---|
| OTU | ||||
| All Amendments | 29 | 0.0001 | 9219 | |
| control x TSP | 10 | 1.1267 | 0.0054 | 460 |
| control x Fe/TSP | 10 | 1.2146 | 0.0027 | 462 |
| control x C/TSP | 10 | 1.1921 | 0.0018 | 461 |
| control x PA | 10 | 1.2206 | 0.0034 | 461 |
| TSP x Fe/TSP | 10 | 1.2157 | 0.0025 | 461 |
| TSP x C/TSP | 10 | 1.2274 | 0.0026 | 458 |
| TSP x PA | 10 | 1.2316 | 0.0017 | 462 |
| Fe/TSP x C/TSP | 10 | 1.1743 | 0.0029 | 459 |
| Fe/TSP x PA | 10 | 1.1689 | 0.002 | 460 |
| C/TSP x PA | 10 | 1.2172 | 0.0015 | 461 |
| Genus | ||||
| All Amendments | 29 | 0.001 | 999 | |
| control x TSP | 10 | 1.1778 | 0.053 | 407 |
| control x Fe/TSP | 10 | 1.3951 | 0.004 | 408 |
| control x C/TSP | 10 | 1.4807 | 0.004 | 421 |
| control x PA | 10 | 1.5277 | 0.003 | 418 |
| TSP x Fe/TSP | 10 | 1.4744 | 0.001 | 401 |
| TSP x C/TSP | 10 | 1.4431 | 0.008 | 403 |
| TSP x PA | 10 | 1.4206 | 0.003 | 414 |
| Fe/TSP x C/TSP | 10 | 1.4467 | 0.003 | 412 |
| Fe/TSP x PA | 10 | 1.541 | 0.002 | 410 |
| C/TSP x PA | 10 | 1.8144 | 0.001 | 414 |
| Family | ||||
| All Amendments | 29 | 0.0001 | 9814 | |
| control x TSP | 10 | 1.202 | 0.028 | 407 |
| control x Fe/TSP | 10 | 1.4994 | 0.006 | 414 |
| control x C/TSP | 10 | 1.7341 | 0.003 | 420 |
| control x PA | 10 | 1.6124 | 0.002 | 412 |
| TSP x Fe/TSP | 10 | 1.4627 | 0.017 | 401 |
| TSP x C/TSP | 10 | 1.438 | 0.005 | 401 |
| TSP x PA | 10 | 1.3524 | 0.01 | 406 |
| Fe/TSP x C/TSP | 10 | 1.5588 | 0.001 | 403 |
| Fe/TSP x PA | 10 | 1.6851 | 0.004 | 416 |
| C/TSP x PA | 10 | 1.8469 | 0.002 | 404 |
| Phylum | ||||
| All Amendments | 29 | 0.0031 | 9909 | |
| control x TSP | 10 | 1.3545 | 0.126 | 410 |
| control x Fe/TSP | 10 | 1.2784 | 0.164 | 413 |
| control x C/TSP | 10 | 1.5367 | 0.066 | 411 |
| control x PA | 10 | 1.842 | 0.013 | 408 |
| TSP x Fe/TSP | 10 | 1.3952 | 0.067 | 421 |
| TSP x C/TSP | 10 | 0.94343 | 0.524 | 402 |
| TSP x PA | 10 | 1.2544 | 0.175 | 414 |
| Fe/TSP x C/TSP | 10 | 1.5042 | 0.054 | 409 |
| Fe/TSP x PA | 10 | 1.5151 | 0.04 | 408 |
| C/TSP x PA | 10 | 1.5619 | 0.05 | 413 |
3.2. Mice fecal microbiome composition
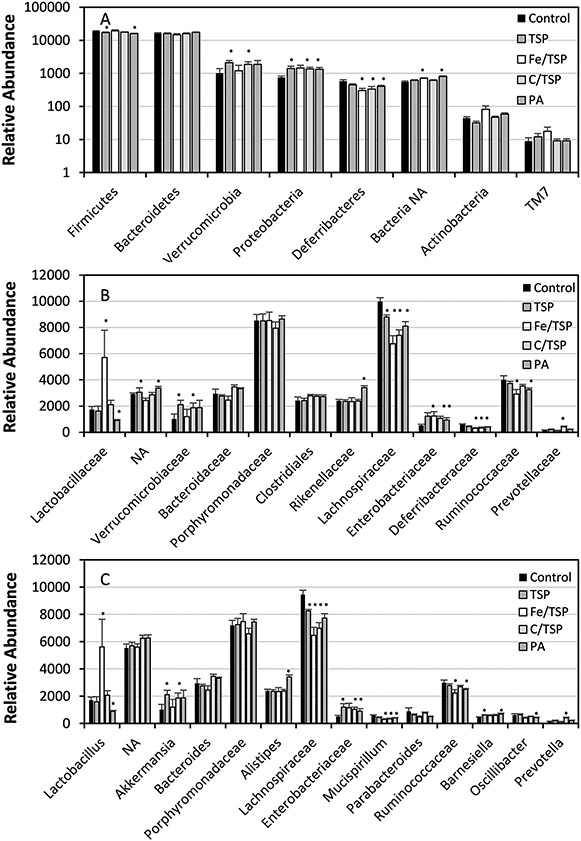
Phyla Firmicutes (44-52%), Bacteroidetes (39-49%), Proteobacteria (2-4%) and Verrucomicrobia (<5%) sequences were prevalent in the feces of mice from all treatments (Fig. 2A), with lower abundances of Deferribacteres (<2%), Actinobacteria (<1%), and TM7 (Candidatus Saccharibacteria; <1%). Relative to control soil, PA had a treatment effect across prevalent phyla; Bacteroidetes and Proteobacteria sequences were higher and Firmicutes and Deferribacteres sequences were lower. When analyzed individually, a treatment effect was observed for Firmicutes (p=0.0435) in feces from mice with all exposures; Bacteroidetes were not significantly different (p=0.3858). Relative abundance of Firmicutes was significantly lower in mice that consumed PA (p=0.0082) treated soil compared to its occurrence in the feces of mice that consumed diets amended with Fe/TSP (p=0.037), C/TSP (p=0.0429) treated soils or with untreated soil (p=0.0082).
Fig. 2.
Relative abundance of dominant phyla (A); families(B); genera (C) (fourth root transformed) in feces of mice receiving dietary amendments of treated and untreated (control) soil. Error bars are standard errors of means. * denotes significant difference of the treatment against the control (p<0.05).
Bacteroidaceae (Bacteroidetes), Lachnospiraceae (Firmicutes), Lactobacillaceae (Firmicutes), Porphyromonadaceae (Bacteroidetes), Rikenellaceae (Bacteroidetes), Ruminococcaceae (Firmicutes), and Verrucomicrobiaceae (Verrucomicrobia) were dominant families found in fecal microbiomes for all treatments (Fig. 2B; Table S2). Also observed were dominant members of the order Clostridiales (Firmicutes) that were not identifiable at the family level. Relative abundances of Enterobacteriaceae were elevated in feces from all treatment groups compared to control; Lachnospiraceae were elevated in mice that that ingested diets amended with Fe/TSP or C/TSP. Compared to effects of consumption of diet amended with untreated soil, consumption of diet amended with Fe/TSP or PA treated soil resulted in increased or reduced Lactobacillaceae, respectively. Consumption of diet amened with TSP, C/TSP and PA treated soil increased Verrucomicrobiaceae relative abundance almost 2-fold increase. Consumption of C/TSP treated soil increased Prevotellaceae (Bacteroidetes) relative abundance compared to untreated and treated soils.
When aggregated at the genus level, Lactobacillus (Firmicutes), Akkermansia (Verrucomicrobia), Bacteroides (Bacteroidetes), Alistipes (Bacteroidetes), Mucispirillum (Deferribacteres), Parabacteroides (Bacteroidetes), Oscillibacter (Firmicutes) dominated in all treatment groups (Fig. 2C, Table S3). Prevalent OTUs in all treatment identifiable at the genus level were Lactobacillus (OTU 1), Akkermansia (OTU 3), Bacteroides (OTU 4), Alistipes (OTU 8), Mucispirillum (OTU 23), Oscillibacter (OTU37), and Parabacteroides (OTU 25) (Table S3). Barnesiella (OTU 31 and/or OTU 35; Bacteroidetes) was identified in the top 20% of microflora from feces of animals that received dietary exposure to amended soils and at lower abundance in control soil. When aggregated at the genus level, these and additional genera contributing to similarity included Dorea (Firmicutes), Allobaculum (Firmicutes), Enterococcus (Firmicutes), Parasutterella (Proteobacteria), Prevotella (Bacteroidetes), Pseudoflavonifractor (Firmicutes), Ruminococcus (Firmiutes), Staphylococcus (Firmicutes), Streptococcus (Firmicutes), and Clostridium (Firmicutes) (Table S4).
3.3. Contributions of bacterial groups to fecal microbiome dissimilarity
An overall statistically significant difference among treatments was observed when the 16S rDNA data was aggregated at the phylum level. Pairwise analysis also revealed varying level of differences in microbiome composition after ingestion of diets amended with untreated, Fe/TSP, or C/TSP, and PA treated soils (Table 3). Verrucomicrobia, Tenericutes, Acidobacteria, TM7, Cyanobacteria, Proteobacteria, Chloroflexi, and an unidentified Bacteria accounted for 70% of the dissimilarity in the microbiomes from mice after consumption of diets amended with PA treated or untreated soils (Table S5). Verrucomicrobia (20.8%) and Tenericutes (20.81%) were the greatest contributors to the dissimilarity observed; similar results were observed when comparing Fe/TSP and C/TSP treated soil amended diets. Verrucomicrobia and Tenericutes were responsible for the dissimilarity in microbiome composition of mice that ingested diet amended PA treated soils and mice that ingested diets amended with Fe/TSP treated (28.7%) or C/TSP treated (33.0%) soil.
Family, genus and OTU level PERMANOVA and pairwise analysis also revealed significant treatment effects (Table 3). Seventy percent of the family contributions to dissimilarity are shown in Table S6. A small number of families contributed to dissimilarities in the fecal microbiome of mice that ingested diets amended with untreated or treated soil; Verrucomicrobiaceae contributions trended the highest and ranged from 2.4% to 4.35%, and Staphylococcaceae (Firmicutes), 1.7% to 2.8%. Relative increases and decreases in average abundance of families detected in all treatment groups are shown in Table S7. Families in the class Rhizobiales (Proteobacteria) accounted for 0.8% to 1.6% in feces from mice treated with Fe/TSP, C/TSP, and PA; the family Deferribacteraceae (Deferribacteres) decreased following dietary exposure to soils treated with TSP, Fe/TSP, and C/TSP; Nocardioidaceae (Actinobacteria) was elevated in Fe/TSP and PA; and Geodermatophilaceae (Actinobacteria) contributed 0.8% and 1.0%, respectively, to dissimilarity in C/TSP and PA treatments compared to control. Average abundance of Gemmatimonadaceae (Gemmatimonadetes) was elevated and contributed 0.90% to dissimilarity in feces of mice treated with TSP treated soil; conversely, when TSP was supplemented with biosolids, consumption of an amended diet reduced the average abundance of Gemmatimonadaceae, contributing 0.8% to dissimilarity.
When aggregated at the genus level for all treatments, primary contributors to dissimilarity included Allobaculum and Ruminococcus, ranging from 2.2% to 3.7% and 2.2% to 2.7% contribution (Table S8). Clostridium contributed 3.1% and 3.0%, respectively, to the fecal microbiome in mice that consumed diet in which TSP was combined with Fe or C treatment. Akkermansia contributed 2.0% to dissimilarity to the fecal microbiome in PA amended soil treated mice compared to control; TSP, Fe/TSP, and C/TSP, 1.7%, 1.7%, and 1.5%, respectively. When comparing to controls, C/TSP treated soil dietary exposure of mice resulted in a two-fold increase in Akkermansia contribution to dissimilarity (2.2%) compared to the other treatments (1.0%).
Data from all OTUs (37,993) indicated that dissimilarity of fecal microbiomes between mice consuming diet amended with treated soils and those with untreated soil ranged from 64.2% to 66.5%. When analysis was restricted to the 50 most abundant OTUs the dissimilarity was reduced to 10.4% to 12. 5%, accounting for only about 3.2% of the total. Within the top ten OTUs, only OTU 34 (Lachnospiraceae) was evident in all treatment types. However, when the range was expanded to the top 50 contributors to dissimilarly comparing the treatment groups to control, 12 OTUs were common across the 4 diets amended with treated soils (Table S9). Of those, 75% were identified as Lachnospiraceae; 17% as Ruminococcaceae; and 8% as Allobaculum. The average abundances of Lachnospiraceae were generally greater in feces of mice that received dietary treated soils compared to control (Fig. 2C).
3.4. Indices of metabolic change: Firmicutes/Bacteroidetes and Bacilli/Clostridia
Intestinal Firmicutes/Bacteroidetes and Bacilli/Clostridia ratios have been linked to physiological and metabolic changes in the host. Therefore, statistical analyses of these phyla, classes, and their ratios were performed. When analyzed together, no overall treatment effect on the relative abundance of fecal Firmicutes and Bacteroidetes was observed in the feces of mice that received the treated soils in their diet (p=0.1616) (Table 4). However, pairwise analysis found significant differences (p=0.0071) in relative abundance of Firmicutes and Bacteroidetes in feces of mice that consumed diets amended with untreated and PA-treated soil. When phyla were examined individually, there was an overall significant difference (p=0.0435) for Firmicutes for all treated soils. Pairwise analysis indicated that PA treated soil reduced abundance of Firmicutes compared to untreated soil (p=0.0082) and decreased the Firmicutes/Bacteroidetes ratio from 1.17 to 0.91. Similarly, TSP amended soil marginally reduced the relative abundance of Firmicutes (p=0.0679), decreasing the Firmicutes/Bacteroidetes ratio to 1.08.
Table 4.
Relative abundance of phyla Firmicutes and Bacteroidetes and classes Bacilli and Clostridia and associated indices. Statistical analyses comparing fecal flora from mice that received treated (TSP, Fe/TSP, C/TSP, PA) or untreated (control) soil in their diet were performed to determine differences in the specific phyla and classes; p<0.05, significant (**); 0.05<p<0.08 marginally significant (*). See Supplemental Table 9 for PERMANOVA and pairwise t-test results.
| Phylum | Relative Abundance | ||||
|---|---|---|---|---|---|
| Control | TSP | Fe/TSP | C/TSP | PA | |
| Firmicutes | 18900 | 17331* | 19403 | 17673 | 15947** |
| Bacteroidetes | 16128 | 16032 | 14818 | 16058 | 17551 |
| Firmicutes/ Bacteroidetes | 1.17 | 1.08 | 1.31 | 1.1 | 0.99** |
| Class | |||||
| Bacilli | 2053 | 1900 | 6321 | 3002 | 1313 |
| Clostridia | 16504 | 15042 | 12727 | 14200 | 14209 |
| Bacilli/Clostrida | 0.12 | 0.13 | 0.5** | 0.21** | 0.09** |
An overall treatment effect was observed for fecal classes Bacilli and Clostridia (p=0.0002; Table S10). Furthermore, consumption of diets amended with Fe/TSP, C/TSP, or PA treated soil had a significant effect on fecal microflora compared to untreated soil group (Table S10). Relative abundance of Clostridia decreased in the three treatment groups; relative abundance of Bacilli was increased when mice received diet amended with Fe/TSP and C/TSP treated soil and decreased with PA treated soil, thus changing the Bacilli/Clostridia ratio from 0.12 to 0.5, 0.21, and 0.09, respectively (Table 4).
4. Discussion
While mammalian factors contribute to Pb transformation, such as dissociation of Pb species in the acidic stomach (Juhasz et al., 2014), the intestinal microbiome is well poised to contribute. Lead is toxic to many microorganisms causing the Pb resistant community to dominate and marginalizing sensitive members, thus altering gut morphology and barrier permeability. These changes can affect the physiological state by modulating carbohydrate, energy, nitrogen and vitamin E and bile acid metabolism and create inflammatory and prooxidant states (Gao et al., 2017; Xia et al., 2018). Modulations in the fecal microflora indicate that the toxic components in the soil matrix may be bioavailable. The shift to a more resistant microbial community may impact Pb bioavailability due to increased expression of resistance mechanisms which may result in Pb adsorption to the gut microbiome and subsequent excretion. Here, we examined effects of ingestion of diets amended with Pb-contaminated soil subjected to different remediation treatments on the microbiome at descending levels of taxonomic organization. The potential health impacts associated with the microflora community shift and their role in transformation of Pb species and bioavailability is discussed.
4.1. Potential health implications of fecal microbiome modulation.
One approach to assessing the effects of toxins on the fecal microbiome is examination of changes in relative abundance at the phylum level. For example, modulation of the Firmicutes/Bacteroidetes ratio has been used as an indicator of obesity, exercise effects, aging, diet, carbohydrate metabolism and inflammation (Breton et al., 2013; Cheng et al., 2018; Denou et al., 2016; Kim et al., 2016; Murphy et al., 2010; Zhang et al., 2021). Similarly, changes in the Bacilli/Clostridia ratio have been linked to an inflammatory response (Breton et al., 2013; Cheng et al., 2018; Denou et al., 2016; Kim et al., 2016; Lee et al., 2020; Murphy et al., 2010; Pearson-Leary et al., 2020). In the present study, the change in the Firmicutes/Bacteroidetes ratio was greatest in mice that consumed diet amended with untreated soil. Smaller changes in the Firmicutes/Bacteroidetes ratio were found in the fecal microbiomes of mice that consumed diets amended with PA or TSP treated soil. This reduction in effect on the ratio may correlate with the lowering of Pb RBA produced by treatment of soils to reduce Pb solubility (Bradham et al., 2018). In a fifteen week drinking water study, treatment with Pb acetate (10, 30 or 100 mg L−1) resulted in an increase in the Bacteroidetes/Firmicutes ratio (i.e. decrease in Firmicutes/Bacteroidetes ratio) which correlated with liver triglyceride, pyruvate and serum triglyceride, total cholesterol and glucose fluctuations (Xia et al., 2018). Interestingly, microflora effects were mediated by the addition of ferric chloride (5 mg L−1) and resulted in changes to the abundance of several antimicrobial resistance genes. Eight weeks of Pb treatment (100 ppm or 500ppm; 1.83 g/L Pb acetate) administered in drinking water revealed no change in fecal Bacteroidetes or Firmicutes relative abundance compared to control, indicating a possible temporal association with their modulation (Breton et al., 2013; Zhai et al., 2017). Differences in the physical and chemical state of Pb in drinking water and soil make it difficult to compare the observed effects on the fecal microbiome. For soil-borne Pb, it may be more appropriate to compare fecal microbiomes found in mice that ingested untreated or treated soils. This approach makes it possible to correlate changes in the RBA of Pb produced by treatment of soils with changes in the composition of the fecal microbiome.
The effect of soil treatment on the fecal microbiome also was examined by comparing the relative abundance differences of classes Bacilli and Clostridia. When compared with the composition of the fecal microbiome from mice that consumed diet amended with untreated soil, there were significant differences (p=0.002) which reflected an increase in Bacilli in microbiomes of mice that consumed diets amended with Fe/TSP or C/TSP treated soil and a decrease in Bacilli in microbiomes of mice that consumed diet amended with PA treated soil (Table 4, Table S10). Similarly, compared with the composition of the fecal microbiome from mice that consumed diet amended with untreated soil, relative abundance of Clostridia was also reduced in fecal microbiomes from mice that consumed diets amended with the three treated soils. Reduced Bacilli to Clostridia ratio is indicative of an elevated inflammatory response (Lee et al., 2020; Pearson-Leary et al., 2020), suggesting that ingestion of diet amended with Fe/TSP or C/TSP treated soil may reduce inflammation below the level found after consumption of diet amended with untreated soil. In contrast, consumption of diet amended with PA treated soil may increase the inflammatory response. If an elevated inflammatory response is considered an adverse effect, then it may be prudent to use fluctuations in Bacilli/Clostridia and Firmicutes/Bacteroidetes in the selection of a soil treatment strategy used in remediation of Pb contaminated soil.
Other differences at the phylum level were seen in fecal microbiomes of mice that consumed diet amended with treated soils. Consumption led to an upward trend in the relative abundance of Verrucomicrobia in fecal microbiomes (Fig. 2A). Notably, this phylum made the largest contribution to dissimilarity between microbiomes from mice that consumed diets amended with untreated or treated soils. Similarly, relative abundance of Tenericutes and candidate phylum TM7 (Candidatus Saccharibacteria) contributed between 6.0% to 13.1% and 6.9% to 9.5%, respectively, to dissimilarity observed in mice that consumed diets amended with untreated or treated soils (Table S5). Compared to consumption of untreated soil, TSP or PA treated soil exhibited reduced relative abundance of Tenericutes; Fe/TSP or C/TSP treated soil increased the relative abundance of Tenericutes. For candidate phylum TM7 the relative abundance was elevated in all mice that consumed diets amended with treated soil (Fig. 2A). Relative abundances of Verrucomicrobia, Tenericutes, and candidate phylum TM7 (Candidatus Saccharibacteria) have been linked to inflammatory response (Kuehbacher et al., 2008). In humans, improved kidney function, evidenced by reduced serum creatinine levels and increased glomerular filtration, is correlated with elevated abundance of candidate phylum TM7 in the feces (Xu et al., 2020). In the horse, levels of Verrucomicrobia and Clostridiales in ileum are associated with induction of regulatory immunity, suggesting a role in modulating inflammatory response (Lindenberg et al., 2019). An eight-week exposure to Pb acetate in drinking water reduced the abundance of Verrucomicrobia which may be indicative of a heightened inflammatory state following Pb exposure (Zhai et al., 2017). Biomarkers of aging and inflammation that are associated with increased levels of IL-6 and IL-17 cytokines and tumor necrosis factor alpha have been reported to accompany decreased abundance of Tenericutes (Huang et al., 2013; Cao et al., 2020; Li et al., 2019). Consumption of diets amended with treated soil marginally increased Proteobacteria abundance in fecal microbiomes (Fig. 2A). The phyla Proteobacteria have been correlated with gut dysbiosis, aging, and inflammation (Huang et al., 2013; Cao et al., 2020; Litvak et al., 2017; Shin et al., 2015). Elevated abundance of Proteobacteria is associated with gut dysbiosis and in aged mice, lower abundance of Proteobacteria is reported (Huang et al., 2013; Cao et al., 2020). An eight-week exposure to Pb acetate in drinking water also reduced Proteobacteria abundance (Zhai et al., 2017). Modulation of the Proteobacteria community associated with consumption of diet amended with a treated soil may be an adverse effect associated with soil remediation. Careful consideration of the full range of effects of consumption of complex treated soils on the composition of the microbiome of the gastrointestinal tract may be part of assessment of treatment and development of a risk assessment framework to evaluate different soil treatments.
Because the contaminated soils are complex matrices containing a wide array of inorganic and organic components, to include metals such as cadmium, chromium, arsenic, and zinc, it is difficult to attribute changes to microbiome composition to a single contaminant, such as Pb. However, evidence from one ecological study suggests a relationship between exposure to Pb in soil and changes in intestinal microflora. A study in deer mice (Peromyscus maniculatus) trapped at a previously remediated site in the former Tri-State Mining District (Missouri, USA) reported to have 270-732 mg kg−1 soil Pb, found significant differences in intestinal microflora community structure compared to P. maniculatus trapped from a reference site (Coolon et al., 2010). In mice that resided on Pb-contaminated soil, Ruminococcaeceae abundance was significantly reduced. Conversely, in a cross sectional study in humans residing near a mining and smelting site who were exposed long term to Pb and other heavy metal contaminated soil, the relative abundances of families Ruminococcaceae, Lachnospiraceae, and Erysipelotrichaceae were elevated in stool (Shao & Zhu, 2020). Intestinal Ruminococcaeceae has been related to health promotion in humans (Hattori & Taylor, 2009); its reduction in mice from Pb contaminated sites may contribute to smaller body size and mass and reduced body fat.
In the present study, ingestion of diets amended with soil for nine days decreased fecal Ruminococcaceae, comparable to the P. maniculatus study and Lachnospiraceae; Erysipelotrichaceae levels declined after consumption of diets amended with Fe/TSP, C/TSP or PA treated soils (Table S2). Verrucomicrobiaceae contributed 2.4% to 4.4% to microbiome dissimilarity between mice consuming diets amended with untreated or treated soil; relative abundance was increased in groups consuming diets amended with treated soils. Verrucomicrobiaceae (as well as genera Akkermansia and Dorea) have been reported to have a causal relationship with inflammatory bowel disease, although the role of these microorganisms in the disease process has not been elucidated (Zhang et al., 2021). Ingestion of diets amended with treated soils were associated with other changes in microbiome composition. Elevated Prevotellaceae levels were observed in mice that consumed diet amended with C/TSP treated soil. Increased levels of Lactobacilaceae, which have been linked with an anti-inflammatory response in the gut (Ojo et al., 2019; Vandana et al., 2020), were found in mice that consumed a diet amended with Fe/TSP treated soil; in these mice Bacteroidaceae levels were marginally decreased. Other studies have reported changes in microflora composition with Pb exposure. Members of the Prevotellaceae family were detected in the feces of mice exposed to 500 ppm of Pb in drinking water for 8 weeks; in contrast, members of this family were absent from the feces of mice not exposed to Pb (Breton et al., 2013). Studies have linked elevated Prevotellaceae levels in humans to a higher genetic risk score associated with body mass index and obesity, suggesting an association with alterations in gut metabolism (Cuevas-Sierra et al., 2020). These results demonstrate the integral role of the microbiome in maintaining gut homeostasis and the complexities in associating function to one or more families.
Aggregating the OTU data at the genus level revealed additional changes in the fecal microbiome of mice that consumed soil-amended diets. For all mice consuming diets amended with treated soils, the relative abundance of Pseudoflavonifractor (Firmicutes) was reduced as compared to its abundance in fecal microbiome of mice that consumed diet amended with untreated soil. Pseudoflavonifractor abundance has been positively correlated with obesity induced Type 2 diabetes and hypertension (Wang et al., 2020; Zuo et al., 2019); reduced Pseudoflavonifractor abundance observed in the current study may be a potential positive health indicator. Consumption of diet amended with C/TSP treated soil increased the relative abundance of Ruminococcus (Firmicutes). Furthermore, Allobaculum (Firmicutes) and Ruminococcus were key contributors to dissimilarity between the microbiomes of mice consuming diets amended with untreated soil and those consuming diets amended with treated soils. Allobaculum abundance has been correlated with a high fat diet and elevated levels of short chain fatty acids, simple carbohydrates, and acetate in the gut (Zhang et al., 2021; Balakrishnan et al., 2021). The mucin degrader Ruminococcus gnavus, which metabolizes complex polysaccharides and is a key symbiont in the mammalian intestinal tract (Crost et al., 2013; Henke et al., 2019), has been associated with Crohn’s disease and inflammation (La Reau & Suen, 2018). Akkermansia (Verrucomicrobia) which contributed approximately 1.7% to dissimilarity in microbiomes in mice consuming TSP +/− Fe and C amended diets was elevated in mice consuming all diets amended with treated soil compared to untreated soil. Therapeutic use of Akkermansia muciniphila, a mucin degrader, has been reported to have beneficial metabolic and immunologic effects including reduced incidence of diabetes mellitus and prevention of age-related colonic mucus layer decline and inflammation (de Vos, 2017; Hänninen et al., 2018; Jayachandran et al., 2020; Van Der Lugt et al., 2019). While not definitive, modulation of the microbiome may be implicated in potential positive or negative health outcomes.
Among the top 50 OTUs contributing to the fecal microbiome dissimilarity between mice consuming diet amended with untreated soil and treated soils, 12 OTUs occurred in all groups consuming diets amended with treated soils (Table S9). Of these, most were either Lachnospiraceae or Ruminococcaceae, which were dominant members of the fecal microflora found in mice receiving diets with treated soils. Notably, OTU 84, identified as Allobaculum, was a prevalent contributor to dissimilarity in fecal microflora found in mice receiving diets with treated soils. Future research on the use of biomarkers of effect to evaluate the influence of ingestion of remediated soils on the composition of the fecal microbiome might focus on use of Allobaculum as candidate biomarker as it is a prevalent contributor at both the OTU and genus level. Additional studies also are needed to determine whether Lachnospiraceae or Ruminococcaceae OTUs can be used as biomarker for changes in the microbiome.
4.2. Relationship of the fecal microbiome to Pb transformation and bioavailability
Microorganisms have evolved survival mechanisms to evade the toxic effects of heavy metals. The related genes are plasmid or chromosomally linked and can co-occur with antibiotic resistance genes in humans and animals (Li et al., 2017). Many phyla, including Proteobacteria, Firmicutes, and Actinobacteria harbor Pb and metal resistance genes (Bharagava et al., 2014; El-Sayed, 2016; Koc et al., 2013). Expression of these genes can result in Pb binding extracellularly to microbial siderophores, exopolysaccharides, and cell phosphoryl-, carboxyl-, or sulfhydryl-groups or sequestration intracellularly by binding to metallothionein (George & Wan, 2020). Hui et al. (2018) demonstrated that Pb resistance genes can sequester Pb in vivo. When a Pb binding protein, PbrR, was engineered to express on the cell surface of Escherichia coli and subsequently introduced into mice, Pb was immobilized at both acidic and neutral pH and correlated with a decrease in bioavailability. Because exposure to Pb and other heavy metals selects for resistance strains, more opportunities for adsorption or precipitation may be available and contribute to reduced Pb bioavailability. Verrucomicrobiaceae and Proteobacteria, which qualitatively increased in relative abundance in mice that received treated soil, have been reported to co-occur with the Pb resistance gene pbrT and other genes associated with heavy metal resistance expression in the environment (Roberto et al., 2019). Members of these phyla could adsorb or bind Pb and make less available for mammalian adsorption. In addition, the higher concentration of available Pb in the untreated soil may have exceeded a microbial Pb resistance threshold, resulting in a reduction of Verrucomicrobiaceae and Proteobacteria populations, which, in turn, resulted in increased mammalian adsorption.
Microbial Pb resistance mechanisms also generate less soluble Pb species such as sulfides, phosphates, and carbonates which have been leveraged in bioremediation strategies to reduce bioavailability (George & Wan, 2020). The untreated soil diet used in this study contained anglesite, galena, organic-Pb, and adsorbed-Pb; no anglesite or galena were detected in the feces and relative amounts of adsorbed and organic Pb were increased (Fig. 3; Bradham et al., 2018). Pb phosphate also was present in the feces of mice provided the untreated soil diet. The low pH in the stomach, or microbial phosphatases or organic acids may have solubilized the Pb species as they transited through the stomach and small intestine. As pH increased in the colon, Pb phosphate may have been formed either abiotically or through phosphate generating microbial processes. In addition, adsorbed and organic Pb also were relatively greater in feces compared to that in the diet which may be a result of Pb adsorbing or binding to exopolysaccharide or the cell surface. Diets prepared with soil treated with PA, TSP, and Fe/TSP contained relatively similar amounts of anglesite, Pb phosphate, pyrophosphate, and organic-Pb; anglesite was not recovered from the feces and relatively more Pb phosphate and organic-Pb and less pyrophosphate was detected, supporting a possible microbiome role. Adsorbed Pb was elevated in feces from mice that received dietary Fe/TSP treated soil compared to other phosphate treatments (TSP, C/TSP, PA) suggesting that Pb may have adsorbed to iron minerals. Feces from mice receiving a diet containing C/TSP had relatively more organic-Pb compared to the other treatments (TSP, Fe/TSP, PA); pyromorphite was relatively less abundant, presumably due to the prevalence of organic associated Pb. Animals that received similar amounts of Pb in their diet from C/TSP and PA treated soils (24.2 ppm), excreted more (184.7 ppm) and less (162.6 ppm) Pb in the feces, respectively (Table S11). Lead excretion was inversely correlated to RBA. Lead was less bioavailable in animals that received C/TSP treated soil (28%) whereas it was more bioavailable (69%) in those that received PA treated soil. Therefore, both the biosolids treatment and the intestinal flora may have adsorbed Pb leading to increased excretion in the feces.
Fig. 3.
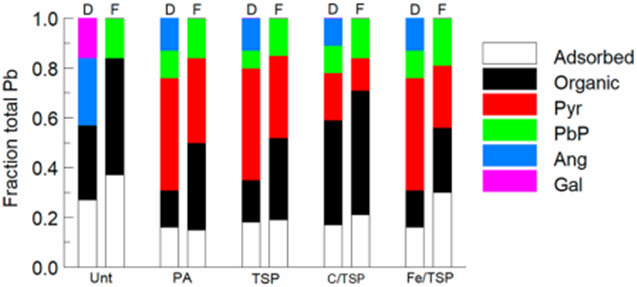
Pb speciation in diet consumed and feces excreted by mice ingesting soil-amended diets. Relative amounts of each Pb species (fraction of total Pb) present in diet (D) and in feces (F) during the standard assay period were calculated using Pb speciation data derived from spectral analysis of diet and feces. Treatments as identified in Table 1. PbP is trilead diphosphate (Pb3(PO4)2), Ang is anglesite (PbSO4), Gal is galena (PbS), Pyr is pyromorphite. Reprinted with permission from Bradham, K. D., Diamond, G. L., Nelson, C. M., Noerpel, M., Scheckel, K. G., Elek, B., Chaney, R. L., Ma, Q, and Thomas, D. J. Long-term in situ reduction in soil Pb bioavailability measured in a mouse model. Environmental Science & Technology, 2018, 52(23), 13908-13913. Copyright 2018 American Chemical Society.
C/TSP soil treatment affected fecal microbiome diversity compared to PA and Fe/TSP treated soils and control when aggregated at the genus and family levels; C/TSP soil treatment fecal microbiome richness was significantly different than that observed for Fe/TSP treated soil and untreated control (Table 2). Compared to control, phyla Actinobacteria (Pseudonocardiaceae), Bacteroidetes (Chitinophagaceae, Cytophagaceae, Prevotellaceae), Firmicutes (Alicyclobacillaceae, Bacillaceae, Clostridiaceae, Enterococcaceae, Lactobacillaceae, Paenibacillaceae, Planococcaceae), Proteobacteria (Enterobacteriaceae, Sutterellaceae), Tenericutes (Anaeroplasmataceae), and Verrucomicrobia (Verrucomicrobiaceae) were elevated in feces from mice that received dietary C/TSP relative to control (Table S7). Members of the associated phyla have been reported to be resistant to high levels of Pb, and therefore have the potential to bind Pb and be excreted in the feces (Adler, Devarajan, Wildi, & Poté, 2016). Feces from mice that received dietary C/TSP treated soil compared to PA treated soil had elevated average abundance of Verrucomicrobia which contributed 19% to the dissimilarity associated with the two treatments and may have more efficient Pb binding. The microbial community differences are suggestive of a microflora role in Pb transformation which results in less Pb bioavailability and higher Pb fecal excretion rate.
5. Conclusions
In summary, this study provides unique insight into the effect of soil remediation alternatives on the murine fecal microbiome and implications for potential health associated outcomes and Pb bioavailability due to change in microbiome equilibrium. Changes in the fecal microbiome may be a consequence of exposure to Pb present in soil. Assessing the significance of those changes in the gastrointestinal tract associated with ingestion of Pb-contaminated soils is an aspect of estimating adverse health effects caused by ingestion of Pb. As shown in the present study, remediation of Pb-contaminated soils by treatment with agents that reduce the solubility and bioavailability of Pb can produce changes in Pb speciation that affects the composition of the fecal microbiome. The consequences of these changes in microbiomes after ingestion of treated soils have not been well characterized, however a link to altered physiology and metabolism and expression of Pb resistant microbial mechanisms can be inferred. Future work is needed to improve our understanding of the relationship between Pb speciation in treated soils and to assess the adverse health effects of these changes. To better delineate treatment effects, future studies will take discrete fecal samples during dietary exposure and while an uncontaminated reference soil may be desirable, its inclusion may introduce another variable to the study because it would be sourced separately. Development of biomarkers of effect to monitor critical changes in microbiome composition will be critical to this work.
Animals
The protocol for use of mice for the determination the bioavailability of Pb in soil was reviewed by the Institutional Animal Care and Use Committee of the US Environmental Protection Agency, Research Triangle Park, North Carolina. These studies reported here conformed to the approved protocol that complied with National Institutes of Health guidelines for the care and use of laboratory animals (NIH Publications No. 8023, revised 1978).
Supplementary Material
Acknowledgements
We would like to thank Dr. Tyler Sowers for his valuable review of earlier versions of this manuscript and Ms. Deborah Vivian, Ms. Mollie Nugent and Ms. Sonya Doten for statistical analysis, data management and literature support. Portions of this work were funded by the U.S. Environmental Protection Agency, Office of Superfund Remediation and Technology Innovation (OSRTI), under Contract 68HERH19D0022. Agency required quality planning documentation was developed in accordance with EPA quality system requirements; therefore, the scientific and technical activities were planned to ensure the research presented is of known and documented quality. The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Declaration of competing interest
No authors have any competing financial and personal relationships with other people or organizations that could inappropriately influence their work.
References
- Adler A, Devarajan N, Wildi W, & Poté J (2016). Metal distribution and characterization of cultivable lead-resistant bacteria in shooting range soils. Soil and Sediment Contamination: An International Journal, 25(4), 378–394. [Google Scholar]
- Anderson MJ (2008). Animal-sediment relationships re-visited: Characterising species' distributions along an environmental gradient using canonical analysis and quantile regression splines. Journal of experimental marine biology and ecology, 366(1-2), 16–27. [Google Scholar]
- Balakrishnan B, Luckey D, Bodhke R, Chen J, Marietta E, Jeraldo P, … Taneja V (2021). Prevotella histicola Protects From Arthritis by Expansion of Allobaculum and Augmenting Butyrate Production in Humanized Mice. Frontiers in Immunology, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta N, Gradwohl R, Snethen K, & Schroder J (2001). Chemical immobilization of Pb, zinc, and cadmium in smelter-contaminated soils using biosolids and rock phosphate. Journal of Environmental Quality, 30(4), 1222–1230. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Kristiansson E, & Larsson DGJ (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology reviews, 42(1). doi: 10.1093/femsre/fux053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharagava RN, Yadav S, & Chandra R (2014). Antibiotic and heavy metal resistance properties of bacteria isolated from the aeration lagoons of common effluent treatment plant (CETP) of tannery industries (Unnao, India). [Google Scholar]
- Bhaskar PV, & Bhosle NB (2006). Bacterial extracellular polymeric substance (EPS): a carrier of heavy metals in the marine food-chain. Environ Int, 32(2), 191–198. doi: 10.1016/j.envint.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Bolan N, Kunhikrishnan A, Thangarajan R, Kumpiene J, Park J, Makino T, … Scheckel K (2014). Remediation of heavy metal (loid) s contaminated soils–to mobilize or to immobilize? Journal of Hazardous Materials, 266, 141–166. [DOI] [PubMed] [Google Scholar]
- Bradham KD, Diamond GL, Nelson CM, Noerpel M, Scheckel KG, Elek B, … Thomas DJ (2018). Long-term in situ reduction in soil lead bioavailability measured in a mouse model. Environmental science & technology, 52(23), 13908–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradham KD, Green W, Hayes H, Nelson C, Alava P, Misenheimer J, … Thomas DJ (2016). Estimating relative bioavailability of soil lead in the mouse. Journal of Toxicology and Environmental Health, Part A, 79(24), 1179–1182. [DOI] [PubMed] [Google Scholar]
- Breton J, Le Clere K, Daniel C, Sauty M, Nakab L, Chassat T, … Foligne B (2013). Chronic ingestion of cadmium and lead alters the bioavailability of essential and heavy metals, gene expression pathways and genotoxicity in mouse intestine. Arch Toxicol, 87(10), 1787–1795. doi: 10.1007/s00204-013-1032-6 [DOI] [PubMed] [Google Scholar]
- Breton J, Massart S, Vandamme P, De Brandt E, Pot B, & Foligne B (2013). Ecotoxicology inside the gut: impact of heavy metals on the mouse microbiome. BMC Pharmacol Toxicol, 14(62), 62. doi: 10.1186/2050-6511-14-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Chaney R, Hallfrisch J, Ryan JA, & Berti WR (2004). In situ soil treatments to reduce the phyto-and bioavailability of lead, zinc, and cadmium. [PubMed] [Google Scholar]
- Cao L, Lee SG, Melough MM, Sakaki JR, Maas KR, Koo SI, & Chun OK (2020). Long-term blackcurrant supplementation modified gut microbiome profiles in mice in an age-dependent manner: An exploratory study. Nutrients, 12(2), 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel SW, Weis CP, Henningsen GM, & Brattin WJ (2006). Estimation of relative bioavailability of lead in soil and soil-like materials using young swine. Environmental health perspectives, 114(8), 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Zhang X, Zhu J, Cheng L, Cao J, Wu Z, … Zheng X (2018). A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols. Food & function, 9(2), 1079–1087. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Tweedley JR, & Valesini FJ (2014). Simple shade plots aid better long-term choices of data pre-treatment in multivariate assemblage studies. Journal of the Marine Biological Association of the United Kingdom, 94(1), 1–16. [Google Scholar]
- Clarke KR, & Warwick R (2001). Change in marine communities. An approach to statistical analysis and interpretation, 2, 1–168. [Google Scholar]
- Cretacci Y, & Parsons PJ (2010). Localized accumulation of lead within and among bones from lead-dosed goats. Environmental research, 110(1), 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, & Juge N (2013). Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PloS one, 8(10), e76341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, Jones KL, Narayanan S, & Wisely SM (2010). Microbial ecological response of the intestinal flora of Peromyscus maniculatus and P. leucopus to heavy metal contamination. Molecular Ecology 19(s1), 67–80. 10.1111/j.1365-294X.2009.04485.x [DOI] [PubMed] [Google Scholar]
- de Vos WM (2017). Microbe Profile: Akkermansia muciniphila: a conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology, 163(5), 646–648. [DOI] [PubMed] [Google Scholar]
- Denou E, Marcinko K, Surette MG, Steinberg GR, & Schertzer JD (2016). High-intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet-induced obesity. American Journal of Physiology-Endocrinology and Metabolism, 310(11), E982–E993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Taylor MP, & Gulson B (2020). A 25-year record of childhood blood lead exposure and its relationship to environmental sources. Environmental research, 186, 109357. [DOI] [PubMed] [Google Scholar]
- Eichler A, Tobler L, Eyrikh S, Malygina N, Papina T, & Schwikowski M (2014). Ice-core based assessment of historical anthropogenic heavy metal (Cd, Cu, Sb, Zn) emissions in the Soviet Union. Environmental science & technology, 48(5), 2635–2642. [DOI] [PubMed] [Google Scholar]
- El-Sayed MH (2016). Multiple Heavy Metala and Antibiotic Resistance of Acinetobacter Baumannii Strain HAF - 13 Isolated from Industrial Effluents. Science & Education Publishing - American journal of Microbiological Research, 4(1), 26–36. doi: 10.12691/ajmr-4-1-3 [DOI] [Google Scholar]
- Freeman GB, Johnson JD, Liao SC, Feder PI, Davis AO, Ruby MV, … Bergstrom PD (1994). Absolute bioavailability of lead acetate and mining waste lead in rats. Toxicology, 91, 151–163. [DOI] [PubMed] [Google Scholar]
- Fry KL, Wheeler CA, Gillings MM, Flegal AR, & Taylor MP (2020). Anthropogenic contamination of residential environments from smelter As, Cu and Pb emissions: Implications for human health. Environmental Pollution, 262, 114235. [DOI] [PubMed] [Google Scholar]
- Gailey AD, Schachter AE, Egendorf SP, & Mielke HW (2020). Quantifying soil contamination and identifying interventions to limit health risks. Current Problems in Pediatric and Adolescent Health Care, 50(1), 100740. [DOI] [PubMed] [Google Scholar]
- Gamiño-Gutiérrez SP, González-Pérez CI, Gonsebatt ME, & Monroy-Fernández MG (2013). Arsenic and lead contamination in urban soils of Villa de la Paz (Mexico) affected by historical mine wastes and its effect on children’s health studied by micronucleated exfoliated cells assay. Environmental Geochemistry and Health, 35(1), 37–51. [DOI] [PubMed] [Google Scholar]
- Gao B, Chi L, Mahbub R, Bian X, Tu P, Ru H, & Lu K (2017). Multi-Omics Reveals that Lead Exposure Disturbs Gut Microbiome Development, Key Metabolites, and Metabolic Pathways. Chem Res Toxicol, 30(4), 996–1005. doi: 10.1021/acs.chemrestox.6b00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SE, & Wan Y (2020). Advances in characterizing microbial community change and resistance upon exposure to lead contamination: Implications for ecological risk assessment. Critical Reviews in Environmental Science & Technology, 50(21), 2223–2270. 10.1080/10643389.2019.1698260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillan DC, Roosa S, Kunath B, Billon G, & Wattiez R (2015). The long-term adaptation of bacterial communities in metal-contaminated sediments: a metaproteogenomic study. Environ Microbiol, 17(6), 1991–2005. doi: 10.1111/1462-2920.12627 [DOI] [PubMed] [Google Scholar]
- Government of South Australia, Department of Health and Wellbeing (2022). Port Pirie Blood Lead Levels. https://www.sahealth.sa.gov.au/wps/wcm/connect/7c8de249-f68e-4f34-8e43-c58a52cf3ce1/Port+Pirie+Blood+Lead+Analysis+for+2021+%281+January+-+31+December+2021%29.pdf?MOD=AJPERES&CACHEID=ROOTWORKSPACE-7c8de249-f68e-4f34-8e43-c58a52cf3ce1-nYZxKaF, retrieved April 12, 2022, 12 pp.
- Hänninen A, Toivonen R, Pöysti S, Belzer C, Plovier H, Ouwerkerk JP, … De Vos WM (2018). Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut, 67(8), 1445–1453. [DOI] [PubMed] [Google Scholar]
- Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, & Clardy J (2019). Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proceedings of the national academy of sciences, 116(26), 12672–12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry H, Naujokas MF, Attanayake C, Basta NT, Cheng Z, Hettiarachchi GM, … Scheckel KG (2015). Bioavailability-Based In Situ Remediation To Meet Future Lead (Pb) Standards in Urban Soils and Gardens. Environmental science & technology, 49(15), 8948–8958. doi: 10.1021/acs.est.5b01693 [DOI] [PubMed] [Google Scholar]
- Hettiarachchi GM, Pierzynski GM, & Ransom MD (2000). In Situ Stabilization of Soil Lead Using Phosphorus and Manganese Oxide. Environmental science & technology, 34(21), 4614–4619. doi: 10.1021/es001228p [DOI] [Google Scholar]
- Hogan K, Marcus A, Smith R, & White P (1998). Integrated Exposure Uptake Biokinetic Model for Lead in Children: Empirical Comparisons with Epidemiologic Data. Environmental health perspectives, 106(Suppl 6), 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, & Chang EB (2013). Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. Journal of Parenteral and Enteral Nutrition, 37(6), 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C, Guo Y, Zhang W, Gao C, Yang X, Chen Y, … Huang X (2018). Surface display of PbrR on Escherichia coli and evaluation of the bioavailability of lead associated with engineered cells in mice. Scientific reports, 8(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynninen A, Touze T, Pitkanen L, Mengin-Lecreulx D, & Virta M (2009). An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol Microbiol, 74(2), 384–394. doi: 10.1111/j.1365-2958.2009.06868.x [DOI] [PubMed] [Google Scholar]
- Jayachandran M, Chung SSM, & Xu B (2020). A critical review of the relationship between dietary components, the gut microbe Akkermansia muciniphila, and human health. Critical reviews in food science and nutrition, 60(13), 2265–2276. [DOI] [PubMed] [Google Scholar]
- Jie S, Li M, Gan M, Zhu J, Yin H, & Liu X (2016). Microbial functional genes enriched in the Xiangjiang River sediments with heavy metal contamination. BMC Microbiol, 16(1), 179. doi: 10.1186/s12866-016-0800-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R, Chambers FM, & Benson-Evans K (1991). Heavy metals (Cu and Zn) in recent sediments of Llangorse Lake, Wales: non-ferrous smelting, Napoleon and the price of wheat - a palaeoecological study. Hydrobiologia, 214, 149–154. [Google Scholar]
- Kang C-H, & So J-S (2016). Heavy metal and antibiotic resistance of ureolytic bacteria and their immobilization of heavy metals. Ecological Engineering, 97, 304–312. doi: 10.1016/j.ecoleng.2016.10.016 [DOI] [Google Scholar]
- Kim K-A, Jeong J-J, Yoo S-Y, & Kim D-H (2016). Gut microbiota lipopolysaccharide accelerates inflamm-aging in mice. BMC microbiology, 16(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc S, Kabatas B, & Icgen B (2013). Multidrug and heavy metal-resistant Raoultella planticola isolated from surface water. Bull Environ Contam Toxicol, 91(2), 177–183. doi: 10.1007/s00128-013-1031-6 [DOI] [PubMed] [Google Scholar]
- Kozlov M, & Zvereva E (2007). Industrial barrens: extreme habitats created by non-ferrous metallurgy. Reviews in Environmental Science and Bio/Technology, 6(1), 231–259. [Google Scholar]
- Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, Schreiber S, & Ott SJ (2008). Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. Journal of Medical Microbiology, 57(12), 1569–1576. doi: 10.1099/jmm.0.47719-0 [DOI] [PubMed] [Google Scholar]
- La Reau AJ, & Suen G (2018). The Ruminococci: key symbionts of the gut ecosystem. journal of microbiology, 56(3), 199–208. [DOI] [PubMed] [Google Scholar]
- Laidlaw MA, Mohmmad SM, Gulson BL, Taylor MP, Kristensen LJ, & Birch G (2017). Estimates of potential childhood lead exposure from contaminated soil using the US EPA IEUBK Model in Sydney, Australia. Environmental research, 156, 781–790. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee S, Mayta A, Mrdjen I, Weghorst C, & Knobloch T (2020). Microcystis toxin-mediated tumor promotion and toxicity lead to shifts in mouse gut microbiome. Ecotoxicology and Environmental Safety, 206, 111204. (Li et al., 2017) [DOI] [PubMed] [Google Scholar]
- Li K, Zhang L, Xue J, Yang X, Dong X, Sha L, … Wang Z (2019). Dietary inulin alleviates diverse stages of type 2 diabetes mellitus via anti-inflammation and modulating gut microbiota in db/db mice. Food & function, 10(4), 1915–1927. [DOI] [PubMed] [Google Scholar]
- Li M-Y, Chen X-Q, Wang J-Y, Wang H-T, Xue X-M, Ding J, … Ma LQ (2021). Antibiotic exposure decreases soil arsenic oral bioavailability in mice by disrupting ileal microbiota and metabolic profile. Environment International, 151, 106444. [DOI] [PubMed] [Google Scholar]
- Li X, Meng D, Li J, Yin H, Liu H, Liu X, … Yan M (2017). Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environmental Pollution, 231, 908–917. doi: 10.1016/j.envpol.2017.08.057 [DOI] [PubMed] [Google Scholar]
- Lindenberg F, Krych L, Fielden J, Kot W, Frøkiær H, Van Galen G, … Hansen A (2019). Expression of immune regulatory genes correlate with the abundance of specific Clostridiales and Verrucomicrobia species in the equine ileum and cecum. Scientific reports, 9(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak Y, Byndloss MX, Tsolis RM, & Bäumler AJ (2017). Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Current opinion in microbiology, 39, 1–6. [DOI] [PubMed] [Google Scholar]
- Marschner B, Welge P, Hack A, Wittsiepe J, & Wilhelm M (2006). Comparison of soil Pb in vitro bioaccessibility and in vivo bioavailability with Pb pools from a sequential soil extraction. Environmental science & technology, 40(8), 2812–2818. [DOI] [PubMed] [Google Scholar]
- Matyar F (2012). Antibiotic and heavy metal resistance in bacteria isolated from the Eastern Mediterranean Sea coast. Bull Environ Contam Toxicol, 89(3), 551–556. doi: 10.1007/s00128-012-0726-4 [DOI] [PubMed] [Google Scholar]
- Matyar F, Gülnaz O, Guzeldag G, Mercimek HA, Akturk S, Arkut A, & Sumengen M (2014). Antibiotic and heavy metal resistance in Gram-negative bacteria isolated from the Seyhan Dam Lake and Seyhan River in Turkey. Annals of microbiology, 64(3), 1033–1040. [Google Scholar]
- Moore JN, & Luoma SN (1990). Hazardous wastes from large-scale metal extraction A case study. Environmental science & technology, 24(9), 1278–1285. [Google Scholar]
- Murphy E, Cotter P, Healy S, Marques TM, O'sullivan O, Fouhy F, … Stanton C (2010). Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut, 59(12), 1635–1642. [DOI] [PubMed] [Google Scholar]
- National Research Council, N. R. (1997). Innovations in ground water and soil cleanup: From concept to commercialization: National Academies Press. [Google Scholar]
- Park JH, Bolan N, Megharaj M, & Naidu R (2011). Concomitant rock phosphate dissolution and lead immobilization by phosphate solubilizing bacteria (Enterobacter sp.). Journal of Environmental Management, 92(4), 1115–1120. [DOI] [PubMed] [Google Scholar]
- Pearson-Leary J, Zhao C, Bittinger K, Eacret D, Luz S, Vigderman AS, … Bhatnagar S (2020). The gut microbiome regulates the increases in depressive-type behaviors and in inflammatory processes in the ventral hippocampus of stress vulnerable rats. Molecular psychiatry, 25(5), 1068–1079. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, … Glöckner FO (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research, 41(D1), D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. [Google Scholar]
- Roberto AA, Van Gray JB, Engohang-Ndong J, & Leff LG (2019). Distribution and co-occurrence of antibiotic and metal resistance genes in biofilms of an anthropogenically impacted stream. Science of The Total Environment, 688, 437–449. [DOI] [PubMed] [Google Scholar]
- Rowan JS, Barnes SJA, Hetherington SL, Lambers B, & Parsons F (1995). Geomorphology and pollution: the environmental impacts of lead mining, Leadhills, Scotland. Journal of Geochemical Exploration, 52, 57–65. [Google Scholar]
- Scheckel KG, Diamond GL, Burgess MF, Klotzbach JM, Maddaloni M, Miller BW, … Serda SM (2013). Amending soils with phosphate as means to mitigate soil lead hazard: a critical review of the state of the science. Journal of Toxicology and Environmental Health, Part B, 16(6), 337–380. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, … Robinson CJ (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23), 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, & Zhu Y (2020). Long-term metal exposure changes gut microbiota of residents surrounding a mining and smelting area. Scientific reports, 10(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N-R, Whon TW, & Bae J-W (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends in biotechnology, 33(9), 496–503. [DOI] [PubMed] [Google Scholar]
- Smith A, Barber J, Davis S, Jones C, Kotra KK, Losada S, … Devlin MJ (2021). Aquatic contaminants in Solomon Islands and Vanuatu: Evidence from passive samplers and Microtox toxicity assessment. Marine Pollution Bulletin, 165, 112118. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Isley CF, & Glover J (2019). Prevalence of childhood lead poisoning and respiratory disease associated with lead smelter emissions. Environment International, 127, 340–352. [DOI] [PubMed] [Google Scholar]
- Teng Z, Shao W, Zhang K, Huo Y, & Li M (2019). Characterization of phosphate solubilizing bacteria isolated from heavy metal contaminated soils and their potential for lead immobilization. Journal of Environmental Management, 231, 189–197. [DOI] [PubMed] [Google Scholar]
- Thompson L, Osuolale O, Knight R, Gilbert J, Jansson J, Ackermann G, … Consortium, E. M. P. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (1994). United States Environmental Protection Agency (USEPA) Guidance Manual for the Integrated Exposure Uptake Biokinetic Model for Lead in Children. EPA/540/R-93/081 1–248. [Google Scholar]
- Van Der Lugt B, Van Beek AA, Aalvink S, Meijer B, Sovran B, Vermeij WP, … Steegenga WT (2019). Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1−/Δ7 mice. Immunity & Ageing, 16(1), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R, Monchy S, Leys N, & Mergeay M (2009). New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek, 96(2), 205–226. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ouyang M, Gao X, Wang S, Fu C, Zeng J, & He X (2020). Phocea, Pseudoflavonifractor and Lactobacillus intestinalis: three potential biomarkers of gut microbiota that affect progression and complications of obesity-induced type 2 diabetes mellitus. Diabetes, metabolic syndrome and obesity: targets and therapy, 13, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GD (2010). Antibiotic resistance in the environment: a link to the clinic? Current opinion in microbiology, 13(5), 589–594. [DOI] [PubMed] [Google Scholar]
- Xia J, Jin C, Pan Z, Sun L, Fu Z, & Jin Y (2018). Chronic exposure to low concentrations of lead induces metabolic disorder and dysbiosis of the gut microbiota in mice. Sci Total Environ, 631-632, 439–448. doi: 10.1016/j.scitotenv.2018.03.053 [DOI] [PubMed] [Google Scholar]
- Xu F, Fu Y, Sun T. y., Jiang Z, Miao Z, Shuai M, … Wang J (2020). The interplay between host genetics and the gut microbiome reveals common and distinct microbiome features for complex human diseases. Microbiome, 8(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Yi H, Wang T, Zhang Y, Zhu X, & Yao J (2017). Application of phosphate solubilizing bacteria in immobilization of Pb and Cd in soil. Environmental Science and Pollution Research, 24(27), 21877–21884. [DOI] [PubMed] [Google Scholar]
- Zartarian V, Xue J, Tornero-Velez R, & Brown J (2017). Children’s lead exposure: A multimedia modeling analysis to guide public health decision-making. Environmental health perspectives, 125(9), 097009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li T, Yu L, Xiao Y, Feng S, Wu J, … Chen W (2017). Effects of subchronic oral toxic metal exposure on the intestinal microbiota of mice. Science Bulletin, 62(12), 831–840. [DOI] [PubMed] [Google Scholar]
- Zhang K, Xue Y, Xu H, & Yao Y (2019). Lead removal by phosphate solubilizing bacteria isolated from soil through biomineralization. Chemosphere, 224, 272–279. doi: 10.1016/j.chemosphere.2019.02.140 [DOI] [PubMed] [Google Scholar]
- Zhang Z-J, Qu H-L, Zhao N, Wang J, Wang X-Y, Hai R, & Li B (2021). Assessment of Causal Direction Between Gut Microbiota and Inflammatory Bowel Disease: A Mendelian Randomization Analysis. Frontiers in Genetics, 12(18). doi: 10.3389/fgene.2021.631061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo K, Li J, Xu Q, Hu C, Gao Y, Chen M, … Yin Q (2019). Dysbiotic gut microbes may contribute to hypertension by limiting vitamin D production. Clinical cardiology, 42(8), 710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.