Abstract
The gene slpA, encoding the S-layer precursor protein in the virulent Clostridium difficile strains C253 and 79–685, was identified. The precursor protein carries a C-terminal highly conserved anchoring domain, similar to the one found in the Cwp66 adhesin (previously characterized in strain 79–685), an SLH domain, and a variable N-terminal domain mediating cell adherence. The genes encoding the S-layer precursor proteins and the Cwp66 adhesin are present in a genetic locus carrying 17 open reading frames, 11 of which encode a similar two-domain architecture, likely to include surface-anchored proteins.
Clostridium difficile is responsible for many cases of antibiotic-associated diarrhea and pseudomembranous colitis in humans (6). It is considered a common microorganism in nosocomial outbreaks (14). Few studies have focused on the importance of virulence factors other than toxins A and B in the pathogenesis of this disease. As in other bacteria, the role of surface proteins in the first step of colonization, i.e., adherence to enterocytes, could be of crucial importance.
Cerquetti et al. have previously described the presence of two superimposed paracrystalline surface layers (S-layers) on different C. difficile strains (4). Each layer is composed of a glycoprotein subunit which slightly varies in molecular mass among the different strains examined. Two S-layer proteins, called P36 and P47, extracted from the toxigenic and virulent C. difficile strain C253, were purified by high-performance liquid chromatography, and their N-terminal sequences were determined. Moreover, properties of adhesiveness to Caco-2 cells were observed for the P36 protein (3).
Karjalainen et al. (9) and Waligora et al. (19) have shown that C. difficile strain 79–685, a toxigenic and virulent isolate, can adhere to various culture cells in vitro and that certain stress factors can increase adherence and expression of some surface proteins. The gene encoding one of these surface proteins (Cwp66, a 66-kDa cell wall protein) has been cloned and characterized (20) (GenBank accession number AF194870). Cwp66 displays a two-domain structure, with an N-terminal cell wall-anchoring domain and a C-terminal surface-exposed domain.
Since genetic manipulation within C. difficile is difficult, thus preventing the construction of isogenic mutants, in the present work we have identified and characterized the slpA gene, encoding the S-layer precursor protein of C. difficile C253 and C. difficile 79–685, by deriving peptide sequences from purified proteins and by analyzing the genome sequence of C. difficile 630, now available (http://www.sanger.ac.uk). This is the first evidence of two S-layer proteins derived from a common precursor through posttranslational processing. A comparative analysis of SlpA and Cwp66 as well as an analysis of the region containing the genes on the genome of C. difficile strain 630 has also been performed.
(This report was presented in part as a poster at Microbial Pathogenesis Session No. 137/B, abstract B-139, of the General Meeting of the American Society for Microbiology, Los Angeles, Calif., 21 to 25 May 2000 [P. Mastrantonio, A.-J. Waligora, M. Cerquetti, A. Sebastianelli, P. Bourlioux, and T. Karjalainen, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. B-139, 2000].)
Characterization of the slpA gene.
In order to search for sequence similarity to other proteins, the two S-layer protein subunits of C. difficile strain C253, P36 and P47, were digested with protease V8 (endoproteinase Glu-C; Sigma-Aldrich, St. Louis, Mo.) and purified using a C18 high-performance liquid chromatography column (Vydac, Hesperia, Calif.). The purified peptides were analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) to determine the molecular weights and sequenced by means of Edman degradation. The sequences obtained for five peptides are presented in Table 1.
TABLE 1.
Amino acid sequences of peptides obtained by digesting C. difficile C253 S-layer proteins with V8 protease
| Peptide | Amino acid sequence |
|---|---|
| P36 | |
| D4 | NH2-AYKAIVALQNDGIE-COOH |
| P36 N-tera | NH2-ATTGTQGYTVVKNDEKKAVKQLQ DGLKDNS-COOH |
| P47 | |
| E1 | NH2-AQLVDALAAAPKAGRF-COOH |
| E2 | NH2-VVNYFVAK-COOH |
| P47 N-ter | NH2-ANDTIASQTPAKVVIKANKLKDLK |
N-ter, N terminal.
When the peptide sequences were submitted to the C. difficile strain 630 genome database using TBLASTN, all peptides could be mapped to the same contig. Analysis of the sequence of this region revealed a unique 2,160-bp gene capable of coding for a protein of 73.4 kDa. To identify the corresponding gene from strain C253, oligonucleotides based on the sequence of C. difficile 630 were synthesized and used for PCR amplification assays (Table 2). The PCR conditions consisted of an initial denaturation at 94°C followed by 30 cycles of 1 min at 94°C, 1 min of annealing at 54°C, and 1 min of extension at 72°C. At the end of the cycling, samples were held at 72°C for 5 min. Amplification was carried out in a final volume of 100 μl with a reaction mixture containing a buffer (10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2), a 200 μM concentration of each deoxynucleoside triphosphate, 100 pmol of each primer, 2.5 U of Takara Ex Taq/TM (Takara Shuzo Co., Ltd., Kyoto, Japan), and 100 ng of template DNA. The complete sequence was obtained by sequencing the overlapping PCR products. The purified PCR products, together with the appropriate oligonucleotide primers, were used as templates for sequencing reactions in a Perkin-Elmer ABI 373A DNA sequencer and an ABI prism dye terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Monza, Italy). All PCR products were sequenced on both strands.
TABLE 2.
Oligonucleotides designed to amplify the slpA genes from C. difficile strains C253 and 79–685
| Primer | Oligonucleotide sequence (5′-3′) | Primer pair (product size in bp) |
|---|---|---|
| C253 | ||
| SL1 | ATA TAA TGT TGG GAG GAA TTT AAG AA | SL1-SL2 (635) |
| SL2 | CTT TAC CAT ACT TAT CCT CTA CAG | |
| SL3 | GCT GCA ACA AAG GCA CTT AAA GTT | SL3-SL4 (929) |
| SL4 | GTC ACT CTT TAA GTT CAT AAC TCT A | |
| SL5 | TAT AGT TGA TGG TCT TGT TGC ATC | SL5-SL6 (930) |
| SL6 | AAA AGA CTT CTC ATG AGA GAA GCC | |
| 79–685 | ||
| SL14 | TAA TAT AAT GTT GGG AGG AAT | SL14-SL15 (2,240) |
| SL15 | TAT TTA AAG TTT TAT TAA AA | |
| SL18 | GCT GTA AGC TTA TAT CCT ATA G | SL14-SL18 (580) |
| SL20 | CTG CTA CTA CTT TAT ATA ATC | SL20-21 (1,177) |
| SL21 | ATA GAG TTA ACT CCA CCA GC | |
| SL22 | CTA TAG GAT ATA AGC TTA CAG CA | SL22-SL21 (890) |
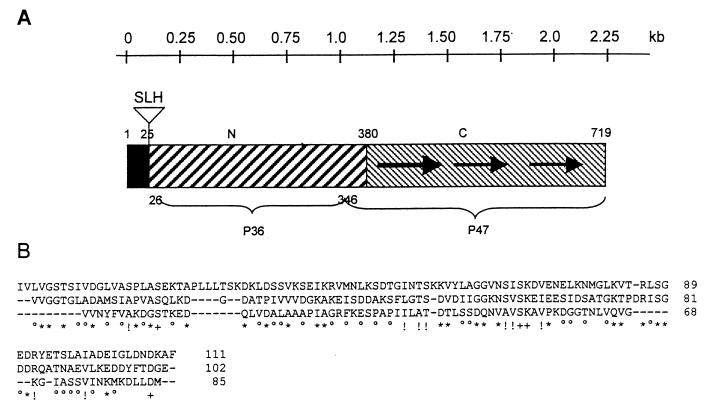
Analysis of the sequence revealed the presence of the 2,160-bp gene also in C. difficile C253 (EMBL accession number AJ291709), exhibiting 100% identity with that found in C. difficile 630 (the sequencing of the gene from strain 630 revealed nucleotide change A→T at position 314 compared to the sequence available on the Internet). This gene was referred to as slpA according to the nomenclature used for other bacteria. The encoded protein shows a characteristic procaryotic signal sequence at the N terminus and a two-domain structure as described previously for the cell wall-anchored adhesin Cwp66 isolated from strain 79–685 (20) (Fig. 1A). The C-terminal domain carries three imperfect intramolecular repeated sequences, a common feature of bacterial surface-exposed proteins (Fig. 1B). The fact that structural analysis of the S-layer of strain C253 revealed the presence of two proteins, P36 and P47, suggests that a specific cleavage of the precursor protein occurs during the maturation process. This cleavage has also been observed for Cwp66; in the cell wall this adhesin is always found in two or even three fragments (20).
FIG. 1.
(A) Diagram of the domain structure of the protein SlpA: a potential leader peptide (black box) is followed by the SLH domain and two larger domains named N and C. The numbers refer to amino acid positions in the protein. The regions corresponding to the P36 and P47 S-layer proteins of C. difficile strain C253 are indicated, as are the positions of repeat sequences (arrows). (B) Alignment of the repeats of the SlpA protein in the C-terminal domain. The length of the C-terminal repeats is between 85 and 111 amino acids. The amino acid positions in the SlpA protein are indicated. ∗ and +, two and three identical amino acids, respectively; ○ and !, two and three functionally identical amino acids (A, S, and T; D and E; N and Q; R and K; I, L, M, and V; F, Y, and W), respectively.
The availability of the N-terminal sequence of the two proteins permitted us to determine that P47 corresponds to the C-terminal part and P36 corresponds to the N-terminal part of the precursor protein (Fig. 1). The molecular masses of the peptides determined by liquid chromatography-mass spectrometry and MALDI-TOF analyses corresponded to those obtained by theoretical digestion of the virtual protein from the contig. Moreover, the molecular masses of the virtual proteins are 34,244 and 39,522 Da for P36 and P47, respectively. These data are in good agreement with the previous experimental results obtained by electrospray ionization-mass spectrometry and MALDI-TOF mass spectrometry (13).
Sequence similarity to other proteins.
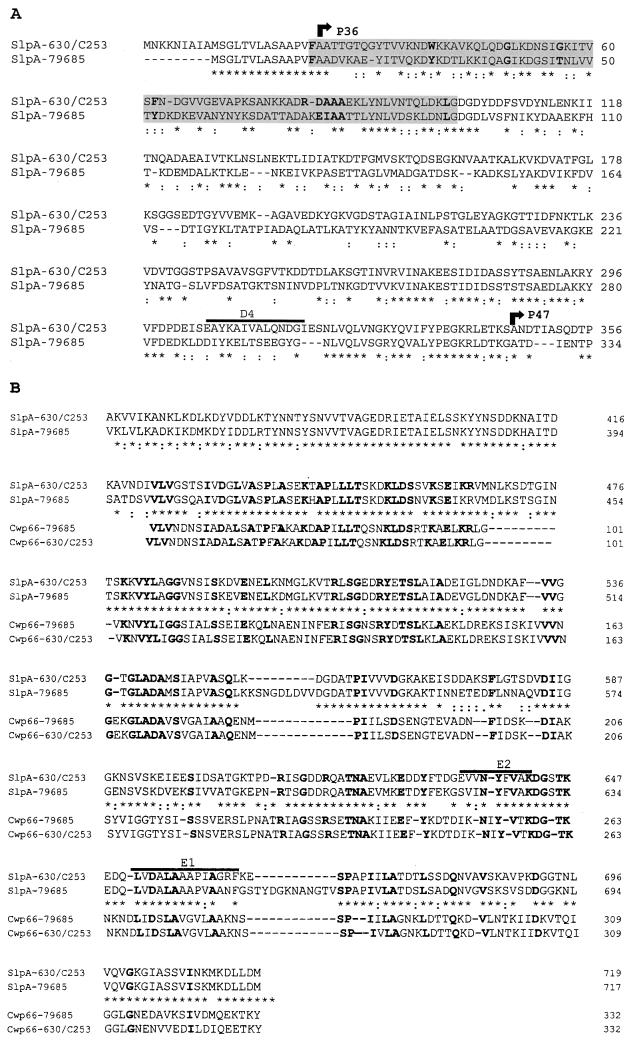
Homology searches were conducted with Fasta3 (16) or Blast 2.0 (1). SlpA carries in its N terminus (Fig. 1A and 2A) a 78-amino-acid sequence showing homology to the SLH (S-layer-homologous) domain, the presence of which has been detected in numerous S-layer proteins (12). This sequence is characterized by several highly conserved residues which are present in SlpA (Fig. 2A) and a particular predicted secondary structure, i.e., two α-helices flanking a β-strand (7) followed by a coiled-coiled domain (11), which has been found in other S-layer proteins (12). Only one of these domains is present in SlpA; in other proteins, several domains can be present, suggesting duplication of a gene fragment. The SLH domains, usually found in proteins that are anchored to the cell wall, were originally proposed to be involved in anchoring the proteins to the peptidoglycan, but more recently, secondary cell wall polymers have been identified as anchoring structures for the SLH domain (5, 8, 15).
FIG. 2.
(A) Amino acid sequence alignment of the N-terminal domains of SlpA from three C. difficile strains, C253, 630, and 79–685. Note the pronounced variability. The SLH domain is shaded in gray; the conserved residues of this domain (12) are in boldface letters. Asterisks or colons denote amino acids that are identical or similar, respectively. The positions of the two peptides of C. difficile C253 (Table 1) are marked above the sequence. The N-terminal amino acids of the P36 and P47 proteins are indicated with arrows. (B) Amino acid sequence alignment of the putative cell wall-anchoring domains of SlpA and Cwp66 from C. difficile strains 630 and C253 and strain 79–685. Asterisks or colons denote amino acids that are identical or similar, respectively, in the SlpA proteins only. Conserved residues in the four proteins are in boldface.
The 339-amino-acid C-terminal domain of SlpA (residues 380 to 719) shows significant homology (29% identity and 51% similarity) to a domain found in N-acetylmuramoyl-l-alanine amidase CwlB of Bacillus subtilis (GenBank accession number Q02114). CwlB is known to be released from the bacteria and reattached to the cell wall peptidoglycan (10) through the domain displaying homology to SlpA (corresponding to the P47 protein). These data suggest that both P36 and P47 carry a motif that could anchor the S-layer proteins to the cell wall, although in the case of P36 the attachment could be indirect through a secondary protein. A similar domain was also found in Cwp66 (20).
Antibodies raised against P47 of C. difficile C253 recognize in other C. difficile strains proteins with molecular masses close to 47 kDa (4). This suggests that this likely cell-anchoring domain is conserved among C. difficile strains. To verify whether this is the case in strain 79–685, the slpA gene was amplified from the strain by PCR as described above using primers listed in Table 2 and its nucleotide sequence was determined. The encoded precursor protein has the same two-domain structure as in strain C253 (Fig. 2). Interestingly, alignments performed with the ClustalW program (18) revealed that one of the domains is well conserved (C-terminal domain) among the three strains studied (77% identity between strains C253 and 630 and strain 79–685) and displays sequence conservation with the cell wall-anchoring N-terminal domain of Cwp66 (Fig. 2B). The second domain is highly variable: only 33% of the residues are identical between strains C253 and 630 and strain 79–685 (Fig. 2A).
P36 (N-terminal domain) shows remote homology (<25% identity) with the SlaP S-layer protein of Bacillus stearothermophilus (P35825), glycosyltransferase-S of Streptococcus mutans (D89979), and various flagellins of Salmonella (e.g., U06197 and U06227). However, glycosyltransferases have been found to mediate adherence in other bacteria (for a review, see reference 17). Even if the N-terminal domain does not show significant homology to known adhesins, preliminary experiments using the Caco-2 cell line and specific antisera have shown it to be involved in the adhesion process (3). Antiserum against whole cells of C. difficile C253 and monospecific antiserum against P36 were raised in adult New Zealand rabbits by following procedures previously described (2). The P36 antiserum, prepared using the antigen obtained by electroelution of the P36 band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis, recognized only P36 when assayed by immunoblot analysis against EDTA- or urea-extracted surface proteins of C. difficile C253 (2, 4). In an inhibition-of-adhesion assay, 3-day-old monolayers of Caco-2 cells infected with C. difficile C253 bacteria preincubated with nonimmune serum, anti-C253 whole-cell serum, and anti-P36 serum showed mean numbers of adherent bacteria per cell ± standard deviations of 43.4 ± 16.42, 0.16 ± 0.20 (P = 0.01), and 3.66 ± 3.23 (P < 0.05), respectively (values were determined from three separate experiments; statistical analysis was performed using Student's t test). Adherence of C. difficile C253 bacteria was specifically blocked by the homologous antiserum and significantly reduced by the P36 antiserum compared to the adherence of bacteria pretreated with a nonimmune serum.
Sequence analysis of SlpA from C. difficile strains 79–685 and C253 (Fig. 2A) revealed pronounced variations in the N-terminal domains suggesting that SlpA is surface exposed and thereby subject to antigenic drift. The surface localization of P36, previously demonstrated by immunofluorescence studies using the P36 antiserum (2), and the above-mentioned data on its role as adhesin may support this hypothesis.
Analysis of a gene cluster carrying slpA and cwp66.
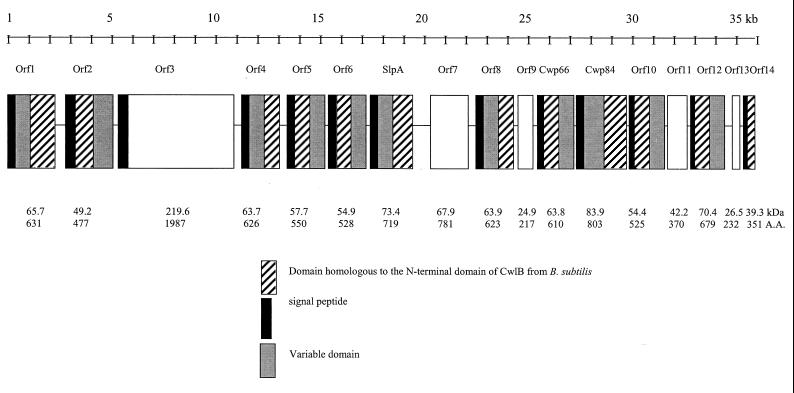
Examination of a 37-kb DNA fragment flanking cwp66 and slpA of the genome sequence of C. difficile strain 630 revealed 17 open reading frames (ORFs) in the same orientation. Eleven of these ORFs encode a domain, present in either the N- or C-terminal part of these putative secreted proteins, showing homology to the cell wall-anchoring domain of the Cwlp autolysin of B. subtilis (Fig. 3). This conserved domain is characterized by the presence of three PII/LL motifs, although the third repeat is less conserved than the first two. The anchoring domain is associated in all cases with a putative peptide signal and with another domain (except for Orf14), whose function appears variable: in Cwp66 and SlpA, this domain carries adhesive function; Cwp84 carries in this domain a cysteine protease active site and has proteolytic activity (our unpublished data); and Orf12 carries an active site found in N-muramyl-l-alanine amidases. The variable domains encoded by the other ORFs display remote homology (<20%) to various bacterial outer membrane proteins. The variable domains of Cwp66 and Orf10 and of Orf5, Orf6, and Orf2 are up to 40% homologous. We suspect that all these proteins could be cell wall anchored through their CwlB-like domain. The presence in multiple genes of sequences encoding the two domains suggests modular evolution and subsequent gene duplication from a common ancestor for B. subtilis cwlB and C. difficile. During evolution, this domain was placed in either the N or C terminus and became associated with another domain as in Cwp66 or many autolysins. Interestingly, the cluster also carries a large gene (orf3) capable of encoding a 220-kDa protein that does not exhibit the two-domain structure. It displays remote homology to various membrane-associated proteins from Plasmodium falciparum.
FIG. 3.
Genetic organization of a 37-kb DNA fragment carrying the slpA and cwp66 genes from the genome of C. difficile 630. The proteins encoded by these genes and their sizes, molecular masses, and domain structures are indicated. Orf7 exhibits significant homology with the SecA protein from B. subtilis, Orf9 with dehydrogenases, Orf12 with N-acetyl-l-alanine-muramyl amidase, and Orf13 with glycosyltransferases. A.A., amino acids.
In conclusion, we report here on the characterization of the slpA gene in C. difficile C253 and 79–685. This gene in C. difficile 630 is located in a cluster including genes encoding proteins with adhesive properties. The isolation of this cluster is an important step in the characterization of the process of colonization by C. difficile: analogously to many other bacteria, C. difficile possesses multiple surface-exposed proteins acting as adhesins. Furthermore, surface proteins are attractive targets for the development of a vaccine against this troublesome pathogen.
Nucleotide sequence accession number.
The DNA sequence of the slpA gene from C. difficile strains C253 and 79–685 have been deposited in the EMBL database under accession number AJ291709 and in the GenBank database under accession number AY004256.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerquetti M, Pantosti A, Stefanelli P, Mastrantonio P. Purification and characterization of an immunodominant 36kDa antigen present on the cell surface of Clostridium difficile. Microb Pathog. 1992;13:271–279. doi: 10.1016/0882-4010(92)90037-o. [DOI] [PubMed] [Google Scholar]
- 3.Cerquetti M, Conti F, Spigaglia P, Mastrantonio P. Adherence of Clostridium difficile C253 to Caco-2 cells. Microb Ecol Health Dis. 1998;10:165. [Google Scholar]
- 4.Cerquetti M, Molinari A, Sebastianelli A, Diociaiuti M, Petruzzelli R, Capo C, Mastrantonio P. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb Pathog. 2000;28:363–372. doi: 10.1006/mpat.2000.0356. [DOI] [PubMed] [Google Scholar]
- 5.Chauvaux S, Matuschek M, Beguin P. Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis cell envelopes. J Bacteriol. 1999;181:2455–2458. doi: 10.1128/jb.181.8.2455-2458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George W L. Antimicrobial agent-associated colitis and diarrhea: historical background and clinical aspects. Rev Infect Dis. 1984;6(Suppl. 1):S208–S213. doi: 10.1093/clinids/6.supplement_1.s208. [DOI] [PubMed] [Google Scholar]
- 7.Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 8.Ilk N, Kosma P, Puchberger M, Egelseer E M, Mayer H F, Sleytr U B, Sára M. Structural and functional analyses of the secondary cell wall polymer of Bacillus sphaericus CCM 2177 that serves as an S-layer-specific anchor. J Bacteriol. 1999;181:7643–7646. doi: 10.1128/jb.181.24.7643-7646.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karjalainen T, Barc M C, Collignon A, Trolle S, Boureau H, Cotte- Laffitte J, Bourlioux P. Cloning of a genetic determinant from Clostridium difficile involved in adherence to tissue culture cells and mucus. Infect Immun. 1994;62:4347–4355. doi: 10.1128/iai.62.10.4347-4355.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda A, Sekiguchi J. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J Bacteriol. 1991;173:7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 12.Lupas A, Engelhardt H, Peters J, Santarius U, Volker S, Baumeister W. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauri P L, Pietta P G, Maggioni A, Cerquetti M, Sebastianelli A, Mastrantonio P. Characterization of surface layer proteins from Clostridium difficile by liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 1999;13:695–703. doi: 10.1002/(sici)1097-0231(19990430)13:8<695::aid-rcm542>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.McFarland L V, Mulligan M E, Kwok R Y, Stamm W E. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 15.Mesnage S, Tosi-Couture E, Fouet A. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol Microbiol. 1999;31:927–936. doi: 10.1046/j.1365-2958.1999.01232.x. [DOI] [PubMed] [Google Scholar]
- 16.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmonds R S, Tompkins G R, George R J. Dental caries and the microbial ecology of dental plaque: a review of recent advances. N Z Dent J. 2000;96:44–49. [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waligora A-J, Barc M-C, Bourlioux P, Collignon A, Karjalainen T. Clostridium difficile cell attachment is modified by environmental factors. Appl Environ Microbiol. 1999;65:4234–4238. doi: 10.1128/aem.65.9.4234-4238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waligora A-J, Hennequin C, Mullany P, Bourlioux P, Collignon A, Karjalainen T. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun. 2001;69:2144–2153. doi: 10.1128/IAI.69.4.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]