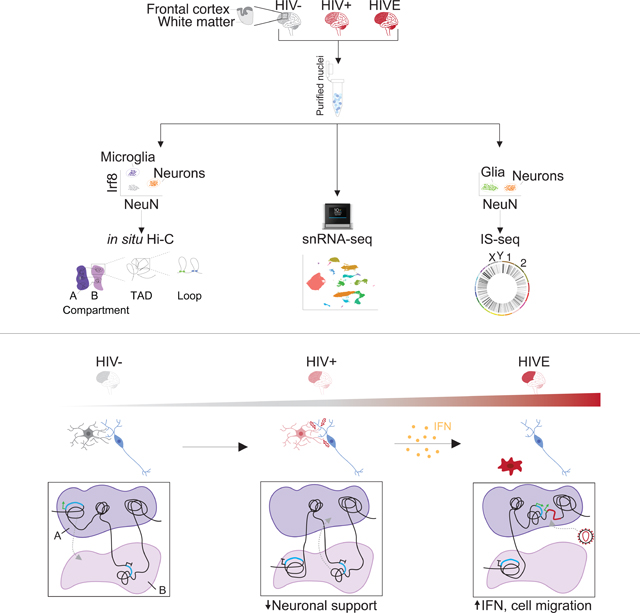
Summary:
To explore genome organization and function in the HIV infected brain, we applied single nuclei transcriptomics, cell-type specific chromosomal conformation mapping, and viral integration site sequencing (IS-seq) to frontal cortex from individuals with encephalitis (HIVE) and without (HIV+). De-repressive changes in 3D genomic compartment structures in HIVE microglia were linked to transcriptional activation of interferon (IFN) signaling and cell migratory pathways, while transcriptional downregulation and repressive compartmentalization of neuronal health and signaling genes occurred in both HIVE and HIV+ microglia. IS-seq recovered 1,221 brain integration sites showing distinct genomic patterns as compared to peripheral lymphocytes, with enrichment for sequences newly mobilized into a permissive chromatin environment after infection. Viral transcription occurred in a subset of highly activated microglia comprising 0.003% of all nuclei in HIVE brain. Our findings point to disrupted microglia-neuronal interactions in HIV and link retroviral integration to remodeling of the microglial 3D genome during infection.
eTOC blurb:
Plaza-Jennings et al. study HIV genome integration patterns in infected brain and link the spatial organization of the chromosomal material in brain immune cells to gene expression changes at single cell resolution, including disruption of neuronal support functions and dynamic interrelations between neuroinflammation and viral insertion into the host genome.
Graphical Abstract
INTRODUCTION
HIV enters the brain within the first two weeks of infection, and biomarkers of central nervous system (CNS) inflammation as well as neurologic symptoms have been observed in acute HIV disease1–3. Considering that roughly 7×109 microglial cells, the primary CNS cell type infected by HIV, reside in an adult human brain4–7, the CNS is potentially a large reservoir site2,8. HIV-associated neurocognitive disorder (HAND) affects 20–50% of the 37 million people living with HIV (PLWH), with the milder forms of HAND predominating in the era of combined antiretroviral therapy (cART)9–11.
Genomic and transcriptomic exploration of the HIV-infected human brain is critical to developing treatments for HAND and HIV cure strategies. However, to date, genomic studies in the HIV infected brain have primarily focused on bulk tissue gene expression profiling and demonstrated neuroinflammation and metabolic alterations12,13. Herein we describe the first cell-type-specific integrative genomics studies, including HIV integration site sequencing, chromosomal conformation (‘3D genome’) mapping, and single nucleus transcription, of frontal lobe tissues from individuals who were HIV-infected with encephalitis and without encephalitis.
RESULTS
Transcriptomic profiling of the HIV-infected brain at a single nucleus resolution
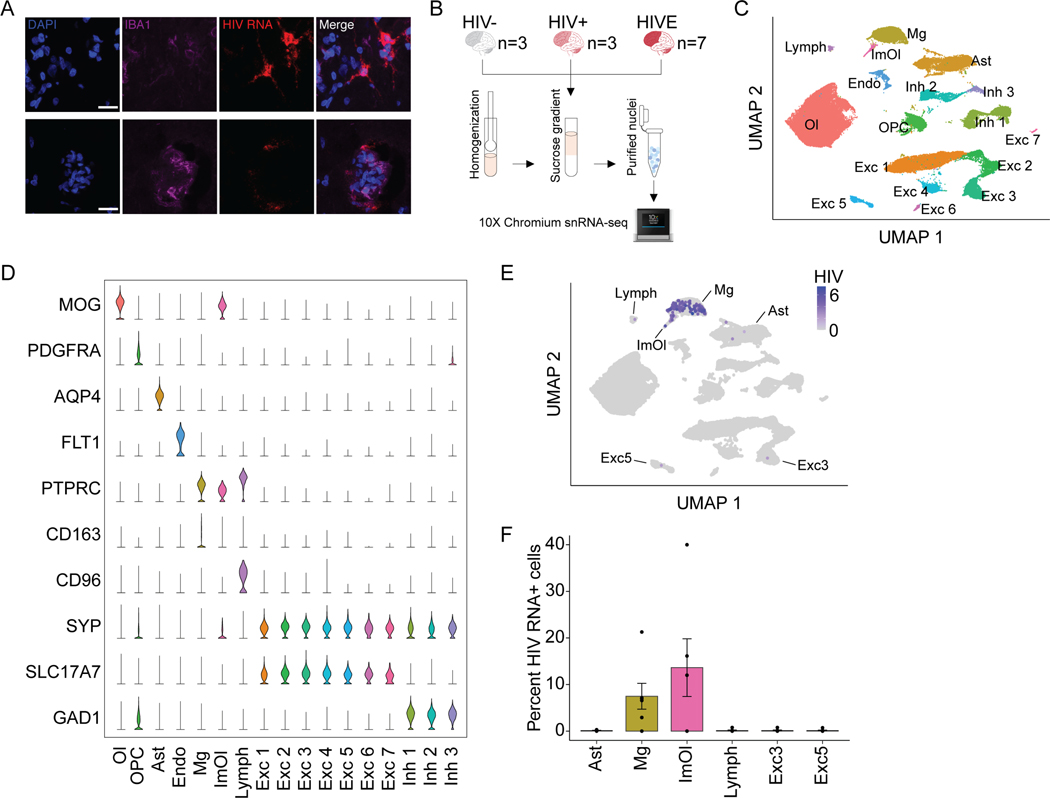
Using dual-labeling RNA fluorescence in situ hybridization (FISH) for HIV transcripts and immunohistochemistry for microglial markers, we first confirmed high levels of HIV RNA expression in encephalitic brain in microglia (Figure 1A). Next, in order to obtain deeper, cell-type specific resolution of transcription in HIV-infected and uninfected brain, we performed 10X Chromium single nucleus RNA-sequencing (snRNA-seq) of frontal lobe, including Frontal Cortex gray matter and underlying subcortical White Matter (FC/WM), of HIV uninfected (HIV-, n=3), HIV infected (HIV+, n=3) and HIV infected with encephalitis (HIVE, n=7) samples (Figure 1B, Table S1). We profiled a total of 69,843 nuclei to an average depth of 119,797 ± 32,766 reads and a median of 2,401 ± 221 expressed genes/nucleus (Supplementary file S1). Consistent with previous studies 14,15, we identified 17 cell type clusters, including excitatory (Exc1–7) and inhibitory neuronal subtypes (Inh1–3), oligodendrocytes (Ol), oligodendrocyte progenitor cells (OPC), astrocytes (Ast), endothelial (Endo), lymphoid cells (Lymph), microglia/CD163+ perivascular macrophages (Mg), and immune oligodendroglia (ImOl; Figure 1C–D, for marker gene expression see Supplementary file S2). ImOl are a previously described, rare cell population with marker gene expression of both oligodendrocyte and immune-related genes16,17.
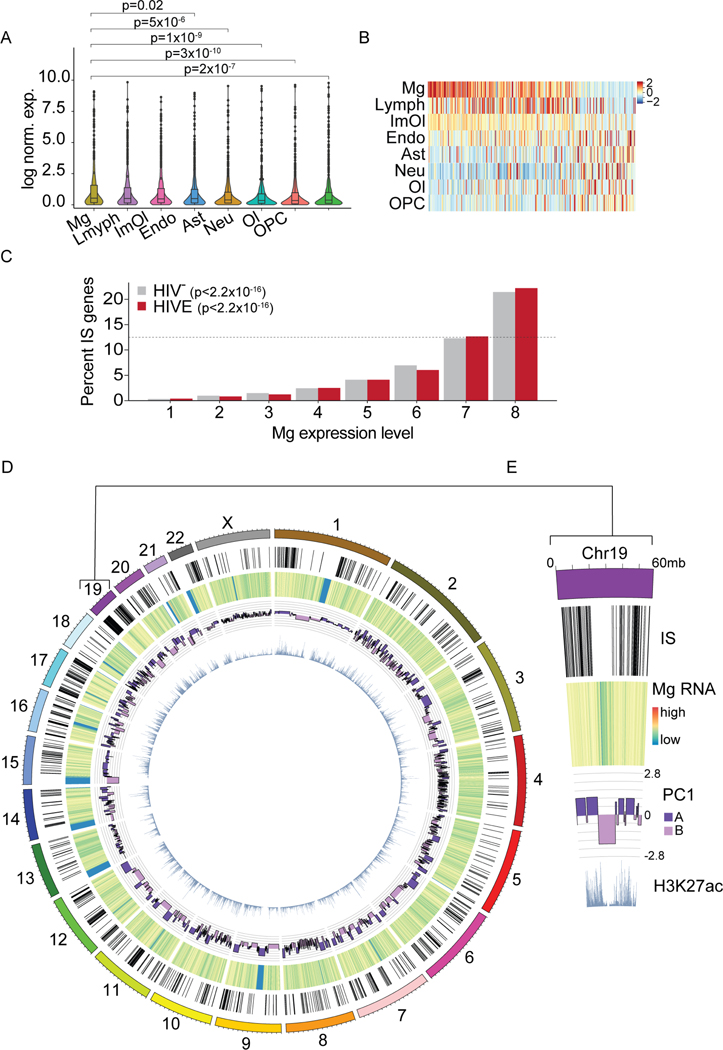
Figure 1. Cell type specificity and prevalence of HIV RNA+ nuclei in HIVE.
(A) Triple stained HIVE brain section with HIV RNA FISH (red), Iba1 (purple), and DAPI (blue). (B) Sample number and workflow of 10X Chromium single nucleus RNA-sequencing. (C-E) All samples (n=13) clustering and gene expression. (C) Uniform Manifold Approximation and Projection (UMAP) plot showing identified cell clusters for n=69,843 total nuclei. Excitatory neurons (Exc1–7), inhibitory neurons (Inh1–3), oligodendrocytes (Ol), oligodendrocyte progenitor cells (OPC), endothelial cells (Endo), astrocytes (Ast), lymphoid cells (Lymph), microglia (Mg), and immune oligodendroglia (ImOl). (D) Marker gene expression for each cluster, each row scaled to the maximum of expression for that gene. (E) UMAP plot displaying log normalized expression of HIV transcripts. (F) Percentage (mean ± S.E.M) of HIV RNA+ HIVE nuclei (n=7 samples, 1 dot = 1 donor) for clusters with HIV RNA+ nuclei. See also Figure S1.
Next, we screened single nuclei for the presence of HIV transcripts and observed HIV-mapping transcripts in all cell types in HIV (Figure S1A). Highly expressed transcripts are associated with low-level contamination in snRNA-seq, potentially accounting for false positive calls in our single-nuclei HIV expression profiles18,19. Thus, we performed snRNA-seq of intermixed HIVE and naïve mouse brain tissue (Figure S1A–E), and thereby determined a stringent read count threshold of >5 HIV-mapping reads required for a nucleus to be considered HIV RNA+, which removed 99.8% of all contaminated nuclei (Figure S1D–F). Overall, 4.8% of nuclei in the HIVE Mg cluster (133/2,756) and in HIVE ImOl (10/209) were HIV RNA+, while the proportion of astrocytes, lymphocytes, and neurons harboring HIV RNA-expressing nuclei was lower by at least one order of magnitude (~0.1 – 0.3%, or 1–3 HIV RNA+ nuclei/cluster) (Figure 1E–F). Consistent with these findings, CD4, encoding the primary viral receptor20, was only detectable in Mg and ImOl clusters. Low expression of CD4 was observed in the lymphoid cluster likely due to low ante-mortem CD4 counts in HIVE donors (Table S1), who comprised 87% of all lymphoid nuclei. Expression of CCR5 and CXCR4, the two major viral co-receptors21, was low across all different FC/WM cell types with the exception of robust CXCR4 expression in the Lymph cluster (Figure S1G). Finally, in contrast to sensitive PCR methods confirming HIV DNA in 3/3 HIV+ brains for which we performed snRNA-seq (Table S1), we did not detect HIV transcripts in any nucleus from HIV+ brain (Figure S1A), while it was readily detectable in 5/7 HIVE brains.
Transcriptomic alterations in HIVE microglia linked to large scale reorganization of chromosomal compartments
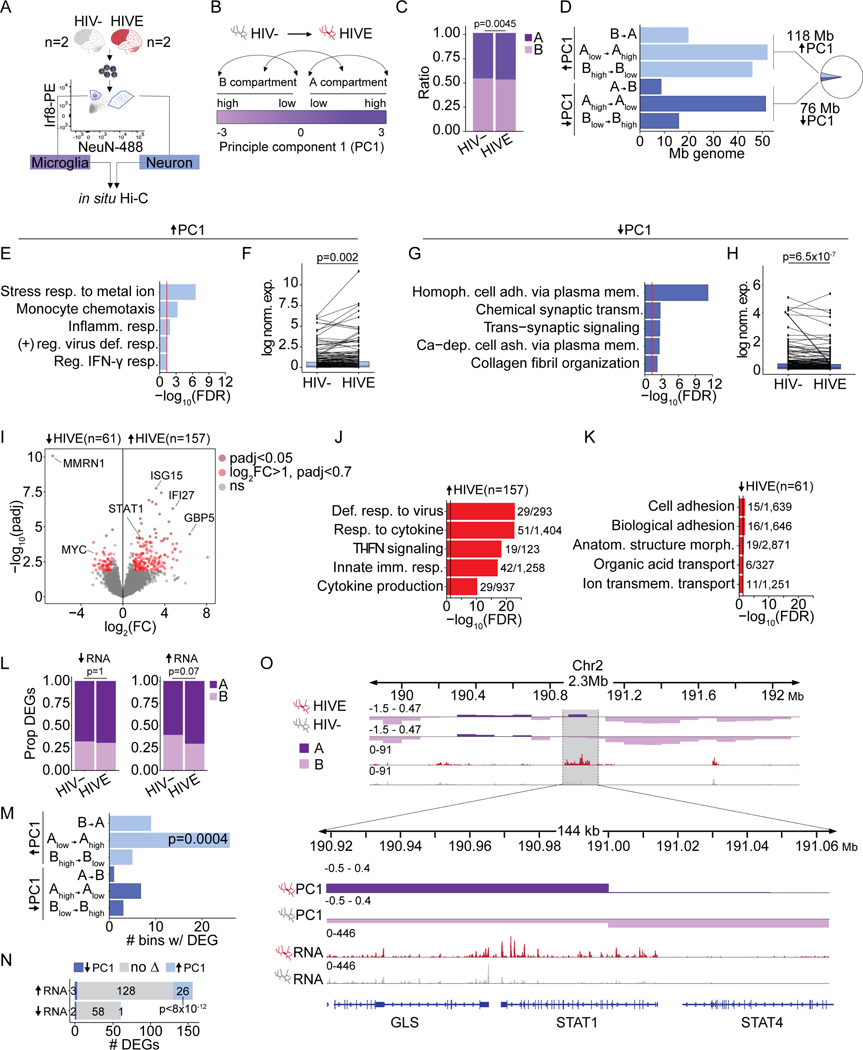
Cell culture models of immune cells undergoing activation have shown broad changes in chromosomal conformation22–26, while 3D reorganization of any immune cell in vivo, including microglia, has not been examined. To explore, we generated genome-scale Hi-C chromosomal contact maps for microglia and neurons from two HIVE brains with confirmed HIV expression by snRNA-seq, together with age-and sex- matched controls (Table S1). Microglial and neuronal nuclei were isolated using fluorescence-activated nuclei sorting (FANS) with microglia- (Interferon regulatory factor 8, Irf8) and neuron-specific (NeuN) markers (Figure 2A, Figure S2A), followed by in situ Hi-C 3D genome profiling with >200M valid intrachromosomal chimeric reads/sample (Supplementary file S1). Pairwise correlations found that the largest Hi-C difference was by cell type followed by diagnosis (Figure S2B). To explore encephalitis-related changes to microglial genome structure, we first focused on megabase-sized (~0.1–10Mb) chromosomal compartments, divided into gene-dense A-compartments with active gene expression and gene-poor B-compartments with low levels of active gene expression (Figure 2B, see Methods). We observed a slight but significant shift in the proportion of the genome found in the A compartment in HIVE (47.3%) vs. HIV- microglia (46.2%, Chi-square p=0.0045, Figure 2C). Analysis also revealed significant changes in A/B compartmentalization (detected by dcHiC, Padj<0.05) encompassing 196Mb, or 6.4% of (haploid) genome in HIVE brain as compared to HIV- (Figure 2D). 118Mb showed a significant increase in the PC1 eigenvector (HIVE > HIV-) corresponding to a switch to a more open conformation in HIVE microglia and 76Mb showed changes in the opposite direction, corresponding to compartment remodeling towards a more closed state (Figure 2D, Supplementary file S3). Of note, there was minimal, negative correlation (R=−0.065) of NeuN+ ΔPC1 and Irf8+ ΔPC1 values for compartment bins undergoing a significant eigenvector change in HIVE vs. HIV- Mg (Figure S2C). Likewise, ΔPC1 values for compartment bins with a significant eigenvector change in both cell types showed a negative correlation (R=−0.29, P = 5.4×10−14) (Figure S2D). Therefore, Hi-C compartment alterations in HIVE are highly specific for microglia and not shared by surrounding neuronal nuclei.
Figure 2. Chromosomal compartment remodeling and transcriptomic alterations in HIVE microglia.
(A) Representative FANS plot for sorted NeuN+/neuronal and Irf8+/microglia nuclei for in situ Hi-C. (B) Schematic A/B Hi-C compartment designation based on the eigenvector value of principle component 1 (PC1). Arrows illustrate the different classes of compartment switches between HIVE and HIV- microglia. (C) Bar plot, proportion of genome in A/B compartments in HIV- and HIVE microglia (P, chi-square). (D) Bar graph summarizing dcHiC genome-wide compartment remodeling in HIVE vs. HIV- microglia. X-axis shows Mb of genome undergoing the compartment switches indicate on the Y-axis. A-compartmentalization (ΔPC1 HIVE-HIV- >0, FDR<0.05) shown in light blue; B-compartmentalization (ΔPC1 HIVE-HIV- <0, FDR<0.05) in dark blue. Inset pie chart shows the total percent of significant A- and B- compartmentalization in the genome. (E, G) Gene ontology enrichments and (F, H) differential expression for genes undergoing A-compartmentalization (E,F) or B-compartmentalization (E,F) in HIVE as compared to HIV- microglia. (F, H) Paired line plots showing average log normalized expression of genes in compartments with significant PC1 increase in HIVE microglia. For visualization, randomly selected subsets of 200 genes are shown. P-value reflects Wilcoxon signed rank test across all genes with expression levels > 25th percentile in either HIV- or HIVE microglia; (F) n=1030, (H) n=566. (I) Volcano plot, -log10(adjust p-value) (Y-axis) and log2(Fold Change) (X-axis) for snRNA-seq differential expression of the microglial cluster (Figure 1C, Mg) from n=3 HIV- and n=7 HIVE brains. Dark red dots mark genes with padj<0.05. Light red dots, genes with log2(FC)>1 and padj<0.7. (J-K) Biological process GO enrichment for up- (J) and downregulated (K) genes with N dataset genes/total GO category genes indicated. Vertical black line marks significance FDR=0.05. (L) Proportion of up- or downregulated microglial DEGs in A and B compartments. P, chi-square. (M) Bar plot, x-axis marks number of 10kb bins undergoing compartment switching that contain a DEG. P, Fisher’s exact test compared to the proportion of the whole genome that underwent the indicated compartment switch. (N) Overlap between up- and downregulated DEGs in HIVE vs. HIV- microglia and all genes that underwent compartment switching. P, hypergeometric test, representation factor 35. (O) Representative browser shot of 144kb window on chromosome 2, showing STAT1/STAT4 gene locus with Hi-C PC1 eigenvector value (top two tracks) at a 10kb resolution, and snRNA-seq normalized read counts for the Mg cluster (bottom two tracks). See also Figures S2, S3 and S5.
Importantly, functional pathway analysis revealed that genes positioned within genomic loci with significantly increased A-compartmentalization in HIVE microglia showed significant enrichment for interferon (IFN) and other cytokine signaling pathways as well as myeloid chemotaxis and migration pathways (Figure 2E, Supplementary file S3–4). Furthermore, regions of increased A compartmentalization in HIVE included a total of 1,940 genes that were expressed at significantly higher levels in HIVE (N=7) as compared to HIV- (N=3) microglia from our snRNA-seq data (Wilcoxon signed rank test, P = 0.002, Figure 2F, Supplementary file S3). Of these, N=335 genes that switched eigenvector direction from B→A showed significant enrichment in immune functions, including cytokine signaling, viral response, inflammatory pathways, and T-cell differentiation pathways (Figure S2E). Another set of N=1,231 genes switched from Alow→Ahigh and were enriched for metal ion stress response pathways due to a group of metallothionein genes involved in the immune response and in mediating monocyte resistance to HIV-induced apoptosis (Figure S2F) 27,28. Genes that switched from Bhigh→Blow (N=308) were also enriched for immune functions, including lymphocyte chemotaxis, IFN, and interleukin pathways (Figure S2G).
In striking contrast to these associations between A-compartmentalization and transcriptional upregulation of IFN and other cytokine and immune signaling genes in the HIVE microglia, gene ontologies of genomic loci undergoing repressive compartment remodeling were enriched for various homeostatic functions, such as cell adhesion, regulation of synaptic signaling, and neurodevelopment (Figure 2G, Figure S2H–I, Supplementary file S4), and were overall expressed at lower levels in HIVE microglia as compared to HIV- (Wilcoxon signed rank test, P = 6.5 × 10−7, Figure 2H).
To further test the link between compartment switching and transcriptional reprogramming in HIVE microglia, we inverted our order of analysis by first calling N=218 differentially expressed genes (DEGs) in the snRNA-seq Mg cluster (DESeq2, log2(Fold change) > 1, Padj < 0.7) and then assigning their respective compartments (Figure 2I, Supplementary file S1). Top functional pathways for the N=157 upregulated genes, as with A-compartmentalized genes, were related to immune signaling, including the interferon (IFN) response with key transcription factors STAT1 and STAT2, and IFN-stimulated genes ISG15, IFI6, IFI27, IFI44, and IFIH1 (Figure 2J, Supplementary file S4). In addition, upregulated expression with A-compartmentalization included several members of the guanylate binding proteins (GBPs), GBP1, 2, 4, and 5, which mediate NFAT-dependent transcription in myeloid cells and interact with the inflammasome to regulate cytokine production and pyroptosis (Figure 2J)29. In contrast, pathway enrichment for the N=61 downregulated and B-compartmentalized genes was limited to cell adhesion genes (Figure 2K, Supplementary file S4).
Of note, from the 157 DEGs upregulated in HIVE Mg, 70% were found in the microglial A compartment in HIVE as compared to 60% in HIV- (Chi-square p=0.07, Figure 2L), with significant preference for Alow→Ahigh compartment switching (Figure 2M) and overall increased A-compartmentalization in HIVE (up-regulated DEGs, 5-fold enrichment over chance, hypergeometric p<8×10–12, Figure 2N). A representative example, the STAT1/STAT4 locus, is shown in Figure 2O. We did not observe significant changes in compartment location of downregulated DEGs (Figure 2L, N). Together, these results suggest that HIV induces open chromatin configuration in microglia that accompanies transcriptomic activation of inflammatory pathways.
3D genome remodeling at IFN-related loci is partially recapitulated by IFN-γ stimulation of cultured microglia
In macrophages, IFN stimulation causes similar changes to 3D genome structure as influenza viral infection30. Because IFN-γ related genes were enriched in regions of A-compartmentalization in our HIVE microglia, we hypothesized that 3D-genome remodeling may be caused in part by IFN stimulation. To explore, we performed in situ Hi-C on HMC3 immortalized microglia stimulated with IFN-γ or a vehicle control (BSA) for 24 hours (Figure S3A–B). Indeed, similarly to HIVE microglia, 7.2% of the HMC3 genome underwent compartment switching upon IFN-γ stimulation, including 130Mb and 93Mb with significant A and B compartmentalization respectively (Figure S3C). ΔPC1 values were modestly but significantly correlated between HMC3 cells and HIVE microglia for regions of compartment switching in HIVE microglia (R=0.17, p=1×10−13) and for regions of significant compartment switching in both HIVE microglia and HMC3 cells (R=0.23, p=0.002; Figure S3D–E). Regions with significant switching in both were enriched for IFN-γ response, stress, and antiviral pathways (Figure S3F). Thus, while global changes to 3D genome structure show only limited correlation between in vivo and in vitro models, IFN stimulation drives changes to 3D genome structure specifically at IFN-regulated loci both in HIVE microglia in vivo and in the HMC3 cell culture model. Illustrative examples include the previously discussed STAT1 locus on chromosome 2 and a large ~2Mb stretch of chromosome 16, containing the immune-related genes NLRC5, CCL22, the metallothionine genes, CX3CL1, HERPUD1, and NUP93, with each of these loci showing a significant increase in PC1 in both HIVE microglia and in IFN-stimulated HMC3 cells (Figure S3G–H).
Microglial plasticity of chromosomal domains and loops linked to altered gene expression
The structural organization of chromosomes is mediated by two main forces – phase separation mechanisms driving the spatial segregation of A and B chromatin compartments and ATP-dependent loop extrusion by CTCF, YY1 and cohesin that furnishes topologically associated domains (TADs) and loops31–33. Peripheral myeloid cells exposed to microbial environments orchestrate a rapid inflammatory response that includes changes both in TAD domain landscapes and compartments34. Therefore, after having seen that HIVE was associated with broad remodeling of compartment architecture, we explored TAD and loops alterations in brain microglia. We first mapped genome-wide TAD insulation scores, a measure comparing the strength of intra-TAD chromosomal contacts in comparison to boundary crossing contacts35. HIVE in comparison to HIV- microglia, showed a significant genome-wide increase in insulation score, indicating weaker domain boundaries (Wilcoxon rank sum P=7×10−11, Figure S4A). Using ENCODE YY1 and CTCF ChIP-seq reference datasets from GM12878 and CD14+ monocytes respectively, we saw significant enrichment of CTCF and YY1 binding specifically at sites with the top 10% largest TAD insulation changes in HIVE Mg, consistent with the role of YY1 and CTCF as regulators of TAD domain insulation and loop formation (Figure S4B) 36–38. The same regions were significantly associated with compartment remodeling, with regions of significant B compartmentalization showing large insulation score increases (Figure S4C–D).
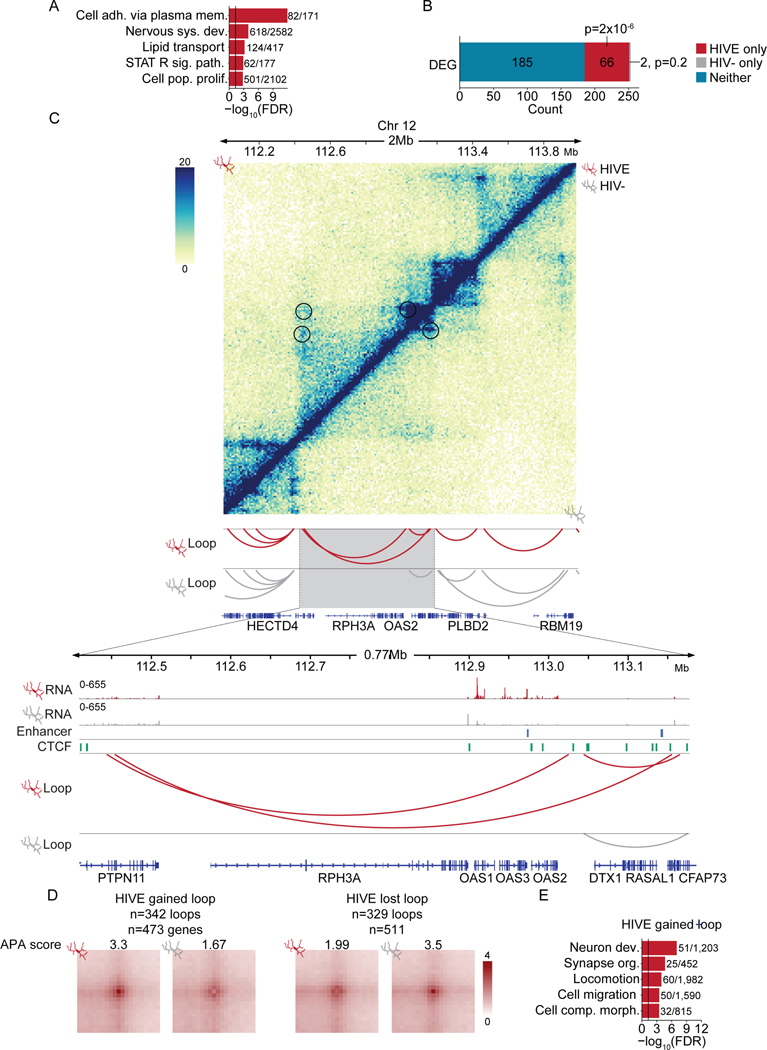
Chromosomal loops, defined as distinct contacts between two loci in the absence of similar interactions in the surrounding sequences39, showed large-scale remodeling genome-wide between groups, with N= 5,959 loops specific to HIVE, N=2,799 loops specific to HIV−, and N=1,493 loops shared between HIVE and HIV− microglia (Supplementary file S5), with highly significant overlap of CTCF binding with loop anchors ( 57% of all HIVE and HIV- loop anchors (Z=272, p<0.001), 54% of differential loop anchors (Z=55, p<0.001, Figure S4E). Genes specific to the span of HIVE loops were enriched for functions including STAT signaling, cell adhesion, and nervous system development (Figure 3A, Supplementary file S4). Furthermore, microglial DEGs, which included multiple oligoadenylate synthetase (OAS) cluster genes on chromosome 12, encompassing OAS1, 2, and 3, defined as IFN-stimulated genes encoding RNases capable of cleaving viral RNAs40, were significantly enriched in HIVE-specific loops (Figure 3B–C). No enrichment was observed for HIV- loops. We performed further loop analysis controlling for read depth41, and identified 342 loops gained and 329 loops lost in HIVE (Figure 3D, Supplementary file S5). HIVE-gained loop anchors contained 473 genes (Supplementary file S5), representing pathways similar to those observed in compartment and differential expression analysis, including synapse organization and cell migration (Figure 3E, Supplementary file S4). These included 9 DEGs (hypergeometric representation factor=4.4, p<2.5×10−4), including SELL, LRRC4C, FLT1 as regulators of cell motility and the long non-coding RNA LINC00513, an important regulator of type I-IFN signaling that promotes STAT1 and STAT2 phosphorylation 42–45. In contrast, HIVE-lost loops, containing 511 genes (Supplementary file S5), lacked significant GO enrichment.
Finally, we also identified multiple loop anchors with conserved loop remodeling in HIVE microglia and IFN-γ treated HMC3 cells (Supplementary file S5), including GBP and other loci showing A compartmentalization and upregulated expression in HIVE microglia (Figure S4F).
The microglial transcriptome and 3D genome in the HIV+ brain without encephalitis
Our studies described above in HIVE microglia revealed transcriptomic reprogramming and widespread reconfiguration of chromosomal conformations encompassing hundreds of megabases, revealing a functional divergence defined by de-repression of immune-related loci, in sharp contrast to the repressive compartmentalization of microglial genes linked to neuronal support functions. We next explored whether similar types of alterations exist in HIV+ brains that had not progressed to encephalitis. We examined the microglial transcriptome in the snRNA-seq datasets from our 3 HIV+ samples that had been processed in parallel to the 10 HIVE and control specimens. Indeed, there was a significant linear correlation (Pearson R=0.41, p=1×10−11) between the log2(fold change) in expression in HIVE and HIV+ vs. HIV- Mg for the 218 DEGs initially identified in HIVE microglia, suggesting that many of the HIVE Mg gene expression changes may be present, to a lesser extent, in HIV+ Mg (Figure S5A). Furthermore, differential expression analysis of HIV+ vs. HIV- microglia, utilizing the same significance threshold used for HIVE vs. HIV- comparisons (log2(FC)>1, padj<0.7), produced 855 DEGs (Figure S5B). 800 genes down-regulated in HIV+ were highly enriched for nervous system and synaptic processes (Figure S5C), gene ontologies closely related to the group of genes showing repressive compartmentalization and decreased expression in HIVE (Figure S5D–E). This included Netrin-G1 ligand LRRC4C/NGL1, which has been implicated in microglial accumulation at axonic compartments of cortico-spinal projection neurons 46. However, in contrast to HIVE, microglial transcriptomes from HIV+ brain lacked upregulation of IFN or other immune-related pathways.
To further corroborate these differences in the genomic response of HIVE and HIV+ Mg, we generated a Hi-C map for Irf8+ Mg nuclei from an HIV+ (non-HIVE) brain. There was a significant positive correlation between ΔPC1 values in HIVE and HIV+ Mg for those regions with a significant compartment switch in HIVE (R=0.38, p<2.2×10−16; Figure S5F), and even a stronger correlation for the 339 100kb bins that underwent a significant compartment switch in both HIVE and HIV+ (R=0.52, p<2.2×10−16; Figure S5G). Remarkably, pathway enrichment analyses revealed that both HIV+ and HIVE microglia exhibit repressive compartment remodeling at BCL2L1, PNOC, and PARKN/PARKIN, which encode broad regulators of auto- and mitophagy implicated in synaptic plasticity and neuronal injury and metabolism (Figure S5H) 47–49. These findings further confirm that the genomic response of Mg residing in the HIV infected brain includes widespread alterations at gene loci regulating microglia-mediated effects on neuronal health and signaling, which are not limited to late-stage disease with encephalitis.
Neuronal and non-neuronal integration site mapping in the HIV infected brain
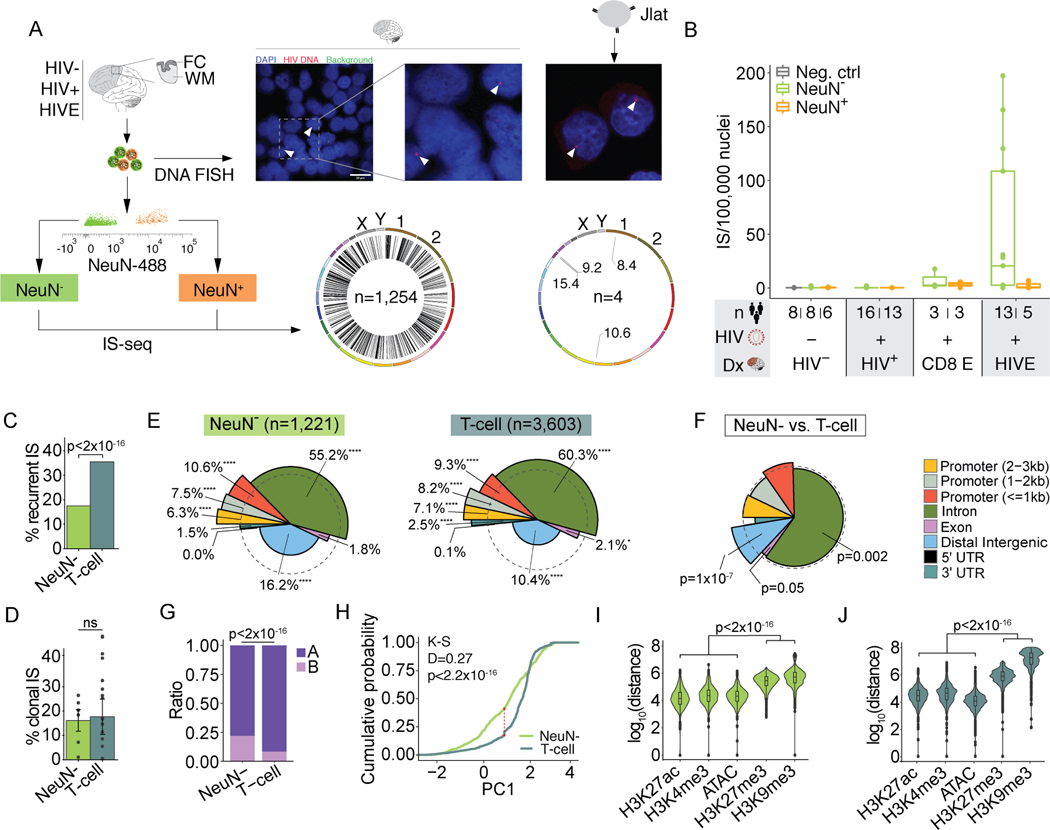
Our single nuclei transcriptome profiling showed that HIV RNA is detectable in a subset of HIVE microglia, with a smaller contribution of other, overwhelmingly non-neuronal, cell types (Figure 1E–F). Therefore, for genome-wide integration site sequencing (IS-seq), we sorted neuronal (NeuN+) and non-neuronal (NeuN-) FC/WM nuclei by FANS (Figure 4A) and performed integration site (IS) sequencing (IS-seq; Figure S6A–B). We validated our IS-seq methods in 4 JLat lymphocyte lines each harboring a single, previously published IS (Figure 4A, Figure S6C). Based on IS-seq from sequential dilutions of JLat clones with uninfected Jurkat DNA, we estimate an overall 1.6% sensitivity of IS detection, a number that is 6-fold higher than previous studies using similar methodology (Figure S6D) 50. Importantly, DNA FISH labeling of nuclei from HIVE brain with an HIV-specific probe showed a single integrated provirus in each infected nucleus, suggesting that surviving, infected cells in the brain are not targeted for repeated infection, similar to what has been observed in peripheral lymphocytes (Figure 4A) 51,52.
Figure 4. Genomic and epigenomic features of IS in human brain.
(A) Representative FANS plot for sorting of NeuN+ and NeuN- nuclei for FC/WM IS-seq and JLat cells for comparison. HIV DNA FISH from HIVE nuclei (left) and JLat cells (right), arrowheads indicate positive signal. Genome-wide IS circos plots (left, brain n=1,254 IS; right, 4 JLat clones n=4 IS). (B) Boxplots show number of IS/100,000 nuclei (y-axis). Diagnosis (Dx) is indicated on the x-axis. CD8 E, CD8 encephalitis; Neg. Ctrl., uninfected Jurkat cell DNA. (C-D) Percent of recurrent (C) and clonal (D) IS for NeuN- and in vivo T-cell IS54–59. Significance testing by chi-square (C) or two-way ANOVA (D). (E) Spie charts comparing proportional IS distribution across 8 genomic features as indicated. Dashed circle demarcates equal (expected) proportions and % denote the IS feature percent. Chi-square test. * 0.01 < p < 0.05, ** 0.001 < p < 0.01, *** 0.0001 < p < 0.001, **** p<0.0001 (Bonferroni multiple comparison factor x 8, significance threshold at p = 0.00625) (F) Spie chart comparing NeuN- IS distribution to T-cell. Chi-square test. (G) Bar plot showing the proportion of NeuN- or in vivo T-cell IS found in A/ B compartments in HIV- microglia or uninfected Jurkat T-Cell Hi-C data respectively. P, Chi-square. (H) Empirical cumulative distribution function graph of NeuN- and T-cell IS for PC1 values. NeuN- IS are compared to HIV- microglia Hi-C data and T-cell IS are compared to uninfected Jurkat Hi-C (two-sample Kolmogorov-Smirnov test, red line Dmax). (I-J) Violin plots showing distance between NeuN- or T-cell IS and the nearest corresponding peak from NeuN- or Jurkat specific open and repressive chromatin marks. Kruskall Wallis rank sum tests. See also Figure S6.
We then generated 53 cell-type specific IS-seq libraries from FC/WM NeuN+ and NeuN- nuclei from 25 unique donors (n=7 HIVE, n=18 HIV+; Table S1), in addition to 20 HIV- control IS-seq libraries (8 from uninfected Jurkat cultures, 12 from HIV- FC/WM sorted nuclei from 6 unique donors). An additional set of 28 libraries, prepared from 14 HIV+, non-encephalitic donors and 8 HIV- samples, failed to produce sufficient library for sequencing, indicating low or absent viral integration (Table S1). We identified n=1,254 IS, the overwhelming majority from NeuN- (n=1,221/1254, 97%, Supplementary file S6). Of note, IS were detected in 85% (6/7) of HIVE and 28% (5/18) of HIV+ donors, including 2 subjects diagnosed with CD8+ T-cell encephalitis, an unusual inflammatory brain disorder affecting cART-medicated and treatment-naïve individuals, and 1 elite controller (Figure 3B)53.
Figure 3. Chromosomal loop remodeling and transcriptomic alterations in HIVE microglia.
(A) Gene ontology enrichment for genes found in HIVE chromosomal loops. Black line indicates significance threshold at FDR=0.05. Numbers indicate the number of genes in the dataset/the number of genes in the GO category. GO categories: cell adh. via plasma mem., cell adhesion via plasma membrane; nervous sys. dev.; nervous system development; STAT R sig. path., STAT receptor signaling pathway; cell pop. prolif., cell population proliferation. (B) Bar graph showing the number of DEGs that are found only in HIVE loops, only in HIV- loops, or in neither HIVE nor HIV- specific loops. Hypergeometric test for the overlap between the DEGs and genes in HIV- or HIVE-specific loops using all genes as a background. (C) Hi-C contact frequency maps of HIVE (top half above diagonal) and HIV- (bottom half below diagonal) microglia and loops shown for a representative locus on chromosome 12. Zoomed in view shows (from top to bottom): normalized read counts from snRNA-seq of HIVE Mg, normalized read counts from snRNA-seq of HIV- Mg, enhancers called from microglial H3K27ac ChIP-seq data, CTCF ChIP peaks from CD14+ monocytes, Hi-C loops from HIVE mg, Hi-C loops from HIV- microglia. (D) Contact frequency maps shown for examples of differential loops that were called on pooled loops from HIVE and HIV-. For all loops, normalized contact frequencies in HIVE and HIV- were queried using a linear regression model and loops with p<0.05 were considered significant. Numbers of loops gained and lost in HIVE and the number of genes located in the anchors of gained/lost loops are indicated. Aggregate peak analysis (APA) scores are shown. (E) Gene ontology enrichment for genes found in HIVE gained loops. Black line indicates significance threshold at FDR=0.05. Numbers indicate the number of genes in the dataset/the number of genes in the GO category. GO categories: Neuron dev., neuron development; synapse org., synapse organization; cell comp. morph., cellular component morphogenesis. See also Figure S4.
We compared the NeuN- IS-seq map with three independent T-cell datasets: 1) n=3,603 compiled IS from patient T-cells infected in vivo54–59, 2) n=687 IS from in vivo infected peripheral blood mononuclear cells (PBMCs) computed from publicly available raw sequencing files58, 3) newly generated n=52,190 IS from in vitro infected Jurkat cells (Supplementary file S6). NeuN- nuclei showed a significantly lower rate of recurrent integration events, defined as >1 separate integration event into the same gene (Figure 4C, Figure S6E). However, clonal integration, indicative of proliferation of infected cells, was of similar magnitude in NeuN- nuclei (16% average across donors) compared to the in vivo T-lymphocyte (18%) and PBMC (12%) datasets. Clonal events in the Jurkat cells, harvested 48h post-infection, were much lower (0.6%) (Figure 4D, Figure S6F).
IS genomic annotations in NeuN- HIVE showed, similar to peripheral T-cells, a preference for proximal promoters, introns, and 3’UTRs (Figure 4E, Figure S6G). However, NeuN- HIVE showed significantly more integration into distal intergenic regions Figure 4F). Consistent with the overall association of intergenic DNA with B-compartment status, we observed significantly higher rates of integration into the B compartment (22% vs. 8%, p<2.2×10−16; Figure 4G) and overall lower PC1 values (Figure 4H) among NeuN- IS compared to T-cell using published Hi-C Jurkat Hi-C60. However, integration in the A compartment was still overall favored in both the brain and T-cells where 46% and 48% of the whole genome were found in the A compartment respectively (Chi-square p<2.2×10−16 for both).
Next, we explored local epigenomic features at viral insertion sites by studying T-cell IS in relation to Jurkat ChIP-seq datasets and similarly, NeuN- IS in relation to NeuN- ChIP-seq datasets (see Methods and Supplementary file S1). These included transcription-facilitative histone H3 lysine 4 trimethylation (H3K4me3) and lysine 27 acetylation (H3K27ac), and repressive H3K9me3 and H3K27me3 marks, together with open chromatin profiles by the Assay for Transposase Accessible Chromatin. Indeed, in both NeuN- and Jurkat, IS were located ~104 bp from the nearest open chromatin region but were 106-108bp from the nearest repressive mark (Figure 4I–J, Figure S6H).
Interestingly, we observed that LEDGF and CPSF6, the two cellular factors involved in mediating integration site selection in T-cells, were expressed at significantly lower levels in microglia as compared to all other cell clusters in our snRNA-seq data (Figure S6I–J). Leveraging ChIP-seq data from U2OS cells (Supplementary file S1) revealed enrichment for CPSF6 binding at T-cell but not NeuN- IS (Figure S6K–L). Of note, a recent IS study in the C20 microglial cell line did not observe the same differences in IS patterns as we observed here compared to T-cells61. However, based on published RNA-seq data, expression of LEDGF and CPSF6 is significantly higher in C20 cells as compared to primary microglia (Figure S6M–N). Both in our snRNA-seq data and published RNA-seq data62, this discrepancy in expression appears to be greatest for LEGDF. Thus, while general IS patterns in the brain are similar to T-cells, we observe differences in the rates of recurrent integration, integration into intergenic and B-compartmentalized regions, and expression and binding of integration targeting host factors.
Viral integration is associated with microglial transcriptional and 3D-genomic features
Of note, genes containing IS (IS genes) were expressed at significantly higher levels in microglia, ImOI, and Lymph clusters as compared to non-immune cell types in the brain (Figure 5A). This is in line with our snRNA-seq data, which showed viral transcription predominantly in the Mg and ImOl clusters (Figure 1E–F). Similarly, microglia showed the highest levels of expression for IS genes most variably expressed across our eight principal cell types (Figure 5B). To examine this further, we grouped all expressed transcripts from HIV-, and separately, from HIVE Mg, into 8 bins of increasing expression (12.5% of expressed genes/bin with bin 8 containing the most highly expressed genes) and determined for each bin the proportion of IS genes it contained. Strikingly, the proportion of IS genes found in each bin showed a steady increase with consecutively higher bin number (Poisson linear model p<2.2×10−16). IS genes were overrepresented in the bin of most highly expressed genes (bin 8, Figure 5C). This effect was seen both in HIV- and HIVE Mg, with only a slight increase in the proportion of IS genes found in bins 7 and 8 in HIVE (69.8% vs 67.4%, Chi-square p=0.3). These findings would be consistent with the hypothesis that microglial genes highly expressed at baseline are at high risk to be targeted for integration when the brain becomes infected with HIV. Global patterns of integration into regions of microglia-specific gene expression and permissive epigenomic features can be observed on a genome-wide scale and are exemplified by chromosome 19, the chromosome with the highest IS density (Figure 5D–E).
Figure 5. Viral integration is associated with microglial transcription and genome structure.
(A) snRNA-seq FC/WM violin plots showing cluster-specific expression of all genes containing IS in HIVE samples (n=7). For cell types composed of more than 1 subcluster, values were averaged across all cells. P, Wilcoxon rank sum (Mg vs. other cell types). (B) Heatmap showing expression centered and scaled by gene for top 300 IS genes most variably expressed across cell types. (C) Graph comparing IS gene expression levels in HIVE and HIV- microglia, divided into 8 equal groups (12.5% of genes/group, ranked by successively higher expression). Dotted line 12.5%. P, Poisson linear regression. (D-E) Genome-wide and chromosome 19 specific circos plot displaying, from the outside in, NeuN- IS, heatmap of log scaled HIVE Mg snRNA-seq read counts, HIVE Mg Hi-C PC1 value, and NeuN- H3K27ac ChIP-seq reads. See also Figure S6.
Viral integration is linked to infection-related reorganization of the microglial genome
In vitro T-cells show 3D genome reorganization in acute immune activation models that impacts integration site selection60. Interestingly, we observed a significant increase in the proportion of IS in the A compartment in HIV+ as compared to HIV- (81% vs. 77%, Chi-square p=0.005, data not shown) and in HIVE as compared to HIV- microglia (82% vs. 78%, Chi-square p=0.02 respectively; Figure 6A). Of those IS found in regions undergoing A compartmentalization in HIVE (N=65) or HIV+ (N=82), N=37 switched in both conditions (Supplementary file 3). These findings indicate that a subset of integration sites may be dependent on disease stage, including presence of encephalitis.
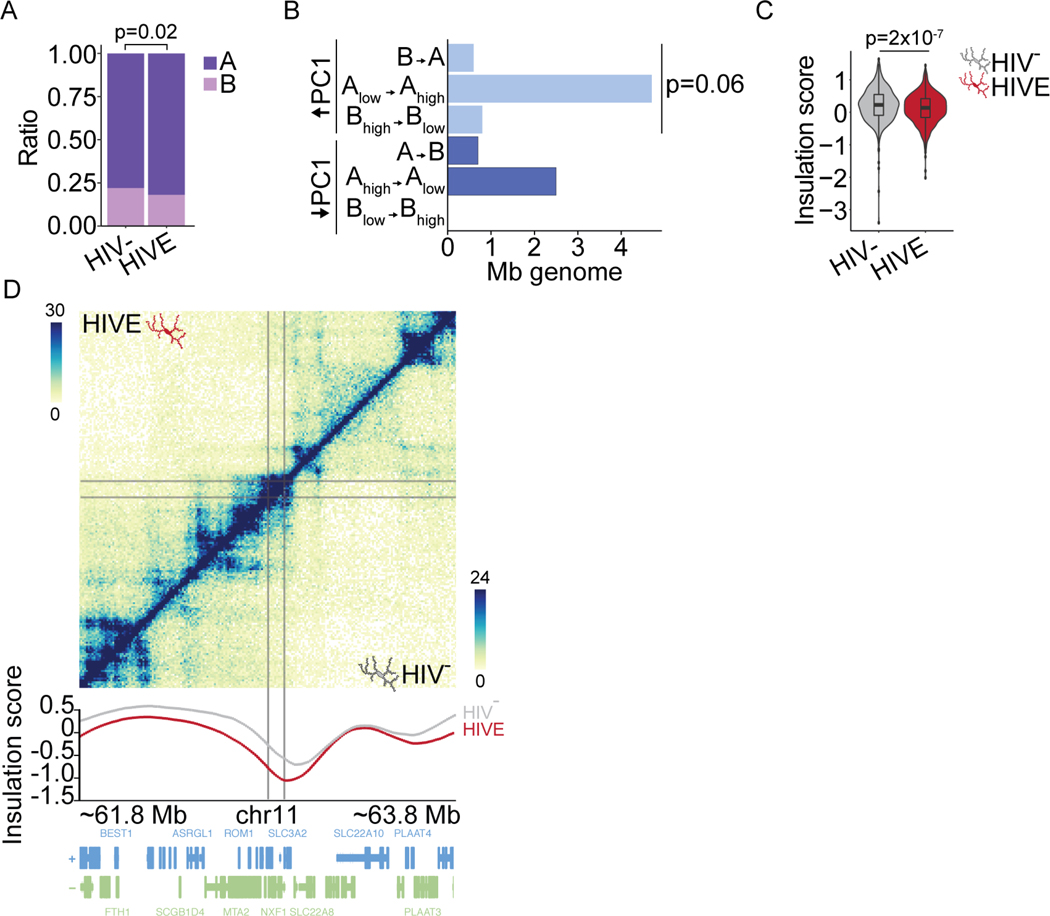
Figure 6. Viral integration is associated with 3D-genomic restructuring in HIVE.
(A) A/B compartment proportions of NeuN- IS in HIV- and HIVE Mg. P, chi-square. (B) Bar graph, dcHiC IS compartment remodeling (HIVE compared to HIV- microglia). X-axis, Mb of genome in 100 kb bins with IS and significantly changed PC1. Y-axis, different compartment switches (HIV- to HIVE), (light blue) A-compartmentalization (DPC1 HIVE-HIV- >0, FDR<0.05), (dark blue) B-compartmentalization (DPC1 HIVE-HIV- <0, FDR<0.05). (C) Violin plot, insulation score at IS in HIV- and HIVE microglia. Wilcoxon rank sum test. (D) Hi-C contact frequency maps of HIVE (top half) and HIV- (bottom half) microglia shown for representative integration sites (gray lines) on chromosome 11. Insulation score plotted below. See also Figure S6.
Overall, 7.6% of IS were found in regions of compartment switching in HIVE (Figure 6B), primarily driven by sequences undergoing significant A-compartmentalization (Chi-square p=0.06, Figure 6B). Furthermore, domains harboring IS showed a significant decrease in insulation score in HIVE as compared to HIV- microglia (Figure 6C–D), an effect extremely specific given that on a genome-wide scale, HIVE Mg showed a significant increase in insulation scores compared to HIV-Mg (Figure S4A). These findings strongly suggest that chromosomal domains undergoing dynamic remodeling in HIVE microglia with high insulation, a potential indicator for open chromatin facilitative to transcription63, are specifically at risk for HIV integration. Furthermore, in good agreement with the aforementioned CTCF and YY1 chromatin occupancies at TAD domains with insulation score alterations in the HIVE vs. HIV- Mg (Figure S4B), loci dually defined by a viral integration site and a robust (top 10th percentile) insulation score change in the HIVE microglia also showed significant enrichment for CTCF and YY1 binding (Figure S6O).
Viral transcription is linked to microglial activation
Integration is a critical step in the HIV life cycle because it allows for efficient transcription of viral genes64,65. Thus, we studied viral transcription using our snRNA-seq data. Surprisingly, HIV was the second most highly expressed transcript in HIV RNA+ microglial nuclei (Figure 7A), suggesting that HIV takes over the cellular transcriptional landscape. Indeed, we observed that HIV RNA+ nuclei tended to cluster together when we performed sub-clustering of HIVE microglia, even though HIV transcripts were excluded when performing the sub-clustering (Figure 7B). Furthermore, principal component analysis of HIV RNA+ and RNA- microglia from each of our 5 HIVE donor samples in which we detected viral transcription, showed a highly consistent shift along PC1 and PC2 for HIV RNA+ microglia (Figure S7A), indicating a shared effect of active viral transcription on cellular gene expression across these different subjects. We counted 69 genes downregulated in HIV RNA+ vs. RNA- Mg nuclei, enriched for cell adhesion and neuronal support functions (Figure 7C–D). Over half of these genes are targets of CTBP2 and SUZ12, both components of Polycomb Repressive Complex 2 (PRC2) that also showed higher levels of transcription in HIV RNA+ microglia (Figure S7B–C). There were 20 significantly upregulated genes in HIV RNA+ microglia, enriched for phagocytic and cell migratory functions (Figure 7C, E).
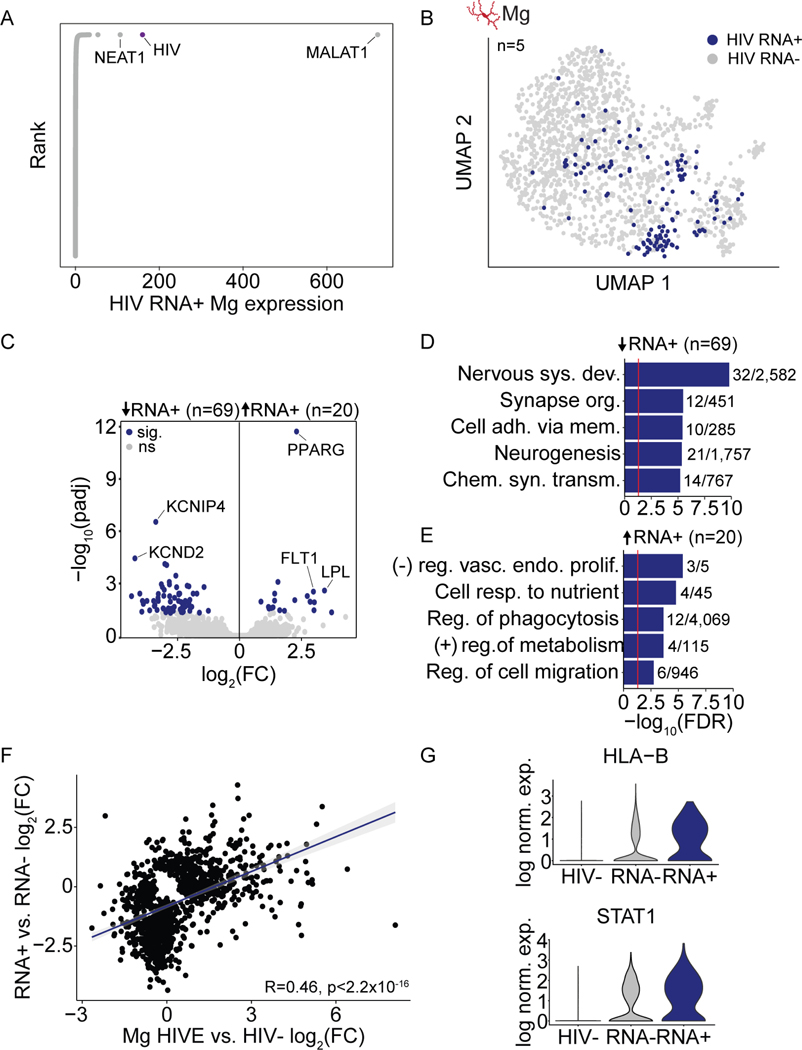
Figure 7. Viral transcription is associated with microglial activation.
(A) Rank plot, log normalized expression of each gene in HIV RNA-expressing (RNA+) HIVE. (B) HIV RNA expression mapped onto sub-clustered microglia from HIVE brains with viral transcription (n=5) (C) Gene expression volcano plot (HIVE HIV RNA+ vs. HIV RNA- nuclei). Blue dots mark genes with padj<0.05. (D-E) Bar plots showing biologic process GO enrichment with N dataset genes/total GO category genes indicated for down- and upregulated genes as indicated. Red line FDR=0.05. (F) Pearson correlation, log2(fold change) in gene expression HIV RNA+ vs. RNA- HIVE microglia (Y-axis) and all HIVE microglia vs. HIV- microglia (X-axis) for genes with unadjusted pval<0.05 in at least one comparison. (G) Violin plots, log normalized expression HLA-B and STAT1 in HIV- microglia, HIVE HIV RNA- microglia, and HIVE HIV RNA+ microglia. See also Figure S7.
Importantly, very similar gene ontologies and functional pathways emerged in our snRNA-seq and Hi-C analyses comparing the entire pool of HIVE brain microglia vs HIV- brain (Figure 2). Thus, we compared gene expression changes in HIV RNA+ vs. HIV RNA- microglia to HIVE vs. HIV- microglia and found a highly significant positive linear relationship (Figure 7F), suggesting that viral transcription occurs in activated microglia that drive differential expression in HIVE. In line with this, for key immune genes, HLA-B and STAT1, we observed a gradient in expression that was lowest in HIV- microglia, intermediate in HIVE microglia not harboring HIV transcript, and highest in HIV RNA+ HIVE microglia (Figure 7G). HIVE HIV RNA- microglia still showed evidence of activation, as there was a high degree of correlation between HIVE HIV RNA- and RNA+ differential expression as compared to HIV- microglia (Figure S7D). However, the magnitude of the fold change in expression of DEGs was higher for HIV RNA+ as compared to RNA- microglia, further reinforcing that HIV RNA+ Mg are the most activated (Figure S7E). Taken together, these findings suggest that viral transcription, detectable on the level of the single nucleus, is linked to a subset of highly activated microglia.
DISCUSSION
Deeper understanding of the invasion and spread of HIV in the brain and its clinical sequelae, including HAND, requires insight into the epigenomic and transcriptomic context of retroviral infection in the human brain. Here, we built an integrative dataset from 79 sequencing files, including snRNA-seq, Hi-C, IS-seq, and ChIP-seq, providing a critically needed neurogenomics resource from postmortem brain of infected donors and controls.
While HIVE is no longer common in the cART era, it is useful as a model to understand how active, unrestricted viral infection impacts the brain, as all individuals undergo a period of unrestricted viral replication prior to initiating cART66,67. In support of this, patients occasionally have neurologic symptoms during acute infection, so it is possible the brain at this stage may resemble more of a HIVE-like state68. It will be interesting to utilize simian immunodeficiency virus (SIV) models or humanized mouse models to assess whether the neurogenomics of acute infection resonates with some of the findings reported here for the HIVE postmortem brain69,70. Moreover, HIVE remains a significant concern for an estimated 9 million PLWH worldwide believed not to be on cART71, given that in the pre-cART era approximately one quarter of patients develop HIVE in the setting of severe immunosuppression72,73.
In any case, our single nuclei profiling, and earlier work in HIVE bulk tissue74,75, all point to elevated expression of interferon (IFN) response and complement cascade genes, and decreased expression of genes related to bioenergetics and neuronal function, indicating an adaptive shift in the microglia of the infected brain from homeostatic, neuronal support functions to inflammation. It is remarkable that non-encephalitic microglia from HIV+ brain show conserved changes in gene expression and 3D genome remodeling at genes related to neuronal health and synaptic function, even in the absences of significant viral integration and transcription. This may represent an early insult that contributes to HAND and to the observed neuronal dysfunction and death in HIV infection in the absence of direct neuronal infection76.
Importantly, according to our microglia-specific Hi-C data from HIVE and non-infected controls, chromosomal conformations at the site of differentially expressed genes undergo a profound reorganization, with altogether 182 megabases of microglial genome—in total length roughly equivalent to the size of human chromosome 5—undergoing a change in A/B compartment structures, together with rewiring of hundreds of contact-specific loops and genome-wide alterations of the kilo-to megabase-scaling TAD domains. We show that 3D genome changes in HIV infected brain are highly cell type-specific and associated with corresponding reprograming of the nuclear transcriptome. Specifically, in encephalitic brain microglia, sites of up-regulated gene expression, including many genes involved in interferon response and viral defense pathways, undergo a reconfiguration to a transcriptionally more permissive and open chromatin environment. At a subset of loci, similar changes were observed in IFN-γ stimulated cultured microglial cells, suggesting that IFN stimulation in part drives 3D remodeling in HIVE.
Critically, these immune activation-associated changes to microglia are linked to viral infection, both in terms of integration and viral transcription. We observed a strong preference for integration into A compartment open chromatin, similar to previous reports in T-cells 54,56,60,77, and found preliminary evidence, pending confirmation in larger cohorts, that a subset of integration sites may be dependent on disease stage, including presence of encephalitis. In addition to activation of immune signaling pathways, HIV-derived transcripts or proteins could impact the cell’s epigenomic organization and chromosomal conformations via additional mechanisms. Illustrative examples exist for other types of viruses. For example, the NS1 protein in influenza virus-infected peripheral macrophages interferes with transcriptional termination, resulting in abnormal read-through of elongating RNA polymerase II across hundreds of kilobases, with the net effects of locus decompaction and chromosomal contact changes 78. Another RNA virus, SARS-CoV-2, causes global 3D genome defects in susceptible lung cells and rewiring of chromosomal contacts and loops at odorant receptor gene loci in the olfactory sensory mucosa 79,80.
We also observed high levels of viral transcription in activated microglia, underscoring the capability of HIV for extremely productive, and ultimately detrimental, seeding and spread in the brain’s susceptible cell population. The presence of a small population of clonal integration sites points to the ability of infected cells to replicate and expand the pool of CNS virus. Regardless of directionality, we saw that immune stimulation and infection are linked, suggesting that microglial activation potentiates infection in some way.
Importantly, we also observed different patterns of IS selection in the brain as compared to T-cells, with lower rates of recurrent integration and higher rates of intergenic integration in brain, which may be caused by lower levels of expression of the integration targeting factors, LEDGF and CPSF6, in brain microglia. In macrophages, rates of genic integration are inversely correlated with expression of LEDGF 81, with LEDGF ablation in macrophages leading to higher rates of intergenic integration82,83. Furthermore, in microglia integration was highly biased for genes in the top 12.5% of most highly expressed genes, while in T-cells preferential integration has been observed in the top ~40% of most highly expressed genes77,84. However, whether this is a biological difference of integration in microglia or the result of nuclear vs. whole cell RNA-seq more accurately capturing the active transcriptional landscape is not clear. Cell-type specific differences integration site targeting have implications for HIV cure strategies as latency reversing agents used in shock-and-kill approaches have different efficacy depending on target site85.
Limitations of the study:
According to the present study, genomic integration sites showed significant changes in 3D structure in HIVE as compared to HIV-. While we cannot determine causality with neurogenomic approaches, the most likely explanation is that infection and inflammation mobilize chromosomal conformations, which in turn increases the susceptibility of genomic loci, primarily those that re-localize into a more open chromatin environment, to become more favored for integration. We believe this is a more likely scenario than specific viral IS driving changes to the 3D genome, considering that the majority of IS are unique, and with our Hi-C method using ensembles of nuclei as input, any single integrant effect is likely to be diluted out. However, whether the 3D genomic changes we observed are present in all microglia or a subset of infected or more activated microglia is unclear. Of note, Irf8, the protein which we used to sort the microglial nuclei, has been shown to be upregulated in reactive microglia, and thus we may have biased towards these cells 86. Future studies, mapping Hi-C chromosomal conformation at single cell resolution, will be required to test these hypotheses conclusively. In addition, much larger cohorts will be required to assess the impact of important comorbidities, including substance use.
In any case, datasets presented here will provide a rich resource to better understand cell-type, sequence- and chromatin status-specific mechanisms that govern HIV integration, expression and eventually, silencing in the human brain.
STAR Methods Text
Resource availability
Lead contact
Requests for further information related to methods and reagents should be directed to the Lead Contact, Schahram Akbarian (schahram.akbarian@mssm.edu).
Materials availability
This study did not generate new or unique reagents.
Data and code availability
Sequencing datasets reported in this paper are available via dbGaP with the accession number phs003080.v1.p1
Original code is available on Mendeley Data at doi:10.17632/ccj4trmsxc.1
Any additional information required for reanalysis of the reported data is available upon request from the Lead Contact.
Experimental model and subject details
Human tissue
Human autopsy brain samples were collected by the Manhattan HIV Brain Bank (MHBB, U24MH100931), using protocols under the supervision of the Icahn School of Medicine at Mount Sinai (ISMMS) Institutional Review Board. Written informed consent was obtained either from decedents or their primary next of kin. Brain processing and histologic assessment were completed as previously described 87. A list of donor IDs and demographic information can be found in Table S1.
Cell lines and cell culture
HMC3 cells were obtained from ATCC (CRL-3304) and grown in EMEM (ATCC) supplemented with 10% FBS (Corning), 100U/mL penicillin, and 100μg/mL streptomycin (Gibco). JLat clones 8.4, 9.2, 10.6, and 15.4 were obtained from the NIH HIV Reagent Program (ARP-9847, −9848, −9849, and −9850). JLat and Jurkat cells were maintained in RPMI 1640 (Gibco) with 10% FBS (Corning) 100U/mL penicillin, and 100μg/mL streptomycin, and 292 μg/mL glutamine (Gibco). HEK293T cells were maintained in DMEM (Gibco) supplemented with 10% FBS (Corning), 100U/mL penicillin, 100μg/mL streptomycin, and 292 μg/mL glutamine (Gibco).
Virus production and infection
HIV strain NL-43 GFP (NLGI) was prepared in HEK293T cells 88. The plasmid was amplified in Stbl3 chemically competent E. Coli (NEB) and isolated using the Qiagen midi-prep kit. HEK293T cells were transformed with the NLGI plasmid using PolyJet (SignaGen) as per the manufacturer’s instructions. Supernatant was harvested from transfected cells after 48 hours, filtered through a 0.2μm syringe filter, and frozen at −80oC until use. Jurkat cells were infected with 75ng p24 equivalent of virus mixed with 8μg/mL Polybrene (Milipore Sigma), they were centrifuged at 1,200g for 90 minutes at 16oC and incubated overnight. Cells were washed and the media was changed the following morning. Jurkat cells were harvested 48 hours post infection.
Method details
Nuclei isolation
Nuclei were isolated from 200–300mg of human postmortem frontal lobe tissue (cortex and underlying white matter) as previously described 89. Briefly, tissue was homogenized in a hypotonic lysis buffer (0.32M sucrose, 5mM CaCl2, 3mM Mg(Ace)2, 0.1mM EDTA, 10mM Tris-HCl pH 8, 1mM DTT, 0.1% Triton X-100). A sucrose gradient (1.8M sucrose, 3mM Mg(Ace)2, 1mM DTT, 10mM Tris-HCl pH8) was layered under the homogenate and nuclei were pelleted via ultracentrifugation at 24,000rpm for 60 minutes at 4°C.
For snRNA-seq nuclei were resuspended in 1mL 0.04% BSA in DPBS. In cases where mouse and human tissue were mixed, cortical tissue from wild-type C57BL/6 mice was combined with human tissue prior to homogenization.
For Hi-C, Tissue was homogenized as described above, and tissue homogenate was fixed in 1% formaldehyde at room temperature for 5 minutes, followed by quenching with 250mM glycine at room temperature for 5 minutes. Fixed nuclei were then spun down and washed once with 50% lysis buffer and 50% sucrose buffer prior to ultracentrifugation. Nuclei were resuspended in 1mL 0.1% BSA in PBS and immunostaining was performed.
For bulk RNA-seq, nuclei were unfixed and isolated as described above except that tissue was first dounced in 1mL lysis buffer with 400U RNase inhibitor (Takara, 2313A). Following ultracentrifugation, nuclei were resuspended in 0.1% BSA prior to immunostaining.
For IS-seq, nuclei were isolated as described above. Following ultracentrifugation, they were resuspended in 0.1% BSA in PBS prior to immunostaining.
Fluorescence in situ hybridization
For DNA FISH of postmortem brain, nuclei extraction was performed as described above. The nuclei pellet was resuspended and fixed in 4% formaldehyde for 10 minutes at room temperature, followed by purification in DPBS. For staining of J-Lat cells, the cells were fixed with 4% formaldehyde for 10 minutes at room temperature and resuspended in DPBS. Approximately 100 μL of either nuclei or cell solution was pipetted onto a charged glass slide before incubation at 60°C for 10 minutes to dehydrate the nuclei. After a brief wash with DPBS, the slides were processed per the RNAscope Multiplex Fluorescent v2 protocol (Advanced Cell Diagnostics). Nuclei were treated with protease, hydrogen peroxide, and incubated with sense DNA probes against the HIV gag-pol gene sequence (Probe-V-HIV1-gagpol, Advanced Cell Diagnostics) for 2 hours at 40°C. Amplifier sequences were polymerized to the probes and treated with Opal 570 dye. Nuclei were briefly incubated in DAPI and mounted in VECTASHIELD antifade medium. Imaging was performed at 63x magnification on a Zeiss LSM780 confocal microscope.
For RNA FISH, human cingulate cortex (approximately 100 mg block) from HIV+ and HIV- tissue was drop-fixed in 4% formaldehyde for 6 hours at 4°C, followed by overnight incubation in 30% sucrose. Sections (20 μm) were cut on a freezing microtome and mounted on charged slides. The tissue was processed as described for DNA FISH, with the exception of a longer protease incubation and use of an RNA-specific probe against gag-pol (Probe-V-HIV1-gagpol-sense-C2, Advanced Cell Diagnostics). Tissue was incubated with an Iba1 antibody (ab178846, Abcam) overnight at 4oC and the following day an Alexa-647 goat anti-rabbit (A-21245, Invitrogen) antibody was incubated for 1 hour at room temperature. Prior to mounting in VECTASHIELD, the slides were incubated for 30 seconds in TrueBlack Lipofuscin Autofluorescence Quencher (Biotium) and rinsed with 1x PBS. Images were acquired at 20x and 63x on a Zeiss LSM780 Confocal microscope.
snRNA-seq
Library preparation
snRNA-seq was performed using the Chromium platform (10× Genomics, Pleasanton, CA) with the 3’ gene expression (3’ GEX) V3.1 kit, using an input of ~16,500 cells. Libraries were sequenced in paired end mode on the NovaSeq (Illumina, San Diego, CA) at an average depth of 176,000 reads per nucleus (range 24,000–200,000).
Data processing
Sequencing data was aligned to a combined pre-mRNA GRCh38 and HXB2 reference genome using the Cell Ranger Single-Cell Software Suite (version 3.0.0, 10x Genomics). Filtered count matrices were loaded into R and empty droplets were detected and removed from each sample with Debris Identification using Expectation Maximization (DIEM) 18. Doublets were detected and removed using DoubletFinder 90. After inspection, two samples which were of low quality were removed (mhbb 154, mhbb 78). This resulted in 69,843 nuclei from 13 samples that passed quality control.
Seurat was used to perform normalization and variance stabilization for each sample prior to merging91,92. We used Harmony to correct for donor effects 93. UMAP dimensional reduction and graph-based clustering were performed on the harmonized dataset in Seurat. After inspection, one cluster that contained low quality nuclei (low read counts and high percent of mitochondrial reads) was removed. Cell cluster identity was assigned based on canonical marker gene expression. We performed further sub-clustering of the microglia cluster. After sub-setting to the cells of interest, HIV was removed as a gene from the dataset. We applied Seurat and Harmony as described above for the full dataset to correct for donor effects and perform graph-based clustering. A cluster that appeared to contain doublets, which expressed canonical neuronal marker genes and had higher average number of genes and UMIs detected per nucleus, was identified and removed.
For visualization of gene expression, read counts were normalized for each nucleus using log normalization (ln(1+feature count/total counts)). Log normalized counts were then used to generate plots using Seurat.
Differential expression analysis
Genes lacking expression in fewer than 10 cells were removed, and HIV was removed as a gene from the data. Read counts were summed within each donor for each cluster to create pseudobulk counts. To determine cluster marker gene expression, DESeq2 was applied with a design of ~ 0 + cluster 94. Each cluster was then compared to all other clusters to determine marker genes. For differential expression between conditions (HIV-, HIV+, HIVE), the count matrix was split into sub-matrices for each cluster. We then used DESeq2 with a model of ~ 0 + condition and identified DEGs by contrasting one condition from another. Differential expression analysis of HIV RNA+ and RNA- microglia was performed similarly. Pseudobulk count matrices were generated within each donor for RNA+ and RNA- microglia and DESeq2 was used with a model of ~ HIV RNA. Gene ontology enrichment was performed using ShinyGO.95
Analysis of mixed human and mouse snRNA-seq
Samples that contained mouse nuclei (mhbb037, mhbb038, mhbb059, mhbb079, mhbb078) were aligned to a combined mm10, GRCh38, and HXB2 reference genome. For each nucleus, the proportions of mm10- and GRCh38-mapping reads were calculated and nuclei with >60% of reads mapping to a single genome were considered to be arising from that species (Figure S1B–C, E). Human nuclei were then filtered using DIEM and DoubletFinder prior to being integrated with the other samples.
For determining HIV read thresholding, nuclei with <600 or >3000 genes detected and >3% of reads mapping to human or mouse mitochondrial genes were removed. Nuclei were identified as human or mouse as described above, and the number of reads mapping to HXB2 in mouse nuclei was determined. For visualization, samples underwent normalization and variance stabilization using SCTransform (Seurat) followed dimensional reduction using UMAP as described above.
Immunostaining and fluorescence-activated nuclei sorting
For Hi-C, fixed nuclei were stained with NeuN-488 (MAB377X, Millipore; 1:1000) and Irf8-PE (U31-644, BD Biosciences, 1:250) antibodies. Nuclei sorted for bulk RNA-seq were stained with NeuN-488, Irf8-PE, and Olig2–647 (ab225100, Abcam, 1:500). For IS-seq, nuclei were stained with NeuN-488 (MAB377X, Millipore; 1:1000). All antibodies were incubated for 1 hour at 4oC protected from light. DAPI was added at a concentration of 5ug/mL just before FANS. Nuclei were sorted using a BD Biosciences FACSAria II machine. For Hi-C, fixed nuclei were sorted into PBS and were pelleted in PBS with 17% sucrose buffer by spinning at 4,000rpm for 5 minutes at 4oC. Nuclei pellets were flash frozen on dry ice and stored at −80oC. For bulk RNA-seq, nuclei were sorted directly into Trizol-LS and snap frozen on dry ice. For IS-seq, nuclei were pelleted in PBS with 17% sucrose buffer prior to lysis and DNA extraction as described below.
Bulk RNA-seq library preparation and analysis
RNA was extracted from nuclei following manufacturer instructions for Trizol-LS. The aqueous phase containing RNA was then further purified using the RNA Clean & Concentrator kit (Zymo, R1013). RNA-seq libraries were prepared using the SMARTer Stranded Total RNA-Seq Kit v2 Pico Input Mammalian (Takara, 634412). Libraries were sequenced on the Illumina MiSeq platform. Shallow sequencing was used as the aim was to confirm cell-type identity. Following sequencing, reads were trimmed using the default settings of TrimGalore (https://github.com/FelixKrueger/TrimGalore) and aligned to GRCh38 using STAR (settings --sjdbOverhang 100 --twopassMode Basic) 96. Reads were counted using featureCounts (settings --ignoreDup -t ‘gene’ -p -O) 97. Read counts were log normalized using the DESeq2 rlog function and log normalized read counts were used to generate heatmaps of marker gene expression 94.
Hi-C
IFN stimulation
HMC3 cells were treated with 200U/mL IFN-γ (Miltenyi) in 0.1% BSA or 0.1% BSA alone (vehicle control) for 24hr before harvesting. For each treatment, three replicates were performed. Cells were removed from the plate and fixed for Hi-C as per the Arima Hi-C Kit protocol (A51008, San Diego, CA). Approximately 50,000 cells were put into Trizol-LS prior to fixation for RNA for qPCR.
qPCR
RNA was extracted from cells in Trizol-LS as per manufacturer instructions. The aqueous phase containing RNA was then further purified using the RNA Clean & Concentrator kit with on column DNase treatment (Zymo, R1013). RNA was reverse transcribed using Superscript RT (Invitrogen, 18091050). qPCR of cDNA was performed using SYBR Green PCR master mix (Applied Biosystems) with primers for GAPDH (Origene), HLA-A98, HLA-Dra99, STAT1, and STAT2100. Data were normalized to GAPDH as a housekeeping gene and were analyzed using the ΔΔCt method.
Library preparation
Nuclei pellets frozen post-FACS and HMC3 cell pellets were thawed, and Hi-C was performed using the Arima-HiC Kit (A51008, San Diego, CA) according to the manufacturer’s instructions. Following enrichment of biotinylated fragments and DNA purification, end repair and adaptor ligation was performed using the Swift Biosciences® Accel-NGS® 2S Plus DNA Library Kit (San Diego, CA), according to the manufacturer’s instructions. DNA libraries were amplified with the Kapa Hyper Prep Kit (07962347001, Wilmington, MA). Libraries were sequenced on the Illumina NovaSeq platform to an average depth of ~212 million valid cis-chimeric reads per sample (San Diego, CA, Supplementary file S1)
Sequencing alignment and contact map generation
We applied HiC-Pro (v2.11.4) to generate contact matrices of HIV- and HIVE microglia 101. Briefly, Bowtie2 (parameters: --very-sensitive -L 30 --score-min L,−0.6,−0.2 --end-to-end --reorder) was used to align Hi-C reads to hg38102. Uniquely mapped read pairs were assigned to restriction enzyme fragments (DpnII, Hinf1; ligation sites = GATCGATC, GANTGATC, GANTANTC, GATCANTC) to filter out invalid ligation products. The resulting valid pairs were then used to generate Hi-C contact maps at 10kb, 40kb, and 100kb resolutions. Hi-C contact maps were normalized using Iterative Correction and Eigenvector decomposition (ICE) via HiC-Pro. Pooled contact maps for HIV- microglia and HIVE microglia were used for the downstream analysis to maximize read depths. We used plotgardener to visualize Hi-C datasets (e.g. contact maps, insulation scores, loops) 103.
Compartment analysis
We applied dcHiC (https://github.com/ay-lab/dcHiC.git) to HIVE and HIV- microglial contact maps at a 100kb resolution. The resulting output contains genomic coordinates, principal component (PC) values for HIVE and HIV- microglia, and significance (p-values and adjusted p-values) of compartment changes between HIVE and HIV- microglia. Genomic regions with positive and negative PC values were classified as compartment A and B, respectively. We also identified regions with differential compartment signatures between HIVE and HIV- microglia using a threshold of padj< 0.05. Regions with a significant increase in PC1 were then further classified as either B→A if there was a complete reversal of eigenvector direction from negative in HIV+ to positive in HIVE, or Alow→Ahigh/Bhigh→Blow when the PC1 value was either positive or negative in both HIV- and HIVE respectively. Regions with a significant decrease in PC1 were similarly classified as either A→B, Ahigh→Alow, or Blow→Bhigh. We intersected differential compartments with gene coordinates (Emsembl v93) to identify genes that are located in regions with differential compartment signatures. We then used ShinyGO to perform gene ontology analysis on genes located in differential compartments.
Insulation score analysis
We calculated insulation scores from contact maps at a 10kb resolution using matrix2insulation.pl from the Github repository: https://github.com/dekkerlab/cworld-dekker.git (parameters: --is 480000 --ids 320000 --im iqrMean --nt 0 --ss 160000 --yb 1.5 --nt 0 --bmoe 0). Insulation scores were then converted to TAD boundaries by insulation2tads.pl in the same Github repository. For each 10kb bin, the absolute difference in insulation was calculated and the bins with the top and bottom 10% largest changes in insulation score were identified. Data were aligned to hg38 using bwa mem with default settings 104. We called peaks using MACS2 with the input ChIP file for background and the settings -B -p .000001 --keep-dup auto 105. We performed overlap permutation tests with regioneR to test for overlap between ChIP peaks and insulation score change regions (n=1000 permutation) 106.
Loop analysis
Loops were called using mustache from contact maps at 10kb resolution of three conditions: HIVE, HIV-, and condition-agnostic (we merged contact maps of HIVE and HIV- to create a chromatin contact map with a higher read depth and resolution) 41. HIVE and HIV- specific loops were identified using the setdiff function in R. To identify genes in HIVE and HIV- specific loops, the entire span of the genome contained within the loop was used. To perform differential loop analysis, we combined loops from HIVE, HIV-, and condition-agnostic. We queried the raw contact frequency of each sample at loop anchors, normalized contact frequencies with the read depth (sum of counts at loop anchors for each sample) and performed differential loop analysis using a linear regression model. Loops with linear regression p < 0.05 were called as differential loops. Difference in contact frequency at differential loops between HIVE and HIV- was further validated by aggregated peak analysis (APA) using hicpeaks (https://pypi.org/project/hicpeaks/). For analysis of genes in differential loops, we identified genes with either of the 10kb anchor regions.
Microglial enhancer identification
We obtained previously published human primary microglia H3K27ac ChIP-seq data from dbGaP (Supplementary file S1) 107. Reads were aligned to hg38 using bwa mem with default settings 104. Enhancers were called by MACS2 using a p-value threshold of p<0.0001 and a narrowPeak setting 105. Super enhancers were called using Rank Ordering of Super-Enhancers (ROSE) with the parameters - s12500 -t2500 108,109.
IS-seq
Library preparation
Pelleted nuclei from FANS were resuspended in nuclear lysis buffer (1X Tris-EDTA pH 8, 0.5% SDS), incubated with 40μg/mL RNaseA (12091–039, Invitrogen) for 15 minutes at 37°C, followed by overnight incubation with 50μg/mL proteinase K at 37°C. Genomic DNA was then isolated using phenol chloroform extraction. DNA from cultured cells was extracted using the Qiagen DNeasy Blood & Tissue Kit (cat. 69504).
Integration site libraries were prepared using a linker-mediated PCR (LM-PCR) approach as previously described 110. This approach allows selective amplification of viral integration sites by using one primer specific for the HIV LTR and another specific for adaptors that have been ligated to fragmented DNA. First, 1μg of DNA was sonicated to an average of 900bp using the Covaris S220 with a peak incident power of 105, duty factor 5% and 200 cycles/burst for 50s at 4–10°C. Sonicated fragments were isolated with SPRIselect beads (Beckman Coulter) at a ratio of 0.7X according to the manufacturer’s instructions. Fragmented DNA was end repaired and A-tailed using the NEBNext Ultra End Repair/dATailing module by incubation at 20°C for 30 minutes followed by 65°C for 30 minutes. End-repaired DNA fragments were ligated with adaptors at a final concentration of 20 μM using the NEBNext Ligation module by incubation at 16oC overnight. Adaptors contained a unique linker sequence, a unique molecular identifier sequence (UMI), and a common linker sequence shared among all samples. Adaptors were partially double stranded at the common linker sequence, while the UMI and unique linker sequences were a single stranded overhang. Adaptors were pre-annealed prior to use by mixing each strand to a final concentration of 20μM and cooling from 95oC to 4oC at a rate of 0.5oC per 90 seconds. Each sample for a given run utilized a different adaptor (see Sherman et al., 2017 for adaptor sequences).
Following adaptor ligation, the DNA was again purified using SPRIselect beads at a ratio of 0.7X and was split into 4 parallel reactions for nested PCR. In each round, one primer was complementary to the adaptor and the other primer was complementary to the HIV LTR sequence to selectively amplify the IS. For both rounds, primers were added at a final concentration of 15μM and a blocking oligo, which prevented amplification from the 5’ HIV LTR, was added at a final concentration of 25μM. PCR was performed using the Advantage 2 polymerase (Clontech) according to the manufacturer’s directions. The first round PCR had the following cycle parameters: 95oC 1 min; 5X [95oC 30 sec, 80oC 30 sec, 70oC 1:00]; 20X [95oC 30 sec, 80oC 30 sec, 67oC 1:00]; 72oC 4 min; 4oC hold. 2ul of the product from the first round PCR were used for the second round amplification, with the parameters: 95°C 1 min; 5X [95oC 30 sec, 80oC 30 sec, 70oC 1:00]; 15X [95oC 30 sec, 80oC 30 sec, 67oC 1:00]; 72°C 4 min; 4oC hold. The adaptor-specific and LTR-specific primers used in the second round PCR reaction introduced the P5 and P7 sequences needed for Illumina sequencing. The LTR-specific second round primer also included a unique sequencing barcode.
Following second round PCR, the 4 parallel reactions for each sample were pooled together and SPRIselect beads were used to clean the DNA at a ratio of 0.7X. Libraries were quantified using the KAPA Library Quantification Kit (KK4873). For each set of libraries prepared, we included a positive control of DNA from infected Jurkat cells and a negative control of DNA from HIV- brain or uninfected Jurkat cells. Positive control libraries were not sequenced, negative control libraries were only sequenced in cases where there was amplification and sufficient library was produced to sequence. Final libraries were submitted for sequencing on either the HiSeq 2500 or NovaSeq platforms (Illumina, San Diego, CA).
Bioinformatic identification of IS
UMIs were first extracted from the reads using UMI-tools, and then reads were trimmed for HIV LTR sequence and the linker sequence using Cutadapt. Only read pairs for which both the LTR end and linker end had exact matches for both trimmed sequences continued to downstream analysis. We aligned trimmed reads to hg38 using Bowtie2 (settings -p 4 -I 150 -X 1000) and converted reads into a bed file that contained the coordinates of the ends of read 1 and read 2 in order to encompass the both the site of integration (where the LTR-genome junction is located) and the sonic breakpoint (coordinate where the linker ligated).
For each library, reads for which the LTR end was within 5 bp and the sonic end was within 15 bp were collapsed as PCR duplicates and the total number of duplicate reads was counted. For analysis of JLat clones, sonic ends were collapsed within 5bp of one another due to the high level of clonality. Only integration sites with >1 read supporting them were considered as valid. Clonal integration sites were defined as those for which the integration site was the same, but the sonic breakpoint was >15 bp apart or >5bp for JLat clones. Any integration sites found in more than one sample, excluding clonal integration sites that came from 2 different assays performed on the same donor, were removed. Any integration sites mapping to non-standard chromosomes were removed prior to analysis.
IS characterization
Brain integration site distributions were compared to 3 different T-cell control sets: 1) a compiled dataset of published in vivo T-cell integration sites 54–59, 2) integration sites we sequenced from infected Jurkat cells, 3) a published in vivo T-cell dataset for which sequencing data was available 58 and we analyzed using our pipeline. We annotated genomic features using ChIPseeker111.
ChIP-seq
Library preparation
Normal healthy control frontal cortex tissue gray matter was used to prepare H3K27me3 (n=3 donors) and H3K9me3 (n=3 donors) ChIP-seq libraries. 200–300mg of tissue was homogenized as described above. For H3K9me3, tissue homogenate was fixed with 1% formaldehyde for 10 minutes and quenched with 125mM glycine for another 10 minutes rotating at room temperature. Nuclei were isolated and FANS was performed with the NeuN antibody as described above. For H3K27me and H3K9me3 ChIP, after FANS, one million NeuN- nuclei were pelleted and resuspended in 300μl of micrococcal nuclease digestion buffer (10mM Tris pH 7.5, 4mM MgCl, and 1mM Ca2+), and digested with 2 uL of MNase (0.2U/μl) for 5 minutes at 28°C to obtain mononucleosomes. The reaction was quenched with 50mM EDTA pH 8. Chromatin was released from nuclei with the addition of hypotonic buffer (0.2mM EDTA pH 8, containing PMSF, DTT, and benzamidine.
All ChIP samples were then incubated with the appropriate antibodies (H3K27me3, Cell Signaling C36B11; H3K9me3, Abcam ab8898) at 4°C overnight. The DNA-protein-antibody complexes were captured by Protein A/G Magnetic Beads (Thermo Scientific 88803) by incubation at 4°C for 2h. Magnetic beads were sequentially washed with low-salt buffer, high-salt buffer, and TE buffer. DNA was eluted from the beads, treated with RNase A, and digested with Proteinase K prior to phenolchloroform extraction and ethanol precipitation. For library preparation, ChIP DNA was end-repaired (End-it DNA Repair kit; Biosearch Technologies) and A-tailed (Klenow Exo-minus; Biosearch Technologies). Adapters (Illumina) were ligated to the ChIP DNA (Fast-Link kit; Biosearch Technologies) and PCR amplified using the Illumina TruSeq ChIP Library Prep Kit. Libraries with the expected size (~275bp) were submitted to the New York Genomics Center for next-generation Illumina sequencing on the HiSeq 2500.
Analysis
A mix of publicly available and newly generated ChIP-seq data were used (see Supplementary file S1). Both publicly available and new generated ChIP-seq data were aligned to hg38 using bwa mem and called peaks using MACS2. Libraries were subsampled to a similar sequencing depth. For H3K27ac, H3K4me3 and ATAC narrow peaks were called; for H3K27me3 and H3K9me3 broad peaks were called. In cases where there was >1 sample, consensus peaks across multiple samples were called as those occurring in at least 2 samples. Blacklist regions were removed. For CTCF and YY1, consensus peaks were downloaded directly from ENCODE.
Quantification and statistical analysis
Statistical analysis
Analyses were performed using R 4.1.2. Statistical tests are indicated in the figure legend. P-values of <0.05 were considered significant for chi-square, ANOVA, and Wilcoxon tests. For snRNA-seq analyses, DESeq2 was used to compare expression across summed, pseudobulk data for comparisons of interest. P-values were obtained using the Wald test and multiple comparison correction was performed using the Benjamini-Hochberg method. For differential expression analysis of HIVE vs. HIV- microglia, genes were considered differentially expressed if they either had an adjusted P-value of <0.05 or if they had a log2(fold change)>1 and an adjusted p-value<0.7 (unadjusted p≤0.16). For differential expression analysis of HIV RNA+ vs HIV RNA- HIVE microglia, genes with an adjusted P-value<0.05 were considered differentially expressed. Gene ontology enrichment utilized the hypergeometric test to derive nominal P-values which were then false discovery rate corrected, and categories with an FDR<0.05 were considered significant. Pearson correlations of differential expression results were performed using the log2(fold change) values from DESeq2 results. Hypergeometric P-values used to compare the overlap between genes sets were obtained using an online tool (http://nemates.org/MA/progs/overlap_stats.html) using a background of 58,219 genes in the human genome. Permutation tests used to study the overlap between genomic coordinates were performed using regioneR with n=1000 permutations and all canonical chromosomes as a genomic background 106.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Recombinant Anti-Iba1 antibody [EPR16588] | Abcam | Cat# ab178846, RRID:AB_2636859 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 | Invitrogen | Cat# A-21245, RRID:AB_2535813 |
| Anti-NeuN Antibody, clone A60, Alexa Fluor®488 conjugated | Millipore Sigma | Cat# MAB377X, RRID:AB_2149209 |
| IRF8 Mouse anti-Human, PE, Clone: U31–644 | BD Biosciences | Cat# 566373, RRID:AB_2739716 |
| Recombinant Alexa Fluor® 647 Anti-Olig2 antibody [EPR2673] | Abcam | Cat# ab225100, RRID:N/A |
| Tri-Methyl-Histone H3 (Lys27) Rabbit mAb | Cell Signaling Technology | Cat# 9733, RRID:AB_2616029 |
| Anti-Histone H3 (tri methyl K9) antibody - ChIP Grade | Abcam | Cat# ab8898, RRID:AB_306848 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemically Competent E. coli | NEB | Cat# C737303 |
| HIV: NL-43 GFP (NLGI) | Cohen et al., 1999 | N/A |
| Biological samples | ||
| Human brain specimens | The Manhattan HIV Brain Bank | https://icahn.mssm.edu/research/manhattan-hiv |
| Chemicals, peptides, and recombinant proteins | ||
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| Fetal Bovine Serum, 500 mL, Regular, USDA approved Origin (Heat Inactivated) | Corning | Cat#35–011-CV |
| Penicillin-Streptomycin-Glutamine (100X) | Gibco | Cat#10378016 |
| Polybrene | Milipore Sigma | Cat#TR1003G |
| Recombinant ribonuclease inhibitor | Takara Bio USA, Inc. | Cat#2313A |
| TrueBlack Lipofuscin Autofluorescence Quencher | Biotium | Cat#23007 |
| PureLink™ RNase A | Invitrogen | Cat#12091039 |
| SPRIselect for Size Selection | Beckman Coulter Life Sciences | Cat#B23318 |
| PolyJet™ In Vitro DNA Transfection Reagent | Cat#SL100688 | |
| Clontech Labs 3P Advantage® 2 Polymerase Mix | Clontech Labs | Cat#639201 |
| Pierce™ Protein A/G Magnetic Beads | Thermo Scientific | Cat#88803 |
| TRIzol™ LS Reagent | Invitrogen | Cat#10296028 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Invitrogen | Cat#D1306 |
| SYBR™ Green PCR Master Mix | Applied Biosystems | Cat#4309155 |
| Human IFN-γ1b research grade | Miltenyi Biotec | Cat#130–096-873 |
| VECTASHIELD Antifade Mounting Medium with Propidium Iodide (PI) | Vector Laboratories | Cat#H-1300–10 |
| Opal 570 Reagent Pack | Akoya Biosciences | Cat#FP1488001KT |
| Critical commercial assays | ||
| Arima-HiC Kit | Arima Genomics | Cat#A51008 |
| Plasmid Midi Kit | Qiagen | Cat#12143 |
| RNA Clean & Concentrator-5 kit | Zymo Research | Cat#R1013 |
| SMARTer® Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian | Takara Bio USA, Inc. | Cat#634412 |
| RNAscope Multiplex Fluorescent v2 Assay | Advanced Cell Diagnostics | Cat#323110 |
| Chromium Next GEM Single Cell 3’ Kit v3.1 | 10x Genomics | Cat#1000268 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | Cat#1000120 |
| Dual Index Kit TT Set A | 10x Genomics | Cat#1000215 |
| SuperScript™ IV First-Strand Synthesis System | Invitrogen | Cat#18091050 |
| Swift Biosciences® Accel-NGS® 2S Plus DNA Library Kit | Swift Biosciences, Inc | Cat#21024 |
| Kapa Hyper Prep Kit | Roche | Cat#07962347001 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69504 |
| NEBNext® Ultra™ II End Repair/dA-Tailing Module | New England Biolabs | Cat#E7546S |
| NEBNext® Ultra™ II Ligation Module | New England Biolabs | Cat#E7595S |
| KAPA Library Quantification Kit | Roche | Cat#KK4873 |
| End-It DNA End-Repair Kit | Biosearch Technologies | Cat#ER0720 |
| Exo-Minus Klenow DNA Polymerase | Biosearch Technologies | Cat#KL11101K |
| Fast-Link kit | Biosearch Technologies | Cat#LK0750H |
| TruSeq ChIP Library Preparation Kit | Illumina | Cat#IP-202–1012 |
| Deposited data | ||
| Raw sequencing data | This paper | dbGaP phs003080.v1.p1 |
| Code | This paper | doi:10.17632/ccj4trmsxc.1 |
| Experimental models: Cell lines | ||
| HMC3 | ATCC | Cat#CRL-3304 |
| Human T Lymphocyte, J-Lat Full Length Cells (8.4) | NIH HIV Reagent Program | Cat#ARP-9847 |
| Human T Lymphocyte, J-Lat Full-Length Cells (9.2) | NIH HIV Reagent Program | Cat#ARP-9848 |
| Human T Lymphocyte, J-Lat Full Length Cells (10.6) | NIH HIV Reagent Program | Cat#ARP-9849 |
| Human T Lymphocyte, J-Lat Full Length Cells (15.4) | NIH HIV Reagent Program | Cat#ARP-9850 |
| Jurkat | NIH HIV Reagent Program | Cat#ARP-177 |
| HEK293T | ATCC | Cat#CRL-3216 |
| Oligonucleotides | ||
| Probe-V-HIV1-gagpol-sense-C2 | Advanced Cell Diagnostics | Cat#317701-C2 |
| Probe-V-HIV1-gagpol | Advanced Cell Diagnostics | Cat#317691 |
| HIV integration site sequencing oligos, see Supplementary Table 1 | Sherman et al., 2017 | N/A |
| Primer: GAPDH Forward: GTCTCCTCTGACTTCAACAGCG | sequence from Origene, ordered from IDT | CAT#: HP205798 |
| Primer: GAPDH Reverse: ACCACCCTGTTGCTGTAGCCAA | sequence from Origene, ordered from IDT | CAT#: HP205798 |
| Primer: HLA-A Forward: CAGACGCCGAGGATGGCC | Huang et al., 2004 | N/A |
| Primer: HLA-A Reverse: CACACAAGGCAGCTGTCTCACA | Huang et al., 2004 | N/A |
| Primer: HLA-DRa Forward: CGAGTTCTATCTGAATCCTG | Wu et al., 1999 | N/A |
| Primer: HLA-DRa Reverse: GTTCTGCTGCATTGCTTTTGC | Wu et al., 1999 | N/A |
| Primer: STAT1 Forward: ACTCAAAATTCCTGGAGCAG | Zuo et al., 2020 | N/A |
| Primer: STAT1 Reverse: ACGCTTGCTTTTCCTTATGTT | Zuo et al., 2020 | N/A |
| Primer: STAT2 Forward: ATGCTGCAGAATCTTGACA | Zuo et al., 2020 | N/A |
| Primer: STAT2 Reverse: TAGTTCAGCTGATCCAAGAAG | Zuo et al., 2020 | N/A |
| Software and algorithms | ||
| Cell Ranger | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Debris Identification using Expectation Maximization | Alvarez et al., 2020 | https://github.com/marcalva/diem |
| DoubletFinder | McGinnis et al., 2019 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Seurat | Butler et al., 2018 | https://github.com/satijalab/seurat |
| Harmony | Broad Institute | https://portals.broadinstitute.org/harmony/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ShinyGO | Ge et al., 2020 | http://bioinformatics.sdstate.edu/go/ |
| TrimGalore | Braham Informatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Bowtie2 | Langmead et al., 2012 | https://github.com/BenLangmead/bowtie2 |
| HiC-Pro | Servant et al., 2015 | https://github.com/nservant/HiC-Pro |
| plotgardener | Kramer et al., 2022 | https://phanstiellab.github.io/plotgardener/ |
| dcHiC | Chakraborty et al., 2022 | https://github.com/ay-lab/dcHiC |
| matrix2insulation.pl | Dekker Lab | https://github.com/dekkerlab/cworld-dekker |
| MACS2 | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| HiCPeaks | Rao et al., 2014 | https://github.com/XiaoTaoWang/HiCPeaks |
| ROSE: Rank Ordering of Super-Enhancers | Young Lab | http://younglab.wi.mit.edu/super_enhancer_code.html |
| Cutadapt | Rahmann Lab | https://cutadapt.readthedocs.io/en/stable/ |
| ChIPseeker | Guangchuang et al., 2015 | https://guangchuangyu.github.io/software/ChIPseeker/ |
Highlights:
Microglial 3D genome remodeling represses neural genes in HIV and HIV encephalitis
Further microglial 3D genome changes in HIV encephalitis are related to IFN stimulation
HIV targets genomic regions that switch to a more open state in HIV infection
HIV is actively transcribed in a subset of highly active microglia
Acknowledgements:
We thank the participants and staff of the Manhattan HIV Brain Bank for their contributions to this work. We thank members of the Akbarian lab, especially Dr. Lucy Bicks, Dr. Jennifer Blaze, Dr. Liz LaMarca, and Dr. Sergio Espeso-Gil for their insightful suggestions and discussions. We thank Dr. Andrew Chess for providing access to his sequencing equipment. We thank the Icahn School of Medicine at Mount Sinai’s Flow Cytometry Core for providing expertise and guidance on nuclei sorting, the Scientific Computing group at the Icahn School of Medicine at Mount Sinai for computational resources and assistance, and the Genomics Core at the Icahn School of Medicine at Mount Sinai for their assistance with single nucleus RNA-sequencing methods.
Diversity and Inclusion:
One or more of the authors of this paper self-identify as an underrepresented ethnic minority in their field of research. One or more of the authors of this paper self-identify as gender minority in their field of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. We support inclusive, diverse and equitable conduct of research.
Funding:
National Institutes of Health grant DP2MH122403 (HW)
National Institutes of Health grant R21DA051921 (HW)
National Institutes of Health grant R61DA048207 (SM, SA)
National Institutes of Health grant DP1DA056018 (SA)
National Institutes of Health grant R01DA054526 (BC, SA)
National Institutes of Health grant R01DA041765 (BC)
National Institutes of Health grant R01NS108801 (SM)
National Institutes of Health grant 5U24MH100931-10 (SM)
Footnotes
Declaration of interests:
The Authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kahn JO, and Walker BD (1998). Acute human immunodeficiency virus type 1 infection. N Engl J Med 339, 33–39. 10.1056/NEJM199807023390107. [DOI] [PubMed] [Google Scholar]
- 2.Davis LE, Hjelle BL, Miller VE, Palmer DL, Llewellyn AL, Merlin TL, Young SA, Mills RG, Wachsman W, and Wiley CA (1992). Early viral brain invasion in iatrogenic human immunodeficiency virus infection. Neurology 42, 1736–1739. 10.1212/wnl.42.9.1736. [DOI] [PubMed] [Google Scholar]
- 3.Longino AA, Paul R, Wang Y, Lama JR, Brandes P, Ruiz E, Correa C, Keating S, Spudich SS, Pilcher C, et al. (2022). HIV Disease Dynamics and Markers of Inflammation and CNS Injury During Primary HIV Infection and Their Relationship to Cognitive Performance. J Acquir Immune Defic Syndr 89, 183–190. 10.1097/QAI.0000000000002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An SF, Groves M, Giometto B, Beckett AA, and Scaravilli F.(1999). Detection and localisation of HIV-1 DNA and RNA in fixed adult AIDS brain by polymerase chain reaction/in situ hybridisation technique. Acta neuropathologica 98, 481–487. [DOI] [PubMed] [Google Scholar]
- 5.Bagasra O., Lavi E., Bobroski L., Khalili K., Pestaner JP., Tawadros R., and Pomerantz RJ. (1996). Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS (London, England) 10, 573–585. 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Thompson KA, Cherry CL, Bell JE, and McLean CA (2011). Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. The American journal of pathology 179, 1623–1629. 10.1016/j.ajpath.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, and Fauci AS (1986). Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233, 1089–1093. 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 8.Valcour V, Chalermchai T, Sailasuta N, Marovich M, Lerdlum S, Suttichom D, Suwanwela NC, Jagodzinski L, Michael N, Spudich S, et al. (2012). Central nervous system viral invasion and inflammation during acute HIV infection. The Journal of infectious diseases 206, 275–282. 10.1093/infdis/jis326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heaton RK, Clifford DB, Franklin DR Jr., Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, et al. (2010). HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75, 2087–2096. 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, Letendre SL, Leblanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, et al. (2011). HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. Journal of neurovirology 17, 3–16. 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggers C., Arendt G., Hahn K., Husstedt IW., Maschke M., Neuen-Jacob E., Obermann M., Rosenkranz T., Schielke E., and Straube E. (2017). HIV-1-associated neurocognitive disorder: epidemiology, pathogenesis, diagnosis, and treatment. Journal of neurology 264, 1715–1727. 10.1007/s00415-017-8503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine AJ, Miller JA, Shapshak P, Gelman B, Singer EJ, Hinkin CH, Commins D, Morgello S, Grant I, and Horvath S.(2013). Systems analysis of human brain gene expression: mechanisms for HIV-associated neurocognitive impairment and common pathways with Alzheimer’s disease. BMC Med Genomics 6, 4. 10.1186/17558794-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canchi S, Swinton MK, Rissman RA, and Fields JA (2020). Transcriptomic analysis of brain tissues identifies a role for CCAAT enhancer binding protein beta in HIV-associated neurocognitive disorder. J Neuroinflammation 17, 112. 10.1186/s12974-0020-01781-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, and Zhang K.(2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nature Biotechnology 36, 70–80. 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch David H, and Kriegstein Arnold R.(2019). Single-cell genomics identifies cell type–specific molecular changes in autism. Science 364, 685–689. 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, and Zhang K.(2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36, 70–80. 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, and Kriegstein AR (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689. 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez M., Rahmani E., Jew B., Garske KM., Miao Z., Benhammou JN., Ye CJ., Pisegna JR., Pietiläinen KH., Halperin E., and Pajukanta P. (2020). Enhancing droplet-based single-nucleus RNA-seq resolution using the semi-supervised machine learning classifier DIEM. Scientific reports 10, 11019. 10.1038/s41598-020-67513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Corbett SE, Koga Y, Wang Z, Johnson WE, Yajima M, and Campbell JD (2020). Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome biology 21, 57. 10.1186/s13059-020-1950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, and Axel R.(1986). The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47, 333–348. 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 21.Wilen CB, Tilton JC, and Doms RW (2012). HIV: cell binding and entry. Cold Spring Harbor perspectives in medicine 2. 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burren OS, Rubio García A, Javierre B-M, Rainbow DB, Cairns J, Cooper NJ, Lambourne JJ, Schofield E, Castro Dopico X, Ferreira RC, et al. (2017). Chromosome contacts in activated T cells identify autoimmune disease candidate genes. Genome biology 18, 165. 10.1186/s13059-017-1285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., McGovern A., Martin P., Duffus K., Ge X., Zarrineh P., Morris AP., Adamson A., Fraser P., Rattray M., and Eyre S. (2020). Analysis of chromatin organization and gene expression in T cells identifies functional genes for rheumatoid arthritis. Nature communications 11, 4402. 10.1038/s41467-020-18180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Yang J, Wei Y, and Wei X.(2020). Epigenetic regulation of macrophages: from homeostasis maintenance to host defense. Cellular & Molecular Immunology 17, 36–49. 10.1038/s41423-019-0315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, and Natoli G.(2012). Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proceedings of the National Academy of Sciences of the United States of America 109, E2865–2874. 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, and Glass CK (2013). Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell 51, 310–325. 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raymond AD, Gekonge B, Giri MS, Hancock A, Papasavvas E, Chehimi J, Kossenkov AV, Nicols C, Yousef M, Mounzer K, et al. (2010). Increased metallothionein gene expression, zinc, and zinc-dependent resistance to apoptosis in circulating monocytes during HIV viremia. J Leukoc Biol 88, 589–596. 10.1189/jlb.0110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi S.(2015). Positive and negative regulators of the metallothionein gene (Review). Mol Med Rep 12, 795–799. 10.3892/mmr.2015.3459. [DOI] [PubMed] [Google Scholar]
- 29.Tretina K, Park E-S, Maminska A, and MacMicking JD (2019). Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. The Journal of experimental medicine 216, 482–500. 10.1084/jem.20182031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. (2018). Transcription Elongation Can Affect Genome 3D Structure. Cell 174, 1522–1536.e1522. 10.1016/j.cell.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, and Mirny LA (2018). Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A 115, E6697–E6706. 10.1073/pnas.1717730115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca NA., Huber W., Haering CH., Mirny L., and Spitz F. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56. 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belaghzal H, Borrman T, Stephens AD, Lafontaine DL, Venev SV, Weng Z, Marko JF, and Dekker J.(2021). Liquid chromatin Hi-C characterizes compartmentdependent chromatin interaction dynamics. Nat Genet 53, 367–378. 10.1038/s41588-02100784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denholtz M, Zhu Y, He Z, Lu H, Isoda T, Döhrmann S, Nizet V, and Murre C.(2020). Upon microbial challenge, human neutrophils undergo rapid changes in nuclear architecture and chromatin folding to orchestrate an immediate inflammatory gene program. Genes & development 34, 149–165. 10.1101/gad.333708.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong Y, Lazaris C, Sakellaropoulos T, Lozano A, Kambadur P, Ntziachristos P, Aifantis I, and Tsirigos A.(2018). Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nature communications 9, 542. 10.1038/s41467-018-03017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. (2017). YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588.e1528. 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., and Mirny LA. (2016). Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038–2049. 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. (2020). New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic acids research 48, D882–d889. 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, and Aiden EL (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakrabarti A, Jha BK, and Silverman RH (2010). New Insights into the Role of RNase L in Innate Immunity. Journal of Interferon & Cytokine Research 31, 49–57. 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roayaei Ardakany A, Gezer HT, Lonardi S, and Ay F.(2020). Mustache: multiscale detection of chromatin loops from Hi-C and Micro-C maps using scale-space representation. Genome biology 21, 256. 10.1186/s13059-020-02167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita Y, Nakanishi T, Ueno M, Itohara S, and Yamashita T.(2020). Netrin-G1 Regulates Microglial Accumulation along Axons and Supports the Survival of Layer V Neurons in the Postnatal Mouse Brain. Cell Reports 31, 107580. 10.1016/j.celrep.2020.107580. [DOI] [PubMed] [Google Scholar]
- 43.Ryu JK., Cho T., Choi HB., Wang YT., and McLarnon JG. (2009). Microglial VEGF receptor response is an integral chemotactic component in Alzheimer’s disease pathology. The Journal of neuroscience : the official journal of the Society for Neuroscience 29, 3–13. 10.1523/JNEUROSCI.2888-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue Z, Cui C, Liao Z, Xia S, Zhang P, Qin J, Guo Q, Chen S, Fu Q, Yin Z, et al. (2018). Identification of LncRNA Linc00513 Containing Lupus-Associated Genetic Variants as a Novel Regulator of Interferon Signaling Pathway. Frontiers in Immunology 9. 10.3389/fimmu.2018.02967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivetic A, Hoskins Green HL, and Hart SJ (2019). L-selectin: A Major Regulator of Leukocyte Adhesion, Migration and Signaling. Front Immunol 10, 1068. 10.3389/fimmu.2019.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita Y, Nakanishi T, Ueno M, Itohara S, and Yamashita T.(2020). Netrin-G1 Regulates Microglial Accumulation along Axons and Supports the Survival of Layer V Neurons in the Postnatal Mouse Brain. Cell Rep 31, 107580. 10.1016/j.celrep.2020.107580. [DOI] [PubMed] [Google Scholar]
- 47.Haque ME, Akther M, Jakaria M, Kim IS, Azam S, and Choi DK (2020). Targeting the microglial NLRP3 inflammasome and its role in Parkinson’s disease. Mov Disord 35, 20–33. 10.1002/mds.27874. [DOI] [PubMed] [Google Scholar]
- 48.Kaneko YS, Nakashima A, Mori K, Nagatsu T, Nagatsu I, and Ota A.(2012). Microglial activation in neuroinflammation: implications for the etiology of neurodegeneration. Neurodegener Dis 10, 100–103. 10.1159/000332936. [DOI] [PubMed] [Google Scholar]
- 49.Zajaczkowska R, Popiolek-Barczyk K, Pilat D, Rojewska E, Makuch W, Wordliczek J, and Mika J.(2018). Involvement of microglial cells in the antinociceptive effects of metamizol in a mouse model of neuropathic pain. Pharmacol Biochem Behav 175, 77–88. 10.1016/j.pbb.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Symons J, Chopra A, Malatinkova E, De Spiegelaere W, Leary S, Cooper D, Abana CO, Rhodes A, Rezaei SD, Vandekerckhove L, et al. (2017). HIV integration sites in latently infected cell lines: evidence of ongoing replication. Retrovirology 14, 2. 10.1186/s12977-016-0325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Josefsson L., King MS., Makitalo B., Brännström J., Shao W., Maldarelli F., Kearney MF., Hu W-S., Chen J., Gaines H., et al. (2011). Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proceedings of the National Academy of Sciences of the United States of America 108, 11199–11204. 10.1073/pnas.1107729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Josefsson L, Palmer S, Faria NR, Lemey P, Casazza J, Ambrozak D, Kearney M, Shao W, Kottilil S, Sneller M, et al. (2013). Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLoS pathogens 9, e1003432-e1003432. 10.1371/journal.ppat.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucas SB, Wong KT, Nightingale S, and Miller RF (2021). HIV-Associated CD8 Encephalitis: A UK Case Series and Review of Histopathologically Confirmed Cases. Frontiers in Neurology 12. 10.3389/fneur.2021.628296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, et al. (2019). Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest 129, 988–998. 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, and Siliciano JD (2004). Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 78, 6122–6133. 10.1128/jvi.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikeda T, Shibata J, Yoshimura K, Koito A, and Matsushita S.(2007). Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. The Journal of infectious diseases 195, 716–725. 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 57.Kok YL., Vongrad V., Shilaih M., Di Giallonardo F., Kuster H., Kouyos R., Günthard HF., and Metzner KJ. (2016). Monocyte-derived macrophages exhibit distinct and more restricted HIV-1 integration site repertoire than CD4+ T cells. Scientific reports 6, 24157. 10.1038/srep24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. (2014). Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179. 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, and Frenkel LM (2014). HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573. 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lucic B, Chen H-C, Kuzman M, Zorita E, Wegner J, Minneker V, Wang W, Fronza R, Laufs S, Schmidt M, et al. (2019). Spatially clustered loci with multiple enhancers are frequent targets of HIV-1 integration. Nature communications 10, 4059–4059. 10.1038/s41467-019-12046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rheinberger M, Costa AL, Kampmann M, Glavas D, Shytaj IL, Penzo C, Tibroni N, Fackler OT, Vlahovicek K, Lucic B, et al. (2022). Genomic profiling of HIV-1 integration in microglia links viral insertions to TAD organization. bioRxiv, 2022.2002.2014.480322. 10.1101/2022.02.14.480322. [DOI] [Google Scholar]
- 62.Rai MA, Hammonds J, Pujato M, Mayhew C, Roskin K, and Spearman P.(2020). Comparative analysis of human microglial models for studies of HIV replication and pathogenesis. Retrovirology 17, 35. 10.1186/s12977-020-00544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ay F., Vu TH., Zeitz MJ., Varoquaux N., Carette JE., Vert JP., Hoffman AR., and Noble WS. (2015). Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics 16, 121. 10.1186/s12864-015-1236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelly J, Beddall MH, Yu D, Iyer SR, Marsh JW, and Wu Y.(2008). Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology 372, 300–312. 10.1016/j.virol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Y, and Marsh JW (2003). Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J Virol 77, 10376–10382. 10.1128/jvi.77.19.1037610382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daar ES, Moudgil T, Meyer RD, and Ho DD (1991). Transient High Levels of Viremia in Patients with Primary Human Immunodeficiency Virus Type 1 Infection. New England Journal of Medicine 324, 961–964. 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- 67.Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, Kroon E, Sawe FK, Sinei S, Sriplienchan S, et al. (2016). Prospective Study of Acute HIV-1 Infection in Adults in East Africa and Thailand. New England Journal of Medicine 374, 2120–2130. 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellmuth J, Fletcher JLK, Valcour V, Kroon E, Ananworanich J, Intasan J, Lerdlum S, Narvid J, Pothisri M, Allen I, et al. (2016). Neurologic signs and symptoms frequently manifest in acute HIV infection. Neurology 87, 148. 10.1212/WNL.0000000000002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Avalos CR, Abreu CM, Queen SE, Li M, Price S, Shirk EN, Engle EL, Forsyth E, Bullock BT, Mac Gabhann F, et al. (2017). Brain Macrophages in Simian Immunodeficiency Virus-Infected, Antiretroviral-Suppressed Macaques: a Functional Latent Reservoir. mBio 8. 10.1128/mBio.01186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Honeycutt JB, and Garcia JV (2018). Humanized mice: models for evaluating NeuroHIV and cure strategies. Journal of neurovirology 24, 185–191. 10.1007/s13365-017-0567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.UNAIDS (2021). Fact Sheet - World AIDS Day 2021.
- 72.Davies J., Everall IP., Weich S., McLaughlin J., Scaravilli F., and Lantos PL. (1997). HIV-associated brain pathology in the United Kingdom: an epidemiological study. AIDS (London, England) 11, 1145–1150. 10.1097/00002030-199709000-00010. [DOI] [PubMed] [Google Scholar]
- 73.Masliah E, DeTeresa RM, Mallory ME, and Hansen LA (2000). Changes in pathological findings at autopsy in AIDS cases for the last 15 years. AIDS (London, England) 14, 69–74. 10.1097/00002030-200001070-00008. [DOI] [PubMed] [Google Scholar]
- 74.Gelman BB, Chen T, Lisinicchia JG, Soukup VM, Carmical JR, Starkey JM, Masliah E, Commins DL, Brandt D, Grant I, et al. (2012). The National NeuroAIDS Tissue Consortium brain gene array: two types of HIV-associated neurocognitive impairment. PLoS One 7, e46178. 10.1371/journal.pone.0046178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanna PP, Repunte-Canonigo V, Masliah E, and Lefebvre C.(2017). Gene expression patterns associated with neurological disease in human HIV infection. PLoS One 12, e0175316. 10.1371/journal.pone.0175316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones G, and Power C.(2006). Regulation of neural cell survival by HIV-1 infection. Neurobiol Dis 21, 1–17. 10.1016/j.nbd.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 77.Kok YL, Vongrad V, Chaudron SE, Shilaih M, Leemann C, Neumann K, Kusejko K, Di Giallonardo F, Kuster H, Braun DL, et al. (2021). HIV-1 integration sites in CD4+ T-cells during primary, chronic, and late presentation of HIV-1 infection. JCI Insight. 10.1172/jci.insight.143940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. (2018). Transcription Elongation Can Affect Genome 3D Structure. Cell 174, 1522–1536 e1522. 10.1016/j.cell.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zazhytska M., Kodra A., Hoagland DA., Frere J., Fullard JF., Shayya H., McArthur NG., Moeller R., Uhl S., Omer AD., et al. (2022). Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell 185, 1052–1064 e1012. 10.1016/j.cell.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang R, Lee JH, Xiong F, Kim J, Hasani LA, Yuan X, Shivshankar P, Krakowiak J, Qi C, Wang Y, et al. (2021). SARS-CoV-2 Restructures the Host Chromatin Architecture. bioRxiv. 10.1101/2021.07.20.453146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, and Bushman FD (2006). HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther 14, 218–225. 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 82.Shun M-C, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, and Engelman A.(2007). LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes & development 21, 1767–1778. 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tchasovnikarova IA, Marr SK, Damle M, and Kingston RE (2022). TRACE generates fluorescent human reporter cell lines to characterize epigenetic pathways. Molecular Cell 82, 479–491.e477. 10.1016/j.molcel.2021.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, and Bushman F.(2002). HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529. 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 85.Chen HC, Martinez JP, Zorita E, Meyerhans A, and Filion GJ (2017). Position effects influence HIV latency reversal. Nature structural & molecular biology 24, 47–54. 10.1038/nsmb.3328. [DOI] [PubMed] [Google Scholar]
- 86.Masuda T, Tsuda M, Yoshinaga R, Tozaki-Saitoh H, Ozato K, Tamura T, and Inoue K.(2012). IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype. Cell reports 1, 334–340. 10.1016/j.celrep.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morgello S., Gelman BB., Kozlowski PB., Vinters HV., Masliah E., Cornford M., Cavert W., Marra C., Grant I., and Singer EJ. (2001). The National NeuroAIDS Tissue Consortium:a new paradigm in brain banking with an emphasis on infectious disease. Neuropathology and Applied Neurobiology 27, 326–335. 10.1046/j.0305-1846.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- 88.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, and Baltimore D.(1999). The Selective Downregulation of Class I Major Histocompatibility Complex Proteins by HIV-1 Protects HIV-Infected Cells from NK Cells. Immunity 10, 661–671. 10.1016/S1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 89.Kundakovic M, Jiang Y, Kavanagh DH, Dincer A, Brown L, Pothula V, Zharovsky E, Park R, Jacobov R, Magro I, et al. (2017). Practical Guidelines for High-Resolution Epigenomic Profiling of Nucleosomal Histones in Postmortem Human Brain Tissue. Biol Psychiatry 81, 162–170. 10.1016/j.biopsych.2016.03.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell systems 8, 329–337.e324. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R.(2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R.(2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh PR., and Raychaudhuri S. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Love MI, Huber W, and Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. 10.1186/s13059014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ge SX, Jung D, and Yao R.(2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liao Y, Smyth GK, and Shi W.(2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 98.Huang J, El-Gamil M, Dudley ME, Li YF, Rosenberg SA, and Robbins PF (2004). T cells associated with tumor regression recognize frameshifted products of the CDKN2A tumor suppressor gene locus and a mutated HLA class I gene product. J Immunol 172, 6057–6064. 10.4049/jimmunol.172.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu Z, Biro PA, Mirakian R, Hammond L, Curcio F, Ambesi-Impiombato FS, and Bottazzo GF (1999). HLA-DMB expression by thyrocytes: indication of the antigen-processing and possible presenting capability of thyroid cells. Clin Exp Immunol 116, 62–69. 10.1046/j.1365-2249.1999.00831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zuo Y., Feng Q., Jin L., Huang F., Miao Y., Liu J., Xu Y., Chen X., Zhang H., Guo T., et al. (2020). Regulation of the linear ubiquitination of STAT1 controls antiviral interferon signaling. Nature communications 11, 1146. 10.1038/s41467-020-14948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, and Barillot E.(2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome biology 16, 259. 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kramer NE, Davis ES, Wenger CD, Deoudes EM, Parker SM, Love MI, and Phanstiel DH (2021). Plotgardener: Cultivating precise multi-panel figures in R. bioRxiv, 2021.2009.2008.459338. 10.1101/2021.09.08.459338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H, and Durbin R.(2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based Analysis of ChIP-Seq (MACS). Genome biology 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gel B, Díez-Villanueva A, Serra E, Buschbeck M, Peinado MA, and Malinverni R.(2016). regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291. 10.1093/bioinformatics/btv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gosselin D., Skola D., Coufal NG., Holtman IR., Schlachetzki JCM., Sajti E., Jaeger BN., O’Connor C., Fitzpatrick C., Pasillas MP., et al. (2017). An environment-dependent transcriptional network specifies human microglia identity. Science 356. 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lovén J, Hoke Heather A., Lin Charles Y., Lau A, Orlando David A., Vakoc Christopher R., Bradner James E., Lee Tong I., and Young Richard A. (2013). Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell 153, 320–334. 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Whyte Warren A., Orlando David A., Hnisz D, Abraham Brian J., Lin Charles Y., Kagey Michael H., Rahl Peter B., Lee Tong I., and Young Richard A. (2013). Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 153, 307–319. 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sherman E, Nobles C, Berry CC, Six E, Wu Y, Dryga A, Malani N, Male F, Reddy S, Bailey A, et al. (2017). INSPIIRED: A Pipeline for Quantitative Analysis of Sites of New DNA Integration in Cellular Genomes. Molecular therapy. Methods & clinical development 4, 39–49. 10.1016/j.omtm.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu G, Wang L-G, and He Q-Y (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing datasets reported in this paper are available via dbGaP with the accession number phs003080.v1.p1
Original code is available on Mendeley Data at doi:10.17632/ccj4trmsxc.1
Any additional information required for reanalysis of the reported data is available upon request from the Lead Contact.