Abstract
Epstein-Barr virus (EBV) immortalized lymphoblastoid cell lines (LCLs) are widely used for banking. This bioresource could be leveraged for creating human iPSC lines to model diseases including CHD. We generated an LCL-derived iPSC line (NCHi001-A) from a patient with congenital aortic valve stenosis. NCHi001-A was EBV and transgenes free, exhibited stem cell-like morphology, expressed pluripotency markers, has a normal karyotype, and could be differentiated into cells of three germ layers in vitro. Relationship inference via a microarray-based analysis showed NCHi001-A is identical to the parental cell line. NCHi001-A can be used for disease modeling, drug discovery, and cell therapy development.
1. Resource Table
| Unique stem cell line identifier | NCHi001-A |
|---|---|
| Alternative name(s) of stem cell line | LCL-iPSC4802 |
| Institution | Nationwide Children’s Hospital |
| Contact information of distributor | Kim.mcbride@nationwidechildrens.org |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info required for human ESC or iPSC | Age:31-year-old |
| Sex: Female | |
| Ethnicity if known: Caucasian | |
| Cell Source | EBV immortalized human lymphoblastoid cell line (LCL) |
| Clonality | Clonal |
| Method of reprogramming | Sendai virus |
| Genetic Modification | NO |
| Type of Genetic Modification | NO |
| Evidence of the reprogramming transgene loss (including genomic copy if applicable) | RT-PCR |
| Associated disease | Congenital aortic valve stenosis |
| Gene/locus | A homozygous NFATC1 variant; chr18:77,153,996 G > A (GRCh37/hg19) |
| Date archived/stock date | 9th March 2022 |
| Cell line repository/bank Ethical approval | https://hpscreg.eu/cell-line/NCHi001-A Genetics of CVD IRB09-00339 |
2. Resource utility
This iPSC line from a patient with congenital aortic valve stenosis containing the homozygous NFATC1 enhancer variant 18:77,153,996 G > A provides an in vitro cellular model to study the role of NFATC1 in the pathogenesis of CHD and potentially facilitates the development of novel therapeutic interventions. Table 1..
Table 1.
Characterization and validation.
| Classification Test Result Data | |||
|---|---|---|---|
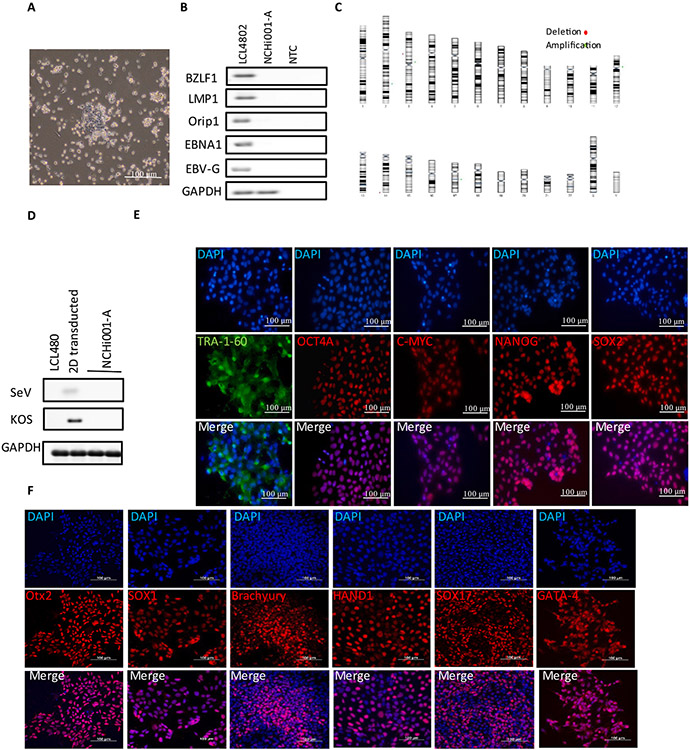
| Morphology | Photography Bright field | Normal | Fig. 1A |
| Phenotype | Qualitative analysis | Expression of pluripotency markers: TRA 1–60, OCT4A, C-MYC, NANOG and SOX2 | Fig. 1E |
| Quantitative analysis | Pluripotent markers positive cells: TRA 1–60: 99 %; OCT4A: 99 %; C-MYC: 99 %; NANOG: 95 % and SOX2: 94 % | Fig. 1E | |
| Genotype | Illumina Infinium CytoSNP-850 K array analysis | Karyotype based microarray analysis confirmed the gain or loss of any significant no more than 5 MB | Fig. 1C |
| Identity | Illumina Infinium CytoSNP-850 K array analysis | Karyotype is normal | Supplementary Fig. 1 |
| Mutation analysis (IF APPLICABLE) | Sequencing | N/A | N/A |
| WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by PCR, Negative | Supplementary Fig. 2 |
| Differentiation potential | Directed differentiation | Positive expression of several lineage specific genes markers assessed with immunocytochemistry. | Fig. 1F |
| List of recommended germ layer markers | Expression of these markers has to be demonstrated at mRNA (RT PCR) or protein (IF) levels, at least 2 markers need to be shown per germ layer | Ectoderm: Otx2 and SOX1, Mesoderm: Brachyury and HAND1 | Fig. 1F |
| Endoderm SOX17 and GATA-4 | |||
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A | |
3. Resource details
Congenital heart disease (CHD) is a common birth defect, affecting 6 to 8 per 1000 live births. Human induced pluripotent stem cells (iPSCs) have opened new approaches to investigate genetic mechanisms of CHD using clinically relevant and patient-specific cardiac cells (e.g., cardiomyocytes, endothelial/endocardial cells) (Lin, McBride et al. 2021).
Human iPSCs have been derived from a variety of cell sources, including EBV-immortalized lymphoblastoid cells lines (LCLs), which are widely used for cell line banking. Bio-banked LCLs are an underutilized resource for generating iPSCs useful for elucidating disease mechanisms of CHD. Here, we generated an iPSC line NCHi001-A, from an EBV-immortalized LCL derived from a 31-year-old female diagnosed with congenital aortic valve stenosis, using the CytoTune™-iPSC 2.0 Sendai Reprogramming kit (Thermo Fisher Scientific).
We reprogrammed LCL4802 with Sendai virus (SeV) containing 4 Yamanaka classical factors: OCT4, KLF4, C-MYC and SOX2 (Zhao et al., 2017; Martineau, Racine et al. 2018). On day 15, iPSCs clones had stem cell-like morphologies with a high nuclear-to-cytoplasmic ratio, and individual colonies were picked and expanded in culture (Fig. 1A). At passage 10, PCR detected no EBV-related genes in the iPSC line NCHi001-A or negative control (NTC), which were present in the parental LCL4802 control (Fig. 1B). In addition, reprogramming transgenes including SeV and KOS were not detected in genomic DNA of NCHi001-A by RT-PCR analysis (Fig. 1D). Immunofluorescent staining was positive for the pluripotent surface antigen TRA-1–60 (99 %) and nuclear pluripotency markers OCT4A (99 %), C-MYC (99 %), NANOG (95 %) and SOX2 (94 %) (Fig. 1E). To evaluate tri-lineage differentiation, iPSCs were differentiated into three germ layers using the Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems). We confirmed the expression of ectoderm (Otx2/SOX1), mesoderm (Brachyury/HAND1), and endoderm (SOX17/GATA-4) markers by immunofluorescence staining (Fig. 1F). These results indicate that these iPSCs have ability to differentiate into cells from all 3 germ layers. We confirmed that NCHi001-A was identical to the parental LCL using KING software (Kinship = 0.5). The karyotype of these iPSCs was normal, and there was no detectable gain or lost (greater than5MB) in all chromosomes (Fig. 1C and Supplementary Fig. 1).
Fig. 1.
Characterization of NCHi001-A iPSC line.
4. Materials and methods
4.1. Cell culture
LCLs isolated from blood were transfected using EBV into immortalized cells using an established protocol (Gilbert and Haines, 2001). EBV-immortalized LCLs were maintained with RPMI1640 (Gibco) containing 20 % FBS at 37 °C, 5 % CO2, and atmospheric O2.
4.2. Reprogramming
EBV-immortalized LCLs were reprogrammed using CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific). On day 0, 100,000 were transfected using Sendai virus. On day 1, cells were reseeded into another well of a 24-well plate. On day 3, cells were transferred into a Matrigel coated 24-well plate pre-loaded with Repro-TeSR™ medium. Medium was refreshed every day. On day 12 to 15, when iPSC clones appeared, mTesR™1 medium was started. After transduction by day 18 to 21, colonies were individually picked and transferred to a new well for expansion. All experiments were performed using iPSCs at least passage 10.
4.3. Rt-pcr
Total RNA was extracted from iPSCs after passage 10 using RNeasy mini-Kit (QIAGEN). Reverse transcription was carried out using Verso cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instruction (Table 2).
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | ||||
|---|---|---|---|---|
| Antibody | Dilution | Company Cat # | RRID | |
| Pluripotency Markers | Mouse anti-TRA-1–60 | 1:100 | Invitrogen Cat # 41–1000 | RRID: AB_2533494 |
| Rabbit anti-Oct-4A | 1:400 | Cell Signaling Cat# 2840P | RRID: AB_2167691 | |
| Rabbit anti-C-MYC | 1:800 | Cell Signaling Cat# 5605P | RRID: AB_1903938 | |
| Rabbit anti-Nanog | 1:200 | Cell Signaling Cat# 4903P | RRID: AB_10559205 | |
| Rabbit anti-Sox2 | 1:400 | Cell Signaling Cat# 3579P | RRID: AB_2195767 | |
| Differentiation Markers | Goat anti-Otx2 | 10 ug/mL | R&D SYSTEMS Cat# AF1979 | RRID: AB_2157172 |
| Goat anti-SOX1 | 10 ug/mL | R&D SYSTEMS Cat# AF3369-SP | RRID: AB_2239879 | |
| Goat anti-Brachyury | 10 ug/mL | R&D SYSTEMS Cat# AF2085 | RRID: AB_2200235 | |
| Goat anti-HAND1 | 10 ug/mL | R&D SYSTEMS Cat# AF3168-SP | RRID: AB_2115853 | |
| Goat anti-SOX17 | 10 ug/mL | R&D SYSTEMS Cat# AF1924 | RRID: AB_355060 | |
| Mouse anti-GATA-4 | 15 ug/mL | R&D SYSTEMS Cat# MAB26061-SP | RRID: Not in database | |
| Secondary antibodies | Rabbit anti-Sox2Alexa Fluor 488 Goat anti Mouse IgG (H + L) | 1:2000 | Invitrogen Cat# A11001 | RRID: AB_2534069 |
| Alexa Fluor 488Goat anti Rabbit IgG (H + L) Alexa Fluor 594 | 1:1000 | Invitrogen Cat# A11012 | RRID: AB_2534079 | |
| Alexa Fluor 594 | ||||
| Donkey anti-Mouse IgG NorthernLights™ NL557-conjugate Antibody | 1:200 | R&D SYSTEMS Cat# NL007 | RRID: AB_663768 | |
| Donkey anti-Goat IgG NorthernLights™ NL557-conjugated Antibody | 1:200 | R&D SYSTEMS Cat# NL001 | RRID: AB_663766 | |
| Primers | Target | Size of band | Forward/reverse primer (5′-3′) | |
| EBV related genes (PCR) | BZLF-1 | 638 bp | TGAAGCAGGCGTGGTTTCAA | |
| 617 bp | CACCTCAACCTGGAGACAAT | |||
| LMP1 | 545 bp | TGAGCAGGATGAGGTCTAGG | ||
| 181 bp | ATGGAACACGACCTTGAGA | |||
| SeV genome and transgene (RT-PCR) | OriP | 528 bp | TCGGGGGTGTTAGAGACAAC | |
| 556 bp | TTCCACGAGGGTAGTGAACC | |||
| SeV | GGA TCA CTA GGT GAT ATC GAG C | |||
| House-Keeping Gene (RT-PCR) | ACC AGA CAA GAG TTT AAG AGA TAT GTA TC | |||
| KOS | ATG CAC CGC TAC GAC GTG AGC GC | |||
| ACC TTG ACA ATC CTG ATG TGG | ||||
| GAPDH | ACCACAGTCCATGCCATCAC TCCACCACCCTGTTGCTGTA | |||
4.4. Immunofluorescence staining
Cells were seeded on 12 mm coverslips coated with Matrigel in 24-well plate. Cells were rinsed twice with DPBS, then fixed using 0.5 mL of 4 % PFA for 10 min at room temperature. PFA was aspirated, and cells were washed with DPBS for 5 min, then gently rinsed with 0.2 M glycine in PBS for 10 min on a shaker. If permeabilization was needed, cells were incubated with 0.5 Triton X-100 plus 1 % protease inhibitor in DPBS for 10 min. Samples were washed three times with DPBS for 10 min on a shaker. For blocking, cells were incubated with 10 % serum from the source species of the secondary antibody raised for 1 h at room temperature. Cells were incubated in 250 μL of primary antibody (Table 2) diluted in blocking buffer in dark overnight at 4 °C. Samples were washed three times with the blocking buffer each for 10 min with shaking. Cells were incubated with the secondary antibody diluted in the blocking buffer for 1 h, unless the primary antibody was conjugated to a fluorochrome. Samples were washed three times with DPBS each for 10 min with shaking. Coverslips were mounted with Invitrogen ™ ProLong ™ Gold Antifade Mountant with DAPI.
4.5. Microscopy
Images were captured using a motorized Nikon Eclipse Ti2-E microscope with a 40X Plan Apo Lambda objective, a Lumencor SOLA LED engine, and a Hamamatsu ORCA Fusion camera.
4.6. Tri-lineage differentiation
The Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems) was used to functionally test the ability of iPSCs to differentiate into cells of three germ layers.
4.7. Karyotyping
Genomic DNA samples were sent to Roswell Park Comprehensive Cancer Center for Illumina Infinium CytoSNP-850 k array analysis. The bioinformatic analysis was performed by using KaryoploteR (version 1.20.0), a Bioconductor package. This R package was used to visualize and identify the distributions of existing variations in the genome on interest.
4.8. Mycoplasma detection
Mycoplasma contamination was evaluated using Venor™GeM Mycoplasma Detection Kit, PCR-based (Sigma-Aldrich) following the manufacturer’s instructions.
Supplementary Material
Acknowledgements
This work was supported by Nationwide Children’s Hospital Foundation. Kim L McBride is supported by Nationwide Children’s Hospital Foundation, Additional Ventures Innovation Fund and NHLBI R01HL109758. This work was partially supported by the American Heart Association (AHA) Career Development Award 18CDA34110293 (M-T.Z.), NIH/NHLBI R01HL155282 (M-T.Z.), R21HL165406 (M-T.Z.), Additional Ventures Innovation Fund (AVIF) and Single Ventricle Research Fund (SVRF) (M-T.Z.).
Abbreviations:
- EBV
Epstein Barr virus
- LCL
lymphoblastoid cell line
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2022.102958.
Data availability
No data was used for the research described in the article.
References
- Gilbert J (2001). “Establishment of permanent cell lines by Epstein-Barr virus transformation.” Current protocols in human genetics / editorial board, Haines Jonathan L. … [et al. ] Appendix 3: Appendix3H. [DOI] [PubMed] [Google Scholar]
- Lin H, McBride KL, Garg V, Zhao MT, 2021. Decoding Genetics of Congenital Heart Disease Using Patient-Derived Induced Pluripotent Stem Cells (iPSCs). Front. Cell Dev. Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau L, et al. , 2018. Lymphoblastoids cell lines - Derived iPSC line from a 26-year-old myotonic dystrophy type 1 patient carrying (CTG)200 expansion in the DMPK gene: CHUQi001-A. Stem Cell Res. 26, 103–106. [DOI] [PubMed] [Google Scholar]
- Zhao MT, et al. (2017). “Molecular and functional resemblance of differentiated cells derived from isogenic human iPSCs and SCNT-derived ESCs.” Proceedings of the National Academy of Sciences 114(52): E11111–E11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.