Abstract
Introduction:
During the second wave of coronavirus disease 2019 (COVID-19), superinfection caused by fungus and multidrug-resistant bacteria worsened the severity of illness in COVID-19 patients. Limited studies from India reported the antimicrobial resistance pattern of secondary infections. In this study, we aim to study the epidemiology of pathogens causing superinfections and genotyping of Gram-negative isolates in COVID-19 patients.
Methods:
This retrospective study was conducted at a dedicated COVID-19 center, India. The identification of bacteria/fungi was done by Vitek2® and matrix-assisted laser desorption/ionization-time of flight mass spectrometry system. Identification of beta-lactamase genes was done using thermal cycler. The diagnosis of mucormycosis was based on 10% potassium hydroxide direct microscopy. Statistical analyses were performed using STATA version 15.1 (StataCorp., College Station, TX, USA). For continuous variables, mean and standard deviation were computed. For comparing proportions of secondary infections across admission location and outcomes, the Chi-squared test of independence was used. To compare the mean and median between intensive care units and outcomes, an independent t-test and a Mann–Whitney test were used.
Results:
Of all the clinical samples, 45.4% of samples were cultured positive for secondary infections. Acinetobacter baumannii (35%) was the most common Gram-negative pathogen, while among Gram positive, it was Enterococcus faecium (40%). Among fungus, Candida spp. (61%) predominates followed by molds. Colistin and tigecycline proved effective against these pathogens. blaNDM was the most prevalent gene followed by the blaOX among the carbapenemase genes.
Conclusions:
The mortality rate among COVID-19 patients with secondary infection was significantly higher compared to the overall mortality rate in COVID-19 patients.
Keywords: Antimicrobial resistance, coronavirus disease 2019, Mucor, NDM
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) had infected 185,906,966 individuals globally, causing an unprecedented 4,019,089 deaths as of July 8, 2021, with India reporting 30,709,557 cases and 405,057 deaths.[1]
As evident from the literature, not all deaths were attributable to SARS-CoV-2 alone, many of them being due to secondary infections.[2,3,4] The same was observed during the influenza pandemic, where secondary/co-infections due to Streptococcus pneumoniae accounted for many deaths. Secondary bacterial infections were also reported in the 2009 swine influenza pandemic,[5] 2002 SARS,[6] and during the 2012 Middle East respiratory syndrome.[7] Co-infections or secondary infections with bacteria, fungi, and viruses in severe respiratory viral infection worsened the severity of the disease and often resulted in fatal outcomes.
Our hospital functioned as a dedicated 285-bedded COVID-19 center with three intensive care units (ICUs), high dependency units (HDUs), COVID-19 operation theaters, dialysis facilities, and wards. We had previously reported a high prevalence of secondary infections in COVID-19 patients due to multidrug-resistant pathogens as part of single and multicentric studies.[8,9]
From March to May 2021, India witnessed a massive surge of cases and death as part of a sudden second wave. As India was coping with the second wave of COVID-19, we also faced a new nemesis in the form of an outbreak of mucormycosis. Mucormycosis is an angio-invasive, potentially life-threatening disease caused by the order Mucorales and requires emergency management.[10] The most common risk factors for mucormycosis include diabetes mellitus, hematopoietic transplants, any immunosuppressive condition, and corticosteroid use.[11,12] The immune system of COVID-19 patients is already under stress; therefore, management may require the use of corticosteroids. Additionally, COVID-19 has been associated with high blood sugar levels and iron overload. All these factors are presumably responsible for the increase in mucormycosis.[13]
COVID-19 patients’ health is known to deteriorate suddenly due to sudden sepsis or secondary infections. Due to the high burden of antimicrobials prescribed to COVID-19 patients in the ICUs, who are also on steroids and invasive devices, it is essential to maintain constant surveillance of the antimicrobial resistance (AMR) profile of organisms causing secondary infections in these cases and be vigilant about fungal infections. To evaluate the burden of secondary infections in context with the severity of COVID-19 patients’ health, we conducted a study in our single center. This study described the overall prevalence of pathogens causing secondary infections for more than a year, the prevalence of AMR genes in Gram-negative pathogens, and the pattern of fungal infections in these patients.
METHODS
We conducted this retrospective study at our 285-bedded COVID-19 center in the capital of India. Data on COVID-19 patients admitted to our center from March 2020 to May 2021 were collected from the laboratory information system software of the microbiology laboratory and analyzed. The data collected included patients’ details, samples’ detail, organism identification, and antimicrobial susceptibility profile. The study included all specimens submitted to the microbiology laboratory for culture. This included a total of 3637 clinical samples including blood, body fluid, respiratory samples, pus, and other samples. We excluded duplicate samples from the study. All clinical specimens were processed in biosafety cabinets with the recommended personal protective equipment, as per standard microbiological techniques. All samples were discarded in accordance with the biomedical waste management guidelines of India.[14]
The samples were processed as per standard microbiological methods. The identification of bacteria/fungi was done by Vitek2® compact identification system (BioMerieux, France) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) system (BioMerieux, France).
Antimicrobial susceptibility test (AST) of the clinical isolates was determined by the Gram-negative, Gram-positive, and yeast Vitek2® AST cards (N235, N280, N281, P628, and YST08) (BioMerieux, France), as per manufacturer's instructions. Minimum inhibitory concentrations (MICs) of antimicrobials were determined and interpreted according to the Clinical and Laboratory Standards Institute-2020 guidelines.[15] The MIC for colistin was determined by the broth microdilution method. Multidrug resistance was defined as resistance to two or more different classes of antimicrobials.
The diagnosis of mucormycosis in our laboratory is based on 10% potassium hydroxide (KOH) direct microscopy, with the addition of calcofluor white as a fluorescent agent to increase sensitivity. Mucorales hyphae are seen as nonseptate or pauci-septate, with a variable width of 6–16 μm. Microscopy is supplemented with the culture of three tubes in sets of two: Sabouraud's dextrose agar (SDA), SDA with chloramphenicol, and SDA with cycloheximide to be incubated at two different temperatures of 25°C and 37°C. The colony characteristic of Mucorales is usually cottony white or grayish black colony. The culture-positive isolates were identified further with the help of the lactophenol cotton blue (LPCB) stain. We also performed MALDI-TOF MS for the identification of isolates using the method of Schwarz et al.[16]
Genotypic profiling
Bacterial genomic DNA was extracted using QIAamp DNA mini kit (Qiagen, Hilden, Germany). Briefly, 200 μl of inoculum was incubated with proteinase K, RNase, and lysis buffer. Ethanol was added followed by centrifugation. Prewash buffer was added to the column and centrifuged. The pellet was washed with wash buffer and again the ethanol treatment was given to improve the yield. The DNA pellet was resuspended in nuclease-free water.
The polymerase chain reaction (PCR)-based identification of beta-lactamase genes was carried out using GenePro Thermal Cycler (Hangzhou Bioer Technology, Bioer, China). A reaction mixture used for PCR is mentioned in Table 1. Table 2 shows the sequence of the primers used for PCR. The PCR conditions included initial denaturation at 94°C for 5 min; the annealing temperature varied according to the primer [Table 2]. It was done for 30 seconds, followed by amplification at 94°C for 30 seconds, elongation at 72°C for 1 min, and final extension at 72°C for 7 min. The PCR product was run on 1.5% agarose gel and stained with ethidium bromide. A 100 base pair ladder was used as a marker to determine the length of the product. The gel was analyzed using a gel documentation system. Table 2 shows the expected length of the PCR product for each gene. The negative control used was nuclease-free water in place of template in a reaction mixture. Klebsiella pneumoniae subsp. pneumoniae (ATCC 700603) (ESBL producer, SHV positive), K. pneumoniae (ATCC BAA-1144) (low-level Amp C producer), Enterobacter cloacae subsp. cloacae (ATCC BAA-1143) (high-level Amp C producer), K. pneumoniae (ATCC BAA1705) (KPC positive), E. cloacae (ATCC BAA-2468) (NDM1 positive), and clinical Gram-negative bacillus strains isolated in the laboratory served as positive controls.
Table 1.
Composition of reaction mixture per sample used for polymerase chain reaction
| Reagents | Volume for 1 reaction (µL) |
|---|---|
| Milli-Q | 14.25 |
| ×10 buffer | 2.5 |
| 25 mM MgCl2 | 1.5 |
| 10 pmole forward primer | 0.5 |
| 10 pmole reverse primer | 0.5 |
| 100 mM dNTPs | 0.5 |
| Taq polymerase (5 U/µL) | 0.25 |
| DNA template | 5 |
Table 2.
The primers used for amplifying different genes and the length of the product obtained after polymerase chain reaction
| Gene | Forward | Reverse | Annealing temperature (°C) | Length of the PCR product (base pairs) |
|---|---|---|---|---|
| ACCM | AACAGCCTCAGCAGCCGGTTA | TTCGCCGCAATCATCCCTAGC | 68.2 | 346 |
| CITM | TGGCCAGAACTGACAGGCAAA | TTTCTCCTGAACGTGGCTGGC | 68.2 | 462 |
| CTXM | AGAATAAGGAATCCCATGGTT | ACCGTCGGTGACGATTTTAG | 52 | 913 |
| DHAM 1and 2 | AACTTTCACAGGTGTGCTGGGT | CCGTACGCATACTGGCTTTGC | 68.2 | 405 |
| EBCM | TCGGTAAAGCCGATGTTGCGG | CTTCCACTGCGGCTGCCAGTT | 68.2 | 302 |
| FOX 1-5B | AACATGGGGTATCAGGGAGATG | CAAAGCCGCTAACCGGATTGG | 68.2 | 190 |
| KPC | ATGTCACTGTATCGCCGTC | AATCCCTCCGAGCGCGAGT | 46.7 | 863 |
| MCR-1 | CGGTCAGTCCGTTTGTTC | CTTGGTCGGTCTGTAGGG | 58 | 309 |
| MCR-2 | TGGTGCTTGTGCCGATTGGA | AGATGGTATTGTTGGTTGCTG | 58 | 567 |
| MCR-3 | TTGGCACTGTATTTTGCATTT | TTAACGAAATTGGCTGGACA | 50 | 542 |
| MCR-4 | ATTGGGATAGTCGCCTTTT | TTACAGCCAGAATCATTATCA | 54 | 487 |
| MCR-5 | ATGCGGTGTCTGCATTTATC | TCATTGTGGTTGTCCTTTTCTG | 50 | 1644 |
| MCR-6 | GTCCGGTCAATCCCTATCTGT | ATCACGGGATTGACATAGCTAC | 55 | 556 |
| MCR-7 | TGCTCAAGCCCTTCTTTTCGT | TTCATCTGCGCCACCTCGT | 55 | 892 |
| MCR-7.1 | AGGGGATAAACCGACCCTGA | TGATCTCGATGTTGGGCACC | 55 | 335 |
| MCR-8 | CCCAAGCTTTTGATTGTCCCTGTCGCCAT | CACCGATAAGAGGAACCAGTGAATTCCGG | 55 | |
| MCR-9 | TTCCCTTTGTTCTGGTTG | GCAGGTAATAAGTCGGTC | 1011 | |
| MOXM | GCTGCTCAAGGAGCACAGGAT | CACATTGACATAGGTGTGGTGC | 68.2 | 520 |
| NDM | GGTGCATGCCCGGTGAAATC | ATGCTGGCCTTGGGGAACG | 61.6 | 660 |
| OXA | CGCAAATGGCACCAGCTTCAAC | TCCTGCACCAGTTTTCCCATACAG | 61.7 | 464 |
| PER | ATGAATGTCATTATAAAAGC | AATTTGGGCTTAGGGCAGAA | 52.3 | 926 |
| SHV | GGGAAACGGAACTGAATGAG | TTAGCGTTGCCAGTGCTCG | 55 | 380 |
| TEM | AGATCAGTTGGGTGCACGAG | TGCTTAATCAGTGAGGCACC | 52 | 750 |
| VEB | GTTAGCGGTAATTAACCAG | TATTCAATAGTAATTCCACG | 59.1 | 820 |
| VIM | AAAGTTATGCCGCACTCACC | TGCAACTTCATGTTATGCCG | 52.3 | 865 |
PCR: Polymerase chain reaction
RESULTS
All the patients (N = 1496) admitted to our center for 1 year (April 2020–May 2021) were diagnosed to be COVID-19 positive based on reverse transcription polymerase chain reaction/cartridge-based nucleic acid amplification test or antigen testing (SD Biosensor, India), as per the diagnostic guideline of the Indian Council of Medical Research, Ministry of Health and Family Welfare.[17]
Out of the 1496 patients, 1240 (82.9%) were critical and admitted to ICU/HDUs while 256 (17.1%) patients were admitted to the wards. Among the 1496 patients, 924 (61.7%) had secondary infections. We included only 924 patients in the analysis. Most of these patients (766, 82.9%) showed severe illness. The overall mortality rate was 678/1496 (45.3). Of the 678 fatalities, 457 (49.5%) were attributed to secondary infections.
Among 924 patients, 585 (62.8%) patients were males; 339 (37.2%) were females. 47.1% of patients were aged between 18 and 60 years; 45.3% were more than 60 years old and 7.6% were <18 years old. The mean ± SD age of the COVID-19-positive patients showing secondary infection was 46.6 ± 19.7 years. Table 3 shows the demographic data of the patients admitted to ICU and wards with their clinical outcome.
Table 3.
Demographic characteristics of coronavirus disease 2019 patients
| Demographics | Overall | Admission | P | Clinical outcome | P* | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Non-ICU | ICU | Alive | Dead | ||||
| Total admissions | 1496 | 256 (17.1) | 1240 (82.9) | 0.994 | 818 (54.6) | 678 (45.3) | 0.048 |
| Patients having secondary infections | 924 | 158 (17.1) | 766 (82.9) | 467 (50.5) | 457 (49.5) | ||
| Age (years), mean±SD (range) | 46.6±19.7 (0-95) | 47.9±19.4 (2-92) | 46.6±19.7 (0-95) | 0.444 | 43.3±20.3 (0-93) | 53.1±18.6 (0-92) | <0.001 |
| 0-16 | 68 (7.6) | 11 (16.2) | 57 (83.8) | 0.949 | 54 (79.4) | 14 (20.6) | <0.001 |
| 16-50 | 433 (47.1) | 73 (16.9) | 360 (83.1) | 239 (55.2) | 194 (44.8) | ||
| >50 | 423 (45.3) | 74 (17.5) | 349 (82.5) | 174 (41.1) | 249 (58.9) | ||
| Gender | |||||||
| Female | 339 (37.2) | 58 (17.1) | 281 (82.9) | 0.995 | 184 (54.3) | 155 (45.7) | 0.084 |
| Male | 585 (62.8) | 100 (17.1) | 485 (82.9) | 283 (48.4) | 302 (51.6) | ||
| Length of stay | |||||||
| Mean±SD | 14.9±11.2 | 8.6±7.6 | 15.8±11.2 | <0.001 | 16.4±11.6 | 13.6±10.7 | <0.001 |
| Median (IQR) | 13 (8-20) | 14 (9-21) | 13 (8-19) | 0.149 | 15 (10-21) | 12 (6-18) | <0.001 |
*Bold-faced P values are significance at α<0.05. SD: Standard deviation, IQR: Interquartile range, ICU: Intensive care unit
During this study period, we received a total of 3637 clinical samples for microscopy/culture and sensitivity testing. Out of these, we included 1652 (45.4%) samples that showed cultured positive for secondary infections with clinically significant pathogens; we did not consider colonizers/insignificant growth in the study. The positive clinical samples included were blood, body fluid, pus and swab, nasal swab/crust/scrap/tissue biopsy, bronchoalveolar lavage (BAL), and endotracheal aspirate (ETA). Among these, pus and swab samples were predominant (431, 26%) followed by blood (405, 25%). Table 4 shows the distribution of different positive clinical samples included in the study.
Table 4.
Distribution of total positive samples for secondary infections
| Sample type | n (%) |
|---|---|
| Pus and swab | 431 (26) |
| Blood | 405 (25) |
| Body fluid | 350 (21) |
| Tracheal | 203 (12) |
| Tissue biopsy/nasal swab | 86 (5) |
| Sputum | 59 (4) |
| BAL | 51 (3) |
| Catheter tip | 24 (1.5) |
| Others | 24 (1.5) |
| CSF | 19 (1) |
| Total | 1652 |
BAL: Bronchoalveolar lavage, CSF: Cerebrospinal fluid
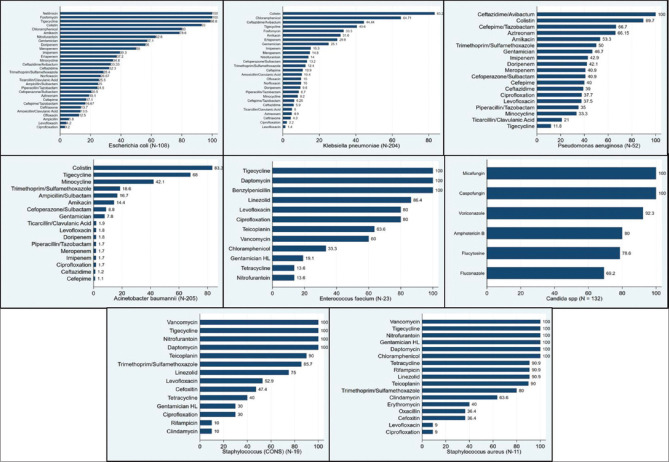
One thousand eight hundred and forty-one pathogens causing secondary infections were isolated from 1652 clinical samples [Table 5]. Bacteria caused 88% (1623/1841) of secondary infections and fungi caused 11.8% (218/1841) of secondary infections. Figure 1 shows the frequency of different pathogens causing secondary infections. Acinetobacter baumannii (205/586, 35%) and K. pneumoniae (204/586, 34.8%) were the most common pathogens among Gram-negative pathogens, followed by Escherichia coli (108/586, 18.4%). Among Gram-positive bacteria (58/1841, 3.2%), Enterococcus faecium isolates were predominant (23/58, 39.7%) followed by 19 isolates (32.8%) of coagulase-negative Staphylococcus aureus and S. aureus (11/58, 19%). The growth of two types of bacteria was observed in 979 cultures (54.8%) [Table 5].
Table 5.
Distribution of pathogens causing secondary infections in various clinical samples of coronavirus disease 2019-positive patients
| Sample type | BAL | Body fluid | Blood | Catheter tip | CSF | Others | Pus and swab | Sputum | Tracheal | Biopsy/nasal swab* | Total (n=1841) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GN organisms (n=586) | |||||||||||
| Acinetobacter baumannii | 14 | 6 | 99 | 0 | 10 | 4 | 32 | 5 | 35 | 0 | 205 |
| Klebsiella oxytoca | 12 | 26 | 106 | 3 | 0 | 1 | 26 | 3 | 27 | 0 | 204 |
| Escherichia coli | 2 | 46 | 20 | 0 | 2 | 3 | 32 | 0 | 3 | 0 | 108 |
| Pseudomonas aeruginosa | 3 | 5 | 8 | 0 | 0 | 3 | 15 | 0 | 16 | 0 | 50 |
| Enterobacter spp. | 1 | 0 | 2 | 0 | 0 | 1 | 3 | 0 | 1 | 0 | 8 |
| Proteus mirabilis | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 4 |
| Pseudomonas spp. | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Salmonella spp. | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Citrobacter koseri | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Serratia ficaria | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Providencia rettgeri | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| GP organisms (n=58) | |||||||||||
| Enterococcus faecium | 0 | 2 | 20 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 23 |
| Staphylococcus (CONS) | 1 | 1 | 14 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 19 |
| Staphylococcus aureus | 0 | 1 | 1 | 0 | 0 | 1 | 8 | 0 | 0 | 0 | 11 |
| Enterococcus faecalis | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Fungus (n=218) | |||||||||||
| Candida spp. | 0 | 116 | 15 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 132 |
| Molds | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 54 | 54 |
| Two types of bacteria | |||||||||||
| Two GP/GN | 18 | 144 | 300 | 19 | 7 | 11 | 307 | 50 | 123 | 0 | 979 |
*Biopsy/nasal swab: Tissue biopsy/nasal scraping/nasal crust/high nasal swab. n: Total number of isolates, GP: Gram-positive, GN: Gram-negative, CONS: Coagulase-negative staphylococcus aureus, CSF: Cerebrospinal fluid, BAL: Bronchoalveolar lavage
Figure 1.
Frequency of different pathogens causing secondary infections in COVID-19 patients. COVID-19: Coronavirus disease 2019
Of the 218 fungal isolates, Candida spp. was 60.5% (132/218) and molds were 39.4% (86/218). We performed KOH mounts and fungal cultures on 96 samples, of which 56 were tissue samples (58.33%), 15 (26.7%) respiratory samples (ETA, BAL, and sputum), 13 (13.54%) nasal/palatal crusts, and 12 (12.5%) nasal swabs. Most samples received between March and May 2021 fell within the second peak. A total of 44 (45.83%) samples had aseptate hyphae, 4 (4.166%) had aseptate hyphae along with septate hyphae, and 6 (6.25%) samples had septate hyphae. Figure 2 shows the KOH mount of broad aseptate hyphae with and without the addition of calcofluor white, growth of Rhizopus arrhizus on SDA tube, and LPCB of R. arrhizus growth. A total of five of the nine samples grew positively in culture (Mucorales in five samples, Mucorales and Aspergillus in three tubes, and Aspergillus from one respiratory sample). We were able to identify the fungal isolates from the culture-positive tubes, based on culture characteristics, followed by teased mounts of the growths on the LPCB mounts.
Figure 2.
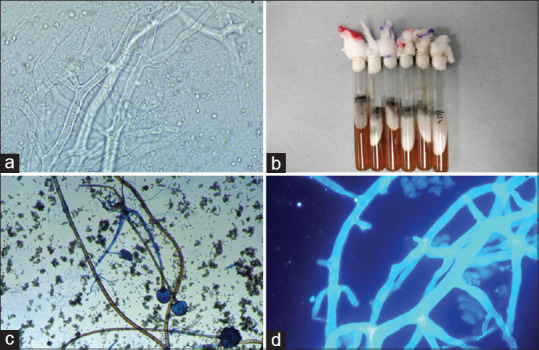
(a) KOH mount (×40) - broad aseptate hyphae, (b) Culture tubes showing growth of Rhizopus arrhizus, (c) Lactophenol cotton blue (×10) showing Rhizopus arrhizus, (d) KOH mount with calcofluor white (×40) - broad aseptate hyphae. KOH: Potassium hydroxide
Almost all the bacterial isolates causing secondary infections in COVID-19 patients showed resistance to more than two classes of antimicrobials and were multidrug-resistant pathogens. All Gram-negative pathogens showed the highest sensitivity to colistin (83%–93%) followed by tigecycline (12%–98%). Among beta-lactamase inhibitors, ceftazidime/avibactam showed the greatest sensitivity (33%–100%). Among carbapenems, meropenem (1.7%–50%) had the highest level of sensitivity in all Gram-negative pathogens, followed by doripenem (2%–46%) and imipenem (2%–43%). All Gram-positive isolates were sensitive to tigecycline and daptomycin. Sensitivity against linezolid and vancomycin was 83% each. There were 36.5% (4/11) methicillin-resistant S. aureus isolated. All candida spp. showed 100% sensitivity against caspofungin and micafungin. Figure 3 shows the antimicrobial sensitivity profile of pathogens causing secondary infections in COVID-19-positive patients.
Figure 3.
Antimicrobial sensitivity profile of pathogens causing secondary infections in COVID-19 patients. COVID-19: Coronavirus disease 2019
Genotypic profile of beta-lactamases
Genotypic profile of 308 isolates recovered from patients admitted to the COVID-19 ICUs was included in this study. The prevalence of beta-lactamases in various isolates is given in Table 6. All three genes’ categories of beta-lactamase genes were found in K. pneumoniae, A. baumannii, E. coli, Pseudomonas aeruginosa, E. cloacae, etc., A high prevalence of ESBL was found in all isolates. The most prevalent ESBL gene was observed to be blaTEM (54.2% positivity) across all the isolates followed by blaSHV (27.3% positivity). The prevalence of blaVEB was the least among the different isolates. AmpC genes were the least prevalent in all the bacterial isolates. blaCITM was the most common AmpC gene among all the isolates forward by blaDHAM-1 and 2. Among carbapenemase genes, blaNDM was most prevalent followed by blaOX gene.
Table 6.
Distribution of beta-lactamase genes in various Gram-negative organisms
| Beta-lactamases | n=308 | Carbapenem-resistant genes | ESBL | AMP C | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||||
| NDM | OXA | KPC | IMP | VIM | TEM | SHV | CTX-M | PER | VEB | CITM | DHAM-1 and amp; 2 | EBCM | MCR 5 | ||
| Acinetobacter baumannii | 101 | 54 (53.5) | 25 (24.8) | 3 (3) | 0 | 0 | 35 (34.7) | 7 (6.9) | 10 (9.9) | 30 (29.7) | 2 (2) | 6 (5.9) | 12 (11.9) | 0 | 0 |
| Enterobacter cloacae | 5 | 3 (60) | 1 (20) | 1 (20) | 0 | 0 | 1 (20) | 1 (20) | 0 | 0 | 0 | 0 | 0 | 1 (20) | 0 |
| Escherichia coli | 59 | 28 (47.5) | 22 (37.3) | 1 (1.7) | 0 | 2 (3.4) | 42 (71.2) | 5 (8.5) | 17 (28.8) | 0 | 0 | 25 (42.4) | 5 (8.5) | 0 | 1 (1.7) |
| Klebsiella pneumoniae | 106 | 66 (62.3) | 26 (24.5) | 25 (23.6) | 3 (2.8) | 1 (0.9) | 79 (74.5) | 66 (62.3) | 45 (42.5) | 1 (0.9) | 0 | 17 (16) | 9 (8.5) | 0 | 1 (0.9) |
| Proteus mirabilis | 1 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 |
| Providencia rettgeri | 1 | 1 (100) | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 29 | 13 (44.8) | 9 (31) | 1 (3.4) | 0 | 0 | 9 (31) | 4 (13.8) | 4 (13.8) | 0 | 2 (6.9) | 2 (6.7) | 4 (13.3) | 0 | 0 |
| Pseudomonas putida | 1 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 |
| Salmonella paratyphi A | 1 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Stenotrophomonas maltophilia | 4 | 2 (50) | 1 (25) | 0 | 0 | 0 | 0 | 0 | 1 (25) | 0 | 0 | 0 | 1 (25) | 0 | 0 |
| Total | 308 | ||||||||||||||
DISCUSSION
It is common to have bacterial or fungal secondary or co-infections in critically ill virus-infected patients.[6,7,8] In our study, we found that more than half of the COVID-19 patients admitted showed secondary infections while 83% of patients admitted to ICU had secondary infections. The secondary infections are more likely to affect critically ill patients. This may be due to severe illness, comorbidities, use of steroids, and the presence of invasive devices which may become colonized with biofilms. A study conducted in patients infected with respiratory viruses showed 11%–35% of secondary infections caused by bacteria.[18] We observed a significant proportion of fungal infections (12%), most of which were in the period of the second COVID-19 wave. Several studies have found bacterial infection as a worrying problem as it complicates the treatment in COVID-19 patients.[8,9] In our study, we found that the mortality rate among COVID-19 patients with secondary infection was significantly higher (50%) compared to the overall mortality rate in COVID-19 patients (45.3%) (P = 0.048). We did not find any significant difference in mortality between males and females. However, the number of male patients with secondary infections was higher than females. Similarly, studies from different countries have reported that male gender is more prone to COVID-19 disease severity.[19,20]
It has been a challenge to detect and distinguish bacterial or fungal co-infection in COVID-19-positive patients based on clinical and radiological findings alone.[21] Thus, including the findings of microbiological investigations in concordance with clinical and radiological findings would help in the better management of patients. Our microbiological test results showed that most of the secondary infections were caused by multidrug-resistant pathogens. A. baumannii (205, 11%) was the predominant multidrug-resistant Gram-negative pathogens causing secondary infections followed by K. pneumoniae (204, 11%) and E. coli (108, 5.86%). Similar results were found in our previous study conducted in 2019.[22] These pathogens showed a high rate of resistance against carbapenem. A. baumannii had the highest resistance (98.3%) followed by K. pneumoniae (84.7%) and E. coli (60.7%) which is in concordance with other studies.[23] Genotypic profiling showed a high prevalence of beta-lactamase genes among all isolates. Many of these were positive for two or more different types of beta-lactamases. ESBL was the most prevalent beta-lactamases followed by carbapenemases and AmpC. Among Gram-positive pathogens, E. faecium isolates showed 40% and 14% resistance against vancomycin and linezolid, respectively. S. aureus showed 36.4% resistance against methicillin. Increased resistance to piperacillin/tazobactam, amoxicillin/clavulanic acid, cefepime/tazobactam, aminoglycosides, and carbapenems was noted in Gram-negative pathogens. Increased resistance was also illustrated for linezolid in E. faecium and S. aureus and against fluconazole in Candida species. The development of mucormycosis in COVID-19 patients is a worrying problem and needs urgent attention.
Mucormycosis is a life-threatening condition; a positive microscopy finding in the setting of high clinical suspicion is sufficient to treat the patient. Culture positivity is seen in only 30%–40% of microscopy-positive cases, and LPCB, MALDI-TOF MS, and PCR can be performed from culture-positive isolates only, making the microscopy more reliable, less expensive, and an easy-to-perform test which can contribute immensely to saving lives of people.[24] The majority of the patients who tested positive for aseptate hyphae had recovered from COVID-19 in the recent past and had developed symptoms like a diminution of vision and periorbital swelling. All of them had a history of having received corticosteroids during their COVID-19 treatment. All our cases were of retro-orbital mucormycosis, it being the most common in India and overall.[12] One was a case of pulmonary aspergillosis aptly supported with the help of radiological and clinical findings.
CONCLUSIONS
We are seeing an increasing number of cases of fungal infections, in particular, molds and antimicrobial-resistant Gram-negative pathogens, which are often highly drug resistant. Continued AMR surveillance for bacteria and prompt detection of fungal pathogens using microscopy will help patients in getting early and appropriate treatment, both definitive and prophylactic. Infection control practices in COVID-19 ICUs need to be improved to prevent cross-transmission of pathogens.
Research quality and ethics statement
The study was approved by the AIIMS Institute Ethics Committee, New Delhi (IEC-556/June 19, 2020). The authors followed applicable EQUATOR Network (http://www.equator-network.org/) guidelines during the conduct of this research project.
Financial support and sponsorship
This work was supported in part through a grant for Center of Excellence on Infection Control from the Indian Council of Medical Research (AMR/193/2019-ECD-II).
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We thank all the laboratory staff, fellow colleagues, and doctors for their constant support and co-operation.
REFERENCES
- 1.India COVID: 34,710,630 Cases and 476,135 Deaths. https://www.worldometers.info/coronavirus/country/india/
- 2.Fattorini L, Creti R, Palma C, Pantosti A. Unit of Antibiotic Resistance and Special Pathogens, Unit of Antibiotic Resistance and Special Pathogens of the Department of Infectious Diseases, Istituto Superiore di Sanità, Rome. Bacterial coinfections in COVID-19: An underestimated adversary. Ann Ist Super Sanita. 2020;56:359–64. doi: 10.4415/ANN_20_03_14. [DOI] [PubMed] [Google Scholar]
- 3.Hughes S, Troise O, Donaldson H, Mughal N, Moore LS. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26:1395–9. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawson TM, Moore LS, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71:2459–68. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris DE, Cleary DW, Clarke SC. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. 2017;8:1041. doi: 10.3389/fmicb.2017.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilder-Smith A, Green JA, Paton NI. Hospitalized patients with bacterial infections: A potential focus of SARS transmission during an outbreak. Epidemiol Infect. 2004;132:407–8. doi: 10.1017/s0950268803001869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memish ZA, Perlman S, Van Kerkhove MD, Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–77. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khurana S, Singh P, Sharad N, Kiro VV, Rastogi N, Lathwal A, et al. Profile of co-infections & Secondary infections in COVID-19 patients at a dedicated COVID-19 facility of a tertiary care Indian hospital: Implication on antimicrobial resistance. Indian J Med Microbiol. 2021;39:147–53. doi: 10.1016/j.ijmmb.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijay S, Bansal N, Rao BK, Veeraraghavan B, Rodrigues C, Wattal C, et al. Secondary infections in hospitalized COVID-19 patients: Indian experience. Infect Drug Resist. 2021;14:1893–903. doi: 10.2147/IDR.S299774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldin C, Soliman SS, Jeon HH, Alkhazraji S, Gebremariam T, Gu Y, et al. PCR-Based approach targeting mucorales-specific gene family for diagnosis of mucormycosis. J Clin Microbiol. 2018;56:e00746–18. doi: 10.1128/JCM.00746-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song G, Liang G, Liu W. Fungal Co-infections associated with global COVID-19 pandemic: A clinical and diagnostic perspective from China. Mycopathologia. 2020;185:599–606. doi: 10.1007/s11046-020-00462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakash H, Chakrabarti A. Epidemiology of mucormycosis in India. Microorganisms. 2021;9:523. doi: 10.3390/microorganisms9030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John TM, Jacob CN, Kontoyiannis DP. When uncontrolled diabetes mellitus and severe COVID-19 converge: The perfect storm for mucormycosis. J Fungi (Basel) 2021;7:298. doi: 10.3390/jof7040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidelines for Handling, Treatment, and Disposal of Waste Generated During Treatment/Diagnosis/Quarantine of COVID-19 Patients – Rev. 4. [Last accessed on 2021 Aug 15]. Available from: https://cpcb.nic.in/uploads/Projects/Bio-Medical-Waste/BMW-GUIDELINES-COVID_1.pdf .
- 15.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Wayne, PA: Supplement M, Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 16.Schwarz P, Guedouar H, Laouiti F, Grenouillet F, Dannaoui E. Identification of mucorales by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Fungi (Basel) 2019;5:56. doi: 10.3390/jof5030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Standard Operating Procedure for Detection of 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases by rRT-PCR: Confirmation Assay. [Last accessed on 2021 Aug 15]. Available from: https://www.icmr.gov.in/pdf/covid/labs/2_SOP_for_Confirmatory_Assay_for_2019_nCoV.pdf .
- 18.Klein EY, Monteforte B, Gupta A, Jiang W, May L, Hsieh YH, et al. The frequency of influenza and bacterial coinfection: A systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10:394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NICE. Managing Suspected or Confirmed Pneumonia COVID-19 Rapid Guideline: Managing Suspected or Confirmed Pneumonia in Adults in the Community. Guidance. 2020. [Last accessed on 2020 Dec 18]. Available from: https://www.nice.org.uk/guidance/ng165/chapter/4-Managing-suspected-or-confirmed-pneumonia .
- 20.Walia K, Madhumathi J, Veeraraghavan B, Chakrabarti A, Kapil A, Ray P, et al. Establishing antimicrobial resistance surveillance & Research network in India: Journey so far. Indian J Med Res. 2019;149:164–79. doi: 10.4103/ijmr.IJMR_226_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P, Liu Z, Chen Y, Xiao Y, Huang X, Fan XG. Bacterial and fungal infections in COVID-19 patients: A matter of concern. Infect Control Hosp Epidemiol. 2020;41:1124–5. doi: 10.1017/ice.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathur P, Khurana S, Kumar S, Gupta D, Aggrawal R, Soni KD, et al. Device associated infections at a trauma surgical Center of India: Trend over eight years. Indian J Med Microbiol. 2021;39:15–8. doi: 10.1016/j.ijmmb.2020.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Balkhair A, Al-Muharrmi Z, Al’Adawi B, Al Busaidi I, Taher HB, Al-Siyabi T, et al. Prevalence and 30-day all-cause mortality of carbapenem-and colistin-resistant bacteraemia caused by Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae: Description of a decade-long trend. Int J Infect Dis. 2019;85:10–5. doi: 10.1016/j.ijid.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clin Microbiol Rev. 2000;13:236–301. doi: 10.1128/cmr.13.2.236-301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]