Abstract
Many host factors of plants are used by viruses to facilitate viral infection. However, little is known about how alpha‐momorcharin (αMMC) counters virus‐mediated attack strategies in tomato. Our present research revealed that the 2b protein of cucumber mosaic virus (CMV) directly interacted with catalases (CATs) and inhibited their activities. Further analysis revealed that transcription levels of catalase were induced by CMV infection and that virus accumulation increased in CAT‐silenced or 2b‐overexpressing tomato plants compared with that in control plants, suggesting that the interaction between 2b and catalase facilitated the accumulation of CMV in hosts. However, both CMV accumulation and viral symptoms were reduced in αMMC transgenic tomato plants, indicating that αMMC engaged in an antiviral role in the plant response to CMV infection. Molecular experimental analysis demonstrated that αMMC interfered with the interactions between catalases and 2b in a competitive manner, with the expression of αMMC inhibited by CMV infection. We further demonstrated that the inhibition of catalase activity by 2b was weakened by αMMC. Accordingly, αMMC transgenic plants exhibited a greater ability to maintain redox homeostasis than wild‐type plants when infected with CMV. Altogether, these results reveal that αMMC retains catalase activity to inhibit CMV infection by subverting the interaction between 2b and catalase in tomato.
Keywords: 2b, alpha‐momorcharin, catalase, cucumber mosaic virus, Solanum lycopersicum
Alpha‐momorcharin directly disrupts the interaction between catalase and 2b by competing with 2b in binding to catalases, altering cellular redox homeostasis, reducing toxic reactive oxygen species production, and augmenting resistance against cucumber mosaic virus infection.
1. INTRODUCTION
In a complex ecological environment, plants often incur various biotic stresses (e.g., viral infections, insect attacks, and fungal diseases) and abiotic stresses (e.g., extreme temperatures, saline soils, and water shortages) throughout their lifetime (Yang & Guo, 2018). Cucumber mosaic virus (CMV), a typical member of the genus Cucumovirus, invades a wide range of plant hosts and causes serious economic losses to vegetable and horticultural crops worldwide (Scholthof et al., 2011). Three positive‐sense RNA strands, designated RNA1, RNA2, and RNA3, constitute the CMV genome. RNA1 encodes the replicase protein 1a, RNA2 encodes the replicase protein 2a, and the 3a movement protein of the virus is encoded by RNA3 (Ding et al., 1994). A subgenome of CMV, designated RNA4, encodes the viral coat protein and is also responsible for coding the 2b protein that plays multiple roles in viral virulence and systemic infection (Chen et al., 2014; Shi et al., 2003). In particular, CMV infection severely hinders the growth and development of tomato crops, resulting in serious disease symptoms such as dwarfism, increased tillering, systemic necrosis, and narrow, tendril‐like leaflets (Di Carli et al., 2010). It is imperative, therefore, that the resistance of tomato plants to CMV infection be improved.
In combating viral infection, plants synthesize compounds to enhance host immunity. Ribosome inactivating proteins (RIPs) are synthesized naturally in a variety of plants, and catalyse inactivating eukaryotic and prokaryotic ribosomes by removing single adenine residues from large rRNAs, which can be divided into three broad groups: type I, type II, and type III (Barbieri et al., 1993). RIPs have been found to function in various aspects of immunity against pathogens in animals (Citores et al., 2021). Moreover, the results of many studies have suggested that expression of RIPs in transgenic plants confers increased tolerance under stressful conditions (Ajji et al., 2016). A growing body of research has proposed the role of RIPs in plant defence against fungi, insect attacks, and viral infections (Fabbrini et al., 2017; Mishra et al., 2022). Although the mechanisms by which RIPs enhance immunity in animals against pathogens are well investigated, their functions in plant responses to viral infection are less well understood.
Alpha‐momorcharin (αMMC) is a typical type I RIP, extracted from Momordica charantia, that removes a specific adenine from 28S rRNA and inhibits protein biosynthesis (Puri et al., 2009). Numerous studies have shown that αMMC exhibits several medicinal properties, including antitumor, antidiabetic, antimicrobial, and antiviral activities, and exhibits an immune‐modulatory effect both in vitro and in vivo (Sur & Ray, 2020 ). Recent studies have found that foliar spraying of αMMC on leaves not only enhanced the defence response of tobacco plants to a variety of pathogens, but also could strengthen crop (M. charantia) resistance to viral infection (Yang et al., 2016; Zhu et al., 2013). Moreover, a study of transgenic tobacco harbouring the αMMC gene showed that the plants increased their immunity to tobacco mosaic virus (TMV) infection (Zhu et al., 2020). Nevertheless, the molecular mechanism by which αMMC contributes to plant resistance in systemic necrosis of plant–virus interactions is largely unknown.
The reactive oxygen species (ROS) burst, including superoxide anion (O2 −), hydrogen peroxide (H2O2), and hydroxyl radical (OH−), is associated with the necrosis of plant tissues caused by viral infections (Díaz‐Vivancos et al., 2008; Yoshioka, 2003). Recent findings have shown that H2O2 and autophagy regulate plant immunity‐related programmed cell death under environmental stresses (Henry et al., 2015; Pérez‐Pérez et al., 2012). Autophagy is a conserved mechanism in eukaryotes that removes harmful or unwanted cellular substances, relies on multiple cell membrane elements, and is regulated by a series of autophagy proteins (ATGs) (Zhuang et al., 2017). Many studies have demonstrated that an accumulation of ROS triggers autophagy (Minibayeva et al., 2012). Furthermore, the evidence of necrosis symptoms in plants indicated that ROS also regulate starvation‐induced autophagy (Scherz‐Shouval & Elazar, 2007). To avoid incurring oxidative damage, plants have evolved defence responses through which antioxidative enzymes metabolize peroxides under viral infection (Das & Roychoudhury, 2014). For example, catalases break down H2O2 into H2O and O2 to overcome oxidative stress, and are found in nearly all organisms (Chelikani et al., 2004). There is mounting research suggesting that catalase participates in plant response to abiotic and biotic stresses (Mathioudakis et al., 2013; Yang et al., 2020).
In this study, we present evidence that αMMC plays a vital role in the transgenic Solanum lycopersicum response to CMV infection. The virus silencing suppressor (VSR) of CMV, 2b, interacted with host factor catalases and inhibited their activities to counter the plant defence response. Using 2b‐overexpressing plants and tobacco rattle virus (TRV)‐induced gene silencing (VIGS) of CAT1 and CAT2 plants, we found that catalase activity was essential for plant defence against CMV infection. To subvert that viral strategy, αMMC interacted with catalases and mitigated the inhibition of their activity by disrupting the interactions between 2b and catalases to prevent the invasion and accumulation of CMV in the host. Moreover, our study demonstrates that catalase has a positive role and is induced as part of the plant immunity response under CMV conditions.
2. RESULTS
2.1. Subcellular localization and spatiotemporal expression of αMMC in tomato
To determine the role of αMMC in the interaction between plants and RNA viruses, transgenic lines of tomato plants overexpressing αMMC were generated (Figure S1). To analyse organ‐specific expression of the αMMC gene in transgenic lines, we measured its transcription levels in tomato roots, stems, and leaves. The results of reverse transcription‐quantitative PCR (RT‐qPCR) suggested that the αMMC gene was expressed in all three examined organs, with the highest expression in roots (Figure 1a). In order to observe the αMMC subcellular location, αMMC‐GFPC was obtained by attaching a green fluorescent protein (GFP) tag at the C‐terminus of αMMC. We found that GFP signals appeared in the cytoplasm of αMMC‐GFPC agro‐infiltrated Nicotiana benthamiana cells (Figure 1b). In contrast, bright fluorescence signals were observed throughout the cell nucleus and cytoplasm in the control plants (35 S:00) (Figure 1b). The αMMC sequence was also analysed through the Web‐server predictors (http://www.csbio.sjtu.edu.cn/bioinf/plant‐multi/). Plant‐mPLoc predicted that αMMC would be located in the cell wall and cytoplasm as it lacks a nuclear localization signal in its sequence (Figure 1c). The αMMC transgenic tomato and wild‐type (WT) tomato had similar phenotypic characteristics and there were no visible differences during the period of growth and reproductive development in a natural environment (Figure S2). These results indicate that the αMMC protein did not affect the growth and development of the transgenic tomato and was located in the cytoplasm of cells.
FIGURE 1.
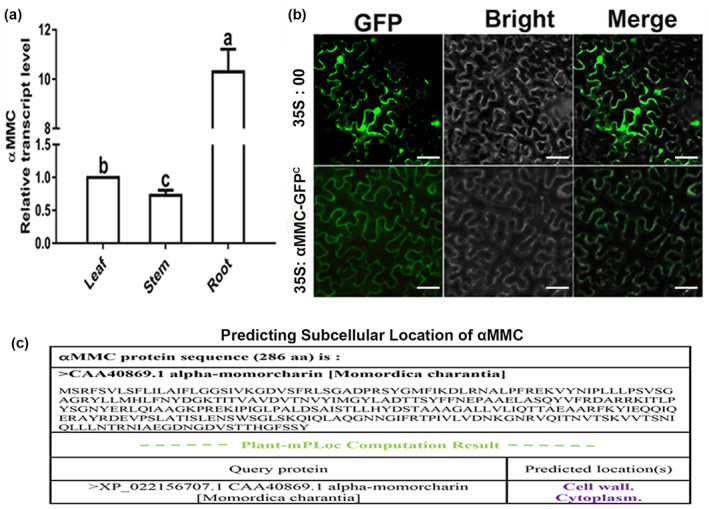
Subcellular localization of αMMC in Nicotiana benthamiana. (a) Organ‐specific expression of the αMMC gene in plants. Expression levels were standardized to Actin, the results of the leaf were set at 1. Values indicate the mean ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05). (b) The 35S:αMMC‐GFPC construct was agro‐infiltrated into N. benthamiana leaves and detected by fluorescence microscopy. The empty vector (35S:00) served as the control. (c) Predicting subcellular location of αMMC. The αMMC sequence was also analysed through the web‐server predictors of plant‐mPLoc
2.2. Transgenic αMMC lines promoted resistance to CMV infection
To evaluate whether αMMC contributes to how tomato responds to infection by CMV, αMMC transgenic seedlings from two independent lines with relative higher expression level of αMMC (i.e., #1 and #8) and WT plants were inoculated with CMV and their symptom severities were examined. At 10 days postinoculation (dpi), the WT plants showed slight dwarfing phenotypes and chlorotic flecks on their leaves, whereas the CMV‐infected systemic leaves had no obvious symptoms in either αMMC line. Furthermore, at 20 dpi, WT plants displayed stronger viral infection symptoms, including severe dwarfing, accelerated leaf senescence, and tissue necrosis, than did the αMMC lines (Figure 2a). Striking symptoms in plants were evidenced by the substantially increased accumulation of CMV in WT plants (Figure 2a–c). The results also showed that the viral coat protein accumulated more in WT plants than that in both αMMC transgenic lines (Figure 2b,c). Moreover, for the 18 individual tomato seedlings of each line inoculated with CMV in three independent experiments, the infection efficiency of transgenic plants was far lower than that of WT plants (Figure 2d). These results together indicate that overexpression of αMMC in tomato plants represses viral infection.
FIGURE 2.
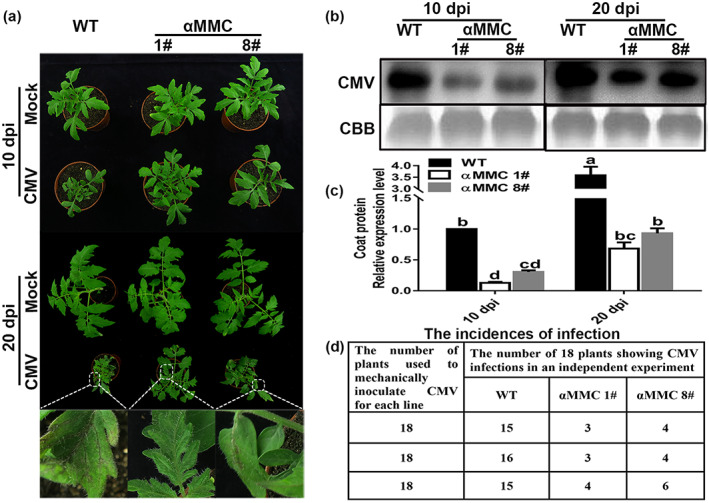
The αMMC‐overexpressing lines promote resistance to CMV infection. (a) Disease symptoms in the wild‐type (WT) plants and the αMMC transgenic plants at 10 days postinoculation (dpi) and 20 dpi. Magnified images of leaves (in white boxes) are shown in the lower panels. Detection of viral coat protein accumulation by (b) western blotting and (c) reverse transcription‐quantitative PCR (RT‐qPCR) analysis. Systemic leaves were harvested for analysis. Coomassie brilliant blue (CBB) staining of RuBisCO protein indicates the loading control. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05). (d) Incidences of CMV infection determined by RT‐qPCR and the visual assessment of disease symptoms of 18 individual plants per line at 40 dpi from three independent experiments
2.3. Involvement of ROS in the αMMC‐regulated CMV defence response
Previous studies reported that virus‐induced host tissue necrosis is associated with ROS bursts (Inaba et al., 2011; Yang et al., 2020), therefore the in situ accumulation of superoxide (O2−) and H2O2 in tomato leaves was detected by nitroblue tetrazolium (NBT) and 3,3′‐diaminobenzidine (DAB) staining, respectively, to investigate whether αMMC affects ROS accumulation. There was greater NBT and DAB staining of WT leaves than of αMMC leaves under CMV‐infected conditions, although it increased in both leaf groups, whereas in the absence of viral infection the staining of these leaves was similar (Figure 3a,b). The measured H2O2 content was also much higher in WT leaves than in αMMC leaves under CMV‐infected conditions (Figure 3c). Oxidative damage is one consequence of stressful conditions incurred by plants, and can be measured by malondialdehyde (MDA) accumulation, relative water content (RWC) of plant leaves, and electrolyte leakage (EL) (Cunha et al., 2016). We found that the MDA content of the αMMC #1and αMMC #8 lines were 49.8% and 37.7% lower, respectively, than those in WT plants when infected with CMV (Figure 3d). The RWC of αMMC leaves resembled that of WT leaves without CMV inoculations (Figure 3e). In contrast, CMV infection led to a reduced RWC, the decline of which in WT plants was greater than that in αMMC #1 or #8 plants (Figure 3e). Conversely, the EL values of plants increased in response to CMV infection, yet EL values were higher in WT plants than in both αMMC lines (Figure 3f). These results imply that αMMC attenuates the accumulation of ROS induced by CMV infection, resulting in less oxidative damage.
FIGURE 3.
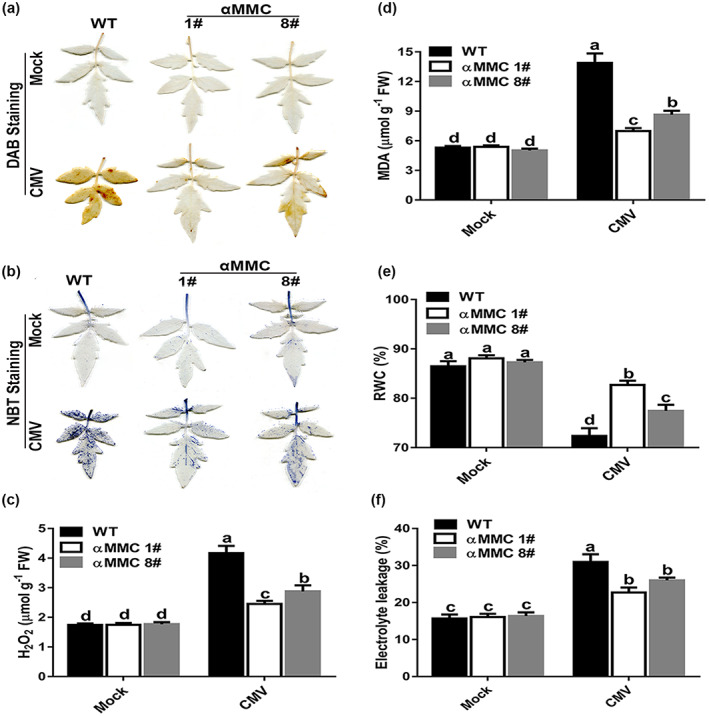
Involvement of reactive oxygen species in the αMMC‐regulated defence response to CMV. (a) H2O2 level evaluated by 3,3′‐diaminobenzidine (DAB) staining. (b) Superoxide level evaluated by nitroblue terazolium (NBT) staining. Five plants were used for each treatment; a picture of one representative leaf is shown. (c) Quantitative detection of H2O2 content. (d) Quantitative measurements of malondialdehyde (MDA) content. Determination of (e) relative water content (RWC) and (f) electrolyte leakage in wild‐type (WT) plants and αMMC transgenic plants. Systemic leaves were harvested for analysis. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05). FW, fresh weight
Autophagy is involved in plant defence responses against stresses, and thus we investigated whether autophagy participates in αMMC‐mediated resistance to CMV infection. The RT‐qPCR analysis showed that transcription levels of ATG genes were up‐regulated in CMV‐inoculated plants compared to mock‐inoculated plants, indicating their involvement in plant defence against CMV infection (Figure S3a–d). In particular, the ATG3, ATG5, ATG8, and ATG16 transcription levels in WT plants were significantly induced when compared with those in αMMC transgenic plants, with transcription levels in αMMC #8 plants exceeding those in #1 plants with CMV infection at 10 dpi (Figure S3a–d). However, under mock inoculation, no distinguishable differences appeared in the expression levels of these genes between αMMC #1 and #8 plants or between both lines and WT plants (Figure S3a–d). Furthermore, to verify the above results, autophagic activity was examined by transmission electron microscopy (TEM). Consistent with the RT‐qPCR results, there was a near total absence of autophagosomes or autophagic bodies in plants without viral infection, whereas the occurrence of both classic double‐membrane autophagosomes and signal‐membrane autophagic bodies increased the most in WT plants infected with CMV, followed by αMMC #8 plants, and increased the least in αMMC #1 plants (Figure S3e). These results demonstrate the involvement of αMMC in regulating the expression of ATG genes and the formation of autophagosome under CMV infection.
2.4. 2b interacted with catalase in vitro and in vivo
Both Inaba et al. (2011) and our previous study demonstrated that a VSR could interact with catalase to promote the accumulation of ROS in necrotic tissues (Yang et al., 2020). In this work, CMV infection induced ROS outbursts, caused autophagy, and elicited disease symptoms accompanied by necrosis on leaves. The catalase of tomato shares a high sequence identity in its nucleotides and deduced amino acid sequences with that of tobacco as well as Arabidopsis (Figure S4). This prompted us to examine whether catalases and the VSR of CMV are involved in facilitating CMV infection. Yeast two‐hybrid assays were carried out to identify interactions between catalases (CAT1 and CAT2) and 2b in vitro. As suspected, the combinations of 2b + CAT1 and 2b + CAT2 in yeast could grow on the four deficient media, but no yeast was detectable in the other negative controls, confirming that tomato catalases could interact with 2b in vitro (Figure 4a). To corroborate the interactions among 2b, CAT1, and CAT2, we performed a glutathione‐S‐transferase (GST) pull‐down assay. The catalases were fused with 6 × His tags, and 2b was fused with a GST tag. In Figure 4b, CAT1 and CAT2 directly bound with GST‐2b in vitro, but did not bind with GST alone (control). To further test the catalase–2b interactions in plants, a bimolecular fluorescence complementation (BiFC) analysis was used. The C‐terminal and N‐terminal fragments of yellow fluorescent protein (YFP) were fused to the 2b protein (2b‐cYFP) and catalase protein (CAT‐nYFP). When 2b‐cYFP was co‐infiltrated with CAT1‐nYFP or CAT2‐nYFP in N. benthamiana leaves, a bright fluorescence signal was detected. However, in CAT1‐nYFP + cYFP, CAT2‐nYFP + cYFP, or nYFP + 2b‐cYFP negative control‐infiltrated leaves, no fluorescence signal was detectable (Figure 4c). In conclusion, these results demonstrate that catalases interact with 2b in vivo and in vitro.
FIGURE 4.
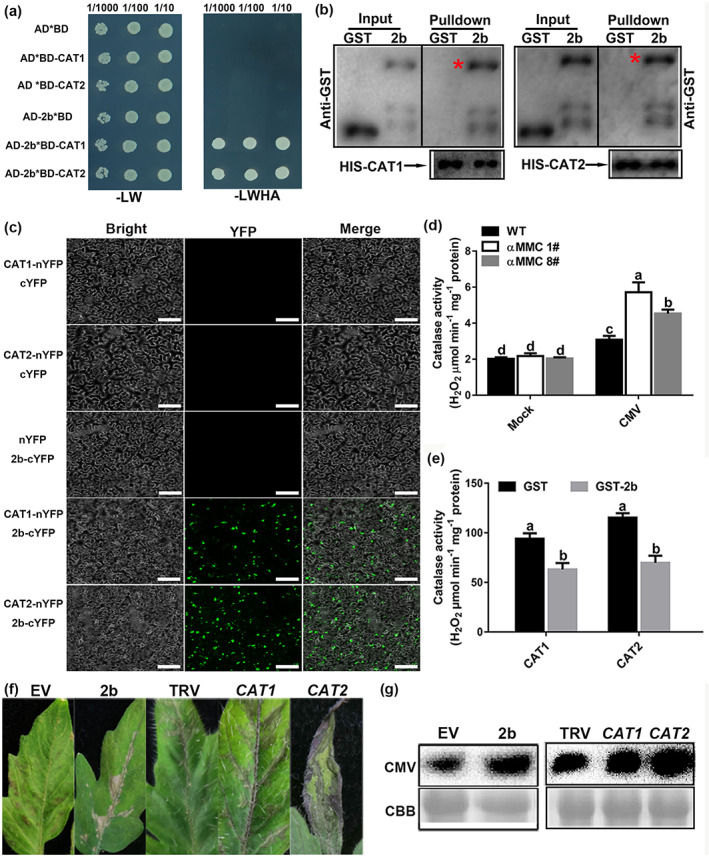
CMV 2b directly interacts with catalases in vivo and inhibits their activities to promote CMV accumulation. (a) CMV 2b interacts with catalases in yeast. Yeast growth on QDO medium lacking Leu, Trp, His, and Ade (−LWHA) illustrated the interaction. (b) Pull‐down assays examining the interactions among 2b, CAT1, and CAT2 in vitro. Purified 2b‐GST or glutathione‐S‐transferase (GST) was incubated with CAT1‐His and CAT2‐His. After immunoprecipitation with His beads, the proteins were monitored by anti‐CAT or anti‐GST antibodies. Red asterisks indicate bands of the target protein. (c) Bimolecular fluorescence complementation (BiFC) analyses of the interactions between catalases and 2b in Nicotiana benthamiana leaves. Green fluorescence illustrates the interaction, bars = 40 μm. (d) Measured catalase activity in wild‐type (WT) and αMMC transgenic plants in response to CMV infection. (e) Detection of CAT1 and CAT2 enzymatic activity in reactions containing 2b‐GST or GST. CAT was incubated with 2b at 37°C. (f) Disease symptoms of CMV infection in the empty vector (EV), 2b‐overexpressing, nonsilenced control (TRV alone), and CAT1‐ or CAT2‐silenced tomato leaves by TRV‐induced gene silencing. (g) Detection of CMV coat protein accumulation in EV, 2b‐overexpressing, TRV‐silenced, CAT1‐silenced, and CAT2‐silenced tomato by western blotting. Systemic leaves were harvested for analysis. Coomassie brilliant blue (CBB) staining of RuBisCO protein indicates the loading control. Values indicate means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05)
Zhang et al. (2015) also reported on catalase activities that were altered by the interaction between catalase and stress factors, which facilitated pathogen infection. As shown in Figure 4d, catalase activity was stronger in CMV‐infected plants than in mock‐infected plants, being significantly increased in αMMC‐overexpressing plants compared with WT plants under CMV infection, especially in line #1. However, under mock‐inoculated conditions, catalase activity differed negligibly between αMMC transgenic plants and WT plants (Figure 4d). The enzymatic activity of catalase protein, as expressed in Escherichia coli BL21 cells and purified in vitro, was also analysed. As shown in Figure 4e, catalase activity in the CAT1 + GST combination surpassed that in the CAT1 + 2b‐GST combination, with similar results obtained for the enzymatic activity of CAT2. To evaluate the effect of catalase activity inhibited by 2b on CMV infection, we measured the accumulation of CMV by silencing the CAT1 or CAT2 gene and transiently overexpressing 2b (Figure S5). In both CAT1‐ and CAT2‐silenced plants, CMV spread faster, tomato manifested stronger disease symptoms, and the level of virus accumulation was higher than that in TRV (control) plants (Figure 4f,g). As shown in Figure 4f,g, the disease symptoms of leaves and western blotting analysis showed that the 2b‐overexpressing tomato plants accumulated more virus than in control (EV) plants and displayed more serious symptoms. Taken together, these results indicated that 2b interacted with catalases and inhibited their activities to facilitate CMV accumulation in host plants.
2.5. αMMC interacted with catalase
A recent study demonstrated that the γb protein of barley stripe mosaic virus interacts with host factors to hasten infection and the onset of disease symptoms by disturbing the ATG7–ATG8 interaction (Yang et al., 2018). In this study, we explored whether αMMC interacted with 2b or catalase to mitigate 2b's inhibition of catalase activity and augment plant resistance against CMV infection. The interaction among αMMC, 2b, and catalase was investigated using the BiFC assay. The YFP fluorescence signals were observed in N. benthamiana leaves transiently expressing the CAT1‐nYFP + αMMC‐cYFP and CAT2‐nYFP + αMMC‐cYFP constructs, but not in leaves expressing αMMC‐nYFP + 2b‐cYFP or the control construct (Figure 5a). Moreover, according to the yeast two‐hybrid analysis, yeast cells expressing αMMC + CAT1 and αMMC + CAT2 grew well on the four deficient media, but yeast growth was suppressed in αMMC + 2b or control transformants, which indicates that αMMC specifically interacted with CAT1 and CAT2 in yeast two‐hybrid assays (Figure 5b). Consistent with the above results, only the catalase proteins were pulled down by αMMC‐MBP, whereas the GST alone (control) could not be pulled down by αMMC‐MBP in the GST pull‐down assays (Figure 5c). These results illustrated that αMMC interacted with catalases, both in vitro and in vivo.
FIGURE 5.
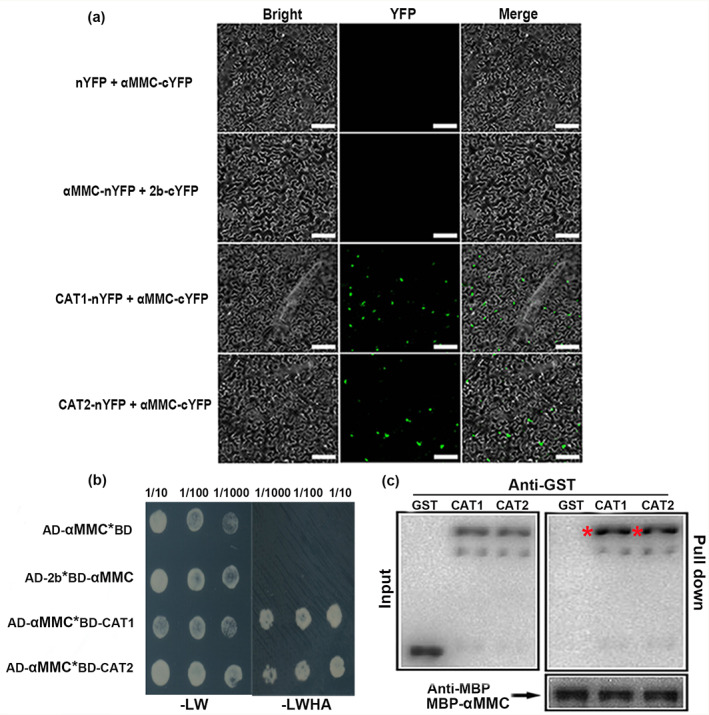
αMMC interacts with catalases. (a) Bimolecular fluorescence complementation analysis of the interactions among catalases, αMMC, and 2b in Nicotiana benthamiana. Green fluorescence illustrates the interaction, bars = 40 μm. (b) Yeast two‐hybrid assay. Yeast cells grown on QDO medium lacking Leu, Trp, His, and Ade (−LWHA) suggested the interaction. AD, GAL4‐activation domain. BD, GAL4 DNA‐binding domain. (c) Pull‐down assay results suggesting that αMMC interacted with CAT1/CAT2 in vitro. The purified CAT1‐GST, CAT2‐GST and glutathione‐S‐transferase (GST) proteins were each incubated with αMMC‐MBP. After immunoprecipitation with maltose‐binding protein (MBP) beads, the proteins were monitored by anti‐MBP or anti‐GST antibodies using western blotting. Red asterisks indicate bands of the target protein
2.6. αMMC interfered with the interactions between 2b and catalases to maintain catalase activity
The mechanism by which αMMC enhances plant defence and suppresses ROS outbursts under CMV‐infected conditions was investigated next. Based on the premise that 2b could interact with catalase directly and that catalase could also interact with αMMC (Figures 4 and 5), whether αMMC competitively interferes with the interaction between catalase and 2b was studied. As shown in Figure 6a, the correlation between 2b‐GST and CAT1‐His weakened with an increasing αMMC‐MBP protein content, while the interaction intensity between CAT1 and αMMC gradually strengthened (Figure 6a). Similarly, αMMC‐MBP also impaired the interaction between 2b‐GST and CAT2‐His in vitro (Figure 6b). A BiFC analysis was used to determine whether αMMC weakens the interactions between catalases and 2b in vivo. As shown in Figure 6c, when CAT1‐nYFP + 2b‐cYFP and CAT2‐nYFP + 2b‐cYFP were infiltrated into N. benthamiana leaves, strong fluorescence signals were detected, which corroborates the earlier descriptive results (Figure 6c). When αMMC‐FLAG was co‐expressed with CAT1‐nYFP + 2b‐cYFP, fluorescence signals were clearly reduced, whereas co‐expression of the empty vector with CAT1‐nYFP + 2b‐cYFP did not decrease fluorescence intensity (Figure 6c,d). When the αMMC‐FLAG protein was co‐expressed with CAT2‐nYFP + 2b‐cYFP in leaves, a parallel result was obtained (Figure 6c,e). The expression of CAT1, CAT2, and 2b exhibited no obvious difference in these combinations (Figure 6f,g). In summary, all the above results demonstrate that αMMC interferes with the interactions between 2b and catalases by binding to catalases.
FIGURE 6.
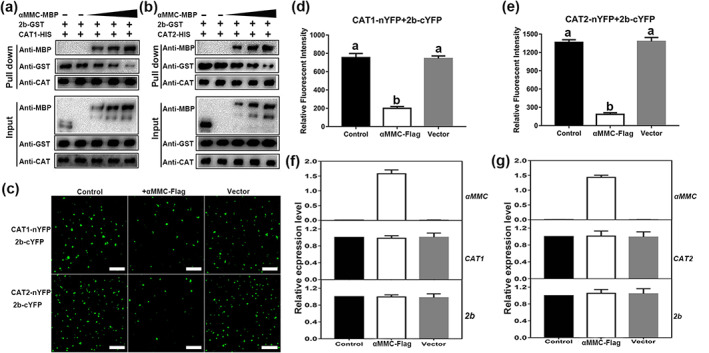
αMMC interferes with the interactions between catalases and 2b. (a, b)Competitive binding assays of 2b and αMMC to catalases in vitro. 2b‐GST and CAT‐His proteins were incubated with increasing concentrations of αMMC‐MBP protein (2.5, 5, or 7.5 μg). Input and pull‐down proteins were detected by western blotting with anti‐GST, ‐MBP, and ‐CAT antibodies. (c) Bimolecular fluorescence complementation assays showing that αMMC interfered with the interaction between 2b and catalases. CAT1‐nYFP or CAT2‐nYFP + 2b‐cYFP (control), αMMC‐FLAG + CAT1‐nYFP/CAT2‐nYFP + 2b‐cYFP (+ αMMC‐FLAG), and empty vector‐FLAG + CAT1‐nYFP/CAT2‐nYFP + 2b‐cYFP (+ vector), bar = 100 μm. (d, e) Quantitative analysis of YFP fluorescence intensity in (c). The fluorescence intensity was estimated by 50 separate fluorescent flecks. (f, g) Measured αMMC, CAT1‐nYFP (CAT1), CAT2‐nYFP (CAT2), and 2b‐cYFP (2b) expression levels in (c) by reverse transcription‐quantitative PCR, for which the results of the control treatment were set to 1. Data are presented as the means ± SD from three independent plants. Lowercase letters represent significant differences (p < 0.05)
Catalase activities were inhibited via 2b interacting with catalases, which promoted the accumulation of CMV. To further explore how αMMC affects catalase activity, we transiently expressed CAT1/CAT2‐FLAG or the empty vector control in N. benthamiana leaf tissues and co‐infiltrated 2b or 2b + αMMC into the same leaf tissues. As shown in Figure S6a, catalase activity in plants transiently expressing CAT1 + 2b was lower than that in plants transiently expressing CAT1. Meanwhile, the catalase activity in plants transiently expressing CAT1 + 2b + αMMC was higher than that in plants transiently expressing CAT1 + 2b but lower than that in plants expressing only CAT1. Similarly, the catalase activity in leaves transiently expressing CAT2 was ranked from low to high as follows: CAT2 + 2b‐expressing plants < CAT2 + 2b + αMMC‐expressing plants < CAT2‐expressing plants (Figure S6b). Consistent with the above results, catalase activity in αMMC‐overexpressing plants was higher than that in WT plants under CMV infection. (Figure 4d). To sum up, these results demonstrate that the αMMC–catalase interaction was essential for maintaining catalase activity in transgenic tomato plants responding to CMV infection.
2.7. αMMC‐induced catalase activity alleviated photosystem damage under CMV infection
To further examine how catalase activity influences αMMC‐mediated CMV resistance, we measured the functioning of photosystem II (PS II), which could be indicated by Fv/Fm (the maximal quantum efficiency of PS II) and ΦPS II (quantum efficiency of PS II), in the CAT‐silenced WT and αMMC #1 plants. High values for Fv/Fm were recorded in all these plants, with no significant differences in Fv/Fm detected between CAT1‐, CAT2‐silenced plants, and TRV control plants under mock‐inoculated conditions (Figure 7a,b). By contrast, CMV infection caused a decline in the Fv/Fm of tomato, and CAT1‐/CAT2‐silenced plants featured significantly lower Fv/Fm values than those in TRV plants. Importantly, compared with TRV‐WT plants, the leaves of TRV‐αMMC plants had a greater Fv/Fm, yet that value was similar between CAT‐silenced WT plants and CAT‐silenced αMMC plants under CMV infection. Furthermore, the Fv/Fm results agreed with the ΦPS II results. Under mock‐inoculated conditions, no differences were detected among any of the plant groups (Figure 7c,d). However, ΦPS II was higher in TRV‐αMMC plants than in TRV‐WT plants under CMV conditions, while the αMMC‐induced resistance to CMV infection, expressed as ΦPS II, was abolished if the genes encoding catalases were silenced. These results indicate that the αMMC‐induced resistance to CMV relies on catalase activity.
FIGURE 7.
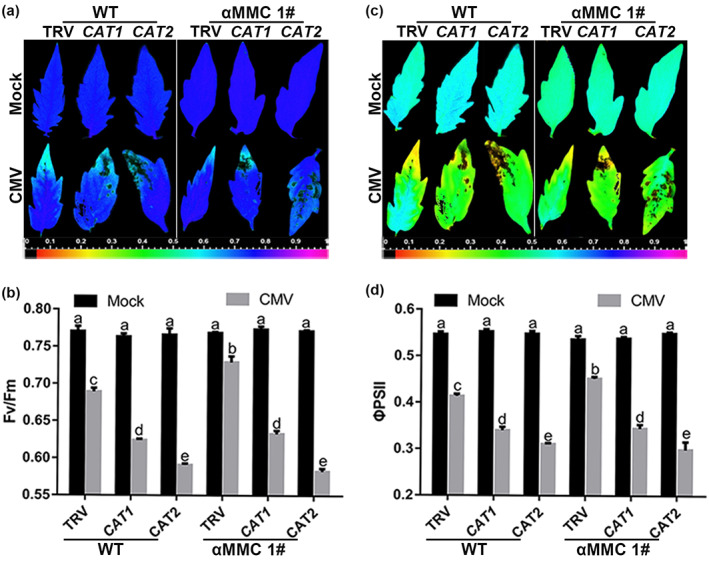
αMMC‐induced catalase activity alleviates photosystem damage under CMV infection conditions. (a, c) Images of the maximal quantum efficiency of PS II (Fv/Fm) (a) and the quantum efficiency of PS II (ΦPS II) (c) in CAT‐silenced wild‐type (WT) and αMMC transgenic plants under CMV infection conditions. (b, d) Average values of Fv/Fm (b) and ΦPS II (d). Systemic leaves were harvested for analysis. Values are the means ± SD from three independent plants. Lowercase letters indicate significant differences (p < 0.05)
2.8. CMV infection altered the expression levels of catalase and αMMC
Systemic leaves were used to estimate whether viral infection altered catalase and αMMC expression patterns in tomato at 10 dpi. The RT‐qPCR results showed that the CAT1 and CAT2 mRNAs were induced in CMV‐infected plants compared with mock‐infected plants (Figure 8a,b). Moreover, as shown in Figure 8c, the greatest increase in catalase protein occurred in WT plants, followed by the αMMC #8 transgenic line, and the lowest increase was in the αMMC #1 transgenic line. Notably, catalase expression was similar between WT plants and the two independent αMMC transgenic lines without CMV infection (Figure 8a–c). Western blotting and RT‐qPCR results suggested a higher level of αMMC expression in line #1 than line #8, which also exhibited similar trends under CMV‐inoculated conditions despite diminished expression of αMMC (Figures 8d and S7). Altogether, these results imply that catalase expression is induced in the plant response to CMV infection, which positively corresponds to virus accumulation in tomato, whereas αMMC expression is down‐regulated by CMV infection.
FIGURE 8.
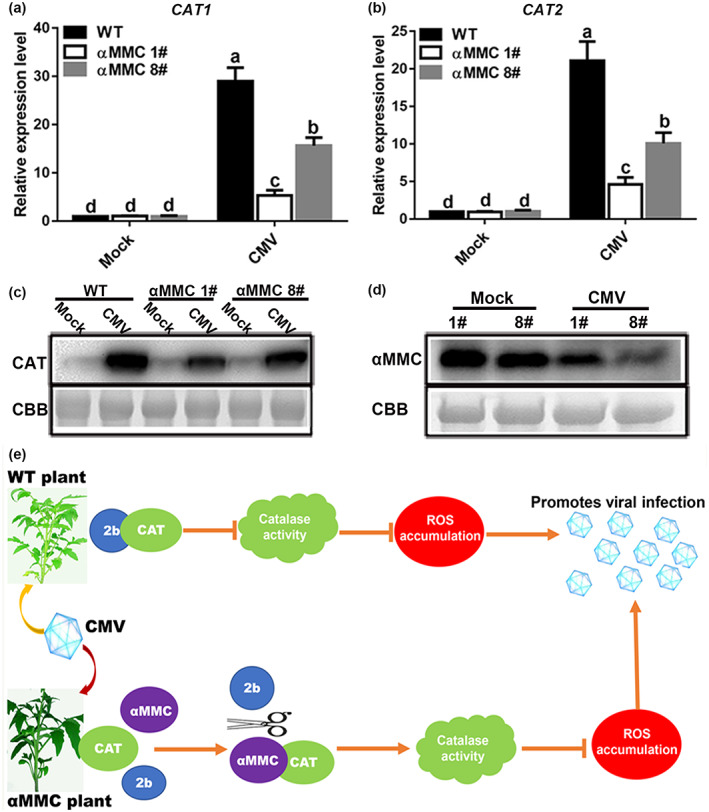
Functional analysis of catalases and αMMC in response to CMV infection in tomato leaves. (a) CAT1 and (b) CAT2 expression levels in wild‐type (WT) plants and αMMC transgenic plants were analysed by reverse transcription‐quantitative PCR at 10 days postinoculation (dpi). Systemic leaves were harvested for analysis. Values indicate the means ± SD from three independent experiments, and lowercase letters represent significant differences (p < 0.05). (c) Catalase protein accumulation in WT plants and αMMC transgenic plants was confirmed by western blotting assay at 10 dpi. (d) Detection of αMMC protein levels at 10 dpi in αMMC transgenic plants using western blotting. Coomassie brilliant blue (CBB) staining of RuBisCO protein indicates the loading control. (e) A working model for αMMC increasing tomato resistance against CMV infection. The model proposes that the CMV‐encoded protein 2b specifically interacts with catalases to repress their enzymatic activity, resulting in virus accumulation, while αMMC interferes with the interactions between 2b and catalases by binding to catalases to preserve catalase enzymatic activity, thus enhancing tomato defence to CMV infection
3. DISCUSSION
In the present work, we explored the mechanism by which αMMC plays an antiviral role during plant–CMV interactions. This study provides evidence for how plant viruses subvert catalase‐regulated antiviral defence to facilitate CMV accumulation in tomato. First, our results show that the transgenic αMMC lines displayed an antiviral phenotype. Second, 2b bound the host factor catalase and inhibited its activity. Third, αMMC contributed to reducing the inhibition of catalase activity mediated by 2b in plants, with catalase expression induced in plant immunity to reduce oxidative stress under CMV conditions. Fourth, the level of αMMC transcription was decreased by CMV infection. Lastly, when αMMC was overexpressed at higher levels in transgenic plants, the plants’ resistance to virus was augmented and their ROS production decreased, as happened in αMMC #1 line compared to #8 line plants under CMV infection. Hence, our work demonstrates that αMMC could lower the inhibition of catalase activity by interfering with the 2b–catalase interaction through its binding to catalase, which lessened toxic accumulation of ROS and enhanced host plant defence against CMV infections.
Viral infections have profound impacts on the growth and productivity of crops around the world. To survive amidst complex environmental stresses, plants rely on intricate defence networks mediated by a variety of host factors (Haxim et al., 2017; Yuan et al., 2017). While αMMC is known to be involved in host immunity responses to various pathogens in mammals (Chen et al., 2019), there are few studies of αMMC in the plant response to pathogens. Our previous study found that αMMC could reduce viral infection in M. charantia plants and that jasmonic acid signalling was involved in their resistance (Yang et al., 2016). In this study, it was further shown that overexpression of αMMC in tomato increased plant resistance to CMV infection, while WT plants were hypersensitive to viral infection, suggesting that αMMC also has a critical function in tomato's defence against CMV infection (Figure 2). Consistent with our research, Qian et al. (2014) showed that αMMC has antifungal activity in rice, and later Zhu et al. (2020) confirmed that overexpression of αMMC in tobacco improved that plant's resistance to TMV infection by mitigating oxidative damage. Moreover, the phenotypic analysis implied that αMMC negligibly impacts plant growth and development (Figures 2a and S2). Collectively, these studies demonstrate that αMMC confers a broad spectrum of plant resistance to disease and could be a promising resistance gene for use in agriculture.
Multiple biotic and abiotic stresses cause ROS bursts in plants, causing oxidative stress that leads to the induction of cell death (Gechev et al., 2005; Jaspers & Kangasjärvi, 2010). Here, our results demonstrated that WT plants suffered more severe oxidative damage than αMMC transgenic lines, because the redox homeostasis was disrupted under CMV conditions. This observation was consistent with the increased incidence of viral infection in WT plants compared with that in αMMC‐overexpressing plants (Figures 2 and 3). Accumulating empirical evidence suggests that autophagy can be activated by ROS, and autophagy, in turn, acts to reduce oxidative damage and increase cell survival in plants under stressful conditions (Gurusamy et al., 2009). Notably, autophagy is an integral part of the plant immune system, and its influence on cell death or cell survival may be on pathogens (Zhou et al., 2018). Mounting studies indicated that autophagy improves resistance and limits disease‐related apoptosis in plants infected with necrotrophic fungi (Lai et al., 2011). In the case of hemibiotrophic pathogens, autophagy is induced in pathogen‐infected plants and inhibits senescence and cell death caused by pathogens to facilitate the reproduction of pathogens and infection (Hafrén et al., 2017). Our results showed that CMV infection was capable of inducing autophagy, and the numbers of autophagosomes and autophagic bodies corresponded well with the ROS levels (Figures 3 and S3). Furthermore, the expression of ATG genes and the abundance of autophagosomes were increased in WT tomato plants in comparison with that in αMMC transgenic tomato plants (Figure S3). All these data suggest that CMV infection induces ROS bursts, which trigger autophagy to degrade the damaged cellular components, and autophagy is therefore repressed in αMMC transgenic plants compared with that in WT plants.
It is well known that antioxidant enzymes contribute to cellular redox homeostasis regulation by minimizing toxic ROS production (Mhamdi et al., 2010). In this study, CMV infection caused ROS accumulation and systemic necrosis symptoms, suggesting that CMV has a counterdefence mechanism to inhibit ROS degradation (Figures 2a and 3). An increasing number of studies have proven that, to facilitate their infection of plants, pathogens use a variety of strategies to evade and suppress host immunity (Jin et al., 2016). For example, the Ageratum leaf curl Sichuan virus‐encoded C4 protein interferes with the gibberellic acid signalling pathway to promote viral infection and symptom development in plants, and the AV2 protein interacts with catalase 2 in tobacco, which accelerates disease development (Li et al., 2022; Roshan et al., 2018). Although certain host factors are used by viruses, and their interaction determines disease phenotypes, the effect of CAT–virus interaction on catalase activity depends on the specific plant–pathogen system. Catalase activities were suppressed by the interactions between catalases and HCPro, a VSR of chilli venial mottle virus, to promote virus accumulation, whereas the triple gene block protein 1 interacted with tomato's CAT1 to increase enzymatic activity, which facilitated viral infection (Mathioudakis et al., 2013; Yang et al., 2020). Our results show that CMV 2b interacts with catalase in vitro and in vivo and inhibits their activities. Silencing of either CAT1 or CAT2 and overexpression of 2b enhanced CMV accumulation and resulted in severe disease symptoms in tomato (Figure 4). These experimental findings clearly suggested that catalase activity might contribute to plant immunity and that CMV 2b limited catalase activity by interacting with catalases to benefit replication, which was consistent with previous studies (Inaba et al., 2011).
Although the interaction of host factors with viral elements could accelerate viral infection, plant immunity also uses some mechanisms to protect the host against stress (Kørner et al., 2013). Recently, Liu et al. (2022) found that the Reduced Dormancy 5 in Arabidopsis thaliana enhanced resistance to CMV by promoting amplification of the virus‐derived small interfering RNAs. Similarly, phosphatidic acid phosphohydrolase 1 negatively regulated replication complex formation of viruses, leading to less of the pathogen accumulating in the host (Zhang et al., 2018). In our study, αMMC interacted only with catalases, but not 2b, to disturb the CAT–2b interaction (Figures 5 and 6). Silencing of CAT1 or CAT2 repressed αMMC‐induced tomato resistance to CMV infection and led to more severe viral symptoms in tomato (Figure 7). Furthermore, catalase activity was induced more strongly in αMMC transgenic lines than that in WT plants under CMV infection, and the catalase activity of plants transiently expressing CAT + 2b was lower than that of plants transiently expressing CAT + 2b + αMMC (Figures 4d and S6). These results suggest that αMMC could alleviate the inhibition of catalase activities by disrupting the interaction between 2b and catalases via binding with catalases, which increases plant tolerance to viral infection. However, molecular experiments showed that CAT expression patterns were activated under CMV inoculation and that the CAT gene expression level in plants was positively correlated with virus accumulation (Figures 2 and 8a–c). Considering the inhibitory effect of 2b on catalase activity, we speculated that plant immunity had to continuously activate the expression of catalase to enhance catalase enzyme activity and eliminate ROS. By measuring the expression level of αMMC, we discovered that CMV infection reduced the expression of αMMC at both the transcriptional and translational level (Figures 8d and S7). These results imply that the virus promotes its infection by inhibiting the expression of αMMC and that plant defence response induces catalase expression to increase its activity to avoid excessive oxidative damage.
In conclusion, this study elucidated the mechanism by which αMMC can foster tomato plant resistance against CMV infection. The possible αMMC‐mediated virus resistance is summarized in the model presented in Figure 8e. On the one hand, CMV 2b interacts with host factor catalases and inhibits their activities to facilitate viral infection. On the other hand, the plant immune mechanism increases catalase expression to reduce ROS accumulation, and αMMC can disrupt the interaction between 2b and catalase to improve plant resistance. Taken together, our work demonstrates that αMMC directly disrupts the interaction between catalase and 2b by competing with 2b for binding to catalases, thereby altering cellular redox homeostasis, reducing toxic ROS production, and augmenting resistance in plants against CMV infection.
4. EXPERIMENTAL PROCEDURES
4.1. Plant growth and virus inoculation
S. lycopersicum ‘Ailsa Craig’ seedlings were grown in a greenhouse at 25°C with a 12‐h light:12‐h dark photoperiod. Seedlings (2–3 weeks old) were selected for their infection with CMV and sodium phosphate buffer via mechanical inoculation, while a phosphate‐buffered saline inoculation served as the mock treatment (control).
4.2. Generation and screening of transgenic tomato
Using specific primers (Table S1), the αMMC gene was amplified from M. charantia cDNA and then ligated to the transformation vector pZP211 by digestion with SalI and BamHI. Transgenic seedlings were generated by Agrobacterium tumefaciens EHA105‐mediated transformation according to previously described methods (Xu et al., 2012), and the ensuing transformed lines were first screened on medium containing kanamycin (50 mg/L). Next, a PCR analysis was performed to detect the αMMC gene and western blot assay was used to test the protein of αMMC. Two independent homozygous lines with stable high expression of αMMC were selected in the study.
4.3. Yeast two‐hybrid analysis
The 2b and αMMC gene fragments were inserted into pGADT7 (Clontech). Meanwhile, the CAT1, CAT2, and αMMC fragments were inserted into pGBKT7 (Clontech). Pairs of plasmid constructs were transferred into yeast cells (AH109) for growth on double dropout (DDO) plates (SD−Leu/−Trp). Finally, yeast cells containing the co‐transformants were transferred from DDO onto quadruple dropout (QDO) plates (SD−Leu/−Trp/−His/−Ade) to determine the interaction between different proteins.
4.4. Detection of BiFC
The BiFC analysis was performed by following previously described methods (Zheng et al., 2019). The coding regions of CAT1, CAT2, and αMMC were inserted into pXY103‐nYFP. The coding regions of αMMC and 2b were inserted into pXY104‐cYFP. The ensuing constructs were used to transform A. tumefaciens; cells carrying the nYFP plasmid were mixed 1:1 with those containing the cYFP plasmid and co‐infiltrated into N. benthamiana leaves for 2 days. The YFP signal was determined by fluorescence microscopy.
4.5. Pull‐down assays
GST pull‐down assays were carried out as described previously (Zhang et al., 2017). To perform the competitive pull‐down assays, a reported methodology was used with slight modifications (Hou et al., 2010). Specifically, 2.5 μg of 2b‐GST protein was mixed with 2.5, 5, or 7.5 μg of αMMC‐maltose‐binding protein (MBP) protein and then combined with beads containing 2.5 μg of CAT1 or CAT2 at 4°C for 2 h using 300 μl of total protein binding buffer (150 mM NaCl, 20 mM Tris, 1 mM PMSF, 0.2% Triton X‐100, 1% protease inhibitor cocktail [pH 8.0]). The beads were washed four times. These proteins were eluted from beads by boiling at 95°C with 30 μl SDS‐PAGE loading buffer and then detected by immunoblotting using anti‐CAT, ‐GST, and ‐MBP antibodies.
4.6. Construction of the VIGS vector and TEM observation
A TRV‐based VIGS approach was used to silence the CAT genes in tomato. The coding‐region fragments of SlCAT1 and SlCAT2 were amplified by PCR, digested by restriction enzymes, and ligated into the linearized pTRV2 vector to generate pTRV2‐CAT1 and pTRV2‐CAT2, respectively (Table S1). The construct for silencing the phytoene desaturase (PDS) gene of tomato was used as positive control to check the method's robustness (Zhu et al., 2018). These plasmids were transferred into A. tumefaciens GV3101 and injected into leaves according to the previously described methodology for the VIGS assays (Zhu et al., 2018), after which tomato plants were infected with CMV at 10 dpi in the VIGS experiment. TEM observation was performed as described by Cao et al. (2015).
4.7. Protein induction and enzymatic activity assays in vitro
The 2b, CAT1, CAT2, and αMMC genes were inserted into the pGEX‐GST, pMAL‐C2X‐MBP, and pET28a‐6 × His vectors. The ensuing fragments were transformed into E. coli BL21 cells to induce protein expression and purification of the expressed protein. CAT activity was measured with CAT assay kit (Visible Light) according to standard procedures. First, 100‐μl samples containing CAT and other proteins were incubated at 37°C for 20 min. Then, these samples (or plant extracts) were fully mixed with a working buffer and allowed to react for 5 min at 37°C; reactions were terminated by adding a stop solution. Based on absorbance at 405 nm, catalase enzymatic activity could be calculated. Here, one unit of catalase activity is defined as the quantity of enzyme catalysing the decomposition of 1 μmol H2O2 per minute. The capacity of the catalase activity was calculated as U/mg = [235.65 × (A 0 − A 1)]/[5 × W], where A 0 and A 1 are the absorbance values of the control reaction and sample reaction, respectively, at 405 nm, and W is the amount of protein in the test sample (mg).
4.8. Total RNA extraction and RT‐qPCR
Total RNA in plants was extracted using the RNeasy Mini Kit (Qiagen) and this was reverse transcribed into cDNA by a M‐MLV reverse transcriptase kit (Invitrogen). Real‐time qPCR was then performed according to the manufacturer's instructions for the TransStart Tip Green qPCR Super Mix Kit (Transgene). The CFX Connect Real‐Time System (Bio‐Rad) was used to analyse the data from RT‐qPCR. Amplifications of the Actin gene in tomato and EF1α in tobacco served as internal references. Finally, relative expression levels of genes were calculated using the comparative 2−∆∆Ct method (Han et al., 2022). All primers used in this study are shown in Table S1.
4.9. Protein extraction and western blotting analysis
Leaves were ground and digested in Eppendorf tubes using 2 × sodium dodecyl sulphate (SDS) sample buffer, then boiled for 10 min and centrifuged at 13,000 × g for 10 min, and the ensuing supernatant retained. For the western blotting analysis, total protein was separated by SDS‐PAGE with the indicated antibodies (Zheng et al., 2019).
4.10. Oxidative damage estimation and chlorophyll fluorescence assay
The relative water content (RWC) of a leaf was defined as RWC (%) = (Fw − Dw)/(Tw − Dw) × 100, where Fw is the fresh leaf weight, Tw is the turgid leaf weight, and Dw is the dry leaf weight. The H2O2 and MDA contents were quantified using H2O2 and MDA kits (Nanjing) as per the manufacturer's protocol. Chlorophyll fluorescence was determined with an imaging pulse amplitude‐modulated fluorometer (IMAG‐MAXI; Heinz Walz), as described previously (Deng et al., 2016). The plants were exposed to darkness for 30 min before being tested.
4.11. Statistical analysis
Samples were analysed in triplicate, and the data presented as the mean ± SD. All statistical analyses were implemented in GraphPad Prism v. 7.0 software using two‐way analysis of variance. A difference at p < 0.05 was considered significant.
CONFLICT OF INTEREST
X.D.H. and Y.T. designed the experiments, Y.T. carried out most of the experiments, and P.Q.D. analysed the data. Y.T. wrote the manuscript, X.D.H. revised the manuscript, and L.H.H. provided some advice and discussion. The authors declare no conflicts of interest.
Supporting information
Figure S1 Identification of αMMC‐overexpressing transgenic tomato. (a) Detection of αMMC expression by reverse transcription (RT)‐PCR in T0 lines. (b) Western blot analysis of αMMC protein level by anti‐αMMC antibody in T0 lines. Coomassie brilliant blue (CBB) staining of RuBisCO protein indicates the loading control. (c) The αMMC gene was examined using PCR analysis in #1T2 lines. (d) The αMMC gene was examined using PCR in #8T2 lines
Figure S2 Phenotype of wild‐type and αMMC transgenic plants in the natural environment
Figure S3 CMV infection induces autophagy in wild‐type and αMMC transgenic plants. The transcription levels of (a) ATG3, (b) ATG5, (c) ATG8, and (d) ATG16 were measured in mock‐ and CMV‐infected tomato at 10 days postinoculation (dpi) by reverse transcription‐quantitative PCR assays. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05). (e) Representative transmission electron microscopy (TEM) images of autophagic structures in wild‐type plants and αMMC transgenic plants at 10 dpi. Autophagic bodies are indicated by red arrows. Bars: 1 μm. Systemic leaves were harvested for analysis
Figure S4 Sequence analysis for CAT1 and CAT2. (a) The nucleic acid sequences of the CAT1 and CAT2 genes of Solanum lycopersicum were compared with homologous genes of Arabidopsis thaliana and Nicotiana tabacum, respectively. (b) Amino acid sequence alignment was identified from A. thaliana, N. tabacum, and S. lycopersicum
Figure S5 Silencing efficiency of CAT1/CAT2 in the wild‐type and the αMMC‐overexpressing plants. (a) Phenotypes of TRV (negative control) and TRV‐PDS (positive control) in tomato and Nicotiana benthamiana. (b) Silencing efficiency of CAT1/CAT2 in the plants
Figure S6 CAT1 and CAT2 enzyme activity assays in empty vector (EV)‐, CAT‐, CAT + 2b‐, or CAT + 2b + αMMC‐overexpressing Nicotiana benthamiana leaves. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05)
Figure S7 Reverse‐transcription quantitative PCR analysis of αMMC expression levels at 10 days postinoculation in αMMC transgenic plants. Systemic leaves were harvested for analysis. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05)
Table S1 A list of primers for the experiments. The highlighted red regions indicate the restriction enzyme cutting site
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (32070167 and 31772131), Fundamental Research Funds for the Central Universities (2022SCUH0006), the International Cooperation Project of Chengdu Science and Technology Bureau (2020‐GH02‐00026‐HZ), and the Hubei Provincial Natural Science Foundation of China (ZRM S2022002103).
Yang, T. , Peng, Q. , Lin, H. & Xi, D. (2023) Alpha‐momorcharin preserves catalase activity to inhibit viral infection by disrupting the 2b–CAT interaction in Solanum lycopersicum . Molecular Plant Pathology, 24, 107–122. Available from: 10.1111/mpp.13279
[Corrections added on 21 November 2022, after first online publication: the sequence of funding projects has been updated in this version.]
DATA AVAILABILITY STATEMENT
The Plant Genome Initiative identifiers for the genes described in this article are as follows: αMMC (X57682), CAT1 (NM_001247898), CAT2 (NM_001247257), ATG3 (XM_004240713), ATG5 (MK189277), ATG8 (XM_015229992), ATG16 (XM_004236251), Actin (NM_001321306), coat protein (MK411768), 2b (MK411766), and NbEF1α (AY206004). The other data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ajji, P.K. , Walder, K. & Puri, M. (2016) Functional analysis of a type‐I ribosome inactivating protein balsamin from Momordica balsamina with anti‐microbial and DNase activity. Plant Foods for Human Nutrition, 71, 265–271. [DOI] [PubMed] [Google Scholar]
- Barbieri, L. , Balteli, M.G. & Stirpe, F. (1993) Ribosome inactivating proteins from plants. Biochimica et Biophysica Acta, 1154, 237–284. [DOI] [PubMed] [Google Scholar]
- Cao, X. , Jin, X. , Zhang, X. , Li, Y. , Wang, C. , Wang, X. et al. (2015) Morphogenesis of endoplasmic reticulum membrane‐invaginated vesicles during beet black scorch virus infection: role of auxiliary replication protein and new implications of three‐dimensional architecture. Journal of Virology, 89, 6184–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelikani, P. , Fita, I. & Loewen, P.C. (2004) Diversity of structures and properties among catalases. Cellular and Molecular Life Science, 61, 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.J. , Deng, X.G. , Peng, X.J. , Zhu, T. , Xi, D.H. & Lin, H.H. (2014) Turnip crinkle virus with nonviral gene cancels the effect of silencing suppressors of P19 and 2b in Arabidopsis thaliana . Physiological and Molecular Plant Pathology, 88, 94–100. [Google Scholar]
- Chen, Y.J. , Zhu, J.Q. , Fu, X.Q. , Su, T. , Li, T. , Guo, H. et al. (2019) Ribosome‐inactivating protein α‐momorcharin derived from edible plant Momordica charantia induces inflammatory responses by activating the NF‐kappaB and JNK pathways. Toxins, 11, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citores, L. , Iglesias, R. & Ferreras, J.M. (2021) Antiviral activity of ribosome‐inactivating proteins. Toxins, 13, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha, J.R. , Neto, M.C.L. , Carvalho, F.E. , Martins, M.O. , Jardim‐Messeder, D. , Margis‐Pinheiro, M. et al. (2016) Salinity and osmotic stress trigger different antioxidant responses related to cytosolic ascorbate peroxidase knockdown in rice roots. Environmental and Experimental Botany, 131, 58–67. [Google Scholar]
- Das, K. & Roychoudhury, A. (2014) Reactive oxygen species (ROS) and response of antioxidants as ROS‐scavengers during environmental stress in plants. Frontiers in Environment Science, 2, 53. [Google Scholar]
- Deng, X.G. , Zhu, T. , Zou, L.J. , Han, X.Y. , Zhou, X. , Xi, D.H. et al. (2016) Orchestration of hydrogen peroxide and nitric oxide in brassinosteroid‐mediated systemic virus resistance in Nicotiana benthamiana . The Plant Journal, 85, 478–493. [DOI] [PubMed] [Google Scholar]
- Di Carli, M. , Villani, M.E. , Bianco, L. , Lombardi, R. , Perrotta, G. , Benvenuto, E. et al. (2010) Proteomic analysis of the plant‐virus interaction in cucumber mosaic virus (CMV) resistant transgenic tomato. Journal of Proteome Research, 9, 5684–5697. [DOI] [PubMed] [Google Scholar]
- Díaz‐Vivancos, P. , Clemente‐Moreno, M.J. , Rubio, M. , Olmos, E. , García, J.A. , Martínez‐Gómez, P. et al. (2008) Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. Journal of Experimental Botany, 59, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. , Anderson, B.J. , Haase, H.R. & Symons, R.H. (1994) New overlapping gene encoded by the cucumber mosaic virus genome. Virology, 198, 593–601. [DOI] [PubMed] [Google Scholar]
- Fabbrini, M.S. , Katayama, M. , Nakase, I. & Vago, R. (2017) Plant ribosome‐inactivating proteins: progesses, challenges and biotechnological applications (and a few digressions). Toxins, 9, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev, T. , Minkov, I. & Hille, J. (2005) Hydrogen peroxide‐induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate‐dependent dioxygenase gene in the cell death process. IUBMB Life, 57, 181–188. [DOI] [PubMed] [Google Scholar]
- Gurusamy, N. , Lekli, I. , Gorbunov, N.V. , Gherghiceanu, M. , Popescu, L.M. & Das, D.K. (2009) Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG‐1 protein. Journal of Cellular and Molecular Medicine, 13, 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén, A. , Macia, J.L. , Love, A.J. , Milner, J.J. , Drucker, M. & Hofius, D. (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1‐mediated targeting of viral capsid protein and particles. Proceedings of the National Academy of Sciences of the United States of America, 114, E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H.Y. , Zou, J.L. , Zhou, J.Y. , Zeng, M.Y. , Zheng, D.C. , Yuan, X.F. et al. (2022) The small GTPase NtRHO1 negatively regulates tobacco defense response to tobacco mosaic virus by interacting with NtWRKY50. Journal of Experimental Botany, 73, 366–381. [DOI] [PubMed] [Google Scholar]
- Haxim, Y. , Ismayil, A. , Jia, Q. , Wang, Y. , Zheng, X.Y. , Chen, T.Y. et al. (2017) Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife, 6, e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, E. , Fung, N. , Liu, J. , Drakakaki, G. & Coaker, G. (2015) Beyond glycolysis: GAPDHs are multi‐functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genetics, 11, e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Lee, L.Y. , Xia, K. , Yan, Y. & Yu, H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental Cell, 19, 884–894. [DOI] [PubMed] [Google Scholar]
- Inaba, J.I. , Kim, B.M. , Shimura, H. & Masuta, C. (2011) Virus‐induced necrosis is a consequence of direct protein‐protein interaction between a viral RNA‐silencing suppressor and a host catalase. Plant Physiology, 156, 2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers, P. & Kangasjärvi, J. (2010) Reactive oxygen species in abiotic stress signaling. Physiologia Plantarum, 138, 405–413. [DOI] [PubMed] [Google Scholar]
- Jin, L. , Qin, Q. , Wang, Y. , Pu, Y. , Liu, L. , Wen, X. et al. (2016) Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathogens, 12, e1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner, C.J. , Klauser, D. , Niehl, A. , Domínguez‐Ferreras, A. , Chinchilla, D. , Boller, T. et al. (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Molecular Plant‐Microbe Interactions, 26, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Lai, Z. , Wang, F. , Zheng, Z. , Fan, B. & Chen, Z. (2011) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. The Plant Journal, 66, 953–968. [DOI] [PubMed] [Google Scholar]
- Li, P. , Guo, L. , Lang, X. , Li, M. , Wu, G. , Wu, R. et al. (2022) Geminivirus C4 proteins inhibit GA signaling via prevention of NbGAI degradation, to promote viral infection and symptom development in N. benthamiana . PLoS Pathogens, 18, e1010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Chen, M. , Li, R. , Li, W.X. , Gal‐On, A. , Jia, Z.Y. et al. (2022) Identification of positive and negative regulators of antiviral RNA interference in Arabidopsis thaliana . Nature Communications, 13, 2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioudakis, M.M. , Veiga, R.S.L. , Canto, T. , Vicente, M. , Dimitris, M. et al. (2013) Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Molecular Plant Pathology, 14, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi, A. , Queval, G. , Chaouch, S. , Vanderauwera, S. , Van Breusegem, F. & Noctor, G. (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress‐mimic models. Journal of Experimental Botany, 61, 4197–4220. [DOI] [PubMed] [Google Scholar]
- Minibayeva, F. , Dmitrieva, S. , Ponomareva, A. & Ryabovol, V. (2012) Oxidative stress‐induced autophagy in plants: the role of mitochondria. Plant Physiology and Biochemistry, 59, 11–19. [DOI] [PubMed] [Google Scholar]
- Mishra, V. , Mishra, R. & Shamra, R.S. (2022) Ribosome inactivating proteins‐an unfathomed biomolecule for developing multi‐stress tolerant transgenic plants. International Journal of Biological Macromolecules, 210, 107–122. [DOI] [PubMed] [Google Scholar]
- Pérez‐Pérez, M.E. , Lemaire, S.D. & Crespo, J.L. (2012) Reactive oxygen species and autophagy in plants and algae. Plant Physiology, 160, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, M. , Kaur, I. , Kanwar, R.K. , Gupta, R.C. , Chauhan, A. & Kanwar, J.R. (2009) Ribosome inactivating proteins (RIPs) from Momordica charantia for antiviral therapy. Current Molecular Medicine, 9, 1080–1094. [DOI] [PubMed] [Google Scholar]
- Qian, Q. , Huang, L. , Yi, R. , Wang, S.Z. & Ding, Y. (2014) Enhanced resistance to blast fungus in rice (Oryza sativa L.) by expressing the ribosome‐inactivating protein alpha‐momorcharin. Plant Science, 217, 1–7. [DOI] [PubMed] [Google Scholar]
- Roshan, P. , Kulshreshtha, A. , Kumar, S. , Purohit, R. & Hallan, V. (2018) AV2 protein of tomato leaf curl Palampur virus promotes systemic necrosis in Nicotiana benthamiana and interacts with host Catalase2. Scientific Reports, 8, 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz‐Shouval, R. & Elazar, Z. (2007) ROS, mitochondria and the regulation of autophagy. Trends in Cell Biology, 9, 422–427. [DOI] [PubMed] [Google Scholar]
- Scholthof, K.B. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. et al. (2011) Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology, 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, B.J. , Miller, J. , Symons, R.H. & Palukaitis, P. (2003) The 2b protein of Cucumoviruses has a role in promoting the cell‐to‐cell movement of pseudo recombinant viruses. Molecular Plant‐Microbe Interactions, 16, 261–267. [DOI] [PubMed] [Google Scholar]
- Sur, S. & Ray, R.B. (2020) Bitter melon (Momordica charantia), a nutraceutical approach for cancer prevention and therapy. Cancers, 12, 2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Yuan, S. , Zhang, D.W. , Lv, X. & Lin, H.H. (2012) The role of alternative oxidase in tomato fruit ripening and its regulatory interaction with ethylene. Journal of Experimental Botany, 63, 5705–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Zhang, Y.L. , Xie, X.L. , Yue, N. , Li, J.J. , Wang, X.B. et al. (2018) Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7–ATG8 interaction. The Plant Cell, 30, 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T. , Meng, Y. , Chen, L.J. , Lin, H.H. & Xi, D.H. (2016) The roles of alpha‐momorcharin and jasmonic acid in modulating the response of Momordica charantia to cucumber mosaic virus. Frontiers in Microbiology, 7, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T. , Qiu, L. , Huang, W. , Xu, Q. , Zou, J. , Peng, Q. et al. (2020) Chilli veinal mottle virus HCPro interacts with catalase to facilitate virus infection in Nicotiana tabacum . Journal of Experimental Botany, 71, 5656–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. & Guo, Y. (2018) Elucidating the molecular mechanisms mediating plant salt‐stress responses. New Phytologist, 217, 523–539. [DOI] [PubMed] [Google Scholar]
- Yoshioka, H. (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans . The Plant Cell, 15, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H.M. , Liu, W.C. & Lu, Y.T. (2017) CATALASE2 coordinates SA‐mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host and Microbe, 21, 143–155. [DOI] [PubMed] [Google Scholar]
- Zhang, K. , Zhang, Y. , Yang, M. , Liu, S. , Li, Z.G. , Wang, X.B. et al. (2017) The Barley stripe mosaic virus γb protein promotes chloroplast‐targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathogens, 13, e1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Li, Q. , Liu, T. , Liu, L. , Shen, D. , Zhu, Y. et al. (2015) Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiology, 167, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , He, G. , Han, G.S. , Zhang, J. , Catanzaro, N. , Diaz, A. et al. (2018) Host pah1p phosphatidate phosphatase limits viral replication by regulating phospholipid synthesis. PLoS Pathogens, 14, e1006988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, T. , Tan, W. , Yang, H. , Zhang, L. , Li, T. , Liu, B. et al. (2019) Regulation of anthocyanin accumulation via MYB75/HAT1/TPL‐mediated transcriptional repression. PLoS Genetics, 15, e1007993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S.M. , Hong, Q. , Lia, Y. , Lia, Q. & Wang, M. (2018) Autophagy contributes to regulate the ROS levels and PCD progress in TMV infected tomatoes. Plant Science, 269, 12–19. [DOI] [PubMed] [Google Scholar]
- Zhu, F. , Zhang, P. , Meng, Y.F. , Xu, F. , Zhang, D.W. , Cheng, J. et al. (2013) Alpha‐momorcharin, a RIP produced by bitter melon, enhances defense response in tobacco plants against diverse plant viruses and shows antifungal activity in vitro. Planta, 237, 77–88. [DOI] [PubMed] [Google Scholar]
- Zhu, F. , Zhu, P.X. , Xu, F. , Che, Y.P. , Ma, Y.M. & Ji, Z.L. (2020) Alpha‐momorcharin enhances Nicotiana benthamiana resistance to tobacco mosaic virus infection through modulation of reactive oxygen species. Molecular Plant Pathology, 21, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T. , Zou, L.J. , Li, Y. , Yao, X.H. , Xu, F. , Deng, X.G. et al. (2018) Mitochondrial alternative oxidase‐dependent autophagy involved in ethylene‐mediated drought tolerance in Solanum lycopersicum . Plant Biotechnology Journal, 16, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X. , Chung, K.P. , Cui, Y. , Lin, W. , Gao, C. , Kang, B.H. et al. (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 114, E426–E435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Identification of αMMC‐overexpressing transgenic tomato. (a) Detection of αMMC expression by reverse transcription (RT)‐PCR in T0 lines. (b) Western blot analysis of αMMC protein level by anti‐αMMC antibody in T0 lines. Coomassie brilliant blue (CBB) staining of RuBisCO protein indicates the loading control. (c) The αMMC gene was examined using PCR analysis in #1T2 lines. (d) The αMMC gene was examined using PCR in #8T2 lines
Figure S2 Phenotype of wild‐type and αMMC transgenic plants in the natural environment
Figure S3 CMV infection induces autophagy in wild‐type and αMMC transgenic plants. The transcription levels of (a) ATG3, (b) ATG5, (c) ATG8, and (d) ATG16 were measured in mock‐ and CMV‐infected tomato at 10 days postinoculation (dpi) by reverse transcription‐quantitative PCR assays. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05). (e) Representative transmission electron microscopy (TEM) images of autophagic structures in wild‐type plants and αMMC transgenic plants at 10 dpi. Autophagic bodies are indicated by red arrows. Bars: 1 μm. Systemic leaves were harvested for analysis
Figure S4 Sequence analysis for CAT1 and CAT2. (a) The nucleic acid sequences of the CAT1 and CAT2 genes of Solanum lycopersicum were compared with homologous genes of Arabidopsis thaliana and Nicotiana tabacum, respectively. (b) Amino acid sequence alignment was identified from A. thaliana, N. tabacum, and S. lycopersicum
Figure S5 Silencing efficiency of CAT1/CAT2 in the wild‐type and the αMMC‐overexpressing plants. (a) Phenotypes of TRV (negative control) and TRV‐PDS (positive control) in tomato and Nicotiana benthamiana. (b) Silencing efficiency of CAT1/CAT2 in the plants
Figure S6 CAT1 and CAT2 enzyme activity assays in empty vector (EV)‐, CAT‐, CAT + 2b‐, or CAT + 2b + αMMC‐overexpressing Nicotiana benthamiana leaves. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05)
Figure S7 Reverse‐transcription quantitative PCR analysis of αMMC expression levels at 10 days postinoculation in αMMC transgenic plants. Systemic leaves were harvested for analysis. Values indicate the means ± SD from three independent experiments and lowercase letters represent significant differences (p < 0.05)
Table S1 A list of primers for the experiments. The highlighted red regions indicate the restriction enzyme cutting site
Data Availability Statement
The Plant Genome Initiative identifiers for the genes described in this article are as follows: αMMC (X57682), CAT1 (NM_001247898), CAT2 (NM_001247257), ATG3 (XM_004240713), ATG5 (MK189277), ATG8 (XM_015229992), ATG16 (XM_004236251), Actin (NM_001321306), coat protein (MK411768), 2b (MK411766), and NbEF1α (AY206004). The other data that support the findings of this study are available from the corresponding author upon reasonable request.


