Abstract
Resveratrol is a phytoalexin that is synthesized by stilbene synthase (STS). Resveratrol in the human diet is known to have beneficial effects on health. We previously identified six novel STS (VqNSTS) transcripts from the transcriptome data of Vitis quinquangularis accession Danfeng‐2. However, the functions of and defensive mechanisms triggered by these VqNSTS transcripts remain unknown. In the present study, we demonstrate that the expression of five of these six novel members, VqNSTS2–VqNSTS6, can be induced by the powdery mildew‐causing fungus Uncinula necator. Additionally, overexpression of VqNSTS4 in the V. vinifera susceptible cultivar Thompson Seedless promoted accumulation of stilbenes and enhanced resistance to U. necator by activating salicylic acid (SA) signalling. Furthermore, our results indicate that the Alfin‐like (AL) transcription factor VqAL4 can directly bind to the G‐rich element (CACCTC) in the VqNSTS4 promoter and activate gene expression. Moreover, overexpression of VqAL4 in Thompson Seedless enhanced resistance to U. necator by promoting stilbene accumulation and activating SA signalling. Conversely, RNA interference‐mediated silencing of VqNSTS4 and VqAL4 resulted in increased susceptibility to U. necator. Collectively, our results reveal that VqNSTS4, regulated by VqAL4, enhances grapevine resistance to powdery mildew by activating SA signalling. Our findings may be useful to improve disease resistance in perennial fruit trees.
Keywords: Chinese wild Vitis quinquangularis , disease resistance, stilbene, VqAL4 transcription factor, VqNSTS
This study demonstrated that the Alfin‐like transcription factor VqAL4 enhances grapevine resistance to powdery mildew by promoting stilbene accumulation and activating salicylic acid signalling.
1. INTRODUCTION
Grapevine (Vitis spp.) is a very important fruit crop of temperate climates that is consumed as table grapes, raisins, grape juice, and wine. The European grapevine, Vitis vinifera, is the predominant species in commercial production because of its superior aroma and flavour characteristics. However, most V. vinifera cultivars are highly susceptible to powdery mildew caused by the ascomycete fungus Uncinula necator, which is an obligate biotrophic fungus (Gadoury et al., 2012; Qiu et al., 2015). Classical disease management methods incur both financial and environmental expenses and can directly lead to the development of fungicide resistance in pathogen populations (Jones et al., 2014; Kunova et al., 2021). Therefore, the focus of current research has shifted to development of disease resistance in the grapes themselves.
The stilbene phytoalexin was discovered from V. vinifera while exploring the natural mechanisms of disease resistance in grapes (Langcake & Pryce, 1977). Resveratrol is the basic unit of stilbene and is a product of the phenylpropanoid pathway that not only exhibits broad‐spectrum resistance to a range of pathogens, including U. necator, Plasmopara viticola, Botrytis cinerea, and Neofusicoccum parvum (Hain et al., 1993; Khattab et al., 2021; Pezet et al., 2004b; Xu et al., 2019), but also exhibits important pharmacological properties relevant to human health, exerting anticancer (Jang et al., 1997) and cardioprotective effects (Barger et al., 2008).
Stilbene synthase (STS) is a key enzyme in the resveratrol synthesis pathway and is encoded by a multigenic family. So far, 48 VvSTS genes have been identified in V. vinifera, of which 32 have potential functions (Parage et al., 2012; Vannozzi et al., 2012). There is considerable interest in the characterization of STS genes as part of a strategy to increase stilbene levels and enhance disease resistance in plants. For example, overexpression of two STS genes from grapevine in tobacco (Nicotiana tabacum) promoted resveratrol accumulation and enhanced resistance to B. cinerea (Hain et al., 1993). The function of the STS genes has also been analysed in tomato (Solanum lycopersicum), rice (Oryza sativa), papaya (Carica papaya), barley (Hordeum vulgare), and wheat (Triticum spp.), and it was shown to improve the plant resveratrol content or resistance to pathogens (Leckband & Lörz, 1998; Stark‐Lorenzen et al., 1997; Thomzik et al., 1997; Zhu et al., 2004).
A detailed study of the transcriptional regulation of STS genes could be important to further enhance disease resistance and the stilbene content of various grape cultivars. Various transcription factors participating in the regulation of STS genes in grapevine have been reported, including MYB, WRKY, ERF, and bZIP transcription factors (Höll et al., 2013; Jiang et al., 2021; Mu et al., 2022; Wang et al., 2019, 2020; Wang & Wang, 2019; Yin et al., 2022). However, it remains unclear whether other transcription factors are involved in the regulation of STS gene expression.
Plant homeodomain (PHD), a type of epigenetic modification‐associated finger protein containing eight conserved metal‐binding residues, Cys4HisCys3, was first reported in Arabidopsis thaliana and parsley (Petroselinum crispum) to participate in plant disease resistance by binding to the promoter of the pr2 gene (Kaadige & Ayer, 2006; Korfhage et al., 1994). The Alfin‐like (AL) protein belongs to the PHD finger family, and was originally identified as a transcription factor family in alfalfa (Medicago sativa) (Bastola et al., 1998). The AL protein family is mainly involved in root development and in responses to abiotic stress, such as salt stress, drought, and low temperatures (Chandrika et al., 2013; Wei et al., 2015; Zhu et al., 2021). There are reports on the functions of AL transcription factors in biotic stress responses. For instance, 10 BrAL genes were found to participate in the response of Brassica rapa to Fusarium oxysporum f. sp. conglutinans infection, and two BoAL genes showed significant expression in Brassica oleracea after inoculation with Pectobacterium carotovorum subsp. carotovorum (Kayum et al., 2015, 2016). However, studies of the grape AL transcription factor have not been conducted.
China is one of the centres of origin of wild grapevines, serving as an exciting resource providing critical disease resistance genes for resistance breeding of grapevine (Wang et al., 1995, 1998). In total, 61 VpSTS genes have been isolated from Chinese wild Vitis pseudoreticulata accession Baihe‐35‐1 (Cao, 2012), and a large number of studies have proved that overexpression of VpSTS genes can enhance resistance to powdery mildew (Dai et al., 2015; Jiao et al., 2016; Wang, 2004; Xu, 2010). In particular, VpSTS29/STS2 overexpression in V. vinifera and A. thaliana significantly increased resistance to powdery mildew (Xu et al., 2019). Another important Chinese wild resource is Vitis quinquangularis accession Danfeng‐2, which has not only high resistance to powdery mildew, but also a high content of stilbenes (Duan, 2002; Shi et al., 2014; Wan et al., 2007). Many VqSTS genes isolated from Danfeng‐2 have been shown to significantly enhance resistance to powdery mildew in the V. vinifera susceptible cultivar Thompson Seedless (Ding et al., 2021; Liu et al., 2019a; Wu et al., 2020; Zhao et al., 2020). For example, VqSTS6, whose expression is highest in ripe fruit, significantly increased the stilbene content and resistance to powdery mildew when overexpressed in V. vinifera, with trans‐piceid contents 16 times higher than in the wild type (Cheng et al., 2016; Liu et al., 2019a).
So far, 41 VqSTS genes have been isolated from Danfeng‐2 using the rapid amplification of cDNA ends (RACE) method (Shi et al., 2014), but it is not clear whether there remain any undiscovered VqSTS genes in Danfeng‐2. To further study the properties of high stilbene content and high resistance to powdery mildew, multiomics analysis of Danfeng‐2 and V. vinifera ‘Cabernet Sauvignon’ was carried out. Interestingly, analysis of Danfeng‐2 transcriptome data (PRJNA306731) revealed many novel transcripts that could not be mapped on the grapevine reference genome (PN40024), including six novel STS transcripts (Li, 2019; Li et al., 2017). However, whether these newly discovered VqNSTS genes contribute to the high levels of resveratrol and the high resistance to U. necator in Danfeng‐2 is still unclear. Furthermore, whether VqNSTS genes have novel regulatory mechanisms is a question worthy of further study. The purpose of this study is to investigate how the VqNSTS genes contribute to resistance to powdery mildew and to reveal the regulatory mechanisms triggered by these genes that control stilbene biosynthesis.
2. RESULTS
2.1. Isolation and characterization of six newly discovered STS genes from V. quinquangularis
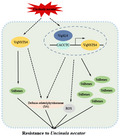
To further explore the relationship between high levels of stilbene and high resistance to powdery mildew in Danfeng‐2, we conducted multiomics analysis on Danfeng‐2 (Li, 2019). Transcriptome data for Danfeng‐2 that were not mapped to a reference genome were assembled, and 112,266 unigenes were generated. Novel transcripts were grouped into 11 clusters. The numbers of transcripts for clusters 1 to 11 were 311, 322, 76, 53, 205, 380, 324, 460, 385, 328, and 6600, respectively (Figure 1a). Six STS transcripts were isolated from cluster 11 with the locus names c50176.graph_c0, c91313.graph_c0, c76190.graph_c0, c56362.graph_c0, c54055.graph_c0, and c11357.graph_c0 (Figure 1b). These newly discovered STS transcripts were cloned from Danfeng‐2 (designated VqNSTS1–VqNSTS6; GenBank: OL589476–OL589481) and they were predicted to be located on chromosome 16 (Table S1).
FIGURE 1.
Identification and expression analysis of novel stilbene synthase transcripts in Vitis quinquangularis accession Danfeng‐2. (a) Expression analysis of novel transcripts in V. quinquangularis accession Danfeng‐2. Novel transcripts are grouped into 11 clusters. The numbers of transcripts from clusters 1 to 11 were 311, 322, 76, 53, 205, 380, 324, 460, 385, 328, and 6600, respectively. Expression of transcripts in cluster 11 was the highest at the ripe stage. (b) Analysis of the expression profiles of novel stilbene synthase transcripts in Danfeng‐2. Six stilbene synthase transcripts were isolated from cluster 11, with locus names c50176.graph_c0, c91313.graph_c0, c76190.graph_c0, c56362.graph_c0, c54055.graph_c0, and c11357.graph_c0. The four developmental stages of Danfeng‐2 berries were represented by DF_GH (green hard stage, 25 days after blooming), DF_BV (before véraison stage, 40 days after blooming), DF_V (véraison stage, 50 days after blooming), and DF_R (ripe stage, 80 days after blooming). (c) Multiple sequence alignment of VqNSTS1–6 amino acid sequences. The red rectangles represent the two conserved motifs. VqNSTS1 is a pseudogene, and VqNSTS5/6 contain mutations in the conserved motif. (d) The phylogenetic tree of VqNSTS1–6 with 41 reported VqSTSs and 48 reported VvSTSs. The 41 reported VqSTSs are from Danfeng‐2. All 48 reported VvSTSs are from V. vinifera ‘Pinot Noir’. They were divided into six subgroups: VqNSTS1, VqNSTS3, VqNSTS5, and VqNSTS6 belong to subgroup II; VqNSTS2 belongs to subgroup V; and VqNSTS4 belongs to subgroup I. (e–i) Reverse transcription‐quantitative PCR (RT‐qPCR) analysis was conducted to determine the relative transcript levels of VqNSTS2–6 after inoculation with Uncinula necator. VqNSTS2/4 showed high expression after U. necator inoculation, especially VqNSTS4. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student's t test). (j–n) RT‐qPCR analysis was conducted to determine the relative transcript levels of VqNSTS2–6 after treatment with 100 μM abscisic acid (ABA), ethylene (Eth), salicylic acid (SA), or methyl jasmonate (MeJA). VqNSTS4 responded significantly to SA treatment. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey's test) was carried out, with asterisks indicating significant differences at *p < 0.05, **p < 0.01.
VqNSTS1 is a 619‐bp pseudogene; VqNSTS4 is a 1035‐bp truncated STS that encodes 345 amino acids, and VqNSTS2, 3, 5, and 6 are all 1179 bp and encoded 393 amino acids (Table S1). Interestingly, two conserved motifs of the STS family, IPNSAGAIAGN and GVLFGFGPGLT, were observed in the amino acid sequences of VqNSTS2–6, but not of VqNSTS1 (Figure 1c). Additionally, the phylogenetic tree of VqNSTSs, with 41 reported VqSTSs and 48 reported VvSTSs, showed that VqNSTS1, VqNSTS3, VqNSTS5, and VqNSTS6 belong to subgroup II, VqNSTS2 belongs to subgroup V, and VqNSTS4 belongs to subgroup I (Figure 1d). Moreover, several sequence alignments of VqNSTSs and reported VqSTSs were carried out to confirm that these VqNSTSs were different from the previously reported VqSTSs. The results showed that although VqNSTSs and the reported VqSTSs were highly conserved, there were amino acid deletions and mutations (Figures [Link], [Link], [Link], [Link], [Link]). For example, the sequence similarity of VqNSTS2 and VqSTS37 was 99.49%, with one amino acid mutation, arginine (R)‐391 to threonine (T) (Figure S1); the sequence similarity of VqNSTS3 and VqSTS33 was 99.24%, with two amino acid mutations, serine (S)‐3 to leucine (L) and serine (S)‐276 to phenylalanine (F) (Figure S2); the sequence similarity of VqNSTS4 and VqSTS1 was 87.53%, with 48 amino acid deletions in the N‐terminus (Figure S3); the sequence similarity of VqNSTS5 and VqSTS33 was 91.35%, with 33 amino acid mutations (Figure S4); and the sequence similarity of VqNSTS6 and VqSTS33 was 93.89%, with 23 amino acid mutations (Figure S5).
The above results demonstrate that VqNSTS1 is a pseudogene caused by the deletion of key motifs; although VqNSTS2–6 and the reported VqSTSs have high homology, VqNSTS2–6 are clearly different from the reported VqSTSs based on their sequence alignments, which suggests that VqNSTS genes are new members of the STS gene family in Danfeng‐2.
2.2. VqNSTS genes respond to U. necator and phytohormones
To determine if the expression of these VqNSTS genes could be induced by U. necator, their expression was determined using reverse transcription‐quantitative PCR (RT‐qPCR) in Danfeng‐2 (Figure 1e–i). Here, VqNSTS1 was not selected for further analysis as it was determined to be a pseudogene, which cannot be translated into a functional protein. The results showed that VqNSTS2 responded most rapidly to U. necator, reaching a maximum at 1 day postinoculation (dpi), 5.3‐fold higher than the control (Figure 1e). In particular, VqNSTS4 showed very high expression levels after U. necator inoculation, with the highest expression at 2 dpi, 8.3‐fold higher than the control (Figure 1g). These results suggest that VqNSTS2 and VqNSTS4 may be involved in the defence response to powdery mildew.
Moreover, many studies have shown that the expression of STS genes can be induced by various phytohormones (Jiao et al., 2016; Ma, 2018; Wu et al., 2020). Therefore, RT‐qPCR analysis was also conducted to investigate the expression patterns of VqNSTS genes after different phytohormone treatments. The results demonstrated that VqNSTS2 was responsive to four phytohormones, particularly ethylene (Eth) and salicylic acid (SA); its transcript levels peaked at 3 h posttreatment (hpt) and 24 hpt, respectively, and its expression was 71.0‐fold and 2.3‐fold higher than the control, respectively (Figure 1j). Upon Eth treatment, VqNSTS3 showed the highest transcript level at 3 hpt, 54.5‐fold higher than the control (Figure 1k). VqNSTS4 responded significantly to SA treatment, with its expression level increasing gradually after 0.5 hpt and reaching a maximum at 48 hpt, 45.5‐fold higher than that of the control (Figure 1l). Upon methyl jasmonate (MeJA) treatment, the VqNSTS5 transcript level increased significantly and peaked at 1 hpt, being 4.9‐fold higher than that of the control (Figure 1m). VqNSTS6 was inducible by Eth and by SA, peaking at 3 hpt and 1 hpt, respectively, being 3.9‐fold and 5.0‐fold higher than in the control (Figure 1n). These results confirmed that expression of VqNSTS2–VqNSTS6 could be induced by U. necator and by phytohormones, with VqNSTS2 and VqNSTS4 being especially inducible.
2.3. VqNSTS4 increases the accumulation of stilbenes and enhances grapevine resistance to U. necator
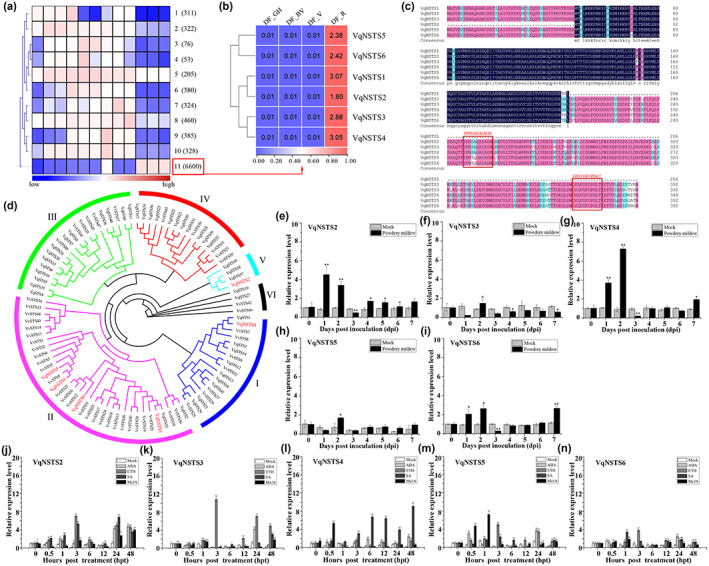
Because VqNSTS4 can be significantly induced by U. necator or SA (Figure 1g,l) despite being truncated, we speculated that it might play an important role in plant resistance to powdery mildew. To test this hypothesis, VqNSTS4 was stably expressed in V. vinifera ‘Thompson Seedless’ through Agrobacterium‐mediated transformation (Figure S6a,b). A total of 56 kanamycin‐resistant grapevine plantlets were obtained and analysed by PCR with specific primers (35S‐GFP‐F/R), of which seven were identified as transgenic grapevines (12.5%) by western blot (Figure S6c,d). Two transgenic lines—OEVqNSTS4‐L1 and OEVqNSTS4‐L2, both with high VqNSTS4‐GFP expression—were selected for further study (Figure S6e). High‐performance liquid chromatography (HPLC) analysis showed that the content of stilbenes was significantly increased in both OEVqNSTS4‐L1 and OEVqNSTS4‐L2, particularly of trans‐piceid, which is the main storage form of stilbene in grapevine (Chong et al., 2009). The contents of trans‐piceid in OEVqNSTS4‐L1 and OEVqNSTS4‐L2 were 2.4‐fold and 1.9‐fold higher than those in the wild‐type Thompson Seedless, respectively (Figure S6f; Table 1). These results suggest that VqNSTS4 can catalyse the synthesis of stilbenes, despite the deletion of 48 amino acids at its N‐terminus.
TABLE 1.
Contents of stilbenes of VqNSTS4 transgenic Vitis vinifera 'Thompson Seedless' lines under natural conditions and 7 days after artificial inoculation with Uncinula necator (PM‐7d)
| Line | trans‐Piceid (μg/g) | trans‐Resveratrol (μg/g) | ε‐Viniferin (μg/g) | Pterostilbene (μg/g) | trans‐Piceatannol (μg/g) |
|---|---|---|---|---|---|
| Wild type (WT) | 125.69 ± 13.23 | – | – | 84.33 ± 9.07 | 17.74 ± 2.33 |
| OEVqNSTS4‐L1 | 305.01 ± 25.83 | 85.58 ± 6.46 | 37.37 ± 7.21 | 102.01 ± 19.29 | 67.84 ± 9.52 |
| OEVqNSTS4‐L2 | 245.06 ± 19.01 | 82.97 ± 5.52 | 38.44 ± 3.94 | 83.29 ± 10.22 | 28.93 ± 7.66 |
| WT‐PM‐7d | 131.09 ± 13.95 | – | 28.66 ± 3.83 | 86.18 ± 7.67 | 14.95 ± 2.33 |
| OEVqNSTS4‐L1‐PM‐7d | 322.08 ± 18.17 | 135.61 ± 7.72 | 74.33 ± 10.20 | 152.54 ± 13.74 | 63.31 ± 10.44 |
| OEVqNSTS4‐L2‐PM‐7d | 564.32 ± 27.57 | 87.93 ± 7.36 | 57.08 ± 6.04 | 85.46 ± 9.15 | 18.34 ± 4.84 |
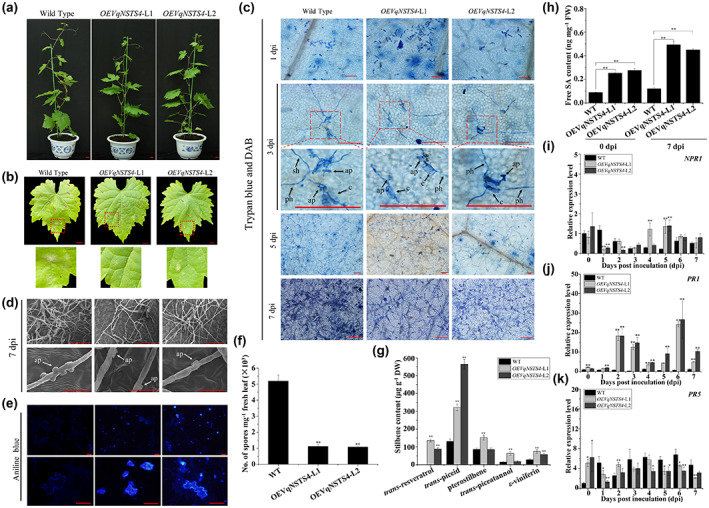
To evaluate disease resistance to powdery mildew, 8‐week‐old OEVqNSTS4‐L1 and OEVqNSTS4‐L2 plants were inoculated with U. necator (Figure 2a). As shown in Figure 2b, while many whitish mildew colonies were seen on the infected leaves of the wild‐type Thompson Seedless, only scattered and sparse mildew colonies were observed on the leaves of VqNSTS4 transgenic grapevines at 7 dpi. Trypan blue staining, scanning electron microscopy, and quantification of the number of spores showed that hyphal growth, sporulation, and appressorium formation on the hyphae were significantly restricted in the VqNSTS4 transgenic lines (Figure 2c,d,f). Also, plentiful callose accumulated in the epidermal cells of the VqNSTS4 transgenic line leaves (Figure 2e). Interestingly, U. necator inoculation promoted the accumulation of trans‐resveratrol, trans‐piceid, and ε‐viniferin in the two VqNSTS4 transgenic lines, while pterostilbene and trans‐piceatannol levels were markedly elevated only in OEVqNSTS4‐L1, at 1.8‐fold and 4.2‐fold higher levels than in wild‐type Thompson Seedless (Figure 2g and Table 1). Taken together, these results demonstrate that overexpression of VqNSTS4 significantly promotes the accumulation of stilbenes and enhances resistance to U. necator in grapevine. To further understand the resistance mechanisms activated by VqNSTS4 overexpression, the content of free SA was measured. As shown in Figure 2h, the content of free SA was 2.8‐fold and 3.1‐fold higher in VqNSTS4 transgenic lines than in wild‐type Thompson Seedless at 0 dpi and 4.1‐fold and 3.8‐fold higher than in the wild type at 7 dpi (Figure 2h). Therefore, we also examined the transcript levels of the SA‐related genes nonexpressor of pathogenesis‐related gene 1 (NPR1), pathogenesis‐related gene 1 (PR1), and PR5 in VqNSTS4 transgenic lines and wild‐type Thompson Seedless after U. necator inoculation. Although the transcript levels of NPR1 and PR5 were not continuously up‐regulated after U. necator inoculation in VqNSTS4 transgenic lines, the transcript level of PR1 was markedly higher than in wild‐type Thompson Seedless (Figure 2i–k). To further study the function of VqNSTS4 in disease resistance, we used RNA interference (RNAi) and investigated the resistance of transiently transformed Danfeng‐2 leaves to U. necator (Figure 3a). Because of the high sequence similarity of VqNSTS4 and VqSTS1, we knocked down VqNSTS4 and VqSTS1 at the same time. However, knockdown with RNAi‐VqNSTS4/STS1 in Danfeng‐2 resulted in the opposite results compared with transgenic overexpression plants (Figure 3b–j, Table 2). Taken together, these results indicate that VqNSTS4 can positively regulate the defence response to U. necator in grapevine by activating the SA signalling pathway.
FIGURE 2.
VqNSTS4 transgenic Vitis vinifera lines exhibit disease resistance resulting from the accumulation of internal stilbene and the expression of resistance genes. (a) Photograph of VqNSTS4 transgenic lines and wild‐type Thompson Seedless before Uncinula necator inoculation. Bars = 1 cm. (b) Photograph of leaves of VqNSTS4 transgenic lines and wild‐type Thompson Seedless at 7 days postinoculation (dpi). Bars = 1 cm. (c) Trypan blue staining and 3,3′‐diaminobenzidine (DAB) staining show the hyphal growth of U. necator and H2O2 accumulation at 1, 3, 5, and 7 dpi. c, conidium; ap, appressorium; ph, primary hypha; sh, secondary hypha. Bars = 100 μm. (d) Scanning electron micrographs of the hyphae and appressoria (ap) of U. necator in VqNSTS4 transgenic lines and wild‐type Thompson Seedless. Upper figures, bars = 100 μm; lower figures, bars = 20 μm. (e) Aniline blue staining showing callose depositions in U. necator‐infected epidermal cells at 7 dpi. Bars = 50 μm. (f) Quantification of spores per mg fresh leaves from VqNSTS4 transgenic lines and wild‐type Thompson Seedless at 7 dpi. (g) Determination of five stilbene contents in the leaves of VqNSTS4 transgenic mutants and wild‐type Thompson Seedless at 7 dpi. (h) Free salicylic acid (SA) content in the leaves of VqNSTS4 transgenic lines and wild‐type Thompson Seedless at 0 dpi and 7 dpi. (i–k) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative transcript levels of SA‐related genes in VqNSTS4 transgenic lines following U. necator inoculation. WT, wild‐type Thompson Seedless. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey's test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01.
FIGURE 3.
Transient silencing of VqNSTS4/STS1 in leaves of Vitis quinquangularis accession Danfeng‐2 reduces resistance to Uncinula necator. (a) Phenotypes of control EV and transiently silenced RNAi‐VqNSTS4/STS1‐GFP Danfeng‐2 leaves at 1 day postinoculation (dpi) and 7 dpi. EV, the empty vector pK7GWIWG2(II)‐35S‐GFP. (b) Trypan blue staining of leaves from EV and transiently silenced RNAi‐VqNSTS4/STS1‐GFP leaves at 1 dpi and 7 dpi. Bars = 100 μm. (c) Reverse transcription‐quantitative PCR (RT‐qPCR) analysis was conducted to determine the relative expression levels of VqNSTS4/STS1 in transiently transformed Danfeng‐2 leaves. (d) Quantification of spores per mg fresh leaves from EV and transiently silenced RNAi‐VqNSTS4/STS1‐GFP leaves at 7 dpi. (e, f) Stilbene contents in EV and transiently silenced RNAi‐VqNSTS4/STS1‐GFP leaves. (g) Free salicylic acid (SA) content in the leaves of EV and transiently silenced RNAi‐VqNSTS4/STS1‐GFP leaves at 7 dpi. (h–j) RT‐qPCR analysis was conducted to determine the relative transcript levels of SA‐related genes in EV and transiently silenced RNAi‐VqNSTS4/STS1‐GFP leaves following U. necator inoculation. The standard deviation (SD) was calculated from three independent replicates. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student's t test).
TABLE 2.
Contents of stilbenes in Vitis quinquangularis accession Danfeng‐2 leaves in which VqNSTS4 is transiently silenced before (PM‐0d) and after artificial inoculation with Uncinula necator (PM‐7d)
| Line | trans‐Piceid (μg/g) | trans‐Resveratrol (μg/g) | ε‐Viniferin (μg/g) |
|---|---|---|---|
| EV‐PM‐0d | 250.64 ± 64.63 | 103.88 ± 22.01 | 15.25 ± 4.83 |
| RNAiVqNSTS4‐PM‐0d | 133.57 ± 27.28 | 79.73 ± 19.59 | 15.49 ± 3.89 |
| EV‐PM‐7d | 309.27 ± 1.04 | 124.02 ± 0.29 | 27.87 ± 0.43 |
| RNAiVqNSTS4‐PM‐7d | 139.38 ± 27.94 | 74.72 ± 21.57 | 18.73 ± 4.88 |
2.4. VqNSTS4 is regulated by the novel transcription factor VqAL4
To identify the transcription factors that regulate the expression of VqNSTS genes, the promoters of VqNSTS2–VqNSTS6 were cloned from Danfeng‐2 and analysed (Figures [Link], [Link], [Link], [Link], [Link]). Several well‐known WRKY and MYB binding sites were identified in the promoters, such as MBS and W‐box elements (Figure 4a, Table S2). The WRKY and MYB transcription factors that have been reported to regulate VqSTS genes directly or indirectly include three WRKY proteins (VqWRKY2, VqWRKY3, and VqWRKY53) (Wang et al., 2020) and three MYB proteins (VqMYB14, VqMYB15, and VqMYB154) (Höll et al., 2013; Jiang et al., 2021). Yeast one‐hybrid (Y1H) analysis was conducted to identify the regulatory relationships between these six transcription factors and VqNSTS promoters (ProVqNSTS). As shown in Figure 4b, VqMYB154 can bind to the promoter of VqNSTS2, VqWRKY3 and VqWRKY53 can bind to the promoter of VqNSTS3, and VqMYB14 can bind to the promoter of VqNSTS6. None of these six VqWRKY/MYB transcription factors was found to bind to the promoters of VqNSTS4 or VqNSTS5 (Figure 4b).
FIGURE 4.
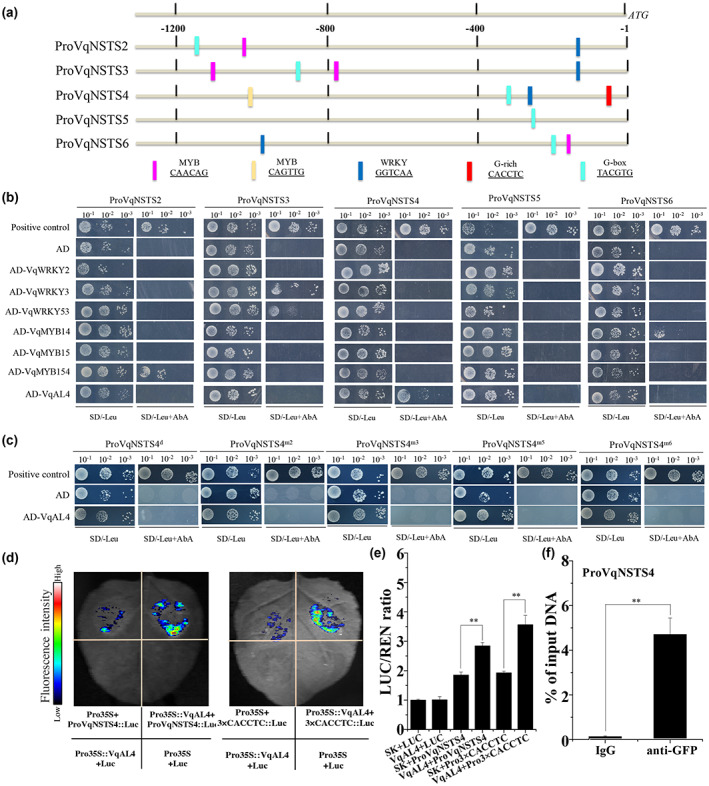
VqNSTS4 has unconventional transcriptional regulation activity and carries specific VqAL4 binding sites. (a) cis‐Regulatory element analysis in the promoters of VqNSTS genes. MYB and WRKY binding elements were found in the ProVqNSTS2, ProVqNSTS3, and ProVqNSTS6 promoters. The ProVqNSTS4 promoter included a G‐rich element in addition to the MYB and WRKY binding elements. (b) Yeast one‐hybrid assays were carried out to determine whether VqWRKYs, VqMYBs, and VqAL4 could bind directly to the promoters of VqNSTS genes. VqMYB154 can bind to ProVqNSTS2, VqWRKY3 and VqWRKY53 can bind to ProVqNSTS3, VqMYB14 can bind to ProVqNSTS6, and VqAL4 can bind to ProVqNSTS4. (c) Yeast one‐hybrid assays were conducted to demonstrate that VqAL4 cannot bind to ProVqNSTS4 m2 , ProVqNSTS4 m3 , ProVqNSTS4 d , ProVqNSTS4 m5 , or ProVqNSTS4 m6 . (d) Luminescence intensity. (e) Ratio of firefly luciferase (LUC) to Renilla luciferase (REN) activity. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey's test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01. (f) Chromatin immunoprecipitation–quantitative PCR assays were carried out to demonstrate that VqAL4 binds to the promoter of VqNSTS4 via the CACCTC/GAGGTG element. The SD was calculated from three independent replicates. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student's t test).
However, a G‐rich element (CACCTC/GAGGTG), which could be recognized by an AL transcription factor, was found in the promoter of VqNSTS4 (Figure 4a) (Bastola et al., 1998; Wei et al., 2017). Meanwhile, coexpression analysis using the transcriptome data of Danfeng‐2 revealed that the Pearson correlation coefficient value between VqNSTS4 and one AL transcription factor (VIT_11s0016g01500) was 0.89. Based on the grapevine gene nomenclature system, this AL transcription factor was designated VqAL4 (Grimplet et al., 2014). To examine whether the VqAL4 transcription factor could directly bind to the VqNSTS4 promoter, Y1H assays were carried out. As shown in Figure 4b, the pGADT7‐VqAL4 plasmid was transferred into the bait strains containing VqNSTS promoters. Only the yeast cells containing ProVqNSTS4 were able to grow normally on SD/−Leu medium containing 200 ng/ml Aureobasidin A (AbA) (Figure 4b). In addition, ProVqNSTS4 could not be bound by the VqAL4 transcription factor after deletion or mutation of the G‐rich element (Figures 4c and S12). These results demonstrate that VqAL4 binds only to the G‐rich element in ProVqNSTS4. Subsequently, dual‐luciferase and chromatin immunoprecipitation (ChIP)‐qPCR assays were conducted to further demonstrate that VqAL4 binds specifically to the G‐rich element in ProVqNSTS4 (Figure 4d–f). The above results indicate that VqNSTS4 is not regulated by reported VqMYB or VqWRKY transcription factors but by a novel transcription factor, VqAL4, through binding to the G‐rich element.
2.5. VqAL4 up‐regulates the expression of VqNSTS4 in grapevine
To analyse the effects of VqAL4 protein accumulation on the transcription of VqNSTS4, a transient expression assay was carried out in Danfeng‐2 leaves using Agrobacterium‐mediated infiltration (Figure 5a–c). As shown in Figure 5d,e, western blot assays demonstrated that 35S‐VqAL4‐GFP and RNAi‐VqAL4‐GFP were expressed in the Danfeng‐2 leaves. The transcript levels of VqAL4 were more elevated in 35S‐VqAL4‐GFP leaves than in those containing empty vector (EV), with 11.4‐fold and 8.7‐fold higher expression, respectively (Figure 5f). Meanwhile, the expression levels of VqNSTS4 in 35S‐VqAL4‐GFP leaves were increased 5.1‐fold and 5.6‐fold compared with those in EV leaves (Figure 5f). However, RNAi‐VqAL4‐GFP leaves of Danfeng‐2 showed the opposite results compared with 35S‐VqAL4‐GFP (Figure 5i). Consistent with the expression of VqNSTS4, the content of stilbenes was increased in 35S‐VqAL4‐GFP leaves and decreased in RNAi‐VqAL4‐GFP leaves, especially for the stilbene associated with strong antifungal activity (Pezet et al., 2004a) ε‐viniferin (Figure 5g,j, Table 3). In addition, the content of free SA was also increased in 35S‐VqAL4‐GFP leaves but decreased in RNAi‐VqAL4‐GFP leaves (Figure 5h,k). These results suggest that VqAL4 can positively regulate the expression of VqNSTS4, thereby promoting the accumulation of stilbene and the synthesis of free SA.
FIGURE 5.
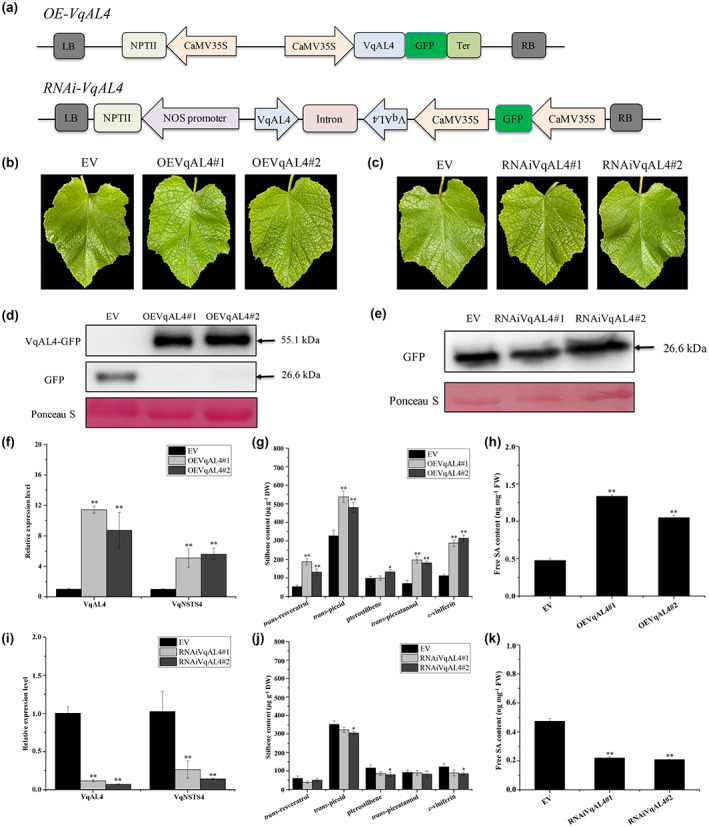
Transient expression of transcription factor VqAL4 in Vitis quinquangularis accession Danfeng‐2 promoted transcription of VqNSTS4 and antifungal stilbene accumulation. (a) Schematic diagram of overexpression and RNAi vectors. (b, c) Transient transformation of 35S‐VqAL4‐GFP and RNAi‐VqAL4‐GFP in Danfeng‐2 leaves using the Agrobacterium vacuum infiltration method. (d) Western blot assays were carried out to determine the expression of 35S‐VqAL4‐GFP in transiently transformed Danfeng‐2 leaves. EV, the empty vector pCAMBIA2300‐35S‐GFP was used as the control. (e) Western blot assays were carried out to determine the expression of RNAi‐VqAL4‐GFP in transiently transformed Danfeng‐2 leaves. EV, the empty vector pK7GWIWG2(II)‐35S‐GFP was used as the control. (f, i) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative expression levels of VqAL4 and VqNSTS4 in transiently transformed Danfeng‐2 leaves. (g, j) Determination of five stilbene contents in transiently transformed Danfeng‐2 leaves. (h, k) Free salicylic acid (SA) content in transiently transformed Danfeng‐2 leaves. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey's test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01.
TABLE 3.
Contents of stilbenes in Vitis quinquangularis accession Danfeng‐2 leaves transiently expressing VqAL4 overexpression (OE) and RNA interference (RNAi) constructs VqAL4
| Line | trans‐Piceid (μg/g) | trans‐Resveratrol (μg/g) | ε‐Viniferin (μg/g) | Pterostilbene (μg/g) | trans‐Piceatannol (μg/g) |
|---|---|---|---|---|---|
| EV | 327.33 ± 31.37 | 53.51 ± 9.53 | 112.24 ± 8.57 | 98.33 ± 11.37 | 70.76 ± 15.90 |
| OEVqAL4#1 | 537.56 ± 31.66 | 187.69 ± 16.06 | 288.05 ± 17.22 | 99.45 ± 11.87 | 197.09 ± 17.98 |
| OEVqAL4#2 | 480.74 ± 27.30 | 131.91 ± 16.43 | 314.60 ± 17.13 | 132.87 ± 12.90 | 181.57 ± 9.11 |
| EV | 352.02 ± 18.50 | 59.94 ± 13.41 | 122.14 ± 17.87 | 116.12 ± 17.80 | 92.12 ± 10.60 |
| RNAiVqAL4#1 | 322.45 ± 14.18 | 38.18 ± 6.83 | 89.38 ± 16.25 | 85.98 ± 11.01 | 89.48 ± 12.70 |
| RNAiVqAL4#2 | 305.26 ± 10.32 | 49.77 ± 9.43 | 85.27 ± 7.84 | 80.31 ± 12.17 | 82.59 ± 16.86 |
2.6. VqAL4 is localized in the nucleus and responds to U. necator and phytohormones
To investigate the function of the transcription factor VqAL4, the structural characteristics and the expression of VqAL4 were analysed. VqAL4 was predicted to be located on chromosome 11, and VqAL4 consists of two functional domains: an Alfin‐like domain (DUF3594) at the N‐terminus and a PHD finger domain at the C‐terminus (Figures 6a and S13). Phylogenetic analysis showed that VqAL4 was clustered into subgroup IV with three VvALs (VvAL1, VvAL3, and VvAL4), one AtAL (AtAL4), and three BrALs (BrAL3, BrAL7, and BrAL10) (Figure 6b). Amino acid sequence comparison between VqAL4 and VvAL4 revealed that the 57th amino acid was changed from leucine in VvAL4 to methionine in VqAL4 (Figure 6c). Furthermore, we performed a subcellular localization assay. The location of the green fluorescence signal from the VqAL4‐GFP fusion protein was identical to the red fluorescence signal from mCherry‐tagged AtHY5, which is a nuclear‐localized protein from Arabidopsis. This indicated that VqAL4 was localized in the nucleus (Figure 6d). A yeast two‐hybrid (Y2H) assay showed that VqAL4 has transcription activation functions in yeast (Figure 6e), suggesting that VqAL4 may function as a transcriptional activator. Additionally, expression analysis showed that the transcript level of VqAL4 was highest in mature leaves, followed by young leaves, and lowest in roots (Figure S14b). Moreover, the expression of VqAL4 could be induced by U. necator and various phytohormones, such as SA and abscisic acid (ABA) (Figure S14c–g). These results indicate that VqAL4 may be involved in powdery mildew resistance in mature leaves as a transcriptional activator.
FIGURE 6.
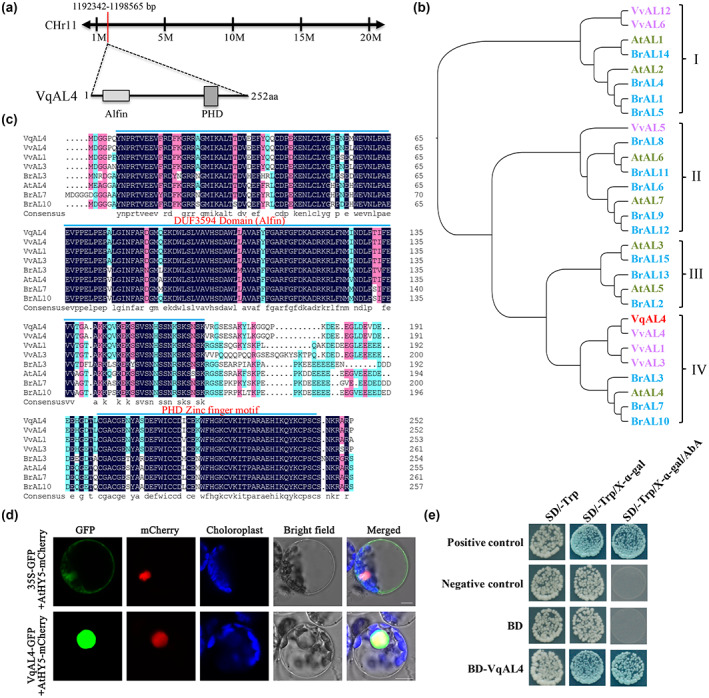
Location and structural analysis of VqAL4 isolated from Vitis quinquangularis accession Danfeng‐2. (a) Chromosomal localization of VqAL4. (b) Phylogenetic analysis of VqAL4 and the Alfin‐like proteins in Vitis vinifera, Brassica rapa, and Arabidopsis thaliana. (c) Multiple sequence alignment of amino acid sequences of VqAL4 and other group IV members, including VvAL4, VvAL1, VvAL3, AtAL4, BrAL3, BrAL7, and BrAL10. (d) Subcellular localization of VqAL4 in A. thaliana protoplasts. (e) Transcriptional activation of VqAL4 in yeast. pGADT7‐T/pGBKT7‐53 was used as a positive control, and pGADT7‐T/pGBKT7‐Lam was used as a negative control.
2.7. VqAL4 enhances resistance to U. necator in grapevine
Based on the finding that VqAL4 expression can be induced by U. necator (Figure S14c), we speculated that VqAL4 is involved in resistance to powdery mildew. Therefore, transgenic VqAL4 overexpression lines were created to evaluate disease resistance (Figure S15a). Two transgenic lines (OEVqAL4‐L1 and OEVqAL4‐L3) were identified from 21 PCR‐positive grapevine plantlets (9.5%) by western blot (Figure S15b). RT‐qPCR analysis showed that VqAL4 was up‐regulated 6.6‐fold in OEVqAL4‐L1 and 8.8‐fold in OEVqAL4‐L3 (Figure S15c). Intriguingly, VvSTS genes were also up‐regulated, and stilbene levels were significantly increased in the VqAL4 transgenic lines (Figure S15c,d, Table 4).
TABLE 4.
Contents of stilbenes in VqAL4 transgenic Vitis vinifera 'Thompson Seedless' lines under natural conditions and 7 days after artificial inoculation with Uncinula necator (PM‐7d)
| Line | trans‐Piceid (μg/g) | trans‐Resveratrol (μg/g) | ε‐Viniferin (μg/g) | Pterostilbene (μg/g) | trans‐Piceatannol (μg/g) |
|---|---|---|---|---|---|
| Wild type (WT) | 126.22 ± 10.03 | – | 23.24 ± 2.86 | 42.91 ± 7.64 | 17.89 ± 2.48 |
| OEVqAL4‐L1 | 156.19 ± 12.01 | 37.64 ± 4.22 | 23.09 ± 2.42 | 37.45 ± 5.90 | 19.08 ± 2.12 |
| OEVqAL4‐L3 | 133.95 ± 9.63 | 64.84 ± 4.36 | 26.46 ± 3.35 | 61.96 ± 8.00 | 37.20 ± 9.19 |
| WT‐PM‐7d | 170.62 ± 11.24 | – | 27.26 ± 5.78 | 47.12 ± 5.69 | 16.71 ± 1.93 |
| OEVqAL4‐L1‐PM‐7d | 242.61 ± 14.34 | 56.72 ± 5.65 | 42.64 ± 5.89 | 43.67 ± 7.58 | 60.69 ± 5.73 |
| OEVqAL4‐L3‐PM‐7d | 215.72 ± 10.76 | 128.69 ± 9.95 | 75.04 ± 7.41 | 74.01 ± 8.46 | 111.78 ± 8.99 |
Eight‐week‐old OEVqAL4‐L1 and OEVqAL4‐L3 were inoculated with U. necator to assess their disease resistance (Figure 7a). The results showed that the infected leaves of the VqAL4 transgenic grapevines were more tolerant to U. necator than the wild‐type Thompson Seedless, as indicated by fewer spores and appressoria, more hypersensitive response (HR)‐like cell death and callose deposition, and higher H2O2 and free SA contents (Figure 7b–h). In addition, HPLC analysis showed that stilbene levels were significantly increased in the two VqAL4 transgenic lines after U. necator inoculation (Figure 7i, Table 4). These results indicate that VqAL4 can enhance resistance to U. necator and promote the accumulation of stilbenes. Given that plentiful H2O2 was detected in the process of resistance to U. necator in the two VqAL4 transgenic grapevine lines (Figure 7g), the expression of respiratory burst oxidase homologue D (RBOHD) and glutathione S‐transferase 1 (GST1) was also examined in addition to NPR1, PR1, and PR5. As shown in Figure 7j–n, the transcript levels of RBOHD, GST1, NPR1, PR1, and PR5 were higher in OEVqAL4‐L1 and OEVqAL4‐L3 than in wild‐type Thompson Seedless. These results demonstrate that overexpression of VqAL4 in grapevine activates the SA signalling pathway. In contrast, transient silencing of VqAL4 in Danfeng‐2 leaves decreased resistance to powdery mildew by preventing SA production (Figure 8).
FIGURE 7.
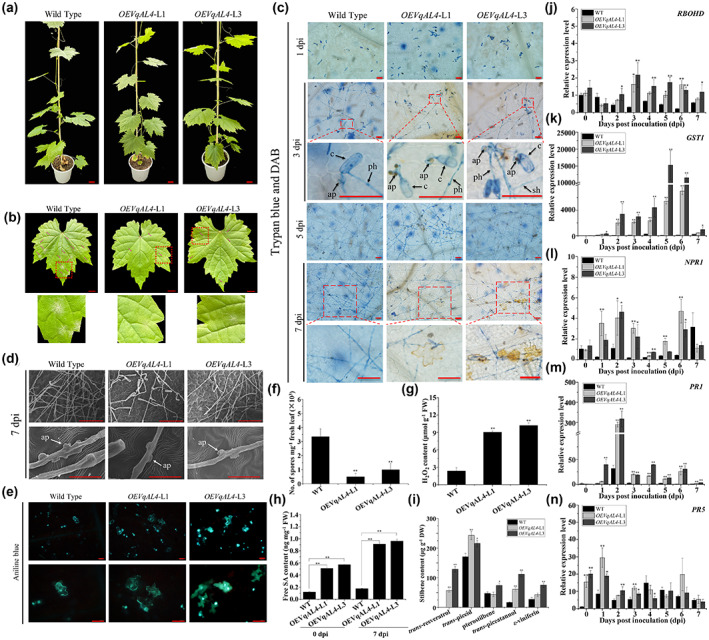
Overexpression of VqAL4 in grapevine enhances resistance to Uncinula necator by promoting stilbene accumulation and activating expression of resistance genes. (a) Photograph of VqAL4 transgenic lines and wild‐type Thompson Seedless before U. necator inoculation. Bars = 1 cm. (b) Photograph of leaves of VqAL4 transgenic lines and wild‐type Thompson Seedless at 7 days postinoculation (dpi). Bars = 1 cm. (c) Trypan blue staining and 3,3′‐diaminobenzidine (DAB) staining show the hyphal growth of U. necator and H2O2 accumulation at 1, 3, 5, and 7 dpi. c, conidium; ap, appressorium; ph, primary hypha; sh, secondary hypha. Bars = 100 μm. (d) Scanning electron micrographs of the hyphae and appressoria (ap) of U. necator in VqAL4 transgenic lines and wild‐type Thompson Seedless. Upper figures, bars = 100 μm; lower figures, bars = 20 μm. (e) Aniline blue staining showing callose depositions in U. necator‐infected epidermal cells at 7 dpi. Bars = 50 μm. (f) Quantification of spores per mg fresh leaves from VqAL4 transgenic lines and wild‐type Thompson Seedless at 7 dpi. (g) H2O2 content of VqAL4 transgenic lines and wild‐type Thompson Seedless leaves at 7 dpi. (h) Free salicylic acid (SA) content in the leaves of VqAL4 transgenic lines and wild‐type Thompson Seedless at 0 dpi and 7 dpi. (i) Determination of five stilbene contents in the leaves of VqAL4 transgenic lines and wild‐type Thompson Seedless at 7 dpi. (j, k) Reverse transcription‐quantitative PCR (RT‐qPCR) analysis was conducted to determine the relative transcript levels of H2O2‐related genes in VqAL4 transgenic lines after U. necator inoculation. WT, wild‐type Thompson Seedless. (l–n) RT‐qPCR analysis was conducted to determine the relative transcript levels of SA‐related genes in VqAL4 transgenic mutants after U. necator inoculation. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey's test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01.
FIGURE 8.
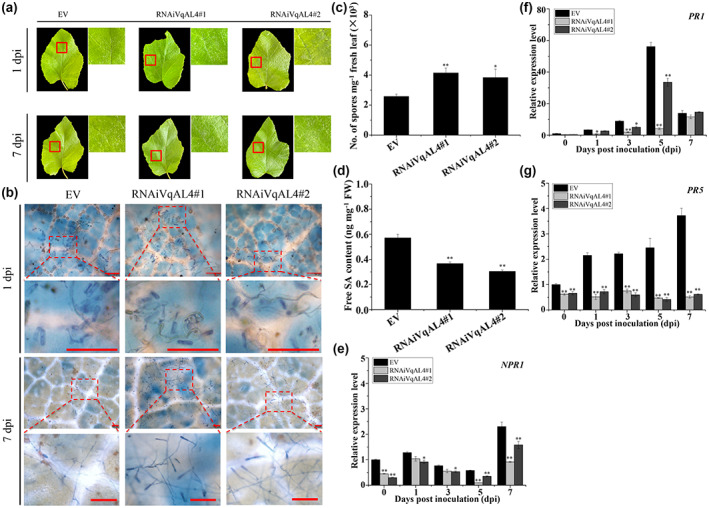
Transient silencing of VqAL4 in leaves of Danfeng‐2 reduces resistance to Uncinula necator. (a) Phenotypes of control EV and transiently silenced RNAi‐VqAL4‐GFP Danfeng‐2 leaves at 1 day postinoculation (dpi) and 7 dpi. EV, the empty vector pK7GWIWG2(II)‐35S‐GFP. (b) Trypan blue staining of leaves from EV and transiently silenced RNAi‐VqAL4‐GFP leaves at 1 dpi and 7 dpi. Bars = 100 μm. (c) Quantification of spores per mg fresh leaves from EV and transiently silenced RNAi‐VqAL4‐GFP leaves at 7 dpi. (d) Free salicylic acid (SA) content in the leaves of EV and transiently silenced RNAi‐VqAL4‐GFP leaves at 7 dpi. (e–g) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative transcript levels of SA‐related genes in EV and transiently silenced RNAi‐VqAL4‐GFP leaves following U. necator inoculation. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey's test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01.
The up‐regulated expression of VvSTS and the increased stilbene content in the VqAL4 transgenic lines compared with wild‐type Thompson Seedless indicate that VqAL4 may also be involved in the regulation of VvSTS genes. We cloned and analysed the promoters of seven VvSTS genes classified in subgroup I along with VqNSTS4 and found a G‐rich element in the promoter of VvSTS32 (Figures 1d, S16a, and [Link], [Link], [Link], [Link], [Link], [Link], [Link]). Y1H, dual‐luciferase, and ChIP‐qPCR assays demonstrated that VqAL4 could bind directly to the VvSTS32 promoter via the CACCTC element (Figure S16b–e). The above results demonstrate that VqAL4 can also up‐regulate VvSTS32 expression by binding to the G‐rich (CACCTC) element.
3. DISCUSSION
3.1. Disease resistance of stilbene synthase genes in grapevines
STS, which exhibits broad‐spectrum resistance and plays defensive roles against a variety of pathogens, is a key enzyme in the synthesis of stilbene (Hain et al., 1993; Khattab et al., 2021; Pezet et al., 2004b; Xu et al., 2019). The Chinese wild V. quinquangularis accession Danfeng‐2 is an important natural resource for investigating the potential relationship between STS and powdery mildew resistance that not only has a high content of stilbene but also exhibits strong disease resistance (Duan, 2002; Shi et al., 2014; Wan et al., 2007). Although 41 VqSTS genes have been cloned from Danfeng‐2 using the RACE method, six novel STS transcripts were identified from the RNA‐seq data of Danfeng‐2 (Li, 2019; Shi et al., 2014). We isolated these six VqNSTS genes from Danfeng‐2 and found that VqNSTS1 was a pseudogene (Figure 1, Table S1). Similarly, several VvSTS genes and VpSTS genes are incomplete or are pseudogenes (Cao, 2012; Parage et al., 2012). Furthermore, we found that the IPNSAGAIAGN conserved motif in VqNSTS5 and VqNSTS6 has been replaced by IPNTQGAIAGN (Figure 1c); the same phenomenon was also observed in VvSTSs (He, 2012). We cannot rule out the possibility that these mutations affect the functions of STS.
Many complete STS genes have been confirmed to have enzymatic activities for resveratrol biosynthesis and to enhance resistance to powdery mildew (Cheng et al., 2016; Liu et al., 2019; Xu et al., 2019). However, in the present study, we identified an N‐terminally truncated STS gene (Figure 1c), VqNSTS4, and showed that its expression was significantly induced by U. necator and SA (Figure 1g,l). In a previous report, it was speculated that two truncated STS genes, VvSTS1 and VvSTS4, may also contribute to stilbene biosynthesis (Vannozzi et al., 2012). Therefore, we created VqNSTS4 transgenic grapevines and demonstrated that VqNSTS4 overexpression significantly promoted the accumulation of stilbenes and enhanced resistance to powdery mildew (Figure 2). Conversely, transient silencing of VqNSTS4 in Danfeng‐2 leaves decreased resistance to powdery mildew (Figure 3). The above results indicate that the deletion of 48 amino acids at the N‐terminus of VqNSTS4 did not affect the disease resistance function of VqNSTS4.
3.2. Regulatory mechanisms of stilbene synthase genes in grapevine
Many studies have focused on the transcriptional regulation of stilbene biosynthesis in grapevine (Orduña et al., 2022; Wong & Matus, 2017). However, most of these focused on two transcription factor families, MYB and WRKY. For example, MYB14 and MYB15 can not only promote the expression of STS genes independently, but also interact with other WRKY transcription factors to regulate the expression of STS genes (Höll et al., 2013; Jiang et al., 2019; Mu et al., 2022; Vannozzi et al., 2018; Wang et al., 2020). In the present study, although several MYB and WRKY binding sites were identified in the promoter of VqNSTS4, VqWRKY2/3/53 and VqMYB14/15/154 transcription factors could not directly regulate VqNSTS4 expression (Figure 4a,b), suggesting that MYB and WRKY transcription factors are specific to the regulation of STS genes.
In addition, other transcription factors have been reported to be involved in the regulation of STS expression. Thus, VqERF114 indirectly regulates the expression of VqSTS genes by interacting with VqMYB35 (Wang & Wang, 2019), while VqbZIP1 interacts with VqSnRK2.4 and VqSnRK2.6 to enhance the regulation of the STS promoters (Wang et al., 2019). In the present study, we also identified a novel transcription factor, VqAL4, that can bind directly to the G‐rich element on the promoter of VqNSTS4 and positively regulate its expression (Figures 4b–f and 5). Similar results were reported in other plants, such as Atriplex hortensis, Glycine max, and A. thaliana, where AL transcription factors could bind to G‐rich elements on target gene promoters to promote their expression (Tao et al., 2018; Wei et al., 2009, 2015). However, it is unclear whether the mechanism of VqNSTS4 regulation by VqAL4 is related to the regulation of STS genes by MYBs and WRKYs. Furthermore, VqAL4 expression was significantly induced by U. necator and VqAL4 enhanced resistance to U. necator in grapevine (Figures 7, 8, and S14c). This is consistent with previous reports that the expression of AL transcription factors is induced by pathogens (Kayum et al., 2015; Kayum et al., 2016). Together, the above findings show that the AL transcription factor in grapevines is involved in disease resistance and reveal a novel regulatory mechanism of STS genes in grapevine.
3.3. Disease resistance pathways activated by stilbene synthase genes in grapevine
Plant defence responses to pathogens are usually viewed as a two‐tiered immune system, constituted by pattern‐triggered immunity (PTI) and effector‐triggered immunity (ETI), which function together as a unified interdependent system that exerts a mutually potentiating effect (Jones & Dangl, 2006; Ngou et al., 2022; Pruitt et al., 2021; Wan et al., 2021). When grapevine is affected by powdery mildew, the two‐tiered immune system is stimulated and a series of typical disease resistance reactions is initiated, including the formation of a haustorium complex and callose‐rich papillae, the accumulation of reactive oxygen species (ROS) and stilbene phytochemicals, and HR‐like cell death (Hu et al., 2021; Qiu et al., 2015; Ramming et al., 2010; Yin et al., 2022). Consistent with previous studies (Liu et al., 2019a; Xu et al., 2019), we observed a large amount of powdery mildew‐induced callose deposition in infected epidermal cells of the leaves of VqNSTS4 transgenic lines after artificial inoculation with U. necator (Figure 2e). Unlike typical disease resistance reactions, in the current and previous studies (Cheng et al., 2016; Liu et al., 2019a), no obvious H2O2 accumulation and HR‐like cell death were observed (Figure 2c). However, overexpression of VqAL4 enhanced grapevine resistance to U. necator and showed typical signs of an antifungal immune response, including a decrease in spore count, an H2O2 burst, HR‐like cell death, and callose accumulation (Figure 7b–g). This difference in the observed strength of the immune reaction may have been due to the difference in the disease resistance pathways activated by different genes as well as the deletion of 48 amino acids at the N‐terminus of VqNSTS4, and should be investigated in greater detail in future studies.
Plant immunity is also often tightly associated with SA signalling (Huang et al., 2020; Zhang & Li, 2019). In the present study, the transcript levels of VqNSTS4 and VqAL4 were significantly up‐regulated after SA treatment (Figures 1l and S14d). Meanwhile, we also observed that the content of free SA and the expression levels of SA‐related genes were significantly increased in transgenic VqNSTS4 and VqAL4 overexpression grapevines (Figures 2h–k and 7h,l–n). In RNAi‐VqNSTS4 and RNAi‐VqAL4 leaves, the content of free SA and the expression levels of SA‐related genes were significantly decreased (Figures 3g–j and 8d–g). These results indicate that VqNSTS4‐mediated or VqAL4‐mediated disease resistance not only promotes the biosynthesis of stilbenes but also activates the SA signalling pathway. Therefore, we speculate that the relationship between the biosynthesis of stilbenes and SA is mutually reinforcing. This is consistent with previous reports that the SA signalling pathway participates in stilbene‐mediated resistance to powdery mildew (Jiao et al., 2016; Xu et al., 2019; Yin et al., 2022). However, the specific cause–consequence relationship between the biosynthesis of stilbenes and SA needs further study.
In conclusion, we have identified novel VqNSTS genes from Danfeng‐2 according to six novel STS transcripts. Among them, VqNSTS4 expression is significantly induced by U. necator, enhancing resistance to U. necator in transgenic overexpression grapevine by activating SA signalling. Additionally, VqAL4 promotes VqNSTS4 expression by binding to the G‐rich element (CACCTC) on the VqNSTS4 promoter, resulting in higher antifungal stilbene levels. Moreover, overexpression of VqAL4 promotes the accumulation of stilbenes and enhances resistance to U. necator by activating SA signalling (Figure 9). These results reveal not only novel disease resistance VqNSTS genes but also a novel transcription factor that promotes their expression. All of these findings prove that VqNSTS4 regulated by VqAL4 improves grapevine resistance to powdery mildew by activating SA signalling. The present study has enriched our knowledge of disease resistance in perennial fruit trees.
FIGURE 9.
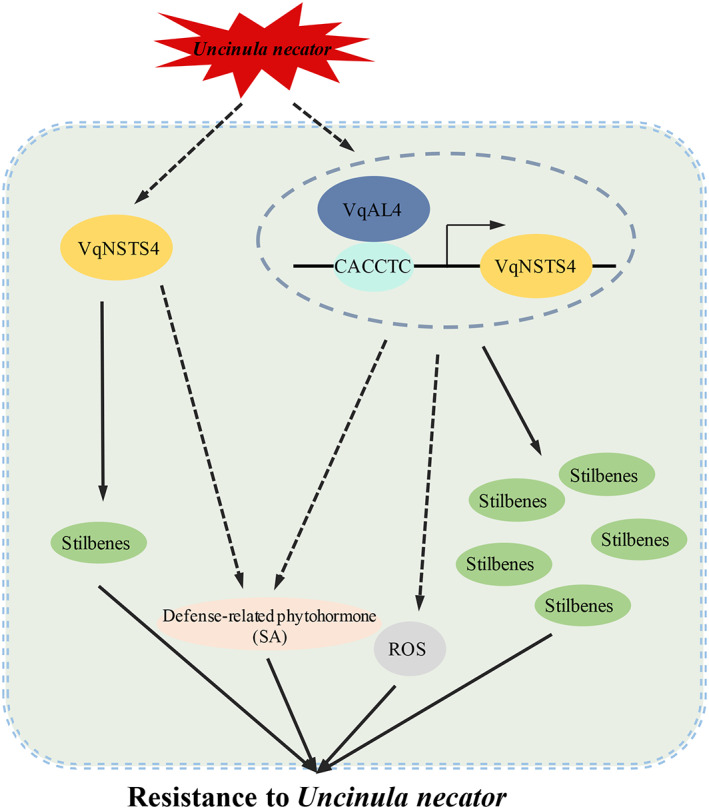
Proposed model of VqNSTS4‐mediated disease resistance to Uncinula necator and its regulatory mechanism. U. necator infection induces expression of VqNSTS4 and VqAL4. VqAL4 positively regulates the transcription of VqNSTS4. VqNSTS4 or VqAL4 transgenic grapevines show enhanced resistance to U. necator by producing more phytoalexin and activating SA signalling. ROS, reactive oxygen species. SA, salicylic acid.
4. EXPERIMENTAL PROCEDURES
4.1. Plant materials
The Chinese wild V. quinquangularis accession Danfeng‐2 was grown in the vineyard of Northwest A&F University, located in Yangling, Shaanxi, China (34°20′ N, 108°24′ E). Proembryonic masses (PEMs) of V. vinifera ‘Thompson Seedless’, to be used as transgenic receptor materials, were cultured on Murashige and Skoog medium supplemented with 1.5 g/L activated carbon at 25°C in the dark. A. thaliana (Col‐0), to be used for subcellular localization, was planted in a chamber at 23/19°C under a light/dark cycle of 16/8 h. Nicotiana benthamiana used for dual‐luciferase assays was grown in a growth chamber at 25°C with a photoperiod of 16 h.
4.2. Identification of novel transcripts in Danfeng‐2
Danfeng‐2 berries were collected at four stages: the green hard stage (DF_GH, 25 days after blooming), the before véraison stage (DF_BV, 40 days after blooming), the véraison stage (DF_V, 50 days after blooming), and the ripe stage (DF_R, 80 days after blooming). Total RNA of grape berries was extracted with a Plant RNA extraction Kit (Omega). Library preparation and sequencing were performed using Illumina HiSeq 2500. After filtering out reads containing adapters, the remaining high‐quality sequencing reads were mapped to the available genome of V. vinifera (Jaillon et al., 2007) using Bowtie2 (Langmead & Salzberg, 2012) with default parameters. Unmapped data were assembled by Trinity (Grabherr et al., 2011). Gene expression levels were calculated and normalized using fragments per kilobase per million mapped reads value (Mortazavi et al., 2008). Cluster analysis was carried out according to gene expression levels in four different developmental stages using MultiExperiment Viewer software (MeV, DFCI Boston).
4.3. Gene cloning and sequence analysis
The coding sequences (CDSs) of VqNSTS genes and VqAL4 were cloned from the cDNA of Danfeng‐2, while the promoters of VqNSTS genes and VvSTS genes were isolated from the genomic DNA of Danfeng‐2 and Thompson Seedless. The cis‐elements in STS promoters were predicted using the Plant‐CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The chromosomal location of VqAL4 was predicted according to the physical location information from the Grape Genome Browser (12X) Database (https://www.genoscope.cns.fr/vitis/). Amino acid sequence alignments were carried out using DNAMAN software v. 7.0.2 (LynnonBiosoft), while phylogenetic trees were constructed with MEGA 5.0 software using the neighbour‐joining approach with 1000 bootstrap replications. The primers used in this study are listed in Table S3.
4.4. Generation of transgenic grapevine using an Agrobacterium‐mediated method
The CDSs of VqNSTS4 and VqAL4 were inserted into the pCAMBIA2300‐GFP expression vector. The recombinant constructs were introduced into Agrobacterium tumefaciens GV3101 and then transformed into PEM of Thompson Seedless according to previously described methods (Zhou et al., 2014). VqNSTS4‐GFP and VqAL4‐GFP transgenic lines were identified by western blot and RT‐qPCR. To confirm the expression of VqNSTS4‐GFP and VqAL4‐GFP fusion proteins, western blot analysis was conducted. Total protein extraction and detection were carried out as described previously (Xu et al., 2019), with the primary antibody anti‐GFP. GFP fluorescence (488 nm) of the fusion protein was examined with a confocal laser scanning microscope (TCS SP8; Leica). For RT‐qPCR, total RNA was extracted using the HiPure Plant RNA Mini Kit (Magen), and 1 μg of RNA was used for cDNA first‐stand synthesis using SumOnetube RT Mixture III (gDNA removal) (SUM 7806). PCR 2× FastSYBR Green Mixture (Cofitt LBQ7505‐05) and an IQ5 RT‐PCR system (Bio‐Rad) were used for qPCR. The PCR mixture was 20 μl, comprised of 0.8 μl of forward primer, 0.8 μl of reverse primer, 1 μl of sample cDNA, 10 μl of SYBR, and 7.4 μl of RNase‐free water. Grape GAPDH (CB973647) was used as a reference gene (Xu et al., 2019). The 2−ΔΔCt method was used to calculate relative expression levels (Livak & Schmittgen, 2001). The primers used for RT‐qPCR are listed in Table S3.
4.5. Artificial inoculation of U. necator and phytohormone treatments
Fully expanded and healthy growing leaves at the third to sixth position of the grape shoot tip were inoculated with U. necator using a previously described method (Wang et al., 1995). Briefly, the surfaces of mature leaves of Danfeng‐2 or transgenic Thompson Seedless grapevines were sprayed with 0.78% glucose solution containing 2 × 105 conidia/ml to the point where tiny liquid droplets appeared but no run‐off occurred. Samples were collected at 0, 1, 2, 3, 4, 5, 6, and 7 dpi. Exogenous ABA (100 μM), Eth (100 μM), SA (100 μM), or MeJA (100 μM) (Wang et al., 2020) was sprayed on the leaves of Danfeng‐2, which were collected at 0, 0.5, 1, 3, 6, 12, 24, and 48 hpt. Mock samples from leaves sprayed with sterile water were collected at the same sampling times as the exogenous treatment samples.
4.6. Y1H assay
Y1H assays were carried out with the Matchmaker Gold Yeast One‐Hybrid System (Clontech). The promoters of VqNSTS genes and VvSTS genes were inserted into pAbAi. The mutations of ProVqNSTS4 (ProVqNSTS4 m2 , ProVqNSTS4 m3 , ProVqNSTS4 d , ProVqNSTS4 m5 , and ProVqNSTS4 m6 ) were inserted into the pAbAi vector. These pAbAi‐baits were linearized using BbsI or BstBI (NEB) and integrated into the genome of the Y1HGold yeast strain. The transformed yeast cells were cultured on SD/−Ura medium to select for positive clones (cultured at 30°C for 3 days). The CDSs of VqAL4, VqWRKY2, VqWRKY3, VqWRKY53, VqMYB14, VqMYB15, and VqMYB154 were inserted into pGADT7 to create the AD‐prey vectors. These AD‐prey vectors were transferred into the bait strains and subsequently growth of varying yeast transformants on SD/−Leu medium supplemented with AbA (200 ng/ml) was examined. pGADT7‐p53/p53‐pAbAi was used as a positive control, and an empty AD vector was used as a negative control. The primers used in this study are listed in Table S3.
4.7. Dual‐luciferase assays
To further demonstrate that transcription factors can activate STS promoters, dual‐luciferase assays were carried out using a previously described method (Wang & Wang, 2019). Briefly, the promoters of VqNSTS4 or VvSTS32 were inserted into the pGreenII 0800‐LUC vector and the CDS of VqAL4 was cloned into the pGreenII 62‐SK vector. These recombinant vectors were then transferred into A. tumefaciens GV3101 and transiently expressed in N. benthamiana leaves by Agrobacterium‐mediated infiltration. The leaves were collected for protein extraction after culture for 3 days. The enzyme activities of firefly luciferase (LUC) and Renilla luciferase (REN) were examined using the Dual Luciferase Reporter Gene Assay Kit (Beyotime).
4.8. ChIP‐qPCR analysis
ChIP‐qPCR analysis was conducted to validate VqAL4 binding to the G‐rich element. For ChIP analysis, 3 g of Danfeng‐2 leaves transiently overexpressing 35S‐VqAL4‐GFP or young leaves from 35S‐VqAL4‐GFP transgenic grapevine were crosslinked in 0.5% formaldehyde for 10 min using vacuum infiltration. Chromatin was isolated using the EpiQuik Plant ChIP Kit (Epigentek) according to the manufacturer's manual and immunoprecipitated by anti‐GFP. The immunoprecipitated and input DNA samples were used for qPCR analysis, and the ChIP‐qPCR results are presented as percentages of the input DNA. The specific primers are listed in Table S3.
4.9. Agrobacterium‐mediated transient expression in grape leaves
Transient transformation in Danfeng‐2 leaves was performed to confirm that VqAL4 regulates VqNSTS4 expression and enhances resistance to U. necator. The A. tumefaciens GV3101 strains containing the fusion vectors 35S‐VqAL4‐GFP, RNAi‐VqAL4‐GFP, and RNAi‐VqNSTS4‐GFP and the empty vectors pCAMBIA2300‐GFP and pK7GWIWG2(II)‐35S‐GFP were cultured in Luria–Bertani (LB) liquid medium with the OD600 value adjusted to 0.6–0.8. Leaves of Danfeng‐2 were immersed in the bacterial solution and held under vacuum for 30 min. These leaves were then cultured in a growth chamber with the petioles wrapped in moist cotton for 72 h and harvested for further analysis. For disease resistance analysis, the transiently transformed leaves of Danfeng‐2 were inoculated with U. necator after 24 h of culture.
4.10. Subcellular localization of the VqAL4 transcription factor
The isolation and transformation of Arabidopsis protoplasts were carried out using a previously described method (Wang et al., 2019). Briefly, 5 μl (1 μg/μl) of 35S‐VqAL4‐GFP and 35S‐AtHY5‐mCherry plasmid were mixed in a 2‐ml centrifuge tube and subsequently transformed into protoplasts using polyethylene glycol–Ca2+‐mediated transformation. The transformed protoplasts were cultured in the dark for 20 h at 25°C, and the expression of fusion protein was examined with a confocal laser scanning microscope (TCS SP8; Leica).
4.11. Y2H assay
For transcriptional activation assays, the full‐length CDS of VqAL4 was inserted into the vector pGBKT7 (BD) to construct the pGBKT7‐VqAL4 (BD‐VqAL4) fusion vector. The plasmid DNA of BD‐VqAL4 was transferred into the Y2HGold yeast strain (Clontech) according to the instructions of the Yeastmaker Yeast Transformation System 2 User Manual (Clontech). pGADT7‐T co‐transformed with pGBKT7‐53 was used as a positive control, and pGADT7‐T co‐transformed with pGBKT7‐Lam was used as a negative control. The transformants were cultured on SD/−Trp, SD/−Trp + X‐α‐gal (40 μg/ml), and SD/−Trp + X‐α‐gal (40 μg/ml) + AbA (200 ng/ml) media at 30°C for 3 days (Wang et al., 2019). The primers used in this study are listed in Table S3.
4.12. Observation of spore growth and development on the leaves of grapevines after artificial inoculation with U. necator
The fungal spores were quantified as previously reported (Weßling & Panstruga, 2012). Briefly, infected leaves were collected at 7 dpi, cut into pieces, and immersed in 5 ml of sterile water containing 0.01% Tween 20. This was shaken for 30 min at 500 rpm and then a haemocytometer was used to count the spores under a microscope. Scanning electron microscopy and aniline blue, trypan blue, and 3,3′‐diaminobenzidine staining were used to visualize hyphae, callose deposition, H2O2 accumulation, and HR‐like cell death (Hu et al., 2021). Samples were taken from three different plants per line for three biological replicates. The extraction and measurement of free SA were carried out using a previously described method (Wang et al., 2020). Briefly, 100 mg of leaves was ground and extracted with 1 ml of extraction solvent (methanol:isopropanol, 20:80 [vol/vol] with 1% of glacial acetic acid) using ultrasonication (4°C). After centrifugation (10,000 rpm for 15 min at 4°C), the supernatant was collected, dried completely under a nitrogen stream, redissolved in 300 μl of methanol, centrifuged (10,000 rpm for 5 min), and filtered through a 0.22‐μm membrane film. Samples were then analysed by HPLC–tandem mass spectrometry with a Shim‐pack XR‐ODS (2.0 mm internal diameter, 75 mm × 1.6 μm) column coupled to a triple quadrupole mass spectrometer (LC‐MS8040; Shimazu) with an electrospray ionization source. A standard sample of free SA (cat. #B21197; Yuanye) was used to confirm the retention time. The H2O2 content was measured by fluorescence spectrophotometry using the Hydrogen Peroxide Assay Kit (Solarbio). All samples were analysed in triplicate.
4.13. Detection and analysis of stilbenes using HPLC
Leaves were collected from different transgenic grapevines and wild‐type Thompson Seedless for stilbene detection using HPLC. Extraction and determination of stilbenes were carried out using a previously described method (Liu et al., 2019a). The leaves were freeze‐dried for 24 h before being extracted in methanol at 4°C for 12 h in the dark. The extracted liquor was filtered through a 0.22‐μm membrane film for HPLC analysis. Standard samples of stilbenes (Sigma) were dissolved in methanol and used to confirm the retention times. HPLC analyses were conducted using a Waters HPLC System (Waters ACQUITY Arc). The gradient elution programme was as follows: 0–1 min, 20% acetonitrile and 80% water; 1–30 min, 20%–75% acetonitrile and 80%–25% water; 30–33 min, 75%–100% acetonitrile and 25%–0% water; 33–36 min, 100% acetonitrile and 0% water; 36–38 min, 100%–20% acetonitrile and 0%–80% water; 38–49 min, 20% acetonitrile and 80% water. Standard samples of trans‐resveratrol (cat. #R5010; Sigma‐Aldrich), trans‐piceid (cat. #15721; Sigma‐Aldrich), pterostilbene (cat. #P1499; Sigma‐Aldrich), trans‐piceatannol (cat. #CFN99024; ChemFaces), and ε‐viniferin (cat. #CFN97067; ChemFaces) were used to confirm the retention times.
CONFLICT OF INTEREST
The authors declare they have no conflicts of interest.
Supporting information
Figure S1 Sequence analysis of VqNSTS2 and VqSTS37. The amino acid sequence homology between VqNSTS2 and VqSTS37 is 99.49%. There was one amino acid mutation: arginine (R)‐391 to threonine (T). The family signature motifs are indicated in the red boxes. The amino acid mutation is indicated in the pink box
Figure S2 Sequence analysis of VqNSTS3 and VqSTS33. The amino acid sequence homology between VqNSTS3 and VqSTS33 is 99.24%. There were two amino acid mutations: serine (S)‐3 to leucine (L) and serine (S)‐276 to phenylalanine (F). The family signature motifs are indicated in the red boxes. The amino acid mutations are indicated in the pink boxes
Figure S3 Sequence analysis of VqNSTS4 and VqSTS1. The amino acid sequence homology between VqNSTS4 and VqSTS1 is 87.53%. There were 48 amino acid deletions in the N‐terminus. The family signature motifs are indicated in red boxes. The amino acid mutations are indicated in the pink box
Figure S4 Sequence analysis of VqNSTS5 and VqSTS33. The amino acid sequence homology between VqNSTS5 and VqSTS33 is 91.35%. There were 33 amino acid mutations. The family signature motifs are indicated in the red boxes. The amino acid mutations are indicated in the pink boxes. There were two amino acid mutations in the first conserved motif: serine (S)‐50 to threonine (T) and alanine (A)‐51 to glutamine (Q)
Figure S5 Sequence analysis of VqNSTS6 and VqSTS33. The amino acid sequence homology between VqNSTS6 and VqSTS33 is 93.89%. There were 23 amino acid mutations. The family signature motifs are indicated in the red boxes. The amino acid mutations are indicated in the pink boxes. There were two amino acid mutations in the first conserved motif: serine (S)‐50 to threonine (T) and alanine (A)‐51 to glutamine (Q)
Figure S6 Creation and identification of VqNSTS4 transgenic grapevines. (a) Schematic diagram of the 35S‐VqNSTS4‐GFP construct. (b) Creation of VqNSTS4 transgenic grapevines via Agrobacterium‐mediated transformation using proembryonic masses of Vitis vinifera ‘Thompson Seedless’ as material. (c) Plantlets of VqNSTS4 transgenic lines in tissue culture vessels. (d) Western blot assays were conducted to determine the expression of VqNSTS4‐GFP in transgenic lines. (e) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative transcript levels of VqNSTS4 in the transgenic lines. (f) Determination of the contents of five stilbenes in the leaves of VqNSTS4 transgenic lines and wild‐type Thompson Seedless. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01
Figure S7 cis‐Regulatory element analysis of ProVqNSTS2. There are 10 cis‐regulatory elements in ProVqNSTS2: one WRKY transcription factor binding site (a W‐box element, TTGACC, −148 to −143 bp), one MYB transcription factor binding site (CAACAG, −1111 to −1106 bp), two abscisic acid‐responsive elements (ABREs, CACGT, −1190 to −1186 bp and −1216 to −1212 bp), three methyl jasmonate (MeJA)‐responsive elements (an ACE element, CATACATGTC, −243 to −234 bp, an I‐box element, GACCTTGTCAT, −483 to −472 bp, and a G‐box element, GTCGTG, −1172 to −1167 bp), and three light‐responsive elements (a Box4 element, ATTAATT, −420 to −414 bp, a Gap‐box element, CAAATGAACAAAGT, −434 to −421 bp, and a GT1 motif, GGTTAA, −571 to −566 bp)
Figure S8 cis‐Regulatory element analysis of ProVqNSTS3. There are 12 cis‐regulatory elements in ProVqNSTS3: one WRKY transcription factor binding site (a W‐box element, TTGACC, −147 to −142 bp), two MYB transcription factor binding sites (CAACAG and CTGTTG, −796 to −791 bp and −1031 to −1026 bp), one abscisic acid‐responsive element (ABRE, CACGT, −891 to −887 bp), four methyl jasmonate (MeJA)‐responsive elements (an ACE element, CATACATGTCT, −241 to −231 bp, an I‐box element, GACCTTGTCAT, −481 to −471 bp, a G‐box element, CGTGGG, −926 to −921 bp, and a TGACG motif, −1203 to −1199 bp), and four light‐responsive elements (two TCT motifs, GTAAGA, −407 to −402 bp and −611 to −606 bp, a Gap‐box element, CAAATGAACAAAGT, −432 to −419 bp, and a GT1 motif, GGTTAA, −569 to −564 bp)
Figure S9 cis‐Regulatory element analysis of ProVqNSTS4. There are 10 cis‐regulatory elements in ProVqNSTS4: one AL transcription factor binding site (a G‐rich element, CACCTC, −76 to −71 bp) (red box), one WRKY transcription factor binding site (a W‐box element, GGTCAA, −285 to −280 bp), one MYB transcription factor binding site (an MBS element, CAGTTG, −1048 to −1043 bp), two abscisic acid‐responsive elements (ABREs, ACGTG and CACGT, −337 to −333 bp and −999 to −995 bp), three methyl jasmonate (MeJA)‐responsive elements (a G‐box element, CACGTG, −338 to −333 bp, a CGTCA motif, TGACG, −942 to −938 bp, and an ACE element, GACACGTATG, −1,001 to −992 bp), and two light‐responsive elements (a TCT motif, GTAAGA, −664 to −659 bp and a Gap‐box element, TGTTCATTTGA, −889 to −879 bp)
Figure S10 cis‐Regulatory element analysis of ProVqNSTS5. There are six cis‐regulatory elements in ProVqNSTS5: one abscisic acid‐responsive element (ABRE, ACGTG, −227 to −223 bp), one methyl jasmonate (MeJA)‐responsive element (a G‐box element, TACGTG, −228 to −222 bp), and four light‐responsive elements (a Gap‐box element, CAAATGAACAATGT, −407 to −394 bp, two TCT motifs, TCTTAC, −691 to −686 bp and −1169 to −1164 bp, and a GT1 motif, GGTTAA, −1002 to −997 bp)
Figure S11 cis‐Regulatory element analysis of ProVqNSTS6. There are nine cis‐regulatory elements in ProVqNSTS6: one MYB transcription factor binding site (an MBS element, CAACTG, −150 to −145 bp), one WRKY transcription factor binding site (a W‐box element, TTGACC, −960 to −955 bp), one abscisic acid‐responsive element (ABRE, CACGT, −907 to −903 bp), four methyl jasmonate (MeJA)‐responsive elements (two G‐box elements, TACGTG and CACGTGCT, −199 to −194 bp and −907 to −900 bp, a CGTCA motif, TGACG, −255 to −251 bp, and an I‐box element, CCTTATCT, −317 to −310 bp), and two light‐responsive elements (a Gap‐box element, CAAATGAACAAGGG, −360 to −347 bp and a TCT motif, TCTTAC, −581 to −576 bp)
Figure S12 The deletions and mutations in the G‐rich element in ProVqNSTS4. ProVqNSTS4: the promoter of VqNSTS4 contains the G‐rich element CACCTC. ProVqNSTS4 m2 : the CACCTC element has mutated to CAACTC, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS2. ProVqNSTS4 m3 : the CACCTC element has mutated to CCAACT, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS3. ProVqNSTS4 d : CACCTC was deleted in the promoter of VqNSTS4. ProVqNSTS4 m5 : the CACCTC element has mutated to CTCTTC, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS5. ProVqNSTS4 m6 : the CACCTC element has mutated to TTTCTC, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS6
Figure S13 The sequence of VqAL4. The coding sequence of VqAL4 is 759 bp long. Initiation and termination codons are marked bold and underlined. The conserved sequences are marked in red. This includes eight sites: TGT (595–597 bp), TGC (604–606 bp), TGC (643–645 bp), TGT (652–654 bp), CAT (667–669 bp), TGT (676–678 bp), TGC (724–726 bp), and TGC (733–735 bp)
Figure S14 Expression analysis of VqAL4 in different organs and under different treatments. (a) Root, stem, young leaf, mature leaf, tendril, inflorescence, young berry, and mature berry of Danfeng‐2 were analysed by reverse transcription‐quantitative PCR (RT‐qPCR). (b) Expression analysis of VqAL4 in different organs. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01. (c) Danfeng‐2 leaves were inoculated with Uncinula necator and sampled for RT‐qPCR analysis. (d–g) Danfeng‐2 leaves were treated with 100 μM salicylic acid (SA), methyl jasmonate (MeJA), abscisic acid (ABA), or ethylene (Eth) and sampled for RT‐qPCR analysis. SD was calculated from three independent replicates. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student’s t test)
Figure S15 Creation and identification of VqAL4 transgenic grapevines. (a) Plantlets of VqAL4 transgenic grapevines in tissue culture vessels. (b) Western blot assays were conducted to determine the accumulation of VqAL4‐GFP fusion protein in VqAL4 transgenic lines. (c) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative transcript levels of VqAL4 and VvSTS genes in the VqAL4 transgenic lines. (d) Determination of the contents of five stilbenes in the leaves of VqAL4 transgenic lines and wild‐type Thompson Seedless. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01
Figure S16 VqAL4 bound to the VvSTS32 promoter and promoted VvSTS32 expression. (a) cis‐Regulatory element analysis of the promoters of VvSTS genes. (b) Yeast one‐hybrid assay demonstrating that VqAL4 binds to the VvSTS32 promoter. (c) Graph showing the luminescence intensity. (d) Ratio of firefly luciferase (LUC) to Renilla luciferase (REN) activity. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01. (e) Chromatin immunoprecipitation (ChIP)‐quantitative PCR analysis of VqAL4 binding to the G‐rich motif. The fragment contains the CACCTC element. The ChIP results are presented as percentages of input DNA. SD was calculated from three independent replicates. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student’s t test)
Figure S17 cis‐Regulatory element analysis of ProVvSTS7. There are three cis‐regulatory elements in ProVvSTS7: one abscisic acid‐responsive element (ABRE, ACGTG, −547 to −543 bp), one methyl jasmonate (MeJA)‐responsive element (a G‐box element, GACGTG, −548 to −543 bp), and one light‐responsive element (a TCT motif, TCTTAC, −356 to −351 bp)
Figure S18 cis‐Regulatory element analysis of ProVvSTS8. There are three cis‐regulatory elements in ProVvSTS8: two MYB transcription factor binding sites (two MBS elements, CAACTG, −158 to −153 bp and −361 to −356 bp) and one light‐responsive element (a GT1 motif, GGTTAA, −501 to −496 bp)
Figure S19 cis‐Regulatory element analysis of ProVvSTS9. There are three cis‐regulatory elements in ProVvSTS9: one WRKY transcription factor binding site (a W‐box element, GGTCCA, −550 to −545 bp), one abscisic acid‐responsive element (ABRE, GACACCTGCG, −732 to −723 bp), and one methyl jasmonate (MeJA)‐responsive element (a TGACG motif, −560 to −556 bp)
Figure S20 cis‐Regulatory element analysis of ProVvSTS25. There are two cis‐regulatory elements in ProVvSTS25: two WRKY transcription factor binding sites (two W‐box elements, TTGACC, −369 to −364 bp and −760 to −755 bp)
Figure S21 cis‐Regulatory element analysis of ProVvSTS27. There are two cis‐regulatory elements in ProVvSTS27: one MYB transcription factor binding site (an MBS element, CAGTTG, −614 to −609 bp) and one light‐responsive element (a GT1 motif, GGTTAA, −727 to −722 bp)
Figure S22 cis‐Regulatory element analysis of ProVvSTS29. There is one cis‐regulatory element in ProVvSTS29: one MYB transcription factor binding site (an MBS element, CAGTTG, −612 to −607 bp)
Figure S23 cis‐Regulatory element analysis of ProVvSTS32. There are four cis‐regulatory elements in ProVvSTS32: one AL transcription factor binding site (a G‐rich element, CACCTC, −77 to −72 bp) (red box), two WRKY transcription factor binding sites (two W‐box elements, GGTCAA, −286 to −281 bp and −534 to −529 bp), and one abscisic acid‐responsive element (ABRE, ACGTG, −338 to −334 bp)
Table S1 Sequence analysis of VqNSTS genes
Table S2 cis‐Regulatory element analysis of ProVqNSTS genes
Table S3 Primers used in this study
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (31872055 and 32272667). We specifically thank Dr Alexander (Sandy) Lang from RESCRIPT Co. (New Zealand) and the company of Cambridge Proofreading (https://proofreading.org/) for useful language editing.
Yan, C. , Yang, N. , Li, R. , Wang, X. , Xu, Y. , Zhang, C. et al. (2023) Alfin‐like transcription factor VqAL4 regulates a stilbene synthase to enhance powdery mildew resistance in grapevine. Molecular Plant Pathology, 24, 123–141. Available from: 10.1111/mpp.13280
Chaohui Yan and Na Yang contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the results are included in this article and its supplementary materials.
REFERENCES
- Barger, J.L. , Kayo, T. , Vann, J.M. , Arias, E.B. , Wang, J. , Hacker, T.A. et al. (2008) A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One, 3, e2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastola, D.R. , Pethe, V.V. & Winicov, I. (1998) Alfin1, a novel zinc‐finger protein in alfalfa roots that binds to promoter elements in the salt‐inducible MsPRP2 gene. Plant Molecular Biology, 38, 1123–1135. [DOI] [PubMed] [Google Scholar]
- Cao, J. (2012) Characterization of stilbene synthase gene and genetic transformation in grape. Master thesis. Yangling, China: Northwest A & F University. [Google Scholar]
- Chandrika, N.N.P. , Sundaravelpandian, K. , Yu, S.‐M. & Schmidt, W. (2013) ALFIN‐LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis . New Phytologist, 198, 709–720. [DOI] [PubMed] [Google Scholar]
- Cheng, S. , Xie, X. , Xu, Y. , Zhang, C. , Wang, X. , Zhang, J. et al. (2016) Genetic transformation of a fruit‐specific, highly expressed stilbene synthase gene from Chinese wild Vitis quinquangularis . Planta, 243, 1041–1053. [DOI] [PubMed] [Google Scholar]
- Chong, J. , Poutaraud, A. & Hugueney, P. (2009) Metabolism and roles of stilbenes in plants. Plant Science, 177, 143–155. [Google Scholar]
- Dai, L. , Zhou, Q. , Li, R. , Du, Y. , He, J. , Wang, D. et al. (2015) Establishment of a picloram‐induced somatic embryogenesis system in Vitis vinifera cv. Chardonnay and genetic transformation of a stilbene synthase gene from wild‐growing Vitis species. Plant Cell Tissue & Organ Culture, 121, 397–412. [Google Scholar]
- Ding, X. , Zhao, K. & Wang, Y. (2021) Expression of stilbene synthase genes from Chinese wild Vitis quinquangularis and its effect on resistance of grape to powdery mildew. Scientia Agricultura Sinica, 54, 310–323. [Google Scholar]
- Duan, C. (2002) Resveratrol content in berries of Chinese wild Vitis species. Master thesis. Yangling, China: Northwest A & F University. [Google Scholar]
- Gadoury, D.M. , Cadle‐Davidson, L. , Wilcox, W.F. , Dry, I.B. , Seem, R.C. & Milgroom, M.G. (2012) Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Molecular Plant Pathology, 13, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. et al. (2011) Full‐length transcriptome assembly from RNA‐seq data without a reference genome. Nature Biotechnology, 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet, J. , Adam‐Blondon, A.‐F. , Bert, P.‐F. , Oliver Bitz, D.C. , Davies, C. , Delrot, S. et al. (2014) The grapevine gene nomenclature system. BMC Genomics, 15, 1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain, R. , Reif, H.J. , Krause, E. , Langebartels, R. , Kindl, H. , Vornam, B. et al. (1993) Disease resistance results from foreign phytoalexin expression in a novel plant. Nature, 361, 153–156. [DOI] [PubMed] [Google Scholar]
- He, M. (2012) Cloning and functional analysis of stilbene synthase and pathogenesis‐related 10 protein genes from Chinese wild grapevines. PhD thesis. Yangling, China: Northwest A & F University. [Google Scholar]
- Höll, J. , Vannozzi, A. , Czemmel, S. , D'Onofrio, C. , Walker, A.R. , Rausch, T. et al. (2013) The R2R3‐MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera . The Plant Cell, 25, 4135–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Cheng, Y. , Yu, X. , Liu, J. , Yang, L. , Gao, Y. et al. (2021) Overexpression of two CDPKs from wild Chinese grapevine enhances powdery mildew resistance in Vitis vinifera and Arabidopsis . New Phytologist, 230, 2029–2046. [DOI] [PubMed] [Google Scholar]
- Huang, W. , Wang, Y. , Li, X. & Zhang, Y. (2020) Biosynthesis and regulation of salicylic acid and N‐hydroxypipecolic acid in plant immunity. Molecular Plant, 13, 31–41. [DOI] [PubMed] [Google Scholar]
- Jaillon, O. , Aury, J.M. , Noel, B. , Policriti, A. , Clepet, C. , Casagrande, A. et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449, 463–467. [DOI] [PubMed] [Google Scholar]
- Jang, M. , Cai, L. , Udeani, G.O. , Slowing, K.V. , Thomas, C.F. , Beecher, C.W.W. et al. (1997) Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science, 275, 218–220. [DOI] [PubMed] [Google Scholar]
- Jiang, J. , Xi, H. , Dai, Z. , Lecourieux, F. , Yuan, L. , Liu, X. et al. (2019) VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. Journal of Experimental Botany, 70, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C. , Wang, D. , Zhang, J. , Xu, Y. , Zhang, C. , Zhang, J. et al. (2021) VqMYB154 promotes polygene expression and enhances resistance to pathogens in Chinese wild grapevine. Horticulture Research, 8, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Xu, W. , Duan, D. , Wang, Y. & Nick, P. (2016) A stilbene synthase allele from a Chinese wild grapevine confers resistance to powdery mildew by recruiting salicylic acid signalling for efficient defence. Journal of Experimental Botany, 67, 5841–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, L. , Riaz, S. , Morales‐Cruz, A. , Amrine, K.C. , McGuire, B. , Gubler, W.D. et al. (2014) Adaptive genomic structural variation in the grape powdery mildew pathogen, Erysiphe necator . BMC Genomics, 15, 1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaadige, M.R. & Ayer, D.E. (2006) The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. Journal of Biological Chemistry, 281, 28831–28836. [DOI] [PubMed] [Google Scholar]
- Kayum, M.A. , Park, J.I. , Ahmed, N.U. , Jung, H.J. , Saha, G. , Kang, J.G. et al. (2015) Characterization and stress‐induced expression analysis of Alfin‐like transcription factors in Brassica rapa . Molecular Genetics and Genomics, 290, 1299–1311. [DOI] [PubMed] [Google Scholar]
- Kayum, M.A. , Park, J.I. , Ahmed, N.U. , Saha, G. , Chung, M.Y. , Kang, J.G. et al. (2016) Alfin‐like transcription factor family: characterization and expression profiling against stresses in Brassica oleracea . Acta Physiologiae Plantarum, 38, 1–14. [Google Scholar]
- Khattab, I.M. , Sahi, V.P. , Baltenweck, R. , Maia‐Grondard, A. , Hugueney, P. , Bieler, E. et al. (2021) Ancestral chemotypes of cultivated grapevine with resistance to Botryosphaeriaceae‐related dieback allocate metabolism towards bioactive stilbenes. New Phytologist, 229, 1133–1146. [DOI] [PubMed] [Google Scholar]
- Korfhage, U. , Trezzini, G.F. , Meier, I. , Hahlbrock, K. & Somssich, I.E. (1994) Plant homeodomain protein involved in transcriptional regulation of a pathogen defense‐related gene. The Plant Cell, 6, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunova, A. , Pizzatti, C. , Saracchi, M. , Pasquali, M. & Cortesi, P. (2021) Grapevine powdery mildew: fungicides for its management and advances in molecular detection of markers associated with resistance. Microorganisms, 9, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langcake, P. & Pryce, R.J. (1977) A new class of phytoalexins from grapevines. Experientia, 33, 151–152. [DOI] [PubMed] [Google Scholar]
- Langmead, B. & Salzberg, S.L. (2012) Fast gapped‐read alignment with bowtie 2. Nature Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband, G. & Lörz, H. (1998) Transformation and expression of a stilbene synthase gene of Vitis vinifera L. in barley and wheat for increased fungal resistance. Theoretical and Applied Genetics, 96, 1004–1012. [Google Scholar]
- Li, R. (2019) Regulation mechanism of stilbenes metabolism in Chinese wild Vitis quinquangularis using multi‐omics analysis. PhD thesis. Yangling, China: Northwest A & F University. [Google Scholar]
- Li, R. , Xie, X. , Ma, F. , Wang, D. , Wang, L. , Zhang, J. et al. (2017) Resveratrol accumulation and its involvement in stilbene synthetic pathway of Chinese wild grapes during berry development using quantitative proteome analysis. Scientific Reports, 7, 9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. , Ma, F. , Wu, F. , Jiang, C. & Wang, Y. (2019) Expression of stilbene synthase VqSTS6 from wild Chinese Vitis quinquangularis in grapevine enhances resveratrol production and powdery mildew resistance. Planta, 250, 1997–2007. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, F. (2018) Expression and regulation of Chinese wild Vitis stilbene synthase genes and their resistance to powdery mildew. PhD thesis. Yangling,: Northwest A & F University. [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. & Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐seq. Nature Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Mu, H. , Li, Y. , Yuan, L. , Jiang, J. , Wei, Y. , Duan, W. et al. (2022) MYB30 and MYB14 form a repressor‐activator module with WRKY8 that controls stilbene biosynthesis in grapevine. The Plant Cell. Available from: 10.1093/plcell/koac1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou, B.P.M. , Ding, P. & Jones, J.D.G. (2022) Thirty years of resistance: zig‐zag through the plant immune system. The Plant Cell, 34, 1447–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orduña, L. , Li, M. , Navarro‐Payá, D. , Zhang, C. , Santiago, A. , Romero, P. et al. (2022) Direct regulation of shikimate, early phenylpropanoid and stilbenoid pathways by subgroup 2 R2R3‐MYBs in grapevine. The Plant Journal, 110, 529–547. [DOI] [PubMed] [Google Scholar]
- Parage, C. , Tavares, R. , Rety, S. , Baltenweck‐Guyot, R. , Poutaraud, A. , Renault, L. et al. (2012) Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiology, 160, 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet, R.G.K. , Gindro, K. , Viret, O. & Richter, H. (2004a) Effects of resveratrol, viniferins and pterostilbene on Plasmopara viticola zoospore mobility and disease development. Vitis, 43, 145–148. [Google Scholar]
- Pezet, R. , Gindro, K. , Viret, O. & Spring, J.L. (2004b) Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiological & Molecular Plant Pathology, 65, 297–303. [Google Scholar]
- Pruitt, R.N. , Gust, A.A. & Nurnberger, T. (2021) Plant immunity unified. Nature Plants, 7, 382–383. [DOI] [PubMed] [Google Scholar]
- Qiu, W. , Feechan, A. & Dry, I. (2015) Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Horticulture Research, 2, 15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramming, D.W. , Gabler, F. , Smilanick, J. , Cadle‐Davidson, M. , Barba, P. , Mahanil, S. et al. (2010) A single dominant locus, Ren4, confers rapid non‐Race‐specific resistance to grapevine powdery mildew. Phytopathology, 101, 502–508. [DOI] [PubMed] [Google Scholar]
- Shi, J. , He, M. , Cao, J. , Wang, H. , Ding, J. , Jiao, Y. et al. (2014) The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera). Plant Physiology and Biochemistry, 74, 24–32. [DOI] [PubMed] [Google Scholar]
- Stark‐Lorenzen, P. , Nelke, B. , Hänßler, G. , Mühlbach, H.P. & Thomzik, J.E. (1997) Transfer of a grapevine stilbene synthase gene to rice (Oryza sativa L.). Plant Cell Reports, 16, 668–673. [DOI] [PubMed] [Google Scholar]
- Tao, J. , Wei, W. , Pan, W. , Lu, L. , Li, Q. , Ma, J. et al. (2018) An Alfin‐like gene from Atriplex hortensis enhances salt and drought tolerance and abscisic acid response in transgenic Arabidopsis . Scientific Reports, 8, 2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomzik, J.E. , Stenzel, K. , Stcker, R. , Schreier, P.H. & Stahl, D.J. (1997) Synthesis of a grapevine phytoalexin in transgenic tomatoes (Lycopersicon esculentum Mill.) conditions resistance against Phytophthora infestans . Physiological and Molecular Plant Pathology, 51, 265–278. [Google Scholar]
- Vannozzi, A. , Dry, I.B. , Fasoli, M. , Zenoni, S. & Lucchin, M. (2012) Genome‐wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biology, 12, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi, A. , Wong, D.C.J. , Holl, J. , Hmmam, I. , Matus, J.T. , Bogs, J. et al. (2018) Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant and Cell Physiology, 59, 1043–1059. [DOI] [PubMed] [Google Scholar]
- Wan, Y. , Schwaninger, H. , He, P. & Wang, Y. (2007) Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis, 46, 132–136. [Google Scholar]
- Wan, J. , He, M. , Hou, Q. , Zou, L. , Yang, Y. , Wei, Y. et al. (2021) Cell wall associated immunity in plants. Stress Biology, 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. (2004) Cloning and analysing for the gene sequences of resistance to Uncinula necator in Chinese wild Vitis species. PhD thesis. Yangling, China: Northwest A & F University. [Google Scholar]
- Wang, L. & Wang, Y. (2019) Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Reports, 38, 1347–1360. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Liu, Y. , He, P. , Chen, J. , Lamikanra, O. & Lu, J. (1995) Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis species. Vitis, 34, 159–164. [Google Scholar]
- Wang, Y. , Liu, Y. , He, P. , Lamikanra, O. & Lu, J. (1998) Resistance of Chinese Vitis species to Elsinoë ampelina (de Bary) Shear. HortScience, 33, 123–126. [Google Scholar]
- Wang, D. , Jiang, C. , Li, R. & Wang, Y. (2019) VqbZIP1 isolated from Chinese wild Vitis quinquangularis is involved in the ABA signaling pathway and regulates stilbene synthesis. Plant Science, 287, 110202. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Jiang, C. , Liu, W. & Wang, Y. (2020) The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. Journal of Experimental Botany, 71, 3211–3226. [DOI] [PubMed] [Google Scholar]
- Wei, W. , Huang, J. , Hao, Y. , Zou, H. , Wang, H. , Zhao, J. et al. (2009) Soybean GmPHD‐type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS One, 4, e7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W. , Zhang, Y.‐Q. , Tao, J.‐J. , Chen, H.‐W. , Li, Q.‐T. , Zhang, W.‐K. et al. (2015) The Alfin‐like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis . The Plant Journal, 81, 871–883. [DOI] [PubMed] [Google Scholar]
- Wei, W. , Tao, J.J. , Chen, H.W. , Li, Q.T. , Zhang, W.K. , Ma, B. et al. (2017) A histone code reader and a transcriptional activator interact to regulate genes for salt tolerance. Plant Physiology, 175, 1304–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling, R. & Panstruga, R. (2012) Rapid quantification of plant‐powdery mildew interactions by qPCR and conidiospore counts. Plant Methods, 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, D.C.J. & Matus, J.T. (2017) Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine: recent applications and future opportunities. Frontiers in Plant Science, 8, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Liu, M. & Wang, Y. (2020) Expression analysis of three stilbene synthase genes from Chinese wild Vitis quinquangularis . Journal of Agricultural Biotechnology, 28, 847–858. [Google Scholar]
- Xu, W. (2010) Cloning and functional analysis of stilbene synthase and its promoter from powdery mildew‐resistant Chinese Vitis pseudoreticulata. PhD thesis. Yangling, China: Northwest A & F University. [Google Scholar]
- Xu, W. , Ma, F. , Li, R. , Zhou, Q. , Yao, W. , Jiao, Y. et al. (2019) VpSTS29/STS2 enhances fungal tolerance in grapevine through a positive feedback loop. Plant Cell and Environment, 42, 2979–2998. [DOI] [PubMed] [Google Scholar]
- Yin, W. , Wang, X. , Liu, H. , Wang, Y. , Nocker, S. , Tu, M. et al. (2022) Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Horticulture Research, 9, uhab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. & Li, X. (2019) Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Current Opinion in Plant Biology, 50, 29–36. [DOI] [PubMed] [Google Scholar]
- Zhao, K. , Ding, X. & Wang, Y. (2020) Research on control of disease resistance by stilbene synthase genes derived from Chinese wild Vitis quinquangularis . Acta Horticulturae Sinica, 47, 1264–1276. [Google Scholar]
- Zhou, Q. , Dai, L. , Cheng, S. , He, J. , Wang, D. , Zhang, J. et al. (2014) A circulatory system useful both for long‐term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant Cell, Tissue and Organ Culture, 118, 157–168. [Google Scholar]
- Zhu, Y.J. , Agbayani, R. , Jackson, M.C. , Tang, C.S. & Moore, P.H. (2004) Expression of the grapevine stilbene synthase gene VST1 in papaya provides increased resistance against diseases caused by Phytophthora palmivora . Planta, 220, 241–250. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Chen, Q. , Liu, X. & Qu, Y. (2021) The Conringia planisiliqua Alfin‐like 2 gene enhances drought and salt tolerance in Arabidopsis thaliana . Theoretical and Experimental Plant Physiology, 33, 427–441. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sequence analysis of VqNSTS2 and VqSTS37. The amino acid sequence homology between VqNSTS2 and VqSTS37 is 99.49%. There was one amino acid mutation: arginine (R)‐391 to threonine (T). The family signature motifs are indicated in the red boxes. The amino acid mutation is indicated in the pink box
Figure S2 Sequence analysis of VqNSTS3 and VqSTS33. The amino acid sequence homology between VqNSTS3 and VqSTS33 is 99.24%. There were two amino acid mutations: serine (S)‐3 to leucine (L) and serine (S)‐276 to phenylalanine (F). The family signature motifs are indicated in the red boxes. The amino acid mutations are indicated in the pink boxes
Figure S3 Sequence analysis of VqNSTS4 and VqSTS1. The amino acid sequence homology between VqNSTS4 and VqSTS1 is 87.53%. There were 48 amino acid deletions in the N‐terminus. The family signature motifs are indicated in red boxes. The amino acid mutations are indicated in the pink box
Figure S4 Sequence analysis of VqNSTS5 and VqSTS33. The amino acid sequence homology between VqNSTS5 and VqSTS33 is 91.35%. There were 33 amino acid mutations. The family signature motifs are indicated in the red boxes. The amino acid mutations are indicated in the pink boxes. There were two amino acid mutations in the first conserved motif: serine (S)‐50 to threonine (T) and alanine (A)‐51 to glutamine (Q)
Figure S5 Sequence analysis of VqNSTS6 and VqSTS33. The amino acid sequence homology between VqNSTS6 and VqSTS33 is 93.89%. There were 23 amino acid mutations. The family signature motifs are indicated in the red boxes. The amino acid mutations are indicated in the pink boxes. There were two amino acid mutations in the first conserved motif: serine (S)‐50 to threonine (T) and alanine (A)‐51 to glutamine (Q)
Figure S6 Creation and identification of VqNSTS4 transgenic grapevines. (a) Schematic diagram of the 35S‐VqNSTS4‐GFP construct. (b) Creation of VqNSTS4 transgenic grapevines via Agrobacterium‐mediated transformation using proembryonic masses of Vitis vinifera ‘Thompson Seedless’ as material. (c) Plantlets of VqNSTS4 transgenic lines in tissue culture vessels. (d) Western blot assays were conducted to determine the expression of VqNSTS4‐GFP in transgenic lines. (e) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative transcript levels of VqNSTS4 in the transgenic lines. (f) Determination of the contents of five stilbenes in the leaves of VqNSTS4 transgenic lines and wild‐type Thompson Seedless. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01
Figure S7 cis‐Regulatory element analysis of ProVqNSTS2. There are 10 cis‐regulatory elements in ProVqNSTS2: one WRKY transcription factor binding site (a W‐box element, TTGACC, −148 to −143 bp), one MYB transcription factor binding site (CAACAG, −1111 to −1106 bp), two abscisic acid‐responsive elements (ABREs, CACGT, −1190 to −1186 bp and −1216 to −1212 bp), three methyl jasmonate (MeJA)‐responsive elements (an ACE element, CATACATGTC, −243 to −234 bp, an I‐box element, GACCTTGTCAT, −483 to −472 bp, and a G‐box element, GTCGTG, −1172 to −1167 bp), and three light‐responsive elements (a Box4 element, ATTAATT, −420 to −414 bp, a Gap‐box element, CAAATGAACAAAGT, −434 to −421 bp, and a GT1 motif, GGTTAA, −571 to −566 bp)
Figure S8 cis‐Regulatory element analysis of ProVqNSTS3. There are 12 cis‐regulatory elements in ProVqNSTS3: one WRKY transcription factor binding site (a W‐box element, TTGACC, −147 to −142 bp), two MYB transcription factor binding sites (CAACAG and CTGTTG, −796 to −791 bp and −1031 to −1026 bp), one abscisic acid‐responsive element (ABRE, CACGT, −891 to −887 bp), four methyl jasmonate (MeJA)‐responsive elements (an ACE element, CATACATGTCT, −241 to −231 bp, an I‐box element, GACCTTGTCAT, −481 to −471 bp, a G‐box element, CGTGGG, −926 to −921 bp, and a TGACG motif, −1203 to −1199 bp), and four light‐responsive elements (two TCT motifs, GTAAGA, −407 to −402 bp and −611 to −606 bp, a Gap‐box element, CAAATGAACAAAGT, −432 to −419 bp, and a GT1 motif, GGTTAA, −569 to −564 bp)
Figure S9 cis‐Regulatory element analysis of ProVqNSTS4. There are 10 cis‐regulatory elements in ProVqNSTS4: one AL transcription factor binding site (a G‐rich element, CACCTC, −76 to −71 bp) (red box), one WRKY transcription factor binding site (a W‐box element, GGTCAA, −285 to −280 bp), one MYB transcription factor binding site (an MBS element, CAGTTG, −1048 to −1043 bp), two abscisic acid‐responsive elements (ABREs, ACGTG and CACGT, −337 to −333 bp and −999 to −995 bp), three methyl jasmonate (MeJA)‐responsive elements (a G‐box element, CACGTG, −338 to −333 bp, a CGTCA motif, TGACG, −942 to −938 bp, and an ACE element, GACACGTATG, −1,001 to −992 bp), and two light‐responsive elements (a TCT motif, GTAAGA, −664 to −659 bp and a Gap‐box element, TGTTCATTTGA, −889 to −879 bp)
Figure S10 cis‐Regulatory element analysis of ProVqNSTS5. There are six cis‐regulatory elements in ProVqNSTS5: one abscisic acid‐responsive element (ABRE, ACGTG, −227 to −223 bp), one methyl jasmonate (MeJA)‐responsive element (a G‐box element, TACGTG, −228 to −222 bp), and four light‐responsive elements (a Gap‐box element, CAAATGAACAATGT, −407 to −394 bp, two TCT motifs, TCTTAC, −691 to −686 bp and −1169 to −1164 bp, and a GT1 motif, GGTTAA, −1002 to −997 bp)
Figure S11 cis‐Regulatory element analysis of ProVqNSTS6. There are nine cis‐regulatory elements in ProVqNSTS6: one MYB transcription factor binding site (an MBS element, CAACTG, −150 to −145 bp), one WRKY transcription factor binding site (a W‐box element, TTGACC, −960 to −955 bp), one abscisic acid‐responsive element (ABRE, CACGT, −907 to −903 bp), four methyl jasmonate (MeJA)‐responsive elements (two G‐box elements, TACGTG and CACGTGCT, −199 to −194 bp and −907 to −900 bp, a CGTCA motif, TGACG, −255 to −251 bp, and an I‐box element, CCTTATCT, −317 to −310 bp), and two light‐responsive elements (a Gap‐box element, CAAATGAACAAGGG, −360 to −347 bp and a TCT motif, TCTTAC, −581 to −576 bp)
Figure S12 The deletions and mutations in the G‐rich element in ProVqNSTS4. ProVqNSTS4: the promoter of VqNSTS4 contains the G‐rich element CACCTC. ProVqNSTS4 m2 : the CACCTC element has mutated to CAACTC, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS2. ProVqNSTS4 m3 : the CACCTC element has mutated to CCAACT, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS3. ProVqNSTS4 d : CACCTC was deleted in the promoter of VqNSTS4. ProVqNSTS4 m5 : the CACCTC element has mutated to CTCTTC, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS5. ProVqNSTS4 m6 : the CACCTC element has mutated to TTTCTC, which is the sequence at the same position (−71 to −76 bp) in ProVqNSTS6
Figure S13 The sequence of VqAL4. The coding sequence of VqAL4 is 759 bp long. Initiation and termination codons are marked bold and underlined. The conserved sequences are marked in red. This includes eight sites: TGT (595–597 bp), TGC (604–606 bp), TGC (643–645 bp), TGT (652–654 bp), CAT (667–669 bp), TGT (676–678 bp), TGC (724–726 bp), and TGC (733–735 bp)
Figure S14 Expression analysis of VqAL4 in different organs and under different treatments. (a) Root, stem, young leaf, mature leaf, tendril, inflorescence, young berry, and mature berry of Danfeng‐2 were analysed by reverse transcription‐quantitative PCR (RT‐qPCR). (b) Expression analysis of VqAL4 in different organs. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01. (c) Danfeng‐2 leaves were inoculated with Uncinula necator and sampled for RT‐qPCR analysis. (d–g) Danfeng‐2 leaves were treated with 100 μM salicylic acid (SA), methyl jasmonate (MeJA), abscisic acid (ABA), or ethylene (Eth) and sampled for RT‐qPCR analysis. SD was calculated from three independent replicates. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student’s t test)
Figure S15 Creation and identification of VqAL4 transgenic grapevines. (a) Plantlets of VqAL4 transgenic grapevines in tissue culture vessels. (b) Western blot assays were conducted to determine the accumulation of VqAL4‐GFP fusion protein in VqAL4 transgenic lines. (c) Reverse transcription‐quantitative PCR analysis was conducted to determine the relative transcript levels of VqAL4 and VvSTS genes in the VqAL4 transgenic lines. (d) Determination of the contents of five stilbenes in the leaves of VqAL4 transgenic lines and wild‐type Thompson Seedless. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01
Figure S16 VqAL4 bound to the VvSTS32 promoter and promoted VvSTS32 expression. (a) cis‐Regulatory element analysis of the promoters of VvSTS genes. (b) Yeast one‐hybrid assay demonstrating that VqAL4 binds to the VvSTS32 promoter. (c) Graph showing the luminescence intensity. (d) Ratio of firefly luciferase (LUC) to Renilla luciferase (REN) activity. The standard deviation (SD) was calculated from three independent replicates. One‐way analysis of variance (Tukey’s test) was carried out. Asterisks indicate significant differences at *p < 0.05, **p < 0.01. (e) Chromatin immunoprecipitation (ChIP)‐quantitative PCR analysis of VqAL4 binding to the G‐rich motif. The fragment contains the CACCTC element. The ChIP results are presented as percentages of input DNA. SD was calculated from three independent replicates. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, Student’s t test)
Figure S17 cis‐Regulatory element analysis of ProVvSTS7. There are three cis‐regulatory elements in ProVvSTS7: one abscisic acid‐responsive element (ABRE, ACGTG, −547 to −543 bp), one methyl jasmonate (MeJA)‐responsive element (a G‐box element, GACGTG, −548 to −543 bp), and one light‐responsive element (a TCT motif, TCTTAC, −356 to −351 bp)
Figure S18 cis‐Regulatory element analysis of ProVvSTS8. There are three cis‐regulatory elements in ProVvSTS8: two MYB transcription factor binding sites (two MBS elements, CAACTG, −158 to −153 bp and −361 to −356 bp) and one light‐responsive element (a GT1 motif, GGTTAA, −501 to −496 bp)
Figure S19 cis‐Regulatory element analysis of ProVvSTS9. There are three cis‐regulatory elements in ProVvSTS9: one WRKY transcription factor binding site (a W‐box element, GGTCCA, −550 to −545 bp), one abscisic acid‐responsive element (ABRE, GACACCTGCG, −732 to −723 bp), and one methyl jasmonate (MeJA)‐responsive element (a TGACG motif, −560 to −556 bp)
Figure S20 cis‐Regulatory element analysis of ProVvSTS25. There are two cis‐regulatory elements in ProVvSTS25: two WRKY transcription factor binding sites (two W‐box elements, TTGACC, −369 to −364 bp and −760 to −755 bp)
Figure S21 cis‐Regulatory element analysis of ProVvSTS27. There are two cis‐regulatory elements in ProVvSTS27: one MYB transcription factor binding site (an MBS element, CAGTTG, −614 to −609 bp) and one light‐responsive element (a GT1 motif, GGTTAA, −727 to −722 bp)
Figure S22 cis‐Regulatory element analysis of ProVvSTS29. There is one cis‐regulatory element in ProVvSTS29: one MYB transcription factor binding site (an MBS element, CAGTTG, −612 to −607 bp)
Figure S23 cis‐Regulatory element analysis of ProVvSTS32. There are four cis‐regulatory elements in ProVvSTS32: one AL transcription factor binding site (a G‐rich element, CACCTC, −77 to −72 bp) (red box), two WRKY transcription factor binding sites (two W‐box elements, GGTCAA, −286 to −281 bp and −534 to −529 bp), and one abscisic acid‐responsive element (ABRE, ACGTG, −338 to −334 bp)
Table S1 Sequence analysis of VqNSTS genes
Table S2 cis‐Regulatory element analysis of ProVqNSTS genes
Table S3 Primers used in this study
Data Availability Statement
The data that support the results are included in this article and its supplementary materials.


