Abstract
Clostridium perfringens enterotoxin is the major virulence factor involved in the pathogenesis of C. perfringens type A food poisoning and several non-food-borne human gastrointestinal illnesses. The enterotoxin gene, cpe, is located on the chromosome of food-poisoning isolates but is found on a large plasmid in non-food-borne gastrointestinal disease isolates and in veterinary isolates. To evaluate whether the cpe plasmid encodes its own conjugative transfer, a C. perfringens strain carrying pMRS4969, a plasmid in which a 0.4-kb segment internal to the cpe gene had been replaced by the chloramphenicol resistance gene catP, was used as a donor in matings with several cpe-negative C. perfringens isolates. Chloramphenicol resistance was transferred at frequencies ranging from 2.0 × 10−2 to 4.6 × 10−4 transconjugants per donor cell. The transconjugants were characterized by PCR, pulsed-field gel electrophoresis, and Southern hybridization analyses. The results demonstrated that the entire pMRS4969 plasmid had been transferred to the recipient strain. Plasmid transfer required cell-to-cell contact and was DNase resistant, indicating that transfer occurred by a conjugation mechanism. In addition, several fragments of the prototype C. perfringens tetracycline resistance plasmid, pCW3, hybridized with pMRS4969, suggesting that pCW3 shares some similarity to pMRS4969. The clinical significance of these findings is that if conjugative transfer of the cpe plasmid occurred in vivo, it would have the potential to convert cpe-negative C. perfringens strains in normal intestinal flora into strains capable of causing gastrointestinal disease.
Clostridium perfringens is an important cause of enteric and histotoxic infections in both humans and animals and produces several potent toxins (22). The alpha-toxin structural gene is invariably located on the chromosome (10), but the genes encoding the beta-, epsilon-, and iota-toxins are located on large plasmids (10, 21).
C. perfringens enterotoxin (CPE) ranks among the most medically important C. perfringens toxins. Although representing only less than 5% of all C. perfringens isolates, CPE-positive type A strains are significant enteric pathogens. In humans, they cause C. perfringens type A food poisoning, which is one of the most common food-borne illnesses in developed countries, and 5 to 20% of all cases of non-food-borne human gastrointestinal (GI) illness, including antibiotic-associated diarrhea and sporadic diarrhea (12, 19). Recent studies (24) have demonstrated that CPE expression is necessary for the pathogenesis of both C. perfringens type A food poisoning and non-food-borne GI disease isolates. In those studies, isogenic cpe knockout mutants were prepared from SM101, a transformable derivative of the food poisoning isolate NCTC8798, which carries a chromosomal cpe gene, and F4969, a non-food-borne GI disease isolate carrying a plasmid cpe gene. Both mutants were constructed by replacing a 0.4-kb segment internal to the cpe gene with the chloramphenicol resistance gene catP. Although sporulating, but not vegetative, culture lysates of both wild-type parents induced significant rabbit ileal loop fluid accumulation and histopathologic damage, neither sporulating nor vegetative culture lysates of the cpe knockout mutants induced either effect. However, virulence was restored when these mutants were complemented with a recombinant plasmid carrying the wild-type cpe gene, confirming that loss of virulence can be specifically attributed to inactivation of the cpe gene and the resultant loss of sporulation-associated CPE production (24).
In human food poisoning isolates, the chromosomal cpe gene is located on what appears to be a transposable genetic element, Tn5565 (8, 9). In contrast, non-food-borne human GI disease isolates and animal isolates carry the cpe gene on large plasmids (11, 13, 27). These plasmids have not yet been fully characterized but appear to vary somewhat in size (average, ≈100 kb) and carry IS elements. Other C. perfringens toxin types occasionally carry cpe plasmids, which may have both IS elements and genes encoding other toxins (15).
There is circumstantial evidence suggesting that the large cpe plasmids are conjugative. Firstly, type A strains carrying the cpe plasmid do not share the same genetic background (12). Secondly, several nonclonal type E isolates have been found to carry a plasmid with a defective but highly conserved copy of the cpe gene as well as the iota-toxin genes iap and ibp (5). The objective of this study was to test the hypothesis that the cpe plasmids are conjugative.
The cpeΩcatP plasmid pMRS4969 is conjugative.
Our approach was to use mixed plate matings to transfer pMRS4969, a plasmid that carries a cpe gene inactivated by deletion of internal cpe sequences and insertion of the catP gene (24). To monitor the transfer of the cpe plasmid, C. perfringens strain MRS4969, which carries the cpeΩcatP plasmid pMRS4969, was initially used as a donor in mixed plate matings (23) with several genetically marked C. perfringens recipient strains. The C. perfringens strains (Table 1) were cultured in various media, as previously described (20, 23). When required, appropriate concentrations of rifampin (1 or 20 μg/ml), nalidixic acid (20 μg/ml), chloramphenicol (5 or 15 μg/ml), or streptomycin (1,000 μg/ml) were added to the medium. Agar plates were grown in a 10% H2–10% CO2–80% N2 atmosphere. For the matings, single colonies of the donor and recipient strains were grown to exponential phase in fluid thioglycollate broth and 100 μl of each culture was mixed and spread on the surface of a nonselective agar plate. After incubation overnight, the cells were resuspended in 3 ml of brain heart infusion broth and 10-fold dilutions were plated onto the appropriate media for selection of the transconjugants.
TABLE 1.
C. perfringens strains
| Strain | Descriptiona | Reference or source |
|---|---|---|
| F4969 | Wild-type CPE+ | 24 |
| MRS4969 | F4969(pMRS4969) Cmr | 24 |
| 13 | Wild type | 18 |
| CW362 | Wlld type | 23 |
| CW504 | CW362 Rifr Nalr | 23 |
| JIR39 | CW362 Chlr Smr | 20 |
| JIR100 | Emr | 4 |
| JIR325 | 13 Rifr Nalr | 17 |
| JIR458 | 13 Smr | T. Bannam and J. Rood |
| JIR4468 | JIR325(pMRS4969) Cmr | Transformant |
| JIR4475 | JIR458(pMRS4969) Cmr | Transconjugant, JIR4468 × JIR458 |
| JIR4479 | JIR325(pMRS4969) Cmr | Transconjugant, JIR4475 × JIR325 |
| JIR4481 | JIR39(pMRS4969) Cmr | Transconjugant, JIR4468 × JIR39 |
Rif, rifampin; Nal, nalidixic acid; Chl, potassium chlorate; Sm, streptomycin; Cm, chloramphenicol; Em, erythromycin.
Unfortunately, MRS4969 produced a substance, presumably a bacteriocin, that killed or inhibited the growth of the potential recipients. Therefore, no transconjugants were obtained in these matings. To move the plasmid into a cleaner background, pMRS4969 was purified from MRS4969 using a cesium chloride-ethidium bromide density gradient (1), without NaCl treatment of the lysate, and used to transform the cpe-negative strain JIR325 to chloramphenicol resistance. Transformation of C. perfringens cells was carried out by electroporation (26) in 0.2-cm Gene Pulser cuvettes (Bio-Rad) at 2.5 kV, 25 μF, and 200 Ω. JIR325 is a rifampin- and nalidixic acid-resistant derivative of strain 13, the transformable C. perfringens strain most commonly used in genetic studies (17). The presence of the cpeΩcatP gene region in the resultant transformant, JIR4468, was confirmed by PCR (Fig. 1) and Southern hybridization analyses (Fig. 2).
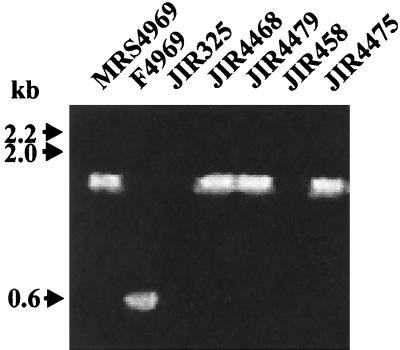
FIG. 1.
PCR analysis of transconjugants. Fresh cells from agar plates were resuspended in 100 μl of H2O and lysed in a microwave oven for 3 min. Three microliters of the supernatant was used in a standard PCR (50-μl volume) in a DNA thermal cycler (Perkin-Elmer) with 30 cycles of 30 s at 94°C, 30 s at 50°C, and 90 s at 72°C. The primers were cpeR (5′-CATCACCTAAGGACTGTTCT-3′) and cpeF (5′-TGTAGAATATGGATTTGGAAT-3′). The resultant products were separated by agarose gel electrophoresis. As shown, wild-type cpe-positive cells yielded a cpe band with a size of 544 bp, whereas a 1.7-kb band was present in strains with the cpeΩcatP region. Molecular sizes of the DNA markers are shown on the left.
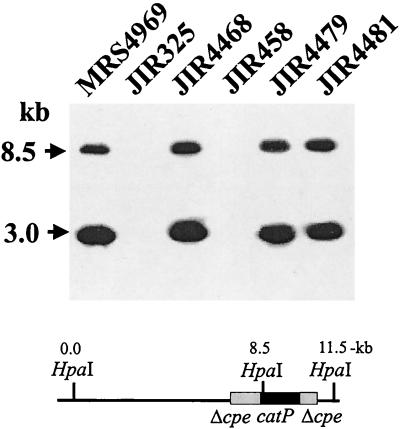
FIG. 2.
Southern blot of parent strains and transconjugants. C. perfringens DNA was isolated as described previously (14), digested with HpaI, separated by electrophoresis on a 1% agarose gel, and transferred by Southern blotting to positively charged nylon membranes (Roche Molecular Biochemicals). The membrane was then hybridized at 68°C overnight with a digoxigenin-labeled cpe probe. To prepare the probe, an ≈1.6-kb fragment containing the cpe gene and the regions ≈300-bp upstream and ≈200-bp downstream, was gel purified from EcoRI- and XbaI-digested pJRC200 DNA (14) and labeled by using a random-primed DNA labeling system (Boehringer Mannheim). After hybridization, the membrane was washed twice for 15 min each in wash solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% sodium dodecyl sulfate) at room temperature. The final washes were twice for 15 min each in 0.5× wash solution (0.5× SSC–0.1% sodium dodecyl sulfate) at 68°C. Hybridized probe was then detected by using a digoxigenin-chemiluminescence detection system with CSPD substrate (Roche Molecular Biochemicals). Molecular sizes of the DNA markers are shown on the left. The map shows the relevant cpeΩcatP region of pMRS4969.
When JIR4468 was mated with JIR458, a streptomycin-resistant derivative of strain 13, chloramphenicol- and streptomycin-resistant transconjugants were observed at consistently high frequencies ranging from 9.0 × 10−2 to 1.5 × 10−3 transconjugants/donor cell. A transconjugant from this mating, JIR4475, was also capable of transferring chloramphenicol resistance in subsequent matings performed by using JIR325 as the recipient and by selecting transconjugants on medium containing chloramphenicol, rifampin, and nalidixic acid. When a JIR325-derived transconjugant from this JIR4475 × JIR325 mating, JIR4479, was used as a donor, the frequency of transfer of chloramphenicol resistance to JIR458 was essentially the same as that observed in the original mating. Matings were also performed with other genetically marked C. perfringens recipients, JIR100, JIR39, and CW504, which have different genetic backgrounds. High-frequency transfer of chloramphenicol resistance was again observed (data not shown). Note that in each of the mating experiments, no colonies were observed on selection plates that contained only the donor or recipient strains. The presence of cpeΩcatP gene region in the resultant transconjugants was confirmed by PCR analysis using cpe primers located on either side of the catP gene (Fig. 1). The recipient strains were negative in this PCR whereas the transconjugants had a PCR band of the same size as that from the donor strain.
The mechanism of transfer of chloramphenicol resistance appeared to involve a conjugation-like mechanism since cell-to-cell contact between the donor and recipient was required. No chloramphenicol- and streptomycin-resistant colonies were obtained when culture supernatant from the donor was mixed with JIR325 cells or when a 0.45-μm-pore-size Millipore membrane was placed between the donor and recipient strains. Furthermore, carrying out the matings in the presence of DNase I (1 mg/plate) had no effect on the frequency of transfer of the chloramphenicol resistance determinant.
Evidence that the transconjugants carry pMRS4969.
Purification of large plasmids from C. perfringens cultures is a difficult task. Therefore, total genomic DNA was prepared from strains MRS4969, JIR325, JIR458, and JIR4468 and two additional independently derived transconjugants, JIR4475 and JIR4481. After digestion with HpaI, Southern blots were hybridized with a cpe-specific probe (Fig. 2). No detectable hybridization signal was observed with HpaI-digested DNA prepared from the cpe-negative recipient strains JIR325 and JIR458; however, as expected from previous observations (24), two hybridizing bands (3.0 and 8.5 kb) were observed with HpaI-digested DNA from the original parent strain MRS4969. An identical hybridization pattern was seen with the pMRS4969 transformant, JIR4468, which was subsequently used as a donor in the matings, as well as the transconjugants JIR4479 and JIR4481 (Fig. 2). These results were as predicted for transconjugant strains derived from the conjugative transfer of pMRS4969.
To confirm these results and to determine whether the intact pMRS4969 plasmid had been transferred, pulsed-field gel electrophoresis (PFGE) was carried out with these strains, and the resultant gel was Southern blotted and hybridized with the cpe-specific probe. In the absence of digestion with restriction endonucleases, only plasmid DNA molecules are able to enter the gel. The results showed that a single plasmid DNA band was detected in MRS4969 (Fig. 3). A hybridizing band of the same size was present in the transconjugants JIR4479 and JIR4481 but not in the recipient strains JIR325 (Fig. 3) and JIR458 (data not shown), providing clear evidence that pMRS4969 has been transferred intact from the donor strains to the recipients in these experiments.
FIG. 3.
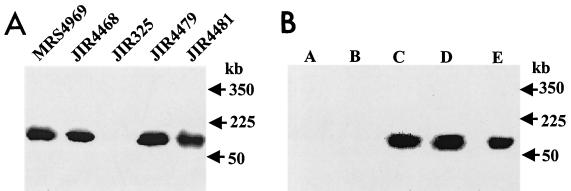
PFGE/Southern hybridization analysis of undigested genomic DNA from parent strains and transconjugants. (A) Total undigested DNA from the strains indicated was separated by pulsed-field gel electrophoresis as previously described (5), with a Bio-Rad CHEF-DR II apparatus, where pulse times were ramped from 50 to 90 s over a period of 22 h. The gel was Southern blotted and probed with a 1.6-kb digoxigenin-labeled cpe-specific probe. Molecular sizes of the DNA markers are shown on the right. (B) Total undigested DNA from transconjugant JIR4479 was separated by pulsed-field gel electrophoresis, and the gels were Southern blotted and separately probed with the five digoxigenin-labeled ClaI fragments (lanes A through E) originally derived from pCW3 (see Fig. 4). For reference, the molecular sizes of the λ DNA standards (Bio-Rad) are shown on the right.
The cpe plasmid pMRS4969 hybridizes to pCW3.
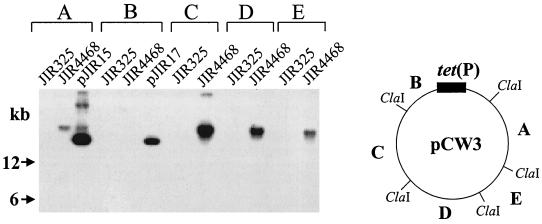
Further hybridization studies were carried out to see if pMRS4969 had any regions of sequence similarity to pCW3 (1, 23), the conjugative tetracycline resistance plasmid that is homologous to all known conjugative C. perfringens plasmids (1–3, 16). Southern blots of BamHI-digested total DNA preparations from JIR325 and JIR4468 were hybridized separately with digoxigenin-labeled probes prepared from separate recombinant plasmids (pJIR15 to pJIR18 and pJIR32), each of which carries one of the five ClaI fragments present in pCW3 (1). None of the five ClaI-derived probes hybridized with DNA from JIR325. However, all of the probes except pJIR17 hybridized to a >20-kb BamHI fragment from JIR4468, with the hybridization of pJIR15 being noticeably weaker (Fig. 4).
FIG. 4.
Hybridization to pCW3-derived probes. Total DNA isolated from strains JIR325 and JIR4468 was digested with BamHI, Southern blotted, and probed separately with the five pCW3-derived ClaI fragments that are present in the recombinant plasmids (1) pJIR15 (A), pJIR17 (B), pJIR16 (C), pJIR18 (D), and pJIR32 (E), as indicated. The pCW3 circular map shows the location of the five ClaI fragments (A through E).
Hybridization analysis of PFGE blots was used to confirm these results. In these experiments, the pJIR16, pJIR18, and pJIR32 probes hybridized to pMRS4969, but no hybridization was observed with pJIR15 or pJIR17 (Fig. 3).
The cpe+ parent plasmid of pMRS4969 represents the first conjugative virulence plasmid to be identified from C. perfringens. All other known conjugative plasmids from C. perfringens confer tetracycline resistance and have substantial similarity to pCW3 (2, 3). Although pMRS4969 does not confer tetracycline resistance, it still has significant similarity to pCW3. The two pCW3-derived ClaI fragments that have little or no similarity to pMRS4969 encode the tetracycline resistance determinant, which is found on pJIR17 (1), and the replication region, which is located on pJIR15 (P. Johanesen, K. Koutsis, D. Lyras, and J. I. Rood, unpublished data). It is tempting to speculate that the regions common to pMRS4969 and pCW3 are involved in conjugation, implying that these plasmids share a common conjugation mechanism. However, detailed DNA sequence analysis and genetic studies of both pCW3 and pMRS4969 need to be carried out before this hypothesis can be confirmed.
Implications for CPE-mediated diseases.
Recent studies (25) suggest that the specific association between chromosomal cpe isolates and C. perfringens type A food poisoning is attributable, at least in part, to the chromosomal cpe isolates being considerably more heat resistant than plasmid cpe isolates, which favors their survival in the cooked foods that constitute the primary vehicle for C. perfringens type A food poisoning (19). The basis for the strong association between plasmid cpe isolates and CPE-associated non-food-borne human GI diseases has remained unclear. However, by demonstrating that the cpe plasmid can be conjugatively transferred between C. perfringens isolates, our present investigation offers a potential explanation for the relationship between isolates that carry the cpe plasmid and CPE-associated non-food-borne human GI diseases. That is, in vivo conjugative transfer of the cpe plasmid to normal intestinal flora isolates of C. perfringens may be important for establishing CPE-associated non-food-borne GI infections. Unlike C. perfringens type A food poisoning, which occurs upon ingestion of food containing massive numbers of C. perfringens cells that have a chromosomal cpe gene (19), CPE-associated non-food-borne human GI diseases appear to result from the ingestion or inhalation of low numbers of cpe-positive C. perfringens spores present in the environment (7). It is possible that the relatively few vegetative cells arising from the germination of these spores have the ability to transfer their cpe plasmid by conjugation to the much more abundant, but naturally cpe-negative, normal flora strains of C. perfringens already present in the GI tract. This process would convert these normal flora isolates to strains capable of causing enteric disease. Since the normal flora are well-adapted for intestinal colonization and growth, this in vivo conversion would hasten disease onset and may even be required to reach the in vivo levels of cpe-positive C. perfringens cells necessary for initiating enteric disease.
In vivo conjugative transfer of the cpe plasmid might also help explain why the GI symptoms derived from the non-food-borne isolates typically are more severe and longer in duration than those of C. perfringens type A food poisoning (6). Since C. perfringens strains in normal flora are proficient in their ability to colonize and persist in the GI tract, conjugative acquisition of the cpe plasmid by these strains may increase the duration of GI infection. One consequence might be more chronic exposure to CPE, which could explain the more severe symptoms associated with CPE-associated non-food-borne diseases relative to those observed during acute C. perfringens type A food poisoning. The latter syndrome is caused by exogenous strains that are not necessarily well adapted to long-term survival in the human GI tract.
Acknowledgments
S. Brynestad and M. R. Sarker have contributed equally to the work and are joint first authors.
This work was supported by research grant 12097/130 to S.B. from the Research Council of Norway, NIH grant 19844-18 and USDA grant 98-02822 to B.A.M., and research grants to J.I.R. from the Australian National Health and Medical Research Council.
We thank Kylie Farrow for assistance with the figures.
REFERENCES
- 1.Abraham L J, Rood J I. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985;13:155–162. doi: 10.1016/0147-619x(85)90038-1. [DOI] [PubMed] [Google Scholar]
- 2.Abraham L J, Rood J I. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J Bacteriol. 1985;161:636–640. doi: 10.1128/jb.161.2.636-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham L J, Wales A J, Rood J I. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985;14:37–46. doi: 10.1016/0147-619x(85)90030-7. [DOI] [PubMed] [Google Scholar]
- 4.Berryman D I, Lyristis M, Rood J I. Cloning and sequence analysis of ermQ, the predominant macrolide-lincosamide-streptogramin B resistance gene in Clostridium perfringens. Antimicrob Agents Chemother. 1994;38:1041–1046. doi: 10.1128/aac.38.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billington S J, Wieckowski E U, Sarkar M R, Bueschel D, Songer J G, McClane B A. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin gene sequences. Infect Immun. 1998;66:4531–4536. doi: 10.1128/iai.66.9.4531-4536.1998. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Borriello S P. Clostridial disease of the gut. Clin Infect Dis. 1995;20(Suppl. 2):S242–S250. doi: 10.1093/clinids/20.supplement_2.s242. [DOI] [PubMed] [Google Scholar]
- 7.Borriello S P, Carman R J. Clostridial diseases of the gastrointestinal tract in animals. In: Borriello S P, editor. Clostridia in gastrointestinal disease. Boca Raton, Fla: CRC Press; 1985. pp. 195–215. [Google Scholar]
- 8.Brynestad S, Granum P E. Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol Lett. 1999;170:281–286. doi: 10.1111/j.1574-6968.1999.tb13385.x. [DOI] [PubMed] [Google Scholar]
- 9.Brynestad S, Synstad B, Granum P E. The Clostridium perfringens enterotoxin gene is on a transposable genetic element in type A human food poisoning strains. Microbiology. 1997;143:2109–2115. doi: 10.1099/00221287-143-7-2109. [DOI] [PubMed] [Google Scholar]
- 10.Canard B, Saint-Joanis B, Cole S T. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992;6:1421–1429. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 11.Collie R, McClane B. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. Anaerobe. 1998;4:69–79. doi: 10.1006/anae.1998.0152. [DOI] [PubMed] [Google Scholar]
- 12.Collie R E, McClane B A. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-foodborne human gastrointestinal diseases. J Clin Microbiol. 1998;36:30–36. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornillot E, Saint-Joanis B, Daube G, Katayama S-I, Granum P E, Canard B, Cole S T. The enterotoxin gene (cpe) of Clostridum perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995;15:639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 14.Czeczulin J R, Collie R E, McClane B A. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect Immun. 1996;64:3301–3309. doi: 10.1128/iai.64.8.3301-3309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama S, Dupuy B, Daube G, China B, Cole S T. Genome mapping of Clostridium perfringens strains with I-CeuI shows many virulence genes to be plasmid-borne. Mol Gen Genet. 1996;251:720–726. doi: 10.1007/BF02174122. [DOI] [PubMed] [Google Scholar]
- 16.Lyras D, Rood J I. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob Agents Chemother. 1996;40:2500–2504. doi: 10.1128/aac.40.11.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyristis M, Bryant A E, Sloan J, Awad M M, Nisbet I T, Stevens D L, Rood J I. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol. 1994;12:761–777. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 18.Mahony D E, Moore T J. Stable L-forms of Clostridium perfringens and their growth on glass surfaces. Can J Microbiol. 1976;22:953–959. doi: 10.1139/m76-138. [DOI] [PubMed] [Google Scholar]
- 19.McClane B A. Clostridium perfringens. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. 2nd ed. 2001. , in press. ASM Press, Washington, D.C. [Google Scholar]
- 20.Rood J I. Transferable tetracycline resistance in Clostridium perfringens strains of porcine origin. Can J Microbiol. 1983;29:1241–1246. doi: 10.1139/m83-193. [DOI] [PubMed] [Google Scholar]
- 21.Rood J I. Virulence genes of Clostridium perfringens. Annu Rev Microbiol. 1998;52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 22.Rood J I, Cole S T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rood J I, Scott V N, Duncan C L. Identification of a transferable resistance plasmid (pCW3) from Clostridium perfringens. Plasmid. 1978;1:563–570. doi: 10.1016/0147-619x(78)90013-6. [DOI] [PubMed] [Google Scholar]
- 24.Sarker R M, Carman R J, McClane B A. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol. 1999;33:946–958. doi: 10.1046/j.1365-2958.1999.01534.x. [DOI] [PubMed] [Google Scholar]
- 25.Sarker R M, Shivers R P, Sparks S G, Juneja V K, McClane B A. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl Environ Microbiol. 2000;66:3234–3240. doi: 10.1128/aem.66.8.3234-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Scott P T, Rood J I. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene. 1989;82:327–333. doi: 10.1016/0378-1119(89)90059-0. [DOI] [PubMed] [Google Scholar]
- 27.Sparks S, Carman R, Sarker M, McClane B A. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J Clin Microbiol. 2001;39:883–888. doi: 10.1128/JCM.39.3.883-888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]