Abstract
Aim
Gastric mucosal changes associated with chronic gastritis are known to be precancerous lesions of gastric cancer. We aimed to identify individuals with a high risk of gastric cancer by detection of microRNAs (miRNA) in the blood as biomarkers.
Methods
Of 1206 individuals screened, 144 who were positive for Helicobacter pylori (H. pylori) by the serum antibody test and who underwent endoscopy were the subjects of this study. For the gross assessment of mucosal inflammation, we applied the Kimura–Takemoto classification, in which normal mucosa was defined as grade 0, and atrophy was categorized as grade 1 (C‐1 and C‐2), grade 2 (C‐3 and O‐1), and grade 3 (O‐2 and O‐3). Serum samples were divided into two phases and used for miRNA microarray profiling. We compared the expression of miRNAs in grade 3 mucosa and other grades. Expression in gastric cancer was confirmed with TCGA data.
Results
miR‐196b‐3p was significantly upregulated, and miR‐92a‐2‐5p was downregulated (P < .05 and q < 0.2). TCGA data showed a high expression of miR‐196b‐3p in gastric cancer cases (P < .001). Comparing grade 3 and the others, the area under the receiver operating characteristic curve using the detected miRNAs was as high as about 0.7. Furthermore, the combination of miRNAs resulted in higher accuracy. In terms of the significance of the combinatory mRNAs, the combination of three miRNAs (miR‐196b‐3p, miR‐92a‐2‐5p, and miR‐6791‐3p) revealed high sensitivity and specificity, with the area under the curve exceeding 0.8.
Conclusion
The identified combinatory miRNAs may represent promising biomarkers of precancerous lesions in gastric cancer.
Keywords: 3‐dimensional microarray, gastric cancer, Kimura–Takemoto classification, miR‐196b, precancerous lesion, serum biomarker
We found that microRNA‐196b‐5p was significantly upregulated in precancerous lesions and that combining it with other microRNAs increased the accuracy of detection. Since endoscopy is associated with risk, the identification of high‐risk groups for gastric cancer is expected to make it possible to clearly identify groups that should undergo endoscopy.

1. INTRODUCTION
Gastric cancer (GC) is one of the essential cancers globally, with more than 1 million new cases and an estimated 783 000 deaths in 2018. 1 Helicobacter pylori (H. pylori) is the leading risk factor for GC, with almost 90% of new cases attributed to this organism. 2 GC is often detected in an advanced stage because it lacks characteristic markers and symptoms. The 5‐y survival rate is 81.6% for Stage I, 59.3% for Stage II, 39.6% for Stage III, and 8.0% for Stage IV. 3 Thus, if we could detect GC at a much earlier stage, we could improve its prognosis and provide patients with curable treatment with minimally invasive surgery, such as endoscopic mucosal resection.
In countries with a high prevalence of GC, such as Japan, China, and Korea, screening is commonly performed. Endoscopy and gastric fluoroscopy are the two most common methods; endoscopy is reported to be more sensitive, 4 and it reduces the cancer mortality rate. 5 As for the GC diagnosis, current medical practice requires observation by endoscopy and pathologic diagnosis by tissue biopsy. Although the risks associated with endoscopy are lower than in the past, it is still a painful and invasive test that requires the documented consent of the patient. The risk of severe complications, such as bleeding, perforation, and infection was 0.78% in the past 3 y in Japan, which is very low. 6 Almost no deaths were associated with endoscopy; unfortunately, however, sedation‐related deaths have been reported. Some reports have shown that sedation is associated with a risk of severe complications. 7 Over‐diagnosis by endoscopy is another problem, and frequent examinations are considered undesirable. 8 Therefore, a screening test is needed to identify individuals with a high risk of GC who would require endoscopic examination.
The gastritis‐metaplasia‐carcinoma sequence is known as the GC carcinogenesis model. 9 Gastric mucosal changes (atrophic changes, including intestinal epithelialization) associated with chronic inflammation are considered precancerous lesions of GC, and inflammation of the gastric mucosa associated with H. pylori is involved in carcinogenesis through this pathway. 10 For the assessment of gastric atrophy, histologic evaluation is the gold standard. However, the Kimura–Takemoto classification, which was established in the 1960s, has been used as an assessment of gross gastric inflammation during endoscopy, and the results are consistent with the histologic diagnosis. 11 This classification is based on detecting atrophic boundaries in the gastric mucosa. Kishino et al 12 and Song et al 13 further proposed scoring for endoscopic atrophic gastritis, and we used that classification in this study. Since the risk of GC correlates with the severity of atrophic gastritis, severe atrophic gastritis is commonly recognized as a precancerous lesion. 14 In studies with post‐detection follow‐up, results showed that the incidence of GC was predominantly higher in the group with a higher gastritis grade. 13
MicroRNAs (miRNAs) are 18–25‐base RNAs that do not encode proteins. 15 Recently, they have attracted attention because they are very stable and easy to detect in blood. 16 They can be identified by collecting blood and performing polymerase chain reaction (PCR) analysis, and the results are reproducible. miRNAs are expected to be biomarkers for some cancers. 16 An association between GC and various miRNAs has been shown. 17 , 18 There have been several reports on miRNA expression in precancerous lesions of GC. 19 , 20 However, the results have not been verified sufficiently, and there are no reports of comprehensive analysis using microarrays.
In this study, we conducted microarray experiments using blood samples and focused on miRNAs characteristically detected in patients with chronic atrophic gastritis as precancerous lesions of GC. We identified cases of severe atrophy of the stomach with H. pylori infection.
2. MATERIALS AND METHODS
2.1. Patients
The study included 1206 people who underwent screening at the USA‐Takata Regional Adult Health Screening Center in 2017. As previously described in a prospective study conducted in the same region, 21 we defined positive cases as those with anti‐H. pylori IgG antibody titers of 3 or higher. In total, 379 patients (32.8%) showed positivity and were subjected to additional examinations. Of the 155 subjects who underwent endoscopy, one patient was found to have GC and was excluded. The study was conducted using blood samples from 144 subjects graded for gastritis (Figure 1). Consent for the study was obtained from all eligible subjects, and information on age and sex was obtained. Residual sera (2 mL) from blood samples collected for physical examination were used for miRNA analysis.
FIGURE 1.
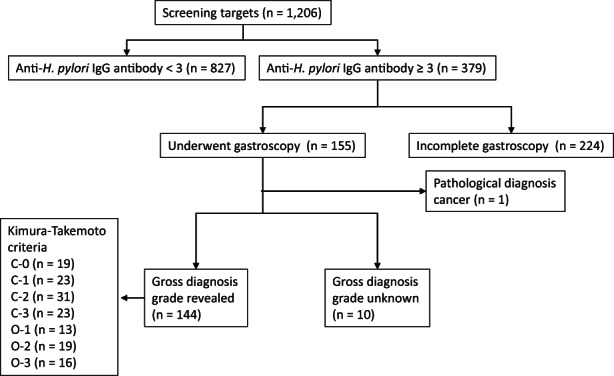
Subject selection. One thousand two hundred six patients underwent screening at the USA‐Takata regional adult health screening center during 2017. There were 379 subjects (32.8%) whose anti‐H. pylori IgG antibodies were three or higher and required further examination. Of the 155 subjects who underwent endoscopy, one was found to have GC and was excluded. The study was conducted using blood samples from 144 subjects graded for gastritis. The Kimura–Takemoto classification was used for visual evaluation. Among the 144 subjects, there were 19 with C‐0, 23 with C‐1, 31 with C‐2, 23 with C‐3, 13 with O‐1, 19 with O‐2, and 16 with 0‐3
The Ethics Committee of the Graduate School of Kyushu University approved the study (Review Number: 597‐00).
2.2. Endoscopic evaluation
The Kimura–Takemoto classification was used for evaluation of the gastric mucosa. 11 This classification is divided into two main types: Closed type (C type) and Open type (O type). The normal gastric mucosa is C‐0, and the two main types are further divided into C‐1,2,3 and O‐1,2,3. 11 The characteristics of each type of atrophy are as follows: C‐1, atrophic mucosa limited to the antrum; C‐2, limited to the gastric angle or the lower corpus; C‐3, limited to the upper canon; O‐1, limited to the surroundings of the gastric cardia, with maintained folds of the greater curvature; O‐3, present in the entire stomach, with lack of folds in the greater curvature as a whole; O‐2, an intermediate type between O‐1 and O‐3. 12 , 13 Additional biopsies were performed if suspicious neoplastic lesions were seen during endoscopy. Four experienced endoscopists reviewed all endoscopic findings. Several meetings were held to minimize interobserver variability.
2.3. miRNA expression microarray analysis
Serum collection and preparation were performed according to the standard operating procedures of the Early Detection Research Network of the National Cancer Institute (https://edrn.nci.nih.gov/). Total RNA was extracted from 300 μL of serum with the 3D‐Gene RNA extraction reagent of a liquid sample kit (Toray Industries, Tokyo, Japan). Comprehensive miRNA expression analysis was performed using a 3D‐Gene miRNA 6iRbase6g kit (Toray Industries) and a 3D‐Gene Human miRNA Oligo chip (Toray Industries), which was designed to detect 2555 miRNA sequences registered in 6iRbase release 20 (http://www.mirbase.org/). If the microarray signal was more significant than the (mean + 2 × standard deviation) signal of the negative control with the top and bottom 5% of the signal intensity removed, respectively, the miRNA was considered to be present. When miRNAs were deemed to be present, the average signal of the negative control from which the top and bottom 5% of the signal intensity were removed was subtracted from the miRNA signal. If the signal value after background subtraction was negative or undetectable, the lowest signal intensity on the microarray was replaced by a matter of minus 0.1 on the bottom two logarithmic scales. The miRNA expression profiles were quantile normalized to normalize the signal between different microarrays, and the ComBat method was applied to remove batch effects.
2.4. miRNA expression data of gastric cancer tissue from a public database
Using the TCGA gastric cancer dataset (https://tcga‐data.nci.nih.gov/tcga/), we compared the miRNA expression in 95 normal gastric mucosal tissues and 441 gastric cancer tissues. We obtained single nucleotide polymorphism (SNP) array data of genes and miRNAs from the Firehose pipeline at the Broad Institute. The copy number of miRNAs was calculated from SNP array data using the ASCAT algorithm and R software version 3.3.2 (The R Foundation, Vienna, Austria). The Broad Institute’s Firehose can be accessed at http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/STAD/20160128/.
2.5. Statistical analysis
The expression profiles of serum miRNAs were compared between the two groups with Fisher’s Exact test. For receiver operating characteristic (ROC) curve analysis, the cutoff value was determined by the point on the ROC curve that was the minimum distance from the upper left corner on the unit square. Fisher’s linear discriminant analysis was performed to calculate the diagnostic sensitivity, specificity, and accuracy. Results with P values less than .05 were considered statistically significant.
3. RESULTS
3.1. Characteristics of the study population
Among the 144 subjects, there were 19 with C‐0, 23 with C‐1, 31 with C‐2, 23 with C‐3, 13 with O‐1, 19 with O‐2, and 16 with 0‐3 (Figure 1). The previous classification was as follows: C‐0 was defined as grade 0, C‐1 and C‐2 as grade 1, C‐3 and O‐1 as grade 2, and O‐2 and O‐3 as grade 3. 12 , 13 Thus, there were 19 subjects with grade 0, 54 subjects with grade 1, 35 subjects with grade 2, and 36 subjects with grade 3. Age and sex were examined for each grade, but no significant differences between groups were observed (Figure 2A). All subjects were divided into the Discovery phase (n = 71) and the Training phase (n = 73; Figure 2B). Grade 3 in this classification is an advanced stage of chronic gastritis, which is defined as a precancerous lesion of GC. 13
FIGURE 2.
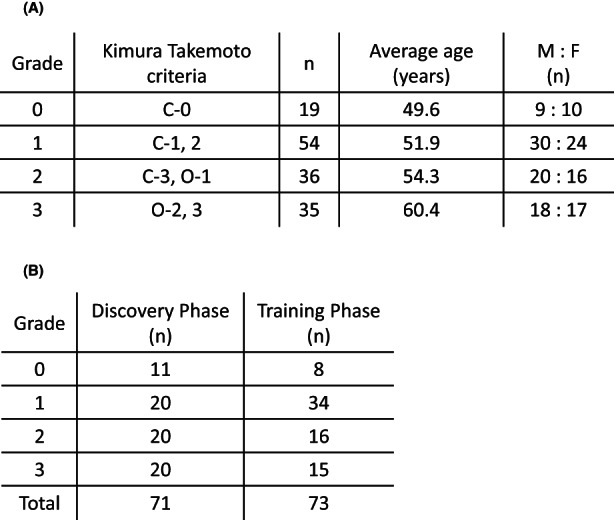
Characteristics of the study population. We divided the subjects into four grades based on the Kimura–Takemoto classification and compared the characteristics of the tissues between each step. C‐0 of the Kimura–Takemoto classification was defined as grade 0, C‐1 and C‐2 as grade 1, C‐3 and O‐1 as grade 2, and O‐2 and O‐3 as grade 3. Thus, there were 19 subjects with grade 0, 54 subjects with grade 1, 35 subjects with grade 2, and 36 subjects with grade 3. Age and sex were examined for each step, but no significant differences were found between groups (A). All subjects were divided into the Discovery phase (n = 71) and the Training phase (n = 73) (B)
3.2. Results of miRNA microarray analysis
We performed a microarray analysis of miRNAs in all subjects, classified them into two clusters in each phase, and compared miRNA expression between grades 0‐2 and 3. Analysis under the condition of P < .05 and q < 0.2 revealed 44 miRNAs in the Discovery phase (Fisher’s Exact test, P = .005; Figure 3A) and 137 miRNAs that showed differential expression in the Training phase (P < .0005; Figure 3B). When we compared the two phases, two common miRNAs were found among the detected miRNAs: miR‐196b‐3p and miR‐92a‐2‐5p. miR‐196b was upregulated in grade 3 (Figure 3C, Discovery phase: q = 0.11, Training phase: q = 0.08), and miR92a‐2‐5p was downregulated (Figure 3D, Discovery phase: q = 0.18, Training phase: q = 0.18). The trend was the same in each phase.
FIGURE 3.
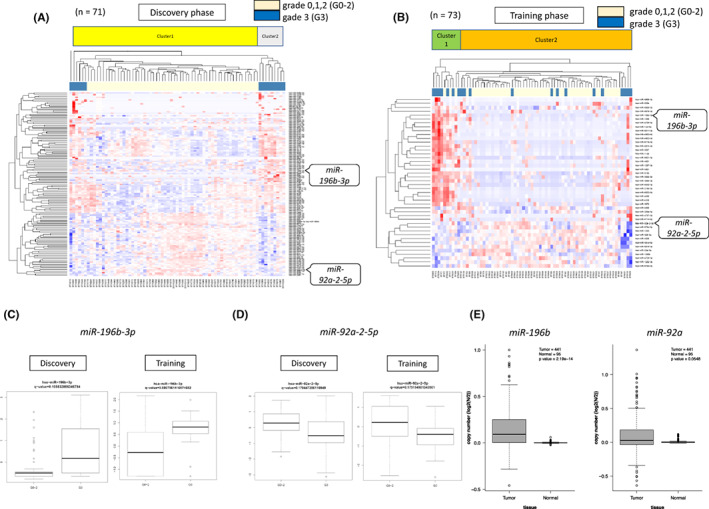
miRNAs detected by microarray analysis. The results of the miRNA microarray analysis are shown. The subjects were divided into two phases: 71 subjects were assigned to the Discovery phase, and 73 subjects were assigned to the Training phase. They were classified into two clusters in each phase, and miRNA expression was compared between grades 0‐2 and 3. Analysis under the condition of P < .05 and q < 0.2 revealed 44 miRNAs in the Dscovery phase (Fisher’s exact test, P = .005) (A). In the Taining phase, 137 miRNAs showed differential expression (P < .0005) (B). The miRNAs satisfying the conditions were miR‐196b‐3p and miR‐92a‐2‐5p. The expression of the two miRNAs was compared in the Discovery phase and Training phase. miR‐196b‐3p was upregulated (Discovery phase: Q = 0.11, Training phase: Q = 0.08) (C), and miR‐92a‐2‐5p was downregulated (Discovery phase: Q = 0.18, Training phase: Q = 0.18) (D) in grade 3 in both phases. In (E), we used the TCGA database to compare the expression of the two miRNAs in GC tissue and normal tissue. The expression of miR‐196b was predominantly highly expressed in GC, while no significant difference in expression was observed between GC and normal tissue for miR‐92a
We investigated the expression status of the above‐detected miRNAs in GC. Using the TCGA database, we performed a comparative analysis of miRNA expression in tissue samples. The results showed that miR‐196b‐3p was highly expressed, with a significant difference in GC tissues compared to normal gastric mucosa (P < .001). Unfortunately, however, miR‐92a‐2‐5p did not show any significant difference between GC tissues and normal gastric mucosa (Figure 3E).
3.3. Analysis using a combination of miRNAs
Comparing grades 0‐2 and grade 3, the areas under the ROC curve (AUCs) of the serum miR‐196b‐3p signature were 0.730 and 0.763 for the two sets of serum samples (Figure 4A). For miR‐92a‐2‐5p, the values were 0.696 and 0.714, respectively (Figure 4B). Since the usefulness of combining miRNAs has been reported, 22 we also analyzed the two detected miRNAs simultaneously. The AUC of the ROC curve showed even higher values in each phase (Figure 4C, Discovery phase: 0.796, Training phase: 0.792).
FIGURE 4.
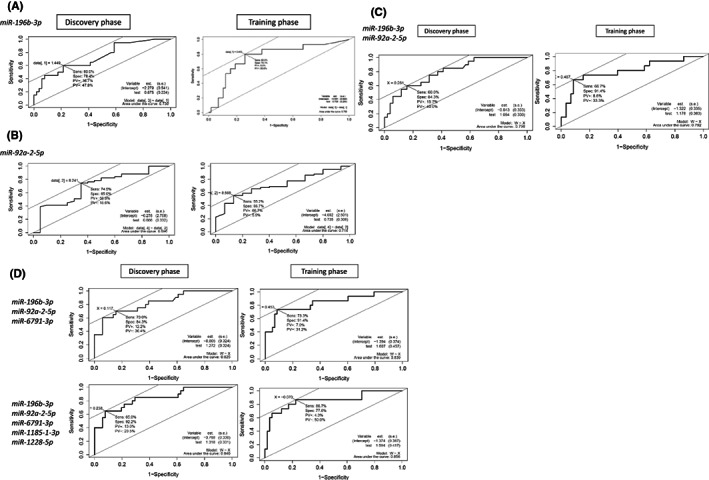
The combination of miRNAs related to grade 3 increased the AUC of the ROC curve. Comparing grades 0‐2 and grade 3, we calculated the area under the receiver operating characteristic (ROC) curve (AUC). First, we calculated the AUC for miR‐196b‐3p alone or miR‐92a2‐5p alone. The results were 0.730 and 0.736 (A) and 0.696 and 0.714 (B) for the Discovery and Training phases, respectively. The combination of miR‐196b‐3p and miR‐92a‐2‐5p increased the AUC (C). When miR‐6791‐3p was added, the AUC increased to 0.825 and 0.839. Furthermore, when miR‐111 and miR‐112 were added, the AUC increased to 0.840 and 0.856 (D)
We tried to increase the number of detected miRNAs by changing the detection conditions: when q < 0.25, miR‐6791‐3p was newly detected as a common miRNA, and when q < 0.3, miR‐1185‐1‐3p and miR‐1228‐5p were newly detected. When we raised q < 0.35, miR‐1185‐1‐3p and miR‐1228‐5p were detected. The AUC value increased when analyzed using three miRNAs with the addition of miR‐6791 (Discovery phase: 0.825, Training phase: 0.839), and further increased when five miRNAs were used (Discovery phase: 0.840, Training phase: 0.856; Figure 4D). However, when we checked the sensitivity and the specificity, we found that the method using three miRNAs was the most suitable for detection, because the specificity in the Discovery phase and the sensitivity in the Training phase decreased in the case of five miRNAs (Table 1).
TABLE 1.
Sensitivity/specificity and AUC for each miRNA and each phase
| miRNA | Discovery phase | Training phase | ||||
|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | AUC | Sensitivity (%) | Specificity (%) | AUC | |
| miR‐196b‐3p (A) | 60 | 78.4 | 0.73 | 80 | 74.1 | 0.763 |
| miR‐92a‐5p (B) | 74.5 | 65 | 0.696 | 55.2 | 86.7 | 0.714 |
| miR‐6791‐3p (C) | 45 | 76.5 | 0.564 | 66.7 | 89.7 | 0.764 |
| miR‐1185‐1‐3p (D) | 95 | 43.1 | 0.678 | 93.3 | 56.9 | 0.757 |
| miR‐1228‐5p (E) | 58.8 | 65 | 0.638 | 63.8 | 80 | 0.683 |
| A and B | 60 | 84.3 | 0.796 | 66.7 | 91.4 | 0.792 |
| A, B, and C | 70 | 84.3 | 0.825 | 73.3 | 91.4 | 0.839 |
| All | 65 | 92.2 | 0.84 | 86.7 | 77.6 | 0.856 |
Note: The sensitivity, specificity, and AUC for the Discovery and Training phases are listed for each of the five miRNAs: miR‐196b‐3p, miR‐92a‐2‐5p, miR‐6791‐3p, miR‐1185‐1‐3p, and miR‐1228‐5p. Below the individual cases, we show where miR‐196b‐3p and miR‐92a‐2‐5p are combined, miR‐6791‐3p is added, and mixed.
Abbreviation: AUC, area under the receiver operating characteristic curve.
4. DISCUSSION
This study identified the serum miRNA, miR‐196b‐3p, in subjects with precancerous lesions of GC by microarray analysis. The miR‐196 gene family (miR‐196a‐1, miR‐196a‐2, miR‐196b) is transcribed from a region harboring three genes, ECRG4, 23 RAD23B, 24 and HOXA10 in the human homeobox (HOX) gene cluster region. miR‐196a‐1 and miR‐196a‐2 have the same mature nucleotide sequence, but mature miR‐196b differs from mature miR‐196A by only one nucleotide. In previous studies, the abundant expression of miR‐196b inhibited ECRG4 and RAD23B. In addition, GC cases with simultaneous overexpression of HOXA10 and miR‐196b had a poor prognosis. 17 Thus, overexpression of miR‐196b and HOXA10 may mediate the development of hematopoietic progenitor cells and gastric cancer with chronic inflammation caused by H. pylori.
In terms of miR‐92a, a former study reported that miR‐92a is downregulated in gastric cancer, warranting further analysis of this miRNA. 18 However, in the current study we found that miR‐92a‐2‐5p did not show cancer‐specific expression compared with that in normal lesions, whereas miR‐92a‐2‐5p was expressed at a lower level in precancerous lesions. In general, the expression of miRNAs in cancerous tissues does not necessarily indicate a sequential or continuous difference from that in precancerous lesions or normal tissues. We assumed that miR‐92a may not be involved in the process of carcinogenesis itself, but in the formation of precancerous lesions, such as chronic inflammation and severe atrophy.
In contrast, several studies have focused on the clinical significance of miRNAs in early‐stage GC lesions. Highly expressed miRNAs in precancerous lesions have been reported in various cancer types. Of those, miR‐421, 19 miR‐19a‐3p, and miR‐483‐5p 25 showed clinical significance, especially in GC cases. Another miRNA, miR‐365, inhibited the progression of precancerous lesions in the stomach. 20 These reports differ from our current study on the following points. They used miRNAs that were already shown to be highly expressed in early and advanced GC, and they confirmed that those miRNAs are also highly expressed in sera from precancerous lesions. In contrast, our study is the first to use 3D microarrays to detect miRNAs highly expressed in atrophic gastritis, a precancerous lesion of GC, compared to patients with other levels of gastritis. Our discovery will support the identification of high‐risk candidates harboring malignant transformation among thousands of patients with gastritis.
The point of the current study is that combining miRNAs increased the detectability of high‐risk cases. Single miRNAs are highly unstable and lack reproducibility. In the current study, there were also large differences in the sensitivity and specificity of specific miRNAs between the two phases. Notably, previous studies have reported that reproducibility is maintained by combining multiple miRNAs. 26 In breast cancer, combining miR‐1246, miR‐1307, miR‐4634, miR‐6861, and miR‐6875 22 upregulated the detectability of cancers. Considering the accuracy and practical clinical benefit, we suggest that the combination of miRNAs should be applied as opposed to one mere miRNA.
As mentioned in the Introduction, endoscopy carries a risk of complications, 6 , 7 and frequent testing may lead to overdiagnosis. 8 Therefore, the current methodology to detect specific serum miRNAs could identify high‐risk cohorts for GC that require fiberscope examination. Performing endoscopy on a limited number of individuals can focus on detecting precancerous lesions, and it is easier to follow the course of subsequent improvement and carcinogenesis. Thus, early cancer detection is possible, and may even lead to a better long‐term prognosis.
A cutoff value of at least 3 for anti‐H. pylori IgG antibodies was used as a criterion for cases in which endoscopy was performed. This cutoff value was set based on a previous report 21 of a prospective study using a population from the same area. Because we did not examine the relationship with H. pylori in this study, the cutoff value was established as an acceptable range for selecting patients who should undergo endoscopic examination. Therefore, it is possible that some patients with antibody levels of <3 also exhibited atrophy. The associations of H. pylori and miRNAs with gastric cancer are well known, 17 and some reports have described fluctuations of miRNAs with H. pylori eradication. 27 However, one major limitation of this study was that we were unable to evaluate these relationships because the history of H. pylori eradication and current infection were not investigated.
In this study, precancerous lesions were defined as grade 3 of gross findings. The rate of carcinogenesis in gastric cancer is known to increase as the grade increases. 13 However, a previous study showed that some cases progressed to carcinogenesis directly from grade 2 or less mild atrophic gastritis without passing through grade 3. 28 Although we were unable to evaluate all cases of carcinogenesis, a previous meta‐analyses reported that the risk associated with grade 3 atrophic gastritis was approximately four times that of other grades. 29 Importantly, in this study, miR‐196b expression increased in a stepwise manner roughly according to the degree of atrophy (Figure S1).
We plan to conduct a functional analysis of the identified miRNAs in future work. Prospective studies on the three miRNAs are also in progress. In this study we were able to identify miRNAs characteristic of precancerous lesions in GC, and we expect that these miRNAs can be applied to select a group of subjects who should undergo endoscopy.
5. CONCLUSION
Three miRNAs (miR‐196b‐3p, miR‐92a‐2‐5p, and miR‐6791‐3p) may be promising biomarkers of precancerous lesions in GC. Identification of these serum miRNAs should identify patients at high risk for GC by using miRNAs in the blood as biomarkers.
DISCLOSURE
Funding: This project was supported by the Japan Society for the Promotion of Science KAKENHI (grant no. 19 K09220), Grant‐in‐Aid for Scientific Research on Innovative Areas (grant no. 15H05912), Priority Issue on Post‐K computer (grant nos. hp170227, hp160219), the Project for Cancer Research and Therapeutic Evolution (grant nos. 19 cm0106504h0004, 21 cm0106475h0002), and a research grant from the Takeda Foundation. This study used the supercomputing resources provided by the Human Genome Center, Institute of Medical Science, University of Tokyo (http://sc.hgc.jp/shirokane.html).
Conflict of Interest: Eiji Oki and Koshi Mimori are Editorial Board Members of the Annals of the Gastroenterological Surgery.
Ethics Approval: The Ethics Committee of the Graduate School of Kyushu University approved the study (review number: 597‐00). Written informed consent for participation in this study was obtained from all patients.
Author Contribution: All of the authors made substantial contributions to the study concept or the data analysis or interpretation, drafted the manuscript or revised it critically for important intellectual content, approved the final version of the manuscript to be published, and agreed to be accountable for all aspects of the work.
Supporting information
Figure S1
ACKNOWLEDGMENT
We thank the members of the Department of Surgery of Kyushu University Beppu Hospital for sample collection and analysis. We also thank M. Kasagi, S. Sakuma, N. Mishima, and T. Kawano for technical assistance.
Otsu H, Nambara S, Hu Q, Hisamatsu Y, Toshima T & Takeishi K et al. Identification of serum microRNAs as potential diagnostic biomarkers for detecting precancerous lesions of gastric cancer. Ann Gastroenterol Surg. 2023;7:63–70. 10.1002/ags3.12610
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans . Infection with Helicobacter pylori . IARC Monogr Eval Carcinog Risks Hum. 1994;61:177–240. [PMC free article] [PubMed] [Google Scholar]
- 3. Monitoring of cancer incidence in Japan ‐ Survival 2009–2011 report (center for Cancer Control And Information Services, National Cancer Center, 2020).
- 4. Choi KS, Suh M. Screening for gastric cancer: the usefulness of endoscopy. Clin Endosc. 2014;47(6):490–6. 10.5946/ce.2014.47.6.490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamashima C, Ogoshi K, Narisawa R, Kishi T, Kato T, Fujita K, et al. Impact of endoscopic screening on mortality reduction from gastric cancer. World J Gastroenterol. 2015;21(8):2460–6. 10.3748/wjg.v21.i8.2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sieg A, Hachmoeller‐Eisenbach U, Eisenbach T. Prospective evaluation of complications in outpatient GI endoscopy: a survey among German gastroenterologists. Gastrointest Endosc. 2001;53(6):620–7. 10.1067/mge.2001.114422 [DOI] [PubMed] [Google Scholar]
- 7. Verschoore T, Vandecandelaere S, Vandecandelaere P, Vanderplancke T, Bergs J. Risk factors for complications and mortality related to endoscopic procedures in adults. Acta Gastroenterol Belg. 2016;79(1):39–46. [PubMed] [Google Scholar]
- 8. Hamashima C. Overdiagnosis of gastric cancer by endoscopic screening. World J Gastrointest Endosc. 2017;9(2):55–60. 10.4253/wjge.v9.i2.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Correa P. The epidemiology of gastric cancer. World J Surg. 1991;15(2):228–34. 10.1007/BF01659057 [DOI] [PubMed] [Google Scholar]
- 10. Ohata H, Kitauchi S, Yoshimura N, Mugitani K, Iwane M, Nakamura H, et al. Progression of chronic atrophic gastritis associated with Helicobacter pylori infection increases risk of gastric cancer. Int J Cancer. 2004;109(1):138–43. 10.1002/ijc.11680 [DOI] [PubMed] [Google Scholar]
- 11. Kimura KTT. An endoscopic recognition of the atrophic border and its significance in chronic gastritis. Endoscopy. 1969;1:87–97. [Google Scholar]
- 12. Kishino M, Nakamura S, Shiratori K. Clinical and endoscopic features of undifferentiated gastric cancer in patients with severe atrophic gastritis. Intern Med. 2016;55(8):857–62. 10.2169/internalmedicine.55.4841 [DOI] [PubMed] [Google Scholar]
- 13. Song JH, Kim SG, Jin EH, Lim JH, Yang SY. Risk factors for gastric tumorigenesis in underlying gastric mucosal atrophy. Gut Liver. 2017;11(5):612–9. 10.5009/gnl16488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sipponen P, Kekki M, Haapakoski J, Ihamaki T, Siurala M. Gastric cancer risk in chronic atrophic gastritis: statistical calculations of cross‐sectional data. Int J Cancer. 1985;35(2):173–7. 10.1002/ijc.2910350206 [DOI] [PubMed] [Google Scholar]
- 15. van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA‐binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–56. 10.1038/nrc3107 [DOI] [PubMed] [Google Scholar]
- 16. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 17. Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, et al. Overexpression of miR‐196b and HOXA10 characterize a poor‐prognosis gastric cancer subtype. World J Gastroenterol. 2013;19(41):7078–88. 10.3748/wjg.v19.i41.7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shin VY, Siu MT, Liu X, Ng EKO, Kwong A, Chu KM. MiR‐92 suppresses proliferation and induces apoptosis by targeting EP4/Notch1 axis in gastric cancer. Oncotarget. 2018;9(36):24209–20. 10.18632/oncotarget.24819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J, Wu L, Sun Y, Yin Q, Chen X, Liang S, et al. Mir‐421 in plasma as a potential diagnostic biomarker for precancerous gastric lesions and early gastric cancer. PeerJ. 2019;7:e7002. 10.7717/peerj.7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang T, Zhang K, Ji K, Zhang C, Jiang Y, Zhang Q, et al. microRNA‐365 inhibits YAP through TLR4‐mediated IRF3 phosphorylation and thereby alleviates gastric precancerous lesions. Cancer Cell Int. 2020;20(1):549. 10.1186/s12935-020-01578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shuto M, Fujioka T, Matsunari O, Okamoto K, Mizukami K, Okimoto T, et al. Association between gastric cancer risk and serum Helicobacter pylori antibody titers. Gastroenterol Res Pract. 2017;2017:1286198. 10.1155/2017/1286198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107(3):326–34. 10.1111/cas.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L, Tang H, Liu G, Xiao S, Liang D, Ma J, et al. MicroRNA‐196b promotes gastric cancer progression by targeting ECRG4. Anticancer Drugs. 2021;32(2):127–37. 10.1097/CAD.0000000000000998 [DOI] [PubMed] [Google Scholar]
- 24. Shen YN, Bae IS, Park GH, Choi HS, Lee KH, Kim SH. MicroRNA‐196b enhances the radiosensitivity of SNU‐638 gastric cancer cells by targeting RAD23B. Biomed Pharmacother. 2018;105:362–9. 10.1016/j.biopha.2018.05.111 [DOI] [PubMed] [Google Scholar]
- 25. Cheng J, Yang A, Cheng S, Feng L, Wu X, Lu X, et al. Circulating miR‐19a‐3p and miR‐483‐5p as novel diagnostic biomarkers for the early diagnosis of gastric cancer. Med Sci Monit. 2020;26:e923444. 10.12659/MSM.923444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, et al. Novel circulating microRNA signature as a potential non‐invasive multi‐marker test in ER‐positive early‐stage breast cancer: a case control study. Mol Oncol. 2014;8(5):874–83. 10.1016/j.molonc.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shiotani A, Uedo N, Iishi H, Murao T, Kanzaki T, Kimura Y, et al. H. Pylori eradication did not improve dysregulation of specific oncogenic miRNAs in intestinal metaplastic glands. J Gastroenterol. 2012;47(9):988–98. 10.1007/s00535-012-0562-7 [DOI] [PubMed] [Google Scholar]
- 28. Quach DT, Hiyama T. Assessment of endoscopic gastric atrophy according to the Kimura–Takemoto classification and its potential application in daily practice. Clin Endosc. 2019;52(4):321–7. 10.5946/ce.2019.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao S, Fan Y, Yin Z, Zhou L. Endoscopic grading of gastric atrophy on risk assessment of gastric neoplasia: a systematic review and meta‐analysis. J Gastroenterol Hepatol. 2021;36(1):55–63. 10.1111/jgh.15177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1


