Abstract
Functional loss of the two major tumor repressors, TP53 and RB1, is frequently involved in the emergence and progression of castration-resistant prostate cancer (CRPC). Inactivating mutations in TP53 and RB1 promote lineage variants that suppress the androgen receptor axis and enhance therapy resistance. The present study provides the first evidence that RB1 loss, and not TP53 loss, is sufficient to activate the master regulator transcription factor ONECUT2 (OC2) in mCRPC. OC2 upregulation is common in CRPC and drives metastasis and lineage plasticity, particularly neuroendocrine differentiation, in model systems. Pharmacologic inhibition of OC2 was reported to suppress established human CRPC metastases in mice. Here we show that RB1 silencing in human and mouse prostate cancer models is sufficient to upregulate OC2, at least in part through epigenetic regulation. OC2 expression downregulated TP53 transcription and inactivated RB1 via phosphorylation. OC2 expression and activation in human CRPC correlated with bi- or single-allelic loss of RB1 and inversely with RB1 expression and activity. A small molecule OC2 inhibitor blocked enzalutamide-induced lineage plasticity in vitro. These findings indicate that activation of OC2 in CRPC occurs in response to RB1 inactivation, and that biomarkers of RB1 activity may be useful for stratifying patients refractory to hormone therapy where OC2 is targeted pharmacologically.
Keywords: ONECUT2, RB1, TP53, lineage plasticity, prostate cancer
Introduction
Prostate cancer (PC) is generally highly treatable, however a subset of patients progress to a therapy-resistant state known as castration-resistant disease (CRPC). About 30,000 men per year die of CRPC in North America [1]. Patients typically respond initially to androgen receptor-(AR)-directed therapies, such as enzalutamide (ENZ), however, therapeutic resistance to all therapies that inhibit AR fail in most patients, consistent with the concept that cancer cell variants emerge that are no longer dependent on activity of the AR [2]. One explanation for progression to treatment resistance is loss of lineage fidelity, such that cancer cell survival and tumor growth is no longer AR-dependent because other mechanisms that govern distinct developmental lineages are activated [3].
The tumor suppressor proteins TP53 and RB1 are well established as playing a major role in cancer progression across a wide spectrum of human malignancies, and their critical role in inhibiting tumorigenesis and progression has been thoroughly investigated [4]. RB1 and TP53 were identified as genomic sites of recurring genetic alterations associated with treatment-emergent neuroendocrine PC (NEPC), a treatment-resistant phenotype that occurs following AR-directed therapy [5]. Considerable evidence supports a role for loss of function of RB1 and TP53 as contributing to treatment resistance. RB1 loss in genetically engineered mouse models (GEMMs) facilitates lineage plasticity and additional loss of TP53 promotes resistance to antiandrogen therapy [6]. Prior studies also demonstrated that combined inactivation of RB1 and TP53 in patients promotes AR-independent lineage plasticity, enhances cell proliferation, and drives stem-like and NEPC phenotypes. Tumor cells with inactivated TP53 and RB1 exhibit drug-resistant phenotypes when challenged with single-agent therapies [4].
We previously reported that the HOX/CUT transcription factor ONECUT2 (OC2) operates as a master regulator protein in some CRPC tumors. OC2 activation suppresses AR activity genome-wide and is a driver of the drug-resistant NEPC phenotype [7]. Because OC2 activation occurs coincident with RB1 and TP53 inactivation in PC, in this study we sought to determine the relationship between active OC2 and the inactivation of these important tumor suppressor genes.
Materials and methods
Public datasets
RNA-Seq data of TP53 and RB1 loss was obtained from the published SU2C Cohort [3]. Knock-down RB1, TP53 and RB1/TP53 RNA-Seq data was downloaded from a published study [4]. Another independent shRB1 dataset was obtained from the GEO database [13]. The RNA-Seq data of the Pten-/- and Pten/RB1-/- mouse models was downloaded from the GEO database [6].
Cell lines
LNCaP (#CRL-1740) was obtained from the American Type Culture Collection (ATCC) and authenticated using the Promega PowerPlex 16 system DNA typing (Laragen). Mycoplasma contamination was routinely monitored using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza #LT07-118). The OC2 overexpression construct was generated by cloning the full-length OC2 cDNA (NM_004852) into the pLenti-C-Myc-DDK-IRES-Puro (Origene) lenti-virus system. Then packing (psPAX2, Addgene #12260), and envelope (pMD2.G, Addgene #12259) plasmids were co-transfected into HEK293T cells to produce lentivirus. Cells were infected with lentivirus supplemented with 10 µg/mL polybrene, then selected by 2 ug/mL puromycin to generate the stable overexpression cells. All cell lines were grown in RPMI-1640 media (Gibco) supplemented with 10% FBS and penicillin/streptomycin.
Cleavage under targets and release using nuclease (CUT&RUN) sequencing
All procedures were performed according to the manufacturer’s protocol from cell signaling technology (CST, #86652). Briefly, 100,000 cells from both vector_control and OC2_Overexpression cells were resuspended in wash buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 0.5 mM spermidine, and protease inhibitor cocktail), concanavalin A-magnetic beads added, then rotated for 10 min at room temperature. Cell-bead conjugates were resuspended in 200 µL of digitonin buffer (wash buffer with 2.5% digitonin solution) containing 2 ug of Anti-Tri-Methyl-Histone H3 (Lys27) (CST, #9733), 2 ug of Anti-Tri-Methyl-Histone H3 (Lys4) (CST, #9751), 5 ug of Anti-ONECUT2 (Proteintech, #21916-1-AP) primary antibody, or rabbit IgG (CST, #66362), rotated overnight at 4°C, resuspended in 250 µL of antibody buffer and 7.5 µL of the pAG-MNase enzyme (#57813, CST), followed by the rotation at 4°C for 1 h. After washing with digitonin buffer, ice-cold 150 µL of digitonin buffer containing CaCl2 was added and incubated on ice for 30 min followed by the addition of 150 µL of stop buffer containing 5 ng S. cerevisiae spike-in DNA used for sample normalization. After incubation at 37°C for 15 min, samples were centrifuged at 16,000 g for 2 min at 4°C. Tubes were placed on a magnetic rack, then supernatants were collected. DNA was purified using DNA purification buffers and spin columns (CST, #14209). The CUT&RUN library was generated with the DNA Library Prep Kit for Illumina (CST, #56795) combined with Multiplex Oligos for Illumina® (Dual Index Primers) (CST, #47538). The adaptor was diluted 1:25 to avoid contamination. The PCR enrichment step run 15 cycles to amplify the adaptor-ligated CUT&RUN DNA.
RNA-seq
RNA concentration, purity, and integrity were assessed by NanoDrop (Thermo Fisher Scientific Inc.) and Agilent Bioanalyzer. RNA-seq libraries were constructed from 1 μg total RNA using the Illumina TruSeq Stranded mRNA LT Sample Prep Kit according to the manufacturer’s protocol. Barcoded libraries were pooled and sequenced on the Illumina HiSeq 2500 generating at least 30 M 100 bp paired-end reads per sample.
RNA-seq data analysis
The RNA-seq data of control and OC2-over expressed cells were generated in the LNCaP cell line. Trim galore was used to remove the adapters. 150 bp paried-end reads were aligned to human reference genome (HG38) using STAR (-alignIntronMin 20-alignIntronMax 1000000-alignSJoverhangMin 8-quantMode GeneCounts) method [14]. Then, gene read counts matrix were used for further analysis.
Western blot analysis
Cell lysates were separated on 4-20% SDS-PAGE (Bio-Rad Laboratories) and transferred to nitrocellulose or PVDF membranes. The membranes were blocked with 5% non-fat dry milk and subsequently incubated with the pertinent primary antibody overnight. Membranes were subsequently washed with TBST (0.1% Tween-20) and incubated with an HRP-conjugated secondary antibody (GE Healthcare Bio-Sciences). After washing with TBST, the protein bands were detected by the Chemidoc MP imaging system (Bio-rad).
Cell viability/proliferation analysis
All procedures were performed according to the XTT cell viability kit protocol (CST, #9095). To assay viability, cells were plated at a density of 2,000 cells/well in triplicate. 48 hours after indicated treatment, viability was assessed at the absorbance of 450 nM. IC50 was generated by a non-linear regression function in Graphpad 9.0. For proliferation assay, the cells were seeded at 2000/well and grown up to 96 hours, then 450 nM absorbance was collected for further analysis.
Luciferase reporter assay
For the TP53 activity assay, the PG13-Luc (containing 13 copies of the p53-binding consensus sequence) plasmid was purchased from Addgene (Addgene, #16442). Lncap wildtype cells were co-transfected with this luciferase construct, and the pRL-SV40 vector (Promega) with Turbofectin 8.0 (Origene, #TF81001). After transfection overnight, cells were washed and treated for 6 hours with CSRM-617 (1 nm/100 nM/10 uM). Luciferase activity was measured using the Dual-Luciferase Assay System (Promega, #1910).
Calculation of OC2, RB1 loss and TP53 signature score
The OC2 signature was published previously [7]. RB1 loss and TP53/RB1 loss data were obtained from a published study [4]. Signature scores of OC2, RB1 loss and TP53/RB1 loss were calculated based on the Z-score method [7].
RB1 and TP53 activity classification analysis
We first ranked tumor samples in the SU2C Cohort based on the expression of RB1, and classified the samples into two groups (top and bottom quartiles), RB1-high and RB1-low. The same classification was also applied in TP53 classification analysis.
CUT&RUN data analysis
OC2, H3K27me3 and H3K4me3 CUT&RUN data were generated in LNCaP cell lines. Briefly, Trim Galore was utilized to remove contaminant adapters and for read-quality trimming (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). 150 bp paired-end reads were aligned to the human reference genome (HG38) using Bowtie2 (v2.2.6) [15]. As spike-in method is commonly used as a control probe in DNA sequencing [16], we did spike-in step and calculated the spike-in mapping rates in each sample. Then, we calculated the scale factor in each sample and applied it in bamCoverage function in deepTools software. Next, Picard MarkDuplicates tool was utilized to mark and remove PCR duplicates in each sample. ENCODE blacklisted regions on HG38 were removed (https://sites.google.com/site/anshulkundaje/projects/blacklists). Finally, a high accuracy peak calling method, SEACR [17], was used to identify significant peaks with the parameters: norm stringent. To visualize the signal in each sample, bamCompare function in DeepTools (v3.1.3) was used to generate Bigwig files with the parameters: -binSize 10-numberofprocessors 5-normalizeUsing CPM-ignoreDuplicates-extendReads 200 [18]. The scale factor was calculated from the spike in step above. Integrative Genomics Viewer (IGV) was used to visualize the bigwig files [19].
Statistical analysis
Data were represented as mean ± SEM wherever necessary. Student’s t test (2-tailed) was used between the data pairs where it is appropriate. Either exact p value or a p value of 0.05 or less was considered significant and have been used. Significance of differential expression was calculated by Wilcoxon Rank-Sum test.
Results
RB1 loss, but not TP53 loss, contributes to OC2 activation
A spectrum of recurrent genomic alterations has been reported to facilitate the development of CRPC. Functional loss of the two major tumor repressor genes, TP53 and RB1, have been shown to be highly involved in the emergence and progression of CRPC [4]. Prior studies showed that TP53 loss-of-function mutations occur in 40-50% of metastatic tumors, while RB1 loss occurs in 12% of cases [4]. As OC2 is highly activated in CRPC patients [7], we next sought to assess the potential association between OC2 and TP53 and RB1 inactivation in CRPC.
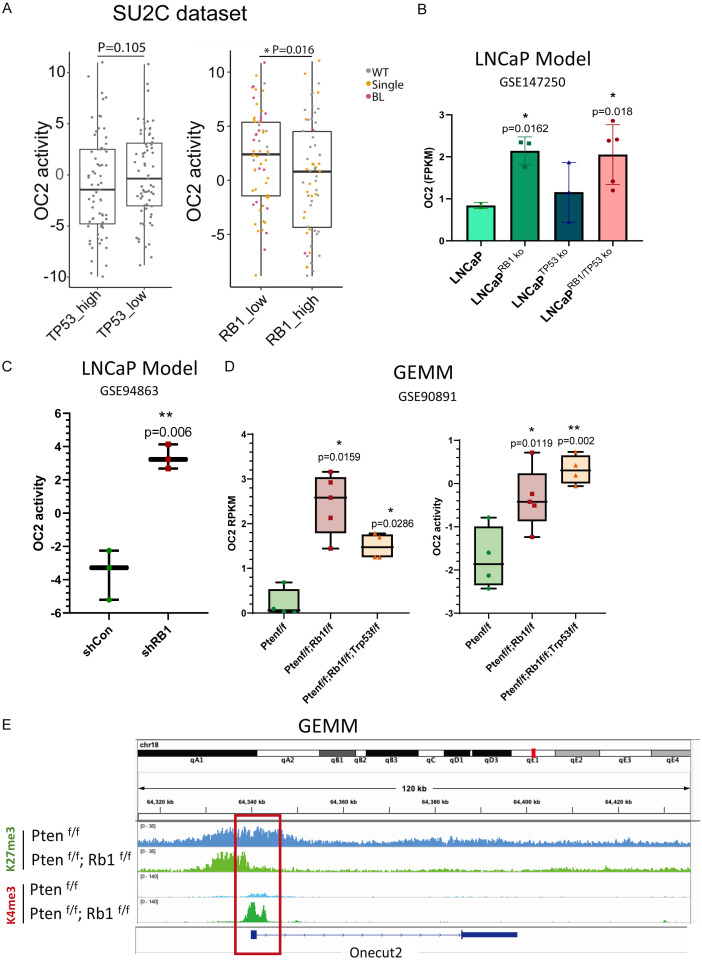
Whole exome, single nucleotide variant (SNV), copy number and matched RNAseq profiles were collected from the SU2C/PCF cohort [3]. OC2 activity was not significantly different when samples were separated into TP53_high (Top 25%) and TP53_low (Bottom 25%) groups. However, when we assessed RB1 loss in the same patient cohorts, OC2 activity was elevated in the RB1-low (Bottom 25% of expression level) group. Next, we applied the copy number information to the matched samples and found the majority of RB1 single copy or biallelic copy loss presented in the RB1-low group (Figure 1A). This observation suggested that in determining whether a tumor is OC2-activated, RB1 copy number detection could be used as a biomarker.
Figure 1.
RB1 loss, but not TP53 loss, contributes to OC2 activation. A. OC2 activity in comparison to TP53 and RB1 expression in SU2C CRPC patient cohorts [3]. All samples were separated into high and low groups based on the transcriptional expression level. The RB1 copy number loss correlates with its expression level and serves as a potential biomarker for stratifying OC2-activated patients. B. LNCaP model with RB1 knockout or combined knockout of RB1/TP53 show increased OC2 expression. TP53 single knockout did not change OC2 expression level. C. Knockdown of RB1 with shRNA activates OC2 in an independent LNCaP model. D. In genetically engineered mouse models (GEMMs), Rb1 knockout and Rb1/Tp53 combined knockout in a Pten-null background elevate both expression and activity of OC2. E. Rb1-null/Pten-null GEMM shows epigenetic activation with loss of repressive mark (K27me3) and gain of activation mark (K4me3) at the Onecut2 promoter compared to Pten-null GEMM.
To validate the causal relationship between OC2 activation and loss of RB1 and/or TP53, LNCaP cells with CRISPR-Cas9 gene knockout (LNCaP TP53-/-; LNCaP RB1-/- and LNCaP RB1-/-; TP53-/-) RNA-seq data were collected from GSE147250. In this dataset, samples with RB1 loss and combined loss of RB1/TP53 have significantly higher OC2 expression compared to wild-type LNCaP cells (Figure 1B). Another independent LNCaP RB1 knockdown cohort, GSE94863, was also analyzed. In this dataset, shRB1 dramatically increased the activity of OC2 in the LNCaP background (Figure 1C). Genomic loss of Rb1 and combined Rb1/Tp53 loss are well-established in GEMMs on the Pten-/- backgound [6]. Compared with Pten-/- models, Rb1-loss and combined Rb1/Tp53 loss significantly contribute to both the expression level and activity of OC2 (Figure 1D). To understand the mechanism of upregulation of OC2 in Rb1 loss models, epigenetic marks at the Onecut2 promoter in an Rb1 loss GEMM were examined by ChIP-sequencing using anti-K27me3 (repressive mark) and anti-K4me3 (activation mark) antibodies. With Rb1 knockout, the K27me3 signal was reduced together with the gain of the K4me3 signal at the promoter region, suggesting that activation of Oc2 by in Rb1 loss was induced by epigenetic regulation (Figure 1E).
OC2 activation induces an RB1 loss and TP53 loss phenotype
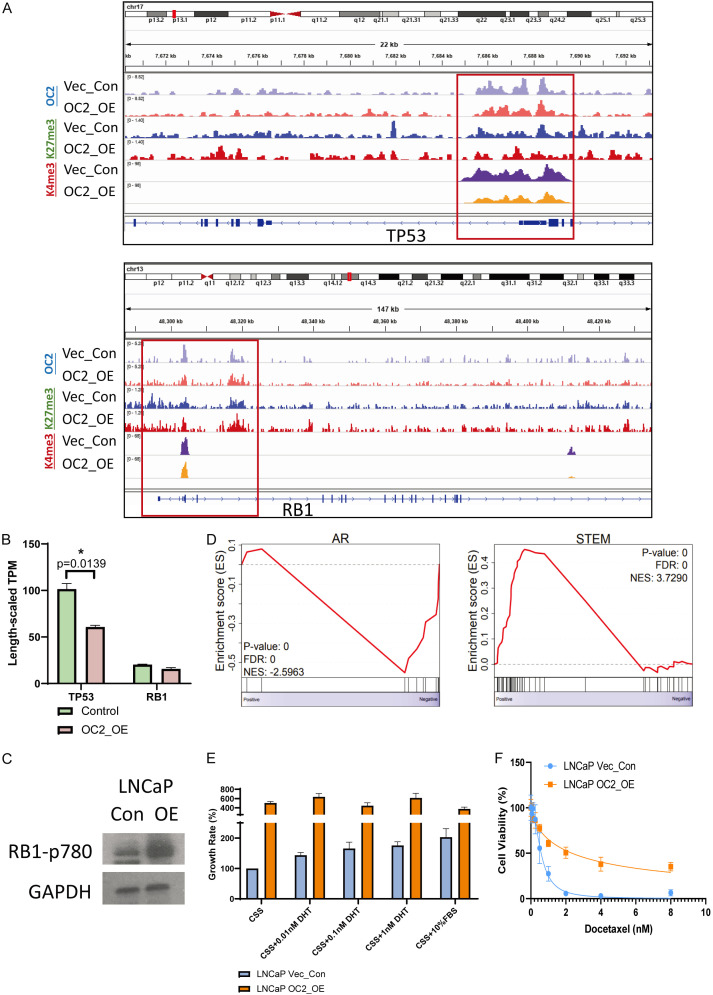
We next evaluated whether OC2 activation affects RB1 or TP53 expression level or signaling activity through enforced expression of OC2 in LNCaP cells. CUT&RUN sequencing with anti-OC2 antibody showed OC2 direct binding to the promoters of both TP53 and RB1, suggesting a role for direct regulation by OC2 of both genes. To further investigate this possibility, we examined the epigenetic marks at both loci. TP53 showed suppression of the K4me3 activation mark, indicating epigenetic repression, while the RB1 promoter did not change (Figure 2A). TP53 expression was significantly repressed under OC2-enforced conditions, while the RB1 mRNA level did not change (Figure 2B). Given the absence of an effect on RB1 expression, we further evaluated the inactivation of RB1 by hyperphosphorylation of the repressive serine phosphorylation site (RB1 p780) [8] in OC2 overexpressed cells. The result showed OC2 activation induced RB1 functional loss through serine 780 phosphorylation (Figure 2C). Collectively, the findings indicate that OC2 activation functionally suppresses both RB1 and TP53, albeit by distinct mechanisms.
Figure 2.
OC2 activation induces an RB1 loss and TP53 loss phenotype. A. OC2 directly binds to the promoter loci of both TP53 and RB1 in LNCaP cells. Enforced OC2 expression suppressed active epigenetic marks at the TP53 promoter while RB1 did not show a significant difference. B. TP53 transcription was downregulated by OC2 activation, while RB1 did not change. C. Enforced OC2 induced functional loss of RB1 through hyperphosphorylation on serine 780. D. Consistent with the RB1/TP53 loss phenotype seen in patient cohorts, gene set enrichment analysis showed that enforced OC2 induced suppression of the AR response transcriptional program and promoted a stem-cell-like program. E. Enforced OC2 overexpression stimulated cell proliferation, however cells a with enforced OC2 expression demonstrated an indifference to AR activation by AR ligand dihydrotestosterone (DHT). CSS = charcoal-stripped serum. FBS = fetal bovine serum. F. Enforced OC2 promoted a docetaxel-resistant phenotype.
Prostate carcinoma with RB1 and TP53 loss-of-function exhibit suppression of the AR transcriptional program and elevation of stem-like features in the cancer cells. The physiological consequence of this lineage plasticity drives AR-dependent cells to become highly proliferative and drug-resistant [4]. Gene set enrichment analysis based on RNA-seq data from LNCaP vector control and OC2-enforced cells demonstrated the repressive AR signaling and elevated stem-like features, consistent with the TP53/RB1 loss phenotype seen in clinical specimens (Figure 2D). OC2-enforced cells also showed 4-fold proliferation rates compared to vector control cells, and the proliferation rates did not change even with stimulation with the AR ligand dihydrotestosterone (DHT) (Figure 2E). These findings suggest OC2-upregulated cells became AR-indifferent. The response of those cells to the first-line drug docetaxel was also evaluated. The IC50 increased from 0.58 nM to 2.35 nM in OC2-enforced cells, and a subset of cells did not exhibit cytotoxicity even at the 8 nM dosage (Figure 2F). Taken together, these findings indicate that OC2 activation in AR-dependent cells drives lineage plasticity and mimics the RB1/TP53 loss-of-function phenotype.
OC2 inhibitor represses the RB1/TP53 loss-induced phenotype
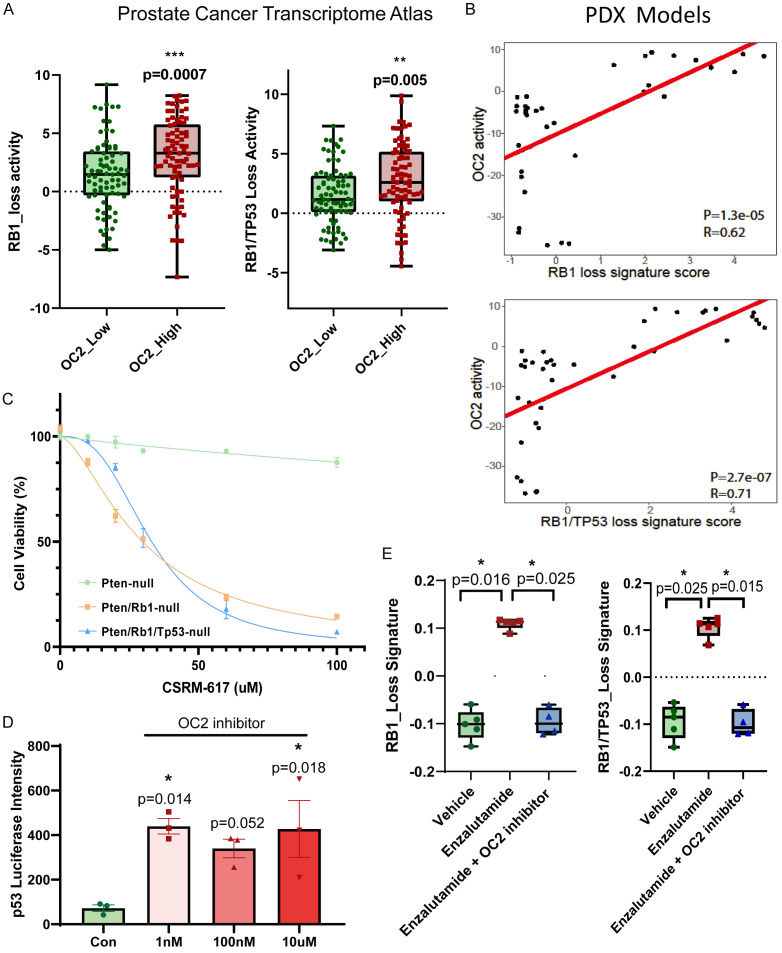
Next, we sought to validate the finding that OC2 activation contributes to the RB1-loss and combined-loss phenotype in clinical cohorts. Transcriptomic profiles collected from 316 CRPC samples in the Prostate Cancer Transcriptome Atlas database [9] (https://www.pcaprofiler.com/) were separated into OC2-high (Top 25%) and OC2-low (Bottom 25%) groups based on OC2 expression level. RB1-loss and combined-loss signatures were calculated by the Z-score algorithm. The OC2-activated patients showed significantly elevated RB1-loss and combined-loss features compared to the OC2-low group (Figure 3A). Prostate cancer PDX models with a CRPC phenotype, developed from clinical specimens, showed a positive correlation of OC2 activity with both RB1 loss (R=0.62) and combined loss (R=0.71) across 39 models (Figure 3B).
Figure 3.
Inhibition of OC2 represses the RB1/TP53 loss-induced phenotype. A. In Prostate Cancer Transcriptome Atlas patient cohorts, the OC2 high group (Top 25%) exhibits higher RB1_loss and RB1/TP53_loss signature activity. B. In LuCaP PDX models (N=39), OC2 activation positively correlates with RB1_loss and RB1/TP53_loss signatures. C. Rb1 and Rb1/Tp53 loss cells from GEMMs are sensitive to the OC2 inhibitor CSRM-617. D. OC2 inhibitor CSRM-617 activates TP53 in TP53 wildtype cells shown by luciferase reporter assay in LNCaP cells with TP53 wildtype phenotype. E. Enzalutamide treatment activates RB1_loss and RB1/TP53_loss signatures in LNCaP cells. Combined treatment with OC2 inhibitor and enzalutamide blocks the activation of the RB1 and RB1/TP53 loss phenotype.
As mentioned above, RB1 knockout and TP53/RB1 knockout activated OC2 in the GEMMs. We previously reported on the development of a small molecule OC2 inhibitor, CSRM-617, which directly targets OC2 and and suppresses established human CRPC metastases in mice [7]. Rb1-loss and Tp53/Rb1-loss GEMM ex vivo cell lines were both sensitive to the OC2 inhibitor, with IC50 at 29.75 uM and 33.06 uM, respectively, whereas the Pten-loss cells did not respond to the inhibitor (Figure 3C) consistent with the findings described above. TP53 activity was measured by luciferase reporter assay with a construct containing 13 copies of the TP53 binding consensus sequence [10]. Compared with the vehicle, even an extremely low dose of CSRM-617 (1 nM) elevated luciferase activity, indicating that inhibition of OC2 activates TP53 activity (Figure 3D).
RB1-loss and RB1/TP53 loss were reported to be associated with resistance to AR signaling inhibitors (ARSIs), such as enzalutamide [11]. In AR-dependent LNCaP cells, treatment with enzalutamide for 7 days significantly activated the RB1-loss and TP53-RB1 combined-loss signatures, and this enzalutamide-induced gene activation effect was blocked when the OC2 inhibitor was applied together with enzalutamide (Figure 3E).
Discussion
RB1 and TP53 are the genes most affected by somatic mutation and copy number loss in mCRPC patients. The combined loss of these tumor suppressors contributes to attenuated AR signaling and the acquisition of a stem-like phenotype, indicating their critical role in maintaining lineage commitment [4]. The present study has provided the first evidence that RB1 loss, and not TP53 loss, is sufficient to activate the master transcription factor ONECUT2 (OC2) in mCRPC. This conclusion is based on data from cell line models, genetically engineered mouse models (GEMM), and clinical samples. In LNCaP cells with RB1 loss, upregulation of OC2 expression and OC2 activation were observed. GEMMs represent conditional mouse models that are genetically engineered to mimic sporadic human cancer. Data from GEMMs with Rb1 and Tp53 knockout demonstrated that Rb1 loss alone, in a Pten-null background, regulates OC2 through epigenetic mechanisms via the removal of suppressive histone marks and replacement with active transcription marks at the Onecut2 promoter. In mCRPC patients, bi- or single-allelic loss of RB1 copy number correlate with reduced RB1 protein and mRNA expression, and activation of OC2 was observed in those specimens. OC2 appears to regulate TP53 and RB1 directly, based on the observation that OC2 binds the TP53 and RB1 promoters. Consistent with this, activation of OC2 repressed TP53 transcription via epigenetic regulation. Evidence for inactivation of RB1 with enforced OC2 was demonstrated through hyperphosphorylation at the repressive serine760 site without an observed change in mRNA expression. Taken together, these findings suggest that OC2 activation drives AR-dependent cells to a functional TP53/RB1 loss-phenotype, which would include AR suppression, stem-like activation, and drug resistance. Consistent with this conclusion, a positive correlation between OC2 activity and RB1-loss/TP53-RB1 combined-loss phenotype was observed in the Prostate Cancer Transcriptome Atlas Database. Treatment of prostate cancer with ARSI results in the emergence of variants no longer dependent on the AR, leading to therapy resistance. Here, we also demonstrated that enzalutamide activates RB1-loss and TP53-RB1 combined loss programs, leading to the emergence of a drug-resistant phenotype. Notably, combination treatment of enzalutamide with the OC2 inhibitor CSRM-617, suppressed the RB1 loss and TP53/RB1 loss-induced transcriptional program.
OC2 was initially identified as a master regulator operating in CRPC from a computational model of transcription factors highly active in lethal human prostate cancer [7]. This model predicted that OC2 was networked with the AR, which was confirmed experimentally. OC2 is a direct regulator of many androgen-regulated genes, including the AR licensing factor FOXA1. OC2 can be expressed in AR-positive cells, but the effect of its activation is largely to suppress the AR transcriptional program. Based on studies of model systems and human prostate cancer, OC2 is predicted to regulate hundreds of other transcription factors, suggesting that its activation in cancer tissues is likely to result in widespread biological effects. Studies by us and others have shown that OC2 promotes a neuroendocrine program and is thus a lineage plasticity driver, consistent with the relationship with RB1 identified in the present study [12].
OC2 is targetable directly with a novel small molecule, CSRM-617, which was shown to suppress established human CRPC metastases in mice. CSRM-617 also inhibits mouse OC2, as demonstrated in vitro (this study) and in vivo, suggesting that the potential for therapeutic targeting of OC2, and its role in lineage plasticity and disease progression, can be studied in autochthonous mouse models. OC2 is expressed widely in CRPC [7], suggesting it is an attractive candidate for a precision therapy approach. Our findings here suggest that OC2 targeting in combination with an ARSI, such as enzalutamide, may be therapeutically beneficial against treatment-refractory disease by suppressing the emergence of AR-indifferent lineage variants. In addition, our findings point to markers of RB1 loss or inactivation as potential biomarkers to identify patients who would benefit from such a therapeutic strategy.
Acknowledgements
We gratefully acknowledge financial support from the NIH/NCI (R01CA271750, P01CA098912, R01CA220327, P50CA092131), the US Army (PC210486, PC180541) and the Urology Care Foundation (to C.Q).
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Nelson PS. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. J. Clin. Oncol. 2012;30:644–646. doi: 10.1200/JCO.2011.39.1300. [DOI] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Sboner A, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 4.Nyquist MD, Corella A, Coleman I, De Sarkar N, Kaipainen A, Ha G, Gulati R, Ang L, Chatterjee P, Lucas J, Pritchard C, Risbridger G, Isaacs J, Montgomery B, Morrissey C, Corey E, Nelson PS. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep. 2020;31:107669. doi: 10.1016/j.celrep.2020.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, Wongvipat J, Ku SY, Gao D, Cao Z, Shah N, Adams EJ, Abida W, Watson PA, Prandi D, Huang CH, de Stanchina E, Lowe SW, Ellis L, Beltran H, Rubin MA, Goodrich DW, Demichelis F, Sawyers CL. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotinen M, You S, Yang J, Coetzee SG, Reis-Sobreiro M, Huang WC, Huang F, Pan X, Yanez A, Hazelett DJ, Chu CY, Steadman K, Morrissey CM, Nelson PS, Corey E, Chung LWK, Freedland SJ, Di Vizio D, Garraway IP, Murali R, Knudsen BS, Freeman MR. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat Med. 2018;24:1887–1898. doi: 10.1038/s41591-018-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies A, Nouruzi S, Ganguli D, Namekawa T, Thaper D, Linder S, Karaoglanoglu F, Omur ME, Kim S, Kobelev M, Kumar S, Sivak O, Bostock C, Bishop J, Hoogstraat M, Talal A, Stelloo S, van der Poel H, Bergman AM, Ahmed M, Fazli L, Huang H, Tilley W, Goodrich D, Feng FY, Gleave M, He HH, Hach F, Zwart W, Beltran H, Selth L, Zoubeidi A. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat Cell Biol. 2021;23:1023–1034. doi: 10.1038/s41556-021-00743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolis M, Bossi D, Vallerga A, Ceserani V, Cavalli M, Impellizzieri D, Di Rito L, Zoni E, Mosole S, Elia AR, Rinaldi A, Pereira Mestre R, D’Antonio E, Ferrari M, Stoffel F, Jermini F, Gillessen S, Bubendorf L, Schraml P, Calcinotto A, Corey E, Moch H, Spahn M, Thalmann G, Kruithof-de Julio M, Rubin MA, Theurillat JP. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nat Commun. 2021;12:7033. doi: 10.1038/s41467-021-26840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, Sunderland K, Kollmannsberger C, Eigl BJ, Finch D, Oja CD, Vergidis J, Zulfiqar M, Azad AA, Nykter M, Gleave ME, Wyatt AW, Chi KN. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 12.Guo H, Ci X, Ahmed M, Hua JT, Soares F, Lin D, Puca L, Vosoughi A, Xue H, Li E, Su P, Chen S, Nguyen T, Liang Y, Zhang Y, Xu X, Xu J, Sheahan AV, Ba-Alawi W, Zhang S, Mahamud O, Vellanki RN, Gleave M, Bristow RG, Haibe-Kains B, Poirier JT, Rudin CM, Tsao MS, Wouters BG, Fazli L, Feng FY, Ellis L, van der Kwast T, Berlin A, Koritzinsky M, Boutros PC, Zoubeidi A, Beltran H, Wang Y, He HH. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat Commun. 2019;10:278. doi: 10.1038/s41467-018-08133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNair C, Xu K, Mandigo AC, Benelli M, Leiby B, Rodrigues D, Lindberg J, Gronberg H, Crespo M, De Laere B, Dirix L, Visakorpi T, Li F, Feng FY, de Bono J, Demichelis F, Rubin MA, Brown M, Knudsen KE. Differential impact of RB status on E2F1 reprogramming in human cancer. J Clin Invest. 2018;128:341–358. doi: 10.1172/JCI93566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fardin P, Moretti S, Biasotti B, Ricciardi A, Bonassi S, Varesio L. Normalization of low-density microarray using external spike-in controls: analysis of macrophage cell lines expression profile. BMC Genomics. 2007;8:17. doi: 10.1186/1471-2164-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meers MP, Tenenbaum D, Henikoff S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019;12:42. doi: 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]