Abstract
Helicobacter bilis is a bacterial pathogen associated with multifocal hepatitis and inflammatory bowel disease in certain strains of mice. This bacterium colonizes the liver, bile, and lower intestine in mice and has also been isolated from a wide spectrum of laboratory animals. In this study, proteins present in the outer membrane preparation (OMP) of four H. bilis strains isolated from a mouse, a dog, a rat, and a gerbil were characterized and compared with that of Helicobacter pylori, a human gastric pathogen. All four H. bilis strains had similar OMP protein profiles that were distinct from those of H. pylori. Immunoblotting demonstrated that OMP proteins from H. bilis and H. pylori have little cross-reactivity, except for their flagellins. Nine major immunogenic polypeptides were present in the H. bilis OMPs. By using two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis, five heat-modifiable proteins with molecular masses of 82, 66, 52, 47 and 37 kDa were identified. The N-terminal sequences of the 46- and 47-kDa OMP proteins had no homology with protein sequences available in public databases. These results indicate that H. bilis has a conserved, unique OMP protein profile that is distinct from those of H. pylori.
Helicobacter bilis is a gram-negative, microaerophilic, spiral bacterium and a member of the rapidly expanding genus Helicobacter (27). This microorganism was originally identified from the bile, livers, and intestines of aged inbred mice and was able to survive in 20% bile in vitro (10). H. bilis has also been isolated from dogs, gerbils, rats, and cats (6, 8, 12; J. G. Fox, F. E. Dewhirst, and B. J. Paster, unpublished data). Natural infection by H. bilis has been associated with an outbreak of colitis in scid mice (23). In addition, experimental infection by H. bilis in scid mice and immunodeficient rats induces inflammatory bowel diseases (IBD) and hepatitis (11, 12, 24). Present evidence indicates that H. bilis has pathogenic potential in a variety of animal species, and it has provided a murine model of human IBD. Recently, H. bilis DNA was amplified from the bile and gallbladder tissues of Chileans with chronic cholecystitis (9), suggesting that this bacterium may play a role in the development of hepatobiliary disease in humans.
Gram-negative bacteria possess an outer membrane (OM) that acts as a selective barrier, controlling the transportation of molecules, such as antibiotics and nutrients, and ions from the environment into the cell (13, 19, 21). In this case, a protein class (termed porins) forms channels and is responsible for facilitating the selective diffusion of hydrophilic molecules across the OM (3, 13). In addition, the OM has an important role in interacting with the host, such as in adhesion and/or invasion of host tissues. Furthermore, many OM proteins from pathogenic bacteria, including porins, are also involved in the modulation of host immune responses, either through inflammation, suppression, or avoidance; these proteins can be important in persistence and/or disease induced by these bacteria.
Given that H. bilis has become an important pathogen in mice and can be utilized in murine models for studying human IBD, it is important to characterize the bacterial components contributing to its colonization, persistence, and pathologic potential. In this study, we identified the profiles, heat modifiability, and immunogenic components of the outer membrane preparation (OMP) from H. bilis strains isolated from four different animal species. In addition, the profiles and the antigenic cross-reactivities of the OMP proteins of H. bilis and Helicobacter pylori were compared.
Conservation of OMP protein profiles among H. bilis strains.
H. bilis strains, including ATCC 51630, from a mouse (10); MIT95-477, from a dog; MIT96-5983, from a gerbil; and MIT96-151N3, from a rat, were used in this study. All of these H. bilis strains were isolated at the Division of Comparative Medicine, Massachusetts Institute of Technology. In addition, H. pylori strains J99 (1), AH244, and Sydney SS1 (18) were used for the preparation of H. pylori OMP. H. bilis and H. pylori were grown on sheep blood agar (Remel, Lenexa, Kans.) for 3 to 4 days under microaerobic conditions (10% CO2, 10% H2, 80% N2). The H. pylori and H. bilis cells were collected and washed three times with 20 ml of 20 mM Tris-HCl (pH. 8.0). The OM proteins were prepared as described previously (5). Briefly, the cells were sonicated on ice (eight times for 30 s each time at intervals of 1 min). Unbroken cells and debris were removed by centrifugation at 4,000 × g for 10 min at 4°C. The supernatants were then centrifuged at 45,000 × g for 20 min at 4°C, and the pellets were washed three times with 20 mM Tris-HCl (pH. 8.0). The pellets were suspended in 0.6% N-lauroyl sarcosine–20 mM Tris-HCl (pH. 8.0) to a final concentration of 1 mg of protein/ml and incubated at room temperature for 20 min. The OMPs were collected by centrifugation at 45,000 × g for 20 min at 4°C. The resulting pellets were dissolved in 20 mM Tris-HCl (pH. 8.0), and the protein concentration was determined by a bicinchoninic acid protein assay (Pierce, Rockford, Ill.) employing bovine serum albumin as a standard. For comparing the OMP profiles among these helicobacters, 10 μg of OMPs were mixed with 2× sample buffer (100 mM Tris-HCl, pH 6.8, 200 mM dithiothreitol, 4% sodium dodecyl sulfate [SDS] 0.2% bromophenol blue, 20% glycerol) and heated at 100°C for 5 min. The samples were centrifuged at 10,000 × g for 10 min. The OMPs of the H. bilis and H. pylori strains were electrophoretically separated on SDS–10 or 12% polyacrylamide gels and visualized with Coomassie blue.
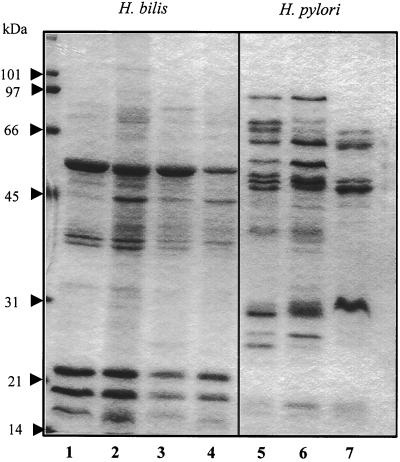
In the OMP samples from four H. bilis strains (ATCC 51630, MIT96-5983, MIT95-477, and MIT96-151N3), approximately 10 polypeptides with apparent molecular masses ranging from 16 to 80 kDa were visualized by Coomassie blue staining on an SDS–12% polyacrylamide gel (Fig. 1, lanes 1 to 4). A conserved pattern of polypeptides was revealed for the four H. bilis strains, with strain-specific differences evident in minor polypeptides. Five of the OM polypeptides, with apparent molecular masses of 16, 18, 22, 36, and 52 kDa, were major components in all strains. An additional two polypeptides with apparent molecular masses of 35 and 44 kDa were also present within the OMPs of all four H. bilis strains, but their expression levels differed significantly between strains. The relative proportion of the OMP proteins made up of a 45-kDa polypeptide in strains MIT95-477 and MIT9615-1N3 (Fig. 1, lanes 2 and 4) was much greater than in strains ATCC 51630 and MIT96-5983 (lanes 1 and 3), whereas the content of an ∼35-kDa polypeptide in ATCC 51630 and MIT95-477 (lanes 1 and 2) was higher than in MIT96-5983 and MIT96-151N3 (lanes 3 and 4).
FIG. 1.
OMP profiles of H. bilis and H. pylori strains on an SDS-10% polyacrylamide gel. H. bilis strains: lane 1, ATCC 51630; lane 2, MIT95-477; lane 3, MIT96-5983; lane 4, MIT96-151N3. H. pylori strains: lane 5, J99; lane 6, AH244; lane 7, SydneySS1. Molecular mass markers are on the left.
The OMP protein profiles of these H. bilis strains were compared to those of three H. pylori strains: J99 (1), AH244, and SydneySS1 (18). Both H. pylori J99 and SydneySS1 are positive for cagA and cagE (also called picB), parts of the H. pylori pathogenicity island (1, 18), whereas AH244 does not contain these two genes (Z. Ge and J. G. Fox, unpublished data). However, the OMP protein profiles in H. pylori strains J99 and AH244 (Fig. 1, lanes 5 and 6) were similar but distinct from that of strain SydneySS1 (lane 7). There was little similarity between the OMP protein profiles of selected H. bilis strains and those of the three H. pylori strains. H. pylori strains AH244 and J99 had more major OMP species (∼10) than H. bilis (∼5). Interestingly, the majority of the H. pylori OMP proteins had masses ranging from 50 to 100 kDa, whereas four of the five major H. bilis OMP proteins had apparent molecular masses of 17 to 36 kDa.
Identification of HMPs in OMP.
The OMP of the H. bilis type strain, ATCC 51630, was used to identify heat-modifiable proteins (HMPs). Twenty-five micrograms of the OMP were mixed with an equal volume of 2× sample buffer, incubated at room temperature for 20 min, and then separated by SDS-polyacrylamide gel electrophoresis (PAGE) using a 10% acrylamide gel. The lane containing the separated OMP proteins was excised, placed in 2× sample buffer, and heated at 100°C for 10 min. The gel strip was then overlaid on an SDS–10% polyacrylamide gel, and the polypeptides were separated electrophoretically. The polypeptides were visualized with Coomassie blue.
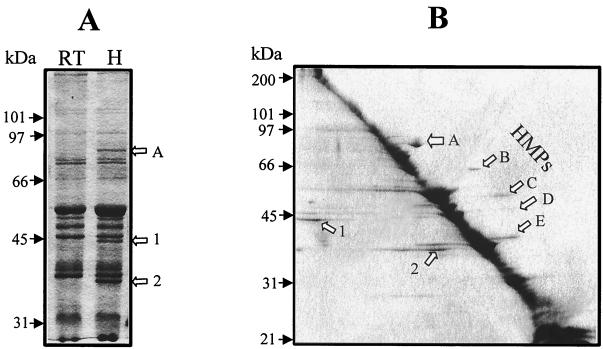
When the heat-denatured and the unboiled OM polypeptides were compared on an SDS–12.5% polyacrylamide gel, three polypeptides in the heat-denatured sample, but not in the unboiled sample, could be identified (corresponding to apparent molecular masses of 35, 44, and 82 kDa) (Fig. 2A). Subsequently, OMP proteins with heat-modifiable electrophoretic mobility were detected by two-dimensional SDS-PAGE. Five polypeptides, which were shown to display slower electrophoretic mobility in the second dimension (Fig. 2B, above the diagonal banding patterns), were identified as HMPs. These proteins were designated OmpA (∼82 kDa), OmpB (∼66 kDa), OmpC (∼52 kDa), OmpD (∼47 kDa), and OmpE (∼37 kDa) (Fig. 2B). Several polypeptides migrated with higher apparent molecular masses in the second-dimensional gel (below the diagonal banding patterns) and likely represent monomers of heat-dissociated high-molecular-weight complexes. Two such polypeptides with apparent molecular masses of 35 and 44 kDa corresponded to the two bands present only in the heated samples in one-dimensional SDS-PAGE (Fig. 2B).
FIG. 2.
OMPs of H. bilis strain ATCC 51630 in first- (A) and second- (B) dimensional SDS-polyacrylamide gels (10%). Five OMPs with heat modifiability are indicated by arrows (A to E). The numbers 1 and 2 denote the heat-dissociated polypeptides from high-molecular-weight complexes. RT and H stand for room temperature and heated, respectively. Molecular mass markers are on the left.
For peptide sequencing of two OMP proteins, 20 μg of the OMP of H. bilis ATCC 51630 were separated on SDS–10% PAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, Calif.). The polypeptides were visualized with Ponceau S solution, and the polypeptide bands of interest were excised with a razor. N-terminal amino acid sequencing was performed by Edman degradation using an Applied Biosystems amino acid sequencer. Two of these OMP polypeptides from the unheated sample, which had approximate molecular masses of 46 and 47 kDa, were sequenced. The N-terminal sequences, AKLSDVINNASIDNF for the 46-kDa protein and KNGADIMEWALQDN for the 47-kDa protein, were not similar to any amino acid sequences available in the public databases Swissprot and Trembl.
Antigenicity of the OMP proteins of H. bilis.
We investigated the in vivo immunogenicities of the H. bilis OMP proteins as well as the immunogenic cross-reactivity between the OMP proteins of H. bilis and those of H. pylori. Anti-H. bilis and anti-H. pylori polyclonal antibodies were obtained from mice experimentally infected with H. bilis strain ATCC 51630 for 24 weeks and from rabbits immunized by cell lysate of strain J99, respectively. For immunoblotting, 2 μg of the respective OMPs was separated on an SDS–12% PAGE minigel apparatus (Bio-Rad, Hercules, Calif.) and then transferred to a nitrocellulose membrane (MSI Westborough, Mass.) in a transblotting buffer (30 mM glycine, 48 mM Tris, 0.0375% [wt/vol] SDS, 20% methanol). Immunoblotting with either anti-H. bilis serum (1 in 2,000 dilution) or anti-H. pylori serum (1 in 10,000 dilution) was performed with the ECL system (Amersham Life Science Inc., Arlington Heights, Ill.) according to the manufacturer's instructions. Reactive polypeptides were visualized on X-ray film (Kodak, Rochester, N.Y.).
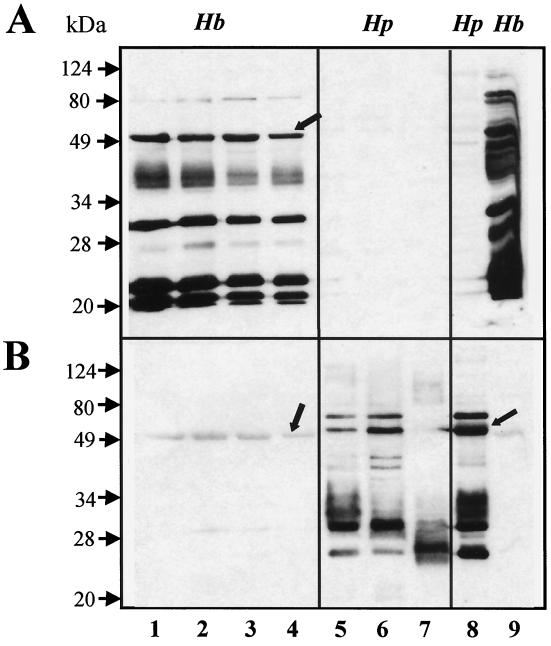
Nine major polypeptide bands with apparent molecular masses of 20, 21, 24, 27, 30, 36, 38, 52, and 80 kDa reacted to the antisera (Fig. 3A, lanes 1 to 4). These immunogenic OMP proteins were highly conserved among the four H. bilis strains. In contrast, whole-cell lysate of the H. bilis type strain, ATCC 51630, contains numerous immunogenic polypeptides (lane 9).
FIG. 3.
Immunoblotting of OMPs from selected H. bilis (Hb) and H. pylori (Hp) strains with the anti-H. bilis (A) and anti-J99 (B) sera. H. bilis strains: lane 1, ATCC 51630; lane 2, MIT95-477; lane 3, MIT96-5983; lane 4, MIT96-151N3. H. pylori strains: lane 5, J99; lane 6, AH244; lane 7, SydneySS1. Cell lysates: lane 8, H. pylori J99; lane 9, H. bilis ATCC 51630. FlaA from H. bilis and H. pylori is denoted by arrows. Molecular mass markers are on the left.
Except for a 52-kDa polypeptide of H. bilis and a 54-kDa polypeptide of H. pylori, there was little intraspecies antigenic cross-reactivity among the OMP proteins (Fig. 3, lanes 1 to 7) or cell lysates (lanes 8 and 9) when either the anti-H. bilis (Fig. 3A) or the anti-H. pylori (Fig. 3B) whole-cell sera were used. This 52-kDa polypeptide in H. bilis OMPs reacted specifically with the monoclonal antibodies to H. pylori FlaA (16). The N-terminal amino acid sequence of the first 15 residues (AFQVNTNINALNAHV) was identical to that of Helicobacter felis FlaA (15) and different by one conservative substitution of Met for a Leu at position 11 in H. pylori, further confirming the identity of this protein as an FlaA homolog.
In addition, both monoclonal and polyclonal antibodies to various OM proteins found in H. pylori were screened. These included monoclonal antibodies specific for HopA, HopC, HopE, JHP438, and JHP117 (5), as well as polyclonal antisera specific for HopB. None of these reacted with either OMs or whole-cell lysates of H. bilis (data not shown).
Due to its likely significance in disease, the composition of the OM of H. pylori, the type species of the genus Helicobacter, has received considerable attention with respect to polypeptide composition and function. The genome of this pathogen encodes a unique protein family comprising 32 OM proteins (1, 14, 25). The members of this protein family, including BabA (15), AlpAB (20), and HopZ (22), have been reported to be necessary for the adhesion of H. pylori to human gastric tissues. Another three members (HopA, HopD, and HopE), as well as HopB and HopC, have been shown to be heat-modified porin proteins (4, 7). It has been proposed that these porins may play a role in H. pylori antibiotic susceptibility and resistance (7, 14). In addition, a 30-kDa OM protein purified from H. pylori was reported to induce the release of cytokines (interleukin 3 [IL-3], IL-4, IL-6, IL-8, interferon gamma, and tumor necrosis factor alpha) from human lymphocytes and monocytes in vitro. This suggested that the surface components of H. pylori, particularly porins, could play an important role in causing the inflammatory and immune responses in the host, possibly leading to gastritis (26).
In this study, the OMP proteins conserved among selected H. bilis isolates showed little similarity to those of three H. pylori strains. The differences in the OM compositions of the two Helicobacter species were further demonstrated by the lack of cross-reactivity with the heterologous species of sera generated to either species as well as the lack of cross antigenicity between the H. bilis OMP proteins and several H. pylori OM proteins (HopA, HopC, HopE, JHP438, and JHP117). A notable exception was the flagellin present in the OMPs of H. bilis. The difference between the OMP protein profiles of the two species as well as the apparent similarities among these selected H. bilis strains may reflect the unique adaptations of this species to its particular gastrointestinal niche. The differences may also indicate that the two microorganisms have different pathogenic mechanisms.
In order to examine the immunorecognition of OM components, sera were obtained from mice infected by H. bilis. Nine OMP proteins with apparent molecular masses ranging from 20 to 80 kDa were found to have strong immunogenicity in vivo. Interestingly, these polypeptides were highly conserved among the four H. bilis strains isolated from mice, gerbils, rats, and dogs, indicating that they could play an essential role in H. bilis colonization, survival, and pathogenesis. It should be noted that except for the 52-kDa reactive polypeptide, FlaA, the identities and biological functions of the immunogenic polypeptides are unknown. Also, it is unclear at present how these OMP proteins induce the production of the corresponding antibodies in their hosts; possible mechanisms include their surface-exposed epitopes, the secreted antigens, or the release of OMP proteins from lysed cells.
Putative porins in the OM of H. bilis were investigated. The sizes of the porins determined by SDS-PAGE are characteristically heat modifiable, migrating more slowly after treatment at 100°C in SDS. This heat modifiability of the porins is due to the presence of extensive β-sheets in their secondary structures (2, 7). Based on this feature, five porins from H. pylori, designated HopA (48 kDa), HopB (49 kDa), HopC (50 kDa), HopD (67 kDa), and HopE (31 kDa), were identified and characterized (4, 7) as members of a larger, functionally important family of proteins. In this study, five HMPs with the apparent molecular masses of 82 (OmpA), 66 (OmpB), 52 (OmpC), 47 (OmpD), and 37 (OmpE) kDa were identified in the OMP of H. bilis. H. bilis OmpB, -C, -D, and -E possessed apparent molecular masses in the range of the H. pylori porins as well as typical bacterial porin sizes (25 to 50 kDa) (4, 7); however, the H. bilis proteins had no cross-reactivity with HopA to -C and HopE, suggesting they are unique. Further investigation of the pore-forming abilities of these HMPs is required in order to elucidate whether they act as porins in vivo. Such studies may lead to the identification of factors that have immunomodulatory activity and thereby play a role in inflammatory processes. Their characterization may also reveal candidates for the development of a vaccine against H. bilis infection.
Acknowledgments
These studies were supported by NIH R01 CA 67529 and NIH R01 DK 52413 and a grant from AstraZeneca.
We thank Gejing Deng for amino acid sequence analysis.
REFERENCES
- 1.Alm R A, Lee L-S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Benz R. Porins from bacterial and mitochondrial outer membranes. Crit Rev Biochem. 1985;19:145–190. doi: 10.3109/10409238509082542. [DOI] [PubMed] [Google Scholar]
- 3.Benz R. Structure and function of porins from gram-negative bacteria. Annu Rev Microbiol. 1988;42:359–393. doi: 10.1146/annurev.mi.42.100188.002043. [DOI] [PubMed] [Google Scholar]
- 4.Doig P, Exner M, Hancock R, Trust T. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J Bacteriol. 1995;177:5447–5452. doi: 10.1128/jb.177.19.5447-5452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doig P, Trust T. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect Immun. 1994;62:4526–4533. doi: 10.1128/iai.62.10.4526-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton K A, Dewhirst F E, Paster B J, Tzellas N, Coleman B E, Paola J, Sherding R. Prevalence and varieties of Helicobacter species in dogs from random sources and pet dogs: animal and public health implications. J Clin Microbiol. 1996;34:3165–3170. doi: 10.1128/jcm.34.12.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Exner M, Doig P, Trust T, Hancock R. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect Immun. 1995;63:1567–1572. doi: 10.1128/iai.63.4.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 9.Fox J G, Dewhirst F E, Shen Z, Feng Y, Taylor N S, Paster B J, Ericson R L, Lau C N, Correa P, Araya J C, Roa I. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 10.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin C L, Riley L K, Livingston R S, Beckwith C S, Besch-Williford C L, Hook J R R. Enterohepatic lesions in SCID mice infected with Helicobacter bilis. Lab Anim Sci. 1998;48:334–339. [PubMed] [Google Scholar]
- 12.Haines D C, Gorelick P L, Battles J K, Pike K M, Anderson R J, Fox J G, Taylor N S, Shen Z, Dewhirst F E, Anver M R, Ward J M. Inflammatory large bowel disease in immunodeficient rats naturally and experimentally infected with Helicobacter bilis. Vet Pathol. 1998;35:202–208. doi: 10.1177/030098589803500305. [DOI] [PubMed] [Google Scholar]
- 13.Hancock R E W. Bacterial outer membranes: evolving concepts. AMS News. 1991;57:175–182. [Google Scholar]
- 14.Hancock R E W, Alm R, Bina J, Trust T J. Helicobacter pylori: a surprisingly conserved bacterium. Nat Biotechnol. 1998;16:216–217. doi: 10.1038/nbt0398-216. [DOI] [PubMed] [Google Scholar]
- 15.Ilver D, Arnqvist A, Ogren J, Frick I M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 16.Josenhans C, Ferrero R L, Labigne A, Suerbaum S. Cloning and allelic exchange mutagenesis of two flagellin genes of Helicobacter felis. Mol Microbiol. 1999;33:350–362. doi: 10.1046/j.1365-2958.1999.01478.x. [DOI] [PubMed] [Google Scholar]
- 17.Kostrzynska M J, Betts D, Austin J W, Trust T J. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J Bacteriol. 1991;173:937–946. doi: 10.1128/jb.173.3.937-946.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee A, O'Rourke J, De Ungria M C, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 19.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- 21.Osborn M J, Wu H C P. Proteins of the outer membrane of gram-negative bacteria. Annu Rev Microbiol. 1980;34:369–422. doi: 10.1146/annurev.mi.34.100180.002101. [DOI] [PubMed] [Google Scholar]
- 22.Peck B, Ortkamp M, Diehl K D, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shomer N H, Dangler C A, Marini R P, Fox J G. Helicobacter bilis/Helicobacter rodentium co-infection associated with diarrhea in a colony of scid mice. Lab Anim Sci. 1998;48:455–459. [PubMed] [Google Scholar]
- 24.Shomer N H, Dangler C A, Schrenzel M D, Fox J G. Helicobacter bilis-induced inflammatory bowel disease in scid mice with defined flora. Infect Immun. 1997;65:4858–4864. doi: 10.1128/iai.65.11.4858-4864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomb J-F, White O, Kerlavage A, Clayton R, Sutton G, Fleischmann R, Ketchum K, Klenk H, Gill S, Dougherty B, Nelson K, Quackenbush J, Zhou L, Kirkness E, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H, Glodek A, McKenney K, Fitzegerald L, Lee N, Adams M, Hickey E, Berg D, Gocayne J, Utterback T, Peterson J, Kelley J, Cotton M, Weidman J, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W, Borodovsky M, Karp P, Smith H, Fraser C, Venter J. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 26.Tufano M A, Rossano F, Catalanotti P, Liguori G, Capasso C, Ceccarelli M T, Marinelli P. Immunobiological activities of Helicobacter pylori porins. Infect Immun. 1994;62:1392–1399. doi: 10.1128/iai.62.4.1392-1399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versalovic J, Fox J G. Helicobacter. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 727–738. [Google Scholar]