Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Vaccines, VLPs
Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) first appeared in Wuhan, China, in December 2019. The 2019 coronavirus disease (COVID-19) pandemic, caused by SARS-CoV-2, has spread to almost all corners of the world at an alarming rate. Vaccination is important for the prevention and control of the COVID-19 pandemic. Efforts are underway worldwide to develop an effective vaccine against COVID-19 using both traditional and innovative vaccine strategies. Compared to other vaccine platforms, SARS-CoV-2 virus-like particles (VLPs )vaccines, as a new vaccine platform, have unique advantages: they have artificial nanostructures similar to natural SARS-CoV-2, which can stimulate good cellular and humoral immune responses in the organism; they have no viral nucleic acids, have good safety and thermal stability, and can be mass-produced and stored; their surfaces can be processed and modified, such as the adjuvant addition, etc.; they can be considered as an ideal platform for COVID-19 vaccine development. This review aims to shed light on the current knowledge and progress of VLPs vaccines against COVID-19, especially those undergoing clinical trials.
1. Introduction
An outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with cases of severe pneumonia of unknown origin has been reported in China (Wuhan, Hubei Province) since December 2019. The 2019 coronavirus disease (COVID-19) outbreak caused by the SARS-CoV-2 coronavirus was declared a global pandemic by the World Health Organization (WHO) on March 11, 2020, and COVID-19 has swept the globe, affecting nearly 208 countries. Globally, as of 16 December 2022 there have been 647,972,911 confirmed cases of COVID-19, including 6,642,832 deaths, and a total of 13,008,560,983 vaccine doses have been administered(https://covid19.who.int). SARS-CoV-2 caused the third viral pandemic of the 21st century, and the third zoonotic coronavirus pandemic outbreak in the past 20 years [1]. SARS-CoV-2 has genetically mutated under genetic evolutionary pressures and will continue to drift mutations. As the SARS-CoV-2 epidemic continues, it will lead to greater transmissibility or higher mortality. Vaccines are an important tool for preventing and controlling outbreaks; therefore, developing a safe and effective SARS-CoV-2 vaccine is today's highest priority and most urgent issue. To date, more than a dozen vaccines have been approved for use by the World Health Organization (WHO) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines). Currently, there are 175 vaccine candidates in clinical trials and 199 vaccine candidates in preclinical trials (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines).
A variety of different vaccine platforms are being used to develop these vaccine candidates: protein-based subunit and virus-like particle vaccines, virus-based attenuated and inactivated vaccines, and gene delivery strategy vaccines such as nucleic acid vaccines and viral vector vaccines (https://www.who.int/publications/). Although traditional vaccines, such as inactivated viruses, live attenuated and subunit vaccines are widely used, they often pose safety, limited cross-protection and immunogenicity issues [2]. In this regard, new vaccine platforms with virus-like particles can highlight their unique advantages.
VLPs are nanoscale particles self-assembled from one or more viral structural proteins with a spatial conformation and antigenic epitopes closer to natural viral particles [3]. These antigenic epitopes are presented in a concentrated and dense manner through VLPs to induce efficient and long-lasting humoral immune responses in vivo. The granular morphology of VLPs makes them more susceptible to phagocytosis by dendritic cells (DCs) and presentation to major histocompatibility complexes class I (MHC I) and class II (MHC II), thus activating an adaptive immune response and inducing strong cellular and humoral immune responses [4]. Furthermore, VLPs do not contain viral genetic material and therefore cannot infect or replicate and have a good safety profile [5]. The immunological advantages of VLPs have made them a hot topic in current vaccine research.
VLPs platforms are not only used for vaccine development but also as vehicles to carry exogenous antigens or deliver adjuvants [6]. VLPs vaccines that have been approved for marketing include Sci-B-Vac™ for hepatitis B, Cervarix®, Gardasil®, Gardasil 9® and Cecolin® for cervical cancer, Mosquirix™ for malaria, and Hecolin® for hepatitis E. In addition, many SARS-CoV and MERS-CoV VLPs vaccines have been reported and some SARS-CoV-2 VLPs vaccines have undergone preclinical and clinical trials.
This review aims to elucidate the current progress and discuss the use of VLPs vaccines against COVID-19.
2. SARS-CoV-2
SARS-CoV-2, a member of the coronavirus family, belongs to the beta-coronavirus, which has a genome size of 26.5–31.7 kb [7]. SARS-CoV-2 contains four structural proteins, a spike protein (S), an envelope protein (E), a nucleocapsid protein (N), and a membrane protein (M) [8]. The S protein is a very important surface protein of the novel coronavirus and is closely related to the infectivity of the virus. S protein contains S1, S2, and RBD (receptor binding domain). It binds to the host cell receptor and mediates the attachment and fusion of the virus and host membrane. The E protein is a component of the viral particle envelope and interacts with the M-protein to form the viral membrane. N protein is abundant in SARS-CoV-2, a highly immunogenic protein involved in the regulation of genome replication and cellular signaling pathways. It binds to the genomic RNA of the virus to form the nucleocapsid. The M protein is also a component of the viral particle envelope. It is involved in the assembly and release of next-generation viral particles and is important for the structural stability and functional expression of other structural proteins (S, E, and N proteins). The M protein determines the viral shape and is the central organizer of coronavirus assembly [9].
SARS-CoV-2 is human-transmissible [10]. The outbreak characteristics and symptoms of COVID-19 share similarities with the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 and the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, both of which are also the β-coronavirus [8], [10]. Although SARS-CoV-2 shares most of the sequence similarities with MERS-CoV, SARS-CoV, however, SARS-CoV-2 shows a longer incubation period, higher infection rate, and faster asymptomatic transmission [11], [12]. From the beginning of the outbreak to the present, SARS-CoV-2 has evolved and mutated, producing more transmissible and virulent variant strains. Some variants have been reported to increase transmissibility and infectivity while evading host antibody recognition. For example, the Alpha (B.1.1.7) variant in the UK, the Beta (B.1.351) variant in South Africa, the Gamma (P.1) variant in Brazil, the Delta (B.1.617.2) variant in India, the Omicron (B.1.1.529) variant in South Africa, the Lambda (C.37) variant in Peru or Chile and the Colombian Mu (B.1.621) variant in Colombia [13], [14], [15], [16], [17], [18]. Since October 11, 2021, there has been a significant increase in confirmed cases worldwide. The properties of SARS-CoV-2 variants have been listed in Table 1 .
Table 1.
Properties of SARS-CoV-2 variants (https://outbreak.info/).
| Lineage | Earliest documented samples | Key mutations of spike protein | Transmissibility |
|---|---|---|---|
| Alpha (B.1.1.7) | UK, September 2020 | DEL69/70, DEL144/145,N501Y | ∼50 % increase in comparison with previously circulating strains [22], [23] |
| Beta (B.1.351) | South Africa, September 2020 | L18F,DEL241/243, K417N,E484K,N501Y | 50 % increase [26], [27], [28] |
| Gamma (P.1) | Brazil, December 2020 | L18F,K417T,E484K,N501Y | 1.4–2.2 times more transmissible [32] |
| Delta(B.1.617.2) | India, December 2020 | G142D,E156G,DEL157/158,L452R,T478K | 97 % increase [33], [34], [35], [36], [37], [38] |
| Omicron (B.1.1.529) | South Africa, November 2021 | DEL143/145,S373P,S375F,K417N,S477N,T478K,E484A,Q493R,Q498R,N501Y,Y505H | Compared with Delta variant, the transmission of Omicron is enhanced [42] |
| Lambda(C.37) | Peru and Chile, April 2021 | T76I, F490S and L452Q | Increased transmissibility relative to that of the parental Wuhan strain [43], [44], [45] |
| Mu (B.1.621) | Colombia, January 2021 | E484K,N501Y,P681H,D614G | Mu is likely to be less transmissible than Delta [46], [47], [48] |
2.1. Alpha (B.1.1.7) variant
The B.1.1.7 mutant strain swept the UK in December 2020 after 2 months of first appearance [19], spread to the US in April 2021, and then rapidly swept the world. The mutant strain belongs to the B.1.1.7 spectrum of SARS-CoV-2 with 17 mutations, 9 of which are on the spike S protein [20], [21]. B.1.1.7 is more infectious than the identified SARS-CoV-2 variants and may cause more severe disease [22], [23].
2.2. Beta (B.1.351) variant
The Beta variant strain, also known as 20H/501Y.V2, includes B.1.351 and its 2 branches, B.1.351.2 and B.1.351.3 [24], [25]. The B.1. 351 variant was first detected in a sample collected in South Africa in December 2020 and became the dominant variant in South Africa within a few weeks [21], [26]. The B.1. 351 variants can alter the conformation of the RBD, increase the affinity for the ACE2 (angiotensin-converting enzyme 2) receptor, and enhances infectivity [27], [28].
2.3. Gamma (P.1) variant
The P.1 variant strain, also known as 20J/501Y.V3, is a branch of the B.1.1.28 spectrum that first appeared in the city of Manaus, Brazil, in December 2020 and caused a massive outbreak throughout the city by mid-January 2021, resulting in approximately 85.4 % of the population being infected with the P.1 variant. This mutant strain has 11 mutations in the spike S protein [29]. There is evidence that some mutations in the P.1 variant strain may affect its transmissibility and antigenicity and may also evade recognition and neutralization of the virus by antibodies produced by prior natural infection or vaccination [30].
2.4. Delta (B.1.617.2) variant
B.1.617.2 first appeared in India in October 2020. In March 2021, this variant triggered a second wave of the COVID-19 epidemic in India, infecting approximately 0.36 % of the population in 2 months, with more than 9.02 million new cases of COVID-19 in India alone as of May 2021, the worst month of the global epidemic since the 2019 epidemic [31], [32]. B.1.617.2 rapidly becoming the most prevalent variant in India and several countries worldwide [33], [34], [35], [36].
2.5. Omicron (B.1.1.529) variant
The first case of B.1.1.529 infection was detected in South Africa on November 24, 2021 [37], and the variant spread rapidly in South Africa, becoming the major epidemic variant and spreading to several countries worldwide. This variant has more than 30 mutations in spike protein S [38], [39]. Omicron's mutation builds on the Delta and Alpha variants and is ultra-transmissible and infectious [40]. As previously reported in China's Health Times, on September 28, 2022, a case of indigenous COVID-19 infection was reported in Hohhot, Inner Mongolia Autonomous Region, which belongs to the evolutionary branch of the COVID-19 Omicron variant strain bf.7. In just a few days, the infection has spread to more than 500 people confirmed. As early as September 23, 2022, the British newspaper Coventry Telegraph reported that the new mutant strain bf.7 was spreading rapidly in Belgium, Germany, France, Denmark, and the United States. The World Health Organization issued a warning that bf.7 may become mainstream globally within weeks.
2.6. Lambda (C.37) variant and Mu (B.1.621) variant
C.37 was first detected in Peru and Chile in April 2021, and in June 2021 it was classified as a virus variant of “concern” by the WHO. The variant subsequently spread to North America, Europe, and the Middle East. The Lambda variant carries a mutation L452Q in the RBD of the Spike protein. The L452R mutation has been reported to be associated with immune evasion and stronger cellular attachment, which can increase viral transmissibility, infectivity, and pathogenicity [41], [42], [43]. B.1.621 is also classified as a virus variant of “concern” by the WHO. Mehul Suthar et al. found that the vaccine had a significantly lower neutralization capacity against the emerging B.1.621 variant than the original strain [44], [45], [46].
Conventional SARS-CoV-2 vaccines generally exert immune protection based on a specific strain s protein. Undoubtedly, as more variants emerge, the protective effect of this vaccine class will be significantly reduced, increasing viral transmissibility and immune escape and posing a significant challenge for COVID-19 control [13]. Rapid mass production of next-generation vaccines against these variants is critical in controlling the COVID-19 pandemic. VLPs are self-assembled biomolecules that closely resemble natural viruses and can be produced on a large scale in short cycles [47]. VLPs exhibit repetitive and dense viral epitopes that can elicit enhanced immunostimulatory effects. VLPs can display SARS-CoV-2 multivalent homologous or variant antigens on their surface and have the potential to become SARS-CoV-2 vaccine candidates that exert broad protection against multiple variants [48].
3. VLPs vaccine platform
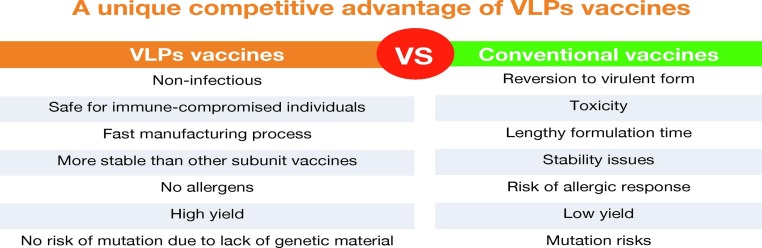
VLPs are highly structured hollow particles, 20–200 nm in diameter, that are structurally identical or similar to natural viral particles, do not contain viral genetic material, and are formed by inserting one or more genes encoding viral structural proteins into an expression vector and then transferring them into a heterologous expression system, such as prokaryotic or eukaryotic cells, for self-assembly [47]. VLPs vaccines are similar to conventional vaccines, the only difference being that VLPs do not contain viral nucleic acids and therefore cannot proliferate [49]. This also means that VLPs vaccines are not infectious and have a higher safety profile. VLPs technology provides an enabling platform for developing effective vaccines against virulent infectious diseases and has been developed in parallel with RNA and viral vector vaccine [50]. VLPs are potentially efficient vaccine candidates due to their unique properties (Fig. 1 ). VLPs are more immunogenic compared to other subunit vaccines. They are potent immunostimulatory molecules that can display repetitive antigenic epitopes on their surface, triggering efficient T and B cell immune responses. The VLPs platform can overcome various side effects associated with conventional vaccines, such as, live attenuated vaccines having a risk of reversion to toxicity and genetic variation, inactivated vaccines having low immunogenicity, unstable toxicity, and low yield, subunit vaccines having a low potential for immune response, poor efficacy, and stability, and some viral vector or RNA vaccines may be associated with complications such as thrombocytopenia and myocarditis [51]. Most VLPs are derived from viral capsid or envelope proteins, and nucleoproteins can also be used [52].
Fig. 1.
The unique advantages of VLPs vaccines compared to conventional vaccines.
VLPs are non-infectious with a good safety profile, as they lack the virus's genetic material and cannot replicate. They are considered safer than conventional viral vaccines [53]. In addition, VLPs are naturally biocompatible and are highly versatile molecules with sizes ranging from 20 to 200 nm. This size range is optimal, allowing them to freely drain into lymph nodes and be more readily taken up by antigen-presenting cells (APCs) [52], [54], particularly DCs, which are then antigenically processed and presented by MHC II molecules [55]. VLPs are highly self-assembling and can self-assemble into different geometrically symmetric conformations, usually icosahedral, helical symmetric, rod-like structures, or spherical, depending on the characteristics of the virus [56]. VLPs can increase their functionality by using different methods such as peptide ligation, gene fusion, and chemical cross-linking, displaying targeted heterologous epitopes, and modifying their external or internal surface proteins [57], [58]. VLPs technology offers significant advantages as it is a more rapid method for vaccine synthesis. New VLPs vaccines against specific strains can be prepared within 12–14 weeks after strain sequencing, compared to the usual 24–32 weeks for conventional vaccines. Different host expression systems (ES) are available for the manufacture of VLPs vaccines, including bacterial, yeast, plant, mammalian, insect, and cell-free expression systems [49].
4. Mechanism of action of VLPs vaccines
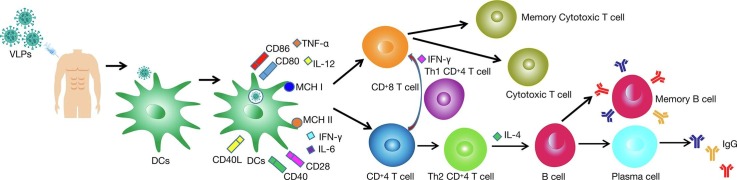
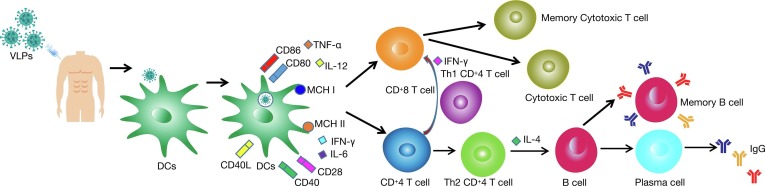
VLPs significantly induced cellular immune responses with the proliferation of CD4+ T and CD8+ T cells, and induced B cell-mediated humoral immune responses [58]. The structure of VLPs is closely related to their efficiency in inducing immune responses in the organism. Studies have shown that the interaction of VLPs with the immune system is mainly related to two factors, VLPs size and their surface geometry [59]. VLPs of appropriate size (20–200 nm) can be drained to lymph nodes through lymphatic vessels. As the injection time increases, VLPs are dispersed to different areas, and myeloid and B cells residing in lymph nodes will carry free VLPs to B-cell follicles [60]. The different distribution pattern of VLPs increases the contact and interaction of particles with immune cells [DCs, macrophages, B cells] in secondary lymphoid organs, facilitating the generation of an immune response against VLPs (Fig. 2 ).
Fig. 2.
Induction of Intrinsic and adaptive immunological responses after immunization with SARS-CoV-2 VLPs vaccines. The administration VLPs stimulates both humoral and cell-mediated immunity. VLPs interact with pattern recognition receptors (PRRs) present on the dendritic cells (DCs), such as the toll-like receptors (TLRs). DCs then engulf the VLPs through phagocytosis. This leads to the maturation of DCs, which then secrete pro-inflammatory cytokines such as TNF-α to IL-12 and recruit more antigen-presenting cells (APCs). VLPs taken up by DCs are subsequently enzymatically cleaved into short peptides that bind to major histocompatibility complexes class I (MHC I) and class II (MHC II) and are translocated to the DCs surface. The short peptide displayed on MHC II, together with CD40 and CD80/86 then interact with T cell receptor (TCR), CD40L, and CD28 presence on the naive helper T cell (Th), respectively. This promotes the proliferation and differentiation of Th cells into type 1 (Th1) and type 2 (Th2) Th cells. Aided by Th1, naive CTL proliferates and differentiates into effector and memory CTLs, providing immediate and long-lasting cellular immunity, respectively. On other hand, aided by Th2, the naive B cell differentiates into plasma B cells which actively secrete antibodies and memory B cells which provide long-lasting humoral immunity.
4.1. Intrinsic immune responses of VLPs
Appropriately sized VLPs can be taken up by DCs through phagocytosis and infiltration mechanisms. VLPs enter DCs and induce their maturation, upregulate the expression levels of cell surface molecules such as CD40, CD80, CD86, and promote the secretion of various chemokines and anti-inflammatory cytokines from DCs [49]. Toll-like receptor (TLR) stimulation with ligands also plays an important role in the VLPs immune response. APC-expressed TLR binds to pathogen-associated pattern molecules and promotes the release of multiple cytokines (IL-12, IL-6, IFN-γ) [61]. Toll-like receptors (TLRs) on B cells promote antibody-type switching. Tian et al. have found that TLR on B cells can bind to TLR ligand-containing QB-VLPs by detecting the immune response of B cells to phage Q-derived VLPs and promote signal transduction by binding to myeloid differentiation protein 88 to cause IFN-γ secretion and IgG2a/c type switching [62]. Nucleic acid molecules contained within the VLPs can also induce TLR-mediated activation. Some studies have shown the possible regulatory role by incorporating the ligand non-coding RNA of TLR in retroviral VLPs and found that non-coding RNA could be effectively encapsulated by VLPs and induce TLR7/8, which in turn activates the immune response [63].
4.2. Adaptive immune responses of VLPs
The highly repetitive surface structure of VLPs facilitates their interaction with the complement system. Studies have shown that IgM or members of the pentapeptide protein family (including C-reactive protein, serum amyloid P, and penetration 3) can bind with high affinity to VLPs with a highly repetitive surface structure and produce a regulatory effect by recruiting C1q, which sequentially activates C1rC1s and triggers a cascade of enzymatic processes in the classical pathway of complement, thereby allowing VLPs entry into cells to activate an adaptive immune response [64].
Adaptive immune responses include both humoral and cellular immune responses, and VLPs can effectively elicit both specific humoral and specific cellular immune responses. As an exogenous antigen, VLPs can be efficiently presented by both MHC I and II. The efficiency of VLPs presentation by MHC I and II was investigated by in vivo experiments, which showed that MHC II cross-presented exogenous antigenic VLPs 10-fold more efficiently than MHC I [65]. Wang et al. [66] induced immune responses in mice with HIV gag-enterovirus 71 (EV71) VP1 VLPs and showed that chimeric VLPs (cVLPs) induced high titer neutralizing antibodies and associated cytokine production, indicating that mixed Th1/Th2 immune response could be generated. Dai et al. [67] successfully packaged Zika virus (Zikavirus, ZIKV) VLPs with an insect baculovirus expression system and performed immune assays in vivo and in vitro, and found that ZIKV VLPs not only induced the production of virus-specific IgG antibodies and Th cytokines (IL-2, IL-4, IL-10) but also stimulated T cells to secrete IFN-γ and induced specific anti-ZIKV T cell immune response.
5. Significance of developing VLPs vaccines against COVID-19
The ongoing SARS-CoV-2 epidemic has caused severe damage to the global economy and has become a concern for every citizen in the world. The COVID-19 global pandemic caused by SARS-CoV-2 was so large that it became the second global pandemic of the 21st century [68]. Currently, three antiviral drugs have been approved by the FDA and WHO for the treatment of humans infected with SARS-CoV-2, including Remdesivir R (Gilead Sciences), Paxlovid R (Pfizer), and Molnupiravir R (Merck and Co.). Based on various considerations of the process and cost of the development of drugs against SARS-CoV-2, researchers are focusing on developing an effective vaccine against COVID-19 for the long term. VLPs against SARS-CoV-2 have played a good role in the prevention and control of the SARS-CoV-2 pandemic. In addition to being a viable vaccine against SARS-CoV-2, VLPs can also be used as a therapeutic agent and a viable diagnostic tool [69]. SARS-CoV-2 must enter the host cell via the endosomal route, and its infection mechanism is slow to evade the immune system effectively, and it typically takes 5–15 days to show symptoms after entering the body. In animal model studies, vaccines against SARS and other coronaviruses (CoVs) strains of Middle East Respiratory Syndrome (MERS) could resist infection with some SARS-CoV-2 strains [70]. Like other CoVs, SARS-CoV-2 consists of four proteins S, N, E, and M, which are structurally conserved in different SARS-CoV-2 strains. The precise contribution of the above four proteins and their associated interaction patterns are essential for the production and assembly of SARS-CoV-2 VLPs [71]. The S protein is common in different coronavirus strains and becomes a potential target for SARS-CoV-2 VLPs vaccine development. Before developing vaccine SARS-CoV-2 VLPs vaccine, they must first be based on previous work on VLPs of closely related viral strains. VLPs of MERS-CoV can be produced by expressing S proteins in insect host cell ES [72], [73]. Previous findings suggested that VLPs produced in rBV ES can effectively induce specific humoral and cellular immune responses to SARS-CoV in mice [74].
As of 13 December 2022, scientists have developed multiple SARS-CoV-2 VLPs vaccine candidates, 19 in the preclinical stage, 6 in the clinical stage, and some in the preliminary stage for use in laboratory animal models (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines). Studies have shown that SARS-CoV-2 VLPs vaccine candidates in these three experimental stages can exhibit good safety and immunogenicity. Due to the flexibility of the virus-like particle vaccine platform, antigens of different SARS-CoV-2 variants can be demonstrated on VLPs. The SARS-CoV-2 VLPs vaccine designed on this basis will show good cross-immunogenicity [75]. Due to the advantages of good safety, short production cycles, and mass production, the VLPs vaccine platform had been used in the preparation of vaccines for many human diseases, among them, VLPs vaccines for hepatitis virus, cervical cancer, and influenza have been marketed one after another. Compared to other platform vaccines on the market, the SARS-CoV-2 VLPs vaccine is expected to be a universal cross-immune candidate against multiple variants and may even provide a basis for the development of vaccine candidates for other life-threatening viruses.
6. Current status of research on VLPs vaccines against COVID-19
VLPs is a self-assembled nanostructures containing key viral structural proteins. VLPs have molecular and morphological characteristics like those of true viruses but are not infectious and non-replicating due to the lack of genetic material. The successful application of VLPs in vaccinology and virology studies has been demonstrated. As a safe and relevant alternative to naturally pathogenic viruses, the construction of SARS-CoV-2 VLPs is necessary for the current fight against the COVID-19 pandemic [76]. Understanding the genomic structure of the SARS-CoV-2 virus is key to developing a COVID-19 vaccine [77], [78]. During viral infection, the body produces antibodies mainly against the N and S proteins. The N protein covers the genome of the virus and is also involved in the release of viral particles from the cell. In contrast, the S protein plays an important role in pathogenesis by binding to host cells through its receptor-binding domain, thus initiating the infection of host cells [79]. S and N proteins have been used as preferred antigens for the development of VLPs vaccines.
6.1. SARS-CoV-2 VLPs based on various expression systems
Currently, researchers in laboratories around the world have utilized conventional host expression systems (ES) for SARS-CoV-2 VLPs vaccine preparation, such as yeast, plant, mammalian, and insect expression systems.
The S, E, and M proteins from the surface capsid of SARS-CoV-2 viruses are important vaccine targets. Saumyabrata et al. [80] reported the recombinant co-expression of three proteins (S, E, and M) in an engineered brewer's yeast platform (D-Crypt™) that self-assembles into virus-like particles (VLPs). This design, as a multi-antigen VLP for SARS CoV-2, has the potential to be a scalable vaccine candidate. Transmission electron microscopy (TEM) images of SARS-CoV-2 along with supporting High-Performance Liquid Chromatography (HPLC), dynamic light scattering (DLS), and correlation analysis data confirmed the VLPs. These images clearly outline the “Corona”-like morphology and uniform size distribution.
Ghorbani et al. [81] used molecular farming biotechnology to synthesize SARS-CoV-2 VLPs vaccines in different plant species, targeting the antigenic epitopes of S proteins and inducing specific immunogenic responses. Moon KB et al. [82] demonstrated that non-infectious SARS-CoV-2 VLPs can be successfully assembled by co-expressing three important viral proteins membrane (M), envelope (E), and nucleocapsid (N) in plants. The shape and size of plant-derived VLPs are similar to native SARS-CoV-2 VLPs without spikes. Although the assembled VLPs do not have S protein spikes, they could be developed as formulations that can improve the immunogenicity of vaccines including S antigens, and further could be used as platforms that can carry S antigens of concern for various mutations.
Xu et al. [83] effectively constructed SARS-CoV-2 VLPs using a mammalian expression system (which has advantages in maintaining the correct protein glycosylation pattern). The results indicate that among the four SARS-CoV-2 structural proteins, the expression of membrane protein (M) and small envelope protein (E) is essential for the efficient formation and release of SARS-CoV-2 VLPs. Furthermore, SARS-CoV-2 VLPs from Vero E6 cells showed a more stable and homogeneous coronal structure compared to SARS-CoV-2 VLPs from HEK-293T cells. The data suggest that SARS-CoV-2 VLPs have the molecular and morphological properties of native viral particles, and these properties confer VLPs as a promising vaccine candidate and a powerful tool for studying SARS-CoV-2. Swann et al. [84] made SARS-CoV-2 VLPs by co-expressing viral proteins S, M, and E in mammalian HEK-293T cells. Such VLPs maintain their structural integrity after being naturally air-dried under environmental conditions, which facilitates the transportation and storage of VLPs for application as vaccines.
Chu et al. [85] developed SARS-CoV-2 virus-like particles (VLPs), which are assembled from full-length spike (S) glycoprotein (S full), S1 or S2, and influenza matrix protein 1 (M1), M1 is the core protein. VLPs expressing full S, S1, and S2, which were successfully constructed by transfecting Sf9 cells. VLPs were confirmed and characterized by Western blot and transmission electron microscopy (TEM). Compared to controls, VLP-immunized mice induced higher levels of spike protein-specific IgG and its subclasses, with IgG2a being the predominant subclass. The intact S and S1 immune sera elicited virus-neutralizing activities, but these were not sufficient to completely inhibit the receptor-ligand binding of SARS-CoV-2. No neutralizing activity was observed for the S2 VLP immune serum. Overall, our results suggest that all-S or S1-containing VLPs can be developed into effective vaccines.
6.2. SARS-CoV-2 VLPs vaccine in clinical trials
The SARS-CoV-2 VLPs vaccines have been developed more rapidly than SARS-CoV and MERS. The average period for vaccine research and commercial development is typically 10 to 15 years, and in response to the SARS-CoV-2 pandemic worldwide, the development time for the SARS-CoV-2 VLPs vaccine has been shortened to 12–18 months. Six SARS-CoV-2 VLPs vaccine candidates are currently in clinical trials (Table 2 ), and one SARS-CoV-2 VLPs vaccine candidate has been commercialized [75].
Table 2.
Overview of the clinical trials of VLPs vaccines against COVID-19 as of November 1, 2022 (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines).
| Developers/Study identifier | Study phase | Type of candidate vaccine | Via | Subjects | Number of subjects | Study location | Project title |
|---|---|---|---|---|---|---|---|
| Radboud University/NCT05329220 | Phase 3 | ABNCoV2 capsid virus-like particle (cVLP) +/- adjuvant MF59 | IM |
Adults (18 Years and older) | 4000 | United States, Georgia | Evaluation of the Immunogenicity, Safety, and Tolerability of a Single Dose of ABNCoV2 Vaccine in Adult Subjects Previously Vaccinated for SARS-CoV-2: a Phase 3 Trial in Two Parts-Randomized, Double-blind, Active Controlled and Open-label, Single-arm |
| Medicago Inc./NCT05040789 | Phase 3 | Coronavirus-Like Particle COVID-19 (CoVLP) MT-2766 Other Name: CoVLP, AS03 adjuvant |
IM | Adults (18–49 years old) (Not yet recruiting) |
900 | Canada, Ontario | A Randomized, Observer-Blind, Multicenter Study to Evaluate the Lot Consistency of a Recombinant Coronavirus-Like Particle COVID-19 Vaccine in Healthy Adults |
| The Scientific and Technological Research Council of Turkey/NCT04962893 | Phase 2 | SARS-CoV-2 VLP: Vaccine/Vaccine-Wuhan; Vaccine-Alpha variant; Vaccine-Wuhan + Alpha variant | SC | Adults (18–59 years old) | 349 | Turkey | Assess the Safety, Efficacy, and Immunogenicity of Authentic SARS-CoV-2 or Alpha Variant Spike Containing VLP Vaccines and Their Combination for the Prevention of COVID-19 in Healthy Adult Volunteers (SAVE STUDY) |
| VBI Vaccines Inc./NCT04773665 | Phase 1/2 | VBI-2902a. An enveloped virus-like particle (eVLP) of SARS-CoV-2 spike (S)/glycoprotein and aluminum phosphate adjuvant | IM |
Adults (18–54 years old) | 114 | Canada, Nova Scotia | A Randomized, Observer-Blind, Placebo-Controlled Study to Evaluate the Safety, Tolerability, and Immunogenicity of the COVID-19 (SARS-CoV-2) Vaccine Candidates VBI-2902a and VBI-2905a in Healthy Adults |
| Serum Institute of India + Accelagen Pty + SpyBiotech/ACTRN12620000817943/ACTRN12620001308987 | Phase 1/2 | RBD SARS-CoV-2 HBsAg VLP/RBD SARS-CoV-2 HBsAg VLP (Adjuvanted with Alum + CpG 1018/Alum alone) | IM/ IM |
Adults (18–79 years old)/Adults (18–79 years old)(Not yet recruiting) | 280/255 | Australia/Australia (Not yet recruiting) | A randomized, placebo-controlled, multi-centre study to evaluate the safety and immunogenicity of COVID-19 Vaccine in Healthy Adults/ A randomized, placebo-controlled, multi-centre study to evaluate the safety and immunogenicity of a novel Receptor Binding Domain (RBD) COVID-19 Vaccine in Healthy Adults |
| Yantai Patronus Biotech Co., Ltd./NCT05125926 | Phase 1 | SARS-CoV-2 Vaccine LYB001, a receptor-binding domain (RBD) from SARS-CoV-2 and virus-like particle (VLP) vector, adjuvanted with aluminum hydroxide. | IM | Adults(18 Years and older) (Not yet recruiting) | 100 | Not yet recruiting |
Safety, Reactogenicity, and Immunogenicity of a SARS-CoV-2 Vaccine LYB001 in Healthy Adults: a Randomized, Double Blinded, Placebo-controlled Phase Ⅰ Trial |
IM, intramuscular; SC, subcutaneous injection.
ABNCoV2 is a clinically tested SARS-CoV-2 VLP vaccine developed by Bavarian Nordic. The vaccine is a Capsid VLP vaccine consisting of a combination of S-RBD antigens expressed by ExpreS2ion Biotechnologies' Drosophila S2 insect cell protein expression system (ExpreS2ion) and AdaptVac's proprietary Capsid VLP (cVLP). The RBD antigen on the cVLP was demonstrated to be targeted and highly reproducible. Preclinical results showed that the vaccine exhibited effective neutralization of SARS-CoV-2 in a mouse model [86]. The virus-neutralizing antibody titers induced by a single injection of the ABNCoV2 vaccine in mice were similar to those detected in patients recovering from SARS-CoV-2 [87]. Currently, A Phase II clinical trial (NCT05077267) has been initiated.
The use of transient plant expression to produce vaccines is a rapid and effective technology in response to pandemic infectious diseases such as SARS-CoV-2. Glaxo SmithKline (GSK), the world's largest vaccine manufacturer, has partnered with Canadian biopharmaceutical company Medicago to develop CoVLP, a SARS-CoV-2 VLP vaccine candidate based on Australian weed plants. The CoVLP vaccine consists of stabilized pre-fusion S protein self-assembled VLPs formulated with Adjuvant System 03(ASO3)and Nonmethylated cytosine phosphate guanine dinucleotide sequence (CpG) as adjuvants. Results of clinical phase I trials have shown that IL-4 and IFN-γ immune responses are well elicited in individuals [88], [89]. CoVLP is currently in phase III clinical trial (NCT04636697) enrolling 30,918 participants in North America and has shown favorable efficacy and safety profiles for CoVLP [90], [91].
Unlike most VLP-based SARS-CoV-2 vaccines undergoing clinical trials, this SARS-CoV-2 VLPs vaccine was developed by the Scientific and Technical Research Council of Turkey containing the M, N, E, and HexaPro S antigens of the virus, supplemented with K-3CpG ODN [92]. The phase I study is designed as a double-blinded, randomized, placebo-controlled, three-armed study composed of two different dose arms against SARS-CoV-2 in dose escalations (first low dose group, followed by high dose group) and placebo arm. Each dose of vaccine will be defined as a cohort, with vaccine administration to 12 participants and placebo administration to 6 participants. The Phase II trial is to evaluate the humoral and cellular immune response of VLP vaccine candidates, as an efficacy criterion. Approximately 330 subjects will be randomized in a 1:1:1 ratio to receive two doses of 40 mcg VLPs vaccines for Wuhan (n = 110) or 40 mcg VLPs vaccines for the Alpha (British) variant (n = 110) or 40 mcg VLP vaccine for Wuhan Alpha variant (n = 110) 21 days apart. The route of administration of this vaccine differs from other clinically tested VLPs vaccines in that this SARS-CoV-2 vaccine is administered subcutaneously [93].
VBI (Viva BioInnovator) Vaccines has also invested in the development of a SARS-CoV-2 VLPs vaccine. Mouse leukemia virus (MLV)-based enveloped virus-like particles (eVLPs) were used to produce the SARS-CoV-2 vaccine candidate, VBI-2900 [94], [95]. VBI-2900 consists of three enveloped virus-like particle (eVLP) vaccine candidates: VBI-2901, a multivalent coronavirus vaccine expressing SARS-CoV-2, SARS-CoV, and MERS-CoV spike proteins. VBI-2902, a monovalent COVID-19 vaccine expressing a modified pre-fusion form of the SARS-CoV-2 ancestral spike protein. VBI-2905, a monovalent COVID-19 vaccine expressing a modified pre-fusion form of the modified Beta variant (also known as B.1.351) spike protein. The vaccine program was developed through a partnership with the National Research Council of Canada (NRC), the Consortium for Epidemic Prevention Innovation (CEPI), and the Government of Canada through its Strategic Innovation Fund. Phase I/II clinical trials of VBI-2902 and VBI-2905, which evaluated safety, tolerability, and immunogenicity, showed that VBI-2902 was well tolerated, had a good safety profile, and induced a robust immune response in all subjects. On October 1, 2021, VBI Vaccines announced the recent initiation of the Phase I clinical study of VBI-2901. The study is expected to enroll approximately 100 adults in Canada and is supported by the Canadian government.
COVIVAXX is a conjugated SpyCatcher: VLPs vaccine that are obtained by fusing the RBD of the S protein of SARS-CoV-2 to the SpyTag peptide using SpyBiotech's SpyCatcher/SpyTag technology to present the RBD on the surface of hepatitis B surface antigen (HBsAg) VLPs. SpyCatcher/SpyTag technology has been widely used in vaccine development to display highly repetitive antigens in specific directions on VLPs through irreversible covalent bonding [96]. HBsAg VLPs is a vaccine that has been licensed by the FDA and has demonstrated high immunogenicity and safety in humans. It can be used as a very attractive universal plug-and-play vector for the presentation of any antigen of interest. Studies have shown that pre-existing immunity to HBsAg does not affect the immunogenicity of VLPs [97]. The COVIVAXX vaccine requires only refrigerated storage at 2–8 degrees Celsius, and the vaccine is currently in phase III clinical trials.
Based on the unique technology platform of VLP, LYB001, a virus-binding particle-like neocrown vaccine developed independently by Yantai PaiNuo Biotechnology Co., Ltd. has been included in the key follow-up vaccine by the State Council's Task Force on Research and Development of Vaccines for Joint Prevention and Control Mechanism. LYB001 is a demonstration of RBD on a VLP vector formulated with aluminum hydroxide. This vaccine has multiple advantages such as high efficiency, broad spectrum, and no genetic material, and is one of the most promising vaccines for blocking new crown mutant strains such as Delta. At a time when Omicron mutants are spreading rapidly, this vaccine is expected to contribute to a more effective solution for the prevention and control of global neocrown outbreaks. It has completed its Phase III clinical trial, the completion of which requires a 360-day safety observation of all participants after the third dose of vaccine. (https://covid19.trackvaccines.org/agency/who/).
Currently, NVX-CoV2372, produced by Novavax, is considered the only marketed VLPs-based SARS-CoV-2 vaccine. Novavax NVX-CoV2373 consists of a full-length SARS-CoV-2 S protein expressed by baculovirus produced in insect cells and Matrix-M1 adjuvant. The vaccine can be prepared on a large scale using an insect cell baculovirus expression system. The vaccine has shown varying degrees of efficacy in different trials in different countries [98], [99], [100]. Novavax claims that the vaccine has some cross-immune protection against the Omicron mutant strain, but is also developing a new generation of Omicron-specific crown vaccines [101].
7. Conclusion and future perspectives
Zoonotic viruses are an unavoidable threat to humans. It always brings new challenges to the prevention and control of related diseases. Prevention is always considered a better approach than treatment in how to control related diseases without causing the loss of human lives. It can be done effectively by vaccinating the population to save human lives.
COVID-19, as a human-animal pathogen, spreads rapidly around the world and poses an unusual threat to global health. With healthcare systems at high risk of collapse in some countries, governments have had to adopt embargoes, including international travel bans and other public containment measures, to try to reduce morbidity and mortality from the virus globally. These actions, while effective, have resulted in significant economic destruction. With the continued emergence of new variants and the alarming increase in confirmed cases of COVID-19 worldwide, there are concerns about the severity of the disease and its global spread, as well as the efficacy of existing vaccines against new variants. Considering this, when undertaking COVID-19 vaccine development, it is important to have a vaccine platform that is flexible enough to allow manufacturers to respond appropriately to new viral variants.
As a novel direction for vaccine research, VLPs have great potential for the development of vaccines against infectious diseases that are similar to SARS-CoV-2 because of their safety, high immunogenicity, and flexibility to be processed and modified according to the needs of manufacturers. Both SARS-CoV-2 vaccines formed using viral structural proteins and SARS-CoV-2 antigens presented by the VLPs platform induce effective humoral and cellular immune responses with the advantage of high safety and mass production. With its unique advantages, the VLPs platform is now widely used in the development of the SARS-CoV-2 vaccine.
Despite the unique advantages of VLPs as a vaccine platform, the following issues still need to be focused on when conducting VLPs vaccine research: target gene selection, manufacturing process research, structural validation, quality control, and immunogenicity evaluation. It is also noteworthy that self-assembled VLPs cannot be applied to the development of all viral vaccines. The viral structure itself, the choice of target genes, and the properties of the expressed structural proteins themselves may affect the success of developing a VLPs vaccine with uniform particle size, stable properties, and good immunogenicity. These need to be fully studied and demonstrated. Improvement and optimization of separation and purification techniques are also important for the successful preparation of VLPs vaccines. The option of the expression system, the improvement of the assembly efficiency of VLPs, and the immune evasion caused by virus mutation are also issues that also must be considered. In addition, some adjuvants may cause side effects and need to be chosen carefully when adding adjuvants for VLPs.
Fortunately, our research experience from SARS-CoV and MERS-CoV VLPs vaccines, as well as our experience in vaccine development from other diseases, has guided us to propose multiple promising SARS-CoV-2 VLPs vaccines. Because VLPs vaccines are highly immunogenic, self-adjuvant, and versatile, VLPs vaccines will be the most potential to become one of the most effective vaccines against coronaviruses if development efforts can be increased.
In conclusion, we hope that countries around the world, regardless of their political ideology, will unite and work together to achieve rapid and successful development of a COVID-19 vaccine shortly to address the COVID-19 pandemic.
Author contributions
XG and YX wrote this article’s manuscript. JL, GL, XL, YX, PL and ZD reviewed, edited and revised the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Natural Science Foundation of China grant (No. 32102751), the Henan Youth Talent Promotion (No. 2021HYTP056), the Key Science and Technology Program of Henan Province (No. 222102310093, No. 222102310701), Key Scientific Research Project of Colleges and Universities of Henan Province (23A320040).
Data availability
No data was used for the research described in the article.
References
- 1.Binot C., Sadoc J.F., Chouard C.H. SARS-COV2, variants, membranes and basic physics. Bull. Acad. Natl. Med. 2022;206(4):445–447. doi: 10.1016/j.banm.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavriatopoulou M., Ntanasis-Stathopoulos I., Korompoki E., Fotiou D., Migkou M., Tzanninis I.G., Psaltopoulou T., Kastritis E., Terpos E., Dimopoulos M.A. Emerging treatment strategies for COVID-19 infection. Clin. Exp. Med. 2021;21(2):167–179. doi: 10.1007/s10238-020-00671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J., Ren Z., Wang H., Zhao Y., Wilker P.R., Yu Z., Sun W., Wang T., Feng N., Li Y., Wang H., Ji X., Li N., Yang S., He H., Qin C., Gao Y., Xia X. Influenza virus-like particles composed of conserved influenza proteins and GPI-anchored CCL28/GM-CSF fusion proteins enhance protective immunity against homologous and heterologous viruses. Int. Immunopharmacol. 2018;63:119–128. doi: 10.1016/j.intimp.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Qian C., Liu X., Xu Q., Wang Z., Chen J., Li T., Zheng Q., Yu H., Gu Y., Li S., Xia N. Recent progress on the versatility of virus-like particles. Vaccines (Basel) 2020;8(1) doi: 10.3390/vaccines8010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu W.Z., Wen Y.C., Lin S.Y., Chen T.C., Chen H.W. Anti-influenza protective efficacy of a H6 virus-like particle in chickens. Vaccines (Basel) 2020;8(3) doi: 10.3390/vaccines8030465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nooraei S., Bahrulolum H., Hoseini Z.S., Katalani C., Hajizade A., Easton A.J., Ahmadian G. Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021;19(1):59. doi: 10.1186/s12951-021-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omoru O.B., Pereira F., Janga S.C., Manzourolajdad A. A Putative long-range RNA-RNA interaction between ORF8 and Spike of SARS-CoV-2. PLoS ONE. 2022;17(9):e0260331. doi: 10.1371/journal.pone.0260331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20(9):e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai C., Zhong Q., Gao G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022;65(2):280–294. doi: 10.1007/s11427-021-1964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowling B.J., Lau M.S., Ho L.M., Chuang S.K., Tsang T., Liu S.H., Leung P.Y., Lo S.V., Lau E.H. The effective reproduction number of pandemic influenza: prospective estimation. Epidemiology. 2010;21(6):842–846. doi: 10.1097/EDE.0b013e3181f20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia Z., Gong W. Will mutations in the spike protein of SARS-CoV-2 lead to the failure of COVID-19 vaccines? J. Korean Med. Sci. 2021;36(18):e124. doi: 10.3346/jkms.2021.36.e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S.E. Galloway, P. Paul, D.R. MacCannell, M.A. Johansson, J.T. Brooks, A. MacNeil, R.B. Slayton, S. Tong, B.J. Silk, G.L. Armstrong, M. Biggerstaff, V.G. Dugan, Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021, MMWR Morb Mortal Wkly Rep 70(3) (2021) 95–99. [DOI] [PMC free article] [PubMed]
- 15.T. Wilton, E. Bujaki, D. Klapsa, M. Majumdar, M. Zambon, M. Fritzsche, R. Mate, J. Martin, Rapid increase of SARS-CoV-2 variant B.1.1.7 detected in sewage samples from England between October 2020 and January 2021, mSystems 6(3) (2021) e0035321. [DOI] [PMC free article] [PubMed]
- 16.McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., De Marco A., Franko N., Tilles S.W., Logue J., Miranda M.C., Ahlrichs M., Carter L., Snell G., Pizzuto M.S., Chu H.Y., Van Voorhis W.C., Corti D., Veesler D. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374(6575):1621–1626. doi: 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- 17.M. McCallum, A.C. Walls, K.R. Sprouse, J.E. Bowen, L. Rosen, H.V. Dang, A. deMarco, N. Franko, S.W. Tilles, J. Logue, M.C. Miranda, M. Ahlrichs, L. Carter, G. Snell, M.S. Pizzuto, H.Y. Chu, W.C. Van Voorhis, D. Corti, D. Veesler, Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants, bioRxiv (2021). [DOI] [PubMed]
- 18.Qin S., Cui M., Sun S., Zhou J., Du Z., Cui Y., Fan H. Genome characterization and potential risk assessment of the novel SARS-CoV-2 variant omicron (B.1.1.529) Zoonoses. 2021;1(1) [Google Scholar]
- 19.K. Roltgen, S.C.A. Nielsen, O. Silva, S.F. Younes, M. Zaslavsky, C. Costales, F. Yang, O.F. Wirz, D. Solis, R.A. Hoh, A. Wang, P.S. Arunachalam, D. Colburg, S. Zhao, E. Haraguchi, A.S. Lee, M.M. Shah, M. Manohar, I. Chang, F. Gao, V. Mallajosyula, C. Li, J. Liu, M.J. Shoura, S.B. Sindher, E. Parsons, N.J. Dashdorj, N.D. Dashdorj, R. Monroe, G.E. Serrano, T.G. Beach, R.S. Chinthrajah, G.W. Charville, J.L. Wilbur, J.N. Wohlstadter, M.M. Davis, B. Pulendran, M.L. Troxell, G.B. Sigal, Y. Natkunam, B.A. Pinsky, K.C. Nadeau, S.D. Boyd, Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination, Cell 185(6) (2022) 1025–1040 e14. [DOI] [PMC free article] [PubMed]
- 20.Dezordi F.Z., Resende P.C., Naveca F.G., do Nascimento V.A., de Souza V.C., Dias Paixao A.C., Appolinario L., Lopes R.S., da Fonseca Mendonca A.C., Barreto da Rocha A.S., Martins Venas T.M., Pereira E.C., Paiva M.H.S., Docena C., Bezerra M.F., Machado L.C., Salvato R.S., Gregianini T.S., Martins L.G., Pereira F.M., Rovaris D.B., Fernandes S.B., Ribeiro-Rodrigues R., Costa T.O., Sousa J.C., Miyajima F., Delatorre E., Graf T., Bello G., Siqueira M.M., Wallau G.L. Unusual SARS-CoV-2 intrahost diversity reveals lineage superinfection. Microb. Genom. 2022;8(3) doi: 10.1099/mgen.0.000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.Y., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenco J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 22.M. Ramanathan, I.D. Ferguson, W. Miao, P.A. Khavari, SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity, Lancet Infect. Dis. 21(8) (2021) 1070. [DOI] [PMC free article] [PubMed]
- 23.M. Ramanathan, I.D. Ferguson, W. Miao, P.A. Khavari, SARS-CoV-2 B.1.1.7 and B.1.351 Spike variants bind human ACE2 with increased affinity, bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 24.P. Wang, M.S. Nair, L. Liu, S. Iketani, Y. Luo, Y. Guo, M. Wang, J. Yu, B. Zhang, P.D. Kwong, B.S. Graham, J.R. Mascola, J.Y. Chang, M.T. Yin, M. Sobieszczyk, C.A. Kyratsous, L. Shapiro, Z. Sheng, Y. Huang, D.D. Ho, Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7, bioRxiv (2021). [DOI] [PubMed]
- 25.Ho D., Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P., Graham B., Mascola J., Chang J., Yin M., Sobieszczyk M., Kyratsous C., Shapiro L., Sheng Z., Nair M., Huang Y. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. Res. Sq. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 26.Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600(7887):21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 27.M. Hoffmann, P. Arora, R. Gross, A. Seidel, B.F. Hornich, A.S. Hahn, N. Kruger, L. Graichen, H. Hofmann-Winkler, A. Kempf, M.S. Winkler, S. Schulz, H.M. Jack, B. Jahrsdorfer, H. Schrezenmeier, M. Muller, A. Kleger, J. Munch, S. Pohlmann, SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies, Cell 184(9) (2021) 2384–2393 e12. [DOI] [PMC free article] [PubMed]
- 28.D. Zhou, W. Dejnirattisai, P. Supasa, C. Liu, A.J. Mentzer, H.M. Ginn, Y. Zhao, H.M.E. Duyvesteyn, A. Tuekprakhon, R. Nutalai, B. Wang, G.C. Paesen, C. Lopez-Camacho, J. Slon-Campos, B. Hallis, N. Coombes, K. Bewley, S. Charlton, T.S. Walter, D. Skelly, S.F. Lumley, C. Dold, R. Levin, T. Dong, A.J. Pollard, J.C. Knight, D. Crook, T. Lambe, E. Clutterbuck, S. Bibi, A. Flaxman, M. Bittaye, S. Belij-Rammerstorfer, S. Gilbert, W. James, M.W. Carroll, P. Klenerman, E. Barnes, S.J. Dunachie, E.E. Fry, J. Mongkolsapaya, J. Ren, D.I. Stuart, G.R. Screaton, Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera, Cell 184(9) (2021) 2348–2361 e6. [DOI] [PMC free article] [PubMed]
- 29.Guo H., Fan Q., Song S., Shen S., Zhou B., Wang H., Cheng L., Ge X., Ju B., Zhang Z. Increased resistance of SARS-CoV-2 Lambda variant to antibody neutralization. J. Clin. Virol. 2022;150–151 doi: 10.1016/j.jcv.2022.105162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T. Funk, A. Pharris, G. Spiteri, N. Bundle, A. Melidou, M. Carr, G. Gonzalez, A. Garcia-Leon, F. Crispie, L. O'Connor, N. Murphy, J. Mossong, A. Vergison, A.K. Wienecke-Baldacchino, T. Abdelrahman, F. Riccardo, P. Stefanelli, A. Di Martino, A. Bella, A. Lo Presti, P. Casaca, J. Moreno, V. Borges, J. Isidro, R. Ferreira, J.P. Gomes, L. Dotsenko, H. Suija, J. Epstein, O. Sadikova, H. Sepp, N. Ikonen, C. Savolainen-Kopra, S. Blomqvist, T. Mottonen, O. Helve, J. Gomes-Dias, C. Adlhoch, C.S. Groups, Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021, Euro Surveill 26(16) (2021). [DOI] [PMC free article] [PubMed]
- 31.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27(7):1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 32.Du M., Liu M., Liu J. Progress in research of epidemiologic feature and control of SARS-CoV-2 Delta variant. Zhonghua Liu Xing Bing Xue Za Zhi. 2021;42(10):1774–1779. doi: 10.3760/cma.j.cn112338-20210808-00619. [DOI] [PubMed] [Google Scholar]
- 33.McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., Sprouse K.R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J.E., Tilles S.W., Pizzuto M.S., Guastalla S.B., Bona G., Pellanda A.F., Garzoni C., Van Voorhis W.C., Rosen L.E., Snell G., Telenti A., Virgin H.W., Piccoli L., Corti D., Veesler D. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373(6555):648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. McCallum, J. Bassi, A. Marco, A. Chen, A.C. Walls, J.D. Iulio, M.A. Tortorici, M.J. Navarro, C. Silacci-Fregni, C. Saliba, M. Agostini, D. Pinto, K. Culap, S. Bianchi, S. Jaconi, E. Cameroni, J.E. Bowen, S.W. Tilles, M.S. Pizzuto, S.B. Guastalla, G. Bona, A.F. Pellanda, C. Garzoni, W.C. Van Voorhis, L.E. Rosen, G. Snell, A. Telenti, H.W. Virgin, L. Piccoli, D. Corti, D. Veesler, SARS-CoV-2 immune evasion by variant B.1.427/B.1.429, bioRxiv (2021).
- 35.W. Ren, X. Ju, M. Gong, J. Lan, Y. Yu, Q. Long, D.J. Kenney, A.K. O'Connell, Y. Zhang, J. Zhong, G. Zhong, F. Douam, X. Wang, A. Huang, R. Zhang, Q. Ding, Characterization of SARS-CoV-2 Variants B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618 by Cell Entry and Immune Evasion, mBio 13(2) (2022) e0009922. [DOI] [PMC free article] [PubMed]
- 36.Kumavath R., Barh D., Andrade B.S., Imchen M., Aburjaile F.F., Ch A., Rodrigues D.L.N., Tiwari S., Alzahrani K.J., Goes-Neto A., Weener M.E., Ghosh P., Azevedo V. The spike of SARS-CoV-2: uniqueness and applications. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.663912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kandeel M., Mohamed M.E.M., Abd El-Lateef H.M., Venugopala K.N., El-Beltagi H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2022;94(4):1627–1632. doi: 10.1002/jmv.27515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren S.Y., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases. 2022;10(1):1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakur V., Ratho R.K. OMICRON (B.1.1.529): a new SARS-CoV-2 variant of concern mounting worldwide fear. J. Med. Virol. 2022;94(5):1821–1824. doi: 10.1002/jmv.27541. [DOI] [PubMed] [Google Scholar]
- 40.Gao S.J., Guo H., Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert! J. Med. Virol. 2022;94(4):1255–1256. doi: 10.1002/jmv.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai M.S., Yang Y.H., Lin Y.S., Chang G.H., Hsu C.M., Yeh R.A., Shu L.H., Cheng Y.C., Liu H.T., Wu Y.H., Wu Y.H., Shen R.C., Wu C.Y. GB-2 blocking the interaction between ACE2 and wild type and mutation of spike protein of SARS-CoV-2. Biomed. Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.112011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moghaddar M., Radman R., Macreadie I. Severity, pathogenicity and transmissibility of delta and Lambda variants of SARS-CoV-2, toxicity of spike protein and possibilities for future prevention of COVID-19. Microorganisms. 2021;9(10) doi: 10.3390/microorganisms9102167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammadi M., Shayestehpour M., Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021;25(4) doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suthar M.S., Arunachalam P.S., Hu M., Reis N., Trisal M., Raeber O., Chinthrajah S., Davis-Gardner M.E., Manning K., Mudvari P., Boritz E., Godbole S., Henry A.R., Douek D.C., Halfmann P., Kawaoka Y., Boyd S.D., Davis M.M., Zarnitsyna V.I., Nadeau K., Pulendran B. Durability of immune responses to the BNT162b2 mRNA vaccine. Med. (N.Y.) 2022;3(1):25–27. doi: 10.1016/j.medj.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruxvoort K.J., Sy L.S., Qian L., Ackerson B.K., Luo Y., Lee G.S., Tian Y., Florea A., Aragones M., Tubert J.E., Takhar H.S., Ku J.H., Paila Y.D., Talarico C.A., Tseng H.F. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. BMJ. 2021;375:e068848. doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acharya A., Pandey K., Thurman M., Klug E., Trivedi J., Sharma K., Lorson C.L., Singh K., Byrareddy S.N. Discovery and evaluation of entry inhibitors for SARS-CoV-2 and its emerging variants. J. Virol. 2021;95(24):e0143721. doi: 10.1128/JVI.01437-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhat T., Cao A., Yin J. Virus-like particles: measures and biological functions. Viruses. 2022;14(2) doi: 10.3390/v14020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., Zhao B., Ding M., Song S., Kang Y., Yu Y., Xu M., Xiang T., Gao L., Feng Q., Zhao Q., Zeng M.S., Krummenacher C., Zeng Y.X. A novel vaccine candidate based on chimeric virus-like particle displaying multiple conserved epitope peptides induced neutralizing antibodies against EBV infection. Theranostics. 2020;10(13):5704–5718. doi: 10.7150/thno.42494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tariq H., Batool S., Asif S., Ali M., Abbasi B.H. Virus-like particles: revolutionary platforms for developing vaccines against emerging infectious diseases. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.790121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lampinen V., Heinimaki S., Laitinen O.H., Pesu M., Hankaniemi M.M., Blazevic V., Hytonen V.P. Modular vaccine platform based on the norovirus-like particle. J. Nanobiotechnol. 2021;19(1):25. doi: 10.1186/s12951-021-00772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharifzadeh M., Mottaghi-Dastjerdi N., Soltany Rezae Raad M. A review of virus-like particle-based SARS-CoV-2 vaccines in clinical trial phases. Iran. J. Pharm. Res. 2022;21(1):e127042. doi: 10.5812/ijpr-127042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Syomin B.V., Ilyin Y.V. Virus-like particles as an instrument of vaccine production. Mol. Biol. 2019;53(3):323–334. doi: 10.1134/S0026893319030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo J., Zhou A., Sun X., Sha W., Ai K., Pan G., Zhou C., Zhou H., Cong H., He S. Immunogenicity of a virus-like-particle vaccine containing multiple antigenic epitopes of toxoplasma gondii against acute and chronic toxoplasmosis in mice. Front. Immunol. 2019;10:592. doi: 10.3389/fimmu.2019.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwarz B., Uchida M., Douglas T. Biomedical and catalytic opportunities of virus-like particles in nanotechnology. Adv. Virus Res. 2017;97:1–60. doi: 10.1016/bs.aivir.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Comas-Garcia M., Colunga-Saucedo M., Rosales-Mendoza S. The role of virus-like particles in medical biotechnology. Mol. Pharm. 2020;17(12):4407–4420. doi: 10.1021/acs.molpharmaceut.0c00828. [DOI] [PubMed] [Google Scholar]
- 56.Fuenmayor J., Godia F., Cervera L. Production of virus-like particles for vaccines. N. Biotechnol. 2017;39(Pt B):174–180. doi: 10.1016/j.nbt.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rupil L.L., Del Carmen Serradell M., Lujan H.D. Production of oral vaccines based on virus-like particles pseudotyped with protozoan-surface proteins. Methods Mol. Biol. 2022;2410:503–537. doi: 10.1007/978-1-0716-1884-4_26. [DOI] [PubMed] [Google Scholar]
- 58.Mohsen M.O., Zha L., Cabral-Miranda G., Bachmann M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 59.Cappelli L., Cinelli P., Giusti F., Ferlenghi I., Utrio-Lanfaloni S., Wahome N., Bottomley M.J., Maione D., Cozzi R. Self-assembling protein nanoparticles and virus like particles correctly display beta-barrel from meningococcal factor H-binding protein through genetic fusion. PLoS ONE. 2022;17(9):e0273322. doi: 10.1371/journal.pone.0273322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Read B.J., Won L., Kraft J.C., Sappington I., Aung A., Wu S., Bals J., Chen C., Lee K.K., Lingwood D., King N.P., Irvine D.J. Mannose-binding lectin and complement mediate follicular localization and enhanced immunogenicity of diverse protein nanoparticle immunogens. Cell Rep. 2022;38(2) doi: 10.1016/j.celrep.2021.110217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson E.A., Lore K. Non-human primates as a model for understanding the mechanism of action of toll-like receptor-based vaccine adjuvants. Curr. Opin. Immunol. 2017;47:1–7. doi: 10.1016/j.coi.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Tian M., Hua Z., Hong S., Zhang Z., Liu C., Lin L., Chen J., Zhang W., Zhou X., Zhang F., DeFranco A.L., Hou B. B cell-intrinsic MyD88 signaling promotes initial cell proliferation and differentiation to enhance the germinal center response to a virus-like particle. J. Immunol. 2018;200(3):937–948. doi: 10.4049/jimmunol.1701067. [DOI] [PubMed] [Google Scholar]
- 63.Sartorius R., Trovato M., Manco R., D'Apice L., De Berardinis P. Exploiting viral sensing mediated by Toll-like receptors to design innovative vaccines. npj Vaccines. 2021;6(1):127. doi: 10.1038/s41541-021-00391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu D., Zhang S., Poteet E., Marin-Muller C., Chen C., Yao Q. Sublingual immunization with chimeric C1q/CD40 ligand/HIV virus-like particles induces strong mucosal immune responses against HIV. Vaccines (Basel) 2021;9(11) doi: 10.3390/vaccines9111236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pishesha N., Harmand T.J., Ploegh H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022 doi: 10.1038/s41577-022-00707-2. [DOI] [PubMed] [Google Scholar]
- 66.Wang X., Dong K., Long M., Lin F., Gao Z., Wang L., Zhang Z., Chen X., Dai Y., Wang H., Zhang H. Induction of a high-titered antibody response using HIV gag-EV71 VP1-based virus-like particles with the capacity to protect newborn mice challenged with a lethal dose of enterovirus 71. Arch. Virol. 2018;163(7):1851–1861. doi: 10.1007/s00705-018-3797-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dai S., Zhang T., Zhang Y., Wang H., Deng F. Zika virus baculovirus-expressed virus-like particles induce neutralizing antibodies in mice. Virol. Sin. 2018;33(3):213–226. doi: 10.1007/s12250-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alvi M.M., Sivasankaran S., Singh M. Pharmacological and non-pharmacological efforts at prevention, mitigation, and treatment for COVID-19. J. Drug Target. 2020;28(7–8):742–754. doi: 10.1080/1061186X.2020.1793990. [DOI] [PubMed] [Google Scholar]
- 69.Chan S.K., Du P., Ignacio C., Mehta S., Newton I.G., Steinmetz N.F. Biomimetic virus-like particles as severe acute respiratory syndrome coronavirus 2 diagnostic tools. ACS Nano. 2021;15(1):1259–1272. doi: 10.1021/acsnano.0c08430. [DOI] [PubMed] [Google Scholar]
- 70.Kaplan R.M., Milstein A. Influence of a COVID-19 vaccine's effectiveness and safety profile on vaccination acceptance. PNAS. 2021;118(10) doi: 10.1073/pnas.2021726118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F.L., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato T., Takami Y., Kumar Deo V., Park E.Y. Preparation of virus-like particle mimetic nanovesicles displaying the S protein of Middle East respiratory syndrome coronavirus using insect cells. J. Biotechnol. 2019;306:177–184. doi: 10.1016/j.jbiotec.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samrat S.K., Tharappel A.M., Li Z., Li H. Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu B., Huang Y., Huang L., Li B., Zheng Z., Chen Z., Chen J., Hu Q., Wang H. Effect of mucosal and systemic immunization with virus-like particles of severe acute respiratory syndrome coronavirus in mice. Immunology. 2010;130(2):254–261. doi: 10.1111/j.1365-2567.2010.03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.C.Y. Yong, W.P.P. Liew, H.K. Ong, C.L. Poh, Development of virus-like particles-based vaccines against coronaviruses, Biotechnol. Prog. (2022) e3292. [DOI] [PMC free article] [PubMed]
- 76.Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., Marra E. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J. Transl. Med. 2020;18(1):222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amawi H., Abu Deiab G.I., Dua K., Tambuwala M.M. COVID-19 pandemic: an overview of epidemiology, pathogenesis, diagnostics and potential vaccines and therapeutics. Ther. Deliv. 2020;11(4):245–268. doi: 10.4155/tde-2020-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiong X., Tortorici M.A., Snijder J., Yoshioka C., Walls A.C., Li W., McGuire A.T., Rey F.A., Bosch B.J., Veesler D. Glycan Shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J. Virol. 2018;92(4) doi: 10.1128/JVI.01628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 80.Mazumder S., Rastogi R., Undale A., Arora K., Arora N.M., Pratim B., Kumar D., Joseph A., Mali B., Arya V.B., Kalyanaraman S., Mukherjee A., Gupta A., Potdar S., Roy S.S., Parashar D., Paliwal J., Singh S.K., Naqvi A., Srivastava A., Singh M.K., Kumar D., Bansal S., Rautray S., Saini M., Jain K., Gupta R., Kundu P.K. PRAK-03202: a triple antigen virus-like particle vaccine candidate against SARS CoV-2. Heliyon. 2021;7(10):e08124. doi: 10.1016/j.heliyon.2021.e08124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghorbani A., Zare F., Sazegari S., Afsharifar A., Eskandari M.H., Pormohammad A. Development of a novel platform of virus-like particle (VLP)-based vaccine against COVID-19 by exposing epitopes: an immunoinformatics approach. New Microbes New Infect. 2020;38 doi: 10.1016/j.nmni.2020.100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moon K.B., Jeon J.H., Choi H., Park J.S., Park S.J., Lee H.J., Park J.M., Cho H.S., Moon J.S., Oh H., Kang S., Mason H.S., Kwon S.Y., Kim H.S. Construction of SARS-CoV-2 virus-like particles in plant. Sci. Rep. 2022;12(1):1005. doi: 10.1038/s41598-022-04883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu R., Shi M., Li J., Song P., Li N. Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol. 2020;8:862. doi: 10.3389/fbioe.2020.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swann H., Sharma A., Preece B., Peterson A., Eldredge C., Belnap D.M., Vershinin M., Saffarian S. Minimal system for assembly of SARS-CoV-2 virus like particles. Sci. Rep. 2020;10(1):21877. doi: 10.1038/s41598-020-78656-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chu K.B., Kang H.J., Yoon K.W., Lee H.A., Moon E.K., Han B.K., Quan F.S. Influenza virus-like particle (VLP) vaccines expressing the SARS-CoV-2 S glycoprotein, S1, or S2 domains. Vaccines (Basel) 2021;9(8) doi: 10.3390/vaccines9080920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Volkmann A., Koopman G., Mooij P., Verschoor E.J., Verstrepen B.E., Bogers W., Idorn M., Paludan S.R., Vang S., Nielsen M.A., Sander A.F., Schmittwolf C., Hochrein H., Chaplin P. A capsid virus-like particle-based SARS-CoV-2 vaccine induces high levels of antibodies and protects rhesus macaques. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.857440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fougeroux C., Goksoyr L., Idorn M., Soroka V., Myeni S.K., Dagil R., Janitzek C.M., Sogaard M., Aves K.L., Horsted E.W., Erdogan S.M., Gustavsson T., Dorosz J., Clemmensen S., Fredsgaard L., Thrane S., Vidal-Calvo E.E., Khalife P., Hulen T.M., Choudhary S., Theisen M., Singh S.K., Garcia-Senosiain A., Van Oosten L., Pijlman G., Hierzberger B., Domeyer T., Nalewajek B.W., Strobaek A., Skrzypczak M., Andersson L.F., Buus S., Buus A.S., Christensen J.P., Dalebout T.J., Iversen K., Harritshoj L.H., Mordmuller B., Ullum H., Reinert L.S., de Jongh W.A., Kikkert M., Paludan S.R., Theander T.G., Nielsen M.A., Salanti A., Sander A.F. Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 2021;12(1):324. doi: 10.1038/s41467-020-20251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ward B.J., Gobeil P., Seguin A., Atkins J., Boulay I., Charbonneau P.Y., Couture M., D'Aoust M.A., Dhaliwall J., Finkle C., Hager K., Mahmood A., Makarkov A., Cheng M.P., Pillet S., Schimke P., St-Martin S., Trepanier S., Landry N. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021;27(6):1071–1078. doi: 10.1038/s41591-021-01370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dube C., Paris-Robidas S., Andreani G., Gutzeit C., D'Aoust M.A., Ward B.J., Trepanier S. Broad neutralization against SARS-CoV-2 variants induced by ancestral and B.1.351 AS03-adjuvanted recombinant plant-derived virus-like particle vaccines. Vaccine. 2022;40(30):4017–4025. doi: 10.1016/j.vaccine.2022.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maharjan P.M., Choe S. Plant-based COVID-19 vaccines: current status, design, and development strategies of candidate vaccines. Vaccines (Basel) 2021;9(9) doi: 10.3390/vaccines9090992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.K.J. Hager, G. Perez Marc, P. Gobeil, R.S. Diaz, G. Heizer, C. Llapur, A.I. Makarkov, E. Vasconcellos, S. Pillet, F. Riera, P. Saxena, P. Geller Wolff, K. Bhutada, G. Wallace, H. Aazami, C.E. Jones, F.P. Polack, L. Ferrara, J. Atkins, I. Boulay, J. Dhaliwall, N. Charland, M.M.J. Couture, J. Jiang-Wright, N. Landry, S. Lapointe, A. Lorin, A. Mahmood, L.H. Moulton, E. Pahmer, J. Parent, A. Seguin, L. Tran, T. Breuer, M.A. Ceregido, M. Koutsoukos, F. Roman, J. Namba, M.A. D'Aoust, S. Trepanier, Y. Kimura, B.J. Ward, V.L.P.S.T. Co, Efficacy and safety of a recombinant plant-based adjuvanted Covid-19 vaccine, N. Engl. J. Med. 386(22) (2022) 2084–2096. [DOI] [PMC free article] [PubMed]
- 92.Motamedi H., Ari M.M., Dashtbin S., Fathollahi M., Hossainpour H., Alvandi A., Moradi J., Abiri R. An update review of globally reported SARS-CoV-2 vaccines in preclinical and clinical stages. Int. Immunopharmacol. 2021;96 doi: 10.1016/j.intimp.2021.107763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abdulla Z.A., Al-Bashir S.M., Al-Salih N.S., Aldamen A.A., Abdulazeez M.Z. A summary of the SARS-CoV-2 vaccines and technologies available or under development. Pathogens. 2021;10(7) doi: 10.3390/pathogens10070788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roy S., Ghani K., de Campos-Lima P.O., Caruso M. A stable platform for the production of virus-like particles pseudotyped with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein. Virus Res. 2021;295 doi: 10.1016/j.virusres.2021.198305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D., Cohen-Gihon I., Lazar S., Gutman H., Glinert I., Cherry L., Vagima Y., Lazar S., Weiss S., Ben-Shmuel A., Avraham R., Puni R., Lupu E., Bar-David E., Sittner A., Erez N., Zichel R., Mamroud E., Mazor O., Levy H., Laskar O., Yitzhaki S., Shapira S.C., Zvi A., Beth-Din A., Paran N., Israely T. A single dose of recombinant VSV-G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11(1):6402. doi: 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alam M.K., Brabant M., Viswas R.S., Barreto K., Fonge H., Ronald Geyer C. A novel synthetic trivalent single chain variable fragment (tri-scFv) construction platform based on the SpyTag/SpyCatcher protein ligase system. BMC Biotech. 2018;18(1):55. doi: 10.1186/s12896-018-0466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marini A., Zhou Y., Li Y., Taylor I.J., Leneghan D.B., Jin J., Zaric M., Mekhaiel D., Long C.A., Miura K., Biswas S. A universal plug-and-display vaccine carrier based on HBsAg VLP to maximize effective antibody response. Front. Immunol. 2019;10:2931. doi: 10.3389/fimmu.2019.02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Formica N., Mallory R., Albert G., Robinson M., Plested J.S., Cho I., Robertson A., Dubovsky F., Glenn G.M., V.S.G. nCo Different dose regimens of a SARS-CoV-2 recombinant spike protein vaccine (NVX-CoV2373) in younger and older adults: a phase 2 randomized placebo-controlled trial. PLoS Med. 2021;18(10):e1003769. doi: 10.1371/journal.pmed.1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masuda T., Murakami K., Sugiura K., Sakui S., Schuring R.P., Mori M. Safety and immunogenicity of NVX-CoV2373 (TAK-019) vaccine in healthy Japanese adults: interim report of a phase I/II randomized controlled trial. Vaccine. 2022;40(24):3380–3388. doi: 10.1016/j.vaccine.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weiss C., Carriere M., Fusco L., Capua I., Regla-Nava J.A., Pasquali M., Scott J.A., Vitale F., Unal M.A., Mattevi C., Bedognetti D., Merkoci A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L.G. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. 2020;14(6):6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.