Abstract
Mutant superoxide dismutase 1 (SOD1) can be constitutively released from motor neurons and transmitted to naïve motor neurons to promote the progression of amyotrophic lateral sclerosis (ALS). However, the biological impacts of this process and the precise mechanisms of SOD1 release remain to be fully resolved. Using biochemical and fluorescent techniques, this study aimed to determine if P2X7 receptor activation could induce mutant SOD1 release from motor neurons and whether this released SOD1 could be transmitted to motor neurons or microglia to mediate effects associated with neurodegeneration in ALS. Aggregated SOD1G93A, released from murine NSC-34 motor neurons transiently transfected with SOD1G93A, could be transmitted to naïve NSC-34 cells and murine EOC13 microglia to induce endoplasmic reticulum (ER) stress and tumour necrosis factor-alpha (TNFα) release, respectively. Immunoblotting revealed NSC-34 cells expressed P2X7. Extracellular ATP induced cation dye uptake into these cells, which was blocked by the P2X7 antagonist AZ10606120, demonstrating these cells express functional P2X7. Moreover, ATP induced the rapid release of aggregated SOD1G93A from NSC-34 cells transiently transfected with SOD1G93A, a process blocked by AZ10606120 and revealing a role for P2X7 in this process. ATP-induced SOD1G93A release coincided with membrane blebbing. Finally, aggregated SOD1G93A released via P2X7 activation could also be transmitted to NSC-34 and EOC13 cells to induce ER stress and TNFα release, respectively. Collectively, these results identify a novel role for P2X7 in the prion-like propagation of SOD1 in ALS and provide a possible explanation for the therapeutic benefits of P2X7 antagonism previously observed in ALS SOD1G93A mice.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11302-022-09863-5.
Keywords: Motor neuron disease, Purinergic receptor, Glia, Neuroinflammation, Protein aggregation, Protein misfolding
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterised by the loss of upper and lower motor neurones and ensuing muscle dysfunction including respiratory paralysis [1]. Neuroanatomically, ALS spreads in a contiguous fashion commencing in a focal region before spreading to neighbouring motor neurons and then other regions of the nervous system [2]. This spreading is thought to involve the prion-like propagation of protein misfolding and aggregation between neighbouring cells [3].
Accumulating evidence suggests a role for mutant superoxide dismutase 1 (SOD1) in the prion-like propagation in ALS [4], a protein long-known to be associated with ALS development [5, 6] and with at least 155 mutations associated with the familial form of this disease [7]. In vitro studies reveal that mutant SOD1 aggregates released from neural cells can be internalised by other neural cells via macropinocytosis to seed the aggregation of intracellular, soluble mutant SOD1 [8], and wild-type SOD1 (SOD1WT) [9]. Once transmission has commenced, SOD1 aggregates are readily transferred from cell to cell in a contact-independent manner [8]. Subsequent studies have confirmed that the internalisation of SOD1 aggregates is mediated via macropinocytosis [10, 11]. Notably, in vivo studies demonstrate the prion-like propagation of SOD1 aggregates initiated by injection of either aggregated recombinant mutant SOD1 or SOD1WT [12, 13] or homogenates from ALS SOD1G85R or SOD1G93A mice [14] or ALS patients [12] into ALS SOD1G85R mice. In vitro studies also show that mutant SOD1 and SOD1WT aggregates can be released from cells via either cell death or exosomes [10], the latter consistent with an earlier report demonstrating mutant SOD1 release via exosomes from murine NSC-34 motor neuron cells [15]. However, other mechanisms of SOD1 release may exist [10] and the precise mechanisms of SOD1 release in vivo and the triggers underpinning these processes are unknown.
In addition to the prion-like propagation of protein misfolding and aggregation, uptake of aggregated mutant SOD1 can initiate cellular events similar to those associated with neurodegeneration in ALS. Uptake of aggregated recombinant mutant SOD1 into human SH-SY5Y neuroblastoma cells induces endoplasmic reticulum (ER) stress [16]. Likewise, uptake of aggregated recombinant mutant SOD1 into murine EOC13 microglia can induce TNFα release [17]. Whether aggregated mutant SOD1 released from cells can induce similar cellular events to aggregated recombinant SOD1 remains unknown.
The P2X7 receptor is a trimeric ligand-gated ion channel/pore activated by extracellular adenosine 5′-triphosphate (ATP) [18, 19] and is present on various immune and neural cell types [20, 21]. The receptor has emerging roles in the development of ALS [22, 23], and its pharmacological blockade can limit disease progression in ALS SOD1G93A mice [24–27]. The role of P2X7 in ALS, however, is yet to be fully established. Notably, P2X7 activation can induce the release of proteins from various cell types [28–30] and in neural cells can induce exosome release [30] and cell death [31]. Collectively, this suggests that P2X7 activation could potentially be involved in the release of aggregated mutant SOD1 from motor neurons.
Using murine NSC-34 motor neurons transfected with human SOD1G93A, this study aimed to determine whether aggregated SOD1G93A released from these cells could be transmitted to naïve NSC-34 cells and murine EOC13 microglia to induce ER stress and TNFα release, respectively. Then, using SOD1G93A-transfected NSC-34 cells, this study aimed to determine if P2X7 activation could induce the release of aggregated SOD1G93A from these cells and whether this released SOD1G93A could also induce ER stress in NSC-34 cells and TNFα release from EOC13 cells.
Materials and methods
Cell culture
Murine neuroblastoma × spinal cord hybrid NSC-34 cells have been described elsewhere [32]. Murine lymphoblast LADMAC, murine microglial EOC13, and human embryonic kidney (HEK)-293 cells were from the American Type Culture Collection (Manassas, VA, USA). NSC-34 and LADMAC cells were maintained in DMEM/F12 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% heat-inactivated FBS (Bovogen Biologicals, Keilor East, Australia) and 2 mM GlutaMAX (Life Technologies) (complete medium). EOC13 cells were maintained in complete medium supplemented with 20% LADMAC conditioned medium. HEK-293 cells were maintained in complete medium supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies). Cells were maintained at 37 °C/5% CO2 and harvested with 0.05% trypsin/0.5 mM EDTA (Life Technologies).
Plasmid purification
pEGFP-N1 expression vectors containing SOD1WT and SOD1G93A cDNAs were kindly provided by Bradley Turner (University of Melbourne, Melbourne, Australia) [33]. pFUW expression vectors containing SOD1G85R and SOD1G127X cDNAs were kindly provided by Edward Pokrishevsky (University of British Columbia, Vancouver, Canada) [9]. Chemically competent DH5α Escherichia coli, kindly provided by Jason McArthur (University of Wollongong, Wollongong, Australia), were transformed with SOD1-containing plasmids using heat shock (42 °C) and colonies resistant to kanamycin (Ameresco, Framingham, MA, USA) selected. Plasmid DNA was purified using the CompactPrep Plasmid Maxi Kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions.
Transfection of NSC-34 cells
NSC-34 cells in complete medium in 6-well plates (7 × 105 cells/well) or 12-well plates (3 × 105 cells/well) were incubated overnight at 37 °C/5% CO2. NSC-34 cells were washed once with DMEM/F12 medium and incubated for 5 h in DMEM/F12 medium containing 0.5 μg plasmid DNA and Lipofectamine 2000 (Life Technologies), or 1.5 μg plasmid DNA and Lipofectamine LTX (Life Technologies), respectively. Cells were then incubated in complete medium for a further 67 h (72 h total). Alternatively, NSC-34 cells were incubated in complete medium containing 2.5 μg plasmid DNA, P3000 reagent, and Lipofectamine 3000 (Life Technologies) for 72 h. Transfection efficiency of EGFP-containing vectors was confirmed by fluorescence microscopy using an Eclipse TE2000 inverted microscope (Nikon, Tokyo, Japan) and was similar between vectors, typically ranging from 17 to 27%. Transiently transfected NSC-34 cells were used at 72 h in subsequent assays.
Constitutive SOD1 release
Conditioned media were collected from transiently transfect NSC-34 cells at 72 h and centrifuged (20,000 × g for 30 min) to obtain pelletable fractions. Pelletable fractions were then resuspended as described for use in subsequent assays.
Detection of cell-associated EGFP by flow cytometry
Pelletable fractions from treated NSC-34 cells (as indicated) were resuspended in 500 µL complete medium and incubated with naïve NSC-34 or EOC13 cells in 24-well plates (5 × 104 cells/well) for 24 h at 37 °C/5% CO2. Cells were visualised by differential interference contrast imaging using an Eclipse TE2000 inverted microscope. Cells were then harvested, washed once with PBS and data acquired using a BD (San Diego, CA, USA) LSR II flow cytometer. The percentage of EGFP+ cells was assessed using FlowJo 8.7.1 (Tree Star, Ashland, OR, USA).
ER stress assay
Pelletable fractions from treated NSC-34 cells (as indicated) were resuspended in 500 µL complete medium and incubated with NSC-34 transiently transfected with X-box-binding protein 1 (XBP1)-tdTomato in 24-well plates (5 × 104 cells/well) for 24 h at 37 °C/5% CO2. Cells were visualised by differential interference contrast and fluorescence imaging using an Eclipse TE2000 inverted microscope with images captured using Image-Pro AMS 6.1 (Media Cybernetics, Rockville, MD, USA) or using an IncuCyte ZOOM Live-cell Analysis System (Essen BioScience, Ann Arbor, MI) with images captured using IncuCyte ZOOM software (Essen BioScience). Cells were then harvested, washed once with PBS and data acquired using a LSR II flow cytometer. The mean fluorescence intensity (MFI) of relative XBP1-tdTomato fluorescence was determined using FlowJo 8.7.1. Alternatively, cells in plates were visualised using a Leica (Wetzlar, Germany) DM IBRE inverted microscope and Leica TCS SP confocal imaging system.
TNFα release assay
Pelletable fractions from treated NSC-34 cells (as indicated) were resuspended in 500 µL complete medium and incubated with naïve EOC13 cells in 24-well plates (5 × 104 cells/well) for 24 h at 37 °C/5% CO2. Media from wells were then centrifuged (20,000 × g for 5 min at 4 °C) and cell-free supernatants stored at − 80 °C until required. The presence of TNFα measured using a Murine TNF-α ELISA Ready-SET-Go! Kit (eBioscience, San Diego, CA, USA) as per the manufacturer’s instructions.
Detection of P2X7 by immunoblotting
Proteins in lysates of NSC-34, EOC13 and P2X7-transfected HEK-293 cells were separated under reducing conditions using Any kD Mini-PROTEAN TGX Stain-Free gels (Bio-Rad, Hercules, CA, USA) and transferred to nitrocellulose membranes (Bio-Rad), and P2X7 detected by immunoblotting using a rabbit anti-murine P2X7 polyclonal antibody (Cat. No. APR-008, Alomone Labs, Jerusalem, Israel) as described [34].
ATP-induced uptake assay
Non-transfected or transiently transfected NSC-34 cells in 6-well plates (7 × 105 cells/mL) were washed twice with DMEM/F12 medium and pre-incubated in the absence or presence of 10 μM AZ10606120 (Tocris Bioscience, Bristol, UK) for 1 h at 37 °C. Cells were then incubated with 25 μM ethidium bromide (Ameresco) in the absence (basal) or presence of 5 mM ATP (Sigma-Aldrich, St Louis, MO, USA) in DMEM/F12 medium for 20 min at 37 °C. Reactions were stopped by the addition of an equal volume of ice-cold DMEM/F12 medium containing 20 mM MgCl2. Cells were then harvested and washed once with DMEM/F12 medium. Data was acquired using a BD LSR II flow cytometer. The MFI of relative ethidium+ uptake was assessed using FlowJo 8.7.1.
ATP-induced SOD1 release assay
Transiently transfected NSC-34 cells in 12-well plates (3 × 105 cells/mL) were washed twice with DMEM/F12 medium and pre-incubated in the absence or presence of 10 µM AZ10606120 in DMEM/F12 medium for 1 h at 37 °C/5% CO2. Cells were then incubated in the absence (basal) or presence of ATP (as indicated) for 20 min at 37 °C. Following incubation in the absence or presence of ATP, medium was collected and centrifuged (20,000 × g for 30 min) to obtain pelletable fractions.
Detection of SOD1 by immunoblotting
Pelletable fractions from treated NSC-34 cells (as indicated), resuspended in equal volumes of sample buffer containing 5% 2-mercaptoethanol (Ameresco) were separated using Any kD Mini-PROTEAN TGX Stain-Free gels and transferred to nitrocellulose membranes. In some experiments, gels were imaged, prior to transfer, using a Bio-Rad Criterion Stain Free Gel Imaging System. Membranes were blocked with either 1% heat denatured casein (Sigma-Aldrich) in PBS (for EGFP) or Tris-buffered saline (TBS; 250 mM NaCl and 50 mM Tris, pH 7.5) containing 0.2% Tween-20 and 5% milk powder (for SOD1) for 1 h. Membranes were incubated with either rabbit anti-EGFP polyclonal antibody (Cat. No. ab290 or ab6556, Abcam, Cambridge, UK) or rabbit anti-SOD1 (Cat. No. abADI-SOD-100, Enzo Life Sciences, Farmingdale, NY, USA) or sheep anti-SOD1 (Cat. No. ab8866, Abcam) polyclonal antibody (depending on availability) in corresponding blocking solution overnight at 4 °C. Membranes were washed three times with PBS containing 0.1% Triton X-100 (for EGFP) or TBS containing 0.2% Tween-20 (for SOD1). Membranes were then incubated with peroxidise-conjugated anti-rabbit (Cat. No. 1706515, Bio-Rad or Cat. No. NA934, Cytiva, Marlborough, MA, USA) or anti-sheep (Cat. No. AB324P, Merck Millipore, Burlington, MA, USA) IgG antibody in corresponding blocking solution for 1 h. Membranes were washed and visualised using SuperSignal West Pico Chemiluminescence Substrate (ThermoFisher Scientific, Waltham, MA, USA). Relative EGFP and SOD1 were quantified from captured images using ImageJ software 1.48 [35].
Detection of SOD1 by transmission electron microscopy
Pelletable fractions (20,000 × g for 30 min) from treated NSC-34 cells (as indicated) were washed and resuspended in PBS. Fractions (2 µL) were then loaded onto carbon-coated nickel grids (ProSciTech, Kirwan, Australia) and incubated for 1 min. Grids were washed 3–5 times with Milli-Q water. In some experiments, grids were blocked for 40 min with 1% heat denatured casein in PBS, followed by incubation with anti-EGFP polyclonal antibody (Cat. No. ab290, Abcam) or an equivalent amount of rabbit IgG (Cat. No. ab171870, Abcam) for 1 h. Grids were washed three times with 1% heat denatured casein in PBS and incubated with 5 nm gold-conjugated anti-rabbit IgG antibody (Cat. No. ab27235, Abcam) 1 h. Grids were washed as above, followed by five washes with Milli-Q water (Merck Millipore). Non-labelled and labelled grids were stained with Milli-Q water containing 2% uranyl acetate (ProSciTech) for 3 min and dried by blotting. Structures were visualised using a JEOL (Akishima, Japan) JEM2011 transmission electron microscope. Images were captured using Gatan Microscopy Suite Version 2.30.542.0 (Pleasanton, CA, USA).
Lactate dehydrogenase release assay
Transiently transfected NSC-34 cells in 6-well plates (7 × 105 cells/well) were washed twice with DMEM/F12 medium and pre-incubated in the absence or presence of 10 µM AZ10606120 in DMEM/F12 medium for 1 h at 37 °C/5% CO2. Cells were then incubated in the absence (basal) or presence of 5 mM ATP in DMEM/F12 medium for 20 min at 37 °C. Cells were visualised by differential interference contrast imaging using an Eclipse TE2000 inverted microscope with images captured using Image-Pro AMS 6.1. The media from wells were collected and the remaining adherent cells lysed with Lysis Solution (Roche, Mannheim, Germany). Lactate dehydrogenase (LDH) release and percent cytotoxicity were determined using the Cytotoxicity Detection KitPLUS (Roche) as per the manufacturer’s instructions using a Molecular Devices (San Jose, CA, USA) SpectraMax Plus 384 Microplate Reader. The percentage of LDH release was determined as the amount of LDH released into the medium over the total amount of LDH released in each sample.
Real-time imaging of ATP-induced SOD1 release
Transiently transfected NSC-34 cells in 8-chamber µ-slides (Ibidi, Grafeling, Germany) (5 × 105 cells/0.2 mL/chamber) were washed twice with DMEM/F12 medium and pre-incubated in DMEM/F12 medium at 37 °C for 1 h. Cells with fluorescent aggregates were identified using a DM IBRE inverted microscope and TCS SP confocal imaging system at 37 °C. Cells were then incubated in the absence (basal) or presence of 5 mM ATP for 20 min at 37 °C with fluorescent and bright-field images captured every 30 s with Leica confocal software.
Detection of cell-associated EGFP by confocal microscopy
Pelletable fractions (20,000 × g for 30 min) from treated NSC-34 cells (as indicated) were resuspended in complete medium and incubated with naïve EOC13 cells in 8-chamber µ-slides (5 × 105 cells/0.2 mL/chamber) for 2 or 24 h at 37 °C/5% CO2. Cells were visualised by a DM IBRE inverted microscope and TCS SP confocal imaging system. Fluorescent and bright-field 30-step z-stacks were captured using Leica confocal software, and either compressed to form single images, or orthogonally sectioned to confirm that fluorescent particles present were intracellular.
Data presentation and statistical analyses
Data are presented as the mean ± SD. Differences between multiple groups were compared using a one-way ANOVA with Tukey’s multiple comparison test. Differences between two groups were compared using a two-tailed unpaired Student’s t-tests. Statistical analyses were performed using GraphPad Version 8.4.0 (San Diego, CA, USA) with p < 0.05 considered statistically significant.
Results
SOD1G93A constitutively released from NSC-34 motor neurons induces ER stress in recipient NSC-34 motor neurons
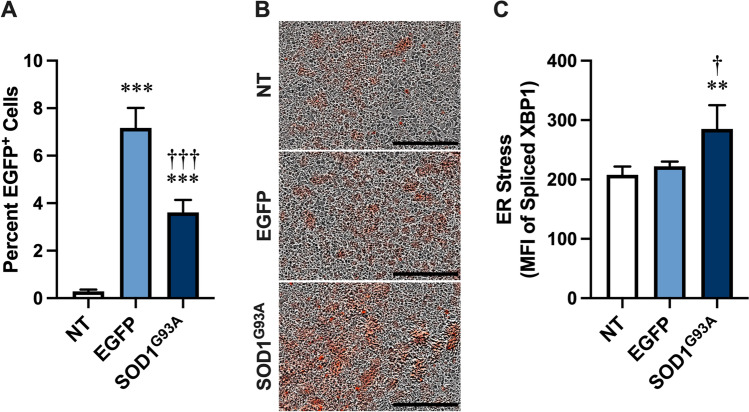
Aggregated SOD1G93A released from NSC-34 motor neurons can be internalised by naïve NSC-34 cells [10]. Furthermore, uptake of aggregated SOD1G93A into human SH-SY5Y neuroblastoma cells can induce ER stress [16]. To determine if uptake of aggregated SOD1G93A released from NSC-34 cells could similarly induce ER stress in naïve NSC-34 cells, NSC-34 cells were transiently transfected with the ER stress reporter, transcription factor XBP1-tdTomato [36, 37], and incubated for 24 h in the presence of pelletable fractions obtained by centrifugation (20,000 × g for 30 min) of conditioned media (72 h) from non-transfected, and EGFP and SOD1G93A-EGFP transfected NSC-34 cells and assessed by flow cytometry and fluorescent microscopy. Flow cytometry revealed that a small proportion of cells incubated with pelletable fractions from EGFP and SOD1G93A-EGFP transfected cells were positive for EGFP, which was significantly greater than in those cells incubated with pelletable fractions from non-transfected cells (Fig. 1A). Fluorescent microscopy demonstrated the presence of a larger proportion of XBP1-tdTomato fluorescent cells following incubation of XBP1-tdTomato-transfected NSC-34 cells with pelletable fractions from EGFP and SOD1G93A-EGFP transfected cells compared to those cells incubated with pelletable fractions from non-transfected cells (Fig. 1B). Notably, flow cytometry revealed a higher amount of fluorescence in cells incubated with pelletable fractions from SOD1G93A-EGFP transfected cells compared to those cells incubated with pelletable fractions from either non-transfected or EGFP transfected cells (Fig. 1C). Together these results indicate that SOD1G93A released from NSC-34 cells can associate with and promote ER stress in recipient NSC-34 cells.
Fig. 1.
SOD1G93A constitutively released from NSC-34 motor neurons induces ER stress in recipient NSC-34 motor neurons. A–C XBP1-tdTomato-transfected NSC-34 cells were incubated with pelletable fractions from conditioned media of non-transfected (NT), or EGFP- or SOD1G93A-EGFP (SOD1G93A)-transfected NSC-34 cells. At 24 h, A the percentage of EGFP+ cells was assessed by flow cytometry, B cellular XBP1-tdTomato fluorescence was visualised by an IncuCyte ZOOM Live-cell Analysis System, and C the MFI of XBP1-tdTomato in cells was assessed by flow cytometry. A, C Data are presented as mean ± SD (n = 4); **p < 0.02, ***p < 0.01 compared to NT, †p < 0.05, †††p < 0.01 compared to EGFP. B Data are representative of n = 4 experiments; bars represent 300 µm
SOD1G93A constitutively released from NSC-34 motor neurons associates with EOC13 microglia to promote TNFα release
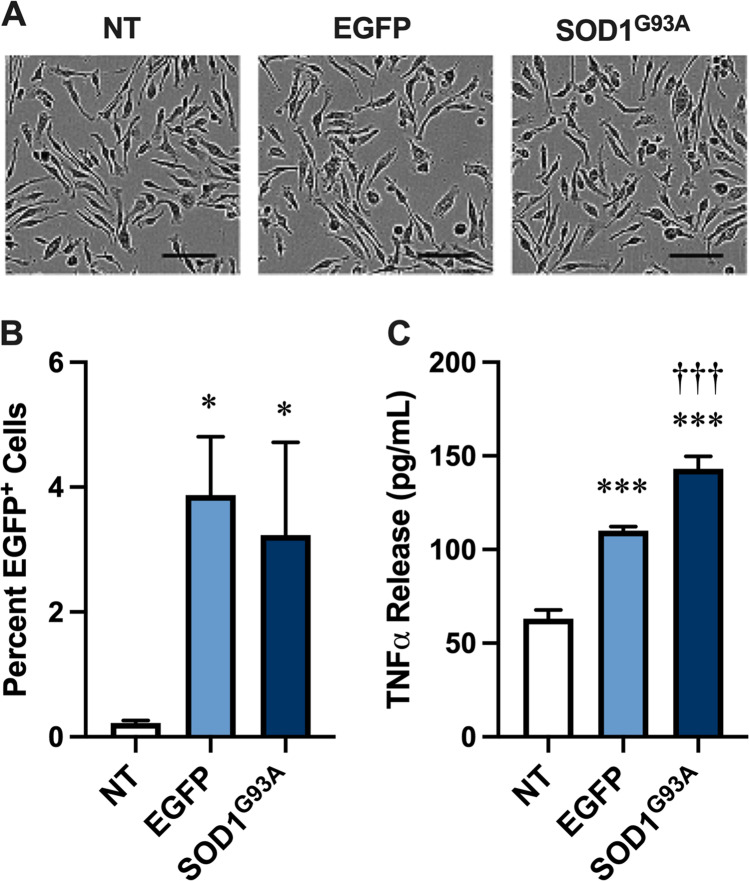
Recombinant SOD1G93A aggregates can be internalised by murine EOC13 microglia and induce TNFα release from these cells over 24 h [17]. Therefore, to determine if SOD1G93A-EGFP released from NSC-34 cells could act in a similar fashion to recombinant SOD1G93A aggregates, EOC13 cells were incubated for 24 h in the presence of pelletable fractions obtained by centrifugation (20,000 × g for 30 min) of conditioned media (72 h) from non-transfected, and EGFP and SOD1G93A-EGFP transfected NSC-34 cells. Phase-contrast microscopy revealed similar morphology of EOC13 cells incubated with each pelletable fraction (Fig. 2A). Flow cytometric analysis revealed that the percent of EOC13 cells positive for EGFP fluorescence was negligible in those incubated with non-transfected pelletable fractions (Fig. 2B). In contrast, a small proportion of EOC13 cells incubated with either EGFP or SOD1G93A-EGFP pelletable fractions were positive for EGFP fluorescence (Fig. 2B) indicating that both EGFP and SOD1G93A-EGFP from NS34 cells associated with EOC13 microglia. Finally, ELISA measurements of media from EOC13 cells revealed that incubation with EGFP and SOD1G93A-EGFP pelletable fractions of conditioned media (72 h) induced significantly greater TNFα release compared to incubation with non-transfected pelletable fractions (Fig. 2C). Notably, TNFα release from EOC13 microglia incubated with SOD1G93A-EGFP pelletable fractions was statistically different than that of cells incubated with EGFP pelletable fractions. TNFα was absent in pelletable fractions from transfected NSC-34 cells (not shown). The relatively small amount of TNFα release from EOC13 microglia incubated with non-transfected pelletable fractions suggests that molecules present within the conditioned medium of non-transfected NSC-34 cells can also stimulate TNFα release from microglia, consistent with the presence of immune mediators in untreated NSC-34 cells [38]. Collectively these results indicate that SOD1G93A released from NSC-34 cells can associate with and promote TNFα release from EOC13 cells.
Fig. 2.
SOD1G93A constitutively released from NSC-34 motor neurons associates with EOC13 microglia to promote TNFα release. A–C EOC13 cells were incubated with pelletable fractions from conditioned media of non-transfected (NT), or EGFP- or SOD1G93A-EGFP (SOD1G93A)-transfected NSC-34 cells. At 24 h, A cells were visualised by differential interference contrast microscopy, B the percentage of EGFP+ cells was assessed by flow cytometry, and C the concentration of TNFα released into cell supernatants was assessed by ELISA. A Data are representative of n = 6 experiments; bars represent 100 µm. B, C Data are presented as mean ± SD (n = 3); *p < 0.05, ***p < 0.01 compared to NT, †††p < 0.01 compared to EGFP
NSC-34 motor neurons express functional P2X7
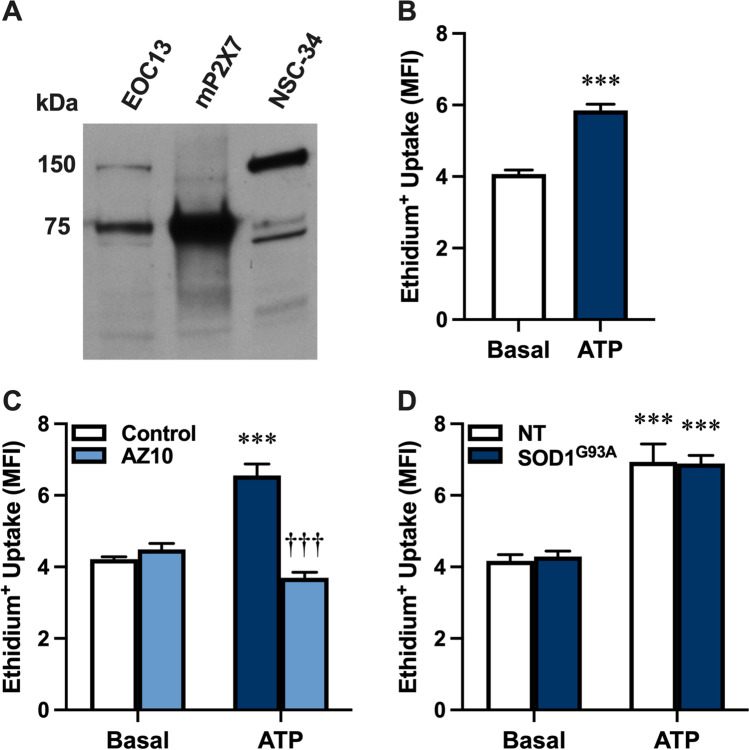
P2X7 has an emerging role in ALS [22, 23], and its activation by extracellular ATP can induce the release of various proteins from different cell types [28–30]. Other studies also indicate that primary rat motor neurons express P2X7 [39, 40]. Thus, it was hypothesised that NSC-34 motor neurons express P2X7, and that activation of this receptor could mediate SOD1G93A release from these cells. To determine if P2X7 is present on NSC-34 motor neuron cells [32], whole lysates of NSC-34 cells, as well as murine EOC13 cells [34] and murine P2X7-transfected HEK-293 cells [41] (as positive controls) were examined by immunoblotting using an anti-P2X7 antibody. Immunoblotting revealed a minor band at 75 kDa in NSC-34 cell lysates, corresponding to the predicted size of glycosylated, monomeric P2X7, as well as major bands at 150 and 65 kDa corresponding to the predicted size of dimeric and non-glycosylated P2X7, respectively (Fig. 3A). Notably, the intensity of the 75 kDa band in NSC-34 cell lysates was lower than that of EOC13 and P2X7-transfected HEK-293 cells.
Fig. 3.
NSC-34 motor neurons express functional P2X7. A P2X7 expression in whole lysates of EOC13, murine P2X7-transfected HEK-293 (mP2X7) and NSC-34 cells was assessed by immunoblotting using an anti-P2X7 antibody. Data is representative of n = 3 experiments. B–D Non-transfected (NT) or D SOD1G93A-EGFP (SOD1G93A)-transfected NSC-34 cells were pre-incubated in the B–D absence or C presence of 10 μM AZ10606120 (AZ10) for 1 h. Cells were then incubated with 25 μM ethidium bromide in the absence (basal) or presence of 5 mM ATP for 20 min, and the reactions were stopped by the addition of 20 mM MgCl2. The MFI of ethidium+ uptake into cells was assessed by flow cytometry. Data are presented as mean ± SD (n = 3); ***p < 0.01 compared to corresponding basal, †††p < 0.01 compared to ATP in the absence of AZ10
To determine whether NSC-34 cells expressed functional P2X7, ATP-induced ethidium+ uptake was assessed by flow cytometry. Incubation with 5 mM ATP induced significant ethidium+ uptake into NSC-34 cells compared to cells incubated in the absence of ATP (basal) (Fig. 3B). Moreover, pre-incubation of NSC-34 cells with 10 µM AZ10606120, a specific P2X7 antagonist [42], prior to incubation with 5 mM ATP completely blocked ATP-induced ethidium+ uptake (Fig. 3C). To determine whether transfection with SOD1 alters P2X7 activity in NSC-34 neurons, cells were transiently transfected with SOD1G93A-EGFP and ATP-induced ethidium+ uptake compared to non-transfected NSC-34 cells. ATP induced similar and significant ethidium+ uptake into both transfected and non-transfected cells (Fig. 3D). Collectively, these results indicate that NSC-34 cells express functional P2X7 and that the activity of this receptor in these cells is not altered by transient transfection with SOD1G93A-EGFP.
P2X7 activation mediates SOD1G93A release from NSC-34 motor neurons
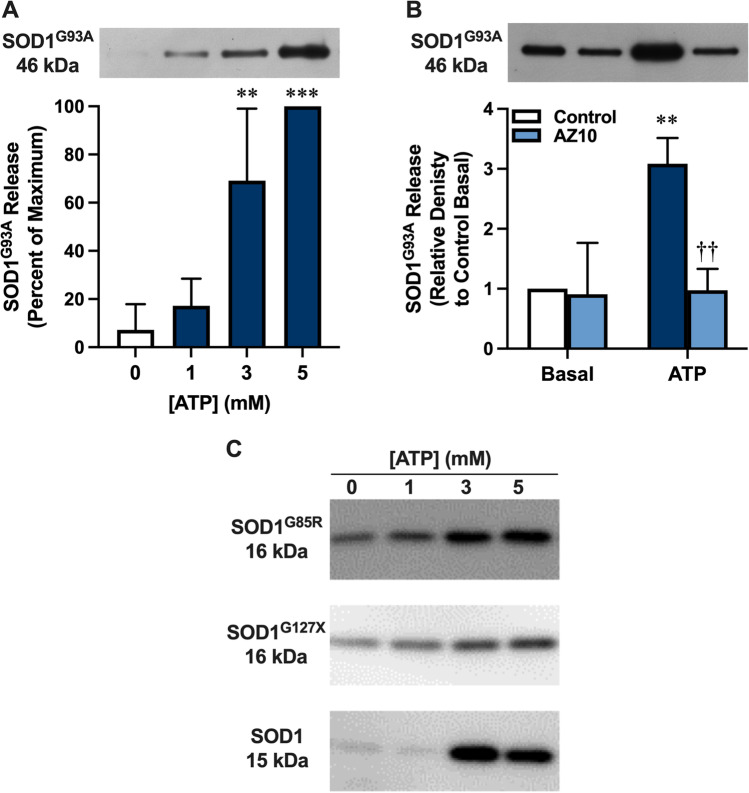
To determine if P2X7 activation could induce SOD1G93A release, NSC-34 cells transiently transfected with SOD1G93A (72 h) were incubated in the absence or presence of ATP (1, 3, or 5 mM) and the presence of SOD1G93A-EGFP in pelletable fractions obtained by centrifugation (20,000 × g for 30 min) examined by immunoblotting using an anti-EGFP antibody. Incubation with either 3 or 5 mM, but not 1 mM, ATP induced significant release of SOD1G93A-EGFP from cells compared to cells incubated in the absence of ATP (Fig. 4A). This ATP-induced release of SOD1G93A-EGFP corresponded to a parallel increase in total protein (Fig. S1), indicating that the ATP effect was not limited to SOD1G93A-EGFP. Pre-incubation of SOD1G93A-EGFP-transfected NSC-34 cells with 10 µM AZ10606120 prior to incubation with 5 mM ATP completely blocked ATP-induced SOD1G93A-EGFP release (Fig. 4B).
Fig. 4.
P2X7 activation mediates SOD1G93A release from NSC-34 motor neurons. A, B SOD1G93A-EGFP (SOD1G93A)-transfected NSC-34 cells, or C non-EGFP SOD1G85R-, non-EGFP SOD1G127X- or non-transfected (SOD1) NSC-34 cells were pre-incubated in the A, C absence or B presence of 10 μM AZ10606120 (AZ10) for 1 h. Cells were then incubated in the absence (0 mM or basal as indicated) or presence of A–C 1, 3 or A–C 5 mM ATP for 20 min and pelletable fractions obtained by centrifugation. A, B The presence of SOD1 in pelletable fractions was assessed by immunoblotting using an anti-EGFP antibody and densitometry. One representative immunoblot (upper image) is shown, and data (lower image) are presented as mean ± SD (n = 3) relative to A maximum release or B release in the absence of AZ10 and ATP; **p < 0.02, ***p < 0.01 compared to 0 mM ATP or corresponding basal, ††p < 0.02 compared to ATP in the absence of AZ10. C The presence of SOD1 in pelletable fractions was assessed by immunoblotting using an anti-SOD1 antibody. Data are representative of n = 2–3 experiments
SOD1G93A can be associated with fibrillar structures that can be macropinocytosed by motor neurons [11] or activate microglia [17]. Therefore, SOD1G93A-EGFP transfected NSC-34 cells were incubated in the absence or presence of 5 mM ATP and the pelletable fractions examined by transmission electron microscopy (TEM). Fibrillar structures, ranging from 100 to 700 nm, were identified in fractions from cells incubated in the presence of ATP but were largely absent in corresponding fractions from cells incubated in the absence of ATP (Fig. S2). To determine if the SOD1G93A-EGFP released following P2X7 activation co-localised with these fibrillar structures, pelletable fractions were examined by TEM and immunogold labelling with an anti-EGFP antibody. SOD1G93A-EGFP, identified by round black gold particles (indicated by arrows), were present alone and in association with fibrillary-like structures (Fig. S3). No immunogold labelling was evident in the corresponding isotype control (Fig. S3).
To determine if ATP could also induce the release of other forms of SOD1, NSC-34 cells, transiently transfected with plasmid DNA encoding non-EGFP-tagged SOD1G85R, non-EGFP-tagged SOD1G127X or murine SOD1, were incubated in the absence or presence of ATP (1, 3, or 5 mM) and the presence of SOD1 in pelletable fractions examined by immunoblotting using an anti-SOD1 antibody. As for SOD1G93A-EGFP, both 3 mM and 5 mM ATP induced the release of SOD1G85R and SOD1G127X from transfected cells, as well as murine SOD1 from non-transfected cells (Fig. 4C). Together, this data indicates that P2X7 activation mediates the release of SOD1G93A (and other forms of SOD1) from NSC-34 cells.
P2X7-mediated SOD1G93A release from NSC-34 motor neurons coincides with membrane blebbing
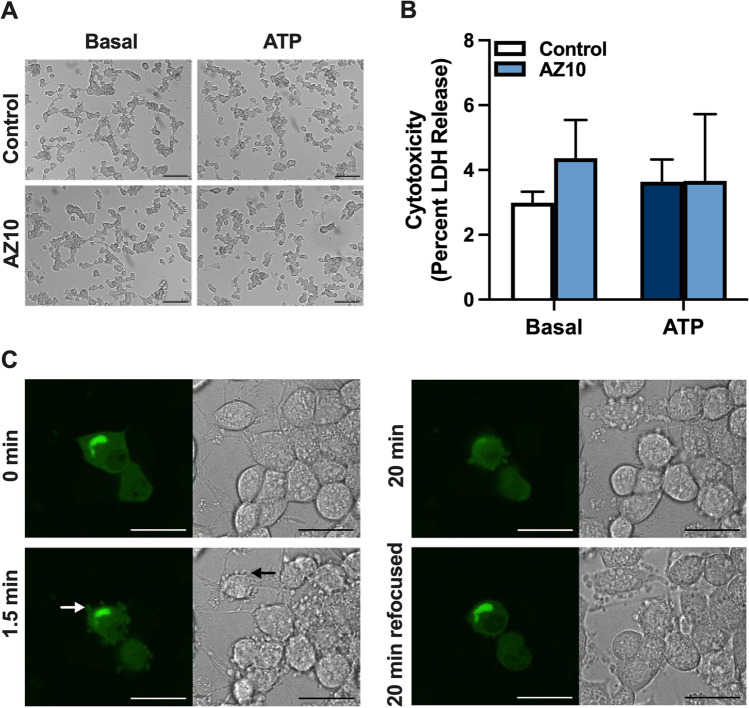
P2X7 activation can induce cytotoxicity under certain circumstances [43] raising the possibility that P2X7-mediated SOD1G93A release from NSC-34 cells occurs via cell death. Therefore, SOD1G93A-EGFP transfected NSC-34 cells were incubated as above and assessed for cell death by microscopy and LDH release. Incubation of cells in the absence (basal) or presence of ATP revealed no morphological differences, with most cells displaying discrete cell bodies with short, spindled shaped processes (Fig. 5A). Moreover, pre-incubation of cells in the absence or presence of AZ10606120 revealed no morphological differences (Fig. 5A). Similarly, cells incubated under the same conditions resulted in no significant differences in LDH release between treatments, with less than 6% of the total cellular LDH released in each treatment group (Fig. 5B).
Fig. 5.
P2X7-mediated SOD1G93A release from NSC-34 motor neurons coincides with membrane blebbing. A–C SOD1G93A-EGFP (SOD1G93A)-transfected NSC-34 cells were pre-incubated in the A–C absence or A, B presence of 10 μM AZ10606120 (AZ10) for 1 h. Cells were then incubated in the absence (basal) or presence of 5 mM ATP for 20 min. A Cell morphology was visualised by differential interference contrast microscopy. Data are representative of n > 20 fields of view; bars represent 100 µm. B Percent of cytotoxicity (LDH release) was determined using the Cytotoxicity Detection KitPLUS. Data are presented as mean ± SD (n = 3). C Membrane blebbing at 0, 1.5, and 20 min was visualised by fluorescent (left images) and bright-field (right images) microscopy. Refocusing was used to examine intracellular SOD1G93A-EGFP at 20 min (20 min refocused). Data are representative of n = 6 experiments; white and black arrows indicate an example of a membrane bleb in transfected and non-transfected cells, respectively; bars represent 25 µm
P2X7 activation can induce membrane blebbing in various cell types [44–46], and membrane blebbing can be associated with protein release [47]. Therefore, to determine a role for membrane blebbing in P2X7-mediated SOD1 release, SOD1G93A-EGFP transfected NSC-34 cells were incubated as above and membrane blebbing examined via confocal microscopy with fluorescent and bright field images collected every 30 s for 20 min (Fig. S4). Morphological changes consistent with membrane blebbing were observed within 1.5 min of ATP addition (Fig. 5C). Membrane blebs ranged in size from 1 to 3 µm and occurred in both SOD1G93A-EGFP expressing (white arrow) and non-expressing (black arrow) cells (Fig. 5C). Moreover, SOD1G93A-EGFP appeared to be released in blebs, evident from the presence of EGFP signal, while the larger mass of cytosolic SOD1G93A-EGFP (identified as EGFP+ foci) appeared to remain inside cells following intracellular refocusing at 20 min (Fig. 5C). Collectively, these results demonstrate that P2X7-mediated SOD1G93A release from NSC-34 motor neurons coincides with membrane blebbing.
SOD1G93A released from NSC-34 motor neurons via P2X7 activation associates with NSC-34 motor neurons and EOC13 microglia to promote ER stress and TNFα release, respectively
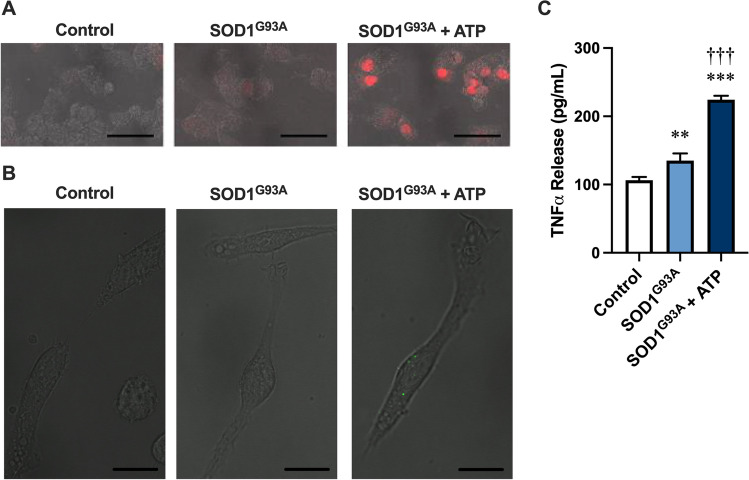
To determine if SOD1G93A-EGFP released from transfected NSC-34 motor neurons via P2X7 activation could associate with naïve NSC-34 cells and induce ER stress, SOD1G93A-EGFP transfected NSC-34 cells were incubated in the absence or presence of 5 mM ATP, and pelletable fractions obtained by centrifugation (20,000 × g for 30 min). Naïve XBP1-tdTomato-transfected NSC-34 cells were then incubated for 24 h in the presence of complete medium (negative control) or pelletable fractions suspended in complete medium. Confocal microscopy revealed a greater proportion of XBP1-tdTomato fluorescence in NSC-34 cells incubated with ATP-induced SOD1G93A-EGFP pelletable fractions compared to cells incubated with complete culture medium or SOD1G93A-EGFP pelletable fractions, both of which had negligible or low amounts of fluorescence, respectively (Fig. 6A).
Fig. 6.
SOD1G93A released from NSC-34 motor neurons via P2X7 activation associates with NSC-34 motor neurons and EOC13 microglia to promote ER stress and TNFα release, respectively. A–C SOD1G93A-EGFP-transfected NSC-34 cells were incubated in the absence (SOD1G93A) or presence of 5 mM ATP (SOD1G93A + ATP) for 20 min, and pelletable fractions obtained by centrifugation and used as follows. A XBP1-tdTomato-transfected NSC-34 cell were incubated with medium alone (Control) or pelletable fractions (SOD1G93A or SOD1G93A + ATP). At 24 h, cellular XBP1-tdTomato fluorescence was visualised by confocal microscopy. Data are representative of n = 2 experiments; bars represent 50 µm. B, C EOC13 cells were incubated with medium alone (control) or pelletable fractions (SOD1G93A or SOD1G93A + ATP). B At 2 h, EGFP fluorescence was visualised by confocal microscopy. Data are representative of n = 6 experiments; bars represent 15 µm. C At 24 h, the concentration of TNFα released into cell supernatants was assessed by ELISA. Data are presented as mean ± SD (n = 3); **p < 0.02, ***p < 0.01 compared to control, †††p < 0.01 compared to SOD1G93A
Finally, to determine if SOD1G93A-EGFP released from transfected NSC-34 cells via P2X7 activation could associate with EOC13 microglia and induce TNFα release, EOC13 cells were incubated for up to 24 h in the presence of complete medium or pelletable fractions as above. Confocal microscopy revealed the presence of EGFP associated with EOC13 cells incubated with ATP-induced SOD1G93A-EGFP pelletable fractions but not in cells incubated with complete medium or SOD1G93A-EGFP pelletable fractions at 2 h (Fig. 6B). ELISA measurements at 24 h revealed that EOC13 cells incubated with ATP-induced SOD1G93A-EGFP pelletable fractions EGFP induced significantly greater TNFα release compared to cells incubated with complete medium or SOD1G93A-EGFP pelletable fractions (Fig. 6C). Together, the above data demonstrate that SOD1G93A released from NSC-34 motor neurons via P2X7 activation associates with both NSC-34 motor neurons and EOC13 microglia to promote ER stress and TNFα release, respectively.
Discussion
Accumulating evidence suggests a role for mutant SOD1 in the prion-like propagation in ALS [4]; however, the biological impacts of this process and the precise mechanisms of SOD1 release remain to be fully resolved. This study demonstrated that aggregated SOD1G93A released from NSC-34 motor neurons could be transmitted to naïve NSC-34 cells and murine EOC13 microglia to induce ER stress and TNFα release, respectively. Moreover, this study demonstrated that NSC-34 cells express P2X7 and that activation of this receptor can induce the release of aggregated SOD1G93A from these cells. Furthermore, this released material could associate with and induce ER stress in NSC-34 cells and TNFα release from EOC13 cells. A limitation regarding the role of P2X7 in NSC-34 cells is that the current study relied on the use of a single P2X7 antagonist, AZ1060120. Although this antagonist is a well-known high affinity, potent and selective P2X7 antagonist [42], which binds the allosteric binding pocket of this receptor [48, 49], additional studies using a second P2X7 antagonist or P2rx7 gene silencing are required to confirm the current observations. Moreover, the current study relied on the use of a single P2X7 agonist, ATP. The P2X7 agonist, 2'(3')-O-(4-benzoylbenzoyl) ATP was not tested due to its potential to activate other P2X receptor subtypes [50].
The major finding of the current study is that P2X7 activation by extracellular ATP induces the release of SOD1G93A aggregates from NSC-34 cells transiently transfected with SOD1G93A, which can be subsequently transmitted to naïve NSC-34 and EOC13 cells to potentially propagate disease. These findings were extended to SOD1G85R and SOD1G127X from transfected cells, as well as murine SOD1 from non-transfected cells, with apparent differences in the amount of SOD1 aggregates released most likely reflecting differences in SOD1 expression or aggregation. Collectively, to the best of our knowledge, this is the first description of a receptor-mediated pathway of aggregated SOD1 release. It is currently thought that SOD1 aggregates are released directly from dead or dying cells or via exosomes from live cells and cells early in the dying process [51]. Thus, P2X7-mediated release provides an alternate but not mutually exclusive mechanism of aggregated SOD1 release. Measurements of LDH release suggest that the P2X7-mediated release of aggregated SOD1 occurred independently of cell death. Similarly, others have shown that P2X7 agonists do not induce death of SOD1G93A- or SOD1WT-transfected NSC-34 cells [52]. However, given that P2X7 activation can induce the death of rat motor neurons [39, 40], it cannot be excluded that P2X7 mediated the release of aggregated SOD1 from cells early in the dying process. In this regard, P2X7 activation on NSC-34 cells transiently transfected with SOD1G93A was associated with membrane blebbing, which can be associated with cell death in some [53] but not all instances [54, 55]. It is also possible that P2X7 activation may mediate SOD1 release to remove excess amounts of this protein, given that P2X7 activation can promote autophagy in SOD1G93A mice [56, 57]. In contrast, it is unlikely that the increase in aggregated SOD1 release following P2X7 activation was due to increased SOD1 synthesis. To the best of our knowledge, the impact of P2X7 activation on SOD1 synthesis is unknown, but the effect of P2X7 activation was studied over a relatively short period (20 min), while the turnover of SOD1 in vitro and in vivo is slow (hours to days) [58, 59]. Other possible mechanisms of aggregated SOD1 release following P2X7 activation include direct exocytosis or exocytosis of extracellular vesicles (ectosomes, exosomes, and/or lysosomes/autophagosomes), mechanisms common to both P2X7 activation [60] and SOD1 release in other contexts [61].
The current study demonstrates that aggregated SOD1G93A released either constitutively or following P2X7 activation can subsequently be transmitted to naïve NSC-34 and EOC13 cells to induce ER stress and TNFα release, respectively. ER stress, resulting from the accumulation of misfolded or unfolded proteins, can stimulate the unfolded protein response, which can eventually result in motor neuron dysfunction and death [62]. Notably, ER stress is found in the motor neurons that degenerate first in SOD1G93A mice, which subsequently influences disease progression [63]. This earlier finding highlighted the contribution of ER stress to the initiation of ALS. However, accumulating evidence also indicates important roles for ER stress in the progression of ALS at later time points [62]. Likewise, TNFα can also contribute to ALS progression, although the contributions of this cytokine to the pathophysiology of ALS remain varied and controversial [64]. Nevertheless, increased release of TNFα may contribute to neuroinflammation, glutamate-mediated excitotoxicity and muscle wasting in this disease [64]. Combined, these observations are consistent with the notion that aggregated SOD1G93A release and transmission to other motor neurons and microglia may facilitate the progression of ALS.
This study demonstrates for the first-time the presence of functional P2X7 on NSC-34 cells. Although the presence of P2X7 on neurons is controversial [65, 66], P2X7 has been observed in mouse and rat motor neurons in situ [67, 68], and activation of these receptors on these cells can induce vesicle exocytosis [67] and, as mentioned above, cell death [39, 40]. However, it should be noted that NSC-34 cells were obtained by fusing N18TG-2 neuroblastoma cells with murine spinal cord cells [32] and that N18TG-2 cells have been shown to express functional P2X7 [69]. Thus, the possibility remains that the presence of P2X7 on NSC-34 cells is acquired from the neuroblastoma cell line rather than spinal cord cells. This caveat notwithstanding, the current study supports the notion that P2X7 activation may mediate the release and transmission of aggregated SOD1 in vivo. Moreover, it is hypothesised that this mechanism may be a target of the central nervous system (CNS)-penetrant P2X7 antagonists JNJ-47965567 [22, 70] or AXX71 [56], which can slow disease progression in ALS SOD1G93A mice in a dose-dependent manner. Whether the poorly CNS-penetrant P2X7 antagonist Brilliant Blue G, which likewise slows disease progression in ALS SOD1G93A mice [71], also impairs P2X7-mediated aggregated SOD1 release remains unknown.
The murine P2rx7 gene is polymorphic, with many mouse strains encoding the well-established loss-of-function polymorphism, P451L [72, 73]. Despite this, the P2rx7 genotype of NSC-34 or EOC13 cells is unknown. EOC13 cells were derived from a C3H mouse [74], a strain that encodes the 451L allele [75]. Thus, EOC13 cells are likely to encode the 451L allele and express P2X7 with reduced activity. However, the P2rx7 genotype of NSC-34 cells is more difficult to ascertain due to their complex origin. As noted above, NSC-34 cells were obtained by fusing N18TG-2 neuroblastoma cells with murine spinal cord cells [32]. This is further complicated with N18TG-2 cells being derived from C1300 tumour cells [76], reportedly from A/J mice [77], a strain that encodes the P451 allele [75]. While the spinal cord cells, used to generate NSC-34 cells, were obtained from CD-1 mice [32], a strain of unknown P2rx7 genotype. CD-1 mice however display P2X7 properties consistent with mice encoding the P451 allele [75]. Thus, NSC-34 cells are likely to encode the P451 allele and express P2X7 with normal activity. Despite the 451L allele encoding for a loss-of-function, the ATP sensitivity of P2X7 containing either a proline or leucine residue at position 451 are similar [78, 79]. Notably, the ATP sensitivity of P2X7 in N18TG-2 cells (EC50 of 1.8 mM) is at least twofold greater [69] than that of recombinant murine P2X7 overexpressed in HEK-293 cells (EC50 of 0.7 mM) [80]. Thus, the aforementioned requirement for relatively high ATP concentrations (≥ 3 mM) to active P2X7 in NCS-34 cells may reflect low P2X7 expression (paralleling the P2X7 immunoblotting data above) or the presence of another, unknown polymorphism in the cells.
In addition to the use of a single P2X7 agonist (ATP) and antagonist (AZ1060120), this study includes other limitations. First, relatively high ATP concentrations (≥ 3 mM) were required to promote aggregated SOD1G93A release from NSC-34 cells. It is unknown if ATP is released and at what concentrations in ALS. However extracellular ATP concentrations are thought to dramatically increase in the CNS under pathological conditions but measuring this directly in vivo remains challenging [81]. Second, there is a possibility that ATP was present in the pelletable fractions obtained following incubation with ATP and this may have mediated the observed effects in naïve NSC-34 or EOC13 cells via activation of P2X7. However, this appears improbable as the pelletable fractions were acquired by centrifugation at 20,000 × g for 30 min, are unlikely to contain residual ATP, a soluble molecule with a molecular weight of 507 Da, with most if not all of the ATP presumably residing in the supernatant. In support of this notion, SOD1 constitutively released from NSC-34 cells caused similar effects in naïve NSC-34 or EOC13 cells to that caused by ATP-induced SOD1. There is the possibility that very low concentrations of ATP, which could activate other P2 receptors potentially present in NSC-34 or EOC13 cells, exist in the pelletable fractions, but the presence of other P2 receptors in these cell types is unknown. Third, the possibility remains that transfection of NSC-34 cells may have induced pro-inflammatory effects to aid the observed effects in naïve NSC-34 or EOC13 cells. For example, others have reported that transfection of NSC-34 cells with mutant SOD1 can cause the release of exosomes containing inflammatory-related microRNAs [82] or the upregulation of cytosolic phospholipase A2α in TNFα-dependent manner [83]. The induction of such pro-inflammatory effects may offer an explanation as to why pelletable fractions from EGFP-transfected NSC-34 caused a small amount of TNFα release from naïve EOC13 cells.
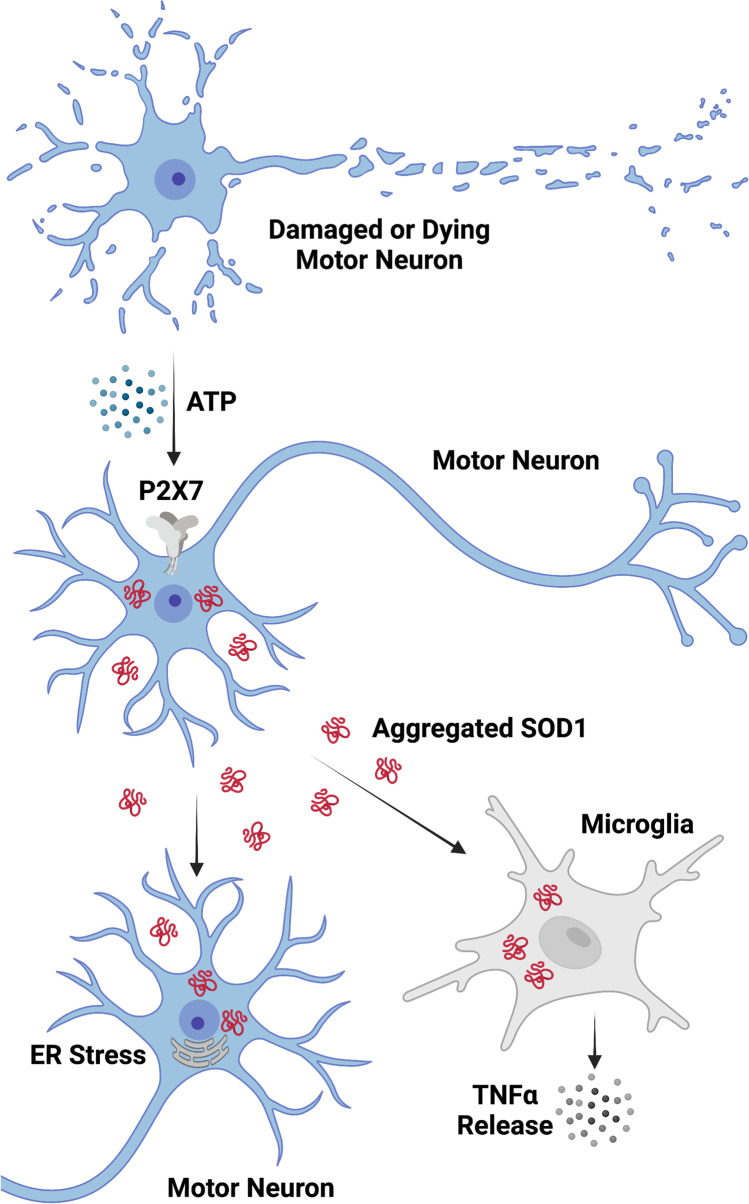
In conclusion, the current study demonstrates that aggregated SOD1G93A can be released constitutively or via P2X7 activation from NSC-34 cells, which can subsequently be transmitted to naïve NSC-34 and EOC13 cells to induce ER stress and TNFα release, respectively (Fig. 7). Collectively, this provides further support for the prion-like propagation of SOD1 in ALS and a possible explanation for the therapeutic benefits observed with P2X7 antagonism in ALS SOD1G93A mice.
Fig. 7.
Role of the ATP-P2X7 signalling axis in the transmission of SOD1 aggregates and progression of ALS. Damaged or dying motor neurons (or activated glia; not shown) in ALS patients release ATP to stimulate P2X7 on neighbouring motor neurons leading to aggregated SOD1 release. Released SOD1 aggregates are internalised by other neighbouring motor neurons or microglia to promote ER stress or TNFα release, respectively, to potentially promote disease progression in ALS patients. Created with BioRender.com
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the administrative and technical support of the Illawarra Health and Medical Research Institute and David Mitchell (University of Wollongong) for assistance with electron microscopy.
Rachael Bartlett
is a Research Fellow in the School of Chemistry and Molecular Bioscience at the University of Wollongong and in the Illawarra Health and Medical Research Institute, Wollongong. She received a BBiotechAdv (Hons I) and a PhD from the University of Wollongong in 2011 and 2016, respectively. Her research to date has focused primarily on neuroinflammation and proteostasis in motor neuron disease. Her other research has focused on molecular mechanisms in psychiatric disorders.
Author contribution
Rachael Bartlett acquired funding, designed and performed experiments, analysed and interpreted data and co-wrote the manuscript. Diane Ly analysed and interpreted data and co-wrote the manuscript. Neil R. Cashman designed experiments, supervised the project, interpreted data, acquired funding and revised the manuscript. Ronald Sluyter conceived the study, acquired funding, designed experiments, supervised the project, interpreted data and wrote the manuscript. Justin J. Yerbury conceived the study, acquired funding, designed experiments, supervised the project, interpreted data and revised the manuscript.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. Rachael Bartlett was a recipient of an Australian Postgraduate Award (University of Wollongong) and a Global Challenges Scholarship (University of Wollongong). Neil R. Cashman received funding from the ALS Society of Canada (ALS Canada), Brain Canada, the R. Howard Webster Foundation, and the Canadian Consortium for Neurodegeneration in Aging (CCNA), and donations from John Tognetti and William Lambert. Ronald Sluyter received funding from MND Australia. Justin J. Yerbury received funding from MND Australia, the National Health and Medical Council of Australia (NHMRC) and FightMND.
Data availability
Data is contained within the article or supplementary material. Data not shown is available from the corresponding authors, upon reasonable request.
Code availability
Not applicable.
Declarations
Ethical approval
This study does not contain any work with animals or human participants performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
All authors give their consent for publication.
Conflicts of interest
Rachael Bartlett declares no conflict of interest.
Diane Ly declares no conflict of interest.
Neil R. Cashman declares no conflict of interest.
Ronald Sluyter declares no conflict of interest.
Justin J. Yerbury declares no conflict of interest.
Footnotes
Ronald Sluyter and Justin J. Yerbury are equal senior authors.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ronald Sluyter, Email: rsluyter@uow.edu.au.
Justin J. Yerbury, Email: jyerbury@uow.edu.au
References
- 1.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 2.Grad LI, Rouleau GA, Ravits J, Cashman NR (2017) Clinical spectrum of amyotrophic lateral sclerosis (ALS). Cold Spring Harb Perspect Med 7:a024117. 10.1101/cshperspect.a024117 [DOI] [PMC free article] [PubMed]
- 3.McAlary L, Plotkin SS, Yerbury JJ, Cashman NR. Prion-like propagation of protein misfolding and aggregation in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:262. doi: 10.3389/fnmol.2019.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAlary L, Yerbury JJ, Cashman NR. The prion-like nature of amyotrophic lateral sclerosis. Prog Mol Biol Transl Sci. 2020;175:261–296. doi: 10.1016/bs.pmbts.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 6.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaur SJ, McKeown SR, Rashid S. Mutant SOD1 mediated pathogenesis of amyotrophic lateral sclerosis. Gene. 2016;577:109–118. doi: 10.1016/j.gene.2015.11.049. [DOI] [PubMed] [Google Scholar]
- 8.Münch C, O'Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc Natl Acad Sci U S A. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grad LI, Guest WC, Yanai A, Pokrishevsky E, O'Neill MA, Gibbs E, Semenchenko V, Yousefi M, Wishart DS, Plotkin SS, Cashman NR. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc Natl Acad Sci U S A. 2011;108:16398–16403. doi: 10.1073/pnas.1102645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grad LI, Yerbury JJ, Turner BJ, Guest WC, Pokrishevsky E, O'Neill MA, Yanai A, Silverman JM, Zeineddine R, Corcoran L, Kumita JR, Luheshi LM, Yousefi M, Coleman BM, Hill AF, Plotkin SS, Mackenzie IR, Cashman NR. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2014;111:3620–3625. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeineddine R, Pundavela JF, Corcoran L, Stewart EM, Do-Ha D, Bax M, Guillemin G, Vine KL, Hatters DM, Ecroyd H, Dobson CM, Turner BJ, Ooi L, Wilson MR, Cashman NR, Yerbury JJ. SOD1 protein aggregates stimulate macropinocytosis in neurons to facilitate their propagation. Mol Neurodegener. 2015;10:57. doi: 10.1186/s13024-015-0053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayers JI, Diamond J, Sari A, Fromholt S, Galaleldeen A, Ostrow LW, Glass JD, Hart PJ, Borchelt DR. Distinct conformers of transmissible misfolded SOD1 distinguish human SOD1-FALS from other forms of familial and sporadic ALS. Acta Neuropathol. 2016;132:827–840. doi: 10.1007/s00401-016-1623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crown A, McAlary L, Fagerli E, Brown H, Yerbury JJ, Galaleldeen A, Cashman NR, Borchelt DR, Ayers JI. Tryptophan residue 32 in human Cu-Zn superoxide dismutase modulates prion-like propagation and strain selection. PLoS ONE. 2020;15:e0227655. doi: 10.1371/journal.pone.0227655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayers JI, Fromholt S, Koch M, DeBosier A, McMahon B, Xu G, Borchelt DR. Experimental transmissibility of mutant SOD1 motor neuron disease. Acta Neuropathol. 2014;128:791–803. doi: 10.1007/s00401-014-1342-7. [DOI] [PubMed] [Google Scholar]
- 15.Gomes C, Keller S, Altevogt P, Costa J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci Lett. 2007;428:43–46. doi: 10.1016/j.neulet.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 16.Sundaramoorthy V, Walker AK, Yerbury J, Soo KY, Farg MA, Hoang V, Zeineddine R, Spencer D, Atkin JD. Extracellular wildtype and mutant SOD1 induces ER-Golgi pathology characteristic of amyotrophic lateral sclerosis in neuronal cells. Cell Mol Life Sci. 2013;70:4181–4195. doi: 10.1007/s00018-013-1385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts K, Zeineddine R, Corcoran L, Li W, Campbell IL, Yerbury JJ. Extracellular aggregated Cu/Zn superoxide dismutase activates microglia to give a cytotoxic phenotype. Glia. 2013;61:409–419. doi: 10.1002/glia.22444. [DOI] [PubMed] [Google Scholar]
- 18.Jiang LH, Caseley EA, Muench SP, Roger S. Structural basis for the functional properties of the P2X7 receptor for extracellular ATP. Purinergic Signal. 2021;17:331–344. doi: 10.1007/s11302-021-09790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sluyter R. The P2X7 Receptor. Adv Exp Med Biol. 2017;1051:17–53. doi: 10.1007/5584_2017_59. [DOI] [PubMed] [Google Scholar]
- 20.Andrejew R, Oliveira-Giacomelli Á, Ribeiro DE, Glaser T, Arnaud-Sampaio VF, Lameu C, Ulrich H. The P2X7 receptor: central hub of brain diseases. Front Mol Neurosci. 2020;13:124. doi: 10.3389/fnmol.2020.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Territo PR, Zarrinmayeh H. P2X7 receptors in neurodegeneration: potential therapeutic applications from basic to clinical approaches. Front Cell Neurosci. 2021;15:617036. doi: 10.3389/fncel.2021.617036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Ruiz C, Calzaferri F, García AG. P2X7 receptor antagonism as a potential therapy in amyotrophic lateral sclerosis. Front Mol Neurosci. 2020;13:93. doi: 10.3389/fnmol.2020.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volonté C, Amadio S, Liguori F, Fabbrizio P. Duality of P2X7 receptor in amyotrophic lateral sclerosis. Front Pharmacol. 2020;11:1148. doi: 10.3389/fphar.2020.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apolloni S, Amadio S, Parisi C, Matteucci A, Potenza RL, Armida M, Popoli P, D'Ambrosi N, Volonté C. Spinal cord pathology is ameliorated by P2X7 antagonism in a SOD1-mutant mouse model of amyotrophic lateral sclerosis. Dis Model Mech. 2014;7:1101–1109. doi: 10.1242/dmm.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett R, Sluyter V, Watson D, Sluyter R, Yerbury JJ. P2X7 antagonism using Brilliant Blue G reduces body weight loss and prolongs survival in female SOD1(G93A) amyotrophic lateral sclerosis mice. PeerJ. 2017;5:e3064. doi: 10.7717/peerj.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cervetto C, Frattaroli D, Maura G, Marcoli M. Motor neuron dysfunction in a mouse model of ALS: gender-dependent effect of P2X7 antagonism. Toxicology. 2013;311:69–77. doi: 10.1016/j.tox.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz-Ruiz C, García-Magro N, Negredo P, Avendaño C, Bhattacharya A, Ceusters M, García AG (2020) Chronic administration of P2X7 receptor antagonist JNJ-47965567 delays disease onset and progression, and improves motor performance in ALS SOD1(G93A) female mice. Dis Model Mech 13:dmm045732. 10.1242/dmm.045732 [DOI] [PMC free article] [PubMed]
- 28.de Torre-Minguela C, Barberà-Cremades M, Gómez AI, Martín-Sánchez F, Pelegrín P. Macrophage activation and polarization modify P2X7 receptor secretome influencing the inflammatory process. Sci Rep. 2016;6:22586. doi: 10.1038/srep22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pupovac A, Geraghty NJ, Watson D, Sluyter R. Activation of the P2X7 receptor induces the rapid shedding of CD23 from human and murine B cells. Immunol Cell Biol. 2015;93:77–85. doi: 10.1038/icb.2014.69. [DOI] [PubMed] [Google Scholar]
- 30.Takenouchi T, Tsukimoto M, Iwamaru Y, Sugama S, Sekiyama K, Sato M, Kojima S, Hashimoto M, Kitani H. Extracellular ATP induces unconventional release of glyceraldehyde-3-phosphate dehydrogenase from microglial cells. Immunol Lett. 2015;167:116–124. doi: 10.1016/j.imlet.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Nishida K, Nakatani T, Ohishi A, Okuda H, Higashi Y, Matsuo T, Fujimoto S, Nagasawa K. Mitochondrial dysfunction is involved in P2X7 receptor-mediated neuronal cell death. J Neurochem. 2012;122:1118–1128. doi: 10.1111/j.1471-4159.2012.07868.x. [DOI] [PubMed] [Google Scholar]
- 32.Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- 33.Turner BJ, Atkin JD, Farg MA, Zang DW, Rembach A, Lopes EC, Patch JD, Hill AF, Cheema SS. Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J Neurosci. 2005;25:108–117. doi: 10.1523/jneurosci.4253-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartlett R, Yerbury JJ, Sluyter R. P2X7 receptor activation induces reactive oxygen species formation and cell death in murine EOC13 microglia. Mediators Inflamm. 2013;2013:271813. doi: 10.1155/2013/271813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 37.Pokrishevsky E, McAlary L, Farrawell NE, Zhao B, Sher M, Yerbury JJ, Cashman NR. Tryptophan 32-mediated SOD1 aggregation is attenuated by pyrimidine-like compounds in living cells. Sci Rep. 2018;8:15590. doi: 10.1038/s41598-018-32835-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keilhoff G, Lucas B, Uhde K, Fansa H. Selected gene profiles of stressed NSC-34 cells and rat spinal cord following peripheral nerve reconstruction and minocycline treatment. Exp Ther Med. 2016;11:1685–1699. doi: 10.3892/etm.2016.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franco MC, Ye Y, Refakis CA, Feldman JL, Stokes AL, Basso M, Fernández M, de Mera RM, Sparrow NA, Calingasan NY, Kiaei M, Rhoads TW, Ma TC, Grumet M, Barnes S, Beal MF, Beckman JS, Mehl R, Estévez AG. Nitration of Hsp90 induces cell death. Proc Natl Acad Sci U S A. 2013;110:E1102–1111. doi: 10.1073/pnas.1215177110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandelman M, Levy M, Cassina P, Barbeito L, Beckman JS. P2X7 receptor-induced death of motor neurons by a peroxynitrite/FAS-dependent pathway. J Neurochem. 2013;126:382–388. doi: 10.1111/jnc.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhaskaracharya A, Dao-Ung P, Jalilian I, Spildrejorde M, Skarratt KK, Fuller SJ, Sluyter R, Stokes L. Probenecid blocks human P2X7 receptor-induced dye uptake via a pannexin-1 independent mechanism. PLoS ONE. 2014;9:e93058. doi: 10.1371/journal.pone.0093058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michel AD, Chambers LJ, Clay WC, Condreay JP, Walter DS, Chessell IP. Direct labelling of the human P2X7 receptor and identification of positive and negative cooperativity of binding. Br J Pharmacol. 2007;151:103–114. doi: 10.1038/sj.bjp.0707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bidula S, Dhuna K, Helliwell R, Stokes L. Positive allosteric modulation of P2X7 promotes apoptotic cell death over lytic cell death responses in macrophages. Cell Death Dis. 2019;10:882. doi: 10.1038/s41419-019-2110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panupinthu N, Zhao L, Possmayer F, Ke HZ, Sims SM, Dixon SJ. P2X7 nucleotide receptors mediate blebbing in osteoblasts through a pathway involving lysophosphatidic acid. J Biol Chem. 2007;282:3403–3412. doi: 10.1074/jbc.M605620200. [DOI] [PubMed] [Google Scholar]
- 45.Verhoef PA, Estacion M, Schilling W, Dubyak GR. P2X7 receptor-dependent blebbing and the activation of Rho-effector kinases, caspases, and IL-1 beta release. J Immunol. 2003;170:5728–5738. doi: 10.4049/jimmunol.170.11.5728. [DOI] [PubMed] [Google Scholar]
- 46.Virginio C, MacKenzie A, North RA, Surprenant A. Kinetics of cell lysis, dye uptake and permeability changes in cells expressing the rat P2X7 receptor. J Physiol. 1999;519(Pt 2):335–346. doi: 10.1111/j.1469-7793.1999.0335m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirov A, Al-Hashimi H, Solomon P, Mazur C, Thorpe PE, Sims PJ, Tarantini F, Kumar TK, Prudovsky I. Phosphatidylserine externalization and membrane blebbing are involved in the nonclassical export of FGF1. J Cell Biochem. 2012;113:956–966. doi: 10.1002/jcb.23425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allsopp RC, Dayl S, Schmid R, Evans RJ. Unique residues in the ATP gated human P2X7 receptor define a novel allosteric binding pocket for the selective antagonist AZ10606120. Sci Rep. 2017;7:725. doi: 10.1038/s41598-017-00732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karasawa A, Kawate T (2016) Structural basis for subtype-specific inhibition of the P2X7 receptor. eLife 5:e22153. 10.7554/eLife.22153 [DOI] [PMC free article] [PubMed]
- 50.Illes P, Müller CE, Jacobson KA, Grutter T, Nicke A, Fountain SJ, Kennedy C, Schmalzing G, Jarvis MF, Stojilkovic SS, King BF, Di Virgilio F. Update of P2X receptor properties and their pharmacology: IUPHAR Review 30. Br J Pharmacol. 2021;178:489–514. doi: 10.1111/bph.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silverman JM, Fernando SM, Grad LI, Hill AF, Turner BJ, Yerbury JJ, Cashman NR. Disease mechanisms in ALS: misfolded SOD1 transferred through exosome-dependent and exosome-independent pathways. Cell Mol Neurobiol. 2016;36:377–381. doi: 10.1007/s10571-015-0294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D'Ambrosi N, Finocchi P, Apolloni S, Cozzolino M, Ferri A, Padovano V, Pietrini G, Carrì MT, Volonté C. The proinflammatory action of microglial P2 receptors is enhanced in SOD1 models for amyotrophic lateral sclerosis. J Immunol. 2009;183:4648–4656. doi: 10.4049/jimmunol.0901212. [DOI] [PubMed] [Google Scholar]
- 53.Ousingsawat J, Wanitchakool P, Kmit A, Romao AM, Jantarajit W, Schreiber R, Kunzelmann K. Anoctamin 6 mediates effects essential for innate immunity downstream of P2X7 receptors in macrophages. Nat Commun. 2015;6:6245. doi: 10.1038/ncomms7245. [DOI] [PubMed] [Google Scholar]
- 54.Mackenzie AB, Young MT, Adinolfi E, Surprenant A. Pseudoapoptosis induced by brief activation of ATP-gated P2X7 receptors. J Biol Chem. 2005;280:33968–33976. doi: 10.1074/jbc.M502705200. [DOI] [PubMed] [Google Scholar]
- 55.Morelli A, Chiozzi P, Chiesa A, Ferrari D, Sanz JM, Falzoni S, Pinton P, Rizzuto R, Olson MF, Di Virgilio F. Extracellular ATP causes ROCK I-dependent bleb formation in P2X7-transfected HEK293 cells. Mol Biol Cell. 2003;14:2655–2664. doi: 10.1091/mbc.02-04-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Apolloni S, Fabbrizio P, Amadio S, Napoli G, Freschi M, Sironi F, Pevarello P, Tarroni P, Liberati C, Bendotti C, Volonté C (2021) Novel P2X7 antagonist ameliorates the early phase of ALS disease and decreases inflammation and autophagy in SOD1-G93A mouse model. Int J Mol Sci 22:10649. 10.3390/ijms221910649 [DOI] [PMC free article] [PubMed]
- 57.Fabbrizio P, Amadio S, Apolloni S, Volonté C. P2X7 receptor activation modulates autophagy in SOD1-G93A mouse microglia. Front Cell Neurosci. 2017;11:249. doi: 10.3389/fncel.2017.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borchelt DR, Guarnieri M, Wong PC, Lee MK, Slunt HS, Xu ZS, Sisodia SS, Price DL, Cleveland DW. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J Biol Chem. 1995;270:3234–3238. doi: 10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- 59.Crisp MJ, Mawuenyega KG, Patterson BW, Reddy NC, Chott R, Self WK, Weihl CC, Jockel-Balsarotti J, Varadhachary AS, Bucelli RC, Yarasheski KE, Bateman RJ, Miller TM. In vivo kinetic approach reveals slow SOD1 turnover in the CNS. J Clin Invest. 2015;125:2772–2780. doi: 10.1172/jci80705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubyak GR. P2X7 receptor regulation of non-classical secretion from immune effector cells. Cell Microbiol. 2012;14:1697–1706. doi: 10.1111/cmi.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cicardi ME, Marrone L, Azzouz M, Trotti D. Proteostatic imbalance and protein spreading in amyotrophic lateral sclerosis. EMBO J. 2021;40:e106389. doi: 10.15252/embj.2020106389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rozas P, Bargsted L, Martínez F, Hetz C, Medinas DB. The ER proteostasis network in ALS: Determining the differential motoneuron vulnerability. Neurosci Lett. 2017;636:9–15. doi: 10.1016/j.neulet.2016.04.066. [DOI] [PubMed] [Google Scholar]
- 63.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 64.Tortarolo M, Lo Coco D, Veglianese P, Vallarola A, Giordana MT, Marcon G, Beghi E, Poloni M, Strong MJ, Iyer AM, Aronica E, Bendotti C. Amyotrophic lateral sclerosis, a multisystem pathology: insights into the role of TNFα. Mediators Inflamm. 2017;2017:2985051. doi: 10.1155/2017/2985051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Illes P, Khan TM, Rubini P. Neuronal P2X7 receptors revisited: do they really exist? J Neurosci. 2017;37:7049–7062. doi: 10.1523/jneurosci.3103-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miras-Portugal MT, Sebastián-Serrano Á, de Diego GL, Díaz-Hernández M. Neuronal P2X7 receptor: involvement in neuronal physiology and pathology. J Neurosci. 2017;37:7063–7072. doi: 10.1523/jneurosci.3104-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moores TS, Hasdemir B, Vega-Riveroll L, Deuchars J, Parson SH. Properties of presynaptic P2X7-like receptors at the neuromuscular junction. Brain Res. 2005;1034:40–50. doi: 10.1016/j.brainres.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Yu Y, Ugawa S, Ueda T, Ishida Y, Inoue K, Kyaw Nyunt A, Umemura A, Mase M, Yamada K, Shimada S. Cellular localization of P2X7 receptor mRNA in the rat brain. Brain Res. 2008;1194:45–55. doi: 10.1016/j.brainres.2007.11.064. [DOI] [PubMed] [Google Scholar]
- 69.Kaiho H, Matsuoka I, Kimura J, Nakanishi H. Identification of P2X7 (P2Z) receptor in N18TG-2 cells and NG108-15 cells. J Neurochem. 1998;70:951–957. doi: 10.1046/j.1471-4159.1998.70030951.x. [DOI] [PubMed] [Google Scholar]
- 70.Ly D, Dongol A, Cuthbertson P, Guy TV, Geraghty NJ, Sophocleous RA, Sin L, Turner BJ, Watson D, Yerbury JJ, Sluyter R. The P2X7 receptor antagonist JNJ-47965567 administered thrice weekly from disease onset does not alter progression of amyotrophic lateral sclerosis in SOD1(G93A) mice. Purinergic Signal. 2020;16:109–122. doi: 10.1007/s11302-020-09692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sluyter R, Bartlett R, Ly D, Yerbury JJ. P2X7 receptor antagonism in amyotrophic lateral sclerosis. Neural Regen Res. 2017;12:749–750. doi: 10.4103/1673-5374.206643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adriouch S, Dox C, Welge V, Seman M, Koch-Nolte F, Haag F. Cutting edge: a natural P451L mutation in the cytoplasmic domain impairs the function of the mouse P2X7 receptor. J Immunol. 2002;169:4108–4112. doi: 10.4049/jimmunol.169.8.4108. [DOI] [PubMed] [Google Scholar]
- 73.Bartlett R, Stokes L, Sluyter R. The P2X7 receptor channel: recent developments and the use of P2X7 antagonists in models of disease. Pharmacol Rev. 2014;66:638–675. doi: 10.1124/pr.113.008003. [DOI] [PubMed] [Google Scholar]
- 74.Walker WS, Gatewood J, Olivas E, Askew D, Havenith CE. Mouse microglial cell lines differing in constitutive and interferon-gamma-inducible antigen-presenting activities for naive and memory CD4+ and CD8+ T cells. J Neuroimmunol. 1995;63:163–174. doi: 10.1016/0165-5728(95)00146-8. [DOI] [PubMed] [Google Scholar]
- 75.Sorge RE, Trang T, Dorfman R, Smith SB, Beggs S, Ritchie J, Austin JS, Zaykin DV, Vander Meulen H, Costigan M, Herbert TA, Yarkoni-Abitbul M, Tichauer D, Livneh J, Gershon E, Zheng M, Tan K, John SL, Slade GD, Jordan J, Woolf CJ, Peltz G, Maixner W, Diatchenko L, Seltzer Z, Salter MW, Mogil JS. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med. 2012;18:595–599. doi: 10.1038/nm.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Augusti-Tocco G, Sato G. Establishment of functional clonal lines of neurons from mouse neuroblastoma. Proc Natl Acad Sci U S A. 1969;64:311–315. doi: 10.1073/pnas.64.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murphy ED (1966) Characteristic Tumours. In: Green EL (ed) Biology of the Laboratory Mouse. Dover Publications, Inc., New York, Ch 27. http://www.informatics.jax.org/greenbook/frames/frame27.shtml
- 78.Donnelly-Roberts DL, Namovic MT, Han P, Jarvis MF. Mammalian P2X7 receptor pharmacology: comparison of recombinant mouse, rat and human P2X7 receptors. Br J Pharmacol. 2009;157:1203–1214. doi: 10.1111/j.1476-5381.2009.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young MT, Pelegrin P, Surprenant A. Identification of Thr283 as a key determinant of P2X7 receptor function. Br J Pharmacol. 2006;149:261–268. doi: 10.1038/sj.bjp.0706880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chessell IP, Simon J, Hibell AD, Michel AD, Barnard EA, Humphrey PP. Cloning and functional characterisation of the mouse P2X7 receptor. FEBS Lett. 1998;439:26–30. doi: 10.1016/s0014-5793(98)01332-5. [DOI] [PubMed] [Google Scholar]
- 81.Beamer E, Kuchukulla M, Boison D, Engel T. ATP and adenosine-two players in the control of seizures and epilepsy development. Prog Neurobiol. 2021;204:102105. doi: 10.1016/j.pneurobio.2021.102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinto S, Cunha C, Barbosa M, Vaz AR, Brites D. Exosomes from NSC-34 Cells Transfected with hSOD1-G93A Are Enriched in miR-124 and Drive Alterations in Microglia Phenotype. Front Neurosci. 2017;11:273. doi: 10.3389/fnins.2017.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malada Edelstein YF, Solomonov Y, Hadad N, Alfahel L, Israelson A, Levy R. Early upregulation of cytosolic phospholipase A2α in motor neurons is induced by misfolded SOD1 in a mouse model of amyotrophic lateral sclerosis. J Neuroinflammation. 2021;18:274. doi: 10.1186/s12974-021-02326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or supplementary material. Data not shown is available from the corresponding authors, upon reasonable request.
Not applicable.