Abstract
Epithelial–mesenchymal transition (EMT) and its reverse mesenchymal–epithelial transition (MET) are critical during embryonic development, wound healing and cancer metastasis. While phenotypic changes during short-term EMT induction are reversible, long-term EMT induction has been often associated with irreversibility. Here, we show that phenotypic changes seen in MCF10A cells upon long-term EMT induction by TGFβ need not be irreversible, but have relatively longer time scales of reversibility than those seen in short-term induction. Next, using a phenomenological mathematical model to account for the chromatin-mediated epigenetic silencing of the miR-200 family by ZEB family, we highlight how the epigenetic memory gained during long-term EMT induction can slow the recovery to the epithelial state post-TGFβ withdrawal. Our results suggest that epigenetic modifiers can govern the extent and time scale of EMT reversibility and advise caution against labelling phenotypic changes seen in long-term EMT induction as ‘irreversible’.
Keywords: epithelial–mesenchymal plasticity, mathematical modelling, chromatin-mediated epigenetic regulation, mesenchymal–epithelial transition (MET), epigenetic memory
1. Introduction
Cellular plasticity—the ability of cells to reversibly alter their phenotypes—is a hallmark of cancer metastasis [1]. One of the most well-investigated axes of cellular plasticity is epithelials–mesenchymal plasticity (EMP) which involves reversible transitions between epithelial (E), mesenchymal (M) and hybrid E/M phenotypes. Aside from metastasis, EMP is also implicated in embryonic development, wound healing, tissue repair and fibrosis [2]. Initially thought of as a binary process, EMP is now understood to encompass many hybrid E/M states and incorporate reversible spontaneous or induced switching among these multiple phenotypes [3]. Studying the dynamics of EMP in cancer cell lines (patient or animal derived) have revealed a phenotypically heterogeneous distribution of E–M states. For instance, the PMC42 breast cancer cells consisted of 80% EpCAMhigh (epithelial) cells and 20% EpCAMlow (mesenchymal) cells. When purified and cultured independently, these two subpopulations recapitulated the phenotypic steady-state distribution of the parental population (80% EpCAMhigh, 20% EpCAMlow) [4]. Similar spontaneous cell-state transitions have been witnessed in other cancer subtypes as well, demonstrating an inherent plasticity [5,6]. The proportion of cells present in E, M and hybrid E/M states in a population at a given time can be regulated by external conditions, including changes in the concentrations of growth factors in culture media and the presence of cytotoxic/cytostatic drugs [6–8]. Further, cell–cell communication through juxtacrine, autocrine and paracrine signalling can also influence cell-state switching and modulate phenotypic heterogeneity [9–11]. All these observations suggest dynamic interconversion among the E, M and hybrid E/M phenotypes.
At an intracellular level, EMP is enabled by a complex interplay of diverse molecules and signalling pathways involved in feedback loops. For instance, epithelial–mesenchymal transition (EMT) is promoted by families of transcription factors such as ZEB, encompassing ZEB1 and ZEB2, the reverse mesenchymal–epithelial transition (MET) is driven by microRNA families such as miR-200, encompassing miRs-200a, -200b, -200c, -141 and -429. ZEB and miR-200 engage in mutually inhibitory feedback loop, wherein the miR-200 family post-transcriptionally inhibits ZEB expression (thus promoting the epithelial state), whereas ZEB represses the miR-200 family transcriptionally [12–15]. Hysteresis, a general term describing the dependence of the state of a system on its history, leads to the lack of recovery to the same state under the same external conditions for a fixed observation time. It has been a useful concept for quantifying the EMT response [16–18]. The phenomenon of hysteresis has often been associated with irreversible EMT, i.e. once cells are induced to attain a M state, it is difficult for them to regain an E state even if the external inducer (e.g. TGFβ) is removed from the culture media. However, recent in vivo and in vitro studies report that not every EMT induction is irreversible. In vivo, during cancer metastasis, reversible EMT has been observed where disseminated cancer cells often regain the epithelial traits of cell–cell adhesion and rapid cell proliferation during metastatic colonization [19,20]. Importantly, the loss of EMT factors such as PRRX1 has been shown to be essential for BT549 cells to form metastases upon extravasation after tail vein injection in immunocompromised mice, thus highlighting the role of reversibility or plasticity in enabling metastasis [20]. Similarly, while epithelial cells can attain a M state within a short-term exposure to EMT inducers in vitro, it is only prolonged exposure to these signals that enables a subpopulation of cells to exhibit seemingly ‘effectively irreversible’ change or a ‘stabilized EMT’ state [21,22]. The extent of EMT reversibility can vary depending on genetic background of the cell, the specific induction factor and/or dose. For instance, long-term treatment of MCF10A cells with TGFβ induced chromatin accessibility changes among genes associated with EMT, apico-basal polarity and stemness. However, most cells also regained certain epithelial traits such as E-cad localization, morphology, and loss in migratory ability in the experimental time window [23]. Thus, further investigation is needed to better understand the dynamics of reversible EMT and MET in vitro and in vivo.
Epigenetic modifications such as gain and loss of DNA methylation and histone-level methylation and acetylation at various promoter and enhancer regions can underlie such effectively ‘irreversible’ changes. When immortalized HMEC cells expressing oncogenic Ras were cultured in 10% serum for multiple passages, the promoter region of CDH1 became increasingly methylated at the DNA level [24]. Similarly, in MDCK cells, autocrine TGFβ signalling maintained promoter methylation of MIR200C, contributing to a stabilized M state [25]. Inhibiting this autocrine signalling reduced the MIR200C promoter methylation levels and thus allowed MET. Consistently, overexpression of miR-200 together with the knockdown of chromatin remodelling protein BRG1 was required to induce a MET in RD sarcoma cells [26]. Together, these reports suggest that chromatin reprogramming can control the reversible dynamics of EMT/MET.
Aside from chromatin-mediated epigenetic aspects, distinct modules of genes whose expression patterns are altered during EMT induction can revert to their pre-treatment levels at varying rates upon withdrawal of the EMT-inducing signal; this has been explicitly seen in prostate cancer cells [27]. Differences in the rate of return to pre-EMT induction expression levels can lead to distinct expression patterns during EMT and MET which can be resolved at the single-cell level. [18,28]. Such variations in expression recovery rates were also observed during the drug holidays after transient 24 h VINC exposure to HL60 leukemia cells and may have implications in rapid switching to a drug-resistant state when cells were re-exposed to drugs [29]. However, it remains unclear whether the observed recovery time differences among genes to their basal expression levels are due to differences in half-lives of mRNAs and/or proteins, slowly evolving and/or accumulating epigenetic modifications, or a combination of these effects.
Here, using joint experiment and mathematical modelling approaches, we show that the time to recover an E state at the population level post-EMT induction depends on induction duration. We used TGFβ to induce EMT in MCF10A cells for two different durations (13 days, 22 days) and measured, at the population level, both gain in epithelial gene products (miR-200b, miR-200c and E-cad) expression and loss of mesenchymal gene (ZEB) expression, up to 18 days post-TGFβ withdrawal (for 13 days induction) and up to 45 days post-withdrawal (for 22 days induction). We hypothesized that the epigenetic repression of miR-200 by ZEB, and consequent accumulation of ‘epigenetic memory’ can prevent MET, thus explaining the time-scale differences in reversibility as seen experimentally for short versus long treatment with TGFβ. We adopted our earlier mathematical modelling formalism describing the chromatin-mediated epigenetic repression of miR-200 by ZEB in a phenomenological manner, i.e. the model accounts for chromatin-mediated epigenetic regulation without delving into molecular-level details of histone modification/DNA methylation [21]. The model assumptions are based on the notion that the long-term EMT-induced changes are indeed of a different character than the those caused by the standard parts of the genetic network, and we speculate that differences are caused by chromatin modifications [30,31]. Our model predicts that while prolonged treatment with an EMT inducer can lead to a slower MET due to differences in signalling activation levels, the accumulation of epigenetic memory appears to be the major determinant of the difference in reversal time scales between short- and long-term EMT induction. Thus, altering the rate of accumulation and/or decay of epigenetic memory, through treatment with various epigenetic modifiers, can govern the extent of reversibility of EMT. Further, our stochastic simulations demonstrate population heterogeneity at a single-cell level by quantifying the time taken to revert to an epithelial state post-withdrawal of the EMT-inducing signal. Overall, our analysis highlights how the time scale of EMT reversibility may depend on the duration of EMT induction and consequent epigenetic changes, and advises caution against mislabelling changes witnessed during long-term EMT induction as ‘irreversible’.
2. Results
2.1. TGFβ-treated MCF10A cells for extended durations require longer withdrawal time to revert to an epithelial state
Earlier experiments in MCF10A cells suggest an effectively ‘irreversible’ switch to a mesenchymal state, by treatment with TGFβ for up to 15 days and withdrawal for 15 days post-treatment [21]. To interrogate the dependence of EMT reversibility on TGFβ induction duration as well as that of withdrawal, we considered two different time scales: (i) short term, i.e. exposure of MCF10A cells to TGFβ for 13 days and under observation for up to 18 days post-withdrawal, and (ii) long term, i.e. exposure of MCF10A cells to TGFβ for 22 days and under observation for up to 45 days post-withdrawal (figure 1a). We found that for the short-term treatment of 13 days, the expression level of epithelial genes (CDH1, miR-200b, miR-200c) and that of mesenchymal marker ZEB1 returned to their pre-treatment levels within roughly 18 days of withdrawal (figure 1b,c). However, the time taken to return to the pre-treatment level was much higher when the EMT induction consisted of 22 days of TGFβ exposure. With this protocol, it took almost 45 days after withdrawal to revert to pre-treatment levels (figure 1b,c). Together, these observations clearly suggested that the time scale of MET—observed through canonical markers measured at the bulk level—depended strongly on the time period of induction, and that MET need not be completely ‘irreversible’ as proposed earlier, even after chronic treatment.
Figure 1.
Time taken to regain basal expression of epithelial and mesenchymal marker (at population level) after short- (13 days) and long-term (22 days) EMT induction. (a) Experimental strategy—MCF10A cells were treated for 13 and 22 days with 5 ng ml−1 TGFβ. After removal of TGFβ, cells were cultured until basal expression levels of epithelial and mesenchymal genes, prior to EMT, were regained. (b,c) Reversal of (b) ZEB1, (c(i)) E-cad, (c(ii)) miR200c, and (c(iii)) miR200c to their basal expression levels (relative to vehicle control) after short- and long-term EMT induction. Data shown are averaged over three independent replicates. For 13 days EMT induction, E-cad and ZEB1 transcripts were quantified at 0, 6, 18 days post-withdrawal and miR-200b-c transcripts were quantified at 0, 18 days post-withdrawal. For 22 days EMT induction, all transcripts above were quantified at 0, 18, 27, 36, 45 days post-withdrawal.
Next, we observed a marked difference in the temporal trajectories defining the recovery of expression levels for CDH1 and ZEB1 mRNAs and miR-200 under 22 days of TGFβ treatment. Here, CDH1 levels did not increase for the first 18 days post-withdrawal, showing an initial silenced phase, but then exhibited a sigmoidal increase trend over the next 27 days, before ultimately saturating (figure 1c(i)). Similar patterns showing an initial lag period were observed for miR-200b and miR-200c levels as well (figure 1c(ii,iii)). ZEB1 levels, on the other hand, followed near-linear decrease for the first 27 days and later plateaued (figure 1b). Together, these trends highlight that different genes may exhibit varying dynamics of recovery, reminiscent of similar observations in LNCaP cells [27].
We further performed gene expression analysis of a specific set of genes associated with EMT, using the NanoString technology. Principal component analysis (PCA) confirmed that cells induced for 13 days showed large variability in gene expression levels during withdrawal periods compared with 22-day induced cells (compare ranges of PC1 values for 13 days (short-term, ST), and 22 days (long-term, LT) cases in electronic supplementary material, figure S1A, B). Upon clustering the genes based on similarities in expression profiles during 18 days of withdrawal, we found three major groups: the first one consists of mesenchymal genes (e.g. VIM, ZEB1; red bar in electronic supplementary material, figure S1C) whose expression decreases during the withdrawal period; the second one consists of epithelial genes whose expression levels increase after 18 days of withdrawal for short-term (13 days) induction but not for 22 days induction (e.g. CDH1, CD24, JAG1; purple bar in electronic supplementary material, figure S1C); and the third one with invariable expression during the withdrawal period after short-term treatment, but highly expressed at 18th day of withdrawal after long-term treatment (cyan bar in electronic supplementary material, figure S1C). These observations suggest that the recovery of epithelial genes is often delayed with an increasing duration of EMT induction, thereby possibly rendering the cell in a hybrid phenotype. Further, during recovery of the epithelial cell-state post inducer treatment, the long-term treated cells take a different trajectory than short-term treated cells by expressing an additional set of genes (cyan bar electronic supplementary material, figure S1C). This difference in trajectory indicates the presence of hysteresis phenomena in long-term treated cells.
2.2. Mathematical model of epigenetic regulation in epithelial–mesenchymal transition
The above-mentioned recovery dynamics of E-cadherin, miR-200b and miR-200c expression levels suggest a transient locking or stabilization of the mesenchymal state observed post-withdrawal (figure 1c). These observations are reminiscent of the durable silencing of miR-200b and miR-200c expression observed in MDCK cells, post TGFβ withdrawal, due to methylated promoter regions [25]. The extent of promoter methylation was shown to increase with the stabilization of mesenchymal state, caused either by the extended duration of TGFβ exposure or prolonged activation of self-sustaining autocrine loops such as those mediated between TGFβ signalling and ZEB1. Therefore, to better understand the role of epigenetic changes in mediating the recovery dynamics upon TGFβ exposure and withdrawal, we adapted our mechanism-based mathematical model of the EMT network [3] to also incorporate epigenetic regulation. In our modelling framework, we consider that upon extended durations of treatment, ZEB1 can elicit methylation of the miR-200 promoter region [32].
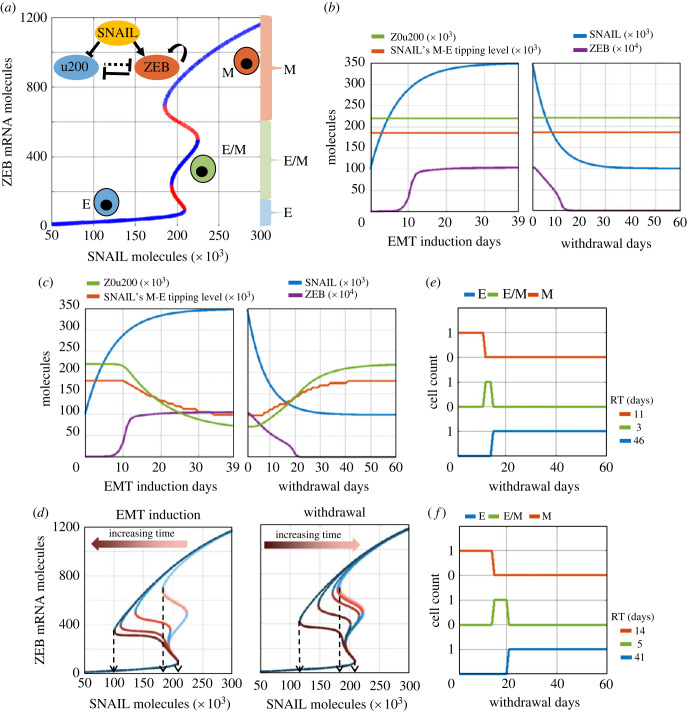
The regulatory network considered in our model includes interactions among miR-200, ZEB and SNAIL at transcriptional and translational levels (figure 2a, inset) [3]. Both SNAIL and ZEB are zinc finger proteins which suppress the expression of epithelial genes such as CDH1 (E-cadherin) and miR-200 by binding to their E-box regions. On the other hand, the miR-200 family binds to the mRNA of ZEB, preventing its translation and consequent EMT. Thus, ZEB and miR-200 inhibit each other [12,13]. SNAIL activates ZEB and inhibits miR-200 indirectly. In this regulatory network, SNAIL represents the cumulative effects of the TGFβ pathway regulating of EMT [16]. In the absence of any epigenetic regulation, the emergent dynamics of interactions among SNAIL, miR-200 and ZEB can allow for (co)existence of multiple cell-states (phenotypes): mesenchymal (M; low miR-200, high ZEB), epithelial (E; high miR-200, low ZEB) and hybrid E/M (medium miR-200, medium ZEB) (figure 2a; blue curves). In this bifurcation diagram, approximately, ZEB mRNA > 600 molecules correspond to a M state, ZEB mRNA < 150 molecules denote an E state, and ZEB mRNA between 150 and 600 molecules show a hybrid E/M state.
Figure 2.
A phenomenological mathematical model to capture epigenetic regulation during EMT. (a) (Inset) Regulatory network incorporating mutual inhibition between epithelial (mir200–u200) and mesenchymal (ZEB) players, with SNAIL as external input. Bifurcation diagram shows equilibrium levels of ZEB mRNA based on SNAIL levels, as resulting from dynamics of the regulatory network. Blue curves are stable equilibria while red ones are unstable. Three distinct coloured braces on the right (red, green and blue) qualitatively represents the ZEB mRNA levels used to assign epithelial (E), mesenchymal (M) or hybrid E/M phenotype. (b) Dynamics of ZEB levels with changing SNAIL levels during EMT induction (left) and withdrawal (right) without considering epigenetic regulation of miR-200 by ZEB. (c) The same as (b) but with incorporating epigenetic regulation. Z0u200 levels and SNAIL M-E tipping levels reflect the extent of epigenetic reprogramming at any time instant. (d) Bifurcation diagrams for ZEB mRNA levels for varying strengths of epigenetic regulation (Z0u200 levels) during EMT induction (left) or withdrawal (right) of EMT-inducing signal, at an interval of 10 days each (0, 10, 20, 30, 39 days post-induction, and 10, 20, 30, 40, 50, 60 days post-withdrawal). Black arrows highlight the SNAIL levels corresponding to E-to-M tipping (right arrow) and M-to-E tipping (left and middle arrows). (e) Changes in cell's phenotype during withdrawal period following EMT induction without epigenetic regulation. (f) Same as (e) but with the impact of epigenetic regulation. RT (residence time) measures the time that the cell spends in each phenotype during withdrawal period of 60 days. Parameters used in (b) and (e): S01 = 100k molecules, S02 = 350k molecules, α = 0, βfor = 1 h, and βrev = 1 h. When considering epigenetic regulation (panels c and f), α = 0.15, βfor = 240 h, and βrev = 240 h; all other parameters as above-mentioned.
First, we investigate the dynamics of this network in absence of any epigenetic influence of ZEB on the miR-200 family promoters. In our simulation framework, the dynamics of SNAIL in a cell is modelled as a deterministic variable which tends to approach a saturation value (equation (4.4) in Material and methods, with zero noise amplitude), and we modulate the SNAIL saturating values to mimic EMT and MET induction in our model. Prior to EMT induction, a cell exhibits SNAIL levels corresponding to an E phenotype (approx. 100k molecules (figure 2a)). During EMT induction, the cellular SNAIL level increases and eventually saturates at a much higher value (approx. 350k molecules), with a corresponding increase in ZEB levels (figure 2b, left), and acquisition of a mesenchymal state (figure 2a). Upon withdrawal of the EMT-inducing signal, the levels of SNAIL and ZEB gradually return to their initial values, thus reflecting MET (figure 2b, right). Thus, in absence of any epigenetic regulation, our model could recapitulate the reversible EMT/MET dynamics.
Next, we examine how incorporating the epigenetic influence on miR-200 mediated by ZEB1 can alter the dynamics of EMT/MET. Experimental data, including ours (figure 1), suggests that the longer a cell stays in the M state, the slower will be its reversibility dynamics following induction withdrawal [24,25]. To obtain these dynamics, we assumed the threshold of ZEB levels needed to suppress miR-200 in the corresponding Hills function (Z0u200) to be a time-dependent function of ZEB levels (Material and methods, equation (4.1)) [21]. Thus, a higher saturating level of ZEB during EMT will continue to decrease the levels of Z0u200, enabling lower levels of ZEB to repress miR-200, and thereby incorporating the impact of epigenetic changes caused by ZEB (figure 2c, left panel green curve versus figure 2b, left panel green curve). The dynamics of SNAIL and ZEB, however, remain unchanged during induction, as expected (compare corresponding blue and violet curves in figure 2c, left panel versus that in figure 2b, left panel).
We further calculated how the temporally varying levels of Z0u200 during EMT induction and withdrawal altered the bifurcation diagram for the EMT network. As the levels of Z0u200 decreased during EMT induction, we saw no observable change in the tipping point levels of SNAIL required for cells to switch from the E to M state, but noticed a complementary decrease in the tipping point for an M to E state switch (dashed arrows in figure 2d). In other words, the epigenetic influence mediated by ZEB can reshape the phenotypic stability landscape such that it becomes more difficult for cells to revert to an epithelial state post-withdrawal. Such changes are not seen in the scenario where epigenetic changes are absent (compare orange curve in figure 2c, left panel versus that in figure 2b, left panel). The longer the EMT induction period, the lower the Z0u200 levels; this trend can explain why short-term EMT induction is expected to have much weaker epigenetic impact as compared with long-term induction (figure 2d). As the EMT-inducing signal is withdrawn, such accumulated epigenetic changes decay slowly, thus leading to recovery of Z0u200 molecules and SNAIL's M to E tipping point to pre-induction levels (figure 2c,d, right). This change ends the lag period in the recovery of levels of EMT/MET regulators (compare orange and green curves in figure 2c, right panel versus that in figure 2b, right panel).
Finally, for comparative analysis, we calculated the recovery time for cells induced to undergo EMT with epigenetic changes versus those without any such changes. We quantified the number of days for which ZEB levels are in the above-mentioned numerical ranges corresponding to the E, M and hybrid E/M states (figure 2a). For the case without any epigenetic changes, the cells revert to an E state 14 days (= 11 days in the M state, followed by 3 days in the hybrid E/M state) post-withdrawal (figure 2e). However, when incorporating epigenetic influence, the cells stay in M and hybrid E/M states longer and return to an E state after 19 days (= 14 days in the M state, followed by 5 days in the hybrid E/M state), thus causing delayed recovery, or in other words, a slower MET (figure 2f). The slower the decay of ‘epigenetic memory’ [33,34] thus accumulated, the higher the delay in cells reverting to an epithelial state post-withdrawal of EMT-inducing signals.
2.3. Longer EMT induction can delay the reversal to an epithelial state post-withdrawal of an EMT-inducing signal
Next, we interrogated how the duration of EMT induction can influence the build-up of ‘epigenetic memory’ and consequently the time scales of reversal to an epithelial state post-withdrawal of the EMT-inducing signal. We consider the response of a cell that is switching between two different values of SNAIL: S01 (prior to EMT induction) and S02 (post EMT-induction; thus S02 > S01). The time scale of epigenetic changes during induction or withdrawal is denoted by the rate of change of Z0u200 levels, given by βfor and βrev respectively. The higher the value of βfor, the slower the reduction in Z0u200 levels and thus the slower the build-up of epigenetic memory. The higher the value of βrev, the slower the return to pre-induction Z0u200 levels and thus the slower the decay of epigenetic memory. We simulated the dynamics of a cell exposed to short-term (13 days) and long-term (39 days) duration of EMT induction, and quantified the reversal time in these two scenarios.
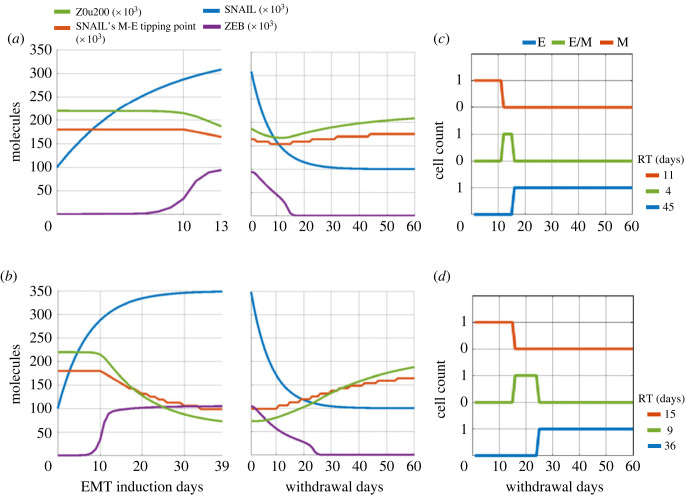
For both short-term and long-term induction cases, EMT was induced by changing SNAIL levels from S01 = 100k molecules to S02 = 350k molecules, with epigenetic regulation time scales taken as βfor = 240 h, and βrev = 720 h. Although ZEB levels increased and then saturated around 15 days of induction, a longer-term induction led to lower Z0u200 levels as compared with short-term induction. Thus, a longer induction conferred a relatively stronger epigenetic memory, as denoted by changes in bifurcation diagrams (electronic supplementary material, figure S2) and thus in tipping points levels of SNAIL required for cells to switch from M to E state (figure 3a versus b; left panels). Consequently, upon withdrawal of the EMT-inducing signal (i.e. reducing SNAIL levels from S02 = 350k molecules to S01 = 100k molecules), the time taken to recover the levels of ZEB and Z0u200 to pre-induction values is slower for long-term induction as compared with short-term induction (figure 3a versus b; right panels).
Figure 3.
Influence of EMT induction time on recovery to an epithelial change post-withdrawal. Simulations for EMT induction for (a) 13 days (short-term), and (b) 39 days (long-term) followed by a withdrawal period of 60 days. Dynamics of ZEB, Z0u200 and the SNAIL levels corresponding to M-to-E tipping point are shown. (c,d) Change in cell's phenotype during withdrawal period after EMT induction for 13 days (a) and 39 days (b). RT (residence time) measures the time that the cell spends in each phenotype during withdrawal period of 60 days. Parameters used in panels a–d: α = 0.15, S01 = 100k molecules, S02 = 350k molecules, βfor = 240 h, and βrev = 720 h.
A quantitative comparative analysis showed the difference in recovery time for cells induced to undergo EMT for different durations. For short-term induction, the cells revert to an E state 15 days (= 11 days in the M state, followed by 4 days in the hybrid E/M state) days post-withdrawal (figure 3c). However, for long-term induction, this return happens after 24 days (= 15 days in the M state, followed by 9 days in the hybrid E/M one), possibly prolonging the residence of cells in hybrid E/M phenotype(s) (figure 3d). These differences are much less prominent if the epigenetic feedback is not considered (electronic supplementary material, figure S3), thus showcasing the impact of a long-term EMT induction on epigenetic-level reprogramming, and increased residence time in M and E/M states.
To assess whether similar results hold for dynamic alterations in other parameters and not just the Hill function threshold (Z0u200), we varied other factors involved in miR200 regulation by ZEB to account for possible epigenetic changes during EMT. First, we made the fold change of miR-200 repression by ZEB into a dynamic variable and a function of ZEB levels (Material and methods, equation (4.2)). This framework provided us with another way to modulate the strength of miR-200 suppression with the increasing duration of EMT induction. However, the fold change () variation did not alter the stability of the M state significantly even after long-term EMT induction, and thus, we observed no difference in epithelial state return times between short- and long-term induction (electronic supplementary material, figures S4 and S5). Next, we considered that increasing the duration of EMT induction could modulate the basal synthesis rate of miR-200 production (Material and methods, equation (4.3)). In this case, the higher the EMT induction time period, the more prominent the change in the bifurcation diagram, thus influencing the SNAIL levels for both M–E and E–M transitions. Hence, we did observe differences in the return time to an epithelial state for an increasing duration of induction (electronic supplementary material, figures S6 and S7), similar to the observations for dynamic Z0u200 levels. Overall, more than one parameter modulation (here, Z0u200 and gμ200) can reflect the impact of epigenetic changes on EMT induction.
2.4. The rate of epigenetic changes during EMT induction and withdrawal determines the time taken to regain an epithelial cell state
The rate and extent of epigenetic reprogramming can depend on multiple factors, such as whether epigenetic changes are mediated via DNA methylation or by histone modification [33]. For instance, GRHL2, a canonical MET inducer, is a pioneering transcription factor capable of directly binding to condensed chromatin to initiate its opening, leading to cell-state changes [35,36]. Such diverse modes of epigenetic regulation can alter the rate of accumulation (βfor) and decay (βrev) of epigenetic memory.
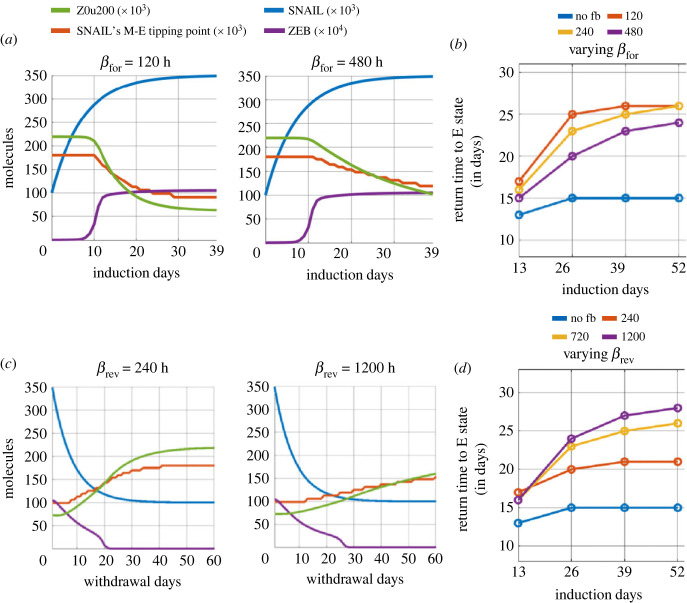
We modulated the values of βfor and βrev to assess their impact on epigenetic reprogramming and the time scales of cell-state transitions. First, we varied βfor (2x = doubled, 0.5x = halved) while maintaining βrev = 720 h. Reduced βfor values (= 120 h) enhanced the rate of decrease of Z0u200 levels (electronic supplementary material, figure S8A), leading to a lower SNAIL's M to E tipping levels at the end of induction period, as compared with the case for larger βfor values (= 480 h) (figure 4a). Higher βfor values result in lower accumulated epigenetic memory and thus a faster reversal to an epithelial state post-withdrawal, irrespective of the induction period (figure 4b, electronic supplementary material, figure S8B). However, the epigenetic memory also saturates to a maximal level, as evident for smaller βfor values (figure 4b). Second, we varied βrev values while maintaining βfor = 240 h. Larger βrev values (= 1200 h) led to a slower recovery from the acquired epigenetic memory accumulated during induction (electronic supplementary material, figure S9A), compared with smaller βrev values (= 240 hrs) (figure 4c), as witnessed by a difference in the slopes of curves of SNAIL's M–E tipping level. The higher the value of βrev, the slower the decay of the accumulated epigenetic memory and thus the longer the delay in reversal to an epithelial state post-withdrawal, irrespective of the induction period (figure 4d, electronic supplementary material, figure S9B).
Figure 4.
Effect of varying rates of acquisition and decay of epigenetic memory. (a) Dynamics of ZEB, and changes in epigenetic regulation measures (Z0u200 and SNAIL levels corresponding to M–E tipping point) during EMT induction with variations in time constant βfor (acquisition rate of epigenetic changes, in units of h). (b) Time to recover to an E state for varied values of βfor and duration of EMT induction. (c) Same as (a) but for varied values of βrev (time scale of decay of epigenetic changes in units of h). (d) Same as (b) but for varied values of βrev and duration of EMT induction. Parameter values, unless specified differently, are: α = 0.15, S01 = 100k molecules, S02 = 350k molecules, βfor = 240 h, and βrev = 720 h, For cases without epigenetic regulation (no fb – no feedback), α = 0.
Again, while considering the fold change () dynamics during EMT induction, variation of both βfor and βrev did not have significant effects on the return time to an epithelial state (observe epithelial return time for a given induction duration with varying βfor and βrev in electronic supplementary material, figure S10A, B). However, significant delays were observed in epithelial return times for smaller βfor and larger βrev while considering dynamic changes in the basal synthesis rate of miR-200 (gμ200) during EMT induction (observe epithelial return time for a given induction duration with varying βfor and βrev in electronic supplementary material, figure S11A, B). Together, these observations suggest that slower response at epigenetic regulation level, either during EMT induction or withdrawal, introduces a latency period for a cell to gain or lose epigenetic memory, thus impacting the rates of cell-state switching.
2.5. Recovery time scales also depend on pre- and post-induction SNAIL levels
The extent of EMT/MET induction in a given cell can depend on multiple factors. These include the dose and duration of the inducing signal, pathways activated by the specific EMT/MET-inducing signal, and variations in the initial cell-state in terms of protein abundance or epigenetic status [26,28]. To represent the impact of these factors, we examined the effects of varying the pre-induction (S01) and post-induction (S02) SNAIL levels on the time taken to recover to epithelial state post-withdrawal. We consider these scenarios both in the presence and absence of epigenetic feedback or memory.
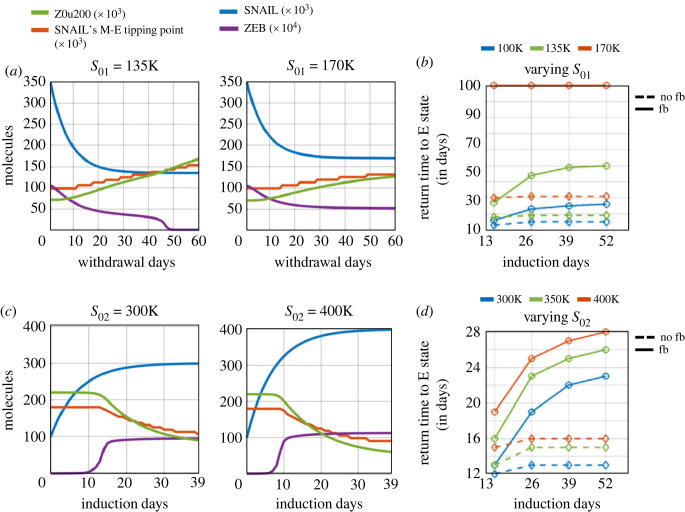
In the absence of epigenetic regulation of miR-200 by ZEB1, we first examined the impact of varying S01. The lower the levels of S01, the faster the recovery dynamics. For S01 = 100k molecules, for 39 days of EMT induction, it takes 14 days (11 days in the mesenchymal state followed by 3 days in the hybrid E/M state) to regain an epithelial phenotype (figure 2b,e). However, at S01 = 135k molecules, the recovery period extends to 18 days, and at S01 = 170k molecules, it extends to 30 days (electronic supplementary material, figure S12). These slower dynamics of SNAIL can be attributed to the difference between post-induction values of SNAIL (S02) and the post-withdrawal values (S01) (equation (4.2)). However, on varying S02, the impact on recovery times is rather small, which can be explained by higher equilibrium levels of SNAIL achieved at the end of the EMT induction period. For S02 = 300k molecules, it takes 12 days to revert to an epithelial state but for S02 = 400k molecules, it increases to 15 days (electronic supplementary material, figure S13). Thus, varying S01 levels had a stronger impact on the time scales of recovery to an epithelial state, than variations in S02 levels.
Next, we incorporated the epigenetic regulation of miR200 by ZEB and considered the case of a high S01 value (= 135k molecules). The build-up of epigenetic memory, as seen before, delayed the time taken for SNAIL's M-E tipping level to increase above the cellular SNAIL levels, and to attain pre-induction values during the period of withdrawal (figure 5a, electronic supplementary material, figure S14A). Consequently, the cell spent more time in M and E/M states and showed delayed recovery dynamics to an epithelial state. At even higher values of S01 (= 170k molecules), the recovery slows further, eventually tending toward the scenario of irreversible EMT (figure 5b, electronic supplementary material, figure S14B).
Figure 5.
Effects of pre-induction (post-withdrawal, S01) and post-induction (S02) SNAIL levels on epithelial recovery time following EMT induction. (a) Dynamics of ZEB, and changes in epigenetic regulation measures—Z0u200 and SNAIL's M–E tipping levels—during withdrawal period with variation in saturating basal SNAIL levels ‘S01’ with epigenetic changes during EMT. EMT induction duration was for 39 days. (b) Time to recover epithelial state with variations in both S01 and EMT induction duration with epigenetic regulation (‘fb’, solid lines) and without epigenetic regulation (‘no fb’, dashed lines). (c) Same as (a) but for varied saturating EMT-induced SNAIL levels ‘S02’ with epigenetic changes during EMT. (d) Same as (c) but for varied S02 and EMT induction duration with epigenetic regulation (solid lines) and without it (dashed lines). Parameter values used, unless otherwise specified, are: S01 = 100k molecules, S02 = 350k molecules, βfor = 240 h, βrev = 720 h, and α = 0.15.
Lastly, we varied the post-induction SNAIL levels (S02) while considering epigenetic changes. With increasing S02, we observed higher steady levels of ZEB mRNA and protein. Also, increasing S02 accelerated EMT, thus stabilizing ZEB in a high state for longer times during the induction period (figure 5c). This prolonged time in a mesenchymal state during induction increased the gap between the tipping point levels of SNAIL for MET and EMT (electronic supplementary material, figure S15), thus acting as a barrier for MET. As expected, the higher the induction time, the stronger the extent of epigenetic memory accumulated, and thus the longer the time required to revert EMT (figure 5d).
We previously observed that varying levels of gμ200 during EMT induction dramatically alters the SNAIL levels corresponding to the M–E and E–M tipping points, which is similar to the response seen with dynamic Z0u200 levels (compare electronic supplementary material, figure S6 with electronic supplementary material, figure S2). Thus, we also expected to observe an increasing return time to the epithelial state with increasing S01 and S02 levels for dynamic gμ200 levels, as was observed with dynamic Z0u200 (observe the epithelial return time for a given induction duration with varying S01 and S02 in electronic supplementary material, figure S11C, D). However, with the dynamic fold change (), increasing S01 values but not S02 values caused greater delay in epithelial state recovery (observe epithelial return time for a given induction duration with varying S01 and S02 in electronic supplementary material, figure S10C, D). This can be explained by observing the slight increase in M state stability caused by dynamic during EMT induction by lowering SNAIL's M–E tipping point (electronic supplementary material, figure S4). Now, as the cell's SNAIL levels settle back to the S01 value during withdrawal, until the time that dynamic SNAIL's M–E tipping point is less than S01, the cell retains its M state.
2.6. Population-level effects of epigenetic changes during EMT
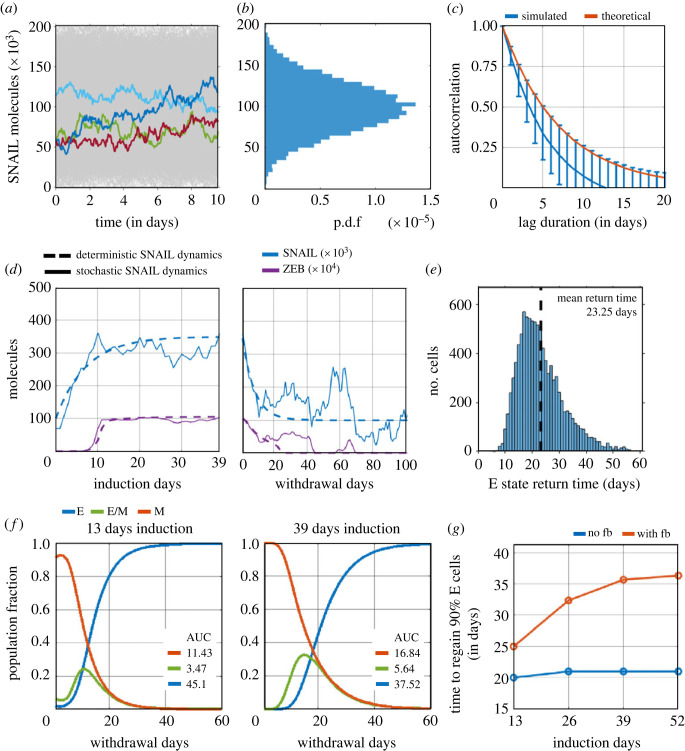
So far, we have examined the influence of epigenetic changes on reversibility towards an epithelial state, through simulating individual cells switching between two discrete levels of SNAIL (S01, S02). These simulations did not consider any stochastic fluctuations in protein levels. However, fluctuations in protein abundance can prevail at the single-cell level [37] due to factors such as stochastic gene expression [38,39] and asymmetry in cell division [40]. To incorporate these factors, we model stochastic fluctuations around the mean level using the Ornstein–Uhlenbeck (OU) process. The OU process determines SNAIL levels by integrating a stochastic differential equation with both deterministic (drift) and stochastic (diffusion) terms (equation (4.2)). Numerical implementation of the OU process provides SNAIL trajectories reflecting distinct cells that are statistically independent within the population (figure 6a). The stochastic fluctuations in SNAIL levels were parameterized based on experimentally estimated values of the coefficient of variation (CV) of distributions of protein levels in a cellular population, and the 50% decorrelation time of single-cell expression levels (figure 6b,c) [37,41].
Figure 6.
Effects of epigenetic changes during EMT on its reversibility in a heterogeneous population. (a) Stochastic fluctuation in SNAIL levels around the population mean (S0 = 100k molecules). Temporal fluctuations in SNAIL levels derived from the total population of 10 000 cells are shown (in grey), of which four stochastic trajectories are coloured. (b) The distribution of aggregate SNAIL levels at any realized time follows a Gaussian distribution; p.d.f. refers to probability density function. (c) Loss of correlation in SNAIL levels of a cell between any two time instants with increasing time separation. Red curve shows theoretical estimation of 50% decorrelation time as 5 days. Blue curve shows mean and standard deviation of decreasing autocorrelation of 10 000 independent fluctuating SNAIL time-series, each being 100 days long. (d) Changes in SNAIL and ZEB levels during EMT induction and withdrawal period for a cell in the population, with and without stochastic fluctuations in SNAIL, shown by solid and dashed curves, respectively. (e) Distribution of return time of cells to epithelial state after undergoing EMT induction for 39 days. Mean return time of population = 23.25 days (dashed line). (f) Change in phenotypic distribution of population after undergoing EMT induction for 13 days and 39 days. ‘AUC’ corresponds to area under the curve and reflects the cumulative fractional share of a phenotype in the population during the withdrawal period of 60 days. The results presented include the mean of three independent simulation runs of 10k cells each. (g) Time taken to regain 90% of epithelial phenotype share in the population for increasing durations of EMT induction. Analysis was performed for scenarios with and without considering epigenetic changes during EMT, denoted by ‘with fb’ and ‘no fb’ respectively. Plots (a–c) were generated from analysing SNAIL time-series data. Time-series was stimulated using a stochastic differential equation (equation (4.4)). Plots (d–g) were obtained using these parameters for simulations: α = 0.15, S01 = 100k molecules, S02 = 350k molecules, βfor = 240 h, βrev = 720 h, for every cell in population (in all simulated cases, population size = 10 000 cells).
The stochastic simulations revealed heterogeneity in the dynamics of SNAIL and ZEB among individual cells within a population during EMT induction, and in the time taken to revert to an epithelial state upon withdrawal (figure 6d,e; electronic supplementary material, figure S16, S17). Despite this heterogeneity, the mean return time (RT) to an epithelial state for a cell in the population remained close to our earlier observations from the deterministic analysis for a 39-day induction period. This similarity was seen for both scenarios—with the accumulation of epigenetic memory (mean RT = 23.25 days in figure 6e, RT = 25 days in figure 4b) as well as without it (mean RT = 14.9 days in electronic supplementary material, figure S17B, RT = 15 days in figure 4b).
Next, we quantified the recovery dynamics to an epithelial phenotype, when a cell population has undergone EMT for varying durations, with and without epigenetic regulation of miR-200 by ZEB. We used two metrics: (i) the cumulative fractional share of a phenotype in the population during the 60-day withdrawal period (figure 6f), estimated by area under the curve (AUC) of phenotypic distribution plot, and (ii) the time taken for epithelial phenotype to comprise 90% of the population (figure 6g). Without considering any epigenetic changes during EMT, a short-term (13 days) induction led to a faster recovery than a long-term (26 days and beyond) one (compare blue curves in electronic supplementary material, figure S18 versus in figure 6f), reminiscent of earlier observations from the deterministic simulations (electronic supplementary material, figure S3). Upon accounting for epigenetic changes during EMT, the differences in recovery times became more pronounced with an increasing induction duration, suggesting a longer residence time of cells in M and hybrid E/M states (figure 6f—compare AUC in left and right panels) and longer delays in regaining 90% epithelial share in the population (figure 6g, red curve).
To ascertain how the rate of epigenetic changes during EMT and the withdrawal period influence the reversibility of EMT at a population level, we varied βfor and βrev for every cell in the population. Increasing βfor lowered the residence time of cells in M and E/M states during withdrawal, thus speeding up the recovery to 90% E share (electronic supplementary material, figure S19A, B). Conversely, increasing βrev enhanced the residence time of cells in M and E/M states during withdrawal, and delayed 90% E share recovery (electronic supplementary material, figure S19C, D). These results corroborated our observations during deterministic analysis (figure 4). Similarities between single-cell and population-level analyses were also seen for varying S01 and S02 levels, where the changes in S01 had a more discernible impact on the dynamics of recovery than variations in S02 (for S01 variation, compare electronic supplementary material, figure S12 and figure 5b with electronic supplementary material, figure S20A, C and electronic supplementary material, figure S14 and figure 5b with electronic supplementary material, figure S20B, C; for S02 variation, compare electronic supplementary material, figure S13 and figure 5d with electronic supplementary material, figure S21A, C and electronic supplementary material, figure S15 and figure 5d with electronic supplementary material, figure S21B, C). Intriguingly and in contrast to the irreversible M state for higher S01 values (170k, figure 5a, right panel, figure 5b) observed for deterministic SNAIL dynamics, stochasticity in SNAIL levels enabled both (i) cell transitions to the E state in the event of considerable dip in cellular SNAIL levels below M–E tipping levels (electronic supplementary material, figure S22), and (ii) spontaneous cell-state switching (electronic supplementary material, figure S23).
Overall, the dynamics of recovery to an epithelial state seen for deterministic SNAIL dynamics at individual cellular level were recapitulated by stochastic simulations for a cellular population whose SNAIL levels fluctuated around and switched between predefined mean SNAIL levels.
3. Discussion
The coexistence of multiple cell-states along the E–M spectrum can be seen as ‘attractors’ or valleys in a gene expression landscape, connected by trajectories that enable cell-state transitions [42,43]. The degree of resolution among distinct cell-states depends on the number of biomarkers used experimentally to identify a cell population [5,6,18,44,45]. These cell-states can transition between each other either spontaneously due to factors such as stochastic gene expression and asymmetric cell division, or under microenvironmental influence such as TGFβ signalling and altered matrix stiffness [28,39,46–48]. Recently, frequency of spontaneous cell-state transition has been shown to depend on the mRNA and protein half-life. Corre et al. showed that cell-state memory can extend up to multiple cell generations for stable transcripts (mRNA and proteins) and that cell-state can occur between two cell divisions for highly unstable transcripts [49]. Similarly, in silico mRNA-microRNA interaction dynamics were shown to give rise to periodic switching in mRNA levels, with oscillation period dependent on mRNA half-life. These periodic oscillations when coupled with noise in mRNA and microRNA transcription process caused asynchronous cell-state transitions [50]. The relative rates of cell-state transitions define the population distribution of cells along the E–M axis, as evident from dynamics of isolated subpopulations in vitro and in vivo [4,5,11]. These rates of transition, and thus the equilibrium state distribution, are determined by the relative stability of each state [51].
Besides transcriptional and translational control, chromatin-based epigenetic regulation of E–M states can influence their relative stability, thus shaping population distributions [52,53]. For instance, the hybrid E/M (EpCAM+ Vim+) and mesenchymal-like (EpCAM- Vim+) cells in the PKV cell line displayed upregulated levels of HMGA2, an epigenetic regulator [6]. Inhibiting HMGA2 using an HDACi (Panobinostat) reduced the mesenchymal fraction of the population. Similarly, ectopic expression of EMT-TFs (SNAIL1, SNAIL2, ZEB1) in MDCK cells conferred a mesenchymal phenotype that included epigenetic silencing of the miR-200 family through DNA methylation [54]. In cells with ectopic expression of SNAIL1, suppression of endogenous ZEB1 expression did not revert cells to an epithelial state; however, SNAIL1 repression led to demethylation of the MIR200C gene, causing MET. In another context, ZEB1 recruited the epigenetic remodelling enzyme BRG1 at the CDH1 promoter and this regulation served as a barrier preventing GRHL2 from inducing MET [26]. Thus, different EMT/MET-TFs can epigenetically control cellular plasticity. Consequently, combinatorial or sequential treatment with epigenetic regulators can govern the patterns of intratumour heterogeneity during metastasis and/or drug treatment [6,55].
Our mathematical model, which captures epigenetic changes during EMT, explains how epigenetic memory can accumulate as a function of the duration of an EMT-inducing signal, and how the reversibility of EMT depends on the rate of decay of this memory and thus the time point of withdrawal at which reversibility is experimentally assessed [23–25]. For instance, the loss in chromatin accessibility of the EPCAM gene was observed upon treatment of MCF10A cells with TGFβ showed recovery to pre-treatment levels for 4 days treatment but not for 10 days [23]. Irreversible chromatin accessibility of many epithelial genes was demonstrated, but the withdrawal period was for 10 days only. Our data suggest that epithelial gene expression can be recovered after long-term EMT induction upon extended withdrawal periods (figure 1), and this is probably reflected in the chromatin state of the cell. Therefore, it is possible that a re-opening of the chromatin state for EPCAM and other epithelial genes would have occurred over longer withdrawal times in the aforementioned study. Different time scales of cell-state reversal can be attributed to the dynamics of heterochromatin. For instance, in embryonic stem cells, a long-term recruitment (4.5 weeks) of heterochromatin protein 1 (HP1) to the Oct4 promoter accumulated both H3K9me3 and DNA methylation that silenced Oct4 expression for multiple generations. However, a short-term recruitment (7 days) accumulated only H3K9me3 and did not silence Oct4 gene expression for long [34]. Consistently, in pluripotent stem cells, the ratio of rate of methylation by DNMTs to the rate of demethylation Nanog-Tet complex determines the stability of the epigenetically silenced Oct gene [56]. Similar differences in time scales of acquiring histone modification and DNA methylation was seen in the CDH1 promoter in HMLE cells grown in 10% serum condition [24].
In our model formalism, the parameters βfor and βrev capture distinct response times resulting from a variety of possible epigenetic regulators to reversibly and/or irreversibly silence a gene. While direct empirical identification of these individual rates is complicated by the fact that diverse epigenetic regulators often act in concert, synthetic biology approaches may be helpful in dissecting the dynamics of epigenetic regulation through different modifications: DNA methylation, histone acetylation and histone methylation. For instance, in CHO-K1 cells, a synthetic genetic circuit was constructed to recruit histone methyl-transferase (EED, KRAB), histone deacetylase (HDAC4) and DNA methyltransferase (DNMT3B) to the fluorescent reporter gene [33]. These diverse regulators caused varied histone modification, with only DNMT3B recruitment leading to DNA methylation at the promoter region. The distribution of gene silencing time at a single-cell level was quite distinct among these epigenetic modifiers after 80 h of recruitment, with silencing due to DNMT3B being the slowest of all epigenetic regulators. While DNMT3B-mediated silencing did not lead to re-activation of expression during 30 days of observation time post-withdrawal, considerable recovery was observed for EEB, KRAB recruitment and full recovery to pre-treatment levels for HDAC4-mediated silencing. Further, increasing the duration of EED, KRAB and HDAC4 recruitment enhanced the fraction of cells showing silenced expression in the population after 30 days of withdrawal [33]. This observation corroborates our findings that the extent and durability of epigenetic changes depend on duration of epigenetic modifier recruitment (in our case, maintenance of high ZEB levels during EMT, and its recruited epigenetic modifiers). It is conceivable that we can capture both the short- and long-term memory effects of epigenetic changes by allowing a cell transition between three gene expression states: (i) active gene expression state, (ii) reversibly silent state, and (iii) irreversibly silent gene state, such that the transition rates among these states are dependent on the dose and/or duration of a specific epigenetic modifier treatment [33,57].
Another factor that can vary the cellular response to external exposure to an EMT inducer (such as TGFβ) is cell-to-cell variability in protein levels. For instance, variation in the concentrations of TGFβ receptor and SMAD TFs among cells dictated their response to TGFβ stimulation in terms of nuclear localization of SMAD2 protein [58]. The cells were clustered into six response classes, whose proportions in the population varied with the concentration of TGFβ treatment. Similarly, cellular variability in levels of multiple proteins (DR4/5 receptors, DISC components, CASP8 and BID) controlled the time of apoptotic event in HeLa cells, in the case of TRAIL-induced apoptosis [59]. Additionally, the inclusion of cellular variability in our simulations, achieved by incorporating the fluctuating dynamics of SNAIL, resulted in certain observable differences when compared with deterministic simulations: (i) individual cells had variable return times to an epithelial state, despite identical exposure dose and duration, and (ii) cells were able to switch phenotype among E, M and hybrid E/M states, establishing a dynamic equilibrium in the phenotypic distribution.
Our simulation results showed that during reduction of SNAIL levels, the cell spends an extended amount of time in a hybrid state post long-term EMT induction (figure 2f). This happens due to reductions in the critical M-to-E tipping point levels for SNAIL in addition to steady-state ZEB mRNA levels (figure 2d). The accumulation of epigenetic memory, as accounted for by lowered Z0u200 levels during long-term EMT, causes a cell to first undergo a continuous transition from an M to hybrid E/M and then a discrete transition from a hybrid E/M to an E state upon SNAIL withdrawal. However, we only see a discrete M to E state transition in a post short-term induction scenario (figure 3, electronic supplementary material, figure S2). Thus, the epigenetic memory helps increase the residence time of cells in the hybrid E/M state. Further, regarding long-term (LT, 22 days) EMT induction, the expression profiles of EMT-related genes during 18 days of withdrawal showed that while epithelial genes (purple bar in electronic supplementary material, figure S1) did not gain expression throughout the withdrawal period, mesenchymal genes expression started to decrease their expression (red bar in electronic supplementary material, figure S1). Therefore, it is possible that the cells attain a hybrid E/M state for an extended duration during inducer withdrawal as is also seen in our simulations (figures 3d and 6f). Similar observations can be made with our qRT-PCR data where the relative expression of ZEB1 mRNA significantly drops from 0 to 18 days and from 18 to 27 days of withdrawal, while we did not see similar fold changes in the expression of E-cad, miR200b and miR-200c (figure 1), which might render the population to be considered as a hybrid state.
Our analysis has many limitations. First, our mathematical model considered a reduced EMT regulatory network comprising a few canonical EMT and MET drivers (SNAIL, ZEB, miR-200), although networks with teams of epithelial and mesenchymal genes have been identified [51]. Second, our model of epigenetic regulation of miR-200 by ZEB is phenomenological and lacks the granularity of including different epigenetic enzymes recruited by one or more EMT/MET-TFs, chromatin status of those EMT/MET-TFs and the dynamics of heterochromatin [60,61]. A few recent models have combined mechanistic dynamics of transcriptional and epigenetic control [56,62], building on previous attempts to explain epigenetic memory [63]. Third, by not considering cell division events, we have excluded the role of cell cycle in epigenetic regulation [64]. Fourth, we do not consider the impact of any extracellular changes during EMT such as increased matrix stiffness, which can give rise to mechanical memory due to mechanochemical feedback loops [65–67]. Fifth, regulatory network of a process, in general, is embedded in a larger pool with other genes/regulatory players, and therefore its dynamics also depend on the local density of interconnections with players not directly involved in the process [68]. Nonetheless, our model was able to (i) provide insight into how the dynamics of epigenetic changes during and post EMT induction can affect the reversibility to an epithelial cell state, corroborating with existing quantitative experimental and theoretical analysis; and (ii) explain experimentally observed time-scale differences for EMT reversibility when cells were exposed to varying duration of TGFβ treatments.
4. Material and methods
4.1. Cell culture
MCF10A cells were a gift from Dr Jeffrey Rosen and cultured as previously described [69]. Cells were treated with 5 ng ml−1 recombinant human TGFβ1 (R&D Systems, 240-B) prepared according to the manufacturer's specifications. Cells were passaged every 3 days during TGFβ1 treatment and after TGFβ1 withdrawal. TGFβ1 was added to fresh growth medium 2 days after passaging and upon passaging.
4.2. RNA isolation and qPCR
For mRNA analysed at days 0, 6 and 18, RNA was extracted from cells using TriZol (ThermoFisher, 15596026) and isolated using the RNeasy Mini Kit (Qiagen, 74104) with RNase-Free DNase (Qiagen, 79254). For mRNA analysed at days 27, 36 and 45 and all miRNAs, RNA was extracted and isolated from cells using the miRNeasy Mini Kit (Qiagen, 217084) and treated with RNase-Free DNase (Qiagen, 79254).
Reverse transcription was performed on 500 ng total RNA using the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (ThermoFisher, 4374966). Power Up SYBR Green Master Mix (ThermoFisher, A25776) was used to perform qPCR using 10 ng cDNA. All qPCR reactions were performed in triplicate. mRNA qPCR primers are included in table 1.
Table 1.
qPCR primer sequences.
| primer | sequence |
|---|---|
| ZEB1 for | 5′ GCACAACCAAGTGCAGAAGA |
| ZEB1 rev | 5′ CATTTGCAGATTGAGGCTGA |
| CDH1 for | 5′ TGCCCAGAAAATGAAAAAGG |
| CDH1 rev | 5′ GTGTATGTGGCAATGCGTTC |
| ACTB for | 5′ CCCTGGCACCCAGCAC |
| ACTB rev | 5′ GCCGATCCACACGGAGTAC |
Reverse transcription and qPCR for miRNAs were performed as previously described [69]. Taqman reverse transcription primers and qPCR probes (ThermoFisher) used were as follows: U6snRNA (001973), miR-200b (002251) and miR-200c (002300). Reverse transcription was performed using the Taqman microRNA Reverse Transcriptase Kit (ThermoFisher, 4366596). qPCR was performed using the TaqMan Fast Advanced Master Mix (ThermoFisher, 4444557).
All qPCR was performed using a CFX96 Touch Real-Time PCR System (BioRad).
4.3. Nanostring analysis
Each time point was analysed in biological duplicates. RNA was extracted from cell cultures using TriZol (ThermoFisher, 15596026) and isolated using the RNeasy Mini Kit (Qiagen, 74104) with RNase-Free DNase (Qiagen, 79254). We used 100 ng RNA as input for probe hybridization to a custom CodeSet. Hybridization was conducted for 16 h at 65°C according to the manufacturer's protocol. Raw counts were normalized to the geometric mean of the count values for the three endogenous control genes, GAPDH, HPRT1 and PKG1. Nanostring nCounter was used to quantify probe detection and assign to transcripts. nSolver 4.0 software was used for clustering of normalized gene counts between samples. We considered only the genes in treatment conditions with average expression levels above 10 for further analysis. The filtered expression data was then z-normalized using the formula Z = (x − μ)/σ where x = the observed value, μ = the mean across all samples, and σ = the standard deviation across all samples. On z-normalized data we performed principal component analysis (PCA) and hierarchical clustering.
4.4. EMT regulatory network
We considered an EMT regulatory network involving interaction between canonical epithelial (miR-200) and mesenchymal (ZEB) markers with miR-200 and ZEB mutually repressing each other (figure 1a, inset). SNAIL transcription factor acts as an input to this network, supressing miR-200 and activating ZEB, and it represents cumulative effects of several EMT-inducing signalling pathways, such as TGFβ, Wnt and Notch [16]. The rate equations capturing the production, degradation and complex interactions between nodes for the network components are as follows:
Here, μ200 = [miR-200], mZ = [ZEB1 mRNA], Z = [ZEB1], and S = [SNAIL1]. [·] represents the concentration of a molecular species within a cell. H is the shifted Hill function.
The functions Yμ, Ym and L describe the post-transcriptional regulation of mRNA activity by micro-RNAs, as described earlier [16].
Here, μ is the microRNA concentration and n is number of micro-RNA binding sites on the mRNA. For the inhibition of ZEB1 mRNA by miR-200, n = 6 and The values of all other kinetic parameters are listed in tables 2 and 3.
Table 2.
Regulatory network parameters—rates of production and degradation; and Hills' coefficient, threshold and fold change for transcriptional regulations. Here, mol. ≡ molecules / cell.
| parameter | value | parameter | value |
|---|---|---|---|
| 3 | |||
| 2 | |||
| 2 | |||
| 2 | |||
| 0.1 | |||
| 0.1 | |||
| 7.5 | |||
| 10.0 | |||
| 10 000 | |||
Table 3.
Parameters for mir200–ZEB mRNA complex translation and degradation.
| no. of miRNA binding sites | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| li | 1.0 | 0.6 | 0.3 | 0.1 | 0.05 | 0.05 | 0.05 |
| 0.0 | 0.04 | 0.2 | 1.0 | 1.0 | 1.0 | 1.0 | |
| 0.0 | 0.005 | 0.05 | 0.5 | 0.5 | 0.5 | 0.5 |
4.5. Framework for incorporating epigenetic regulation of miR-200 expression from ZEB
Epigenetic regulation of miR-200 can happen through increasingly methylation of its upstream promoter region, as noted during continued maintenance of mesenchymal state [25]. We phenomenologically incorporate this epigenetic regulation into our modelling framework using following three ways:
-
(1) Strengthening the miR-200 suppression by ZEB depending on the duration for which ZEB is maintained at high levels. This is achieved either by making:
-
(a) Threshold of repression of miR-200 by ZEB a dynamic variable whose rate of change depended on ZEB levels [21], as shown below,
4.1 -
(b) Fold change of repression of miR-200 by ZEB a dynamic variable whose rate of change depended on ZEB levels, as shown below,
4.2
-
(a)
-
(2) Reduce the basal rate of expression of miR-200 depending on the duration for which ZEB is maintained at high levels. This is achieved by making basal rate a dynamics variable, as shown below,
4.3
Here, Z0u200 : threshold for transcriptional repression (variable parameter); Z0u2000 : basal threshold (constant parameter); fold change of transcriptional repression (variable parameter); basal fold change (constant parameter); gμ200 : basal rate miR-200 synthesis (variable parameter); basal rate miR-200 synthesis (constant parameter); Z : ZEB levels; β: time constant (constant parameter); α : epigenetic regulation strength (constant parameter).
In simulations where Z0u200 is variable parameter, the basal threshold Z0u2000 = 220 × 103 molecules [16], and the epigenetic regulation strength parameter, α = 0.15, corresponding to a strong epigenetic regulation. α values greater than 0.15 give a negative value for Z0u200 levels within relevant range of SNAIL levels (0–600 × 103 molecules), thus becoming biologically inappropriate. The time constant β scales the response time of threshold Z0u200 to changes in ZEB levels. To take into account any possible differences in the molecular mechanisms and/or reaction rates of epigenetic changes during EMT and its reversal, we consider two independent β values: (i) βfor, during induction, and (ii) βrev, during withdrawal. To analyse the cellular response without epigenetic regulation, we set α = 0, βfor = 1 h and βrev = 1 h.
For the variable case, and α = 5 × 10−8; and when gμ200 is variable, molecules/hour and α = 0.0013. Note again that α value is adjusted for each case so that variable parameter (/gμ200) remains positive throughout simulation.
4.6. EMT induction and withdrawal simulation set-up
SNAIL levels are being used to control the induction and reversal of EMT, based on bifurcation diagram (figure 2a). These levels can be affected by its stochastic gene expression, variability in upstream signalling pathway activity, and varying microenvironmental cues [58]. To model the temporal variability of SNAIL levels, we represented its dynamics using Ornstein–Uhlenbeck (OU) process, as mentioned earlier [37],
| 4.4 |
Here, γSNAIL : return rate, S0 : mean cellular SNAIL level, W(t) : Weiner process, σn : standard deviation of noise. With the above equation, SNAIL levels follow a stochastic trajectory whose statistical characteristics at stationary state are highlighted in table 4.
Table 4.
Statistical characteristic of stochastic SNAIL dynamics (at stationary state). Where, N is a normalizing factor, and τ is lag duration. τ1/2 represents the average time at which a cell's SNAIL level changes by 50% due to stochasticity in its expression.
| characteristics | expression |
|---|---|
| mean, μ | S0 |
| variance, σ2 | |
| coefficient of variation, CV | |
| probability density function, p.d.f. | |
| autocorrelation function, A () | ) |
| 50% decorrelation time, |
In the simulations:
- (1)
-
(2) S0 attains two values:
-
(a) S01 (pre-induction/post-withdrawal levels): mean SNAIL level of a cell in the population prior to EMT induction and to which it returns after a prolonged withdrawal period.
-
(b) S02 (post-induction levels): mean SNAIL level of a cell in the population after long duration of EMT induction.
-
(a)
-
(3)
τ1/2 = 120 h (5 days) following experimental studies which report mixing times of proteins in human cancer cell lines [37,41]. We considered τ1/2 to remain conserved irrespective of the mean SNAIL level of a cell (S0).
-
(4) With the above constraint, the return rate (γSNAIL) in SNAIL's dynamics gets defined
and
-
(5) For stochastic (noisy) SNAIL dynamics, the choice of standard deviation of noise in SNAIL dynamics, σn, is made by considering the relation between CV, τ1/2, S0 and σn values derived by eliminating αSNAIL from CV and τ1/2 expressions (table 4) as shown below,
-
(6) Note that the CV of SNAIL levels is inversely proportional to square root of S0 (table 3). Here, CV value is taken as 0.3 at S0 = 150 000 molecules. This choice of CV lies within the biological observed range for CVs of several proteins from variety of pathways [41]. Substituting numerical values of CV = 0.3, S0 = 150 000 molecules, and τ1/2 = 120 h above, we get
-
(7)
The above value σn is used during both EMT induction and withdrawal time points for all stochastic population-level simulations.
For population-level analysis (figure 6 and corresponding electronic supplementary material figures), we simultaneously generate 10 000 independent trajectories of SNAIL (equation (4.2)), each representing an individual cell (figure 6a).
4.6.1. Simulation procedure
A cell in the model is described by six variables: {miR-200, ZEB mRNA, ZEB, SNAIL, Z0u200 and S0}. The ZEB mRNA levels, as per bifurcation diagram, are used to assign phenotypes (table 5).
Table 5.
ZEB mRNA ranges and cell's phenotype.
| phenotype | ZEB mRNA range (in molecules) |
|---|---|
| epithelial (E) | <160 |
| hybrid (E/M) | > = 160 & < = 568 |
| mesenchymal (M) | >568 |
4.6.2. For deterministic single-cell EMT induction and withdrawal
-
(1)
Initialize model parameters: assign values to α, τ1/2, βfor, βrev, γSNAIL,for, γSNAIL,rev, S01 and S02 either using the user inputs or relations described in section 3. Set σn = 0.
-
(2)
Initialize cellular network components: set SNAIL = S01, Z0u200 = Z0u2000 = 220×103 molecules, mir200 = 0, ZEB mRNA = 0, and ZEB = 0. Simulate the model for an arbitrary long time so that variables settle to their steady values for the given SNAIL level.
-
(3)
Define the time for EMT induction and SNAIL withdrawal.
-
(4)
For EMT induction, assign S0 = S02, β = βfor, and γSNAIL = γSNAIL,for. Then simulate the model for defined number of induction days.
-
(5)
When induction duration ends, assign S0 = S01, β = βrev, and γSNAIL = γSNAIL,rev. Then simulate the model for defined number of withdrawal days.
4.6.3. For stochastic population-level EMT induction and withdrawal
-
(1)
Initialize model parameters: assign values to α, CV, S0CV#, τ1/2, βfor, βrev, γSNAIL,for, γSNAIL,rev, S01, S02 and σn either using the user inputs or relations described in section 3.
-
(2)
Generate 10k normally distributed SNAIL samples centred around S0 = S01 with variance, . Assign one SNAIL sample to each cell with the values of the other cellular network components as mir200 = 0, ZEB mRNA = 0, ZEB = 0, Z0u200 = Z0u2000, and S0 = sampled SNAIL value. Simulate the model for 1000 h with σn = 0 so that variables settle to their steady values for the given SNAIL levels.
-
(3)
Again, for 1000 h, run the system while considering noise in SNAIL dynamics (use σn as determined above).
-
(4)
Define the time for EMT induction and SNAIL withdrawal.
-
(5)
For EMT induction, assign S0 = S02 for every cell, β = βfor, and γSNAIL = γSNAIL,for. Then simulate the model for defined number of induction days.
-
(6)
When induction duration ends, assign S0 = S01 for every cell, β = βrev, and γSNAIL = γSNAIL,rev. Then simulate the model for defined number of withdrawal days.
#S0CV : S0 value for which CV value is initialized
For simulating scenarios with the and parameter variation, replace Z0u200 and Z0u2000 in the above steps with and and and ), respectively.
Data accessibility
The gene expression data using NanoString technology and codes used for simulation in this study can be accessed at: https://github.com/Paras-Jain20/EMT_Epigenetic_Decay.
Supplementary material is available online [70].
Authors' contributions
P.J.: formal analysis, writing—original draft; S.C.: formal analysis; K.M.: formal analysis; S.S.: formal analysis; S.R.: formal analysis; J.T.G.: methodology, writing—review and editing; H.L.: methodology, writing—review and editing; J.T.: supervision, writing—review and editing; M.T.: conceptualization, supervision, writing—review and editing; M.K.J.: conceptualization, funding acquisition, resources, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
M.K.J. was supported by Ramanujan Fellowship (SB/S2/RJN-049/2018) awarded by Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India. M.T. was supported by Faculty Development Awards and Provost Awards funded by Widener University. J.T. was supported by the Susan G. Komen Foundation Career Catalyst Research Grant (grant no. CCR18548469) and NIH NIGMS (grant no. R15AI156742-01A1). J.T.G. is a CPRIT Scholar in Cancer Research and supported by grant RR210080.
References
- 1.Welch DR, Hurst DR. 2019. Defining the hallmarks of metastasis. Cancer Res. 79, 3011-3027. ( 10.1158/0008-5472.CAN-19-0458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J, et al. 2020. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341-352. ( 10.1038/s41580-020-0237-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripathi S, Levine H, Jolly MK. 2020. The physics of cellular decision-making during epithelial-mesenchymal transition. Annu. Rev. Biophys. 49, 1-18. ( 10.1146/annurev-biophys-121219-081557) [DOI] [PubMed] [Google Scholar]
- 4.Bhatia S, Monkman J, Blick T, Pinto C, Waltham A, Nagaraj SH, Thompson EW. 2019b. Interrogation of phenotypic plasticity between epithelial and mesenchymal states in breast cancer. J. Clin. Med. 8, 893. ( 10.3390/jcm8060893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastushenko I, et al. 2018. Identification of the tumour transition states occurring during EMT. Nature 556, 463-468. ( 10.1038/s41586-018-0040-3) [DOI] [PubMed] [Google Scholar]
- 6.Ruscetti M, et al. 2016. HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene 35, 3781-3795. ( 10.1038/onc.2015.444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. 2011. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633-644. ( 10.1016/j.cell.2011.07.026) [DOI] [PubMed] [Google Scholar]
- 8.Suhas Jagannathan N, Ihsan MO, Kin XX, Welsch RE, Clément MV, Tucker-Kellogg L. 2020. Transcompp: understanding phenotypic plasticity by estimating Markov transition rates for cell state transitions. Bioinformatics 36, 2813-2820. ( 10.1093/bioinformatics/btaa021) [DOI] [PubMed] [Google Scholar]
- 9.Boareto M, Jolly MK, Goldman A, Pietilä M, Mani SA, Sengupta S, Ben-Jacob E, Levine H, Onuchic JN. 2016. Notch-Jagged signalling can give rise to clusters of cells exhibiting a hybrid epithelial/mesenchymal phenotype. J. R. Soc. Interface 13, 20151106. ( 10.1098/rsif.2015.1106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neelakantan D, et al. 2017. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat. Commun. 8, 15773. ( 10.1038/ncomms15773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto M, et al. 2017. Intratumoral bidirectional transitions between epithelial and mesenchymal cells in triple-negative breast cancer. Cancer Sci. 108, 1210-1222. ( 10.1111/cas.13246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582-589. ( 10.1038/embor.2008.74) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S-MM, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894-907. ( 10.1101/gad.1640608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian X-J, Zhang H, Xing J. 2013. Coupled reversible and irreversible bistable switches underlying TGFβ-induced epithelial to mesenchymal transition. Biophys. J. 105, 1079-1089. ( 10.1016/j.bpj.2013.07.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe K, Panchy N, Noguchi S, Suzuki H, Hong T. 2019. Combinatorial perturbation analysis reveals divergent regulations of mesenchymal genes during epithelial-to-mesenchymal transition. npj Syst. Biol. Appl. 5, 21. ( 10.1038/s41540-019-0097-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. 2013. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc. Natl Acad. Sci. USA 110, 18 144-18 149. ( 10.1073/pnas.1318192110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celià-Terrassa T, et al. 2018. Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat. Commun. 9, 5005. ( 10.1038/s41467-018-07538-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karacosta LG, Anchang B, Ignatiadis N, Kimmey SC, Benson JA, Shrager JB, Tibshirani R, Bendall SC, Plevritis SK. 2019. Mapping lung cancer epithelial-mesenchymal transition states and trajectories with single-cell resolution. Nat. Commun. 10, 5587. ( 10.1101/570341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beerling E, et al. 2016. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 14, 2281-2288. ( 10.1016/J.CELREP.2016.02.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ocaña OH, Córcoles R, Fabra Á, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. 2012. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709-724. ( 10.1016/J.CCR.2012.10.012) [DOI] [PubMed] [Google Scholar]
- 21.Jia W, Deshmukh A, Mani SA, Jolly MK, Levine H. 2019. A possible role for epigenetic feedback regulation in the dynamics of the epithelial-mesenchymal transition (EMT). Phys. Biol. 16, 066004. ( 10.1088/1478-3975/ab34df) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsuno Y, Meyer DS, Zhang Z, Shokat KM, Akhurst RJ, Miyazono K, Dernyck R. 2019. Chronic TGF-β exposure drives stabilized EMT, tumor stemness, and cancer drug resistance with vulnerability to bitopic mTOR inhibition. Sci. Signal. 12, eaau8544. ( 10.1126/scisignal.aau8544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson KS, et al. 2022. CTCF expression and dynamic motif accessibility modulates epithelial–mesenchymal gene expression. Cancers (Basel). 14, 209. ( 10.3390/cancers14010209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumont N, Wilson MB, Crawford YG, Reynolds PA, Sigaroudinia M, Tlsty TD. 2008. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc. Natl Acad. Sci. USA 105, 14 867-14 872. ( 10.1073/pnas.0807146105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregory PA, et al. 2011. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 22, 1686-1698. ( 10.1091/mbc.E11-02-0103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somarelli JA, et al. 2016. Mesenchymal-epithelial transition in sarcomas is controlled by the combinatorial expression of MicroRNA 200s and GRHL2. Mol. Cell. Biol. 36, 2503-2513. ( 10.1128/MCB.00373-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stylianou N, et al. 2019. A molecular portrait of epithelial–mesenchymal plasticity in prostate cancer associated with clinical outcome. Oncogene 38, 913-934. ( 10.1038/s41388-018-0488-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook DP, Vanderhyden BC. 2020. Context specificity of the EMT transcriptional response. Nat. Commun. 11, 2142. ( 10.1038/s41467-020-16066-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisco AO, Brock A, Zhou J, Moor A, Mojtahedi M, Jackson D, Huang S. 2013. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat. Commun. 4, 1-11. ( 10.1038/ncomms3467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eichelberger L, et al. 2020. Maintenance of epithelial traits and resistance to mesenchymal reprogramming promote proliferation in metastatic breast cancer. bioRxiv 998823. ( 10.1101/2020.03.19.998823) [DOI]
- 31.Zhang Y, et al. 2022. Genome-wide CRISPR screen identifies PRC2 and KMT2D-COMPASS as regulators of distinct EMT trajectories that contribute differentially to metastasis. Nat. Cell Biol. 24, 554-564. ( 10.1038/s41556-022-00877-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. 2012. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062-2074. ( 10.1038/onc.2011.383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, Oshimura M, Elowitz MB. 2016. Dynamics of epigenetic regulation at the single-cell level. Science 351, 720-724. ( 10.1126/science.aab2956) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. 2012. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447-1460. ( 10.1016/j.cell.2012.03.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balsalobre A, Drouin J. 2022. Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Mol. Cell Biol. 23, 449-464. ( 10.1038/s41580-022-00464-z) [DOI] [PubMed] [Google Scholar]
- 36.Chen AF, Liu AJ, Krishnakumar R, Freimer JW, DeVeale B, Blelloch R. 2018. GRHL2-dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell 23, 226-238. ( 10.1016/j.stem.2018.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, Wennborg A, Aurell E, Dekel E, Zou JZ, Xu Y, Huang S, Ernberg I. 2016. Dynamics inside the cancer cell attractor reveal cell heterogeneity, limits of stability, and escape. Proc. Natl Acad. Sci. USA 113, 2672-2677. ( 10.1073/pnas.1519210113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balázsi G, Van Oudenaarden A, Collins JJ. 2011. Cellular decision making and biological noise: from microbes to mammals. Cell 144, 910-925. ( 10.1016/j.cell.2011.01.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Hu J, Li Y, Guo M. 2021. Volumetric compression develops noise-driven single-cell heterogeneity. Proc. Natl Acad. Sci. USA 118, e2110550118. ( 10.1073/pnas.2110550118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huh D, Paulsson J. 2011. Random partitioning of molecules at cell division. Proc. Natl Acad. Sci. USA 108, 15 004-15 009. ( 10.1073/pnas.1013171108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigal A, et al. 2006. Variability and memory of protein levels in human cells. Nature 444, 643-646. ( 10.1038/nature05316) [DOI] [PubMed] [Google Scholar]
- 42.Burkhardt DB, San Juan BP, Lock JG, Krishnaswamy S, Chaffer CL. 2022. Mapping phenotypic plasticity upon the cancer cell state landscape using manifold learning. Cancer Discov. OF1–OF13 12, 1847-1859. ( 10.1158/2159-8290.cd-21-0282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S, Enrberg I, Kauffman SA. 2009. Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Semin. Cell Dev. Biol. 20, 869-876. ( 10.1016/j.semcdb.209.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhatia S, Monkman J, Blick T, Duijf PH, Nagaraj SH, Thompson EW. 2019a. Multi-omics characterization of the spontaneous mesenchymal–epithelial transition in the PMC42 breast cancer cell lines. J. Clin. Med. 8, 1253. ( 10.3390/jcm8081253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pillai M, Jolly MK. 2021. Systems-level network modeling deciphers the master regulators of phenotypic plasticity and heterogeneity in melanoma. iScience 24, 103111. ( 10.1016/j.isci.2021.103111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain P, Bhatia S, Thompson EW, Jolly MK. 2022. Population dynamics of epithelial-mesenchymal heterogeneity in cancer cells. Biomolecules 12, 348. ( 10.3390/biom12030348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matte BF, Kumar A, Placone JK, Zanella VG, Martins MD, Engler AJ, Lamers ML. 2019. Matrix stiffness mechanically conditions EMT and migratory behavior of oral squamous cell carcinoma. J. Cell Sci. 132, jcs224360. ( 10.1242/jcs.224360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tripathi S, Chakraborty P, Levine H, Jolly MK. 2020. A mechanism for epithelial-mesenchymal heterogeneity in a population of cancer cells. PLoS Comput. Biol. 16. ( 10.1371/journal.pcbi.1007619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corre G, et al. 2014. Stochastic fluctuations and distributed control of gene expression impact cellular memory. PLoS ONE 9, e115574. ( 10.1371/journal.pone.0115574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nordick B, Yu PY, Liao G, Hong T. 2022. Nonmodular oscillator and switch based on RNA decay drive regeneration of multimodal gene expression. Nucleic Acids Res. 50, 3693-3708. ( 10.1093/nar/gkac217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hari K, Ullanat V, Balasubramanian A, Gopalan A, Jolly MK. 2022. Landscape of epithelial–mesenchymal plasticity as an emergent property of coordinated teams in regulatory networks. eLife 11, 1110. ( 10.7554/eLife.76535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown M, Abdollahi B, Wilkins O. 2022. Phenotypic heterogeneity driven by plasticity of the intermediate EMT state governs disease progression and metastasis in breast cancer. Sci. Adv. 8, eabj8002. ( 10.1126/sciadv.abj8002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam WL, Weinberg RA. 2013. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19, 1438-1449. ( 10.1038/nm.3336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diaz-Martin J, Diaz-Lopez A, Moreno-Bueno G, Castilla MÁ, Rosa-Rosa JM, Cano A, Palacios J. 2014. A core microRNA signature associated with inducers of the epithelial-to-mesenchymal transition. J. Pathol. 232, 319-329. ( 10.1002/path.4289) [DOI] [PubMed] [Google Scholar]
- 55.Sharma SV, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69-80. ( 10.1016/j.cell.2010.02.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen T, Ali Al-Radhawi M, Sontag ED. 2021. A mathematical model exhibiting the effect of DNA methylation on the stability boundary in cell-fate networks. Epigenetics 16, 436-457. ( 10.1080/15592294.2020.1805686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukund A, Bintu L. 2022. Temporal signaling, population control, and information processing through chromatin-mediated gene regulation. J. Theor. Biol. 535, 110977. ( 10.1016/j.jtbi.2021.110977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strasen J, Sarma U, Jentsch M, Bohn S, Sheng C, Horbelt D, Knaus P, Legewie S, Loewer A. 2018. Cell-specific responses to the cytokine TGFβ are determined by variability in protein levels. Mol. Syst. Biol. 14, e7733. ( 10.15252/msb.20177733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. 2009. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428-432. ( 10.1038/nature08012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Al-Radhawi MA, Tripathi S, Zhang Y, Sontag ED, Levine H. 2022. Epigenetic factor competition reshapes the EMT landscape. Proc. Natl Acad. Sci. USA 119, e2210844119. ( 10.1073/PNAS.2210844119/-/DCSUPPLEMENTAL) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D'Alessio AC, Young RA, Weinberg RA. 2013. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61-74. ( 10.1016/j.cell.2013.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Folguera-Blasco N, Pérez-Carrasco R, Cuyàs E, Menendez JA, Alarcón T. 2019. A multiscale model of epigenetic heterogeneity-driven cell fate decisionmaking. PLoS Comput. Biol. 15, e1006592. ( 10.1371/journal.pcbi.1006592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dodd IB, Micheelsen MA, Sneppen K, Thion G. 2007. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 129, 813-822. ( 10.1016/j/.cell.2007.02.053) [DOI] [PubMed] [Google Scholar]
- 64.Nashun B, Hill PW, Hajkova P. 2015. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 34, 1296-1308. ( 10.15252/embj.201490649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng Y, Chakraborty P, Jolly MK, Levine H. 2021. A theoretical approach to coupling the epithelial-mesenchymal transition (EMT) to extracellular matrix (ECM) stiffness via LOXL2. Cancers (Basel). 13, 1609. ( 10.3390/cancers13071609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar S, Das A, Sen S. 2014. Extracellular matrix density promotes EMT by weakening cell-cell adhesions. Mol. Biosyst. 10, 838-850. ( 10.1039/c3mb70431a) [DOI] [PubMed] [Google Scholar]
- 67.Price CC, Mathur J, Boerckel JD, Pathak A, Shenoy VB. 2021. Dynamic self-reinforcement of gene expression determines acquisition of cellular mechanical memory. Biophys. J. 120, 5074-5089. ( 10.1016/j.bpj.2021.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harlapur P, Duddu AS, Hari K, Kulkarni P, Jolly MK. 2022. Functional resilience of mutually repressing motifs embedded in larger networks. Biomolecules 12, 1842. ( 10.3390/biom12121842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toneff MJ, et al. 2016. The Z-cad dual fluorescent sensor detects dynamic changes between the epithelial and mesenchymal cellular states. BMC Biol. 14, 47. ( 10.1186/s12915-016-0269-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain P, et al. 2023. Epigenetic memory acquired during long-term EMT induction governs the recovery to the epithelial state. Figshare. ( 10.6084/m9.figshare.c.6365740) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Jain P, et al. 2023. Epigenetic memory acquired during long-term EMT induction governs the recovery to the epithelial state. Figshare. ( 10.6084/m9.figshare.c.6365740) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The gene expression data using NanoString technology and codes used for simulation in this study can be accessed at: https://github.com/Paras-Jain20/EMT_Epigenetic_Decay.
Supplementary material is available online [70].