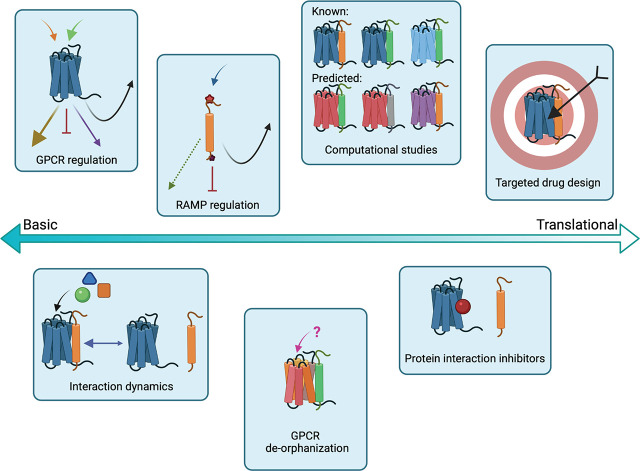
Abstract
G protein-coupled receptors (GPCRs) are known to interact with several other classes of integral membrane proteins that modulate their biology and pharmacology. However, the extent of these interactions and the mechanisms of their effects are not well understood. For example, one class of GPCR-interacting proteins, receptor activity-modifying proteins (RAMPs), comprise three related and ubiquitously expressed single-transmembrane span proteins. The RAMP family was discovered more than two decades ago, and since then GPCR–RAMP interactions and their functional consequences on receptor trafficking and ligand selectivity have been documented for several secretin (class B) GPCRs, most notably the calcitonin receptor-like receptor. Recent bioinformatics and multiplexed experimental studies suggest that GPCR–RAMP interactions might be much more widespread than previously anticipated. Recently, cryo-electron microscopy has provided high-resolution structures of GPCR–RAMP–ligand complexes, and drugs have been developed that target GPCR–RAMP complexes. In this review, we provide a summary of recent advances in techniques that allow the discovery of GPCR–RAMP interactions and their functional consequences and highlight prospects for future advances. We also provide an up-to-date list of reported GPCR–RAMP interactions based on a review of the current literature.
Significance Statement
Receptor activity-modifying proteins (RAMPs) have emerged as modulators of many aspects of G protein-coupled receptor (GPCR)biology and pharmacology. The application of new methodologies to study membrane protein–protein interactions suggests that RAMPs interact with many more GPCRs than had been previously known. These findings, especially when combined with structural studies of membrane protein complexes, have significant implications for advancing GPCR-targeted drug discovery and the understanding of GPCR pharmacology, biology, and regulation.
I. Introduction
Receptor activity-modifying proteins (RAMPs) and their roles in modulating G protein-coupled receptor (GPCR) cell biology and pharmacology were first described nearly 25 years ago. GPCR–RAMP complexes have now been targeted therapeutically, and high-resolution structures have been reported. In addition, it is now clear that many more GPCRs may interact with RAMPs than previously anticipated. An updated comprehensive review about the role of RAMPs is timely because the rate of discovery of new GPCR–RAMP complexes is accelerating, and the techniques used to study GPCRs and RAMPs are rapidly evolving. Recently, bioinformatics, multiplexed proteomics screens, and genetics studies in animal and cell-based models have dramatically expanded the known GPCR–RAMP interactome. In addition, molecular dynamics (MD) simulations and cryoelectron microscopy (cryo-EM) with single particle reconstruction have been used to study known GPCR–RAMP interactions for the first time. In this review, we first provide an overview of the main facets of RAMP research, highlighting key previous reviews for more exhaustive coverage when applicable. Next, we explore the current state of methodologies for identification of GPCR–RAMP complexes and for their functional characterization. We delve into the findings from these new avenues of investigation and critique the pros and cons of different approaches. We conclude by pointing out gaps in our knowledge and future potential avenues for investigation.
A. G Protein-Coupled Receptors
The G protein-coupled receptors (GPCRs) superfamily comprises approximately 800 distinct receptor genes, of which approximately 400 are nonolfactory receptors (Fredriksson et al., 2003). GPCRs respond to diverse classes of agonist ligands and can trigger or modulate a wide range of intracellular responses. For example, activated GPCR signaling cascades can induce changes in second-messenger levels, activate cellular kinases and other regulatory enzymes, regulate ion channels, and alter gene transcription (Pierce et al., 2002; Hauser et al., 2017). While all GPCRs display the canonical seven helical transmembrane (TM) structure, different GPCR classes have substantial differences in other aspects of their architecture, especially at their extracellular N-terminal tails, intracellular C-terminal tails, and intracellular loops. Post-translational modifications (PTMs) of GPCRs include glycosylation, tyrosine sulfation, serine and threonine phosphorylation, and acylation. GPCRs derive their name from the ability to bind heterotrimeric (αβγ) guanine nucleotide-binding regulatory proteins (G proteins) to cause guanine-nucleotide exchange. The active GTP-bound form of the G protein α subunit or the free G protein βγ heterodimer subunit can then interact with downstream cellular effector enzymes or channels. G proteins are classified according to conserved primary structures of the Gα subunits (αi/o, αs, α12/13, and αq/11) and generally initiate different signaling cascades (Wu et al., 2019). Canonical GPCR activation of Gαs is associated with generation of the second messenger cAMP, whereas Gαi activation reduces cAMP levels. Gαq activation is associated with generation of inositol 1,4,5-trisphosphate (IP3) and release of intracellular calcium (Ca2+). Several effectors downstream of GPCR activation can mediate activation of extracellular signal-regulated kinase (ERK) signaling pathways. Phosphorylation of the C-terminal tail of the active GPCR by GPCR kinases (GRKs) creates a substrate for the binding of adaptor/signaling molecules called nonvisual arrestins (β-arrestins). β-arrestin binding turns off G protein signaling and, in some cases, initiates separate signaling cascades.
Bias in signaling between G protein and β-arrestin pathways can be observed pharmacologically when a given ligand preferentially promotes signaling along one or the other pathway (Kolb et al., 2022). Arrestin binding to active phosphorylated GPCRs also drives arrestin-mediated trafficking and receptor internalization pathways (Luttrell and Lefkowitz, 2002; Gurevich and Gurevich, 2019). Internalized receptors, which in some cases remain competent to signal even when removed from the plasma membrane (PM), are either recycled back to the cell membrane or degraded. The role of RAMPs and other accessory proteins in trafficking has emerged for several GPCRs. More study will be needed as additional GPCR–RAMP interacting pairs are discovered, especially since the behavior of specific receptors can vary depending on cellular context.
1. GPCR Classification Based on Phylogeny
Based on phylogenetic analysis of genomic sequences or primary structures, human GPCRs are grouped into five main receptor families—termed the glutamate, rhodopsin, adhesion, Frizzled (Fzd)/sweet taste receptor (TAS2), and secretin families—in the so-called GRAFS system (Fredriksson et al., 2003). The structural hallmarks of GPCRs within each family generally include unique N-terminal tail domains. For example, rhodopsin receptors generally have relatively short (< 50 amino acid residues) N-terminal tails and an orthosteric ligand-binding site within the seven-helical TM core of the receptor. The molecular composition of agonist ligands that bind to GPCRs varies widely, especially among members of the rhodopsin receptor family (Fredriksson et al., 2003). One reason for this observation is that the rhodopsin GPCR family is the largest family, with a total of approximately 700 members (Fig. 1). The rhodopsin family is also referred to as class A. The class A–F system is a homology-based classification that is designed to encapsulate GPCRs in both vertebrates and invertebrates (Attwood and Findlay, 1994). Class A receptors correspond to the rhodopsin family, class B receptors to a subfamily of the secretin and adhesion receptor family, and class C receptors to the glutamate receptor family. Classes D, E, and F include some receptors not found in humans. Here, we will primarily use the GRAFS classification system, which includes only human GPCRs. Within the rhodopsin GPCR family, many discrete receptors respond to diverse odorant molecules and are termed olfactory GPCRs. Nonolfactory, rhodopsin family GPCRs respond to small molecules, including amines, purines, and lipids, as well as peptides and larger glycoproteins. The secretin family is composed of 15 GPCRs that share intermediate-size (approximately 150 amino acid residues) N-terminal hormone-docking/binding domains, which play a pivotal role in the binding of medium-length (approximately 30 or more amino acid residues) peptide ligands. Metabotropic glutamate receptors (mGluRs) on the other hand have long (> 600 amino acid residue) N-terminal tails that comprise a venus-flytrap domain, which includes the orthosteric ligand-binding domain and a cysteine-rich domain. The mGluRs form functional dimers (Pin and Bettler, 2016). The adhesion family of GPCRs is characterized by long N-terminal tails (approximately 200–2800 amino acid residues) with multiple O- and S-glycosylation sites, as well as epidermal growth factor-binding domains and proteolytic sites that are important for their ability to facilitate cellular adhesion. The Fzd receptors have intermediate-length N-terminal tails (approximately 200 amino acid residues), which include a Cys-rich domain and the ligand-docking/binding domain. The Fzd receptors respond to secreted glycoproteins named Wingless-related integration sites and are instrumental in embryonic development and cellular proliferation pathways (Nusse and Clevers, 2017). The TAS2 receptors are interesting because they are like mGluRs in that they form functional dimers and have a venus-flytrap domain that includes the orthosteric ligand-binding site.
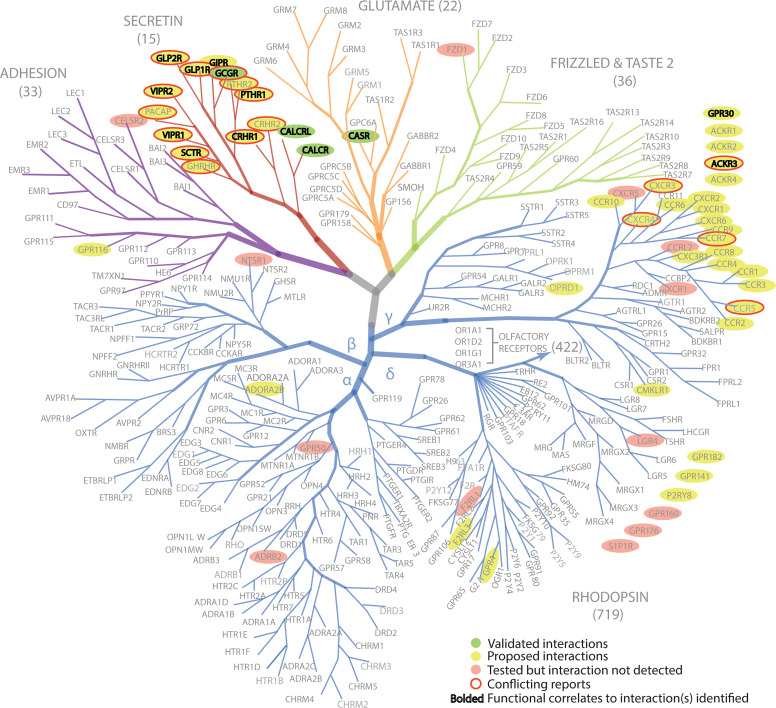
Fig. 1.
GPCR classifications based on phylogenetic analysis. The phylogenetic tree positions of GPCRs tested for RAMP interactions are indicated. Adapted from Lv et al. (2016).
2. GPCR Protein–Protein Interactions and Oligomerization
While GPCRs were originally thought to function simply as monomeric ligand-activated binary switches for various intracellular signaling events, it is now known that GPCR signaling and trafficking involves many oligomeric components that undergo allosteric regulation (Pierce et al., 2002). A key element of this complexity arises from protein–protein interactions (PPIs) between GPCRs or between GPCRs and allosteric modulators (Maurice et al., 2011). Many GPCRs form homo- and hetero-oligomers with other GPCRs. For example, mGluRs homodimerize, and it has been shown that some mGluRs such as mGluR-2 and mGLuR-4 can heterodimerize (Moreno Delgado et al., 2017). Another recent study solved several cryo-EM structures of mGluR-2 and mGluR-7 homodimers and heterodimers (Du et al., 2021). The GPCR Interaction database, GPCR-HetNet (http://www.gpcr-hetnet.com) indicates a total of 537 pairwise interactions between GPCRs, encompassing 183 GPCRs (Borroto-Escuela et al., 2014). The functional consequences of GPCR dimerization and higher-order oligomerization vary and are unknown in some cases. Some GPCR oligomers are disease specific. For example, oligomerization between the GPCR dopamine D1 receptor and the ion channel N-methyl D-aspartate (NMDA) receptor (which is not a GPCR) has been shown to play a role in L-DOPA-induced dyskinesia pathology (Fiorentini et al., 2003). The dopamine D1 receptor-NMDA receptor PPI also highlights the diversity in GPCR-interacting proteins, which is explored further in the next section.
3. GPCR Accessory Proteins
GPCRs can also interact with other non-GPCR membrane proteins, such as RAMPs, receptor transporting proteins (RTPs), receptor expression-enhancing proteins (REEPs), melanocortin receptor accessory proteins (MRAPs), and receptor-component protein (RCP) (Roux and Cottrell, 2014). RTPs are a family of transmembrane proteins that facilitate cell-surface trafficking and ligand-induced responses of odorant receptors (Yu et al., 2017). REEPs mediate the traffic of odorant receptors through modulation of the endoplasmic reticulum (ER) cargo capacity (Björk et al., 2013). MRAPs differentially modulate the expression, trafficking, and signaling of melanocortin receptor 2 (MCR2) and the adrenocorticotropic receptor, with important implications for diseases such as obesity (Berruien and Smith, 2020). More specifically, MRAP1 is required for MCR2 trafficking and function, while MRAP2 interacts with several MCRs by mechanisms that are more poorly understood (Sebag and Hinkle, 2007; 2009a, 2009b; Chung et al., 2008). RCP is a peripheral membrane protein that selectively promotes coupling of a specific GPCR–RAMP complex to Gs (Evans et al., 2000; Routledge et al., 2020).
B. Receptor Activity-Modifying Proteins
Receptor activity-modifying protein (RAMP)1, RAMP2, and RAMP3 are single TM spanning proteins that are ubiquitously expressed in human tissues and unique to vertebrates, indicating that they are likely to be a relatively recent evolutionary development (McLatchie et al., 1998; Parameswaran and Spielman, 2006; GTEx Consortium, 2015; Uhlén et al., 2015; Klein et al., 2016). They have structured extracellular N-terminal tails and short intracellular C-terminal tails. The three RAMPs share only about 30% primary structure homology (McLatchie et al., 1998; Parameswaran and Spielman, 2006; Klein et al., 2016). The discovery of RAMPs resulted from the search for the GPCR that signals in response to the peptide calcitonin-gene related peptide (CGRP) (McLatchie et al., 1998). In their milestone discovery study, McLatchie et al. found that RAMP1 interacts with the secretin family GPCR called calcitonin (CT) receptor-like receptor (CALCRL). CALCRL is the gene that encodes the CALCRL receptor, which is also sometimes unofficially abbreviated as CRLR or CLR. Here, we will refer to the calcitonin receptor-like receptor as CALCRL, which is also how it appears in the original GRAFS system publication (Fredriksson et al., 2003). The CALCRL-RAMP1 complex, but not CALCRL alone, is activated by CGRP, while the CALCRL-RAMP2 and the CALCRL-RAMP3 complexes signal primarily in response to distinct peptides, adrenomedullin (AM) or adrenomedullin 2 (AM2, also referred to as intermedin), respectively. CALCRL-RAMP3 can signal in response to both AM and AM2. The complex of CALCRL-RAMP1 is called the CGRP receptor (CGRPR), whereas the complexes of CALCRL-RAMP2 or CALCRL-RAMP3 are called the AM 1 receptor (AM1R) and AM 2 receptor (AM2R), respectively.
CGRP, AM, and AM2 belong to the calcitonin family of peptides. CGRP is a 37-amino acid residue neuropeptide that is primarily secreted by sensory neurons but is found throughout the central and peripheral nervous system. CGRP-mediated signaling is important in the pathophysiology of diseases, including migraine, which is discussed in more detail in Section D. For reviews focused on CGRP signaling and physiology, we recommend Russell et al. (2014); Kim and Granstein (2021); and Argunhan and Brain (2022). Hay et al. have also reviewed CGRP with a broader focus on CT/CGRP peptide family pharmacology (Hay et al., 2018). AM is a 52-amino acid residue peptide synthesized by adipocytes and a few other cell types. AM is a potent vasodilator and also has other regulatory functions. AM2 is closely related to AM and is widely expressed in the nervous system and peripheral tissue (Hong et al., 2012).
1. Comparisons of the 3 RAMPs
The three RAMPs have the same topology, which includes an extracellular domain, a TM α-helix, and a nine-amino acid cytoplasmic C-terminal tail. The extracellular N-terminal tail is approximately 90 to 100 amino acid residues in length and contains a three-helix bundle, with RAMP2 having an extracellular domain (ECD) that is 26 amino acids longer than that of RAMP1 or RAMP3, which have similar ECD structures (Parameswaran and Spielman, 2006). Bioinformatics analysis suggests that RAMP1 and RAMP3 share a higher sequence similarity than either of them do with RAMP2. The same study also found that RAMP1 and RAMP3 coevolved with a set of GPCRs distinct from the set of GPCRs that evolved with RAMP2 (Barbash et al., 2017a).
The RAMPs have several known and putative sites of PTM. RAMP2 and RAMP3 have one and four predicted N-link glycosylation sites within their ECDs, respectively. A recent paper reports that RAMP1 contains a consensus motif WXXW for C-mannosylation (Mizuta et al., 2022). C-mannosylation is a rare PTM consisting of a carbon-carbon bond linking a single α- or β-D-mannopyranose to the pyrrole ring of the first tryptophan residue in the WXXW motif (Crine and Acharya, 2021). The C-mannosylation of RAMP1 at tryptophan 56 enhances protein stability. RAMP2 and RAMP3 also contain WXXW and WXWC motifs, respectively, but they were not analyzed further in this study.
The C-terminal tails of the RAMPs contain a conserved serine-lysine (SK) motif. For RAMP1 only, this motif is embedded within the ER retention signal QRSKT, which interestingly is overridden upon association with CALCRL (Steiner et al., 2002). The role of the SK motif in RAMPs has not yet been elucidated definitively. For example, Kuwasako et al. studied C-terminal truncation mutants of RAMP1, 2, and 3 coexpressed with CALCRL and concluded that the SK motif in RAMP3 negatively regulates receptor internalization, whereas the SK motif in RAMP2 is involved in the forward trafficking of CALCRL to the PM. They also show that the complete removal of the RAMP3 C-terminal tail does not diminish the maximum extent of internalization and hypothesize about the potential regulatory roles of the SK motif of the other RAMPs. The limitation of this study was that truncation mutants, and not site-specific amino acid substitutions, were employed (Kuwasako et al., 2006). The RAMP C-terminal tails were also studied in the context of RAMP interaction with the calcitonin receptor (CALCR). Udawela et al. used RAMP C-terminal deletion constructs to show that the RAMP C-termini are important for the CALCR-RAMP ligand-binding phenotype in splice isoform A of CALCR. The RAMP C-terminal tail did not appear to be directly involved in CALCR-RAMP signaling. However, the authors found that the RAMPs may interact with other cellular components via their C-terminal tails to facilitate G protein coupling to the receptor (Udawela et al., 2006a). Udewala and colleagues followed up on their findings by applying the same approach to study splice isoform B of CALCR, with similar results (Udawela et al., 2008).
Interestingly, RAMP3 contains a type 1 PSD-95/Discs-large/ZO-1 (PDZ) recognition site in its C-terminal tail. PDZ is an acronym derived from the names of the first three protein structures in which this protein scaffolding domain was observed. RAMP3 has been shown to affect the trafficking of an associated GPCR, such as CALCRL, after receptor activation through RAMP3 interaction with Na+/H+ exchanger regulatory factor (NHERF) or N-ethylmaleimide-sensitive factor (NSF), a vesicle-fusing ATPase (Bomberger et al., 2005a, 2005b; Klein et al., 2016). The RAMPs have putative phosphorylation sites within their cytoplasmic tails, but Hilairet et al. showed that agonist stimulation of the CALCRL-RAMP1 complex with CGRP leads to phosphorylation of CALCRL, but not RAMP1, in the complex (Hilairet et al., 2001). RAMPs also have putative ubiquitination sites, although RAMP ubiquitination has not been directly demonstrated. On the other hand, RAMPs affect GPCR ubiquitination, and it has been shown that AM-induced activation of CALCRL-RAMP2 promotes CALCRL ubiquitination, whereas CGRP-induced activation of CALCRL-RAMP1 does not (Cottrell et al., 2007; Roux et al., 2017).
2. RAMPs in Other Species
As discussed in the review of Klein et al., RAMPs have been identified in 53 species, including many model organisms (Foord et al., 2005; Klein et al., 2016). According to a phylogenetic analysis by Klein and colleagues based on a database of the European Bioinformatics Institute called TreeFam (TF333286), most organisms have three distinct RAMP genes, as is the case for Homo sapiens, but there are a few fish species that have two RAMP1-like and two RAMP2-like genes, thereby encoding five RAMPs in total (Ruan et al., 2008; Guindon et al., 2010; Klein et al., 2016). A different study, which included a tissue expression analysis, coevolution analysis, and phylogenetic comparison of GPCRs and RAMPs, used data from the Orthologous Matrix and identified 44 species with at least one orthologous GPCR and RAMP gene (Barbash et al., 2017a).
There have been several studies of the roles of RAMPs in fish and invertebrates. For example, Nag et al. identified and characterized CALCRLs and RAMPs in pufferfish. The authors then went on to show that, in pufferfish, RAMP1 affects CALCRL glycosylation and trafficking and that some RAMPs can be expressed as multimers on the surface (Nag et al., 2006; Nag et al., 2012). Sekiguchi and colleagues identified three CT/CGRP family peptides, one CALCR/CALCRL, and three RAMP-like proteins in the basal chordate amphioxus (Branchiostoma floridae). Their work is the first molecular and functional characterization of a CT/CGRP family receptor and of RAMPs from invertebrates (Sekiguchi et al., 2016). Two reviews by Sekiguchi provide a summary of the CT/CGRP family peptides and their receptors in mammals and invertebrate deuterostomes, highlighting teleosts, urochordates, cephalochordates, and invertebrate chordates, including ascidians and amphioxi (Sekiguchi, 2018, 2022). Moreover, putative CT/CGRP family peptides are identified in cartilaginous fish based on genomic data analysis.
3. RAMP Localization and Homodimerization
RAMPs were recently shown to be allosteric modulators of GPCR function (Gingell et al., 2016; Lee et al., 2016; Pioszak and Hay, 2020). They are also known to be chaperones for GPCRs that promote receptor translocation from the ER to the PM. Interestingly, RAMP1 has been shown to interact with tubulin (Kunz et al., 2007). RAMP1 has also been shown to colocalize in the Golgi as a disulfide-linked homodimer, suggesting additional possible roles for the RAMPs apart from affecting the biology of GPCRs (Hilairet et al., 2001). These findings were further corroborated by a bioluminescence resonance energy transfer (BRET)-based study of CALCRL and RAMP1, which showed that RAMP1 and CALCRL may both homodimerize (Héroux et al., 2007). There is additional evidence that RAMP1 homodimers may be disrupted by complex formation with CALCRL or the vasoactive intestinal peptide (VIP) and pituitary adenylyl cyclase-activating protein (PACAP) receptor 1 (VIPR1) (Udawela et al., 2004). Another study also applied a BRET-based method and showed that there is potential RAMP dimerization in intracellular biosynthetic compartments that can be disrupted by expression of the secretin receptor (SCTR) (Harikumar et al., 2009). However, the presence, regulation, and function of RAMP homodimers has not been well characterized.
C. Overview of How RAMPs Affect GPCR Biology and Pharmacology
RAMPs have been reported to interact with up to 46 GPCRs (Table 1; Fig. 1). Here, we define an interaction as: (i) formation of a relatively stable and long-lasting physical bimolecular complex; (ii) a transient physical complex formation that has some functional consequence; or (iii) indirect effects mediated by complex formation, either stable and long-lasting, or transient with another relevant regulatory protein. RAMPs can exert a range of effects on an interacting GPCR (Fig. 2), including a chaperone function to facilitate the transport of a receptor to the cell surface. For example, in the absence of RAMPs, CALCRL is poorly localized to the cell membrane. RAMPs have been shown to act as forward trafficking chaperones for several additional GPCRs, including corticotropin-releasing hormone (CRH) receptor 1 (CRHR1), G protein-coupled estrogen receptor 1 (GPR30), and calcium-sensing receptor (CaSR) (Bouschet and Henley, 2005; Bouschet et al., 2008a; Lenhart et al., 2013; Wootten et al., 2013). RAMPs can also modulate ligand selectivity, affect the downstream signaling of an activated receptor, or alter receptor recycling after agonist stimulation (Fig. 2). Receptor-RAMP interactions that affect GPCR cellular trafficking and recycling can result in apparent alterations of receptor expression levels. How a RAMP affects a particular GPCR must be determined experimentally, and it is currently not possible to predict from theoretical or structural considerations. In addition, it is possible that signaling molecules can induce RAMP expression, as was reported in the case in which parathyroid hormone (PTH) induced RAMP3 expression in osteoblasts (Phelps et al., 2005). Co-regulated expression of GPCRs and RAMPs was also suggested in a study that looked at concordant GPCR and RAMP mRNA levels using multiplexed error-correcting fluorescence in situ hybridization (MERFISH) (Barbash et al., 2019). Modes of RAMP-mediated regulation of GPCR function have been reviewed earlier (Hay and Pioszak, 2016), so the aim here is to highlight some key individual studies.
TABLE 1.
Reports of GPCRs tested for RAMP interaction, followed by summary statistics GPCRs are grouped by family and sorted alphabetically within each family. Key references are provided.
| GPCR | Abbreviation | Uniprot | Family | RAMP |
|---|---|---|---|---|
| Adhesion G-protein coupled receptor F5 | ADGRF5 | Q8IZF4 | Adhesion | RAMP3 (Lorenzen et al., 2019) |
| Cadherin EGF LAG seven-pass G-type receptor 2 | CELSR2 | Q9HCU4 | Adhesion | None (only RAMP2 tested) (Barbash et al., 2019) |
| Frizzled family receptor 1 | FZD1 | Q9UP38 | Frizzled | None (only RAMP2 tested) (Barbash et al., 2019) |
| Calcium-sensing receptor | CaSR | P41180 | Glutamate | RAMP1 and 3 (Bouschet et al., 2008; Bouschet et al., 2005; Desai et al., 2014) |
| Pituitary adenylate-cyclase activating polypeptide type 1 | ADYCAP1R1 | P41586 | Secretin | RAMP1, 2, and 3 (Lorenzen et al., 2019) RAMP2 and 3 (M. Harris et al., preprint, DOI: https://doi.org/10.1101/2021.04.08.436756) |
| Calcitonin receptor-like receptor | CALCRL | Q16602 | Secretin | RAMP1, 2 and 3 (McLatchie et al., 1998) and many others (including solved structures) |
| Calcitonin receptor | CALCR | P30988 | Secretin | RAMP1, 2 and 3 (Armour et al., 1999; Christopoulos et al., 1999) and many others |
| Corticotropin-releasing hormone receptor 1 | CRHR1 | P34998 | Secretin | None (Tasma et al., 2020) RAMP2 (Wootten et al., 2013; Wootten et al., 2013;Bailey et al., 2019) RAMP3 (Lorenzen et al., 2019) RAMP2,3 (M. Harris et al., preprint) |
| Corticotropin-releasing hormone receptor 2 | CRHR2 | Q13324 | Secretin | None (Lorenzen et al., 2019; Tasma et al., 2020) None (only RAMP2 tested) (Bailey et al., 2019) RAMP2,3 (M. Harris et al., preprint) |
| Glucagon receptor | GCGR | P47871 | Secretin | RAMP2 (Christopoulos et al., 2003; Weston et al., 2015; Cegla et al., 2017; McGlone et al., 2021) RAMP1,3 (Lorenzen et al., 2019) RAMP1,2,3 (Shao et al., 2022; M. Harris et al., preprint) |
| Growth hormone-releasing hormone | GHRHR | Q02643 | Secretin | None (Christopoulos et al., 2003) RAMP2,3 (Lorenzen et al., 2019) RAMP1,2,3 (Shao et al., 2022; M. Harris et al., preprint) RAMP1,2 with splice variant 1 of GHRHR (Shao et al., 2022) |
| Gastric inhibitory polypeptide receptor | GIPR | P48546 | Secretin | RAMP1,2,3 (Lorenzen et al., 2019; Shao et al., 2022; M. Harris et al., preprint) |
| Glucagon-like peptide 1 receptor | GLP1R | P43220 | Secretin | None (Christopoulos et al., 2003; Wootten et al., 2013) RAMP2,3 (Shao et al., 2022) RAMP1,2,3 (Lorenzen et al., 2019) (M. Harris et al., preprint) |
| Glucagon-like peptide 2 receptor | GLP2R | O95838 | Secretin | None (Christopoulos et al., 2003) RAMP1,2,3 (Lorenzen et al., 2019; M. Harris et al., preprint) RAMP3 (Shao et al., 2022) |
| Parathyroid hormone 1 receptor | PTH1R | Q03431 | Secretin | RAMP2 (Christopoulos et al., 2003; Nemec et al., 2022) RAMP1,3 (Lorenzen et al., 2019) RAMP2,3 (M. Harris et al., preprint) RAMP3 (Phelps et al., 2005) |
| Parathyroid hormone 2 receptor | PTH2R | P49190 | Secretin | RAMP3 (Christopoulos et al., 2003) RAMP1,2 (Lorenzen et al., 2019) RAMP1,2,3 (M. Harris et al., preprint) |
| Secretin receptor | SCTR | P47872 | Secretin | RAMP3 (Harikumar et al., 2009) RAMP1,2 (Lorenzen et al., 2019) RAMP1,2,3 (Shao et al., 2022; M. Harris et al., preprint) |
| VIP and PACAP receptor 1 | VIPR1 | P32241 | Secretin | RAMP1,2,3 (Christopoulos et al., 2003) (M. Harris et al., preprint) RAMP2,3 (Lorenzen et al., 2019) |
| VIP and PACAP receptor 2 | VIPR2 | P41587 | Secretin | RAMP1,2,3 (Wootten et al., 2013; (M. Harris et al., preprint) RAMP2,3 (Lorenzen et al., 2019) |
| Atypical chemokine receptor 1 | ACKR1 | Q16570 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| Atypical chemokine receptor 2 | ACKR2 | O00590 | Rhodopsin | RAMP1,3 (Mackie et al., 2019) |
| Atypical chemokine receptor 3 | ACKR3 | P25106 | Rhodopsin | RAMP2,3 (Lorenzen et al., 2019) RAMP1,2,3 (Mackie et al., 2019) |
| Atypical chemokine receptor 4 | ACKR4 | Q9NPB9 | Rhodopsin | RAMP2,3 (Mackie et al., 2019) |
| Atypical chemokine receptor 5 | ACKR5 | O00421 | Rhodopsin | None (Mackie et al., 2019) |
| Adenosine A2B receptor | ADORA2B | P29275 | Rhodopsin | RAMP2 (Barbash et al., 2019) |
| Beta 2 adrenergic receptor | B2ADR | P07550 | Rhodopsin | None (Mackie et al., 2019; Shao et al., 2022; M. Harris et al., preprint) |
| C-C Chemokine receptor type 1 | CCR1 | P32246 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 2 | CCR2 | P41597 | Rhodopsin | RAMP2,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 3 | CCR3 | P51677 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 4 | CCR4 | P51679 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 5 | CCR5 | P51681 | Rhodopsin | None (Lorenzen et al., 2019) RAMP2,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 6 | CCR6 | P51684 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 7 | CCR7 | P32248 | Rhodopsin | None (Lorenzen et al., 2019) RAMP3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 8 | CCR8 | P51685 | Rhodopsin | RAMP3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 9 | CCR9 | P51686 | Rhodopsin | RAMP1,3 (Mackie et al., 2019) |
| C-C Chemokine receptor type 10 | CCR10 | P46092 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| Chemokine-like receptor 1 | CMKLR1 | Q99788 | Rhodopsin | RAMP2,3 (Mackie et al., 2019) |
| Chemokine C-X3-C receptor 1 | CX3CR1 | P49238 | Rhodopsin | RAMP1,2,3 (Mackie et al., 2019) |
| C-X-C chemokine receptor type 1 | CXCR1 | P25024 | Rhodopsin | RAMP1,2 (Mackie et al., 2019) |
| C-X-C chemokine receptor type 2 | CXCR2 | P25025 | Rhodopsin | RAMP2,3 (Mackie et al., 2019) |
| C-X-C chemokine receptor type 3 | CXCR3 | P49682 | Rhodopsin | None (Lorenzen et al., 2019) RAMP3 (Mackie et al., 2019) |
| C-X-C chemokine receptor type 4 | CXCR4 | P61073 | Rhodopsin | None (Lorenzen et al., 2019) RAMP1,3 (Mackie et al., 2019) |
| C-X-C chemokine receptor type 5 | CXCR5 | P32302 | Rhodopsin | None (Mackie et al., 2019) |
| C-X-C chemokine receptor type 6 | CXCR6 | O00574 | Rhodopsin | RAMP3 (Mackie et al., 2019) |
| Proteinase-activated receptor 2 | F2RL1 | P55085 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| Proteinase-activated receptor 2 | F2RL3 | Q96RI0 | Rhodopsin | RAMP2 (Barbash et al., 2019) |
| G-protein coupled receptor 4 | GRP4 | P46093 | Rhodopsin | RAMP1,2,3 (Lorenzen et al., 2019) |
| G protein-coupled estrogen receptor 1 | GPR30 | Q99527 | Rhodopsin | RAMP3 (Lenhart et al., 2013) |
| Melatonin-related receptor | GPR50 | Q15385 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| Probably G protein-coupled receptor 141 | GPR141 | Q7Z602 | Rhodopsin | RAMP2 (Barbash et al., 2019) |
| Probably G protein-coupled receptor 141 | GPR160 | Q9UJ42 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| G protein-coupled receptor 176 | GPR176 | Q80WT4 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| G-protein coupled receptor 182 | GRP182 | O15218 | Rhodopsin | RAMP1,2,3 (Lorenzen et al., 2019) |
| Leucine-rich repeat-containing G-protein coupled receptor 4 | LGR4 | Q9BXB1 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| P2Y purinoceptor 8 | P2RY8 | Q86VZ1 | Rhodopsin | RAMP2 (Barbash et al., 2019) |
| Neurotensin receptor type 1 | NTSR1 | P30989 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| δ-type opioid receptor | OPRD1 | P41143 | Rhodopsin | RAMP2 (Barbash et al., 2019) |
| Sphingosine 1-phosphate receptor 1 | S1PR1 | P21453 | Rhodopsin | None (only RAMP2 tested) (Barbash et al., 2019) |
| Chemokine XC receptor 1 | XCR1 | P46094 | Rhodopsin | None (Mackie et al., 2019) |
| Summary statistics | RAMP1 | RAMP2 | RAMP3 | ||
|---|---|---|---|---|---|
| No. of GPCR interactors (at least one study) | 28 | 30 | 41 | ||
| Secretin | Rhodopsin | Glutamate | Adhesion | Frizzled | |
| No. of GPCRs that interact with any RAMP(s) (based on at least one report) | 15 | 29 | 1 | 1 | — |
| No. of GPCRs that don’t interact with any RAMP (based on at least one report) | 5 | 8a | — | 1 | 1 |
| No. of GPCRs in the family | 15 | 719 | 22 | 33 | 11 |
Key references provided.
aAnd 7 additional instances where the GPCR does not appear to interact with a RAMP, but only RAMP2 was tested.
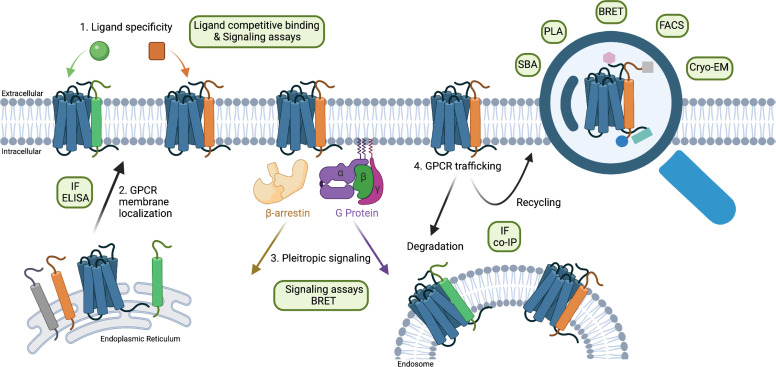
Fig. 2.
Summary of how RAMPs modulate GPCR biology. The four major regulatory effects of RAMPs on RAMP-interacting GPCRs are shown, with techniques commonly used to interrogate the regulatory effect or the presence of the complex in green text bubbles.
1. Ligand Selectivity
Potential GPCR–RAMP interactions that affect the ligand specificity and selectivity for the GPCR in the complex are particularly interesting. So far, this effect has only been well documented for the receptors CALCRL (McLatchie et al., 1998; Hilairet et al., 2001; Husmann et al., 2003) and CALCR (Armour et al., 1999; Christopoulos et al., 1999; Hay et al., 2005; Udawela et al., 2006a, 2006b; Morfis et al., 2008; Gingell et al., 2014). The ligand selectivity of the complexes formed between CALCRL and the three RAMPs was discussed at the start of section B. CALCR in complex with any one of the three RAMPs binds amylin and forms the amylin receptors 1-3 (AMY1-3). AMY1-3 bind amylin with high affinity and CT with low affinity. CALCR in the absence of interaction with RAMPs has the opposite phenotype: it binds CT and is also capable of binding amylin but with lower affinity than for CT. Amylin is a 37-amino acid residue peptide hormone that is cosecreted with insulin from pancreatic β-cells and plays a role regulating food intake and glucose metabolism (Hay et al., 2015). CT is a 32-amino acid residue peptide hormone produced by parafollicular C-cells in the thyroid and among other functions regulates calcium metabolism. Notably, the RAMP1-based amylin receptor complex, AMY1, binds CGRP with high affinity and may be a dual receptor for CGRP and amylin (Hay et al., 2018). It is therefore important to interpret studies with RAMP1 in the context of both these peptides.
The degree of ligand discrimination is not very selective in the case of other RAMP-interacting GPCRs. Moreover, the species from which the receptor or RAMP are derived can influence these pharmacological profiles, and it has been shown recently that there are differences between mouse and human CT and CALCRL (Garelja et al., 2022). There are conflicting results regarding whether RAMPs affect the ligand selectivity of the glucagon receptor (GCGR). Weston et al. showed that RAMP2 increases glucagon potency and efficacy for activating the GCGR, while Cegla et al. and Shao et al. found that RAMP2 does not alter glucagon binding to or activation of the GCGR (Weston et al., 2015; Cegla et al., 2017; Shao et al., 2022). One example where RAMP-mediated ligand selectivity effects have not been demonstrated is the receptor VIPR1, for which RAMPs have been shown not to affect binding of the VIP ligand (Christopoulos et al., 2003). It has been suggested that the mechanism of peptide-hormone binding for CALCRL and CALCR is unique compared with peptide binding to other secretin family GPCRs. The hormones that bind selectively to GPCR–RAMP complexes have been hypothesized to retain a partial N-terminal α-helix motif in solution that extends upon binding. Solution structures of other peptide ligands that bind to secretin family GPCRs tend to be more disorganized and form an extended α-helix only upon binding. This difference in the ability of a receptor to induce secondary structure in peptide ligands upon binding may help to explain why the RAMPs only appear to act as ligand-binding selectivity switches for CALCRL-RAMP or CALCR-RAMP complexes (Liang et al., 2020b; Deganutti et al., 2021).
2. GPCR Trafficking
The chaperone activities of RAMPs that affect GPCR cellular trafficking and localization have been examined for CALCRL (McLatchie et al., 1998; Kuwasako et al., 2000; Bomberger et al., 2012), CALCR (Hay et al., 2006; Morfis et al., 2008), VIPR1 (Christopoulos et al., 2003), CaSR (Bouschet et al., 2005; Bouschet et al., 2008) , SCTR (Harikumar et al., 2009), VIP and PACAP receptor 2 (VIPR2) (Wootten et al., 2013), GPR30 (Lenhart et al., 2013), CRHR1 (Wootten et al., 2013;Bailey et al., 2019), glucagon-like peptide 1 receptor (GLP1R) (Wootten et al., 2013), GCGR (Cegla et al., 2017; McGlone et al., 2021), atypical chemokine receptor 3 (ACKR3) and other chemokine receptors (Mackie et al., 2019), and gastric-inhibitory polypeptide (GIP) receptor (GIPR) (M. Harris et al., preprint, DOI: https://doi.org/10.1101/2021.04.08.436756).
It is important to note that, unlike the case of CALCRL, many other RAMP-interacting GPCRs, such as CALCR, traffic to the cell surface even in the absence of RAMP coexpression. Thus, both RAMP-free and RAMP-associated GPCRs are present on the cell surface, potentially confounding the measurements of RAMP-specific pharmacological effects, which are discussed later. RAMPs may also have the inverse effect of decreasing apparent GPCR surface expression, as evidenced by the flow cytometry-based surface expression screen for chemokine receptors reported by Mackie and colleagues (Mackie et al., 2019). Another example is GCGR, which demonstrated increased internalization upon coexpression of RAMP2, in both basal and agonist-stimulated conditions, when assayed in Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK) 293T cells (Cegla et al., 2017; McGlone et al., 2021). There is also evidence for the presence of CALCRL-RAMP1 complexes in endosomal compartments that are signaling competent (Yarwood et al., 2017).
RAMPs can also affect the movement of an interacting receptor from the PM after agonist stimulation. The RAMP-specific regulation of receptor desensitization has been studied most extensively for CALCRL. CALCRL-RAMP1 and CALCRL-RAMP2 internalization has been shown to be β-arrestin dependent, whereas RAMP3 mediates CALCRL internalization through PPIs between the PDZ domain of RAMP3 and NSF and NHERF, which were introduced in Section B, Subsection 1 (Hilairet et al., 2001; Bomberger et al., 2005a, 2005b; Kuwasako et al., 2006; Héroux et al., 2007). Recently, RAMP3 has been shown to be required for the rapid recycling of ACKR3 (Mackie et al., 2019). The PDZ motif of RAMP3 has also been implicated in GIPR localization (McGlone et al., 2021) (M. Harris et al., preprint, DOI: https://doi.org/10.1101/2021.04.08.436756).
There is some overlap between the discussion of the effect of RAMPs on GPCR trafficking after agonist stimulation and on the ability of RAMPs to modulate β-arrestin recruitment, which is an element of the GPCR desensitization process. Therefore, some of the studies referenced here are discussed in more detail in the section on G protein and β-arrestin-mediated signaling.
3. G Protein- and β-Arrestin-Mediated Signaling
The effects of RAMPs on downstream GPCR signaling pathways are pleiotropic, and thus far, no clear patterns have emerged. Depending on the receptor, RAMPs can either augment or inhibit GPCR signaling through G protein- and β-arrestin-mediated signaling pathways. The effect of RAMP association on receptor pharmacology and signaling is discussed further later, where we provide select examples that are not meant to be a comprehensive review of all instances of RAMP effect on G protein- and β-arrestin-mediated signaling. The modulatory effects of the RAMPs on G protein-mediated signaling has been reviewed for several secretin family receptors (Hay and Pioszak, 2016; Klein et al., 2016).
There have been several studies on β-arrestin recruitment to CALCRL-RAMP complexes (Hilairet et al., 2001; Kuwasako et al., 2006; Héroux et al., 2007; Schönauer et al., 2015; Kuwasako et al., 2016; Gingell et al., 2020). For example, Héroux et al. showed that when CALCRL is coexpressed with RAMP1, there is much higher β-arrestin recruitment to CALCRL compared with the case where CALCRL is expressed alone (Héroux et al., 2007). More recently, Pearce and colleagues carried out a complete characterization of β-arrestin1 and β-arrestin2 recruitment to all three CALCRL-RAMP complexes. The authors also characterized the effect of the GRKs on CALCRL-RAMP complexes, and the effects of RAMPs on agonist-dependent and agonist-independent trafficking (Pearce et al., 2022). Using a BRET-based approach they found that CALCRL-RAMP1 recruits both β-arrestins more potently than CALCRL-RAMP2 and CALCRL-RAMP3 and that the three complexes have different internalization and recycling pathways. Characterization of the effect of GRK expression on CALCRL-RAMP signaling revealed that GRK5 and GRK6, out of the six GRKs tested, had the strongest effects on the surface expression of CALCRL-RAMP complexes.
In studies of GCGR, Cegla et al. used a non-BRET-based β-arrestin recruitment assay to study GCGR with RAMP2, and in contrast to the Héroux et al. and Pearce et al. results for CALCRL-RAMP1, showed that coexpression of GCGR with RAMP2 abolished β-arrestin recruitment to GCGR (Cegla et al., 2017). Recently, McGlone et al. showed that RAMP2-GCGR coexpression enhanced GCGR internalization in both basal and stimulated conditions compared with GCGR expressed alone. GCGR was shown to colocalization with an early endosome marker and an increase in ligand-stimulated cAMP production was measured upon RAMP2 coexpression. RAMP2 did not seem to affect G protein subtype bias for GCGR. The authors argued that the spaciotemporal pattern of GCGR signaling was altered due to RAMP2 coexpression, although they were not able to connect their findings to an in vivo phenotype in mice with hepatic RAMP2 overexpression (McGlone et al., 2021).
Shao and colleagues interrogated the effect of RAMPs on both G protein- and β-arrestin-mediated signaling of the glucagon family receptors and showed that RAMPs affect receptor signaling in a RAMP-, GPCR-, and ligand-dependent manner. For example, cAMP production was decreased upon stimulation with GIP when GIPR was coexpressed with RAMP3 compared with the case where GIPR was expressed alone. However, recruitment of β-arrestin1 or β-arrestin2 was not significantly affected. In contrast, RAMP3 coexpression with glucagon-like peptide 2 (GLP2) receptor (GLP2R) resulted in a decrease of Gq activation, and both β-arrestin1 and β-arrestin2 coupling, while GLP2-mediated cAMP production was not significantly affected. In all the cases tested, RAMP2 only seemed to affect the β-arrestin recruitment to an interacting receptor, and for some GPCRs, like growth hormone-releasing hormone receptor (GHRHR), none of the RAMPs seemed to have any effect on cAMP production, Gq activation, or β-arrestin1/2 recruitment (Shao et al., 2022). The authors posited that the modulatory effects of the RAMPs, or lack thereof for some receptors, may be cell line-dependent and that a RAMP may be affecting other aspects of GPCR biology that were not measured, such as receptor internalization and degradation.
In a preprint from April 2021, Harris et al. describe how RAMPs regulate the signaling bias and internalization of GIPR (Harris et al., preprint, DOI: https://doi.org/10.1101/2021.04.08.436756). The authors reported that GIPR can activate multiple G protein effectors, not just the “classically activated” Gs subtype. They then studied the effect of GIPR-RAMP coexpression on GIP signaling and proposed that RAMP3 association impairs GIPR-mediated activation of Gs and therefore reduces cAMP accumulation. In contrast, RAMP1 and RAMP2 association with GIPR is linked to reduced Gq, G11, and G15 activation, and therefore attenuated Ca2+ mobilization and ERK1/2 phosphorylation. However, many of the effects observed are relatively subtle.
4. GPCR Activation Dynamics
In recent study, Nemec and colleagues investigated the class B GPCR–RAMP interaction of PTH 1 receptor (PTH1R) and RAMP2 (Nemec et al., 2022). They used a Förster resonance energy transfer (FRET) experiment with C-terminally tagged PTH1R (mCitrine, acceptor) and RAMPs (mTurquoise2, donor) to show that PTH1R interacts with RAMP2, but not with RAMP1 or RAMP3. Using a PTH1R FRET biosensor, a rapid superfusion system, and a circularly permuted green-fluorescent protein-based PTH1R biosensor, they draw two conclusions. First, coexpression of RAMP2 may promote a preactivated conformation of PTH1R, given that PTH1R is activated by PTH twice as quickly in the presence of RAMP2 than alone. Second, there are no RAMP2-dependent changes on PTH1R activation by PTHrP, which is another endogenous agonist for the receptor.
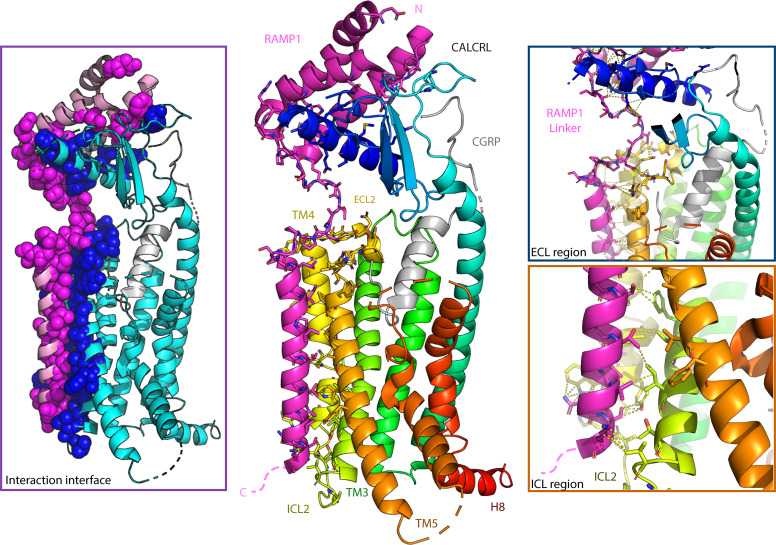
Looking at events downstream of PTH1R activation with BRET- and FRET-based assays, Nemec et al. found that RAMP2 coexpression accelerated Gs activation and increased the potency of Gi3 activation upon PTH stimulation compared with PTH1R expressed alone. However, the G protein activation profile for PTHrP-stimulated PTH1R was not affected. RAMP2 coexpression also increased β-arrestin2 recruitment to both PTH- and PTHrP-stimulated PTH1R but did not affect GRK2 recruitment or ERK activation. Modeling a PTH1R-PTH-RAMP2-Gs complex based on the previously solved CALCRL-CGRP-RAMP1-Gs structure indicated that the RAMP2 linker and ECD make important contacts with PTH1R ECD and extracellular loop (ECL) 2, which connects TM4 and TM5 (Liang et al., 2018). These PHT1R-RAMP2 contacts may promote or stabilize significant preactivation conformational changes, thereby providing a structure-based explanation for the observed effects of RAMP2 on PTH1R activation. Overall, the findings suggest that RAMP2 promotes a unique “partially preactivated state” of PTH1R. The effects of RAMP2 were ligand specific, suggesting that endogenous ligands can be regulated differently. The authors posit that their findings can be exploited to advance treatments to increase bone density, since RAMP2 affects β-arrestin2 recruitment to PTH1R, which may in turn upregulate PTH1R-mediated effects on bone mass.
D. Physiologic Relevance of RAMPs
Shortly after the discovery of the three RAMPs in human-derived cell lines, they were also identified in rodents (Husmann et al., 2000). In the decades following, many groups have investigated the effects of modulating RAMP expression on phenotypes in mice and in human cells, and more have hypothesized the potential roles of RAMP dysfunction in disease. The review by Serafin and colleagues (Serafin et al., 2020) focuses on in vivo RAMP studies, so here we first highlight some foundational studies and then summarize key recent findings from about 2019 onwards.
1. Physiologic Studies of RAMPs in Mice
Physiologic studies in mouse models have mostly focused on the interaction between RAMPs and just a subset of interacting GPCRs, largely secretin (class B) receptors such as CALCRL. Different RAMP transgenic mouse models demonstrate distinctive phenotypes, and it is important to note that several GPCRs could contribute to the phenotypes of RAMP transgenic mice. Since the exact number of RAMP-interacting GPCRs has not yet been determined, the effects observed in global knockout (KO) mouse studies might ultimately be attributable to GPCRs other than the intended target. Therefore, at present it is difficult to assign a phenotype associated with a global RAMP KO transgenic model to a specific RAMP-interacting GPCR, as there may be many different GPCR–RAMP complexes contributing to the observed phenotypes.
RAMP1 KO mice are viable but show dysfunction in the vascular system as well as an alteration in inflammatory responses. Kurashige et al. generated RAMP1 KO mice that exhibited suppressed wound-induced angiogenesis, lymphangiogenesis, and healing compared with wild-type (WT). In particular, the KO mice showed reduced expression of vascular endothelial growth factor (VEGF)-A, VEGF-C, and VEGFR-3 and suppressed formation of lymphatic vessels for draining interstitial fluids (Kurashige et al., 2014). The role of RAMP1 in lymphangiogenesis was studied in the context of a mouse model of secondary lymphedema in RAMP1 KO mice. The RAMP1 KO mice displayed sustained lymphedema, suppressed lymphangiogenesis, and reduced expression of VEGF-C and VEGFR-3 distal to lymphatic lesions, suggesting that RAMP1 plays a role in accelerated lymphangiogenesis associated with reduced recruitment of proinflammatory macrophages (Mishima et al., 2017).
Recently, Yin and colleagues studied the role of RAMP1 in wound healing using a skin wound mouse model and mouse skin fibroblast cell lines. They found that RAMP1 expression levels were altered during skin wound healing. Moreover, RAMP1 overexpression (OE) promoted cell proliferation and was associated with increased yes-associated protein expression and altered expression patterns of G proteins (Yin et al., 2022). Interestingly, CALCRL-RAMP1 has been shown to have mechanoresponsive properties and is involved in mechanical force transduction in macrophages in mice, thereby pointing to a role for RAMP1 in innate immunity (Muschter et al., 2019). A role for RAMP1 in CGRP sensory nerve regulation of chondroitin sulfate synthesis in the context of extracellular matrix homeostasis of intervertebral discs was also reported (Hu et al., 2022). The precise mechanism for mechanical force transduction effects mediated by GPCR complexes remains to be elucidated.
Although both CALCRL-RAMP2 and CALCRL-RAMP3 complexes form a receptor for AM, KO mice of RAMP2 and RAMP3 have revealed distinct roles of these two RAMP isoforms. Genetic loss of RAMP2 causes embryonic lethality due to defects in vascular development and cardiac mitochondrial dysregulation. Consistent with the role of RAMP2 in vasculature, endothelial restoration of RAMP2 expression rescues Ramp2−/− lethality, but mice still exhibit cardiomyopathy. Haploinsufficient RAMP2 mice survive to birth but demonstrate increased vascular permeability (Dackor et al., 2007; Barrick et al., 2012; Yoshizawa et al., 2013; Yamauchi et al., 2014). In a rodent model of renal dysfunction with deficiency in vascular endothelium RAMP2, there were elevated levels of exogenous AM in the plasma, pointing to a role of RAMP2 in AM distribution (Hosoda et al., 2022).
In contrast to the situation with RAMP2, Ramp3−/− mice have no major abnormalities but exhibit higher blood pressure and reduced lymphatic vessel function, and therefore RAMP3 may be involved in regulation of draining through lymphatic vessels (Yamauchi et al., 2014; Shindo et al., 2022). Interestingly, the phenotypes described here are recapitulated by genetic loss of Calcrl or AM. RAMP3 KO mice exhibit an age-dependent weight decrease phenotype compared with control. Other RAMP3 null mice studies have pointed to a role for RAMP3 in negatively regulating bone adaptation (Dackor et al., 2007). A recent study on the effect of RAMP3 on skeletal growth and development showed that Ramp3−/− young mice have increased bone volume, osteoblast numbers, and bone apposition rate compared with WT mice. RAMP3 may act interdependently with RAMP1 in this context. Pacharne and colleagues showed that there are correlations between the mRNA levels of RAMP3 with RAMP1 but not with RAMP2 in osteoblasts cultured from the Ramp3−/− mice (Pacharne et al., 2022).
Studies of RAMP OE in the context of CALCRL have also been reported. In mice with neural OE of RAMP1, increased sensitivity to CGRP caused increased neurogenic inflammation (Tsujikawa et al., 2007; Li et al., 2014; Pawlak et al., 2017). Mouse models with RAMP1 OE in the central nervous system have been generated and show that neuronal RAMP1 is positively correlated with energy expenditure and is involved in modulating the brain actions of amylin and CGRP (Zhang et al., 2011). As RAMP2 KO is lethal, some studies use OE mouse models to interrogate RAMP2 function. Tam et al. generated transgenic mice with RAMP2 OE in smooth muscles to study the role of RAMP2 in blood pressure and vascular function. The authors found that RAMP2 plays a key role in the sensitivity and potency of AM-induced hypotensive response (Tam et al., 2006).
Studies of CALCRL-RAMP interactions and their functional consequences have dominated the landscape of in vivo–focused RAMP studies. However, the effects of RAMPs on several other GPCRs have also been studied in mouse model systems. Wootten et al. showed a loss of responsiveness to CRH in RAMP2+/− mice (Wootten et al., 2013). McGlone et al. studied the effect of RAMP2 on GCGR trafficking in the liver with experiments that employed cell lines and mouse models. Although they observed an effect of RAMP2 on GCGR cellular localization and signaling in HEK 293T and mouse embryonic fibroblast (MEF) cells, they did not see phenotypic changes or differences in glucose tolerance, glycemic response, or insulin tolerance in lean and obese mice with hepatic RAMP2 upregulation compared with control mice. The authors speculated that the lack of a readily observable effect of RAMP2 upregulation in hepatocytes on carbohydrate metabolism indicates that there is a compensatory mechanism involved (McGlone et al., 2021).
Liu et al., showed that female RAMP3 homozygous KO mice had a decreased glucose tolerance (Liu et al., 2018). Using RAMP1/RAMP3 KO mice, Lutz et al. showed that RAMP1 and RAMP3 are involved in amylin (also known as islet amyloid polypeptide) signaling in the brain (Lutz et al., 2018). The study also showed that amylin may negatively regulate its receptor by affecting the downregulation of RAMP1 and RAMP3 mRNA levels. Another study employing global KO RAMP1, RAMP3, and RAMP1+RAMP3 mice investigated the effect of RAMPs in the context of food intake, energy balance, and amylin receptor function (Coester et al., 2020). The authors found that RAMP1 has a role in mediating fat utilization, whereas RAMP3 is likely involved in glucose homeostasis. Notably, mice with the RAMP1+RAMP3 double KO that were on a high-fat diet had higher food intake, weight gain, and leptin levels compared with WT mice fed the same diet. RAMP1+RAMP3 KO mice also displayed amylin insensitivity. These results extend upon the findings of the previous report from the same authors that RAMP1+RAMP3 KO mice were insensitive to the effects of amylin on eating or of leptin on food intake. RAMP1 KO effects were sex dependent, suggesting the possibility of some influence of female sex hormones (Coester et al., 2020).
In addition to its reported roles in metabolism and skeletal growth and development, RAMP3 may also be involved in cardiovascular physiology. RAMP3 acts as a chaperone and essential regulator of GPR30 function. Genetic loss of RAMP3 eliminated the cardioprotective effects of GPR30 activation in chronic hypertension and cardiac hypertrophy mouse model (Lenhart et al., 2013) . Studies in mice have also pointed to a potential role of ACKR3 and its regulation by RAMP3 in cardiovascular disease (Duval et al., 2022). Mackie et al. showed that ACKR3 and RAMP3 form a complex that can scavenge AM, which in turn reduces AM bioavailability and decreases signaling through the CALCRL-RAMP3 complex, a pathway thought to be involved in angiogenesis (Mackie et al., 2019). Additional studies also reported ACKR3 interaction with RAMP3 and showed that RAMP3 does not affect β-arrestin recruitment in response to AM, although the CALCRL-RAMP2 and CALCRL-RAMP3 complexes seemed to play roles as AM scavengers (Szpakowska et al., 2018; Meyrath et al., 2021). Additional work is needed to ascribe AM as a physiologic ligand for the ACKR3-RAMP3 complex and to dissect the precise regulatory role for ACKR3 in AM signaling.
2. Small Molecules and Biologics Targeting RAMP-Interacting GPCRs
CGRP, which signals primarily through the CALCRL-RAMP1 complex, plays an important role in the pathophysiology of migraine. Small molecules and monoclonal antibodies (mAbs) have been developed to inhibit the effects of CGRP by blocking the interaction between CGRP and CALCRL-RAMP1 (Reuter et al., 2018; Tepper, 2018; Wattiez et al., 2020). Erenumab was the first Food and Drug Administration (FDA)–approved therapeutic mAb targeting the RAMP1-CALCRL complex. There are three other FDA-approved mAbs that target the CGRP peptide instead of the receptor indicated for treatment of migraine: eptinezumab (approved February 2020), fremanezumab (approved September 2018), and galcanezumab (approved September 2018), which were all in clinical trials at the same time as erenumab. The small molecule ubrogepant, which is a CALCRL-RAMP1 antagonist, was approved in December 2019 after showing promising clinical trial results (Edvinsson et al., 2018). Two additional small molecules, rimegepant and atogepant have been approved recently (Scuteri et al., 2022). The CGRP antagonists olcegepant and telcagepant bind to CALCRL and RAMP1 directly, with the RAMP affecting the selectivity of the small molecules, an effect that is reviewed in more detail in Sexton et al. (Sexton et al., 2009). Another orally dosed compound called MK-3207 was developed, but it and tecalgepant have been discontinued due to side effects on the liver associated with chronic dosing (Bell et al., 2010; Hewitt et al., 2011; Bucknell et al., 2020).
More anti-migraine drugs targeting the CALCRL-RAMP1 complex are currently in the pipeline. Uniquely, zavegepant is an intranasal small molecule CALCRL-RAMP1 antagonist that has recently undergone a phase II/III trial, with promising results (Chaturvedula et al., 2013; Croop et al., 2021). Using olcegepant as a starting point, Bucknell et al. have used a structure-activity relationship-based approach and developed a new CALCRL-RAMP1 antagonist, called HTL22562 (Bucknell et al., 2020). An in silico drug repurposing study using molecular docking has identified the FDA-approved compounds pentagastrin and leuprorelin as potential antagonists of CALCRL-RAMP1 (Aksoydan and Durdagi, 2022).
Novel classes of antimigraine therapeutics targeting CALCRL-RAMP1 are also being developed. Jamaluddin et al. have developed and tested the activity of lipidated peptide analogs based on CGRP (Jamaluddin et al., 2022). Cansfield et al. have developed novel macrocycle antagonists for CALCRL-RAMP1 and solved two crystal structures of the extracellular portion of the complex bound to each of two different macrocycles (Cansfield et al., 2022).
AM and the CALCRL-RAMP2 and CALCRL-RAMP3 complexes have been implicated in tumor progression. Selective CALCRL-RAMP2 antagonists have recently been developed as anti-tumor therapeutics, although they are still in the preclinical stage of development (Avgoustou et al., 2020; Jailani et al., 2022).
Shifting from therapeutics targeting CALCRL to those targeting CALCR, several amylin analogs, notably pramlintide and cagrilintide, have been developed. Pramlintide is a synthetic amylin analog that is FDA approved for use with insulin to treat patients with type 1 and type 2 diabetes mellitus (Ratner et al., 2004; Ryan et al., 2005). Cagrilintide is an amylin analog and dual amylin and calcitonin receptor agonist (DACRA) that has shown promising results in weight reduction in overweight and obese individuals in a phase II clinical trial (Fletcher et al., 2021; Kruse et al., 2021; Lau et al., 2021). Salmon CT, which is also a DACRA, has been tested in a phase III clinical trial for postmenopausal osteoporosis and showed some benefit over placebo (Binkley et al., 2012). The synthetic peptide DACRA called KPB-088 has shown promising results in preclinical studies for weight loss and improving key metabolic parameters (Larsen et al., 2020). A preclinical study has also identified an amylin receptor peptide antagonist called AC253 that may confer protection from Alzheimer’s disease progression (Soudy et al., 2019).
Overall, RAMP regulation is correlated with various disease states (Jacob et al., 2012), information that can potentially be leveraged with tissue-specific GPCR expression to identify functional consequences of GPCR–RAMP interactions relevant to the pathophysiology of disease. Key information needed to make targeting GPCR–RAMP complexes viable includes a strong understanding of how RAMPs regulate GPCR biology at the cell and system level. These insights can, in turn, be applied to rational drug design, usually in combination with cell-based screening strategies to create targeted therapeutics with minimal off-target effects.
3. Other Potential Associations Between RAMPs and Human Disease
CALCRL upregulation has been identified in promoting treatment resistance and increased stemness in transformed cells in acute myeloid leukemia (AML). As discussed in Grandits et al., there are conflicting reports as to whether AM, CGRP, or both are the relevant ligands contributing to the observed effect (Grandits and Wieser, 2021). Therefore, the expression and regulation of the three RAMPs in AML, and in different systems of studying AML such as cell lines and animal models, remains to be validated (Grandits and Wieser, 2021; Larrue et al., 2021).
Interestingly, a long noncoding RNA was found to be encoded on the antisense strand of RAMP2 and was denoted RAMP2-AS1. RAMP2-AS1 regulates endothelial cell homeostasis and may also play a role in cancer-related angiogenesis (Cheng et al., 2020; Hassani et al., 2021; Lai et al., 2021; Song et al., 2021; Li et al., 2022).
In a study on migration of CR6-interacting factor 1 (CRIF1)-deficient endothelial cells, CRIF1 expression was inversely correlated with mRNA levels of RAMP2, RAMP3, and AM2. Addition of exogenous AM2 led to increased expression of RAMP2, RAMP3, and AM2 in human umbilical vein endothelial cells (HUVECs). The authors posited that these findings could represents a mechanism by which AM2 compensates for CRIF1 deficiency (Nagar et al., 2021). Clark and colleagues focused on endogenous CALCRL in human cardiomyocytes and HUVECs and demonstrated that CALCRL exhibits RAMP-dependent signaling bias using multiple cellular readouts (Clark et al., 2021).
Though not studying the RAMPs directly, Han et al. have identified CALCRL as a biomarker for low-grade glioma prognostic risk and developed a model in which the expression of the gene for CALCRL was noted to be inversely correlated to risk score (Han et al., 2021). A different study, which also was not focused on the RAMPs directly, implicated AM in progression of severe acute respiratory syndrome coronavirus 2 infection. Kita and Kitamura described that AM administration correlated with reduction of inflammation in rodent models and reviewed clinical trials for coronavirus disease pandemic 2019 focused on therapeutics related to AM (Kita and Kitamura, 2022). AM-based therapeutics are also currently being investigated for treatment of irritable bowel disorder (Ashizuka et al., 2021). Patel et al. have created a compound mouse model of genetically depleted CALCR in an Alzheimer’s disease predisposition background to identify whether amylin receptor activation or blockage might be beneficial to treat or prevent Alzheimer’s disease. Their work suggests that expression of the amylin receptor is inversely correlated with spatial memory. Although the mechanism underlying this observation is not known, the authors posit a few explanations, such as the known connection between Alzheimer’s development and glia and brain vasculature alterations and the connection between vasculature and the amylin receptor. The results support the potential utility of developing amylin receptor antagonists as Alzheimer’s disease therapeutic agents (Patel et al., 2021).
A small cohort study on posttraumatic headache revealed correlations between headache burden postconcussion injury and particular single-nucleotide polymorphisms (SNPs) in the genes for RAMP1 and CGRP (La Fountaine et al., 2022). In an exome sequencing study of sporadic primary open angle glaucoma, six different point mutations in RAMP2 were identified in a cohort of 398 cases (Gong et al., 2019). The RAMP2 protein variants corresponding to the six somatic mutations were tested for localization and CALCRL-RAMP2 signaling in African green monkey kidney (COS)-7 cells in culture. If RAMP2 forms complexes with other receptors in retinal ganglion cells, then the RAMP2 point mutations might be affecting those interactions and contributing to the pathology through mechanisms that are not CALCRL-dependent. Prakash et al. looked for correlations between five SNPs in and around the RAMP3 gene in cohorts of 25- and 75-year-old women to try to identify correlations between RAMP3 and age-related body composition phenotypes (Prakash et al., 2019). They found that RAMP3 SNPs may play a minor role in increased age-related fracture risk and fat mass. The authors did not find any RAMP3-related differences in bone density.
II. Strategies to Identify GPCR–RAMP Interactions
Several methods have been developed to identify and quantify direct PPIs. However, measuring functionally relevant interactions between and among membrane proteins presents unique challenges (Fig. 3). While not exhaustive, the following section provides summaries of key studies and methodologies directly related to the problem of identifying GPCR–RAMP interactions (Table 2).
Fig. 3.
Schematic highlighting the most common methodologies used to identify GPCR–RAMP interactions.
TABLE 2.
Summary of experimental methods used to identify GPCR–RAMP interactions Studies are sorted chronologically.
| Study | Method | Looked for | GPCR–RAMP tested | Also looked at |
|---|---|---|---|---|
| McLatchie et al. (1998) | Expression cloning | Signaling in response to CGRP | Discovered CALCRL–RAMP1,2,3 | Ligand binding, FACS for RAMP and CALCRL surface expression, immunoblot (crosslinking, radioligand labeling, glycosylase treatment) |
| Christopoulos et al. (1999) | Radioligand binding | Increased binding of amylin and salmon CT upon RAMP expression | Discovered CALCR–RAMP1,2,3 | Competition assay for peptide radioligand binding, cAMP response, immunoblot (crosslinking), IF for RAMP1 localization |
| Christopoulos et al. (2003) | IF | Increased RAMP surface expression | Interacting: VIPR1–RAMP1,2,3 GCGR–RAMP2 PTH1R–RAMP2 PTH2R–RAMP3 Noninteracting: GHRH, VIPR2, GLP1R, GLP2R |
VIPR1 signaling (cAMP, PI), radioligand binding |
| Bouschet et al. (2005) | IF | Increased GPCR surface expression GPCR–RAMP colocalization |
Interacting: CaSR–RAMP1,3 Noninteracting: CaSR–RAMP2 |
Surface biotinylation, siRNA KD of RAMP1 (look at CaSR surface expression), co-IP, IF of GPCR with different cell compartment markers, GPCR glycosylation changes |
| Harikumar et al. (2009) | Fluorescence, BRET | Increased RAMP surface expression Saturating BRET signal that increases then plateaus. |
Interacting: SCTR–RAMP3 Noninteracting: SCTR–RAMP1,2 |
Bimolecular fluorescence complementation (look for fluorescence at the PM), assays with truncation and chimeric mutants of SCTR with WT or truncation mutants of RAMP, SCTR signaling (cAMP, Ca2+ flux, ERK1/2 phosphorylation) |
| Wootten et al. (2013) | ELISA | Increased RAMP surface expression Increased GPCR surface expression |
Interacting: VIPR2–RAMP1,2,3, CRHR1–RAMP2 Noninteracting: GLP1R |
G-protein binding to GPCR, GPCR signaling (cAMP, Ca2+ flux), in vivo experiment (measured plasma levels of adrenocorticotropic hormone in RAMP2+/− mice) |
| Lenhart et al. (2013) | IF BRET |
Increased RAMP surface expression and colocalization with GPCR, Saturating BRET signal that increases then plateaus |
Interacting: GPR30–RAMP3 | Co-IP, expression changes in vivo, localization, changes in vivo, in vivo experiment (studied cardiac fibrosis and left ventricular hypertrophy in RenTgMk; RAMP3+/+ and RAMP3−/− mice) |
|
Cegla et al. (2017) (Note: GPCR–RAMP interaction was previously published) |
IF | Changes in GPCR surface expression GPCR–RAMP co-localization |
GCGR–RAMP2 | Radioligand binding, GPCR signaling with RAMP2 overexpression or siRNA KD (cAMP, Ca2+ flux, β-arrestin1 recruitment) |
| Barbash et al. (2019) | MERFISH | Significant changes in GPCR expression at the mRNA level upon RAMP2 co-expression | 14 GPCRs: ADORA2B, S1PR1, NTSR1, OPRD1, F2RL3, GPR50, GPR141, GPR160, GPR176, LGR4, P2YR8, CELSR2, FZD1 (tested with RAMP2 only) | Bioinformatics comparison with phylogenic correlation coefficient |
| Bailey et al. (2019) | FACS ELISA |
Increased RAMP surface expression (FACS) Increased GPCR surface expression (ELISA) |
Interacting: CRHR1α, CRHR1β–RAMP2 Noninteracting: CRHR2β–RAMP2 |
GPCR signaling (cAMP), GPCR and RAMP expression (mRNA), molecular modeling of interaction interface |
| Lorenzen et al. (2019) | SBA | High median fluorescence intensity signal relative to control | Overview: 15 Secretin GPCRs plus ACKR3, ADGRF5, CCR5, CCR7, CXCR3, CXCR4, GPR4, GPR182 (with all 3 RAMPs) | PLA |
| Mackie et al. (2019) | BRET FACS |
Saturating BRET signal that increases then plateaus Increased RAMP surface expression |
Overview: 24 Chemokine GPCRs (with all 3 RAMPs) | PLA, co-IF with different biomarkers, ACKR3 signaling (cAMP), coculture scavenging activity assay, in vivo experiment (retinal angiogenesis in Admhi/hi, Adm+/+, Ackr3+/−, Ackr3−/−, RAMP3+/+, and RAMP3−/− mice) |
| Shao et al. (2022) | BRET IF |
Saturating BRET signal that increases then plateaus Altered RAMP surface expression and colocalization, |
Overview: 7 glucagon family GPCRs (with all 3 RAMPs) | GPCR signaling (cAMP, Gαq activation, β-arrestin1 and β-arrestin2 recruitment) |
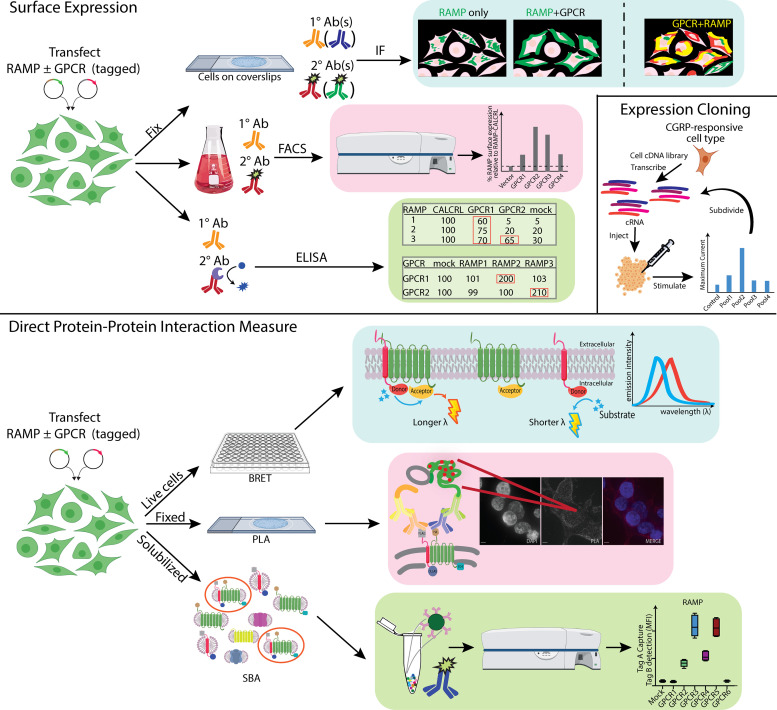
A. Expression Cloning
A cell-based expression cloning approach was used to identify the GPCR that signals in response to CGRP stimulation. This work led to the discovery of RAMP1, which was needed to form functional complexes with CALCRL (McLatchie et al., 1998). SK-N-MC cells are known to respond to CGRP stimulation, so to identify the CGRP-responsive receptor, SK-N-MC cRNA pools were injected into Xenopus oocytes and the oocytes’ response to CGRP stimulation was measured. The cRNA pool that corresponded to an elevated response was subdivided and tested further until RAMP1 was identified. No direct binding assays between CALCRL and RAMP1 were performed (although CGRP binding was measured with radionuclide-labeled ligand). However, McLatchie et al. observed that coexpression of CALCRL and RAMP1 led to a dose-dependent response to CGRP, increased trafficking of both proteins to the cell surface, complex formation as detected by immunoblot, and a change in the glycosylation pattern of CALCRL (terminal glycosylation) (McLatchie et al., 1998). Public database searches revealed the existence of RAMP2 and RAMP3. The authors then showed that RAMP2 and RAMP3 could each form an AM receptor when coexpressed with CALCRL.
B. Methods to Detect Changes in Surface Expression
The rationale behind using indirect, surface expression-focused methods to identify RAMP-interacting GPCRs is that the RAMPs, RAMP1 and RAMP2 in particular, have poor cell-surface expression on their own. The N-linked glycosylation patterns of the RAMPs vary. As introduced previously, RAMP1 has no N-glycosylation sites within its extracellular domain and the ER retention signal QSKRT within its C-terminal tail (Steiner et al., 2002). RAMP2 has one N-glycosylation site, whereas RAMP3 has four. RAMP2 and RAMP3 lack the QSKRT motif. Therefore, it is important to note that some GPCR-independent RAMP3 surface expression is possible and has been observed (Parameswaran and Spielman, 2006). As RAMP1 is not glycosylated, it is probably not translocated to the cell surface without interacting with another protein that is properly glycosylated (Bomberger et al., 2012). However, glycosylation can merely be a marker that a protein has traveled through the various intracellular compartments necessary for subsequent surface expression. It may not play a role in the function of a mature surface-expressed receptor, and it has been shown that functional GPCRs without any glycosylation can be generated (Reeves et al., 2002). Therefore, any potential functional role of glycosylation in RAMP surface trafficking must also be interpreted carefully. Overall, the different N-linked glycosylation patterns, and therefore abilities of the three RAMPs to translocate independently to the cell surface, must be considered when analyzing RAMP cell surface expression data.
The most commonly used techniques to identify GPCR–RAMP interactions that correlate with changes in surface expression are immunofluorescence (IF), fluorescence tag-based microscopy, ELISA, fluorescence-activated cell sorting (FACS), and immunoprecipitation (IP). Although these experimental techniques are the foundation for many GPCR–RAMP interaction discoveries, details such as the tags used and whether localization of the GPCR or RAMP or both are tracked vary widely. The traditional approach of monitoring changes in tagged RAMP surface expression upon coexpression with a GPCR is indirect but is still relevant in recent work. In some cases, co-overexpression of the RAMP and the RAMP-interacting GPCR changes RAMP surface localization, as illustrated by the examples described later. Most studies that report on RAMP surface localization do not simultaneously monitor the GPCR and therefore do not provide direct evidence for GPCR–RAMP complex formation.
Many of the first examples demonstrating a particular GPCR–RAMP interaction tested for changes in GPCR or RAMP surface expression and validated the putatively identified complex with other approaches. Christopoulos et al. transfected c-myc- or hemagglutinin (HA)- tagged RAMP ± GPCR, where the GPCR was either VIPR1, PTH1R, PTH 2 receptor (PTH2R), or GCGR, and looked for an increase in RAMP surface localization by IF. They noted that cellular background is an important factor (Christopoulos et al., 2003). Earlier, Christopoulos and colleagues identified the CALCR-RAMP1/2/3 interactions with a combination of experimental methods including radioligand binding assay (each of the three RAMPs) and IF of c-myc-tagged RAMP expressed in the presence or absence of CALCR (RAMP1 only) (Christopoulos et al., 1999). Cegla et al. studied GCGR in more detail using an ELISA-based method to measure GCGR surface expression in CHO cells stably expressing GCGR alone or with RAMP2 (Cegla et al., 2017). Bouschet et al. studied the glutamate family (class C) receptor CaSR with all three RAMPs using myc- (RAMP1) or HA- (RAMP2/3) tagged RAMPs. Uniquely, they used a pH-sensitive fluorescently tagged CaSR to monitor changes in both RAMP and CaSR surface expression with IF. Surface biotinylation assays were used to quantitate GPCR surface expression (Bouschet et al., 2005). IP was then used to validate the findings and provide additional information about trafficking. Harikumar and colleagues used COS cells transfected with yellow fluorescent protein (YFP)-tagged RAMPs, expressed with or without SCTR, and monitored changes in RAMP surface expression with fluorescence microscopy. They followed up with BRET-based studies and functional assays to provide additional evidence for the SCTR-RAMP3 interaction (Harikumar et al., 2009). Lenhart et al. studied whether RAMP3 interacts with GPR30 using HA-RAMP3 and FLAG-GPR30 and the IF technique, looking for GPCR–RAMP colocalization at the cell surface (Lenhart et al., 2013). They complemented their results from the microscopy experiments with other techniques including BRET. The authors also carried out IP and immunoblot experiments on fractionated cardiac lysates of mice and compared the amount of GPR30 in the membrane and cytosolic fractions of RAMP3+/+ and RAMP3−/− mice with a heart-disease prone genetic background.COS)
Wootten et al. transfected FLAG-tagged RAMPs ± HA-tagged GPCRs and looked at whether GPCR coexpression increased RAMP surface expression by ELISA for the receptors CRHR1β, GLP1R, and VIPR2. They also checked whether the RAMP increased GPCR surface expression, which is less commonly investigated (Wootten et al., 2013). In a study from 2019, Bailey et al. used HA-tagged versions of different CRHR1 subtypes, CRHR1α and β, and measured increases in FLAG-RAMP surface expression by FACS (Bailey et al., 2019). CRH receptor 2 (CRHR2) was also included in the study but was not epitope tagged. Examining RAMP2 cell surface expression, they showed that CRHR1α and CRHR1β coexpression with RAMP2 increased RAMP2 surface expression, suggesting that the receptor and RAMP interact. They also tested whether RAMP surface expression decreased after agonist stimulation using an ELISA method. Mackie et al. used FACS for “hit validation” after a BRET-based screen for RAMP-interacting chemokine receptors. With flow cytometry they measured changes in HA- and FLAG-tagged RAMP1, 2, and 3 surface expression upon GPCR coexpression (Mackie et al., 2019). The FACS-based RAMP surface expression data did not correlate well with the results from the BRET-based screen, suggesting that monitoring surface expression changes, by FACS and in general, may not be the most informative approach for identifying GPCR–RAMP interacting pairs (Mackie et al., 2019).
C. Recent Applications of Methods to Map the GPCR–RAMP Interactome
Unlike earlier studies that primarily focused on studying one GPCR at a time, recent reports have applied screening approaches to identify GPCR–RAMP interactions across entire subfamilies of GPCRs. The BRET-based screening approach noted previously was used to map the chemokine GPCR–RAMP interactome (Mackie et al., 2019) and the glucagon family GPCR–RAMP interactome (Shao et al., 2022). In a recent preprint (Harris et al., preprint, DOI: https://doi.org/10.1101/2021.04.08.436756) a BRET screen has also been used to map the secretin family GPCR–RAMP interactome. Applying a different technique, Lorenzen et al. developed a multiplexed immunoassay using a suspension bead array (SBA) for mapping the secretin family GPCR–RAMP interactome (Lorenzen et al., 2019). Notably, the SBA and BRET assays enable direct measurement of GPCR–RAMP interactions, and their respective strength and limitations are described in greater detail later. The results provide an insightful direct comparison of both approaches. In general, there is good but not perfect agreement in the results of BRET- and SBA-based approaches across the studies, which can serve as a cross-validation of the results. In the case of both BRET and SBA screens, the results should be interpreted as “high probability hits” that require validation by other methods.
Collectively, the GPCR–RAMP interactome screens reported so far have used FACS, IF, and proximity ligation assay (PLA) for hit validation. Within this subsection we discuss the in situ PLA, an orthogonal immunoassay used for hit validation. The PLA enables direct detection of GPCR–RAMP complexes in cell membranes and is semiquantitative. As mentioned, Mackie et al. also use a flow cytometry validation method to look at changes of surface expression of RAMP upon GPCR coexpression but, as discussed previously, measuring changes in surface expression is an indirect method with results that must be interpreted very carefully because a true GPCR–RAMP interaction might not always increase RAMP surface expression. For example, a RAMP may cause the retention of an interacting GPCR, or, for the case of RAMP3, there may already be RAMP present on the surface. Shao et al. looked at colocalization and changes in RAMP surface expression with IF to validate their findings (Shao et al., 2022).
1. Bioluminescence Resonance Energy Transfer
The bioluminescence resonance energy transfer (BRET) assay has been developed and improved extensively since it was described in 1999 (Xu et al., 1999). The most recent generation of BRET assay, called NanoBRET, uses a 19-kilo-Daltons (kDa) subunit of nanoluciferase (Nluc) that is more stable and yields brighter signal than other versions of luciferase (Hall et al., 2012; Machleidt et al., 2015; Dale et al., 2019; El Khamlichi et al., 2019). The principle behind BRET is that two proteins of interest are ectopically expressed in cells, with one protein fused to a luminescence “donor” and the other to a fluorescence “acceptor” that absorbs the resonance energy that is emitted by the donor. If the two proteins are sufficiently close and a chemiluminescence substrate is added, there will be a detectable signal of the “acceptor” fluorescence. Quantitation of BRET signal depends on the Förster distance that defines resonance energy transfer efficiency from the donor to the acceptor fluorophore and generally must be less than 10nm, which is the same magnitude as the size of a typical protein. BRET-based methods to study GPCRs were pioneered two decades ago, shortly after the development of the BRET assay (El Khamlichi et al., 2019; Angers et al., 2000).
Mackie et al. used a BRET-based screen to identify GPCR–RAMP interactions among the rhodopsin family (class A) chemokine receptors (Mackie et al., 2019). Twenty-four chemokine receptors were screened for interactions with each of the three RAMPs, with CALCRL-RAMP1/2/3 used as a positive control and the β2-adrenergic receptor (β2ADR)-RAMP1/2/3 as a negative control. A constant amount of an expression construct encoding for GPCR fused to Renilla luciferase (Rluc, donor) was transfected along with increasing amounts of an expression construct encoding RAMP fused to YFP (acceptor). Then, the BRET ratio was measured upon addition of coelenterazine h substrate. A BRET ratio threshold was applied to exclude noninteracting receptors with BRET ratios below the cutoff. Hits were further parsed by applying a best-fit comparison and classifying interactions that yielded BRET data that could be well described by a hyperbolic curve as true hits. Although Mackie and colleagues were the first to use BRET to screen for tens of GPCR–RAMP interactions, BRET assays have been applied previously for identifying and studying specific GPCR–RAMP interactions and are mentioned earlier in this review. For example, Lenhart et al. used BRET assays to show the interaction between GPR30 and RAMP3 (Lenhart et al., 2013). Earlier on, Harikumar et al. used BRET assays to test for interactions between SCTR and RAMP3 (Harikumar et al., 2009). Héroux and colleagues applied their expertise in BRET assays to study CALCRL-RAMP1 complexes (Héroux et al., 2007).
The most recent peer-reviewed BRET-based GPCR–RAMP interaction screen was conducted by Shao and colleagues and focused on seven glucagon family GPCRs: GCGR, GHRHR, splice variant 1 of GHRHR (SV1), GIPR, GLP1R, GLP2R, and SCTR. Like the earlier work, the authors transfected a constant amount of GPCR-Rluc8 (donor) with increasing amounts of RAMP fused to a variant of the acceptor YFP, called Ypet, and measured the BRET ratio upon addition of coelenterazine h substrate. Applying a similar hit classification criterion as Mackie et al., the authors postulated that out of the 21 possible GPCR–RAMP complexes tested, there were only four likely negative hits: GLP2R-RAMP1, GLP2R-RAMP2, GLP1R-RAMP1, and SV1-RAMP3. Shao et al., proposed an underlying structural mechanism for why GLP2R does not interact with RAMP1, as amino acids on GLP2R that are likely to be RAMP-interacting, based on the CALCRL-RAMP structures, would form repulsive interactions with RAMP1. Interestingly, the authors qualitatively validated the complexes identified with IF using HA-tagged receptor and FLAG-RAMP. Consistent with the previous discussion on the limitations of identifying GPCR–RAMP complexes through monitoring changes in surface expression, Shao et al. observed that in most cases RAMP surface expression was not significantly altered upon coexpression of an interacting GPCR, with the exceptions of SCTR-RAMP1, and SCTR, GCGR, or SV1 in complex with RAMP3. When SCTR was coexpressed with RAMP1, RAMP1 surface expression increased. However, when SCTR, GCGR, and SV1 were coexpressed with RAMP3, RAMP3 surface expression decreased compared with its surface expression when expressed alone (Shao et al., 2022).
Overall, there is good agreement between the BRET screen results described here and those obtained using the SBA method as described in more detail later (Lorenzen et al., 2019). An example of an exception to the good agreement is the putative complex between GHRHR-RAMP1, which was not identified by SBA. Table 1 lists the reported RAMP interactions for individual receptors across studies, enabling direct comparison of agreement across studies on a receptor-by-receptor basis. Although Shao and colleagues detected the GHRHR-RAMP1 interaction by BRET, they did not identify any functional consequences for complex formation in terms of GPCR-mediated cAMP production, Gq activation, or β-arrestin recruitment.
In the previously introduced Harris et al. preprint, the authors conducted a BRET-based screen to test for secretin family GPCR–RAMP interactions and showed that GIPR interacts with all three RAMPs. Like in the Mackie et al. study, Harris and colleagues also used flow cytometry to measure the surface expression of each RAMP coexpressed with the GPCRs to validate the BRET results (Mackie et al., 2019; Harris et al., preprint, DOI: https://doi.org/10.1101/2021.04.08.436756). The authors identified several GPCR–RAMP interactions either through the BRET screen, flow cytometry, or both but focused on GIPR for subsequent investigation. GIPR exhibited complex formation with the three RAMPs as determined by BRET-based assay. Further, coexpression of GIPR and each RAMP correlated with a significant increase of RAMP surface localization compared with RAMP expressed alone, assessed by flow cytometry. As in the other studies, the BRET-based assay results were not always in agreement with the flow cytometry data, again highlighting that RAMPs can modulate an interacting GPCR in many ways. The RAMPs are not necessarily always acting as chaperones from the ER to the PM for an interacting GPCR, as they are for CALCRL. Harris et al. suggest that not all GPCR–RAMP complexes traffic to the surface because they may be instead targeted for degradation or reside intracellularly. Still, the results from the BRET-based portion of the screen from this preprint are added to the list of published GPCR–RAMP interactions (Table 1). Here, we again see a generally good but not perfect agreement between the complexes identified by this BRET screen compared with those previously identified by SBA (Lorenzen et al., 2019).
BRET assays have multiple advantages, including their capacity to accommodate medium to high throughput analysis. As a BRET assay measures protein proximity and binding, it can, at least in theory, also be used to study the kinetics and dynamics of GPCR–RAMP complex formation and the effect of a ligand on complex stability. Moreover, BRET assays can be adapted to study cell membrane-specific interactions, as was done by Harris and colleagues. BRET-based assays are conducted in live cells, are scalable, and do not depend on the availability of validated antibodies (Abs) for every new target. One disadvantage of BRET assays is that they require ectopic expression of engineered RAMPs and GPCRs. It is possible that adding a C- or N-terminal BRET donor or acceptor, especially one of a larger molecular weight, might interfere with endogenous interactions or cause some artifacts, which is a concern for the Mackie et al. and Shao et al. studies that used YFP/YPet and Rluc. Effects of the luciferase tag on function have been somewhat ameliorated by the emergence of Nluc, since it is about one-half the size of Rluc8 (19.1 kDa vs. 36 kDa) and, depending on the chemiluminescent substrate used, up to 100-fold brighter. Moreover, Nluc-tagged GPCRs have been shown to traffic to the surface normally (Stoddart et al., 2015). Another disadvantage of BRET in general is that very high OE levels of the proteins being studied may cause artifacts. A recent paper has reported an application of CRISPR to facilitate “endogenous” BRET, which would address some of these pitfalls, but this approach is not yet in widespread use (White et al., 2017).
2. Multiplexed Suspension Bead Array
To circumvent the limitations in throughput of other approaches, Lorenzen et al. developed a multiplexed assay using the suspension bead array (SBA) platform and performed a proof-of-concept study to detect GPCR–RAMP PPIs on a larger scale than had been attempted earlier (Lorenzen et al., 2019). The SBA immunoassay detects GPCR–RAMP binding in a multiplexed format and is based on magnetic, color-coded beads that can be coupled to anti-RAMP or anti-GPCR specific Abs and subsequently read-out using a Luminex flow cytometer. In a single experiment, the SBA assay enables the determination of three modalities: Ab specificity, quantitation of target protein expression levels, and quantitative detection of the presence of GPCR–RAMP complexes. In this context, Ab specificity refers to affinity for the target receptor and lack of cross-reactivity with other receptors in the same subfamily, which tend to have the highest homology. Using dual epitope-tagged GPCR and RAMP constructs and mAbs targeting the four different tags allows for the measurement of a single interaction using up to eight different capture-detection schemes. This strategy serves as an immediate internal validation and increases the confidence in the results obtained.
Lorenzen et al. first used 23 dual-epitope-tagged secretin, adhesion, and rhodopsin subfamily GPCRs and three dual-tagged RAMPs to validate anti-GPCR Abs from the Human Protein Atlas (HPA) against 19 of the receptors studied (Uhlén et al., 2015). Developing specific anti-GPCR Abs presents a significant challenge: (i) it can be very difficult to purify high-quality, functional GPCRs to use as immunogen; (ii) most of a typical GPCR is hydrophobic and occluded in the plasma membrane or in a detergent-lipid micelle; (iii) the ECD of GPCRs can be poorly immunogenic; (iv) there is high homology among human and mouse GPCRs; and (v) there is high homology between closely related GPCRs such that anti-GPCR Abs tend to have high cross-reactivity (Hutchings et al., 2017). The HPA adopted a unique pipeline approach to systematically develop approximately 2,400 Abs for more than 600 GPCRs. The HPA uses 50 to 150 amino acid-residue long peptide immunogens to generate polyclonal Abs in rabbits (Uhlén et al., 2005). Lorenzen et al. used one of the SBA modalities to validate the selectivity of 55 anti-GPCR HPA Abs and found low cross-reactivity against all other tested overexpressed GPCRs for 31 of the Abs (Lorenzen et al., 2019).
Lorenzen and colleagues then used the SBA approach to study GPCR–RAMP interactions and showed that RAMP-interacting GPCRs generally either form complexes with all three RAMPs, or with RAMP2 and RAMP3. These findings are in line with previous bioinformatics work that suggested that RAMP1 and RAMP3 coevolved with a similar set of GPCRs that is distinct from RAMP2 and that RAMP1 and RAMP3 evolved less than RAMP2 (Benítez-Páez and Cárdenas-Brito, 2008; Barbash et al., 2017). The GPCR–RAMP complexes detected by the SBA are consistent with most of the earlier “indirect approach” findings. The SBA also revealed that there are several additional secretin receptors, rhodopsin family orphan receptors, and chemokine GPCRs that can form complexes with RAMPs. As previously noted, the data from a single experiment is highly multiplexed; therefore, it is possible to generate very high confidence “hits” from just one microtiter plate of expressed GPCRs and RAMPs. The SBA can achieve multiple aims within the same experiment, including detection of PPIs and simultaneous validation of Abs. The validated Abs can then be used to detect specific GPCR–RAMP interactions by SBA without depending on epitope tags. While Lorenzen et al. used ectopically expressed GPCR and RAMP constructs, which represents a potential limitation to the SBA, once Abs are validated, the SBA can be applied to study endogenous GPCR–RAMP interactions in cells and tissues if sensitivity is adequate. The flip side to the potential strength of the SBA approach is that validated Abs are required for endogenous PPI studies. Conversely, using epitope-tagged constructs, while cumbersome, can enable the capture of a whole library of tagged GPCRs onto SBA beads. The SBA approach is also scalable to high throughput.
3. Proximity Ligation Assay
The proximity ligation assay (PLA) is an immunolocalization assay that was successfully used to verify the results of the aforementioned SBA assay with five GPCR–RAMP pairs (Lorenzen et al., 2019). The PLA was also used to validate a newly identified interaction between ACKR3 and RAMP3 (Mackie et al., 2019). The PLA is an immunoassay with stringent distance constraints. It relies on special oligonucleotide-conjugated Ab probes that bind to two primary Abs from different species, and that in turn are bound to two potentially interacting proteins (Söderberg et al., 2006). Previously, PLA has been applied to detect endogenous GPCR heterodimers in both cells and tissues with primary Abs targeting the native GPCR (Gomes et al., 2016). Lorenzen et al. used the PLA with Abs targeting FLAG and HA N-terminal epitope tags engineered onto the respective receptor and RAMP constructs and validated five GPCR–RAMP2 complexes detected by SBA. Mackie et al. also used the same basic PLA approach to validate the ACKR3-RAMP3 interaction in situ. The DuoLink PLA detects PPIs that are up to 40-nm apart, a maximum distance determined by the size of the oligonucleotide-conjugated Abs and the length of the connector oligonucleotide that serves as part of the template for rolling circle amplification (sigmaaldrich.com). For comparison, the interreceptor distance between two GPCRs in a dimeric complex is about 4.5 nm, and an IgG Ab has a diameter of about 10 nm (Gurevich and Gurevich, 2008). PLA can be applied to study endogenously expressed proteins if there are verified Abs available for both protein targets. PLA assay kits with the proprietary DuoLink probes have been used for both the Lorenzen et al. and Mackie et al. GPCR–RAMP screening studies. Recently, the company Navinci Diagnostics (www.navinci.se) has developed its own proprietary system with an additional reaction step that is purported to confer superior sensitivity (Klaesson et al., 2018).
One potential disadvantage of the PLA is low throughput. Individual samples such as cells or tissue slices must be mounted on coverslips for PLA processing and imaged with confocal or deconvolution-based fluorescent microscopy. In theory, PLA is amenable to flow cytometry, and DuoLink does offer a flow-adapted PLA kit. Scaling up through use of micro-titer plates could also be possible. A 384-well-based PLA screen was recently performed to identify compounds that affected integration of tau and bridging integrator 1, a genetic risk factor for Alzheimer’s disease (Mendes et al., 2020). However, there are some barriers to implementation, and this level of throughput for PLA is not common so far. Moreover, multiplexing the PLA with many colors is limited by the nature of the PLA probes. The PLA probes and reaction components are proprietary, which could also be considered a disadvantage to the technique. The PLA requires specific and functional Abs against the target proteins, in this case the GPCRs and RAMPs, to study the interactions between endogenous proteins. The use of the SBA approach to validate anti-RAMP and anti-GPCR Abs could enable the parallel advancement of PLA strategies, as Abs validated by SBA assay could potentially be applied to “endogenous” PLA in cells or tissues expressing potential GPCR–RAMP pairs of interest.
D. Computational Approaches to Identify GPCR–RAMP Interactions
There have been relatively few studies using computational approaches to identify or validate hypothesized RAMP-interacting GPCRs. Using a global coexpression and coevolution analysis, Barbash and colleagues showed that GPCRs and RAMPs are globally coexpressed and likely coevolved, suggesting that GPCR–RAMP interactions should be widespread among the superfamily of GPCRs (Barbash et al., 2017). The authors did not discuss hypotheses about mechanisms of specific GPCR–RAMP interactions, but their work clearly points to specific GPCR subfamilies that might be most likely to interact with RAMPs. In a follow-up study, Barbash et al. selected 14 GPCRs based on their original phylogenetic analysis and measured changes in GPCR mRNA levels upon RAMP2 coexpression using MERFISH (Barbash et al., 2019). The results agreed with the original bioinformatics analysis, thereby strengthening the hypothesis of widespread GPCR–RAMP interactions.
III. Molecular Characterization of GPCR–RAMP Interactions
There have been numerous recent reports of structures of GPCR–RAMP complexes (Table 3). There are also several recently solved structures of GPCRs that are now known to interact with RAMPs, although the RAMP is not a part of the reported structure. Published structures inform MD simulations and other computational investigations. Static structures provide useful information but do not give the whole picture, as there is mounting evidence that RAMPs affect key aspects of GPCR structural dynamics. These themes are discussed in more detail next. Mutagenesis continues to be particularly valuable where elements of a receptor structure are poorly resolved.
TABLE 3.
Published structures with RAMPs Asterisk (*) indicates structure obtained by cryo-electron microscopy. Otherwise, structures obtained by X-ray crystallography. All structures marked Full include the ECD and TM domains of the RAMP, with no or very poor C-terminal density observed. Stabilizing mutations made to the RAMP and/or GPCR are not listed but may be present.
| GPCR | RAMP | Full length? | Relevant molecules present | Other molecules present | PDB Ref | Ref |
|---|---|---|---|---|---|---|
| None | RAMP1 | ECD only | MSE | 2YX8 | Kusano et al. (2008) | |
| None | RAMP2 | ECD only | Ca+2 | 2XVT | Not published (Quigley, 2010) | |
| None | RAMP2 | ECD only | MSE | 3AQE | Kusano et al. (2012) | |
| CALCRL | RAMP1 | ECD (both RAMP and GPCR) | Olcegepant | 3N6, 3N7, sulfate ion | 3N7S | ter Haar et al. (2010) |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | AM variant | Maltose | 5V6Y | Booe et al. (2018) |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | AM2 | Maltose, sodium ion | 6D1U | Roehrkasse et al. (2018) |
| CALCRL | RAMP1 | ECD + TM (no C terminus), CALCRL full length | CGRP, Gs heterotrimer | Nb35 | 6E3Y* | Liang et al. (2018a) |
| CALCRL | RAMP1 | ECD (both RAMP and GPCR) | Telcagepant | 3N6, N7R | 3N7R | ter Haar et al. (2010) |
| CALCRL | RAMP1 | ECD (both RAMP and GPCR) | (unliganded) | sulfate ion, MSE | 3N7P | ter Haar et al. (2010) |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | CGRP analog | Maltose, magnesium ion | 4RWG | Booe et al. (2015) |
| CALCRL | RAMP1 | ECD (both RAMP and GPCR) | Erenumab (Fab) | 6UMG | Garces et al. (2020) | |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | Olcegepant | SN6, tetraethyl glycol (PG4), maltose, alpha-D-glucopyranose | 6ZIS | Bucknell et al. (2020) |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | HTL22562 | tetraethyl glycol (PG4), maltose, alpha-D-glucopyranose | 6ZHO | Bucknell et al. (2020) |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | HTL0028125 (macrocycle) | Unknown (PDB entry on hold) | 7P0F | Cansfield et al. (2022) |
| CALCRL | RAMP1 | MBP–RAMP1 ECD–CALCRL ECD fusion | Macrocycle compound 13 | Unknown (PDB entry on hold) | 7P0I | Cansfield et al. (2022) |
| CALCRL | RAMP1 | ECD + TM (no C terminus), CALCRL full length | Detergent micelle | 7KNT* | Josephs et al. (2021) | |
| CALCRL | RAMP1 | ECD + TM (no C terminus), CALCRL full length | CGRP | Detergent micelle | 7KNU* | Josephs et al. (2021) |
| CALCRL | RAMP2 | ECD only | MSE | 3AQF | Kusano et al. (2012) | |
| CALCRL | RAMP2 | MPB-RAMP2 ECD–CALCRL ECD fusion | AM | Maltose, 1,2-ethanediol | 4RWF | Booe et al. (2015) |
| CALCRL | RAMP2 | MPB–RAMP2 ECD–CALCRL ECD fusion | high-affinity AM (37-52) S45R/K46L/S48G/Q50W | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose, formic acid, amino group | 6V2E | Booe et al. (2020) |
| CALCRL | RAMP2 | Full | AM, Gs heterotrimer | Nb35 | 6UUN* | Liang et al. (2020a) |
| CALCRL | RAMP3 | Full | AM2, Gs heterotrimer | Nb35 | 6UVA* | Liang et al. (2020a) |
| CALCRL | RAMP3 | Full | AM, Gs heterotrimer | Nb35 | 6UUS* | Liang et al. (2020a) |
| CALCR | RAMP1 | Full | Rat amylin, Gs heterotrimer | Nb35, P42, phosphatidylethanolamine, cholesterol hemisuccinate, palmitic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose | 7TYF* | Cao et al. (2022) |
| CALCR | RAMP1 | Full | Salmon CT, Gs heterotrimer | Nb35, cholesterol hemisuccinate, palmitic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose | 7TYW* | Cao et al. (2022) |
| CALCR | RAMP2 | Full | Rat amylin, Gs heterotrimer | Nb35, cholesterol hemisuccinate, palmitic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose | 7TYX* | Cao et al. (2022) |
| CALCR | RAMP2 | Full | Salmon CT, Gs heterotrimer | Nb35, cholesterol hemisuccinate, palmitic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose | 7TYY* | Cao et al. (2022) |
| CALCR | RAMP2 | Full | CT, Gs heterotrimer | Nb35 | 7TYH* | Cao et al. (2022) |
| CALCR | RAMP3 | Full | Rat amylin, Gs heterotrimer | Nb35, P42, cholesterol hemisuccinate, palmitic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose | 7TZF* | Cao et al. (2022) |
MBP, maltose binding protein; MSE, selenomethionine, an L-peptide linking molecule; Nb35, Nanobody 35.
A. Mutagenesis Approaches to Study GPCR–RAMP Interactions
Site-directed mutagenesis was frequently used to determine key amino acid residues in RAMPs that are responsible for their functional effects on GPCRs (Gingell et al., 2010). The technique is now mostly used to complement other approaches, such as cryo-EM structural determination and MD simulations. For example, Qi et al. used in silico alignment of the RAMPs followed by functional characterization of mutants to determine that position 74 in RAMP1 and RAMP3 is important for AM pharmacology (Qi et al., 2008). Woolley et al. used targeted CALCRL mutagenesis along with MD simulations to determine residues that affect signaling of the CRGPR (Woolley et al., 2017). They determined which CALCRL alanine mutations affected CALCRL-RAMP1 expression, CGRP or AM ligand binding, and G protein-mediated signaling.
Gingell et al. coupled alanine mutagenesis functional studies with modeling of the AMY1 receptor, which is discussed in more detail later, to identify key residues for ligand potency and selectivity (Gingell et al., 2016). Lee and colleagues performed mutagenesis on RAMP2 and an amylin analog called AC413 with a fluorescence polarization readout to provide evidence for the interaction between specific residues on the peptide and RAMP2 (Lee and Pioszak, 2020). Sun et al. used mutagenesis to study GLP2R, and Liang et al. complemented their recently solved structures of CALCRL-RAMP2 and CALCRL-RAMP3 with mutagenesis studies of the RAMP linker regions (Liang et al., 2020a; Sun et al., 2020). Another recent cryo-EM “companion paper” focused on alanine scanning mutagenesis of AM (Garelja et al., 2020). Pham et al. studied the AMY3 receptor with alanine scanning mutagenesis of the CALCR ECL2 and ECL3 (Pham et al., 2019). Functional readouts of surface expression, peptide binding, cAMP accumulation, and ERK phosphorylation revealed peptide ligand-dependent differences in the roles of the loops.
Unnatural amino acid incorporation is another mutagenesis approach that may yield insights into important sites and residues of a GPCR. This strategy is also known as genetic code expansion and is based on amber codon suppression using an orthogonal tRNA and amino acyl-tRNA synthetase pair engineered to recognize an amber codon introduced into a transfected gene of interest and incorporate a particular unnatural amino acid. Ye et al. first employed the strategy for studies of GPCRs (Ye et al., 2008). Specific unnatural amino acids can be introduced to facilitate “targeted photo-crosslinking” or bio-orthogonal covalent labeling with small-molecule fluorophores to create FRET or BRET conformational sensors (Koole et al., 2017; Kowalski-Jahn et al., 2021). Simms and colleagues employed a targeted photo-cross-linking strategy to study the ECL2 of CALCRL and identified two major contact points for CGRP, I284 and L291 (Simms et al., 2018). For earlier, more complete reviews that have a focus on RAMP mutagenesis, we recommend these sources Qi and Hay (2010) and Hay and Pioszak (2016).
B. Recent GPCR–RAMP Complex Structures and Molecular Dynamics Simulations
1. Insights From Recently Solved Three-Dimensional Structures
Over the past decade, high-resolution X-ray crystallography and cryo-EM with single-particle reconstruction, along with high-performance computational approaches, including long-time-scale all-atom MD simulations, have provided significant insights into the molecular mechanism of signaling by GPCRs. Several GPCR structures have been reported in complex with RAMPs, and these provide important validations for the existence of specific GPCR–RAMP complexes. All currently available RAMP structures and costructures are listed, along with references and PDB accession codes, in Table 3. Ideally, structures would provide insights about GPCR–RAMP subtype specificity determinants. GPCR–RAMP structures might also be expected to reveal a mechanism to explain how certain RAMPs affect ligand-binding specificity and selectivity (Fig. 4). Overall, the structures available to date show that the presence of the RAMP has only relatively subtle effects on the structure of the GPCR in its respective GPCR–RAMP complex. Additional structural studies will help to reveal more about the mechanism of regulatory effects of RAMPs on GPCR pharmacology. Earlier studies with chimeric ECD proteins showed that both RAMP1 and RAMP2 have ECDs with a three-helix bundle fold and have similar interactions with CALCRL. Because RAMPs make only minimal contacts to a given agonist, the molecular mechanism by which RAMPs affect ligand binding to CALCRL, based on current information, is mostly through allosteric shaping of CALCRL conformation.
Fig. 4.
Structure of CALCRL in complex with RAMP1 and CGRP obtained from cryo-EM (PDB code: 6E3Y) (Liang et al., 2018). Center panel: CALCRL, rainbow color gradation from deep blue (N-terminal tail) to deep red (C-terminal tail); RAMP1, magenta; CGRP, gray. The original published structure also included the bound G protein, which has been removed here for clarity. The N-terminal tails and the ECDs of CALCRL and RAMP1 are at the top, while their C-terminal tails and intracellular domains (ICDs) are at the bottom. The C-terminal residues of RAMP1 that stretch beyond the membrane bilayer are not resolved (dashed line). All residues of RAMPs that interact with CALCRL as determined by all recent cryo-EM structures are shown as sticks in the center and right-side insets (Liang et al., 2018; Liang et al., 2020a; Josephs et al., 2021). Similarly, residues of CALCRL that may interact with either RAMP1, 2, or 3 are shown as sticks in the center panel and right-side insets. In the right-side insets, any atoms between CALCRL and RAMP1 that are within 4 Å of each other are marked with yellow dashed lines. The left-side boxed structure shows interacting residues as spheres to highlight the potential CALCRL–RAMP interaction interface. All RAMP amino acid residues with potential interactions with CALCRL are shown as magenta spheres. Amino acid residues in the TM, ECL, and ECD regions of CALCRL that potentially interact with RAMPs are shown as blue spheres.
A cryo-EM structure at a global resolution of 3.3 Å of a complex including RAMP1, CALCRL, CGRP, and a Gs protein heterotrimer defined the interaction of the TM domains and confirmed findings from previous crystal structures of the ECDs (Liang et al., 2018). RAMP1 forms extensive contacts with CALCRL, causing approximately 23% of the RAMP1 surface to be buried. The TM domain of RAMP1 nestles into an interface between TM helices 3, 4, and 5 of CALCRL, and the ECD of RAMP1 interacts with the ECD and ECL2 of CALCRL. As seen in prior crystal structures, there are minimal contacts between RAMP1 and the agonist CGRP. Based on the structural models and complementary MD simulations, which are discussed in more detail in the next section, the authors postulated that RAMP1 is stabilizing the ECD and ECL2 of CALCRL, which promotes CGRP binding to the complex. This conclusion, which is relatively self-evident from the structure itself, requires further refinement to provide more general insights about the effects of RAMPs on GPCR ligand binding characteristics.
The full-length cryo-EM structures of RAMP2 in complex with CALCRL, AM, and Gs, and two structures of CALCRL-RAMP3-Gs, with either AM or AM2 bound to the activated complex, have also been solved recently (Liang et al., 2020a). Across all structures, there is minimal contact between any of the ligands and any of the RAMPs, again highlighting that RAMPs modulate GPCR biology and pharmacology allosterically (Pioszak and Hay, 2020). The authors found that the identity of the complexed RAMP affects the orientation of the ECD of CALCRL relative to the receptor core and that CALCRL-RAMP2 had greater motion of its ECD overall compared with that of CALCRL-RAMP3. The RAMP also alters the kinking of TM6 in CALCRL, the conformation of its intracellular loop (ICL) 2, and the positioning of its ECL3. CALCRL-RAMP1 (active, CGRP bound) exhibited the most different and dramatic ECD rotation of the complexes, although the ECDs across all structures were highly dynamic and therefore of lower resolution.
The use of so-called cryoSPARC software to perform multivariate analysis of the cryo-EM data suggested that the different RAMPs affected the GPCR ECD mobilities in subtly different ways. Since a component of the motion of the GPCR ECD and the bound G protein occur in a coordinated manner, the RAMP may be indirectly influencing GPCR-G protein interactions. Interestingly, it appears that the C-terminal tail of RAMP3, but not RAMP2, makes transient contacts with the G protein. In both the 2018 and 2020 Liang et al. studies there was limited, or no density observed for the C-terminal tail of CALCRL and the RAMP. Considering other known CALCRL PPIs, the authors suggested that one implication of RAMP-dependent ICL2 orientation might be differences in the CALCRL-RCP interaction and therefore G protein signaling. The authors proposed a critical role for the RAMP “linker” region, which connects its TM and extracellular N-terminal domain, for exerting RAMP-specific stabilizing effects on the CALCRL extracellular regions. To test this hypothesis experimentally, they created a series of chimeric RAMPs, exchanging different portions of the linker regions in the three RAMPs, and then tested CALCRL-RAMP G protein-mediated signaling in response to CGRP, AM, and AM2. Linker exchange affected signaling to varying degrees, with the results indicating that the RAMP linker contributes to the allosteric modulation imparted by the RAMPs, perhaps through different intracellular interactions that alter receptor dynamics.
A companion paper to Liang et al. (2020a) focused on extensive alanine scanning mutations of AM (Garelja et al., 2020). The authors characterized CALCRL-RAMP signaling profiles for the unmodified AM peptide and peptides with single alanine substitutions, revealing AM residues that are critical for function. Good agreement between known ligand–receptor interactions and functional aberrations upon mutation of an involved amino acid residue highlights that mutagenesis and functional characterization studies are an important way to confirm and contextualize structure-based findings.
To complement the active, ligand-bound structures obtained by Liang and colleagues in 2018 and 2020, Josephs et al. recently published the cryo-EM structure of unmodified apo-CALCRL-RAMP1 and unmodified CGRP-bound CALCRL-RAMP1 without transducer protein bound. To assess the conformational dynamics of the complexes, the authors conducted a cryoSPARC multivariate analysis on the cryo-EM data and performed hydrogen-deuterium exchange mass spectrometry (HDX-MS) experiments (Josephs et al., 2021).
In many published structural studies, the protein of interest is commonly modified to increase stability at the expense of other potential native constraints, thereby altering native dynamics. As highlighted by the previously discussed publications, RAMP-mediated effects on GPCR dynamics are important to consider, so obtaining cryo-EM data on unmodified GPCR–RAMP complex represents a key advance in understanding native complex dynamics. Josephs and colleagues aimed to provide insights into RAMP-CALCRL activation and showed that the effect of RAMP1 on CALCRL dynamics plays an important role in initiating the activation process after CGRP C-terminal tail binding. Comparing the two CALCRL-RAMP1 structures, the apo complex with the CGRP bound-inactive complex, revealed that the ECD of RAMP1 differed significantly in relative position in the apo structure compared with that in the CGRP-bound structure but the ECD of CALCRL did not. Interactions between residues in the RAMP1 linker and CALCRL ECL2 are stronger in the CGRP-bound structure, suggesting that the RAMP is stabilizing active or active-like conformations.
Akin to the previous cryo-EM structures, there was low resolution or no density for the CALCRL and RAMP1 C-terminal tails and portions of the CALCRL ECD, ECL3, and ICL3. Notably, the density of the RAMP1 linker region was too low for confident side chain assignment. HDX-MS studies of apo and CGRP-bound complex dynamics agreed with the complementary three-dimensional variance analysis of the cryo-EM data. Interestingly, both approaches showed that the RAMP1 ECD and C-terminal tail were highly dynamic in the apo structure and that the RAMP ECD was largely stabilized upon CGRP binding, whereas the C-terminal tail increased in mobility. The authors proposed a model for CGRP binding and activation in which binding of the peptide C-terminal tail stabilized the dynamic RAMP1 ECD, promoting the interaction of RAMP1 with the ECL3 of CALCRL. The resulting stabilization of ECL3 promotes the dynamic motion of the intercellular facing portion of CALCRL, facilitating G protein binding. Engagement of the transducer promotes the numerous structural rearrangements associated with a fully active CALCRL-RAMP1 complex and binding of the CGRP N-terminus deep within the TM7 cavity.
The cryo-EM structures of the three AMY receptors (CALCR-RAMP1/2/3) have been solved in complex with Gs and either rat amylin (CALCR-RAMP1/2/3), salmon CT (CALCR-RAMP1/2), or human CT (CALCR-RAMP2) (Cao et al., 2022). These six structures have been reported alongside three structures of CALCR (no RAMP) bound to Gs and each of the abovementioned three peptide ligands. This study supports the findings that amylin and CT agonists bind and activate CALCR through distinct mechanisms, and the differences may be attributed to characteristics of the peptide agonists and to allosteric modulation by the RAMPs.
Overall, the presence of the RAMP had little effect on the CALCR core in rat amylin-bound structures. In all three rat amylin-bound AMY receptors there was a 12 Å rigid-body translation of the CALCR ECD when compared with the location of the ECD in CT-bound CALCR with no RAMP present. The RAMPs did not have a pronounced effect on the TMD and ECLs of CALCR, but there were subtle differences in the conformation of ICL2. Accompanying analysis of structure dynamics showed that all three RAMPs made contacts with the αN helix of Gαs, but with varied strength, dynamics, and relative positioning to ICL2. Therefore, the RAMPs may differentially affect G protein coupling efficiency.
The structural and dynamic consequences of RAMP2 complex formation with CALCRL were most distinct from those of RAMP1 and RAMP3, thereby potentially explaining the different ligand-binding characteristics and selectivity of each AMY subtype. RAMP1 and RAMP3 formed a much more robust TM interface with CALCR than RAMP2, thereby conferring a higher degree of stability to the CALCR ECD for AMY1 and AMY3. Further, RAMP1 and RAMP3, but not RAMP2, form stabilizing interactions with CALCR ECD loop 5, which may in turn stabilize the proximal rat amylin residues. The authors also use dynamics analysis to show that the weak RAMP2 TM–CALCR TMD interaction contributes to decreased complex stability within both the TMD and ECD, weaker amylin potency, and stronger CT potency. Conversely, RAMP2 and RAMP3, but not RAMP1, may form transient polar contacts with tyrosine 37 of rat amylin.
The identity of the peptide ligand did not have a pronounced effect on the CALCR-RAMP interface, but the RAMPs allosterically modulated ligand selectivity of CALCR. The selective agonist rat amylin preferentially bound CALCR when its ECD was stabilized by the RAMPs, as this enabled the peptide to adopt a unique so-called bypass motif. Conversely, salmon CT, a nonselective agonist, appeared to stabilize the CALCR-RAMP interface. Cao and colleagues postulate that the increased stability of AMY1 and AMY3 corresponds to a higher activation energy that salmon CT must overcome to bind and activate the complex, and therefore it has higher potency for CALCR and AMY2 (Cao et al., 2022).
Similar to the CALCRL-RAMP structures, there were no direct interactions observed between each RAMP and the peptide N-terminus but some hydrophobic interactions with the C-terminus. As is the case with CALCRL-RAMP complexes, the RAMP interacts with TM3, 4, and 5 and make extensive contact with ECL2 of CALCR. Unlike the CALCRL-RAMP structures, in which each unique complex had different GPCR and RAMP ECD orientations and locations, CALCR-RAMP complexes exhibited only subtle differences in RAMP ECD orientation relative to the CALCR ECD. Another contrast is that the RAMP linker region was more stabilized in the AMY receptors than in CALCRL-RAMP complexes. As in the CALCRL-RAMP structures, there was limited or no density for the RAMP linker region and portions of the C-terminal tail. The effect of each RAMP on conformational dynamics is a key contributor to the allosteric modulation imparted by the RAMPs on both CALCRL and CALCR.
Cryo-EM has enabled the determination of multiple different CALCRL-RAMP and CALCR-RAMP structures in the span of just a few years. This powerful technique has also highlighted the importance of understanding the dynamics of GPCR–RAMP interactions, which play a vital role in how the RAMPs affect the pharmacology of interacting receptors.
2. Computational-Based Insights Into GPCR–RAMP Dynamics
Multiple groups have carried out MD simulations and other computational approaches, taking advantage of solved structures to generate hypotheses regarding how RAMPs may alter GPCR dynamics. Liang and colleagues used the CALCRL-RAMP1 cryo-EM structure as the basis for simulations that indicated that RAMP1 is stabilizing the positioning of the CALCRL ECD, thereby increasing the stability of the C-terminal region of CGRP (Liang et al., 2018). In general, they observed that the regions of lower resolution or lacking cryo-EM density (i.e., RAMP C-terminus, CALCRL ECL3) in their solved structure are predicted to have high mobility in MD simulations, indicating that dynamics play an important role in GPCR–RAMP interactions and complex stability. Modeling predicted that the C-terminal tail of RAMP1 interacts transiently with ICL2 of CALCRL and the αN helix of the Gαs subunit. There were no persistent interactions observed between RAMP1 and the ligand CGRP. Simulations of CGRP-CALCRL without bound RAMP1 revealed increased ECD dynamics and decreased persistence of key intermolecular interactions within CALCRL that are thought to contribute to signal propagation.
Bower and colleagues modeled a full-length AMY1 receptor with amylin bound to test the importance of the C-terminal amino acid sequence of amylin for binding to CALCR in the presence of RAMP. Their simulations showed that the ligand-interacting residues of CALCR were RAMP1-dependent and that RAMP1 affected the number and persistence of intermolecular interactions within CALCR and between CALCR and amylin. The presence of RAMP1 also affected amino acid bond angle values for CALCR and amylin. The amylin-CALCR binding pathway was stabilized by RAMP1, especially for the C-terminal amide form of the peptide (Bower et al., 2018). Overall, there is good agreement between the findings of Bower et al. and the recently solved AMY1 structure.
As introduced previously, Bailey et al. studied the secretin family GPCRs CRHR1 and CRHR2 and experimentally identified putative CRHR1-RAMP2 interactions. The authors then identified a potential contact interface between RAMP2 and CRHR by molecular modeling of the extracellular portions, thus supporting their experimental findings (Bailey et al., 2019). On the other hand, a study by Tasma et al. investigating biased signaling mediated by ligands of CRH receptors found that CRHR1 or CRHR2 expression had no effect on RAMP1 or RAMP2 surface expression (Tasma et al., 2020).
In line with these studies, others have shown that the presence of a RAMP affects GPCR flexibility and dynamics. To highlight a few of these works, Weston and colleagues focused on the RAMP-dependent G protein-signaling bias (Gs vs. Gi versus Gq/11) of activated CALCRL-RAMP complexes (Weston et al., 2016). Gingell et al. conducted MD simulations showing that the CALCR N-terminal tail as well as the ECD loop 4 and EDC loop 5 are more flexible in the presence of RAMP1. The decreased rigidity of the receptor may be exploited by ligands such as amylin (Gingell et al., 2016). The results of this study are consistent with those from later work by Deganutti and colleagues, which drew on the multitude of recently published cryo-EM structures of secretin family GPCRs to computationally interrogate ligand binding to CALCRL (Deganutti et al., 2021). Deganutti et al. used a combination of supervised MD and classic MD simulations to study the second step in the proposed “two-step binding mechanism” of a ligand to a class B GPCR, namely the binding of the peptide ligand N-terminal tail to the receptor TM domain. Their study was the first example of dynamic docking of CGRP and the small molecule antagonist telcagepant to CALCRL-RAMP1. The study identified residues in CALCRL ECD loop 4 as playing an important role in peptide association, and the authors speculated that different RAMPs may promote divergent ECD loop 4 states, thereby allosterically modulating the affinity of CALCRL to different peptides. Consistent with previously published structures, very few interactions were formed between CGRP and RAMP1 in the docking. Also consistent with recent findings, differences in the dynamics of the RAMP linkers may affect selectivity.
As mentioned, Woolley et al. also used a combined experimental and computational approach to study the activation of the CALCRL-RAMP1 complex (Woolley et al., 2017). CALCRL mutants that had a significant difference in a parameter such as signaling compared with WT were then analyzed by MD simulation. The authors found that mutation of certain residues at the extracellular face of the TM bundle affected signaling in a ligand- and RAMP-dependent manner and that tighter CALCRL TM packing correlated with higher ligand potency. They proposed that the ECLs of CALCRL play an important role in the ligand binding and the subsequent activation process, a hypothesis that is further supported by the recent structure and dynamics studies discussed here. The authors also found that certain CALCRL residues were involved in receptor activation in a RAMP-independent manner, again consistent with later insights into the CALCRL-RAMP1 interface (Liang et al., 2018). Woolley and colleagues also used computational modeling to compare their modeled CALCRL-RAMP to the cryo-EM structure of CALCR solved in the absence of RAMP (the CALCRL-RAMP1 structure was not solved yet at that time). It appears that the RAMP caused reorganization of TM1, TM6, and TM7 to restrict the outward movement of the top of TM6, and therefore ECL3, in CALCRL-RAMP1 relative to CALCR.
The previously mentioned study by Pham et al. of the AMY3 receptor included MD simulations that accompanied the mutagenesis experiments (Pham et al., 2019). The simulations showed that ECL2 and ECL3 loop dynamics are highly dependent on RAMP3, further supporting an allosteric mechanism by which RAMPs regulate an interacting receptor. Overall, computation modeling suggests that the dynamics of CALCRL ECL2, ECL3, and ECD loop 4 are highly RAMP-dependent.
IV. Perspectives and Future Directions
A. Drug Discovery and GPCR–RAMP Pharmacology
Several outstanding gaps in knowledge about GPCR–RAMP interactions hamper our full understanding of GPCR-mediated pharmacology. Filling these gaps is important for both basic and translational research, especially since many GPCRs remain orphan. Identification of more RAMP-interacting GPCRs may enhance deorphanization efforts since ligands could be identified for GPCR–RAMP complexes that do not bind to the same GPCR in the absence of a RAMP. Although approximately one-third of current FDA-approved drugs target GPCRs, many difficult or refractory GPCR drug targets remain undrugged. Small molecules targeting GPCRs still have a relatively high failure rate in advanced clinical trials (Hauser et al., 2017). Advancing our understanding of how RAMPs regulate GPCRs might help to address this problem. One notable set of examples illustrating the importance of RAMPs to drug discovery targeting GPCRs are the recently approved antimigraine therapeutics that target the CALCRL-RAMP1 complex (Scuteri et al., 2022). The success of drugs targeting CALCRL-RAMP1 indicates the importance of considering the RAMP for a RAMP-interacting GPCR in GPCR-targeted drug design. Drug screens that involve both GPCRs and interacting RAMPs can potentially yield hits with a higher chance of clinical success. Elucidating the full breadth of GPCR–RAMP interactions can inform drug screen design and hopefully decrease the pipeline failure rate. Targeting GPCR–RAMP PPIs, for example by targeting the interaction interface to develop inhibitors, or conversely, bivalent ligand “glues,” might be another approach to modulate GPCR–RAMP pharmacology. Chemical tools developed in the course of drug discovery programs could also be leveraged to address many basic research questions in the future (Chang and Hsu, 2019; Hendrikse et al., 2020).
B. Elucidating Endogenous GPCR–RAMP Interactions
A combination of evidence from bioinformatics, coexpression analysis, and multiplexed direct binding assays suggests that GPCR–RAMP interactions are widespread and are likely to be identified across different classes of GPCRs. We do not yet know the functional effects of all known and potential GPCR–RAMP interactions. However, the array of functional studies on GPCR–RAMP pairs also suggests that RAMPs can exert a diverse range of effects on GPCR biology and pharmacology. Recent developments in the field of affinity proteomics can be leveraged to study GPCR–RAMP complexes. For example, SBA on endogenous tissues and other approaches to study endogenous GPCR–RAMP interactions should be attempted. Such studies may reveal which GPCR–RAMP interactions are most prominent in different tissues and cell types. Cross-linking mass spectrometry proteomics also holds the potential to identify GPCR–RAMP interactions in native systems. Additionally, RNAseq or single-cell RNAseq and related mRNA profiling strategies could be used to measure transcriptional effects of RAMP-dependent GPCR signaling pathways in endogenous systems. Such studies might address the question of how RAMPs are regulated and to what extent their expression is interdependent.
Some work along these lines was reported earlier (Pondel and Mould, 2005; Jacob et al., 2012). However, it is still unknown whether global deletion of a single RAMP alters the expression of other RAMPs (Coester et al., 2020). Is there some sort of negative feedback loop that downregulates one RAMP upon increased expression of another? Is there some redundancy and thus compensatory mechanisms for expressing the three RAMPs, and, if so, what are they? Globally and on a single-cell level, mRNA can usually be detected for more than one RAMP (DepMap.org). However, what about the cellular RAMP composition at the protein level?
C. Future Prospects for Structural Studies
High resolution CALCRL- and CALCR-RAMP structures are now available to help provide insights about GPCR–RAMP interactions and their functional correlates. However, data are lacking for the cytoplasmic, C-terminal-tail portions of CALCRL, CALCR, and the interacting RAMP. In addition, there is a lack of structural information to date about CALCRL-RAMP complexes bound to β-arrestin, or bound transducer protein other than Gs if they are in the active form. The full-length, inactive state structure of GLP1R was recently solved, and multiple different active-conformation structures have also been published. These structures will enable a more detailed comparison of those structures with inactive and active GLP1R structures bound to RAMP1, 2, or 3, once those structures become available (Jazayeri et al., 2017; Liang et al., 2017; Zhang et al., 2017; Wu et al., 2020). The structures of two other RAMP-interacting secretin family GPCRs have also been solved recently, that of PTH1R and GCGR, such that now there are structures available for all 15 secretin family (class B) receptors (Zhang et al., 2018; Zhao et al., 2019; Cong et al., 2022). A structure of one of these GPCRs in complex with a RAMP would prove to be very interesting. For example, it has been postulated that peptide interaction with TM7/ECL3/TM6 are involved in biased agonism (Lei et al., 2018) and a structure of a receptor with and without an interacting RAMP would help to test this hypothesis.
A GPCR–RAMP complex structure for a rhodopsin family and/or glutamate family receptor is another step in expanding our understanding of how the RAMPs regulate GPCRs of different families. As discussed in the first section, class A, or rhodopsin family, receptors traditionally lack the large extracellular domains common to class B, or secretin family, GPCRs, so there will be a need to explore whether the RAMP linker-GPCR ECL2/3 interactions plays a similar role in activation of rhodopsin family GPCR–RAMP complexes as it does in CGRP-mediated CALCRL-RAMP1 activation. A full-length structure may reveal whether a particular RAMP affects a rhodopsin family receptor allosterically, or whether the RAMP adopts a conformation that enables direct contacts with a ligand.
Liang and colleagues determined that the consensus structures of CALCRL-RAMP are largely similar and hypothesized that the unique influence of each RAMP on GPCR dynamics is the main driver of distinct GPCR–RAMP phenotypes (Liang et al., 2020a). Moving forward it will be necessary to consider the dynamics of a complex when analyzing a new structure. Cryo-EM is particularly well suited to this future direction, as it can reveal receptor dynamics through analysis of the different conformations captured. However, additional methods to assess dynamics, such as HDX-MS, and perhaps also FRET and double electron-electron resonance may provide complementary insights. Previous MD simulations have provided insights that were supported by structures obtained subsequently. Therefore, MD simulation continues to be an important tool to understand GPCR–RAMP dynamics but one that is intricately tied to the availability of some structural information.
There is significant potential in applying other computational approaches to study GPCR–RAMP interactions. A few examples of other approaches include: (i) homology modeling to identify potential RAMP-interacting GPCRs, (ii) machine learning using known interactions to predict other pairs, and (iii) MD simulations to design drugs targeting GPCR–RAMP complexes. Coarse-grain simulations have not yet been successfully applied to demonstrate the biophysical basis for GPCR–RAMP complex formation, but this strategy should prove promising, especially when structures of additional complexes become available (Periole et al., 2012).
D. Dynamics of GPCR–RAMP Complex Formation
How does a RAMP select a GPCR partner in cells with multiple RAMPs and GPCRs present? There are many open questions pertaining to confirming the direct physiologic and pathophysiological relevance of specific GPCR–RAMP pairs. Arguably, extremely detailed analysis has been carried out for only the CALCRL-RAMP1 complex, in part because of the challenges of working in cellular systems containing 10s of GPCRs and all three RAMPs. As highlighted within the sections here, new methods, including structural biology using cryo-EM and single particle reconstruction methods, are beginning to address the question of how RAMPs affect GPCR activation. A related question is: Once formed, are GPCR–RAMP interactions stable and long lasting or transient? Do interaction dynamics vary for each RAMP receptor pair, or are there common shared features? Do interaction dynamics vary with cell membrane compartment?
RAMP1 has been shown to localize primarily in the ER and the Golgi and to interact with tubulin (Hilairet et al., 2001; Kunz et al., 2007), so perhaps the RAMPs play some sort of more generalized chaperoning role inside the cell, where they interact with non-GPCR proteins. Moreover, within the context of modulating GPCR biology, do RAMPs exert any effects that have so far not been reported or characterized? For example, RAMPs may regulate GPCRs by disrupting hetero- or homodimerization. RAMP3 has been shown to interact with ACKR3, a receptor that can heterodimerize with CXCR4 (Levoye et al., 2009; Mackie et al., 2019). The CXCR4-ACKR3 heterodimer has a distinct signaling profile relative to CXCR4 alone (Décaillot et al., 2011). Therefore, the RAMP3 interaction with ACKR3 may regulate heterodimer formation by competition for binding at the TM3, 4, and 5 interface and thereby affect CXCR4 signaling.
Our understanding of the PTMs of RAMPs is incomplete. There are putative phosphorylation and ubiquitination sites on the C-terminal tails of the RAMPs, which have not yet been confirmed experimentally. It is possible that RAMP phosphorylation regulates dynamics of GPCR–RAMP interaction?
E. RAMPs in Human Disease
The precise role of RAMPs in human disease states remains to be determined. Additional mouse genetic models should prove useful to create and study relevant disease models. Genome sequencing studies focused on identifying RAMP variants and their correlation with SNPs or somatic mutations in their interacting GPCRs, especially in patient cohorts with a particular pathology, could identify novel disease connections for all three RAMPs. Genome sequencing studies across different large populations might reveal mutations in RAMPs that correlate with various predispositions for disease. For example, a recent genome-wide association study of migraine, expanding upon previous work, identified 86 novel loci associated with migraine, and 123 loci in total. One previously unknown locus was that for the genes CALCA and CALCB, which encode the two isoforms of CGRP. However, CALCRL and RAMP1 genes did not show a statistically comparable association with migraine (Hautakangas et al., 2022).
The role of anti-GPCR auto Abs in human immune disorders and post-infections syndromes has recently been discovered (Cabral-Marques et al., 2018; Skiba and Kruse, 2021). However, the potential role for RAMPs in the pathophysiology of autoimmune syndromes has not been considered. It will be interesting to test serum samples from patients with systemic sclerosis and other disorders for anti-RAMP Ab activity and to determine whether the presence of RAMPs plays a role in the immunogenicity of GPCRs associated with auto-immune diseases.
Acknowledgments
The authors thank the dedicated scientists from their laboratories and those of their collaborators, who contributed to studies on GPCRs and RAMPs, in particular Shahar Barbash, Annika Bendes, Émilie Ceraudo, Leo Dahl, Tea Dodig-Crnković, Thomas Huber, Manija A. Kazmi, Kevin Kemelmakher-Liben, Cassandra Koole, Torbjörn Persson, Elisa Pin, Marcus Saarinen, Roger D. Vaughan, and Harriet Watkins. The authors also thank Alexander Payne for his help with the creation of Figure 4. The authors acknowledge the support of Mathias Uhlèn, who pioneered the creation of the Human Protein Atlas.
Abbreviations
- Ab
antibody
- ACKR3
atypical chemokine receptor 3
- AM
adrenomedullin
- AM2
adrenomedullin2
- AM1R
adrenomedullin 1 receptor
- AM2
adrenomedullin 2 receptor
- AML
acute myeloid leukemia
- AMY
amylin receptor
- β-arrestin
non-visual arrestin
- β2ADR
β2-adrenergic receptor
- BRET
bioluminescence resonance energy transfer
- Ca 2+
calcium
- CALCR
calcitonin receptor
- CALCRL
calcitonin receptor-like receptor
- CaSR
calcium-sensing receptor
- CGRP
calcitonin-gene related peptide
- CGRPR
calcitonin-gene related peptide receptor
- CHO
Chinese hamster ovary
- COS
African green monkey kidney
- CRH
corticotropin-releasing hormone
- CRHR1
corticotropin-releasing hormone receptor 1
- CRHR2
corticotropin-releasing hormone receptor 2
- CRIF1
CR6-interacting factor 1
- cryo-EM
cryo-electron microscopy
- CT
calcitonin
- DACRA
dual amylin and calcitonin receptor agonist
- ECD
extracellular domain
- ECL
extracellular loop
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated kinase
- FACS
fluorescence-activated cell sorting
- FDA
Food and Drug Administration
- FRET
Förster resonance energy transfer
- G protein
heterotrimeric (αβγ) guanine-nucleotide regulatory protein
- GCGR
glucagon receptor
- GHRHR
growth hormone-releasing hormone receptor
- GIP
gastric inhibitory polypeptide
- GIPR
gastric inhibitory polypeptide receptor
- GLP1R
glucagon-like peptide 1 receptor
- GLP2
glucagon-like peptide 2
- GLP2R
glucagon-like peptide 2 receptor
- GPCR
G protein-coupled receptor
- GRAFS
glutamate, rhodopsin, adhesion, Frizzled (Fzd)/sweet taste receptor (TAS2), and secretin families
- GRK
GPCR kinase
- GPR30
G protein-coupled estrogen receptor 1
- HA
hemagglutinin
- HDX-MS
hydrogen-deuterium exchange mass spectrometry
- HEK
human embryonic kidney
- HPA
Human Protein Atlas
- HUVEC
human umbilical vein endothelial cells
- ICL
intracellular loop
- IP3
inositol 1, 4, 5-trisphosphate
- IF
immunofluorescence
- IP
immunoprecipitation
- IP3
inositol 1, 4, 5-trisphosphate
- kDa
kilo-Daltons
- KO
knockout
- mAb
monoclonal antibody
- mAbs
molecules and monoclonal antibodies
- MCR2
melanocortin receptor 2
- MD
molecular dynamics
- MEF
mouse embryonic fibroblast
- MERFISH
multiplexed error-correcting fluorescence in situ hybridization
- mGluRs
metabotropic glutamate receptor
- MRAP
melanocortin receptor accessory protein
- NHERF
Na+ /H+ exchanger regulatory factor
- Nluc
nanoluciferase
- NMDA
N-methyl D-aspartate
- NSF
N-ethylmaleimide-sensitive factor
- OE
overexpression
- PACAP
pituitary adenylyl cyclase-activating protein
- PDZ
PSD-95/Discs-large/ZO-1
- PLA
proximity ligation assay
- PM
plasma membrane
- PPI
protein–protein interaction
- PTH
parathyroid hormone
- PTH1R
parathyroid hormone 1 receptor
- PTH2R
parathyroid hormone 2 receptor
- PTM
post-translational modification
- RAMP
receptor activity-modifying protein
- RCP
receptor-component protein
- REEP
receptor expression-enhancing proteins
- Rluc
Renilla luciferase
- RTP
receptor transporting proteins
- SBA
suspension bead array
- SCTR
secretin receptor
- SK
serine-lysine
- SNP
single-nucleotide base polymorphism
- SV1
splice variant 1 of GHRHR
- TAS2
sweet taste receptor GPCR family
- TM
transmembrane
- VEGF
vascular endothelial growth factor
- VIP
vasoactive and intestinal peptide
- VIPR1
VIP and PACAP receptor 1
- VIPR2
VIP and PACAP receptor 2
- YFP
yellow fluorescent protein
- WT
wild-type.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Kotliar, Lorenzen, Schwenk, Hay, Sakmar.
Footnotes
Financial support for studies of GPCR–RAMP interactions was provided by the KTH Center for Applied Precision Medicine, the Erling-Persson Family Foundation, and grants for Science for Life Laboratory. The Human Protein Atlas was funded by the Knut and Alice Wallenberg Foundation. Additional funding was provided by the Nicholson Short-Term Exchange, the Albert Cass Fellowship, and the Alexander Mauro Fellowship. This work was also supported by National Institutes of Health National Institute of General Medical Sciences [Grant T32-GM136640], a Rothschild postdoctoral fellowship, Bengt Winblad’s Foundation for Young Scientists, the Margaretha af Ugglas Foundation, the Gun och Bertil Stohnes Foundation, the Robertson Therapeutic Development Fund, the Danica Foundation, and the Denise and Michael Kellen Foundation through Kellen Women’s Entrepreneurship Fund.
No author has an actual or perceived conflict of interest with the contents of this article.
References
- Aksoydan B, Durdagi S (2022) Virtual drug repurposing study for the CGRPR identifies pentagastrin and leuprorelin as putative candidates. J Mol Graph Model 116:108254. [DOI] [PubMed] [Google Scholar]
- Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M (2000) Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc Natl Acad Sci USA 97:3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argunhan F, Brain SD (2022) The vascular-dependent and -independent actions of calcitonin gene-related peptide in cardiovascular disease. Front Physiol 13:833645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour SL, Foord S, Kenakin T, Chen WJ (1999) Pharmacological characterization of receptor-activity-modifying proteins (RAMPs) and the human calcitonin receptor. J Pharmacol Toxicol Methods 42:217–224. [DOI] [PubMed] [Google Scholar]
- Ashizuka S, Kita T, Inatsu H, Kitamura K (2021) Adrenomedullin: a novel therapeutic for the treatment of inflammatory bowel disease. Biomedicines 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Findlay JB (1994) Fingerprinting G-protein-coupled receptors. Protein Eng 7:195–203. [DOI] [PubMed] [Google Scholar]
- Avgoustou P, Jailani ABA, Zirimwabagabo JO, Tozer MJ, Gibson KR, Glossop PA, Mills JEJ, Porter RA, Blaney P, Bungay PJ, et al. (2020) Discovery of a first-in-class potent small molecule antagonist against the adrenomedullin-2 receptor. ACS Pharmacol Transl Sci 3:706–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Harris M, Barkan K, Winfield I, Harper MT, Simms J, Ladds G, Wheatley M, Poyner D(2019) Interactions between RAMP2 and CRF receptors: The effect of receptor subtypes, splice variants and cell context. Biochim Biophys Acta Biomembr 1861:997–1003. [DOI] [PubMed] [Google Scholar]
- Bailey S, Harris M, Barkan K, Winfield I, Harper MT, Simms J, Ladds G, Wheatley M, Poyner D(2019) Interactions between RAMP2 and CRF receptors: The effect of receptor subtypes, splice variants and cell context. Biochim Biophys Acta Biomembr 1861:997–1003. [DOI] [PubMed] [Google Scholar]
- Barbash S, Lorenzen E, Persson T, Huber T, Sakmar TP(2017) GPCRs globally coevolved with receptor activity-modifying proteins, RAMPs. Proc Natl Acad Sci USA 114:12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash S, Persson T, Lorenzen E, Kazmi MA, Huber T, Sakmar TP (2019) Detection of concordance between transcriptional levels of GPCRS and receptor-activity-modifying proteins. iScience 11: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick CJ, Lenhart PM, Dackor RT, Nagle E, Caron KM (2012) Loss of receptor activity-modifying protein 3 exacerbates cardiac hypertrophy and transition to heart failure in a sex-dependent manner. J Mol Cell Cardiol 52:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell IM, Gallicchio SN, Wood MR, Quigley AG, Stump CA, Zartman CB, Fay JF, Li CC, Lynch JJ, Moore EL, et al. (2010) Discovery of MK-3207: a highly potent, orally bioavailable CGRP receptor antagonist. ACS Med Chem Lett 1:24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez-Páez A, Cárdenas-Brito S (2008) Dissection of functional residues in receptor activity-modifying proteins through phylogenetic and statistical analyses. Evol Bioinform Online 4:153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruien NNA, Smith CL (2020) Emerging roles of melanocortin receptor accessory proteins (MRAP and MRAP2) in physiology and pathophysiology. Gene 757:144949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkley N, Bolognese M, Sidorowicz-Bialynicka A, Vally T, Trout R, Miller C, Buben CE, Gilligan JP, Krause DS; Oral Calcitonin in Postmenopausal Osteoporosis (ORACAL) Investigators (2012) A phase 3 trial of the efficacy and safety of oral recombinant calcitonin: the Oral Calcitonin in Postmenopausal Osteoporosis (ORACAL) trial. J Bone Miner Res 27:1821–1829. [DOI] [PubMed] [Google Scholar]
- Björk S, Hurt CM, Ho VK, Angelotti T (2013) REEPs are membrane shaping adapter proteins that modulate specific g protein-coupled receptor trafficking by affecting ER cargo capacity. PLoS One 8:e76366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger JM, Parameswaran N, Hall CS, Aiyar N, Spielman WS (2005a) Novel function for receptor activity-modifying proteins (RAMPs) in post-endocytic receptor trafficking. J Biol Chem 280:9297–9307. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Parameswaran N, Spielman WS (2012) Regulation of GPCR trafficking by RAMPs. Adv Exp Med Biol 744:25–37. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Spielman WS, Hall CS, Weinman EJ, Parameswaran N (2005b) Receptor activity-modifying protein (RAMP) isoform-specific regulation of adrenomedullin receptor trafficking by NHERF-1. J Biol Chem 280:23926–23935. [DOI] [PubMed] [Google Scholar]
- Booe JM, Walker CS, Barwell J, Kuteyi G, Simms J, Jamaluddin MA, Warner ML, Bill RM, Harris PW, Brimble MA, et al. (2015) Structural basis for receptor activity-modifying protein-dependent selective peptide recognition by a G protein-coupled receptor. Mol Cell 58:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booe JM, Warner ML, Pioszak AA (2020) Picomolar affinity antagonist and sustained signaling agonist peptide ligands for the adrenomedullin and calcitonin gene-related peptide receptors. ACS Pharmacol Transl Sci 3:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booe JM, Warner ML, Roehrkasse AM, Hay DL, Pioszak AA (2018) Probing the mechanism of receptor activity-modifying protein modulation of GPCR ligand selectivity through rational design of potent adrenomedullin and calcitonin gene-related peptide antagonists. Mol Pharmacol 93:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Brito I, Romero-Fernandez W, Di Palma M, Oflijan J, Skieterska K, Duchou J, Van Craenenbroeck K, Suárez-Boomgaard D, Rivera A, et al. (2014) The G protein-coupled receptor heterodimer network (GPCR-HetNet) and its hub components. Int J Mol Sci 15:8570–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet T, Henley JM (2005) Calcium as an extracellular signalling molecule: perspectives on the calcium sensing receptor in the brain. C R Biol 328:691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet T, Martin S, Henley JM (2005) Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci 118:4709–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet T, Martin S, Henley JM(2008) Regulation of calcium-sensing-receptor trafficking and cell-surface expression by GPCRs and RAMPs. Trends Pharmacol Sci 29:633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower RL, Yule L, Rees TA, Deganutti G, Hendrikse ER, Harris PWR, Kowalczyk R, Ridgway Z, Wong AG, Swierkula K, et al. (2018) Molecular signature for receptor engagement in the metabolic peptide hormone amylin. ACS Pharmacol Transl Sci 1:32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucknell SJ, Ator MA, Brown AJH, Brown J, Cansfield AD, Cansfield JE, Christopher JA, Congreve M, Cseke G, Deflorian F, et al. (2020) Structure-based drug discovery of N-((R)-3-(7-methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2′-oxo-1′,2′-dihydrospiro[piperidine-4,4′-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): a calcitonin gene-related peptide receptor antagonist for acute treatment of migraine. J Med Chem 63:7906–7920. [DOI] [PubMed] [Google Scholar]
- Cabral-Marques O, Marques A, Giil LM, De Vito R, Rademacher J, Günther J, Lange T, Humrich JY, Klapa S, Schinke S, et al. (2018) GPCR-specific autoantibody signatures are associated with physiological and pathological immune homeostasis. Nat Commun 9:5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansfield AD, Ator MA, Banerjee J, Bestwick M, Bortolato A, Brown GA, Brown J, Butkovic K, Cansfield JE, Christopher JA, et al. (2022) Novel Macrocyclic antagonists of the calcitonin gene-related peptide receptor: design, realization, and structural characterization of protein-ligand complexes. ACS Chem Neurosci 13:751–765. [DOI] [PubMed] [Google Scholar]
- Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, et al. (2022) A structural basis for amylin receptor phenotype. Science 375:eabm9609. [DOI] [PubMed] [Google Scholar]
- Cegla J, Jones BJ, Gardiner JV, Hodson DJ, Marjot T, McGlone ER, Tan TM, Bloom SR (2017) RAMP2 influences glucagon receptor pharmacology via trafficking and signaling. Endocrinology 158:2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Hsu SYT (2019) Development of chimeric and bifunctional antagonists for CLR/RAMP receptors. PLoS One 14:e0216996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedula PV, Mercer SE, Pin SS, Thalody G, Xu C, Conway CM, Keavy D, Signor L, Cantor GH, Mathias N, et al. (2013) Discovery of (R)-N-(3-(7-methyl-1H-indazol-5-yl)-1-(4-(1-methylpiperidin-4-yl)-1-oxopropan-2-yl)-4-(2-oxo-1,2-dihydroquinolin-3-yl)piperidine-1-carboxamide (BMS-742413): a potent human CGRP antagonist with superior safety profile for the treatment of migraine through intranasal delivery. Bioorg Med Chem Lett 23:3157–3161. [DOI] [PubMed] [Google Scholar]
- Cheng C, Zhang Z, Cheng F, Shao Z (2020) Exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells promotes angiogenesis through miR-2355-5p/VEGFR2 axis. OncoTargets Ther 13:3291–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos A, Christopoulos G, Morfis M, Udawela M, Laburthe M, Couvineau A, Kuwasako K, Tilakaratne N, Sexton PM (2003) Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem 278:3293–3297. [DOI] [PubMed] [Google Scholar]
- Christopoulos G, Perry KJ, Morfis M, Tilakaratne N, Gao Y, Fraser NJ, Main MJ, Foord SM, Sexton PM(1999) Multiple Amylin Receptors Arise from Receptor Activity-Modifying Protein Interaction with the Calcitonin Receptor Gene Product. Mol Pharmacol 56:235–242. [DOI] [PubMed] [Google Scholar]
- Chung TT, Webb TR, Chan LF, Cooray SN, Metherell LA, King PJ, Chapple JP, Clark AJ (2008) The majority of adrenocorticotropin receptor (melanocortin 2 receptor) mutations found in familial glucocorticoid deficiency type 1 lead to defective trafficking of the receptor to the cell surface. J Clin Endocrinol Metab 93:4948–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Mullooly N, Safitri D, Harris M, de Vries T, MaassenVanDenBrink A, Poyner DR, Gianni D, Wigglesworth M, Ladds G (2021) CGRP, adrenomedullin and adrenomedullin 2 display endogenous GPCR agonist bias in primary human cardiovascular cells. Commun Biol 4:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coester B, Pence SW, Arrigoni S, Boyle CN, Le Foll C, Lutz TA (2020) RAMP1 and RAMP3 differentially control amylin’s effects on food intake, glucose and energy balance in male and female mice. Neuroscience 447:74–93. [DOI] [PubMed] [Google Scholar]
- Cong Z, Liang YL, Zhou Q, Darbalaei S, Zhao F, Feng W, Zhao L, Xu HE, Yang D, Wang MW (2022) Structural perspective of class B1 GPCR signaling. Trends Pharmacol Sci 43:321–334. [DOI] [PubMed] [Google Scholar]
- Cottrell GS, Padilla B, Pikios S, Roosterman D, Steinhoff M, Grady EF, Bunnett NW (2007) Post-endocytic sorting of calcitonin receptor-like receptor and receptor activity-modifying protein 1. J Biol Chem 282:12260–12271. [DOI] [PubMed] [Google Scholar]
- Crine SL, Acharya KR (2021) Molecular basis of C-mannosylation—a structural perspective. FEBS J DOI: 10.1111/febs.16265 [online ahead of print] [DOI] [PubMed] [Google Scholar]
- Croop R, Madonia J, Conway C, Thiry A, Forshaw M, Murphy A, Jensen C, Dubowchik G, Coric V, Lipton R (2021) Intranasal zavegepant is effective and well tolerated for the acute treatment of migraine: a phase 2/3 dose-ranging clinical trial. Neurology 96:4976. [Google Scholar]
- Dackor R, Fritz-Six K, Smithies O, Caron K (2007) Receptor activity-modifying proteins 2 and 3 have distinct physiological functions from embryogenesis to old age. J Biol Chem 282:18094–18099. [DOI] [PubMed] [Google Scholar]
- Dale NC, Johnstone EKM, White CW, Pfleger KDG (2019) NanoBRET: the bright future of proximity-based assays. Front Bioeng Biotechnol 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Décaillot FM, Kazmi MA, Lin Y, Ray-Saha S, Sakmar TP, Sachdev P (2011) CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem 286:32188–32197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deganutti G, Atanasio S, Rujan RM, Sexton PM, Wootten D, Reynolds CA (2021) Exploring ligand binding to calcitonin gene-related peptide receptors. Front Mol Biosci 8:720561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai AJ, Roberts DJ, Richards GO, Skerry TM (2014) Role of receptor activity modifying protein 1 in function of the calcium sensing receptor in the human TT thyroid carcinoma cell line. PLoS One 9:e85237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Wang D, Fan H, Xu C, Tai L, Lin S, Han S, Tan Q, Wang X, Xu T, et al. (2021) Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature 594:589–593. [DOI] [PubMed] [Google Scholar]
- Duval V, Alayrac P, Silvestre JS, Levoye A (2022) Emerging roles of the atypical chemokine receptor 3 (ACKR3) in cardiovascular diseases. Front Endocrinol (Lausanne) 13:906586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvinsson L, Haanes KA, Warfvinge K, Krause DN (2018) CGRP as the target of new migraine therapies—successful translation from bench to clinic. Nat Rev Neurol 14:338–350. [DOI] [PubMed] [Google Scholar]
- El Khamlichi C, Reverchon-Assadi F, Hervouet-Coste N, Blot L, Reiter E, Morisset-Lopez S (2019) Bioluminescence resonance energy transfer as a method to study protein-protein interactions: application to G protein coupled receptor biology. Molecules 24:PMC6384791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BN, Rosenblatt MI, Mnayer LO, Oliver KR, Dickerson IM (2000) CGRP-RCP, a novel protein required for signal transduction at calcitonin gene-related peptide and adrenomedullin receptors. J Biol Chem 275:31438–31443. [DOI] [PubMed] [Google Scholar]
- Fiorentini C, Gardoni F, Spano P, Di Luca M, Missale C (2003) Regulation of dopamine D1 receptor trafficking and desensitization by oligomerization with glutamate N-methyl-D-aspartate receptors. J Biol Chem 278:20196–20202. [DOI] [PubMed] [Google Scholar]
- Fletcher MM, Keov P, Truong TT, Mennen G, Hick CA, Zhao P, Furness SGB, Kruse T, Clausen TR, Wootten D, et al. (2021) AM833 is a novel agonist of calcitonin family G protein-coupled receptors: pharmacological comparison with six selective and nonselective agonists. J Pharmacol Exp Ther 377:417–440. [DOI] [PubMed] [Google Scholar]
- Foord SM, Topp SD, Abramo M, Holbrook JD (2005) New methods for researching accessory proteins. J Mol Neurosci 26:265–276. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin L-G, Schiöth HB (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63:1256–1272. [DOI] [PubMed] [Google Scholar]
- Garces F, Mohr C, Zhang L, Huang CS, Chen Q, King C, Xu C, Wang Z (2020) Molecular insight into recognition of the cgrpr complex by migraine prevention therapy Aimovig (erenumab). Cell Rep 30:1714–1723.e1716. [DOI] [PubMed] [Google Scholar]
- Garelja ML, Au M, Brimble MA, Gingell JJ, Hendrikse ER, Lovell A, Prodan N, Sexton PM, Siow A, Walker CS, et al. (2020) Molecular mechanisms of class B GPCR activation: insights from adrenomedullin receptors. ACS Pharmacol Transl Sci 3:246–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garelja ML, Bower RL, Brimble MA, Chand S, Harris PWR, Jamaluddin MA, Petersen J, Siow A, Walker CS, Hay DL (2022) Pharmacological characterisation of mouse calcitonin and calcitonin receptor-like receptors reveals differences compared with human receptors. Br J Pharmacol 179:416–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingell JJ, Burns ER, Hay DL (2014) Activity of pramlintide, rat and human amylin but not Aβ1-42 at human amylin receptors. Endocrinology 155:21–26. [DOI] [PubMed] [Google Scholar]
- Gingell JJ, Qi T, Bailey RJ, Hay DL (2010) A key role for tryptophan 84 in receptor activity-modifying protein 1 in the amylin 1 receptor. Peptides 31:1400–1404. [DOI] [PubMed] [Google Scholar]
- Gingell JJ, Rees TA, Hendrikse ER, Siow A, Rennison D, Scotter J, Harris PWR, Brimble MA, Walker CS, Hay DL (2020) Distinct patterns of internalization of different calcitonin gene-related peptide receptors. ACS Pharmacol Transl Sci 3:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingell JJ, Simms J, Barwell J, Poyner DR, Watkins HA, Pioszak AA, Sexton PM, Hay DL (2016) An allosteric role for receptor activity-modifying proteins in defining GPCR pharmacology. Cell Discov 2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes I, Sierra S, Devi LA (2016) Detection of receptor heteromerization using in situ proximity ligation assay. Curr Protoc Pharmacol 75:2.16.11–12.16.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Zhang H, Huang L, Chen Y, Shi Y, Tam PO-S, Zhu X, Huang Y, Lei B, Sundaresan P, et al. (2019) Mutant RAMP2 causes primary open-angle glaucoma via the CRLR-cAMP axis. Genet Med 21:2345–2354. [DOI] [PubMed] [Google Scholar]
- Grandits AM, Wieser R (2021) Gene expression changes contribute to stemness and therapy resistance of relapsed acute myeloid leukemia: roles of SOCS2, CALCRL, MTSS1, and KDM6A. Exp Hematol 99:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2015) Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV (2008) GPCR monomers and oligomers: it takes all kinds. Trends Neurosci 31:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV (2019) GPCR signaling regulation: the role of GRKs and arrestins. Front Pharmacol 10:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Zuo Z, Qu M, Zhou Y, Li Q, Wang H (2021) Comprehensive analysis of inflammatory response-related genes, and prognosis and immune infiltration in patients with low-grade glioma. Front Pharmacol 12:748993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar KG, Simms J, Christopoulos G, Sexton PM, Miller LJ (2009) Molecular basis of association of receptor activity-modifying protein 3 with the family B G protein-coupled secretin receptor. Biochemistry 48:11773–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani B, Taheri M, Asgari Y, Zekri A, Sattari A, Ghafouri-Fard S, Pouresmaeili F (2021) Expression analysis of long non-coding RNAs related with FOXM1, GATA3, FOXA1 and ESR1 in breast tissues. Front Oncol 11:671418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautakangas H, Winsvold BS, Ruotsalainen SE, Bjornsdottir G, Harder AVE, Kogelman LJA, Thomas LF, Noordam R, Benner C, Gormley P, et al. ; International Headache Genetics Consortium; HUNT All-in Headache; Danish Blood Donor Study Genomic Cohort (2022) Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat Genet 54:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DL, Chen S, Lutz TA, Parkes DG, Roth JD (2015) Amylin: pharmacology, physiology, and clinical potential. Pharmacol Rev 67:564–600. [DOI] [PubMed] [Google Scholar]
- Hay DL, Christopoulos G, Christopoulos A, Poyner DR, Sexton PM (2005) Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol Pharmacol 67:1655–1665. [DOI] [PubMed] [Google Scholar]
- Hay DL, Garelja ML, Poyner DR, Walker CS (2018) Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Br J Pharmacol 175:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DL, Pioszak AA (2016) Receptor activity-modifying proteins (RAMPs): new insights and roles. Annu Rev Pharmacol Toxicol 56:469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DL, Poyner DR, Sexton PM (2006) GPCR modulation by RAMPs. Pharmacol Ther 109:173–197. [DOI] [PubMed] [Google Scholar]
- Hendrikse ER, Liew LP, Bower RL, Bonnet M, Jamaluddin MA, Prodan N, Richards KD, Walker CS, Pairaudeau G, Smith DM, et al. (2020) Identification of small-molecule positive modulators of calcitonin-like receptor-based receptors. ACS Pharmacol Transl Sci 3:305–320.</jHeHrn> [DOI] [PMC free article] [PubMed] [Google Scholar]
- Héroux M, Breton B, Hogue M, Bouvier M (2007) Assembly and signaling of CRLR and RAMP1 complexes assessed by BRET. Biochemistry 46:7022–7033. [DOI] [PubMed] [Google Scholar]
- Hewitt DJ, Aurora SK, Dodick DW, Goadsby PJ, Ge YJ, Bachman R, Taraborelli D, Fan X, Assaid C, Lines C, et al. (2011) Randomized controlled trial of the CGRP receptor antagonist MK-3207 in the acute treatment of migraine. Cephalalgia 31:712–722. [DOI] [PubMed] [Google Scholar]
- Hilairet S, Bélanger C, Bertrand J, Laperrière A, Foord SM, Bouvier M (2001) Agonist-promoted internalization of a ternary complex between calcitonin receptor-like receptor, receptor activity-modifying protein 1 (RAMP1), and beta-arrestin. J Biol Chem 276:42182–42190. [DOI] [PubMed] [Google Scholar]
- Hong Y, Hay DL, Quirion R, Poyner DR (2012) The pharmacology of adrenomedullin 2/intermedin. Br J Pharmacol 166:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda H, Nakamura T, Yoshihara F (2022) Plasma clearance of intravenously infused adrenomedullin in rats with acute renal failure. Biomolecules 12:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Lv X, Wei L, Wang Y, Zheng G, Yang C, Zang F, Wang J, Li J, Wu X, et al. (2022) Sensory nerve maintains intervertebral disc extracellular matrix homeostasis via CGRP/CHSY1 axis. Adv Sci (Weinh) DOI: 10.1002/advs.202202620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husmann K, Born W, Fischer JA, Muff R (2003) Three receptor-activity-modifying proteins define calcitonin gene-related peptide or adrenomedullin selectivity of the mouse calcitonin-like receptor in COS-7 cells. Biochem Pharmacol 66:2107–2115. [DOI] [PubMed] [Google Scholar]
- Husmann K, Sexton PM, Fischer JA, Born W (2000) Mouse receptor-activity-modifying proteins 1, -2 and -3: amino acid sequence, expression and function. Mol Cell Endocrinol 162:35–43. [DOI] [PubMed] [Google Scholar]
- Hutchings CJ, Koglin M, Olson WC, Marshall FH (2017) Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. Nat Rev Drug Discov 16:787–810. [DOI] [PubMed] [Google Scholar]
- Jacob A, Wu R, Wang P (2012) Regulation of RAMP expression in diseases. Adv Exp Med Biol 744:87–103. [DOI] [PubMed] [Google Scholar]
- Jailani ABA, Bigos KJA, Avgoustou P, Egan JL, Hathway RA, Skerry TM, Richards GO (2022) Targeting the adrenomedullin-2 receptor for the discovery and development of novel anti-cancer agents. Expert Opin Drug Discov 17:839–848. [DOI] [PubMed] [Google Scholar]
- Jamaluddin A, Chuang CL, Williams ET, Siow A, Yang SH, Harris PWR, Petersen JSSM, Bower RL, Chand S, Brimble MA, et al. (2022) Lipidated Calcitonin gene-related peptide (CGRP) peptide antagonists retain CGRP receptor activity and attenuate CGRP action in vivo. Front Pharmacol 13:832589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Rappas M, Brown AJH, Kean J, Errey JC, Robertson NJ, Fiez-Vandal C, Andrews SP, Congreve M, Bortolato A, et al. (2017) Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546:254–258. [DOI] [PubMed] [Google Scholar]
- Josephs TM, Belousoff MJ, Liang YL, Piper SJ, Cao J, Garama DJ, Leach K, Gregory KJ, Christopoulos A, Hay DL, et al. (2021) Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science 372:eabf7258. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Granstein RD (2021) Roles of calcitonin gene-related peptide in the skin, and other physiological and pathophysiological functions. Brain Behav Immun Health 18:100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita T, Kitamura K (2022) Adrenomedullin therapy in moderate to severe COVID-19. Biomedicines 10:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaesson A, Grannas K, Ebai T, Heldin J, Koos B, Leino M, Raykova D, Oelrich J, Arngården L, Söderberg O, et al. (2018) Improved efficiency of in situ protein analysis by proximity ligation using UnFold probes. Sci Rep 8:5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KR, Matson BC, Caron KM (2016) The expanding repertoire of receptor activity modifying protein (RAMP) function. Crit Rev Biochem Mol Biol 51:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb P, Kenakin T, Alexander SPH, Bermudez M, Bohn LM, Breinholt CS, Bouvier M, Hill SJ, Kostenis E, Martemyanov K, Neubig RR, Onaran HO, Rajagopal S, Roth BL, Selent J, Shukla AK, Sommer ME, Gloriam DE (2022) Community guidelines for GPCR ligand bias: IUPHAR review 32. Br J Pharmacol 179: 3651–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koole C, Reynolds CA, Mobarec JC, Hick C, Sexton PM, Sakmar TP (2017) Genetically encoded photocross-linkers determine the biological binding site of exendin-4 peptide in the N-terminal domain of the intact human glucagon-like peptide-1 receptor (GLP-1R). J Biol Chem 292:7131–7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski-Jahn M, Schihada H, Turku A, Huber T, Sakmar TP, Schulte G (2021) Frizzled BRET sensors based on bioorthogonal labeling of unnatural amino acids reveal WNT-induced dynamics of the cysteine-rich domain. Sci Adv 7:eabj7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Hansen JL, Dahl K, Schäffer L, Sensfuss U, Poulsen C, Schlein M, Hansen AMK, Jeppesen CB, Dornonville de la Cour C, et al. (2021) Development of cagrilintide, a long-acting amylin analogue. J Med Chem 64:11183–11194. [DOI] [PubMed] [Google Scholar]
- Kunz TH, Mueller-Steiner S, Schwerdtfeger K, Kleinert P, Troxler H, Kelm JM, Ittner LM, Fischer JA, Born W (2007) Interaction of receptor-activity-modifying protein1 with tubulin. Biochim Biophys Acta 1770:1145–1150. [DOI] [PubMed] [Google Scholar]
- Kurashige C, Hosono K, Matsuda H, Tsujikawa K, Okamoto H, Majima M (2014) Roles of receptor activity-modifying protein 1 in angiogenesis and lymphangiogenesis during skin wound healing in mice. FASEB J 28:1237–1247. [DOI] [PubMed] [Google Scholar]
- Kusano S, Kukimoto-Niino M, Akasaka R, Toyama M, Terada T, Shirouzu M, Shindo T, Yokoyama S (2008) Crystal structure of the human receptor activity-modifying protein 1 extracellular domain. Protein Sci 17:1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano S, Kukimoto-Niino M, Hino N, Ohsawa N, Okuda K, Sakamoto K, Shirouzu M, Shindo T, Yokoyama S (2012) Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding. Protein Sci 21:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K, Cao YN, Chu CP, Iwatsubo S, Eto T, Kitamura K (2006) Functions of the cytoplasmic tails of the human receptor activity-modifying protein components of calcitonin gene-related peptide and adrenomedullin receptors. J Biol Chem 281:7205–7213. [DOI] [PubMed] [Google Scholar]
- Kuwasako K, Sekiguchi T, Nagata S, Jiang D, Hayashi H, Murakami M, Hattori Y, Kitamura K, Kato J (2016) Inhibitory effects of two G protein-coupled receptor kinases on the cell surface expression and signaling of the human adrenomedullin receptor. Biochem Biophys Res Commun 470:894–899. [DOI] [PubMed] [Google Scholar]
- Kuwasako K, Shimekake Y, Masuda M, Nakahara K, Yoshida T, Kitaura M, Kitamura K, Eto T, Sakata T (2000) Visualization of the calcitonin receptor-like receptor and its receptor activity-modifying proteins during internalization and recycling. J Biol Chem 275:29602–29609. [DOI] [PubMed] [Google Scholar]
- La Fountaine MF, Hohn AN, Leahy CL, Weir JP, Testa AJ (2022) Observations from a prospective small cohort study suggest that CGRP genes contribute to acute posttraumatic headache burden after concussion. Front Neurol 13:947524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CH, Chen AT, Burns AB, Sriram K, Luo Y, Tang X, Branciamore S, O’Meally D, Chang SL, Huang PH, et al. (2021) RAMP2-AS1 regulates endothelial homeostasis and aging. Front Cell Dev Biol 9:635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrue C, Guiraud N, Mouchel PL, Dubois M, Farge T, Gotanègre M, Bosc C, Saland E, Nicolau-Travers ML, Sabatier M, et al. (2021) Adrenomedullin-CALCRL axis controls relapse-initiating drug tolerant acute myeloid leukemia cells. Nat Commun 12:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen AT, Sonne N, Andreassen KV, Karsdal MA, Henriksen K (2020) Dose frequency optimization of the dual amylin and calcitonin receptor agonist KBP-088: long-lasting improvement in food preference and body weight loss. J Pharmacol Exp Ther 373:269–278. [DOI] [PubMed] [Google Scholar]
- Lau DCW, Erichsen L, Francisco AM, Satylganova A, le Roux CW, McGowan B, Pedersen SD, Pietiläinen KH, Rubino D, Batterham RL (2021) Once-weekly cagrilintide for weight management in people with overweight and obesity: a multicentre, randomised, double-blind, placebo-controlled and active-controlled, dose-finding phase 2 trial. Lancet 398:2160–2172. [DOI] [PubMed] [Google Scholar]
- Lee S, Pioszak AA (2020) Molecular interaction of an antagonistic amylin analog with the extracellular domain of receptor activity-modifying protein 2 assessed by fluorescence polarization. Biophys Chem 267:106477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Hay DL, Pioszak AA (2016) Calcitonin and amylin receptor peptide interaction mechanisms: insights into peptide-binding modes and allosteric modulation of the calcitonin receptor by receptor activity-modifying proteins. J Biol Chem 291:8686–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S, Clydesdale L, Dai A, Cai X, Feng Y, Yang D, Liang YL, Koole C, Zhao P, Coudrat T, et al. (2018) Two distinct domains of the glucagon-like peptide-1 receptor control peptide-mediated biased agonism. J Biol Chem 293:9370–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart PM, Broselid S, Barrick CJ, Leeb-Lundberg LMF, Caron KM(2013) G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. J Mol Endocrinol 51:191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B (2009) CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood 113:6085–6093. [DOI] [PubMed] [Google Scholar]
- Li L, Gan YP, Peng H (2022) RAMP2-AS1 inhibits CXCL11 expression to suppress malignant phenotype of breast cancer by recruiting DNMT1 and DNMT3B. Exp Cell Res 416:113139. [DOI] [PubMed] [Google Scholar]
- Li M, Wetzel-Strong SE, Hua X, Tilley SL, Oswald E, Krummel MF, Caron KM (2014) Deficiency of RAMP1 attenuates antigen-induced airway hyperresponsiveness in mice. PLoS One 9:e102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y-LKhoshouei MDeganutti GGlukhova AKoole CPeat TSRadjainia MPlitzko JMBaumeister WMiller LJet al. (2018) Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature 561:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y-L, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, et al. (2017) Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546:118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Belousoff MJ, Fletcher MM, Zhang X, Khoshouei M, Deganutti G, Koole C, Furness SGB, Miller LJ, Hay DL, et al. (2020a) Structure and dynamics of adrenomedullin receptors AM1 and AM2 reveal key mechanisms in the control of receptor phenotype by receptor activity-modifying proteins. ACS Pharmacol Transl Sci 3:263–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Belousoff MJ, Zhao P, Koole C, Fletcher MM, Truong TT, Julita V, Christopoulos G, Xu HE, Zhang Y, et al. (2020b) Toward a structural understanding of class B GPCR peptide binding and activation. Mol Cell 77:656–668.e5. [DOI] [PubMed] [Google Scholar]
- Liu T, Kamiyoshi A, Tanaka M, Iida S, Sakurai T, Ichikawa-Shindo Y, Kawate H, Hirabayashi K, Dai K, Cui N, et al. (2018) RAMP3 deficiency enhances postmenopausal obesity and metabolic disorders. Peptides 110:10–18. [DOI] [PubMed] [Google Scholar]
- Lorenzen E, Dodig-Crnković T, Kotliar IB, Pin E, Ceraudo E, Vaughan RD, Uhlèn M, Huber T, Schwenk JM, Sakmar TP (2019) Multiplexed analysis of the secretin-like GPCR–RAMP interactome. Sci Adv 5:eaaw2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Lefkowitz RJ (2002) The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci 115:455–465. [DOI] [PubMed] [Google Scholar]
- Lutz TA, Coester B, Whiting L, Dunn-Meynell AA, Boyle CN, Bouret SG, Levin BE, Le Foll C (2018) Amylin selectively signals onto POMC neurons in the arcuate nucleus of the hypothalamus. Diabetes 67:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Liu J, Shi Q, Tan Q, Wu D, Skinner JJ, Walker AL, Zhao L, Gu X, Chen N, et al. (2016) In vitro expression and analysis of the 826 human G protein-coupled receptors. Protein Cell 7:325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machleidt T, Woodroofe CC, Schwinn MK, Méndez J, Robers MB, Zimmerman K, Otto P, Daniels DL, Kirkland TA, Wood KV (2015) NanoBRET—a novel BRET platform for the analysis of protein-protein interactions. ACS Chem Biol 10:1797–1804. [DOI] [PubMed] [Google Scholar]
- Mackie DI, Nielsen NR, Harris M, Singh S, Davis RB, Dy D, Ladds G, Caron KM(2019) RAMP3 determines rapid recycling of atypical chemokine receptor-3 for guided angiogenesis. Proc Natl Acad Sci USA 116:24093–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice P, Guillaume J-L, Benleulmi-Chaachoua A, Daulat AM, Kamal M, Jockers R (2011) GPCR-interacting proteins, major players of GPCR function. Adv Pharmacol 62:349–380. [DOI] [PubMed] [Google Scholar]
- McGlone ER, Manchanda Y, Jones B, Pickford P, Inoue A, Carling D, Bloom SR, Tan T, Tomas A (2021) Receptor activity-modifying protein 2 (RAMP2) alters glucagon receptor trafficking in hepatocytes with functional effects on receptor signalling. Mol Metab 53:101296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM (1998) RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393:333–339. [DOI] [PubMed] [Google Scholar]
- Mendes T, Herledan A, Leroux F, Deprez B, Lambert JC, Kilinc D (2020) High-content screening for protein-protein interaction modulators using proximity ligation assay in primary neurons. Curr Protoc Cell Biol 86:e100. [DOI] [PubMed] [Google Scholar]
- Meyrath M, Palmer CB, Reynders N, Vanderplasschen A, Ollert M, Bouvier M, Szpakowska M, Chevigné A (2021) Proadrenomedullin N-terminal 20 peptides (PAMPs) are agonists of the chemokine scavenger receptor ACKR3/CXCR7. ACS Pharmacol Transl Sci 4:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Ito Y, Nishizawa N, Amano H, Tsujikawa K, Miyaji K, Watanabe M, Majima M (2017) RAMP1 signaling improves lymphedema and promotes lymphangiogenesis in mice. J Surg Res 219:50–60. [DOI] [PubMed] [Google Scholar]
- Mizuta H, Takakusaki A, Suzuki T, Otake K, Dohmae N, Simizu S (2022) C-mannosylation regulates stabilization of RAMP1 protein and RAMP1-mediated cell migration. FEBS J DOI: 10.1111/febs.16592 [online ahead of print] [DOI] [PubMed] [Google Scholar]
- Moreno Delgado D, Møller TC, Ster J, Giraldo J, Maurel D, Rovira X, Scholler P, Zwier JM, Perroy J, Durroux T, et al. (2017) Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. eLife 6 DOI: 10.7554/eLife.25233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfis M, Tilakaratne N, Furness SG, Christopoulos G, Werry TD, Christopoulos A, Sexton PM (2008) Receptor activity-modifying proteins differentially modulate the G protein-coupling efficiency of amylin receptors. Endocrinology 149:5423–5431. [DOI] [PubMed] [Google Scholar]
- Muschter D, Beiderbeck AS, Späth T, Kirschneck C, Schröder A, Grässel S (2019) Sensory neuropeptides and their receptors participate in mechano-regulation of murine macrophages. Int J Mol Sci 20:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag K, Kato A, Nakada T, Hoshijima K, Mistry AC, Takei Y, Hirose S (2006) Molecular and functional characterization of adrenomedullin receptors in pufferfish. Am J Physiol Regul Integr Comp Physiol 290:R467–R478. [DOI] [PubMed] [Google Scholar]
- Nag K, Sultana N, Hirose S (2012) Calcitonin receptor-like receptor (CLR) influences posttranslational events of receptor activity-modifying proteins (RAMPs). Biochem Biophys Res Commun 418:824–829. [DOI] [PubMed] [Google Scholar]
- Nagar H, Kim S, Lee I, Choi SJ, Piao S, Jeon BH, Shong M, Kim CS (2021) CRIF1 deficiency suppresses endothelial cell migration via upregulation of RhoGDI2. PLoS One 16:e0256646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec K, Schihada H, Kleinau G, Zabel U, Grushevskyi EO, Scheerer P, Lohse MJ, Maiellaro I (2022) Functional modulation of PTH1R activation and signaling by RAMP2. Proc Natl Acad Sci USA 119:e2122037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Clevers H (2017) Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169:985–999. [DOI] [PubMed] [Google Scholar]
- Pacharne S, Livesey M, Kadmiel M, Wang N, Caron KM, Richards GO, Skerry TM (2022) Accelerated development with increased bone mass and skeletal response to loading suggest receptor activity modifying protein-3 as a bone anabolic target. Front Endocrinol (Lausanne) 12:807882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran N, Spielman WS (2006) RAMPs: the past, present and future. Trends Biochem Sci 31:631–638. [DOI] [PubMed] [Google Scholar]
- Patel A, Kimura R, Fu W, Soudy R, MacTavish D, Westaway D, Yang J, Davey RA, Zajac JD, Jhamandas JH (2021) Genetic depletion of amylin/calcitonin receptors improves memory and learning in transgenic Alzheimer’s disease mouse models. Mol Neurobiol 58:5369–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak JB, Wetzel-Strong SE, Dunn MK, Caron KM (2017) Cardiovascular effects of exogenous adrenomedullin and CGRP in Ramp and Calcrl deficient mice. Peptides 88:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce A, Redfern-Nichols T, Harris M, Poyner DR, Wigglesworth M, Ladds G (2022) Determining the effects of differential expression of GRKs and β-arrestins on CLR-RAMP agonist bias. Front Physiol 13:840763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T (2012) Structural determinants of the supramolecular organization of G protein-coupled receptors in bilayers. J Am Chem Soc 134:10959–10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V, Zhu Y, Dal Maso E, Reynolds CA, Deganutti G, Atanasio S, Hick CA, Yang D, Christopoulos A, Hay DL, et al. (2019) Deconvoluting the molecular control of binding and signaling at the amylin 3 receptor: RAMP3 alters signal propagation through extracellular loops of the calcitonin receptor. ACS Pharmacol Transl Sci 2:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps E, Bezouglaia O, Tetradis S, Nervina JM (2005) Parathyroid hormone induces receptor activity modifying protein-3 (RAMP3) expression primarily via 3′,5′-cyclic adenosine monophosphate signaling in osteoblasts. Calcif Tissue Int 77:96–103. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ (2002) Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3:639–650. [DOI] [PubMed] [Google Scholar]
- Pin JP, Bettler B (2016) Organization and functions of mGlu and GABAB receptor complexes. Nature 540:60–68. [DOI] [PubMed] [Google Scholar]
- Pioszak AA, Hay DL (2020) RAMPs as allosteric modulators of the calcitonin and calcitonin-like class B G protein-coupled receptors. Adv Pharmacol 88:115–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondel MD, Mould R (2005) Cloning and transcriptional analysis of the mouse receptor activity modifying protein-1 gene promoter. BMC Mol Biol 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash J, Herlin M, Kumar J, Garg G, Akesson KE, Grabowski PS, Skerry TM, Richards GO, McGuigan FEA (2019) Analysis of RAMP3 gene polymorphism with body composition and bone density in young and elderly women. Gene X 2:100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Christopoulos G, Bailey RJ, Christopoulos A, Sexton PM, Hay DL (2008) Identification of N-terminal receptor activity-modifying protein residues important for calcitonin gene-related peptide, adrenomedullin, and amylin receptor function. Mol Pharmacol 74:1059–1071. [DOI] [PubMed] [Google Scholar]
- Qi T, Hay DL (2010) Structure-function relationships of the N-terminus of receptor activity-modifying proteins. Br J Pharmacol 159:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner RE, Dickey R, Fineman M, Maggs DG, Shen L, Strobel SA, Weyer C, Kolterman OG (2004) Amylin replacement with pramlintide as an adjunct to insulin therapy improves long-term glycaemic and weight control in Type 1 diabetes mellitus: a 1-year, randomized controlled trial. Diabet Med 21:1204–1212. [DOI] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA 99:13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter U, Goadsby PJ, Lanteri-Minet M, Wen S, Hours-Zesiger P, Ferrari MD, Klatt J (2018) Efficacy and tolerability of erenumab in patients with episodic migraine in whom two-to-four previous preventive treatments were unsuccessful: a randomised, double-blind, placebo-controlled, phase 3b study. Lancet 392:2280–2287. [DOI] [PubMed] [Google Scholar]
- Roehrkasse AM, Booe JM, Lee SM, Warner ML, Pioszak AA (2018) Structure-function analyses reveal a triple β-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design. J Biol Chem 293:15840–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge SJ, Simms J, Clark A, Yeung HY, Wigglesworth MJ, Dickerson IM, Kitchen P, Ladds G, Poyner DR (2020) Receptor component protein, an endogenous allosteric modulator of family B G protein coupled receptors. Biochim Biophys Acta Biomembr 1862:183174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux BT, Bauer CC, McNeish AJ, Ward SG, Cottrell GS (2017) The role of ubiquitination and hepatocyte growth factor-regulated tyrosine kinase substrate in the degradation of the adrenomedullin type I receptor. Sci Rep 7:12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux BT, Cottrell GS (2014) G protein-coupled receptors: what a difference a “partner” makes. Int J Mol Sci 15:1112–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, Hériché JK, Hu Y, Kristiansen K, Li R, et al. (2008) TreeFam: 2008 update. Nucleic Acids Res 36:D735–D740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell FA, King R, Smillie SJ, Kodji X, Brain SD (2014) Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev 94:1099–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan GJ, Jobe LJ, Martin R (2005) Pramlintide in the treatment of type 1 and type 2 diabetes mellitus. Clin Ther 27:1500–1512. [DOI] [PubMed] [Google Scholar]
- Schönauer R, Kaiser A, Holze C, Babilon S, Köbberling J, Riedl B, Beck-Sickinger AG (2015) Fluorescently labeled adrenomedullin allows real-time monitoring of adrenomedullin receptor trafficking in living cells. J Pept Sci 21:905–912. [DOI] [PubMed] [Google Scholar]
- Scuteri D, Tonin P, Nicotera P, Bagetta G, Corasaniti MT (2022) Real world considerations for newly approved CGRP receptor antagonists in migraine care. Expert Rev Neurother 22:221–230. [DOI] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM (2007) Melanocortin-2 receptor accessory protein MRAP forms antiparallel homodimers. Proc Natl Acad Sci USA 104:20244–20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM (2009a) Opposite effects of the melanocortin-2 (MC2) receptor accessory protein MRAP on MC2 and MC5 receptor dimerization and trafficking. J Biol Chem 284:22641–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM (2009b) Regions of melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor trafficking and signaling. J Biol Chem 284:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T (2018) The calcitonin/calcitonin gene-related peptide family in invertebrate deuterostomes. Front Endocrinol (Lausanne) 9:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T (2022) Evolution of calcitonin/calcitonin gene-related peptide family in chordates: identification of CT/CGRP family peptides in cartilaginous fish genome. Gen Comp Endocrinol 328:114123. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Kuwasako K, Ogasawara M, Takahashi H, Matsubara S, Osugi T, Muramatsu I, Sasayama Y, Suzuki N, Satake H (2016) Evidence for conservation of the calcitonin superfamily and activity-regulating mechanisms in the basal chordate Branchiostoma floridae: insights into the molecular and functional evolution in chordates. J Biol Chem 291:2345–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafin DS, Harris NR, Nielsen NR, Mackie DI, Caron KM (2020) Dawn of a new RAMPage. Trends Pharmacol Sci 41:249–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton PM, Poyner DR, Simms J, Christopoulos A, Hay DL (2009) Modulating receptor function through RAMPs: can they represent drug targets in themselves? Drug Discov Today 14:413–419. [DOI] [PubMed] [Google Scholar]
- Shao L, Chen Y, Zhang S, Zhang Z, Cao Y, Yang D, Wang MW (2022) Modulating effects of RAMPs on signaling profiles of the glucagon receptor family. Acta Pharm Sin B 12:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T, Tanaka M, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, Sakurai T (2022) Receptor activity modifying protein RAMP sub-isoforms and their functional differentiation, which regulates functional diversity of adrenomedullin. Biology (Basel) 11:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms J, Uddin R, Sakmar TP, Gingell JJ, Garelja ML, Hay DL, Brimble MA, Harris PW, Reynolds CA, Poyner DR (2018) Photoaffinity cross-linking and unnatural amino acid mutagenesis reveal insights into calcitonin gene-related peptide binding to the calcitonin receptor-like receptor/receptor activity-modifying protein 1 (CLR/RAMP1) complex. Biochemistry 57:4915–4922. [DOI] [PubMed] [Google Scholar]
- Skiba MA, Kruse AC (2021) Autoantibodies as endogenous modulators of GPCR Signaling. Trends Pharmacol Sci 42:135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. [DOI] [PubMed] [Google Scholar]
- Song Z, Zhang Y, Chen Z, Zhang B (2021) Identification of key genes in lung adenocarcinoma based on a competing endogenous RNA network. Oncol Lett 21:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudy R, Kimura R, Patel A, Fu W, Kaur K, Westaway D, Yang J, Jhamandas J (2019) Short amylin receptor antagonist peptides improve memory deficits in Alzheimer’s disease mouse model. Sci Rep 9:10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Muff R, Gujer R, Fischer JA, Born W (2002) The transmembrane domain of receptor-activity-modifying protein 1 is essential for the functional expression of a calcitonin gene-related peptide receptor. Biochemistry 41:11398–11404. [DOI] [PubMed] [Google Scholar]
- Stoddart LA, Johnstone EKM, Wheal AJ, Goulding J, Robers MB, Machleidt T, Wood KV, Hill SJ, Pfleger KDG (2015) Application of BRET to monitor ligand binding to GPCRs. Nat Methods 12:661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Chen LN, Zhou Q, Zhao LH, Yang D, Zhang H, Cong Z, Shen DD, Zhao F, Zhou F, et al. (2020) A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor. Cell Res 30:1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpakowska M, Meyrath M, Reynders N, Counson M, Hanson J, Steyaert J, Chevigné A (2018) Mutational analysis of the extracellular disulphide bridges of the atypical chemokine receptor ACKR3/CXCR7 uncovers multiple binding and activation modes for its chemokine and endogenous non-chemokine agonists. Biochem Pharmacol 153:299–309. [DOI] [PubMed] [Google Scholar]
- Tam CW, Husmann K, Clark NC, Clark JE, Lazar Z, Ittner LM, Götz J, Douglas G, Grant AD, Sugden D, et al. (2006) Enhanced vascular responses to adrenomedullin in mice overexpressing receptor-activity-modifying protein 2. Circ Res 98:262–270. [DOI] [PubMed] [Google Scholar]
- Tasma Z, Wills P, Hay DL, Walker CS (2020) Agonist bias and agonist-dependent antagonism at corticotrophin releasing factor receptors. Pharmacol Res Perspect 8:e00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper SJ (2018) History and review of anti-calcitonin gene-related peptide (CGRP) therapies: from translational research to treatment. Headache 58(Suppl 3):238–275. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Koth CM, Abdul-Manan N, Swenson L, Coll JT, Lippke JA, Lepre CA, Garcia-Guzman M, Moore JM (2010) Crystal structure of the ectodomain complex of the CGRP receptor, a class-B GPCR, reveals the site of drug antagonism. Structure 18:1083–1093. [DOI] [PubMed] [Google Scholar]
- Tsujikawa K, Yayama K, Hayashi T, Matsushita H, Yamaguchi T, Shigeno T, Ogitani Y, Hirayama M, Kato T, Fukada S, et al. (2007) Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc Natl Acad Sci USA 104:16702–16707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udawela M, Christopoulos G, Morfis M, Christopoulos A, Ye S, Tilakaratne N, Sexton PM (2006a) A critical role for the short intracellular C terminus in receptor activity-modifying protein function. Mol Pharmacol 70:1750–1760. [DOI] [PubMed] [Google Scholar]
- Udawela M, Christopoulos G, Morfis M, Tilakaratne N, Christopoulos A, Sexton PM (2008) The effects of C-terminal truncation of receptor activity modifying proteins on the induction of amylin receptor phenotype from human CTb receptors. Regul Pept 145:65–71. [DOI] [PubMed] [Google Scholar]
- Udawela M, Christopoulos G, Tilakaratne N, Christopoulos A, Albiston A, Sexton PM (2006b) Distinct receptor activity-modifying protein domains differentially modulate interaction with calcitonin receptors. Mol Pharmacol 69:1984–1989. [DOI] [PubMed] [Google Scholar]
- Udawela M, Hay DL, Sexton PM (2004) The receptor activity modifying protein family of G protein coupled receptor accessory proteins. Semin Cell Dev Biol 15:299–308. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Björling E, Agaton C, Szigyarto CA-K, Amini B, Andersen E, Andersson A-C, Angelidou P, Asplund A, Asplund C, et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4:1920–1932. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347:1260419. [DOI] [PubMed] [Google Scholar]
- Wattiez AS, Sowers LP, Russo AF (2020) Calcitonin gene-related peptide (CGRP): role in migraine pathophysiology and therapeutic targeting. Expert Opin Ther Targets 24:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C, Lu J, Li N, Barkan K, Richards GO, Roberts DJ, Skerry TM, Poyner D, Pardamwar M, Reynolds CA, et al. (2015) Modulation of glucagon receptor pharmacology by receptor activity-modifying protein-2 (RAMP2). J Biol Chem 290:23009–23022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C, Winfield I, Harris M, Hodgson R, Shah A, Dowell SJ, Mobarec JC, Woodlock DA, Reynolds CA, Poyner DR, et al. (2016) Receptor activity-modifying protein-directed G protein signaling specificity for the calcitonin gene-related peptide family of receptors. J Biol Chem 291:21925–21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CW, Vanyai HK, See HB, Johnstone EKM, Pfleger KDG (2017) Using nanoBRET and CRISPR/Cas9 to monitor proximity to a genome-edited protein in real-time. Sci Rep 7:3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley MJ, Reynolds CA, Simms J, Walker CS, Mobarec JC, Garelja ML, Conner AC, Poyner DR, Hay DL (2017) Receptor activity-modifying protein dependent and independent activation mechanisms in the coupling of calcitonin gene-related peptide and adrenomedullin receptors to Gs. Biochem Pharmacol 142:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D, Lindmark H, Kadmiel M, Willcockson H, Caron KM, Barwell J, Drmota T, Poyner DR (2013) Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. Br J Pharmacol 168:822–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Yang L, Hang K, Laursen M, Wu L, Han GW, Ren Q, Roed NK, Lin G, Hanson MA, et al. (2020) Full-length human GLP-1 receptor structure without orthosteric ligands. Nat Commun 11:1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu V, Yeerna H, Nohata N, Chiou J, Harismendy O, Raimondi F, Inoue A, Russell RB, Tamayo P, Gutkind JS (2019) Illuminating the Onco-GPCRome: novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J Biol Chem 294:11062–11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Piston DW, Johnson CH (1999) A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc Natl Acad Sci USA 96:151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi A, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, Igarashi K, Toriyama Y, Tanaka M, Liu T, Xian X, et al. (2014) Functional differentiation of RAMP2 and RAMP3 in their regulation of the vascular system. J Mol Cell Cardiol 77:73–85. [DOI] [PubMed] [Google Scholar]
- Yarwood RE, Imlach WL, Lieu T, Veldhuis NA, Jensen DD, Klein Herenbrink C, Aurelio L, Cai Z, Christie MJ, Poole DP, et al. (2017) Endosomal signaling of the receptor for calcitonin gene-related peptide mediates pain transmission. Proc Natl Acad Sci USA 114:12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Köhrer C, Huber T, Kazmi M, Sachdev P, Yan ECY, Bhagat A, RajBhandary UL, Sakmar TP (2008) Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem 283:1525–1533. [DOI] [PubMed] [Google Scholar]
- Yin S, Song R, Ma J, Liu C, Wu Z, Cao G, Liu J, Zhang G, Zhang H, Sun R, et al. (2022) Receptor activity-modifying protein 1 regulates mouse skin fibroblast proliferation via the Gαi3-PKA-CREB-YAP axis. Cell Commun Signal 20:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, Iesato Y, Koyama T, Uetake R, Yang L, Yamauchi A, et al. (2013) Novel regulation of cardiac metabolism and homeostasis by the adrenomedullin-receptor activity-modifying protein 2 system. Hypertension 61:341–351. [DOI] [PubMed] [Google Scholar]
- Yu T, Su X, Pan Y, Zhuang H (2017) Receptor-transporting protein (RTP) family members play divergent roles in the functional expression of odorant receptors. PLoS One 12:e0179067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qiao A, Yang L, Van Eps N, Frederiksen KS, Yang D, Dai A, Cai X, Zhang H, Yi C, et al. (2018) Structure of the glucagon receptor in complex with a glucagon analogue. Nature 553:106–110. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun B, Feng D, Hu H, Chu M, Qu Q, Tarrasch JT, Li S, Sun Kobilka T, Kobilka BK, et al. (2017) Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546:248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Morgan DA, Kuburas A, Thedens DR, Russo AF, Rahmouni K (2011) Neuronal receptor activity-modifying protein 1 promotes energy expenditure in mice. Diabetes 60:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Ma S, Sutkeviciute I, Shen DD, Zhou XE, de Waal PW, Li CY, Kang Y, Clark LJ, Jean-Alphonse FG, et al. (2019) Structure and dynamics of the active human parathyroid hormone receptor-1. Science 364:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]