Abstract
RNA-binding proteins are important regulators of T cell activation, proliferation, and cytokine production. The zinc finger protein 36 (ZFP36) family genes (Zfp36, Zfp36l1, and Zfp36l2) encode RNA-binding proteins that promote the degradation of transcripts containing AU-rich elements. Numerous studies have demonstrated both individual and shared functions of the ZFP36 family in immune cells, but their collective function in T cells remains unclear. Here, we found a redundant and critical role for the ZFP36 proteins in regulating T cell quiescence. T cell-specific deletion of all three ZFP36 family members in mice resulted in early lethality, immune cell activation, and multiorgan pathology characterized by inflammation of the eyes, central nervous system, kidneys, and liver. Mice with T cell-specific deletion of any two Zfp36 genes were protected from this spontaneous syndrome. Triply-deficient T cells overproduced proinflammatory cytokines, including IFNγ, TNF, and GM-CSF, due to increased mRNA stability of these transcripts. Surprisingly, T cell-specific deletion of both Zfp36l1 and Zfp36l2 rendered mice resistant to experimental autoimmune encephalomyelitits due to failed priming of antigen-specific CD4+ T cells. ZFP36L1 and ZFP36L2 double-deficient CD4+ T cells had poor proliferation during in vitro T helper cell polarization. Thus, the ZFP36 family redundantly regulates T cell quiescence at homeostasis, but ZFP36L1 and ZFP36L2 are specifically required for antigen-specific T cell clonal expansion.
One Sentence Summary:
T cell homeostasis depends redundantly on the ZFP36 family of RNA-binding proteins while T cell priming depends on ZFP36L1 and ZFP36L2.
INTRODUCTION
Post-transcriptional regulation of messenger RNAs is an important mechanism to regulate protein production. A variety of molecules, including microRNAs, long-noncoding RNAs, and RNA-binding proteins, positively and negatively regulate mRNA turnover and translation. These molecules recognize specific mRNA secondary structures or linear sequences, such as adenine-uridine-rich elements (AREs), to bind target transcripts (1). Many cytokine and chemokine mRNAs contain AREs within their 3’ untranslated regions which control their stability, including the mRNAs expressed from the Tnf and Ifng loci. When the AREs of Tnf or Ifng are genetically deleted, resulting mice display excessive production of these cytokines, leading to immunopathology (2, 3).
T cells depend on individual or redundant members of RNA-binding protein families, such as the Regnase family (4-8) and the Roquin-1/−2 family (6, 9-13) to negatively regulate their activation and prevent autoreactivity. The broadly expressed mammalian zinc finger protein 36 (ZFP36) family, composed of Zfp36 (encoding the protein commonly called tristetraprolin or TTP), Zfp36l1, and Zfp36l2, binds AREs to negatively regulate mRNA transcript production into protein. Rodents additionally contain a fourth family member, Zfp36l3, which is only expressed in the placenta and yolk sac (14-16). The ZFP36 family can recruit deadenylase complexes to promote target mRNA decay or inhibit target mRNA translation in a decay-independent manner (17-24). Global deficiency in mice of any one family member is either developmentally lethal (Zfp36l1 or Zfp36l2) (25, 26) or results in a severe inflammatory condition with excessive TNF and IL-23 production leading to cachexia and arthritis (Zfp36) (17, 27-29). While Zfp36l1 and Zfp36l2 redundantly control early thymic development of T cells (30, 31), there is little understanding of how the ZFP36 family collectively coordinates effector T cell responses. ZFP36 has been shown to negatively regulate mRNA targets in T cell pathways of activation and proliferation (32) and negatively regulate T cell IL-17A production in aged mice (33). ZFP36L2 was shown to be an important translational repressor of many transcripts (including Ifng) in memory CD4+ and CD8+ T cells (24). However, the full spectrum of ZFP36 family activity in T cells, both individually and redundantly, remains unexplored.
We investigated the collective functions of the ZFP36 family of genes in T cells using a conditional deletion strategy in mice. T cell-specific deletion of all three family members resulted in a lethal, multiorgan inflammatory syndrome characterized by markedly increased T cell cytokine production. Mice deficient in any combination of two ZFP36 family members did not exhibit this syndrome, revealing functional redundancy within the family in the regulation of T cell quiescence. Together, the ZFP36 family negatively regulated activation, proliferation, and cell death pathways in T cells. Using ZFP36 family single- and double-deficient mice, we found that T cells surprisingly rely on ZFP36L1 and ZFP36L2 for CD4+ T cell priming in experimental autoimmune encephalomyelitis (EAE). Taken together, our studies reveal that T cell homeostasis is critically maintained by the three ZFP36 family members acting redundantly, while, in contrast, an antigen-specific autoimmune response is specifically supported by ZFP36L1 and ZFP36L2.
RESULTS
The ZFP36 family acts redundantly in T cells for the prevention of a lethal inflammatory syndrome
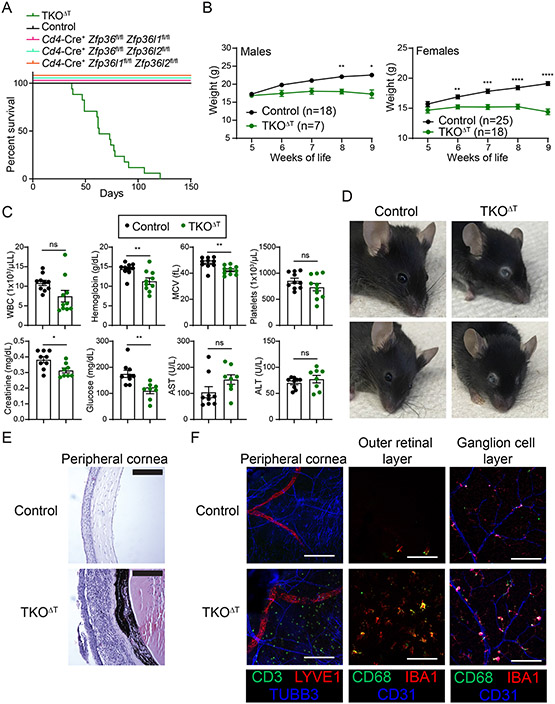
As global knockout mice for individual ZFP36 family members have severe or lethal phenotypes (17, 25-29), we crossed mice with floxed alleles of Zfp36, Zfp36l1, and Zfp36l2 to Cd4-Cre mice to conditionally delete individual and multiple family members in T cells. Mice with T cell-specific conditional deletion of all three family members, Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl Zfp36l2fl/fl (hereafter referred to as TKOΔT), died beginning at 6 weeks of age with a median survival of approximately 10 weeks (Fig. 1A). Mice with T cell-specific deletion of any two ZFP36 family members (broadly referred to as DKOΔT mice; Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl, Cd4-Cre+ Zfp36fl/fl Zfp36l2fl/fl, or Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl) did not experience this early lethality, indicating full redundancy within the ZFP36 family in T cells to prevent lethality. TKOΔT male and female mice failed to gain weight compared to Cd4-Cre− Zfp36fl/fl Zfp36l1fl/fl Zfp36l2fl/fl littermate controls (hereafter Controls) (Fig. 1B). TKOΔT mice displayed microcytic anemia, reduced serum creatinine levels, hypoglycemia, decreased arterial blood pressure, and bradycardia compared to Control mice (Fig. 1C and data file S1).
Figure 1: TKOΔT mice exhibit spontaneous mortality and ocular pathology.
A, Survival curves in the indicated mice at homeostasis (n=6-18/group). B, Control and TKOΔT mice were weighed weekly (n=7-25/group). C, Blood and sera were collected from Control and TKOΔT mice for CBCs and the indicated chemistry tests (pooled from 2 experiments, n=8-10/group). D, Photographs of the gross ocular phenotype in TKOΔT mice. Two representative mice from each genotype are shown. E, H&E staining on corneas from Control and TKOΔT eyes. Scale bar is 50 μm. F, Immunofluorescent staining of different eye regions of Control and TKOΔT mice. The peripheral cornea was stained with anti-CD3 (green), anti-LYVE1 (red), and anti-TUBB3 (blue), and the outer retinal layer and ganglion cell layer were stained with anti-CD68 (green), anti-IBA1 (red), and anti-CD31 (blue). Scale bar is 100 μm. Data in B,C are mean ± s.e.m. Two-way ANOVA with Šídák’s multiple comparisons test (B); unpaired two-sided Student’s t-test (C). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
One notable aspect of the syndrome seen in TKOΔT mice was severe ocular inflammation (Fig. 1D). H&E staining revealed infiltration of immune cells in multiple regions of the eyes of TKOΔT mice, including the cornea (Fig. 1E and fig. S1A). Immunofluorescent imaging in TKOΔT mice showed T cell infiltration of the peripheral cornea and myeloid cell activation in multiple ocular regions, including the cornea, choroid, and retina (Fig. 1F and fig. S1B).
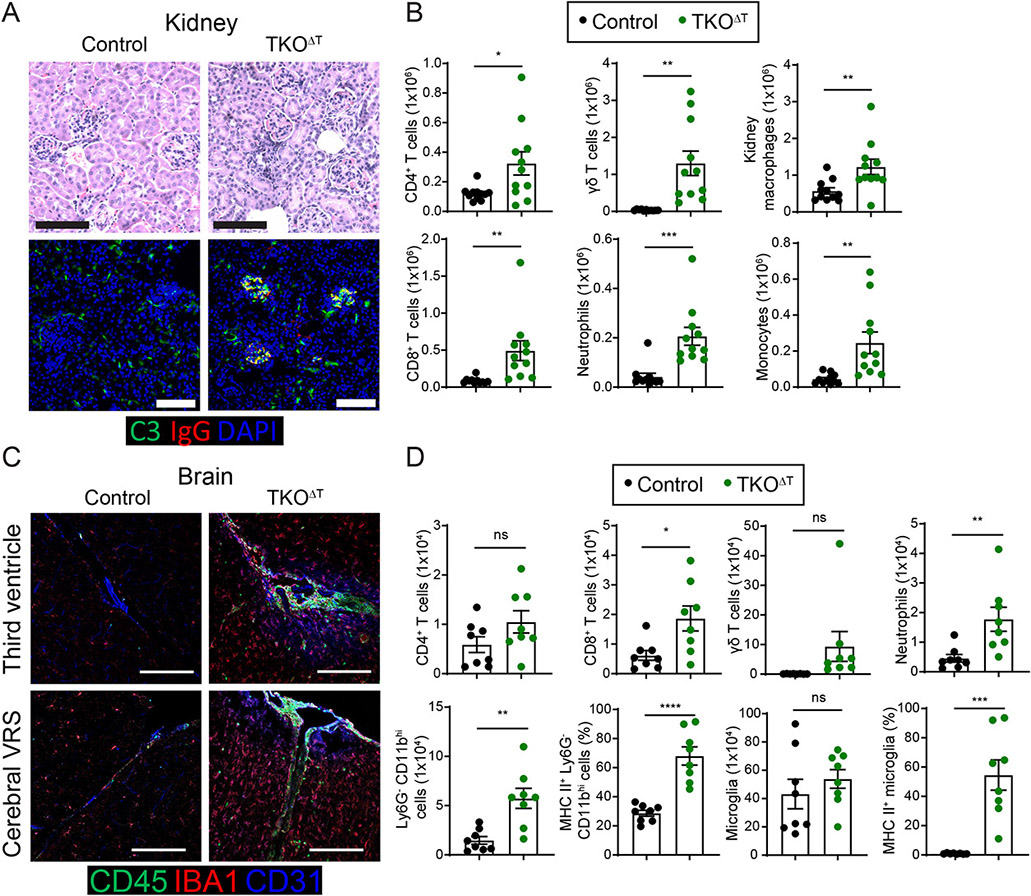
An initial necropsy performed on a 9 week-old TKOΔT mouse revealed marked inflammation of the kidneys and liver. Subsequent studies showed that kidneys of TKOΔT mice consistently displayed extraglomerular inflammation (Fig. 2A) and in some cases glomerulonephritis. We found evidence of complement C3 protein and IgG deposition in the glomeruli of kidneys from TKOΔT mice but not Control mice (Fig. 2A). Using flow cytometry, we characterized an increase in immune cell populations in the kidneys of TKOΔT mice, including CD4+ T cells, CD8+ T cells, γδ T cells, neutrophils, monocytes, and kidney resident macrophages (Fig. 2B and fig. S2C). Livers of TKOΔT mice displayed periportal inflammation and immune cell infiltration as determined by flow cytometry (fig. S2, A-B, E).
Figure 2: Broad immune cell activation and infiltration into peripheral organs of TKOΔT mice.
A, Top: H&E staining of kidney sections from Control and TKOΔT mice. Bottom: Complement C3 (green), mouse IgG (red), and DAPI (blue) immunofluorescent staining of kidneys from Control and TKOΔT mice. B, Quantitation of kidney CD4+ T cells (F4/80− Ly6Clow CD11blow TCRγδ− TCRβ+ CD4+), CD8+ T cells (F4/80− Ly6Clow CD11blow TCRγδ− TCRβ+ CD8α+), γδ T cells (F4/80− Ly6Clow CD11blow TCRβ− TCRγδ+), neutrophils (F4/80− Ly6Chi CD11bint Ly6G+), kidney macrophages (F4/80+ Ly6C− MHC II+), and monocytes (F4/80− Ly6Cint CD11b+) in Control and TKOΔT mice (pooled from 6 experiments, n=11/group). C, CD45 (green), IBA1 (red) and CD31 (blue) immunofluorescent staining of brains from Control and TKOΔT mice at the third ventricle (top) and a cerebral Virchow-Robin space (VRS) (bottom). Scale bar is 100 μm. D, Quantitation of brain CD4+ T cells (CD45+ CD11blow TCR γδ− TCRβ+ CD4+), CD8+ T cells (CD45+ CD11blow TCR γδ− TCRβ+ CD8α+), γδ T cells (CD45+ CD11blow TCRβ− TCR γδ+), neutrophils (CD45+ CD11bhi Ly6G+), Ly6G− CD11bhi cells (CD45+ CD11bhi Ly6G−), MHC II+ Ly6G− CD11bhi cells, microglia (CD45int CD11bint), and MHC II+ microglia in Control and TKOΔT mice (pooled from 4 experiments, n=8/group). Data in B,D are mean ± s.e.m. Unpaired two-sided Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
We noticed an impaired gait and righting reflex in some TKOΔT mice and therefore analyzed their central nervous system (CNS). TKOΔT mice had increased CD45+ cells in brain interface regions, such as the third ventricle and cerebral Virchow-Robin spaces (VRS), relative to Control mice (Fig. 2C). Flow cytometric analysis of brains of TKOΔT mice showed increased CD8+ T cells, γδ T cells, neutrophils, and Ly6G− CD11bhi myeloid cells (Fig. 2D and fig. S2D). There was also a higher percentage of MHC II+ cells amongst Ly6G− CD11bhi myeloid cells and microglia (Fig. 2D). In summary, TKOΔT mice develop a multiorgan inflammatory syndrome involving the eyes, kidneys, liver and brain.
T cell lymphopenia and activation in TKOΔT mice
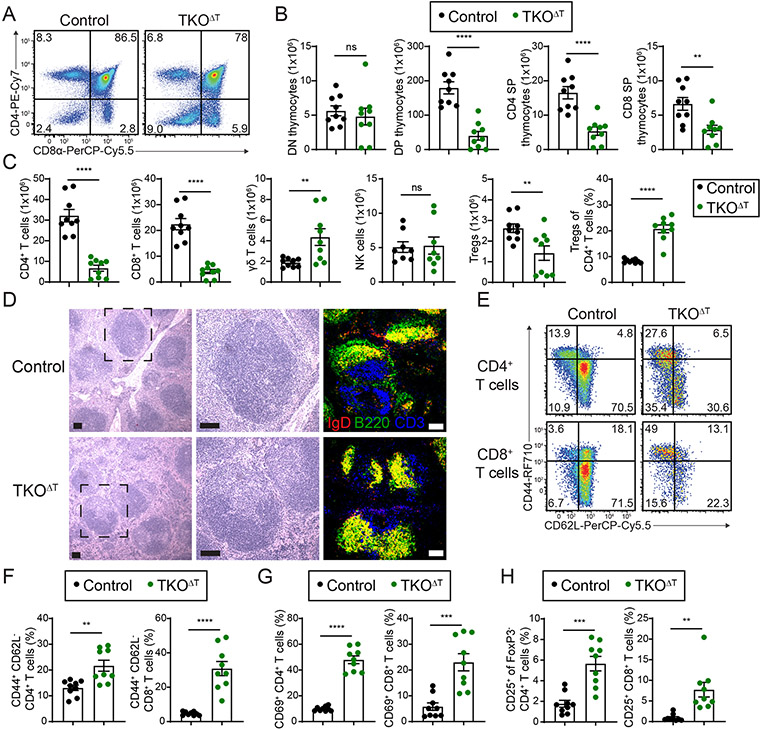
As Cd4-Cre-mediated recombination of target genes is largely complete by the double positive thymocyte stage (34), we examined the thymi of TKOΔT mice. Compared to Control mice, TKOΔT mice experienced a defect in T cell development starting at the double negative (DN) 4 stage of development, with robust numerical reductions of double positive (DP), CD4 single positive (SP), and CD8 SP thymocytes (Fig. 3, A and B, and fig. S3, A and B). Additionally, splenic CD4+ and CD8+ T cells failed to develop from TKOΔT bone marrow in mixed bone marrow chimeras, indicating a cell-intrinsic T cell developmental disadvantage (fig. S3, C and D). Despite these developmental defects, TKOΔT T cells did not have a dramatically altered TCR repertoire, with only minor differences in the usage of TCR Vβ gene segments compared to Control T cells (fig. S3E).
Figure 3: T cell lymphopenia and T cell activation in TKOΔT mice.
A, CD4 and CD8α staining of thymocytes (pre-gated as Ly6G− NK1.1− CD19− TCRγδ− CD11b−) from Control and TKOΔT mice. B, Quantitation of CD4− CD8α− (double negative, DN), CD4+ CD8α+ (double positive, DP), CD4+ CD8α− (CD4 single positive, CD4 SP), and CD4− CD8α+ (CD8α single positive, CD8 SP) thymocytes (pooled from 5 experiments, n=9/group). C, Quantitation of the indicated splenic populations in Control and TKOΔT mice: CD4+ T cells, CD8+ T cells, γδ T cells, NK cells (NK1.1+ TCRβ−), and the number and frequency of Tregs (TCRβ+ CD4+ CD25+ FoxP3+) (pooled from 5 experiments, n=8-9/group). D, H&E and immunofluorescent staining of spleens from Control and TKOΔT mice. Left: low magnification view of H&E stained spleen. Middle: high magnification view of the dashed black box. Right: Immunofluorescent staining for CD3 (blue), B220 (green), and IgD (red) of spleens from Control and TKOΔT mice. Scale bar is 100 μm. E, CD44 and CD62L staining of splenic CD4+ and CD8+ T cells from Control and TKOΔT mice. F-H, Quantitation of the percentage of (F) CD44+ CD62L− CD4+ and CD8+ T cells, (G) CD69+ CD4+ and CD8+ T cells, and (H) CD25+ FoxP3− CD4+ and CD25+ CD8+ T cells in the spleens of indicated mice (pooled from 5 experiments, n=9/group). Data in B,C,F-H are mean ± s.e.m; unpaired two-sided Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
T cells were found in the periphery of TKOΔT mice, though mature CD4+ and CD8+ T cells were severely reduced in number in the spleen and blood compared to Control mice and there was a concomitant increase in γδ T cells and neutrophils with no change in NK cells (Fig. 3C and fig. S3F). Splenic regulatory T cells (Tregs) were slightly decreased in TKOΔT mice versus Control mice, though they were increased as a percentage of the CD4+ T cell population (Fig. 3C). H&E and immunofluorescent staining of splenic sections from TKOΔT and Control mice confirmed that white pulp follicles in TKOΔT mice contained reduced CD3+ T cells and revealed an absence of B220+ IgD− marginal zone B cells (Fig. 3D). We confirmed a significant reduction in splenic marginal zone B cells (B220+ CD19+ CD21hi CD23low) and in total blood B cells in TKOΔT mice compared to Control mice (fig. S3, F and G). Total splenic B cells (B220+ CD19+), follicular B cells (B220+ CD19+ CD21int CD23hi), and serum immunoglobulin levels were not significantly reduced in TKOΔT mice (fig. S3, G and H). Given that immune complexes were observed in the glomeruli of TKOΔT kidneys (Fig. 2A), we screened Control, TKOΔT, and DKOΔT sera for the presence of autoantibodies using a microarray panel of autoantigens. Serum from TKOΔT mice contained autoantibodies, especially of the IgM isotype, specific for a number of nuclear autoantigens, including dsDNA and several ribonucleoproteins (RNPs) (fig. S4A). Control and DKOΔT sera were largely devoid of IgM autoantibodies and contained variable levels of IgG autoantibodies as measured by this microarray assay.
Splenic CD4+ and CD8+ T cells in TKOΔT mice displayed an activated phenotype, with a higher percentage of CD44+ CD62L−, CD69+, and CD25+ cells compared to Control T cells (Fig. 3, E to H). With the exception of Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl T cells which showed a partially activated phenotype, DKOΔT CD4+ and CD8+ T cells showed minimal signs of activation (fig. S4B). To examine the possibility that defective Treg function could contribute to the increased activation of conventional TKOΔT T cells, we phenotyped splenic Tregs from Control and TKOΔT mice (fig. S4C). TKOΔT Tregs expressed similar or greater levels of several markers associated with Treg functionality, including ICOS, CTLA-4, PD-1, TIGIT, and CD44, compared to Control Tregs (fig. S4D). In addition, a similar proportion of Control and TKOΔT Tregs expressed Helios, suggesting normal Treg development. Nevertheless, TKOΔT Tregs were deficient compared to Control Tregs in suppressing proliferation of wild type CD4+ T cells in an in vitro suppression assay (fig. S4E). Taken together, the absence of the ZFP36 proteins in T cells resulted in T cell lymphopenia, T cell activation, absence of marginal zone B cells, and autoreactive immunoglobulins.
TKOΔT T cells overproduce proinflammatory cytokines
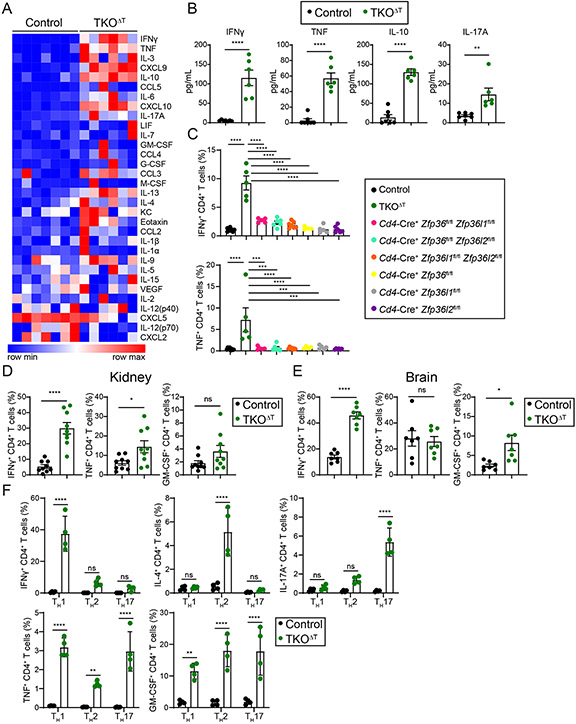
Knowing that the ZFP36 family proteins are important negative regulators of T cell cytokine production (24, 32, 33), we examined whether hypercytokinemia was a feature of TKOΔT mice. Sera of TKOΔT mice contained elevated cytokines and chemokines compared to Control mice, especially IFNγ, TNF, IL-10, and IL-17A (Fig. 4, A and B). When bulk splenocytes were cultured without stimulation for 4 h in the presence of Brefeldin A, TKOΔT CD4+ T cells produced excessive IFNγ and TNF (Fig. 4C). This secretion was not seen in either Control CD4+ T cells or CD4+ T cells deficient in one or two ZFP36 family members. A similar phenomenon was seen in TKOΔT CD8+ T cells, though one double deletion strain, Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl, had an intermediate phenotype in IFNγ production (fig. S5A). To determine whether non-stimulated production of IFNγ and TNF from TKOΔT T cells required de novo transcription and/or translation, bulk splenocytes were cultured in the presence of Brefeldin A with or without a transcription inhibitor (Actinomycin D) or a translation inhibitor (cycloheximide, CHX). In CD4+ TKOΔT T cells, IFNγ production was only partially reliant on new transcription, while TNF production was dependent on new transcription (fig. S5B). In CD8+ TKOΔT T cells, IFNγ production was not reliant on new transcription, while TNF was only partially reliant on new transcription (fig. S5B).
Figure 4: Hypercytokinemia in TKOΔT mice.
A, Heat map of serum cytokines and chemokines from Control and TKOΔT mice (n=6-7/group). B, Concentration of IFNγ, TNF, IL-10, and IL-17A in the sera of Control and TKOΔT mice (n=6-7/group). C, Quantitation of IFNγ+ and TNF+ CD4+ T cells from the indicated mice determined by ICS of bulk splenocytes cultured for 4 h with Brefeldin A (pooled from 4 experiments, n=5-8/group). D,E, Quantitation of IFNγ+, TNF+, and GM-CSF+ CD4+ T cells from the (D) kidney (pooled from 5 experiments, n=9/group) and (E) brain (pooled from 4 experiments, n=7/group) of Control and TKOΔT mice. F, CD4+ splenic T cells were polarized in different TH conditions. On day 4, cells were cultured for 4 h with Brefeldin A before ICS for IFNγ, IL-4, IL-17A, TNF, and GM-CSF (pooled from 2 experiments, n=4/group). Data in B-F are mean ± s.e.m. Unpaired two-sided Student’s t-test (B,D,E); one-way ANOVA with Tukey’s multiple comparisons test (C), two-way ANOVA with Šídák’s multiple comparisons test (F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
T cells isolated from infiltrated peripheral organs of TKOΔT mice overproduced proinflammatory cytokines upon stimulation with PMA and ionomycin. There was an increased percentage of IFNγ+ and TNF+ CD4+ T cells, as well as a higher percentage of IFNγ+ CD8+ T cells in the kidneys of TKOΔT mice (Fig. 4D and fig. S5C). In the brains of TKOΔT mice, there was an increase in the proportion of IFNγ+ and GM-CSF+ CD4+ T cells, with no difference in cytokine production from TKOΔT CD8+ T cells (Fig. 4E and fig. S5D). Infiltrating CD4+ and CD8+ TKOΔT T cells in the liver primarily overproduced IFNγ (fig. S5E).
As TKOΔT CD4+ T cells overproduced cytokines ex vivo, we hypothesized that polarization to different TH subsets would result in greater cytokine production. After 4 days of polarization and without re-stimulation, TKOΔT TH1 cells overproduced IFNγ, TKOΔT TH2 cells overproduced IL-4, TKOΔT TH17 cells overproduced IL-17A, and all three subsets overproduced TNF and GM-CSF compared to Control T cells (Fig. 4F). When the cells were re-stimulated, most cytokine production was equivalent between TKOΔT and Control TH subsets, except for GM-CSF, which was markedly increased in all TKOΔT TH subsets (fig. S5F). In summary, TKOΔT T cells overproduced several proinflammatory cytokines ex vivo and had a greater propensity for cytokine production after TH polarization.
Transcriptional analysis reveals dysregulation of proliferation and cell death in TKOΔT T cells
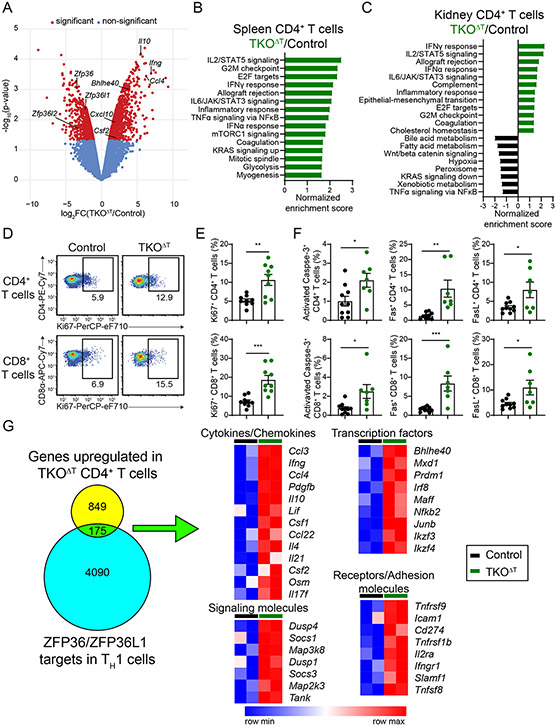
We sorted CD4+ and CD8+ T cells from the spleens and kidneys of Control and TKOΔT mice for RNA-sequencing. In splenic TKOΔT CD4+ T cells, we found 750 downregulated genes and 1024 upregulated genes compared to Control splenic CD4+ T cells, with several upregulated genes encoding for cytokines (Il10, Ifng, Ccl4, Cxcl10, and Csf2) (Fig. 5A and data file S2). Gene set enrichment analysis revealed pathways enriched in TKOΔT CD4+ T cells including activation pathways (IL2/STAT5 signaling, IFNγ response, Allograft rejection, IL6/JAK/STAT3 signaling, Inflammatory response, TNF signaling via NFκB, and IFNα response), proliferative pathways (G2M checkpoint, E2F targets, Mitotic spindle), and metabolic pathways (Glycolysis, mTORC1 signaling) (Fig. 5B). No pathways were statistically enriched in Control splenic CD4+ T cells. Similar activation and proliferation pathways were found to be enriched in kidney TKOΔT CD4+ T cells, while several metabolic pathways (Bile acid metabolism, Fatty acid metabolism, Xenobiotic metabolism) were enriched in Control CD4+ T cells (Fig. 5C).
Figure 5: RNA-sequencing reveals inflammatory and proliferative pathways enriched in TKOΔT T cells.
A-C, RNA-sequencing of splenic and kidney CD4+ T cells from Control and TKOΔT mice (n=2/group). A, Volcano plot identifying differentially expressed genes between Control and TKOΔT splenic CD4+ T cells. Red dots indicate genes significantly differentially expressed (P-value < 0.05 and fold change ≥ 2). B,C, Hallmark pathways enriched in TKOΔT (B) splenic CD4+ T cells and (C) kidney CD4+ T cells by GSEA. If more than 15 pathways were statistically enriched, only the top 15 pathways are shown. D, Ki67 staining of splenic CD4+ and CD8+ T cells. E, Quantitation of Ki67+ CD4+ and CD8+ splenic T cells from Control and TKOΔT mice (pooled from 4 experiments, n=9/group). F, Quantitation of activated Caspase-3+, Fas+, and FasL+ CD4+ and CD8+ splenic T cells from Control and TKOΔT mice (pooled from 4 experiments, n=7-10/group). G, Left: Overlap of the 1024 genes upregulated in splenic CD4+ TKOΔT T cells with published ZFP36/ZFP36L1 mRNA targets in murine TH1 cells (GSE96074) (32). Right: Heat maps of gene expression of selected genes from the overlapping set. Data in E,F are mean ± s.e.m. Unpaired two-sided Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
There were 1085 downregulated genes and 1386 upregulated genes in splenic TKOΔT CD8+ T cells compared to Control splenic CD8+ T cells (fig. S6A and data file S2). Hallmark pathways enriched in splenic TKOΔT CD8+ T cells were similar to those enriched in splenic TKOΔT CD4+ T cells, including many of the same activation and proliferative pathways, but also pathways that might indicate a stress response (Apoptosis, Reactive oxygen species) (fig. S6B). Apoptosis was also enriched in splenic TKOΔT CD4+ T cells, though it was not within the top 15 pathways. Kidney TKOΔT CD8+ T cells were enriched in pathways similar to those enriched in splenic TKOΔT CD8+ T cells (fig. S6C).
Because proliferative pathways were enriched in CD4+ and CD8+ TKOΔT T cells, we stained for Ki67, a nuclear marker of proliferation. Spleens of TKOΔT mice contained a two-fold increase in the frequency of Ki67+ CD4+ and CD8+ T cells compared to Control mice (Fig. 5, D and E). The Hallmark pathway Apoptosis was enriched in CD4+ and CD8+ TKOΔT T cells, with increased expression of cell death genes, including Casp3, Fas, Fasl, Casp7, and Bcl2l1 (data file S2). To validate some of these differences, we stained for activated Caspase-3, Fas, and FasL. We found increased percentages for all three markers in splenic CD4+ and CD8+ TKOΔT T cells compared to Control T cells (Fig. 5F). Our data revealed that TKOΔT T cells were more proliferative and apoptotic, potentially related to activation-induced cell death.
We compared genes upregulated in our RNA-sequencing dataset with a list of mRNA transcripts bound by ZFP36 and/or ZFP36L1 in murine TH1 cells (32). Of the 1024 genes significantly upregulated in spleen CD4+ TKOΔT T cells, 175 of those (17%) were identified as putative direct targets of the ZFP36 family in T cells (Fig. 5G). These putative targets included different classes of transcripts, including cytokines and chemokines (Ccl3, Ifng, Il10, Csf2), transcription factors (Bhlhe40, Prdm1, Irf8, Ikzf3), signaling molecules (Dusp4, Socs1, Map3k8), and receptors and adhesion molecules (Tnfrsf9, Icam1, Tnfrsf1b, Il2ra, and Tnfsf8). Of the 1386 genes significantly upregulated in splenic CD8+ TKOΔT T cells, 246 of those (18%), were potentially directly regulated by the ZFP36 family (fig. S6D). There were some putative direct mRNA targets that increased in both CD4+ and CD8+ TKOΔT T cells (Ifng, Ccl3, Bhlhe40, Prdm1, Ikzf3, Il2ra, and Tnfrsf9). While our data are consistent with direct regulation of some bound mRNA targets of the ZFP36 family that were upregulated in TKOΔT T cells in vivo, many more dysregulated genes are likely indirect targets downstream of directly regulated genes.
We also performed RNA-sequencing of splenic CD4+ and CD8+ T cells from the three strains of DKOΔT mice. We found that DKOΔT T cells did not display the same transcriptional dysregulation as TKOΔT T cells, although CD4+ T cells from Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice showed an intermediate upregulation of many genes (fig. S7A). Genes that were increased at both the RNA and protein levels in TKOΔT T cells, such as Ifng, Tnf, and Il2ra, were not increased in T cells from any of the DKOΔT strains (fig. S7B), revealing that any one ZFP36 family member is able to negatively regulate gene expression in T cells and maintain T cell quiescence.
Activated TKOΔT CD4+ T cells have increased mRNA stability
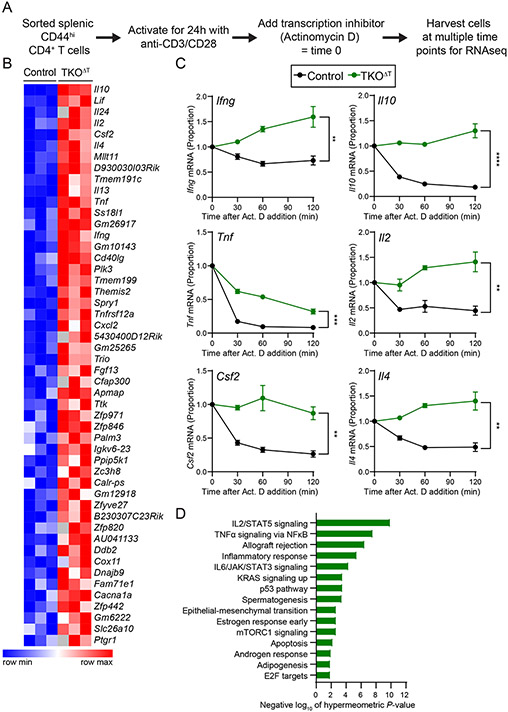
To address whether the ZFP36 family collectively regulate the mRNA decay rates of target transcripts, we performed global mRNA decay assays in activated TKOΔT and Control CD4+ T cells. To account for the increased activation state of TKOΔT CD4+ T cells, we sorted CD44hi splenic CD4+ T cells from Control and TKOΔT mice and activated them for 24 h on anti-CD3/anti-CD28 coated plates. We then treated the cells with Actinomycin D (a transcription inhibitor), harvested cells at different time points after this treatment, and performed RNA-sequencing. Decay curves were generated for individual transcripts and the area under the curve (AUC) was calculated (Fig. 6A).
Figure 6: Increased cytokine gene transcript stability in TKOΔT CD4+ T cells.
A, Experimental schema. CD44hi CD4+ splenic T cells were sorted from Control and TKOΔT mice (n=3/group). Cells were then activated for 24 h on anti-CD3/CD28 coated cell culture plates at which point some cells were harvested (time 0) and some cells were treated with Actinomycin D (Act. D). At different times post-addition (30, 60, 120 min), cells were harvested for RNA-sequencing. B, Heat map representation of area under the curve (AUC) values for the top 50 most significant genes with increased AUC (P-value < 0.05) in TKOΔT CD4+ T cells compared to Control CD4+ T cells. C, mRNA decay curves for Ifng, Il10, Tnf, Il2, Csf2, and Il4. All time points are graphed as a fraction of the time 0 counts per million reads mapped. D, Enriched Hallmark pathways (FDR q-value < 0.05) in genes with increased AUC (P-value < 0.05) in TKOΔT CD4+ T cells compared to Control CD4+ T cells. Data in C are mean ± s.e.m. Unpaired two-sided Student’s t-test computed on the AUC. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
Two hundred fifty-two transcripts from TKOΔT CD4+ T cells had an increased AUC, indicating increased mRNA stability (data file S3). Many of the top 50 transcripts with an increased AUC in TKOΔT CD4+ T cells compared to Control CD4+ T cells were transcripts encoding cytokines, including Il10, Lif, Il24, Il2, Csf2, Il4, Il13, Tnf, Ifng, and Cxcl2 (Fig. 6, B and C). Hallmark pathways enriched in the genes with increased AUC in TKOΔT CD4+ T cells included inflammatory pathways (IL2/STAT5 signaling, TNFα signaling via NFκB, Allograft rejection, Inflammatory response, IL6/JAK/STAT3 signaling) and proliferative pathways (p53 pathway, E2F targets) (Fig. 6D). Overall, we conclude that many proinflammatory genes are dysregulated via increased mRNA stability in TKOΔT CD4+ T cells.
Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice are resistant to EAE
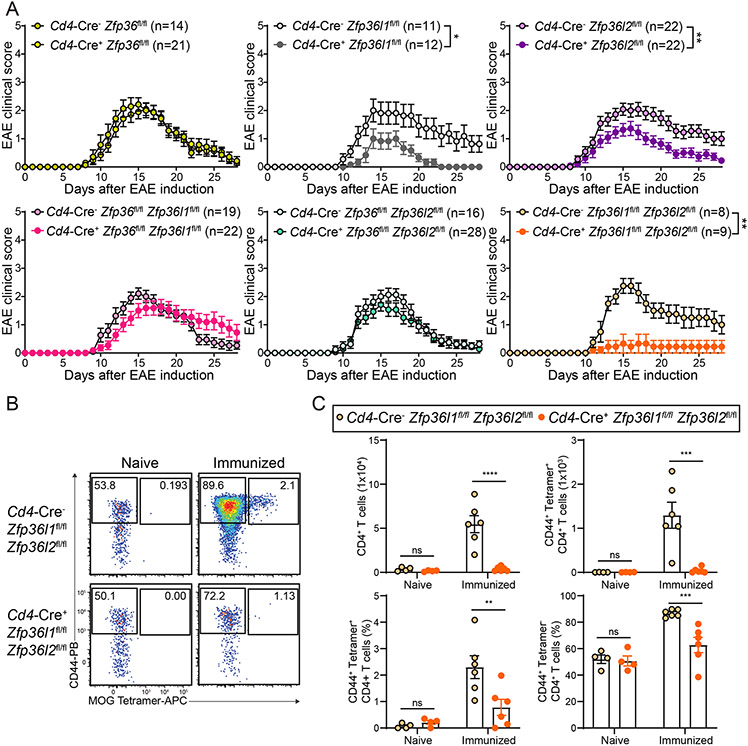
As TKOΔT mice develop neuroinflammation as part of their spontaneous syndrome, we decided to test our single and double deletion strains of the ZFP36 family, which have largely normal splenic T cell numbers (fig. S8A), in the experimental autoimmune encephalitomyelitis (EAE) model of induced neuroinflammation. Cd4-Cre+ Zfp36fl/fl, Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl, and Cd4-Cre+ Zfp36fl/fl Zfp36l2fl/fl mice had no difference in clinical disease compared to their Cd4-Cre− controls (Fig. 7A). However, to our surprise, Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice were almost completely resistant to EAE induction compared to their Cd4-Cre− controls, indicating a specific role for ZFP36L1 and ZFP36L2 in positively regulating T cell encephalitogenicity. Mice singly deficient in either of these two genes (Cd4-Cre+ Zfp36l1fl/fl or Cd4-Cre+ Zfp36l2fl/fl) had modest protection from EAE clinical symptoms. The resistance to EAE seen in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice was likely not due to any T cell abnormalities in naïve mice, as this strain had no impairment in thymic T cell development nor in their populations of splenic CD4+ and CD8+ T cells (fig. S8, B and C).
Figure 7: Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice are protected from EAE.
A, Clinical EAE scoring in the indicated strains of mice. B, CD44 and MOG38-49-I-Ab Tetramer staining on CD4+ T cells from the CNS of naïve or MOG35-55 peptide immunized (day 14) Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl and Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice. C, Quantitation of CD4+ cells, CD44+ MOG38-49-I-Ab Tetramer+ CD4+ T cells, the frequency of CD44+ MOG38-49-I-Ab Tetramer+ CD4+ T cells, and the frequency of CD44+ MOG38-49-I-Ab Tetramer− CD4+ T cells in the CNS of naïve (pooled from 2 experiments, n=4/group) or immunized (day 14) Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl and Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice (pooled from 2 experiments, n=6/group). Data in A,C are mean ± s.e.m. Mann-Whitney U test between area under the curve for individual mice (A); two-way ANOVA with Šídák’s multiple comparisons test (C). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
To examine whether the normal EAE susceptibilities seen in Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl and Cd4-Cre+ Zfp36fl/fl Zfp36l2fl/fl mice were dependent on compensation by the remaining Zfp36 family member (either ZFP36L2 or ZFP36L1, respectively), we generated the following mice: Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl Zfp36l2WT/fl and Cd4-Cre+ Zfp36fl/fl Zfp36l1WT/fl Zfp36l2fl/fl, creating haploinsufficiency at the one remaining Zfp36 family gene. This resulted in decreased gene expression of the remaining family member in CD4+ T cells relative to DKOΔT CD4+ T cells (fig. S9A). Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl Zfp36l2WT/fl and Cd4-Cre+ Zfp36fl/fl Zfp36l1WT/fl Zfp36l2fl/fl mice were either partially or completely protected from EAE (fig. S9B), indicating that the normal disease susceptibility in Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl and Cd4-Cre+ Zfp36fl/fl Zfp36l2fl/fl mice was due to compensation by ZFP36L2 or ZFP36L1 in a gene dosage-dependent manner. As expected, Cd4-Cre+ Zfp36WT/fl Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells had decreased Zfp36 expression and mice of this genotype were protected from EAE, similar to their Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl counterparts (Fig. 7A).
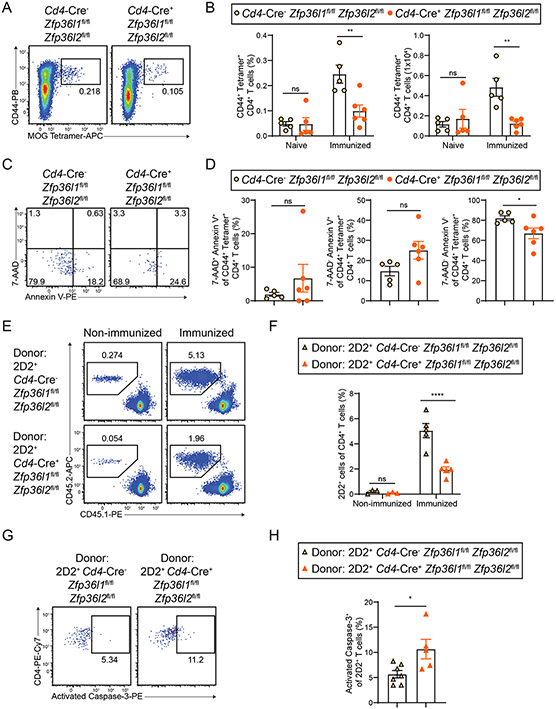
To investigate the immunological basis for the resistance to EAE in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice, we examined CNS-infiltrating immune cells following EAE induction. At the peak of EAE clinical disease, Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CNS had reduced signs of neuroinflammation, including less MHC II expression on microglia and fewer and less activated infiltrating myeloid cells (CD45hi Ly6G− CD11bhi), compared to Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl controls (fig. S10, A and B). The number of CNS neutrophils (CD45hi Ly6G+ CD11bhi) in each group of mice was equivalent (fig. S10B). In addition, fewer total and myelin oligodendrocyte glycoprotein (MOG)-specific (MOG38-49-I-Ab Tetramer+ CD44+) CD4+ T cells infiltrated the CNS of Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice compared to their Cd4-Cre− controls (Fig. 7, B and C). There was also a decreased frequency of MOG-specific CD4+ T cells and non-MOG-specific activated CD4+ T cells in the CNS of Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice. CNS-infiltrating CD4+ T cells from Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl mice produced proinflammatory cytokines such as GM-CSF and IFNγ upon PMA/ionomcyin re-stimulation, and this was diminished in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice (fig. S10, C and D). There was no difference in the frequency of IL-17A+ CD4+ T cells between the two genotypes. There was also an increased frequency of IL-10-producing CD4+ T cells in the CNS from naïve Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice compared to Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl mice and a trend towards greater IL-10 production during EAE from Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells compared to Cd4-Cre− CD4+ T cells.
Impaired CD4+ T cell responses in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice
Given the impaired MOG-specific CD4+ T cell response in the CNS on day 14 post-EAE immunization, we assessed whether Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells responded normally during in vitro polarization. Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells failed to expand to the same extent as Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells in TH1 and TH2 conditions but expanded normally in TH17 conditions (fig. S11A). CD4+ T cells from Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl and Cd4-Cre+ Zfp36fl/fl Zfp36l2fl/fl mice did not have expansion defects in TH1 and TH2 conditions and had enhanced expansion in TH17 conditions (fig. S11, B and C). During polarization, Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells proliferated less than their Cd4-Cre− controls, as determined by a CFSE dilution assay (fig. S11, D and E).
In light of these data, we next investigated MOG-specific T cell priming in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice. We immunized mice with MOG35-55 and examined draining lymph nodes 7 days later. Immunization of Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice induced fewer MOG38-49-I-Ab Tetramer+ CD44+ CD4+ T cells, both by frequency and number, compared to their Cd4-Cre− controls (Fig. 8, A and B). MOG-specific CD4+ T cells that developed in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice appeared more susceptible to cell death than their Cd4-Cre− controls, as seen by a decrease in live (7-AAD− Annexin V−) CD4+ T cells in the draining lymph nodes of immunized mice (Fig. 8, C and D).
Figure 8: Deficient CD4+ T cell priming in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice.
A, CD44 and MOG38-49-I-Ab Tetramer staining on CD4+ T cells from the popliteal lymph nodes of MOG35-55 peptide immunized mice (day 7). B, Quantitation of the frequency and number of CD44+ MOG38-49-I-Ab Tetramer+ CD4+ T cells from the inguinal lymph nodes from naïve mice (pooled from 2 experiments, n=5/group) and the popliteal lymph nodes of MOG35-55 peptide immunized mice (day 7) (pooled from 2 experiments, n=5-6/group). C, 7-AAD and Annexin V staining on CD44+ MOG38-49-I-Ab Tetramer+ CD4+ T cells from the popliteal lymph nodes of MOG35-55 peptide immunized mice (day 7). D, Quantitation of necrotic (7-AAD+ Annexin V+), apoptotic (7-AAD− Annexin V+), or live (7-AAD− Annexin V−) CD4+ T cells from the popliteal lymph nodes of Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl and Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice 7 days after MOG35-55 peptide immunization (pooled from 2 experiments, n=5-6/group). E, Staining of popliteal lymph nodes to identify donor 2D2+ T cells (CD45.2) and endogenous CD4+ T cells (CD45.1) gated on TCRβ+ CD4+ T cells on day 5 post-MOG35-55 hock immunization or in non-immunized mice. F, Quantitation of the frequency of popliteal lymph node donor 2D2+ T cells on day 5 post-MOG35-55 hock immunization or in non-immunized mice (pooled from 2 experiments, n=3-5/group). G, Activated Caspase-3 staining of popliteal lymph node donor 2D2+ T cells on day 3 post-MOG35-55 hock immunization. H, Quantitation of activated Caspase-3+ popliteal lymph node donor 2D2+ T cells on day 3 post-MOG35-55 hock immunization (pooled from 2 experiments, n=5-7). Data in B,D,E,H are mean ± s.e.m. Two-way ANOVA with Šídák’s multiple comparisons test (B,F); unpaired two-sided Student’s t-test (D,H). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001; ns, not significant.
To determine whether these T cell defects were entirely cell intrinsic and to track MOG-specific cells more easily, we crossed 2D2 TCR transgenic mice with Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice as a source of MOG-specific donor T cells. We transferred CFSE-labeled 2D2+ Cd4-Cre− or Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells (CD45.2) to congenically marked recipients (CD45.1) one day prior to MOG35-55 immunization. At day 5 post-immunization, markedly fewer 2D2+ Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells were recovered from the draining lymph nodes compared to 2D2+ Cd4-Cre− Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells (Fig. 8, E and F). At this time point, 2D2+ cells of both genotypes showed maximal CFSE dilution (fig. S12, A and B). At day 3 post-immunization, despite no increase in the frequency of 2D2+ cells of either genotype relative to non-immunized mice (fig. S12C), cells of both genotypes were highly proliferative in immunized mice (fig. S12, D and E), suggesting that this time point is optimal for examining early T cell priming. 2D2+ Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CD4+ T cells showed increased apoptosis as determined by activated Caspase-3 staining on day 3 post-immunization (Fig. 8, G and H). Overall, our in vitro and in vivo data indicate that CD4+ T cells require ZFP36L1 and ZFP36L2 cell-intrinsically to maintain T cell fitness during clonal expansion.
DISCUSSION
Here we find that the ZFP36 family of RNA-binding proteins critically and redundantly regulates T cell homeostasis. Mice with T cell-specific deletion of the three ZFP36 family members develop a spontaneous inflammatory syndrome likely driven by excessive cytokine production from triply-deleted T cells. Data from DKOΔT mice indicate that any two ZFP36 family proteins are sufficient to maintain T cell quiescence and prevent autoinflammation. The kidney and liver inflammation seen in TKOΔT mice resembles that seen in mice with excessive TNF (TnfΔARE) or IFNγ (IfngΔARE) production, though these strains of mice lack the neuroinflammation seen in TKOΔT mice (2, 3). In addition, TnfΔARE mice experience ileitis (2) while TKOΔT mice lack gut pathology. Mice with induced overexpression of GM-CSF from CD4+ T cells experience neurological defects and immune cell infiltration into the CNS, but lack the lethality seen in TKOΔT mice (35). It will be informative to cross TKOΔT mice with mice deficient in IFNγ, TNF, or GM-CSF to see which aspects of the spontaneous syndrome, if any, are alleviated in the absence of specific cytokines. TKOΔT mice lacked significant immunopathology in their lungs, heart, skin, and joints. It has been reported that aged Cd4-Cre+ Zfp36fl/fl mice develop dermatitis (33); however, a small group of Cd4-Cre+ Zfp36fl/fl mice from our colony aged to 12 months did not develop overt skin pathology, a discrepancy that may be caused by microbiota differences between mouse facilities.
TKOΔT Tregs expressed normal markers of Treg function but were less efficient at suppression of effector CD4+ proliferation compared to Control Tregs, indicating that ZFP36 family-deficient Tregs may contribute to the TKOΔT syndrome. Lower suppression efficiency could be due to poor suppression on a per cell basis or poor survival of TKOΔT Tregs in vitro. Despite the functional deficiencies of TKOΔT Tregs, TKOΔT mice differ from Scurfy mice, which completely lack Tregs due to mutations in Foxp3 (36). Similar to the TKOΔT syndrome, Scurfy mice develop multiorgan pathology but with additional features not seen in TKOΔT mice, including lymphadenopathy, dermatitis, and pulmonary inflammation (36-39). Scurfy mice succumb to their disease more quickly with a median survival of 4 weeks compared to a median survival of 10 weeks in TKOΔT mice. The precise cause of death in TKOΔT mice remains uncertain but their cachexia, anemia, bradycardia, and hypotension likely contribute to progressive organ dysfunction.
We found that the ZFP36 family members collectively destabilized cytokine transcripts in activated CD4+ T cells. ZFP36 and ZFP36L2 have been shown to individually inhibit translation of cytokine transcripts in T cells (24, 32). While prior work demonstrated that memory T cells from Cd4-Cre+ Zfp36l2fl/fl mice did not have a difference in the decay of Ifng transcript (24), these results are likely explained by compensation by the remaining ZFP36 family members. In the future, TKOΔT T cells could be used to obtain a more complete list of mRNA targets negatively regulated by the ZFP36 family at the translational level. It will be interesting to see if specific mRNA targets are preferentially regulated by mRNA decay, decay-independent translational inhibition, or both.
Given the spontaneous neuroinflammation involving cytokine-producing T cells in TKOΔT mice, we hypothesized that single and double T cell deletions of ZFP36 family members might result in increased severity of EAE. Unexpectedly, Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice were markedly protected from EAE. This finding is of interest as single nucleotide polymorphisms in non-coding regions of human ZFP36L1 and ZFP36L2 have been linked to increased risk for multiple sclerosis in genome-wide association studies (40-42). Given the intermediate protection from EAE seen in Cd4-Cre+ Zfp36l1fl/fl and Cd4-Cre+ Zfp36l2fl/fl mice, it was initially surprising that Cd4-Cre+ Zfp36fl/fl Zfp36l1fl/fl and Cd4-Cre+ Zfp36fl/fl Zfp36l2fl/fl mice were not also protected from EAE. However, when these DKOΔT strains were made haploinsufficient in their single remaining Zfp36 family member, they were protected from EAE. We interpret these data to indicate that there is a minimum cell-intrinsic expression of either Zfp36l1 or Zfp36l2 needed for CD4+ T cell priming and ultimately encephalitogenicity in this model.
Our results indicate that ZFP36L1 and ZFP36L2 specifically regulate mRNA targets that are induced upon and required for T cell activation. Lack of ZFP36L1 and ZFP36L2 led to decreased T cell fitness in vivo and in vitro, which may be due to increased cell death, decreased cell cycle progression, impaired metabolic reprogramming, or some combination of these processes. In contrast to our data, it has been suggested that ZFP36L1 and ZFP36L2 are negative regulators of proliferation (31, 43-45) and positive regulators of cell death (46, 47), although this has not been rigorously tested in T cells. Interestingly, recent data analyzing the combined loss of ZFP36 and ZFP36L1 in T cells found increased proliferation relative to control T cells in the context of anti-CD3 stimulation (48). These data in combination with our studies of T cells lacking ZFP36L1 and ZFP36L2 suggest that different ZFP36 family member pairs may cooperate in T cells to achieve opposite outcomes (i.e. suppression or support of proliferation).
It remains unclear why some biological processes are redundantly or non-redundantly regulated by ZFP36 family members. Global knockout of any one Zfp36 family member in mice results in distinct phenotypes (17, 25-29); whether this is due to a lack of functional compensation by the remaining Zfp36 family members or merely a lack of expression of a functionally redundant remaining Zfp36 family member in the relevant cell type(s) remains unclear. ZFP36L1 and ZFP36L2 are more similar to each other in amino acid sequence than they are to ZFP36 (55) and have been shown to be functionally redundant in some cell types for certain processes (30, 31, 43, 56). A direct comparison of mRNA targets bound by individual members of the ZFP36 family in the same cell type and context (e.g. during activation) would be an important advance to clarify redundant and non-redundant targets. In addition, many other non-ZFP36 family member RNA-binding proteins also bind to AREs, such as ELAV1 (also known as HuR, an mRNA stabilizer) and HNRNPD (also known as AUF1, a stabilizer or de-stabilizer depending on the protein isoform). It is unclear how these other RNA-binding proteins participate in either the TKOΔT syndrome or in T cell priming in Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice. HuR, which acts in opposition to the ZFP36 family, interacts with many of the same mRNA targets with similar but sometimes non-overlapping binding sites and distinct affinities (57, 58). How other RNA-binding proteins function at newly available binding sites upon loss of the ZFP36 family members is an interesting area of future investigation.
In summary, we found a crucial role for the ZFP36 family proteins in regulating T cell quiescence in the steady state. The ZFP36 proteins collectively promoted cytokine transcript decay and limited protein production such that without this regulation, triply-deficient T cells induced a lethal inflammatory syndrome. In contrast to full redundancy of the ZFP36 family during T cell homeostasis, ZFP36L1 and ZFP36L2 together played a shared role in promoting antigen-specific clonal expansion of CD4+ T cells in an induced model of neuroinflammation that could not be compensated for by ZFP36. Thus, the ZFP36 proteins are critical regulators of T cell homeostasis and autoimmunity, displaying complex redundancies depending on the immunological context.
MATERIALS AND METHODS
Study Design
The overall objective of this study was to determine some of the redundant and distinct roles of the ZFP36 family proteins in murine T cells. This was achieved by generating T cell-specific deletion of the ZFP36 family genes (Zfp36, Zfp36l1, and Zfp36l2) and studying these strains of mice during homeostasis and the EAE model. Outcomes in homeostasis were survival, changes in immune cell populations in lymphoid and peripheral organs, and T cell function (cytokine production, gene expression by RNA-sequencing, and mRNA stability measurements). Outcomes in the EAE model were clinical disease scores, changes in immune cell populations in the CNS, and T cell priming in the draining lymph nodes. In EAE experiments, mice were randomly assigned to the naïve or immunized groups. No sample size calculation was performed, but previous experience and pilot experiments led us to typically use greater than or equal to 4 mice per group to disprove statistically significant differences. The study was unblinded. One sample from the mRNA decay-sequencing experiment (TKOΔT #3 time60) was excluded based on global gene expression differences from the other replicates. This non pre-established exclusion was based on an unbiased assessment of global gene expression differences. Exclusions of animals were rare, and only due to evidence of unrelated injury or sickness. Exclusions of flow cytometry samples were rare and only due to technical difficulties. This is a pre-established policy in our lab. All data, excluding blood leukocyte quantitation by flow cytometry using CountBright beads, RNA-sequencing experiments, RT-qPCR experiments, and serum cytokine multiplex analysis, are pooled from at least two independent experiments, and numbers of experimental repeats are indicated in the figure legends.
Mice
Zfp36fl/fl, Zfp36l1fl/fl, and Zfp36l2fl/fl mice on the C57BL/6 background have been previously described (59-61). These mice were crossed to Cd4-Cre mice (Jackson Laboratory, 022071) to generate single-, double-, and triple-conditionally deleted strains. C57BL/6NTac (B6) and B6.SJL mice were purchased from Taconic (4007) or Jackson Laboratory (002014). Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl mice were crossed to 2D2 TCR transgenic mice (C57BL/6-Tg(Tcra2D2,Tcrb2D2)1Kuch/J, Jackson Laboratory, 006912) (62). All strains were bred and maintained in our specific pathogen-free animal facility. Both sexes were analyzed. Littermates were used for experiments whenever possible, although in some cases, mice from multiple litters were used in a single experiment. In some cases, mice were weighed weekly. All animal experiments were approved by the Institutional Animal Care and Use Committee of Washington University in St. Louis.
Complete blood counts and blood chemistry
Blood from the submandibular vein was collected in EDTA-coated tubes (BD Microtainer 365974). For blood chemistry analysis, sera were obtained from blood collected into Serum Separator Tubes (BD Microtainer 365956). Complete blood counts (CBCs) and blood chemistry measurements were performed by the Division of Comparative Medicine (DCM) Research Animal Diagnostic Laboratory at Washington University School of Medicine.
Blood pressure and heart rate measurements
Blood pressure and heart rates were measured as previously described (63). Briefly, mice were anesthetized with 2% isoflurane and maintained at 37 °C on a heating pad. The right common carotid artery was exposed and a Millar pressure transducer (model SPR-1000) was introduced and advanced to the ascending aorta after which isoflurane anesthesia was reduced to 1.5%. Systolic blood pressure, diastolic blood pressure, and heart rate were recorded (PowerLab data acquisition system, ADInstruments). Data from the average of a 3 min period of stable recording was analyzed using LabChart 8 (ADInstruments). Heart weights were normalized to mouse body weight and reported as a fraction.
Generation of mixed bone marrow chimeric mice
B6 x B6.SJL F1 mice (CD45.1/CD45.2) were lethally irradiated (1,000 rads) and injected intravenously with 16 million total bone marrow cells (8 million B6.SJL CD45.1 cells plus either 8 million Control (CD45.2) or TKOΔT (CD45.2) cells). Mice were provided drinking water containing trimethoprim-sulfamethoxazole (Pharmaceutical Associates, Inc. 0121-0854-16) (trimethoprim at 0.26 mg/mL and sulfamethoxazole at 1.3 mg/mL) for 2 weeks following transplantation and were allowed to reconstitute for at least 12 weeks.
EAE induction
Mice were subcutaneously immunized with 50 μg MOG35-55 peptide (CS Bio Co.) in Complete Freund’s Adjuvant (CFA) (made with 5 mg/mL heat-killed Mycobacterium tuberculosis H7Ra (BD Difco) in Incomplete Freund’s Adjuvant (BD Difco)) in each axilla. 300 ng of pertussis toxin (PTX, List Biological Laboratories) was given intraperitoneally on the day of immunization and two days later. Mice were observed for clinical symptoms for at least 28 days using a standard 5-point clinical scoring system (0, no clinical symptoms; 1, limp tail; 2, altered gait or impaired righting reflex; 3, hind limb paralysis; 4, forelimb paralysis; 5, moribund or deceased) (64). For analysis of T cell priming, mice were subcutaneously injected in the hock (65) with 10 nanomoles MOG35-55 peptide emulsified in CFA at day 0. Mice were subsequently injected with 300 ng PTX i.p. on days 0 and 2. Popliteal lymph nodes from immunized mice and inguinal lymph nodes from naïve mice were harvested on day 7.
2D2+ T cell transfer
CD4+ T cells were purified from 2D2+ Cd4-Cre− or Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl (CD45.2) spleens and inguinal lymph nodes (MojoSort Mouse CD4 T Cell Isolation kit) and stained with 1 μM CFSE for 10 min at RT. 2x106 live CFSE-labeled cells were transferred i.p. to B6.SJL (CD45.1) recipient mice. One day later (day 0), recipient mice were subcutaneously injected in the hock with 10 nanomoles of MOG35-55 peptide emulsified in CFA and given 300 ng PTX i.p. on days 0 and 2. Popliteal lymph nodes from immunized and non-immunized mice were harvested on days 3 and 5. 2D2+ cells were gated as TCRβ+ CD4+ CD45.2+ CD45.1− and examined for CFSE dilution and/or activated Caspase-3.
Tissue processing
Spleens and thymi were passed through a 70-μm cell strainer. Red blood cells were lysed with ACK lysis buffer. When harvesting non-lymphoid organs, mice were transcardially perfused with 30 mL of PBS. The following tissues were digested in complete Iscove’s Modified Dulbecco’s Media (cIMDM: 10% FBS, sodium pyruvate, L-glutamine, penicillin/streptomycin, nonessential amino acids, and 2-mercaptoethanol) by incubation with stirring for 1 h at 37 °C in the following digestion conditions: lymph nodes (250 μg/mL Collagenase B, Sigma and 30 U/mL DNase I, Sigma), kidneys (4 mg/mL Collagenase D, Sigma), and liver (4 mg/mL Collagenase D and 30 U/mL DNase I). Enzymatic digestion was halted with EDTA (Corning, 5mM final). Brains and spinal cords were digested in Hank’s Buffered Salt Solution + 10% FBS with Collagenase IV (0.1 mg/mL, Sigma) for 40 min at 37 °C with rocking before being passed through a 70-μm cell strainer. For the CNS tissues, cell suspensions from a single mouse in 10.5 mL PBS were mixed with 4.5 mL isotonic Percoll (GE Healthcare 17-0891-01) and spun at 4 °C at 900 g with no brake. The top layer was discarded, and pelleted cells were resuspended in PBS. In all cases, cells were passed through a 70-μm cell strainer and counted with a hemocytometer before analysis.
Flow cytometry
Surface staining of cells was conducted in sterile PBS with 0.5% BSA, 2 mM EDTA, and 0.02% sodium azide (FACS buffer) after blocking with anti-CD16/32 (clone 2.4G2, BioXCell) for 5 min at 4 °C. For intracellular cytokine staining (ICS) and cytoplasmic antigens (activated Caspase-3 and CTLA-4), the BD Cytofix/Cytoperm kit was used (BD 554714). For ICS, cells were cultured for 4 h at 37 °C in the presence of brefeldin A (BFA) (Enzo Life Sciences, 1 μg/mL) with or without PMA (Enzo Life Sciences, 50 ng/mL) and ionomycin (Enzo Life Sciences, 1 μM). In some experiments, Actinomycin D (Sigma A9415, 10 μg/mL) or cycloheximide (CHX) (Sigma 01810, 10 μg/mL) was added in the presence of BFA for 4 h of cell culture. Post surface staining, cells were fixed with either 4% methanol-free paraformaldehyde (Electron Microscopy Sciences, if left overnight) or the provided BD Fixation/Permeabilization solution (if proceeding through ICS staining) according to the manufacturer’s protocol. Cells were permeabilized with the provided BD Perm/Wash, and ICS staining was performed for 20 min at 4 °C.
For nuclear antigens (Ki67, FoxP3, and Helios), the True-Nuclear Transcription Factor Buffer Set (BioLegend 424401) was used. Post surface staining, cells were fixed with 1X Fix Concentrate buffer in the provided Fix Diluent for 45 min at room temperature (RT). Cells were washed with FACS buffer and stored overnight at 4 °C followed by nuclear permeabilization and nuclear staining the next day. For detection of MOG-specific CD4+ T cells, APC-conjugated MOG38-49-I-Ab tetramers (National Institutes of Health Tetramer Core Facility) were used at a 1:200 dilution in media. Lymph node or CNS cells were stained for 30 min at 37 °C, followed by normal surface staining for other markers.
For Annexin V and 7-AAD staining, the Annexin V Apoptosis Detection Kit was used (BioLegend 640930) with azide-free FACS buffer and 7-AAD and anti-Annexin V antibodies were used at a 1:20 dilution. To screen for TCR Vβ gene usage, the BD Pharmingen Mouse Vβ TCR Screening Panel was used according to the manufacturer’s protocol (BD Biosciences 557004). CountBright absolute counting beads (Life Technologies C36950) were used to enumerate concentrations of blood leukocytes according to the manufacturer’s protocol. Flow cytometry was performed on an LSRFortessa instrument (BD) with FlowJo software (Treestar) used for analysis. Antibodies and fluorescent dyes used in this study are listed in data file S4.
T helper cell polarization
TH polarization was performed as previously described with minor modifications (66). Briefly, CD4+ T cells (for experiments using TKOΔT mice: Invitrogen Dynabeads FlowComp Mouse CD4) or naïve CD4+ T cells (for experiments using DKOΔT mice: EasySep Mouse Naïve CD4+ T Cell Isolation Kit or MojoSort Mouse CD4 Naïve T Cell Isolation Kit) were isolated from spleens and cultured in cIMDM at 37 °C/8% CO2 with plate-bound anti-CD3e (Leinco, clone 145-2C11, 2 μg/mL) and anti-CD28 (BioXCell, clone 37.51, 2 μg/mL) in the presence of neutralizing antibodies and cytokines as follows. TH1: anti-IL-4 (10 μg/mL) and IL-12 (10 ng/mL); TH2: anti-IFNγ (5 μg/mL) and IL-4 (10 ng/mL); TH17: anti-IFNγ (5 μg/mL), anti-IL-4 (10 μg/mL), TGFβ1 (2 ng/mL), IL-6 (25 ng/mL), IL-23 (10 ng/mL), and IL-1β (10 ng/mL). Cells were split on day 3 and harvested and counted on day 4 for ICS. In some experiments, naïve CD4+ T cells were labeled with 10 μM CFSE for 20 min at RT for analysis of CFSE dilution on day 4 of TH culture. In different experiments, TH cell fold expansion was calculated by determining the number of cells on day 4 relative to the starting number of cells upon initiation of culture.
Treg suppression assay
CD25+ CD4+ T cells were isolated from the spleens of Control or TKOΔT mice (CD45.2) using the MojoSort Mouse CD4+ CD25+ Regulatory T Cell Isolation Kit and Treg purity was confirmed by CD25 and FoxP3 staining. Tconventional (Tconv) cells were purified from B6.SJL mice (CD45.1) using the MojoSort Mouse CD4 Naïve T Cell Isolation Kit and labeled with 10 μM CFSE as described above. Splenocytes from B6 x B6.SJL F1 mice (CD45.1/.2) were irradiated (3400 rads) and, along with soluble anti-CD3e (Leinco, clone 145-2C11, 2.5 ng/μL) were used to stimulate T cells. Tregs and Tconv cells were plated at various Treg:Tconv ratios. Tconv cells were gated as FSChi CD4+ CD45.1+ and analyzed on day 3 of co-culture for CFSE dilution. Percent suppression was calculated using the geometric mean fluorescence intensity (MFI) of CFSE and the following formula: .
ELISAs and autoantigen microarrays
For quantitation of serum immunoglobulins, Nunc Maxisorp plates were coated with isotype-specific capture antibodies from the SBA Clonotyping System (SouthernBiotech 5300-05B) in carbonate buffer (34.88 mM NaHCO3, 15.00 mM Na2CO3, pH 9.7) at 4 °C overnight. Following blocking with PBS + 0.5% BSA (1 h at RT), diluted serum was added and incubated for 1 h at RT, followed by incubation with HRP-conjugated anti-mouse immunoglobulin isotype-specific antibodies for 1 h at RT. Substrate solution (BD OptEIA, 555214) was added, reactions were stopped with 1M H3PO4 (Alfa Aesar), and absorbance was read at OD450 on an iMark Microplate Reader (Bio-Rad). Standard curves were generated with purified mouse immunoglobulin standards from the C57BL/6 Mouse Immunoglobulin Panel (SouthernBiotech 5300-01B). For serum cytokine and chemokine analysis, a mouse 31-plex array was performed (Eve Technologies, Calgary, Canada). Mouse sera were screened for autoreactive IgM and IgG on autoantigen microarrays at the UT Southwestern Medical Center Genomics & Microarray Core Facility. Heatmaps represent normalized net signal intensity after removing antigens that had a signal-to-noise ratio greater than 3 in fewer than 10% of the samples and were created using Phantasus (https://ctlab.itmo.ru/phantasus/).
Microscopy
Initial necropsies on Control and TKOΔT mice were performed by the DCM Animal Research Diagnostic Laboratory at Washington University School of Medicine. Tissues were fixed in 10% neutral buffered formalin and embedded in paraffin blocks prior to sectioning and H&E staining. For immunofluorescent staining of spleens and kidneys, organs were frozen in O.C.T. compound (Fisher 4585) prior to sectioning. Cut sections were fixed in acetone (Alfa Aesar) and blocked with CAS-Block (Invitrogen). Staining was performed with the antibodies listed in data file S4 all diluted in CAS-Block. Sections were mounted with Abcam Fluoroshield Mounting Medium with DAPI. Images were captured with a Nikon Eclipse E800 microscope and MicroPublisher 5.0 RTV or EXi Blue cameras (QImaging) using QCapture software. Images were merged and leveled in Adobe Photoshop.
Immunofluorescence (IF) staining of whole-mounted tissues from eyeballs was performed as previously described (67). In brief, eyeballs were gently enucleated and fixed in 4% paraformaldehyde (PFA) for 30 min at RT. Corneas, retinas, and RPE-choroid-sclera complexes were dissected from the eyeballs and fixed in 1% PFA for 1 h at RT. Brains were fixed in 4% PFA overnight at 4 °C, dehydrated in 20% sucrose solution overnight at 4°C, embedded and frozen in Tissue-Tek O.C.T. compound (Sakura), and cut into 14-μm sections using a cryostat (Leica, CM1950). All samples were blocked with 5% goat or donkey serum in PBST (0.3% Triton X-100 in PBS) for 30 min, incubated in primary antibodies at 4°C overnight, washed in PBST, and incubated in secondary antibodies at RT for 2 h. Samples were washed in PBST and mounted on microscope slides with Vectashield (Vector Laboratories, H-1200) or FluoroSave (Millipore 345789). Staining was performed with the antibodies listed in data file S4. IF images were acquired using a confocal microscope (Zeiss, LSM 800 with Airyscan). Confocal images of whole-mount tissues are maximum intensity projections of tiled z-stack images taken at the optimal interval through the entire thickness of tissues, which were all taken at a resolution of 1024 × 1024 pixels with the Plan-Apochromat 20×/0.8 NA M27 lens with multichannel scanning in the frame. ZEN 3.0 (blue edition; Zeiss) and Fiji (https://imagej.net/Fiji) were used to acquire and process images.
RT-qPCR
For analysis of Zfp36 family member gene expression, splenic naïve CD4+ T cells were purified using EasySep Mouse Naïve CD4+ T Cell Isolation Kits or MojoSort Mouse CD4 Naïve T Cell Isolation Kits. RNA was isolated using the E.Z.N.A. MicroElute Total RNA Kit (OMEGA Bio-Tek R6831), and quantitated using a Nanodrop 2000 spectrophotometer (Thermo Fisher). cDNA was synthesized with 50 ng of RNA using the High Capacity RNA-to-cDNA kit (Applied Biosciences, 4387406). qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosciences, 4367659) on a CFX Connect Real-Time System (Bio-Rad). Gene expression was determined relative to Hprt by the ΔCT method. The following primers were used: Hprt forward 5’-TCAGTCAACGGGGGACATAAA-3’, Hprt reverse 5’-GGGGCTGTACTGCTTAACCAG-3’, Zfp36 forward 5’-CCACCTCCTCTCGATACAAGA-3’, Zfp36 reverse 5’-GCTTGGCGAAGTTCACCCA-3’, Zfp36l1 forward 5’-CCTATCAGATGGAGAGGTGCTGTC-3’, Zfp36l1 reverse 5’-TGTAGTTGAGCATCTTGTTACCCTGG-3’, Zfp36l2 forward 5’-GAGGGCACCTCCCAACCT-3’, Zfp36l2 reverse 5’-TGACAGAAGTGTGGTCGACATTT-3’.
RNA-sequencing
Bulk CD4+ and CD8+ T cells were sorted on an BD Aria II cytometer from spleens (Control, TKOΔT, and DKOΔT mice) and kidneys (Control and TKOΔT mice) as CD45+ Ly6G− NK1.1− CD11b− TCRγδ− B220− 7-AAD− TCRβ+ CD8α− CD4+ and CD45+ Ly6G− NK1.1− CD11b− TCRγδ− B220− 7-AAD− TCRβ+ CD4− CD8α+ cells, respectively. RNA was isolated using the Quick-DNA/RNA Microprep Plus Kit (Zymo Research D7005). RNA was submitted to the Genome Technology Access Center at the McDonnell Genome Institute at Washington University in St. Louis for cDNA synthesis (Clonetech SMARTer Ultra Low RNA kit) followed by RNA-sequencing on a NovaSeq 6000 (Illumina) with paired-end reads extending 150 bases. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq software and a custom python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 primary assembly with STAR version 2.5.1a (68). Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.6-p5 (69). All gene counts were then imported into the R/Bioconductor package EdgeR (70) and trimmed mean of M values (TMM) normalization size factors were calculated to adjust for samples with differences in library size.
Genes with an expression value of greater than 1 count per million (CPM) in any sample were considered expressed. Phantasus (https://ctlab.itmo.ru/phantasus/) was used for log2 transformation and quantile normalization of the data, differential gene expression analysis (Limma), and to generate volcano plots and heat maps. Genes were considered statistically differentially expressed with a P-value < 0.05 and fold change ≥ 2. Gene Set Enrichment Analysis (GSEA) of all expressed genes was performed using GSEA software (https://www.gsea-msigdb.org/gsea/index.jsp) (71, 72) and Hallmark gene sets (73). Pathways were considered statistically enriched with an FDR of < 25% and a nominal P-value < 5%. For comparison of RNA-sequencing data to published ZFP36 and ZFP36L1 target mRNAs, HITS-CLIP data from GSE96074 (32) (CD4+ T cells activated in TH1 conditions for 4 h) was compared to genes statistically upregulated in TKOΔT CD4+ or CD8+ splenic T cells. Venn diagrams were generated with the Venn Diagram Plotter tool (Pacific Northwest National Laboratories, omics.pnl.gov).
mRNA decay assay
CD44hi CD4+ T cells (CD19− TCRγδ− CD8α− CD62L− CD25− 7-AAD− CD4+ CD44hi) were sorted on an BD Aria II cytometer from the spleens of Control and TKOΔT mice (n=3/group) after pre-enrichment using Invitrogen Dynabeads FlowComp Mouse CD4 kit. Cells were activated on anti-CD3/CD28 coated plates overnight (see TH cell polarization section above). At approximately 24 h post-activation, one well of cells were harvested (time 0). For the remaining cells, Actinomycin D was added (10 μg/mL) and cells were harvested at different time points (30 min, 60 min, and 120 min). Cells were lysed in TRK lysis buffer and RNA was isolated using the E.Z.N.A. MicroElute Total RNA Kit (OMEGA Bio-Tek R6831). RNA was submitted for RNA-sequencing as described above; only genes with a CPM value of ≥1 in Control time 0 samples were analyzed. The area under the curve (AUC) in mRNA decay experiments was calculated as the trapezoidal area for each tested time interval using Microsoft Excel. The average of 3 biological replicate AUC values for a given mRNA transcript was compared between genotypes using an unpaired two-sided Student’s t-test. Genes with significantly increased AUC in TKOΔT CD4+ T cells (P-value < 0.05) were cross-referenced to the Hallmark gene sets (73) in the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
Statistical analysis
All data, excluding blood leukocyte quantitation by flow cytometry using CountBright beads, RNA-sequencing experiments, RT-qPCR experiments, and serum cytokine multiplex analysis, are from at least two independent experiments. Data were analyzed by either Student’s t-tests, one-way ANOVA, two-way ANOVA, or Mann-Whitney U, as indicated in the figure legends using Prism (GraphPad Software, Inc.). For relevant comparisons where no P-value is shown the P-value was >0.05. Horizontal bars represent the mean and error bars represent the standard error of the mean (s.e.m.).
Supplementary Material
Data file S5: Raw data.
Data file S1: Blood pressure and heart rate measurements in Control and TKOΔT mice.
Fig. S1: Additional characterization of the ocular phenotype in TKOΔT mice.
Fig. S2: Liver inflammation in TKOΔT mice and representative flow cytometric gating for peripheral organs.
Fig. S3: Further analysis of primary and secondary lymphoid organs in TKOΔT mice.
Fig. S4: Identification of autoantibodies, evaluation of T cell activation in DKOΔT mice, and phenotyping of TKOΔT Tregs.
Fig. S5: Further analysis of cytokine production from TKOΔT T cells.
Fig. S6: RNA-sequencing from TKOΔT splenic and kidney CD8+ T cells.
Fig. S7: RNA-sequencing from DKOΔT splenic CD4+ and CD8+ T cells.
Fig. S8: Single and double deletion strains are normal during homeostasis.
Fig. S9: Clinical EAE in DKOΔT mice made haploinsufficient for their remaining Zfp36 family gene.
Fig. S10: Analysis of Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CNS on day 14 post-EAE induction.
Fig. S11: ZFP36L1 and ZFP36L2 are uniquely required for optimal TH cell expansion.
Fig. S12: Further analysis of day 5 and day 3 lymph node responses after 2D2+ T cell transfer.
Data file S3: mRNA decay-sequencing from activated Control and TKOΔT CD4+ T cells.
Data file S4: Antibodies and dyes.
Data file S2: RNA-sequencing data from Control and TKOΔT spleen and kidney CD4+ and CD8+ T cells.
Acknowledgements:
We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant UL1TR002345 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. We acknowledge the NIH Tetramer Core Facility for provision of MOG38-49-I-Ab tetramers. We thank E. Lantelme, P. Akitani, and A. Cullen for help with cell sorting. We thank Debbie Stumpo for mice and advice concerning their genotyping and husbandry. We thank C.-C. Lin for initial help conceiving of the study. We thank N. Jarjour for critical reading of the manuscript. We thank G. Wu and G. Fox for insightful feedback, and we thank the members of the D. Lenschow laboratory (especially L. Fox, M. Locke, and Y.-C. Perng) for reagents and helpful discussions about this project.
Funding:
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) (R01AI113118) and the National Multiple Sclerosis Society (RG-2111-38724) (B.T.E.). M.E.C. was supported by the National Science Foundation Graduate Research Fellowship program (DGE-1745038) and by grant T32AI007163 from the NIAID. J.T. was supported by the National Institute of General Medical Sciences (F31GM146361). This work was also supported in part by the Intramural Research Program of the NIEHS (P.J.B.). R.S.A was supported by NIH grant R01EY019287, P30EY02687 (Vision Core Grant), the Jeffery T. Fort Innovation Fund, the Starr Foundation, Retina Associates of St. Louis Research Fund, and an unrestricted grant from Research to Prevent Blindness to the John F. Hardesty, MD Department of Ophthalmology and Visual Sciences at Washington University School of Medicine.
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability: The RNA-sequencing and mRNA decay sequencing data have been deposited in the GEO repository under accession code GSE192956. Mice with floxed alleles of Zfp36, Zfp36l1, and Zfp36l2 are available from Dr. Perry Blackshear under a material transfer agreement with the National Institute of Environmental Health Sciences. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Anderson P, Post-transcriptional control of cytokine production. Nat Immunol 9, 353–359 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G, Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Hodge DL, Berthet C, Coppola V, Kastenmüller W, Buschman MD, Schaughency PM, Shirota H, Scarzello AJ, Subleski JJ, Anver MR, Ortaldo JR, Lin F, Reynolds DA, Sanford ME, Kaldis P, Tessarollo L, Klinman DM, Young HA, IFN-gamma AU-rich element removal promotes chronic IFN-gamma expression and autoimmunity in mice. J Autoimmun 53, 33–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S, Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, Tsujimura T, Rakugi H, Isaka Y, Takeuchi O, Akira S, Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Jeltsch KM, Hu D, Brenner S, Zöller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL, Gloury R, Martin N, Lohs C, Lech M, Stehklein JE, Geerlof A, Kremmer E, Weber A, Anders HJ, Schmitz I, Schmidt-Supprian M, Fu M, Holtmann H, Krappmann D, Ruland J, Kallies A, Heikenwalder M, Heissmeyer V, Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol 15, 1079–1089 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Garg AV, Amatya N, Chen K, Cruz JA, Grover P, Whibley N, Conti HR, Hernandez Mir G, Sirakova T, Childs EC, Smithgall TE, Biswas PS, Kolls JK, McGeachy MJ, Kolattukudy PE, Gaffen SL, MCPIP1 Endoribonuclease Activity Negatively Regulates Interleukin-17-Mediated Signaling and Inflammation. Immunity 43, 475–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minagawa K, Wakahashi K, Kawano H, Nishikawa S, Fukui C, Kawano Y, Asada N, Sato M, Sada A, Katayama Y, Matsui T, Posttranscriptional modulation of cytokine production in T cells for the regulation of excessive inflammation by TFL. J Immunol 192, 1512–1524 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, Roberts IS, Copley RR, Bell JI, Cornall RJ, Goodnow CC, A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, Goodnow CC, Vinuesa CG, Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 450, 299–303 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Bertossi A, Aichinger M, Sansonetti P, Lech M, Neff F, Pal M, Wunderlich FT, Anders HJ, Klein L, Schmidt-Supprian M, Loss of Roquin induces early death and immune deregulation but not autoimmunity. J Exp Med 208, 1749–1756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, Zöller J, Warth SC, Hoefig KP, Lohs C, Neff F, Kremmer E, Schick J, Repsilber D, Geerlof A, Blum H, Wurst W, Heikenwälder M, Schmidt-Supprian M, Heissmeyer V, Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity 38, 655–668 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Pratama A, Ramiscal RR, Silva DG, Das SK, Athanasopoulos V, Fitch J, Botelho NK, Chang PP, Hu X, Hogan JJ, Maña P, Bernal D, Korner H, Yu D, Goodnow CC, Cook MC, Vinuesa CG, Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 38, 669–680 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Blackshear PJ, Phillips RS, Ghosh S, Ramos SB, Richfield EK, Lai WS, Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod 73, 297–307 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Gingerich TJ, Stumpo DJ, Lai WS, Randall TA, Steppan SJ, Blackshear PJ, Emergence and evolution of Zfp36l3. Mol Phylogenet Evol 94, 518–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stumpo DJ, Trempus CS, Tucker CJ, Huang W, Li L, Kluckman K, Bortner DM, Blackshear PJ, Deficiency of the placenta- and yolk sac-specific tristetraprolin family member ZFP36L3 identifies likely mRNA targets and an unexpected link to placental iron metabolism. Development 143, 1424–1433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carballo E, Lai WS, Blackshear PJ, Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281, 1001–1005 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ, Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 19, 4311–4323 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ, Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275, 17827–17837 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Lai WS, Kennington EA, Blackshear PJ, Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 23, 3798–3812 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franks TM, Lykke-Andersen J, TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21, 719–735 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J, Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol 31, 256–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N, Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 20, 735–739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salerno F, Engels S, van den Biggelaar M, van Alphen FPJ, Guislain A, Zhao W, Hodge DL, Bell SE, Medema JP, von Lindern M, Turner M, Young HA, Wolkers MC, Translational repression of pre-formed cytokine-encoding mRNA prevents chronic activation of memory T cells. Nat Immunol 19, 828–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y, Blackshear PJ, Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol Cell Biol 24, 6445–6455 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, Shelley WC, Richfield EK, Ray MK, Yoder MC, Aplan PD, Blackshear PJ, Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114, 2401–2410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ, A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Carballo E, Blackshear PJ, Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98, 2389–2395 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, Van Simaeys G, Blackshear PJ, Leo O, Goriely S, Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J Exp Med 210, 1675–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, Ferrando AA, Grutz G, Turner M, Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol 11, 717–724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel KU, Bell LS, Galloway A, Ahlfors H, Turner M, The RNA-Binding Proteins Zfp36l1 and Zfp36l2 Enforce the Thymic β-Selection Checkpoint by Limiting DNA Damage Response Signaling and Cell Cycle Progression. J Immunol 197, 2673–2685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore MJ, Blachere NE, Fak JJ, Park CY, Sawicka K, Parveen S, Zucker-Scharff I, Moltedo B, Rudensky AY, Darnell RB, ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng H, Ning H, Wang Q, Lai J, Wei L, Stumpo DJ, Blackshear PJ, Fu M, Hou R, Hoft DF, Liu J, Tristetraprolin Regulates T(H)17 Cell Function and Ameliorates DSS-Induced Colitis in Mice. Front Immunol 11, 1952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawada S, Scarborough JD, Killeen N, Littman DR, A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Spath S, Komuczki J, Hermann M, Pelczar P, Mair F, Schreiner B, Becher B, Dysregulation of the Cytokine GM-CSF Induces Spontaneous Phagocyte Invasion and Immunopathology in the Central Nervous System. Immunity 46, 245–260 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Godfrey VL, Wilkinson JE, Russell LB, X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol 138, 1379–1387 (1991). [PMC free article] [PubMed] [Google Scholar]
- 37.Godfrey VL, Wilkinson JE, Rinchik EM, Russell LB, Fatal lymphoreticular disease in the scurfy (sf) mouse requires T cells that mature in a sf thymic environment: potential model for thymic education. Proc Natl Acad Sci U S A 88, 5528–5532 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F, Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27, 68–73 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Hadaschik EN, Wei X, Leiss H, Heckmann B, Niederreiter B, Steiner G, Ulrich W, Enk AH, Smolen JS, Stummvoll GH, Regulatory T cell-deficient scurfy mice develop systemic autoimmune features resembling lupus-like disease. Arthritis Res Ther 17, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Multiple Sclerosis Genetics Consortium (IMSGC), Wellcome Trust Case Control Consortium 2, Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, Edkins S, Gray E, Booth DR, Potter SC, Goris A, Band G, Oturai AB, Strange A, Saarela J, Bellenguez C, Fontaine B, Gillman M, Hemmer B, Gwilliam R, Zipp F, Jayakumar A, Martin R, Leslie S, Hawkins S, Giannoulatou E, D'Alfonso S, Blackburn H, Martinelli Boneschi F, Liddle J, Harbo HF, Perez ML, Spurkland A, Waller MJ, Mycko MP, Ricketts M, Comabella M, Hammond N, Kockum I, McCann OT, Ban M, Whittaker P, Kemppinen A, Weston P, Hawkins C, Widaa S, Zajicek J, Dronov S, Robertson N, Bumpstead SJ, Barcellos LF, Ravindrarajah R, Abraham R, Alfredsson L, Ardlie K, Aubin C, Baker A, Baker K, Baranzini SE, Bergamaschi L, Bergamaschi R, Bernstein A, Berthele A, Boggild M, Bradfield JP, Brassat D, Broadley SA, Buck D, Butzkueven H, Capra R, Carroll WM, Cavalla P, Celius EG, Cepok S, Chiavacci R, Clerget-Darpoux F, Clysters K, Comi G, Cossburn M, Cournu-Rebeix I, Cox MB, Cozen W, Cree BA, Cross AH, Cusi D, Daly MJ, Davis E, de Bakker PI, Debouverie M, D'Hooghe M B, Dixon K, Dobosi R, Dubois B, Ellinghaus D, Elovaara I, Esposito F, Fontenille C, Foote S, Franke A, Galimberti D, Ghezzi A, Glessner J, Gomez R, Gout O, Graham C, Grant SF, Guerini FR, Hakonarson H, Hall P, Hamsten A, Hartung HP, Heard RN, Heath S, Hobart J, Hoshi M, Infante-Duarte C, Ingram G, Ingram W, Islam T, Jagodic M, Kabesch M, Kermode AG, Kilpatrick TJ, Kim C, Klopp N, Koivisto K, Larsson M, Lathrop M, Lechner-Scott JS, Leone MA, Leppä V, Liljedahl U, Bomfim IL, Lincoln RR, Link J, Liu J, Lorentzen AR, Lupoli S, Macciardi F, Mack T, Marriott M, Martinelli V, Mason D, McCauley JL, Mentch F, Mero IL, Mihalova T, Montalban X, Mottershead J, Myhr KM, Naldi P, Ollier W, Page A, Palotie A, Pelletier J, Piccio L, Pickersgill T, Piehl F, Pobywajlo S, Quach HL, Ramsay PP, Reunanen M, Reynolds R, Rioux JD, Rodegher M, Roesner S, Rubio JP, Rückert IM, Salvetti M, Salvi E, Santaniello A, Schaefer CA, Schreiber S, Schulze C, Scott RJ, Sellebjerg F, Selmaj KW, Sexton D, Shen L, Simms-Acuna B, Skidmore S, Sleiman PM, Smestad C, Sørensen PS, Søndergaard HB, Stankovich J, Strange RC, Sulonen AM, Sundqvist E, Syvänen AC, Taddeo F, Taylor B, Blackwell JM, Tienari P, Bramon E, Tourbah A, Brown MA, Tronczynska E, Casas JP, Tubridy N, Corvin A, Vickery J, Jankowski J, Villoslada P, Markus HS, Wang K, Mathew CG, Wason J, Palmer CN, Wichmann HE, Plomin R, Willoughby E, Rautanen A, Winkelmann J, Wittig M, Trembath RC, Yaouanq J, Viswanathan AC, Zhang H, Wood NW, Zuvich R, Deloukas P, Langford C, Duncanson A, Oksenberg JR, Pericak-Vance MA, Haines JL, Olsson T, Hillert J, Ivinson AJ, De Jager PL, Peltonen L, Stewart GJ, Hafler DA, Hauser SL, McVean G, Donnelly P, Compston A, Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International Multiple Sclerosis Genetics Consortium (IMSGC), Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, Oturai A, Saarela J, Fontaine B, Hemmer B, Martin C, Zipp F, D'Alfonso S, Martinelli-Boneschi F, Taylor B, Harbo HF, Kockum I, Hillert J, Olsson T, Ban M, Oksenberg JR, Hintzen R, Barcellos LF, Agliardi C, Alfredsson L, Alizadeh M, Anderson C, Andrews R, Søndergaard HB, Baker A, Band G, Baranzini SE, Barizzone N, Barrett J, Bellenguez C, Bergamaschi L, Bernardinelli L, Berthele A, Biberacher V, Binder TM, Blackburn H, Bomfim IL, Brambilla P, Broadley S, Brochet B, Brundin L, Buck D, Butzkueven H, Caillier SJ, Camu W, Carpentier W, Cavalla P, Celius EG, Coman I, Comi G, Corrado L, Cosemans L, Cournu-Rebeix I, Cree BA, Cusi D, Damotte V, Defer G, Delgado SR, Deloukas P, di Sapio A, Dilthey AT, Donnelly P, Dubois B, Duddy M, Edkins S, Elovaara I, Esposito F, Evangelou N, Fiddes B, Field J, Franke A, Freeman C, Frohlich IY, Galimberti D, Gieger C, Gourraud PA, Graetz C, Graham A, Grummel V, Guaschino C, Hadjixenofontos A, Hakonarson H, Halfpenny C, Hall G, Hall P, Hamsten A, Harley J, Harrower T, Hawkins C, Hellenthal G, Hillier C, Hobart J, Hoshi M, Hunt SE, Jagodic M, Jelčić I, Jochim A, Kendall B, Kermode A, Kilpatrick T, Koivisto K, Konidari I, Korn T, Kronsbein H, Langford C, Larsson M, Lathrop M, Lebrun-Frenay C, Lechner-Scott J, Lee MH, Leone MA, Leppä V, Liberatore G, Lie BA, Lill CM, Lindén M, Link J, Luessi F, Lycke J, Macciardi F, Männistö S, Manrique CP, Martin R, Martinelli V, Mason D, Mazibrada G, McCabe C, Mero IL, Mescheriakova J, Moutsianas L, Myhr KM, Nagels G, Nicholas R, Nilsson P, Piehl F, Pirinen M, Price SE, Quach H, Reunanen M, Robberecht W, Robertson NP, Rodegher M, Rog D, Salvetti M, Schnetz-Boutaud NC, Sellebjerg F, Selter RC, Schaefer C, Shaunak S, Shen L, Shields S, Siffrin V, Slee M, Sorensen PS, Sorosina M, Sospedra M, Spurkland A, Strange A, Sundqvist E, Thijs V, Thorpe J, Ticca A, Tienari P, van Duijn C, Visser EM, Vucic S, Westerlind H, Wiley JS, Wilkins A, Wilson JF, Winkelmann J, Zajicek J, Zindler E, Haines JL, Pericak-Vance MA, Ivinson AJ, Stewart G, Hafler D, Hauser SL, Compston A, McVean G, De Jager P, Sawcer SJ, McCauley JL, Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45, 1353–1360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.International Multiple Sclerosis Genetics Consortium, Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galloway A, Saveliev A, Łukasiak S, Hodson DJ, Bolland D, Balmanno K, Ahlfors H, Monzón-Casanova E, Mannurita SC, Bell LS, Andrews S, Díaz-Muñoz MD, Cook SJ, Corcoran A, Turner M, RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science 352, 453–459 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Suk FM, Chang CC, Lin RJ, Lin SY, Liu SC, Jau CF, Liang YC, ZFP36L1 and ZFP36L2 inhibit cell proliferation in a cyclin D-dependent and p53-independent manner. Sci Rep 8, 2742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, Li PJ, Diolaiti ME, Ashworth A, Marson A, Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell 175, 1958–1971.e1915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebner F, Sedlyarov V, Tasciyan S, Ivin M, Kratochvill F, Gratz N, Kenner L, Villunger A, Sixt M, Kovarik P, The RNA-binding protein tristetraprolin schedules apoptosis of pathogen-engaged neutrophils during bacterial infection. J Clin Invest 127, 2051–2065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zekavati A, Nasir A, Alcaraz A, Aldrovandi M, Marsh P, Norton JD, Murphy JJ, Post-transcriptional regulation of BCL2 mRNA by the RNA-binding protein ZFP36L1 in malignant B cells. PLoS One 9, e102625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petkau G, Mitchell TJ, Chakraborty K, Bell SE, V DA, Matheson L, Turner DJ, Saveliev A, Gizlenci O, Salerno F, Katsikis PD, Turner M, The timing of differentiation and potency of CD8 effector function is set by RNA binding proteins. Nat Commun 13, 2274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patial S, Curtis AD 2nd, Lai WS, Stumpo DJ, Hill GD, Flake GP, Mannie MD, Blackshear PJ, Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc Natl Acad Sci U S A 113, 1865–1870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks DG, Walsh KB, Elsaesser H, Oldstone MB, IL-10 directly suppresses CD4 but not CD8 T cell effector and memory responses following acute viral infection. Proc Natl Acad Sci U S A 107, 3018–3023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sedlyarov V, Fallmann J, Ebner F, Huemer J, Sneezum L, Ivin M, Kreiner K, Tanzer A, Vogl C, Hofacker I, Kovarik P, Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol Syst Biol 12, 868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newman R, Ahlfors H, Saveliev A, Galloway A, Hodson DJ, Williams R, Besra GS, Cook CN, Cunningham AF, Bell SE, Turner M, Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat Immunol 18, 683–693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loh XY, Sun QY, Ding LW, Mayakonda A, Venkatachalam N, Yeo MS, Silva TC, Xiao JF, Doan NB, Said JW, Ran XB, Zhou SQ, Dakle P, Shyamsunder P, Koh AP, Huang RY, Berman BP, Tan SY, Yang H, Lin DC, Koeffler HP, RNA-Binding Protein ZFP36L1 Suppresses Hypoxia and Cell-Cycle Signaling. Cancer Res 80, 219–233 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Wang E, Zhou H, Nadorp B, Cayanan G, Chen X, Yeaton AH, Nomikou S, Witkowski MT, Narang S, Kloetgen A, Thandapani P, Ravn-Boess N, Tsirigos A, Aifantis I, Surface antigen-guided CRISPR screens identify regulators of myeloid leukemia differentiation. Cell Stem Cell 28, 718–731.e716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu M, Blackshear PJ, RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 17, 130–143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bye AJH, Pugazhendhi D, Woodhouse S, Brien P, Watson R, Turner M, Pell J, The RNA-binding proteins Zfp36l1 and Zfp36l2 act redundantly in myogenesis. Skelet Muscle 8, 37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR, HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J Biol Chem 276, 47958–47965 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Bhandare S, Goldberg DS, Dowell R, Discriminating between HuR and TTP binding sites using the k-spectrum kernel method. PLoS One 12, e0174052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu LQ, Stumpo DJ, Blackshear PJ, Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol 188, 5150–5159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hyatt LD, Wasserman GA, Rah YJ, Matsuura KY, Coleman FT, Hilliard KL, Pepper-Cunningham ZA, Ieong M, Stumpo DJ, Blackshear PJ, Quinton LJ, Mizgerd JP, Jones MR, Myeloid ZFP36L1 does not regulate inflammation or host defense in mouse models of acute bacterial infection. PLoS One 9, e109072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dumdie JN, Cho K, Ramaiah M, Skarbrevik D, Mora-Castilla S, Stumpo DJ, Lykke-Andersen J, Laurent LC, Blackshear PJ, Wilkinson MF, Cook-Andersen H, Chromatin Modification and Global Transcriptional Silencing in the Oocyte Mediated by the mRNA Decay Activator ZFP36L2. Dev Cell 44, 392–402.e397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK, Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med 197, 1073–1081 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin M, Roth RA, Kozel BA, Mecham RP, Halabi CM, Loss of Angiotensin II Type 2 Receptor Improves Blood Pressure in Elastin Insufficiency. Front Cardiovasc Med 8, 782138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stromnes IM, Goverman JM, Active induction of experimental allergic encephalomyelitis. Nat Protoc 1, 1810–1819 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Kamala T, Hock immunization: a humane alternative to mouse footpad injections. J Immunol Methods 328, 204–214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin CC, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, Cook LE, Egawa T, Taneja R, Murphy TL, Russell JH, Edelson BT, Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun 5, 3551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Kim YH, Park DY, Bae H, Lee DH, Kim KH, Hong SP, Jang SP, Kubota Y, Kwon YG, Lim DS, Koh GY, YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. J Clin Invest 127, 3441–3461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S5: Raw data.
Data file S1: Blood pressure and heart rate measurements in Control and TKOΔT mice.
Fig. S1: Additional characterization of the ocular phenotype in TKOΔT mice.
Fig. S2: Liver inflammation in TKOΔT mice and representative flow cytometric gating for peripheral organs.
Fig. S3: Further analysis of primary and secondary lymphoid organs in TKOΔT mice.
Fig. S4: Identification of autoantibodies, evaluation of T cell activation in DKOΔT mice, and phenotyping of TKOΔT Tregs.
Fig. S5: Further analysis of cytokine production from TKOΔT T cells.
Fig. S6: RNA-sequencing from TKOΔT splenic and kidney CD8+ T cells.
Fig. S7: RNA-sequencing from DKOΔT splenic CD4+ and CD8+ T cells.
Fig. S8: Single and double deletion strains are normal during homeostasis.
Fig. S9: Clinical EAE in DKOΔT mice made haploinsufficient for their remaining Zfp36 family gene.
Fig. S10: Analysis of Cd4-Cre+ Zfp36l1fl/fl Zfp36l2fl/fl CNS on day 14 post-EAE induction.
Fig. S11: ZFP36L1 and ZFP36L2 are uniquely required for optimal TH cell expansion.
Fig. S12: Further analysis of day 5 and day 3 lymph node responses after 2D2+ T cell transfer.
Data file S3: mRNA decay-sequencing from activated Control and TKOΔT CD4+ T cells.
Data file S4: Antibodies and dyes.
Data file S2: RNA-sequencing data from Control and TKOΔT spleen and kidney CD4+ and CD8+ T cells.