Abstract
Wolbachia are the most widely distributed intracellular bacteria, and their most common effect on host phenotype is cytoplasmic incompatibility (CI). A variety of models have been proposed to decipher the molecular mechanism of CI, among which the host modification (HM) model predicts that Wolbachia effectors play an important role in sperm modification. However, owing to the complexity of spermatogenesis and testicular cell-type heterogeneity, whether Wolbachia have different effects on cells at different stages of spermatogenesis or whether these effects are linked with CI remains unknown. Therefore, we used single-cell RNA sequencing to analyse gene expression profiles in adult male Drosophila testes that were infected or uninfected by Wolbachia. We found that Wolbachia significantly affected the proportion of different types of germ cells and affected multiple metabolic pathways in germ cells. Most importantly, Wolbachia had the greatest impact on germline stem cells, resulting in dysregulated expression of genes related to DNA compaction, and Wolbachia infection also influenced the histone-to-protamine transition in the late stage of sperm development. These results support the HM model and suggest that future studies on Wolbachia-induced CI should focus on cells in the early stages of spermatogenesis.
Keywords: Wolbachia, Drosophila melanogaster, cytoplasmic incompatibility, host modification, spermatogenesis
1. Introduction
The intracellular symbiotic bacteria of the genus Wolbachia are widely distributed in arthropods and nematodes and are mainly located in the testes and ovaries of arthropod hosts [1,2]. Wolbachia can manipulate host reproduction in several ways to enhance their own maternal transmission, and the best-studied host phenotype induced by Wolbachia is cytoplasmic incompatibility (CI), in which mating between Wolbachia-infected males and uninfected females leads to embryonic lethality [3]. Numerous studies have discovered that CI is related to zygotic interphase defects in de novo nucleosome assembly and replication, leading to delayed activation of the cell cycle kinase Cdk1 and improper chromosome condensation [4–6].
Although the molecular mechanism of CI has not been fully deciphered, a series of models have been proposed. The earliest proposed CI model was the ‘mod-resc’ (modification/rescue) model, which hypothesizes that CI is caused by the modification of sperm by a factor of Wolbachia and that another factor derived from Wolbachia-infected eggs can rescue this modification [7]. Over the past few years, Wolbachia-encoded CI factors (CifA and CifB) have been identified that appear to be the key to explaining the processes of Wolbachia-induced CI and rescue [8,9]. Correspondingly, a variety of improved CI models based on the ‘mod-resc’ model have been proposed, including the HM (host modification) [5,10] and TA (toxin–antidote) [11] models. The HM model assumes that Cif proteins can modify sperm and that this modification leads to the segregation failure of paternally derived chromosomes and the phenotype of CI in fertilized eggs, unless this modification is reversed or removed by CifA derived from Wolbachia-infected females [10]. The TA model assumes that in embryos, CifB from the male gamete plays the role of toxin, which can lead to the CI phenotype, while these defects can be rescued by the CifA antidote binding CifB in the embryo [11].
Some insightful results about CI have emerged recently, providing evidence supporting one or more predictions of these models. Some evidence seems to support the theory of toxin–antidote binding in the TA model, such as in vitro pull-down experiments showing that CifA can bind to CifB [8], and high-resolution structures of CifA and CifB also support the binding of the two proteins [12]. Studies have shown that CifB is sufficient to induce CI in males, and this CifB-induced sterility is rescued by CifA expression in females [13,14]. However, whether this rescue is due to the binding of CifA and CifB in the embryo under the assumption of the TA model is still unclear as no in vivo data in the host embryos has yet been obtained. Alternatively, the co-expression of CifA and CifB may be the key to inducing CI, according to the two-by-one or HM model [10,15]. For example, in some cases, co-expression of CifA and CifB is necessary for the induction of CI in transgenic male flies [8,9,16], and transmission of the same Wolbachia strain into different hosts may induce CI phenotypes to different degrees [17,18], which suggests that host factors play an important role in Wolbachia-induced CI. Moreover, many studies have indicated the importance of the establishment of Wolbachia modification in the testes during the induction of CI, consistent with the HM model. For example, for some CI-inducing Wolbachia strains, the host sperm cells in the pre-fertilization stages show abnormal morphology or lower competitiveness, suggesting that the sperm cells have been modified [19–21]. Importantly, a paper recently published by Kaur et al. [22] strongly supported the HM model by providing evidence that Cif proteins localize to the nuclear DNA of the host in the process of spermatogenesis and cause abnormal histone retention in elongating spermatids and protamine deficiency in mature sperm.
Based on the HM model, many researchers have carried out various omics studies on the putative modification of testes by Wolbachia and identified multiple host genes and metabolic pathways that may be closely related to CI [23–30]. However, owing to the complexity of spermatogenesis and the heterogeneity of cell types in the testes, we still do not know whether Wolbachia have different effects on cells at different stages of spermatogenesis or whether these effects may be associated with CI. For example, studies have shown that the distribution pattern of Wolbachia in Drosophila testicular cells is uneven, Wolbachia are present in developing spermatocytes and spermatogonia but absent in mature sperm, so the CI-related effects of Wolbachia may start occurring early in sperm development [31–34]. Furthermore, in the parasitic wasp Nasonia vitripennis, Wolbachia-induced CI is close to 100%, but Wolbachia are found in only 28% of developing sperm, suggesting that Wolbachia may also modify uninfected germ cells by secreting effectors [34]. Based on the above evidence, we speculate that the effects of Wolbachia on testicular cells at different stages of spermatogenesis are complex, and the bacteria may have different modification effects on different cell types.
While previous studies have shown the complex impacts of Wolbachia on spermatogenesis that can result in downstream sperm defects [17], a deeper investigation of these defects is needed to confirm their connection to CI. In this study, we employed single-cell RNA sequencing (scRNA-seq) of 1-day-old adult male Drosophila melanogaster testes infected or uninfected with Wolbachia. We attempted to compare the effects of Wolbachia on gene expression profiles in different cell types of Drosophila testes in order to identify the cell type that Wolbachia mainly affects and to explore the host metabolic pathway and gene alterations. Our results showed that Wolbachia have the strongest effect on cells in the early stage of spermatogenesis, and the alteration in the process of DNA compaction in the early stage of sperm development may be related to the establishment of HM in the mechanism of CI.
2. Methods
(a) . Fruit fly rearing
Drosophila melanogaster naturally infected Wolbachia strain wMel was available in our laboratory; Wolbachia-uninfected D. melanogaster strains were established following the previously reported method [28]. All flies were reared on standard Drosophila cornflour medium at 25°C with a photoperiod of 14 h L : 12 h D (light : dark) and 40% relative humidity [35]. Each strain had been cultivated for more than 40 generations before this experiment.
(b) . Embryo hatch rate experiment
We performed the CI test as previously described by Yamada et al. [36]. We crossed 3-day-old virgin female flies (Wolbachia-infected or uninfected) with 1-, 3- and 5-day-old male flies (Wolbachia-infected or uninfected) to examine the effect of male age on the strength of CI. We used agarose/apple juice medium and smeared fresh yeast on the surface for flies to lay eggs. First, a single 3-day-old virgin female was put into each plastic vial containing medium. After 4 h of acclimation, a single virgin male was added to each vial. After mating for approximately 6 h, the females were transferred to a new vial containing fresh medium and incubated at 25°C for 24 h for egg laying. Then, the females were removed, and the eggs were counted immediately; females that laid fewer than five eggs were discarded from the experiment. All vials were incubated at 25°C for another 48 h, and the number of remaining unhatched eggs was counted. Embryo hatch rates were calculated by determining the ratio of the number of hatched eggs to the total number of eggs.
(c) . Microbiome analysis
One-day-old Wolbachia-infected and uninfected male flies were used for intestinal genome extraction and 16S rRNA gene amplicon sequencing (see electronic supplementary material, text for details). FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for raw read quality control, and fastp was used to quality filter the raw data with the following parameters: fastp -q 20 -u 20 -n 5 -c. We used DADA2 (v.1.24.0) to produce amplicon sequence variants (ASVs) based on denoising algorithms [37], used the silver138 database to annotate ASV sequences, used the R package 'phyloseq' to normalize the ASV table to eliminate any bias due to differences in the sampling sequencing depth, and used the sintax_summary function from ‘usearch’ to determine the frequencies of each named taxon at different ranks. We performed principal component analysis (PCA) using the ‘prcomp’ R package. We used ‘vegdist’ to calculate Bray–Curtis dissimilarities and performed principal coordinates analyses (PCoA) by using the ‘ape’ R package. Permutational multivariate analysis of variance (PERMANOVA) was conducted using the adonis2 function in the ‘vegan’ R package to analyse the significance of the community differences. Data link: https://doi.org/10.5061/dryad.stqjq2c5w.
(d) . Preparation of single-cell suspensions
One-day-old Wolbachia-infected and uninfected males were used. For each group, 20–30 pairs of adult male testes were dissected one by one in cold EBSS (NaCl 6.8 g l−1, NaH2PO4 0.112 g l−1, KCl 0.4 g l−1, d-glucose 1 g l−1, NaHCO3 2.2 g l−1, without calcium, magnesium and phenol red). Cells were dissociated by lysis solution (EBSS containing 4 mg ml−1 elastase and 2.5 mg ml−1 type IV collagenase) and filtered for purification. Libraries were prepared with the 10X Genomics chromium 3′ kit, and sequenced on a MGISEQ2000 instrument (BGI, Shenzhen, China) using 100 nucleotide (nt) paired-end sequencing, with at least 7 million reads obtained per library. A detailed description of the methods can be found in electronic supplementary material, methods.
(e) . Downstream single-cell RNA sequencing analysis
The FASTQ file of each sample (WinfM and WuninfM) was used for Cell Ranger (v.6.1; https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest) analysis. This output was then imported into the Seurat (v.4.0.1) R package [38,39] for quality control and downstream analysis of our scRNA-seq data. Data link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA788731?reviewer=n7svq5k71ohhepvg14k41k4hhs.
(f) . Identification of cell types in single-cell RNA sequencing data
Specific markers in each cluster were identified by the ‘FindAllMarkers’ function in the 'Seurat' package' (options: only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25). All predicted marker genes for each cell type are shown in electronic supplementary material, table S1. We used well-known reported marker genes for cell type identification [40,41].
(g) . Functional enrichment analysis of differentially expressed genes
We identified differentially expressed genes (DEGs) for each cell type between Wolbachia-infected and uninfected samples through the 'FindMarkers' function in ‘Seurat’ (logfc.threshold = 0.25, min.pct = 0.25 and test.use = wilcox), and |log2FC| > 0.25 and p-value < 0.05 were considered statistically significant. For the DEGs of each cell type, the ‘clusterProfiler’ (v.3.18.1, [42]) was used with enrichGO for functional analysis, with Fisher's exact tests (two-sided) performed, and p-value adjusted by the Benjamini–Hochberg (BH) procedure, with adjusted p-value (p-adjust) < 0.05 defined as statistically significant.
(h) . Pseudotime inference analysis and identification of differential gene expression patterns
The Monocle3 package (v.1.0.0) [43] was used to analyse single-cell trajectories to reveal differential gene expression patterns associated with cell-state transitions. We reconstructed the expression pattern of some selected genes of interest involved in spermatogenesis or CI in germ cells of WuninfM and WinfM samples along pseudotime, and genes with significant divergence in expression dynamics are displayed. Details are provided in electronic supplementary material, text.
(i) . Distribution of Wolbachia reads in single-cell RNA sequencing data and annotation of Wolbachia genes
To detect the presence of Wolbachia genes in scRNA-seq data, we first selected Wolbachia reads from scRNA-seq data of the WinfM sample and calculated the density of Wolbachia reads by matching each read with a 10× barcode to each cell type. Then, we used Trinity (v.2.8.5; https://github.com/trinityrnaseq/trinityrnaseq/releases) to assemble and eggNOG-mapper (http://eggnog-mapper.embl.de/) to annotate Wolbachia reads in each cell type. As a control, we also mapped scRNA-seq reads of the WuninfM sample to the Wolbachia wMel reference genome, and no Wolbachia reads were detected.
(j) . cif gene expression assays
To identify cif genes in the Wolbachia genome, DNA was extracted from Wolbachia-infected male flies using a QIAGEN DNeasy Blood & Tissue DNA kit (Qiagen, Germany), with uninfected males as the negative control. cif-specific primers were used to amplify cif gene fragments (WD0631-F: ATAAAGGCGTTTCAGCAGGA, WD0632-R: TTGCCAGCCATCATTCATAA) [44]. The PCR mixture contained forward primer (1.25 µl), reverse primer (1.25 µl), Q5 High-Fidelity 2X Master Mix (12.5 µl; New England Biolabs, Ipswich, MA, USA), DNA template (2 µl) and nuclease-free water (8 µl). The PCR programme used was as follows: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 54°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR products were detected with 1% agarose gel electrophoresis and Sanger sequencing.
Quantitative reverse transcription PCR (qRT-PCR) was performed to determine the relative expression dynamics of cif genes with increasing Drosophila male age. One-, 3- and 5-day-old virgin male flies were collected for RNA extraction (siblings from hatch rate assays). The male testis tissues were dissected in cold phosphate-buffered saline (PBS), and 10 pairs of testes were placed into a single 1.5 ml tube with 500 μl of TRIzol. We used the TransZol Up Plus RNA Kit (TransGen, Beijing, China) to extract and purify RNA. First-strand synthesis of cDNA was completed by using TransScript First-Strand cDNA Synthesis SuperMix with random primers (TransGen, Beijing, China). The specific primers used for the Wolbachia cif genes for qPCR were as described by Shropshire et al. [33], and all the primers are listed in electronic supplementary material, table S2. qPCR was performed using a CFX96 system (Bio-Rad) with PerfectStart Green qPCR SuperMix (TransGen, Beijing, China). The qPCR programme was as follows: 50°C for 2 min, 95°C for 2 min, followed by 39 cycles of 95°C for 15 s, 54°C for 15 s and 72°C for 20 s, and a final dissolution step. All samples were tested in triplicate. The Wolbachia ftsZ and D. melanogaster β-spec genes were used as reference genes for normalization. The fold change was calculated as 2–ΔΔCq.
3. Results
(a) . Wolbachia induced the strongest cytoplasmic incompatibility in 1-day-old males
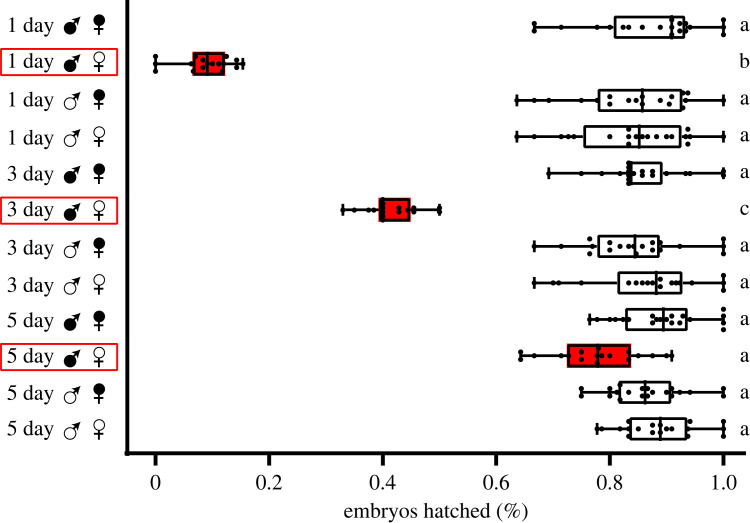
To test the CI strength of the Wolbachia strain wMel, we performed a hatch rate experiment on male flies of different ages. The results showed that the strength of CI induced by Wolbachia was closely related to male age, since Wolbachia induced almost 100% CI in 1-day-old males, while 5-day-old males expressed no CI (figure 1). We thus focused on 1-day-old males for subsequent studies. p-values are reported in electronic supplementary material, table S3.
Figure 1.
Embryo hatch rate analyses showing the effect of Wolbachia-induced cytoplasmic incompatibility (CI), which decreased with male age. The horizontal axis shows the hatch rate, and the vertical axis shows different cross groups (days represent male age, black solid circles represent Wolbachia-infected samples, and open circles represent uninfected samples). CI crosses are coloured red, boxplots represent median and interquartile ranges. Letters to the right represent statistically significant differences based on α = 0.05 calculated by Kruskal–Wallis test followed by Dunn's test with corrections for multiple comparisons between all groups; crosses with different letters are significantly different. These data demonstrate that 1-day-old males show the strongest CI.
(b) . The gut bacterial community structures of male flies were highly correlated with Wolbachia infection
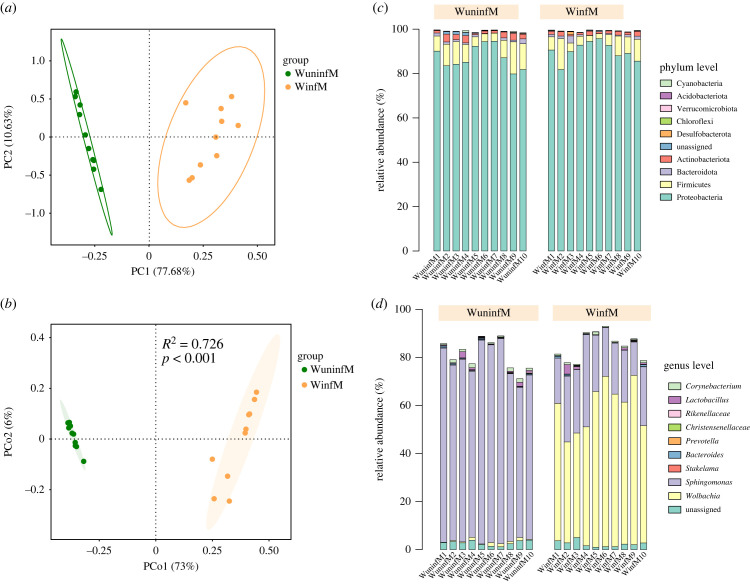
Uninfected Drosophila strains are mainly obtained through antibiotic treatment. However, many studies have shown that antibiotic treatment can significantly change the host's microbiome [45–48], which may lead to changes in host gene expression. To detect changes in the microbiome of the uninfected Drosophila strain after recovery from antibiotic treatment for more than 40 generations, we performed microbiome analyses. PCA showed that Wolbachia-infected and uninfected samples were well divided into two clusters. Consistent with this, the two Drosophila strains were also significantly separated in the PCoA (PERMANOVA: R2 = 0.728 and p < 0.001) (figure 2a,b). To further determine which bacterial group in Drosophila was changed owing to the long-term differences in Wolbachia infection and non-infection, we compared the differences in the bacterial community based on the top 10 bacterial groups at the phylum and genus levels. There was no significant difference in abundance between the groups at the phylum level (figure 2c). However, at the genus level, Wolbachia was the dominant genus in the Wolbachia-infected samples, and Wolbachia was also the genus with the largest difference between the two groups (figure 2d). Therefore, we speculate that the change in host gene expression is mainly related to Wolbachia infection.
Figure 2.
Gut bacterial diversity analyses of Wolbachia-infected (WinfM) and uninfected (WuninfM) Drosophila males. (a) Principal component analysis (PCA) of 16S rRNA gene-sequencing data of gut microbes obtained from Wolbachia-infected and uninfected male flies. (b) Principal coordinate analysis (PCoA) based on Bray–Curtis distances between both groups, with R2 and p-values derived from the adjusted PERMANOVA models. (c,d) Top 10 relative abundances of bacteria, showing bacterial community structure at the phylum and genus levels.
(c) . Single-cell expression atlas and cell typing in adult Drosophila testes infected and uninfected with Wolbachia
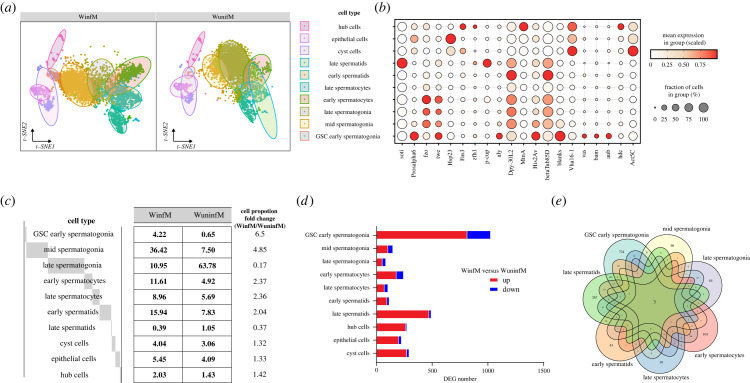
To understand the modification effects of Wolbachia on different cell types in testes, we performed scRNA-seq on Wolbachia-infected (WinfM) and uninfected (WuninfM) one-day-old male Drosophila testes. After the initial quality control step (see Methods), we acquired a total of 8372 high-quality cells, with 4385 cells from WinfM and 3987 from WuninfM samples. To explore the cellular compositions, we applied PCA to DEGs across all cells for dimensionality reduction. We further used unsupervised graph clustering to partition the cells into 10 clusters and visualized them via t-distributed stochastic neighbour embedding (t-SNE) (figure 3a). We generated cluster-specific marker genes via differential gene expression analysis and used previously reported marker genes to identify the cell types. These well-known cell type markers, such as soti for late spermatids, prosalpha, aly and bam for germline stem cells (GSCs), hdc for hub cells, and Act5C for somatic cells [36,37], were used to determine the cellular identity of the clusters (figure 3b). We finally identified a total of 10 cell types, with seven germ cell types, including GSC early spermatogonia, mid spermatogonia, late spermatogonia, early spermatocytes, late spermatocytes, early spermatids and late spermatids, and three somatic cell types, hub, cyst and epithelial cells. The relative expression of markers in all cell types is shown in electronic supplementary material, table S1.
Figure 3.
Diverse cell types identified in Drosophila testes from 1-day-old males by single-cell RNA sequencing (scRNA-seq) analysis. (a) A t-distributed stochastic neighbour embedding (t-SNE) visualization and clustering of WuninfM and WinfM samples. (b) Dot plot of scaled expression of marker genes in each cell type. For each dot, the size refers to the proportion of cells expressing a gene, and the colour represents the calculated scaled expression value. (c) Proportions of each cell type in the WinfM and WuninfM samples and cell proportion fold changes between the two samples. (d) Numbers of differentially expressed genes (DEGs) between WinfM and WuninfM samples in each cell type (|log2-fold change| ≥ 0.25 and p-value < 0.05); up: upregulated in WinfM, down: downregulated in WinfM. (e) Venn diagram showing only three coregulated DEGs among all germ cell types.
Following initial cell-type characterization, we calculated the percentage of each cell type in both samples. In the WuninfM sample, the vast majority of cells (63.78%) were in the late spermatogonia stage, and the cell type with the lowest proportion (0.65%) was GSC early spermatogonia, while in the WinfM sample, the cell type with the highest proportion (36.42%) was mid spermatogonia and the lowest proportion was observed in late spermatids (0.39%) (figure 3c). We further compared the differences in the proportion of each cell type between the two samples and found that the proportions of most cell types varied greatly upon Wolbachia infection. For germ cells, the proportions of GSC early spermatogonia, late spermatogonia, mid spermatogonia, early spermatocytes and late spermatids differed greatly between the WinfM and WuninfM samples, among which the proportions of GSC early spermatogonia, early spermatocytes and mid spermatogonia were higher in the WinfM sample, and the proportions of late spermatogonia and late spermatids were lower in the WinfM sample, than in the WuninfM sample (figure 3c). For the three somatic cell types, the proportions in the WinfM sample were slightly higher than those in the WuninfM sample, and the difference was not as significant as those of germ cells (figure 3c). All these findings indicate that several cell types are affected by Wolbachia infection, suggesting that the cell-type-specific effects of Wolbachia deserve further exploration.
(d) . Cell-type-specific aberrant gene expression in Wolbachia-infected samples
Next, we compared transcriptome profiles between WinfM and WuninfM samples for each cell type (electronic supplementary material, table S4). We found that the number of upregulated genes was higher than the number of downregulated genes in all cell types in the WinfM sample compared with the WuninfM sample; moreover, the number of DEGs varied greatly among different types of germ cells. GSC early spermatogonia contained the largest number of DEGs among all germ cells, followed by late spermatids and early spermatocytes, suggesting that Wolbachia might have a greater effect on these cell types (figure 3d). In addition, a total of 1653 DEGs were found in all germ cells; however, only three of them (CG43800, hydra and Sclp) displayed differential expression in all germ cells (figure 3e), implying that the majority of the DEGs were cell-type-specific. These results indicate the cell-type-specific impact of Wolbachia on gene expression in Drosophila testes.
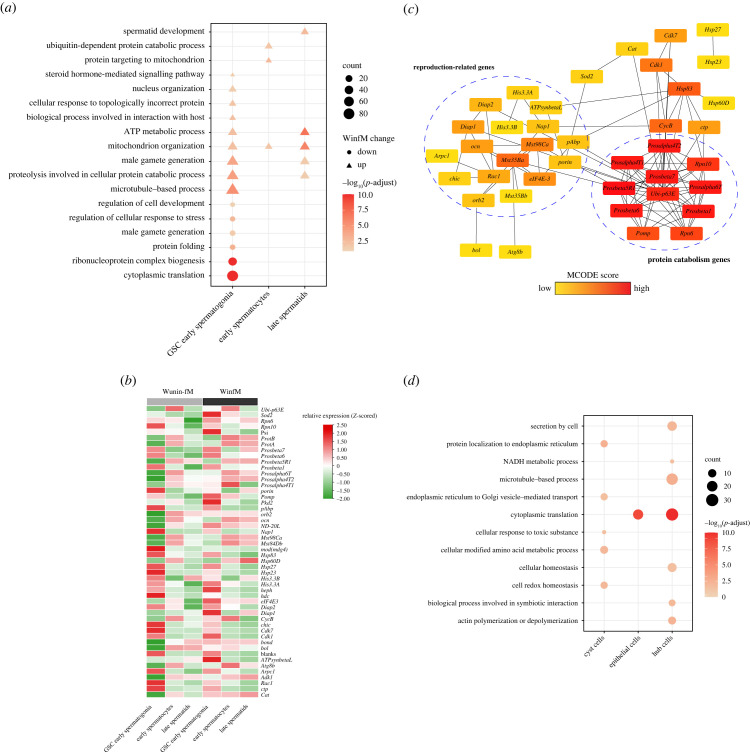
(e) . Functional enrichment analyses of cell-type-specific differentially expressed genes revealed differential effects of Wolbachia on different cell types
To further clarify the function of DEGs between WinfM and WuninfM samples, we performed functional enrichment analysis on DEGs of each cell type (electronic supplementary material, table S5). Among germ cells, GSC early spermatogonia, late spermatids and early spermatocytes showed larger changes in cell proportion and more DEGs than other cell types, which led us to focus on these three germ cell types in subsequent analyses. For the three germ cell types, we found that the upregulated DEGs in the infected samples were mostly enriched in microtubule-based, ubiquitin-dependent protein catabolic, proteolysis involved in protein catabolic, reproduction-related and ATP metabolic processes. The downregulated genes in GSC early spermatogonia were significantly enriched in translation, ribonucleoprotein complex biogenesis, regulation of cellular response to stress, nucleus organization and male gamete generation processes (figure 4a).
Figure 4.
Functional analysis of the cell-type-specific differentially expressed genes (DEGs) between WinfM and WuninfM samples. (a) Enriched biological processes of DEGs in the three germ cell types. Triangles represent the enriched GO terms of upregulated genes in WinfM, while dots represent the enriched GO terms of downregulated genes in WinfM; Fisher's exact test (two-sided) performed. The p-values were adjusted by the Benjamini–Hochberg (BH) method, with adjusted p-value (p-adjust) < 0.05 defined as statistically significant. (b) Relative expression of selected genes in the three germ cell types. (c) Protein–protein interaction network analysis of selected DEGs, showing only connected nodes in the network. The nodes represent the genes, and edges indicate interaction associations between nodes. The MCODE (Molecular Complex Detection) plugin from Cytoscape was utilized to identify highly interconnected clusters from the network. Colours of the dots represent the connectivity index derived from MCODE scores. (d) Enriched biological processes of DEGs in the three somatic cell types; Fisher's exact test (two-side) performed. The p-values were adjusted by the BH method, with adjusted p-value (p-adjust) < 0.05 defined as statistically significant.
To further target the key genes in the above biological processes, we selected genes with three criteria (fold change, p-value, previously reported related to CI) and determined their differential expression patterns in the three types of cells; these genes were mainly differentially expressed in GSC early spermatogonia (figure 4b). Then, we performed protein–protein interaction network analysis on these genes and found that the interaction network of these genes is mainly divided into two clusters: protein catabolism-related genes (such as multiple testis-specific proteolysis genes) and reproduction-related genes (such as His3.3A, His3.3B, ProtA, ProtB, Nap1 and ATPsynbetaL). Therefore, the DEGs involved in these two biological processes in germ cells, especially in GSCs, may be the key host factors for Wolbachia-mediated modification of sperm (figure 4c).
Interestingly, the functional enrichment results of DEGs in the three somatic cell types were quite different from those of germ cells. Somatic DEGs were mainly involved in transcription, cellular secretion, cell redox homeostasis, NADH metabolism, cellular response to toxic substances and other processes (figure 4d). These results indicate that Wolbachia have distinct interactions with somatic and germ cells.
(f) . Wolbachia infection caused various differentially expressed genes to exhibit different dynamic expression patterns during spermatogenesis
Different genes tend to have different expression patterns at different stages of sperm development. To decipher the transcriptomic dynamics during spermatogenesis, we performed pseudotime analysis on the germ cells of the WinfM and WuninfM samples using Monocle3 and found that both samples had similar cell development trajectories (electronic supplementary material, figure S1).
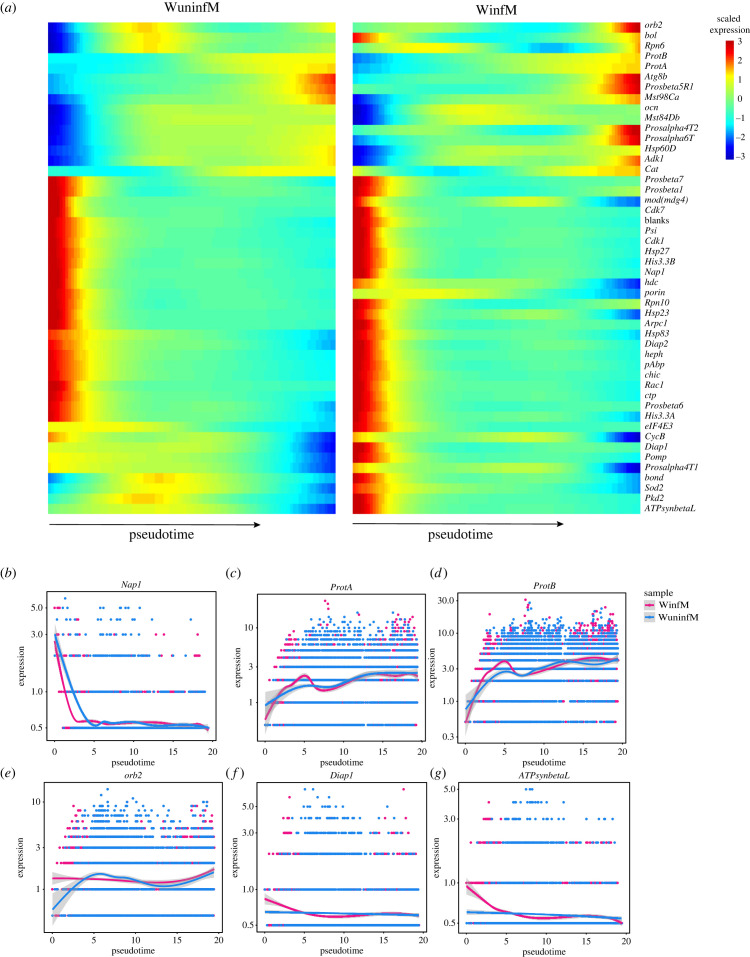
We reconstructed the dynamic expression patterns for the selected DEGs as shown in figure 4b. We found that multiple genes, such as Nap1, ProtA, ProtB, CycB, bol, Diap1, Pomp, bond, sod2, orb2, pAbp, ATPsynbetaL, Pkd2, Prosalpha4T2, Mst84Db and eIF4E3, exhibited significant dynamic differences in expression between the two samples (figure 5a and electronic supplementary material, figure S2). Among them, six candidate genes, Nap1, ProtA, ProtB, orb2, Diap1 and ATPsybetaL, were selected according to two independent criteria—the differences in expression patterns along the trajectories and the fold changes in expression levels between the two samples. These genes were related to male gamete generation; they showed significant dynamic expression differences between WinfM and WuninfM samples in early spermatogenesis; however, in late spermatogenesis, the expression patterns converged (figure 5b–g). These patterns suggest that Wolbachia have a greater effect on the early stage of spermatogenesis. In addition, we found that the expression levels of ProtA and ProtB were significantly increased at the later stage of sperm development (figure 5c,d).
Figure 5.
Cell trajectory analysis of spermatogenesis-related genes. (a) Heatmap showing the expression pattern of selected differentially expressed genes (DEGs) across single cells along pseudotime in WuninfM and WinfM samples. (b–g) Smoothed expression curves of several candidate genes along the trajectories of spermatogenesis in both samples. The blue and pink lines represent the WuninfM and WinfM samples, respectively, and the shaded area around the line represents the 95% confidence interval.
(g) . The presence of active Wolbachia genes in different cells suggested differential interaction of Wolbachia with different cell types
Based on the HM model that Wolbachia-induced CI depends on the modification of sperm by Wolbachia effectors, such as CifA and CifB, we attempted to determine the expression pattern of Wolbachia genes from the scRNA-seq data. We assumed that the number of detected Wolbachia sequences was positively correlated with the number of active Wolbachia cells. Based on this assumption, we discovered that the distribution of Wolbachia among different cells was uneven. The highest Wolbachia density was in somatic cells, mostly in hub cells, while among germ cells, the highest density was in GSC early spermatogonia, and there was a decreasing trend in Wolbachia density during the process of spermatogenesis (electronic supplementary material, figure S3a). Then, we performed assembly and functional annotation of the Wolbachia sequences in each cell type, with a total of 38 Wolbachia genes annotated (cifA and cifB were not detected). We found that the number of Wolbachia genes annotated in different cells varied, with the most genes annotated in the hub cells (electronic supplementary material, figure S3b,c). In hub cells, the annotated Wolbachia genes were involved in translation, amino acid transport, vesicular transport, cell membrane/cell wall synthesis and other processes (electronic supplementary material, figure S3d), implying that Wolbachia proliferation occurs primarily in hub cells. Interestingly, in several cell types, the iscU gene was annotated; this gene encodes a component of the iron–sulfur (Fe–S) cluster scaffold, and Fe–S clusters are important cofactors in the functioning of diverse enzymes involved in iron homeostasis and oxidative stress response. WD_1250 expressed in hub cells is also associated with redox homeostasis. Thus, the expression of Wolbachia genes with different functions detected in different cell types suggests that Wolbachia genes interact differently with different cell types in Drosophila testes.
Since we did not detect any expression of cif genes from the scRNA-seq data, to further confirm whether there are cif genes in the Wolbachia genome and to measure the expression of cif genes, we performed amplification, sequencing and qPCR expression analyses on cifs. The cif sequences of Wolbachia obtained by amplification and sequencing were completely consistent with the cif gene reference sequences of Wolbachia wMel (electronic supplementary material, figure S4a,b). The expression of cifA and cifB had no significant correlation with male age, but the expression ratio of cifA and cifB decreased with male age (not statistically significant) (electronic supplementary material, figure S4c). Therefore, the expression of cif genes does not sufficiently explain the variation in CI strength, which has been confirmed in previous studies [33].
4. Discussion
The Wolbachia-induced reproductive regulation phenotype of CI has been extensively studied in a variety of insects; however, the molecular mechanism of CI remains controversial. In the present study, through microbiome analysis, we first determined that changes in host gene expression may be mainly associated with Wolbachia infection. Next, through single-cell RNA sequencing, our results revealed that Wolbachia have the greatest effect on cells in the early stages of sperm development from three perspectives. First, the proportion of early spermatogenesis cells, such as GSC early spermatogonia, in the WinfM sample was significantly higher than that in the WuninfM sample, suggesting that in the Drosophila testes, Wolbachia infection causes more cells to be retained in the early stages of sperm development. Second, the number of DEGs in GSC early spermatogonia was the largest among all cell types, suggesting that Wolbachia may cause more modifications in early spermatogenesis, such as in GSCs. Finally, in the analysis of gene expression dynamics, multiple DEGs related to spermatogenesis or male gamete generation, such as Nap1, Diap1, ProtB, ProtA, orb2 and ATPsynbetaL, also showed differential expression patterns along the developmental trajectories between samples of WinfM and WuninfM, since these genes showed a pattern of significant differential expression in the early stage of spermatogenesis and were more similar in the late stages, suggesting that Wolbachia mainly affect the early stages of spermatogenesis.
Wolbachia not only affect processes related to reproduction in germ cells but also significantly affect the processes of mitochondrial energy metabolism and proteolysis. On the one hand, the upregulated DEGs in the three germ cells were significantly enriched in biological processes related to ATP metabolic processes, and studies have shown that Wolbachia infection can significantly enhance host energy consumption, resulting in excessive oxidative stress and intracellular DNA damage, which in turn affects sperm development [49–51]. On the other hand, the ubiquitin proteolysis pathway plays a key role in chromosomal remodelling and is involved in the conversion of histones to protamine during the later stages of spermatogenesis [52], and mutations in some testis-specific proteolysis genes may even lead to defects in sperm nuclear morphology, with histones being not effectively removed [53]. In our results, multiple testis-specific proteolysis genes, including Prosalpha4T1, Prosbeta5R, Prosalpha3, Prosbeta6, Prosbeta7, Rpn12 and Rpn6, were upregulated in germ cells, especially in GSC early spermatogonia and late spermatids. We speculate that Wolbachia can modify the protein catabolic process and mitochondrial energy metabolic process, leading to abnormal sperm cell production.
In addition to the effect on germ cells, the influence of Wolbachia on somatic cells cannot be ignored, since hub cells and cyst cells also play a crucial role in the development of germ cells by secreting various signalling molecules [54,55]. Here, we found that Wolbachia induced more differential gene expression between WinfM and WuninfM samples in hub cells and cyst cells than in most germ cells, and in these somatic cells, the DEGs were significantly enriched in processes of translation, cellular response to toxic substances, cellular homeostasis, cell redox homeostasis, NADH metabolic process and secretion by cells, significantly different from the results for germ cells. In addition, we found that the density of Wolbachia sequences was the highest in hub cells, indicating that Wolbachia metabolic activity may be more active in hub cells, therefore resulting in alterations in many pathways involved in metabolism and cellular homeostasis in these cells.
According to the HM model, Wolbachia factors can modify host factors [10], indicating that host factors play an important role in CI. A recent study has shown that Wolbachia modifies developing sperm at the canoe stage [22]. Here, we found that the cells of the sperm developmental stage most affected by Wolbachia were GSC early spermatogonia and that Wolbachia-induced downregulated genes in GSC early spermatogonia were significantly enriched in male gamete generation. Importantly, three genes involved in chromosome condensation, including Nap1, His3.3B and mod(mdg4), were only downregulated in GSC early spermatogonia, among which Nap1 encodes a host protein closely related to CI [56,57]. Therefore, we speculate that the DNA compaction process in early spermatogonia is affected by Wolbachia. These results are also highly consistent with the newly discovered results showing that the CifA and CifB proteins are localized to nuclear DNA in the early stage of spermatogenesis, indicating that Wolbachia-mediated modification of sperm may be established in the early stage of sperm development [22]. Our results also indicate the influence of Wolbachia in the later stage of spermatogenesis. During sperm development, histones bind to the nucleosomes in the early stage, being replaced by protamine at later stages for the tight packaging of DNA in the nucleus within the sperm head [58]. We found that the expression levels of the Protamine A (ProtA) and Protamine B (ProtB) genes were significantly higher in late spermatids than in early germ cells and were significantly upregulated in Wolbachia-infected samples. ProtA and ProtB play important roles in chromosome remodelling during the transition from histones to protamine in the late stage of sperm development, where they are integrated into the sperm nucleus, contributing to the formation of condensed sperm chromatin, and mutations in ProtA and ProtB genes can cause improper nuclear morphology [58–60]. The new results of Kaur et al. [22] show that Cifs can cause abnormal histone retention in elongating spermatids and protamine deficiency in mature sperm. Our results showing higher expression of the protamine genes ProtA and ProtB in late spermatids in Wolbachia-infected testes also indicate that the modification of sperm by Wolbachia is also closely related to this chromosome remodelling stage.
5. Conclusion
Overall, we thoroughly analysed the effects of Wolbachia on gene expression in Drosophila testes at the single-cell transcriptome level. We identified the metabolic pathways most affected by Wolbachia in germ cells, such as reproduction and protein degradation. We also revealed that early spermatogenesis cells are the cell type strongly impacted by Wolbachia and recapitulate previous finding that the sperm DNA compaction is the key target for CI modification. Thus, our data support the hypothesis that Wolbachia factors can modify sperm cells prior to fertilization and suggest that further studies of the establishment of CI should focus on cells in the early stages of sperm development and in the cellular stages of chromosome remodelling.
Acknowledgements
We would like to thank Dr Haoyuan Hu for donating fruit flies.
Contributor Information
Dawei Huang, Email: huangdw@nankai.edu.cn.
Jinhua Xiao, Email: xiaojh@nankai.edu.cn.
Data accessibility
The accession number for the scRNA sequencing data reported in this paper is SRA: SAMN24018191, SAMN24018192. These data have been deposited in the Sequence Read Archive under BioProject: PRJNA788731. Data link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA788731?reviewer=n7svq5k71ohhepvg14k41k4hhs. 16S sequencing data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.stqjq2c5w [61]. R scripts used in this manuscript are available in electronic supplementary material, software file [62].
Authors' contributions
W.D.: formal analysis, methodology, visualization, writing—original draft; B.S.: funding acquisition, methodology, software; Y.M.: writing—review and editing; D.H.: funding acquisition, resources, supervision; J.X.: conceptualization, funding acquisition, investigation, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed herein.
Conflict of interest declaration
The authors declare no competing interests.
Funding
This work was supported by the National Science Foundation of China (grant nos 31830084, 31970440 and 32170497), and also by ‘the Fundamental Research Funds for the Central Universities’, Nankai University (grant nos 96172158, 96173250 and 91822294).
References
- 1.Kaur R, Shropshire JD, Cross KL, Leigh B, Mansueto AJ, Stewart V, Bordenstein SR, Bordenstein SR. 2021. Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host Microbe 29, 879-893. ( 10.1016/j.chom.2021.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741-751. ( 10.1038/nrmicro1969) [DOI] [PubMed] [Google Scholar]
- 3.Werren JH, O'Neill SL. 1997. The evolution of heritable symbionts. In Influential passengers: inherited microorganisms and arthropod reproduction (eds O'Neill SL, Hoffmann AA, Werren JH), pp. 1-41. Oxford, UK: Oxford University Press. [Google Scholar]
- 4.Lassy CW, Karr TL. 1996. Cytological analysis of fertilization and early embryonic development in incompatible crosses of Drosophila simulans. Mechanisms Dev. 57, 47-58. ( 10.1016/0925-4773(96)00527-8) [DOI] [PubMed] [Google Scholar]
- 5.Tram U, Sullivan W. 2002. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296, 1124-1126. ( 10.1126/science.1070536) [DOI] [PubMed] [Google Scholar]
- 6.Landmann F, Orsi GA, Loppin B, Sullivan W. 2009. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 5, e1000343. ( 10.1371/journal.ppat.1000343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werren JH. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42, 587-609. ( 10.1146/annurev.ento.42.1.587) [DOI] [PubMed] [Google Scholar]
- 8.Beckmann JF, Ronau JA, Hochstrasser M. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 17007. ( 10.1038/nmicrobiol.2017.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LePage DP, et al. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243-247. ( 10.1038/nature21391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shropshire JD, Leigh B, Bordenstein SR, Duplouy A, Riegler M, Brownlie JC, Bordenstein SR. 2019. Models and nomenclature for cytoplasmic incompatibility: caution over premature conclusions – a response to Beckmann et al. Trends Genet. 35, 397-399. ( 10.1016/j.tig.2019.03.004) [DOI] [PubMed] [Google Scholar]
- 11.Beckmann JF, Bonneau M, Chen H, Hochstrasser M, Poinsot D, Mercot H, Weill M, Sicard M, Charlat S. 2019. The toxin–antidote model of cytoplasmic incompatibility: genetics and evolutionary implications. Trends Genet. 35, 175-185. ( 10.1016/j.tig.2018.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Y, et al. 2021. Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors. Proc. Natl Acad. Sci. USA 118, e2107699118. ( 10.1073/pnas.2107699118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams KL, Abernathy DG, Willett BC, Selland EK, Itoe MA, Catteruccia F. 2021. Wolbachia cifB induces cytoplasmic incompatibility in the malaria mosquito vector. Nat. Microbiol. 6, 1575-1582. ( 10.1038/s41564-021-00998-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun G, Zhang M, Chen H, Hochstrasser M. 2022. The CinB nuclease from wNo Wolbachia is sufficient for induction of cytoplasmic incompatibility in Drosophila. mBio 13, e0317721. ( 10.1128/mbio.03177-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shropshire JD, Bordenstein SR. 2019. Two-by-one model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 15, e1008221. ( 10.1371/journal.pgen.1008221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Ronau JA, Beckmann JF, Hochstrasser M. 2019. A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl Acad. Sci. USA 116, 22 314-22 321. ( 10.1073/pnas.1914571116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shropshire JD, Leigh B, Bordenstein SR. 2020. Symbiont-mediated cytoplasmic incompatibility: what have we learned in 50 years? eLife 9, e61989. ( 10.7554/eLife.61989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baião GC, Janice J, Galinou M, Klasson L. 2021. Comparative genomics reveals factors associated with phenotypic expression of Wolbachia. Genome Biol. Evol. 13, evab111. ( 10.1093/gbe/evab111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tram U, Fredrick K, Werren JH, Sullivan W. 2006. Paternal chromosome segregation during the first mitotic division determines Wolbachia-induced cytoplasmic incompatibility phenotype. J. Cell Sci. 119, 3655-3663. ( 10.1242/jcs.03095) [DOI] [PubMed] [Google Scholar]
- 20.Riparbelli MG, Giordano R, Callaini G. 2007. Effects of Wolbachia on sperm maturation and architecture in Drosophila simulans Riverside. Mechanisms Dev. 124, 699-714. ( 10.1016/j.mod.2007.07.001) [DOI] [PubMed] [Google Scholar]
- 21.Champion de Crespigny FE, Wedell N. 2006. Wolbachia infection reduces sperm competitive ability in an insect. Proc. R. Soc. B 273, 1455-1458. ( 10.1098/rspb.2006.3478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur R, Leigh BA, Ritchie IT, Bordenstein SR. 2022. The Cif proteins from Wolbachia prophage WO modify sperm genome integrity to establish cytoplasmic incompatibility. PLoS Biol. 20, e3001584. ( 10.1371/journal.pbio.3001584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xi Z, Gavotte L, Xie Y, Dobson SL. 2008. Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genom. 9, 1. ( 10.1186/1471-2164-9-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biwot JC, Zhang HB, Liu C, Qiao JX, Yu XQ, Wang YF. 2020. Wolbachia-induced expression of kenny gene in testes affects male fertility in Drosophila melanogaster. Insect Sci. 27, 869-882. ( 10.1111/1744-7917.12730) [DOI] [PubMed] [Google Scholar]
- 25.Zug R, Hammerstein P. 2015. Wolbachia and the insect immune system: what reactive oxygen species can tell us about the mechanisms of Wolbachia–host interactions. Front. Microbiol. 6, 1201. ( 10.3389/fmicb.2015.01201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang HB, Cao Z, Qiao JX, Zhong ZQ, Pan CC, Liu C, Zhang LM, Wang YF. 2021. Metabolomics provide new insights into mechanisms of Wolbachia-induced paternal defects in Drosophila melanogaster. PLoS Pathog. 17, e1009859. ( 10.1371/journal.ppat.1009859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Wang JL, Zheng Y, Xiong EJ, Li JJ, Yuan LL, Yu XQ, Wang YF. 2014. Wolbachia-induced paternal defect in Drosophila is likely by interaction with the juvenile hormone pathway. Insect Biochem. Mol. Biol. 49, 49-58. ( 10.1016/j.ibmb.2014.03.014) [DOI] [PubMed] [Google Scholar]
- 28.Dou W, Miao Y, Xiao J, Huang D. 2021. Association of Wolbachia with gene expression in Drosophila testes. Microb. Ecol. 82, 805-817. ( 10.1007/s00248-021-01703-0) [DOI] [PubMed] [Google Scholar]
- 29.Bing XL, Lu YJ, Xia CB, Xia X, Hong XY. 2019. Transcriptome of Tetranychus urticae embryos reveals insights into Wolbachia-induced cytoplasmic incompatibility. Insect Mol. Biol. 29, 193-204. ( 10.1111/imb.12620) [DOI] [PubMed] [Google Scholar]
- 30.Zheng Y, Wang JL, Liu C, Wang CP, Walker T, Wang YF. 2011. Differentially expressed profiles in the larval testes of Wolbachia infected and uninfected Drosophila. BMC Genom. 12, 595. ( 10.1186/1471-2164-12-595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark ME, Veneti Z, Bourtzis K, Karr TL. 2002. The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila. Mechanisms Dev. 111, 3-15. ( 10.1016/s0925-4773(01)00594-9) [DOI] [PubMed] [Google Scholar]
- 32.Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. 2008. The genetics and cell biology of Wolbachia–host interactions. Annu. Rev. Genet. 42, 683-707. ( 10.1146/annurev.genet.41.110306.130354) [DOI] [PubMed] [Google Scholar]
- 33.Shropshire JD, Hamant E, Cooper BS. 2021. Male age and Wolbachia dynamics: investigating how fast and why bacterial densities and cytoplasmic incompatibility strengths vary. mBio 12, e0299821. ( 10.1128/mBio.02998-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark ME, Bailey-Jourdain C, Ferree PM, England SJ, Sullivan W, Windsor DM, Werren JH. 2008. Wolbachia modification of sperm does not always require residence within developing sperm. Heredity (Edinb.) 101, 420-428. ( 10.1038/hdy.2008.71) [DOI] [PubMed] [Google Scholar]
- 35.Ashburner M, Golic KG, Hawley RS. 1989. Drosophila: a laboratory handbook. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 36.Yamada R, Floate KD, Riegler M, O'Neill SL. 2007. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics 177, 801-808. ( 10.1534/genetics.106.068486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581-583. ( 10.1038/nmeth.3869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. ( 10.1038/nbt.4096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. ( 10.1038/nbt.3192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White-Cooper H. 2012. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2, 11-22. ( 10.4161/spmg.19088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witt E, Benjamin S, Svetec N, Zhao L. 2019. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife 8, e47138. ( 10.7554/eLife.47138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284-287. ( 10.1089/omi.2011.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J, et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496-502. ( 10.1038/s41586-019-0969-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindsey ARI, Rice DW, Bordenstein SR, Brooks AW, Bordenstein SR, Newton ILG. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol. Evol. 10, 434-451. ( 10.1093/gbe/evy012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Audsley MD, Seleznev A, Joubert DA, Woolfit M, O'Neill SL, McGraw EA. 2018. Wolbachia infection alters the relative abundance of resident bacteria in adult Aedes aegypti mosquitoes, but not larvae. Mol. Ecol. 27, 297-309. ( 10.1111/mec.14436) [DOI] [PubMed] [Google Scholar]
- 46.Duan XZ, et al. 2020. Recent infection by Wolbachia alters microbial communities in wild Laodelphax striatellus populations. Microbiome 8, 104. ( 10.1186/s40168-020-00878-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simhadri RK, Fast EM, Guo R, Schultz MJ, Vaisman N, Ortiz L, Bybee J, Slatko BE, Frydman HM. 2017. The gut commensal microbiome of Drosophila melanogaster is modified by the endosymbiont Wolbachia. mSphere 2, e00287-17. ( 10.1128/mSphere.00287-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye YH, Seleznev A, Flores HA, Woolfit M, McGraw EA. 2017. Gut microbiota in Drosophila melanogaster interacts with Wolbachia but does not contribute to Wolbachia-mediated antiviral protection. J. Invertebr. Pathol. 143, 18-25. ( 10.1016/j.jip.2016.11.011) [DOI] [PubMed] [Google Scholar]
- 49.Brennan LJ, Haukedal JA, Earle JC, Keddie B, Harris HL. 2012. Disruption of redox homeostasis leads to oxidative DNA damage in spermatocytes of Wolbachia-infected Drosophila simulans. Insect Mol. Biol. 21, 510-520. ( 10.1111/j.1365-2583.2012.01155.x) [DOI] [PubMed] [Google Scholar]
- 50.Walczak-Jedrzejowska R, Wolski JK, Slowikowska-Hilczer J. 2013. The role of oxidative stress and antioxidants in male fertility. Cent. Eur. J. Urol. 66, 60-67. ( 10.5173/ceju.2013.01.art19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur R, Shropshire D, Leigh B, Bordenstein S. 2022. Nuclease proteins CifA and CifB promote spermatid DNA damage associated with symbiont-induced cytoplasmic incompatibility. bioRxiv, 2022.04.04.487029. ( 10.1101/2022.04.04.487029) [DOI]
- 52.Sassone-Corsi P. 2002. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176-2178. ( 10.1126/science.1070963) [DOI] [PubMed] [Google Scholar]
- 53.Zhong L, Belote JM. 2007. The testis-specific proteasome subunit Prosα6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development 134, 3517-3525. ( 10.1242/dev.004770) [DOI] [PubMed] [Google Scholar]
- 54.Spradling A, Fuller MT, Braun RE, Yoshida S. 2011. Germline stem cells. Cold Spring Harb. Perspect. Biol. 3, a002642. ( 10.1101/cshperspect.a002642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voog J, et al. 2014. Escargot restricts niche cell to stem cell conversion in the Drosophila testis. Cell Rep. 7, 722-734. ( 10.1016/j.celrep.2014.04.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. 2010. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37, 834-842. ( 10.1016/j.molcel.2010.01.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beckmann JF, Sharma GD, Mendez L, Chen H, Hochstrasser M. 2019. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine–histone exchange factors. eLife 8, e50026. ( 10.7554/eLife.50026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanippayoor RL, Alpern JH, Moehring AJ. 2013. Protamines and spermatogenesis in Drosophila and Homo sapiens: a comparative analysis. Spermatogenesis 3, e24376. ( 10.4161/spmg.24376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura S, Loppin B. 2016. The Drosophila chromosomal protein Mst77F is processed to generate an essential component of mature sperm chromatin. Open Biol. 6, 160207. ( 10.1098/rsob.160207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tirmarche S, Kimura S, Sapey-Triomphe L, Sullivan W, Landmann F, Loppin B. 2014. Drosophila protamine-like Mst35Ba and Mst35Bb are required for proper sperm nuclear morphology but are dispensable for male fertility. G3 4, 2241-2245. ( 10.1534/g3.114.012724) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dou W, Sun B, Miao Y, Huang D, Xiao J. 2023. Data from: Single-cell transcriptome sequencing reveals Wolbachia-mediated modification in early stages of Drosophila spermatogenesis. Dryad Digital Repository. ( 10.5061/dryad.stqjq2c5w) [DOI]
- 62.Dou W, Sun B, Miao Y, Huang D, Xiao J. 2023. Single-cell transcriptome sequencing reveals Wolbachia-mediated modification in early stages of Drosophila spermatogenesis. Figshare. ( 10.6084/m9.figshare.c.6350567) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession number for the scRNA sequencing data reported in this paper is SRA: SAMN24018191, SAMN24018192. These data have been deposited in the Sequence Read Archive under BioProject: PRJNA788731. Data link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA788731?reviewer=n7svq5k71ohhepvg14k41k4hhs. 16S sequencing data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.stqjq2c5w [61]. R scripts used in this manuscript are available in electronic supplementary material, software file [62].