Abstract
Human T-lymphotropic virus type I (HTLV-I) infects CD4+ T-cells resulting in a latent, life-long infection in patients. Crosstalk between oncogenic viral factors results in the transformation of the host cell into an aggressive cancer, adult T-cell leukemia/lymphoma (ATL). ATL has a poor prognosis with no currently available effective treatments, urging the development of novel therapeutic strategies. Recent evidence exploring those mechanisms contributing to ATL highlights the viral anti-sense gene HTLV-I bZIP factor (HBZ) as a tumor driver and a potential therapeutic target. In this work, a series of zinc-finger protein (ZFP) repressors were designed to target within the HTLV-I promoter that drives HBZ expression at highly conserved sites covering a wide range of HTLV-I genotypes. ZFPs were identified that potently suppressed HBZ expression and resulted in a significant reduction in the proliferation and viability of a patient-derived ATL cell line with the induction of cell cycle arrest and apoptosis. These data encourage the development of this novel ZFP strategy as a targeted modality to inhibit the molecular driver of ATL, a possible next-generation therapeutic for aggressive HTLV-I associated malignancies.
Graphical Abstract
Graphical Abstract.
A targeted zinc-finger protein (ZFP) repressor to the 3′-LTR promoter of HTLV-I suppresses the HBZ oncogene resulting in anti-proliferative effects in adult T-cell leukemia (ATL).
INTRODUCTION
Human T-lymphotropic virus type I (HTLV-I), a retrovirus, is transmitted by bodily fluids and establishes a life-long infection in patients. The virus infects primarily CD4+ T-cells in which the reverse transcribed genome integrates within the host cell to form a provirus. Viruses are predicted to cause about 15% of known cancers world-wide (1), and HTLV-I is the established etiological agent involved in the development of a group of blood-borne malignances. Through a complex interplay between viral factors over an extended incubation time, the virus has been linked to the transformation of CD4+ T-cells into a tumor state, resulting in acute T-cell leukemia/lymphoma (ATL). In its most aggressive form, acute ATL, the prognosis for the overall survival rate is ∼9 months. There remains no vaccine or treatment for HTLV-I, and, furthermore, ATL is refractory to chemotherapy and radiation therapy with no effective, FDA approved alternative cancer treatment. The C-C Motif Chemokine Receptor 4 (CCR4) is upregulated on the surface of most ATLs (2), and a monoclonal antibody, mogamulizumab, has been used in clinical trials in CCR4-positive ATL patients with limited improvement in disease outcomes (3). However, a sub-class of ATLs with gain-of-function CCR4 mutations substantially improved the antibody's treatment response (4). Nonetheless, the overall lack of effective approaches to inhibit ATL urges the development of novel therapeutic strategies.
HTLV-I has ∼9 kb genome flanked by long terminal repeats (LTRs) at the 5′ and 3′ ends that serve as promoters to drive sense and anti-sense expression, respectively. The HTLV-I transactivator protein Tax is expressed from the 5′ LTR, along with other accessory and structural genes involved in productive viral replication, and is a well-established factor in clonal expansion and oncogenic transformation (5). However, Tax is highly immunogenic resulting in cytotoxic CD8+ T-cell clearance of Tax-positive cells, and in ATL is generally lowly expressed or silent as a result of gene mutation, 5′LTR truncation, or promoter epigenetic hypermethylation (6).
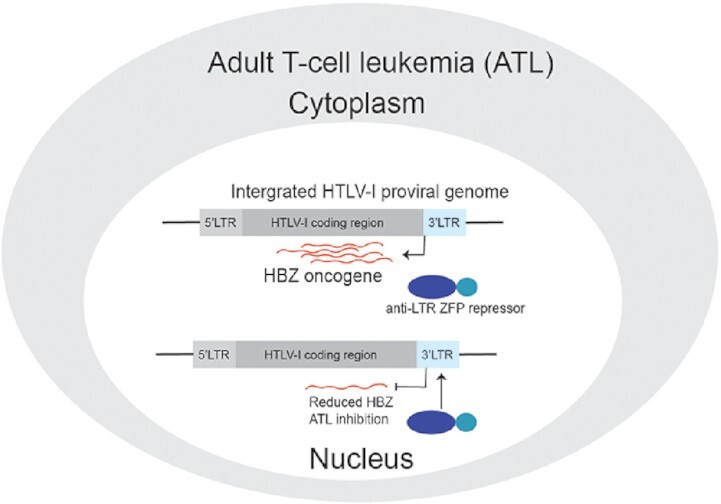
Recently, the anti-sense HTLV-I bZIP factor (HBZ) gene expressed from the 3′LTR has been realized as playing an underappreciated role in oncogenesis as it suppresses apoptosis (7), induces genetic instability (8), and causes T-cell lymphomas in HBZ transgenic mice (9). Importantly, the HBZ RNA and protein have been implicated in various proliferative and pathological roles in ATL (10), such as the up-regulation of CCR4 that augments the tumor's migration and proliferation (11). Furthermore, all primary ATL samples are positive for HBZ expression (12), and the selective inhibition of HBZ reduced proliferation in a range of HTLV-I cell lines (13,14), presenting a potential common molecular target for cancer intervention.
The cys2his2 zinc-finger proteins (ZFPs) are abundant endogenous regulatory proteins that bind specific DNA motifs to control gene expression. As a result of well-characterized rules for DNA motif recognition, custom zinc-finger arrays can be generated to target unique sequences and artificially regulate a gene of interest (15). Here, the development of a novel ZFP-based therapy is described that suppresses HBZ expression as a means to inhibit ATL proliferation. A series of HTLV-I ZFP repressors were designed to target the LTR at highly conserved sequences across various HTLV-I genotypes. Potent anti-HBZ ZFP inhibitors were identified which were further modified to improve anti-HBZ activity, and then tested in a patient-derived ATL cell line, resulting in the significant reduction of proliferation markers. This study demonstrates the utility of a novel anti-HTLV-I molecular approach to inhibit ATL.
MATERIALS AND METHODS
Vectors and cell culture
For details on vector generation, see Supplemental Materials and Methods. The HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, MA, USA). The TL-Om1, Jurkat cells or Jurkat-LTR-HBZ-IRES-GFP-Puro cells were maintained in RPMI media supplemented with 10% fetal bovine serum. The TL-Om1 cells were kindly provided by Prof. Kazuo Sugamura (16). All cell lines were cultured at 37°C and 5% CO2.
To generate the Jurkat-LTR-HBZ-IRES-GFP-Puro cell line, the LTR-HBZ-IRES-GFP-puro vector (Supplemental Materials and Methods) was linearized, purified, and 1 μg of DNA was electroporated using the Neon® transfection system into a Jurkat cell line using the electroporation conditions below. The media was then supplemented with 1.5 μg/ml puromycin (Gibco, Thermo Fisher Scientific, MA, USA).
In vitro mRNA synthesis and electroporation
The ZFP templates were linearized by digestion with XbaI and purified with the Zymo DNA Clean & Concentrator-25 kit (Zymo Research, CA, USA) and 1 μg of template was used for mRNA production with the T7 mScript™ Standard mRNA Production System according to instructions (Cellscript, WI, USA). The integrity and molecular weight of the mRNA was confirmed using PAGE loaded on to 6% Novex™ TBE-Urea Gels (Thermo fisher scientific, MA, USA), and visualised with ethidium bromide staining.
For the proliferation assays, a total of 5 × 104 TL-Om1 or Jurkat cells were electroporated with 2 or 4 μg of mRNA using the 10 μl Neon® transfection system. The electroporation conditions were as follows: ATL55T(+) and TL-Om1 cells: 1325 V, 10 ms, 3 pulse; Jurkat cells: 1450 V, 10 ms, 3 pulse. For the experiments using DNA vectors, 1 μg of expression vector was electroporated into 2×105 TL-Om1 cells with the same described conditions. For the qPCR, western blot, and apoptosis assays, 1 × 106 TL-Om1 cells were electroporated with the described amount of mRNA. For the LTR-GFP knockdown assays, 1 × 106 Jurkat-LTR-HBZ-IRES-GFP-Puro cells were electroporated with the described amount of mRNA. For the cell cycle arrest assays, 2 × 106 TL-Om1 cells were electroporated with 4 μg of mRNA. The electroporated cells were added to 1 ml of pre-warmed complete media in a 48-well plate and processed for further analysis at the described timepoints.
Transfections and luciferase assays
For the reporter assays, HEK293 cells were seeded at 1.2 × 105 cells per well, and 24 h later were transfected using Lipofectamine 3000® (Thermo fisher scientific, MA, USA) with 250 ng of HBZ luciferase reporter vector (Rluc-HTLV-I-LTR-Fluc or Rluc(splice)-HTLV-I-LTR-Fluc) and 250 ng of the ZFP expression vector. At 48 h post-transfection, the levels of Rluc and Fluc were assessed using a Dual-luciferase® Reporter Assay and activity detected on the Glomax® Explorer system (Promega, WI, USA). For the detection of HBZ RNA and protein, transfections were performed with the pcDNA-LTR-HBZ-3xFLAG vector as described above. At 48 h post-transfection, the samples were processed for either the RT-qPCR or western blot assays as described below.
RT-qPCR assay
After treatment, at the specific time points, RNA was extracted from the HEK293 or TL-Om1 cells using the Promega Maxwell™ RSC simplyRNA Tissue Kit (Promega, WI, USA). One-microgram of HEK293 RNA or 200 ng of TL-Om1 or ATL55T(+) RNA was reverse transcribed using the QuantiTect® Reverse Transcription Kit (Qiagen, Hilden, Germany), and 4 μl of RT-template was amplified in a LightCycler® 96 (Roche, Basel, Switzerland) using the KAPA Sybr® Fast qPCR Master Mix (Sigma-aldrich, MO, USA) with the following conditions: initial denaturation: 95°C for 3 min, denaturation 95°C for 5 s, annealing/extension at 60°C for 20 s. The data was analysed with the LightCycler® 96 software (V1.1.0.1320). The primers used to detect the various expressed and endogenous targets are described in Supplementary Table S1.
Western blots
After treatment, the cells were lysed in M-PER™ Mammalian Protein Extraction Reagent supplemented with Halt™ Protease Inhibitor Cocktail and the protein concentration determined by Pierce™ BCA Protein Assay Kit according to manufacturer protocols (Thermo Fisher Scientific, MA, USA). Equal amounts of protein from each sample was loaded onto a 4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, CA, USA) and transferred using the Trans-Blot® Turbo™ Transfer System with Trans-Blot® Turbo™ Mini Nitrocellulose Transfer Packs (Bio-Rad, CA, USA). The membrane was blocked with 3% BSA TBS-T and subsequently probed with the following antibodies: α-FLAG mouse mAb Anti-Flag® M2 (Cat. No. F1804; Milliporesigma, CA, USA), α-myc mouse mAb 9B11 (Cat. No. 2276; Cell Signalling Technology, MA, USA), or α-alpha tubulin rabbit polyclonal (Cat. No. 4074; Abcam, Cambridge, United Kingdom). Secondary antibodies used were the HRP-conjugated α-Mouse IgG goat antibody (Cat. No. 1705047; Bio-Rad, CA, USA) or Immun-Star™ Goat Anti-Rabbit (GAR)-HRP Conjugate (Cat. No. 170546; Bio-Rad, CA, USA), and exposed using a Pierce™ SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, MA, USA). The Bio-Rad Chemidoc™ Touch Gel-Imaging System was used to detect the signal and analysed using the Bio-rad Image Lab™ Software V6.1.0. All antibodies were diluted in blocking buffer.
Cell proliferation assays
After treatment of the TL-Om1 cells or Jurkat cells, an alamarBlue™ Cell Viability assay was performed at the designated time points (Thermo Fisher Scientific, MA, USA) and fluorescent signal measured on the Glomax® Explorer system (Promega, WI, USA). To measure cell viability and counts, trypan blue staining was performed and the cells assessed on the Countess® II Automated Cell Counter (Thermo Fisher Scientific, MA, USA).
Cell cycle assay
At 24 h post-treatment, the TL-Om1 cells were collected, washed twice with PBS, and the fixed with ice-cold 70% ethanol for 30 min at 4°C. The cells were pelleted by centrifugation at 850 g for 5 min, washed twice with PBS, and resuspended in FxCycle™ PI/RNase Staining Solution (Thermo Fisher Scientific, MA, USA). Single cells were then counted to 10000 events on a BD Accuri™ C6 and cell cycle phase analysed using the FlowJo vX5.0 software.
Apoptosis assays
To assess apoptosis, Annexin V and propidium iodide (PI) staining was performed. One-hundred thousand TL-Om1 cells were electroporated with the described amount of ZFP mRNA and the cells were harvested at 24 or 48 h. The cells were washed twice with ice-cold PBS, the pellet resuspend in 100 μl of 1× Annexin V Binding Buffer (Cat. No. 51-66121E; BD Biosciences, NJ, USA), and then 1 μl of anti-Annexin V-FITC (Cat. No. 556419; BD Biosciences, NJ, USA) and 2 μl of PI stain was added (Cat. No. P3566; Thermo Fisher Scientific, MA, USA) and incubated for 15 min in the dark at RT. Four-hundred microliters of 1x Annexin V Binding Buffer was added and 10 000 events were assessed on a BD Accuri™ C6 flow cytometer and analysed using the FlowJo vX5.0 software
To assess Caspase 3/7 activity, the TL-Om1 cells electroporated with mRNA as described above were assessed using the Caspase-Glo® 3/7 Assay System according to manufacturer instructions (Promega, WI, USA) and the signal detected on the Glomax® Explorer system (Promega, WI, USA).
FACS analysis of CCR4 surface expression
For the detection of the CCR4 receptor, TL-Om1 cells after electroporation were centrifuged at 1000 rpm for 5 min and resuspened in 45 μl of PBS with 1% bovine serum albumin (BSA) and incubated with 5 μl of a mouse PE anti-human CD194 L291H4 (Cat. No. 359411; Biolegend, CA, USA) for 30 min at RT in the dark. Five-hundred microliters of PBS with 1% BSA was added, the cells washed, and resuspended in 100 μl of PBS with 1% BSA. Single cells were counted to a total of 10000 events using the BD Accuri™ C6 and analysed on the FlowJo vX5.0 software.
ATAC-seq and analysis
ATAC-seq analysis was performed by the City of Hope integrative genomic core. A previously published OMNI ATAC-Seq protocol (17) was used for cell lysis, tagmentation, and DNA purification. The Tn5 treated DNA was amplified with 10 cycles of PCR in 50 μl reaction volumes. 1.8× AmpurXP beads purification was used for the PCR product clean-up. The libraries were validated with Agilent Bioanalyzer DNA High Sensitivity Kit, and quantified with qPCR. ATAC-seq libraries were sequenced on Illumina NovaSeq6000 with S4 Reagent v1.5 kit (Illumina, Cat 20028312) at Tgen with the sequencing length of 2 × 101. Real-time analysis (RTA) 3.4.4 software was used to process the image analysis. Raw sequencing reads were filtered using the fastp (https://github.com/OpenGene/fastp) (18) and aligned against a reference genome with HTLV sequence in chromosome 1 into the hg38 genome using HISAT2 V2.1.0 (19) aligner with its very-sensitive default parameters. Furthermore, aligned reads with a mapping quality less than 20 along with PCR duplicates were filtered out using samtools v1.6 (20). Detection of open chromatin areas was performed with the MACS2 v2.2.5 peak calling tool using the paired-end alignment information setup (–BAMPE parameter), after which the peaks detected within the promoter regions of protein coding genes defined as 3 kb upstream from the Transcription Start Site (TSS) were selected for analysis. The peaks are annotated using ChIPseeker (https://bioconductor.org/packages/release/bioc/html/ChIPseeker.html) and UCSC genome hg38 with default settings. The pathway enrichments were done using ReactomePA package (https://bioconductor.org/packages/release/bioc/html/ReactomePA.html), including three canonical pathway databases, KEGG (https://www.genome.jp/kegg/), Reactome (https://reactome.org/), and Biocarta (https://maayanlab.cloud/Harmonizome/resource/Biocarta). The node sizes represent the number of genes overlapped with the pathway genes while the heatmap represent the statistical significance. The peaks are reannotated with narrower genomic regions (tssRegion = c(-1000, 1000)). The R/Bioconductor package csaw (21) was used to detect differential accessibility among groups.
Statistical analysis
Graphing and statistical analyses was performed using GraphPad Prism version 8 (V8.1.2).
RESULTS
Screening of potent ZFP repressors of the HTLV-I LTR promoter
The 3′ LTR of the HTLV-I drives the expression of the anti-sense HBZ RNA and protein, implicated in ATL proliferation and pathology (Figure 1). Using the ZF Tools Ver 3.0 software (22), a series of nine ZFPs were generated to target the LTR of HTLV-I, each recognizing a unique 18 nucleotide DNA motif (Figure 1 and Supplementary Table S1 and S2). The ZFP coding sequence was inserted into a cytomegalovirus (CMV) expression vector and fused to a nuclear localization signal (NLS) and well-known krüppel-associated box (KRAB) repressor domain, ZFP10/KOX1 (23) (Supplementary Figure S1A). To assess if the ZFPs affected HTLV-I promoter expression of the HBZ transcript, the ZFPs were co-transfected with a bi-directional expression vector containing the HTLV-LTR driving Firefly (Fluc) and Renilla (Rluc) luciferase in the sense and anti-sense direction, respectively (Figure 2A). The HBZ intron was maintained so that the 5′ HBZ sequence located within the LTR spliced to Rluc, which provides for luciferase activity to serve as an indicator of spliced HBZ transcript expression. At 48 hrs post-transfection the ZFP3-KRAB and ZFP5-KRAB demonstrated a strong reduction in Rluc levels (>99%) compared to a control ZFP known to target the LTR of human immunodeficiency virus (ZFP-HIV-KRAB) (Figure 2A) (24). Furthermore, the ZFP5-KRAB was able to potently inhibit sense Fluc activity and ZFP3-KRAB demonstrated ∼50% inhibition. To assess if the ZFP affected basal LTR promoter activity, the ZFP expression vectors were transfected into HEK293 cells with a bi-directional expression vector without the spliced intron and, likewise, ZFP3-KRAB and ZFP5-KRAB showed a comparable level of luciferase suppression to their activity against the spliced vector, suggesting the ZFPs functionally augmented promoter activity to affect HBZ reporter expression (Supplementary Figure S1B).
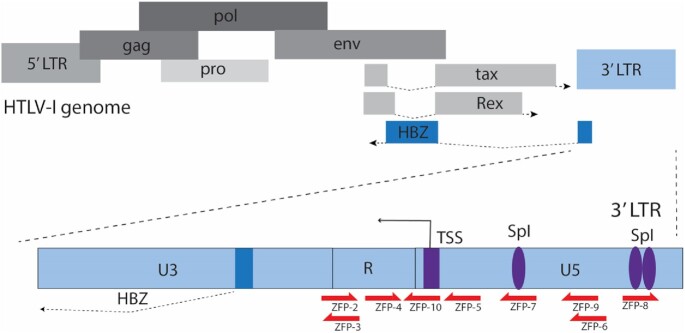
Figure 1.
Schematic of the HTLV-I genome and ZFP target sites. A schematic depicting the 5′ LTR and 3′ LTRs flanking the ∼9 kb integrated HTLV-I genome with the 3′ LTR (light blue) driving the expression of the anti-sense HBZ gene. The representative target sites of a series of ZFP within the LTR are indicated (red arrows, ZFP2-10). Transcription factor SpI binding sites and the transcription start site (TSS) in the 3′ LTR are shown in purple. The HBZ coding sequence is highlighted in dark blue.
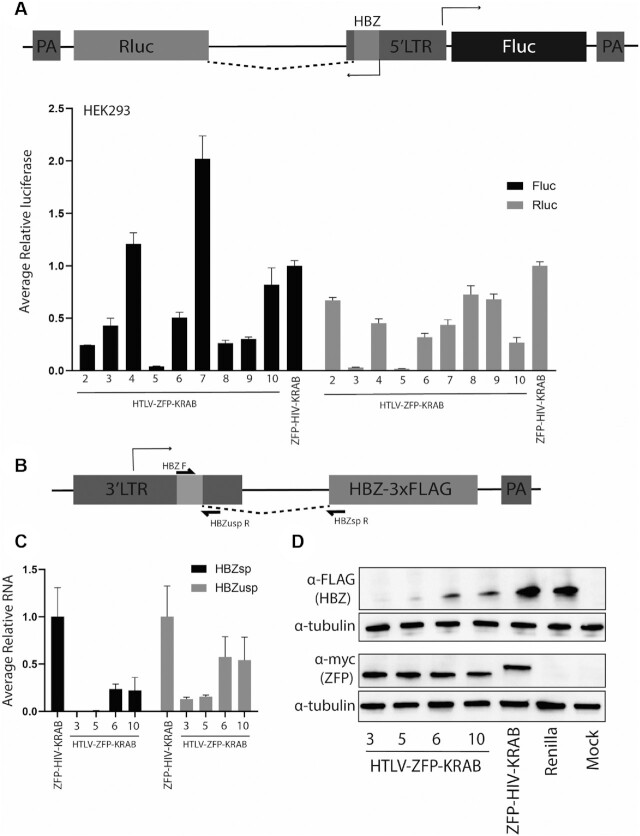
Figure 2.
Screening of ZFP repressors that inhibit HTLV-I LTR expression. (A) HEK293 cells were transfected with a vector that contains a HTLV-I LTR bidirectionally driving the expression Rluc (anti-sense) and Fluc (sense) luciferase. A mutated Rluc translational start ensures that expression of Rluc only occurs if the 5′ HBZ sequence within the LTR is spliced onto the reporter. A series of HTLV-I ZFP-KRAB repressors (2–10) were transfected with the reporter vector and 48 hrs post-transfection the levels of luciferase were determined. (B) Schematic of the HBZ exogenous vector. (C) HEK293 cells were transfected with a vector containing the HTLV-I 3′-LTR driving the expression of the HBZ-3xFLAG with the ZFP vectors, and 48 hrs post-transfection the levels of HBZ RNA were assessed. Both spliced (HBZsp) and nascent HBZ RNA (HBZusp) was detected. For (A) and (C), error bars represent standard deviation from samples treated in triplicate from two independent experiments. The levels of luciferase or HBZ RNA was made relative to a ZFP-HIV-KRAB control, set a 100%. (D) HEK293 cells were transfected as described in (C) and the HBZ-3xFLAG and ZFPs were detected through their Flag and myc tags, respectively. A Rluc expression vector or untreated cells (mock) were included as ZFP and HBZ detection controls, respectively. Alpha-tubulin was detected as a loading control.
To assess if the ZFP repressors reduced HBZ RNA and protein expression, an exogenous vector containing the 3′LTR driving the expression of the HBZ transcript was generated (LTR-HBZ) (Figure 2B) (25). The four top ZFP-KRAB repressors (ZFP3,5,6 and 10) were transfected into HEK293 cells with the LTR-HBZ vector, and the expression of the spliced and nascent HBZ RNAs and ZFP mRNAs were readily detectable by RT-qPCR (Supplementary Figure S1C and D, respectively) as well as the FLAG-tagged HBZ or myc-tagged ZFP proteins (Figure 2D). When compared to the ZFP-HIV-KRAB control, potent suppression of the spliced HBZ RNA was observed with ZFP3- and ZFP5-KRAB (>99%) (Figure 2C) and HBZ protein (Figure 2D), which corroborated the luciferase reporter data (Figure 2A). The ZFP5-KRAB had no significant effect on the levels of HBZ from a CMV-HBZ vector (Supplementary Figure S2A). However, upon electroporation into a Jurkat cell line, the ZFP3-KRAB had a non-specific restrictive effect on growth (Supplementary Figure S2B) and, as a result, the ZFP5-KRAB was selected as the lead candidate.
Modified ZFP repressors inhibits LTR activity of all circulating genotypes
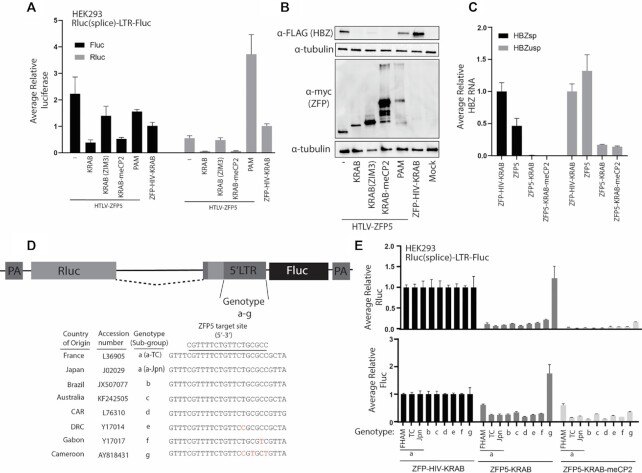
To further enhance repressor activity of the ZFPs, new versions of the ZFP5 with alternative repressor domains were generated. These domains have been described to have more potent activity which included a novel KRAB repressor ZIM3 (26), the current KOX1 KRAB fused to an additional methyl CpG binding protein 2 (meCP2) (27), or replacing the KRAB with a recently described fusion repressor from this group, PAM (28) (Supplementary Figure S3A). The ZFP5 variants were transfected into HEK293 cells with the HBZ spliced Rluc reporter or LTR-HBZ vectors, and the ZFP5-KRAB-meCP2 showed comparable suppressive activity to the ZFP5-KRAB when detecting HBZ spliced Rluc levels (Figure 3A), protein (Figure 3B), and RNA levels (Figure 3C and Supplementary Figure S3B). A ZFP5 without a KRAB domain was also tested to determine if steric hinderance at the promoter was causing HBZ suppression. The ZFP5 alone slightly suppressed promoter activity by ∼50%, but potent suppression required the KRAB domain, demonstrating a domain-specific effect (Figure 3A–C). THE ZFP5-KRAB (ZIM3) suppressed activity by ∼50%, suggesting the ZIM3 KRAB was not contributing to suppression, and the ZFP5-PAM was ineffective, likely from poor expression of the fusion protein (Figure 3A and B). Overall, in these assays, the ZFP5-KRAB-meCP2 demonstrated comparable activity to the ZFP5-KRAB and was therefore selected for further characterization.
Figure 3.
Anti-HTLV-I ZFP repressors inhibit the LTRs from multiple HTLV-I genotypes. (A) HEK293 were transfected with a vector containing the HTLV-1 LTR bi-directional reporter to measure Fluc (sense) or the HBZ(spliced)-Rluc (anti-sense) activity with the modified ZFP5 vectors, and at 48 h post-transfection the levels of luciferase activity were assessed. The modified vectors were generated by fusing the ZFP5 to a KRAB(ZIM3), KRAB-meCP2 or PAM. A ZFP5 without a KRAB domain was also included (–). The levels of HBZ (B) protein or (C) RNA were determined after transfecting HEK293 cells with an LTR-HBZ and the modified ZFP5 vectors. (D) A schematic of the HTLV-I LTR bidirectional vector with the LTR upstream of the HBZ translation start replaced with sequences from different HTLV-I genotypes (a-g). The country of viral origin, accession numbers, genotypes and ZFP5 target site sequences are indicated. Mismatches are highlighted in red. (E) HEK293 cells were transfected with an LTR(a-g) spliced reporter vector with the ZFP5-KRAB and ZFP5-KRAB-meCP2 vectors, and 48 h post-transfection the levels of Rluc and Fluc luciferase was determined. For (A), (C) and (E), the ZFP5 vectors were made relative to a control ZFP-HIV-KRAB, which was set a 100%. Error bars represent standard deviation from samples treated in triplicate. For (B), the HBZ and ZFPs were detected through a FLAG tag and myc tag, respectively. Untreated cells (mock) were included as ZFP and HBZ detection controls and α-tubulin as a loading control.
The ZFPs were designed to target conserved sites within the LTR to ensure activity against a wide-range of HTLV-I genotypes. The reference LTR sequence of each global circulating genotype (a–g) was inserted upstream of the HBZ start site in the spliced Rluc luciferase reporter vector (Figure 3D). The ZFP5 target site is fully conserved within genotypes a–d, single mismatches in genotypes e and f, and a triple mismatch in genotype g. To test the repressive activity, the ZFP5 vectors were transfected into HEK293 cells with the spliced Rluc luciferase reporter vectors of each genotype, and the ZFP5-KRAB successfully knocked down each genotype, except for the triple mismatch genotype g, while the ZFP5-KRAB-meCP2 inhibited luciferase expression from all genotypes (Figure 3E). These data suggest that the ZFPs should affect HBZ expression in a wide range of circulating HTLV-I genotypes and the ZFP5-KRAB-meCP2 may have enhanced functionality compared to unmodified ZFP5.
Transient HBZ suppression by a ZFP repressor significantly inhibits ATL cell line proliferation
To determine if the ZFP repressors inhibited the proliferation of ATL, the ZFP vectors were tested in a patient-derived TL-Om1 cells. These cells have been well-characterized to have a single HTLV-I proviral integrant (16), and positive for HBZ but negative for Tax expression (29) (Supplementary Figure S4A) as a result of hypermethylation of the 5′LTR (30). As all primary ATLs express HBZ (12), these features make the TL-Om1 cells a representative model for studying the anti-proliferative effects through HBZ repression.
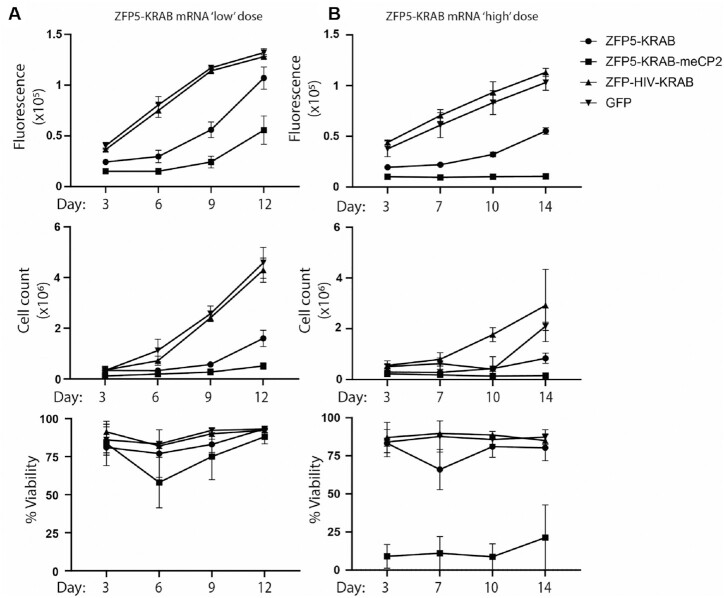
The TL-Om1 cells were generally negatively affected by the introduction of DNA vectors into the cells; however, transient expression of the ZFPs would be preferable for therapeutic development as mRNA is emerging as the nucleic acid of choice for such applications. Accordingly, the ZFP5-KRAB was generated as mRNA and electroporated in the TL-Om1 cells, which was efficiently delivered and well-tolerated. In the cells electroporated with ZFP5-KRAB mRNA at a ‘low’ dose, a clear reduction in TL- Om1 proliferation was observed compared to controls, although no effect on cell viability was observed (Figure 4A and Supplementary Figure S4B). The ZFP5-KRAB-meCP2 mRNA showed higher inhibition of proliferation compared to the ZFP5-KRAB. Although there was no significant effect on viability between the treated groups, there were fluctuations in the viability in the ZFP5-KRAB-meCP2 treated cells at day 6 at the ‘low’ dose. Increasing the amount ZFP5-KRAB-meCP2 mRNA to a ‘high’ dose resulted in a potent anti-proliferative effect and marked reduction in viability compared to the ZFP5-KRAB (Figure 4B and Supplementary Figure S4B). Of interest, the ZFP5-KRAB showed similar fluctuations in viability at day 7 at the ‘high’ dose, further suggesting a dosing effect.
Figure 4.
Anti-proliferative effects of the anti-HBZ ZFP repressors. TL-Om1 cells were electroporated with (A) 2 μg ‘low’ dose or (B) 4 μg ‘high’ dose of mRNA expressing the ZFP5-KRAB or ZFP5-KRAB-meCP2 and outgrowth was assessed through proliferation (top panel), viability (middle panel), or cell count (bottom panel). The ZFP-HIV-KRAB or GFP mRNAs were included as negative controls. Error bars represent standard deviation from samples treated in triplicate.
Lastly, to confirm these anti-proliferative effects were specific to a HTLV-I leukemia cell line, these conditions were repeated in Jurkats cells, a non-HTLV-I leukemia cell line. The ZFP5 repressors had no effect on the proliferation, cell count, or viability of these cells (Supplementary Figure 5A and B). Furthermore, to assess if the ZFP could affected the LTRs from other retroviral vectors, the HTLV-targeted ZFPs were transfected into a reporter cell line with the HIV-1 LTR driving the expression of GFP and no effect on reporter levels was observed (Supplementary Figure S5C). Furthermore, suppression of the HTLV-I LTR in another ATL cell lines, ATL55T(+), likewise resulted in a reduction in HBZ RNA levels and proliferation (Supplementary Figure S5D-F). These data demonstrate that the anti-proliferative effects from the ZFP5 repressors were specific to HTLV-I.
The ZFP repressors affected HBZ levels and downstream induced CCR4
Next, the effect the ZFP5 repressors had on HBZ expression in TL-Om1 cells was assessed. The ZFP5-KRAB and ZFP5-KRAB-meCP2 mRNA electroporated cells showed a comparable reduction in HBZ RNA levels (Figure 5A and Supplementary Figure S7A). As expected, the detected ZFP5 repressor mRNA and protein rapidly reduced over a 72 or 48 h period, respectively, (Supplementary Figure 6A–C), and the declined in ZFP mRNA was mirrored by a concordant increase in HBZ RNA levels (Supplementary Figure S6C), confirming the ZFPs were affecting HBZ expression within its genomic context.
Figure 5.
Anti-HTLV-I ZFPs reduce HBZ-induced CCR4 levels. TL-Om1 cells were electroporated with 2 μg of ZFP5-KRAB or ZFP5-KRAB-meCP2 mRNA, and the levels of (A) HBZ spliced RNA, (B) CCR4 RNA, (C) or surface CCR4 receptor was assessed at 24 and 48 h post-electroporation. Cells treated with a ZFP-HIV-KRAB mRNA or untreated cells (mock) were included as negative controls. Error bars represent standard deviation from samples treated in triplicate and P-values were determined by one-way ANOVA analysis (Dunnett's post-test) when compared to the ZFP-HIV-control (*P < 0.05, **P < 0.01 ***P < 0.001, ****P < 0.0001).
The HBZ RNA and protein affects a number of host genes in ATL and both upregulate CCR4 expression (11). To confirm if the ZFPs were affecting HBZ-induced host proteins, the CCR4 mRNA levels were assessed, and a significant reduction of about 50% was observed at 24 h but only in the ZFP5-KRAB-meCP2 treated cells (Figure 5B). Even though CCR4 mRNA levels were re-established at 48 h, the amount of surface CCR4 detected by flow cytometry was reduced at 24 and 48 h (Figure 5C). Increasing the amount of ZFP mRNA to the ‘high’ dose did not improve the reduction of HBZ or CCR4 levels (Supplementary Figure S7A–C).
The ZFP5-KRAB and ZFP5-KRAB-meCP2 showed comparable levels of HBZ suppression in the TL-Om1 cells (Figure 5A), but only ZFP5-KRAB-meCP2 was able to affect CCR4 levels (Figure 5B and C), suggesting that the ZFP5-KRAB-meCP2 was a more potent effector. Considering this observation, we surmised that the anti-proliferative effects were masking the extent of HBZ suppression. To assess the anti-HBZ effects of the ZFPs in the absence of proliferative factors, the ZFP mRNAs were electroporated into a Jurkat cell line engineered with an LTR-HBZ in-frame with a GFP reporter (Supplementary Figure S8A). In the absence of confounding anti-proliferative effects, the ZFP5-KRAB-meCP2 had a more rapid decline in GFP signal than the ZFP5-KRAB, demonstrating the ZFP5-KRAB-meCP2 was a more potent repressor (Supplementary Figure S8B). Overall, these data demonstrate that the ZFPs reduced HBZ mRNAs levels in TL-Om1 cells, and the ZFP5-KRAB-meCP2 was a more potent effector that can affect a downstream HBZ-induced gene.
The ZFP repressors induce cell cycle arrest and activate apoptotic pathways
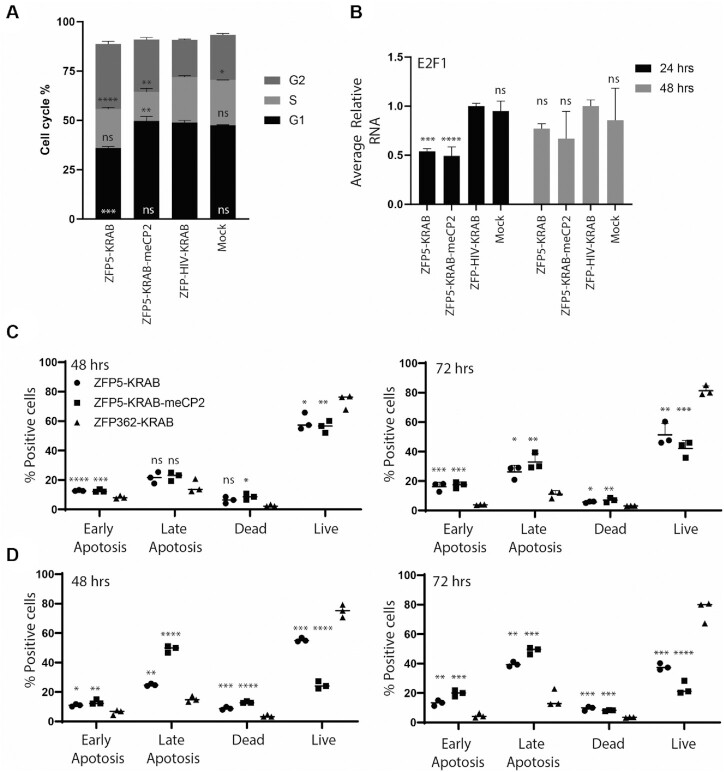
To better understand the mechanisms behind the anti-proliferative effects, a cell cycle arrest assay was performed. At 24 h post-electroporation, the ZFP5-KRAB was able to induce an increase in G2/M phase and a reduction in G1, suggesting the inhibition of HBZ was causing G2/M arrest in the TL-Om1 cells (Figure 6A). Notably, the ZFP5-KRAB-meCP2 resulted in a different arrest profile, resulting in a likewise increase in G2 phase, although to a lesser extent than the ZFP5-KRAB, and a reduction of cells in S phase. The HBZ RNA is known to upregulate the transcription factor E2F1, which is a well-known driver of cell cycle progression (31). The levels of E2F1 mRNA were reduced at 24 hrs in the ZFP treated TL-Om1 cells (Figure 6B), further demonstrating the ZFPs were affecting cell cycle factors induced by HBZ.
Figure 6.
Anti-HBZ ZFPs cause cell cycle arrest and apoptosis. (A) TL-Om1 cells were electroporated with 2 μg of mRNA expressing the ZFP5-KRAB or ZFP5-KRAB-meCP2 and the percentage of cell cycle phase was assessed at 24 hrs post-electroporation. (B) The levels of E2F1 mRNA were assessed at 24 h and 48 hrs post-electroporation. Cells treated with a ZFP-HIV-KRAB mRNA or untreated (mock) were included as negative controls. For (C), the samples were made relative to the ZFP-HIV-KRAB set at 100%. To assess the induction of apoptosis, TL-Om1 cells were electroporated with a (C) 2 μg ‘low’ dose or (D) 4 μg ‘high’ dose of mRNA and Annexin V and PI detected at 48 and 72 h post-electroporation. ZFP-HIV-KRAB was used as a negative control. For (A) and (B), error bars represent standard deviation from samples treated in triplicate. For (C) and (D), the line represents the mean from samples treated in triplicate. The P-values were determined by one-way ANOVA analysis (Dunnett's post-test) when compared against ZFP-HIV-control (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
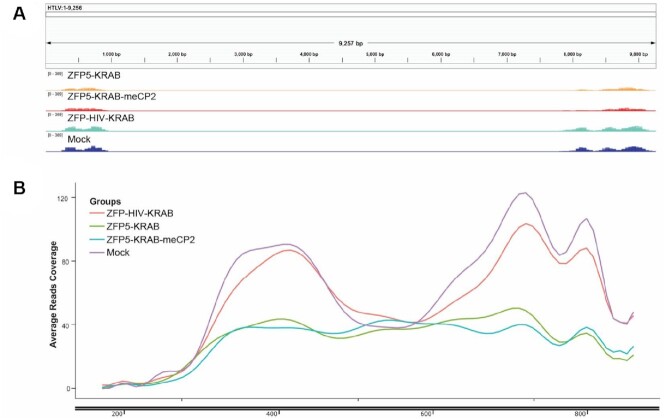
Next, induction of apoptosis by the ZFPs was assessed. When determining the activation caspase 3/7 activity, the ZFP repressors induced activity in the TL-Om1 cells to comparable levels even when using a ‘low’ or ‘high’ dose, with no effect observed with the ZFP-HIV control (Supplementary Figure S9). Annexin V/PI staining revealed that at the ‘low’ dose, both ZFP repressors induced a modest but equitable induction of late-stage apoptosis at 48 and 72 h (Figure 6C). However, at the ‘high’ dose, TL-Om1 cells receiving the ZFP-KRAB-meCP2, strongly induced late-stage apoptosis at 48 h compared to the ZFP5-KRAB (Figure 6D), which was comparable at the 72-h time point. Collectively, these data demonstrate the anti-HBZ ZFPs induced anti-proliferative effects through cell cycle arrest and the induction of apoptosis in a ATL cell line. Lastly, to demonstrate a mechanism of chromatin remodelling at the LTR by the anti-HTLV ZFPs, treated TL-Om1 cells were subjected to ATAC-seq (32). Briefly, chromatin is exposed to Tn5 transposase and euchromatin regions at transcriptional active genomic sites are more accessible to transposase tagmentation. Treatment with the ZFP5-KRAB or ZFP5-KRAB-meCP2 resulted in reduced reads across the HTLV-I genome (Figure 7A) and a reduction in nucleosome-free regions in the LTR (Figure 7B). Furthermore, pathway analysis was performed for differential chromatin accessibility across TSS sites. P53 is functionally inhibited by HBZ and a top hit was genes associated with p53 transcription regulation in the ZFP treated samples, which was not observed in the ZFP-HIV-KRAB treated cells (Supplementary Figure S10), suggesting anti-HBZ ZFPs are affecting genes downstream of p53.
Figure 7.
LTR-targeted ZFP repressors reduce chromatin accessibility. TL-Om1 cells were electroporated with 4 μg of mRNA expressing the ZFP5-KRAB or ZFP5-KRAB-meCP2 and at 24 h the cells were subjected to ATAC-seq to assess chromatin accessibility. (A) IGV of the HTLV-I genome displaying accessibility. (B) Enrichment plot of nucleosome-free regions across HTLV-I’s LTR. The read counts are the average of triplicate treated cells.
DISCUSSION
Current approved treatments for ATL have limited improvements on patient survival, and ATL is considered refractory to chemotherapy and radiation therapy, promoting the development of novel therapeutics. Here we describe a novel molecular therapy against a potential gene driver of ATL, the anti-sense HBZ gene, which is functional against the LTRs of a broad range of HTLV-I genotypes. Other knockdown studies have shown that a reduction in HBZ results in reduced proliferation in the TL-Om1 cells as well as a number of in vitro HTLV-I transformed cells (MT-1, SLB-1, PBLACH) (13,14), suggesting that the anti-HBZ ZFP repressors will affect a wide range of ATL samples.
A zinc-finger nuclease that introduces mutations into the LTR through nuclease activity has been shown to reduce HTLV-I associated tumor growth in vitro and in vivo (33). However, no further characterization of the mechanism of inhibition was performed. In the knockdown studies, reduced proliferation in HTLV-I cell lines was observed (13,14), but no reduction in viability (13). The ZFP repressors showed a rapid and strong induction of late-stage apoptosis and, at the ‘high’ dose, the ZFP5-KRAB-meCP2 resulted in a stark reduction in viability (Figure 4 and 6). These different observations may reflect the potency of HBZ inhibition, where previous studies knocking down the HBZ RNA and protein levels was limited, and may be insufficient to induce cell death (13). However, some of the cell lines in these studies also expressed a functional Tax oncogene, which may affect anti-proliferative effects and additional studies will be needed to determine the threshold of ATL oncogene suppression required to induce cell death.
Still, it is unclear why ZFP5-KRAB-meCP2 reduced viability at the ‘high’ dose as caspase 3/7 activation was similar to the ZFP5-KRAB (Supplementary Figure S9), but a more potent and rapid induction of late-stage apoptosis was observed (Figure 6D). Furthermore, the ZFP5-KRAB-meCP2 caused S-phase arrest which has been linked to apoptosis, and these observations may suggest this modality is more effective at committing the TL-Om1 cells to programmed death. Furthermore, the ZFP5-KRAB-meCP2 at the ‘high’ dose was the only system that substantially reduced viability (Figure 4B), affecting an HBZ downstream factor (Figure 5B and C), suggesting a possible threshold for reversing HBZ-induced factors involved in maintaining the tumor state. The HBZ protein has proapoptotic function while the HBZ RNA has pro-survival effects (10), and this apparent threshold may support the ‘oncogenic shock’ model for this viral oncogene (34), where the reduction of the oncogene's pro-survival signals are outbalanced by the proapoptotic signals, ultimately driving the cell down a cellular death pathway. Further studies elucidating this mechanism would assist in a more rational design of anti-HBZ modalities.
An alternative explanation may be warranted. The ZFP5-KRAB-meCP2 was selected as the meCP2 component may elicit epigenetic changes at the target promoter (35), allowing for sustained, if not permanent, silencing. The ‘high’ dose ZFP5-KRAB-meCP2 may elicit a sustained suppressive effect on HBZ, resulting in cell death. Further studies should explore this possibility, and, if so, epigenetic modulators like ‘block and lock’ strategies for the silencing of the LTR of HIV (28,36), could be applied to the inhibition of HBZ as an ATL treatment approach. Regardless, whether the effect was through potency or duration, the unique observation presented here suggests that the ablation of HBZ expression may prove to be a viable means to eliminate HBZ-driven malignances.
HBZ has been implicated in a wide range of pathological features of ATL. The upregulation of CCR4 is known to enhance ATL proliferation and trafficking (11), especially migration to the skin (2). The reduction in CCR4 surface levels by the anti-HBZ ZFPs was low (Figure 6C) and is unclear if this will be enough to affect pro-migratory and proliferative effects but warrants further investigation. Furthermore, the HBZ protein is associated with bone degeneration through the RANKL/c-Fos pathway (37), and the HBZ RNA is known to augment Survivin (10), a factor involved in chemoresistance and a feature of ATL (38,39). Therefore, targeting HBZ with the ZFP repressors may be a means to modify a spectrum of ATL disease features.
HTLV-I has been associated with another disorder, HTLV-I associated myelopathy/tropical spastic paraparesis (HAM/TSP), which is a progressive, chronic neurological disorder that has been associated with HBZ and Tax expression (40). Furthermore, there are currently no therapeutics that can suppress Tax-mediated productive infection in active HTLV-I. The ZFP5 did affect 5′LTR activity in reporter assays (Figure 2A), however, we observed no significant suppression of Tax transcripts in the ATL55T(+) cells, demonstrating the repressive activity of the ZFPs is 3′ LTR specific. Still, novel ZFP repressors specifically designed to inhibit the 5′ LTR could be developed to affect Tax expression, an important factor in active infection and HAM/TSP.
Delivery of gene therapies remains a challenge within the field. Although viral vectors, such as T-cell tropic adeno-associated viral vectors (AAVs), may be an option, the sustained expression of the ZFPs in potential off-target tissues through systemic administration would not be advisable and antibody responses to the vector would preclude repeat dosing. There has been significant interest in the current development of mRNA and lipid nanoparticle (LNP) formulations because of the success of the COVID-19 LNP-based vaccine. Currently, T-cell delivery in vivo with LNPs is limited; however, there has been recent success with T-cell delivery in vivo (41), and the combination of the ZFP mRNA with an LNP formulation to target ATL cells could be a feasible approach. More innovative solutions could explore extracellular vesicles (EVs) as an emerging delivery platform. EVs are a broad group of small, membraned nano-size products derived from the cell, which are biocompatible and non-immunogenic, and are being developed as a delivery system for therapeutic cargo (42). Recent work has demonstrated that ZFP activators can be transferred to recipient cells to activate an endogenous gene (43) as well as deliver a ZFP repressor targeted to HIV’s LTR resulting in epigenetic repression of HIV after systematic administration in a humanized mouse model (28). Therefore, potential platforms compatible with systemic administration are available that could prove to be a viable, druggable, approach for clinical application of this novel modality.
In conclusion, described here is a novel ZFP repressor that can target the HTLV-I LTR and suppress the HBZ gene, resulting in the reduced proliferation of a patient derived ATL cell line. These data not only add to the growing body of evidence establishing HBZ a molecular driver and potential target in ATL, but encourages the further development of this modality to potentially treat HTLV-I associated malignancies.
DATA AVAILABILITY
The authors declare that all data of this study are available within the article and the Supplementary Information.
Supplementary Material
ACKNOWLEDGEMENTS
Thank you to Prof. Kazuo Sugamura who kindly provided us with the TL-Om1 cell line. Thank you to Dr Ye and Dr Maeda for kindly supplying the ATL55T(+) cell line. Thank you to Dr Zaia and Dr Cardoso for reviewing the manuscript and providing valuable input. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Author contributions: T.S. and K.V.M. conceived and designed the experiments; T.S., C.S. and R.R. conducted the experimentation; T.S. and K.V.M. wrote the main manuscript and prepared the tables/figures. All authors reviewed and edited the manuscript.
Contributor Information
Tristan A Scott, Center for Gene Therapy, City of Hope – Beckman Research Institute and Hematological Malignancy and Stem Cell Transplantation Institute at the City of Hope. 1500 E. Duarte Rd., Duarte, CA 91010, USA.
Citradewi Soemardy, Center for Gene Therapy, City of Hope – Beckman Research Institute and Hematological Malignancy and Stem Cell Transplantation Institute at the City of Hope. 1500 E. Duarte Rd., Duarte, CA 91010, USA.
Roslyn M Ray, Center for Gene Therapy, City of Hope – Beckman Research Institute and Hematological Malignancy and Stem Cell Transplantation Institute at the City of Hope. 1500 E. Duarte Rd., Duarte, CA 91010, USA.
Kevin V Morris, Menzies Health Institute Queensland, School of Pharmacy and Medical Science, Griffith University, Gold Coast Campus 4222, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
National Institute of Health, National Institute of Mental Health [R01 113407-01]; National Institute of Health Allergy and Infectious Disease [R56AI147684-01 to K.V.M.].
Conflict of interest statement. K.V.M. and T.A.S. have submitted a provisional patent application based on this technology (048440-836P01US).
REFERENCES
- 1. Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 2006; 118:3030–3044. [DOI] [PubMed] [Google Scholar]
- 2. Yoshie O. Expression of CCR4 in adult T-cell leukemia. Leuk. Lymphoma. 2005; 46:185–190. [DOI] [PubMed] [Google Scholar]
- 3. Adrienne A.P., Paul A.F., Olivier H., Juan C.R., Brady E.B., Juliana P., Farooq W., Tatyana F., Graham P.T., Ahmed S. et al. Mogamulizumab versus investigator's choice of chemotherapy regimen in relapsed/refractory adult T-cell leukemia/lymphoma. Haematologica. 2019; 104:993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sakamoto Y., Ishida T., Masaki A., Murase T., Yonekura K., Tashiro Y., Tokunaga M., Utsunomiya A., Ito A., Kusumoto S. et al. CCR4 mutations associated with superior outcome of adult T-cell leukemia/lymphoma under mogamulizumab treatment. Blood. 2018; 132:758–761. [DOI] [PubMed] [Google Scholar]
- 5. Giam C.Z., Jeang K.T. HTLV-1 Tax and adult T-cell leukemia. Front. Biosci. 2007; 12:1496–1507. [DOI] [PubMed] [Google Scholar]
- 6. Takeda S., Maeda M., Morikawa S., Taniguchi Y., Yasunaga J.-i., Nosaka K., Tanaka Y., Matsuoka M. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer. 2004; 109:559–567. [DOI] [PubMed] [Google Scholar]
- 7. Tanaka-Nakanishi A., Yasunaga J.-i., Takai K., Matsuoka M. HTLV-1 bZIP factor suppresses apoptosis by attenuating the function of FoxO3a and altering its localization. Cancer Res. 2014; 74:188–200. [DOI] [PubMed] [Google Scholar]
- 8. Vernin C., Thenoz M., Pinatel C., Gessain A., Gout O., Delfau-Larue M.-H., Nazaret N., Legras-Lachuer C., Wattel E., Mortreux F. HTLV-1 bZIP factor HBZ promotes cell proliferation and genetic instability by activating OncomiRs. Cancer Res. 2014; 74:6082–6093. [DOI] [PubMed] [Google Scholar]
- 9. Satou Y., Yasunaga J.-i., Zhao T., Yoshida M., Miyazato P., Takai K., Shimizu K., Ohshima K., Green P.L., Ohkura N. et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011; 7:e1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitobe Y., Yasunaga J.-i., Furuta R., Matsuoka M. HTLV-1 bZIP factor RNA and protein impart distinct functions on T-cell proliferation and survival. Cancer Res. 2015; 75:4143–4152. [DOI] [PubMed] [Google Scholar]
- 11. Sugata K., Yasunaga J.-i., Kinosada H., Mitobe Y., Furuta R., Mahgoub M., Onishi C., Nakashima K., Ohshima K., Matsuoka M. HTLV-1 viral factor HBZ induces CCR4 to promote T-cell migration and proliferation. Cancer Res. 2016; 76:5068. [DOI] [PubMed] [Google Scholar]
- 12. Kataoka K., Nagata Y., Kitanaka A., Shiraishi Y., Shimamura T., Yasunaga J.-i., Totoki Y., Chiba K., Sato-Otsubo A., Nagae G. et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 2015; 47:1304–1315. [DOI] [PubMed] [Google Scholar]
- 13. Arnold J., Zimmerman B., Li M., Lairmore M.D., Green P.L. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood. 2008; 112:3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Satou Y., Yasunaga J.-i., Yoshida M., Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Nat. Acad. Sci. USA. 2006; 103:720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Papworth M., Kolasinska P., Minczuk M. Designer zinc-finger proteins and their applications. Gene. 2006; 366:27–38. [DOI] [PubMed] [Google Scholar]
- 16. Kuramitsu M., Okuma K., Yamagishi M., Yamochi T., Firouzi S., Momose H., Mizukami T., Takizawa K., Araki K., Sugamura K. et al. Identification of TL-Om1, an adult T-Cell leukemia (ATL) cell line, as reference material for quantitative PCR for human T-lymphotropic virus 1. J. Clin. Microbiol. 2015; 53:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017; 14:959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M. et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021; 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lun A.T.L., Smyth G.K. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2015; 44:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mandell J.G., Barbas C.F. III Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006; 34:W516–W523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003; 4:231–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scott T.A., O’Meally D., Grepo N.A., Soemardy C., Lazar D.C., Zheng Y., Weinberg M.S., Planelles V., Morris K.V. Broadly active zinc finger protein-guided transcriptional activation of HIV-1. Mol. Ther.–Methods Clin. Dev. 2021; 20:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sugamura K., Fujii M., Kannagi M., Sakitani M., Takeuchi M., Hinuma Y. Cell surface phenotypes and expression of viral antigens of various human cell lines carrying human T-cell leukemia virus. Int. J. Cancer. 1984; 34:221–228. [DOI] [PubMed] [Google Scholar]
- 26. Alerasool N., Segal D., Lee H., Taipale M. an efficient KRAB domain for CRISPRi applications in human cells. Nat. Methods. 2020; 17:1093–1096. [DOI] [PubMed] [Google Scholar]
- 27. Yeo N.C., Chavez A., Lance-Byrne A., Chan Y., Menn D., Milanova D., Kuo C.-C., Guo X., Sharma S., Tung A. et al. an enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods. 2018; 15:611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shrivastava S., Ray R.M., Holguin L., Echavarria L., Grepo N., Scott T.A., Burnett J., Morris K.V. Exosome-mediated stable epigenetic repression of HIV-1. Nat. Commun. 2021; 12:5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanaka Y., Mizuguchi M., Takahashi Y., Fujii H., Tanaka R., Fukushima T., Tomoyose T., Ansari A.A., Nakamura M. Human T-cell leukemia virus type-I Tax induces the expression of CD83 on T cells. Retrovirology. 2015; 12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koiwa T., Hamano-Usami A., Ishida T., Okayama A., Yamaguchi K., Kamihira S., Watanabe T. 5'-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 2002; 76:9389–9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawatsuki A., Yasunaga J.I., Mitobe Y., Green P.L., Matsuoka M. HTLV-1 bZIP factor protein targets the Rb/E2F-1 pathway to promote proliferation and apoptosis of primary CD4(+) T cells. Oncogene. 2016; 35:4509–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan F., Powell D.R., Curtis D.J., Wong N.C. from reads to insight: a hitchhiker's guide to ATAC-seq data analysis. Genome Biol. 2020; 21:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tanaka A., Takeda S., Kariya R., Matsuda K., Urano E., Okada S., Komano J. a novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia. 2013; 27:1621–1627. [DOI] [PubMed] [Google Scholar]
- 34. Sharma S.V., Fischbach M.A., Haber D.A., Settleman J. “Oncogenic shock”: explaining oncogene addiction through differential signal attenuation. Clin. Cancer Res. 2006; 12:4392s–4395s. [DOI] [PubMed] [Google Scholar]
- 35. Fuks F., Hurd P.J., Wolf D., Nan X., Bird A.P., Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003; 278:4035–4040. [DOI] [PubMed] [Google Scholar]
- 36. Vansant G., Bruggemans A., Janssens J., Debyser Z. Block-and-lock strategies to cure HIV infection. Viruses. 2020; 12:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiang J., Rauch D.A., Huey D.D., Panfil A.R., Cheng X., Esser A.K., Su X., Harding J.C., Xu Y., Fox G.C. et al. HTLV-1 viral oncogene HBZ drives bone destruction in adult T cell leukemia. JCI Insight. 2019; 4:e128713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garg H., Suri P., Gupta J.C., Talwar G.P., Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016; 16:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. El Hajj H., Tsukasaki K., Cheminant M., Bazarbachi A., Watanabe T., Hermine O. Novel treatments of adult T cell leukemia lymphoma. Front. Microbiol. 2020; 11:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Enose-Akahata Y., Vellucci A., Jacobson S. Role of HTLV-1 tax and HBZ in the pathogenesis of HAM/TSP. Front. Microbiol. 2017; 8:2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rurik J.G., Tombácz I., Yadegari A., Fernández P.O.M., Shewale S.V., Li L., Kimura T., Soliman O.Y., Papp T.E., Tam Y.K. et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022; 375:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O’Brien K., Breyne K., Ughetto S., Laurent L.C., Breakefield X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020; 21:585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Villamizar O., Waters S.A., Scott T., Grepo N., Jaffe A., Morris K.V. Mesenchymal stem cell exosome delivered zinc finger protein activation of cystic fibrosis transmembrane conductance regulator. J. Extracell. Vesicles. 2021; 10:e12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data of this study are available within the article and the Supplementary Information.