Abstract
The tumor suppressor p53 and phosphoinositide 3-kinase (PI3K)-Akt pathway have fundamental roles in regulating cell growth and apoptosis, and they are frequently mutated in cancer. Here, we show that genotoxic stress induces nuclear Akt activation by a p53-dependent mechanism that is distinct from the canonical membrane-localized PI3K-Akt pathway. Upon genotoxic stress, a nuclear PI3K binds to p53 in the non-membranous nucleoplasm to generate a p53-PI3,4,5P3 complex, which recruits Akt and PDK1 and mTORC2 that are required to activate Akt, and phosphorylates FOXOs, thereby inhibiting DNA damage-induced apoptosis. Wild-type p53 activates nuclear Akt in an on/off fashion upon stress, whereas mutant p53 dose dependently stimulates high basal Akt activity. The p53-PI3,4,5P3 complex is dephosphorylated to p53-PI4,5P2 by the phosphatase and tensin homolog (PTEN) to inhibit Akt activation. The nuclear p53-phosphoinositide signalosome is distinct from the canonical membrane-localized pathway and insensitive to PI3K inhibitors currently in the clinic, underscoring its therapeutic relevance.
Keywords: p53; Akt; PI3K; phosphoinositide; PI4,5P2; PI3,4,5P3; nucleus
In brief
p53 assembles a PI3K-PI3,4,5P3-Akt pathway that regulates nuclear Akt activation independent of the canonical pathway on membranes.
INTRODUCTION
Receptor activation of phosphoinositide (PI) 3-kinase (PI3K) generates PI3,4,5P3 that binds to and activates the serine/threonine protein kinases Akt at membranes1, 2 and is a key regulator of cell growth, survival, and metabolism. This pathway is often hyperactivated in cancer3–5. In the canonical membrane-localized PI3K-Akt pathway, PI3,4,5P3 is required for recruiting PDK1, mTORC2, and Akt to membranes via their respective pleckstrin homology (PH) domains5, 6, enabling active PDK1 to phosphorylate T308 on Akt7. Subsequent phosphorylation of Akt primarily by mTORC2 on S473 stimulates full Akt activity with continuous PI3,4,5P3 binding5, 8, 9. Active Akt is present in the cytoplasm2 and nucleus10. Reports suggest translocation of active Akt from the cytoplasm to the nucleus11–13 and de novo nuclear activation of Akt14–16. However, the underlying mechanisms of nuclear Akt activation and the functional role of Akt in the nucleus are poorly understood.
The tumor suppressor p53 maintains genome integrity in response to cellular stress and is the most mutated gene in cancer17, 18. We have previously demonstrated that p53 interacts with the type I phosphatidylinositol phosphate (PIP) kinase PIPKIα (encoded by PIP5K1A), generating PI4,5P2 that forms a complex with p53 by binding to the C-terminus of p53. The p53-PI4,5P2 complex then recruits small heat shock proteins (sHSPs), which bind and stabilize p5319, but the functions of the p53-PI4,5P2 complex are unknown. Intriguingly, the nuclear PI3K inositol polyphosphate multikinase (IPMK)20 and the 3-phosphatase PTEN21 also interact with p53. Moreover, IPMK and PTEN regulate the interconversion of PI4,5P2 and PI3,4,5P3 bound to the nuclear receptor steroidogenic factor 1 (SF-1), controlling its transcriptional activity22, 23. Akt was also identified as a potential p53 interactor24. These diverse findings suggest that p53 may be modified by multiple phosphoinositide kinases and phosphatases to produce p53-PI3,4,5P3 and regulate Akt activation in the nucleus independently of the canonical membrane-localized pathway.
Here, we demonstrate that genotoxic stress activates nuclear Akt by a p53-dependent mechanism. p53-PI4,5P2 is converted to p53-PI3,4,5P3 in response to genotoxic stress by the nuclear PI3K IPMK, and the 3-phosphatase PTEN reverses this reaction. PI3,4,5P3 binding stimulates the assembly of PDK1, mTORC2, Akt, and FOXOs on p53, leading to nuclear activation of Akt and phosphorylation of FOXOs. Inhibition of PIPKIα or IPMK, but not class I PI3Ks, blocks nuclear p53-PI3,4,5P3 formation and Akt activation. The p53-PI3,4,5P3-Akt complex is regulated by cellular stress and attenuates DNA damage and FOXO-regulated cell death. Our findings establish a PI3K-Akt pathway that dynamically assembles on p53 to control nuclear Akt activation. This p53-phosphoinositide signalosome is independent of the canonical membrane-localized PI3K-Akt pathway, underscoring its therapeutic relevance.
RESULTS
p53 interacts with nuclear Akt and regulates its activation
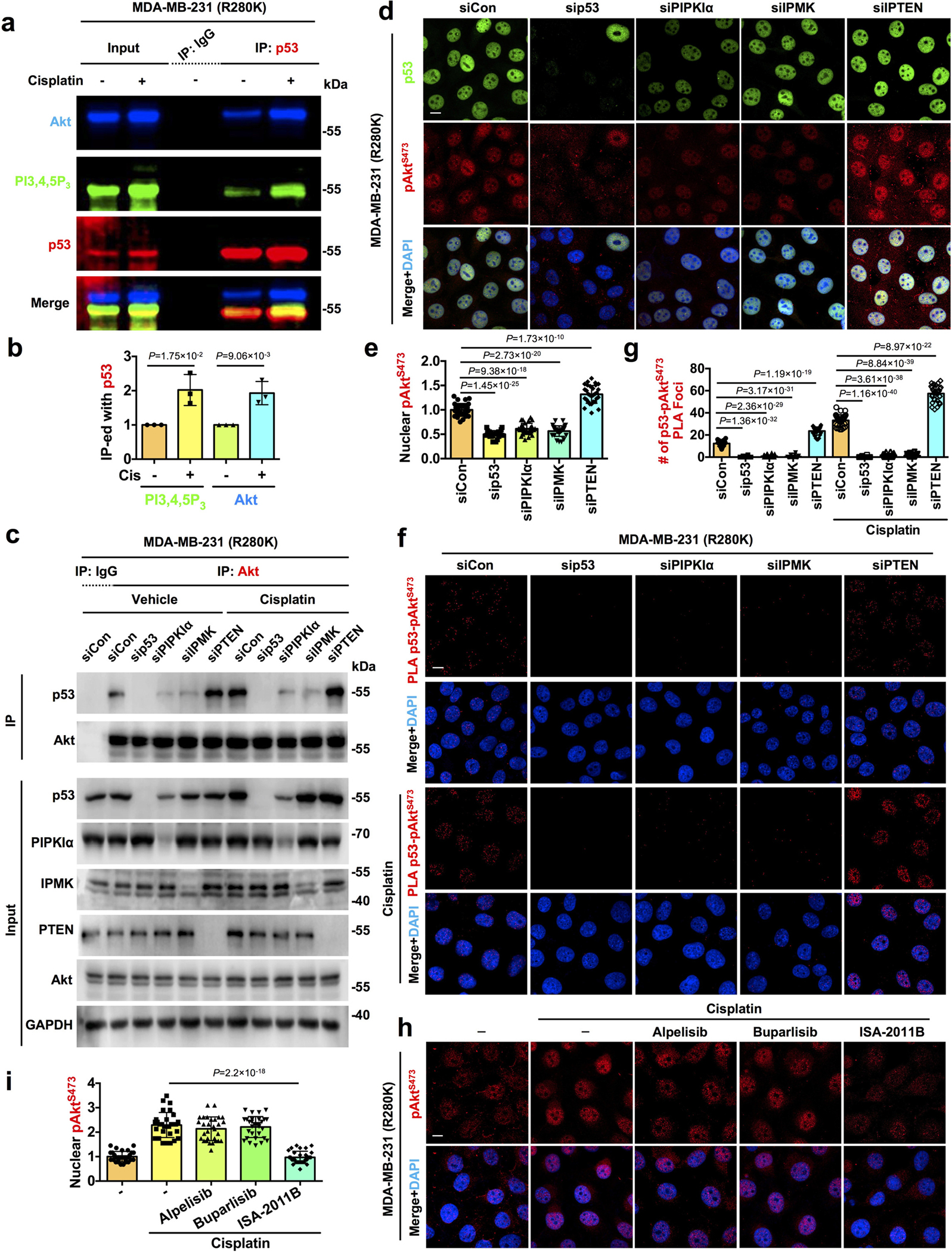
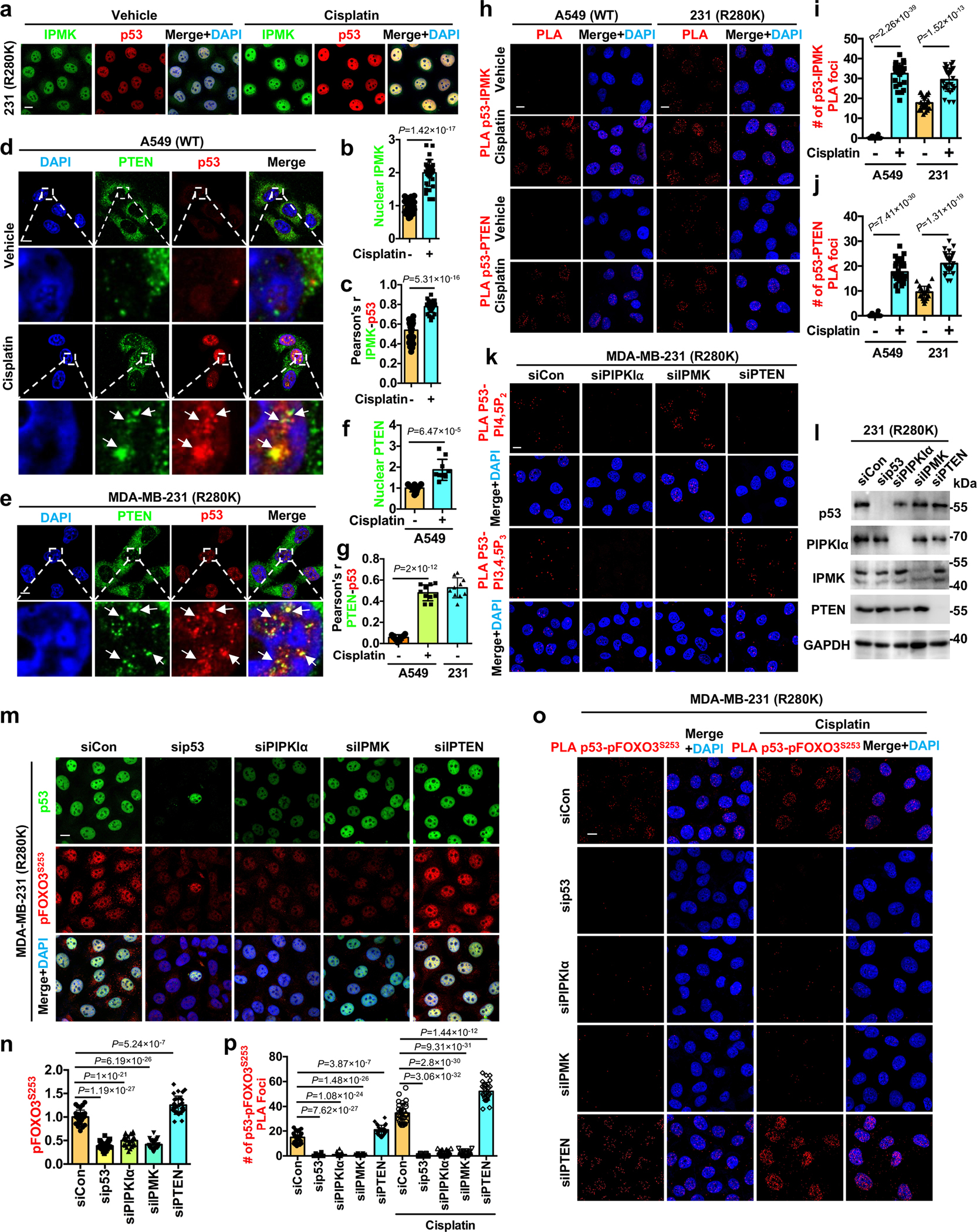
Although Akt is activated in the nucleus by genotoxic stress and active Akt is observed in the nucleus of cancer cells10, 14, 15, 25, the mechanisms of nuclear Akt activation and its function remain enigmatic. To gain mechanistic insights into nuclear Akt activation, we examined the subcellular distribution of phosphorylated Akt under normal and stressed conditions by immunofluorescent (IF) staining. Consistent with prior reports15, 16, genotoxic stress increased the nuclear levels of two active Akt phospho-forms (pAktT308 and pAktS473) but not total Akt in the membrane-free nucleoplasm (Fig. 1a,b and Extended Data Fig. 1a,b). The stress-induced nuclear Akt activation was abolished upon mTOR or Akt inhibition, which also eliminated EGF-stimulated Akt phosphorylation (Extended Data Fig. 1c–g), supporting the specificity of nuclear Akt activation. Given the critical role of class I PI3K-mediated PI3,4,5P3 production in Akt activation in the canonical membrane-localized PI3K-Akt pathway3, 4, we postulated a functional role of class I PI3Ks in stress-induced nuclear Akt activation. However, the increase in nuclear pAktS473 levels by cisplatin treatment was not attenuated by the class I PI3K inhibitors alpelisib and buparlisib26 at concentrations that suppressed EGF-stimulated Akt activation (Extended Data Fig. 1h–j). These results are consistent with a previous report that the stress-induced nuclear PI3,4,5P3 generation, the PIPn-messenger required for Akt activation3, 4, was depleted upon IPMK knockdown (KD), but not Class I PI3K KD27, indicating that nuclear Akt activation is independent of the canonical PI3K-Akt pathway.
Figure 1. p53 interacts with nuclear Akt and regulates its activation.
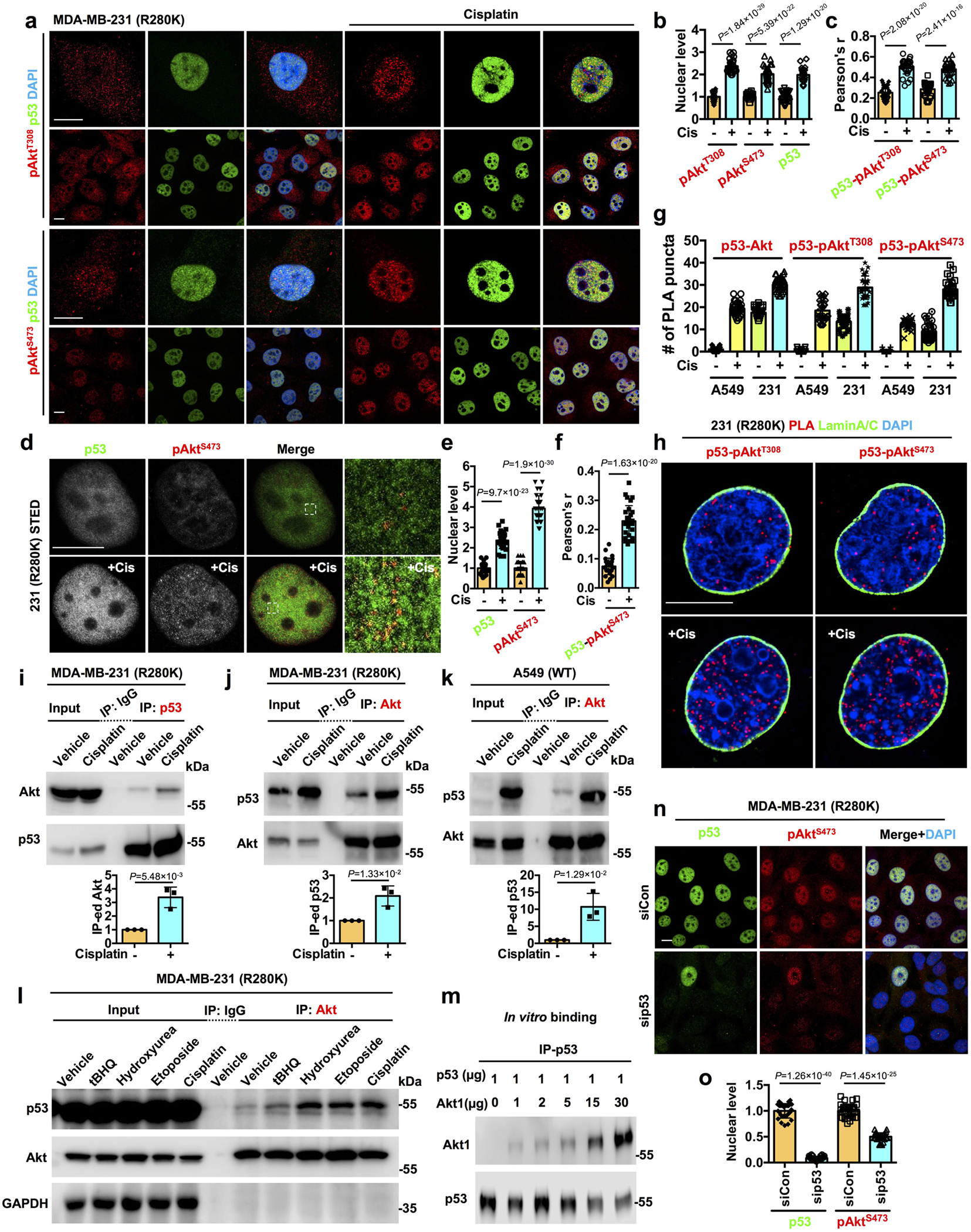
a-c, Confocal images of IF staining against pAktT308/pAktS473 and p53 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear levels of pAktT308/pAktS473 and p53 normalized to vehicle treated cells (b) and their colocalization determined by Pearson’s correlation coefficient (c) were quantified. N=30 cells from representative experiments of three repeats.
d-f, STED super-resolution images of IF staining against pAktS473 and p53 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear levels of pAktS473 and p53 normalized to vehicle treated cells (e) and their colocalization (f) were quantified. N=30 cells from representative experiments of three repeats.
g, Quantification of nuclear PLA complexes of p53-Akt/pAktT308/pAktS473 in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. N=30 cells from representative experiments of three repeats. See PLA images in Extended Data Fig. 2a.
h, PLA of p53-pAktT308/pAktS473 overlaid with the nuclear envelope marker Lamin A/C in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h.
i-k, Co-immunoprecipitation (IP) of endogenous p53 and Akt from MDA-MB-231 and A549 cells treated with control vehicle or 30 μM cisplatin for 24 h. The Akt IP-ed by mutant p53 (i), mutant p53 IP-ed by Akt (j), and wild-type p53 IP-ed by Akt (k) were analyzed by WB. N=3.
l, Co-IP of p53 and Akt from MDA-MB-231 cells treated with 100 μM tert-butylhydroquinone (tBHQ), 100 μM hydroxyurea, 100 μM etoposide, 30 μM cisplatin, or control vehicle for 24 h. Representative data of three independent experiments are shown.
m, In vitro binding of recombinant p53 with Akt1. A constant amount of p53 immobilized on anti-p53 agarose was incubated with the indicated increasing amounts of Akt1. p53 was IP’ed and p53-bound Akt1 was analyzed by WB. Representative data of three independent experiments are shown.
n-o, IF staining of p53 and pAktS473 in MDA-MB-231 cells 48 h after transient transfection with control siRNAs or siRNAs against p53. The nuclear p53 and pAktS473 levels normalized to siCon treated cells were quantified (o). N=30 cells from representative experiments of three repeats.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Akt was identified as a putative p53 interacting protein24 and as p53 interacts with the PIP 5-kinase PIPKIα and its product PI4,5P219, the PI3,4,5P3 precursor, this suggested a regulatory link between nuclear Akt and p53. Cisplatin-induced genotoxic stress increased the colocalization of the active Akt isoforms (pAktT308 and pAktS473) with mutant p53R280K (Fig. 1a–c). Multiple cell stressors augmented nuclear Akt activation and colocalization of pAktS473 with mutant p53 (Extended Data Fig. 1k–n). Stimulated-depletion-emission (STED) microscopy confirmed that genotoxic stress enhanced the colocalization of mutant p53 and pAktS473 in the nucleus, although not all p53 and pAktS473 colocalized (Fig. 1d–f). Genotoxic stress also increased nuclear levels of wild-type p53 and pAktS473 and their nuclear colocalization, but not cytosolic colocalization (Extended Data Fig. 1o–u). Cisplatin stimulated the association of wild-type p53 (p53WT) and mutant p53R280K with total Akt, pAktT308, and pAktS473 in the nucleus as quantified by proximity ligation assay (PLA). Importantly, only mutant p53 interacted with Akt and pAkt under unstressed conditions (Fig. 1g and Extended Data Fig. 2a). Multiple cellular stressors increased the levels of the p53-pAktS473 complex in the nucleus (Extended Data Fig. 2b, c). The mutant p53R280K-pAkt complexes identified by PLA localized to the non-membranous nucleoplasm and were stimulated by genotoxic stress (Fig. 1h). 3D section images and reconstitution confirmed the nuclear localization of the p53-pAktS473 complex to membrane-free regions (Extended Data Fig. 2d and Extended Data Videos 1–2). The interactions of p53WT and p53R280K with Akt were quantified by co-IP and were stimulated by cisplatin (Fig. 1i–k and Extended Data Fig. 2e). Multiple stress stimuli increased mutant p53 binding to Akt (Fig. 1l). Direct binding of recombinant p53 and Akt1 proteins was quantified by pull down (Fig. 1m) and by microscale thermophoresis (MST) that demonstrated a 10±2 nM affinity interaction (Extended Data Table 1). To determine the functional role of p53 in nuclear Akt activation, p53 was KD’ed and quantification of p53R280K and pAktT308/S473 levels showed a loss of nuclear pAktT308/S473 upon p53 KD (Fig. 1n, o and Extended Data Fig. 2f–h). Together, these data indicate that p53 interacts with Akt in the nucleus and promotes its activation.
p53 recruits upstream Akt-activating kinases
To elucidate the mechanisms by which p53 activates nuclear Akt, we examined the association of p53 with upstream kinases required for cytosolic Akt activation. Consistently, genotoxic stress increased nuclear levels of active and nuclear-retained PDK1 (pPDK1S241 and pPDK1S396)28, 29, their colocalization with p53R280K (Fig. 2a–c), and the association of p53WT and p53R280K with PDK1 (total PDK1, pPDK1S241, and pPDK1S396) and the Sin1 subunit of mTORC230, as quantified by PLA (Fig. 2d,e and Extended Data Fig. 3a). The data indicate de novo nuclear PDK1 and mTORC2 activation on p53 as there were no changes in total nuclear PDK1 and Sin1 upon genotoxic stress (Extended Data Fig. 3b–d). The p53-pPDK1 and p53-Sin1 complexes localized to the non-membranous nucleoplasm (Fig. 2f). The interaction of p53WT and p53R280K with PDK1 and Sin1 was confirmed by co-IP and stimulated by genotoxic stress (Fig. 2g–j and Extended Data Fig. 2e). Additionally, p53 directly bound to PDK1 and Sin1 with high affinity, 6±0.2 and 20±1 nM, respectively (Extended Data Table 1). Collectively, these data establish a robust, stress-induced association of p53 with Akt and its activating kinases in the nucleus (Fig. 2k).
Figure 2. p53 recruits upstream Akt-activating kinases in the nucleus.
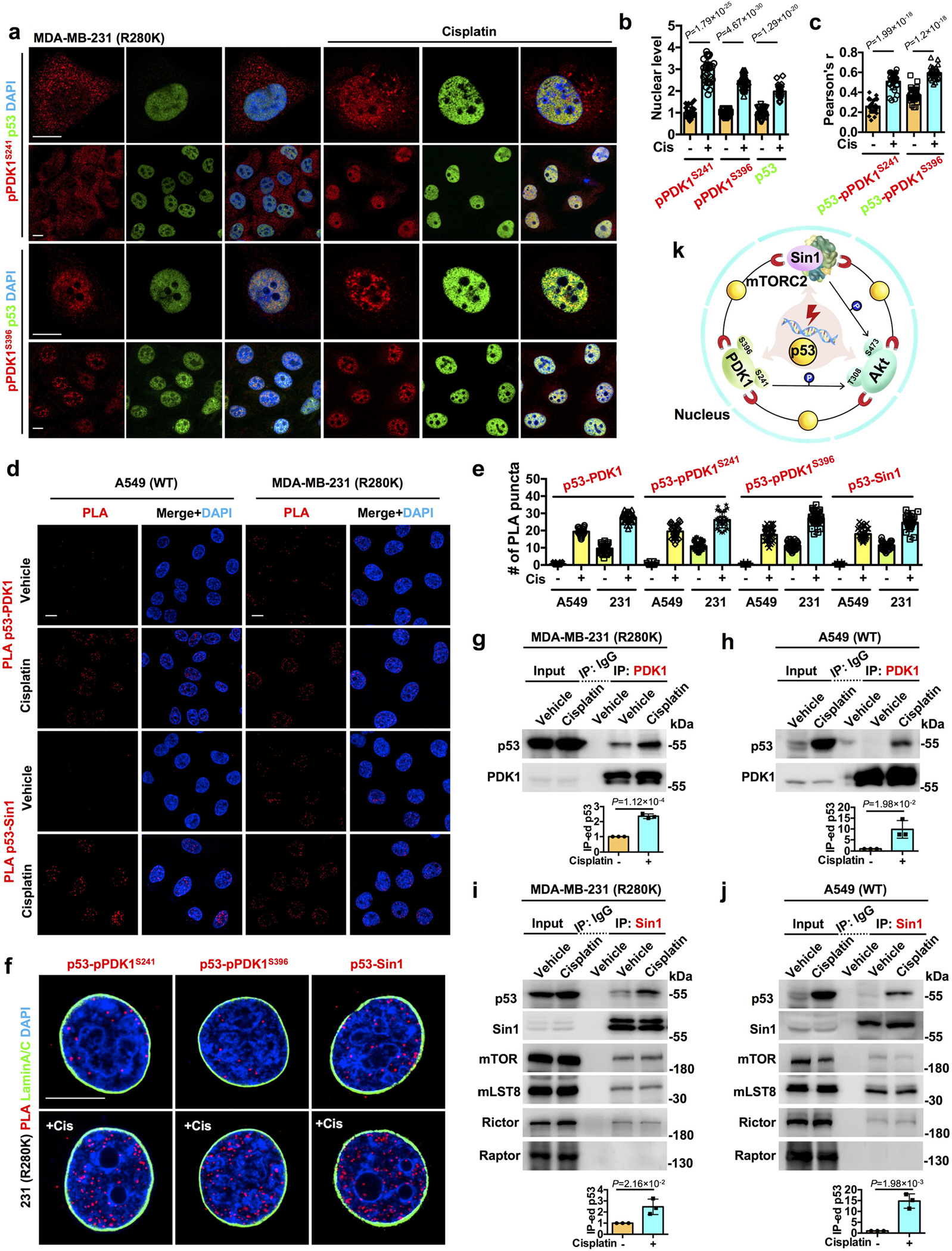
a-c, Confocal IF staining against pPDK1S241/pPDK1S396 and p53 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear levels of pPDK1S241/pPDK1S396 and p53 normalized to vehicle treated cells (b) and their colocalization (c) were quantified. N=30 cells from representative experiments of three repeats.
d-e, PLA of p53-PDK1/Sin1 in A549 and MDA-MB-231 treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear PLA foci of p53-PDK1/pPDK1S241/pPDK1S396/Sin1 were quantified (e). See images of PLA p53-pPDK1S241/pPDK1S396 in Extended Data Fig. 3a.
f, PLA of p53-pPDK1S241/pPDK1S396/Sin1 overlaid with the nuclear envelope marker Lamin A/C in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h.
g-j, Co-IP of endogenous p53 and PDK1/Sin1 from MDA-MB-231 and A549 cells treated with control vehicle or 30 μM cisplatin for 24 h. The mutant and wild-type p53 IP-ed by PDK1 (g,h), and the mutant, wild-type p53, mTORC2 components (mTOR, mLST8, and Rictor) IP-ed by Sin1 (i,j) were analyzed by WB. The mTORC1 component Raptor was probed as the negative control (i,j). N=3. See also Extended Data Fig. 2e.
k, Schematic diagram summarizes the nuclear stress-induced p53 complex that activates Akt.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
The nuclear p53-Akt complex targets FOXOs
Akt phosphorylates multiple downstream effectors, including tuberous sclerosis complex 2 (TSC2), glycogen synthase kinase (GSK3), and members of the forkhead box O (FOXO) family4. FOXOs are reported to also have functional interactions with p5331–33. FOXOs directly bind p5332, and p53 regulates MDM2 ubiquitination and degradation of FOXOs33. The FOXO family, FOXO1, FOXO3, FOXO4, and FOXO6 in humans, regulates cell senescence and apoptosis in response to genotoxic stress and other stimuli5, 34. Akt phosphorylates FOXO proteins at three residues (Fig. 3a), resulting in nuclear exclusion and degradation of FOXOs to promote cell survival5. FOXO1/3, FOXO fragments, and phosphorylated forms co-IP’ed with Akt and p53R280K and were increased by cisplatin (Fig. 3b–d).
Figure 3. The nuclear p53-Akt complex targets FOXOs.
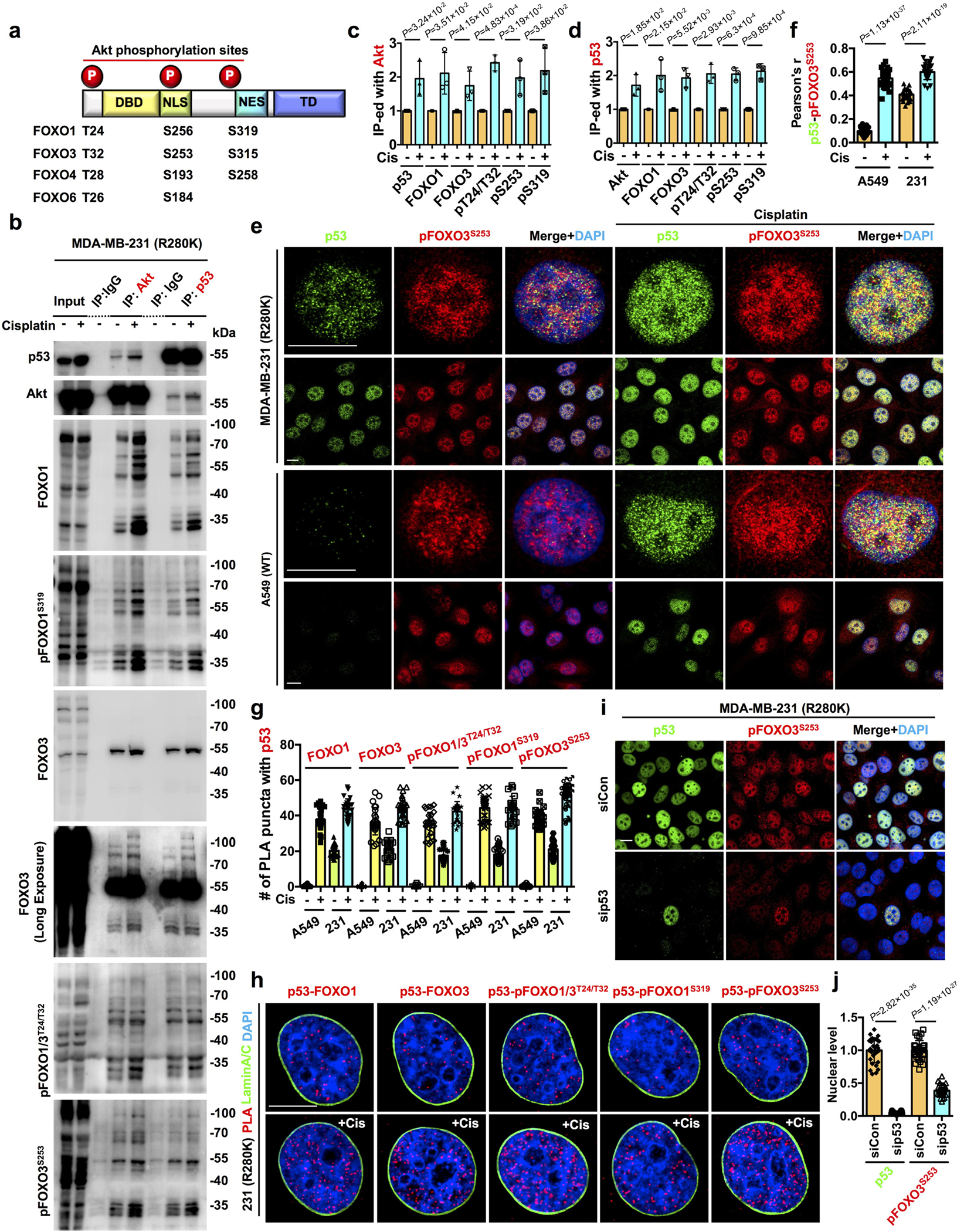
a, Domain map of FOXO family members and their phosphorylation sites by Akt.
b-d, IP analysis of total and phosphorylated forms of FOXOs in association with Akt and p53 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The FOXOs in association with Akt (c) and p53 (d) normalized to vehicle treated cells were quantified. N=3.
e-f, Confocal IF staining of p53 and pFOXO3S253 in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear colocalization of p53 and pFOXO3S253 was quantified (f). N=30 cells from representative experiments of three repeats. See quantification of nuclear pFOXO3S253 in Extended Data Fig. 3f.
g, Quantification of nuclear PLA complexes of p53 with FOXOs in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. N=30 cells from representative experiments of three repeats. See PLA images in Extended Data Fig. 3g.
h, PLA of p53-FOXOs overlaid with the nuclear envelope marker Lamin A/C in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h.
i-j, Confocal IF staining of p53 and pFOXO3S253 in MDA-MB-231 cells 48 h after transient transfection with control siRNAs or siRNAs against p53. The nuclear p53 and pFOXO3S253 levels normalized to siCon treated cells were quantified (j). N=30 cells from representative experiments of three repeats.
For all panels, data are represented as mean ±SD, p value denotes t-test. Scale bar: 5 μm.
Nuclear pFOXO3S253 colocalized with p53WT and p53R280K, and these colocalizations were enhanced by genotoxic stress (Fig. 3e,f). Genotoxic stress induced nuclear FOXO3 phosphorylation and modestly reduced total nuclear FOXO3 (Fig. 3e and Extended Data Fig. 3b, e–f), consistent with prior reports that phosphorylated FOXOs are degradated35, 36. FOXO1/3 and their phosphorylated forms associated with p53 and this was stress-regulated (Fig. 3g,h and Extended Data Fig. 3g). KD of p53R280K diminished nuclear pFOXO3S253 levels (Fig. 3i,j) and total pFOXO3S253 with little impact on cellular FOXO3 levels (Extended Data Fig. 3h). These findings indicate that the p53-Akt complex recruits and phosphorylates FOXOs in the nucleus, thereby underscoring the functional role of this complex in nuclear Akt and p53 signaling.
p53 binds and forms a stable complex with PI3,4,5P3
PI3,4,5P3 directly binds Akt and continuous binding appears to be required for Akt activation3, 4, 9. Thus, we postulated that stress-induced PI3,4,5P3 linked to p53 may serve as a signal to recruit and activate Akt. As PI4,5P2 forms a stable complex with p5319, we investigated if PI3,4,5P3 is also associated with p53. The PI4,5P2 and PI3,4,5P3 epitopes were detected by fluorescent immunoblotting (IB) of FLAG-immunoprecipitated (IP), ectopically expressed p53WT or p53175H. Notably, the interaction of PI4,5P2 and PI3,4,5P3 with p53WT and p53175H was attenuated by the PIPKIα inhibitor ISA-2011B37 (Fig. 4a). PI4,5P2 and PI3,4,5P3 interacted constitutively with multiple p53 mutant proteins and with p53WT in response to cisplatin treatment in all cancer cell lines tested (Fig. 4b). PI3,4,5P3 binding to p53R280K was enhanced by cisplatin and abolished by ISA-2011B37. In contrast, the class I PI3K inhibitors alpelisib and buparlisib did not block p53R280K-PI3,4,5P3 complex formation (Fig. 4c) at concentrations that inhibited EGF-induced Akt phosphorylation (Extended Data Fig. 1h).
Figure 4. p53 binds PI3,4,5P3 and IPMK and PTEN regulate the interconversion of nuclear p53-PI complexes.
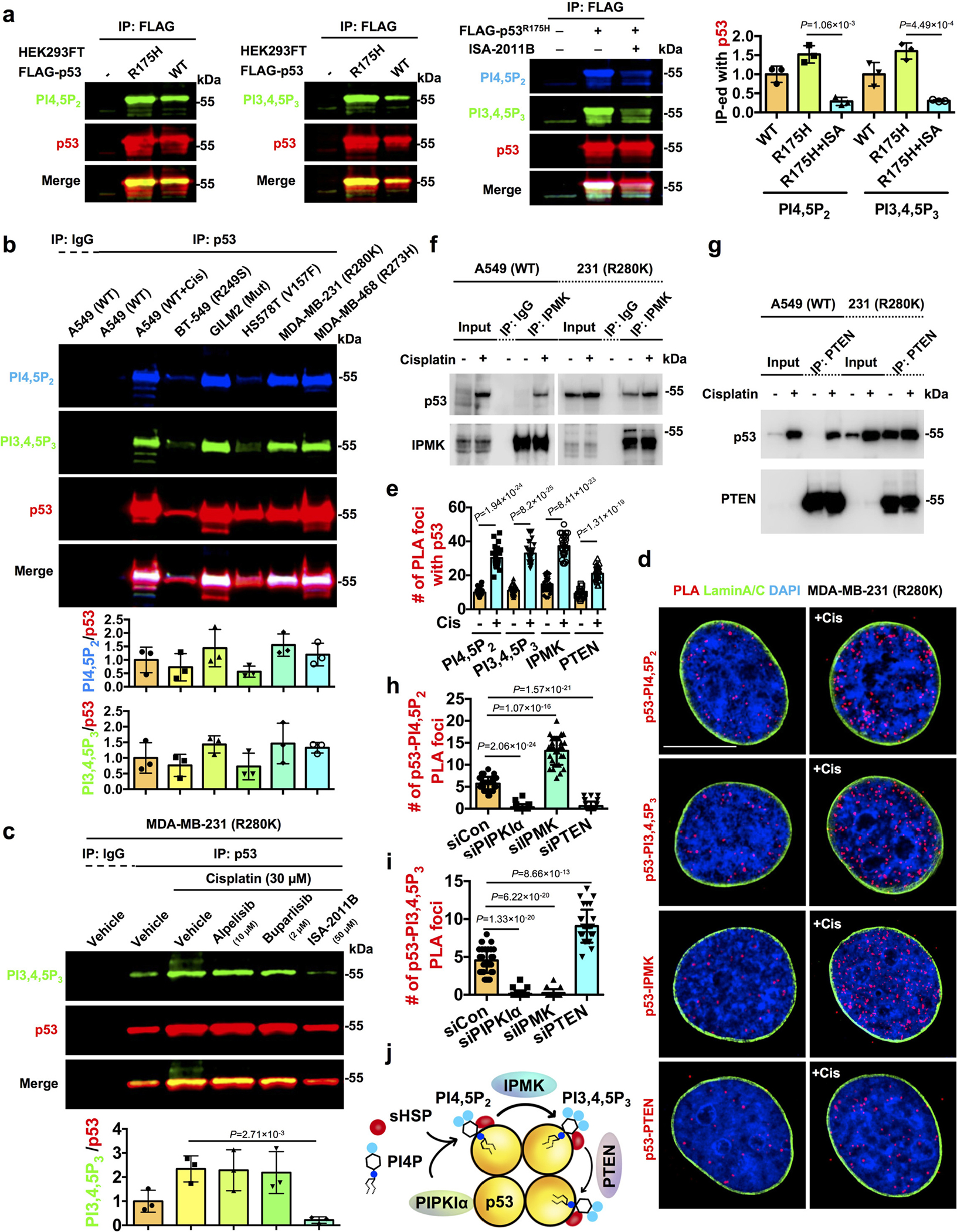
a, Fluorescent IP-WB detects on-site PI4,5P2 and PI3,4,5P3 association with epitope-tagged p53 downstream of PIPKIα. FLAG-tagged mutant (R175H) or wild-type p53 were transiently transfected into HEK293FT cells for 24 h. Then the cells were treated with 50 μM PIPKIα inhibitor ISA-2011B (ISA) for 24 h. The ectopically expressed p53 was IP-ed with FLAG antibody and analyzed by WB using fluorescent antibodies detecting PI4,5P2, PI3,4,5P3, and p53 simultaneously. N=3.
b, Fluorescent IP-WB detects on-site PI4,5P2 and PI3,4,5P3 association with endogenous mutant p53 and stress-induced wild-type p53 in a panel of cancer cells. A549 cells expressing wild-type p53 were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for IP against p53. BT-549, GILM2, HS578T, MDA-MB-231, and MDA-MB-468 cells expressing mutant p53 were directly processed for IP against mutant p53. The on-site PI4,5P2 and PI3,4,5P3 association with p53 was analyzed simultaneously by WB using fluorescent antibodies. N=3.
c, Fluorescent IP-WB detects stress-induced PI3,4,5P3 association with endogenous mutant p53 downstream of PIPKIα but independent from class I PI3Ks. MDA-MB-231 cells were treated with control vehicle or 30 μM cisplatin with or without the presence of the α-specific PI3K inhibitor alpelisib (10 μM), the pan-PI3K inhibitor buparlisib (2 μM), or the PIPKIα inhibitor ISA-2011B (50 μM) for 24 h. Then the cells were processed for IP against p53 and analyzed by WB using fluorescent antibodies detecting PI3,4,5P3 and p53 simultaneously. N=3.
d-e, PLA of p53-PI4,5P2/PI3,4,5P3/IPMK/PTEN overlaid with the nuclear envelope marker Lamin A/C in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear PLA foci were quantified (e). N=30 cells from representative experiments of three repeats.
f, Co-IP of IPMK with wild-type and mutant p53 from A549 and MDA-MB-231 cells respectively treated with 30 μM cisplatin or control vehicle for 24 h. Representative data of three independent experiments are shown.
g, Co-IP of PTEN with wild-type and mutant p53 from A549 and MDA-MB-231 cells respectively treated with 30 μM cisplatin or control vehicle for 24 h. Representative data of three independent experiments are shown.
h-i, Quantification of the nuclear PLA foci of p53-PI4,5P2/PI3,4,5P3 in MDA-MB-231 cells 48 h after transient transfection with control siRNAs or siRNAs against PIPKIα, IPMK, and PTEN. N=30 cells from representative experiments of three repeats. See PLA images in Extended Data Fig. 5k and knockdown validation by WB in Extended Data Fig. 5l.
j, Schematic illustration of p53 in a complex with PI4,5P2 and PI3,4,5P3 downstream of PIPKIα and their interconversion by IPMK and PTEN.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Genotoxic stress increased nuclear PI3,4,5P3 levels and PI3,4,5P3 colocalization with p53R280K as determined by IF (Extended Data Fig. 4a–c). PLA of p53 with PI4,5P2 and PI3,4,5P3 revealed that cisplatin greatly increased the p53WT-PI4,5P2 and p53WT-PI3,4,5P3 complexes, while these phosphoinositides associated constitutively with mutant p53 but were enhanced by cell stress (Extended Data Fig. 4d–h). The p53-PI3,4,5P3 complexes were abolished by ISA-2011B but not by alpelisib or buparlisib (Extended Data Fig. 4i,j). Moreover, the p53-PI4,5P2 and p53-PI3,4,5P3 complexes were localized in the nucleoplasm in regions devoid of membranes (Fig. 4d,e). 3D section images further confirmed the nuclear localization of the p53-PI3,4,5P3 complexes in membrane-free regions (Extended Data Fig. 4k). The interaction of p53 with PI, PI4,5P2, and PI3,4,5P3 was quantified by MST: PI4,5P2 and PI3,4,5P3, but not PI, bound to recombinant p53 (Extended Data Table 1). These data indicate that PI4,5P2 and PI3,4,5P3 form a stable nuclear complex with stress-activated p53WT and mutant p53 that is dependent on PIPKIα and insensitive to class I PI3K inhibitors.
IPMK and PTEN regulate the interconversion of nuclear p53-PIPn complexes
The PI3K IPMK and the 3-phosphatase PTEN associate with p5320, 21. We confirmed the interactions of IPMK and PTEN with stress-induced p53WT and mutant p53 by co-IP (Fig. 4f, g and Extended Data Fig. 3i). IF staining revealed that genotoxic stress enhanced the content of nuclear IPMK and PTEN and their colocalization with p53 (Extended Data Fig. 5a–g). Both IPMK and PTEN associated with stress-activated p53WT and p53R280K in the nucleus in regions distinct from the nuclear envelop as detected by PLA (Fig. 4d, e and Extended Data Fig. 5h–j). IPMK and PTEN directly bound to p53 with 42±13 nM and 124±22 nM affinity, respectively (Extended Data Table 1). To reveal the functional role of PIPKIα, IPMK, and PTEN on potential p53-PI4,5P2⇆p53-PI3,4,5P3 interconversion, we silenced the expression of each enzyme individually and then quantified cellular content of p53R280K-PI4,5P2/PI3,4,5P3 complexes by PLA (Fig. 4h, i and Extended Data Fig. 5k, l). PIPKIα KD prevented the formation of both p53-PI4,5P2 and p53-PI3,4,5P3 complexes. IPMK silencing inhibited the formation of p53-PI3,4,5P3 complexes, while increasing p53-PI4,5P2 complexes. PTEN KD increased p53-PI3,4,5P3, consistent with its 3-phosphatase activity, and diminished the p53-PI4,5P2 complex. These data indicate that IPMK and PTEN dynamically regulate the interconversion of p53-PI4,5P2 and p53-PI3,4,5P3 complexes downstream of PIPKIα generation of p53-PI4,5P2 (Fig. 4j).
The p53-PI3,4,5P3 complex recruits and activates the nuclear Akt pathway
PDK1, Sin1, and Akt are all PI3,4,5P3 effectors that require binding to PI3,4,5P3 for their activities30, 38, 39. These data suggest that the p53-PI3,4,5P3 complex serves as a PI3,4,5P3 reservoir for activating the Akt pathway in the nucleus. Consistently, PI3,4,5P3 and Akt co-IP’ed with p53R280K and these interactions were enhanced by cisplatin (Fig. 5a, b), indicating that p53, PI3,4,5P3, and Akt are present in one complex. KD of p53, PIPKIα, or IPMK decreased the interaction of p53R280K with Akt as measured by co-IP under basal or stressed conditions, while PTEN KD enhanced this interaction, underscoring the functional role of PI3,4,5P3 in the formation of the p53-Akt complex (Fig. 5c). Furthermore, KD of p53R280K, PIPKIα, or IPMK reduced the nuclear pAktS473 and pFOXO3S253 content quantified by IF, whereas PTEN KD enhanced nuclear pAktS473 and pFOXO3S253 levels (Fig. 5d,e and Extended Data Fig. 5m,n). Additionally, p53R280K-pAktS473 and p53R280K-pFOXO3S253 complexes measured by PLA were diminished by KD of p53, PIPKIα, or IPMK, but enhanced by PTEN KD under basal and stressed conditions (Fig. 5f,g and Extended Data Fig. 5o,p). Notably, cisplatin-induced nuclear Akt activation was suppressed by the PIPKIα inhibitor ISA-2011B, but not by the class I PI3K inhibitors (Fig. 5h,i). These data indicate that p53-PI3,4,5P3 recruits and activates Akt in the nucleus. Significantly, the activity of this nuclear p53-PI3,4,5P3 signalosome is dynamically regulated by the nuclear PI3K IPMK that is distinct from the class I PI3Ks in the canonical membrane-localized pathway.
Figure 5. p53-PI3,4,5P3 recruits and activates the nuclear Akt pathway.
a-b, Triple fluorescent IP-WB detects stress-induced Akt association with the p53-PI3,4,5P3 complex. MDA-MB-231 cells were treated with 30 μM cisplatin or control vehicle for 24 before being processed for IP against p53 and analyzed by WB using fluorescent antibodies detecting Akt, PI3,4,5P3, and p53 simultaneously. The p53 associated PI3,4,5P3 and Akt levels normalized to vehicle treated cells were quantified (b). N=3.
c, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for IP against Akt. The Akt-associated p53 and the input of whole cell lysates were analyzed by WB. Representative images of three independent experiments are shown.
d-e, Confocal IF staining of p53 and pAktS473 in MDA-MB-231 cells 48 h after transient transfection with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. The nuclear pAktS473 levels normalized to siCon treated cells were quantified (e). N=30 cells from representative experiments of three repeats.
f-g, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before processing for PLA between p53 and pAktS473. The nuclear PLA foci were quantified (g). N=30 cells from representative experiments of three repeats.
h-i, Confocal IF staining of pAktS473 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h with or without the presence of the PI3Kα inhibitor alpelisib (10 μM), the pan-PI3K inhibitor buparlisib (2 μM), or the PIPKIα inhibitor ISA-2011B (50 μM). The nuclear pAktS473 levels normalized to vehicle treated cells were quantified (i). N=30 cells from representative experiments of three repeats.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
The p53-PI signalosome links the nuclear PI3K-Akt pathway to DNA-repair and cell survival
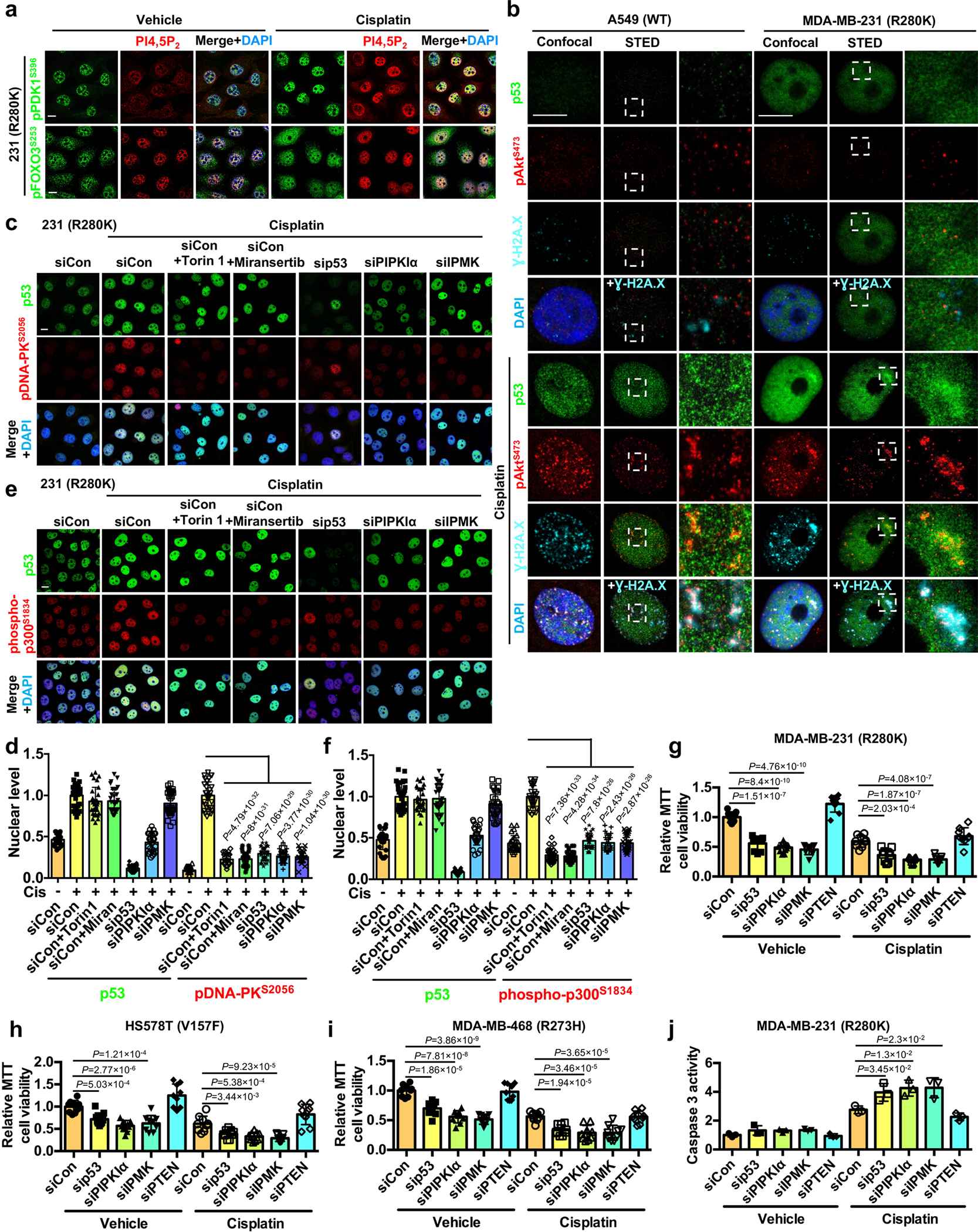
Nuclear pPDK1S396 and pFOXO3S253 were localized to nuclear speckle foci in response to cellular stress (Fig. 2a and 3e). These foci are enriched in PI4,5P2 (Fig. 6a,b and Extended Data Fig. 6a) and PIP kinases and are distinct from known membrane structures19, 40–42. Genotoxic stress stimulation of the p53-PI3,4,5P3-Akt-FOXO complex assembly suggests a role in DNA damage repair and/or apoptosis. Indeed, p53, Akt, FOXOs, and phosphoinositides have all been implicated in the DNA damage response18, 19, 25, 27, and the p53-PI4,5P2 complex has been localized to sites of DNA damage19. Using the phosphorylated histone H2A.X variant, γ-H2A.X, as a marker for sites of DNA double-strand breaks (DSBs)43, STED revealed a specific enrichment of pAktS473 colocalized with p53WT or p53R280K at DNA damage sites (Fig. 6c, d and Extended Data Fig. 6b). KD of p53R280K, PIPKIα, or IPMK increased the cisplatin-induced levels, while PTEN silencing diminished γ-H2A.X levels (Fig. 6e, f), indicating that the p53-PI3,4,5P3 complex and activated Akt facilitate DNA repair.
Figure 6. The p53-PI signalosome links the nuclear PI3K-Akt pathway to DNA-repair and cell survival.
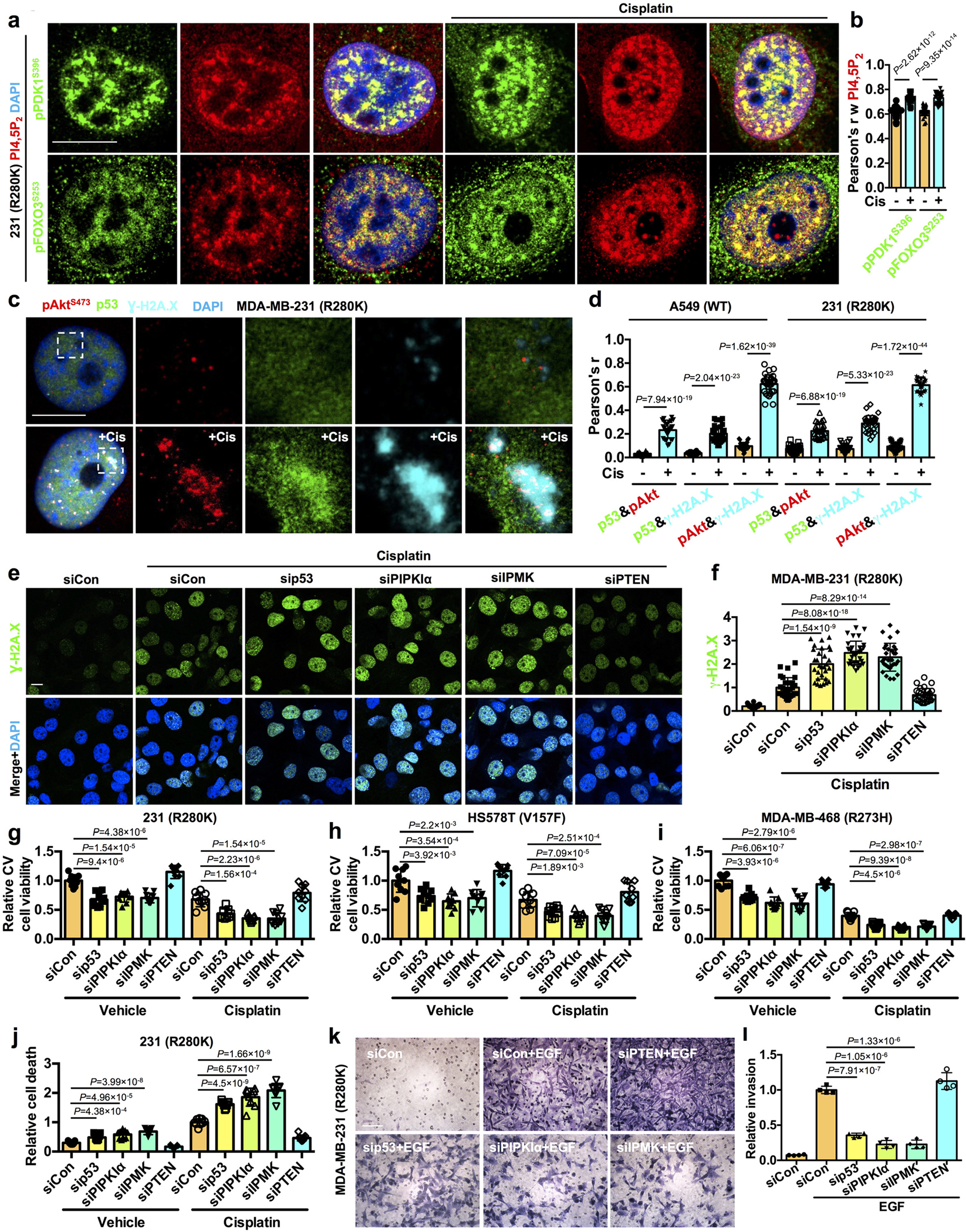
a-b, MDA-MB-231 cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for IF staining against PI4,5P2 and pPDK1S396/pFOXO3S253. The nuclear colocalization of PI4,5P2 and pPDK1S396/pFOXO3S253 was quantified (B). N=30 cells from representative experiments of three repeats. Scale bar: 5 μm. See Extended Data Fig. 6a for extended images.
c-d, STED super-resolution images of IF staining against p53, pAktS473, and the DNA DSB marker Ɣ-H2A.X in MDA-MB-231 or A549 cells treated with 30 μM cisplatin or control vehicle for 24 h. The nuclear colocalization of p53, pAktS473, and Ɣ-H2A.X was quantified (d). N=30 cells from representative experiments of three repeats. Scale bar: 5 μm. See Extended Data Fig. 6b for extended images.
e-f, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for IF staining against Ɣ-H2A.X. The nuclear level of Ɣ-H2A.X normalized to siCon transfected cells treated with cisplatin was quantified (f). N=30 cells from representative experiments of three repeats. Scale bar: 5 μm.
g-i, MDA-MB-231, HS578T, MDA-MB-468 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for crystal violet (CV) viability assay. Cell viability was normalized to siCon transfected cells treated with vehicle. N=3 from three independent experiments, each in triplicate.
j, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for cell death detection ELISA. Cell death was normalized to siCon transfected cells treated with vehicle. N=3 from three independent experiments, each in triplicate.
k-l, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were serum-starved for 24 h and then scored for invasion through Laminin-coated transwell inserts with 8 μm pores using a 10 ng/ml EGF chemoattractant for 16 h. The invading cells at the insert bottom were stained with crystal violet, imaged, and quantified based on the extracted dye using a plate reader. Invasion was normalized to siCon transfected cells treated with EGF. N=4. Scale bar: 100 μm.
For all panels, data are represented as mean ± SD, p value denotes t-test.
Nuclear Akt has been reported to regulate DNA damage repair pathways44–48. This includes promoting the kinase activity of DNA-dependent protein kinase (DNA-PK) by stimulating its phosphorylation at Ser 2056, which facilitates efficient DNA double-strand breaks (DSB) repair in the non-homologous end-joining (NHEJ) pathway48. Akt also phosphorylates p300 at Ser 1834 to increase its histone acetyltransferase activity to enable initial damage recognition factors to access damaged DNA in the nucleotide excision repair (NER) pathway49. Both Torin 1 (mTOR inhibitor) and Miransertib (Akt inhibitor) attenuate the stress-induced nuclear levels of pDNA-PKS2056 and phosphop-300S1834 (Extended Data Fig. 6c–f), supporting that nuclear Akt controls phosphorylation of DNA-PK and p300 as previously reported44, 48, 49. Similarly, p53 KD suppressed the stress-induced nuclear levels of pDNA-PKS2056 and phospho-p300S1834, supporting the dual role for both p53 and Akt in regulating the nuclear activity of DNA-PK and p300 in DNA damage repair (Extended Data Fig. 6c–f). Consistently, KD of PIPKIα and IPMK individually mimicked the effects of p53 KD in reducing the genotoxic stress-induced content of pDNA-PKS2056 and phospho-p300S1834 (Extended Data Fig. 6c–f), indicating a role for PIPn isomer synthesis and the p53-Akt complex in regulating nuclear DNA-PK and p300 in DNA damage repair.
We next examined the functional role of the nuclear p53-Akt complex in DNA damage-induced apoptosis. KD of mutant p53, PIPKIα, or IPMK sensitized cancer cells to cisplatin treatment and increased caspase-3 activity, whereas PTEN KD conferred resistance to genotoxic stress as determined by multiple cell death assays (Fig. 6g–j and Extended Data Fig. 6g–j). Moreover, KD of mutant p53, PIPKIα, or IPMK reduced cell invasion, an early step in metastasis, while PTEN KD enhanced invasion (Fig. 6k–l). Collectively, these results support a pro-survival role of the nuclear p53-PI3,4,5P3 complex that recruits and activates Akt at sites of DNA damage to regulate genotoxic stress.
The interaction of Akt with p53 is PIPn-dependent and regulated by the C-terminus of p53
To gain additional mechanistic insights, we first examined which Akt isoforms (Akt1, Akt2, and Akt3)5 interacted with p53. p53 co-IP’d with all three Akt isoforms, albeit most robustly with Akt1 (Fig. 7a). Akt1, Akt2, and Akt3 are activated by PI3,4,5P3 and PI3,4P250, suggesting that p53-PI3,4,5P3 plays a role in their nuclear activation (Fig. 4). Consistent with this idea, Akt binding to p53 was dependent on the PI3,4,5P3-binding PH domain of Akt as the PI3,4,5P3-binding defective Akt R25C mutant (both full-length Akt and Akt PH domain) was markedly impaired in p53 binding (Fig. 7b–c). We previously demonstrated that p53 associates with PI4,5P2 via a polybasic PIPn-binding motif in the p53 C-terminal regulatory domain (CTD)19. Using a series of p53 truncation mutants, the Akt and Sin1 (mTORC2 subunit) binding domains were mapped to the C-terminus of p53, which includes the PIPn-binding motif (Fig. 7d–e). As PI3,4,5P3 is generated from PI4,5P2, we determined if the PIPn-binding motif in p53 is required for PI3,4,5P3 binding using p53 mutants (6Q and R379Q) that are impaired in PI4,5P2 binding19. The p53 6Q and R379Q mutants markedly disrupted PI4,5P2 and PI3,4,5P3 binding (Fig. 7f,g and Extended Data Fig. 7a). Akt1 interacted with p53WT, p53R175H, and p53R248Q, but these interactions were lost in the corresponding p53 6Q and R379Q p53 mutants (Fig. 7h–i and Extended Data Fig. 7b–c), indicating that the PIPn binding to p53 regulates assembly of the p53-Akt nuclear complex.
Figure 7. The interaction of p53 with Akt is PIPn-dependent and mediated by the C-terminus of p53.
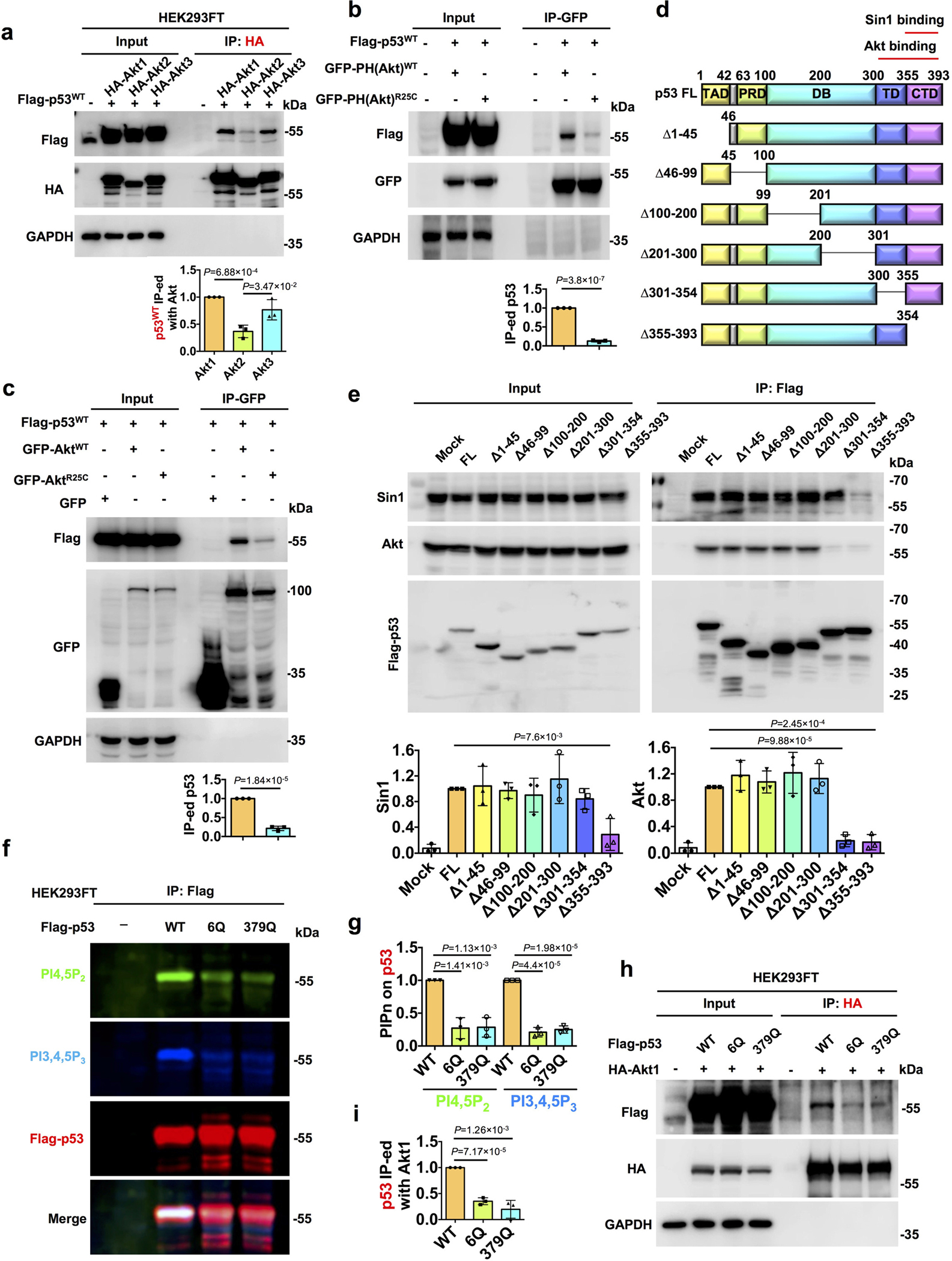
a, p53 co-IP-ed with all three Akt isoforms (Akt1, Akt2, and Akt3). FLAG-tagged wild-type p53 and HA-tagged Akt1/2/3 were co-transfected into HEK293FT cells for 48 h. The ectopically-expressed Akt1/2/3 were IP-ed with HA antibody, and analyzed by WB. The co-IP-ed p53 level was normalized to the level obtained by Akt1 IP. N=3.
b, The interaction of the Akt PH with p53 is regulated by PIPn binding to the Akt PH. FLAG-tagged wild-type p53 and GFP-tagged Akt PH domain (WT or PIPn binding-defective R25C mutant) were co-transfected into HEK293FT cells for 48 h. The ectopically expressed WT or R25C mutant Akt PH domain were IP-ed with GFP antibody, and then analyzed by WB. N=3.
c, The interaction of full-length Akt with p53 is regulated by PIPn-binding to the Akt PH. FLAG-tagged p53 and GFP-tagged full-length Akt (WT or R25C mutant) were co-transfected into HEK293FT cells for 48 h. The ectopically expressed WT or R25C mutant Akt were IP-ed with GFP antibody, and then analyzed by WB. N=3.
d, A schematic representation of p53 truncation mutants used in the study. DB, DNA-binding domain; FL, full length; PRD, proline-rich domain; TAD, transactivation domain; TD, tetramerization domain; CTD, C-terminal regulatory domain. The Akt and Sin1 binding domains identified in (e) are indicated.
e, Flag-tagged p53 truncation mutants were transiently transfected into HEK293FT cells for 48 h. The ectopically expressed p53 was IP-ed with FLAG antibody, and the associated endogenous Sin1 and Akt were analyzed by WB. N=3. Binding was normalized to that of FL p53.
f-g, Fluorescent IP-WB detects on-site PI4,5P2 and PI3,4,5P3 association with p53 that is dependent on its PIPn-binding motif. FLAG-tagged p53 (WT or PIPn-binding defective 6Q or 379Q mutants) were transiently transfected into HEK293FT cells for 48 h. The ectopically expressed p53 was IP-ed with FLAG antibody and analyzed by WB using fluorescent antibodies detecting PI4,5P2, PI3,4,5P3, and p53 simultaneously. N=3.
h-i, The interaction of Akt with p53 requires the PIPn-binding motif of p53. HA-tagged Akt1 and FLAG-tagged WT p53 or mutant p53 (6Q or 379Q) were co-transfected into HEK293FT cells for 48 h. The ectopically expressed Akt1 was IP-ed with HA antibody, and analyzed by WB. N=3.
For all panels, data are represented as mean ± SD, p value denotes t-test.
Regulation of the nuclear Akt pathway by p53 is dependent on the PIPn-binding motif of p53
Wild-type p53 protein is normally expressed at low levels but is dramatically induced by stress, whereas mutant p53 is expressed at high basal levels due to enhanced protein stability17, 18. There are stochastic variations in the levels of wild-type and mutant p53 in cells (Fig. 8a–c and Extended Data Fig. 7d–k). To assess the relationship between p53 and active nuclear Akt levels, we quantified the correlation between nuclear p53 and pAktS473. Under basal and stressed conditions, there were strong correlations between nuclear p53WT and pAktS473 levels with Pearson’s r > 0.751. While the stochastic levels of p53WT segregated the nuclear active pAkt levels in an on/off mode, p53R280K levels correlated with nuclear pAkt expression in a dose-dependent fashion (Fig. 8a–c). Nuclear Akt phosphorylated pFOXO3S253 levels also correlated with p53R280K expression under basal and stressed conditions (Fig. 8d–g). In addition, the nuclear levels of pAktS473 and pFOXO3S253 in p53-high cells were significantly greater than those in p53-low cells. To refine this observation, we ectopically expressed Flag-tagged p53R175H in A549 cells and observed that p53R175H enhanced pAktS473 levels in the nucleus under basal conditions, while nuclear pAktS473 levels were further increased by genotoxic stress. The nuclear content of pAktS473 in p53R175H expressing cells was significantly greater than that in p53R175H-negative cells (Extended Data Fig. 7l–o). These results demonstrate that p53 activates nuclear Akt in a dose-dependent fashion and this dose-dependence likely underlies the requirement of cellular stress to stabilize p53WT to assemble the p53-PI3,4,5P3-Akt complex and activate nuclear Akt, while the hyper-stabilized mutant p53 protein17, 18 constitutively activates this pathway with a modest increase by stress.
Figure 8. Regulation of the nuclear Akt pathway by p53 is dependent on the PIPn-binding motif of p53.
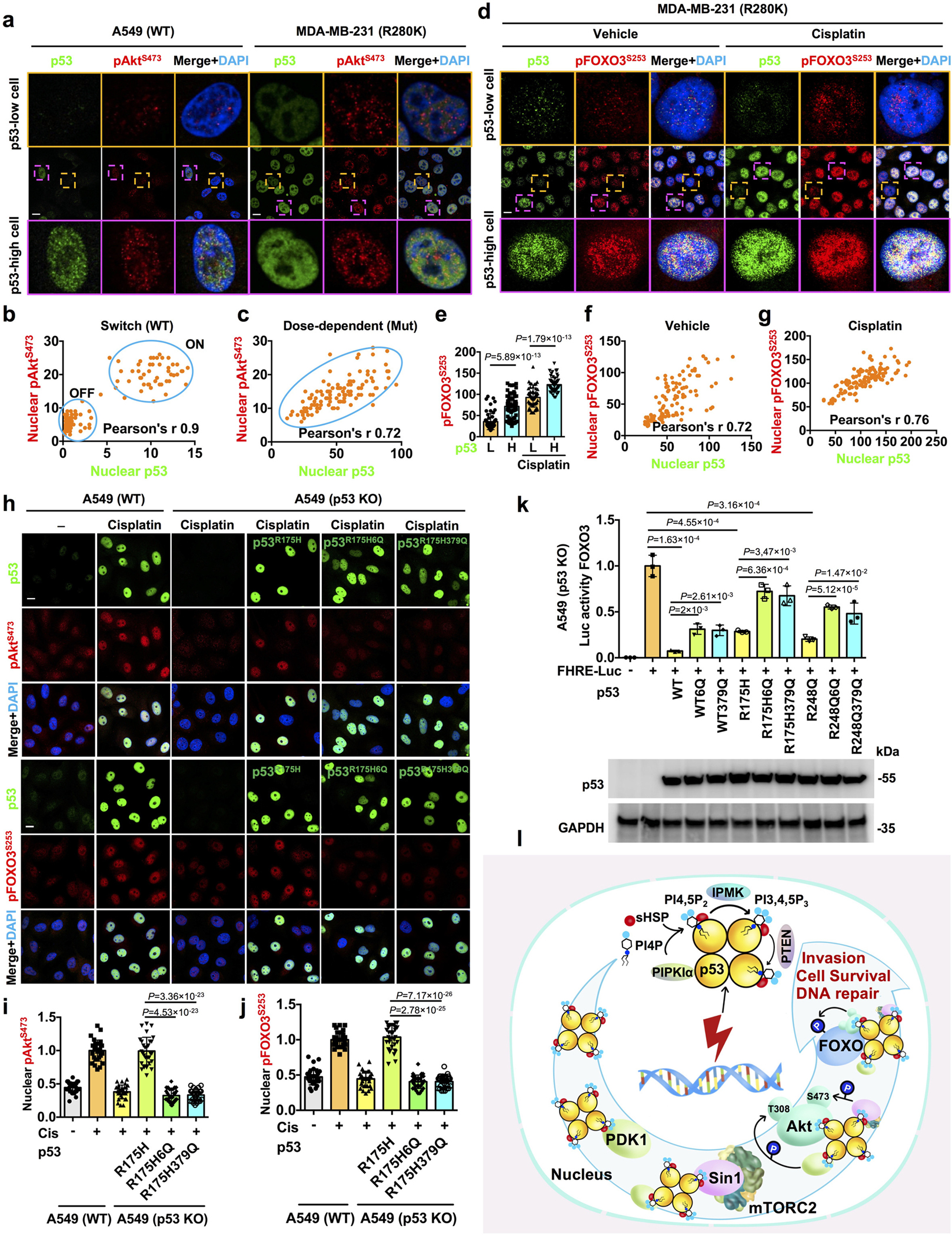
a-c, A549 and MDA-MB-231 cells after 24 h of treatment with control vehicle were fixed and processed for IF staining for p53 and pAktS473. The nuclear p53 and pAktS473 levels were quantified and the correlation between the nuclear p53 and pAktS473 was calculated by Pearson’s r (b,c). N=120 cells from representative experiments of three repeats. See extended images and quantification in Extended Data Fig. 7d–k.
d-g, MDA-MB-231 cells were treated with 30 μM cisplatin or control vehicle for 24 h and were then fixed and processed for IF staining of p53 and pFOXO3S253. The nuclear levels of p53 and pFOXO3S253 were quantified. The low (L) and high (H) levels of nuclear p53 were determined based on the average of the p53 levels in the corresponding control vehicle or cisplatin-treated groups (e). The correlation between the nuclear p53 and pFOXO3S253 was determined by Pearson’s r (f,g). N=120 cells from representative experiments of three repeats.
h-j, A549 p53 KO cells transiently transfected with Flag-tagged mutant p53R175H or p53R175H 6Q/379Q mutant and then 24 h later treated with 30 μM cisplatin for 24h. A549 WT cells were treated with 30 μM cisplatin or control vehicle for 24h. The cells were then fixed and processed for IF staining of p53 and pAktS473/pFOXO3S253. The nuclear pAktS473/pFOXO3S253 levels in A549 WT cells, p53 KO cells, and Flag-tagged mutant p53-positive cells were quantified and normalized to levels in A549 WT cells treated with cisplatin as indicated (i,j). N=30 cells from representative experiments of three repeats.
k, p53 inhibits the transcriptional activity of FOXO3 by a PIPn-dependent mechanism. A549 p53 KO cells were co-transfected with constructs expressing WT p53, mutant p53, or their corresponding 6Q or 379Q mutants and a FOXO3 responsive element with luciferase readout (FHRE-Luc) as indicated for 48 h. Cells were then lysed and processed for luciferase assay to measure the transcriptional activity of FOXO3. Luciferase activity was normalized to that in cells transfected only with FHRE-Luc. The expression levels of different p53 constructs were examined by WB. N=3.
l, A schematic model of the stress-induced nuclear p53-phosphoinositide signalosome that regulates Akt activation, DNA-repair, cell survival, and invasion.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
We next quantitated the effect of p53 and its PIPn-binding defective mutants on nuclear Akt activation in p53 knockout (KO) A549 cells (Extended Data Fig. 7p). Transfection of p53R175H enhanced stress-induced nuclear Akt activation (pAktT308 and pAktS473) and the phosphorylation of DNA-PK and p300 in the p53 KO A549 cells, while the PIPn-binding defective p53 6Q and R379Q mutants were markedly impaired in these activities (Fig. 8h–i, and Extended Data Fig. 8a–c). These results establish that p53 activates the nuclear Akt pathway and components of the DNA repair machinery (DNA-PK and p300) by a mechanism that requires the p53-PIPn complex.
As the p53-Akt nuclear complex binds and phosphorylates FOXOs (Fig. 3 and Extended Data Fig. 3), we examined whether this activity depends on PIPn binding to p53. Extopic expression of p53R175H enhanced stress-induced nuclear Akt activation and pFOXO3S253 levels in p53 KO A549 cells, and these functions were markedly impaired in the corresponding p53 6Q and R379Q mutants (Fig. 8h–j and Extended Data Fig. 8a). Additionally, p53WT, p53R175H, and p53R248Q inhibited the transcriptional activity of FOXO3 as previously reported52 (Fig. 8k), but the corresponding p53 6Q and R379Q mutants attenuated FOXO3 transcriptional inhibition (Fig. 8k). Together, these data link the p53-PIPn complexes to their ability to negatively regulate FOXO3 transcriptional activity, providing additional evidence for the functional relevance of the p53-PIPn complex.
We also examined the functional role of PIPn binding to p53 in the cytoprotective and pro-invasive activities of mutant p53. Ectopic expression of p53R175H or p53R248Q in p53 KO A549 cells enhanced cell viability upon cisplatin treatment (Extended Data Fig. 8d) and increased invasion upon EGF-stimulation (Extended Data Fig. 8e–f), while these activities were diminished by the p53 6Q and R379Q mutations. Collectively, these data support the dynamic regulation of nuclear Akt activation by p53-PIPn complexes in response to genotoxic stress that activate multiple downstream targets to confer stress-resistance.
DISCUSSION
Since the discovery of the nuclear generation of PIPs53 in regions spatially distinct from known membrane compartments42, key components of the canonical membrane-localized cytoplasmic PI3K-Akt pathway have been shown to be present in the nucleus40. Nuclear PI signaling enables the synthesis of PIPns and their dynamic interconversions by kinases and phosphatases with no detectable association with membrane structures, implying a distinct nuclear mechanism independent from the cytoplasmic pathway40, 54. Accumulating evidence points to nuclear PDK1-mTORC2-Akt activation distinct from the classic membrane-localized cytosolic pathway10, 44, 55–57. The nucleus contains all the machinery necessary to activate Akt, including PIP kinases, PI3-kinases, PI4,5P2, PI3,4,5P3, PDK1, mTORC2, and Akt10, 40–42. In the canonical membrane-localized PI3K-Akt pathway, PI3,4,5P3 recruits PDK1, mTORC2, and Akt to membranes via their PH domains30, 38, 39. The cytoplasmic PI3K pathway is assembled by the IQGAP1 scaffold and is compartmentalized to endosomes1, 58, 59.
Here, we report the discovery of a novel compartmentalized nuclear PI3K-Akt pathway independent of the membrane-localized pathway that is scaffolded on p53 (Fig. 8l). This nuclear p53-PIPn signalosome dynamically binds to PIPKIα, IPMK, and PTEN to enable stress-regulated modifications of the p53-PIPn complexes with spatial and temporal specificity. IQGAP1 in the cytosol membranes scaffolds the full Akt pathway components1, the p53-PI3,4,5P3 complex has an analogous role in the nucleus by recruiting Akt, PDK1, and mTORC2 leading to activation of Akt. Yet, the p53 scaffold is separate from membrane suggesting a different structure and compartmentalization that nevertheless recruits and regulates the Akt pathway components that also function in a membrane-based compartment. A key distinction is the pathway specific PI3Ks. In the cytosol, class I PI3Ks are incorporated into the IQGAP1 scaffold, whereas on the nuclear p53 scaffold, IPMK generates the p53-PI3,4,5P3 complex.
The nuclear p53-PI3,4,5P3-Akt complex also recruits, phosphorylates, and suppresses the transcriptional activity of FOXOs, known Akt substrates with roles in proliferation and DNA damage-induced apoptosis25, 27, 36, 60. The resulting inactivating phosphorylation of FOXOs mitigates DNA damage and apoptosis, underscoring the cytoprotective function of this complex. The localization of the p53-PI3,4,5P3-Akt complex at sites of DNA DSBs identified by γ-H2A.X foci and its phosphorylation of DNA-PK and p300 downstream of active nuclear Akt in the NHEJ and NER DNA repair pathways, respectively48, 49, supports the function role of the p53-PIPn complex in the DNA damage response.
The discovery of a direct link between p53 and Akt and the dose-dependent induction of nuclear Akt activation by p53 point to a new function of p53. In the absence of stress, p53WT is stochastically expressed, activating nuclear Akt in an on/off mode, whereas mutant p53 is highly expressed and activates nuclear Akt constitutively, reflecting the hyperstability of mutant p53 relative to p53WT17, 18. Indeed, the loss of transcriptional regulation by mutant p53 and its gain of Akt activation may be underlying mechanisms for its survival, proliferative and oncogenic roles in cancer. Consistent with this idea, knock-in of mutant p53 in mice leads to elevated Akt signaling and tumorigenesis61 and expression levels of p53, a surrogate marker for p53 mutational status62, 63, correlate with active Akt in human colorectal carcinomas64.
The p53-PI signalosome is spatially distinct from the membrane-localized PI3K-Akt pathway and utilizes a distinct PI3K, the nuclear PI3K IPMK that binds p5320 and generates p53-PI3,4,5P3. Notably, the p53-PI signalosome and nuclear Akt activation are not impacted by class I PI3K inhibitors, including the α-specific inhibitor alpelisib65. As such, the discovery of the nuclear p53-PI signalosome has significant therapeutic implications for cancers driven by mutant p53. Finally, the discovery that PIPns tightly associate with key proteins such as p53 and the nuclear poly(A) polymerase Star-PAP66 suggests that this may represent a “third messenger” system whereby second messengers in the canonical membrane-localized PI signaling pathway become tightly associated with key regulatory proteins to confer new functional activities.
METHODS
Cell culture and constructs
A549, BT-549, Cal33, HCT116, HS578T, MDA-MB-231, MDA-MB-468, SUM1315, HEK293FT, and MCF-10A cells were purchased from ATCC. GILM2 cells were described previously67. A549 p53 KO cells were purchased from Abcam (#ab276092). MCF10A cells were cultured as described1 and the other cells were maintained in DMEM (#10-013-CV, Corning) supplemented with 10% fetal bovine serum (#SH30910.03, Hyclone) and 1% penicillin/streptomycin (#15140-122, Gibco). The cell lines used in this study were routinely tested for mycoplasma contamination, and mycoplasma negative cells were used. None of the cell lines used in this study is listed in the database of commonly misidentified cell lines maintained by ICLAC. The p53 constructs used for this work were described previously19. Constructs were transfected in mammalian cells by the lipid-based delivery system from Invitrogen (Lipofectamine™3000, #L3000015, Thermo Fisher Scientific) according to the manufacturer’s instructions. Typically, 2–5 μg of DNA and 6–10 μl of lipid were used for transfecting cells in 6-well plates. Cells that had at least 80% transfection efficiency were used for further analysis.
Antibodies and reagents
Monoclonal antibodies against p53 (clone DO-1, #SC-126, Santa Cruz Biotechnology), p53 (clone 7F5, #2527, Cell Signaling), Sin1 (clone D7G1A, #12860, Cell Signaling), Akt (clone C67E7, #4691, Cell Signaling), pAktS473 (clone 193H12, #4058, Cell Signaling), pAktT308(clone D25E6, #13038, Cell Signaling), PTEN (clone 17.A, #MA5-12278, Invitrogen), FOXO1 (clone C29H4, #2880, Cell Signaling), FOXO3 (clone D19A7, #12829, Cell Signaling), pDNA-PKS2056 (clone E9J4G, #68716, Cell Signaling), mTOR (Clone 7C10, #2983, Cell Signaling), Rictor (Clone H-11, #sc-271081, Santa Cruz Biotechnology), Raptor (Clone 24C12, #2280, Cell Signaling), HA-tag (clone C29F4, #3724, Cell Signaling), GFP-tag (clone B-2, #sc-9996, Santa Cruz Biotechnology), Flag-tag (clone D6W5B, #14793, Cell Signaling), GAPDH (clone 0411, #sc-47724, Santa Cruz Biotechnology), and polyclonal antibodies against PIPKIα (PIP5K1A, #9693, Cell Signaling), PDK1 (#3062, Cell Signaling), pPDK1S241 (#ab131098, Abcam), pPDK1S396 (#PA5-12888, Invitrogen), pFOXO1T24/FOXO3T32 (#9464, Cell Signaling), pFOXO1S319 (#2486, Cell Signaling), pFOXO3S253 (#PA5-37578, Invitrogen), phospho-p300S1834 (#PA5-105823, Invitrogen), mLST8 (#HPA041841, MilliporeSigma) were used in this study. Polyclonal antibodies against PIPKIα and IPMK were produced as described previously1. For conventional immunostaining and PLA analysis of phosphoinositides, anti-PI4,5P2 (#Z-P045) and PI3,4,5P3 (#Z-P345) antibodies were purchased from Echelon Biosciences. For immunoblotting analyses, antibodies were diluted at a 1:1000 ratio except for p53 (clone DO-1, 1:5000) and GAPDH (clone 0411, 1:5000). For immunoprecipitation, antibody-conjugated agarose was purchased from Santa Cruz Biotechnology, including agarose-conjugated antibodies against p53 (#sc-126AC), PDK1 (#sc-17765AC), Sin1 (#sc-393166AC), Akt (#sc-5298AC), HA-tag (#sc-7392AC), GFP-tag (#sc-9996AC) and Flag-tag (#sc-166355AC). For immunostaining analyses and proximity ligation assay (PLA), antibodies were diluted at a 1:100 ratio. Nuclear envelope (Alexa Fluor®488 Lamin A/C, clone 4C11, #8617, Cell Signaling, 1:200) and DNA damage site (Alexa Fluor®488-ɣ-H2A.X, clone EP854(2)Y, #ab195188, Abcam, 1:200) markers were used to examine subnuclear regions. The constructs of GFP-PH(Akt)WT (#51465), GFP-PH(Akt)R25C (#51466), GFP-AktWT (#39531), GFP-AktR25C (#39532), HA-Akt1(#73408), HA-Akt2 (#16000), HA-Akt3 (#9017), and FHRE-Luc (#1789) were purchased from Addgene. The Flag-tagged p53 constructs and corresponding PIPn-binding defective 6Q and R379Q mutant, and p53 domain deletion mutants were described previously19. For recombinant protein production, the DNA constructs for His-tagged p53, PIPKIα, IPMK, and PTEN were purchased from Genscript and the recombinant proteins were purified as previously described19. Recombinant Akt1 (#ab79792) and PDK1 (also known as PDPK1, #ab60834) were purchased from Abcam, and the recombinant Sin1 (also known as MAPKAP1, #TP311745) was purchased from Origene. For the knockdown (KD) experiments, pooled-siRNAs targeting the 3’UTR of IPMK (sense 5’-CCAAGAGAGCUGGAAUUCUAUAAUA-3’ and antisense 5’-UAUUAUAGAAUUCCAGCUCUCUUGG-3’; sense 5’-CAAAGGACAACUGUCAGACACAGAA-3’ and antisense 5’-UUCUGUGUCUGACAGUUGUCCUUUG-3’) were purchased from Thermo Fisher Scientific. The ON-TARGETplus siRNA SMARTpool with 4 siRNAs in combination against human p53, PIPKIα, and PTEN was purchased from Dharmacon. Non-targeting siRNA (Dharmacon) was used as a control. siRNAs were delivered to cells by RNAiMAX reagent (#13778150, Thermo Fisher Scientific), and KD efficiency was determined by immunoblotting. KD efficiency greater than 80% was required to observe phenotypic changes in the study. The PIPKIα inhibitor ISA-2011B (#HY-16937), mTOR inhibitor KU-0063794 (#HY-50710), Akt inhibitors Miransertib (#HY-19719) and Capivasertib (#HY-15431) were purchased from MedChemExpress. Cisplatin (#S1166), class I PI3K inhibitors alpelisib (BYL719, #S2814) and buparlisib (BKM120, #S2247), and Akt inhibitors MK-2206 (#S1078), Tricirbine (#S1117), and Ipatasertib (#S2808) were purchased from Sellekchem. The mTOR inhibitors Torin 1 (#A11587) was purchased from Adooq Bioscience and Palomid 529 (#SML0801) was purchased from MilliporeSigma.
Immunoprecipitation and immunoblotting
Cells were removed from the medium after the indicated treatment, washed once with ice-cold PBS, and lysed in an ice-cold RIPA lysis buffer system (#sc-24948, Santa Cruz Biotechnology) with 1 mM Na3VO4, 5 mM NaF, and 1x protease inhibitor cocktail (#11836153001, Roche). The cell lysates were then sonicated at 15% amplitude for 15 s. After sonication, the cell lysates were incubated at 4°C with continuous rotation for 1 h and subsequently centrifuged at maximum speed for 10 min to collect the supernatant. The protein concentration in the supernatant was measured by the Bradford protein assay (#5000201, BIO-RAD) according to the manufacturer’s instructions. Equal amounts of protein were used for further analysis. All antibodies were diluted at a 1:1000 ratio for immunoblotting. For immunoprecipitation, 0.5–1 mg of protein was incubated with 20 μl of antibody-conjugated agarose (Santa Cruz Biotechnology) at 4°C for 24 h. After washing five times with lysis buffer, the protein complex was eluted with SDS sample buffer. For immunoblotting, 5–20 μg of protein were loaded. For immunoblotting of immunoprecipitated complexes, horseradish peroxidase (HRP)-conjugated antibodies were used to avoid non-specific detection of immunoglobulin in the immunoprecipitated samples. HRP-conjugated p53 (#sc-126HRP), PDK1 ((#sc-17765HRP), and Sin1 (#sc-393166HRP) antibodies were purchased from Santa Cruz Biotechnology. Immunoblots were developed by Odyssey Imaging System (LI-COR Biosciences) and the intensity of protein bands was quantified using ImageJ. The unsaturated exposure of immunoblot images was used for quantification with the appropriate loading controls as standards. Statistical analysis of the data was performed with Microsoft Excel, using data from at least three independent experiments.
Fluorescent IP-WB
Cells were lysed in a RIPA lysis buffer system after the indicated treatment and quantified for protein concentration as described above. For endogenous p53 or FLAG-tagged p53 immunoprecipitation, 0.5–1 mg of cell lysates were incubated with 20 μl anti-p53 (#sc-126 AC, Santa Cruz Biotechnology) or anti-FLAG (#sc-166355AC, Santa Cruz Biotechnology) mouse monoclonal IgG antibody-conjugated agarose at 4°C for 24 h. Normal immunoglobulin (IgG)-conjugated agarose was used as a negative control (#sc-2343, Santa Cruz Biotechnology). After washing 5 times with PBST (PBS with 0.1% Tween 20), the protein complex was eluted with SDS sample buffer. The sample was then boiled at 95°C for 10 min. For immunoblotting, 5–20 μg of protein were loaded. The protein complexes associated with p53 were resolved by SDS-PAGE and transferred onto a PVDF membrane (#IPVH00010, MilliporeSigma). The membrane was blocked with 3% BSA in PBS for 1 h at room temperature. For double fluorescent IP-WB detecting p53-PI4,5P2/PI3,4,5P3 complex, anti-p53 rabbit monoclonal IgG antibody (clone 7F5, #2527, Cell Signaling) at 1:2000 dilution and anti-PI4,5P2 mouse monoclonal IgM antibody (#Z-P045, Echelon Biosciences) or PI3,4,5P3 mouse monoclonal IgM antibody (#Z-P345, Echelon Biosciences) at 1:2000 dilution were mixed together in blocking buffer with 0.02 % Sodium Azide and incubated with the membrane at 4°C overnight. The next day, the membrane was washed three times with PBST for 10 min each time. For the secondary antibody incubation, goat anti-rabbit IgG antibody conjugated with IRDye 800CW fluorophore (#926-32211, LI-COR) detectable on the 800 nm wavelength channel of the Odyssey Fc Imaging System (LI-COR Biosciences) and goat anti-mouse IgM antibody conjugated with IRDye 680RD fluorophore (#926-68180, LI-COR) detectable on the 700 nm wavelength channel at 1:10000 dilution were mixed together in blocking buffer with 0.01 % SDS and 0.1% Tween 20 and incubated with the membrane at room temperature for 2 h. The membrane was then washed three times with PBST for 10 min each time. The images were subsequently acquired using the 700 and 800 nm wavelength channels simultaneously on the Odyssey Fc Imaging System (LI-COR Biosciences). The p53-associated PI4,5P2/PI3,4,5P3 complex was visualized by overlapping the 700 and 800 nm channels. For triple fluorescent IP-WB, Alexa fluor 594 Dye (detectable on the 600 nm channel), IRDye 680RD (detectable on the 700 nm channel), and IRDye 800CW (detectable on the 800 nm channel) conjugated antibodies were used together to visualize the complex by overlapping the 600, 700, and 800 nm channels. Statistical analysis of the data was performed with Microsoft Excel, using data from at least three independent experiments.
In vitro binding assay
His-tagged p53 protein was expressed in BL-21(DE3) E. coli (#EC0114, Thermo Fisher Scientific), lysed with 1% Brij58, sonicated, and purified with Ni-NTA-agarose (#166038887, Qiagen) as previously described19. The eluates were buffer-exchanged into PBS using a dialysis cassette (#66380, Thermo Fisher Scientific), flash-frozen, and stored at −80 °C. Recombinant Akt1 protein was purchased from Abcam (#ab79792). The binding assay was performed in PBST by incubating a constant amount of His-tagged p53 with an increasing amount of Akt1 in the presence of 20 μl anti-p53 antibody-conjugated agarose (#sc-126AC, Santa Cruz). After incubating overnight at 4°C, unbound proteins were removed by washing three times with PBST, and the protein complex was analyzed by immunoblotting.
Microscale Thermophoresis (MST) Assay
The MST assay was used to measure the binding affinity of purified recombinant proteins in vitro as described previously68. The target protein was fluorescently labeled by Monolith Protein Labeling Kit RED-NHS 2nd Generation (#MO-L011, Nano Temper) following the manufacturer’s instruction. A sequential titration of unlabeled ligand proteins, PI-PolyPIPosomes, or PI-micelles was made in a Tris-based MST buffer containing 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 80 mM KCl, and 0.05% Tween-20 and mixed with an equal volume of fluorescently labeled target protein prepared at 10 nM concentration in the same MST buffer, making the final target protein at a constant concentration of 5 nM and the ligand protein or lipid as the gradient. The PI-PolyPIPosomes for MST were purchased from Echelon Biosciences, including PI PolyPIPosomes (#Y-P000), PI4,5P2 PolyPIPosomes (#Y-P045), and PI3,4,5P3 PolyPIPosomes (#Y-P039). The synthetic PIs for MST were also purchased from Echelon Biosciences, including PI diC16 (#P-0016), PI4,5P2 diC16 (#P-4516), and PI3,4,5P3 diC16 (#P-3916), which were dissolved in the MST buffer with 5 min sonication to prepare the PI-micelles. The target-ligand mixtures were loaded into Monolith NT.115 Series capillaries (#MO-K022, Nano Temper) and the MST traces were measured by Monolith NT.115 pico, and the binding affinity was auto-generated by MO. Control v1.6 software.
Immunofluorescence (IF), Confocal and STED Microscopy
For immunofluorescence studies, cells were grown on coverslips coated with 0.2 % gelatin (#G9391, MilliporeSigma). Cells were fixed with 4% paraformaldehyde (PFA) (#sc-281692, Santa Cruz Biotechnology) for 20 min at room temperature followed by washing three times with PBS. Next, the cells were permeabilized with 0.3% Triton-X100 for 10 min and rewashed three times with PBS. The cells were then blocked with 3% BSA in PBS for one hour at room temperature. After blocking, cells were incubated with a primary antibody overnight at 4°C. The cells were then washed three times with PBS and incubated with fluorescent-conjugated secondary antibodies (Molecular Probes) for 1 hour at room temperature. For STED super-resolution microscopy, secondary antibodies conjugated with Abberior® STAR RED/580 dyes were used at 1:200 dilution (#41699 and #52405, MilliporeSigma). After secondary antibody incubation, the cells were washed three times with PBS and nuclei counterstained with 1 μg/ml 4’,6-diamidino-2-phenylindole (DAPI) (#D3571, Invitrogen) in PBS for 30 min at room temperature. The cells were subsequently washed three times with PBS and mounted in Prolong™ Glass Antifade Mountant media (#P36984, Thermo Fisher Scientific). The images were taken by Leica SP8 3xSTED Super-Resolution Microscope, which is both a point scanning confocal and 3xSTED super-resolution microscope. The Leica SP8 3xSTED microscope was controlled by LASX software (Leica Microsystems). All images were acquired using the 100X objective lens (N.A. 1.4 oil). The z-stack images were taken with each frame over a 0.2 μm thickness. For quantification, the mean fluorescent intensity of channels in each cell was measured by LASX. The colocalization of double staining channels was quantified by LASX using Pearson’s correlation coefficient (Pearson’s r), ranging between 1 and −1. A value of 1 represents perfect correlation, 0 means no correlation, and −1 means perfect negative correlation69. Pearson’s r greater than 0.7 suggests a strong correlation51. The quantitative graph was generated by GraphPad Prism. The images were processed using ImageJ.
Proximity Ligation Assay (PLA)
PLA was utilized to detect in situ protein-protein/PI interaction as previously described19, 69, 70. After fixation and permeabilization, cells were blocked before incubation with primary antibodies as in routine IF staining. The cells were then processed for PLA (#DUO92101, MilliporeSigma) according to the manufacturer’s instruction and previously published69, 70. The slides post-PLA were further processed for immunofluorescent staining against the nuclear membrane marker (Lamin A/C, #8617, Cell Signaling). The slides were mounted with Duolink® In Situ Mounting Medium with DAPI (#DUO82040, MilliporeSigma). The Leica SP8 confocal microscope detected PLA signals as discrete punctate foci and provided the intracellular localization of the complex. ImageJ was used to quantify the nuclear PLA foci.
MTT assay
In 96-well plates, 5×104 cells/well were transfected with control siRNAs or siRNAs targeting p53, PIPKIα, IPMK, and PTEN for 48 h. The cells were then treated with the control vehicle or 30 μM cisplatin for 20 h. Next, the cells were incubated with 100 μl of fresh medium plus 10 μl of the 12 mM MTT stock solution from the Vybrant® MTT cell proliferation assay kit (#V13154, Thermo Fisher Scientific) in the presence of the control vehicle or 30 μM cisplatin. After incubation for 4 hours at 37°C, all but 25 μl of the medium was removed from each well and 50 μl of DMSO was added. The mixture was incubated at 37°C for 10 min, and the absorbance was read at 540 nm using a Synergy HTX Multi-Mode Microplate reader (BioTek Instruments, Inc.).
Crystal Violet Cell Viability assay
In 96-well plates, 5×104 cells/well of MDA-MB-231, HS578T, or MDA-MB-468 cells were transfected with control siRNAs or siRNAs targeting p53, PIPKIα, IPMK, and PTEN for 48 h. A549 p53 KO cells were transfected with Flag-tagged p53 constructs for 24h. The cells were then treated with the control vehicle or 30 μM cisplatin for 24 h. The cells were then fixed with 4% PFA and stained with 0.2% Crystal Violet. After washing the cells 3 times with PBS, the dye was extracted from the cells with 10% acetic acid and quantified by measuring the absorbance at 570 nm using a Synergy HTX Multi-Mode Microplate reader.
Cell Death Detection ELISA
In 96-well plates, 5×104 cells/well were transfected with control siRNAs or siRNAs targeting p53, PIPKIα, IPMK, and PTEN for 48 h. The cells were then treated with the control vehicle or 30 μM cisplatin for 24 h. Next, the cells were lysed and processed for cell death detection ELISA assay by following the manufacturer’s instruction (#11774425001, MilliporeSigma). At the end of the assay, the absorbance was read at 405 nm using a Synergy HTX Multi-Mode Microplate reader.
Caspase-3 Activity Assay
In 6-well plates, 1×106 cells/well were transfected with control siRNAs or siRNAs targeting p53, PIPKIα, IPMK, and PTEN for 48 h. The cells were then treated with the control vehicle or 30 μM cisplatin for 24 h. Next, the cells were lysed and processed for Caspase-3 activity assay by following the manufacturer’s instruction (#E-13183, Thermo Fisher Scientific). At the end of the assay, the fluorescence was read at excitation/emission 342/441 nm using a Synergy HTX Multi-Mode Microplate reader.
Luciferase Assay
A549 p53 KO cells were co-transfected with constructs expressing wild-type, mutant p53, and or p53 6Q or R379Q mutants with FOXO3 responsive element with luciferase readout (FHRE-Luc) (#1789, Addgene) for 48 h. The cells were then lysed, and the cell lysates were mixed with the luciferase assay reagent to measure the light produced by a Synergy HTX Multi-Mode Microplate reader following the luciferase assay system manufacturer’s instructions (#E1500, Promega).
Transwell Invasion Assay
The bottom polycarbonate filter surface of a 6.5 mm diameter insert with 8 μm pores in a 24-well plate (#3422, Corning) was coated with 10 μg/ml of Laminin (#CC095, MilliporeSigma) diluted in PBS for 3 h at 37°C. MDA-MB-231 cells were transiently transfected with control siRNAs or siRNAs targeting p53, PIPKIα, IPMK, and PTEN, then serum-starved for 24 h. A549 p53 KO cells were transiently transfected with mutant p53 (R175H or R248Q) or their corresponding 6Q or R379Q mutants, and then serum-starved for 24 h. Next, 5×104 transfected cells were suspended in 200 μl serum-free medium containing 0.5% BSA and then were plated in the upper insert chamber in 500 μl serum-free medium with 0.5% BSA. 10 ng/ml EGF was added to the lower chamber. Cells were allowed to invade for 16 h at 37°C. Cells on the bottom of the filter were then fixed with 4% PFA and stained with 0.2% Crystal Violet. The stained cells were imaged using a Leica MC170 HD microscope controlled by the LAS EZ software (Leica). At the end of the assay, the dye was extracted by 10% acetic acid from the cells and quantified by measuring the optical density at 570 nm using a Synergy HTX Multi-Mode Microplate reader.
Statistics and Reproducibility
Two-tailed unpaired t-tests were used for pair-wise significance unless otherwise indicated. We note that no power calculations were used. Sample sizes were determined based on previously published experiments where significant differences were observed 19. Each experiment was repeated at least three times independently, and the number of repeats is defined in each figure legend. We used at least three independent experiments or biologically independent samples for statistical analysis.
Resource and Data Availability
All data supporting the findings of this study are available from the corresponding authors on reasonable request.
Extended Data
Extended Data Fig. 1. p53 associates with stress-induced active Akt in the nucleus.
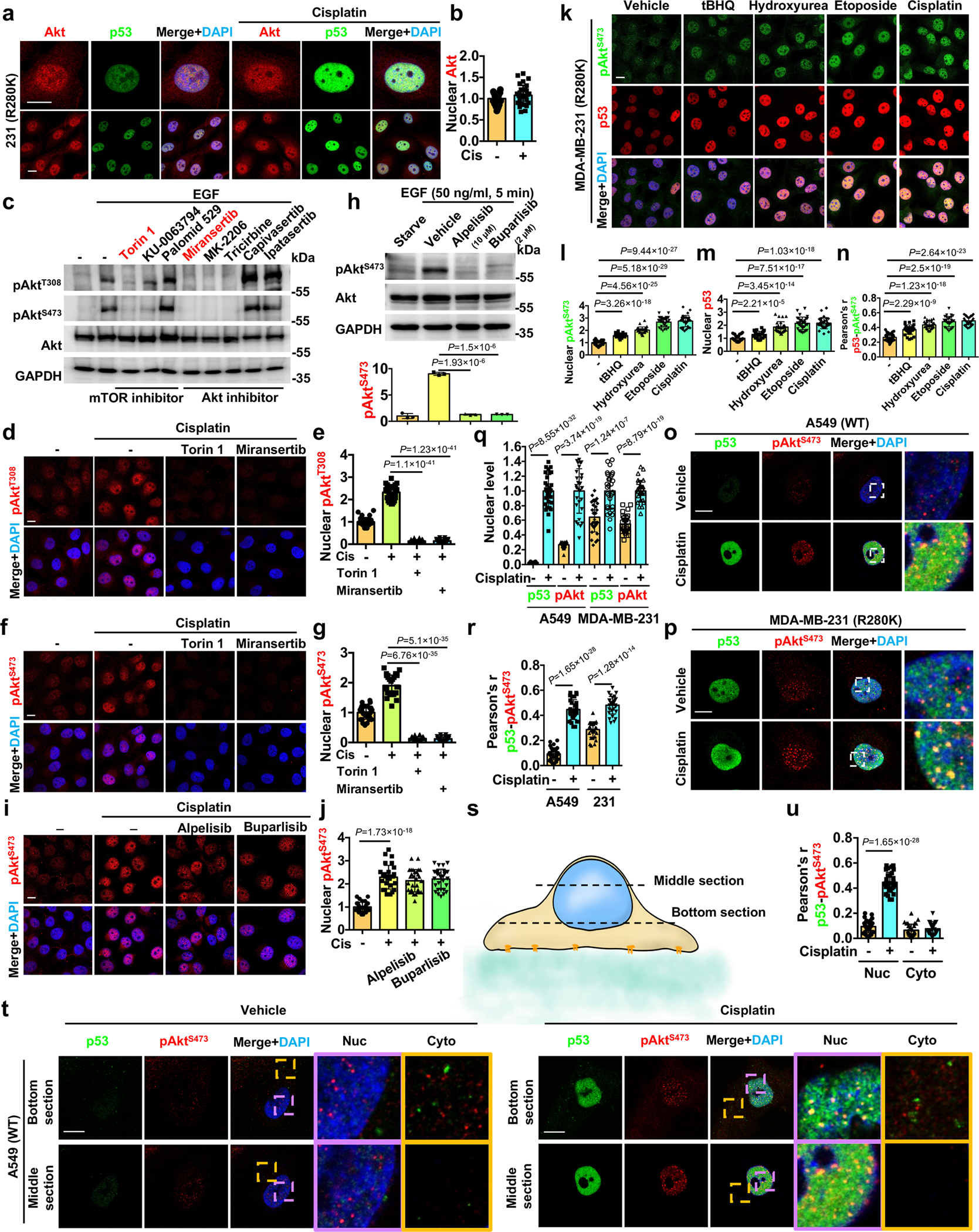
a-b, Confocal images of IF staining against Akt and p53 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear levels of Akt (b) normalized to vehicle treated cells were quantified. N=30 cells from representative experiments of three repeats.
c, WB analysis of pAktT308/S473 in MDA-MB-231 cells after 24 h starvation and treatment with 10 μM mTOR inhibitor Torin 1/KU-0063794/Palomid 529 or Akt inhibitor Miransertib/MK-2206/Tricirbine/Capivasertib/Ipatasertib, followed by 5 min stimulation with 50 ng/ml EGF. Representative images of three independent experiments are shown.
d-g, Confocal IF staining of pAktT308/S473 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h with or without the mTOR inhibitor Torin 1 (10 μM) or the Akt inhibitor Miransertib (10 μM). The nuclear pAktT308/S473 levels normalized to vehicle treated cells were quantified (e,g). N=30 cells from representative experiments of three repeats.
h, WB analysis of pAktS473 in MDA-MB-231 cells after 24 h starvation and treatment with 10 μM PI3Kα inhibitor alpelisib, 2 μM pan-PI3K inhibitor buparlisib, or control vehicle, followed by 5 min stimulation with 50 ng/ml EGF. N=3.
i-j, Confocal IF staining of pAktS473 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h with or without the presence of the PI3Kα inhibitor alpelisib (10 μM) or the pan-PI3K inhibitor buparlisib (2 μM). The nuclear pAktS473 levels normalized to vehicle treated cells were quantified (j). N=30 cells from representative experiments of three repeats.
k-n, Confocal IF staining of p53 and pAktS473 in MDA-MB-231 cells treated with 100 μM tBHQ, 100 μM hydroxyurea, 100 μM etoposide, 30 μM cisplatin, or control vehicle for 24 h. The nuclear levels of pAktS473 (l) and p53 (m) normalized to vehicle treated cells and their colocalization (n) were quantified. N=30 cells from representative experiments of three repeats.
o-r, Confocal IF staining of p53 and pAktS473 in A549 and MDA-MB-231 cells treated with 30 μM cisplatin or control vehicle for 24 h. The nuclear levels of p53 and pAktS473 normalized to cisplatin treated cells (q) and their colocalization (r) were quantified. N=30 cells from representative experiments of three repeats.
s-u, Two-panel section of confocal IF staining against p53 and pAktS473 in A549 cells treated with 30 μM cisplatin or control vehicle for 24 h. The middle section focusing on the nucleus and the bottom section focusing on the cytosol are indicated in the model (s). The nuclear and cytosolic colocalization of p53 and pAktS473 was quantified (u). N=30 cells from representative experiments of three repeats.
For all panels, data are represented as mean ±SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 2. p53 associates with the stress-induced nuclear Akt pathway.
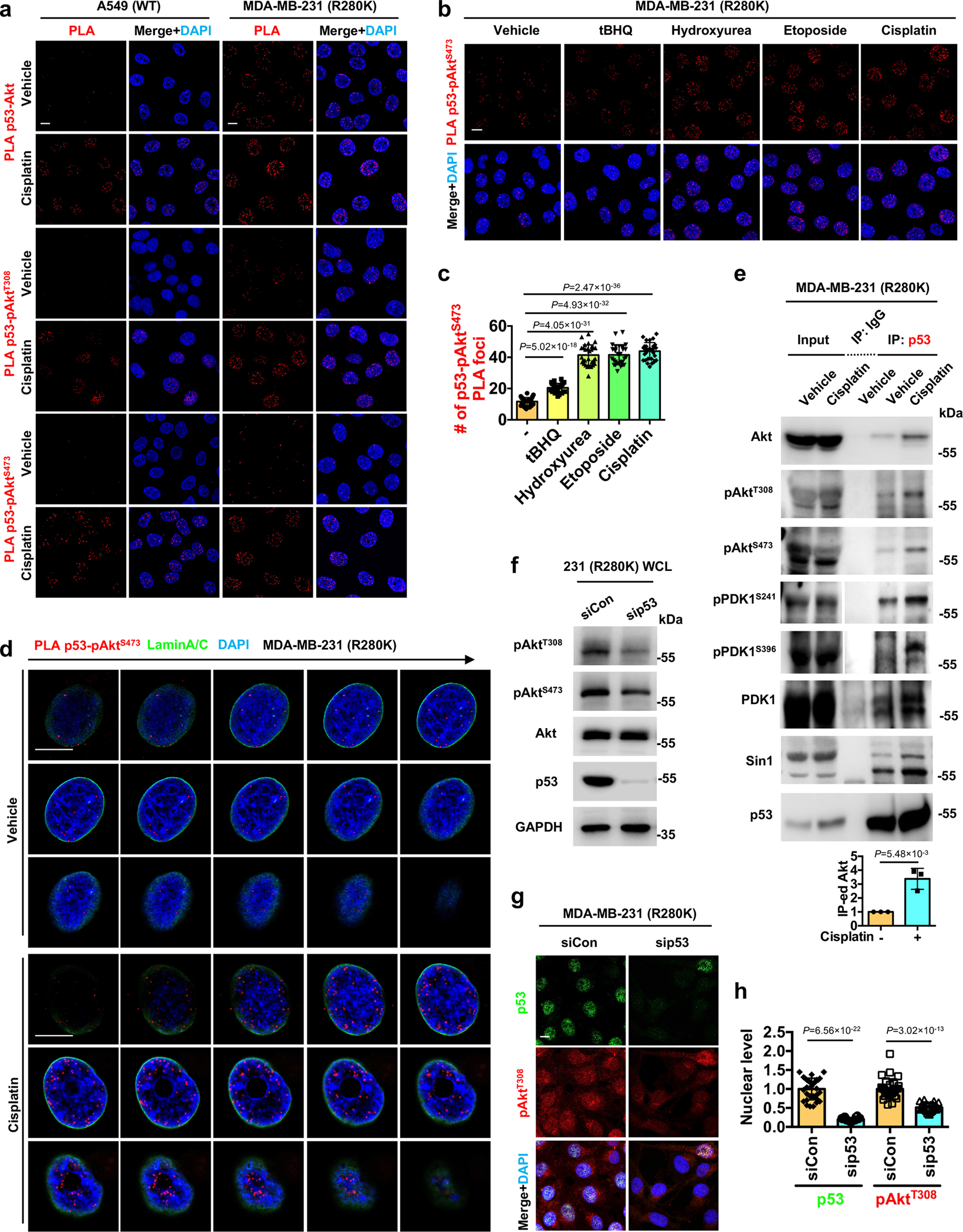
a, PLA of p53-Akt/pAktT308/pAktS473 in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. See quantification in Fig. 1g.
b-c, PLA of p53-pAktS473 in MDA-MB-231 cells treated with 100 μM tBHQ, 100 μM hydroxyurea, 100 μM etoposide, 30 μM cisplatin, or control vehicle for 24 h. The nuclear PLA foci were quantified (c). N=30 cells from representative experiments of three repeats.
d, 3D section of PLA between p53 and pAktS473 overlaid with Lamin A/C in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. Each frame of the 3D sections was over a 0.2 μm thickness.
e, Co-IP of p53 and Akt pathway components from MDA-MB-231 cells treated with 30 μM cisplatin or control vehicle for 24 h. N=3.
f, MDA-MB-231 cells were transient-transfected with control siRNAs or siRNAs against p53 for 48 h. Then the cell lysates were analyzed by WB for p53/Akt/pAktT308/pAktS473. Representative data of three independent experiments are shown.
g-h, Confocal IF staining of p53 and pAktT308 in MDA-MB-231 cells 48 h after transient transfection with control siRNAs or siRNAs against p53. The nuclear p53 and pAktT308 levels normalized to siCon transfected cells were quantified (h). N=30 cells from representative experiments of three repeats.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 3. p53 associates with PDK1, FOXOs, and IPMK.
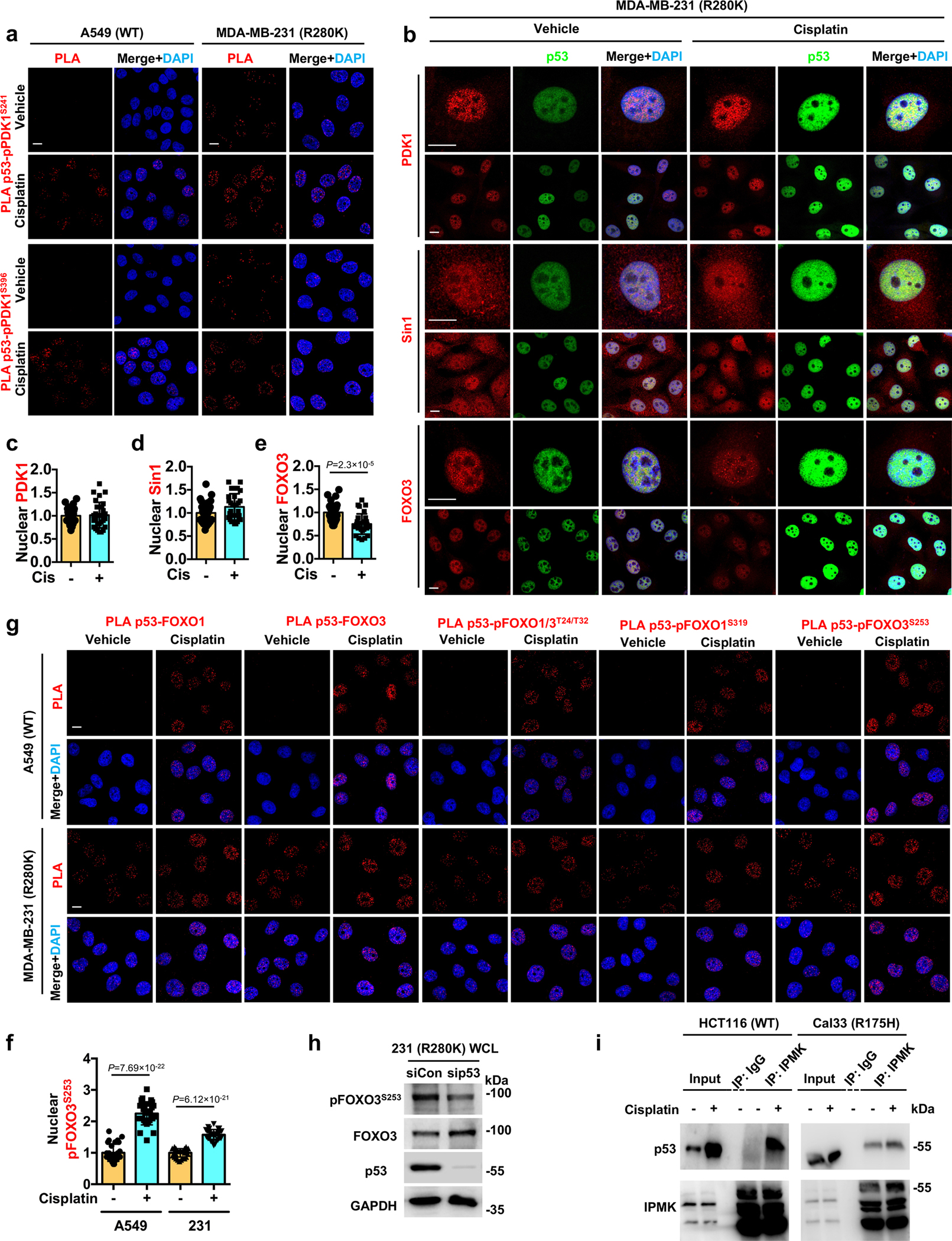
a, PLA of p53-pPDK1S241/pPDK1S396 in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. See quantification in Fig. 2e.
b-e, Confocal images of IF staining against PDK1/Sin1/FOXO3 and p53 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear levels of PDK1 (c), Sin1 (d), FOXO3 (e) normalized to vehicle treated cells were quantified. N=30 cells from representative experiments of three repeats.
f, Quantification of nuclear pFOXO3S253 in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h (levels normalized to vehicle treated cells). N=30 cells from representative experiments of three repeats. Representative images are shown in Fig. 3e.
g, PLA of p53-FOXOs in A549 and MDA-MB-231 cells treated with vehicle or 30 μM cisplatin for 24 h. See quantification in Fig. 3g.
h, MDA-MB-231 cells were transient transfected with control siRNAs or siRNAs against p53 for 48 h. Then the cell lysates were analyzed by WB for p53/FOXO3/pFOXO3S253. Representative data of three independent experiments are shown.
i, Co-IP of IPMK with wild-type and mutant p53 from HCT116 and Cal33 cells respectively treated with 30 μM cisplatin or control vehicle for 24 h. Representative data of three independent experiments are shown.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 4. p53 associates with PI4,5P2 and PI3,4,5P3 in the nucleus.
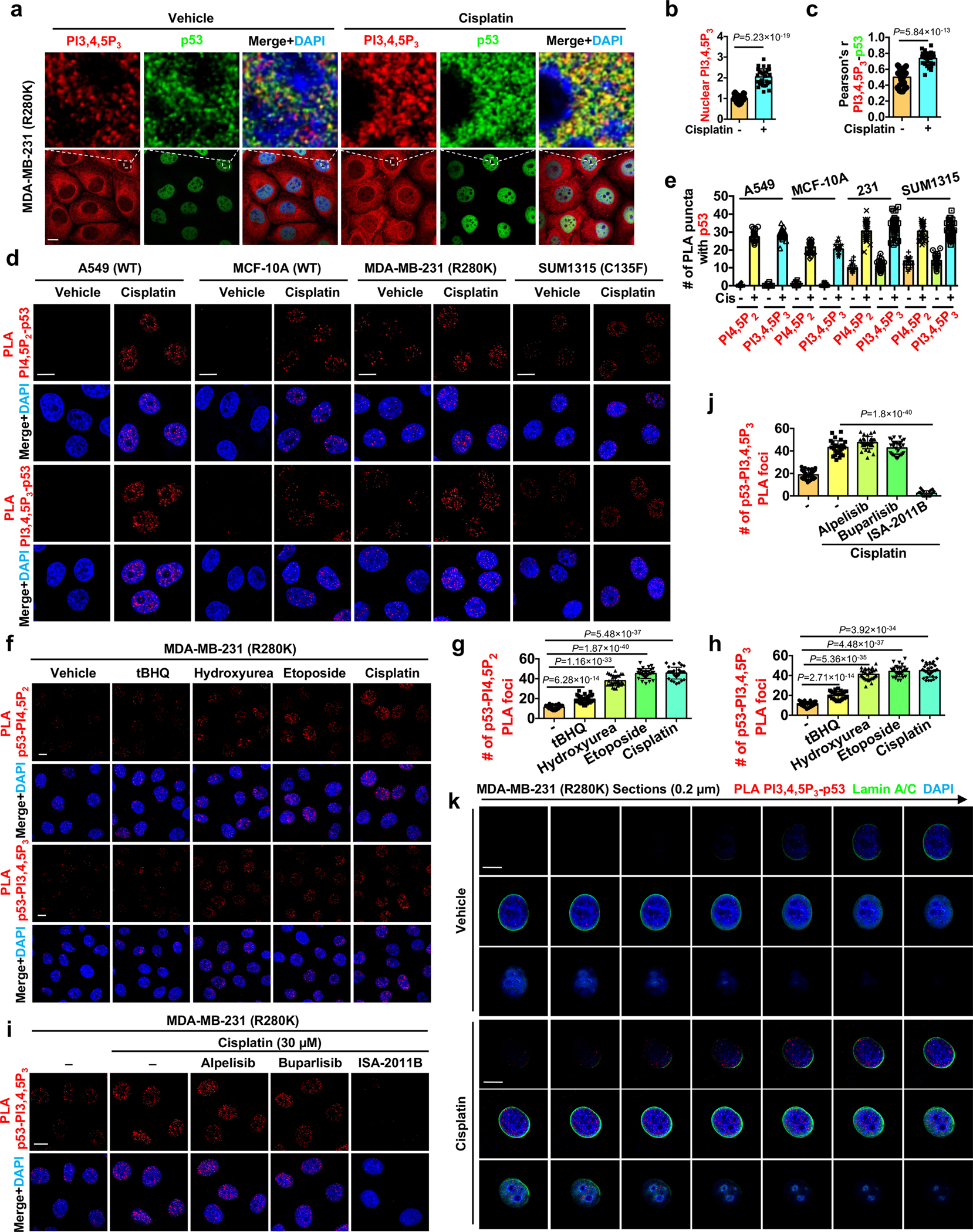
a-c, Confocal IF staining against p53 and PI3,4,5P3 in MDA-MB-231 cells treated with 30 μM cisplatin or control vehicle for 24 h. The nuclear level of PI3,4,5P3 normalized to vehicle treated cells (b) and its colocalization with p53 (c) were quantified. N=30 cells from representative experiments of three repeats.
d-e, PLA of p53-PI4,5P2/PI3,4,5P3 in A549, MCF-10A, MDA-MB-231, and SUM1315 cells treated with 30 μM cisplatin or control vehicle for 24 h. The nuclear PLA foci were quantified (e). N=30 cells from representative experiments of three repeats.
f-h, PLA of p53-PI4,5P2/PI3,4,5P3 in MDA-MB-231 cells treated with 100 μM tBHQ, 100 μM hydroxyurea, 100 μM etoposide, 30 μM cisplatin, or control vehicle for 24 h. The nuclear PLA foci were quantified (g,h). N=30 cells from representative experiments of three repeats.
i-j, PLA of p53-PI3,4,5P3 in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin with or without the presence of the PI3Kα inhibitor alpelisib (10 μM), the pan-PI3K inhibitor buparlisib (2 μM), or the PIPKIα inhibitor ISA-2011B (50 μM) for 24 h. The nuclear PLA foci were quantified (j). N=30 cells from representative experiments of three repeats.
k, 3D section of PLA between p53 and PI3,4,5P3 overlaid with Lamin A/C in MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin treatment for 24 h. Each frame of the 3D sections was over a 0.2 μm thickness.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 5. IPMK and PTEN associate with p53 and regulate the interconversion of nuclear p53-bound PI4,5P2 and PI3,4,5P3 and FOXO phosphorylation.
a-c, Confocal IF staining against p53 and IPMK in MDA-MB-231 cells treated with 30 μM cisplatin or control vehicle for 24 h. The nuclear level of IPMK normalized to vehicle treated cells (b) and its colocalization with p53 (c) were quantified. N=30 cells from representative experiments of three repeats.
d-g, Confocal IF staining against p53 and PTEN in A549 or MDA-MB-231 cells treated with 30 μM cisplatin or control vehicle for 24 h. The nuclear level of PTEN normalized to vehicle treated cells (f) and its colocalization with p53 (g) were quantified. N=10 cells from representative experiments of three repeats.
h-j, PLA of p53-IPMK/PTEN in A549 and MDA-MB-231 cells treated with control vehicle or 30 μM cisplatin for 24 h. The nuclear PLA foci were quantified (i,j). N=30 cells from representative experiments of three repeats.
k-l, PLA of p53-PI4,5P2/PI3,4,5P3 in MDA-MB-231 cells 48 h after of transient transfection with control siRNAs or siRNAs against PIPKIα, IPMK, and PTEN (k). See quantification in Fig. 4h,i. The knockdown efficiency was validated by WB (l).
m-n, Confocal IF staining of p53 and pFOXO3S253 in MDA-MB-231 cells 48 h after transient transfection with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. The nuclear pFOXO3S253 levels normalized to siCon transfected cells were quantified (n). N=30 cells from representative experiments of three repeats.
o-p, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for PLA between p53 and pFOXO3S253. The nuclear PLA foci were quantified (p). N=30 cells from representative experiments of three repeats.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 6. The nuclear p53-PI signalosome regulates DNA damage repair and cell survival.
a, Confocal IF staining against PI4,5P2 and pPDK1S396/pFOXO3S253 in MDA-MB-231 cells treated with 30 μM cisplatin or control vehicle for 24 h. See Fig. 6a,b for the nuclear region and quantification of the nuclear colocalization of PI4,5P2 and pPDK1S396/pFOXO3S253.
b, Confocal and STED super-resolution images of IF staining against p53, pAktS473, and the DNA DSB marker Ɣ-H2A.X in MDA-MB-231 and A549 cells treated with 30 μM cisplatin or control vehicle for 24 h. See quantification in Fig. 6d.
c-f, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, and IPMK. 24 h later, cells were treated with 30 μM cisplatin, 10 μM mTOR inhibitor Torin1, 10 μM Akt inhibitor Miransertib or control vehicle for 24 h before being processed for IF staining of p53 and pDNA-PKS2056/phosphor-p300S1834. The nuclear levels of p53/pDNA-PKS2056/phosphor-p300S1834 normalized to vehicle treated cells were quantified (d,f). N=30 cells from representative experiments of three repeats.
g-i, MDA-MB-231, HS578T, MDA-MB-468 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for MTT cell viability assay. Cell viability was normalized to siCon transfected cells. N=3 from three independent experiments, each in triplicate.
j, MDA-MB-231 cells were transfected with control siRNAs or siRNAs against p53, PIPKIα, IPMK, and PTEN. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for 24 h before being processed for Caspase-3 activity assay. Activity was normalized to siCon transfected cells. N=3.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 7. p53 interaction with Akt requires the p53 C-terminal PIPn-binding motif and correlates with nuclear Akt activation.
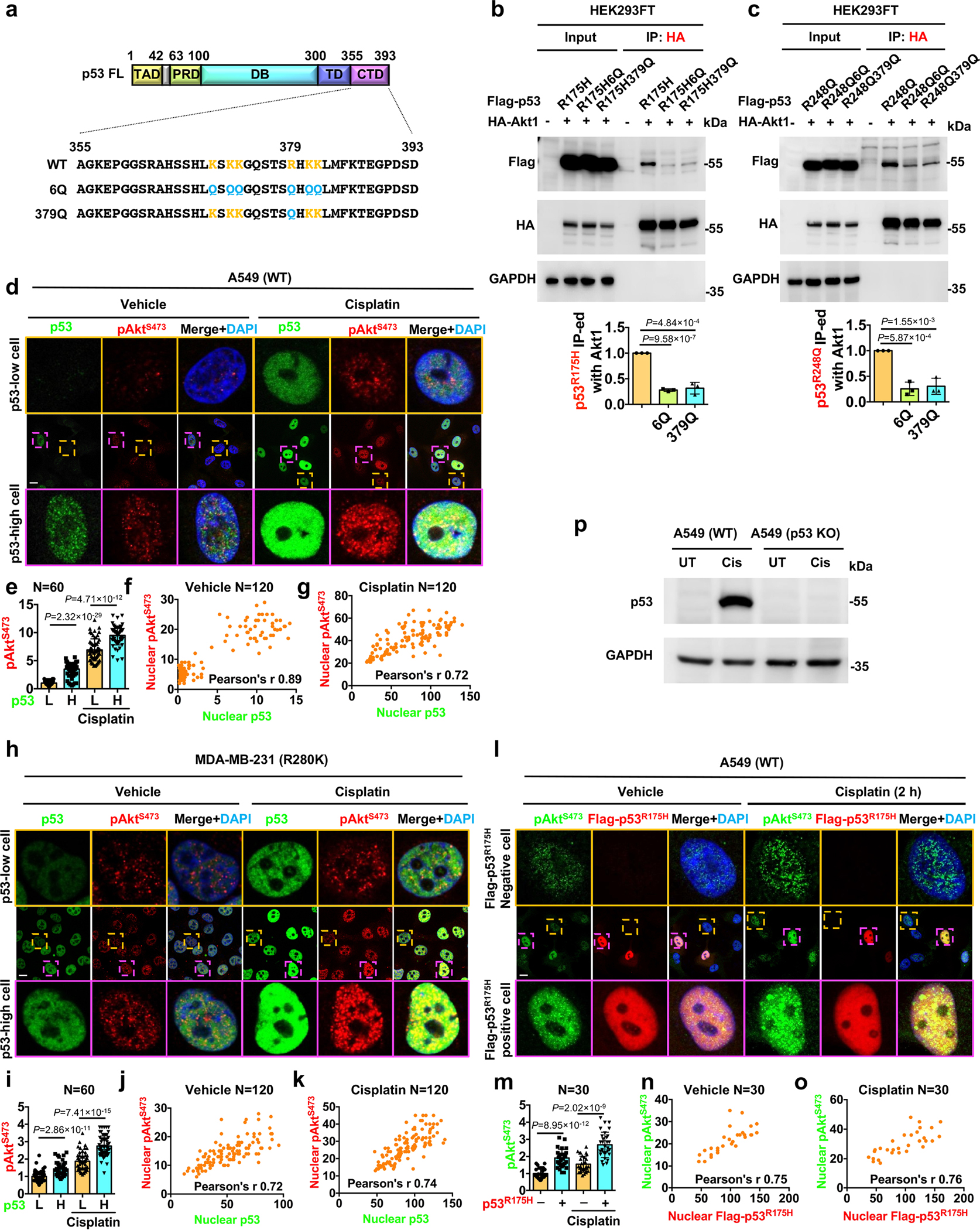
a, Schematic representation of p53 domains (top). 1 or 6 basic residues in the CTD were mutated to glutamine (Q) to generate the PIPn-binding defective mutant 6Q or 379Q, respectively. TAD, transactivation domain; PRD, proline-rich domain; DBD, DNA-binding domain; TD, tetramerization domain; CTD, C-terminal regulatory domain.
b-c, The interaction of Akt with mutant p53 is regulated by the PIPn-binding motif of p53. HA-tagged Akt1 and FLAG-tagged mutant p53R175H, p53R248Q, or their corresponding 6Q or 379Q mutants, were co-transfected into HEK293FT cells for 48 h. The ectopically expressed Akt1 was IP-ed with HA antibody, and then analyzed by WB. N=3.
d-g, A549 cells were treated with 30 μM cisplatin or control vehicle for 24 h and then were fixed and processed for confocal IF staining of p53 and pAktS473. The nuclei were counter-stained by DAPI. The nuclear levels of p53 and pAktS473 were quantified. The low (L) and high (H) levels of nuclear p53 were determined based on the average p53 levels in the corresponding control vehicle- or cisplatin-treated groups (e). The correlation between nuclear p53 and pAktS473 was determined by Pearson’s r (f,g). N=120 cells from representative experiments of three repeats.
h-k, MDA-MB-231 cells were treated with 30 μM cisplatin or control vehicle for 24 h and then fixed and processed for IF staining against p53 and pAktS473. The nuclei were counter-stained by DAPI. The nuclear levels of p53 and pAktS473 were quantified. The low (L) and high (H) levels of nuclear p53 were determined based on the average of the p53 levels in the corresponding control vehicle or cisplatin-treated groups (i). The correlation between the nuclear p53 and pAktS473 was determined by Pearson’s r (j,k). N=120 cells from representative experiments of three repeats.
l-o, A549 cells were transiently transfected with Flag-tagged mutant p53R175H and then 24 h later treated with 30 μM cisplatin or control vehicle for 4 h. The cells were then fixed and processed for IF staining of Flag-tagged mutant p53 and pAktS473. The nuclear pAktS473 levels in Flag-tagged mutant p53R175H-negative and -positive cells were quantified (m). The correlation between the nuclear levels of Flag-tagged mutant p53R175H and pAktS473 was determined by Pearson’s r (n,o). N=30 cells from representative experiments of three repeats.
p, A549 wild-type and p53 KO cells were treated with 30 μM cisplatin or control vehicle for 24 h. The p53 KO was validated by WB.
For all panels, data are represented as mean ± SD, p value denotes t-test. Scale bar: 5 μm.
Extended Data Fig. 8. Stress-induced Akt, DNA-PK, and p300 phosphorylation by p53 is dependent on the PIPn-binding motif of p53.
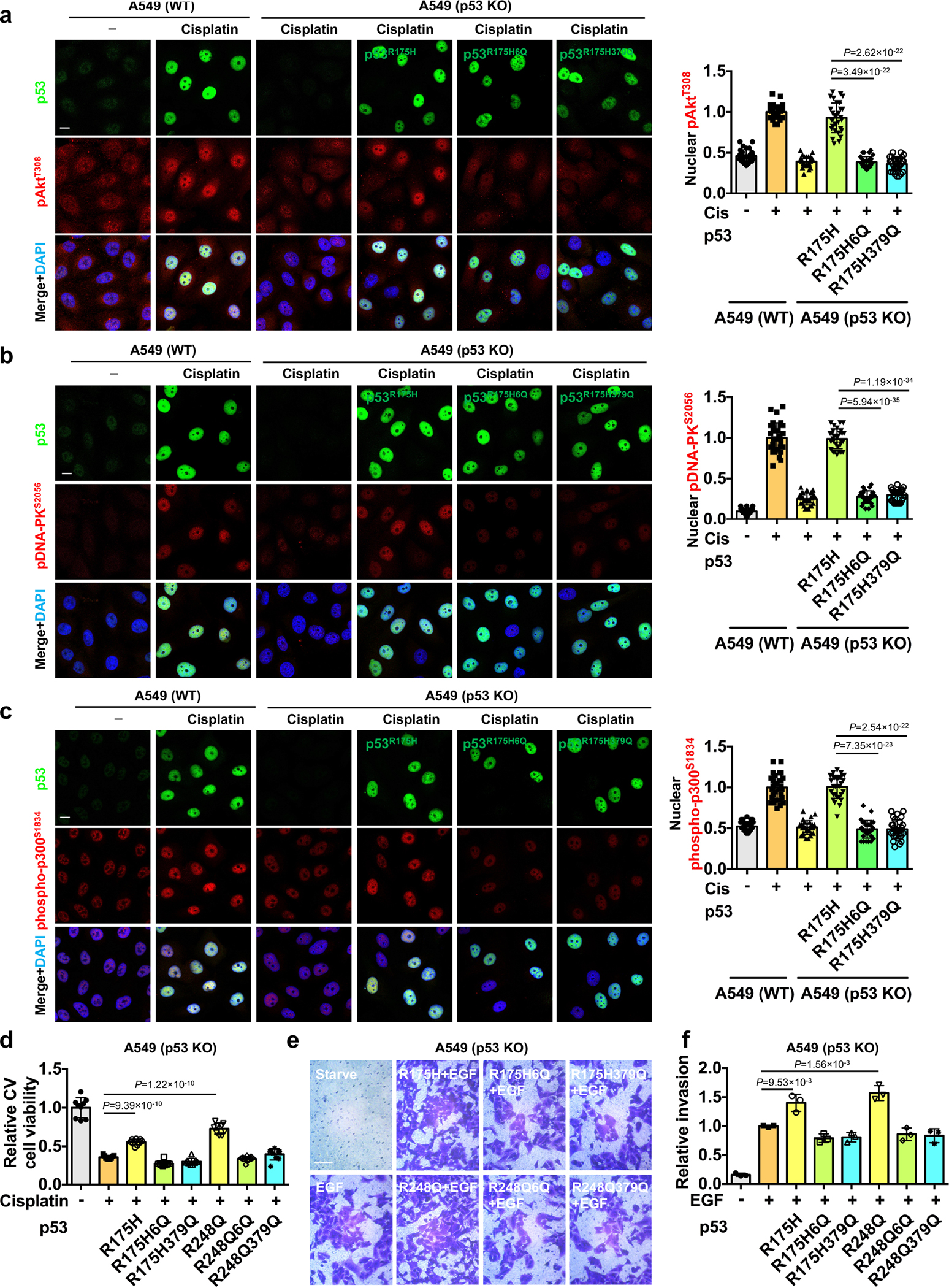
a, A549 p53 KO cells were transiently transfected with Flag-tagged mutant p53R175H or its corresponding 6Q or 379Q mutant for 24 h and then treated with 30 μM cisplatin for an additional 24 h. A549 WT cells were treated with 30 μM cisplatin or control vehicle for 24 h. The cells were then fixed and processed for IF staining of p53 and pAktT308. The nuclear pAktT308 levels in A549 WT cells, p53 KO cells, and Flag-tagged mutant p53-positive cells normalized to A549 WT cells treated with cisplatin were quantified as indicated. N=30 cells from representative experiments of three repeats. Scale bar: 5 μm.
b, A549 p53 KO cells were transiently transfected with Flag-tagged mutant p53R175H or its corresponding 6Q or 379Q mutant for 24 h and then treated with 30 μM cisplatin for an additional 24 h. A549 WT cells were treated with 30 μM cisplatin or control vehicle for 24 h. The cells were then fixed and processed for IF staining of p53 and pDNA-PKS2056. The nuclear pDNA-PKS2056 levels in A549 wild-type cells, p53 KO cells, and Flag-tagged mutant p53-positive cells normalized to A549 WT cells treated with cisplatin were quantified as indicated. N=30 cells from representative experiments of three repeats. Scale bar: 5 μm.
c, A549 p53 KO cells were transiently transfected with Flag-tagged mutant p53R175H or its corresponding 6Q or 379Q mutant for 24 h and then treated with 30 μM cisplatin for an additional 24 h. A549 wild-type cells were treated with 30 μM cisplatin or control vehicle for 24 h. The cells were then fixed and processed for confocal IF staining of p53 and phospho-p300S1834. The nuclear phospho-p300S1834 levels in A549 wild-type cells, p53 KO cells, and Flag-tagged mutant p53-positive cells normalized to A549 WT cells treated with cisplatin were quantified as indicated. N=30 cells from representative experiments of three repeats. Scale bar: 5 μm.
d, A549 p53 KO cells were transiently transfected with Flag-tagged mutant p53R175H, p53R248Q or their corresponding 6Q or 379Q mutant. 24 h later, cells were treated with 30 μM cisplatin or control vehicle for an additional 24 h before being processed for crystal violet (CV) viability assay. Cell viability was normalized to untransfected cells treated with vehicle. N=3 from three independent experiments, each in triplicate.
e-f, A549 p53 KO cells were transiently transfected with Flag-tagged mutant p53R175H, p53R248Q or their corresponding 6Q or 379Q mutant. 24 h later, cells were serum-starved for an additional 24 h and then scored for invasion through Laminin-coated transwell inserts with 8 μm pores using a 10 ng/ml EGF chemoattractant for 16 h. The invading cells at the insert bottom were stained with crystal violet, imaged, and quantified based on the extracted dye using a plate reader. Invasion was normalized to untransfected cells treated with EGF. N=3. Scale bar: 100 μm.
For all panels, data are represented as mean ± SD, p value denotes t-test.
Extended Data Table 1.
The binding affinity of the nuclear p53-PI signalosome components with p53
| Binding partners | Binding affinity with p53 (Kd) measured by MST |
|---|---|
| Akt1 | 10±2 nM |
| PDK1 | 6±0.2 nM |
| Sin1 | 20±1 nM |
| PIPKIa | 86±34 nM |
| IPMK | 42±13 nM |
| PTEN | 124±22 nM |
| PI (PolyPIPosome) | No binding detected |
| PI4,5P2 (PolyPIPosome) | 150±12 nM |
| PI3,4,5P3 (PolyPIPosome) | 1497±150 nM |
| PI (C16 Micelles) | No binding detected |
| PI4,5P2 (C16 Micelles) | 83±7 nM |
| PI3,4,5P3 (C16 Micelles) | 670±92 nM |
The interaction of recombinant fluorescently labeled p53 and non-labeled signalosome components was quantitated by microscale thermophoresis (MST) assay. A constant concentration of fluorescently labeled p53 (target, 5 nM) was incubated with increasing concentrations of non-labeled ligands to calculate the binding affinity. The binding affinity determined by MST are shown as indicated Kd values. MST was performed using a Monolith NT.115 pico, and the binding affinity was auto-generated by MO. Control v1.6 software. Data are represented as mean ± SD; n=3.
Supplementary Material
MDA-MB-231 cells were treated with control vehicle for 24 h before being processed for PLA between p53 and pAktS473 (Red). Lamin A/C stained the nuclear envelopes (Green), and the nuclei were counter-stained with DAPI (Blue). The z-stack images were taken with a Leica SP8 confocal microscope with each frame over a 0.2 μm thickness. The video of the 3D reconstitution of a representative cell from three independent experiments was generated by LASX.
MDA-MB-231 cells were treated with 30 μM cisplatin for 24 h before being processed for PLA between p53 and pAktS473 (Red). Lamin A/C stained the nuclear envelopes (Green), and the nuclei were counter-stained with DAPI (Blue). The z-stack images were taken with a Leica SP8 confocal microscope with each frame over a 0.2 μm thickness. The video of the 3D reconstitution of a representative cell from three independent experiments was generated by LASX.
ACKNOWLEDGMENTS
We thank Drs. Anjon Audhya, Michael Sussman, and Wolfgang Peti for discussions and comments, and Lance Rodenkirch for technical support. This work was supported in part by a National Institutes of Health grant R35GM134955 (R.A.A.), Department of Defense Breast Cancer Research Program grants W81XWH-17-1-0258 (R.A.A.), W81XWH-17-1-0259 (V.L.C.) and W81XWH-21-1-0129 (V.L.C.), and a grant from the Breast Cancer Research Foundation (V.L.C.).
Footnotes
DECLARATION OF INTERESTS
Authors declare that they have no competing interests.
REFERENCES
- 1.Choi S et al. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol 18, 1324–1335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thapa N, Horn HT & Anderson RA Phosphoinositide spatially free AKT/PKB activation to all membrane compartments. Adv Biol Regul 72, 1–6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fruman DA et al. The PI3K Pathway in Human Disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoxhaj G & Manning BD The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer 20, 74–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokoe D et al. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277, 567–570 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Alessi DR et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7, 261–269 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Sarbassov DD, Guertin DA, Ali SM & Sabatini DM Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Ebner M, Lucic I, Leonard TA & Yudushkin I PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell 65, 416–431 e416 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Martelli AM et al. The emerging multiple roles of nuclear Akt. Biochim Biophys Acta 1823, 2168–2178 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Meier R, Alessi DR, Cron P, Andjelkovic M & Hemmings BA Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bbeta. J Biol Chem 272, 30491–30497 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Andjelkovic M et al. Role of translocation in the activation and function of protein kinase B. J Biol Chem 272, 31515–31524 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Borgatti P et al. Threonine 308 phosphorylated form of Akt translocates to the nucleus of PC12 cells under nerve growth factor stimulation and associates with the nuclear matrix protein nucleolin. J Cell Physiol 196, 79–88 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Wang R & Brattain MG AKT can be activated in the nucleus. Cell Signal 18, 1722–1731 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Bozulic L, Surucu B, Hynx D & Hemmings BA PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell 30, 203–213 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Boehme KA, Kulikov R & Blattner C p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci U S A 105, 7785–7790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller PA & Vousden KH p53 mutations in cancer. Nat Cell Biol 15, 2–8 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Kastenhuber ER & Lowe SW Putting p53 in Context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S, Chen M, Cryns VL & Anderson RA A nuclear phosphoinositide kinase complex regulates p53. Nat Cell Biol 21, 462–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu R et al. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal 6, ra22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman DJ et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 3, 117–130 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Blind RD, Suzawa M & Ingraham HA Direct modification and activation of a nuclear receptor-PIP(2) complex by the inositol lipid kinase IPMK. Sci Signal 5, ra44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blind RD et al. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc Natl Acad Sci U S A 111, 15054–15059 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y et al. Global tumor protein p53/p63 interactome: making a case for cisplatin chemoresistance. Cell Cycle 11, 2367–2379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Turner KM, Alfred Yung WK, Chen K & Zhang W Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro Oncol 16, 1313–1323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer IA & Arteaga CL The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med 67, 11–28 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Wang YH et al. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat Commun 8, 2118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casamayor A, Morrice NA & Alessi DR Phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo. Biochem J 342 ( Pt 2), 287–292 (1999). [PMC free article] [PubMed] [Google Scholar]
- 29.Moon Z, Wang Y, Aryan N, Mousseau DD & Scheid MP Serine 396 of PDK1 is required for maximal PKB activation. Cell Signal 20, 2038–2049 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Liu P et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov 5, 1194–1209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung YM et al. FOXO3 signalling links ATM to the p53 apoptotic pathway following DNA damage. Nat Commun 3, 1000 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F et al. Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53. J Mol Biol 384, 590–603 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Fu W et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem 284, 13987–14000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb AE & Brunet A FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci 39, 159–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H & Tindall DJ Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim Biophys Acta 1813, 1961–1964 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Tang N, Hadden TJ & Rishi AK Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 1813, 1978–1986 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Semenas J et al. The role of PI3K/AKT-related PIP5K1alpha and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc Natl Acad Sci U S A 111, E3689–3698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Currie RA et al. Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem J 337 ( Pt 3), 575–583 (1999). [PMC free article] [PubMed] [Google Scholar]
- 39.Calleja V et al. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol 5, e95 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M et al. The nuclear phosphoinositide response to stress. Cell Cycle 19, 268–289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barlow CA, Laishram RS & Anderson RA Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol 20, 25–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boronenkov IV, Loijens JC, Umeda M & Anderson RA Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell 9, 3547–3560 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma A, Singh K & Almasan A Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol 920, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Liu L et al. DNA-PK promotes activation of the survival kinase AKT in response to DNA damage through an mTORC2-ECT2 pathway. Sci Signal 15, eabh2290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plo I et al. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res 68, 9404–9412 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Liu P et al. Akt-mediated phosphorylation of XLF impairs non-homologous end-joining DNA repair. Mol Cell 57, 648–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu N, Lao Y, Zhang Y & Gillespie DA Akt: a double-edged sword in cell proliferation and genome stability. J Oncol 2012, 951724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toulany M et al. Akt promotes post-irradiation survival of human tumor cells through initiation, progression, and termination of DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Res 10, 945–957 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Huang WC & Chen CC Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol Cell Biol 25, 6592–6602 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu SL et al. Quantitative Lipid Imaging Reveals a New Signaling Function of Phosphatidylinositol-3,4-Bisphophate: Isoform- and Site-Specific Activation of Akt. Mol Cell 71, 1092–1104 e1095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schober P, Boer C & Schwarte LA Correlation Coefficients: Appropriate Use and Interpretation. Anesth Analg 126, 1763–1768 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Miyaguchi Y, Tsuchiya K & Sakamoto K P53 negatively regulates the transcriptional activity of FOXO3a under oxidative stress. Cell Biol Int 33, 853–860 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Cocco L et al. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem J 248, 765–770 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis WJ, Lehmann PZ & Li W Nuclear PI3K signaling in cell growth and tumorigenesis. Front Cell Dev Biol 3, 24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikani CK et al. Proliferative and antiapoptotic signaling stimulated by nuclear-localized PDK1 results in oncogenesis. Sci Signal 5, ra80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiraishi I et al. Nuclear targeting of Akt enhances kinase activity and survival of cardiomyocytes. Circ Res 94, 884–891 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Fu W & Hall MN Regulation of mTORC2 Signaling. Genes (Basel) 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thapa N et al. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4. Nat Cell Biol 22, 1357–1370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen M et al. The Specificity of EGF-Stimulated IQGAP1 Scaffold Towards the PI3K-Akt Pathway is Defined by the IQ3 motif. Sci Rep 9, 9126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzivion G, Dobson M & Ramakrishnan G FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta 1813, 1938–1945 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Hanel W et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ 20, 898–909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iggo R, Gatter K, Bartek J, Lane D & Harris AL Increased expression of mutant forms of p53 oncogene in primary lung cancer. Lancet 335, 675–679 (1990). [DOI] [PubMed] [Google Scholar]
- 63.Yemelyanova A et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol 24, 1248–1253 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Muller PA et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Andre F et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Mellman DL et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451, 1013–1017 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Malin D, Chen F, Schiller C, Koblinski J & Cryns VL Enhanced metastasis suppression by targeting TRAIL receptor 2 in a murine model of triple-negative breast cancer. Clin Cancer Res 17, 5005–5015 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Asmari M, Ratih R, Alhazmi HA & El Deeb S Thermophoresis for characterizing biomolecular interaction. Methods 146, 107–119 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Costes SV et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86, 3993–4003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen M, Horn HT, Wen T, Cryns VL & Anderson RA Assessing In Situ Phosphoinositide-Protein Interactions Through Fluorescence Proximity Ligation Assay in Cultured Cells. Methods Mol Biol 2251, 133–142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MDA-MB-231 cells were treated with control vehicle for 24 h before being processed for PLA between p53 and pAktS473 (Red). Lamin A/C stained the nuclear envelopes (Green), and the nuclei were counter-stained with DAPI (Blue). The z-stack images were taken with a Leica SP8 confocal microscope with each frame over a 0.2 μm thickness. The video of the 3D reconstitution of a representative cell from three independent experiments was generated by LASX.
MDA-MB-231 cells were treated with 30 μM cisplatin for 24 h before being processed for PLA between p53 and pAktS473 (Red). Lamin A/C stained the nuclear envelopes (Green), and the nuclei were counter-stained with DAPI (Blue). The z-stack images were taken with a Leica SP8 confocal microscope with each frame over a 0.2 μm thickness. The video of the 3D reconstitution of a representative cell from three independent experiments was generated by LASX.


