Summary
The Calvin–Benson–Bassham (CBB) cycle is arguably the most important pathway on earth, capturing CO2 from the atmosphere and converting it into organic molecules, providing the basis for life on our planet. This cycle has been intensively studied over the 50 yr since it was elucidated, and it is highly conserved across nature, from cyanobacteria to the largest of our land plants. Eight out of the 11 enzymes in this cycle catalyse the regeneration of ribulose‐1‐5 bisphosphate (RuBP), the CO2 acceptor molecule. The potential to manipulate RuBP regeneration to improve photosynthesis has been demonstrated in a number of plant species, and the development of new technologies, such as omics and synthetic biology provides exciting future opportunities to improve photosynthesis and increase crop yields.
Keywords: biotechnology, Calvin–Benson–Bassham Cycle, modelling, multigene, photosynthesis, transgenic
| Contents | ||
|---|---|---|
| Summary | 350 | |
| I. | Introduction | 350 |
| II. | Transgenic manipulation of RuBP regeneration | 352 |
| III. | Modelling to take a more predictive approach to identify targets to improve the Calvin–Benson–Bassham cycle? | 353 |
| IV. | Gaps in our knowledge about the Calvin–Benson–Bassham cycle | 354 |
| V. | Conclusions | 354 |
| Acknowledgements | 355 | |
| References | 355 |
I. Introduction
The Calvin–Benson–Bassham (CBB) cycle is the primary photosynthetic pathway for assimilation of atmospheric CO2 in over 85% of terrestrial plants, which are named C3 species as the first stable product of this cycle is a three‐carbon compound, glycerate3‐phosphate (Geiger & Servaites, 1994; Sharkey, 2019). The CBB cycle involves 11 enzymes, and the biochemical steps have been divided into three stages: carboxylation carried out by 1,5‐bisphosphate carboxylase/oxygenase (Rubisco), reduction, and RuBP regeneration (Fig. 1). Under light saturating and CO2‐limiting conditions Rubisco activity is the major determinant of the efficiency of carbon fixation, but as CO2 levels rise and light intensity decreases, this balance shifts towards both the reductive and regenerative phases of the CBB cycle that catalyse the synthesis of the CO2 acceptor molecule, RuBP (Fig. 1). Improving photosynthesis has been identified as a target to increase crop yield based on theory, modeling and empirical studies (Box 1). A major focus of efforts to improve photosynthesis is still the enzyme Rubisco, through the application of protein engineering strategies and also via manipulation of its expression in transgenic plants (Parry et al., 2012; Zhou & Whitney, 2019; Yoon et al., 2020; Makino, 2021). However, manipulating the expression of other enzymes of the CBB cycle can also enhance photosynthesis and growth. The aim of this insight will be to highlight the current status and future potential to improve the processes leading to regeneration of RuBP to deliver a step change in photosynthesis and boost crop yield.
Fig. 1.
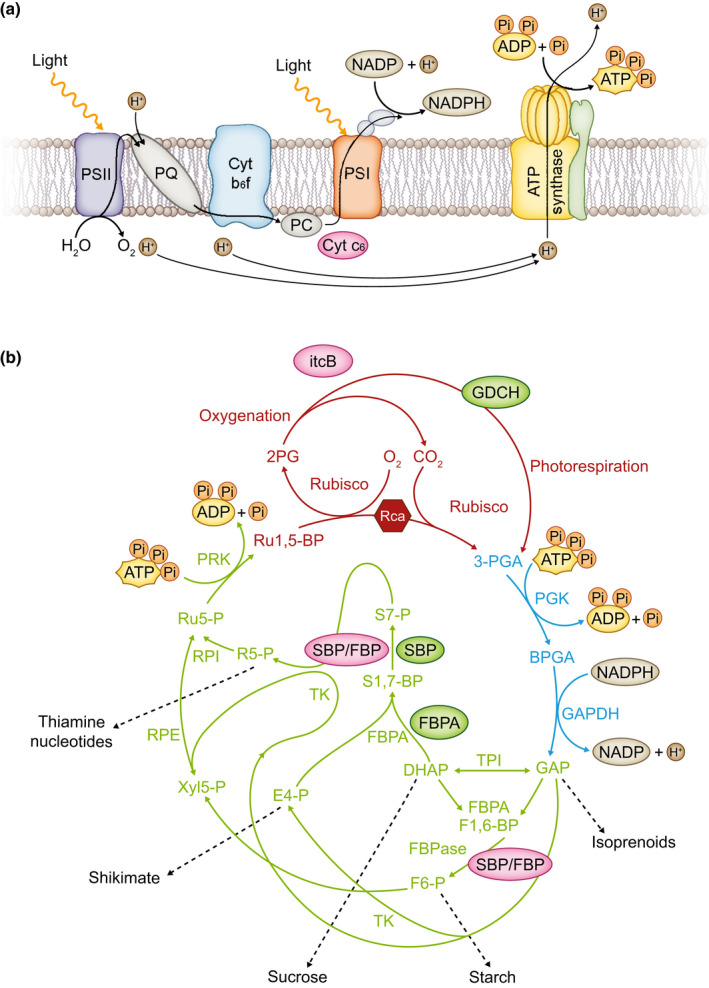
The Calvin–Benson–Bassham (CBB) cycle. (a) Energy in the form of ATP and NADPH needed to drive the CBB cycle is produced in the thylakoid membrane‐located electron transport chain. (b) The first step in the CBB cycle is catalysed by ribulose 1,5‐bisphosphate carboxylase/oxygenase (Rubisco) resulting in the formation of 3‐phosphoglycerate (3‐PGA). The next two reactions form the reductive phase and are catalysed by phosphoglycerate kinase (PGK), forming glycerate 1,3‐bisphosphate (BPGA) using ATP and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) which forms glyceraldehyde 3‐phosphate (GAP) consuming NADPH. Triose phosphate isomerase (TPI) catalyses the production of dihydroxyacetone phosphate (DHAP) and together with GAP enters the regenerative phase of the cycle, catalysed by fructose 1,6‐bisphosphate/sedoheptulose 1,7‐bisphosphate aldolase (FBPA), forming sedoheptulose 1,7‐bisphosphate (S1,7‐BP) and fructose 1,6‐bisphosphate (F1,6‐BP). Sedoheptulose 1,7‐bisphosphatase (SBPase) and FBPase (fructose 1,6‐bisphosphatase) then produce sedoheptulose 7‐phosphate (S7‐P) and fructose 6‐phosphate (F6‐P), which are converted to 5C compounds in reactions catalysed by transketolase (TK), ribose 5‐P isomerase (RPI) and ribulose 5‐phosphate epimerase (RPE), resulting in the formation of ribulose 5‐P (Ru5P). The final step in the cycle is catalysed by ribulose 5‐phosphate kinase (PRK), producing the CO2 acceptor molecule ribulose 1,5‐bisphosphate (Ru1,5‐BP). The three phases of the CBB cycle are shown: (1) carboxylation (red), (2) reduction (blue) and (3) regeneration (green). The products of the CBB cycle are exported to a number of biosynthetic pathways (grey dashed lines): isoprenoid, starch, sucrose, shikimate, thiamine and nucleotide. Rubisco has a competing oxygenase reaction which results in the formation of 2‐phosphoglycerate, which enters the photorespiratory pathway. The manipulations related to ribulose‐1‐5 bisphosphate (RuBP) regeneration discussed in this paper are in the electron transport chain algal cytochrome C6 (CytC6), the photorespiratory cycle H‐subunit of glycine decarboxylate (GDCH), the putative transporter from an alga (ictB), the endogenous SBPase and FBPA enzymes and the cyanobacterial bifunctional sedoheptulose 1,7‐bisphosphatase/fructose 1,6‐bisphosphatase (SBPase/FBPase) enzyme. Overexpression of endogenous proteins is shown in green and foreign proteins in pink.
Box 1. Why target photosynthesis to increase crop yields?
There is a pressing need to increase the yield of crop plants in order to feed our growing population, and this needs to be achieved within the next 20 yr without increasing land area used, or increasing water or nutrient inputs. The yield potential is the maximum yield attainable from a crop when the best adapted crop variety is grown, in optimal conditions with no biotic or abiotic stress. Yield potential is determined by a combination of the availability of light, the ability to capture the available light, conversion of the fixed energy into biomass, and plant architecture in terms of harvest index. Energy conversion is the only one of these four components that is well below its potential maximum, and this parameter is determined by photosynthetic efficiency (Zhu et al., 2010). The significance of photosynthetic efficiency for yield can be described by the following equation:
W h, harvested yield; S, solar energy; e i, light interception efficiency; e c, energy conversion efficiency; e p, harvest index.
Although there have been some doubts expressed about the strategy of targeting photosynthesis to improve yield, evidence that increased yield can be obtained by improving photosynthetic CO2 assimilation (the Calvin–Benson–Bassham (CBB) cycle) comes from studies with a range of plants grown in field conditions under elevated CO2 (Ainsworth & Long, 2021). Further evidence has come from transgenic manipulation of photosynthesis that showed under field conditions that an increase in photosynthesis and biomass was obtained (South et al., 2019). An increase in grain yield in rice grown in paddy fields was also found in plants with increased levels of ribulose 1,5‐bisphosphate carboxylase/oxygenase (Rubisco) (Yoon et al., 2020), and increased biomass and water use efficiency was observed in field‐grown tobacco plants expressing the cyanobacterial sedoheptulose 1,7‐bisphosphatase/fructose 1,6‐bisphosphatase (SBPase/FBPase) and the algal cytochrome C6 (see Fig. 2).
II. Transgenic manipulation of RuBP regeneration
Synthesis of the CO2 acceptor molecule, RuBP, requires the two steps in the reductive phase of the cycle to produce the C3 molecule glyceraldehyde 3‐phosphate (GAP) utilizing ATP and NADPH from the light reactions. The biochemical steps in the regenerative phase of the cycle use this GAP, and through the action of eight enzymes catalysing 10 reactions produce RuBP. In the 1990s antisense technology was used as an empirical approach to identify which of these enzymes exert control over the flow of CO2 in this pathway (Fig. 1). These studies demonstrated that no one enzyme had complete control over CO2 assimilation under all environmental conditions and, over and above Rubisco, identified sedoheptulose 1,7‐bisphosphatase (SBPase), fructose 1,6‐bisphosphate aldolase (FBPA) and transketolase (TK) as promising targets for overexpression and improvement of photosynthesis (Stitt & Schulze, 1994; Raines, 2003). Based on these empirical studies, a transgenic overexpression approach has shown that increasing the levels of SBPase can improve photosynthesis and growth in algae and a number of plant species including tobacco (in the field and glasshouse), wheat, rice, and Arabidopsis (Lefebvre et al., 2005; Simkin et al., 2015; Driever et al., 2017; Suzuki et al., 2019). Furthermore, tomato plants with increased SBPase activity were found to be more chilling tolerant with increased photosynthetic capacity (Ding et al., 2017). Overexpression of FBPA in tobacco also resulted in positive effects on photosynthesis and biomass (Uematsu et al., 2012; Simkin et al., 2015), and in tomato an increase in seed weight under both optimal and sub‐optimal temperatures was observed (Cai et al., 2022). Introduction of the bifunctional cyanobacterial CBB cycle enzyme sedoheptulose 1,7‐bisphosphatase/fructose 1,6‐bisphosphatase (SBPase/FBPase) into tobacco plants, lettuce and soybean (under elevated CO2), has also resulted in improved CO2 assimilation and growth (Miyagawa et al., 2001; Ichikawa et al., 2010; Kohler et al., 2017; Benes et al., 2020).
Given the central role of the CBB cycle in primary carbon metabolism, improvements in RuBP regeneration can also be realized through the combined introduction of proteins that function outside of the CBB cycle (Fig. 1). Two examples of this approach are as follows: the putative transporter ictB when introduced into tobacco in combination with SBPase and FBPA resulted in a further improvement in photosynthesis and growth over single gene manipulations, and overexpression of the H subunit of glycine decarboxylase together with SBPase and FBPA in Arabidopsis also resulted in additional positive effects when compared to the single manipulations (Simkin et al., 2015; Simkin et al., 2017). RuBP regeneration can also be limited by the availability of the ATP and NADPH produced by light reactions. To remove this potential bottleneck, plants were produced with a combination of overexpression of either the endogenous SBPase enzyme or bifunctional cyanobacterial SBPase/FBPase together with the algal cytochrome C6 protein, which rapidly transfers electrons from the cytochrome b6/f complex to photosystem I. Interestingly, tobacco plants carrying these manipulations were shown to exhibit not only improved photosynthesis and yield but also improved water use efficiency when grown in field conditions (Fig. 2) (Lopez‐Calcagno et al., 2020). Another more recent example is a study in which the co‐overexpression of SBPase and cytosolic FBPase showed a synergistic effect in transgenic tobacco plants, resulting in improvements in biomass, plant height, stem diameter and pod weight (Li et al., 2022). Additional combinations of targets for improving RuBP regeneration have been proposed. For example, overexpression of triose phosphate isomerase (TPI) in conjunction with other CBB cycle enzymes may provide further enhancements in carbon assimilation, by removing triose phosphate limitation (Suzuki et al., 2022). The expression of a group of CBB cycle genes (FBA1, RCA1, FBP5 and PGK1) was increased when the expression of the Brassinole resistant 1 transcription factor (BZR1) was increased and enhanced photosynthetic capacity was observed, suggesting that simultaneous overexpression of these proteins may stimulate the CBB cycle (Yin et al., 2022). These new findings, together with advancements in modeling, open up opportunities to re‐engineer the CBB cycle to maximize improvements.
Fig. 2.
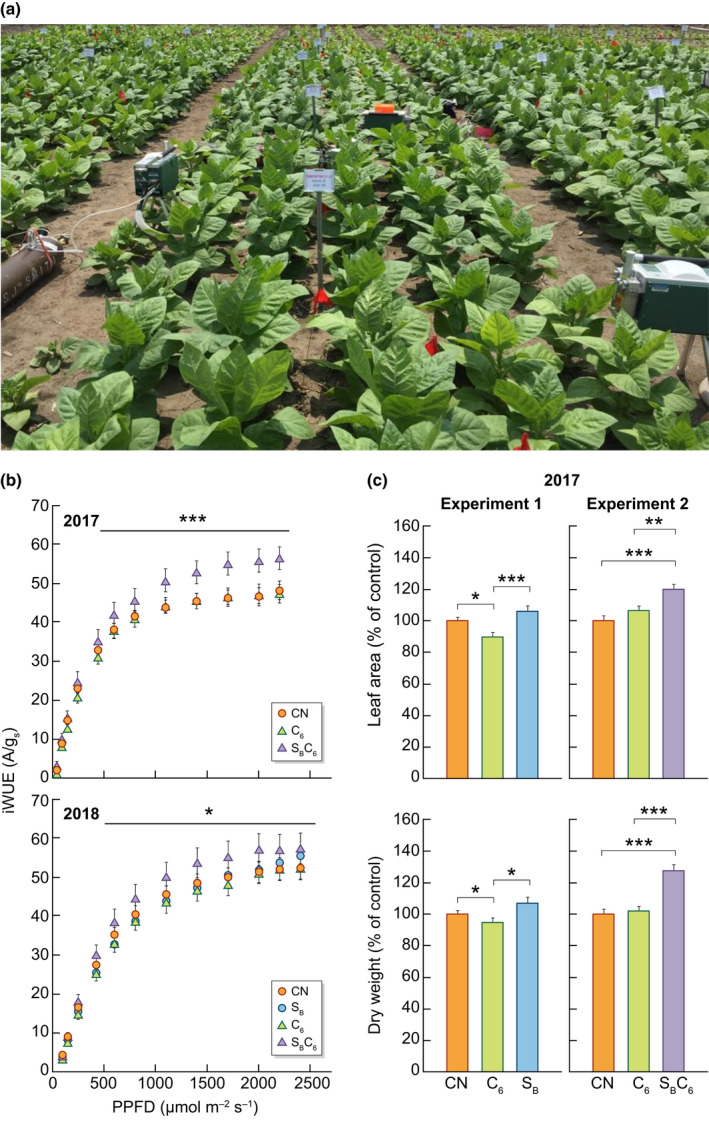
Expression of sedoheptulose 1,7‐bisphosphatase/fructose 1,6‐bisphosphatase and cytochrome C6 in transgenic tobacco improves water use efficiency and biomass in field‐grown plants. (a) Transgenic tobacco plants in field plots in Illinois. (b) Intrinsic water use efficiency and (c) biomass are increased in field‐grown transgenic plants expressing the bifunctional sedoheptulose 1,7‐bisphosphatase/fructose 1,6‐bisphosphatase (SBPase/FBPase) enzyme together with the algal cytochrome C6. CN, wild‐type and azygous controls; A/gs, assimilation rate over stomatal conductance; C, cytochrome C6; iWUE, intrinsic water use efficiency; PPFD, photosynthetic photon flux density; SB, SBPase/FBPase (Lopez‐Calcagno et al., 2020). In (b), asterisks indicate significant differences between the transgenic plants and the controls, using a linear mixed‐effects model and type III analysis of variance (ANOVA) and contrast analysis (*, P < 0.05; **, P < 0.01; ***, P < 0.001); in (c), asterisks indicate significance between transgenic plants and controls, or between genotypes, using ANOVA with Tukey's post‐hoc test (*, P < 0.05; **, P < 0.01, ***, P < 0.001). This figure is modified from figures presented in Lopez‐Calcagno et al. (2020).
III. Modelling to take a more predictive approach to identify targets to improve the Calvin–Benson–Bassham cycle?
A number of models incorporating the CBB cycle have been established, and the major strength of these mathematical models is that they offer a tool with which to study a range of scenarios and simulations that would not be possible experimentally. Kinetic models of the CBB cycle raised the hypothesis that SBPase was a major control point in the CBB cycle and that Rubisco is not always the sole limiting factor (Janasch et al., 2018). A model using an evolutionary algorithm suggested that simultaneous overexpression of SBP and FBPA could lead to significant improvement in CO2 assimilation rates (Zhu et al., 2007). Experimental evidence supporting this hypothesis came from a study showing that the combined over‐expression of SBPase and FBPA in tobacco resulted in a cumulative increase in biomass (Simkin et al., 2015).
A recent modelling study has hypothesised that sub‐cycles may exist within the CBB cycle and proposed that excessive increases in the activity of individual enzymes in one sub‐cycle could impact on the availability of metabolites for other sub‐cycles. The impact of this metabolite imbalance would be to lower reaction rates and the overall CBB cycle flux (Zhao et al., 2020). Interestingly, increasing the TK levels in tobacco resulted in a negative growth phenotype, reduced rates of photosynthesis and partial thiamine auxotrophy, suggesting that this manipulation of the CBB cycle had caused an imbalance in flux out of the cycle (Khozaei et al., 2015).
A model of the CBB cycle that included the electron transport chain has revealed the importance of balancing supply and demand reactions in order to ensure the efficiency of the CBB cycle. This merged model raised the hypothesis that a ‘standby’ mode during light–dark transitions may be essential to allow the CBB cycle to restart under increasing light, and a role for the oxidative pentose phosphate pathway was proposed (Matuszynska et al., 2019). The outputs from this model are supported by experimental studies showing that the CBB cycle relies on carbon influx from anaplerotic reactions to compensate for the depletion of intermediates, particularly under the fluctuating light conditions found in natural environments (Makowka et al., 2020; Xu et al., 2022).
IV. Gaps in our knowledge about the Calvin–Benson–Bassham cycle
Although the CBB cycle is ubiquitous and highly conserved between species, there remains a number of significant gaps in our knowledge, including the following.
The extent of natural variation that exists in the kinetic parameters and regulation of individual enzymes is unknown
Individual CBB cycle enzymes from different C3 species can exhibit diversity in their primary protein sequences but, with the exception of Rubisco, the functional implication of this diversity has not been studied systematically, and to date detailed data on the catalytic properties of any individual enzyme of the cycle is available for only a very few species. Extending our knowledge of the natural diversity of these enzymes will allow a better understanding of the relationship between the structure/function of the CBB cycle enzymes and the specific catalytic roles of the conserved and nonconserved amino acids. To achieve this the catalytic diversity needs to be analysed alongside the CBB cycle enzyme sequences, to identify potential catalytic switches for improving photosynthesis and productivity. Four enzymes involved in the CBB cycle, GAPDH, SBPase, FBPase and PRK, are regulated by light via redox changes in the chloroplast, through the thioredoxin (trx) system and the CP12 protein. Increasing the expression of these regulators in conjunction with their CBB cycle targets has yet to be explored as a strategy but could be a viable option given the indications from overexpression of trx on its own (Nikkanen & Rintamäki, 2019; Gurrieri et al., 2021).
Genetic factors regulating the coordinated expression of the C3 cycle gene are not known for even one species
The RNA abundance of CBB cycle enzymes changes during plant development, in response to light conditions and the accumulation of sugars; however, the detailed molecular mechanisms underlying the regulation of individual genes have not been elucidated, and even less is known about the coordination of expression of the whole pathway (Wang et al., 2017). The availability of whole genome sequences, including those of crop plants, together with the advent of omics technologies, eQTL and bioinformatics advances has enabled new approaches. A study of Populus tomentosa identified 40 transcription factors with potential roles in the regulation of 46 CBB cycle genes (Wang et al., 2019). Ten of these transcription factors were explored in more detail using a combination of metabolic analysis and outputs from gene regulatory network analysis. Interestingly, half of the SNPs identified were located in the promoter regions of the genes. Promoter scanning results revealed that 121 cis‐motifs co‐occurred in 80% of promoters of genes involved in the CBB cycle. The value of this approach is that it can provide insight into common regulatory mechanisms that would enable multitarget nontransgenic (gene editing) approaches to be incorporated into strategies to improve RuBP regeneration and CO2 fixation.
The regulation of the allocation of carbon through and from the C3 cycle to adjacent pathways has not been addressed holistically
Metabolite profiling of CBB cycle intermediates from C3 and C4 species revealed specificity and diversity in the CBB cycle between C3 species (Stitt et al., 2021). A comparative study between Arabidopsis and rice showed that these two C3 species prioritise different reactions when exposed to changes in irradiance (Borghi et al., 2019). The implication of these findings is that strategies to improve photosynthesis will need to be tailored depending on the crop, highlighting the need for systematic analyses of target species and cultivars within the same species. It is unlikely that metabolic profiling could be used to screen as a high‐throughput, automated approach. This type of study is likely to be most beneficial to target individual species in different environments or to provide data that can be built into models.
V. Conclusions
Advances in kinetic flux and multiscale modelling have provided novel predictions on how to further enhance RuBP regeneration. Testing these outputs will require the application of rapid high‐throughput and iterative approaches to identify the best candidates with which to achieve improvements in photosynthesis (Benes et al., 2020). At the same time, new approaches enabling identification of genetic factors and mechanisms involved in regulating the expression of CBB cycle genes will underpin the application of gene‐editing technologies to modify this pathway. Excitingly, it may even be possible to use synthetic biology to build a completely synthetic, more efficient CO2 fixation pathway to operate in parallel with the endogenous cycle (Erb & Zarzycki, 2016; Schwander et al., 2016; Löwe & Kremling, 2021) or to introduce improved enzymes to operate within the existing cycle. The advent of these new technologies provides future researchers with an exciting toolbox with which to exploit the full potential that improvements in RuBP regeneration can contribute to increasing photosynthetic performance and crop yield.
Acknowledgements
I would like to thank Professor Tracy Lawson and Dr Amanda Cavanagh for helpful comments on drafts of this paper. Thanks to Dr Patricia Lopez‐Calcagno and Dr Shellie Wall for preparing Figs 1 and 2. This study was supported by the Realising Improved Photosynthetic Efficiency (RIPE) initiative awarded to CAR by the University of Illinois, USA. RIPE was possible through support from the Bill & Melinda Gates Foundation, DFID and FFAR, grant. This work was also supported by the Biotechnology and Biological Sciences Research Council (BBSRC) grants BB/J004138/1, BB/H01960X/1 and BB/N021045/1 and the European Union's Horizon2020 research and innovation programme (No.862201) project CAPITALISE.
References
- Ainsworth EA, Long SPP. 2021. 30 years of free‐air carbon dioxide enrichment (FACE): what have we learned about future crop productivity and its potential for adaptation? Global Change Biology 27: 27–49. [DOI] [PubMed] [Google Scholar]
- Benes B, Guan K, Lang M, Long SP, Lynch JP, Marshall‐Colon A, Peng B, Schnable J, Sweetlove LJ, Turk MJ. 2020. Multiscale computational models can guide experimentation and targeted measurements for crop improvement. The Plant Journal 103: 21–31. [DOI] [PubMed] [Google Scholar]
- Borghi GL, Moraes TA, Günther M, Feil R, Mengin V, Lunn JE, Stitt M, Arrivault S. 2019. Relationship between irradiance and levels of Calvin–Benson cycle and other intermediates in the model eudicot Arabidopsis and the model monocot rice. Journal of Experimental Botany 70: 5809–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai BB, Ning Y, Li Q, Li QY, Ai XZ. 2022. Effects of the chloroplast fructose‐1,6‐bisphosphate aldolase gene on growth and low‐temperature tolerance of tomato. International Journal of Molecular Sciences 23: 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Wang M, Zhang S. 2017. Overexpression of a Calvin cycle enzyme SBPase improves tolerance to chilling‐induced oxidative stress in tomato plants. Scientia Horticulturae 214: 27–33. [Google Scholar]
- Driever SM, Simkin AJ, Alotaibi S, Fisk SJ, Madgwick PJ, Sparks CA, Jones HD, Lawson T, Parry MAJ, Raines CA. 2017. Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 372: 20160384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb TJ, Zarzycki J. 2016. Biochemical and synthetic biology approaches to improve photosynthetic CO2‐fixation. Current Opinion in Chemical Biology 34: 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger DR, Servaites JC. 1994. Diurnal regulation of photosynthetic carbon metabolism in C3 Plants. Annual Review of Plant Physiology and Plant Molecular Biology 45: 235–256. [Google Scholar]
- Gurrieri L, Fermani S, Zaffagnini M, Sparla F, Trost P. 2021. Calvin–Benson cycle regulation is getting complex. Trends in Plant Science 26: 898–912. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Tamoi M, Sakuyama H, Maruta T, Ashida H, Yokota A, Shigeoka S. 2010. Generation of transplastomic lettuce with enhanced growth and high yield. GM Crops 1: 322–326. [DOI] [PubMed] [Google Scholar]
- Janasch M, Asplund‐Samuelsson J, Steuer R, Hudson EP. 2018. Kinetic modeling of the Calvin cycle identifies flux control and stable metabolomes in Synechocystis carbon fixation. Journal of Experimental Botany 70: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozaei M, Fisk S, Lawson T, Gibon Y, Sulpice R, Stitt M, Lefebvre SC, Raines CA. 2015. Overexpression of plastid transketolase in tobacco results in a thiamine auxotrophic phenotype. Plant Cell 27: 432–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler IH, Ruiz‐Vera UM, VanLoocke A, Thomey ML, Clemente T, Long SP, Ort DR, Bernacchi CJ. 2017. Expression of cyanobacterial FBP/SBPase in soybean prevents yield depression under future climate conditions. Journal of Experimental Botany 68: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Lawson T, Fryer M, Zakhleniuk OV, Lloyd JC, Raines CA. 2005. Increased sedoheptulose‐1,7‐bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiology 138: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y‐Y, Guo L‐N, Liang C‐Z, Meng Z‐G, Tahira S, Guo S‐D, Zhang R. 2022. Overexpression of Brassica napus cytosolic fructose‐1,6‐bisphosphatase and sedoheptulose‐1,7‐bisphosphatase genes significantly enhanced tobacco growth and biomass. Journal of Integrative Agriculture 21: 49–59. [Google Scholar]
- Lopez‐Calcagno PE, Brown KL, Simkin AJ, Fisk SJ, Vialet‐Chabrand S, Lawson T, Raines CA. 2020. Stimulating photosynthetic processes increases productivity and water‐use efficiency in the field. Nature Plants 6: 1054. [DOI] [PubMed] [Google Scholar]
- Löwe H, Kremling A. 2021. In‐depth computational analysis of natural and artificial carbon fixation pathways. BioDesign Research 2021: 9898316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A. 2021. Photosynthesis improvement for enhancing productivity in rice. Soil Science and Plant Nutrition 67: 513–519. [Google Scholar]
- Makowka A, Nichelmann L, Schulze D, Spengler K, Wittmann C, Forchhammer K, Gutekunst K. 2020. Glycolytic shunts replenish the Calvin–Benson–Bassham cycle as anaplerotic reactions in cyanobacteria. Molecular Plant 13: 471–482. [DOI] [PubMed] [Google Scholar]
- Matuszynska A, Saadat NP, Ebenhoh O. 2019. Balancing energy supply during photosynthesis ‐ a theoretical perspective. Physiologia Plantarum 166: 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S. 2001. Overexpression of a cyanobacterial fructose‐1,6‐/sedoheptulose‐1,7‐bisphosphatase in tobacco enhances photosynthesis and growth. Nature Biotechnology 19: 965–969. [DOI] [PubMed] [Google Scholar]
- Nikkanen L, Rintamäki E. 2019. Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochemical Journal 476: 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Andralojc PJ, Scales JC, Salvucci ME, Carmo‐Silva AE, Alonso H, Whitney SM. 2012. Rubisco activity and regulation as targets for crop improvement. Journal of Experimental Botany 64: 717–730. [DOI] [PubMed] [Google Scholar]
- Raines CA. 2003. The Calvin cycle revisited. Photosynthesis Research 75: 1–10. [DOI] [PubMed] [Google Scholar]
- Schwander T, Borzyskowski LSV, Burgener S, Cortina NS, Erb TJ. 2016. A synthetic pathway for the fixation of carbon dioxide in vitro . Science 354: 900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD. 2019. Discovery of the canonical Calvin–Benson cycle. Photosynthesis Research 140: 235–252. [DOI] [PubMed] [Google Scholar]
- Simkin AJ, Lopez‐Calcagno PE, Davey PA, Headland LR, Lawson T, Timm S, Bauwe H, Raines CA. 2017. Simultaneous stimulation of sedoheptulose 1,7‐bisphosphatase, fructose 1,6‐bisphophate aldolase and the photorespiratory glycine decarboxylase‐H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnology Journal 15: 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, McAusland L, Headland LR, Lawson T, Raines CA. 2015. Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. Journal of Experimental Botany 66: 4075–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR. 2019. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Luca Borghi G, Arrivault S. 2021. Targeted metabolite profiling as a top‐down approach to uncover interspecies diversity and identify key conserved operational features in the Calvin–Benson cycle. Journal of Experimental Botany 72: 5961–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Schulze D. 1994. Does rubisco control the rate of photosynthesis and plant‐growth ‐ an exercise in molecular ecophysiology. Plant, Cell & Environment 17: 465–487. [Google Scholar]
- Suzuki Y, Ishiyama K, Yoon DK, Takegahara‐Tamakawa Y, Kondo E, Suganami M, Wada S, Miyake C, Makino A. 2022. Suppression of chloroplast triose phosphate isomerase evokes inorganic phosphate‐limited photosynthesis in rice. Plant Physiology 188: 1550–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Wada S, Kondo E, Yamori W, Makino A. 2019. Effects of co‐overproduction of sedoheptulose‐1,7‐bisphosphatase and Rubisco on photosynthesis in rice. Soil Science and Plant Nutrition 65: 36–40. [Google Scholar]
- Uematsu K, Suzuki N, Iwamae T, Inui M, Yukawa H. 2012. Increased fructose 1,6‐bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. Journal of Experimental Botany 63: 3001–3009. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie J, Du Q, Song F, Xiao L, Quan M, Zhang D. 2019. Transcription factors involved in the regulatory networks governing the Calvin–Benson–Bassham Cycle. Tree Physiology 39: 1159–1172. [DOI] [PubMed] [Google Scholar]
- Wang P, Hendron RW, Kelly S. 2017. Transcriptional control of photosynthetic capacity: conservation and divergence from Arabidopsis to rice. New Phytologist 216: 32–45. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wieloch T, Kaste JAM, Shachar‐Hill Y, Sharkey TD. 2022. Reimport of carbon from cytosolic and vacuolar sugar pools into the Calvin Benson cycle explains photosynthesis labeling anomalies. Proceedings of the National Academy of Sciences, USA 119: e2121531119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Tang M, Xia X, Yu J. 2022. BRASSINAZOLE RESISTANT 1 mediates brassinosteroid‐induced calvin cycle to promote photosynthesis in tomato. Frontiers in Plant Science 12. doi: 10.3389/fpls.2021.811948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon D‐K, Ishiyama K, Suganami M, Tazoe Y, Watanabe M, Imaruoka S, Ogura M, Ishida H, Suzuki Y, Obara M et al. 2020. Transgenic rice overproducing Rubisco exhibits increased yields with improved nitrogen‐use efficiency in an experimental paddy field. Nature Food 1: 134–139. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tang Q, Chang T, Xiao Y, Zhu X‐G. 2020. Why an increase in activity of an enzyme in the Calvin–Benson cycle does not always lead to an increased photosynthetic CO2 uptake rate?—a theoretical analysis. In Silico Plants 3: diaa009. [Google Scholar]
- Zhou Y, Whitney S. 2019. Directed evolution of an improved rubisco; in vitro analyses to decipher fact from fiction. International Journal of Molecular Sciences 20: 5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, de Sturler E, Long SP. 2007. Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: a numerical simulation using an evolutionary algorithm. Plant Physiology 145: 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR. 2010. Improving photosynthetic efficiency for greater yield. Annual Review of Plant Biology 61: 235–261. [DOI] [PubMed] [Google Scholar]


