Abstract
Striped large-eye bream, Gnathodentex aureolineatus (Lacepède, 1802), is of high economic value and has important ecological functions in coral reefs. However, the genetic information of this species is quite limited, and there is taxonomical difficulty in the family Lethrinidae. Here, we present the complete mitochondrial genome of G. aureolineatus obtained with a long PCR approach and Sanger sequencing. The mitogenome was 16,940 bp in length, consisting of 37 genes (13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes) and two non-coding regions. Both maximum-likelihood and Bayesian inference phylogenetic trees placed the genus Gnathodentex sister to Monotaxis within Lethrinidae. These results contribute toward the taxonomy, conservation, and phylogeny of Lethrinidae.
Keywords: Striped large-eye bream, mitogenome, phylogenetics
Striped large-eye bream, Gnathodentex aureolineatus (Lacepède, 1802), is one of the most common coral reef fishes widely distributed in the Indo-Pacific (Francis and Randall 1993). In China, G. aureolineatus has been recorded in South China Sea coral reefs (Shen et al. 1993; Li et al. 2020; Zhang et al. 2021). As one of the major predators in coral reefs (Carpenter and Allen 1989; Skinner et al. 2020), they usually form aggregations and travel on subtidal reef flats, lagoon platforms, or the upper edge of seaward reefs (Lieske and Myers 1994; Carpenter 1997). However, the genetic information is quite limited for G. aureolineatus, since only several of their nuclear and mitochondrial genes have been used in phylogenetic analyses (Lautredou et al. 2013; Chen and Borsa 2020; Fabian et al. 2021). In addition, the family Lethrinidae has been considered a taxonomically difficult group (Chen and Borsa 2020; Ramesh et al. 2020; Zhang et al. 2021). To establish genetic data for species identification, conservation, and evolutionary clarification, we aimed to sequence the complete mitogenome of G. aureolineatus and constructed the phylogenetic relationships in Lethrinidae.
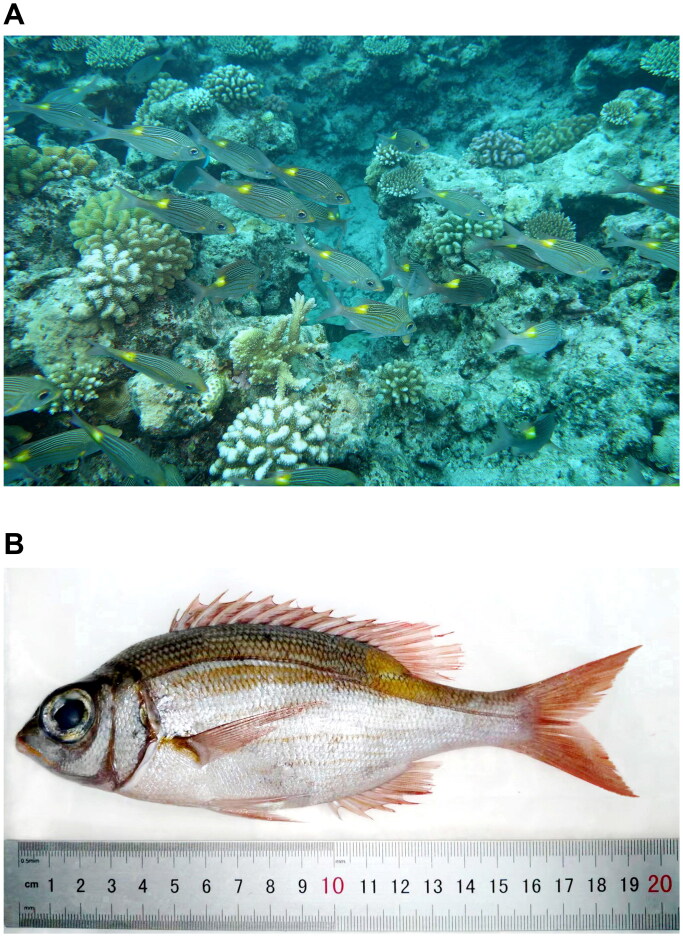
Three individuals of G. aureolineatus were collected in Triton Island (15°47′N 111°12′E), Xisha, China, on the same survey approved by the Animal Care and Ethical Committee of the South China Sea Institute of Oceanology, Chinese Academy of Sciences (Guo et al. 2016). According to WORMS (https://www.marinespecies.org/) and FishBase (https://www.fishbase.de/), we conducted species identification, along with the collection of the dorsal muscles for total genomic DNA extraction. Then, three specimens (Figure 1) were deposited at the South China Sea Tropical Marine Biology Collection, Chinese Academy of Sciences (Minglan Guo, guominglan@scsio.ac.cn) under voucher numbers: SCSTMBC030980-030982.
Figure 1.
Specimen of Gnathodentex aureolineatus (Lacepède, 1802). (A) G. aureolineatus found in the coral reefs off Triton Island, Xisha, China (JH Yang and YL Gao). (B) Fixed specimen (ML Guo).
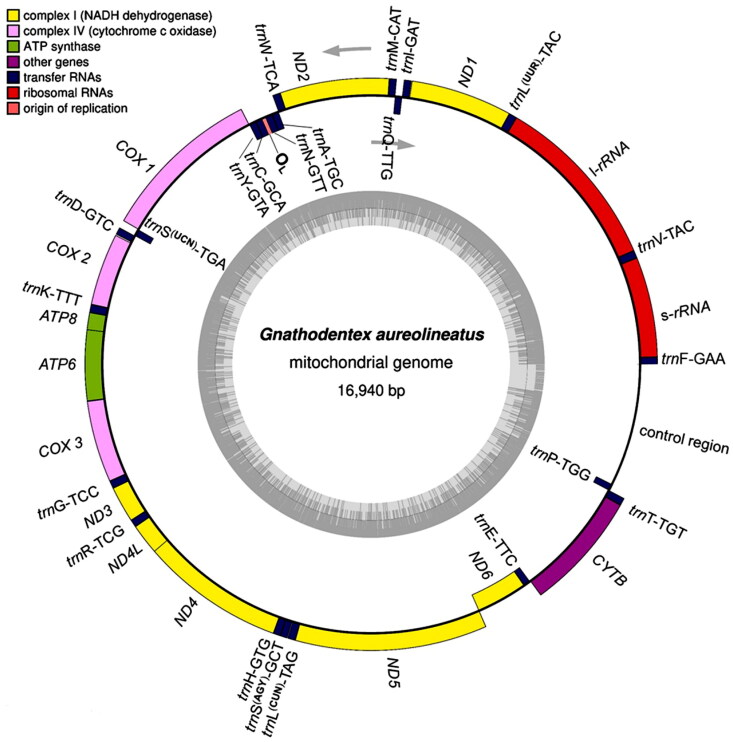
Based on the mitogenome sequences of Monotaxis grandoculis (AP009166), Lethrinus laticaudis (KU530221), and L. obsoletus (AP009165) in Lethrinidae, we designed five pairs of primers (Table 1) to amplify the genes using long PCR for Sanger sequencing (Guo et al. 2016). All sequences were assembled using DNAman software, and the overlaid regions were checked using Sanger sequencing with primers designed from the obtained sequences. The assembled mitogenome (GenBank accession no: OM302214 or NC_063714) was annotated using MITOS (Bernt et al. 2013), aligned with Blastn, and checked with tRNAscan-SE (Chan and Lowe 2019). It was 16,940 bp in length, containing 37 genes (13 protein-coding genes, two rRNA genes, and 22 tRNA genes), the origin of light-strand replication (OL), and the control region (D-loop) (Table 2). The GenBank file was used to construct the mitogenome circle map on OrganellarGenomeDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Figure 2).
Table 1.
Sequences of primers used in this study.
| Primers | Sequence (5’-3’) |
|---|---|
| GA F1/12S | CACAAAGGTTTGGTCCTGACTTTAC |
| GA R1/16S | GGGCTCTGCCACCTTAACATGY |
| GA F2/16S | CTAGTACGAAAGGACCGAGAAGATGA |
| GA R2/COI | TGTGTGATAAGGRGGMGGGCAGC |
| GA F3/COI | CGACGRTACTCAGACTACCCHGA |
| GA R3/ND4L | TTGTAGGAGRTTTARGCTYTGVAGG |
| GA F4/ND4L | CACCGAACCCACCTWCTCTCYGC |
| GA R4/Cytb | GCAATTTTTAGTARKGGGTGKGTTTT |
| GA F5/Cytb | GCCAGGACTYTAACCAGGACTAATG |
| GA R5/12S | GCGGTGGCTGGCACGAGTTTTAC |
The five pairs of primers (GA F1/12S and GA R1/16S, GA F2/16S and GA R2/COI, GA F3/COI and GA R3/ND4L, GA F4/ND4L and GA R4/Cytb, GA F5/Cytb and GA R5/12S) were designed for the mitogenome amplification and Sanger sequencing.
Table 2.
Characteristics of the mitochondrial genome of Gnathodentex aureolineatus.
| Locus (abbreviation) | size |
Codon |
Anti-codon | Intergenic nucleotidea | Strandb | ||
|---|---|---|---|---|---|---|---|
| Nucleotide (position) | Amino acid | Start | Stop | ||||
| tRNAPhe (trnF) | 68 (1–68) | GAA | 0 | H | |||
| 12S rRNA (s-rRNA) | 958 (69–1027) | 0 | H | ||||
| tRNAVal (trnV) | 74 (1028–1101) | TAC | 0 | H | |||
| 16S rRNA (l-rRNA) | 1698 (1102–2800) | 0 | H | ||||
| tRNALeu(UUR) (trnL) | 75 (2801–2875) | TAA | 0 | H | |||
| ND1 (ND1) | 975 (2876–3850) | 324 | ATG | TAA | 4 | H | |
| tRNAIle (trnl) | 70 (3855–3924) | GAT | 0 | H | |||
| tRNAGln (trnQ) | 71 (3995–3925) | TTG | −1 | L | |||
| tRNAMet (trnM) | 69 (3995–4063) | CAT | 0 | H | |||
| ND2 (ND2) | 1046 (4064–5109) | 348 | ATG | TA– | 0 | H | |
| tRNATrp (trnW) | 74 (5110–5183) | TCA | 0 | H | |||
| tRNAAla (trnA) | 69 (5252–5184) | TGC | 1 | L | |||
| tRNAAsn (trnN) | 73 (5326–5254) | GTT | 1 | L | |||
| OL | 40 (5328–5367) | −3 | – | ||||
| tRNACys (trnC) | 68 (5432–5365) | GCA | 0 | L | |||
| tRNATyr (trnY) | 70 (5502–5433) | GTA | 1 | L | |||
| COX1 (COX1) | 1551 (5504–7054) | 516 | GTG | TAG | 1 | H | |
| tRNASer(UCN) (trnS) | 71 (7126–7056) | TGA | 1 | L | |||
| tRNAAsp (trnD) | 72 (7128–7199) | GTC | 8 | H | |||
| COX2 (COX2) | 694 (7208–7901) | 231 | ATG | T— | 0 | H | |
| tRNALys (trnK) | 73 (7902–7976) | TTT | 1 | H | |||
| ATP8 (ATP8) | 168 (7978–8145) | 55 | ATG | TAA | −10 | H | |
| ATP6 (ATP6) | 683 (8136–8818) | 227 | ATG | TA– | 0 | H | |
| COX3 (COX3) | 785 (8819–9603) | 261 | ATG | TA– | 0 | H | |
| tRNAGly (trnG) | 72 (9604–9675) | TCC | 0 | H | |||
| ND3 (ND3) | 349 (9676–10,024) | 116 | ATG | T— | 0 | H | |
| tRNAArg (trnR) | 70 (10,025–10,094) | TCG | 0 | H | |||
| ND4L (ND4L) | 297 (10,095–10,391) | 98 | ATG | TAA | −7 | H | |
| ND4 (ND4) | 1381 (10,385–11,765) | 460 | ATG | T— | 0 | H | |
| tRNAHis (trnH) | 69 (11,766–11,834) | GTG | 0 | H | |||
| tRNASer(AGY) (trnS) | 68 (11,835–11,902) | GCT | 4 | H | |||
| tRNALeu(CUN) (trnL) | 73 (11,907–11,979) | TAG | 0 | H | |||
| ND5 (ND5) | 1839 (11,980–13,818) | 612 | ATG | TAA | −4 | H | |
| ND6 (ND6) | 522 (14,336–13,815) | 173 | ATG | TAG | 1 | L | |
| tRNAGlu (trnE) | 69 (14,406–14,338) | TTC | 5 | L | |||
| CYTB (CTYB) | 1141 (14,412–15,552) | 380 | ATG | T— | 1 | H | |
| tRNAThr (trnT) | 73 (15,554–15,626) | TGT | −1 | H | |||
| tRNAPro (trnP) | 69 (15,694–15,626) | TGG | 0 | L | |||
| Control region | 877 (15,695–16,940) | – | |||||
aNumbers correspond to the nucleotides separating different genes. Negative numbers mean overlapping nucleotides between adjacent genes. bH and L indicate genes encoded on the heavy and light strands, respectively.
Figure 2.
Mitochondrial genome map of Gnathodentex aureolineatus. Genes on the heavy and light strands were shown outside and inside the outer circle, respectively. The inner grey ring indicates the GC content. tRNA genes were abbreviated and linked with anti-codons.
The gene arrangement in the mitogenome of G. aureolineatus was identical to that of the above species in Lethrinidae. ND6 and eight tRNA genes were transcribed on the light strand, while the other 28 genes were transcribed on the heavy strand. There were two forms of codon recognition in both tRNALeu (UUR and CUN) and tRNASer (UCN and AGY) (Figure 2). Moreover, intergenic nucleotides occurred between protein-coding genes and/or tRNA genes. Except for COX 1, all protein-coding genes began with the typical start codon ATG. The incomplete stop codon T— appeared in COX2, ND3, ND4, and CYTB, and TA– appeared in ND2, ATP6, and COX3 (Table 2).
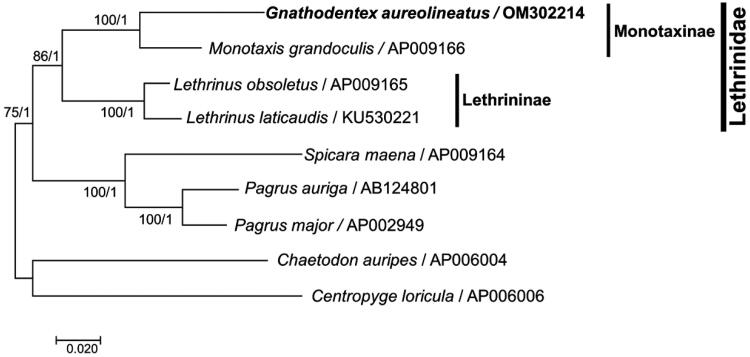
We explored the phylogenies of nine suborder Percoidei mitogenomes available in the NCBI database, including those of G. aureolineatus and two outgroup species Chaetodon auripes and Centropyge loricula (Miya et al. 2001; Yamanoue et al. 2007; Taillebois et al. 2016). All concatenated amino acid sequences of 12 protein-coding genes on the heavy strand were aligned with code constraint under Clustal X (v2.1) (http://www.clustal.org/clustal2/). Then, bootstrapped maximum-likelihood (ML) with 1000 replications and Bayesian Inference (BI) trees were constructed using MEGA 7 (Kumar et al. 2016) and MrBayes v3.2.7 (Huelsenbeck and Ronquist 2001), respectively, based on the result of multiple sequence alignment. The subfamily Monotaxinae (Gnathodentex and Monotaxis) was separated from Lethrininae (Lethrinus) in Lethrinidae (Figure 3). This confirmed the phylogeny in Lethrinidae reported using morphology (Carpenter and Johnson 2002) and nuclear or mitochondrial genes (Chen and Borsa 2020; Lautredou et al. 2013; Fabian et al. 2021).
Figure 3.
Maximum-likelihood and Bayesian Inference phylogenetic trees constructed with the mitogenomes of nine fish species in the suborder Percoidei, including Gnathodentex aureolineatus. The numbers above branches indicate the values of the Maximum-likelihood and Bayesian Inference phylogenetic trees, respectively.
This study provides a new mitogenome and phylogenetic relationship in Lethrinidae. The mitogenome will be useful in delimitating problematic groups in taxonomy. These data will offer genome resources for the conservation of G. aureolineatus and a reference for species delimitation and evolutionary research in Lethrinidae.
Acknowledgments
The authors would like to thank JH Yang for sample collection and Meng Qu for phylogenetic analysis. We thank the reviewers for their constructive comments which greatly improved this manuscript.
Funding Statement
This work was supported by Hainan Province Science and Technology Special Fund [ZDKJ2019011], Natural Science Foundation of Guangdong Province, China [2018A0303130173], Fund of Key Laboratory of Aquatic Product Processing, Ministry of Agriculture and Rural Affairs, China [NYJG201509], Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences [ISEE2018PY01], Science and Technology Planning Project of Guangdong Province, China [2020B1212060058].
Author contributions
MG conceived, designed, and performed the experiments, analyzed the data, prepared the figure, authored the draft, prepared the funding, and approved the final draft. Y G conceived and designed the experiments, collected the samples, revised drafts, and approved the final draft. HH conceived and designed the experiments, interpreted the data, reviewed the draft, prepared the funding, and approved the final draft. All authors commented on the draft, gave final consent for publication, and agreed to be accountable for all aspects of this work.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The mitogenome sequence data supporting this study are openly available in GenBank with accession no. OM302214 (https://www.ncbi.nlm.nih.gov/nuccore/OM302214.1/) and corresponding RefSeq NC_063714 (https://www.ncbi.nlm.nih.gov/nuccore/NC_063714.1/). The associated BioProject number is PRJNA845703.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Carpenter KE, Allen GR.. 1989. FAO Species Catalogue. Vol.9. Emperor fishes and large-eye breams of the world (family Lethrinidae). An annotated and illustrated catalogue of lethrinid species known to date. FAO Fish. Synop. Rome: FAO. 125(9):118. [Google Scholar]
- Carpenter KE, Johnson GD.. 2002. A phylogeny of sparoid fishes (Perciforms, Percoidei) based on morphology. Ichthyol Res. 49(2):114–127. [Google Scholar]
- Carpenter KE. 1997. Lethrinidae. Emperor sanppers. In: Carpenter KE, Niem V, editors. FAO identification guide for fishery purposes. The Western Central Pacific. Rome: Food and Agriculture Organization of the United Nation; p. 3017. [Google Scholar]
- Chan PP, Lowe TM.. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Borsa P.. 2020. Diversity, phylogeny, and historical biogeography of large-eye seabreams (Teleostei: lethrinidae). Mol Phylogenet Evol. 151:106902. [DOI] [PubMed] [Google Scholar]
- Fabian V, Houk P, Lemer S.. 2021. Phylogeny of Micronesian emperor fishes and evolution of trophic types. Mol Phylogenet Evol. 162:107207. [DOI] [PubMed] [Google Scholar]
- Francis MP, Randall JE.. 1993. Further additions to the fish faunas of Lord Howe and Norfolk Islands, Southwest Pacific Ocean. Pac Sci. 47(2):118–135. [Google Scholar]
- Guo M, Huang H, Gao Y.. 2016. Complete mitochondrial genome of darkfin hind Cephalopholis urodeta (Perciformes, Epinephelidae). Mitochondrial DNA B Resour. 1(1):913–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautredou A-C, Motomura H, Gallut C, Ozouf-Costaz C, Cruaud C, Lecointre G, Dettai A.. 2013. New nuclear markers and exploration of the relationships among Serraniformes (Acanthomorpha, Teleostei): the importance of working at multiple scales. Mol Phylogenet Evol. 67(1):140–155. [DOI] [PubMed] [Google Scholar]
- Li J, Sun C, Zhang L, Ding J, Jiang F, Wang Z, Wang Z, Fu L.. 2020. Current distribution characteristics of trace elements in the coral-reef systems of Xisha Islands, China. Mar Pollut Bull. 150:110737. [DOI] [PubMed] [Google Scholar]
- Lieske E, Myers R.. 1994. Collins pocket guide. In: Coral reef fishes. Indo-Pacific & Caribbean including the Red Sea. New York: Haper Collins Publishers; p. 400. [Google Scholar]
- Miya M, Kawaguchi A, Nishida M.. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009. [DOI] [PubMed] [Google Scholar]
- Ramesh CH, Koushik S, Shunmugaraj T, Ramana Murthy MV.. 2020. Current biodiversity of Mandapam group of Islands in Gulf of Mannar Marine Biosphere Reserve, Southeast coast of Tamil Nadu, India. Reg Stud Mar Sci. 39:101429. [Google Scholar]
- Shen SC, Lee SC, Shao KT, Mok HK, Chen CT, Chen CH, Tzeng CS.. 1993. Fishes of Taiwan. Taipei: Department of Zoology, National Taiwan University; p. 960. [Google Scholar]
- Skinner C, Mill AC, Newman SP, Alsagoff SN, Polunin NVC.. 2020. The importance of oceanic atoll lagoons for coral reef predators. Mar Biol. 167(2):19. [Google Scholar]
- Taillebois L, Crook DA, Saunders T, Williams SM, Ovenden JR.. 2016. The complete mitochondrial genome of the grass emperor, Lethrinus laticaudis (Perciformes: lethrinidae). Mitochondrial DNA B Resour. 1(1):277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanoue Y, Miya M, Matsuura K, Yagishita N, Mabuchi K, Sakai H, Katoh M, Nishida M.. 2007. Phylogenetic position of tetraodontiform fishes within the higher teleosts: Bayesian inferences based on 44 whole mitochondrial genome sequences. Mol Phylogenet Evol. 45(1):89–101. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang J, Shi D, Chen Z.. 2021. Assessment of coral reef fish stocks from the Nansha Islands, South China Sea, using length-based Bayesian biomass estimation. Front Mar Sci. 7:610707. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mitogenome sequence data supporting this study are openly available in GenBank with accession no. OM302214 (https://www.ncbi.nlm.nih.gov/nuccore/OM302214.1/) and corresponding RefSeq NC_063714 (https://www.ncbi.nlm.nih.gov/nuccore/NC_063714.1/). The associated BioProject number is PRJNA845703.