Abstract
We report the discovery of a novel genetic locus within Staphylococcus aureus that encodes a cluster of at least five exotoxin-like proteins. Designated the staphylococcal exotoxin-like genes 1 to 5 (set1 to set5), these open reading frames have between 38 and 53% homology to each other. All five proteins contain consensus sequences that are found in staphylococcal and streptococcal exotoxins and toxic shock syndrome toxin 1 (TSST-1). However, the SETs have only limited overall sequence homology to the enterotoxins and TSST-1 and thus represent a novel family of exotoxin-like proteins. The prototypic gene in this cluster, set1, has been cloned and expressed. Recombinant SET1 stimulated the production of interleukin-1β, interleukin-6, and tumor necrosis factor alpha by human peripheral blood mononuclear cells. PCR analysis revealed that set1 was distributed among other strains of S. aureus but not in the other staphylococcal species examined. Sequence analysis of the set1 genes from different strains revealed at least three allelic variants. The protein products of these allelic variants displayed a 100-fold difference in their cytokine-inducing potency. The distribution of allelic variants of the set genes among strains of S. aureus may contribute to differences in the pathogenic potential of this bacterium.
Staphylococcus aureus is a major human pathogen that causes a broad and diverse range of diseases including the life-threatening toxic shock syndrome and less serious conditions such as boils and food poisoning. Such human diseases are believed to be caused by a disparate group of exoproteins secreted by S. aureus. The exoproteins fall into two main groups: (i) enzymes such as hyaluronidase, proteases, and nucleases, and (ii) exotoxins including leukocidins, exfoliative toxins, enterotoxins, and toxic shock syndrome toxin 1 (TSST-1). However, some exoproteins, for instance the hemolysins which possess both enzymatic and toxin activities, cannot be molded into this artificial classification. The enzymatic exoproteins probably function primarily to provide nutrients for bacterial growth and in so doing contribute to malaise, while the exotoxins are the major cause of disease.
Among the numerous staphylococcal exotoxins, those that have received the most attention are the enterotoxins and TSST-1. There are currently 11 staphylococcal enterotoxins (SEs), SEA, SEB, SEC1, SEC2, SEC3, SED, SEE, SEG, SEH, SEI and SEJ), and these are believed to be the main causative agents of staphylococcal food poisoning (2, 3). Although the SEs also cause toxic shock, TSST-1 is the predominant causative agent (17). These exotoxins are also superantigens (14, 37, 38) and, together with the exotoxins of Streptococcus pyogenes, have been termed pyrogenic toxins because, in addition to a number of shared biological activities including the ability to stimulate T-cell proliferation, they are pyrogenic (4, 5). The pyrogenicity of these toxins is due to their potent immunostimulatory properties (16). These immunostimulatory activities are not confined to actions on T cells but are also, in part, due to the stimulation of proinflammatory cytokine production by major histocompatibility complex class II-bearing cells such as monocytes (7, 9, 19). Although the staphylococcal and streptococcal pyrogenic exotoxins are grouped together because of their shared biological activities, these exotoxins also possess distinct activities; for instance, only the SEs cause emesis and only the streptococcal exotoxins are cardiotoxic.
In addition to having primary-structure homology, crystal structures for SEA (30), SEB (33), SEC2 (22), SED (32), TSST-1 (1), and streptococcal pyrogenic exotoxin C (SPEC) (28) demonstrate that these proteins have similar tertiary and quaternary structures. They all possess an N-terminal β-barrel globular domain (OB fold) and a C-terminal β-grasp motif composed of five β strands.
Of the clinical isolates of S. aureus, 40% produce one or more of the known exotoxins (31), and it has been suggested that there are as yet unidentified exotoxin genes (20, 27). We describe the identification of a new exotoxin-like gene, set1. Cloning of this gene has revealed that it forms part of a gene cluster containing a novel family of at least five staphylococcal exotoxin-like genes, designated set1 to set5. The distribution of the prototypic gene from this cluster, set1, among selected staphylococcal species was examined. The capacity of recombinant SET1 to induce proinflammatory cytokine production by human peripheral blood mononuclear cells (PBMCs) was also examined.
MATERIALS AND METHODS
Identification of set1.
A fragment of the set1 gene was initially cloned using a screening method incorporating alkaline phosphatase (PhoA) as a reporter molecule to identify secreted proteins, as previously described for Streptococcus pneumoniae (23). This system utilizes an alkaline phosphatase-negative strain of Escherichia coli CC118 (PhoA−) and plasmid pHRM104, which contains a truncated E. coli phoA gene lacking its signal sequence. Thus, for the secretion of functional alkaline phosphatase activity, genes encoding signal sequences must be cloned in frame with the phoA gene in the reporter system. A 5-μg sample of S. aureus NCTC 6571 genomic DNA was digested with Sau3A, fragments of 1 to 4 kb were ligated to pHRM104 which had been linearized with BamHI, and the ligations were used to transform E. coli CC118. Transformants exhibiting alkaline phosphatase activity were detected by growth on nutrient agar containing 5-bromo-4-chloro-3-indolylphosphate. Plasmid DNA was prepared from all positive clones, and the sequences of the inserts were determined. One of these clones, pExp5, contained a 237-bp DNA fragment of set1.
Cloning of the full-length set1 gene and identification of the novel set gene cluster.
A set1-specific probe (253 bp) was PCR amplified using pExp5 as template DNA and the primer pair 5′-GGGACAGAATAATACTATGAAATTAAAAACG and 5′-ATCTTTTTGGTTAAAGCGTAC. Labeling of the set1 probe DNA with digoxigenin-dUTP (DIG-dUTP), hybridization, and subsequent immunological detection were carried out as specified by the manufacturer (Roche Diagnostics Ltd.). Southern blots of genomic DNA digested with an array of restriction endonucleases and probed with the set1 probe revealed that this gene was present on a 5.2-kb EcoRI fragment. Chromosomal DNA was digested with EcoRI and ligated to pUC19. The ligation mix was transformed into E. coli TOP10. Transformants were transferred to Hybond-N membranes (Amersham, Little Chalfont, United Kingdom) and screened for the set1 gene using the 253-bp set1 probe. One clone, pSET16, had a 5.2-kb insert containing the set1 gene. The 5.2-kb insert was sequenced by primer walking using a BigDye terminator kit as specified by the manufacturer (ABI Perkin-Elmer). The cycle-sequencing reactions were run on an ABI 310 genetic analyzer. The template was sequenced in both directions in duplicate.
Computer-aided modeling of protein structures.
The SET sequences, excluding SET2, for which we did not have complete sequence data, were modeled against two target proteins, TSST-1 and SPEC, using a Silicon Graphics computer. Coordinate files for SPEC (1an8) and TSST-1 (2tss) were downloaded from the Brookhaven Protein Database. We used MODELLER software, which optimizes spatial restraints derived from the sequence alignments, to build models of the SET proteins (29). Protein models thus derived were viewed using MOLSCRIPT. Based on sequence alignments using CLUSTAL_X Windows 1 (35), the SET3 and SET4 sequences were modeled using TSST-1 (2tss.pdb) as template while the SET1 and SET5 sequences were modeled using SPEC (1an8.pdb).
Distribution of the set1 gene among staphylococci.
Staphylococcal strains were screened for set1 by PCR. Chromosomal DNA from S. aureus NCTC 6571, NCTC 8325-4, FRI326, and eight clinical isolates (obtained from Geoffrey Scott, University College London Hospital) and S. epidermidis NCTC 11964 and NCTC 11047 were used as templates. A total of 30 cycles of amplification were performed using the forward primer 5′-GAATTCAGATTGGGAGAATAATACTATG and the reverse primer 5′-AGATCTCAACGTTTCATCGTTAAGCTGC.
Dot blot analysis of set1 mRNA production.
Overnight cultures of S. aureus grown in 1% casein hydrolysate–2.5% yeast extract were diluted (1:50) in fresh broth and incubated at 37°C with aeration. Aliquots were taken at various time points, and the cells were harvested and washed with 0.5% Tween 80. The cell pellets were resuspended in 0.5 ml of lysis solution (consisting of 9.6 ml of Divolab no. 1 detergent [DiversyLever Ltd.], 24 ml of 500 mM sodium acetate [pH 4], 66.4 ml of distilled water), 0.5 ml of acid-phenol (pH 4), and 0.1 ml of chloroform-isoamyl alcohol (24:1) and lysed using a Ribolyser (Hybaid). After a 10-min incubation on ice followed by centrifugation, the supernatants were extracted with phenol-chloroform and the RNA was precipitated. RNA (2 μg) was blotted onto a nylon membrane and probed using a DIG-labeled set1 probe. The hybridization and subsequent chemiluminescent detection were carried out as specified by the manufacturer (Roche Diagnostics Ltd.).
Cloning of set1 into an N-terminal polyhistidine expression vector.
The oligonucleotide 5′-GGATCCGCAGAAAAACAAGAGAGAGTAC and 5′-GTCGACAACGTTTCATCG-TTAAGCTGCC were designed to amplify the 677-bp set1 gene minus the DNA encoding the N-terminal signal peptide and also contained recognition sequences for the restriction enzymes BamHI and SalI (underlined), respectively. The PCR fragment was initially cloned into PCR2.1-TOPO and transformed into TOP10 (Invitrogen, Leek, The Netherlands). The set1 gene was extracted from PCR2.1-TOPO on a BamHI-SalI fragment and ligated to BamHI-SalI digested pQE30 (Qiagen Ltd., Crawley, United Kingdom). The ligation mix was transformed into E. coli JM109(pREP4), and transformants were selected by growth at 30°C on Luria-Bertani agar containing 100 μg of ampicillin per ml and 25 μg of kanamycin per ml.
Expression of set1 and purification of recombinant SET1.
For gene expression, positive clones were grown overnight in Terrific broth, diluted 1:25 in fresh broth, and incubated for a further 1 h at 37°C. Gene expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 28°C. Cells were harvested by centrifugation at 6,000 × g for 30 min and then resuspended and lysed for 20 min in B-PER protein extraction reagent (Pierce & Warriner Ltd.) containing 20 mM imidazole, 1.5 μM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 10 μM leupeptin. Lysates were clarified by centrifugation at 23,000 × g for 15 min. Recombinant proteins were purified using Ni-nitrilotriacetic acid-agarose columns under native conditions as specified by the manufacturer (Qiagen Ltd.), except that after the lysates were loaded onto the column, an additional column wash, consisting of 2.5 mg of polymyxin B per ml in wash buffer, was performed to remove contamination with lipopolysaccharide.
Finally, recombinant SET1 (rSET1) was further purified by gel filtration chromatography using a Superdex75 column preequilibrated in phosphate-buffered saline and attached to a Pharmacia SMART system (Amersham-Pharmacia Biotech).
Assay of cytokine production by PBMCs.
PBMCs were prepared as previously described (34). Cells were plated at a density of 2 × 106 cells/ml in 24-well plates and stimulated with graded concentrations of protein. SEB (Sigma-Aldrich Ltd.) was used as a positive control. Cell supernatants were assayed for the presence of interleukin-1β (IL-1β), IL-6, and tumor necrosis factor alpha (TNF-α) using two-site enzyme-linked immunosorbent assays as previously described (26, 34).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported here have been deposited in the GenBank database under accession numbers AF094826 (set gene cluster fragment), AF188835 (NCTC 6571 set1 gene), AF188836 (FRI326 set1 gene), and AF188837 (NCTC 8325-4 set1 gene).
RESULTS
Identification of set1.
In a previous study we employed an alkaline phosphatase fusion strategy to identify proteins exported by S. aureus. Using this system, we isolated one clone, pExp5, that contained DNA encoding the N-terminal 79 amino acids of a secreted protein fused to phoA. Comparison of the nucleotide sequence of pExp5 with those in the Oklahoma University sequencing project of S. aureus NCTC 8325 (http://www.genome.ou.edu/staph.html) revealed homology (91%) to contig 386 (designation as of July 1998). Contig 386 contained a partial open reading frame encoding a putative 191-amino-acid protein (ORF386), and a search of the SwissProt database revealed that ORF386 had approximately 25% identity to a number of staphylococcal and streptococcal exotoxins. The homology included two regions that were similar to the consensus staphylococcal enterotoxin/streptococcal exotoxin signatures (PROSITE and PCOC00250) (12, 16). The first of these signatures is a well-conserved region found in the majority of the staphylococcal enterotoxins but not TSST-1, while the second signature is a more diffuse sequence which is present in both the enterotoxins and TSST-1. This similarity suggested that ORF386 may code for a novel exotoxin. The DNA sequence identified in pExp5 as encoding an N-terminal fragment (including the signal peptide sequence) of an unknown protein and that of ORF386 were very similar although not identical, and we hypothesized that the gene within pExp5 may also encode a novel exotoxin-like protein. We have designated this open reading frame set1 (for staphylococcal exotoxin-like gene 1).
Cloning of the full-length set1 gene and identification of the novel exotoxin-like gene cluster.
The set1 gene was mapped to a 5.2-kb EcoRI S. aureus genomic DNA fragment by using a 253-bp probe designed to target the 5′ end of set1 (data not shown). To clone this fragment, a complete S. aureus NCTC 6571 genomic DNA-pUC19 EcoRI library was constructed in E. coli and screened using the 253-bp set1 probe. One positive clone, pSET16, containing a 5,175-bp fragment was used for further analysis. The complete DNA sequence of this 5,175-bp fragment was determined and has been deposited in GenBank (accession no. AF094826). Analysis of the DNA sequence of pSET16 revealed four complete open reading frames together with two partial open reading frames at the 5′ and 3′ extremities of the DNA. A BLAST search of the SwissProt database indicated that the four complete open reading frames and one of the partial open reading frames (5′ end) had homology, although low, to those encoding a number of staphylococcal and streptococcal exotoxins, including TSST-1. The second partial open reading frame at the 3′ extremity encoded the start of a possible host specificity determinant methylase (HsdM)-like protein (54% identity over 26 amino acids to Klebsiella pneumoniae HsdM). The novel exotoxin-like genes have been designated set1 to set5 based on the order of their discovery.
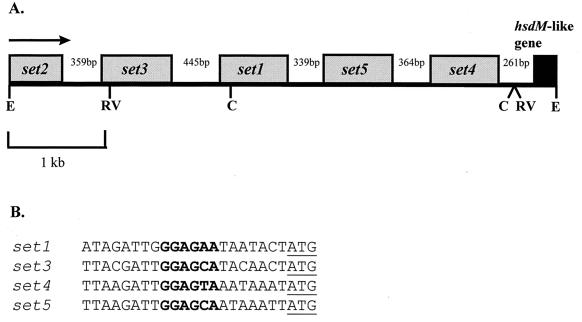
The organization of the set genes is shown in Fig. 1A. The distance between each of the set genes varies from 339 to 446 bp, while the shorter intergenic region between set4 and the hsdM-like gene is 261 bp. Apart from the set2 gene (for which the upstream region has not been sequenced), all genes are preceded by putative ribosome-binding sites 7 bp upstream of their start codons (Fig. 1B).
FIG. 1.
(A) Arrangement of the set genes in S. aureus NCTC 6571. The direction of transcription of all of the genes is shown by the arrow. The numbers refer to the sizes of the intergenic spaces. The letters refer to sites cut by the restriction endonucleases: C, ClaI; E, EcoRI; RV, EcoRV. (B) Putative ribosome-binding sites (bold) upstream of the set genes. The start codons are underlined.
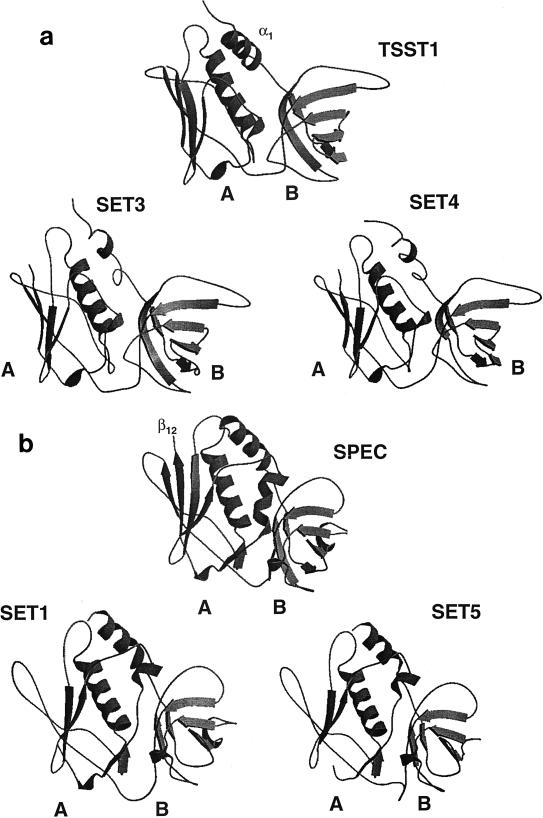
The four complete set genes encode proteins of between 227 and 234 amino acids, which have 38 to 53% homology to each other. The partial set2 gene encodes the C-terminal 172 amino acids of this protein. An alignment of the amino acid sequences of the SET proteins is shown in Fig. 2. Each of the SET proteins (apart from SET2, for which the N-terminal region has not been sequenced) possesses putative N-terminal signal sequences, as determined using the SignalP algorithm for gram-positive bacteria (21), indicating that these are secreted proteins. The mature extracellular proteins are predicted to consist of 201 (SET1), 204 (SET3), 202 (SET4), and 207 (SET5) amino acids. Although the similarity between the SET proteins and the known staphylococcal and streptococcal exotoxins is low, being around 25%, each of the SET proteins possesses the staphylococcal/streptococcal exotoxin consensus signature 2 (K-X2-[LIV]-X2-[LIV]-D-X3-R-X2-L-X5-[LIV]-Y) as determined by searches of the PROSITE database (11) (Fig. 2). In addition, the SET proteins have an amino acid sequence which is similar to the exotoxin consensus signature 1 (PROSITE, PCOC00250) (Fig. 2). SET1, SET2, and SET5 have the greatest homology to SPEC (25, 26, and 25% sequence identity, respectively), while SET3 and SET4 are most closely related to TSST-1 (25 and 27% identity, respectively). Based on this homology, the SET sequences were modeled against two target protein structures, TSST-1 (PDB file 2tss) and SPEC (PDB file 1an8). The protein models of SET1 and SET5 depict two domain structures joined by a central α-helix (Fig. 3a). Domain B has topology equivalent to the oligosaccharide- and oligonucleotide-binding fold (OB-fold) found in SPEC and other unrelated toxins. However, the A domain of SET1 and SET5 differs from that of SPEC in that it has one less β-strand in the β-grasp motif. The missing β-strand corresponds to β12 in the SPEC model and is important in zinc binding and in providing biological activity. The protein models of SET3 and SET4 also depict a two-domain structure joined by a central α helix (Fig. 3b). Once again, the B domain forms a β-barrel structure. The β-grasp motif of the A domain in SET3 and SET4 is similar to that of TSST-1. However, the short N-terminal α-helix (α1) of TSST-1 is not maintained in SET3 and SET4. Although there is low amino acid sequence identity between the SET proteins and those of the target protein structures, these theoretical models demonstrate that the SET proteins could adopt typical superantigen-type structures.
FIG. 2.
Multiple alignment of the SET proteins. Highly conserved residues which are present in at least three of the proteins are shown in boldface type, and the putative N-terminal signal peptides are underlined. The staphylococcal and streptococcal exotoxin consensus signatures 1 and 2 are shown underneath the alignment at positions 121 to 129 and 162 to 185, respectively.
FIG. 3.
Schematic diagram of the modeled structures for SET proteins, TSST-1, and SPEC. The SET sequences were modeled against two target protein structures, TSST-1 (PDB file 2tss) and SPEC (PDB file 1an8) on a Silicon Graphics computer. All models showed the potential to form two domains: in domain A, the C terminus has a five-strand β-sheet surrounding a long central α-helix, and in the smaller domain B, the N terminus shows β-strands folding into a barrel creating the OB-fold. (a) The A domain of SET3 and SET4 has one less β-strand than the structure for TSST-1. (b) SET1 and SET5 differ from SPEC in having one less β-strand in the A domain. The missing strand corresponds to β12 in the SPEC model.
Distribution of the set1 gene among staphylococci.
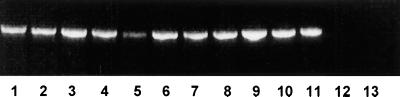
Ten strains of S. aureus, FRI326, NCTC 8325-4, and eight clinical isolates, together with two strains of S. epidermidis (NCTC 11964 and NCTC 11047) were analyzed by PCR for the presence of the set1 gene. Products were generated for all of the S. aureus strains but not for either of the S. epidermidis strains (Fig. 4). Cloning and sequencing of the set1 genes from three of the S. aureus strains (FRI326, NCTC 6571, and NCTC 8325-4) revealed that the NCTC 6571 gene had 92 and 88% identity to the FRI326 and NCTC 8325-4 set1 genes, respectively. The respective GenBank accession numbers for the set1 genes from FRI326, NCTC 8325-4, and NCTC 6571 are AF188836, AF188837, and AF188835. The predicted protein sequences differed in 30 (FRI326) and 34 (NCTC 8325-4) positions from the NCTC 6571 SET1 sequence, corresponding to 87 and 84% identity, respectively, at the amino acid level (Fig. 5).
FIG. 4.
PCR analysis for the presence of the set1 gene in staphylococci. Lanes: 1 to 8, clinical S. aureus isolates; 9, S. aureus NCTC 6571; 10, S. aureus FRI326; 11, S. aureus NCTC 8325-4; 12, S. epidermidis NCTC 11964; 13, S. epidermidis NCTC 11047.
FIG. 5.
Alignment of the SET1 amino acid sequences from S. aureus NCTC 6571, S. aureus NCTC 8325-4, and S. aureus FRI326. Differences between the NCTC 8325-4 and FRI326 SET1 sequences compared to the NCTC 6571 sequence are shown in bold.
Dot blot analysis of set1 mRNA production.
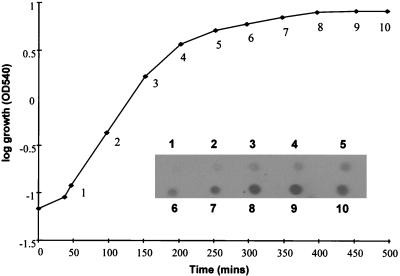
To determine if set1 is expressed in S. aureus, total cellular RNA extracted at various growth stages from S. aureus NCTC 6571 was probed with a probe specific to set1. Figure 6 shows that the expression of set1 was growth phase dependent, with the highest level of expression in the stationary phase.
FIG. 6.
Expression of the set1 gene by S. aureus NCTC 6571. Total cellular RNA was extracted at the time points shown, and 2 μg of total RNA was probed with a DIG-labeled set1 DNA fragment. OD540, optical density at 540 nm.
Cloning and expression of set1.
The set1 gene encoding the mature protein minus the signal peptide was PCR amplified from pSET16 DNA and inserted into the pQE30 N-terminal histidine tag fusion vector. This construct was introduced into E. coli JM109. Protein production was found to be most efficient when bacteria were grown at 28°C after induction (data not shown), giving yields of approximately 20 mg of soluble protein per liter. Analysis of SET1 by gel filtration chromatography revealed that it existed as a monomeric protein of about 23 to 24 kDa. The set1 genes from NCTC 8325-4 and FRI326 were also cloned and expressed using this system and found to encode monomeric proteins with molecular masses identical to that of SET1 from NCTC 6571. All rSET proteins were homogenous as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and migrated with molecular masses of about 23 to 24 kDa.
Capacity of rSET1 to stimulate production of the proinflammatory cytokines IL-1β, IL-6, and TNFα.
All characterized staphylococcal enterotoxins are able to stimulate proinflammatory cytokines. The ability of rSET1 from strain NCTC 6571 to stimulate the production of the cytokines IL-1β, IL-6 and TNF-α by PBMCs was compared to that of SEB. To establish the purity and to ensure that similar concentrations of each protein preparation were used, rSET1 and commercial SEB were compared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Stimulation of PBMCs with rSET1 resulted in the production of all three of the proinflammatory cytokines tested (Fig. 7 to 9).
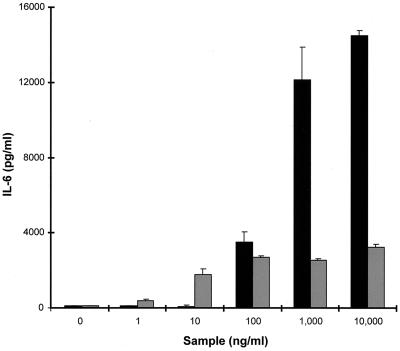
FIG. 7.
Production of IL-6 by PBMCs in response to stimulation with graded concentrations of either rSET1 (■) or SEB (░⃞). The figure shows the results from one representative experiment of at least three, expressed as the mean and standard deviation of three replicate cultures.
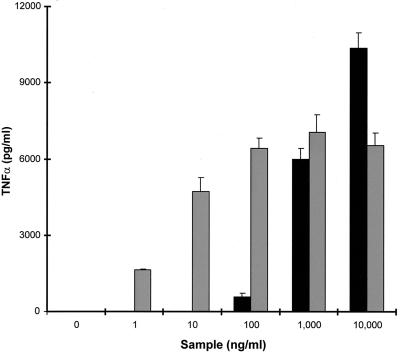
FIG. 9.
Production of TNF-α by PBMCs in response to stimulation with graded concentrations of either rSET1 (■) or SEB (░⃞). The figure shows the results from one representative experiment of at least three; the data are the means and standard deviations of three replicate cultures.
rSET1 stimulated the secretion of IL-6 by PBMCs over the dose range of 100 ng/ml to 10 μg/ml, compared to SEB, which was active at inducing IL-6 production over the dose range of 10 ng/ml to 10 μg/ml. However, the maximal levels of IL-6 produced in response to rSET1 were almost 10-fold greater than those induced by SEB (Fig. 7). Heating of rSET1 at 100°C for 20 min reduced its ability to stimulate IL-6 production by 69%.
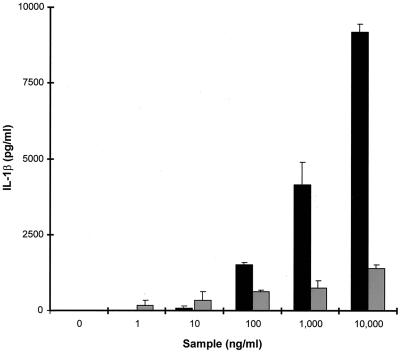
SEB was a very poor inducer of IL-1β secretion by PBMCs. rSET1, on the other hand, showed a similar dose response to that produced for IL-6 secretion, being active over the range of 100 ng/ml to 10 μg/ml. The maximal level of IL-1β produced by PBMCs stimulated with rSET1 was, again, almost 10-fold greater than that induced by SEB (Fig. 8).
FIG. 8.
Production of IL-1β by PBMCs in response to stimulation with graded concentrations of either rSET1 (■) or SEB (░⃞). The figure shows the results from one representative experiment of at least three; the data are the means and standard deviations of three replicate cultures.
rSET1 was not a potent inducer of TNF-α secretion, being active only over the concentration range from 1 to 10 μg/ml. In contrast, SEB was a potent inducer of TNF-α secretion by PBMCs, having activity over the dose range of 1 ng/ml to 10 μg/ml. Once again, the maximum level of cytokine secretion was produced in response to rSET1 and not to SEB; however, the levels were only slightly higher (Fig. 9).
Ability of allelic variants of SET1 to stimulate the production of IL-6 by PBMCs.
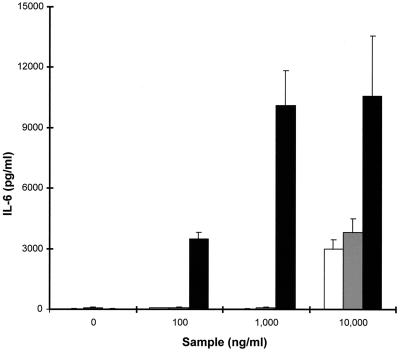
As described above, we have identified at least three allelic variants of set1 corresponding to the genes from strains NCTC 6571, FRI326, and NCTC 8325-4. Allelic variants of other exotoxins such as SPEA have different biological activities (15). To determine if the different alleles of SET1 have different biological activities, we cloned and expressed the genes for these proteins in E. coli and compared the ability of the rSET1 proteins to stimulate IL-6 secretion by PBMCs. All three alleles were able to stimulate PBMCs to secrete IL-6 (Fig. 10). However, the alleles from strains FRI326 and NCTC 8325-4 were active only at 10 μg/ml. In contrast, the SET1 from strain NCTC 6571 was active at 100 ng/ml and thus was the most potent allele, being 100-fold more potent than the other SET1 alleles. Additionally, the levels of IL-6 induced by 100 ng of SET1 from NCTC 6571 per ml were equivalent to those produced by 10 μg of the SET1s from strains FRI326 and NCTC 8325-4 per ml (Fig. 10).
FIG. 10.
Production of IL-6 by PBMCs in response to stimulation with graded concentrations of rSET1 encoded by S. aureus strains NCTC 6571 (■), FRI326 (░⃞), and NCTC 8325-4 (□). The figure shows the results from one representative experiment of at least three, and the data are the means and standard deviations of three replicate cultures.
DISCUSSION
The staphylococcal exotoxins are a heterogeneous group of bacterial toxins that include the enterotoxins, which cause food poisoning, and, together with TSST-1, are responsible for toxic shock syndrome. Although at least 11 enterotoxins have been identified, it is widely believed that more remain to be discovered, since the known enterotoxin genes do not account for all cases of food poisoning and toxic shock syndrome. Some of the exotoxins are also superantigens, and it has been suggested that they may even play a role in autoimmune diseases (8, 18). For instance, the enterotoxins have been suspected of triggering symptoms resembling arthritis in patients with staphylococcal toxic shock syndrome (6, 16).
In a previous study, we attempted to identify novel secreted proteins from S. aureus by using an alkaline phosphatase gene fusion technique. This method allowed us to identify a clone that contained an insert encoding the N-terminal 79 amino acids of a putative novel exotoxin-like protein, SET1. In this study, we have cloned the entire open reading frame of set1 from S. aureus NCTC 6571, which was found on a 5.2-kb EcoRI fragment and was flanked by further exotoxin-like genes. In all, we have identified four complete and one partial novel exotoxin-like genes. We have designated these genes set1 to set5, since they encode staphylococcal exotoxin-like proteins. All of the genes possess sequences similar to ribosome-binding sites immediately upstream of their start codons. Apart from set2 (for which the complete DNA sequence has not yet been determined), the set genes encode putative extracellular proteins which have typical gram-positive bacterial N-terminal signal peptides made up of between 25 and 30 amino acids. The predicted mature SET proteins are of a similar size to the staphylococcal enterotoxins and TSST-1, consisting of between 201 and 207 amino acids with predicted molecular masses ranging from 23 to 24 kDa.
To our knowledge, only two other genetic loci encoding contiguous staphylococcal toxin genes have previously been described. The first of these contains the leukocidin genes hlg2, lukS, and lukF (36). The second locus contains the Panton-Valentine leukocidin genes lukS-PV and lukF-PV and occurs in only a few strains of S. aureus (24). The genetic locus encoding the Panton-Valentine leukocidins resides on the recently sequenced bacteriophage wPVL (13). The genetic locus that we describe in this report represents the third such locus. While we have not completely determined the boundaries of this putative exotoxin cluster, we have determined that these genes are flanked at the 3′ end by an hsdM-like gene. Interestingly, we have recently identified a fourth genetic locus that contains a cluster of contiguous staphylococcal virulence genes (unrelated to the exotoxin genes described in this report) which are also flanked at the 3′ end by an identical hsdM-like gene (S. P. Nair, R. J. Williams, and B. Henderson, unpublished data). At present the relevance of the hsdM-like gene and its position in these virulence clusters is unclear.
We are at present unsure whether the set genes are expressed as part of a polycistronic transcript or individually. Most of the staphylococcal enterotoxin genes are expressed individually, the exception being seg, which is transcribed on a large (6.7-kb) transcript, although the identities of the flanking genes are unknown (20). The intergenic regions between each of the set genes are large (approximately 400 bp), making it unlikely that these genes are expressed as a polycistronic transcript. We have shown that, as with the majority of the staphylococcal exotoxin genes, set1 is expressed during the postexponential phase of growth.
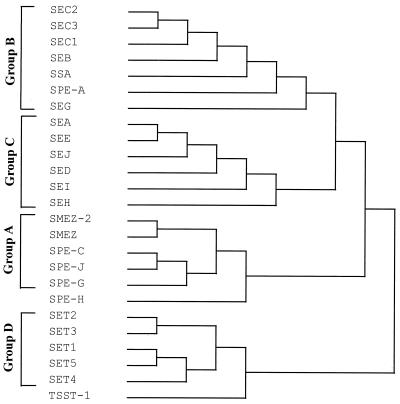
The staphylococcal and streptococcal pyrogenic exotoxins have a reasonable degree of homology at the amino acid level and, according to their primary sequences, can at present be divided into three groups (A to C). However, two of the pyrogenic toxins, SPEH and TSST-1, cannot be placed in any of these groups because of their low sequence homology. Phylogenetic comparison of the SET proteins with the known pyrogenic exotoxins has revealed that they form a distinct family of exotoxin-like proteins, group D. The SET family lies between the group A exotoxins and TSST-1 (Fig. 11). Although there is little primary structure conservation between TSST-1 and the other families of pyrogenic exotoxins, they do have common tertiary and quaternary structural features. All of the pyrogenic exotoxins for which crystal structure data are available consist of two domains (A and B). The minimalist structure found in common is a large A domain consisting of a five-strand β-sheet surrounding a long central α-helix and a smaller B domain composed of β-strands folded into a barrel or claw motif. Computer-aided modeling of the SET proteins has demonstrated that these proteins could adopt a structure composed of two domains joined by a central α-helix, as is found in the other pyrogenic exotoxins. However, while the B domain β-barrel or claw motif of the pyrogenic exotoxins is maintained in the models of the SET proteins, there are structural differences in the A domain. SET1 and SET5 have one less β-strand in the β-grasp motif of domain A. SET3 and SET4 also differ from TSST-1 in the A domain. The high degree of similarity between the structures of the pyrogenic exotoxins and the differences found in the modeled structures of the SET proteins suggests that SETs form a distinct family of exotoxin-like proteins.
FIG. 11.
Phylogenetic tree showing the relationship between the known streptococcal pyrogenic exotoxins, the staphylococcal enterotoxins, TSST-1, and the novel exotoxins described in this report. The multiple alignment was constructed by DNAsis software using the Higgins and Sharp algorithm (10).
We have cloned and expressed the prototypical gene in this cluster, set1, and demonstrated that rSET1 has the ability to stimulate the production of the cytokines IL-1β, IL-6, and TNF-α by PBMCs. SEB also stimulates the production of these three cytokines; however, the two toxins have differing patterns of efficacies and potencies. Given the similarities of the immunomodulatory effects of rSET1 to those produced by the known pyrogenic exotoxins, it is likely that this protein may play an important role in the pathology of staphylococcal diseases. In addition, set1 is flanked by at least four other set genes and, given the sequence similarities between SET1 and the other SET proteins, it is probable that their protein products may also have similar or more potent biological activities, and thus they may act in concert. Previous studies have shown that slight amino acid differences between the members of the pyrogenic toxin family can result in greatly altered biological properties. In one of the most recent discoveries, the 17-amino-acid difference between the streptococcal mitogenic exotoxin z (SMEZ) isolated from two Streptococcus pyogenes strains led to different superantigenic potencies on human PBMCs and, indeed, to different Vβ specificity profiles (25). During this study, we have found the set1 gene in three different S. aureus strains and also in eight clinical S. aureus isolates, although, admittedly, we do not know if the set1 gene is expressed in all S. aureus strains. Interestingly, the proteins from S. aureus FRI326 and NCTC 8325-4 have less than 90% amino acid sequence identity to SET1 from NCTC 6571. These allelic variants of SET1 were found to have different capacities to stimulate PBMCs to secrete IL-6. The SET1 proteins encoded by strains FRI326 and NCTC 8325-4 were relatively inactive compared to that from strain NCTC 6571. At present we have identified three allelic variants of the set1 gene. It is possible that there are other SET1 alleles and that these code for proteins that are even more potent at inducing proinflammatory cytokine production by PBMCs. Thus, the allelic form of set1 possessed by a particular strain of S. aureus may have a major impact on the virulence of this organism. There are four other exotoxin genes in the SET virulence cassette, whose protein products remain to be characterized. If these proteins also have the ability to stimulate proinflammatory cytokine production by PBMCs, this virulence cluster may represent one of the most important pathogenicity determinants in S. aureus.
ACKNOWLEDGMENT
We are grateful to the Arthritis Research Campaign for funding R.J.W. (grant N0514).
REFERENCES
- 1.Acharya K R, Passalacqua E F, Jones E Y, Harlos K, Stuart D I, Brehm R D, Tranter H S. Structural basis of superantigen action inferred from crystal structure of toxic-shock syndrome toxin-1. Nature. 1994;367:94–97. doi: 10.1038/367094a0. [DOI] [PubMed] [Google Scholar]
- 2.Bergdoll M. Enterotoxins. In: Easmon C S F, Adlam C, editors. Staphylococci and staphylococcal infections. New York, N.Y: Academic Press, Inc.; 1983. pp. 550–598. [Google Scholar]
- 3.Bergdoll M S. Staphylococcus aureus. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 463–523. [Google Scholar]
- 4.Bohach G A, Fast D J, Nelson R D, Schlievert P M. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990;17:251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 5.Bohach G A, Stauffacher C V, Ohlendorf D H, Chi Yi, Vath G M, Schlievert P M. The staphylococcal and streptococcal pyrogenic toxin family. Adv Exp Med Biol. 1996;391:131–154. doi: 10.1007/978-1-4613-0361-9_8. [DOI] [PubMed] [Google Scholar]
- 6.Chesney P J. Clinical aspects and spectrum of illness of toxic shock syndrome. Overview. Rev Infect Dis. 1989;11:S1–S7. [PubMed] [Google Scholar]
- 7.Fast D J, Schlivert P M, Nelson R D. Toxic shock syndrome-associated staphylococcal and streptococcal pyrogenic toxins are potent inducers of tumor necrosis factor production. Infect Immun. 1989;57:291–294. doi: 10.1128/iai.57.1.291-294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman S M, Posnett D N, Tumang J R, Cole B C, Crow M K. A potential role for microbial superantigens in the pathogenesis of systemic autoimmune disease. Arthritis Rheum. 1991;34:468–480. doi: 10.1002/art.1780340412. [DOI] [PubMed] [Google Scholar]
- 9.Hackett S P, Stevens D L. Streptococcal toxic shock syndrome synthesis of tumor necrosis factor and interleukin-1 by monocytes stimulated with pyrogenic exotoxin A and streptolysin O. J Infect Dis. 1992;165:879–885. doi: 10.1093/infdis/165.5.879. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iandolo J J. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu Rev Microbiol. 1989;43:375–402. doi: 10.1146/annurev.mi.43.100189.002111. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko J, Kimura T, Narita S, Tomita T, Kamio Y. Complete nucleotide sequence and molecular characterisation of the temperate staphylococcal bacteriophage wPVL carrying Panton-Valentine leukocidin genes. Gene. 1998;215:57–67. doi: 10.1016/s0378-1119(98)00278-9. [DOI] [PubMed] [Google Scholar]
- 14.Kappler J, Kotzin B, Herron L, Gelfand E W, Bigler R D, Boylston A, Carrel S, Posnett D N, Choi Y, Marrack P. Vβ-specific stimulation of human T cells by staphylococcal toxins. Science. 1989;244:811–813. doi: 10.1126/science.2524876. [DOI] [PubMed] [Google Scholar]
- 15.Kline J B, Collins C M. Analysis of the superantigenic activity of mutant and allelic forms of streptococcal pyrogenic exotoxin A. Infect Immun. 1996;64:861–869. doi: 10.1128/iai.64.3.861-869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 17.Miethke T, Duschek K, Wahl C, Heeg K, Wagner H. Pathogenesis of the toxic shock syndrome: T cell mediated lethal shock caused by the superantigen TSST-1. Eur J Immunol. 1993;23:1494–1500. doi: 10.1002/eji.1830230715. [DOI] [PubMed] [Google Scholar]
- 18.Mourad W, Scholl P, Diaz A, Geha R, Chatila T. The staphylococcal toxic shock syndrome toxin 1 triggers B cell proliferation and differentiation via major histocompatibility complex-unrestricted cognate T/B cell interaction. J Exp Med. 1989;170:2011–2022. doi: 10.1084/jem.170.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller-Alouf H, Alouf J E, Gerlach J H, Ozegowski Fitting C, Cavaillon J M. Comparative study of cytokine release by human peripheral blood mononuclear cells stimulated with Streptococcus pyogenessuperantigenic erythrogenic toxins, heat-killed streptococci, and lipopolysaccharide. Infect Immun. 1994;62:4915–4921. doi: 10.1128/iai.62.11.4915-4921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson S H, Tremaine M T, Betley M J, Welch R A. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen H, Engelbrecht J, Brunak K, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Papageorgiou A V, Acharya K R, Shapiro R, Passalacqua E F, Brehm R D, Tranter H S. Crystal structure of the superantigen enterotoxin C2 from Staphylococcus aureusreveals a zinc-binding site. Structure. 1995;3:769–779. doi: 10.1016/s0969-2126(01)00212-x. [DOI] [PubMed] [Google Scholar]
- 23.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 24.Prevost G, Cribier B, Couppie P, Petiau P, Supersac G, Finck-Babancon V, Monteil H, Piemont Y. Panton-Valentine leukocidin and gamma-hemolysin from Staphylococcus aureusATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995;63:4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proft T, Moffatt S L, Berkahn C J, Fraser J D. Identification and characterization of novel superantigens from Streptococcus pyogenes. J Exp Med. 1999;189:89–101. doi: 10.1084/jem.189.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddi K, Nair S P, White P A, Hodges S, Tabona P, Meghji S, Poole S, Wilson M, Henderson B. Surface-associated material from the bacterium Actinobacillus actinomycetemcomitanscontains a peptide which, in contrast to lipopolysaccharide, directly stimulates fibroblast interleukin-6 gene transcription. Eur J Biochem. 1996;236:871–876. doi: 10.1111/j.1432-1033.1996.00871.x. [DOI] [PubMed] [Google Scholar]
- 27.Ren K, Bannan J D, Panchi V, Cheung A L, Robbins J C, Fischetti V A, Zabrskie J B. Characterisation and properties of a new staphylococcal exotoxin. J Exp Med. 1994;180:1675–1683. doi: 10.1084/jem.180.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussel A, Anderson B F, Baker H M, Fraser J D, Baker E N. Crystal structure of the streptococcal superantigen SPE-C: dimerization and zinc binding suggests a novel mode of interaction with MHC class II molecules. Nat Struct Biol. 1997;4:635–643. doi: 10.1038/nsb0897-635. [DOI] [PubMed] [Google Scholar]
- 29.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Schad E M, Zaitseva I, Zaitsev V N, Dohlsten M, Kalland T, Schlievert P M, Ohlendorf D H, Svensson L A. Crystal structure of the superantigen staphylococcal enterotoxin type A. EMBO J. 1995;14:3293–3301. doi: 10.1002/j.1460-2075.1995.tb07336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz F J, Steiert M, Hofmann B, Verhoef J, Hadding U, Heinz H P, Kohrer K. Development of a multiplex-PCR for direct detection of the genes for enterotoxin B and C, and toxic shock syndrome toxin-1 in Staphylococcus aureusisolates. J Med Microbiol. 1998;47:335–340. doi: 10.1099/00222615-47-4-335. [DOI] [PubMed] [Google Scholar]
- 32.Sundstrom M, Abrahmsen L, Antonsson P, Mehindate K, Mourad W, Dolsten M. The crystal structure of staphylococcal enterotoxin type D reveals Zn2+-mediated homodimerization. EMBO J. 1996;15:6832–6840. [PMC free article] [PubMed] [Google Scholar]
- 33.Swaminathan S, Furey W, Pletcher J, Sax M. Crystal structure of staphylococcal enterotoxin B, a superantigen. Nature. 1992;359:801–806. doi: 10.1038/359801a0. [DOI] [PubMed] [Google Scholar]
- 34.Tabona P, Reddi K, Khan S, Nair S P, Crean StJ, Meghji S, Wilson M, Preuss M, Miller A D, Poole S, Carne S, Henderson B. Homogeneous Escherichia colichaperonin 60 induces IL-1β and IL-6 gene expression in human monocytes by a mechanism independent of protein conformation. J Immunol. 1998;161:1414–1421. [PubMed] [Google Scholar]
- 35.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomita T, Kamio Y. Molecular biology of the pore-forming cytolysins from Staphylococcus aureusα- and γ-hemolysins and leukocidin. Biosci Biotechnol Biochem. 1997;61:565–572. doi: 10.1271/bbb.61.565. [DOI] [PubMed] [Google Scholar]
- 37.Wen R, Cole G A, Surman S, Blackman M A, Woodland D L. Histocompatibility complex class II-associated peptides control the presentation of bacterial superantigens to T-cells. J Exp Med. 1996;183:1083–1092. doi: 10.1084/jem.183.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White J, Herman A, Pullen A M, Kubo R, Kappler J W, Marrack P. The Vβ-specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell. 1989;56:27–35. doi: 10.1016/0092-8674(89)90980-x. [DOI] [PubMed] [Google Scholar]