Abstract
Lipid nanoparticle (LNP)–based mRNA delivery holds promise for the treatment of inherited retinal degenerations. Currently, LNP-mediated mRNA delivery is restricted to the retinal pigment epithelium (RPE) and Müller glia. LNPs must overcome ocular barriers to transfect neuronal cells critical for visual phototransduction, the photoreceptors (PRs). We used a combinatorial M13 bacteriophage–based heptameric peptide phage display library for the mining of peptide ligands that target PRs. We identified the most promising peptide candidates resulting from in vivo biopanning. Dye-conjugated peptides showed rapid localization to the PRs. LNPs decorated with the top-performing peptide ligands delivered mRNA to the PRs, RPE, and Müller glia in mice. This distribution translated to the nonhuman primate eye, wherein robust protein expression was observed in the PRs, Müller glia, and RPE. Overall, we have developed peptide-conjugated LNPs that can enable mRNA delivery to the neural retina, expanding the utility of LNP-mRNA therapies for inherited blindness.
Peptide-guided LNPs enable mRNA delivery to photoreceptors, expanding the utility of LNP-mRNA therapies for inherited blindness.
INTRODUCTION
Lipid nanoparticles (LNPs) are the most clinically advanced nonviral platform for mRNA delivery. Their worldwide dissemination as part of the coronavirus disease 2019 vaccine has proven their safety and efficacy (1). Our efforts have focused on using LNP mRNA delivery for the treatment of inherited retinal degenerations (IRDs). IRDs are a complex group of genetic disorders that arise from mutations in the over 300 different genes associated with retinal pathology (2, 3). These genes are critical for photoreceptor (PR) and retinal pigment epithelium (RPE) function and when compromised, progressive cell death leads to blindness (4, 5).
Gene augmentation, editing, and silencing are the most attractive forms of clinical care for these patients as they correct the causative genomic malfunction. Luxturna, which uses an adeno-associated virus (AAV2) to deliver a normal copy of the RPE65 gene to the RPE, is U.S. Food and Drug Administration (FDA)–approved for patients with biallelic RPE65 Leber congenital amaurosis (6). Luxturna has established the safety and efficacy of gene augmentation via subretinal delivery, and, now, many different AAV gene therapies are under preclinical and/or clinical development (7, 8). In addition to subretinal administration, intravitreal delivery of AAVs, for X-linked retinoschisis, Leber hereditary optic neuropathy, and mutation-independent optogenetic strategies, is also demonstrating safety and efficacy in various clinic trials (9–12). AAVs have facilitated the advancement of gene editing in the retina, as the first in vivo CRISPR genome-editing medicine, EDIT-101, was administered to patients with CEP290 Leber congenital amaurosis (13). While both subretinal and intravitreal AAV gene therapy strategies are benefiting patients (6, 12), AAVs have three main limitations, including limited DNA packaging capacity (<5 kb), immunogenicity (14), and the ability to constitutively express Cas9 nucleases (15, 16), which highlight the critical need to develop next-generation gene delivery vehicles for the retina.
LNPs have a well-established safety profile in the clinic (17–19). These modular systems can encapsulate large size cargos, and their synthetic biodegradable chemistries alleviate the pressures of neutralizing antibodies and sustained immune responses (20, 21). Our long-term goal is to develop LNP gene editing platforms for IRDs. Ideally, gene editors would be delivered in the form of mRNA, allowing for robust and transient expression of nucleases, mitigating off-target effects. We know that LNPs can deliver mRNA-based cargo, which leads to rapid protein production, in hard-to-transfect nondividing cells with no risk of genomic integration (22, 23). However, before replacing their viral counterparts, LNPs must be able to transfect the neuronal cells, which harbor many of the mutations associated with IRDs. Our previous work demonstrates that regardless of compositional modifications, protein expression in the retina is predominately restricted to the phagocytic RPE cells and Müller glia (24–28). Either through subretinal or intravitreal delivery, LNPs have been unable to penetrate the neural retina, limiting their ability to deliver genes or gene editors to one of the most important cellular targets, the PRs.
We postulated that chemically decorating LNPs with a short 7-nucleotide oligomer peptide would allow them to permeate into the neural retina. Peptides are sequences of amino acids with varying lengths that can be naturally occurring or chemically synthesized. A given peptide chain can fulfill both a structural or bioactive role depending on charge densities, hydrophobicity, hydrophilicity, structural conformations, and chemical modifications (29, 30). By crossing biological barriers, peptides have been able to enhance drug delivery, imaging agents, and nanoparticle drug targeting (31, 32). We used a diverse M13 bacteriophage–based, heptameric peptide library to identify peptide sequences that bind the neural retina in vivo. Then, chemically synthesized, peptide conjugates of these hits were used to confirm localization to our cell of interest. Decoration of these peptides on the surface of LNPs with varying surface densities resulted in successful delivery of mRNA to the neural retina in a mouse model. These results translated to the more clinically relevant nonhuman primate (NHP), where robust protein expression was observed in the PRs, Müller glia, and RPE. Overall, we have identified peptide-conjugated LNPs that can facilitate the delivery of mRNA to the neural retina, expanding the utility of LNP-mRNA therapies for inherited blindness.
RESULTS
In vivo bacteriophage biopanning
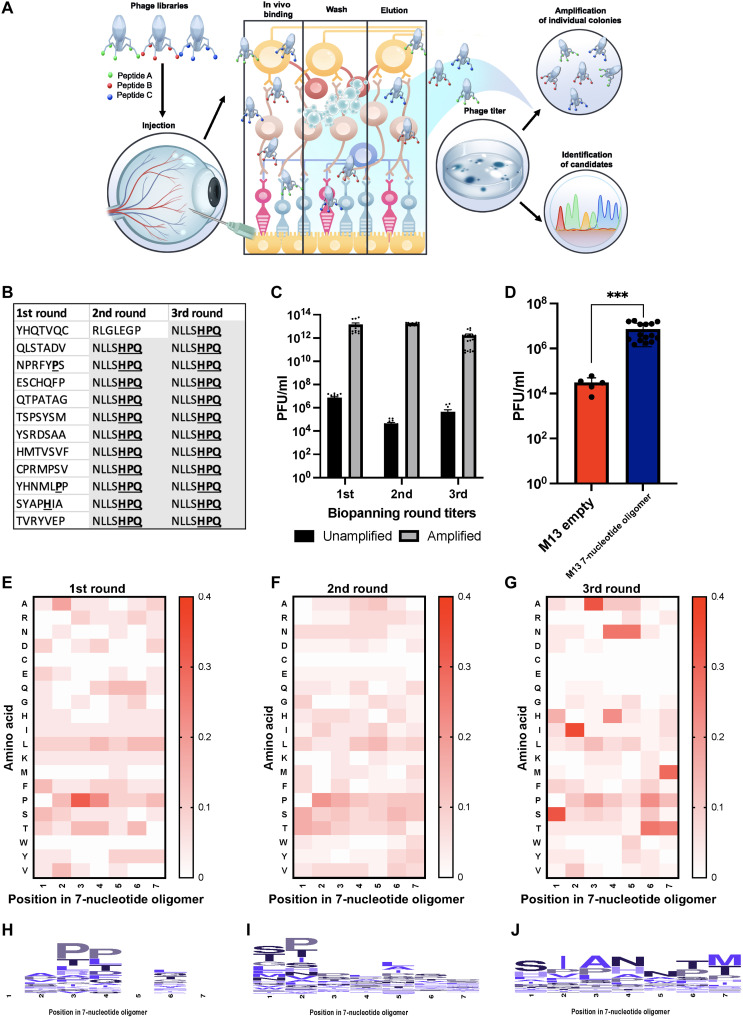
To elucidate retina-specific peptide sequences, we intravitreally injected a naïve heptameric M13 bacteriophage–based library in mice. At 6 hours after injection, the neural retina was harvested and washed, and bound bacteriophage-peptide fusions were eluted off the tissue. Collected bacteriophage-peptide fusions were either amplified for another round of injections (enriched library) or sequenced to elucidate the specific bacteriophage-peptide fusions that bound to the neural retina after intravitreal delivery (Fig. 1A). The initial validation experiment using the naïve M13 bacteriophage library against streptavidin showed that three iterative rounds of biopanning were necessary to isolate a known target sequence (Fig. 1B). Thus, when using the M13 bacteriophage library in vivo, BALB/c mice were injected intravitreally for three iterative rounds of biopanning. Titers were measured after each round, quantifying the amount of unamplified bacteriophage-peptide fusions isolated from the tissue, and then amplified in culture (enriched library) for subsequent panning round injection. Titers measured after each round of in vivo biopanning were detected within the expected ranges (Fig. 1C). An empty, M13 bacteriophage library, delivered at an equivalent concentration, was used as a negative control. Titers retrieved from the neural retina after the first round of biopanning were significantly higher when using the full M13 bacteriophage library compared to the empty M13 bacteriophage library (Fig. 1D). A notable 100-fold higher bacteriophage-peptide count from the naïve library biopanning suggests that the displayed peptides were facilitating the binding of the neural retina, as opposed to other proteins on the surface of the empty bacteriophage itself (Fig. 1D). After each round of biopanning, eluted unamplified phage plaques were Sanger sequenced, and the normalized occurrence of each amino acid at each position in the heptameric moiety was displayed to elucidate the enrichment across the three rounds. After the third round of biopanning, SIA(N/H)NT(M/T) appeared as a motif, which is shown in the heatmaps and accompanying enrichment plots (Fig. 1, E to J). Overall, after the third round of biopanning, over 150 phage plaques were sequenced, and 30 unique sequences were identified for retinal targeting showcasing the attained enrichment (table S1).
Fig. 1. In vivo development of peptide phage display biopanning.
(A) In vivo schematic. Heptameric phage libraries were injected intravitreally into BALB/c mice and exposed for 6 hours, followed by retina extraction. Loosely bound phages were subjected to washes and elution to be used in titering and amplification rounds for subsequent biopanning rounds. The process was carried out three times and enriched specific phage-peptide binders were isolated, followed by DNA extraction and Sanger sequencing. (B) Streptavidin-targeted phage biopanning positive control. (C) In vivo phage titers after each round of biopanning. (D) In vivo peptide-targeting validation. Empty M13 bacteriophages were used as a negative control compared to peptide-containing library at the same concentration injected intravitreally in mice using an ordinary one-way analysis of variance (ANOVA). n = 3; means ± SEM. ***P ≤ 0.001. (E to G) Heatmaps depicting phage biopanning enrichment after rounds 1 to 3 in vivo of peptide sequences isolated for each round. For biopanning rounds, n = 3 with at least five technical replicates. (H to J) Consensus sequence for each amino acid in the heptameric sequence isolated after biopanning rounds 1 to 3 in vivo.
Differential binding of in vivo–isolated phage-displayed peptides
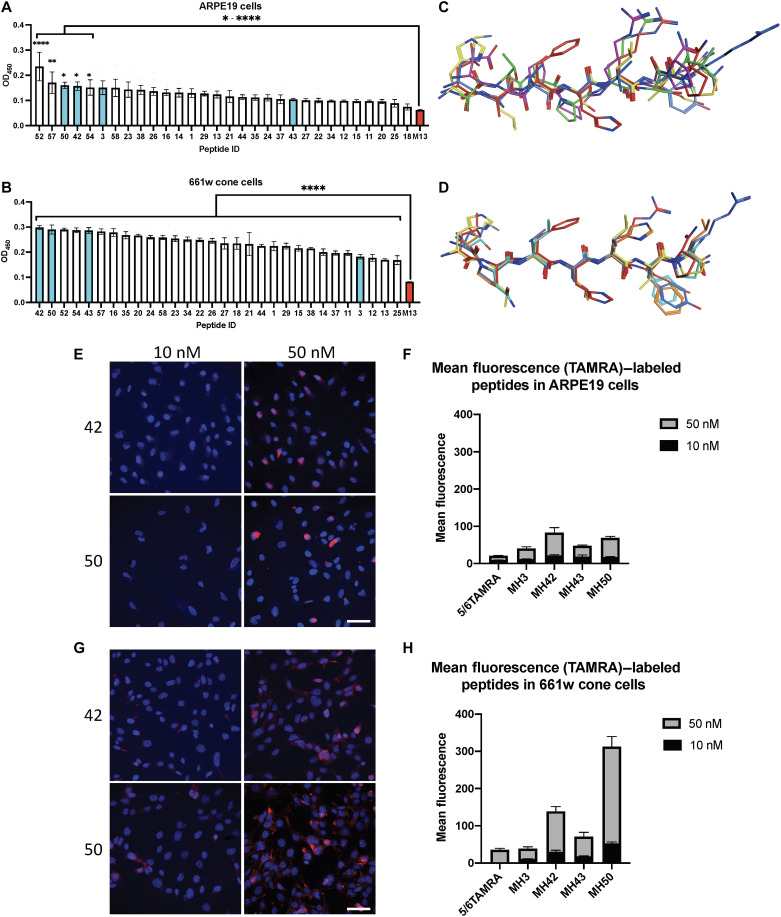
After identifying 30 peptide sequences, we worked to identify top-performing candidates that can target LNPs to the neural retina. Since bacteriophages were eluted from the entire neural retina, we sought to prescreen the candidates for their ability to bind to our cell of interest, the PRs. Single bacteriophage peptides were amplified in Escherichia coli, and a cell-based enzyme-linked immunosorbent assay (ELISA) was developed against the 661w mouse cone PR cell line and the hARPE19 cell line as a control. As expected, for most of the bacteriophage-peptide candidates, binding affinities were weaker when tested against RPE cells compared to 661w cone cells. However, in hARPE19 cells, peptides MH52, MH57, MH50, MH42, MH54, and MH3 showed significantly higher binding to RPE cells compared to control (*P ≤ 0.05 and ****P ≤ 0.0001; Fig. 2A). In the 661w cone cells, all 30 phage peptides tested exhibited significantly increased binding affinities compared to empty bacteriophage control, highlighting their neural retina affinity (****P ≤ 0.0001; Fig. 2B). Some phage-peptide fusions exhibited high binding across both of the ocular cell lines, such as peptides MH42, MH50, MH52, and MH57. However, the binding affinity, as measured by optical density at 450 nm, was close to twofold higher when binding to 661w cone cells compared to hARPE19 cells. After confirming binding affinity to our cell of interest, we continued our analysis with five peptides that had the highest affinity for 661w cells (42, 43, 50, 52, and 54) and ARPE19 cells (3, 50, 52, 54, and 57) as a control (figs. S1 and S2).
Fig. 2. In vitro binding, structural analysis, and internalization of in vivo–isolated peptide candidates.
Single phage-peptide differential binding properties were elucidated using cell-based ELISA against model cell lines of desired target tissues in neural retina. Three high-performing hits and a low binder were selected from ELISA results for further analysis and validation across both cell lines. (A and B) Cell-based ELISA results for ARPE19 cells and 661w cells, respectively, with selected hits highlighted in cyan. Empty M13 bacteriophage used as negative control (red). (C and D) Molecular operating environment (MOE) structural superposition of selected candidates for ARPE19 and 661w cells, respectively. (E and G) Confocal microscopy images of TAMRA-labeled peptides 42 and 50 cell internalization after 30-min incubation with ARPE19 and 661w cells, respectively. Scale bars, 50 μm. (F and H) Mean fluorescence intensity quantification of confocal images. ELISA binding experiments performed in duplicate with six technical replicates; means ± SEM. An ordinary one-way ANOVA, with Tukey’s correction for multiple comparisons test was used for comparisons between treatments. *P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001.
Pharmaceutically relevant properties and three-dimensional superposition of peptide sequences tested
The physicochemical properties of the peptide sequences and their structure governed by amino acid composition were analyzed using the QikProp module of Schrödinger drug discovery suite and Molecular Operating Environment (MOE) software. Pharmaceutically relevant physicochemical properties were computed and reported for all of the candidate peptide sequences tested (table S1), while those that had highest binding against 661w cone cells and hARPE19 cells are highlighted (figs. S1A and S2A). As expected, for hARPE19 top performers, there were no conserved structural patterns of pertinence in the structural superposition analysis (Fig. 2C). For 661w cone cell top candidates, the structural superposition of physiologically relevant conformations showed a high conservation in three-dimensional (3D) models across all five peptides (Fig. 2D). In general, the top peptides were made up of mostly neutral, slightly basic amino acid residues, which were preferable given their compatibility with our LNP carriers and required endosomal escape–prone profile (figs. S1B and S2). In contrast, MH3, which showed reduced binding affinity in the 661w cone cells, contained slightly more acidic amino acid residues (fig. S2B).
Cell internalization of fluorescently-labeled peptides
Although differential binding results were promising, before LNP conjugation, we wanted to confirm cellular internalization of peptide ligands. Four peptides were chosen from the 661w cone cell–based ELISA results: three high binders (MH50, MH43, and MH42) and one low binder (MH3) to serve as negative control (Fig. 2, A and B). These peptides were synthesized with a carboxy-end linker, -GGGS (table S1), followed by a C-terminal cysteine that was conjugated to a 5-(and-6)-carboxytetramethylrhodamine (TAMRA) dye.
TAMRA-conjugated peptides were added to hARPE19 and 661w cells at concentrations of 10 and 50 nM to evaluate their internalization (Fig. 2, E to H, and fig. S3, A and B). After 30-min incubation at 37°C, cells were fixed and 4′,6-diamidino-2-phenylindole (DAPI)–stained for confocal imaging. We observed 2.3× higher uptake in 661w cone cells when compared to RPE cells as measured by mean fluorescence intensity across peptides tested at concentrations of 10 and 50 nM (Fig. 2, F and H). Peptides MH50 and MH42 elicited 4.1- and 1.7-fold higher cellular uptake compared to the negative control MH3 in 661w cells, respectively. MH42 and MH50 were also the top 2–performing peptides in the 661w differential binding affinity ELISA. Together, these in vitro validation studies suggest that these peptides can bind to and be internalized into our cells of interest (Fig. 2).
In vivo validation of labeled peptides with conjugated TAMRA dye
After verifying binding and internalization in vitro, we proceeded to test penetrative properties and targeting capabilities of our TAMRA-labeled peptide candidates in BALB/c mice. First, we needed to elucidate the pharmacokinetic profile of the peptides candidates. All peptides conjugated to TAMRA dye were injected intravitreally, and eyes were harvested at 1, 6, and 24 hours after administration (Fig. 3 and fig. S4). TAMRA dye was visible in the retina at 1 hour after injection; however, maximum levels of labeled peptide were observed at the 6-hour time point. Peptides were cleared by 24 hours after injection (Fig. 3 and fig. S4). In vivo fundus imaging after MH42 injection support these findings and show the highest TAMRA fluorescence intensity at 6 hours and whole clearance by 24 hours (Fig. 3, A to D). With maximum localization of labeled peptide observed at 6 hours, all quantification was performed at the 6-hour time point. In addition to intravitreal delivery, TAMRA-labeled peptides were also injected subretinally and harvested at the 6-hour time point (Fig. 3 and fig. S4).
Fig. 3. In vivo injections of TAMRA-conjugated peptide candidates in BALB/c mice.
Representative images of the clearance kinetics and in vivo targeting of TAMRA-labeled peptides injected intravitreally and subretinally into BALB/c mice and extracted at specific time points after injection. (A to D) In vivo fundus images of peptide MH42 intravitreally and subretinally delivered at the corresponding time points. Top: Bright-field images of the eye. Bottom: Red fluorescence demonstrating localization of the peptide. (E to H and I to L) Confocal images of 12-μm cryosections following intravitreal and subretinal injections of peptides MH42 and MH50, respectively, to validate peptide targeting. Scale bars, 25 μm. (M) Schematic identifying the RPE/choroid and PR layers used for quantification. For the analysis, these layers were manually segmented in ImageJ. (N) Intravitreal and (O) subretinal mean fluorescence intensity quantification of confocal images for localized fluorescence in PR or RPE/choroid layers. An ordinary one-way ANOVA, with Tukey’s correction for multiple comparisons test was used for comparisons between groups. n = 4 to 8 eyes per group; means ± SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001. TAMRA dye injected as negative control. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer.
Both MH42 and MH50 were localized to PRs including the outer nuclear layer and inner/outer segments as well as RPE (Fig. 3, E to L). For quantification, the PR and RPE/choroid layers were segmented to obtain mean fluorescent intensity (Fig. 3M). When injected intravitreally, MH42 demonstrated 2.2- and 3.2-fold increase in PR and RPE/choroid localization compared to TAMRA dye injection control, respectively (**P ≤ 0.01; Fig. 3N and fig. S4C). With MH42 subretinal administration, increased PR and RPE/choroid localization was observed by 5.1- and 7.9-fold, respectively (**P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; Fig. 3O and fig. S4D). For MH50, PR and RPE/choroid localization was increased by 1.8- and 2.6-fold after intravitreal administration and by 4.4- and 6.6-fold after subretinal administration (*P ≤ 0.05 and **P ≤ 0.01;Fig. 3, N and O, and fig. S4, C and D). As expected, the negative control, MH3, showed poor accumulation in RPE and PRs at all time points tested after injection. In the case of peptide MH43, affinity toward the PRs was observed at the 6-hour time point after intravitreal injection but not after subretinal injection (fig. S4, A and B).
In vivo validation of targeting peptides on surface of Cre mRNA–loaded LNPs in Ai9 mice
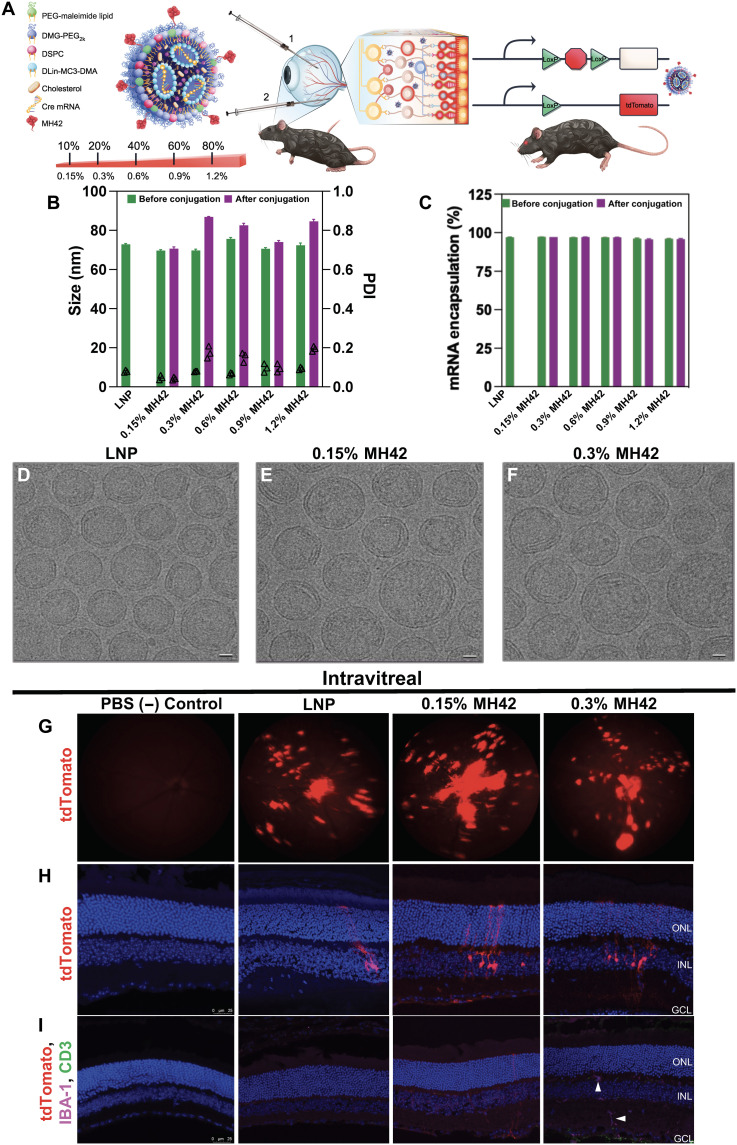
Encouraged with the localization of TAMRA dye–conjugated peptide candidates in BALB/c mice, we used a gene delivery reporter mouse model to visualize cell-based gene editing. Ai-9 mice stably express a floxed stop codon upstream of a tdTomato cassette in all cells, which is only translated after successful Cre-recombinase (Cre) delivery recombination (Fig. 4A). We encapsulated Cre mRNA inside of LNPs formulated with cholesterol, (6Z,9Z,28Z,31Z)-heptatriacont-6,9,28,31-tetraene-19-yl 4-(dimethylamino)butanoate (DLin-MC3-DMA), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2K) as the ionizable, structural, and PEG lipid, respectively (Fig. 4A). For LNP-peptide conjugates, molar equivalents of the PEG-lipid component were substituted for varying amounts of functionalized PEG lipid, in this case, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG2K-maleimide), for direct conjugation to the peptides via their substituted carboxy-end cysteine. The carboxy end of the peptide ligand’s terminal cysteine was amide capped to mimic the physicochemical properties of the original peptide obtained from the screenings. The free ─SH group on the cysteine side chain facilitated direction conjugation onto the surface of our LNP with the PEG lipid via thio-ester conjugation. We explored a wide range of targeting ligand surface densities, evaluating a gradient from 0.15 to 1.2% of the 1.5% total PEG-lipid content, equivalent to 10 to 80% of total PEG (fig. S5A). LNPs with varying levels of ligand on surface were formulated with Cre mRNA and subsequently injected into Ai-9 mice. Unconjugated LNPs loaded with Cre-mRNA served as nontargeted baseline comparison in our studies. LNP characterization using dynamic light scattering and nanoparticle tracking analysis (NTA) measured LNP size as 70 to 76 nm in diameter before peptide conjugation with a uniform polydispersity index (PDI) of 0.1 indicating a homogeneous preparation. After conjugation, LNP-MH42 conjugates displayed slightly higher diameters (71 to 87 nm) and an increased PDI likely due to the inclusion of peptides on their surface (Fig. 4B). mRNA encapsulation ranged between 95 and 99% for all LNPs ensuring almost complete encapsulation of cargo across all formulations tested irrespective of ligand conjugation amount (Fig. 4C). Cryo–electron microscopy (EM) of LNPs with and without peptides corroborated once again that LNP morphology was undisturbed by the conjugation strategy (Fig. 4, D to F). To quantify the amount of peptide conjugated on surface of nanoparticle, we used a fluorometric absorbance kit that detects free maleimide groups before and after conjugation. The calculated total amount (w/v) of peptide was 376 and 492 pg for 0.15 and 0.3% MH42 per microliter of LNP solution, respectively.
Fig. 4. Peptide-conjugated LNP characterization and in vivo injections in Ai9-tdTomato mice with increasing peptide surface density.
Particle characterization and representative confocal images of MH42 peptide–conjugated, Cre mRNA–loaded LNPs intravitreally administered to Ai9-Rosa mice. (A) Schematic of LNP formulation and conjugation with peptide via maleimide-thiol chemistry and Cre mouse model depicting both routes of administration trialed. (B and C) Graphs depicting size, polydispersity, and mRNA encapsulation efficiency of the LNPs with varying ratios of MH42 peptide conjugation. % refers to molar percent of peptide-functionalized PEG in the formulation. (D to F) Cryo–transmission EM images of the unconjugated control LNP, 0.15% MH42, and 0.3% MH42-LNP. Scale bars, 20 nm. (G) Representative fundus images showing in vivo tdTomato expression after intravitreal delivery of unconjugated and conjugated LNPs. (H) Confocal microscopy images of tdTomato expression following intravitreal uptake and translation of Cre mRNA LNPs. (I) Confocal images showing no T cell infiltration (CD3) or microglia activation (IBA-1) associated with intravitreal delivery and expression. Microglia were restricted to the plexiform layers (arrowheads). Confocal images taken at ×20. n = 6 eyes per group. Scale bars, 50 μm.
At 7 days after intravitreal or subretinal administration of LNPs with and without MH42, tdTomato expression was visualized in vivo with fundus imaging and postmortem with confocal microscopy of retinal cryosections. We initially tested untargeted LNP and five different conjugated preparations, spanning 10 to 80% of total 1.5% PEG content, intravitreally (Fig. 5, G to I, and fig. S5B). Formulations with over 40% substitution with PEG-maleimide showed no detectable levels of transfection (fig. S5B). However, MH42 at 0.15% (10% of total PEG), 0.3% (20% of total PEG), and 0.6% (40% of total PEG) showed Müller glia transfection after intravitreal administration (Fig. 5, G and H, and fig. S5B). Müller glia transfection was not associated with any signs of an immune response or retinal toxicity. There was no T cell infiltration as evident by CD3 staining and the microglia, labeled with IBA-1, were restricted to the plexiform layers (Fig. 4I, arrowheads). Positive controls for IBA-1 and CD3 stains are located in the Supplementary Materials (fig. S6).
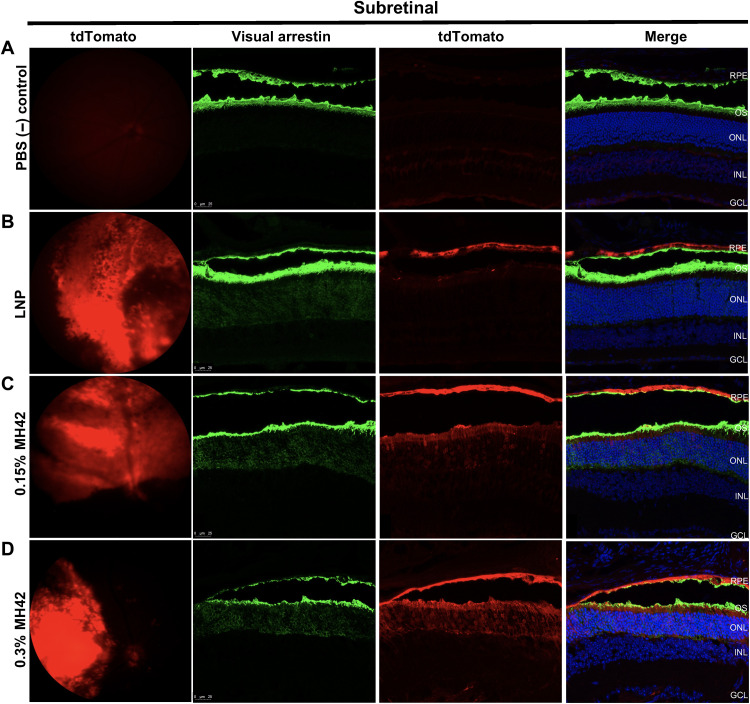
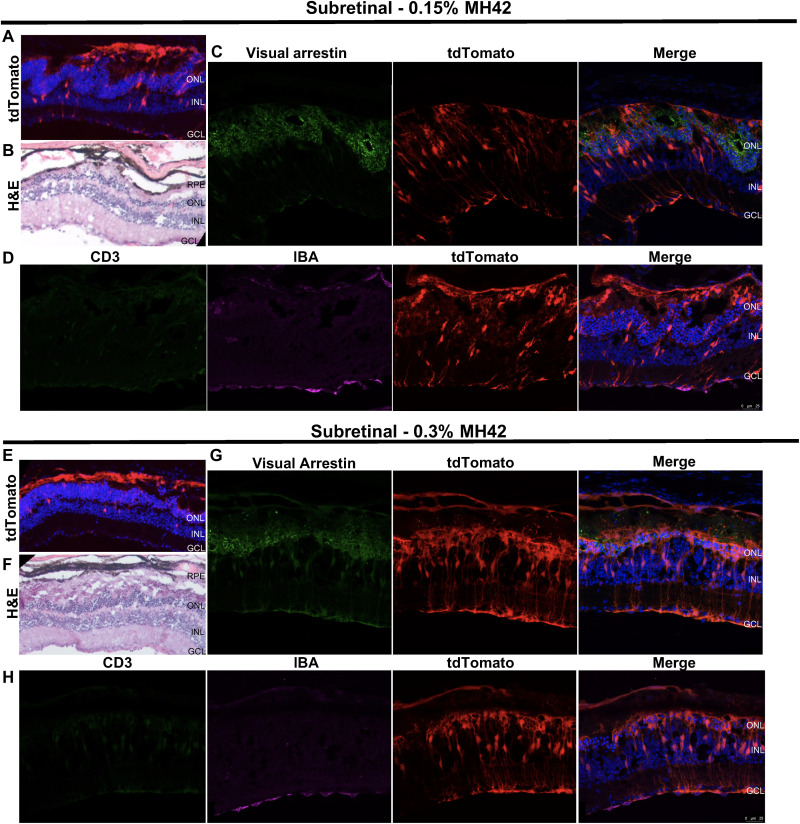
Fig. 5. MH42-conjugated LNPs mediate PR expression after subretinal administration.
Representative fundus images showing in vivo tdTomato expression combined with ×40 confocal images of retinal cross sections expressing tdTomato (red) and stained with visual arrestin (green; rods and cones) and DAPI (blue) for (A) phosphate-buffered saline (PBS), (B) untargeted LNPs (n = 2), (C) 0.15% MH42 LNPs (n = 6), and (D) 0.3% MH42 LNPs (n = 4).
After subretinal administration of untargeted LNPs, transfection was predominately observed in the RPE, as previously published (24, 25) (Fig. 5B). However, with the addition of MH42 at 0.15 and 0.3%, tdTomato expression was observed in the RPE and PRs. Retinal cross sections were costained with visual arrestin, which labels the cytoplasm of rod and cone cell bodies as well as the entire outer segments (Fig. 5, A to D). Visual arrestin labeling aligned with the tdTomato expression, confirming PR delivery by MH42 LNP (Fig. 5, C and D). Areas of MH42 LNP–injected retinas contained disrupted retinal morphology corresponding with the loss of the PRs (Fig. 6, A, B, E, and F). More specifically, there was a loss of PR inner/outer segments and the formation of outer nuclear layer rosettes (Fig. 6, C and G). There was still presence of PR transfection, but, more notably, there was an increase in Müller glia transfection in these areas (Fig. 6, C and G). Retinal cross sections were stained with IBA-1 and CD3 to detect an immune response corresponding to the retinal damage (Fig. 6, D and H). T cell infiltration was not detected and there was no evidence of microglia activation (Fig. 6, D and H). Even with these retinal disruptions, overall, we determined 0.15 and 0.3% peptide substitution to be optimal for mRNA delivery of ligand-functionalized LNPs. (Figs. 4 to 6).
Fig. 6. Retinal toxicity associated with subretinal administration of MH42 LNPs.
Top: Representative images from retinas injected with 0.15% MH42 LNPs (n = 6). Bottom: Representative images from retinas injected with 0.3% MH42 LNPs (n = 4). (A and E) Confocal images at ×10 containing tdTomato expression (red) and labeled with DAPI (blue). (B and F) Hematoxylin and eosin (H&E) images show the retinal morphology in some areas of tdTomato expression. (C and G) Confocal images at ×40 demonstrating tdTomato expression (red) and stained with visual arrestin (green) and DAPI (blue). (D and H) Confocal images at ×40 showing tdTomato expression (red) and stained with CD3 (green; T cells), IBA-1 (microglia; magenta), and DAPI (blue).
Subretinal delivery to NHP retina
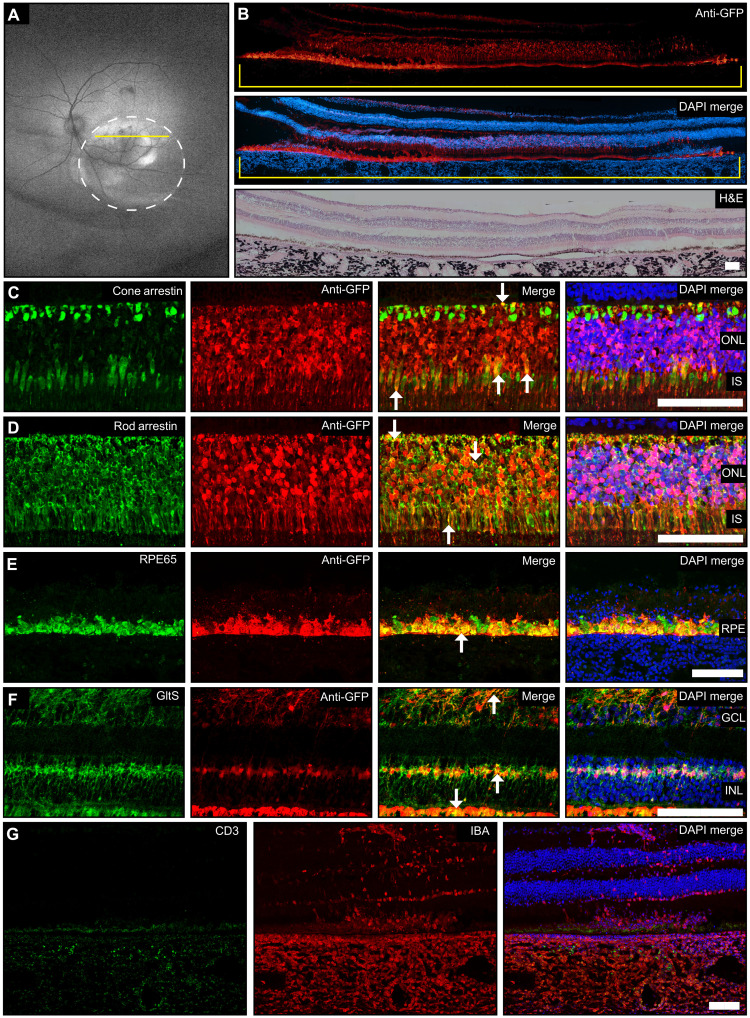
Since subretinal delivery with our formulations showed PR transfection, we explored the translatability of 0.15% MH42 LNP–mediated mRNA delivery in a highly relevant NHP model, the rhesus macaque. Green fluorescent protein (GFP) mRNA LNPs were formulated using cholesterol, DLin-MC3-DMA, DSPC, and DMG-PEG2K. For direct conjugation to the peptides, 0.15% molar equivalent of the PEG-lipid component was substituted for functionalized PEG, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carboxy(polyethylene glycol)-2000, N-hydroxysuccinimide ester (DSPE-PEG2K-carboxy-NHS). MH42 was modified by adding a C-terminal lysine for conjugation via the primary amine on its side chain. The α-NH2 was acetylated to prevent conjugation with functionalized LNP, while the carboxy end was left unchanged with amide capping to maintain the physicochemical properties of the initially screened original peptide to react with primary amine on the lysine side chain, thereby directionally conjugating onto the surface of the LNP. Formulated nanoparticles were 91.6 nm in diameter with a PDI value of less than 0.05. Encapsulation efficiency was 98.5%. At 48 hours after subretinal delivery of 50 μg of GFP mRNA in a 100-μl volume, wide-field fundus autofluorescence images showed GFP expression within the margins of the bleb (Fig. 7A). Confocal images of retinal cross sections immunolabeled with an antibody specific to GFP showed robust expression in the PRs and RPE throughout the bleb (Fig. 7B). Hematoxylin and eosin (H&E) imaging demonstrated reattachment of the neural retina to the RPE within the bleb, 48 hours after injection (Fig. 7B). Further staining showed GFP expression colabeled with cone arrestin, rod arrestin, RPE65, and glutamine synthetase, verifying that expression was localized to PRs, RPE, and Müller glia, respectively (Fig. 7, C to F). Arrows highlight places of colocalization for each cell type. For both rod and cone arrestin, colocalization with GFP was observed in the synapses and inner segments. In addition, the rod arrestin and glutamine synthetase antibodies labeled the cytoplasm of the cell bodies, while the GFP expression was observed in the nucleus of the cell bodies. Therefore, much of the colocalization has the appearance of the antibody surrounding the GFP expression (Fig. 7, D and E). We evaluated the immune response using CD3 and IBA-1 markers. Imaging showed microglia activation and, to a lesser degree, T cell infiltration in the choroid, corroborating immune-related inflammation (Fig. 7G). These data provide additional evidence of successful delivery of mRNA to the neural retina with the utilization of 0.15% MH42 peptide–conjugated LNP.
Fig. 7. MH42-conjugated LNPs mediate expression in the neural retina after subretinal administration in the NHP.
(A) Wide-field fundus autofluorescence imaging 48 hours after subretinal delivery of MH42 LNPs (n = 1). Circle demarks the location of the bleb, and yellow line indicates cross section that corresponds to immunohistochemistry. (B) Montaged ×10 confocal images of primate retinal cross sections labeled with anti-GFP antibody (red) and DAPI (blue). A montaged ×10 H&E image shows the retinal morphology in areas of GFP expression. (C to F) Confocal images (×40) of retinal cross sections costained with anti-GFP and cell-specific antibodies cone arrestin (cones), rod arrestin (rods and s-cones), RPE65 (RPE), and glutamine synthetase (Müller glia). Arrowheads represent co-localization. (C to G) Confocal cross sections (×20) labeled with CD3 (T cells; green) and IBA-1 (microglia; red) to observe the elicited immune response. GltS, glutamine synthetase.Scale bars represent 100μM.
DISCUSSION
Gene therapies are quickly advancing to tackle genetic diseases of the retina as evidenced by recent landmark achievements such as the first FDA-approved gene therapy for a genetic condition (6) or the first-in-human delivery of CRISPR-Cas components to the retina for in vivo gene editing (13). Concurrent with the success of mRNA vaccines, nonviral LNPs have gained enormous momentum in the gene editing field. For example, systemic administration of LNP mRNA encoding for RNA-guided Cas9 has led to knockdown of misfolded transthyretin (TTR) protein in six patients with amyloidosis, with over 90% reduction in TTR protein (33). LNP-encapsulated CRISPR-Cas components have also shown tolerability and preliminary efficacy in a clinical trial addressing hereditary angioedema after a single systemic administration (Intellia). In addition, LNPs have been deployed in primates to deliver mRNA-encoded nucleases for base editing of proprotein convertase subtilisin/kexin type 9, a protein implicated in low-density lipoprotein–cholesterol management tied to ischemic heart disease (34).
As LNPs have become the principal nonviral gene delivery vehicle, our goal is to translate this platform into gene-editing therapeutics for the diverse forms of IRD. The main limitation of LNPs with regard to retinal delivery is their inability to penetrate the neural retina. We have shown that LNPs have restricted expression to the RPE when delivered subretinally (24), and after intravitreal delivery, they get sequestered in the vitreous, leading to minimal Müller glia expression (25). Targeting of the neural retina, and, in particular, the PRs, is required to advance LNP therapeutics for IRDs. Here, we show that multiple rounds of in vivo biopanning with a bacteriophage library containing 1.28 × 109 unique peptides elucidated 7-nucleotide oligomer peptide sequences that had increased binding affinity to 661w cone PR cells and increased PR localization after subretinal administration. Furthermore, we describe conjugation methods that allows for the generation of peptide-conjugated LNPs with ideal size, encapsulation, and morphology. When injected subretinally, peptide-conjugated LNPs mediated expression in PRs, RPE, and Müller glia in both rodents and NHPs. This is our first report showing LNP-mediated transfection in the PRs and successful translation to the NHP, which overall demonstrates the advancement of retinal LNP-mRNA delivery.
We initially hypothesized that these peptide-conjugated LNPs would increase neural retina penetration from the vitreous and would possibly offer cell-specific expression. We reasoned that injecting the bacteriophage library intravitreally would increase our chances of identifying penetrating ligands that could function with both intravitreal and subretinal delivery. For instance, any peptides that would be able to cross the vitreous humor and the inner limiting membrane to get to the neural retina would, in principle, also be able to penetrate the neural retina from the subretinal space. Although neuronal cells were not transfected from the vitreous, after subretinal delivery, successful transfection of the PRs was achieved. Results were consistent across species and conjugation strategies. First, we observed PR localization of the TAMRA-dye conjugated peptides in BALB/c mice. We determined the optimal surface density of MH42 onto the LNPs that lead to efficient gene delivery in Ai9 mouse. It is likely that intermediate surface density of these peptides provided either penetrative properties or interacted with still-undetermined cell receptors that mediated more efficient gene delivery; similar optimized MH42 LNP led to PR transfection in NHPs. In both the Ai9 mice and the NHP, the RPE and Müller glia were transfected as well. Most likely because of their highly phagocytic nature (24, 25). Future studies will use the use of RPE- and Müller glia–specific microRNAs to silence off-target expression (35).
QikProp analysis showed that MH42 has the highest predicted potential for .globular formation with low binding to serum albumin across peptides tested. It also has a histidine residue not found in the other candidates selected. PR transfection after subretinal delivery could have been a result of improved interaction with cell membranes or extracellular matrix leading to increased uptake and/or escape of nucleic acid cargo into the cytoplasmic compartment. Slightly basic amines have been postulated to be ideal in generating endosomal rupture events via proton sponge effect and osmotic swelling; therefore, it is possible this peptide’s physicochemical properties are aiding in release from endosomal vesicles and increased expression (36). Future studies examining MH42 LNP uptake, internalization, and trafficking in vivo will enable understanding of the mechanism of action.
We attempted to assess the immune response associated with the MH42 LNP delivery. In mice, we observed areas of the retina that had PR cell loss. These areas had disrupted/swollen RPE, a thinned outer nuclear layer, and increased Müller glia transfection. In these same areas, there was no detectable T cell infiltration or microglia activation. It is likely that the PR cell loss in the rodents is due to dose-related RPE toxicity from LNP overload/accumulation or robust tdTomato expression, which is known to be toxic (37). Once the RPE is compromised, they are unable to support the PRs, which leads to cell death. In contrast, in the NHP, we did not see any PR cell loss, but in areas of disrupted RPE, there was T cell infiltration in the choroid and microglia activation in the retina. The NHP was dosed at a 100× higher concentration than the rodents, which could account for the observable immune response. In addition, NHPs are known to have increased immunoreactivity compared to rodents (38). Overall, we suspect that the toxicity and immunogenicity are associated with LNP dose and these effects can be mitigated by reducing LNP dose. We will need to fully characterize the immune response associated with LNP-mRNA delivery to the retina, which would include performing a dose escalation study and assessing the kinetics of local and systemic responses, with and without immunosuppression. In addition to dose, novel biodegradable systems or new materials designed for retinal delivery can mitigate toxic effects. The current LNP formulation contains the MC3 ionizable lipid, which was developed for liver targeting and has been associated with increased toxicity as compared to biodegradable counterparts (21). Lowering the dose in combination with retina-targeting lipids could pave the way toward safe and effective LNP-mRNA therapeutics.
One main limitation of the study is that we eluted peptides from the entire neural retina after intravitreal delivery of the bacteriophage library. It is possible that if PRs were isolated and phage was eluted from an isolated PR population, then ligands may have improved PR-specific uptake. In addition, continued modifications of our elucidated peptide ligands, whether by means of circularization, chemical, or residue substitution strategies, have the potential to further enhance penetrative and targeting abilities. Further work is also needed to reveal the receptors within the retina that enable in vivo uptake, internalization, and endosomal escape of MH42 LNPs (39). In conclusion, this study shows that peptide-conjugated LNPs can be powerful tools to advance mRNA-based therapeutics in the retina. The translation of our results into the NHP eye highlights the advancement of LNP-mRNA delivery toward clinical application for IRDs.
MATERIALS AND METHODS
LNP and mRNA materials
Cre recombinase (Cre) and GFP mRNA fully substituted with 5-methoxyuridine were acquired from TriLink BioTechnologies (L-7211 and L7203, respectively). DLin-MC3-DMA was purchased from BioFine International Inc. (BC, Canada). DSPC was acquired from Avanti Polar Lipids Inc. (Alabaster, AL). DMG-PEG2K and cholesterol were obtained from Sigma-Aldrich (St. Louis, MO). For peptide-conjugated formulations, DMG-PEG2K was substituted with DSPE-PEG2K-maleimide for Ai9 mice and DSPE-PEG2K-carboxy-NHS for NHP.
LNP formulation, conjugation, and characterization
LNPs were formulated by microfluidic mixing using a previously described method (39). Briefly, ethanol solutions containing DLin-MC3-DMA, cholesterol, DSPC, and DMG-PEG2K, at molar ratios of 50:38.5:10:1.5, were mixed with 50 mM citrate buffer containing mRNA using a microfluidic mixer at a ratio of 1:3. LNPs were dialyzed two times using phosphate-buffered saline (PBS; pH 7.4) and concentrated with 10-kDa Amicon Ultra centrifuge filters (Millipore, Burlington, MA). For peptide-conjugated LNPs, DSPE-PEG2K-maleimide– and DSPE-PEG2K-carboxy-NHS–functionalized LNPs were incubated at 4°C overnight in PBS with peptide ligands at a molar excess of 10:1 (peptide:PEG) and subsequently concentrated via centrifugation. Size distribution and PDI of LNPs were determined via dynamic light scattering using a Zetasizer Nano ZSP (Malvern Instruments, UK), and NTA was done using ZetaView TWIN equipped with video microscope PMX-220 (Ammersee, Germany). mRNA encapsulation efficiency was determined using Quant-iT RiboGreen RNA reagent before and after peptide conjugation (Invitrogen, Carlsbad, CA). Conjugated peptide ligand concentration was determined using fluorometric maleimide assay kit (Sigma-Aldrich, catalog no. MAK167) as per the manufacturer’s protocol. This assay was used to measure the peptide content on DSPE-PEG-maleimide LNPs. It was not used for DSPE-PEG2Kcarboxy-NHS–functionalized LNPs.
Cell culture
Cell culture media and reagents were purchased from Thermo Fisher Scientific (Waltham, MA). 661w cone cells were provided by Muayyad Al-Ubaidi, University of Houston, Houston, TX. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) high glucose (catalog no. 11965175) + 10% fetal bovine serum (FBS) + progesterone (20 μg/ml) and Hydro-21 Heme at 37°C and 5% CO2 and split at a ratio of 1:6 once per week. hARPE19 cells (CRL-2302, American Type Culture Collection) were cultured in DMEM/F12 (50:50 mix; catalog no. 11320033) + 10% FBS at 37°C and 5% CO2 and split at a ratio of 1:10 once per week.
Mouse models
Albino BALB/c and Ai9-Rosa mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Male and female mice aged 2 to 4 months were used in experiments. All the experimental procedures followed the protocols approved by the Institutional Animal Care and Use Committee at Oregon Health and Science University and were in adherence to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Phage library and phage reagents
The Ph.D.-7 phage display peptide library was purchased from New England Biolabs (Ipswich, MA, USA). This pentavalent display library contained phages [approximately 1 × 1013 plaque-forming units (PFU)/ml] with a clonal diversity of 1.28 × 109 unique peptide sequences. All reagents necessary for phage display incubation, isolation, amplification, and purification prepared as described in the Ph.D.-7 manufacturer’s manual. Empty M13K phage vector (catalog no. E8101, New England Biolabs) was used to produce M13 phage without any peptides displayed on surface and this was used as a negative control.
Enzyme-linked immunosorbent assays
M13 phage coat protein monoclonal antibody (E1), biotin conjugated (catalog no. MA1-34468), and poly–horseradish peroxidase (HRP) streptavidin secondary antibody (catalog no. N200) were purchased from Thermo Fisher Scientific.
Peptide synthesis
Peptides with added linker were conjugated with TAMRA fluorophore at the C-terminal cysteine’s side chain. The peptides were labeled, synthesized, and purified using 9-fluorenyl methoxycarbonyl solid-phase peptide synthesis and reversed-phase high-performance liquid chromatography purification by Thermo Fisher Scientific. Peptides used have the following sequences: MH3, DGPPRKPGGGSC; MH42, SPALHFLGGGSC; MH43, SNLAAFPGGGSC; and MH50, MPVAVYRGGGSC.
In vivo biopanning
Before intravitreal injections, mice were topically administered 0.5% proparacaine, 1% tropicamide, and 2.5% phenylephrine and anesthetized with ketamine (100 mg/kg)/xylazine (10 mg/kg). To begin injection, 2.5% hypromellose was placed over the eye, and a 30-gauge needle was used to make an incision in the limbus. Going through the scleral incision in the limbus, using a Hamilton syringe with a 33-gauge 20° beveled needle, 1.5 μl of Ph.D.-7 phage library (1 × 1011 PFU/ml) was administered to the intravitreal space for each of the three panning rounds performed. An empty, M13 phage library was administered as a negative control for the first round of panning. A 2% fluorescein solution was added to the phage libraries to observe and confirm successful intravitreal delivery. Six hours after administration, the neural retina was extracted from mouse eyes and washed 10 times with 0.1% Tris-buffered saline with Tween 20 (TBST), followed by 200 μl of 0.2 M glycine-HCl (pH 2.2) and bovine serum albumin (BSA; 1 mg/ml) elution for 10 min at 4°C with gentle rocking. Thirty microliters of 1 M tris-HCl (pH 9.1) neutralization buffer was added to the solution. Ten microliters of the resulting neutralized, eluted phage solution was used for phage titering, while the rest was used for amplification in E. coli K12 ER2738, followed by phage purification according to the manufacturer. Serial dilutions of phages were prepared in Luria-Bertani medium. Single-stranded DNA was extracted and purified from individual phage clones picked from titering plates for Sanger sequencing by capillary electrophoresis (Applied Biosystems, 3730xl DNA Analyzer) to elucidate candidate peptides after each unamplified round [the sequencing primer, 5′-CCCTCATAGTTAGCGTAACG-3′ (96 g of III)].
Heatmaps normalized occurrence (% occurrence)
A Python script was written to take in a “.txt” file containing tab-delimited sample labels and the corresponding 7-nucleotide oligomer peptide. The script parses the input.txt file, counting the total number of peptide sequences, and adds each sequence to a list. A 20 by 7 integer matrix was initialized with each element having a value of 0, the 20 rows corresponding to the 20 amino acids and the seven columns corresponding to the positions in the 7-nucleotide oligomer. The list of sequences was then analyzed, each character in each sequence was checked against a list of the one letter amino acid codes to ensure that it was a valid amino acid, if the check passed the element in the final 20 by 7 matrix corresponding to the amino acid of the character and the position the character in the sequence was iterated by 1. Once all sequences were analyzed, the final 20 by 7 matrix contained counts of each amino acid at each position in the sequence. The counts were then divided by the total number of sequences to yield a “normalized occurrence” or % occurrence of amino acids in each position of the sequence. The 20 by 7 matrix of normalized occurrence values was written to an output.txt file, tab delimited, where it was exported to prism and the heatmaps were then finalized. The script was run on all three rounds of in vivo–enriched phage.
Cell-based ELISA
ARPE19 and 661w cells were seeded a day before the experiment on 96-well plates (104 cells per well). On the day of the experiment, individual phage clones were incubated for 1 hour at room temperature in PBS with gentle rocking. Cells were then fixed in 4% paraformaldehyde solution at room temperature for 15 min. This was followed by three washes with PBS, and then the cells were blocked for 30 min at room temperature with 1% BSA in PBS. Biotin-conjugated, mouse anti-M13 phage coat protein primary antibody was added (catalog no. MA1-34468, Thermo Fisher Scientific) at 1:5000 dilution in block buffer with 0.1% TBST and incubated for 2 hours at room temperature with gentle rocking. The wells were washed three more times with 0.1% TBST and subsequently incubated with poly-HRP streptavidin secondary antibody (catalog no. N200, Thermo Fisher Scientific) at 1:10,000 dilution in block buffer with 0.1% TBST for 1 hour at room temperature. Wells were then washed three times with 0.1% TBST; 3,3′,5,5′-tetramethylbenzidine (Thermo Fisher Scientific) was added to the wells and incubated at room temperature for 5 to 10 min; the reaction was stopped with 0.2 M sulfuric acid; and absorbance was read at 450 nm using TECAN Infinite200 Pro spectrophotometer (Tecan Group Ltd., Switzerland).
Cell internalization and image analysis
Approximately 50,000 cells were seeded per well of 8-well μ-slide (Ibidi, Fitchburg, WI) and incubated with 10 and 50 nM TAMRA-labeled peptides for 30 min at 37°C, followed by washing with PBS and fixation in 4% paraformaldehyde for 10 min at room temperature. Cells were then washed three times, DAPI-stained, and cover-slipped for confocal microscopy imaging. Fluorescence confocal images of cell internalization studies were captured with the same exposure settings and were analyzed for fluorescence intensity using ImageJ (version 1.45; National Institutes of Health, Bethesda, MD). First, the hARPE or 661w cells were outlined, and ImageJ calculated the pixel intensity of the different cell treatments with different peptides. At least three images were analyzed for each peptide tested.
Structural analysis superposition of peptides using MOE
Using the molecular builder module in MOE (Chemical Computing Group, Quebec, Canada), the peptide structure was drawn. The peptide side chain, N terminus, and the C terminus were assigned appropriate charges (depending on their ionization state at physiological pH). The peptide was then energy minimized using the Assisted Model Building with Energy Refinement module in MOE. The same process was repeated for all the peptides. The top peptide candidates were superposed using the default superposition module in MOE software.
Pharmaceutically relevant physicochemical properties of peptides isolated
The peptide database prepared in MOE software was exported as Schrödinger compatible file (.mae). The database was manually checked for structure correctness. Pharmaceutically relevant absorption, distribution, metabolism, and excretion properties were calculated using QikProp module in the Schrödinger software (Schrödinger Inc., New York, NY).
Mouse injections
For subretinal injections, mice were topically administered 0.5% proparacaine, 1% tropicamide, and 2.5% phenylephrine and anesthetized with ketamine (100 mg/kg)/xylazine (10 mg/kg). To initiate the injection, 2.5% hypromellose was used to cover the eye and a 30-gauge needle was used to make an incision in the limbus. A glass coverslip was then placed over the eye to allow for visualization of the retina. Going through the scleral incision in the limbus, using a Hamilton syringe with a 33-gauge blunt needle, 1 μl of PBS, peptide, or LNP-Cre were delivered to the subretinal space. A 2% fluorescein solution was added to the PBS and LNPs so retinal detachment could be confirmed. For most injections, scleral incisions in the limbus were created nasally, and PBS or LNPs were delivered temporally. Intravitreal injections were performed as previously described elsewhere (25). For LNP-Cre subretinal injections, 200 ng (1 μl and 200 ng/μl) was delivered. For intravitreal injections, 1.1 μg (1.5 μl and 741 ng/μl) was injected.
In vivo validation with TAMRA-labeled peptides
Top peptide candidates were synthesized and subsequently conjugated with TAMRA (Thermo Fisher Scientific) at the carboxy cysteine and injected either subretinally or intravitreally into BALB/c mice aged 2 to 3 months following the same procedure as described earlier for phage in vivo biopanning. Following peptide injection, ophthalmic fundus imaging was conducted to preemptively gauge the retina distribution of the top-performing TAMRA-labeled peptide candidate injected. At specified time points, mouse eyes were enucleated and fixed using 4% paraformaldehyde in PBS overnight at 4°C. Eyes were cryopreserved in 30% sucrose solution for 2 hours before embedding in optimal cutting temperature (OCT) medium, followed by cryosectioning and confocal imaging.
In vivo validation with LNP-conjugated peptides
Ai9 mice were injected intravitreally and subretinally with LNPs and LNP peptides loaded with Cre mRNA. At 7 days after injection, ophthalmic fundus imaging was performed to observe in vivo tdTomato distribution in the retina. Mouse eyes were then enucleated and fixed using 4% paraformaldehyde in PBS overnight at 4°C. Eyes were cryopreserved in 30% sucrose solution for 2 hours before embedding in OCT medium. Retinal cryosections were 12 μm in thickness. Slides were stain with antibodies specific for visual arrestin (rod and cone PRs; catalog no. sc-166383, Santa Cruz Biotechnology, Dallas, TX), IBA-1 (microglia; catalog no. 019-19741, Wako Chemicals, Richmond, VA), and CD3 (T cells; catalog no. sc20047, Santa Cruz Biotechnology, Dallas, TX). Primary antibodies were used at a concentration of 1:100, 1:500, and 1:50, respectively. Detection of primary antibodies was achieved using Alexa Fluor secondary antibodies at a concentration of 1:300; DAPI was used as a nuclear marker counterstain. Confocal imaging was performed using TCS SP8 X (Leica Microsystems, Buffalo Grove, IL). Z-stacks (spanned 10 μm with 1-μm interval) were collected using a 40× objective, and maximum intensity projections were used for further analysis. Slides were additionally stained with H&E and viewed on a Leica DMI3000 B microscope (Leica Microsystems GmbH, Wetzlar, Germany). All images were taken at a magnification of ×10.
Fundus imaging
In vivo retinal imaging was performed with the Micron IV (Phoenix Research Laboratories, Pleasanton, CA). To observe general retinal morphology, bright-field images were acquired. To capture tdTomato, we used a 534/42-nm BrightLine single-band bandpass filter (Semrock, Rochester, NY). Light intensity, exposure, and gain were kept consistent across all Red Fluorescent Protein images.
Confocal microscopy
All confocal imaging was performed using TCS SP8 X (Leica Microsystems, Buffalo Grove, IL). Z-stacks (spanned 10 μm with 1-μm interval) were collected using a 20× or 40× objective, and maximum intensity projections were used for further analysis. For the main Fig. 3, mean fluorescent intensity measurements were performed on region of interest–gated images for RPE/choroid and PR cell layers specifically using ImageJ.
NHP in vivo delivery and imaging
One male rhesus macaque, aged 10 years old, was used for this study. All protocols involving NHPs were approved by the Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee and conducted in accordance with the National Institutes of Health (NIH) Guidelines for the Care and Use of Laboratory Animals and Animal Research:Reporting of In Vivo Experiments guidelines. Pupils were dilated to a minimum of 8 mm using phenylephrine (2.5%; Bausch and Lomb, Rochester, NY, USA) and tropicamide (1% tropicacyl; Akorn, Lake Forest, IL, USA) eye drops. MH42 LNPs were administered into the subretinal space through a 27G/38G subretinal cannula (#5194, Microvision, Redmond, WA, USA) using the Alcon vitrectomy machine and a pars plana transvitreal approach. First, a pre-bleb of 30 μl of balanced salt solution was generated in the posterior pole. Then, 100 μl of MH42 LNP (500 ng/μl) was delivered within and expanded the bleb. After the injection, dexamethasone (0.5 ml and 10 mg/ml) and cefazolin (0.5 ml and 125 mg/ml) were administered subconjunctivally. There were no complications noted during surgery. The animal received comprehensive multimodal retinal imaging before injection (baseline) and at 48 hours after injection. For each imaging session, the animal was anesthetized by an intramuscular injection of telazol (1:1 mixture of tiletamine hydrochloride and zolazepam hydrochloride, 3.5 to 5.0 mg/kg) and maintained with ketamine (1 to 2 mg/kg) as required. Heart rate and peripheral blood oxygen saturation were monitored by pulse oximetry. Rectal temperature was maintained between 37.0° and 38.0°C by water-circulating heated pads. For image acquisition, animals were positioned prone with the head supported by a chinrest; the pupils were dilated to a minimum of 8 mm using phenylephrine (2.5%; Bausch and Lomb, Rochester, NY, USA) and tropicamide (1% tropicacyl; Akorn, Lake Forest, IL, USA) eye drops. Imaging included wide-field color fundus and autofluorescence (Optos Inc., Marlborough, MA). Following imaging, the contact lenses and eyelid specula were removed, and erythromycin ointment was applied to each eye.
NHP immunohistochemistry and immunofluorescence imaging
After the animal was euthanized, eyes were collected and immersion fixed in 4% paraformaldehyde in PBS for 24 to 48 hours. Dissected eyes were then cryoprotected in increasing sucrose gradients (up to 30%), embedded in OCT compound, and frozen in an embedding mold. Frozen blocks were sectioned at 16 μm using a cryostat (CM1850, Leica, Wetzlar, Germany). Sections were collected throughout the bleb. H&E were examined for subretinal bleb injection site retinotomy, trans-scleral injection site, evidence of immunological reaction, and any signs of pathology. Following examination of the stained sections, adjacent slides were used for immunohistochemistry. LNP expression was identified by colocalization of GFP (anti-GFP; catalog no. ab290, Abcam, Cambridge, UK) with glutamine synthetase (catalog no. sc74430, Santa Cruz Biotechnologies, CA), rod, and cone arrestin (provided by W. Clay Smith, PhD, University of Florida), and RPE65 (catalog no. ab13826, Abcam). Immune infiltrates were identified using antibodies against IBA-1 (microglia; catalog no. 019-19741 Wako Chemicals, Richmond, VA) and CD3 (T cells; catalog no. sc20047, Santa Cruz Biotechnologies), respectively. All primary antibodies were used at a concentration of 1:500, except for anti-CD3 and anti-rod arrestin which were used at 1:50. Detection of primary antibodies was achieved using Alexa Fluor secondary antibodies at a concentration of 1:300; DAPI was used as a nuclear marker counterstain.
Statistical analysis
For TAMRA quantification from peptide injections, regions of interest were created for RPE/choroid and PR cell layer for mean fluorescent intensity values. Fold change measurements were compared to TAMRA-only controls. An ordinary one-way analysis of variance (ANOVA), with Tukey’s correction for multiple comparisons test, was used for comparisons between groups (Prism 8 software, GraphPad Software, La Jolla, CA). Data are presented as means ± SEM. A P < 0.05 was considered as statistically significant.
Cryo–electron microscopy
Vitrification of samples was performed by dispensing 2 μl of LNPs onto a glow-discharged 300-mesh lacey carbon film–coated copper grid using Vitribot Mark IV (FEI). Grids were blotted with filter paper for 3 s at 22°C and 100% relative humidity; thereafter, grids were plunged into the copper cup containing liquid ethane cooled by liquid nitrogen. Any defect in frozen grids was carefully checked, clipped, and assembled into cassettes. After vitrification, frozen grids were maintained at a temperature below −170°C using liquid nitrogen. Imaging was performed in 300-keV Glacios cryo-EM equipped with Falcon III and K3 Summit camera (Gatan) with Direct Electron Detector in counting mode with a magnification of ×45,000 at an electron dose of ∼15 to 20 e−/Å2. The captured images were then processed and analyzed manually using ImageJ software.
Acknowledgments
We thank A. Zlitni for input into conjugation and characterization of LNP peptides and C. Beinat for access to computational software. We thank J. (David) Kim for help with preparation of initial batch of LNPs tested. We thank T. Chiba for help developing peptide analytics scripts. We thank I. Fries for assisting with animal ocular injections and cryosectioning. We thank L. Renner and R. Spears for assisting with NHP animal handling and S. Shubert for assistance with immunohistochemistry.
Funding This project was supported through funding from the National Eye Institute 1R21EY031066 (to G.S.), 1R01EY033423-01A1 (to G.S.), ONPRC Pilot Grant (to R.C.R.), PhRMA Predoctoral Drug Delivery Fellowship (to M.L.), ONPRC Core Grant P51 OD011092 and S10RR024585, Casey Eye Institute Core Grant P30 EY010572 from the NIH, and unrestricted departmental funding from Research to Prevent Blindness.
Author contributions: Conceptualization and supervision: G.S. Research design: G.S., M.H.-B., and R.C.R. Methodology: M.H.-B., R.C.R., M.Ga., C.A., W.T., N.J., J.S., R.R., and A.K.L. Contributed analytical tools: M.H.-B., M.Gu., A.J., and M.L. Data analysis: M.H.-B., R.C.R., M.L., C.S., J.S., and G.S. Designed graphical abstract: M.H.-B., T.K., and O.T. Writing–original draft, review, and editing: M.H.-B., R.C.R., and G.S., with input from all authors.
Competing interests: G.S., M.H., and R.C.R. are inventors on a patent application to this work filed by the Oregon State University (provisional patent application no. 63/292,824). G.S. is a stockholder and cofounder of EnterX Bio. EnterX Bio has a scientific research agreement with OSU. G.S. has a conflict management plan at OSU. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The peptides can be provided by G.S. pending scientific review and a completed material transfer agreement. Requests for the peptides should be submitted to sahay@ohsu.edu and advantage@oregonstate.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Other Supplementary Material for this : manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.N. Chaudhary, D. Weissman, K. A. Whitehead, mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 817–838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. V. Garafalo, A. V. Cideciyan, E. Héon, R. Sheplock, A. Pearson, C. WeiYang Yu, A. Sumaroka, G. D. Aguirre, S. G. Jacobson, Progress in treating inherited retinal diseases: Early subretinal gene therapy clinical trials and candidates for future initiatives. Prog. Retin. Eye Res. 77, 100827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RetNet–Retinal Information Network (14 February 2022).
- 4.S. Veleri, C. H. Lazar, B. Chang, P. A. Sieving, E. Banin, A. Swaroop, Biology and therapy of inherited retinal degenerative disease: Insights from mouse models. Dis. Model. Mech. 8, 109–129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Makin, Four technologies that could transform the treatment of blindness. Nature 10.1038/d41586-019-01107-8 (2019). [DOI] [PubMed] [Google Scholar]
- 6.S. Russell, J. Bennett, J. A. Wellman, D. C. Chung, Z. F. Yu, A. Tillman, J. Wittes, J. Pappas, O. Elci, S. McCague, D. Cross, K. A. Marshall, J. Walshire, T. L. Kehoe, H. Reichert, M. Davis, L. Raffini, L. A. George, F. P. Hudson, L. Dingfield, X. Zhu, J. A. Haller, E. H. Sohn, V. B. Mahajan, W. Pfeifer, M. Weckmann, C. Johnson, D. Gewaily, A. Drack, E. Stone, K. Wachtel, F. Simonelli, B. P. Leroy, J. F. Wright, K. A. High, A. M. Maguire, Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: A randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Georgiou, K. Fujinami, M. Michaelides, Inherited retinal diseases: Therapeutics, clinical trials and end points—A review. Clin. Experiment. Ophthalmol. 49, 270–288 (2021). [DOI] [PubMed] [Google Scholar]
- 8.I. Vázquez-Domínguez, A. Garanto, R. W. J. Collin, Molecular therapies for inherited retinal diseases—Current standing, opportunities and challenges. Genes 10, 654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D. Dhurandhar, N. K. Sahoo, I. Mariappan, R. Narayanan, Gene therapy in retinal diseases: A review. Indian J. Ophthalmol. 69, 2257–2265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. Amato, A. Arrigo, E. Aragona, M. P. Manitto, A. Saladino, F. Bandello, M. Battaglia Parodi, Gene therapy in inherited retinal diseases: An update on current state of the art. Front. Med. 8, 750586 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J.-A. Sahel, E. Boulanger-Scemama, C. Pagot, A. Arleo, F. Galluppi, J. N. Martel, S. D. Esposti, A. Delaux, J.-B. de Saint Aubert, C. de Montleau, E. Gutman, I. Audo, J. Duebel, S. Picaud, D. Dalkara, L. Blouin, M. Taiel, B. Roska, Partial recovery of visual function in a blind patient after optogenetic therapy. Nat. Med. 27, 1223–1229 (2021). [DOI] [PubMed] [Google Scholar]
- 12.N. J. Newman, P. Yu-Wai-Man, V. Carelli, M. L. Moster, V. Biousse, C. Vignal-Clermont, R. C. Sergott, T. Klopstock, A. A. Sadun, P. Barboni, A. A. DeBusk, J. F. Girmens, G. Rudolph, R. Karanjia, M. Taiel, L. Blouin, G. Smits, B. Katz, J. A. Sahel, C. Vignal, R. Hage, C. B. Catarino, C. Priglinger, S. Priglinger, S. Thurau, B. von Livonius, D. Muth, A. Wolf, J. al-Tamami, A. Pressler, C. Schertler, M. Hildebrandt, M. Neuenhahn, G. Heilweil, I. Tsui, G. B. Hubbard, A. Hendrick, M. Dattilo, J. Peragallo, E. Hawy, L. DuBois, Med, D. Gibbs, A. F. Filho, J. Dobbs, M. Carbonelli, L. di Vito, M. Contin, S. Mohamed, C. la Morgia, S. Silvestri, J. Acheson, M. Eleftheriadou, S. Esposti, M. Gemenetzi, L. Leitch-Devlin, W. R. Tucker, N. Jurkute, M. SantaMaria, H. Tollis, J. A. Haller, M. Massini, Efficacy and safety of intravitreal gene therapy for leber hereditary optic neuropathy treated within 6 months of disease onset. Ophthalmology 128, 649–660 (2021). [DOI] [PubMed] [Google Scholar]
- 13.H. Ledford, CRISPR treatment inserted directly into the body for first time. Nature 579, 185 (2020). [DOI] [PubMed] [Google Scholar]
- 14.K. Bucher, E. Rodríguez-Bocanegra, D. Dauletbekov, M. D. Fischer, Immune responses to retinal gene therapy using adeno-associated viral vectors—Implications for treatment success and safety. Prog. Retin. Eye Res. 83, 100915 (2021). [DOI] [PubMed] [Google Scholar]
- 15.A. Kantor, M. E. McClements, R. E. MacLaren, CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 21, 6240 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Kantor, M. E. McClements, C. F. Peddle, L. E. Fry, A. Salman, J. Cehajic-Kapetanovic, K. Xue, R. E. MacLaren, CRISPR genome engineering for retinal diseases. Prog. Mol. Biol. Transl. Sci. 182, 29–79 (2021). [DOI] [PubMed] [Google Scholar]
- 17.D. Adams, A. Gonzalez-Duarte, W. D. O’Riordan, C. C. Yang, M. Ueda, A. V. Kristen, I. Tournev, H. H. Schmidt, T. Coelho, J. L. Berk, K. P. Lin, G. Vita, S. Attarian, V. Planté-Bordeneuve, M. M. Mezei, J. M. Campistol, J. Buades, T. H. Brannagan III, B. J. Kim, J. Oh, Y. Parman, Y. Sekijima, P. N. Hawkins, S. D. Solomon, M. Polydefkis, P. J. Dyck, P. J. Gandhi, S. Goyal, J. Chen, A. L. Strahs, S. V. Nochur, M. T. Sweetser, P. P. Garg, A. K. Vaishnaw, J. A. Gollob, O. B. Suhr, Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 18.F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, Frenck RW Jr, L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen, W. C. Gruber; C4591001 Clinical Trial Group , Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. R. Baden, H. M. el Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller, T. Zaks; COVE Study Group , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis, R. van der Meel, The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 16, 630–643 (2021). [DOI] [PubMed] [Google Scholar]
- 21.S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova, J. J. Senn, A. Lynn, A. Bulychev, I. McFadyen, J. Chan, Ö. Almarsson, M. G. Stanton, K. E. Benenato, A novel amino lipid series for mRNA delivery: Improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. R. Cullis, M. J. Hope, Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25, 1467–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.X. Hou, T. Zaks, R. Langer, Y. Dong, Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. Patel, R. C. Ryals, K. K. Weller, M. E. Pennesi, G. Sahay, Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release Off. J. Control. Release Soc. 303, 91–100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. C. Ryals, S. Patel, C. Acosta, M. McKinney, M. E. Pennesi, G. Sahay, The effects of PEGylation on LNP based mRNA delivery to the eye. PLOS ONE 15, e0241006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. Bejarano-Escobar, H. Sánchez-Calderón, J. Otero-Arenas, G. Martín-Partido, J. Francisco-Morcillo, Müller glia and phagocytosis of cell debris in retinal tissue. J. Anat. 231, 471–483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.F. Mazzoni, H. Safa, S. C. Finnemann, Understanding photoreceptor outer segment phagocytosis: Use and utility of RPE cells in culture. Exp. Eye Res. 126, 51–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. Reichenbach, A. Bringmann, New functions of Müller cells. Glia 61, 651–678 (2013). [DOI] [PubMed] [Google Scholar]
- 29.D. Tesauro, A. Accardo, C. Diaferia, V. Milano, J. Guillon, L. Ronga, F. Rossi, Peptide-based drug-delivery systems in biotechnological applications: Recent advances and perspectives. Molecules 24, 351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Liu, X. Fang, Y. Yang, C. Wang, Peptide-enabled targeted delivery systems for therapeutic applications. Front. Bioeng. Biotechnol. 9, 701504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. Albertini, V. Mathieu, N. Iraci, M. van Woensel, A. Schoubben, A. Donnadio, S. M. L. Greco, M. Ricci, A. Temperini, P. Blasi, N. Wauthoz, Tumor targeting by peptide-decorated gold nanoparticles. Mol. Pharm. 16, 2430–2444 (2019). [DOI] [PubMed] [Google Scholar]
- 32.U. Hennrich, K. Kopka, Lutathera®: The first FDA- and EMA-approved radiopharmaceutical for peptide receptor radionuclide therapy. Pharmaceuticals 12, 114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana, M. L. Maitland, J. Seitzer, D. O’Connell, K. R. Walsh, K. Wood, J. Phillips, Y. Xu, A. Amaral, A. P. Boyd, J. E. Cehelsky, M. D. McKee, A. Schiermeier, O. Harari, A. Murphy, C. A. Kyratsous, B. Zambrowicz, R. Soltys, D. E. Gutstein, J. Leonard, L. Sepp-Lorenzino, D. Lebwohl, CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 34.K. Musunuru, A. C. Chadwick, T. Mizoguchi, S. P. Garcia, J. E. DeNizio, C. W. Reiss, K. Wang, S. Iyer, C. Dutta, V. Clendaniel, M. Amaonye, A. Beach, K. Berth, S. Biswas, M. C. Braun, H. M. Chen, T. V. Colace, J. D. Ganey, S. A. Gangopadhyay, R. Garrity, L. N. Kasiewicz, J. Lavoie, J. A. Madsen, Y. Matsumoto, A. M. Mazzola, Y. S. Nasrullah, J. Nneji, H. Ren, A. Sanjeev, M. Shay, M. R. Stahley, S. H. Y. Fan, Y. K. Tam, N. M. Gaudelli, G. Ciaramella, L. E. Stolz, P. Malyala, C. J. Cheng, K. G. Rajeev, E. Rohde, A. M. Bellinger, S. Kathiresan, In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021). [DOI] [PubMed] [Google Scholar]
- 35.H. Quintero, M. Lamas, microRNA expression in the neural retina: Focus on Müller glia. J. Neurosci. Res. 96, 362–370 (2018). [DOI] [PubMed] [Google Scholar]
- 36.S. Patel, J. Kim, M. Herrera, A. Mukherjee, A. V. Kabanov, G. Sahay, Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev. 144, 90–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.C.-J. Zhang, H. Mou, Z.-B. Jin, Fluorescent protein tdTomato is deleterious to the retina. Invest. Ophthalmol. Vis. Sci. 63, 4128–F0365 (2022). [Google Scholar]
- 38.Z. B. Bjornson-Hooper, G. K. Fragiadakis, M. H. Spitzer, H. Chen, D. Madhireddy, K. Hu, K. Lundsten, D. R. McIlwain, G. P. Nolan, A Comprehensive atlas of immunological differences between humans, mice, and non-human primates. Front. Immunol. 13, 867015 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Herrera, J. Kim, Y. Eygeris, A. Jozic, G. Sahay, Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery. Biomater. Sci. 9, 4289–4300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1