Abstract
Parvovirus B19 (B19V) infects human erythroid progenitor cells (EPCs) and causes several hematological disorders and fetal hydrops. Amino acid (aa) 5–68 of minor capsid protein VP1 (VP1u5–68aa) is the minimal receptor binding domain for B19V to enter EPCs. Here, we carried out a genome-wide CRISPR-Cas9 guide RNA screen and identified tyrosine protein kinase receptor UFO (AXL) as a proteinaceous receptor for B19V infection of EPCs. AXL gene silencing in ex vivo expanded EPCs remarkably decreased B19V internalization and replication. Additions of the recombinant AXL extracellular domain or a polyclonal antibody against it upon infection efficiently inhibited B19V infection of ex vivo expanded EPCs. Moreover, B19V VP1u interacted with the recombinant AXL extracellular domain in vitro at a relatively high affinity (KD = 103 nM). Collectively, we provide evidence that AXL is a co-receptor for B19V infection of EPCs.
AXL interacts with parvovirus B19 VP1 unique region and plays an important role in virus entry of human erythroid progenitors.
INTRODUCTION
Human parvovirus B19 (B19V) belongs to the species Primate erythroparvovirus 1 in the genus Erythroparvovirus of the family Parvoviridae (1). As a small nonenveloped virus, the genome of B19V is a single-stranded DNA of ~5.6 kb that is flanked by two identical inverted terminal repeats (2). Besides being known as a causative agent of the common fifth disease (erythema infectiosum), B19V infection can cause several hematological diseases, including transient aplastic crisis, chronic anemia, and nonimmune hydrops fetalis. In patients with increased destruction of erythrocytes and a high demand for erythrocyte production, such as sickle cell disease and hereditary spherocytosis patients, B19V infection causes transient aplastic crisis (3, 4). B19V infection can be life-threatening to sickle cell disease patients (5). In immunocompromised patients, such as organ transplant recipients, acquired immunodeficiency syndrome patients, or post-chemotherapy patients, persistent B19V infections of bone marrow can cause chronic anemia associated with pure red cell aplasia (2, 5). B19V infection of pregnant women during early pregnancy can cause severe complications with the fetus, including miscarriage, hydrops fetalis, and even sometimes intrauterine fetal death (5, 6).
In bone marrow and fetal liver, a fraction of primitive multipotent hematopoietic stem cells (HSCs) commit to the erythroid lineage, a process called erythropoiesis. These stem cells give rise to burst-forming unit erythroid (BFU-E) and colony-forming unit erythroid (CFU-E) progenitor cells, pro-normoblasts, erythroblasts, reticulocytes, and ultimately mature erythrocytes (7). B19V infection shows a remarkable tropism for erythroid progenitor cells (EPCs), including late BFU-E, CFU-E, pro-normoblasts, and early erythroblasts (2, 8). The hematological diseases caused by B19V infection (transient aplastic crisis, pure red cell aplasia, chronic anemia, and hydrops fetalis) are directly related to the cytotoxicity of B19V infection (5, 9). Ex vivo expanded CD36+ EPCs are fully permissive to B19V infection (10–12). In vitro, a few erythroid or megakaryoblastoid leukemia cell lines, e.g., UT7/Epo-S1 cells, can also be infected by B19V (13).
The B19V capsid has a T = 1 icosahedral structure and is composed of 60 structural proteins, of which ~5% are VP1 of 83 kDa and ~95% are VP2 of 58 kDa (14, 15). VP1 is identical to VP2 except for an additional N-terminal 227–amino acid (aa) residue, referred to as the VP1 unique region (VP1u). In contrast to other parvoviruses, e.g., adeno-associated virus (AAV) and minute virus of mice (MVM) (16, 17), B19V VP1u is important for eliciting an efficient immune response (18–21), as the N terminus (aa1–80) of VP1u is rich in neutralizing epitopes (18, 19). Upon infection, when the virus interacts with the cells, B19V VP1u is exposed out of the capsid and becomes accessible to neutralization antibodies (22, 23). The structural dynamics of B19V VP1u is dependent on an optimal condition at pH 7.4 (24), rather than at pH <5.5 (16, 17). The externalized N-terminal aa5–80 of VP1u is responsible for cellular binding and internalization (25), and aa5–68 (VP1u5–68aa) was further defined as the minimal domain of VP1u for B19V entry of EPCs through its participation in virus binding and internalization (26). The N-terminally green fluorescent protein–fused VP1u5–68aa (GFP-VP1u5–68aa) enters EPCs nearly five times more efficiently than the full-length VP1u; thus, VP1u5–68aa represents the minimal receptor binding domain (26, 27).
The cellular receptors that mediate B19V entry of EPCs are still obscure, and several candidates, including erythrocyte P antigen (globside) (28, 29), Ku80 autoantigen (30), and integrin α5β1 (31), have been proposed. P-antigen has long been thought of as the primary attachment receptor of B19V (28, 29), which mediates virus attachment to cells (32). However, it has recently been argued that P-antigen is dispensable for B19V entry, but it is essential for productive infection at a post-entry level (33). After internalization, B19V interacts with P-antigen exclusively under acidic conditions in the endosome, which plays an important role in endocytic trafficking of the virus (34). EPCs barely express Ku80 on the cell surface (35), and the essential role of Ku80 in B19V infection is attributed to the function of the DNA-PK complex in the infection-induced DNA damage response that plays a role in B19V DNA replication (36). In addition, neither native integrin α5β1 nor recombinant Ku80 blocked B19V infection of EPCs (see Discussion). Therefore, our current understanding of B19V cell entry cannot fully explain the importance of the extended VP1u during virus entry.
To identify the proteinaceous receptor that mediates B19V infection through interacting with VP1u, we used a genome-wide CRISPR-Cas9 knockout screen based on the internalization of GFP-VP1u5–68aa determined by flow cytometry. The tyrosine protein kinase receptor UFO (AXL) gene had the most noticeable enriched single-guide RNA (sgRNA) reads among genes encoding cell membrane–associated proteins, depending on the enrichment over two rounds of selections. AXL is a member of the TAM receptors, a unique family of receptor tyrosine kinases that also includes Tyro3 and Mer. Binding of the TAM receptor with the ligand growth arrest–specific gene 6 (Gas6) or protein S (a blood anticoagulant cofactor) plays an essential role in phagocytosis of apoptotic cells, in the regulation of innate immunity, in the signaling of autoimmune disease, and in mediating virus infection (37). AXL is a transmembrane receptor with a molecular weight of 100 to 140 kDa that contains the extracellular domain at the N terminus and an intracellular tyrosine kinase domain at the C terminus (38). The extracellular domain contains tandem immunoglobulin-like repeats that mediate the binding of ligand Gas6 or protein S and two fibronectin type III motifs. In this study, AXL was further investigated for its role in VP1u-mediated entry and B19V infection of EPCs.
RESULTS
Genome-wide CRISPR-Cas9 knockout screen identifies host cell entry factors for B19V VP1u internalization
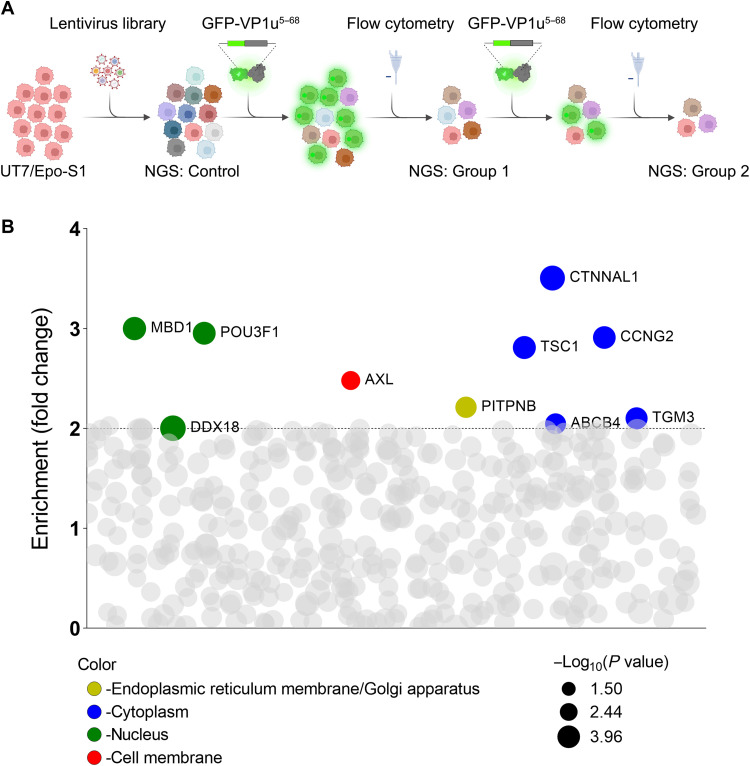
To identify the host cell entry factors of B19V VP1u, recombinant GFP-VP1u5–68aa protein served as a selection reporter to mimic the entry of B19V (26). A CRISPR-Cas9 knockout pooled lentiviral library (39) was transduced to B19V permissive cell line UT7/Epo-S1 cells, followed by two-round selections based on the internalization of GFP-VP1u5–68aa using flow cytometry (Fig. 1A). sgRNAs present in the GFP-negative (GFP−) cell population were identified by next-generation sequencing (NGS) (Fig. 1A), and 10 genes were identified with more than eightfold enriched at the first round of selection and further enriched by twofold at the second round of selection (Fig. 1B and tables S1 to S4). Further gene ontology analysis demonstrated that tyrosine protein kinase receptor UFO (AXL) was the only hit among the cell membrane–associated genes (Fig. 1B). Other hits included phosphatidylinositol transfer protein β (PITPNB) in the endoplasmic reticulum membrane/Golgi apparatus; catenin α-like 1 (CTNNAL1), TSC complex subunit 1 (TSC1), cyclin-G2 (CCNG2), ATP binding cassette subfamily B member 4 (ABCB4), and transglutaminase 3 (TGM3) in the cytoplasm; and methyl-CpG binding domain protein 1 (MBD1), DEAD-Box helicase 18 (DDX18), and POU class 3 homeobox 1 (POU3F1) in the nucleus. We decided to further investigate the cell membrane–associated protein AXL.
Fig. 1. A pooled genome-scale CRISPR screen identifies AXL as an important host protein for B19V VP1u entry.
(A) Schematic diagram of the CRISPR-Cas9 gRNA screen. UT7/Epo-S1 cells were transduced with a CRISPR knockout pooled lentiviral library, followed by two-round selections based on the internalization of GFP-expressing VP1u5–68aa (GFP-VP1u5–68aa) by flow cytometry. The diagram was created with BioRender. (B) Enrichment of gene gRNAs across iteratively sorted cells. sgRNAs presented in the GFP− cell population were identified by NGS. Ten genes, as depicted, that have enriched sgRNAs by >8-fold at the first screen, which were further enriched at the second screen, were identified depending on the enrichment fold from the second to the first screen. The data were log2-transformed. The log2 fold changes were calculated by comparison with the control groups (tables S1 and S2). The size of the circle represents the P values at log10 scale as indicated. The Y axis represents the enriched fold changes by comparison of the log2 changes between round 2 and round 1. The color of each circle represents the function of the genes, and the bubble size is proportional to the statistical significance [−log10(P value)] of the enrichment [relative to the unselected control group (tables S3 and S4)].
AXL is required for efficient internalization of GFP-VP1u5–68aa
To validate the role of AXL in B19V VP1u internalization, we used lentiviral vector-based AXL-specific small hairpin (sh)RNA (shAXL) to silence AXL expression in UT7/Epo-S1 cells. The same strategy was used later to silence AXL expression in primary CD36+ EPCs. After selection with puromycin, expression of both AXL protein and mRNA was nearly eliminated in shAXL-transduced UT7/Epo-S1 cells (S1shAXL) compared to scramble RNA-expressing lentivirus (shScr)–transduced control cells (S1shScr) (fig. S1, A and B). Cell surface AXL expression was remarkably reduced (fig. S1C). We analyzed the cell proliferation of the parent UT7/Epo-S1 cells, S1shScr and S1shAXL, using a bromodeoxyuridine (BrdU) incorporation assay. The results showed no significant difference in cell populations of the G2-M, S, and G0-G1 phases, confirming that the silenced expression of AXL had a negligible influence on the cell cycle (fig. S1, D and E).
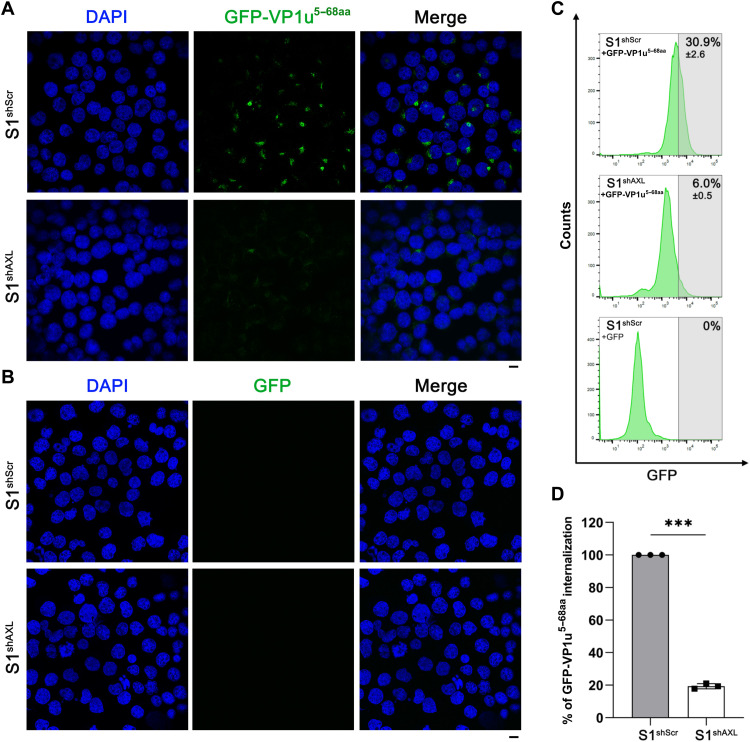
The internalization of GFP-VP1u5–68aa was assessed in both S1shScr and S1shAXL cells. A concentration of 2 μM GFP-VP1u5–68aa or GFP was incubated with the cells to perform an internalization assay. After incubation at 37°C for 1.5 hours, the cells were analyzed by immunofluorescence assay and flow cytometry. Notably, the internalization of GFP-VP1u5–68aa was substantially decreased in the S1shAXL cells compared to that in the S1shScr cells (Fig. 2A). Flow cytometry analysis indicated that the percentage of the S1shAXL cells that internalized GFP-VP1u5–68aa was detected to be at least >4 times less than that in the S1shScr cells (Fig. 2C). As a control, no GFP was internalized in both S1shScr and S1shAXL cells (Fig. 2B). Collectively, these results demonstrated that AXL silencing severely blocks VP1u5–68aa entry of UT7/Epo-S1 cells.
Fig. 2. Silencing of AXL expression in UT7/Epo-S1 cells inhibits internalization of GFP-VP1u5–68aa.
(A and B) GFP-VP1u5–68aa internalization assay. Lentiviral vector–based shRNA shAXL or shScr-transduced UT7/Epo-S1 cells (S1shAXL or S1shScr) were incubated with GFP-VP1u5–68aa (A) or control GFP protein (B) at 37°C for 1.5 hours, followed by washing with PBS three times. The cells were then cytospun onto slides, fixed, permeabilized, and visualized under a confocal microscope. (C and D) Flow cytometry. After incubation at 37°C for 1.5 hours, the internalization of GFP-VP1u5–68 or control GFP protein was analyzed by flow cytometry. A reference line was drawn on the basis of the GFP protein control for a cutoff between the GFP− and GFP+ populations (C). The numbers represented the percentage of the cells in the indicated range of fluorescence density. The percentage (mean ± SD) obtained from S1shAXL was normalized to S1shScr, which is arbitrarily set up to 100% (D). Nuclei were stained with DAPI (blue). Scale bars, 10 μm.
AXL is an important host factor for B19V entry
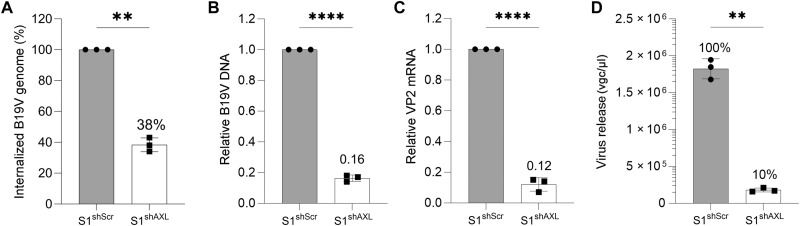
We next evaluated the internalization and replication of B19V in S1shScr and S1shAXL cells. To this end, we infected the cells with B19V, and the internalized B19V genome was quantified by quantitative polymerase chain reaction (qPCR). The results showed that the internalized B19V genome in the S1shAXL cells only reached ~40% of those in the control S1shScr cells (Fig. 3A). We then analyzed B19V DNA replication and viral (VP2) mRNA expression at 2 days post-infection (dpi) in S1shScr and S1shAXL cells, respectively. The results showed that B19V DNA replication and VP2 mRNA expression in S1shAXL cells were significantly decreased by ~6- and ~7-fold, respectively, compared to those in the S1shScr cells (Fig. 3, B and C). In addition, the released B19V was decreased by 10 times in the S1shAXL cells versus that in the S1shScr cells (Fig. 3D).
Fig. 3. Silencing of AXL expression efficiently inhibits B19V infection of UT7/Epo-S1 cells.
(A) Quantification of internalized B19V. Lentivirus-transduced UT7/Epo-S1 cells (S1shAXL or S1shScr) were incubated with B19V at an MOI of 1000 at 37°C for 1.5 hours. After treatment with trypsin and benzonase, the cells were washed with PBS three times, and viral DNA was extracted for quantification by qPCR. (B to D) Quantification of B19V replication and egress. S1shAXL or S1shScr cells were infected with B19V at an MOI of 1000. At 2 dpi, the cells were collected for quantification of B19V replication by relative viral DNA levels versus mitochondrial DNA (B), relative VP2 mRNA versus β-actin mRNA (C), or virus egress (D). The values (mean ± SD) obtained from S1shAXL are normalized to those from the S1shScr cells, which are arbitrarily set up to 100% [(D), S1shScr]. All the experiments were performed in triplicate, and the data were analyzed by Student’s t test (**P < 0.01 and ****P < 0.0001).
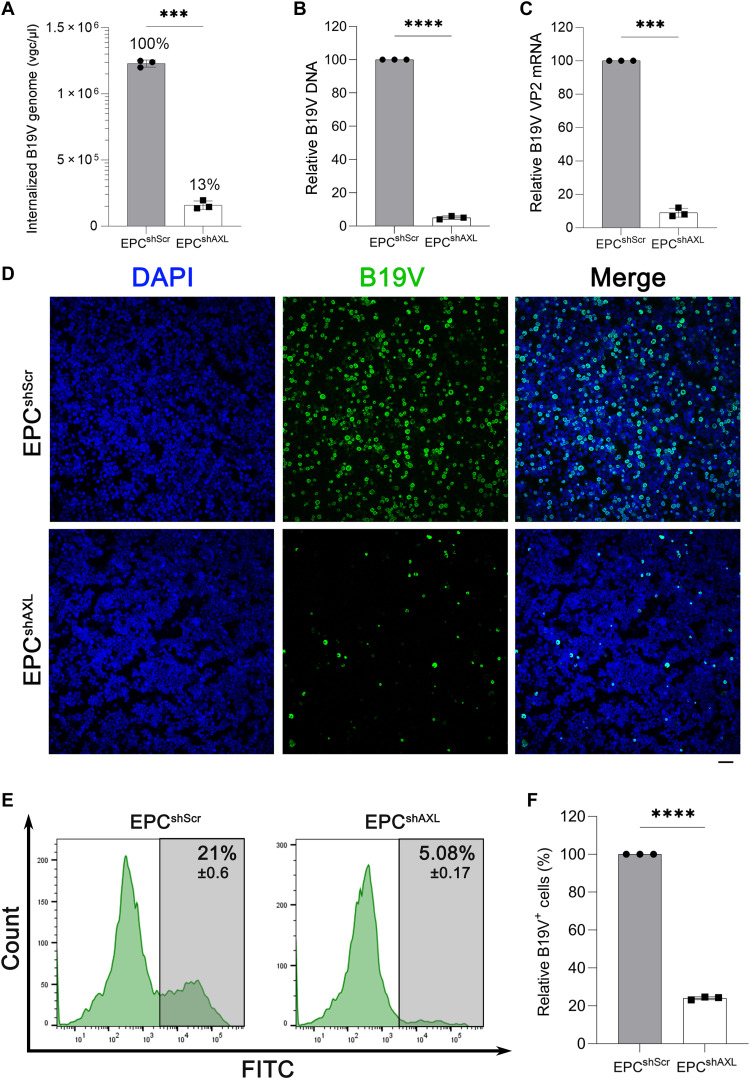
Next, the function of AXL in B19V entry was explored in ex vivo expanded CD36+ EPCs. The lentiviral shAXL silenced the expression of total AXL in EPCs and on the cell surface (fig. S2, A to C) while showing negligible influence on the cell cycle of EPCs (fig. S2, D and E). The entry and replication of B19V were evaluated in shAXL-transduced EPCs (EPCshAXL) and the shScr-transduced EPCs control (EPCshScr), respectively. B19V entry of EPCshAXL was decreased by ~90% compared to that of the EPCshScr control (Fig. 4A). At 2 dpi, levels of replicated viral DNA (Fig. 4B) and VP2 mRNA expression (Fig. 4C) were also significantly decreased in EPCshAXL compared to those of the EPCshScr control. We next used an anti-intact B19V capsid monoclonal antibody (clone 521-5D) (40) to perform immunofluorescence assay and flow cytometry analyses of B19V infection. The results demonstrated a drastic decrease in B19V capsid–expressing cells (B19V+) in EPCshAXL compared to that of the EPCshScr control (Fig. 4, D and E), and the amount of B19V+ cells decreased to a fifth in EPCshAXL compared to that of the EPCshScr control (Fig. 4F). Together, the silencing of AXL significantly decreased B19V entry, replication, and capsid expression in both UT7/Epo-S1 cells and ex vivo expanded CD36+ EPCs.
Fig. 4. AXL gene silencing of CD36+ EPCs inhibits B19V infection.
Lentivirus-transduced CD36+ EPCs were incubated with B19V at an MOI of 1000 at 37°C for 1.5 hours. (A) Quantification of internalized B19V. Cells were treated with trypsin and benzonase, followed by washing with PBS. The internalized B19V was quantified by qPCR. (B and C) Quantification of B19V replication. At 2 dpi, the indicated cells were collected and treated with benzonase, followed by quantification of B19V DNA using qPCR (B) and VP2 mRNA using RT-qPCR (C). (D) Immunofluorescence assay. At 2 dpi, cells were collected and resuspended in DMEM and cytospun onto slides, followed by fixation, permeabilization, and sequentially staining with anti-B19V capsid (clone 521-5D) and an FITC-conjugated secondary antibody (green). Confocal images were taken at a magnification of ×20. Nuclei were stained with DAPI (blue). Scale bar, 50 μm. (E and F) Flow cytometry. At 2 dpi, infected EPCs were fixed, permeabilized, and stained with anti-B19V capsid (#clone 521-5D) and an FITC-conjugated secondary antibody, sequentially, followed by flow cytometry (E). All the experiments were performed in triplicate (F), and the data shown were analyzed by Student’s t test (***P < 0.001 and ****P < 0.0001).
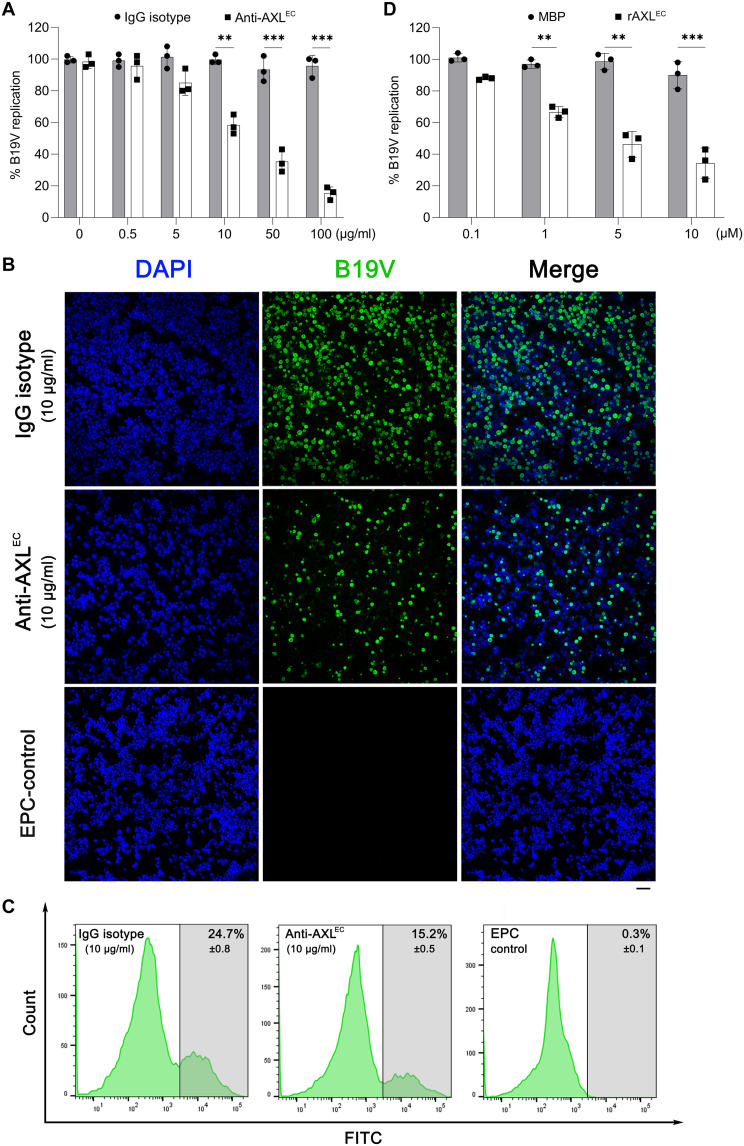
Both recombinant AXL extracellular domain and a polyclonal antibody against the domain inhibit B19V infection of EPCs
To examine whether AXL plays a role in the early steps of B19V infection, we used a recombinant protein of the extracellular domain of AXL (rAXLEC) and a polyclonal antibody against this domain to inhibit B19V entry of EPCs. An anti-AXLEC antibody or rAXLEC were incubated with the cells during B19V infection to block cell surface AXL and neutralize the virus, respectively. At 2 dpi, B19V DNA replication in anti-AXLEC–incubated EPCs was decreased in a dose-dependent manner when compared to the cells treated with a control immunoglobulin G (IgG) isotype (Fig. 5A). At 10 μg/ml of anti-AXLEC, B19V+ cells were decreased by ~40% compared with the IgG control (Fig. 5, B and C), and at 100 μg/ml, B19V DNA replication was further decreased by more than 80% (Fig. 5A). The incubation of rAXLEC from 1 to 10 μM also significantly inhibited B19V replication in CD36+ EPCs in a dose-dependent manner when compared with the control of a nonspecific protein [maltose-binding protein (MBP)] (Fig. 5D). Cells treated with antibodies or proteins exhibited no differences in cell proliferation. Overall, these results demonstrated that both the anti-AXLEC antibody and rAXLEC markedly inhibited B19V infection of CD36+ EPCs, strongly supporting the important role of AXL during the entry of B19V into CD36+ EPCs.
Fig. 5. Both anti-AXL antibody and AXL protein efficiently inhibit B19V infection of CD36+ EPCs.
(A and D) Quantification of B19V DNA replication. (A) CD36+ EPCs were incubated with an anti-AXL or an IgG isotype control (at indicated concentrations) at 4°C before B19V infection (MOI = 1000). (D) CD36+ EPCs were incubated with soluble rAXLEC or MBP (0.1 to 10 μg/ml) during B19V infection. At 2 dpi, the cells were collected and quantified for replicated B19V genome and mitochondrial DNA using qPCR. The number of replicated B19V genomes of each group is standardized with the mitochondrial DNA numbers. Data shown are relative to the mock-treated cell group, which is arbitrarily set up as 100%. (B and C) Immunofluorescence assay and flow cytometry. CD36+ EPCs were incubated with anti-AXL or IgG isotype control (10 μg/ml) at 4°C before B19V infection. At 2 dpi, the cells were collected and stained with an anti-capsid antibody (clone 521-5D), followed by immunofluorescence assay (B) and flow cytometry (C), respectively. Scale bar, 50 μm.
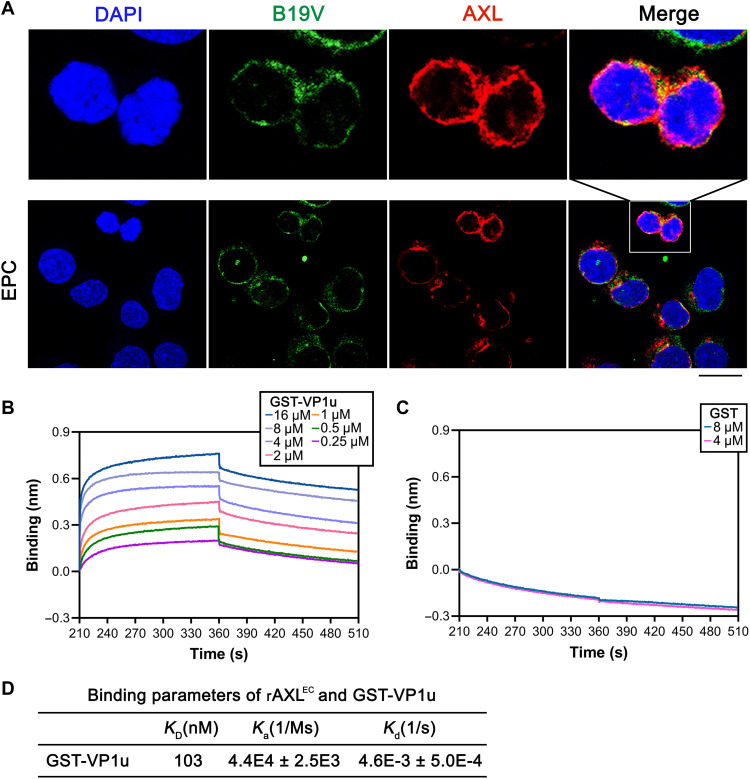
AXL colocalizes with B19V VP1u in vivo and binds to B19V VP1u in vitro
We next asked whether AXL interacts with B19V VP1u. To this end, EPCs were infected with B19V. After incubation at 4°C for 1 to 2 hours, the localization of AXL and B19V virions in the cells was imaged by immunofluorescence assays with specific antibodies. The results showed colocalization of signals of AXL and B19V virions (Fig. 6A). Moreover, we analyzed the distribution of AXL expression and the VP1u binding on cell surface using flow cytometry. GFP-VP1u and control GFP proteins were incubated with CD36+ EPCs at 4°C for 1 hour. Following cell surface staining of AXL, the flow cytometry results showed that GFP-VP1u was distributed to the subset of AXL-positive CD36+ EPCs (fig. S3A). For the GFP control, we detected AXL expression, but no GFP signal, on the cell surface (fig. S3B). These results substantiated the association of B19V VP1u with AXL on the surface of B19V permissive cells.
Fig. 6. AXL interacts with GST-VP1u.
(A) Immunofluorescent assay for colocalization of AXL with B19V virions. CD36+ EPCs cells were infected with B19V at an MOI of ~5000 vgc per cell for 1.5 hours at 4°C. The cells were then washed with ice-cold PBS, then cytospun onto slides, fixed, permeabilized, and stained with an anti-B19V capsid antibody (clone MAB860-55D). Confocal images were taken under a confocal microscope. Nuclei were stained with DAPI (blue). Scale bar, 10 μm. (B to D) Kinetics of the in vitro interaction between AXL and VP1u using Ni-NTA biosensors. The binding kinetics depicts the associations and dissociations of 4 μM rAXLEC with GST-VP1u at the indicated concentrations (B). Binding kinetics of 4 μM AXLHis with 4 and 8 μM GST was used as a control (C). The binding parameters of AXL and GST-VP1u are shown (D). Equilibrium dissociation constant KD value represents the ratio of dissociation [Kd (1/s)] and association [Ka (1/Ms)] computed from the real-time binding curves of rAXLEC to GST-VP1u. The values are shown with means ± SDs based on at least three repeated experiments.
To further measure a direct interaction between AXL and VP1u, a biolayer interferometry (BLI) assay was performed to determine the binding kinetics of rAXLEC with VP1u. A concentration of 4 μM rAXLEC was loaded to the assays for the association and dissociation with different concentrations of glutathione S-transferase (GST)–VP1u and GST only (as a control). A notable shift was observed in the binding kinetics between rAXLEC and GST-VP1u, which was exhibited in a dose-dependent manner (Fig. 6B). As a control, GST only showed no binding at 4 or 8 μM (Fig. 6C). An equilibrium dissociation constant (KD) value of 103 nM was calculated from Ka (association rate constant; 1/Ms) and Kd (dissociation rate constant; 1/s) derived from the binding kinetics assays (Fig. 6D), indicating a high binding affinity between AXL and B19V VP1u. Collectively, AXL colocalized with B19V virions on the surface of CD36+ EPCs, and rAXLEC directly interacted with B19V VP1u at a relatively high affinity (KD = 103 nM) in vitro compared with the KD values of most ligand and receptor binding (41).
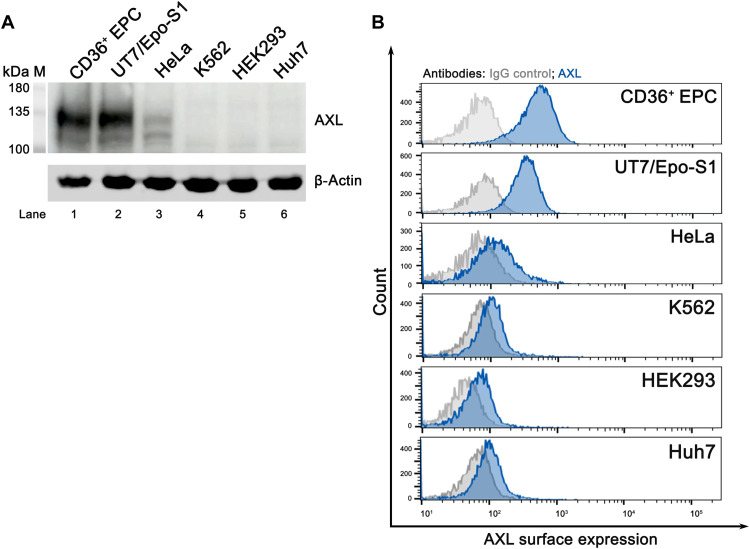
AXL is expressed on the cell surface of B19V permissive cells
Because B19V infection exhibits a high tropism for EPCs (2), we then asked whether the surface expression of AXL contributes to the tropism. To this end, the expression of AXL in B19V permissive cells (CD36+ EPCs and UT7/Epo-S1) and nonpermissive cells [HeLa, K562, human embryonic kidney (HEK) 293, and Huh7] was probed by Western blotting for whole-cell expression and by flow cytometry for cell surface expression. While AXL was overall highly expressed in the B19V permissive cells (EPCs and UT7/Epo-S1 cells), no AXL was detectable in the B19V nonpermissive cells (K562, HEK293, and Huh7 cells), except for a low level in HeLa cells (Fig. 7A). However, flow cytometry analysis showed that AXL was much less expressed on the cell surface of HeLa cells compared to that on CD36+ EPCs or UT7/Epo-S1 cells (Fig. 7B). In addition, we checked the AXL expression on the surface of CD36+ EPCs during the progression of differentiation. The results showed that AXL cell surface expression peaked at day 9 of differentiation, and much less was expressed on cells differentiated for 12 and 15 days (fig. S4), echoing the best window of differentiated EPCs for B19V infection (11).
Fig. 7. AXL is abundantly expressed on the cell surface of B19V permissive cells but not on B19V nonpermissive cells.
(A) Western blotting detection of AXL expression. Whole-cell lysates from CD36+ EPC, UT7/Epo-S1, HeLa, K562, HEK293, and Huh7 cells were probed with anti-AXL antibody. β-Actin was probed as a loading control. (B) Analysis of cell surface expression of AXL by flow cytometry. CD36+ EPC, UT7/Epo-S1, HeLa, K562, HEK293, and Huh7 cells were directly stained with an anti-AXL antibody, followed by incubation with Texas Red–conjugated secondary antibody. Cell surface expression of AXL was quantified by flow cytometry. Results shown are cell surface expression of AXL in the indicated cell types.
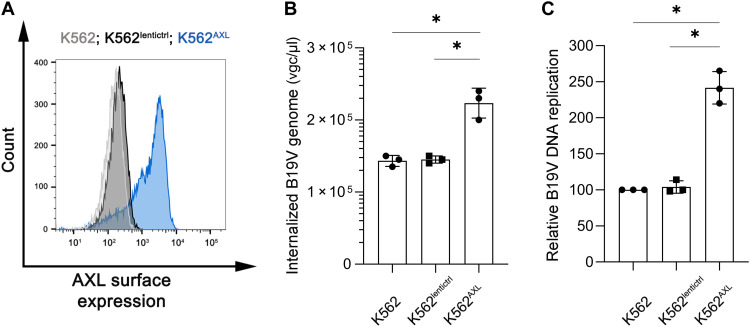
We then asked whether the expression of AXL helped B19V infection. AXL-expressing lentivirus was transduced into K562 cells to express AXL on the cell surface (K562AXL) (Fig. 8A). We then infected K562AXL cells with B19V. The expression of AXL on K562 cells resulted in ~2-fold augmentation of B19V entry and infection compared to the K562lentictrl control cells (Fig. 8, B and C). Together, these results suggested that the expression of AXL on the cell surface is correlated to B19V entry.
Fig. 8. Overexpression of AXL on K562 cells increases B19V infection.
(A) Cell surface expression of AXL. K562 cells were transduced with lentivirus-expressing AXL protein to express AXL on the cell surface (K562AXL). Blank lentivirus vector serves as control (K562lentictrl). Flow cytometry was used to analyze cell surface expression of AXL. (B and C) Overexpression of AXL on K562 cells increases B19V entry and infection. K562, K562AXL, and K562lentictrl cells were infected with B19V at an MOI of ~2000 at 37°C for 1.5 hours. (B) Internalized B19V virions were quantified by qPCR. (C) At 2 dpi, total DNA was extracted and analyzed for the replicated viral DNA and mitochondrial DNA by qPCR. The numbers shown are mitochondrial DNA–standardized B19V genome levels of the infected K562AXL and K562lentictrl cells related to that of the control K562 cells.
DISCUSSION
In this study, using a genome-wide CRISPR-Cas9 gRNA library screen based on B19V VP1u entry, we identified that AXL is a co-receptor for B19V. VP1u directly interacts with AXL at a relatively high affinity (KD = 103 nM), and AXL colocalizes with B19V virions during early infection. Silencing of AXL expression significantly decreased both VP1u and B19V entry and B19V replication of ex vivo expanded human EPCs. Both the recombinant AXL extracellular domain and its specific antibody markedly blocked B19V infection. Thus, we provide strong evidence that AXL is a co-receptor for B19V infection of human EPCs.
AXL has been reported to be an entry factor for several viruses, including Zika virus (ZIKV), Ebola and Marburg viruses, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (42–44). However, rather than having a direct interaction with the virus (45), AXL serves as a docking site for Gas6- or protein S–bound virus particles (37), and the tyrosine kinase activity is required for AXL potentiation of infection by Ebola (46) and ZIKV (47). AXL has been reported to directly bind to the N-terminal domain of the SARS-CoV-2 spike (S) protein but at a relatively low affinity (KD = 882 nM) (44). Notably, AXL directly interacts with VP1u with a KD of 103 nM, ~8 times higher than that with the S protein, which is higher than the binding affinity between most viruses with their cognate receptors (44, 48). R428, a pharmacological inhibitor of AXL autophosphorylation of its tyrosine kinase domain (49), at 1 μM, significantly impaired ZIKV infection (47) but not B19V infection of EPCs, while EPCs retained excellent viability (fig. S5), indicating that the kinase activity of AXL is not necessary for B19V entry. The recombinant extracellular domain of the human AXL protein (Glu33-Pro449; accession #NP_001690.2) and a polyclonal antibody raised against it significantly inhibited B19V infection of EPCs. The receptor/co-receptors identified for B19V entry were unsubstantiated (28–31, 33–35). Neither purified recombinant Ku80 nor native α5β1 blocked B19V infection of CD36+ EPCs (fig. S6), strongly arguing against their roles in mediating B19V entry. Considering the cell surface expression of AXL, its direct interaction with externalized VP1u on the capsid, and its essential role in virus entry and infection, we propose that AXL is a bona fide B19V proteinaceous co-receptor.
AXL is highly expressed in hematopoietic tissues, including erythroid progenitors and proerythroblasts during erythropoiesis (50, 51). Although the TAM receptors, AXL and Mer, play a critical role in erythropoiesis in mouse models, mice lacking AXL or Mer exhibited normal erythropoiesis (51). On the other hand, we observed that near abolishment of AXL did not influence the cell cycle progression of the CD36+ EPCs, echoing the finding in the mouse knockout model. Thus, the decrease in B19V infection of the AXL expression–silenced EPCs and UT7/Epo-S1 cells was due to the function of AXL in mediating virus entry rather than affecting the cell cycle progression on which B19V DNA replication relies (52, 53).
K562 cells, a human erythroleukemia cell line, can be induced to differentiate into the megakaryocytic pathway by treatment with phorbol esters [e.g., phorbol 12-myristate 13-acetate (PMA)] or into the erythrocytic pathway using hemin (50). PMA-treated/differentiated K562 cells render AXL expression (50). PMA-differentiated K562 cells have been shown to permit B19V infection, although it was claimed to be due to the up-regulated integrin α5β1 expression (31). Here, we showed that AXL expression on the cell surface of K562 cells rendered an ~2-fold increase in internalization of B19V into K562 cells and an ~2-fold increase in virus replication, suggesting that P antigen or other receptors/coreceptors may play a cooperative role in B19V entry. The high tropism of B19V infection in EPCs is also controlled by the erythropoietin (Epo)/Epo receptor signaling (35), in particular, the activation of the signal transducer and activator of the transcription 5 (STAT5) pathway (54), which explains why we could not turn on the AXL-expressing K652 cells to become highly permissive to B19V infection.
Although B19V can transmit via the blood and from pregnant mothers to the fetus (2), the major transmission route is respiratory (55). Droplet transmission has been confirmed after intranasal inoculation of volunteers (55, 56), and during the prodrome stage, B19V DNA can be detected in the upper airways (55). Nevertheless, it is still unknown how B19V overcomes the airway epithelial barrier to eventually reach the bone marrow for its replication. The B19V natural host cells, human erythroid progenitors, and B19V permissive UT7/Epo-S1 cells express a high level of AXL on the cell surface, while B19V nonpermissive HEK293 cells, K562, and HeLa cells express negligible AXL on the cell surface, which echoes the high tropism of erythroid lineage cells of B19V. AXL is also differentially expressed on the surface of EPCs under erythroid lineage differentiation. The day 9 EPCs expressed the AXL on cell surface the highest, which also have been shown to be the most permissive cells to B19V infection (11). Notably, AXL is widely expressed in human pulmonary epithelial cells (44), indicating that AXL may facilitate B19V infection of human airways. Apparently, further investigations are required to assure a role of AXL in B19V transmission through the respiratory tract.
In summary, using an unbiased genome-wide CRISPR-Cas9 gRNA screen with VP1u entry of a B19V permissive cell line, we identified AXL as a proteinaceous co-receptor for B19V entry of human EPCs. Thus, AXL could serve as a potential target for antiviral development of B19V infection–caused diseases.
MATERIALS AND METHODS
Primary cells and cell lines
Primary human CD36+ EPCs
Primary human CD36+ EPCs were expanded from CD34+ HSCs ex vivo as previously described (57). Briefly, HSCs were cultured in Wong medium (11) under normoxia (5% CO2 and 21% O2) up to day 4 (the day of procurement was designated as day 0) and frozen in liquid nitrogen. In each experiment, day 4 cells were seeded under normoxia for 2 to 3 days and transferred to a hypoxic condition (1% O2 and 5% CO2) (12) for 2 days, which was designated as day 9 EPCs. These cells express erythroid cell marker CD36+ (35) and were named CD36+ EPCs.
UT7/Epo-S1 cell line
UT7/Epo-S1 cells, which are a human megakaryoblastoid cell line, were obtained from K. Brown (National Heart, Lung, and Blood Institute, National Institutes of Health) with permission from K. Sugamura (13). The UT7/Epo-S1 cells were cultured under normoxic conditions in Dulbecco’s modified Eagle’s medium (DMEM; #SH30022.01, Cytiva Life Science, Marlborough, MA) containing 10% fetal bovine serum (FBS; #F0926, MilliporeSigma, St. Louis, MO), Epo (2 U/ml; Amgen, Thousand Oaks, CA), penicillin (50 IU/ml), and streptomycin (50 μg/ml).
Other cell lines
HEK293 cells [#CRL-1573, American Type Culture Collection (ATCC), Manassas, VA], HEK293T cells (#CRL-3216, ATCC), HeLa cells (#CRM-CCL-2, ATCC), and Huh7 cells [JCRB0403,The Japanese Collection of Research Bioresources(JCRB) Cell Bank] were cultured in DMEM supplemented with 10% FBS at 37°C with a 5% CO2 atmosphere. K562 cells (#CCL-243, ATCC) were cultured with RPMI 1640 medium (#SH30027, Cytiva) with 10% FBS.
Virus infection and quantification
Virus infection
Plasma samples containing B19V at ~5 × 1012 virus genomic copies (vgc)/ml, as quantified by real-time qPCR (35), were provided by ViraCor Eurofins Laboratories (Lenexa, KS). No anti-B19V–specific IgG and IgM antibodies were detected in the plasma samples using enzyme-linked immunosorbent assay (ELISA) kits (DRG International Inc., Springfield, NJ). Day 9 CD36+ EPCs were infected with B19V at a multiplicity of infection (MOI) of 1000 or 5000 vgc per cell.
Virus/replication quantification
For quantification of internalized virions, cells were infected with B19V at 37°C for 1 hour and washed, followed by trypsinization and benzonase treatment to remove the attached virus and degrade DNA/RNA, respectively. For the quantification of egressed virus, at 2 dpi, the supernatant from indicated cell medium was collected for treatment with benzonase followed by DNA extraction using the Quick-DNA/RNA Pathogen Kit (#R1042, Zymo Research Co., Irvine, CA). Extracted DNA was quantified using a probe-based qPCR for B19V DNA as previously described (57). For quantification of viral DNA replication, at 2 dpi, the viral DNA of the infected cells was extracted using the Quick-DNA/RNA Pathogen Kit. Extracted DNA was quantified using a probe-based qPCR for B19V DNA and mitochondrial DNA as previously described (57, 58). For quantification of VP2 mRNA, at 2 dpi, total RNA was extracted from infected cells using TRIzol reagent (Thermo Fisher Scientific). After reverse transcription (RT) of the total RNA with a random primer, both VP2 and β-actin mRNAs were quantified using probe-based multiplex qPCR as previously described (57, 58).
GFP-VP1u internalization assay
GFP-VP1u internalization assay was performed as previously described (26). Briefly, 5 × 105 of UT7/Epo-S1 or CD36+ EPCs were harvested, washed, and resuspended in phosphate-buffered saline (PBS) buffer (pH 7.4) at a final volume of 500 μl. The cells were then incubated with purified GFP-VP1u5–68aa protein (or purified GST protein as control) at 2 μM in a final volume of 500 μl for 1.5 hours at 37°C with gentle rotation. After being trypsinized for 3 min at 37°C and washed three times with PBS, the cells were spun onto the slides using Cytospin 4 (Thermo Fisher Scientific) for immunofluorescent assays or were subjected to flow cytometry. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole).
Genome-wide CRISPR gRNA screen
CRISPR-Cas9 gRNA screen
An amount of 1.0 × 108 UT7/Epo-S1 cells were transduced with the Human Brunello CRISPR Knockout Pooled Library (#73179-LV, Addgene, Watertown, MA), which targets 19,114 human genes with 76,441 gRNAs (~4 gRNAs for each gene), at an MOI of 0.3 (39). Two days after transduction, puromycin (#P8833, Sigma-Aldrich) was added to a final concentration of 2.5 μg/ml, and the cells were selected in puromycin for 7 days. When library-transduced cells reached 2.0 × 108, half of the cells were pelleted, and genomic DNA (gDNA) was extracted using the Blood & Cell Culture DNA Maxi Kit (#13362, Qiagen, Germantown, MD) according to the manufacturer’s instructions and used for NGS as a baseline (NGS0). The GFP-VP1u5–68aa internalization assay was performed by using the other 1.0 × 108 cells. The GFP− cells were sorted with a three-laser flow cytometer (LSR II, BD Biosciences, San Jose, CA). When the GFP− cells reached 2.0 × 108, half of the cells were taken for gDNA extraction (NGS1nd) and the other half were used to perform a second round of GFP-VP1u5–68aa internalization assay to get the second round of GFP− gDNA (NGS2nd).
NGS and data analysis
gDNA was subjected to PCR-based amplification of guide sequences and indexed according to the protocol from the Broad Institute of MIT and Harvard (59) at GeneGoCell Inc., San Diego, CA. The samples were sequenced with Illumina NovaSeq 6000 platform, and the sgRNAs were analyzed by using software package MaGeCK (60). The resulting data were log2-transformed to generate log2-norm raw data, and the abundance of perturbations was calculated to obtain log2 fold change values by comparing with the baseline log2-normalized values. Bubble plots of screening data were generated to display the enriched genes during the NGS2nd versus NGS1nd, where the bubble size represents the P values on the log10 scale, and the y axis represents the fold change of all significantly enriched genes.
Lentivirus production, transduction, and quantification of AXL gene silencing
shRNA-expressing lentiviruses, shAXL (#TRCN0000001040) and shScramble (shScram; #SHC016V), were purchased from MilliporeSigma. AXL-expressing lentivirus (Lenti-AXL) and control lentivirus (LentiCtrl) were made using Addgene plasmid (#116714) according to previously described methods (35). UT7/Epo-S1 cells, CD36+ EPCs, or K562 cells were transduced with the lentivirus at an MOI of ~5 (57).
To quantify the silencing efficiency of the AXL gene expression, total RNA was extracted from stable shRNA-expressing cells by using TRIzol (Thermo Fisher Scientific). After RT using a random primer (#48190011, Invitrogen), the relative level of AXL mRNA was quantified with qPCR using 6-carboxyfluorescein (FAM)–labeled AXL probe (5′6-FAM-AGGAGCCTGACGAAATCCTCTATGTCA-IABkFQ3′) and AXL-specific primers (forward AXL: 5′-CTGCGGGAAGATTTGGAGAA-3′; reverse AXL: 5′-AGGATAACCTCCACCCTCAT-3′), with quantification of β-actin mRNA as an internal control in a multiplex format (61).
Antibody/protein inhibition of B19V infection
For antibody inhibition, an anti-AXL polyclonal (goat IgG) antibody (#AF154, R&D Systems, Minneapolis, MN), which was generated against human AXL (Glu33-Pro440), or IgG isotype control (#AB-108-C, R&D Systems) was incubated with day 9 CD36+ EPCs at concentrations ranging from 0.5 to 100 μg/ml in Wong medium for 1 hour at 4°C.
For protein inhibition, purified proteins—GST-VP1uFlag, rAXLEC, Ku80, α5β1, and GSTFlag or MBP control—were added to 1 × 106 hypoxia-cultured day 9 CD36+ EPCs at various concentrations. The cells were then infected with B19V at an MOI of 1000. At 2 dpi, B19V replication was measured by the quantification of viral DNA using qPCR and the observation and quantification of B19V capsid–expressing cells by immunofluorescence assay and flow cytometry, respectively. All data presented are representative of three independent experiments.
Western blotting
Western blotting was carried out as previously described (57). Briefly, the cell lysates were separated on SDS–polyacrylamide electrophoresis gels and transferred onto a polyvinylidene difluoride membrane (#IPVH00010, MilliporeSigma). After blocking with 5% nonfat milk, the membrane was incubated with indicated primary and secondary antibodies sequentially. Signals were visualized by enhanced chemiluminescence with a Fuji LAS 4000 imaging system (Cytiva) or LI-COR Odyssey imaging system (LI-COR Corporate, Lincoln, NE).
Cell proliferation and cell viability assays
A BrdU incorporation assay was performed to analyze the cell proliferation (cycle) of UT7/Epo-S1 cells and CD36+ EPCs, followed by flow cytometry analysis, as described previously (57). The cell viability assay was performed as previously described using a CytoTox-Glo cytotoxicity assay kit (#G9290, Promega) (58).
Flow cytometry
B19V-infected (B19V+/capsid-positive) or GFP-, GFP-VP1u5–68aa-, and GFP-VP1u(1-227aa)–treated cells were analyzed using flow cytometry as previously described (35, 57, 58). Briefly, the infected or treated cells were collected, fixed, and permeabilized. A mouse anti-B19V intact capsid monoclonal antibody (clone 521-5D) (40) was incubated with the cells, followed by staining with a fluorescein isothiocyanate (FITC)–conjugated anti-mouse secondary antibody. The cells were further stained with DAPI before flow cytometry analysis on the three-laser flow cytometer (LSR II, BD Biosciences, San Jose, CA), and data were analyzed using FACSDiva software (BD Biosciences) or FlowJo software (FlowJo LLC, Ashland, OR).
For staining of AXL on the cell surface, indicated samples were incubated with goat anti-AXL antibody at room temperature for 1 hour. After washing three times with PBS–fetal calf serum (2%), the samples were incubated with fluorochrome-labeled second antibody at room temperature for 1 hour. The samples were then washed three times with PBS-FBS (2%) and fixed with 2% paraformaldehyde at room temperature for 15 min before analysis.
Immunofluorescence assay and confocal microscopy
Immunofluorescence assay was carried out as described previously (26, 58). Briefly, 1 × 105 of treated cells were cytospun onto the slide and fixed. The fixed cells were then incubated with an anti-B19V capsid antibody [#MAB8292/clone 521-5D (40) or #MAb860-55D (25, 62)] to detect intact B19V virions or costained with anti-AXL antibody (#AF154, R&D Systems). The slide was incubated with corresponding secondary antibodies, and DAPI was used to stain the nuclei. After washing, the slide was mounted with Mowiol and a cover glass. Confocal images were taken using a confocal microscope (Leica TCS SP8).
BLI assay
BLI kinetics analysis was performed using BLItz BLI system (FortéBio/Sartorius, Bohemia, NY) as previously described (63). Briefly, the Ni-NTA biosensors (#18-5101, FortéBio/Sartorius) were first hydrated in kinetic buffer [50 mM tris-HCl (pH 7.4) and 150 mM NaCl] for 10 min. rAXLHis (4 μM) in the kinetic buffer was mounted on Ni-NTA biosensors to generate the baseline. The mounted Ni-NTA biosensors were then dipped into binding buffer containing the indicated concentration of GST-VP1u or control GST to determine the binding parameters. Last, the biosensor was dipped into the kinetic buffer to complete a final dissociation step. Ka and Kd were determined using BLItz Pro software. The binding assays were performed at least three times for the calculation of KD (mean ± SD).
Antibodies, proteins, and inhibitors used in the study
Primary antibodies
The following primary antibodies were purchased: mouse monoclonal antibody of anti-B19V capsid (#MAB8292, clone 521-5D) and mouse anti–β-actin (#A5441) from MilliporeSigma; mouse monoclonal antibody against B19V conformational epitope in VP2 (MAB 860-55D) from MIKROGEN (Neuried, Germany); goat anti-AXL antibody (#AF154) from R&D Systems Minneapolis, MN); rabbit anti-AXL (#8661S, clone C89E7), anti-Ku80 (#2180), and anti-integrin α5 (#98204) from Cell Signaling Technology; and mouse anti-Flag (#200-301-B13) and rabbit anti-BrdU (#600-401-C29) from Rockland, Limerick, PA. Anti-B19V VP1u antibody was in-house prepared.
Secondary antibodies
Horseradish peroxidase (HRP)–conjugated anti-rabbit IgG (#A0545), anti-goat IgG (AP106P), and HRP-conjugated anti-mouse IgG (#A4416) were purchased from MilliporeSigma. DyLight 800–conjugated anti-rabbit IgG (#5151S) and DyLight 800–conjugated anti-mouse IgG (#5257S) were purchased from Cell Signaling Technology. FITC-conjugated anti-mouse IgG (#715-095-151), Texas Red–conjugated anti-goat IgG (#705-075-003), Cy-5–conjugated anti-goat IgG (#705-175-003), and HRP-conjugated anti-rat IgG (#112-035-003) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Control antibodies
Normal goat IgG control (#AB-108-C) and normal rabbit IgG control (#AB-105-C) were purchased from R&D Systems.
Purified proteins
Recombinant AXL protein was purchased from Sino Biological (#10279-H08H, Wayne, PA) or ABclonal (#RP00087, Woburn, MA). Recombinant human Ku80 protein and native integrin α5β1 complex were purchased from Abcam (#ab132946, Boston, MA) and R&D Systems (#3230-A5, Minneapolis, MN), respectively. Recombinant B19V VP1u5–68aa, GST-VP1u(1-227aa), GST, and MBP proteins were purified as previously described (26).
Pharmacological inhibitors
AXL kinase inhibitor R428 was purchased from APExBio (#A8329, Boston, MA).
Research standards
Ethics statement
B19V-containing plasma samples were obtained from ViraCor Eurofins Laboratories (Lenexa, KS). Primary human CD34+ HSCs used in this study were isolated from the bone marrow of healthy human donors and were purchased from AllCells LLC (Alameda, CA). Both the cell and virus samples were deidentified, and therefore, institutional review board (IRB) reviews were waived.
Statistics analysis
Statistical analysis was performed by using GraphPad Prism version 9.3, and error bars show means and SDs. Statistical significance P values were determined by using Student’s t test. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05 were regarded as statistically significant, and n.s. was regarded as statistically insignificant.
Acknowledgments
We thank the members of the J.Q. laboratory for their technical support and valuable discussions. We thank S. McFarlin for the critical reading of this manuscript. We are indebted to K. Brown and K. Sugamura for providing the UT7/Epo-S1 cells. We are grateful to the Confocal Microscopy Core Laboratory and the Flow Cytometry Core Laboratory of the University of Kansas Medical Center.
Funding: This study was supported by the National Institutes of Health (R21AI130613). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions: K.N., W.Z., P.X., and F.C. designed, conducted, and analyzed experiments. E.Y.Z. and A.Z.-C. contributed to the bioinformatics analysis. S.K. contributed essential reagents. J.Q. designed and supervised the experiments and acquired funds. K.N. and J.Q. wrote the manuscript draft. All authors contributed to the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data are available from the National Center for Biotechnology Information Sequencing Read Archive (SRA) under accession numbers SRR19919803, SRR19940445, and SRR19926120 and BioProject under accession number PRJNA854912.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.S. F. Cotmore, M. Agbandje-McKenna, M. Canuti, J. A. Chiorini, A.-M. Eis-Hubinger, J. Hughes, M. Mietzsch, S. Modha, M. Ogliastro, J. J. Pénzes, D. J. Pintel, J. Qiu, M. Soderlund-Venermo, P. Tattersall, P. Tijssen; Ictv Report Consortium , ICTV virus taxonomy profile: Parvoviridae. J. Gen. Virol. 100, 367–368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. Qiu, M. Söderlund-Venermo, N. S. Young, Human parvoviruses. Clin. Microbiol. Rev. 30, 43–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. R. Serjeant, J. M. Topley, K. Mason, B. E. Serjeant, J. R. Pattison, S. E. Jones, R. Mohamed, Outbreak of aplastic crises in sickle cell anaemia associated with parvovirus-like agent. Lancet 2, 595–597 (1981). [DOI] [PubMed] [Google Scholar]
- 4.J. R. Pattison, S. E. Jones, J. Hodgson, L. R. Davis, J. M. White, C. E. Stroud, L. Murtaza, Parvovirus infections and hypoplastic crisis in sickle-cell anaemia. Lancet 1, 664–665 (1981). [DOI] [PubMed] [Google Scholar]
- 5.N. S. Young, K. E. Brown, Parvovirus B19. N. Engl. J. Med. 350, 586–597 (2014). [DOI] [PubMed] [Google Scholar]
- 6.A. Riipinen, E. Vaisanen, M. Nuutila, M. Sallmen, R. Karikoski, M. L. Lindbohm, K. Hedman, H. Taskinen, M. Söderlund-Venermo, Parvovirus b19 infection in fetal deaths. Clin. Infect. Dis. 47, 1519–1525 (2008). [DOI] [PubMed] [Google Scholar]
- 7.B. E. Torbett, J. S. Friedman, Erythropoiesis: An overview, in Erythropoietins, Erythropoietic Factors and Erythropoiesis, S. G. Elliott, M. A. Foote, G. Molineux, Eds. (Birkhauser Verlag, 2009), pp. 3–18. [Google Scholar]
- 8.K. Ozawa, G. Kurtzman, N. Young, Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233, 883–886 (1986). [DOI] [PubMed] [Google Scholar]
- 9.H. Chisaka, E. Morita, N. Yaegashi, K. Sugamura, Parvovirus B19 and the pathogenesis of anaemia. Rev. Med. Virol. 13, 347–359 (2003). [DOI] [PubMed] [Google Scholar]
- 10.N. Sol, J. J. Le, I. Vassias, J. M. Freyssinier, A. Thomas, A. F. Prigent, B. B. Rudkin, S. Fichelson, F. Morinet, Possible interactions between the NS-1 protein and tumor necrosis factor alpha pathways in erythroid cell apoptosis induced by human parvovirus B19. J. Virol. 73, 8762–8770 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Wong, N. Zhi, C. Filippone, K. Keyvanfar, S. Kajigaya, K. E. Brown, N. S. Young, Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J. Virol. 82, 2470–2476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Y. Chen, S. Kleiboeker, J. Qiu, Productive Parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIF alpha. PLOS Pathog. 7, e1002088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.E. Morita, K. Tada, H. Chisaka, H. Asao, H. Sato, N. Yaegashi, K. Sugamura, Human parvovirus B19 induces cell cycle arrest at G(2) phase with accumulation of mitotic cyclins. J. Virol. 75, 7555–7563 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. F. Cotmore, V. C. McKie, L. J. Anderson, C. R. Astell, P. Tattersall, Identification of the major structural and nonstructural proteins encoded by human parvovirus B19 and mapping of their genes by procaryotic expression of isolated genomic fragments. J. Virol. 60, 548–557 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.K. Ozawa, N. Young, Characterization of capsid and noncapsid proteins of B19 parvovirus propagated in human erythroid bone marrow cell cultures. J. Virol. 61, 2627–2630 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. Kronenberg, B. Böttcher, C. W. von der Lieth, S. Bleker, J. A. Kleinschmidt, A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79, 5296–5303 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. Mani, C. Baltzer, N. Valle, J. M. Almendral, C. Kempf, C. Ros, Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 80, 1015–1024 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.T. Saikawa, S. Anderson, M. Momoeda, S. Kajigaya, N. S. Young, Neutralizing linear epitopes of B19 parvovirus cluster in the VP1 unique and VP1-VP2 junction regions. J. Virol. 67, 3004–3009 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Anderson, M. Momoeda, M. Kawase, S. Kajigaya, N. S. Young, Peptides derived from the unique region of B19 parvovirus minor capsid protein elicit neutralizing antibodies in rabbits. Virology 206, 626–632 (1995). [DOI] [PubMed] [Google Scholar]
- 20.G. Kurtzman, N. Frickhofen, J. Kimball, D. W. Jenkins, A. W. Nienhuis, N. S. Young, Pure red-cell aplasia of 10 years’ duration due to persistent parvovirus B19 infection and its cure with immunoglobulin therapy. N. Engl. J. Med. 321, 519–523 (1989). [DOI] [PubMed] [Google Scholar]
- 21.S. Kajigaya, H. Fujii, A. Field, S. Anderson, S. Rosenfeld, L. J. Anderson, T. Shimada, N. S. Young, Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc. Natl. Acad.Sci. U.S.A. 88, 4646–4650 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Bonsch, C. Kempf, C. Ros, Interaction of parvovirus B19 with human erythrocytes alters virus structure and cell membrane integrity. J. Virol. 82, 11784–11791 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C. Bonsch, C. Zuercher, P. Lieby, C. Kempf, C. Ros, The globoside receptor triggers structural changes in the B19 virus capsid that facilitate virus internalization. J. Virol. 84, 11737–11746 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R. V. Lakshmanan, J. A. Hull, L. Berry, M. Burg, B. Bothner, R. McKenna, M. Agbandje-McKenna, Structural dynamics and activity of B19V VP1u during the pHs of cell entry and endosomal trafficking. Viruses 14, 1922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. Leisi, N. Ruprecht, C. Kempf, C. Ros, Parvovirus B19 uptake is a highly selective process controlled by VP1u, a novel determinant of viral tropism. J. Virol. 87, 13161–13167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.W. Zou, K. Ning, P. Xu, X. Deng, F. Cheng, S. Kleiboeker, J. Qiu, The N-terminal 5-68 amino acids domain of the minor capsid protein VP1 of human parvovirus B19 enters human erythroid progenitors and inhibits B19 infection. J. Virol. 95, e00466-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. Ros, J. Bieri, R. Leisi, The VP1u of human parvovirus B19: A multifunctional capsid protein with biotechnological applications. Viruses 12, 1463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.K. E. Brown, S. M. Anderson, N. S. Young, Erythrocyte P antigen: Cellular receptor for B19 parvovirus. Science 262, 114–117 (1993). [DOI] [PubMed] [Google Scholar]
- 29.K. E. Brown, J. R. Hibbs, G. Gallinella, S. M. Anderson, E. D. Lehman, P. McCarthy, N. S. Young, Resistance to parvovirus B19 infection due to lack of virus receptor (erythrocyte P antigen). N. Engl. J. Med. 330, 1192–1196 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Y. Munakata, T. Saito-Ito, K. Kumura-Ishii, J. Huang, T. Kodera, T. Ishii, Y. Hirabayashi, Y. Koyanagi, T. Sasaki, Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood 106, 3449–3456 (2005). [DOI] [PubMed] [Google Scholar]
- 31.K. A. Weigel-Kelley, M. C. Yoder, A. Srivastava, Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: Requirement of functional activation of beta1 integrin for viral entry. Blood 102, 3927–3933 (2003). [DOI] [PubMed] [Google Scholar]
- 32.K. A. Weigel-Kelley, M. C. Yoder, A. Srivastava, Recombinant human parvovirus B19 vectors: Erythrocyte P antigen is necessary but not sufficient for successful transduction of human hematopoietic cells. J. Virol. 75, 4110–4116 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Bieri, C. Ros, Globoside is dispensable for Parvovirus B19 entry but essential at a postentry step for productive infection. J. Virol. 93, e00972-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. Bieri, R. Leisi, C. Bircher, C. Ros, Human parvovirus B19 interacts with globoside under acidic conditions as an essential step in endocytic trafficking. PLOS Pathog. 17, e1009434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Y. Chen, W. Guan, S. Lou, Z. Liu, S. Kleiboeker, J. Qiu, Role of erythropoietin receptor signaling in Parvovirus B19 replication in human erythroid progenitor cells. J. Virol. 84, 12385–12396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Y. Luo, S. Lou, X. Deng, Z. Liu, Y. Li, S. Kleiboeker, J. Qiu, Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J. Virol. 85, 8046–8055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.G. Lemke, Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol. 5, ea009076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.C. M. Gay, K. Balaji, L. A. Byers, Giving AXL the axe: Targeting AXL in human malignancy. Br. J. Cancer 116, 415–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. G. Doench, N. Fusi, M. Sullender, M. Hegde, E. W. Vaimberg, K. F. Donovan, I. Smith, Z. Tothova, C. Wilen, R. Orchard, H. W. Virgin, J. Listgarten, D. E. Root, Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.N. Zhi, I. P. Mills, J. Lu, S. Wong, C. Filippone, K. E. Brown, Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J. Virol. 80, 5941–5950 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.W. Ma, L. Yang, L. He, Overview of the detection methods for equilibrium dissociation constant K(D) of drug-receptor interaction. J. Pharm. Anal. 8, 147–152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. P. Strange, B. Jiyarom, Z. N. Pourhabibi, X. Xie, C. Baker, H. Sadri-Ardekani, P. Y. Shi, S. Verma, Axl promotes Zika virus entry and modulates the antiviral state of human sertoli cells. MBio 10, e01372-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Shimojima, A. Takada, H. Ebihara, G. Neumann, K. Fujioka, T. Irimura, S. Jones, H. Feldmann, Y. Kawaoka, Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 80, 10109–10116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Wang, Z. Qiu, Y. Hou, X. Deng, W. Xu, T. Zheng, P. Wu, S. Xie, W. Bian, C. Zhang, Z. Sun, K. Liu, C. Shan, A. Lin, S. Jiang, Y. Xie, Q. Zhou, L. Lu, J. Huang, X. Li, AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 31, 126–140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. A. Brindley, C. L. Hunt, A. S. Kondratowicz, J. Bowman, P. L. Sinn, P. B. McCray Jr., K. Quinn, M. L. Weller, J. A. Chiorini, W. Maury, Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology 415, 83–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Shimojima, Y. Ikeda, Y. Kawaoka, The mechanism of Axl-mediated Ebola virus infection. J. Infect. Dis. 196, S259–S263 (2007). [DOI] [PubMed] [Google Scholar]
- 47.S. Liu, L. J. DeLalio, B. E. Isakson, T. T. Wang, AXL-mediated productive infection of human endothelial cells by Zika Virus. Circ. Res. 119, 1183–1189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. Pillay, N. L. Meyer, A. S. Puschnik, O. Davulcu, J. Diep, Y. Ishikawa, L. T. Jae, J. E. Wosen, C. M. Nagamine, M. S. Chapman, J. E. Carette, An essential receptor for adeno-associated virus infection. Nature 530, 108–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. J. Holland, A. Pan, C. Franci, Y. Hu, B. Chang, W. Li, M. Duan, A. Torneros, J. Yu, T. J. Heckrodt, J. Zhang, P. Ding, A. Apatira, J. Chua, R. Brandt, P. Pine, D. Goff, R. Singh, D. G. Payan, Y. Hitoshi, R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 70, 1544–1554 (2010). [DOI] [PubMed] [Google Scholar]
- 50.A. Neubauer, A. Fiebeler, D. K. Graham, J. P. O’Bryan, C. A. Schmidt, P. Barckow, S. Serke, W. Siegert, H. R. Snodgrass, D. Huhn, E. T. Liu, Expression of axl, a transforming receptor tyrosine kinase, in normal and malignant hematopoiesis. Blood 84, 1931–1941 (1994). [PubMed] [Google Scholar]
- 51.H. Tang, S. Chen, H. Wang, H. Wu, Q. Lu, D. Han, TAM receptors and the regulation of erythropoiesis in mice. Haematologica 94, 326–334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.W. Zou, Z. Wang, M. Xiong, A. Y. Chen, P. Xu, S. S. Ganaie, Y. Badawi, S. Kleiboeker, H. Nishimune, S. Q. Ye, J. Qiu, Human parvovirus B19 utilizes cellular DNA replication machinery for viral DNA replication. J. Virol. 92, e01881-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. S. Ganaie, J. Qiu, Recent advances in replication and infection of human parvovirus B19. Front. Cell. Infect. Microbiol. 8, 166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.S. S. Ganaie, W. Zou, P. Xu, X. Deng, S. Kleiboeker, J. Qiu, Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLOS Pathog. 13, e1006370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. J. Anderson, P. G. Higgins, L. R. Davis, J. S. Willman, S. E. Jones, I. M. Kidd, J. R. Pattison, D. A. Tyrrell, Experimental parvoviral infection in humans. J. Infect. Dis. 152, 257–265 (1985). [DOI] [PubMed] [Google Scholar]
- 56.C. G. Potter, A. C. Potter, C. S. Hatton, H. M. Chapel, M. J. Anderson, J. R. Pattison, D. A. Tyrrell, P. G. Higgins, J. S. Willman, H. F. Parry, Variation of erythroid and myeloid precursors in the marrow and peripheral blood of volunteer subjects infected with human parvovirus (B19). J. Clin. Invest. 79, 1486–1492 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Wang, S. S. Ganaie, F. Cheng, P. Xu, K. Ning, X. Wang, S. Kleiboeker, S. Cheng, J. Qiu, RNA binding motif protein RBM45 regulates expression of the 11-kilodalton protein of parvovirus B19 through binding to novel intron splicing enhancers. MBio 11, e00192-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. Ning, A. Roy, F. Cheng, P. Xu, S. Kleiboeker, C. R. Escalante, J. Wang, J. Qiu, High throughput screening identifies inhibitors for parvovirus B19 infection of human erythroid progenitor cells. J. Virol. 96, e01326-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.K. R. Sanson, R. E. Hanna, M. Hegde, K. F. Donovan, C. Strand, M. E. Sullender, E. W. Vaimberg, A. Goodale, D. E. Root, F. Piccioni, J. G. Doench, Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.W. Li, H. Xu, T. Xiao, L. Cong, M. I. Love, F. Zhang, R. A. Irizarry, J. S. Liu, M. Brown, X. S. Liu, MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.W. Guan, F. Cheng, Y. Yoto, S. Kleiboeker, S. Wong, N. Zhi, D. J. Pintel, J. Qiu, Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 82, 9951–9963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. Gigler, S. Dorsch, A. Hemauer, C. Williams, S. Kim, N. S. Young, S. Zolla-Pazner, H. Wolf, M. K. Gorny, S. Modrow, Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J. Virol. 73, 1974–1979 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.K. Ning, Z. Wang, F. Cheng, Z. Yan, J. Qiu, The small nonstructural protein NP1 of human bocavirus 1 directly interacts with Ku70 and RPA70 and facilitates viral DNA replication. PLOS Pathog. 18, e1010578 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S4