Abstract
Recent proteome and transcriptome profiling of Alzheimer's disease (AD) brains reveals RNA splicing dysfunction and U1 small nuclear ribonucleoprotein (snRNP) pathology containing U1-70K and its N-terminal 40-KDa fragment (N40K). Here we present a causative role of U1 snRNP dysfunction to neurodegeneration in primary neurons and transgenic mice (N40K-Tg), in which N40K expression exerts a dominant-negative effect to downregulate full-length U1-70K. N40K-Tg recapitulates N40K insolubility, erroneous splicing events, neuronal degeneration and cognitive impairment. Specifically, N40K-Tg shows the reduction of GABAergic synapse components (e.g., the GABA receptor subunit of GABRA2), and concomitant postsynaptic hyperexcitability that is rescued by a GABA receptor agonist. Crossing of N40K-Tg and the 5xFAD amyloidosis model indicates that the RNA splicing defect synergizes with the amyloid cascade to remodel the brain transcriptome and proteome, deregulate synaptic proteins, and accelerate cognitive decline. Thus, our results support the contribution of U1 snRNP-mediated splicing dysfunction to AD pathogenesis.
Introduction
RNA splicing is a fundamental regulatory process in eukaryotic gene expression, which is particularly important to expand molecular diversity in the nervous system1. Mutations of RNA splicing genes are linked to numerous neurodegenerative disorders2-4, such as SMN1 in spinal muscular atrophy5, TARDBP (TDP-43 protein), FUS, TAF15, MATR3, TIA1, hnRNPA1, and hnRNPA2B1 in amyotrophic lateral sclerosis and frontotemporal dementia6,7. Mutant FUS can induce neuronal toxicity, which may be mediated by its interaction with U1 snRNP, an essential complex in RNA splicing8-10. In addition, genetic linkage of RNA splicing components to neurodegeneration has been found in a chemical mutagenesis screen of mice11, in which a mutant U2 snRNA gene causes alternative splicing defects, leading to ataxia and cerebellar neurodegeneration. Thus, the deregulation of RNA splicing is a key molecular mechanism linked to neurodegenerative disease.
Dysfunction of RNA splicing has been implicated in the development of Alzheimer’s disease, as TDP-43 aggregates were detected in up to ~50% of AD brains12. While amyloid plaques and neurofibrillary tangles are the hallmarks of AD pathology13,14, abnormal synaptic changes are correlated with progressive cognitive decline, indicating that synaptic dysfunction is a crucial factor contributing to cognitive impairment15,16. More recently, the development of mass spectrometry (MS) enables comprehensive analysis of proteome directly from human clinical specimens17,18. Unbiased profiling of aggregated brain proteome discovered the deposition of U1 snRNP and Prp5/DDX46, an RNA helicase connecting the U1 and U2 snRNP complexes19. Further profiling of the AD whole proteome and phosphoproteome identified the alterations of other RNA binding factors and splicing components20,21. Global structural profiling of AD cases also suggested conformational changes of RNA splicing components, such as the U2 snRNP subunit SF3B, and heterogeneous nuclear ribonucleoproteins (hnRNP H2, M and U)22. Consistently, transcriptomic profiling of human brain tissues confirmed reproducible, aberrant RNA splicing events in multiple AD cohorts19,23.
U1 snRNP is highly accumulated in the AD aggregated proteome, only behind Aβ, tau and complement proteins19,24,25. The U1 snRNP complex is comprised of U1 snRNA, U1-70K, U1A, U1C, and seven Sm proteins (B/B’, D1, D2, D3, E, F and G)26. Immunohistochemical staining and electron microscopy confirmed that the U1 snRNP forms a new type of cytoplasmic tangle-like fibril in sporadic and familial AD, as well as in trisomy 21 cases19,24,27,28. The presence of U1 snRNA in the aggregates was also shown by quantitative PCR and immunofluorescence studies27. Moreover, The core subunit U1-70K was found to be cleaved to a N-terminal fragment, ~40 kDa with ~300 amino acids named N40K29. The aggregation of U1 snRNP occurs as early as in the stage of mild cognitive impairment19, and is correlated with the neuropathological hallmarks of Aβ plaques and tau tangles during AD progression24. Interestingly, the U1 snRNP pathology was not found in other examined neurodegenerative diseases, including Parkinson’s disease, amyotrophic lateral sclerosis, frontotemporal lobar degeneration, and corticobasal degeneration19. Taken together, these findings strongly support a unique U1 snRNP pathology in AD. However, the post-mortem human brain studies only uncover disease-correlated proteins, which do not indicate causation, especially considering that molecular changes may appear years earlier than the onset of AD clinical symptoms. So, it is not clear whether the observed U1 snRNP pathology in human samples plays a causative role in the development of AD.
In this study, we present a mouse model (N40K-Tg) of U1 snRNP dysfunction to investigate its role in AD pathogenesis. The function of U1 snRNP is impaired by the dominant-negative effect of the N40K fragment, which competes with U1-70K to assemble the U1 snRNP complex, resulting in a significant loss of full length U1-70K protein and splicing defects in primary neurons and mice. We have thoroughly characterized the neuron-specific N40K-Tg mice by multi-omics, biochemical, histopathological, and electrophysiological approaches, along with mouse behavior studies. The N40K-Tg mice display protein insolubility, neurodegeneration and abnormality in memory tests, as well as splicing defects enriched in synaptic signaling. In particular, GABAergic synapse deregulation and neuronal hyperexcitability are identified in the N40K-Tg mice. Finally, the double transgenic model from N40K-Tg and 5xFAD mice shows a strong synergy between RNA splicing defects and the amyloid cascade.
Results
U1 snRNP dysfunction elicits excitatory toxicity in neurons
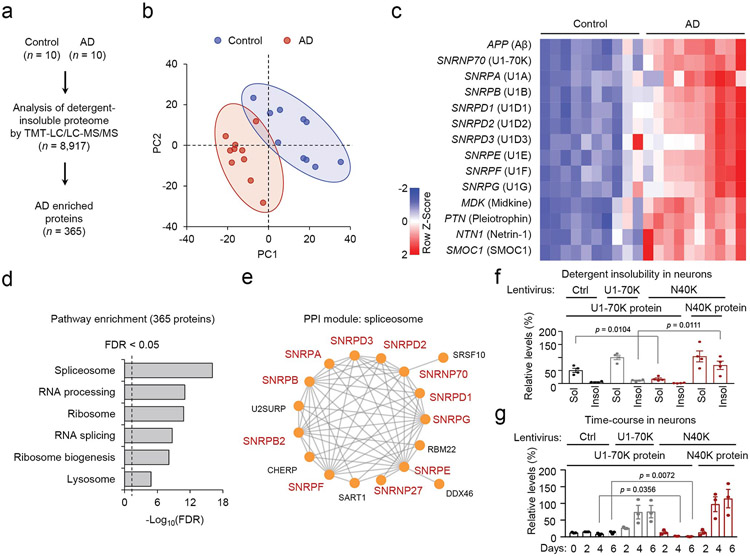
Multiple U1 snRNP components are aggregated in AD brain, which were previously detected in detergent-insoluble fractions by semi-quantitative label-free mass spectrometry in two studies (n = 4,216 proteins and 2,711 proteins)19,24, as aggregated proteins often have low solubility and are partially resistant to detergent extraction30. With the development of the latest tandem-mass-tag (TMT) strategy, two-dimensional liquid chromatography (LC/LC) and tandem mass spectrometry (MS/MS) approach31,32, we re-profiled 8,917 proteins in the detergent-insoluble proteome from 10 control and 10 AD brains (Supplementary Tables 1, 2), which increased the coverage by more than 2-fold. We found that 365 proteins were significantly accumulated in the AD cases (Fig. 1a, Extended Data Fig. 1a, 1b, FDR < 0.05), including Aβ, MDK, PTN, NTN1, SMOC1, and U1-70K, as well as other U1 snRNP subunits (U1A, Sm proteins B/B’, D1, D2, D3, E, F and G, Fig. 1b, Extended Data Fig. 1c). MDK, PTN, NTN1 and SMOC1 have recently been identified among the most increased proteins in human AD brains and they are colocalized in amyloid plaques18. Spliceosome is the most enriched pathway and protein-protein interaction module in the AD detergent-insoluble proteome (Extended Data Fig. 1d, 1e, FDR < 0.05), in agreement with the previous proteomic finding19,24.
Figure 1. Tissue proteomics confirms U1 snRNP aggregation and N40K shows detergent insolubility and dominant negative effects to deplete U1-70K through proteasomal degradation.
a, Volcano plot of detergent-insoluble proteome by deep TMT-LC/LC-MS/MS analysis (10 AD and 10 non-dementia control cases). Among 8,917 identified proteins, 365 proteins were accumulated in the AD samples. Dashed lines indicate the cutoffs (FDR < 0.05, log2(AD/Ctrl) > 0.30, equivalent to 2-fold of the standard deviation). b, Diagram of the U1 snRNP complex. c, Diagrams of U1-70K and N40K domains: RRM: RNA recognition motif, LC: low complexity domain. d, Aggregation analysis of U1-70K and N40K. Control EGFP (Ctrl), U1-70K-FLAG, and N40K-FLAG were expressed in mouse cortical neurons (DIV 12) by lentiviruses (MOI 10) and extracted with 1% sarkosyl to produce detergent soluble (Sol) and insoluble (Insol) fractions for immunoblotting. The experiment was repeated 4 times and quantified (Extended Data Fig. 1f). e, The dominant-negative effect of N40K in a time course analysis. Cortical neurons were infected with lentiviruses for the analysis (DIV 12, MOI 10). The experiment was triplicated with quantification (Extended Data Fig. 1g). f, Endogenous U1-70K mRNA quantitated by qRT-PCR in the control and N40K-expressing neurons (3 replicates, mean ± SEM, Student’s t-test, ns: not significant). g, The model of U1-70K downregulation induced by N40K expression. h, U1-70K or N40K assembled in U1 snRNP. Control EGFP (Ctrl), U1-70K-FLAG or N40K-FLAG was expressed in HEK 293T cells and immunoprecipitated by FLAG Ab for SDS-PAGE, followed by Ponceau S staining and immunoblotting by U1A and U1C antibodies. i, U1-70K downregulation is mediated by the ubiquitin-proteasome pathway. Neurons were infected with the N40K lentivirus for 2 days (DIV 12, MOI 10), then treated with solvent DMSO 0.1% (Ctrl), the proteasome inhibitor MG132 (10 μM) or the autophagy inhibitor bafilomycin A (Baf, 10 nM) for 1 or 2 days, and harvested for immunoblotting. The experiment was repeated 4 times and the U1-70K levels were quantified (two-way ANOVA and Tukey's multiple comparison test). Data are shown as mean ± SEM. Full statistical information is in Source Data Statistics.
Moreover, U1-70K was found to be cleaved to a N-terminal fragment, ~40 kDa with ~300 amino acids named N40K in the AD detergent-insoluble proteome19,29. Both U1-70K and N40K contain disordered low complexity (LC) domains involved in aggregation (Fig. 1c)33,34. When human U1-70K or N40K is ectopically expressed in mouse primary cortical neurons and subjected to extraction with 1% sarkosyl detergent, an anionic surfactant, N40K protein clearly shows higher insolubility than U1-70K protein (Fig. 1d), suggesting that N40K is prone to protein aggregation.
In addition to N40K insolubility, we found that N40K exerts a strong dominant-negative effect to downregulate native U1-70K protein in neurons. First, in AD brain, N40K appears to be inversely correlated with full length U1-70K in protein abundance29. Second, in human N40K lentivirus infected mouse neurons, endogenous U1-70K protein is substantially downregulated (Fig. 1d, Extended Data Fig. 1f), and this downregulation occurs in a time-dependent manner after the infection of N40K virus (Fig. 1e, Extended Data Fig. 1g). In contrast, the RNA level of native U1-70K remains unchanged (Fig. 1f), indicating of a posttranscriptional mechanism. As U1-70K N-terminus interacts with other subunits in the U1 snRNP complex35, we propose that N40K can compete with U1-70K to assemble U1 snRNP, and free U1-70K is then degraded by the proteasome (Fig. 1g). To validate this hypothesis, we performed immunoprecipitation analysis to indicate that either U1-70K or N40K can pull down U1A and U1C equally (Fig. 1h). Moreover, when applying two chemicals to inhibit N40K-induced U1-70K degradation, we found that U1-70K can be rescued by the treatment of MG132 (a proteasomal inhibitor) but not bafilomycin A1 (an autophagy inhibitor) (Fig. 1i), supporting that U1-70K is degraded by the ubiquitin-proteasome system.
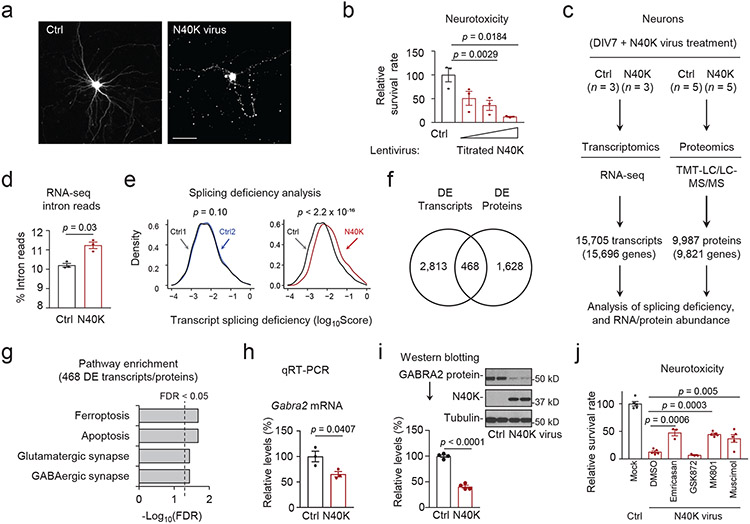
We further observed morphological degeneration and cell death caused by N40K expression in primary neurons (Fig. 2a, 2b), like our previous report of N40K neurotoxicity29. To investigate the mechanism of neurotoxicity, we performed deep RNA-seq and MS-based proteomics experiments (Supplementary Tables 1, 3) to profile 15,705 transcripts and 9,987 proteins in N40K-expressing neurons, identifying N40K-induced changes in RNA splicing, RNA abundance, and protein levels (Fig. 2c). Consistent with splicing dysfunction in AD19, N40K-expressing neurons display a higher global intron read percentage than controls (Fig. 2d). For individual transcripts, we computed a splicing deficiency score, the ratio between length-normalized intron and exon reads19. A total of 2,874 transcripts exhibit significant splicing changes (Supplementary Table 4, FDR < 0.05), showing an obvious shift in the distribution of their splicing deficiency scores in N40K-expressing neurons (Fig. 2e). Subsequently, the abundance of 3,281 transcripts and 2,096 proteins are altered in these neurons (Supplementary Tables 5, 6, FDR < 0.05), in which 468 RNAs/proteins show consistent changes (Fig. 2f). Interestingly, these overlapped RNAs/proteins are enriched in numerous pathways, including ferroptosis, apoptosis, glutamatergic synapse, and GABAergic synapse (Fig. 2g, Supplementary Table 7, FDR < 0.05). Thus, these large-scale molecular profiling clearly reveals that N40K expression has a significant impact on RNA splicing, the transcriptome and the proteome of neurons, especially on the components in synaptic function.
Figure 2. Comprehensive analysis of transcriptome and proteome reveals synaptic pathway in N40K-induced neuron death.
a, Morphology of representative cultured neurons with lentiviral expression of control EGFP (Ctrl) and N40K (with EGFP for visualization). Scale bar: 20 μm. The experiment was repeated three times independently. b, Neuron survival rate under dose-dependent N40K expression. Mouse cortical neurons were transduced with the control or N40K lentivirus at MOI 7, 20, or 60 for 5 days followed by the CellTiter-Glo luminescent cell viability assay (n=3),). Cortical neurons were transduced with control or N40K lentivirus at MOI 10 for 4 days and harvested for c-I. c, Transcriptomic and proteomic profiling. Sample sizes are indicated. To simplify data processing, we analyzed the major transcripts without considering alternative splicing forms. d, Mapped intronic reads increase in N40K-expressing neurons. e, Histograms of splicing deficiency scores of individual transcripts in N40K-expressing neurons analyzed by Kolmogorov-Smirnov test (left: comparison of Ctrl1 and Ctrl2 is shown as a null (p = 0.1), right: comparison of average score of control and N40K neurons (n = 3, p < 2 .2 x 10−16). f, Overlapping of DE RNAs (FDR < 0.05) and proteins (FDR < 0.05). g, Pathway enrichment of the overlapped DE transcripts/proteins (selected from Supplementary Table 7). FDR was derived from p values (Fisher's exact test) by the BH procedure. h, Quantitative RT-PCR of Gabra2 mRNA (n=3). i, Western blotting of GABRA2 and N40K in cultured neurons transduced by control and N40K lentivirus. The analysis was repeated with 4 different wells of neurons. j, Relative survival rate of pharmacological inhibitors on N40K transduced neurons. Cortical neurons were transduced with vector (Ctrl) or N40K lentivirus for 2 days followed with pharmacological treatments for 3 days: 100 μM emricasan (apoptosis inhibitor), 50 μM GSK872 (necrosis inhibitor), 10 μM MK801 (NMDAR antagonist), or 50 μM muscimol (GABAAR agonist). All replicates were from different wells of neuron culture. Data presented as means ± SEM, p values as indicated by two-tailed Student’s t-test (d, h, i) or one-way ANOVA followed by Tukey's multiple comparison test (b, j). Full statistical information is in Source Data Statistics.
We then focused on synaptic proteins and found that Gabra2, one of the alpha subunits in GABAA receptors36, was among the altered proteins in N40K-expressing neurons (Supplementary Table 6). We confirmed the decrease of Gabra2 RNA and GABRA2 protein by quantitative RT-PCR (Fig. 2h) and western blotting (Fig. 2i), respectively. Since GABAA receptors play a central role to inhibit synaptic activities, GABRA2 downregulation may lead to synaptic hyperactivity, resulting in N40K-induced cell death. To examine this hypothesis, we performed a rescue assay by treating the N40K cellular model with different pathway inhibitors. Indeed, neuronal toxicity can be markedly reduced by either muscimol (an agonist of inhibitory GABAA receptor) or MK801 (an antagonist of excitatory NMDA receptor) in the primary culture (Fig. 2j), while the toxicity is partially rescued by emricasan (a patented apoptosis inhibitor), but not GSK872 (a necroptosis inhibitor). These results suggest that the N40K neurotoxicity is mediated, at least partially, by elevated synaptic activities and apoptosis. It should be mentioned that necroptosis, ferroptosis and excitotoxicity are implicated in AD37,38. Our cellular model recapitulates the excitotoxicity but not the necroptosis, which may be due to its difference from the neurodegenerative condition in AD brain. Overall, this cellular model experiments indicate that N40K expression induces U1-70K depletion, compromises U1 snRNP function, impairs RNA splicing, and alters synaptic proteins and activities, leading to neurotoxicity.
N40K-Tg displays splicing defects and cognitive impairment
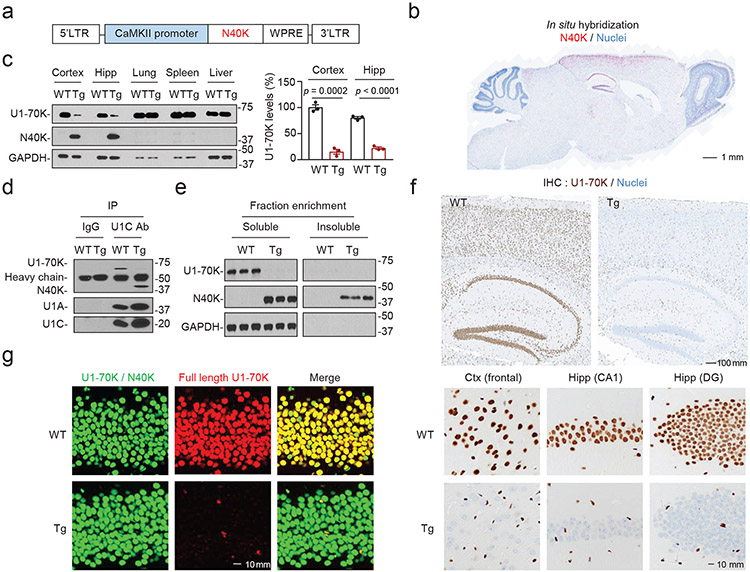
Beyond the N40K cellular model, we generated an N40K-Tg transgenic mouse model to determine whether N40K accumulation and U1-70K depletion may be causative of AD symptoms. In the Tg, human N40K gene is expressed under a neuron-specific CaMKIIα promotor39 to produce two lines: Tg396 and Tg318 with the transgene inserted in chromosomes 18 and 10, respectively (Fig. 3a-b, Extended Data Fig. 2a, 2b). We fully characterized the Tg396 mouse line and confirmed major conclusions in the second Tg318 line. In both lines, N40K is expressed in a brain region-specific pattern at a ~2-fold level of native U1-70K in wild type (WT) mice (Fig. 3c, and Extended Data Fig. 2c, 2d). As anticipated, N40K expression results in significant U1-70K reduction in cortex and hippocampus (Fig. 3c). Immunoprecipitation analysis confirms that N40K competes with U1-70K for assembling U1 snRNP (Fig. 3d). In protein extraction assay, a significant portion of N40K is present in the detergent-insoluble fraction (Fig. 3e). Comprehensive proteomic analysis of the detergent-insoluble fraction reveals the accumulation of N40K, other U1 snRNP subunits (U1A, U1C, and Sm proteins), and some other RNA binding proteins (Extended Data Fig. 3a-e and Supplementary Table 8, FDR < 0.05). The MS analysis profiled a total of 8,509 proteins in the detergent-insoluble fractions, revealing 90 proteins highly enriched in N40K-Tg, when compared to the WT littermates. These 90 proteins are almost exclusively enriched in RNA related pathways. Furthermore, U1-70K immunostaining validates its dramatic neuron-specific reduction in cortical and hippocampal regions in N40K-Tg compared with WT littermates (Fig. 3f, 3g). Together, the N40K-Tg model exhibits N40K insolubility and U1-70K downregulation.
Figure 3. Biochemical and cellular characterization of the N40K-Tg mouse model.
a, Tg construct expressing N40K specifically in neurons, including CaMKIIα (CaMKII) promoter, a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) and long terminal repeats (LTR). b, N40K mRNA expression by in situ hybridization. Tg brain slides were stained with RNAscope probe specific for human N40K (Red) and hematoxylin for nuclei (blue). Scale bar: 1 mm. c, Western blotting to confirm N40K expression and loss of U1-70K specifically in the brain. Ctx: cortex; Hipp: hippocampus. The experiments were performed with three replicates and the loss of U1-70K by western blotting was quantified (mean ± SEM, Student’s t-test, two-tailed). d, N40K-assembled U1 snRNP complex in the Tg brain occludes the binding of U1-70K. Immunoprecipitation of U1C Ab was followed by immunoblotting (3 replicates). e, Aggregation analysis of N40K in the Tg brain. The mouse brain tissues were differentially extracted to generate detergent soluble (Sol) and insoluble (Insol) fractions, followed by immunoblotting (3 replicates). f, Immunostaining of WT and Tg brain tissue with U1-70K C-terminal Ab (brown) that recognized only full length U1-70K but not N40K. Slides were counter-stained with hematoxylin for nuclei (blue). DG: dentate gyrus. Scale bar, 100 μm (upper), 10 μm (lower) (n = 3 replicates). g, Confocal images of immunofluorescence staining (hippocampal DG region, green: U1-70K and N40K detected by U1-70K N-terminal Ab, red: full length U1-70K detected by U1-70K C-terminal Ab. Scale bar, 10 μm. In b, f, g, each assay was repeated from three mice. Full statistical information is in Source Data Statistics.
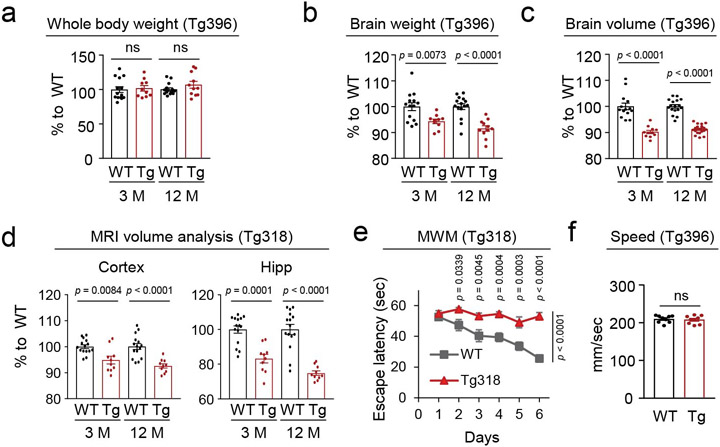
Next, we examined whether N40K-Tg displays neurodegeneration in the brain. Compared with WT littermates, the Tg mice show no visible change in whole body weight (Extended Data Fig. 4a) but display a small decrease (<10%) in brain weight and volume in 3- and 12-month-old mice (Extended Data Fig. 4b, 4c). Magnetic resonance imaging (MRI) shows a comparable decrease of cortical and hippocampal volumes in N40K-Tg (Fig. 4a and Extended Data Fig. 4d, both Tg lines). Counting of neuronal marker NeuN-stained cells indicates a significant decrease (~30%) in neuronal density within cerebral cortex and hippocampus (Fig. 4b). These experiments support age-dependent neuronal loss in the N40-Tg mice.
Figure 4. N40K-Tg mice display neuron loss and cognitive impairment.
Tg396 was used in these studies. a, Volume change of mouse whole brain, cortex and hippocampus (Hipp) measured by MRI at different ages (1-month-old: WT n = 11, Tg n = 8; 3-month-old: WT n = 14, Tg n = 11; 12-month-old: WT n = 17, Tg n = 20; two-way ANOVA and Sidak's multiple comparison test). b, Quantification of NeuN+ neurons in 12-month-old mouse cortex, CA1 and DG of hippocampus (WT n = 3-4, Tg n = 3-4, Student's t-test, two-tailed). c, Novel object recognition test (3-month-old: WT n = 14, Tg n = 11; 12-month-old: WT n = 17, Tg n = 10, Student's t-test, two-tailed). d, Percent of correct choice in Y-maze (3-month-old: WT n = 12, Tg n = 12; 12-month-old: WT n = 15, Tg n =12, Student’s t-test, two-tailed). e, Morris water maze task (12-month-old: WT n = 18, Tg n = 14, two-way ANOVA and Sidak's multiple comparison test). f, Morris water maze probe test. At day 6, the hidden platform was removed to measure distance traveled in the target quadrant (12-month-old: WT n = 14, Tg n = 12, two-way ANOVA and Sidak's multiple comparison test). g, Visual test of 12-month-old WT and N40K-Tg mice (12-month-old: WT n = 9, Tg n =9, Student's t-test, two-tailed). h, Open field test. Left: total traveling distance. Right: center/total ratio (12-month-old: WT n = 11, Tg n = 12, Student's t-test, two-tailed). Data are shown as mean ± SEM. Full statistical information is in Source Data Statistics.
We then asked whether N40K expression perturbs brain cognitive function. Compared with WT, N40K-Tg mice (12-month-old) display a decline in novel object recognition (Fig. 4c), and significant defects of working memory in Y-maze test (Fig. 4d) and spatial memory in Morris water maze (Fig. 4e, 4f, and Extended Data Fig. 4e, both Tg lines). The deficit appears to be limited to memory tasks, as the Tg animals display normal performance in swimming speed in Morris water maze (Extended Data Fig. 4f), visual acuity test (Fig. 4g) and locomotor activity in open field test (Fig. 4h). These results demonstrate impairments of recognition and spatial memory in the aged N40K-Tg mice.
GABAergic synapses mediate hyperexcitability in N40K-Tg
To investigate how N40K expression contributes to cognitive impairment, we performed RNA-seq analysis of WT and N40K-Tg mouse brain at 3 and 12 months of age (n = 15,705 transcripts, Supplementary Tables 3, 9). Compared with WT, N40K-Tg mice display an increase in intron reads (Fig. 5a and Supplementary Table 3). While 416 transcripts show statistically significant splicing changes in the 3-month-old Tg mice, the number increases to 4, 428 in the 12-month-old Tg mice (Supplementary Table 9, FDR < 0.05), illustrated by a shift of the distribution of splicing deficiency scores (Fig. 5a). We also re-analyzed RNA transcripts (n = 18,957) in human brain tissues previously reported23 (ROS/MAP cohort, filtered by RNA integrity number (RIN) of at least 8, accepting 18 control and 35 AD cases), and found higher intron reads and splicing deficiency of 2,673 transcripts in AD (Fig. 5b, Supplementary Tables 3, 10, FDR < 0.05). Cross-species comparison reveals 984 overlapped transcripts (Fig. 5c and Supplementary Table 11), significantly enriched in multiple pathways, such as glutamatergic, GABAergic, cholinergic, dopaminergic, and serotonergic synapses, long-term potential (LTP), and autophagy (Fig. 5d, Supplementary Table 12, FDR < 0.05). The transcripts/proteins were further analyzed by protein-protein interaction (PPI) network21, also revealing PPI modules enriched in glutamatergic synapse, GABAergic synapse, LTP, and autophagy (Fig. 5e, Supplementary Table 13, FDR < 0.05). Strikingly, these PPI modules contain 15 transcripts/proteins identified in the inhibitory postsynaptic density (iPSD), including GABRA2, GABRA3, GABRA5, GABRB2, and GABRB3 (Fig. 5e)40. It should be aware of that these iPSD proteins can locate on the surface of excitatory neurons, which is expected from the cell type specific expression of N40K under the CaMKIIα promotor.
Figure 5. N40K-Tg mice exhibit AD-related splicing defects enriched in synaptic function.
a, Age-dependent increase of mapped intronic reads and histogram of splicing deficiency score of individual transcripts in Tg mice (Tg396, 3-month-old: WT n = 3, and Tg n = 3, 12-month-old: WT n = 3, and Tg n = 3 biologically independent mouse samples). We quantified the percentage of intron reads (Student's t-test, two-tailed) and transcript splicing deficiency (Kolmogorov–Smirnov test). b, Increase of mapped intron reads and histograms of splicing deficiency scores of individual transcripts in ROS/MAP human AD cases23 (Ctrl n = 18, AD n = 35, left: Student's t-test, two-tailed; right: Kolmogorov–Smirnov test). Each point represents data of individual human cases c, Venn diagram of mouse and human splicing-defective transcripts. d, Pathway enrichment analysis of shared splicing-defective transcripts in the Tg and AD, showing selected pathways from Supplementary Table 12. FDR was derived from p values (Fisher's exact test) by the BH procedure. e, Four examples of enriched PPI modules (Supplementary Table 13), with the proteins in iPSD highlighted in red. Each dot represents a protein, whereas the interaction is indicated by connected lines. f, Validation of intron retention in selected transcripts by the density of RNA reads (ex: exon; in: intron) and RT-PCR. Red boxes show genomic regions selected for quantifying RNA-seq reads (Student's t-test, two-tailed). Scale bar, 10 kb. g, qRT-PCR analysis of Gabra2 transcripts (Student's t-test). Data are shown as mean ± SEM. in a, f, g. each point represents a data point of one mouse (WT n = 3, and Tg n = 3). Full statistical information is in Source Data Statistics.
To verify the splicing deficiency of individual transcripts in N40K-Tg mice, we used the RNA-seq data to define intron retention events and analyzed specific intron retention by RT-PCR. We focused on 4 transcripts involved in synaptic regulation, including Gabra2 (introns 4 and 6), Gng7 (intron 2, a G protein subunit that modulate neurotransmission41,42), Kcnh1 (intron 9, potassium channel involved in epilepsy43) and Camk1d (intron 5, the neurosignaling kinase linked to AD44) (Fig. 5f, Extended Data Fig. 5a, 5b). In addition, splicing deficiency may lead to downregulation of mRNA level, as exemplified by Gabra2 through quantitative RT-PCR (Fig. 5g). Thus, our data support that the N40K-Tg mice display splicing defects throughout the transcriptome, especially in the transcripts associated with synaptic function, consistent with splicing defects identified in human AD cases19,23.
Considering that a large number of iPSD transcripts exhibit splicing deficiency in N40K-Tg and in human AD cases, we next analyzed the level of GABRA2 protein in the Tg mice and human AD brains, as well as the relevant synaptic activity. The GABRA2 protein level dramatically decreases in the Tg, as early as in 3-month-old mice (Fig. 6a). The GABRA2 protein also decreases in human AD brain tissues (Fig. 6b), reminiscent of a previous report of reduced GABAA receptor inhibitory function in AD45. As GABRA2 reduction is likely to de-repress synaptic activity, we performed electrophysiological experiments in the perforant pathway (Fig. 6c) which plays a central role in learning and is altered early and severely by neuropathology in AD46,47. Indeed, N40K-Tg mice show significant impairments in long-term potentiation in the perforant pathway (Fig. 6d). We then examined the presynaptic and postsynaptic activities. Although the frequency of presynaptic neurotransmitter release was normal (Fig. 6e), excessive postsynaptic strength was observed in response to stimuli, and this postsynaptic hyperexcitability was largely rescued by the administration of the GABAA receptor agonist muscimol (Fig. 6f, 6g). Thus, our results strongly support that N40K-induced splicing defects lead to downregulation of GABAergic synapse components and neuronal hyperexcitability, contributing to the deterioration of memory and cognition in mice.
Figure 6. N40K-Tg mice show GABRA2 reduction, post-synaptic hyperexcitation and LTP impairment.
a, GABRA2 protein reduction in N40K-Tg by western blotting (n = 5 mice in each group, Student’s t-test). b, GABRA2 protein in AD cases by western blotting (Ctrl n = 8, and AD n = 8, Student’s t-test). c, Synaptic activity assay in perforant pathway, stimulating from entorhinal cortex and recording on dentate gyrus (DG). d, LTP was shown as field excitatory postsynaptic potential (fEPSP) slope versus recording time, showing impairment in Tg396 mice. Representative traces before and after Theta-burst stimulation (TBS) were shown (WT animals n = 5, slices n = 7; Tg animals n = 5, Tg slices n = 11). The data of the last 20 min were used for statistical analysis (two-way ANOVA). e, Paired-pulse ratios that quantify presynaptic function, showing no significant difference between WT and Tg mice. two-way ANOVA). f-g, The plot of fEPSP slope against stimulus intensity shows a larger excitability in Tg compared with WT, which were alleviated upon the treatment with 2.5 μM muscimol (an agonist of GABAA receptor). Data are shown as mean ± SEM. Statistical significance was analyzed by two-way ANOVA: all four conditions together (vertical line), or any of the two conditions. (WT animals n = 5, slices n = 7; Tg animals n = 5, Tg slices n = 11, WT + muscimol animals n = 5, slices n = 5; Tg + muscimol animals n = 5, slices n = 5 in d, e, f, g). Full statistical information is in Source Data Statistics.
Splicing dysfunction synergizes with the amyloid cascade
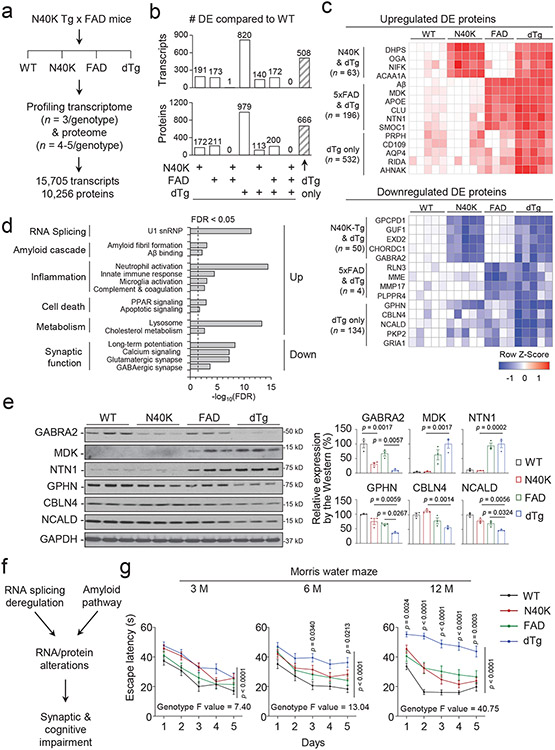
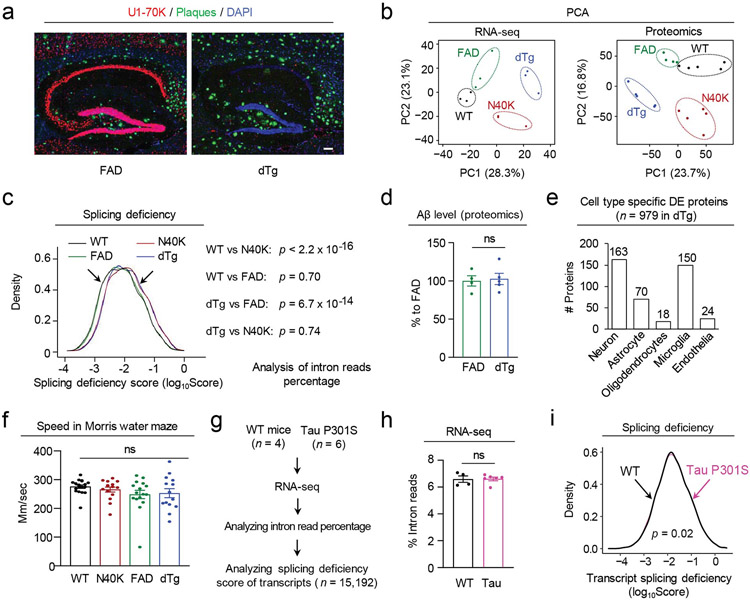
One central question is whether splicing dysfunction interacts with the amyloid cascade in AD pathogenesis, as the U1 snRNP and plaque pathologies are correlated but distinct in the brain of AD patients19,24. To address this question, we crossed N40K-Tg with 5xFAD, an AD model of amyloidosis, to generate double transgenic mouse model (dTg) that presents both pathologies (Extended Data Fig. 6a). To dissect the interaction between splicing dysfunction and amyloid cascade in molecular level, we profiled 15,705 transcripts and 10,256 proteins in the four genotypes (WT, N40K-Tg, 5xFAD and dTg, 6-month-old, Fig. 7a and Extended Data Fig. 6b). Compared with WT by splicing deficiency scores, dTg and N40K-Tg show the same severity of splicing defect, while 5xFAD has no detectable splicing defect (Extended Data Fig. 6c, and Supplementary Table 14). Conversely, the N40K-Tg in the 5xFAD background can induce U1-70K loss but does not significantly affect the Aβ level (Extended Data Fig. 6d).
Figure 7. Synergistic effects of human N40K and amyloid pathway on synaptic deregulation and cognitive deficiency.
a, N40K-Tg mice bred with 5xFAD mice to obtain WT, N40K (N40K-Tg, Tg396), FAD (5xFAD), and double transgenic mice (dTg) to profile transcriptome (3 mice/genotype, 6-month-old) and proteome (5 mice/genotype except 4 mice for FAD, 6-month-old). To simplify data processing, we only analyzed the major transcripts without considering alternative splicing forms. b, Numbers of transcripts and proteins differentially expressed (DE, FDR < 0.05) in N40K, FAD or dTg mice alone and in any combinations of these three genotypes. c, Heatmaps of selected DE proteins to show synergistic effects. d, Pathway enrichment of the DE proteins in dTg (selected from Supplementary Table 17). FDR was derived from p values (Fisher's exact test) by the BH procedure. e, Expression levels of GABRA2, MDK, NTN1, GPHN, CBLN4, and NCALD in 6-month-old WT, N40K, FAD, and dTg mice (3 mice/genotype) were analyzed by western blotting and quantified. Data are shown as mean ± SEM. Statistical significance was analyzed by one-way ANOVA followed by Tukey's multiple comparison test. The ANOVA overall p value is significant for each protein. dTg vs FAD or dTg vs N40K comparisons: f, The diagram of synergistic effect of splicing dysfunction and amyloid pathway. g, Morris water maze test of 4-genotyped mice at three different ages (3-month-old: WT n = 18, N40K n = 19, FAD n = 19, dTg n = 19; 6-month-old: WT n = 18, N40K n = 19, FAD n = 19, dTg n = 18; 12-month-old: WT n = 18, N40K n = 16, FAD n = 15, dTg n = 14). Statistical significance at each age was analyzed by two-way ANOVA. The effect of genotype in all three ages is significant, and the F value of the genotype increases with ages, consistent with larger difference at the older ages. The comparison of FAD and dTg at each training day using Tukey's multiple comparison test, the difference was not significant at 3-month-old, but became significant at 6-month-old (3 and 5 days) and 12-month-old (all five individual days). Full statistical information is in Source Data Statistics.
We further analyzed differentially expressed (DE) transcripts/proteins in three Tg genotypes versus WT. In N40K-Tg, 5xFAD and dTg mice, 191 transcripts/172 proteins, 173 transcripts/211 proteins, and 820 transcripts/979 proteins are significantly changed, respectively (Fig. 7b, Supplementary Tables 15, 16, FDR < 0.05). As expected, N40K-Tg and 5xFAD mice share only a few DE components, but both genotypes share their majority of DE components with dTg. Strikingly, the dTg mice displays more than 50% unique DE components (508 transcripts/666 proteins, Fig. 7b) that are not present in either N40K-Tg or 5xFAD. This synergy is shown in both upregulated and downregulated proteins (Fig. 7c). The DE proteins in dTg mice are enriched in diverse pathways (Fig. 7d, Supplementary Table 17, FDR < 0.05); for example, the downregulated proteins are highly enriched in synaptic function. The dTg DE list also contains a large number of cell type specific proteins, including 163 in neurons, 70 in astrocytes, 18 in oligodendrocytes, 150 in microglia, and 24 in endothelia, implicating the contribution of different cell types (Extended Data Fig. 6e, Supplementary Table 18). We validated some DE proteins in the mouse brain by western blotting, including GABRA2, MDK/midkine and NTN1/netrin 1 (two Aβ-corrected proteins in human AD brain21), GPHN/gephyrin (a scaffold at inhibitory synapses to cluster glycine and GABA receptors48), CBLN4/cerebellin 4 (an inhibitory transmission regulator reducing Aβ toxicity49), and NCALD/neurocalcin-delta (a neuronal calcium sensor related to SMA50) (Fig. 7e). Additionally, we performed deep RNA-seq analysis of a tau mouse model (P301S)51, and did not find difference in the percentage of total intron reads, but a slight defect in splicing deficiency, compared with wild type littermates (Extended Data Fig. 6g-I, Supplementary Table 19). We did not cross N40K-Tg and P301S mice in this study. Overall, the transcriptomic and proteomic data raise a new concept that the splicing and amyloid pathways can synergize to amplify molecular alterations in the dTg mouse brain (Fig. 7f).
In addition, we compared the DE proteins in the three mouse models (N40K-Tg, 5xFAD and dTg) with the human AD proteome, motivated by recent transcriptomic comparison between AD human brains and mouse models52. A meta-analysis of seven deep AD proteomic datasets revealed 2,698 DE proteins in the brain18, in which 2,381 proteins have orthologues in mice. When overlapping the 2,381-protein list with the 172 (N40K-Tg), 211 (5xFAD) and 979 (dTg) DE proteins, 15, 80 and 296 proteins displayed consistent changes, respectively (Extended Data Fig. 7a, 7b). These overlapped proteins are enriched in numerous pathways and protein-protein interaction modules, such as RNA splicing, amyloid cascade, cytoskeleton and extracellular matrix, endocytosis and exocytosis, growth and development, inflammation, lipid metabolism, protein degradation and synaptic function (Extended Data Fig. 7c, 7d, Supplementary Tables 20, 21). Importantly, during this mouse-human comparison, the synergistic effect in the dTg is observed in almost all pathways listed above, and the enriched pathways in the dTg are consistent with those identified in AD proteomic studies21,53.
To investigate whether splicing dysfunction can synergize with the amyloid cascade in cognition impairment, we performed Morris water maze with littermate animals of the four genotypes (WT, N40K-Tg, 5xFAD, and dTg) at three different ages (3-, 6- and 12-month-old) (Fig. 7g). Compared with N40K-Tg or 5xFAD, dTg mice show a trend of incapability at 3-month-old, statistically significant memory impairment at 6-month-old, and much more severe impairment at 12-month-old, although swimming speed in the test is similar for all mice (Extended Data Fig. 6f). These age-dependent results suggest that the synergistic interaction of the splicing and amyloid pathways accelerates the development of cognitive impairment.
In addition, as TDP-43 inclusions display in up to half of AD cases12, we examined the TDP-43 pathology (stage 0 to 3) in the ROS/MAP cases23 (Supplementary Tables 3, RIN > 8, n = 53), and found a weak correlation between TDP-43 stages and intron read percentage in the RNA-seq analysis (Extended Data Fig. 8a, r = 0.12). Although direct intron read comparison between different TDP-43 stages did not yield results of statistical significance (Extended Data Fig. 8b), the splicing deficiency scores of individual transcripts revealed expected splicing alteration in the cases with high TDP-43 pathology (Extended Data Fig. 8c), implicating a role of TDP-43 in splicing dysfunction, which might co-exist with the impact of U1 snRNP pathology in some AD cases. We further explored if TDP-43 proteinopathy is induced in the mouse models of N40K-Tg, 5xFAD and tau P301S mice, but found negative results in these models (Extended Data Fig. 8d).
Discussion
A mouse model to recapitulate RNA splicing dysfunction in AD
U1 snRNP proteinopathy has recently been identified by proteomics and validated by immunohistochemistry in AD patient brain19,24,27-29, together with widespread RNA splicing dysfunction detected by transcriptomics19,23, but its contribution to AD progression was not well studied due to the challenge of recapitulating mRNA splicing dysfunction in AD mouse models (e.g. the 5xFAD amyloidosis model). Since U1 snRNP is central in the function of RNA splicing, U1 snRNP genes are essential for viability of human cell lines in a genome-wide CRISPR screen54, and knocking out U1-70K gene in Drosophila also results in lethality55. During our study of N40K toxicity, unexpectedly, we found a potent dominant-negative effect of N40K to reduce the U1-70K protein level in neurons. We then utilized this effect to impair U1 snRNP-mediated splicing function in a Tg mouse model. This model provides a general approach to perturb RNA splicing mechanisms in the mouse brain.
In N40K-Tg mice, neuron-specific expression of N40K is driven by the CaMKIIα promotor39, and N40K impacts neuronal function by potential loss- and gain-of-function mechanisms. Clearly, N40K expression causes a loss-of-function effect by depleting full length U1-70K protein. The depletion is dependent on the expression level of N40K, which is approximately 2-fold higher than that of native U1-70K in the WT mice. As a result, the majority of U1-70K in the cortex and hippocampus is depleted to perturb the normal process of RNA splicing. Moreover, N40K aggregation might exhibit a role of gain-of-function. The disordered low complexity (LC) domains in N40K are liable to protein aggregation33,34, possibly through a process of liquid-liquid phase separation4,56. Although we did not find obvious tangle formation in N40K-Tg, we detected evident accumulation of N40K in detergent-insoluble fraction of mouse brain by western blotting and by large-scale mass spectrometry. In addition, other RNA-binding components (e.g. UT14A, and RRP1) are also sequestrated in the detergent-insoluble fraction, which may restrict their physiological functions in neurons, similar to the toxic roles of many known misfolded proteins in neurodegeneration57. In addition, protein aggregation might lead to the activation of unfolded protein response and ER stress58,59. We then examined the known genes/proteins activated by the ER stress in the transcriptomic and proteomic analyses of WT and N40K-Tg mice. Although we detected the expression of six genes/proteins in three ER stress pathways (i.e., XBP1/JNK, PERK/EIF2A/ATF4, ATF6), none were significantly changed in N40K-Tg, suggesting that the ER stress may not contribute to the defects observed in N40K-Tg, in accordance with the marginal overexpression of N40K. Based on our collected data, although we cannot completely rule out the role of other pleiotropic effects of genetic perturbation, RNA splicing dysfunction is believed to be the major event detected in the N40K-Tg mice.
How can the N40K-Tg mice survive with the loss of the majority of U1-70K in neurons? We believe that the truncated N40K protein replaces U1-70K in spliceosome and can partially function in mRNA splicing. Structural studies of human U1 snRNP complex reveal that the N-terminus and RRM domain in N40K are indispensable for U1 snRNP assembly35, supporting that N40K has ability to stabilize the complex of U1 snRNP. A genetic experiment in Drosophila strongly supports this partially functional hypothesis of N40K, in which expressing fly N40K homologous portion rescues the embryonic lethality of the U1-70K knockout fly55. However, without the C-terminus of U1-70K, the function of the N40K-assemblied U1snRNP complex is compromised, demonstrated by impaired splicing efficiency in the cellular and mouse models.
More excitingly, the N40K-Tg model mimics U1 snRNP proteinopathy, RNA splicing defects and neuronal loss observed in AD brain. The N40K-Tg results are highly similar in two independent mouse lines (Tg396 and Tg318) that have different gene integration sites in mouse chromosomes. Remarkably, splicing dysfunction in N40K-Tg mice alters a variety of synaptic components, which may contribute to the imbalance between excitatory and inhibitory pathways60. In particular, GABRA2 decreases significantly in the N40K-Tg, which is also confirmed in examined human AD cases by western blotting and is supported by recent meta-analysis of AD proteomics18, and single-cell RNA profiling of human AD samples61. The downregulation of GABAergic pathway proteins is in agreement with synaptic hyperexcitability and LTP impairment, demonstrated by our electrophysiological study. The synaptic hyperexcitability may also be attributed to neuronal loss observed in the N40K-Tg mouse brain.
Interaction of U1 snRNP, amyloid, tau and TDP-43 pathologies
We provide clear evidence that U1 snRNP dysfunction synergizes with the amyloid pathway to accelerate cognitive impairment, although Aβ and U1 snRNP pathologies appear to be independent in mice. It should be noted that the human mutated APP was expressed as a recombinant protein in 5xFAD mice, bypassing the processing of RNA splicing. Therefore, the 5xFAD model is not suitable for studying the direct impact of splicing dysfunction on human APP transcripts. Nevertheless, our double transgenic mice offer a model to study the synergistic effect downstream of Aβ and U1 snRNP pathologies. Through temporal spatial memory test by water maze, we determined the synergetic effect in cognitive decline as early as in 6-month-old mice. Moreover, proteomics profiling of the same age mice identified aggravated alterations of numerous synaptic components (e.g., Gabra2 and gephyrin) in double Tg mice, compared with N40K-Tg or 5xFAD mice. The results suggest the synergetic effect can occur to reduce inhibitory neurotransmission, which orchestrates neuronal networks underlying the cognitive process48, although the synergy is also observed in other pathways. The result is consistent with the known function of Aβ-induced synaptic hyperexcitability60, leading to cognitive impairment by abnormal patterns of neuronal activity62. Together, Aβ may exacerbate synaptic hyperexcitability together with dysfunctional U1 snRNP, providing a possible explanation to the synergistic effect.
Unlike amyloid plaques, tau tangles appears to be partially overlapped with aggregated U1-70K by immunohistochemistry in human AD brain19, and U1 snRNP complex and/or other RNA splicing proteins (e.g. TIA1 and SRRM2) were detected to interact with tau in AD brain tissues and/or tau mouse models63-65. The U1 snRNP pathology, however, has not been identified in the human cases of non-AD tauopathies, such as corticobasal degeneration and frontotemporal lobar degeneration (FTLD)-tau19, implicating that tau pathology alone cannot induce U1-70K aggregation. Furthermore, our RNA-seq comparison of a tau mouse model (P301S) and wild type did not show statistically significant difference in the percentage of total intron reads, although a minor change of splicing deficiency was observed. Therefore, the N40K-Tg model is the only mouse model currently recapitulating widespread splicing defects in AD. Nevertheless, it is still highly possible that U1 snRNP complex may co-aggregate with tau in neurodegenerative cases, reminiscent of enhanced fibrillization of tau and alpha-synuclein in the inclusion formation66.
In addition, TDP-43 inclusions have been reported in a significant portion (up to ~50%) of AD cases12, and are emphasized by a recently proposed disease entity of limbic-predominant age-related TDP-43 encephalopathy (LATE)67. Our correlation analysis of RNA splicing and TDP-43 pathology suggest that TDP-43 and U1 snRNP pathologies may occur simultaneously to affect RNA splicing in some AD cases, but the TDP-43 proteinopathy cannot be induced in N40K-Tg, 5xFAD and tau P301S mouse models. The data reiterate the limitation of the mouse models, none of which recapitulates the full spectrum of complex pathologies in human AD brains. Each mouse model, however, allows the detailed investigation of selected mechanism, and different models (e.g., N40K-Tg and 5xFAD) may be crossed to dissect the interaction of multiple pathways.
The N40K-Tg model has some limitations. First, the expression of human N40K in the mice is controlled under a short CaMKIIα promotor39 due to the package capacity of recombinant Lentivirus. It is possible that N40K-induced splicing defects might perturb brain development. For instance, downregulation of GABRA2 protein is detected in young N40K-Tg mice (i.e., 3-month-old). This caveat may be alleviated by N40K inducible mouse models in the future. Moreover, N40K expression in neurons results in the depletion of full length U1-70K (loss-of-function) and the co-aggregation of N40K and other proteins (gain-of-function), increasing the complexity for data interpretation. Finally, some of the splicing-related molecular changes in N40K-Tg mice are not expected to occur in human cases, because of the species difference in genomic intron-exon architecture. Future development of human iPSC models would partially address this concern.
In summary, we thoroughly studied the toxic mechanism of AD-associated N40K and splicing defects in primary cultured neurons and mouse models. Our findings lead to critical mechanistic insights into understanding how RNA splicing dysfunction causes neural hyperexcitation, cognitive abnormalities, and neurodegeneration in these disease models. Our studies strongly suggest that U1 snRNP proteinopathy and splicing disruption may play an unexpected role in AD pathogenesis, implicating a novel pathway for therapeutic targeting.
Methods
All experiments conducted in this study comply with all relevant ethical regulations. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at St Jude Children’s Research Hospital. Human postmortem brain tissue samples (frontal gyrus) were provided by the Brain and Body Donation Program at Banner Sun Health Research Institute, in which the studies were approved and written consent for sample collection was obtained. Clinical and pathological diagnoses were based on the established criteria68.
Statistics and reproducibility.
No statistical methods were used to predetermine sample sizes, but the sample sizes were similar to those reported in previous publications69-71. Data distribution was assumed to be normal, but this was not formally tested. Statistical methods were applied to estimate p values followed by the analysis of false discovery rate. No animals were excluded from the study. The experiments were not randomized, in which animals were used when available. The investigators were blinded to the genotype information during data acquisition for experiments of MRI, stereology, electrophysiology and behavior tests. In small-scale analyses, two-tailed unpaired Student’s t-test was used for two-sample comparisons, while one-way or two-way ANOVA (Graphpad Prism 8.0.2) was used for group comparisons followed by Tukey's or Sidak’s post hoc correction. The Kolmogorov-Smirnov test was used to analyze statistical significance of splicing deficiency of the comparison between mouse models and human cases.
Primary mouse neuronal culture.
Mouse cortical neurons were isolated from mouse embryos (C57BL/6, Charles River) at E18, and cultured in completed Neurobasal A (Life Technologies, Cat# 10888022) supplemented with B27 complex (Thermo Fisher, Cat# 17504044) and GlutaMAX (Thermo Fisher, Cat# 35050-061). Half of the medium was replaced by new medium every two days. Lentiviral infection was performed at DIV 7-12. For time-dependent assay: neurons were infected at MOI 10 for 2, 4, and 6 days. For inhibition of protein degradation, after neuronal infection with N40K lentivirus (MOI 10) for 2 days, MG132 or Bafilomycin A1 were added for 1-2 days, followed by harvest and immunoblotting.
Mouse model.
All mice (C57BL/6J) were housed under a 12 hour: 12 hour (light: dark) cycle at 22-25 °C and 40% - 60% humidity. N40K-Tg mouse lines were generated in St Jude Children’s Research Hospital by lentiviral transgenic method as described72. N40K lentiviral vector (FCK1.3GW-N40K, 1x108 titer) was microinjected into perivitelline space of fertilized eggs from C57BL/6J. Genetic background was confirmed by C57BL/6 substrain characterization panel (The Jackson Laboratory). The founder line (Tg396) and another line (Tg318) showed high N40K expression and was backcrossed to C57BL/6J for at least two generations. Hemizygous Tg mice and non-transgenic wild type littermates were used in experiments. 5xFAD mice were purchased from The Jackson Laboratory (MMRRC_034848-JAX). We crossed N40K-Tg with 5xFAD, to generate double transgenic mouse model (dTg). In all mouse experiments, similar numbers of male and female animals were used.
Lentiviral vectors.
Human U1-70K or N40K coding sequences were cloned into BamHI/EcoRI cites of FCK(1.3)GW vectors (Addgene, plasmid #27230), driven by a modified mouse CaMKIIα promoter which shows strong expression in pyramidal neurons39.
Cell culture and immunoprecipitation (IP).
HEK 293T cells (ATCC, CRL-3216) were cultured in DMEM media (Thermo Fisher Scientific, Cat# 11995-065) supplemented with 10% FBS (Thermo Fisher Scientific, Cat# 16140-071). Cells were transfected with plasmids of pCDNA3.1, pCNDA3.1-U1-70K-FLAG, or pCDNA3.1-N40K-FLAG using X-tremeGENE 9 DNA Transfection Reagent (Millipore Sigma, Cat# XTG9-RO), following the manual with 1:3 ratio (DNA/transfection reagent). After two days, transfected cells were harvested and extracted with the lysis buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP40, 1 mM EDTA, 1 mM EGTA, and 1x protease inhibitor cocktail (Roche, Cat# 11836170001). IP was performed with anti-FLAG M2 magnetic beads (Millipore Sigma, Cat# M8823) following the manufacturer’s protocol.
In the IP experiment from mouse brain, the cortex of 6-month-old WT or N40K-Tg mice was homogenized in the hypotonic buffer (10 mM HEPES, pH 8.0, 0.2 mM PMSF, 0.5 mM DTT, phosphatase inhibitor, protease inhibitor cocktail and RNAse inhibitor) at 5:1 ratio (w/w) in Dounce homogenizer, followed by the addition of other components to adjust the final concentrations to 10 mM HEPES, pH 8.0, 1.5 mM MgCl2, 600 mM NaCl, and 0.5% Triton X-100. The homogenates were further lysed by glass bead beating in Bullet Blender at 4 C for 5 min at speed 6. The lysates were centrifuged at 21,000 x g for 15 min to remove debris. The supernatants were dialyzed against the low salt buffer containing 10 mM HEPES, pH 8.0, 1.5 mM MgCl2, 150 mM NaCl and 0.5% Triton X-100. IP was performed with rabbit anti-U1C Ab (Santa Cruz, Cat# sc-374428) coupled to Dynabeads (Thermo Fisher Scientific, Cat# 10003D) overnight at 4 C. Then the beads were washed three times with the cold low salt buffer and spliceosome complexes were eluted by the sample loading buffer for SDS-PAGE and western blotting.
Neuron survival assay.
Neuron survival/death was determined by Cell-Titer Glo assay (Promega, Cat# G7572) for viability. The cells were equilibrated to 21 °C for 20 min, and then an equal volume of Cell-Titer Glo was added to each well. After incubation for 30 min, luminescence was detected with an Envision 2102 Multilabel Plate Reader (PerkinElmer Life Sciences). For dose-dependent toxicity assay of N40K, mouse cortical neurons at DIV 7 were infected with lentiviral particles at MOI 7, 20, 60 for 5 days. For pharmacological inhibition assay, neuron infected with lentivirus at MOI 20 for 2 days, and then inhibitor compounds were added for 3 more days.
Detergent extraction.
The analysis was adapted from a previously published protocol19. Human brains or primary neurons were extracted with 1% sarkosyl (N-lauroyl-sarcosine) in Lysis Buffer (10 μM Tris, pH 7.5, 5 mM EDTA, 1 mM DTT, 10% sucrose, and protease inhibitor cocktail). Primary neurons (DIV 12) were infected with lentiviral vector expressing EGFP, U1-70K-FLAG, and N40K-FLAG by CaMKIIα promoter (MOI 10) for 4 days. Mouse hippocampi were extracted with RIPA buffer (10 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 10% glycerol, and protease inhibitor cocktail, 10 μl buffer per mg tissue). The insoluble pellet was resuspended in 8 M Urea with 2% SDS for Western blotting or proteome profiling.
Mass spectrometry-based proteomics.
We used an optimized protocol of TMT-LC/LC-MS/MS for deep proteome profiling31,32. Protein samples were lysed by homogenization in the lysis buffer (50 mM HEPES, pH 8.5, 8 M urea, and 0.5% sodium deoxycholate), and their concentrations were measured by the BCA assay (Thermo Fisher, Cat# 23227) and confirmed by Coomassie-stained short SDS gels 73. Quantified protein samples (~0.1 mg per TMT channel) were digested with Lys-C (Wako, distributor 121-05063, 1:100 w/w) for 2 h at 21 °C, followed by dilution to decrease urea to 2 M and trypsin digestion (Promega, Cat# V5113, 1:50 w/w) overnight at 21 °C. Cys residues were reduced and alkylated by iodoacetamide. The proteolysis was terminated by adding trifluoroacetic acid to 1%. The resulting peptides were desalted with the Sep-Pak C18 cartridges (Waters), TMT-labeled (Thermo Fisher, Cat# A34808 and A44520), and pooled equally. The pooled peptides were resolved by basic pH reverse phase LC on an XBridge C18 column (3.5 μm beads, 4.6 mm x 25 cm, Waters; buffer A: 10 mM ammonium formate, pH 8.0; buffer B: 95% acetonitrile, 10 mM ammonium formate, pH 8.0, ~2 h gradient, 40-80 concatenated fractions collected)74. Each fraction was analyzed by acidic pH LC-MS/MS (75 μm x ~20 cm, 1.9 μm C18 resin from Dr. Maisch GmbH, buffer A: 0.2% formic acid, 5% DMSO; buffer B: buffer A plus 65% acetonitrile, ~1.5 h gradient). The settings of Q Exactive HF Orbitrap MS (Thermo Fisher) included the MS1 scan (~410-1600 m/z, 60,000 resolution, 1 x 106 AGC and 50 ms maximal ion time) and 20 data-dependent MS2 scans (fixed first mass of 120 m/z, 60,000 resolution, 1 x 105 AGC, ~110 ms maximal ion time, HCD, 32-35% normalized collision energy, ~1.0 m/z isolation window with 0.3 m/z offset, and ~15 s dynamic exclusion)32.
The raw MS data were searched against protein database by the JUMP software (v1.13.4)75, which utilizes both pattern matching and de novo tag scoring to improve the sensitivity and specificity. A composite target/decoy database was used to evaluate FDR in peptide identification76,77. The protein target database combined downloaded Swiss-Prot, TrEMBL, and UCSC databases (human: 83,955 entries; mouse: 59,423 entries). Search parameters included precursor/product ion mass tolerance (± 10 ppm), full trypticity, static mass shift (TMT tags of 229.16293 and Cys carbamidomethylation of 57.02146 on cysteine), dynamic mass shift (Met oxidation of 15.99491), two maximal miscleavage sites, and three maximal modification sites. Peptide-spectrum matches (PSMs) were filtered by matching scores and mass accuracy to limit protein FDR below 1%.
Proteins were quantified from TMT reporter ions based on our published method78. Briefly, TMT reporter ion intensities were extracted for each PSM, corrected by isotopic distribution of TMT reagents, filtered to remove poor PSMs (e.g., minimum intensity of 1,000), and adjusted to eliminate sample pooling bias. The relative protein intensities were averaged from all assigned PSMs after removing outliers (e.g., Dixon’s Q-test or generalized extreme Studentized deviate test). Finally, the absolute protein intensities were derived by multiplying the relative intensities by the grand mean of top three abundant PSMs.
The analysis of differential expressed proteins in proteomic datasets essentially followed the limma R package (v3.48.3)79 in multiple steps: (i) obtain the protein quantification data from MS as described above; (ii) perform a log transformation of the data80; (iii) calculate the p values by moderated t-test, and FDR values by the Benjamini-Hochberg procedure, using limma R package; (iv) calculate the mean for each protein under different conditions and derive log2(fold change); and (v) fit log2(fold change) data of all proteins to Gaussian distribution to generate a “global” standard deviation value. Statistically significant changes were usually based on FDR cutoff (0.05) and log2(fold change) cutoff (two standard deviations).
RNA-seq analysis.
For primary neurons, mouse cortical neurons (DIV7) were infected with lentiviral vectors expressing EGFP or N40K driven by CaMKIIα promoter at MOI 20 for 3 days. Total RNA was extracted by RNAeasy Mini kit (Qiagen, Cat# 74104). For mouse hippocampus or cortex, total RNA was extracted by RNeasy Lipid Tissue Mini Kit (Qiagen, Cat# 74804) as described in the manual, including with DNAse I digestion to remove contaminated DNA. Total RNA was eluted with RNase-free water and stored at −80°C. RNA quality was examined by 2100 Bioanalyzer RNA 6000 Nano assay (Agilent, Cat# 5067-1511). RNA sequencing libraries were prepared with the TruSeq Stranded mRNA Library Prep Kit (Illumina, Cat# 20020594), including poly(A) RNA purification. Libraries were then quantified using the Quant-iT PicoGreen dsDNA assay (Thermo Fisher, Cat# P11496) and MiSeq Reagent Kit Nano v2 (Illumina, Cat# MS-102-2001). One hundred cycle paired-end sequencing (100 bp) was performed on an Illumina HiSeq 2500. Library preparation and RNA-seq were performed at the Genome Sequencing Core at the St Jude Children’s Research Hospital. An average of 130 million reads were obtained for each sample. For human cortex samples, we obtained the RNA-seq raw reads from the ROS/MAP study23. Individual cases were first filtered by RNA integrity (RIN > 8) and then categorized by the cognitive COGDX score and the neuritic plaque CERAD score, resulting in 18 control samples (COGDX = 1 and CERAD = 4) and 35 AD samples (COGDX ≥ 4 and CERAD of 1-3).
Reads from RNA-seq were mapped by our in-house mapping pipeline ‘Strongarm’ (v1.0)81. Reads were aligned to the following four database files using BWA (v0.5.5) aligner: (i) the (mouse mm9) human hg19 reference sequence; (ii) RefSeq; (iii) a sequence file representing all possible combinations of non-sequential pairs in RefSeq exons; and (iv) AceView database flat file downloaded from UCSC representing transcripts constructed from ESTs. The mapping results were aligned to the reference genome coordinates and the final BAM file was constructed by selecting the best alignment in the four databases. The quantification of mapped transcripts was estimated with htseq-count82.
To extract the reads mapped to whole genes, including exons and introns, we converted the BAM to BED files using bedtools (http://code.google.com/p/bedtools, bamToBed, v2.16.1), and used Table Browser program (http://genome.ucsc.edu) to generate three BED files: (i) Reference Sequence Genes (Whole Gene), (ii) Reference Sequence Genes (Exons) and (iii) Reference Sequence Genes (Introns). Finally, we used intersectBed (v2.16.1) to define the intersections between mapped reads and the three RefSeq BED files.
Differential gene expression in transcriptomic datasets was performed using moderated t-test in the limma R package and voom83 (inside limma v3.48.3) with the raw read counts. The p values were converted to FDR values (cutoff of 0.05) by the Benjamini-Hochberg procedure.
Splicing deficiency analysis.
The global splicing deficiency was first evaluated by the percentage of reads mapped to introns, after normalized to the total reads mapped to whole genes19. Moreover, we defined a splicing deficiency score for each gene as the ratio of length-corrected read counts aligned to introns and exons:
A high splicing deficiency score indicates low splicing efficiency. For comparative analysis, p values were calculated by moderated t-test in the limma R package and followed by the Benjamini-Hochberg procedure to derive FDR values (cutoff of 0.05).
RT-PCR for validating intron retention.
RT-PCR was performed by a two-step method. Total RNA from each sample was converted to cDNA using random primers [High-Capacity Reverse Transcription Kit (Thermo Fisher, Cat# 4374966). The products are then subjected to PCR analysis using primers (Supplementary Table 23). RT-PCR products were resolved by gel electrophoresis followed by SYBR Safe DNA Gel Stain. Gel bands were quantified by Image J. Intron retention was calculated by
Quantitated RT-PCR.
mRNA levels of mouse endogenous U1-70K and human N40K levels were quantitated with TaqMan assay (Mouse U1-70K Mm0120544, Human N40K Hs01091623, Thermo Fisher Scientific) Relative expression levels were calculated by ΔΔCT method with using 18S as the reference gene.
Pathway enrichment by KEGG and gene ontology databases.
Pathway enrichment analysis was carried out by the JUMPn software (v1.13.0)84 to identify the biological functions of dysregulated genes/proteins in a given dataset. The analysis was performed using Fisher’s exact test against the Gene Ontology (GO) biological process, molecular function, and cellular component annotations, and KEGG pathway database, separately. The homologous genes between human and mouse were used as the background. The p values derived from Fisher’s exact test were further adjusted into FDR using the Benjamini-Hochberg procedure for multiple testing. Enriched pathways with FDR values < 0.05 were considered statistically significant.
Protein-protein interaction (PPI) network analysis.
The analysis was performed based on our previously published protocol85 with modifications. DE genes/proteins were superimposed onto a composite PPI database by combining STRING (v11)86, BioPlex (v3.0)87, and InWeb_IM (v2016_09_12)88. The BioPlex database was developed by the method of affinity purification and mass spectrometry, whereas the STRING and InWeb contain information from various sources. Due to this heterogeneity of PPI interactions, the STRING and InWeb databases were further filtered by the edge score to ensure high quality, with the following rules: (i) only edges with evidence of physical interactions (e.g. through co-IP or yeast two-hybrid) were considered; (ii) edges of high confidence, as filtered by the edge score, with the cutoff determined by best fitting the log-log degree distribution using the scale free criteria89. The finally accepted STRING and InWeb databases were combined with BioPlex and the inhibitory postsynaptic density interactome40 to construct a composite PPI database, which includes 20,485 proteins and 1,152,607 PPI connections. PPI modules were then defined by a three-step procedure: (i) extracting a subnetwork by retaining PPI between two proteins if both were from the DE protein list; (ii) calculating a topologically overlapping measure (TOM)90 between each pair of proteins for the resulting PPI subnetwork, and (iii) dividing this network into individual modules based on the TOM clustering using the hybrid dynamic tree-cutting method91. The biological functions of each PPI module were further obtained using the proteins in each module as the input to perform the pathway enrichment analysis as described above.
Fluorescence in situ hybridization of mouse chromosomes.
Purified FCK(1.3)GW-U170K plasmid DNA was labeled with a red-dUTP (Alexa Fluor 594, Thermo Fisher, Cat# C11400) by nick translation. The labeled transgene probe was combined with sheared mouse cot DNA and hybridized to metaphase and interphase nuclei derived from the transgenic mouse lung fibroblast culture. Specific chromosome control probes labeled with green-dUTP (Alexa Fluor 488, Thermo Fisher, Cat# C11397) were used to confirm the localization. Chromosome 18 control probe (RP24-255J24) for Tg396 and chromosome 10 control probe (RP24-360A19/10A1)) for Tg318 were used. The chromosomes were stained with 4,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific, Cat# D21490) and analyzed.
mRNA in situ hybridization.
In situ hybridization for human N40K mRNA in formalin-fixed paraffin embedded (FFPE) tissues92 was completed on a Discovery Ultra automation system platform (Ventana Medical Systems) using RNAscope® VS Reagent Kit – RED (Advanced Cell Diagnostics). An oligoprobe set specific for N40K (not for U1-70K) (Advanced Cell Diagnostics, Cat# 760-234) was applied according to the manufacturer’s instruction. Hybridization signals were detected by chromogenic development with Fast Red, followed by counterstaining with hematoxylin. Each sample was quality controlled for RNA integrity with RNAscope oligoprobes for PPIB RNA, and for non-specific background staining with oligoprobes for bacterial dapB RNA. The specific RNA staining signal was identified as intracellular red punctate dots.
Immunohistochemistry.
The staining was performed essentially as previously reported21. Briefly, brain tissue samples were fixed with 10% formalin, and embedded in paraffin. 5-10 μm sections were deparaffinized, rehydrated and rinsed with water. Antigen retrieval was performed with 10 mM citric buffer (pH 6.0) with boiled water bath (20 min) and cool down to room temperature. Endogenous peroxidase activity is blocked by 1% H2O2 in PBS buffer (10 min). After rinsed by PBS, sections were then blocked with 5% donkey serum in PBS with 0.3% Triton X-100 (PBST) for 30 min at room temperature followed with primary antibodies diluted with PBST plus 2% BSA and 0.2% skim milk for overnight at 4°C. Resource and dilution of primary antibodies are listed in Supplementary Table 22. After washing with PBS, sections were then incubated with secondary antibodies (Jackson ImmunoResearch Laboratories) with 1:500 dilution, 1 hour at room temperature. For fluorescence staining, fluorescent dye-conjugated secondary antibodies were used with nuclear counter stain by DAPI (Thermo Fisher Scientific, Cat# D21490) or followed with amyloid plaque stain by 0.02% thioflavine S (Sigma-Aldrich, Cat# T1892) for 5 min. For chromogenic immunodetection, biotin-conjugated secondary antibodies were used followed with ABC reaction by Vectastain ABC kit (Vector Laboratories, Cat# PK-4000) and developed by 3,3-diaminobenzidine solution (Vector Laboratories, Cat# SK-4100). Images were captured by Zeiss LSM 780 confocal microscopy or Zeiss Axioscan.Z1.
Quantitation of brain cells by stereology.
Design-based stereology (thick section, 3-dimensional) was carried out in an investigator blinded manner. Brain tissue was cryopreserved followed by serially sagittal cryosectioning at 40 μm. Every 5th section was immunostained with anti-NeuN antibodies followed with chromogenic development of DAB. NeuN+ neurons were counted in the cortical regions using the optical fractionator probe (StereoInvestigator). Counts of NeuN+ cells were estimated using Microbrightfield Stereo Investigator (MBF Biosciences, Williston, VT) and the optical fractionator method93 using a Olympus BX-51 microscope and 40 X objective. The density of cortical NeuN+ neurons were calculated by total number of NeuN+ neurons divided by the estimated volume.
In vivo magnetic resonance imaging (MRI) and brain volume analysis.
Preclinical MRI studies were performed using a 7 T Bruker ClinScan system (Bruker BioSpin MRI GmbH) equipped with a 12 S gradient coil. A mouse head volume coil (Bruker BioSpin) was used for high resolution brain imaging. Animals were anesthetized and maintained with 1.5-2% isoflurane during MRI sessions. Transverse T2-weighted turbo spin echo images were acquired for volume measurements (TR/TE = 3840/50 ms, FOV = 25 x 25, matrix = 320 x 320, NEX = 1, Thickness = 0.4 mm, scan time = 11.5 min). Volume measurements (brain, hippocampus and cortex) were obtained by manually segmenting the regions and computing volumes using OsiriX (v5.7, Pixmeo, Switzerland).
Behavior studies.
Spatial working memory was conducted by “Y-maze” with delayed non-matching to position task94. Y-maze apparatus consisted of a start arm (47 x 10 x 10 cm) leading to two goal arms (34 x 10 x 10 cm). Each mouse was first familiarized individually, with running in the maze and presentation with a positive reinforcer of a drop of sweet milk. Habituation was performed on the first three days of testing, in which mice were released from the start location to freely investigate the maze, with the positive reinforcer placed in the stem and in the reward wells. After habituation and demonstration of consumption of milk reward, the experiment trials were initiated. Each trial consisted of a run from a start arm to a sample arm, a return to the start arm with 15 s interval, and subsequent free choice of goal arm. Each mouse choosing the non-sample arm was counted as correct response, and the mouse was rewarded. Sample arm was determined randomly, with an upper limit of three consecutive same sample arms. A total of 12 trials were given over a period of 3 days. Water restriction was performed through the study, in which mice were allowed 2 h of free access to water per day after testing.
General locomotor and exploratory activities were measured by “open field test”. The apparatus consisted of a clear square arena with blue plastic floor measuring 16 inches × 16 inches with walls 16 inches tall (San Diego Instruments). Testing was performed under white light condition. Mice were brought into the testing room and allowed to habituate for 1 h before the test. The activity of the mice was video recorded and scored using visual tracking software (CleverSys, TopScan Suite v3). Locomotor activity was determined by allowing the mice to freely investigate the testing arena for 15 min. The surfaces of the field were wiped between each animal with 70% alcohol to avoid olfactory cues.
Long-term recognition memory was measured by “novel object recognition test (NOR)”95. NOR was conducted in the square locomotion chamber as open field under normal light. Each mouse was habituated to the testing arena for 10 min and returned to home cage for 5-10 min. On the day of testing, the mice were habituated to the testing arena for 3-10 min and then returned to home cages for 5-10 min. Next the mice were placed back in the testing arena for 8 min with two identical objects (T1). The objects were plastic, custom fabricated (4cm in height, 4cm in diameter). The mice were then placed back into home cages. 1 h later, each mouse was returned to the testing arena for 8 min and exposed to one familiar object (T1) and one novel object (T2). The activity of the mice was video recorded and scored using visual tracking software (CleverSys, TopScan Suite v3). The time each mouse spent intentionally touching the object and the time the mouse was within 1 cm of the object facing it were recorded as touch time. NOR% was calculated as the percent of time each animal explored the novel object relative to all objects. The surfaces of the field were cleaned with 70% alcohol after each trial.
Long-term spatial memory was measured by “Morris water maze”96. The maze consisted of a 122-cm-diameter pool filled with water clouded with white, nontoxic, water-based paint. The maze was virtually divided into four quadrants, with one containing a hidden platform located 0.8 cm below the water surface. Each mouse was trained to find the platform, orienting by extra maze cues placed asymmetrically as spatial references. Individually, the mouse was placed into the water of a quadrant in a random fashion to prevent strategy learning. Each mouse was given 60 s to find the hidden platform. If the mouse failed to find the platform, it was placed onto the platform. Each mouse was allowed to stay on the platform for 20 s and was then dried and put back to home cage. Each animal was trained four trials per day (an inter-trial latency of at least 60 sec) for five consecutive days. Spatial probe trials, were conducted 24 h after the last training session (day 6), the platform was removed, and mouse was allowed to swim for 60 sec. The drop position was in the first quadrant, with the mouse facing the wall at start. For the probe trial, data were given as the traveled distance in quadrant Q3, representing the quadrant where the platform had been located, and compared to the averaged distance the animal has traveled in the remaining quadrants. We use the same batch of mice (WT, N40K, 5xFAD, dTg littermates) for MWM at 3, 6, or 12 months of age.
Visual acuity was measured using the OptoMotry System (CerebralMechanics) as described97. All tests were performed in bright-light conditions to measure cone function.
Electrophysiology.
Mice were rapidly sacrificed and acute transverse hippocampal slices (400 μm) were processed as previously described98. Field recordings were made using an 8-submerged recording chamber setup (Campden Instruments), Digidata digitizers (Molecular Devices), computer-controlled microelectrode amplifiers (Molecular Devices), STG2004 stimulators (Multichannel Systems), and pClamp software (Molecular Devices, Clampex v10.3.2.1, Multiclamp 700B v2.1.0.16/Axoclamp 900A v1.1.0.1, and MC Stimulus v3.4.4), with each chamber recorded independently. We examined the medial perforant pathway which projects from entorhinal cortex to dentate gyrus in hippocampus, as it plays a critical role in learning and is affected early and severely by neuropathology in AD46,47. Field excitatory postsynaptic potentials (fEPSPs) from the molecular layer of the dentate gyrus were recorded by using an extracellular glass pipette (3–5 MΩ), while the medial portion the perforant path was stimulated using a bipolar tungsten electrode (FHC). Input-Output curves were generated for each slice and stimulus strength set at the magnitude that elicited 50% maximal output in the input-output curve. Paired-pulse response was then assessed at 20-, 50-, 100-, 200-, and 1000 ms intervals, any slices that did not exhibit paired-pulse depression were not used. Baseline responses were recorded for at least 20 min and LTP was induced using three rounds of theta burst-stimulation. Each round was separated by 5 min and contained six trains with 10 s between each train. Each train was five bursts separated by 200 ms, with each burst being four 100 Hz pulses.
Extended Data
Extended Data Fig. 1. Deep TMT profiling of detergent insoluble proteome in AD and quantitative analysis of western blotting.
a, Proteomic profiling of detergent-insoluble proteome of 10 control and 10 AD cases by TMT-LC/LC-MS/MS. A total of 8,917 proteins were identified, out of which 365 proteins were found to be increased in AD detergent insoluble proteome. b, Principal component analysis of the insoluble proteome in control and AD cases. c, Heatmap of selected top proteins and U1 snRNP components enriched in AD. d, Pathway enrichment analysis of the proteins enriched in the AD insoluble proteome (Fisher's exact test and the BH procedure to generate FDR values). e, Enriched protein-protein interaction module of spliceosome. f, Quantified data of western blotting (4 replicates) in Fig. 1d (Student’s t-test). g, Quantified data of western blotting (triplicates) in Fig. 1e to show that the N40K expression leads to the depletion of endogenous U1-70K in neurons (Student’s t-test). Data are shown as mean ± SEM. * p < 0.05, ** p < 0.01.
Extended Data Fig. 2. Generation of multiple N40K Tg lines with N40K expression and U1-70K downregulation.
a, Strategy for producing N40K Tg lines by the injection of N40-expressing lentivirus. b, Chromosomal localization of Tg determined by fluorescence in situ hybridization (FISH) analysis. Left: Chr18 (green), N40K (red) in Tg396 line; Right: Chr10 (green), N40K (red) in Tg318 line. c, Western blotting of U1-70K and N40K in Tg396 and Tg318 Tg lines. N40K expression led to similar depletion of U1-70K protein in the hippocampus of both lines. d, Quantitation of relative N40K levels in cortex and hippocampus of Tg396 lines by western blotting. Titrated Tg proteins produce a linear response curve (R2 = 0.98). According to the curve equation, N40K is at a ~2-fold level of native U1-70K as in WT mice (Student t-test). Data are shown as mean ± SEM. * p < 0.05.
Extended Data Fig. 3. Proteomics analysis of insoluble fraction in WT and N40K-Tg mice.
a, Proteomic profiling of detergent-insoluble proteome of WT and N40K-Tg mice (12-month-old, WT n = 3, Tg n = 3). b, Principal component analysis of the insoluble proteome of WT and N40K. c, Heatmap of differentially expressed proteins in N40K-Tg insoluble fraction. d, Pathway enrichment analysis of differentially expressed proteins in N40K-Tg insoluble fraction (Fisher's exact test) followed by FDR analysis by the BH procedure. e, Enriched PPI module of spliceosomal snRNP complex (Fisher's exact test and the BH procedure, FDR < 0.05). Each dot represents a protein, whereas the interaction is indicated by connected lines.
Extended Data Fig. 4. N40K Tg mice exhibit brain weight loss and brain volume reduction, but with normal locomotive activities.
a-c, N40K Tg mice showed reduction of brain weight and brain volume, whereas the body weight had no change at different ages (3-month-old: WT n = 14, Tg n = 10-11; 12-month-old: WT n = 14-16, Tg n = 11-19, Student t-test). d, Volume change of Tg318 mouse in cortex, and hippocampus (Hipp) measured by MRI at different ages (3-month-old: WT n = 15, Tg n = 10; 12-month-old: WT n = 15, Tg n = 10, two-way ANOVA followed by Sidak's multiple comparison test). e, Morris water maze task for Tg318 mice. Compared to WT control, Tg318 mice showed significant difference at day 3-6 (two-way ANOVA followed by Sidak's multiple comparison test). f, Speed in the probe trial of Morris water maze at day 6 showed no significant difference between WT and Tg mice in Figure 4g (12-month-old: WT n = 9, Tg n = 9, Student t-test). Data are shown as mean ± SEM, ns (not significant); * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
Extended Data Fig. 5. Validation of splicing deficient transcripts of synaptic pathway in N40K-Tg mice.
a-b, Quantitation of intron accumulation at the selected region of Gabra2, Gng7, Kcnh1, and Camk1d. IGV software was used to display RNA read density with same scale range in each gene from RNA-seq (ex: exon; in: intron). Red boxes show selected regions for quantification. Intron reads % was analyzed by RNA-seq ((intron-exon junctions + introns)/(intron-exon junctions + introns + exons + exon-exon junctions)). Intron retention % was analyzed by RT-PCR ((intron-containing PCR band intensity)/(intron-containing PCR band intensity + exon-exon PCR band intensity)). Gabra2: intron 6 vs exon 6 and 7; Gng7: intron 2 vs exon 2 and 3, Kcnh1: intron 9 vs exon 9 and 10, and Camk1d: intron 5 vs exon 5 and 6. 12-month-old: WT n = 3, Tg n = 3. Scale bar, 10 kb. Data are shown as mean ± SEM. Statistical significance was analyzed by Student's t-test. Data are shown as mean ± SEM, * p < 0.05; ** p < 0.01; *** p < 0.001.
Extended Data Fig. 6. Profiling analysis of dTg and tau mice.
a, Representative co-immunofluorescence staining of 5xFAD and dTg brain slides with Thioflavin S for plaques and the U1-70K C-terminal antibody for U1-70K depletion. Scale bar, 100 μm. b, PCA for RNA-seq and proteomics studies. c, Distribution of splicing deficiency scores of mapped transcripts. Statistical comparisons between different genotypes are shown (Kolmogorov–Smirnov test). d, Relative Aβ level in 5xFAD and dTg by the proteomics analysis (mean ± SEM, Student’s t-test, ns: not significant). e, Cell type enriched DE proteins in the dTg mice. The 979 DE proteins were overlapped with the cell type expression data from RNA-seq analysis96. f, Swimming speed of WT, N40K Tg, 5xFAD, dTg in the Morris water maze experiment (mean ± SEM, one-way ANOVA, ns: not significant). g, RNA-seq analysis of WT and Tau (P301S) mice. h, The percentage of mapped intron reads in all transcripts from the cortices of WT (n = 5) and Tau P301S (n = 6) mice (mean ± SEM, Student’s t-test, ns: not significant). i, Distribution of splicing deficiency scores of mapped transcripts (Kolmogorov–Smirnov test).
Extended Data Fig. 7. Proteomic comparison of the three mouse models and AD.
a, The workflow to examine the consistent changes in the three mouse models and human AD cases by overlapping differentially expressed proteins. b, DE protein numbers in the mouse models, human cases, and the overlapped portions. c, Enriched pathways of the human-overlapped DE proteins in three genotypes (selected from supplementary table 19, Fisher's exact test and the BH procedure to derive FDR, FDR cutoff of 0.05). d, Enriched protein-protein interaction modules (selected from supplementary table 20, Fisher's exact test and the BH procedure to derive FDR). FDR cutoff is 0.05 except the Aβ binding module, which is selected due to biological significance.
Extended Data Fig. 8. Analysis of the role of TDP-43 in human cases and the mouse models.
a, The weak correlation between the percentage of intron reads and the stages of TDP-43 pathology. Pearson correlation coefficient (r) is shown. b, The percentage of mapped intron reads in all transcripts from human cases of different TDP-43 stages (mean ± SEM, Student’s t-test, ns: not significant). As the sample size was small, we merged stages 0-1 and stages 2-3 in the analysis. c, Distribution of splicing deficiency scores of mapped transcripts (Kolmogorov–Smirnov test). d, Staining of plaques, Tau, TDP-43, and nuclei in a human AD brain sample (positive controls) and in the mouse models (cortex, ~12-month-old). Phosphorylated Tau and TDP-43 antibodies were used.
Supplementary Material
Acknowledgments
We thank all other lab and center members for discussion and technical support. We thank St. Jude Shared Resources and Core Facilities, including Animal Research Center, Transgenic/Gene Knockout, In Vivo Imaging and Therapeutics, Veterinary Pathology, Cytogenetics, Genome Sequencing, Proteomics and Metabolomics, Applied Bioinformatics, and Cell and Tissue Imaging. We also thank J. Jankowsky for providing plasmids, I. Bayazitov for electrophysiology, F. Zheng for the discussion of behavior studies, A. Hemphill for Western blots of replicates during revision and I. Chen for helpful discussion and guidance. This work was partially supported by National Institutes of Health grants R01AG047928 (J.P.), R01AG053987 (J.P.), RF1AG068581 (J.P.), RF1AG064909 (G.Y. and J.P.), U54NS110435 (J.P.), U19AG069701 (J.P.), R01MH095810 (S.S.Z.), American Lebanese Syrian Associated Charities (ALSAC). The Banner Sun Health Research Institute Brain and Body Donation Program was supported by National Institutes of Health grants U24NS072026, P30AG072980, P30AG19610, the Arizona Department of Health Services, the Arizona Biomedical Research Commission and the Michael J. Fox Foundation for Parkinson’s Research.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
ROSMAP data can be download from this website (https://adknowledgeportal.synapse.org/) and mapped to human transcriptome (hg19). All mouse RNA-seq data were deposited in the GEO database: GSE115177 (mouse brain) and GSE196873 containing two sub-series IDs: GSE196871 (mouse brain) and GSE196872 (mouse neuronal culture) and mapped to mouse transcriptome (mm9). The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD031587 (human aggregated proteome), PXD031581 (mouse aggregated proteome), and PXD031546 (mouse neuronal culture), PXD023395 and PXD031545 (mouse brain). Both hg19 and mm9 can be download from UCSC website (https://genome.ucsc.edu/index.html). Other data underlying the paper are provided as source data files or available from the corresponding author upon reasonable request.
Code availability
The source codes are available at https://github.com/PengLabStJude/N40K_model.
References
- 1.Raj B & Blencowe BJ Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 87, 14–27 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Cooper TA, Wan L & Dreyfuss G RNA and disease. Cell 136, 777–793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scotti MM & Swanson MS RNA mis-splicing in disease. Nat Rev Genet 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathieu C, Pappu RV & Taylor JP Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science 370, 56–60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefebvre S et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie IR, Rademakers R & Neumann M TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 9, 995–1007 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Nguyen HP, Van Broeckhoven C & van der Zee J ALS Genes in the Genomic Era and their Implications for FTD. Trends Genet 34, 404–423 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Jutzi D et al. Aberrant interaction of FUS with the U1 snRNA provides a molecular mechanism of FUS induced amyotrophic lateral sclerosis. Nat Commun 11, 6341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y et al. U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Res 43, 3208–3218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun 6, 6171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Y, Mu JC & Ackerman SL Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 148, 296–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josephs KA et al. Staging TDP-43 pathology in Alzheimer's disease. Acta Neuropathol 127, 441–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman BT et al. National Institute on Aging-Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease. Alzheimers Dement 8, 1–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeTure MA & Dickson DW The neuropathological diagnosis of Alzheimer’s disease. Mol Neurodegener 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terry RD et al. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol 30, 572–580 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Selkoe DJ Alzheimer's disease is a synaptic failure. Science 298, 789–791 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Aebersold R & Mann M Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Bai B et al. Proteomic landscape of Alzheimer's Disease: novel insights into pathogenesis and biomarker discovery. Molecular neurodegeneration 16, 55 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai B et al. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc Natl Acad Sci U S A 110, 16562–16567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson ECB et al. Deep proteomic network analysis of Alzheimer's disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Molecular neurodegeneration 13, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai B et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer’s Disease Progression. Neuron 105, 975–991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu K et al. Global Profiling of Lysine Accessibility to Evaluate Protein Structure Changes in Alzheimer's Disease. J Am Soc Mass Spectrom 32, 936–945 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raj T et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer's disease susceptibility. Nat Genet 50, 1584–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hales CM et al. Changes in the detergent-insoluble brain proteome linked to amyloid and tau in Alzheimer's Disease progression. Proteomics 16, 3042–3053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz BM & Peng J Deep Profiling of the Aggregated Proteome in Alzheimer's Disease: From Pathology to Disease Mechanisms. Proteomes 6, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J & Nagai K Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458, 475–480 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hales CM et al. Aggregates of small nuclear ribonucleic acids (snRNAs) in Alzheimer's disease. Brain Pathol 24, 344–351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hales CM et al. U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer's disease due to autosomal dominant genetic mutations and trisomy 21. Molecular neurodegeneration 9, 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai B et al. Integrated Approaches for Analyzing U1-70K Cleavage in Alzheimer's Disease. Journal of proteome research 13, 4526–4534 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy J & Selkoe DJ The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Bai B et al. Deep Profiling of Proteome and Phosphoproteome by Isobaric Labeling, Extensive Liquid Chromatography, and Mass Spectrometry. Methods Enzymol 585, 377–395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z et al. 27-plex Tandem Mass Tag Mass Spectrometry for Profiling Brain Proteome in Alzheimer's Disease. Anal Chem 92, 7162–7170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diner I et al. Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. The Journal of biological chemistry 289, 35296–35313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishof I et al. RNA-binding proteins with basic-acidic dipeptide (BAD) domains self-assemble and aggregate in Alzheimer's disease. The Journal of biological chemistry 293, 11047–11066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo Y, Oubridge C, van Roon AM & Nagai K Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition. Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigel E & Steinmann ME Structure, function, and modulation of GABA(A) receptors. The Journal of biological chemistry 287, 40224–40231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan J, Amin P & Ofengeim D Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat Rev Neurosci 20, 19–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo T et al. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer's disease. Mol Neurodegener 15, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dittgen T et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A 101, 18206–18211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uezu A et al. Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oleskevich S, Leck KJ, Matthaei K & Hendry IA Enhanced serotonin response in the hippocampus of Galphaz protein knock-out mice. Neuroreport 16, 921–925 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Schwindinger WF et al. Loss of G protein gamma 7 alters behavior and reduces striatal alpha(olf) level and cAMP production. The Journal of biological chemistry 278, 6575–6579 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Simons C et al. Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat Genet 47, 73–77 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Floudas CS, Um N, Kamboh MI, Barmada MM & Visweswaran S Identifying genetic interactions associated with late-onset Alzheimer's disease. BioData Min 7, 35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Limon A, Reyes-Ruiz JM & Miledi R Loss of functional GABA(A) receptors in the Alzheimer diseased brain. Proc Natl Acad Sci U S A 109, 10071–10076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hyman BT, Van Hoesen GW, Kromer LJ & Damasio AR Perforant pathway changes and the memory impairment of Alzheimer's disease. Ann Neurol 20, 472–481 (1986). [DOI] [PubMed] [Google Scholar]
- 47.Scheff SW & Price DA Alzheimer's disease-related alterations in synaptic density: neocortex and hippocampus. J Alzheimers Dis 9, 101–115 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Tyagarajan SK & Fritschy JM Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci 15, 141–156 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Chacon PJ et al. Cerebellin 4, a synaptic protein, enhances inhibitory activity and resistance of neurons to amyloid-beta toxicity. Neurobiol Aging 36, 1057–1071 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Upadhyay A et al. Neurocalcin Delta Knockout Impairs Adult Neurogenesis Whereas Half Reduction Is Not Pathological. Front Mol Neurosci 12, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshiyama Y et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Wan YW et al. Meta-Analysis of the Alzheimer's Disease Human Brain Transcriptome and Functional Dissection in Mouse Models. Cell Rep 32, 107908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson ECB et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salz HK et al. The Drosophila U1-70K protein is required for viability, but its arginine-rich domain is dispensable. Genetics 168, 2059–2065 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xue S et al. Low-complexity domain of U1-70K modulates phase separation and aggregation through distinctive basic-acidic motifs. Sci Adv 5, eaax5349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winklhofer KF, Tatzelt J & Haass C The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J 27, 336–349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheper W & Hoozemans JJ The unfolded protein response in neurodegenerative diseases: a neuropathological perspective. Acta Neuropathol 130, 315–331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hetz C The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13, 89–102 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Ranasinghe KG et al. Neurophysiological signatures in Alzheimer's disease are distinctly associated with TAU, amyloid-beta accumulation, and cognitive decline. Sci Transl Med 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grubman A et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type-specific gene expression regulation. Nat Neurosci 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Palop JJ & Mucke L Amyloid-beta-induced neuronal dysfunction in Alzheimer's disease: from synapses toward neural networks. Nat Neurosci 13, 812–818 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh YC et al. Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer's Disease. Cell Rep 29, 301–316 e310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Apicco DJ et al. Dysregulation of RNA Splicing in Tauopathies. Cell Rep 29, 4377–4388 e4374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lester E et al. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron 109, 1675–1691 e1679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giasson BI et al. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science 300, 636–640 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Nelson PT et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 142, 1503–1527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beach TG et al. Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology 35, 354–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kosse C & Burdakov D Natural hypothalamic circuit dynamics underlying object memorization. Nat Commun 10, 2505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Achilly NP, Wang W & Zoghbi HY Presymptomatic training mitigates functional deficits in a mouse model of Rett syndrome. Nature 592, 596–600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai B et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer's Disease Progression. Neuron 105, 975–991 e977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lois C, Hong EJ, Pease S, Brown EJ & Baltimore D Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Xu P, Duong DM & Peng J Systematical Optimization of Reverse-Phase Chromatography for Shotgun Proteomics. J Proteome Res 8, 3944–3950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H et al. Systematic optimization of long gradient chromatography mass spectrometry for deep analysis of brain proteome. J Proteome Res 14, 829–838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X et al. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Mol Cell Proteomics 13, 3663–3673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng J, Elias JE, Thoreen CC, Licklider LJ & Gygi SP Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2, 43–50 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Niu M et al. Extensive Peptide Fractionation and y1 Ion-Based Interference Detection Method for Enabling Accurate Quantification by Isobaric Labeling and Mass Spectrometry. Anal Chem 89, 2956–2963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mertens BJA Transformation, Normalization, and Batch Effect in the Analysis of Mass Spectrometry Data for Omics Studies. Statistical Analysis of Proteomics, Metabolomics, and Lipidomics Data Using Mass Spectrometry, 1–21 (2016). [Google Scholar]
- 81.Rusch M et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat Commun 9, 3962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Law CW, Chen Y, Shi W & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vanderwall D et al. JUMPn: A Streamlined Application for Protein Co-Expression Clustering and Network Analysis in Proteomics. J Vis Exp (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan H et al. Integrative Proteomics and Phosphoproteomics Profiling Reveals Dynamic Signaling Networks and Bioenergetics Pathways Underlying T Cell Activation. Immunity 46, 488–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huttlin EL et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li T et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 14, 61–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barabasi AL & Oltvai ZN Network biology: understanding the cell's functional organization. Nat Rev Genet 5, 101–113 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Ravasz E, Somera AL, Mongru DA, Oltvai ZN & Barabasi AL Hierarchical organization of modularity in metabolic networks. Science 297, 1551–1555 (2002). [DOI] [PubMed] [Google Scholar]
- 91.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.West MJ, Slomianka L & Gundersen HJ Unbiased stereological estimation of the total number of neurons in thesubdivisions of the rat hippocampus using the optical fractionator. Anat Rec 231, 482–497 (1991). [DOI] [PubMed] [Google Scholar]
- 94.Chapman PF et al. Impaired synaptic plasticity and learning in aged amyloid precursor protein transgenic mice. Nat Neurosci 2, 271–276 (1999). [DOI] [PubMed] [Google Scholar]
- 95.Uslaner JM et al. The muscarinic M1 receptor positive allosteric modulator PQCA improves cognitive measures in rat, cynomolgus macaque, and rhesus macaque. Psychopharmacology (Berl) 225, 21–30 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Heneka MT et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prusky GT, Alam NM, Beekman S & Douglas RM Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci 45, 4611–4616 (2004). [DOI] [PubMed] [Google Scholar]
- 98.Bayazitov IT, Richardson RJ, Fricke RG & Zakharenko SS Slow presynaptic and fast postsynaptic components of compound long-term potentiation. J Neurosci 27, 11510–11521 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ROSMAP data can be download from this website (https://adknowledgeportal.synapse.org/) and mapped to human transcriptome (hg19). All mouse RNA-seq data were deposited in the GEO database: GSE115177 (mouse brain) and GSE196873 containing two sub-series IDs: GSE196871 (mouse brain) and GSE196872 (mouse neuronal culture) and mapped to mouse transcriptome (mm9). The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD031587 (human aggregated proteome), PXD031581 (mouse aggregated proteome), and PXD031546 (mouse neuronal culture), PXD023395 and PXD031545 (mouse brain). Both hg19 and mm9 can be download from UCSC website (https://genome.ucsc.edu/index.html). Other data underlying the paper are provided as source data files or available from the corresponding author upon reasonable request.
The source codes are available at https://github.com/PengLabStJude/N40K_model.