Abstract
The cellular recycling process of autophagy is essential for survival, development, and homeostasis. Autophagy also plays an important role in aging and has been linked to longevity in many species, including the nematode C. elegans. Study of the physiological roles of autophagy during C. elegans aging requires methods for the spatiotemporal analysis of autophagy. Here we describe a method for assessing autophagic flux in multiple tissues of C. elegans by quantifying the pool of autophagic vesicles using fluorescently labelled Atg8/LGG-1 reporters upon autophagy inhibition using bafilomycin A1 (BafA). This methodology has revealed that autophagic activity varies in different cell types of C. elegans during aging.
Keywords: Autophagic flux, Aging, Bafilomycin A, C. elegans
1. Introduction
Macroautophagy (hereafter referred to as autophagy) facilitates degradation and recycling of cytosolic components, referred to as cargo, in response to nutrient deprivation or other stresses. Autophagy is initiated by the nucleation of a double membrane, which forms the phagophore. Upon completion and maturation, autophagosomes fuse with acidic lysosomes, resulting in the degradation of the sequestered content by hydrolases [1]. Autophagy is essential for survival, development, and homeostasis and may protect against pathologies, including neurodegeneration and aging [2, 3].
Autophagic vesicles are commonly monitored using fluorescently tagged Atg8/LC3. During autophagy induction, Atg8 is cleaved, conjugated to phosphatidylethanolamine, and inserted into the autophagosomal membrane [4]. GFP-tagged Atg8 visualizes phagophores, autophagosomes and amphisomes (collectively referred to below as autophagosomal vesicles) as fluorescent punctae. The C. elegans homolog of Atg8 [5], LGG-1, is expressed in various tissues of the adult animal, including neurons [6]. A GFP-tagged LGG-1/Atg8 reporter under the control of its endogenous promoter visualizes autophagosomal structures in hypodermal seam cells, intestinal cells, body-wall muscle and pharynx [6,7], whereas a GFP-tagged LGG-1/Atg8 reporter under the control of a neuronal promoter (rgef-1p) specifically visualizes pan-neuronal autophagosomal structures [8]. However, quantifying the number of autophagosomes does not conclusively inform on the status of autophagy, which is a dynamic, multistep process [4]. An increase in the number of GFP::LGG-1/Atg8 punctae could result from increased formation of autophagosomes (i.e., increased autophagic activity), or a block of downstream steps of the autophagy process (i.e., decreased autophagic activity), respectively [9–11]. To distinguish between these two scenarios, acute inhibition of autophagy is performed using “autophagy flux” assays [4, 11]. In such assays, autophagy is inhibited by using chemical inhibitors such as bafilomycin A1 (BafA), which blocks autophagosomal turnover by inhibiting V-ATPase activity and preventing lysosomal acidification [9, 11]. Autophagosome numbers are compared between BafA inhibition and the control conditions. A change in the number of autophagosomes upon BafA addition indicates that autophagy is active, whereas no change indicates that the cell/tissue is experiencing a block in autophagy (Fig. 1). Using this assay, we recently assessed autophagic activity in a tissue-specific manner of animals of different ages and found that autophagic activity declines with age in the intestine, pharynx, body-wall muscle, and nerve-ring neurons of wild-type C. elegans, albeit with tissue-specific differences [9, 10]. Furthermore, our analyses of long-lived animals revealed that they were capable of maintaining autophagic activity in distinct tissues with age [9, 10]. The tissue-specific analysis of autophagic activity with age is an important step in understanding the role of autophagy in lifespan and healthspan determination.
Fig. 1.
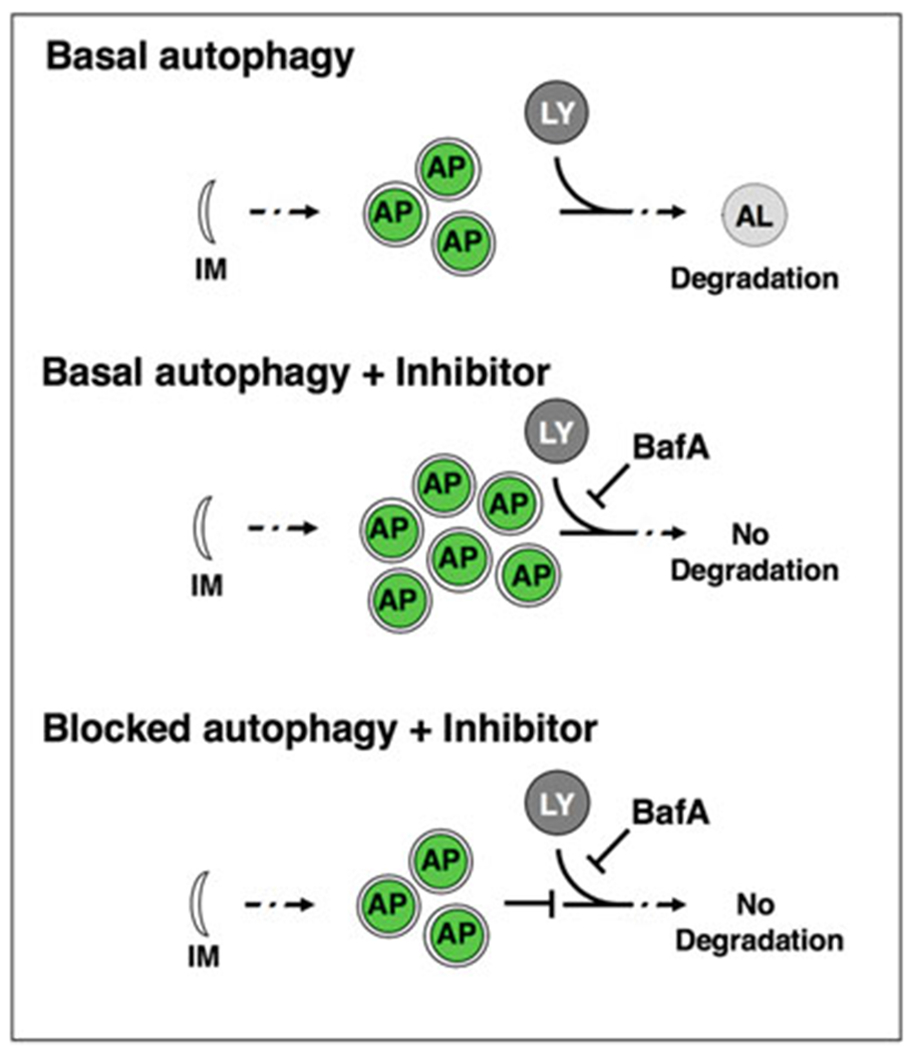
Autophagy flux assay. Basal autophagy is initiated with the nucleation of a double membrane (isolation membrane, IM), which forms the phagofore. The phagofore expands into an autophagosome (AP) that sequesters cytosolic material (also referred to as cargo) destined for degradation. Autophagosomes fuse with acidic lysosomes (LY) to form autolysosomes (AL), in which the cargo is subsequently degraded. In autophagy flux assays, the chemical autophagy inhibitor Bafilomycin A (BafA) can be used to block autophagic turnover. In situations where autophagy is active, the addition of BafA will change the apparent number of autophagosomes. If autophagy is blocked, BafA addition will not change the number of autophagosomes compared to control conditions
Here we describe our autophagy flux assay that can be used to assess autophagic activity in distinct tissues of adult C. elegans of different ages [9, 11–13].
2. Materials
2.1. C. elegans Strains
Strains expressing GFP-tagged LGG-1 (see Note 1).
adIs2122[lgg-1p::gfp::lgg-1 + rol-6] (DA2123) [6, 7] to assess autophagic flux in hypodermal seam cells, intestine, pharynx, and body-wall muscle.
-
sqIs24[rgef-1p::gfp::lgg-1 + unc-122p::rfp] (MAH242) [8] to assess autophagic flux in nerve-ring neurons.
Both C. elegans strains can be obtained from the Caenorhabditis Genetic Center (CGC).
2.2. C. elegans Maintenance
Worm pick: Cut ~3 cm of platinum wire (90% platinum, 10% iridium wire, 30 G, e.g., Genesee Scientific). Break off the thin part of a glass Pasteur pipette (5¾ in., e.g., Fisherbrand). Melt the glass at the site of breakage on a Bunsen burner and attach the platinum wire. Flatten the end of the platinum wire using small forceps or bend the platinum wire to create a small loop. The platinum wire pick should be sterilized over a flame while using.
Normal Growth Media (NGM) agar plates: Mix 2.5 g Bacto peptone, 3 g NaCl, and 20 g agar in 975 ml H2O. After autoclaving, allow to cool down to ~55 °C, and add 1 ml of 1 M CaCl2, 1 ml of 1 M MgSO4, 1 ml of 5 mg/ml cholesterol in ethanol, and 25 ml of 1 M KPO4 buffer (pH 6.0). Dispense into 6 cm dishes.
E. coli OP50 is used as a food source and can be obtained from the CGC. Culture OP50 in LB medium. Apply about 80–100 μl of OP50 culture to NGM plates and allow a lawn to grow before transferring animals.
C. elegans strains can be maintained and cultured under standard conditions at 20 °C [14]. For aging experiments, animals are synchronized by transferring eggs manually using a platinum wire worm pick (see Note 2). Eggs are allowed to hatch on NGM plates seeded with OP50 bacteria. Once reproductive (3 days of incubation at 20 °C), adult animals need to be transferred away from progeny daily until they reach the desired day of adulthood. Preferably, animals of all ages should be imaged on the same day, in which case eggs are picked on different days to achieve appropriate staging on the day of the experiment. (We note that a large number of experimental conditions may make it impossible to image all animals on the same day. Best practices should be used to ensure reproducibility.) Animals should always be well-fed, since autophagy activity is sensitive to the nutritional status of the animals.
2.3. Reagents for Injections
Dimethyl sulfoxide (DMSO, e.g., Sigma).
Water.
1 mg of Bafilomycin A1 (BafA) (from BioViotica Rare Active Natural Products, Product number BVT-0252). Prepare a 25 mM BafA stock solution by dissolving 1 mg of BafA in 64 μl of DMSO. The stock concentration needs to be this high to keep the final DMSO concentration of 0.2% (w/v). Make aliquots of 5 μl and store BafA stock solution at −20 °C.
Dextran, Texas Red, 3000 MW Lysine Fixable (Thermo Fisher Scientific, Product number D3328). To control for successful injection into the tissue of interest, a dye like Dextran Texas Red that is visible under a fluorescent microscope and of a molecular weight larger than BafA is mixed with the BafA and DMSO. Prepare a 25 mg/ml stock solution by resuspending the coinjection dye in water. Make aliquots of 10–20 μl and store at −20 °C.
M9 buffer: 6 g Na2HPO4, 3 g KH2PO4, 5 g NaCl, and 0.25 g MgSO4·7H2O in 1 L H2O. Sterilize by autoclaving.
Halocarbon oil 700 (e.g., Sigma) for mounting of the animals to be injected.
2.4. Injections
Needle puller (e.g., Flaming/Brown Micropipette Puller Model P-94, Sutter Instruments).
Injection needles: Injection needles are produced from thin glass capillaries, such as Kwik-Fil Borosilicate Glass Capillaries (1.0 mm, 4 in., e.g., World Precision Instruments, Inc.).
Pipette tips for loading needles (e.g., Microloader tips (e.g., Eppendorf) 0.5–20 μl 100 mm) (see Note 3).
Injection scope (e.g., Zeiss Axio) with needle micromanipulator (e.g., Eppendorf) and joystick (e.g., Transferman NK2 (Eppendorf)).
Electronic microinjector that provides the pressure for injecting small volumes via the attached needle (e.g., Femtojet (Eppendorf)).
Microscope slide coverslips 22 × 50 mm with Thickness #1.5 (e.g., Slip-rite coverslip (Thermo)).
Injection pads are used to immobilize animals for the injection procedure. An injection pad is a glass coverslip (22 × 50 mm) (e.g., Slip-rite coverslip (Thermo)) with a dried layer of 2% (w/v) agarose (in water) on the center of it. Prepare boiling 2% agarose in water (usually 10 ml of water with 0.2 g of agarose), use a glass Pasteur pipette (5¾ in., e.g., Fisherbrand) to place 1–4 drops (~50 μl) onto the center of a 22 × 50 mm glass coverslip, immediately flatten the drop with another 22 × 50 mm coverslip. Let the pads dry for a few minutes and then slide the coverslip off. Let the slides dry for at least 1 day before use. Label the right side of the upward-facing coverslip with an “R” (optional).
2.5. Fluorescence Microscopy
Confocal microscope (e.g., Zeiss LSM 710) or Zeiss Imager Z1 including apotome.2.
Sodium azide is used to anesthetize live animals for imaging. Prepare M9 buffer containing 150 mM NaN3 (add 1.5 ml of 1M NaN3 (dissolved in M9) to 8.5 ml ofM9 buffer) (see Note 4).
Microscope slides (25 × 75 × 1 mm, e.g., Premium Microscope Slides Superfrost (Fisher Scientific)) and coverslips (22 × 50 mm, Thickness #1.5, e.g. Slip-rite coverslip (Thermo)).
Imaging slides are used to immobilize worms for imaging. Prepare boiling 2% agarose in M9 buffer and add NaN3 to a final concentration of 150 mM (usually 8.5 ml of water with 0. 2 g of agarose and 1.5 ml of 1 M NaN3 in M9). This solution can be kept at 65 °C for repeated use. Using a glass Pasteur pipette (5¾ in. e.g., Fisherbrand), place 1–4 drops (~50 μl of this solution to the center of a microscope slide (25 × 75 × 1 mm) and immediately flatten the drop with another microscope slide. Let the pads dry for a few minutes and then separate the microscope slides. Imaging pads should be made fresh and should not be allowed to dry out.
3. Methods
3.1. Preparing Solutions for Injection
The solutions for injections should be prepared fresh before the injections.
BafA solution: Make a 100 μM BafA working solution by adding 1 μl of the 25 mM BafA stock solution to 249 μl of water. The working solution should not be reused. Make the BafA injection solution by mixing 5 μl of 100 μM BafA with 1 μl of the 25 mg/ml injection dye stock with 4 μl of water. The BafA concentration in the injection solution is 50 μM.
DMSO solution: Make a 0.4% DMSO (w/v) working solution by adding 1 μl of DMSO (e.g., Sigma) to 249 μl of water. Make the DMSO injection solution by mixing 5 μl of 0.4% DMSO with 1 μl of the 25 mg/ml injection dye stock with 4 μl of water. The DMSO concentration in the injection solution is 0.2%.
3.2. Preparing Injection Needles
The individual settings for the needle-pulling program have to be established individually and depend on the filament, the capillaries, and the needle-pulling machine. The parameters that determine the needle shape are typically heat, pull, velocity and time. The pulled needles should be sealed at the tip and have a taper of about 6 mm but will need to be adjusted depending on injection set-up. Prepulled injection needles can also be purchased from several companies (e.g., Femtotips II (Eppendorf)). See Note 5 for needle-pulling protocol using Micropipette Puller model P-94 from Sutter Instruments.
3.3. Injections
Animals should be injected with BafA at least 2 h before imaging to achieve steady-state levels of autophagosomes after the treatment with the inhibitor; these levels do not appear to change over the next 24 h [9].
3.3.1. Loading the Needles
Use a 20 μl pipette with a 0.5–20 μl 100 mm Microloader pipette tip (Eppendorf) to load 2–4 μl of the BafA or DMSO solution into the needle (see Note 3).
3.3.2. Breaking the Needle Tip
After placing the needle in the micromanipulator, it is necessary to physically break its tip to open the capillary and allow the flow of the solution. The needle can be broken on the side of a small coverslip (18 × 18 mm, e.g., micro cover glass (e.g., VWR)) attached to a larger cover slide (22 × 50 mm, e.g., Slip-rite coverslip (Thermo)). Place a drop of microinjection oil (Halocarbon oil 700) on a 22 × 50 mm coverslip and place the smaller 18 × 18 mm cover glass on top, so that the edge of the smaller cover glass is in the oil. Place this coverslip on the microscope and bring into focus using the 10× objective. Move the needle so that the needle tip is illuminated. Dip the needle tip into the oil and position it at the same level of the side of the small coverslip and then change to 40 or 100× objective. Using the needle manipulator joystick (e.g., Transferman NK2 (Eppendorf)), move the needle into the side of the coverslip to break off the tip. The tip needs to be small and sharp enough to penetrate the animal without injury. If the needle has been successfully broken, you should be able to see the solution coming out of the needle by pressing the inject button (tip: the best needles have a beveled tip, which helps piercing the animal’s cuticle).
3.3.3. Injection Settings for FemtoJet (Eppendorf)
The injection pressure (Pi) should be set to 1500–2000 hPa for C. elegans injections. The compensation pressure (Pc), which prevents anything from being sucked into the needle, should be set to ~125 hPa.
3.3.4. Injection Procedure
Mounting animals for injections: Take an injection pad and put a drop of microinjection oil on the dried agarose drop on the injection pad. Dip the worm pick into the Halocarbon oil and pick an animal of the desired age from its plate (should be well-fed). Avoid transferring large amounts of bacteria to the injection pad. Transfer the animal into the oil on the injection pad and gently press the animal onto the agarose to aid in immobilization of the animal. Make sure that the animal is submerged in enough oil to slow down desiccation, without preventing the animals from adhering to the agarose.
With the needle well above the slide, bring the animal into focus using the 10× objective. Bring the needle into the same level as the animal by gently lowering the needle until it comes into focus (tip: the needle should be next to the animal as it is lowered and not on top of it). Change to the 40× or 100× objective and focus on the anterior region of the animal.
-
To assess autophagic flux in intestine, pharynx, and muscle, DA2123 (adIs2122[lgg-1p::gfp::lgg-1 + rol-6]) (Fig. 2a) [6, 7] animals are injected into the anterior area of the animal. The injection dye should penetrate all desired tissues if injected into anterior intestinal cells, the extracellular space near posterior pharyngeal bulb, or into the intestinal lumen.
To assess autophagic flux in nerve-ring neurons, MAH242 (sqIs24[rgef-1p::gfp::lgg-1 + unc-122p::rfp]) (Fig. 2e) [8] animals are injected in the space surrounding the pharynx but be careful not to damage the nerve-ring.
Move the needle into the animal and press either the left mouse button or the foot pedal.
Once the animal has been injected, move the needle first out of the animal and then way above the injection pad (tip: once the needle is out of the animals, it is easiest to just move the needle up, then it will already be in the right position for the next animal and will only have to be lowered).
Recover the injected animal by placing a drop of M9 buffer on top of the animal to make it float and use the worm pick to lift it up and place it on an NGM plate with E. coli OP50. The animals should be incubated for at least 2 h at the cultivation temperature.
Fig. 2.
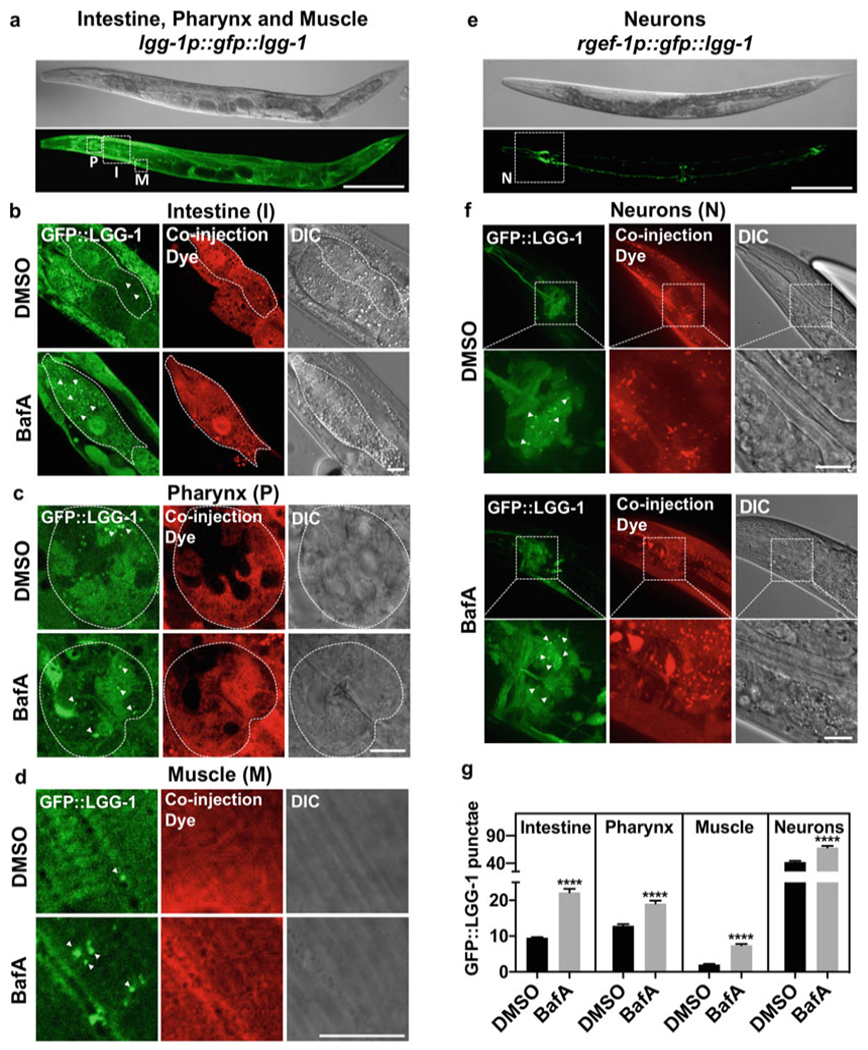
Autophagic flux analysis in select tissues of adult C. elegans. (a) Representative image of strain DA2123 (adIs2122[lgg-1p::gfp::lgg-1 + rol-6]) used to assess autophagic flux in intestine (I), pharynx (P), and muscle (M), outlined by dashed boxes. Scale bar 200 μm. (b–d) Representative images of GFP::LGG-1-positive punctae (green, some indicated by arrowheads) in lgg-1p::gfp::lgg-1-expressing animals (DA2123) injected with DMSO (DMSO) or Bafilomycin A (BafA) with coinjection dye (Dextran, Texas Red) and differential interference contrast (DIC) images are shown. Scale bar 10 μm. Animals shown were injected on day 1 of adulthood. Tissues examined included the intestine (b), pharynx (c), and body-wall muscle (d). (e) Representative image of strain MAH242 (sqIs24[rgef-1p::gfp::lgg-1 + unc-122p::rfp]) used to assess autophagic flux in nerve-ring neurons (N), outlined by a dashed box. Scale bar 200 μm. (f) Representative images of GFP::LGG-1-positive punctae (green) in regf-1p::gfp::lgg-1-expressing animals (MAH242) injected with DMSO (DMSO) or Bafilomycin A (BafA) with coinjection dye (Dextran, Texas Red) and differential interference contrast (DIC) images are shown. Scale bar 10 μm. Animals shown were injected on day 1 of adulthood. (g) Quantification of GFP::LGG-1 punctae in animals injected with DMSO (DMSO) or Bafilomycin A (BafA) on day 1 of adulthood. Data are the mean ± SEM of ≥25 animals combined from at least three experiments. ****p < 0.0001 by Student’s t-test. The data in this figure were published in Fig. 3, Supplement 3 of [9]
3.4. Imaging
3.4.1. Mounting Animals for Imaging
Prepare a fresh imaging slide (see Subheading 2.5) and place a drop (~10 μl) of M9 buffer containing 150 mM NaN3 onto the flattened agarose. Manually transfer the living and injected worms onto the pad. Cover the slide with a 22 × 50 mm coverslip (Thickness #1.5, e.g., Slip-rite coverslip (Thermo)) and image immediately. Imaging pads should be made fresh and should not be allowed to dry out.
3.4.2. Imaging
Imaging can be performed using a confocal microscope (e.g., Zeiss LSM 710) or Zeiss Imager Z1 including apotome.2.
Using the 10× objective determine whether the animals have been properly injected by monitoring the coinjected dye using the red channel. The tissue of interest should be positive for the dye as well as the GFP signal.
To assess autophagic flux in the anterior intestine, use the 100× objective and position the animal ideally with the terminal pharyngeal bulb in one of the corners and the rest of the animal positioned along the diagonal. Images can be acquired as a Z-stack or as a single snap of the anterior intestinal cells (for analysis the best slices of the Z-stack are chosen). When focusing on the intestinal cells, ideally, the nuclei should be visible. Note that the intestine is much dimmer than the pharynx and the gain or exposure time will have to be adjusted accordingly (Fig. 2b).
To assess autophagic flux in the terminal pharyngeal bulb, use the 100× objective and position the animals with the terminal pharyngeal bulb in the center of the view field, or in one of the corners and the animal positioned along the diagonal, if the anterior intestine will be imaged as well. Make sure that the coinjection dye has penetrated the pharyngeal bulb; as we found that the pharynx is more resistant to dye-filling than other tissues [9]. Focus on the terminal pharyngeal bulb so that the pharyngeal opening is visible and acquire an image (Fig. 2c).
To assess autophagic flux in the body-wall muscle, two separate regions can be chosen. Use the 100× objective and focus either on the body-wall muscle in the head region, or on the body-wall muscle region above the germ line arm, rather than above the intestine (in contrast to the intestine, the germ line does not have gfp::lgg-1 expression making focusing on the body-wall muscle easier). Body-wall muscle striation should be visible when acquiring this image (Fig. 2d).
To assess autophagic flux in the nerve-ring neurons use the 100× objective and focus on the area between the pharyngeal bulbs. For imaging of the nerve-ring neurons, images should be acquired as a Z-stack (~20–25 slices with 1 μm intervals). Collapse the images to a stack using maximal orthogonal projection (this step can also be performed during image analysis using Image J (National Institute of Health) (Fig. 2f).
Differential interference contrast (DIC) images can be acquired if desired or if unsure about the tissues.
Collect and save the acquired images.
3.4.3. Image Analysis and Statistical Analysis
Open images in Image J (National Institute of Health).
Use the cell counter function to count the GFP::LGG-1-positive punctae.
To assess autophagic flux in the terminal pharyngeal bulb, count the green GFP::LGG-1-positive punctae in the terminal pharyngeal bulb.
To assess autophagy flux in the anterior intestinal cell, count the green GFP::LGG-1-positive punctae in the individual cells that are visible in the image.
To assess autophagic flux in the body-wall muscle, draw a box that is 1000 μm2 using the polygon tool. The size of the area can be determined using the measure tool and can be saved as an ROI. Count the green GFP::LGG-1-positive punctae in the 1000 μm2 area.
To assess autophagic flux in the nerve-ring neurons, collapse the images to a stack using maximal projection (this can be done either with the microscope software or using Image J). The nerve-ring will be clearly visible, count the green GFP:: LGG-1-positive punctae therein (i.e., between the two pharyngeal bulbs).
Document the obtained data for each tissue for DMSO-injected animals and BafA-injected animals (Fig. 2g).
3.4.4. Statistical Analysis
Statistical analysis of the number of GFP::LGG-1-positive punctae can be done using Student’s t-test if DMSO and BafA conditions are compared at a single timepoint (as in Fig. 2g). If more than one condition or strain is analyzed at a single timepoint, two-way ANOVA should be performed. Analysis of multiple timepoints need to be compared using extended statistical analysis, that is, Poisson regression [9].
Acknowledgments
The nematode strains used in this work can be obtained by the Caenorhabditis Genetics Center (University of Minnesota), which is supported by the NIH—Office of Research Infrastructure Programs (P40 OD010440). This work was funded by the Julie Martin Mid-Career Award in Aging Research from The Ellison Medical Foundation/AFAR (to M.H.), and by NIH grants AG058038 (to C.K.) and AG028664 (to M.H.).
4 Notes
Strains expressing dual-tagged LGG-1, that is, mCherry:: GFP::LGG-1, can also be used for BafA injections [9]:
qIs11[lgg-1p::mcherry::GFP::lgg-1 + rol-6] (MAH215) to assess autophagy flux in intestine, pharynx, and body-wall muscle.
sqEx67[rgef-1p::mcherry::GFP::lgg-1 + rol-6] (MAH508) (pick animals with Rol phenotype to maintain) to assess autophagy flux in nerve-ring neurons.
Both strains can be obtained from the Caenorhabditis Genetic Center (CGC).
These reporters inform on the pool size of autophagosomes (punctae positive for green and red fluorescence) as well as the pool size of autolysosomes (punctae positive for red fluorescence, since the fluorescence of pH-sensitive GFP will be quenched in the acidic environment of the lysosomes [15]). If autophagy is active, BafA injection may cause changes in the pools of either autophagosomes (punctae positive for green and red fluorescence) or autolysosomes (punctae positive for red fluorescence), or both. If autophagy is blocked, BafA injections should not change the pool sizes of autophagosomes or autolysosomes [9]. We note that the intensity of the red fluorescence compared to green is stronger and in order to see mCherry-positive punctae clearly, the gain of the red channel needs to be set lower than for the green channel. If the intensity of the red channel is increased to effectively overexpose red punctae, the cytoplasmic mCherry::GFP::LGG-1 signal will appear yellow [9]. The number of autolysosomes is calculated as the total number of punctae positive in the red channel subtracted by the number of punctae positive in the green channel, with the latter number of punctae representing the number of autophagosomes. We acknowledge that red punctae can also represent amphisomes, resulting from the fusion of autophagosomes with acidic endosomes [4].
The coinjection dye to use for BafA injection of animals carrying the tandem LGG-1 reporter is Dextran, Cascade Blue 3000 MW, Anionic Lysine Fixable (Thermo Fisher Scientific, Product number D7132).
Animals can be synchronized by hypochlorous acid treatment. Hypochlorous acid solution is made by mixing 150 ml of 5% (v/v) NaOCl (sodium hypochlorite, bleach) with 75 ml 5 N KOH and 275 ml H2O to a final volume of 500 ml. The solution is mixed and sterile filtered. The bleach is light sensitive, and the solution should be stored covered in aluminum foil at 4 °C. For hypochlorous acid treatment, gravid animals are washed off NGM plates using a 5–10 ml M9 solution and collected in a 15 ml conical tube. Centrifuge the 15 ml conical tube at low speed 1500–2000 × g for 30 s to gently pellet the animals. Aspirate the M9 medium leaving 200–500 μl of M9, depending on the size of the worm pellet. Add 10 ml of hypochlorous acid solution. Mix by inverting or rocking. Monitor under a stereoscope until worms release the eggs and dissolve. Centrifuge at 1500–2000 × g for 30 s. A white or clear egg pellet should be visible. Aspirate as much of the hypochlorous acid solution as possible without disturbing the egg pellet. Quickly add 10 ml of M9 medium and mix to remove the hypochlorous acid solution. Centrifuge at 1500–2000 × g for 30 s and aspirate most of the solution. Repeat the washing step one more time. Transfer the eggs onto an NGM media plate with OP50 bacteria immediately after bleaching or transfer eggs into a fresh 15 ml conical tube containing 3-5 ml M9 medium for overnight hatching. Transfer the hatched L1 larvae onto NGM plates with OP50 bacteria the next day.
Injection needles can be loaded with injection solution by capillary forces. Pipet 2–4 μl of the injection solution on the unpulled end of the injection needle and allow 5–7 min for collection of the solution in the needle tip. The disadvantage of this method is that air bubbles can be trapped in the needle tip, which will prevent injection.
The gfp::lgg-1 reporter is sensitive to the type of anesthetics used to mount the transgenic animals for imaging. In contrast to 150 mM sodium azide, 0.2–2 mM of Levamisole or Tetramisole hydrochloride leads to the formation of different sized GFP-positive punctae 5–10 min after mounting [11].
- Turn on the needle puller. Place the capillary on the ridge on the left side and slide toward and past the filament without touching the filament. Once the capillary has passed through the filament, tighten the screw.
- Push both levers toward the filament.
- Loosen the left screw just enough so that the capillary is still is held in the ridge but can slide along the ridge. Push the capillary further through the filament so that the middle of the capillary is right in the filament and the two pulled needles will have the same length.
- Tighten both screws and close the lid.
- Initiate needle pulling and the filament will heat up and melt the capillary. Once the glass is molten, the levers will snap back, resulting in two needles.
- Take out the needles without touching the tips and place them into a needle holder box (an empty 15 cm petri dish with a strip of modeling clay or wax to hold the needles).
- Check the needles under a dissecting scope to see whether they look correct.
The original version of this chapter was revised. The correction to this chapter is available at https://doi.org/10.1007/978-1-0716-0592-9_24
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451 (7182):1069–1075. 10.1038/nature06639.nature06639 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinsztein DC, Marino G, Kroemer G (2011) Autophagy and aging. Cell 146 (5):682–695. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- 3.Hansen M, Rubinsztein DC, Walker DW (2018) Autophagy as a promoter of longevity: insights from model organisms Nat Rev Mol Cell Biol 19:579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12 (1):1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manil-Segalen M, Lefebvre C, Jenzer C, Trichet M, Boulogne C, Satiat-Jeunemaitre B, Legouis R (2014) The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev Cell 28(1):43–55. 10.1016/j.devcel.2013.11.022 [DOI] [PubMed] [Google Scholar]
- 6.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301 (5638):1387–1391 [DOI] [PubMed] [Google Scholar]
- 7.Kang C, You YJ, Avery L (2007) Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev 21 (17):2161–2171. 10.1101/gad.1573107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelino S, Chang JT, Kumsta C, She X, Davis A, Nguyen C, Panowski S, Hansen M (2016) Intestinal autophagy improves healthspan and longevity in C. elegans during dietary restriction. PLoS Genet 12(7):e1006135. 10.1371/journal.pgen.1006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M (2017) Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife 6. 10.7554/eLife.18459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JT, Hansen M (2018) Age-associated and tissue-specific decline in autophagic activity in the nematode C. elegans. Autophagy 14 (7):1276–1277. 10.1080/15548627.2018.1445914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Chang JT, Guo B, Hansen M, Jia K, Kovacs AL, Kumsta C, Lapierre LR, Legouis R, Lin L, Lu Q, Melendez A, O’Rourke EJ, Sato K, Sato M, Wang X, Wu F (2015) Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy 11 (1):9–27. 10.1080/15548627.2014.1003478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumsta CCJ, Schmalz J, Hansen M (2017) Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat Commun 8:14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A, Hansen M (2015) Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol Cell 57(1):55–68. 10.1016/j.molcel.2014.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77(1):71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3 (5):452–460. 4451 [pii] [DOI] [PubMed] [Google Scholar]


