Abstract
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is the most common monogenic disorder of the cerebral small blood vessels. It is caused by mutations in the NOTCH3 gene on chromosome 19, and more than 280 distinct pathogenic mutations have been reported to date. CADASIL was once considered a very rare disease with an estimated prevalence of 1.3–4.1 per 100,000 adults. However, recent large-scale genomic studies have revealed a high prevalence of pathogenic NOTCH3 variants among the general population, with the highest risk being among Asians. The disease severity and age at onset vary significantly even among individuals who carry the same NOTCH3 mutations. It is still unclear whether a significant genotype–phenotype correlation is present in CADASIL. The accumulation of granular osmiophilic material in the vasculature is a characteristic feature of CADASIL. However, the exact pathogenesis of CADASIL remains largely unclear despite various laboratory and clinical observations being made. Major hypotheses proposed so far have included aberrant NOTCH3 signaling, toxic aggregation, and abnormal matrisomes. Several characteristic features have been observed in the brain magnetic resonance images of patients with CADASIL, including subcortical lacunar lesions and white matter hyperintensities in the anterior temporal lobe or external capsule, which were useful in differentiating CADASIL from sporadic stroke in patients. The number of lacunes and the degree of brain atrophy were useful in predicting the clinical outcomes of patients with CADASIL. Several promising blood biomarkers have also recently been discovered for CADASIL, which require further research for validation.
Keywords: CADASIL, NOTCH3 mutation, magnetic resonance imaging, biomarker
INTRODUCTION
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is the most common Mendelian disorder of the cerebral small blood vessels.1,2,3 It is caused by mutations of the NOTCH3 gene located on the short arm of chromosome 19 between bands 13.2 and 13.1.4,5 The human NOTCH3 gene consists of 33 exons that encode the single-pass transmembrane receptors comprising 2,321 amino acids.6 Most pathogenic variants are located in exons 2–24 that correspond to the 34 epidermal growth factor-like repeats (EGFRs) and lead to an odd number of cysteines within a given EGFR.3 CADASIL is microscopically characterized by abnormal accumulation of the NOTCH3 extracellular domain (NECD) around vascular smooth-muscle cells (VSMCs) and pericytes.7 It is still unclear whether impairment of canonical Notch signaling is responsible for inducing the disease process of CADASIL. On the other hand, accumulation of various extracellular matrix (ECM) proteins along with NECD has been suggested as a potential cause of small-vessel pathology in CADASIL.8 Unfortunately, the exact mechanism of the disorder has not yet been elucidated despite the causative gene being identified more than 25 years ago.
Until recently, CADASIL was regarded as a rare genetic disorder with an estimated prevalence of 1.3–4.1 per 100,000 among Caucasians.9,10,11,12 However, large-scale human genomic studies found a very high prevalence of pathogenic NOTCH3 variants in the general population (3.4 per 1,000 individuals), with the risk being highest among Asians.13,14 The ready availability of genetic diagnosis for the general population has resulted in more patients with CADASIL being identified, and several studies have investigated its clinical manifestations with the aim of predicting its clinical course. Unfortunately, it is still unclear whether a significant genotype–phenotype correlation is present in CADASIL because of large variations in disease onset and severity even within those with the same mutation or from the same family.15,16,17
Brain magnetic resonance imaging (MRI) is the most useful brain imaging tool for differential diagnoses, disease severity assessments, and outcome predictions for patients with CADASIL.18,19,20 Several characteristic MRI features can help distinguish CADASIL from sporadic ischemic stroke, and the number of lacunes and the degree of brain atrophy were very useful in predicting the clinical outcomes of patients with CADASIL. In cases when brain MRI is not available for various reasons, blood biomarkers may be very useful for screening subjects at a high risk of developing CADASIL, diagnosis of clinically probable patients without genetic evidence, clinical outcome and prognosis predictions, and serving as surrogate endpoints in clinical trials on CADASIL.21,22 Several promising blood biomarkers for CADASIL have been discovered recently, and further research is needed to validate their clinical usefulness.23,24
This review presents the recent developments in the epidemiology, genetics, clinical manifestations, pathogenesis, and biomarkers of CADASIL.
EPIDEMIOLOGY AND GENETICS
Historical overview: from pedigrees to the causal gene
The history of CADASIL can be traced back to 1977. Since then, an autosomal dominant disease presenting with relapsing strokes and gradually evolving into severe dementia in relatively young adults from several families has been reported using different names such as “hereditary multi-infarct dementia” and “chronic familial vascular encephalopathy.”25,26 In the early 1990s, Tournier-Lasserve et al.27 characterized CADASIL as an autosomal dominantly inherited disorder with recurrent subcortical stroke-like episodes and leukoencephalopathy based on an analysis of 45 family members from a four-generation pedigree in France. Autopsy of one of the affected family members revealed arteriopathy with concentric thickening of the vascular walls, granular osmiophilic material (GOM) deposition in the media and adventitia, and duplication of the internal elastic lamella.28 Based on the findings of these two pioneering studies,27,28 the disease is now known as CADASIL.
In 1996, several mutations in the NOTCH3 that encodes the NOTCH3 receptor were identified as the cause of CADASIL.5,29 The NOTCH3 gene encodes single-pass transmembrane proteins with extracellular domains composed of 34 tandem EGFRs. Each EGFR contains six cysteine residues that form three disulfide bonds (https://www.uniprot.org/uniprot/Q9UM47). More than 280 distinct pathogenic mutations of NOTCH3 have been reported to date. Most of these mutations are referred to as cysteine-altering, corresponding to mutations that cause an odd number of cysteine residues and consequently disrupt the conformation of EGFRs.6,30 In certain rare circumstances, missense variants that affect residues other than cysteine (i.e., “cysteine-sparing mutations”) and loss-of-function variants in NOTCH3 are considered as pathogenic.4,31,32,33 Detection of GOM deposits in skin biopsy34,35 and/or mutation–phenotype cosegregation in multiple members of the same family are prerequisites of supporting the pathogenicity of cysteine-sparing and loss-of-function variants in NOTCH3.
Disease prevalence and frequency of NOTCH3 mutations
CADASIL was once considered to be a very rare disease. Early epidemiological studies estimated that CADASIL prevalence in northeast England, west Scotland, Glasgow, and central Italy were 1.32, 1.98, 4.6, and 4.1 per 100,000 adults, respectively,9,10,11,12 and the frequencies of mutation carriers in these populations were predicted to range from 4.1 to 10.7 per 100,000 adults. These numbers may be underestimations since the studies were conducted retrospectively by reviewing registered cases, most of which were diagnosed during a time of restricted access to comprehensive genetic testing for NOTCH3.
Following advances in next-generation sequencing techniques, exome or genome databases have been established in many countries and provide excellent opportunities to assess the true prevalence of pathogenic NOTCH3 mutations in general populations (Table 1). Rutten et al.13 found that 3.4 per 1000 persons among 60,706 unrelated individuals in the Exome Aggregation Consortium (ExAC) database harbored a cysteine-altering mutation of NOTCH3 suggesting that the prevalence of CADASIL could be 100-fold higher than previously predicted. Grami et al.14 subsequently analyzed the 18 genes responsible for inherited stroke in 101,635 individuals from the Genome Aggregation Database, the upgraded version of the ExAC database, and found that the frequency of cysteine-altering mutations of NOTCH3 ranged from 0.67 to 11.78 per 1,000 persons among participants from 1 of 7 different ethnicities (Table 1). In line with these findings, 2.2 per 1,000 persons among 200,632 unrelated individuals in the UK Biobank and 1.4 per 1,000 persons among 92,456 participants in the MyCode Initiative cohort carried cysteine-altering mutations of NOTCH3.36,37
Table 1. Frequencies of cysteine-altering NOTCH3 mutations in different populations.
| Source | Regions or populations | Number of subjects (mutation carriers/number of screened individuals) | Frequency of mutation carrier* | |
|---|---|---|---|---|
| All cysteine-altering NOTCH3 mutations | Most frequent variant | |||
| gnomAD (Genome Aggregation Database)14 | South Asian | NA | 0.01178 | p.Arg1231Cys (0.0054) |
| East Asian | NA | 0.01117 | p.Arg544Cys (0.0037) | |
| Ashkenazi Jewish | NA | 0.00067 | p.Arg951Cys (0.00017) | |
| African/African-American | NA | 0.00087 | p.Arg717Cys (0.00013) | |
| Non-Finnish European | NA | 0.002 | p.Arg1231Cys (0.00032) | |
| Finnish European | NA | 0.00117 | p.Arg767Cys (0.0004) | |
| Latino/admixed American | NA | 0.00285 | p.Arg1231Cys (0.00066) | |
| UK Biobank36 | UK | 443/200,632 | 0.0022 | p.Arg1231Cys (0.0006) |
| The MyCode Initiative37 | Pennsylvania, USA | 131/92,456 | 0.0014 | p.Arg1231Cys (0.00091) |
| KRGDB (Korean Reference Genome Database)39 | South Korea | 2/1,722 | 0.0012 | p.Arg544Cys (0.0012) |
| Korea1K (The Korean Genome Project)39 | South Korea | 4/1,094 | 0.0044 | p.Arg544Cys (0.0044) |
| Jeju Island39 | South Korea | 10/1,000 | 0.01 | p.Arg544Cys (0.009) |
| Taiwan38 | Taiwan | 61/7,038 | 0.0087 | p.Arg544Cys (0.0085) |
*Frequency of mutation carriers=number of subjects carrying NOTCH3 cysteine-altering mutations/overall number of screened individuals.
NA, not available.
The frequencies of pathogenic NOTCH3 mutations in Asian populations appear to be even higher than those observed in European populations. Lee et al.38 analyzed 7,038 healthy adults in Taiwan and found that a single NOTCH3 mutation, p.Arg544Cys, was carried by 0.85% of the general population. Kang et al.39 found that the p.Arg544Cys mutation was also common in Jeju Island, South Korea, being found at a rate of 9 per 1,000 local residents. However, for South Korean people living outside Jeju Island, the estimated frequency of NOTCH3 mutation carriers was slightly lower: 1.2–4.4 per 1,000 persons.39 Considering the strikingly high global and regional frequencies of pathogenic NOTCH3 mutations, CADASIL should no longer be recognized as a rare disease. In fact, NOTCH3 mutations have been identified in 5.6%–6.5% of Taiwanese patients with small-vessel-occlusion stroke,38,40 3.5% of Japanese patients with acute lacunar infarction,41 4% of patients with acute ischemic stroke in Jeju Island, South Korea,42 and 0.5% of Caucasian patients younger than 70 years with lacunar infarction.43 More attention should therefore be paid to NOTCH3 mutations as important risk factors for stroke in general populations.
Mutation hotspots and founder effects
There are 33 exons in NOTCH3, but most of the pathogenic mutations that cause CADASIL are located on exons 2–24, which encode the 34 EGFR domains. In particular, exons 2–6 are considered mutation hotspots, though there are ethnic variations. In patients with CADASIL from France, UK, and Germany, 55%–72.9% of the mutations were located in exon 4 of NOTCH3, with exons 3, 5, and 6 being the next most common locations in order.6,44,45 For patients with CADASIL from Dutch families, half of the mutations were detected in exon 4% and 15% in exon 11.46 The most frequently detected pathogenic NOTCH3 mutations in Japan were p.Arg133Cys and p.Arg182Cys in exon 4, and p.Arg75Pro in exon 3, which contributed to 23%, 13%, and 13% of the patients with CADASIL, respectively.47 NOTCH3 p.Arg607Cys in exon 11 was the mutation hotspot of eastern China and accounted for one-fifth of the patients with CADASIL.48 In central Italy, the most frequently observed NOTCH3 mutation was p.Arg1006Cys in exon 19 (16.1%), followed by p.Arg1231Cys in exon 22 (12.2%), p.Arg607Cys in exon 11 (7.4%), and p.Arg528Cys in exon 10 (7.4%).10 The optimal strategy for performing mutation analysis of NOTCH3 is therefore population-specific, and it is crucial to investigate the mutation profiles of patients with CADASIL from different ethnic groups and geographic areas.
A single NOTCH3 mutation, p.Arg544Cys in exon 11, accounted for 90.3%, 70.5%, and 15.5% of the CADASIL pedigrees in Jeju Island, Taiwan, and southeastern China, respectively.16,48,49 The homozygous p.Arg544Cys mutation has been reported in a Japanese patient with CADASIL with consanguineous parents50 and four Taiwanese patients with CADASIL from three different families.16,51 Patients from Taiwan and Fujian that carried the p.Arg544Cys NOTCH3 mutation shared an identical haplotype and could be descendants of a common ancestor.16,48 A founder effect has also been demonstrated on the west coast of Finland, where 85.7% of the families with CADASIL harbored the p.Arg133Cys mutation.52 It was particularly interesting that a founder effect of p.Arg133Cys was also observed for most of the p.Arg133Cys-mutation carriers with CADASIL living in Kyushu, Japan.47 Moreover, the p.Arg75Pro NOTCH3 mutation has only been identified in patients with CADASIL from Japan, South Korea, and China, and not in those from European countries,39,47,53,54,55 implying that a founder effect of these mutations may exist in East Asia.
CLINICAL MANIFESTATIONS
Heterogeneity in clinical manifestations
The classical presentations of CADASIL comprise four main symptoms that occur in succession or in isolation: 1) migraine with aura that has an average age at onset of 30 years, 2) recurrent subcortical ischemic stroke with an average age at onset of 40–59 years, 3) mood and psychiatric disturbance, and 4) cognitive impairment with an average age at onset of 50–59 years.3,17,56,57 Gait disturbance, urinary incontinence, and pseudobulbar palsy are also frequently reported. Epileptic seizure occurs in 5%–10% of patients with CADASIL,17,57 while an acute-onset reversible encephalopathy called “CADASIL coma” affects 10% of patients.58,59 These classical symptoms were described based on observations of European patients with CADASIL in earlier studies. Subsequent studies involving East Asian patients found a higher incidence of intracerebral hemorrhage (ICH), a lower incidence of migraine, and an higher age at symptom onset.15,16,54,60,61 ICH has been reported in only 0.5%–2.4% of European patients,62,63 but was found in 17%–25% and 21% of South Korean and Taiwanese patients, respectively, most of whom carried the p.Arg544Cys mutation.15,60,61 Cerebral microbleeds (CMBs) on MRI were also more common in Asian than Caucasian patients (66%–73.5% vs. 25%–60%).60,61,62,64,65,66 The presence of CMBs in the brain stem and a total CMB count of >10 were independently associated with an increased risk of ICH in CADASIL.61 In contrast, the prevalence of migraine was markedly lower in Asian patients with CADASIL than in Caucasian patients (2.7%–28.4% vs. 42%–75%).10,16,44,48,54,67,68 One recent study screened 2,884 Taiwanese patients with migraine, including 324 with migraine with aura, and 3,502 controls for NOTCH3 p.Arg544Cys mutation. The study identified 32 (1.1%) patients with migraine and 36 (1.0%) controls who carried this mutation,69 suggesting that p.Arg544Cys does not increase the risk of migraine with aura or of migraine as a whole. Both ethnicity and different NOTCH3 mutations might contribute to the clinical heterogeneity of CADASIL.
Variable disease severity
It has been noted that disease severity and age at the first symptom onset may vary dramatically among individuals who carry different NOTCH3 mutations (Fig. 1). In earlier European studies,3,17,56,57 the average ages of the first ischemic event were 41.2–49.3 years, with a range between 20 and 70 years for patients with CADASIL who mostly carried a NOTCH3 mutation in exons 2–6. However, subsequent reports indicated that ischemic events in CADASIL can be very subtle, with the first event often occurring at an extremely advanced age. For example, Pescini et al.70 reported a patient with CADASIL carrying the p.Cys1131Trp mutation who experienced their first minor stroke at 79 years old. Lee et al.71 analogously reported a male who carried the p.Arg544Cys mutation and had experienced their first lacunar stroke symptom at 86 years old, and three asymptomatic p.Arg544Cys carriers who only presented subclinical leukoencephalopathy on MRI at 59–67 years old.38 There is increasing evidence that late-onset CADASIL with a mild phenotype is not rare, and the phenotypic spectrum of NOTCH3 mutations has now expanded to include mild cerebral small-vessel disease (SVD), an attenuated and delayed-onset CADASIL phenotype, and the classical CADASIL with middle-age-onset stroke and dementia.13,37,72,73
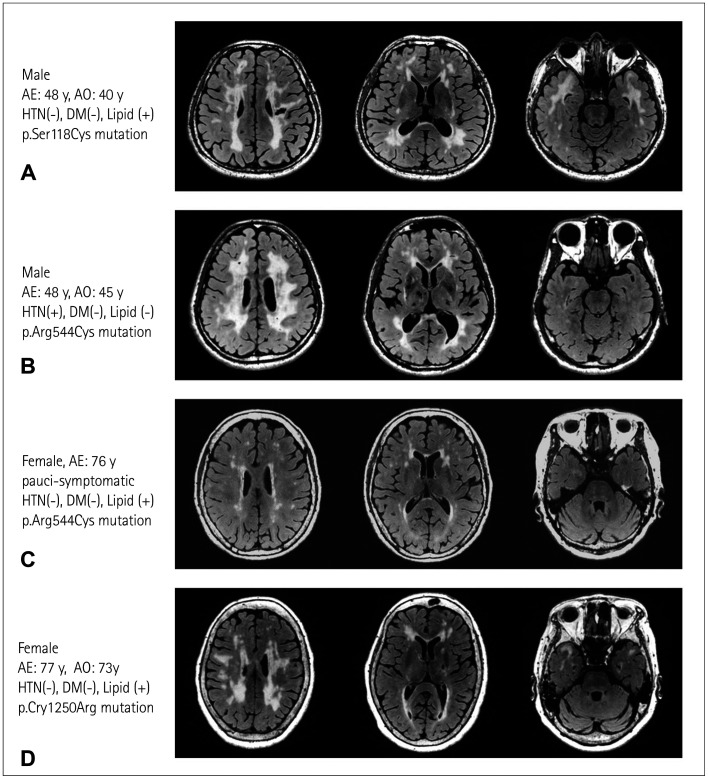
Fig. 1. Variable neuroimaging severity among patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) carrying different NOTCH3 mutations. Representative fluid-attenuated inversion recovery (FLAIR) images showing (A) typical magnetic resonance imaging (MRI) features including lacunar infarct, diffuse white-matter hyperintensities (WMHs) involving the corona radiata, periventricular regions, and anterior temporal poles in a patient with CADASIL carrying a heterozygous NOTCH3 p.Ser118Cys mutation located within epidermal growth factor-like repeat (EGFR)2 of the NOTCH3 protein; (B and C) variable severity of WMHs without anterior temporal pole involvement in a middle-age-onset patient with CADASIL and an elderly paucisymptomatic individual, both of whom carried a heterozygous p.Arg544Cys NOTCH3 mutation that altered the amino acid residue between EGFR13 and EGFR14; and (D) typical MRI features of CADASIL in a late-onset patient with a heterozygous p.Cys1250Arg mutation located within the EGFR32. AE, age at examination; AO, age at stroke onset; HTN, hypertension; DM, diabetes mellitus; lipid, hypercholesterolemia.
Genotype–phenotype correlations have been observed in a group of disparate NOTCH3 mutations. For example, the p.Arg544Cys mutation seems to be associated with a mild disease severity and late disease onset. Compared with patients carrying other NOTCH3 mutations, Taiwanese patients with CADASIL carrying the p.Arg544Cys mutation had a 9.1-year delay to the first symptom onset and a lower percentage of white-matter hyperintensities (WMHs) in the anterior temporal pole.16 Similar findings were also obtained in the cysteine-sparing NOTCH3 mutation p.Arg75Pro. The age at symptom onset was much higher among Japanese patients with CADASIL harboring p.Arg75Pro than those with other NOTCH3 mutations (53.6 years vs. 44.2 years), and anterior temporal pole was involved in fewer p.Arg75Pro carriers than in subjects with other mutations (29% vs. 77%).47
The theory of a “NOTCH3 mutation position effect” recently emerged in an attempt to explain the vast phenotypic variations in CADASIL. This theory was deduced from a study in the Netherlands that found that patients with CADASIL and pathogenic NOTCH3 mutations affecting one of the first six EGFR domains had an earlier stroke onset by 12 years, a shorter survival time of 8.4 years, and a higher WMH volume on MRI when compared with patients who carried mutations altering other EGFR domains.73 A similar trend was also observed in Japanese patients with CADASIL, in whom the median age at first stroke/transient ischemic attack was significantly lower in patients who harbored NOTCH3 cysteine-altering mutations in EGFRs 1–6 than in subjects who carried mutations in EGFRs 7–34 (52 years vs. 55 years).74 The milder aggregation properties of mutant NOTCH3 proteins might be one of the factors leading to a milder phenotype in patients with CADASIL who carry the cysteine-altering mutations in EGFRs 7–34, who were found to have less extracellular NOTCH3 aggregation and GOM deposits.75
While the positions of NOTCH3 mutations may substantially contribute to the severity of CADASIL, there is still an obvious phenotypic variation among patients who carry the same NOTCH3 mutation, even among those from the same ethnic group. Choi et al.15 examined 15 South Korean patients with CADASIL carrying the p.Arg544Cys mutation from Jeju Island, and found a greatly varied disease onset age range of 43–66 years. The 79 patients with the p.Arg544Cys mutation in a Taiwanese CADASIL cohort also presented a wide range of ages at the first ischemic event, of 25–86 years.16 The large variation in onset ages in patients with the same ethnic background carrying the identical mutation imply that some other genetic or environmental factors could modify the severity of CADASIL. Further studies that target actionable factors that may modify the disease course and alleviate the symptoms of CADASIL will be valuable.
PATHOGENESIS OF CADASIL
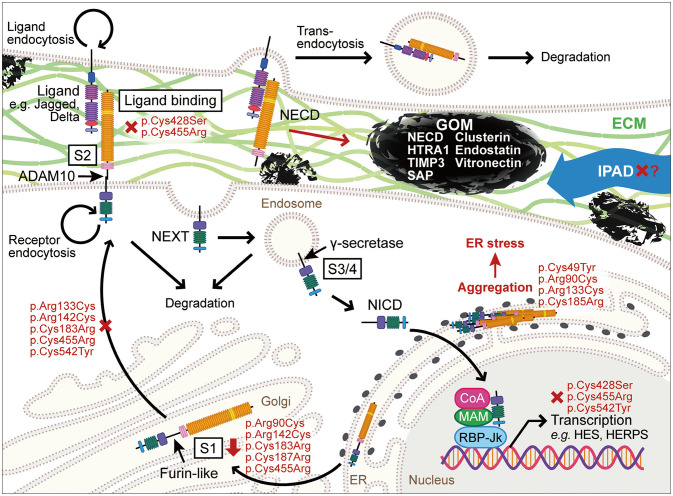
The pathogenesis of CADASIL remains largely obscure despite various hints and clues provided by cell culture and animal model studies as well as clinical observations. Major hypotheses proposed so far have included aberrant NOTCH3 signaling, toxic NECD aggregation, and matrisomes (Fig. 2).
Fig. 2. Pathogenesis of CADASIL-causing NOTCH3 mutations. There is currently still no consensus on the pathogenesis of CADASIL. Major hypotheses proposed so far are aberrant NOTCH3 signaling, toxic NOTCH3 extracellular domain (NECD) aggregation, and matrisomes. ECM, extracellular matrix; ER, endoplasmic reticulum; GOM, granular osmiophilic material; IPAD, intramural periarterial drainage; NEXT, Notch3 extracellular truncation; NICD, Notch3 intracellular domain.
Aberrant NOTCH3 signaling
The NOTCH receptor comprises a large NECD with 34 EGFR and 3 cysteine-rich Lin12/Notch repeats, a transmembrane domain, and the Notch3 intracellular domain (NICD).76 Translated NOTCH3 is cleaved by furin-like convertase at the S1 site to form a heterodimer before presentation at the cell surface.77 Ligand binding induces a proteolytic cleavage at S2 by ADAM10 and transendocytosis of the NECD.78 Cleavage by γ-secretase releases NICD, which then enters the nucleus to activate target genes. In humans, the expressions of five ligands (Jagged [Jag]-1, Jag-2, Delta-like [Dll]-1, Dll-3, and Dll-4) differ significantly depending on cell type, and Jag-1 is primarily expressed in VSMCs.79
Since most of the mutations that cause CADASIL alter the number of cysteine residues in the NECD, which leads to a three-dimensional conformational change, it was reasonable to suspect that loss of NOTCH3 signaling activity could play a role. Various studies have examined the receptor processing and function of mutant NOTCH3, but so far they have failed to reach a consistent conclusion (Table 2). NOTCH3 signaling activity was significantly reduced by impairments of ligand binding due to the mutations in the ligand-binding domain (p.Cys428Ser and p.Cys455Arg) and receptor presentation (p.Cys542Tyr).80,81 However, many other mutations presented with no abnormalities in the overall, maturation, ligand-interaction, and signal-transduction processes.80,81,82,83 Several studies have detected altered S1 cleavage of mutant NOTCH3/Notch3 (p.Arg133Cys, p.Cys183Arg, and mouse p.Arg142Cys and p.Cys187Arg).81,84,85 The reduced ratio of S1-cleaved mutant (p.Arg142Cys) receptors to full-length receptors may be caused by impaired receptor trafficking, which leads to intracellular aggregation and reduced cell-surface presentation, although the ligand-induced signaling itself remains intact.84 In the case of the rat Notch3 p.Cys187Arg mutation, receptor presentation and ligand binding were unaffected, but mutant Notch3 induced increased signaling activation by Jag-1 and reduced activation by Dll-1 compared with wild-type Notch3.85 To complicate the matter, the NOTCH3-activating mutation p.Leu1515Pro in the juxtamembrane region near S1 has been found in CADASIL-like cerebral SVD without GOM deposits nor NECD immunoreactivity in the vasculature.86 The mutant NOTCH3 reduced S1 cleavage but destabilized the NOTCH3 heterodimer, resulting in increased S2 cleavage and NECD shredding in a ligand-independent manner.
Table 2. Effects of NOTCH3 mutations on the NOTCH3 signaling pathway.
| EGFR | Mutation | Receptor processing | Receptor presentation | Ligand binding | Signal transduction |
|---|---|---|---|---|---|
| 2 | p.Arg90Cys76,79 | ○ | ○ | ○Jag-1 | ○RBP-Jk (Jag-1) |
| 3 | p.Arg133Cys77,79 | ○/S1↓ | ○ or delayed | ○Jag-1 | ○RBP-Jk (Jag-1) |
| 3 | Mouse p.Arg142Cys (human p.Arg141Cys)80 | S1↓ | x | ○ | ○RBP-Jk (Dll-1, Jag-1) |
| 4 | Rat p.Arg171Cys78 | ○ | ○ | ○Dll-1 | - |
| 4 | p.Cys183Arg77 | S1↓ | delayed | ○Jag-1 | ○RBP-Jk (Jag-1) |
| 4 | Rat p.His184Cys78 | ○ | ○ | ○Dll-1 | - |
| 4 | p.Cys185Arg79 | ○ | ○ | ○ | ○ |
| 4 | Rat p.Cys187Arg (human p.Cys185Arg)81 | S1↓ | ○ | ○Dll-1, Dll-4, Jag-1 | CBF1 (Dll-1↓, Jag-1↑) |
| 5 | p.Cys212Ser76 | ○ | ○ | ○Jag-1 | ○RBP-Jk (Jag-1) |
| 10 | p.Cys428Ser76 | ○ | ○ | x | ↓RBP-Jk (Jag-1) |
| 11 | p.Arg449Cys79 | ○ | ○ | ○ | ○ |
| 11 | p.Cys455Arg77 | S1↓ | delayed | x | ↓RBP-Jk (Jag-1) |
| 13 | p.Cys542Tyr76 | ○ | x | ○Jag-1 | ↓RBP-Jk (Jag-1) |
| 13 | Rat p.Cys544Tyr78 | ○ | ○ | ○Dll-1 | - |
| 14 | Rat p.Arg560Cys78 | ○ | ○ | ○Dll-1 | - |
| 26 | p.Arg1006Cys76 | ○ | ○ | ○Jag-1 | ○RBP-Jk (Jag-1) |
| p.Leu1515Pro82 | S1↓, S2↑ | - | - | ↑RBP/Jk (ligand independent) |
о and x denote normal and impaired, respectively. S1, S1 cleavage; Jag-1, Jagged -1; Dll-1, Delta -1; Dll-4, Delta -4.
EGFR, epidermal growth factor-like repeat.
These conflicting data suggest that small-vessel pathology is at least partially attributable to the disruption of tightly controlled NOTCH3 signaling: timely and swift on-and-off switching of NOTCH3 signaling with a sufficient (but not excessive) signal strength would be key to forming and maintaining healthy arteries and arterioles. However, CADASIL pathogenesis cannot be fully explained by uncontrolled NOTCH3 signaling alone, which has led researchers to shift their focus to GOM deposition.
Toxic NECD aggregation
The accumulation of GOM deposition in the vasculature is a prominent and characteristic feature of CADASIL. The components of GOM had long been a mystery until Ishiko et al.7 demonstrated the presence of NECD but not NICD in GOM. Several studies have found that mutant NOTCH3 is prone to forming multimeric complexes partially due to the formation of disulfide bridges between unpaired cysteine residues in mutant NECD.87,88,89 Young et al.88 found that some of the NOTCH3 mutations in the first three EGFRs result in an increased probability of unpaired cysteines at a few specific positions near the mutation site, indicating that the unpaired cysteine interferes with the other normal disulfide pairing to produce multiple unpaired cysteines, further contributing to multimerization. Intracellular clearance of these multimers was impaired, partly due to the dysfunction of the autophagylysosomal pathway, which induces endoplasmic reticulum (ER) stress and eventually cell death.90,91,92 ER stress caused by mutant NOTCH3 also induces RhoA/Rho kinase activation and leads to abnormal VSMC growth and cytoskeletal reorganization, which may underlie the reduced vasodilation and vasoreactivity of arteries in CADASIL.93
In addition to the direct toxicity of intracellular NECD accumulation, extracellular NECD multimers (GOM), may also disrupt VSMC function by repressing NOTCH3-regulated smooth-muscle transcripts in a dominant negative manner.90 However, a study involving mutant NOTCH3 transgenic mice found that morphological abnormalities of VSMCs preceded GOM deposition and that there was no correlation between the presence/amount of GOM and degenerating VSMC,94 which questions the magnitude of their contribution to the abnormality in VSMC. Another possibility is that GOM contributes to the pathogenesis of CADASIL by physically blocking intramural periarterial drainage (IPAD), just as Aβ aggregates do in Alzheimer’s disease and cerebral amyloid angiopathy. IPAD is a key fluid drainage pathway that removes harmful metabolites from the interstitial fluid along capillary and artery walls to the cervical lymph nodes at the base of skull.95 NECD-positive GOM is found in the tunica media and basement membrane of the capillaries, arterioles and meningeal arteries, which are IPAD routes.96,97 The enlarged perivascular spaces in CADASIL may indicate IPAD disruption.98 Nonetheless, considering that the driving force of IPAD is considered to be artery pulsation,99,100 impaired vasoreactivity may be the primary cause of vascular GOM deposition, which then reduces the efficiency of IPAD and results in white-matter degeneration.
Matrisome theory
It was particularly interesting that high-temperature requirement protein A1 (HTRA1), the gene mutation of which is responsible for CARASIL (cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy), is also present in GOM deposits.101,102 HTRA1 is a serine protease that represses transforming growth factor-β (TGF-β) signaling pathway by degrading pro-TGF-β and type-II TGF-β receptor.103,104 HTRA1 mutations in CARASIL are considered to be loss-of-function or hypomorphic mutations, leading to the activation of TGF-β signaling that causes rarefaction of the mural cells in the capillaries and arteries.102,105,106 HTRA1 substrate accumulation in the CADASIL vasculature and increased TGF-β expression in VSMC suggest that recruitment of HTRA1 to GOM may interfere with the normal function of and results in HTRA1 loss-of-function phenotype just like in CARASIL.101,107,108,109 The other components of GOM, namely clusterin, endostatin, vitronectin, tissue inhibitors of metalloproteinases 3 (TIMP3), serum amyloid P component (SAP), and latent transforming growth factor binding protein 1 (LTBP-1),110,111,112,113,114 are also related to the ECM and TGF-β signaling pathway, or so-called matrisome.115 Matrisome is a term that encompasses core ECM proteins (collagens, proteoglycans, and glycoproteins) and ECM-associated proteins (secreted factors, ECM regulators, and ECM-affiliated proteins).116 Several other hereditary diseases with small-vessel pathology (e.g., COL4A1/A2-related SVD and Fabry disease), have also been directly or indirectly linked to matrisome abnormality, which implies its involvement in the development of small-vessel pathology.8,115 Regardless of whether ECM protein deposition is the primary cause or an adaptive consequence, the resulting arterial stenosis certainly exacerbates the cerebral hypoperfusion and contributes to the increased susceptibility to ischemic insult.
Environmental factors
The phenotypes and severity of CADASIL can vary in a family with the same mutation and even with the same genetic background, which indicates the importance of environmental factors in its pathogenesis.117,118 Prospective cohort studies have identified hypertension and smoking as factors associated with increased risks of stroke and dementia.67,119 Both hypertension and smoking are vascular risk factors linked to arterial stiffness and impaired vascular reactivity.120,121 Arterial stiffness was previously classically recognized as a consequence of ECM deposition in the artery walls, but VSMC phenotypic modulation/dedifferentiation has recently been considered to be another contributing factor.122 A few studies have found that hypertension and smoking can cause VSMC to change from a contractile to a synthetic-like phenotype.123,124 Such phenotypic change would further exacerbate already malfunctioning IPAD and accelerate the disease progression.
Collectively, these data suggest that the CADASIL pathogenesis is difficult to explain by a single clear-cut mechanism. Considering the time course of CADASIL development, it is plausible that impaired vasoreactivity is the primary cause of its pathology, although the mechanism–abnormal NOTCH3 signaling or intracellular NECD accumulation–still needs to be investigated further.
BIOMARKERS FOR CADASIL
Brain imaging
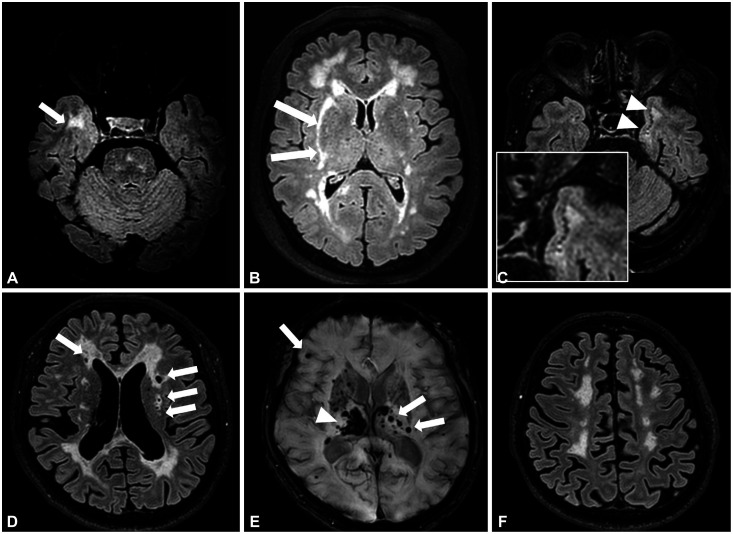
CADASIL is a genetic model of pure subcortical vascular dementia. Brain MRI markers for cerebral SVD such as lacunes, WMHs, enlarged perivascular space, CMBs, and brain atrophy are frequently found in patients with CADASIL, and their topographic characteristics are important in distinguishing CADASIL from sporadic SVD (Fig. 3).19,125,126,127 Of the SVD markers on imaging, WMHs in the anterior temporal lobe or external capsule are more characteristic of CADASIL than are sporadic ischemic WMHs. The sensitivity and specificity of the anterior temporal lobe lesions, which are present even in young patients aged 20–30 years, were 89%–90% and 86%–100% for a CADASIL diagnosis, respectively.19,44,127 Compared to WMHs in other regions, those found in the anterior temporal areas were more severe even in young patients. External capsule involvement exhibited a sensitivity of 93% and specificity of 45%. However, studies on East Asians found lesser involvement of anterior temporal WMHs (20.0%–70.5%).15,16,47,48,128 Subcortical lacunar lesions refer to linearly arranged groups of round circumscribed lesions just below the cortex at the junction between the gray matter and the white matter with a signal intensity that was identical to that of the cerebrospinal fluid on all pulse sequences, and they were first identified in a brain MRI study of 54 Dutch patients with CADASIL.129 The subcortical lacunar lesions were found in 59% of those patients and were frequently observed in the anterior temporal lobes and in areas where diffuse WMHs expanded into arcuate fibers. In contrast, these lesions were not observed in other conditions associated with SVD. The specificity and sensitivity of subcortical lacunar lesions for CADASIL diagnosis were 100% and 59%, respectively.129 The sensitivity and specificity of subcortical lacunar lesions have unfortunately not been validated in other study populations.
Fig. 3. Characteristic brain MRI features of CADASIL. Axial FLAIR MRI images of a 59-year-old male present prominent WMHs in the right anterior temporal lobe (arrow) (A). WMHs involving right external capsule (arrows) were noted in a 66-year-old female (B). Axial FLAIR MRI images of a 68-year-old male presented subcortical lacunar lesions at the junction between the white matter and the gray matter in the left anterior temporal lobe (arrowheads and the enlarged figure in the white box) (C). Multiple lacunar infarctions (arrows) at the bilateral subcortical white matter (D). Several cerebral microbleeds (arrows) with a right thalamic hematoma (arrowhead) (E) and marked brain atrophy (F). All patients had the same p.Arg544Cys mutation.
Cross-sectional studies of brain SVD markers have found the number of lacunes to be independently associated with disability and cognitive dysfunction in patients with CADASIL, while the lesion load of WMHs and CMBs were not associated with cognitive dysfunction after correcting for the effect of age.18,130 Brain atrophy is a strong predictor of dementia in patients with subcortical ischemic vascular disease and it was also found to be independently associated with age and lacunar lesion volume but not with WMHs or CMBs in CADASIL.131,132 The degree of brain atrophy was strongly correlated with cognitive dysfunction and disability in patients with CADASIL in cross-sectional analyses.132,133
In a longitudinal study, the number of lacunes and brain parenchymal fraction (an indicator of brain atrophy) at baseline independently predicted incident stroke during 3 years of follow-up in 290 patients with CADASIL.20 The volume of lacunes was also a strong predictor of 3-year changes in cognitive dysfunction and disability.134 In the same study, brain parenchymal fraction also predicted incident stroke, dementia, and disability in patients with CADASIL. However, the volume of WMHs or CMBs was not associated with incident stroke or dementia. In a study on 369 patients from the Paris-Munich cohort with a median follow-up time of 39 months, CMB presence was associated with a 1.87-fold increased risk of incident stroke (95% confidence interval=1.10–3.26) after adjusting for demographic and clinical variables.62 However, the presence of CMBs was not a significant predictor of incident stroke after further adjusting for other MRI markers including the volume of lacunes and brain parenchymal fraction.
There are several limitations to using brain MRI to research CADASIL. Firstly, the repeated examinations required in longitudinal follow-ups are expensive. Secondly, quantitative MRI measurements that are useful for monitoring CADASIL severity are often time-consuming and require specialized software. Thirdly, MRI examinations cannot be performed on patients who are uncooperative due to advanced disease or with a pacemaker or claustrophobia.
Blood biomarkers
Several promising blood biomarkers have been identified in patients with CADASIL and in a mouse model of CADASIL (Table 3).23,24,135,136,137 Neurofilaments are the main element of the neuroaxonal cytoskeleton and consist of light, intermediate, and heavy chains with different sizes. They are released into the extracellular fluid when axons are damaged, and they can be measured in either the cerebrospinal fluid or blood.138,139 Serum neurofilament light chain (NfL) was significantly elevated in patients with CADASIL compared with healthy controls, and its level was correlated with age in both patients with CADASIL and healthy controls.23 The area under the curve for differentiating CADASIL from controls was 0.75, which increased to 0.85 after adjusting for age. The age-related increase in NfL level was more prominent in patients with CADASIL than in healthy controls. In particular, there was a sharp increase in serum NfL levels from 40 years old, which is when lacunes often start to develop in patients with CADASIL.23 The serum NfL level had a strong correlation with brain MRI markers for SVD even after adjusting for age.136 It was also associated with the presence of ischemic or hemorrhagic stroke, neurological deficit, cognitive dysfunction, and overall disability at baseline, and was useful for predicting incident stroke, cognitive decline, disability, or even mortality in patients with CADASIL.23,140 However, the NfL level was also elevated in sporadic cerebral SVD and other various neurological disorders, and it is therefore not specific to CADASIL.141,142
Table 3. Biomarkers found in blood or tissue in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL).
| Name | Origin | Findings in CADASIL | Biological function | |
|---|---|---|---|---|
| Blood biomarkers | ||||
| Neurofilament light chain23,132,136 | Patients with CADASIL | Elevated in serum Associated with disability Predicted stroke and cognitive decline |
Main element of neuroaxonal cytoskeleton | |
| NECD24,133 | Mouse model of CADASIL and patients with CADASIL | Decreased in blood Correlated with white-matter hyperintensities |
Unknown | |
| HTRA124 | Mouse model of CADASIL | Elevated | Serine protease transforming-growth-factor signaling | |
| Endostatin24 | Mouse model of CADASIL | Elevated | Angiogenesis inhibitor | |
| Biomarkers found in proteomic analysis of brain vessels | ||||
| Serum amyloid P component106,109,110 | Postmortem or biopsied brain vessels of patients with CADASIL | Elevated and colocalized with NOTCH3 | Amyloid formation | |
| TIMP3110 | Postmortem brain vessels of patients with CADASIL | Elevated and colocalized with NOTCH3 | Matrix metalloproteinases inhibitor | |
| Vitronectin106,109,110 | Postmortem or biopsied brain vessels of patients with CADASIL | Elevated and colocalized with NOTCH3 | Cell adhesion | |
| Neurofilament110 | Postmortem brain vessels of patients with CADASIL | Decreased | Main element of neuroaxonal cytoskeleton | |
| Neurofascin110 | Postmortem brain vessels of patients with CADASIL | Decreased | Links the ECM to the intracellular cytoskeleton | |
| Internexin α110 | Postmortem brain vessels of patients with CADASIL | Decreased | Neuronal intermediate filament protein | |
| Solute carrier family 4110 | Postmortem brain vessels of patients with CADASIL | Decreased | Membrane transport | |
| Smooth-muscle myosin heavy chain110 | Postmortem brain vessels of patients with CADASIL | Elevated | Smooth-muscle contraction | |
| Other ECM proteins106,109,110 | Postmortem or biopsied brain vessels of patients with CADASIL | Elevated | ECM components | |
ECM, extracellular matrix; HTRA, high-temperature requirement protein A1; NECD, NOTCH3 extracellular domain; TIMP3, tissue inhibitors of metalloproteinases 3.
In a mouse model of CADASIL (p.Cys455Arg), the plasma NECD level was significantly lower in mutant than control mice.24 NECD was also detected in human blood, and its level was significantly higher in the serum than the plasma. In an earlier in vitro study, monoclonal antibody treatment, which activates Notch3 receptors independently from ligand binding, caused a significant increase in the NECD released in the supernatant of the cells that expressed wild-type or mutant Notch3.143 Low serum NECD levels in a CADASIL animal model could therefore be suggestive of impaired Notch signaling. However, low serum NECD could be caused by impaired NECD clearance in CADASIL, because it is well known that mutant NECD tend to accumulate in aggregates between VSMCs, and the mutant NOTCH3 displayed significantly enhanced multimerization with less solubility and delayed clearance compared with wild-type NOTCH3 in previous in vitro studies.87,90 Gao et al.137 recently found that the NOTCH3 protein level in plasma exosomes was significantly lower in 30 patients with CADASIL than in 30 age-matched healthy controls. They also reported that the plasma exosome NOTCH3 protein levels were positively correlated with WMH severity measured on the Fazekas scale. However, it was not correlated with cognitive scores in patients with CADASIL. In addition to NECD, the mouse model also indicated significantly increased plasma levels of endostatin and HTRA1 compared with control mice.24 Endostatin is a proteolytic fragment of collagen18α1 and has been identified as a component of GOM.114 The clinical utility of those potential blood biomarkers needs to be validated in large-scale human CADASIL studies.
Biomarkers found in brain vessels
Proteomic analyses of human brain arteries have disclosed several important proteins with levels that differ significantly between patients with CADASIL and controls. The molecular functions or biological processes of the proteins included ECM constituents, cytoskeleton, protein processing and vesicular traffic, and cell adhesion.144 In a comparative postmortem proteome profiling analysis of two brain arteries in patients with the p.Arg1031Cys NOTCH3 mutation, Arboleda-Velasquez et al.114 identified 19 proteins whose levels differed significantly between patients with CADASIL and controls. Most of them were ECM proteins that were significantly increased in patients with CADASIL such as collagen 1α2, collagen12α1, collagen14α1, collagen18α1, laminin α5, laminin γ1, lactadherin, clusterin, vinculin, leucine-rich repeat proteoglycan, and perlecan. Other proteins that were decreased in patients with CADASIL were neurofilament, neurofascin, internexin α, and solute carrier family 4.
As described above, several proteins have been identified as components of GOM deposits. In the brain vessels from seven postmortem French patients with CADASIL (mean age=63 years), 72 proteins that were mostly ECM proteins were only present in CADASIL samples.110 To investigate the proteins that were expressed in the early disease stage, those authors also performed a proteomic analysis of brain arteries from 10- to 12-month-old transgenic mice. In both human and mice samples enriched with NECD, TIMP3 and vitronectin were present in CADASIL and were almost absent in controls.110 TIMP3 and vitronectin were colocalized with NECD on immunohistochemical staining. In brain vessels, TIMP3 activity was significantly increased in postmortem patients with CADASIL compared with controls. Serum TIMP3 and related matrix metalloproteinase levels have not yet been investigated in patients with CADASIL. SAP, annexin A2, and periostin were also significantly elevated in the GOM-enriched leptomeningeal or superficial temporal arteries of three Japanese patients with CADASIL compared with controls.113 On immunofluorescence staining, only SAP was colocalized with NOTCH3 in the leptomeningeal arteries, which was also observed in subcutaneous blood vessels in patients with CADASIL, while there was weak staining of SAP in the perivascular region of the control subject.
CONCLUSIONS AND FUTURE PERSPECTIVES
Recent large-scale genomic studies have found the prevalence of pathogenic NOTCH3 mutations to be much higher than previously thought, with the risk being highest among Asians. Pathogenic NOTCH3 mutations frequently found in general populations were located in EGFRs 7–34, while most of these mutations found in patients with CADASIL were located in EGFRs 1–6. Nonetheless, the pathogenic NOTCH3 mutation frequently found in general populations was associated with an increased SVD burden on brain MRI and a higher stroke risk. Despite the increasing awareness of the disorder, the exact pathomechanism of CADASIL has not yet been elucidated. Active research using large-scale human CADASIL cohorts as well as cell culture and mouse model studies are underway to understand how NOTCH3 mutations lead to damage of the cerebral small blood vessels. In addition to brain MRI, several promising blood biomarkers have been identified that can be useful in screening high-risk populations and monitoring the disease severity in the future.
Footnotes
- Conceptualization: Jay Chol Choi, Masafumi Ihara, Yi-Chung Lee.
- Methodology: Jay Chol Choi, Masafumi Ihara, Yi-Chung Lee, Yi-Chu Liao, Yumi Yamamoto.
- Visualization: Jay Chol Choi, Yumi Yamamoto, Yi-Chung Lee, Yi-Chu Liao.
- Writing—original draft: Jay Chol Choi, Masafumi Ihara, Yi-Chung Lee, Yi-Chu Liao, Yumi Yamamoto.
- Writing—review & editing: Jay Chol Choi, Masafumi Ihara, Yi-Chung Lee, Yi-Chu Liao, Yumi Yamamoto.
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Funding Statement: This work was supported by the 2022 education, research and student guidance grant funded by Jeju National University.
Availability of Data and Material
Data sharing not applicable to this article as no datasets were generated or analyzed during the study.
References
- 1.Choi JC. Genetics of cerebral small vessel disease. J Stroke. 2015;17:7–16. doi: 10.5853/jos.2015.17.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi JC. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: a genetic cause of cerebral small vessel disease. J Clin Neurol. 2010;6:1–9. doi: 10.3988/jcn.2010.6.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. Cadasil. Lancet Neurol. 2009;8:643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 4.Tikka S, Mykkänen K, Ruchoux MM, Bergholm R, Junna M, Pöyhönen M, et al. Congruence between NOTCH3 mutations and GOM in 131 CADASIL patients. Brain. 2009;132(Pt 4):933–939. doi: 10.1093/brain/awn364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 6.Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssière C, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–1515. doi: 10.1016/S0140-6736(97)08083-5. [DOI] [PubMed] [Google Scholar]
- 7.Ishiko A, Shimizu A, Nagata E, Takahashi K, Tabira T, Suzuki N. Notch3 ectodomain is a major component of granular osmiophilic material (GOM) in CADASIL. Acta Neuropathol. 2006;112:333–339. doi: 10.1007/s00401-006-0116-2. [DOI] [PubMed] [Google Scholar]
- 8.Joutel A, Haddad I, Ratelade J, Nelson MT. Perturbations of the cerebrovascular matrisome: a convergent mechanism in small vessel disease of the brain? J Cereb Blood Flow Metab. 2016;36:143–157. doi: 10.1038/jcbfm.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razvi SS, Davidson R, Bone I, Muir KW. The prevalence of cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL) in the west of Scotland. J Neurol Neurosurg Psychiatry. 2005;76:739–741. doi: 10.1136/jnnp.2004.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi S, Zicari E, Carluccio A, Di Donato I, Pescini F, Nannucci S, et al. CADASIL in central Italy: a retrospective clinical and genetic study in 229 patients. J Neurol. 2015;262:134–141. doi: 10.1007/s00415-014-7533-2. [DOI] [PubMed] [Google Scholar]
- 11.Moreton FC, Razvi SS, Davidson R, Muir KW. Changing clinical patterns and increasing prevalence in CADASIL. Acta Neurol Scand. 2014;130:197–203. doi: 10.1111/ane.12266. [DOI] [PubMed] [Google Scholar]
- 12.Narayan SK, Gorman G, Kalaria RN, Ford GA, Chinnery PF. The minimum prevalence of CADASIL in northeast England. Neurology. 2012;78:1025–1027. doi: 10.1212/WNL.0b013e31824d586c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutten JW, Dauwerse HG, Gravesteijn G, van Belzen MJ, van der Grond J, Polke JM, et al. Archetypal NOTCH3 mutations frequent in public exome: implications for CADASIL. Ann Clin Transl Neurol. 2016;3:844–853. doi: 10.1002/acn3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grami N, Chong M, Lali R, Mohammadi-Shemirani P, Henshall DE, Rannikmäe K, et al. Global assessment of Mendelian stroke genetic prevalence in 101 635 individuals from 7 ethnic groups. Stroke. 2020;51:1290–1293. doi: 10.1161/STROKEAHA.119.028840. [DOI] [PubMed] [Google Scholar]
- 15.Choi JC, Kang SY, Kang JH, Park JK. Intracerebral hemorrhages in CADASIL. Neurology. 2006;67:2042–2044. doi: 10.1212/01.wnl.0000246601.70918.06. [DOI] [PubMed] [Google Scholar]
- 16.Liao YC, Hsiao CT, Fuh JL, Chern CM, Lee WJ, Guo YC, et al. Characterization of CADASIL among the Han Chinese in Taiwan: distinct genotypic and phenotypic profiles. PLoS One. 2015;10:e0136501. doi: 10.1371/journal.pone.0136501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dichgans M, Mayer M, Uttner I, Brüning R, Müller-Höcker J, Rungger G, et al. The phenotypic spectrum of CADASIL: clinical findings in 102 cases. Ann Neurol. 1998;44:731–739. doi: 10.1002/ana.410440506. [DOI] [PubMed] [Google Scholar]
- 18.Liem MK, van der Grond J, Haan J, van den Boom R, Ferrari MD, Knaap YM, et al. Lacunar infarcts are the main correlate with cognitive dysfunction in CADASIL. Stroke. 2007;38:923–928. doi: 10.1161/01.STR.0000257968.24015.bf. [DOI] [PubMed] [Google Scholar]
- 19.O’Sullivan M, Jarosz JM, Martin RJ, Deasy N, Powell JF, Markus HS. MRI hyperintensities of the temporal lobe and external capsule in patients with CADASIL. Neurology. 2001;56:628–634. doi: 10.1212/wnl.56.5.628. [DOI] [PubMed] [Google Scholar]
- 20.Chabriat H, Hervé D, Duering M, Godin O, Jouvent E, Opherk C, et al. Predictors of clinical worsening in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: prospective cohort study. Stroke. 2016;47:4–11. doi: 10.1161/STROKEAHA.115.010696. [DOI] [PubMed] [Google Scholar]
- 21.Jickling GC, Sharp FR. Biomarker panels in ischemic stroke. Stroke. 2015;46:915–920. doi: 10.1161/STROKEAHA.114.005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilar-Bergua A, Riba-Llena I, Nafría C, Bustamante A, Llombart V, Delgado P, et al. Blood and CSF biomarkers in brain subcortical ischemic vascular disease: involved pathways and clinical applicability. J Cereb Blood Flow Metab. 2016;36:55–71. doi: 10.1038/jcbfm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravesteijn G, Rutten JW, Verberk IMW, Böhringer S, Liem MK, van der Grond J, et al. Serum neurofilament light correlates with CADASIL disease severity and survival. Ann Clin Transl Neurol. 2019;6:46–56. doi: 10.1002/acn3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Primo V, Graham M, Bigger-Allen AA, Chick JM, Ospina C, Quiroz YT, et al. Blood biomarkers in a mouse model of CADASIL. Brain Res. 2016;1644:118–126. doi: 10.1016/j.brainres.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Sonninen V, Savontaus ML. Hereditary multi-infarct dementia. Eur Neurol. 1987;27:209–215. doi: 10.1159/000116158. [DOI] [PubMed] [Google Scholar]
- 26.Stevens DL, Hewlett RH, Brownell B. Chronic familial vascular encephalopathy. Lancet. 1977;1:1364–1365. doi: 10.1016/s0140-6736(77)92576-4. [DOI] [PubMed] [Google Scholar]
- 27.Tournier-Lasserve E, Iba-Zizen MT, Romero N, Bousser MG. Autosomal dominant syndrome with strokelike episodes and leukoencephalopathy. Stroke. 1991;22:1297–1302. doi: 10.1161/01.str.22.10.1297. [DOI] [PubMed] [Google Scholar]
- 28.Baudrimont M, Dubas F, Joutel A, Tournier-Lasserve E, Bousser MG. Autosomal dominant leukoencephalopathy and subcortical ischemic stroke. A clinicopathological study. Stroke. 1993;24:122–125. doi: 10.1161/01.str.24.1.122. [DOI] [PubMed] [Google Scholar]
- 29.Ducros A, Nagy T, Alamowitch S, Nibbio A, Joutel A, Vahedi K, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, genetic homogeneity, and mapping of the locus within a 2-cM interval. Am J Hum Genet. 1996;58:171–181. [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuno T, Mizuta I, Watanabe-Hosomi A, Mukai M, Koizumi T. Clinical and genetic aspects of CADASIL. Front Aging Neurosci. 2020;12:91. doi: 10.3389/fnagi.2020.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muiño E, Gallego-Fabrega C, Cullell N, Carrera C, Torres N, Krupinski J, et al. Systematic review of cysteine-sparing NOTCH3 missense mutations in patients with clinical suspicion of CADASIL. Int J Mol Sci. 2017;18:1964. doi: 10.3390/ijms18091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moccia M, Mosca L, Erro R, Cervasio M, Allocca R, Vitale C, et al. Hypomorphic NOTCH3 mutation in an Italian family with CADASIL features. Neurobiol Aging. 2015;36:547.e5–547.e11. doi: 10.1016/j.neurobiolaging.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 33.Pippucci T, Maresca A, Magini P, Cenacchi G, Donadio V, Palombo F, et al. Homozygous NOTCH3 null mutation and impaired NOTCH3 signaling in recessive early-onset arteriopathy and cavitating leukoencephalopathy. EMBO Mol Med. 2015;7:848–858. doi: 10.15252/emmm.201404399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruchoux MM, Maurage CA. CADASIL: cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. J Neuropathol Exp Neurol. 1997;56:947–964. [PubMed] [Google Scholar]
- 35.Joutel A, Favrole P, Labauge P, Chabriat H, Lescoat C, Andreux F, et al. Skin biopsy immunostaining with a Notch3 monoclonal antibody for CADASIL diagnosis. Lancet. 2001;358:2049–2051. doi: 10.1016/S0140-6736(01)07142-2. [DOI] [PubMed] [Google Scholar]
- 36.Cho BPH, Nannoni S, Harshfield EL, Tozer D, Gräf S, Bell S, et al. NOTCH3 variants are more common than expected in the general population and associated with stroke and vascular dementia: an analysis of 200,000 participants. J Neurol Neurosurg Psychiatry. 2021;92:694–701. doi: 10.1136/jnnp-2020-325838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hack RJ, Rutten JW, Person TN, Li J, Khan A, Griessenauer CJ, et al. Cysteine-altering NOTCH3 variants are a risk factor for stroke in the elderly population. Stroke. 2020;51:3562–3569. doi: 10.1161/STROKEAHA.120.030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YC, Chung CP, Chang MH, Wang SJ, Liao YC. NOTCH3 cysteine-altering variant is an important risk factor for stroke in the Taiwanese population. Neurology. 2020;94:e87–e96. doi: 10.1212/WNL.0000000000008700. [DOI] [PubMed] [Google Scholar]
- 39.Kang CH, Kim YM, Kim YJ, Hong SJ, Kim DY, Woo HG, et al. Pathogenic NOTCH3 variants are frequent among the Korean general population. Neurol Genet. 2021;7:e639. doi: 10.1212/NXG.0000000000000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang SC, Chen YR, Chi NF, Chen CH, Cheng YW, Hsieh FI, et al. Prevalence and clinical characteristics of stroke patients with p.R544C NOTCH3 mutation in Taiwan. Ann Clin Transl Neurol. 2018;6:121–128. doi: 10.1002/acn3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada T, Washida K, Irie K, Saito S, Noguchi M, Tomita T, et al. Prevalence and atypical clinical characteristics of NOTCH3 mutations among patients admitted for acute lacunar infarctions. Front Aging Neurosci. 2020;12:130. doi: 10.3389/fnagi.2020.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi JC, Lee KH, Song SK, Lee JS, Kang SY, Kang JH. Screening for NOTCH3 gene mutations among 151 consecutive Korean patients with acute ischemic stroke. J Stroke Cerebrovasc Dis. 2013;22:608–614. doi: 10.1016/j.jstrokecerebrovasdis.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Kilarski LL, Rutten-Jacobs LC, Bevan S, Baker R, Hassan A, Hughes DA, et al. Prevalence of CADASIL and Fabry disease in a cohort of MRI defined younger onset lacunar stroke. PLoS One. 2015;10:e0136352. doi: 10.1371/journal.pone.0136352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markus HS, Martin RJ, Simpson MA, Dong YB, Ali N, Crosby AH, et al. Diagnostic strategies in CADASIL. Neurology. 2002;59:1134–1138. doi: 10.1212/wnl.59.8.1134. [DOI] [PubMed] [Google Scholar]
- 45.Opherk C, Peters N, Herzog J, Luedtke R, Dichgans M. Long-term prognosis and causes of death in CADASIL: a retrospective study in 411 patients. Brain. 2004;127(Pt 11):2533–2539. doi: 10.1093/brain/awh282. [DOI] [PubMed] [Google Scholar]
- 46.Oberstein SA. Diagnostic strategies in CADASIL. Neurology. 2003;60:2020. [PubMed] [Google Scholar]
- 47.Ueda A, Ueda M, Nagatoshi A, Hirano T, Ito T, Arai N, et al. Genotypic and phenotypic spectrum of CADASIL in Japan: the experience at a referral center in Kumamoto University from 1997 to 2014. J Neurol. 2015;262:1828–1836. doi: 10.1007/s00415-015-7782-8. [DOI] [PubMed] [Google Scholar]
- 48.Chen S, Ni W, Yin XZ, Liu HQ, Lu C, Zheng QJ, et al. Clinical features and mutation spectrum in Chinese patients with CADASIL: a multicenter retrospective study. CNS Neurosci Ther. 2017;23:707–716. doi: 10.1111/cns.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi JC, Song SK, Lee JS, Kang SY, Kang JH. Diversity of stroke presentation in CADASIL: study from patients harboring the predominant NOTCH3 mutation R544C. J Stroke Cerebrovasc Dis. 2013;22:126–131. doi: 10.1016/j.jstrokecerebrovasdis.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Mukai M, Mizuta I, Ueda A, Nakashima D, Kushimura Y, Noto YI, et al. A Japanese CADASIL patient with homozygous NOTCH3 p.Arg-544Cys mutation confirmed pathologically. J Neurol Sci. 2018;394:38–40. doi: 10.1016/j.jns.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 51.Soong BW, Liao YC, Tu PH, Tsai PC, Lee IH, Chung CP, et al. A homozygous NOTCH3 mutation p.R544C and a heterozygous TREX1 variant p.C99MfsX3 in a family with hereditary small vessel disease of the brain. J Chin Med Assoc. 2013;76:319–324. doi: 10.1016/j.jcma.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Mykkänen K, Savontaus ML, Juvonen V, Sistonen P, Tuisku S, Tuominen S, et al. Detection of the founder effect in Finnish CADASIL families. Eur J Hum Genet. 2004;12:813–819. doi: 10.1038/sj.ejhg.5201221. [DOI] [PubMed] [Google Scholar]
- 53.Kim YE, Yoon CW, Seo SW, Ki CS, Kim YB, Kim JW, et al. Spectrum of NOTCH3 mutations in Korean patients with clinically suspicious cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Neurobiol Aging. 2014;35:726.e1–726.e6. doi: 10.1016/j.neurobiolaging.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Shindo A, Tabei KI, Taniguchi A, Nozaki H, Onodera O, Ueda A, et al. A nationwide survey and multicenter registry-based database of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy in Japan. Front Aging Neurosci. 2020;12:216. doi: 10.3389/fnagi.2020.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Y, Sun Q, Zhou Y, Yi F, Tang H, Yao L, et al. NOTCH3 variants and genotype-phenotype features in Chinese CADASIL patients. Front Genet. 2021;12:705284. doi: 10.3389/fgene.2021.705284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chabriat H, Vahedi K, Iba-Zizen MT, Joutel A, Nibbio A, Nagy TG, et al. Clinical spectrum of CADASIL: a study of 7 families. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Lancet. 1995;346:934–939. doi: 10.1016/s0140-6736(95)91557-5. [DOI] [PubMed] [Google Scholar]
- 57.Desmond DW, Moroney JT, Lynch T, Chan S, Chin SS, Mohr JP. The natural history of CADASIL: a pooled analysis of previously published cases. Stroke. 1999;30:1230–1233. doi: 10.1161/01.str.30.6.1230. [DOI] [PubMed] [Google Scholar]
- 58.Schon F, Martin RJ, Prevett M, Clough C, Enevoldson TP, Markus HS. “CADASIL coma”: an underdiagnosed acute encephalopathy. J Neurol Neurosurg Psychiatry. 2003;74:249–252. doi: 10.1136/jnnp.74.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drazyk AM, Tan RYY, Tay J, Traylor M, Das T, Markus HS. Encephalopathy in a large cohort of British cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy patients. Stroke. 2019;50:283–290. doi: 10.1161/STROKEAHA.118.023661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JS, Ko K, Oh JH, Park JH, Lee HK, Floriolli D, et al. Cerebral microbleeds, hypertension, and intracerebral hemorrhage in cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Front Neurol. 2017;8:203. doi: 10.3389/fneur.2017.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao YC, Hu YC, Chung CP, Wang YF, Guo YC, Tsai YS, et al. Intracerebral hemorrhage in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: prevalence, clinical and neuroimaging features and risk factors. Stroke. 2021;52:985–993. doi: 10.1161/STROKEAHA.120.030664. [DOI] [PubMed] [Google Scholar]
- 62.Puy L, De Guio F, Godin O, Duering M, Dichgans M, Chabriat H, et al. Cerebral microbleeds and the risk of incident ischemic stroke in CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) Stroke. 2017;48:2699–2703. doi: 10.1161/STROKEAHA.117.017839. [DOI] [PubMed] [Google Scholar]
- 63.Nannucci S, Rinnoci V, Pracucci G, MacKinnon AD, Pescini F, Adib-Samii P, et al. Location, number and factors associated with cerebral microbleeds in an Italian-British cohort of CADASIL patients. PLoS One. 2018;13:e0190878. doi: 10.1371/journal.pone.0190878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lesnik Oberstein SA, van den Boom R, van Buchem MA, van Houwelingen HC, Bakker E, Vollebregt E, et al. Cerebral microbleeds in CADASIL. Neurology. 2001;57:1066–1070. doi: 10.1212/wnl.57.6.1066. [DOI] [PubMed] [Google Scholar]
- 65.Dichgans M, Holtmannspötter M, Herzog J, Peters N, Bergmann M, Yousry TA. Cerebral microbleeds in CADASIL: a gradient-echo magnetic resonance imaging and autopsy study. Stroke. 2002;33:67–71. doi: 10.1161/hs0102.100885. [DOI] [PubMed] [Google Scholar]
- 66.Chung CP, Chen JW, Chang FC, Li WC, Lee YC, Chen LF, et al. Cerebral microbleed burdens in specific brain regions are associated with disease severity of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. J Am Heart Assoc. 2020;9:e016233. doi: 10.1161/JAHA.120.016233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adib-Samii P, Brice G, Martin RJ, Markus HS. Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: study in 200 consecutively recruited individuals. Stroke. 2010;41:630–634. doi: 10.1161/STROKEAHA.109.568402. [DOI] [PubMed] [Google Scholar]
- 68.Choi JC, Song SK, Lee JS, Kang SY, Kang JH. Headache among CADASIL patients with R544C mutation: prevalence, characteristics, and associations. Cephalalgia. 2014;34:22–28. doi: 10.1177/0333102413497598. [DOI] [PubMed] [Google Scholar]
- 69.Wang YF, Liao YC, Tzeng YS, Chen SP, Lirng JF, Fuh JL, et al. Mutation screening and association analysis of NOTCH3 p.R544C in patients with migraine with or without aura. Cephalalgia. 2022;42:888–898. doi: 10.1177/03331024221080891. [DOI] [PubMed] [Google Scholar]
- 70.Pescini F, Bianchi S, Salvadori E, Poggesi A, Dotti MT, Federico A, et al. A pathogenic mutation on exon 21 of the NOTCH3 gene causing CADASIL in an octogenarian paucisymptomatic patient. J Neurol Sci. 2008;267:170–173. doi: 10.1016/j.jns.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 71.Lee YC, Yang AH, Soong BW. The remarkably variable expressivity of CADASIL: report of a minimally symptomatic man at an advanced age. J Neurol. 2009;256:1026–1027. doi: 10.1007/s00415-009-5048-z. [DOI] [PubMed] [Google Scholar]
- 72.Rutten JW, Hack RJ, Duering M, Gravesteijn G, Dauwerse JG, Overzier M, et al. Broad phenotype of cysteine-altering NOTCH3 variants in UK Biobank: CADASIL to nonpenetrance. Neurology. 2020;95:e1835–e1843. doi: 10.1212/WNL.0000000000010525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rutten JW, Van Eijsden BJ, Duering M, Jouvent E, Opherk C, Pantoni L, et al. The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1-6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7-34 pathogenic variant. Genet Med. 2019;21:676–682. doi: 10.1038/s41436-018-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukai M, Mizuta I, Watanabe-Hosomi A, Koizumi T, Matsuura J, Hamano A, et al. Genotype-phenotype correlations and effect of mutation location in Japanese CADASIL patients. J Hum Genet. 2020;65:637–646. doi: 10.1038/s10038-020-0751-9. [DOI] [PubMed] [Google Scholar]
- 75.Gravesteijn G, Hack RJ, Mulder AA, Cerfontaine MN, van Doorn R, Hegeman IM, et al. NOTCH3 variant position is associated with NOTCH3 aggregation load in CADASIL vasculature. Neuropathol Appl Neurobiol. 2022;48:e12751. doi: 10.1111/nan.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bellavia D, Checquolo S, Campese AF, Felli MP, Gulino A, Screpanti I. Notch3: from subtle structural differences to functional diversity. Oncogene. 2008;27:5092–5098. doi: 10.1038/onc.2008.230. [DOI] [PubMed] [Google Scholar]
- 77.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Groot AJ, Habets R, Yahyanejad S, Hodin CM, Reiss K, Saftig P, et al. Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol Cell Biol. 2014;34:2822–2832. doi: 10.1128/MCB.00206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hofmann JJ, Iruela-Arispe ML. Notch signaling in blood vessels: who is talking to whom about what? Circ Res. 2007;100:1556–1568. doi: 10.1161/01.RES.0000266408.42939.e4. [DOI] [PubMed] [Google Scholar]
- 80.Joutel A, Monet M, Domenga V, Riant F, Tournier-Lasserve E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am J Hum Genet. 2004;74:338–347. doi: 10.1086/381506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peters N, Opherk C, Zacherle S, Capell A, Gempel P, Dichgans M. CADASIL-associated Notch3 mutations have differential effects both on ligand binding and ligand-induced Notch3 receptor signaling through RBP-Jk. Exp Cell Res. 2004;299:454–464. doi: 10.1016/j.yexcr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 82.Haritunians T, Boulter J, Hicks C, Buhrman J, DiSibio G, Shawber C, et al. CADASIL Notch3 mutant proteins localize to the cell surface and bind ligand. Circ Res. 2002;90:506–508. doi: 10.1161/01.res.0000013796.73742.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Low WC, Santa Y, Takahashi K, Tabira T, Kalaria RN. CADASIL-causing mutations do not alter Notch3 receptor processing and activation. Neuroreport. 2006;17:945–949. doi: 10.1097/01.wnr.0000223394.66951.48. [DOI] [PubMed] [Google Scholar]
- 84.Karlström H, Beatus P, Dannaeus K, Chapman G, Lendahl U, Lundkvist J. A CADASIL-mutated Notch 3 receptor exhibits impaired intracellular trafficking and maturation but normal ligand-induced signaling. Proc Natl Acad Sci U S A. 2002;99:17119–17124. doi: 10.1073/pnas.252624099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haritunians T, Chow T, De Lange RP, Nichols JT, Ghavimi D, Dorrani N, et al. Functional analysis of a recurrent missense mutation in Notch3 in CADASIL. J Neurol Neurosurg Psychiatry. 2005;76:1242–1248. doi: 10.1136/jnnp.2004.051854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fouillade C, Chabriat H, Riant F, Mine M, Arnoud M, Magy L, et al. Activating NOTCH3 mutation in a patient with small-vessel-disease of the brain. Hum Mutat. 2008;29:452. doi: 10.1002/humu.9527. [DOI] [PubMed] [Google Scholar]
- 87.Opherk C, Duering M, Peters N, Karpinska A, Rosner S, Schneider E, et al. CADASIL mutations enhance spontaneous multimerization of NOTCH3. Hum Mol Genet. 2009;18:2761–2767. doi: 10.1093/hmg/ddp211. [DOI] [PubMed] [Google Scholar]
- 88.Young KZ, Rojas Ramírez C, Keep SG, Gatti JR, Lee SJ, Zhang X, et al. Oligomerization, trans-reduction, and instability of mutant NOTCH3 in inherited vascular dementia. Commun Biol. 2022;5:331. doi: 10.1038/s42003-022-03259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duering M, Karpinska A, Rosner S, Hopfner F, Zechmeister M, Peters N, et al. Co-aggregate formation of CADASIL-mutant NOTCH3: a single-particle analysis. Hum Mol Genet. 2011;20:3256–3265. doi: 10.1093/hmg/ddr237. [DOI] [PubMed] [Google Scholar]
- 90.Meng H, Zhang X, Yu G, Lee SJ, Chen YE, Prudovsky I, et al. Biochemical characterization and cellular effects of CADASIL mutants of NOTCH3. PLoS One. 2012;7:e44964. doi: 10.1371/journal.pone.0044964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takahashi K, Adachi K, Yoshizaki K, Kunimoto S, Kalaria RN, Watanabe A. Mutations in NOTCH3 cause the formation and retention of aggregates in the endoplasmic reticulum, leading to impaired cell proliferation. Hum Mol Genet. 2010;19:79–89. doi: 10.1093/hmg/ddp468. [DOI] [PubMed] [Google Scholar]
- 92.Hanemaaijer ES, Panahi M, Swaddiwudhipong N, Tikka S, Winblad B, Viitanen M, et al. Autophagy-lysosomal defect in human CADASIL vascular smooth muscle cells. Eur J Cell Biol. 2018;97:557–567. doi: 10.1016/j.ejcb.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 93.Neves KB, Harvey AP, Moreton F, Montezano AC, Rios FJ, Alves-Lopes R, et al. ER stress and Rho kinase activation underlie the vasculopathy of CADASIL. JCI Insight. 2019;4:e131344. doi: 10.1172/jci.insight.131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruchoux MM, Domenga V, Brulin P, Maciazek J, Limol S, Tournier-Lasserve E, et al. Transgenic mice expressing mutant Notch3 develop vascular alterations characteristic of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Am J Pathol. 2003;162:329–342. doi: 10.1016/S0002-9440(10)63824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carare RO, Aldea R, Agarwal N, Bacskai BJ, Bechman I, Boche D, et al. Clearance of interstitial fluid (ISF) and CSF (CLIC) group-part of vascular professional interest area (PIA): cerebrovascular disease and the failure of elimination of amyloid-β from the brain and retina with age and Alzheimer’s disease-opportunities for therapy. Alzheimers Dement (Amst) 2020;12:e12053. doi: 10.1002/dad2.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamamoto Y, Craggs LJ, Watanabe A, Booth T, Attems J, Low RW, et al. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol. 2013;72:416–431. doi: 10.1097/NEN.0b013e31829020b5. [DOI] [PubMed] [Google Scholar]
- 97.Weller RO, Djuanda E, Yow HY, Carare RO. Lymphatic drainage of the brain and the pathophysiology of neurological disease. Acta Neuropathol. 2009;117:1–14. doi: 10.1007/s00401-008-0457-0. [DOI] [PubMed] [Google Scholar]
- 98.Yamamoto Y, Ihara M, Tham C, Low RW, Slade JY, Moss T, et al. Neuropathological correlates of temporal pole white matter hyperintensities in CADASIL. Stroke. 2009;40:2004–2011. doi: 10.1161/STROKEAHA.108.528299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aldea R, Weller RO, Wilcock DM, Carare RO, Richardson G. Cerebrovascular smooth muscle cells as the drivers of intramural periarterial drainage of the brain. Front Aging Neurosci. 2019;11:1. doi: 10.3389/fnagi.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schley D, Carare-Nnadi R, Please CP, Perry VH, Weller RO. Mechanisms to explain the reverse perivascular transport of solutes out of the brain. J Theor Biol. 2006;238:962–974. doi: 10.1016/j.jtbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 101.Zellner A, Scharrer E, Arzberger T, Oka C, Domenga-Denier V, Joutel A, et al. CADASIL brain vessels show a HTRA1 loss-of-function profile. Acta Neuropathol. 2018;136:111–125. doi: 10.1007/s00401-018-1853-8. [DOI] [PubMed] [Google Scholar]
- 102.Hara K, Shiga A, Fukutake T, Nozaki H, Miyashita A, Yokoseki A, et al. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N Engl J Med. 2009;360:1729–1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- 103.Graham JR, Chamberland A, Lin Q, Li XJ, Dai D, Zeng W, et al. Serine protease HTRA1 antagonizes transforming growth factor-β signaling by cleaving its receptors and loss of HTRA1 in vivo enhances bone formation. PLoS One. 2013;8:e74094. doi: 10.1371/journal.pone.0074094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shiga A, Nozaki H, Yokoseki A, Nihonmatsu M, Kawata H, Kato T, et al. Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-β1 via cleavage of proTGF-β1. Hum Mol Genet. 2011;20:1800–1810. doi: 10.1093/hmg/ddr063. [DOI] [PubMed] [Google Scholar]
- 105.Shiga A, Nozaki H, Nishizawa M, Onodera O. [Molecular pathogenesis of cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy] Brain Nerve. 2010;62:595–599. Japanese. [PubMed] [Google Scholar]
- 106.Kato T, Sekine Y, Nozaki H, Uemura M, Ando S, Hirokawa S, et al. Excessive production of transforming growth factor β1 causes mural cell depletion from cerebral small vessels. Front Aging Neurosci. 2020;12:151. doi: 10.3389/fnagi.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.An E, Sen S, Park SK, Gordish-Dressman H, Hathout Y. Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest Ophthalmol Vis Sci. 2010;51:3379–3386. doi: 10.1167/iovs.09-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beaufort N, Scharrer E, Kremmer E, Lux V, Ehrmann M, Huber R, et al. Cerebral small vessel disease-related protease HtrA1 processes latent TGF-β binding protein 1 and facilitates TGF-β signaling. Proc Natl Acad Sci U S A. 2014;111:16496–16501. doi: 10.1073/pnas.1418087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Panahi M, Yousefi Mesri N, Samuelsson EB, Coupland KG, Forsell C, Graff C, et al. Differences in proliferation rate between CADASIL and control vascular smooth muscle cells are related to increased TGFβ expression. J Cell Mol Med. 2018;22:3016–3024. doi: 10.1111/jcmm.13534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Monet-Leprêtre M, Haddad I, Baron-Menguy C, Fouillot-Panchal M, Riani M, Domenga-Denier V, et al. Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain. 2013;136(Pt 6):1830–1845. doi: 10.1093/brain/awt092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kast J, Hanecker P, Beaufort N, Giese A, Joutel A, Dichgans M, et al. Sequestration of latent TGF-β binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol Commun. 2014;2:96. doi: 10.1186/s40478-014-0096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delaney CE, Weagant BT, Addison CL. The inhibitory effects of endostatin on endothelial cells are modulated by extracellular matrix. Exp Cell Res. 2006;312:2476–2489. doi: 10.1016/j.yexcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 113.Nagatoshi A, Ueda M, Ueda A, Tasaki M, Inoue Y, Ma Y, et al. Serum amyloid P component: a novel potential player in vessel degeneration in CADASIL. J Neurol Sci. 2017;379:69–76. doi: 10.1016/j.jns.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 114.Arboleda-Velasquez JF, Manent J, Lee JH, Tikka S, Ospina C, Vanderburg CR, et al. Hypomorphic Notch 3 alleles link Notch signaling to ischemic cerebral small-vessel disease. Proc Natl Acad Sci U S A. 2011;108:E128–E135. doi: 10.1073/pnas.1101964108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yamamoto Y, Ihara M. Disruption of transforming growth factor-β superfamily signaling: a shared mechanism underlying hereditary cerebral small vessel disease. Neurochem Int. 2017;107:211–218. doi: 10.1016/j.neuint.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 116.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11:M111.014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mykkänen K, Junna M, Amberla K, Bronge L, Kääriäinen H, Pöyhönen M, et al. Different clinical phenotypes in monozygotic CADASIL twins with a novel NOTCH3 mutation. Stroke. 2009;40:2215–2218. doi: 10.1161/STROKEAHA.108.528661. [DOI] [PubMed] [Google Scholar]
- 118.Liu X, Zuo Y, Sun W, Zhang W, Lv H, Huang Y, et al. The genetic spectrum and the evaluation of CADASIL screening scale in Chinese patients with NOTCH3 mutations. J Neurol Sci. 2015;354:63–69. doi: 10.1016/j.jns.2015.04.047. [DOI] [PubMed] [Google Scholar]
- 119.Singhal S, Bevan S, Barrick T, Rich P, Markus HS. The influence of genetic and cardiovascular risk factors on the CADASIL phenotype. Brain. 2004;127(Pt 9):2031–2038. doi: 10.1093/brain/awh223. [DOI] [PubMed] [Google Scholar]
- 120.Safar ME, Asmar R, Benetos A, Blacher J, Boutouyrie P, Lacolley P, et al. Interaction between hypertension and arterial stiffness. Hypertension. 2018;72:796–805. doi: 10.1161/HYPERTENSIONAHA.118.11212. [DOI] [PubMed] [Google Scholar]
- 121.Rogers RL, Meyer JS, Shaw TG, Mortel KF, Thornby J. The effects of chronic cigarette smoking on cerebrovascular responsiveness to 5 per cent CO2 and 100 per cent O2 inhalation. J Am Geriatr Soc. 1984;32:415–420. doi: 10.1111/j.1532-5415.1984.tb02215.x. [DOI] [PubMed] [Google Scholar]
- 122.Lacolley P, Regnault V, Segers P, Laurent S. Vascular smooth muscle cells and arterial stiffening: relevance in development, aging, and disease. Physiol Rev. 2017;97:1555–1617. doi: 10.1152/physrev.00003.2017. [DOI] [PubMed] [Google Scholar]
- 123.Yoshiyama S, Chen Z, Okagaki T, Kohama K, Nasu-Kawaharada R, Izumi T, et al. Nicotine exposure alters human vascular smooth muscle cell phenotype from a contractile to a synthetic type. Atherosclerosis. 2014;237:464–470. doi: 10.1016/j.atherosclerosis.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 124.Arribas SM, González C, Graham D, Dominiczak AF, McGrath JC. Cellular changes induced by chronic nitric oxide inhibition in intact rat basilar arteries revealed by confocal microscopy. J Hypertens. 1997;15(12 Pt 2):1685–1693. doi: 10.1097/00004872-199715120-00073. [DOI] [PubMed] [Google Scholar]
- 125.Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12:822–838. doi: 10.1016/S1474-4422(13)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chabriat H, Levy C, Taillia H, Iba-Zizen MT, Vahedi K, Joutel A, et al. Patterns of MRI lesions in CADASIL. Neurology. 1998;51:452–457. doi: 10.1212/wnl.51.2.452. [DOI] [PubMed] [Google Scholar]
- 127.van den Boom R, Lesnik Oberstein SA, Ferrari MD, Haan J, van Buchem MA. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: MR imaging findings at different ages--3rd-6th decades. Radiology. 2003;229:683–690. doi: 10.1148/radiol.2293021354. [DOI] [PubMed] [Google Scholar]
- 128.Kim Y, Choi EJ, Choi CG, Kim G, Choi JH, Yoo HW, et al. Characteristics of CADASIL in Korea: a novel cysteine-sparing Notch3 mutation. Neurology. 2006;66:1511–1516. doi: 10.1212/01.wnl.0000216259.99811.50. [DOI] [PubMed] [Google Scholar]
- 129.van Den Boom R, Lesnik Oberstein SA, van Duinen SG, Bornebroek M, Ferrari MD, Haan J, et al. Subcortical lacunar lesions: an MR imaging finding in patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Radiology. 2002;224:791–796. doi: 10.1148/radiol.2243011123. [DOI] [PubMed] [Google Scholar]
- 130.Viswanathan A, Gschwendtner A, Guichard JP, Buffon F, Cumurciuc R, O’Sullivan M, et al. Lacunar lesions are independently associated with disability and cognitive impairment in CADASIL. Neurology. 2007;69:172–179. doi: 10.1212/01.wnl.0000265221.05610.70. [DOI] [PubMed] [Google Scholar]
- 131.Jouvent E, Viswanathan A, Mangin JF, O’Sullivan M, Guichard JP, Gschwendtner A, et al. Brain atrophy is related to lacunar lesions and tissue microstructural changes in CADASIL. Stroke. 2007;38:1786–1790. doi: 10.1161/STROKEAHA.106.478263. [DOI] [PubMed] [Google Scholar]
- 132.Fein G, Di Sclafani V, Tanabe J, Cardenas V, Weiner MW, Jagust WJ, et al. Hippocampal and cortical atrophy predict dementia in subcortical ischemic vascular disease. Neurology. 2000;55:1626–1635. doi: 10.1212/wnl.55.11.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peters N, Holtmannspötter M, Opherk C, Gschwendtner A, Herzog J, Sämann P, et al. Brain volume changes in CADASIL: a serial MRI study in pure subcortical ischemic vascular disease. Neurology. 2006;66:1517–1522. doi: 10.1212/01.wnl.0000216271.96364.50. [DOI] [PubMed] [Google Scholar]
- 134.Jouvent E, Duchesnay E, Hadj-Selem F, De Guio F, Mangin JF, Hervé D, et al. Prediction of 3-year clinical course in CADASIL. Neurology. 2016;87:1787–1795. doi: 10.1212/WNL.0000000000003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pescini F, Donnini I, Cesari F, Nannucci S, Valenti R, Rinnoci V, et al. Circulating biomarkers in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy patients. J Stroke Cerebrovasc Dis. 2017;26:823–833. doi: 10.1016/j.jstrokecerebrovasdis.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 136.Duering M, Konieczny MJ, Tiedt S, Baykara E, Tuladhar AM, Leijsen EV, et al. Serum neurofilament light chain levels are related to small vessel disease burden. J Stroke. 2018;20:228–238. doi: 10.5853/jos.2017.02565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao D, Shang J, Sun R, Shi Y, Jiang H, Ma M, et al. Changes in the morphology, number, and protein levels of plasma exosomes in CADASIL patients. J Alzheimers Dis. 2021;81:221–229. doi: 10.3233/JAD-210101. [DOI] [PubMed] [Google Scholar]
- 138.Petzold A. Neurofilament phosphoforms: surrogate markers for axonal injury, degeneration and loss. J Neurol Sci. 2005;233:183–198. doi: 10.1016/j.jns.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 139.Teunissen CE, Khalil M. Neurofilaments as biomarkers in multiple sclerosis. Mult Scler. 2012;18:552–556. doi: 10.1177/1352458512443092. [DOI] [PubMed] [Google Scholar]
- 140.Chen CH, Cheng YW, Chen YF, Tang SC, Jeng JS. Plasma neurofilament light chain and glial fibrillary acidic protein predict stroke in CADASIL. J Neuroinflammation. 2020;17:124. doi: 10.1186/s12974-020-01813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lu CH, Macdonald-Wallis C, Gray E, Pearce N, Petzold A, Norgren N, et al. Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. Neurology. 2015;84:2247–2257. doi: 10.1212/WNL.0000000000001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bergman J, Dring A, Zetterberg H, Blennow K, Norgren N, Gilthorpe J, et al. Neurofilament light in CSF and serum is a sensitive marker for axonal white matter injury in MS. Neurol Neuroimmunol Neuroinflamm. 2016;3:e271. doi: 10.1212/NXI.0000000000000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Machuca-Parra AI, Bigger-Allen AA, Sanchez AV, Boutabla A, Cardona-Vélez J, Amarnani D, et al. Therapeutic antibody targeting of Notch3 signaling prevents mural cell loss in CADASIL. J Exp Med. 2017;214:2271–2282. doi: 10.1084/jem.20161715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Muiño E, Fernández-Cadenas I, Arboix A. Contribution of “omicΠ studies to the understanding of Cadasil. A systematic review. Int J Mol Sci. 2021;22:7357. doi: 10.3390/ijms22147357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the study.