Abstract
Viable Escherichia coli and Staphylococcus aureus bacteria elicited markedly different in vitro tumor necrosis factor alpha (TNF-α) responses when placed in coculture with peritoneal murine macrophages. These include quantitative differences in TNF-α mRNA expression and corresponding protein product secretion as well as kinetic differences in the profiles of the TNF-α responses. Further, lipopolysaccharide (from E. coli) is a major contributing factor to these differences, as revealed by comparative experiments with endotoxin-responsive (C3Heb/FeJ) and endotoxin-hyporesponsive (C3H/HeJ) macrophages. Nevertheless, the eventual overall magnitude of the TNF-α secretion of macrophages in response to S. aureus was at least equivalent to that observed with E. coli, while appearing at time periods hours later than the E. coli-elicited TNF-α response. Both the magnitude and kinetic profile of the TNF-α responses were found to be relatively independent of the rate of bacterial proliferation, at least to the extent that similar results were observed with both viable and paraformaldehyde-killed microbes. Nevertheless, S. aureus treated in culture with the carbapenem antibiotic imipenem manifests markedly altered profiles of TNF-α response, with the appearance of an early TNF-α peak not seen with viable organisms, a finding strikingly similar to that recently reported by our laboratory from in vivo studies (R. Silverstein, J. G. Wood, Q. Xue, M. Norimatsu, D. L. Horn, and D. C. Morrison, Infect. Immun. 68:2301–2308, 2000). In contrast, imipenem treatment of E. coli-cocultured macrophages does not significantly alter the observed TNF-α response either in vitro or in vivo. In conclusion, our data support the concept that the host inflammatory response of cultured mouse macrophages in response to viable gram-positive versus gram-negative microbes exhibits distinctive characteristics and that these distinctions are, under some conditions, altered on subsequent bacterial killing, depending on the mode of killing. Of potential importance, these distinctive in vitro TNF-α profiles faithfully reflect circulating levels of TNF-α in infected mice. These results suggest that coculture of peritoneal macrophages with viable versus antibiotic-killed bacteria and subsequent assessment of cytokine response (TNF-α) may be of value in clarifying, and ultimately controlling, related host inflammatory responses in septic patients.
Despite appropriate antimicrobial therapy and intensive supportive treatment, septic shock remains a major cause of morbidity and mortality in the United States and worldwide. The mortality rate can, under some circumstances, exceed 40 to 50%, and only a slight reduction in mortality from septic shock has been reported within the last 3 decades (7, 11, 12, 18). Septic shock is now recognized to be associated with multiple critical organ failure, usually attributable to the uncontrolled release of multiple proinflammatory and anti-inflammatory cytokines and biologically active oxygen radicals. Bacterial endotoxin and other bacterial wall components, such as peptidoglycan, lipoteichoic acid, and, more recently identified, bacterial DNA (6, 9, 10, 12–14, 19, 25, 27, 28) are among the microbial factors that have been strongly implicated as initiating these responses. Among those cytokines most often identified in septic patients, tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1), IL-6, IL-8, gamma interferon, IL-12, and nitric oxide (NO) have been widely investigated as potentially pivotal causative factors in gram-negative and gram-positive sepsis (1, 9, 18, 26).
Early efforts to treat this disease focused primarily on gram-negative microbe-induced septic shock. More recently, emphasis has been increasingly shifted to gram-positive sepsis inasmuch as gram-positive pathogens now account for up to 50% of severe cases of all sepsis. In addition, the mortality from gram-positive sepsis is significant and often exceeds the observed mortality from gram-negative sepsis (7, 18, 26). To date, the precise pathogenic mechanisms responsible for gram-positive and gram-negative bacterial sepsis have not been fully identified. In this respect, these mechanisms may well be significantly different as to their active roles in septic shock in terms of the involvement of both bacterial factors (cell wall components and soluble and secreted bacterial products) and host factors (e.g., susceptibility and innate immune response) (18, 23, 24, 26). In any case, activation of the innate immune responses in gram-positive and gram-negative sepsis is generally accepted to be important and largely responsible for observed experimental and clinical symptoms of sepsis including in vitro studies, animal models, and clinical trials.
Many studies have documented kinetically earlier and quantitatively increased levels of proinflammatory cytokines such as TNF-α, IL-1, and IL-6 during gram-negative sepsis in comparison with those documented during gram-positive sepsis (1, 4, 16, 23), whereas relatively few studies have suggested the equivalent involvement of cytokines in gram-positive and gram-negative sepsis (27, 32). Recent studies from our own laboratory have demonstrated that intraperitoneal injection of viable gram-positive Staphylococcus aureus in mice triggers levels of circulating TNF-α production in blood at least as high as those resulting from gram-negative Escherichia coli infection; however the appearance of the TNF-α peak induced by S. aureus treatment occurred at least 6 h later than that from E. coli treatment. In contrast, antibiotic imipenem treatment of S. aureus-infected mice converts this response to a quantitatively smaller TNF-α peak response, which can be detected much earlier than that documented in the absence of the antibiotic and which is kinetically similar to that induced by E. coli (both viable and antibiotic killed) (24).
In order to better understand whether or not different mechanisms may be involved in the host innate immune response in gram-positive and gram-negative bacterial sepsis, we have initiated a series of in vitro studies using viable E. coli and viable S. aureus in coculture with mouse macrophages in efforts to reproduce as closely as possible the in vivo host macrophage inflammatory response. These studies have been predicated on the concept that the inflammatory macrophage can be a major source of proinflammatory cytokines during sepsis. We demonstrate here that the introduction of viable S. aureus into a macrophage culture will induce TNF-α release with a magnitude comparable to that seen with viable E. coli treatment. In addition, as was earlier shown in vivo, the appearance of elevated TNF-α levels in the supernatant of macrophages cocultured with microbes in response to viable S. aureus is significantly delayed relative to that noted during the in vitro macrophage response to E. coli. Furthermore, in vitro addition of the antibiotic imipenem to S. aureus-macrophage cocultures results in an early but quantitatively reduced induction of TNF-α not detectable with viable S. aureus organisms. In contrast, little change of TNF-α release stimulated by either viable or antibiotic-killed E. coli is observed. We conclude from these studies that the qualitative and quantitative differences in circulating serum TNF-α responses in mice to viable versus antibiotic-killed S. aureus and E. coli reported earlier can be faithfully reproduced at the level of the cultured peritoneal macrophage, as assessed by levels of TNF-α expression and release into macrophage culture supernatants.
MATERIAL AND METHODS
Animals.
Female C3Heb/FeJ and C3H/HeJ mice were purchased from the Jackson Laboratory (Bar Harbor, Maine). Animals were maintained in the American Association for Accreditation of Laboratory Animal Care-certified University of Kansas Medical Center animal facility with food and water provided ad libitum. Eight- to 12-week-old mice were used for isolation of peritoneal macrophages.
Reagents.
RPMI 1640 cell culture medium and low-endotoxin heat-inactivated fetal bovine serum (FBS) were purchased from Sigma (St. Louis, Mo.). Imipenem was obtained from Merck & Co. (West Point, Pa.). TRIzol reagent was purchased from Life Technologies (Grand Island, N.Y.). GeneAmp RNA PCR kits were obtained from Perkin-Elmer (Norwalk, Conn.). TNF-α enzyme-linked immunosorbent assay kits were purchased from R&D Systems, Inc. Minneapolis, Minn.
Bacteria.
E. coli strain O111:B4 was originally obtained from List Biological Laboratories, Campbell, Calif. S. aureus M was generously provided by Chia Y. Lee, Department of Microbiology, Molecular Genetics, and Immunology, University of Kansas Medical Center.
Bacterial growth.
Several colonies from a streaked plate of E. coli or S. aureus grown overnight on Trypticase soy agar were used to initiate bacterial growth. Bacteria were inoculated into 2 ml of Trypticase soy broth in a 10-ml culture tube and were aerated by mechanical shaking overnight at 37°C. Then 200 μl of the overnight culture was subcultured in 10 ml of RPMI 1640 culture medium (without antibiotics) supplemented with 10% FBS in order to adapt bacteria to the macrophage culture medium. Cells were grown with aeration until mid-log phase as monitored by light scattering at 650 nm.
Isolation and culture of peritoneal macrophages.
Peritoneal exudate macrophages were obtained from C3Heb/FeJ or C3H/HeJ mice that had been injected intraperitoneally 5 days earlier with 1.5 ml of 4% sterile thioglycolate broth (Difco Laboratories, Detroit, Mich.). Peritoneal exudate cells were harvested by three cycles of peritoneal lavage with 5 ml of RPMI 1640 culture medium, and the cells obtained were then washed once with Hanks balanced salt solution (HBSS), followed by centrifugation at 800 × g for 10 min. Cells were resuspended in RPMI 1640 culture medium supplemented with 10% FBS. Cells were counted and plated into either 24-well cell culture plates (Costar, Cambridge, Mass.) or 6-well cell culture plates (Costar) at approximate densities of 5 × 105 and 2.5 × 106 cells per well, respectively. Cells were incubated in a 5% CO2 humidified culture incubator at 37°C for 2 h to allow macrophages to adhere to the plates. Nonadherent cells were removed by washing two times with HBSS solution. Since viable bacteria had been used to treat macrophages, no penicillin or streptomycin was added to the culture medium during the isolation, washing, or subsequent culturing period.
RNA isolation and reverse transcription-PCR (RT-PCR).
The E. coli or S. aureus cells were added to the established primary cultures of peritoneal macrophages in six-well plates and cocultured for various lengths of time as specified in Results. Following coculture, TRIzol reagent was added directly to the plate wells and total RNA was extracted from the cells directly from the culture plates by following the instructions provided by manufacturer. Equal amounts of total RNA (0.5 to 1.0 μg) for each sample were reverse-transcribed, and cDNA was amplified using GeneAmp RNA PCR kits by following the manufacturer's suggested protocol (Perkin-Elmer). The primers used were as follows: for mouse β-actin, the sense primer was TGTGATGGTGGGAATGGGTCG and the antisense primer was TTTGATGTCACGCACGATTTCC; for mouse TNF-α, the sense primer was GGCAGGTCTACTTTGGAGTCATTGC and the antisense primer was ACATTCGAGGCTCCAGTGAATTCGG). Amplified samples were subjected to electrophoresis on 1.2% agarose gels, stained with ethidium bromide, and photographed.
Quantitative TNF-α analysis.
Peritoneal macrophages were treated with various doses of E. coli or S. aureus for different times, and culture supernatants were filtered to remove the remaining bacteria. In most instances, culture supernatant were frozen at −70°C until analyzed. TNF-α was measured using mouse enzyme-linked immunosorbent assay kits by following manufacturers' instructions (Genzyme, Inc., Cambridge, Mass.; R&D Systems, Inc.). As a standard, recombinant mouse TNF-α, with a detection limit of 15 pg/ml, was used for the assay.
Statistics.
TNF-α secretion and accumulation in macrophage-bacterium coculture supernatants and the level of visible bacteria are presented as means ± standard errors of the means (SEM), with differences between groups assessed for significance via Student's t-test method (5). A calculated P value of <0.05 was considered significant.
RESULTS
Viable E. coli and viable S. aureus evoke different patterns of TNF-α release from mouse peritoneal macrophages.
One of the primary aims of these studies was to establish whether or not viable E. coli and/or viable S. aureus could elicit innate immune responses as reflected by TNF-α production and/or release in vitro and whether the kinetic patterns would approximate those observed earlier in an experimental mouse animal model (24). To pursue this aim, cultures of C3Heb/FeJ mouse peritoneal macrophages were cocultured with 5 × 105 CFU of viable E. coli or viable S. aureus/ml and macrophage secretion of TNF-α into cell culture medium was then measured in supernatants collected at fixed time intervals through 6 h of incubation. Macrophage viability during this same time frame was assessed by a 3-[4,5-dimethyl thiazol-2-yl]-2,5-diphenyltetrazolium bromide spectrophotometric assay (15). When macrophages were cocultured with E. coli, percentages of macrophage viability relative to that seen in the absence of bacteria, as determined in quadruplicate experiments, were 113% ± 8% at 1 h, 97% ± 11% at 2 h, 109% ± 13% at 3 h, and 130% ± 6% at 6 h. The corresponding percentages with S. aureus were 121% ± 11%, 97% ± 8%, 114% ± 19%, and 90% ± 16%. Thus, there are no apparent or systematic differences in macrophage viability during the 6-h incubation period of the experiment with either of these bacteria.
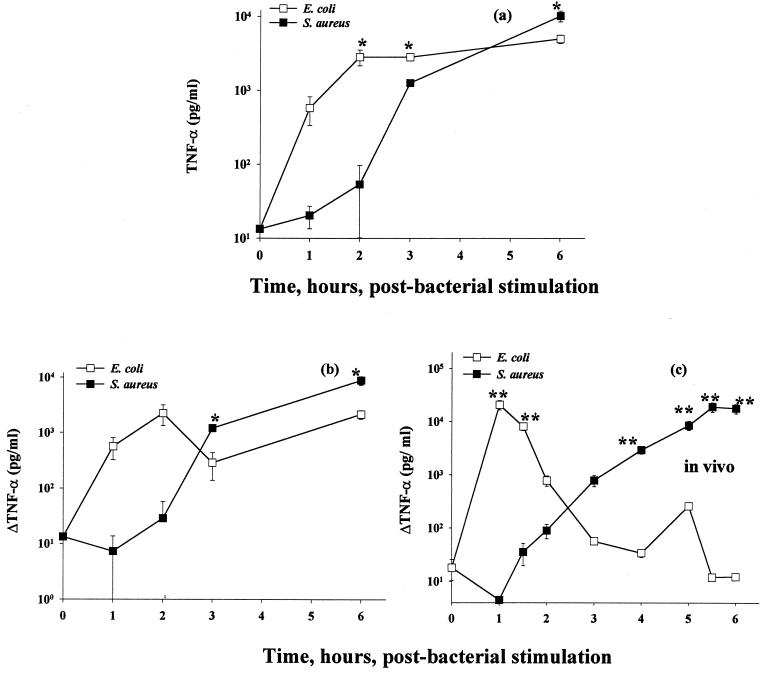
As shown by the data in Fig. 1a, E. coli induced substantially higher levels of TNF-α in culture supernatants during the first 2 h than the almost undetectably low levels of TNF-α induced by cultured macrophages stimulated with S. aureus during the same time period. At later times, however, S. aureus-stimulated macrophages stimulated production of substantial amounts of TNF-α. In fact, by 6 h, the quantitative magnitude of TNF-α release was found to be higher than that detected in culture supernatants from E. coli-stimulated macrophages.
FIG. 1.
TNF-α secretion from mouse macrophage–viable-bacterium cocultures. Thioglycolate-elicited C3Heb/FeJ mouse macrophages were cocultured with 5 × 105 CFU of E. coli O111:B4 or S. aureus M/ml. (a) TNF-α was assayed in supernatants collected at 0 to 6 h, as described in Materials and Methods. (b) Secondary plot of the data in panel a. Specifically, from each datum shown in panel a, the experimental value for TNF-α determined at the immediately preceding experimental time point has been subtracted. (c) Serum TNF-α data taken from reference 24, shown here on a log scale, from the trunk blood of outbred (CF-1) mice challenged intraperitoneally with the same proliferating bacterial strains as those used in the present study. For the in vitro data, three independent experiments were performed; for the in vivo data, two experiments were performed, with five mice per datum per experiment. Data are means ± SEM. ∗, P < 0.05; ∗∗, P < 0.005. For the data lacking error bars, variation was too small to indicate.
In order to more closely compare these TNF-α profiles with those earlier reported in vivo (from serum) (24), it would be of value to take into account the fact that TNF-α is usually rapidly cleared from the circulation, with a half-life of 6 to 7 min (2). In this regard, therefore, we reevaluated the data shown in Fig. 1a as the net increase in accumulated TNF-α, as shown in Fig. 1b. For comparison, earlier-reported in vivo responses of mice to E. coli versus S. aureus, as assessed by measuring circulating levels of TNF-α, are shown in Fig. 1c. These data collectively support the conclusion that the in vitro profiles of TNF-α secretion induced by S. aureus and E. coli do, in fact, reflect the findings earlier reported following in vivo infection (24).
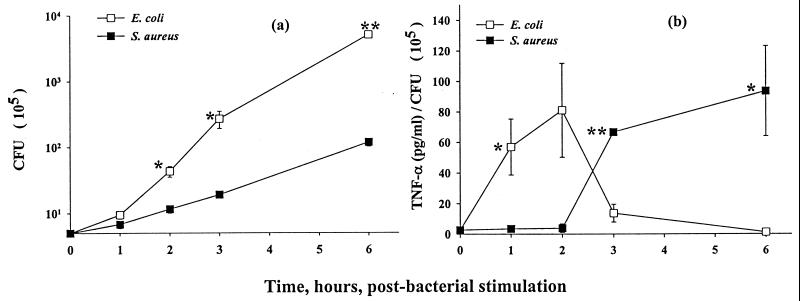
Given that these in vitro experiments resulted in ongoing bacterial growth, we have considered the question of whether the observed differences in TNF-α secretion occurred independently of the rate of bacterial proliferation. To assess this factor, we monitored bacterial growth in the macrophage cultures exposed to E. coli or S. aureus over the interval of coculture. As illustrated by the data shown in Fig. 2a, E. coli replicates much more rapidly than does S. aureus in the primary macrophage cell cultures. Approximately 14-fold and 43-fold differences in total CFU between E. coli and S. aureus in macrophage cultures were detected at 3 and 6 h, respectively.
FIG. 2.
Bacterial proliferation and normalized TNF-α secretion from mouse macrophage–viable-bacterium cocultures. Thioglycolate-elicited C3Heb/FeJ mouse macrophages were cocultured with 5 × 105 CFU of E. coli O111:B4 or S. aureus M/ml. (a) Bacterial proliferation was determined at the times indicated; (b) TNF-α concentration from Fig. 1a normalized to bacterial number from panel a. Data are means ± SEM. ∗, P < 0.05; ∗∗, P < 0.005. Three independent experiments were performed. For the data lacking error bars, variation was too small to indicate.
In order to put these data in perspective, we have also calculated the ratio of TNF-α to the actual number of live bacteria in the cell culture medium (as estimated by CFU), and these data are illustrated in Fig. 2b. The calculated ratio (Fig. 2b) reflects the levels of TNF-α secreted by macrophages relative to the concentration of activating stimulus (live bacteria at 105 CFU/ml) at various times of coculture. The kinetic profiles of the ratio of TNF-α levels to bacterial counts in macrophages continued to parallel the kinetics of TNF-α secretion (Fig. 2b versus Fig. 1b) as well as the in vivo profiles (Fig. 2b versus Fig. 1c). The calculated ratio during the first 2 h for coculture of E. coli with macrophags was higher than that for coculture of S. aureus-treated macrophages. Between 3 and 6 h, however, the ratio declined to much lower levels in macrophages treated with E. coli whereas, in contrast, the ratio increased substantially by 3 h and remained higher at 6 h in S. aureus-treated macrophages. These data indicate that the different patterns of TNF-α secretion from mouse peritoneal macrophages in response to viable E. coli versus viable S. aureus are most likely not directly related to the different bacterial growth rates; rather they reflect a true difference in the activating potential of these prototype gram-positive and gram-negative microorganisms.
The different in vitro profiles of TNF-α secretion are related to intrinsic structural differences between E. coli and S. aureus.
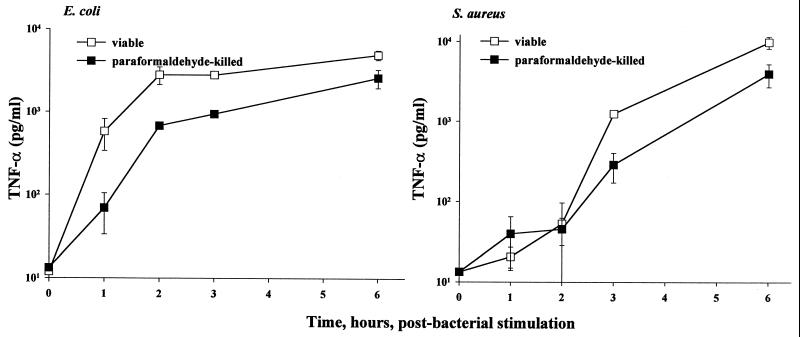
In order to further ascertain whether or not the observed differences in TNF-α release by primary macrophage cultures treated with viable E. coli and S. aureus are, in fact, due to inherent differences between these bacteria, we incubated macrophages with paraformaldehyde-killed bacteria for various times and measured the TNF-α released into culture supernatants in comparison with that released by cocultures of viable organisms with macrophages. As demonstrated by the data shown in Fig. 3, macrophages treated with paraformaldehyde-killed bacteria generated TNF-α kinetic profiles qualitatively similar to those generated by viable bacteria-treated macrophages (data from Fig. 1a shown in Fig. 3). Both viable and paraformaldehyde-killed E. coli induced early TNF-α secretion from macrophages compared to S. aureus. The similar profiles of TNF-α secretion from macrophages stimulated by paraformaldehyde-killed bacteria and viable bacteria-treated macrophages strengthen the hypothesis that fundamental differences between E. coli and S. aureus gave rise to the observed differences in macrophage innate immune responses to these microorganisms.
FIG. 3.
Comparisons of TNF-α secretions from paraformaldehyde-killed versus viable E. coli- and S. aureus-stimulated mouse macrophages from 0 to 6 h. Macrophage preparation and the TNF-α assay were as described for Fig. 1. E. coli O111:B4 and S. aureus M concentrations: paraformaldehyde-killed, 107 CFU/ml; viable, 5 × 105 CFU/ml. Data are means ± SEM for three independent experiments. For the data lacking error bars, variation was too small to indicate.
LPS is the major component from E. coli that stimulates macrophages to release TNF-α.
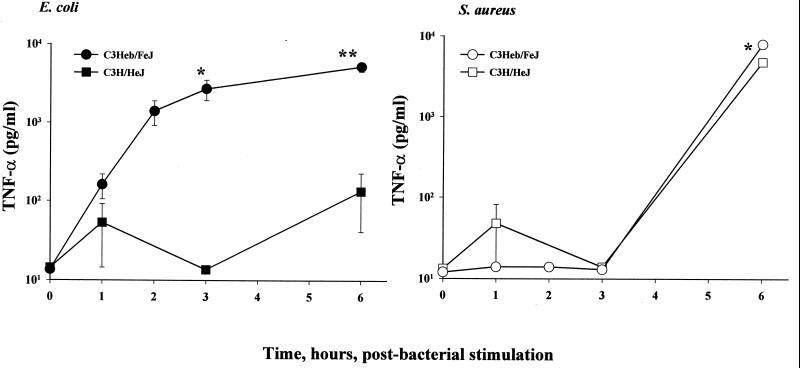
One major structural difference between gram-negative and gram-positive microbes is, of course, that the former, but not the latter, contain biologically active endotoxin (lipopolysaccharide [LPS]) in an intact membrane bilayer in the outer leaflet exterior to the cell wall. In order to determine the extent to which the different abilities of E. coli and S. aureus to induce macrophage TNF-α may be related to the presence or absence of LPS, we utilized macrophages derived from C3H/HeJ mice to conduct experiments comparable to those described above with C3Heb/FeJ macrophages. C3H/HeJ mice possess a genetic lesion that renders them significantly less responsive to LPS. Cultures of C3H/HeJ (LPS-hyporesponsive) and C3Heb/FeJ (LPS-normoresponsive) mouse peritoneal exudate macrophages were treated with 5 × 104 CFU of either E. coli or S. aureus/ml for 0 to 6 h, and the TNF-α released into the cell culture medium was then assayed from the supernatants taken at fixed times. As shown by the data in Fig. 4, only very low levels of soluble TNF-α were detectable in culture in supernatants from viable E. coli-stimulated C3H/HeJ mouse macrophages, in contrast to the magnitude of the TNF-α concentrations found in coculture supernatants from C3Heb/FeJ mouse macrophages. In contrast, following S. aureus stimulation, the TNF-α concentrations detectable in the supernatants markedly increased between 3 and 6 h and were essentially independent of whether or not the stimulated macrophages had been originally harvested from C3Heb/FeJ mice or from C3H/HeJ mice. Thus, LPS responsiveness appears to be a major determinant for the TNF-α response following macrophage stimulation by viable E. coli but not by S. aureus.
FIG. 4.
Comparisons of TNF-α secretion from C3Heb/FeJ (endotoxin-normoresponsive) versus C3H/HeJ (endotoxin-hyporesponsive) mouse macrophages cocultured with viable bacteria for 0 to 6 h. Macrophage and bacterial preparations and the TNF-α assay were as described for Fig. 1, except that each bacterial concentration was 10-fold lower: E. coli O111:B4 concentration, 5 × 104 CFU/ml; S. aureus M concentration, 5 × 104 CFU/ml. Data are means ± SEM for three independent experiments. For the data lacking error bars, variation was too small to indicate.
Transcriptional differences in TNF-α response are manifest by macrophages stimulated by viable E. coli versus viable S. aureus.
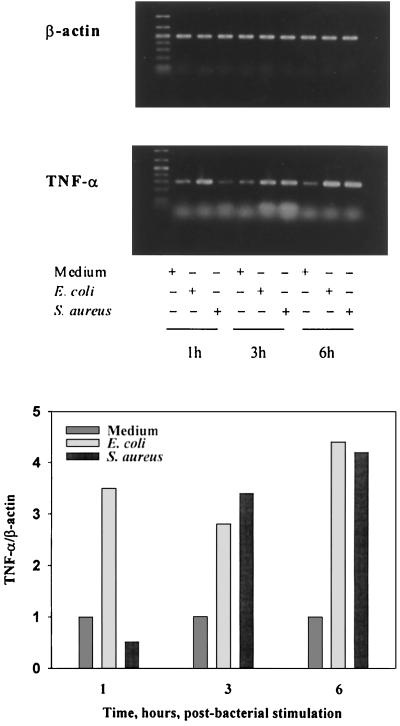
We next queried whether or not the differences in TNF-α secretion from macrophages that we had demonstrated using viable E. coli and S. aureus as stimuli would also be reflected at the level of gene transcription. For this purpose, we used RT-PCR to examine TNF-α mRNA expression in C3Heb/FeJ mouse peritoneal macrophages that had been treated with 5 × 104 CFU of live E. coli or S. aureus/ml for fixed time periods from 0 to 6 h. In order to normalize the RT-PCR levels, we also measured cellular levels of mRNA of the housekeeping gene for β-actin. As shown by the data in Fig. 5, E. coli stimulation induced an early (1 h poststimulation) upregulation of approximately three- to fourfold in TNF-α mRNA expression in the stimulated macrophages. Comparable induction of TNF-α mRNA by S. aureus was also observed, but only after 3 h of coculture. These findings are consistent with the temporal differences in TNF-α release shown earlier in Fig. 1 to 4. Together, these findings support the concept that the differential responses of macrophages to E. coli and S. aureus are mediated by early events in cell activation. Qualitatively similar temporal differences in IL-1β, IL-10, and inducible nitric oxide synthase mRNA expression were also observed (data not shown), suggesting a global character to the observed distinctions between macrophages stimulated by viable gram-positive versus gram-negative bacteria.
FIG. 5.
Expression of β-actin and TNF-α mRNA in C3Heb/FeJ mouse macrophages cocultured with viable bacteria. Total RNA from macrophages cocultured with 5 × 104 CFU of viable E. coli O111:B4 or S. aureus M for 1 to 6 h was isolated. An equal amount was reverse transcribed, and cDNAs were amplified using PCR. (Upper panel) Samples were then subjected to electrophoresis and stained with ethidium bromide. (Lower panel) Densitometric scanning of amplified TNF-α mRNA normalized against the β-actin mRNA control for the medium, E. coli, and S. aureus. Macrophage preparation was as described for Fig. 1.
Bacterial killing by the carbapenem antibiotic imipenem selectively alters the kinetics and magnitude of TNF-α secretion from macrophages stimulated by S. aureus versus E. coli.
The new data shown in Fig. 1 to 5 are indication of marked temporal differences in TNF-α secretion in macrophages cocultured with viable E. coli versus S. aureus. We have also shown that viability would not necessarily serve as an absolute requirement for such differences, inasmuch as paraformaldehyde-killed bacteria yielded similar outcomes (Fig. 3). Nevertheless, there have been several earlier studies from our laboratory (3, 17, 24; D. C. Morrison, Editorial, J. Endotoxin Res. 3:171, 1996) and from other laboratories (8, 11, 20–22, 29–31) establishing that killing or otherwise inhibiting proliferation of bacteria through the use of efficacious antibiotic treatments can significantly alter inflammatory responses depending on the bacteria and specific antibiotic(s). We therefore investigated the effect of one such antibiotic, imipenem, which effectively kills both gram-positive and gram-negative microbes in vivo and in vitro.
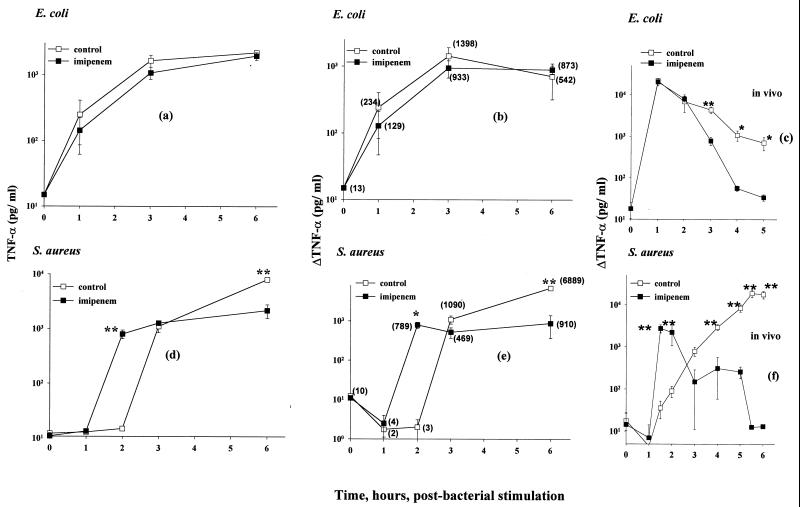
C3Heb/FeJ mouse macrophages were separately treated with live E. coli or S. aureus in coculture, with or without imipenem (50 μg/ml). TNF-α concentrations in isolated culture supernatants were then assayed. Bacterial killing was established as being complete by 2 h, with 99.9% of E. coli cells and 90% of S. aureus cells being killed within the first hour (data not shown). With E. coli, the imipenem treatment had little apparent effect on either the magnitude or the kinetics of TNF-α appearance following release from the macrophages. By contrast, and at the same bacterial concentrations, the macrophage response to S. aureus was found to be markedly affected by the imipenem treatment. Thus, as shown by the results in Fig. 6e, between 1 and 2 h following the imipenem treatment, the magnitude of TNF-α release increased from 4 to 789 pg/ml, whereas in the absence of the imipenem treatment, the corresponding differences were negligible. At later times, it was only in the absence of the imipenem treatment that the TNF-α response continued to rise, to 1,090 pg/ml between 2 and 3 h and to a difference increasing to 6.9 ng/ml between 3 and 6 h. These observations are qualitatively comparable to those previously reported (24) from in vivo experiments in which mice were challenged with these same proliferating bacterial strains, with and without the imipenem killing. For purposes of direct comparison, these published data are reproduced here (in logarithmic form) in Fig. 6c (E. coli) and Fig. 6f (S. aureus). The early increase in TNF-α was once again seen with S. aureus following imipenem treatment, as well as a marked and continuing later increase in TNF-α when the antibiotic treatment was omitted. As with the in vitro experiments, neither of these effects was apparent with the E. coli, even though the imipenem was also confirmed to be effective at killing these organisms in vitro and in vivo.
FIG. 6.
Effect of imipenem treatment on TNF-α secretion from macrophages stimulated by E. coli versus S. aureus. C3Heb/FeJ mouse macrophages were cocultured with 5 × 105 CFU of viable E. coli O111:B4 or viable S. aureus M/ml, with (■) and without (□) imipenem (50 μg/ml), for 0 to 6 h. (a and d) TNF-α in supernatants collected at 0 to 6 h, assayed as described in Materials and Methods. (b and e) Secondary plots of the data in panels a and d, respectively. Specifically, from each datum shown in panels a and d, the experimental value for TNF-α determined at the immediately preceding experimental time point has been subtracted. (c and f) Serum TNF-α data taken from reference 24, shown here on a log scale, from the trunk blood of outbred (CF-1) mice challenged intraperitoneally with the same proliferating bacterial strains as those used in the present study and concomitantly with a second intraperitoneal injection of 20 mg of imipenem per kg of body weight. For the in vitro data, three independent experiments were performed; for the in vivo data, two experiments were performed, with five mice per datum per experiment. Data are means ± SEM, ∗ P < 0.05; ∗∗, P < 0.005. For the data lacking error bars, variation was too small to indicate.
DISCUSSION
Marked temporal differences in TNF-α secretion from peritoneal macrophages are readily detected following coculture with viable prototypic gram-positive (S. aureus) and gram-negative (E. coli) bacteria. These differences are also found when TNF-α mRNA expression in macrophage cultures is measured. When each of the microbes was rapidly killed with the broad-spectrum antibiotic imipenem, the effects ofTNF-α secretion in response to S. aureus were significantly altered in comparison to that in response to control untreated S. aureus. In contrast, responses to E. coli were not measurably distinguishable, as assessed by measuring TNF-α levels in culture supernatants with and without imipenem treatment. Collectively, these data accurately reflect the results of earlier published serum TNF-α kinetic profiles in mice in response to viable E. coli and S. aureus, and the effects of antibiotic chemotherapy.
It is noteworthy that paraformaldehyde killing of both S. aureus and E. coli, by contrast, did not evoke TNF-α profiles different from those seen with corresponding viable bacteria. Further evidence for a dominant role for structural differences in eliciting these differences and, in particular, a key role for endotoxin is the fact that the macrophage TNF-α responses to E. coli but not to S. aureus are affected when cells from the C3H/HeJ endotoxin-hyporesponsive mouse are substituted for C3Heb/FeJ macrophages. That other microbial factors most likely are involved as well, however, is strongly supported by observations, both from published (24) and unpublished data from our laboratory, that the cell wall-active antibiotics imipenem and ceftazidime behave qualitatively similarly in both in vitro and in vivo models of mediator TNF-α production, even though differences in the abilities of these antibiotics to elicit endotoxin-mediated release are well documented (D. C. Morrison, Editorial, J. Endotoxin Res. 3:171, 1996). Also, the early TNF-α release associated with imipenem (or ceftazidime) killing of the S. aureus could arise from the release of one or more bacterial components, already identified as being capable of eliciting host inflammatory responses. These would include lipoteichoic acid and peptidoglycan (6, 30) or microbial DNA or some combination of these microbial factors.
The present study, like previous studies from our laboratory (17, 23, 24), has employed S. aureus and E. coli as prototype gram-positive and gram-negative bacteria. The data reported here not only further establish a distinct temporal difference between E. coli and S. aureus in eliciting macrophages to release TNF-α but also provide a reliable and highly reproducible in vitro macrophage model. These results, by essentially mirroring our earlier published results in vivo with each of these bacteria (24), support the systematic extension of the concept to other gram-positive and gram-negative microbes and modes of bactericidal action.
We have previously reported that treatment of gram-negative bacteria with cell wall-active antibiotics can induce the release of biologically active endotoxin (3; D. C. Morrison, Editorial, J. Endotoxin Res. 3:171, 1996). To our knowledge, however, there are few, if any, prior reports establishing that treatment of gram-positive organisms with antibiotics can promote the effective release of microbial factors and corresponding host inflammatory response. Given the widespread use of antibiotics, and particularly cell wall-active antimicrobials, the potential relevance of this finding to treatment of the gram-positive septic patient should not be entirely discounted in the consideration of treatment options. Whether or not different antibiotics elicit different (either qualitatively or quantitatively) profiles of cytokine production remains to be established and is currently under active investigation in our laboratory.
ACKNOWLEDGMENTS
This work was supported by NIH grant NI 23447, grants from the Jane Harley Estate Fellowship and Ernest F. Lied Foundation, and an unrestricted medical grant from Merck & Company.
We thank Jian Jun Gao, Donald C. Johnson, MeiGuey Lei, Alexander Shnyra, and Qiao Xue for their helpful advice and Chia Lee for generously providing S. aureus.
REFERENCES
- 1.Anderson J, Nagy S, Bjork L, Abrams J, Holm S, Anderson U. Bacterial toxin-induced cytokine production studied at the single-cell level. Immunol Rev. 1992;127:69–96. doi: 10.1111/j.1600-065x.1992.tb01409.x. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B A, Milsark I W, Cerami A. Cachectin/tumor necrosis factor: production, distribution, and metabolic fate in vivo. J Immunol. 1985;6:3973–3977. [PubMed] [Google Scholar]
- 3.Bucklin S, Morrison D C. Bacteremia versus endotoxemia in experimental mouse leukopenia—role of antibiotic chemotherapy. J Infect Dis. 1996;174:1249–1254. doi: 10.1093/infdis/174.6.1249. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J, Abraham E. Microbiological findings and correlations with serum tumor necrosis factor-α in patients with severe sepsis and septic shock. J Infect Dis. 1999;180:116–121. doi: 10.1086/314839. [DOI] [PubMed] [Google Scholar]
- 5.Croxton F E. Elementary statistics with application in medicine, Prentice-Hall, Englewood Cliffs, N.J. 1953. pp. 235–239. and 326–327. [Google Scholar]
- 6.De Kimpe S J, Kengatharan M, Thiemermann C, Vane J R. The cell wall components peptidoglycan and lipoteichoic acid from Staphylococcus aureus act in synergy to cause shock and multiple organ failure. Proc Natl Acad Sci USA. 1995;92:10359–10363. doi: 10.1073/pnas.92.22.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman G, Silva E, Vincent J L. Has the mortality of septic shock changed with time? Care Med. 1998;26:2078–2086. doi: 10.1097/00003246-199812000-00045. [DOI] [PubMed] [Google Scholar]
- 8.Frieling J T M, Mulder J A, Hendriks T, Curfs J H A J, van der Linden C J, Sauerwein R W. Differential induction of pro- and anti-inflammatory cytokines in whole blood by bacteria: effects of antibiotic treatment. Antimicrob Agents Chemother. 1997;41:1439–1443. doi: 10.1128/aac.41.7.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J J, Zuvanich E G, Xue Q, Horn D L, Silverstein R, Morrison D C. Bacterial DNA and lipopolysaccharide act in synergy in inducing nitric oxide production in RAW 264.7 macrophages. J Immunol. 1999;163:4095–4099. [PubMed] [Google Scholar]
- 10.Heumann D, Barras C, Severin A, Glauser M P, Tomasz A. Gram-positive cell walls stimulate synthesis of tumor necrosis factor alpha and interleukin-6 by human monocytes. Infect Immun. 1994;62:2715–2721. doi: 10.1128/iai.62.7.2715-2721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn D L, Opal S M, Lomastro E. Antibiotics, cytokines, and endotoxin: a complex and evolving relationship in gram-negative sepsis. Scand J Infect Dis. 1996;101(Suppl.):9–13. [PubMed] [Google Scholar]
- 12.Horn, D. L., D. C. Morrison, S. M. Opal, R. Silverstein, K. Visvanathan, and J. B. Zabriskie. What are the microbial components implicated in the pathogenesis of sepsis? Report on a symposium. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 13.Kengatharan K M, De Kimpe S, Robson C, Foster S J, Thiemermann C. Mechanism of Gram-positive shock: identification of peptidoglycan and lipoteichoic acid moieties essential in the induction of nitric oxide synthase, shock and multiple organ failure. J Exp Med. 1998;188:305–315. doi: 10.1084/jem.188.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattsson E, Rollof J, Verhoef J, van Dijk H, Fleer A. Serum-induced potentiation of tumor necrosis factor alpha production by human monocytes in response to staphylococcal peptidoglycan: involvement of different serum factors. Infect Immun. 1994;62:3837–3843. doi: 10.1128/iai.62.9.3837-3843.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mossmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 16.Muller-Alouf H, Alouf J E, Gerlach D, Ozegowski J H, Fitting C, Cavaillon J-M. Comparative study of cytokine release by human peripheral blood mononuclear cells stimulated with Streptococcus pyogenes superantigenic erythrogenic toxins, heat-killed streptococci, and lipopolysaccharide. Infect Immun. 1994;62:4915–4921. doi: 10.1128/iai.62.11.4915-4921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norimatsu M, Morrison D C. Correlation of antibiotic-induced endotoxin release and cytokine production in Escherichia coli-inoculated mouse whole blood ex vivo. J Infect Dis. 1997;177:1302–1307. doi: 10.1086/515291. [DOI] [PubMed] [Google Scholar]
- 18.Opal S M, Cohen J. Clinical gram-positive sepsis: does it fundamentally differ from gram-negative bacterial sepsis? Crit Care Med. 1999;27:1608–1616. doi: 10.1097/00003246-199908000-00039. [DOI] [PubMed] [Google Scholar]
- 19.Opal S M, Cross A C. Clinical trials for severe sepsis: past failures and future hopes. Infect Dis Clin N Am. 1999;13:285–298. doi: 10.1016/s0891-5520(05)70075-1. [DOI] [PubMed] [Google Scholar]
- 20.Periti P, Mazzei T. Antibiotic-induced release of bacterial cell wall components in the pathogenesis of sepsis and septic shock: a review. J Chemother. 1998;10:427–448. doi: 10.1179/joc.1998.10.6.427. [DOI] [PubMed] [Google Scholar]
- 21.Prins J M. Endotoxin, antibiotics, and inflammation in gram-negative infections. In: Brade H, Opal S M, Vogel S N, Morrison D C, editors. Endotoxin in health and disease. New York, N.Y: Marcel Dekker, Inc.; 1999. p. 887. [Google Scholar]
- 22.Schneider C M, Huzly D, Vetter C, von Specht B U, Daschner F D. Tumor necrosis factor alpha and interleukin 6 release induced by antibiotic killing of Pseudomonas aeruginosa and Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1997;16:467–471. doi: 10.1007/BF02471914. [DOI] [PubMed] [Google Scholar]
- 23.Silverstein R, Norimatsu M, Morrison D C. Fundamental differences during gram-positive versus gram-negative sepsis become apparent during bacterial challenge of D-galactosamine-treated mice. J Endotox Res. 1997;4:173–181. [Google Scholar]
- 24.Silverstein R, Wood J G, Xue Q, Norimatsu M, Horn D L, Morrison D C. Differential host inflammatory responses to viable versus antibiotic-killed bacteria in experimental microbial sepsis. Infect Immun. 2000;68:2301–2308. doi: 10.1128/iai.68.4.2301-2308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sparwasser T, Miethke T, Lipford G, Borschert G K, Häcker H, Heeg K, Wagner H. Bacterial DNA causes septic shock. Nature. 1997;386:336–337. doi: 10.1038/386336a0. [DOI] [PubMed] [Google Scholar]
- 26.Sriskandan S, Cohen J. Gram-positive sepsis mechanisms and differences from gram-negative sepsis. Infect Dis Clin N Am. 1999;13:397–412. doi: 10.1016/s0891-5520(05)70082-9. [DOI] [PubMed] [Google Scholar]
- 27.Timmerman C P, Mattsson E, Martinez L, De Graaf L, van Strijp J A G, Verbrugh H A, Verhoef J, Fleer A. Induction of release of tumor necrosis factor from human monocytes by staphylococci and staphylococcal peptidoglycan. Infect Immun. 1993;61:4167–4172. doi: 10.1128/iai.61.10.4167-4172.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallejo J G, Baker C J, Edwards M. Roles of the bacterial cell wall and capsule in induction of tumor necrosis factor alpha by type III group B streptococci. Infect Immun. 1996;64:5042–5046. doi: 10.1128/iai.64.12.5042-5046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Langevelde P, Kwappenberg K M C, Groeneveld P H P, Mattie H, van Dissel J P. Antibiotic-induced lipopolysaccharide (LPS) release from Salmonella typhi: delay between killing by ceftazidime and imipenem and release of LPS. Antimicrob Agents Chemother. 1998;42:739–743. doi: 10.1128/aac.42.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Langevelde P, van Dissel J P, Ravensbergen E, Appelmelk B J, Schrijver I A, Groeneveld P H P. Antibiotic-induced release of lipoteichoic acid and peptidoglycan from Staphylococcus aureus: quantitative measurements and biological reactivities. Antimicrob Agents Chemother. 1998;42:3073–3078. doi: 10.1128/aac.42.12.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Langevelde P, Ravensbergen E, Grashoff P, Beekhuizen H, Groeneveld P H P, van Dissel J P. Antibiotic-induced cell wall fragments of Staphylococcus aureus increase endothelial chemokine secretion and adhesiveness for granulocytes. Antimicrob Agents Chemother. 1999;43:2984–2989. doi: 10.1128/aac.43.12.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakabayashi G, Gelfand J A, Jung W K, Connolly R J, Burke J F, Dinarello C A. Staphylococcus epidermidis induces complement activation, tumor necrosis factor and interleukin-1, a shock-like state and tissue injury in rabbits without endotoxemia. Comparison to Escherichia coli. J Clin Investig. 1991;87:1925–1935. doi: 10.1172/JCI115218. [DOI] [PMC free article] [PubMed] [Google Scholar]