Abstract
Outer membrane protein E (OMP E) is a 50-kDa protein of Moraxella catarrhalis which has several features that suggest that the protein may be an effective vaccine antigen. To assess the conservation of OMP E among strains of M. catarrhalis, 22 isolates were studied with eight monoclonal antibodies which recognize epitopes on different regions of the protein. Eighteen of 22 strains were reactive with all eight antibodies. The sequences of ompE from 16 strains of M. catarrhalis were determined, including the 4 strains which were nonreactive with selected monoclonal antibodies. Analysis of sequences indicate a high degree of conservation among strains, with sequence differences clustered in limited regions of the gene. To assess the stability of ompE during colonization of the human respiratory tract, the sequences of ompE of isolates collected from patients colonized with the same strain for 3 to 9 months were determined. The sequences remained unchanged. These results indicate that OMP E is highly conserved among strains of M. catarrhalis, and preliminary studies indicate that the gene which encodes OMP E remains stable during colonization of the human respiratory tract.
Moraxella catarrhalis is a common and important human respiratory tract pathogen. It is the third most common cause of otitis media in children, accounting for approximately 3.5 million episodes of otitis media annually in the United States (6, 11, 15, 20, 25). Adults with chronic obstructive pulmonary disease (COPD) experience recurrent lower respiratory tract infections, often called exacerbations. M. catarrhalis is a common cause of these infections (15, 18, 23, 27, 30). Exacerbations of COPD lead to substantial morbidity and mortality and increased health care costs worldwide (14, 19, 22). In view of the impact of M. catarrhalis infections, there is considerable interest in developing a vaccine to prevent infections caused by M. catarrhalis. Infants would be immunized to prevent otitis media, with particular emphasis on preventing otitis media in otitis-prone children who experience recurrent episodes of otitis media. The second population which would benefit from such a vaccine would be adults with COPD.
Outer membrane protein E (OMP E) is a 50-kDa heat-modifiable OMP which has characteristics indicating that it may be an effective vaccine antigen (3, 4). The protein is expressed in all strains of M. catarrhalis studied thus far (2, 4, 16). Three independent lines of experiments indicate that OMP E contains epitopes on the bacterial surface; these include adsorption studies with polyclonal antisera raised to whole bacterial cells (16), immunofluorescence microscopy with polyclonal antisera raised to purified OMP E (4), and flow cytometry with monoclonal antibodies (MAbs) (4, 17).
An important consideration in evaluating an OMP as a potential vaccine is the extent to which the protein is conserved among strains of the species. An ideal vaccine candidate would be highly conserved among strains so that immunization with the protein from one strain would generate protective antibodies to all or most strains of the species. Studies of OMP E with four MAbs and analysis of PCR restriction fragment length polymorphisms of ompE suggest that the protein and gene show conservation among strains (3, 4). The goal of the present study was to more rigorously assess the degree of conservation of OMP E by analysis of strains with eight MAbs and to determine the sequences of ompE of 16 selected strains. In addition, the stability of ompE in isolates which colonize the human respiratory tract was assessed preliminarily.
(This work was presented in part at the 101st General Meeting of the American Society for Microbiology, 20 to 24 May 2001, Orlando, Fla.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Twenty-one clinical isolates of M. catarrhalis were recovered from sputum (15), middle ear fluid (2), the nasopharynx (2), the adenoid (1), and sinus aspirate (1). The geographic sources of the isolates were Buffalo, N.Y. (15), Birmingham, United Kingdom (2), Houston, Tex. (1), Philadelphia, Pa. (1), Mountain Home, Tenn. (1), and Utrecht, The Netherlands (1). Ten of the sputum isolates were recovered from the sputum of adults monitored in a prospective COPD study clinic (see below). M. catarrhalis strain ATCC 25240 was obtained from the American Type Culture Collection. Strains were grown on brain heart infusion agar at 35°C under 5% CO2.
COPD Study Clinic.
Isolates of M. catarrhalis were recovered from adults enrolled in a prospective study of COPD at the Buffalo Veterans Affairs Medical Center. To be included in the COPD Study Clinic, a patient must have chronic bronchitis as defined by the American Thoracic Society (1) and be willing to visit the clinic monthly and at the time of a suspected exacerbation. At each clinic visit, the patient undergoes a clinical evaluation and serum and sputum samples are collected. For this study, an equal volume of 6.5 mM dithiothreitol in phosphate-buffered saline was added to the sputum. The sputum was mixed by vortexing and incubated at 37°C for 20 min. After an aliquot of sputum was removed for culture, the mixture was centrifuged at 27,000 × g for 30 min at 4°C. The sputum supernatants were stored at −80°C. M. catarrhalis was identified using standard methods, and isolates were stored at −80°C in Mueller-Hinton broth containing 10% glycerol.
Cell envelope preparation.
Bacterial strains were grown overnight on brain heart infusion plates. Bacteria from one plate were harvested by suspension in 2 ml of 0.01 M HEPES, pH 7.4. Cells were pelleted by centrifugation at 1,000 × g for 15 min at 4°C. The pellet was resuspended in 0.5 ml of HEPES buffer and sonicated three times for 10 s with a Branson Sonifier (small tip, setting 4). After transfer of the entire suspension to a fresh tube, cell envelopes were pelleted by centrifugation at 16,000 × g for 45 min at 4°C. The cell envelopes were suspended in ∼0.5 ml of HEPES buffer and stored at −20°C until use.
MAbs.
Seven MAbs which recognize epitopes on OMP E have been described previously (4, 17). MAb 2E11 was developed from a fusion in which mice were immunized subcutaneously with 50 μg of purified recombinant OMP E from M. catarrhalis strain ATCC 25240 (4) with incomplete Freund's adjuvant on days 0 and 14. On day 28 the mice received approximately 108 CFU of M. catarrhalis strain ATCC 25240 intraperitoneally without adjuvant. On day 31, splenocytes were fused with SP2/0-Ag-14 plasmacytoma cells by a modification of the procedure of Kennett (10) and another previously described method (21).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot assay.
Whole-bacterial-cell lysates and cell envelope preparations were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot assay as previously described (17).
ELISA.
Levels of immunoglobulin to OMP E were assayed in human serum and sputum supernatants by enzyme-linked immunosorbent assay (ELISA) as previously described (4).
PFGE.
Strains of M. catarrhalis recovered from the sputum of adults monitored in the COPD Study Clinic were subjected to pulsed-field gel electrophoresis (PFGE) as previously described (12). Isolates were determined to be identical to one another when identical PFGE patterns were observed following the restriction of genomic DNA separately with both SpeI and NheI in the pairs of isolates.
Determination of sequences of ompE.
Primers GCGCGCGGATCCGCAGGCCTGGATCGCTC and ATATATGAATTCTTTGGCGTGATAAGCAAG were used to amplify ompE by PCR from genomic DNA prepared using a genomic DNA kit (Promega). The PCR mixture consisted of 10 ng of genomic DNA, 100 ng of each primer, 1 μl of 10 mM deoxynucleoside triphospate, 5 μl of Thermopol buffer (New England Biolabs), 0.5 μl of VentI polymerase (New England Biolabs), and 40.5 μl of water (total volume, 50 μl). After an initial denaturation for 3 min at 94°C, 30 cycles of the following program were carried out: 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. This was followed by incubation at 72°C for 3 min.
The 1,300-bp amplicon was either sequenced directly or cloned into pCR Blunt (Invitrogen) using the manufacturer's directions. For cloning into the plasmid, three separate PCRs were performed and the amplicons were cloned individually into pCR Blunt. The sequences were determined for two of the clones. If these two sequences differed, then the third clone would have been subjected to sequencing. For all isolates, the sequences of the first two clones were identical to one another.
RESULTS
Strain specificity of MAs.
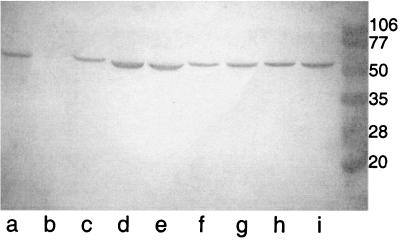
The 22 isolates (21 clinical isolates and 1 American Type Culture Collection strain) of M. catarrhalis were assayed with each of the eight MAbs in an immunoblot assay using either whole-bacterial-cell lysates or cell envelope preparations. Figure 1 shows the results of an immunoblot assay with MAb 1B3 for nine clinical isolates of M. catarrhalis. Eight of the clinical isolates were reactive with MAb 1B3, whereas strain 534 (lane b) was nonreactive. The eight MAbs recognize at least six different epitopes based on the results of the present study in combination with assays of overlapping recombinant fusion peptides containing sequences which span OMP E as reported previously (17). Table 1 shows the regions of OMP E which contain the epitopes recognized by each of the MAbs. MAb 2E11 was reactive with full-length OMP E in native form (whole-cell lysates of M. catarrhalis) and in recombinant form (purified OMP E with an amino-terminal histidine tag) (4), but it was nonreactive with each of five overlapping recombinant fusion peptides which span the sequence of the protein. Therefore, MAb 2E11 recognizes a conformational epitope on OMP E.
FIG. 1.
Immunoblot assay with MAb 1B3. Lanes contain whole-bacterial-cell lysates of strains 93P3B1 (a), 534 (b), 55P26B1 (c), 47P31B1 (d), 19P7B1 (e), 14P15B1 (f), 12P6B1 (g), 10P28B1 (h), and 3P3B1 (i). Molecular mass standards are noted in kilodaltons on the right.
TABLE 1.
Summary of reactivities of eight MAbs to OMP E from 22 strains of M. catarrhalis
| Result | No. of strains with result with MAba
|
|||||
|---|---|---|---|---|---|---|
| 1B3 and 9G10d (aa 80–180) | 12D5 (aa 160–260) | 4C11 (aa 160–260) | 5B3 and 14E10 (aa 240–340) | 9E3 (aa 240–340) | 2E11 | |
| Reactive | 18 | 21 | 22 | 18 | 22 | 22 |
| Nonreactive | 4 | 1 | 0 | 4 | 0 | 0 |
Region of OMP E recognized by MAbs based on amino acid (aa) sequence of mature protein (17).
Three of the MAbs (4C11, 9E3, and 2E11) recognized epitopes on all 22 strains (Table 1). Eighteen of the 22 isolates expressed the epitopes recognized by MAbs 1B3, 9G10d, 5B3, and 14E10. Of the four nonreactive isolates, two were sputum isolates from Buffalo, N.Y., one was a sputum isolate from Birmingham, United Kingdom, and one was a nasopharyngeal isolate from Utrecht, The Netherlands. MAb 12D5, which recognizes an epitope in the region of amino acids 160 to 260, was reactive with 21 of 22 isolates. The nonreactive isolate was the same sputum isolate from Birmingham, which was nonreactive with four other MAbs.
Sequence analysis of ompE.
The nucleotide sequence of the gene which encodes OMP E was determined for 16 strains of M. catarrhalis, including the four isolates which were nonreactive with some of the eight MAbs. Table 2 shows that OMP E displays a substantial degree of sequence conservation among strains of M. catarrhalis. Analysis of the amino acid sequences reveals that six different sequences of OMP E are represented by the 16 strains. Seven strains which are reactive with all eight MAbs have an identical amino acid sequence (Table 2, top row). Single amino acid differences among strains were identified at five positions, as noted in Table 2. In three additional positions in the protein, differences in two to six amino acids were noted. These include positions 77 and 78 (VQ versus IK), positions 112 to 117 (LAYKS versus FTYRRA) and positions 137 and 138 (IV versus TL). Overall, nucleotide sequences were 95.7 to 100% homologous with ompE of strain ATCC 25240. Amino acid sequences were 96.6 to 100% identical to OMP E of strain ATCC 25240.
TABLE 2.
Amino acid differences and MAb reactivities of OMP E from strains of M. catarrhalis
| Strain(s) | Amino acid(s) at position(s)
|
MAb reactivitya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 37 | 65 | 77–78 | 95 | 112–117 | 137–138 | 202 | 269 | Reactive | Nonreactive | |
| ATCC 25240, 7169, 15P4B1, 3614, 3583, 14P15B1, 14P23B1 | A | I | VQ | I | LAYKS | IV | K | A | All 8 | None |
| 8185 | S | V | VQ | I | LAYKS | IV | K | A | All 8 | None |
| 5P7B1, 5P10B1, 15P1B1 | S | V | IK | I | LAYKS | IV | K | A | All 8 | None |
| 4608 | A | V | IK | V | FTYRRA | TL | K | V | 9E3, 12D5, 4C11, 2E11 | 1B3, 9G10d, 5B3, 14E10 |
| 15P9B1, 15P12B1, 3.215 | S | V | IK | V | FTYRRA | TL | K | V | 9E3, 12D5, 4C11, 2E11 | 1B3, 9G10d, 5B3, 14E10 |
| 534 | S | V | IK | V | FTYRRA | TL | R | V | 9E3, 4C11, 2E11 | 1B3, 9G10d, 12D5, 5B3, 14E10 |
Reactivities with eight MAbs noted in Table 1.
Preliminary epitope analysis.
Since the regions of OMP E containing epitopes recognized by MAbs are known, correlating patterns of the reactivity of MAbs with amino acid sequences allows predictions regarding the location of epitopes recognized by the MAbs. MAbs 1B3 and 9G10d recognize the same, or a closely related, epitope on the bacterial surface in the region represented by amino acids 80 to 180 (4, 17). Amino acid sequences of OMP E of strains which were reactive with MAbs 1B3 and 9G10d were compared with sequences of nonreactive strains. Comparison of sequences in the region of amino acids 80 to 180 reveals that amino acid 95 (I), amino acids 112 to 116 (LAYKS), and/or amino acids 137 and 138 (IV) are important for reactivity of MAbs 1B3 and 9G10d (Table 2). In an effort to determine whether the MAbs recognize a linear epitope in the region of amino acids 112 to 116 and/or amino acids 137 and 138, individual recombinant fusion peptides representing amino acid sequences 107 to 121, 102 to 126, 130 to 145, and 107 to 145 were expressed and assayed by immunoblotting. MAbs 1B3 and 9G10d were nonreactive with these fusion peptides but reactive with the fusion peptide corresponding to amino acids 80 to 180 as reported previously (data not shown) (17).
MAbs 5B3 and 14E10 recognized an epitope in the nonreduced form of OMP E in the region of amino acids 240 to 340 (17). All 11 sequenced strains which were reactive with MAbs 5B3 and 14E10 had an alanine at position 269, while all 5 sequenced strains which were nonreactive has a valine at position 269 (Table 2). The amino acid sequence in this region of OMP E was otherwise identical among all strains. This observation indicates that the alanine at position 269 is important for the reactivity of MAbs 5B3 and 14E10.
Strain 534 is the only strain which is nonreactive with MAb 12D5, which recognizes an epitope in the region of amino acids 160 to 260 (17). Strain 534 is also the only one of the 16 sequenced strains which shows a difference in this region of the protein (Table 2). Therefore, the lysine at position 202 is critical for the reactivity of MAb 12D5.
Stability of ompE in the human respiratory tract.
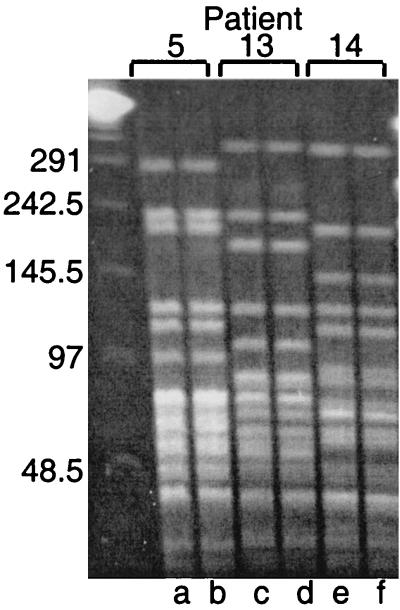
Three pairs of isolates recovered from the sputum of adults with COPD who were colonized by M. catarrhalis for durations of 3, 4, and 9 months were studied. PFGE on the prospectively collected isolates established that the patients were colonized by the same strain during these periods (Fig. 2). The gene which encodes OMP E was amplified by PCR and the sequences were determined. The sequences of ompE from all three sets of isolates were identical in every nucleotide from the beginning and end of the colonizing periods. This result indicates that the sequence of ompE was stable during colonization of the human respiratory tract in these three pairs of isolates.
FIG. 2.
Ethidium bromide-stained pulsed-field gel of genomic DNA cut with SpeI. The brackets at the top show the pairs of isolates and patient identification numbers from the COPD Study Clinic. Lanes contain DNA from strains 5P7B1 (a), 5P10B1 (b), 13P3B1 (c), 13P7B1 (d), 14P15B1 (e), and 14P23B1 (f). Molecular size standards are noted on the left in kilobases.
Human antibody response to OMP E.
To determine the extent to which OMP E was subjected to immune selective pressure while colonizing the human respiratory tracts of the three patients who were persistently colonized for 3 to 9 months, antibodies to OMP E were measured in supernatants of serum and sputum from these patients. Table 3 shows the results of quantitative ELISAs of serum immunoglobulin G (IgG) and sputum supernatant IgA at the beginning of the period of colonization. All three patients had detectable levels of serum IgG, and one of the three patients had detectable IgA in sputum supernatant. Assays of serum and sputum supernatants from before and after the colonization revealed that the level of antibody did not change during the period of colonization by M. catarrhalis.
TABLE 3.
Levels of immunoglobulin to OMP E in supernatants of serum and sputum from three adults with chronic bronchitis colonized by M. catarrhalis
| Patient identification no. | Serum pair | Serum IgG (μg/ml) | Sputum IgA (μg/ml) |
|---|---|---|---|
| 5 | 5E6-5E11 | 0.54 | 0.7 |
| 13 | 13E2-13E8 | 0.25 | <0.2 |
| 14 | 14E14-14E24 | 1.88 | <0.2 |
DISCUSSION
Previous work on OMP E suggested that the protein is conserved among strains of M. catarrhalis. This preliminary conclusion was based on several observations. (i) Adsorption assays with rabbit polyclonal antiserum showed that OMP E contains surface-exposed determinants that are shared among 17 of 20 strains (16). (ii) Analysis of PCR restriction fragment length polymorphisms with two restriction enzymes revealed identical patterns in 20 strains (3). (iii) Four MAbs recognized epitopes in all of the 19 strains studied (4). The present study was undertaken to rigorously assess the degree of sequence conservation of OMP E among strains of M. catarrhalis. Analysis of sequences of ompE from 16 strains revealed that the gene is well conserved among strains, showing 95.7 to 100% homology in nucleotide sequence. The protein sequences show 96.6 to 100% identity in comparison to the previously sequenced gene in strain ATCC 25240 (GenBank accession number L31788).
The sequences of OMP E appear to fall into two clusters based on sequence differences at positions 95, 112 to 117, 137 and 138, and 269 (Table 2). Analysis of the genetic relationship among strains of M. catarrhalis has recently been undertaken by several groups with various methods (5, 13, 26, 28, 29). In general, a high degree of genetic diversity exists among strains. Of interest, two studies have observed that complement-resistant strains form a distinct subpopulation among the strains (5, 26). Future studies will determine whether clusters of strains defined by the sequence of OMP E are associated with subpopulations of M. catarrhalis.
Immunoblot assays of clinical isolates with a battery of eight MAbs identified a small number of strains which did not react with selected MAbs. Comparing the amino acid sequences of reactive and nonreactive strains allows the prediction of epitopes recognized by the antibodies, since the MAbs are known to recognize specific regions of OMP E. MAbs 1B3 and 9G10d are especially interesting because they recognize an epitope on the surface of the intact bacterial cell (4). They may recognize the same epitope since they both bind a fusion peptide corresponding to amino acids 80 to 180 (17) and they inhibit the binding of one another to OMP E in a competitive ELISA (4). Comparison of sequences from amino acids 80 to 180 in reactive and nonreactive strains reveals differences at three sites, including amino acids 95, 112 to 117, and 137 and 138 (Table 2). Any or all three of these sites may be part of the epitope recognized by MAbs 1B3 and 9G10d. Fusion proteins corresponding to sequences in and around the region of amino acids 107 to 145 did not react with the MAbs, suggesting either that the epitope is located around amino acid 95 or that a larger peptide is required to reproduce the conformation of the epitope.
Comparison of sequences and patterns of reactivity with MAbs reveals that single amino acid differences account for reactivity with selected MAbs. For example, MAb 12D5 recognizes amino acids 160 to 260 (17). Strain 534 is nonreactive with 12D5 and differs from reactive strains in a single amino acid in that amino acid range, indicating that the lysine at position 202 is critical for the epitope recognized by MAb 12D5. Similarly, MAbs 5B3 and 14E10 recognize amino acids 240 to 340. Reactive and nonreactive strains differ by a single amino acid in this region of OMP E. The valine in place of alanine at position 269, a difference of a single methyl group, accounts for the lack of reactivity with MAbs 5B3 and 14E10.
The gene which encodes protein P2, the major outer membrane protein of nontypeable Haemophilus influenzae, undergoes nonsynonymous point mutations under immune selective pressure and undergoes horizontal transfer between strains in the human respiratory tract (7, 8, 24). These are thought to be mechanisms of immune evasion, thereby facilitating persistent colonization of the respiratory tract. As a preliminary assessment of the stability of OMP E of M. catarrhalis in the human respiratory tract, the sequence of ompE was determined in sets of isolates which colonized the respiratory tracts of adults with COPD continuously for 3 to 9 months. Immunoassays showed that all three patients had serum IgG to OMP E prior to colonization, and no change in the level was observed during colonization (Table 3). The genes showed identical nucleotides at all positions, indicating that the gene did not undergo changes during persistent colonization. A similar observation has been made with OMP CD of M. catarrhalis (9).
OMP E has several characteristics which indicate that it may be an effective vaccine antigen. The protein is present in all strains tested (2, 3). It is abundantly expressed on the bacterial surface based on results of immunofluorescence assays and flow cytometry with MAbs (4). OMP E is immunogenic in animals (4, 17). Some adults with COPD have serum IgG to OMP E, and the majority have mucosal IgA to OMP E (4). The present study establishes that OMP E is highly conserved among strains of M. catarrhalis, an important characteristic for an effective vaccine antigen.
ACKNOWLEDGMENT
This work was supported by grant AI28304 from the National Institute of Allergy and Infectious Diseases and the Department of Veterans Affairs.
REFERENCES
- 1.American Thoracic Society. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. Am Rev Respir Dis. 1987;136:225–244. doi: 10.1164/ajrccm/136.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Bartos L C, Murphy T F. Comparison of the outer membrane proteins of 50 strains of Branhamella catarrhalis. J Infect Dis. 1988;158:761–765. doi: 10.1093/infdis/158.4.761. [DOI] [PubMed] [Google Scholar]
- 3.Bhushan R, Craigie R, Murphy T F. Molecular cloning and characterization of outer membrane protein E of Moraxella (Branhamella) catarrhalis. J Bacteriol. 1994;176:6636–6643. doi: 10.1128/jb.176.21.6636-6643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhushan R, Kirkham C, Sethi S, Murphy T F. Antigenic characterization and analysis of the human immune response to outer membrane protein E of Branhamella catarrhalis. Infect Immun. 1997;65:2668–2675. doi: 10.1128/iai.65.7.2668-2675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bootsma H J, van der Heide H G, van de Pas S, Schouls L M, Mooi F R. Analysis of Moraxella catarrhalis by DNA typing: evidence for a distinct subpopulation associated with virulence traits. J Infect Dis. 2000;181:1376–1387. doi: 10.1086/315374. [DOI] [PubMed] [Google Scholar]
- 6.Brook I, Gober A E. Microbiologic characteristics of persistent otitis media. Arch Otolaryngol Head Neck Surg. 1998;124:1350–1352. doi: 10.1001/archotol.124.12.1350. [DOI] [PubMed] [Google Scholar]
- 7.Duim B, van Alphen L, Eijk P, Jansen H M, Dankert J. Antigenic drift of non-encapsulated Haemophilus influenzae major outer membrane protein P2 in patients with chronic bronchitis is caused by point mutations. Mol Microbiol. 1994;11:1181–1189. doi: 10.1111/j.1365-2958.1994.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 8.Groeneveld K, van Alphen L, Voorter C, Eijk P P, Jansen H M, Zanen H C. Antigenic drift of Haemophilus influenzae in patients with chronic obstructive pulmonary disease. Infect Immun. 1989;57:3038–3044. doi: 10.1128/iai.57.10.3038-3044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiao C B, Sethi S, Murphy T F. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from the human respiratory tract. Microb Pathog. 1995;19:215–225. doi: 10.1016/s0882-4010(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 10.Kennett R H. Cell fusion. Methods Enzymol. 1979;58:345–359. doi: 10.1016/s0076-6879(79)58149-x. [DOI] [PubMed] [Google Scholar]
- 11.Klein J O. Otitis media. Clin Infect Dis. 1994;19:823–833. doi: 10.1093/clinids/19.5.823. [DOI] [PubMed] [Google Scholar]
- 12.Klingman K L, Pye A, Murphy T F, Hill S L. Dynamics of respiratory tract colonization by Moraxella (Branhamella) catarrhalis in bronchiectasis. Am J Respir Crit Care Med. 1995;152:1072–1078. doi: 10.1164/ajrccm.152.3.7663786. [DOI] [PubMed] [Google Scholar]
- 13.Martinez G, Ahmed K, Zheng C H, Watanabe K, Oishi K, Nagatake T. DNA restriction patterns produced by pulsed-field gel electrophoresis in Moraxella catarrhalis isolated from different geographical areas. Epidemiol Infect. 1999;122:417. doi: 10.1017/s0950268899002381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miravitlles M, Espinosa C, Fernandez-Laso E, Martos J A, Maldonado J A, Gallego M Study Group of Bacterial Infection in COPD. Relationship between bacterial flora in sputum and functional impairment in patients with acute exacerbations of COPD. Chest. 1999;116:40–46. doi: 10.1378/chest.116.1.40. [DOI] [PubMed] [Google Scholar]
- 15.Murphy T F. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol Rev. 1996;60:267–279. doi: 10.1128/mr.60.2.267-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy T F, Bartos L C. Surface-exposed and antigenically conserved determinants of outer membrane proteins of Branhamella catarrhalis. Infect Immun. 1989;57:2938–2941. doi: 10.1128/iai.57.10.2938-2941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy T F, Brauer A L, Yuskiw N, Hiltke T J. Antigenic structure of outer membrane protein E of Moraxella catarrhalis and construction and characterization of mutants. Infect Immun. 2000;68:6250–6256. doi: 10.1128/iai.68.11.6250-6256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy T F, Sethi S. Bacterial infection in chronic obstructive pulmonary disease. Am Rev Respir Dis. 1992;146:1067–1083. doi: 10.1164/ajrccm/146.4.1067. [DOI] [PubMed] [Google Scholar]
- 19.Murphy T F, Sethi S. A national strategy for research in chronic obstructive pulmonary disease. JAMA. 1997;277:1596. [PubMed] [Google Scholar]
- 20.Ruuskanen O, Heikkinen T. Otitis media: etiology and diagnosis. Pediatr Infect Dis J. 1994;13:S23–S26. [PubMed] [Google Scholar]
- 21.Sarwar J, Campagnari A A, Kirkham C, Murphy T F. Characterization of an antigenically conserved heat-modifiable major outer membrane protein of Branhamella catarrhalis. Infect Immun. 1992;60:804–809. doi: 10.1128/iai.60.3.804-809.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seneff M G, Wagner D P, Wagner R P, Zimmerman J E, Knaus W A. Hospital and 1-year survival of patients admitted to intensive care units with acute exacerbation of chronic obstructive pulmonary disease. JAMA. 1995;274:1852–1857. [PubMed] [Google Scholar]
- 23.Sethi S, Murphy T F. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state of the art review. Clin Microbiol Rev. 2001;14:336–363. doi: 10.1128/CMR.14.2.336-363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith-Vaughan H C, Sriprakash K S, Mathews J D, Kemp D J. Nonencapsulated Haemophilus influenzae in aboriginal infants with otitis media: prolonged carriage of P2 porin variants and evidence for horizontal P2 gene transfer. Infect Immun. 1997;65:1468–1474. doi: 10.1128/iai.65.4.1468-1474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Hare G F, Shurin P A, Marchant C D, Cartelli N A, Johnson C E, Fulton D, Carlin S, Kim C H. Acute otitis media caused by Branhamella catarrhalis: biology and therapy. Rev Infect Dis. 1987;9:16–27. doi: 10.1093/clinids/9.1.16. [DOI] [PubMed] [Google Scholar]
- 26.Verduin C M, Kools-Sijmons M, van der Plas J, Vlooswijk J, Tromp M, van Dijk H, Banks J, Verbrugh H, van Belkum A. Complement-resistant Moraxella catarrhalis forms a genetically distinct lineage within the species. FEMS Microbiol Lett. 2000;184:1–8. doi: 10.1111/j.1574-6968.2000.tb08981.x. [DOI] [PubMed] [Google Scholar]
- 27.Verghese A, Roberson D, Kalbfleisch J H, Sarubbi F. Randomized comparative study of cefixime versus cephalexin in acute bacterial exacerbations of chronic bronchitis. Antimicrob Agents Chemother. 1990;34:1041–1044. doi: 10.1128/aac.34.6.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vu-Thien H, Dulot C, Moissenet D, Fauroux B, Garbarg-Chenon A. Comparison of randomly amplified polymorphic DNA analysis and pulsed-field gel electrophoresis for typing of Moraxella catarrhalis strains. J Clin Microbiol. 1999;37:450–452. doi: 10.1128/jcm.37.2.450-452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker E S, Preston R A, Post J C, Ehrlich G D, Kalbfleisch J H, Klingman K L. Genetic diversity among strains of Moraxella catarrhalis: analysis using multiple DNA probes and a single-locus PCR-restriction fragment length polymorphism method. J Clin Microbiol. 1998;36:1977–1983. doi: 10.1128/jcm.36.7.1977-1983.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson R. The role of infection in COPD. Chest. 1998;113:242S–248S. doi: 10.1378/chest.113.4_supplement.242s. [DOI] [PubMed] [Google Scholar]