Abstract
The coronavirus disease 2019 (COVID-19) pandemic is ongoing because of the repeated emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, highlighting the importance of developing vaccines for variants that may continue to emerge. In the present review, we discuss humoral immune responses against SARS-CoV-2 with a focus on the antibody breadth to the variants. Recent studies have revealed that the temporal maturation of humoral immunity improves the antibody potency and breadth to the variants after infection or vaccination. Repeated vaccination or infection further accelerates the expansion of the antibody breadth. Memory B cells play a central role in this phenomenon, as the reactivity of the B-cell antigen receptor (BCR) on memory B cells is a key determinant of the antibody potency and breadth recalled upon vaccination or infection. The evolution of memory B cells remarkably improves the reactivity of BCR to antigenically distinct Omicron variants, to which the host has never been exposed. Thus, the evolution of memory B cells toward the variants constitutes an immunological basis for the durable and broad control of SARS-CoV-2 variants.
Keywords: Immunological memory, Vaccine, B cell, Antibody, SARS-CoV-2
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative virus of coronavirus disease 2019 (COVID-19), is currently threatening the lives of people worldwide. COVID-19 presents with mild to severe respiratory symptoms, and more than 500 million cases with more than 6 million deaths have been reported globally as of August 31, 2022 (https://covid19.who.int/). Since the initial case was identified in December 2019, several vaccines that confer protection against SARS-CoV-2 have rapidly been developed and authorized for emergency use. COVID-19 vaccines efficiently confer protection against SARS-CoV-2 infection, hospitalization, and death [1, 2].
Immunological memory responses that are formed and maintained in response to primary antigen exposure protect the host from subsequent viral infection. Immunological memory responses are of particular importance for vaccination, as they are the basis for conferring protection against re-infection by a variety of pathogens [3]. Immunological memory involves two classes of responses, humoral and cellular immunity, driven by B and T cells, respectively. The present review will focus on the humoral immune system, while cellular immunity to SARS-CoV-2 has been reviewed elsewhere [4].
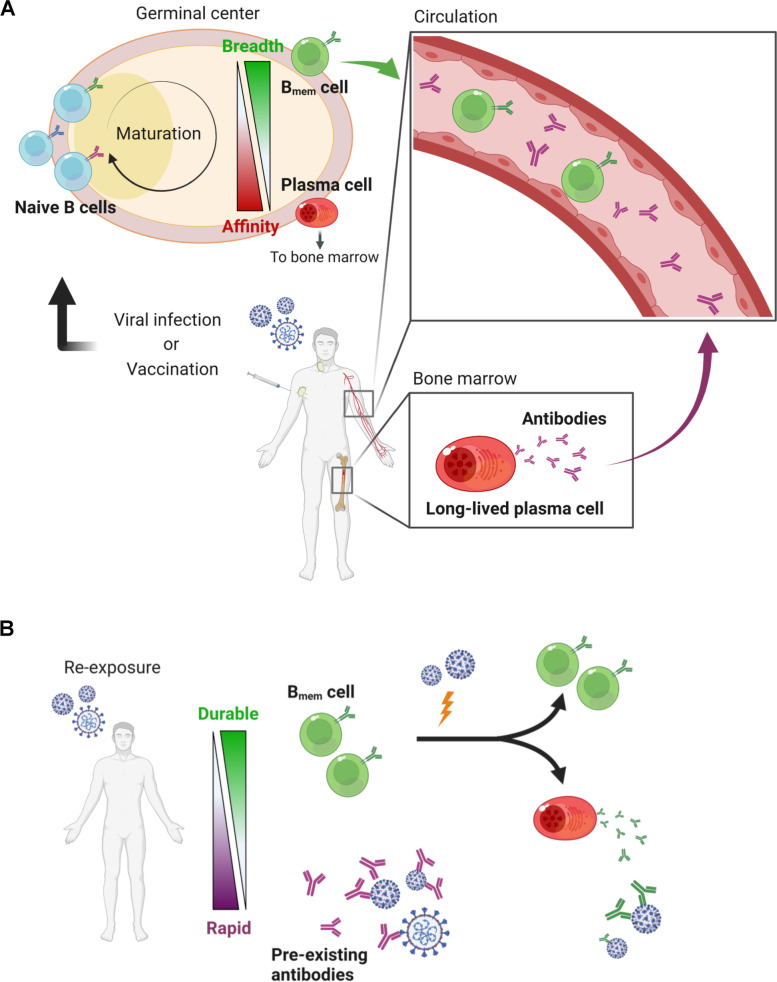
Humoral memory responses to secondary or further antigen exposure involve two defense walls: long-lived plasma and memory B (Bmem) cells. Both long-lived B cells largely develop within germinal centers (GCs), where those with high-affinity B cell receptors (BCRs) to the antigens emerge with somatic hypermutations and are clonally selected (Fig. 1A). However, the two long-lived B cells are functionally different from each other: plasma cells mediate prompt and effective protection via pre-existing antibodies as the first line of defense, and Bmem cells serve as the backup of pre-existing antibodies by robustly supplying plasma cells in response to re-invading antigens (Fig. 1B). In cases of acute viral infection, the circulating antibody level declines in a biphasic manner after its peak: a rapid decline in the early phase (< 3 months) followed by a gradual decline in the late phase (< 1 year) [5]. The kinetics are compatible with that of plasma cells in the bone marrow, which decline from its peak to the baseline within 1 year [5]. By stark contrast, Bmem cells, circulating in the peripheral blood, increase in numbers in 2–3 weeks and are stably maintained for up to 1 year [6, 7]. Further differences between plasma cells and Bmem cells have been suggested in their reactivity to variants [8–10]. Plasma cells are highly specific to homologous antigens with high affinity, while Bmem cells tend to have BCRs with low affinity but relatively broad reactivity to heterologous antigens of variants [11–13].
Fig. 1.
Humoral immune memory responses. A Humoral immune responses to a primary antigen exposure are depicted. Viral infection or vaccination induces Bmem and plasma cells through germinal center reaction, conferring immunological memory. B Memory responses to antigen re-exposure are described. Pre-existing antibodies secreted from long-lived plasma cells rapidly eliminate the viruses, whereas Bmem cells function as a backup reservoir for plasma cells. The figure was created with BioRender.com
In-depth immune profiling on SARS-CoV-2 convalescent and vaccinated individuals have greatly advanced our understanding of humoral immunity in relation to the breadth to virus variants. In this review, we discuss the recent progress in humoral memory responses to SARS-CoV-2. A comprehensive view of these responses will provide more insights into developing vaccines for the effective and broad control of SARS-CoV-2 variants that are likely to emerge in the future.
Main text
Neutralizing antibodies as an immune correlate of protection
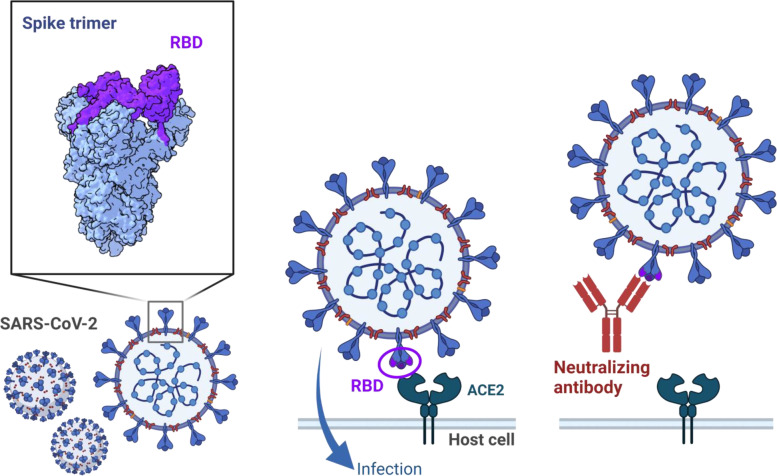
SARS-CoV-2 contains structural proteins, including spike, envelope, membrane, and nucleocapsid proteins, and non-structural proteins encoded by several open reading frames [14]. The spike protein binds to host angiotensin-converting enzyme 2 (ACE2) through a receptor-binding domain (RBD), which is critical for viral entry into host cells [15–18] (Fig. 2). Although SARS-CoV-2 infection elicits antibodies targeting a variety of viral proteins, including structural and non-structural proteins [19], antibodies against spike RBD are particularly important for protective immunity, as they exhibit potent neutralizing activity [20–26]. Despite detection of neutralizing activity by non-RBD antibodies [27, 28], the majority of neutralizing antibodies circulating in the blood are known to target RBD [23–25]. The fact that all monoclonal antibody therapeutics in licensure recognize RBD epitopes [29–31] also illustrates the immunodominant nature of RBD for neutralizing antibodies. Furthermore, the successful use of neutralizing antibodies as an immune correlate of protection in vaccinees supports the functional role in preventing symptomatic diseases [32–34].
Fig. 2.
Antibody-dependent neutralization of SARS-CoV-2 entry through RBD. SARS-CoV-2 infection through RBD of spike proteins and neutralization by a protective antibody are depicted. The structural analysis on trimeric spike protein (upper left) is visualized with UCSF ChimeraX software (Refs. [35, 36]) using data from PDB (PDB ID: 6VSB, https://doi.org/10.2210/pdb6VSB/pdb, Ref. [37]). The figure was created with BioRender.com
SARS-CoV-2 variants escape the neutralizing antibody with RBD mutations
SARS-CoV-2 continuously changes its genome through mutations or viral recombination, and some emerging variants have acquired the ability to escape from neutralizing antibodies [38]. Antibody neutralization is one of the driving forces for selecting viral escape mutations, as observed during viral incubation in vitro under the pressure of neutralizing plasma [39–41]. Virus evolution has also been demonstrated in convalescent plasma treatment of an immunocompromised patient with persistent SARS-CoV-2 infection [42].
Variants with increased transmissibility, virulence, or immune escape are threats to global public health. Multiple SARS-CoV-2 variants with mutations in their spike RBD have emerged since the ancestral strain was identified in December 2019. The latest Omicron variant, which was detected in November 2021, subsequently diverged into a number of sublineages, e.g., BA.1, BA.5, BQ.1.1, and XBB. Each sublineage has 15 or more amino acid substitutions in its RBD [43]. The number of RBD mutations was greater than that of pre-Omicron variants with no more than three mutations. With numerous RBD mutations, Omicron variants strongly escape neutralizing antibodies [44–59]. A few antibodies retain neutralizing activity against a widely circulating Omicron subvariant, BA.5, by binding to conserved sites of the RBD between the ancestral and Omicron strains [57, 59].
Temporal maturation of plasma antibodies induced by SARS-CoV-2 infection or vaccination
Serum antibodies induced by infection or vaccination act as the first wall of defense against future SARS-CoV-2 infections. Neutralizing antibodies of COVID-19 convalescent plasma peaks around 1–2 months after the infection, followed by a biphasic decrease, namely, early rapid (< 6 months), and late gradual (< 12 months) [60–62], roughly compatible with the kinetics of an influenza vaccine [5]. The titer declines but is still detectable even at late time points (5–12 months after infection). Indeed, spike-specific plasma cells are also detected in the bone marrow of convalescent individuals 11 months after infection [62].
The antibody titers induced by COVID-19 vaccination have essentially the same kinetics. COVID-19 vaccines have been designed to activate B cells specific to the SARS-CoV-2 spike, as the spike is a target of protective antibodies, as described above. Several modalities of COVID-19 vaccines have been developed, such as mRNA, viral-vector, inactivated whole virus, and protein vaccines. Among these, immune responses induced by mRNA vaccines, which were approved for human use for the first time, have been intensely analyzed. Vaccination induces neutralizing antibodies to SARS-CoV-2 in the plasma, which reaches its peak at approximately 1 month after the 2nd vaccination and then declines. The neutralizing activity is low but still detectable 5 months after the 2nd dose [63–66]. In line with this, spike-specific plasma cells can be detected at 5 months after vaccination in the draining lymph nodes and bone marrow [67]. The antibody response induced by an influenza vaccine persists up to 5 years above detectable levels [68]; however, it remains unclear how long the neutralizing activity induced by COVID-19 vaccine would remain above detectable levels.
Although the antibody quantity decreases, plasma antibodies mature in terms of neutralizing potency and breadth over time. Neutralization potency, which has been reported to correlate with the prognosis of patients [69], increased from the early to the late time points in convalescent plasma [61] and vaccinated plasma (unpublished data). Interestingly, neutralizing breadth to the SARS-CoV-2 variants also increases over time, despite individuals being exposed only to the ancestral strain [60, 61, 63, 65, 66]. The qualitative maturation of plasma antibodies after antigen exposure suggests gradual changes in antibody repertoires in long-lived plasma cells, which continue to replenish circulating antibodies over time [67].
Repeated antigen exposures further improve humoral immune responses to the Omicron variants
Although plasma antibodies mature as mentioned above, the Omicron variants strongly escape vaccine-induced or convalescent serum polyclonal antibodies [44–48, 51, 53, 58, 59, 65, 70–77]. In some vaccinees, the neutralization titer for the Omicron variants after the 2nd dose of vaccination is not detectable throughout the time course. In other vaccinees with detectable titers against the Omicron variants, the Omicron neutralization titer after two doses of the vaccine has been reported to be 10 to > 120 times lower than that of the ancestral Wuhan strain.
Meanwhile, current studies on vaccinated cohorts have revealed that humoral immunity improves reactivity to Omicron variants even after the 3rd vaccination by the ancestral Wuhan-based strain [45, 46, 48, 77–79]. Neutralization titers to the Omicron variants after the 3rd dose of vaccines are still 5–10 times lower than those to the ancestral strain, but the difference is smaller than those after the 2nd dose from the same cohort. Improved neutralization to the Omicron variants has also been observed in breakthrough infections of pre-Omicron strains after two doses of vaccine [80]. Collectively, the three exposures to non-Omicron antigens induce neutralizing antibodies to the Omicron variants, indicating that repeated antigen exposure, even upon the ancestral strain, expands the breadth to the variants.
Memory B cells are durably maintained with the breadth to variants
Exposure to the 3rd antigen with vaccination or breakthrough infection boosts the neutralizing titer to the same or higher level compared with the 2nd vaccination [45, 46, 48, 77–82]. Antibodies are largely supplied by plasma cells that originate from reactivated Bmem cells. As observed in influenza vaccine studies [6, 7], Bmem cells specific to the spike are stably maintained or even increase in number from 1 to 6 months after the 2nd vaccination [63–65]. These Bmem cell numbers further increase after the 3rd vaccination [79, 81].
Bmem cells exhibit extended cross-reactivity to the virus variants compared to antibodies secreted by plasma cells. Even after the 2nd dose of the vaccine, 30–50% of Wuhan RBD-reactive Bmem cells are cross-reactive to the Omicron variants, and their numbers increase over time [65, 81, 83]. A fraction of the Omicron-reactive Bmem cells retain BCRs which neutralize Omicron variants [65, 79, 83]. Thus, the 3rd vaccination and breakthrough infection reactivate the Omicron-neutralizing Bmem cells, which differentiate into plasma cells secreting Omicron-neutralizing antibodies. Indeed, a fraction of Omicron-reactive Bmem cells exhibit an activated phenotype after the 3rd dose [81].
The interval between antigen exposures is also a key factor in the cross-reactivity of humoral immune responses. A long interval between the 1st and 2nd doses of COVID-19 mRNA vaccines enhances the plasma neutralization titer against the Alpha, Beta, Gamma, and Delta variants compared with a short interval [84, 85]. At the breakthrough infection of non-Omicron variants after the 2nd vaccination, a longer interval from the vaccination induced a higher neutralization titer of the Omicron variant [80]. The longer interval may enable Bmem cells to undergo maturation for a longer time to increase cross-reactivity before reactivation.
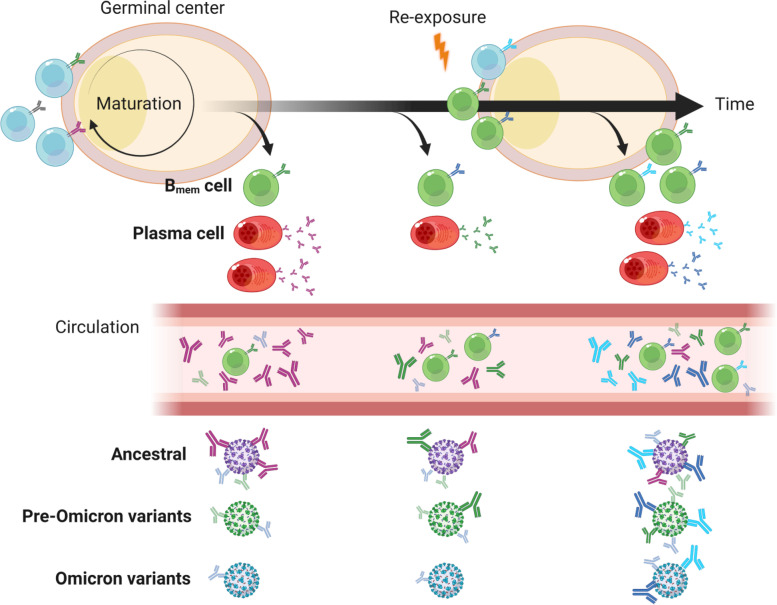
Collectively, Bmem cells have evolved their BCR repertoire to acquire broad reactivity to the variants over time after ancestral strain-based vaccination (Fig. 3). Hence, Bmem cells may play a pivotal role in both the durability and breadth of protection against emerging SARS-CoV-2 variants.
Fig. 3.
Evolution of humoral immune responses to SARS-CoV-2. Bmem cells evolve their BCRs over time to acquire the breadth to pre-Omicron variants. The BCR breadth is further expanded to Omicron variants after re-exposures by vaccination or infection. These processes may play a pivotal role in durable and broad protection against SARS-CoV-2 variants that emerge in the future. The figure was created with BioRender.com
Memory B cell selection underlying the evolution
Bmem cells encompass phenotypically and functionally heterogeneous subsets [3, 86, 87]. After the 2nd vaccination, the subset composition of Bmem cells changes over time, skewing toward CD21+CD27+ resting Bmem cell dominance from the early to late time points [65]. The accumulation of resting Bmem cells is due to an increase in the number of cross-reactive resting Bmem cells but not due to the Bmem cells that are reactive only to the ancestral strain, resulting in the evolution of reactivity in the resting Bmem cells. CD27+ Bmem cells, including the resting subset, are intrinsically programmed to enhance survival and rapidly respond to restimulation [88]. Hence, the increase in the evolved cross-reactive resting Bmem cells may contribute to a rapid cross-reactive antibody response after antigen re-exposure, as observed at the 3rd vaccination [81].
Antigen exposure, such as infection or vaccination, shapes the B cell repertoire through BCR-dependent selection and maturation. The antigen activates antigen-reactive naïve B cells and Bmem cells, which in turn undergo Bmem/plasma cell differentiation or further maturation in GCs in lymphoid tissues, where B cells diversify their BCRs and undergo selection [8, 89–91]. Throughout the GC reaction, a fraction of GC B cells differentiate into Bmem and plasma cells. These processes diversify and continuously change the BCR repertoire, which may contribute to the evolution of Bmem and plasma cell BCRs induced by SARS-CoV-2 infection and vaccination. Indeed, GCs have been detected in the draining lymph nodes of mRNA vaccinees and persist for months [67, 92, 93]. The Bmem cells after vaccination accumulate mutations in their BCRs over time [63, 79], indicating that the vaccine-induced Bmem cells are derived from the GCs. In addition, COVID-19 convalescent individuals also sustain Bmem cells with mutated BCRs and bone marrow plasma cells, which is indicative of their GC-dependent development [60, 62, 94]. Although patients with severe COVID-19 harbor dysfunctional responses at acute phase [95, 96], the higher number of circulating follicular helper T cells and comparable affinity maturation of plasma IgG titer in the convalescent phases suggest the involvement of functional GC responses at a different time or location [97, 98]. Otherwise, non-GC pathway could be involved in severe cases [96]. Bmem cell evolution may involve B-cell selection in GCs over time. The prolonged interval between antigen exposures may result in a longer selection of Bmem cells before reactivation, which in turn confers more evolved humoral antibody responses, as mentioned above. In COVID-19 convalescent individuals, Bmem cells also evolve their reactivity against variant strains [60, 99]. The evolved cross-reactivity of a Bmem cell pool can be achieved in two ways: mutation-dependent maturation of pre-existing Bmem cell clones, and expansion of newly emerging clones with cross-reactivity. Bmem cells after the 3rd vaccination include both persistent and newly emerged Bmem cell clones [79], indicating the contribution of both pre-existing and new Bmem cells to the evolving reactivity of the BCR repertoire. The BCRs of the newly emerging clones bind distinct epitopes from those of the other clones, thereby changing the epitope distribution of the overall Bmem cell repertoire.
One possible mechanism underlying the change in paratope distribution is a process called antibody feedback. In this process, the antibodies in circulation mask the antigenic epitopes for B-cell selection and suppress B cell activation toward the same epitopes [100–104]. COVID-19 vaccines or active SARS-CoV-2 infection predominantly activate spike-specific B cells toward immunodominant epitopes [23, 105]. Activated B cells expand and differentiate into GC B, Bmem, and plasma cells, which in turn secrete antibodies. The predominant antibodies produced at the early stage may dampen the selection of B cells specific to the predominant epitopes at the later stage and the next vaccination, resulting in a change in the predominant epitopes of the B cell repertoire over time. A study demonstrated that antibody feedback by monoclonal antibodies alters epitopes of Bmem cells induced by COVID-19 vaccination [106].
Whereas repeated COVID-19 vaccination improves cross-reactivity to variant strains, repeated vaccination with homologous flu hemagglutinin (HA) skews antibody response towards the more specific HA head domain of the targeted strain [107]. Concordantly, compared to plasmablasts, a single dose of HA vaccination induces GC B cells with more specific reactivity against the vaccinated HA, which may be involved in later humoral responses [108]. Some of those GC B cells recognize different epitopes on the HA from those plasma antibodies bind. Thus, homologous flu HA protein and COVID-19 mRNA vaccines both induce the evolution of B cell responses, but to opposite directions, specific and broad reactivity, respectively. The discrepancy in the direction may be related to the properties of cross-reactive antibodies and their epitopes, since most of flu cross-reactive antibodies which target well-conserved stalk region of HA are polyreactive [107]. This implies a selective disadvantage of the flu cross-reactive B cells due to immune tolerance. Collectively, the Bmem cell evolution is induced independent of antigens and modality, while the direction of the evolution may vary by antigens.
Further investigation of the mechanism underlying the evolution of broadly reactive Bmem cells, induced by COVID-19 vaccination, is indispensable for the development of universal vaccines that confer durable and broad protection against future SARS-CoV-2 variants and other pandemic viruses.
Conclusion
The repeated emergence of SARS-CoV-2 variants escaping vaccine-induced immune responses has emphasized the importance of developing universal vaccines that induce protective immune memory toward a variety of variants, ideally including future escaping variants. Recent intense cohort studies on COVID-19-convalescent and vaccinated individuals have revealed that Bmem cells play a pivotal role in durable and broad humoral memory responses. Bmem cells evolve their reactivity to the variants over time, which are reactivated to secrete antibodies into circulation in response to subsequent antigen exposure. In summary, persistent maturation and repeated reactivation are the two key events involved in the evolution of humoral immune responses to SARS CoV-2. Although most COVID-19 vaccine studies have focused on responses to mRNA vaccines, some studies have reported that vaccines with other modalities induce Bmem cells of similar quality [109, 110]. In addition to these modalities, switching to Omicron-adapted vaccines from the ancestral strain is considered and discussed in the current situation of the Omicron outbreak. However, the cross-reactivity of Bmem cells induced by the Omicron-adapted vaccine booster is comparable to that of the ancestral booster in nonhuman primates [111]. Further analyses of B-cell responses to vaccines with various modalities and antigens will provide more insights into the evolution of Bmem cells, which may be pivotal for the development of universal vaccines.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing. Molecular graphics and analyses were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- BCR
B-cell antigen receptor
- Bmem Cell
Memory B cell
- COVID-19
Coronavirus disease 2019
- GC
Germinal center
- HA
Hemagglutinin
- RBD
Receptor-binding domain
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
Authors’ contributions
R.K. wrote the manuscript’s initial draft. S.M. and Y.T. revised the manuscript. All the authors have read and approved the final manuscript.
Funding
The present work was supported by JSPS KAKENHI Grant Numbers JP22K16383 (to R.K.) and Japan Agency for Medical Research and Development grants JP20fk0108516j0001 (to S.M.), JP20fk0108104 (to Y.T.), JP20fk0108534 (to Y.T.), and JP22fk0108141 (to Y.T.).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Saya Moriyama, Email: sayamrym@niid.go.jp.
Yoshimasa Takahashi, Email: ytakahas@niid.go.jp.
References
- 1.Self WH, Tenforde MW, Rhoads JP, Gaglani M, Ginde AA, Douin DJ, et al. Comparative effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) vaccines in preventing COVID-19 hospitalizations among adults without immunocompromising conditions - United States, March-August 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1337–1343. doi: 10.15585/mmwr.mm7038e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botton J, Semenzato L, Jabagi M-J, Baricault B, Weill A, Dray-Spira R, et al. Effectiveness of Ad26.COV2.S vaccine vs BNT162b2 vaccine for COVID-19 hospitalizations. JAMA Netw Open. 2022;5:e220868. doi: 10.1001/jamanetworkopen.2022.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGrath JJC, Li L, Wilson PC. Memory B cell diversity: insights for optimized vaccine design. Trends Immunol. 2022;43:343–354. doi: 10.1016/j.it.2022.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 5.Davis CW, Jackson KJL, McCausland MM, Darce J, Chang C, Linderman SL, et al. Influenza vaccine-induced human bone marrow plasma cells decline within a year after vaccination. Science. 2020;370:237–241. doi: 10.1126/science.aaz8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews SF, Chambers MJ, Schramm CA, Plyler J, Raab JE, Kanekiyo M, et al. Activation dynamics and immunoglobulin evolution of pre-existing and newly generated human memory B cell responses to influenza hemagglutinin. Immunity. 2019;51:398–410.e5. doi: 10.1016/j.immuni.2019.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Inoue T, Shinnakasu R, Kurosaki T. Generation of high quality memory B cells. Front Immunol. 2021;12:825813. doi: 10.3389/fimmu.2021.825813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akkaya M, Kwak K, Pierce SK. B cell memory: building two walls of protection against pathogens. Nat Rev Immunol. 2020;20:229–238. doi: 10.1038/s41577-019-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laidlaw BJ, Cyster JG. Transcriptional regulation of memory B cell differentiation. Nat Rev Immunol. 2021;21:209–220. doi: 10.1038/s41577-020-00446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J Exp Med. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leach S, Shinnakasu R, Adachi Y, Momota M, Makino-Okamura C, Yamamoto T, et al. Requirement for memory B-cell activation in protection from heterologous influenza virus reinfection. Int Immunol. 2019;31:771–779. doi: 10.1093/intimm/dxz049. [DOI] [PubMed] [Google Scholar]
- 13.Adachi Y, Onodera T, Yamada Y, Daio R, Tsuiji M, Inoue T, et al. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J Exp Med. 2015;212:1709–1723. doi: 10.1084/jem.20142284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JF-W, Kok K-H, Zhu Z, Chu H, To KK-W, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 16.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 20.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–7. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piccoli L, Park Y-J, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W-T, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreano E, Nicastri E, Paciello I, Pileri P, Manganaro N, Piccini G, et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–35.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Muecksch F, Cho A, Gaebler C, Hoffmann H-H, Ramos V, et al. Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins. Immunity. 2022;55:998–1012.e8. doi: 10.1016/j.immuni.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–47.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi H, Liu B, Wang X, Zhang L. The humoral response and antibodies against SARS-CoV-2 infection. Nat Immunol. 2022;23:1008–1020. doi: 10.1038/s41590-022-01248-5. [DOI] [PubMed] [Google Scholar]
- 30.Gruell H, Vanshylla K, Weber T, Barnes CO, Kreer C, Klein F. Antibody-mediated neutralization of SARS-CoV-2. Immunity. 2022;55:925–944. doi: 10.1016/j.immuni.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang R, Wang Y, Wang L, Deng X, Huo S, Jiang S, et al. Neutralizing monoclonal antibodies against highly pathogenic coronaviruses. Curr Opin Virol. 2021;53:101199. doi: 10.1016/j.coviro.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 33.Earle KA, Ambrosino DM, Fiore-Gartland A, Goldblatt D, Gilbert PB, Siber GR, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert PB, Montefiori DC, McDermott AB, Fong Y, Benkeser D, Deng W, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc Natl Acad Sci U S A. 2021;118:e2103154118. doi: 10.1073/pnas.2103154118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt F, Weisblum Y, Rutkowska M, Poston D, DaSilva J, Zhang F, et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600:512–516. doi: 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia S, Wang L, Zhu Y, Lu L, Jiang S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct Target Ther. 2022;7:241. doi: 10.1038/s41392-022-01105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.VanBlargan LA, Errico JM, Halfmann PJ, Zost SJ, Crowe JE, Jr, Purcell LA, et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28:490–5. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, Iketani S, Guo Y, Chan JF-W, Wang M, Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–81. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell. 2022;185:447–56.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iketani S, Liu L, Guo Y, Liu L, Chan JF-W, Huang Y, et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–6. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou T, Wang L, Misasi J, Pegu A, Zhang Y, Harris DR, et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B11529. Science. 2022;376:eabn8897. doi: 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du W, Hurdiss DL, Drabek D, Mykytyn AZ, Kaiser FK, González-Hernández M, et al. An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern. Sci Immunol. 2022;7:eabp9312. doi: 10.1126/sciimmunol.abp9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, Jia Z, Bao L, Wang L, Cao L, Chi H, et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature. 2022;603:919–925. doi: 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–66.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gruell H, Vanshylla K, Tober-Lau P, Hillus D, Schommers P, Lehmann C, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 56.Takashita E, Kinoshita N, Yamayoshi S, Sakai-Tagawa Y, Fujisaki S, Ito M, et al. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. N Engl J Med. 2022;386:1475–7. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–33.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.22.1, BA.4 and BA.5. Nature. 2022;608:603–8. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Muecksch F, Schaefer-Babajew D, Finkin S, Viant C, Gaebler C, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moriyama S, Adachi Y, Sato T, Tonouchi K, Sun L, Fukushi S, et al. Temporal maturation of neutralizing antibodies in COVID-19 convalescent individuals improves potency and breadth to circulating SARS-CoV-2 variants. Immunity. 2021;54:1841–52.e4. doi: 10.1016/j.immuni.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner JS, Kim W, Kalaidina E, Goss CW, Rauseo AM, Schmitz AJ, et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- 63.Cho A, Muecksch F, Schaefer-Babajew D, Wang Z, Finkin S, Gaebler C, et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotaki R, Adachi Y, Moriyama S, Onodera T, Fukushi S, Nagakura T, et al. SARS-CoV-2 Omicron-neutralizing memory B-cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci Immunol. 2022;7:eabn8590. doi: 10.1126/sciimmunol.abn8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pegu A, O’Connell SE, Schmidt SD, O’Dell S, Talana CA, Lai L, et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science. 2021;373:1372–1377. doi: 10.1126/science.abj4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim W, Zhou JQ, Horvath SC, Schmitz AJ, Sturtz AJ, Lei T, et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141–145. doi: 10.1038/s41586-022-04527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trieu M-C, Jul-Larsen Å, Sævik M, Madsen A, Nøstbakken JK, Zhou F, et al. Antibody responses to influenza A/H1N1pdm09 virus after pandemic and seasonal influenza vaccination in healthcare workers: a 5-year follow-up study. Clin Infect Dis. 2019;68:382–392. doi: 10.1093/cid/ciy487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–88.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collie S, Champion J, Moultrie H, Bekker L-G, Gray G. Effectiveness of BNT162b2 Vaccine against Omicron variant in South Africa. N Engl J Med. 2022;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin A, Kawabata H, et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 73.Quandt J, Muik A, Salisch N, Lui BG, Lutz S, Krüger K, et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci Immunol. 2022;7:eabq2427. doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hachmann NP, Miller J, Collier A-RY, Ventura JD, Yu J, Rowe M, et al. Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. 2022;387:86–8. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qu P, Faraone J, Evans JP, Zou X, Zheng Y-M, Carlin C, et al. Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 subvariants. N Engl J Med. 2022;386:2526–8. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muik A, Lui BG, Wallisch A-K, Bacher M, Mühl J, Reinholz J, et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375(11):678–80. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, et al. SARS-CoV-2 breakthrough infections elicit potent, broad and durable neutralizing antibody responses. Cell. 2022;185:872–80.e3. doi: 10.1016/j.cell.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muecksch F, Wang Z, Cho A, Gaebler C, Ben Tanfous T, DaSilva J, et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature. 2022;607:128–134. doi: 10.1038/s41586-022-04778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyamoto S, Arashiro T, Adachi Y, Moriyama S, Kinoshita H, Kanno T, et al. Vaccination-infection interval determines cross-neutralization potency to SARS-CoV-2 Omicron after breakthrough infection by other variants. Med. 2022;3:249–261. doi: 10.1016/j.medj.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goel RR, Painter MM, Lundgreen KA, Apostolidis SA, Baxter AE, Giles JR, et al. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. Cell. 2022;185:1875–7.e8. doi: 10.1016/j.cell.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Servellita V, Syed AM, Morris MK, Brazer N, Saldhi P, Garcia-Knight M, et al. Neutralizing immunity in vaccine breakthrough infections from the SARS-CoV-2 Omicron and Delta variants. Cell. 2022;185:1539–48.e5. doi: 10.1016/j.cell.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sokal A, Broketa M, Barba-Spaeth G, Meola A, Fernández I, Fourati S, et al. Analysis of mRNA vaccination-elicited RBD-specific memory B cells reveals strong but incomplete immune escape of the SARS-CoV-2 Omicron variant. Immunity. 2022;55:1096–104.e4. doi: 10.1016/j.immuni.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hall VG, Ferreira VH, Wood H, Ierullo M, Majchrzak-Kita B, Manguiat K, et al. Delayed-interval BNT162b2 mRNA COVID-19 vaccination enhances humoral immunity and induces robust T cell responses. Nat Immunol. 2022;23:380–385. doi: 10.1038/s41590-021-01126-6. [DOI] [PubMed] [Google Scholar]
- 85.Payne RP, Longet S, Austin JA, Skelly DT, Dejnirattisai W, Adele S, et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184:5699–714.e11. doi: 10.1016/j.cell.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhattacharya D. Instructing durable humoral immunity for COVID-19 and other vaccinable diseases. Immunity. 2022;55:945–964. doi: 10.1016/j.immuni.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sanz I, Wei C, Jenks SA, Cashman KS, Tipton C, Woodruff MC, et al. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Front Immunol. 2019;10:2458. doi: 10.3389/fimmu.2019.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Good KL, Avery DT, Tangye SG. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. J Immunol. 2009;182:890–901. doi: 10.4049/jimmunol.182.2.890. [DOI] [PubMed] [Google Scholar]
- 89.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2022;40:413–442. doi: 10.1146/annurev-immunol-120419-022408. [DOI] [PubMed] [Google Scholar]
- 90.Cyster JG, Allen CDC. B Cell Responses: Cell interaction dynamics and decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takahashi Y, Kelsoe G. Role of germinal centers for the induction of broadly-reactive memory B cells. Curr Opin Immunol. 2017;45:119–125. doi: 10.1016/j.coi.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner JS, O’Halloran JA, Kalaidina E, Kim W, Schmitz AJ, Zhou JQ, et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mudd PA, Minervina AA, Pogorelyy MV, Turner JS, Kim W, Kalaidina E, et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185:603–13.e15. doi: 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaneko N, Kuo H-H, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adamo S, Chevrier S, Cervia C, Zurbuchen Y, Raeber ME, Yang L, et al. Profound dysregulation of T cell homeostasis and function in patients with severe COVID-19. Allergy. 2021;76:2866–2881. doi: 10.1111/all.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Wu Q, Liu Z, Wang Q, Wu J, Hu Y, et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat Microbiol. 2021;6:51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- 99.Goel RR, Apostolidis SA, Painter MM, Mathew D, Pattekar A, Kuthuru O, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grantham WG, Fitch FW. The role of antibody feedback inhibition in the regulation of the secondary antibody response after high and low dose priming. J Immunol. 1975;114:394–398. doi: 10.4049/jimmunol.114.1_Part_2.394. [DOI] [PubMed] [Google Scholar]
- 101.Karlsson MCI, Wernersson S, de Ståhl TD, Gustavsson S, Heyman B. Efficient IgG-mediated suppression of primary antibody responses in Fcγ receptor-deficient mice. Proc Natl Acad Sci. 1999;96:2244–2249. doi: 10.1073/pnas.96.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Meyer-Hermann M, George LA, Figge MT, Khan M, Goodall M, et al. Germinal center B cells govern their own fate via antibody feedback. J Exp Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McNamara HA, Idris AH, Sutton HJ, Vistein R, Flynn BJ, Cai Y, et al. Antibody feedback limits the expansion of B cell responses to malaria vaccination but drives diversification of the humoral response. Cell Host Microbe. 2020;28:572–85.e7. doi: 10.1016/j.chom.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 104.Ellebedy AH, Nachbagauer R, Jackson KJL, Dai Y-N, Han J, Alsoussi WB, et al. Adjuvanted H5N1 influenza vaccine enhances both cross-reactive memory B cell and strain-specific naive B cell responses in humans. Proc Natl Acad Sci U S A. 2020;117:17957–17964. doi: 10.1073/pnas.1906613117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abbott RK, Crotty S. Factors in B cell competition and immunodominance. Immunol Rev. 2020;296:120–131. doi: 10.1111/imr.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schaefer-Babajew D, Wang Z, Muecksch F, Cho A, Loewe M, Cipolla M, et al. Antibody feedback regulates immune memory after SARS-CoV-2 mRNA vaccination. Nature. 2022 doi: 10.1038/s41586-022-05609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turner JS, Zhou JQ, Han J, Schmitz AJ, Rizk AA, Alsoussi WB, et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature. 2020;586:127–132. doi: 10.1038/s41586-020-2711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Z, Mateus J, Coelho CH, Dan JM, Moderbacher CR, Gálvez RI, et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–51.e17. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cho A, Muecksch F, Wang Z, Ben Tanfous T, DaSilva J, Raspe R, et al. Antibody evolution to SARS-CoV-2 after single-dose Ad26.COV2.S vaccine in humans. J Exp Med. 2022;219:e20220732. doi: 10.1084/jem.20220732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gagne M, Moliva JI, Foulds KE, Andrew SF, Flynn BJ, Werner AP, et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron. Cell. 2022;185:1556–71.e18. doi: 10.1016/j.cell.2022.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.