Abstract
Pathogenic germline variants in the CHEK2 gene have been shown to cause a moderate increased risk of breast cancer. Here, we present a striking CHEK2 family with a biallelic carrier of two frameshift pathogenic variants, to draw attention and to encourage a comprehensive genetic and cancer risk education for biallelic carriers of CHEK2 pathogenic variants.
Keywords: biallelic, breast cancer, checkpoint kinase 2, CHEK2, familial, genetic, heterozygous
The biallelic germline pathogenic variants of CHEK2 gene confer a high risk of early‐onset breast cancer development to their female carriers that calls in for their more aggressive management and surveillance.
1. INTRODUCTION
Pathogenic germline mutations in the Checkpoint kinase 2 (CHEK2) gene have been shown to cause a moderate increased risk of breast cancer, prostate cancer, and colon cancer. 1 , 2 , 3 , 4 , 5 Like most cancer predisposition genes, published cancer risk estimates are intended for heterozygous carriers who carry one pathogenic variant. In other, better studied cancer predisposition genes such as BRCA2, ATM, and PALB2, individuals who carry two pathogenic variants in these genes (homozygous carriers or compound heterozygous carriers) are known to have more severe cancer phenotypes and can also have other clinical features such as bone marrow failure, skeletal problems, and ataxia. 6 , 7 Unlike these and other syndromes, CHEK2 is distinguished by not having a defined recessive phenotype 8 and there are little data about cancer risks for individuals who are biallelic carriers of CHEK2 pathogenic variants, meaning they carry two pathogenic variants in CHEK2.
In this report, we present a striking CHEK2 family with compound heterozygous frameshift variants in CHEK2.
2. CASE REPORT
The proband (indicated by the arrow on the pedigree, Figure 1) is a 37‐year‐old female with past medical history significant for gastroesophageal reflux disease (GERD), fatty liver, leiomyoma, history of ovarian cysts, anxiety, recurrent erosion of the cornea, and splenomegaly. She presented to her primary care provider with the complaint of noting that her left breast “felt different” without feeling a discrete mass, since about a month prior to presentation. A mammogram was ordered and noted suspicious bilateral abnormalities and suspicious left intramammary and axillary lymph nodes. She underwent bilateral biopsies and left lymph node biopsy. Core biopsy of her right breast lesion showed ductal carcinoma in situ (DCIS), high nuclear grade with comedonecrosis and microcalcifications; Estrogen receptor (ER) 95% Positive, progesterone receptor (PR) 93% Positive. Left breast biopsy showed invasive carcinoma with ductal and lobular features, preliminary modified Bloom‐Richardson grade 2; ER 87% positive, PR 2% positive, human epidermal growth factor receptor 2 (HER2) (3+) positive. Fine‐needle aspiration of her left intramammary lymph node was positive for adenocarcinoma. Her diagnosis was left Stage IIIA – T0‐2 N2 M0 or T3 N1‐2 M0 infiltrating ductal carcinoma, grade two (Intermediate) and right DCIS.
FIGURE 1.
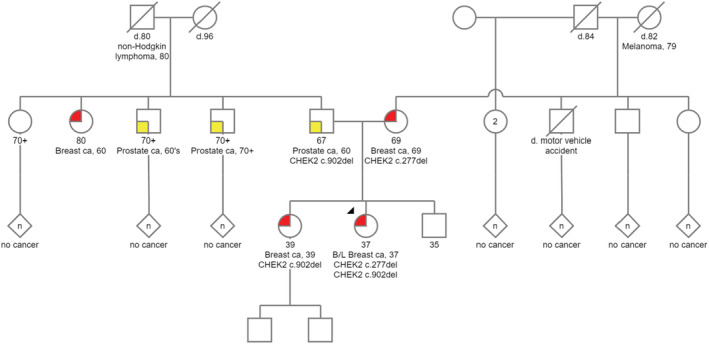
Family pedigree. Proband is marked with arrow head.
Given her young age at diagnosis, the patient underwent germline genetic testing via a 15 breast cancer gene panel (ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, FH, NBN, NF1, PALB2, PTEN, RAD50, STK11, TP53). The patient was found to carry biallelic CHEK2 pathogenic variants: c.277del (p.Trp93Glyfs*17) and c.902del (p.Leu301Trpfs*3). Maternal ancestry reported as German, English, and Welsh, and paternal ancestry reported as Russian and Western European.
She completed neoadjuvant chemotherapy with dose‐dense doxorubicin and cyclophosphamide followed by paclitaxel with pertuzumab and trastuzumab. Patient then underwent bilateral mastectomies with left axillary lymph node dissection (ALND) and right sentinel lymph node biopsy (SLNB) and immediate tissue expander placement. Pathology showed complete pathologic response with <0.1 cm of residual DCIS in both breasts. She then completed radiation therapy to the left chest wall/reconstructed breast and regional lymph nodes. She is planned to continue on pertuzumab and trastuzumab for total of 1 year of therapy and adjuvant endocrine therapy with ovarian suppression.
Proband's sister is a 39‐year‐old female with past medical history of corneal abrasions, acquired hypothyroidism, endometriosis who presented for a screening mammogram at age 39 after her sister was diagnosed with breast cancer. She had imaging with biopsy that showed right grade 3 DCIS, multicentric – cTis, cN0, cM0 ER 91%–100% positive, PR 91%–100% positive. Patient elected to have bilateral mastectomy. Patient underwent germline testing for the two pathogenic CHEK2 variants identified in her sister and was found to only carry the c.902del variant, not the c.277del variant.
Proband's father is a 67‐year‐old male with a past medical history of recurrent erosion of cornea, senile nuclear sclerosis, and hypertension. He was diagnosed with prostate adenocarcinoma at 60 years old, Gleason score 6 and was treated by a robot assisted laparoscopic prostatectomy. He also had a colonoscopy with tubular adenoma. He underwent cascade germline testing for both familial CHEK2 variants that revealed only the c.902del variant.
Proband's mother is a 69‐year‐old female with a past medical history of type II diabetes and hypertension who presented with a right breast mass and nipple discharge after the diagnosis of breast cancer in her two daughters. Imaging and biopsy were performed. She was diagnosed with invasive ductal carcinoma, stage IA (pT1c, pN1a, cM0) grade one; ER 91%–100% positive, PR 91%–100% positive, HER2 negative. Patient underwent right mastectomy with ALND; pathology showed invasive ductal carcinoma, 16 mm with background DCIS. 2/20 LNs involved. Oncotype score was 13; no chemotherapy recommended. Adjuvant radiation to the chest wall and supraclavicular and infraclavicular lymph nodes was completed. Adjuvant endocrine therapy was initiated. She underwent cascade germline testing for both familial CHEK2 variants that revealed only the c.277del variant.
The proband's 35‐year‐old brother has not undergone germline testing and reportedly has not been diagnosed with any cancers at the time of this paper.
3. DISCUSSION
In this report, we have presented a striking CHEK2 family with compound heterozygous frameshift mutations in CHEK2, c.277del (p.Trp93Glyfs*17) and c.902del (p.Leu301Trpfs*3).
The majority of current research and literature about CHEK2 pathogenic variants is on heterozygous frameshift variants, specifically the c.1100delC variant. Frameshift variants are thought to impose a lifetime risk of 20%–40% for a first breast cancer and an increased risk of a second primary breast cancer being up to 29%. 5 , 9 This cancer risk estimation is thought to be due to the type of variant of CHEK2, a patient's ancestry, a family history of breast cancer, and the age at cancer diagnosis in the family. 5 , 10 The c.1100delC is a founder variant and is present in at least 1.1% of individuals of Northern and Eastern European ancestry. 11 Missense variants of CHEK2 (such as the p.I157T variant) are less clear but seem to be of lower penetrance. Specifically, the risk for female breast cancer appears to be lower. 12 Other frameshift variants, like the ones reported in this family, are assumed to have similar cancer risks as the c.1100delC variant.
There are little data about cancer risks for individuals who are biallelic carriers of CHEK2 pathogenic variants, especially the biallelic non‐c.1100delC carriers. Paperna et al. have previously reported three patients with homozygous pathogenic variant of CHEK2. One of these patients had presented with multiple different neoplasms of different origins. 13 In a study, evaluating 2554 Dutch independent familial non‐BRCA1/2 breast cancer cases, Adank et al. identified 8 homozygotes carriers of CHEK2*1100delC. They demonstrated that the female homozygotes carriers of CHEK2*1100delC had a greater than twofold increased breast cancer risk compared to familial heterozygotes CHEK2*1100delC carriers. 14 Another study found that biallelic carriers of pathogenic variants of CHEK2 were significantly more likely to have one primary breast cancer, be diagnosed with breast cancer at or before age 50, and have a second breast cancer diagnosis as compared to monoallelic carriers. Of note, in this latter study, 16 out of 31 were trans c.1100delC. Additionally, the authors suggest that biallelic carriers of CHEK2 pathogenic variants could have breast cancer risks higher than those associated with pathogenic variants in in BRCA1 or BRCA2. 15 In present study, the proband, who is the biallelic carriers of two non‐c.1100delC CHEK2 pathogenic variants, presented at a younger age and more advanced stage compared to her family members.
In our present study, prior to the proband and her sister's diagnosis of breast cancer, only their paternal family history met the current criteria set by NCCN guidelines for germline testing for hereditary prostate cancer. Their maternal family history was not suspicious for hereditary cancer and did not meet the criteria for germline testing for hereditary cancer, per available information provided by the patient. Therefore, further research regarding the risk stratification for biallelic carriers is essential due to the possible significant implications for treatment of an initial breast malignancy and cancer surveillance for unaffected biallelic carriers. Current breast cancer screening guidelines in NCCN include breast mammography with consideration of tomosynthesis and consideration of breast MRI starting by age 40 are stated for monoallelic heterozygous carriers only. Unfortunately, there has not been enough information to incorporate a risk stratification and surveillance guidelines for biallelic carriers. If the risk of developing breast cancer for biallelic carriers of CHEK2 pathogenic variants is truly in the high‐risk range, similar to that of a BRCA gene mutation, these women are likely not being offered appropriate high‐risk breast screening young enough, nor being offered risk reducing surgery including bilateral mastectomies. Cybulski et al. have estimated the lifetime risk of breast cancer for heterozygote carriers of CHEK2 pathogenic variants to be 20% for a woman with no family history of breast cancer, and 28%, 34%, and 44% for a woman with one second‐degree relative affected, with one first‐degree relative affected, and with both a first‐ and second‐degree relative affected, respectively. 10 Therefore, they recommended addition of magnetic resonance imaging (MRI) for screening as well as chemoprevention with Tamoxifen for carriers of pathogenic variants of CHEK2 with a family history of breast cancer. 10
Another interesting finding in the review of this family pedigree is the occurrence of cutaneous melanoma at the age of 79 in maternal grandmother of the proband and non‐Hodgkin lymphoma at the age of 80 in her paternal grandfather. The maternal grandmother of the proband could have been a carrier of the pathogenic variant c.277del. A few previous studies have evaluated the risk of cutaneous melanoma in CHEK2 pathogenic variant carriers. Of the six different studies that were reviewed by Bui et al. 16 in 2020, one large study has demonstrated a statistically significant twofold risk of melanoma in individuals with the CHEK2*1100delC variant but the other five other studies did not show an association. Association between the pathogenic variant c.277del and malignant melanoma requires further investigations. Previous studies have suggested association between CHEK2 variants and analyzed hereditary CHEK2 variants and non‐Hodgkin lymphoma. A study in 2015 evaluated the coding sequence of the CHEK2 gene in a large group of non‐Hodgkin lymphoma patients, and found that CHEK2 variants could modify the risk and clinical course of non‐Hodgkin lymphoma. 17 Specifically, they showed that the I157T variant had statistically significant association with an increased risk, and the c.319 + 43dupA alteration was the only variant associated with a significant decrease in risk of non‐Hodgkin lymphoma.
Furthermore, to our knowledge, there is no published information regarding prostate cancer risks for biallelic carriers of CHEK2 pathogenic variants. Additionally, other cancers have been reported in association with CHEK2 pathogenic variants, but have not been statistically significant in their risk correlation to be added to current screening guideline recommendations by NCCN for CHEK2 heterozygous carriers. 8 , 18 , 19 Cancer risk management per national practice guidelines is based on lifetime cancer risks, and further research is needed to understand the interactions of moderate penetrance genes and family history on lifetime cancer risk. When providing a patient with an accurate cancer risk assessment with moderate risk genes, the family history of cancer should be assessed to guide risk assessment and screening for the family.
Regarding the patients that were presented in this report, the following surveillance has been recommended at this time; The proband and her sister were recommended to have colonoscopies every 5 years and annual chest wall examinations (since both had bilateral mastectomies). Mother was recommended to have colonoscopies every 5 years and her surveillance for breast cancer is pending her completion of her breast cancer treatment. Father was recommended to have annual PSA and colonoscopies every 5 years.
4. CONCLUSION
This report draws attention to compound heterozygous carriers of pathogenic variant CHEK2 and to encourage a comprehensive genetic and cancer risk education to give patients the ability to choose the most appropriate treatment and surveillance options for them given the many confounding factors at work in this assessment.
AUTHOR CONTRIBUTIONS
Tahereh Soleimani: Conceptualization; validation; writing – original draft; writing – review and editing. Corrie Bourdon: Conceptualization; data curation; investigation; validation; writing – original draft; writing – review and editing. Jacquelyn Davis: Conceptualization; writing – original draft; writing – review and editing. Thais Fortes: Conceptualization; supervision; validation; writing – review and editing.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
FUNDING INFORMATION
No grants or fundings have been received for this research.
ETHICS STATEMENT
Ethics approval obtained from Sparrow Hospital Ethics committee.
CONSENT
Written informed consent was obtained from the patient to publish this report in accordance with the journal's patient consent policy.
ACKNOWLEDGEMENT
None.
Soleimani T, Bourdon C, Davis J, Fortes T. A case report of biallelic CHEK2 heterozygous variant presenting with breast cancer. Clin Case Rep. 2023;11:e06820. doi: 10.1002/ccr3.6820
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer‐specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30(35):4308‐4316. doi: 10.1200/jco.2012.42.7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weischer M, Bojesen SE, Ellervik C, Tybjaerg‐Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta‐analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008;26(4):542‐548. doi: 10.1200/jco.2007.12.5922 [DOI] [PubMed] [Google Scholar]
- 3. Kurian AW, Hughes E, Handorf EA, et al. Breast and ovarian cancer penetrance estimates derived from germline multiple‐gene sequencing results in women. JCO Precis Oncol. 2017;1:1‐12. doi: 10.1200/po.16.00066 [DOI] [PubMed] [Google Scholar]
- 4. CHEK2 Breast Cancer Case‐Control Consortium . CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74(6):1175‐1182. doi: 10.1086/421251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stolarova L, Kleiblova P, Janatova M, et al. CHEK2 germline variants in cancer predisposition: stalemate rather than checkmate. Cell. 2020;9(12):2675. doi: 10.3390/cells9122675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44(1):1‐9. doi: 10.1136/jmg.2006.043257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katsuki Y, Takata M. Defects in homologous recombination repair behind the human diseases: FA and HBOC. Endocr Relat Cancer. 2016;23(10):T19‐T37. doi: 10.1530/erc-16-0221 [DOI] [PubMed] [Google Scholar]
- 8. Rahman N, Scott RH. Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum Mol Genet. 2007;16 Spec No 1:R60‐R66. doi: 10.1093/hmg/ddm026 [DOI] [PubMed] [Google Scholar]
- 9. Kriege M, Hollestelle A, Jager A, et al. Survival and contralateral breast cancer in CHEK2 1100delC breast cancer patients: impact of adjuvant chemotherapy. Br J Cancer. 2014;111(5):1004‐1013. doi: 10.1038/bjc.2014.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cybulski C, Wokołorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29(28):3747‐3752. doi: 10.1200/jco.2010.34.0778 [DOI] [PubMed] [Google Scholar]
- 11. Meijers‐Heijboer H, van den Ouweland A, Klijn J, et al. Low‐penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55‐59. doi: 10.1038/ng879 [DOI] [PubMed] [Google Scholar]
- 12. Dorling L, Carvalho S, Allen J, et al. Breast cancer risk genes – association analysis in more than 113,000 women. N Engl J Med. 2021;384(5):428‐439. doi: 10.1056/NEJMoa1913948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paperna T, Sharon‐Shwartzman N, Kurolap A, et al. Homozygosity for CHEK2 p.Gly167Arg leads to a unique cancer syndrome with multiple complex chromosomal translocations in peripheral blood karyotype. J Med Genet. 2020;57(7):500‐504. doi: 10.1136/jmedgenet-2018-105824 [DOI] [PubMed] [Google Scholar]
- 14. Adank MA, Jonker MA, Kluijt I, et al. CHEK2*1100delC homozygosity is associated with a high breast cancer risk in women. J Med Genet. 2011;48(12):860‐863. doi: 10.1136/jmedgenet-2011-100380 [DOI] [PubMed] [Google Scholar]
- 15. Rainville I, Hatcher S, Rosenthal E, et al. High risk of breast cancer in women with biallelic pathogenic variants in CHEK2. Breast Cancer Res Treat. 2020;180(2):503‐509. doi: 10.1007/s10549-020-05543-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bui AN, LeBoeuf NR, Nambudiri VE. Skin cancer risk in CHEK2 mutation carriers. J Eur Acad Dermatol Venereol. 2021;35(2):353‐359. doi: 10.1111/jdv.16729 [DOI] [PubMed] [Google Scholar]
- 17. Havranek O, Kleiblova P, Hojny J, et al. Association of Germline CHEK2 gene variants with risk and prognosis of non‐Hodgkin lymphoma. PLoS One. 2015;10(10):e0140819. doi: 10.1371/journal.pone.0140819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiang HP, Geng XP, Ge WW, Li H. Meta‐analysis of CHEK2 1100delC variant and colorectal cancer susceptibility. Eur J Cancer. 2011;47(17):2546‐2551. doi: 10.1016/j.ejca.2011.03.025 [DOI] [PubMed] [Google Scholar]
- 19. Zlowocka‐Perlowska E, Narod SA, Cybulski C. CHEK2 alleles predispose to renal cancer in Poland. JAMA Oncol. 2019;5(4):576. doi: 10.1001/jamaoncol.2019.0022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.


