Abstract
Cancer is generally regarded as a localised disease, with the well-established role of the tumour microenvironment. However, the realm of cancer goes beyond the tumour microenvironment, and cancer should also be regarded as a systemic and environmental disease. The exposome (i.e., the totality of exposures), which encompasses diets, supplements, smoking, alcohol, other lifestyle factors, medications, etc., likely alters the microbiome (inclusive of bacteria, viruses, archaea, fungi, parasites, etc.) and immune system in various body sites and influences tumour phenotypes. The systemic metabolic / inflammatory status, which is likely influenced by exposures and intestinal physiological changes, may affect tissue microenvironment of colorectum and any other organs. Germline genomic factors can modify disease phenotypes via gene-by-environment interactions. Although challenges exist, it is crucial to advance not only basic experimental research that can analyse the effects of exposures, microorganisms, and microenvironmental components on tumour evolution but also interdisciplinary human population research that can dissect the complex pathogenic roles of the exposome, microbiome, and immunome. Metagenomic, metatranscriptomic, and metabolomic analyses should be integrated into well-designed population research combined with advanced methodologies of artificial intelligence and molecular pathological epidemiology. Ideally, a prospective cohort study design that enables biospecimen (such as stool) collection before disease detection should be considered to address reverse causation and recall biases. Robust experimental and observational research together can provide insights into dynamic interactions between environmental exposures, microbiota, tumour, and immunity during carcinogenesis processes, thereby helping us develop precision prevention and therapeutic strategies to ultimately reduce the cancer burden.
Keywords: biobank, bioinformatics, computational biology, microbiology, precision medicine
Introduction and Purpose of the Article
While a tumour evolves with the accumulation of genomic and epigenomic aberrations in neoplastic cells, it generates its intrinsic microenvironment, where neoplastic cells interact with immune and other non-neoplastic cells. Among various components of the tumour microenvironment, increasing attention has been devoted to microorganisms that encompass viruses, bacteria, fungi, archaea, etc. Microorganisms, which are ubiquitously present in and around the human body and particularly abundant in digestive tracts, not only influence oncogenesis in various organs but also shape the host’s antitumour immunity in the local and systemic environment.1–4
To better understand cancer, we should also account for the exposome, i.e., the totality of exposures including diets, supplements, alcohol, smoking, medications, microorganisms, etc. Many exposures have been established as either risk or protective factors for cancer. In addition, various exposures may influence tumour development through alterations of the tumour microenvironment.5 Systemic physiological statuses such as immune, inflammatory, metabolic, and hormonal conditions are also influenced by exposures (including the microbiota), and in turn, influence local tumour development.5,6 Taken together, cancer can be regarded as a microenvironmental, systemic, and environmental disease (Figure 1). Therefore, we need to examine not only tumour cells and the surrounding microenvironment but also the effects of various exposures and systemic factors on tumours. However, there have been technical and practical hurdles to performing such integrative analyses of these factors in human populations. To date, large-scale human population studies have rarely been conducted to elucidate the complex interactions between the exposome, microbiota, and cancer.
Figure 1.
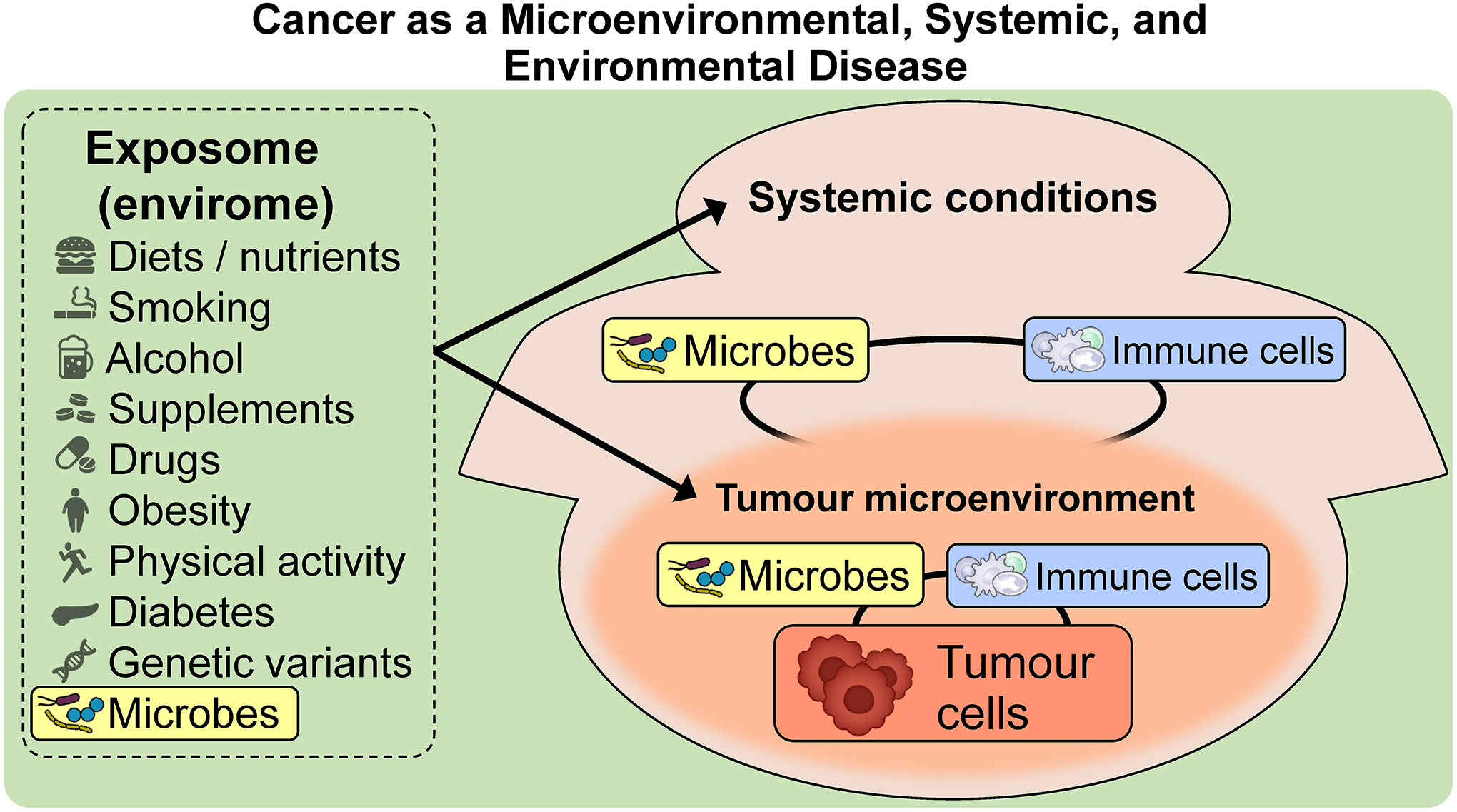
Cancer as a microenvironmental, systemic, and environmental disease. Tumour generates its intrinsic microenvironment, where tumour cells interact with immune cells, microbes, and other cells as well as non-cellular components. Notably, there are no clear boundaries of the tumour microenvironment, which rather blends into tissue outside of the tumour. Systemic conditions, especially systemic immunity, affect the local tumour microenvironment and contribute to tumourigenic processes. The exposome, which encompasses diets, alcohol, medications, lifestyle factors, etc., influences tumour phenotypes by modulating systemic conditions and the tumour microenvironment. Microbes, which may exist in the tumour microenvironment, in distant organs, or around the human body, potentially influence tumour phenotypes directly or indirectly by modulating the host’s local and systemic antitumour immunity.
This article aims to provide a viewpoint that cancer is an environmental, systemic, and microenvironmental disease where the microbiome plays a pivotal role in the interplay of various factors. We emphasise the increasing need for transdisciplinary multi-modal research to assess the interactive effects of the exposome, microbiome, and the tumour microenvironment on tumours, using experimental models and human populations. Such integrative research approaches can help us develop better strategies for precision cancer prevention and therapeutics.
This article uses the standardised nomenclature system for genes and gene products according to the Human Genome Organisation Gene Nomenclature Committee (HGNC),7 to increase clarity and reduce ambiguity associated with colloquial protein names.
Tumour Microenvironment
Tumour arises through a dynamic network
Tumour arises with the accumulation of cellular genomic and epigenomic alterations. Emerging evidence indicates that the expansion of clones harbouring somatic mutations in cancer-associated genes is common in non-neoplastic tissues, especially in aging individuals or individuals with chronic inflammation.8 Somatic driver mutations for colorectal cancer (CRC) were documented in approximately 1% of normal colorectal crypts in middle-aged individuals.9 Most newly-forming clones are destined to be well controlled or eliminated.10 Some mutant clones in phenotypically normal epithelium may purge other clones through cell competition, thereby exerting an antitumourigenic effect and preserving tissue integrity.10 Eventually, one of these mutant clones may proliferate enough to form a benign neoplasm. A benign tumour may further accumulate genomic and epigenomic alterations and progress to malignancy.
A neoplasm generates its microenvironment, where tumour cells, microorganisms, immune cells, other cells, and extracellular matrix components (e.g., collagen, fibronectin) interact via dynamic crosstalks.11,12 The intercellular communications are regulated by direct cell-to-cell contact and through biomolecules (e.g., cytokines, chemokines, growth factors). Tumours harbour distinct microbial communities, which vary by tumour types13–15 and may persist during the metastatic process.16 As such, polymorphic microbiomes have recently been noted as a new cancer hallmark.17 Intracellular bacteria may exist within tumour cells and/or immune cells.14 Peptides derived from intracellular bacteria can be presented by tumour cells and influence immune responses.18 Intratumoural microbes interact with tumour cells, immune cells, and other cells / components. In mice bearing inflammation-induced colon tumours, neutrophil depletion increased intratumoural microbes, induced IL17A-mediated inflammatory response, and promoted tumour growth.19 Certain microbes may promote carcinogenesis through producing tumourigenic molecules or inducing a pro-inflammatory microenvironment, whereas other microbes may exert tumour-suppressive properties through maintaining vigourous antitumour immune responses.1,3,20
Microbes involved in tumourigenesis across various organs
Accumulating evidence supports the involvement of specific microbes in tumourigenesis of various organs (Table 1).1–4 Herein, we discuss not only bacteria but also fungi, archaea, and viruses that have been implicated in gastroenterological tumours. Presumably, enormous amounts of data on the pro/anti-carcinogenic roles of microorganisms and microbial dysbiosis will emerge in the next decade.
Table 1.
Specific microorganisms implicated in gastroenterological tumourigenesis by organ site
| Specific microbes in a spectrum of organs | Findings |
|---|---|
| Colorectal cancer | |
| Atopobium parvulum Actinomyces odontolyticus |
Atopobium parvulum and Actinomyces odontolyticus were characteristically enriched for multiple polypoid colorectal adenomas and intramucosal colorectal carcinomas.90 |
| Bacteroides fragilis | Enterotoxigenic Bacteroides fragilis (ETBF) is enriched in colorectal serrated lesions and adenoma as well as colorectal cancer.208 ETBF and pks+ E. coli appear to synergistically induce colorectal tumourigenesis.103 |
| Escherichia coli | Polyketide synthetase (pks)-producing E. coli appear to inhabit colorectal tissues in approximately 20% of healthy individuals, 40% of patients with inflammatory bowel disease, and 60% of patients with familial adenomatous polyposis or colorectal cancer.103–105 The pks+ E. coli appears to directly induce DNA damage and mutagenesis.102 |
| Fusobacterium nucleatum | F. nucleatum is implicated in the initiation and progression of colorectal cancer.21–26 Intratumoural F. nucleatum appears to promote colorectal carcinogenesis by activating anti-inflammatory myeloid cells,23 suppressing NK and T cells,24,25 and activating the CTNNB1 (beta-catenin)-WNT signalling pathway.26 The presence of F. nucleatum in colorectal cancer is associated with decreased CD3+CD4+CD45RO (PTPRC)+ cells30 and increased tumour-infiltrating macrophages31 in the tumour microenvironment. Colorectal cancer containing F. nucleatum is characterised by proximal (caecal) tumour localisation, BRAF mutations, microsatellite instability-high status, CpG island methylator phenotype-high status, and poor prognosis.27–29 |
| Viral microbiome (virome) | Faecal specimens of patients with colorectal cancer were characterised by increased viral diversity and enrichment of bacteriophages (e.g., Siphoviridae, Myoviridae, Inovirus, Tunalikevirus) that were expected to be bacterium-virus community network hubs.37,38 |
| Fungal microbiome (mycobiome) | Faecal specimens of patients with colorectal cancer were characterised by increased Basidiomycota:Ascomycota ratio, high amount of Malasseziomycetes, low amount of Saccharomycetes and Pneumocystidomycetes, increased co-occurring fungal intrakingdom correlations, and disappearance of some co-occurring bacterial–fungal correlations (e.g., fungal Ascomycota and bacterial Proteobacteria).39 |
| Archaeal microbiome | Faecal specimens of patients with colorectal cancer had enrichment of halophilic and depletion of methanogenic archaea.40 The halophilic Natrinema species J7-2 increased progressively in faecal specimens of healthy individuals, to patients with colorectal adenomas, to patients with colorectal cancer.40 |
| Multi-kingdom microbiome | Faecal 16 multi-kingdom microbiome markers, including 11 bacterial (e.g., F. nucleatum, Parvimonas micra, Gemella morbillorum), 4 fungal (e.g., Talaromyces islandicus, Aspergillus rambellii), and 1 archaeal (Pyrobaculum arsenaticum) feature, achieved good performance in diagnosing patients with colorectal cancer.34 Bacterial–fungal interactions may contribute to CRC pathogenesis via upregulation of D-arginine and D-ornithine and stimulation of the butanoate metabolism pathways. |
| Gastric cancer | |
| Helicobacter pylori | Chronic infection with H. pylori is a leading cause of gastric cancer.41–44,209 H. pylori induces multistep carcinogenesis, namely progression from chronic gastritis, tissue atrophy, intestinal metaplasia, and benign tumour to carcinoma.42 The eradication of H. pylori is an established strategy to prevent gastric cancer.187 |
| Epstein-Barr virus (EBV) | EBV-associated gastric cancer is characterised by male predominance, early-onset, proximal tumour localisation, less tumour differentiation, poorly differentiated morphology with marked lymphocytic infiltration, PIK3CA and ARID1A mutations, lack of TP53 mutation, 9p24.1 amplification, hypermethylation of CDKN2A promoter, CD274 overexpression.46 EBV-encoded microRNAs BART11 and BART17-3p inhibit FOXP1 and PBRM1, respectively, and enhance the transcription of CD274, resulting in the promotion of tumour immune evasion.47 |
| Liver cancer | |
| Bacterial microbiome | The composition of intratumoural microbiota in hepatocellular carcinoma differed according to aetiological factors.56,57 Ruminococcus gnavus was characteristically enriched for virus-related hepatocellular carcinoma.57 Four proteogenomic subgroups of intrahepatic cholangiocarcinoma had distinct intratumoural microbiota diversity, composition, and functions.58 |
| Faecal microbiome | Butyrate-producing bacterial genera (e.g., Ruminococcus) were decreased, while lipopolysaccharide-producing genera (e.g., Klebsiella) were increased in faecal samples from patients with early-stage hepatocellular carcinoma than those from controls.210
Gram-negative commensal gut bacteria induced hepatocytes to generate an immunosuppressive environment by recruiting CXCR2+ polymorphonuclear myeloid-derived suppressor cells through TLR4-dependent CXCL1 production, ultimately promoting the development of intrahepatic cholangiocarcinoma.54 Enterococcus faecalis was abundant in faecal samples from patients with hepatitis C virus-related chronic liver disease.55 GelE-positive E. faecalis appears to promote liver carcinogenesis by expressing the metallopeptidase gelE, which increased gut permeability, leading to elevated plasma lipopolysaccharide and activation of TLR4-MYD88 proliferative signalling in hepatocytes.55 |
| Oesophageal cancer | |
| Fusobacterium nucleatum | The presence of F. nucleatum in oesophageal cancer is associated with the low efficacy of chemotherapy.166 Intratumoural F. nucleatum may contribute to chemoresistance by promoting autophagy.166 |
| Pancreatic cancer | |
| Bacterial microbiome | Pancreatic cancer tissue contains more bacteria than normal pancreatic tissue.48 Proteobacteria, Bacteroidetes, and Firmicutes were highly colonised and prevalent bacterial phyla in pancreatic cancer.48 A signature of three intratumoural bacterial genera (Pseudoxanthomonas, Streptomyces, Saccharopolyspora) and high alpha-diversity of intratumoural bacteria were associated with better patient outcomes. Proteobacteria, Bacteroidetes, and Firmicutes were highly colonised and prevalent bacterial phyla in pancreatic cancer.48 |
| Fungal microbiome (mycobiome) | Pancreatic cancer tissue contains more fungi than normal pancreatic tissue.49 Malassezia promoted tumour progression by attaching to mannose-binding lectin, thereby activating the complement cascade.49 In response to the intratumoural mycobiome, pancreatic cancer cells produced IL33 as a chemoattractant for type 2 immune cells, leading to tumour progression.51 |
| Faecal microbiome | Veillonella atypica, Fusobacterium nucleatum/hwasookii, and Alloscardovia omnicolens were enriched in faeces of patients with pancreatic adenocarcinoma, whereas Romboutsia timonensis, Faecalibacterium prausnitzii, Bacteroides coprocola, and Bifidobacterium bifidum species clusters were depleted.52 Enrichments of Streptococcus species and Veillonella species (V. parvula and V. atypica) and a depletion of Faecalibacterium prausnitzii were common signatures for faecal specimens of patients with pancreatic adenocarcinoma.53 |
Abbreviations: EBV, Epstein-Barr virus; ETBF, enterotoxigenic Bacteroides fragilis; pks, polyketide synthase.
The colorectum hosts the largest load and diversity of bacterial species among all organs; therefore, the dysregulated microbiota has been examined extensively in the development of colorectal diseases, including CRC. Metagenomic analyses demonstrated enrichment of Fusobacterium nucleatum in CRC tissues compared to adjacent normal tissues.21,22 F. nucleatum appears to exert carcinogenic effects on the colorectal epithelium by activating myeloid-derived suppressor cells,23 suppressing NK and T cells via interaction with TIGIT and CEACAM1 inhibitory immunoreceptors,24,25 and activating the CTNNB1 (beta-catenin)-WNT signalling pathway via ANXA1 (annexin A1) upregulation.26 CRC containing F. nucleatum is characterised by proximal tumour localisation, BRAF mutation, high-level microsatellite instability, high-level CpG island methylator phenotype,27–29 decreased CD3+CD4+CD45RO(PTPRC)+ cells,30 and increased tumour-associated macrophages.31 Specifically, F. nucleatum subspecies animalis may play a role in most of these associations.32
In addition to bacteria, non-bacterial microorganisms, including viruses, fungi, archaea, and parasites, likely play pathogenic roles in various cancer types, including CRC.33,34 Viruses represent an essential component of the intestinal microbial community and have been implicated in inflammatory bowel diseases35,36 and CRC.34,37,38 The faecal virome of CRC patients appeared more diverse than that of CRC-free individuals and enriched for bacteriophages that are expected to be bacterium-virus community hubs,37,38 suggesting a role of the virome in colorectal carcinogenesis via its modulating effect on the bacterial community. Regarding the mycobiome, CRC patients exhibited faecal fungal dysbiosis with an increased Basidiomycota:Ascomycota ratio.39 Additionally, the faecal microbiota was characterised by increased co-occurring fungal intrakingdom correlations and disappearance of co-occurring bacterial–fungal correlations (e.g., fungal Ascomycota and bacterial Proteobacteria), indicating that synergistic intrafungal and antagonistic bacterial–fungal associations may contribute to colorectal carcinogenesis.39 The faecal archaeome of CRC patients was characterised by enrichment of halophilic archaea (e.g., Natrinema species J7-2) and depletion of methanogenic archaea.40 Multi-kingdom microbiota analyses of CRC metagenomic datasets identified 16 microbial biomarkers (including 11 bacterial, 4 fungal, and 1 archaeal feature) that achieved better performance than single-kingdom markers in diagnosing CRC patients.34 Moreover, exploration of the metagenomic functions indicated that bacterial–fungal interactions might contribute to colorectal carcinogenesis via upregulation of D-arginine and D-ornithine and stimulation of butanoate metabolism.34
Chronic infection with Helicobacter pylori is a leading cause of gastric cancer.41–43 H. pylori typically resides in the gastric mucus layer and promotes chronic inflammation, mucosal atrophy, and intestinal metaplasia.42,44 H. pylori infection can induce infiltrations of immune cells that produce inflammatory mediators such as TGFB1 (transforming growth factor-β), thereby contributing to gastric tumourigenesis.45
Epstein-Barr virus (EBV) is another pathogenic microbe associated with certain forms of gastric cancer. EBV-associated gastric cancer, which comprises 7–10% of gastric cancer cases, is characterised by male predominance, young-onset, proximal tumour localisation, abundant tumour-infiltrating lymphocytes, PIK3CA and ARID1A mutations, CDKN2A promoter hypermethylation, and CD274 (PD-L1) overexpression.46 EBV-encoded microRNAs BART11 and BART17-3p appear to promote immune escape by increasing the enhancer-mediated CD274 transcription.47
Pancreatic cancer tissue harbours greater amounts of bacteria and fungi than normal pancreatic tissue.48,49 Intratumoural microbes in pancreatic cancer may have migrated from the gastrointestinal tract via the pancreatic duct system, as illustrated by the observation that fluorescently labelled bacteria and fungi migrated into the pancreas in a retrograde manner.48,49 Bacterial translocation to the pancreas may be caused by the biliary infection, as Enterococcus species were commonly detected in bile juice and pancreatic cancer tissue.50 Pancreatic intratumoural microbes may create an immunosuppressive microenvironment by activating distinct Toll-like receptors (TLRs) in monocytic cells.48 In mice, bacterial ablation decreased myeloid-derived suppressor cells and increased antitumour M1 macrophages.48 Intratumoural bacterial composition in pancreatic cancer influences patient outcomes, as indicated by observations that a signature of three bacterial genera (Pseudoxanthomonas, Streptomyces, Saccharopolyspora) and high alpha-diversity of intratumoural bacteria were both associated with better patient outcomes. Intratumoural fungi also play a crucial role in pancreatic carcinogenesis. Intratumoural Malassezia appears to augment the progression of pancreatic cancer by attaching to mannose-binding lectin and thereby activating the complement cascade.49 Furthermore, in response to the intratumoural mycobiome, pancreatic cancer cells appear to produce IL33 (interleukin 33) as a chemoattractant for type 2 immune cells, which can stimulate tumour growth by secreting pro-tumourigenic cytokines.51
Emerging evidence suggests a feasibility of non-invasive faecal microbiota-based screening for the early detection of pancreatic cancer.52,53 Pancreatic adenocarcinoma could be predicted robustly and accurately by metagenomic classifiers based on faecal microbial species.52,53 Veillonella species (e.g., V. atypica) and Streptococcus species were enriched, and Faecalibacterium prausnitzii was depleted in faecal samples of pancreatic adenocarcinoma patients.52,53
The liver is chronically exposed to intestinal microbes and their metabolites because of its anatomical connection with the gut via the portal vein and bile duct systems. The microbes and their metabolites may produce pro-inflammatory or immunosuppressive conditions, which may result in liver carcinogenesis. Gram-negative commensal gut bacteria can induce hepatocytes to form a tumour-promoting environment by recruiting immunosuppressive CXCR2+ polymorphonuclear myeloid-derived suppressor cells through TLR4-dependent CXCL1 production, eventually promoting the development of intrahepatic cholangiocarcinoma.54 Enterococcus faecalis, a species enriched in faecal samples of patients with HCV (hepatitis C virus)-related chronic hepatitis, appears to promote liver carcinogenesis via the expression of the metallopeptidase gelE.55 In mice, gelE-positive E. faecalis promoted liver carcinogenesis in a TLR4-dependent manner by increasing gut permeability via its gelatinase activity and elevating plasma lipopolysaccharide that acts on hepatocytes.55 The composition of intratumoural microbiota in hepatocellular carcinoma (HCC) differs according to aetiological factors.56,57 Ruminococcus gnavus was characteristically enriched for virus-related HCCs.57 As for intrahepatic cholangiocarcinoma, four subgroups characterised by proteogenomic profiling had distinct intratumoural microbiota diversity, compositions, and functions.58 As viral exposure history differs between HCC patients and HCC-free individuals, a viral exposure signature, determined by serological profiling, could identify HCC prior to a clinical diagnosis.59 Exposure to HBV (hepatitis B virus), HCV, two influenza strains (H1N1 and H3N2), and cytomegalovirus correlated with increased HCC risk.59
Systemic Conditions as a Component of Tumour
Cancer should be recognised as a systemic disease, as systemic conditions can influence a tumour and vice versa. Persistent local and systemic inflammation is a hallmark of cancer. Systemic immune, inflammatory, metabolic, and hormonal statuses may contribute to oncogenesis through their effects on cellular genomic and epigenomic aberrations as well as local tissue microenvironment. Systemic antitumour immunity suppresses tumour initiation, progression, and metastasis.60–62 Diabetes mellitus, a metabolic syndrome characterised by hyperglycaemia, hyperinsulinaemia, and insulin resistance, increases cancer risk.63–65 Higher levels of GDF15, CRP, IL6, and TNFRSF1B (HGNC:11917; TNF receptor superfamily 1B) and lower levels of ADIPOQ (HGNC:13633; adiponectin) and 25-hydroxyvitamin D in blood have been associated with cancer risk and mortality.66–73 Vitamin D is an immunomodulator that helps maintain immune homeostasis and induces tumour-suppressive immune responses.74,75 The inverse association of vitamin D levels with CRC risk appeared stronger for tumours exhibiting higher lymphocytic infiltrates.76
Conversely, localised or metastatic cancer can alter systemic immune and metabolic conditions.60 In preclinical models, TP53 loss in breast carcinoma cells induced the secretion of WNT ligands that stimulate tumour-associated macrophages to produce IL1B, thereby causing systemic inflammation and tumour metastasis.77 In an analysis of The Cancer Genome Atlas (TCGA) cohorts of 33 cancer types, unique microbial communities were detected in tumour tissues and blood samples, indicating that microbes may migrate between tumour tissue and bloodstream.13 Intratumoural microbes may move to distant organs, creating a microenvironment (“premetastatic niche”) where tumour cells can implant, survive, and proliferate.78 In a preclinical study using CRC-bearing mice, intratumoural Escherichia coli disrupted the gut vascular barrier and created a premetastatic niche in the liver, promoting CRC metastasis.78 Tumour microenvironment may favour local bacterial implantation and growth from circulating microbes.4 Rapidly formed vasculature due to tumour growth is characterised by irregular organisation and leakiness, which may permit microbial migration between the tumour microenvironment and bloodstream.
Exposome as a Component of Tumour
The exposome (the totality of exposures), which includes the microbiome, influences tumour phenotypes via its complex effects on neoplastic cells, tumour microenvironment, and systemic physiological states. The systemic conditions are also conceptually a part of the exposome. Certain exposures may predispose individuals to cancer development as well as influence its disease course and outcomes.63,79,80 Cigarette smoking is the leading cause of cancer.79,81 Inhaled carcinogens in cigarette smoke directly damage DNA and produce mutations in epithelial cells.81 Smoking also appears to induce colorectal carcinogenesis via its modulating effects on systemic and local immune reactions.81,82 The association between smoking and CRC incidence was stronger for tumours containing fewer T cells and macrophages, supporting immunosuppressive effects of smoking.80,83 Higher physical activity was associated with decreased CRC incidence and mortality through its influences on energy balance, cellular prostaglandin biosynthesis, and systemic inflammatory statuses.84 The beneficial association of exercise with CRC prognosis was stronger in CRC with fewer tumour-infiltrating CD3+ lymphocytes, supporting interactive effects of physical activity and immune response on clinical outcomes.85 Essentially, the exposome, which is one of the determinants of tumour evolution and phenotypes, can be regarded as an extended component of the tumour. This notion is helpful in increasing the recognition of exposure modifications as effective preventative and therapeutic strategies for cancer.
Microbiota as a Pivot of Interplay of the Exposome and Tumour
Emerging evidence indicates a mediating and modifying role of the microbiota (which is itself a component of the exposome) in the effects of other exposures on tumour cells and the microenvironment (Figure 2). The gut microbiota plays a pivotal role in the association between diets and cancer.86–89 High intake of red meat and low dietary fibre intake were correlated with enrichment of Fusobacterium in faeces of healthy individuals.90 The abundance of CRC-related bacteria or bile acid-metabolising bacteria (e.g., Bilophila wadsworthia) was correlated with a high intake of red meat and a low intake of fruits and vegetables.91 Processed and animal-derived foods were associated with Firmicutes, Ruminococcus species of the Blautia genus, and endotoxin synthesis pathways.92 In contrast, plant foods and fish were linked to short-chain fatty acid (SCFA)-producing microbes and nutrient metabolism pathways. These diet-microbiota associations are consistent across healthy individuals and patients with chronic inflammatory bowel diseases (e.g., Crohn’s disease, ulcerative colitis).92 Further research has characterised metabolomic and metagenomic profiles of stool specimens from patients with colorectal tumours.90,93 Compared to plant-based foods, animal-based foods contain abundant taurine, which increases taurocholic acid in the liver and gut.94 Taurocholic acid is metabolised to genotoxic H2S by B. wadsworthia and tumour-promoting deoxycholic acid by Clostridium scindens.95 H2S-producing pathways were upregulated in CRC patients based on faecal examinations.90 Notably, African Americans harbour higher amounts of sulfidogenic bacteria and B. wadsworthia than non-Hispanic Whites in the U.S., suggesting that these microbial differences might explain the higher incidence of CRC in African Americans.96
Figure 2.
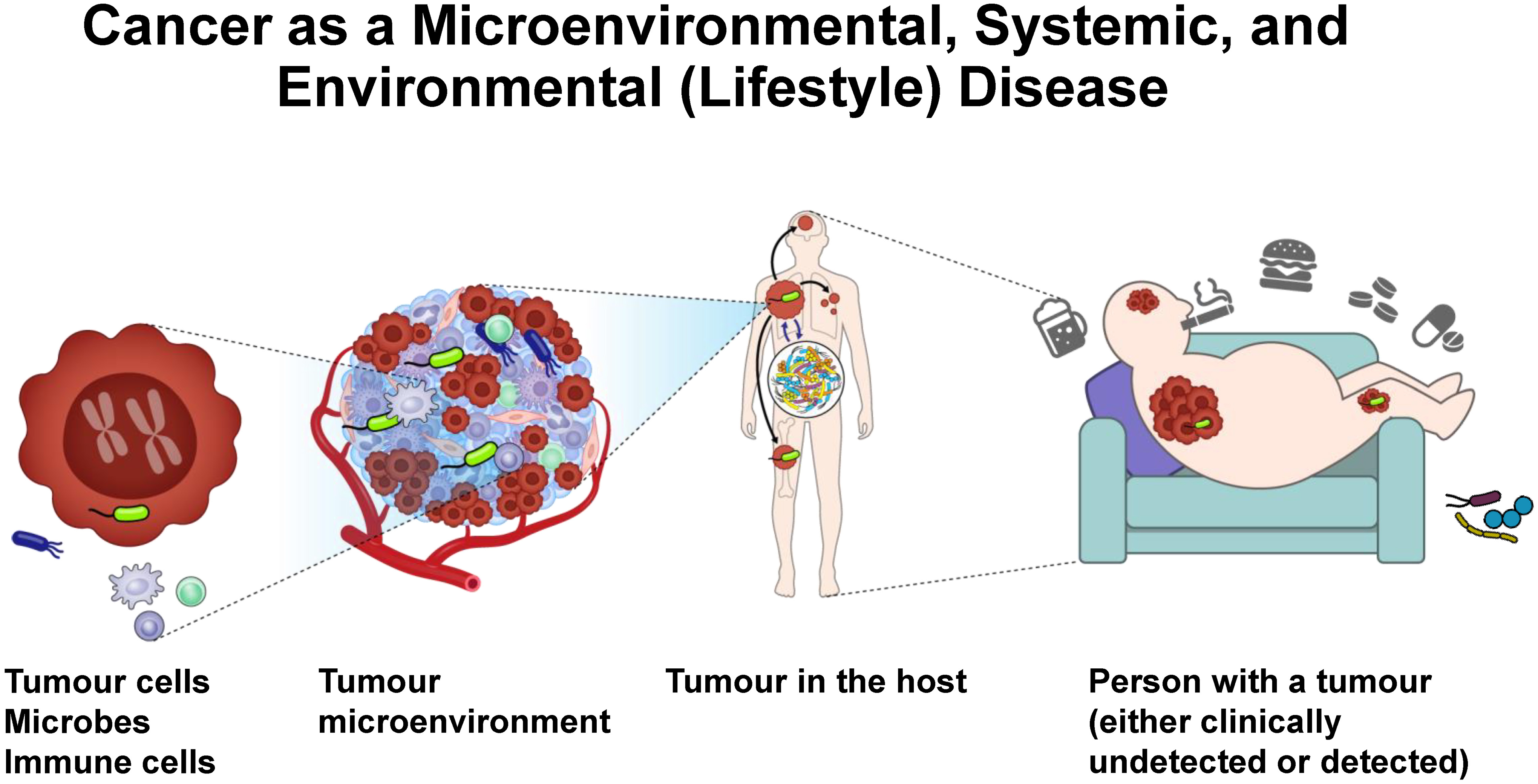
Multi-level perspectives of cancer-microbe associations. Tumour cells, microbes, and immune cells are major constituents of the tumour microenvironment. Cancer may metastasise to other organs. Microbiota, especially the gut microbiota, influences cancer phenotypes via systemic host-tumour-microbiome interactions. The exposome (e.g., diets, smoking, alcohol, supplements, medications, obesity, physical inactivity) influences tumour phenotypes and clinical outcomes of cancer patients via its complex effects on tumour cells, tumour microenvironment, and systemic conditions.
Physical and physiological conditions (e.g., obesity, surgery) can influence the gut microenvironment, thereby promoting tumourigenesis.86 Gastrointestinal surgery influenced the gut microbiota with increased species diversity and enrichment of deoxycholic acid and branched-chain amino acids.97 Metagenomic and metabolomic analyses of faecal samples suggested that patients with a history of gastrectomy had higher amounts of oral microbes, aerobes, or facultative anaerobes, as well as higher levels of deoxycholic acid and branched-chain amino acids in the intestine.97 Interestingly, patients with a history of total gastrectomy had abundant CRC-related bacteria (e.g., F. nucleatum),97 which might explain an increased risk of developing metachronous CRC in those patients.98
Role of Basic Experimental Research
Studies based on epidemiological or clinical cohorts have demonstrated associations of specific bacterial taxa with clinical and molecular characteristics of tumours.27,28,30,31,99–101 Functional analyses using preclinical model systems (e.g., organoids, animal models) can provide biological evidence for tumourigenic roles of specific bacterial species or communities and thereby support findings of population-based studies. Such experimental studies are essential to moving the cancer-microbiome research from a discovery phase to a translation phase, and finally to an implementation phase for cancer prevention, diagnosis, and treatment.
Organoids
Patient-derived organoids are ex vivo tissue cultures that form three-dimensional organ-like structures. Organoids can be genetically manipulated to recapitulate specific genetic mutations observed in patient tumours. Certain E. coli strains harbour the genetic island pks (polyketide synthase), which encodes a set of enzymes required for colibactin synthesis. The pks+ E. coli appear to inhabit the colorectal tissues in approximately 20% of healthy individuals, 40% of patients with inflammatory bowel diseases, and 60% of patients with familial adenomatous polyposis or CRC.102–105 A study using human intestinal organoids and CRC tissues indicated that pks+ E. coli might directly contribute to specific mutational signatures of CRC.102 These pks+ E. coli-induced mutational signatures were closely matched with those in healthy human colon crypts and suggested to be induced during early childhood,9 implying that the exposure to pks+ E. coli may occur during early childhood and predispose individuals to CRC development in later life.
Mouse models
Genetically engineered mouse models (GEMMs) serve as a valuable tool in preclinical cancer research and allow for microbiome studies of conventional microbiota (conventionally housed or specific pathogen-free) or those of gnotobiotic GEMMs. In particular, germ-free mice facilitate examinations of animals without microbes or gnotobiotic animals exclusively colonised by defined microbial species or communities.106
In a study using ApcMin/+ mice,23 oral administration of F. nucleatum increased intestinal tumour formation, supporting its carcinogenic role. However, exposure to other F. nucleatum strains did not increase tumour formation in germ-free or specific pathogen-free ApcMin/+ and ApcMin/+;IL10−/− mice,107 suggesting the existence of tumour-promoting virulence factors in specific F. nucleatum strains. A study utilising patient-derived xenografts of CRC demonstrated that Fusobacterium and co-existing cancer-specific microbes persist following serial implantation,16 suggesting that the microbiota is an intrinsic component of the tumour microenvironment. This study also serves as a proof-of-principle work of microbe-targeted treatment, demonstrating that bacterial ablation reduced tumour burden in mice harbouring Fusobacterium-positive human tumours.16
A synergistic role of microbial members in the initiation of hereditary CRC was assessed by examining colonic mucosal biofilms composed of enterotoxigenic Bacteroides fragilis (ETBF) and pks+ E. coli in GEMMs.103 Co-colonisation of pks+ E. coli and ETBF led to faster tumour onset, greater mortality, and higher levels of colonic inflammation than infection with either bacterial strain alone. ETBF enhanced pks+ E. coli colonisation through mucus degradation and subsequently increased cellular DNA damage and IL17A production with the aid of pks+ E. coli. Interestingly, mucosal biofilms from CRC patients or even healthy individuals were tumourigenic in germ-free ApcMinΔ850/+;IL10−/− or ApcMinΔ850/+ and specific pathogen-free ApcMinΔ716/+ mice, suggesting a carcinogenic potential of bacterial biofilms.108
Population-based data indicate associations of the intratumoural microbiota with clinical outcomes of pancreatic cancer patients.109 To validate these associations, antibiotic-pretreated C57BL/6 mice which received faecal microbiota transplantation (FMT) from pancreatic cancer patients were orthotopically implanted with KPC (Pdx1-Cre;LSL-Krasp.G12D/+;LSL-Tp53p.R172H/+) pancreatic cancer cells. FMT from long-term survivors resulted in decreased tumour burden with increased antitumour T cell infiltrates.109 Collectively, intratumoural and intestinal microbiome data at the time of therapy initiation may guide treatment strategies, including microbial manipulation.
Challenges in basic experimental research
A major challenge of the current preclinical models is the difficulty in accurately recapitulating the complexity of tumour microenvironment along with varieties of microbial populations and immune cells in humans. Bacterial culture has been a fundamental method of analysing microbes, which enables the reproduction of microorganisms in a predetermined culture medium under controlled laboratory conditions. However, approximately 70–80% of the intestinal bacterial species cannot be cultured.110 It has been difficult to examine bacterial populations in the human body, retarding our understanding of complex microbial communities in humans. To overcome these challenges, mechanistic approaches should be sophisticated at both reductionist and community levels.
Human Population Research with Innovative Microbiomics Technologies
To overcome the limitations of the conventional microbiology assays, including bacterial culture, next-generation sequencing (NGS) emerged as a culture-free technology in the early 2000s. NGS-based high-throughput technologies allow for analyses of unculturable or previously unidentified microbes and thereby facilitate examinations of an entire spectrum of microbial populations at the nucleic acid level (i.e., the metagenome).111 Metagenomic approaches have been increasingly utilised to assess taxonomic and functional characteristics of the microbiota.112
In recent meta-analyses,93,113 CRC-related microbial alterations were noted consistently across three continents, despite considerable differences not only in environmental, dietary, and lifestyle factors between the populations but also in approaches of taxonomic profiling and statistical analyses.93,113 A core set of intestinal microorganisms was strongly associated with CRC (e.g., F. nucleatum, Parvimonas micra, Gemella morbillorum). Most of those core species decreased dramatically after tumour removal.90
Metabolomics analysis is another omics approach that has expanded our understanding of complex interactions between microbiota, metabolites, and the host. SCFAs play pivotal roles in modulating inflammation and tumourigenesis.114 Amounts of metabolites can be measured comprehensively using mass spectrometry and/or nuclear magnetic resonance spectroscopy. Especially, various charged metabolites can be quantified using capillary electrophoresis time-of-flight-mass spectrometry.115 These assays have revealed dynamic alterations of metabolomic profiles during colorectal tumourigenesis, including increased levels of deoxycholic acids and branched-chain amino acids in early-stage CRC.90 Secondary bile acids (e.g., deoxycholic acids) might promote colorectal carcinogenesis through generating reactive oxygen and nitrogen species, which potentially damage DNA and promote resistance to apoptosis.116 Stool specimens from CRC patients showed increased amino acids and decreased SCFAs.117 Recent advances in metabolomics technologies have provided novel insights into intestinal metabolic dynamics in the complex microbial ecosystem.1
Integration of Microbiomics into Exposome Research
Epidemiology is a fundamental scientific field that studies the aetiology and consequence of a disease of interest in human populations. However, the importance of epidemiological studies in addressing the complex roles of tumour microenvironment has not been fully recognised. A substantial gap remains between basic experimental and epidemiological research.
It is conceivable that risk factor exposures may contribute to tumourigenesis at least some time points during possibly decades-long latency from normal cells to clinically-detectable cancer.118,119 Certain exposures (e.g., strong radiation), even for a short time, can directly cause cellular alterations and increase cancer risks for a lifetime. Other exposures (e.g., obesity) may help altered cells clonally expand and accumulate additional molecular changes over a long time period via mechanisms including promotion of cellular proliferation and suppression of antitumour immunity.120 Therefore, it is desirable to examine various exposures in each individual longitudinally over time. Furthermore, the relationships between exposures and tumour development may differ by tumoural characteristics. Based on this notion, molecular pathological epidemiology (MPE) research examines associations of exposures with disease subtypes classified by molecular and pathological signatures.121,122 For microbiome-related cancer, utilising microbiomic data from biospecimens (tumour/normal tissue, stool, blood, saliva, etc.), MPE research can provide evidence for the association of an exposure with development and consequence of tumour subtypes with specific microbial features (Figure 3). For example, the inverse relationship of fibre-rich “prudent diets” with CRC incidence appeared stronger for tumours containing abundant F. nucleatum.101 Hence, the MPE approach can link prudent diets with specific intratumoural bacteria, thereby supporting causality and uncovering a strong association that is otherwise masked in the traditional epidemiological analysis of overall CRC. Another study showed that a positive association between inflammatory diets and CRC incidence was stronger for tumours containing abundant F. nucleatum.100 These findings suggest that the F. nucleatum-rich tumour subtype may be affected by dietary factors and that dietary interventions may help control microbiota-related CRC.122,123 Moreover, it is of particular interest to examine microbial features in the intestine or other organs (as exposures) in relation to tumours subtyped by intratumoural microbial characteristics. With its unique strengths, MPE research can contribute to developing microbe-targeted strategies for cancer prevention and treatment.
Figure 3.
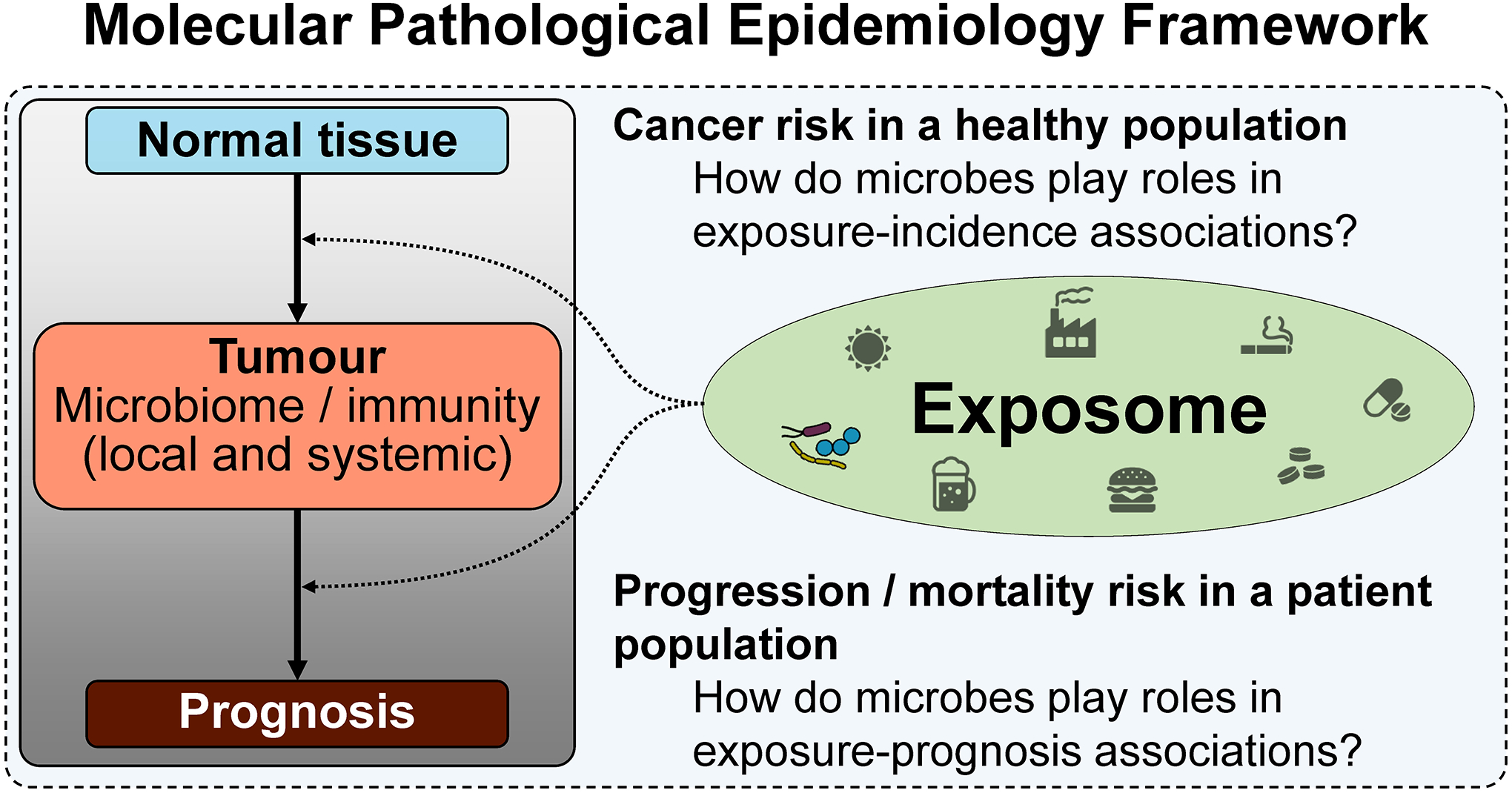
Analytical framework of molecular pathological epidemiology (MPE) in cancer-microbiome research. MPE research examines associations of an exposure of interest with the development and consequence of tumour subtypes with specific microbial / immune features, potentially providing evidence for complex interactions between exposome and tumour during cancer development and progression. Notably, the “tumour” in this figure may be a benign (premalignant) or malignant tumour, which can be analysed for its microbial, immune, and other characteristics.
Translational Potentials for Microbe-targeted Preventive and Therapeutic Strategies
Microbiome-modulating strategies
Substantial evidence supports microbial manipulation as a promising strategy for cancer prevention and treatment.124–127 Potential microbial interventions include dietary modifications, pre/probiotics, antimicrobial agents, FMT, and bacterial cocktails. Microbial interventions can be applied as an adjunct to traditional cancer therapeutics or stand-alone therapy and may mitigate the adverse effects of anticancer therapies.128,129
Evidence supports dietary alterations as a microbiota-modifying intervention.130–132 High-fibre, less western-style diets can prevent gut microbes from consuming mucous glycoproteins, and strengthen the mucus barrier function.133–135 Mediterranean diets may induce SCFA production and exert anti-inflammatory properties, potentially reducing the risk of chronic inflammation-related diseases, including CRC.136
Oral administrations of pre/probiotics are easy-to-implement ways to modulate microbial populations.137 Prebiotics are non-viable substances that facilitate the growth or activity of certain bacterial species, whereas probiotics are individual or combinations of bacteria. Administration of probiotics containing Bifidobacterium lactis and Lactobacillus acidophilus to CRC patients increased butyrate-producing bacteria and decreased CRC-related bacteria in the intestine.138
Antimicrobial agents, including antibiotics, have an appreciable influence on the development and consequence of cancer by modulating the microbiota. Antibiotics may eliminate favourable microbes and decrease microbial diversity, resulting in microbial dysbiosis. Developing narrow spectrum antimicrobial or anti-virulence approaches with little disruption to the human microbial ecosystem would be an ideal strategy for cancer control. For example, fidaxomicin selectively targets Clostridium difficile with minimal effects on gut commensals, as supported by findings that a fidaxomicin-binding determinant of RNA polymerase is present in C. difficile but is absent in intestinal bacteria.139 Another example is sequence-specific antimicrobials based on programmed CRISPR-Cas13a packaged into a bacteriophage capsid, which potentially target antimicrobial-resistant bacteria.140 This technology would facilitate the development of antimicrobials that can selectively eliminate carcinogenic microbes with minimal dysregulation of the commensal microbial flora for cancer control. The duration of trials investigating antimicrobial approaches for cancer prevention and treatment needs to be determined based on data on numbers of outcome and adverse events in exposed and unexposed populations in previous studies.
The FMT approach, where favourable microbial ecosystem of a donor is transplanted to a recipient, has been investigated in C. difficile infection141–143 and ulcerative colitis.144,145 Emerging evidence suggests the effectiveness of FMT for cancer control.146 This approach may be more beneficial than the administration of limited microbial species, given that an appropriate microbial ecosystem comprising various microbes enables the host to maintain normal physiological function and homeostasis.125,147 Recent studies have reported utilities of FMT to enhance immunotherapeutic efficacy.148–154 However, FMT has some risks, as illustrated by a report of antibiotic-resistant bacteraemia after receiving FMT in patients with C. difficile infection155 and a preclinical study that showed inflammation-associated carcinogenesis in FMT-treated mice.156
Bacterial cocktails are a mixture of purified bacteria with a presumably better safety profile compared to FMT. Bacterial cocktails, including Firmicutes species extracted from the stool of healthy human donors, relieved symptoms of C. difficile infection as effectively as FMT.157 Bacterial cocktails can also enhance immunotherapeutic efficacy. For example, a mixture of 11 bacterial strains could expand the populations of CD8+IFNG (interferon-γ)+ T cells and boost their activity to kill tumour cells, thereby augmenting the efficacy of anti-PDCD1 (PD-1) or anti-CTLA4 treatment.158 Most healthy individuals did not harbour these beneficial microbes, and in a minority of individuals who did, the microbial abundance was low.158 This study emphasises that highly abundant microbes in certain niches are not necessarily functionally important and that specific microbes with experimentally proven functions should be targeted for cancer control.
Microbial interventions for cancer therapeutics
Microbiota potentially affects the efficacy of anticancer therapies. Several studies have reported differential effects of cancer therapeutics by the microbiota (Table 2).159–163 Certain microbes appear to enhance chemotherapeutic efficacy, while others may have the opposite effect.159–163 The antitumour effect of oxaliplatin was reduced without the innate gut microbiota that stimulates tumour-infiltrating myeloid-derived cells to produce reactive oxygen species.164 In CRC, F. nucleatum may provoke tumour resistance to oxaliplatin and 5-fluorouracil by upregulating ULK1 and ATG7 expressions.165 Similarly, in oesophageal cancer, intratumoural F. nucleatum may promote autophagy by increasing ATG7 levels, thereby conferring chemoresistance to oesophageal cancer.166 Cyclophosphamide impairs the intestinal mucosal barrier and promotes bacterial translocation to the spleen and lymph nodes, where the microbes provoke antitumour Th17-mediated immune responses.167 In tumour-bearing mice, ablation of gram-positive bacteria decreased Th17 cells and induced resistance to cyclophosphamide.167 Through their drug-metabolising activities,159 gut microbes can influence chemotherapeutic efficacy. In pancreatic cancer, intratumoural bacteria directly provoke gemcitabine resistance by metabolising gemcitabine into its inactive form, and antibiotics (ciprofloxacin) can reverse the chemoresistance and facilitate the apoptosis of cancer cells when administered with gemcitabine.168 The microbial profile also influences chemotherapeutic toxicity. Indeed, several microbes increase the risk of chemotherapy-induced adverse events by producing drug-metabolising enzymes, and elimination of those microbes ameliorates the adverse effects.125,159 Irinotecan is detoxified in the liver and transfers through the bile duct to the intestine, where microbe-derived beta-glucuronidases can reactivate it and thereby cause diarrhoea and other toxicities.169,170
Table 2.
Studies investigating microbial manipulation in relation to the effectiveness of cancer therapeutics with microbial data
| Microbial intervention | Preclinical / clinical | Findings | |
|---|---|---|---|
| Chemotherapy | |||
| Colon cancer211 | Antibiotics | Preclinical | In mice, antibiotics dysregulated the gut microbiome and reduced anti-tumour effect of 5-fluorouracil. |
| Colon cancer, lymphoma, and melanoma164 | Antibiotics | Preclinical | Antibiotics-treated or germ-free mice responded poorly to CpG-oligonucleotide immunotherapy or oxaliplatin with reduced production of reactive oxygen species and immune-related cytokines. |
| Hepatocellular carcinoma212 | Antibiotics | Clinical | Shorter progression-free / overall survival times were observed among patients receiving carbapenem before or during systemic interferon injection and hepatic arterial infusion of 5-fluorouracil with or without cisplatin. Antianaerobic drugs were also associated with poor prognosis. The abundance of Blautia in faecal microbiota was associated with favourable survival outcomes. |
| Lung cancer167 | Antibiotics | Preclinical | In antibiotics-treated or germ-free mice, tumours were resistant to cyclophosphamide with reduced reactivity of T helper 17 cells. Transfer of T helper 17 cells partially restored the antitumour effect of cyclophosphamide. |
| Immunotherapy | |||
| Colon cancer176 | Engineered probiotics | Preclinical | In mice, an engineered Escherichia coli strain colonised in the tumour and increased local L-arginine levels, resulting in increased tumour-infiltrating T cells and synergistic effects with anti-CD274 (PD-L1) therapy. |
| Colon cancer175 | Probiotics | Preclinical | In mice, Bifidobacterium accumulated within the tumour, when administered systemically or orally, and restored tumour response to anti-CD47 therapy through upregulation of the STING1 signalling. |
| Colon cancer213 | Antibiotics | Preclinical | In mice, antibiotics enhanced antitumour immune responses and antitumour effects of neoantigen cancer vaccines delivered by DNA electroporation. |
| Colon cancer and melanoma214 | Antibiotics and probiotics | Preclinical | In mice, antibiotics attenuated antitumour effect of the HAVCR2 (Tim-3) blockade. The therapeutic efficacy was restored by oral gavage of faecal bacteria. |
| Colon cancer, melanoma, NSCLC, and sarcoma148 | Antibiotics and FMT | Preclinical and clinical | In mice and patients, T cell response to Bacteroides thetaiotaomicron or Bacteroides fragilis was correlated with the effectiveness of ipilimumab (anti-CTLA4 antibody). Antibiotics-treated mice did not respond to the CTLA4 blockade. In mice, FMT from patients with gut microbiota related to a good response to ipilimumab promoted the outgrowth of B. fragilis and enhanced the antitumour effect. |
| Melanoma177 | Diets and oral probiotics | Clinical | In patients, high fibre consumption was associated with better response to anti-PDCD1 (PD-1)-based therapy, with the most pronounced benefit observed in patients with sufficient dietary fibre and no probiotic use. |
| Melanoma150 | FMT | Preclinical and clinical | Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium were more frequently identified in stool samples of patients responding to anti-PDCD1 (PD-1) or anti-CTLA4 therapy. In mice, FMT from the responders restored T cell-mediated immune response and antitumour effect of anti-CD274 (PD-L1) therapy. |
| Melanoma153,154 | FMT | Clinical | In patients with melanoma refractory to anti-PDCD1 (PD-1) therapy, FMT from responders restored tumour sensitivity to the PDCD1 blockade. The FMT resulted in favourable immune and microbial profiles in the gut and tumour microenvironment. |
| Melanoma and bladder cancer149 | Oral probiotics | Preclinical | In mice, oral Bifidobacterium administration improved tumour control and, when combined with the CD274 (PD-L1) blockade, almost eliminated the tumour. The key mediators were enhanced dendritic cell function and recruitment of CD8+ T cells to the tumour microenvironment. |
| NSCLC, renal cell carcinoma, and urothelial carcinoma151 | Antibiotics and FMT | Preclinical and clinical | In patients with advanced cancer, antibiotics use was associated with poor response to anti-PDCD1 (PD-1) or anti-CD274 (PD-L1) therapy. In mice transplanted with sarcoma cells, FMT from responding patients improved the effectiveness of the immune checkpoint blockade. |
| Pancreatic cancer48 | Antibiotics | Preclinical | In mice, antibiotics upregulated PDCD1 (PD-1) expression in CD4+ cells and CD8+ cells in the tumour and synergised with anti-PDCD1 therapy in tumour suppression. |
| Hormone therapy | |||
| Prostate cancer215 | Antibiotics | Preclinical | In mice, antibiotics delayed castration resistance of the tumour. FMT from patients with hormone-sensitive prostate cancer and Prevotella stercorea administration promoted tumour suppression. |
Abbreviations: FMT, faecal microbiota transplantation; NSCLC, non-small cell lung carcinoma.
Recent studies support the effects of the intestinal microbiota on responses to immune checkpoint inhibitors such as anti-PDCD1 (PD-1), anti-CD274 (PD-L1), and anti-CTLA4 monoclonal antibodies.148–152,163,171–173 The gut microbiota may influence immunotherapeutic efficacy through its complex interactions with the host, which modulate antitumour immunity.125,172,173 Administration of specific microbes to tumour-bearing mice enhanced the efficacy of anti-CTLA4 treatment by triggering Th1-dependent immune reactions in tumour-draining lymph nodes and shifting dendritic cells towards a pro-inflammatory state.148 Similar enhancement was observed in the blockade of the CD274-PDCD1 axis with increased CD8+ cytotoxic T cells and decreased FOXP3+ regulatory T cells (Tregs) in the tumour microenvironment.149,151 Patients with favourable microbiota who responded to immune checkpoint inhibition had higher levels of effector CD4+ and CD8+ T cells and lower levels of Tregs and myeloid-derived suppressor cells in blood, compared to non-responding patients with unfavourable microbiota.152 In mouse models of various tumour types, FMT or oral administration of favourable bacteria enhanced the efficacy of immune checkpoint blockades.148–151 Furthermore, increased diversity of the gut microbiota may augment the effectiveness of this treatment strategy.151,152 Benefits from antibiotics in patients receiving immunotherapy depend on tumour types. Antibiotic treatment reduced the efficacy of immune checkpoint inhibition in patients with lung, kidney, or bladder cancer,151,174 while patients with pancreatic cancer benefitted from antibiotic administration.48 Microbes can travel to the distant tumour microenvironment and, on-site, enhance immunotherapeutic efficacy. Tumour microenvironment is often hypoxic, and therefore may enable anaerobic microbes to preferentially proliferate.175 In a preclinical model, systemic or oral administration of Bifidobacterium, an anaerobic commensal gut bacterium, led to its accumulation in the tumour microenvironment and enhanced the local effect of anti-CD47 immunotherapy via STING1 signalling.175 Engineered microbial therapies may enable metabolic modulation in the tumour microenvironment, leading to enhanced immunotherapeutic efficacy. In mice, colonisation of tumours with probiotic E. coli Nissle 1917 strain increased intratumoural arginine concentrations and tumour-infiltrating T cells, enhancing the efficacy of PDCD1 (PD-1) blockade.176 Diets and over-the-counter probiotic supplements may have differential effects on immunotherapeutic efficacy.177 In melanoma patients, high fibre consumption was associated with a better response to anti-PDCD1 (PD-1)-based therapy, with the most pronounced benefit observed in patients with sufficient dietary fibre and no probiotic use.177
The gastrointestinal microbiota influences adverse events of immune checkpoint inhibition.128,129,178,179 Certain microbes elicit immunotherapy-related toxicity, whereas others counteract it.128,129 In melanoma patients, the abundance of Bacteroidetes species was associated with decreased risk of CTLA4 blockade-induced colitis.180 In patients with urological cancer, FMT ameliorated immunotherapy-induced refractory colitis with decreased CD8+ T cells and increased anti-inflammatory FOXP3+ Tregs in colonic mucosa.181
Preclinical in vivo studies have demonstrated that FMT can enhance immunotherapeutic efficacy.148–152 Two first-in-human trials reported the safety and feasibility of FMT combined with anti-PDCD1 (PD-1) therapy for therapy-refractory metastatic melanoma.153,154 Both studies suggested that FMT might help overcome immunotherapy resistance by increasing antitumour immune responses locally and systemically.153,154
Despite accumulating evidence indicating immune- and microbiome-modulating effects of various exposures, it remains unknown whether (and if so, how) the association between the microbiota and immunotherapeutic efficacy (or toxicity) is modified by exposures. Integrative MPE research strategies are needed to address this research gap. Previous studies have suggested differential effects of aspirin use, vitamin D level, physical activity, cigarette smoking, and coffee intake on clinical outcomes of CRC patients by levels of lymphocytic reaction or tumour CD274 (PD-L1) expression.85,182–185 Investigations of the effects of modifiable exposures on clinical outcomes and underlying mechanisms (presumably through microbiota and immunity) can have substantial implications in the development of precision medicine.
Microbial interventions for cancer prevention
Growing evidence for the role of microbiota in oncogenesis supports microbial manipulation as a promising strategy for cancer prevention. For instance, pharmacological eradication of H. pylori can decrease gastric cancer risk and is recommended as a preventative treatment for gastric diseases.186,187 While the gastric microbiota resembles the oral microbiota in H. pylori-uninfected persons, H. pylori, when present, dominates the gastric microbial population and reduces the microbial diversity.188 In a population-based study, successful elimination of H. pylori restored gastric microbiota to a similar status as found in uninfected individuals.189
Chronic infection with HCV causes chronic hepatitis, cirrhosis, and HCC. Recent advances in combination therapy with direct-acting antiviral drugs have provided a dramatic increase in the rate of sustained virologic response.190 Despite adverse effects associated with the antiviral drugs, this treatment strategy has no substantial effects on bacterial communities in the body. Therefore, these classes of agents would be a good candidate for selective antimicrobial strategies for cancer prevention and treatment.
F. nucleatum is a potential target to prevent colorectal carcinogenesis. Prudent and anti-inflammatory diets have been associated with a lower incidence of F. nucleatum-enriched CRC,100,101 suggesting the usefulness of diet-modifying preventative strategies. In a preclinical model, aspirin effectively killed F. nucleatum strain Fn7-1 and inhibited F. nucleatum-potentiated colonic tumourigenesis.191 In humans, daily aspirin intake was associated with a lower abundance of F. nucleatum in colonic adenoma tissues.191
Studies also pointed to a link between sulfur microbial diets (associated with abundant sulfur-reducing bacteria in stool) and the development of CRC192 and early-onset colorectal adenomas.193 Decreasing animal fat consumption appeared to suppress detrimental Bacteroidetes species,194 while high-fibre diets increased beneficial SCFA-producing bacteria.195 Therefore, diet-induced microbial alteration may influence the carcinogenesis processes and serve as effective cancer prevention strategies.
Two randomised controlled trials assessed the effect of probiotics and prebiotics on preventing colorectal tumours.196,197 One trial of 80 participants revealed that administration of synbiotics (i.e., combined prebiotics and probiotics) changed faecal microbiota and blood IL2 and IFNG levels.196 Another trial of 380 participants observed adenoma risk reduction by probiotics use.197 However, large-scale trials investigating microbial interventions for cancer prevention have not been conducted. Therefore, costs and potential adverse events should be considered when designing trials using healthy populations (Table 3).
Table 3.
Consideration and challenges ahead to modulate the microbiome for cancer prevention in healthy populations
| Intervention type | Cost | Potential disadvantages and considerations | Considerations across all intervention types | |
|---|---|---|---|---|
| Diets, nutrient supplements | Low | Varied effects by individuals; low compliance |
|
|
| Prebiotics (i.e., food ingredients that support beneficial microbes in the gut) | Relatively low | Varied effects by individuals; unknown pernicious effects | ||
| Probiotics (i.e., microorganisms that provide health benefits) | Relatively low | Variation of quality control, bioavailability, and standardisation; known and unknown pernicious effects | ||
| Antibiotics | Relatively high | Depletion of commensal / symbiotic bacteria; introduction of antibiotics-resistant bacteria; short-term and long-term side effects | ||
| Medication | Relatively high | Short-term and long-term side effects | ||
| Faecal microbial transplantation | High | Potential to affect the risk of other diseases; procedural risks; unknown safety profiles; donor selection is crucial but difficult |
Challenges and Future Directions
Technical difficulties exist in microbiomic profiling of clinical specimens, including tissue, stool, blood, saliva, urine, etc.198,199 As pre-analytical and analytical factors change microbial compositions, it is essential to standardise methods of specimen collection, processing, storage, and analyses. Although formalin-fixed paraffin-embedded (FFPE) tissue specimens are often used in human population studies, microbial profiles differ between FFPE and fresh tissue specimens. Intratumoural heterogeneity of microbial populations may pose another challenge.200 Multiple biopsies from each tumour should be conducted when feasible.200 In situ approaches (e.g., immunofluorescence) and spatial transcriptomic profiling enable spatial analyses of specific microbes in relation to tumour, immune, and other cell types.16,201,202
A gap remains between microbiomic analyses and epidemiological research. To address this gap, prospective cohort studies that examine the microbiome are needed. Most investigations that assessed the microbiome and cancer risk have used case-control or cross-sectional study designs, which have inherent limitations (Table 4). In typical case-control or cross-sectional studies, exposure information and biospecimens are collected at or after disease diagnosis. However, disease processes often influence individuals’ physiological states and exposures, likely altering biospecimen analytes (microbiome, metabolome, etc.). Hence, a difference in any analyte between cases and controls may be a consequence rather than a cause of the disease. Because of this phenomenon (i.e., “reverse causation”), results from case-control studies may not be helpful for aetiological inference or future risk assessments. Therefore, such results need to be tested in prospective cohort studies or randomised controlled trials that collected biospecimens long before disease detection.
Table 4.
Comparison of case-control versus prospective cohort study designs for the study of cancer aetiologies
| Case-control study design | Prospective cohort study design | |
|---|---|---|
| Source of bias | ||
| 1. Selection bias due to differential background populations that have given rise to cancer cases from controls | Yes (D) | No (A) |
| 2. Referral bias | Often large (D) | Small to moderate (A) |
| 3. Recall bias in long-term exposure measurements | Often large (D) | Small if the information is repeatedly obtained during follow-up (A) |
| 4. Differential recall bias between cancer cases and controls | Yes (D) | No (A) |
| Sample size of cancer cases | Can be large; enables to study rare cancer types (A) | Determined by cohort size (D) |
| Measurement of the incidence rate across different generations | Difficult (D) | Possible (A) |
| Collection of detailed clinical information | Relatively easy (A) | Relatively difficult (D) |
| Collection of biospecimens including tumour tissue | Relatively easy (A) | Relatively difficult (D) |
| Collection of biospecimens years before cancer diagnosis | Difficult or impossible (D) | Possible (A) |
| Time needed for follow-up | No (A) | Yes (D) |
| Overall cost | Relatively low (A) | Very high (D) |
| Generalisability (given the same statistical power) | Low to moderate (D) | Moderate to high (A) |
Abbreviations: A, advantage; D, disadvantage.
Given the multi-factorial processes of tumour development and progression, it is imperative to comprehensively integrate and analyse diverse types of data on exposures, the microbiome, and immune status in research on human cancer. Although conducting such comprehensive analyses is challenging, there is no alternative way. Experimental research using model systems under controlled environment is important and can shed light on pathogenic mechanisms. However, ultimately, we need to validate experimental findings from model systems in real human tumours. Otherwise, we cannot fully understand cancer as microenvironmental, systemic, and environmental diseases. Despite the enormous amounts of resources that have been invested in cancer research, our understanding of cancer currently remains limited because most research efforts have been focused on short-term goals with rather limited data collection and analyses, which cannot adequately decipher cancer (as depicted in Figure 4). It is time to shift our minds to judiciously invest our finite resources for well-designed studies with comprehensive data collection and integrated analyses.
Figure 4.
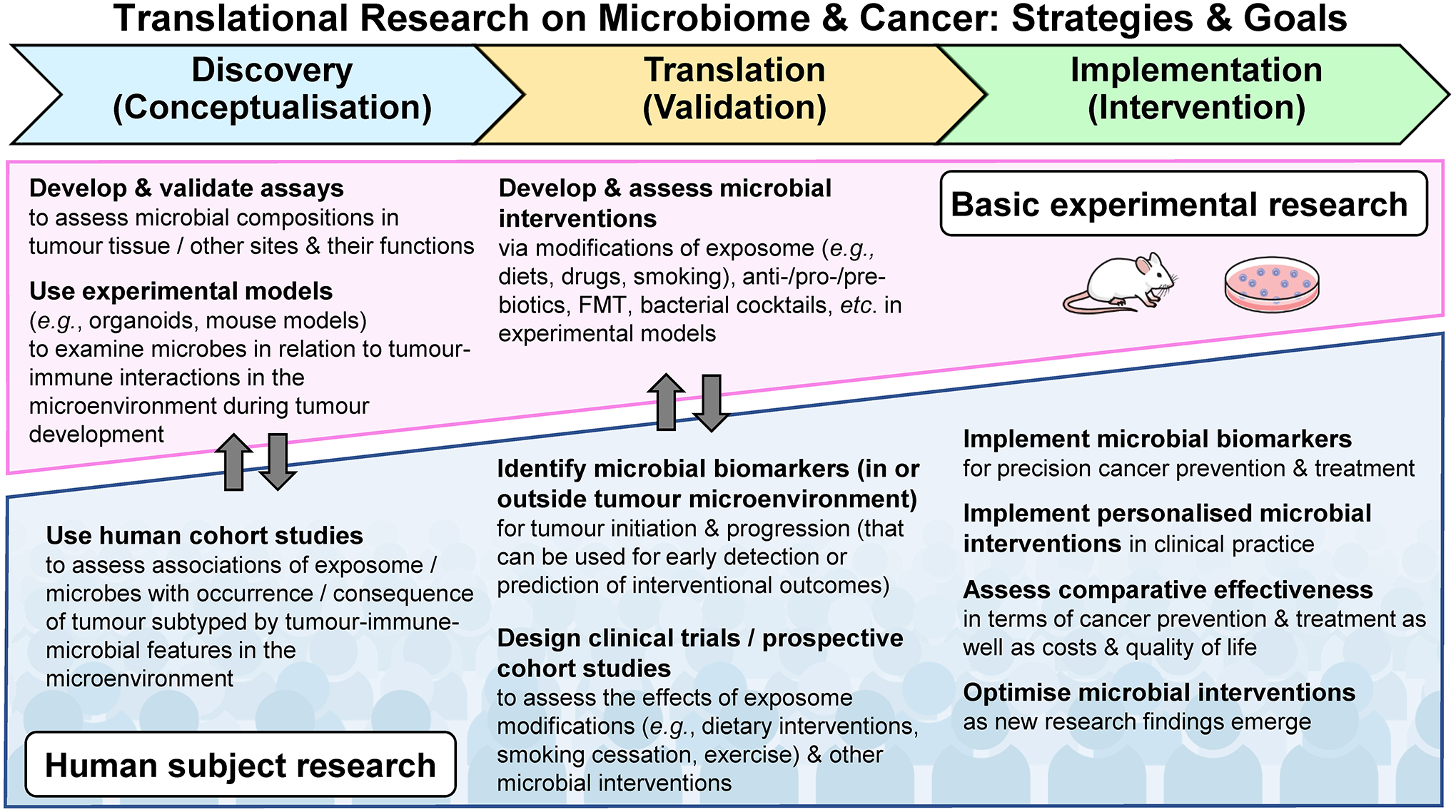
Roadmap of transdisciplinary cancer-microbiome research from a discovery phase to a translation phase and a final implementation phase for targeted cancer prevention and treatment. The exposome represents the totality of exposures (including but not limited to diets, drugs, and smoking), which can be examined individually or collectively in relation to cancer development and progression. In contrast to research on non-neoplastic diseases, cancer research is characterised by the availability of tumour tissue specimens for examinations of the microenvironment where tumour cells, immune cells, and microorganisms form a dynamic interactive network. Tumour tissue research plays a key role in discovering and validating new insights into the mechanism through which microorganisms may influence cancer initiation and progression by interacting with the exposome and immune cells. FMT, faecal microbiota transplantation.
Mounting evidence indicates a significant role of gene-by-environment interaction in various diseases including cancer.203 Hence, integrative multi-level analyses of genetic, environmental, systemic, and tumoural factors combined with microbiomic analyses of various biospecimens will drastically increase the values of microbiome studies.
A limited number of transdisciplinary education / training programmes has led to a paucity of investigators with transdisciplinary expertise. Conducting integrative microbiomic research necessitates expertise in various scientific disciplines including microbiology, immunology, pathology, epidemiology, bioinformatics, genetics, statistics, etc. Transdisciplinary research approaches will contribute to the generation of new research ideas and may lead to new fields of investigation.204,205
In recent decades, the incidence of various early-onset cancer types (tumours arising in bone marrow, breast, colorectum, endometrium, extrahepatic bile duct, gallbladder, head and neck, kidney, liver, oesophagus, pancreas, prostate, stomach, and thyroid) among individuals under age 50 has been increasing worldwide.206,207 Among these, the rise of prostate and thyroid cancers appears to be largely attributable to increased screening and early detection. Notably, eight of the remaining 12 early-onset cancer types arise in the aerodigestive system, implying the critical pathogenic role of the microbiome in this phenomenon. Integrative microbiomic research will likely shed light on the aetiologies of rising early-onset cancer.
Conclusions
Cancer is a complex condition that should be recognised as an environmental, systemic, and microenvironmental disease. A neoplasm evolves under the influence of various exposures that affect the local and systemic status of immune and microbial activities in the host. Therefore, effective research approaches should account for the interplay between the exposome, microorganisms, immune and other host cell populations, and neoplastic cells. Taking advantage of mechanistic evidence from basic experimental research, integrative microbiomic research can serve as a unique methodological framework and potentially provides novel insights into the host-tumour-microbiome interactions, thereby guiding microbe-targeted strategies for cancer control. Given the increasing availability of multi-omics analysis platforms to interrogate tumour, microbial, and immune signatures, the integrative approach would improve our understanding of the complex cancer pathogenesis. Despite the substantial challenges, there are ample opportunities for integrative microbiomic research to advance cancer science and ultimately reduce the cancer burden through effective precision prevention and treatment.
Key messages.
Cancer is a complex condition that should be recognised as an environmental, systemic, and microenvironmental disease.
The exposome (the totality of exposures including diets, supplements, smoking, alcohol, medications, obesity, physical activity, etc.) influences tumour phenotypes via its complex effects on tumour cells, tumour microenvironment, microorganisms, and systemic conditions.
Microorganisms, which ubiquitously exist in the tumour microenvironment and around the whole human body, play a pivotal role in shaping tumour phenotypes via complex host-tumour-microbiome interactions.
Transdisciplinary research integrating analyses of the exposome, microbiome, and tumour microenvironment based on experimental models and human populations is needed to examine the dynamic interplay of these factors and develop targeted cancer prevention and therapeutics.
Funding:
This work was supported by a U.S. National Institutes of Health (NIH) grants (R35 CA197735 to S.O. and R01 CA248857 to S.O.), and by Cancer Research UK Grand Challenge Award (OPTIMISTICC [C10674/A27140] to S.O.). K.I. was supported by grants from JSPS KAKENHI (JP22H02930), the Takeda Science Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Ichiro Kanehara Foundation, Grant for Lung Cancer Research, Suzuki Foundation for Urological Medicine, Foundation for Promotion of Cancer Research in Japan, and the Yakult Bio-Science Foundation. T.H. was supported by grants from JSPS KAKENHI (JP19K08362) and the Takeda Science Foundation. T.U. was supported by grants from the Mishima Kaiun Memorial Foundation, Japan Society for the Promotion of Science (201960541), and Prevent Cancer Foundation. S.Y. was supported by grants from the National Cancer Center Research and Development Fund (2020-A-4), JSPS KAKENHI (JP20H033620), Practical Research for Innovative Cancer Control from the Japan Agency for Medical Research and Development (AMED) (JP21ck0106546), Project for Cancer Research and Therapeutic Evolution (P-CREATE) from AMED (JP22cm0106477); Project for Promotion of Cancer Research and Therapeutic Evolution (P-PROMOTE) from AMED (JPama221404), United States-Japan Cooperative Medical Science Program from AMED (JP20jk0210009); AIP Accelerated Program from JST (21-191029679); Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Joint Research Project of the Institute Medical Science, the University of Tokyo, the Takeda Science Foundation, the Yasuda Medical Foundation, the Mitsubishi Foundation, and the Princess Takamatsu Cancer Research Fund. The funding source had no role in the decision to submit the manuscript to publication or preparation, review, and approval of the manuscript.
Conflicts of interest:
K.I. received research grants from Konica Minolta, Inc. and Daiichi Sankyo Co., Ltd. outside the submitted work. S.B. is a co-inventor on a U.S. Provisional Patent Application no. 62/534,672, that covers targeting of Fusobacterium for treatment of colorectal cancer. S.B. has consulted for glaxosmithkline (GSK) and BiomX, and is currently on the cancer program scientific advisory board for BiomX. This study was not funded by any of these companies. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
Abbreviations:
- aka
also known as
- CRC
colorectal cancer
- EBV
Epstein-Barr virus
- ETBF
enterotoxigenic Bacteroides fragilis
- FFPE
formalin-fixed paraffin-embedded
- FMT
faecal microbiota transplantation
- GEMM
genetically engineered mouse model
- HCC
hepatocellular carcinoma
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HGNC
Human Genome Organisation Gene Nomenclature Committee
- MPE
molecular pathological epidemiology
- NGS
next-generation sequencing
- NSCLC
non-small cell lung carcinoma
- PD-1
programmed cell death 1
- PD-L1
programmed cell death 1 ligand 1
- PI3K
phosphatidylinositol-4,5-bisphosphonate 3-kinase
- pks
polyketide synthase
- SCFA
short-chain fatty acid
- Treg
regulatory T cell
Footnotes
Use of standardised official symbols: We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes and gene products, including ADIPOQ, ANXA1, APC, ARID1A, ATG7, BRAF, CD3, CD4, CD8, CD47, CD274, CDKN2A, CEACAM1, CRP, CTLA4, CTNNB1, CXCL1, CXCR2, FOXP1, FOXP3, GDF15, HAVCR2, IFNG, IL1B, IL2, IL33, IL6, IL10, IL17A, JAK2, KRAS, MYD88, NOTCH1, PBRM1, PDCD1, PDX1, PIK3CA, PTPRC, STING1, TGFB1, TIGIT, TLR, TLR4, TNFRSF1B, TP53, ULK1, and WNT; all of which are described at www.genenames.org. The official gene symbols are italicised to differentiate from non-italicised gene product names (and other colloquial names). Names of non-human genes and gene products have the first Capital letter followed by small case letters.
Note added in proof
A recent study has shown a positive relationship between long-term Western-style diets and increased incidence of colorectal cancer containing high amounts of pks+ E. coli,216 which further attests to the power of the MPE approach in deciphering dietary influences on tumourigenic processes via the microbe.
References
- 1.Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science 2021;371:eabc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell 2021;39:1317–41. [DOI] [PubMed] [Google Scholar]
- 3.LaCourse KD, Johnston CD, Bullman S. The relationship between gastrointestinal cancers and the microbiota. Lancet Gastroenterol Hepatol 2021;6:498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An Y, Zhang W, Liu T, Wang B, Cao H. The intratumoural microbiota in cancer: new insights from inside. Biochim Biophys Acta Rev Cancer 2021;1876:188626. [DOI] [PubMed] [Google Scholar]
- 5.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA Jr., et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018;67:1168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cani PD, Jordan BF. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nat Rev Gastroenterol Hepatol 2018;15:671–82. [DOI] [PubMed] [Google Scholar]
- 7.Fujiyoshi K, Bruford EA, Mroz P, Sims CL, O’Leary TJ, Lo AWI, et al. Opinion: Standardizing gene product nomenclature-a call to action. Proc Natl Acad Sci U S A 2021;118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kakiuchi N, Ogawa S. Clonal expansion in non-cancer tissues. Nat Rev Cancer 2021;21:239–56. [DOI] [PubMed] [Google Scholar]
- 9.Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 2019;574:532–7. [DOI] [PubMed] [Google Scholar]
- 10.Colom B, Herms A, Hall MWJ, Dentro SC, King C, Sood RK, et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature 2021;598:510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008;27:5904–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal 2020;18:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368:973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greathouse KL, Stone JK, Harris CC. Cancer-Type-Specific Bacteria: Freeloaders or Partners? Cancer Cell 2020;38:158–60. [DOI] [PubMed] [Google Scholar]
- 16.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358:1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D Hallmarks of Cancer: New Dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 18.Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021;592:138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triner D, Devenport SN, Ramakrishnan SK, Ma X, Frieler RA, Greenson JK, et al. Neutrophils Restrict Tumor-Associated Microbiota to Reduce Growth and Invasion of Colon Tumors in Mice. Gastroenterology 2019;156:1467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parida S, Sharma D. The Microbiome and Cancer: Creating Friendly Neighborhoods and Removing the Foes Within. Cancer Res 2021;81:790–800. [DOI] [PubMed] [Google Scholar]
- 21.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015;42:344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology 2019;8:e1581531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep 2019;20:e47638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol 2016;7:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gethings-Behncke C, Coleman HG, Jordao HWT, Longley DB, Crawford N, Murray LJ, et al. Fusobacterium nucleatum in the Colorectum and Its Association with Cancer Risk and Survival: A Systematic Review and Meta-analysis. Cancer Epidemiol Biomarkers Prev 2020;29:539–48. [DOI] [PubMed] [Google Scholar]
- 30.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol 2015;1:653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch 2017;471:329–36. [DOI] [PubMed] [Google Scholar]
- 32.Borozan I, Zaidi SH, Harrison TA, Phipps AI, Zheng J, Lee S, et al. Molecular and pathology features of colorectal tumors and patient outcomes are associated with Fusobacterium nucleatum and its subspecies animalis. Cancer Epidemiol Biomarkers Prev 2022;31:210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol 2019;16:690–704. [DOI] [PubMed] [Google Scholar]
- 34.Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang AJ, et al. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat Microbiol 2022;7:238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019;68:1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarris G, de Rougemont A, Charkaoui M, Michiels C, Martin L, Belliot G. Enteric Viruses and Inflammatory Bowel Disease. Viruses 2021;13:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 2018;155:529–41.e5. [DOI] [PubMed] [Google Scholar]
- 38.Hannigan GD, Duhaime MB, Ruffin MTt, Koumpouras CC, Schloss PD. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. mBio 2018;9:e02248–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019;68:654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coker OO, Wu WKK, Wong SH, Sung JJY, Yu J. Altered Gut Archaea Composition and Interaction With Bacteria Are Associated With Colorectal Cancer. Gastroenterology 2020;159:1459–70.e5. [DOI] [PubMed] [Google Scholar]
- 41.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984;1:1311–5. [DOI] [PubMed] [Google Scholar]
- 42.Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes 2020;11:1220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 2020;8:e180–e90. [DOI] [PubMed] [Google Scholar]
- 44.Peek RM Jr., Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2002;2:28–37. [DOI] [PubMed] [Google Scholar]
- 45.Baj J, Korona-Głowniak I, Forma A, Maani A, Sitarz E, Rahnama-Hezavah M, et al. Mechanisms of the Epithelial-Mesenchymal Transition and Tumor Microenvironment in Helicobacter pylori-Induced Gastric Cancer. Cells 2020;9:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito M, Kono K. Landscape of EBV-positive gastric cancer. Gastric Cancer 2021;24:983–9. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Ge J, Wang Y, Xiong F, Guo J, Jiang X, et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat Commun 2022;13:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019;574:264–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maekawa T, Fukaya R, Takamatsu S, Itoyama S, Fukuoka T, Yamada M, et al. Possible involvement of Enterococcus infection in the pathogenesis of chronic pancreatitis and cancer. Biochem Biophys Res Commun 2018;506:962–9. [DOI] [PubMed] [Google Scholar]
- 51.Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell 2022;40:153–67.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kartal E, Schmidt TSB, Molina-Montes E, Rodríguez-Perales S, Wirbel J, Maistrenko OM, et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut 2022:doi: 10.1136/gutjnl-2021-324755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagata N, Nishijima S, Kojima Y, Hisada Y, Imbe K, Miyoshi-Akiyama T, et al. Metagenomic identification of microbial signatures predicting pancreatic cancer from a multinational study. Gastroenterology 2022:doi: 10.1053/j.gastro.2022.03.054. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Ma C, Duan Y, Heinrich B, Rosato U, Diggs LP, et al. Gut Microbiome Directs Hepatocytes to Recruit MDSCs and Promote Cholangiocarcinoma. Cancer Discov 2021;11:1248–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iida N, Mizukoshi E, Yamashita T, Yutani M, Seishima J, Wang Z, et al. Chronic liver disease enables gut Enterococcus faecalis colonization to promote liver carcinogenesis. Nat Cancer 2021;2:1039–54. [DOI] [PubMed] [Google Scholar]
- 56.Chakladar J, Wong LM, Kuo SZ, Li WT, Yu MA, Chang EY, et al. The Liver Microbiome Is Implicated in Cancer Prognosis and Modulated by Alcohol and Hepatitis B. Cancers (Basel) 2020;12:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komiyama S, Yamada T, Takemura N, Kokudo N, Hase K, Kawamura YI. Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci Rep 2021;11:10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong L, Lu D, Chen R, Lin Y, Zhu H, Zhang Z, et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell 2022;40:70–87.e15. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Tang W, Budhu A, Forgues M, Hernandez MO, Candia J, et al. A Viral Exposure Signature Defines Early Onset of Hepatocellular Carcinoma. Cell 2020;182:317–28.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat Rev Cancer 2021;21:345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zitvogel L, Perreault C, Finn OJ, Kroemer G. Beneficial autoimmunity improves cancer prognosis. Nat Rev Clin Oncol 2021;18:591–602. [DOI] [PubMed] [Google Scholar]
- 62.Finn OJ. The dawn of vaccines for cancer prevention. Nat Rev Immunol 2018;18:183–94. [DOI] [PubMed] [Google Scholar]
- 63.Renehan AG, Zwahlen M, Egger M. Adiposity and cancer risk: new mechanistic insights from epidemiology. Nat Rev Cancer 2015;15:484–98. [DOI] [PubMed] [Google Scholar]
- 64.Lega IC, Lipscombe LL. Review: Diabetes, Obesity, and Cancer-Pathophysiology and Clinical Implications. Endocr Rev 2020;41:bnz014. [DOI] [PubMed] [Google Scholar]
- 65.Vigneri P, Frasca F, Sciacca L, Pandini G, Vigneri R. Diabetes and cancer. Endocr Relat Cancer 2009;16:1103–23. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto T, Kawada K, Obama K. Inflammation-Related Biomarkers for the Prediction of Prognosis in Colorectal Cancer Patients. Int J Mol Sci 2021;22:8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harlid S, Gunter MJ, Van Guelpen B. Risk-Predictive and Diagnostic Biomarkers for Colorectal Cancer; a Systematic Review of Studies Using Pre-Diagnostic Blood Samples Collected in Prospective Cohorts and Screening Settings. Cancers (Basel) 2021;13:4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song M, Mehta RS, Wu K, Fuchs CS, Ogino S, Giovannucci EL, et al. Plasma Inflammatory Markers and Risk of Advanced Colorectal Adenoma in Women. Cancer Prev Res (Phila) 2016;9:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mehta RS, Chong DQ, Song M, Meyerhardt JA, Ng K, Nishihara R, et al. Association Between Plasma Levels of Macrophage Inhibitory Cytokine-1 Before Diagnosis of Colorectal Cancer and Mortality. Gastroenterology 2015;149:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehta RS, Song M, Bezawada N, Wu K, Garcia-Albeniz X, Morikawa T, et al. A prospective study of macrophage inhibitory cytokine-1 (MIC-1/GDF15) and risk of colorectal cancer. J Natl Cancer Inst 2014;106:dju016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bao Y, Giovannucci EL, Kraft P, Stampfer MJ, Ogino S, Ma J, et al. A prospective study of plasma adiponectin and pancreatic cancer risk in five US cohorts. J Natl Cancer Inst 2013;105:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inamura K, Song M, Jung S, Nishihara R, Yamauchi M, Lochhead P, et al. Prediagnosis Plasma Adiponectin in Relation to Colorectal Cancer Risk According to KRAS Mutation Status. J Natl Cancer Inst 2016;108:djv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carlberg C, Muñoz A. An update on vitamin D signaling and cancer. Semin Cancer Biol 2022;79:217–30. [DOI] [PubMed] [Google Scholar]
- 74.Pandolfi F, Franza L, Mandolini C, Conti P. Immune Modulation by Vitamin D: Special Emphasis on Its Role in Prevention and Treatment of Cancer. Clin Ther 2017;39:884–93. [DOI] [PubMed] [Google Scholar]
- 75.Dou R, Ng K, Giovannucci EL, Manson JE, Qian ZR, Ogino S. Vitamin D and colorectal cancer: molecular, epidemiological and clinical evidence. Br J Nutr 2016;115:1643–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song M, Nishihara R, Wang M, Chan AT, Qian ZR, Inamura K, et al. Plasma 25-hydroxyvitamin D and colorectal cancer risk according to tumour immunity status. Gut 2016;65:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wellenstein MD, Coffelt SB, Duits DEM, van Miltenburg MH, Slagter M, de Rink I, et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 2019;572:538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell 2021;39:708–24.e11. [DOI] [PubMed] [Google Scholar]
- 79.Grando SA. Connections of nicotine to cancer. Nat Rev Cancer 2014;14:419–29. [DOI] [PubMed] [Google Scholar]
- 80.Hamada T, Nowak JA, Masugi Y, Drew DA, Song M, Cao Y, et al. Smoking and Risk of Colorectal Cancer Sub-Classified by Tumor-Infiltrating T Cells. J Natl Cancer Inst 2019;111:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer 2003;3:733–44. [DOI] [PubMed] [Google Scholar]
- 82.Domagala-Kulawik J Effects of cigarette smoke on the lung and systemic immunity. J Physiol Pharmacol 2008;59 Suppl 6:19–34. [PubMed] [Google Scholar]
- 83.Ugai T, Väyrynen JP, Haruki K, Akimoto N, Lau MC, Zhong R, et al. Smoking and Incidence of Colorectal Cancer Subclassified by Tumor-Associated Macrophage Infiltrates. J Natl Cancer Inst 2021:djab142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oruç Z, Kaplan MA. Effect of exercise on colorectal cancer prevention and treatment. World J Gastrointest Oncol 2019;11:348–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koh H, Hamada T, Song M, Liu L, Cao Y, Nowak JA, et al. Physical Activity and Colorectal Cancer Prognosis According to Tumor-Infiltrating T Cells. JNCI Cancer Spectr 2018;2:pky058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020;158:322–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 2016;13:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hullar MA, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer Treat Res 2014;159:377–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nat Immunol 2017;18:843–50. [DOI] [PubMed] [Google Scholar]
- 90.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 2019;25:968–76. [DOI] [PubMed] [Google Scholar]
- 91.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 92.Bolte LA, Vich Vila A, Imhann F, Collij V, Gacesa R, Peters V, et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021;70:1287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 2019;25:667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trefflich I, Dietrich S, Braune A, Abraham K, Weikert C. Short- and Branched-Chain Fatty Acids as Fecal Markers for Microbiota Activity in Vegans and Omnivores. Nutrients 2021;13:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ridlon JM, Wolf PG, Gaskins HR. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016;7:201–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yazici C, Wolf PG, Kim H, Cross TL, Vermillion K, Carroll T, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017;66:1983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Erawijantari PP, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Influence of gastrectomy for gastric cancer treatment on faecal microbiome and metabolome profiles. Gut 2020;69:1404–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eom BW, Lee HJ, Yoo MW, Cho JJ, Kim WH, Yang HK, et al. Synchronous and metachronous cancers in patients with gastric cancer. J Surg Oncol 2008;98:106–10. [DOI] [PubMed] [Google Scholar]
- 99.Kosumi K, Hamada T, Koh H, Borowsky J, Bullman S, Twombly TS, et al. The Amount of Bifidobacterium Genus in Colorectal Carcinoma Tissue in Relation to Tumor Characteristics and Clinical Outcome. Am J Pathol 2018;188:2839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu L, Tabung FK, Zhang X, Nowak JA, Qian ZR, Hamada T, et al. Diets That Promote Colon Inflammation Associate With Risk of Colorectal Carcinomas That Contain Fusobacterium nucleatum. Clin Gastroenterol Hepatol 2018;16:1622–31.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol 2017;3:921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 2020;580:269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018;359:592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013;8:e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kennedy EA, King KY, Baldridge MT. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front Physiol 2018;9:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tomkovich S, Yang Y, Winglee K, Gauthier J, Mühlbauer M, Sun X, et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res 2017;77:2620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest 2019;129:1699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019;178:795–806.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. New insights from uncultivated genomes of the global human gut microbiome. Nature 2019;568:505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mizutani S, Yamada T, Yachida S. Significance of the gut microbiome in multistep colorectal carcinogenesis. Cancer Sci 2020;111:766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019;25:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014;12:661–72. [DOI] [PubMed] [Google Scholar]
- 115.Soga T, Igarashi K, Ito C, Mizobuchi K, Zimmermann HP, Tomita M. Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Anal Chem 2009;81:6165–74. [DOI] [PubMed] [Google Scholar]
- 116.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol 2009;15:3329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013;8:e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015;347:78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ogino S, Nowak JA, Hamada T, Milner DA Jr., Nishihara R. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol 2019;14:83–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol 2017;52:265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mima K, Kosumi K, Baba Y, Hamada T, Baba H, Ogino S. The microbiome, genetics, and gastrointestinal neoplasms: the evolving field of molecular pathological epidemiology to analyze the tumor-immune-microbiome interaction. Hum Genet 2021;140:725–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hamada T, Nowak JA, Milner DA Jr., Song M, Ogino S Integration of microbiology, molecular pathology, and epidemiology: a new paradigm to explore the pathogenesis of microbiome-driven neoplasms. J Pathol 2019;247:615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng WY, Wu CY, Yu J. The role of gut microbiota in cancer treatment: friend or foe? Gut 2020;69:1867–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Inamura K Gut microbiota contributes towards immunomodulation against cancer: New frontiers in precision cancer therapeutics. Semin Cancer Biol 2021;70:11–23. [DOI] [PubMed] [Google Scholar]
- 126.Derosa L, Routy B, Desilets A, Daillère R, Terrisse S, Kroemer G, et al. Microbiota-Centered Interventions: The Next Breakthrough in Immuno-Oncology? Cancer Discov 2021:2396–412. [DOI] [PubMed] [Google Scholar]
- 127.Sieow BF, Wun KS, Yong WP, Hwang IY, Chang MW. Tweak to Treat: Reprograming Bacteria for Cancer Treatment. Trends Cancer 2021;7:447–64. [DOI] [PubMed] [Google Scholar]
- 128.Andrews MC, Duong CPM, Gopalakrishnan V, Iebba V, Chen WS, Derosa L, et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat Med 2021;27:1432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sullivan RJ, Weber JS. Immune-related toxicities of checkpoint inhibitors: mechanisms and mitigation strategies. Nat Rev Drug Discov 2021:doi: 10.1038/s41573-021-00259-5. [DOI] [PubMed] [Google Scholar]
- 130.Weir TL, Trikha SRJ, Thompson HJ. Diet and cancer risk reduction: The role of diet-microbiota interactions and microbial metabolites. Semin Cancer Biol 2021;70:53–60. [DOI] [PubMed] [Google Scholar]
- 131.Ocvirk S, Wilson AS, Appolonia CN, Thomas TK, O’Keefe SJD. Fiber, Fat, and Colorectal Cancer: New Insight into Modifiable Dietary Risk Factors. Curr Gastroenterol Rep 2019;21:62. [DOI] [PubMed] [Google Scholar]
- 132.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016;167:1339–53.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, et al. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 2018;23:27–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol 2019;16:35–56. [DOI] [PubMed] [Google Scholar]
- 136.Beam A, Clinger E, Hao L. Effect of Diet and Dietary Components on the Composition of the Gut Microbiota. Nutrients 2021;13:2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cunningham M, Azcarate-Peril MA, Barnard A, Benoit V, Grimaldi R, Guyonnet D, et al. Shaping the Future of Probiotics and Prebiotics. Trends Microbiol 2021;29:667–85. [DOI] [PubMed] [Google Scholar]
- 138.Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol 2017;4:e000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cao X, Boyaci H, Chen J, Bao Y, Landick R, Campbell EA. Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile. Nature 2022;604:541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kiga K, Tan XE, Ibarra-Chávez R, Watanabe S, Aiba Y, Sato’o Y, et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun 2020;11:2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 2011;53:994–1002. [DOI] [PubMed] [Google Scholar]
- 142.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013;368:407–15. [DOI] [PubMed] [Google Scholar]
- 143.Juul FE, Garborg K, Bretthauer M, Skudal H, Oines MN, Wiig H, et al. Fecal Microbiota Transplantation for Primary Clostridium difficile Infection. N Engl J Med 2018;378:2535–6. [DOI] [PubMed] [Google Scholar]
- 144.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015;149:102–9.e6. [DOI] [PubMed] [Google Scholar]
- 145.Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, et al. Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial. JAMA 2019;321:156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bullman S, Eggermont A, Johnston CD, Zitvogel L. Harnessing the microbiome to restore immunotherapy response. Nature Cancer 2021;2:1301–4. [DOI] [PubMed] [Google Scholar]
- 147.McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. The Lancet Oncology 2019;20:e77–e91. [DOI] [PubMed] [Google Scholar]
- 148.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- 152.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021;371:602–9. [DOI] [PubMed] [Google Scholar]
- 154.Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021;371:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, et al. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N Engl J Med 2019;381:2043–50. [DOI] [PubMed] [Google Scholar]
- 156.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017;153:1621–33.e6. [DOI] [PubMed] [Google Scholar]
- 157.Ratner M Microbial cocktails raise bar for C. diff. treatments. Nat Biotechnol 2020;38:1366–7. [DOI] [PubMed] [Google Scholar]
- 158.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019;565:600–5. [DOI] [PubMed] [Google Scholar]
- 159.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019;570:462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 2019;363:eaat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pryor R, Martinez-Martinez D, Quintaneiro L, Cabreiro F. The Role of the Microbiome in Drug Response. Annu Rev Pharmacol Toxicol 2020;60:417–35. [DOI] [PubMed] [Google Scholar]
- 162.Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut 2020;69:1510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Inamura K Roles of microbiota in response to cancer immunotherapy. Semin Cancer Biol 2020;65:164–75. [DOI] [PubMed] [Google Scholar]
- 164.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017;170:548–63.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu Y, Baba Y, Ishimoto T, Tsutsuki H, Zhang T, Nomoto D, et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer 2021;124:963–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357:1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh AS, Keefe DM. Faecal microflora and beta-glucuronidase expression are altered in an irinotecan-induced diarrhea model in rats. Cancer Biol Ther 2008;7:1919–25. [DOI] [PubMed] [Google Scholar]
- 170.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010;330:831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018;359:1366–70. [DOI] [PubMed] [Google Scholar]
- 172.Matson V, Chervin CS, Gajewski TF. Cancer and the Microbiome-Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology 2021;160:600–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhou CB, Zhou YL, Fang JY. Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer 2021;7:647–60. [DOI] [PubMed] [Google Scholar]
- 174.Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol 2018;29:1437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med 2020;217:e20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 2021;598:662–6. [DOI] [PubMed] [Google Scholar]
- 177.Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021;374:1632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Postow MA, Sidlow R, Hellmann MD. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med 2018;378:158–68. [DOI] [PubMed] [Google Scholar]
- 179.Chang AE, Golob JL, Schmidt TM, Peltier DC, Lao CD, Tewari M. Targeting the Gut Microbiome to Mitigate Immunotherapy-Induced Colitis in Cancer. Trends Cancer 2021;7:583–93. [DOI] [PubMed] [Google Scholar]
- 180.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med 2018;24:1804–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak JA, Yang J, et al. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol 2017;35:1836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hamada T, Liu L, Nowak JA, Mima K, Cao Y, Ng K, et al. Vitamin D status after colorectal cancer diagnosis and patient survival according to immune response to tumour. Eur J Cancer 2018;103:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fujiyoshi K, Chen Y, Haruki K, Ugai T, Kishikawa J, Hamada T, et al. Smoking Status at Diagnosis and Colorectal Cancer Prognosis According to Tumor Lymphocytic Reaction. JNCI Cancer Spectr 2020;4:pkaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ugai T, Haruki K, Väyrynen JP, Borowsky J, Fujiyoshi K, Lau MC, et al. Coffee Intake of Colorectal Cancer Patients and Prognosis According to Histopathologic Lymphocytic Reaction and T-Cell Infiltrates. Mayo Clin Proc 2022;97:124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ford AC, Yuan Y, Moayyedi P. Helicobacter pylori eradication therapy to prevent gastric cancer: systematic review and meta-analysis. Gut 2020;69:2113–21. [DOI] [PubMed] [Google Scholar]
- 187.Liou JM, Malfertheiner P, Lee YC, Sheu BS, Sugano K, Cheng HC, et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut 2020;69:2093–112. [DOI] [PubMed] [Google Scholar]
- 188.Serrano C, Harris PR, Smith PD, Bimczok D. Interactions between H. pylori and the Gastric Microbiome: Impact on Gastric Homeostasis and Disease. Curr Opin Physiol 2021;21:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, et al. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut 2020;69:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Brennan CA, Nakatsu G, Gallini Comeau CA, Drew DA, Glickman JN, Schoen RE, et al. Aspirin Modulation of the Colorectal Cancer-Associated Microbe Fusobacterium nucleatum. mBio 2021;12:e00547–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nguyen LH, Ma W, Wang DD, Cao Y, Mallick H, Gerbaba TK, et al. Association Between Sulfur-Metabolizing Bacterial Communities in Stool and Risk of Distal Colorectal Cancer in Men. Gastroenterology 2020;158:1313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nguyen LH, Cao Y, Hur J, Mehta RS, Sikavi DR, Wang Y, et al. The Sulfur Microbial Diet Is Associated With Increased Risk of Early-Onset Colorectal Cancer Precursors. Gastroenterology 2021;161:1423–32.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 195.Makki K, Deehan EC, Walter J, Backhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018;23:705–15. [DOI] [PubMed] [Google Scholar]
- 196.Rafter J, Bennett M, Caderni G, Clune Y, Hughes R, Karlsson PC, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr 2007;85:488–96. [DOI] [PubMed] [Google Scholar]
- 197.Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, Takeyama I, et al. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int J Cancer 2005;116:762–7. [DOI] [PubMed] [Google Scholar]
- 198.Wilkinson JE, Franzosa EA, Everett C, Li C, Hu FB, Wirth DF, et al. A framework for microbiome science in public health. Nat Med 2021;27:766–74. [DOI] [PubMed] [Google Scholar]
- 199.Morales E, Chen J, Greathouse KL. Compositional Analysis of the Human Microbiome in Cancer Research. Methods Mol Biol 2019;1928:299–335. [DOI] [PubMed] [Google Scholar]
- 200.Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, et al. Microbial Community Heterogeneity Within Colorectal Neoplasia and its Correlation With Colorectal Carcinogenesis. Gastroenterology 2021;160:2395–408. [DOI] [PubMed] [Google Scholar]
- 201.Zhang W, Svensson Akusjärvi S, Sönnerborg A, Neogi U. Characterization of Inducible Transcription and Translation-Competent HIV-1 Using the RNAscope ISH Technology at a Single-Cell Resolution. Front Microbiol 2018;9:2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol 2020;38:586–99. [DOI] [PubMed] [Google Scholar]
- 203.McAllister K, Mechanic LE, Amos C, Aschard H, Blair IA, Chatterjee N, et al. Current Challenges and New Opportunities for Gene-Environment Interaction Studies of Complex Diseases. Am J Epidemiol 2017;186:753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu L, Nevo D, Nishihara R, Cao Y, Song M, Twombly TS, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol 2018;33:381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Campbell PT, Ambrosone CB, Nishihara R, Aerts H, Bondy M, Chatterjee N, et al. Proceedings of the fourth international molecular pathological epidemiology (MPE) meeting. Cancer Causes Control 2019;30:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gupta S, Harper A, Ruan Y, Barr R, Frazier AL, Ferlay J, et al. International Trends in the Incidence of Cancer Among Adolescents and Young Adults. J Natl Cancer Inst 2020;112:1105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Akimoto N, Ugai T, Zhong R, Hamada T, Fujiyoshi K, Giannakis M, et al. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol 2021;18:230–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zamani S, Taslimi R, Sarabi A, Jasemi S, Sechi LA, Feizabadi MM. Enterotoxigenic Bacteroides fragilis: A Possible Etiological Candidate for Bacterially-Induced Colorectal Precancerous and Cancerous Lesions. Front Cell Infect Microbiol 2019;9:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Meng W, Bai B, Sheng L, Li Y, Yue P, Li X, et al. Role of Helicobacter pylori in gastric cancer: advances and controversies. Discov Med 2015;20:285–93. [PubMed] [Google Scholar]
- 210.Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019;68:1014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yuan L, Zhang S, Li H, Yang F, Mushtaq N, Ullah S, et al. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed Pharmacother 2018;108:184–93. [DOI] [PubMed] [Google Scholar]
- 212.Iida N, Mizukoshi E, Yamashita T, Terashima T, Arai K, Seishima J, et al. Overuse of antianaerobic drug is associated with poor postchemotherapy prognosis of patients with hepatocellular carcinoma. Int J Cancer 2019;145:2701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lione L, Salvatori E, Petrazzuolo A, Massacci A, Maggio R, Confroti A, et al. Antitumor efficacy of a neoantigen cancer vaccine delivered by electroporation is influenced by microbiota composition. Oncoimmunology 2021;10:1898832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lee B, Lee J, Woo MY, Lee MJ, Shin HJ, Kim K, et al. Modulation of the Gut Microbiota Alters the Tumour-Suppressive Efficacy of Tim-3 Pathway Blockade in a Bacterial Species- and Host Factor-Dependent Manner. Microorganisms 2020;8:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pernigoni N, Zagato E, Calcinotto A, Troiani M, Mestre RP, Calì B, et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 2021;374:216–24. [DOI] [PubMed] [Google Scholar]
- 216.Arima K, Zhong R, Ugai T, Zhao M, Haruki K, Akimoto N, et al. Western-style diet, pks island-carrying Escherichia coli, and colorectal cancer: analyses from two large prospective cohort studies. Gastroenterology (online ahead of print. doi: 10.1053/j.gastro.2022.06.054). [DOI] [PMC free article] [PubMed] [Google Scholar]


