Abstract
One prerequisite for the virulence of Yersinia pestis, causative agent of bubonic plague, is the yersiniabactin (Ybt) siderophore-dependent iron transport system that is encoded within a high-pathogenicity island (HPI) within the pgm locus of the Y. pestis chromosome. Several gene products within the HPI have demonstrated functions in the synthesis or transport of Ybt. Here we examine the roles of ybtU and ybtT. In-frame mutations in ybtT or ybtU yielded strains defective in siderophore production. Mutant strains were unable to grow on iron-deficient media at 37°C but could be cross-fed by culture supernatants from a Ybt-producing strain of Y. pestis. The ybtU mutant failed to express four indicator Ybt proteins (HMWP1, HMWP2, YbtE, and Psn), a pattern similar to those for other ybt biosynthetic mutants. In contrast, strains carrying mutations in ybtT or ybtS (a previously identified gene required for Ybt biosynthesis) produced all four proteins at wild-type levels under iron-deprived conditions. To assess the effects of ybtT, -U, and -S mutations on transcription of ybt genes, reporter plasmids with ybtP or psn promoters controlling lacZ expression were introduced into these mutants. Normal iron-regulated β-galactosidase activity was observed in the ybtT and ybtS mutants, whereas a significant loss of expression occurred in the ΔybtU strain. These results show that ybtT and ybtU genes are involved in the biosynthesis of the Ybt siderophore and that a ybtU mutation but not ybtT or ybtS mutations affects transcription from the ybtP and psn promoters.
To cause infections, pathogenic bacteria must be able to remove iron, an essential trace nutrient, from host iron- and/or heme-chelating proteins (12, 43, 65). Yersinia pestis, the causative agent of bubonic and pneumonic plague, possesses an ATP-binding cassette (ABC) hemoprotein transport system (Hmu) that allows it to use a variety of host hemoproteins (36, 64). The organism also contains one putative and two known inorganic iron transport systems. The Yfu system was identified by a search of the Y. pestis KIM10+ genome database (www.genome.wisc.edu) and belongs to a family of ABC iron transporters present in Yersinia enterocolitica (Yfu), Neisseria spp. (Fbp), Haemophilus influenzae (Hit), Actinobacillus pleuropneumoniae (Afu), and Serratia marcescens (Sfu) (7). Whether this system is functional in Y. pestis is currently unknown. The Y. pestis Yfe system belongs to a family of cation-transporting ABC systems and transports both iron and manganese. This system appears to function to acquire iron during the later stages of plague (3, 4).
The third inorganic iron transport system synthesizes the siderophore yersiniabactin (Ybt), which is composed of a phenolate, a thiazoline, and a thiazolidine ring (15, 19, 47) and which has considerable similarity to the siderophores pyochelin and anguibactin produced by Pseudomonas aeruginosa and Vibrio anguillarum, respectively (18, 38). The Ybt iron acquisition system is essential for the virulence of Y. pestis during the early stages of infection in mice (3). Ybt biosynthetic, regulatory, and transport genes are encoded within a high-pathogenicity island (HPI) that is present in highly pathogenic isolates of Y. pestis, Yersinia pseudotuberculosis, and Y. enterocolitica, as well as several types of pathogenic Escherichia coli (8, 9, 13, 29, 33, 50, 56). In Y. pestis, HPI resides within the pgm locus, a 102-kb region of chromosomal DNA subject to high-frequency deletion (9, 10, 26, 29, 32, 41).
The genes encoding the Ybt systems of Y. pestis (see Fig. 1) and Y. enterocolitica have been completely sequenced and show >97% sequence identity (10, 29, 46, 50). One notable exception to this sequence identity is the unique insertion of a 125-bp ERIC sequence (enterobacterial repetitive intergenic consensus sequence; also called an intergenic repeated unit) within the promoter region of ybtA of Y. enterocolitica (50). ybtA encodes a transcriptional activator of ybt genes (22). Although this suggests possible differences in regulatory responses, some of the Ybt biosynthetic genes appear to be functionally interchangeable among the three pathogenic yersiniae: Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica (14, 47).
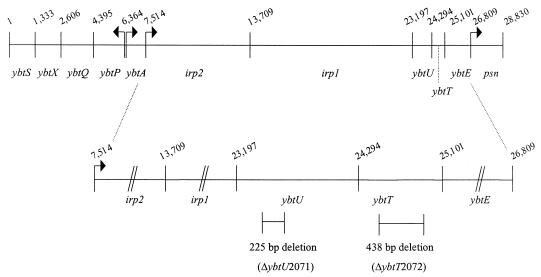
FIG. 1.
Genetic organization of the Y. pestis Ybt operons. Arrows, putative promoters for the operons. Gene designations are given below the lines, while nucleotide numbers are shown above the lines. Numbering corresponds to that used for GenBank accession no. AF091251 (29). An expanded version of the irp1-irp2-ybtU-ybtT-ybtE operon is shown; irp1, irp2, and ybtE genes are not drawn to scale in the expanded version. The size and location of the ybtU and ybtT deletions are indicated on the expanded map.
Iron from Fe-Ybt is transported into the cell via an outer membrane (OM) receptor (termed Psn in Y. pestis and FyuA in Y. enterocolitica) in conjunction with an ABC transport system encoded by ybtP and ybtQ. Psn, which likely binds the Fe-Ybt complex, also binds the bacteriocin pesticin (23–25, 34, 40, 51). Translocation of the substrate across the OM is TonB dependent (21, 34). YbtP and YbtQ resemble inner-membrane permeases that are each fused to an ATP-binding domain. Both proteins are required for use of iron from Ybt. No periplasmic binding protein has been identified for the Ybt system (23, 29).
Ybt production occurs via a mixed polyketide synthase-nonribosomal peptide synthetase (NRPS) strategy that assembles the siderophore in modular fashion from salicylate, a linker group derived from malonyl-coenzyme A, three molecules of cysteine, and three methyl groups donated by S-adenosylmethionine (29). The requirement of three gene products (high-molecular-weight protein 2 [HMWP2], YbtE, and YbtS) for Ybt synthesis has been clearly demonstrated genetically. YbtS is likely required for the final steps in salicylate biosynthesis (2, 29). YbtE adenylates salicylate and transfers this activated compound to HMWP2 (30). HMWP2, encoded by irp2, possesses domains involved in nonribosomal peptide synthesis and likely participates in the initial cyclization and condensation reactions involving salicylate and two cysteine molecules (29–31, 62). HMWP1, encoded by irp1, contains polyketide/fatty acid synthase and modified NRPS domains that add the branched isobutyryl-alcohol linker and the last thiazoline moiety. Phosphopantetheinylation of a peptidyl carrier protein domain of HMWP1 (PCP3) has been demonstrated (29). The roles of the remaining ybt genes (ybtX, ybtT, and ybtU) contained within the HPI were undetermined or uncertain. The product encoded by ybtX is predicted to be extremely hydrophobic. However, a strain carrying a deletion in ybtX had no discernible in vitro phenotype (23). While YbtT contains a thioesterase domain (2, 29), a definitive role for YbtT in Ybt biosynthesis has not been demonstrated. YbtU has no significant homology to proteins in the database that have defined enzymatic or regulatory functions (29), nor has a function for YbtU in Ybt production or utilization been determined. In this study, we report that in-frame deletions in ybtU and ybtT abolish Ybt siderophore synthesis. Similar to mutations in other Ybt biosynthetic genes, the ybtU mutation downregulates expression of ybt genes; however, mutations in ybtT and ybtS did not have this regulatory effect.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All relevant characteristics of strains used in this study are presented in Table 1. All the Y. pestis strains used in this study were derived from KIM6+, an avirulent strain that possesses all of the known Y. pestis virulence determinants except for pCD1, a 70.5-kb plasmid encoding the low-calcium response (Lcr) stimulon (26, 59). The Lcr virulence regulon is unrelated to the Pgm+ phenotype and has no demonstrable role in iron metabolism (48, 49).
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Cloning host | 1 |
| DH5α (λpir) | Strain for propagating plasmids with R6K origins, derived from DH5α | S. C. Straley |
| Y. pestis | ||
| KIM6+ | Pgm+ (Hms+ Ybt+) Lcr− | 24 |
| KIM6-2046.1 | Hms+ Ybt− (irp2::kan2046.1) Lcr− Kmr | 47 |
| KIM6-2046.3 | Hms+ Ybt− (Δirp2-2046.3) Lcr− | 2 |
| KIM6-2070.1 | Hms+ Ybt− (ybtS::kan2070.1) Lcr− Kmr | 29 |
| KIM6-2071 | Hms+ Ybt− (ΔybtU2071) Lcr− | This study |
| KIM6-2072 | Hms+ Ybt− (ΔybtT2072) Lcr− | This study |
| Plasmids | ||
| pACYC184 | 4.2-kb cloning vector; Cmr Tcr | 16 |
| pBluescript II KS+ | 2.9-kb cloning vector; Apr | Stratagene |
| pEUPP1 | 15.4-kb low-copy-number psn::lacZ reporter plasmid; Spcr; iron-, Fur-, and YbtA-regulated expression of β-galactosidase | 22 |
| pEUYbtP | 15.4-kb low-copy-number ybtP::lacZ reporter plasmid; Spcr; iron-, Fur-, and YbtA-regulated expression of β-galactosidase | 23 |
| pKNG101 | 6.8-kb suicide vector; sacB+; R6K origin; Smr | 39 |
| pPROEX-1 | 4.7-kb protein expression vector; Apr | Gibco/BRL |
| pPSN3 | 9.9-kb SalI fragment from pSDR498.4 ligated into pBGL2; Apr | 25 |
| pPSN345 | 3.45-kb SphI/EcoRI fragment from pPSN3 ligated into pUC18; ybtU+ ybtT+; Apr | This study |
| pSDR498.1 | ∼4.6-kb BamHI/EcoRI fragment from pSDR498 ligated into pHC79; ybtS+; Apr | 29, 35 |
| pSUC1 | 4.7-kb suicide vector; sacB+; R6K origin; Apr | 23 |
| pUC18 | 2.7-kb cloning vector; Apr | 68 |
| pYbtS-H6 | 1,305-bp PCR fragment from pSDR498.1 ligated into pPROEX-1; IPTG-regulated expression of YbtS-H6; Apr | This study |
| pYbtT-H6 | 800-bp PCR fragment from pPSN345 ligated into pPROEX-1; IPTG-regulated expression of YbtT-H6; Apr | This study |
| pYbtU-H6 | 1,100-bp PCR fragment from pPSN345 ligated into pPROEX-1; IPTG-regulated expression of YbtU-H6; Apr | This study |
| pYbtTU1 | ∼3.9-kb SphI fragment containing ybtT and ybtU from pPSN3 ligated into pACYC184; Cmr | This study |
| pYbtT1 | pYbtTU1 with 438-bp PvuI fragment deleted; Cmr | This study |
| pYbtT1.1 | 3.5-kb SphI fragment from pYbtT1 ligated into pSUC1; ΔybtT2072 (deletion of 808 bp), sacB+; R6K origin; Apr | This study |
| pYbtU1 | 526-bp EagI/EcoRV fragment from pYbtTU1 ligated into pBluescript II KS+; Apr | This study |
| pYbtU2 | 467-bp PvuII fragment from pYbtTU1 ligated into the EcoRV site of pYbtU1; Cmr | This study |
| pYbtU2.1 | ∼1.0-kb SacI/SalI fragment from pYbtU2 ligated into pSUC1; ΔybtU2071 (deletion of 225 bp), sacB+; R6K origin; Apr | This study |
| pYbtU2.2 | ∼1.0-kb SalI/SmaI fragment from pYbtU2.1 ligated into pKNG101; ΔybtU (deletion of 225 bp); sacB+; R6K origin; Smr | This study |
Y. pestis strains with a plus sign possess an intact 102-kb pgm locus containing the genes for hemin storage (hms) and the Ybt system. All other Y. pestis strains contain a mutation within the pgm locus due to either a deletion or insertion of an antibiotic resistance cassette. Strains synthesizing the siderophore yersiniabactin are designated Ybt+, while those affected in yersiniabactin production are Ybt−. Lcr− indicates the absence of the low-calcium response virulence plasmid pCD1. Apr, Kmr, Spcr, Smr, Tcr, and Cmr, resistance to ampicillin, kanamycin, spectinomycin, streptomycin, tetracycline, and chloramphenicol, respectively.
All strains were stored at −20°C in phosphate-buffered glycerol. Y. pestis cells were grown routinely at 30°C on Congo red agar (63) from glycerol stocks and then grown in heart infusion broth (Difco Laboratories) or on tryptose-blood agar base (Difco). For iron-deficient growth, Y. pestis cells were grown in the chemically defined medium PMH, which had been extracted prior to use with Chelex 100 resin (Bio-Rad Laboratories) (61). The residual iron that was not removed from deferrated PMH by the resin was precipitated by the addition of 0.5 mM NaCO3–0.01 mM MnCl2–4.0 mM CaCl2 (PMH-S) or chelated by supplementation with 2,2′-dipyridyl (PMH-DIP) at a concentration of 100 μM. PMH-S and PMH-DIP plates were solidified with 1% agarose. PMH-S and PMH-DIP plates were subsequently used in cross-feeding experiments or to determine the growth characteristics of the ybt mutants at 37°C as previously described (24). For iron-replete growth, Y. pestis strains were cultivated in PMH supplemented with 10 μM FeCl3.
All glassware used for iron-restricted studies was soaked overnight in chromic/sulfuric acid (46.3 g of K2Cr2O7 per liter of 12 M sulfuric acid) or ScotClean (OWL Scientific, Inc.) to remove contaminating iron and copiously rinsed in deionized water. E. coli cells were grown in Luria broth. Where appropriate, ampicillin (100 μg/ml), spectinomycin (100 μg/ml), tetracycline (6.25 μg/ml), streptomycin (50 μg/ml), and kanamycin (50 μg/ml) were added to cultures.
Plasmids, sequencing, and recombinant DNA techniques.
All the plasmids used in this study are listed in Table 1. Plasmids were purified from overnight cultures by alkaline lysis (6) and further purified when necessary by polyethylene glycol precipitation (37). Standard cloning and recombinant DNA methods (53) were used to construct the various plasmids in Table 1. A standard CaCl2 procedure was used to introduce plasmids into E. coli (53). Y. pestis cells were transformed by electroporation as previously described (24). Plasmid DNA was sequenced by the dideoxynucleotide chain termination method (54) using Sequenase, version 2.0 (Amersham Pharmacia Biotech), 35S-dATP (New England Nuclear/Dupont), and 7-deaza-dGTP. Synthetic oligonucleotide primers purchased from Integrated DNA Technologies were used to extend the sequence. Samples were electrophoresed at 70 W on 6% polyacrylamide gels containing Tris-borate-EDTA buffer and 8.3 M urea. Gels were fixed in a 10% ethanol–10% acetic acid solution, dried, and exposed to Kodak BioMax MR film at room temperature.
Generating ybtU and ybtT mutant strains and expression plasmids.
All Y. pestis mutant strains were generated by homologous recombination using mutated DNA fragments cloned into suicide vectors carrying the sacB gene and an R6K origin of replication. For construction of an in-frame ΔybtT strain (KIM6-2072), we first subcloned an ∼3.9-kb SphI fragment of plasmid pPSN3 into the SphI site of pACYC184 to yield pYbtTU1. Removal of a 438-bp PvuI fragment from pYbtTU1 resulted in plasmid pYbtT1. Subcloning an ∼3.5-kb SphI fragment from pYbtT1 into the SphI site of the suicide plasmid pSUC1 generated pYbtT1.1.
To generate the in-frame ΔybtU mutant (strain KIM6-2071), a 526-bp EagI/EcoRV fragment of plasmid pYbtTU1 was subcloned into the corresponding sites in pBluescript II KS+ (Stratagene) to yield pYbtU1. A 467-bp PvuII fragment was removed from pYbtTU1 and ligated into the EcoRV site of the plasmid pYbtU1, generating pYbtU2. The orientation of the insert was determined by DNA sequencing. We then subcloned an ∼1-kb SacI/SalI fragment of pYbtU2 into the SacI/SalI sites in pSUC1 to yield pYbtU2.1. Gene replacement attempts with pYbtU2.1 did not yield the desired chromosomal integrants. Therefore, we constructed an alternate suicide plasmid, pYbtU2.2, by subcloning a SalI/SmaI fragment of pYbtU2.1 into the SalI/SmaI sites in pKNG101.
To generate hexahistidine fusion proteins, the ybtT and ybtU gene coding regions were amplified with Pfu polymerase (Stratagene) from pPSN345 by PCR using primers ybtT-1 (5′-TGATGGCGCCTCTGTGACGCAATCTGCAATG-3′) and M13 reverse (5′-AGCGGATAACAATTTCA-3′) and ybtU-1 (GGAATTCTTATGATGCCGTCCGCCTCC) and ybtU-2 (CGGGATCCTCACAGCGCCTCCTTATC), respectively. Reaction mixtures contained 0.2 mM deoxynucleoside triphosphates and 0.2 μM primers, and reactions consisted of 20 s at 94°C, 20 s at 50°C, and 90 s at 72°C for 30 cycles, followed by a single cycle at 72°C for 10 min for ybtT. Amplification conditions for ybtU were 45 s at 94°C, 30 s at 50°C, and 2 min at 72°C for 30 cycles followed by a single cycle at 72°C for 10 min. The ybtS coding region was amplified with Pfu polymerase (Stratagene) from pSDR498.1 using primers ybtS-1 (5′-GGAATTCTTATGAAAATCAGTGAATTT-3′) and ybtS-2 (5′-CGGGATCCCTACACCATTAAATAGGG-3′). Amplification conditions for ybtS consisted of 45 s at 94°C, 30 s at 45°C, and 2 min at 72°C for 30 cycles followed by a single cycle at 72°C for 10 min. The 827-, 1,305-, and 1,100-bp products of ybtT, ybtS, and ybtU, respectively, were purified from low-melting-point agarose and digested with KasI/EcoRI (for ybtT) or with BamHI/EcoRI (for ybtS and ybtU), followed by ligation into the corresponding sites of pPROEX1, yielding pYbtT-H6, pYbtS-H6, and pYbtU-H6. Ligated products were transformed into DH5α cells. Positive clones containing the desired inserts were identified by restriction enzyme digests and verified by PCR using a set of nested primers specific for the ybtS, ybtT, and ybtU coding regions. Expression of all three gene products was verified in the positive clones by IPTG (isopropyl-β-d-thiogalactopyranoside) induction of Luria broth minicultures followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (data not shown). pYbtT-H6, pYbtS-H6, and pYbtU-H6 were electroporated into ΔybtT, ΔybtS, and ΔybtU mutants, respectively. Growth stimulation in these three strains was tested by bioassay on PMH-DIP as described above.
Protein analyses.
To label cellular proteins, whole cells of Y. pestis strains acclimated to growth under iron-deficient or iron-sufficient conditions by serial passage in PMH, with or without FeCl3 (10 μM), for a total of approximately six generations, were labeled with 35S-amino acids (DuPont NEN Research Products) for 1 h as previously described (25). To analyze the effect of Ybt on protein synthesis, purified Ybt from Y. pestis KIM6+ was added to cells, acclimated to iron starvation, at the same time as 35S-amino acids. An equivalent number of counts was electrophoresed on 7.5% polyacrylamide gels containing SDS. Dried gels were exposed to Kodak BioMax MR film at room temperature.
Ybt bioassay.
Culture supernatants were obtained from ybt mutants inoculated into deferrated PMH and grown for a total of six to nine generations at 37°C as previously described (24). The cells were pelleted by centrifugation, and the supernatant was filtered through a 0.45-μm-pore-size filter. For growth responses, PMH-S or PMH-DIP plates were overlayered with 0.04 optical density (at 620 nm) units of KIM6-2046.1 (irp2::kan2046.1) cells grown in deferrated PMH and 25 μl of filtered supernatants from iron-deficient ybt mutant cultures were added to wells in the plates.
The ybtU and ybtT mutants were also tested for their ability to promote the growth of KIM6-2046.1 at 37°C by streaking the mutants adjacent to KIM6-2046.1 on PMH-S or PMH-DIP plates. Prior to streaking, the mutants were adapted to iron-deficient growth conditions as described above. Y. pestis strains that do not produce Ybt are unable to grow on PMH-S or PMH-DIP at 37°C but can be cross-fed by Ybt-producing strains.
β-Galactosidase assays.
Lysates were prepared from cells carrying the ybtP::lacZ or psn::lacZ reporter plasmid and grown in PMH in the presence or absence of iron through two transfers for a total of approximately six generations, as previously described (60). β-Galactosidase activities were measured spectrophotometrically following cleavage of ONPG (4-nitro-phenyl-β-d-galactopyranoside). Activity is expressed in Miller units (44).
Ybt detection and purification.
Y. pestis KIM6+, KIM6-2071, and KIM6-2072 were grown at 37°C in 100 ml of deferrated PMH for approximately eight generations. The supernatant of each culture was filtered through a 0.45-μm-pore-size filter. The presence of the Ybt siderophore was determined using methods modified from Chambers et al. (15) and Drechsel et al. (19) as previously described (47). Three C18-SEP-PAK cartridges were used in a preliminary purification step. The sample, in 50% methanol, was then applied to an analytical C-18 high-pressure liquid chromatography (HPLC) column and eluted with 100% methanol. The Ybt siderophore was detected by its absorbance maximum at 385 nm (19) and by bioassay.
RESULTS
Mutation of ybtU or ybtT causes loss of siderophore production.
Most biosynthetic enzymes for the synthesis of Ybt are encoded within a large, putative operon of five genes: irp2, irp1, ybtU, ybtT, and ybtE (Fig. 1). The likely functions of HMWP1, HMWP2, and YbtE in the synthesis of Ybt have been described previously (29, 30, 62). To study the functions of YbtT and YbtU in Ybt production or utilization, we constructed in-frame deletions in both genes (Fig. 1). We have been unable to identify the YbtU and YbtT products by SDS-PAGE. However, when grown under appropriate conditions, both the YbtT− and YbtU− mutants express YbtE (Fig. 2), the product of the final, downstream gene of the operon, indicating that both mutations are in-frame deletions.
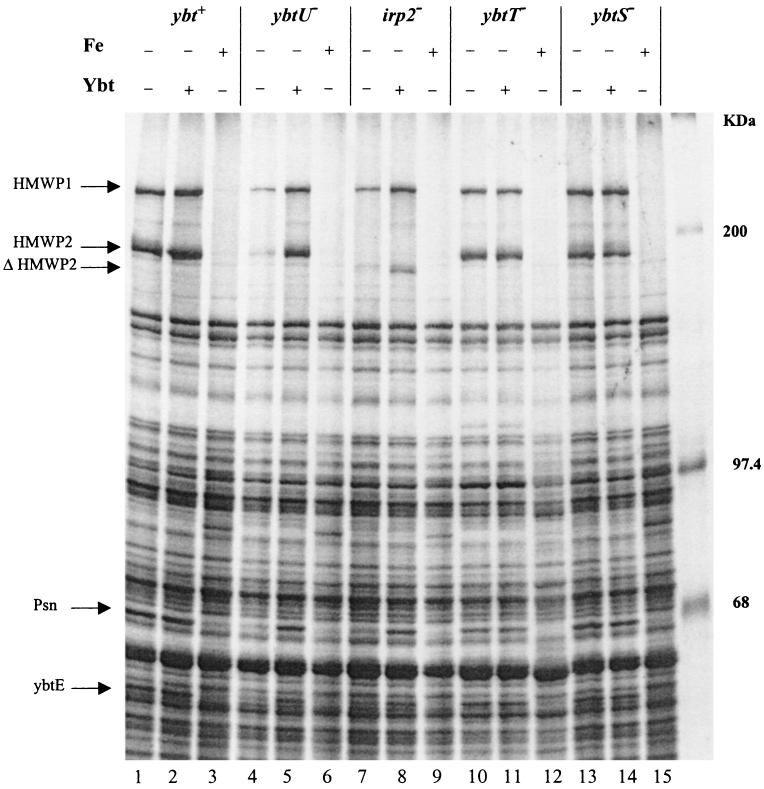
FIG. 2.
SDS-PAGE analysis of whole-cell proteins from Y. pestis strains grown in PMH in the presence (+) or absence (−) of FeCl3 (10 μM) or Ybt. Ybt was added since previous studies showed induction of Ybt protein expression when the siderophore was added to Ybt biosynthetic mutants (2, 22). Cultures from Y. pestis KIM6+ (lanes 1 to 3), KIM6-2071 (ΔybtU2071; lanes 4 to 6), KIM6-2046.3 (Δirp2-2046.3; lanes 7 to 9), KIM6-2072 (ΔybtT2072; lanes 10 to 12), and KIM6-2070.1 (ybtS::kan2070.1; lanes 13 to 15) were incubated with 35S-labeled amino acids for 1 h. Total cellular proteins were separated on a 7.5% polyacrylamide gel and visualized by autoradiography. Arrows, iron-regulated proteins HMWP1 (240 kDa), HMWP2 (190 kDa), Psn (68 kDa), and YbtE (56 kDa) and the truncated irp2 gene product (ΔHMWP2) expressed by the Y. pestis in-frame deletion mutant KIM6-2046.3 (Δirp2-2046.3).
Two different media (PMH-S and PMH-DIP) were used to examine the ability of the mutants to grow at 37°C under iron-chelated conditions. Neither KIM6-2071 (ΔybtU) nor KIM6-2072 (ΔybtT) grew at 37°C on the iron-chelated media (Table 2), indicating that the mutated strains lost the ability to either synthesize or utilize Ybt. Supernatants from iron-deficient cultures of KIM6-2071 and KIM6-2072 were unable to stimulate the growth of a Y. pestis strain (KIM6-2046.1) defective in Ybt synthesis (Table 2). However, culture supernatant from KIM6+, a yersiniabactin-producing strain of Y. pestis, allowed the growth of KIM6-2046.1 as well as the YbtU− and YbtT− mutants under these conditions. Similar results were obtained with KIM6-2070.1 (ΔybtS::kan2070.1) (Table 2), a strain previously demonstrated to be defective in Ybt synthesis (29). These results suggest that the YbtU− and YbtT− mutants are defective in synthesis of the Ybt siderophore. To clearly demonstrate that Ybt was not produced at significant levels, we attempted to purify Ybt from 100 ml of iron-deficient culture supernatant of each mutant, as well as the KIM6+ parental strain. Although a significant amount of Ybt was produced by KIM6+ as determined by HPLC analysis, we were not able to detect any Ybt production by the YbtU−, YbtT−, and YbtS− mutants (data not shown).
TABLE 2.
Growth of Y. pestis KIM6+ and ybt mutants on PMH-S and PMH-DIP
| Strain | Relevant characteristics | Growth on PMH-DIP and PMH-Sa | Growth stimulation on PMH-DIP and PMH-S
|
|
|---|---|---|---|---|
| of KIM6-2046.1b | by KIM6+c | |||
| KIM6+ | ybt+ | + | + | NDd |
| KIM6-2071 | ΔybtU2071 | − | − | + |
| KIM6-2072 | ΔybtT2072 | − | − | + |
| KIM6-2070.1 | ybtS::kan2070.1 | − | − | + |
| KIM6-2071(pYbtU-H6) | ΔybtU2071 ybtU-H6 | + | + | ND |
| KIM6-2072(pYbtT-H6) | ΔybtT2072 ybtT-H6 | + | + | ND |
| KIM6-2070.1(pYbtS-H6) | ΔybtS::kan2070.1 ybtS-H6 | + | + | ND |
| KIM6-2046.1 | irp2::kan2046.1 | − | − | + |
The presence (+) or absence (−) of growth on PMH-S and PMH-DIP plates at 37°C.
Each strain was tested for its ability to promote the growth of KIM6-2046.1 (irp2::kan2046.1) at 37°C on PMH-S and PMH-DIP either by streaking adjacent to KIM6-2046.1 or by spent-culture supernatants.
Each strain was tested for its ability to use the exogenous Ybt siderophore at 37°C by streaking adjacent to KIM6+ on PMH-S and PMH-DIP at 37°C.
ND, not determined.
For complementation analyses, ybtT, ybtU, and ybtS were amplified by PCR and cloned into an IPTG-inducible His tag expression vector generating pYbtT-H6, pYbtU-H6, and pYbtS-H6. The recombinant plasmids restored the ability of the respective mutants to grow on iron-chelated plates at 37°C. In addition, culture supernatant from each of the complemented strains was able to promote the growth of KIM6-2046.1 (irp2::kan2046.1) on PMH-DIP plates (Table 2). Although the genes encoding the recombinant proteins are inducible by IPTG in E. coli, there is no direct evidence for IPTG induction in Y. pestis. SDS-PAGE analysis showed that YbtU-H6 expression was induced by IPTG in Y. pestis but to a much lesser extent than in E. coli (data not shown). However, in the experiments described above, IPTG addition was not necessary for complementation in Y. pestis, indicating that there was sufficient production of the fusion proteins in the absence of IPTG to restore Ybt synthesis. Furthermore, the additional amino acid residues on the His-tagged fusion proteins apparently did not perturb their ability to function in Ybt synthesis.
The ΔybtU2071 mutation results in decreased expression of ybt operons.
Total 35S-labeled proteins synthesized by cells grown under iron-sufficient and iron-deficient conditions were analyzed by SDS-PAGE to determine the effect, if any, of the ΔybtU2071 mutation on the expression of other iron-regulated proteins. Previously, we showed that mutations causing loss of siderophore production lower expression of other Ybt biosynthetic genes (those encoding HMWP1, HMWP2, and YbtE) as well as the Ybt receptor (Psn) (2, 22). The analysis of whole-cell extracts of strains grown in iron-deficient media revealed that the YbtU− mutant had a pattern of protein expression similar to that of a Δirp2-2046.3 mutant that is defective in siderophore biosynthesis. In the absence of iron, the levels of expression of HMWP1, HMWP2, YbtE, and Psn proteins were greatly reduced in KIM6-2071 (ΔybtU2071) and KIM6-2046.3 (Δirp2-2046.3) cells (Fig. 2, lanes 4 and 7) compared to those in the parental strain, KIM6+ (Fig. 2, lane 1). Cells grown in the presence of iron do not express detectable levels of these proteins (Fig. 2, lanes 3, 6, 9, 12, and 15).
We have previously shown that addition of purified Ybt or supernatant containing Ybt to Y. pestis Ybt biosynthetic mutants restores expression of HMWP1, HMWP2, YbtE, and Psn (2, 22, 47). Similarly, when purified Ybt was added to iron-deficient cultures, both the YbtU− and ΔHMWP2 mutants expressed wild-type levels of HMWP1, HMWP2, Psn, and YbtE (Fig. 2, lanes 5 and 8).
To test the effect of the ybtU2071 mutation on gene transcription, we used two well-characterized reporter plasmids with promoters that have been demonstrated to be regulated by Fur, iron, YbtA, and the Ybt siderophore: pEUYbtP, a ybtP promoter fusion to lacZ, and pEUPP1, a psn promoter fusion to lacZ. Both reporter genes were cloned into the low-copy-number plasmid pEU730 (2, 22, 23, 27). Because our Y. pestis strains are phenotypically β-galactosidase negative, any β-galactosidase activity is due to the presence of the reporter plasmid (61). The β-galactosidase activities of cells bearing these plasmids and grown in deferrated PMH, in the presence or absence of added iron, are presented in Table 3. As expected for KIM6+, which contains all the genes needed for Ybt synthesis and utilization, expression of lacZ from the ybtP promoter is iron regulated; there is a 23-fold repression of β-galactosidase activity in cells grown in the presence of surplus iron compared to those cultured under iron-deficient conditions. Expression of the ybtP::lacZ reporter in the YbtU− mutant, KIM6-2071, was still somewhat iron regulated (12-fold repression); however the overall expression was greatly reduced (Table 3). The ybtP promoter was more active than the psn promoter during iron deficiency in the Ybt+ strain: a 23-fold versus a 14-fold induction. However, the effects of the ybtU mutation on the two promoters were similar, reducing expression to ∼2,000 Miller units, a 16-fold reduction for the ybtP promoter and an 11-fold reduction for the psn promoter (Table 3). These studies suggest that loss of expression of HMWP1, HMWP2, YbtE, and Psn in the YbtU− mutant results from decreased transcription from the relevant ybt promoters.
TABLE 3.
β-Galactosidase activities of Y. pestis strains containing either a ybtP::lacZ or a psn::lacZ reporter plasmida
| Strain | β-Galactosidase activityb of cells grown:
|
Ratio
|
|||
|---|---|---|---|---|---|
| −Fe | +Fe | −Fe/+Fe | Wild type/mutant
|
||
| −Fe | +Fe | ||||
| With pEUYbtP (ybtP::lacZ) | |||||
| KIM6+ (ybt+) | 30,356 ± 4,196 | 1,308 ± 162 | 23.2 ± 2.6 | ||
| KIM6-2071 (ΔybtU2071) | 1,926 ± 394 | 165 ± 18 | 11.6 ± 2.3 | 15.8 ± 3.8 | 7.9 ± 0.7 |
| KIM6-2072 (ΔybtT2072) | 29,541 ± 8,474 | 1,324 ± 317 | 22.3 ± 8.4 | 1.0 ± 0.2 | 1.0 ± 0.4 |
| KIM6-2070.1 (ybtS::kan2070.1) | 29,670 ± 2,637 | 1,106 ± 105 | 29.2 ± 3.5 | 1.0 ± 0.2 | 1.2 ± 0.1 |
| With pEUPP1 (psn::lacZ) | |||||
| KIM6+ (ybt+) | 22,199 ± 3,388 | 1,595 ± 400 | 13.9 ± 5.5 | ||
| KIM6-2071 (ΔybtU2071) | 2,042 ± 326 | 138 ± 37 | 14.8 ± 2.0 | 10.9 ± 1.6 | 11.5 ± 6.0 |
| KIM6-2072 (ΔybtT2072) | 23,125 ± 3,351 | 949 ± 311 | 24.4 ± 9.1 | 1.0 ± 0.1 | 1.7 ± 0.5 |
| KIM6-2070.1 (ybtS::kan2070.1) | 22,349 ± 3,370 | 1,386 ± 585 | 16.1 ± 5.5 | 1.0 ± 0.2 | 1.2 ± 0.3 |
| With no reporter | |||||
| KIM6+ (ybt+) | 0 | 0 | |||
Cells were harvested during exponential growth at 37°C after approximately six generations in PMH containing either no added iron (−Fe) or 10 μM FeCl3 (+Fe).
Enzyme activities are expressed in Miller units (44). The values are averages ± standard deviations of three or four individual reactions from two or more independent cultures.
The level of expression from the psn::lacZ reporter in KIM6+ is higher here (Table 3) than in previously reported results (22). Although several factors, including the level of residual iron remaining in different preparations of PMH, could help account for this, we have found that mutations occurred within the promoter region of pEUPP1 after its construction. In other reporter strains, we have identified a single base change in the putative −10 region that reduces its similarity to the consensus and could decrease overall transcriptional efficiency (data not shown). A similar mutation might have caused the lower expression values reported previously (22). The pEUPP1 plasmid used in this study does not contain this mutation.
The ybtT2072 and ybtS2070.1 mutations do not affect expression of ybt operons.
We also performed analyses of iron-repressible protein expression by YbtT− and YbtS− mutants of Y. pestis. Expression of HMWP1, HMWP2, YbtE, and Psn by these two mutants was iron repressible with patterns similar to that of the parental strain, KIM6+ (Fig. 2). In contrast to the YbtU− and ΔHMWP2 mutants, the YbtT− and YbtS− mutants produced all four proteins at wild-type levels under iron-deprived conditions (Fig. 2, lanes 10 and 13) and the addition of purified Ybt during growth of these mutants did not increase expression of these four proteins (Fig. 2, lanes 11 and 14).
We also tested the effects of the ybtT and ybtS mutations on transcription of the ybtP and psn promoter fusions to lacZ. The β-galactosidase activities from strains KIM6-2072 (ΔybtT) and KIM6-2070.1 (ΔybtS) carrying either pEUYbtP or pEUPP1 were essentially the same as that observed with KIM6+ cells carrying these reporter plasmids (Table 3). These results indicate that the ΔybtT2072 and ΔybtS2070.1 mutations do not affect the transcription of the ybtPQXS or psn operons or expression of Ybt proteins. This is in sharp contrast to the previous three Ybt biosynthetic mutants, which all decreased expression of the Ybt system (this study; 2, 22, 23, 47).
DISCUSSION
While there are some differences among the HPIs of the pathogenic yersiniae, all encode an essentially identical and interchangeable siderophore-iron transport system (Ybt). The ybt genes appear to be organized into four operons: (i) psn, (ii) irp2-irp1-ybtU-ybtT-ybtE, (iii) ybtA, and (iv) ybtP-ybtQ-ybtX-ybtS (10, 23, 24, 29, 46, 47) (Fig. 1). In Y. pestis, it has previously been shown that the OM receptor Psn and an ABC transporter composed of YbtP and YbtQ are essential for use of the Ybt siderophore (23, 24). YbtA is an AraC-type activator for the other ybt operons and represses its own transcription (22). The role of YbtX is unclear since a mutation disrupting ybtX had no demonstrable effect on Ybt synthesis or iron uptake (23). HMWP1 and HMWP2 (encoded by irp1 and irp2, respectively), YbtE, and YbtS have demonstrated roles in siderophore synthesis (2, 29, 30). Here we have determined that the two remaining genes, ybtU and ybtT, also appear to be required for Ybt siderophore synthesis. Thus, cells with mutations in ybtU or ybtT were unable to grow on iron-deficient medium at 37°C and culture supernatants from these mutants could not stimulate the growth of a Ybt biosynthetic mutant and did not contain detectable levels of siderophore. However, the ybtU and ybtT mutants could grow on iron-deficient medium at 37°C when provided with exogenous Ybt. These results suggest that the ybtU and ybtT mutants can use Ybt but are unable to produce the siderophore.
While our results indicate that YbtT and YbtU are required for Ybt synthesis, the precise role of these proteins in the production of the siderophore is unknown. YbtT has a thioesterase domain with the G(Y/W/H)SxG signature motif (amino acids [aa] 102 to 106 in YbtT) and a distal GxH conserved sequence (aa 134 to 136 in YbtT) (2, 29, 58). The four highest homologies to YbtT in the database (3 January 2000) are to external putative thioesterases; NrpT in Proteus mirabilis is part of a NRPS/polyketide synthase system involved in swarming (39.7% identity and 56.6% similarity), AngT is encoded within the anguibactin siderophore gene cluster in V. anguillarum (36.7% identity and 57.7% similarity), PikAV is required for macrolide antibiotic biosynthesis in Streptomyces venezuelae (33% identity and 50.9% similarity), and Srf4 (or SrfA-TE; 26.2% identity and 50.6% similarity) of Bacillus subtilis is needed for surfactin biogenesis (17, 20, 28, 66, 67). PchC, a putative thioesterase of the pyochelin siderophore system of P. aeruginosa (57), has 29.6% identity and 47.2% homology to YbtT. Many bacterial NRPS systems possess a C-terminal thioesterase domain as part of the NRPS in addition to a separate gene encoding an external thioesterase. In most systems, the biochemical function of the external thioesterase is unknown. However, it is thought to serve an editing function by removing aberrant structures on mischarged NRPSs caused by nonspecific thioesterification (11, 42, 55). Thus, loss of AngT expression reduced anguibactin synthesis to 30 to 40% of wild-type levels (66) and deletion of the gene encoding SrfA-TE caused a sixfold drop in synthesis of surfactin (55). Deletion of tylO, encoding an external putative thioesterase in Streptomyces fradiae, reduced tylosin synthesis by 85% (11). Our bioassay detects Ybt in iron-deficient culture supernatants diluted 1:16 (data not shown). Thus, if any Ybt siderophore is produced by the YbtT− mutant, it is at less than 6% of wild-type levels. Therefore, loss of the YbtT putative external thioesterase appears to have a slightly greater effect on product synthesis than do mutations in external thioesterase genes in other systems. In this regard the YbtT− mutant resembles the S. venezuelae pikAV mutant where biosynthesis of three macrolide antibiotics was reduced to less than 5%. The S. venezuelae macrolide biosynthesis gene cluster has no product containing an internal thioesterase domain (67). A thioesterase domain, present in the C terminus of HMWP1, is hypothesized to release the completed Ybt siderophore from the enzyme complex (29). Experiments to test the role of the HMWP1 thioesterase domain in Ybt production are in progress.
Our results show that a ΔybtU mutation affects Ybt synthesis and regulation. The YbtU− mutant had greatly reduced transcription from the psn and ybtP promoters and reduced levels of HMWP1, HMWP2, YbtE, and Psn proteins. However, it is unlikely that YbtU is a direct regulator; the regulatory effects of the ΔybtU mutation and polar mutations in the upstream genes irp1 and irp2 are corrected by the addition of the Ybt siderophore (2, 47). This suggests that the regulatory effects in these mutants are caused by loss of siderophore synthesis. The role of YbtU in siderophore synthesis is unknown. Protein motif and homology searches have not provided insight into the function of this protein. TopPred2 and DAS (http://www/biohemi.su.se) predict that YbtU has one potential transmembrane domain with a significant score (Fig. 3). YbtU is predicted by PSORT (45) to be an inner membrane protein. The two highest homologies in the database (3 January 2000) are to PchG of P. aeruginosa (pyochelin biosynthesis) and NrpU of P. mirabilis (swarming phenotype) (28, 57) (Fig. 3 shows the alignment), whose functions are also unknown.
FIG. 3.
Amino acid sequence alignment of YbtU from Y. pestis, PchG from P. aeruginosa, and NrpU from P. mirabilis. Black background, residues with identity to YbtU; gray background, conservative amino acid substitutions. The consensus line shows identical residues in all three proteins (capital letters), identical residues in two proteins (lowercase letters), and conservative amino acids in two proteins (colons). A putative transmembrane region is labeled and overlined.
In Y. pestis, we proposed that Ybt functions as a signal molecule by binding to the AraC-type regulator YbtA. The Ybt-YbtA complex then activates transcription of other genes in the Ybt system and represses transcription of ybtA. Thus mutants in ybtA had reduced β-galactosidase activity from a psn::lacZ reporter plasmid but elevated expression from a ybtA::lacZ reporter (22). In addition, strains bearing mutations in irp2 or ybtE, both genes encoding products involved in Ybt synthesis, had significantly reduced expression of HMWP1, HMWP2, and Psn (2). Curiously, mutations in psn (encoding the OM receptor for Ybt) and ybtP (encoding a putative ABC transporter involved in Ybt uptake) do not affect regulation of gene transcription or the levels of Ybt proteins (2, 22, 24, 47). This is in contrast to results obtained with Y. enterocolitica, where a strain with a mutant OM receptor, designated FyuA, exhibited reduced levels of HMWP2, suggesting the involvement of FyuA in regulation. However, despite a reduction in HMWP2, a key component in Ybt biosynthesis, this mutant has been described as a Ybt overproducer (46, 51). Psn and FyuA are 98% identical at the deduced amino acid level, making it unlikely that sequence differences would account for the observed regulatory discrepancies between Y. enterocolitica and Y. pestis (24, 51). An ERIC sequence is present in the ybtA promoter region of Y. enterocolitica IB strains (52) but absent from Y. pestis KIM6+ (29). The effect of this insertion on expression of YbtA has not been experimentally determined (52). Alternatively, the dissimilarity between Y. pestis and Y. enterocolitica Ybt receptor mutants could be due to differential permeation of Ybt through the OM. Bengoechea et al. (5) concluded that the OM of Y. pestis was more permeable to small hydrophobic molecules than the OM of Y. enterocolitica. Thus Y. enterocolitica FyuA may be required to translocate Ybt to the periplasm because of the increased OM impermeability.
The effects of a ΔybtU2071 mutation on expression of ybt genes are similar to those observed with other Ybt biosynthetic mutants (irp1, irp2, and ybtE) and support the hypothesis that the siderophore functions as a regulatory molecule. However, the lack of regulatory effects caused by ΔybtT2072 and ybtS::kan2070.1 appears to contradict this since neither mutant made detectable levels of Ybt. YbtT is a potential proofreading thioesterase, while YbtS is hypothesized to synthesize salicylate, the first moiety activated to initiate chain elongation and Ybt synthesis (29). YbtE catalyzes the adenylation of salicylate and transfer of this activated group to the N-terminal aryl carrier protein domain of HMWP2. Albeit at a much lower efficiency, YbtE also adenylates 2,3-dihydroxybenzoate (30). Thus, in the YbtS− mutant YbtE may activate 2,3-dihydroxybenzoate or another phenolate compound to initiate synthesis of an aberrant Ybt molecule. The low efficiency of YbtE-catalyzed activation or chain elongation from the altered phenolate moiety may lead to low levels of the altered siderophore that might interact with YbtA and thus allow normal regulation of the Ybt system. Similarly, YbtT− mutants may produce an aberrant compound(s) that can function as an inducer(s) in concert with YbtA. The altered Ybt molecule must either be produced at low levels or be retained within the bacterial cells since this molecule was not detected by our HPLC analysis. Alternatively, it is possible that small amounts of Ybt are synthesized by the YbtT− mutant and are sufficient for normal regulation but not for growth stimulation. Further experiments will be necessary to determine the nature of the signal molecule in Ybt+ cells as well as in YbtT− and YbtS− mutants.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI42738 from the National Institutes of Health.
We thank Scott W. Bearden for assistance with some sequence analyses and construction of pPSN345.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 2.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearden S W, Perry R D. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 4.Bearden S W, Staggs T M, Perry R D. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol. 1998;180:1135–1147. doi: 10.1128/jb.180.5.1135-1147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengoechea J-A, Brandenburg K, Seydel U, Díaz R, Moriyón I. Yersinia pseudotuberculosis and Yersinia pestis show increased outer membrane permeability to hydrophobic agents which correlates with lipopolysaccharide acyl-chain fluidity. Microbiology. 1998;144:1517–1526. doi: 10.1099/00221287-144-6-1517. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun V, Hantke K, Köster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 8.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 9.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchrieser C, Rusniok C, Frangeul L, Couve E, Billault A, Kunst F, Carniel E, Glaser P. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect Immun. 1999;67:4851–4861. doi: 10.1128/iai.67.9.4851-4861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler A R, Bate N, Cundliffe E. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem Biol. 1999;6:287–292. doi: 10.1016/S1074-5521(99)80074-X. [DOI] [PubMed] [Google Scholar]
- 12.Byers B R, Arceneaux E L. Microbial iron transport: iron acquisition by pathogenic microorganisms. Met Ions Biol Syst. 1998;35:37–66. [PubMed] [Google Scholar]
- 13.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carniel E, Guiyoule A, Guilvout I, Mercereau-Puijalon O. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol Microbiol. 1992;6:379–388. doi: 10.1111/j.1365-2958.1992.tb01481.x. [DOI] [PubMed] [Google Scholar]
- 15.Chambers C E, McIntyre D D, Mouck M, Sokol P A. Physical and structural characterization of yersiniophore, a siderophore produced by clinical isolates of Yersinia enterocolitica. Biometals. 1996;9:157–167. doi: 10.1007/BF00144621. [DOI] [PubMed] [Google Scholar]
- 16.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosmina P, Rodriguez R, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 18.Cox C D, Rinehart K L, Jr, Moore M L, Cook C J., Jr Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1981;78:4256–4260. doi: 10.1073/pnas.78.7.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drechsel H, Stephan H, Lotz R, Haag H, Zähner H, Hantke K, Jung G. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann. 1995;1995:1727–1733. [Google Scholar]
- 20.Farrell D H, Mikesell P, Actis L A, Crosa J H. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P22 cro and regulation by iron. Gene. 1990;86:45–51. doi: 10.1016/0378-1119(90)90112-5. [DOI] [PubMed] [Google Scholar]
- 21.Ferber D M, Fowler J M, Brubaker R R. Mutations to tolerance and resistance to pesticin and colicins in Escherichia coli. J Bacteriol. 1981;146:506–511. doi: 10.1128/jb.146.2.506-511.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 23.Fetherston J D, Bertolino V J, Perry R D. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32:289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 24.Fetherston J D, Lillard J W, Jr, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 26.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 27.Froehlich B, Husmann L, Caron J, Scott J R. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaisser S, Hughes C. A locus coding for putative non-ribosomal peptide/polyketide synthase functions is mutated in a swarming-defective Proteus mirabilis strain. Mol Gen Genet. 1997;253:415–427. doi: 10.1007/s004380050339. [DOI] [PubMed] [Google Scholar]
- 29.Gehring A M, DeMoll E, Fetherston J D, Mori I, Mayhew G F, Blattner F R, Walsh C T, Perry R D. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 30.Gehring A M, Mori I, Perry R D, Walsh C T. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 31.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hare J M, McDonough K A. High-frequency RecA-dependent and -independent mechanisms of Congo red binding mutations in Yersinia pestis. J Bacteriol. 1999;181:4896–4904. doi: 10.1128/jb.181.16.4896-4904.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hare J M, Wagner A K, McDonough K A. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol Microbiol. 1999;31:291–303. doi: 10.1046/j.1365-2958.1999.01172.x. [DOI] [PubMed] [Google Scholar]
- 34.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65 000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 35.Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 36.Hornung J M, Jones H A, Perry R D. The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 37.Humphreys G O, Willshaw G A, Anderson E S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975;383:457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- 38.Jalal M A F, Hossain M B, van der Helm D, Sanders-Loehr J, Actis L A, Crosa J H. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J Am Chem Soc. 1989;111:292–296. [Google Scholar]
- 39.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 40.Kutyrev V V, Filippov A A, Oparina O S, Protsenko O A. Analysis of Yersinia pestis chromosomal determinants Pgm+ and PstS associated with virulence. Microb Pathog. 1992;12:177–186. doi: 10.1016/0882-4010(92)90051-o. [DOI] [PubMed] [Google Scholar]
- 41.Lucier T S, Brubaker R R. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2078–2086. doi: 10.1128/jb.174.7.2078-2086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marahiel M A, Stachelhaus T, Mootz H D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 43.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 44.Miller J H. A short course in bacterial genetics. A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 45.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 46.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry R D, Balbo P B, Jones H A, Fetherston J D, DeMoll E. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology. 1999;145:1181–1190. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- 48.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakin A, Noelting C, Schubert S, Heesemann J. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect Immun. 1999;67:5265–5274. doi: 10.1128/iai.67.10.5265-5274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 52.Rakin A, Schubert S, Pelludat C, Brem D, Heesemann J. The high-pathogenicity island of yersiniae. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 77–90. [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider A, Marahiel M A. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch Microbiol. 1998;169:404–410. doi: 10.1007/s002030050590. [DOI] [PubMed] [Google Scholar]
- 56.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serino L, Reimann C, Visca P, Beyeler M, Della Chiesa V, Haas D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J Bacteriol. 1997;179:248–257. doi: 10.1128/jb.179.1.248-257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw-Reid C, Kelleher N, Losey H C, Gehring A M, Berg C, Walsh C T. Assembly line enzymology by multimodular nonribosomal peptide synthetases: the thioesterase domain of Escherichia coli EntF catalyzes both elongation and cyclolactonization. Chem Biol. 1999;6:385–400. doi: 10.1016/S1074-5521(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 59.Sikkema D J, Brubaker R R. Resistance to pesticin, storage of iron, and invasion of HeLa cells by yersiniae. Infect Immun. 1987;55:572–578. doi: 10.1128/iai.55.3.572-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staggs T M, Fetherston J D, Perry R D. Pleiotropic effects of a Yersinia pestis fur mutation. J Bacteriol. 1994;176:7614–7624. doi: 10.1128/jb.176.24.7614-7624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Staggs T M, Perry R D. Identification and cloning of a fur regulatory gene in Yersinia pestis. J Bacteriol. 1991;173:417–425. doi: 10.1128/jb.173.2.417-425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suo Z, Walsh C T, Miller D A. Tandem heterocyclization activity of the multidomain 230 kDa HMWP2 subunit of Yersinia pestis yersiniabactin synthetase: interaction of the 1-1382 and 1383-2035 fragments. Biochemistry. 1999;38:14023–14035. doi: 10.1021/bi991574n. [DOI] [PubMed] [Google Scholar]
- 63.Surgalla M J, Beesley E D. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol. 1969;18:834–837. doi: 10.1128/am.18.5.834-837.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson J M, Jones H A, Perry R D. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinberg E D, Weinberg G A. The role of iron in infection. Curr Opin Infect Dis. 1995;8:164–169. [Google Scholar]
- 66.Wertheimer A M, Verweij W, Chen Q, Crosa L M, Nagasawa M, Tolmasky M E, Actis L A, Crosa J H. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect Immun. 1999;67:6496–6509. doi: 10.1128/iai.67.12.6496-6509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue Y, Zhao L, Liu H, Sherman D H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]