Abstract
Antibiotics, among the most used medications in children, affect gut microbiome communities and metabolic functions. These changes in microbiota structure can impact host immunity. We hypothesized that early-life microbiome alterations would lead to increased susceptibility to allergy and asthma. To test this, mouse pups between postnatal days 5 – 9 were orally exposed to water (control) or to therapeutic doses of azithromycin or amoxicillin. Later in life, these mice were sensitized and challenged with a model allergen, house dust mite (HDM), or saline. Mice with early-life azithromycin exposure that were challenged with HDM had increased IgE and IL-13 production by CD4+ T cells compared to unexposed mice; early-life amoxicillin exposure led to fewer abnormalities. To test that the microbiota contained the immunological cues to alter IgE and cytokine production after HDM challenge, germ-free mice were gavaged with fecal samples of the antibiotic-perturbed microbiota. Gavage of adult germ-free mice did not result in altered HDM responses, however, their offspring, which acquired the antibiotic-perturbed microbiota at birth showed elevated IgE levels and CD4+ cytokines in response to HDM, and altered airway reactivity. These studies indicate that early-life microbiota composition can heighten allergen-driven Th2/Th17 immune pathways and airway responses in an age-dependent manner.
Keywords: azithromycin, amoxicillin, microbiome, perinatal, immunity, development, house dust mite
Introduction
Global asthma incidence has increased over the past five decades, with 300 million people currently affected 1. Asthma is clinically diverse and categorized into different endotypes. The type 2 (T2) high endotype has increased type 2 cytokine responses, characterized by production of Interleukin (IL)-4, IL-5 and IL-13 by either T helper 2 (Th2) cells or type 2 innate lymphocytes. Asthmatic children without these T2 signatures are classified as the T2-low endotype 2, 3. T2-high asthma, including early-onset allergic, late-onset eosinophilic, and exercise-induced asthma, mostly responds to medications targeting T2 immune responses 2–4. The T2-low endotype includes neutrophilic and obesity-related forms of asthma, which are characterized by type 17 immune responses and neutrophil recruitment that mediate pulmonary inflammation, and have fewer available therapies 2, 5.
Early-life antibiotic use has been associated with increased asthma risk in epidemiological studies 6–9, and reducing community-wide antibiotic exposure has been associated with diminished asthma incidence 10. Asthma is part of an allergic continuum also including atopic dermatitis, food allergy, and allergic rhinitis; early-life antibiotic exposure has been associated with increased risk of all of these disorders 11–15. These observations support the hypothesis that early-life antibiotic use is causally involved in asthma predisposition, but direct evidence has been lacking.
Amoxicillin and azithromycin are the two most commonly prescribed pediatric antibiotics in the U.S. and globally 6, 16, 17. The developing early-life microbiota is perturbed by antibiotics, with effects related to antibiotic class, dose, duration, and host age during exposure 18–20. Early-life microbiome perturbation can change microbial metabolites and alter microbiome resilience and colonization resistance, altering host immune and metabolic development 21–26.
We now report studies of early-life administration of azithromycin and amoxicillin to mice, at levels prescribed in children, that highlight interactions of microbiome perturbation with host age, and downstream inflammatory and immune consequences. We provide direct evidence that azithromycin perturbation of the early-life microbiota enhances the immunological and physiological features of allergen-driven responses.
Results
Early-life antibiotic exposures alter responses to allergen challenge.
To assess whether a clinically relevant antibiotic exposure alters susceptibility to allergic airway inflammation, C57BL/6 select-pathogen-free (SPF) pups were orally dosed from postnatal day (P)5 for 5 days with azithromycin or amoxicillin at pharmacological levels for children [amoxicillin, 100mg/kg of bodyweight/day; azithromycin, 30mg/kg of bodyweight/day] or water as a control (Figure 1A). Following antibiotic exposure, on P46, mice were sensitized intranasally with HDM extract, or PBS as a negative control, and subsequently challenged intranasally three times with HDM or PBS. Mice were euthanized at P60 and immune responses to HDM were compared across the groups, 50 days after the antibiotic exposure had ended. We first asked whether the antibiotic exposures affected inflammatory responses to HDM antigen. Analysis of bronchoalveolar lavage (BAL), collected to assess leukocytic airway infiltration, indicated that mice exposed to azithromycin or amoxicillin during early-life, and subsequently sensitized and challenged with HDM, compared to HDM-sensitized and -challenged control mice, led to no significant changes in BAL cellularity (Figure S1A–C). However, azithromycin-HDM mice had increased serum (Figure S1D) and lung (Figure S1E) total IgE levels, which were strongly correlated (Figure S1F). In the PBS-challenged mice, prior antibiotic exposure did not affect BAL cellularity, eosinophilia, or IgE levels (Figure S1A–F). In total, these results indicate that early-life antibiotic exposures to azithromycin led to increased IgE responses following antigen challenge.
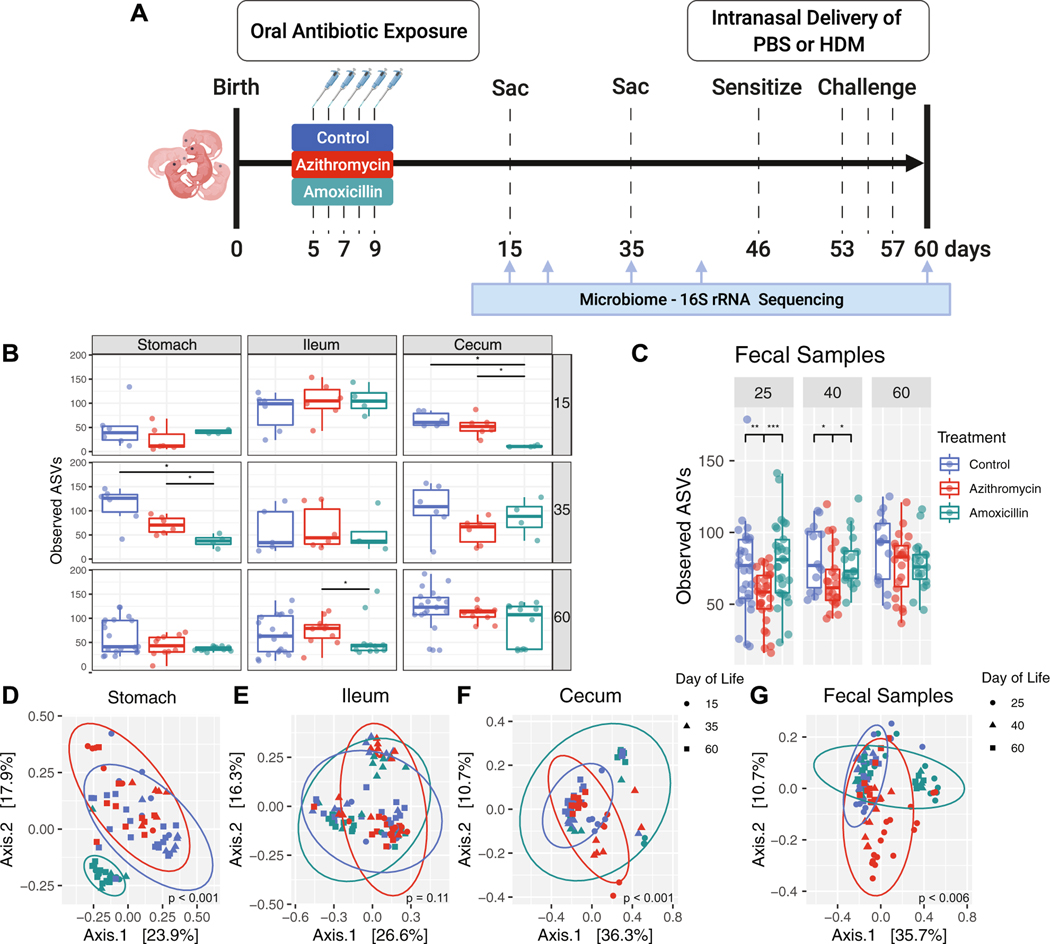
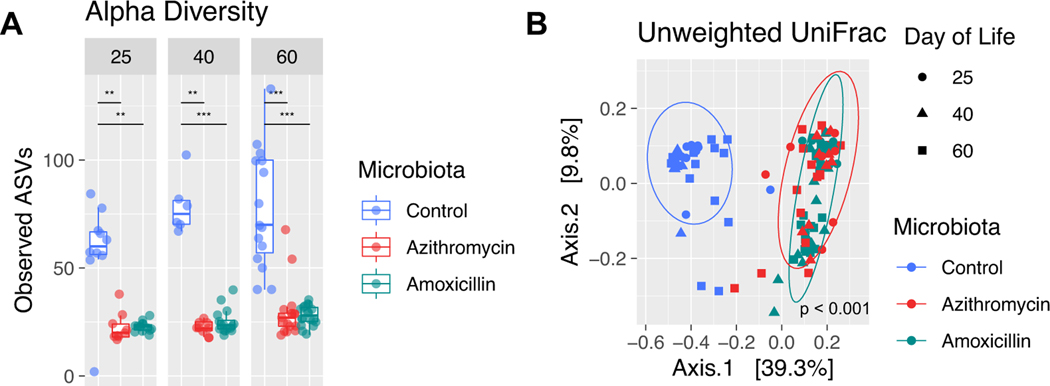
Figure 1: Evaluation of microbiota composition in antibiotic-exposed mice.
A, mice received early-life treatment with azithromycin, amoxicillin, or not—control, between P5–9 by direct oral administration; dams were unexposed. For each independent experiment, there were at least 2 or 3 litters per treatment group. At P46, mice were intranasally sensitized with 1μg of HDM antigen or PBS as a negative control for HDM sensitization. One week later, mice received three separate intranasal challenges with 10μg of HDM antigen, or again with PBS as a negative control. All mice were sacrificed 3 days after the last challenge at P60. DNA extracted from mouse stomach tissue, ileal tissue with contents, colon tissue with contents and fecal pellets and was subjected to 16S rRNA sequencing. Tissue samples were collected at sacrifice on days 15, 35 and 60, while fecal samples were collected on days 25, 40, and 60. Analysis of alpha diversity (Observed ASVs) (B,C) and beta diversity (D-G) (unweighted UniFrac) are from one representative experiment. Significance for alpha diversity was determined by a nonparametric one-way ANOVA (*p<0.05; **p<0.01; ***p<0.001) and for beta diversity, a Permutational Multivariate Analysis of Variance was performed to compare antibiotic exposure, day of life, and the interaction of both factors (a p-value for each comparison is reported on the UniFrac plot). Results are shown for: D, stomach (18.8% variance explained by treatment, 7.2% by day of life, and 9.3% by both); E, ileum (4.9% variance explained by treatment, 7.7% by day of life, and 6.8% by both); F, cecum (9.8% variance explained by treatment, 17.9% by day of life, and 6.3% by both); and G, fecal pellets (13.4% variance explained by treatment, 7.4% by day of life, and 9.3% by both).
We then asked whether differences in Th2 or Th17 cytokines would explain the IgE patterns observed in mice that had early-life antibiotic exposure. First, compared to unexposed mice, azithromycin-exposed mice had an increase in lung CD4+ T cell infiltration when sensitized and challenged with HDM (Figure S1H). Among the mice that were HDM-sensitized/challenged, those exposed to azithromycin had higher pulmonary IL-13 production at P60 than the unexposed controls (Figure S1G,I), consistent with higher IgE titers (Figure S1D,E). Mice that had been exposed to amoxicillin and were then subsequently challenged with HDM had a higher frequency of IL-17A-producing CD4+ Th cells than untreated controls (Figure S1J). There were no significant differences in IFNγ production in any experimental group (Figure S1K). We next sought to determine if these cytokine changes in antibiotic-exposed mice affected lung inflammation. Histological examination of lungs showed increased inflammatory scores and PAS+ cells related to the HDM challenge, but no significant differences with respect to prior antibiotic exposure (Figure S2A–E). As there were no differences in lung inflammation (Figure S2A–C), we did not expect any deficits in FoxP3+ regulatory T cells (Tregs) and peripherally induced (p)Treg populations in the lung tissue in response to antibiotic exposure, and this was indeed the case (Figure S2F). In total, these findings indicate that the early-life immunological perturbations were mostly associated with azithromycin exposure, and limited to heighted IgE responses, and increased lung CD4+ T cell infiltration and IL-13 production.
Role of the microbiota in altering immunological responses.
We then sought to determine if the differences observed in the antibiotic-exposed mice were due to direct effects of the drugs on the murine immune system, or whether they were transduced via perturbation of the microbiome. To address this, germ-free pups were treated with antibiotics, or not, and later sensitized and challenged with HDM, exactly as outlined in Figure 1A. At P60, no differences in BAL cellularity or granulocyte frequency were found between the antibiotic-exposed or unexposed germ-free mice (Figures S3A). Flow cytometric analysis of lung tissue also showed no significant differences in lung CD4+ T cell abundance or frequencies with respect to antibiotic exposure (Figure S3B). As observed in the SPF mice (Figure S1B, S1G–J), the proportions of eosinophils and Th cells rose in mice challenged with HDM compared with PBS, but there were no significant differences in relation to the antibiotic exposure (Figures S3A–C). Thus, in the absence of a microbiota, antibiotic treatment had no discernable impact on immune responses to allergen challenge. These results provide evidence that the observed antibiotic-driven effects on the immune responses to HDM are microbiota-dependent and do not result from the direct drug interactions with murine tissues, consistent with prior results 19.
Identification of taxa perturbed by the early-life antibiotic exposures.
To determine whether antibiotic-exposure led to microbial differences in the experimental mice, we examined gastrointestinal and pulmonary samples using 16S rRNA sequencing. Antibiotic exposure led to significantly reduced representation of amplicon sequence variants (ASVs) in the gastric microbiota at P35 (Figure 1B). The fecal microbiota of azithromycin-exposed mice showed significantly reduced ASVs at P25 and P40 (Figure 1C). These findings indicate a reduction in taxon richness for at least a month following the antibiotic exposure with partial recovery, later in life (Figure 1C). Utilizing unweighted UniFrac analysis across multiple timepoints (Figure 1D–G) and plots of taxonomic abundance at P60 (Figure S4), amoxicillin, but not azithromycin, altered gastric microbiota composition consistently at all timepoints (Figure 1D, S4A). There were no differences in the ileal microbiota (Figure 1E, S4B), the cecal microbiota was altered by exposure to either antibiotic across all timepoints (Figure 1F, S4C). Both antibiotic regimens changed colonic community structure, determined by analysis of serially obtained fecal samples (Figure 1G, S4D).
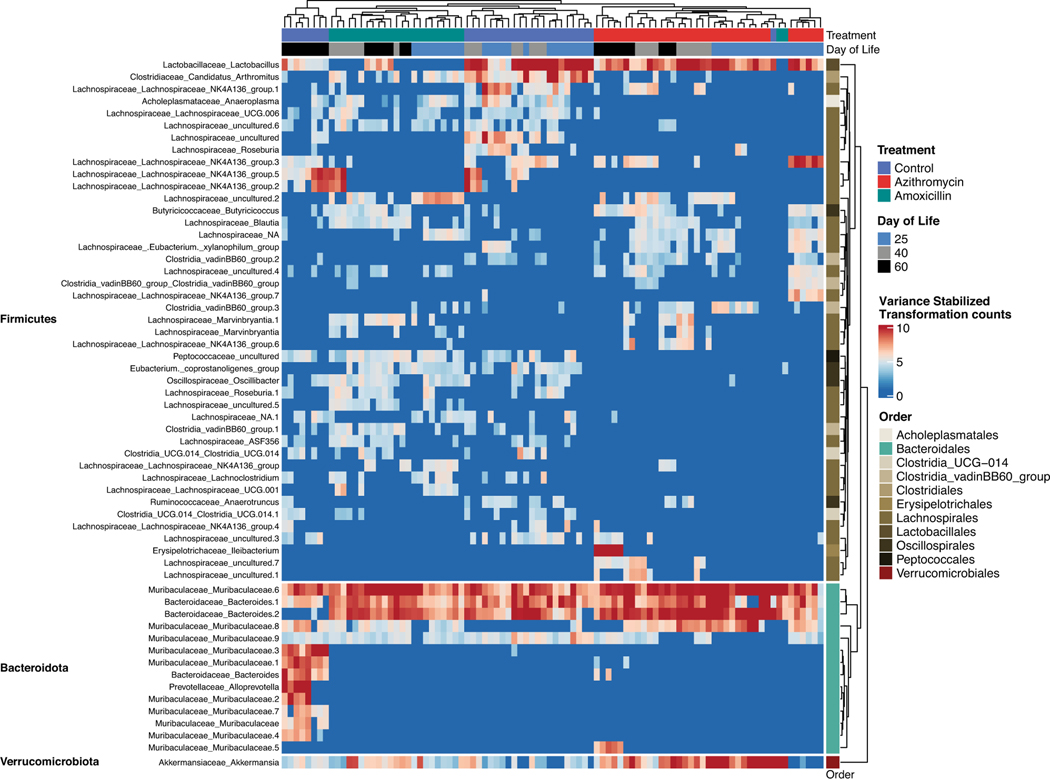
Each of the antibiotic exposures led to significantly altered colonic representation of multiple ASVs, with unsupervised hierarchical clustering of differentially abundant ASVs showing that colonic samples from azithromycin-exposed mice were distinct from the other treatment groups (Figure 2). By P60, control mice had high abundances of Bacteroidota (especially Muribaculaceae species), which did not occur in mice with early-life antibiotic exposure. To assess changes in Muribaculaceae, the abundance of all Muribaculaceae strains, at the genus level, was evaluated in fecal samples over time (Figure S5). Using a mixed linear effects model, we found that Muribaculaceae changed significantly over time in all groups, and was significantly different when comparing control and azithromycin (p-value = 0.020). Further, the change over time in the azithromycin group significantly differed from the change in abundance over time in control mice (p-value = 0.015). Both groups of antibiotic-exposed mice had increased Verrucomicrobia (Akkermansia), and the azithromycin-exposed mice had unique losses within the Firmicutes, mostly Lachnospiraceae species. Azithromycin-exposed mice showed a unique enrichment of Ileibacterium in P60 samples that was not observed in amoxicillin or control samples. Samples from the amoxicillin-exposed mice clustered more closely with control samples until P60 but showed reduced Lactobacillus abundances compared to both other groups (Figure 2).
Figure 2: Identifying significantly altered ASVs between control and antibiotic-exposed mice.
Unsupervised hierarchical clustering of differentially abundant ASVs from 92 fecal samples (columns) from 42 individual mice across multiple timepoints, identified using DESeq2, with false discovery rate (FDR) set to p < 0.01. Samples are color-coded by treatment, day of life, and body site. Rows indicate statistically significant taxa (n = 58), taxa annotation at the genus level (species level information also included when available, annotated as: genus_species).
We then assessed whether there were antibiotic-induced compositional differences in the lungs. At P60, but not before, the mice exposed to azithromycin early in life had reduced representation of pulmonary ASVs (Figure S6A), but there were few other differences in pulmonary microbiota compositions between antibiotic-exposed and control mice (Figure S6B, S6C). In total, these data indicate persistent effects on the host microbiome with effects being either conserved across both antibiotics, or antibiotic-specific with the most substantial effects on the colon, cecum, and stomach microbiota composition. Given the antibiotic-induced effects on the host gastrointestinal bacterial communities, we sought to test if altered microbiota composition is involved in host immunomodulation in response to HDM sensitization and challenge.
Role of the altered microbiome in the immunological phenotypes of adult mice.
We next asked whether the compositional differences in the intestinal microbiota directly led to an altered immune response to HDM. To address this question, we experimentally transferred microbiota collected from P25 mice (15 days after the antibiotic exposure had ceased) into 6–8-week-old germ-free mice (Figure 3A). P25 was selected as the timepoint of transfer to ensure that fecal pellets did not contain any residual antibiotics, and that it was early enough in development to permit a maturation of the transferred microbial community in the new murine host. The fecal pellets used for transfer were either perturbed by the prior antibiotic exposure or from control mice (no antibiotic exposure). Consistent with the timeline of the initial studies, 45 days post-transfer, the mice were sensitized and challenged with HDM or PBS and sacrificed at post-transplant day 60 to evaluate immune responses. Apart from a rise in BAL cellularity in the mice that received the amoxicillin-perturbed microbiota, there were no significant differences observed in serum IgE, BAL cellular composition, or cytokine production by CD4+ T cells in mice that received different microbiota (Figure S7). These results indicate that conventionalization of germ-free adult mice with perturbed microbiota was not sufficient to transfer any phenotype previously observed in SPF-conventional mice exposed to antibiotics during the neonatal period (Figure S1). One explanation for this finding is that there is an age-window during which the immune system is most sensitive to perturbations in intestinal microbiota.
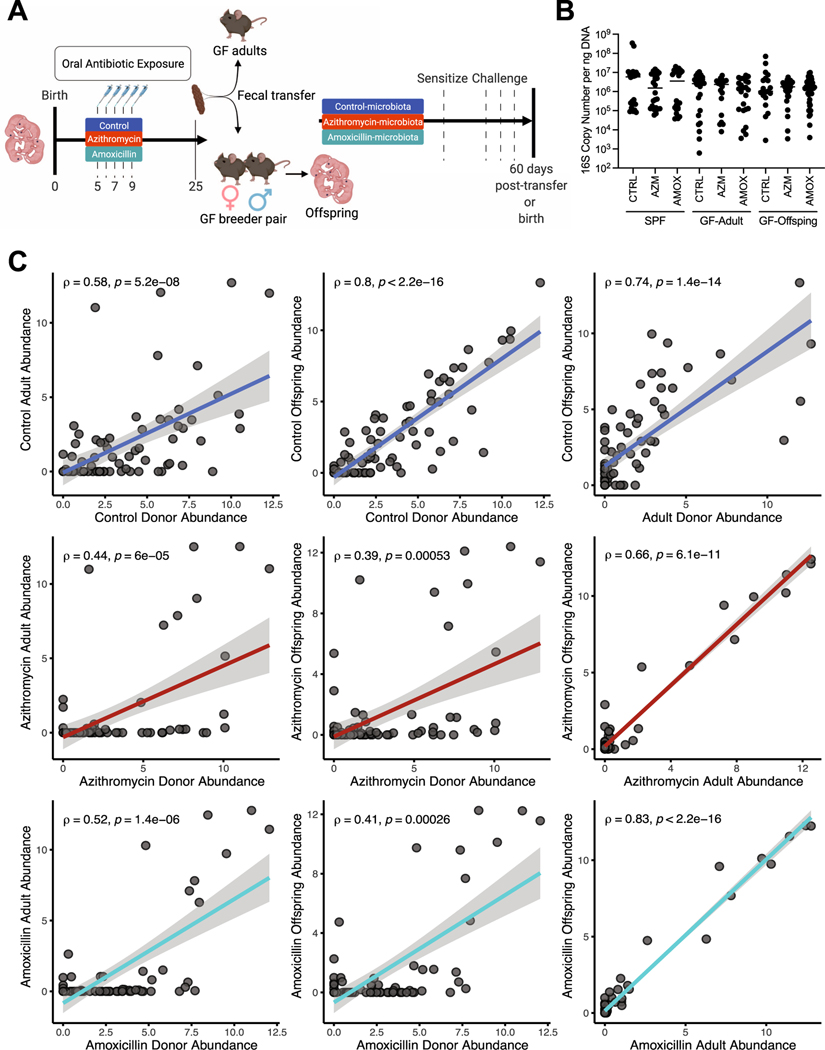
Figure 3: Differential microbiota in experiments conventionalizing germ-free mice.
A. Adult (6–8-week-old) germ-free mice were conventionalized with a suspension of fecal microbiota prepared from frozen P25 fecal samples collected from SPF mice varying in antibiotic-exposure (Figures 1–3). A subset (n = 6–8) of these mice were evaluated for immune responses to HDM through sensitization and challenge with antigen or to PBS. Other conventionalized mice (n = 3 per donor) were used as breeder pairs to provide progeny exposed to the conventionalized microbiota from birth. B. 16S rRNA copy number was determined using qPCR, and copy number was normalized to the total DNA concentration of the sample and reported as copy number per ng of DNA. Fecal samples for day of life/day post gavage 25, 40, and 60 were analyzed for copy number. C. Spearman correlations between donor SPF mice, adult germ-free mice gavaged with stool and conventionalized offspring mice (3A) for all three experimental groups. Each point represents the log2 abundance of a unique taxon at the genus level within each experimental group of mice. All identified genera were included (n = 74). The correlations strength (Spearman rho) was positive (ρ > 0.4 in all cases), and significant (p < 0.001) in all comparisons. Sequencing data was pooled from three sequencing runs, and multiple experiments.
Role of the altered microbiome in the immunological phenotypes of mice exposed from birth.
To address the possibility of an age-dependent variable, we assessed immunological responses to HDM in mice exposed to the perturbed microbiota from birth (Figure 3A). From within the original group of conventionalized germ-free adult mice, some animals were used for antigen challenge (described above), but others were used for breeding. As such, their offspring acquired either a perturbed or control fecal microbiota from birth via vertical transmission. First, to determine if there were any differences in bacterial abundance based on mode of microbiota acquisition, we quantified 16S copy number in the fecal samples from the original SPF cohort, the germ-free adults that were gavaged with fecal samples, and the progeny of the germ-free mice (Figure 3B). We found no differences in 16S copy number between the experimental groups. To assess the compositional fidelity of the transferred perturbed or control microbiota, we compared the relative abundances of all annotated genera between the SPF donor, germ-free adult recipient, and the resultant offspring mice in each group (Figure 3C). These analyses showed significant correlation of the taxa present in donor and gavaged adult mice, and donor and offspring mice, indicating high fidelity transfer (Figure 3C), and that the correlation strength (Spearman rho) was favorable (ρ > 0.4) in all cases. These analyses were conducted on samples from multiple experiments and sequencing runs, subject to batch effects which could skew the comparison between samples. The greatest fidelity between the experimental generations involved transfer of the control microbiota, and less so for the antibiotic-perturbed microbiota. This may be an indication of reduced community resiliency and stability, confirming prior observations 27. This reduced resiliency may factor into the diversity loss observed in the transfer of the antibiotic-perturbed microbiota within fecal samples.
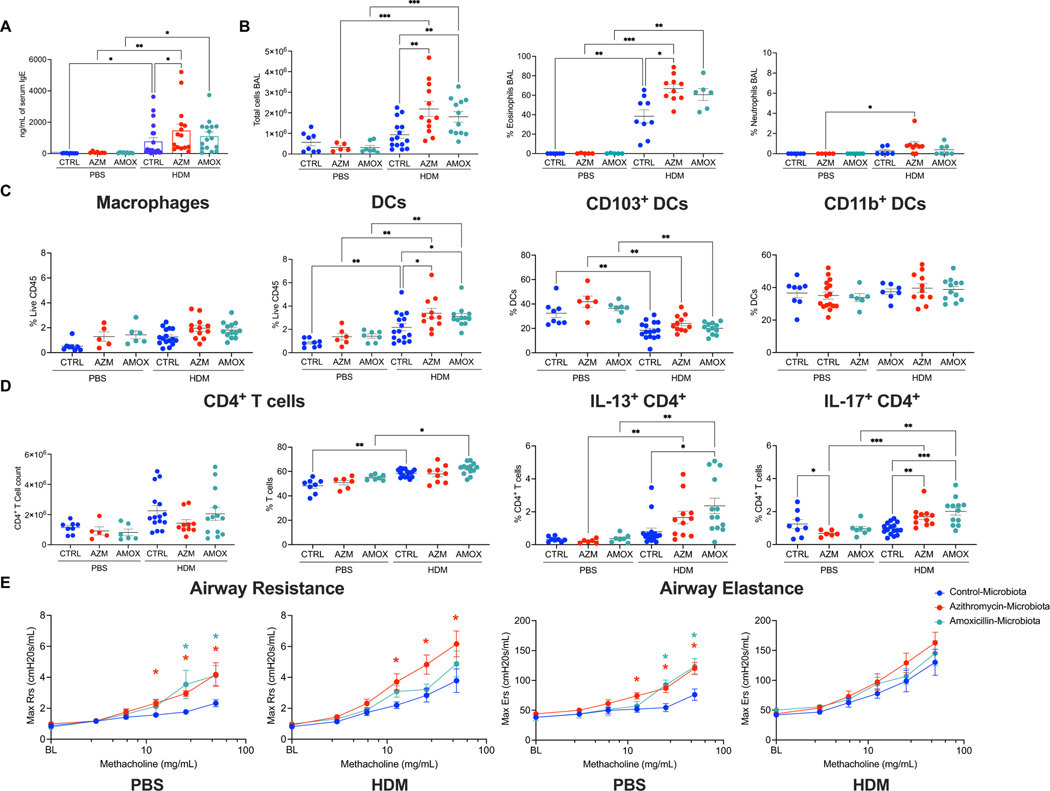
Based on these transfers, we had a way to generate offspring mice that had not received antibiotics themselves but were exposed to the perturbed microbiota from birth (Figures 3A). Using the same HDM allergen model and measurements of immunity, we assessed if these animals would mirror the effects of directly administered antibiotics in terms of heightened IgE and IL-13 responses. The mice that had an azithromycin-altered microbiota from birth had higher serum IgE concentrations than control mice, after HDM sensitization and challenge (Figure 4A) which also was observed in the SPF mice following the early-life azithromycin exposure (Figure S1D). Following HDM challenge, the mice colonized with the azithromycin-perturbed microbiota also had increased BAL cellularity and eosinophil and neutrophil proportions (Figure 4B) and had higher proportions of dendritic cells (Figure 4C). We did not observe changes in infiltration or frequencies of CD4+ T cells associated with receipt of either antibiotic-perturbed-microbiota (Figure 4D). We did observe significant increases in CD4+ T cells producing IL-17A in mice that received either azithromycin or amoxicillin perturbed microbiota, and IL-13 in mice that received the amoxicillin-perturbed microbiota (Figures 4D). The mice exposed to the amoxicillin-perturbed microbiota from birth did not have altered IgE levels, like the amoxicillin-exposed SPF mice, but had increased total number of BAL cells and increased proportions of pulmonary dendritic cells (Figures 4A–B, Figure S1D). In total, this experiment demonstrates that acquiring an antibiotic-perturbed microbiota from birth leads to allergen-induced immunological hyperactivity.
Figure 4: Phenotypes in the progeny of parental germ-free mice conventionalized with antibiotic-perturbed microbiota.
Progeny of germ-free mice that had been conventionalized with the fecal microbiota of antibiotic-exposed or control SPF mice [see Figure 3A] were evaluated at sacrifice for immune responses to HDM antigen. A. Serum IgE levels were determined by IgE-specific ELISA. B. BAL was evaluated for total cell counts by trypan blue staining, or after BAL samples were cytocentrifuged and differentially stained, for proportion of eosinophils, or neutrophils. Lung single cell suspensions were isolated and evaluated by flow cytometry for differences in the C. myeloid compartment or D. stimulated ex vivo using PMA and ionomycin. Data was collected from three independent experiments; each individual experiment yielded the same trend. Subsets of the offspring mice were evaluated for differences in lung function by methacholine challenge assay, using a Scireq FlexiVent apparatus. Offspring mice sensitized and challenged with HDM or PBS were evaluated for E. airway resistance and airway elastance in response to increasing methacholine doses (PBS, n = 5–8 per group and HDM, n = 10–12 per group, data is from three independent experiments). *p<0.05; **p<0.01; ***p<0.001, ****p<0.0001; nonparametric one-way ANOVA.
Physiologic consequences of exposure to an antibiotic-perturbed microbiota.
Finally, we sought to determine whether the observed immunological changes had physiological consequences related to asthma risk or severity. We addressed this question using experimental methacholine challenges to assess bronchial airway reactivity in the mice exposed to control or the altered microbiota from birth and subjected to HDM or PBS challenge (Figure 3A). The mice exposed from birth to either of the two antibiotic-perturbed microbiota and challenged with PBS showed higher airway reactivity measured by both maximal resistance and elastance (Figure 4E). As expected 28, both airway resistance and elastance in response to increasing doses of methacholine were significantly greater in all mice that had prior HDM challenge than in those PBS-challenged. Among the HDM-challenged mice, those exposed to the azithromycin-perturbed microbiota from birth had significantly greater airway resistance than mice exposed to the control microbiota; exposure to the amoxicillin-perturbed microbiota had little effect (Figure 4E). There were no significant differential effects on airway elastance among the HDM sensitized and challenged mice, but the antibiotic-perturbed microbiota mice had higher airway elastance relative to control among the PBS challenged mice. (Figure 4E). In total, these experimental findings provide evidence that exposure to antibiotic-perturbed microbiota from birth affects pulmonary physiological function with and without allergen challenge. These results experimentally link antibiotic-perturbed early-life microbiota to allergic predisposition, heightened immunological responses to allergen, and clinical features of allergic asthma.
Microbiota composition associated with altered immune phenotypes.
To characterize differences in microbiota composition of the mice that were born with control or antibiotic-perturbed microbiota, 16S rRNA sequencing was employed. Analysis of observed ASVs (Figure 5A) revealed that mice with the antibiotic-perturbed microbiota had significantly reduced alpha diversity relative to control animals and did not show the increases in observed ASVs with maturation that were seen in controls. An unweighted UniFrac analysis showed clear and significant differences in fecal community composition between control-microbiota mice and antibiotic-perturbed microbiota mice (Figure 5B). Differentially abundant ASVs are highlighted in Figure S8, with the mice exposed to the antibiotic-perturbed microbiota having increased abundances of Akkermansia, Muribaculum, Bacteroides, Lachnostridium, and Blautia. Many of these ASVs are consistent with differentially abundant taxa identified in the SPF experiments (Figure 2). To further highlight consistently altered colonic ASVs associated with antibiotic exposure in these experiments, ASVs that were differentially abundant between controls and azithromycin-perturbed mice from both the SPF experiments (Figure 1A) and the transfer experiments (Figure 3A) from P25 and P60 are identified in Tables S1 and S2, respectively. Table S1 identifies potentially important ASVs during early-life (P25) which include Segmented filamentous bacteria (SFB; Candidatus Arthomitus), Akkermansia, Oscillospirales, and Muribaculaceae species. All apart from Akkermansia, were enriched in control mice. Later at P60, Muribaculaceae and Oscillospirales continue to be present in increased abundance in control mice, and Lachnospiraceae and Clostridia also show higher representation in control mice relative to the azithromycin-perturbed microbiota mice. Parabacteroides was the only ASV overrepresented in azithromycin-microbiota mice at P60 in both sets of experiments. These data provide evidence that the mice that were exposed from birth to antibiotic-perturbed microbiota had clear compositional differences, and reduced diversity relative to control-microbiota animals.
Figure 5: Altered alpha and beta diversity in mice that had an antibiotic-perturbed or control microbiota from birth.
A. Alpha diversity across P25, P40 and P60 for each group of mice based on birth-microbiota. Significance for alpha diversity was determined by a nonparametric one-way ANOVA (*p<0.05; **p<0.01; ***p<0.001) B. Beta diversity, based on unweighted UniFrac analysis of P25, P40, P65 fecal pellets. Each sample is color-coded by microbiota group and symbols are used to differentiate day of life. Permutational Multivariate Analysis of Variance was performed to compare microbiota, day of life, and the interaction of both factors (p-value < 0.001); 41% of variance could be attributed to the founding microbiota source; 3.5% of variance could be attributed to day of life, and 3.3% of variance was attributed to both microbiota source and day of life.
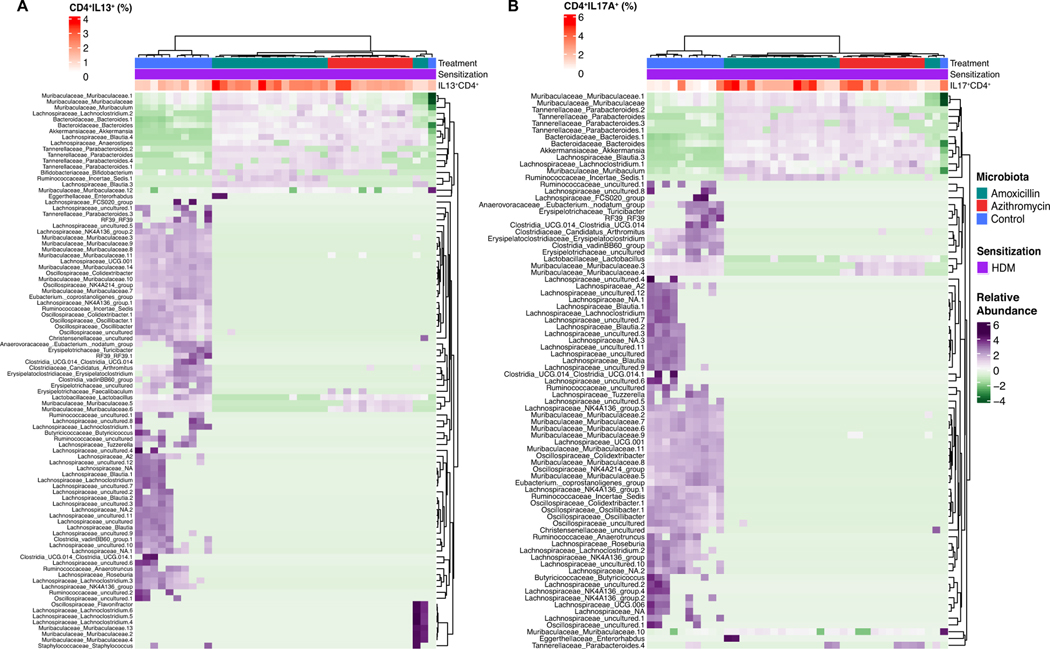
Next, to determine ASVs that may be associated with the immune phenotypes observed in the microbiota transfer experiments, we correlated lung IL-13 and IL-17A production by CD4+ T cells to ASVs that significantly differed between recipient mice that received control or antibiotic-perturbed microbiota. Non-hierarchical clustering of significantly correlated ASVs (p<0.05) revealed that there are distinct microbial signatures in the control and antibiotic-perturbed microbiota groups that correlate with both IL-13 and IL-17A production (Figure 6A, 6B). Many Lachnospiracae and Muribaculaceae strains were enriched in control mice and negatively correlated with cytokine production. Akkermansia and some Tannerellaceae strains had higher relative abundance in the mice receiving the antibiotic-perturbed microbiota and significantly correlated with elevated cytokine production (Figure 6A, 6B). We also tested correlations between ASV relative abundance and total BAL counts, lung eosinophilia, and serum IgE without identifying any significant associations or clear compositional differences. In total, these data indicate that there are compositional differences associated with the CD4+ T cell phenotypes in this study. Further, the germ-free experiments permitted the transfer of compositional microbiome differences found in SPF antibiotic-exposed mice into new hosts, and showed that germ-free mice that developed with the perturbed microbiota from birth had altered immune and physiological responses to HDM.
Figure 6: ASVs in the progeny mice conventionalized with antibiotic-perturbed microbiota correlate with elevated CD4+ T cell production of IL-13 and IL-17A.
Spearman correlations were used identify the colonic ASVs from the offspring-recipient mice sensitized and challenged with HDM (Figure 4A) that significantly correlate with lung CD4+ T cell production of A. IL-13 and B. IL-17A. Each row is an ASV that is significantly correlated with the frequency of cytokine produced by pulmonary Th cells. ASVs were considered based on nominal significance (p<0.05). The heatmap includes 39 colonic samples from 22 unique mice all of which were sensitized and challenged with HDM. Samples are annotated by source of microbiota, sensitization, timepoint at which the sample was collected, and the body site (colon or fecal pellet). The heatmap is colored by scaled (z-score) ASV relative abundance.
Discussion
Our data indicate that direct administration of either of the two most commonly prescribed antibiotics in pediatric practice 6, 16, 17 to murine pups causes sustained differences in gastrointestinal microbial communities, extending prior studies with antibiotic administration in drinking water that exposes both dams and pups 18, 19, 26. This was evident by the unweighted UniFrac analysis of stomach, cecum, and fecal samples of the antibiotic-treated mice which indicated long-term compositional differences; alpha-diversity was largely restored by the time of sacrifice (Figure 1B–E). Interestingly, although dams remained untreated in our studies, we still found sustained differences in the gastrointestinal microbiota of the antibiotic-exposed pups into adulthood. This reveals that there was not a substantial restoration of the pup microbiome from their mom to resemble an untreated mouse. This is consistent with prior work in which co-housing control and antibiotic exposed mice did not rescue microbiota-dependent immune phenotypes or restore microbiome composition 26, 29. This work provides experimental evidence consistent with the epidemiological data that early-life antibiotic use is associated with increased risk of asthma and related allergic diseases in children later in their life 6–8, 30. We found that azithromycin exposure during early-life heightened IgE and lung CD4+ T cell IL-13 production. That treatment of germ-free mice with antibiotics had no effect on allergen-specific immune responses indicates that the antibiotic effects require the presence of a microbiota as an intermediate. This is consistent with earlier studies, which found that treatment with the macrolide tylosin altered the immunological status of mice, but when germ-free mice were treated with tylosin, no immunological effects were observed 19. The lack of effect seen when transplanting the antibiotic-perturbed microbiota into adult germ-free mice, when taken together with the strong observed influence this microbial composition had on mice exposed to this microbiota from birth, provides evidence for a critical age window in which host microbial composition exerts immunomodulatory effects. This is in line with prior observations showing that human infant microbiota composition during the first 100 days of life is a critical determinant of asthma susceptibility 31 and adds to understanding of an early ontogenic window of opportunity for impacts of the intestinal microbiota on the immune landscape of the lung. This experiment also provides mechanistic evidence for a direct role of the perturbed microbiota in modulating allergy risk and severity, and further rules out direct effects of the antibiotic on host tissues in playing any substantial role.
Although broad spectrum super-therapeutic antibiotic regimens perturb immune dynamics in the lung and elsewhere 32–34, to our knowledge, no other studies have shown that early-life exposure to a single measured antibiotic at therapeutic levels alter the lung cytokine environment following antigen challenge. Similarly, no other studies have shown that clinically relevant doses, durations, and routes of antibiotic exposures were sufficient to alter gut microbial composition in mice for periods >6 weeks, and that microbiota composition was sufficient to alter the immune landscape and pulmonary function. Previous murine studies have shown that super-therapeutic antibiotics resulted in reduced Tregs in the lymph nodes draining the lungs 34. In our studies, there were no observed decreases in frequency or abundance of Tregs or pTregs in the lung tissue of the antibiotic-exposed mice before or after HDM challenge. Our results provide evidence that the antibiotic-driven pulmonary immune changes are not modulated by altered pulmonary Treg numbers but do not rule out functional differences of these T cells, or contributions of gastrointestinal tract-derived Tregs.
Although our studies indicate effects of a perturbed intestinal microbiota as a causal element, we did not examine contributions of viral, bacterial, or fungal infections, which lead to antibiotic prescriptions for children 4, 35, 36. We also did not find significant changes in the pulmonary microbiota, although it is known that airway microbial changes may influence asthma pathogenesis in humans 42 and mice 37, 38. The lower airway microbiota has also been associated with the development of wheeze in children 39, and pulmonary microbial signatures were found to cluster based on asthma endotype in adults 40. Despite no experimental evidence of the pulmonary microbiota influencing our observations, the contributions of the altered intestinal microbiota to the enhanced allergic/inflammatory phenomena were both substantial and consistent in the models we used.
A brief early-life amoxicillin course uniquely and persistently altered the gastric microbiota. Such effects can potentially influence host immunity, paralleling antibiotic abrogation of Helicobacter pylori immunomodulatory properties related to asthma 14, 41–43. Amoxicillin exposure altered IL-17 CD4+ T cell HDM responses, with phenotypes consistent with neutrophilic asthma 44. Amoxicillin-exposed mice had notable decreases in gastric and colonic Lactobacillus and Lachnospiraceae strains relative to controls. In prior murine studies, oral Lactobacillus administration reduced inflammation in models of antigen-driven lung inflammation 45, 46. Lachnospiraceae produce the short-chain fatty acid butyrate 47, which was shown to have anti-inflammatory effects in the lungs of OVA-sensitized and challenged mice 32, 48, 49. The mechanism for the amoxicillin-induced IL-17-dominanted immune responses to HDM may involve reductions of these important gastric taxa, potentially impacting gastric-derived pTregs, which have been reported to reduce HDM-associated pulmonary inflammation in murine models of H. pylori infection 50. In our own data, we found that increased colonic abundance of Lachnospiraceae was negatively associated with lung CD4+ T cell IL-17 production.
Mice that developed from birth in the presence of the azithromycin-perturbed microbiota had heightened Type 2 and Type 17 immune responses to HDM; the heightened Type 2 responses were also observed in SPF mice exposed to azithromycin during early-life. Azithromycin exposure reduced intestinal alpha-diversity until P40, a finding consistent with the association between reduced 1st-year of life bacterial diversity and higher subsequent asthma risk in human children 51. Germ-free mice have hyper-IgE responses and increased susceptibility to anaphylaxis; however this can be abrogated through conventionalization with a diverse microbiota during early-life but not later 52. These data support the hypothesis of a critical window of time for host-microbiota interactions which are dependent on both composition and age. ASVs that significantly correlated with CD4+ T cell cytokine production in the lung tissue of the mice that received antibiotic-perturbed microbiota (Figure 6) included Muribaculaceae, a family within the phylum Bacteroidetes, and Lachnospiraceae a family within the phylum Firmicutes, which both were reduced. In prior studies, exposure to these phyla over the first 3 years of life negatively correlated with atopy and atopic wheeze in children 53. Both phyla also contribute to short-chain fatty acid production 54, metabolites that reduce inflammation in other murine asthma models 32, 48, 49. Children with the highest levels of butyrate and propionate at one year of age are less likely to develop asthma during early childhood 48. Azithromycin-exposed mice had reduced SFB levels during early-life and enrichment for Akkermansia. SFB potently induce Th17 cells 55, loss of this taxon could lead to dysregulation of T cell differentiation and T cell immunity. Akkermansia has been shown to bloom following antibiotic treatment in both mice 18, 19, and humans 56, and to degrade mucins and generate metabolites including acetate and propionate 57. Although Akkermansia is a biomarker of recent antibiotic exposure, we are not able to conclude from these experiments the functional role of its antibiotic-induced bloom. These data from humans and mice suggest a time window during early-life in which protection against allergic diseases is associated with particular microbial compositions and metabolites. Our transfers to germ-free mice provide direct evidence that this developmental window is a critical variable.
In a Danish cohort, children diagnosed with asthma by age 7 already had decreased air flow and increased methacholine responsiveness at age 1 month compared to children who did not develop asthma later 58. Similarly, an abnormal microbiota at age 1 year was significantly associated with risk of asthma developing by age 6 59. Airway resistance in azithromycin-perturbed microbiota mice differing from controls (Figure 4E) without antigen challenge provides evidence for predisposition to altered lung physiology that is microbiome-dependent. This supports observations in these childhood cohorts 58, 59.
In total, our studies indicate a window during early-life, with conserved cross-talk between the assembling gastrointestinal microbiota and the host immune system that supports healthy development 60, but which can be abrogated by therapeutic antibiotic regimens that substantially affect microbiome composition. Despite their many clinical benefits, antibiotics disrupt the conserved early-life mutualism between host and microbes. Further understanding of the mechanisms by which perturbation of this microbiota-immune axis influences predisposition to asthma and other allergic disorders will inform methods in the clinic to mitigate or reverse these processes.
Materials and Methods
Mice.
C57BL/6J mice were purchased from Jackson Laboratories at New York University School of Medicine, or from Javier at the University of Zürich and acclimated to the animal facility for one week prior to breeding. C57BL/6 germ-free mice were obtained from Taconic Biosciences, and maintained in a laminar flow isolator at a germ-free facility at NYU School of Medicine. Mice were maintained on a 12-hour light/dark cycle and allowed ad libitum access to rodent standard lab chow and water. For germ-free mice, all food and water were autoclaved. All mouse experiments were approved by the New York University Langone Institutional Animal Care and Use Committee (IACUC protocol IA16-00785) and complied with federal and institutional regulations. All animal experimentation at the University of Zurich was reviewed and approved by the Zurich Cantonal Veterinary Office (licenses ZH170/2014 and ZH086/2020).
Antibiotic treatment.
Between P5–9, mice received either sterile water, or an oral suspension of 30mg/kg of active azithromycin (Teva, Fairfield NJ) or 100mg/kg of amoxicillin (Sigma, St. Louis MO). Oral suspensions were prepared in sterile water. Mouse pups were weighed, and the volume of the suspension adjusted for each mouse to achieve the desired daily dose, with a new suspension prepared every 48 hours. Dams remained untreated in these experiments. For antibiotic treatment of germ-free pups, autoclaved water was used to prepare the antibiotic suspension in a hood with laminar flow, and the suspension was filter-sterilized. New antibiotic suspensions were prepared for each germ-free treatment.
House dust mite (HDM) antigen model.
Lyophilized HDM antigen was purchased from Greer (XPB82D3A2.5, Cambridge MA) and dissolved in sterile PBS (Corning, Oneonta NY) at a concentration of 2mg/mL based on total protein concentration. At P45, offspring mice were sensitized intranasally with 1ug of HDM in 50ul PBS, or with PBS alone as a negative control. On P53, P55 and P57, mice were challenged intranasally with 10ug of HDM in 50ul PBS, or with PBS alone as a negative control. On P60, mice were euthanized, blood was collected by cardiac puncture, and the lungs perfused with PBS. Gastrointestinal tract samples were snap-frozen on dry ice and stored at −80°C.
BAL collection.
After tracheotomy and insertion of an 18-gauge cannula, BAL was collected. Total viable cells were counted by trypan blue dye exclusion, and differential cell counts were performed on cytocentrifuged samples stained with Shandon Kwik-Diff Stains (Thermo Scientific, Springfield Township NJ).
Lung histology.
For histopathology, lungs were fixed by means of inflation and immersion in 4% formalin and embedded in paraffin. Tissue sections were stained with hematoxylin and eosin (H&E) or periodic acid–Schiff (PAS). Images were evaluated blindly using a BX40 Olympus microscope (Olympus, Center Valley PA). Peribronchial inflammation was scored from 0–4. PAS-positive goblet cells were quantified per millimeter of basal membrane on at least three different representative airways on PAS-stained slides. More detailed information on scoring has been described previously 61.
IgE ELISA.
The left lung was collected and homogenized in T-PER tissue protein extraction reagent (Thermo Scientific) supplemented with Halt protease and phosphatase inhibitors (Thermo Scientific) to extract total protein. ELISA MAX Deluxe set mouse total IgE (BioLegend, San Diego CA) was used to quantify IgE levels in both serum and lung tissue homogenate. Lung tissue homogenate immunoglobulin concentrations were normalized to the total protein levels in the sample, quantitated by BCA assay (Pierce, Springfield Township NJ).
Flow Cytometry.
At mouse sacrifice, the middle lung lobe was perfused with PBS, excised, minced into small pieces using dissection scissors and submerged in DMEM (Corning) supplemented with 10% fetal calf serum (FCS) (Corning) and penicillin and streptomycin (Corning). Lungs were enzymatically digested in DMEM supplemented with 10% FCS, 0.5mg of Collagenase 1A (Sigma, United States) and 0.05mg of DNase I (Worthington Biochemical, Lakewood, NJ) for 35 minutes at 37°C with shaking and were subsequently mechanically disrupted. Lung suspensions were passed through a 70-micron filter (Milltenyi, Sommerville MA). For stimulation experiments, lung cell suspensions were cultured for 4 hours at 37°C in RPMI supplemented with PMA (Sigma), ionomycin (Sigma), golgi plug (BD), 10% FCS, beta mercaptoethanol, and penicillin/streptomycin. Cell suspensions were stained with fluorescently tagged antibodies (BioLegend San Diego, CA; BD Franklin Lakes, NJ; and eBiosciences San Diego, CA). All data was acquired on a BD LSRII flow cytometer at the NYU School of Medicine Flow Cytometry Core or a BD Fortessa at the University of Zurich Flow Cytometry Core. All data was analyzed using FlowJo (v10.6.1, BD). Count bright beads (Invitrogen) were used to determine cell counts in flow cytometry experiments.
FlexiVent and methacholine challenge.
Mice were anesthetized with a cocktail of ketamine and xylazine, tracheotomized and a 20-gauge metal cannula was inserted into the trachea and connected to the computer-operated animal ventilator (FlexiVent, Scireq, Montreal, Canada). Mice were mechanically ventilated and baseline measurements recorded based on standard Scireq protocols 62. A nebulizer was used to aerosolize PBS or increasing doses of methacholine (Sigma) were delivered to the mouse, and airway resistance measured. A dose response curve was generated using methacholine at doubling doses from 3.125mg/ml to 50mg/ml.
DNA isolation and 16S rRNA sequencing.
DNA was extracted from gastrointestinal samples with the DNeasy PowerSoil HTP 96 Kit (Qiagen, Germantown MD) or DNeasy PowerLyzer Powersoil kit (Qiagen). DNA was extracted from lung lobes using a DNeasy Blood and Tissue Kit (Qiagen). PBS, HDM, and no sample controls also were extracted to assess for background bacterial contamination of lung samples. The V4 region of the bacterial 16S rRNA gene was amplified in triplicate, using barcoded fusion primers (F515/R806) 63, then pooled, and DNA was quantified with Quant-iT PicoGreen (Invitrogen, Springfield Township, NJ). A maximum of 96 samples were then pooled in equal quantities of 300ng, and then purified with a QIAquick PCR purification kit (Qiagen) and quantified with a Qubit Fluorometer (Life Technologies, Springfield Township, NJ). Finally, these samples were pooled at an equal molar concentration (50nM) and sequenced on an Illumina MiSeq platform at the New York University School of Medicine Genome Technology Center. All data were processed and analyzed using Quantitative Insights into Microbial Ecology (QIIME2, version 2020.02) 64. Sequencing reads were trimmed and denoised using DADA2 65 and aligned using MAFFT 66. Taxonomy was assigned using Silva 138 (released December 2019), cyanobacteria, mitochondrial, and chloroplast sequences were removed, and only ASVs that were present in ≥10 samples and had a total count >15 across all samples were included in subsequent analyses. For the SPF experiments, 416 samples were included at a sequencing depth of 5.8 million reads with an average number of reads per sample of 13,527 (range 5042 – 84746). For the GF-transfer experiments, 285 samples were included at a sequencing depth of 5.6 million reads, at an average number of reads per sample of 18,773 (range 8962–99359). Beta diversity (unweighted UniFrac) analyses, rarefied at 5000 reads, were performed using the QIIME2 pipeline, and plots were generated in R using ggPlot2. Alpha diversity (Observed ASVs) and taxonomic abundance plots were generated using the Phyloseq package, and differential abundance plots and heatmaps were generated using the DESeq2 and Complex Heatmap packages in R 67, 68. Lung 16S rRNA samples were subject to background subtraction prior to analysis using the prevalence method of the Decontam open source R package using the conservative threshold of 0.5 69. The pipeline was used to remove contaminant ASVs found in background control samples (extraction blank, PCR negative control, PBS and HDM samples) from true lung samples. 16S copy number was determined in DNA samples by qPCR using SyberGreen (Applied Biosystems) and universal 16S primers 785F/907R, and normalized to the DNA concentration determined by the Quant-iT PicoGreen assay. A standard curve using a plasmid that contains the 16S rRNA gene from Propionibacterium acnes was used to determine 16S rRNA copy number.
Fecal transfer to germ-free mice.
Selected frozen mouse fecal pellets (25mg) were suspended in 1mL of Beef Broth medium (10.0g Beef Extract, 10.0g Peptone, 5.0g NaCl in 1.0L of water, pH 7.2), then disrupted by passage through a 1mL sterile syringe without needle attached, vortex-mixed, and immediately used for gavage into recipient mice. Sterile disposable 18G gavage needles (Fine Science Tools, Foster City CA) were used to transfer fecal suspensions to conventionalize germ-free mice. Fecal samples were collected periodically to determine engraftment and stability of the community over time.
Statistical Analyses.
Statistical analysis was performed with Prism 9.2.0 (GraphPad Software). Statistical comparisons between groups were performed using a Kruskal-Wallis test (one-way nonparametric ANOVA). To determine statistical significance between HDM sensitized and challenged mice, the matching PBS group was used as the reference for comparison: control vs. control; azithromycin vs. azithromycin; amoxicillin vs. amoxicillin. This was based on the a priori assumption that antibiotic treatment alone is sufficient to disrupt the immune status of HDM naïve mice 19. Differences were considered statistically significant when p < 0.05. Statistical significance between community structures was determined using an Adonis test and reported as significant when p < 0.01. Differentially abundant ASVs were determined by contrasting the relevant treatment groups and selecting for ASVs that were significant using a Benjamini Hochberg false discovery rate (FDR) of p < 0.01. Spearman correlations between all detectable genera between the donor, adult and offspring mice were used to detect the fidelity of microbiota transfer. This analysis considers all detectable bacteria genera and their group average relative abundance (e.g., the mean of each genus in each experimental group of mice). Correlations of the relative abundance of the genera were then conducted between donor and transfer groups to allow for evaluation of transfer strength (|ρ| > 0.4) and significance (p<0.05).
Supplementary Material
Key Messages.
Mice that acquired an azithromycin-perturbed microbiota from birth had heightened Th2 and Th17 responses, and airway reactivity to methacholine
The altered HDM phenotype could only be transferred to a new murine host when those mice acquired the perturbed microbiome at birth
Acknowledgements
We thank Dr. Leopoldo Segal, Dr. Yonghua Li, Dr. Juan Lafaille, Dr. Shruti Naik, and Angela Fallegger for scientific input and support for these studies. Dr. Elitza Ivanova for technical assistance with the methacholine challenge assay. And Drs Huilin Li and Kelly Ruggles for their input on data analysis. Study designs were generated using BioRender.
Funding
TCB was supported by the National Institutes of Health (TL1TR001447, T32ES007324, T32AI007180). This work was supported by NIH U01AI22285 and the Leducq and Sergei Zlinkoff foundations for M.J.B. and the Swiss National Science Foundation (BSCGIO 462 157841/1) for A.M. Flow cytometry technologies were provided by the NYU Langone Cytometry and Cell Sorting Laboratory, and 16SrRNA sequencing provided by the NYU Genome Technology Center and were supported in part by grant P30CA016087 from the National Institutes of Health/National Cancer Institute. Work in the SBK lab was supported by NIH R01HL125816. CT was support by the German Research Foundation (DFG) (Ta 275/7-1); SR was supported by the German Research Foundation (DFG) (RE 3652/4-1).
Data Availability
The data is available at the following link https://qiita.ucsd.edu/study/description/14608
Footnotes
Disclosure
The authors have declared that no conflicts of interest exist.
Disclosure
The authors declare no competing financial or conflicts of interest.
References
- 1.Braman SS. The global burden of asthma. Chest 2006; 130(1): 4S–12S. [DOI] [PubMed] [Google Scholar]
- 2.Svenningsen S, Nair P. Asthma Endotypes and an Overview of Targeted Therapy for Asthma. Front Med (Lausanne) 2017; 4: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med 2012; 18(5): 716–725. [DOI] [PubMed] [Google Scholar]
- 4.Sugita K, Sokolowska M, Akdis CA. Key points for moving the endotypes field forward. Implementing precision medicine in best practices of chronic airway diseases. Elsevier, 2019, pp 107–114. [Google Scholar]
- 5.Choy DF, Hart KM, Borthwick LA, Shikotra A, Nagarkar DR, Siddiqui S et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Science Translational Medicine 2015; 7(301): 301ra129. [DOI] [PubMed] [Google Scholar]
- 6.Metsälä J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Prenatal and post‐natal exposure to antibiotics and risk of asthma in childhood. Clinical & Experimental Allergy 2015; 45(1): 137–145. [DOI] [PubMed] [Google Scholar]
- 7.Aversa Z, Atkinson EJ, Schafer MJ, Theiler RN, Rocca WA, Blaser MJ et al. Association of Infant Antibiotic Exposure with Childhood Health Outcomes. Mayo Clinic Proceedings 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozyrskyj AL, Ernst P, Becker AB. Increased risk of childhood asthma from antibiotic use in early life. Chest 2007; 131(6): 1753–1759. [DOI] [PubMed] [Google Scholar]
- 9.Risnes KR, Belanger K, Murk W, Bracken MB. Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am J Epidemiol 2011; 173(3): 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patrick DM, Sbihi H, Dai DL, Al Mamun A, Rasali D, Rose C et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. The Lancet Respiratory Medicine 2020. [DOI] [PubMed] [Google Scholar]
- 11.Ahmadizar F, Vijverberg SJ, Arets HG, de Boer A, Lang JE, Garssen J et al. Early‐life antibiotic exposure increases the risk of developing allergic symptoms later in life: a meta-analysis. Allergy 2018; 73(5): 971–986. [DOI] [PubMed] [Google Scholar]
- 12.Metsälä J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology 2013: 303–309. [DOI] [PubMed] [Google Scholar]
- 13.Sun W, Svendsen ER, Karmaus WJ, Kuehr J, Forster J. Early-life antibiotic use is associated with wheezing among children with high atopic risk: a prospective European study. Journal of Asthma 2015; 52(7): 647–652. [DOI] [PubMed] [Google Scholar]
- 14.Borbet TC, Zhang X, Muller A, Blaser MJ. The role of the changing human microbiome in the asthma pandemic. J Allergy Clin Immunol 2019; 144(6): 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas M, Custovic A, Woodcock A, Morris J, Simpson A, Murray CS. Atopic wheezing and early life antibiotic exposure: a nested case–control study. Pediatric allergy and immunology 2006; 17(3): 184–188. [DOI] [PubMed] [Google Scholar]
- 16.Chai G, Governale L, McMahon AW, Trinidad JP, Staffa J, Murphy D. Trends of outpatient prescription drug utilization in US children, 2002–2010. Pediatrics 2012; 130(1): 23–31. [DOI] [PubMed] [Google Scholar]
- 17.Hales CM, Kit BK, Gu Q, Ogden CL. Trends in Prescription Medication Use Among Children and Adolescents-United States, 1999–2014. JAMA 2018; 319(19): 2009–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 2015; 6: 7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz VE, Battaglia T, Kurtz ZD, Bijnens L, Ou A, Engstrand I et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat Commun 2017; 8(1): 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016; 8(343): 343ra382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemppainen KM, Vehik K, Lynch KF, Larsson HE, Canepa RJ, Simell V et al. Association Between Early-Life Antibiotic Use and the Risk of Islet or Celiac Disease Autoimmunity. JAMA Pediatr 2017; 171(12): 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX et al. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell host & microbe 2018; 23(5): 653–660. e655. [DOI] [PubMed] [Google Scholar]
- 23.Roubaud-Baudron C, Ruiz VE, Swan AM, Vallance BA, Ozkul C, Pei Z et al. Long-Term Effects of Early-Life Antibiotic Exposure on Resistance to Subsequent Bacterial Infection. mBio 2019; 10(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X-S, Li J, Krautkramer KA, Badri M, Battaglia T, Borbet TC et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. Elife 2018; 7: e37816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nature microbiology 2019; 4(12): 2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Borbet TC, Fallegger A, Wipperman MF, Blaser MJ, Müller A. An Antibiotic-Impacted Microbiota Compromises the Development of Colonic Regulatory T Cells and Predisposes to Dysregulated Immune Responses. Mbio 2021; 12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulfer AF, Battaglia T, Alvarez Y, Bijnens L, Ruiz VE, Ho M et al. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol 2018; 3(2): 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piyadasa H, Altieri A, Basu S, Schwartz J, Halayko AJ, Mookherjee N. Biosignature for airway inflammation in a house dust mite-challenged murine model of allergic asthma. Biology open 2016; 5(2): 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulfer AF, Schluter J, Zhang Y, Brown Q, Pathmasiri W, McRitchie S et al. The impact of early-life sub-therapeutic antibiotic treatment (STAT) on excessive weight is robust despite transfer of intestinal microbes. The ISME journal 2019; 13(5): 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmadizar F, Vijverberg SJ, Arets HG, de Boer A, Turner S, Devereux G et al. Early life antibiotic use and the risk of asthma and asthma exacerbations in children. Pediatric Allergy and Immunology 2017; 28(5): 430–437. [DOI] [PubMed] [Google Scholar]
- 31.Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science Translational Medicine 2015; 7(307): 307ra152–307ra152. [DOI] [PubMed] [Google Scholar]
- 32.Cait A, Hughes M, Antignano F, Cait J, Dimitriu P, Maas K et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal immunology 2018; 11(3): 785–795. [DOI] [PubMed] [Google Scholar]
- 33.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences 2011; 108(13): 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adami AJ, Bracken SJ, Guernsey LA, Rafti E, Maas KR, Graf J et al. Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatr Res 2018; 84(3): 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almqvist C, Wettermark B, Hedlin G, Ye W, Lundholm C. Antibiotics and asthma medication in a large register-based cohort study – confounding, cause and effect. Clinical & Experimental Allergy 2012; 42(1): 104–111. [DOI] [PubMed] [Google Scholar]
- 36.Arrieta M-C, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. Journal of Allergy and Clinical Immunology 2018; 142(2): 424–434. e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor SL, Leong LE, Choo JM, Wesselingh S, Yang IA, Upham JW et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. Journal of Allergy and Clinical Immunology 2018; 141(1): 94–103. e115. [DOI] [PubMed] [Google Scholar]
- 38.Hanashiro J, Muraosa Y, Toyotome T, Hirose K, Watanabe A, Kamei K. Schizophyllum commune induces IL-17-mediated neutrophilic airway inflammation in OVA-induced asthma model mice. Scientific Reports 2019; 9(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L, Shen C, Chen Y, Yang X, Luo X, Hang C et al. Follow-up study of airway microbiota in children with persistent wheezing. Respir Res 2021; 22(1): 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Aziz MI, Brinkman P, Vijverberg SJH, Neerincx AH, Riley JH, Bates S et al. Sputum microbiome profiles identify severe asthma phenotypes of relative stability at 12 to 18 months. J Allergy Clin Immunol 2021; 147(1): 123–134. [DOI] [PubMed] [Google Scholar]
- 41.Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest 2011; 121(8): 3088–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyburz A, Fallegger A, Zhang X, Altobelli A, Artola-Boran M, Borbet T et al. Transmaternal Helicobacter pylori exposure reduces allergic airway inflammation in offspring through regulatory T cells. J Allergy Clin Immunol 2019; 143(4): 1496–1512 e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler DB, Reuter S, van Wijck Y, Urban S, Kyburz A, Maxeiner J et al. Effective treatment of allergic airway inflammation with Helicobacter pylori immunomodulators requires BATF3-dependent dendritic cells and IL-10. Proc Natl Acad Sci U S A 2014; 111(32): 11810–11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020; 52(2): 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proceedings of the National Academy of Sciences 2014; 111(2): 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu C-T, Chen P-J, Lee Y-T, Ko J-L, Lue K-H. Effects of immunomodulatory supplementation with Lactobacillus rhamnosus on airway inflammation in a mouse asthma model. Journal of Microbiology, Immunology and Infection 2016; 49(5): 625–635. [DOI] [PubMed] [Google Scholar]
- 47.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS microbiology letters 2009; 294(1): 1–8. [DOI] [PubMed] [Google Scholar]
- 48.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019; 74(4): 799–809. [DOI] [PubMed] [Google Scholar]
- 49.Yip W, Hughes MR, Li Y, Cait A, Hirst M, Mohn WW et al. Butyrate Shapes Immune Cell Fate and Function in Allergic Asthma. Frontiers in Immunology 2021; 12(299). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altobelli A, Bauer M, Velez K, Cover TL, Muller A. Helicobacter pylori VacA Targets Myeloid Cells in the Gastric Lamina Propria To Promote Peripherally Induced Regulatory T-Cell Differentiation and Persistent Infection. MBio 2019; 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abrahamsson T, Jakobsson H, Andersson AF, Björkstén B, Engstrand L, Jenmalm M. Low gut microbiota diversity in early infancy precedes asthma at school age. Clinical & Experimental Allergy 2014; 44(6): 842–850. [DOI] [PubMed] [Google Scholar]
- 52.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 2013; 14(5): 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. Journal of Allergy and Clinical Immunology 2014; 134(3): 593–601.e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444(7122): 1027–1031. [DOI] [PubMed] [Google Scholar]
- 55.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139(3): 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubourg G, Lagier J-C, Armougom F, Robert C, Audoly G, Papazian L et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. International Journal of Antimicrobial Agents 2013; 41(2): 149–155. [DOI] [PubMed] [Google Scholar]
- 57.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. International journal of systematic and evolutionary microbiology 2004; 54(5): 1469–1476. [DOI] [PubMed] [Google Scholar]
- 58.Bisgaard H, Jensen SM, Bønnelykke K. Interaction between asthma and lung function growth in early life. American journal of respiratory and critical care medicine 2012; 185(11): 1183–1189. [DOI] [PubMed] [Google Scholar]
- 59.Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 2018; 9(1): 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Research 2020: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reuter S, Lemmermann NAW, Maxeiner J, Podlech J, Beckert H, Freitag K et al. Coincident airway exposure to low-potency allergen and cytomegalovirus sensitizes for allergic airway disease by viral activation of migratory dendritic cells. PLOS Pathogens 2019; 15(3): e1007595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGovern TK, Robichaud A, Fereydoonzad L, Schuessler TF, Martin JG. Evaluation of respiratory system mechanics in mice using the forced oscillation technique. JoVE (Journal of Visualized Experiments) 2013; (75): e50172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A et al. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2016; 1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology 2019; 37(8): 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13(7): 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30(4): 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014; 15(12): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wipperman MF, Fitzgerald DW, Juste MAJ, Taur Y, Namasivayam S, Sher A et al. Antibiotic treatment for Tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci Rep 2017; 7(1): 10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018; 6(1): 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available at the following link https://qiita.ucsd.edu/study/description/14608