Abstract
To investigate the contribution of the TonB protein to high-affinity iron acquisition in Pseudomonas aeruginosa, we constructed tonB-inactivated mutants from strain PAO1 and its derivative deficient in producing the siderophores pyoverdin and pyochelin. The tonB mutants could not grow in a free-iron-restricted medium prepared by apotransferrin addition, even though the medium was supplemented with each purified siderophore or with a heme source (hemoglobin or hemin). The tonB inactivation was shown to make P. aeruginosa unable to acquire iron from the transferrin with either siderophore. Introduction of a plasmid carrying the intact tonB gene restored growth of the tonB mutant of PAO1 in the free-iron-restricted medium without any supplements and restored growth of the tonB mutant of the siderophore-deficient derivative in the medium supplemented with pyoverdin, pyochelin, hemoglobin, or hemin. In addition, animal experiments showed that, in contrast to PAO1, the tonB mutant of PAO1 could not grow in vivo, such as in the muscles and lungs of immunosuppressed mice, and could not kill any of the animals. The in vivo growth ability and lethal virulence were also restored by introduction of the tonB-carrying plasmid in the tonB mutant. These results indicate clearly that the intact tonB gene—and, therefore, the TonB protein encoded by it—is essential for iron acquisition mediated by pyoverdin and pyochelin and via heme uptake in P. aeruginosa and suggest that the TonB-dependent iron acquisition may be essential for P. aeruginosa to infect the animal host.
Iron is one of the essential elements for almost all bacteria, and the ability of pathogenic bacteria to acquire iron in hosts is essential for their growth and infection (7, 23). In animal hosts, iron is usually bound to proteins such as transferrin, lactoferrin, and ferritin and bound as heme to hemoglobin (Hb) and various enzymes (26, 41). To utilize such complexes as iron sources, bacteria possess some sophisticated mechanisms, including an iron uptake system mediated by high-affinity iron chelators called siderophores and a heme uptake system, which involve specific receptors (21, 26, 41). In these systems of gram-negative bacteria, a cytoplasmic membrane protein known as TonB is generally accepted to play a crucial role.
Although the tonB gene encoding the TonB protein has been identified in many gram-negative bacteria, the molecular location and functions of the protein have been primarily demonstrated by studies of Escherichia coli (reviewed in reference 6). The TonB protein is anchored via its N-terminal region to and associated with ExbB and ExbD proteins in the cytoplasmic membrane and in large part extends to the periplasm. The TonB protein is thought to change its conformation in response to the electrochemical potential (proton motive force) of the cytoplasmic membrane and thereby to interact with outer membrane receptor proteins (gated channels) for internalizing bound ligands. By using tonB mutants, it has been shown that uptake of iron-siderophore complexes and utilization of iron sources found in animal hosts, including heme, Hb, transferrin, and lactoferrin, are TonB-dependent processes in various bacteria (4, 12, 15, 16, 18, 33). As a consequence, it is likely that the TonB protein may contribute to the in vivo growth and virulence of pathogenic gram-negative bacteria. However, there are only a few reports that have addressed this point based on experimental facts (16, 36).
Pseudomonas aeruginosa, a ubiquitous gram-negative rod, is considered to be an important opportunistic pathogen and highly pathogenic for individuals with compromised immunity (5). This organism is able to acquire iron by means of siderophores and to utilize heme compounds as iron sources (19, 24, 27, 34). This bacterium also possesses a homolog of the TonB protein (29).
P. aeruginosa produces siderophores pyoverdin (Pvd) and pyochelin (Pch) (9, 10). It can use not only them but also heterologous siderophores, including enterobactin, to acquire iron (11, 27). Outer membrane proteins FpvA (28), FptA (1), and PfeA (11) were characterized as receptors for iron complexes of Pvd, Pch, and enterobactin, respectively. Initially, their TonB-dependencies in internalizing the ligands were speculated to be based on the homology between their amino acid sequences as deduced from the genes and those of other known TonB-dependent receptors. Thereafter, when the tonB gene in P. aeruginosa was identified by Poole et al. (29), it was shown that growth of Pvd-deficient tonB mutants in an iron-restricted medium was not observed even in the presence of Pvd or enterobactin. This finding suggested that iron acquisition via FpvA and PfeA might be a TonB-dependent process. Pch-mediated iron acquisition via FptA might also be a TonB-dependent process, but it has not been shown experimentally as yet. This point should be examined and clarified in order to understand fully the contribution of the TonB protein to the iron acquisition, ability to grow, and virulence of P. aeruginosa, because Pch shows a certain impact on P. aeruginosa infections (8, 34). In our previous study, the virulence of a Pch- and Pvd-deficient mutant derived from wild-type strain PAO1 was significantly attenuated in immunosuppressed mice in comparison to an isogenic Pvd-deficient mutant (34), indicating the contribution of Pch to the virulence.
In addition, it has not been established yet whether the heme utilization in P. aeruginosa is a TonB-dependent process or not. Heme utilization was also suggested to play an important role in P. aeruginosa infections (34). One of the heme uptake systems known to exist in this organism is the system mediated by an extracellular heme-binding protein, HasA, which was identified as a homolog of that in Serratia marcescens (19). A receptor responsible for the heme-HasA complex, HasR, in P. aeruginosa was also recently identified (25). Moreover, another heme uptake system in this organism was recently shown to be expressed from the phu locus, which consisted of the PhuR receptor gene and the phuSTUVW operon, which encodes a typical ATP binding cassette transporter (25). Although these receptors have also been assigned to the TonB-dependent family (R. E. W. Hancock laboratory website [http://www.cmdr.ubc.ca/bobh/TonBfamily.html]) based on the sequence homology determined from the gene analysis, their TonB dependencies have not been experimentally confirmed.
The purpose of the present study was to clarify whether iron acquisition mediated by the siderophores Pvd and Pch and heme utilization are TonB-dependent processes in P. aeruginosa and, furthermore, whether the TonB protein would be required for the infectivity of P. aeruginosa in the animal host. To achieve this purpose, we constructed P. aeruginosa tonB-inactivated mutants from wild-type strain PAO1 and its Pvd- and Pch-deficient derivative by allelic exchange and examined them in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. The P. aeruginosa mutants PAD08 and PAD14, plasmid pMMBD, and plasmids in the pHT series were generated in this study as described below. E. coli strains DH5α (TOYOBO, Tokyo, Japan) and S17-1 (32) were utilized as hosts for plasmid multiplication and donors for conjugal transfer and mobilization of plasmids, respectively. Media used were Luria (L) broth; Vogel-Bonner (VB) minimal medium (39), which is selective for P. aeruginosa; and succinate minimal medium (22) containing 0.2% Casamino Acids (SMMCA; the concentration of contaminating iron in this medium, measured with Fe-750 reagents [Eiken Chemical Co., Ltd., Tokyo, Japan], was less than 1 μM). Solid media were prepared by addition of agar (1.5%). Where appropriate, selective agents included in media were as follows: ampicillin, 100 μg/ml for E. coli; tetracycline, 10 μg/ml for E. coli and 50 μg/ml for P. aeruginosa; streptomycin, 500 μg/ml for P. aeruginosa; chloramphenicol, 200 μg/ml for P. aeruginosa; and carbenicilin, 400 μg/ml for P. aeruginosa. Unless otherwise stated, bacteria were cultured at 37°C. For conjugal transfer and mobilization of plasmids from E. coli to P. aeruginosa, the recipient cells were grown overnight at 43°C (38). For iron acquisition assays and animal experiments, P. aeruginosa strains were grown in SMMCA containing 10 or 40 μM FeSO4 for 16 h, harvested by centrifugation, and suspended and incubated in SMMCA containing 1 μM FeCl3 for an additional 4 h.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| Pseudomonas aeruginosa | ||
| PAO1 | Prototroph | |
| PAD06 | PAO1 ΔpvdA::ΩSm/Sp | 34 |
| PAD07 | PAO1 ΔpchD::Tc ΔpvdA::ΩSm/Sp | 34 |
| PAD08 | PAO1 ΔtonB::Tc | This study |
| PAD14 | PAO1 ΔpchD::Tc ΔpvdA::ΩSm/Sp ΔtonB | This study |
| Escherichia coli | ||
| DH5α | recA1 endA1 gylA96 thi-1 hsdR17 supE44 Δ(lac)U169 (φ80dlacΔM15) | TOYOBO |
| S17-1 | pro thi recA hsdR Tpr Smr; chromosomally integrated RP4-2-Tc::Mu-Km::Tn7; mobilizer of plasmids carrying the R68-derived Mob region | 32 |
| Plasmids | ||
| pMT5059 | Apr; pBR322 derivative carrying multiple cloning sites and a NotI site | 38 |
| pMT5071 | Kmr Cmr; pMOB3 derivative carrying NotI-flanked Mob cassette with ΩCm gene cartridge | 37 |
| pMT5056 | Apr Tcr; pBend2 derivative carrying Tcr gene cartridge flanked with multiple restriction enzyme sites | 38 |
| pMMB67EH | Apr Cbr; multi-host range tacP expression vector | 13 |
| pMMBD | Apr Cbr; pMMB67EH derivative in which PvuII-EcoRI fragment including lacIq and tac promoter is deleted | This study |
| pHT007 | Apr; pMT5059 carrying tonB gene in EcoRI-BamHI site | This study |
| pHT011 | Apr Tcr; pMT5059::ΔtonB::Tc | This study |
| pHT018 | Apr; pMT5059::ΔtonB | This study |
| pHT014 | Apr Cbr; pMMBD carrying tonB gene in EcoRI-BamHI site | This study |
Tpr, trimethoprim resistant; Smr, streptomycin resistant; Mob, plasmid mobilization; Apr, ampicillin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Tcr, tetracycline resistant; Cbr, carbenicilin resistant.
Recombinant DNA techniques.
Established procedures were used for preparation of plasmids, DNA manipulation, agarose gel electrophoresis, and transformation of E. coli (20). Competent cells of E. coli were prepared as described elsewhere (30). Plasmids and DNA fragments were purified with a commercially available kit (Prep-A-Gene DNA purification systems; Bio-Rad).
PCR and gene cloning.
Bacterial chromosomal DNA was extracted with TRIzol LS Reagent (Life Technologies) as described by the manufacturer. The tonB gene (on a DNA fragment corresponding to base positions 5 to 1165 of GenBank sequence U23764) was amplified from the chromosomal DNA of P. aeruginosa strain PAO1 by PCR with synthesized primers 5′-CGGAATTCGCGGAATGATCCGCCAAGGT-3′ (sense) and 5′-GAAGATCTGCGCGGCTCTTTTCGTTGTC-3′ (antisense); the fragment contains 101 nucleotides upstream of the tonB start codon to 31 nucleotides downstream of the tonB stop codon. The 5′ region of the sense primer was artificially flanked with two additional bases, CG, plus EcoRI sequence (GAATTC), and that of the antisense primer was artificially flanked with two additional bases, GA, plus BglII sequence (AGATCT). The PCR was performed under the same conditions as described previously (34) except for the addition of 10% dimethyl sulfoxide to the reaction mixture (29) in the present case. The amplified tonB gene was separated by agarose gel electrophoresis and purified from the gel. After digestion with EcoRI and BglII, it was cloned into the EcoRI-BamHI site of pMT5059 (38), resulting in pHT007. The purified tonB gene, which included the original promoter region, was also cloned into the EcoRI-BamHI site of pMMBD which was generated from pMMB67EH (13) by deletion of the PvuII-EcoRI fragment containing lacIq and tac promoter, resulting in pHT014.
Construction of mutants.
Allelic exchange mutagenesis of the P. aeruginosa chromosome was carried out with a system already established (31, 34, 38). The pMT5059 derivative carrying the tonB gene, pHT007, was digested with NaeI for deletion of an internal part of the gene (approximately 0.3 kb); into this plasmid was inserted a StuI-flanked tetracycline-resistant gene cartridge (Tcr; 1.6 kb) excised from pMT5056 (38), resulting in pHT011. An 8.5-kb NotI fragment containing the mobilization cassette derived from pMT5071 (37) was subsequently inserted into the NotI site of pHT011 carrying ΔtonB::Tc. The plasmid thus constructed was conjugally mobilized from E. coli strain S17-1 to P. aeruginosa strain PAO1. Then, P. aeruginosa transconjugants were selected on VB agar plates containing tetracycline. A colony of tetracycline-resistant transconjugants was next spread onto L agar plates containing tetracycline, 5% sucrose, and 40 μM FeSO4 for selection of an allelic exchange mutant, PAD08. When chromosomal DNA of this mutant was subjected to PCR under the same conditions as those for amplification of the normal tonB gene (1.2 kb), a size change was observed in the PCR product as expected (2.5 kb), indicating that the expected allelic exchange had successfully occurred in the mutant obtained. On the other hand, the NaeI-digested plasmid pHT007 was subjected to self-ligation, resulting in pHT018 carrying a partially deleted tonB gene. The mobilization cassette was subsequently inserted into the NotI site of pHT018 carrying ΔtonB. The generated plasmid was, similar to that described above, introduced into the Pvd- and Pch-deficient mutant, PAD07 (PAO1 ΔpchD::Tc ΔpvdA::ΩSm) (34); then, transconjugants were selected on VB agar plates containing chloramphenicol. A colony of chloramphenicol-resistant transconjugants was next spread onto L agar plates containing 5% sucrose and 40 μM FeSO4. Sucrose-resistant colonies were screened by PCR for those possessing only the tonB (0.9-kb) deletion in the chromosomal DNA, and a positive clone was selected as an allelic exchange mutant, PAD14. It was also confirmed by PCR under the conditions described previously (34) that mutations in pchD and pvdA were maintained in this tonB mutant (data not shown). The mutants generated in the present study, as well as PAO1, were all susceptible to carbenicillin and chloramphenicol (the MICs of carbenicillin and chloramphenicol for the mutants in SMMCA supplemented with 40 μM FeSO4 were 64 and 16 μg/ml, respectively), indicating that the vector plasmid which mediated the allelic exchange did not remain in the mutants.
Transformation of P. aeruginosa.
P. aeruginosa strains PAD08 and PAD14 were transformed with pMMBD and pHT014 by conjugal mobilization from E. coli strain S17-1 carrying the plasmid. P. aeruginosa transformants were selected on L agar plates containing tetracycline, carbenicillin, and 40 μM FeSO4.
Purification of siderophores.
Pvd was purified from the supernatant (2 liter) of a 2-day culture of P. aeruginosa strain PAO1 in succinate minimal medium. The culture supernatant was concentrated to approximately 60 ml by evaporation and treated with ethyl acetate (60 ml). Then, the collected aqueous phase was applied to a column (2.5 by 30 cm) of ion-exchange resin (DIAION HP20; Mitsubishi Chemical Co., Tokyo, Japan) which had been equilibrated with distilled water. After elution of nonbinding substances with water, the bound material was fractionated by elution with water-ethanol (1:1). Fractions corresponding to a major peak with absorption at 280 nm were collected and evaporated. Dried substances were dissolved again in distilled water and lyophilized. The final extract (ca. 140 mg) was stored as Pvd at −20°C and dissolved in distilled water just before use. Pch was extracted and purified as described below, based on previous reports (2, 10). The supernatant (1 liter) of an overnight culture of PAD06, a Pvd-deficient mutant derived from strain PAO1 (34), in SMMCA containing 1 μM FeCl3 was made acidic (pH 2 to 3) with HCl and treated with ethyl acetate (500 ml). The organic phase was evaporated, dissolved again in a small volume of chloroform, and applied onto a preparative silica thin-layer plate for chromatography in chloroform-acetic acid-ethanol (19:1:1). A fluorescent band corresponding to an Rf of 0.35 to 0.40 under UV light was scraped from the plate, and the fluorescent substance was eluted with dichloromethane-ethanol (1:1) and evaporated. After weighing the final extract (ca. 5 mg) as Pch, it was dissolved again in the dichloromethane-ethanol, divided into several vials, evaporated again, and stored at −20°C. Pch was dissolved in ethanol just before use. Purified Pvd and Pch were identified by analysis with 1H-nuclear magnetic resonance and measurements of absorption spectra and specific fluorescence (2, 9, 10) (data not shown).
In vitro growth assays.
Assays were performed in 96-well round-bottom plates as described previously (34). A free-iron-restricted medium used was made by addition of 25 μM apotransferrin (apoTsf; from bovines; Life Technologies) and 20 mM sodium bicarbonate to SMMCA containing 10 μM FeCl3. When required, the medium was supplemented with twofold serial dilution of FeSO4, Pvd, Pch, Hb (from bovines; Sigma), or hemin (Hm) (Sigma). After inoculation of bacteria at approximately 105 CFU/ml, assay plates were incubated without shaking under 5% CO2 and 95% air for 20 h. Bacterial growth of the cultures was measured as the optical density at 590 nm (OD590).
Preparation of [59Fe]transferrin.
The apoTsf was dissolved at 100 μM in nitrogen-free succinate minimal medium (nf-SMM) containing 20 mM sodium bicarbonate, and 1 ml of the apoTsf solution was mixed with 10 μl of 59FeCl3 solution (0.257 mg/ml in 0.5 N HCl; specific activity, 13.8 mCi/mg; NEN Life Science Products). An hour after incubation at 37°C under 5% CO2 and 95% air, the mixture was filtered with centrifugal filter units (Ultrafree C3-LTK; nominal molecular-weight limit, 30,000; Millipore) and [59Fe]transferrin recovered on the filter was dissolved again in the same medium to its initial volume. This process was repeated three times to remove free iron.
Iron acquisition assay.
Bacterial suspension (approximately 109 CFU/ml in the sodium bicarbonate-containing nf-SMM) was dispensed into wells (50 μl/well) of a 96-well filtration plate (0.45-μm Durapore type; MultiScreen; Millipore) and mixed with Pvd or Pch dissolved at various concentrations in the sodium bicarbonate-containing nf-SMM (40 μl/well). The assay was initiated by addition of the [59Fe]transferrin solution (10 μl/well) and lasted for 1 h at 37°C under 5% CO2 and 95% air. Bacterial cells were then harvested on the filters of the well bottoms and washed twice with 1 mM ethylenediamine di(o-hydroxyphenylacetic acid) solution by filtration with a vacuum system (MultiScreen filtration system vacuum manifold; Millipore). After drying of the filters and addition of scintillation cocktail (Microscint O; Packard Instrument), the radioactivity associated with bacteria was measured as 59Fe uptake by bacteria with a scintillation counter for multiwell plates (TopCount; Packard Instrument).
Animal experiments.
Animal experiments were performed as described previously (34). Briefly, male ddY mice (5 to 6 weeks of age; Japan SLC Ltd., Hamamatsu, Japan), which had received intraperitoneal injections of cyclophosphamide (3 mg/mouse) for immunosuppression, received intramuscular or intranasal inoculations with bacteria suspended in saline. At various times after the bacterial inoculation, the muscles or lungs were collected and assayed for viable bacteria. L agar plates supplemented with 40 μM FeSO4 were used for the assays. In the animal studies, we followed the animal experimentation guidelines of the Daiichi Pharmaceutical Co., Ltd., Animal Care and Use Committee.
RESULTS
Construction of P. aeruginosa tonB mutants and their demand for free iron for growth.
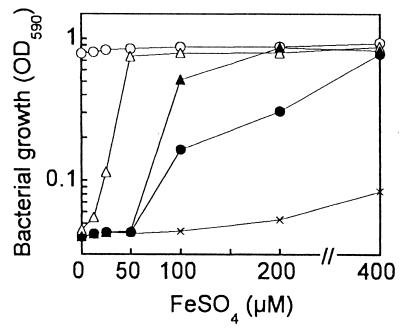
Before this study, it was reported that P. aeruginosa tonB-inactivated mutants could not be obtained from Pvd-producing strains (29). However, by an established allelic-exchange procedure (31, 34, 38), we succeeded in generating a tonB mutant, PAD08, from the wild-type strain PAO1 capable of producing both Pvd and Pch. In fact, PAD08 was confirmed to produce the siderophores, when examined as described previously (34) (data not shown). In addition, another tonB mutant, PAD14, was constructed from a PAO1-derived mutant, PAD07 (34), deficient in both Pvd and Pch production. To obtain these tonB mutants, we modified the medium used for final selection; namely, we supplemented the L agar containing 5% sucrose with a substantial amount of ferrous salt (40 μM FeSO4). The modification was made on the hypothesis that, if the tonB inactivation resulted in impairment of some high-affinity iron acquisition systems, much iron in the medium would be needed for efficient growth (colony formation) of the tonB mutant.
Neither of the P. aeruginosa tonB mutants obtained could grow in a free-iron-restricted medium prepared by apoTsf addition (25 μM apoTsf in SMMCA containing 10 μM ferric iron) (34), in which the iron was expected to exist as transferrin. The tonB mutants grew, however, when the medium was supplemented with excess FeSO4 (more than 100 μM) (Fig. 1). As shown previously (34) and in Fig. 1, the Pvd- and Pch-deficient strain PAD07 hardly grew in the apoTsf-added medium, but this strain fully grew in the medium supplemented with FeSO4 at 50 μM (Fig. 1). The feature of PAD07 must be inherited by its tonB mutant, PAD14. However, compared with PAD07, PAD14 required more free iron for growth in the medium (Fig. 1). Additionally, the siderophore-producing tonB mutant PAD08 required more free iron for growth than PAD14 did (Fig. 1). Thus, the tonB mutants showed high demand for free iron for their growth.
FIG. 1.
Effects of ferrous salt on growth of P. aeruginosa strain PAO1 and its mutants in a free-iron-restricted medium, SMMCA containing 10 μM ferric iron together with 20 mM sodium bicarbonate and 25 μM apoTsf. Bacterial growth was measured as the OD590 of the culture 20 h after inoculation of the bacteria at approximately 105 CFU/ml. Symbols: ○, strain PAO1; ●, tonB mutant of PAO1, PAD08; ▵, Pvd- and Pch-deficient mutant of PAO1, PAD07; ▴, tonB mutant of PAD07, PAD14; ×, no inoculum (medium control).
Requirement of the P. aeruginosa tonB gene for siderophore-mediated iron acquisition.
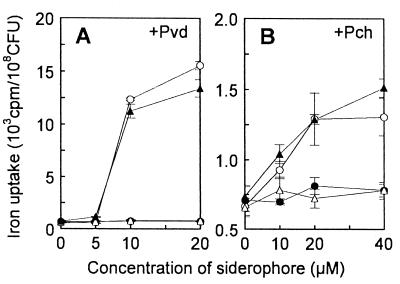
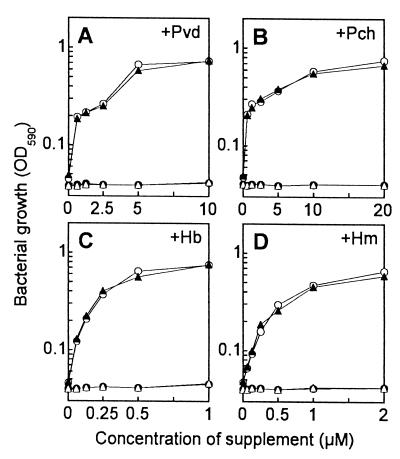
The inability of the siderophore-producing tonB mutant PAD08 to grow in the apoTsf-added medium without excess FeSO4 (Fig. 1) suggested impairment of iron acquisition from transferrin mediated by both Pvd and Pch. The mutant PAD08 could not grow in the medium even after supplementation with either purified siderophore (data not shown). Accordingly, we examined the iron acquisition from [59Fe]transferrin by PAD08. Iron uptake by this mutant was not promoted in the presence of purified Pvd or Pch, whereas the uptake by the parental strain PAO1 was promoted in a manner dependent on increased concentrations of each siderophore supplemented (Fig. 2), indicating that the tonB inactivation made P. aeruginosa unable to acquire iron with either siderophore. On the other hand, by introduction of a plasmid carrying the intact tonB gene (pHT014), but not by that of the vector plasmid (pMMBD), the ability of PAD08 to acquire iron in the presence of Pvd (Fig. 2A) and Pch (Fig. 2B) and to grow in the apoTsf-added medium without any supplements (data not shown) was restored. In contrast, the ability of the siderophore-deficient tonB mutant PAD14 to grow in the medium was restored neither by introduction of the tonB-carrying plasmid nor by the siderophore supplementation to the medium (Fig. 3A and B). However, PAD14 transformed with the tonB-carrying plasmid [PAD14(pHT014)] grew as well as PAD07, the parental strain of PAD14, in the medium supplemented with Pvd (Fig. 3A) or Pch (Fig. 3B), depending on increased concentrations of each siderophore. These results indicate that the tonB gene is essential for the iron acquisition mediated by Pvd and Pch in P. aeruginosa.
FIG. 2.
Effects of Pvd (A) and Pch (B) on iron acquisition from [59Fe]transferrin by P. aeruginosa strain PAO1 (○), its tonB mutant PAD08 (●), and transformants of PAD08 with a plasmid carrying the intact tonB gene (pHT014) (▴) or the vector plasmid (pMMBD) (▵). Iron uptake by bacteria was measured as radioactivity (counts per minute) associated with bacterial cells after incubation for 1 h and was shown as the radioactivity corrected by dividing by the count (CFU) of viable cells subjected to the assay. Each datum point represents the mean ± standard deviation (error bar) for triplicate determinations.
FIG. 3.
Effects of Pvd (A), Pch (B), Hb (C), and Hm (D) on growth of P. aeruginosa PAO1-derived Pvd- and Pch-deficient mutant PAD07 (○), its tonB mutant PAD14 (●), and PAD14 transformed with a plasmid carrying the intact tonB gene (pHT014) (▴) or the vector plasmid (pMMBD) (▵) in a free-iron-restricted medium, SMMCA containing 10 μM ferric iron together with 20 mM sodium bicarbonate and 25 μM apoTsf. Bacterial growth was measured as the OD590 of the culture 20 h after inoculation of the bacteria at approximately 105 CFU/ml.
Requirement of the P. aeruginosa tonB gene for heme utilization.
The influence of tonB inactivation on heme utilization in P. aeruginosa was also examined by the bacterial growth assay. As results, even though Hb or Hm was supplemented as a heme source into the apoTsf-added free-iron-restricted medium, no growth was observed for the tonB mutants PAD08 (data not shown) and PAD14 (Fig. 3C and D). However, the transformant of PAD14 with the tonB-carrying plasmid [PAD14(pHT014)] grew as well as PAD07, the parental strain of PAD14, in the presence of Hb (Fig. 3C) or Hm (Fig. 3D). It was notable that the concentrations of the heme sources (0.5 to 2 μM) that supported fully the bacterial growth were much lower than those of FeSO4 (more than 100 μM; Fig. 1) and siderophores (5 to 20 μM; Fig. 3A and B). These results indicate that the tonB gene is essential for the high-affinity heme utilization in P. aeruginosa.
Requirement of the P. aeruginosa tonB gene for in vivo bacterial growth and virulence.
Furthermore, to investigate the contribution of the tonB gene to the infectivity of P. aeruginosa, we inoculated wild-type strain PAO1, its tonB mutant PAD08, and transformants of the mutant into immunosuppressed mice. Bacterial growth was evaluated as the increase in the number of viable cells at sites (the calf muscles or lungs) where bacteria had been introduced. Virulence of the bacteria was assessed as lethality in the mice. The tonB mutant PAD08, in contrast to PAO1, did not show growth or rather showed a decrease in the number of viable bacteria with time in both the muscles after intramuscular inoculation (Table 2) and the lungs after intranasal inoculation (Table 3). Irrespective of the inoculation route, the tonB mutant could not kill the mice at all, whereas PAO1 killed almost all of the animals within a day (Tables 2 and 3). Even though PAD08 was intramuscularly inoculated at an approximately 10-fold-greater inoculum size compared with that of PAO1, it was defective in its growth and virulence expression (Table 2). On the other hand, by the transformation with the tonB-carrying plasmid (pHT014), PAD08 was completely restored to growth in the lungs and expression of the lethality in the mice, but not by that with the vector plasmid (pMMBD) (Table 3). These results indicate that the tonB gene is required for the growth ability and virulence of P. aeruginosa during infection in the animal host.
TABLE 2.
Growth and virulence of P. aeruginosa strain PAO1 and its tonB mutant PAD08 in immunosuppressed mice after intramuscular inoculation
| Inoculum
|
Count of viable bacteriaa in muscles at time (hpi):
|
Lethalityb | |||
|---|---|---|---|---|---|
| Strain | Log10 CFU | 16 | 40 | 64 | |
| PAO1 | 6.32 | 9.28 ± 0.13 | NDc | ND | 5/5 |
| PAD08 | 6.40 | 4.54 ± 0.09 | 3.63 ± 0.20 | 3.48 ± 0.42 | 0/5 |
| 7.40 | 7.39 ± 0.18 | 6.17 ± 0.40 | 3.94 ± 0.05 | 0/5 | |
Samples were collected from three mice per group at the indicated times (hours postinoculation [hpi]) and assayed for viable bacteria. Values are mean log10 CFU ± standard deviations.
Number of dead mice at 24 h postinfection/number of mice inoculated for observation of bacterial virulence.
ND, not done because of the death of all mice inoculated.
TABLE 3.
Growth and virulence of P. aeruginosa strain PAO1 and its tonB mutant PAD08 in immunosuppressed mice after intramuscular inoculation
| Strain inoculated | Count of viable bacteriaa in the lungs at time (hpi):
|
Lethalityb | |||
|---|---|---|---|---|---|
| 1 | 12 | 36 | 60 | ||
| PAO1 | 5.73 ± 0.37 | 8.01 ± 0.41 | NDc | ND | 6/7 |
| PAD08 | 6.28 ± 0.09 | 5.33 ± 0.30 | <2.50 ± 0.19 | <2.30 | 0/7 |
| PAD08(pMMBD) | 6.42 ± 0.10 | 5.50 ± 0.23 | 2.69 ± 0.48 | <2.30 | 0/7 |
| PAD08(pHT014) | 6.39 ± 0.23 | 8.16 ± 0.33 | ND | ND | 6/7 |
Samples were collected from three or five mice per group at the indicated times (hours postinoculation [hpi]) and assayed for viable bacteria. Values are mean log10 CFU ± standard deviations. The lowest limit of bacterial detection was 2.30; the limit value was included for calculation of the mean if data were below the limit.
Number of dead mice at 24 h postinfection/number of mice inoculated for observation of bacterial virulence.
ND, not done because of the death of almost all mice inoculated.
DISCUSSION
We clarified that the tonB gene—and, therefore, the TonB protein encoded by it—is essential for Pch-mediated iron acquisition and heme utilization in P. aeruginosa. Pvd-mediated iron acquisition was also confirmed to be a TonB-dependent process as reported previously (29). Furthermore, we demonstrated the requirement of the P. aeruginosa tonB gene for infection.
In the present study, we used two types of P. aeruginosa tonB mutants, which were constructed by allelic exchange from wild-type strain PAO1 and its Pvd- and Pch-deficient mutant. A tonB mutant (PAD08) derived from a Pvd-producing strain of P. aeruginosa (PAO1) is for the first time reported here. Our success in obtaining such a mutant, we believe, is attributable to our inclusion of a considerable amount of ferrous salt in the medium for final selection. It was reported that a similar approach resulted in the successful generation of a Xanthomonas campestris pv. campestris tonB mutant (40). As we speculated before and confirmed after generating P. aeruginosa tonB mutants, they showed increased demand for free iron for their growth (Fig. 1). A higher free-iron demand of the Pvd- and Pch-deficient tonB mutant PAD14, compared with the demand of parental strain PAD07, suggests that besides iron acquisition systems mediated by Pvd an Pch, there might be another TonB-dependent mechanism unknown but related to the iron assimilation in P. aeruginosa. In this regard, the heme uptake system is not included, because the medium used did not originally contain heme sources. In addition, a higher free-iron demand of the siderophore-producing tonB mutant PAD08, compared with the demand of PAD14, suggests that the siderophore production might negatively influence the iron utilization by the tonB mutant. We assume that Pvd secretion may result in retention of some level of the free-iron-restricted condition in the apoTsf-added medium even after ferrous salt supplementation, for Pvd is able to bind and oxidize ferrous ion, as reported recently (42). When a P. aeruginosa tonB mutant, such as PAD08, senses iron restriction, the mutant probably falls into a “dilemma” between an accelerated production of siderophores, resulting in iron chelation, and the inability to take up iron-siderophore complexes. This speculation may explain the comparative difficulty in generating the tonB mutant from a Pvd-producing strain, which was previously pointed out by Poole et al. (29).
Our results showing the requirement of the tonB gene for iron acquisition mediated by Pvd and Pch and heme utilization in P. aeruginosa (Fig. 2 and 3) strongly support the TonB dependency of outer membrane receptors involved in such high-affinity iron acquisition, which has heretofore been primarily speculative based on the genetic information. However, receptor proteins FpvA and FptA, responsible for ferric Pvd and Pch, respectively, are known to lack the amino acid sequence corresponding to the so-called TonB box (1, 28), which is usually found at the N terminus of TonB-dependent receptor proteins and has been supposed to interact with the TonB protein in E. coli (6), whereas another receptor protein of P. aeruginosa, PfeA for ferric enterobactin, contains the TonB box (11). Consequently, the so-called TonB box is not thought to be always required for the cooperation of receptors with the TonB protein in P. aeruginosa. It is possible that the secondary structure corresponding to the so-called TonB box, rather than its primary sequence, might be crucial and/or that the direct contact site of the TonB-dependent receptor with the TonB protein might be present at regions different from the so-called TonB box.
One of the important results obtained in the present study is that the P. aeruginosa tonB mutant was unable to grow and express virulence in immunosuppressed mice (Tables 2 and 3). The findings in vivo, combined with those in vitro, imply that TonB-dependent iron acquisition may be essential for P. aeruginosa infection of the animal host. Earlier we demonstrated that the production of both Pvd and Pch—and, therefore, iron acquisition with these siderophores—has a considerable impact on the bacterial growth and virulence of P. aeruginosa in mice (34). At the same time, based on a certain virulence and the heme utilization ability of a Pvd- and Pch-deficient mutant (PAD07), we proposed an important role of heme uptake as non-siderophore-mediated iron acquisition in P. aeruginosa infections (34). Our present data further support this proposal, since the P. aeruginosa tonB mutant (PAD08) which was defective in heme utilization in addition to the siderophore-mediated iron acquisition was defective in experimental infections. Additionally, as we have speculated on the presence of another TonB-dependent mechanism undefined but related to iron assimilation, it is possible that such a mechanism might contribute, in part, to P. aeruginosa growth in vivo.
Furthermore, it is also possible as a reason for the reduced infectivity of the tonB mutant that the tonB inactivation might affect some mechanisms, besides the iron acquisition system, related to bacterial growth ability and virulence in vivo. Vitamin B12 uptake is a well-known TonB-dependent process in E. coli (3). In Aeromonas hydrophilia, a TonB-like protein, ExeB, is suggested to function in an exotoxin secretion system (14). If the TonB protein played a similar role in secretion of some virulence factors, the tonB inactivation would attenuate bacteria for the infection. For a much better understanding of the contribution of the TonB protein to the infectivity of P. aeruginosa, possible functions or roles of the protein in this organism must be further investigated in the future.
The tonB gene of Haemophilus influenzae type b is known to be essential for the virulence expression in infections of infant rats induced by intraperitoneal and intranasal inoculations (16). In this organism, the utilization of heme and transferrin-bound iron was demonstrated to be a TonB-dependent process (15, 16). On the other hand, as tonB inactivation did not attenuate Salmonella enterica serovar Typhimurium for infection of mice by the intraperitoneal route (36), the TonB protein is not always required for virulence. When inoculated by the intragastric route, the serovar Typhimurium tonB mutant was certainly attenuated and at a disadvantage during colonization of Peyer's patches and mesenteric lymph nodes in mice but not during colonization of the intestinal lumen, liver, and spleen (36). Thus, the contribution of TonB-dependent systems to the bacterial infection appears to vary among bacterial genera and infection sites of the host.
Under the anaerobic conditions found in the intestine, soluble ferrous iron may be available for bacteria. The Feo system, independent of the TonB protein for its function, is known to be responsible for ferrous iron uptake by enteric bacteria, E. coli and serovar Typhimurium (17, 36). The presence of Feo homologs in P. aeruginosa has been recently supposed based on the whole genome analysis (Pseudomonas Genome Project website [http://www.pseudomonas.com/]). In serovar Typhimurium, the feo mutation reduced an ability of the bacterium to colonize the mouse intestine, and the double mutation in feo and tonB resulted in further reduction of this ability (36). Since P. aeruginosa is able to cause a variety of infections, including intestinal disorders and intestine-derived sepsis, in immunocompromised hosts (5, 35), it is of interest to determine whether the TonB-dependent system contributes to such infections by this organism. In addition, molecular and physiological characterization of the Feo-like system in P. aeruginosa is needed in order to advance our understanding of the iron acquisition mechanisms in this organism and the relationship between them and bacterial pathogenesis.
ACKNOWLEDGMENTS
We thank N. Gotoh and M. Tsuda for their gifts of bacterial strains and plasmids and M. Takemura and Y. Ishida for their contribution to the siderophore purification processes.
ADDENDUM
While the present work was being reviewed, a paper by Zhao and Poole (43) was published. The paper (43) showed that the tonB1 gene, which is the same gene as the tonB gene we have focused on in our study, was essential for heme utilization in P. aeruginosa. This is consistent with our results.
REFERENCES
- 1.Ankenbauer R G, Quan H N. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J Bacteriol. 1994;176:307–319. doi: 10.1128/jb.176.2.307-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankenbauer R G, Toyokuni T, Staley A, Rinehart K, Jr, Cox C D. Synthesis and biological activity of pyochelin, a siderophore of Pseudomonas aeruginosa. J Bacteriol. 1988;170:5344–5351. doi: 10.1128/jb.170.11.5344-5351.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassford P J, Jr, Bradbeer C, Kadner R J, Schnaitman C A. Transport of vitamin B12 in tonB mutants of Escherichia coli. J Bacteriol. 1976;128:242–247. doi: 10.1128/jb.128.1.242-247.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas G D, Anderson J E, Sparling P F. Cloning and functional characterization of Neisseria gonorrhoeae tonB, exbB and exbD genes. Mol Microbiol. 1997;24:169–179. doi: 10.1046/j.1365-2958.1997.3421692.x. [DOI] [PubMed] [Google Scholar]
- 5.Bodey G P, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 6.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 7.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 8.Cox C D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect Immun. 1982;36:17–23. doi: 10.1128/iai.36.1.17-23.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox C D, Adams P. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect Immun. 1985;48:130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox C D, Graham R. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J Bacteriol. 1979;137:357–364. doi: 10.1128/jb.137.1.357-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean C R, Poole K. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J Bacteriol. 1993;175:317–324. doi: 10.1128/jb.175.2.317-324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost G E, Rosenberg H. Relationship between the tonB locus and iron transport in Escherichia coli. J Bacteriol. 1975;124:704–712. doi: 10.1128/jb.124.2.704-712.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fruste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 14.Howard S P, Meiklejohn H G, Shivak D, Jahagirdar R. A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol Microbiol. 1996;22:595–604. doi: 10.1046/j.1365-2958.1996.d01-1713.x. [DOI] [PubMed] [Google Scholar]
- 15.Jarosik G P, Maciver I, Hansen E J. Utilization of transferrin-bound iron by Haemophilus influenzae requires an intact tonB gene. Infect Immun. 1995;63:710–713. doi: 10.1128/iai.63.2.710-713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarosik G P, Sanders J D, Cope L D, Muller-Eberhard U, Hansen E J. A functional tonB gene is required for both utilization of heme and virulence expression by Haemophilus influenzae type b. Infect Immun. 1994;62:2470–2477. doi: 10.1128/iai.62.6.2470-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kammler M, Schon C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koebnik R, Baumler A J, Heesemann J, Braun V, Hantke K. The TonB protein of Yersinia enterocolitica and its interactions with TonB-box proteins. Mol Gen Genet. 1993;237:152–160. doi: 10.1007/BF00282796. [DOI] [PubMed] [Google Scholar]
- 19.Létoffé S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 21.Marinez J L, Delgado-Iribarren A, Baquero F. Mechanisms of iron acquisition and bacterial virulence. FEMS Microbiol Rev. 1990;75:45–56. doi: 10.1111/j.1574-6968.1990.tb04085.x. [DOI] [PubMed] [Google Scholar]
- 22.Meyer J M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 23.Neilands J B. Iron absorption and transport in microorganisms. Annu Rev Nutr. 1981;1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 24.Noya F, Arias A, Fabiano E. Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J Bacteriol. 1997;179:3076–3078. doi: 10.1128/jb.179.9.3076-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 26.Otto B R, Verweij-van Vudht A M J J, MacLaren D M. Transferrin and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 27.Poole K, Dean C, Heinrichs D, Neshat S, Krebs K, Young L, Kilburn L. Siderophore-mediated iron transport in Pseudomonas aeruginosa. In: Nakazawa T, editor. Molecular biology of Pseudomonas. Washington, D.C.: American Society for Microbiology; 1996. pp. 371–383. [Google Scholar]
- 28.Poole K, Neshat S, Krebes K, Heinrichs D E. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J Bacteriol. 1993;175:4597–4604. doi: 10.1128/jb.175.15.4597-4604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole K, Zhao Q, Neshat S, Heinrichs D E, Dean C R. The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology. 1996;142:1449–1458. doi: 10.1099/13500872-142-6-1449. [DOI] [PubMed] [Google Scholar]
- 30.Schweizer H P. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 32.Simon R, O'Connell M, Labe M, Puhler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 33.Stojiljkovic I, Srinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in Neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834–1839. [DOI] [PMC free article] [PubMed]
- 35.Tancrede C H, Andremont A O. Bacterial translocation and gram-negative bacteremia in patients with hematological malignancies. J Infect Dis. 1985;152:99–103. doi: 10.1093/infdis/152.1.99. [DOI] [PubMed] [Google Scholar]
- 36.Tsolis R M, Baumler A J, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuda M. Use of a transposon-encoded site-specific resolution system for construction of large and defined deletion mutations in bacterial chromosome. Gene. 1998;207:33–41. doi: 10.1016/s0378-1119(97)00601-x. [DOI] [PubMed] [Google Scholar]
- 38.Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–431. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel H J, Bonner D M. Acetylornithase of E. coli: partial purification and properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 40.Wiggerich H G, Klauke B, Koplin R, Priefer U B, Puhler A. Unusual structure of the tonB-exb DNA region of Xanthomonas campestris pv. campestris: tonB, exbB, and exbD1 are essential for ferric iron uptake, but exbD2 is not. J Bacteriol. 1997;179:7103–7110. doi: 10.1128/jb.179.22.7103-7110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 42.Xiao R, Kisaalita W S. Fluorescent pseudomonad pyoverdines bind and oxidize ferrous ion. Appl Environ Microbiol. 1998;64:1472–1476. doi: 10.1128/aem.64.4.1472-1476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Q, Poole K. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett. 2000;184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]