This is a correction to Yogananda CG, Shah BR, Nalawade SS, et al. MRI-based deep-learning method for determining glioma MGMT promoter methylation status. AJNR Am J Neuroradiol 2021;42:845–52 [10.3174/ajnr.A7029] [33664111]
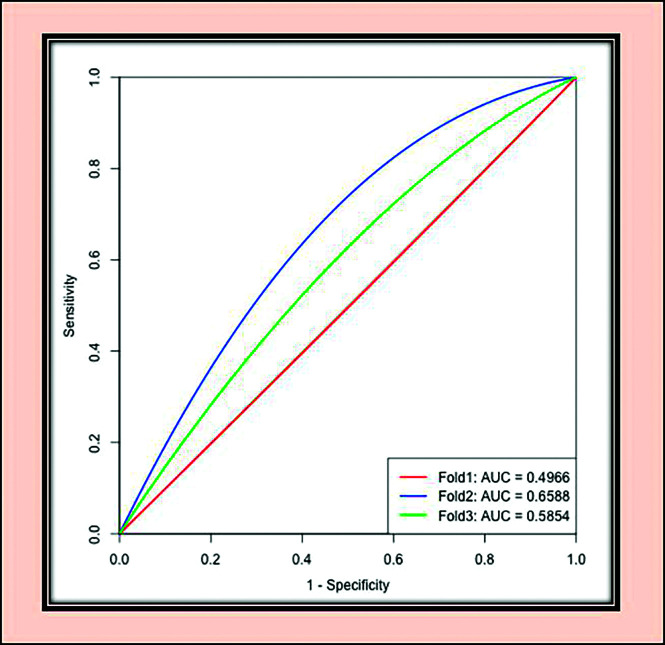
There was an error in the Python code for the 3-fold cross-validation procedure. This resulted in the use of the training cases instead of the set-aside test cases for the testing procedure for molecular marker accuracy. This caused our reported accuracies from the TCIA/TCGA data set to be artificially inflated. The corrected accuracies for the Table (computed using nnU-Net1), along with the updated receiver operating characteristic (ROC) curve for Fig 3 are provided here. The updated accuracies do not outperform other reported methods for MGMT molecular marker prediction using MR imaging.
Cross-validation results
| Fold Description |
MGMT-Net |
||
|---|---|---|---|
| % Accuracy | AUC | Dice Score | |
| Fold no. | |||
| Fold 1 | 59.75 | 0.4966 | 0.7906 |
| Fold 2 | 73.49 | 0.6588 | 0.7725 |
| Fold 3 | 64.63 | 0.5854 | 0.7874 |
| Average | 65.95 (SD, 0.06) | 0.5802 (SD, 0.081) | 0.7835 (SD, 0.009) |
FIG 3.
ROC analysis for MGMT-net. Separate curves are plotted for each cross-validation fold along with corresponding area under the curve (AUC) values.
Reference
- 1.Isensee F, Jaeger PF, Kohl SAA, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203–11 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]