Abstract
Background
Microphthalmia-associated transcription factor (MITF) is a master regulator of melanogenesis and is mainly expressed in melanoma cells. MITF has also been reported to be expressed in non-pigmented cells, such as osteoclasts, mast cells, and B cells. However, the roles of MITF in immunosuppressive myeloid cells, including myeloid-derived suppressor cells (MDSCs), remain unclear. Here, we investigated the role of MITF in the differentiation process of MDSCs during tumor development.
Methods
In vitro-generated murine MDSCs and primary MDSCs from breast cancer-bearing mice or lung carcinoma-bearing mice were used to determine the expression level of MITF and the activity of MDSCs. Additionally, we investigated whether in vivo tumor growth can be differentially regulated by coinjection of MDSCs in which MITF expression is modulated by small molecules. Furthermore, the number of MITF+ monocytic (MO)-MDSCs was examined in human tumor tissues or tumor-free lymph nodes by immunohistochemistry (IHC).
Results
The expression of MITF was strongly increased in MO-MDSCs from tumors of breast cancer-bearing mice compared with polymorphonuclear MDSCs. We found that MITF expression in MDSCs was markedly induced in the tumor microenvironment (TME) and related to the functional activity of MDSCs. MITF overexpression in myeloid cells increased the expression of MDSC activity markers and effectively inhibited T-cell proliferation compared with those of control MDSCs, whereas shRNA-mediated knockdown of MITF in myeloid cells altered the immunosuppressive function of MDSCs. Modulation of MITF expression by small molecules affected the differentiation and immunosuppressive function of MDSCs. While increased MITF expression in MDSCs promoted breast cancer progression and CD4+ or CD8+ T-cell dysfunction, decreased MITF expression in MDSCs suppressed tumor progression and enhanced T-cell activation. Furthermore, IHC staining of human tumor tissues revealed that MITF+ MO-MDSCs are more frequently observed in tumor tissues than in tumor-free draining lymph nodes obtained from patients with cancer.
Conclusions
Our results indicate that MITF regulates the differentiation and function of MDSCs and can be a novel therapeutic target for modulating MDSC activity in immunosuppressive TMEs.
Keywords: myeloid-derived suppressor cells; tumor microenvironment; immunomodulation; biomarkers, tumor
WHAT IS ALREADY KNOWN ON THIS TOPIC
Microphthalmia-associated transcription factor (MITF) is a key regulator of melanogenesis and modulates proliferation and development of melanocytes. MITF controls differentiation of other cell types, such as osteoclasts, mast cells, B cells, and natural killer cells. However, its role on the expression of MITF in immunosuppressive myeloid cells, such as myeloid-derived suppressor cells (MDSCs), as well as its role in MDSCs, has not been reported.
WHAT THIS STUDY ADDS
MITF is strongly expressed in monocytic (MO)-MDSCs in the tumor microenvironment. Suppression of MITF expression with shRNAs or inhibitor reduced the expression of MDSC activity markers. Moreover, we observed that the modulation of MITF expression in MDSCs affects T-cell activity and tumor progression in both of in vitro and in vivo.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our findings unveil an important role of MITF in immunoregulatory function of MDSCs, suggesting that targeting of MITF in MDSCs could be a novel strategy for improving therapeutic efficacy in cancer.
Introduction
Anticancer drugs mainly induce cancer cell regression and T-cell activity. However, tumor progression proceeds through a complicated network between diverse immune cells and cancer cells. Although numerous studies have attempted to increase T-cell function, there are no effective clinical therapies to overcome drug resistance in patients with cancer. Tumor progression and resistance are related to myeloid-derived suppressor cell (MDSC) accumulation in tumor tissues to enhance tumor immune evasion.1 2 In recent studies, combination therapies of antitumor drugs, immune checkpoint inhibitors (ICIs), and molecules targeting immune suppressive myeloid cells, such as MDSCs, have been used to overcome disease progression and drug resistance in cancers, including breast cancer, lung cancer, and melanoma.3 4
MDSCs endow the tumor microenvironment (TME) with a strong immunosuppressive function and promote tumor progression; these cells can be classified into two subsets in mice, either polymorphonuclear (PMN)-MDSCs (Ly6G+Ly6ClowCD11b+) or monocytic (MO)-MDSCs, Ly6G−Ly6ChighCD11b+), depending on various stimulating factors, such as granulocyte–macrophage colony-stimulating factor (GM-CSF), vascular endothelial growth factor (VEGF), interleukin (IL)-6, and IL-1β.5 6 In humans, PMN-MDSCs express CD15+CD14−HLA-DR−CD11b+, and MO-MDSCs are defined as CD15−CD14+HLA-DR−CD11b+. However, phenotypical markers in humans and mice are not sufficient to distinguish MDSCs from neutrophils and monocytes. Therefore, it is essential to develop specific biomarkers that can discriminate between MDSCs and other myeloid cells.7 8 Activated MDSCs participate in extensive crosstalk with various cells, including CD4/CD8 T cells through arginase 1 (Arg1), inducible nitric oxide synthase (iNOS or NOS2), and indoleamine-2,3-dioxygenase, and suppress the immune response to tumor cells.9 10 Tumor-associated MDSCs are stimulated by hypoxic conditions and induce the expression of hypoxia-inducible factor-1α (HIF-1α). HIF-1α increases the expression of Arg1, iNOS, and programmed death-ligand 1 (PD-L1), which enhances the suppressive activity of MDSCs on T cells and interrupts ICI therapy.11 Numerous studies have explored the mechanisms that disrupt MDSC activation, but there has been little success thus far in inhibiting suppressive crosstalk with various immune cells. Thus, further studies are required to determine how to alter or block the mechanisms of MDSC activation.
Microphthalmia-associated transcription factor (MITF) is a master regulator of the expression of various genes related to survival, metastasis, cell cycle arrest, and differentiation. The MITF promoter is mainly targeted by diverse transcription factors, such as cyclic AMP regulatory element-binding protein, SRY-related high-mobility group box 10, and paired box 3 (PAX3).12 13 In addition, MITF was shown to target HIF-1α, which is known to be increased by tumor conditions and enhances the expression of Arg1 through binding to promoter site HIF-1α.14 15 Interestingly, MITF is expressed in other cell types, including osteoclasts, mast cells, B cells, and natural killer (NK) cells.16–18 Altered MITF expression in osteogenesis regulates osteoclast differentiation. The role of MITF in the maintenance of the mature and resting B-cell state has also been demonstrated. Dysfunction of MITF expression induces the differentiation of B cells into plasma cells and increases autoantibody production via inhibition of IRF4 expression.16 19 However, the effect of MITF on other myeloid cells is poorly understood.
In the present study, we demonstrate the functional role of MITF in MDSCs in the TME. An increase in MITF expression was observed in tumor-associated MDSCs in vivo, and MITF expression in bone marrow (BM)-MDSCs was markedly induced by tumor cell-conditioned medium (TCCM) used to mimic the TME. We found that MITF regulates the immunosuppressive function of MDSCs obtained from mouse breast and lung cancer models and the antitumor effect of T cells in a breast cancer model. In addition, MITF was able to modulate the expression of Arg1 via HIF-1α. Furthermore, MITF expression was correlated with poor survival in patients with breast and lung cancers, and elevated numbers of MITF+ MO-MDSCs were located in the tumor tissues of patients with lung and head and neck (H&N) cancers compared with tumor-draining lymph nodes without metastasis. Therefore, our findings suggest that MITF can modulate the immunosuppressive activity of MDSCs in the TME and can be a novel target for developing biomarkers of MDSCs in the TME.
Materials and methods
Mice and tumor models
Female Balb/c mice at 6–8 weeks of age were purchased from Daehan Biolink (Eumseong, Chungcheongbuk-Do, South Korea). Tumor models were generated by subcutaneous injection of 5×105 4T1 cells/mouse (or PBS alone as a control) into the mammary fat pad. Tumor growth was measured using digital calipers at the indicated time points (A×B2)/2 (mm3) (A>B). Tumor-bearing mice were sacrificed humanely within a month.
Differentiation of murine BM-derived MDSCs in vitro
BM cells were obtained from mouse femurs. BM cells were treated with red blood cell (RBC) lysing buffer (Sigma-Aldrich, St. Louis, Missouri, USA) for the depletion of RBCs. For the depletion of lymphocytes, two monoclonal antibodies (GK1.5 and J11d.2) were added to the cells for 30 min at 4℃ and cytotoxicity medium containing Low Tox-M Rabbit Complement (Cedarlane Laboratory, Hornby, Canada) was used to remove the lymphocytes. Next, the remaining cells were cultured in RPMI 1640 with 10% heat-inactivated FBS in the presence of 10 ng/mL GM-CSF (PeproTech, Rocky Hill, New Jersey, USA), in the absence or presence of 10 ng/mL IL-6 (PeproTech) and 30% TCCM in an atmosphere of 5% CO2 in a 37°C humidified incubator. The cells were collected on day 4 for cell analysis.
Isolation of MDSCs
Single cell suspensions from spleens and tumor tissues of tumor-bearing mice were stained with antibodies (online supplemental table 1), and then the CD45+CD11b+Gr1+ (MDSCs), CD45+CD11b+Ly6ChighLy6G− (MO-MDSCs) or CD45+CD11b+Ly6ClowLy6G+ (PMN-MDSCs) populations were sorted by a S3 Cell Sorter (Bio-Rad Laboratories, Hercules, California, USA) with purities of >85%.
jitc-2022-005699supp001.pdf (6.9MB, pdf)
For analysis of the effects of MDSCs on tumor growth, MDSCs were purified from the spleens of tumor-bearing mice using a MACS MDSC Isolation Kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Splenic MDSCs (5×105 cells/mL) were cultured either alone (DMSO) or with compounds (3-isobutyl-1-methylxanthine (IBMX)) and ML-329) in complete medium for 48 hours.
In vivo bioluminescence imaging
ML-329- or DMSO-treated MDSCs (4×105 cells/mouse) were coinjected subcutaneously with 4T1-luc2 cells (4×105 cells/mouse) into Balb/c mice. In another in vivo experiment, 4T1-luc2 cells (2×105 cells/mouse) were subcutaneously injected into Balb/c mice with or without IBMX- or DMSO-treated MDSCs (4×105 /mouse). Tumor-bearing mice were given an intraperitoneal injection of 3 mg/mouse D-luciferin (Promega, Madison, Wisconsin, USA) dissolved in saline. After the injection, the mice were anesthetized with isoflurane (2% in 1 L/min oxygen), and bioluminescence images were acquired using the IVIS Lumina III XRMS (PerkinElmer, Waltham, Massachusetts, USA). Tumor growth was monitored every 2–3 days.
Transfection assay
The control and MITF shRNAs were purchased from Sigma-Aldrich. pBabe-empty and pBabe-MITF vectors were obtained from Addgene (Watertown, Massachusetts, USA). All plasmid DNAs were transfected into myeloid cells using Lipofectamine LTX & Plus reagents (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. The transfected cells were cultured in complete medium with 10 ng/mL GM-CSF for 4 days. MITF expression was confirmed by western blot analysis or real-time PCR on day 3.
T-cell proliferation assay
Splenic CD3+ cells obtained from Balb/c mice were sorted using a S3 Cell Sorter (Bio-Rad Laboratories). Sorted CD3+ T cells with high purity (> 95%) using an antibody (145–2 C11, eBioscience) were stained with 2.5 µM CFSE for 7 min at room temperature using a CellTrace CFSE Cell Proliferation Kit (Invitrogen). An equal volume of filtered-FBS was added to CD3+ T cells and incubated for 3 min. T cells were stimulated with 3 µg/mL anti-CD3 (145–2 C11, eBioscience) and 1 µg/mL anti-CD28 (37.51, eBioscience) antibodies and cocultured with MDSCs in complete medium for 72 hours. Then, CD8+ T-cell proliferation was analyzed by flow cytometry.
Patients and histological evaluation
We retrospectively collected tissue from 20 patients with lung cancer and H&N cancer who underwent surgery at Seoul National University Hospital (Seoul, South Korea) in 2019. None had received chemotherapy before surgery or had distant metastasis at the time of diagnosis. Patients with non-small cell lung cancer (n=10) and H&N squamous cell carcinoma (n=10) were included in this cohort.
Representative formalin-fixed paraffin-embedded blocks of tumor and matched tumor-draining lymph nodes (TDLNs) without metastasis were collected in each case. Serial sections of 4 µm were cut from the paraffin blocks. Mouse monoclonal anti-human MITF (clone C5/D5; RTU, Ventana Medical Systems, Tucson, Arizona, USA), mouse monoclonal anti-human CD11b (CL1719; 1:1000, ATLAS Antibodies AB, Täby, Sweden), and mouse monoclonal anti-human CD14 (CL1638, 1:1000, ATLAS Antibodies AB) antibodies were used to identify MO-MDSCs. All immunohistochemistry (IHC) studies were performed using the Ventana Benchmark XT automated staining system (Ventana Medical Systems) according to the manufacturer’s protocol.
Nuclear immunoreactivity for MITF and membranous to cytoplasmic staining for CD14 and CD11b were evaluated. Normal human tonsil tissue was used as a positive control. Serial sections were used to evaluate MDSCs with CD14, CD11b, and MITF. MO-MDSCs were defined as CD14+, CD11b+, and MITF+ cells in the same location. Three high-power fields (HPFs) were evaluated in each case, and the absolute numbers of triple-positive cells were calculated per HPF. The mean number of MO-MDSCs per HPF in tumors and lymph nodes of each case was recorded.
Statistical analysis
Statistical analysis was performed using Student’s t-test and one-way analysis of variance (Tukey’s post hoc test for multiple comparisons) with GraphPad PRISM software V.9 (GraphPad Software, San Diego, California, USA). All results are presented as the mean±SEM; p values of <0.05 were considered to be statistically significant.
Other materials and methods are provided in the online supplemental materials and methods of this paper.
Results
MITF expression is associated with increased MDSC infiltration into tumors
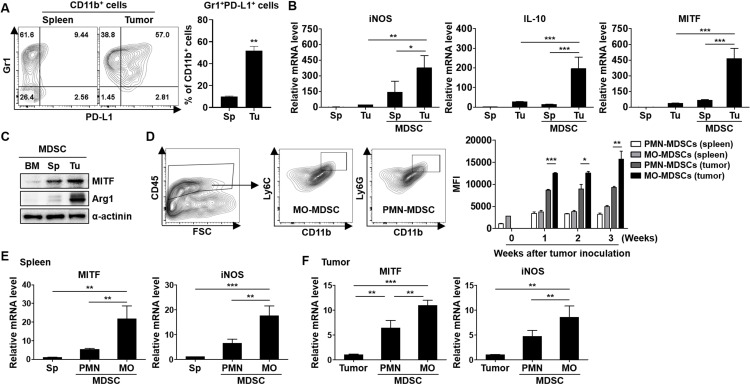
When the MDSC population in the TME was measured, the number of MDSCs was increased in the splenocytes and tumor tissues at the indicated time points (online supplemental figure S1A). We found that MITF expression was gradually increased in the tumor-exposed MDSCs, depending on tumor development (online supplemental figure S1B). To elucidate the interaction between MITF expression and the immunosuppressive activity in MDSCs, we initially investigated whether an increased expression level of PD-L1, an activation marker of MDSCs,20 is observed in tumor-associated MDSCs compared with splenic MDSCs from mice bearing 4T1 tumors (figure 1A). Concurrently, the elevation of iNOS, IL-10, and MITF mRNA levels was observed in the tumor-exposed MDSCs compared with the splenic MDSCs (figure 1B). An increase in the protein expression of Arg1 and MITF was also detected in the tumor-exposed MDSCs compared with the splenic or BM-derived MDSCs (figure 1C). These data indicate that MITF expression may be related to MDSC infiltration into tumors and the immunosuppressive activity of tumor-associated MDSCs. Next, we compared the expression levels of MITF in myeloid cells from splenocytes and tumor tissues of the 4T1-bearing mice at various time points. Interestingly, MITF expression was markedly increased in MDSCs compared with dendritic cells and macrophages, depending on tumor progression (online supplemental figure S2A, B). To investigate the origin of MITF expression in myeloid cells, we analyzed MITF+ cells from tumors. MITF+ cells were mainly observed in CD11b+Gr1+ cells and were related to tumor development, but this correlation was not detected in MITF− cells (online supplemental figure S2C). We then explored whether MITF expression was differentially expressed in different subsets of MDSCs. The expression was strongly increased in the MO-MDSCs from tumors of the 4T1-bearing mice compared with PMN-MDSCs (figure 1D). The expression levels of iNOS and MITF mRNAs in the splenic MDSCs or tumor-associated MDSCs were significantly increased in the MO-MDSCs (figure 1E, F). These findings suggest that MITF expression in MDSCs is induced by tumor conditions, and that elevated MITF expression may promote the activation and differentiation of MDSCs, especially MO-MDSCs.
Figure 1.
Expression of MDSC activation markers and MITF in Sp MDSCs or Tu-associated MDSCs of Tu-bearing mice. (A) Flow cytometry analysis of single cells isolated from spleen and Tu tissues from the 4T1 Tu-bearing mice on day 21 after Tu inoculation. The expression levels of CD45, CD11b, Gr1, and PD-L1 were analyzed (n=4–5). (B) mRNA expression in Sp MDSCs and Tu-MDSCs obtained from the mice bearing 4T1 Tus was detected by real-time PCR. (C) MITF and Arg1 expression in BM (BM-derived MDSCs), Sp (Spc MDSCs), and Tu (Tu-MDSCs) was evaluated by Western blot analysis. (D) The gating strategy for MO-MDSC and PMN-MDSCs in Tus followed by staining with Ly6C and Ly6G antibodies is shown. The splenocytes and Tu cells were stained with CD11b, Ly6C, Ly6G, CD45, and MITF antibodies and analyzed by flow cytometry. The graphs represent MITF expression in the subtype of MDSCs from Tu-free or Tu-bearing mice at various time points (n=3–4). (E, F) mRNA expression levels of MITF in MDSCs obtained from splenocytes and Tu cells were measured by real-time PCR. The experiments were independently repeated at least three times. *P<0.05, **P<0.01, ***P<0.001. Arg1, arginase 1; BM, bone marrow; IL, interleukin; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; MO, monocytic; PMN, polymorphonuclear; Sp, splenic; Tu, tumor.
TME induces MITF expression in MDSCs
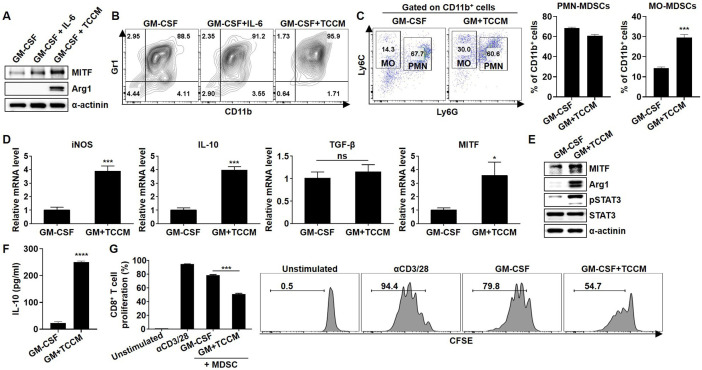
Previous studies have reported that IL-6 or the TME induces immunosuppressive activity in MDSCs.8 Therefore, we investigated whether MITF expression in BM-MDSCs can be induced by IL-6 or TCCM. Indeed, the expression of MITF and Arg1 was increased in the TCCM-treated BM-MDSCs (figure 2A). Consequently, although we found that MITF expression was strongly induced by IL-6 treatment, experiments were usually performed using MDSC-like cells (MDSC-LCs) after treatment with GM-CSF only. Next, we explored whether MDSC subpopulations are differentially regulated by the TME, and the proportion of MO-MDSCs was significantly increased after TCCM treatment compared with that of PMN-MDSCs (figure 2B, C). Increases in the expression levels of MDSC activation-related factors, such as Arg1, pSTAT3, iNOS, and IL-10,6 were detected in BM-MDSCs after TCCM treatment, and MITF expression was also significantly induced in the TCCM group (figure 2D, E). Consistent with the mRNA expression results, the amount of secreted IL-10 was markedly increased in the TCCM-treated BM-MDSCs (figure 2F). MDSCs have been shown to play a central role in the inhibition of T-cell proliferation through various mechanisms.21 Intriguingly, CD8+ T-cell proliferation was more strongly inhibited after incubation with TCCM-treated BM-MDSCs than after incubation with GM-CSF-treated BM-MDSCs (figure 2G), suggesting a change in MITF expression in BM-MDSCs by molecules present in the TME. In fact, IL-18 or IL-10 treatment of BM-MDSCs, which has been shown to promote suppressive activity of MDSCs,22 23 elevated the expression levels of MDSC-inhibitory factors and MITF in the presence of GM-CSF, depending on the exposure time and concentration (online supplemental figure S3A–H). Furthermore, when BM cells were cultured in the presence of IL-18 for 96 hours, the IL-10 level was increased after treatment with 50 ng/mL IL-18 (online supplemental figure S3I). In addition, IL-10 production in 4T1 cells was confirmed after TCCM treatment (online supplemental figure S3J). Accordingly, these data suggest that MITF is markedly induced by soluble factors present in the TME, indicating that increased MITF expression can be associated with the immunosuppressive function of MDSCs.
Figure 2.
Induction of BM-MDSC activation and MITF expression by TCCM. BM cells were obtained from the femurs of Balb/c mice, and then lymphocytes were depleted. The cells were cultured in fresh medium with 10 ng/mL GM-CSF in the absence or presence of 10 ng/mL IL-6 or 30% TCCM for 96 hours. (A) MITF and Arg1 in BM-MDSCs were determined by western blot analysis. (B) BM-MDSCs were stained with CD11b and Gr1 antibodies and analyzed by flow cytometry. (C) Cells were stained with CD11b, Ly6C, and Ly6G antibodies and analyzed by flow cytometry. (D) The mRNA expression of iNOS, IL-10, TGF-β, and MITF in BM-MDSCs was measured by real-time PCR. (E) MITF, pSTAT3, and Arg1 were evaluated by western blot analysis. (F) BM cells were treated with TCCM for 96 hours and then cultured in serum-free medium. After 48 hours, the supernatant was collected and used to detect IL-10 by ELISA. (G) Splenic CD3+ T cells were labeled with 2.5 µM CFSE. CFSE-labeled T cells were stimulated with 3 µg/mL plate-bound anti-CD3 mAb and 1 µg/mL soluble anti-CD28 mAb for 2 hours. BM-MDSCs were cocultured with CFSE-labeled CD3+ T cells for 72 hours, and then CD8+ T-cell proliferation was measured by flow cytometry at an MDSC:T-cell ratio of 1:2. All experiments were independently repeated at least three times. *P<0.05, ***P<0.001, ****P<0.0001. Arg1, arginase 1; BM, bone marrow; GM, granulocyte–macrophage; GM-CSF, granulocyte–macrophage colony-stimulating factor; IL, interleukin; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; MO, monocytic; PMN, polymorphonuclear; TCCM, tumor cell-conditioned medium; TGF-β, transforming growth factor beta.
MDSC activity is affected by the MITF expression level
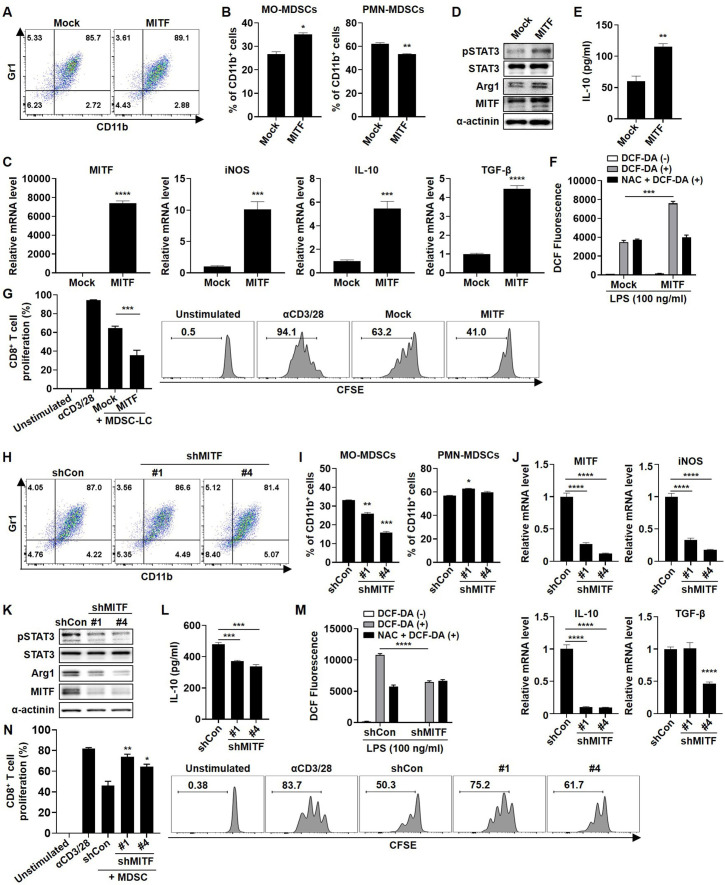
To validate whether MITF plays a role as a regulator of suppressive activity in MDSCs, we transfected myeloid cells from BM with MITF plasmid DNA. As shown figure 3A, >85% of the population in both transfected groups showed expression of MDSC differentiation markers. Interestingly, MITF overexpression induced the MO-MDSC-LC population (figure 3B) and markedly increased the mRNA and protein expression of MDSC activation markers (figure 3C, D). Similarly, IL-10 secretion was markedly increased by MITF overexpression (figure 3E). Moreover, MITF overexpression in MDSC-LCs induced reactive oxygen species (ROS) production, but treatment with N-acetylcysteine (NAC), which inhibits ROS production, reduced ROS levels (figure 3F). Consistent with previous data, CD8+ T-cell proliferation was more strongly inhibited by MITF overexpression than the control vector transfection (figure 3G). In contrast, the inhibition of MITF expression by MITF shRNA transfection attenuated MO-MDSC differentiation and suppressed the expression of MDSC-inhibitory factors and ROS production (figure 3H–M). Intriguingly, the T-cell proliferation assay showed that CD8+ T-cell proliferation was markedly restored by the inhibition of MITF expression (figure 3N). Thus, our results indicate that knockdown of MITF regulates the MO-MDSC differentiation and immunosuppressive functions of MDSCs, leading to restoration of T-cell proliferation in parallel with suppression of activation marker expression and ROS generation.
Figure 3.
MITF expression in MDSCs regulates the immunosuppressive function. Myeloid cells depleted of lymphoid cells were transfected with pBabe-empty or pBabe-MITF (A–G) or control shRNA or MITF shRNAs (clones 1 and 4) (H–N), and then cells were cultured with fresh complete medium containing 10 ng/mL GM-CSF (A–G) or GM-CSF+TCCM (H–N). (A, H) After 4 days, the cells were stained with CD11b and Gr1 antibodies and analyzed by flow cytometry. (B, I) Cells were stained with CD11b, Ly6C, and Ly6G antibodies and analyzed by flow cytometry. (C, J) mRNA expression in BM-MDSCs under each condition was evaluated by real-time PCR. (D, K) The expression levels of MITF and activity markers in MDSCs were determined by western blot analysis. (E, L) Transfected cells were cultured in serum-free medium for 48 hours. IL-10 secretion in the supernatant was determined by ELISA. (F, M) The cells were pretreated with 10 mM NAC and 100 ng/mL LPS for 24 hours. Then, the cells were stained with 10 µM 2ʹ,7ʹ-dichlorofluorescin diacetate (DCF-DA) for 30 min. The ROS levels of harvested cells were measured by flow cytometry and analyzed by FlowJo software. (G, N) The transfected BM-MDSCs were cocultured with CFSE-labeled CD3+ T cells for 72 hours, and then CD8+ T-cell proliferation was measured by flow cytometry at an MDSC:T-cell ratio of 1:2. All experiments were independently repeated at least three times. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. IL, interleukin; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; MO, monocytic; PMN, polymorphonuclear; TGF-β, transforming growth factor beta.
ML-329, an inhibitor of MITF, abrogates the immunosuppressive capacity of MDSCs
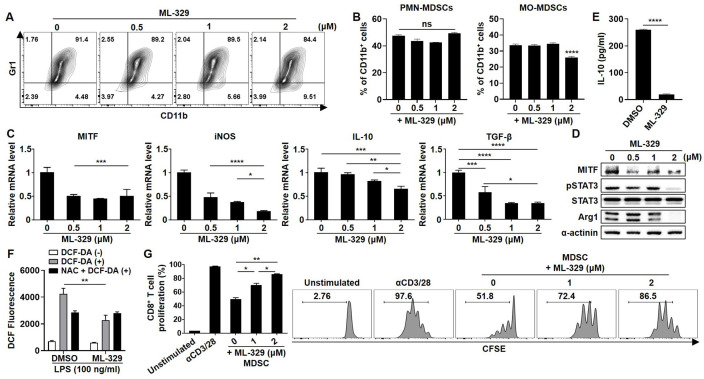
As MITF might be a promising therapeutic biomarker of MDSCs, we sought to identify the small molecules that can regulate MITF expression and determine their effects on the immunosuppressive capacity of MDSCs. ML-329 was reported to inhibit the expression of MITF in dendritic cells.24 As expected, MITF inhibition by treatment with ML-329 in BM cells resulted in a gradual decline in MO-MDSC differentiation (figure 4A, B). In addition, MITF inhibition by ML-329 treatment was found to decrease the expression levels of activation markers in MDSCs, and IL-10 and ROS production (figure 4C–F). Finally, CD8+ T-cell proliferation was gradually reversed by ML-329 treatment in BM-MDSCs in a dose-dependent manner (figure 4G). These observations suggest the idea that MITF inhibitors could be used therapeutically to target MDSCs by regulating ROS production as well as T-cell proliferation.
Figure 4.
Effect of an MITF inhibitor on the activation and differentiation of MDSCs. Myeloid cells were cultured in complete medium containing 10 ng/mL GM-CSF and TCCM. The indicated concentration of ML-329 was added to the cells for 96 hours. (A) BM-MDSCs were stained with CD11b and Gr1 antibodies and analyzed by flow cytometry. (B) Cells were stained with CD11b, Ly6C, and Ly6G antibodies and analyzed by flow cytometry. (C, D) The expression levels of MITF and activity markers in MDSCs were evaluated by real-time PCR and western blot analysis. (E) IL-10 secretion in the supernatant was determined by ELISA. (F) The cells were pretreated with 10 mM NAC and 100 ng/mL LPS for 24 hours. Then, the cells were stained with 10 µM DCF-DA for 30 min. The ROS levels of harvested cells were measured by flow cytometry and analyzed by FlowJo software. (G) BM-MDSCs were cocultured with CFSE-labeled CD3+ T cells for 3 days. CD8+ T-cell proliferation was measured by flow cytometry at an MDSC:T-cell ratio of 1:1. All experiments were independently repeated at least three times. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. BM, bone marrow; GM-CSF, granulocyte–macrophage colony-stimulating factor; IL, interleukin; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; MO, monocytic; PMN, polymorphonuclear; TCCM, tumor cell-conditioned medium; TGF-β, transforming growth factor beta.
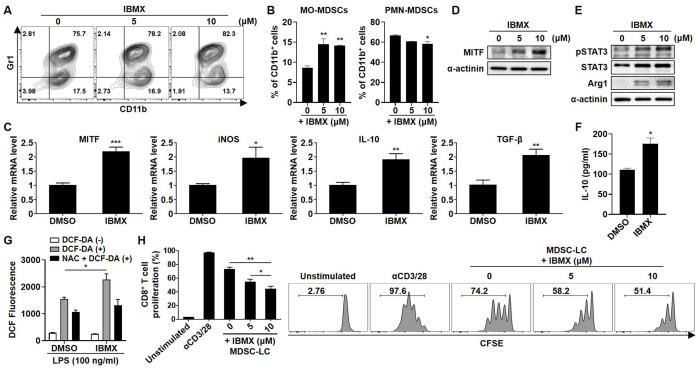
IBMX induces MITF expression in MDSC-LCs and enhances their immunoregulatory activity
Next, we tested IBMX, which is well known as an inducer of MITF through increases in cAMP,25 to regulate MITF expression in BM cells. The number of CD11b+Gr1+ MDSC-LCs and MO-MDSC-LCs was increased by IBMX treatment (figure 5A, B). Furthermore, IBMX induced the expression of activation markers, IL-10 production, and ROS generation in MDSC-LCs (figure 5C–G). Moreover, IBMX treatment significantly suppressed CD8+ T-cell proliferation in a dose-dependent manner (figure 5H). Taken together, these findings suggest that MITF inducers, such as IBMX, enhance the expression of MITF and MDSC activation markers and promote the immunosuppressive activity of MDSCs.
Figure 5.
Effect of IBMX on the immunosuppressive activity of MDSC-LCs. Myeloid cells were cultured in complete medium containing 10 ng/mL GM-CSF. The indicated concentration of IBMX was added to the cells for 96 hours. (A) BM-MDSCs were stained with CD11b and Gr1 antibodies and analyzed by flow cytometry. (B) Cells were stained with CD11b, Ly6C, and Ly6G antibodies and analyzed by flow cytometry. (C–E) The expression levels of MITF and activity markers in MDSCs were evaluated by real-time PCR and western blot analysis. (F) IL-10 secretion in the supernatant was determined by ELISA. (G) The cells were pretreated with 10 mM NAC and 100 ng/mL LPS for 24 hours. Then, the cells were stained with 10 µM DCF-DA for 30 min. The ROS levels of harvested cells were measured by flow cytometry and analyzed by FlowJo software. (H) BM-MDSCs were cocultured with CFSE-labeled CD3+ T cells for 3 days. CD8+ T-cell proliferation was measured by flow cytometry at an MDSC:T-cell ratio of 1:2. All experiments were independently repeated at least three times. *P<0.05, **P<0.01, ***P<0.001. BM, bone marrow; IL, interleukin; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; MO, monocytic; PMN, polymorphonuclear.
MITF upregulates immune suppressive activity in MDSC-LCs via HIF-1α
In the TME, HIF-1α expression increases in MDSCs due to hypoxia, a characteristic feature of the TME.26 A recent study revealed that HIF-1α promotes the differentiation of MDSCs in the TME. Moreover, HIF-1α regulates the function of MDSCs via upregulation of arginase and iNOS15 and promotes MDSC accumulation via ENTPD2.27 However, the regulator of HIF-1α in MDSCs has not been identified. MITF directly binds to the HIF-1α promoter and induces HIF-1α expression.14 We hypothesized that MITF increases the expression of MDSC activation markers through HIF-1α. As expected, TCCM treatment upregulated HIF-1α expression in BM-MDSCs (online supplemental figure 4A, B). IBMX increased HIF-1α protein levels in a dose-dependent manner, whereas TCCM-enhanced HIF-1α mRNA expression was suppressed by the MITF inhibitor (online supplemental figure 4C–E). To assess whether MITF induces the activation markers of MDSCs via HIF-1α, we added the HIF-1α inhibitor, 2-methoxyestradiol (2-ME2), to BM cells treated with IBMX. Predictably, IBMX-induced Arg1 expression was attenuated by an HIF-1α inhibitor (online supplemental figure 4F). Last, CD8+ T-cell proliferation was restored by MDSC-LCs treated with IBMX+2-ME2 compared with those treated with IBMX alone, indicating that MITF can induce the immunosuppressive function of MDSC-LCs via HIF-1α (online supplemental figure 4G). These findings indicate that MITF might increase the immunosuppressive effect of MDSCs through upregulation of HIF-1α expression.
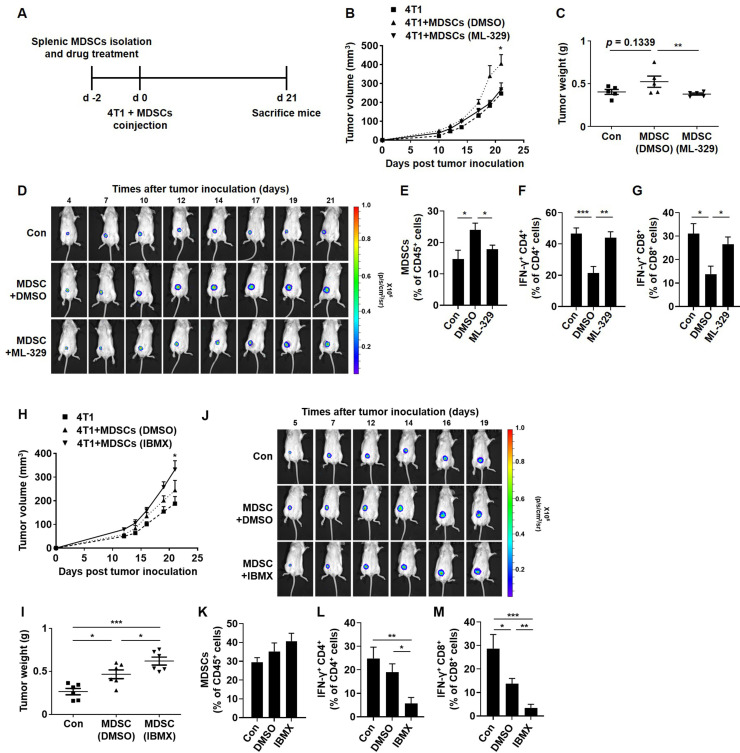
MITF regulates the tumor progression and immunosuppressive activity of MDSCs from tumor-bearing mice
Next, we assessed whether in vivo tumor growth can be differentially regulated by coinjection of MDSCs that are cultured in the absence or presence of ML-329. MDSCs obtained from the splenocytes of tumor-bearing mice were treated with ML-329 for 48 hours and coinjected with 4T1-luc2 cells into mice (figure 6A). As shown in figure 6B, tumor volume was significantly increased in the mice coinjected with 4T1-luc2 cells and the DMSO-treated MDSCs (DMSO-MDSCs). Interestingly, the 4T1-luc2 cells coinjected with the ML-329-treated MDSCs (ML-329-MDSCs) resulted in attenuated tumor growth, leading to a tumor size similar to that of 4T1-luc2 cells. The tumor weight and luciferase intensity of 4T1 cells were reduced in the mice coinjected with 4T1-luc2 cells and ML-329-MDSCs (figure 6C, D). To evaluate the immune cell proportion, we measured tumor-associated MDSCs in tumor tissues formed by coinjection with DMSO-MDSCs, and their proportion was significantly decreased in the mice coinjected with ML-329-MDSCs (figure 6E). Furthermore, as interferon gamma (IFN-γ) is an activity marker for T cells,28 tumor-infiltrating T cells were intracellularly stained with an anti-IFN-γ antibody and analyzed by flow cytometry. The accumulation of IFN-γ-positive CD4 or CD8 T cells was hampered in DMSO-MDSCs, whereas ML-329-MDSCs elicited markedly improved T-cell activity comparable to the level in the 4T1-luc2 cells coinjected with non-treated MDSCs (figure 6F, G). In contrast, the tumor volume, weight, and luciferase intensity of 4T1 cells were markedly increased in the mice coinjected with 4T1-luc2 cells and IBMX-treated MDSCs (IBMX-MDSCs) (figure 6H–J). Unexpectedly, when the tumor-associated MDSC population was analyzed in tumor tissues, the percentage of MDSCs did not reach statistical significance (figure 6K). However, the IBMX-MDSC group showed remarkably suppressed T-cell activity compared with the DMSO-MDSC group (figure 6L, M). Collectively, these observations indicate that increased MITF expression in MDSCs may promote tumor progression and suppress the immune response mediated by CD4 or CD8 T cells in the TME.
Figure 6.
MITF expression in MDSCs regulates tumor progression in vivo. (A) The experimental design for the mouse in vivo model. (B–M) MDSCs from the splenocytes of 4T1-bearing mice were isolated by magnetic-activated cell sorting (MACS). Splenic MDSCs were cultured in complete medium containing 10 ng/mL GM-CSF and TCCM or GM-CSF only and then treated with ML-329 (2 µM) or IBMX (10 µM) for 48 hours. (B, H) To evaluate the effect of MITF modulation on tumor development, Balb/c mice were subcutaneously injected with 4T1-luc2 cells alone or coinjected at a 1:1 or 1:2 ratio with DMSO-treated, ML-329-treated or IBMX-treated MDSCs into the mammary fat pad. The tumor volume of the 4T1-bearing mice from five mice/group (B) and six mice/group (H) was measured by a vernier caliper and recorded for the indicated time points. (C,I) Tumor weight was measured on day 21. (D, J) The 4T1-bearing mice were intraperitoneally injected with 3 mg D-luciferin per mouse. After the injection, the mice were anesthetized with isoflurane, and representative bioluminescence images were analyzed using IVIS. (E, K) The tumor-associated immune cells from the 4T1-bearing mice were stained with CD11b, Gr1, and CD45 antibodies and analyzed by flow cytometry. (F, G, L, M) To evaluate the effect of MDSCs on the activity of T cells, harvested tumor-associated cells were stained with IFN-γ, CD4, CD8, and CD45 antibodies. IFN-γ-positive tumor-infiltrating CD4 (F, L) or CD8 (G, M) cells were evaluated by flow cytometry. *P<0.05, **P<0.01, ***P<0.001. GM-CSF, granulocyte–macrophage colony-stimulating factor; IFN-γ, interferon gamma; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; TCCM, tumor cell-conditioned medium.
Enhanced MITF expression in MDSCs is linked to tumor development in a murine lung carcinoma model
To investigate the correlation between MITF expression and MDSC activity in other cancer types such as murine Lewis lung cancer, we established an LLC1-bearing mouse model. Consistent with previous results obtained from the 4T1 breast tumor model, the expression levels of MITF were gradually increased in MDSCs from tumor tissues, depending on tumor development (online supplemental figure 5A). Next, we compared the mRNA expression of MITF and iNOS in MDSCs from splenocytes and tumor tissues. The expression levels of MITF and iNOS were strongly increased in the tumor-exposed MDSCs compared with the splenic MDSCs with or without tumors (online supplemental figure 5B). To examine the expression of MITF in MDSCs from primary lung tumor tissues, we injected intravenously LLC1 cells into mice. As expected, the tumor-infiltrated MDSC proportion was increased in lung tissues at the indicated time points, and an elevation of MITF expression in the tumor-exposed MDSCs coincided with tumor development (online supplemental figure 5C). To examine the effect of the TME on the regulation of MITF expression, we generated murine BM-MDSCs using LLC1-TCCM. The expression of MDSC activation markers and MITF was markedly increased by TCCM in a percentage-dependent manner (online supplemental figure 5D, E). These results suggest that MITF expression in tumor-associated MDSCs plays an important role in MDSC differentiation and activation not only in breast cancer models but also in lung carcinoma models.
High MITF expression is associated with poor survival in patients with breast and lung cancers
To investigate whether MITF expression is associated with the survival of patients with cancer, we searched the Cancer Genome Atlas (TCGA) dataset and found that patients with low MITF expression had longer overall survival and disease-specific survival rates than those with high MITF expression in the Kaplan-Meier plot of breast cancer (online supplemental figure 6A, B). Similarly, to investigate the overall survival in lung cancer type, we analyzed the dataset by Q-omics software (http://qomics.sookmyung.ac.kr) and found that patients with low MITF expression had longer overall survival rates in the Kaplan-Meier plot of patients with lung squamous cell carcinoma (online supplemental figure 6C). These results suggest that MITF is a potential prognostic marker for survival of patients with breast and lung cancers.
MITF expression is elevated in tumor-infiltrating MDSCs from patients with cancer
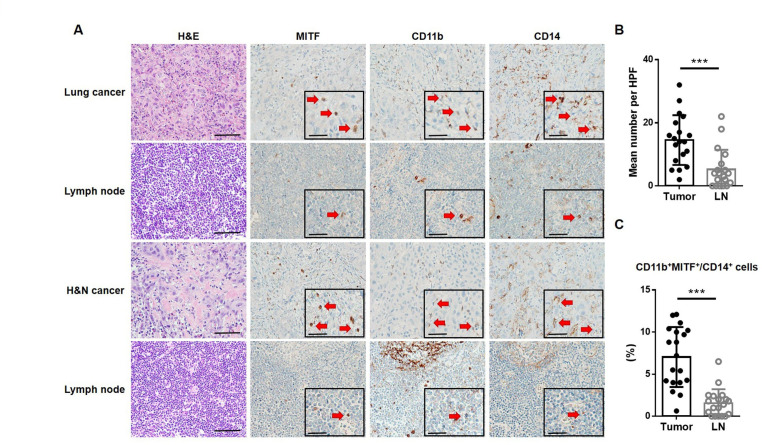
To analyze the presence of MITF+ MO-MDSCs in human primary lung or H&N cancer and TDLN, we used three IHC markers, MITF, CD11b, and CD14, in this study to identify MITF+ MO-MDSCs. Serial sections of tumor and lymph node slides from patients with lung cancer (n=10) or H&N cancer (n=10) were stained for the expression of each marker with H&E (figure 7A). Histological examination was performed under a light microscope in an HPF. The mean number of MITF+ MO-MDSCs in tumor tissues was significantly higher than that in TDLNs (figure 7B). The proportion of CD11b+MITF+ cells among the total myeloid cells, which are represented by cells with CD14 expression, was calculated. The proportion was also significantly higher in tumors than TDLNs (figure 7C). These data suggest that MITF+ MO-MDSCs are found at significantly higher levels in tumor tissues than TDLNs.
Figure 7.
MITF expression is elevated in tumor-infiltrating MO-MDSCs from patients with cancer. (A) representative IHC images of H&E staining and immunostaining with anti-MITF, anti-CD11b, and anti-CD14 antibodies for detecting MO-MDSCs in lung or H&N cancers and matched LNs. Inlet pictures show positive cells of each marker (red arrows) in a representative area at higher magnification (scale bar of 50 µm). (B) The number of MITF+ MO-MDSCs (MITF+, CD11b+, and CD14+ cells using IHC) per HPF in tumors and LNs from patients with lung or H&N cancer. (C) The percentage of MITF+ and CD11b+ cells among CD14+ cells in tumors and LNs from patient with lung or H&N cancer. ***P<0.001. H&N, head and neck; HPF, high-power field; IHC, immunohistochemistry; LN, lymph node; MDSC, myeloid-derived suppressor cell; MITF, microphthalmia-associated transcription factor; MO, monocytic.
Discussion
MDSCs consist of two main populations, PMN-MDSCs and MO-MDSCs. However, MDSCs are morphologically and phenotypically similar to neutrophils or monocytes. Due to the heterogeneity of MDSCs and the lack of specific biomarkers, the development of molecules targeting MDSC suppressive functions is progressing relatively slowly. MDSCs can enable tumor cells to evade immune surveillance by diverse mechanisms and accelerate tumor progression. MDSCs suppress the T cell-mediated immune response through Arg1, iNOS, IL-10, and transforming growth factor beta (TGF-β).29 30 MDSCs also induce the development of regulatory T cells via secretion of IL-1031 and inhibit NK cells through interaction between the NK-cell receptor NKp30 and MDSC membrane-bound TGF-β.32 33 Therefore, these molecules are thought to play a crucial role in immunological homeostasis, and the inhibition of MDSC expansion and activation can enhance the efficacy of anticancer therapy.
In our previous study, we reported that IRF4 regulates the differentiation and function of MDSCs.34 In addition, glycoprotein non-metastatic B (GPNMB) expressed by MDSCs was reported to inhibit T-cell activation.35 Interestingly, both IRF4 and GPNMB are known to be transcriptional targets regulated by MITF. MITF suppresses IRF4 expression and induces GPNMB expression.19 36 In previous studies, MITF has been reported to be a master regulator of melanogenesis by binding to promoter sites. Intriguingly, MITF is also expressed in immune cells, such as osteoclasts, B cells, mast cells, and NK cells.17 18 37 However, the role of MITF in MDSCs has not been reported. Additionally, in osteoclast differentiation, MITF plays an important role in transcriptional activation by another transcription factor, PU.1.38 PU.1 is critically important in hematopoietic development.39 High PU.1 expression promotes HSC differentiation into myeloid cells.40 These reports suggest that MITF contributes to MDSC differentiation. Thus, further studies are needed on the effect of MITF on MDSC differentiation.
Here, we found that MITF expression is induced in MDSCs and is related to tumor development. Compared with splenic MDSCs, tumor-associated MDSCs strongly expressed MITF and activation markers of MDSCs. MITF expression in splenic MDSCs or tumor-MDSCs was significantly increased in MO-MDSCs compared with PMN-MDSCs. Moreover, we observed that soluble factors used for mimicking the TME can induce MDSC activation and MITF expression. These results demonstrate that MITF could elevate the immunosuppressive function of MDSCs during tumor development, indicating that an increase in MITF expression can be associated with MDSC activation. Previous studies have shown that PAX3 is activated by STAT3 and that MITF expression is directly regulated by PAX3 at the transcriptional level.41 Given that STAT3 plays a crucial role in the activation and differentiation of MDSCs,42 these reports indicate that MITF expression can be regulated by STAT3 in MDSCs and correlated with the activity of MDSCs in the TME. Moreover, this study showed that MITF expression can be regulated in various ways, such as transfection of plasmid vectors or treatment with IBMX and ML-329. As expected, induction of MITF expression elicited the immunosuppressive activity of MDSCs. Conversely, disruption of MITF expression reduced the differentiation and immunoregulatory functions of MDSCs to restore T-cell proliferation by inhibiting MDSC activation markers and ROS generation. ML-329 was shown to decrease the expression of MITF and multiple MITF target genes in melanoma cell lines and dendritic cells.24 43 However, its effect on MDSC differentiation and activation has not been reported. We observed that MITF expression and the immunosuppressive activity of MDSCs were effectively inhibited by ML-329. Thus, the effects of ML-329 on cancer therapy should be further investigated with respect to safety and toxicity studies in vivo.
A previous report showed that the TME induces HIF-1α, which enhances iNOS and Arg1 expression in MDSCs.15 We found that HIF-1α mRNA and protein levels were increased in MDSCs treated with TCCM in vitro. In a previous study, it was reported that HIF-1α is a direct transcriptional target of MITF.14 Consistent with this report, ML-329, which is known as an MITF inhibitor, suppressed TCCM-induced HIF-1α expression in MDSCs. In addition, HIF-1α in MDSC-LCs was increased by IBMX-induced MITF. Predictably, IBMX-induced Arg1 expression was attenuated by a HIF-1α inhibitor, and CD8+ T-cell proliferation was significantly restored by an HIF-1α inhibitor. As HIF-1α is known to be a direct regulator of PD-L1, MITF may regulate PD-L1 expression in tumor conditions.20 Thus, further study is required to investigate whether MITF upregulates PD-L1 expression in MDSCs and attenuates the efficacy of anticancer therapy in combination with ICIs. If MITF regulates PD-L1 expression via HIF-1α, the regulation of MITF expression in MDSCs will be very important in ICI therapy. Notably, a recent report showed that CCAAT/enhancer binding protein alpha (C/EBPα) has a negative role in protumor functions including Arg1 and iNOS of MDSCs.44 MITF binds the C/EBPα promoter to suppress the transcription of C/EBPα in mast cell differentiation,37 suggesting that MITF inhibits C/EBPα expression in MDSCs.
The results of the present study showed that coinjection of cancer cells and ML-329-treated MDSCs suppressed tumor growth and improved T-cell activation. Conversely, coinjection of cancer cells and IBMX-treated MDSCs enhanced tumor progression and abrogated the activities of CD4/CD8 T cells. However, the proportion of MDSCs in tumor tissues was not significantly affected by IBMX-treated MDSCs. Previous studies have reported the effect of the TME on the differentiation of MO-MDSCs into tumor-associated macrophages (TAMs). Moreover, HIF-1α is involved in MO-MDSC-to-TAM differentiation and the accumulation of TAMs under hypoxic conditions. Although the mechanisms by which MITF enhances MO-MDSC-to-TAM differentiation remain unclear, there is growing evidence that it is possible to promote the accumulation of TAMs in tumor tissues by inducing MITF expression in MDSCs via HIF-1α.15 45 These results suggest that inhibition of MITF expression in MDSCs overcomes immune tolerance in the TME and improves the efficacy of anticancer therapy. Finally, we observed that MITF expression is induced in tumor-associated MDSCs and plays a crucial role not only in a breast cancer model but also in a lung carcinoma model. Consistent with these findings in mice, the proportion of MITF+ MO-MDSCs was significantly higher in the tumor tissues than in TDLNs from patients with human cancer. Notably, MITF+ MO-MDSCs were observed in the TDLNs, although their numbers were much lower than those in the TME. A recent study demonstrated that MO-MDSCs home to TDLNs via the CCR2/CCL20-dependent pathway and are highly suppressive.46 Therefore, the present study suggested that MITF+ MO-MDSCs in the TME may be recruited to and may elicit immune suppressive functions in TDLNs.
In this study, MITF was highly expressed in tumor-associated MDSCs and MO-MDSCs, compared with PMN-MDSCs, and could affect CD8+ T-cell proliferation. Furthermore, inhibition of MITF expression reduced the proportion of MO-MDSCs in the TME and downregulated the suppressive molecules, such as iNOS and Arg1. Nevertheless, some evidence indicated that MITF also influenced the suppressive activity of PMN-MDSCs, because ROS generation was reduced by MITF inhibition. In fact, it cannot be excluded that the differences shown are actually due to a direct effect of MITF on MO-MDSC suppressive activity rather than a reduction/increase of MO-MDSC frequency within the bulk in vitro population. Therefore, further investigation is needed to explain the exact mechanisms by which MITF is involved in the regulation of MDSC population and immunosuppressive function of MDSCs.
In summary, this study describes a novel role of MITF as a master regulator of the differentiation and immunoregulatory functions of MDSCs in tumors, and provides strategies for improving therapeutic efficacy by inhibiting MITF as a novel approach to restore T-cell activity and hamper proliferation in cancer.
Footnotes
Correction notice: This article has been corrected since it was first published online. The conclusion of the abstract has been updated to: "Our results indicate that MITF regulates the differentiation and function of MDSCs and can be a novel therapeutic target for modulating MDSC activity in immunosuppressive TMEs. Figure 6 has also been updated.
Contributors: AL, HP, and J-SL designed research, acquired the data, prepared the figures, and wrote the paper. SL and JL analyzed and interpreted the data. JK and YKJ acquired the data using tumor tissues, and YY and M-SL provided the experimental materials and facilities. J-SL is responsible for the overall content as the guarantor.
Funding: This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea (NRF-2016R1A5A1011974 and 2019R1A2C1088825). JL was supported by a grant from the NRF (2022R1I1A1A01063959).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee, after approval by the institutional ethical committee of Sookmyung Women’s University (approval number SMWU-IACUC-1410-018-01). This study followed the World Medical Association Declaration of Helsinki recommendations and was approved by the institutional review board of Seoul National University Hospital (numbers 2106-028-1225 and 2007-150-1143).
References
- 1.Gabrilovich DI. Myeloid-Derived suppressor cells. Cancer Immunol Res 2017;5:3–8. 10.1158/2326-6066.CIR-16-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng J, Fleming JB. Inflammation and myeloid cells in cancer progression and metastasis. Front Cell Dev Biol 2021;9:759691. 10.3389/fcell.2021.759691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, Liu T, Zhu W, et al. Targeting MDSC for immune-checkpoint blockade in cancer immunotherapy: current progress and new prospects. Clin Med Insights Oncol 2021;15:117955492110355. 10.1177/11795549211035540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim J, Lee A, Lee HG, et al. Modulation of immunosuppression by oligonucleotide-based molecules and small molecules targeting myeloid-derived suppressor cells. Biomol Ther 2020;28:1–17. 10.4062/biomolther.2019.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groth C, Hu X, Weber R, et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer 2019;120:16–25. 10.1038/s41416-018-0333-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009;9:162–74. 10.1038/nri2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stromnes IM, Greenberg PD, Hingorani SR. Molecular pathways: myeloid complicity in cancer. Clin Cancer Res 2014;20:5157–70. 10.1158/1078-0432.CCR-13-0866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronte V, Brandau S, Chen S-H, et al. Recommendations for myeloid-derived suppressor cell Nomenclature and characterization standards. Nat Commun 2016;7:12150. 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Ding Y, Deng Y, et al. Role of myeloid-derived suppressor cells in the promotion and immunotherapy of colitis-associated cancer. J Immunother Cancer 2020;8:e000609. 10.1136/jitc-2020-000609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raber P, Ochoa AC, Rodríguez PC. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest 2012;41:614–34. 10.3109/08820139.2012.680634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber R, Riester Z, Hüser L, et al. IL-6 regulates CCR5 expression and immunosuppressive capacity of MDSC in murine melanoma. J Immunother Cancer 2020;8:e000949. 10.1136/jitc-2020-000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 2006;12:406–14. 10.1016/j.molmed.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 13.Kawakami A, Fisher DE. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab Invest 2017;97:649–56. 10.1038/labinvest.2017.9 [DOI] [PubMed] [Google Scholar]
- 14.Buscà R, Berra E, Gaggioli C, et al. Hypoxia-inducible factor 1{alpha} is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J Cell Biol 2005;170:49–59. 10.1083/jcb.200501067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corzo CA, Condamine T, Lu L, et al. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med 2010;207:2439–53. 10.1084/jem.20100587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meadows NA, Sharma SM, Faulkner GJ, et al. The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. J Biol Chem 2007;282:1891–904. 10.1074/jbc.M608572200 [DOI] [PubMed] [Google Scholar]
- 17.Shahlaee AH, Brandal S, Lee Y-N, et al. Distinct and shared transcriptomes are regulated by microphthalmia-associated transcription factor isoforms in mast cells. J Immunol 2007;178:378–88. 10.4049/jimmunol.178.1.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Yue X, Yu J, et al. MITF Regulates Downstream Genes in Response to Vibrio parahaemolyticus Infection in the Clam Meretrix Petechialis. Front Immunol 2019;10:1547. 10.3389/fimmu.2019.01547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin L, Gerth AJ, Peng SL. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J Exp Med 2004;200:115–22. 10.1084/jem.20040612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noman MZ, Desantis G, Janji B, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med 2014;211:781–90. 10.1084/jem.20131916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monu NR, Frey AB. Myeloid-derived suppressor cells and anti-tumor T cells: a complex relationship. Immunol Invest 2012;41:595–613. 10.3109/08820139.2012.673191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K, Kassem S, Cleynen A, et al. Dysregulated IL-18 is a key driver of immunosuppression and a possible therapeutic target in the multiple myeloma microenvironment. Cancer Cell 2018;33:634–48. 10.1016/j.ccell.2018.02.007 [DOI] [PubMed] [Google Scholar]
- 23.Yaseen MM, Abuharfeil NM, Darmani H, et al. Mechanisms of immune suppression by myeloid-derived suppressor cells: the role of interleukin-10 as a key immunoregulatory cytokine. Open Biol 2020;10:200111. 10.1098/rsob.200111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutknecht M, Geiger J, Joas S, et al. The transcription factor MITF is a critical regulator of GPNMB expression in dendritic cells. Cell Commun Signal 2015;13:19. 10.1186/s12964-015-0099-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park H-Y, Wu C, Yonemoto L, et al. MITF mediates cAMP-induced protein kinase C-beta expression in human melanocytes. Biochem J 2006;395:571–8. 10.1042/BJ20051388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu DK-C, Xu IM-J, Lai RK-H, et al. Hypoxia induces myeloid-derived suppressor cell recruitment to hepatocellular carcinoma through chemokine (C-C motif) ligand 26. Hepatology 2016;64:797–813. 10.1002/hep.28655 [DOI] [PubMed] [Google Scholar]
- 27.Chiu DK-C, Tse AP-W, Xu IM-J, et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun 2017;8:517. 10.1038/s41467-017-00530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gocher AM, Workman CJ, Vignali DAA. Interferon-γ: teammate or opponent in the tumour microenvironment? Nat Rev Immunol 2022;22:158–72. 10.1038/s41577-021-00566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Ye Y, Liu P, et al. Suppression of T cells by myeloid-derived suppressor cells in cancer. Hum Immunol 2017;78:113–9. 10.1016/j.humimm.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 30.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park M-J, Lee S-H, Kim E-K, et al. Interleukin-10 produced by myeloid-derived suppressor cells is critical for the induction of Tregs and attenuation of rheumatoid inflammation in mice. Sci Rep 2018;8:3753. 10.1038/s41598-018-21856-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoechst B, Voigtlaender T, Ormandy L, et al. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology 2009;50:799–807. 10.1002/hep.23054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Han Y, Guo Q, et al. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol 2009;182:240–9. 10.4049/jimmunol.182.1.240 [DOI] [PubMed] [Google Scholar]
- 34.Nam S, Kang K, Cha JS, et al. Interferon regulatory factor 4 (IRF4) controls myeloid-derived suppressor cell (MDSC) differentiation and function. J Leukoc Biol 2016;100:1273–84. 10.1189/jlb.1A0215-068RR [DOI] [PubMed] [Google Scholar]
- 35.Chung J-S, Tamura K, Akiyoshi H, et al. The DC-HIL/syndecan-4 pathway regulates autoimmune responses through myeloid-derived suppressor cells. J Immunol 2014;192:2576–84. 10.4049/jimmunol.1301857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ripoll VM, Meadows NA, Raggatt L-J, et al. Microphthalmia transcription factor regulates the expression of the novel osteoclast factor GPNMB. Gene 2008;413:32–41. 10.1016/j.gene.2008.01.014 [DOI] [PubMed] [Google Scholar]
- 37.Qi X, Hong J, Chaves L, et al. Antagonistic regulation by the transcription factors C/EBPα and MITF specifies basophil and mast cell fates. Immunity 2013;39:97–110. 10.1016/j.immuni.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey HA, Hildreth BE, Samuvel DJ, et al. Eomes partners with PU.1 and MITF to regulate transcription factors critical for osteoclast differentiation. iScience 2019;11:238–45. 10.1016/j.isci.2018.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mak KS, Funnell APW, Pearson RCM, et al. PU.1 and haematopoietic cell fate: dosage matters. Int J Cell Biol 2011;2011:1–6. 10.1155/2011/808524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 1998;12:2403–12. 10.1101/gad.12.15.2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong L, Li Y, Cao J, et al. FGF2 regulates melanocytes viability through the STAT3-transactivated Pax3 transcription. Cell Death Differ 2012;19:616–22. 10.1038/cdd.2011.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Y-L, Banerjee S, White SV, et al. STAT3 in tumor-associated myeloid cells: multitasking to disrupt immunity. Int J Mol Sci 2018;19:19. 10.3390/ijms19061803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faloon PW, Bennion M, Weiner WS, et al. A small molecule inhibitor of the MITF molecular pathway. In: Probe reports from the NIH molecular libraries program. Bethesda, MD: National Center for Biotechnology Information (US), 2010. [PubMed] [Google Scholar]
- 44.Mackert JR, Qu P, Min Y, et al. Dual negative roles of C/EBPα in the expansion and pro-tumor functions of MDSCs. Sci Rep 2017;7:14048. 10.1038/s41598-017-12968-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trillo-Tinoco J, Sierra RA, Mohamed E, et al. AMPK Alpha-1 intrinsically regulates the function and differentiation of tumor myeloid-derived suppressor cells. Cancer Res 2019;79:5034–47. 10.1158/0008-5472.CAN-19-0880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lahmar Q, Schouppe E, Morias Y, et al. Monocytic myeloid-derived suppressor cells home to tumor-draining lymph nodes via CCR2 and locally modulate the immune response. Cell Immunol 2021;362:104296. 10.1016/j.cellimm.2021.104296 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005699supp001.pdf (6.9MB, pdf)
Data Availability Statement
Data are available upon reasonable request.