Abstract
Dinoflagellates of the genus Alexandrium are responsible for harmful algal blooms and produce paralytic shellfish toxins (PSTs). Their very large and complex genomes make it challenging to identify the genes responsible for toxin synthesis. A family-based genomic association study was developed to determine the inheritance of toxin production in Alexandrium minutum and identify genomic regions linked to this production. We show that the ability to produce toxins is inheritable in a Mendelian way, while the heritability of the toxin profile is more complex. We developed the first dinoflagellate genetic linkage map. Using this map, several major results were obtained: 1. A genomic region related to the ability to produce toxins was identified. 2. This region does not contain any polymorphic sxt genes, known to be involved in toxin production in cyanobacteria. 3. The sxt genes, known to be present in a single cluster in cyanobacteria, are scattered on different linkage groups in A. minutum. 4. The expression of two sxt genes not assigned to any linkage group, sxtI and sxtG, may be regulated by the genomic region related to the ability to produce toxins. Our results provide new insights into the organization of toxicity-related genes in A. minutum, suggesting a dissociated genetic mechanism for the production of the different analogues and the ability to produce toxins. However, most of the newly identified genes remain unannotated. This study therefore proposes new candidate genes to be further explored to understand how dinoflagellates synthesize their toxins.
Keywords: Alexandrium minutum, Genetic linkage map, Paralytic Shellfish Toxins (PSTs), sxtG, sxtI, Toxin inheritance
Data Summary
The RNA-Seq sequences of the different strains can be found at this address: https://www.ebi.ac.uk/ena/browser/view/PRJEB52370 (ENA accession number: PRJEB52370). The reference transcriptome is available on Seanoe dataportal: https://doi.org/10.17882/45445.
Impact Statement.
Dinoflagellates are micro-eukaryotes of primary ecological importance. Of special interest, some species produce toxins that bio-accumulate in the trophic network. Being non-model with huge genomes, very few genomic resources and studies have been developed for these organisms. By developing the first genetic linkage map and genomic association study based on sexual crosses in dinoflagellates, the present study takes a major step toward a better understanding of the genetic basis of Paralytic Shellfish Toxin (PST) production. So far, PST production in dinoflagellates has been investigated by focusing on candidate genes homolog to genes known to be involved in PST production in cyanobacteria (sxt genes). Here, we show that the ability to produce toxins is inherited in a Mendelian manner and that the types of toxins produced follows a more complex pattern. We identified a genomic region associated to PST production and genes potentially regulated by this region. Altogether, we identified new candidate genes related to toxin production and discussed the importance of some genes considered as markers of toxicity.
Introduction
Genetic association studies aim to link phenotype variations to genomic regions. The first attempts to understand the genetic bases of phenotypic variability were based on linkage maps developed after characterizing inheritance between parents and F1 offspring resulting from artificial sexual crosses [1, 2]. Following the development of molecular markers in the 1980s, quantitative trait locus (QTL) mapping studies were developed, mostly in plants, where numerous offspring may be obtained from a couple of parents [3–7]. In micro-eukaryotes, such approaches were limited by both the ability to perform sexual crosses in vitro and to phenotype the individuals, yeasts being a notable exception [8–10]. Since the explosion of genomic resources, the focus shifted from family- to population-wide association studies, mostly in humans and pathogenic bacteria [11–13].
However, in non-model micro-eukaryotes, genetic association studies using either linkage map or genome sequences are still scarce. For instance, the SAR supergroup (a clade containing Stramenopiles, Alveolata, and Rhizaria) is a deep branching eukaryote clade encompassing brown macro-algae, as well as numerous micro-eukaryotes such as ascomycetes, ciliates, radiolarians, and dinoflagellates. Within this clade, about ten genetic maps have been developed, mostly in parasites [14–23]. There is no genetic map available for dinoflagellates, which may be explained by the difficulties of obtaining hundreds of descendants [24] to analyse quantitative traits. However, a smaller number of offspring may be sufficient to investigate the genetic linkage of many qualitative traits.
Dinoflagellates are aquatic microorganisms displaying numerous peculiar genetic and molecular characteristics. They have multiple copies of genes arranged in tandem [25], numerous chromosomes in a permanent semi-condensed state and lack histones [26–28]. Only a few of their genes seem to be transcriptionally regulated [29–33], and transcription appears to be driven by a trans-splicing mechanism controlled by a specific sequence, the splice leader [34, 35]. Moreover, dinoflagellates have very large genomes (3 to 245 Gbp [36]), strongly limiting the feasibility of sequencing the entire genome. As a result, only genomes a few Gbp long have been sequenced, e.g. the coral symbiont Breviolum minutum [37] and Polarella glacialis [38].
Among dinoflagellates, four genera are known to produce Paralytic Shellfish Toxins (PSTs): Gymnodinium, Pyrodinium, Centrodinium, and finally Alexandrium, for which at least 11 species are known to be PST-producers [39, 40]. PSTs are non-protein toxins, produced by cyanobacteria and dinoflagellates, responsible for the Paralytic Shellfish Poisoning (PSP) syndrome in humans following the consumption of contaminated seafood. PSTs correspond to more than 57 analogues of saxitoxin (STX) classified into different groups, based on the nature of the substitutions on radicals R1 to R5 (Fig. 1; Table 1; [41]).
Fig. 1.
Intermediates involved in the PST biosynthetic pathway in dinoflagellates [45, 111]. The sxt gene products involved in the biosynthetic pathway in cyanobacteria, and potentially involved in dinoflagellates, are indicated by dotted circles.
Table 1.
Radicals of different saxitoxin analogues
|
Type of molecule |
Analogue |
R1 |
R2 |
R3 |
R4 |
R5 |
Type of toxin |
|---|---|---|---|---|---|---|---|
|
Non-sulphated |
STX |
H |
H |
H |
OCONH2 |
OH |
Carbamate |
|
Mono-sulphated |
GTX1 |
OH |
H |
OSO3- |
OCONH2 |
OH |
|
|
GTX2 |
H |
H |
OSO3- |
OCONH2 |
OH |
||
|
GTX3 |
H |
OSO3- |
H |
OCONH2 |
OH |
||
|
GTX4 |
OH |
OSO3- |
H |
OCONH2 |
OH |
||
|
GTX5 |
H |
H |
H |
OCONHSO3- |
OH |
N-sulfocarbamoyl |
|
|
GTX6 |
OH |
H |
H |
OCONHSO3- |
OH |
||
|
Di-sulphated |
C1 |
H |
H |
OSO3- |
OCONHSO3- |
OH |
|
|
C2 |
H |
OSO3- |
H |
OCONHSO3- |
OH |
||
|
C3 |
OH |
H |
OSO3- |
OCONHSO3- |
OH |
||
|
C4 |
OH |
OSO3- |
H |
OCONHSO3- |
OH |
||
|
Decarbamoylated |
dcSTX |
H |
H |
H |
OH |
OH |
Decarbamoyl |
|
dcGTX1 |
OH |
H |
OSO3- |
OH |
OH |
||
|
dcGTX2 |
H |
H |
OSO3- |
OH |
OH |
||
|
dcGTX3 |
H |
OSO3- |
H |
OH |
OH |
||
|
dcGTX4 |
OH |
OSO3- |
H |
OH |
OH |
The first identified precursors of PSTs in cyanobacteria and dinoflagellates were arginine, acetate, and S-adenosyl methionine (SAM) [42] and several biosynthetic intermediates were confirmed experimentally by LC-MS analyses [43]. Some intermediates were also found in the dinoflagellate Alexandrium tamarense [44, 45], notably the two first products of the cyanobacterial biosynthetic pathway Int-A' and Int-C’2 [46]. The ability to produce toxins (qualitatively and quantitatively) shows both intra- and inter-specific polymorphisms in dinoflagellates, as well as cyanobacteria [47, 48].
So far, the genetic basis of PST production has mainly been studied in cyanobacteria. The first gene identified as involved in PST production was a carbamoyltransferase, named sxtI, whose role is to transfer a carbamoyl group to STX precursors in the PST biosynthetic pathway [49]. Using a genome walking approach upstream and downstream of sxtI, a cluster of 26 genes was identified as responsible for the synthesis, regulation, modification, and transport of these toxins in several species of cyanobacteria [49–51]. The genes sxtA, sxtG, sxtH, and sxtI were systematically present in PST-producing cyanobacteria [43]. The first intermediate in the PST biosynthetic pathway, Int-A', is thought to be produced by SxtA, and Int-C’2 by SxtG and SxtB. The various core genes (sxtDIJKHTSUVW) lead to saxitoxin formation and its analogues are obtained by the action of tailoring genes (sxtLNXO, as well as sxtSUL and GxtA), while the remaining sxt genes are dedicated to toxin regulation and transport [43].
Following these pioneer studies, homologues of cyanobacterial sxt genes were identified in dinoflagellates, with a particular focus on sxtA and sxtG [52–55]. In dinoflagellates, sxtA consists of four catalytic domains and has two isoforms: the short form, consisting of domains A1 to A3, and the long form, consisting of domains A1 to A4. Although the gene sxtG and the sxtA4 domain first appeared to be present in PST-producing strains only, they were also identified in non-producing strains of species that usually produce PST [47, 53, 56]. Conversely, there is no published data showing a PST-producing strain lacking sxtA4, as reported in [57, 57]. In addition, a strain of A. affine, a usually non-toxic species, producing low levels of PST has not been investigated for the presence of sxt genes [58].
The functional role of cyanobacterial SxtA, GxtA, SxtSUL, SxtT, SxtH, SxtN in the biosynthesis of STX and analogues was recently confirmed in vitro [59–61], and in vivo after SxtA was expressed in E. coli [62]. In dinoflagellates, sxt genes have indeed been found [52–55], but experimental evidence of their implication in toxin production is still lacking. Current knowledge about this biosynthetic pathway in dinoflagellates is summarized in Fig. 1.
So far, a single study specifically investigated PST synthesis inheritance from parents to recombinant offspring in dinoflagellates [63]. The authors crossed A. catenella strains with different toxin profiles and obtained offspring. After analysing the toxin profile and mating type of the offspring, the authors concluded that the genes responsible for toxin synthesis are encoded in the nucleus and that the toxin profile is inherited in a Mendelian manner, i.e. the F1s inherit the toxin composition of one of the parent strains.
The present study aimed at identifying genomic regions associated with the ability to produce PSTs in the dinoflagellate A. minutum, one of the PST-producing species of the genus Alexandrium, responsible for harmful algal blooms (HABs) worldwide. A classic genetic approach, involving sexual crosses of four A. minutum clonal strains (i.e. two PST-producing and two non-producing) was performed in order to obtain F1 offpsring. The PST quota and profile of parental strains and F1s were determined to investigate the inheritance of PST production and test the hypothesis of a Mendelian segregation suggested by Sako et al. [63] in terms of: 1. ability to produce the toxins, 2. types of analogues produced and 3. quantity of toxins. The genome size of A. minutum (24 to 28 pg [47]) precludes whole genome sequencing and assembly. As an alternative, RNA-Seq was used to focus on the coding regions of the expressed genes of the genome and to identify thousands of SNPs among parental strains [64]. A genetic map based on the SNP markers recombination in the F1s was developed. Using these genomic resources, we investigated: 1. whether sxt genes form a gene cluster, 2. the genomic regions associated with toxicity and if they contain sxt genes, 3. the genes, whose expression seems to be controlled by the genomic regions linked to toxicity and if these genes correspond to sxt.
Methods
Strains, crosses, and cell cultures
The dinoflagellate A. minutum has a haplontic life cycle [39] with alternance of asexual haploid cell multiplication and sexual reproduction, leading to the formation of a diploid resting cyst. This cyst enters a dormancy phase after which germination associated with meiosis may occur, leading to new haploid vegetative cells [65]. The dinoflagellate A. minutum appears to be heterothallic, which means that sexual reproduction is only possible between strains with a different mating-type [65, 66]. Four clonal parental strains (Table 2; Table S1, available in the online version of this article) were crossed considering previously established sexual types [33]: mt+ strains were crossed with mt− strains, resulting in four crosses, namely T1xT2, NT1xNT2, T1xNT1, and T2xNT2.
Table 2.
Characteristics of the four parental strains
|
T1 |
T2 |
NT1 |
NT2 |
|
|---|---|---|---|---|
|
Sampling site |
Bay of Brest, France |
Bay of Cork, Ireland |
Bay of Concarneau, France |
Bay of Concarneau, France |
|
Year of isolation |
2013 |
2010 |
2010 |
2010 |
|
Mating Type |
– |
+ |
+ |
– |
|
Toxinic profile |
C1/C2/GTX3/GTX2/dcGTX2/dcGTX3/STX |
GTX3/GTX2/STX |
Non toxic |
Non toxic |
Unless stated otherwise, all strains were grown in K medium containing Na2SiO3 [67], referred hereafter as ‘K medium’, and under a 12/12 L:D cycle, 18 °C and 80 µmol photons m−2 s−1.
Parental strains were grown for 10 days and diluted in their own culture filtrate (0.2 µM) to obtain an initial cell density of 5 000 cells per millilitre. Pairwise crosses were performed by mixing 1 ml of each strain in 24 well plates. Resting cysts resulting from gamete fusion produced mucus and bound to the bottom of the plates. After ten days, microplates were washed twice using sterile sea water (to remove vegetative cells), and stored at 4 °C in the dark for 8 months. After this dormancy period, microplates were washed with sterile sea water and placed for 5 days in an algal incubator after addition of 1 ml of K/2 (K diluted in A. minutum filtrate (0.2 µM)) per well. The presence of vegetative cells was confirmed and quantified under a microscope. Single cell isolation was then performed using serial dilution in K medium. Growth was assessed regularly and culture volume progressively increased to 30 ml of K medium, leading to a total of 62 F1 strains that were confirmed to be recombinant and monoclonal using SNP markers (see below).
Toxin extraction and analysis
Toxin quantification was performed across three sets of experiments (PST1, PST2, and PST3). The PST1 analysis was performed in 2020 on 52 strains collected at the end of the exponential phase, including all four parents, with one biological replicate per strain. The PST2 analysis was performed later in 2020, on three parent strains (NT1, NT2, T1), 18 F1s from the T2xNT2 cross and two F1s from the T1xNT1 cross, all harvested in stationary phase, with one biological replicate per strain. The PST3 analysis was performed in 2021, focusing on the T2xNT2 cross, with five biological replicates harvested at the end of the exponential phase. Cells were pelleted and stored at −20 °C before extraction.
The samples were prepared and analysed according to Shin et al. [40] but from non-lyophilised cell pellets. Briefly, they were resuspended in 1 ml of 1 % acetic acid and lysed in an ultrasonic bath for 15 min before SPE clean-up on ENVI-carb cartridges (Supelco, Bellefonte, Pennsylvania, USA), diluted 1/4 in acetonitrile and finally analysed by LC-MS/MS. The method includes 19 PST analogues, and limits of detection and quantification are provided in Shin et al. [40].
RNA extraction, library preparation and sequencing
For mRNA extraction, parental and F1 strains were grown for 10 days in 50 ml and cells were pelleted by centrifugation (4500 r.p.m. for 8 min at 4 °C). Just after centrifugation, cell pellets were sonicated on ice for 30 s in LBP buffer (Macherey-Nagel, Duren, Germany). Extraction was performed using NucleoSpin RNA Plus kit (Macherey-Nagel) following the manufacturer’s protocol. Extracted RNA was quantified using a Biotek Epoch. Library preparation was performed using the Illumina mRNA TruSeq stranded kit starting from 0.5 µg of total RNA. Library quality was assessed on a Bioanalyzer using high-sensitivity DNA analysis chips. Paired-end sequencing was performed using 2×150 bp cycles on Illumina Hiseq 3000 at the GeT-PlaGe France Genomics sequencing platform (Toulouse, France).
Bioinformatics analysis
Alignment and genotyping
Ambiguous, low quality reads, sequencing adapters and spliced-leader sequences were trimmed from raw reads using Trimmomatic version 0.33 [68] with parameters ILLUMINACLIP:Adapters.fasta : 2:30:10, LEADING : 3, TRAILING : 3, MAXINFO : 80:0.8, and MINLEN : 70.
Trimmed reads were aligned to a reference transcriptome developed previously [64] using BWA-MEM (Burrows-Wheeler Alignment Tool [69]). Only reads with a mapping score >10 were retained. Pairs for which the two reads did not map concordantly on the same transcript were removed. Samtools was used to sort and index bam files, as well as to generate a raw count table for gene expression analysis [70]. Freebayes [71] was used to genotype the strains using 457 368 SNPs previously validated [64]. Freebayes was first run enforcing diploidy. As A. minutum vegetative cells are haploid, monoclonal F1 strains had to be homozygotes at the various SNP positions. Non-monoclonal strains were identified and discarded based on the presence of numerous heterozygous SNPs. Freebayes was then run enforcing haploidy. Nucleotide divergence between the parental strains was calculated by dividing for each pair of strains the number of polymorphic SNP sites by the total number of sites covered by more than ten reads using BEDtools [72]. The genotype of the F1 strains was compared to the parental strains to infer the recombination pattern of the parental strains. Out of the 79 initial F1 cultures, 62 strains were identified as clonal and recombinant (Fig. 2), and used to build the genetic linkage map. Among them, ten strains were lost, thus 52 strains were considered to assess toxicity (Table S1).
Fig. 2.
SNP parental contribution for each F1 of NT1xNT2 (a), T1xNT1 (b), T2xNT2 (c), T1xT2 (d) crosses.
Annotation
The reference transcriptome was initially annotated based on homology with the UNIPROT/SWISSPROT database [64]. To improve the annotation of sxt genes, a tblastn [73] analysis was performed against the reference transcriptome using as queries sxt genes from dinoflagellates and cyanobacteria extracted from GenBank (sxt A to Z and sxtSUL, DIOX, PER, ACT) (e-value <1e−10). An additional blastx was performed with the transcripts identified by tblastn against the nr database (evalue <1e−5) with these candidate transcripts. TransDecoder version 5.5.0 [74] was used on the transcriptome to select the longest ORFs and generate the associated protein sequences. These sequences were searched against the HMM library pfam-A.hmm [75] using the hmmsearch function of the HMMER programme (version 3.1b2 [76]).
Genetic linkage map
Transcripts presenting genetic variability (polymorphic at one or several SNP positions) among the four parents were considered to develop a genetic linkage map (38 690 transcripts). Transcripts presenting multiple recombination events in the F1 strains, possibly reflecting chimeric transcripts (1798 transcripts), as well as transcripts displaying missing data in at least one strain (23 172 transcripts) were removed from the initial dataset. The remaining 13 720 transcripts were used to develop a genetic linkage map using the mstmap function of the R package Asmap [77–79], with default parameters except for bychr parameter (set to ‘false’, no preconception about the partition of transcripts in different chromosomes) and a p-value threshold set to 1e−7.
Genetic association and eQTL analysis of toxicity
Hereafter, a strong focus is made on the T2xNT2 cross. The genetic linkage between toxicity (considered as a qualitative trait: toxic, non-toxic) and the 13 720 transcripts used to build the map was tested using Fisher’s exact test, focusing on the T2xNT2 cross only, followed by a false discovery rate (FDR) correction, with significance set at 0.05. Transcripts displaying genetic variation between T2 and NT2, but not among the four parental strains and displaying a recombination pattern matching the identified region of interest were manually assigned to this region. Such an approach only focuses on genes present in the genomes of all the strains, but displays genetic variation among them.
An eQTL analysis was performed to identify SNP variants associated with gene expression. Raw read counts of all F1 RNA-Seq libraries were normalized using the vsd function of the Deseq2 package [80]. Still focusing on the T2xNT2 cross, these expression data were used to perform the eQTL analysis on the 31 transcripts of the gene cluster linked to toxicity (see Results) using the R package MatrixEQTL [81]. This analysis could indirectly identify genes underlying the ability to produce toxins due to a presence/absence pattern, as such genes would not be expressed if absent from the genome of a given strain.
Results
Recombinant F1 strains
Four parental strains (T1, T2, NT1, and NT2; Table 2, Table S1) differing genetically (pairwise nucleotide divergence ranging from 0.24–0.35 % based on 15e+06 bp) and in terms of toxin profiles were crossed. A total of 62 monoclonal F1s were obtained; 27 from the NT1xNT2 cross (named I to XXVII, Table S1), 23 from the T2xNT2 cross (named A to W, Table S1), seven from T1xT2 (named α, β, γ, δ, ε, Φ, and φ) and five from T1xNT1 (named a, b, c, e, and w). Considering the genetic peculiarities of dinoflagellates and since recombination patterns have never been investigated in A. minutum, the parental contribution was calculated for each of the 62 clonal F1s (Fig. 2). The mean SNP contribution of each parent did not differ from a 0.5/0.5 expectation (t-test: p-values between 0.27 and 0.71), confirming that all F1s are recombinant from the parental genotypes. The contribution averaged 0.503/0.497 overall (standard deviation 0.038), and 0.506/0.494, 0.518/0.482, 0.505/0.495, 0.505/0.495, for NT1xNT2 cross (Fig. 2a), T1xNT1 cross (Fig. 2b), T2xNT2 (Fig. 2c), and T1xT2 (Fig. 2d), respectively (standard deviations: 0.03, 0.03, 0.05, and 0.03, respectively). Moreover, the contribution of the two parents did not differ between crosses (Fisher Exact test, p-values=1).
Toxin inheritance
Toxin profiles and quotas were assessed at different growth phases. The analogues detected for a given strain are the same across analyses and replicates, except for analogues present in very small quantities, which are not always detected. The quantities of toxins produced vary between replicates (Table S2).
T1 parental strain produced eight toxins at a total toxin content of 40 pg per cell (Fig. 3a) and composed of 62 % of C1 and C2 analogues, 30 % of GTX3, 5 % GTX2 and traces of dcGTX2, dcGTX3, GTX5, and STX (<1 %). T2 parental strain produced only three analogues: GTX3 (55 %), GTX2 (26 %), and STX (19 %) for a toxin content of 1.4 pg per cell. The two parental strains NT1 and NT2 did not produce any of the 19 screened toxins at a detectable level. The toxin content in toxic F1s varied from 0.025 to 10 pg/cell.
Fig. 3.
Quantities of different PST analogues measured in parents (a) and offspring of T1xT2 (b), T1xNT1 (c), and T2xNT2 (d) crosses in pg/cell. NT1xNT2 offspring are not represented as they are all non-toxic.
No toxin was detected in any of the 27 F1s from the NT1xNT2 cross (Table S2). All the F1s from T1xT2 cross produced toxins (Fig. 3b). Of the four T1xNT1 F1s, only one, strain e, produced toxins (Fig. 3c). Of the 18 F1s from the T2xNT2 cross, 13 produced toxins (Fig. 3d). To test whether the ability to produce toxins followed Mendelian proportions in the progeny, a Fisher test was performed considering F1s of toxic x non-toxic crosses. This trait did not deviate significantly from the expected value of 50 % of toxic F1s (p-value 0.54 for both crosses taken together, p-value=0.3 for the T2xNT2 cross).
When considering analogues individually, C1 and C2 were produced in large amounts (32 and 31 %, respectively) by T1 but were never found in its progeny. The same applied to GTX5, although this analogue was produced by T1 at low amount. STX was present in the two toxic parents (albeit at trace level in T1, i.e. 0.2 % of toxin content) but not always detected, neither in T1xT2, T2xNT2 nor T1xNT1 F1s. In the T1xNT1 cross, dcGTX2 and dcGTX3 were the only analogues detected, in low amounts (0.08 and 0.03 pg/cell, respectively), in a single descendant (strain e), while the profile of the toxic parent consisted of eight toxins. These two analogues were also found in four toxic F1s (strains C, D, K and V) of the T2xNT2 cross, whereas they were not detected in either parent. These results showed a complex toxin profile inheritance that cannot be explained by Mendelian segregation at a single locus.
Linkage map
A linkage map was obtained by analysing the recombination pattern of 13 720 transcripts in the 62 clonal recombinant F1s. These transcripts were clustered into 1685 bins (a bin encompassing a set of co-localized markers) distributed across 139 linkage groups (Fig. 4), corresponding to a total length of 5727 centimorgans (cM). Linkage groups had an average length of 41. 2 cM (minimum 0, maximum 122. 09 cM). The smallest linkage group contained one transcript and the largest 275, distributed in 28 bins. The density (cM/number of transcripts) per transcript averaged 0 .43 cM/transcript (minimum 0, maximum 1.88). The distance between two adjacent markers averaged 3 61 cM (minimum 1.61, maximum 16. 7 cM) and this distance was below 5 cM in 82.9 % of cases.
Fig. 4.
Genetic map: 139 linkage groups ordered according to their size in cM. Orange area represents the toxicity-related gene cluster (l62). Blue areas are not on scale and each one represents the location of one sxt homologue in the transcriptome.
Genetic linkage
To investigate the possibility of a genomic region associated with the ability to produce toxins (qualitative trait), the focus was placed on the 18 F1s (five non-toxic, 13 toxic) from the T2xNT2 cross. A region correlated to this toxin-producing ability was identified (Fig. 5a). This region was located on the L62 linkage group, between positions 0 and 16.2 cM (Fig. 5b). It consisted of 31 transcripts. This region was then manually enriched (see Methods) up to 50 transcripts, of which 25 could be annotated (Table 3, Table S3 for all pfam domain hits and GO terms). These include, among others, a sulfotransferase, two potassium channels and a caspase domain. None of the transcripts present in L62 matched any sxt homologue.
Fig. 5.
Genomic association of the ability to produce toxins (a) Results of the Fisher test associating linkage groups with the toxin-producing capacity of F1s from the T2xNT2 cross; (b) Focus on linkage group 62 containing the region (0 to 16.2 cM) significantly related to the ability to produce toxins in T2xNT2 F1s.
Table 3.
L62 transcript annotation. Only the pfam domain with the best overall score was shown for each transcript. The list of all pfam domains, as well as the list of GO terms associated with each transcript, are available in Table S3
|
Transcript |
Position in L62 |
Putative function |
Best pfam domain hit |
Pfam evalue |
Uniprot hit |
Uniprot evalue |
|---|---|---|---|---|---|---|
|
comp104658_c0_seq1 |
0 cM |
Sulfotransferase |
Sulfotransfer_3 |
1.4e-06 |
/ |
/ |
|
comp99374_c0_seq1 |
0 cM |
Diphthamide biosynthesis / Chaperone DnaJ |
DnaJ |
1.7e-13 |
DPH4_ASHGO |
1,00e-08 |
|
comp103051_c0_seq1 |
0 cM |
/ |
/ |
/ |
/ |
/ |
|
comp111463_c0_seq2 |
0 cM |
/ |
/ |
/ |
/ |
/ |
|
comp32533_c0_seq1 |
0 cM |
/ |
/ |
/ |
/ |
/ |
|
comp65252_c0_seq1 |
0 cM |
/ |
/ |
/ |
/ |
/ |
|
comp74963_c0_seq1 |
0 cM |
/ |
/ |
/ |
/ |
/ |
|
comp128798_c0_seq1 |
3.2 cM |
EGF-like domain / Tyrosine protein-kinase |
Ephrin_rec_like |
3.6e-06 |
SCUB1_HUMAN |
3,00e-06 |
|
comp106553_c0_seq1 |
3.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp111972_c0_seq1 |
4.8 cM |
EF-hand |
EF-hand_1 |
0.019 |
/ |
/ |
|
comp117087_c1_seq1 |
4.8 cM |
Tetratricopeptide repeat |
TPR_12 |
1.8e-21 |
TTC28_MOUSE |
1,00e-09 |
|
comp130842_c0_seq1 |
4.8 cM |
EamA-like transporter family |
EamA |
6.4e-06 |
/ |
/ |
|
comp94939_c0_seq1 |
4.8 cM |
/ |
/ |
/ |
/ |
/ |
|
comp105001_c0_seq1 |
6.4 cM |
Leucyl/phenylalanyl-tRNA--protein transferase |
Leu_Phe_trans |
8.8e-24 |
/ |
/ |
|
comp121729_c0_seq1 |
6.4 cM |
Ankyrin repeat |
Ank_4 |
1.9e-24 |
ANR50_HUMAN |
3,00e-13 |
|
comp125039_c0_seq1 |
6.4 cM |
Glycosyltransferase |
Glyco_tranf_2_3 |
0.0079 |
/ |
/ |
|
comp129241_c1_seq1 |
6.4 cM |
/ |
/ |
/ |
/ |
/ |
|
comp131122_c0_seq1 |
6.4 cM |
/ |
/ |
/ |
/ |
/ |
|
comp101486_c0_seq1 |
16.2 cM |
Potassium channel / Ion channel |
Ion_trans_2 |
7.7e-23 |
TOK1_YEAST |
8,00e-11 |
|
comp102705_c1_seq4 |
16.2 cM |
NAD-binding domain |
NAD_binding_10 |
2.5e-10 |
/ |
/ |
|
comp102814_c0_seq1 |
16.2 cM |
GTP-ase |
MMR_HSR1_Xtn |
3.3e-34 |
DRG2_ARATH |
9,00e-125 |
|
comp109406_c0_seq1 |
16.2 cM |
Chlorophyll A-B binding protein |
Chloroa_b-bind |
2.4e-23 |
FCPA_MACPY |
6,00e-13 |
|
comp115397_c0_seq1 |
16.2 cM |
Methyltransferase |
Methyltransf_21 |
1.6e-05 |
/ |
/ |
|
comp116297_c0_seq1 |
16.2 cM |
Peptidase C14 (Caspase/Metacaspase) |
Peptidase_C14 |
4.5e-17 |
MCA2_ARATH |
2,00e-17 |
|
comp118522_c0_seq1 |
16.2 cM |
Potassium channel / Ion channel |
Ion_trans_2 |
1.5e-21 |
KCO5_ARATH |
3,00e-09 |
|
comp123175_c0_seq1 |
16.2 cM |
Amine oxidase |
Amino_oxidase |
5.6e-36 |
PPOC_TOBAC |
7,00e-121 |
|
comp123308_c0_seq2 |
16.2 cM |
Alpha/beta hydrolase |
Abhydrolase_3 |
6.9e-12 |
/ |
/ |
|
comp124989_c0_seq1 |
16.2 cM |
Pyridine nucleotide-disulphide oxidoreductases |
Pyr_redox_2 |
3.3e-51 |
/ |
/ |
|
comp125738_c0_seq1 |
16.2 cM |
Cysteine dioxygenase |
CDO_I |
3.3e-53 |
CDO1_HUMAN |
3,00e-27 |
|
comp125984_c0_seq1 |
16.2 cM |
WEB protein/Apolipoprotein |
Apolipoprotein |
0.002 |
Y5673_ARATH |
4,00e-04 |
|
comp129241_c0_seq1 |
16.2 cM |
Protein kinase / Cyclic nucleotide-binding domain |
cNMP_binding |
1.1e-11 |
KAPR_ASPNG |
5,00e-14 |
|
comp24034_c0_seq1 |
16.2 cM |
Galactosyltransferase |
Galactosyl_T |
2,00E-06 |
/ |
/ |
|
comp74512_c0_seq1 |
16.2 cM |
Plastid lipid-associated protein |
PAP_fibrillin |
0.00057 |
/ |
/ |
|
comp125555_c0_seq9 |
16.2 cM |
Myosin-2 heavy chain |
/ |
/ |
MYS2_DICDI |
3,00e-05 |
|
comp100745_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp101484_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp113111_c3_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp118918_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp121267_c1_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp122552_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp124016_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp124374_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp129321_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp130665_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp131282_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp25504_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp26886_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp29037_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp29558_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
|
comp30077_c0_seq1 |
16.2 cM |
/ |
/ |
/ |
/ |
/ |
In the T1xNT1 cross, the distribution of SNPs in the L62 region did not match the toxicity pattern observed in the offspring (Table S8): F1s b, c and e all had the SNP of the non-toxic parent in 43 transcripts; offspring ‘a’ had the SNP of the toxic parent in only 16 transcripts (from 0 to 6.4 cM); finally, in seven transcripts there was no polymorphic SNP between the two parents.
Identifying sxt homologues
A total of 68 transcripts homologues to cyanobacterial sxt genes were identified in the reference transcriptome (Table S4). Of these, 29 are monomorphic and do not display any SNP, 35 do not form a gene cluster and are distributed on 30 different linkage groups (Fig. 4).
Four linkage groups contained two sxt genes: region L1 contained one sxtA and one sxtU in positions 74.6 and between 90.8 and 92.4 cM respectively; region L11 contained two sxtU in positions 79.6 and between 58.3 and 59.9 cM; region L28 contained two sxtU in position 3.23 cM; region L67 contains one sxtU and one sxtA between positions 13.1 and 19.5 cM and in position 26.02 cM respectively. Only three of the 12 putative sxtA homologues could be assigned to linkage groups. Neither of them was in the same linkage group. Finally, four sxt homologues could not be assigned to a particular linkage group, despite having differentiated SNPs between parents. Among them, an sxtA -long isoform- (comp112540_c0_seq1) and an sxtU (comp108760_c0_seq1) could not be genotyped for all offspring (Table S5) but displayed a recombination pattern incompatible with any linkage group (including L62). The other two appeared to be chimeric transcripts.
eQTL
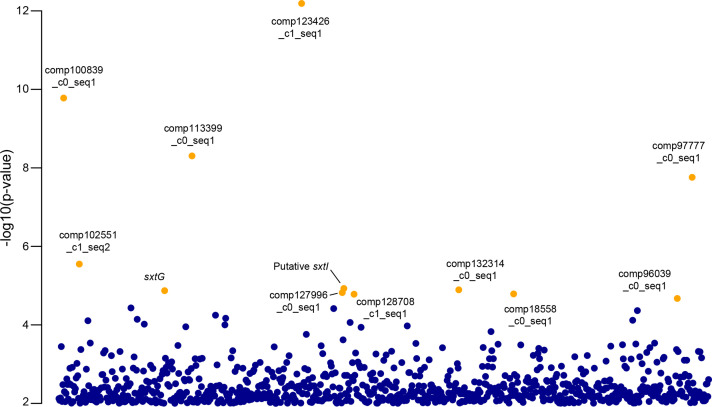
The genetic linkage analysis allowed us to detect inherited differences on coding sequences. However, genetic differences between strains may be carried by non-coding elements, such as regulatory sequences, to which our genetic linkage analysis does not give access (as only the expressed sequences are considered). An eQTL analysis was performed to link the L62 region associated with toxin production to gene expression. This analysis revealed 12 transcripts, whose expression is linked to the 0–16.2 region of the L62 linkage group (Fig. 6, FDR<0.2). One, comp97777_c0_seq1, belonged to the toxicity-related L62 cluster, six belonged to other linkage groups, and five monomorphic transcripts could not be assigned to any linkage group (Table S6).
Fig. 6.
eQTL analysis associating the L62 region (0 to 16.2 cM) with the expression of certain transcripts (in orange the transcripts, whose expression is significantly linked to this region).
Six of these 12 transcripts are annotated, two of which correspond to sxt homologues. The first one, comp110949_c1_seq1 (Fig. 6, Fig. 7b) is a sxtG homologue (Table S4) catalysing the second step of the PST biosynthetic pathway in cyanobacteria. The second one, comp127996_c2_seq1 (Fig. 6, Fig. 7a) corresponds to the sxtI gene, a carbamoyltransferase involved in the last step leading to complete formation of STX in cyanobacteria. Both of these transcripts are overexpressed in toxic F1s compared to non-toxic ones. The other identified transcripts are poorly annotated (Table S6).
Fig. 7.
Normalized expression of transcripts found by eQTL analysis, in F1s of all crosses according to their phenotype (toxic or non-toxic).
The expression of the 12 transcripts identified by the eQTL analysis was then examined in all F1s of the four crosses (Fig. 7). The sxtI homologue (Fig. 7a) and comp127996_c0_seq1 (Fig. 7f) showed the same expression pattern, with a clear demarcation between toxic and non-toxic individuals. The only notable exception being the toxic T1xNT1 e strain, displaying contrasting expression levels compared to the other toxic strains. It should be noted that this specific strain displayed both very low toxicity levels and a singular toxinic profile (Fig. 3c). The sxtG (Fig. 7b) homologue, comp123426_c1_seq1 (Fig. 7d), comp100839_c0_seq1 (Fig. 7e) and comp102551_c1_seq2 (Fig. 7g) followed the same pattern of demarcation between toxic and non-toxic F1s, but with a less clear delimitation than sxtI and comp127996_c0_seq1. The six remaining transcripts displayed differential expression only in the T2xNT2 F1s with high variability for the NT1xNT2 F1s (Fig. 7c and h-l).
Discussion
The present study developed the first genomic association study in dinoflagellates to identify genetic regions associated with toxin production. Following crossing experiments and F1 phenotyping, the ability to produce PST appeared inherited as a simple Mendelian trait compatible with a single locus genetic determinism. However, toxin profile inheritance, i.e. the types of PST analogues produced, was not compatible with Mendelian inheritance and was probably governed by a more complex genetic determinism involving several genes. SNP genotyping and subsequent recombination analyses were used to obtain the first dinoflagellate genetic map. Contrasting with PST-producing cyanobacteria, in which the sxt genes responsible for toxin production were grouped in a single gene cluster, dinoflagellate sxt genes appeared scattered in numerous linkage groups. In accordance with the simple Mendelian inheritance hypothesis, a single genomic region was associated with the ability to produce PSTs. This genomic region did not carry any polymorphic sxt gene. An eQTL analysis identified several transcripts, including two sxt, whose expression seemed to be controlled by the genomic region associated with the ability to produce PSTs. These results are discussed below.
Linkage map and genomic association study in dinoflagellates
Very few linkage maps were developed for members of the SAR clade. Previous maps were obtained from species with genome sizes ranging from 23 to 560 Mbp [17, 20–23, 82], while A. minutum genome is estimated to be ~25 Gbp [47]. Despite this huge difference in genome size, the A. minutum linkage map was only slightly larger than previous ones, with 139 linkage groups spanning 5727 cM, compared to fewer than 60 linkage groups spanning 1782.7 to 2883. 9 cM in previous studies. The average distance between two markers is 3.6 cM, compared to the mean distance ranging from 0.3 to 6.6 cM in previous studies, indicating that the A. minutum map is resolutive. Finally, although linkage groups do not necessarily correspond to chromosomes, as a matter of comparison, 143 chromosomes were observed in A. fundyense [83].
Recent genomic association studies tend to focus on quantitative traits, requiring hundreds or even thousands of individuals [24]. However, traits of biological interest are not necessarily quantitative. This is especially the case for the ability to produce toxins or disease susceptibility [84]. The present study clearly demonstrates that a few tens of offspring may be sufficient to be resolutive for genomic association studies focusing on such qualitative traits.
Toxicity inheritance
Mendelian inheritance of the ability to produce toxins
In agreement with previous studies [85–88], the present study showed that the PST profile, i.e. the ability to produce toxins and the types of analogues produced, may be considered as a character rather stable for a given strain. The only marginal exceptions were for analogues produced at concentrations close to the detection limits, which may rather reflect technical limitations (already noticed in [88]). The higher variability of quantities and ratio of analogues is also perfectly in agreement with previous studies showing that environmental factors (e.g. biotic, abiotic, nutrients, physiological state) can modulate the toxin level [89–94].
None of the F1s obtained from crosses between the two non-toxic strains were toxic, all of the F1s obtained from crosses between the two toxic strains were toxic and the ability to produce toxin did not deviate from the 50 % expectation when toxic and non-toxic parental strains were crossed. As vegetative dinoflagellates cells are haploids, there is no dominance involved. These results suggest that the ability to produce toxins may be inherited as a simple mono locus Mendelian trait. This finding was in agreement with the genomic association analysis that identified a single genomic region correlating with the ability to produce toxins. Martins et al. [95] reported a culture of A. minutum that lost its ability to produce toxins, while a subculture was still able to produce PSTs. Since this loss was a non-transient feature, a mutation in the PST biosynthetic pathway was considered rather than differential regulation at the gene level. This is consistent with this study, where the ability to produce is indeed controlled by a single genetic region in T2xNT2 cross. However, there was no association between the distribution of SNPs and the ability to produce toxins in the four offspring of the T1xNT1 cross, suggesting an alternative mechanism for this cross.
Multigenic bases of the toxin profiles
When examining the types of analogues produced, inheritance tends to be more complex. It deviated from Mendelian expectations and probably involved several loci. The best illustration concerned C1 and C2 analogues, produced in large amounts by T1, but never found in F1s. If the transmission was indeed Mendelian, the probability of finding C1 and C2 analogues in the five F1s of the T1xT2 cross would be 1/2 per F1, leading to a probability of finding no F1 with the T1 profile of 3 %. These results indicated that the types of analogues produced have a multigenic basis. The partial inheritance of the toxin profiles contrasted with previous work [63] in A. catenella that highlighted a strict Mendelian inheritance of the toxinic profiles of the parental strains in F1, without any modification of the parental profiles. The strains used by Sako et al. [63] belonged to a different species and displayed toxinic profiles extremely different from the ones analysed in the present study. Altogether, these results illustrate that depending on the genetic make-up of the parental strains and on the produced analogues, parental profiles may sometimes be inherited as a whole and sometimes strongly modified by sexual reproduction.
This finding is perfectly compatible with the sxt genes being spread-out in the genomes of dinoflagellates rather than regrouped in a single cluster as in cyanobacteria. Considering that sxt genes are involved in PST production in dinoflagellates, this may have major consequences regarding the ability to produce the various PST analogues and on gene expression. First, in perfect accordance with the complex pattern of PST analogues inheritance identified in the present study, meiosis following gamete fusion would lead to new combinations of alleles with potential impacts on the ability to produce the various PST analogues. Second, in gene clusters the same cis module may co-regulate several genes, while independent cis modules would be required to regulate the expression of sxt genes spread out in various genomic regions. Since each of these independent modules can diverge genetically, this would imply the possibility of independent regulation of all these genes, even with a single trans-acting regulator.
In line with this hypothesis, a major result was the absence of genetically variable sxt genes in the region of interest. Indeed, such genes have long been associated with toxin production in cyanobacteria [43, 50, 51], and the identification of numerous homologues in dinoflagellates ([52]; present study) make them ideal candidates to explain toxin production in dinoflagellates. Here, 68 sxt homologues were identified. None of the 39 sxt homologues displaying genetic variability among parental strains was present in the genomic region associated with toxicity. The 29 remaining were genetically monomorphic in the parental strains and could not be assigned to a linkage group. As these genes are monomorphic, genetic variation of their coding region cannot be involved in the ability to produce toxins. However, genetic variation in the cis regulatory region could translate into gene expression differences. Out of the 29 monomorphic sxt homologues, two displayed over-expression in the toxic strains. The two sxt homologues, whose expression were upregulated in toxic strains, were sxtG and sxtI. These two genes were previously reported as candidates involved in toxicity in dinoflagellates. The sxtG homologues were reported in PST-producing dinoflagellate species only [56] and sxtG expression was upregulated in experimental conditions favouring toxin production [96]. Similarly, sxtI expression was upregulated in conditions favouring toxin production, and downregulated when toxin synthesis was stopped [96, 97]. Although identifying the molecular bases of the production of the various analogues is beyond the scope of the present study, the case of the strain e (T1xNT1 cross) might be worth being mentioned. In this strain, only dcGTX2 and three analogues are present and sxtI expression is extremely low. Based on the dinoflagellate putative biosynthetic pathway [98], carbamoylated analogues (GTX1 to 6, C1 to C4, STX, and NeoSTX) seem to be catalysed by SxtI, whereas decarbamoylated analogues (‘dc’-type analogues) appear to be produced from dc-STX without involving SxtI. The absence of C1 and C2 in strain e could be explained by such a mechanism.
Gene expression regulation of sxtI and sxtG by the toxicity-related L62 region adds additional evidence that these two genes are related to toxin production in dinoflagellates and could be compatible with the above-mentioned hypothesis of trans-regulation by L62 of the genes responsible for the toxin profile.
Candidate genes
No evidence for sxtA implication in the ability to produce PST
The sxtA gene is by far the most studied when investigating the production of PSTs. This gene presents two isoforms in dinoflagellates: a short form comprising three domains (A1 to A3) homologue to cyanobacterial sxtA domains and a fourth domain without any known homology [64] and a long form with the four cyanobacterial domains (A1 to A4 [53]). It was postulated that this long form would be the cause of toxicity in dinoflagellates as it was repeatedly found in toxic strains, and rarely in non-toxic ones [52, 53, 56, 99, 100]. A recent study identified the presence of the long isoform in both toxic and non-toxic parental strains used in the present study among others [47]. At least two homologues of the long form of sxtA were identified in the present study. Neither of them appeared to be related to the ability to produce toxins. One could be assigned to the L1 linkage group. The other one could not be assigned to any group, but distribution of parental markers in the progeny was clearly incompatible with L62. Similarly, sxtA expression was not controlled by this region. This is consistent with Wiese et al. [101] who showed that sxtA4 expression in A. catenella does not vary when the amount of PSTs changes between different growth phases. However, in the same species, sxtA4 expression has been shown to vary between toxic and non-toxic mutants [100]. In addition, a strain of A. catenella, containing both toxic and non-toxic cells, has been studied multiple times to elucidate the mechanisms responsible for the loss of toxicity of part of these cells [102–105]. In the non-toxic subclone, sxtA4 expression was significantly reduced. It was postulated that changes in the 3′UTR region of sxtA4 were responsible for the loss of toxicity via post-transcriptional regulatory mechanisms [105]. In our study, the mechanism is probably distinct, as sxtA4 expression did not change between toxic and non-toxic individuals. However, a distinct post-transcriptional or translational mechanism, potentially regulated by the L62 region, cannot be excluded to explain the loss of toxicity in F1 strains.
At an evolutionary scale, it has been shown that of the three sxtA1 isoforms detected in dinoflagellates, only one is thought to be specific to STX-producing species [56]. In the present study, genetic variation of the sxtA1 isoforms could not be related to toxicity. The evolution of sxtA by acquisition of the specific domain sxtA4 has also been suggested in toxin-producing cyanobacteria and dinoflagellates [106]. In the present study, and as mentioned above, the polymorphism of sxtA4 does not correspond to the variations in toxicity.
Altogether, our study and previous ones suggest that depending on the precise genetic make-up of the strains, there may be multiple ways to lose the ability to produce PSTs in dinoflagellates.
Other genes involved in toxin production
The implication of L62 or of differentially expressed genes without homology with sxt in PST production cannot be excluded. It is however difficult to identify clear candidates. A major reason is the lack of homology with genes of identified functions for a great majority of dinoflagellate genes [64]. In the L62 region of interest, only 25/50 genes had a putative homology with a protein domain. The putative functions may be attributed to many different cellular processes (e.g. methyltransferase-, carboxylesterase- domains). Nevertheless, it is worth mentioning some of the identified homologues since their functions have previously been associated with toxin production in different marine dinoflagellates. For example, comp104658_c0_seq1 displays a domain homologous to a sulfotransferase (L62, 0 cM). In Gymnodinium catenatum, O-sulfotransferases are responsible for the transformation of STX into GTX 2 and 3 [107].
A second candidate might be a gene encoding a metacaspase (L62, 16 cM). This metacaspase is a specific arginine/lysine protease. Caspases, as well as their counterparts in bacteria and plants, orthocaspases and metacaspases, are involved in programmed cell death, however these enzymes might have other functions in plants, algae and cyanobacteria such as cell homeostasis, culture ageing [108] or even roles in survival pathways and cell cycle progression as suggested in A. tamarense [109]. A study on metacaspases from environmental samples from the Baltic Sea also indicates that several metacaspases were in the same expression cluster than the genes related to toxin production in Nodularia spumigena , suggesting a link between toxin synthesis, metacaspase expression and programmed cell death [110].
Finally, of the 12 transcripts identified by eQTL analysis, two were not annotated, including comp127996_c0_seq1, for which the expression pattern is identical to that of sxtI. Further studies are needed to discover or confirm the putative functions of the identified genes, and their implication in the PST biosynthetic pathway in dinoflagellates, e.g. by genetic knock-out, long read sequencing of part of the genome (L62 region for example), or by screening natural populations or a larger number of offspring.
Conclusion
The present study showed that the ability to produce toxins in A. minutum is an inheritable Mendelian character and seems to be controlled by a single genetic region. This region does not contain homologues of sxt genes identified in cyanobacteria. However, it seems to control the expression of sxtI and sxtG gene homologues. Unfortunately, it was impossible to determine whether these two homologues are physically present in the genomic region associated with toxicity, if they displayed genetic polymorphism in their regulatory region (cis-regulation), or if they are in another linkage group regulated by a gene present in L62 (trans-regulation). A starting point to test these hypotheses would require the development of specific sequencing strategies targeting the cis regulatory or the entire L62 regions.
The present study also showed that toxin profiles seem to be controlled by several genes, since these profiles are not transmitted as a whole in progeny. This multigenetic origin of the profiles would be in accordance with the sxt genes being spread out in multiple linkage groups.
The sxtA homologues were not found to be related to toxin production in this study, as they were absent from the identified genetic region and were not differentially expressed between toxic and non-toxic strains. Caution has nevertheless to be taken, as gene expression regulation in dinoflagellates is believed to be mainly driven by post-transcriptional mechanisms involving trans-splicing [34]. Finally, most of the newly discovered candidate genes are not annotated and therefore represent new perspectives to unravel the mechanisms that govern the synthesis of PSTs in dinoflagellates.
Supplementary Data
Funding information
This work was financially supported by the Bretagne Region and Ifremer Research Institute.
Acknowledgements
The authors acknowledge Pascale Malestroit for preliminary work that lead to the obtention of the F1 strains, Laure Guillou for initially providing the parental strains, Sébastien Artigaud for insightful comments, the Pôle de Calcul et de Données Marines (PCDM) for access to the DATARMOR calculator, the Sebimer team for bioinformatic assistance, the GetPlage sequencing facility for sequencing, and the reviewers for their insightful comments.
Author contribution
Conceptualization: M.L.G., Data curation, Formal analysis: L.M., M.L.G., Funding acquisition: M.L.G., Investigation: J.Q., M.L., G.A.R., D.R., M.L.G., Methodology, Project administration, Resources, Software, Supervision: M.L.G., D.R., H.H., Validation, Visualization: L.M., Writing – original draft: L.M., Writing – review & editing: M.L.G., L.M., D.R., H.H.
Conflicts of interest
The author(s) declare that there are no conflicts of interest.
Footnotes
Abbreviations: eQTL, expression quantitative trait loci; HAB, Harmful algal bloom; PSP, paralytic shellfish poisoning; PST, paralytic shellfish toxin; SNP, single nucleotide polymorphism; STX, saxitoxin.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary tables are available with the online version of this article.
References
- 1.Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Zool. 1913;14:43–59. doi: 10.1002/jez.1400140104. [DOI] [Google Scholar]
- 2.Emerson RA, Beadle GW, Fraser AC. A Summary of Linkage Studies in Maize: Memoir. Ithaca, New York: Cornell University, Agricultural Experiment Station; 1935. p. 84. p. [Google Scholar]
- 3.Ingvarsson PK, Street NR. Association genetics of complex traits in plants. New Phytol. 2011;189:909–922. doi: 10.1111/j.1469-8137.2010.03593.x. [DOI] [PubMed] [Google Scholar]
- 4.Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana . Proc Natl Acad Sci. 1999;96:4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juenger T, Purugganan M, Mackay TFC. Quantitative trait loci for floral morphology in Arabidopsis thaliana . Genetics. 2000;156:1379–1392. doi: 10.1093/genetics/156.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne M, Murrell JC, Owen JV, Williams ER, Moran GF. Mapping of quantitative trait loci influencing frost tolerance in Eucalyptus nitens . Theor Appl Genet. 1997;95:975–979. doi: 10.1007/s001220050650. [DOI] [Google Scholar]
- 7.Frewen BE, Chen TH, Howe GT, Davis J, Rohde A, et al. Quantitative trait loci and candidate gene mapping of bud set and bud flush in populus. Genetics. 2000;154:837–845. doi: 10.1093/genetics/154.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 9.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 10.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, et al. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 12.Kobras CM, Fenton AK, Sheppard SK. Next-generation microbiology: from comparative genomics to gene function. Genome Biol. 2021;22:123. doi: 10.1186/s13059-021-02344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet. 2017;18:41–50. doi: 10.1038/nrg.2016.132. [DOI] [PubMed] [Google Scholar]
- 14.Shirley MW, Harvey DA. A genetic linkage map of the apicomplexan protozoan parasite Eimeria tenella . Genome Res. 2000;10:1587–1593. doi: 10.1101/gr.149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Lee T, Testa A, Robold A, van’t Klooster J, Govers F. High-density genetic linkage maps of Phytophthora infestans reveal trisomic progeny and chromosomal rearrangements. Genetics. 2004;167:1643–1661. doi: 10.1534/genetics.104.029652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinelli A, Hunt P, Fawcett R, Cravo PVL, Walliker D, et al. An AFLP-based genetic linkage map of Plasmodium chabaudi chabaudi. Malar J. 2005;4:11. doi: 10.1186/1475-2875-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blake DP, Oakes R, Smith AL. A genetic linkage map for the apicomplexan protozoan parasite Eimeria maxima and comparison with Eimeria tenella . Int J Parasitol. 2011;41:263–270. doi: 10.1016/j.ijpara.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Katzer F, Lizundia R, Ngugi D, Blake D, McKeever D. Construction of a genetic map for Theileria parva: identification of hotspots of recombination. Int J Parasitol. 2011 May;41(6):669–675. doi: 10.1016/j.ijpara.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Pattaradilokrat S, Zhu F, Jiang H, Liu S, et al. Linkage maps from multiple genetic crosses and loci linked to growth-related virulent phenotype in Plasmodium yoelii . Proc Natl Acad Sci. 2011;108:E374–82. doi: 10.1073/pnas.1102261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heesch S, Cho GY, Peters AF, Le Corguillé G, Falentin C, et al. A sequence-tagged genetic map for the brown alga Ectocarpus siliculosus provides large-scale assembly of the genome sequence. New Phytol. 2010;188:42–51. doi: 10.1111/j.1469-8137.2010.03273.x. [DOI] [PubMed] [Google Scholar]
- 22.Shan T, Pang S, Li J, Li X, Su L. Construction of a high-density genetic map and mapping of a sex-linked locus for the brown alga Undaria pinnatifida (Phaeophyceae) based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Genomics. 2015;16:902. doi: 10.1186/s12864-015-2184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N, Zhang L, Tao Y, Guo L, Sun J, et al. Construction of a high density SNP linkage map of kelp (Saccharina japonica) by sequencing Taq I site associated DNA and mapping of a sex determining locus. BMC Genomics. 2015;16:189. doi: 10.1186/s12864-015-1371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vales MI, Schön CC, Capettini F, Chen XM, Corey AE, et al. Effect of population size on the estimation of QTL: a test using resistance to barley stripe rust. Theor Appl Genet. 2005;111:1260–1270. doi: 10.1007/s00122-005-0043-y. [DOI] [PubMed] [Google Scholar]
- 25.Bachvaroff TR, Place AR. From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae . PLoS ONE. 2008;3:e2929. doi: 10.1371/journal.pone.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzo PJ. Those amazing dinoflagellate chromosomes. Cell Res. 2003;13:215–217. doi: 10.1038/sj.cr.7290166. [DOI] [PubMed] [Google Scholar]
- 27.Livolant F, Bouligand Y. New observations on the twisted arrangement of Dinoflagellate chromosomes. Chromosoma. 1978;68:21–44. doi: 10.1007/BF00330370. [DOI] [Google Scholar]
- 28.Rill RL, Livolant F, Aldrich HC, Davidson MW. Electron microscopy of liquid crystalline DNA: direct evidence for cholesteric-like organization of DNA in dinoflagellate chromosomes. Chromosoma. 1989;98:280–286. doi: 10.1007/BF00327314. [DOI] [PubMed] [Google Scholar]
- 29.Johnson JG, Morey JS, Neely MG, Ryan JC, Van Dolah FM. Transcriptome remodeling associated with chronological aging in the dinoflagellate, Karenia brevis. Mar Genomics. 2012;5:15–25. doi: 10.1016/j.margen.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Erdner DL, Anderson DM. Global transcriptional profiling of the toxic dinoflagellate Alexandrium fundyense using massively parallel signature sequencing. BMC Genomics. 2006;7:88. doi: 10.1186/1471-2164-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moustafa A, Evans AN, Kulis DM, Hackett JD, Erdner DL, et al. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. aziz RK, editor. PLoS ONE. 2010;5:e9688. doi: 10.1371/journal.pone.0009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin S. Genomic understanding of dinoflagellates. Res Microbiol. 2011;162:551–569. doi: 10.1016/j.resmic.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Seveno J, Even Y, Le Gac M. Strong constitutive expression divergence among strains but no evidence of differential expression associated with sexual reproduction in Alexandrium minutum . Harmful Algae. 2020;100:101940. doi: 10.1016/j.hal.2020.101940. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, et al. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lidie KB, van Dolah FM. Spliced leader RNA-mediated trans-splicing in a dinoflagellate, Karenia brevis . J Eukaryot Microbiol. 2007;54:427–435. doi: 10.1111/j.1550-7408.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 36.Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS ONE. 2009;4:e6978. doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 38.Stephens TG, González-Pech RA, Cheng Y, Mohamed AR, Burt DW, et al. Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions. BMC Biol. 2020;18:56. doi: 10.1186/s12915-020-00782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson DM, Alpermann TJ, Cembella AD, Collos Y, Masseret E, et al. The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae. 2012;14:10–35. doi: 10.1016/j.hal.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin HH, Li Z, Réveillon D, Rovillon G-A, Mertens KN, et al. Centrodinium punctatum (Dinophyceae) produces significant levels of saxitoxin and related analogs. Harmful Algae. 2020;100:101923. doi: 10.1016/j.hal.2020.101923. [DOI] [PubMed] [Google Scholar]
- 41.Wiese M, D’Agostino PM, Mihali TK, Moffitt MC, Neilan BA. Neurotoxic alkaloids: saxitoxin and its analogs. Mar Drugs. 2010;8:2185–2211. doi: 10.3390/md8072185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu Y. Microalgal metabolites. Chem Rev. 1993;93:1685–1698. doi: 10.1021/cr00021a002. [DOI] [Google Scholar]
- 43.Kellmann R, Mihali TK, Jeon YJ, Pickford R, Pomati F, et al. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl Environ Microbiol. 2008;74:4044–4053. doi: 10.1128/AEM.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchiya S, Cho Y, Konoki K, Nagasawa K, Oshima Y, et al. Synthesis and identification of proposed biosynthetic intermediates of saxitoxin in the cyanobacterium Anabaena circinalis (TA04) and the dinoflagellate Alexandrium tamarense (Axat-2) Org Biomol Chem. 2014;12:3016–3020. doi: 10.1039/c4ob00071d. [DOI] [PubMed] [Google Scholar]
- 45.Tsuchiya S, Cho Y, Konoki K, Nagasawa K, Oshima Y, et al. Biosynthetic route towards saxitoxin and shunt pathway. Sci Rep. 2016;6:20340. doi: 10.1038/srep20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho Y, Tsuchiya S, Yoshioka R, Omura T, Konoki K, et al. Column switching combined with hydrophilic interaction chromatography-tandem mass spectrometry for the analysis of saxitoxin analogues, and their biosynthetic intermediates in dinoflagellates. J Chromatogr A. 2016;1474:109–120. doi: 10.1016/j.chroma.2016.10.065. [DOI] [PubMed] [Google Scholar]
- 47.Geffroy S, Lechat MM, Le Gac M, Rovillon GA, Marie D, et al. From the sxtA4 gene to saxitoxin production: what controls the variability among Alexandrium minutum and Alexandrium pacificum strains? Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.613199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Agostino VC, Krock B, Degrati M, Sastre V, Santinelli N, et al. Occurrence of toxigenic microalgal species and phycotoxin accumulation in mesozooplankton in Northern Patagonian Gulfs, Argentina. Environ Toxicol Chem. 2019;38:2209–2223. doi: 10.1002/etc.4538. [DOI] [PubMed] [Google Scholar]
- 49.Kellmann R, Mihali TK, Michali TK, Neilan BA, Neilan BA. Identification of a saxitoxin biosynthesis gene with a history of frequent horizontal gene transfers. J Mol Evol. 2008;67:526–538. doi: 10.1007/s00239-008-9169-2. [DOI] [PubMed] [Google Scholar]
- 50.Mihali TK, Kellmann R, Neilan BA. Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 2009;10:8. doi: 10.1186/1471-2091-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mihali TK, Carmichael WW, Neilan BA. In: PLoS ONE. Hofmann A, editor. Vol. 6. 2011. A putative gene cluster from a Lyngbya wollei bloom that encodes paralytic shellfish toxin biosynthesis; e14657. vol. p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hackett JD, Wisecaver JH, Brosnahan ML, Kulis DM, Anderson DM, et al. Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol Biol Evol. 2013;30:70–78. doi: 10.1093/molbev/mss142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stüken A, Orr RJS, Kellmann R, Murray SA, Neilan BA, et al. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE. 2011;6:e20096. doi: 10.1371/journal.pone.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orr RJS, Stüken A, Murray SA, Jakobsen KS. Evolutionary acquisition and loss of saxitoxin biosynthesis in dinoflagellates: the second “core” gene, sxtG. Appl Environ Microbiol. 2013;79:2128–2136. doi: 10.1128/AEM.03279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perini F, Galluzzi L, Dell’Aversano C, Iacovo ED, Tartaglione L, et al. SxtA and sxtG gene expression and toxin production in the Mediterranean Alexandrium minutum (Dinophyceae) Mar Drugs. 2014;12:5258–5276. doi: 10.3390/md12105258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray SA, Diwan R, Orr RJS, Kohli GS, John U. Gene duplication, loss and selection in the evolution of saxitoxin biosynthesis in alveolates. Mol Phylogenet Evol. 2015;92:165–180. doi: 10.1016/j.ympev.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 57.Li Z, Mertens KN, Nézan E, Chomérat N, Bilien G, et al. Discovery of a new clade nested within the genus Alexandrium (Dinophyceae): morpho-molecular characterization of Centrodinium punctatum (Cleve) F.J.R. Taylor. Protist. 2019;170:168–186. doi: 10.1016/j.protis.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen-Ngoc L. An autecological study of the potentially toxic dinoflagellate Alexandrium affine isolated from Vietnamese waters. Harmful Algae. 2004;3:117–129. doi: 10.1016/S1568-9883(03)00062-3. [DOI] [Google Scholar]
- 59.Chun SW, Hinze ME, Skiba MA, Narayan ARH. Chemistry of a unique polyketide-like synthase. J Am Chem Soc. 2018;140:2430–2433. doi: 10.1021/jacs.7b13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lukowski AL, Ellinwood DC, Hinze ME, DeLuca RJ, Du Bois J, et al. C-H hydroxylation in paralytic shellfish toxin biosynthesis. J Am Chem Soc. 2018;140:11863–11869. doi: 10.1021/jacs.8b08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lukowski AL, Denomme N, Hinze ME, Hall S, Isom LL, et al. Biocatalytic detoxification of paralytic shellfish toxins. ACS Chem Biol. 2019;14:941–948. doi: 10.1021/acschembio.9b00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soeriyadi AH, Mazmouz R, Pickford R, Al-Sinawi B, Kellmann R, et al. Heterologous expression of an unusual ketosynthase, SxtA, leads to production of saxitoxin intermediates in Escherichia coli . Chembiochem. 2021;22:845–849. doi: 10.1002/cbic.202000675. [DOI] [PubMed] [Google Scholar]
- 63.Sako Y, Kim CH, Ishida Y. Mendelian inheritance of paralytic shellfish poisoning toxin in the marine dinoflagellate Alexandrium catenella . Biosci Biotechnol Biochem. 1992;56:692–694. doi: 10.1271/bbb.56.692. [DOI] [PubMed] [Google Scholar]
- 64.Le Gac M, Metegnier G, Chomérat N, Malestroit P, Quéré J, et al. Evolutionary processes and cellular functions underlying divergence in Alexandrium minutum . Mol Ecol. 2016;25:5129–5143. doi: 10.1111/mec.13815. [DOI] [PubMed] [Google Scholar]
- 65.Figueroa RI, Garcés E, Bravo I. Comparative study of the life cycles of Alexandrium tamutum and Alexandrium minutum (gonyaulacales, dinophyceae) in culture 1. J Phycol. 2007;43(5):1039–1053. [Google Scholar]
- 66.Anderson DM. In: The Physiological Ecology of Harmful Algal Blooms. Anderson DM, Cembella AD, Hallegraeff GM, editors. Heidelberg, Germany: Springer-Verlag; 1998. Physiology and bloom dynamics of toxic Alexandrium species, with emphasis on life cycle transitions; pp. 29–48. [Google Scholar]
- 67.Keller MD, Selvin RC, Claus W, Guillard RRL. Media for the culture of oceanic ultraphytoplankton. J Phycol. 1987;23:633–638. doi: 10.1111/j.1529-8817.1987.tb04217.x. [DOI] [Google Scholar]
- 68.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing arXiv:12073907 [q-bio] [Internet] [ July 20; 2012 ]. http://arxiv.org/abs/1207.3907 n.d. accessed.
- 72.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.Haas B, Papanicolaou A. TransDecoder (find coding regions within transcripts) [Internet] Available from. 2016. https://github.com/TransDecoder/TransDecoder/wiki
- 75.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor J, Butler D. R package asmap: efficient genetic linkage map construction and diagnosis. J Stat Soft. 2017;79 doi: 10.18637/jss.v079.i06. [DOI] [Google Scholar]
- 78.Team RC. R: A language and environment for statistical computingg [Internet] Available from. 2020. https://www.R-project.org/
- 79.Team Rs. Boston, MA: RStudio, PBC; 2019. RStudio: Integrated Development for R [Internet]http://www.rstudio.com/ [Google Scholar]
- 80.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye N, Zhang X, Miao M, Fan X, Zheng Y. Saccharina genomes provide novel insight into kelp biology. Nat Commun. 2015;6:6986. doi: 10.1038/ncomms7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: a remarkable evolutionary experiment. Am J Bot. 2004;91:1523–1534. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- 84.Траспов АА, Костюнина ОВ, Белоус АА, Карпушкина ТВ, Свеженцева НА, et al. Whole-genome association studies of distribution of developmental abnormalities and other breeding-valuable qualitative traits in offspring of the Russian large-white boars. Vavilovskii Zhurnal Genet Selektsii. 2020;24:185–190. doi: 10.18699/VJ20.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cembella AD, Sullivan JJ, Boyer GL, Taylor FJR, Andersen RJ. Variation in paralytic shellfish toxin composition within the Protogonyaulax tamaronsis/catenella species complex; red tide dinoflagellates. Biochemical Systematics and Ecology. 1987;15:171–186. doi: 10.1016/0305-1978(87)90018-4. [DOI] [Google Scholar]
- 86.Boyer GL, Sullivan JJ, Andersen RJ, Taylor FJR, Harrison PJ, et al. Use of high-performance liquid chromatography to investigate the production of paralytic shellfish toxins by Protogonyaulax spp. in culture. Marine Biology. 1986;93:361–369. doi: 10.1007/BF00401103. [DOI] [Google Scholar]
- 87.Oshima Y, Blackburn SI, Hallegraeff GM. Comparative study on paralytic shellfish toxin profiles of the dinoflagellate Gymnodinium catenatum from three different countries. Marine Biology. 1993;116:471–476. doi: 10.1007/BF00350064. [DOI] [Google Scholar]
- 88.Parkhill JP, Cembella AD. Effects of salinity, light and inorganic nitrogen on growth and toxigenicity of the marine dinoflagellate Alexandrium tamarense from northeastern Canada. J Plankton Res. 1999;21:939–955. doi: 10.1093/plankt/21.5.939. [DOI] [Google Scholar]
- 89.Jean N, Perié L, Dumont E, Bertheau L, Balliau T, et al. Metal stresses modify soluble proteomes and toxin profiles in two Mediterranean strains of the distributed dinoflagellate Alexandrium pacificum . Sci Total Environ. 2022;818:151680. doi: 10.1016/j.scitotenv.2021.151680. [DOI] [PubMed] [Google Scholar]
- 90.Hansen G, Daugbjerg N, Franco JM. Morphology, toxin composition and LSU rDNA phylogeny of Alexandrium minutum (Dinophyceae) from Denmark, with some morphological observations on other European strains. Harmful Algae. 2003;2:317–335. doi: 10.1016/S1568-9883(03)00060-X. [DOI] [Google Scholar]
- 91.Grzebyk D. Effects of salinity and two coastal waters on the growth and toxin content of the dinoflagellate Alexandrium minutum . J Plankton Res. 2003;25:1185–1199. doi: 10.1093/plankt/fbg088. [DOI] [Google Scholar]
- 92.Hamasaki K. Variability in toxicity of the Ddinoflagellate Alexandrium tamarense isolated from Hiroshima Bay, Western Japan, as a reflection of changing environmental conditions. J Plankton Res. 2001;23:271–278. doi: 10.1093/plankt/23.3.271. [DOI] [Google Scholar]
- 93.Lee TCH, Kwok OT, Ho KC, Lee FWF. Effects of different nitrate and phosphate concentrations on the growth and toxin production of an Alexandrium tamarense strain collected from Drake Passage. Mar Environ Res. 2012;81:62–69. doi: 10.1016/j.marenvres.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 94.Selander E, Kubanek J, Hamberg M, Andersson MX, Cervin G, et al. Predator lipids induce paralytic shellfish toxins in bloom-forming algae. Proc Natl Acad Sci. 2015;112:6395–6400. doi: 10.1073/pnas.1420154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martins CA, Kulis D, Franca S, Anderson DM. The loss of PSP toxin production in a formerly toxic Alexandrium lusitanicum clone. Toxicon. 2004;43:195–205. doi: 10.1016/j.toxicon.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 96.Hii KS, Lim PT, Kon NF, Takata Y, Usup G, et al. Physiological and transcriptional responses to inorganic nutrition in a tropical Pacific strain of Alexandrium minutum: Implications for the saxitoxin genes and toxin production. Harmful Algae. 2016;56:9–21. doi: 10.1016/j.hal.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 97.Zhang SF, Zhang Y, Lin L, Wang DZ. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella at different stages of toxin biosynthesis during the cell cycle. Mar Drugs. 2018;16:E491. doi: 10.3390/md16120491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Verma A, Barua A, Ruvindy R, Savela H, Ajani PA, et al. The genetic basis of toxin biosynthesis in dinoflagellates. Microorganisms. 2019;7:E222. doi: 10.3390/microorganisms7080222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murray SA, Wiese M, Stüken A, Brett S, Kellmann R, et al. sxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl Environ Microbiol. 2011;77:7050–7057. doi: 10.1128/AEM.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Zhang SF, Lin L, Wang DZ. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar Drugs. 2014;12:5698–5718. doi: 10.3390/md12115698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wiese M, Murray SA, Alvin A, Neilan BA. Gene expression and molecular evolution of sxtA4 in a saxitoxin producing dinoflagellate Alexandrium catenella . Toxicon. 2014;92:102–112. doi: 10.1016/j.toxicon.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 102.Omura T, Onodera H, Ishimaru T, Oshima Y. La Mer. 2003. Non-toxic mutational subclones in the paralytic shellfish poisoning causative dinoflagellates, Alexandrium spp; pp. 86–93. [Google Scholar]
- 103.Cho Y, Hiramatsu K, Ogawa M, Omura T, Ishimaru T, et al. Non-toxic and toxic subclones obtained from a toxic clonal culture of Alexandrium tamarense (Dinophyceae): Toxicity and molecular biological feature. Harmful Algae. 2008;7:740–751. doi: 10.1016/j.hal.2008.02.008. [DOI] [Google Scholar]
- 104.Cho Y, Tsuchiya S, Yoshioka R, Omura T, Konoki K, et al. The presence of 12β-deoxydecarbamoylsaxitoxin in the Japanese toxic dinoflagellate Alexandrium determined by simultaneous analysis for paralytic shellfish toxins using HILIC-LC–MS/MS. Harmful Algae. 2015;49:58–67. doi: 10.1016/j.hal.2015.09.003. [DOI] [Google Scholar]
- 105.Cho Y, Hidema S, Omura T, Koike K, Koike K, et al. SxtA localizes to chloroplasts and changes to its 3’UTR may reduce toxin biosynthesis in non-toxic Alexandrium catenella (Group I)✰ . Harmful Algae. 2021;101:101972. doi: 10.1016/j.hal.2020.101972. [DOI] [PubMed] [Google Scholar]
- 106.Thi Nhu Bui Q, Kim H, Wang H, Ki JS. Unveiling the genomic structures and evolutionary events of the saxitoxin biosynthetic gene sxtA in the marine toxic dinoflagellate Alexandrium . Mol Phylogenet Evol. 2022;168:107417. doi: 10.1016/j.ympev.2022.107417. [DOI] [PubMed] [Google Scholar]
- 107.Yoshida T, Sako Y, Uchida A, Kakutani T, Arakawa O, et al. Purification and characterization of sulfotransferase specific to O-22 of 11-hydroxy saxitoxin from the toxic dinoflagellate Gymnodinium catenatum (dinophyceae) Fisheries Sci. 2002;68:634–642. doi: 10.1046/j.1444-2906.2002.00471.x. [DOI] [Google Scholar]
- 108.Klemenčič M, Funk C. Structural and functional diversity of caspase homologues in non-metazoan organisms. Protoplasma. 2018;255:387–397. doi: 10.1007/s00709-017-1145-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jauzein C, Erdner DL. Stress-related responses in Alexandrium tamarense cells exposed to environmental changes. J Eukaryot Microbiol. 2013;60:526–538. doi: 10.1111/jeu.12065. [DOI] [PubMed] [Google Scholar]
- 110.Asplund-Samuelsson J, Sundh J, Dupont CL, Allen AE, McCrow JP, et al. Diversity and expression of bacterial metacaspases in an aquatic ecosystem. Front Microbiol. 2016;7:1043. doi: 10.3389/fmicb.2016.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cho Y, Tsuchiya S, Omura T, Koike K, Oikawa H, et al. Metabolomic study of saxitoxin analogues and biosynthetic intermediates in dinoflagellates using 15N-labelled sodium nitrate as a nitrogen source. Sci Rep. 2019;9:11. doi: 10.1038/s41598-019-39708-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.