Abstract
Enterococcus faecium is a lactic acid bacterium that confers beneficial health effects in humans. However, lately, a number of E. faecium strains have been linked to the spread of nosocomial infections in the hospital environment. Therefore, any potential commercial usage of E. faecium isolates should be preceded by an assessment of infection risk. In the current study, the genomes of two novel E. faecium strains Am1 (larval isolate) and Bee9 (adult bee isolate) isolated from the gut of Apis mellifera L. (honeybee) were sequenced to allow evaluation of their safety. In particular, their genomes were screened for antibiotic-resistance and virulence genes. In addition, their potential to spread resistance in the environment was evaluated. The analysis revealed that Am1 and Bee9 possess 2832 and 2844 protein-encoding genes, respectively. In each case, the genome size was 2.7 Mb with a G+C content of 37.9 mol%. Comparative analysis with probiotic, non-pathogenic and pathogenic enterococci revealed that there are variations between the two bee E. faecium isolates and pathogenic genomes. They were, however, closely linked to the probiotic comparison strains. Phenotypically, the Am1 and Bee9 strains were susceptible to most antibiotics tested, but showed intermediate sensitivity towards erythromycin, linezolid and trimethoprim/sulfamethoxazole. Notably, no genes associated with antibiotic resistance in clinical isolates (e.g. vancomycin resistance: vanA, vanB, vanS, vanX and vanY) were present. In addition, the insertion sequences (IS16, ISEfa11 and ISEfa5), acting as molecular pathogenicity markers in clinically relevant E. faecium strains, were also absent. Moreover, the analysis revealed the absence of three key pathogenicity-associated genes (acm, sgrA, ecbA) in the Am1 and Bee9 strains that are found in the prominent clinical isolates DO, V1836, Aus0004 and Aus0085. Overall, the findings of this investigation suggest that the E. faecium isolates from the bee gut have not suffered any recent clinically relevant antibiotic exposure. It also suggests that E. faecium Am1 and Bee9 are safe potential probiotic strains, because they lack the phenotypic and genetic features associated with strains eliciting nosocomial infections.
Keywords: Apis mellifera, antibiotic resistance, comparative genomics, Enterococcus faecium, lactic acid bacteria (LAB), virulence
Data Summary
For Enterococcus faecium Am1, the whole-genome shotgun project has been deposited at GenBank/ENA/DDBJ under the accession number JAHLTJ000000000. The version described in this paper is version JAHLTJ010000000. For E. faecium Bee9, this whole-genome shotgun project has been deposited at GenBank/ENA/DDBJ under the accession number JAHLXG000000000. The version described in this paper is version JAHLXG010000000. The raw sequencing data are available in the Sequence Read Archive (SRA) database under the accession numbers SRR15238610 and SRR15245957, respectively.
Impact Statement.
Enterococcus species are widespread indigenous bacteria that inhabit the gastrointestinal tract of humans, insects and animals. Although Enterococcus species have not yet obtained generally regarded as safe (GRAS) status from the US Food and Drug Administration (FDA), probiotic strains have been made commercially available. Thus, comprehensive assessment of their safety characteristics is necessary to distinguish between beneficial and pathogenic Enterococcus strains. Whole-genome sequencing (WGS) provides a valuable tool for assessing the safety of Enterococcus strains. Thus, we have used WGS to provide insight into the virulence and antibiotic-resistance properties of two Enterococcus faecium strains (Am1 and Bee9) isolated from honeybee gut. Our combined WGS, in silico and phenotypic analyses indicates that the two E. faecium isolates are stable and safe, and of low microbial risk.
Introduction
Enterococci are facultative anaerobic, non-spore-forming, Gram-positive lactic acid bacteria (LAB). They are characterized by the different relationships that they may establish with their hosts, where they may act as commensals, probiotics or opportunistic pathogens. Therefore, enterococci are one of the most controversial groups of LAB. The majority of enterococci have been isolated from humans, animals, plants and insects as commensal LAB. For example, some Enterococcus faecium strains have beneficial probiotic qualities. They also have an impact on intestinal flora balance and modulate the immune system of humans [1]. However, other strains have emerged as opportunistic human pathogens with a high propensity for spreading antibiotic-resistance and virulence genes via horizontal gene transfer (HGT) [2]. Given the paucity of treatment alternatives, vancomycin-resistant Enterococcus , in particular, is considered a persistent global problem affecting both developed and developing nations [3].
Before considering application of any Enterococcus strain as a safe starter culture in the food or feed sectors, a thorough examination is required to ensure that such strains do not represent any risk to health. For instance, one of the main challenges is determining potential E. faecium strains that are most likely to have generally recognized as safe (GRAS) status while avoiding pathogenic ones. Adopting next-generation sequencing (NGS) technology to analyse the bacterial genomes of Enterococcus sp. strains, with special emphasis on antibiotic-resistance and virulence genes, is one of the currently recommended techniques [4]. Whole-genome sequencing (WGS) is becoming the method of choice for identifying and characterizing bacterial genomes. A recent survey conducted on enterococci isolated from different environments confirmed that conventional molecular approaches are less effective than WGS in providing a detailed analysis of antimicrobial-resistance genes (ARGs) and virulence genes [5].
The gut microbiota of social insects, Apis mellifera L. (honeybee), is one of the E. faecium habitats that has recently been explored. A broad range of bacteria, including many LAB, may be found in the gut of bees [6]. By competing with pathogens for resources and production of organic acids, antimicrobial peptides and bacteriocins, the gut microbiota of A. mellifera protect their hosts from pathogenic attacks [6, 7].
Therefore, in the current study, we adopted WGS technology to identify the antibiotic-resistance and virulence genes encoded by two honeybee E. faecium strains (Am1 and Bee9). Furthermore, we conducted a comparative study of their genomes versus other E. faecium strains isolated from hospitalized patients, milk and commercial probiotics. Overall, the findings of this study support the safe use of E. faecium Am1 and Bee9 as these strains lack phenotypic and genotypic characteristics associated with the emergence of nosocomial infections.
Methods
LAB isolation from bee gut
LAB were isolated from A. mellifera larvae and adults. The larvae were provided by the Department of Applied Entomology and Zoology, Faculty of Agriculture, Alexandria University, Egypt. The adult bees were collected from Kafr El-Dawar governorate (Egypt) in early Summer 2019. Briefly, every bee was surface sterilized using 96 % ethanol and dissected on the surface of a sterilized wax plate or glass slide to separate the insect’s gut. The separated gut was kept in De Man–Rogosa–Sharpe (MRS; HiMedia) broth for enrichment at 37 °C, for 24 h, under microaerophilic conditions generated by a candle jar. For purification, each liquid culture was serially diluted and plated on the surface of MRS agar under the same incubation conditions for 24–48 h. Finally, individual phenotypically unique colonies were picked for a further two rounds of purification using the streak-plate method. Pure cultures were stored at −20 °C in MRS broth supplemented with 50 % (v/v) glycerol for further investigation.
Morphological and biochemical characterization
Two bacterial isolates, one isolated from the A. mellifera larval stage (Am1) and one from the adult stage (Bee9), were morphologically and biochemically characterized. The two bacterial isolates' morphology was checked using scanning electron microscopy (SEM; JSM-IT 200; JEOL), at the EM Unit, Alexandria University, Egypt. The bacterial isolates were examined for Gram staining and catalase reaction, followed by full biochemical characterization using the VITEK 2 GP ID card (bioMérieux; https://www.biomerieux.com). The haemolytic activity was evaluated according to procedures described elsewhere [8]. E. faecium cultures were streaked onto 5 % (v/v) blood agar plates and incubated for 24 h at 37 °C.
Genomic DNA extraction
Genomic DNA was isolated from 1.5 ml of 18 h overnight grown bacterial culture in MRS broth at 37 °C using a GeneJET genomic DNA purification kit (Thermo Fisher Scientific), following the manufacturer’s instructions for Gram-positive bacterial DNA isolation. In the final step, the DNA was eluted in nuclease-free water (Thermo Fisher Scientific), instead of an elution buffer, for subsequent library preparation. The concentration and purity of the extracted DNA were checked using a NanoDropND-2000 spectrophotometer (Thermo Fisher Scientific) and 1 % (w/v) agarose gel electrophoresis.
Genome sequencing, assembly and quality assurance
Genomic DNA libraries were prepared using the Nextera XT library prep kit (Illumina), following the manufacturer’s protocol. DNA quantification and library preparation were carried out on a Hamilton Microlab STAR automated liquid handling system (Hamilton Bonaduz). Pooled libraries were quantified using the Kapa Biosystems library quantification kit for Illumina. WGS was performed by MicrobesNG (http://microbesng.uk) on a HiSeq platform (Illumina), using a 250 bp paired-end protocol with 30× sequence coverage. The adapters were trimmed using Trimmomatic (version 0.30) with a sliding cut-off of Q15 [9]. De novo assembly was performed with SPAdes software (version 3.7.0) [10]. The quality of the genome assemblies was assessed using the Quality Assessment Tool for Genome Assemblies (quast) [11].
Genome annotation
Genome annotation was performed using Prokka software (version 1.11) [12], the Pathosystems Resource Integration Center (PATRIC; version 3.6.12) server (https://www.bv-brc.org/) [13] and the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (pgap) [14]. The contigs were recorded and reoriented relative to the reference genome based on a MUMmer whole-genome alignment [15]. Total numbers of coding sequences (CDSs), tRNA and rRNA genes from the genomes were predicted using PATRIC. Subsystem categories and features distribution of all E. faecium genomes included in this study was assessed using Rapid Annotations using Subsystems Technology (annotation scheme: RASTtk) [16].
Phylogenetic analysis using 16S rRNA gene sequences
For phylogenetic analysis of the two E. faecium isolates, the 16S rRNA sequences of Enterococcus type strains were downloaded from the NCBI database, followed by use of ClustalW 2.1 for alignment, Gblocks 0.91b for alignment refinement, phylogeny using two programs (PhyML 3.1/3.0 aLRT and MrBayes) and tree rendering by TreeDyn 198.3. The previously listed programs were used through phylogeny.fr [17]. Finally, FigTree (version 1.4.4) (available through http://tree.bio.ed.ac.uk/software/figtree/) was used as a tree graphical viewer.
Comparative genomic analyses
A comparative genomic analysis of E. faecium Am1 and Bee9 WGS assemblies was performed to reveal significant similarities and differences between these two bee isolates and other published E. faecium strains, namely: SM21 (bee gut isolate); DO, V1836, Aus0004, Aus0085 (clinical isolates); T110, FS86 (commercial probiotic); and NRRL_B-2354 (milk and dairy utensils isolate). These comparator strains were selected as representative examples of probiotic, non-pathogenic and pathogenic isolates. The chromosomal genome sequences were downloaded from the GenBank database and accession numbers are listed in Table 1. Command-line search tools such as blast were used for comparison of sequences locally through the web server. blast Atlas was generated to align query genomes using E. faecium DO as a reference. GView server (https://server.gview.ca) [18] was used with a cut-off value 1×10−10 and percentage identity cut-off values of 80 %.
Table 1.
General genome features of E. faecium strains Am1 and Bee9 against other selected E. faecium genomes
|
Strain |
Source |
Geographical location |
Accession no.* |
Genome length (bp) |
Total number of genes (CDSs) |
tRNA |
rRNA |
G+C (mol%) |
Reference |
|---|---|---|---|---|---|---|---|---|---|
|
Am1 |
Bee gut – larvae |
Egypt |
2 732 750 |
2832 |
62 |
6 |
37.99 |
This study |
|
|
Bee9 |
Bee gut – adult |
Egypt |
2 734 007 |
2844 |
56 |
6 |
37.97 |
This study |
|
|
SM21 |
Bee gut |
Argentina |
2 544 244 |
2477 |
60 |
10 |
38.10 |
[7] |
|
|
NRRL_B-2354 |
Milk and dairy utensils |
USA |
2 635 572 |
2832 |
48 |
18 |
37.84 |
[48] |
|
|
T110 |
Commercial probiotic product |
India |
2 693 877 |
2522 |
66 |
18 |
38.47 |
[61] |
|
|
FS86 |
Commercial probiotic product |
Russia |
2 685 395 |
2634 |
57 |
16 |
38.40 |
[62] |
|
|
DO |
Clinical isolate |
USA |
2 698 137 |
2788 |
31 |
3 |
37.8 |
[63] |
|
|
V1836 |
Clinical isolate |
Denmark |
2 884 831 |
2885 |
67 |
18 |
37.95 |
[64] |
|
|
Aus0004 |
Clinical isolate |
Australia |
2 955 294 |
3006 |
47 |
18 |
38.36 |
[65] |
|
|
Aus0085 |
Clinical isolate |
Australia |
2 994 661 |
3089 |
76 |
18 |
38.32 |
[65] |
*All accession numbers are from GenBank.
Identification of ARGs and antimicrobial-susceptibility testing (AST)
ARGs were identified in the E. faecium genomes using the Genome Annotation Service in PATRIC by deploying the k-mer-based ARG detection method, which use PATRIC’s curated collection of representative antibiotic-resistance variants [19] and assigns to each ARG functional annotation, broad mechanism of antibiotic resistance, drug class and specific antibiotic it confers resistance to.
ARG screening results were validated using minimum inhibitory concentration (MIC) and antibiotic-sensitivity tests. Bacterial suspensions equivalent to 0.5 McFarland turbidity were prepared and 3 ml suspension equivalent to 107 c.f.u. ml−1 was subjected to AST. The test was performed using the GP susceptibility card AST-P592 (bioMérieux) and the VITEK 2 system (version 9.02), according to the manufacturer’s guidelines. MIC interpretation was performed according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI). The antibiotic-sensitivity test was carried out on Muller Hinton agar (MHA; HiMedia) plates containing antibiotic discs. The plates were incubated at 37 °C for 24 h and the results were expressed in millimetres of inhibition. The isolates were classified as susceptible (S), intermediate (I) or resistant (R) based on CLSI recommendations [20].
Screening for virulence genes
The VFanalyzer platform (https://www.mgc.ac.cn), available through the Virulence Factor Database (VFDB), was used to detect virulence factors in the genomes under investigation. E. faecium DO was specified as a representative genome for this analysis. The DO strain was selected for this analysis because it has a complete genome sequence and has been used in multiple pathogenesis studies [21, 22]. The statistics (as of 4th August 2021) of the genus Enterococcus in the VFDB are the following: number of species and strains is 2 and 13, respectively. Moreover, the number of virulence factors and genes is 25 and 113, respectively. For validation of virulence-gene screening results, the presence/absence of two virulence genes (collagen-binding, ace [23], and surface protein, esp [8]) was examined using conventional PCR. Enterococcus faecalis 24FS (NCBI accession no. PGCH01000000) was used as a positive control.
Detection of genetic mobile elements and antimicrobial peptide genes
Genes recently acquired by HGT were predicted with IslandViewer 4 (https://www.pathogenomics.sfu.ca/islandviewer/) using the IslandPath-DIMOB method [24] and E. faecium DO as a reference. Insertion sequences (ISs) and transposons were predicted with the ISfinder server (https://www-is.biotoul.fr) [25] using blastn (version 2.2.31+). Integrative and conjugative elements (ICEs) were identified by the ICEfinder web-based tool [26], with an E value of 1×10−150. Plasmids were detected by the PlasmidFinder (version 2.0) online tool [27] provided by the Center for Genomic Epidemiology (https://www.genomicepidemiology.org). Clustered regularly interspaced short palindromic repeats (CRISPRs) were predicted with the CRISPRFinder tool [28]. Prophage sequences were identified using the PHAge Search Tool Enhanced Release web server (PHASTER) (https://phaster.ca) [29]. The fasta sequence format of the E. faecium Am1 and Bee9 genome contigs were mined for bacteriocin-encoding genes using the BAGEL4 server (http://bagel4.molgenrug.nl) [30].
Examination of LAB characteristics
The antagonistic activity of Am1 and Bee9 neutralized cell-free supernatant (CFS) was determined by the agar well diffusion assay [31] against Gram-negative bacteria, Escherichia coli (ATCC 8739), Pseudomonas aeruginosa (ATCC 27853), and Gram-positive bacteria, Bacillus subtilis (ATCC 6633), Staphylococcus aureus (ATCC 25923). Am1 and Bee9 were also investigated for auto-aggregation properties and in vitro cell surface hydrophobicity [32, 33]. Lactiplantibacillus plantarum 10CH (NCBI accession no. CP023728) was used as the control strain.
Results
Morphological and biochemical characterization using the VITEK 2 system
Two stable Gram-positive and catalase-negative bacterial isolates, namely Am1 and Bee9, were identified by 99 % probability as being E. faecium species using the VITEK 2 system. Furthermore, the VITEK 2 biochemical characterization revealed that the two isolates were able to grow on different carbon sources and had the ability to tolerate a high concentration of NaCl (6.5 %). The detailed results of this biochemical characterization are reported in Table S1 (available with the online version of this article). Neither isolate displayed blood haemolysis activity on the surface of blood agar and, thus, were identified as γ-haemolytic bacteria. SEM revealed that the two strains possess a coccoidal cell morphology (Fig. 1).
Fig. 1.
Growth of E. faecium Am1 (a) and Bee9 (c) on the surface of blood agar after 24 h incubation at 37 °C. (b) and (d) demonstrate SEM examinations of Am1 and Bee9 cells, respectively.
Genome assembly features of E. faecium Am1 and Bee9
WGS of Am1 and Bee9 yielded 595 821 and 407 492 reads, respectively. De novo assembly was performed using SPAdes yielding 81 and 76 contigs of over 1000 bp (Table S2). The genomes had 2832 and 2844 predicted protein CDSs, respectively. The general genomic features of Am1 and Bee9 indicated total genome sizes of 2.7 Mb and a G+C content of 37.9 mol%, as predicted using the PATRIC server.
Subsystem analysis
The genome annotations obtained using the RASTk server were used to generate an overview of the subsystem categories and feature distribution of genomes under investigation. The distribution of Gene Ontology (GO) categories was similar among the tested E. faecium isolates, including Am1 and Bee9. For metabolism categories, genes for carbohydrate and protein metabolism were the most abundant, followed by genes for DNA and RNA metabolism. Furthermore, genes related to cell wall and capsule biogenesis were among the most abundant, especially in Am1 and Bee9. The genomes also revealed genes involved in biosynthesis of essential amino acids (glycine, valine, methionine, isoleucine, arginine, serine, lysin and tryptophan) and vitamins (B6, biotin, thiamine, riboflavin and folate). An overview of the genomes subsystem annotations is provided in Fig. 2.
Fig. 2.
Subsystem categories and features distribution of the E. faecium genomes based on the RASTk annotation server. For each E. faecium strain, the number of genes identified in each category is displayed. The colour scale represents the number of genes found in each category; the deeper the colour, the more genes were detected in that category.
Phylogenetic analysis
Phylogenetic analysis based on the 16S rRNA gene sequence using two different methods (Fig. 3) revealed the evolutionary relationships for the two bee-gut isolates with respect to other closely related species of Enterococcus . The two isolates, regardless of the phylogenetic analysis method used, were grouped together in the same clade with E. faecium strain ATCC 19434 (NCBI accession no. NR_115764) and E. faecium strain SM21 (NCBI accession no. EU428012.1). The SM21 strain was isolated from the intestinal tract of A. mellifera.
Fig. 3.
Phylogenetic analyses of E. faecium Am1 and Bee9 with closely related homologues inferred using the maximum-likelihood method (a) and Bayesian inference (b). The evolutionary analyses were carried out in phylogeny.fr (available at https://www.phylogeny.fr/), with tree building using PhyML and MrBayes programs, respectively. The evolutionary trees were graphically displayed using the FigTree program (available at http://tree.bio.ed.ac.uk/software/figtree/).
Comparative genome analysis
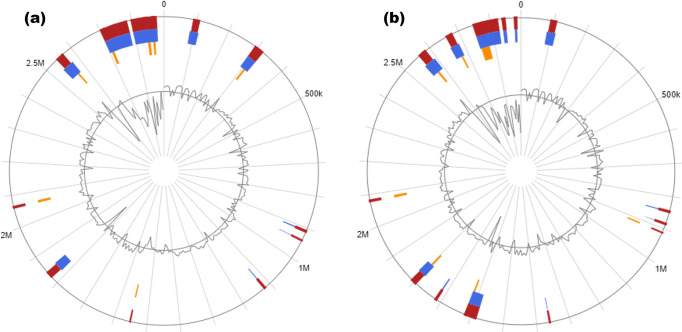
The blast Atlas presentation of the gene content revealed common characteristics between the two bee E. faecium isolates and the aligned genomes. Regions of significant similarity were identified among the two strains (Am1 and Bee9), non-clinical E. faecium (NRRL B-2354) and probiotic strains (T110 and FS86), illustrating their similarity in genomic content (Fig. 4). However, two pathogenic islands including most of the virulence and antibiotic-resistance genes predicted in clinical strains (DO, V1836, Aus0004 and Aus0085) were absent in both probiotic strains and the two bee isolates.
Fig. 4.

blast Atlas overview of E. faecium (Am1 and Bee9) and selected genomes from the NCBI database based on blastn. The outermost circles demonstrate the two E. faecium , Am1 (red) and Bee9 (brown), and the genomes of strains used in comparison: SM21 (orange), T110 (dark green), FS86 (light green), NRRL B-2354 (purple), Aus0085 (blue), Aus0004 (red violet), V1836 (light purple). Strain E. faecium DO (navy blue) was used as a reference genome. The innermost circles represent the G+C content (black), G+C skew curve (violet) and Clusters of Orthologous Genes (COG) categories. The genome sequences were analysed using the GView server using both alignment length and percent identity cut-off values of 80 %.
ARG analysis and AST
ARG analysis of the two bee isolates' genomes revealed genes conferring resistance to low-level aminoglycosides [aac(6’)-li], fusidane (ef-G, ef-TU; translation elongation factor), peptides (liaR, liaS; transcriptional regulatory protein), (efrB; ABC multidrug efflux pump), erythromycin and other macrolide antibiotics (msrC; macrolide-resistance gene), (efmA; multidrug efflux pump), (dfrE; dihydrofolate reductase); (adeC; efflux pump) (clsA; cardiolipin synthase A). The genomes did not contain the genes for vancomycin resistance (vanA, vanB, vanS, vanX and vanY), which are responsible for vancomycin-resistant Enterococcus infections in the human population (Fig. 5).
Fig. 5.
Heat map representation of virulence genes/factors and antibiotic-resistance genes identified in E. faecium Am1 and Bee9 using the VFanalyzer platform and the k-mer-based ARG detection method available through PATRIC, respectively. The dark and light blue colours represent present and absent genes, respectively.
AST profiles, determined over a ~12 h period by VITEK 2 analysis, provided MIC values for clinically relevant antibiotics (Table 2). Both AST profile and the disc diffusion method (Table S3) revealed that the isolates were susceptible to the following antibiotics: ampicillin, imipenem, gentamicin, streptomycin, ciprofloxacin, teicoplanin, vancomycin, tetracycline and tigecycline. Isolate Am1 showed intermediate sensitivity to erythromycin and linezolid, while isolate Bee9 was susceptible to linezolid in the AST profile. Furthermore, the disc diffusion assay revealed that the two isolates were resistant to penicillin G, cefoxitin, azithromycin and colistin sulphate.
Table 2.
MIC (µg ml−1) interpretation of E. faecium Am1 and Bee9 detected by VITEK 2 as per CLSI guidelines
|
Antimicrobial |
MIC value (interpretation*) |
|
|---|---|---|
|
Am1 |
Bee9 |
|
|
Ampicillin |
≤ 2 (S) |
≤ 2 (S) |
|
Imipenem |
≤ 1 (S) |
≤ 1 (S) |
|
Gentamicin high level (synergy) |
SYN-S (S) |
SYN-S (S) |
|
Streptomycin high level (synergy) |
SYN-S (S) |
SYN-S (S) |
|
Ciprofloxacin |
≤ 0.5 (S) |
≤ 0.5 (S) |
|
Erythromycin |
1 (I) |
1 (I) |
|
Linezolid |
4 (I) |
2 (S) |
|
Teicoplanin |
≤ 0.5 (S) |
≤ 0.5 (S) |
|
Vancomycin |
≤ 0.5 (S) |
≤ 0.5 (S) |
|
Tetracycline |
≤ 1 (S) |
≤ 1 (S) |
|
Tigecycline |
≤ 0.12 (S) |
≤ 0.12 (S) |
*Interpretation of MIC values S (susceptible) and I (intermediate) according to the CLSI [20].
Virulence-gene analysis
Screening for virulence factors using the VFanalyzer platform revealed that the two bacterial genomes possess six adherence-associated genes (ebpA, ebpB, ebpC, strC, efaA and scm), three specifying antiphagocytosis (cpsA/uppS, cpsB/cdsA and cpsC), one biofilm gene (bopD) and four immune evasion-associated factors. Interestingly, the comparative analysis with the other E. faecium strains (SM21, NRRL B-2354, T110, FS86, Aus0004, Aus0085, DO and V1836) included in our analyses revealed that two of the immune evasion factors (eps3 and wchJ) were unique to E. faecium Am1 and Bee9 (Fig. 5). The protein products of eps3 and wchJ share similarity with exopolysaccharide biosynthesis protein and UDP-N-acetylglucosamine-LPS N-acetylglucosamine transferase, respectively. Interestingly, three virulence genes (acm, sgrA, ecbA) were uniquely absent in Am1 and Bee9 strains in comparison to the clinical isolates (DO, V1836, Aus0004 and Aus0085). However, three genes were detected in all E. faecium strains regardless the source of isolation, namely bopD, cpsA/upps and cpsB/cdsA. PCR validation confirmed the absence of ace and esp virulence genes (Fig. S1).
Genomic islands (GIs)
The analysis of the Am1 and Bee9 genomes revealed 9 and 12 GIs, respectively, based on the IslandPath prediction method (Fig. 6). The GIs were well distributed over the genome, and each GI encoded proteins of both hypothetical and known function (Tables S4 and S5). GIs included ribosomal proteins, transposable elements (IS3, IS4, IS6, IS30, IS256, IS200/IS605, IS630), prophage integrase (phiRv2), tyrosine recombinase (xerC, xerD) and conserved virulence factor B (cvfB).
Fig. 6.
Analysis of the GIs of E. faecium Am1 (a) and Bee9 (b) predicted by IslandViewer web server. The predicted GIs are shown in different colours within the circular image based on the tools used: sigi-hmm, which predicts GIs based on a hidden Markov model (orange); IslandPath-DIMOB, which predicts GIs based on features associated with GIs (blue); and an integration of three methods (red).
Genetic mobile elements (insertion sequences, ICEs, plasmids, CRISPRs)
Insertion sequence elements, major genetic mobile elements in E. faecium isolates, were predicted with ISfinder and blastn analysis. Seventy-two IS elements were predicted in E. faecium Am1, while in E. faecium Bee9 48 IS elements were identified. Most IS elements in Am1 and Bee9 belong to the IS3 family (Tables S6 and S7). The isolates showed an absence of IS16, ISEfa11 and ISEfa5 insertion sequences, known as molecular pathogenicity markers in clinically relevant E. faecium strains [34–36]. Although several IS elements were found and pathogenicity related IS elements were absent in the genomes of Am1 and Bee9, further analysis using different approaches would support this in silico finding.
ICEs were detected with the ICEfinder web-based tool. In E. faecium Am1, two putative ICE elements (IMEs; integrative and mobilizable elements) of 25 368 and 38 627 bp length were detected that contained integrase and relaxase genes. The E. faecium Bee9 genome also revealed the presence of two putative IMEs, of 34 956 and 27 880 bp. Larval isolate Am1 contained a 214 319 bp plasmid (pNB2354_1) with a repUS15 replicon (accession no. CP004064) and 8 347 bp plasmid (pGL) with a rep29 replicon (accession no. HQ696461). Isolate Bee9 contained only one plasmid (pNB2354_1). The detected plasmids lack the critical virulence factors present in pathogenic E. faecium strains. The CRISPR finder tool identified one confirmed CRISPR element (199 bp) in isolate Bee9 with four direct repeats of 24 bp and three spacers, with one associated with a cas-family gene.
Prophages
The phaster tool identified one intact (PHAGE_Entero_IME_EFm5, 17.3 kb) and three incomplete (PHAGE_Escher_RCS47, 15.3 kb; PHAGE_Entero_phiFL3A, 15.3 kb; and PHAGE_Lister_LP_030_3, 9 kb) prophage genomes in E. faecium Am1. The Bee9 isolate carried one intact (PHAGE_Entero_IME_EFm5, 23.9 kb) and three incomplete prophage regions (PHAGE_Entero_phiFL3A, 18 kb; PHAGE_Staphy_SPbeta_like, 15.1 kb; and PHAGE_Lister_A118, 10.1 kb).
Bacteriocin genes
The bacteriocinogenic potential of Am1 and Bee9 was assessed using the BAGEL4 server. The data revealed that the predicted bacteriocins corresponded to a class II (small heat-stable non-lantibiotic peptides) bacteriocins designated enterocin P (EntP; 5 735 Da) and a class III (large thermolabile bacteriolysins) bacteriocin designated enterolysin A (EnlA; 43 155 Da) (Fig. 7).
Fig. 7.
Putative bacteriocin-related gene clusters using the BAGEL4 web server. (a, b) Predicted enterocin P (EntP) and enterolysin A (EnlA) in E. faecium Am1 genome; (c, d) Predicted EntP and EnlA in E. faecium Bee9 genome.
Hydrophobicity, autoaggregation and antagonistic characteristics
The affinity of Am1 and Bee9 toward the hydrophobic agent xylene was investigated and the results revealed 54.78 and 54.58 % hydrophobicity, respectively. The ability of the two isolates to form autoaggregates was determined as 45.44 and 44.65 % for Am1 and Bee9, respectively. Am1 and Bee9 neutralized CFS revealed antagonistic activity against S. aureus (ATCC 25923) and Escherichia coli (ATCC 8739) (Fig. 8).
Fig. 8.
Examination of some LAB characteristics. Histograms in (a) and (b) show the hydrophobicity and autoaggregation percentage achieved by E. faecium Am1 and Bee9. The histogram shows the percentages obtained by L. plantarum strain 10CH and L. plantarum laboratory isolate (+ve C). The results of the antibiotic disc diffusion method for Am1 and Bee9 are shown in (c) and (d), respectively. The (e), (f), (g) and (h) plates demonstrate the antagonistic activity of Am1 and Bee9 neutralized CFS against S. aureus (ATCC 25923), Escherichia coli (ATCC 8739), P. aeruginosa (ATCC 27853) and B. subtilis (ATCC 6633), respectively.
Discussion
In the current study, we utilized WGS technology to assess two E. faecium isolates, Am1 and Bee9, originally isolated from A. mellifera larval and adult guts, respectively, in terms of their antibiotic and virulence-gene profiles. Moreover, a comparative genomic study was conducted to identify unique and common characteristics between these two isolates and other probiotic, non-pathogenic and pathogenic E. faecium strains.
The analysis of antibiotic-resistance genes revealed the absence of vancomycin- and teicoplanin-associated-resistance genes in Am1 and Bee9 isolates. This result was further confirmed using the VITEK 2 system and the disc diffusion method, which indicated a sensitive (S) status for these clinically relevant antibiotics. Sensitivity to vancomycin is essential for the safe application of Enterococcus strains intended for food or feed consumption. The reason for this is that vancomycin is regarded as a ‘drug of last resort’ commonly used in the treatment of enterococcal infections [37].
The unique absence of three key virulence genes (acm, sgrA, ecbA) in Am1 and Bee9, in contrast to clinical isolates, indicates the safety of these bee-gut isolates. These three genes were reported from previous studies to be associated with enhancing the virulence of clinical isolates. For instance, sgrA and ecbA along with esp were reported as important markers enriched in hospital-associated E. faecium isolates and ~70–100 % of E. faecium hospital-associated isolates possess sgrA and ecbA genes. Furthermore, the presence of esp was found to enhance the ability of E. faecium and E. faecalis to form biofilms that facilitate the colonization of medical devices [38–40]. Therefore, these genes may play an important role in adhesion, colonization, biofilm formation and pathogenesis. Hence, it is highly recommended to include hospital-associated E. faecium strains in comparative genomic studies against commensal ones as it pinpoints important genes [41, 42]. However, the two bee isolates were found to have two distinct Exopolysaccharide (EPS)-related genes, eps3 and wchJ. Although the production of exopolysaccharide (EPS) may be seen as virulent, it may also aid in the mobility of these non-motile bacteria. It also improves their capacity to endure stressful conditions such as high pH, osmolarity and temperature. It might also explain the two isolates' antagonistic properties toward other bacterial species, since EPS can accumulate metabolites that are toxic to other bacteria [1].
When ingesting a meal or feed contaminated with glycopeptide-antibiotic-resistant or virulent enterococci, there are two main dangers. First, there is the risk of the resistant bacteria being established in the host gut. Second, by conjugation, resistance or virulence genes from glycopeptide-resistant enterococci may transfer to other gut bacteria [37, 43, 44]. It is confirmed that enterococci can pass on their resistance towards glycopeptide antibiotics to other enterococci [45] and even other Gram-positive bacteria [46]. Therefore, it is critical to ensure the absence of these transferable resistances before use of any new Enterococcus strain in food or feed applications. However, the presence of glycopeptide-resistance or virulence genes does not imply that these genes will be easily transferred to other bacteria. As a result, SCAN (Scientific Committee on Animal Nutrition) advises checking whether these genes are transferable [43]. Screening of Am1 and Bee9 genomes for IS elements revealed that they harboured different numbers of IS families. IS elements are considered the smallest transposable elements that play an important role in shaping bacterial genomes [47]. Notably, both strains revealed the absence of IS16 insertion sequences, which are used as markers for hospital-acquired multidrug-resistant (MDR) E. faecium strains [35]. The Am1 and Bee9 genomes also lacked the ISEfa5 and ISEfa11 insertion sequences that are associated with the vancomycin-resistance genes vanS (sensor histidine kinase), vanX (dipeptidase) and vanY (carboxypeptidase) [34]. The number of IS elements was significantly higher in the adult (Bee9) than the larval bee (Am1) isolate. This might be related to the exposure of adult bees to different ecological niches as they move freely without restrictions. Further investigations are required to verify these findings.
Additionally, the Am1 and Bee9 genomes were mined for other mobile genetic elements, including plasmids, transposons, CRISPR-cas and prophages. Mega plasmid pNB2354_1 was predicted in both bee isolates. This plasmid was previously reported in the non-clinical E. faecium NRRL B-2354 strain [48]. ICE and transposon gene induction leads to the potential transfer of DNA to appropriate recipients through processes of HGT [49]. Unlike clinical strains, the genomes of Am1 and Bee9 lacked transposons Tn1547, Tn1549 and Tn5382, which are responsible for the transfer of vanB resistance alleles [50–52]. CRISPR functions in defence against bacteriophage and foreign DNA [47]. According to genomic data, the larval isolate Am1 has no CRISPR elements, but the adult bee isolate (Bee9) has one CRISPR locus that may offer phage defence in the adult bee gut. The biotic and abiotic complexity of the adult insect’s gut ecosystem, where multiple bacterial species and individuals coexist, as well as variation in phage and plasmids, may explain how this CRISPR element evolved and was maintained [53].
The RAST-based annotation suggested an abundance of carbohydrate metabolism subsystems. This apparent high capacity for carbohydrate utilization agrees with the ability of E. faecium strains to utilize a wide variety of saccharides [54]. The ability to utilize various saccharides plays a role in biofilm formation by Gram-positive bacteria [55]. Gut colonization through biofilm formation is required for the interaction of bacterial cells with the host and supports resistance to gastrointestinal tract conditions [56]. The metabolic diversity in bee-gut associated E. faecium is vital in the adaptation of these strains to the host bee gut under various environmental and stress responses. Overall, enterococci often require rich and complex nutrients to support growth as a result of their fastidious nature [57].
Am1 and Bee9 strains were found to possess two bacteriocin gene clusters specifying enterocin P (entP) and enterolysin A (enlA). Enterocin P is a class II bacteriocin that displays a strong inhibitory activity against food pathogenic Listeria monocytogenes by permeabilizing the membrane, causing an ionic imbalance and leakage of inorganic ions [58]. Enterolysin A is a class III bacteriocin that inhibits growth of bacteria by the cleavage of the peptide bonds within the stem peptide, as well as in the interpeptide bridge, of Gram-positive bacterial cell walls [59]. The presence of bacteriocin genes in bee-gut-associated E. faecium genomes suggests a protective activity for these bacterial strains against host pathogens that would support their colonization of the gastro-intestinal tract. Finally, Am1 and Bee9 isolates revealed hydrophobicity and autoaggregation potential, which are beneficial characteristics for LAB. These characters are necessary for adhesion to intestinal cells and to form a barrier that prevents the colonization by pathogens [60].
Conclusion
The genomes of Am1 and Bee9 isolated from larval and adult bee gut were sequenced and followed by their incorporation in a comparative genome analysis. The assembled genomes have provided insights into the virulence and antibiotic resistance in bee-gut E. faecium and the data from this study would support the use of bee-gut E. faecium as a safe and of low microbial risk source of LAB.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
We gratefully acknowledge Professor Simon Colin Andrews, Professor of Molecular Microbiology, School of Biological Sciences, University of Reading, UK, for his kind help in revising and improving the English language in the manuscript. We would also like to thank Professor Dr Ahlam A. Alfazairy, Department of Applied Entomology and Zoology, Faculty of Agriculture, Alexandria University, Egypt, for her kind help with the bee dissection step.
Author contributions
H.A.H.Z. and N.M.E. contributed significantly to design experimental work, data acquisition analysis and interpretation in all aspects. Furthermore, both authors took part in drafting, revising and critically reviewing the article and gave final approval of this version.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ARG, antimicrobial-resistance gene; AST, antimicrobial-susceptibility testing; CDS, coding sequence; CFS, cell-free supernatant; CLSI, Clinical and Laboratory Standards Institute; CRISPR, clustered regularly interspaced short palindromic repeat; GI, genomic island; HGT, horizontal gene transfer; ICE, integrative and conjugative element; LAB, lactic acid bacteria; MIC, minimum inhibitory concentration; NCBI, National Center for Biotechnology Information; PATRIC, Pathosystems Resource Integration Center; SEM, scanning electron microscopy; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and seven supplementary tables are available with the online version of this article.
References
- 1.Krawczyk B, Wityk P, Gałęcka M, Michalik M. The many faces of Enterococcus spp. – commensal, probiotic and opportunistic pathogen. Microorganisms. 2021;9:1900. doi: 10.3390/microorganisms9091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida-Santos AC, Novais C, Peixe L, Freitas AR. Enterococcus spp. as a producer and target of bacteriocins: a double-edged sword in the antimicrobial resistance crisis context. Antibiotics. 2021;10:1215. doi: 10.3390/antibiotics10101215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimov SG, Guyrova A, Vladimirova A, Dimitrov M, Peykov S, et al. WGS-based characterization of the potentially beneficial Enterococcus faecium EFD from a beehive. Mol Biol Rep. 2020;47:6445–6449. doi: 10.1007/s11033-020-05663-5. [DOI] [PubMed] [Google Scholar]
- 5.Rogers LA, Strong K, Cork SC, McAllister TA, Liljebjelke K, et al. The role of whole genome sequencing in the surveillance of antimicrobial resistant Enterococcus spp.: a scoping review. Front Public Health. 2021;9:599285. doi: 10.3389/fpubh.2021.599285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson KE, Sheehan TH, Mott BM, Maes P, Snyder L, et al. Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera) PLoS One. 2013;8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carina Audisio M, Torres MJ, Sabaté DC, Ibarguren C, Apella MC. Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol Res. 2011;166:1–13. doi: 10.1016/j.micres.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 13.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brettin T, Davis JJ, Disz T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI Performance Standards for Antimicrobial Susceptibility Testing, supplement M100, 27th edn. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 21.Shaw TD, Fairley DJ, Schneiders T, Pathiraja M, Hill RLR, et al. The use of high-throughput sequencing to investigate an outbreak of glycopeptide-resistant Enterococcus faecium with a novel quinupristin-dalfopristin resistance mechanism. Eur J Clin Microbiol Infect Dis. 2018;37:959–967. doi: 10.1007/s10096-018-3214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanderson H, Ortega-Polo R, Zaheer R, Goji N, Amoako KK, et al. Comparative genomics of multidrug-resistant Enterococcus spp. isolated from wastewater treatment plants. BMC Microbiol. 2020;20:20. doi: 10.1186/s12866-019-1683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004;53:13–20. doi: 10.1099/jmm.0.05353-0. [DOI] [PubMed] [Google Scholar]
- 24.Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing Group. Lau BY, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 26.Liu M, Li X, Xie Y, Bi D, Sun J, et al. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019;47:D660–D665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Heel AJ, de Jong A, Song C, Viel JH, Kok J, et al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maragkoudakis PA, Zoumpopoulou G, Miaris C, Kalantzopoulos G, Pot B, et al. Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J. 2006;16:189–199. doi: 10.1016/j.idairyj.2005.02.009. [DOI] [Google Scholar]
- 32.Zommiti M, Cambronel M, Maillot O, Barreau M, Sebei K, et al. Evaluation of probiotic properties and safety of Enterococcus faecium isolated from artisanal Tunisian meat “Dried Ossban.”. Front Microbiol. 2018;9:1685. doi: 10.3389/fmicb.2018.01685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rokana N, Singh BP, Thakur N, Sharma C, Gulhane RD, et al. Screening of cell surface properties of potential probiotic lactobacilli isolated from human milk. J Dairy Res. 2018;85:347–354. doi: 10.1017/S0022029918000432. [DOI] [PubMed] [Google Scholar]
- 34.López M, Sáenz Y, Alvarez-Martínez MJ, Marco F, Robredo B, et al. Tn1546 structures and multilocus sequence typing of vanA-containing enterococci of animal, human and food origin. J Antimicrob Chemother. 2010;65:1570–1575. doi: 10.1093/jac/dkq192. [DOI] [PubMed] [Google Scholar]
- 35.Mikalsen T, Pedersen T, Willems R, Coque TM, Werner G. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis [published correction appears in BMC Genomics 2015;16:689] BMC Genomics. 2015;16:282. doi: 10.1186/s12864-015-1407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghattargi VC, Gaikwad MA, Meti BS, Nimonkar YS, Dixit K, et al. Comparative genome analysis reveals key genetic factors associated with probiotic property in Enterococcus faecium strains. BMC Genomics. 2018;19:652. doi: 10.1186/s12864-018-5043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondi M, Laukova A, de Niederhausern S, Messi P, Papadopoulou C, et al. Controversial aspects displayed by enterococci: probiotics or pathogens? Biomed Res Int. 2020;2020:9816185. doi: 10.1155/2020/9816185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vu J, Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol. 2011;6:357–366. doi: 10.1007/s11515-011-1167-x. [DOI] [Google Scholar]
- 39.Biswas PP, Dey S, Adhikari L, Sen A. Virulence markers of vancomycin resistant enterococci isolated from infected and colonized patients. J Glob Infect Dis. 2014;6:157–163. doi: 10.4103/0974-777X.145242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strateva T, Atanasova D, Savov E, Petrova G, Mitov I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz J Infect Dis. 2016;20:127–133. doi: 10.1016/j.bjid.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendrickx APA, van Wamel WJB, Posthuma G, Bonten MJM, Willems RJL. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189:8321–8332. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanchi H, Mottawea W, Sebei K, Hammami R. The genus Enterococcus: between probiotic potential and safety concerns – an update. Front Microbiol. 2018;9:1791. doi: 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein G. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int J Food Microbiol. 2003;88:123–131. doi: 10.1016/s0168-1605(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 44.Egan SA, Kavanagh NL, Shore AC, Mollerup S, Samaniego Castruita JA, et al. Genomic analysis of 600 vancomycin-resistant Enterococcus faecium reveals a high prevalence of ST80 and spread of similar vanA regions via IS1216E and plasmid transfer in diverse genetic lineages in Ireland. J Antimicrob Chemother. 2022;77:320–330. doi: 10.1093/jac/dkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimoto Y, Kita I, Suzuki M, Hirakawa H, Ohtaki H, et al. First report of the local spread of vancomycin-resistant enterococci ascribed to the interspecies transmission of a vanA gene cluster-carrying linear plasmid. mSphere. 2020;5:e00102-20. doi: 10.1128/mSphere.00102-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morroni G, Brenciani A, Litta-Mulondo A, Vignaroli C, Mangiaterra G, et al. Characterization of a new transferable MDR plasmid carrying the pbp5 gene from a clade B commensal Enterococcus faecium . J Antimicrob Chemother. 2019;74:843–850. doi: 10.1093/jac/dky549. [DOI] [PubMed] [Google Scholar]
- 47.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopit LM, Kim EB, Siezen RJ, Harris LJ, Marco ML. Safety of the surrogate microorganism Enterococcus faecium NRRL B-2354 for use in thermal process validation. Appl Environ Microbiol. 2014;80:1899–1909. doi: 10.1128/AEM.03859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/AAC.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Launay A, Ballard SA, Johnson PDR, Grayson ML, Lambert T. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob Agents Chemother. 2006;50:1054–1062. doi: 10.1128/AAC.50.3.1054-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López M, Hormazábal JC, Maldonado A, Saavedra G, Baquero F, et al. Clonal dissemination of Enterococcus faecalis ST201 and Enterococcus faecium CC17-ST64 containing Tn5382-vanB2 among 16 hospitals in Chile. Clin Microbiol Infect. 2009;15:586–588. doi: 10.1111/j.1469-0691.2009.02741.x. [DOI] [PubMed] [Google Scholar]
- 53.Westra ER, Levin BR. It is unclear how important CRISPR-Cas systems are for protecting natural populations of bacteria against infections by mobile genetic elements. Proc Natl Acad Sci USA. 2020;117:27777–27785. doi: 10.1073/pnas.1915966117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manero A, Blanch AR. Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol. 1999;65:4425–4430. doi: 10.1128/AEM.65.10.4425-4430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pillai SK, Sakoulas G, Eliopoulos GM, Moellering RC, Murray BE, et al. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis . J Infect Dis. 2004;190:967–970. doi: 10.1086/423139. [DOI] [PubMed] [Google Scholar]
- 56.Nishiyama K, Sugiyama M, Mukai T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms. 2016;4:E34. doi: 10.3390/microorganisms4030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.APHA-AWWA-WEF Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association, American Water Works Association and Water Environment Federation; 2005. [Google Scholar]
- 58.Herranz C, Chen Y, Chung HJ, Cintas LM, Hernández PE, et al. Enterocin P selectively dissipates the membrane potential of Enterococcus faecium T136. Appl Environ Microbiol. 2001;67:1689–1692. doi: 10.1128/AEM.67.4.1689-1692.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nilsen T, Nes IF, Holo H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol. 2003;69:2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baccouri O, Boukerb AM, Farhat LB, Zébré A, Zimmermann K, et al. Probiotic potential and safety evaluation of Enterococcus faecalis OB14 and OB15, isolated from traditional Tunisian testouri cheese and rigouta, using physiological and genomic analysis. Front Microbiol. 2019;10:881. doi: 10.3389/fmicb.2019.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Natarajan P, Parani M. First complete genome sequence of a probiotic Enterococcus faecium strain T-110 and its comparative genome analysis with pathogenic and non-pathogenic Enterococcus faecium genomes. J Genet Genomics. 2015;42:43–46. doi: 10.1016/j.jgg.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Buzikov RM, Piligrimova EG, Shadrin AM. Complete genome sequence of Enterococcus faecium FS86, used for propagation of bacteriophages with therapeutic potential. Microbiol Resour Announc. 2020;9:e00776-20. doi: 10.1128/MRA.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin X, Galloway-Peña JR, Sillanpaa J, Roh JH, Nallapareddy SR, et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012;12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubin IMC, Pedersen MS, Mollerup S, Kaya H, Petersen AM, et al. Association between vancomycin-resistant Enterococcus faecium colonization and subsequent infection: a retrospective WGS study. J Antimicrob Chemother. 2020;75:1712–1715. doi: 10.1093/jac/dkaa074. [DOI] [PubMed] [Google Scholar]
- 65.Lam MMC, Seemann T, Bulach DM, Gladman SL, Chen H, et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol. 2012;194:2334–2341. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.