Abstract
Spinal cord ischemia/reperfusion injury (SCII) is a severe complication driven by apoptosis and neuroinflammation. An increase in the expression of c‐Fos, a member of the AP‐1 family, is known as a neuronal activation marker in SCII. The AP‐1 family is composed of Jun, Fos, and is associated with the regulation of cytokines expression and apoptosis. Fra‐1 is a member of the Fos family, however, the contribution of Fra‐1 to SCII is still unclear. In our study, Fra‐1 was highly upregulated especially in neurons and microglia and promoted apoptosis by changing the expression of Bax/Bcl‐2 after SCII. Furthermore, we found that Fra‐1 directly regulated the transcription expression of S100A8. We demonstrated that knockdown of Fra‐1 alleviated S100A8 mediated neuronal apoptosis and inflammatory factor release, thus improved motor function after SCII. Interestingly, we showed that administration of TAK‐242, the TLR4 inhibitor, to the ischemia/reperfusion (I/R) injury induced rats suppressed the activation of the ERK and NF‐κB pathways, and further reduced Fra‐1 expression. In conclusion, we found that Fra‐1‐targeted S100A8 was expressed the upstream of Fra‐1, and the Fra‐1/S100A8 interaction formed a feedback loop in the signaling pathways activated by SCII.
Keywords: apoptosis, feedback loop, ischemia/reperfusion, neuroinflammation, oxygen–glucose deprivation/reoxygenation, spinal cord ischemia/reperfusion injury
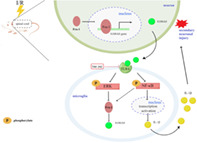
Schematic illustration depicting the mechanism by which Fra‐1‐mediated S100A8/TLR4/NF‐κB and ERK signaling in the regulation of apoptosis and neuroinflammation during spinal I/R.
1. INTRODUCTION
Spinal cord ischemia/reperfusion injury (SCII) is a severe complication after spinal or thoracic surgery that often triggers a variety of pathophysiological reactions, including the release of excitatory amino acids, inflammatory reactions and neuronal apoptosis [1, 2]. The reactions caused by SCII can result in paralysis or paraplegia and even threaten the life of patients [3]. Although various studies on SCII have been reported, the mechanisms of this injury are not sufficiently clear, and few effective protective measures are used in the clinic.
The AP‐1 complex is a dimer consisting of Jun (c‐Jun, JunB and JunD) and Fos (c‐Fos, FraB, Fra‐1, and Fra‐2) family proteins and other bZIP transcription factors [4]. AP‐1 transcription factors are signal activated, and they regulate apoptosis and stress responses [5,6]. Increasing evidence has indicated that c‐Fos and c‐Jun can be induced to high expression levels after ischemia injury and that this upregulation moderates apoptosis [7, 8]. Nevertheless, the role of Fra‐1 after ischemic injury has rarely been studied. Some reports have indicated that Fra‐1 controls cell proliferation, survival, and metastasis, but the therapeutic effect of Fra‐1 depends on the cancer cell context [9]. In addition, Fra‐1 has been shown to suppress apoptosis [10, 11, 12]. However, Fra‐1 has also been reported to promote apoptosis, indicating that the role of Fra‐1 in apoptosis might vary with the type of cell and stimulation [13, 14]. Therefore, understanding the function of Fra‐1 may help identify novel potential targets for therapeutic intervention after SCII.
S100A8 has attracted long‐standing interest as a therapeutic target in myocardial ischemia [15, 16]. However, the role of S100A8 after SCII has been unknown. In the previous study, S100A8 has been found to be expressed in microglia and neurons in the central nervous system and is secreted by neuronal cells under hypoxic conditions, which promotes activation of the Toll‐like receptor 4(TLR4)/ nuclear factor‐κB (NF‐κB) pathway and production of interleukin‐1β (IL‐1β), resulting in neuroinflammation and apoptosis [17]. Multiple lines of evidence support the idea that S100A8, a known ligand of TLR4, can induce downstream TLR4 genes [18, 19, 20]. Besides, phosphorylated extracellular regulated protein kinases (p‐ERK) and phosphorylated NF‐κB (p‐NB‐κB) in TLR4 downstream pathways, act as important factors contributing to inflammation and apoptosis [17, 21, 22], especially after SCII [23, 24]. Moreover, extracellular regulated protein kinases (ERK) and NF‐κB signaling regulates the abundance of Fra‐1 via transcriptional induction and posttranslational stabilization [25, 26, 27, 28].
In this study, we first investigated Fra‐1 and its target gene S100A8, and we found that in conjunction, they promoted apoptosis and neuroinflammation through the TLR4/ERK and NF‐κB pathways after SCII. Notably, the feedback loop formed by Fra‐1/S100A8 aggravates the ischemic/reperfusion cascade reaction, so comprehensive identification of the feedback loop can help us to understand the pathophysiological process of SCII.
2. MATERIALS AND METHODS
2.1. Animals
Male Sprague–Dawley rats (200–250 g) were obtained from the Animal Center of China Medical University, Shenyang (China). All rats were housed for at least 1 week to allow their adaptation to the environment, and they had free access to water and food. The rats were randomly assigned to control and experimental groups.
2.2. Drugs and reagents
Recombinant rat S100A8 (rS100A8, E. coli derived, with purity >95%) was purchased from Med Chem Express (China, cat. no. HY‐P71275). TAK‐242 (purity >98%) was purchased from Absin (Shanghai, China, cat. no. abs814050). The following antibodies were used for Western Blotting: primary antibodies against Fra‐1 (sc‐376148, 1:1000), TLR4 (sc‐293072, 1:1000) and IL‐1β (sc‐12742, 1:1000) were purchased from Santa Cruz Biotechnology; primary antibodies against S100A8 (15792‐1‐AP, 1:1000), Bcl‐2 (60178‐1‐Ig, 1:2000), Bax (60267‐1‐Ig, 1:2000) and secondary antibodies goat anti‐mouse IgG (SA00001‐1, 1:10000) and goat anti‐rabbit IgG (SA00001‐2, 1:10000) were purchased from the PROTEINTECH; primary antibodies to cleaved caspase‐3 (9661, 1:1000), ERK1/2 (4695, 1:1000), p‐ERK1/2 (4370, 1:1000), NF‐κB (8242, 1:1000), and p‐NF‐κB (3033, 1:1000) were purchased from Cell Signaling Technology. The following antibodies were used for Immunofluorescence are: a primary antibody against Fra‐1 (sc‐376148, 1:100) was purchased from Santa Cruz Biotechnology; a primary antibody against cleaved caspase‐3 (9661, 1:200) was purchased from Cell Signaling Technology; primary antibodies against NeuN (26975‐1‐AP, 1:200), GFAP (16825‐1‐AP, 1:200), and Iba‐1 (10904‐1‐AP, 1:200) were purchased from PROTEINTECH; and secondary antibodies donkey anti‐rabbit IgG (H + L, GB22403, 1:200) and goat anti‐mouse IgG (H + L, GB21301, 1:200) were purchased from Servicebio.
2.3. Intrathecal administration
Intrathecal administration was operated as described previously [1]. In summary, after receiving anesthesia, each rat was placed in the prone position with the back was arched. A 25 μl microliter syringes needle (Gaoge Co, Shanghai, China) was placed in the subarachnoid space between segments L4‐L6, and the drug was slowly injected in 1 min. Successful puncture was evident when, after the needle entered the subarachnoid space, the rat tail laterally flicked. Ten microliters of 2 × 1010 vg/ml synthetic adeno‐associated virus (AAV) targeting Fra‐1 and S100A8 along with the respective negative controls (purchased from GeneChem Co., Ltd., China) was intrathecally injected into the rats. The sequences were as follows:
AAV‐sh‐Fra‐1 sequence (5′‐ACCGGCGAAAGAGTAGCAGCAGCACTCGAGTGCTGCTGCTACTCTTTCGTTTT‐3′);
AAV‐sh‐S100A8 sequence (5′‐ACCGGCCACAATTATTCTGGTATACTCGAGTATACCAGAATAATTGTGGTTTT‐3′).
2.4. Drug administration to animals
Recombinant S100A8 (rS100A8) protein was diluted using normal saline. The TLR4 inhibitor, TAK‐242 was dissolved in 1% dimethyl sulfoxide (DMSO). The rS100A8 (10 μg in 10 μl) was intrathecally injected before induction of spinal ischemia. Sham rats were injected with 10 μl of normal saline. The process was performed following a previously reported method [20, 29]. TAK‐242 was intraperitoneally injected (3 mg/kg) 30 min before the injection of rS100A8 [20, 29, 30]. The rS100A8 and TAK‐242 can permeate the blood–brain barrier [20, 29, 30].
2.5. Rat models
The SCII rat models were established as previously described [31]. In summary, we anesthetized the rats with pentobarbital sodium (50 mg/kg), and the rats were endotracheally intubated and mechanically ventilated. Then, through the cervicothoracic incision, between the left common carotid artery and the left subclavian artery the aortic arch was cross‐clamped to establish the spinal cord ischemia model. After 14 min, we released the occlusion to start reperfusion. We performed the same procedure without clamping for rats in the sham group.
2.6. Tarlov score
Tarlov score was used to assess the motor function of the hind limbs of rats [32]. The scores of motor function were observed and recorded by two experimentals who were blind to the grouping. The score was expressed as: 0—no movement of lower limbs; 1—weak movement of lower limbs; 2—joint of lower limb can move, but unable to stand; 3—abnormal standing but unable to walk; 4—Normal.
2.7. Motor functional assessment
The postoperative motor function of the rats was assessed by toe footprint analysis [33, 34]. In summary, the rats were allowed to walk into a dark compartment on a gangway (100 cm × 12 cm × 10 cm). The rats did not need to be pretrained or conditioned before completing this task. The rat hind paws were dipped in two different nontoxic dyes. A ruler was attached to the gangway, and clear footprints were obtained with the imaging equipment, which was placed under the transparent bottom. The walking posture and toe hold of the rats were clearly visible on the transparent surface. After the rats walked across the gangway, the dyes on the toes were washed off. Differences in foot posture between the SCII group and the treatment group were analyzed.
2.8. Cell culture and treatment
We purchased VSC4.1 motor neurons from Beina Chuanglian Biotechnology Institute (BNCC, China). They were cultured in an incubator (5% CO2; 37°C) in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS500‐S, AusGeneX, Australia) and 1% penicillin/streptomycin(Solarbio, China) as previously described [35].
After placing 5 × 105 cells/well in a 6‐well plate, short interfering RNA (siRNA) against Fra‐1 (si‐Fra‐1; 50 nmol/L) or the plasmid used for Fra‐1 overexpression (OE‐Fra‐1; 2 μg) was added to 200 μl of jet PRIMETM Buffer, and mixed with 4 μl jet PRIMETM reagent and then added to cell cultures. After 4–6 h, the culture solution was replaced with DMEM medium. After 48 h of transfection, western blotting was performed to measure the protein levels in the cells of each group. The Fra‐1 OE vector (OE‐Fra‐1, EX‐Rn24015‐M90) and its negative control plasmid (EX‐NEG‐M90) were purchased from GeneCopoeia (China).
The si‐Fra‐1‐1 sequence (forward, 5′‐CCGACAAGUUGGAGGAUGATT‐3′, and reverse, 5′‐UCAUCCUCCAACUUGUCGGTT‐3′);
si‐Fra‐1‐2 sequence (forward, 5′‐ GGAUGGUGCAGCCUCAUUUTT‐3′, and reverse, 5′‐AAAUGAGGCUGCACCAUCCTT‐3′);
si‐Fra‐1‐3 sequence (forward, 5′‐CGAAAGAGUAGCAGCAGCATT‐3′, and reverse, 5′‐UGCUGCUGCUACUCUUUCGTT‐3′) were purchased from GENERAL BIOL (China).
2.9. Oxygen–glucose deprivation/reoxygenation (OGD/R) model
We established the OGD/R model as described previously [36]. VSC4.1 neurons were washed twice using Hank's balanced salt solution (HBSS), cultured in glucose‐free DMEM (Gibco) and then incubated in a hypoxic chamber (94% N2, 5% CO2, and 1% O2; 37°C)for 6 h. After two washes with HBSS, the cultured cells were transferred to initial medium and incubated in an incubator (5% CO2; 37°C) for 24 h to induce reoxygenation. The control cells were cultured in normal medium in the incubator (5% CO2; 37°C).
2.10. Detection of cell viability
The vitality values of the OGD/R‐induced cells were detected and compared with those of the non‐oxygen–glucose deprived group. As in the previous study [37], the cells were added into 96‐well plates with 100 μl cell suspension in each well, and the cell density was 5 × 103/well. When the cell density reached 5 × 104/well, the OGD/R model was established, and the 96‐well plates were removed from the hypoxia chamber at 0.5, 1, 2, 4, 6, and 8 h after oxygen and glucose deprivation. Then 10uL CCK‐8 reagent (MedChemExpress, China) was added to each well and incubated at 37°C for 2 h in dark. And the cells were detected at 12, 24, and 36 h after reoxygenation, and the same volume of CCK‐8 reagent was added to each well at the same incubation conditions. We set up the blank control well, which contained no cells, only medium and reagent. The absorbance of cells was detected with a microplate reader (BioTek, USA) and the wavelength was set at 450 nm.
2.11. Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA from L4‐L6 spinal cord specimens and cells was obtained with TRIzol reagent (TaKaRa, Japan). Then, cDNA was obtained by reverse transcription of the total RNA using the PrimeScript RT reagent kit (RR047A, Takara). Finally, qRT‐PCR was performed with a TB Green Premix Ex TaqII kit (RR820A, Takara) on an ABI 7500 qRT‐PCR instrument (Applied Biosystems). Relative expression of Fra‐1 and S100A8 mRNA was normalized to that of GAPDH. The following primers (Sangon Biotech, China) were used: Fra‐1 (forward, 5′‐CACAGACAGTCCAGCAGCAGAAG‐3′, and reverse, 5′‐GATAGGTCAGAGGTCGGGGATAGC‐3′), S100A8 (forward, 5′‐AGTGCCCTCAGTTTGTGCAG‐3′, and reverse, 5′‐CGCCCACCCTTATCACCAAC‐3′).
2.12. Western blotting
The protein of spinal cord tissues (L4‐L6 segments) or cells was lysed and extracted. Then, we used the BCA protein assay kit (Beyotime, Biotechnology) to detect the protein concentrations. Next, we used 8 ~ 15% SDS‐PAGE gels to separate the protein samples, which were then transferred to PVDF membranes (Millipore, Temecula, CA). After blocking with 5% skim milk (1 h), the membranes were incubated with primary antibodies (overnight; 4°C). Subsequently, after two washes with Tris‐buffered saline‐Tween 20 (TBST) buffer,the membranes incubated with secondary antibodies (1 h; 37°C). Finally, we visualized the membranes using an ECL Western HRP kit (17046, ZEN BIO, China), and the band intensity was measured by an imaging system (Bio‐Rad). The expression of β‐actin was the reference to the expression levels of the proteins.
2.13. Hematoxylin and eosin (H&E) staining
The L4‐L6 segments of rat spinal cords were fixed in 4% paraformaldehyde (G1101, Servicebio, China). After fixation for 24 h, the samples were embedded in paraffin. Then, the blocks were sliced into 4 μm slices, deparaffinized, and rehydrated. Finally, the slices were stained with H&E staining kit reagent (Servicebio G1005, China). The stained sections were examined by two experimenters who were blind to the grouping using an optical microscope (Nikon, Japan). Normal neurons are typically considered to contain a round or ovoid clear nucleus [2]. The average number of normal neurons was calculated with Image J software (download from the NIH website).
2.14. Double immunofluorescence (IF)
As previously described [2], for the in vivo experiment, spinal cord slices were deparaffinized and rehydrated. Then, we performed the antigen retrieval by immersing the slices in heated sodium citrate. After blocking with QuickBlock (P0260, Beyotime; 15 min; room temperature), the slices were incubated with primary antibodies (overnight; 4°C). Then, the slices were incubated with secondary antibodies (1 h; room temperature) in the dark. For the experiment in vitro, the neurons were fixed with 4% paraformaldehyde (20 min; 4°C). Then, the neurons were blocked with QuickBlock (15 min; room temperature). Similarly, the neurons were incubated with primary antibodies (overnight; 4°C) and then with secondary antibodies (1 h; room temperature) in the dark. We counterstained the nuclei with 4, 6‐diamidino‐2‐ phenylindole (DAPI; G1012, Servicebio; 10 min; room temperature). Finally, we captured the images with a fluorescence microscope (Nikon, Japan), and we measured the integrated fluorescence densities by Image J software.
2.15. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
Spinal cord slices were stained with TMR (red) TUNEL apoptosis detection kit (G1502, Servicebio) following the manufacturers's instructions. We stained the nuclei with DAPI (G1012, Servicebio; 10 min) in the dark. Finally, we captured images by a fluorescence microscope (Nikon, Japan).The apoptosis rate (the ratio of TUNEL‐positive cells to total nuclei) was calculated by two investigators who were blinded to the grouping using Image J software.
2.16. Chromatin immunoprecipitation coupled with qPCR (ChIP‐qPCR)
A total of 1 × 107 VSC4.1 cells obtained after OGD/R induction were used to perform ChIP experiments as described previously [38]. Briefly, the cells were cross‐linked with 1% formaldehyde. Then, the cells were lysed to obtain the ultrasonically fragmented chromatin. Chromatin was immunoprecipitated using a ChIP‐grade antibody against Fra‐1 (5841, Cell Signaling Technology). Then, the cross‐links in the protein‐DNA complexes were broken, and the DNA was purified. Finally, qRT‐PCR was performed with the obtained DNA products. The sequences of the S100A8 primers were as follows: S100A8‐1 (forward, 5′‐GCATGGCAGAAGGAGCAAGAG‐3′, and reverse, 5′‐GGAACTCAGACTCATCACGAAGAC‐3′); S100A8‐2 (forward, 5′‐AACGGAGGACAGGGCTAAGAC‐3′, and reverse, 5′‐CAGGGAAGACATTTACCAGACAGG‐3′); and S100A8‐3 (forward, 5′‐CATCCTTCTTCTGCTCCGTCTTC‐3′, and reverse, 5′‐GCTCCACACTGTTCCACTTCC‐3′).
2.17. Statistical analysis
We used GraphPad Prism 8.0 software for data analysis. The data at normal distribution are shown as the mean ± SEM. Kruskal–Wallis test was used for comparison among multiple groups of data that did not conform to normal distribution. For the data conforming to a normal distribution, the comparison between two groups was analyzed with the unpaired t‐test and the one‐way or two‐way ANOVA were used for differences among multiple groups followed by Tukey's test. p values <0.05 were considered to be statistically significant.
3. RESULTS
3.1. The expression of Fra‐1 at different time points after spinal I/R
To investigate the Fra‐1 expression at different time points after spinal I/R, the spinal cord ischemia model was established and sampled at different time points after reperfusion (Figure 1A). As shown in Figure 1B–D, the mRNA and protein levels of Fra‐1 were measured, and the results showed that the expression of Fra‐1 increased over time and peaked 48 h after I/R. Considering that Fra‐1 affects the apoptosis response under some conditions [10, 11, 12, 13, 14], we measured the protein expression of cleaved caspase‐3 and found that Fra‐1 and cleaved caspase‐3 were positively correlated, as shown in Figure 1C–F. These data indicated that Fra‐1 may influence the apoptosis response activated by I/R.
FIGURE 1.
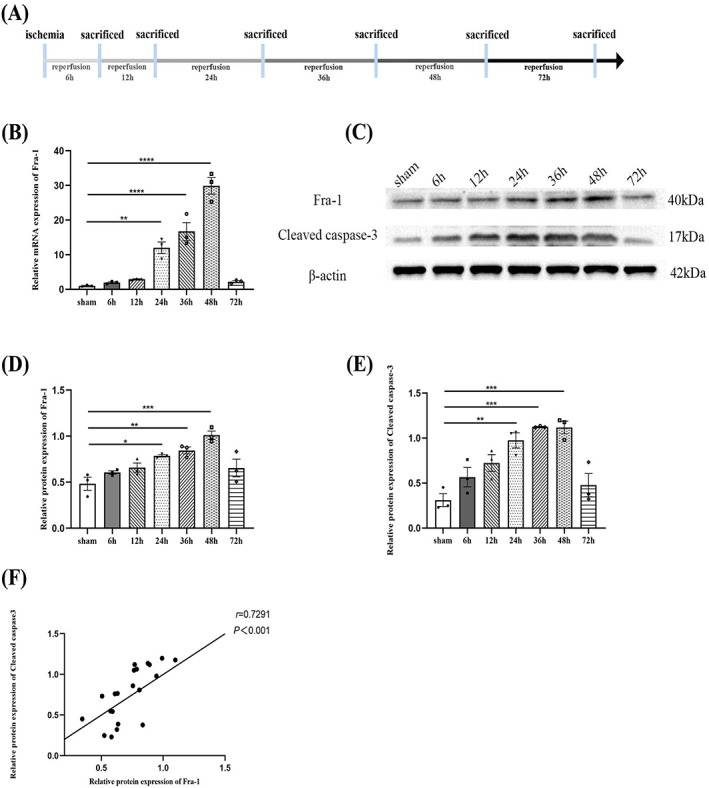
The expression of Fra‐1 at different time points after spinal ischemia/reperfusion (I/R). (A) Schematic of the experimental design shows the timeline for grouping rats. At different time points (6, 12, 24, 36, 48 and 72 h) after I/R induction, the L4‐L6 segments of spinal cords were harvested from the rats. (B) qRT‐PCR‐assisted detection of the relative mRNA level of Fra‐1 after I/R (n = 3 rats/group). (C–E) Western blot‐assisted analysis of the protein expression levels of Fra‐1 and cleaved caspase‐3 after I/R (n = 3 rats/group). (F) Correlation analysis of Fra‐1 and cleaved caspase‐3 protein expression levels. All data represent the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.2. Cellular distribution of Fra‐1 expression after spinal I/R
To identify cell types with upregulated Fra‐1 expression after I/R, according to the previous results, spinal cord ischemia model was established and sampled 48 h after reperfusion (Figure 2A). We performed IF assays with markers represented NeuN, Iba‐1, and GFAP cell type. As shown in Figure 2B–D, in the anterior horn of spinal cord, Fra‐1 was increased in both the NeuN‐positive and Iba‐1‐positive cells, and that the expression level of Fra‐1was higher in neurons 48 h after I/R. The ventral horn of spinal cord injury is associated with motor outcome [39]. Therefore, we speculated that Fra‐1 might affect motor function of rats after I/R. At the same time, according to the results we observed Fra‐1 was also highly expressed in microglia, which may be involved in microglia‐related inflammation induced by I/R.
FIGURE 2.
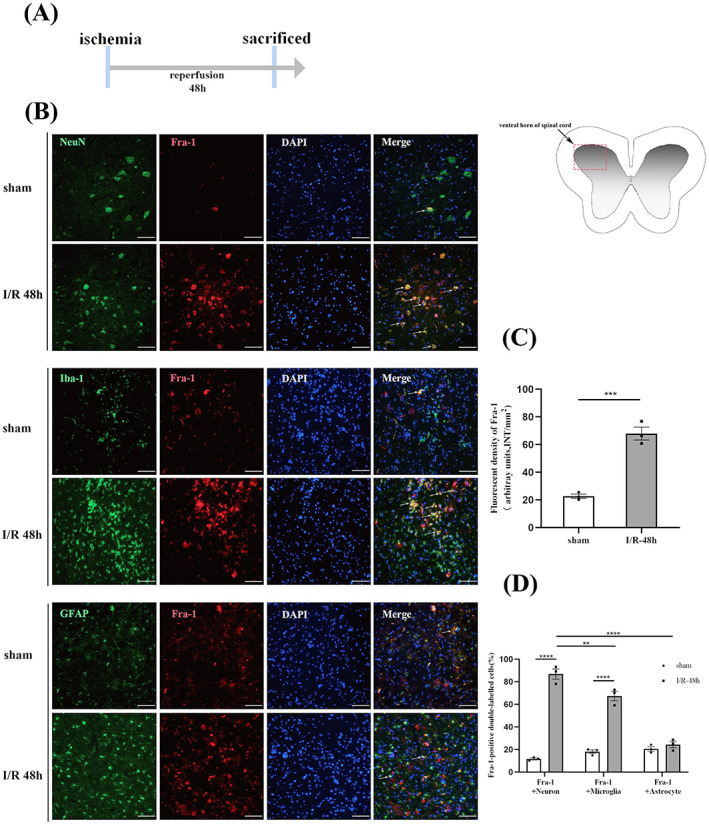
Cellular distribution of Fra‐1 expression after spinal I/R (A) Schematic of the experimental design shows the timeline for grouping rats. Spinal cord ischemia model was established. After 48 h of I/R, tissue samples were collected. (B) In the ventral horn of the spinal cord, the distribution and expression of Fra‐1 (red) were detected by double immunofluorescence (IF) analysis with NeuN (green, neuron marker), Iba‐1 (green, microglial marker) and GFAP (green, astrocyte marker) 48 h after I/R (n = 3 rats/group). Scale bar = 100 μm. (C) Quantified Fra‐1 fluorescence intensity (INT/mm2) after I/R. (D) Quantified Fra‐1‐positive double‐labeled neurons, microglia and astrocytes in the spinal cord 48 h after I/R. All data represent the mean ± SEM. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3.3. Fra‐1 regulates apoptosis response in OGD/R‐stressed cells
In previous experiments, Fra‐1 was found to be positively correlated with cleaved caspase‐3 and colocalize with neurons in the ventral horn of spinal cord, and therefore, the role of Fra‐1 in neurons after SCII was further assessed. Because OGD/R‐induced neuronal injury can mimic I/R injury in vitro [35], the OD values of VSC4.1 cells at 0.5, 1, 2, 4, 6, and 8 h after oxygen and glucose deprivation were detected. Compared with the non‐oxygen–glucose deprived group, the cell activity at 0.5, 1, 2, 4, 6, and 8 h after oxygen and glucose deprivation was significantly decreased, and the cell activity at 6 and 8 h after oxygen and glucose deprivation was the lowest (Figure 3A). The OD values of VSC4.1 cells at 12, 24, and 36 h after reoxygenation were detected. Compared with cells at 6 h after oxygen and glucose deprivation, the cell activity was significantly increased, and the activity was highest at 24 and 36 h after reoxygenation (Figure 3B). The VSC4.1 neurons were used in vitro after OGD/R and transfected with the synthetic si‐Fra‐1‐1, si‐Fra‐1‐2, si‐Fra‐1‐3, and the plasmid overexpressing Fra‐1 (OE‐Fra‐1). The western blot results revealed that Fra‐1 expression was significantly downregulated by si‐Fra‐1 and upregulated by OE‐Fra‐1, respectively, relative to its expression in the OGD/R group (Figure 3C,D,G,I). The Bcl‐2 and Bax are involved in mitochondrial apoptosis pathways [40]. Our results showed that, cleaved caspase‐3 and Bax were significantly decreased, but Bcl‐2 was increased in VSC4.1 cells after OGD/R following knockdown of Fra‐1 (Figure 3E,F). In contrast, overexpression of Fra‐1 had the opposite effects (Figure 3G,I). As shown in Figure 3H,J, quantitative results of cell IF assays revealed that the expression levels of Fra‐1 and cleaved caspase‐3 were notably increased after OGD/R compared with that in the control group, but knocking down Fra‐1 significantly reduced the expression of these proteins. So the conclusion that can be drawn from the results is that after OGD/R, Fra‐1 upregulated Bax, downregulated Bcl‐2, activated caspase‐3, and promoted neuronal apoptosis with the mitochondrial pathway.
FIGURE 3.
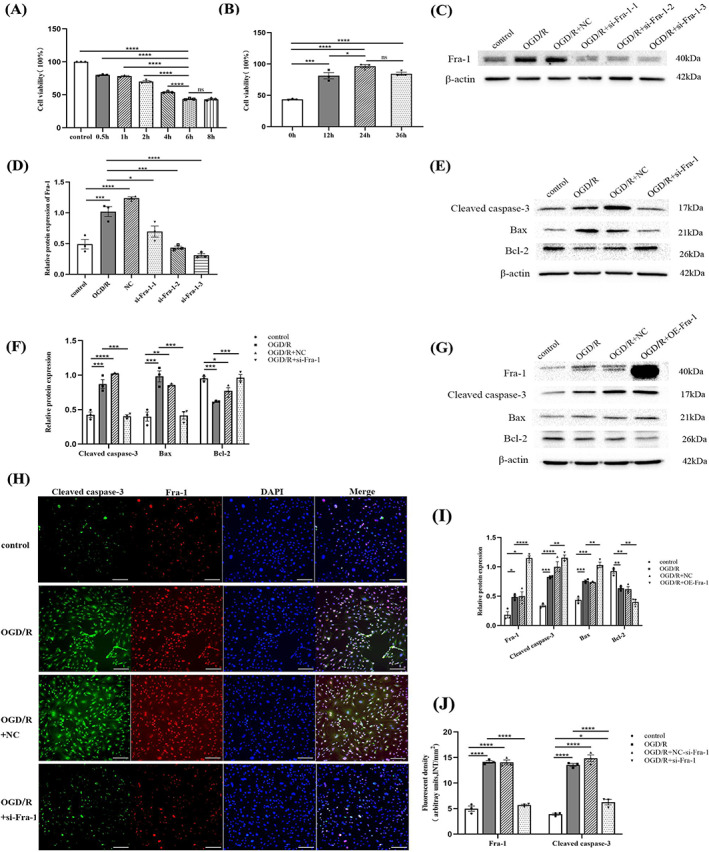
Fra‐1 regulates apoptosis response in oxygen–glucose deprivation/reoxygenation (OGD/R)‐stressed VSC4.1 neurons. (A,B) CCK‐8‐assisted analysis of cell viability after OGD/R. (C,D) Western blot‐assisted analysis of the relative protein levels of Fra‐1 after transfection with si‐Fra‐1‐1, si‐Fra‐1‐2, si‐Fra‐1‐3 under OGD/R. (E,F) Western blot‐assisted analysis of the relative protein levels of cleaved caspase‐3, Bax and Bcl‐2 in VSC4.1 neurons after transfection with si‐Fra‐1 under OGD/R. (G,I) Western blot‐assisted analysis of the relative protein levels in VSC4.1 neurons after transfection with the plasmid used for Fra‐1 overexpression (OE‐Fra‐1) under OGD/R. (H) Immunofluorescence (IF) assays of cleaved caspase‐3 and Fra‐1 after OGD/R were performed. Scale bar = 100 μm. (J) Quantification of the cleaved caspase‐3 and Fra‐1 fluorescence intensity (INT/mm2). All data represent the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3.4. Knockdown of Fra‐1 alleviates spinal I/R induced injury in vivo
To investigate the effect of Fra‐1 after I/R, at 7 days before the establishment of spinal cord ischemia model, sh‐Fa‐1 were intrathecally injected, and samples were collected 48 h after reperfusion (Figure 4A). H&E staining showed a reduction in normal neurons in the ventral horn of the spinal cord after I/R. Whereas in I/R + sh‐Fra‐1 group, the number of normal neurons increased (Figure 4B,C). In addition, as observed in vitro neuronal experiments, decreased cleaved caspase‐3 and Bax but increased Bcl‐2 were detected in the spinal cord samples taken from rats with injection of sh‐Fra‐1 after I/R as shown by western blotting (Figure 4D,E). The motor function of the hind limbs of the rats was determined by Tarlov score and visualizing their paw patterns. Tarlov score showed that the scores of the rats in I/R group and NC group were significantly lower than those of sham group. After administration of sh‐Fra‐1, Tarlov score was significantly increased compared with the I/R group (Figure 4F). Paw prints were observed in three rats from each group of 10 rats in Tarlov score experiment. Compared with those in the sham group, the rats subjected to I/R were unable to stand on their lower limbs and walked mainly by the fore limbs. After injection of sh‐Fra‐1, the hind limb motor function of the I/R injured rats improved, and the stride lengths were significantly increased (Figure 4G,H).
FIGURE 4.
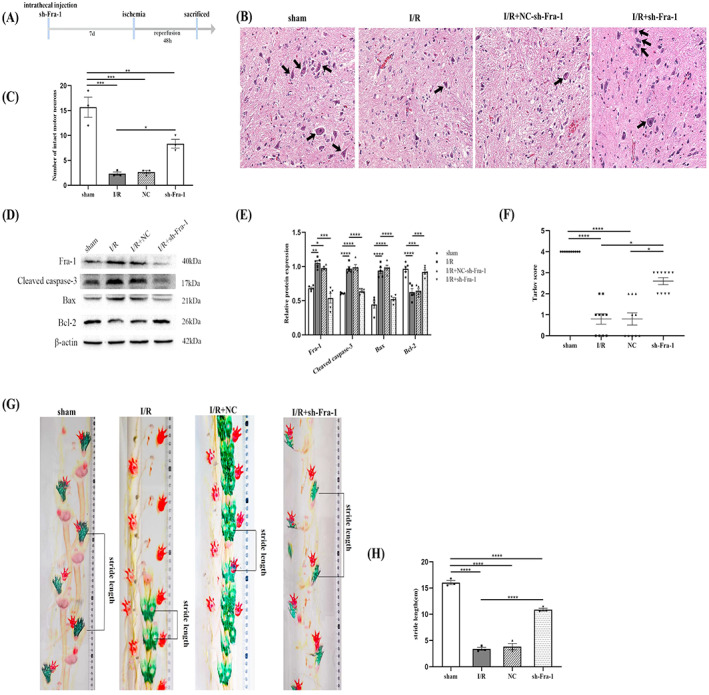
Knockdown of Fra‐1 alleviates spinal I/R induced injury in vivo. (A) Schematic of the experimental design shows the timeline for grouping rats. The rats were intrathecally injected sh‐Fra‐1 7 days before I/R induction. After 48 h of I/R, tissue samples were collected. (B,C) H&E staining‐assisted detection of the number of normal neurons in the ventral horn of the spinal cord (n = 3 rats/group). The black arrows indicate normal neurons. Scale bar = 100 μm. (D,E) Western blot‐assisted analysis of the relative levels of the Fra‐1,cleaved caspase‐3,Bax,Bcl‐2 after I/R (n = 5 rats/group). (F) Tarlov score analysis was used to evaluate neuromotor function in different groups of rats (n = 10 rats/group). Kruskal–Wallis test was used for comparison among multiple groups of data. (G) Limb motor function of rats was determined by examining paw patterns and stride length (n = 3 rats/group). (H) The stride length of the rats in different groups was analyzed. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3.5. S100A8 is a direct target of Fra‐1
To investigate the S100A8 expression after SCII, we examined the samples taken at different time points after establishing the ischemia/reperfusion model (Figure 5A). As previously reported, the S100A8 protein level is significantly upregulated after post‐traumatic brain injury [20]. RNA sequencing results by qRT‐PCR showed S100A8 was increased 48 h after SCII [41]. Similarly, we found that the S100A8 protein level was increased and peaked 36 and 48 h after I/R (Figure 5B,C). Moreover, Pearson's analysis showed that S100A8 protein expression was significantly related to Fra‐1 expression after I/R (Figure 5D). Because Fra‐1 can activate its target genes by directly binding to respective promoters [9, 42], we sought to determine whether S100A8 is a direct target of Fra‐1. We chose three putative Fra‐1‐binding sites (P1‐P3) in the S100A8 promoter on the basis of the JASPAR database and then designed and synthesized S100A8 primers (Figure 5E). The ChIP‐qPCR results showed that Fra‐1 was highly enriched in the P3 area of the S100A8 gene promoter (Figure 5F). These results suggest that S100A8 is a direct target of Fra‐1 in neurons under OGD/R. Moreover, intrathecal injection of sh‐Fra‐1 significantly decreased S100A8 mRNA and protein expressions in rat models subjected to SCII (Figure 5H–J). Therefore, these results proved that after SCII, Fra‐1 upregulated S100A8 expression at the transcriptional level.
FIGURE 5.
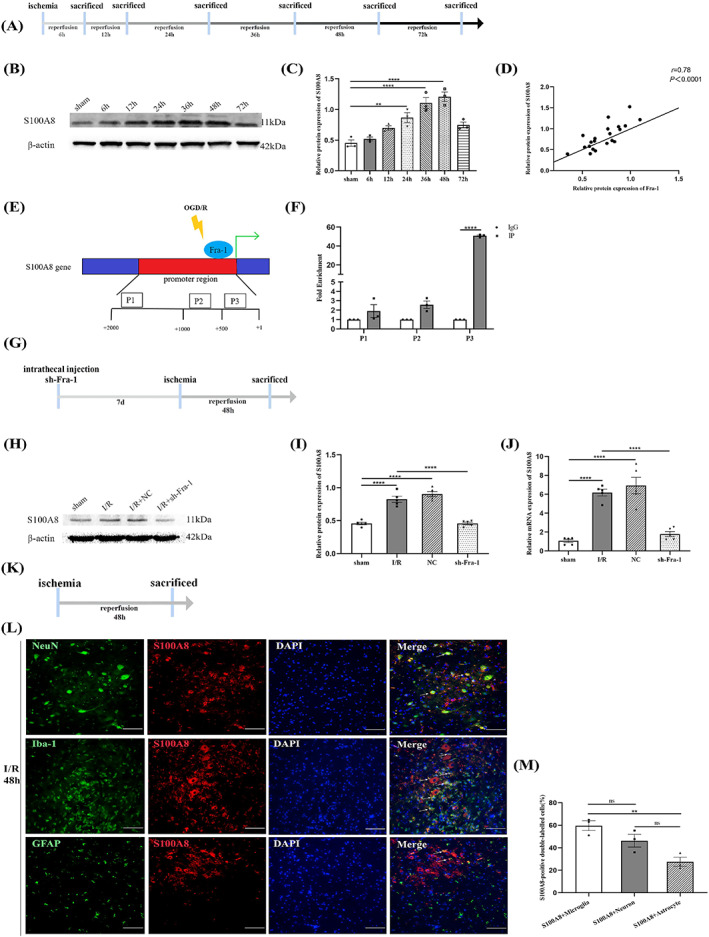
S100A8 is a direct target of Fra‐1. (A) Schematic of the experimental design shows the timeline for grouping rats. At different time points (6, 12, 24, 36, 48, and 72 h) after I/R induction, tissue samples were collected. (B,C) Western blot‐assisted analysis and quantification of S100A8 at different time points after I/R (n = 3 rats/group). (D) Correlation analysis of Fra‐1 and S100A8 protein expression. (E) We selected three putative Fra‐1 binding sites of the S100A8 promoter (P1–P3). (F) ChIP‐qPCR‐assisted analysis of S100A8 promoter fragments containing the binding site for Fra‐1. (G) The rats were intrathecally injected sh‐Fra‐1 7 days before I/R induction. After 48 h of I/R, tissue samples were collected. (H,I) Western blot‐assisted analysis and quantification of S100A8 protein expression after intrathecal injection of sh‐Fra‐1 (n = 5 rats/group). (J) qRT‐PCR‐assisted detection of the relative mRNA level of S100A8 in rat spinal cord samples (n = 5 rats/group). (K) Spinal cord ischemia model was established. After 48 h of I/R, tissue samples were collected. (L) In the ventral horn of the spinal cord, the distribution and expression of S100A8 (red) were detected by double immunofluorescence (IF) analysis with NeuN (green, neuron marker), Iba‐1 (green, microglial marker) and GFAP (green, astrocyte marker) after I/R (n = 3 rats/group). Scale bar = 100 μm. (M) Quantified S100A8‐positive double‐labeled neurons, microglia and astrocytes in the spinal cord after I/R. All data represent the mean ± SEM. **p < 0.01, ****p < 0.0001, ns, not significant.
To identify cell types with upregulated S100A8 expression after SCII, according to the previous results, we examined the samples taken 48 h after ischemia/reperfusion (Figure 5K). In immunofluorescence detection, the Neun, Iba‐1 and GFAP were used for cell markers. As shown in Figure 5L,M, in the anterior horn of spinal cord, compared with GFAP‐positive cells S100A8 was higher in microglia 48 h after I/R. SCII‐induced elevated S100A8 may be involved in microglia‐associated inflammation.
3.6. Fra‐1/S100A8‐mediated TLR4 signaling promotes ERK1/2 and NF‐κB pathways activation and IL‐1β production after spinal I/R
To determine the functional relevance of S100A8 after I/R, sh‐S100A8 was injected intrathecally 7 days before the establishment of spinal cord ischemia model, and samples were collected 48 h after reperfusion (Figure 6A). The result showed that S100A8 was increased after I/R, and decreased in the I/R + sh‐S100A8 group compared with I/R group by western blot (Figure 6F,I). Accumulating evidence has previously revealed that S100A8 can induce cell inflammation and apoptosis via the TLR4/NF‐κB and ERK1/2 pathways [17, 19, 20]. Because p‐NF‐κB, p‐ERK1/2, and IL‐1β are key players in spinal I/R‐induced apoptosis and inflammation [2, 23, 24], we sought to determine whether the production or activation of these molecules is triggered by S100A8 after SCII. Consistent with our hypothesis, the results revealed that the protein levels of cleaved caspase‐3, TLR4, IL‐1β, p‐ERK/ERK and p‐NF‐κB/NF‐κB were increased after I/R as shown by western blot (Figure 6F,I,J). In the I/R + sh‐S100A8 group, these protein levels decreased compared with I/R group (Figure 6F,I,J). Hence, S100A8 promoted apoptosis and inflammation by increasing TLR4 expression and activating the downstream ERK1/2 and NF‐κB pathways.
FIGURE 6.
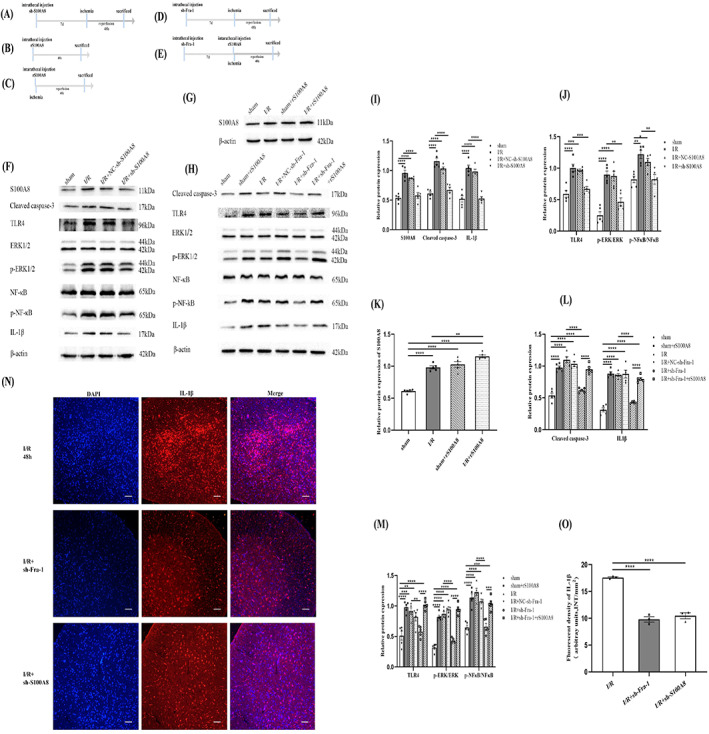
Fra‐1/S100A8‐mediated TLR4 signaling promotes ERK1/2 and NF‐κB pathways activation and IL‐1β production after spinal I/R. (A) Schematic of the experimental design shows the timeline for grouping rats. The rats were intrathecally injected sh‐S100A8 7 days before I/R induction. After 48 h of I/R, tissue samples were collected. (B) The rats were intrathecally injected rS100A8, and tissue samples were collected 48 h later. (C) The rats were intrathecally injected rS100A8 before I/R induction. After 48 h of I/R, tissue samples were collected. (D) The rats were intrathecally injected sh‐Fra‐1 7 days before I/R induction. After 48 h of I/R, tissue samples were collected. (E) The rats were intrathecally injected sh‐Fra‐1 7 days before administration of rS100A8 and intrathecally injected rS100A8 before I/R induction. After 48 h of I/R, tissue samples were collected. (F‐M) Western blot‐assisted analysis and quantification of the protein levels of S100A8, cleaved caspase‐3, TLR4, p‐ERK/ERK, p‐NF‐κB/NF‐κB and IL‐1β (n = 5 rats/group). (N) Immunofluorescence (IF) assays of IL‐1β expression in different groups were performed (n = 3 rats/group). Scale bar =100 μm. (O) Quantification of the IL‐1β fluorescence intensity (INT/mm2). All data represent the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
In order to detect the regulation of S100A8 protein by recombinant S100A8 (rS100A8), the rats were divided into different groups. The rS100A8 was intrathecally injected to rats, and tissue samples were collected 48 h later (Figure 6B). The rS100A8 was intrathecally injected to the rats before I/R induction. After 48 h of I/R, tissue samples were collected (Figure 6C). The results of western blot revealed that, compared with that in the sham group, S100A8 was significantly upregulated in the rats treated with rS100A8 (Figure 6G,K). Compared with that in I/R group, S100A8 was significantly intensified in the rS100A8‐treated rats after I/R induction (Figure 6G,K). To further investigate whether Fra‐1 promotes apoptosis and inflammation by upregulating the target gene S100A8, we examined the rats injected with sh‐Fra‐1 before ischemia and sampled at 48 h after reperfusion (Figure 6D). And the rats in the other group were intrathecally injected sh‐Fra‐1 7 days before administration of rS100A8 and intrathecally injected rS100A8 before I/R induction and tissue samples were collected after 48 h of reperfusion (Figure 6E). As expected, compared with those in the sham group rats, the protein expression levels of cleaved caspase‐3, TLR4, p‐ERK/ERK, p‐NF‐κB/NF‐κB, and IL‐1β were upregulated in the rS100A8‐treated rats (Figure 6H,L,M). In the I/R + sh‐Fra‐1 group, these protein levels deceased compared with those in the I/R group rats (Figure 6H,L,M). Besides, the results of I/R + sh‐Fra‐1 group and I/R + sh‐Fra‐1 + rS100A8 group showed that elevated level of S100A8 reversed the reduction in cleaved caspase‐3, TLR4, p‐ERK/ERK, p‐NF‐κB/NF‐κB, and IL‐1β protein levels caused by intrathecal injection with sh‐Fra‐1 (Figure 6H,L,M). The immunofluorescence results showed that IL‐1β expression was significantly decreased in the I/R + sh‐Fra‐1 and I/R + sh‐S100A8 groups compared with the I/R group (Figure 6N,O). Therefore, Fra‐1 may involve in promoting apoptosis and inflammation responses after spinal I/R by regulating transcription of S100A8.
3.7. TAK‐242 attenuates S100A8‐induced apoptosis and activation of ERK1/2 and NF‐κB pathways and IL‐1β production after spinal I/R
To identify the association between TLR4 expression and apoptosis or inflammation, the spinal samples taken 48 h after intrathecal injection of rS00A8 were examined (Figure 7A). And the rats in the other group were intraperitoneally injected TAK‐242 30 min before administration of rS100A8 and intrathecally injected rS100A8 before I/R induction, and tissue samples were collected after 48 h of reperfusion (Figure 7B). As shown in Figure 7C,D, compared with the sham group, the number of TUNEL‐positive cells in the ventral horn of the spinal cord increased significantly in the rS100A8 and I/R groups. The results of the I/R + rS100A8 + TAK‐242 group versus I/R groups showed that the number of TUNEL‐positive cells was significantly decreased (Figure 7C,D). Besides, our results revealed that the protein levels of p‐ERK/ERK, p‐NF‐κB/NF‐κB and IL‐1β were significantly decreased in the I/R + rS100A8+ TAK‐242 group compared with that in the I/R group rats (Figure 7E,F). These results indicated that S100A8 promoted apoptosis and inflammation by activating the ERK1/2 and NF‐κB pathways downstream of TLR4, and TAK‐242 inhibited apoptosis induced by S100A8. IL‐1β increased the expression of activated caspase‐3 and apoptosis within infarcted forebrain area [43]. Taken together, the results from these in vivo and in vitro analyses led us to hypothesize that microglia was activated by S100A8 secreted from neurons, promoted the production of IL‐1β in turn aggravated neuronal apoptosis under spinal I/R. Besides, Fra‐1‐induced apoptosis and inflammation after SCII were mediated by S100A8 and that the mechanism of action depended on TLR4 pathway activation.
FIGURE 7.
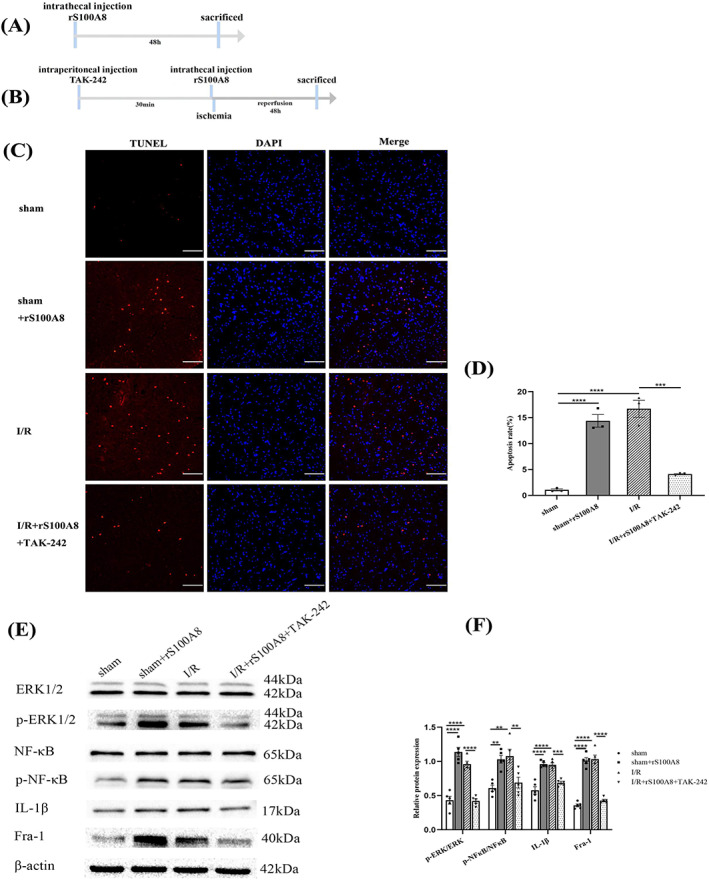
TAK‐242 attenuates S100A8‐induced apoptosis and activation of ERK1/2 and NF‐κB pathways and IL‐1β production, and activated TLR4/NF‐κB and ERK1/2 pathways promote Fra‐1 expression after I/R. (A) Schematic of the experimental design shows the timeline for grouping rats. The rats were intrathecally injected rS100A8, and tissue samples were collected 48 h later. (B) The rats were intraperitoneally injected TAK‐242 30 min before administration of rS100A8 and intrathecally injected rS100A8 before I/R induction. After 48 h of I/R, tissue samples were collected. (C) TUNEL staining of the ventral horn of the spinal cord showing apoptotic cells (n = 3 rats/group). Scale bar = 100 μm. (D) Quantification of TUNEL‐positive cells after I/R. (E,F) Western blot‐assisted analysis and quantification of the protein levels of p‐ERK/ERK, p‐NF‐κB/NF‐κB, IL‐1β, and Fra‐1 (n = 5 rats/group). All data represent the mean ± SEM. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3.8. Activated TLR4/NF‐κB and ERK1/2 pathways promote Fra‐1 expression after I/R
It has been reported that under lipopolysaccharide (LPS) stimulation in alveolar macrophages, both the NF‐κB and ERK1/2 signaling pathways promote transcriptional activation of Fra‐1 [28]. We conducted further experiments with rats treated with rS100A8 and TAK‐242 to elucidate whether the TLR4/ERK and NF‐κB pathways promote the expression of Fra‐1 after SCII. Interestingly, we found that Fra‐1 was increased in the rS100A8‐treated rats compared with its expression in the sham group rats (Figure 7E,F). TAK‐242 was administered to I/R‐injured rats before they were injected rS100A8 (Figure 7B), and Fra‐1 expression was found to be decreased in the I/R + rS100A8 + TAK‐242 group compared with the I/R group (Figure 7E,F).
These findings from our study raised the intriguing possibility that activated TLR4 mediated ERK and NF‐κB pathways activation, which in turn amplified Fra‐1 and S100A8 expression to accelerate the apoptosis rate and inflammation signaling cascades after SCII (Figure 8).
FIGURE 8.
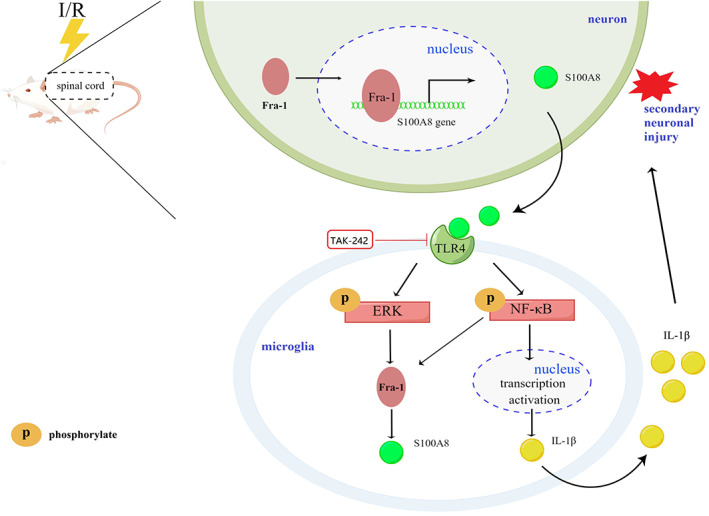
Schematic illustration of a proposed working model wherein Fra‐1‐mediated S100A8/TLR4/NF‐κB and ERK signaling and the generated feedback loop in the regulation of apoptosis and neuroinflammation during spinal I/R. I/R stress promotes Fra‐1 expression mainly in neurons of spinal cord. Fra‐1, as the transcription factor, translocates into the nucleus and binds S100A8 gene to activate S100A8 transcription. Upon recognition of S100A8 that possibly secreted by neurons following spinal I/R insult, microglia initiate the TLR4 signaling that activates NF‐κB and ERK pathways. NF‐κB pathway induces the production of the inflammatory cytokine IL‐1β. The NF‐κB and ERK pathways, in turn, promotes Fra‐1 expression. TAK‐242, as a TLR4 receptor inhibitor, inhibits the responses activated by S100A8 binding with the TLR4 receptor, which rescues NF‐κB and ERK activity as well as the interactions of microglia with neuron. Therefore, Fra‐1, through S100A8/TLR4 pathway, forms a feedback loop to promote sustained activation of microglia and induces microglia‐mediated neuroinflammation and apoptosis after spinal I/R.
4. DISCCUSSION
Neuronal apoptosis and inflammation are the key causes of motor dysfunction after SCII. Here, we not only firstly demonstrate a crucial role for Fra‐1 in rats after SCII but also confirm, for the first time, that Fra‐1 is highly expressed in neurons and microglia and promotes apoptosis and inflammatory responses by directly regulating S100A8 transcription. In addition, further evidence show that upregulated S100A8 induces activation of the TLR4 pathways, phosphorylation of ERK1/2 and NF‐κB and production of IL‐1β. Moreover, the activated ERK1/2 and NF‐κB pathways in turn enhanced Fra‐1 expression. Together, these data are the first to reveal a mechanism by which feedback regulation is established through the Fra‐1 signaling pathway after SCII.
The Jun(c‐Jun, JunB and JunD) family and Fos (c‐Fos, FraB, Fra‐1, and Fra‐2) family proteins constitute the AP‐1 transcription complex [4]. The transcription factors in the AP‐1 family participate in the regulation of various signaling pathway, especially the pathways involved in stress and apoptosis responses [5, 6]. The c‐jun from Jun family is highly expressed after cerebral ischemia injury and plays a role in initiating apoptosis, but JunD from Jun family alleviates cerebral ischemia/reperfusion injury by inhibiting IL‐1β production [8, 44]. In various types of cells and stimuli, Fos family members perform different functions, although they have similar structures [45]. Previous studies have shown that c‐Fos, a member of the Fos family, promotes apoptosis by regulating miR‐27a expression after myocardial I/R injury [7]. Fra‐1 is a member of the Fos family, with members that dimerize with Jun family members [4], but the function of Fra‐1 after I/R has been unclear. Results from different studies have shown that Fra‐1 has antiapoptotic or proapoptotic functions under different conditions [10, 11, 12, 13, 14]. For example, Fra‐1 potentially promotes neuronal apoptosis in rats after traumatic brain injury [46]. Bcl‐2 and Bax are important signaling molecules that regulate apoptosis in mitochondrial apoptosis pathway [47]. Bcl‐2 has anti‐apoptotic effect, while Bax has pro‐apoptotic effect. The imbalance between Bcl‐2 and Bax may lead to apoptosis [48]. In our study, we similarly found that after SCII, Fra‐1 was primarily upregulated in neurons and promoted neuronal apoptosis by changing the Bax/Bcl‐2 (pro‐/anti‐apoptotic) expression. So we demonstrate that Fra‐1 promotes neuronal apoptosis through the mitochondrial pathway after spinal cord ischemia/reperfusion injury.
In a previous study, after bleomycin‐induced acute lung injury, Fra‐1 deletion in mice was shown to decrease the expression levels and activity of certain genes, such as S100A8, indicating a potential role for Fra‐1 in cytokine and chemokine signaling [49]. This finding agrees with our results of a ChIP‐qPCR assay showing that S100A8 is a direct target of Fra‐1. Furthermore, knocking down Fra‐1 leads to decreased levels of S100A8 expression in spinal cord tissues obtained from I/R‐injured rats. S100A8, a Ca2+ regulatory protein, can be actively secreted or passively released by specific cells. For example, S100A8 is specifically released by activated phagocytes during the interaction between phagocytes and activated endothelial cells in the context of inflammation [50]. Besides, it has been found that S100A8 is expressed in microglia and neurons in the central nervous system. And S100A8 can be secreted by neurons under hypoxia condition, which promotes activation of the TLR4/ NF‐κB pathway in microglia and generates IL‐1β, leading to neuroinflammation and apoptosis [17]. Indeed, increased S100A8 has been previously shown to be an important factor in the pathogenesis produced by peripheral blood neutrophils after myocardial infarction or stroke [15, 16], and S100A8 acts as an endogenous ligand of TLR4 [18, 19]. Specifically, after traumatic brain injury, S100A8 mediates TLR4 pathway activation to induce inflammation [20]. However, S100A8 is rarely studied in spinal cord ischemia/reperfusion injury. In our study, we find that Fra‐1 promotes transcriptional upregulation of S100A8 in neurons after OGD/R. Activated NF‐κB and ERK1/2 are signaling pathways downstream of TLR4 and participate in apoptosis and inflammatory responses after ischemic injury [23, 24, 51]. The production of proinflammatory cytokines, primarily IL‐1β, is caused by activation of NF‐κB and ERK signaling [52]. Herein, we describe multiple lines of evidence showing that S100A8 is highly expressed in the spinal cords of rats after I/R and that knocking down S100A8 expression reduces cleaved caspase‐3, p‐ERK/ERK, p‐NF‐κB/NF‐κB, and IL‐1β expression. Besides, we show that upregulation of S100A8 by rS100A8 increases p‐ERK/ERK, p‐NF‐κB/NF‐κB, and IL‐1β expression, subsequently promoting apoptosis and inflammation. TAK‐242 can disrupt the interaction between TLR4 and the ligand, thereby inhibiting TLR4 signal transduction and its downstream signal activation [53, 54]. A previous study reported that TAK‐242 treatment alleviated S100A8‐induced neurological deficits and brain edema [20] also NF‐κB phosphorylation and proinflammatory cytokine production [29]. In the study about hypoxic–ischemic brain injury, NF‐κB signaling can regulate microglia‐mediated neuroinflammation [55]. Consistent with previous results, our study shows that as TAK‐242 inhibits S100A8 binding to TLR4, the activation of ERK1/2 and NF‐κB pathways and the production of IL‐1β are prevented. Study has shown that IL‐1β activates caspase‐3‐mediated neuronal apoptosis under hypoxia‐ischemia [43]. Herein, IL‐1β produced by the S100A8/TLR4 pathway in turn leads to secondary neuronal injury, that is, exacerbates neuronal apoptosis and promotes inflammatory responses.
Feedback regulation of transcription factors plays an essential role in the regulation of perturbations or diseases [56, 57]. Previous studies have indicated that the ERK and NF‐κB pathways can regulate Fra‐1 expression [26, 27, 28]. Besides, based on the conclusion that Fra‐1 was also elevated in microglia after SCII, we further studied the role of Fra‐1 in the inflammatory response associated with activated microglia following I/R injury. In our study, we confirm that Fra‐1 shows targeted binding with S100A8. Surprisingly, in turn, treatment with rS100A8 was shown to increase Fra‐1 expression. Inhibition of S100A8 induced TLR4 pathway activation by TAK‐242 decreased ERK and NF‐κB phosphorylation and Fra‐1 expression.
However, there are some limitations in this study. Our study only analyzed cells and rat samples, not clinical data. In future studies, we will add the collection and analysis of clinical samples to make the conclusions of this study more evidence‐based. Our results suggest that Fra‐1 may be involved in regulation of the signaling pathway at the transcriptional level and thus may activate apoptosis and promote inflammation after SCII. But our study only provided data on the changes in protein expression of the feedback loop formed by Fra‐1/S100A8 in the condition of SCII, and specific data of the mechanism on the interaction between neurons and microglia were not very sufficient. We need to further investigate the ischemia mechanism of Fra‐1 in more detail.
5. CONCLUSIONS
In conclusion, our data demonstrate that elevation of Fra‐1 expression promotes apoptotic and inflammatory responses after SCII by activating the TLR4/NF‐κB and ERK1/2 pathways through enhanced S100A8 transcriptional activity. In addition, S100A8 controls Fra‐1 expression via the TLR4 pathway. TAK‐242, however, inhibits TLR4/NF‐κB and ERK1/2 pathways activation triggered by S100A8 to attenuate apoptosis and neuroinflammation after SCII. Our findings provide novel insights into SCII, showing a feedback loop through which apoptosis and inflammation is regulated by Fra‐1. On the basis of these findings, the specific function and mechanism of action of Fra‐1 need to be further studied because additional findings may lead to new ideas for spinal I/R therapy in the clinic.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China (grant number 81771342).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The animal experiments were approved by the Animal Care and Utilization Committee of China Medical University, Shenyang (China).
Chen Y, Dong Y, Zhang Z‐L, Han J, Chen F‐S, Tong X‐Y, et al. Fra‐1 induces apoptosis and neuroinflammation by targeting S100A8 to modulate TLR4 pathways in spinal cord ischemia/reperfusion injury. Brain Pathology. 2023;33(1):e13113. 10.1111/bpa.13113
Funding information National Natural Science Foundation of China, Grant/Award Number: 81771342
DATA AVAILABILITY STATEMENT
The data analyzed during the current study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Wang D, Chen F, Fang B, Zhang Z, Dong Y, Tong X, et al. MiR‐128‐3p alleviates spinal cord ischemia/reperfusion injury associated neuroinflammation and cellular apoptosis via SP1 suppression in rat. Front Neurosci. 2020;14:609613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mi J, Yang Y, Yao H, Huan Z, Xu C, Ren Z, et al. Inhibition of heat shock protein family A member 8 attenuates spinal cord ischemia‐reperfusion injury via astrocyte NF‐kappaB/NLRP3 inflammasome pathway: HSPA8 inhibition protects spinal ischemia‐reperfusion injury. J Neuroinflammation. 2021;18:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miranda V, Sousa J, Mansilha A. Spinal cord injury in endovascular thoracoabdominal aortic aneurysm repair: prevalence, risk factors and preventive strategies. Int Angiol. 2018;37:112–26. [DOI] [PubMed] [Google Scholar]
- 4. Huang CK, Dai D, Xie H, Zhu Z, Hu J, Su M, et al. Lgr4 governs a pro‐inflammatory program in macrophages to antagonize post‐infarction cardiac repair. Circ Res. 2020;127:953–73. [DOI] [PubMed] [Google Scholar]
- 5. Na J, Bak DH, Im SI, Choi H, Hwang JH, Kong SY, et al. Antiapoptotic effects of glycosaminoglycans via inhibition of ERK/AP1 signaling in TNFalphastimulated human dermal fibroblasts. Int J Mol Med. 2018;41:3090–8. [DOI] [PubMed] [Google Scholar]
- 6. Takeuchi K, Motoda Y, Ito F. Role of transcription factor activator protein 1 (AP1) in epidermal growth factor‐mediated protection against apoptosis induced by a DNA‐damaging agent. FEBS J. 2006;273:3743–55. [DOI] [PubMed] [Google Scholar]
- 7. Bao Y, Qiao Y, Yu H, Zhang Z, Yang H, Xin X, et al. miRNA‐27a transcription activated by c‐Fos regulates myocardial ischemia‐reperfusion injury by targeting ATAD3a. Oxid Med Cell Longev. 2021;2021:2514947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao P, Liu XW, Zhao NN, Fang R, Wen Q, Zeng KX, et al. Correlations of neuronal apoptosis with expressions of c‐Fos and c‐Jun in rats with post‐ischemic reconditioning damage. Eur Rev Med Pharmacol Sci. 2018;22:2832–8. [DOI] [PubMed] [Google Scholar]
- 9. Talotta F, Casalino L, Verde P. The nuclear oncoprotein Fra‐1: a transcription factor knocking on therapeutic applications' door. Oncogene. 2020;39:4491–506. [DOI] [PubMed] [Google Scholar]
- 10. Li R, Che W, Liang N, Deng S, Song Z, Yang L. Silent FOSL1 enhances the radiosensitivity of glioma stem cells by Down‐regulating miR‐27a‐5p. Neurochem Res. 2021;46:3222–46. [DOI] [PubMed] [Google Scholar]
- 11. Yang Y, You B, Dong S, Zhou C. FRA‐1 suppresses apoptosis of helicobacter pylori infected MGC‐803 cells. Mol Biol Rep. 2021;48:611–21. [DOI] [PubMed] [Google Scholar]
- 12. Zhong G, Chen X, Fang X, Wang D, Xie M, Chen Q. Fra‐1 is upregulated in lung cancer tissues and inhibits the apoptosis of lung cancer cells by the P53 signaling pathway. Oncol Rep. 2016;35:447–53. [DOI] [PubMed] [Google Scholar]
- 13. Zhang M, Liang L, He J, He Z, Yue C, Jin X, et al. Fra‐1 inhibits cell growth and the Warburg effect in cervical cancer cells via STAT1 regulation of the p53 signaling pathway. Front Cell Dev Biol. 2020;8:579629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu X, Yang X, Zhu R, Dai M, Zhu M, Shen Y, et al. Involvement of Fra‐1 in retinal ganglion cell apoptosis in rat light‐induced retina damage model. Cell Mol Neurobiol. 2017;37:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sreejit G, Abdel‐Latif A, Athmanathan B, Annabathula R, Dhyani A, Noothi SK, et al. Neutrophil‐derived S100A8/A9 amplify granulopoiesis after myocardial infarction. Circulation. 2020;141:1080–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo D, Zhu Z, Xu T, Zhong C, Wang A, Xie X, et al. Plasma S100A8/A9 concentrations and clinical outcomes of ischemic stroke in 2 independent multicenter cohorts. Clin Chem. 2020;66:706–17. [DOI] [PubMed] [Google Scholar]
- 17. Ha JS, Choi HR, Kim IS, Kim EA, Cho SW, Yang SJ. Hypoxia‐induced S100A8 expression activates microglial inflammation and promotes neuronal apoptosis. Int J Mol Sci. 2021;22:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagareddy PR, Kraakman M, Masters SL, Stirzaker RA, Gorman DJ, Grant RW, et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 2014;19:821–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee JS, Lee NR, Kashif A, Yang SJ, Nam AR, Song IC, et al. S100A8 and S100A9 promote apoptosis of chronic eosinophilic leukemia cells. Front Immunol. 2020;11:1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He GY, Zhao CH, Wu DG, Cheng H, Sun LA, Zhang DL, et al. S100A8 promotes inflammation via toll‐like receptor 4 after experimental traumatic brain injury. Front Neurosci. 2020;14:616559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh MC, Lo YS, Chuang YC, Lin CC, Ho HY, Hsieh MJ, et al. Dehydrocrenatidine extracted from Picrasma quassioides induces the apoptosis of nasopharyngeal carcinoma cells through the JNK and ERK signaling pathways. Oncol Rep. 2021;46:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu L, Ge F, Hu Y, Yu Y, Guo K, Miao C. Sevoflurane postconditioning attenuates hepatic ischemia‐reperfusion injury by limiting HMGB1/TLR4/NF‐κB pathway via modulating microRNA‐142 in vivo and in vitro. Front Pharmacol. 2021;12:646307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen F, Li X, Li Z, Zhou Y, Qiang Z, Ma H. The roles of chemokine (C‐X‐C motif) ligand 13 in spinal cord ischemia‐reperfusion injury in rats. Brain Res. 2020;1727:146489. [DOI] [PubMed] [Google Scholar]
- 24. Sun Z, Zhao T, Lv S, Gao Y, Masters J, Weng H. Dexmedetomidine attenuates spinal cord ischemia‐reperfusion injury through both anti‐inflammation and anti‐apoptosis mechanisms in rabbits. J Transl Med. 2018;16:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lau EY, Lo J, Cheng BY, Ma MK, Lee JM, Ng JK, et al. Cancer‐associated fibroblasts regulate tumor‐initiating cell plasticity in hepatocellular carcinoma through c‐met/FRA1/HEY1 signaling. Cell Rep. 2016;15:1175–89. [DOI] [PubMed] [Google Scholar]
- 26. Basbous J, Chalbos D, Hipskind R, Jariel‐Encontre I, Piechaczyk M. Ubiquitin‐independent proteasomal degradation of Fra‐1 is antagonized by Erk1/2 pathway‐mediated phosphorylation of a unique C‐terminal destabilizer. Mol Cell Biol. 2007;27:3936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang C, Li Z, Shao F, Yang X, Feng X, Shi S, et al. High expression of collagen triple helix repeat containing 1 (CTHRC1) facilitates progression of oesophageal squamous cell carcinoma through MAPK/MEK/ERK/FRA‐1 activation. J Exp Clin Cancer Res. 2017;36:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mishra RK, Potteti HR, Tamatam CR, Elangovan I, Reddy SP. c‐Jun is required for nuclear factor‐κB‐dependent, LPS‐stimulated Fos‐related antigen‐1 transcription in alveolar macrophages. Am J Respir Cell Mol Biol. 2016;55:667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gong H, Su WJ, Cao ZY, Lian YJ, Peng W, Liu YZ, et al. Hippocampal Mrp8/14 signaling plays a critical role in the manifestation of depressive‐like behaviors in mice. J Neuroinflammation. 2018;15:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu YP, Tao YN, Wu YP, Zhang J, Jiao W, Wang YH, et al. Sleep deprivation aggravates brain injury after experimental subarachnoid hemorrhage via TLR4‐MyD88 pathway. Aging (Albany NY). 2021;13:3101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Z, Li X, Chen F, Li Z, Wang D, Ren X, et al. Downregulation of LncRNA Gas5 inhibits apoptosis and inflammation after spinal cord ischemia‐reperfusion in rats. Brain Res Bull. 2021;168:110–9. [DOI] [PubMed] [Google Scholar]
- 32. Jia H, Li Z, Chang Y, Fang B, Zhou Y, Ma H. Downregulation of long noncoding RNA TUG1 attenuates MTDH‐mediated inflammatory damage via targeting miR‐29b‐1‐5p after spinal cord ischemia reperfusion. J Neuropathol Exp Neurol. 2021;80:254–64. [DOI] [PubMed] [Google Scholar]
- 33. Klapdor K, Dulfer BG, Hammann A, van der Staay FJ. A low‐cost method to analyse footprint patterns. J Neurosci Methods. 1997;75:49–54. [DOI] [PubMed] [Google Scholar]
- 34. Singh U, Upadhya M, Basu S, Singh O, Kumar S, Kokare DM, et al. Transient receptor potential Vanilloid 3 (TRPV3) in the cerebellum of rat and its role in motor coordination. Neuroscience. 2020;424:121–32. [DOI] [PubMed] [Google Scholar]
- 35. Wang H, Chen FS, Zhang ZL, Zhou HX, Ma H, Li XQ. MiR‐126‐3p‐enriched extracellular vesicles from hypoxia‐preconditioned VSC 4.1 neurons attenuate Ischaemia‐reperfusion‐induced pain hypersensitivity by regulating the PIK3R2‐mediated pathway. Mol Neurobiol. 2021;58:821–34. [DOI] [PubMed] [Google Scholar]
- 36. Wang H, Yu Q, Zhang ZL, Ma H, Li XQ. Involvement of the miR‐137‐3p/CAPN‐2 interaction in ischemia‐reperfusion‐induced neuronal apoptosis through modulation of p35 cleavage and subsequent Caspase‐8 Overactivation. Oxid Med Cell Longev. 2020;2020:2616871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou HJ, Wang LQ, Zhan RY, Zheng XJ, Zheng JS. lncRNA MEG3 restrained the M1 polarization of microglia in acute spinal cord injury through the HuR/A20/NF‐κB axis. Brain Pathol. 2022;e13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ying Y, Ma X, Fang J, Chen S, Wang W, Li J, et al. EGR2‐mediated regulation of m(6)a reader IGF2BP proteins drive RCC tumorigenesis and metastasis via enhancing S1PR3 mRNA stabilization. Cell Death Dis. 2021;12:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huber E, David G, Thompson AJ, Weiskopf N, Mohammadi S, Freund P. Dorsal and ventral horn atrophy is associated with clinical outcome after spinal cord injury. Neurology. 2018;90:e1510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu YP, Yuan H, Xu Y, Liu RM, Luo Y, Xiao JH. Protective effects of Ligularia fischeri root extracts against ulcerative colitis in mice through activation of Bcl‐2/Bax signalings. Phytomedicine. 2022;99:154006. [DOI] [PubMed] [Google Scholar]
- 41. Zhou Z, Han B, Jin H, Chen A, Zhu L. Changes in long non‐coding RNA transcriptomic profiles after ischemia‐reperfusion injury in rat spinal cord. PeerJ. 2020;8:e8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vallejo A, Erice O, Entrialgo‐Cadierno R, Feliu I, Guruceaga E, Perugorria MJ, et al. FOSL1 promotes cholangiocarcinoma via transcriptional effectors that could be therapeutically targeted. J Hepatol. 2021;75:363–76. [DOI] [PubMed] [Google Scholar]
- 43. Savard A, Brochu ME, Chevin M, Guiraut C, Grbic D, Sébire G. Neuronal self‐injury mediated by IL‐1β and MMP‐9 in a cerebral palsy model of severe neonatal encephalopathy induced by immune activation plus hypoxia‐ischemia. J Neuroinflammation. 2015;12:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diaz‐Cañestro C, Reiner MF, Bonetti NR, Liberale L, Merlini M, Wüst P, et al. AP‐1 (activated Protein‐1) transcription factor JunD regulates ischemia/reperfusion brain damage via IL‐1β (interleukin‐1β). Stroke. 2019;50:469–77. [DOI] [PubMed] [Google Scholar]
- 45. Hannemann N, Cao S, Eriksson D, Schnelzer A, Jordan J, Eberhardt M, et al. Transcription factor Fra‐1 targets arginase‐1 to enhance macrophage‐mediated inflammation in arthritis. J Clin Invest. 2019;129:2669–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu X, Jiang R, Gong P, Liu Q, Chen Y, Hou S, et al. Up‐regulation of FOS‐like antigen 1 contributes to neuronal apoptosis in the cortex of rat following traumatic brain injury. Metab Brain Dis. 2018;33:115–25. [DOI] [PubMed] [Google Scholar]
- 47. Yao J, Cao X, Zhang R, Li YX, Xu ZL, Zhang DG, et al. Protective effect of Baicalin against experimental colitis via suppression of oxidant stress and apoptosis. Pharmacogn Mag. 2016;12:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Edlich F. BCL‐2 proteins and apoptosis: recent insights and unknowns. Biochem Biophys Res Commun. 2018;500:26–34. [DOI] [PubMed] [Google Scholar]
- 49. Rajasekaran S, Reddy NM, Zhang W, Reddy SP. Expression profiling of genes regulated by Fra‐1/AP‐1 transcription factor during bleomycin‐induced pulmonary fibrosis. BMC Genomics. 2013;14:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pruenster M, Kurz AR, Chung KJ, Cao‐Ehlker X, Bieber S, Nussbaum CF, et al. Extracellular MRP8/14 is a regulator of β2 integrin‐dependent neutrophil slow rolling and adhesion. Nat Commun. 2015;6:6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao H, Chen Z, Xie LJ, Liu GF. Suppression of TLR4/NF‐κB signaling pathway improves cerebral ischemia‐reperfusion injury in rats. Mol Neurobiol. 2018;55:4311–9. [DOI] [PubMed] [Google Scholar]
- 52. Holló K, Ducza L, Hegyi Z, Dócs K, Hegedűs K, Bakk E, et al. Interleukin‐1 receptor type 1 is overexpressed in neurons but not in glial cells within the rat superficial spinal dorsal horn in complete Freund adjuvant‐induced inflammatory pain. J Neuroinflammation. 2017;14:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sha T, Iizawa Y, Ii M. Combination of imipenem and TAK‐242, a toll‐like receptor 4 signal transduction inhibitor, improves survival in a murine model of polymicrobial sepsis. Shock. 2011;35:205–9. [DOI] [PubMed] [Google Scholar]
- 54. Matsunaga N, Tsuchimori N, Matsumoto T, Ii M. TAK‐242 (resatorvid), a small‐molecule inhibitor of Toll‐like receptor (TLR) 4 signaling, binds selectively to TLR4 and interferes with interactions between TLR4 and its adaptor molecules. Mol Pharmacol. 2011;79:34–41. [DOI] [PubMed] [Google Scholar]
- 55. Li B, Dasgupta C, Huang L, Meng X, Zhang L. MiRNA‐210 induces microglial activation and regulates microglia‐mediated neuroinflammation in neonatal hypoxic‐ischemic encephalopathy. Cell Mol Immunol. 2020;17:976–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Inoue K. Feedback enrichment analysis for transcription factor‐target genes in signaling pathways. Biosystems. 2020;198:104262. [DOI] [PubMed] [Google Scholar]
- 57. Marbach D, Lamparter D, Quon G, Kellis M, Kutalik Z, Bergmann S. Tissue‐specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016;13:366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed during the current study are available from the corresponding author upon reasonable request.


