Abstract
Glioblastoma is the most common primary malignant brain tumor in adults with an overall survival of only 14.6 months. Hypoxia is known to play a role in tumor aggressiveness but the influence of hypoxia on the immune microenvironment is not fully understood. The aim of this study was to investigate the expression of immune‐related proteins in normoxic and hypoxic tumor areas by digital spatial profiling. Tissue samples from 10 glioblastomas were stained with a panel of 40 antibodies conjugated to photo‐cleavable oligonucleotides. The free oligo‐tags from normoxic and hypoxic areas were hybridized to barcodes for digital counting. Differential expression patterns were validated by Ivy Glioblastoma Atlas Project (GAP) data and an independent patient cohort. We found that CD44, Beta‐catenin and B7‐H3 were upregulated in hypoxia, whereas VISTA, CD56, KI‐67, CD68 and CD11c were downregulated. PD‐L1 and PD‐1 were not affected by hypoxia. Focusing on the checkpoint‐related markers CD44, B7‐H3 and VISTA, our findings for CD44 and VISTA could be confirmed with Ivy GAP RNA sequencing data. Immunohistochemical staining and digital quantification of CD44, B7‐H3 and VISTA in an independent cohort confirmed our findings for all three markers. Additional stainings revealed fewer T cells and high but equal amounts of tumor‐associated microglia and macrophages in both hypoxic and normoxic regions. In conclusion, we found that CD44 and B7‐H3 were upregulated in areas with hypoxia whereas VISTA was downregulated together with the presence of fewer T cells. This heterogeneous expression should be taken into consideration when developing novel therapeutic strategies.
Keywords: digital spatial profiling, glioblastoma, hypoxia, immune checkpoints
1. INTRODUCTION
Glioblastoma (GBM) is the most frequent and malignant primary brain tumor. The current treatment options include surgery followed by radiotherapy and concomitant chemotherapy with temozolomide. Despite this, disease recurrence is inevitable and GBM patients have a poor prognosis with a median survival range from 12 to 15 months [1]. Tumor cells secrete chemo‐attractants that recruit the immunosuppressive cells including tumor‐associated microglia and macrophages (TAMs), myeloid derived suppressor cells and regulatory T cells, all helping to reduce the immune response [2, 3]. TAMs infiltrate the tumor tissue comprising up to 30% of the total cell population [4] and contribute to tumor progression by stimulating angiogenesis, cancer cell proliferation and extracellular matrix remodeling [5, 6]. Moreover, they suppress the immune response by producing immunosuppressive cytokines and expressing programmed death‐ligand 1 (PD‐L1) [7]. Targeting of immune checkpoint molecules has improved survival in other cancers like lung cancer and melanoma [8]. GBM tumor cells, but also TAMs, are known to express PD‐L1 and infiltrating T cells express programmed cell death protein 1 (PD‐1) [9, 10]. These factors add to the potential benefit of immune checkpoint therapy in GBMs [11].
Hypoxia is known to play a role in tumor aggressiveness by stimulating cell proliferation, tumor invasion, and therapy resistance leading to tumor recurrence [2, 12]. The hypoxic niche has also been associated with increased expression of GBM stem cell‐like markers and production of pro‐angiogenic factors [13]. Hypoxia has by itself been suggested to induce immunosuppression by impairing T cell proliferation and effector function in vitro [14]. Of importance, hypoxia has been suggested to increase expression of immune checkpoint molecules such as PD‐L1 but also the amount of extracellular adenosine, a potent immunosuppressive metabolite increasing PD‐1 on tumor‐specific T cells in colon carcinoma in vitro [15] and cytotoxic T‐lymphocyte‐associated protein 4 (CTLA‐4) levels on T cells in vitro [16]. Hypoxia also appears to support the recruitment and polarization towards a more pro‐tumorigenic phenotype of TAMs in GBM and breast cancer models [17, 18], thereby contributing to the immunosuppressive features in the microenvironment.
The influence of hypoxia on the immune system is not fully understood and uncovering this area has potential implications for novel therapeutic strategies. In this study, the aim was to investigate and compare the immune landscape in normoxic and hypoxic tumor areas. We especially wanted to focus on checkpoint‐related molecules that have gained increased attention in the treatment of GBMs. Nanostrings Digital spatial profiling (DSP) technology combines standard immunofluorescence with a digital barcoding technology, where antibodies are coupled to photo‐cleavable oligonucleotide tags. This allows spatial simultaneous measurement of multiple protein markers. Using this technology, we investigated the expression of immune‐related protein markers covering immune checkpoints/immune activating molecules, immune cell markers and immune signaling pathways. We validated our results using Ivy Glioblastoma Atlas Project RNA sequencing data from five laser microdissected GBM compartments that include pseudopalisading cells (PC), central tumor (CT), infiltrating tumor (IT), leading edge (LE) and microvascular proliferation (MP). In addition, we used immunohistochemical staining and quantitative image analysis in an independent cohort consisting of 19 GBMs. We have previously shown that the use of quantitative image analysis is of great advantage by producing continuous measurements and eliminating intra‐observer variability [19, 20].
2. MATERIALS AND METHODS
2.1. Patient material
All samples were obtained from the well characterized Region of Southern Denmark glioma cohort [19, 20]. The cohort was immunohistochemically stained for hypoxia‐inducible factor 1α (Hif‐1α). Tissue from 10 GBM patients containing both positive and negative Hif‐1α regions was included in the study. All patients were diagnosed with a primary glioma between 2005 and 2009 (Table 1). Additionally, 19 IDH‐wild‐type GBMs with the same characteristics as described above were included for validation. All samples were evaluated by two pathologists and reclassified according to the 2016 World Health Organization Classification of Tumors of the Central Nervous System.
TABLE 1.
Patient characteristics
| Variables | No. |
|---|---|
| Patients (n) | 10 |
| Age | |
| Median | 64 |
| Range | 40–76 |
| Gender | |
| Female | 4 |
| Male | 6 |
| WHO 2016 | |
| GBM IDH‐wildtype | 9 |
| GBM IDH‐mutant | 1 |
| Performance status | |
| 0–1 | 5 |
| 2–4 | 5 |
| Post‐surgical treatment | |
| No post‐surgical treatment | 2 |
| Less than Stupp | 2 |
| Stupp | 6 |
| Survival (months) | |
| Mean | 17.4 |
| Range | 2–59.9 |
| MGMT status | |
| Methylated | 4 |
| Unmethylated | 6 |
| IDH1 | |
| WT | 9 |
| Mutated | 1 |
Abbreviations: GBM, glioblastoma; IDH1, isocitrate dehydrogenase 1; MGMT, O‐6‐methylguanine‐DNA methyltransferase.
2.2. Digital spatial profiling
DSP analysis of 40 immune‐related protein markers (Immuno‐oncology panel, Nanostring) was performed on 5 micrometer formalin‐fixed paraffin‐embedded (FFPE) sections from 10 GBM patients. The slides were stained with a cocktail of primary oligo‐conjugated antibodies (Table 2). Regions of interest (ROI) were selected based on a Hif‐1α immunohistochemically stained and scanned sections merged with scanned sections stained with glial fibrillary acidic protein (GFAP) and ionized calcium‐binding adapter molecule 1 (Iba1) to ensure presence of tumor cells and TAMs, respectively. The ROIs comprised circles (diameter from 200 to 600 μm) and rectangles (200 μm × 400 μm and 500 μm × 500 μm). Ultraviolet exposure of ROIs released the oligonucleotides from bound antibodies. The free tags were captured and hybridized to NanoString optical barcodes for ex situ digital counting in the nCounter analysis system (NanoString Technologies, Seattle). The obtained digital counts were normalized using three positive controls correcting for system variation and subsequently according to area using three house‐keeping controls (Histone H2, S6 and pS6). Pan‐cytokeratin was part of the panel but of minor biological importance and therefore not further investigated.
TABLE 2.
Immune oncology panel from Nanostring
| Signaling pathways and survival molecules | Immune cell checkpoint and immune activating molecules | Pan‐immune cell markers |
|---|---|---|
| AKT | CD44 | CD45 |
| p‐AKT | GZMB | CD45RO |
| PTEN | B‐2‐microglobulin | CD3 |
| B‐catenin | B7‐H3 | CD4 |
| STAT3 | PD‐1 | CD8A |
| p‐STAT3 | PD‐L1 | CD14 |
| BCL2 | VISTA | CD68 |
| HER2 | OX40L | CD11C |
| Ki‐67 | IDO‐1 | CD56 |
| ICOS | CD163 | |
| B7‐H4 | CD66B | |
| HLA‐DR | FOXP3 | |
| STING |
To obtain a total score of the level of visualization markers, GFAP, Iba1 and Hif‐1α was scored according to the average percentage of positive cells from 0 to 4: Score 0: 0% positive cells, 1: 1%–25%, 2: 25%–50%, 3: 50%–75%, 4: 75%–100%. A mean score was calculated for hypoxic and normoxic ROIs in each tumor. The scoring system and scoring procedure was made under supervision of a neuropathologist.
2.3. Immunohistochemical staining
Fresh tissue was fixed in 4% neutral buffered formaldehyde and paraffin embedded and 3 μm sections were cut on a microtome. Immunohistochemical staining was carried out on an automated immunostainer (BenckMark. Discovery). The sections were dewaxed in xylene and rehydrated with ethanol. The standardized protocol included epitope retrieval for 32–48 min at 100°C. Endogen peroxidase activity was blocked by OptiView peroxidase inhibitor. Afterwards, the sections were incubated with primary antibody (Table 3). Amplification step includes standard OptiView amplification from Ventana (Ref: 760‐099) with H2O2 + OptiView amplifier for 4 min and OptiView multimer for 4 min. The antigen–antibody complex was visualized using Optiview‐DAB according to manufactures recommendations. The slides were scanned on a Hamamatsu digital slide scanner. Specific staining reaction was ensured by using a tissue microarray with more than 40 different normal and cancer tissues. Omission of primary antibody abolished all staining reaction.
TABLE 3.
Antibodies included in the study
| Antibody | Clone/product code | Concentration | Species/type | Epitope retrieval | Detection/instrument |
|---|---|---|---|---|---|
| Hif‐1α | 54 BD biosciences | 1:100 32 min at 36°C + amplification | Mice monoclonal | HIER: CC1 for 48 min at 100°C | Optiview Bench Mark Discovery |
| CD44 | DF1485 Novus biologicals | 1:200 32 min at 36°C | Mice monoclonal | HIER: CC1 for 32 min at 100°C | Optiview Benck Mark Discovery |
| VISTA | D1L2G Cell Signaling Technology | 1:100 32 min at 36°C | Rabbit monoclonal | HIER: CC1 for 48 min at 100°C | Optiview Benck Mark Discovery |
| B7‐H3 | AF1027 R&D systems | 1:800 32 min at 36°C | Goat polycloncal | CC1 for 32 min at 100°C HIER: | Optiview Benck Mark Discovery |
| Iba1 | 019–19,741 Wako Pure Chemical | 1:2000 16 min at 36°C | Rabbit polyclonal | HIER: CC1 for 32 min at 100°C | Optiview Bench Mark Discovery |
| CD4 | SP35 Ventana Medical Systems | Ready‐to‐use 24 min at 36°C | Rabbit monoclonal | HIER: CC1 for 32 min at 100°C | Optiview Bench Mark Discovery |
| CD8 | C8/144B | 1:100 32 min at 36°C | Mice monoclonal | HIER: CC1 for 32 min at 100°C | Optiview Bench Mark Discovery |
| FOXP3 | 236A/E7 | 1:40 16 min at 36°C + amplification | Mice monoclonal | HIER: CC1 for 64 min at 100°C | Optiview Bench Mark Discovery |
Note: Cell conditioning solution (CC1 Tris‐EDTA buffer, pH 7.8).
Abbreviation: HIER, heat‐induced epitope retrival.
2.4. Digital image analysis
Automated digital image analysis and quantitation were performed using Visiopharm Image Analysis Software, version 2018.4. ROIs were manually outlined containing at least three hypoxic regions with high Hif‐1α expression and three normoxic regions with no Hif‐1α expression (circles with a diameter between 600‐1000 μm). Sample images of ROIs were collected using a 20× objective. The expression was quantified by using a pixel‐based algorithm to detect positive staining. We used a threshold‐based classifier (VISTA, B7‐H3, and Iba1) or a membrane classification (CD44) using a DAB (HDAB‐DAB) feature. The threshold‐based classifier measured the area of positive immune staining. The threshold had been set so only clearly positive staining was measured. For each of the four marker, the area fraction (AF) (defined as the positive area/total area of the ROI) was quantified. For the T cell markers (CD4, CD8, and FOXP3), a cell‐based classification was used identifying all nuclei using the RGB‐R feature. Positive T cells were then manually differentiated from the negative cells based on their membrane expression of CD4, CD8, or FOXP3 and morphology (for CD4). For each of the T cell markers, the AF of positive cells (defined as the positive nuclei area/total nuclei area) was quantified.
2.5. Patient dataset analysis
From the Ivy Glioblastoma Atlas Project (IvyGAP), RNA data from microlaser dissected areas in GBM tissue (n = 122) was used to examine the mRNA levels of CD44, B7‐H3 and VISTA. The dataset was exported directly from the GlioVIS database and expressed in log2 transcripts (fragments per kilobase per million [FPKM]).
2.6. Statistics
The Student's paired t‐test was applied to calculate the significance of the difference in protein expression between hypoxic and normoxic regions. All p‐values <0.05 was considered statistically significant. The protein expression of the significant differentially expressed proteins, was subsequently standardized to have a mean count per area of 0 and a standard deviation of 1, and used to generate heatmaps using the heatmap2 function from the gplots R‐package to visualize the associated protein levels. Cluster analysis was performed by principal component analysis, K‐means clustering and hierarchical clustering. Principal component analysis was performed and visualized using the prcomp and biplot functions from the stats R‐package. K‐means cluster calculation was performed using the kmeans and clusplot from the stats and cluster R‐packages, respectively. The hierarchical clustering was performed using the pvclust function embedded in the pvclust R‐package with correlation as distance measure and the average agglomerative method as clustering. One thousand bootstrap replications were used to assess the bootstrap support for each bifurcation node in the resulting tree. Multiple comparisons were analyzed with one‐way ANOVA and Bonferroni post‐test using Graph Pad Prism 5.0 (GraphPad software, San Diego, CA). Statistical significance was defined as *p < 0.5, **p < 0.01, ***p < 0.001.
3. RESULTS
3.1. Digital spatial profiling
We investigated the expression of 40 immune‐related protein markers in hypoxic and normoxic tumor areas using DSP technology (Table 2). ROIs were selected based on the presence of Hif‐1α expression (hypoxia) (Figure 1A,C) or absence (normoxia) (Figure 1A,B). The Hif‐1α stained slide was merged with a slide stained with double immunofluorescence ensuring presence of TAMs (Iba1) (Figures 1E–G and S1) and tumor cells (GFAP) (Figures 1I–K and S1) in the outlined ROIs. The positive staining in each ROI was scored. Hif‐1α (Figure 1D) was only expressed in hypoxic regions (mean score = 3.0) and not in normoxic regions (mean score = 0). Iba1 (Figure 1H) and GFAP (Figure 1L) expression was high in both normoxia (GFAP: mean score = 3.5, Iba1: mean score = 2) and hypoxia (GFAP: Mean score = 3.5, Iba1: mean score = 1.9).
FIGURE 1.
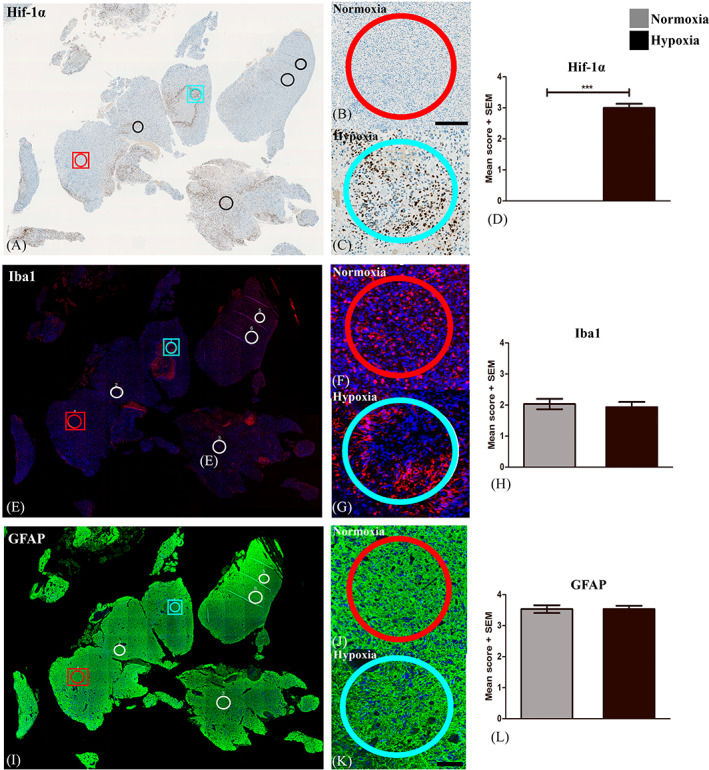
Selection of Regions of interest (ROIs). (A) ROIs were outlined in areas with no or high Hif‐1α expression and defined as (B) normoxic and (C) hypoxic ROIs, respectively. Double immunofluorescence staining with Iba1 and GFAP ensured the presence of (E‐G) tumor‐associated microglia and macrophages and (I‐K) tumor cells, respectively. The expression of each marker was scored and (D) Hif‐1α was significantly higher expressed in hypoxic ROIs as compared to normoxic, whereas (H) Iba1 and (L) GFAP was expressed equally in normoxia and hypoxia. Statistical significance was defined as *p < 0.5, **p < 0.01, ***p < 0.001. Abbreviations: Hypoxia‐inducible factor 1α (Hif‐1α), glial fibrillary acidic protein (GFAP), ionized calciumbinding adapter molecule 1 (Iba1).
In a hierarchal cluster analysis (Figure 2A), measurements from ROIs within the same tumor tended to cluster rather than regions with hypoxia or normoxia across tumors. We included one patient with a known IDH1 mutation (Tumor 3) and found that the protein profiling pattern suggested a low expression of most immune‐related molecules. In general, we found relatively high counts for PD‐L1, AKT, CD56, STAT3, CD44, Beta‐catenin and Beta‐2 microglobulin in all ROIs, whereas CD11c, CD4, CD68, PTEN, KI‐67, VISTA had a medium range expression (Figures 2 and S2). When comparing the protein expression between hypoxia and normoxia CD44 (p = 0.006, 33%), Beta‐catenin (p = 0.04, 18%) and B7‐H3 (p = 0.02, 27%) were all upregulated in hypoxia (Figure 2B–G), whereas VISTA (p = 0.02, −41%), CD56 (p = 0.01, −24%), KI‐67 (p < 0.0001, −91%), CD68 (p = 0.01, −35%) and CD11c (p = 0.001, −39%) were all downregulated in hypoxia (Figure 2H–Q). The observed association between the deregulated markers and the frequencies of T cells was not found for all tumors (Figure S5). Interestingly, immune‐related checkpoint molecules PD‐L1 and PD‐1 were not affected by hypoxia (Figure S2).
FIGURE 2.
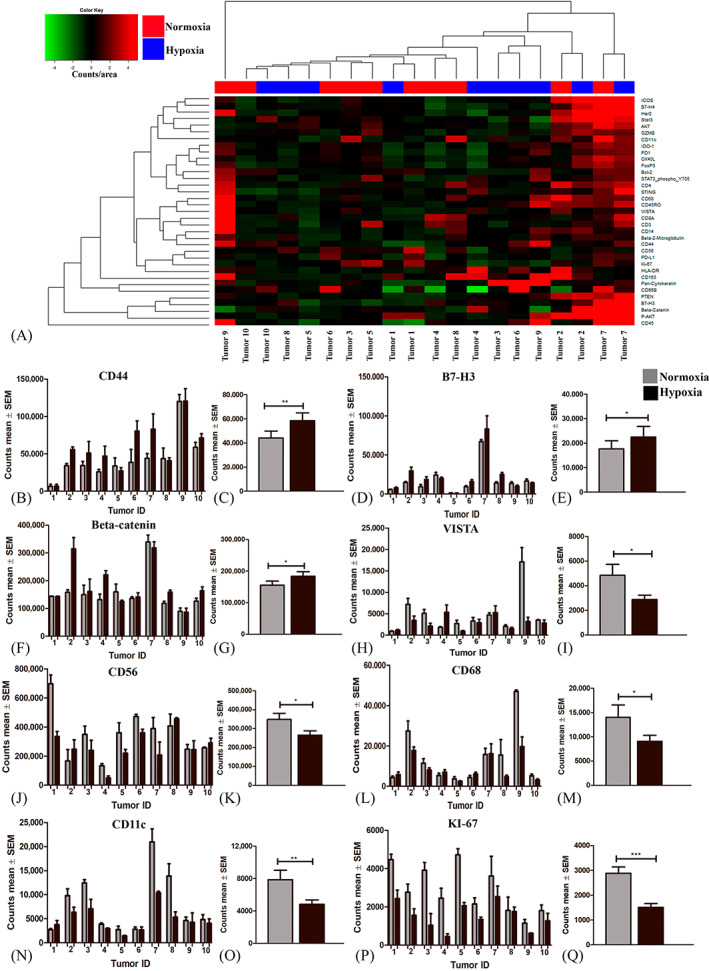
Differences in protein expression between hypoxic and normoxic tumor regions acquired from Digital Spatial Profiling (DSP). (A) Unsupervised cluster analysis of protein profiling results. (B, D, F, H, J, L, N, P) The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and (C, E, G, I, K, M, O, P) the mean count across all tumors are illustrated in the figure. (B‐C) CD44, (D‐E) B7‐H3 and (F‐G) Beta‐catenin were significantly upregulated in areas with hypoxia, whereas (H‐I) VISTA, (J‐K) CD56, (L‐M) CD68, (N‐O) CD11c, (P‐Q) KI‐67 were downregulated. Statistical significance was defined as *p < 0.5, **p < 0.01 = **, ***p < 0.001. Abbreviations: Regions of interest (ROI).
3.2. IvyGAP validation
In the validation we focused on the checkpoint‐related markers CD44, B7‐H3 and VISTA. Using the IvyGap database RNA sequencing data from laser microdissected tissue, we found that CD44 mRNA expression in areas with PC was significantly higher compared to CT (p < 0.001, 20%), IT (p < 0.001, 19%), LE (p < 0.001, 64%) and MP (p < 0.001, 23%) (Figure 3A,B). B7‐H3 mRNA expression was significantly higher in areas with MP (p < 0.001, 22%) compared to PC (Figure 3A,C). VISTA mRNA expression was significantly lower in areas with PC compared to CT (p < 0.001, 255%), IT (p < 0.001, 480%), LE (p < 0.001, 565%) and MP (p < 0.001, 399%) (Figure 3A,D). Thus, mRNA expression in PC correlated with protein hypoxia data for CD44 and VISTA identified by DSP.
FIGURE 3.
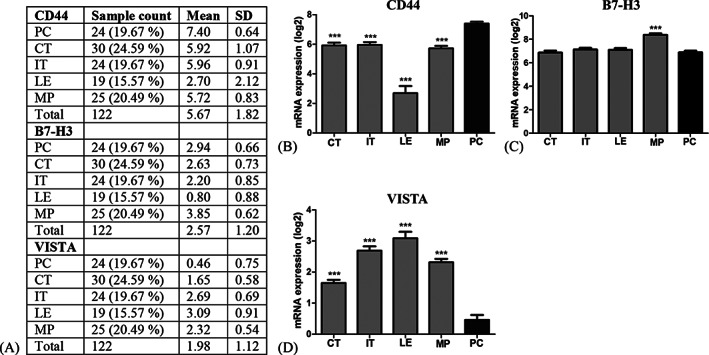
IvyGAP database mRNA sequencing data for CD44 (A and B), B7‐H3 (A and C) and VISTA (A and D). (A) Sample characteristics from 122 RNA samples generated from laser‐microdissected areas from 10 GBMs. CD44 was upregulated and VISTA downregulated in PC compared to the other types of areas. B7‐H3 was upregulated in MP areas as compared to PC areas. Statistical significance was defined as *p < 0.5, **p < 0.01, ***p < 0.001. Abbreviations: Ivy Glioblastoma atlas project (IvyGAP), glioblastoma (GBM), pseudopalisading cells (PC), central tumor (CT), infiltrating tumor (IT), leading edge (LE), microvascular proliferation (MP).
3.3. Immunohistochemical validation of CD44, B7‐H3 and VISTA
Expression of the deregulated markers CD44, B7‐H3 and VISTA were evaluated in the same normoxic and hypoxic areas as outlined for immune cell quantitation. In the immunohistochemical staining we found that CD44 was expressed in the membrane of cells with tumor cell morphology and the amount of CD44 positive cells was high in regions with necrosis and pseudopalisades (Figure 4B) as compared to normoxic areas (Figure 4A). B7‐H3 was expressed in the membrane of tumor cells as well as TAMs (Figure 4E,F). B7‐H3 expression was moderate in areas with normoxia (Figure 4E) whereas expression increased in and around pseudopalisades and areas with necrosis (Figure 4F). VISTA was expressed in the membrane of cells with TAM morphology and in ramifications of infiltrating cells with characteristic microglia morphology (Figure 4I,J). VISTA expression was reduced or absent in areas with necrosis and pseudopalisades (Figure 4J). Vessels exhibited high expression of all markers and these were excluded from analysis. When applying the software‐based classifiers, the quantitative estimates of AF CD44 (p = 0.052) (Figure 4C,D,M) and B7‐H3 (p = 0.005) (Figure 4G,H,N) showed a significant increase in hypoxia, whereas VISTA decreased in hypoxia (p = 0.054) (Figure 4K,L,O).
FIGURE 4.
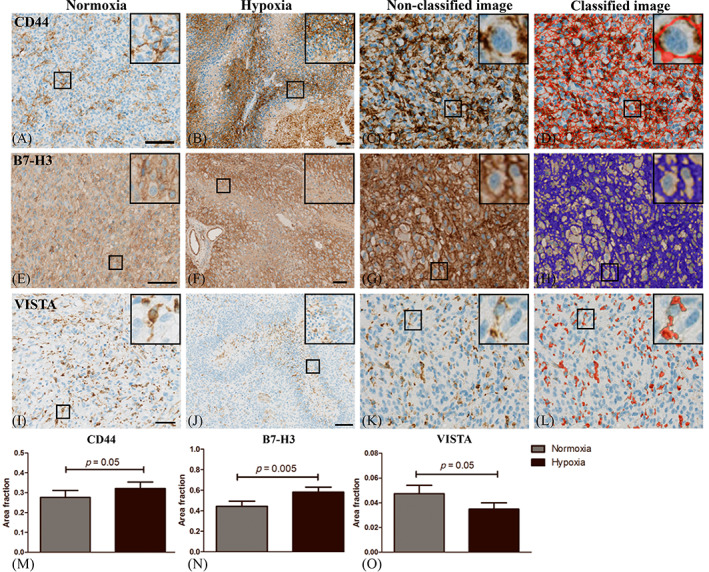
Immunohistochemical CD44, B7‐H3 and VISTA staining patterns in glioblastoma (GBM). (A) CD44 was moderately expressed in areas with normoxia whereas (B) hypoxic areas contained high CD44 expression especially in pseudopalisading tumor cells. (E) B7‐H3 was expressed by tumor‐ and microglia cells (F) and the expression increased around pseudopalisades and necrotic areas. (I) VISTA was expressed in the membrane and ramifications of microglia but (J) the expression was absent or low in areas with necrosis and pseudopalisades. Three areas with hypoxia and normoxia per tumor were manually outlined and quantitation of (C‐D) CD44, (G‐H) B7‐H3 and (K‐L) VISTA was carried out using a trained pixel‐based classifier to detect the area of membrane staining. Scalebar 100 μm. (M‐N) The area fraction (AF) of CD44 and B7‐H3 was significantly higher in tumor regions with hypoxia whereas (O) AF VISTA decreased in areas with hypoxia. Statistical significance was defined as *p < 0.5, **p < 0.01, ***p < 0.001.
3.4. Hypoxia and immune cell composition
DSP analysis revealed relatively high counts of CD4 and CD11c. CD11c was significant lower in hypoxic areas (Figure 2) whereas CD4 tended to decrease in hypoxia (Figure S3). To further explore the immune cell composition in hypoxic areas TAMs (Iba1), T‐helper cells (CD4), cytotoxic T cells (CD8) and regulatory T cells (FOXP3) were investigated in an additional patient material consisting of 19 GBMs (Figure 5). As expected TAMs were the most frequently occurring immune cells, but with no significant differences in AF Iba1 between hypoxic and normoxic areas (p = 0.2) (Figure 5U). Iba1+ cells comprised of up to 0.33 (median = 0.14) of the total hypoxic area (Figure 5I,M). In the normoxic areas Iba1 comprised up to 0.35 (median = 0.15) of the total tumor area (Figure 5A,E). AF CD4 indicated no differences (p = 0.2, 31%) between hypoxia (Figure 5J,N,V) and normoxia (Figure 5B,F). Interestingly, AF CD8 was significantly decreased in hypoxia (p = 0.04, −69%) (Figure 5K,O,X) as compared to normoxia (Figure 5C,G). AF FOXP3 was similar (Figure 5Y) (p = 0.8, 7%) between hypoxic (Figure 5L,P) and normoxic areas (Figure 5D,H).
FIGURE 5.
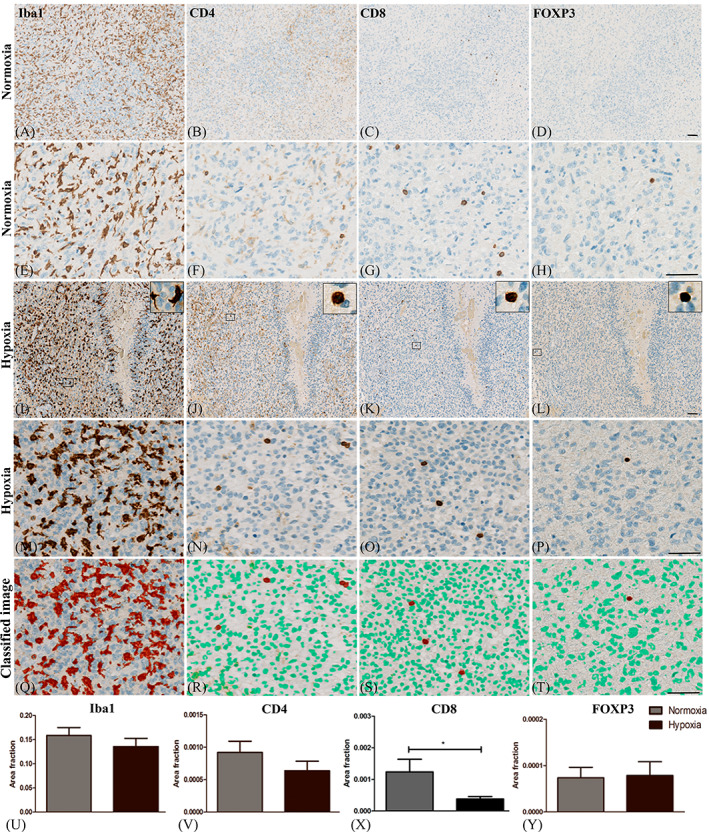
Presence of immune cells in hypoxic and normoxic regions. (A) The amount of tumor‐associated microglia and macrophages (TAMs), (B) T‐helper cells (C) Cytotoxic T cells and (D) regulatory T cells were evaluated in (I‐P) hypoxic and (A‐H) normoxic regions. (M, Q, N, R, O, S, P, T). A pixel‐based classifier was trained to detect expression of each marker. AF (U) Iba1, (V) CD4 and (Y) FOXP3 did not differ between regions with hypoxia and normoxia. (X) AF CD8 was significantly decreased in areas with hypoxia as compared to normoxia. Scalebar: 100μm. Statistical significance was defined as *p < 0.5, **p < 0.01, ***p < 0.001. Abbreviations: Area fraction (AF) and ionized calcium‐binding adapter molecule 1 (Iba1).
4. DISCUSSION
In this study we found differential expression of the immune checkpoint‐related markers CD44, B7‐H3 and VISTA in normoxic and hypoxic GBM regions. This was found by DSP and validated with IvyGAP project data and by immunohistochemistry.
We found that CD44 was upregulated in hypoxic areas. CD44 is a cell surface glycoprotein receptor and involved in the progression and metastasis of cancer cells [21]. Using immunohistochemistry, we detected high CD44 expression in GBM tumor cells, especially in pseudopalisades and other areas around necrosis. The antibody used in the DSP approach recognizes residues around proline 210 and is therefore separate from the splice site and not affected by splicing. Digital quantitation of CD44 protein expression and RNA sequencing data from IvyGAP confirmed CD44 upregulation in hypoxic regions. CD44 and osteopontin (CD44 ligand secreted from myeloid‐ and tumor cells) was proposed as a promising immune checkpoint in human colon carcinoma [22] and moreover implicated in the pathogenesis of several cancer types including GBMs [23]. GBM cell‐derived osteopontin increased tumor invasion and radiation resistance in vitro and in vivo [24, 25]. In microglia and macrophages, osteopontin enhanced activation and induced a pro‐tumorigenic phenotype [26]. Interestingly, the expression profile of human and murine GBM‐associated microglia suggested osteopontin as one of the most upregulated genes compared to resting microglia [27, 28]. CD44 was included in the DSP panel caused by its role as immune checkpoint marker but was also associated with stem‐cell properties. The hypoxic/perinecrotic niche is known for stem‐like cell enrichment [29] and CD44 suggested as a stem cell marker [30], although its function remains controversial [31]. The high expression of CD44 in hypoxic areas may be explained by a GBM in vitro study showing hypoxia‐induced binding of CD44 to Hif‐2α [32]. Similar results have been reported in breast‐ and gastric cancer with CD44 binding to Hif‐1α [33, 34]. However, cells in hypoxic areas will most likely be difficult to target caused by poor vascularization and impaired drug delivery, consequently with high expression of immune checkpoints, including CD44, contributing to immune evasion.
We found that B7‐H3 was upregulated in areas with hypoxia. B7‐H3 (CD276) is a co‐stimulator that belongs to the B7‐CD28 family and it works as a ligand and has an inhibitory function on T cell activation and proliferation [35]. B7‐H3 is overexpressed in different cancer types like acute leukemia and ovarian cancer and associated with poor prognosis [36, 37], including GBMs [38]. Large‐scale analysis using The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) database found that B7‐H3 expression was associated with malignancy grade and shorter survival in GBM [39]. This study also proposed that B7‐H3 was upregulated in microglia and macrophages and may facilitate immune evasion. The role of B7‐H3 remains controversial and some studies suggest that it works as an immune co‐stimulator [40], while other studies suggest it works as a co‐inhibitor [41]. A previous study showed that B7‐H3 regulate aerobic glycolysis in a Hif‐1α dependent pathway in breast cancer cells [42], whereas a colorectal cancer study found no association [43]. Therapeutic targeting of B7‐H3 with a monoclonal antibody inhibited tumor growth in murine renal and bladder xenografts [44]. This was extended to a phase 1 clinical trial with monotherapy (NCT02982941) or combined with anti PD‐1 (NCT02475213) and anti CTLA‐4 therapy (NCT02381314) [45] in solid tumors and with no published results yet. We found a high expression of B7‐H3 in GBM, which may suggest that therapeutic targeting of B7‐H3 in GBM is relevant. However, our results at the same time showed increased expression of B7‐H3 in hypoxic GBM areas and the same goes for CD44. The increased expression of checkpoint markers suggests a potential mechanism for immune evasion in these areas caused by decreased blood flow and limited drug delivery.
This study suggested that VISTA was downregulated in areas with hypoxia. We found intense VISTA staining of microglia in GBM tissue. VISTA (V‐domain immunoglobulin suppressor of T cell activation) has dual activities and it works as a stimulatory ligand for antigen presenting cells inducing immune activation and also as a negative ligand suppressing T cell activation [46]. Previously VISTA has been found to be abundantly expressed by microglia and differentially regulated in different central nervous system pathologies [47]. VISTA acts as an immune checkpoint, suppressing T cell proliferation and enhancing the conversion of naive T cells into regulatory T cells [48, 49]. In line with this VISTA depleted mice were more resistant to glioma growth [50] and no tumors developed in 20% of the VISTA depleted mice. In a fibrosarcoma model, VISTA overexpression reduced anti‐tumor immunity in mice [51]. In contrast to our results other studies suggested that hypoxia in the tumor microenvironment is associated with increased VISTA expression in tumor infiltrating inflammatory cells [52, 53]. Deng et al. showed that VISTA and Hif‐1α activity correlated in colon cancer patients and that myeloid derived suppressor cells showed increased VISTA expression in hypoxic tumor regions [54]. The divergence between our study and the mentioned results may be explained by the different type of cancers including their different microenvironment. Importantly, a major determinant of the immune biology in GBMs is TAMs, which infiltrate the tumor tissue and comprise up to 30% of the total cell population. Different amounts and expression levels of TAMs in areas with and without hypoxia may therefore influence the VISTA expression level together with different influence of hypoxia on the polarization state of these cells.
PD‐1 and PD‐L1 are immune checkpoint proteins. PD‐1 is a cell surface receptor and binding to the ligand PD‐L1 results in negative regulation of the T cell response [10]. We found and validated that hypoxia did not affect PD‐L1 expression in GBM patient material. The ROIs investigated in our study were selected based on the expression of Hif‐1α and the cell type markers GFAP (astrocytes) and IBA1 (microglia and macrophages) and we ensured that the tissue was vital. However, since the ROIs in our study covered regions with different cell types, the sensitivity in terms of changes occurring e.g. exclusively in tumor cells is not optimal. We will be able to obtain more precise measurements in future studies measuring e.g. PD‐L1 level exclusively in tumor cells by using a tumor cell specific morphology marker. This may explain the different results obtained in our study and the studies discussed below. Accordingly, a recent study identified upregulation of PD‐L1 in glioma cell lines exposed to hypoxia but the lines were commercial lines (U251 and U343) [55] and not low‐passage patient‐derived GBM lines. Another previous study suggested that PD‐L1 was enriched in perinecrotic and pseudopalisading niches of GBMs, but this finding was obtained using IvyGAP mRNA data [56] and did not report on the protein level. In melanoma and lung cancer in vivo tumor models—having a different microenvironment compared to glioblastoma—Noman et al. showed that PD‐L1 expression in hypoxic tumor areas was increased [57]. Although PD‐L1 has a hypoxia responsive element in its promotor region which directly binds Hif‐1α [58], this pathway might be affected by different regulators in vivo and in different cancers and PD‐L1 expression may be regulated independent of CD44, B7‐H3 and VISTA. This is in line with, another study proposing that VISTA worked from a pathway different from the PD‐L1 providing a rationale to concurrently target the proteins [59]. However, VISTA regulation of PD‐L1 has been found in other cancer types [60]. VISTA immune cell expression has thus been associated with high PD‐L1 in gastric tumor cells suggesting a dual synergistic mechanism for tumor evasion [60]. Further multiplexing studies are needed to investigate potential association of expression of checkpoint markers.
In hierarchal cluster analysis we did not see two major hypoxic or normoxic cluster but a more mixed pattern. DSP data suggested no major differences in T cell populations between normoxia and hypoxia, although there was a trend towards decreased CD4 and CD8 counts in hypoxic regions. Using immunohistochemistry we found a significant decrease in CD8+ T cells in hypoxic regions and this has to our knowledge not been shown in GBM before. We found no differences in regulatory T cells or CD4+ T cells, though CD4 tended to decrease in hypoxia. This is in line with in vitro findings suggesting that hypoxia induce immunosuppression by impairing T cell proliferation and function [16, 61] including impaired cytotoxic T cell induction [62].
Our immunohistochemical analysis showed high levels of TAMs in GBMs but we were not able to demonstrate a significant difference in the frequency of these cells in normoxia and hypoxia. Macrophages are known to promote cancer growth by being immunosuppressive and secreting factors increasing angiogenesis and tumor growth [6] and some studies indicate that macrophages exposed to hypoxia increase production of inflammatory mediators [63, 64, 65] while other suggest hypoxia as an unfavorable growth condition leading to decreased phagocytic abilities [65, 66]. Although we found a similar frequency of TAMs in normoxic and hypoxic regions our results with different expression of checkpoint markers in these cells were in line with influence of hypoxia on the cell phenotype.
The high expression of CD44 and B7‐H3 might explain the decreased number of T cells (CD8+ T cells) in hypoxic regions. The combination of few T cells, high amounts of TAMs and increased expression of checkpoint‐related molecules suggests that hypoxic areas will be difficult to target with immune therapy. Furthermore, hypoxia is associated with abnormal vascularization resulting in reduced oxygen supply but also impaired delivery of drugs. We found that VISTA expression was low and even absent in some hypoxic areas hereby reducing the risk of VISTA‐mediated escape from VISTA targeted therapy in hypoxic areas with impaired delivery of checkpoint‐blockers. Hence, VISTA might be novel promising target in GBMs.
Currently there are no FDA‐approved immune therapies for GBM patients. Despite this several new immunotherapies are currently being tested in clinical trials [67]. One trial with VISTA monotherapy (NCT02671955) is ongoing, although the expression of VISTA in the tumor microenvironment is unknown. GBM is a highly immunosuppressive tumor containing only few infiltrating T cells [68]. In addition, most GBM patients are treated with steroids to control peri‐tumoral edema and potentially decreasing the efficacy of immunotherapies [69, 70]. Our study suggests that both normoxic and hypoxic tumor regions contained high amounts of TAMs. Increased knowledge about the microenvironment, including the influence of hypoxia on the expression of checkpoint markers could be the way forward.
In conclusion, we found that CD44 and B7‐H3 were upregulated in hypoxic GBM areas whereas VISTA was downregulated together with the presence of fewer T cells. This heterogeneous expression of checkpoint‐related markers should be taken into consideration when developing new immune‐related targeted therapies.
AUTHOR CONTRIBUTIONS
The DSP analysis was performed by Nanostring and ROI selection was done by Stine Asferg Petterson and Bjarne Winther Kristensen. Stine Asferg Petterson carried out the experiments and Mia Dahl Sørensen, Signe Regner Michaelsen and Stine Asferg Petterson carried out the digital image analysis. Mark Burton, Mads Thomassen and Torben A. Kruse performed the hierarchal cluster analysis. Stine Asferg Petterson and Bjarne Winther Kristensen designed the study, interpreted the results and drafted the manuscript. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ETHICS STATEMENT
This study was approved by the Regional Committee on Health Research Ethics for Southern Denmark (Project‐ID S‐20150148) as well as the Danish Data Protection Agency (file number: 16/11065). The use of tissue was not prohibited by any patient according to the Danish Tissue Application Register.
Supporting information
Appendix S1. Supporting Information.
Figure S1. Images of Iba1, GFAP and HIF‐1α staining in tumors and outlined ROIs. Overview scans of the whole tissue sections and focused images of each outlined ROI from fluorescent staining of Iba1 and GFAP, as well as from HIF‐1α staining of neighbor tissue sections. ROI, regions of interest.
Figure S2. Immune cell checkpoints and immune activating molecules. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and the mean count across all tumors are illustrated in the figure. ROI, regions of interest.
Figure S3. Pan‐immune cell markers. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and the mean count across all tumors are illustrated in the figure. ROI, regions of interest.
Figure S4. Signaling pathways and survival molecules. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and the mean count across all tumors are illustrated in the figure. ROI, regions of interest.
Figure S5. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in individual tumors are illustrated in the figure. ROI, regions of interest.
ACKNOWLEDGMENTS
We would like to thank Helle Wohlleben for assistance with the immunohistochemical stainings. The project was supported by the Danish Medical Research Council 4183‐00183.
Petterson SA, Sørensen MD, Burton M, Thomassen M, Kruse TA, Michaelsen SR, et al. Differential expression of checkpoint markers in the normoxic and hypoxic microenvironment of glioblastomas. Brain Pathology. 2023;33(1):e13111. 10.1111/bpa.13111
Funding information Danish Medical Research Council, Grant/Award Number: 4183‐00183
Contributor Information
Stine Asferg Petterson, Email: stine.asferg.petterson@rsyd.dk.
Bjarne Winther Kristensen, Email: bjarne.winther.kristensen.01@regionh.dk.
DATA AVAILABILITY STATEMENT
All data underlying the findings reported in the manuscript has been submitted as part of the article. The data that support the findings of this study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella‐Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. [DOI] [PubMed] [Google Scholar]
- 2. Colwell N, Larion M, Giles AJ, Seldomridge AN, Sizdahkhani S, Gilbert MR, et al. Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem‐like cells. Neuro Oncol. 2017;19(7):887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Otvos B, Silver DJ, Mulkearns‐Hubert EE, Alvarado AG, Turaga SM, Sorensen MD, et al. Cancer stem cell‐secreted macrophage migration inhibitory factor stimulates myeloid derived suppressor cell function and facilitates glioblastoma immune evasion. Stem Cells. 2016;34(8):2026–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yi L, Xiao H, Xu M, Ye X, Hu J, Li F, et al. Glioma‐initiating cells: a predominant role in microglia/macrophages tropism to glioma. J Neuroimmunol. 2011;232(1–2):75–82. [DOI] [PubMed] [Google Scholar]
- 5. Laoui D, Van Overmeire E, Movahedi K, Van den Bossche J, Schouppe E, Mommer C, et al. Mononuclear phagocyte heterogeneity in cancer: different subsets and activation states reaching out at the tumor site. Immunobiology. 2011;216(11):1192–202. [DOI] [PubMed] [Google Scholar]
- 6. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bloch O, Crane CA, Kaur R, Safaee M, Rutkowski MJ, Parsa AT. Gliomas promote immunosuppression through induction of B7‐H1 expression in tumor‐associated macrophages. Clin Cancer Res. 2013;19(12):3165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bersanelli M, Buti S. From targeting the tumor to targeting the immune system: transversal challenges in oncology with the inhibition of the PD‐1/PD‐L1 axis. World J Clin Oncol. 2017;8(1):37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berghoff AS, Kiesel B, Widhalm G, Rajky O, Ricken G, Wohrer A, et al. Programmed death ligand 1 expression and tumor‐infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heiland DH, Haaker G, Delev D, Mercas B, Masalha W, Heynckes S, et al. Comprehensive analysis of PD‐L1 expression in glioblastoma multiforme. Oncotarget. 2017;8(26):42214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Romani M, Pistillo MP, Carosio R, Morabito A, Banelli B. Immune checkpoints and innovative therapies in glioblastoma. Front Oncol. 2018;8:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23(9):3265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aderetti DA, Hira VVV, Molenaar RJ, van Noorden CJF. The hypoxic peri‐arteriolar glioma stem cell niche, an integrated concept of five types of niches in human glioblastoma. Biochim Biophys Acta Rev Cancer. 2018;1869(2):346–54. [DOI] [PubMed] [Google Scholar]
- 14. Ohta A, Madasu M, Subramanian M, Kini R, Jones G, Chouker A, et al. Hypoxia‐induced and A2A adenosine receptor‐independent T‐cell suppression is short lived and easily reversible. Int Immunol. 2014;26(2):83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Allard B, Pommey S, Smyth MJ, Stagg J. Targeting CD73 enhances the antitumor activity of anti‐PD‐1 and anti‐CTLA‐4 mAbs. Clin Cancer Res. 2013;19(20):5626–35. [DOI] [PubMed] [Google Scholar]
- 16. Ohta A, Kini R, Ohta A, Subramanian M, Madasu M, Sitkovsky M. The development and immunosuppressive functions of CD4(+) CD25(+) FoxP3(+) regulatory T cells are under influence of the adenosine‐A2A adenosine receptor pathway. Front Immunol. 2012;3:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo X, Xue H, Shao Q, Wang J, Guo X, Chen X, et al. Hypoxia promotes glioma‐associated macrophage infiltration via periostin and subsequent M2 polarization by upregulating TGF‐beta and M‐CSFR. Oncotarget. 2016;7(49):80521–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tripathi C, Tewari BN, Kanchan RK, Baghel KS, Nautiyal N, Shrivastava R, et al. Macrophages are recruited to hypoxic tumor areas and acquire a pro‐angiogenic M2‐polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget. 2014;5(14):5350–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petterson SA, Dahlrot RH, Hermansen SK, K A Munthe S, Gundesen MT, Wohlleben H, et al. High levels of c‐Met is associated with poor prognosis in glioblastoma. 2015;122(3):517–27. [DOI] [PubMed] [Google Scholar]
- 20. Sorensen MD, Dahlrot RH, Boldt HB, Hansen S, Kristensen BW. Tumour‐associated microglia/macrophages predict poor prognosis in high‐grade gliomas and correlate with an aggressive tumour subtype. Neuropathol Appl Neurobiol. 2018;44(2):185–206. [DOI] [PubMed] [Google Scholar]
- 21. Liu WH, Lin JC, Chou YC, Li MH, Tsai JT. CD44‐associated radioresistance of glioblastoma in irradiated brain areas with optimal tumor coverage. Cancer Med. 2020;9(1):350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klement JD, Paschall AV, Redd PS, Ibrahim ML, Lu C, Yang D, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest. 2018;128(12):5549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subraman V, Thiyagarajan M, Malathi N, Rajan ST. OPN—revisited. J Clin Diagn Res. 2015;9(6):ZE10–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henry A, Nokin MJ, Leroi N, Lallemand F, Lambert J, Goffart N, et al. New role of osteopontin in DNA repair and impact on human glioblastoma radiosensitivity. Oncotarget. 2016;7(39):63708–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamour V, Henry A, Kroonen J, Nokin MJ, von Marschall Z, Fisher LW, et al. Targeting osteopontin suppresses glioblastoma stem‐like cell character and tumorigenicity in vivo. Int J Cancer. 2015;137(5):1047–57. [DOI] [PubMed] [Google Scholar]
- 26. Ellert‐Miklaszewska A, Wisniewski P, Kijewska M, Gajdanowicz P, Pszczolkowska D, Przanowski P, et al. Tumour‐processed osteopontin and lactadherin drive the protumorigenic reprogramming of microglia and glioma progression. Oncogene. 2016;35(50):6366–77. [DOI] [PubMed] [Google Scholar]
- 27. Szulzewsky F, Arora S, de Witte L, Ulas T, Markovic D, Schultze JL, et al. Human glioblastoma‐associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia. 2016;64(8):1416–36. [DOI] [PubMed] [Google Scholar]
- 28. Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, et al. Glioma‐associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2):e0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia‐inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown DV, Daniel PM, D'Abaco GM, Gogos A, Ng W, Morokoff AP, et al. Coexpression analysis of CD133 and CD44 identifies proneural and mesenchymal subtypes of glioblastoma multiforme. Oncotarget. 2015;6(8):6267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang HH, Liao CC, Chow NH, Huang LL, Chuang JI, Wei KC, et al. Whether CD44 is an applicable marker for glioma stem cells. Am J Transl Res. 2017;9(11):4785–806. [PMC free article] [PubMed] [Google Scholar]
- 32. Johansson E, Grassi ES, Pantazopoulou V, Tong B, Lindgren D, Berg TJ, et al. CD44 interacts with HIF‐2alpha to modulate the hypoxic phenotype of perinecrotic and perivascular glioma cells. Cell Rep. 2017;20(7):1641–53. [DOI] [PubMed] [Google Scholar]
- 33. Liang G, Li S, Du W, Ke Q, Cai J, Yang J. Hypoxia regulates CD44 expression via hypoxia‐inducible factor‐1alpha in human gastric cancer cells. Oncol Lett. 2017;13(2):967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryu D, Ryoo IG, Kwak MK. Overexpression of CD44 standard isoform upregulates HIF‐1alpha signaling in hypoxic breast cancer cells. Biomol Ther (Seoul). 2018;26(5):487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Picarda E, Ohaegbulam KC, Zang X. Molecular pathways: targeting B7‐H3 (CD276) for human cancer immunotherapy. Clin Cancer Res. 2016;22(14):3425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu Y, Lv X, Wu Y, Xu J, Wang L, Chen W, et al. Expression of costimulatory molecule B7‐H3 and its prognostic implications in human acute leukemia. Hematology (Amsterdam, Netherlands). 2015;20(4):187–95. [DOI] [PubMed] [Google Scholar]
- 37. Zang X, Sullivan PS, Soslow RA, Waitz R, Reuter VE, Wilton A, et al. Tumor associated endothelial expression of B7‐H3 predicts survival in ovarian carcinomas. Mod Pathol. 2010;23(8):1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang X, Zhao S, Zhang Y, Wang Y, Zhang Z, Yang M, et al. B7‐H3 as a novel CAR‐T therapeutic target for glioblastoma. Mol Ther Oncolytics. 2019;14:279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang C, Zhang Z, Li F, Shen Z, Qiao Y, Li L, et al. Large‐scale analysis reveals the specific clinical and immune features of B7‐H3 in glioma. Onco Targets Ther. 2018;7(11):e1461304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo L, Chapoval AI, Flies DB, Zhu G, Hirano F, Wang S, et al. B7‐H3 enhances tumor immunity in vivo by costimulating rapid clonal expansion of antigen‐specific CD8+ cytolytic T. Cell. 2004;173(9):5445–50. [DOI] [PubMed] [Google Scholar]
- 41. Leitner J, Klauser C, Pickl WF, Stockl J, Majdic O, Bardet AF, et al. B7‐H3 is a potent inhibitor of human T‐cell activation: no evidence for B7‐H3 and TREML2 interaction. Eur J Immunol. 2009;39(7):1754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lim S, Liu H, Madeira da Silva L, Arora R, Liu Z, Phillips JB, et al. Immunoregulatory protein B7‐H3 reprograms glucose metabolism in cancer cells by ROS‐mediated stabilization of HIF1alpha. Cancer Res. 2016;76(8):2231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi T, Ma Y, Cao L, Zhan S, Xu Y, Fu F, et al. B7‐H3 promotes aerobic glycolysis and chemoresistance in colorectal cancer cells by regulating HK2. Cell Death Dis. 2019;10(4):308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loo D, Alderson RF, Chen FZ, Huang L, Zhang W, Gorlatov S, et al. Development of an Fc‐enhanced anti‐B7‐H3 monoclonal antibody with potent antitumor activity. Clin Cancer Res. 2012;18(14):3834–45. [DOI] [PubMed] [Google Scholar]
- 45. Burugu S, Dancsok AR, Nielsen TO. Emerging targets in cancer immunotherapy. Semin Cancer Biol. 2018;52(Pt 2):39–52. [DOI] [PubMed] [Google Scholar]
- 46. Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O'Connell S, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74(7):1924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Borggrewe M, Grit C, Den Dunnen WFA, Burm SM, Bajramovic JJ, Noelle RJ, et al. VISTA expression by microglia decreases during inflammation and is differentially regulated in CNS diseases. Glia. 2018;66(12):2645–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen L, Flies DB. Molecular mechanisms of T cell co‐stimulation and co‐inhibition. Nat Rev Immunol. 2013;13(4):227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nowak EC, Lines JL, Varn FS, Deng J, Sarde A, Mabaera R, et al. Immunoregulatory functions of VISTA. Immunol Rev. 2017;276(1):66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Flies DB, Han X, Higuchi T, Zheng L, Sun J, Ye JJ, et al. Coinhibitory receptor PD‐1H preferentially suppresses CD4(+) T cell‐mediated immunity. J Clin Invest. 2014;124(5):1966–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208(3):577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mahoney K, Freeman GJNI. Acidity changes immunology: a new VISTA pathway. Nat Immunol. 2020;21:13–6. [DOI] [PubMed] [Google Scholar]
- 53. Yuan L, Tatineni J, Mahoney KM, Freeman GJ. VISTA: a mediator of quiescence and a promising target in cancer immunotherapy. Trends Immunol. 2021;42(3):209–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deng J, Li J, Sarde A, Lines JL, Lee YC, Qian DC, et al. Hypoxia‐induced VISTA promotes the suppressive function of myeloid‐derived suppressor cells in the tumor microenvironment. Cancer Immunol Res. 2019;7(7):1079–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ding XC, Wang LL, Zhang XD, Xu JL, Li PF, Liang H, et al. The relationship between expression of PD‐L1 and HIF‐1alpha in glioma cells under hypoxia. J Hematol Oncol. 2021;14(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ricklefs FL, Alayo Q, Krenzlin H, Mahmoud AB, Speranza MC, Nakashima H, et al. Immune evasion mediated by PD‐L1 on glioblastoma‐derived extracellular vesicles. Sci Adv. 2018;4(3):eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, et al. PD‐L1 is a novel direct target of HIF‐1alpha, and its blockade under hypoxia enhanced MDSC‐mediated T cell activation. J Exp Med. 2014;211(5):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barsoum IB, Koti M, Siemens DR, Graham CH. Mechanisms of hypoxia‐mediated immune escape in cancer. Cancer Res. 2014;74(24):7185–90. [DOI] [PubMed] [Google Scholar]
- 59. Liu J, Yuan Y, Chen W, Putra J, Suriawinata AA, Schenk AD, et al. Immune‐checkpoint proteins VISTA and PD‐1 nonredundantly regulate murine T‐cell responses. Proc Natl Acad Sci U S A. 2015;112(21):6682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Böger C, Behrens H‐M, Krüger S, Röcken C. The novel negative checkpoint regulator VISTA is expressed in gastric carcinoma and associated with PD‐L1/PD‐1: a future perspective for a combined gastric cancer therapy? Onco Targets Ther. 2017;6(4):e1293215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Naldini A, Carraro F, Silvestri S, Bocci V. Hypoxia affects cytokine production and proliferative responses by human peripheral mononuclear cells. J Cell Physiol. 1997;173(3):335–42. [DOI] [PubMed] [Google Scholar]
- 62. Caldwell CC, Kojima H, Lukashev D, Armstrong J, Farber M, Apasov SG, et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. Journal of immunology. Baltimore, Md: 1950. 2001;167(11):6140–9. [DOI] [PubMed] [Google Scholar]
- 63. Ghezzi P, Dinarello CA, Bianchi M, Rosandich ME, Repine JE, White CW. Hypoxia increases production of interleukin‐1 and tumor necrosis factor by human mononuclear cells. Cytokine. 1991;3(3):189–94. [DOI] [PubMed] [Google Scholar]
- 64. Scannell G, Waxman K, Kaml GJ, Ioli G, Gatanaga T, Yamamoto R, et al. Hypoxia induces a human macrophage cell line to release tumor necrosis factor‐alpha and its soluble receptors in vitro. J Surg Res. 1993;54(4):281–5. [DOI] [PubMed] [Google Scholar]
- 65. Yun JK, McCormick TS, Villabona C, Judware RR, Espinosa MB, Lapetina EG. Inflammatory mediators are perpetuated in macrophages resistant to apoptosis induced by hypoxia. Proc Natl Acad Sci U S A. 1997;94(25):13903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leeper‐Woodford SK, Mills JW. Phagocytosis and ATP levels in alveolar macrophages during acute hypoxia. Am J Respir Cell Mol Biol. 1992;6(3):326–34. [DOI] [PubMed] [Google Scholar]
- 67. McGranahan T, Therkelsen KE, Ahmad S, Nagpal S. Current state of immunotherapy for treatment of glioblastoma. Curr Treat Options Oncol. 2019;20(3):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24(9):1459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Arbour KC, Mezquita L, Long N, Rizvi H, Auclin E, Ni A, et al. Impact of baseline steroids on efficacy of programmed cell death‐1 and programmed death‐ligand 1 blockade in patients with non‐small‐cell lung cancer. J Clin Oncol. 2018;36(28):2872–8. [DOI] [PubMed] [Google Scholar]
- 70. Reardon DA, Neuberg DS, Keskin DB, Tirosh I, Anandappa A, Mathewson ND, et al. Effect of dexamethasone in glioblastoma (GBM) patients on systemic and intratumoral T‐cell responses induced by personalized neoantigen‐targeting vaccine. 2018;36(15_suppl):2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Figure S1. Images of Iba1, GFAP and HIF‐1α staining in tumors and outlined ROIs. Overview scans of the whole tissue sections and focused images of each outlined ROI from fluorescent staining of Iba1 and GFAP, as well as from HIF‐1α staining of neighbor tissue sections. ROI, regions of interest.
Figure S2. Immune cell checkpoints and immune activating molecules. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and the mean count across all tumors are illustrated in the figure. ROI, regions of interest.
Figure S3. Pan‐immune cell markers. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and the mean count across all tumors are illustrated in the figure. ROI, regions of interest.
Figure S4. Signaling pathways and survival molecules. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in each individual tumor and the mean count across all tumors are illustrated in the figure. ROI, regions of interest.
Figure S5. Differences in protein expression between hypoxic and normoxic tumor regions acquired from digital spatial profiling (DSP). The average count for hypoxic and normoxic ROIs for each marker in individual tumors are illustrated in the figure. ROI, regions of interest.
Data Availability Statement
All data underlying the findings reported in the manuscript has been submitted as part of the article. The data that support the findings of this study are available from the corresponding author on reasonable request.


