Abstract
Background
Nematode-trapping fungi are a highly specialised group in fungi and are essential regulators of natural nematode populations. At present, more than 130 species have been discovered in Zygomycota (Zoopagaceae), Basidiomycota (Nematoctonus), and Ascomycota (Orbiliaceae). Amongst them, nematode-trapping fungi in Orbiliaceae have become the research focus of carnivorous fungi due to their abundant species. During the investigation of carnivorous fungi in Yunnan, China, four fungal strains isolated from burned forest soil were identified as two new nematode-trapping species in Drechslerella (Orbiliaceae), based on multigene phylogenetic analysis and morphological characters.
New information
Drechslerelladaliensis sp. nov. is characterised by its ellipsoid, 1–2-septate macroconidia, clavate or bottle-shaped, 0–1-septate microconidia and unbranched, simple conidiophores. D.xiaguanensis sp. nov. is characterised by fusiform or spindle-shaped, 2–4-septate conidia and unbranched, simple conidiophores. Both of them produce constricting rings to capture nematodes. The phylogenetic analysis, based on combined ITS, TEF1-α and RPB2 sequences, determined their placement in Drechslerella. D.daliensis forms a basal lineage closely nested with D.hainanensis (YMF1.03993). D.xiaguanensis forms a sister lineage with D.bembicodes (1.01429), D.aphrobrocha (YMF1.00119) and D.coelobrocha (FWY03-25-1).
Keywords: carnivorous fungi, constricting rings, new species, Orbiliaceae, taxonomy
Introduction
Nematode-trapping fungi are important predators that capture nematodes by specialised trap structures (Barron 1977, Li et al. 2006, Swe et al. 2011, Zhang and Hyde 2014). They play vital roles in maintaining energy balance and nutrient cycles in soil ecosystems and exhibit great potential for biocontrol application in agricultural management (Cooke 1962, Ulzurrun and Hsueh 2018, Zhang et al. 2020a). Most nematode-trapping fungi belong to Orbiliaceae, which have been extensively studied due to their abundant species and sophisticated trapping devices (Linford et al. 1938, Jaffee et al. 1993, Wolstrup et al. 1996, Jaffee et al. 1998, Morton et al. 2003, Liu et al. 2009, El-Borai et al. 2011, Kumar et al. 2011, Swe et al. 2011, Vilela et al. 2012). Currently, 116 predatory species in Orbiliaceae have been reported (Glockling and Dick 1994, Li et al. 2006, Wu et al. 2012, Li et al. 2013, Liu et al. 2014, Zhang and Hyde 2014, Quijada et al. 2020, Zhang et al. 2020, Zhang et al. 2020b, Zhang et al. 2022). They are classified into three genera according to their types of trapping structure: 1) Arthrobotrys (67 species), catching nematodes using adhesive networks; 2) Dactylellina (34 species), capturing nematodes by adhesive knobs, adhesive branches and non-constricting rings and 3) Drechslelrella (15 species), trapping nematodes with constricting rings (Scholler et al. 1999, Li et al. 2005, Yang et al. 2007, Zhang and Hyde 2014).
Drechslerella was established by Subramanian (1963) with the type species D.acrochaeta (Drechsler) Subram. It is a small genus separated from Monacrosporium, based on conidia producing filamentous appendages at the apex, which are lacking in Monacrosporium. However, filamentous appendages are usually produced when conidia germinate and are also commonly found in some species of Arthrobotrys. Therefore, Liu and Zhang (1994) treated Drechslerella as a synonym of Monacrosporium, based on their similar conidial morphology. Subsequently, the generic concept of nematode-trapping fungi in Orbiliaceae was revised, based on molecular phylogenetic analysis. Drechslerella is characterised by producing constricting rings to capture nematodes (Liou and Tzean 1997, Pfister 1997, Ahren et al. 1998, Scholler et al. 1999, Li et al. 2005). Drechslerella currently includes 15 accepted species, 13 of which have been reported in China (Zhang and Mo 2006, Zhang and Hyde 2014). They mainly occur in the soil or sediment of various ecosystems such as forests, mangroves, freshwater, brackish water, heavy metal polluted areas and even in tree trunks and animal faeces (Jansson and Autery 1961, Hao et al. 2005, Mo et al. 2006, Su et al. 2007, Swe et al. 2009, Zhang and Hyde 2014, She et al. 2020, Zhang et al. 2020). In soil, Drechslerella species are mainly distributed in the upper litter and humus layer and closely related to the density of soil nematodes (Burges and Raw 1967, Gray and Bailey 1985, Zhang and Hyde 2014). Drechslerella species lack nematodes mainly by the rapid expansion of the three cells that make up the constricting ring. This method of trapping nematodes mainly by mechanical force is significantly different from that of species in Arthrobotrys and Dactylellina (capture nematodes mainly with adhesive material) (Zhang and Mo 2006, Zhang and Hyde 2014). Therefore, Drechslerella is the most special genus amongst Orbiliaceae NTF and it is also a key group in studying the origin and evolution of carnivorous fungi.
The studies of nematode-trapping fungi have been poorly addressed in extreme habitats (Onofri and Tosi 1992, Mo et al. 2008, Swe et al. 2008). Our previous research investigated the succession of nematode-trapping fungi after fire disturbance in forests in China (She et al. 2020). Four strains were isolated and identified as two new nematode-trapping fungi in Orbiliaceae. The aim of this study is to introduce these two new species, D.daliensis and D.xiaguanensis, based on morphology and phylogenetic evidence. The discovery of these two species increased the diversity of nematode-trapping fungi and provided more valuable materials for studying the evolution and origin of carnivorous fungi, as well as more potential species for the biological control of plant and animal parasitic nematodes.
Materials and methods
Samples collection, isolation and morphology
The soil samples were collected from a burned forest in Cangshan Mountain, Dali City, Yunnan Province, China (100°07’44”N, 25°45’49”E). The sampling site information has been described by She et al. (2020). About 100 g of soil was collected from 10–20 cm depth using a 35 mm-diameter soil borer. The soil sample was placed into a zip lock bag and samples were brought back to the laboratory and stored at 4°C until processing.
The soil samples were sprinkled on corn meal agar (CMA) plates with sterile toothpicks. Free-living nematodes (Panagrellusredivivus Goodey) were added as bait to promote the germination of nematode-trapping fungi. After three weeks of incubation at 26°C, the plates were observed under a stereomicroscope to find the spores of nematode-trapping fungi. A single spore was transferred to a fresh CMA plate using a sterile toothpick, repeating this step until the pure culture was obtained.
Fungal isolates were transferred to fresh potato dextrose agar plate (PDA) using a sterile toothpick and incubated at 26°C for colony characteristics observation. The pure cultures were transferred to fresh CMA observation plates (an observation well of 2×2 cm was made by removing the agar from the centre of the CMA plate) and incubated at 26°C. When the mycelium overspread the observation well, about 500 nematodes (P.redivivus) were added to the well to induce the formation of trapping devices. The types of trapping devices were checked using a stereomicroscope. All morphological characters were captured and measured with an Olympus BX53 microscope (Olympus Corporation, Japan).
DNA extraction, PCR amplification and sequencing
The genomic DNA was extracted from the mycelium grown on PDA plates according to the method described by Jeewon et al. (2002). The primer pairs ITS4-ITS5 (White et al. 1990), 526F-1567R (O'Donnell et al. 1998) and 6F-7R (Liu et al. 1999) were used to amplify the ITS, TEF1-α and RPB2 genes, respectively. The PCR amplification was performed as follows: 4 minutes of pre-denaturation at 94°C, followed by 35 cycles of 45 seconds of denaturation at 94°C, 1 minute of annealing at 52°C (ITS), 55°C (TEF1-α), 54°C (RPB2), 1.5-2 minutes of extension at 72°C and a final extension of 10 minutes at 72°C. The PCR products were purified with a DiaSpin PCR Product Purification Kit (Sangon Biotech Company, Limited, Shanghai, China). ITS and RPB2 genes were sequenced in forward and reverse directions using PCR primers and the TEF1-α region was sequenced using the 247F-609R primer pair (Yang et al. 2007) (BioSune Biotech Company, Limited, Shanghai, China).
Phylogenetic analysis
The sequences generated in this study were compared against the NCBI GenBank database using BLASTn (BLASTn; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome; accessed on 16 July 2022). The morphological and BLASTn search results placed these two species in the genus Drechslerella. Drechslerella were searched in the Index Fungorum (http://www.indexfungorum.org; accessed on 16 August 2022) and Species Fungorum (http://www.speciesfungorum.org; accessed on 16 August 2022) and all relevant records were checked individually according to the relevant documents to ensure that all Drechslerella taxa were considered in this study (Li et al. 2013, Zhang and Hyde 2014). All reliable ITS, TEF1-α and RPB2 sequences of Drechslerella taxa were downloaded from the GenBank database (Table 1). The three genes datasets (including our two new species) were aligned using MAFFT online version (Madeira et al. 2022, https://www.ebi.ac.uk/Tools/msa/mafft/), then manually adjusted and linked via BioEdit v.7.2.3 (Hall 1999) and MEGA6.0 (Tamura et al. 2013). Dactylaria sp. YNWS02-7-1 and Vermisporafusarina YXJ02-13-5 were selected as outgroups (Yang et al. 2007). Phylogenetic trees were inferred with Maximum Likelihood (ML), Maximum Parsimony (MP) and Bayesian Inference analyses (BI).
Table 1.
GenBank accession numbers of isolates included in this study. The type strains are marked with T at the end of the strain number. The newly-generated sequences are indicated in bold.
| Taxa | Strain numbers | GenBank accession numbers | Reference | ||
| ITS | TEF1-α | RPB2 | |||
| Arthrobotrysconoides | YMF1.00009 | MF948387 | MF948544 | MF948468 | Unpublished |
| Arthrobotrysguizhouensis | YMF1.00014T | MF948390 | MF948547 | MF948471 | Unpublished |
| Arthrobotrysshizishanna | YMF1.00022 | MF948392 | MF948549 | MF948473 | Unpublished |
| Dactylaria sp. | YNWS02-7-1 | AY773457 | AY773399 | AY773428 | Yang et al. (2007) |
| Dactylellinaappendiculata | CBS 206.64T | AF106531 | DQ358227 | DQ358229 | Hagedorn and Scholler (1999) |
| Dactylellinacopepodii | CBS 487.90T | U51964 | DQ999835 | DQ999816 | Liou and Tzean (1997) |
| Dactylellinamammillata | CBS229.54T | AY902794 | DQ999843 | DQ999817 | Li et al. (2006) |
| Dactylellinayushanensis | CGMCC 3.19713T | MK372061 | MN915113 | MN915112 | Zhang et al. (2020) |
| Drechslerellaanchonia | CBS109.37 | AY965753 | —— | —— | Li et al. (2006) |
| Drechslerellaaphrobrocha | YMF1.00119 | MF948397 | —— | MF948477 | Unpublished |
| Drechslerellabembicodes | 1.01429 | MH179731 | —— | MH179835 | Unpublished |
| Drechslerellabrochopaga | 701 | AY773456 | AY773398 | AY773427 | Yang et al. (2007) |
| Drechslerellabrochopaga | 1.01829 | MH179750 | —— | MH179852 | Unpublished |
| Drechslerellabrochopaga | CBS218.61 | U51950 | —— | —— | Liou and Tzean (1997) |
| Drechslerellabrochopaga | ATCC 96710 | EF445987 | —— | —— | Smith and Jaffee (2009) |
| Drechslerellabrochopaga | DHP 212 | U72609 | —— | —— | Pfister (1997) |
| Drechslerellabrochopaga | BCRC 34361 | FJ380936 | —— | —— | Zhang et al. (2020b) |
| Drechslerellabrochopaga | H.B.9925 | KT222412 | —— | —— | Zhang et al. (2020b) |
| Drechslerellabrochopaga | H.B.9965 | KT380104 | —— | —— | Zhang et al. (2020b) |
| Drechslerellabrochopaga | 6178 | DQ656615 | —— | —— | Zhang et al. (2020b) |
| Drechslerellacoelobrocha | FWY03-25-1 | AY773464 | AY773406 | AY773435 | Yang et al. (2007) |
| Drechslerellacoelobrocha | 1.0148 | MH179744 | MH179847 | Unpublished | |
| Drechslerelladactyloides | 1.00031 | MH179690 | MH179554 | MH179799 | Unpublished |
| Drechslereladactyloides | expo-5 | AY773463 | AY773405 | AY773434 | Yang et al. (2007) |
| Drechslerelladactyloides | 1.00131 | MH179705 | —— | MH179813 | Unpublished |
| Drechslerelladaliensis | CGMCC 3.20131 | MT592896 | OK556701 | OK638157 | This study |
| Drechslerelladaliensis | DLU22-1 | OK643974 | OK556700 | OK638158 | This study |
| Drechslerelladoedycoides | YMF1.00553 | MF948401 | —— | MF948481 | Unpublished |
| Drechslerelladoedycoides | CBS 586.91 | MH862283 | —— | —— | Vu et al. (2019) |
| Drechslerelladoedycoides | CBS175.55 | MH857432 | —— | —— | Liou and Tzean (1997) |
| Drechslerellaeffusa | YMF1.00583 | MF948405 | MF948557 | MF948484 | Unpublished |
| Drechslerellaeffusa | CBS 774.84 | MH861835 | —— | —— | Vu et al. (2019) |
| Drechslerellahainanensis | YMF1.03993 | KC952010 | —— | —— | Li et al. (2013) |
| Drechslerellaheterospora | YMF1.00550 | MF948400 | MF948554 | MF948480 | Unpublished |
| Drechslerellapolybrocha | CBS 319.56 | MH857657 | —— | —— | Vu et al. (2019) |
| Drechslerellapolybrocha | CCRC 32872 | U51973 | —— | —— | Vu et al. (2019) |
| Drechslerellapolybrocha | DHP 133 | U72606 | —— | —— | Zhang et al. (2020b) |
| Drechslerellapolybrocha | H.B. 8317 | KT222361 | —— | —— | Unpablished |
| Drechslerellastenobrocha | YNWS02-9-1 | AY773460 | AY773402 | AY773431 | Yang et al. (2007) |
| Drechslerellaxiaguanensis | CGMCC 3.20132 | MT592900 | OK556699 | OK638159 | This study |
| Drechslerellaxiaguanensis | DLU23-1 | OK643975 | OK556698 | OK638160 | This study |
| Drechslerellayunnanensis | 1.01863 | MH179759 | —— | MH179861 | Unpublished |
| Drechslerellayunnanensi | YMF1.03216 | HQ711927 | —— | —— | Yu et al. (2009) |
| Vermisporafusarina | YXJ02-13-5 | AY773447 | AY773389 | AY773418 | Yang et al. (2007) |
SYM+I+G, GTR+I+G and GTR+I+G models were selected as best-fit optimal substitution models for ITS, TEF1-α and RPB2, respectively, via jModelTest v.2.1.10 (Posada 2008) under the Akaike Information Criterion (AIC).
MrBayes v. 3.2.6. (Huelsenbeck and Ronquist 2001) was used to perform the Bayesian Inference (BI) analysis. The multiple sequence alignment file was converted into the MrBayes compatible NEXUS file via Fasta Convert (Hall 2005). The dataset was partitioned and the optimal substitution models of each gene were equivalently replaced to conform to the setting of MrBayes. Six simultaneous Markov Chains were run for 10,000,000 generations and trees were sampled every 100 generations (a total of 100,000 trees). The first 25% of trees were discarded and the remaining trees were used to calculate the posterior probabilities (PP) in the majority rule consensus tree. All the above parameters are edited into the MrBayes block in the NEX file.
IQ-Tree v.1.6.5 (Nguyen et al. 2014) was used to perform the Maximum Likelihood (ML) analysis. The dataset was partitioned and each gene was analysed with its corresponding model. The rapid bootstrapping method with 1000 replicates (Felsenstein 1985) was used to compute the bootstrap support values (BS).
Maximum Parsimony (MP) analysis was performed via the web CIPRES Science Gateway v. 3.3 (Miller et al. 2010, https://www.phylo.org) by PAUP 4. a168 on XSEDE using the heuristic search option with 1000 random sequence additions. Max-trees were set up at 5000 and no increase. Clade stability was assessed using a bootstrap analysis with 1,000 replicates (Felsenstein 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated for all trees generated under different optimality criteria. All the above parameters are edited into the PAUP block in the NEX file.
The trees were visualised with FigTree v.1.3.1 (Rambaut 2009). The backbone tree was edited and reorganised by Microsoft PowerPoint (2013) and Adobe Photoshop CS6 software (Adobe Systems, USA). Sequences derived from this study were deposited in GenBank (Table 1).
Taxon treatments
Drechslerella daliensis
Fa Zhang, Xiao-Yan Yang, Kevin D Hyde sp. nov.
25550C67-0C86-508A-9865-0630E2CAEDBB
http://www.indexfungorum.org/Names:IF558120
Facesoffungi number:FOF 10565
Materials
Type status: Holotype. Occurrence: occurrenceRemarks: Isolated from burned forest soil; occurrenceID: 82BE156C-BBA2-57F5-B468-EA13407B9F19; Taxon: scientificName: Drechslerelladaliensis; kingdom: Fungi; phylum: Ascomycota; class: Orbiliomycetes; order: Orbiliales; family: Orbiliaceae; genus: Drechslerella; specificEpithet: daliensis; taxonRank: species; scientificNameAuthorship: Fa Zhang, Xiao-Yan Yang, Kevin D. Hyde; Location: country: China; countryCode: CHN; stateProvince: Yunnan; county: Dali; locationRemarks: China, Yunnan Province, Dali City, Cangshan Mountain, burned forest soil, 25 July 2017; Identification: identifiedBy: Fa Zhang; Record Level: language: English; collectionID: CGMCC3.20131
Type status: Isotype. Occurrence: occurrenceRemarks: Isolated from burned forest soil; occurrenceID: 9A5F7D25-49A6-5CC9-925B-595C9BB01673; Taxon: scientificName: Drechslerelladaliensis; kingdom: Fungi; phylum: Ascomycota; class: Orbiliomycetes; order: Orbiliales; family: Orbiliaceae; genus: Drechslerella; specificEpithet: daliensis; taxonRank: Species; Location: country: China; countryCode: CHN; stateProvince: Yunnan Province; county: Dali; locationRemarks: China, Yunnan Province, Dali City, burned forest soil; Identification: identifiedBy: Fa Zhang; Record Level: language: English; collectionID: DLU22-1
Description
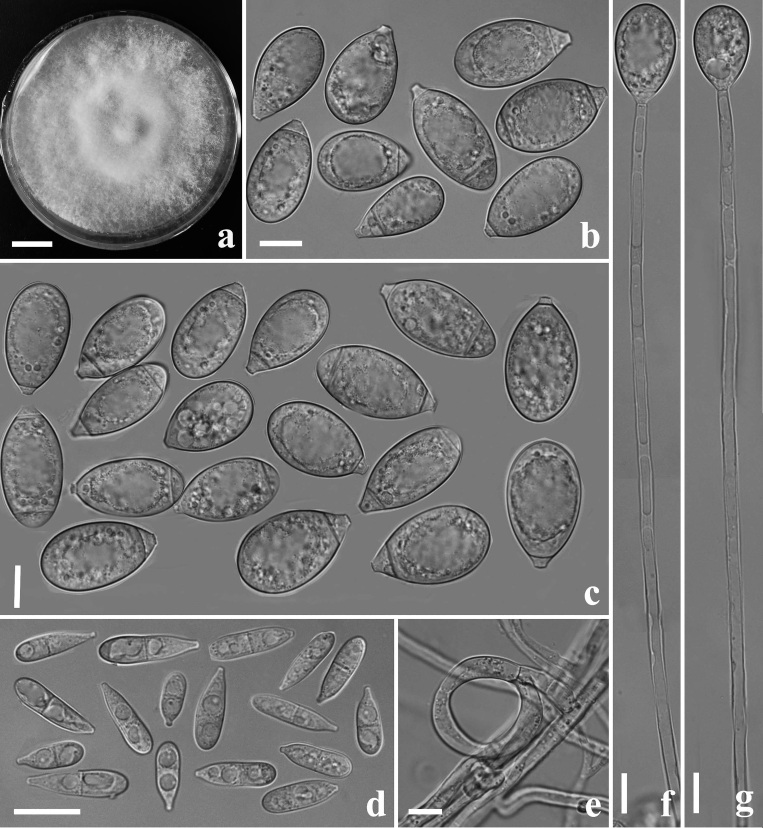
Colonies white, cottony, slow-growing on PDA medium, reaching 50 mm diameter after 18 days at 26°C. Mycelium hyaline, septate, branched, smooth. Conidiophores 125–335 µm (x̅ = 216.5 µm, n = 50) long, 3–6.5 µm (x̅ = 4.5 µm, n = 50) wide at the base, 2–3.5 µm (x̅ = 3 µm, n = 50) wide at the apex, hyaline, erect, septate, unbranched, bearing a single conidium at the apex. Conidia two types: Macroconidia 20–49.5 × 8.5–15 µm (x̅ = 38.5–12 µm, n = 50), hyaline, smooth, ellipsoid, broadly rounded at the apex, truncate at the base, 1–2-septate, mostly 2-septate. Microconidia 6.5–22 × 3.5–7 µm (x̅ = 15.5–5 µm, n = 50), hyaline, smooth, clavate or bottle-shaped, broadly rounded at the apex, truncate at the base, 0–1-septate. Chlamydospores not observed. Capturing nematodes with three-celled constricting rings, in the non-constricted state, the outer diameter is 21–32 µm (x̅ = 26 µm, n = 50), the inner diameter is 12–21 µm (x̅ = 15.5 µm, n = 50), stalks 5.5–11 µm (x̅ = 8.5µm, n = 50) long and 4–6.5 µm (x̅ = 5µm, n = 50) wide (Fig. 1).
Figure 1.
Drechslerelladaliensis (holotype, CGMCC3.20131). a Culture colony; b, c Macroconidia; d Microconidia; e Constricting rings; f, g Conidiophores. Scale bars: a = 1 cm; b–g = 10 µm.
Diagnosis
D.daliensis differs from D.hainanensis by its thinner macroconidia and shorter microconidia.
Etymology
The species name “daliensis” refers to the locality (Dali) of the type specimen.
Distribution
China, Yunnan Province, Dali City, from burned forest soil.
Drechslerella xiaguanensis
Fa Zhang, Xiao-Yan Yang, Kevin D. Hyde sp. nov.
5D097623-BB9F-5713-85A4-A28DD1A7E6A1
http://www.indexfungorum.org/Name:IF558121
Facesoffungi number: FOF10566
Materials
Type status: Holotype. Occurrence: occurrenceRemarks: Isolated from burned forest soil; occurrenceID: 7D732B1B-4091-549C-97B3-64CC0D42FFC0; Taxon: scientificName: Drechslerellaxiaguanensis; kingdom: Fungi; phylum: Ascomycota; class: Orbiliomycetes; order: Orbiliales; family: Orbiliaceae; genus: Drechslerella; specificEpithet: xiaguanensis; taxonRank: Species; scientificNameAuthorship: Fa Zhang, Xiao-Yan Yang, Kevin D. Hyde; Location: country: China; countryCode: CHN; stateProvince: Yunnan; county: Dali; locationRemarks: China, Yunnan Province, Dali City, Cangshan Mountain, burned forest soil, 25 July 2017; Identification: identifiedBy: Fa Zhang; Record Level: language: English; collectionID: CGMCC3.20132
Type status: Isotype. Occurrence: occurrenceRemarks: Isolated from burned forest soil; occurrenceID: A14AF229-0901-5266-92D8-8950A34DCCDF; Taxon: scientificName: Drechslerellaxiaguanensis; kingdom: Fungi; phylum: Ascomycota; class: Orbiliomycetes; order: Orbiliales; family: Orbiliaceae; genus: Drechslerella; specificEpithet: xiaguanensis; taxonRank: Species; Location: country: China; countryCode: CHN; stateProvince: Yunnan Province; county: Dali; locationRemarks: China, Yunnan Province, Dali City, Cangshan Mountain, burned forest soil; Identification: identifiedBy: Fa Zhang; Record Level: language: English; collectionID: DLU23-1
Description
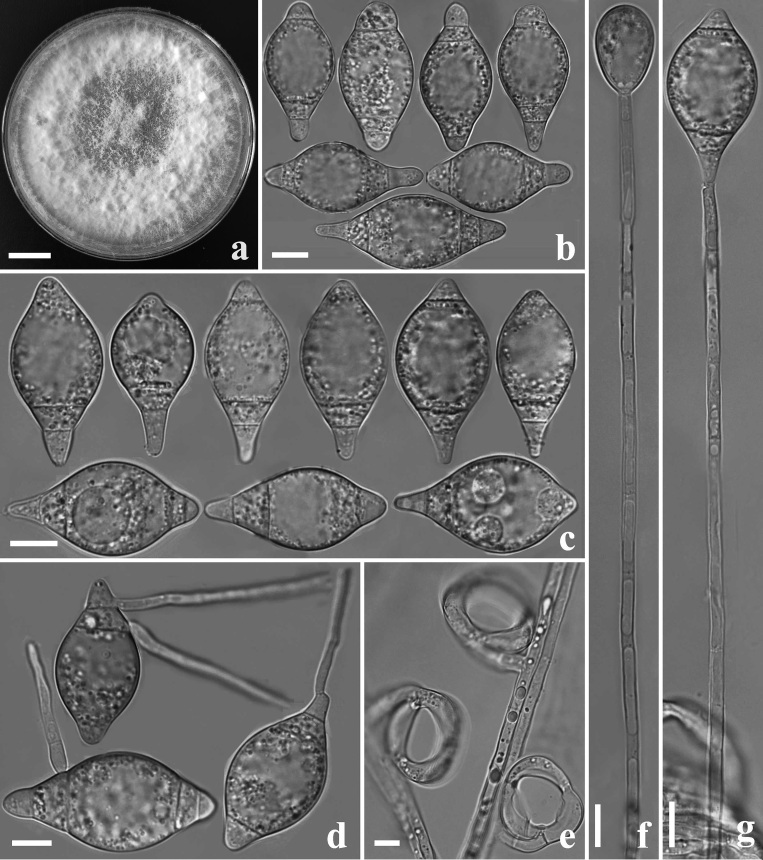
Colonies white, cottony, slow-growing on PDA medium, reaching 50 mm diameter after 15 days at 26°C. Mycelium hyaline, smooth, septate, branched. Conidiophores 145–315 µm (x̅ = 208.5 µm, n = 50) long, 3–6 µm (x̅ = 4 µm, n = 50) wide at the base, 2–3 µm (x̅ = 2.5 µm, n = 50) wide at the apex, hyaline, erect, septate, unbranched, bearing a single conidium at the apex. Conidia 33–52 × 9.5–28 µm (x̅ = 42.5–15.5 µm, n = 50), hyaline, smooth, fusiform, spindle-shaped, rounded and swollen at the both ends, 2–4-septate, mostly 3-septate, germinating tubes produced from both ends. Chlamydospores not observed. Capturing nematodes with three-celled constricting rings, in the non-constricted state, the outer diameter is 19–27.5 µm (x̅ = 24 µm, n = 50), the inner diameter is 12.5–20.5 µm (x̅ = 17 µm, n = 50), stalks 5–11.5 µm (x̅ = 9 µm, n = 50) long and 4.5–6 µm (x̅ = 5 µm, n = 50) wide (Fig. 2).
Figure 2.
Drechslerellaxiaguanensis (holotype, CGMCC3.20132). a Culture colony; b, c Conidia; d Germinating conidia; e Constricting rings; f, g Conidiophore. Scale bars: a = 1 cm; b–g = 10 µm.
Diagnosis
D.xiaguanensis differs from D.aphrobrocha by its smaller conidia and swollen cells at both ends of conidia.
Etymology
The species name “xiaguanensis” refers to the locality (Xiaguan) of the type specimen.
Distribution
China, Yunnan Province, Dali City, Cangshan Mountain, from burned forest soil.
Analysis
Phylogenetic analyses
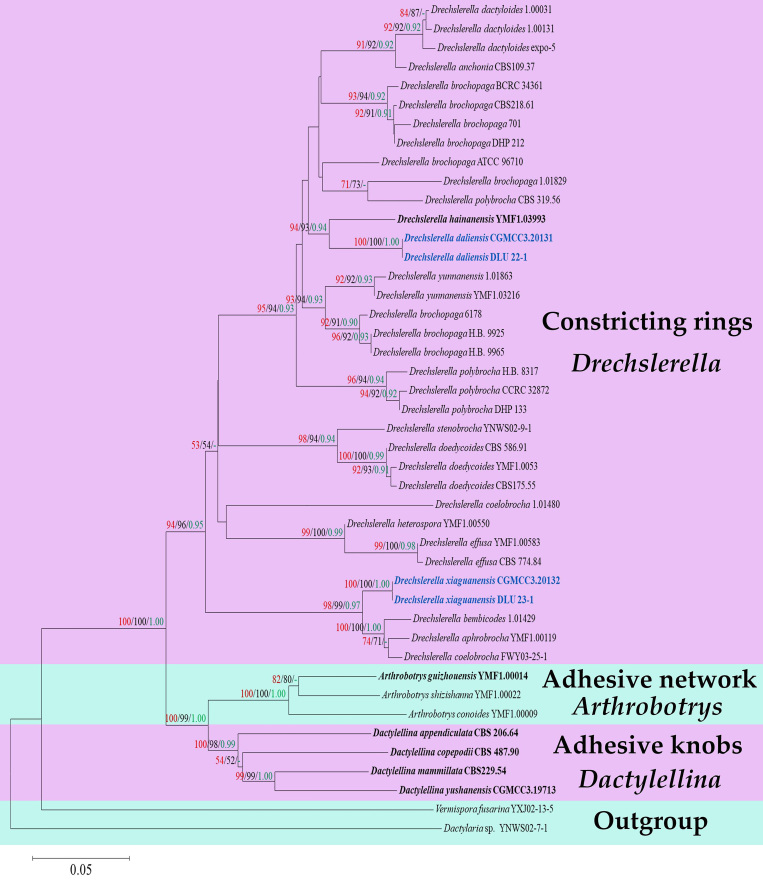
A total of 15 Drechslerella related taxa were listed in Index Fungorum (http://www.indexfungorum.org; accessed on 16 August 2022) and Species Fungorum (http://www.speciesfungorum.org; accessed on 16 August 2022), representing 15 valid Drechslerella species. Amongst them, 13 species have available molecular data. The combined ITS, TEF1-α and RPB2 sequence dataset contained 42 nematode-trapping taxa in Orbiliaceae (3 Arthrobotrys species, 4 Dactylellina species and 35 Drechslerella taxa representing 15 species). The final dataset comprised 1939 characters (ITS = 591, TEF1-α = 534 and RPB2 = 814), including 807 conserved characters, 1072 variable characters and 748 parsimony-informative characters. After Maximum Likelihood (ML) analysis, a best-scoring likelihood tree was obtained with a final ML optimisation likelihood value of -7146.589745. For Bayesian analysis (BI), the first 25% of trees were discarded in a burn-in period, the consensus tree was calculated with the remaining trees and the Bayesian posterior probabilities were evaluated with a final average standard deviation of the split frequency of 0.009547. Within Maximum Parsimony (MP) analysis, a strict consensus tree was obtained from the two equally most parsimonious trees (TL = 2817, CI = 0.471, RI = 0.514, RC = 0.296, HI = 0.404). The trees inferred by ML, MP and BI showed similar topologies. Therefore, the best-scoring ML tree was selected for presentation (Fig. 3).
Figure 3.
Maximum Likelihood tree, based on combined ITS, TEF1-α and RPB2 sequence data from 42 nematode-trapping taxa in Orbiliaceae. Bootstrap support values for Maximum Parsimony (red) and Maximum Likelihood (black) equal or greater than 50% and Bayesian posterior probabilities values (green) greater than 0.90 are indicated above the nodes. New isolates are in blue, ex-type strains are in bold.
The phylogram inferred from the ITS+TEF1-α+RPB2 dataset clustered 42 Orbiliaceae nematode-trapping fungi into two large clades according to their mechanisms for catching nematodes: 1) The genus Drechslerella that captures nematodes by mechanical force (Zhang and Hyde 2014); 2) The genera Arthrobotrys and Dactylellina capture nematode by adhesive material (Zhang and Hyde 2014). Our two new species D.daliensis and D.xiaguanensis clustered in Drechslerella with high statistical support. D.daliensis forms a basal lineage closely nested with D.hainanensis (YMF1.03993) with 94% MPBS, 93% MLBS and 0.94 BYPP support. D.xiaguanensis forms a sister lineage with D.bembicodes (1.01429), D.aphrobrocha (YMF1.00119) and D.coelobrocha (FWY03-25-1) with 98% MPBS, 99% MLBS and 0.97 BYPP support (Fig. 3).
Discussion
Drechslerelladaliensis and D.xiaguanensis produce constricting rings to capture nematodes, which is consistent with the genus Drechslerella (Zhang and Hyde 2014). The multi-genes phylogenetic analysis also confirmed that they are members of Drechslerella.
Phylogenetically, D.daliensis (CGMCC3.20131) forms a sister lineage to D.hainanensis (YMF 1.03993) with 97% MLBS, 96% MPBS and 0.95 BYPP support (Fig. 3). A comparison of ITS nucleotide shows 10.15% difference (60/591 bp) between them. Morphologically, amongst 17 species in Drechslerella (plus our two new species), D.daliensis, D.effusa, D.hainanensis and D.heterospora produce ellipsoid 0–3 septate conidia (Li et al. 2013, Zhang and Hyde 2014). The difference between D.daliensis and D.effusa is that the conidiophores of D.daliensis produce only a single conidium at the apex, while the conidiophores of D.effusa usually bear two or more conidia (Zhang and Hyde 2014). D.daliensis can be easily distinguished from D.heterospora by their microconidia size and the apex characteristic of conidiophore: the microconidia of D.daliensis are significantly smaller than those of D.heterospora (6.5–22 × 3.5–7 µm vs. 23–40 × 5.3–8 µm), the conidiophores of D.heterospora usually swollen and spherical at the apex, while those of D.daliensis are not swollen. In addition, D.daliensis does not produce chlamydospores, while D.heterospora produces chlamydospores in chains (Zhang and Hyde 2014). It is challenging to distinguish D.daliensis and D.hainanensis according to their shape characteristics. The difference between them is that the macroconidia of D.daliensis are thinner than those of D.hainanensis (20–49.5 × 8.5–15 µm vs. 32.5–43 × 17–25 µm) and the microconidia are shorter than those of D.hainanensis (6.5–22 × 3.5–7 µm vs. 18.2–22.8 × 4.2–5.3 µm) (Li et al. 2013).
In the phylogenetic analysis, D.xiaguanensis (CGMCC3.20131) forms a sister lineage to D.bembicodes (1.01429), D.aphrobrocha (YMF1.00119) and D.coelobrocha (FWY03-25-1) with 100% MLBS, 100% MPBS and 1.00 BYPP support (Fig. 3). Comparison of ITS nucleotide shows 2.6% (15/577 bp), 5.2% (30/577 bp) and 3.6% (20/556 bp) between D.xiaguanensis and D.bembicodes, D.aphrobrocha and D.coelobrocha, respectively. Morphologically, they can be distinguished by their conidia size: the conidia of D.xiaguanensis are thinner than those of D.bembicodes, shorter than those of D.coelobrocha and smaller than those of D.aphrobrocha (D.xiaguanensis 33–52 (42.5) × 9.5–28 (15.5) µm vs. D.bembicodes 36–43.2 (40) × 16.8–21.6 (20.5) µm vs. D.coelobrocha 45.6–55.2 (49.5) × 16.8–21.6 (19.8) µm vs. D.aphrobrocha 40–57.5 (51) × 15.5–35 (24.6) µm). In addition, the cells at both ends of some conidia of D.xiaguanensis are swollen, while D.bembicodes, D.aphrobrocha and D.coelobrocha are not (Drechsler 1950, Zhang and Mo 2006, Zhang and Hyde 2014). Based on the above, we propose D.daliensis and D.xiaguanensis as two new species of Drechslerella.
Amongst nematode-trapping fungi, species in Arthrobotrys are the dominant group in most ecosystems due to their strong reproductive and saprophytic ability, while the species in Dactylellina and Drechslerella, with weaker competitive abilities were rare (Jaffee et al. 1998, Hao et al. 2005, Elshafie et al. 2006, Su et al. 2007, Mo et al. 2008, Yang et al. 2008, Swe et al. 2009, Wachira et al. 2009, Yang et al. 2011). However, many species of Dactylellina and Drechslerella have been isolated from the burning forest in Cangshan, Yunnan (She et al. 2020). Amongst them, two new Dactylellina species (Zhang et al. 2020) and two new Drechslerella species (this paper) have been identified. We speculate that the reasons for this unusual phenomenon may be as follows: in normal habitat, Arthrobotrys species usually occupy the main living resources and are mainly distributed in the upper soil where humus, air and space are abundant due to their strong reproductive and saprophytic ability, while those species of Dactylellina and Drechslerella are mainly distributed in the lower soil where humus is scarce. When a fire occurs, Arthrobotrys species distributed in the upper soil are more vulnerable to the fire and are wiped out and then the habitat plaques form. In contrast, the rare species distributed in the lower layer are protected by the upper soil and preserved. In the subsequent recovery stage, these species can grow in large numbers and occupy the habitat plaque to form the dominant population in the area. Based on the above, we speculate that we would find more rare nematode-trapping fungi in burned forests. In addition, according to this principle, we speculate that other saprophytic fungi also have similar laws. Further research is underway and will be reported later.
Supplementary Material
Acknowledgements
This research was supported by the National Natural Science Foundation Program of P.R. China [Project ID: 31360013, 31460015], the National Natural Science Foundation Program-Yunnan union fund [Project ID:U1602262].
Funding Statement
This research was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) [Grant No. 2019QZKK0402], the National Natural Science Foundation Program-Yunnan union fund [Project ID: U1602262].
References
- Ahren D, Ursing BM, Tunlid A. Phylogeny of nematode-trapping fungi based on 18S rDNA sequences. FEMS Microbiology Letters. 1998;158(2):179–184. doi: 10.1111/j.1574-6968.1998.tb12817.x. [DOI] [PubMed] [Google Scholar]
- Barron GL. The nematode-destroying fungi. Canadian Biological Publications Ltd; Guelph: 1977. 45 [Google Scholar]
- Burges A, Raw F. Soil Biology. Academic Press; London: 1967. [Google Scholar]
- Cooke R. C. The ecology of nematode‐trapping fungi in the soil. Annals of Applied Biology. 1962;50(3):507–513. doi: 10.1111/j.1744-7348.1962.tb06045.x. [DOI] [Google Scholar]
- Drechsler C. Several species of Dactylellina and Dactylaria that capture free-living nematode. Mycologia. 1950;42:1–79. doi: 10.1080/00275514.1950.12017816. [DOI] [Google Scholar]
- El-Borai F. E., Campos-Herrera R., Stuart R. J., Duncan L. W. Substrate modulation, group effects and the behavioral responses of entomopathogenic nematodes to nematophagous fungi. Journal of Invertebrate Pathology. 2011;106(3):347–356. doi: 10.1016/j.jip.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Elshafie A. E., Al-Mueini R, Al-Bahry S. N., Akindi A. Y., Mahmoud I, Al-Rawahi S. H. Diversity and Trapping Efficiency of Nematophagous Fungi from Oman. Phytopathologia mediterranea. 2006;45:266–270. doi: 10.1400/56490. [DOI] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Glockling S. L., Dick M. W. Dactylellamegalobrocha, a new species of nematophagous fungus with constricting rings. Mycological Research. 1994;98(8):845–853. doi: 10.1016/S0953-7562(09)80252-9. [DOI] [Google Scholar]
- Gray N. F., Bailey F. Ecology of nematophagous fungi: vertical distribution in a deciduous woodland. Plant and Soil. 1985;86(2):217–223. doi: 10.1007/bf02182896. [DOI] [Google Scholar]
- Hagedorn G., Scholler M. A reevaluation of predatory orbiliaceous fungi. I. phylogenetic analysis using rDNA sequence data. Sydowiahorn. 1999;51:27–48. [Google Scholar]
- Hall BG. Phylogenetic trees made easy: a how-to manual, 2nd ed. Journal of Heredity. 2005;96(4):469–470. doi: 10.1093/jhered/esi046. [DOI] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Aymposium Aeries. 1999;41(41):95–98. [Google Scholar]
- Hao Y. E., Mo M. H., Su H. Y., Zhang K. Q. Ecology of aquatic nematode-trapping hyphomycetes in southwestern China. Aquatic Microbial Ecology. 2005;40(2):175–181. doi: 10.3354/ame040175. [DOI] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jaffee B., Tedford E., Muldoon A. Tests for density-dependent parasitism of nematodes by nematode-trapping and endoparasitic fungi. Biological Control. 1993;3(4):329–336. doi: 10.1006/bcon.1993.1043. [DOI] [Google Scholar]
- Jaffee B., Ferris H., Scow K. Nematode-trapping fungi in organic and conventional cropping systems. Phytopathology. 1998;88(4):344–350. doi: 10.1094/PHYTO.1998.88.4.344. [DOI] [PubMed] [Google Scholar]
- Jansson H. B., Autery C. L. An Arthrobotrys from brackish water. Mycologia. 1961;53:432–433. doi: 10.2307/3756586. [DOI] [Google Scholar]
- Jeewon R., Liew E. C., Hyde K. D. Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Molecular Phylogenetics and Evolution. 2002;25(3):378–392. doi: 10.1016/S1055-7903(02)00422-0. [DOI] [PubMed] [Google Scholar]
- Kumar N., Singh R., Singh K. Occurrence and colonization of nematophagous fungi in different substrates, agricultural soils and root galls. Archives of Phytopathology and Plant Protection. 2011;44(12):1182–1195. doi: 10.1080/03235408.2010.484945. [DOI] [Google Scholar]
- Li J., Qian W., Qiao M., Bai Y., Yu Z. A new Drechslerella species from Hainan, China. Mycotaxon. 2013;125:183–188. doi: 10.5248/125.183. [DOI] [Google Scholar]
- Linford M., Yap F., Oliveira J. M. Reduction of soil populations of the root-knot nematode during decomposition of organic matter. Soil Science. 1938;45(2):127–142. doi: 10.1097/00010694-193802000-00004. [DOI] [Google Scholar]
- Liou G. Y., Tzean S. S. Phylogeny of the genus Arthrobotrys and allied nematode-trapping fungi based on rDNA sequences. Mycologia. 1997;89(6):876–884. doi: 10.1080/00275514.1997.12026858. [DOI] [Google Scholar]
- Liu SR, Su HY, Su XJ, Zhang F, Liao GH, Yang XY. Arthrobotrysxiangyunensis, a novel nematode-trapping taxon from a hot-spring in Yunnan Province, China. Phytotaxa. 2014;174(2):89–96. doi: 10.11646/phytotaxa.174.2.3. [DOI] [Google Scholar]
- Liu X. Z., Zhang K. Q. Nematode-trapping species of Monacrosporium with special reference to two new species. Mycological Research. 1994;98(8):862–868. doi: 10.1016/S0953-7562(09)80255-4. [DOI] [Google Scholar]
- Liu X. Z., Xiang M., Che Y. The living strategy of nematophagous fungi. Mycoscience. 2009;50(1):20–25. doi: 10.1007/S10267-008-0451-3. [DOI] [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16(12):1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Li Y., Hyde K. D., Jeewon R., Cai L., Vijaykrishna D., Zhang K. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia. 2005;97(5):1034–1046. doi: 10.1111/j.1574-6968.1998.tb12817.x. [DOI] [PubMed] [Google Scholar]
- Li Yan, Jeewon Rajesh, Hyde Kevin D, Mo Ming-He, Zhang Ke-Qin. Two new species of nematode-trapping fungi: relationships inferred from morphology, rDNA and protein gene sequence analyses. Mycological Research. 2006;110(Pt 7):790–800. doi: 10.1016/j.mycres.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Madeira F, Pearce M, Tivey A. R.N, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Research. 2022;50 doi: 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop. 2010:1–8. doi: 10.1109/GCE.2010.5676129. [DOI]
- Mo M. H., Chen W. M., Su H. Y., Zhang K. Q., Duan C. Q., He D. M. Heavy metal tolerance of nematode-trapping fungi in lead-polluted soils. Applied Soil Ecology. 2006;31(1–2):11–19. doi: 10.1016/j.apsoil.2005.04.008. [DOI] [Google Scholar]
- Mo M. H., Chen W. M., Yang H. R., Zhang K. Q. Diversity and metal tolerance of nematode-trapping fungi in Pb-polluted soils. The Journal of Microbiology. 2008;46(1):16. doi: 10.1007/s12275-007-0174-8. [DOI] [PubMed] [Google Scholar]
- Morton C. O., Mauchline T. H., Kerry B. R., Hirsch P. R. PCR-based DNA fingerprinting indicates host-related genetic variation in the nematophagous fungus Pochoniachlamydosporia. Mycological Research. 2003;107(2):198–205. doi: 10.1017/S0953756203007251. [DOI] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., Haeseler A., Minh B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2014;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences. 1998;95(5):2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofri S., Tosi S. Arthrobotrysferox sp. nov., a springtail-capturing hyphomycete from continental Antarctica. Mycotaxon. 1992;44(2):445–451. [Google Scholar]
- Pfister D. H.Castor. Pollux and life histories of fungi. Mycologia. 1997;89(1):1–23. doi: 10.1080/00275514.1997.12026750. [DOI] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Quijada L, Baral HO, Beltran-Tejera E, Pfister DH. Orbiliajesu-laurae (Ascomycota, Orbiliomycetes), a new species of neotropical nematode-trapping fungus from Puerto Rico, supported by morphology and molecular phylogenetics. Willdenowia. 2020;50(2):241–251. doi: 10.3372/wi.50.50210. [DOI] [Google Scholar]
- Rambaut A. FigTree v1. 3.1. http://tree. bio. ed. ac. uk/software/figtree/ Molecular evolution, phylogenetics and epidemiology. 2009
- Scholler M., Hagedorn G., Runner A. A reevaluation of predatory orbiliaceous fungi. II. A new generic concept. Sydowiahorn. 1999;51:89–113. [Google Scholar]
- She R., Zhou X. J., Wang H. Q., Zhang F., Yang X. Y., Xiao W. Succession of soil nematode-trapping fungi following fire disturbance in forest. Journal of Forest Research. 2020;25(6):433–438. doi: 10.1080/13416979.2020.1793465. [DOI] [Google Scholar]
- Smith M. E., Jaffee B. A. PCR primers with enhanced specificity for nematode-trapping fungi (Orbiliales) Microbial Ecology. 2009;58(1):117–128. doi: 10.1007/s00248-008-9453-0. [DOI] [PubMed] [Google Scholar]
- Subramanian C. V. Dactylella, Monacrosporium and Dactylina. Journal of the Indian Botanical Society. 1963;42:291–300. [Google Scholar]
- Su H. Y, Hao Y. E, Mo M,H, Zhang K. Q. The ecology of nematode-trapping hyphomycetes in cattle dung from three plateau pastures. Veterinary Parasitology. 2007;144:293–298. doi: 10.1016/j.vetpar.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Swe A., Jeewon R., Hyde K. D. Nematode-trapping fungi from mangrove habitats. Cryptogamie. 2008;29(4):333. [Google Scholar]
- Swe Aung, Jeewon Rajesh, Pointing S. B., Hyde K. D. Diversity and abundance of nematode-trapping fungi from decaying litter in terrestrial, freshwater and mangrove habitats. Biodiversity and Conservation. 2009;18(6):1695–1714. doi: 10.1007/s10531-008-9553-7. [DOI] [Google Scholar]
- Swe A., Li J., Zhang K. Q., Pointing S. B., Jeewon R., Hyde K. D. Nematode-trapping fungi. Current Research in Environmental and Applied Mycology. 2011;1(1):1–26. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulzurrun G., Hsueh Y. P. Predator-prey interactions of nematode-trapping fungi and nematodes: both sides of the coin. Applied Microbiol Biotechnol. 2018;102:3939–3949. doi: 10.1007/s00253-018-8897-5. [DOI] [PubMed] [Google Scholar]
- Vilela VLR, Feitosa TF, Braga FR, Araujo JV, Oliveira SDV, Silva SHE, Athayde ACR. Biological control of goat gastrointestinal helminthiasis by Duddingtoniaflagrans in a semi-arid region of the northeastern Brazil. Veterinary Parasitology. 2012;188(1-2):127–133. doi: 10.1016/j.vetpar.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Vu D, Groenewald M, Vries M De, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PE, Robert V, Verkley GJM. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachira P., Mibey R., Okoth S., Kimenju J., Kiarie J. Diversity of nematode destroying fungi in Taita Taveta, Kenya. Fungal Ecology. 2009;2(2):60–65. doi: 10.1016/j.funeco.2008.11.002. [DOI] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;18(1):315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- Wolstrup J., Nansen P., Gronvold J., Henriksen S., Larsen M. Toward practical biological control of parasitic nematodes in domestic animals. Journal of Nematology. 1996;28(2):129. [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Kim D. G., Ryu Y. H., Zhou X. B. Arthrobotryskoreensis, a new nematode-trapping species from Korea. Sydowia. 2012;64(1):129–136. [Google Scholar]
- Yang H., Yang D, Zhou J. Diversity of nematode-trapping fungi in soil of eucalyptus. Journal-Yunnan university natural sciences. 2008;30(1):101. [Google Scholar]
- Yang X. Y, Liu L. P, Su X. J, Ye Y. B, Huang A. Y, Su H. Y. Study on the biological diversity of nematode-trapping fungi in Erhai Lake. Agricultural Science & Technology. 2011;12(8):1100–1102. [Google Scholar]
- Yang Y., Yang E., An Z., Liu X. Z. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proceedings of the National Academy of sciences. 2007;104(20):8379–8384. doi: 10.1073/pnas.0702770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Qin L., Zhang Y., Qiao M., Kong Y., Zhang K. A new Drechslerella species isolated from Orbiliacf.orientalis. Mycotaxon. 2009;110(1):253–259. doi: 10.5248/110.253. [DOI] [Google Scholar]
- Zhang F., Zhou X. -J., Monkai J., Li F. T., Liu S. -R., Yang X. -Y., Wen X., Hyde K. D. Two new species of nematode-trapping fungi (Dactylellina, Orbiliaceae) from burned forest in Yunnan, China. Phytotaxa. 2020;452(1):65–74. doi: 10.11646/phytotaxa.452.1.6. [DOI] [Google Scholar]
- Zhang F., Boonmee S., Bhat J. D., Xiao W., Yang X. Y. New Arthrobotrys nematode-trapping species (Orbiliaceae) from terrestrial soils and freshwater sediments in China. Journal of Fungi. 2022;8(7):671. doi: 10.3390/jof8070671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. X, Wei Z. Y, Zhang J, Liu X. F. Classification of dendrocola nematode-trapping fungi. Journal of Forestry Research. 2020;32(3):1295–1304. doi: 10.1007/s11676-020-01159-x. [DOI] [Google Scholar]
- Zhang K. Q., Mo M. H. Flora fungorum sinicorum (Vol. 33): Arthrobotrys et gengra cetera cognata. Science Press; Beijin: 2006. 97-104. Chinese. [Google Scholar]
- Zhang K. Q., Hyde K. D. Nematode-trapping fungi. Springer Science & Business; Berlin: 2014. [DOI] [Google Scholar]
- Zhang Y., Li S., Li H., Wang R., Zhang K. -Q., Xu J. Fungi-nematode interactions: diversity, ecology, and biocontrol prospects in agriculture. Journal of Fungi. 2020;6(4) doi: 10.3390/jof6040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qiao M., Baral H. O., Xu J., Zhang K. Q., Yu Z. F. Morphological and molecular characterization of Orbiliapseudopolybrocha and O.tonghaiensis, two new species of Orbiliaceae from China. International Journal of Systematic and Evolutionary Microbiology. 2020;70(4):2664–2676. doi: 10.1099/ijsem.0.004088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.