Abstract
Background
Freshwater fungi play an indispensable role in the ecosystem and have great research value. Based on morphological and phylogenetic analyses of a concatenated dataset of ITS, LSU and SSU sequences, a new species, Phaeoisarialaianensis, was introduced as a freshwater hyphomycete from Anhui Province, China.
New information
Phaeoisarialaianensis was morphologically described as erect, rigid, dark brown to black, velvety synnemata which has macronematous, septate, branched, brown to dark brown, parallel adpressed conidiophores with polyblastic, integrated, terminal, hyaline to pale brown, smooth, denticulate, sympodial conidiogenous cells and ellipsoidal to obovoid, rounded at the apex, obtuse and tapering towards base, septate, guttulate conidia. Based on molecular and morphological characteristics, it is confirmed to be a new species. All illustrations and descriptions have been provided.
Keywords: Ascomycota, Phaeoisaria , morphology, phylogenetic anaysis, taxonomy
Introduction
Phaeoisaria (Pleurotheciales) was established by Höhnel (1909) to accommodate Phaeoisariabambusae as the type species, a hyphomycetous taxon isolated from a bamboo substrate. This genus is characterised by indeterminate synnemata with parallel adpressed conidiophores with numerous sympodially extending denticulate conidiogenous cells and aseptate or septate ellipsoidal, obovoidal, fusiform-cylindrical to falcate, hyaline conidia (Höhnel 1909, Réblová et al. 2016, Hyde et al. 2018, Luo et al. 2018, Boonmee et al. 2021). Nevertheless, indeterminate synnemata have not been observed in some species, such as P.curvata (de Hoog and Papendorf 1976), P.glauca (de Hoog and Papendorf 1976), P.loranthacearum (Crous et al. 2015), P.fasciculata (Réblová et al. 2016), P.annesophieae (Crous et al. 2017) and P.dalbergiae (Crous et al. 2021).
In the past decades, an increasing number of new species was assigned to Phaeoisaria by distinguishing characters (Crous et al. 2017, Hyde et al. 2018, Hyde et al. 2019, Luo et al. 2019, Boonmee et al. 2021, Crous et al. 2021). Until now, 26 species have been accepted in the genus Phaeoisaria (http://www.speciesfungorum.org/Names/Names.asp). These species are relatively common and have a worldwide distribution, while only four of them have been recorded in China. Moreover, there are presently only 15 species having the molecular data in Phaeoisaria. In this study, we depicted a new species, Phaeoisarialaianensis, from submerged wood in Anhui Province of China, with both morphological examination and molecular phylogenetic analysis.
Materials and methods
Samples collection, specimen examination and isolation
Submerged rotting wood samples were gathered from Laian County, Anhui Province, China and were brought back to the laboratory to be incubated in plastic boxes at room temperature. Fungi on the host surface were observed with a Nikon SMZ-1270 microscope (Nikon Corporation, Japan) and morphologically photographed with a Nikon ECLIPSE Ni-U compound microscope (Nikon Corporation, Japan), which was equipped with a Nikon DS-Fi3 camera. The structure of fungi was determined by PhotoRuler 1.1.3.0 (The Genus Inocybe, Hyogo, Japan) and figures were processed by Adobe Photoshop 2020 (Adobe Systems, USA). According to the method of Li et al. (2021), single spore isolation and pure culture were carried out. Fungal specimens were deposited in the Fungus Herbarium, Jiangxi Agricultural University, Nanchang, China.
DNA extraction, PCR amplification and sequencing
By using the improved CTAB method (Doyle and Doyle 1987), fungal total genomic DNA was extracted from fresh mycelium. Three gene regions (ITS, LSU and SSU), were respectively amplified by polymerase chain reaction (PCR) using the primers of ITS1/ITS4 (White et al. 1990), LROR/LR7 (Hopple and Vilgalys 1999) and NS1/NS4 (White et al. 1990), with 25 μl of the final volume including 9.5 μl ddH2O, 12.5 μl 2× Taq PCR MasterMix (Qingke, Changsha, China), 1 μl of DNA template and 1 μl of each primer (10 μM). Then amplifications were conducted under the PCR conditions described by Zhai et al. 2022. The PCR products were purified and the sequencing reactions were commercially conducted with the corresponding forward and reverse primers by QingKe Biotechnology Co. (Changsha, China). All sequences were edited with SeqMan v. 7.1.0 (DNASTAR, lnc, Madison, WI) and were deposited in the NCBI GenBank database.
Phylogenetic analysis
The sequences of 69 strains were retrieved from recent articles (Luo et al. 2018, Hyde et al. 2019, Boonmee et al. 2021) and downloaded from GenBank (Table 1). Each matrix of ITS, LSU and SSU was aligned using the online service of MAFFT v.7 (http://mafft.cbrc.jp/alignment/server/large.html, Katoh et al. 2019) and then the sequences of three regions were concatenated by PhyloSuite v.1.2.2 (Zhang et al. 2020). By using RAxML v.7.2.6 (Stamatakis and Alachiotis 2010), Maximum Likelihood (ML) analysis was performed, which used a GTRGAMMA substitution model with 1000 bootstrap replicates. The Markov Chain Monte Carlo (MCMC) method in MrBayes was used to estimate the posterior probabilities (PP) (Zhaxybayeva and Gogarten 2002) and it was set as four chains (2 hot chains and 2 cold chains) running 2,000,000 generations synchronously, resulting in 40002 trees in total. Based on the initial 25% of sampled data being cut off as burn-in, PhyloSuite v.1.2.2 (Zhang et al. 2020) was used to infer Bayesian inference phylogeny under the JC+I+G+F model of the concatenation of ITS, LSU and SSU. After visualisation by FigTree v.1.4.4 (Rambaut 2018), the phylogenetic tree was edited and illustrated using Adobe Illustrator 2020 (Adobe Systems Inc., USA). The aligned matrices and trees were submitted to TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S29791).
Table 1.
Sequences used in this study. Note: Ex-type strains are in bold. The sequences of new species are indicated as underlined and unavailable sequences in GenBank are indicated by hyphen "-".
| Taxonomy | Strain | GenBank accession numbers | ||
| ITS | LSU | SSU | ||
| Adelosphaeriacatenata | CBS 138679 | KT278721 | KT278707 | KT278692 |
| Ascotaiwaniafusiformis | MFLUCC 15-0625 | – | KX550894 | KX550898 |
| Ascotaiwaniafusiformis | MFLU 15-1156 | MG388215 | NG–057114 | – |
| Ascotaiwanialignicola | NIL 00005 | HQ446341 | HQ446364 | HQ446284 |
| Ascotaiwaniasawadae | SS00051 | HQ446340 | HQ446363 | HQ446283 |
| Bactrodesmiastrumobovatum | FMR 6482 | FR870264 | FR870266 | – |
| Bactrodesmiastrumpyriforme | FMR 10747 | FR870263 | FR870265 | – |
| Brachysporiellasetosa | HKUCC 3713 | – | AF132334 | – |
| Canalisporiumexiguum | SS 00809 | GQ390296 | GQ390281 | GQ390266 |
| Canalisporiumgrenadoideum | BCC 20507 | – | GQ390267 | GQ390252 |
| Canalisporiumpulchrum | SS03982 | GQ390292 | GQ390277 | GQ390262 |
| Conioscyphalignicola | CBS 335.93 | – | AY484513 | JQ437439 |
| Conioscyphaminutispora | CBS 137253 | – | MH878131 | – |
| Conioscyphaperuviana | CBS 137657 | – | KF781539 | – |
| Conioscyphavaria | CBS 113653 | – | AY484512 | AY484511 |
| Fuscosporellapyriformis | MFLUCC 16-0570 | MG388217 | KX550896 | KX550900 |
| Helicoonfarinosum | DAOM 241947 | JQ429145 | JQ429230 | – |
| Leotialubrica | AFTOLID 1 | DQ491484 | AY544644 | AY544746 |
| Melanotrigonumovale | CBS 138815 | KT278722 | KT278711 | KT278698 |
| Melanotrigonumovale | CBS 138744 | KT278725 | KT278710 | KT278697 |
| Melanotrigonumovale | CBS 138743 | KT278724 | KT278709 | KT278696 |
| Melanotrigonumovale | CBS 138742 | KT278723 | KT278708 | KT278695 |
| Microglossumrufum | OSC100641 | – | DQ470981 | DQ471033 |
| Mucisporaobscuriseptata | MFLUCC 15-0618 | MG388218 | KX550892 | KX550897 |
| Parafuscosporellamoniliformis | MFLUCC 15-0626 | MG388219 | KX550895 | KX550899 |
| Phaeoisariaannesophieae | CBS 143235 | MG022180 | MG022159 | – |
| Phaeoisariaannesophieae | MFLU190531 | MT559109 | MT559084 | – |
| Phaeoisariaaquatica | MFLUCC 16-1298 | MF399237 | MF399254 | – |
| Phaeoisariaclematidis | MFLUCC 16-1273 | MF399229 | MF399246 | – |
| Phaeoisariaclematidis | MFLUCC 17-1341 | MF399230 | MF399247 | MF399216 |
| Phaeoisariaclematidis | MFLUCC 17-1968 | MG837022 | MG837017 | MG837027 |
| Phaeoisariaclematidis | DAOM 226789 | JQ429155 | JQ429231 | JQ429243 |
| Phaeoisariadalbergiae | CPC 39540 | OK664703 | – | – |
| Phaeoisariafasciculata | CBS 127885 | KT278719 | KT278705 | KT278693 |
| Phaeoisariafasciculata | DAOM 230055 | KT278720 | KT278706 | KT278694 |
| Phaeoisariafiliformis | MFLUCC 18-0214 | MK878381 | MK835852 | MK834785 |
| Phaeoisariaguttulata | MFLUCC 17-1965 | MG837021 | MG837016 | MG837026 |
| Phaeoisarialaianensis | CCTCC AF 2022069 | ON937559 | ON937557 | ON937562 |
| Phaeoisarialaianensis | CCTCC AF 2022073 | ON937560 | ON937561 | ON937558 |
| Phaeoisarialoranthacearum | CBS 140009 | KR611888 | MH878676 | – |
| Phaeoisarialoranthacearum | BYCDW25 | MG820097 | – | – |
| Phaeoisarialoranthacearum | BYCDW24 | MG820098 | – | – |
| Phaeoisariamicrospora | MFLUCC 16-0033 | MF671987 | MF167351 | – |
| Phaeoisariapseudoclematidis | MFLUCC 11-0393 | KP744457 | KP744501 | KP753962 |
| Phaeoisariasedimenticola | CGMCC3.14949 | JQ074237 | JQ031561 | – |
| Phaeoisariasedimenticola | S-908 | MK878380 | MK835851 | – |
| Phaeoisariasiamensis | MFLUCC 16-0607 | MK607610 | MK607613 | MK607612 |
| Phaeoisariasparsa | FMR 11939 | – | HF677185 | – |
| Phaeoisariasynnematica | NFCCI 4479 | MK391494 | MK391492 | – |
| Phragmocephalastemphylioides | KAS 4277 | KT278730 | KT278717 | – |
| Plagiascomafrondosum | CBS 139031 | – | KT278713 | KT278701 |
| Pleurotheciellacentenaria | DAOM 229631 | JQ429151 | JQ429234 | JQ429246 |
| Pleurotheciellarivularia | CBS 125237 | JQ429161 | JQ429233 | JQ429245 |
| Pleurotheciellarivularia | CBS 125238 | JQ429160 | JQ429232 | JQ429244 |
| Pleurotheciellauniseptata | KUMCC 15-0407 | MF399231 | MF399248 | – |
| Pleurotheciumaquaticum | MFLUCC 17-1331 | MF399245 | MF399263 | – |
| Pleurotheciumaquaticum | MFLUCC 21-0148 | OM654775 | OM654772 | OM654807 |
| Pleurotheciumfloriforme | MFLUCC 15-0628 | KY697281 | KY697277 | KY697279 |
| Pleurotheciumobovoideum | CBS 209.95 | EU041784 | EU041841 | – |
| Pleurotheciumpulneyense | MFLUCC 16-1293 | – | MF399262 | MF399228 |
| Pleurotheciumrecurvatum | CBS 138747 | KT278728 | KT278714 | KT278703 |
| Pleurotheciumrecurvatum | CBS 131272 | JQ429149 | JQ429237 | JQ429251 |
| Pleurotheciumrecurvatum | CBS 101581 | JQ429148 | AF261070 | JQ429248 |
| Pleurotheciumsemifecundum | CBS 131482 | JQ429158 | JQ429239 | JQ429253 |
| Pleurotheciumsemifecundum | CBS 131271 | JQ429159 | JQ429240 | JQ429254 |
| Savoryellalongispora | SAT 00322 | HQ446359 | HQ446380 | HQ446302 |
| Savoryellapaucispora | SAT 00866 | – | HQ446381 | HQ446303 |
| Savoryellaverrucosa | SS 00052 | HQ446353 | HQ446374 | HQ446296 |
| Sterigmatobotrysmacrocarpa | DAOM 230059 | JQ429154 | GU017316 | – |
| Sterigmatobotrysmacrocarpa | PRM 915682 | JQ429153 | GU017317 | JQ429255 |
| Sterigmatobotrysrudis | DAOM 229838 | JQ429152 | JQ429241 | JQ429256 |
Taxon treatments
Phaeoisaria laianensis
Y. Liu, G.P. Xu, X.Y. Yan, D.M. Hu & Z.J. Zhai sp. nov.
2EE81D2F-85F9-51D4-9628-863AEF4186AB
844773
Materials
Type status: Holotype. Occurrence: recordedBy: Yu Liu; occurrenceID: 99B9C819-CA87-5634-AB08-7E7A79E1ADE0; Taxon: scientificName: Phaeoisarialaianensis; acceptedNameUsage: Phaeoisarialaianensis Y. Liu, D.M. Hu & Z.J. Zhai; kingdom: Fungi; phylum: Ascomycota; class: Sordariomycetes; order: Pleurotheciales; family: Pleurotheciaceae; genus: Phaeoisaria; specificEpithet: laianensis; taxonRank: species; verbatimTaxonRank: species; scientificNameAuthorship: Y. Liu, D.M. Hu & Z.J. Zhai; Location: continent: Asia; country: China; stateProvince: Anhui; county: Laian; locality: Wawuzhuang; verbatimElevation: 35; locationRemarks: Label transliteration; verbatimCoordinates: 32.66 N, 118.65 E; verbatimLatitude: 32.66; verbatimLongitude: 118.65; Identification: identifiedBy: Yu Liu and Zhi-jun Zhai; dateIdentified: 2021; Event: samplingProtocol: collecting; eventDate: 06-05-2021; year: 2021; month: 5; day: 6; habitat: Freshwater; Record Level: type: PhysicalObject; language: en; rightsHolder: Dian-Ming Hu and Zhi-jun Zhai; institutionID: HFJAU10040; collectionID: LKJ17; institutionCode: the Herbarium of Fungi, Jiangxi Agricultural University (HFJAU); collectionCode: Fungi; ownerInstitutionCode: HFJAU
Description
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: Undetermined. Asexual morph: Colonies effuse, solitary, scattered, dark brown to black, hairy, covered by white conidial mass. Mycelium partly superficial, partly immersed. Synnemata 290–848 × 9.3–30.7 µm (x̅ = 532 × 18.6, SD = 159 × 5, n = 20), erect, rigid, dark brown to black, velvety, smooth, composed of compactly and parallel adpressed conidiophores. Conidiophores 116.2–491.1 × 2–3.2 µm (x̅ = 276.1 × 2.4, SD = 96.7 × 0.5, n = 10), macronematous, synnematous, septate, branched, brown to dark brown, smooth. Conidiogenous cells 8.3–27.5 × 2.3–3.8 µm (x̅ = 17.1 × 2.7, n = 10), polyblastic, integrated, terminal, hyaline to pale brown, smooth, denticulate, sympodial, each with several denticulate conidiogenous loci, 0.8–1.6 × 0.4–0.8 µm (x̅ = 1.3 × 0.7, n = 10). Conidia 5–7.2 × 1.7–2.9 µm (x̅ = 5.9 × 1.7, SD = 0.5 × 0.3, n = 50), ellipsoidal to obovoid, straight, rounded at the apex, obtuse and tapering towards base, hyaline, aseptate, guttulate, smooth-walled. (Fig. 1).
Figure 1.

Phaeoisarialaianensis (HFJAU 10040, Holotype) a, b Colonies on wood; c, d Conidiophores; e, f Conidiogenous cells with conidia; g Germinating conidium; h Conidia; i, j Colony on PDA for 26 days from above and reverse. Scale bars: a, b = 100 µm, c, d = 50 µm, e–h = 10 µm.
Culture characteristics: Conidia germinated within 24 h in which germ tubes were produced from both ends or sides at 28℃ on PDA. The colony on PDA grows up slowly and reaches 24.5 mm in 26 days, periphery grey, surface folded, middle grey-green to black, raised with mycelium in the centre, covered with lots of white conidia, powdery, reverse grey to black.
Material examined: China, Anhui Province, alt. 35 m, near 32.66°N, 118.65°E, on decaying wood submerged in a freshwater stream, 6 May 2021, Y. Liu, G.P. Xu and Z.J. Zhai, LKJ17 (HFJAU 10040, holotype), ex-type living culture, CCTCC AF 2022069 = CCTCC AF 2022073.
Etymology
The name reflects the district where this fungus was found.
Notes
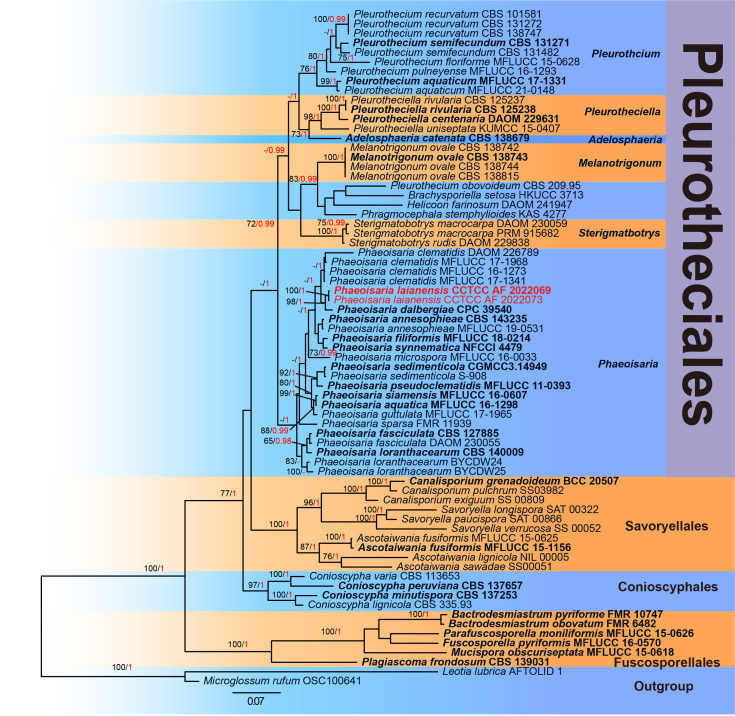
Phylogenetic analysis shows that Phaeoisarialaianensis is a phylogenetically-distinct species, most closely related to P.dalbergiae and then to P.clematidis (Fig. 2). However, P.laianensis is easily distinguished from P.dalbergiae by its ellipsoidal to obovoid, rounded at the apex and tapering towards base conidia (Crous et al. 2021). In addition, P.laianensis has synnemata, which is absent in P.dalbergiae (Crous et al. 2021), also in P.curvata, P.glauca (de Hoog and Papendorf 1976), P.loranthacearum (Crous et al. 2015), P.annesophieae and P.fasciculata (Réblová et al. 2016) (Table 2). The new species is similar to P.clematidis in having resembling synnemata or conidia (Hughes 1958, Luo et al. 2018), while the former has shorter synnemata (290–848 µm vs. 1000–1500 µm) and smaller conidia (5–7.2 µm wide vs. 4–10 µm wide) than P.clematidis (Table 2). Likewise, P.laianensis has longer synnemata than P.siameneis (290–848 µm vs. 330–380 µm), smaller conidiophores than P.guttulata (Hyde et al. 2018) and P.aquatica (116.2–491.1 × 2–3 µm vs. 480–700 × 2–5 µm and 1028–1262 × 3.5–4.5 µm) (Luo et al. 2018) and smaller conidia (5–7.2 × 1.7–2.9 µm) than P.annesophiae (4.5–9 × 2–3.5 µm) (Crous et al. 2017), P.synnematica (4–11 µm long) (Boonmee et al. 2021) and P.siamensis (3–4 µm wide) (Table 2). In addition, it can be differentiated from P.filiforms by the indeterminate asexual morph of the latter species (Luo et al. 2019).
Figure 2.
Phylogenetic tree of Bayesian analysis, based on a concatenated alignment of ITS, LSU and SSU sequences. Branch support is shown at the nodes, Maximum Likelihood bootstrap support (BS, black) ≥ 60% and Bayesian posterior probability (PP, red) ≥ 0.95. Leotialubrica (AFTOLID 1) and Microglossumrufum (OSC100641) are selected as the outgroup taxa. The new species is marked in red and ex-type strains are in bold.
Table 2.
Synopsis 1 of asexual morphological characteristics of Phaeoisaria species. Note: Hyphens “-” are indeterminate or unavailable data.
| Species | Synnemata (µm) | Synnemata characteristics | Conidiophores (µm) | Conidiophores characteristics | Conidia (µm) | References |
| Phaeoisarialaianensis | 290‒848 × 9.3‒30.7 | Erect, rigid, dark brown to black, velvety, smooth, composed of compactly and parallel adpressed conidiophores | 116.2‒491.1 × 2‒3.2 | Macronematous, synnematous, septate, branched, brown to dark brown, smooth | 5‒7.2 × 1.7‒2.9 | This study |
| P.aguilerae | - | - | - | - | 18–29.5 × 4–5 | Castañeda Ruiz et al. (2002) |
| P.annesophieae | - | - | Conidiophores indeterminate | Sometimes grouping in strands of 2–4 hyphae, a rising from aerial hyphae, cylindrical, hyaline to pale brown | 4.5–9 × 2–3.5 | Crous et al. (2017) |
| P.aquatica | - | Erect, rigid, dark brown to black, velvety, smooth | 1028–1262 × 3.5–4.5 | Macronematous, synnematous, brown to dark brown, smooth | 6.5–7.5 × 2.5–3.5 | Luo et al. (2018) |
| P.bambusae | - | Erect, rigid, dark brown toblack, velvety, smooth | - | Macronematous, synnematous, brown to dark brown, smooth | - | Höhnel (1909), Hyde et al. (2019), Luo et al. (2019), Réblová et al. (2016) |
| P.caffra | - | Synnemata composed of at least 10 adpressed hyphae | - | Conidiophores not tuberculate | 7.5–12 × 2.5–3.5 | Castañeda Ruiz et al. (2002), de Hoog and Papendorf (1976) |
| P.clavulata | - | Stiff synnemata, composed of parallel hyphae, packed with slender, curved conidiogenous cells with very thin, fragile conidiogenous rachides | - | - | 1–2 long | Castañeda Ruiz et al. (2002), de Hoog and Papendorf (1976), Mason and Ellis (1953) |
| P.clematidis | 1000–1500 × 20–80 | Conidiomata scattered, indeterminate, erect, rigid, superficial, dark brown composed of compact appressed conidiophores | 312–568 × 2.5–3.5 | Macronematous, septate, branched, brown to dark brown, smooth | 4–10 × 1.5–2.5 | Castañeda Ruiz et al. (2002), Hughes (1958), Luo et al. (2018) |
| P.curvata | - | - | Conidiophores indeterminate | - | (4–)6–8(–11) × (1–)2–3 | de Hoog and Papendorf (1976) |
| P.dalbergiae | - | - | 10–50 × 1.5–2.5 | Indeterminate, erect, subcylindrical, hyaline, smooth, 0–2-septate, unbranched or branched at apex | 0.5 µm diam, (5 –)6–7 × (1.5–)2 | Crous et al. (2021) |
| P.fasciculata | - | Synnemata absent | 25–65 × 3.0–3.5 | Macronematous, arising from brown, thick-walled cells, cylindrical, pale brown, subhyaline towards the apex, unbranched, smooth-walled | 6.0–8.0 (–9.0) × 2.0 | Réblová et al. (2016) |
| P.filiformis | - | - | - | - | - | Luo et al. (2019) |
| P.glauca | - | - | Conidiophores indeterminate | - | 2.5–3.5 × 1.6–2.2 | de Hoog and Papendorf (1976) |
| P.guttulata | - | Erect, rigid, dark brown to black, velvety, smooth, composed of compactly and parallel adpressed conidiophores | 480–700 × 2–5 | Macronematous, synnematous, erect, septate, smooth, mid-brown to dark brown | 3.5–5.5 × 2.5–4.8 | Hyde et al. (2018) |
| P.infrafertilis | - | Synnemata narrow, composed of only 5-6 brown adpressed hyphae | - | - | 19.5–22 × 2–3 | de Hoog and Papendorf (1976), Sutton and Hodges (1976) |
| P.loranthacearum | - | - | 10–30 × 2–3 | Arising from superficial hyphae, erect, solitary, branched at base or not, subcylindrical, straight to geniculate-sinuous, 1–3-septate, hyaline | (5)7– 8(9) × (1.5) 2(3) | Crous et al. (2015) |
| P.magnifica | - | Synnemata brush-like, synnemata with flaring hyphae at the tip | - | Growing well away from the column in the apical portion | 5–6.5 × 4–4.5 | de Hoog and Papendorf (1976), Deighton (1974) |
| P.microspora | 35–238 µm long, 4–31 µm wide at the base, 5–35 µm wide at the apex | Erect, straight or flexuous, dark brown at base, pale brown at apex | 25–225 × 1–3 | Macronematous, synnematous, septate, branched at the apex, smooth, pale to dark brown | 4.5–6.9 × 1.3–3.1 | Hyde et al. (2017) |
| P.muscariformis | - | - | - | - | 12–22 × 4 | Castañeda Ruiz et al. (2002), Siboe et al. (1999) |
| P.pseudoclematidis | 200–500 µm long, 40–80 µm wide at the base, 40– 60 µm wide in the middle, 20–30 µm wide at the apex | Erect, rigid, dark brown, velvety, smooth, composed of compactly and parallel adpressed conidiophores | 50–500 × 2–3 | Macronematous, synnematous, brown to dark brown, septate, branched, smooth | 5–8.5 × 3–4 | Liu et al. (2015) |
| P.sedimenticola | up to 4000 µm high or sometimes longer, 70– 90 µm wide | Erect, cylindrical to subulate, consisting of very regular, parallel, brown hyphae | aseptate (3.5–)4.5–5.5(–7.5) × (2.5–)3–4(–4.5) 1-septate (4.5–)5.5–6.5(–9) × (2–)2.5–3.5(–4.5) |
Cheng et al. (2014) | ||
| P.siamensis | 330–380 × 20–25(–30) | Conidiomata scattered, indeterminate, erect, rigid, superficial, dark brown composed of compactly appressed conidiophores | 2–2.5(–3) µm wide | Macronematous, in synnematous conidiomata, scattered, synnemata subulate or cylindrical, indeterminate, at the base 13–15 µm beneath the fertile portion with conidiogenous cells, composed of medium to dark brown, smooth, septate parallel hyphae, splaying out at the middle to apex | 5–8 × 3–4 | Hyde et al. (2019) |
| P.sparsa | - | Synnemata composed of at least 10 adpressed hyphae | - | Not tuberculate | 10–15.5 × 2.5–3.5 | de Hoog and Papendorf (1976), Sutton (1973) |
| P.sparsavar.cubensis | - | - | - | - | (4–)7– 11(–17) ×(1.5–) 2– 3(–4) | Mercado-Sierra et al. (1997), Mel’nik (2012) |
| P.synnematica | 399–960 × 12–30 | Synnematal, erect, rigid, dark brown to olivaceous brown, composed of compactly parallel appressed conidiophores, cylindrical to clavate | 1.5–960 × 1–3.5 | Macronematous to semi- macronematous, highly geniculate, dark brown to olivaceous brown, synnematous, simple to dichotomously branched, emerging out at the apex and along the sides of the upper half or two thirds of each synnema, dark brown at the base, brown to pale brown | 4–11 × 2–5 | Boonmee et al. (2021) |
| P.tuberculata | - | Synnemata composed of at least 10 adpressed hyphae | - | Conspicuously tuberculate | 8–13.5 × 1.5–2 | Castañeda Ruiz et al. (2002), Sutton (1993) |
| P.uniseptata | - | - | - | - | (3.5–) 5.5– 7.5 (–10) × 1.5–3 | de Hoog and Papendorf (1976), Mercado-Sierra (1984), Mel’nik (2012) |
| P.vietnamensis | 330–380 µm high, 20–25(– 30) µm wide at the base | - | 2–2.5(–3) µm wide | Macronematous, in synnematous conidiomata, scattered, synnemata subulate or cylindrical, indeterminate composed of medium to dark brown, smooth, septate parallel hyphae | 18.5– 23.5 × 4.5–5 | Mel’nik (2012) |
Analysis
Phylogenetic analysis
The aligned matrix for the combined analysis, ITS+LSU+SSU had 3105 bp, including ITS = 509 bp, LSU = 1172 bp and SSU = 1424 bp. No topological conflict exists between the tree generated by ML analysis and the Bayesian tree. The Bayesian tree is shown with BS and PP in Fig. 2. All 15 Phaeoisaria species in our analyses form a monophyletic group (BS/PP = 59/1.00). Most importantly, the two collections of Phaeoisarialaianensis form an independent lineage with strong support (BS/PP = 100/1.00). This lineage groups with P.dalbergiae into a highly supported clade (BS/PP = 98/1.00), which is sister to P.clematidis (BS/PP = 54/1.00). After searching of NCBIs GenBank nucleotide database based on a megablast, the ITS sequence of P.laianensis was found to share 97.46% similarity with P.dalbergiae (CPC 39540) and 96.35% similarity with P.clematidis (DAOM 226789). In addition, the sequence has nine different loci from that of P.dalbergiae and 15 different loci from that of P.clematidis.
Discussion
In our molecular phylogenetic tree, Phaeoisaria consists of 15 species and is supported as a monophyletic group (BS/PP = 59/1.00, Fig. 2). The low ML bootstrap might be due to a large number of unavailable sequences for 13 species in Phaeoisaria. However, the independent lineage of P.laianensis (BS/PP = 100/1.00, Fig. 2) is established and groups with P.dalbergiae into a highly supported clade (BS/PP = 98/1.00, Fig. 2). This clade is sister to the four collections of P.clematidis although with lower support (BS/PP = 54/1.00, Fig. 2). In addition, the morphological characters of P.laianensis can be effortlessly distinguished from P.dalbergiae and P.clematidis and other species in Phaeoisaria (Tables 2, 3). Notably, our results favour P.laianensis as a new species in the genus. However, molecular data for Phaeoisaria species require enriching to clarify more species relationships in the genus.
Table 3.
Synopsis 2 of asexual morphological characteristics of Phaeoisaria species. Note: Hyphens "-" are indeterminate or unavailable data.
| Species | Conidia septation | Conidia characteristics | Host | District | References |
| Phaeoisarialaianensis | Aseptate | Ellipsoidal to obovoid, straight, rounded at the apex, obtuse and tapering towards base, hyaline, guttulate, smooth-walled | Decaying wood | China, Anhui Province | This study |
| P.aguilerae | 1-septate, rarely 2–3-septate | Clavate or cylindrical, curved, with obtuse, rounded apex, slightly uncinate, and truncate base, hyaline, smooth | Decaying twig submerged in river | Cuba | Castañeda Ruiz et al. (2002) |
| P.annesophieae | Aseptate | Ellipsoidal to obovoid, straight or slightly curved, rounded at the ends or sometimes tapering towards the base, hyaline, guttulate, smooth-walled | Isolated from soil | The Netherlands, Geldermalsen | Crous et al. (2017) |
| P.aquatica | Aseptate | Ellipsoidal to obovoidal, rounded at the apex, hyaline, with two guttules smooth-walled | Decaying wood submerged in Jinsha River | China, Yunnan Province | Luo et al. (2018) |
| P.bambusae | aseptate or septate | Ellipsoidal to obovoidal, fusiform-cylindrical to falcate, hyaline, straight, guttulate, smooth-walled | Unidentified submerged bamboo | Indonesia | Höhnel (1909), Hyde et al. (2019), Luo et al. (2019), Réblová et al. (2016) |
| P.caffra | Aseptate, rarely 1-sepate | Conidia straight, ellipsoidal to clavate, obovoid, not attenuated at the apex, pale yellow brown, smooth | On decaying leaf of Podocarpus | Cape Province | Castañeda Ruiz et al. (2002), de Hoog and Papendorf (1976) |
| P.clavulata | Aseptate | Broadly ellipsoidal to ± spherical, subspherical, smooth, hyaline | On rotten decorticated wood | Great Britain | Castañeda Ruiz et al. (2002), de Hoog and Papendorf (1976), Mason and Ellis (1953) |
| P.clematidis | Aseptate | Obovoidal, rounded at the apex, obtuse and tapering towards base, hyaline, smooth-walled | Decaying wood submerged in Lancang River | China, Yunnan Province | Castañeda Ruiz et al. (2002), Hughes (1958), Luo et al. (2018) |
| P.curvata | Aseptate | Smooth, thin-walled, hyaline, clavate to obovoid and pointed at base, curved, occasionally sickle-shaped | Rotten leaves of Parinaricapensis | South West Africa | de Hoog and Papendorf (1976) |
| P.dalbergiae | Aseptate | Solitary, hyaline, smooth, thin-walled, guttulate, subcylindrical to obovoid, tapering towards both ends, apex subobtuse, base with truncate hilum | On bark of Dalbergiaarmata | South africa, Northern Province | Crous et al. (2021) |
| P.fasciculata | Aseptate | Ellipsoidal to obovoid, straight, rounded at the apex, obtuse and tapering towards base, hyaline, smooth-walled | Decorticated wood of Sambucusnigra | Canada, Ontario, Goulbourn Twp | Réblová et al. (2016) |
| P.filiformis | - | - | Decaying wood submerged in freshwater stream | Thailand, Sai khu Waterfall | Luo et al. (2019) |
| P.glauca | Aseptate | Smooth, thin-walled, hyaline, guttuliform to ellipsoidal,with pointed base, occasionally sickle-shaped | On rotten wood of Quercus sp. | America, Newfield | de Hoog and Papendorf (1976) |
| P.guttulata | Aseptate | Globose to obovoid, hyaline, smooth-walled, guttulate | Decaying wood submerged in Suoluo River | China, Guizhou Province | Hyde et al. (2018) |
| P.infrafertilis | Aseptate, rarely 1-sepate | Conidia falcate,hyaline | On dead leaves of Eucalyptus | Brazil | de Hoog and Papendorf (1976), Sutton and Hodges (1976) |
| P.loranthacearum | - | Solitary, hyaline, smooth, fusoidal-ellipsoidal with obtuse ends, straight to falcate, guttulate | On twigs of Loranthuseuropaeus | Germany | Crous et al. (2015) |
| P.magnifica | Aseptate | Straight, ellipsoidal to obovo1d, clavate, very pale olivaceous, smooth | On Bambusa | New Caledonia | de Hoog and Papendorf (1976), Deighton (1974) |
| P.microspora | Aseptate | Solitary, fusiform, straight, smooth-walled, guttulate, hyaline | On decaying wood | Thailand, Krabi, Wat ThumSua | Hyde et al. (2017) |
| P.muscariformis | 3-sepate | Cylindrical-fusiform, subhyaline, smooth | On leaves of Tiliacorakenyensis | Kenya | Castañeda Ruiz et al. (2002), Siboe et al. (1999) |
| P.pseudoclematidis | Aseptate | Cylindrical-ovate, straight, hyaline, smooth-walled, guttulate | On dead culm of bamboo (Bambusae) | Thailand, Chiang Rai | Liu et al. (2015) |
| P.sedimenticola | Aseptate, 1-septate | Smooth-walled, hyaline, with a pointed base, usually aseptate when attached to the conidiogenous cells, 0–1-septate after release; aseptate conidia, obovoid to ellipsoidal; 1-septate conidia, obovoid, slightly constricted at septum | Isolated from surface of marine sediment in intertidal zone | China, Shandong Province | Cheng et al. (2014) |
| P.siamensis | Aseptate | Globose to subglobose, hyaline | Saprobic on decaying fruits | Thailand, Chiang Mai Province | Hyde et al. (2019) |
| P.sparsa | 0-3-septate | Fusiform to clavate, conidia straight, ellipsoidal to fusiform, hyaline, not attenuated at the apex | On bark of Acerspicatum | Saskatchewan | de Hoog and Papendorf (1976), Sutton (1973) |
| P.sparsavar.cubensis | 0–1(–4)-septate | Fusiform, cylindrical or clavate, hyaline, sometimes slightly curved | On dead branch | Cuba | Mercado-Sierra et al. (1997), Mel’nik (2012) |
| P.synnematica | 0–1-septate | Dimorphic, clavate to ellipsoidal, cylindrical to falcate, base narrowly truncate, tip obtuse, variable in size, sometimes constricted near septa, 1–2-guttulate, hyaline, smooth-walled | Dead bark of Azadirachtaindica (Meliaceae) | India, Maharashtra | Boonmee et al. (2021) |
| P.tuberculata | Asepate, rarely 1-sepate | Conidia fusiform, straight, the apex attenuated, hyaline, smooth, guttulate | On Labiatae | Malawi | Castañeda Ruiz et al. (2002), de Hoog and Papendorf (1976), Sutton (1993) |
| P.uniseptata | Mostly with a median septum | Two-celled, fusiform, ellipsoid, hyaline, cylindrical or clavate | On dead branch | Cuba | de Hoog and Papendorf (1976), Mercado-Sierra (1984), Mel’nik (2012) |
| P.vietnamensis | A single median septum | Fusiform-subcylindrical to short obovoid-subclavate, somewhat attenuated towards the base, apex obtuse, straight to slightly curved, not constricted, hyaline, smooth, often guttulate | On bark of a living unidentified liane | South Vietnam, Dong Nai Province | Mel’nik (2012) |
Phaeoisaria predominantly occurs on leaves, barks, decaying wood and twigs of plants from the freshwater or terrestrial habitats (Table 3), while some are isolated from surface marine sediments (e.g. P.sedimenticola, Cheng et al. 2014), some from soil (e.g. P.annesophieae, Crous et al. 2017) and some from saprobic decaying fruits (e.g. P.siameneis, Hyde et al. 2019). Consequently, the habitats of Phaeoisaria are various. In this research, we introduce another lignicolous freshwater fungus, P.laianensis, discovered in China and it is noteworthy that the freshwater in which this species exists has been somewhat polluted. Phaeoisaria is thought to play an important role in nutrient and carbon cycling, biological diversity and ecosystem functioning of freshwater ecosystems, for their ability to decompose lignocellulose in woody litter, softening the wood and releasing nutrients (Bucher et al. 2004, Vijaykrishna et al. 2005, Hyde et al. 2016, Luo et al. 2018). Nonetheless, some Phaeoisaria species are pathogenic to humans, for example, it has been reported that P.clematidis and Phaeoisaria sp. can cause corneal inflammation of the eye (keratitis) (Guarro et al. 2000, Chew et al. 2010) and the former species is saprotrophic, which is similar to P.laianensis. What is the role of P.laianensis in ecosystem functioning? Is this species also pathogenic to humans? Such questions are waiting to be investigated by researchers.
Supplementary Material
Acknowledgements
We are grateful to Deng-Mei Fan (Agricultural college, Jiangxi Agricultural University) for very helpful comments on earlier drafts of this manuscript. This study was supported by the National Natural Science Foundation of China (NSFC 32070023 and NSFC 32060014), the Natural Science Foundation of Jiangxi Province (20151BAB214002) and Science and Technology Plan Project of Jiangxi Province (GJJ160417).
Funding Statement
Natural Science Foundation of China (NSFC 32070023 and NSFC 32060014), the Natural Science Foundation of Jiangxi Province (20151BAB214002) and Science and Technology Plan Project of Jiangxi Province (GJJ160417)
Contributor Information
Dian-Ming Hu, Email: hudianming1@163.com.
Zhi-Jun Zhai, Email: zhjzh002@163.com.
References
- Boonmee S., Wanasinghe D. N., Calabon M. S., Huanraluek N., Chandrasiri S. K. U., Jones G. E. B., Rossi W., Leonardi M., Singh S. K., Rana S., Singh P. N., Maurya D. K., Lagashetti A. C., Choudhary D., Dai Y. C., Zhao C. L., Mu Y. H., Yuan H. S., He S. H., Phookamsak R., Jiang H. B., Martín M. P., Dueñas M., Telleria M. T., Kałucka I. L., Jagodziński A. M., Liimatainen K., Pereira D. S., Phillips A. J. L., Suwannarach N., Kumla J., Khuna S., Lumyong S., Potter T. B., Shivas R. G., Sparks A. H., Vaghefi N., Abdel-Wahab M. A., Abdel-Aziz F. A., Li G. J., Lin W. F., Singh U., Bhatt R. P., Lee H. B., Nguyen T. T. T., Kirk P. M., Dutta A. K., Acharya K., Sarma V. V., Niranjan M., Rajeshkumar K. C., Ashtekar N., Lad S., Wijayawardene N. N., Bhat D. J., Xu R. J., Wijesinghe S. N., Shen H. W., Luo Z. L., Zhang Z. Y., Sysouphanthong P., Thongklang N., Bao D. F., Aluthmuhandiram J. V. S., Abdollahzadeh J., Javadi A., Dovana F., Usman M., Khalid A. N., Dissanayake A. J., Telagathoti A., Probst M., Peintner U., Garrido-Benavent I., Bóna L., Merényi Z., Boros L., Zoltán B., Stielow J. B., Jiang N., Tian C. M., Shams E., Dehghanizadeh F., Pordel A., Javan-Nikkhah M., Denchev T. T., Denchev C. M., Kemler M., Begerow D., Deng C. Y., Harrower E., Bozorov T., Kholmuradova T., Gafforov Y., Abdurazakov A., Xu J. C., Mortimer P. E., Ren G. C., Jeewon R., Maharachchikumbura S. S. N., Phukhamsakda C., Mapook A., Hyde K. D. Fungal diversity notes 1387-1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity. 2021;111:1–335. doi: 10.1007/s13225-021-00489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher V. V. C., Pointing S. B., Hyde K. D., Reddy C. A. Production of wood decay enzymes, loss of mass, and lignin solubilization in wood by diverse tropical freshwater fungi. Microbial Ecology. 2004;48:331–337. doi: 10.1007/s00248-003-0132-x. [DOI] [PubMed] [Google Scholar]
- Castañeda Ruiz R. F., Velasquez S., Cano J., Saikawa M., Guarro J. Phaeoisariaaguilerae anam. sp. nov. from submerged wood in Cuba with notes and reflections in the genus Phaeoisaria. Cryptogamie Mycologie. 2002;23(1):9–18. [Google Scholar]
- Cheng X., Li W., Zhang T. A new species of Phaeoisaria from intertidal marine sediment collected in Weihai, China. Mycotaxon. 2014;127(1):17–24. doi: 10.5248/127.17. [DOI] [Google Scholar]
- Chew H. F., Jungkind D. L., Mah D. Y., Raber I. M., Toll A. D., MJ Tokarczyk, Cohen E. J. Post-traumatic fungal keratitis caused by Carpoligna sp. Cornea. 2010;29(4):449–452. doi: 10.1097/ICO.0b013e3181af3954. [DOI] [PubMed] [Google Scholar]
- Crous P. W., Schumacher R. K., Wingfield M. J., Lombard L., Giraldo A., Christensen M., Gardiennet A., Nakashima C., Pereira O. L., AJ Smith, Groenewald J. Z. Fungal systematics and evolution: FUSE 1. Sydowia. 2015;67:81–118. doi: 10.12905/0380.sydowia67-2015-0081. [DOI] [Google Scholar]
- Crous P. W., Wingfield M. J., Burgess T. I., Carnegie A. J., Hardy G. E.S.J., Smith D., Summerell B. A., Cano-Lira J. F., Guarro J., Houbraken J., Lombard L., Martín M. P., Sandoval-Denis M., Alexandrova A. V., Barnes C. W., Baseia I. G., Bezerra J. D.P., Guarnaccia V., May T. W., Hernández-Restrepo M., Stchigel A. M., Miller A. N., Ordoñez M. E., Abreu V. P., Accioly T., Agnello C., Agustincolmn A., Albuquerque C. C., Alfredo D. S., Alvarado P., Araújo-Magalhães G. R., Arauzo S., Atkinson T., Barili A., Barreto R. W., Bezerra J. L., Cabral T. S., Camello R. F., Cruz R. H.S.F., Daniëls P. P., Da Silva B. D.B., De Almeida D. A.C., De Carvalho Júnior A. A., Decock C. A., Delgat L., Denman S., Dimitrov R. A., Edwards J., Fedosova A. G., Ferreira R. J., Firmino A. L., Flores J. A., García D., Gené J., Giraldo A., Góis J. S., Gomes A. A.M., Gonçalves C. M., Gouliamova D. E., Groenewald M., Guéorguiev B. V., Guevara-Suarez M., Gusmão L. F.P., Hosaka K., Hubka V., Huhndorf S. M., Jadan M., Jurjević Ž, B Kraak, V Kučera, TKA Kušan, I Lacerda, Lamlertthon S., Lisboa W. S., Loizides M., Luangsa-Ard J. J., Lysková P., Maccormack W. P., Macedo D. M., Machado A. R., Malysheva E. F., Marinho P., Matočec N., Meijer M., Mešić A., Mongkolsamrit S., Moreira K. A., Morozova O. V., Nair K. U., Nakamura N., Noisripoom W., Olariaga I., Oliveira R. J.V., Paiva L. M., Pawar P., Pereira O. L., Peterson S. W., Prieto M., Rodríguez-Andrade E., Rojodeblas C., Roy M., Santos E. S., Sharma R., Silva G. A., Souza-Motta C. M., Takeuchi-Kaneko Y., Tanaka C., Thakur A., Smith M. T., Tkalčec Z., Valenzuela-Lopez N., Vanderkleij P., Verbeken A., Viana M. G., Wang X. W., Groenewald J. Z. Fungal Planet description sheets: 625-715. Persoonia: Molecular Phylogeny and Evolution of Fungi. 2017;39(1):270–467. doi: 10.3767/persoonia.2017.39.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P. W., Osieck E. R., Jurjević Ž., Boers J., Iperen A. L., Starink-Willemse M., Dima B., Balashov S., Bulgakov T. S., Johnston P. R., Morozova O. V., Pinruan U., Sommai S., Alvarado P., Decock C. A., Lebel T., McMullan-Fisher S., Moreno G., Shivas R. G., Zhao L., Abdollahzadeh J., Abrinbana M., Ageev D. V., Akhmetova G., Alexandrova A. V., Altés A., Amaral A. G.G., Angelini C., Antonín V., Arenas F., Asselman P., Badali F., Baghela A., Bañares A., Barreto R. W., Baseia I. G., Bellanger J. M., Berraf-Tebbal A., Biketova A. Yu., Bukharova N. V., Burgess T. I., Cabero J., Câmara M. P.S., Cano-Lira J. F., Ceryngier P., Chávez R., Cowan D. A., de Lima A. F., Oliveira R. L., Denman S., Dang Q. N., Dovana F., Duarte I. G., Eichmeier A., Erhard A., Esteve-Raventós F., Fellin A., Ferisin G., Ferreira R. J., Ferrer A., Finy P., Gaya E., Geering A. D.W., Gil-Durán C., Glässnerová K., Glushakova A. M., Gramaje D., Guard F. E., Guarnizo A. L., Haelewaters D., Halling R. E., Hill R., Hirooka Y., Hubka V., Iliushin V. A., Ivanova D. D., Ivanushkina N. E., Jangsantear P., Justo A., Kachalkin A. V., Kato S., Khamsuntorn P., Kirtsideli I. Y., Knapp D. G., Kochkina G. A., Koukol O., Kovács G. M., Kruse J., Kumar T. K. A., Kušan I., Læssøe T., Larsson E., Lebeuf R., Levicán G., M Loizides, P Marinho, JJ Luangsa-ard, EG Lukina, Magaña-Dueñas V., Maggs-Kölling G., Malysheva E. F., Malysheva V. F., Martín B., Martín M. P., Matočec N., McTaggart A. R., Mehrabi-Koushki M., Mešić A., Miller A. N., Mironova P., Moreau P. A., Morte A., Müller K., Nagy L. G., Nanu S., Navarro-Ródenas A., Nel W. J., Nguyen T. H., Nóbrega T. F., Noordeloos M. E., Olariaga I., Overton B. E., Ozerskaya A. M., Palani P., Pancorbo F., Papp V., Pawłowska J., Pham T. Q., Phosri C., Popov E. S., Portugal A., Pošta A., Reschke K., Reul M., Ricci G. M., Rodríguez A., Romanowski J., Ruchikachorn N., Saar I., Safi A., Sakolrak B., Salzmann F., Sandoval-Denis M., Sangwichein E., Sanhueza L., Sato T., Sastoque A., Senn-Irlet B., Shibata A., Siepe K., Somrithipol S., Spetik M., Sridhar P., Stchigel A. M., Stuskova K., Suwannasai N., Tan Y. P., Thangavel R., Tiago I., Tiwari S., Tkalčec Z., Tomashevskaya M. A., Tonegawa C., Tran H. X., Tran N. T., Trovão J., Trubitsyn V. E., Van Wyk J., Vieira W. A.S., Vila J., Visagie C. M., Vizzini A., Volobuev S. V., Vu D. T., Wangsawat N., Yaguchi T., Ercole E., Ferreira B. W., de Souza A. P., Vieira B. S., Groenewald J. Z. Fungal Planet description sheets: 1284-1382. Persoonia: Molecular Phylogeny and Evolution of Fungi. 2021;47(1):178–374. doi: 10.3767/persoonia.2021.47.06. [DOI] [Google Scholar]
- de Hoog G. S., Papendorf M. C. The genus Phaeoisaria. Persoonia: Molecular Phylogeny and Evolution of Fungi. 1976;8(4):407–414. [Google Scholar]
- Deighton F. C. Four synnematous hyphomycetes. Transactions of British Mycological Society. 1974;62:243–252. doi: 10.1016/S0007-1536(74)80033-1. [DOI] [Google Scholar]
- Doyle J. J., Doyle J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Guarro J., Vieira L. A., de Freitas D., Gené J., Zaror L., Hofling-Lima A. L., Fischman O., Zorat-Yu C., Figueras M. J. Phaeoisariaclematidis as a cause of keratomycosis. Journal of Clinical Microbiology. 2000;38(6):2434–2437. doi: 10.1128/JCM.38.6.2434-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhnel F. V. Fragmente zur Mykologie (VI. Mitteilung, Nr. 182 bis 288) Sitzungsberichten der Kaiserliche Akademie der Wissenschaften in Wien Mathematische-Naturwissenschaftliche Klasse, Abt I. 1909;118(182):275–452. [Google Scholar]
- Hopple J. S.J., Vilgalys R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups, and monophyly. Molecular Phylogenetics and Evolution. 1999;13(1):1–19. doi: 10.1006/mpev.1999.0634. [DOI] [PubMed] [Google Scholar]
- Hughes S. J. Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Canadian Journal of Botany. 1958;36(6):727–836. doi: 10.1139/b58-067. [DOI] [Google Scholar]
- Hyde K. D., Fryar S., Tian Q., Bahkali A. H., Xu J. C. Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecology. 2016;19:190–200. doi: 10.1016/j.funeco.2015.07.002. [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Abreu V. P., Bazzicalupo A. L., Chethana K. W. Thilini, Clericuzio M., Dayarathne M. C., Dissanayake A. J., Ekanayaka A. H., He M. Q., Hongsanan S., Huang S. K., Jayasiri S. C., Jayawardena R. S., Karunarathna A., Konta S., Kušan I., Lee H. G., Li J., Lin CG., Liu N. G., Lu Y. Z., Luo Z. L., Manawasinghe I. S., Mapook A., Perera R. H., Phookamsak R., Phukhamsakda C., Siedlecki I., Soares A. M., Tennakoon D. S., Tian Q., Tibpromma S., Wanasinghe D. N., Xiao Y. P., Yang J., Zeng X. Y., Abdel-Aziz F. A., Li W. J., Senanayake I. C., Shang Q. J., Daranagama D. A., de Silva N. I., Thambugala K. M., Abdel-Wahab M. A., Bahkali A. H., Berbee M. L., Boonmee S., Bhat D. J., Bulgakov T. S., Buyck B., Camporesi E., Castañeda-Ruiz R. F., Chomnunti P., Doilom M., Dovana F., Gibertoni T. B., Jadan M., Jeewon R., Jones E. B.G., Kang J. C., Karunarathna S. C., Lim Y. W., Liu J. K., Liu Z. Y., Plautz H. L., Lumyong S., Maharachchikumbura S. S.N., Matočec N., McKenzie E. H.C., Mešić A., Miller D., Pawłowska J., Pereira O. L., Promputtha I., Romero A. I., Ryvarden L., Su H. Y., Suetrong S., Tkalčec Z., Vizzini A., Wen T. C., Wisitrassameewong K., Wrzosek M., Xu J. C., Zhao Q., Zhao R. L., Mortimer P. E. Fungal diversity notes 603-708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity. 2017;87(1):1–235. doi: 10.1007/s13225-017-0391-3. [DOI] [Google Scholar]
- Hyde K. D., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K. W.T., Dayarathne M. C., Silva N. I., Dissanayake A. J., Ekanayaka A. H., Hongsanan S., Huang S. K., Jayasiri S. C., Jayawardena R. S., Jiang H. B., Karunarathna A., Lin C. G., Liu J. K., Liu N. G., Lu Y. Z., Luo Z. L., Maharachchimbura S. S.N., Manawasinghe I. S., Pem D., Perera R. H., Phukhamsakda C., Samarakoon M. C., Senwanna C., Shang Q. J., Tennakoon D. S., Thambugala K. M., Tibpromma S., Wanasinghe D. N., Xiao Y. P., Yang J., Zeng X. Y., Zhang J. F., Zhang S. N., Bulgakov T. S., Bhat D. J., Cheewangkoon R., Goh T. K., Jones E. B.G., Kang J. C., Jeewon R., Liu Z. Y., Lumyong S., Kuo C. H., McKenzie E. H.C., Wen T. C., Yan J., Zhao Q. Mycosphere notes 169-224. Mycosphere. 2018;9(2):271–430. doi: 10.5943/mycosphere/9/2/8. [DOI] [Google Scholar]
- Hyde K. D., Tennakoon D. S., Jeewon R., Bhat D. J., Maharachchikumbura S. S. N., Rossi W., Leonardi M., Lee H. B., Mun H. Y., Houbraken J., Nguyen T. T. T., Jeon S. J., Frisvad J. C., Wanasinghe D. N., Luücking R., Aptroot A., Cáceres M. E. S., Karunarathna S. C., Hongsanan S., Phookamsak R., de Silva N. I., Thambugala K. M., Jayawardena R. S., Senanayake I. C., Boonmee S., Chen J., Luo Z. L., Phukhamsakda C., Pereira O. L., Abreu V. P., Rosado A. W.C., Bart B., Randrianjohany E., Hofstetter V., Gibertoni T. B., da Silva Soares A. M., Plautz Jr. H. L., Sotão H. M. P., Xavier W. K. S., Bezerra J. D. P., de Oliveira T. G. L., de Souza Motta C. M., Magalhães O. M. C., Bundhun D., Harishchandra D., Manawasinghe I. S., Dong W., Zhang S. N., Bao D. F., Samarakoon M. C., Pem D., Karunarathna A., Lin C. G., Yang J., Perera R. H., Kumar V., Huang S. K., Dayarathne M. C., Ekanayaka A. H., Jayasiri S. C., Xiao Y., Konta S., Niskanen T., Liimatainen K., Dai Y. C., Ji X. H., Tian X. M., Mešić A., Singh S. K., Phutthacharoen K., Cai L., Sorvongxay T., Thiyagaraja V., Norphanphoun C., Chaiwan N., Lu Y. Z., Jiang H. B., Zhang J. F., Abeywickrama P. D., Aluthmuhandiram J. V.S., Brahmanage R. S., Zeng M., Chethana T., Wei D., Réblová M., Fournier J., Nekvindová J., do Nascimento Barbosa R., dos Santos J. E.F., de Oliveira N. T., Li G. J., Ertz D., Shang Q. J., Phillips A. J. L., Kuo C. H., Camporesi E., Bulgakov T. S., Lumyong S., Jones E. B. G., Chomnunti P., Gentekaki E., Bungartz F., Zeng X. Y., Fryar S., Tkalčec Z., Liang J., Li G., Wen T. C., Singh P. N., Gafforov Y., Promputtha I., Yasanthika E., Goonasekara I. D., Zhao R. L., Zhao Q., Kirk P. M., Liu J. K., Yan J., Mortimer P. E., Xu J. C. Fungal diversity notes 1036-1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity. 2019;96(1):1–242. doi: 10.1007/s13225-019-00429-2. [DOI] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. K., Hyde K. D., Jones E. B. G., Ariyawansa H. A., Bhat D. J., Boonmee S., Maharachchikumbura S. S., McKenzie E. H. C., Phookamsak R., Phukhamsakda C., Shenoy B. D., Abdel-Wahab M. A., Buyck B., Chen J., Chethana K. W. T., Singtripop C., Dai D. Q., Dai Y. C., Daranagama D. A., Dissanayake A. J., Doilom M., D’souza M. J., Fan X. L., Goonasekara I. D., Hirayama K., Hongsanan S., Jayasiri S. C., Jayawardena R. S., Karunarathna S. C., Li W. J., Mapook A., Norphanphoun C., Pang K. L., Perera R. H., Peršoh D., Pinruan U., Senanayake I. C., Somrithipol S., Suetrong S., Tanaka K., Thambugala K. M., Tian Q., Tibpromma S., Udayanga D., Wijayawardene N. N., Wanasinghe D., Wisitrassameewong K., Zeng X. Y., Abdel-Aziz F. A., Adamčík S., Bahkali A. H., Boonyuen N., Bulgakov T. S., Callac P., Chomnunti P., Greiner K., Hashimoto A., Hofstetter V., Kang J. C., Lewis D., Li X. H., Liu X. Z., Liu Z. Y., Matsumura M., Mortimer P. E., Rambold G., Randrianjohany E., Sato G., Sriindrasutdhi V., Tian C. M., Verbeken A., Brackel W. V., Wang Y., Wen T. C., Xu J. C., Yan J. Y., Zhao R. L., Camporesi E. Fungal diversity notes 1-110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- Li X. H., Liu Y. L., Song H. Y., Hu D. M., Gao Y., Hu H. J., Zhou J. P. Sporidesmiellalignicola sp. nov., a new hyphomycetous fungus from freshwater habitats in China. Biodiversity Data Journal. 2021;9:77414. doi: 10.3897/BDJ.9.e77414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. L., Hyde K. D., Bhat D. J., Maharachchikumbura S. S.N., Bao D. F., Li W. L., Su X. J., Yang X. Y. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress. 2018;17(5):511–530. doi: 10.1007/s11557-018-1377-6. [DOI] [Google Scholar]
- Luo Z. L., Hyde K. D., Liu J. K., Maharachchikumbura S. S.N., Jeewon R., Bao D. F., Bhat D. J., Lin C. G., Li W. L., Yang J., Liu N. G., Lu Y. Z., Jaya-wardena R. S., Li J. F., Su H. Y. Freshwater sordariomycetes. Fungal Diversity. 2019;99:451–660. doi: 10.1007/s13225-019-00438-1. [DOI] [Google Scholar]
- Mason E. W., Ellis M. B. British species of Periconia. Mycological Paper. 1953;56:1–127. [Google Scholar]
- Mel’nik V. A. Phaeoisariavietnamensis sp. nov. and P.clematidis (hyphomycetes) from Vietnam. Mycosphere. 2012;3(6):957–960. doi: 10.5943/mycosphere/3/6/10. [DOI] [Google Scholar]
- Mercado-Sierra A. Nuevas especis de Deightoniella, Phaeoisaria, Sporidesmium, Y Taeniolella (Hyphomycetes) de Cuba. Acta Botánica Cubana. 1984;21:1–10. [Google Scholar]
- Mercado-Sierra A., Figueras M. J., Gene J. New or rare hyphomycetes from Cuba Ⅷ. Species of Lylea, Phaeisaria, Arxiella, Graphium, Periconia, and Ramichloridium. Mycotaxon. 1997;63:369–375. [Google Scholar]
- Rambaut A. FigTree v1.4.4: Tree figure drawing tool. https://github.com/rambaut/figtree/releases 2018
- Réblová M., Seifert K. A., Fournier J., Štěpánek V. Newly recognized lineages of perithecial ascomycetes: the new orders Conioscyphales and Pleurotheciales. Persoonia: Molecular Phylogeny and Evolution of Fungi. 2016;37(1):57–81. doi: 10.3767/003158516X689819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboe G. M., Kipk P. M., Cannon P. F. New dematiaceous hyphomycetes from Kenyan rare plants. Mycotaxon. 1999;73:283–302. [Google Scholar]
- Stamatakis A., Alachiotis N. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics. 2010;26(12):132–139. doi: 10.1093/bioinformatics/btq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton B. C. Hyphomycetes from Manitoba and Saskatchewan, Canada. Mycological Papers. 1973;132:1–143. [Google Scholar]
- Sutton B. C., Hodges C. S. Eucalyptus micofungi. Microdochium and Phaeoisaria species from Brazil. Nova Hedwigia. 1976;27:215–222. [Google Scholar]
- Sutton B. C. Mitosporic fungi from Malawi. Mycological Papers. 1993;167:1–93. [Google Scholar]
- Vijaykrishna D., Jeewon R., Hyde K. D. Fusoidisporaaquatica: a new freshwater ascomycete from Hong Kong based on morphology and phylogeny inferred from rDNA gene sequences. Sydowia. 2005;57(2):267–280. [Google Scholar]
- White T. J., Bruns T., Lee S. J.W.T., Taylor J. In: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Innis M., Gelfand D., Shinsky J., White T., editors. Academic Press; 1990. PCR protocols: a guide to methods and applications. Academic Press, New York. [DOI] [Google Scholar]
- Zhai Z. J., Yan J. Q., Li W. W., Gao Y., Hu H. J., Zhou J. P., Song H. Y., Hu D. M. Three novel species of Distoseptispora (Distoseptisporaceae) isolated from bamboo in Jiangxi Province, China. MycoKeys. 2022;88:35–54. doi: 10.3897/mycokeys.88.79346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W. X., Wang G. T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources. 2020;20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O., Gogarten J. P. Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC genomics. 2002;3(1):1–15. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.