Abstract
Species delimitation and species diagnosis must remain separate operations to avoid constructing taxonomies comprised of non-monophyletic species based on morphological similarity as opposed to phylogenetic propinquity. This is particularly true for highly specialized species such as the range-restricted upland taxa in the Cyrtodactylusbrevipalmatus group of Indochina where strong selection pressure for an arboreal lifestyle has contributed to morphologically similar but distantly related species. This in turn, has resulted in a history of erroneous taxonomies that have actually obscured rather than revealed the diversity within this group. A Bayesian phylogeny of the C.brevipalmatus group recovered at least 15 putative species-level lineages, at least seven of which are undescribed, and of which four are described herein. A total evidence morphological data set comprised of 16 normalized morphometric, 15 meristic, and seven categorical characters scored across 51 individuals was subjected to a multiple factor analysis (MFA) and an analysis of variance (ANOVA) to diagnose the putative species. These new species descriptions contribute to focusing attention to the unrealized diversity in upland tropical ecosystems, which due to climate change, are some of the most impearled ecosystems on the planet. Thus, it is paramount that taxonomies do not conflate species identities and underrepresent true diversity.
Keywords: geckos, genetics, morphology, Southeast Asia, taxonomy
Introduction
A cornerstone of biodiversity conservation is a phylogenetic-based taxonomy where the names of the component species are consistent with the patterns and processes by which they evolved. Taxonomies constructed from paraphyletic or polyphyletic species misrepresent history, thus obscuring true diversity and potentially countermanding the effectiveness of conservation efforts. Before a newly discovered population can be given a new name, or the name of an existing species be successfully challenged, it should be properly delimited and diagnosed. Delimiting and diagnosing species are independent operations used together to construct taxonomies that reflect, and are consistent with evolutionary history. Unfortunately, these two operations are often conflated when analyses to diagnose species, which are most often rooted in morphological similarity, are equated with analyses to delimit species, which are rooted in phylogenetic propinquity. The unfortunate consequence of this is that taxonomies may be constructed using non-monophyletic species, thus obscuring rather than revealing the group’s actual diversity. This is especially true for taxonomies comprised of highly specialized cryptic species where, in the absence of a phylogeny, morphological convergence can be mistaken for common ancestry (see Grismer 2008; Grismer et al. 2020, 2022). Despite methodological improvements over the years that have increased the efficacy of both delimitation and diagnostic procedures, they have had little effect on operational decisions to not conflate them.
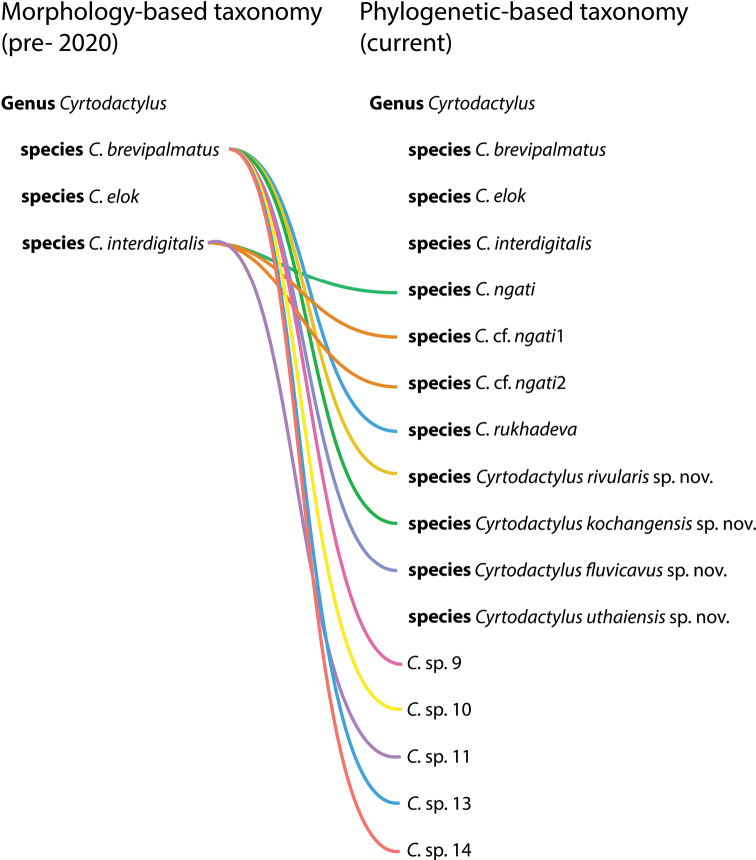
Convergent morphology results from the specific resource requirements that necessitate a particular functional morphology (Ricklefs 2012; Dehling et al. 2016; Baulechner et al. 2020). In such cases, phylogenetic analysis becomes paramount to disentangle convergence from common ancestry in order to construct proper phylogenetic taxonomies. This is especially true of clades containing highly specialized species such as the Cyrtodactylusbrevipalmatus group (sec. Grismer et al. 2021a, b, c). This group currently contains four nominal species that range from northern Vietnam and Laos, southward through Thailand to southern Peninsular Malaysia (Fig. 1). Nearly all members of this adaptive radiation are highly specialized for an arboreal lifestyle and all have a prehensile tail carried in a tightly coiled position, a cryptic color pattern of different shades of brown that closely match the substrate of their microhabitat, and generally slow, deliberate “chameleon-like” movements. Presumably, the selection pressures for such a specialized lifestyle have contributed to morphological convergence within this relatively small group (Grismer et al. 2022). Thus, as demonstrated by Grismer (2008) and Grismer et al. (2021c, 2022), previous morphology-based operations intending to delimit species resulted in reoccurring taxonomies composed of non-monophyletic species (Smith 1935; Welch et al. 1990; Ulber 1993; Manthey and Grossmann 1997; Stuart 1999; Nabhitabhata et al. 2004; Nabhitabhata and Chan-ard 2005; Pauwels and Chan-ard 2006; Ellis and Pauwels 2012) that underrepresented the group’s diversity. Grismer et al. (2021c, 2022) were the first to employ phylogenetic analyses to the brevipalmatus group to delimit species and statistically based morphological analyses to diagnose those species. As such, by treating these analyses as independent operations and their results as corroborating evidence, a revised taxonomy consistent with the group’s evolutionary history was disentangled from a convergence-based taxonomy.
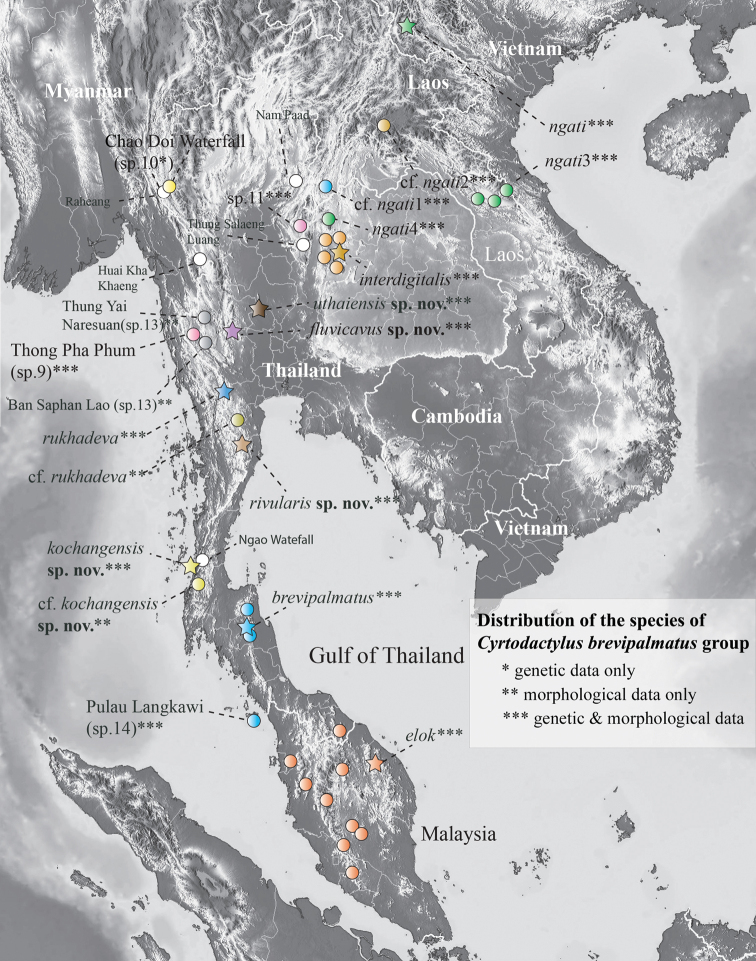
Figure 1.
Distribution of nominal species and unnamed populations and specimens of the Cyrtodactylusbrevipalmatus group. Stars denote type localities. White circles are literature localities from which specimens were not examined and remain unidentified. Locality data for all material examined is in Table 1.
The phylogenetic-based taxonomy of Grismer et al. (2022) is expanded here with significantly increased morphological and genetic sampling from new localities in Thailand at Ko (= island) Chang (ZMKU R 00945) and Khlong Nakha Wildlife Sanctuary (THNHM 01667), Ranong Province; Hua Hin, Prachuap Khiri Khan Province (ZMKU R 00946–47); Si Sawat (ZMKU R 00958–64) and Thong Pha Phum (ZMKU R 00950–57), Kanchanaburi Province; and an isolated population from a small hilly region at Lan Sak, Uthai Thani Province (ZMKU R 00949) in the Chao Phraya River Basin. In this analysis, we discuss the high degree of morphological similarity throughout the phylogeny despite the relatively high degrees of genetic divergence. The analyses also recovered the existence of 4–10 new populations we hypothesize to be distinct species, four of which are described herein.
Materials and methods
Species delimitation
The general lineage concept (GLC: de Queiroz 2007) adopted herein proposes that a species constitutes a population of organisms evolving independently from other such populations owing to a lack of, or limited gene flow. By “independently,” it is meant that new mutations arising in one species cannot spread readily into another species (Barraclough et al. 2003; de Queiroz 2007). Under the GLC implemented herein, molecular phylogenies recovered monophyletic mitochondrial lineages of individuals (populations) that were used to develop initial species-level hypotheses, the grouping stage of Hillis (2019). Discrete color pattern data and univariate and multivariate analyses of morphological data were then used to search for characters and morphospatial patterns consistent with the tree-designated species-level hypotheses, the construction of boundaries representing the hypothesis-testing step of Hillis (2019), –thus providing independent diagnoses to complement the molecular analyses. In this way, delimiting (phylogeny) and diagnosing (taxonomy) species are not conflated (Frost and Hillis 1990; Frost and Kluge 1994; Hillis 2019).
Species boundaries were cross-checked using a Bayesian Poisson Tree Process for species delimitation (bPTP; Zhang et al. 2013), thus providing an independent framework to complement the empirically based thresholds of the morphological and molecular analyses. This method delimits species from single-locus gene trees with low population samples (Lin et al. 2018) by detecting genetic clustering beyond the expected levels of a null hypothesis which infers that all individuals of a population form a genetically, interacting nexus. A Markov Chain Monte Carlo (MCMC) was run for 10,000 generations on the bPTP web server at https://species.h-its.org/ptp/ and checked for convergence. Delimitation accuracy was based on the normalized mutual information criterion (NMI) (Vinh et al. 2010). The model relies on the prediction that independent evolution leads to the appearance of distinct genetic clusters, separated by relatively longer internal branches (Barraclough et al. 2003; Acinas et al. 2004). Such groups therefore diverge into discrete clades of genetic variation. These clades are then diagnosed by statistically defensible morphological analyses.
Genetic data
Methods for DNA extraction, sequencing, and editing followed Grismer et al. (2022) and resulted in a 1,386 base pair segment of the mitochondrial NADH dehydrogenase subunit 2 gene (ND2) and adjacent tRNAs. All material examined is listed in Table 1 along with GenBank accession numbers for the new and published genetic materials.
Table 1.
Cyrtodactylus specimens examined in this study. Institutional abbreviations follow Sabaj (2020) except that YC = Yodchaiy Chuaynkern from Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen, Thailand.
| Species | Location | Catalog no. | GenBank no. |
|---|---|---|---|
| C.brevipalmatus | Thailand, no data | LSUHC 1899 | not in tree |
| C.brevipalmatus | Thailand, Nakhon Si Thammarat Province, Nopphitam District, Khao Nan National Park, Huay Lak Protected Unit | THNHM 10670 | not in tree |
| C.brevipalmatus | Thailand, Nakhon Si Thammarat Province, Lan Saka District, Khao Luang National Park | THNHM 14112 | not in tree |
| C.brevipalmatus | Thailand, Nakhon Si Thammarat Province, Khao Ram Mt. | AUP-00573 | OK626313 |
| C.cf.brevipalmatus | Peninsular Malaysia, Kedah State, Pulau Langkawi, Gunung Raya | LSUHC 11788 | not in tree |
| C.cf.brevipalmatus | Peninsular Malaysia, Kedah State, Pulau Langkawi, Gunung Raya | LSUHC 15076 | not in tree |
| C.elok | Peninsular Malaysia, Pahang State, Fraser’s Hill, the Gap | ZRC 2.6091/LSUHC 6471 | JQ889180 |
| C.elok | Peninsular Malaysia, Negeri Sembilan State | LSUHC 8238 | not in tree |
| C.elok | Peninsular Malaysia, Pahang State, near Cameron Highlands | LSUHC 12180 | not in tree |
| C.elok | Peninsular Malaysia, Pahang State, near Cameron Highlands | LSUHC 12181 | not in tree |
| C.elok | Malaysian pet trade, no data | ZMMU R-16144 | not in tree |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00958 paratype | OP620036 |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00959 holotype | OP620037 |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00960 paratype | OP620038 |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00961 paratype | OP620039 |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00962 paratype | OP620040 |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00963 paratype | OP620041 |
| Cyrtodactylusfluvicavus sp. nov. | Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park | ZMKU R 00964 paratype | OP620042 |
| C.interdigitalis | Thailand, Phetchabun Province, Nam Nao National Park, Tham Yai Nam Nao | THNHM 20226 paratype | not in tree |
| C.interdigitalis | Thailand, Phetchabun Province, Nam Nao National Park, Tham Yai Nam Nao | THNHM 20227 paratype | not in tree |
| C.interdigitalis | Thailand, Phetchabun Province, Nam Nao National Park, Tham Yai Nam Nao | THNHM 20228 paratype | not in tree |
| C.interdigitalis | Thailand, Phetchabun Province, Nam Nao National Park, Tham Yai Nam Nao | THNHM 20229 paratype | not in tree |
| C.interdigitalis | Thailand, Phetchabun Province, Nam Nao National Park, Tham Yai Nam Nao | YC000952 | ON055281 |
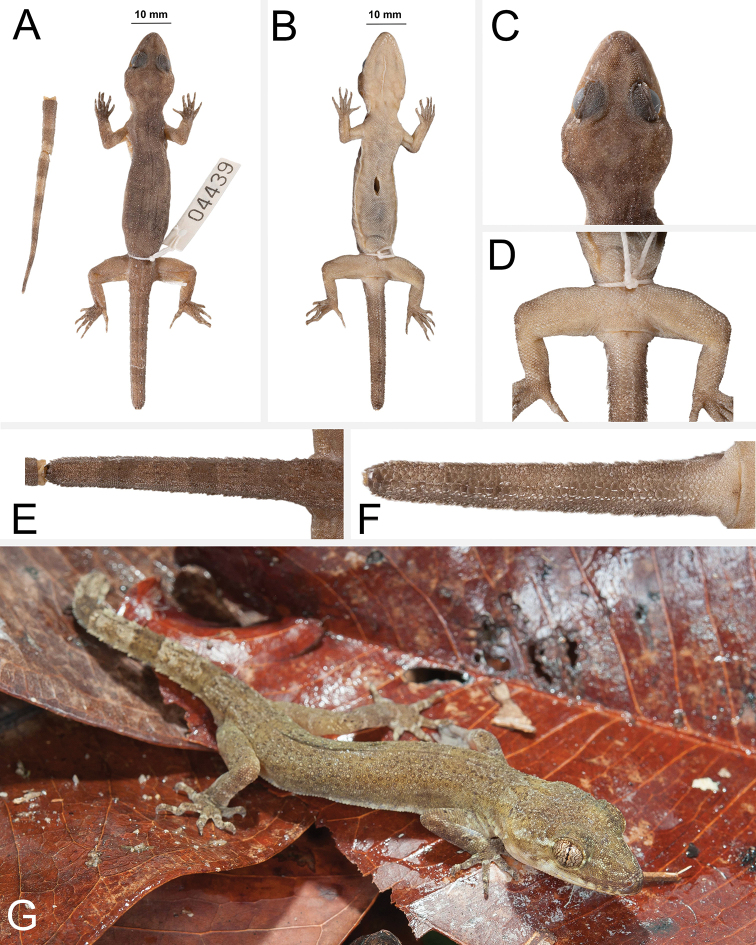
| Cyrtodactyluskochangensis sp. nov. | Thailand, Ranong Province, Mueng Ranong District, Ko Phayam Subdistrict, Ko Chang | ZMKU R 00945 holotype | OP620023 |
| C.cf.kochangensis sp. nov. | Thailand, Ranong Province, Khlong Nakha Wildlife Sanctuary | THNHM 01667 | not in tree |
| C.ngati | Vietnam, Dien Bien Province, Dien Bien District, Pa Thom Commune, Pa Xa Lao Village, karst forest near Pa Thom Cave | HNUE-R00111 holotype | ON411220 |
| C.ngati | Vietnam, Dien Bien Province, Dien Bien District, Pa Thom Commune, Pa Xa Lao Village, karst forest near Pa Thom Cave | HNUE-R00112 paratype | not in tree |
| C.ngati | Vietnam, Dien Bien Province, Dien Bien District, Pa Thom Commune, Pa Xa Lao Village, karst forest near Pa Thom Cave | IEBR 4829 paratype | OK626318 |
| C.ngati | Vietnam, Dien Bien Province, Dien Bien District, Pa Thom Commune, Pa Xa Lao Village, karst forest near Pa Thom Cave | VNUF R.2020.12 paratype | OK626319 |
| C.ngati3 | Laos, Khammouane Province | VNUF R.2014.50 | ON411221 |
| C.ngati3 | Laos, Khammouane Province, Phou Hin Poun National Biodiversity Conservation Area | FMNH 255454 | JQ889181 |
| C.ngati3 | Laos, Khammouane Province, Phou Hin Poun National Biodiversity Conservation Area | FMNH 270492 | OK626315 |
| C.ngati3 | Laos, Khammouane Province, Phou Hin Poun National Biodiversity Conservation Area | FMNH 270493 | not in tree |
| C.ngati4 | Thailand, Loei Province, Nam San Noi River, Phu Luang Wildlife Sanctuary | FMNH 265806 | JX519471 |
| C.cf.ngati1 | Laos, Xaignabouli Province, Ban Pha Liep, Houay Liep Stream | NCSM 79472 | OK626316 |
| C.cf.ngati2 | Laos, Vientiane Province | ZMMU R-14917 | not in tree |
| C.cf.ngati2 | Laos, Vientiane Province, tributary of Nam Pha River, Houay Wan Stream | NCSM 80100 | OK626317 |
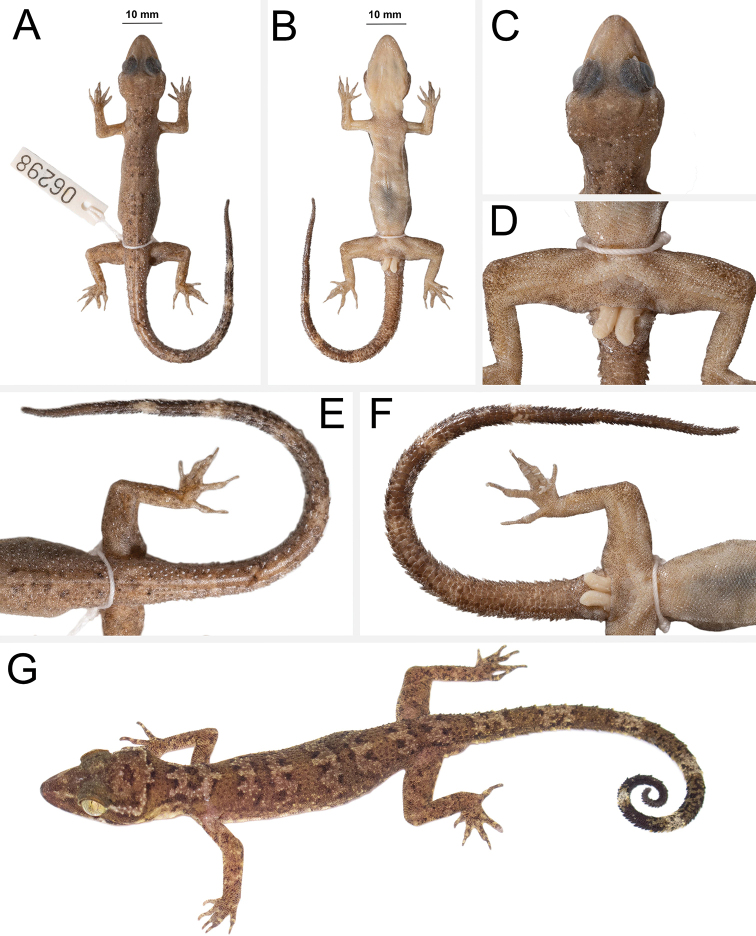
| Cyrtodactylusrivularis sp. nov. | Thailand, Prachuap Khiri Khan Province, Hua Hin District, Huai Sat Yai Subdistrict, Kaeng Krachan National Park, Pa La-U Waterfall | ZMKU R 00946 paratype | OP620024 |
| Cyrtodactylusrivularis sp. nov. | Thailand, Prachuap Khiri Khan Province, Hua Hin District, Huai Sat Yai Subdistrict, Kaeng Krachan National Park, Pa La-U Waterfall | ZMKU R 00947 holotype | OP620025 |
| C.rukhadeva | Thailand, Ratchaburi Province, Suan Phueng District, Khao Laem Mountain | ZMMU R-16851 holotype | OK626320 |
| C.rukhadeva | Thailand, Ratchaburi Province, Suan Phueng District, Hoop Phai Tong | ZMMU R-16852 paratype | not in tree |
| C.rukhadeva | Thailand, Ratchaburi Province, Suan Phueng District, Suan Phueng Subdistrict | ZMKU R 00948 | OP620026 |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 01807 | not in tree |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 03251 | not in tree |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 03252 | not in tree |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 03253 | not in tree |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 03254 | not in tree |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 24622 | not in tree |
| C.cf.rukhadeva | Thailand, Phetchaburi Province, Kaeng Krachan National Park | THNHM 24838 | not in tree |
| Cyrtodactylusuthaiensis sp. nov. | Thailand, Uthai Thani Province, Lan Sak District, Thung Na Ngam Subdistrict | ZMKU R 00949 holotype | OP620027 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Thong Pha Phum National Park | AUP-01715 | MT468909 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00950 | OP620028 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00951 | OP620029 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00952 | OP620030 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00953 | OP620031 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00954 | OP620032 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00955 | OP620033 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00956 | OP620034 |
| C. sp.9 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Pilok Subdistrict, Thong Pha Phum National Park | ZMKU R 00957 | OP620035 |
| C. sp.10 | Thailand, Tak Province, Tha Song Yang District, Mae Moei National Park, Chao Doi Waterfall | AUP-00680 | MT468902 |
| C. sp.11 (C.cf.interdigitalis) | Thailand, Phitsanulok Province, Phu Hin Rong Kla National Park | ZMMU R-16492 | MW792061 |
| C. sp.13 | Thailand, Tak Province, Umphang District, Thung Yai Naresuan Wildlife Sanctuary | THNHM 00104 | not in tree |
| C. sp.13 | Thailand, Kanchanaburi Province, Thong Pha Phum District, Ban Saphan Lao | THNHM 27821 | not in tree |
| C. sp.14 (C.cf.brevipalmatus) | Peninsular Malaysia, Kedah State, Pulau Langkawi, Gunung Raya | USMHC 2555 | OK626314 |
Morphological data
Morphological data included both meristic and morphometric characters. To reduce the degree of researcher bias, data were taken using the protocol of Le et al. (2021) and Grismer et al. (2022) and where possible, double checked by LLG using high resolution digital photographs and/or the actual specimens. All data were taken on the left side of the body (when possible) and measured to the nearest 0.1 mm using digital calipers under a Nikon SMZ745 stereomicroscope. Morphometric data taken were: snout-vent length (SVL), taken from the tip of the snout to the vent; tail length (TL), taken from the vent to the tip of the tail–original or partially regenerated; tail width (TW), taken at the base of the tail immediately posterior to the postcloacal swelling; humeral length (HumL), taken from the proximal end of the humerus at its insertion point in the glenoid fossa to the distal margin of the elbow while flexed 90°; forearm length (ForL), taken on the ventral surface from the posterior margin of the elbow while flexed 90° to the inflection of the flexed wrist; femur length (FemL), taken from the proximal end of the femur at its insertion point in the acetabulum to the distal margin of the knee while flexed 90°; tibia length (TibL), taken on the ventral surface from the posterior margin of the knee while flexed 90° to the base of the heel; axilla to groin length (AG), taken from the posterior margin of the forelimb at its insertion point on the body to the anterior margin of the hind limb at its insertion point on the body; head length (HL), the distance from the posterior margin of the retroarticular process of the lower jaw to the tip of the snout; head width (HW), measured at the angle of the jaws; head depth (HD), the maximum height of head measured from the occiput to base of the lower jaw posterior to the eyes; eye diameter (ED), the greatest horizontal diameter of the eye-ball; eye to ear distance (EE), measured from the anterior edge of the ear opening to the posterior edge of the bony orbit; eye to snout distance or snout length (ES), measured from anteriormost margin of the bony orbit to the tip of snout; eye to nostril distance (EN), measured from the anterior margin of the bony orbit to the posterior margin of the external nares; interorbital distance (IO), measured between the dorsomedial-most edges of the bony orbits; internarial distance (IN), measured between the external nares across the rostrum; and ear length (EL), greatest oblique length across the auditory meatus.
Meristic characters evaluated were the number of supralabial scales (SL), counted from the largest scale at the corner of the mouth or posterior to the eye, to the rostral scale; infralabial scales (IL), counted from termination of enlarged scales at the corner of the mouth to the mental scale; number of paravertebral tubercles (PVT) between the limb insertions counted in a straight line immediately left of the vertebral column; number of longitudinal rows of body tubercles (LRT) counted transversely across the body midway between the limb insertions from one ventrolateral body fold to the other; number of longitudinal rows of ventral scales (VS) counted transversely across the abdomen midway between limb insertions from one ventrolateral fold to the other; number of transverse rows of ventral scales (VSM) counted along the midline of the body from the postmentals to just anterior to the cloacal opening, stopping where the scales become granular; number of expanded subdigital lamellae on the fourth toe proximal to the digital inflection (TL4E) counted from the base of the first phalanx where it contacts the body of the foot to the largest scale on the digital inflection–the large contiguous scales on the palmar and plantar surfaces were not counted; number of small, generally unmodified subdigital lamellae distal to the digital inflection on the fourth toe (TL4U) counted from the digital inflection to the claw including the claw sheath; total number of subdigital lamellae (TL4T) beneath the fourth toe (i.e. TL4E + TL4U = TL4T); number of expanded subdigital lamellae on the fourth finger proximal to the digital inflection (FL4E) counted the same way as with TL4E; number of small generally unmodified subdigital lamellae distal to the digital inflection on the fourth finger (FL4U) counted the same way as with TL4U; total number of subdigital lamellae (FL4T) beneath the fourth toe (i.e. FL4E + FL4U = FL4T); total number of enlarged femoral scales (FS) from each thigh combined as a single metric; number of enlarged precloacal scales (PCS);number of precloacal pores (PP) in males; the number of femoral pores (FP) in males from each thigh combined as a single metric; and the number of dark body bands (BB) between the dark band on the nape and the hind limb insertions on the body. A post-sacral or sacral band when present, was not counted. Categorical characters evaluated were the presence or absence of tubercles on the flanks (FKT); single enlarged, unmodified, medial subcaudal scales (SC2); enlarged medial subcaudals intermittent, medially furrowed, posteriorly emarginated (SC3); slightly enlarged medial subcaudals (SC1); large or small dorsolateral caudal tubercles (DCT) forming a narrow or wide ventrolateral caudal fringe (VLF1); ventrolateral caudal fringe scales generally homogenous or not (VLF2); and the cross-section of the tail round or square (TLcross).
Phylogenetic analyses
An input file implemented in BEAUti (Bayesian Evolutionary Analysis Utility) v. 2.4.6 was run in BEAST (Bayesian Evolutionary Analysis Sampling Trees) v. 2.4.6 (Drummond et al. 2012) on CIPRES (Cyberinfrastructure for Phylogenetic Research; Miller et al. 2010) in order to generate a BEAST phylogeny, employing a lognormal relaxed clock with unlinked site models and linked trees and clock models. bModelTest (Bouckaert and Drummond 2017), implemented in BEAST, was used to numerically integrate over the uncertainty of substitution models while simultaneously estimating the phylogeny using Markov chain Monte Carlo (MCMC). MCMC chains were run using a Yule prior for 40,000,000 generations and logged every 4,000 generations. The BEAST log file was visualized in Tracer v. 1.7.0 (Rambaut et al. 2018) to ensure effective sample sizes (ESS) were clearly above 200 for all parameters. A maximum clade credibility tree using mean heights at the nodes was generated using TreeAnnotator v. 1.8.0 (Rambaut and Drummond 2013) with a burn-in of 1,000 trees (10%). Nodes with Bayesian posterior probabilities (BPP) of 0.95 and above were considered strongly supported (Huelsenbeck et al. 2001; Wilcox et al. 2002). Uncorrected pairwise sequence divergences were calculated in MEGA 11 (Tamura et al. 2021) using the complete deletion option to remove gaps and missing data from the alignment prior to analysis.
Statistical analyses
All statistical analyses were conducted using R Core Team (2018). Morphometric characters used in statistical analyses were SVL, AG, HumL, ForL, FemL, TibL, HL, HW, HD, ED, EE, ES, EN, IO, EL, and IN. Tail metrics were not used due to the high degree of incomplete sampling (i.e., regenerated, broken, or missing). In order to most successfully remove the effects of allometry (sec. Chan and Grismer 2022), size was normalized using the following equation: Xadj = log(X)-β[log(SVL)-log(SVLmean)], where Xadj = adjusted value; X = measured value; β = unstandardized regression coefficient for each population; and SVLmean = overall average SVL of all populations (Thorpe 1975, 1983; Turan 1999; Lleonart et al. 2000), accessible in the R package GroupStruct (available at https://github.com/chankinonn/GroupStruct). The morphometrics of each species were normalized separately and then concatenated so as not to conflate potential intra- with interspecific variation (Reist 1986; McCoy et al. 2006). The juvenile Cyrtodactylusngati (HNUE-R00112) was removed from the data so as not to skew the normalization results. All data were scaled to their standard deviation to ensure they were analyzed on the basis of correlation and not covariance. Meristic characters analyzed were SL, IL, PVT, LRT, VS, VSM, TL4E, TL4U, TL4T, FL4E, FL4U, FL4T, FS, PCS, and BB. Precloacal and femoral pores were omitted from the multivariate analyses due to their absence in females. Categorical characters analyzed were DCT, VLF1, VLF2, TLcross, SC1, SC2, and SC3.
Small sample sizes (N = 1 or 2) for some of the species/populations precluded them from statistical analyses. A Levene’s test for the normalized morphometric and meristic characters was conducted to test for equal variances across all groups. Characters with equal variances were analyzed with an analysis of variance (ANOVA) and TukeyHSD post hoc test for mean comparisons involving more than three groups. Those with unequal variances were subjected to Welch’s F-test and Games-Howell post hoc test to test for mean comparisons involving more than three groups.
Morphospatial clustering and positioning among the species/populations was analyzed using multiple factor analysis (MFA) on a concatenated data set comprised of 15 meristic characters, 16 normalized morphometric characters, and seven categorical characters (Suppl. material 1). For this test, it was not necessary to remove populations represented by small sample sizes. The MFA was implemented using the mfa () command in the R package FactorMineR (Husson et al. 2017) and visualized using the Factoextra package (Kassambara and Mundt 2017). MFA is a global, unsupervised, multivariate analysis that incorporates qualitative and quantitative data (Pagès 2015) simultaneously, making it possible to analyze different data types in a nearly total evidence environment. In an MFA, each individual is described by a different set of variables (i.e., characters) which are structured into different data groups in a global data frame, in this case quantitative data (i.e., meristics and normalized morphometrics) and categorical data (i.e., scale, tubercle, and caudal morphology). In the first phase of the analysis, separate multivariate analyses are carried out for each set of variables: principal component analyses (PCA) for each quantitative data set and a multiple correspondence analysis (MCA) for the categorical data. The data sets are then normalized separately by dividing all their elements by the square root of their first eigenvalue. For the second phase of the analysis, these normalized data sets are concatenated into a single matrix for a final global PCA of the normalized data. Standardizing the data in this manner prevents one data type from overleveraging another. In other words, the normalization of the data in the first phase prevents data types with the greatest number of characters or the greatest amount of variation from outweighing other data types in the second phase. This way, the contributions of each data type to the overall variation in the data set are scaled to define the morphospatial distance between individuals as well as calculating each data type’s contribution to the overall variation in the analysis (Pagès 2015; Kassambara and Mundt 2017).
A non-parametric permutation multivariate analysis of variance (PERMANOVA) from the vegan package 2.5–3 in R (Oksanen et al. 2020) was used to determine if the centroid locations and group clustering of each species/population in the MFA were statistically different from one another (Skalski et al. 2018). The analysis was based on the calculation of a Gower (dis)similarity matrix using 50,000 permutations based on the loadings of the first four dimensions of the MFA. A pairwise post hoc test calculates the differences between all combinations of population pairs, generating a p-value, a Bonferroni-adjusted p-value, and a pseudo-F ratio (F statistic). A p < 0.05 is considered significant and larger F statistics indicate more pronounced group separation. A rejection of the null hypothesis (i.e., centroid positions and/or the spread of the data points [i.e. clusters] are no different from random) signifies a statistically significant difference between species/populations.
Phylomorphospace
The BEAST phylogeny was projected onto the first two dimensions of the MFA plot using the phylomorphospace () command from the R package phytools (Revell 2012). This allows one to map the history of a group’s morphological diversification and infer the magnitude and direction of shape change along branches of the phylogeny (Sidlauskas 2008). Outgroups and species from the phylogenetic analyses not represented in the morphological dataset were excluded. To eliminate a potential bias caused by the highly derived Cyrtodactyluselok (see Grismer et al. 2022), it was not included.
Some of the populations examined had only genetic or morphological data. However, only phylogenetically delimited populations bearing morphological differences from other populations were described as new species. In some cases, populations represented by only morphological data but in close geographic proximity to named species from which they could not be separated morphologically, were considered conferre (cf.) to the named species pending further investigation.
Results
Phylogenetic analysis
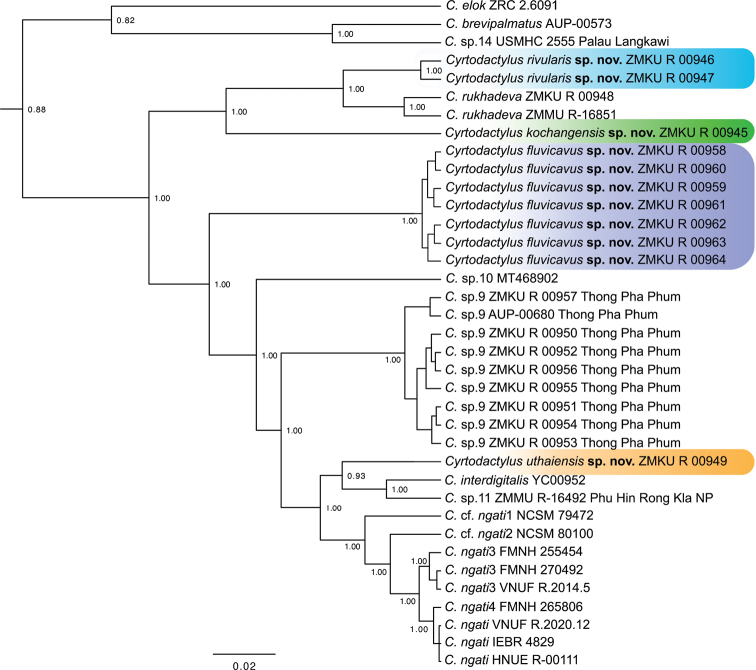
The BEAST analysis recovered 8–10 new species and two major clades within the brevipalmatus group: a weakly supported clade (BPP = 0.82) from the southernmost portion of the Thai-Malay Peninsula south of the Isthmus of Kra comprised of Cyrtodactyluselok, C.brevipalmatus, and C. sp.14 and a well-supported clade (1.00) comprised of all other species north of the Isthmus of Kra (Figs 1, 2). Within the latter, a well-supported (1.00) more exclusive clade containing the sister species from Hua Hin, Prachuap Khiri Khan (Cyrtodactylusrivularis sp. nov. [see below]) and C.rukhadeva with the Ko Chang population (Cyrtodactyluskochangensis sp. nov. [see below]) being sister to these. All relationships within this clade have strong nodal support (1.00) and its taxa occur only in the northernmost portion of the Thai-Malay Peninsula (Fig. 1). The population from Si Sawat (Cyrtodactylusfluvicavus sp. nov. [see below]), C. sp.10 from Chao Doi Waterfall, Mae Moei National Park, Tak Province, and C. sp.9 from Thong Pha Phum are sequentially related to a strongly supported (1.00) clade containing the remaining species. Within that latter clade, C.interdigitalis is the strongly supported (1.00) sister species to C. sp.11 from Phu Hin Rong Kla National Park and together they compose the well-supported (0.93) sister lineage to the Uthai Thani population (Cyrtodactylusuthaiensis sp. nov. [see below]). The remaining species all form a strongly supported (1.00) sequentially related clades with C.ngati.
Figure 2.
Maximum clade credibility BEAST phylogeny of the Cyrtodactylusbrevipalmatus group highlighting the new species described herein. Bayesian posterior probabilities (BPP) are listed at the nodes.
Uncorrected pairwise sequence divergences within the brevipalmatus group range from 2.84–22.84% (Table 2). Ranges for those of the new species described below are as follows: Cyrtodactylusrivularis sp. nov. ranges from 3.61–21.81%, being most similar to C.rukhadeva and most distant to C.elok; Cyrtodactylusfluvicavus sp. nov. ranges from 9.55–20.26%, being most similar to the Thong Pha Phum population (C. sp.9) and most distant to C.elok; Cyrtodactyluskochangensis sp. nov. ranges from 12.00–20.90%, being most similar to Cyrtodactylusrivularis sp. nov. and most distant to C.elok; and Cyrtodactylusuthaiensis sp. nov. ranges from 5.81–21.16%, being most similar to C.cf.ngati1 and most distant to C.elok.
Table 2.
Mean (minimum–maximum) percentages of uncorrected pairwise sequence divergence (p-distances) among the putative species of the Cyrtodactylusbrevipalmatus group based on 1,386 base pairs of mitochondrial NADH dehydrogenase subunit 2 gene (ND2) and adjacent tRNAs. Intraspecific p-distance are in bold font. n/a = data not applicable.
| C.brevipalmatus | C.cf.ngati1 | C.cf.ngati2 | C.elok | Cyrtodactylusfluvicavus sp. nov. | C.interdigitalis | Cyrtodactyluskochangensis sp. nov. | C.ngati, C.ngati3 and C.ngati4 | Cyrtodactylusrivularis sp. nov. | C.rukhadeva | C. sp.9 (Thong Pha Phum) | C. sp.10 | C. sp.11 | C. sp.14 | Cyrtodactylusuthaiensis sp. nov. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C.brevipalmatus N = 1 | n/a | ||||||||||||||
| C.cf.ngati1 N = 1 | 21.03 | n/a | |||||||||||||
| C.cf.ngati2 N = 1 | 21.68 | 4.39 | n/a | ||||||||||||
| C.elok N = 1 | 20.77 | 22.58 | 21.42 | n/a | |||||||||||
| Cyrtodactylusfluvicavus sp. nov. N = 7 | 18.86 | 10.64 | 11.02 | 20.15 | 0.10 | ||||||||||
| (18.84–18.97) | (10.58–10.84) | (10.97–11.23) | (20.13–20.26) | (0.00–0.26) | |||||||||||
| C.interdigitalis N = 1 | 20.77 | 6.97 | 9.16 | 22.84 | 12.02 | n/a | |||||||||
| (12.00–12.13) | |||||||||||||||
| Cyrtodactyluskochangensis sp. nov. N = 1 | 19.35 | 14.58 | 14.71 | 20.90 | 12.31 | 15.23 | n/a | ||||||||
| (12.26–12.31) | |||||||||||||||
| C.ngati, C.ngati3 and C.ngati4 N = 7 | 20.70 | 3.30 | 3.71 | 21.11 | 11.34 | 8.13 | 14.58 | 0.84 | |||||||
| (20.65–20.90) | (2.84–4.00) | (3.35–4.26) | (20.90–21.42) | (11.10–11.87) | (7.74–8.65) | (14.45–14.84) | (0.00–1.55) | ||||||||
| Cyrtodactylusrivularis sp. nov. N = 2 | 20.00 | 15.87 | 15.03 | 21.61 | 12.57 | 15.48 | 12.26 | 15.03 | 0.52 | ||||||
| (19.74–20.26) | (15.61–16.13) | (14.84–15.23) | (21.42–21.81) | (12.26–13.03) | (15.23–15.74) | (12.00–12.52) | (14.71–15.48) | ||||||||
| C.rukhadeva N = 2 | 20.65 | 15.42 | 15.48 | 21.61 | 12.25 | 16.00 | 13.10 | 15.23 | 4.65 | 1.55 | |||||
| (20.13–21.16) | (14.84–16.00) | (14.84–16.13) | (21.16–22.06) | (11.61–13.03) | (15.35–16.65) | (12.52–13.68) | (14.19–16.23) | (3.61–5.68) | |||||||
| C. sp.9 (Thong Pha Phum) N = 9 | 20.34 | 7.93 | 9.51 | 22.02 | 9.75 | 8.96 | 13.22 | 8.81 | 13.12 | 13.25 | 0.22 | ||||
| (20.13–20.65) | (7.74–8.00) | (9.42–9.55) | (21.81–22.32) | (9.55–9.94) | (8.77–9.03) | (13.03–13.29) | (8.13–9.68) | (12.77–13.42) | (12.52–13.94) | (0.00–0.52) | |||||
| C. sp.10 N = 1 | 19.87 | 9.29 | 10.84 | 21.94 | 10.12 | 10.19 | 13.68 | 10.21 | 13.94 | 14.32 | 8.06 | n/a | |||
| (10.06–10.32) | (10.06–10.45) | (13.68–14.19) | (13.68–14.97) | (7.87–8.13) | |||||||||||
| C. sp.11 N = 1 | 20.39 | 7.23 | 8.90 | 22.19 | 11.12 | 3.87 | 14.58 | 8.28 | 15.35 1) | 15.61 | 8.96 | 10.45 | n/a | ||
| (11.10–11.23) | (8.00–8.65) | (15.10–15.6 | (14.97–16.26) | (8.77–9.03) | |||||||||||
| C. sp.14 N = 1 | 6.45 | 20.90 | 20.65 | 20.00 | 18.34 | 20.13 | 19.10 | 20.52 | 19.74 | 20.00 | 19.60 | 18.84 | 19.61 | n/a | |
| (18.32–18.45) | (20.26–20.65) | (19.48–20.00) | (19.48–20.52) | (19.48–19.87) | |||||||||||
| Cyrtodactylusuthaiensis sp. nov. N = 1 | 19.74 | 5.81 | 8.13 | 21.16 | 10.12 | 7.1 | 13.94 | 6.97 | 13.94 | 13.94 | 7.80 | 8.39 | 6.58 | 19.48 | n/a |
| (10.06–10.32) | (6.58–7.61) | (13.68–14.19) | (13.29–14.58) | (7.61–7.87) | |||||||||||
Bayesian Poisson Tree Process (bPTP)
The bPTP species delimitation analysis recovered 16 putative species within the brevipalmatus group with varying degrees of support (Table 3). The newly acquired material in this study recovered as distinct species are Cyrtodactylusfluvicavus sp. nov., Cyrtodactyluskochangensis sp. nov., Cyrtodactylusrivularis sp. nov., the holotype of C.rukhadeva (ZMMU R-16851), C.rukhadeva (ZMKU R 00948), Cyrtodactylusuthaiensis sp. nov., C. sp.9 from Thong Pha Phum, and C. sp.14 from Pulau Langkawi. However, Cyrtodactylusrivularis sp. nov. and C. sp.9 were not recovered with strong support (NMI 0.575 and 0.565, respectively) despite them being genetically distinct (Table 2) and morphologically diagnosable (Tables 4–6). The analysis also separated the two specimens of C.rukhadeva from the same locality which only differ by a genetic distance of 1.55% and are similar in morphology. Sukumaran and Knowles (2017) demonstrated that many species delimitation analyses recover clades not species and that a wider range of empirical data are necessary to interpret these boundaries (Coyne and Orr 1998; Feulner et al. 2007; Fontaneto et al. 2007; Knowles and Carstens 2007; Leach et al. 2009) as was done here (see below).
Table 3.
Species delimited by the bPTP analysis.
| Species | NMI |
|---|---|
| C.brevipalmatus | 1 |
| C.cf.ngati1 | 1 |
| C.cf.ngati2 | 0.967 |
| C.elok | 1 |
| Cyrtodactylusfluvicavus sp. nov. | 0.957 |
| C.interdigitalis | 0.997 |
| Cyrtodactyluskochangensis sp. nov. | 1 |
| C.ngati | 0.841 |
| Cyrtodactylusrivularis sp. nov. | 0.575 |
| C.rukhadeva ZMKU R 00948 | 0.947 |
| C.rukhadeva ZMMU R-16851 | 0.947 |
| Cyrtodactylusuthaiensis sp. nov | 1 |
| C. sp.9 (Thong Pha Phum) | 0.565 |
| C. sp.10 | 1 |
| C. sp.11 | 0.997 |
| C. sp.14 | 1 |
Table 4.
Summary statistics of the normalized morphometric data for the putative species of the Cyrtodactylusbrevipalmatus group. SD = ± standard deviation. Character abbreviations are listed in the Materials and methods.
| Species | SVL | AG | HumL | ForL | FemL | TibL | HL | HW | HD | ED | EE | ES | EN | IO | EL | IN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C.brevipalmatus (N = 5) | ||||||||||||||||
| Mean | 1.82 | 1.49 | 0.96 | 0.96 | 1.06 | 1.02 | 1.27 | 1.10 | 0.87 | 0.67 | 0.75 | 0.86 | 0.73 | 0.68 | 0.07 | 0.32 |
| SD | 0.020 | 0.027 | 0.031 | 0.023 | 0.024 | 0.036 | 0.012 | 0.006 | 0.017 | 0.041 | 0.025 | 0.008 | 0.019 | 0.043 | 0.058 | 0.041 |
| Lower | 1.80 | 1.46 | 0.92 | 0.92 | 1.02 | 0.97 | 1.25 | 1.10 | 0.86 | 0.63 | 0.71 | 0.85 | 0.70 | 0.62 | -0.02 | 0.25 |
| Upper | 1.85 | 1.52 | 0.99 | 0.98 | 1.09 | 1.07 | 1.29 | 1.11 | 0.89 | 0.73 | 0.77 | 0.87 | 0.75 | 0.73 | 0.13 | 0.36 |
| C.elok (N = 4) | ||||||||||||||||
| Mean | 1.91 | 1.59 | 0.80 | 1.05 | 1.14 | 1.13 | 1.34 | 1.20 | 1.00 | 0.71 | 0.83 | 0.95 | 0.79 | 0.70 | 0.19 | 0.43 |
| SD | 0.016 | 0.012 | 0.341 | 0.026 | 0.022 | 0.024 | 0.001 | 0.012 | 0.008 | 0.017 | 0.020 | 0.008 | 0.010 | 0.070 | 0.062 | 0.033 |
| Lower | 1.89 | 1.57 | 0.34 | 1.02 | 1.11 | 1.10 | 1.34 | 1.18 | 0.98 | 0.68 | 0.81 | 0.94 | 0.78 | 0.61 | 0.15 | 0.40 |
| Upper | 1.93 | 1.60 | 1.09 | 1.08 | 1.16 | 1.15 | 1.34 | 1.21 | 1.00 | 0.72 | 0.85 | 0.95 | 0.80 | 0.76 | 0.28 | 0.48 |
| Cyrtodactylusfluvicavus sp. nov. (N = 7) | ||||||||||||||||
| Mean | 1.86 | 1.52 | 0.95 | 1.01 | 1.11 | 1.03 | 1.31 | 1.14 | 0.91 | 0.69 | 0.78 | 0.92 | 0.79 | 0.73 | 0.20 | 0.38 |
| SD | 0.027 | 0.009 | 0.013 | 0.017 | 0.018 | 0.012 | 0.007 | 0.011 | 0.011 | 0.007 | 0.016 | 0.008 | 0.011 | 0.006 | 0.04 | 0.02 |
| Lower | 1.82 | 1.51 | 0.94 | 0.98 | 1.10 | 1.01 | 1.30 | 1.13 | 0.90 | 0.68 | 0.76 | 0.91 | 0.78 | 0.72 | 0.14 | 0.35 |
| Upper | 1.89 | 1.54 | 0.98 | 1.04 | 1.15 | 1.05 | 1.32 | 1.16 | 0.93 | 0.70 | 0.81 | 0.94 | 0.81 | 0.74 | 0.24 | 0.40 |
| C.interdigitalis (N = 4) | ||||||||||||||||
| Mean | 1.86 | 1.50 | 0.98 | 1.01 | 1.10 | 1.08 | 1.30 | 1.12 | 0.80 | 0.72 | 0.76 | 0.90 | 0.77 | 0.68 | 0.13 | 0.33 |
| SD | 0.060 | 0.016 | 0.032 | 0.015 | 0.020 | 0.007 | 0.012 | 0.006 | 0.187 | 0.026 | 0.026 | 0.024 | 0.034 | 0.0307 | 0.033 | 0.035 |
| Lower | 1.78 | 1.48 | 0.94 | 0.99 | 1.07 | 1.07 | 1.28 | 1.12 | 0.56 | 0.69 | 0.73 | 0.88 | 0.74 | 0.66 | 0.10 | 0.29 |
| Upper | 1.91 | 1.52 | 1.01 | 1.02 | 1.12 | 1.09 | 1.31 | 1.13 | 0.96 | 0.75 | 0.79 | 0.94 | 0.81 | 0.73 | 0.17 | 0.38 |
| Cyrtodactyluskochangensis sp. nov. (N = 1) | ||||||||||||||||
| Value | 1.78 | 1.51 | 1.00 | 0.95 | 1.05 | 1.02 | 1.30 | 1.14 | 0.91 | 0.69 | 0.76 | 0.91 | 0.78 | 0.68 | 0.05 | 0.36 |
| C.cf.kochangensis sp. nov. (N = 1) | ||||||||||||||||
| Value | 1.85 | 1.50 | 1.02 | 0.94 | 1.08 | 1.08 | 1.27 | 1.09 | 0.90 | 0.72 | 0.70 | 0.88 | 0.75 | 0.60 | 0.12 | 0.36 |
| C.ngati (N = 3) | ||||||||||||||||
| Mean | 1.83 | 1.47 | 0.91 | 0.99 | 1.06 | 1.05 | 1.31 | 1.08 | 0.85 | 0.57 | 0.76 | 0.86 | 0.81 | 0.74 | -0.12 | 0.43 |
| SD | 0.009 | 0.002 | 0.004 | 0.007 | 0.000 | 0.006 | 0.001 | 0.003 | 0.010 | 0.035 | 0.016 | 0.015 | 0.004 | 0.009 | 0.019 | 0.008 |
| Lower | 1.82 | 1.47 | 0.91 | 0.98 | 1.06 | 1.04 | 1.31 | 1.08 | 0.84 | 0.55 | 0.74 | 0.85 | 0.80 | 0.73 | -0.13 | 0.42 |
| Upper | 1.84 | 1.47 | 0.91 | 1.00 | 1.06 | 1.05 | 1.31 | 1.09 | 0.86 | 0.61 | 0.77 | 0.88 | 0.81 | 0.75 | -0.09 | 0.43 |
| C.ngati3 (N = 3) | ||||||||||||||||
| Mean | 1.88 | 1.58 | 0.94 | 1.00 | 1.12 | 1.07 | 1.32 | 1.12 | 0.95 | 0.69 | 0.81 | 0.93 | 0.80 | 0.77 | 0.08 | 0.41 |
| SD | 0.039 | 0.001 | 0.002 | 0.02 | 0.001 | 0.017 | 0.007 | 0.002 | 0.014 | 0.003 | 0.004 | 0.007 | 0.006 | 0.022 | 0.016 | 0.01 |
| Lower | 1.85 | 1.58 | 0.93 | 0.98 | 1.12 | 1.05 | 1.31 | 1.12 | 0.94 | 0.69 | 0.80 | 0.92 | 0.79 | 0.74 | 0.07 | 0.40 |
| Upper | 1.92 | 1.58 | 0.94 | 1.02 | 1.12 | 1.09 | 1.33 | 1.12 | 0.96 | 0.69 | 0.81 | 0.94 | 0.80 | 0.78 | 0.10 | 0.42 |
| C.ngati4 (N = 1) | ||||||||||||||||
| Value | 1.87 | 1.50 | 0.87 | 1.02 | 1.13 | 1.07 | 1.33 | 1.11 | 0.91 | 0.72 | 0.79 | 0.94 | 0.83 | 0.75 | 0.51 | 0.43 |
| C.cf.ngati1 (N = 1) | ||||||||||||||||
| Value | 1.89 | 1.59 | 0.96 | 1.03 | 1.13 | 1.12 | 1.34 | 1.12 | 0.95 | 0.85 | 0.76 | 0.96 | 0.82 | 0.73 | 0.28 | 0.49 |
| C.cf.ngati2 (N = 2) | ||||||||||||||||
| Mean | 1.92 | 1.59 | 1.01 | 1.02 | 1.17 | 1.1 | 1.34 | 1.15 | 0.95 | 0.72 | 0.80 | 0.94 | 0.80 | 0.65 | 0.03 | 0.37 |
| SD | 0.035 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Lower | 1.89 | 1.59 | 1.01 | 1.02 | 1.17 | 1.10 | 1.34 | 1.15 | 0.95 | 0.72 | 0.80 | 0.94 | 0.80 | 0.65 | 0.03 | 0.37 |
| Upper | 1.94 | 1.59 | 1.01 | 1.02 | 1.17 | 1.10 | 1.34 | 1.15 | 0.95 | 0.72 | 0.80 | 0.94 | 0.80 | 0.65 | 0.03 | 0.37 |
| Cyrtodactylusrivularis sp. nov. (N = 2) | ||||||||||||||||
| Mean | 1.85 | 1.53 | 0.89 | 0.98 | 1.04 | 1.03 | 1.30 | 1.16 | 0.91 | 0.76 | 0.80 | 0.91 | 0.77 | 0.75 | 0.05 | 0.34 |
| SD | 0.025 | 0.01 | 0.002 | 0.007 | 0.007 | 0.005 | 0.005 | 0.001 | 0.013 | 0.007 | 0.024 | 0.005 | 0.004 | 0.025 | 0.019 | 0.016 |
| Lower | 1.83 | 1.53 | 0.89 | 0.97 | 1.03 | 1.03 | 1.29 | 1.16 | 0.91 | 0.75 | 0.79 | 0.91 | 0.77 | 0.74 | 0.04 | 0.33 |
| Upper | 1.87 | 1.54 | 0.90 | 0.98 | 1.04 | 1.04 | 1.30 | 1.16 | 0.92 | 0.76 | 0.82 | 0.91 | 0.78 | 0.77 | 0.06 | 0.35 |
| C.rukhadeva and C.cf.rukhadeva (N = 10) | ||||||||||||||||
| Mean | 1.85 | 1.49 | 1.01 | 0.95 | 1.05 | 1.02 | 1.30 | 1.14 | 0.92 | 0.71 | 0.75 | 0.91 | 0.77 | 0.67 | 0.09 | 0.35 |
| SD | 0.026 | 0.028 | 0.055 | 0.029 | 0.027 | 0.023 | 0.009 | 0.018 | 0.025 | 0.036 | 0.034 | 0.014 | 0.015 | 0.087 | 0.069 | 0.022 |
| Lower | 1.79 | 1.45 | 0.91 | 0.90 | 0.99 | 0.97 | 1.29 | 1.10 | 0.87 | 0.62 | 0.70 | 0.89 | 0.74 | 0.46 | 0.00 | 0.32 |
| Upper | 1.88 | 1.54 | 1.08 | 1.00 | 1.09 | 1.05 | 1.32 | 1.16 | 0.95 | 0.75 | 0.80 | 0.92 | 0.79 | 0.73 | 0.23 | 0.38 |
| Cyrtodactylusuthaiensis sp. nov. (N = 1) | ||||||||||||||||
| Value | 1.76 | 1.80 | 0.95 | 0.99 | 1.09 | 1.02 | 1.28 | 1.10 | 0.76 | 0.72 | 0.74 | 0.88 | 0.75 | 0.68 | 0.19 | 0.32 |
| C. sp.9 (Thong Pha Phum) (N = 8) | ||||||||||||||||
| Mean | 1.86 | 1.53 | 0.91 | 0.97 | 1.08 | 1.02 | 1.30 | 1.16 | 0.89 | 0.71 | 0.77 | 0.90 | 0.77 | 0.74 | 0.09 | 0.34 |
| SD | 0.024 | 0.016 | 0.034 | 0.021 | 0.032 | 0.019 | 0.014 | 0.017 | 0.008 | 0.015 | 0.004 | 0.012 | 0.007 | 0.012 | 0.061 | 0.023 |
| Lower | 1.81 | 1.50 | 0.86 | 0.93 | 1.02 | 0.99 | 1.27 | 1.14 | 0.88 | 0.69 | 0.76 | 0.89 | 0.76 | 0.72 | 0.00 | 0.30 |
| Upper | 1.88 | 1.55 | 0.95 | 1.00 | 1.12 | 1.05 | 1.32 | 1.19 | 0.90 | 0.74 | 0.78 | 0.93 | 0.78 | 0.76 | 0.18 | 0.38 |
| C. sp.11 (N = 1) | ||||||||||||||||
| Value | 1.83 | 1.53 | 1.01 | 0.99 | 1.13 | 1.09 | 1.30 | 1.14 | 0.75 | 0.70 | 0.78 | 0.92 | 0.78 | 0.61 | 0.08 | 0.35 |
| C. sp.13 (N = 2) | ||||||||||||||||
| Mean | 1.83 | 1.45 | 0.94 | 0.94 | 1.07 | 1.01 | 1.27 | 1.11 | 0.91 | 0.67 | 0.75 | 0.88 | 0.76 | 0.74 | 0.15 | 0.34 |
| SD | 0.040 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Lower | 1.80 | 1.45 | 0.94 | 0.94 | 1.07 | 1.01 | 1.27 | 1.11 | 0.91 | 0.67 | 0.75 | 0.88 | 0.76 | 0.74 | 0.15 | 0.34 |
| Upper | 1.86 | 1.45 | 0.94 | 0.94 | 1.07 | 1.01 | 1.27 | 1.11 | 0.91 | 0.67 | 0.75 | 0.88 | 0.76 | 0.74 | 0.15 | 0.34 |
Table 6.
Significant p-values from the results of the ANOVA analyses comparing all combinations of OTU pairs of the Cyrtodactylusbrevipalmatus group. SVL and TL4U are not listed because no species pairs differed significantly from one another. * = Results based on a Games-Howell post hoc test. Character abbreviations are listed in the Materials and methods.
| Morphometric characters | AG* | HumL* | ForL | FemL | TibL | HL | HW | HD* | ED* | EE* | ES | EN* | IO | EL | IN |
| Cyrtodactylusfluvicavus sp. nov. vs. C.brevipalmatus | 0.00 | 0.01 | < 0.001 | 0.001 | 0.048 | < 0.001 | 0.004 | 0.002 | 0.004 | ||||||
| C.interdigitalis vs. C.brevipalmatus | 0.00 | 0.00 | 0.00 | < 0.001 | |||||||||||
| C.ngati vs. C.brevipalmatus | 0.00 | 0.003 | 0.001 | < 0.001 | |||||||||||
| C.ngati3 vs. C.brevipalmatus | 0.014 | 0.03 | 0.01 | < 0.001 | 0.008 | 0.038 | < 0.001 | 0.004 | 0.000 | ||||||
| C.rukhadeva vs. C.brevipalmatus | < 0.001 | 0.001 | 0.022 | < 0.001 | 0.021 | ||||||||||
| C. sp.9 vs. C.brevipalmatus | 0.01 | < 0.001 | < 0.001 | 0.03 | |||||||||||
| C.interdigitalis vs. Cyrtodactylusfluvicavus sp. nov. | 0.01 | ||||||||||||||
| C.ngati vs. Cyrtodactylusfluvicavus sp. nov. | < 0.001 | 0.002 | 0.04 | < 0.001 | 0.005 | < 0.001 | < 0.001 | ||||||||
| C.ngati3 vs. Cyrtodactylusfluvicavus sp. nov. | < 0.001 | 0.025 | 0.038 | ||||||||||||
| C.rukhadeva vs. Cyrtodactylusfluvicavus sp. nov. | 0.049 | < 0.001 | < 0.001 | 0.004 | |||||||||||
| C. sp.9 vs. Cyrtodactylusfluvicavus sp. nov. | 0.0 | 0.007 | 0.013 | 0.023 | 0.007 | ||||||||||
| C.ngati vs. C.interdigitalis | 0.007 | 0.031 | 0.010 | < 0.001 | 0.000 | ||||||||||
| C.ngati3 vs. C.interdigitalis | 0.011 | 0.003 | |||||||||||||
| C.rukhadeva vs. C.interdigitalis | 0.0 | 0.01 | 0.00 | 0.044 | |||||||||||
| C. sp.9 vs. C.interdigitalis | 0.00 | 0.007 | |||||||||||||
| C.ngati3 vs. C.ngati | < 0.001 | 0.019 | 0.006 | < 0.001 | 0.001 | ||||||||||
| C.rukhadeva vs. C.ngati | 0.003 | < 0.001 | < 0.001 | 0.046 | < 0.001 | 0.001 | < 0.001 | < 0.001 | 0.001 | ||||||
| C. sp.9 vs. C.ngati | < 0.001 | < 0.001 | 0.042 | 0.007 | < 0.001 | < 0.001 | 0.000 | ||||||||
| C.rukhadeva vs. C.ngati3 | < 0.001 | 0.021 | 0.0 | 0.00 | 0.007 | 0.02 | 0.006 | ||||||||
| C. sp.9 vs. C.ngati3 | 0.001 | 0.01 | 0.03 | 0.003 | 0.043 | 0.001 | 0.019 | 0.019 | 0.003 | ||||||
| C. sp.9 vs. C.rukhadeva | 0.02 | 0.004 | 0.02 | 0.033 | |||||||||||
| Meristic characters | SL | IL* | PVT* | LRT | VS | VSM | TL4E | TL4T | FL4E | FL4U* | FL4T* | FS | PCS* | BB* | |
| Cyrtodactylusfluvicavus sp. nov. vs. C.brevipalmatus | 0.037 | < 0.001 | 0.05 | ||||||||||||
| C.interdigitalis vs C.brevipalmatus | < 0.001 | 0.000 | 0.002 | 0.044 | |||||||||||
| C.ngati vs C.brevipalmatus | < 0.001 | 0.028 | < 0.001 | ||||||||||||
| C.ngati3 vs C.brevipalmatus | 0.017 | 0.029 | |||||||||||||
| C.rukhadeva vs C.brevipalmatus | < 0.001 | < 0.001 | |||||||||||||
| C. sp.9 vs C.brevipalmatus | < 0.001 | 0.003 | 0.022 | < 0.001 | 0.05 | ||||||||||
| C.interdigitalis vs Cyrtodactylusfluvicavus sp. nov. | 0.000 | 0.000 | 0.021 | < 0.001 | 0.000 | < 0.001 | |||||||||
| C.ngati vs Cyrtodactylusfluvicavus sp. nov. | 0.037 | 0.002 | < 0.001 | < 0.001 | 0.033 | 0.000 | 0.002 | < 0.001 | |||||||
| C.ngati3 vs Cyrtodactylusfluvicavus sp. nov. | 0.005 | ||||||||||||||
| C.rukhadeva vs Cyrtodactylusfluvicavus sp. nov. | 0.000 | 0.001 | 0.001 | ||||||||||||
| C. sp.9 vs Cyrtodactylusfluvicavus sp. nov. | < 0.001 | < 0.001 | 0.001 | 0.004 | 0.020 | ||||||||||
| C.ngati vs C.interdigitalis | 0.001 | 0.029 | < 0.001 | < 0.001 | 0.029 | ||||||||||
| C.ngati3 vs C.interdigitalis | 0.029 | 0.025 | 0.005 | ||||||||||||
| C.rukhadeva vs C.interdigitalis | 0.004 | 0.011 | 0.001 | 0.044 | 0.001 | 0.002 | |||||||||
| C. sp.9 vs C.interdigitalis | 0.003 | < 0.001 | 0.005 | 0.043 | 0.01 | < 0.001 | |||||||||
| C.ngati3 vs C.ngati | 0.0267 | 0.001 | < 0.001 | ||||||||||||
| C.rukhadeva vs C.ngati | < 0.001 | < 0.001 | 0.008 | < 0.001 | 0.008 | 0.03 | < 0.001 | ||||||||
| C. sp.9 vs C.ngati | 0.003 | 0.016 | 0.011 | 0.000 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | |||||||
| C.rukhadeva vs C.ngati3 | |||||||||||||||
| C. sp.9 vs C.ngati3 | 0.001 | 0.042 | < 0.001 | 0.001 | |||||||||||
| C. sp.9 vs C.rukhadeva | 0.029 | < 0.001 | 0.002 | 0.001 |
Statistical analyses
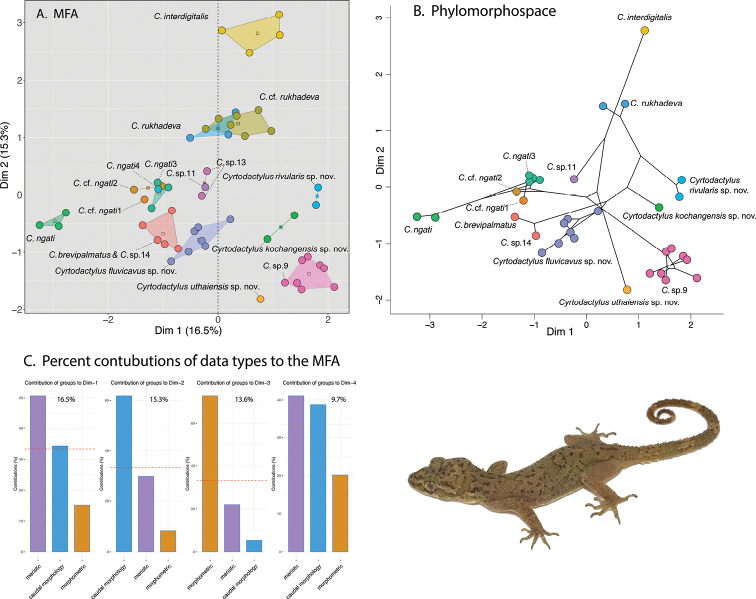
The ANOVA and TukeyHSD post hoc analyses of the adjusted morphometric and meristic characters were consistent with the phylogenetic and pairwise distance data in recovering a number of statistically significant differences between the Cyrtodactylusfluvicavus sp. nov. and C.interdigitalis as well as others (Table 6). Cyrtodactylusfluvicavus sp. nov. plotted separately in the MFA, only slightly overlapping with C.brevipalmatus (Fig. 3A). Cyrtodactyluskochangensis sp. nov., Cyrtodactylusrivularis sp. nov. and Cyrtodactylusuthaiensis sp. nov. plotted separately from each other and all other populations. A PERMANOVA analysis recovered several instances of various combinations of statistically significant differences among all the populations/species numbering more than two samples in regards to their clustering and centroid placement (Table 7). The contributions of each data type are shown in Fig. 3C.
Figure 3.
AMFA of the species-level lineages based on the BEAST phylogeny (Fig. 2) B phylomorphospace projection of the BEAST phylogeny C percent contribution of each data type to Dim 1–4 of the MFA. The percentage score at the top of each graph is the percent contribution of that dimension to the overall variation in the data set.
Table 7.
Summary statistics from the PERMANOVA analysis of the OTUs and proposed morphogroups of the Cyrtodactylusbrevipalmatus group.
| OTU pairs | F model | R2 | p-value | p-adjusted |
|---|---|---|---|---|
| C.rukhadeva vs. C.cf.ngati2 | 6.8474 | 0.4064 | 0.015 | 0.544 |
| C.rukhadeva vs. C.ngati3 | 8.8824 | 0.4467 | 0.003 | 0.122 |
| C.rukhadeva vs. C.interdigitalis | 3.3630 | 0.2189 | 0.006 | 0.201 |
| C.rukhadeva vs. C.ngati | 10.4580 | 0.4874 | 0.003 | 0.114 |
| C.rukhadeva vs. C.brevipalmatus | 6.5983 | 0.3367 | 0.000 | 0.012 |
| C.rukhadeva vs. Cyrtodactylusfluvicavus sp. nov. | 6.6357 | 0.3067 | 0.000 | 0.004 |
| C.rukhadeva vs. C. sp.9 | 3.8646 | 0.1945 | 0.001 | 0.033 |
| C.cf.ngati2 vs. C.brevipalmatus | 15.4818 | 0.7559 | 0.048 | 1.000 |
| C.cf.ngati2 vs. Cyrtodactylusfluvicavus sp. nov. | 15.9186 | 0.6946 | 0.027 | 0.967 |
| C.cf.ngati2 vs. C. sp.9 | 19.0130 | 0.7038 | 0.022 | 0.804 |
| C.ngati3 vs. C.interdigitalis | 4.4753 | 0.4723 | 0.029 | 1.000 |
| C.ngati3 vs. C.brevipalmatus | 14.9425 | 0.7135 | 0.018 | 0.643 |
| C.ngati3 vs. Cyrtodactylusfluvicavus sp. nov. | 8.7953 | 0.5237 | 0.009 | 0.317 |
| C.ngati3 vs. C. sp.9 | 14.7978 | 0.6218 | 0.006 | 0.226 |
| C.interdigitalis vs. C.ngati | 9.8976 | 0.6644 | 0.029 | 1.000 |
| C.interdigitalis vs. C.brevipalmatus | 4.5646 | 0.3947 | 0.008 | 0.278 |
| C.interdigitalis vs. Cyrtodactylusfluvicavus sp. nov. | 6.7120 | 0.4272 | 0.003 | 0.124 |
| C.interdigitalis vs. C. sp.9 | 5.6585 | 0.3614 | 0.002 | 0.067 |
| C.ngati vs. C.brevipalmatus | 7.4818 | 0.5550 | 0.018 | 0.643 |
| C.ngati vs. Cyrtodactylusfluvicavus sp. nov. | 22.8234 | 0.7405 | 0.008 | 0.283 |
| C.ngati vs. C. sp.9 | 17.0146 | 0.6540 | 0.006 | 0.227 |
| C.brevipalmatus vs. Cyrtodactylusfluvicavus sp. nov. | 17.8585 | 0.6410 | 0.001 | 0.048 |
| C.brevipalmatus vs. C. sp.9 | 9.3960 | 0.4607 | 0.001 | 0.025 |
| Cyrtodactylusfluvicavus sp. nov. vs. C. sp.9 | 8.2047 | 0.3869 | 0.000 | 0.005 |
Morphological and phylogenetic discordance
The phylomorphospace analysis illustrates that the morphological similarities among the species/populations in the MFA are discordant with their phylogenetic placement in the tree (Fig. 3A, B). In other words, species/populations that are similar in morphology are not necessarily each other’s closest relatives. For example, Cyrtodactylusinterdigitalis is morphologically more similar to the distantly related C.rukhadeva than it is to C. sp.11 or Cyrtodactylusuthaiensis sp. nov. to which it is closely related. Similarly, C. sp.9 is morphologically similar to the distantly related Cyrtodactyluskochangensis sp. nov. than to the closely related clade containing Cyrtodactylusuthaiensis sp. nov., C.interdigitalis, C. sp.11, C.cf.ngati1, C.cf.ngati2, C.ngati3, C.ngati4, and C.ngati. Other examples of morphological and phylogenetic discordance can be seen by comparing Fig. 3A, B.
Taxonomy
Given the phylogenetic delimitation of the Si Sawat, Prachuap Khiri Khan, Uthai Thani, and Ko Chang populations (Figs 2, 3), the statistically significant diagnostic placement of the Si Sawat population in morphospace (Fig. 3A; Table 7) and its statistically significant diagnostic morphological differences (Table 6), we describe them below as new species. The phylogenetic delimitation and statistically significant morphological differences among C. sp.9 from Thong Pha Phum and C. sp.11 from Phu Hin Rong Kla National Park will require descriptions at a future date.
. Cyrtodactylus fluvicavus sp. nov.
C909A4C7-64EF-5999-9A31-19E7EB8BF7B6
https://zoobank.org/B65469C6-55A1-401B-9EC3-FAFF151A2973
Figs 4 , 5 Suggested Common Name: Tham Than Lot Bent-toed Gecko
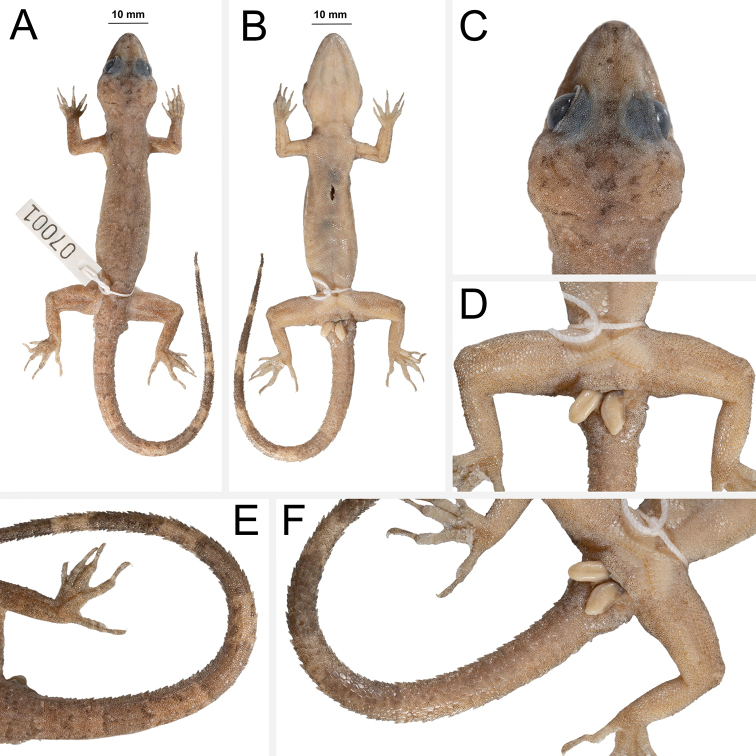
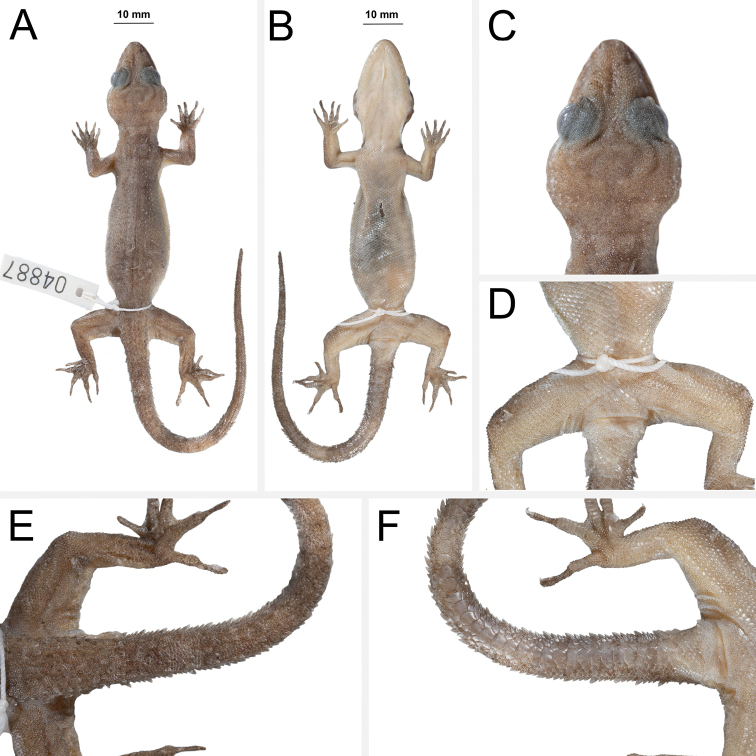
Figure 4.
Holotype of Cyrtodactylusfluvicavus sp. nov. ZMKU R 00959 (field no. AA 07001) from Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park A dorsal view B ventral view C dorsal view of head D ventral view of femoral and precloacal regions E dorsal view of tail F ventral view of tail.
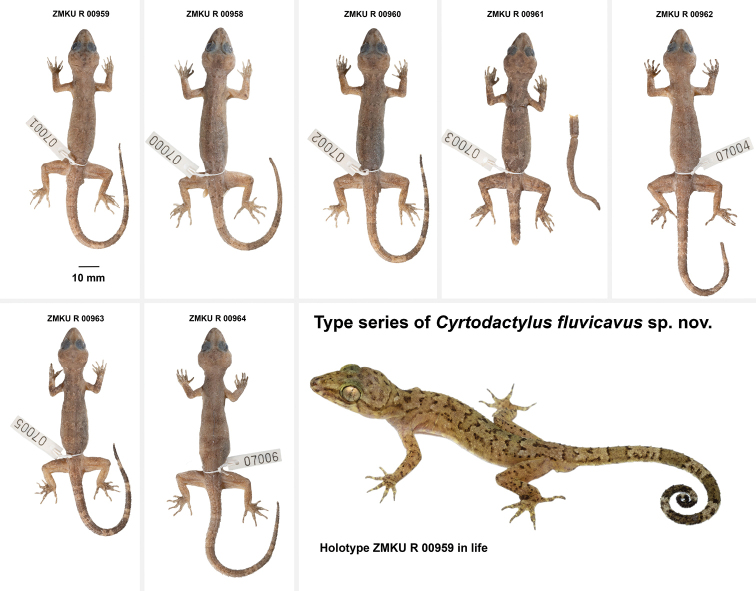
Figure 5.
Type series of Cyrtodactylusfluvicavus sp. nov. from Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park.
Cyrtodactylus sp. Yodthong, Rujirawan, Stuart, Grismer, Aksornneam, Termprayoon, Ampai & Aowphol, 2022: 161.
Holotype.
Adult male ZMKU R 00959 from Tham Than Lot Noi-Tham Than Lot Yai Nature Trail, Chaloem Rattanakosin National Park, Khao Chot Subdistrict, Si Sawat District, Kanchanaburi Province, Thailand (14.66930°N, 99.29060°E, 526 m a.s.l.), collected by Korkhwan Termprayoon, Akrachai Aksornneam, Natee Ampai, and Siriporn Yodthong on 20 April 2019.
Paratypes.
Adult males ZMKU R 00958 and ZMKU R 00960 and adult females ZMKU R 00961–64 bear the same collection site as the holotype.
Diagnosis.
Cyrtodactylusfluvicavus sp. nov. can be separated from all other species of the brevipalmatus group by the combination of having 11–13 supralabials, 9 or 10 infralabials, 26–30 paravertebral tubercles, 14–18 rows of longitudinally arranged tubercles, 30–39 transverse rows of ventrals, 154–175 longitudinal rows of ventrals, 9–11 expanded subdigital lamellae on the fourth toe, 10–13 unexpanded subdigital lamellae on the fourth toe, 19–22 total subdigital lamellae on the fourth toe; 7–9 expanded subdigital lamellae on the fourth finger, 9–11 unexpanded subdigital lamellae on the fourth finger, 17–19 total subdigital lamellae on the fourth finger; 9–12 total enlarged femoral scales, 8–11 total femoral pores in males; 14 or 15 precloacal pores in males; 14 or 15 enlarged precloacals; enlarged femorals and enlarged precloacals not continuous; proximal femorals less than one-half the size of the distal femorals; small tubercles on forelimbs and flanks; small dorsolateral caudal tubercles and narrow ventrolateral caudal fringe; ventrolateral caudal fringe composed scales of different size; tail circular in cross-section; slightly enlarged unpaired medial subcaudals not posteromedially furrowed; maximum SVL 78.2 mm; three dark transverse body bands (Tables 4–6).
Description of holotype
(Fig. 4). Adult male SVL 72.5 mm; head moderate in length (HL/SVL 0.28), width (HW/HL 0.70), depth (HD/HL 0.42), distinct from neck, triangular in dorsal profile; lores concave slightly anteriorly, weakly inflated posteriorly; prefrontal region concave; canthus rostralis rounded; snout elongate (ES/HL 0.42), rounded in dorsal profile; eye large (ED/HL 0.25); ear opening obliquely elongate, small; eye to ear distance greater than diameter of eye; rostral rectangular, divided by a deep furrow, bordered posteriorly by large left and right supranasals and one small azygous internasal, bordered laterally by first supralabials; external nares bordered anteriorly by rostral, dorsally by large supranasal, posteriorly by two smaller postnasals, bordered ventrally by first supralabial; 12R/12L rectangular supralabials, second through seventh supralabials nearly same size as first, then tapering abruptly below eye; 10R/10L infralabials tapering smoothly to just below and slightly past posterior margin of eye; scales of rostrum and lores flat to domed, larger than granular scales on top of head and occiput; scales of occiput intermixed with distinct, small tubercles; superciliaries subrectangular, largest dorsally; mental triangular, bordered laterally by first infralabials and posteriorly by large left and right trapezoidal postmentals contacting medially for 50% of their length posterior to mental; one row of slightly enlarged, elongate sublabials extending posteriorly to sixth(L) and fifth(R) infralabial; gular and throat scales small, granular, grading posteriorly into slightly larger, flatter, smooth, imbricate, pectoral and ventral scales.
Body relatively short (AG/SVL 0.46) with well-defined ventrolateral folds; dorsal scales small, granular interspersed with larger, conical, semi-regularly arranged, weakly keeled tubercles; tubercles extend from occipital region onto base of tail and slightly beyond as paravertebral rows; smaller tubercles extend anteriorly onto nape and occiput, diminishing in size anteriorly; approximately 17 longitudinal rows of tubercles at midbody; approximately 30 paravertebral tubercles; small tubercles on flanks; 34 longitudinal rows of flat, imbricate, ventral scales much larger than dorsal scales; 155 transverse rows of ventral scales; 15 large, pore-bearing, precloacal scales; no deep precloacal groove or depression; and two rows of post-precloacal scales on midline.
Forelimbs moderate in stature, relatively short (ForL/SVL 0.14); granular scales of forearm slightly larger than those on body, interspersed with large tubercles; palmar scales rounded, slightly raised; digits well-developed, relatively short, inflected at basal interphalangeal joints; digits narrower distal to inflections; subdigital lamellae wide, transversely expanded proximal to joint inflections, narrower transverse lamellae distal to joint inflections; claws well-developed, claw base sheathed by a dorsal and ventral scale; 8R/8L expanded and 10R/10L unexpanded lamellae beneath the fourth finger; hind limbs larger and thicker than forelimbs, moderate in length (TibL/SVL 0.16), covered dorsally by granular scales interspersed with moderately sized, conical tubercles dorsally and posteriorly and anteriorly by flat, slightly larger, subimbricate scales; ventral scales of thigh flat, subimbricate, larger than dorsals; subtibial scales flat, imbricate; one row of 5R/6L enlarged pore-bearing femoral scales not continuous with enlarged pore-bearing precloacal scales, terminating distally at knee; proximal femoral scales smaller than distal femorals, the former forming an abrupt union with much smaller, rounded, ventral scales of posteroventral margin of thigh; plantar scales flat; digits relatively long, well-developed, inflected at basal interphalangeal joints; 9R/9L wide, transversely expanded subdigital lamellae on fourth toe proximal to joint inflection extending onto sole, and 11R/11L unexpanded lamellae beneath the fourth toe; and claws well-developed, claw base sheathed by a dorsal and ventral scale.
Tail original, 97.6 mm long (TL/SVL 1.34), 5.2 mm in width at base, tapering to a point; sub-circular or nearly round in cross-section; dorsal scales flat, square bearing tubercles forming paravertebral rows and small tubercles forming a dorsolateral longitudinal row; slightly larger, posteriorly directed, semi-spinose tubercles forming narrow but distinct ventrolateral caudal fringe; larger scales of ventrolateral fringe occur at regular intervals; medial subcaudals slightly enlarged but not paired, distinctly enlarged single medial subcaudals absent; subcaudals, larger than dorsal caudals; base of tail bearing hemipenial swellings; 3R/2L conical postcloacal tubercles at base of hemipenial swellings; and postcloacal scales flat, imbricate.
Coloration in life
(Fig. 5). Ground color of the head, body, limbs, and tail brown; faint, diffuse mottling on the top of the head; thin, dark brown postorbital stripe; ventral portion of lores and suborbital region dark brown; nuchal band faint, bearing two dark-colored posterior projections; paired dark brown paravertebral blotches on nape; three wide faint irregularly shaped body bands edged in dark brown between limb insertions; band interspaces bearing irregularly shaped dark-colored markings; dark-colored speckling on limbs and digits; digits bearing pale-colored bands; eight wide dark-colored caudal bands separated by seven pale-colored bands; first six dark-colored and seven pale-colored caudal bands encircle tail; all ventral surfaces beige, generally immaculate; iris orange-gold in color.
Variation
(Fig. 5). Individuals of the type series are very similar in overall coloration and pattern. TL and TW of complete original tails (ZMKU R 00959–00960, ZMKU R 00963–00964) are 79.7–97.6 mm (mean 91.4 ± 8.2 mm; N = 4) and 4.0–5.2 mm (mean 4.5 ± 0.5; N = 4), respectively. The posterior one-third of the tail of ZMKU R 00958 is regenerated (TL 72.0 mm, TW 4.9 mm); that of ZMKU R 00962 is missing (TL 83.8 mm, TW 4.4 mm); and that of ZMKU R 00961 is broken one-third of the way passed the base and nearly all of the broken section is regenerated (TL 73.6 mm, TW 4.07 mm). Regenerated tail sections lack a color pattern. ZMKU R 00963–64 have seven dark-colored and six pale-colored caudal bands as opposed to eight and seven bands, respectively, in the holotype. ZMKU R 00958, ZMKU R 00960, ZMKU R 00962, ZMKU R 00964 are slight less boldly marked than the holotype. Meristic and morphometric differences are listed in Table 5.
Table 5.
Sex and raw meristic, categorical, and morphometric data used in the analyses from specimens in the Cyrtodactylusbrevipalmatus group. m = male; f = female; R/L = right/left; / = data unavailable.
| Species | C.brevipalmatus | C.cf.brevipalmatus | C.cf.brevipalmatus | C.brevipalmatus | C.brevipalmatus | C.elok | C.elok | C.elok | C.elok | C.interdigitalis | C.interdigitalis | C.interdigitalis | C.interdigitalis | C. sp.11 | C.ngati | C.ngati | C.ngati | C.ngati | C.ngati3 | C.ngati3 | C.ngati3 | C.ngati4 | C.cf.ngati1 | C.cf.ngati2 | C.cf.ngati2 | C.rukhadeva | C.rukhadeva | C.rukhadeva | C.cf.rukhadeva | C.cf.rukhadeva | C.cf.rukhadeva | C.cf.rukhadeva | C.cf.rukhadeva | C.cf.rukhadeva | C.cf.rukhadeva | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactylusfluvicavus sp. nov. | Cyrtodactyluskochangensis sp. nov | C.cf.kochangensis | Cyrtodactylusuthaiensis sp. nov. | Cyrtodactylusrivularis sp. nov. | Cyrtodactylusrivularis sp. nov. | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.9 Thong Pha Phum | C. sp.13 | C. sp.13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Institutional catalog number | LSUHC 1899 | LSUHC 15076 | LSUHC 11788 | THNHM 10670 | THNHM 14112 | LSUHC 8238 | LSUHC 12180 | LSUHC 12181 | ZMMU R-16144 | THNHM 20226 paratype | THNHM 20228 paratype | THNHM 20229 paratype | THNHM 20227 paratype | ZMMU R-16492 | HNUE-R00111 holotype | IEBR 4829 paratype | VNUF R.2020.12 paratype | HNUE-R00112 paratype | FMNH 255454 | FMNH 270493 | FMNH 270492 | FMNH 265806 | NCSM 79472 | ZMMU R-14917 | NCSM 80100 | ZMMU R-16851 holotype | ZMMU R-16852 paratype | ZMKU R 00948 topotype | THNHM 24622 | THNHM 24838 | THNHM 03251 | THNHM 03252 | THNHM 03253 | THNHM 03254 | THNHM 01807 | ZMKU R 00959 holotype | ZMKU R 00958 paratype | ZMKU R 00960 paratype | ZMKU R 00961 paratype | ZMKU R 00962 paratype | ZMKU R 00963 paratype | ZMKU R 00964 paratype | ZMKU R 00945 holotype | THNHM 01667 | ZMKU R 00949 holotype | ZMKU R 00947 holotype | ZMKU R 00946 paratype | ZMKU R 00950 | ZMKU R 00951 | ZMKU R 00952 | ZMKU R 00953 | ZMKU R 00954 | ZMKU R 00955 | ZMKU R 00956 | ZMKU R 00957 | THNHM 00104 | THNHM 27821 |
| Sex | m | f | f | f | f | f | m | m | f | f | f | f | f | m | m | f | f | f | f | m | m | m | f | f | f | m | f | f | m | f | m | m | f | m | m | m | m | m | f | f | f | f | f | m | m | f | f | f | m | f | m | m | f | m | f | f | f |
| Meristic data | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Supralabials (SL) | 11 | 12 | 10 | 14 | 12 | 11 | 8 | 13 | 9 | 14 | 12 | 11 | 12 | 11 | 10 | 10 | 10 | 10 | 13 | 13 | 13 | 10 | 14 | 9 | 12 | 11 | 9 | 14 | 11 | 13 | 13 | 11 | 12 | 13 | 12 | 12R/12L | 13R/12L | 13R/12L | 11R/12L | 12R/12L | 13R/12L | 12R/11L | 12R/13L | 12 | 13R/15L | 13R/12L | 13R/12L | 12 | 13 | 13 | 14 | 13 | 13 | 13 | 13 | 12 | 15 |
| Infralabials (IL) | 8 | 10 | 9 | 11 | 11 | 11 | 8 | 11 | 9 | 9 | 8 | 8 | 7 | 9 | 9 | 9 | 9 | 9 | 10 | 9 | 11 | 8 | 11 | 10 | 12 | 10 | 11 | 9 | 10 | 10 | 10 | 10 | 10 | 11 | 10 | 10R/10L | 10R/10L | 9R/10L | 10R/10L | 10R/10L | 10R/10L | 10R/10L | 9R/9L | 10 | 10R/11L | 11R/10L | 10R/9L | 8 | 8 | 10 | 10 | 9 | 10 | 10 | 9 | 10 | 11 |
| Paravertebral tubercles (PVT) | 39 | 37 | 38 | 37 | 37 | 0 | 0 | 0 | 0 | 32 | 33 | 33 | 33 | 30 | 39 | 40 | 38 | 40 | 28 | 27 | 26 | 27 | 28 | 32 | 29 | 27 | 30 | 30 | 26 | 28 | 27 | 27 | 30 | 30 | 26 | 30 | 28 | 27 | 27 | 28 | 26 | 28 | 34 | 29 | 33 | 34 | 33 | 32 | 33 | 34 | 34 | 36 | 36 | 30 | 30 | 33 | 29 |
| Longitudinal rows of tubercles (LRT) | 15 | 16 | 17 | 16 | 14 | 6 | 7 | 4 | 4 | 19 | 20 | 19 | 19 | 18 | 18 | 18 | 17 | 22 | 19 | 18 | 17 | 19 | 18 | 24 | 19 | 19 | 20 | 19 | 18 | 19 | 18 | 18 | 19 | 19 | 19 | 17 | 17 | 14 | 16 | 17 | 18 | 16 | 14 | 19 | 17 | 20 | 18 | 21 | 19 | 20 | 20 | 21 | 21 | 19 | 19 | 18 | 20 |
| Ventral scales (VS) | 38 | 38 | 38 | 36 | 39 | 45 | 45 | 47 | 36 | 42 | 40 | 42 | 43 | 34 | 38 | 36 | 35 | 32 | 37 | 36 | 36 | 33 | 33 | 36 | 35 | 34 | 43 | 38 | 38 | 36 | 37 | 37 | 39 | 34 | 35 | 34 | 37 | 33 | 30 | 36 | 37 | 39 | 35 | 34 | 36 | 34 | 37 | 34 | 33 | 33 | 34 | 30 | 33 | 32 | 33 | 37 | 36 |
| Ventral scales along middle of the body (VSM) | 176 | 170 | 182 | 154 | 160 | 190 | 225 | 234 | 192 | 187 | 170 | 187 | 178 | 160 | 168 | 164 | 178 | 158 | 159 | 166 | 156 | 158 | 164 | 166 | 165 | 154 | 152 | 165 | 162 | 158 | 157 | 159 | 168 | 160 | 161 | 155 | 154 | 155 | 172 | 164 | 175 | 170 | 172 | 159 | 159 | 160 | 166 | 173 | 158 | 156 | 166 | 159 | 159 | 150 | 169 | 159 | 165 |
| Expanded subdigital lamellae on 4th toe (TL4E) | 7 | 8 | 9 | 8 | 8 | 10 | 9 | 9 | 9 | 12 | 10 | 10 | 11 | 9 | 8 | 10 | 9 | 9 | 10 | 10 | 8 | 10 | 9 | 8 | 10 | 9 | 9 | 9 | 8 | 9 | 9 | 10 | 9 | 10 | 10 | 9R/9L | 10R/10L | 9R/9L | 9R/9L | 10R/11L | 9R/10L | 9R/9L | 9R/8L | 8 | 8R/(broken)L | 9R/9L | 9R/9L | 9 | 10 | 9 | 8 | 10 | 8 | 9 | 9 | 9 | 7 |
| Unmodified subdigital lamellae on 4th toe (TL4U) | 13 | 11 | 11 | 11 | 12 | 11 | 10 | 11 | 9 | 14 | 13 | 12 | 14 | 10 | 11 | 10 | 11 | 10 | 11 | 11 | 11 | 11 | 12 | 10 | 10 | 11 | 11 | 12 | 11 | 13 | 12 | 12 | 15 | 13 | 13 | 11R/11L | 12R/11L | 10R/10L | 12R/12L | 11R/11L | 10R/10L | 12R/13L | 12R/11L | 13 | 12R/(broken)L | 13R/13L | 12R/13L | 12 | 14 | 13 | 12 | 13 | 12 | 11 | 13 | 12 | 12 |
| Total subdigital lamellae 4th toe (TL4T) | 20 | 19 | 20 | 19 | 20 | 21 | 19 | 20 | 18 | 26 | 23 | 22 | 23 | 19 | 13 | 16 | 17 | 16 | 21 | 21 | 19 | 21 | 21 | 18 | 20 | 20 | 18 | 21 | 19 | 22 | 21 | 22 | 14 | 23 | 23 | 20R/20L | 22R/21L | 19R/19L | 21R/21L | 21R/22L | 19R/20L | 22R/22L | 21R/19L | 21 | 20 | 22R/22L | 21R/22L | 21 | 24 | 22 | 20 | 23 | 20 | 20 | 22 | 21 | 19 |
| Expanded subdigital lamellae on 4th finger (FL4E) | 8 | 8 | 8 | 7 | 8 | 8 | 9 | 9 | 9 | 9 | 8 | 9 | 9 | 10 | 6 | 6 | 7 | 6 | 8 | 8 | 8 | 8 | 9 | 7 | 9 | 9 | 8 | 8 | 7 | 8 | 8 | 8 | 8 | 8 | 8 | 8R/8L | 8R/8L | 8R/8L | 8R/8L | 7R/7L | 8R/9L | 7R/7L | 8R/8L | 8 | 7R/7L | 8R/8L | 8R/8L | 8 | 7 | 7 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Unmodified subdigital lamellae on 4th finger (FL4U) | 9 | 11 | 10 | 10 | 10 | 12 | 13 | 9 | 8 | 12 | 11 | 12 | 13 | 9 | 9 | 9 | 9 | 9 | 10 | 10 | 10 | 10 | 8 | 9 | 10 | 10 | 9 | 11 | 10 | 11 | 10 | 10 | 12 | 12 | 12 | 10R/10L | 10R/10L | 10R/9L | 11R/11L | 10R/10L | 9R/9L | 10R/10L | 10R/10L | 12 | 11R/11L | 11R/10L | 12R/12L | 10 | 12 | 12 | 11 | 12 | 12 | 11 | 12 | 11 | 10 |
| Total subdigital lamellae 4th finger (FL4T) | 17 | 19 | 18 | 17 | 18 | 20 | 22 | 18 | 17 | 21 | 21 | 21 | 22 | 19 | 15 | 15 | 18 | 15 | 18 | 18 | 18 | 18 | 17 | 16 | 19 | 19 | 17 | 19 | 17 | 17 | 18 | 18 | 20 | 20 | 20 | 18R/18L | 18R/18L | 18R/17L | 19R/19L | 17R/17L | 17R/18L | 17R/17L | 18R/18L | 20 | 18R/18L | 19R/18L | 20R/20L | 18 | 19 | 19 | 19 | 20 | 20 | 19 | 20 | 19 | 18 |
| Enlarged femoral scales (R/L) | 0 | 0 | 0 | 8R/8L | 7R/7L | 0 | 0 | 0 | 0 | 11R/8L | 10R/9L | 8R/8L | 9R/10L | 9R/8L | 10R/10L | 9R/8L | 10R/9L | 8R/9L | 9R/7L | 8R/9L | 9R/9L | 8R/8L | 9R/8L | 7R/8L | 7R/8L | 9R/8L | 8R/8L | 9R/8L | 9R/L | 9R/9L | 9R/7L | 7R/7L | 6R/7L | 5R/8L | 7R/7L | 5R/6L | 4R/5L | 5R/6L | 6R/6L | 5R/6L | 5R/6L | 6R/6L | 6R/6L | 7R/7L | 8R/8L | 8R/8L | 6R/8L | 5R/7L | 8R/8L | 8R/8L | 7R/8L | 8R/8L | 7R/8L | 7R/6L | 8R/8L | 9R/9L | 7R/10L |
| Total enlarged femoral scales (FS) | 16 | 10 | 11 | 16 | 14 | 0 | 0 | 0 | 0 | 14 | 19 | 19 | 19 | 17 | 20 | 17 | 19 | 17 | 16 | 17 | 18 | 16 | 17 | 15 | 15 | 17 | 16 | 17 | 18 | 18 | 16 | 14 | 13 | 13 | 14 | 11 | 9 | 11 | 12 | 11 | 11 | 12 | 12 | 14 | 16 | 16 | 14 | 12 | 16 | 16 | 15 | 16 | 15 | 13 | 16 | 18 | 17 |
| Total femoral pores in males (FP) | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 14 | 0 | 0 | 0 | 0 | 14 | 15 | 13 | 0 | 0 | 0 | 17 | 0 | 0 | 14 | 0 | 12 | 13 | 0 | 11 | 13 | 11 | 8 | 10 | 0 | 0 | 0 | 0 | 0 | 14 | 12 | 0 | 0 | 0 | 16 | 0 | 14 | 15 | 14 | 12 | 0 | 0 | 0 |
| Enlarged precloacal scales (PCS) | 7 | 7 | 7 | 8 | 7 | 8 | 8 | 8 | 7 | 14 | 15 | 13 | 19 | 13 | 13 | 13 | 13 | 13 | 15 | 13 | 13 | 13 | 12 | 13 | 13 | 17 | 13 | 15 | 15 | 15 | 14 | 13 | 15 | 15 | 14 | 15 | 14 | 14 | 15 | 14 | 15 | 15 | 12 | 16 | 14 | 15 | 15 | 17 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 14 | 16 |
| Precloacal pores in males (PP) | 7 | 0 | 0 | 0 | 0 | 0 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 13 | 13 | 13 | 13 | 0 | 0 | 0 | 17 | 13 | 0 | 15 | 0 | 14 | 13 | 0 | 15 | 14 | 15 | 14 | 14 | 0 | 0 | 0 | 0 | 0 | 16 | 14 | 0 | 0 | 0 | 15 | 0 | 15 | 15 | 15 | 15 | 0 | 0 | 0 |
| Postcloacal tubercles (PCT) | 3 | 3 | 2 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 2 | 3 | 4 | 3 | 2 | 2R/3L | 3 | 2 | 3 | 2 | 2 | 3 | 2 | 3R/2L | 3R/2L | 3R/3L | 1R/1L | 3R/2L | 3R/3L | 2R/2L | 1R/1L | 3 | 3R/3L | 2R/2L | 3R/3L | 2 | 2R/3L | 3 | 3 | 2R/3L | 2R/3L | 3 | 2 | 3 | 3 |
| Body bands (BB) | 4 | 6 | 3 | 5 | 5 | 5 | 5 | 3 | 3 | 5 | 5 | 5 | 5 | 3 | 6 | 6 | 6 | 6 | 3 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | / | / | 5 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 5 | 5 | 6 | 3 | 4 | 3 | 4 | 3 | 4 | 3 | 5 | 4 | 4 | 3 | / |
| Categorical data | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Small tubercles on flank (FKT) | present | present | present | present | present | absent | absent | absent | absent | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present | present |
| Dorsolateral caudal tubercles (DCT) | small | small | small | / | small | large | large | large | large | small | / | small | small | large | small | small | small | small | small | small | small | small | small | small | small | small | small | small | small | small | small | small | small | small | / | small | small | small | small | small | small | small | large | large | large | large | large | large | large | large | large | large | large | / | large | small | small |
| Ventrolateral caudal fringe narrow or wide (VLF1) | narrow | narrow | narrow | / | narrow | wide | wide | wide | wide | narrow | / | narrow | narrow | wide | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | narrow | / | narrow | narrow | narrow | narrow | narrow | narrow | narrow | wide | wide | wide | wide | wide | wide | wide | wide | wide | wide | wide | / | wide | narrow | narrow |
| Ventrolateral caudal fringe scales generally homogenous (VLF2) | no | no | no | / | no | no | no | no | no | yes | yes | yes | yes | yes | no | no | no | no | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | / | no | no | no | no | no | no | no | no | no | no | yes | yes | no | no | no | no | no | no | / | no | yes | yes |
| Tail cross-section (TLcross) | circular | circular | circular | / | circular | square | square | square | square | circular | / | circular | circular | square | circular | circular | circular | circular | circular | circular | circular | circular | circular | circular | circular | square | square | square | square | square | square | square | square | square | / | circular | circular | circular | circular | circular | circular | circular | square | / | circular | square | square | square | square | square | square | square | square | / | square | circular | circular |
| Slightly enlarged medial subcaudals (SC1) | present | present | present | / | absent | absent | absent | absent | absent | absent | / | absent | absent | present | present | present | present | present | / | present | present | present | present | present | present | absent | absent | absent | absent | absent | absent | absent | absent | absent | / | present | present | present | present | present | present | present | present | present | present | absent | absent | present | present | present | present | present | present | / | present | present | present |
| Single enlarged medial subcaudal (SC2) | absent | absent | absent | / | absent | absent | absent | absent | absent | absent | / | absent | absent | absent | absent | absent | absent | absent | / | absent | absent | absent | absent | absent | absent | present | present | present | present | present | present | present | present | present | / | absent | absent | absent | absent | absent | absent | absent | absent | absent | absent | present | present | absent | absent | absent | absent | absent | absent | / | absent | absent | absent |
| Enlarged medial subcaudals intermittent, medially furrowed, posteriorly emarginate (SC3) | no | no | no | / | no | no | no | no | no | yes | / | yes | yes | no | no | no | no | no | / | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | no | yes | no | no | no | no | no | no | yes | no | / | no | no | no |
| Morphometric data | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SVL | 68.8 | 70.8 | 64.1 | 65.95 | 63.79 | 80.2 | 78.2 | 84.8 | 78.6 | 81.19 | 74.80 | 78.56 | 59.70 | 68.1 | 66.5 | 68.1 | 69.3 | 46.6 | 83.6 | 70.2 | 74.1 | 73.8 | 78.0 | 87.1 | 77.7 | 74.9 | 71.7 | 71.6 | 68.3 | 71.8 | 73.6 | 75.3 | 74.7 | 73.2 | 61.5 | 72.5 | 72.0 | 69.6 | 68.4 | 76.8 | 65.7 | 78.2 | 60.1 | 70.2 | 58.1 | 73.9 | 68.1 | 73.1 | 73.5 | 73.7 | 73.2 | 64.4 | 76.6 | 76.6 | 74.2 | 63.7 | 72.9 |
| AG | 35.7 | 33.4 | 30.1 | 30.0 | 26.5 | 39.7 | 37.8 | 41.5 | 36.2 | 34.5 | 33.7 | 32.7 | 24.6 | 34.6 | 28.8 | 29.8 | 30.2 | 19.7 | 41.3 | 35.4 | 37.0 | 31.3 | 38.2 | 41.9 | 36.8 | 34.6 | 32.6 | 33.9 | 27.3 | 29.9 | 30.9 | 31.3 | 32.2 | 30.3 | 26.2 | 33.4 | 33.6 | 32.0 | 30.4 | 35.6 | 30.6 | 38.1 | 29.0 | 31.5 | 26.6 | 34.8 | 33.2 | 34.8 | 33.9 | 35.4 | 33.6 | 28.5 | 37.1 | 33.2 | 35.1 | 25.8 | 30.6 |
| HumL | 9.7 | 9.3 | 8.0 | 9.6 | 9.5 | 10.2 | 9.1 | 10.1 | 1.7 | 9.8 | 10.2 | 11.2 | 7.4 | 10.3 | 7.9 | 8.1 | 8.5 | 5.6 | 8.6 | 8.7 | 8.6 | 6.9 | 8.7 | 11.5 | 9.2 | 10.7 | 10.4 | 7.9 | 9.8 | 8.3 | 12.2 | 11.3 | 11.8 | 11.0 | 10.1 | 9.1 | 8.8 | 9.0 | 8.0 | 10.0 | 7.5 | 10.1 | 6.5 | 10.2 | 7.0 | 8.1 | 7.6 | 8.4 | 7.2 | 9.0 | 9.0 | 7.2 | 8.0 | 8.1 | 8.6 | 7.6 | 10.1 |
| ForL | 9.9 | 9.8 | 8.9 | 8.2 | 8.7 | 11.5 | 11.7 | 11.8 | 10.2 | 10.6 | 10.5 | 11.1 | 8.4 | 8.5 | 9.2 | 10.0 | 10.1 | 6.5 | 10.2 | 9.3 | 10.4 | 10.0 | 10.3 | 10.4 | 10.7 | 8.6 | 7.9 | 9.6 | 8.7 | 8.5 | 9.0 | 10.6 | 9.6 | 9.2 | 7.9 | 10.5 | 10.3 | 10.5 | 10.1 | 11.1 | 8.8 | 10.8 | 7.6 | 8.6 | 8.3 | 9.7 | 9.1 | 9.5 | 9.1 | 9.2 | 9.8 | 9.2 | 10.0 | 8.6 | 9.8 | 8.1 | 9.6 |
| FemL | 12.0 | 12.6 | 11.5 | 11.7 | 9.8 | 12.9 | 14.2 | 14.6 | 13.1 | 14.7 | 13.2 | 12.7 | 10.2 | 12.6 | 11.5 | 11.5 | 11.5 | 7.6 | 13.7 | 12.7 | 13.0 | 13.1 | 13.1 | 15.2 | 14.2 | 12.6 | 11.8 | 10.5 | 10.8 | 10.9 | 11.5 | 10.2 | 11.9 | 12.1 | 9.5 | 13.1 | 12.5 | 12.5 | 13.5 | 14.1 | 11.5 | 13.9 | 10.4 | 12.1 | 10.0 | 11.4 | 10.4 | 12.8 | 11.6 | 12.3 | 12.5 | 10.9 | 13.7 | 10.8 | 12.5 | 10.7 | 12.8 |
| TibL | 11.6 | 12.2 | 10.5 | 9.7 | 8.2 | 13.5 | 14.0 | 13.8 | 12.3 | 13.1 | 11.9 | 12.9 | 10.2 | 11.4 | 10.8 | 11.1 | 11.8 | 7.8 | 12.5 | 11.8 | 11.2 | 11.1 | 12.8 | 12.6 | 12.7 | 10.1 | 9.3 | 11.2 | 9.7 | 10.7 | 10.9 | 11.7 | 11.3 | 11.1 | 9.1 | 11.3 | 10.6 | 10.2 | 9.9 | 11.2 | 9.4 | 12.3 | 8.4 | 11.8 | 8.4 | 11.2 | 10.3 | 10.5 | 10.1 | 10.6 | 10.6 | 9.9 | 11.1 | 10.0 | 11.4 | 10.1 | 10.2 |
| HL | 19.3 | 19.3 | 19.0 | 17.9 | 18.2 | 21.8 | 21.6 | 21.9 | 21.7 | 20.8 | 19.9 | 21.7 | 16.7 | 18.4 | 20.1 | 20.4 | 20.7 | 16.1 | 21.7 | 20.6 | 20.3 | 20.7 | 21.2 | 22.1 | 21.4 | 20.2 | 19.2 | 19.7 | 19.7 | 19.9 | 20.8 | 21.3 | 20.8 | 21.5 | 17.9 | 20.1 | 20.5 | 19.7 | 20.1 | 21.2 | 18.6 | 21.3 | 17.3 | 18.3 | 16.1 | 20.3 | 19.3 | 19.9 | 20.9 | 20.1 | 20.0 | 17.6 | 20.4 | 19.3 | 20.0 | 17.6 | 19.9 |
| HW | 13.2 | 13.8 | 12.3 | 12.3 | 12.0 | 15.6 | 16.1 | 15.9 | 15.1 | 14.0 | 13.4 | 14.2 | 11.4 | 13.1 | 12.6 | 12.0 | 11.8 | 8.8 | 13.8 | 12.5 | 13.0 | 12.3 | 12.7 | 14.8 | 13.5 | 14.6 | 13.4 | 14.0 | 13.1 | 13.9 | 14.9 | 15.0 | 13.1 | 14.1 | 11.8 | 14.0 | 13.4 | 12.9 | 13.0 | 14.9 | 13.0 | 15.4 | 11.6 | 12.1 | 10.9 | 14.9 | 13.7 | 14.5 | 14.3 | 15.7 | 13.9 | 12.8 | 14.7 | 14.4 | 14.1 | 11.9 | 13.8 |
| HD | 8.0 | 7.6 | 7.6 | 7.3 | 7.0 | 9.6 | 9.8 | 10.4 | 9.8 | 3.4 | 8.6 | 8.7 | 6.6 | 8.3 | 7.4 | 7.2 | 6.6 | 5.1 | 9.2 | 8.4 | 9.1 | 7.6 | 8.3 | 8.7 | 9.2 | 9.2 | 8.5 | 8.3 | 7.3 | 8.9 | 8.2 | 8.2 | 8.1 | 8.9 | 7.5 | 8.5 | 8.1 | 8.3 | 7.9 | 8.1 | 7.8 | 8.3 | 6.5 | 7.8 | 6.3 | 8.2 | 8.2 | 7.8 | 7.7 | 7.9 | 7.7 | 7.0 | 8.2 | 7.8 | 7.6 | 7.7 | 8.4 |
| ED | 5.2 | 4.5 | 4.3 | 5.3 | 4.4 | 4.8 | 5.0 | 5.7 | 5.0 | 5.3 | 5.5 | 5.9 | 4.4 | 4.4 | 3.8 | 4.1 | 3.4 | 2.6 | 4.9 | 4.9 | 4.9 | 4.8 | 6.5 | 4.6 | 6.0 | 4.6 | 4.3 | 5.5 | 4.9 | 5.1 | 5.8 | 5.4 | 5.0 | 5.5 | 4.7 | 5.0 | 5.0 | 4.9 | 4.7 | 5.1 | 4.5 | 5.3 | 4.2 | 5.2 | 4.6 | 5.8 | 5.6 | 5.0 | 5.1 | 5.0 | 5.0 | 4.8 | 5.6 | 5.3 | 4.9 | 4.1 | 5.3 |
| EE | 5.7 | 5.9 | 4.9 | 5.7 | 5.7 | 6.4 | 7.1 | 7.0 | 6.8 | 5.8 | 6.2 | 6.4 | 4.8 | 6.2 | 5.8 | 5.5 | 5.9 | 4.4 | 6.9 | 6.1 | 6.2 | 5.7 | 5.3 | 6.5 | 6.2 | 6.2 | 6.2 | 5.8 | 5.1 | 6.2 | 5.6 | 5.7 | 5.4 | 6.2 | 4.3 | 6.5 | 5.9 | 5.7 | 5.8 | 6.1 | 5.4 | 6.5 | 5.0 | 4.9 | 4.7 | 6.5 | 6.2 | 5.9 | 5.9 | 6.0 | 5.9 | 5.3 | 6.1 | 6.0 | 6.0 | 4.9 | 6.3 |
| ES | 7.4 | 7.6 | 7.0 | 7.0 | 7.2 | 8.6 | 8.7 | 9.5 | 8.6 | 8.3 | 7.8 | 9.1 | 6.8 | 7.7 | 7.5 | 7.6 | 6.9 | 5.0 | 9.0 | 8.3 | 8.3 | 8.2 | 8.7 | 8.8 | 8.4 | 8.3 | 7.7 | 7.9 | 7.4 | 8.1 | 8.4 | 8.8 | 8.1 | 8.6 | 7.3 | 8.5 | 8.3 | 8.2 | 8.1 | 9.2 | 7.3 | 9.3 | 6.9 | 7.5 | 6.4 | 8.3 | 7.9 | 7.9 | 8.5 | 7.9 | 7.9 | 7.3 | 8.2 | 7.9 | 7.9 | 7.2 | 8.0 |
| EN | 5.7 | 5.4 | 4.9 | 5.3 | 5.4 | 6.0 | 6.2 | 6.5 | 6.2 | 6.0 | 5.5 | 6.8 | 5.1 | 5.5 | 6.7 | 6.3 | 6.2 | 4.5 | 6.5 | 6.2 | 6.1 | 6.2 | 6.2 | 6.6 | 6.0 | 6.3 | 5.7 | 5.8 | 5.4 | 6.0 | 6.2 | 6.4 | 5.8 | 6.2 | 5.3 | 6.5 | 6.2 | 5.9 | 6.1 | 6.6 | 5.6 | 6.5 | 5.2 | 5.5 | 4.9 | 6.1 | 5.8 | 6.0 | 6.1 | 6.0 | 5.8 | 5.4 | 6.1 | 6.0 | 5.9 | 5.6 | 5.9 |
| IO | 5.4 | 4.7 | 4.7 | 4.2 | 5.2 | 5.7 | 5.4 | 5.4 | 3.9 | 4.8 | 4.7 | 5.5 | 4.3 | 2.9 | 5.6 | 5.4 | 5.6 | 4.2 | 6.6 | 5.6 | 5.4 | 5.1 | 4.9 | 3.5 | 5.7 | 3.3 | 3.1 | 5.6 | 4.5 | 4.7 | 5.6 | 5.7 | 5.7 | 5.6 | 4.2 | 5.5 | 5.4 | 5.3 | 5.1 | 5.6 | 5.0 | 5.6 | 4.2 | 4.0 | 4.3 | 5.8 | 5.5 | 5.4 | 5.5 | 5.8 | 5.5 | 4.9 | 5.7 | 5.6 | 5.3 | 4.8 | 6.1 |
| EL | 1.0 | 1.4 | 1.1 | 1.3 | 1.0 | 1.9 | 1.4 | 1.5 | 1.4 | 1.3 | 1.3 | 1.6 | 1.2 | 0.9 | 0.8 | 0.8 | 0.7 | 0.3 | 1.3 | 1.1 | 1.2 | 1.0 | 1.5 | 1.2 | 0.9 | 1.2 | 1.0 | 1.4 | 1.6 | 1.5 | 1.2 | 1.3 | 1.2 | 1.2 | 0.9 | 1.4 | 1.5 | 1.7 | 1.4 | 1.8 | 1.6 | 1.8 | 1.0 | 1.3 | 1.5 | 1.1 | 1.1 | 1.1 | 1.5 | 1.5 | 1.2 | 1.2 | 1.0 | 1.2 | 1.3 | 1.4 | 1.4 |
| IN | 1.7 | 2.1 | 2.3 | 2.1 | 2.2 | 2.7 | 2.6 | 2.5 | 3.1 | 2.1 | 2.2 | 2.5 | 1.8 | 2.3 | 2.8 | 2.6 | 2.6 | 2.0 | 2.8 | 2.5 | 2.5 | 2.3 | 2.7 | 2.2 | 2.5 | 2.2 | 2.1 | 2.1 | 2.0 | 2.2 | 2.4 | 2.5 | 2.4 | 2.3 | 2.0 | 2.3 | 2.4 | 2.5 | 2.3 | 2.3 | 2.3 | 2.6 | 1.9 | 2.2 | 1.8 | 2.3 | 2.0 | 2.3 | 2.4 | 2.2 | 2.0 | 2.0 | 2.3 | 2.2 | 2.2 | 2.1 | 2.3 |
Distribution.
Cyrtodactylusfluvicavus sp. nov. is currently known from the type locality at Tham Than Lot Noi-Tham Than Lot Yai Nature Trail in Chaloem Rattanakosin National Park, Si Sawat District, Kanchanaburi Province, western Thailand (Fig. 1).
Etymology.
The specific epithet fluvicavus comes from the Latin fluvius, meaning stream, river, or flow and the Latin cavus, meaning hollow or hole and refers to a landmark cave in the Chaloem Rattanakosin National Park which has a stream that flows through it.
Comparisons.
Cyrtodactylusfluvicavus sp. nov. is the sister species to a clade composed of ten lineages in a phylogenetic sequence of C. sp.9, C. sp.10, Cyrtodactylusuthaiensis sp. nov., C. sp.11, C.interdigitalis, C.cf.ngati1, C.cf.ngati2, C.ngati3, and the sister lineages C.ngati4 and C.ngati (Fig. 2). Cyrtodactylusfluvicavus sp. nov. differs from those lineages by an uncorrected pairwise sequence divergence of 9.55–12.13% and from all members of the brevipalmatus group by 11.61–20.26% (Table 2). It differs categorically from C.elok by having as opposed to lacking paravertebral tubercles and femoral pores, and by having 14–18 as opposed to 4–7 longitudinal rows of tubercles. It differs from C.brevipalmatus, C.interdigitalis, C.ngati, C.ngati3, C.rukhadeva, and C. sp.9 in having statistically significant different mean values of the morphometric characters of AG, HumL, ForL, FemL, TibL, HL, HW, HD, EE, ES, EN, EL and IN (Table 6). It differs further from C.brevipalmatus, C.interdigitalis, C.ngati, C.ngati3, C.rukhadeva, and C. sp.9 in having statistically significant different mean values of the meristic characters of SL, PVT, LRT, VS, VSM, TL4E, TL4T, FL4E, FL4U, FL4T, FS, PCS, and BB. Statistically significant and discrete differences between Cyrtodactylusfluvicavus sp. nov. and all other species and populations are presented in Tables 4–6.
Natural history.
All individuals were found in karst forests bearing mixed deciduous and dry evergreen trees amidst rocky streams and a nearby waterfall (Fig. 6). This area is surrounded by agricultural fields and residential areas. Specimens (N = 7) were collected at night (1900–2100 h) during the dry season (April) on the tree trunks or palm tree leaves (57.1%; N = 4), twigs of shrubs (14.3%; N = 1), karst walls (14.3%; N = 1), and a wooden bridge (14.3%; N = 1) at 526 m elevation with a temperature of 31.9 °C and relative humidity of 56.9%. The holotype (ZMKU R 00959) and two specimens (ZMKU R 00960, ZMKU R 00962) were found on tree trunks ≤ 100 cm above ground level. One specimen (ZMKU R 00964) was found on a palm tree branch approximately 50 cm above the ground. Another specimen (ZMKU R 00963) was found on the twig of a shrub. Another specimen (ZMKU R 00961) was found on a karst wall approximately 3 m above the ground, and another (ZMKU R 00958) on a wooden bridge over a stream. Given these observations, this species appears to be an arboreal habitat generalist. The new species was found to co-occur with two other species of gekkonid lizards, Cyrtodactylusmonilatus Yodthong, Rujirawan, Stuart, Grismer, Aksornneam, Termprayoon, Ampai & Aowphol, 2022 and Dixoniussiamensis (Boulenger, 1899).
Figure 6.
Habitat of Cyrtodactylusfluvicavus sp. nov. at Thailand, Kanchanaburi Province, Si Sawat District, Khao Chot Subdistrict, Chaloem Rattanakosin National Park.
. Cyrtodactylus rivularis sp. nov.
2BBE9CD2-58DC-5F5B-991D-225EF3E44746
https://zoobank.org/B3381A9D-0049-4C5B-BEB1-DB4D60509F0E
Figs 7 , 8 Suggested Common Name: Pa La-U Bent-toed Gecko
Figure 7.
Holotype of Cyrtodactylusrivularis sp. nov. ZMKU R 00947 (field no. AA 04887) from Thailand, Prachuap Khiri Khan Province, Hua Hin District, Huai Sat Yai Subdistrict, Kaeng Krachan National Park, Pa La-U Waterfall A dorsal view B ventral view C dorsal view of head D ventral view of femoral and precloacal regions E dorsal view of tail F ventral view of tail.
Figure 8.
Type series of Cyrtodactylusrivularis sp. nov. from Thailand, Prachuap Khiri Khan Province, Hua Hin District, Huai Sat Yai Subdistrict, Kaeng Krachan National Park, Pa La-U Waterfall A holotype ZMKU R 00947 B paratype ZMKU R 00946.
Holotype.
Adult female ZMKU R 00947 from Pa La-U Waterfall, Kaeng Krachan National Park, Huai Sat Yai Subdistrict, Hua Hin District, Prachuap Khiri Khan Province, Thailand (12.53685°N, 99.45972°E, 368 m a.s.l.), collected by Attapol Rujirawan, Siriporn Yodthong, Korkhwan Termprayoon, Natee Ampai, and Piyawan Puanprapai on 15 September 2017.
Paratype.
Adult female ZMKU R 00946 bearing the same data as the holotype.
Diagnosis.
Cyrtodactylusrivularis sp. nov. can be separated from all other species of the brevipalmatus group by the combination of having 12 or 13 supralabials, 9–11 infralabials, 33 or 34 paravertebral tubercles, 18–20 rows of longitudinally arranged tubercles, 34–37 transverse rows of ventrals, 160–166 longitudinal rows of ventrals, nine expanded subdigital lamellae on the fourth toe, 12 or 13 unexpanded subdigital lamellae on the fourth toe, 21 or 22 total subdigital lamellae on the fourth toe; eight expanded subdigital lamellae on the fourth finger, 10–12 unexpanded subdigital lamellae on the fourth finger, 18–20 total subdigital lamellae on the fourth finger; 14–16 total enlarged femoral scales; 15 enlarged precloacals; enlarged femorals and enlarged precloacals not continuous, and lacking pores; proximal femorals less than one-half the size of the distal femorals; small tubercles on forelimbs and flanks; large dorsolateral caudal tubercles and wide ventrolateral caudal fringe; ventrolateral caudal fringe composed generally homogeneous scales; tail square in cross-section; single enlarged unpaired medial subcaudals not posteromedially furrowed; maximum SVL 73.9 mm; three or four dark transverse body bands (Tables 4, 5).
Description of holotype
(Fig. 7). Adult female SVL 73.9 mm; head moderate in length (HL/SVL 0.27), width (HW/HL 0.73), depth (HD/HL 0.40), distinct from neck, triangular in dorsal profile; lores concave slightly anteriorly, weakly inflated posteriorly; prefrontal region concave; canthus rostralis rounded; snout elongate (ES/HL 0.41), rounded in dorsal profile; eye large (ED/HL 0.29); ear opening horizontally elongate, small; eye to ear distance greater than diameter of eye; rostral rectangular, divided dorsally by a deep furrow, bordered posteriorly by large left and right supranasals and one slightly smaller azygous internasal, bordered laterally by first supralabials; external nares bordered anteriorly by rostral, dorsally by large supranasal, posteriorly by two smaller postnasals, bordered ventrally by first supralabial; 13R/12L rectangular supralabials, first two largest, then tapering abruptly below eye; 11R/10L infralabials tapering smoothly to just below eye and then more rapidly beyond posterior margin of eye; scales of rostrum and lores flat to domed, larger than granular scales on top of head and occiput; scales of occiput intermixed with distinct, small tubercles; superciliaries subrectangular, largest anteriorly; mental triangular, bordered laterally by first infralabials, posteriorly by large left and right elongate postmentals contacting medially for approximately 40% of their length posterior to mental; one row of two (R) and four (L) slightly enlarged sublabials extending posteriorly to third(L) and second(R) infralabial, subsequent sublabials much smaller; gular and throat scales small, granular, grading posteriorly into slightly larger, flatter, smooth, imbricate, pectoral and ventral scales.
Body relatively short (AG/SVL 0.47) with well-defined ventrolateral folds; dorsal scales small, granular, interspersed with larger conical, semi-regularly arranged, weakly keeled tubercles; tubercles extend from occipital region onto base of tail and slightly beyond as paravertebral rows; tubercles of nape and occiput small; approximately 20 longitudinal rows of tubercles at midbody; approximately 34 paravertebral tubercles; tubercles on flanks nearly same size as those on dorsum; 34 longitudinal rows of flat, imbricate, ventral scales much larger than dorsal scales; 160 transverse rows of ventral scales; no pore-bearing, precloacal scales; 15 enlarged precloacal scales; no deep precloacal groove or depression; and three rows of post-precloacal scales on midline.
Forelimbs moderate in stature, relatively short (ForL/SVL 0.13); granular scales of forearm slightly larger than those on body, interspersed with tubercles; palmar scales rounded, slightly raised; digits well-developed, relatively short, inflected at basal interphalangeal joints; digits narrower distal to inflections; subdigital lamellae wide, transversely expanded proximal to joint inflections, narrower transverse lamellae distal to joint inflections; claws well-developed, claw base sheathed by a dorsal and ventral scale; 8R/8L expanded and 11R/11L unexpanded lamellae beneath the fourth finger; hind limbs larger and thicker than forelimbs, moderate in length (TibL/SVL 0.15), covered dorsally by granular scales interspersed with moderately sized, conical tubercles dorsally and posteriorly and anteriorly by flat, slightly larger, subimbricate scales; ventral scales of thigh flat, subimbricate, larger than dorsals; subtibial scales flat, imbricate; no pore-bearing femoral scales; 8R/8L enlarged femoral scales; enlarged femoral scales not contiguous with enlarged precloacal scales, terminating distally at knee; proximal femoral scales smaller than distal femoral scales, the former forming an abrupt union with much smaller, rounded, ventral scales of posteroventral margin of thigh; plantar scales flat; digits relatively long, well-developed, inflected at basal interphalangeal joints; 9R/9L wide, transversely expanded subdigital lamellae on fourth toe proximal to joint inflection that extend onto sole, and 13R/13L unexpanded lamellae beneath the fourth toe; and claws well-developed, sheathed by a dorsal and ventral scale at base.
Posterior one-half of tail regenerated, tail long 91.5 mm (TL/SVL 1.24), 4.8 mm in width at base, tapering to a point; nearly square in cross-section; dorsal scales flat, square bearing large tubercles forming a discontinuous dorsolateral longitudinal row; slightly larger, posteriorly directed, semi-spinose tubercles forming large distinct ventrolateral caudal fringe; scales of ventrolateral fringe generally homogeneous; single medial subcaudals enlarged but not paired; subcaudal scales, larger than dorsal caudal scales; base of tail bearing hemipenial swellings; 2R/2L conical postcloacal tubercles at base of hemipenial swellings; and postcloacal scales flat, imbricate.
Coloration in life
(Fig. 8). Ground color of the head, body, limbs, and tail pale brown; faint, diffuse mottling on rostrum; lores darkly colored; wide, distinct, pale-colored post-orbital stripe; nuchal band faint, bearing two posterior projections; three very faint, wide irregularly shaped body bands edged in slightly darker brown between limb insertions; band interspaces bearing irregularly shaped, faint, dark-colored markings; dark-colored speckling on limbs and digits; digits bearing pale-colored bands; four wide faint dark-colored caudal bands separated by three pale-colored bands on original portion of tail; all caudal bands encircle tail; all ventral surfaces beige, generally immaculate; iris orangish to coppery in color.
Variation
(Fig. 8). The paratype (ZMKU R 00946) closely approximates the holotype in overall coloration and pattern except that it is more boldly marked. It has four dark-colored body bands as opposed to three and a complete original tail (TL 89.0mm, TW 4.1 mm) bearing eight dark-colored and seven pale-colored bands. The pale-colored postorbital stripe is slightly thinner and less distinct. Meristic and morphometric differences are listed in Table 5.
Distribution.
Cyrtodactylusrivularis sp. nov. is currently known from the type locality at Pa La-U Waterfall, Kaeng Krachan National Park, Huai Sat Yai Subdistrict, Hua Hin District Prachuap Khiri Khan Province, Thailand.
Etymology.
The specific epithet rivularis is derived from the Latin rivus, meaning stream, brook, or creek refers to rocky brook or stream habitat of the new species.
Comparisons.
Cyrtodactylusrivularis sp. nov. is the sister species to C.rukhadeva (Fig. 2) from which it differs by an uncorrected pairwise sequence divergence of 3.61% (Table 2). It differs from C.rukhadeva and C.cf.rukhadeva by having large versus small dorsolateral caudal tubercles and a wide versus narrow ventrolateral fringe. Although no statistical mean differences were recovered between Cyrtodactylusrivularis sp. nov. and C.rukhadeva which we attribute to the small sample sizes of both species (N = 2), they do respectively differ discretely (at this point) in their ranges of SL (12 or 13 vs. 9–11), PVT (33–34 vs. 27–30), VSM (160–166 vs. 152–154), and TL4T (21–22 vs. 18–20) and the morphometric characters of HumL, ForL, FemL, TibL, HD, ED, and IO (Table 5). Discrete differences among Cyrtodactylusrivularis sp. nov. and all other species and populations are presented in Tables 4, 5.
Natural history.
The holotype and paratype were collected at night (1900–2055 h) on granite boulders by a rocky stream dry evergreen forest at 368 m in elevation (Fig. 9) with a temperature of 25.3 °C and relative humidity of 86.9%. The new species was found to co-occur with two other species of gekkonid lizards, Cyrtodactylusoldhami (Theobald, 1876) and Gehyramutilata (Wiegmann, 1834).
Figure 9.
Habitat of Cyrtodactylusrivularis sp. nov. at Thailand, Prachuap Khiri Khan Province, Hua Hin District, Huai Sat Yai Subdistrict, Kaeng Krachan National Park, Pa La-U Waterfall.
Comments.
Cyrtodactylusrivularis sp. nov. and C.cf.rukhadeva occur on separate branches of the phylogeny, thus neither is embedded within one another’s branch indicating there is no evidence of gene flow between them. Both species are reported to occur within the boundaries of Kaeng Krachan National Park (Grismer et al. 2022). Kaeng Krachan National Park is the largest national park in Thailand, encompassing 2,914.7 km2 across five districts through the provinces of Phetchaburi and Prachuap Khiri Khan and extending approximately 300 km in latitude. As such, it harbors a wide range of habitats from lowland forests near sea level to cloud forests at its highest peak of 1,513 m in elevation. The seven specimens of C.cf.rukhadeva (THNHM 01807, 24622, 24838, 03251–54) were obtained by various collectors between 11 March 1991 and 25 October 2015 with no specific collection data other than Phetchaburi Province. The locality reported by Grismer et al. (2021c) was an estimate obtained from Ulber (1993) from the central portion of the park in Phetchaburi Province who referred to a specimen he examined (THNHM 24838) as C.brevipalmatus. Cyrtodactylusrivularis sp. nov. however, is known only from the Park’s southernmost limit in Prachuap Khiri Khan Province. Additional collecting and DNA sequencing will be required to establish the approximate distribution limits of each population.
. Cyrtodactylus kochangensis sp. nov.
7E67190E-06B0-564C-B4C8-E5A323705A73
https://zoobank.org/96DF655B-BBB7-4C06-9418-744EBCD14703
Fig. 10 Suggested Common Name: Ko Chang Bent-toed Gecko
Figure 10.
Holotype of Cyrtodactyluskochangensis sp. nov. ZMKU R 00945 (field no. AA 04439) from Thailand, Ranong Province, Mueng Ranong District, Ko Phayam Subdistrict, Ko Chang A dorsal view B ventral view C dorsal view of head D ventral view of femoral and precloacal regions E dorsal view of tail F ventral view of tail G holotype in life.
Holotype.
Adult female ZMKU R 00945 from Ko Chang Island, Ko Phayam Subdistrict, Mueang Ranong District, Ranong Province, Thailand (9.82411°N, 98.43999°E, 36 m a.s.l.), collected by Siriporn Yodthong, Natee Ampai, Attapol Rujirawan, and Piyawan Puanprapai on 8 July 2017.
Additional material examined.
Cyrtodactyluscf.kochangensis sp. nov. adult male THNHM 01667 from Khlong Naka Wildlife Sanctuary, Suk Samran District, Ranong Province, Thailand (~N 9.4596, E 98.5044, elevation unknown), collected by Yodchaiy Chuaynkern between 28 December 2000 and 2 January 2001.
Diagnosis
(based on the holotype).Cyrtodactyluskochangensis sp. nov. can be separated from all other species of the brevipalmatus group by the combination of having 12 or 13 supralabials, nine infralabials, 34 paravertebral tubercles, 14 rows of longitudinally arranged tubercles, 35 transverse rows of ventrals, 172 longitudinal rows of ventrals, 8 or 9 expanded subdigital lamellae on the fourth toe, 11 or 12 unexpanded subdigital lamellae on the fourth toe, 19–21 total subdigital lamellae on the fourth toe; eight expanded subdigital lamellae on the fourth finger, ten unexpanded subdigital lamellae on the fourth finger, 18 total subdigital lamellae on the fourth finger; 12 total enlarged femoral scales; 12 enlarged precloacal scales; enlarged femoral and enlarged precloacal scales not continuous; proximal femoral scales less than one-half the size of the distal femorals; small tubercles on forelimbs and flanks; large dorsolateral caudal tubercles and a wide ventrolateral caudal fringe not composed homogeneous scales; tail square in cross-section; slightly enlarged paired medial subcaudals not posteromedially furrowed; maximum SVL 60.1 mm; five dark transverse body bands (Tables 4, 5).
Description of holotype
(Fig. 10). Adult female SVL 60.1 mm; head moderate in length (HL/SVL 0.29), width (HW/HL 0.67), depth (HD/HL 0.38), distinct from neck, triangular in dorsal profile; lores flat anteriorly, weakly inflated posteriorly; prefrontal region slightly concave; canthus rostralis rounded; snout elongate (ES/HL 0.40), rounded in dorsal profile; eye large (ED/HL 0.24); ear opening subcircular, small; eye to ear distance greater than diameter of eye; rostral rectangular, furrowed dorsally, bordered posteriorly by large left and right supranasals and one slightly smaller azygous internasal, bordered laterally by first supralabials; external nares bordered anteriorly by rostral, dorsally by large supranasal, posteriorly by two slightly smaller postnasals, bordered ventrally by first supralabial; 12R/13L rectangular supralabials tapering smoothly to below eye; 9R/9L infralabials tapering smoothly to below eye; scales of rostrum and lores domed, slightly larger than granular scales on top of head and occiput; scales of occiput intermixed with distinct, small tubercles; superciliaries subrectangular, largest anteriorly; mental triangular, bordered laterally by first infralabials and posteriorly by large left and right trapazoidal postmentals contacting medially for approximately 40% of their length posterior to mental; one row of six (R,L) slightly enlarged sublabials extending posteriorly fifth infralabials, subsequent sublabials much smaller; gular and throat scales small, granular, grading posteriorly into slightly larger, flatter, smooth, imbricate, pectoral and ventral scales.
Body relatively short (AG/SVL 0.48) with well-defined ventrolateral folds; dorsal scales small, granular, interspersed with larger conical, semi-regularly arranged, weakly keeled tubercles; tubercles extend from occipital region onto base of tail and slightly beyond as paravertebral rows; tubercles of nape and occiput smaller than those on body; approximately 14 longitudinal rows of tubercles at midbody; approximately 34 paravertebral tubercles; tubercles on upper flanks smaller than those on dorsum; approximately 35 longitudinal rows of flat, imbricate, ventral scales much larger than dorsal scales; approximately 172 transverse rows of ventral scales; no pore-bearing, precloacal scales; 12 enlarged precloacal scales; no deep precloacal groove or depression; and approximately three rows of post-precloacal scales on midline.
Forelimbs moderate in stature, relatively short (ForL/SVL 0.13); granular scales of forearm slightly larger than those on body, interspersed with tubercles; palmar scales rounded, slightly raised; digits well-developed, relatively short, inflected at basal interphalangeal joints; digits narrower distal to inflections; subdigital lamellae wide, transversely expanded proximal to joint inflections, narrower transverse lamellae distal to joint inflections; claws well-developed, claw base sheathed by a dorsal and ventral scale; 8R/8L expanded and 10R/10L unexpanded lamellae beneath the fourth finger; hind limbs larger and thicker than forelimbs, moderate in length (TibL/SVL 0.14), covered dorsally by granular scales interspersed with moderately sized, conical tubercles dorsally and posteriorly and anteriorly by flat, slightly larger, subimbricate scales; ventral scales of thigh flat, subimbricate, larger than dorsals; subtibial scales flat, imbricate; no pore-bearing femoral scales; 6R/6L enlarged femoral scales; enlarged femoral scales not contiguous with enlarged precloacal scales, terminating distally at knee; proximal femoral scales smaller than distal femorals, the former forming an abrupt union with much smaller, rounded, ventral scales of posteroventral margin of thigh; plantar scales flat; digits relatively long, well-developed, inflected at basal interphalangeal joints; 9R/8L wide, transversely expanded subdigital lamellae on the fourth toe proximal to joint inflection that extend onto sole, and 12R/11L unexpanded lamellae beneath the fourth toe; and claws well-developed, sheathed by a dorsal and ventral scale at base.
Tail original (but in two pieces), long 80.3 mm (TL/SVL 1.34), 4.0 mm in width at base, tapering to a point; nearly square in cross-section; dorsal scales flat, square bearing tubercles forming paravertebral rows and large tubercles forming a dorsolateral longitudinal row; slightly larger, posteriorly directed, semi-spinose tubercles forming wide distinct ventrolateral caudal fringe; scales of ventrolateral fringe generally interspersed at regular intervals with larger spinose scales; medial subcaudal scales paired, slightly enlarged; subcaudals, larger than dorsal caudal scales; base of tail bearing hemipenial swellings; one conical postcloacal tubercle at base of hemipenial swellings; and postcloacal scales flat, imbricate.
Coloration in life
(Fig. 10). Ground color of the head, body, limbs, and tail pale brown; faint, diffuse mottling on rostrum; lores darkly colored; wide, distinct, pale-colored postorbital stripe; nuchal band faint, bearing two posterior projections; three very faint, wide, irregularly shaped body bands between limb insertions edged in slightly darker brown; band interspaces bearing irregularly shaped, faint, dark-colored markings; dark-colored speckling on limbs and digits; digits bearing pale-colored bands; four wide, faint, dark-colored caudal bands separated by three pale-colored bands on original portion of tail; all caudal bands encircle tail; all ventral surfaces beige, generally immaculate; and iris orangish to coppery in color.
Variation.
The additional specimen (THNHM 01667) closely approximates the holotype in overall coloration and pattern except that it is more boldly marked. It has four dark-colored body bands as opposed to three and a complete original tail bearing eight dark-colored and seven pale-colored bands. The pale-colored postorbital stripe is slightly thinner and less distinct. Meristic and morphometric differences are listed in Table 5. Given its overall morphological and color pattern similarities and close geographic proximity to the holotype (~ 40 km), we consider this individual as C.cf.kochangensis sp. nov. pending genetic data.
Distribution.
Cyrtodactyluskochangensis sp. nov. is currently known only from the type locality at Ko Chang Island, Ko Phayam Subdistrict, Mueang Ranong District, Ranong Province, Thailand. The additional population of pending species status occurs in the Khlong Naka Wildlife Sanctuary, Suk Samran District Ranong Province.
Etymology.
The specific epithet kochangensis is in reference to the type locality, Ko Chang, Ranong Province, Thailand
Comparisons
(based on the holotype). Cyrtodactyluskochangensis sp. nov. forms a clade with the sister species Cyrtodactylusrivularis sp. nov. and C.rukhadeva (Fig. 2) from which it differs by an uncorrected pairwise sequence divergence of 12.00–12.52% and 12.52–13.68%, respectively (Table 2) and it and C.cf.kochangensis sp. nov. are separated from C.rukhadeva and Cyrtodactylusrivularis sp. nov. by geographic distance of no less than ~280–470 km (Fig. 1). The small sample size (N = 1) precludes it from statistical analyses, however at this point, it differs from C.rukhadeva and C.cf.rukhadeva in having 34 PVT versus 26–30; 14 LRT versus 18–20; 172 VSM versus 152–165; 12 FS versus 13–18; 12 PCS versus 13–17 and 1 PCT versus 2 or 3, collectively. From C.rukhadeva by having 12 FS versus 16 or 17; and five BB versus three. From Cyrtodactylusrivularis sp. nov. it differs in having 14 LRT versus 18–20; 172 VSM versus 160–166; 12 FS versus 14–16; and five BB versus three or four. Discrete differences between Cyrtodactyluskochangensis sp. nov. and C.cf.kochangensis sp. nov. and all other species and populations are presented in Tables 4, 5.
Natural history.
The holotype (ZMKU R 00945) was collected at night (2107 h) among branches of a small tree approximately 100 cm above the ground at 36 m elevation with a temperature of 28.6 °C and relative humidity of 83.9%. The surrounding habitat was dry evergreen forest with a rocky stream nearby (Fig. 11). The new species was found to co-occur with two other species of gekkonid lizards, Cyrtodactylusoldhami (Theobald, 1876) and Gekkotokehos (Grismer, Wood, Grismer, Quah, Thy, Phimmachak, Sivongxay, Seateun, Stuart, Siler, Mulcahy, Anamza & Brown, 2019).
Figure 11.
Habitat of Cyrtodactyluskochangensis sp. nov. at Thailand, Ranong Province, Mueng Ranong District, Ko Phayam Subdistrict, Ko Chang.
. Cyrtodactylus uthaiensis sp. nov.
333ED315-A455-5578-9D5E-3A3BC20D8AEB
https://zoobank.org/9422F241-3A46-4FAD-97C4-D6A6AF16B405
Fig. 12 Suggested Common Name: Uthai Thani Bent-toed Gecko
Figure 12.
Holotype of Cyrtodactylusuthaiensis sp. nov. ZMKU R 00949 (field no. AA 06298) from Thailand, Uthai Thani Province, Lan Sak District, Thung Na Ngam Subdistrict A dorsal view B ventral view C dorsal view of head D ventral view of femoral and precloacal regions E dorsal view of tail F ventral view of tail G holotype in life.
Holotype.
Adult male ZMKU R 00949 from Thung Na Ngam Subdistrict, Lan Sak District, Uthai Thani Province, Thailand (15.37649°N, 99.63426°E, 106 m a.s.l.), collected by Attapol Rujirawan, Siriporn Yodthong, Korkhwan Termprayoon, and Natee Ampai on 18 June 2018.
Diagnosis.
Cyrtodactylusuthaiensis sp. nov. can be separated from all other species of the brevipalmatus group by the combination of having 13–15 supralabials, 10–11 infralabials, 33 paravertebral tubercles, 17 rows of longitudinally arranged tubercles, 36 transverse rows of ventrals, 159 longitudinal rows of ventrals, eight expanded subdigital lamellae on the fourth toe, 12 unexpanded subdigital lamellae on the fourth toe, 20 total subdigital lamellae on the fourth toe; seven expanded subdigital lamellae on the fourth finger, 11 unexpanded subdigital lamellae on the fourth finger, 18 total subdigital lamellae on the fourth finger; 16 total enlarged femoral scales, 12 total femoral pores; 14 enlarged pore-bearing precloacals; enlarged femorals and enlarged precloacals not continuous; proximal femorals less than one-half the size of the distal femorals; small tubercles on forelimbs and flanks; large dorsolateral caudal tubercles and wide ventrolateral caudal fringe; ventrolateral caudal fringe composed scales of different size; tail circular in cross-section; slightly enlarged medial subcaudals intermittent, medially furrowed, posteriorly emarginated; maximum SVL 58.1 mm; and six dark transverse body bands (Tables 5, 6).
Description of holotype
(Fig. 12). Adult male SVL 58.1 mm; head moderate in length (HL/SVL 0.28), width (HW/HL 0.68), depth (HD/HL 0.39), distinct from neck, triangular in dorsal profile; lores concave slightly anteriorly, weakly inflated posteriorly; prefrontal region slightly concave; canthus rostralis rounded; snout elongate (ES/HL 0.40), rounded in dorsal profile; eye large (ED/HL 0.29); ear opening elliptical, small; eye to ear distance greater than diameter of eye; rostral rectangular, dorsally furrowed, bordered posteriorly by large left and right supranasals, bordered laterally by first supralabials; external nares bordered anteriorly by rostral, dorsally by large supranasal, posteriorly by two smaller postnasals, ventrally by first supralabial; 13R/15L rectangular supralabials tapering smoothly to posterior margin of eye; 10R/11L infralabials tapering smoothly to posterior margin of eye; scales of rostrum and lores flat to domed, slightly larger than granular scales on top of head and occiput; scales of occiput intermixed with distinct, small tubercles; superciliaries subrectangular, largest dorsally and anteriorly; mental triangular, bordered laterally by first infralabials and posteriorly by large left and right trapezoidal postmentals contacting medially for approximately 40% of their length posterior to mental; one row of slightly enlarged, elongate sublabials extending posteriorly to fifth(L) and seventh(R) infralabial; gular and throat scales small, granular, grading posteriorly into slightly larger, flatter, smooth, imbricate, pectoral and ventral scales.
Body relatively short (AG/SVL 0.46) with well-defined ventrolateral folds; dorsal scales small, granular, interspersed with larger, conical, semi-regularly arranged, weakly keeled tubercles; tubercles extend from occipital region onto base of tail and slightly beyond as paravertebral rows; smaller tubercles extend anteriorly onto nape and occiput, diminishing in size anteriorly; approximately 17 longitudinal rows of tubercles at midbody; approximately 33 paravertebral tubercles; small tubercles on flanks; 36 longitudinal rows of flat, imbricate, ventral scales much larger than dorsal scales; 159 transverse rows of ventral scales; 16 total large femoral scales; 12 total femoral pores; 14 enlarged pore-bearing precloacals; no deep precloacal groove or depression; and two rows of post-precloacal scales on midline.
Forelimbs moderate in stature, relatively short (ForL/SVL 0.14); granular scales of forearm slightly larger than those on body, interspersed with large tubercles; palmar scales rounded, slightly raised; digits well-developed, relatively short, inflected at basal interphalangeal joints; digits narrower distal to inflections; subdigital lamellae wide, transversely expanded proximal to joint inflections, narrower transverse lamellae distal to joint inflections; claws well-developed, claw base sheathed by a dorsal and ventral scale; 7R/7L expanded and 11R/11L unexpanded lamellae beneath the fourth finger; hind limbs larger and thicker than forelimbs, moderate in length (TibL/SVL 0.14), covered dorsally by granular scales interspersed with moderately sized, conical tubercles dorsally and posteriorly and anteriorly by flat, slightly larger, subimbricate scales; ventral scales of thigh flat, subimbricate, larger than dorsals; subtibial scales flat, imbricate; one row of 6R/6L of enlarged pore-bearing femoral scales not continuous with enlarged pore bearing precloacal scales, terminating distally at knee; 8R/8L enlarged femoral scales; proximal femoral scales smaller than distal femorals, the former forming an abrupt union with much smaller, rounded, ventral scales of posteroventral margin of thigh; plantar scales flat; digits relatively long, well-developed, inflected at basal interphalangeal joints; 8R/(broken)L wide, transversely expanded subdigital lamellae on fourth toe proximal to joint inflection that extend onto sole, 12R/(broken)L unexpanded lamellae beneath first toe; and claws well-developed, sheathed by a dorsal and ventral scale at base.
Tail original, long 76.7 mm (TL/SVL 1.32), 4.1 mm in width at base, tapering to a point; sub-circular or nearly round in cross-section; dorsal scales flat, square bearing tubercles forming paravertebral rows and large tubercles forming a dorsolateral longitudinal row; slightly larger, posteriorly directed, semi-spinose tubercles forming small but distinct ventrolateral caudal fringe; larger scales of ventrolateral fringe occur at regular intervals; slightly enlarged medial subcaudals intermittent, medially furrowed, posteriorly; single enlarged medial subcaudals absent; subcaudal scales, larger than dorsal caudal scales; base of tail bearing hemipenial swellings; 3R/3L conical postcloacal tubercles at base of hemipenial swellings; and postcloacal scales flat, imbricate.
Coloration in life
(Fig. 12). Ground color of the head, body, limbs, and tail pale-brown; dark, diffuse mottling on interorbital region and snout; dark blotch on top of head; wide, pale-colored postorbital stripe irregularly edged in dark brown extends from posterior margin of one eye across nape to posterior margin of other eye; ventral portion of lores, suborbital region, and supralabials darkly mottled; wide, dark brown nuchal band, bearing two posterior projections; six irregularly shaped darkly edged body bands extending between forelimb and hind limb insertions followed by one dark sacral band; paired dark brown paravertebral blotches on nape; band interspaces bearing irregularly shaped, dark-colored markings; dark-colored speckling on limbs and digits; digits bearing pale-colored bands; seven wide dark-colored caudal bands separated by six pale-colored bands; caudal bands encircle tail resulting in heavily mottled subcaudal region; all other ventral surfaces beige, generally immaculate; and iris gold in color.
Distribution.
Cyrtodactylusuthaiensis sp. nov. is currently known from the type locality at Thung Na Ngam Subdistrict, Lan Sak District, Uthai Thani Province, Thailand.
Etymology.
The specific epithet uthaiensis refers to the type locality, Uthai Thani Province, Thailand.
Comparisons.
Cyrtodactylusuthaiensis sp. nov. is the sister species to a clade comprised the sister species C.interdigitalis and C. sp.11 (Fig. 2). Together, these taxa form the sister lineage to C.cf.ngati1, C.cf.ngati2, and all other C.ngati. Cyrtodactylusuthaiensis sp. nov. differs from those lineages by an uncorrected pairwise sequence divergence of 5.81–8.13% (Table 2). We are aware that any comparison based on morphometric and meristic characters are preliminary being that there is only one sample of Cyrtodactylusuthaiensis sp. nov. and that additional sampling may preclude some characters and being diagnostic just as it may reveal that other characters are diagnostic (see Table 5). Therefore, at this point we rely on the invariable categorical characters to separate Cyrtodactylusuthaiensis sp. nov. from other species in the brevipalmatus group. Cyrtodactylusuthaiensis sp. nov. differs from C.brevipalmatus, Cyrtodactylusfluvicavus sp. nov., C.interdigitalis, C.ngati, C.ngati3, C.cf.ngati1, C.cf.ngati2, C.rukhadeva, C.cf.rukhadeva and C. sp.13 by having large dorsolateral caudal tubercles (DCT) forming a wide ventrolateral caudal fringe (VLF1). Cyrtodactylusuthaiensis sp. nov. is further differentiated from C.ngati3, C.cf.ngati1, C.cf.ngati2, C.interdigitalis, C.rukhadeva, C.cf.rukhadeva, and sp.13 by having a ventrolateral fringe not homogenous (VLF2). It differs from Cyrtodactyluskochangensis sp. nov., Cyrtodactylusrivularis sp. nov., C.rukhadeva, C.cf.rukhadeva, and C. sp.11 by having tail that is more circular in cross-section than square (TLcross). From Cyrtodactylusrivularis sp. nov., C.rukhadeva and C.cf.rukhadeva, it differs by having enlarged, unmodified, medial subcaudal scales (SC1). From Cyrtodactylusrivularis sp. nov., C.rukhadeva and C.cf.rukhadeva, it differs by lacking single, enlarged, medial subcaudal scales (SC2). From all species in the brevipalmatus group except C.interdigitalis it differs by having posteriorly emarginated, medial subcaudals bearing a median furrow (SC3).
Natural history.
Cyrtodactylusuthaiensis sp. nov. is the only species of the brevipalmatus group that occurs in an isolated hilly area within the Chao Phraya River Basin (Fig. 1). The holotype (ZMKU R 00949) was collected at night (2055 h) on a bamboo twig approximately 170 cm above ground level at 106 m elevation. The habitat was isolated karst formations within a mixed deciduous forest. This area was surrounded by agricultural fields (plantations and rice fields) and human residential areas (Fig. 13). The new species was found to co-occur with a gekkonid lizard, Dixoniussiamensis (Boulenger, 1899).
Figure 13.
Habitat of Cyrtodactylusuthaiensis sp. nov. at Thailand, Uthai Thani Province, Lan Sak District, Thung Na Ngam Subdistrict.
Discussion
A taxonomy consistent with evolutionary history is paramount to any downstream comparative analyses employed to address the evolution of other features of the group be they ecology, behavior, or habitat preference. By delimiting new and existing species using a phylogenetic analysis and then diagnosing those species using univariate and multivariate statistical analyses of their morphological data, phylogenetic history will not conflated with convergent evolution (Fig. 14). This is extremely important for conservation and natural resource management programs so that efforts can be vectored towards all recognized species without missing the species that were previously masquerading among the synonymies of an erroneous taxonomy. This is especially true for range-restricted, highly specialized species, whose general morphological similarity often does not align with their phylogenetic history as exemplified in the brevipalmatus group (Fig. 3). Given that upland tropical regions are currently some of the most vulnerable to climate change (Gehrig-Fasel et al. 2007; Myers-Smith et al. 2011; Scridel et al. 2018), this argument becomes especially germane for the entire brevipalmatus group— the majority of whose species are upland populations and potentially range-restricted.
Figure 14.
Comparison of a morphology-based taxonomy pre-2020 and the current phylogenetic-based taxonomy illustrating the misrepresentation of the diversity of the Cyrtodactylusbrevipalmatus group by the former. Colored lines connecting species on the left to species on the right indicate under which name the former were masquerading as the latter.
The description of the four new species, Cyrtodactylusfluvicavus sp. nov., Cyrtodactyluskochangensis sp. nov., Cyrtodactylusrivularis sp. nov., and Cyrtodactylusuthaiensis sp. nov. adds to the rapidly growing list of new species of Cyrtodactylus from Thailand. Many of the poorly sampled populations reported here such as C. sp.9 from Thong Pha Phum National Park, Kanchanaburi Province; C. sp.10 from Chao Doi Waterfall, Tak Province; C. sp.11 from Phu Hin Rong Kla National Park, Phitsanulok Province; C. sp.13 from Ban Saphan Lao, Kanchanaburi Province and Thung Yai Naresuan Wildlife Sanctuary, Tak Provinces; C. sp.14 from Langkawi Island, Peninsular Malaysia; C.cf.ngati1 and C.cf.ngati2 from Xaignabouli and Vientiane Provinces, respectively, Laos (Figs 1, 2), will likely be described as new species when new material becomes available. Additionally, many populations listed in various non-technical websites as C.interdigitalis or C.brevipalmatus based on overall similarity or general distribution, may also be recovered as new species or delegated to some of the more recently described species. This research continues to highlight the unrealized biodiversity in climatically imperiled upland ecosystems throughout Southeast Asia.
Supplementary Material
Acknowledgements
This work was financially supported by Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (grant no. RGNS 64-038). NA was supported by Srinakharinwirot University Research Grant (no. 596/2564). This research was reviewed and approved by the Institutional Animal Care and Use Committee of Faculty of Science, Kasetsart University (ACKU64-SCI-006) and the Department of National Parks, Wildlife and Plant Conservation, Thailand provided the research permission. We would like to thank Suchai Horradee (Chaloem Rattanakosin National Park), Charoen Jaichon (Thong Pha Phum National Park) and Mana Phermpool (Kaeng Krachan National Park) for facilitating the fieldwork. Wachara Sanguansombat and Sunchai Makchai (Thailand Natural History Museum) made specimens in their care available for study. Piyawan Puanprapai assisted with fieldwork. We would like to thank Bryan L. Stuart for suggestions on this research and Truong Nguyen and an anonymous reviewer improved the manuscript.
Citation
Grismer LL, Aowphol A, Yodthong S, Ampai N, Termprayoon K, Aksornneam A, Rujirawan A (2022) Integrative taxonomy delimits and diagnoses cryptic arboreal species of the Cyrtodactylus brevipalmatus group (Squamata, Gekkonidae) with descriptions of four new species from Thailand. ZooKeys 1129: 109–162. https://doi.org/10.3897/zookeys.1129.90535
Funding Statement
Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (grant no. RGNS 64-038)
Supplementary materials
Data frame for the multiple factor analysis of the putative species of the Cyrtodactylusbrevipalmatus group
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
L. Lee Grismer, Anchalee Aowphol, Siriporn Yodthong, Natee Ampai, Korkhwan Termprayoon, Akrachai Aksornneam, Attapol Rujirawan
Data type
morphological data
Explanation note
Data frame for the multiple factor analysis of the putative species of the Cyrtodactylusbrevipalmatus group.
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. (2004) Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430(6999): 551–554. 10.1038/nature02649 [DOI] [PubMed] [Google Scholar]
- Barraclough TG, Birky Jr CW, Burt A. (2003) Diversification in sexual and asexual organisms. Evolution 57(9): 2166–2172. 10.1111/j.0014-3820.2003.tb00394.x [DOI] [PubMed] [Google Scholar]
- Baulechner D, Jauker F, Neubauer TA, Wolters V. (2020) Convergent evolution of specialized generalists: Implications for phylogenetic and functional diversity of carabid feeding groups. Ecology and Evolution 10(20): 11100–11110. 10.1002/ece3.6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR, Drummond AJ. (2017) bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evolutionary Biology 17(1): 42. 10.1186/s12862-017-0890-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KO, Grismer LL. (2022) GroupStruct: An R package for allometric size correction. Zootaxa 5124(4): 471–482. 10.11646/zootaxa.5124.4.4 [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. (1998) The evolutionary genetics of speciation. Philosophical Transactions of the Royal Society of London Series B, Biological science 353(1366): 287–305. 10.1098/rstb.1998.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz K. (2007) Species concepts and species delimitation. Systematic Biology 56(6): 879–886. 10.1080/10635150701701083 [DOI] [PubMed] [Google Scholar]
- Dehling DM, Jordano P, Schaefer HM, Böhning-Gaese K, Schleuning M. (2016) Morphology predicts species’ functional roles and their degree of specialization in plant–frugivore interactions. Proceedings of the Royal Society B, Biological Sciences 283(1823): 1–7. 10.1098/rspb.2015.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012) Bayesian phylogenetics with BEAUti and BEAST 1.7. Molecular Biology and Evolution 29(8): 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Pauwels OSG. (2012) The bent-toed geckos (Cyrtodactylus) of the caves and karst of Thailand. Cave Karst Science 39(1): 16–22. [Google Scholar]
- Feulner PGD, Kirschbaum F, Mamonekene V, Ketmaier V, Tiedemann R. (2007) Adaptive radiation in African weakly electric fish (Teleostei: Mormyridae: Campylomormyrus): a combined molecular and morphological approach. Journal of Evolutionary Biology 20(1): 403–414. 10.1111/j.1420-9101.2006.01181.x [DOI] [PubMed] [Google Scholar]
- Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. (2007) Independently evolving species in asexual bdelloid rotifers. PLoS Biology 5(4): e87. 10.1371/journal.pbio.0050087 [DOI] [PMC free article] [PubMed]
- Frost DR, Hillis DM. (1990) Species in concept and practice: Herpetological application. Herpetologica 46(1): 86–104. [Google Scholar]
- Frost DR, Kluge AG. (1994) A consideration of the epistemology in systematic biology, with special reference to species. Cladistics 10(3): 259–294. 10.1111/j.1096-0031.1994.tb00178.x [DOI] [Google Scholar]
- Gehrig-Fasel J, Guisan A, Zimmermann NE. (2007) Tree line shifts in the Swiss Alps: Climate change or land abandonment? Journal of Vegetation Science 18(4): 571–582. 10.1111/j.1654-1103.2007.tb02571.x [DOI]
- Grismer LL. (2008) On the distribution and identification of Cyrtodactylusbrevipalmatus Smith, 1923 and Cyrtodactyluselok Dring, 1979. The Raffles Bulletin of Zoology 56(1): 177–179. [Google Scholar]
- Grismer LL, Wood Jr PL, Le MD, Quah ESH, Grismer JL. (2020) Evolution of habitat preference in 243 species of Bent‐toed geckos (Genus Cyrtodactylus Gray, 1827) with a discussion of karst habitat conservation. Ecology and Evolution 10(24): 13717–13730. 10.1002/ece3.6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grismer LL, Wood Jr PL, Poyarkov NA, Le MD, Kraus F, Agarwal I, Oliver PM, Nguyen SN, Nguyen TQ, Karunarathna S, Welton LJ, Stuart BL, Luu VQ, Bauer AM, O’Connell KA, Quah ESH, Chan KO, Ziegler T, Ngo H, Nazarov RA, Aowphol A, Chomdej S, Suwannapoom C, Siler CD, Anuar S, Tri NV, Grismer JL. (2021a) Phylogenetic partitioning of the third-largest vertebrate genus in the world, Cyrtodactylus Gray, 1827 (Reptilia; Squamata; Gekkonidae) and its relevance to taxonomy and conservation. Vertebrate Zoology 71: 101–154. 10.3897/vertebrate-zoology.71.e59307 [DOI] [Google Scholar]
- Grismer LL, Wood Jr PL, Poyarkov NA, Le MD, Karunarathna S, Chomdej S, Suwannapoom C, Qi S, Liu S, Che J, Quah ESH, Kraus F, Oliver PM, Riyanto A, Pauwels OSG, Grismer JL. (2021b) Karstic landscapes are foci of species diversity in the World’s Third-Largest Vertebrate genus Cyrtodactylus Gray, 1827 (Reptilia: Squamata; Gekkonidae). Diversity 13(5): 183. 10.3390/d13050183 [DOI] [Google Scholar]
- Grismer LL, Suwannapoom C, Pawangkhanant P, Nazarov RA, Yushchenko PV, Naiduangchan M, Le MD, Luu VQ, Poyarkov NA. (2021c) A new cryptic arboreal species of the Cyrtodactylusbrevipalmatus group (Squamata: Gekkonidae) from the uplands of western Thailand. Vertebrate Zoology 71: 723–746. 10.3897/vz.71.e76069 [DOI] [Google Scholar]
- Grismer LL, Rujirawan A, Yodthong S, Stuart BL, Le MD, Le DT, Chuaynkern Y, Wood Jr PL, Aowphol A. (2022) The taxonomy and phylogeny of the Cyrtodactylusbrevipalmatus group (Squamata: Gekkonidae) with emphasis on C.interdigitalis and C.ngati. Vertebrate Zoology 72: 245–269. 10.3897/vz.72.e80615 [DOI] [Google Scholar]
- Hillis DM. (2019) Species delimitation in herpetology. Journal of Herpetology 53(1): 3–12. 10.1670/18-123 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294(5550): 2310–2314. 10.1126/science.1065889 [DOI] [PubMed] [Google Scholar]
- Husson F, Josse J, Le S, Mazet J. (2017) FactoMine R: explortory data analysis and data mining. R package, version 1.36.
- Kassambara A, Mundt F. (2017) Factoextra: extract and visualize the result of multivariate data analyses. R package, version 1.0.5.999.
- Knowles LL, Carstens BC. (2007) Delimiting species without monophyletic gene trees. Systematic Biology 56(6): 887–895. 10.1080/10635150701701091 [DOI] [PubMed] [Google Scholar]
- Le DT, Sitthivong S, Tran TT, Grismer LL, Nguyen TQ, Le MD, Ziegler T, Luu VQ. (2021) First record of the Cyrtodactylusbrevipalmatus group (Squamata: Gekkonidae) from Vietnam with description of a new species. Zootaxa 4969(3): 492–510. 10.11646/zootaxa.4969.3.3 [DOI] [PubMed] [Google Scholar]
- Leach AD, Koo MS, Spencer CL, Papenfuss TJ, Fisher RN, McGuire JA. (2009) Quantifying ecological, morphological, and genetic variation to delimit species in the coast horned lizard species complex (Phrynosoma). Proceedings of the National Academy of Sciences of the United States of America 106(30): 12418–12423. 10.1073/pnas.0906380106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XL, Stur E, Ekrem T. (2018) Exploring species boundaries with multiple genetic loci using empirical data from non-biting midges. Zoologica Scripta 47(3): 325–341. 10.1111/zsc.12280 [DOI] [Google Scholar]
- Lleonart J, Salat J, Torres GJ. (2000) Removing allometric effects of body size in morphological analysis. Journal of Theoretical Biology 205(1): 85–93. 10.1006/jtbi.2000.2043 [DOI] [PubMed] [Google Scholar]
- Manthey U, Grossmann W. (1997) Amphibien and Reptilien Südostasiens. Natur und Tier Verlag, Münster, Germany, 512 pp. [Google Scholar]
- McCoy MW, Bolker BM, Osenberg CW, Miner BG, Vonesh JR. (2006) Size correction: Comparing morphological traits among populations and environments. Oecologia 148(4): 547–554. 10.1007/s00442-006-0403-6 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE), New Orleans (USA), November 2010, IEEE, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Myers-Smith IH, Forbes BC, Wilmking M, Hallinger M, Lantz T, Blok D, Tape KD, Macias-Fauria M, Sass-Klaassen U, Levesque E, Boudreau S, Ropars P, Hermanutz L, Trant A, Collier LS, Weijers S, Rozema J, Rayback SA, Schmidt NM, Schaepman-Strub G, Wipf S, Rixen C, Menard CB, Venn S, Goetz S, Andreu-Hayles L, Elmendorf S, Ravolainen V, Welker J, Grogan P, Epstein HE, Hik DS. (2011) Shrub expansion in tundra ecosystems: Dynamics, impacts and research priorities. Environmental Research Letters 6(4): 045509. 10.1088/1748-9326/6/4/045509 [DOI] [Google Scholar]
- Nabhitabhata J, Chan-ard T. (2005) Thailand Red Data: Mammals, Reptiles and Amphibians. Office of Natural Resources and Environmental Policy and Planning, Bangkok, Thailand, 234 pp. [Google Scholar]
- Nabhitabhata J, Chan-ard T, Chuaynkern Y. (2004) Checklist of Amphibians and Reptiles in Thailand. Office of Environmental Policy and Planning, Bangkok, Thailand, 152 pp. [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. (2020) Package ‘vegan’. Version 2.5-7. https://cran.r-project.org/web/packages/vegan/
- Pagès J. (2015) Multiple Factor Analysis by Example Using R. CRC Press, New York, USA, 272 pp. 10.1201/b17700 [DOI] [Google Scholar]
- Pauwels OSG, Chan-ard T. (2006) Reptiles of Kaeng Krachan National Park, western Thailand. The Natural History Bulletin of the Siam Society 54(1): 89–108. [Google Scholar]
- R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna. http://www.R-project.org [accessed 1 June 2022]
- Rambaut A, Drummond AJ. (2013) TreeAnnotator v1.8.0 MCMC Output Analysis. https://beast.community/treeannotator
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67(5): 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reist JD. (1986) An empirical evaluation of coefficients used in residual and allometric adjustment of size covariation. Canadian Journal of Zoology 64(6): 1363–1368. 10.1139/z86-203 [DOI] [Google Scholar]
- Revell LJ. (2012) Phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3(2): 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Ricklefs RE. (2012) Species richness and morphological diversity of passerine birds. Proceedings of the National Academy of Sciences of the United States of America 109(36): 14482–14487. 10.1073/pnas.1212079109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabaj MH. (2020) Codes for natural history collections in ichthyology and herpetology. Copeia 108(3): 593–669. 10.1643/ASIHCODONS2020 [DOI] [Google Scholar]
- Scridel D, Brambilla M, Martin K, Lehikoinen A, Iemma A, Matteo A, Jähnig S, Caprio E, Bogliani G, Pedrini P, Rolando A, Arlettaz R, Chamberlain D. (2018) A review and meta-analysis of the effects of climate change on Holarctic mountain and upland bird populations. The Ibis 160(3): 489–515. 10.1111/ibi.12585 [DOI] [Google Scholar]
- Sidlauskas B. (2008) Continuous and arrested morphological diversification in sister clades of characiform fishes: A phylomorphospace approach. Evolution 62(12): 3135–3156. 10.1111/j.1558-5646.2008.00519.x [DOI] [PubMed] [Google Scholar]
- Skalski JR, Richins SM, Townsend R. (2018) A statistical test and sample size recommendations for comparing community composition following PCA. PLoS ONE 13(10): e0206003. 10.1371/journal.pone.0206033 [DOI] [PMC free article] [PubMed]
- Smith MA. (1935) The fauna of British India, including Ceylon and Burma. Reptilia and Amphibia. Vol. II. Sauria. Taylor and Francis, London, 440 pp. [Google Scholar]
- Stuart BL. (1999) Amphibians and Reptiles. In: Duckworth JW, Salter RE, Khounboline K. (Eds) Wildlife in Lao PDR: 1999 Status Report.IUCN-The World Conservation Union/ Wildlife Conservation Society/Centre for Protected Areas and Watershed Management, Vientiane, Laos, 43–67.
- Sukumaran J, Knowles LL. (2017) Multispecies coalescent delimits structure, not species. Proceedings of the National Academy of Sciences of the United States of America 114(7): 1607–1612. 10.1073/pnas.1607921114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38(7): 3022–3027. 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe RS. (1975) Quantitative handling of characters useful in snake systematics with particular reference to intraspecific variation in the Ringed Snake Natrixnatrix (L.). Biological Journal of the Linnean Society 7(1): 27–43. 10.1111/j.1095-8312.1975.tb00732.x [DOI] [Google Scholar]
- Thorpe RS. (1983) A review of the numerical methods for recognising and analysing racial differentiation. In: Felsenstein J. (Ed.) Numerical Taxonomy.NATO ASI Series, Volume 1. Springer, Berlin, Heidelberg, 404–423. 10.1007/978-3-642-69024-2_43 [DOI]
- Turan C. (1999) A note on the examination of morphometric differentiation among fish populations: The Truss System. Turkish Journal of Zoology 23(3): 259–263. [Google Scholar]
- Ulber T. (1993) Bemerkungen über cyrtodactyline Geckos aus Thailand nebst Beschreibungen von zwei neuen Arten (Reptilia: Gekkonidae). Mitteilungen aus dem Museum für Naturkunde in Berlin 69(2): 187–200. 10.1002/mmnz.19930690202 [DOI] [Google Scholar]
- Vinh NX, Epps J, Bailey J. (2010) Information theoretic measures for clusterings comparison: Variants, properties, normalization and correction for chance. Journal of Machine Learning Research 11: 2837–2854. [Google Scholar]
- Welch KRG, Cooke PS, Wright AS. (1990) Lizards of the Orient: A Checklist. Robert E. Krieger Publishing Company, Malabar, Florida, USA, 372 pp. [Google Scholar]
- Wilcox TP, Zwickl DJ, Heath TA, Hillis DM. (2002) Phylogenetic relationships of the Dwarf Boas and a comparison of Bayesian and bootstrap measures of phylogenetic support. Molecular Phylogenetics and Evolution 25(2): 361–371. 10.1016/S1055-7903(02)00244-0 [DOI] [PubMed] [Google Scholar]
- Yodthong S, Rujirawan A, Stuart BL, Grismer LL, Aksornneam A, Termprayoon K, Ampai N, Aowphol A. (2022) A new species in the Cyrtodactylusoldhami group (Squamata, Gekkonidae) from Kanchanaburi Province, western Thailand. ZooKeys 1103: 139–169. 10.3897/zookeys.1103.84672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Kapli P, Pavlidis P, Stamatakis A. (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29(22): 2869–2876. 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data frame for the multiple factor analysis of the putative species of the Cyrtodactylusbrevipalmatus group
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
L. Lee Grismer, Anchalee Aowphol, Siriporn Yodthong, Natee Ampai, Korkhwan Termprayoon, Akrachai Aksornneam, Attapol Rujirawan
Data type
morphological data
Explanation note
Data frame for the multiple factor analysis of the putative species of the Cyrtodactylusbrevipalmatus group.