Abstract
A family of abundant surface proteins (Vpmas [variable proteins of Mycoplasma agalactiae]) undergoing phase variation in M. agalactiae has been characterized using monoclonal antibodies and specific polyclonal sera. Two expressed members of 39 kDa (Vpma39) and 34 kDa (Vpma34), which varied in expression between clones of a lineage, shared a common amino-terminal sequence but were immunologically distinct. An amino-terminal oligonucleotide probe identified multiple vpma genes which were clustered within a 14-kb ClaI genomic fragment. Rearrangements were found to have occurred within the vpma locus between clones which correlated with changes in their Vpma phenotype. Two neighboring vpma genes were cloned and sequenced from one M. agalactiae clonal variant expressing Vpma39. The two genes, vpmaX and vpmaY, were orientated divergently and shared highly homologous 5′ untranslated regions, 25-amino-acid (aa) lipoprotein leader sequences, and amino-terminal sequences. The vpmaY gene coded for 346 aa and 84% of the open reading frame, comprised of 1.5 units of a large repeat of 186 aa. Although the sequence for an entire second vpmaY repeat was present, it was prematurely terminated by insertion of two nucleotides. The vpmaX gene encoded 221 aa and possessed 102 aa of the 186-aa repeat of vpmaY. Many of the features in common between the vpma genes were also found to be shared by the vsp genes of M. bovis, which also undergo DNA rearrangements concomitant with phenotypic changes. Since M. bovis is the closest phylogenetic relative to M. agalactiae, the vpma and vsp gene families most probably represent homologous systems.
Mycoplasma agalactiae, a cell wall-less bacterium belonging to the class Mollicutes, is considered the classical agent of the syndrome contagious agalactia in sheep and goats. It primarily affects the mammary glands, the joints and eyes, and, to a lesser extent, the respiratory tract, causing various clinical manifestations such as mastitis, arthritis, conjunctivitis, and pneumonia (4, 12). In most cases, infections in the udder of susceptible lactating females result in a rapid drop in milk production, shedding of the pathogen in the milk, and possibly spread to secondary sites and other animals. During subsequent lactations, infected animals may present minor to no clinical symptoms, yet M. agalactiae can still be shed in the milk and other body fluids. These asymptomatic carriers contribute to the prevalence of the disease since they can transmit the pathogen to susceptible animals for up to 4 years after the initial infection (4). Because contagious agalactia is worldwide and affects all breeds of sheep and goats, expensive measures are necessary for diagnosis and control both in regions where the disease is enzootic and in those with sporadic outbreaks.
As for many mycoplasma pathogens, antigenic variation of M. agalactiae surface proteins is believed to play a major role in the survival and dissemination of this organism within and between hosts. During their evolution, successful pathogenic bacteria have evolved a wide range of mechanisms to multiply and survive within their complex immunocompetent hosts (13, 21, 37). Mycoplasmas are no exception, and accumulating evidence has shown the presence in these minimal prokaryotes of various sophisticated genetic systems that generate extensive phenotypic variation within populations derived from single organisms (9, 36, 52). Despite the limited coding capacity of mycoplasma genomes (22), most of these systems often involve families of multiple but distinct single-copy genes, clustered on the chromosome and coding for major surface components. The overall function of these systems is to provide the mycoplasma with a highly versatile surface architecture drawn randomly from its gene pool in order to compensate for the lack of regulatory systems (22) that would allow the pathogen to modulate its interaction within the host. In an ultimate step, these systems may participate in establishing and maintaining successful infections, the latter being particularly pertinent for mycoplasma infections, as they result in diseases which are often characterized by their chronicity. The phenomenon of antigenic diversity via phase or size variation is driven by reversible, stochastic, high-frequency mutational events that affect the ON↔OFF expression (phase variation) and/or the structure (size variation) of each member of a multigene family. Remarkably, all highly mutable genes, silent or expressed, maintain functional complete open reading frames (ORFs) as if their coding sequences were under a high selection pressure (6, 10, 24, 26, 27), which may be due to periodic expression in the host.
One mode for controlling the expression of individual genes within multigene families is the random but high-frequency insertions or deletions of nucleotides in a repeated DNA sequence located within or near the promoter region of each member so that a particular repeat length dictates whether a gene is transcribed or not. This includes the Vlp system in M. hyorhinis [mutation in the number of adenosines in a poly(A) tract] (53), and the pMGA system in M. gallisepticum (mutation in the number of GAA repeats) (18, 19). Alternatively, ON↔OFF switching of genes can occur as a result of DNA rearrangement(s) within a multigene locus to link an ORF to an active promoter such as for the V-1 (or Vsa) system in M. pulmonis (6) and is speculated for the Vsp system in M. bovis (24). Several single-copy genes, such as the vaa gene in M. hominis (55) and the gene coding for p78 in M. fermentans (46), undergo ON↔OFF expression via frameshift translational control. Individual genes in some of the above-mentioned systems can further increase surface diversity by varying their size via insertion/deletion of reiterated coding units as shown for the Vlps (53), the Vsps (23), the single-copy vaa genes of M. pulmonis (54), and the gene coding for the MB antigen of Ureaplasma urealyticum (56).
All multigene families so far characterized in mycoplasmas encode surface lipoproteins which are highly immunogenic in their host and, most important, are structurally different from one mycoplasma species to the other. Although the molecular mechanisms by which several systems generate surface antigenic variation have been deciphered in mycoplasmas (6, 19, 53), the exact functions of these surface components remain to be fully assessed; this effort has been hampered by the difficulty in isolating or distinguishing one member from another in a given family.
Variability between M. agalactiae strains and isolates has been shown to exist at the protein level (5, 44, 49); however, little is known about the genetic systems that generate surface variation in this species. In a previous study to develop M. agalactiae-specific monoclonal antibodies (MAbs) for diagnostic purposes, four MAbs were shown to immunostain M. agalactiae colonies, revealing typical sectorial staining associated with the presence of variable epitopes (5). In this study, we used two such MAbs to characterize a new family of abundant surface proteins (Vpmas [variable proteins of M. agalactiae]) in M. agalactiae type strain PG2, which undergo phase variation via high-frequency DNA rearrangements. Examination of the vpma system revealed many features in common with the vsp system of M. bovis, a mycoplasma that is phylogenetically related to M. agalactiae and induces similar clinical signs in cattle (33).
MATERIALS AND METHODS
Mycoplasma culture and derivation of clonal lineages.
M. agalactiae type strain PG2 was isolated from a sheep in Spain (44). Mycoplasmas were grown in standard medium according to Aluotto et al. (1) at 37°C. Clonal lineages were obtained from PG2 as follows. Optimal concentrations of M. agalactiae type strain PG2 cells were inoculated onto solid agar plates; after 3 days at 37°C, colonies were lifted onto nitrocellulose membranes and immunostained with MAb 3B3, using standard methods as previously described (39). One sectored and one negative clone (clones 55 and 60, respectively) were picked and grown in 1 ml of liquid growth medium at 37°C for 3 days. These cultures were again grown on solid growth medium, and colonies were immunostained as before to obtain uniformly staining first-generation clones. This procedure was repeated on selected clones to obtain second and, for the clone 60 lineage, third-generation clones as shown in Fig. 1A.
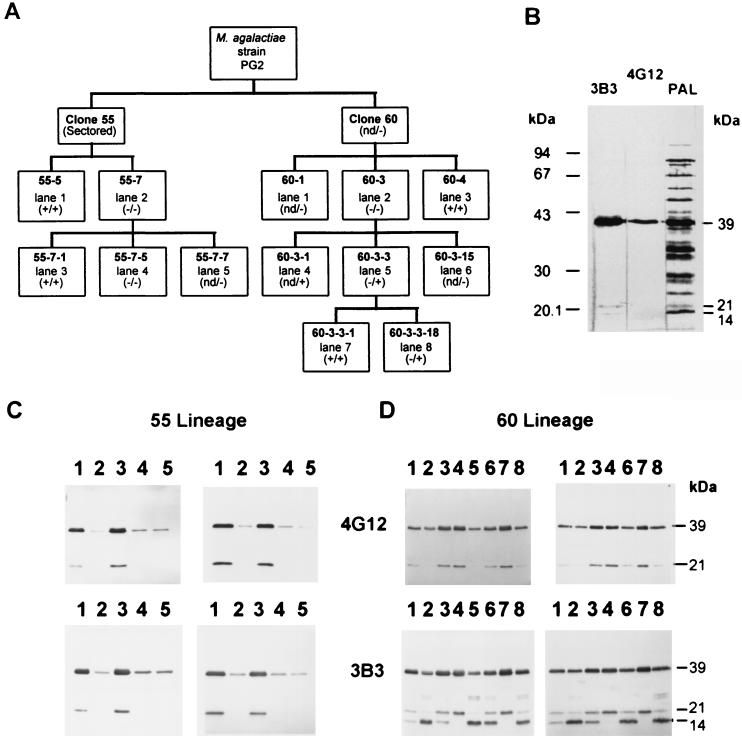
FIG. 1.
(A) Outline showing clones of the 55 (from clone 55) and 60 (from clone 60) lineages, derived from M. agalactiae type strain PG2 based on colony immunostaining positive (+) or negative (−) with MAb 3B3. These clones were also immunostained at the colony level with MAb 4G12 except for those marked “nd” (not determined). (−/+) indicates 4G12-negative and 3B3-positive colonies; (−/−) indicates 4G12- and 3B3-negative colonies. (B) Western blot analysis of Triton X-114-phase material from the parental M. agalactiae type strain PG2 using 3B3, 4G12, or polyclonal anti-M. agalactiae sheep serum (PAL) as indicated above each lane. (C and D) Western blot analysis of colony clones from the 55 (C) and 60 (D) lineages. Whole-cell lysates (left panels) and cell material partitioned into the Triton X-114 detergent phase (right panels) were immunostained with 4G12 (top panels) and 3B3 (bottom panels). Lanes are labeled according to the lane number shown in panel A for the corresponding lineage.
Antibodies and immunostaining.
MAbs 3B3 and 4G12 (both immunoglobulin G1 [IgG1] isotype) were produced as part of a collaborative research project of the AFSSA-Lyon (Lyon, France) and the Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia (Brescia, Italy), using M. agalactiae type strain PG2 as the whole-cell antigen source, and characterized as previously described (5). Sheep anti-M. agalactiae serum (PAL), with a high anti-M. agalactiae antibody titer in enzyme-linked immunosorbent assay, was obtained from a naturally infected ewe in the French Pyrenees. Rabbit sera specific to the 39-kDa protein from clone 55-5 (Vpma39) and to the 34-kDa protein from clone 55-7 (Vpma34) were produced as previously described (18, 19). Briefly, proteins of clones 55-5 and 55-7 that partitioned in the Triton X-114 detergent phase were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. The proteins of interest were then located according to size by Ponceau S staining, excised, pulverized into fine particles by sonication, and injected into New Zealand White rabbits using a protocol previously described (19). Western blot and colony blot analyses were performed as previously described (39). Rabbit antiserum was diluted 1/1,000. Goat anti-mouse IgG (Fc fragment specific) conjugated to horseradish peroxidase (Accurate Chemical and Scientific Corporation) and swine anti-rabbit Ig-horseradish peroxidase conjugate (DAKO) were used at dilutions of 1/1,000 to 1/2,000. Membranes were developed using 4-chloro-1-naphthol or 3,3′-diaminobenzidine tetrahydrochloride (DAB tablets; D-5905; Sigma). Low-molecular-weight protein standards (Amersham Pharmacia Biotech) were used for Coomassie blue-stained SDS-polyacrylamide gels, which were prestained with Remazol brilliant blue R (Sigma); prestained standards from Bio-Rad were used for Western blots.
Amino-terminal sequencing.
Proteins partitioned into the Triton X-114 phase from clones were subjected to reducing SDS-PAGE and transferred to Immobilon-P (Millipore) membranes. Protein bands of interest were identified by staining with Ponceau S, excised, and sequenced at the Institute of Biochemistry, University of Vienna, Vienna, Austria, using an Applied Biosystems model 476A automated protein sequencer.
Protease treatment of intact mycoplasmas.
Intact cells were treated with increasing concentrations of trypsin or carboxypeptidase Y as previously described (39). Briefly, cells from late-logarithmic-phase culture were harvested by centrifugation and washed three times with phosphate-buffered saline (PBS). Suspended cells from 200 μl (for trypsin digest) or 167 μl (carboxypeptidase Y) of original culture were incubated at 37°C for 1 h in 30 μl of PBS at pH 8 (trypsin) or 2 h in 150 μl of PBS at pH 7.2 (carboxypeptidase Y) containing either no enzyme, 20, 40, 80, or 160 U of trypsin (Sigma T-1426), or 7.6, 19, 28.5, 38, or 76 U of carboxypeptidase Y (Sigma C-3888). Cells were then washed once in PBS and lysed in 25 μl of reducing SDS-PAGE loading buffer; equivalent amounts were subjected to SDS-PAGE and Western blot analysis.
Quantitation of Vpma protein abundance in culture medium supernatants.
Cells were grown in liquid growth medium at 37°C, and 8-ml aliquots were taken after 2, 4, and 7 days of growth; 7 ml of each aliquot was centrifuged at 24,000 × g for 20 min at 4°C, and a portion of the remainder was diluted for CFU determination. The cell pellets were washed once in PBS and finally resuspended in 200 μl of PBS. The culture supernatants were centrifuged again at 24,000 × g for 20 min at 4°C and then passed through one 0.22-μm-pore-size filter followed by successive filtering through three 0.1 μm-pore-size filters. A 50-μl aliquot from each supernatant was grown on solid growth medium and confirmed to contain no viable mycoplasma cells. Tenfold dilutions of the supernatant and the corresponding cell fraction were subjected to reducing SDS-PAGE and Western blot analysis using rabbit anti-Vpma34 and rabbit anti-Vpma39 sera, respectively. The dilution before which no immunostaining protein band could be detected was used to calculate the abundance of the Vpma protein in the culture supernatant as a percentage of that present in the cell fraction for the same volume of original culture medium. This dilution was chosen to ensure that the staining was not at saturation levels. Protein band abundances were calculated from scanned Western blots (UMAX model UC1260 scanner [Umax Data System Inc.], with identical settings for all blots), using ImageMaster 1D software from Pharmacia Biotech. The values indicated were calculated as the raw volume, which represents the sum of the intensities of every pixel in the stained protein band.
Oligonucleotide sequences.
The A3F oligonucleotide sequence, 5′-AA(A/G)TG(T/C)GG(A/T)GG(A/T)AC(A/T)A(A/C)(A/T)(A/G)A-3′, was designed from the common amino-terminal sequence between Vpma39 and Vpma34, KCGGTKD/E, and degeneracies were reduced based on mycoplasmas having a bias for A or T in the third position of degenerate codons. The vspS-2 oligonucleotide sequence, 5′-AATTCCCTTTGTAGCAGCT-3′ (see Fig. 6), was designed from the leader sequence of vspA from M. bovis type strain PG45, GenBank accession number L81118 (23). Specific primers for the vpmaY gene were Y2F (5′-ccggaattCCCTAAAACTCCAGCAGAAGGCAG-3′) and Y2R (5′-gctctagACATCAGACATCGCTGACGTTAAG-3′) (capital letters represent vpmaY-specific sequences, and lowercase letters are nucleotides introduced to facilitate further cloning).
FIG. 6.
(A) Clustal W DNA alignment of the 5′ noncoding and coding regions of vspA, vpmaY, and vpmaX. An asterisk represents identity between all three nucleotide sequences, and a dash represents a gap introduced to optimize the alignment. The translated amino acid sequences are aligned above the DNA sequences. A putative ribosomal binding sequence (SD) is overlined, and an 11-nucleotide imperfect inverted repeat is marked by an arrowed line above the sequence. A box encompasses the vspA sequence used to design the leader oligonucleotide vspS-2.
Genomic DNA isolation, Southern blotting PCR, and DNA sequencing.
Mycoplasma genomic DNA was prepared as previously described (26). For Southern blot analysis, 1 μg of DNA was digested with the appropriate restriction enzyme (Promega) and subjected to agarose gel electrophoresis. DNA fragments were then transferred to H-bond N+ membranes (Amersham) using a standard protocol (43), prehybridized under previously described conditions (20), hybridized overnight with a digoxigenin (DIG)-labeled probe, and washed under stringent conditions followed by nonradioactive detection performed according to the manufacturer's recommendations. The probe was stripped from the membrane by treating the membrane twice with 0.2 M NaOH–0.1% (wt/vol) SDS for 10 min at room temperature. Oligonucleotides were labeled at the 3′ end using DIG-ddUTP and terminal transferase (DIG oligonucleotide tailing kit; Boehringer Mannheim) according to the manufacturer's instructions. Labeled A3F was hybridized with the membrane overnight at 50°C and washed two times in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 50°C for 10 min. Labeled vspS-2 was hybridized with the membrane overnight at 45°C and washed two times in 6× SSC–0.1% SDS at 45°C for 10 min.
PCR amplifications were performed in a total volume of 50 μl using 0.25 U of Taq DNA polymerase (Promega) in 1× buffer supplied by the company, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 0.4 μM primer, and 100 ng of genomic DNA template. Cycling conditions were 1 cycle at 94°C for 1 min, 29 cycles of 94°C for 30 s, 50°C (A3F) or 64°C (Y2F and Y2R) for 45 s, and 72°C for 5 min (A3F) or 1 min 15 s (Y2F and Y2R), followed by 1 cycle at 72°C for 7 min, performed in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler. PCR products were cloned into the T-tailed vector pGEM-Teasy (Promega) and used as a template for DIG labeling by PCR using a DIG-11-dUTP (Boehringer Mannheim)/dATP ratio of 1:19 and the above PCR conditions. PCR probes were hybridized with the membrane overnight at 50°C (PCR9 and/or PCR91 from A3F) or 56°C (PCRY2 from Y2F and Y2R) and washed in 0.5× SSC–0.5% SDS at 50°C (PCR9 and/or PCR91) or 0.2× SSC–0.5% SDS at 56°C (PCRY2) for 2 h.
Sequencing of DNA fragments cloned into plasmid vectors was done by VBC-Genomics Bioscience Research GmbH, Vienna, Austria, using IRD 700 or IRD 800 dye-labeled sequencing primers, dideoxy PCR, and a Li-COR model 4200 DNA sequencer.
Construction and screening of an M. agalactiae genomic library.
Genomic DNA from M. agalactiae clone 55-5 was digested with HindIII, extracted with phenol (43), precipitated with ethanol, and resuspended in water; 1 μg was ligated with 50 ng of pUC18, prepared with HindIII and bacterial alkaline phosphatase (Amersham Pharmacia Biotech), in a final volume of 5 μl with 0.5 U of ligase (Boehringer Mannheim) and 1× buffer supplied by the company. Escherichia coli DH10B transformants were screened by hybridization of colonies on H-bond N+ membranes (Amersham), using a mix of two vpma-specific PCR fragments (PCR9 and PCR91) as probes. Probe-positive recombinant clones were purified from the genomic library by three rounds of screening.
Database searches and DNA analysis.
Advanced BLASTp and BLASTx searches were used at the web site for the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) for amino acid or translated DNA sequences against all nonredundant sequences in the databases: GenBank CDS translation, PDB, SwissProt, PIR, and PRF for protein query sequences; GenBank, EMBL, DDBJ, and PDB for DNA query sequences. DNA alignments were performed using Clustal W version 1.7 (48). Translations and calculations of molecular mass and pI of proteins were performed using the programs at the Expert Protein Analysis System proteomics server (http://www.expasy.ch/).
Nucleotide sequence accession number.
The nucleotide sequences described here have been assigned GenBank accession number AF248865.
RESULTS
Related surface proteins (Vpmas) in M. agalactiae clonal lineages undergo phase variation.
M. agalactiae type strain PG2 expressed a major 39-kDa protein and several lower-molecular-weight minor proteins which were detected by MAbs 3B3 and 4G12 (Fig. 1B) and are recognized by antibodies elicited during a natural infection (Fig. 1B). In an earlier study (5), the epitopes detected by these MAbs were suspected of undergoing phase variation; to investigate this further, we obtained two clonal lineages, designated 55 and 60, consisting of MAb-negative or MAb-positive immunostaining colonies from M. agalactiae type strain PG2 (Fig. 1A). Western blot analysis of the 55 lineage (Fig. 1C, left panels) revealed that all clones with uniformly MAb-staining colonies expressed 39- and 21-kDa proteins that carried both 3B3 and 4G12 epitopes (Fig. 1C, lanes 1 and 3). MAb-negative clones of the 55 lineage were essentially negative in Western blot analysis (Fig. 1C, lanes 2, 4, and 5), showing only a faint trace of the 39-kDa product which was further shown by colony blot immunostaining to represent a small percentage of cells that had turned expression of the 39-kDa protein back to the ON phenotype. The frequency of this phenotypic back switching for clone 55-7 was calculated to be between 10−3 to 10−2 per cell per generation and was even higher for some other clones. The ability to isolate a MAb-positive clone (55-7-1, expressing the 39- and 21-kDa proteins) from a negative clone (55-7) demonstrated that these proteins were undergoing phase variation, and they were designated Vpmas.
Interestingly, although Western blot analysis of clones of the 60 lineage resulted in MAb 4G12-staining profiles that were similar to those for clones of the 55 lineage, the 3B3-staining profiles were more complex and revealed the presence of additional minor Vpmas of 27 kDa (doublet) and 14 kDa (Fig. 1D, lanes 1, 2, 3, 5, 6, and 8). Interestingly, clone 60-3, which was both 4G12 and 3B3 negative after colony immunostaining, displayed in Western blot analysis a 3B3 immunoprofile identical to those of clones 60-3-3 and 60-3-3-18, which were also 4G12 negative but 3B3 positive after colony immunostaining (Fig. 1A and D). Whether this represents evidence of a masking phenomenon in clone 60-3, as observed for several surface proteins in M. fermentans (47), was not further investigated.
The reactivity of the Vpmas in colony blotting with 4G12 and 3B3 suggested their association with the cell membrane. This was confirmed by exclusive partitioning of the 39-kDa and all minor proteins (27, 21, and 14 kDa) into the Triton X-114 detergent phase (Fig. 1C and D, right panels; Fig. 2A) and by protease treatment of intact cells using either trypsin or carboxypeptidase Y. For both enzymes, the amount of Vpma39 and Vpma21 was reduced and the amount of degradation products increased in 55-5 as the concentration of protease increased. Carboxypeptidase Y resulted in the reduction of Vpma39 and the appearance of two smaller degradation products of approximately 20 kDa (both 3B3 and 4G12 positive) and 14 kDa (only 4G12 positive), indicating that the 4G12 epitope was localized closer to the membrane (or amino terminus) relative to 3B3. With respect to 55-5, Vpma39 was observed to be the most abundant membrane protein, while the 21-kDa protein was present in only a relatively minor amount (Fig. 2A) which correlated with the intensity of immunostaining (Fig. 1C).
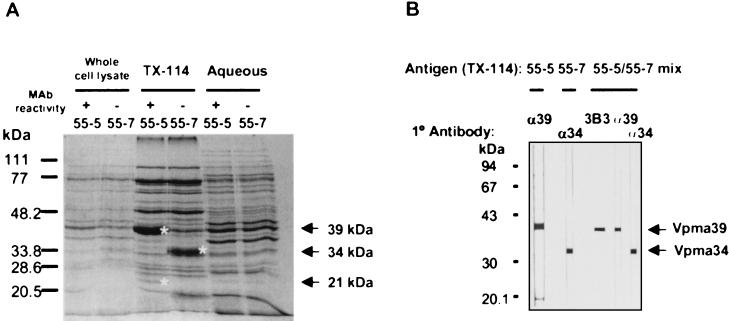
FIG. 2.
A Coomassie blue-stained reducing SDS-polyacrylamide gel of whole-cell lysate, Triton X-114 partitioning material (TX-114), and Triton X-114 aqueous-phase material from two clones of the 55 lineage, 55-5 and 55-7, as indicated. MAb reactivity refers to whether the clones do (+) or do not (−) bind 4G12 and 3B3. Differences between clones are indicated by asterisks at the right of the protein bands. (B) Western blot analysis on strips containing Triton X-114-partitioned antigen from clones indicated at the top. The primary antibody used is indicated above each strip (α39, rabbit anti-Vpma39 serum; α34, rabbit anti-Vpma34 serum).
Attempts to obtain clones expressing a single product failed. We do not yet know whether these variable minor proteins are encoded by individual genes or whether they simply represent proteolytic cleavage products of the higher-molecular-mass 39-kDa protein(s). Due to the complex Vpma phenotype displayed by clones of the 60 lineage, we concentrated on analysis of the 55 lineage.
Characterization of a new member of the Vpma family, Vpma34.
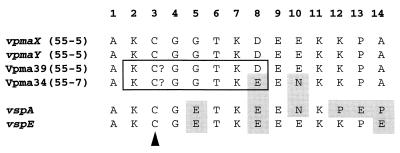
Comparison of Coomassie blue- and/or silver-stained SDS-polyacrylamide gels of Triton X-114-partitioning proteins from clones expressing Vpma39 (55-5 and 55-7-1) with those that did not (55-7, 55-7-5, and 55-7-7) revealed the presence of a new abundant 34-kDa protein in the Vpma39-negative clones (shown in Fig. 2A for 55-5 and 55-7). In contrast to Vpma39 (and Vpma21), this 34-kDa protein did not react with either 3B3 or 4G12. Alignment of the amino-terminal sequences of the 34-kDa protein from 55-7 and Vpma39 from 55-5 (Fig. 3) showed that they were identical in their first 7 amino acids (aa) and overall, having only two differences out of a total of 14 aa (86% identity). A phenylthiohydantoin amino acid peak for both proteins was absent in cycle 3 and probably corresponded to a cysteine residue. Most importantly, this observation indicated that these proteins were related, and hence the 34-kDa protein was formally redesignated Vpma34. Specific rabbit antisera raised to Vpma39 (from 55-5) and Vpma34 (from 55-7) were shown in Western blot analysis to be specific for the Vpma protein to which each was raised (Fig. 2B), indicating that Vpma39 and Vpma34 are immunologically distinct. Immunostaining of all clones (60-3-3 not included) with anti-Vpma34 confirmed abundant expression of Vpma34 in all MAb-negative clones except 60-3 and 60-3-3-18 (data not shown), indicating that there was not always coordinate expression between Vpma39 and Vpma34.
FIG. 3.
Alignment of all determined Vpma amino-terminal sequences either by amino-terminal sequencing (Vpma39 and Vpma34) or translated from the DNA sequence (vpmaX and vpmaY) and from two homologous vsp genes from M. bovis. GenBank accession numbers are L81118 for VspA and AF162139 for VspE (23, 24). Amino acids which differ from the top sequence (vpmaX) are shaded. The sequences boxed were used to design the vpma gene-specific oligonucleotide 3AF. The arrowhead indicates the proposed cysteine residues involved in lipid attachment.
Abundance of Vpmas in the culture medium.
Close examination of colonies immunostained with 3B3 revealed the presence of a halo surrounding colonies expressing the 3B3 target epitope. This prompted us to examine whether the Vpma components were released into the culture medium. Vpma39 and Vpma34 were found to be present in the growth medium after 2, 4, and 7 days growth at 37°C. In Table 1, the estimated amounts of Vpma proteins in the culture supernatants are compared with the abundance present in cells for the same volume of culture medium after 2 and 4 days of growth. Supernatant Vpma abundances were not calculated at 7 days of growth, as viable cell counts had fallen to below 106/ml. Vpma levels in the supernatants were high after 2 days of growth (56 to 59% of the amount in cells for the same original culture volume); although still high after 4 days growth, levels of full-length Vpma were reduced (38 to 36%). Degraded Vpma intermediates were detected only for Vpma34. Only approximately 10% of full-length Vpma39 and Vpma34 in culture supernatants after 2 days of growth were able to partition into the Triton X-114 detergent phase.
TABLE 1.
Relative abundances of Vpma39 and Vpma34 in mycoplasma culture supernatants in various cell fractions
| Culture | Day | CFU/ml (109) | Relative abundance of Vpmas (% of cell fraction) |
|---|---|---|---|
| 55-5 | 2 | 2.6 | 56a |
| 4 | 2.1 | 38a | |
| 55-7 | 2 | 2.3 | 59b |
| 4 | 2.6 | 36b | |
| 2 | 2.3 | 298c | |
| 4 | 2.6 | 421c |
Anti-Vpma39 serum was used to detect and calculate values for full-length Vpma39.
Anti-Vpma34 serum was used to detect and calculate values for full-length Vpma34.
Anti-Vpma34 serum was used to detect and calculate values for degraded products of Vpma34.
DNA rearrangement as a basis for Vpma phase variation.
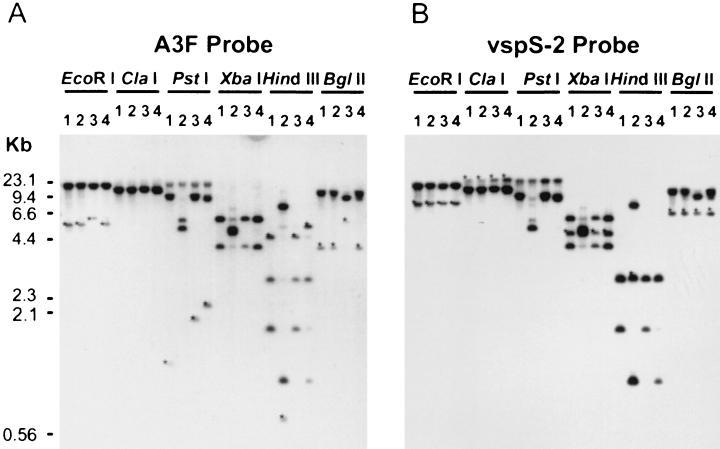
To estimate the number of vpma genes present within the M. agalactiae genome, a vpma-specific oligonucleotide (A3F) was designed based on the common amino-terminal sequence between Vpma39 and Vpma34 (Fig. 3) and used as a probe in Southern blot analysis. Results obtained with restricted genomic DNA from clonal variants 55-5 (MAb positive), 55-7 (MAb negative), and two other clones from the 60 lineage, 60-3 (MAb negative) and 60-4 (MAb positive), are illustrated in Fig. 4A. Assuming that each A3F amino-terminal sequence represents a single, distinct vpma gene, the hybridization of the A3F probe with four HindIII DNA fragments in clone 60-4 (Fig. 4A, lane 4) indicated that there could be at least four vpma genes clustered within an approximate 14-kb ClaI fragment (Fig. 4A, lane 4). Each clone also exhibited a unique hybridization pattern when several restriction enzyme digests were compared (Fig. 4A).
FIG. 4.
Southern blot analysis of genomic DNA from clones 55-5 (lanes 1), 55-7 (lanes 2), 60-3 (lanes 3), and 60-4 (lanes 4). The membrane was first hybridized with DIG-labeled A3F oligonucleotide (A) and then stripped and hybridized with DIG-labeled vspS-2 oligonucleotide based on the leader sequence of vspA from M. bovis type strain PG45 (B). Restriction digests of the genomic DNA are indicated above the lane sets. Asterisks show DNA fragments that are detected with only one probe.
Molecular characterization of two vpma genes, vpmaX and vpmaY.
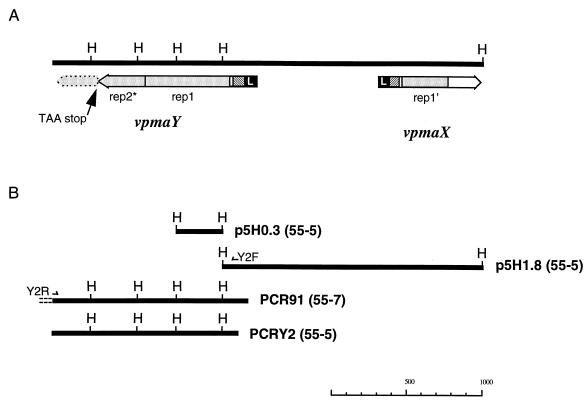
To better define the nature of the vpma genes, a HindIII chromosomal DNA library from 55-5 was screened with the A3F oligonucleotide probe. However, because of the low signal-to-background ratio obtained with this probe, clones of interest could not be definitively identified; the screening was then performed with two PCR fragments containing vpma-specific coding regions, PCR75 and PCR91, as probes. These were obtained using the 3AF primer to generate PCR products from 55-7 DNA on the basis that vpma genes occurred in a convergent orientation by analogy with the well-characterized system of vsa genes in M. pulmonis (6). In Southern blot analysis, the PCR75 probe hybridized with the 4.7- and 2.9-kb A3F-specific HindIII fragments from 55-5, while the PCR91 probe hybridized to the 2.9- and 1.8-kb HindIII fragments, with only weak cross-hybridization with the 4.7-kb A3F-specific HindIII fragments from 55-5. The PCR91 probe additionally recognized a 0.3-kb HindIII fragment which was not detected by 3AF (Fig. 4A, lane 1). Using a mix of PCR75 and PCR91 probes to screen the 55-5 HindIII DNA library, recombinant clones containing either the 0.3-kb (p5H0.3) or the 1.8-kb (p5H1.8) HindIII fragment were obtained. DNA sequence from the 1.8-kb HindIII fragment revealed one complete and one partial ORF in a divergent orientation which represented vpma genes; these were designated vpmaX and vpmaY, respectively (Fig. 5A). The remainder of the vpmaY ORF was obtained from the sequence at one end of PCR91 and from PCRY2, whose sequence was identical to that of PCR91 (Fig. 5B).
FIG. 5.
(A) Schematic HindIII (H) genomic DNA map showing the organization and orientation of the ORFs of vpmaX and vpmaY. The ORFs begin with a homologous leader sequence (L; shaded black) followed by a short nonrepeated homologous region (diagonal pattern). Unique sequences between genes are not shaded. Stippled shading represent homologous repeated sequences within the ORFs. The vpmaY rep1 is a complete repeat of 186 aa, rep2* is a partial repeat which is prematurely terminated by a TAA stop codon (indicated by an arrow), while the remainder of the repeat is present but in a different reading frame and indicated by a broken-line boundary. The vpmaX rep1′ is also partial, consisting of 102 aa. (B) Fragments used to deduce the map shown in panel A. The 0.3-kb HindIII fragment (p5H0.3), the 1.8-kb HindIII fragment (p5H1.8), and the PCR fragment PCRY2 were all from clone 55-5; PCR fragment PCR91 was from clone 55-7.
The ORF of vpmaY was 346 aa in length, beginning with a 25-aa leader sequence that ended in an acylation/peptidase II cleavage motif of AAKC (Fig. 6). Assuming that the VpmaY protein would be processed at its mature amino terminus as for Vpma39 and Vpma34 (cleaved between the two alanine residues of the motif), then the calculated molecular mass of the mature protein would be 35 kDa (324 aa), with a theoretical pI of 8.27. Mature VpmaY contained a high percentage of charged amino acids (34%) which were almost balanced to give an overall net charge of +2. Following the leader sequence of vpmaY there are 31 aa containing four proline residues; the remainder of the ORF comprises of one complete repeat of 186 aa followed immediately by a second partial repeat comprising 104 aa of the amino-terminal part of the first repeat. Although the sequence is present to encode a complete second repeat of 186 aa, an insertion of two nucleotides (TA), located after the 104th codon of the second repeat, immediately introduces a TAA stop codon to truncate the second repeat by 44%.
The ORF of vpmaX is 221 aa and also contained a 25-aa leader sequence which was highly homologous to that of vpmaY, having only 3 amino acid and 11 nucleotide differences and ending with an identical acylation/peptidase II cleavage motif (Fig. 6). The mature polypeptide was predicted to be 22.4 kDa (199 aa) and contained an even higher proportion of charged amino acids (42%), with a net charge of −1, and a lower pI of 6.33 compared with VpmaY. Although vpmaX had no repetitive sequences, it did possess a truncated version of the vpmaY repeat, sharing 87% aa identity (100% similarity) with 102 aa of the amino-terminal end of the repeat. The region between this partial repeat and the cysteine of the leader (27 aa) was homologous to the equivalent region in vpmaY (74% amino acid identity), particularly at the amino-proximal end, with the first 12 aa being identical (Fig. 3 and 6). For the remainder of the vpmaX ORF, following the partial repeat, there is little homology to vpmaY and no evidence for continuation of the repeat. Overall, the ORFs of vpmaX and vpmaY shared 74% identity at the DNA level and 63% identity (79% similarity) at the amino acid level.
In Southern blot analysis (Fig. 4A), the A3F probe bound to three HindIII fragments from 55-5 DNA and four HindIII fragments from 60-4 DNA. Since the 1.8-kb HindIII fragment from 55-5 contained two vpma genes, it is feasible that other, larger probe-positive fragments in this and other clones also contain more than one vpma gene; thus, the total number of genes is most probably more than four.
Relationship of the Vpmas to the Vsps of M. bovis.
Comparing the first 14 aa of Vpma39 and Vpma34 to known mycoplasma protein sequences, Vpma34 was found to have good identity to VspA of M. bovis, 10 out of 14 aa (Fig. 3), while Vpma39 had 8 out of 14 aa matches to VspA. Later, when more vsp genes had been published (24), database searches showed that Vpma39 had an even greater match to VspE (11 out of 14 aa [Fig. 3]). An oligonucleotide (vspS-2) designed from the leader sequence of vspA bound essentially to the same restriction fragments in clones 55-5, 55-7, 60-3, and 60-4 as detected by the vpma common probe, A3F (Fig. 4B). However, the 4.7-kb HindIII fragment from 55-5, which hybridized with the 3AF probe, did not hybridize with vspS-2, possibly because the vpma genes residing on the 4.7-kb HindIII fragment have more mismatches to the vspS-2 oligonucleotide. The 25-aa lipoprotein leader sequences (including the cysteine residue) deduced from both vpmaY and vpmaX were identical to that of vspA, having only one and two amino acid differences, respectively (Fig. 6). There were 6 out of 11 amino acid identities following the cysteine residue of their leader sequences (Fig. 6) but no significant homology to vspA or any other vsp gene within the remainder of the coding sequence. The 5′ untranslated regions of both vpmaX and vpmaY also possessed high homology to the equivalent region of vspA (Fig. 6) and to other published vsp gene sequences for this region, vspB, vspE, vspF, and vspH (3, 24), where the identity ranged between 86 and 94% over 78 bp upstream from the start codon, with the highest being 94% identity between vpmaY and vspA and 94% identity between vpmaX and vspB.
DISCUSSION
Data collected in this study have revealed the presence in M. agalactiae of a multigene family encoding variable abundant surface proteins (Vpmas) that vary in expression. Analysis of this system, at the protein and molecular levels, predicts that the vpma gene family consists of more than four members that have several features in common with multigene families in other Mycoplasma species (6, 10, 24, 26, 27, 53). Increasing evidence for the presence of these large gene families in pathogenic mycoplasmas, which are considered to have a minimal-sized genome, highlights the importance of these gene products as they have been retained (or acquired) during the regressive evolution of the genomes of these bacteria.
Based on their amino-terminal sequences, both vpmaX and vpmaY could be likely candidates to encode Vpma39 in clone 55-5 (Fig. 3), and it may be possible that other vpma genes also possess this sequence. The amino-terminal sequence of Vpma34 from 55-7 is different from that of VpmaX and VpmaY and must therefore represent the gene product of a yet uncharacterized vpma gene. To definitively assign the genes for Vpma39 and Vpma34, it will be necessary to obtain the sequences for all vpma genes and to design gene-specific probes for use in Northern analysis with total RNA isolated from clones 55-5 and 55-7.
The characterization of two Vpma proteins, Vpma39 and Vpma34, and two vpma genes, vpmaX and vpmaY, has revealed that the genes are related to the vsp genes of M. bovis (24). Many of the features shared by vpmaX and vpmaY were also found in all known vsp genes of M. bovis (24). These include the high identity within the 5′ untranslated regions between vpmaX, vpmaY, and four vsp genes for which this region has been published (3, 23, 24), the leader sequences, and a short region of approximately 10 amino-terminal aa for all genes; however, there is little homology within the remainder of vpma and vsp coding sequences besides a high proportion of charged amino acids. All vpma and vsp genes also exhibit the same lipoprotein motif (AAKC), which is consistent with the observation that this motif is conserved within members of the same family in mycoplasmas (AISC for Vlps except TISC for VlpF [10], AASC for pMGAs [26, 27], LIAC for Vsas [6], and AAKC for Vsps [24], where C is mandatory for lipid attachment). Due to the close phylogenetic relationship between M. agalactiae and M. bovis (32, 35), we suggest that the vpma and vsp multigene families were acquired and evolved from a common ancestor.
The acylation/peptidase II cleavage motif of vpma genes (AAKC), like that of vsp genes, is atypical, as documented prokaryotic lipoprotein signal sequences do not contain a charged residue (lysine) at the amino-terminal side of the cysteine residue (11, 45, 50). We have chemically verified for Vpma39 and Vpma34 that Vpmas also have an unusual proteolytic processing of their leader sequences and are cleaved between the two alanine (A) residues (i.e., A↓AKC) instead of the lysine (K) and cysteine (C) residues (i.e., AAK↓C) as predicted from all other known prokaryote lipoproteins. If indeed the Vpmas, like the Vsps in M. bovis, are covalently attached to a membrane lipid moiety via their amino-terminal cysteine residue, then it represents evidence of an alternative lipoprotein signal processing in prokaryotes. Interestingly, another lipoprotein from M. agalactiae, P48, has recently been characterized (38) whose gene has homology to the M. fermentans malp gene, encoding the MALP-404 lipoprotein and MALP-2 lipopeptide, which possesses a potent macrophage-stimulatory activity (8, 30). Like the Vpmas, the P48 lipoprotein is cleaved two amino acids amino terminal to the cysteine to give a mature protein of ASC…even though it possesses the typical lipoprotein motif AASC. In contrast, the same lipoprotein motif in all members of the pMGA family of M. gallisepticum is cleaved as a typical lipoprotein between the serine and cysteine residues (20, 25). These data argue against the hypothesis that a specific atypical lipoprotein motif sequence dictates altered cleavage specificity; rather, they suggest that the peptidase II-like enzyme of M. agalactiae possesses an altered cleavage specificity. Whether this is shared by lipoproteins from other phylogenetically related species, such as M. bovis, is not known but could define either a new species or phylogenetic cluster trait.
Both vpma and vsp genes contain repeated sequences; however, the 558-bp (186-aa) repetitive element that is repeated 1.5 times in the vpmaY gene is more than twice the size of the largest vsp coding repeat (vspN, 261 bp, 87 aa). It is more common for vsp genes to contain multiple tandemly repeated sequences of between 18 and 36 bp (6 to 12 aa) (24), whose in-frame insertions or deletions have been responsible for the observed size variations in VspA and VspB (2, 23). Repeated efforts to isolate size variants of Vpma39 or Vpma34, or other Vpmas bearing 4G12 and/or 3B3 epitopes, were unsuccessful, suggesting that the corresponding vpma genes do not contain small (≥8-aa) coding repeats. Another difference in this respect is that vsp genes do not contain partial repeats, as is the case for vpmaX and vpmaY, both of which possess homologous and almost identically sized truncated repeats 0.5 unit in length.
The structural differences and low homology within the bulk of the coding sequences between the two vpma genes identified in this study and the 13 vsp genes from M. bovis raises the question of their function. Recently the Vsps have been shown to be involved in host adhesion (41), and small synthetic peptides of 3 to 7 aa have precisely identified the epitopes involved in cytadhesion to embryonic bovine lung cells (42). These adherence epitopes were also found to be encoded by vpmaX (one GTK and one KEK epitope) and vpmaY (one GTK, one NDL, and two KEK epitopes). As mentioned above, M. agalactiae and M. bovis species are closely related, inducing very similar clinical symptoms in distinct hosts (4, 33). It is possible that this host adherence function of Vsps is also possessed by the Vpmas of M. agalactiae and that the two related families, Vpmas and Vsps, have similar functions during the disease process. The fact that structural differences exist between vpma and vsp genes and that there is no significant homology between the bulk of their coding sequences may play a role in their different host specificities. Also, VpmaX and VpmaY possess an unusually high proportion of charged amino acids, a trait shared by the Vsps of M. bovis (24), the Vaa lipoprotein of M. hominis, involved in host binding (54), and the Vsa lipoproteins of M. pulmonis, implicated in hemadsorption (51). Whether Vpmas of M. agalactiae participate in host adhesion and whether the attribute of possessing a high proportion of charged amino acids contributes to this adherence has yet to be elucidated and may be only one facet of a more multifunctional role for both Vpmas and Vsps.
A recent study has reported the polymorphism among M. agalactiae strains of DNA fragments hybridizing with a specific probe to the conserved 5′ region between vsp genes and to the entire vspA gene of M. bovis (14). This may reflect DNA rearrangements that have occurred within the vpma locus of these strains or, alternatively, that they possess different repertoires of vpma genes. The former interpretation is based on our results showing that the vpma genes undergo high-frequency DNA rearrangements within isogenic lineages derived from a single strain propagated in vitro. These mutational events occur spontaneously within the vpma locus and were shown to correlate with ON and OFF switching in Vpma expression. The exact genetic mechanism underlying this phenomenon remains to be fully assessed but may be similar to what has been shown for the vsa genes of M. pulmonis (6) and has been speculated for control of vsp gene expression in M. bovis (24), where rearrangements align the ORF of a silent gene with an active promoter. The vpma DNA rearrangements occur at such high frequencies that they most probably involve an active mechanism such as a site-specific recombinase. This high frequency also exemplifies one type of mutator mechanism for “contingency loci” proposed by Moxon et al. (28, 29), which have been found in bacteria to allow a large repertoire of phenotypic variation within a population while minimizing deleterious effects on fitness.
The relative abundance of Vpma39 and Vpma34 in the cell-free culture supernatant relative to that in the cells for the same volume was remarkably high (up to 59%). It is interesting to speculate as to whether this phenomenon would also occur in the host and what pathological function this would play. A similar phenomenon has been observed for two membrane proteins of Actinobacillus pleuropneumoniae, a transferrin-binding protein and an outer membrane lipoprotein, which were both present in membrane blebs (15, 16). When used as recombinant immunogens, these proteins were found to protect animals from a homologous challenge (15, 40). These extracellular bleb structures have also been observed in M. penetrans (17, 31); however, it is unlikely that the majority of Vpmas in culture supernatants exist within blebs, as only 10% partition into the Triton X-114 detergent phase. We observed that injection of the 55-5 culture supernatant into the teat canal of lactating ewes induced a transient hypogalactia whereas control media did not alter milk production (data unpublished). Whether a cell-free source of Vpmas specifically induces this phenomenon is currently under investigation.
Although switching ON and OFF members of a gene family encoding surface proteins in mycoplasma would aid the organism in avoiding the host immune response, it is still unknown whether it functions in additional ways to optimize pathogen-host interactions. Gene-specific and protein-specific Vpma reagents will hopefully provide essential tools for the investigation of their role in disease pathogenesis in vivo. These reagents will also provide invaluable diagnostic and epidemiological tools as have equivalent reagents specific for Vsps of M. bovis in recent studies (3, 7, 34).
ACKNOWLEDGMENTS
This work was supported in part by grant P12545-GEN from the Fonds zur Förderung der wissenschaftlichen Forschung (to C. Citti and R. Rosengarten).
We thank Karin Siebert, Thomas Beier, and Joachim Spergser for their assistance and Marc Marenda for critical reading of the manuscript.
REFERENCES
- 1.Aluotto B B, Wittler R G, Williams C O, Faber J E. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int J Syst Bacteriol. 1970;20:35–58. [Google Scholar]
- 2.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier T, Hotzel H, Lysnyansky I, Grajetzki C, Heller M, Rabeling B, Yogev D, Sachse K. Intraspecies polymorphism of vsp genes and expression profiles of variable surface protein antigens (Vsps) in field isolates of Mycoplasma bovis. Vet Microbiol. 1998;63:189–203. doi: 10.1016/s0378-1135(98)00238-7. [DOI] [PubMed] [Google Scholar]
- 4.Bergonier D, Berthelot X, Poumarat F. Contagious agalactia of small ruminants: current knowledge concerning epidemiology, diagnosis and control. Rev Sci Tech Off Int Epizoot. 1997;16:848–873. doi: 10.20506/rst.16.3.1062. [DOI] [PubMed] [Google Scholar]
- 5.Bergonier D, De Simone F, Russo P, Solsona M, Lambert M, Poumarat F. Variable expression and geographic distribution of Mycoplasma agalactiae surface epitopes demonstrated with monoclonal antibodies. FEMS Microbiol Lett. 1996;143:159–165. doi: 10.1111/j.1574-6968.1996.tb08475.x. [DOI] [PubMed] [Google Scholar]
- 6.Bhugra B, Voelker L L, Zou N, Yu H, Dybvig K. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 7.Brank M, Le Grand D, Poumarat F, Bezille P, Rosengarten R, Citti C. Development of a recombinant antigen for antibody-based diagnosis of Mycoplasma bovis infection in cattle. Clin Diagn Lab Immunol. 1999;6:861–867. doi: 10.1128/cdli.6.6.861-867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calcutt M J, Kim M F, Karpas A B, Mühlradt P F, Wise K S. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans. Infect Immun. 1999;67:760–771. doi: 10.1128/iai.67.2.760-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citti C, Rosengarten R. Mycoplasma genetic variation and its implication for pathogenesis. Wien Klin Wochenschr. 1997;109:562–568. [PubMed] [Google Scholar]
- 10.Citti C, Watson-McKown R, Droesse M, Wise K S. Gene families encoding phase- and size-variable surface lipoproteins of Mycoplasma hyorhinis. J Bacteriol. 2000;182:1356–1363. doi: 10.1128/jb.182.5.1356-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleavinger C M, Kim M F, Im J H, Wise K S. Identification of mycoplasma membrane proteins by systematic TnphoA mutagenesis of a recombinant library. Mol Microbiol. 1995;18:283–293. doi: 10.1111/j.1365-2958.1995.mmi_18020283.x. [DOI] [PubMed] [Google Scholar]
- 12.Cottew G S. Caprine-ovine mycoplasmas. In: Barile M F, Razin S, Tully J G, Whitcomb R F, editors. The mycoplasmas. New York, N.Y: Academic Press; 1979. pp. 103–132. [Google Scholar]
- 13.Deitsch K W, Moxon E R, Wellems T E. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol Mol Biol Rev. 1997;61:281–293. doi: 10.1128/mmbr.61.3.281-293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flitman-Tene R, Levisohn S, Rosenbusch R, Rapoport E, Yogev D. Genetic variation among Mycoplasma agalactiae isolates detected by the variant surface lipoprotein gene (vspA) of Mycoplasma bovis. FEMS Microbiol Lett. 1997;156:123–128. doi: 10.1111/j.1574-6968.1997.tb12716.x. [DOI] [PubMed] [Google Scholar]
- 15.Gerlach G F, Anderson C, Klashinsky S, Rossi-Campos A, Potter A A, Willson P J. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect Immun. 1993;61:565–572. doi: 10.1128/iai.61.2.565-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlach G F, Anderson C, Potter A A, Klashinsky S, Willson P J. Cloning and expression of a transferrin-binding protein from Actinobacillus pleuropneumoniae. Infect Immun. 1992;60:892–898. doi: 10.1128/iai.60.3.892-898.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giron J A, Lange M, Baseman J B. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect Immun. 1996;64:197–208. doi: 10.1128/iai.64.1.197-208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glew M D. Gene expression studies of pMGA, a surface protein of Mycoplasma gallisepticum. Ph.D. thesis. Melbourne, Victoria, Australia: University of Melbourne; 1997. [Google Scholar]
- 19.Glew M D, Baseggio N, Markham P F, Browning G F, Walker I D. Expression of the pMGA genes of Mycoplasma gallisepticum is controlled by variation in the GAA trinucleotide repeat lengths within the 5′ noncoding regions. Infect Immun. 1998;66:5833–5841. doi: 10.1128/iai.66.12.5833-5841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glew M D, Markham P F, Browning G F, Walker I D. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology. 1995;141:3005–3014. doi: 10.1099/13500872-141-11-3005. [DOI] [PubMed] [Google Scholar]
- 21.Henderson I R, Owen P, Nataro J P. Molecular switches—the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 22.Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lysnyansky I, Sachse K, Rosenbusch R, Levisohn S, Yogev D. The vsp locus of Mycoplasma bovis: gene organization and structural features. J Bacteriol. 1999;181:5734–5741. doi: 10.1128/jb.181.18.5734-5741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markham P F, Glew M D, Brandon M R, Walker I D, Whithear K G. Characterization of a major hemagglutinin protein from Mycoplasma gallisepticum. Infect Immun. 1992;60:3885–3891. doi: 10.1128/iai.60.9.3885-3891.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markham P F, Glew M D, Browning G F, Whithear K G, Walker I D. Expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect Immun. 1998;66:2845–2853. doi: 10.1128/iai.66.6.2845-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markham P F, Glew M D, Sykes J E, Bowden T R, Pollocks T D, Browning G F, Whithear K G, Walker I D. The organisation of the multigene family which encodes the major cell surface protein, pMGA, of Mycoplasma gallisepticum. FEBS Lett. 1994;352:347–352. doi: 10.1016/0014-5793(94)00991-0. [DOI] [PubMed] [Google Scholar]
- 28.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 29.Moxon E R, Thaler D S. Microbial genetics. The tinkerer's evolving tool-box. Nature. 1997;387:659–662. doi: 10.1038/42607. [DOI] [PubMed] [Google Scholar]
- 30.Mühlradt P F, Kiess M, Meyer H, Sussmuth R, Jung G. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med. 1997;185:1951–1958. doi: 10.1084/jem.185.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neyrolles O, Brenner C, Prevost M C, Fontaine T, Montagnier L, Blanchard A. Identification of two glycosylated components of Mycoplasma penetrans: a surface-exposed capsular polysaccharide and a glycolipid fraction. Microbiology. 1998;144:1247–1255. doi: 10.1099/00221287-144-5-1247. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson B, Uhlén M, Johansson K-E. Phylogeny of some mycoplasmas from ruminants based on 16S rRNA sequences and definition of a new cluster within the hominis group. Int J Syst Bacteriol. 1996;46:1093–1098. doi: 10.1099/00207713-46-4-1093. [DOI] [PubMed] [Google Scholar]
- 33.Pfützner H, Sachse K. Mycoplasma bovis as an agent of mastitis, pneumonia, arthritis and genital disorders in cattle. Rev Sci Tech Off Int Epizoot. 1996;15:1477–1494. doi: 10.20506/rst.15.4.987. [DOI] [PubMed] [Google Scholar]
- 34.Poumarat F, Le Grand D, Solsona M, Rosengarten R, Citti C. Vsp antigens and vsp-related DNA sequences in field isolates of Mycoplasma bovis. FEMS Microbiol Lett. 1999;173:103–110. doi: 10.1111/j.1574-6968.1999.tb13490.x. [DOI] [PubMed] [Google Scholar]
- 35.Razin S, Freundt E A. The mycoplasmas. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: Williams & Wilkins; 1984. pp. 740–770. [Google Scholar]
- 36.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson B D, Meyer T F. Genetic variation in pathogenic bacteria. Trends Genet. 1992;8:422–427. doi: 10.1016/0168-9525(92)90325-x. [DOI] [PubMed] [Google Scholar]
- 38.Rosati S, Pozzi S, Robino P, Montinaro B, Conti A, Fadda M, Pittau M. P48 major surface antigen of Mycoplasma agalactiae is homologous to a malp product of Mycoplasma fermentans and belongs to a selected family of bacterial lipoproteins. Infect Immun. 1999;67:6213–6216. doi: 10.1128/iai.67.11.6213-6216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosengarten R, Wise K S. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J Bacteriol. 1991;173:4782–4793. doi: 10.1128/jb.173.15.4782-4793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi-Campos A, Anderson C, Gerlach G F, Klashinsky S, Potter A A, Willson P J. Immunization of pigs against Actinobacillus pleuropneumoniae with two recombinant protein preparations. Vaccine. 1992;10:512–518. doi: 10.1016/0264-410x(92)90349-o. [DOI] [PubMed] [Google Scholar]
- 41.Sachse K, Grajetzki C, Rosengarten R, Hänel I, Heller M, Pfützner H. Mechanisms and factors involved in Mycoplasma bovis adhesion to host cells. Zentbl Bakteriol. 1996;284:80–92. doi: 10.1016/s0934-8840(96)80157-5. [DOI] [PubMed] [Google Scholar]
- 42.Sachse K, Helbig J H, Lysnyansky I, Grajetzki C, Müller W, Jacobs E, Yogev D. Epitope mapping of immunogenic and adhesive structures in repetitive domains of Mycoplasma bovis variable surface lipoproteins. Infect Immun. 2000;68:680–687. doi: 10.1128/iai.68.2.680-687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Solsona M, Lambert M, Poumarat F. Genomic, protein homogeneity and antigenic variability of Mycoplasma agalactiae. Vet Microbiol. 1996;50:45–58. doi: 10.1016/0378-1135(95)00200-6. [DOI] [PubMed] [Google Scholar]
- 45.Sutcliffe I C, Russell R R. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theiss P, Wise K S. Localized frameshift mutation generates selective, high-frequency phase variation of a surface lipoprotein encoded by a mycoplasma ABC transporter operon. J Bacteriol. 1997;179:4013–4022. doi: 10.1128/jb.179.12.4013-4022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theiss P M, Kim M F, Wise K S. Differential protein expression and surface presentation generate high-frequency antigenic variation in Mycoplasma fermentans. Infect Immun. 1993;61:5123–5128. doi: 10.1128/iai.61.12.5123-5128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tola S, Idini G, Manunta D, Casciano I, Rocchigiani A M, Angioi A, Leori G. Comparison of Mycoplasma agalactiae isolates by pulsed field gel electrophoresis, SDS-PAGE and immunoblotting. FEMS Microbiol Lett. 1996;143:259–265. doi: 10.1111/j.1574-6968.1996.tb08490.x. [DOI] [PubMed] [Google Scholar]
- 50.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 51.Watson H L, Zheng X, Cassell G H. Structural variations and phenotypic switching of mycoplasmal antigens. Clin Infect Dis. 1993;17(Suppl.):S183–S186. doi: 10.1093/clinids/17.supplement_1.s183. [DOI] [PubMed] [Google Scholar]
- 52.Wise K S. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1993;1:59–63. doi: 10.1016/0966-842X(93)90034-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yogev D, Rosengarten R, Watson-McKown R, Wise K S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, Wise K S. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64:2737–2744. doi: 10.1128/iai.64.7.2737-2744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q, Wise K S. Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma. Mol Microbiol. 1997;25:859–869. doi: 10.1111/j.1365-2958.1997.mmi509.x. [DOI] [PubMed] [Google Scholar]
- 56.Zheng X, Teng L J, Watson H L, Glass J I, Blanchard A, Cassell G H. Small repeating units within the Ureaplasma urealyticum MB antigen gene encode serovar specificity and are associated with antigen size variation. Infect Immun. 1995;63:891–898. doi: 10.1128/iai.63.3.891-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]