Abstract
mRNA vaccines have been established as a safe and effective modality, thanks in large part to the expedited development and approval of COVID-19 vaccines. In addition to the active, full-length mRNA transcript, mRNA fragment species can be present as a byproduct of the cell-free transcription manufacturing process or due to mRNA hydrolysis. In the current study, mRNA fragment species from BNT162b2 mRNA were isolated and characterized. The translational viability of intact and fragmented mRNA species was further explored using orthogonal expression systems to understand the risk of truncated spike protein or off-target antigen translation. The study demonstrates that mRNA fragments are primarily derived from premature transcriptional termination during manufacturing, and only full-length mRNA transcripts are viable for expression of the SARS-CoV-2 spike protein antigen.
Keywords: mRNA, Vaccine, COVID-19, HPLC, Comirnaty
Introduction
The COVID-19 pandemic triggered a need for swift development and production of safe and effective vaccines and therapies targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The potential for rapid construct design and manufacturing positioned mRNA-based vaccines as the leading vaccine platform.1 The potential of mRNA vaccines was soon realized with the monumental achievement of delivering a COVID-19 vaccine in just nine months.2 mRNA-based COVID-19 vaccines from Pfizer/BioNTech (BNT162b2; Comirnaty) and Moderna (mRNA-1273; Spikevax) proved to be safe and > 90% efficacious in preventing COVID-19 infection,3 and BNT162b2 became the first mRNA vaccine to receive emergency use authorization and full FDA approval.4 In the first year of vaccination, it is estimated that up to 20 million deaths were averted globally.5 One study estimated that COVID-19 vaccination prevented approximately 27 million infections, 1.6 million hospitalizations, and 237,000 deaths in adults in the US in the first ten months of vaccine availability.6
Development of mRNA for use in vaccines and other therapeutics progressed for decades prior to the COVID-19 pandemic.7, 8, 9, 10 Key developments enabling the success of mRNA vaccines include the finding that modified uridine incorporated into mRNA can help evade the innate immune response11 and the identification of optimized 5’-cap and poly(A) tail structures.12 , 13 The 5’-cap, 3’-poly-adenylation tract (poly(A) tail), and an integral mRNA transcript of the target antigen sequence are critical quality attributes for the mRNA component in the approved COVID-19 vaccines.14, 15, 16 These features ensure transcript stability and translational efficiency to produce the intended antigen protein. The mRNA component is co-formulated with four structural & functional lipids, which protect the mRNA from degradation and facilitate cellular entry.17 While the pandemic created an opportunity to implement this relatively new vaccine technology, the platform is now established to develop new vaccines and other therapies using mRNA technology.
The mRNA component in BNT162b2 encodes the SARS-CoV-2 spike protein, which is comprised of the S1 domain (containing the ACE2 Receptor Binding Domain, RBD) and the S2 domain.18 The mRNA drug substance is manufactured using a cell-free transcription process commonly called in vitro transcription (IVT), where linear template DNA encoding the SARS-CoV-2 spike protein is transcribed into mRNA using T7 RNA polymerase, nucleotide triphosphates, and other necessary cofactors.19 DNase I and proteinase K digest the DNA template and enzyme components of the IVT reaction, respectively, followed by a number of purification and filtration steps, resulting in a highly pure mRNA.14 , 15 A robust control strategy (including process controls, routine testing, non-routine characterization, and product and process understanding) ensures process- and product-related impurities are well-characterized and appropriately cleared in the manufacturing process to ensure product safety. mRNA fragment species are a product-related impurity that can be generated during the IVT process by abortive transcription or through hydrolytic degradation of the full-length mRNA transcript.20 While mRNA fragments are not considered a high risk to vaccine safety or efficacy, high levels of mRNA fragments would correspond to lower relative RNA integrity (intact RNA), which could impact efficacy at extreme levels. mRNA degradation is controlled in the BNT162b2 drug substance and drug product through stabilizing formulation components and suitable long-term storage conditions. mRNA fragments generated during the IVT process can be present in the drug substance; however, fragment levels are controlled by an appropriate RNA integrity acceptance criterion. Beyond the potential to impact efficacy at elevated levels (i.e., outside the established control limits), it is also important to consider the potential for mRNA fragments to translate truncated or off-target (e.g., frame-shifted) proteins to ensure product consistency.
In the current study, BNT162b2 fragment species were isolated and characterized, and their potential to express truncated spike proteins or other off-target proteins was evaluated. The data demonstrate that BNT162b2 fragments originate predominantly from premature transcriptional stops during manufacturing and do not have the capacity for protein translation, as evidenced by orthogonal protein expression systems. Thus, no undesired protein byproducts derived from these fragments can be expected for the BNT162b2 vaccine.
Materials & Methods
Materials
BNT162b2 mRNA (active substance) is synthesized from linear DNA via an in vitro transcription (IVT) process. The mRNA then undergoes subsequent purification and filtration prior to freezing for long-term storage. Transcripts lacking 5’-cap were generated by the same process, with the omission of the cap analog in the reaction. Full-length mRNA lacking the poly(A) tail was generated as a control sample using a modified linear DNA template that lacked the poly(A) encoding sequence.
Purification of Fragments and Intact RNA by Ion-Pair Reversed Phase High Performance Liquid Chromatography (IP-RP-HPLC)
Separation of mRNA was performed using ion pair reversed phase high performance liquid chromatography (IP-RP-HPLC) using an Alliance 2695 HPLC system (Waters) with a UV detector and a DNAPac RP column (DNAPac RP, 4 µm, 3.0 × 100 mm) (Thermo Fisher Scientific) held at 65 °C. The chromatography was performed at a flow rate of 0.25 mL/min. Mobile phase A consists of 0.1% triethylamine (TEA), 0.85% 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP), and 5% methanol. Mobile phase B consisted of 0.1% TEA, 0.85% HFIP, and 50% methanol and mobile phase C contained 50% methanol and 30% 2-propanol (IPA). Approximately 30 µg of a 0.5 mg/mL (diluted in DEPC water) mRNA was injected onto the column and UV absorbance at 260 nm was monitored.
Peaks 1 and 2 were purified individually by manual collection from multiple runs, partially dried by vacuum centrifugation, and pooled to generate sufficient material for downstream analyses. The mRNA concentration of each purified peak was determined using absorbance at 260 nm.
Capillary Electrophoresis
Sample separation was performed on a Fragment Analyzer (Agilent Technologies), an automated multiplexed capillary electrophoresis (CE) instrument equipped with high intensity light emitting diode (LED) excitation light source that is focused across the capillary array detection window and imaged onto a sensitive, two-dimensional charge coupled device (CCD) detector. Separation is achieved by applying an electric field through a narrow bore (50 μm i.d.) fused silica capillary array filled with a conductive, sieving gel matrix. RNA was analyzed using the RNA Analysis Kit (Agilent Technologies). The RNA separation gel was mixed with an intercalating dye at a v/v ratio of 10,000:1. RNA was mixed with the RNA diluent marker (15 nt), denatured at 70°C for 2 min, and cooled on ice prior to analysis. Denatured RNA samples were electrokinetically injected at 5 kV for 6 s, and electrophoresis was performed for 60 min at 8 kV. An RNA ladder was similarly analyzed as a calibrator for nucleotide size. By monitoring the relative fluorescence unit (RFU) intensity as a function of time during the CE separation, digital electropherograms representative of the RNA content of samples are collected. Data analysis was performed using ProSize (Agilent Technologies).
5’-Cap Analysis
The quantitation of 5’-Cap was accomplished by first treating the mRNA material with RNase H after it has been annealed to a probe designed to complement the 5’ end. The 5’ end related species were then isolated and separated by reversed-phase high-performance liquid chromatography (RP-HPLC) using an Agilent 1260 system with quaternary pumps and DAD detector. UV absorbance at 260 nm was monitored. An ACQUITY UPLC Oligonucleotide BEH C18 Column (130Å, 1.7 µm, 2.1 mm X 150 mm, 1K - 30K) (Waters) held at 75 °C was used.
Relative quantification of the 5’ end cap level (5’-Cap) was determined from relative peak areas. The percentage of species that are capped was determined by dividing the percent area for the capped species by the total area of all 5’ end related species.
Poly(A) Tail Analysis
Confirmation of the 3’ Poly(A) tail length of mRNA was achieved by digesting mRNA with RNase T1 and RNase A followed IP-RP-HPLC. RNase T1 specifically cleaves single-stranded RNA on the 3’-side of guanosine (G) residues, and RNase A cleaves single-stranded RNA on the 3’-side of uridine (U) and cytidine (C) residues. The resulting digestion product includes the two expected poly(A) tracts that are encoded by the linear DNA template. The poly(A) tracts were then separated by IP-RP-HPLC/UV using an Agilent 1260 system with quaternary pumps and DAD detector and a DNAPac RP column (DNAPac RP, 4 µm, 3.0 × 100 mm) (Thermo Fisher Scientific) held at 60 °C. The chromatography was evaluated using UV detection at 260 nm.
Digital droplet polymerase chain reaction (ddPCR) technology was used to quantify the poly(A) tail content. The ddPCR method uses a two-step reverse transcription (RT) for the quantitation of mRNA with a poly(A) tail. First, an Oligo-dT primer which is target-specific for the poly A tail of the mRNA was used to generate complementary deoxyribonucleic acid (cDNA) by RT from the test sample. Second, cDNA is diluted to a final theoretical total input copy number in the ddPCR reaction. A set of primers and a probe which are target-specific for the cDNA sequence at the 3’ end was used for digital droplet polymerase chain amplification. Fluorescence in the droplets was measured at the end of amplification. Poly(A) tail content was determined as a percent of the copy number quantified for cDNA by ddPCR with 3’ end over the theoretical total input cDNA copy number.
Antigen Expression Analyses
To monitor cell-based antigen expression, HEK-293 cells (Cell biolabs) were seeded into a 24-well clear flat bottom cell culture plate and incubated at 37°C, 5% CO2 for ∼24 hours. An RNA sample dilution series is prepared and mixed with 1x Lipofectamine MessengerMax solution (Thermo Fisher Scientific). The sample dilution series was then added to the cell assay plate, centrifuged, and incubated in a 37°C, 5% CO2 incubator for 20 to 28 hours. The cell lysates were analyzed with the specific antibodies to detect the SARS-CoV-2 spike proteins using ProteinSimple technology. 12-230 kDa Wes Separation Module and 25 capillary cartridges were used. SARS-CoV-2 Spike S1 subunit mouse antibody (R&D systems, Catalog No. MAB105403) and SARS-CoV-2 Spike S2 subunit mouse antibody (R&D systems, Catalog No. MAB10557) were used. mRNA sample results are reported as ProteinSimple Wes assay run lane images.
A cell-free in-vitro translation system was used to translate mRNA samples comprising different RNA integrity levels into corresponding protein or peptides. For sample preparation, mRNA samples were subjected to thermal degradation using elevated temperature (90°C) and incubation times of 2, 4, 6, 9, 10, and 20 minutes. Resulting RNA integrity levels are measured by CE as described above. For in-vitro translation, an expression system using rabbit reticulocyte lysate (nuclease treated; Promega) was used based on manufacturer´s recommendations containing biotinylated lysine tRNA (via a so called transcend RNA; Promega), which is incorporated into the resulting proteins and peptides. After translation the proteins and peptides were separated via SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) at 150 V. For Western Blot analysis the separated samples were transferred to a nitrocellulose membrane followed by detection of the biotinylated species using streptavidin alkaline phosphatase (Promega).
Results
mRNA Fragment Isolation
The BNT162b2 mRNA drug substance is comprised of full length, integral mRNA encoding the SARS-CoV-2 spike protein. In addition, it consists of 5’- and 3’-untranslated regions (UTR), a 7-methylguanosine-derived cap (m7GpppNmN) on the 5’-end, and a 3’ polyadenylation region.21 The 5’ cap, poly(A) tail, and other drug substance attributes are well-characterized and routinely tested for each batch to ensure a consistently high-quality vaccine.
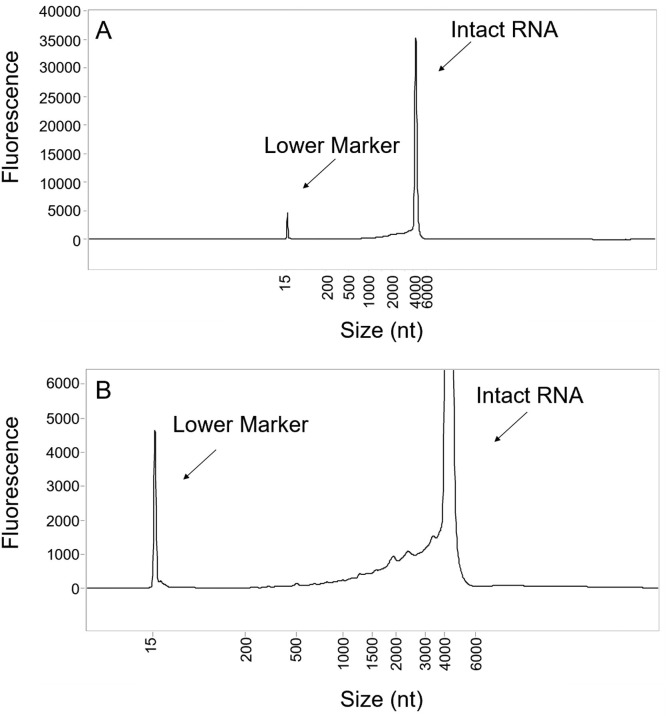
RNA integrity is a measure of the intact RNA transcript and is measured using capillary electrophoresis (CE). The BNT162b2 electropherogram (Fig. 1 ) reveals a single predominant species migrating at the expected size, with low-level species migrating to the left of the intact RNA peak, consistent with smaller mRNA fragments. Based on manufacturing process knowledge and the directional 5’-to-3’ polymerase transcription, these low-level species were hypothesized to be 5’-end BNT162b2 fragments generated from premature transcription stops during the IVT reaction. In addition, these could be derived from hydrolysis of full-length transcripts.
Fig. 1.
BNT162b2 RNA integrity electropherogram (A) and zoomed view (B) of fragment species. The capillary electropherogram shows a main species corresponding to the intact mRNA (∼4300 nucleotides, nt). A lower molecular weight marker is included in the sample preparation and migrates at a size of ∼15 nt. Fragment species migrate between the main peak and the lower marker peak.
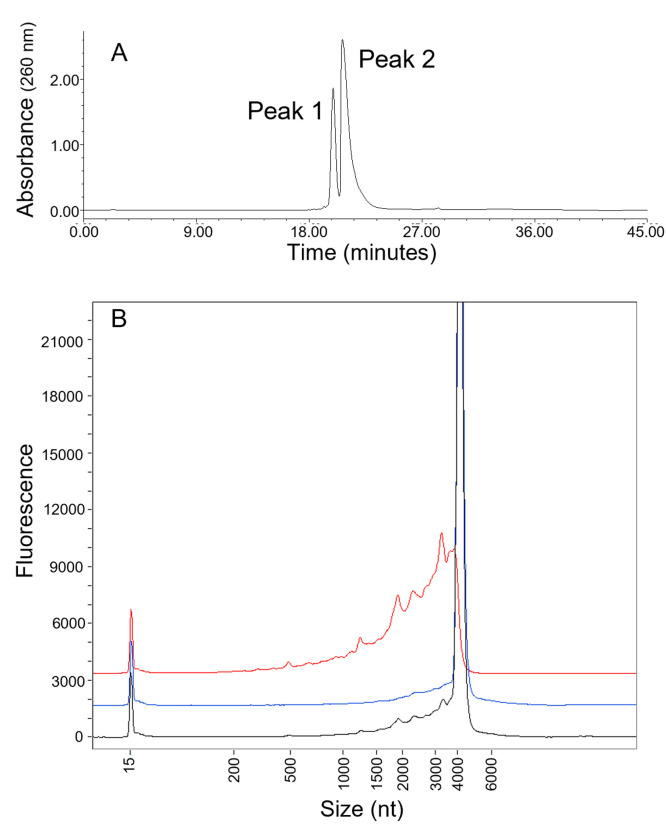
In an effort to isolate and characterize these low-level fragment species, an ion-pair reversed phase HPLC (IP-RP-HPLC) method was developed. The resulting chromatogram reveals two main peaks (Fig. 2 A). The peaks were collected and analyzed using CE for further characterization. The resulting electropherograms (Fig. 2B) reveal that the first IP-RP-HPLC peak (Peak 1, red trace) is comprised of fragment species, with little or no intact mRNA species observed. Further, the electrophoretic profile of “Peak 1” shows that the most abundant fragment peaks are consistent with those observed in the starting, unfractionated material (black trace), indicating that the IP-RP-HPLC method maintains the overall composition of material during species enrichment and fractionation. CE electropherograms additionally demonstrate that IP-RP-HPLC “Peak 2” (blue trace) is comprised predominantly of intact mRNA, with only trace-level fragment species present.
Fig. 2.
Separation of two species (Peak 1 and Peak 2 in panel A) was achieved using ion pairing RP-HPLC. (B) RNA integrity analysis of the isolated Peak 1 and Peak 2 materials demonstrates that Peak 1 consists of fragmented species (red trace), and Peak 2 is predominantly intact mRNA (blue trace). The unfractionated starting material electropherogram is shown in the black trace.
Characterization of Fragment and Intact mRNA Species
In addition to RNA integrity, the 5’-cap and poly(A) tail are critical quality attributes for mRNA vaccines, due to their roles in improving transcript stability and translational capability.22 , 23 In an effort to characterize mRNA fragment species, the IP-RP-HPLC peaks were isolated and evaluated for 5’-cap and poly(A) tail attributes, compared with the input material.
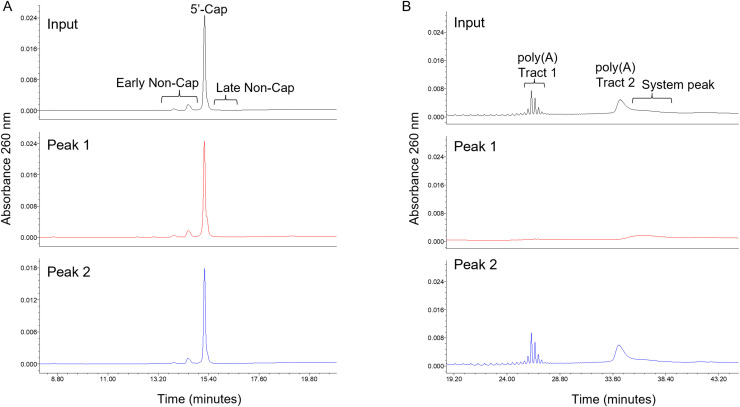
5’-cap content is evaluated similar to the method described previously,24 with the RP-HPLC separation of capped and non-capped species adapted for UV detection. The resulting material is chromatographically separated using RP-HPLC to evaluate the relative abundance of capped and uncapped mRNA (Fig. 3 A). The resulting chromatogram for the input (unfractionated) material shows a predominant peak, which corresponds to the 5’-capped species, flanked by minor/trace level non-capped species. The identities of all species have been confirmed by mass spectrometry analysis (data not shown). After fractionation and 5’-cap evaluation, both “Peak 1” fragment species and “Peak 2” enriched intact mRNA samples were observed to be predominantly capped at their 5’-end, and at levels consistent with the input (unfractionated) material. The results indicate that a vast majority of intact and fragmented 5’-end species are capped at their 5’-end, as would be expected from the in vitro transcription process.
Fig. 3.
(A) 5’-Cap analysis demonstrates that Peak 1 (middle) and Peak 2 (bottom) materials contain the expected capping structure at the 5’ end, consistent with the capping structure observed in the input BNT162b2 material (top). Relative quantitation of the capped content demonstrates comparable levels of capped/non-capped species across all three samples. (B) Poly(A) tail analysis reveals that the fragment species in Peak 1 (middle) lack the poly(A) tracts that are observed in the BNT162b2 input (top) and intact species enriched in the Peak 2 sample (bottom).
The poly(A) tail attribute was evaluated both for length of the poly-adenylation tracts and for content (the percentage of transcripts containing the poly(A) tail). First, a chromatographic method was developed to evaluate the approximate length of the poly(A) tail. A second method using droplet digital PCR (ddPCR) provides orthogonal information on the poly(A) tail content in each sample.
The poly(A) tail chromatogram of the input (unfractionated) material shows the expected length and distribution of the two poly(A) tracts in BNT162b2 (Fig. 3B). In contrast to the 5’-cap attribute described above, the poly(A) tail was only observed in the input material and in “Peak 2”, the latter of which corresponds to predominantly intact mRNA. The poly(A) tail region was not observed by RP-HPLC in the fragment-enriched “Peak 1” sample. Further, quantitation of poly(A) tail content by ddPCR demonstrated high levels (>85%) of transcripts containing poly(A) tail in the input (unfractionated) material and in the intact mRNA species enriched in “Peak 2”. Consistent with the observations from the chromatographic method, the poly(A) tail content in the fragment-enriched “Peak 1” sample was below the limit of quantitation for the ddPCR method (<4%).
The observation that BNT162b2 fragments are predominantly capped at their 5’-end and lack the poly(A) tail is consistent with fragment species forming mainly as a result of premature transcriptional stops during the in vitro transcription process. We next evaluated the role of 5’-cap and poly(A) tail on protein expression and the propensity for fragmented species to express truncated or off-target antigens.
Assessment of the Potential for Translation Into Truncated S1S2 or Other Off-Target Proteins/Peptides
The BNT162b2 mRNA encodes the SARS-CoV-2 spike protein, including S1 and S2 domains.21 To assess the potential for translation of mRNA fragments into truncated spike proteins/peptides or other proteins/peptides, two orthogonal protein expression systems were utilized and translated proteins were evaluated by Western blot.
Protein Expression in Transfected HEK-293 Cells
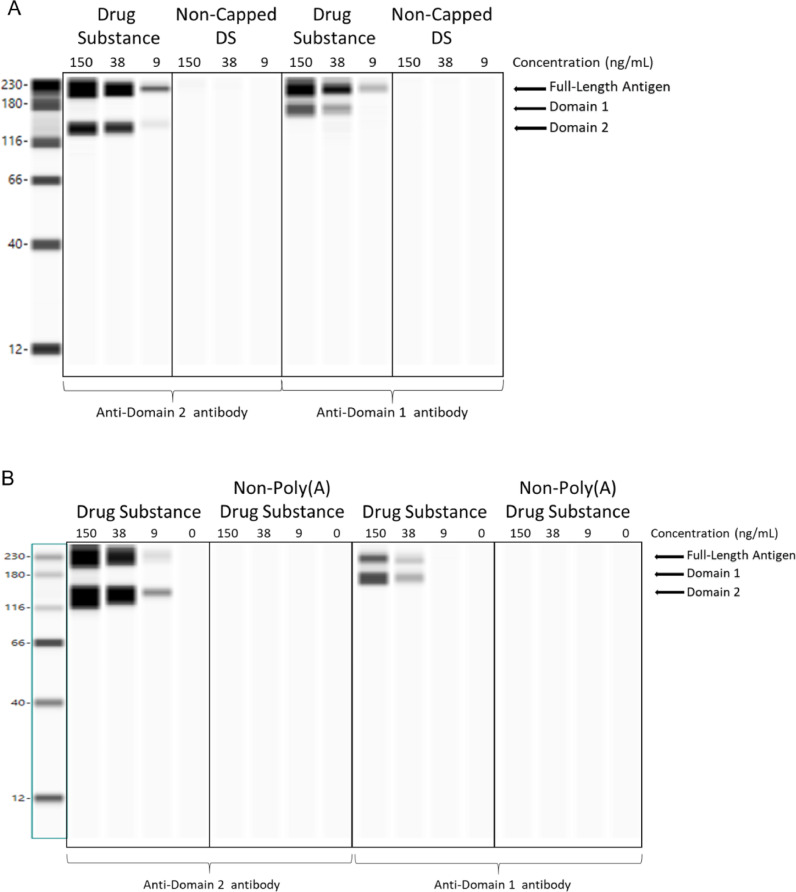
To characterize the expressed spike protein, BNT162b2 mRNA was mixed with Lipofectamine MessengerMax and then transfected into HEK-293 cells. Following incubation, cell lysates were evaluated for the expressed protein antigen by Western blot using antibodies specific for the SARS-CoV-2 spike protein S1 and S2 domains. As shown in Fig. 4 , BNT162b2 mRNA containing 5’-cap and poly(A) tail elements (“Drug Substance” in Fig. 4) is translated into proteins detected by both S1 and S2 domain antibodies. The full-length spike protein antigen and cleaved S1 and S2 domain proteins are observed by Western blot at sizes expected for the glycosylated proteins. The concentrations shown for each sample lane correspond to the amounts of mRNA transfected per well of HEK-293 cells, and a correlation was observed between the amount of mRNA transfected and expressed protein band intensity.
Fig. 4.
Expression analysis of mRNA transfected HEK-293 cells demonstrates the requirement for 5’-cap and poly(A) tail for protein translation. BNT162b2 drug substance and control mRNAs lacking 5’-cap (panel A), or poly(A) tail (panel B) were transfected into HEK-293 cells at 9, 38, or 150 ng/mL. Western blots from cell lysates were probed with an anti-S1 domain antibody (right half of each blot) or anti-S2 domain antibody (left half of each blot), revealing expression of the full length (S1S2) spike protein from the drug substance mRNA. Individual S1 and S2 domains are also observed and likely arise from S1S2 cleavage in the cell lysate. In contrast, protein expression is not observed after transfection with non-capped mRNA (panel A) or non-polyadenylated mRNA (panel B).
To demonstrate that transcripts require both 5’-cap and poly(A) tail for protein translation, full-length transcripts lacking either 5’-cap or poly(A) tail were produced and transfected into HEK-293 cells, and the resulting cell lysates were analyzed by Western blot using S1 and S2 detection antibodies (Fig. 4). In contrast to BNT162b2 mRNA, which contains 5’-cap and poly(A) tail elements, spike protein expression was not observed for the control transcripts lacking either the 5’-cap (Fig. 4A) or the poly(A) tail (Fig. 4B). This control experiment using full-length transcripts demonstrates that both 5’-cap and poly(A) tail are required for spike protein expression in HEK-293 cells.
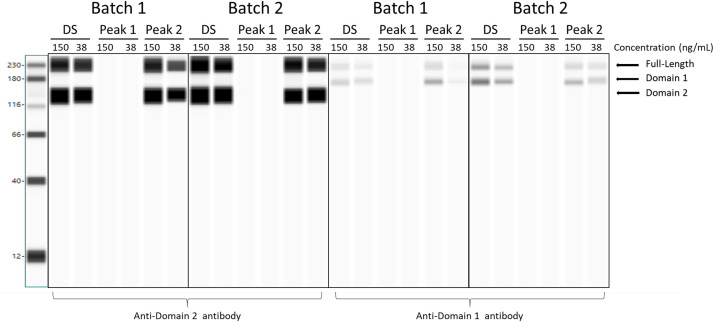
Next, HEK-293 cells were transfected with the input (unfractionated) mRNA, purified fragment species (“Peak 1”), or enriched intact mRNA (“Peak 2”) characterized above, using two different starting mRNA batches. Consistent with the control test results described above using full-length mRNA transcripts, cells transfected with the unfractionated mRNA or with enriched intact mRNA (“Peak 2”) material show the expected spike protein expression by Western blot, whereas cells transfected with “Peak 1” material comprised of fragmented species do not show evidence of full-length or truncated spike protein expression (Fig. 5 ). The results are consistent across batches, and the samples containing intact, capped, and polyadenylated mRNA show a dose-dependent expression response.
Fig. 5.
Evaluation of fragmented mRNA species’ translation. BNT162b2 drug substance (DS), “Peak 1” fragment species, and “Peak 2” enriched intact mRNA species were transfected into HEK-293 cells at 150 or 38 ng/mL. Cell lysates were probed by Western blot for spike protein expression using anti-S1 domain (right half of each blot) or anti-S2 domain (left half of each blot) antibodies. DS and enriched intact mRNA samples show the expected expression of full-length spike protein and the presence of S1 and S2 domains. Spike protein (full-length or truncated forms) expression was not observed from cells transfected with “Peak 1” fragment species, which lack the poly(A) tail.
Cell-Free Protein Expression
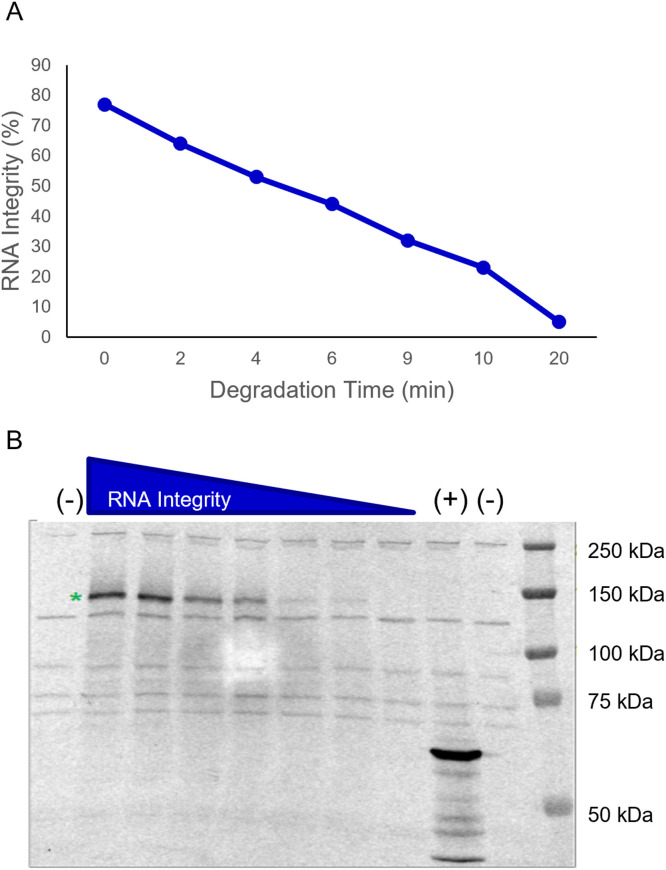
In a separate experiment, the potential for transcript fragments to produce aberrant protein/peptides was evaluated using a cell-free in vitro expression system. BNT162b2 drug substance was intentionally degraded by exposure to elevated temperature, generating samples with various levels of fragmented species (Fig. 6 A). Fragment species in thermally degraded samples are generated by hydrolysis, which is a relevant degradation pathway subsequent to in-vitro transcription. Unlike the enriched fragmented species described above, which are thought to be formed during transcription, thermally degraded samples offer additional insight into the translational viability of 3’-end fragments, which would lack 5’-cap, but retain the poly(A) tail.
Fig. 6.
Evaluation of translation from degraded mRNA species. (A) Full-length mRNA was thermally degraded to generate samples with diminishing levels of intact mRNA (and increased fragments). (B) Expression was evaluated in a cell-free expression system that incorporates a biotinylated lysine into newly translated proteins. SARS-CoV-2 spike protein expression is observed at the expected size for the non-glycosylated form (designated with a “*”) and at levels correlating with RNA integrity. No additional protein expression (i.e., truncated or off-target proteins) is observed beyond the species expressed in the negative control (-). “(+)” represents the positive control, “(-)” represents a negative control.
The starting and degraded materials were subjected to in vitro expression using rabbit reticulocyte lysate, wherein biotinylated lysine is incorporated into newly translated proteins and peptides for detection by Western blot. This approach enables detection of both truncated spike and other non-spike proteins/peptides, if present, without the need for protein/peptide-specific detection antibodies. After in vitro expression, the non-degraded BNT162b2 sample produced a protein of approximately 140 kDa, which is consistent with the expected size of the aglycosylated spike protein (denoted with a “*” in Fig. 6B). For degraded samples, Western blot band intensity for the full-length protein correlated with RNA integrity; however, no truncated or other protein species were detected beyond the background bands observed in the negative control sample. These results further demonstrate that BNT162b2 fragments, either generated as part of the in vitro transcription process or due to hydrolysis, lack translational viability.
Discussion
In the present study, mRNA fragment species generated during the BNT162b2 in vitro transcription process were isolated, characterized, and evaluated for potential translational viability. We demonstrated that fragmented mRNA species contain the expected 5’-cap structure but lack a poly(A) tail. We showed through control experiments using full-length transcripts that 5’-cap and poly(A) tail are required for translation of the BNT162b2 transcript into spike protein, and that fragmented transcript species lacking a poly(A) tail are not viable for protein translation. Lastly, we demonstrated that thermally degraded mRNA transcripts, which presumably include fragments lacking 5’-cap, poly(A) tail, or both, are only viable for translating the full-length spike protein at levels commensurate with the full-length (non-degraded) mRNA transcript.
The newly developed IP-RP-HPLC method enabled isolation of fragment species (“Peak 1”) and enrichment of full-length, intact mRNA (“Peak 2”). The similar CE fragment profiles of isolated fragment species and the starting material indicate that separation and fractionation are non-destructive to the mRNA. However, low-level residual fragment species are observed in the enriched, full-length mRNA sample (Fig. 2). Low level fragments observed in “Peak 2” are thought to either originate from carry-over during “Peak 1” fractionation or from low levels of mRNA hydrolysis that could occur during sample handling. Because full-length, intact mRNA and fragment species differ in transcript length and the presence of the poly(A) tail, these two attributes likely contribute to the mode of IP-RP-HPLC separation that enables isolation of fragment species.
Translation of mRNA to protein was evaluated using two orthogonal systems, one cell-based and one cell-free. In HEK-293 cells, proteins translated from mRNA transcripts can be post-translationally modified, and this cell-based translation system reveals the spike protein's capacity for glycosylation,25 , 26 which could be important for immunogenicity.27 However, extensive glycosylation introduces challenges in accurately characterizing protein size by Western blot. In contrast to HEK-293 cell transfection, the cell-free translation system lacks the cellular machinery needed for glycosylation, which in turn offers a more accurate determination of the translated spike protein size by Western blot.
The Western blot used to evaluate HEK-293 cell-based translation used spike-protein specific detection antibodies, providing specificity in confirming the identity of the expressed protein(s). In addition to the full-length spike protein, S1 and S2 protein domains were observed in the Western blot of transfected cell lysates. These individual domains are not thought to be expressed as truncated proteins from mRNA fragments, but rather are likely proteolytically clipped from the full-length protein in the cell lysate. This is supported by the demonstration that 5’-cap and poly(A) tail attributes (the latter of which is lacking in fragment species) are required for translation and that the individual S1 and S2 protein domains are not observed in the orthogonal cell-free expression system.
The cell-free system incorporates a biotinylated lysine residue into newly translated proteins, and therefore Western blot detection is not dependent on protein/antigen-specific detection antibodies. While lacking specificity for the spike protein, the detection of all newly translated proteins was central to demonstrating that BNT162b2 mRNA fragments lack translational viability. The results of this study indicate that BNT162b2 fragment species predominantly arise from premature transcriptional stoppage during in vitro transcription, and the additional characterization of thermally degraded materials in the cell-free system further demonstrates that fragments generated either during in vitro transcription or via mRNA hydrolysis lack capacity for protein translation.
The active mRNA ingredient in BNT162b2 is well-characterized and gives rise to a single, homogeneous polypeptide antigen of the expected size. In addition to improving BNT162b2 product knowledge, the present study strengthens in vitro transcriptional process understanding and is relevant for other mRNA transcripts manufactured from a similar process. It is, however, important to assess the off-target protein translation risk for new construct designs, for example to evaluate novel sequence elements (e.g. alternative poly(A) tail designs13) or the potential introduction of hidden open reading frames within the gene of interest or during construct design and optimization.28 In the formulated vaccine drug product, the lipid nanoparticle protects the mRNA from hydrolytic fragmentation; however, additional mRNA degradation mechanisms (e.g., through interactions with lipid excipients in the formulated drug product29 , 30) can potentially affect antigen translation. While not in scope of the present study, these non-hydrolytic degradation mechanisms should be explored and appropriately controlled in the final drug product.
The COVID-19 pandemic brought an unprecedented urgency for a vaccine targeting SARS-CoV-2, and mRNA technology proved key to rapidly meeting the global need for a safe and efficacious vaccine without compromising product and process understanding.4 Heightened characterization studies, routine product testing, and a well-controlled manufacturing process are part of a robust, holistic control strategy that ensures consistently high-quality vaccine production.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Himakshi K. Patel, Kun Zhang, Rachael Utegg, Elaine Stephens, Shauna Salem, Heidi Welch, Jeff Ryczek, David J. Cirelli, and Thomas F. Lerch are full-time employees and may be shareholders of Pfizer Inc. Svenja Grobe, Julia Schlereth, and Andreas N. Kuhn are full-time employees and may be shareholders of BioNTech SE.
Acknowledgments
Funding
This study was sponsored by Pfizer Inc and BioNTech SE.
Acknowledgements
The authors thank our fellow colleagues at BioNTech and Pfizer, including the Vaccine R&D team at Pfizer for generating control transcripts. We also thank the clinical trial participants and their families, investigators, sites, and staff, as well as governments and regulatory authorities worldwide; healthcare workers; first responders; teachers and other essential workers; vendors, suppliers, and other support agencies and teams.
References
- 1.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourla A. 1st ed. HarperCollins Publishers; 2022. Moonshot. [Google Scholar]
- 3.Thompson MG, Burgess JL, Naleway AL, Tyner HL, Yoon SK, Meece J, Olsho LEW, Caban-Martinez AJ, Fowlkes A, Lutrick K, Kuntz JL, Dunnigan K, Odean MJ, Hegmann KT, Stefanski E, Edwards LJ, Schaefer-Solle N, Grant L, Ellingson K, Groom HC, Zunie T, Thiese MS, Ivacic L, Wesley MG, Lamberte JM, Sun X, Smith ME, Phillips AL, Groover KD, Yoo YM, Gerald J, Brown RT, Herring MK, Joseph G, Beitel S, Morrill TC, Mak J, Rivers P, Harris KM, Hunt DR, Arvay ML, Kutty P, Fry AM, Gaglani M. Interim estimates of vaccine effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines in preventing SARS-CoV-2 infection among health care personnel, first responders, and other essential and frontline workers - eight U.S. Locations, December 2020-March 2021. MMWR Morb Mortal Wkly Rep. 2021;70(13):495–500. doi: 10.15585/mmwr.mm7013e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis LM, Badkar AV, Cirelli D, Combs R, Lerch TF. The race to develop the Pfizer-BioNTech COVID-19 vaccine: from the pharmaceutical scientists' perspective: race to the COVID-19 vaccine. J Pharm Sci. 2022 doi: 10.1016/j.xphs.2022.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson OJ, Barnsley G, Toor J, Hogan AB, Winskill P, Ghani AC. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect Dis. 2022;22(9):1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steele MK, Couture A, Reed C, Iuliano D, Whitaker M, Fast H, Hall AJ, MacNeil A, Cadwell B, Marks KJ, Silk BJ. Estimated number of COVID-19 infections, hospitalizations, and deaths prevented among vaccinated persons in the US, December 2020 to September 2021. JAMA Netw Open. 2022;5(7) doi: 10.1001/jamanetworkopen.2022.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolgin E. The tangled history of mRNA vaccines. Nature. 2021;597(7876):318–324. doi: 10.1038/d41586-021-02483-w. [DOI] [PubMed] [Google Scholar]
- 8.Kreiter S, Diken M, Selmi A, Tureci O, Sahin U. Tumor vaccination using messenger RNA: prospects of a future therapy. Curr Opin Immunol. 2011;23(3):399–406. doi: 10.1016/j.coi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Pascolo S. Vaccination with messenger RNA. Methods Mol Med. 2006;127:23–40. doi: 10.1385/1-59745-168-1:23. [DOI] [PubMed] [Google Scholar]
- 10.Vallazza B, Petri S, Poleganov MA, Eberle F, Kuhn AN, Sahin U. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. Wiley Interdiscip Rev RNA. 2015;6(5):471–499. doi: 10.1002/wrna.1288. [DOI] [PubMed] [Google Scholar]
- 11.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn AN, Beibetaert T, Simon P, Vallazza B, Buck J, Davies BP, Tureci O, Sahin U. mRNA as a versatile tool for exogenous protein expression. Curr Gene Ther. 2012;12(5):347–361. doi: 10.2174/156652312802762536. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn AN, Diken M, Kreiter S, Vallazza B, Tureci O, Sahin U. Determinants of intracellular RNA pharmacokinetics: Implications for RNA-based immunotherapeutics. RNA Biol. 2011;8(1):35–43. doi: 10.4161/rna.8.1.13767. [DOI] [PubMed] [Google Scholar]
- 14.European Medicines Agency. February 19, 2021. European public assessment report (EPAR). ed.
- 15.European Medicines Agency. Last Updated July 28, 2022. Comirnaty - Summary of Product Characteristics. ed.
- 16.European Medicines Agency. March 11, 2021. European Public Assessment Report (Spikevax). ed.
- 17.Hald Albertsen C, Kulkarni JA, Witzigmann D, Lind M, Petersson K, Simonsen JB. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv Drug Deliv Rev. 2022;188 doi: 10.1016/j.addr.2022.114416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine. 2021;39(16):2190–2200. doi: 10.1016/j.vaccine.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott D, Ladomery M. Oxford University Press; 2016. Molecular Biology of RNA. [Google Scholar]
- 21.Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Guler A, Loschko J, Maddur MS, Ota-Setlik A, Tompkins K, Cole J, Lui BG, Ziegenhals T, Plaschke A, Eisel D, Dany SC, Fesser S, Erbar S, Bates F, Schneider D, Jesionek B, Sanger B, Wallisch AK, Feuchter Y, Junginger H, Krumm SA, Heinen AP, Adams-Quack P, Schlereth J, Schille S, Kroner C, de la Caridad Guimil, Garcia R, Hiller T, Fischer L, Sellers RS, Choudhary S, Gonzalez O, Vascotto F, Gutman MR, Fontenot JA, Hall-Ursone S, Brasky K, Griffor MC, Han S, Su AAH, Lees JA, Nedoma NL, Mashalidis EH, Sahasrabudhe PV, Tan CY, Pavliakova D, Singh G, Fontes-Garfias C, Pride M, Scully IL, Ciolino T, Obregon J, Gazi M, Carrion R, Jr., Alfson KJ, Kalina WV, Kaushal D, Shi PY, Klamp T, Rosenbaum C, Kuhn AN, Tureci O, Dormitzer PR, Jansen KU, Sahin U. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592(7853):283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 22.Geall AJ, Mandl CW, Ulmer JB. RNA: the new revolution in nucleic acid vaccines. Semin Immunol. 2013;25(2):152–159. doi: 10.1016/j.smim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Sahin U, Kariko K, Tureci O. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 24.Beverly M, Dell A, Parmar P, Houghton L. Label-free analysis of mRNA capping efficiency using RNase H probes and LC-MS. Anal Bioanal Chem. 2016;408(18):5021–5030. doi: 10.1007/s00216-016-9605-x. [DOI] [PubMed] [Google Scholar]
- 25.Sanda M, Morrison L, Goldman R. N- and O-Glycosylation of the SARS-CoV-2 spike protein. Anal Chem. 2021;93(4):2003–2009. doi: 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng T, Li T, Chen G, Zhu Y, Xu L, Lin Y, Sun H, Zhang H, Fang Q, Hong J, Wu D, Gao S, Li S, Wang Y, Zhang T, Chen Y, Yuan Q, Zheng Q, Yu H, Zhao Q, Zhang J, Li S, Xia N, Gu Y. Characterization and immunogenicity of SARS-CoV-2 spike proteins with varied glycosylation. Vaccine. 2022;40(47):6839–6848. doi: 10.1016/j.vaccine.2022.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beaudoin CA, Bartas M, Volna A, Pecinka P, Blundell TL. Are there hidden genes in DNA/RNA vaccines? Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.801915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oude Blenke E, Ornskov E, Schoneich C, Nilsson GA, Volkin DB, Mastrobattista E, Almarsson O, Crommelin DJA. The storage and in-use stability of mRNA vaccines and therapeutics: not a cold case. J Pharm Sci. 2022 doi: 10.1016/j.xphs.2022.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Packer M, Gyawali D, Yerabolu R, Schariter J, White P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat Commun. 2021;12(1):6777. doi: 10.1038/s41467-021-26926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]