Abstract
With the introduction of large-scale molecular profiling methods and high-throughput sequencing technologies, the genomic features of most lymphoid neoplasms have been characterized at an unprecedented scale. Although the principles for the classification and diagnosis of these disorders, founded on a multidimensional definition of disease entities, have been consolidated over the past 25 years, novel genomic data have markedly enhanced our understanding of lymphomagenesis and enriched the description of disease entities at the molecular level. Yet, the current diagnosis of lymphoid tumors is largely based on morphological assessment and immunophenotyping, with only few entities being defined by genomic criteria. This paper, which accompanies the International Consensus Classification of mature lymphoid neoplasms, will address how established assays and newly developed technologies for molecular testing already complement clinical diagnoses and provide a novel lens on disease classification. More specifically, their contributions to diagnosis refinement, risk stratification, and therapy prediction will be considered for the main categories of lymphoid neoplasms. The potential of whole-genome sequencing, circulating tumor DNA analyses, single-cell analyses, and epigenetic profiling will be discussed because these will likely become important future tools for implementing precision medicine approaches in clinical decision making for patients with lymphoid malignancies.
Complementing the recently published Blood articles outlining the 2022 International Consensus Classifications for hematological malignancies (Vol. 140, Issue 11), this pair of Special Reports illustrates how molecular pathology can be applied to precision medicine. de Leval and colleagues summarize the potential of DNA sequencing of tumors and cell-free plasma, epigenetic profiling, and single-cell analyses to inform clinical decision-making about diagnosis, prognosis, and treatment for patients with lymphoid neoplasms. Similarly, Duncavage and colleagues cover genomic profiling for myeloid neoplasms and the acute leukemias, focusing principally on somatic changes but also with emphasis on the emerging importance of germline gene mutations in certain diseases. Both articles provide up-to-date references for how to apply genomic information to practice.
Introduction
Genetics is an integral part of the contemporary classification of lymphoid neoplasms.1,2 Recurrent chromosomal alterations, discovered by cytogenetics,3 were instrumental in defining certain lymphoma entities and, in select tumors, represent a cornerstone for diagnosis in complement to morphological and immunophenotypic analyses. Some rearrangements lead to either dysregulation of oncogenic proteins or expression of gene fusions. Fluorescence in situ hybridization (FISH) is most often used to detect chromosomal aberrations (Figure 1), with rearrangements detected using either fusion or break-apart probes. Clonality assessment of immunoglobulin (IG) and TR loci rearrangements using PCR-based analyses4 or, more recently, high-throughput sequencing (HTS)5 is often useful in the assessment of lymphoid proliferations. However, the finding of clonal rearrangements is not always synonymous with lymphoid neoplasms because dominant clones can be seen in reactive conditions as well, highlighting the importance of appropriate integration with all other pathologic features.4
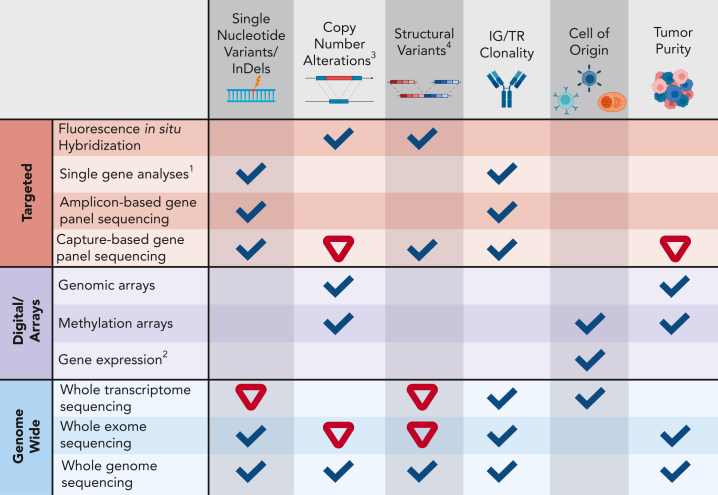
Figure 1.
Detection capacity of genomic aberrations with different technologies.1Includes various technologies that may interrogate single nucleotide changes through to the sequence of the entire gene (AS-PCR, fragment analysis, Sanger sequencing, and others). 2Includes gene expression arrays, NanoString, and RT-MLPA assays. 3Most technologies, except FISH, cannot detect subclonal CNAs (<20%) with high confidence. 4Including gene fusions. Ticks indicate good capacity to determine a certain aberration/feature, whereas an inverted red triangle indicates a limited/insufficient detection capacity. AS-PCR, allele-specific oligonucleotide polymerase chain reaction; CNA, copy number aberration; IG, immunoglobulin; indel, insertion-deletion; RT-MLPA, reverse transcriptase multiplex ligation–dependent probe amplification; TR, T-cell receptor locus. Created with BioRender.com.
With the introduction of HTS-based technologies over the past 10 to 15 years, the genomic landscapes of many lymphoid neoplasms were characterized at an unprecedented scale.6 Although a predominant gene mutation was identified in only a few lymphoma entities, such as the MYD88L265P mutation in lymphoplasmacytic lymphoma (LPL) and BRAFV600E mutation in hairy cell leukemia,7,8 in most lymphoid neoplasms, a much more diverse pattern is observed with only a small number of variably frequent aberrations followed by a long tail of uncommonly mutated genes.9, 10, 11 These studies have also disentangled the diverse (sub)clonal architecture of lymphoid neoplasms, including early drivers, later alterations linked to clinical aggressiveness, and passenger mutations.12, 13, 14 Despite the heterogeneous mutation landscapes between distinct diseases, there are also common themes of affected cellular processes and signaling pathways (supplemental Figure 1 and supplemental Table 1 [available on the Blood website]). Based on newly acquired knowledge, clinically relevant genomic aberrations have been identified with diagnostic, prognostic, and predictive impact in different entities.15,16 Although the number of alterations that facilitate diagnosis and risk stratification is increasing, relatively few are currently linked to prediction of therapeutic response.17,18
HTS-based technologies range from targeted sequencing of a limited number of genes (gene panels) to whole-exome sequencing (WES) for the assessment of coding regions of genes or whole-genome sequencing (WGS). These methods have different capacities to detect somatic aberrations because targeted approaches typically have a higher sequence depth than genome-wide technologies and, therefore, detect subclonal alterations with greater sensitivity and are more robust to lower tumor purity. In amplicon-based sequencing panels, a limited number of genes or hotspot regions are generally included (∼20-50), and only single-nucleotide variants (SNVs) and indels or specific gene fusions are detected (Figure 1).19 Capture-based panels enable simultaneous interrogation of SNVs and indels, copy-number aberrations (CNAs) (ie, deletions and amplifications), and structural variants (SVs, including rearrangements).20,21 These comprehensive panels can include sequencing of DNA and/or RNA and assessment of other more complex markers, such as IG and TR rearrangements and DNA methylation. Recently developed “all-in-one” capture-based panels can detect the most relevant types of genomic aberrations associated with lymphoproliferations.22, 23, 24
Gene expression profiling (GEP) and DNA methylation analyses have been pivotal in identifying lymphoma subgroups and “cell-of-origin” signatures.25, 26, 27, 28, 29 Subsequently, selective targeted approaches have been developed to detect differential expression of key genes that inform on these subgroups.29, 30, 31 Whole-transcriptome sequencing (WTS; commonly referred to as RNA-seq), an alternative unbiased method, may have future routine applications in clinical diagnostic laboratories.32 Apart from tumor genetics, the tumor microenvironment (TME) plays a key role in shaping lymphoma development and response to treatment.33 Advances in single-cell analysis (SCA) methodologies, along with tools for in silico deconvolution of bulk tissue WTS,34 are leading to a better understanding of tumor heterogeneity within its TME landscape.35
The application of clinical molecular diagnostics to lymphoid proliferations is currently constrained by several practical considerations. The optimal source consists of nucleic acids extracted from fresh surgical biopsy specimens or liquid samples (blood or bone marrow), but clinical assays must be adapted to formalin-fixed paraffin-embedded (FFPE) tissues, which is the main diagnostic material, and to limited samples (eg, needle biopsies). Currently, targeted gene panels WES and WTS are feasible for FFPE material, but WGS remains more challenging.36 For HTS-based assays, important parameters include tumor cell content, technical performance (eg, sequence coverage/depth, background artifacts), the need for unique molecular identifiers, and turnaround time. Key aspects related to variant interpretation and reporting include variant classification systems used,37, 38, 39 variants of uncertain significance, and the presence of clonal hematopoiesis (CH). For patients experiencing relapse, the most recent sample is usually preferentially analyzed, but comparison of sequential biopsies may be necessary depending on the clinical question posed.
Current classification of lymphoid tumors remains largely based on morphological assessment and immunophenotyping, but it is likely that future schemes will further integrate genomic-based features to characterize and define (sub)entities and direct therapies.2 This paper, which accompanies the International Consensus Classification of mature lymphoid neoplasms,2 will address how genomic testing already complements existing criteria and provides a novel lens on disease classification. More specifically, its contributions to diagnostic refinement, risk stratification, and therapy prediction will be considered for the main categories of lymphoid neoplasms (Tables 1 and 2), along with its value in helping resolve potentially challenging differential diagnoses (Table 3). Histiocytic and dendritic cell neoplasms, being of myeloid or mesenchymal derivation, have traditionally been discussed with lymphomas, given overlapping clinical presentation, and will be addressed in a similar fashion. Finally, how WGS, analysis of circulating tumor DNA (ctDNA), or liquid biopsy specimens, epigenetic profiling, and single-cell analyses may become important tools for implementing precision medicine approaches in clinical decision making of patients with lymphoid malignancies in the near future will be envisioned.
Table 1.
Clinical impact of genomic testing in B-cell neoplasms
| Entity | Genetic alteration: test | Diagnostic use | Clinical impact | Future assays |
|---|---|---|---|---|
| B-cell neoplasms | IG gene rearrangement: PCR-based assays with fragment analysis or HTS | Useful in certain circumstances to demonstrate monoclonality of B-cell lymphoproliferations to establish a diagnosis; mandatory in certain entities (eg, pediatric-type FL) | WGS for the detection of CNAs and SVs WTS to detect microenvironment signatures |
|
| Chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) | IGHV mutation status∗: IGHV sequencing | Prognostic and predictive. IGHV gene mutational status remains stable through the disease course and only needs to be performed once | Determining BcR stereotypy and IGLV3-21R110 mutation status for risk stratification; tracking of resistance mutations (BTK, PLCG2, and BCL2; supplemental Table 3) WGS for mutations, CNAs, SVs, and complex karyotype determination MRD testing using HTS to guide therapy decisions |
|
| del(11q), +12, del(13q), del(17p)∗: FISH | Prognostic and del(17p) is predictive. FISH testing should be performed before each new course of therapy | |||
| TP53 mutations∗: HTS | Prognostic and predictive. TP53 sequencing should be performed before each new course of therapy unless already demonstrated | |||
| Detection of complex karyotype (≥5 abnormalities): cytogenetics∗ or SNP arrays | Prognostic | |||
| Hairy cell leukemia | BRAF V600E mutation: sequencing or IHC | Useful to support the diagnosis on biopsy samples and in cases with uncommon presentations463 | ||
| Follicularlymphoma (FL) | BCL2 rearrangement†: FISH (or cytogenetics) | Consider if BCL2 IHC is negative. Further workup of BCL2-R–negative FL shown in scenario 1B in Table 3 | ||
| EZH2 mutation†: HTS | EZH2 mutation is predictive of response to EZH2 inhibition.81 Tazemetostat is approved by the FDA for use in patients with EZH2-mutated FL (detected by an FDA-approved test) who have received at least 2 prior lines of systemic therapy (and all adult patients, including with wt EZH2 with relapsed/refractory disease and no other satisfactory alternative treatment options) | |||
| Marginal zone lymphomas (MZL) |
BCL2 and CCND1 rearrangements: FISH† MYD88 L265 mutation†: AS-PCR or HTS |
Detection prompts considering a diagnosis of other entities; see scenarios 1 and 2 in Table 3 and supplemental Figure 3 | ||
| Extranodal MZL of mucosa associated lymphoid tissue (MALT lymphoma) |
MALT1, BCL10, FOXP1 rearrangements†: FISH +3, +1888: cytogenetics and FISH |
Detection is useful in certain circumstances to support the diagnosis | ||
| t(11;18) BIRC3::MALT1∗: FISH in H pylori–positive gastric MALT lymphoma | MALT1 rearrangements are associated with lack of antibiotic response in H pylori–positive gastric MALT lymphoma91 | |||
| SplenicMZL | del(7q)†, +3, +1888: cytogenetics and FISH KLF2, NOTCH2 mutations88: HTS |
Detection is useful in certain circumstances to support the diagnosis | ||
| NodalMZL | +3, +1888: cytogenetics and FISH KLF2, NOTCH2, PTPRP88 mutations: HTS |
Detection is useful in certain circumstances to support the diagnosis | ||
| Mantle cell lymphoma | CCND1 rearrangement†: FISH | Consider if CCND1 IHC is negative | MRD testing using HTS to guide treatment decisions WTS or targeted gene expression panel for proliferation and signatures of nnMCL vs cMCL |
|
| CCND2 and CCND3 rearrangement†: FISH | Consider in CCND1-R–negative tumors | |||
| TP53 mutation∗: HTS‡ | Prognostic and guide management111 | |||
|
Multiple myeloma (MM) MM-NOS MM with recurrent genetic abnormality MM with CCND family translocation MM with MAF family translocation MM with NSD2 translocation MM with hyperdiploidy |
t(4;14) NSD2::IGH; t(14;16) IGH::MAF; t(11;14) CCND1::IGH;∗,§ gain of odd numbered chromosomes: FISH on bone marrow plasma cells (CD138-positive selected sample strongly recommended)∗ | Diagnostic of the ICC subtypes of MM | t(11;14) predictive of response to venetoclax134 | WGS for subtype assignment, risk stratification, and decision making MRD using HTS for decision making |
| t(4;14) NSD2::IGH; t(14;16) IGH::MAF; amp(1q); del(1p), del(17p)∗; TP53 mutations464 For SMM: t(4;14) NSD2::IGH; t(14;16) IGH::MAF; 1q gain/amplification; del(13)145 and MYC rearrangement139: FISH and HTS |
Risk stratification at diagnosis and relapse | The adverse prognosis of high-risk genetics is partially overcome by the addition of a proteasome inhibitor131 and/or anti-CD38 MoAb132 to first-line therapy | ||
| Lymphoplasmacytic lymphoma | MYD88 L265 mutation: AS-PCR testing on bone marrow∗ (or other highly sensitive HTS-based method: consider AS-PCR as a reflex test if negative) | Diagnostic. Aids in the differential with small B-cell lymphomas; see scenario 2A in Table 3 | HTS methods for sensitive mutation detection | |
| CXCR4 mutations†: highly sensitive HTS-based method | Predictive of primary resistance to ibrutinib therapy160 | |||
|
Diffuse large B-cell lymphoma, NOS Germinal center B-cell subtype Activated B-cell subtype |
MYC, BCL2, and/or BCL6 rearrangement (latter two can be performed concurrently or only if MYC rearrangement is detected): FISH∗ | Required to exclude HGBCL-DH-BCL2 and HGBCL-DH-BCL6 | See “High-grade B-cell lymphoma” | Genetic subtype assignment (eg, LymphGen187) by panel, exome or WGS and BCL2 and BCL6 rearrangement detection and WTS or targeted gene expression panels (DHITsig29/MHG signature199) HTS-based ctDNA testing465 for response-adapted management |
| COO determination: GEP or widely used IHC surrogates∗ | Required to assign DLBCL, NOS gene expression subtypes | Prognostic for outcomes following R-CHOP (GEP)466; predictive of response to treatment at relapse177 | ||
|
High-grade B-cell lymphomas (HGBCL) HGBCL with MYC and BCL2 rearrangement (HGBCL-DH-BCL2) HGBCLwithMYCandBCL6rearrangement (HGBCL-DH-BCL6) HGBCL, NOS |
MYC, BCL2, and/or BCL6 rearrangement (latter two can be performed concurrently or only if MYC rearrangement is detected): FISH∗ | Required for the diagnosis of HGBCL-DH-BCL2 and HGBCL-DH-BCL6 | Prognostic and predictive: HGBCL-DH-BCL2 has poor prognosis with R-CHOP and likely benefits from treatment intensification467 | Rearrangement detection and MYC partner determination by HTS HTS analysis of HGBCL, NOS tumors to assign these tumors to definitive disease categories |
| Burkitt lymphoma | MYC, BCL2, and/or BCL6 rearrangement (latter two can be performed concurrently or only if MYC rearrangement is detected): FISH∗ | Required to exclude HGBCL-DH-BCL2 and HGBCL-DH-BCL6 | ||
| Pediatric lymphomas | ||||
| Pediatric-type FL Pediatric nodal MZL |
BCL2 or BCL6 rearrangements†: FISH IRF8, MAP2K1 TNFRSF14 mutations†: HTS B-cell clonality testing |
Useful in certain circumstances for diagnosis; see also scenario 3A in Table 3. Of note, pediatric-type FL and pediatric nodal MZL are not readily distinguishable by genomic features | Detection of CNAs and SVs using HTS | |
| Large B-cell lymphoma with11qaberration | 11q aberration: SNP array or FISH | Required for diagnosis of LBCL-11q | ||
| Large B-cell lymphoma withIRF4rearrangement |
IRF4 rearrangement: FISH CARD11, IRF4 mutations†: HTS |
FISH required for diagnosis of LBCL-IRF4 rearrangement Useful in certain circumstances for diagnosis; see also scenario 3A in Table 3. |
||
| Classic Hodgkin lymphoma | ctDNA for the detection of genetic aberrations in the Hodgkin/Reed-Sternberg cells and for response-adapted therapy Detection of amplification of 9p24.1 by FISH as a favorable biomarker for PD1 inhibitors in relapsed/refractory CHL248 |
AS-PCR, allele-specific polymerase chain reaction; BcR, B-cell receptor; BL, Burkitt lymphoma; BTK, Bruton’s tyrosine kinase; CHL, classic Hodgkin lymphoma; cMCL, conventional MCL; CLL, chronic lymphocytic leukemia; COO, cell-of-origin; ctDNA, circulating tumor DNA; DLBCL, diffuse large B-cell lymphoma; FDA, Food and Drug Administration; FL, follicular lymphoma; HGBCL, high-grade B-cell lymphoma; IGHV, immunoglobulin heavy variable; IHC, immunohistochemistry; LBCL-IRF4, large B-cell lymphoma with IRF4 rearrangement; MALT, mucosa-associated lymphoid tissue; MCL, mantle cell lymphoma; MHG, molecular high grade; MM, multiple myeloma; MRD, measurable residual disease; MZL, marginal zone lymphoma; NMZL, nodal MZL; NMM, newly diagnosed multiple myeloma; nnMCL, non-nodal MCL; NOS, not otherwise specified; R-CHOP, rituximab in combination with cyclophosphamide, doxorubicin, vincristine, and prednisone; SLL, small lymphocytic lymphoma; SMM, smoldering multiple myeloma; SMZL, splenic MZL; SNP, single nucleotide polymorphism; wt, wild-type.
Required/strongly recommended in the National Comprehensive Cancer Network 2022 guidelines.
Useful in certain circumstances in the National Comprehensive Cancer Network 2022 guidelines.
IHC for TP53 has reported 82% sensitivity for TP53 missense mutations.468
IGH break-apart FISH can be used to screen before the other FISH assays are performed.
Table 2.
Clinical impact of genomic testing in T-cell neoplasms
| Entity | Genetic alteration: test | Diagnostic use | Clinical impact | Future assays |
|---|---|---|---|---|
| T-cell neoplasms | TRG and/or TRB gene rearrangements∗,†: PCR-based assays with fragment analysis or HTS | Demonstration of monoclonal TCR gene rearrangement is (1) recommended to support a diagnosis of T-cell lymphoma, especially when morphology and immunophenotyping are not fully conclusive for T-cell lymphoma/leukemia, and to diagnose clonal T-LPD; (2) useful in the assessment of atypical T-cell populations and establishing lineage in phenotypically ambiguous malignancies; and (3) helping in the distinction between T and NK origin | Accurate diagnosis of a neoplastic T-cell proliferation | WTS or targeted gene expression assays to determine T-cell repertoire and disease classification and detect driver fusions282,469 WGS to detect CNAs and SVs ctDNA assays for disease monitoring |
| Mutations and small indels in genes recurrently altered: HTS Various gene fusions: HTS or FISH |
Useful in certain circumstances to establish clonality or to support the diagnosis of a specific entity | Mechanism of actionable alterations and how they could be targeted clinically is displayed in Figure 5 | ||
| ALCL, ALK-positive | ALK gene fusions†: IHC, FISH, or transcript detection | Mandatory to establish the diagnosis of ALK-positive ALCL | Use of ALK inhibitors | HTS to guide second-/third-generation ALK inhibitors in cases of resistance to ALK inhibitors470 |
| ALCL, ALK-negative | DUSP22-IRF4 (6p25.3) rearrangement†: FISH; TP63 (3q28) rearrangement†: FISH | DUSP22-R defines a subtype of ALK- ALCL2; see scenario 4E in Table 3 | Treatment may be adapted according to genomic configuration with (possibly) less aggressive therapy in patients with DUSP22-R ALCL‡ | |
| TFHL angioimmunoblastic type; follicular type; NOS | TET2, DNMT3A, IDH2, RHOA mutations†: HTS (or PCR-based for RHOAG17V and IDH2R172) | Useful in certain circumstances to support the diagnosis; see scenario 4B in Table 3 | DNMT3A hotspot mutation may be predictive of nonresponse to standard chemotherapy and associated with adverse prognosis471 | |
| PTCL, NOS | Mutations and small indels in genes recurrently altered: HTS | Demonstration of genomic alterations useful in certain circumstances to establish clonality and support the diagnosis | Adverse prognostic impact of higher mutation load, complex genomic imbalances, TP53 mutations, and Th2 molecular subgroup280,281,284 | WGS, cytogenetics or array-based determination of SVs Gene expression–based subtyping469 (or IHC surrogate472) for risk stratification and patient selection |
| HSTCL | I(7q), trisomy 8†: FISH or cytogenetics INO80, PIK3CD, SETD2, STAT5B, STAT3, TET3, SMARCA2 mutations†: HTS |
Useful in certain circumstances to support the diagnosis; see scenario 4C in Table 3 | ||
| ENKTCL, nasal type | CD274 SVs and amplifications: HTS | Useful in certain circumstances for prediction of response to PD1 inhibitors329, 330, 331, 332 | Integrated HTS and TME analysis for disease stratification and guiding treatment decisions326,333 | |
| Adult T-cell leukemia/lymphoma | Clonal HTLV-1 integration: HTS | Useful in certain circumstances to support the diagnosis in HTLV-1 carriers | Disease follow-up and clonal evolution340,473 | HTS to assess risk of transformation in HTLV-1 carriers and guide treatment decisions340 |
| Mutations in genes related to immune function, signaling, cell cycle: HTS | Useful in certain circumstances for prognostic or predictive value. CCR4 mutations predictive of response to mogamulizumab.344,345 Some alterations indicative of unfavorable prognosis (TP53 or PRKBC mutations; TcR/NF-κB pathway alterations in the indolent subtype)340,346,474 |
|||
| T-LGLL and NK-LGLL | STAT3 and STAT5B mutations†: HTS | Useful in certain circumstances to support the diagnosis; see scenario 4C in Table 3 | STAT3 mutations relate with neutropenia | |
| T-cell prolymphocytic leukemia | inv(14)(q11q32), t(14;14)(q11;q32), t(X;14)(q28;q11), trisomy 8: FISH (TCL1A or MCTP1) or cytogenetics∗ |
Strongly recommended for establishing the diagnosis; see scenario 4C in Table 3 | Prognosis: complex karyotype (≥3 aberrations) indicative of less favorable prognosis366 |
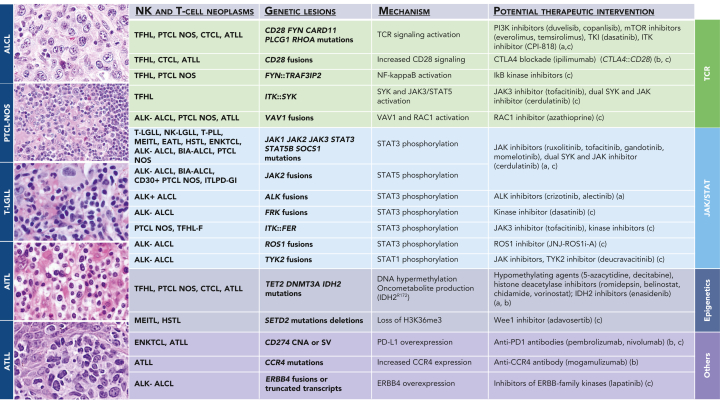
Figure 5 shows the potential therapeutic targeting of specific genetic alterations that may be common to several T/NK-cell neoplastic entities.
ALCL, anaplastic large-cell lymphoma; ALK, anaplastic lymphoma kinase; ENKTCL, extranodal NK/T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; HTLV, human T-lymphotropic virus; LPD, lymphoproliferative disorder; NK-LGLL, chronic lymphoproliferative disorder of natural killer cells; TFHL, follicular helper T-cell lymphoma; T-LGLL, T-cell large granular lymphocytic leukemia.
Required/strongly recommended in the National Comprehensive Cancer Network 2022 guidelines.
Useful in certain circumstances in the National Comprehensive Cancer Network 2022 guidelines.
National Comprehensive Cancer Network 2022 treatment guidelines.
Table 3.
Utility of genomic testing in selected diagnostic settings
| Diagnostic scenario | Genomic testing |
|---|---|
| Scenario 1: Small B-cell lymphomas | |
| 1A: CD5-positivesmall B-cell lymphoma: SLL/CLL; MCL; CD5-positive MZLs | Demonstration of CCND1, CCND2, or CCND3 rearrangement establishes the diagnosis of MCL; demonstration of BCL2 rearrangement is rare in SLL/CLL and favors FL. Overlapping and heterogeneous mutational landscapes; mutations in the following genes have the most discriminant value: ATM, BIRC3, MEF2B (favor MCL); BRAF, KLF2, NOTCH2, and PTPRD (favor MZLs), NOTCH1, SF3B1, XPO1 (favor SLL/CLL) |
| 1B: CD5-negative, CD10-negative,BCL2-R–negative small B-cell lymphoma: MZLs (including pediatric type); BCL2-R–negative, CD23-positive follicle center lymphoma; FL (without BCL2-R); hairy cell leukemia (tumor presentation) | Demonstration of BCL6 rearrangement or 1p36 deletion favors FL. Overlapping and heterogeneous mutational landscapes; mutations in the following genes have the most discriminant value: KLF2, NOTCH2, PTPRD, CARD11, IRF8, MAP2K1 (favor MZLs and pediatric-type MZL); CREBBP, EZH2, TNFRSF14 (in FLs), STAT6 (favor BCL2-R–negative, CD23-positive follicle center lymphoma); BRAF (in virtually all hairy cell leukemias, also in some MZLs) |
| 1C: Cutaneous involvement by follicular B-cell lymphoma: primary cutaneous follicle center lymphoma; systemic FL | Demonstration of BCL2 rearrangement favors systemic FL but does not exclude primary cutaneous follicle center lymphoma. Mutational landscapes overlap with less frequent incidence of mutations in BCL2, CREBBP, EP300, EZH2, KMT2D more frequent mutations in TNFAIP3, and similar occurrences of TNFRSF14 mutations or 1p36 deletions in primary cutaneous vs systemic cases |
| Scenario 2: B-cell neoplasms with plasmacytic differentiation and plasma cell neoplasms | |
| 2A: Small B-cell lymphoma with plasmacytic differentiation: LPL; nodal MZLs; splenic MZL; extranodal MZL (MALT lymphoma); FL | Demonstration of BCL2 rearrangement supports the diagnosis of FL. Demonstration of trisomies of chromosomes 3 and 18 or del(7q) supports the diagnosis of MZL. Translocations of MALT1, FOXP1, and BCL10 are specific for MALT lymphomas. MYD88L265P mutation is highly suggestive of LPL but not entirely specific because it is also found in a subset of other small B-cell lymphomas. Coexisting CXCR4 mutation further increases the specificity for LPL. Overlapping and heterogeneous mutational landscapes; mutations in the following genes have the most discriminant value: MYD88 and CXCR4 (favor LPL); BRAF, KLF2, NOTCH2, PTPRD, TNFAIP3 (favor MZLs); CREBBP, EZH2, TNFRSF14 (favor FL) |
| 2B: Bone marrow withIgM-secretingneoplasm: IgM MGUS, plasma cell type; IgM MGUS, NOS; LPL; IgM plasmacytoma; IgM plasma cell myeloma | Demonstration of translocations of CCND or MAF family genes or NSD2 indicates a plasma cell neoplasm. Mutational landscapes are distinct with MYD88L265P mutation present in most LPL and MGUS, NOS; other discriminant mutations involve ARID1A, CD79B, CXCR4, KMT2D (in lymphoplasmacytic neoplasms) and BRAF, DIS3, KRAS, NRAS, TENT5C, and TRAF3 (in plasma cell neoplasms). Genomic testing does not resolve the differential diagnosis of MGUS vs lymphoma or myeloma |
| 2C: Small B-cell lymphoma, with spleen, bone marrow, or blood involvement: splenic MZL; hairy cell leukemia; splenic diffuse red pulp small B-cell lymphoma; hairy cell leukemia variant; MCL | Demonstration of CCND1 rearrangement establishes the diagnosis of MCL. Detection of del(7q) is not discriminant in this context. Mutational landscapes are distinct with BRAFV600E mutation being a highly diagnostically sensitive marker for hairy cell leukemia, although not entirely specific; other mutations supportive of diagnosis in this context include MAP2K1 mutations (favor hairy cell leukemia variant); those in KLF2 and NOTCH2 (favor splenic MZL); and those in BCOR and CCND3 (favor splenic diffuse red pulp small B-cell lymphoma) |
| 2D:EBV-negativeplasmablastic neoplasm: plasmablastic lymphoma; plasmablastic MM; ALK-positive DLBCL | Demonstration of translocations of CCND or MAF families or NSD2 indicates an MM; ALK translocations (generally substituted by IHC) define ALK-positive DLBCL. Demonstration of MYC rearrangement while supporting the diagnosis of plasmablastic lymphoma does not exclude plasmablastic MM. Overlapping and heterogeneous mutational landscapes; mutations in the following genes more frequent in plasmablastic lymphoma: EP300, MYC, SOCS1, STAT3, TET2, and TP53 |
| Scenario 3: LBCLs | |
| 3A:Nodal-basedfollicular B-cell lymphoproliferations with a predominance of large cells in the pediatric population: pediatric-type FL; follicular hyperplasia; LBCL-IRF4 rearrangement; in adults: FL grade 3A; FL grade 3B; LBCL-IRF4 rearrangement | Demonstration of monoclonal IG gene rearrangement is useful to establish the diagnosis of lymphoma over reactive hyperplasia, in particular in pediatric conditions. Demonstration of BCL2 rearrangement favors grade 3A over grade 3B FL and excludes pediatric entities. BCL6 rearrangement occurs in both grade 3A and 3B cases, more commonly in 3B, but not in pediatric-type FL Demonstration of IRF4 (or IGH, IGK or IGL) rearrangements is essential for supporting LBCL-IRF4 rearrangement; demonstration of one or several IRF4 mutations in exon 1-2 is a strong indicator of IRF4 rearrangement including cryptic translocation. IRF4 rearrangement can be present in association with other rearrangement(s) (BCL2 or MYC) in DLBCLs, and these do not qualify for LBCL-IRF4. Overlapping and heterogeneous mutational landscapes; mutations in the following genes have the most discriminant value: IRF8 and MAP2K1 (pediatric-type FL; note that the same mutations are found in pediatric nodal MZL); IRF4 and MYC (LBCL-IRF4); CARD11 (LBCL-IRF4 and FL, not in pediatric-type FL); BCL2, CREBBP, EZH2, and KMT2D (FL) |
| 3B: Aggressive mature B-cell lymphomas: BL; LBCL with 11q aberration; HGBCL (NOS; with MYC and BCL2 rearrangements; with MYC and BCL6 rearrangements); DLBCL, NOS | Demonstration or exclusion of MYC, BCL2, and/or BCL6 rearrangements or 11q aberrations, are essential in this differential diagnosis and should be applied according to the algorithm presented in Figure 4. Mutations in ID3 and TCF3 favor BL whereas B2M, CREBBP, EZH2, MYD88L265P, SOCS1, and TNFRSF14 mutations favor other aggressive B-cell entities. Similarly, BCL2 mutations imply the presence of IGH::BCL2, thereby favoring entities other than BL |
| 3C: LBCL involving mediastinum: PMBCL; DLBCL, NOS involving mediastinum; mediastinal gray-zone lymphoma | Demonstration of BCL2 or BCL6 rearrangement favors DLBCL, NOS, as these uncommonly occur in PMBCL; conversely, CIITA rearrangement, CD274 rearrangement or CNV are typical of primary mediastinal lymphomas. Mutations in IL4R, ITPKB, NFKBIE, SOCS1, STAT6, and XPO1 are characteristic of PMBCL, while several genes often mutated in DLBCL, NOS, such as CD79B, CREBBP, KMT2D, MYD88, PIM1, and others, are not altered in PMBCL. Mediastinal gray-zone lymphoma has genomic features closer to PMBCL than to DLBCL, NOS, but distinctive genomic features between mediastinal gray-zone lymphoma and PMBCL are not described. Gene expression–based tests differentiate PMBCL from DLBCL, NOS |
| 3D: Cyclin D1–positive blastoid or pleomorphic B-cell neoplasm: MCL; DLBCL, NOS positive for cyclin D1 expression; DLBCL, NOS with CCND1 rearrangement | Demonstration of CCND1 translocation indicates MCL or DLBCL with CCND1 rearrangement. Demonstration of additional BCL2, BCL6, or MYC rearrangement is common in DLBCL with CCND1 translocation. Blastoid MCL may harbor secondary MYC rearrangement or TP53 mutations. Mutations in ATM, BIRC3, NSD2, and UBR5 support mantle cell lymphoma |
| Scenario 4: T-cell lymphoproliferations | |
| 4A: Hodgkin/Reed-Sternberg(–like) cells in a T-cell background: CHL; nodular lymphocyte-predominant B-cell lymphoma; T-cell/histiocyte-rich LBCL; TFHL; PTCL NOS. | Clonality testing for IG and TR rearrangements is useful in the differential diagnosis because a monoclonal TR rearrangement supports a diagnosis of T-cell lymphoma and argues against CHL or B-cell lymphomas; conversely, monoclonal IG rearrangements may be variably demonstrated in CHL, nodular lymphocyte-predominant B-cell lymphoma, and T-cell/histiocyte-rich LBCL as well as in PTCLs with an associated B-cell component (more often present in TFHLs). Demonstration of mutations in genes commonly mutated in T-cell lymphomas (CARD11, CD28, DNMT3A, IDH2, PLCG1, RHOA, STAT3, and TET2) supports that diagnosis; caution is required when interpreting mutations present only in TET2 and/or DNMT3A, which can be related to CH |
| 4B: Expansions of T cells with follicular helper phenotype: reactive TFH cells in benign lymphadenopathies; reactive TFH cells in small B-cell lymphomas; early involvement by TFHL | Demonstration of a monoclonal TR gene rearrangement or somatic mutations in other genes is useful in the distinction between reactive vs neoplastic expansions of TFH cells. Demonstration of mutations in genes commonly mutated in TFHL (most specific: IDH2 and RHOA; others: CARD11, CD28, DNMT3A, PLCG1, and TET2) supports TFHL; caution is required when interpreting mutations present only in TET2 and/or DNMT3A, which can be related to CH and are not per se indicative of a T-cell neoplasm; in cases of reactive TFH expansions, the presence of mutations in genes related to B-cell lymphomas favor MZLs or FLs |
| 4C:EBV-negativecytotoxic T-lymphocytosis in blood, bone marrow, or spleen: T-LGLL; HSTCL; reactive T-cell expansions | Monoclonal TR gene rearrangements or somatic mutations (PIK3CD, SETD2, STAT3, STAT5B, and TNFAIP3) favor neoplasia over reactive expansions. Isochromosome 7q is characteristic of HSTCL. Mutations in the following genes may help differentiating between HSTCL (CD8−/+ Tαβ or Tγδ) and CD8+Tαβ or Tγδ−LGLL: SETD2 (exclusive to HSTCL), STAT3 (less common in HSTCL than in T-LGLL), STAT5B (less common in T-LGLL than in HSTCL) |
| 4D: Intestinal T-cell lymphoproliferations: RCDII; EATL; MEITL; intestinal T-cell lymphoma, NOS; indolent gastrointestinal lymphoproliferative disorders | Demonstration of a monoclonal TR rearrangement is useful in the distinction of (type I refractory) celiac disease and RCDII, as well as for distinguishing indolent clonal T-lymphoproliferative disorders from prominent inflammatory infiltrates. T-cell or NK-cell lymphoproliferations are further supported by somatic mutations or fusions (STAT3, JAK3, JAK2::STAT3, others). Most discriminant mutated genes between EATL and MEITL are JAK1 and STAT3 (more commonly mutated in EATL) and GNAI2, JAK3, SETD2, and STAT5B (more commonly mutated in MEITL) |
| 4E: Lymphoproliferations of large CD30-positiveT cells: ALCL, ALK-positive; ALCL, ALK-negative; BIA-ALCL; PTCL, NOS; primary cutaneous CD30-positive lymphoproliferative disorders; transformed mycosis fungoides; subsets of EATL, or ENKTCL | Demonstration of ALK rearrangement (generally substituted by IHC) defines ALCL, ALK-positive. Demonstration of DUSP22 rearrangement in ALK-negative CD30-positive large-cell lymphoproliferations establishes the diagnosis of ALCL, ALK-negative, over PTCL, NOS, but does not discriminate between primary cutaneous vs systemic ALCL, ALK-negative. VAV1 and TP63 rearrangements occur in small subsets of ALCL, ALK-negative but are not specific for that entity. Demonstration of ALK, DUSP22, or TP63 translocations exclude BIA cases, whereas chromosome 20q loss is characteristic of that entity. Overlapping and heterogeneous mutational landscapes, including mutations in STAT3 and JAK1, are common to several entities |
| Scenario 5: Successive neoplasms | |
| Clonal relationship between successive hematologic neoplasms | Analysis of IG or TR gene rearrangements helps to distinguish between clonally related and clonally unrelated neoplasms and to establish transdifferentiation in cases of secondary histiocytic/dendritic cell neoplasms; interpretation may be ambiguous in cases of clonal evolution; sequencing-based clonality assays provide more precise results in that setting. Analysis of somatic mutations provides information on linear vs divergent evolution and secondary genomic alterations |
Refer to supplemental Figure 1 and supplemental Table 1 for prevalence of genetic aberrations in the major entities.
BIA, breast implant–associated; CHL, classic Hodgkin lymphoma; EATL, enteropathy-associated T-cell lymphoma; EBV, Epstein-Barr virus; MEITL, monomorphic epitheliotropic intestinal T-cell lymphoma; MGUS, monoclonal gammopathy of undetermined significance; PMBCL, primary mediastinal large B-cell lymphoma; PTCL, peripheral T-cell lymphoma; RCDII, type II refractory celiac disease.
Mature B-cell neoplasms
Chronic lymphocytic leukemia/small lymphocytic lymphoma
Molecular genetic characterization guides management of newly diagnosed patients with CLL/SLL. The somatic hypermutation (SHM) status of the clonally rearranged immunoglobulin heavy variable (IGHV) gene should be determined once as it remains constant through the disease course.40,41 Compared with IGHV-mutated (M) CLL (<98% identity compared with germ line sequence), IGHV-unmutated (U) CLL has shorter time-to-therapy initiation, remission duration with therapy, and overall survival (OS).42 In patients without TP53 aberration, chemoimmunotherapy remains a reasonable first-line treatment option for patients with M-CLL, whereas initial targeted therapy (BTK inhibitors [BTKis], BH3 mimetic with anti-CD20 antibody) has become the standard of care for U-CLL.43, 44, 45, 46, 47 Stereotyped B-cell receptors (BcRs) occur in 41% of cases, with some subsets having distinctive outcomes (supplemental Table 2).48, 49, 50, 51, 52, 53
Among CNAs routinely detected by FISH (del(11q), trisomy 12, del(13q), and del(17p)),54 del(17p) confers a poorer prognosis and predicts suboptimal responses to chemoimmunotherapy. Complex karyotype (≥5 abnormalities) confers poor outcome, even with targeted therapies,55 and can be detected by cytogenetics or SNP arrays.56,57
Driver mutations differ in U-CLL and M-CLL (supplemental Figure 2) and affect cellular signaling pathways: BcR (IGLV3-21R110),58,59 TLR (MYD88), NF-κB (BIRC3), NOTCH (NOTCH1, FBXW7), DNA damage response (ATM, TP53), RNA processing (SF3B1, XPO1), and chromatin modification (H1-4, ZMYM3, CHD2).52,60 Although TP53 aberrations are typically biallelic (ie, del(17p) and TP53 mutation), isolated TP53 mutations can also occur in the absence of del(17p). Both clonal (variant allele frequency > 10%) and subclonal (<10%) TP53 mutations61,62 are associated with poor chemoimmunotherapy response. TP53 aberrations can arise at relapse/progression and thus, if not previously identified, should be evaluated before each course of therapy. ATM mutations are associated with poor, nondurable responses to DNA-damaging chemotherapy.63 Mutations of BTK, PLCG2, and CARD1164,65 and of BCL266 have been associated with resistance to BTKi and venetoclax, respectively (supplemental Table 3).
Epigenetic alterations can also affect CLL outcomes.67 However, none are routinely tested clinically. HTS and flow cytometry have been applied to the quantification of measurable residual disease (MRD) and might guide therapy in the future.68,69 Consensus recommendations were recently published.70
Follicular lymphoma
The classic form of FL, in situ follicular neoplasia, and duodenal-type FL are all characterized by the t(14;18)(q32;q21) IGH::BCL2 translocation deregulating BCL2 expression.71 FL pathogenesis involves a complex network of genetic, epigenetic, and microenvironmental factors, driven by (1) recurrent mutations in genes encoding, in particular, several epigenetic regulators (eg, CREBBP, KMT2D, EZH2), as well as transcription factors (eg, MEF2B, FOXO1, STAT6) and components of the mechanistic target of rapamycin (mTOR) signaling pathway (eg, RRAGC, ATP6V1B2); and (2) perturbations in interactions with their immune environment (eg, TNFRSF14 inactivation, N-glycosylation sites in the IGV genes).71, 72, 73, 74, 75, 76, 77, 78 However, identifying these lesions and combinations thereof,79 along with GEP signatures,80 has not yet entered routine testing, given the lack of reproducible prognostic or predictive value at diagnosis for patients treated with standard chemoimmunotherapy. FDA approval mandates EZH2 mutation detection for the treatment with the EZH2 inhibitor tazemetostat81 in patients having received at least 2 previous systemic therapies, but this is not required for those lacking alternative options in later treatment lines.
Molecular analyses may assist in the differential diagnosis of BCL2-translocation–negative FLs,82 which share genetic alterations with nodal FL although at different frequencies (BCL2R-negative CD23+ follicle center lymphoma, primary cutaneous follicle center lymphoma, pediatric-type FL, testicular FL, and large B-cell lymphoma with IRF4 rearrangement [LBCL-IRF4]) and in distinguishing primary vs secondary cutaneous disease83,84 (Table 3).
Phylogenetic analyses of spatial and temporally acquired mutations in t(14;18)-positive cells revealed a marked heterogeneity inferring the existence of a long-lived common mutated precursor B-cell population that is capable of evading treatment and seeding new episodes of disease.85, 86, 87 Current challenges include characterizing this precursor B cell, identifying molecular predictors of early relapse/histologic transformation, and recognizing better stratification factors in the context of a rapidly evolving therapeutic landscape.
Marginal zone lymphomas
Extranodal (MALT), nodal (N), and splenic (S) MZLs have distinct genetic changes. Nevertheless, they commonly affect signaling pathways central to the homeostasis of normal MZ B cells, including BcR, NF-κB, and NOTCH.88
MALT lymphomas have distinct genomic alterations according to their primary anatomic site.89,90 The t(11;18)(q21;q21) BIRC3::MALT1 fusion occurs most often in gastric and pulmonary MALT lymphomas.89 This rearrangement is more common in Helicobacter pylori–negative gastric MALT lymphomas and is associated with a lack of antibiotic response in H pylori–positive cases.91 The t(14;18)(q32;q21) IGH::MALT1 translocation is usually found in lung and ocular adnexa MALT lymphomas.89 The t(3;14)(p14.1;q32) FOXP1::IGH translocation associates with thyroid and ocular adnexa MZL and primary cutaneous marginal zone lymphoproliferative disorder (LPD).89 The t(1;14)(p22;q32) BCL10::IGH translocation is found in gastric and lung MALT lymphomas and skin MZL-LPD.89 Mutations of TNFAIP3 are reported in all types of MZL, but enriched in ocular adnexa MALT lymphoma. Mutations of FAS are enriched in primary cutaneous MZ-LPDs.90
MALT lymphoma translocations are lacking in SMZL and NMZL. SMZL shows hemizygous deletion of 7q31-3292 and, rarely, translocations juxtaposing CDK6 to IG loci.93 SMZL and NMZL have a common genetic background characterized by mutations of NOTCH genes (eg, NOTCH2, NOTCH1, SPEN), genes involved in noncanonical NF-κB signaling (eg, BIRC3, TRAF3), and KLF2, a master regulator of both NOTCH and NF-κB signaling.94 Among MZLs, BRAF and PTPRD mutations are nearly exclusive to NMZL.95,96 SMZL comprises 2 main genetic clusters, characterized by mutations affecting NF-κB, NOTCH, and KLF2 (NNK), or by mutations of TP53, MAPK, and TLR (DMT).97 Cytogenetic and molecular features can assist in the differential diagnosis of MZL and other small B-cell lymphomas (Table 3; supplemental Figure 3).
Mantle cell lymphoma
MCL includes 2 subtypes: conventional MCL (cMCL) and the less common leukemic non-nodal MCL (nnMCL). Both share rearrangements involving CCND1, and less frequently CCND2 or CCND3, mainly with IGH or IG light chain loci.98,99 FISH break-apart probes are recommended for the detection of these rearrangements, although CCND1 immunohistochemistry (IHC) typically obviates the need for CCND1 FISH. Identification of uncommon cryptic translocations requires specific probes or HTS analysis.99, 100, 101 cMCL derives from naïve-like B cells, carries unmutated IGHV genes, and has a different expression profile with typically high SOX11 levels. nnMCL originates in memory-like B cells, carries mutated IGHV genes, and is typically SOX11 negative.102,103 In both subtypes, CCND1 rearrangement is acquired in B-cell precursors mediated by RAG activity, although in a minority of cases, it occurs in mature B cells by IG class-switch and AID-driven mechanisms.104 cMCL carries frequent (>15%) mutations in ATM, KMT2D, TP53, BIRC3, and the 3′ untranslated region (3′-UTR) of CCND1 leading to higher oncogene expression. Less common mutations (5%-15%) occur in NSD2, NOTCH1/2, HNRNPH1, CARD11, SP140, and SMARCA1, among others. The most common mutations in nnMCL are CCND1 in the 5′-region (mediated by AID) and TP53.104, 105, 106, 107, 108 MCL, particularly blastoid/pleomorphic of both subtypes, accumulates numerous and complex genomic structural alterations that worsen the prognosis, with TP53, CDKN2A deletions, and MYC rearrangements being of particular impact.104,109,110 TP53 aberrations are associated with poor prognosis in patients undergoing chemoimmunotherapy and autologous stem cell transplant; future studies should focus on this very-high-risk group.103,111, 112, 113, 114, 115, 116 The proliferation signature defines patients with different clinical trajectories.117, 118, 119 Resistance to BTK or BCL2 inhibitors due to acquired BTK or BCL2 mutations is uncommon in MCL but may involve alterations in other genes and transcriptome reprogramming with overexpression of OxPhos, MYC, alternative NF-κB, and mTOR pathways.120, 121, 122, 123
Multiple myeloma
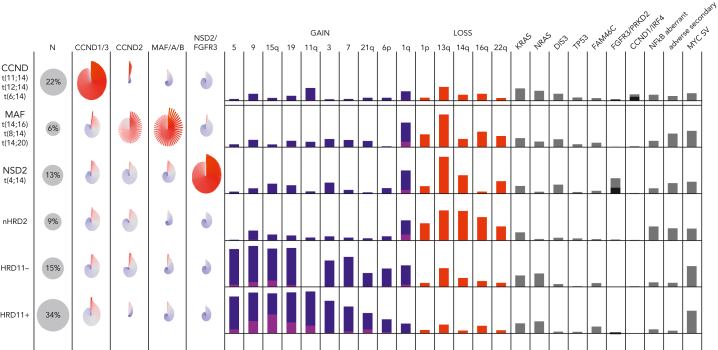
Classification of MM is based on primary abnormalities invariant through disease progression from monoclonal gammopathy of uncertain significance (MGUS) to smoldering (S)MM to MM.2 Moreover, genomic profiling is important for risk stratification in which adverse genetic events may be acquired during disease progression. There are 5 nonoverlapping disease subgroups: (1) CCND family translocation, (2) MAF family translocation, (3) NSD2 translocation, (4) hyperdiploid (gains of chromosome 3, 5, 7, 9, 11, 15, 19, and 21), and (5) MM-NOS, lacking all the preceding features.124, 125, 126, 127 In the future, the hyperdiploid group will likely be further subdivided, for instance based on the presence of trisomy 11 and CCND1 expression (Figure 2).124,125 Disease classification currently relies on FISH assays (Table 1) but can be achieved more comprehensively using GEP and/or WGS.124,126, 127, 128
Figure 2.
The molecular classification of MM. Data from the COMMpass study (clinical trial identifier: NCT0145297) are summarized, showing the 5 nonoverlapping subgroups and their associated gene expression, CNVs, SVs, and SNVs. The pinwheels show the expression of CCND, MAF, NSD2, and FGFR3 for individual patients in each group. Gains of chromosomes (or arms) are shown in blue (1 copy) or purple (>1 copy). For illustration, the hyperdiploid subgroup is further subdivided into those with (HRD11-positive) and without (HRD11-negative) trisomy 11, and the patients without translocations or hyperdiploidy are labeled nHRD2 (MM, NOS). Mutations of FGFR3 (black) and PRKD2 (gray) are common in NSD2, whereas mutations of CCND1 (black) and IRF4 (gray) are common in CCND. A variety of different mutations can activate NF-κB (TRAF3, BIRC2/3, and others). Adverse secondary events include biallelic inactivation of CDKN2C, TP53, or RB1. MYC SVs are most common in hyperdiploid MM.
Adverse risk is associated with specific primary genetic events (t(4;14)(p16;q32) NSD2::IGH, t(14;16)(q32;q23) IGH::MAF) and, beyond these subgroups, secondary genetic events (1q gain/amplification, del(1p), del(17p), and TP53 mutation).129,130 Not all therapies have been shown to benefit patients with high-risk genetics; however, a prolongation of progression-free survival (PFS) is seen with the addition of a proteosome inhibitor for patients with t(4;14) or del(17p) or of daratumumab or tandem stem cell transplant for these patients.131, 132, 133 These data strongly support the use of a quadruplet regimen (an anti-CD38 antibody, a proteosome inhibitor, a thalidomide analog, and a glucocorticoid) for the treatment of newly diagnosed high-risk MM. Genetics can also help guide therapy for standard-risk patients. For example, relapsed patients with t(11;14) benefit from treatment with venetoclax, an effect not observed in the cohort overall.134 Much more prognostic information can be obtained from high-risk scores based on GEP (GEP70, EMC92), reflecting important biological aspects of the disease, such as proliferation.135,136 WGS adds information about clonal heterogeneity, focal CNAs, chromothripsis, and important SVs, such as those involving MYC, present in almost half of newly diagnosed MM.137, 138, 139, 140, 141, 142 This may perhaps be most relevant for patients with SMM, where the presence of genetic events associated with adverse prognosis in newly diagnosed MM (t(4;14), t(14;16), add(1q), del(17p)), as well as others that are not (del(13q), NRAS and KRAS mutations, MYC rearrangements) are associated with more rapid progression to symptomatic MM and may become the basis for a genetic definition of MM requiring treatment.139,143, 144, 145, 146, 147
Lymphoplasmacytic lymphoma
Somatic mutations in MYD88 (MYD88Mut) and CXCR4 (CXCR4Mut) occur in 95% to 97% and 30% to 40% of patients with IgM-secreting LPL (Waldenström macroglobulinemia; WM/LPL), respectively,148 and in 50% to 90% and 10% to 20% of patients with IGM MGUS, respectively.148, 149, 150, 151 In IgM MGUS, NOS, nearly all individuals with MYD88Mut will progress to WM/LPL.150 Up to 80% of non–IgM-secreting LPL also harbor MYD88Mut.152 Nearly all MYD88Mut IgM and non-IgM LPL and IgM MGUS, NOS cases express the L265P variant, though rarely non-L265P variants have been identified.148,150, 151, 152 MYD88Mut triggers BTK-directed NF-κB prosurvival signaling, whereas CXCR4 mutations trigger extracellular signal-regulated kinase (ERK) and protein kinase B (AKT) signaling relevant to drug resistance, particularly BTKi.148 In LPL/WM, MYD88 and CXCR4 mutations affect disease presentation, prognosis, time-to-treatment initiation, and/or treatment outcome.148,153 Patients with wild-type MYD88 (MYD88WT) have NF-κB pathway activating mutations overlapping those found in DLBCL, are at higher risk of disease transformation and/or death, and show decreased response activity and/or shorter PFS following treatment with BTKi and bendamustine/rituximab.148,153, 154, 155, 156, 157 Zanubrutinib shows major response activity in MYD88WT and can be considered.158 Other B-cell malignancies, including IgM-secreting MM, can be confused with MYD88WT WM and should be ruled out because management can differ.155 MYD88 mutation status should ideally be determined by allele-specific (AS) PCR-based diagnostics because HTS may miss up to one-third of MYD88Mut WM/LPL, particularly those with low bone marrow disease burden.159 CXCR4 mutations are typically subclonal and affect the depth of response, time to attainment of major responses, and/or PFS following ibrutinib or zanubrutinib.148,149,160, 161, 162 Over 40 nonsense and frameshift variants of CXCR4Mut are described.149,153,163 Nonsense variants (most commonly CXCR4S338X), are particularly associated with high serum IgM levels, symptomatic hyperviscosity, shorter time-to-treatment initiation, lower response activity and shorter PFS on ibrutinib, and shorter OS.153,164,165 Up to two-thirds of CXCR4Mut may be missed by HTS, particularly those with low disease burden and low variant allele frequencies.166 CXCR4 antagonists are being investigated in WM/LPL. Heterozygous loss of 6q, present in up to half of the patients with WM, is mutually exclusive of CXCR4Mut and includes regulatory genes of BTK (IBTK), BCL2 (BCLAF1), NF-κB (HIVEP2, TNFAIP3), and apoptosis (FOXO3).148,167 Following ibrutinib treatment, tumor evolution leading to biallelic del(6q) has been observed.168 The BTKC481 mutation has been observed in patients with WM/LPL with acquired resistance to ibrutinib, particularly those with CXCR4Mut.169 TP53 mutations are rare in WM/LPL and are associated with poor outcomes, though patients carrying these mutations respond to ibrutinib.170,171
Diffuse large B-cell lymphoma
The molecular subclassification of DLBCL is key to understanding therapeutic efficacy. Currently, LBCL should be evaluated for rearrangements, typically by FISH (though imperfect172), to identify high-grade B-cell lymphoma (HGBCL) with MYC and BCL2 rearrangements, which responds poorly to R-CHOP chemotherapy, and the provisional entity HGBCL with MYC and BCL6 rearrangements.2 The activated B-cell–like (ABC) and germinal center B-cell–like (GCB) DLBCL COO subtypes25 should be distinguished by GEP,173 or approximated by IHC,174 providing useful prognostic information.26,175 ABC-DLBCLs typically rely upon BcR-dependent NF-κB signaling for survival, engendering sensitivity to BTK inhibition.176, 177, 178, 179 Younger, newly diagnosed patients with ABC-DLBCL may benefit from the addition of a BTKi to R-CHOP,180,181 although this requires validation. However, recent clinical studies suggest that the binary COO classification is insufficiently granular to predict the efficacy of all precision medicine strategies.182,183
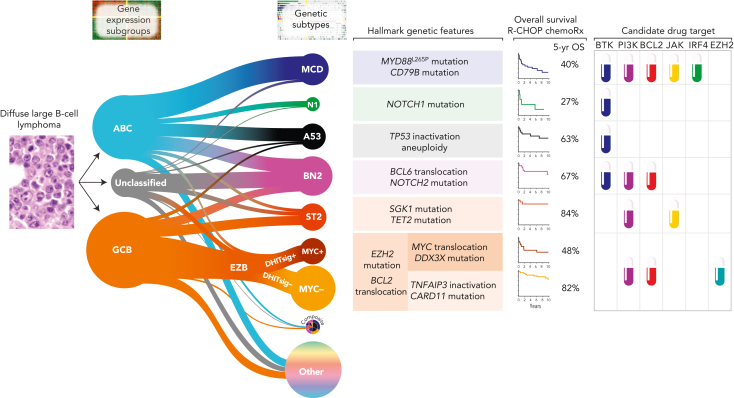
An important refinement and extension of the DLBCL COO classification emerged from 3 independent studies184, 185, 186 that used multiplatform genomic profiling to detect patterns of co-occurring genetic alterations, converging on ∼7 subtypes with recurrent biological features (Figure 3). Several DLBCL genetic subtypes share core genomic alterations with indolent B-cell lymphomas, suggesting that some apparently de novo DLBCL may arise from clinically occult indolent lymphomas and that the evolutionary paths of DLBCL and indolent lymphomas share key driver events at their inception. The MCD DLBCL subtype encompasses genetically related primary extranodal entities, including primary DLBCL of the central nervous system and of the testis, among others, reflecting shared biology typified by BcR signaling and escape from immune recognition.187 The genetic subtypes, with distinct outcomes following R-CHOP, reveal oncogenic pathways that suggest therapeutic vulnerabilities, providing a framework for future drug development. For example, the addition of a BTKi to R-CHOP may be particularly beneficial in the MCD and N1 genetic subtypes.181
Figure 3.
Genetic subgroups of DLBCL illustrated using the LymphGen algorithm. The relationships between COO and the probabilistic assignments to genetics-based subgroups are shown. The size of the subgroup circles approximates the proportions of patients in each group, with the prevalence based on Schmitz et al,185 adjusted for a population-based distribution of COO subgroups. Tumors assigned with high confidence to ≥2 subgroups are assigned to the composite group, while ∼37% of tumors are not assigned to any subgroup with sufficient confidence (other). The hallmark genetic features are those frequent within that subgroup but are not required for that assignment. OS following R-CHOP chemoimmunotherapy along with inferred drug targets are shown. GCB, germinal center B-cell–like.
One publicly available approach to assign individual DLBCL tumors to genetic subtypes is the LymphGen algorithm, which performed comparably in 4 independent DLBCL cohorts.187,188 One subtype, EZB, is further subdivided into MYC+ and MYC– subtypes based on a GEP signature29 that reflects germinal center dark vs light zone origin and MYC target gene expression.187 LymphGen classifies ∼63% of DLBCL tumors, with ∼6% assigned to more than 1 subtype, indicating a compound pathogenesis. A key task ahead is to understand how to categorize the remaining 37% of DLBCLs that are unassigned using LymphGen. Some may represent rare, undescribed subtypes, whereas others may be classifiable into existing subtypes using WGS, GEP, epigenetic profiling, and analysis of the TME.
Given the above, efficient progress toward precision medicine for DLBCL will require the incorporation of genetic profiling in future clinical trials. At a minimum, this would entail WES (or WGS), analysis of MYC, BCL2, and BCL6 rearrangements, and WTS to gauge the phenotype of the malignant cells and TME, both of which provide prognostic information.26,34,175,189 Initially, this molecular profiling will likely be performed retrospectively, but our deepening understanding of the therapeutic vulnerabilities of each genetic subtype will foster clinical trials that use genomic profiling to stratify patients into treatment arms.
High-grade B-cell lymphomas
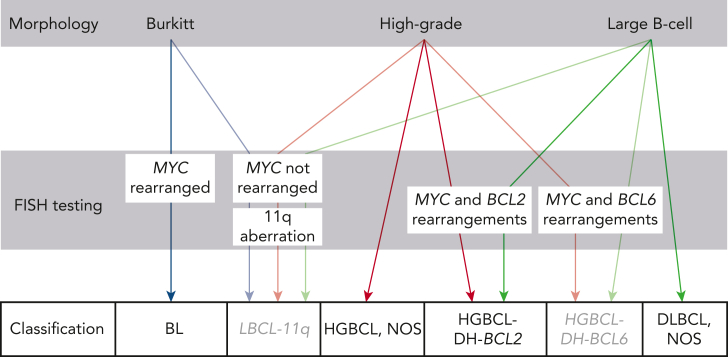
HGBCL with MYC and BCL2 (with or without BCL6) rearrangement (HGBCL-DH-BCL2) is diagnosed by detecting these rearrangements in tumors with high-grade or large B-cell morphology (Figure 4).2 This is typically achieved using break-apart FISH probes, although up to 20% of diagnoses may be missed using this approach.172 The partner gene for MYC is an IG locus in approximately half of HGBCL-DH-BCL2.190,191 The existence of “cryptic” rearrangements and the potential prognostic implication of partner loci190,191 may lead to capture-based rearrangement detection supplanting FISH.192,193
Figure 4.
Approach to diagnosing HGBCL. Lymphomas that potentially fall into the HGBCL categories can have high-grade (blastoid or intermediate [between BL and large-cell]) morphology or resemble DLBCL. Tumors with morphology resembling BL and other HGBCL are assigned to the provisional entity LBCL with 11q aberration (LBCL-11q) if they lack MYC rearrangement and have 11q aberration. The full morphological spectrum of cases with this aberration requires further study. Other cases in this category present with large-cell morphology. Tumors should not be assigned to LBCL-11q if they harbor concurrent MYC and BCL2 or MYC and BCL6 rearrangements. Tumors with morphology resembling BL and an immunophenotype consistent with BL, lacking both MYC rearrangement and 11q aberration, are likely diagnosed as HGBCL, NOS, acknowledging that rare MYC rearrangements cryptic to FISH have been observed.
The mutational landscape of HGBCL-DH-BCL2 is relatively homogeneous, with frequent mutations in BCL2, KMT2D, CREBBP, TNFRSF14, and EZH2.194, 195, 196 These mutations are frequent in FL, suggesting that these tumors either arise from (occult) FL or an FL-like precursor. In contrast, albeit based on modest numbers, the mutational landscape of HGBCL-DH-BCL6 is heterogeneous.194,196 Coupled with ∼30% harboring t(3;8)(q27;q24) BCL6::MYC (ie, “pseudo-double hit”),197 the upcoming World Health Organization classification has removed this category,198 although it is retained as a provisional category in the International Consensus Classification,2 encouraging further investigation.
HGBCL-DH-BCL2 and BL share a common GEP signature, “molecular high grade”199 or “double-hit signature” (DHITsig).29 These signatures, observed in a larger group of aggressive tumors (including EZB-MYC+187), encompass germinal center dark-zone programs. The biology of these poor prognosis “dark-zone” lymphomas requires exploration to determine whether shared targetable biology warrants defining a future lymphoma entity broader than that identified by gene rearrangements alone.
HGBCL, NOS, remains a rare category defined by morphology and lack of defining genomic rearrangements.2 The molecular landscape is very diverse200 and, where available, molecular assessment is recommended to determine whether individual tumors can be aligned or reassigned to defined entities.
Burkitt lymphoma
BL is characterized by MYC translocations, which almost exclusively involve an IG partner. MYC mutations due to aberrant activity of AID (somatic hypermutation [SHM]) are typically found, but most of them are likely inconsequential. BL in malaria-endemic regions is generally EBV positive, whereas this is less common in sporadic cases found elsewhere (adult 20%, pediatric 6%).201 BL risk is higher in patients with immunodeficiency, including individuals with HIV infection.202 Pathogenesis may vary according to the EBV status, as reflected by the higher prevalence of some driver mutations in EBV-negative tumors.203, 204, 205, 206 However, currently, EBV status and genomic characteristics do not influence treatment decisions. Potential prognostic associations have been reported for TP53 mutations, and this could eventually improve risk stratification.207 Patients with disease refractory to standard therapies represent an unmet clinical need,202 and genomic analysis has identified potential therapeutic vulnerabilities.205 Many genes recurrently mutated in BL are also drivers in other lymphomas originating from germinal center B cells, with driver mutations more prevalent in BL, highlighting distinguishing oncogenic mechanisms. These include mutations that inactivate the protein translation factor DDX3X, thereby buffering the proteotoxic stress caused by dysregulated MYC expression,208 and mutations in either TCF3 or its negative regulator ID3.205 TCF3 promotes constitutive BcR signaling that activates PI3 kinase and is essential to BL survival.205 In contrast, EZH2, CREBBP, and KMT2D mutations are rarely observed in BLs, though common in GCB DLBCL.203,204 Establishing the presence of such mutations could ultimately be combined with current criteria to improve the robustness of BL diagnosis and identify potential therapeutic targets.205
Pediatric B-cell lymphomas
Several types of B-cell lymphoma that typically occur in pediatric and young adult populations have characteristic genomic aberrations. Pediatric-type FL (PTFL) presents as localized disease, has pure follicular morphology, high proliferation, and lacks BCL2 expression and/or rearrangement. Molecular confirmation of monoclonality is crucial.4,209 Lack of cytogenetic complexity and detection of TNFRSF14 alterations and/or MAP2K1 or IRF8 mutations, in the absence of mutations in histone modifier genes, favor this diagnosis.209, 210, 211, 212, 213 The presence of IRF4, MYC, or BCL6 rearrangement exclude pediatric-type FL. Of note, pediatric nodal MZL and PTFL share clinical and morphological features, low genetic complexity, and similar mutational and methylation profiles, indicating that they are probably part of a single disease with differences in the histological spectrum.214,215
LBCL-IRF4 frequently involves the head and neck or gastrointestinal tract. Tumors are composed of large cells, with or without follicular component expressing germinal center phenotype, and moderate/high levels of MUM1/IRF4. IRF4 rearrangements are detectable by FISH break-apart probes.216,217 Rearrangements of BCL6 but not BCL2 may be observed. Frequently mutated genes include IRF4, most likely by juxtaposition to IG loci, BCL6, and NF-κB pathway genes (CARD11, CD79B, MYD88).216,218 Losses of 17p and 11q12-qter gains are characteristic.216,218,219 In tumors with consistent pathological and clinical features but FISH-negative for IRF4 rearrangement, the demonstration of IG rearrangement in the absence of BCL2, BCL6, and MYC rearrangements, and/or the presence of IRF4 somatic mutations, could support inclusion in this diagnostic category.218,220 LBCL-IRF4 also presents rarely in adults. Moreover, IRF4 rearrangement may be observed in other LBCLs in association with BCL2 and/or MYC rearrangements, and these tumors should not be classified as LBCL-IRF4.221
LBCL with 11q aberration (LBCL-11q) should be considered in tumors with high-grade/large-cell morphology, germinal center phenotype, and very high proliferation (>90%) without MYC rearrangement (Figure 4). Most LBCL-11q carry the prototypical 11q23.2-q23.3 gain/11q24-qter loss, but some have a single terminal loss or proximal gains together with terminal copy neutral loss of heterozygosity (CN-LOH).222, 223, 224, 225 In cytogenetic studies, the gained region is usually inverted.223,226 Irrespective of aberration patterns, ETS1 and FLI1 genes are included in the minimally deleted region or CN-LOH, differently to other 11q aberrations observed in DLBCL.227,228 The commercially available 11q FISH assay has limitations in detecting gain/CN-LOH and 11q-inverted-gain alteration patterns. Further genomic SV analyses to confirm the LBCL-11q diagnosis may be helpful in those cases. LBCL-11q cases have recurrent mutations in ETS1, GNA13, BTG2, and NFRKB genes and lack typical BL alterations.224,225
Hodgkin and mediastinal lymphomas
Classic Hodgkin lymphoma (CHL), primary mediastinal large B-cell lymphoma (PMBCL), and mediastinal gray-zone lymphoma (MGZL) are related diseases that share common genetic alterations, phenotypes, and clinical features, including anterior mediastinal involvement.2 The current classification does not incorporate molecular diagnostics, but several assays can be considered to increase diagnostic precision and aid biomarker development. GEP assays suitable for FFPE biopsy specimens have been developed to differentiate DLBCL from PMBCL.229, 230, 231 WES has revealed the contrasting mutational landscape of PMBCL to DLBCL and CHL (supplemental Figure 1).232,233 GEP and WES studies in MGZL confirmed the presence of genetic and phenotypic features shared with, and intermediate between, CHL and PMBCL. The predominance of T-cell and macrophage-rich TME suggests a closer relationship to HL234 and further studies are needed to refine borderline cases.235 WES analysis helped distinguish mutational profiles of MGZL (eg, B2M, TNFAIP3, GNA13 mutations) from extramediastinal (“nonthymic”) cases (supplemental Figure 1),236 the latter of which are no longer included in “gray-zone” lymphomas.1,2
GEP studies have been reported in CHL with the goal to predict outcomes after standard-of-care treatments.237,238 Overall, testing at diagnosis in adult CHL is disappointing with a lack of validation in treatment-intense239 and response-adapted240,241 trials. Outcome prediction models in relapsed CHL and pediatric patients await further validation.242, 243, 244 Although the mutational landscape of CHL is established,245, 246, 247 mutational testing for clinical purposes is hampered by the scarcity of the malignant Hodgkin Reed-Sternberg cells. Recent studies suggest the clinical utility of FISH-determined 9p24.1 amplification (harboring CD274, PDCD1LG2, and JAK2) as a favorable predictive biomarker in patients with relapsed/refractory CHL treated with PD1 inhibitors.248 ctDNA-based assessments of remission status and MRD show promise for dynamic disease monitoring with potential implications for response-adapted therapy.246,249
Mature T-cell and NK-cell neoplasms
Anaplastic large cell lymphomas
ALCL comprises 4 clinically, pathologically, and genetically distinct subtypes: 2 systemic forms (ALCL, ALK-positive and ALCL, ALK-negative) and 2 site-specific forms (primary cutaneous ALCL [pcALCL] and breast implant-associated [BIA] ALCL).2 Accurate diagnosis of ALCL requires integration of histologic, immunophenotypic, genetic, and clinical data. Genetic and molecular characterization additionally aids in prognosis and potential therapeutic targets (Table 2; Figure 5).
Figure 5.
Recurrent genetic lesions in mature NK-cell and T-cell neoplasms with potential therapeutic intervention. Representative histology of entities with frequent genetic lesions potentially amenable to therapeutic intervention are shown on the left. The genetic lesions are presented according to functional groups related to TcR signaling, JAK/STAT pathway, epigenetics, or others. Therapeutic efficacy is supported by clinical trial (a); case reports, small case series, or retrospective analyses (b); or experimental or in silico data (c). AITL, Angioimmunoblastic T-cell lymphoma; ATLL, adult T-leukemia/lymphoma; CTCL, cutaneous T-cell lymphoma; ITLPD-GI, indolent clonal T-cell LPD of the gastrointestinal tract; TFHL-F, TFHL, follicular type; T-PLL, T-cell prolymphocytic leukemia. Sources referenced: 268, 270, 278, 284, 291, 293, 330, 338, 339, 341, 345, 475, 476, 477, 478, 479, 480, 481, 482, 483, 484, 485, 486, 487.
Most ALCLs have clonally rearranged TR genes.250 ALCL, ALK-positive is defined by the presence of ALK fusions encoding oncogenic proteins, typically identified by IHC.251 ALK rearrangement is occasionally seen in cases otherwise resembling pcALCL.252,253 The partner is NPM1 in >80% of cases. ALK tyrosine kinase inhibitors have efficacy in some clinical settings.254 NOTCH pathway activation, resulting from recurrent NOTCH1 mutations or ALK fusions, represents another candidate therapeutic target.255
ALCL, ALK-negative is genetically heterogeneous.256 DUSP22 rearrangement, seen in 19% to 30% of cases, defines a distinct genetic subtype associated with mutations of MSC2; prognosis is generally favorable but high-risk cases occur.256, 257, 258, 259 DUSP22-R also occurs in pcALCL and lymphomatoid papulosis.260, 261, 262, 263, 264 ALCL, ALK-negative with TP63 rearrangement appears largely chemorefractory,256,257,265 and the losses of TP53 and/or PRDM1 are associated with inferior outcome.266 pcALCL with TP63 rearrangement may also follow an aggressive course.265 Rare cases with dual DUSP22/TP63 rearrangements exist.257,267 A subset of ALCL, ALK-negative expresses potentially targetable truncated ERBB4.268
ALCL, ALK-positive and about two-thirds of ALCL, ALK-negative share STAT3-mediated oncogenesis; genetic alterations driving STAT3 activation in ALCL, ALK-negative include JAK1 and STAT3 mutations, and rearrangement involving ROS1, TYK2, FRK, and JAK2.261,269, 270, 271, 272, 273 These findings also may be seen in pcALCL.270,274 BIA-ALCL shows activating JAK/signal transducer and activator of transcription (JAK/STAT) alterations as well as epigenetic modifier mutations and loss of chromosome 20q13.13.275, 276, 277 Therapies targeting the JAK/STAT pathway are being explored.278 TP53 mutations are detected in a small subset of systemic and BIA-ALCLs.273,276
TFH lymphoma and peripheral T-cell lymphoma, NOS
In follicular helper T-cell lymphoma (TFHL) and peripheral T-cell lymphoma (PTCL), not otherwise specified (NOS), most common genetic abnormalities, including SNVs, CNAs, and rearrangements, affect genes of epigenetic regulators (eg, TET2, DNMT3A, IDH2), T-cell receptor (TcR) signaling and activation (eg, RHOA, VAV1, CD28, ICOS, FYN, LCK), phosphatidylinositol 3-kinase/protein kinase B pathway, and tumor suppressor genes (eg, TP53, CDKN2A, ATM, PTEN, RB1).279, 280, 281, 282, 283, 284 (supplemental Figure 1) Genetic testing of newly diagnosed nodal PTCL for commonly reported alterations, ideally using HTS-based panels targeting tumor DNA with high depth and, if necessary, RNA, may be clinically useful as the genomic profile may have implications for accurate diagnosis, risk stratification, and therapy selection (Table 2; Figure 5).
The diagnosis of PTCL integrates clonality assessment, which is performed by TR rearrangement analysis. Although these methods are sensitive, false-positive results may occur in reactive conditions.4 HTS-based gene panels may provide higher specificity for clonality in PTCL while preserving sensitivity comparable to TR rearrangement–based analyses. Therefore, these panels may have broader diagnostic utility by providing both evidence of clonality and characteristic mutational profile.285,286
Some genetic aberrations, including tyrosine kinase gene fusions, are broadly seen across different types of nodal PTCL,282,283 whereas others are more characteristic of phenotypic subtypes. Specifically, TFHLs frequently carry mutations of TET2, DNMT3A, RHOA, and IDH2, rarely seen in combination in other PTCL,280,287 thus providing diagnostic utility. In PTCL, NOS, 2 molecular subgroups, namely PTCL-TBX21 and PTCL-GATA3, show distinct genetic profiles. PTCL-GATA3 demonstrates high genomic complexity characterized by biallelic deletion/mutation of TP53, CDKN2A/B, or RB1. Meanwhile, PTCL-TBX21 shows low genomic complexity and few recurrent specific genetic changes, such as chromosome 5 gain and focal 14q32 gain, including the BCL11B locus.280
TET2 and DNMT3A mutations, often seen in TFHL but also less commonly in other PTCL, NOS, are also the most frequent mutations seen in CH.288 Emerging evidence suggests that in TFHL, bone marrow myeloid precursors may also carry identical mutations, indicating a clonal link/filiation.289,290 The background CH appears to be the source of myeloid neoplasms seen in patients with TFHL, particularly after cytotoxic therapy.290 Therefore, genomic analysis of marrow for CH clones at diagnosis and during disease monitoring may be required to assess the risk of development of a secondary myeloid neoplasm and ensure early diagnosis.290 When interpreting mutational profiles, special attention should be given to avoid misinterpretation of background CH as tumor-specific mutations.
The mutational profile may also provide prognostic information. Mutations leading to loss of tumor suppressor genes, such as TP53 and CDKN2A, have been associated with adverse outcomes in PTCL, NOS.280,281 TFHL, which frequently carries mutations in genes regulating the epigenetic machinery, have a higher response rate to hypomethylating agents such as 5-azacytidine and histone deacetylase inhibitors such as romidepsin.291, 292, 293 However, the predictive value of individual gene mutations has not been clearly established, and whether there are implications in PTCLs not fitting the diagnostic criteria of TFHL is unknown.
Extranodal PTCLs
Extranodal T-cell and NK-cell lymphoma entities derive mostly from innate cells, are relatively organ-specific, and often portend poor outcomes. Although their recognition relies primarily on morphological and immunophenotypic criteria and considering clinical features, genomic traits may be diagnostically useful. Frequent oncogenic activation of the JAK/STAT signaling pathway may be an attractive therapeutic target (Figure 5).278,294,295
Distinctive genomic features help differentiate between enteropathy-associated T-cell lymphoma (EATL), monomorphic epitheliotropic T-cell lymphoma (MEITL), and indolent T/NK LPDs of the gastrointestinal tract (Table 3). Alterations in the JAK/STAT pathway genes primarily target STAT3 and JAK1 in EATL and STAT5B and JAK3 in MEITL; a recurrent deletion in JAK3 characterizes some indolent gastrointestinal NK LPDs,296 and a proportion of indolent clonal T-cell LPDs of the gastrointestinal tract harbor hotspot STAT3 mutations or JAK2::STAT3 fusion.297, 298, 299 Deleterious lesions of SETD2 gene, translating into reduced H3K36 trimethylation, are almost constant in MEITL, rare in EATL, and not found in indolent gastrointestinal T/NK LPDs.298,300, 301, 302, 303 Conversely, KMT2D and TET2 are frequently mutated in EATL and gastrointestinal T-cell LPDs.298,304,305 Detection of somatic mutations in indolent T/NK LPDs supports the neoplastic nature of these processes. Because EATL-associated mutations or add(1q) are frequently present in type II refractory celiac disease (RCDII), HTS or FISH help assess intestinal intraepithelial lymphocyte proliferations and risk of transformation from RCDII to EATL.304,306,307
Hepatosplenic T-cell lymphoma (HSTCL) must be distinguished from T-cell large granular lymphocytic leukemia (T-LGLL), from reactive expansions of γδ T cells or florid γδ T-cell lymphoproliferations causing splenomegaly, with or without association to primary immune deficiency.308,309 Diagnostic confirmation is supported by HSTCL-associated genomic imbalances (isochromosome 7q,310,311 trisomy 8312) or mutations (INO80, PIK3CD, SETD2, TET3, SMARCA2; and STAT5B or STAT3, also found in T-LGLL).294,313,314
Extranodal NK/T-cell lymphoma, nasal type (ENKTCL) has a heterogeneous derivation from NK or T cells.315 Germ line single-nucleotide polymorphisms (SNPs) associated with increased risk of ENKTCL316,317 or with patient survival318 have been described. Among the genomic landscape of ENKTCL,319, 320, 321, 322, 323, 324 mutations in DDX3X, TP53, and KMT2D reportedly confer a worse prognosis.325,326 Chronic active EBV disease of T-cell or NK-cell type may harbor mutations in genes altered in ENKTCL,327 and the constellation of mutations found in aggressive NK-cell leukemia is similar to those in ENKTCL.328 A large integrative multiomics analysis of ENKTCL biopsies defined 3 molecular subtypes with different biology and vulnerabilities: tumor suppressor/immune modulator (TSIM); MYC-related, having the worst outcome; and histone epigenetic altered, having the best outcome.326 Tumors harboring SVs or amplification of CD274 may show greater sensitivity to immune checkpoint inhibitors.329, 330, 331, 332 Four TME subgroups defined by expression profiling alone may represent immunotherapy biomarkers.333
CTCLs comprise a collection of diseases, with heterogeneous genomic portraits overlapping those of other T-cell lymphomas with particularly frequent CNAs. Germline or somatic mutations in HAVCR2 are specifically associated with subcutaneous panniculitis–like T-cell lymphoma and are associated with more severe clinical presentation and a higher risk of hemophagocytic syndrome.334, 335, 336
Leukemic/disseminated NK and T-cell neoplasms
Adult T-cell lymphoma/leukemia is a virally driven neoplasm in which a single HTLV-1–positive clone expands, outcompeting thousands of other infected cells and undergoing malignant transformation.337 The neoplastic cells harbor frequent gain-of-function alterations in TcR/NF-κB signaling, including activating mutations in PLCG1 and PRKCB, CTLA4/ICOS::CD28 fusions, and REL truncations.338, 339, 340 Recurrent alterations targeting immune-related molecules are also observed, including SVs involving the 3′-UTR of CD274, resulting in programmed death-ligand 1 (PD-L1) overexpression.341 Other commonly targeted pathways include transcriptional regulation (alterations in the CIC-ATXN1 complex and IKZF2 intragenic deletions), T-cell trafficking (CCR4 and CCR7 truncating mutations), tumor suppression (TP53), and epigenetic modification (ARID2, EP300).338,339,342 Aggressive subtypes show more genetic alterations, whereas STAT3 mutations are more frequent in indolent subtypes.343 Retrospective data have suggested that gain-of-function CCR4 mutations are associated with significantly improved survival when treated with mogamulizumab344,345 and that SNVs and CNAs of TP53 are associated with inferior OS, regardless of treatment strategies.346
In T-LGLL, mutations in STAT3 and STAT5B are the most common gain-of-function mutations.347, 348, 349 In particular, STAT3 mutations are a feature of CD8+ T-LGLL (∼45%) and some T-γ/δ LGLL, whereas STAT5B mutations are mostly associated with the indolent CD4+ T-LGLL form (∼60%)350 or with the rare aggressive variant of CD8+ T-LGLL.351, 352, 353 The presence of a STAT3 mutation is strongly linked to CD8+ T-LGLL characterized by neutropenia and the CD16+/CD56− phenotype.352,354,355 Other genes have been found recurrently (TNFAIP3) and occasionally (eg, BCL11B, FLT3, PTPN23) mutated in patients with T-LGLL.353
Mutations of STAT3 (∼30%)356, TET2 (∼25%), and CCL22 (27%)357 have been detected in NK-LGLL, while this disorder appears to be devoid of STAT5B genetic lesions.358, 359, 360 TNFAIP3 mutation has been found in ∼6% of NK-LGLL.360,361
T-cell prolymphocytic leukemia (T-PLL) is characterized by chromosomal inversions or translocations involving TCL1 family genes, best demonstrated by FISH,362 resulting in constitutive overexpression of TCL1A or MCTP1, and found in virtually all cases.363, 364, 365 Complex karyotypic abnormalities, present in >70% of cases, portend a poor prognosis.366 Monoallelic deletions and/or mutations of ATM are common.367, 368, 369 Up to 75% of patients harbor mutations in STAT5B, JAK1, or JAK3.370,371
Histiocytic and dendritic cell neoplasms (HDCNs)
In myeloid-derived HDCNs (Langerhans cell histiocytosis [LCH], Erdheim-Chester disease [ECD], juvenile xanthogranuloma, Rosai-Dorfman-Destombes disease [RDDD]), mutually exclusive recurrent mutations in MAPK (BRAF, ARAF, NRAS, KRAS, MAPK1/2) and, less frequently, in phosphatidylinositol-3-kinase (PIK3CA) pathways have been reported.372, 373, 374, 375, 376, 377, 378, 379, 380, 381, 382, 383, 384, 385, 386, 387, 388 None of these mutations are specific for HDCNs because they can occur in many tumors of different histogenesis. However, in HDCNs, these somatic alterations arise in the setting of relatively few other mutations.382,389, 390, 391 BRAFV600E is identified in the majority of LCH and ECD cases, and the bone marrow may represent the primary tumor cell reservoir, given detection of BRAFV600E in hematopoietic stem cells.372, 373, 374, 375 In LCH, the severity of disease is associated with the ability to detect BRAFV600E (or other MAPK activating mutations) in myeloid precursors in bone marrow and peripheral blood.372,392 In keeping with their hematopoietic origin, they can occur in association with myeloid, as well as B-cell and T-cell neoplasms, with evidence of a shared clonal origin.393,394 In adults, ECD lesions can bear evidence of mutations arising from CH.395 In LCH-associated neurodegeneration (LCH-ND), BRAFV600E has been detected in peripheral blood and brain biopsies/autopsy of patients with BRAFV600E+ lesions, suggesting potential for shared clonal hematopoietic origins of systemic disease and LCH-ND.396 ALK-positive histiocytosis is characterized by the fusion of ALK with different partners (typically KIF5B), leading to activation of signaling pathways and sensitivity to ALK inhibitors.388 RDDD likely represents a more diverse spectrum of biological conditions with a common phenotype with recurrent MAPK pathway mutations identified in RDDD, although at a lower frequency than in other histiocytic diagnoses. Histiocytic sarcoma shows a history of lymphoid neoplasm in more than 20% of cases and frequently carries mutations of CDKN2A and TP53.397 In most instances, mutations involving at least 1 gene in the MAPK pathway (most commonly BRAF) are also detected.398 By contrast, follicular dendritic/reticular cell sarcomas and EBV+ inflammatory follicular dendritic cell/fibroblastic reticular cell tumors are of mesenchymal origin, unrelated to a hematopoietic precursor.399 Follicular dendritic cell sarcoma (FDCS) shows mutations affecting CDKN2A, NFKBIA, TP53, and BIRC3.397,398 GEP studies and immunohistochemical analyses have revealed constitutive overexpression of PD-L1 in LCH and FDCS, which might represent a target for immune checkpoint inhibitors.386,387,399
Identification of somatic alterations is clinically important in histiocytic disorders, not only to confirm diagnosis but also to inform risk stratification and therapy; for example, BRAFV600E is associated with an increased risk of relapse and CNS disease in LCH. Determining mutations is also required to determine the suitability of specific inhibitors (eg, ALK, BRAFV600E, second-generation RAF or MEK). Vemurafenib is approved by the FDA for front-line therapy for ECD,400 and cobimetinib has breakthrough designation for study in adult histiocytic disorders.401 Near-universal responses are reported in pediatric patients with LCH treated with MAPK inhibition in retrospective studies, and prospective pediatric trials are in progress.402,403 Finally, genomic characterization of histiocytic lesions is helpful to support identification of mutated cells in blood or bone marrow aspirate, which informs the extent of disease and persistence of precursors.389,392,404,405 In patients treated with MAPK inhibitors, BRAFV600E typically remains detectable in peripheral blood and bone marrow, and high relapse/progression rates are associated with cessation of inhibitor therapy.402,403,406 Systematic molecular investigations of these orphan neoplasms are warranted to discover novel effective therapeutic targets; their treatment still represents an unmet clinical need.
Technologies poised to enter clinical practice
WGS: ongoing opportunities for discovery
Although WES has clearly informed on the diverse protein-coding mutations relevant to individual cancers, WGS interrogates the understudied regions, allowing SVs, CNAs, and noncoding mutations to be detected.10,104,105,204,339,407,408 Therefore, it represents an opportunity for identifying gaps in our understanding of the etiology of cancers and the shortcomings of current clinical assays (Figure 1). Some of the emerging genetic subgrouping systems for lymphomas rely on the presence of specific driver mutations (including SVs and CNAs) and patterns of SHM.187,409, 410, 411
SVs involving an oncogene and an active regulatory element causing ectopic oncogene expression (eg, BCL2) or those forming a functional fusion gene (eg, NPM1::ALK) can be diagnostic, prognostic, or predictive for targeted therapies. For oncogenes having promiscuous rearrangement partners (eg, MYC, BCL6), the identity of partners may have a differential influence on prognosis.191 Atypical examples of common SVs can arise from cassette-like insertions of oncogenes or enhancers or from complex rearrangements, and these can be cryptic to FISH.172,412,413 Functional MYC rearrangements can also reside distal to the gene,192 making their detection through targeted sequencing panels challenging.414
Mutations affecting U1 spliceosomal RNA have been found in CLL and other cancers, causing broad perturbation of splicing.415 There is also a growing list of noncoding mutations that alter splicing in cis by creating novel protein isoforms10,339 or influencing the abundance of wild-type protein.105 3′-UTR SVs or mutations are known to increase the expression of multiple oncogenes such as CCND1,416 CD274,341 and NFKBIZ.407 Such events are not readily detected by standard assays but could have therapeutic implications.330
ctDNA and lymphoma liquid biopsies
Circulating tumor DNA (ctDNA) represents the fraction of cell-free DNA released by tumor cells into body fluids (ie, blood plasma, cerebrospinal fluid).417 Therefore, ctDNA is an easily accessible source of tumor DNA amenable to serial minimally invasive sampling for the genotyping or monitoring of diverse malignancies.418 HTS-based assays applied to ctDNA can detect IG and TR rearrangements, multiple classes of gene mutations, fusions, and CNAs.21 Amplicon-based assays can also track single mutations at known loci.21 The sensitivity of HTS assays incorporating molecular barcodes and/or bioinformatics that suppress error rates can even surpass amplicon-based PCR approaches, with monitoring detection limits approaching 10−7.419 However, most currently available commercial noninvasive tumor genotyping methods seldom achieve detection of actionable genotypes below ∼0.5% allelic levels.420
Given its high positive predictive value, ctDNA genotyping represents a potential tool for supporting lymphoma diagnosis in certain clinical situations, such as inaccessible tumor sites, and to overcome sampling biases.420 Genotyping of ctDNA can provide information that may complement or potentially replace genomic interrogation of tissue biopsies and inform on newly acquired genetic changes following treatment. This may be relevant if actionable mutations are predictive biomarkers for treatment tailoring.417 In addition to this use of baseline liquid biopsies for genotyping and subtype classification,24 ctDNA measurement at baseline allows for measurement of tumor burden,421 and serial measurements allow for dynamic monitoring of tumor response and residual disease422 (Figure 6).
Figure 6.
Applications of ctDNA in lymphoma. Schematic illustrates the potential applications of liquid biopsy assessment, as used for the identification of clinically actionable adverse-risk features in lymphomas at different disease milestones. A lymphoid tumor (left of vessel) is imagined as being accessible through blood plasma by analysis of ctDNA fragments. ctDNA is represented by purple double-stranded DNA molecules, and yellow double strands represent non–tumor-derived cell-free DNA molecules. The patient is evaluated by ctDNA profiling during various disease milestones over time (diagnosis, treatment, and relapse).488 During this temporal sequence, ctDNA can inform risk at diagnosis, during therapy, immediately after induction therapy, in surveillance of disease, and at progression or disease transformation,424 as illustrated in the panels associated with each milestone. At diagnosis, profiling of tumor DNA obtained from either tissue biopsies or noninvasively through genotyping of plasma (depicted as blood collection tubes),24 allows for the identification of patients with high tumor burden,246,421 histological subtypes,489 and prediction of outcomes.422 Assessment of ctDNA during and after lymphoma treatment facilitates the detection of both emerging resistance mutations and MRD before progression,419 with potential for noninvasive prediction of relapse and transformation.490 Tumor evolution in an anecdotal patient with DLBCL is illustrated, showing tumor response and clonal evolution over the course of the disease (detectable subclones at diagnosis are shown in blue/yellow; an emergent subclone after therapy is shown in red). EMR, early molecular response; MMR, major molecular response; TMTV, total metabolic tumor volume.
These applications likely allow ctDNA to complement and enhance conventional imaging for staging and response assessments.423 Nevertheless, clinical translation of ctDNA analysis in the management of lymphoma requires further understanding of the (1) pathophysiology of cell-free DNA across lymphomas; (2) impact of preanalytics on ctDNA assay results; (3) technical validity and real-time feasibility of state-of-the-art ctDNA assays; and (4) clinical utility of ctDNA assays to guide diagnosis, treatment tailoring, and residual disease identification.424
Single cell analyses
SCA is a breakthrough technology that directly addresses the challenge of complex heterogeneous cell populations in cancer, including immune cells of the TME. Currently, diverse SCA approaches exist, differing on the basis of throughput and data type, from genome, transcriptome, and epigenome to proteome analysis (supplemental Table 4).425 Ongoing efforts aim at integrating multiple data platforms at the individual cell level. This emerging technology has already enabled the functional characterization of cellular identity (including new immune cell types),35,426 deconvolution of cell heterogeneity,427,428 tracking of tumor, and immune cell clonal dynamics at unprecedented resolution,429,430 and has challenged the COO dogma toward a highly plastic view of cancer where dynamic transitions of cell states coexist within the tumor bulk.431,432
Although SCA is presently insufficiently mature to supply specific recommendations for clinical practice, the field is rapidly developing with new tools for data generation/analysis alongside an avalanche of new biologic insights, together with processing and cost streamlining. In particular, new workflows for spatial visualization compatible with FFPE tissues are anticipated to coalesce the expertise of pathologists, molecular biologists, and cytometrists, with high potential for affecting clinical decision making. Thus, across blood malignancies, routine application of SCA can be envisioned for the purposes of diagnosis; for neoplastic and immune population monitoring while on treatment; for monitoring of MRD; and for guiding treatment decisions upon relapse.433,434
Incorporation of SCA within the clinical arena will require the maturation of integrative multiomics analyses, access to appropriate (fresh/live-frozen) longitudinal specimens, including those from clinical trials (supplemental Table 4), and robust and standardized practices for biospecimen collection and computational analyses.
DNA methylation and chromatin profiling
Epigenetic mechanisms play a critical role in lymphomagenesis and have significant clinical diagnostic and outcome implications. Lymphoid tumors maintain a DNA methylation imprint of their cellular origin, which is useful for diagnostic and patient stratification purposes.27,28,102,435, 436, 437, 438 On the other hand, aberrant cytosine methylation patterning is a universal finding in lymphoid neoplasms.439,440 Mechanisms driving this process include the hypermethylation effect of epigenetic modifier mutations such as in TET2,441, 442, 443, 444, 445, 446 the hypomethylating bystander effect of AID where methylcytosine is replaced with unmethylated nucleotides,447 and lymphoma proliferative history associated with gradual accumulation of DNA methylation changes in repressed/heterochromatic regions.430,435 These factors contribute to lymphomagenesis, generate intraclonal heterogeneity, and have significant clinical impact.435,448, 449, 450, 451 Lymphoid neoplasms harbor recurrent hypermethylation of specific genes, including the canonical tumor suppressor gene CDKN2A, related to disease progression,452 and SMAD1, which is a biomarker for chemotherapy resistance453 that can be reversed using DNA methyltransferase inhibitors454 and is currently under validation in a phase 2/3 clinical trial. It is warranted to bring at least some of these findings, including epigenetic biomarkers for COO, proliferative history, and key genes, into clinical practice.
Aberrant histone modifications are also critically relevant to lymphomagenesis. Recent genome-wide chromatin profiling studies have uncovered extensive changes in the activity of regulatory elements, which are targets of drugs such as BET inhibitors.455, 456, 457, 458 Aberrant chromatin patterns are caused by mutations in epigenetic modifiers439 and aberrant transcription factor function. For example, gain-of-function mutations in EZH2 cause profound spreading of the H3K27me3 promoter repressive mark, which is reversed by EZH2 inhibitors459, 460, 461; KMT2D loss-of-function mutations cause loss of enhancer-activating H3K4me1 marks and may be reverted through inhibition of histone demethylases; and the loss of gene body H3K36me3 due to SETD2 mutations that causes activation-induced cytidine deaminase–induced genomic instability.462
Conclusion
The ultimate goal of disease classification is to provide a biologically and clinically relevant framework, reflecting pathogenetic paths and encompassing therapeutically targetable alterations and vulnerabilities. The quality/depth and the amount of data massively generated by newer technologies encompass groundbreaking opportunities to refine classification and define useful structure within “not otherwise specified” disease entities. The diagnostic value of genomic characteristics and measurable impact on clinical management in many lymphoma entities still needs to be addressed, likely best achieved by retrospective and prospective genomic testing in clinical trials. Importantly, given identical DNA alterations, similar pathway alterations or expression signature being observed across pathologically and clinically distinct entities, morphology remains critical in the diagnostic process. Finally, tension is generated by the ideal that any classification should be applicable in a global fashion, including sites where access to resources and technologies are limited. The degree to which genomics will be further integrated into classification in the coming years will depend on defining clinically useful distinctions supported by widely available supportive diagnostics.
Conflict-of-interest disclosure: L.d.L. reports consultancy for AbbVie, Bayer, Bio Ascend, Lunaphore, Novartis. A.A.A. reports consultancy for Celgene, Chugai, Genentech, Gilead, Janssen, Pharmacyclics, and Roche; research funding from Celgene and Pfizer; and ownership interests CiberMed, ForeSight, and FortySeven. E.C. reports consultancy for Genmab, Illumina, NanoString, and Takeda; educational honoraria from AstraZeneca, EUSA Pharma, Janssen, and Takeda; is a named inventor on a patent describing the use of gene expression to subtype aggressive B-cell lymphomas, one of which is licensed to NanoString Technologies, and an author in a protected bioinformatic pipeline (IgCaller) licensed to Diagnostica Longwood. A. Dogan reports consultancy for EUSA Pharma, Incyte, and Loxo; and research funding from Roche and Takeda. S.M.H. reports consultancy for Cimieo Therapeutics, Daiichi Sankyo, Kyowa Hakko Kirin, ONO Pharmaceuticals, SecuraBio, Shoreline Biosciences, Takeda, Tubulis, and Yingli Pharma; and research funding from ADC Therapeutics, Affimed, C4, Celgene, Crispr Therapeutics, Daiichi Sankyo, Kyowa Hakko Kirin, Millennium/Takeda, Seattle Genetics, and Verastem/SecuraBio. A.M.M. reports consulting for AstraZeneca, Epizyme, Exo Therapeutics, Janssen, and Treeline Biosciences; and research funding from AstraZeneca, Epizyme, and Janssen. R.R. reports consultancy for AbbVie, AstraZeneca, Illumina, Janssen, and Roche. D.R. reports consultancy for AbbVie, AstraZeneca, BeiGene, BMS, and Janssen; and research funding from AbbVie, AstraZeneca, BeiGene, BMS, and Janssen. C.S. reports consultancy for AbbVie, Bayer, and Seattle Genetics; and research funding from Trillium Therapeutics, BMS, and Epizyme. S.P.T. reports consultancy for BeiGene, Janssen, and Pharmacyclics; and research funding from BeiGene, BMS, Eli Lilly, Janssen, Pharmacyclics, and X4 Pharmaceuticals. A.D.Z. reports consultancy for Amgen, AstraZeneca, BeiGene, BMS/Celgene/JUNO, Genentech/Roche, Gilead/Kite, Janssen, MEI Pharma, Novartis, and Pharmacyclics/AbbVie; scientific advisory board for Adaptive Biotechnologies and Lymphoma Research Foundation; and research funding from AbbVie, BeiGene, Genentech/Roche, and MEI Pharma. C.E.A. reports consultancy for Electra and Sobi; and research support from Roche/Genetech. W.C.C. is a named inventor on a patent describing the use of gene expression to subtype aggressive B-cell lymphomas, one of which is licensed to NanoString Technologies. L.C. reports advisory boards for AbbVie and Roche. F.d’A. reports advisory board for Kyova Kirin; and research funding from Servier. S.D. reports consultancy for Incyte, Roche, and Takeda. M.D. reports speakers’ honoraria from Amgen, AstraZeneca, Bayer, BMS/Celgene, Gilead/Kite, Incyte, Janssen, Novartis, and Roche; scientific advisory board for AstraZeneca, Bayer, BMS/Celgene, Genmab, Gilead/Kite, Incyte, Janssen, Lilly/Loxo, MorphoSys, Novartis, and Roche; and research funding (institutional) from AbbVie, Bayer, BMS/Celgene, Gilead/Kite, Janssen, and Roche. K.D. reports scientific advisory board for AbbVie, ADC Therapeutics, AstraZeneca, BeiGene, Daiichi Sankyo, Genentech, Genmab, Incyte, and MorphoSys. A.L.F. reports research funding from Seattle Genetics; and is a named inventor on technology for which Mayo Clinic holds unlicensed patents. F.F. reports advisory boards or speaker honoraria from EUROPharma, Roche, and Stemline; and research funding from Stemline. P. Gaulard reports consultancy for Gilead and Takeda; and research funding from Alderan, Innate Pharma, Sanofi, and Takeda. P. Ghia reports consultancy for AbbVie, AstraZeneca, BeiGene, BMS, Janssen, Lilly/Loxo, MSD, and Roche; and research funding from AbbVie, AstraZeneca, and Janssen. J.G.G. reports honoraria from AbbVie, Amgen, AstraZeneca, BMS, Gilead/Kite, Janssen, and Novartis; and research funding from AstraZeneca, BMS, and Janssen. O.H. reports holding stock in AB science; is a cofounder of AB science and Inatherys; and reports research funding from AB science, AbbVie, Alexion, BMS, Inatherys, Innate Pharma, and Takeda. D.J.H. reports research funding from AstraZeneca. E.D.H. reports consultancy for Abcon, Astellas, Cytomx, and Novartis; and research funding from AbbVie, Eli Lilly, and Virtuoso. K. Karube reports speaker honoraria from AstraZeneca, Chugai Pharmaceutical, Eisai, Kyowa Kirin, Janssen, Otsuka Pharmaceutical, Takeda, Symbio, and Meiji Seika Pharma; and research funding from Takeda. K. Kataoka reports honoraria from Astellas Pharma, AstraZeneca, Chugai Pharmaceutical, Eisai, Kyowa Kirin, Janssen, Novartis, Ono Pharmaceutical, Otsuka Pharmaceutical, Sumitomo Dainippon Pharma, and Takeda; and research funding from Asahi Kasei Pharma, Chugai Pharmaceutical, Chordia Therapeutics, Eisai, Kyowa Kirin, Japan Blood Products Organization, JCR Pharmaceutical, Mochida Pharmaceutical, Nippon Shinyaku, Ono Pharmaceutical, Otsuka Pharmaceutical, Shionogi, Sumitomo Dainippon Pharmaceutical, Takeda, and Teijin Pharma; is a named inventor on a patent for genetic alterations as a biomarker in T-cell lymphomas and for PD-L1 abnormalities as a predictive biomarker for immune checkpoint blockade therapy; and reports stocks in Asahi Genomics. W.S.K. reports research funding from BeiGene, Boryong, Kyowa Kirin, Merck, Roche, and Sanofi. G.L. reports consultancy for AbbVie, ADC, AstraZeneca, Bayer, BMS, Celgene, Constellation, Gilead, Genmab, Incyte, Janssen, Karyopharm, MorphoSys, NanoString, Novartis, Roche, and Takeda; speakers’ honoraria from AbbVie, Bayer, Celgene, Janssen, and Roche; and research funding from AstraZeneca, Bayer, Celgene, Gilead, Janssen, MorphoSys, Novartis, and Roche. L.P. reports research funding from AstraZeneca. M.A.P. reports speaker honoraria and advisory board fees from Celgene, Gilead, Janssen, Kyowa Kirin, Millennium/Takeda, and NanoString. S.J.R. reports research funding from BMS and Merck. G.A.S. reports consulting and advisory boards for AbbVie, Bayer, BeiGene, BMS/Celgene, Epizyme, Genentech/Roche, Genmab, Incyte, Janssen, Kite/Gilead, Loxo, Miltenyi, Molecular Partners, MorphoSys, Nordic Nanovector, Novartis, Rapt, Regeneron, and Takeda; and is a shareholder of Owkin. J.S.-M. reports consultancy and advisory boards for AbbVie, Amgen, BMS, Celgene, GSK, Haemalogix, Janssen-Cilag, Karyopharm, MSD, Novartis, Regeneron, Roche, Sanofi, SecuraBio, and Takeda. K.J.S. reports consultancy for BMS, Janssen, Kyowa, Merck, Novartis, and Seattle Genetics; and research funding from BMS. L.H.S. reports consulting and honoraria from AbbVie, Acerta, Amgen, Apbiologix, AstraZeneca, Celgene, Chugai, Gilead, Incyte, Janssen, Kite, Karyopharm, Lundbeck, Merck, MorphoSys, Roche/Genentech, Sandoz, Seattle Genetics, Servier, Takeda, Teva, TG Therapeutics, and Verastem; and research funding from Roche/Genentech and Teva. L.M.S. is one of the developers of the LymphGen software, and NCI has applied for copyright protection for this software; and is an inventor on NCI patents relevant to cell-of-origin classification of DLBCL. C.S.T. reports honoraria from AbbVie, BeiGene, and Janssen. J.T. reports research funds (institutional) from BeiGene, BMS, Cellectar, Janssen, Pharmacyclics, Roche, and Takeda. C.J.W. reports equity in BioNTech; research funds from Pharmacyclics. P.L.Z. reports consultancy for MSD TG Therapeutics, EUSAPharma, and Novartis; advisory board and speakers’ bureau for BeiGene, BMS, Celltrion, EUSAPharma, Gilead, Incyte, Janssen-Cilag, Kyowa Kirin, MSD TG Therapeutics, Novartis, Roche, Servier, and Takeda; and advisory board for ADC Therapeutics, Sandoz, and Secura Bio. D.W.S. reports consultancy for AbbVie, AstraZeneca, Incyte, and Janssen; research funds from Janssen and Roche/Genentech; and is a named inventor on a patent describing the use of gene expression to subtype aggressive B-cell lymphomas, one of which is licensed to NanoString Technologies. The remaining authors declare no competing financial interests.
Acknowledgment
Laura Hilton contributed to the production of figures.
Authorship
Contribution: L.d.L. and D.W.S. conceived the structure of the manuscript, coordinated the writing, and wrote and edited the manuscript; and all authors wrote or contributed to the contents of the manuscript and approved it.
Footnotes
Data are available on request from the corresponding authors, Laurence de Leval (laurence.deleval@chuv.ch) and David W. Scott (dscott8@bccancer.bc.ca).
The online version of this article contains a data supplement.
Contributor Information
Laurence de Leval, Email: laurence.deleval@chuv.ch.
David W. Scott, Email: dscott8@bccancer.bc.ca.
Supplementary Material
References
- 1.Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of the Haematopoietic and Lymphoid Tissues. Revised 4th ed. IARC; 2017. [Google Scholar]
- 2.Campo E, Jaffe ES, Cook JR, et al. The international consensus classification of mature lymphoid neoplasms: a report from the Clinical Advisory Committee. Blood. 2022;140(11):1229–1253. doi: 10.1182/blood.2022015851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zech L, Haglund U, Nilsson K, Klein G. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int J Cancer. 1976;17:47–56. doi: 10.1002/ijc.2910170108. [DOI] [PubMed] [Google Scholar]
- 4.Langerak AW, Groenen PJTA, Brüggemann M, et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia. 2012;26(10):2159–2171. doi: 10.1038/leu.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Brand M, Rijntjes J, Mobs M, et al. Next-generation sequencing-based clonality assessment of Ig gene rearrangements: a multicenter validation study by EuroClonality-NGS. J Mol Diagn. 2021;23(9):1105–1115. doi: 10.1016/j.jmoldx.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Rosenquist R, Bea S, Du MQ, Nadel B, Pan-Hammarstrom Q. Genetic landscape and deregulated pathways in B-cell lymphoid malignancies. J Intern Med. 2017;282(5):371–394. doi: 10.1111/joim.12633. [DOI] [PubMed] [Google Scholar]
- 7.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treon SP, Xu L, Yang G, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puente XS, Bea S, Valdes-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 11.Parry M, Rose-Zerilli MJJ, Ljungström V, et al. Genetics and prognostication in splenic marginal zone lymphoma: revelations from deep sequencing. Clin Cancer Res. 2015;21(18):4174–4183. doi: 10.1158/1078-0432.CCR-14-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenquist R, Rosenwald A, Du MQ, et al. Clinical impact of recurrently mutated genes on lymphoma diagnostics: state-of-the-art and beyond. Haematologica. 2016;101(9):1002–1009. doi: 10.3324/haematol.2015.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buhler MM, Martin-Subero JI, Pan-Hammarstrom Q, Campo E, Rosenquist R. Towards precision medicine in lymphoid malignancies. J Intern Med. 2022;292(2):221–242. doi: 10.1111/joim.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansouri L, Thorvaldsdottir B, Laidou S, Stamatopoulos K, Rosenquist R. Precision diagnostics in lymphomas – recent developments and future directions. Semin Cancer Biol. 2022;84:170–183. doi: 10.1016/j.semcancer.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Davis AR, Stone SL, Oran AR, et al. Targeted massively parallel sequencing of mature lymphoid neoplasms: assessment of empirical application and diagnostic utility in routine clinical practice. Mod Pathol. 2021;34(5):904–921. doi: 10.1038/s41379-020-00720-7. [DOI] [PubMed] [Google Scholar]
- 19.Sutton LA, Ljungström V, Mansouri L, et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach will facilitate implementation in a clinical setting. Haematologica. 2015;100(3):370–376. doi: 10.3324/haematol.2014.109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Intlekofer AM, Joffe E, Batlevi CL, et al. Integrated DNA/RNA targeted genomic profiling of diffuse large B-cell lymphoma using a clinical assay. Blood Cancer J. 2018;8(6):60. doi: 10.1038/s41408-018-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherer F, Kurtz DM, Diehn M, Alizadeh AA. High-throughput sequencing for noninvasive disease detection in hematologic malignancies. Blood. 2017;130(4):440–452. doi: 10.1182/blood-2017-03-735639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart JP, Gazdova J, Darzentas N, et al. Validation of the EuroClonality-NGS DNA capture panel as an integrated genomic tool for lymphoproliferative disorders. Blood Adv. 2021;5(16):3188–3198. doi: 10.1182/bloodadvances.2020004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navrkalova V, Plevova K, Hynst J, et al. Lymphoid NeXt-generation sequencing (LYNX) panel: a comprehensive capture-based sequencing tool for the analysis of prognostic and predictive markers in lymphoid malignancies. J Mol Diagn. 2021;23(8):959–974. doi: 10.1016/j.jmoldx.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Scherer F, Kurtz DM, Newman AM, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8:364ra155. doi: 10.1126/scitranslmed.aai8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alizadeh AA, Eisen MB, David RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 26.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 27.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 28.Oakes CC, Seifert M, Assenov Y, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48(3):253–264. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ennishi D, Jiang A, Boyle M, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2019;37(3):190–201. doi: 10.1200/JCO.18.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33(26):2848–2856. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotty R, Hu K, Stevenson K, et al. Simultaneous identification of cell of origin, translocations, and hotspot mutations in diffuse large B-cell lymphoma using a single RNA-sequencing assay. Am J Clin Pathol. 2020;155(5):748–754. doi: 10.1093/ajcp/aqaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skaftason A, Qu Y, Abdulla M, et al. Transcriptome sequencing of archived lymphoma specimens is feasible and clinically relevant using exome capture technology. Genes Chromosomes Cancer. 2022;61(1):27–36. doi: 10.1002/gcc.23002. [DOI] [PubMed] [Google Scholar]
- 33.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14(8):517–534. doi: 10.1038/nrc3774. [DOI] [PubMed] [Google Scholar]
- 34.Steen CB, Luca BA, Esfahani MS, et al. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell. 2021;39(10):1422–1437.e1410. doi: 10.1016/j.ccell.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoki T, Chong LC, Takata K, et al. Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov. 2020;10(3):406–421. doi: 10.1158/2159-8290.CD-19-0680. [DOI] [PubMed] [Google Scholar]
- 36.Meggendorfer M, Jobanputra V, Wrzeszczynski KO, et al. Analytical demands to use whole-genome sequencing in precision oncology. Semin Cancer Biol. 2022;84:16–22. doi: 10.1016/j.semcancer.2021.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mateo J, Chakravarty D, Dienstmann R, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2018;29(9):1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horak P, Griffith M, Danos AM, et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC) Genet Med. 2022;24(5):986–998. doi: 10.1016/j.gim.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 41.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 42.Agathangelidis A, Chatzidimitriou A, Chatzikonstantinou T, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: the 2022 update of the recommendations by ERIC, the European Research Initiative on CLL. Leukemia. 2022;36:1961–1968. doi: 10.1038/s41375-022-01604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woyach JA, Ruppert AS, Heerema NA, et al. Ibrutinib regimens Versus chemoimmunotherapy in older patients with untreated CLL. N Engl J Med. 2018;379(26):2517–2528. doi: 10.1056/NEJMoa1812836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer K, Al-Sawaf O, Bahlo J, et al. Venetoclax and obinutuzumab in patients with CLL and coexisting conditions. N Engl J Med. 2019;380(23):2225–2236. doi: 10.1056/NEJMoa1815281. [DOI] [PubMed] [Google Scholar]
- 45.Moreno C, Greil R, Demirkan F, et al. Ibrutinib plus obinutuzumab versus chlorambucil plus obinutuzumab in first-line treatment of chronic lymphocytic leukaemia (iLLUMINATE): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20(1):43–56. doi: 10.1016/S1470-2045(18)30788-5. [DOI] [PubMed] [Google Scholar]
- 46.Shanafelt TD, Wang XV, Kay NE, et al. Ibrutinib–rituximab or chemoimmunotherapy for chronic lymphocytic leukemia. N Engl J Med. 2019;381(5):432–443. doi: 10.1056/NEJMoa1817073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Sawaf O, Zhang C, Tandon M, et al. Venetoclax plus obinutuzumab versus chlorambucil plus obinutuzumab for previously untreated chronic lymphocytic leukaemia (CLL14): follow-up results from a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2020;21(9):1188–1200. doi: 10.1016/S1470-2045(20)30443-5. [DOI] [PubMed] [Google Scholar]
- 48.Maura F, Cutrona G, Fabris S, et al. Relevance of stereotyped B-cell receptors in the context of the molecular, cytogenetic and clinical features of chronic lymphocytic leukemia. PLoS One. 2011;6(8):e24313. doi: 10.1371/journal.pone.0024313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatopoulos K, Belessi C, Moreno C, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood. 2006;109(1):259–270. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 50.Agathangelidis A, Darzentas N, Hadzidimitriou A, et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012;119(19):4467–4475. doi: 10.1182/blood-2011-11-393694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.ten Hacken E, Gounari M, Ghia P, Burger JA. The importance of B cell receptor isotypes and stereotypes in chronic lymphocytic leukemia. Leukemia. 2019;33(2):287–298. doi: 10.1038/s41375-018-0303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nadeu F, Diaz-Navarro A, Delgado J, Puente XS, Campo E. Genomic and epigenomic alterations in chronic lymphocytic leukemia. Annu Rev Pathol. 2020;15(1):149–177. doi: 10.1146/annurev-pathmechdis-012419-032810. [DOI] [PubMed] [Google Scholar]
- 53.Agathangelidis A, Chatzidimitriou A, Gemenetzi K, et al. Higher-order connections between stereotyped subsets: implications for improved patient classification in CLL. Blood. 2021;137(10):1365–1376. doi: 10.1182/blood.2020007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 55.Rigolin GM, Del Giudice I, Bardi A, et al. Complex karyotype in unfit patients with CLL treated with ibrutinib and rituximab: the GIMEMA LLC1114 phase 2 study. Blood. 2021;138(25):2727–2730. doi: 10.1182/blood.2021011883. [DOI] [PubMed] [Google Scholar]
- 56.Schweighofer CD, Coombes KR, Majewski T, et al. Genomic variation by whole-genome SNP mapping arrays predicts time-to-event outcome in patients with chronic lymphocytic leukemia: a comparison of CLL and HapMap genotypes. J Mol Diagn. 2013;15(2):196–209. doi: 10.1016/j.jmoldx.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133(11):1205–1216. doi: 10.1182/blood-2018-09-873083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nadeu F, Royo R, Clot G, et al. IGLV3-21R110 identifies an aggressive biological subtype of chronic lymphocytic leukemia with intermediate epigenetics. Blood. 2021;137(21):2935–2946. doi: 10.1182/blood.2020008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maity PC, Bilal M, Koning MT, et al. IGLV3-21∗01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling. Proc Natl Acad Sci U S A. 2020;117(8):4320–4327. doi: 10.1073/pnas.1913810117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landau DA, Wu CJ. Chronic lymphocytic leukemia: molecular heterogeneity revealed by high-throughput genomics. Genome Med. 2013;5(5):47. doi: 10.1186/gm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bomben R, Rossi FM, Vit F, et al. TP53 mutations with low variant allele frequency predict short survival in chronic lymphocytic leukemia. Clin Cancer Res. 2021;27(20):5566–5575. doi: 10.1158/1078-0432.CCR-21-0701. [DOI] [PubMed] [Google Scholar]
- 62.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139–2147. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Austen B, Powell JE, Alvi A, et al. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood. 2005;106(9):3175–3182. doi: 10.1182/blood-2004-11-4516. [DOI] [PubMed] [Google Scholar]
- 64.Smith CIE, Burger JA. Resistance mutations to BTK inhibitors originate from the NF-κB but not from the PI3K-RAS-MAPK arm of the B cell receptor signaling pathway. Front Immunol. 2021;12:689472. doi: 10.3389/fimmu.2021.689472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang E, Mi X, Thompson MC, et al. Mechanisms of resistance to noncovalent Bruton’s tyrosine kinase inhibitors. N Engl J Med. 2022;386(8):735–743. doi: 10.1056/NEJMoa2114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blombery P. Mechanisms of intrinsic and acquired resistance to venetoclax in B-cell lymphoproliferative disease. Leuk Lymphoma. 2020;61(2):257–262. doi: 10.1080/10428194.2019.1660974. [DOI] [PubMed] [Google Scholar]
- 67.Mansouri L, Wierzbinska JA, Plass C, Rosenquist R. Epigenetic deregulation in chronic lymphocytic leukemia: clinical and biological impact. Semin Cancer Biol. 2018;51:1–11. doi: 10.1016/j.semcancer.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 68.Del Giudice I, Raponi S, Della Starza I, et al. Minimal residual disease in chronic lymphocytic leukemia: a new goal? Front Oncol. 2019;9:689. doi: 10.3389/fonc.2019.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fürstenau M, De Silva N, Eichhorst B, Hallek M. Minimal residual disease assessment in CLL: ready for use in clinical routine? HemaSphere. 2019;3(5) doi: 10.1097/HS9.0000000000000287. e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wierda WG, Rawstron A, Cymbalista F, et al. Measurable residual disease in chronic lymphocytic leukemia: expert review and consensus recommendations. Leukemia. 2021;35(11):3059–3072. doi: 10.1038/s41375-021-01241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carbone A, Roulland S, Gloghini A, et al. Follicular lymphoma. Nat Rev Dis Primers. 2019;5(1):83. doi: 10.1038/s41572-019-0132-x. [DOI] [PubMed] [Google Scholar]
- 72.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okosun J, Wolfson RL, Wang J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48(2):183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheung KJ, Johnson NA, Affleck JG, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70(22):9166–9174. doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- 77.Boice M, Salloum D, Mourcin F, et al. Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell. 2016;167(2):405–418.e413. doi: 10.1016/j.cell.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu D, McCarthy H, Ottensmeier CH, Johnson P, Hamblin TJ, Stevenson FK. Acquisition of potential N-glycosylation sites in the immunoglobulin variable region by somatic mutation is a distinctive feature of follicular lymphoma. Blood. 2002;99(7):2562–2568. doi: 10.1182/blood.v99.7.2562. [DOI] [PubMed] [Google Scholar]
- 79.Pastore A, Jurinovic V, Kridel R, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015;16(9):1111–1122. doi: 10.1016/S1470-2045(15)00169-2. [DOI] [PubMed] [Google Scholar]
- 80.Huet S, Tesson B, Jais J-P, et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: a retrospective training and validation analysis in three international cohorts. Lancet Oncol. 2018;19(4):549–561. doi: 10.1016/S1470-2045(18)30102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morschhauser F, Tilly H, Chaidos A, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020;21(11):1433–1442. doi: 10.1016/S1470-2045(20)30441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nann D, Ramis-Zaldivar JE, Müller I, et al. Follicular lymphoma t(14;18)-negative is genetically a heterogeneous disease. Blood Adv. 2020;4(22):5652–5665. doi: 10.1182/bloodadvances.2020002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barasch NJK, Liu YC, Ho J, et al. The molecular landscape and other distinctive features of primary cutaneous follicle center lymphoma. Hum Pathol. 2020;106:93–105. doi: 10.1016/j.humpath.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 84.Zhou XA, Yang J, Ringbloom KG, et al. Genomic landscape of cutaneous follicular lymphomas reveals 2 subgroups with clinically predictive molecular features. Blood Adv. 2021;5(3):649–661. doi: 10.1182/bloodadvances.2020002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okosun J, Bodor C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112(10):E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossi D, Bertoni F, Zucca E. Marginal-zone lymphomas. N Engl J Med. 2022;386(6):568–581. doi: 10.1056/NEJMra2102568. [DOI] [PubMed] [Google Scholar]
- 89.Zucca E, Bertoni F. The spectrum of MALT lymphoma at different sites: biological and therapeutic relevance. Blood. 2016;127(17):2082–2092. doi: 10.1182/blood-2015-12-624304. [DOI] [PubMed] [Google Scholar]
- 90.Vela V, Juskevicius D, Dirnhofer S, Menter T, Tzankov A. Mutational landscape of marginal zone B-cell lymphomas of various origin: organotypic alterations and diagnostic potential for assignment of organ origin. Virchows Arch. 2022;480(2):403–413. doi: 10.1007/s00428-021-03186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu H, Ruskon-Fourmestraux A, Lavergne-Slove A, et al. Resistance of t(11;18) positive gastric mucosa-associated lymphoid tissue lymphoma to Helicobacter pylori eradication therapy. Lancet. 2001;357(9249):39–40. doi: 10.1016/S0140-6736(00)03571-6. [DOI] [PubMed] [Google Scholar]
- 92.Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood. 2011;117(5):1595–1604. doi: 10.1182/blood-2010-01-264275. [DOI] [PubMed] [Google Scholar]
- 93.Gailllard B, Cornillet-Lefebvre P, Le QH, et al. Clinical and biological features of B-cell neoplasms with CDK6 translocations: an association with a subgroup of splenic marginal zone lymphomas displaying frequent CD5 expression, prolymphocytic cells, and TP53 abnormalities. Br J Haematol. 2021;193(1):72–82. doi: 10.1111/bjh.17141. [DOI] [PubMed] [Google Scholar]
- 94.Clipson A, Wang M, de Leval L, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177–1185. doi: 10.1038/leu.2014.330. [DOI] [PubMed] [Google Scholar]
- 95.Pillonel V, Juskevicius D, Ng CKY, et al. High-throughput sequencing of nodal marginal zone lymphomas identifies recurrent BRAF mutations. Leukemia. 2018;32(11):2412–2426. doi: 10.1038/s41375-018-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spina V, Khiabanian H, Messina M, et al. The genetics of nodal marginal zone lymphoma. Blood. 2016;128(10):1362–1373. doi: 10.1182/blood-2016-02-696757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonfiglio F, Bruscaggin A, Guidetti F, et al. Genetic and phenotypic attributes of splenic marginal zone lymphoma. Blood. 2022;139(5):732–747. doi: 10.1182/blood.2021012386. [DOI] [PubMed] [Google Scholar]
- 98.Puente XS, Jares P, Campo E. Chronic lymphocytic leukemia and mantle cell lymphoma: crossroads of genetic and microenvironment interactions. Blood. 2018;131(21):2283–2296. doi: 10.1182/blood-2017-10-764373. [DOI] [PubMed] [Google Scholar]
- 99.Martín-Garcia D, Navarro A, Valdés-Mas R, et al. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1− mantle cell lymphoma. Blood. 2019;133(9):940–951. doi: 10.1182/blood-2018-07-862151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sander B, Quintanilla-Martinez L, Ott G, et al. Mantle cell lymphoma—a spectrum from indolent to aggressive disease. Virchows Arch. 2016;468(3):245–257. doi: 10.1007/s00428-015-1840-6. [DOI] [PubMed] [Google Scholar]
- 101.Peterson JF, Baughn LB, Ketterling RP, et al. Characterization of a cryptic IGH/CCND1 rearrangement in a case of mantle cell lymphoma with negative CCND1 FISH studies. Blood Adv. 2019;3(8):1298–1302. doi: 10.1182/bloodadvances.2019031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Queiros AC, Beekman R, Vilarrasa-Blasi R, et al. Decoding the DNA methylome of mantle cell lymphoma in the light of the entire B cell lineage. Cancer Cell. 2016;30(5):806–821. doi: 10.1016/j.ccell.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clot G, Jares P, Giné E, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood. 2018;132(4):413–422. doi: 10.1182/blood-2018-03-838136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nadeu F, Martin-Garcia D, Clot G, et al. Genomic and epigenomic insights into the origin, pathogenesis, and clinical behavior of mantle cell lymphoma subtypes. Blood. 2020;136(12):1419–1432. doi: 10.1182/blood.2020005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pararajalingam P, Coyle KM, Arthur SE, et al. Coding and noncoding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood. 2020;136(5):572–584. doi: 10.1182/blood.2019002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hill HA, Qi X, Jain P, et al. Genetic mutations and features of mantle cell lymphoma: a systematic review and meta-analysis. Blood Adv. 2020;4(13):2927–2938. doi: 10.1182/bloodadvances.2019001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yi S, Yan Y, Jin M, et al. Genomic and transcriptomic profiling reveals distinct molecular subsets associated with outcomes in mantle cell lymphoma. J Clin Invest. 2022;132(3):e153283. doi: 10.1172/JCI153283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Müller H, Walter W, Hutter S, et al. Aberrant somatic hypermutation of CCND1 generates non-coding drivers of mantle cell lymphomagenesis. Cancer Gene Ther. 2022;29:484–493. doi: 10.1038/s41417-022-00428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bea S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(45):18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jain P, Zhang S, Kanagal-Shamanna R, et al. Genomic profiles and clinical outcomes of de novo blastoid/pleomorphic MCL are distinct from those of transformed MCL. Blood Adv. 2020;4(6):1038–1050. doi: 10.1182/bloodadvances.2019001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eskelund CW, Dahl C, Hansen JW, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017;130(17):1903–1910. doi: 10.1182/blood-2017-04-779736. [DOI] [PubMed] [Google Scholar]
- 112.Delfau-Larue MH, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood. 2015;126(5):604–611. doi: 10.1182/blood-2015-02-628792. [DOI] [PubMed] [Google Scholar]
- 113.Halldórsdóttir AM, Lundin A, Murray F, et al. Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia. 2011;25(12):1904–1908. doi: 10.1038/leu.2011.162. [DOI] [PubMed] [Google Scholar]
- 114.Ferrero S, Rossi D, Rinaldi A, et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: a FIL study. Haematologica. 2020;105(6):1604–1612. doi: 10.3324/haematol.2018.214056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eskelund CW, Albertsson-Lindblad A, Kolstad A, et al. Lenalidomide plus bendamustine-rituximab does not overcome the adverse impact of TP53 mutations in mantle cell lymphoma. Haematologica. 2018;103(11):e541–e543. doi: 10.3324/haematol.2018.194399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aukema SM, Hoster E, Rosenwald A, et al. Expression of TP53 is associated with the outcome of MCL independent of MIPI and Ki-67 in trials of the European MCL Network. Blood. 2018;131(4):417–420. doi: 10.1182/blood-2017-07-797019. [DOI] [PubMed] [Google Scholar]
- 117.Scott DW, Abrisqueta P, Wright GW, et al. New molecular assay for the proliferation signature in mantle cell lymphoma applicable to formalin-fixed paraffin-embedded biopsies. J Clin Oncol. 2017;35(15):1668–1677. doi: 10.1200/JCO.2016.70.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Holte H, Beiske K, Boyle M, et al. The MCL35 gene expression proliferation assay predicts high-risk MCL patients in a Norwegian cohort of younger patients given intensive first line therapy. Br J Haematol. 2018;183(2):225–234. doi: 10.1111/bjh.15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Croci GA, Hoster E, Beà S, et al. Reproducibility of histologic prognostic parameters for mantle cell lymphoma: cytology, Ki67, p53 and SOX11. Virchows Arch. 2020;477(2):259–267. doi: 10.1007/s00428-020-02750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang L, Yao Y, Zhang S, et al. Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci Transl Med. 2019;11(491):eaau1167. doi: 10.1126/scitranslmed.aau1167. [DOI] [PubMed] [Google Scholar]
- 121.Zhang S, Jiang VC, Han G, et al. Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma. Nat Commun. 2021;12(1):2877. doi: 10.1038/s41467-021-22872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thompson ER, Nguyen T, Kankanige Y, et al. Single-cell sequencing demonstrates complex resistance landscape in CLL and MCL treated with BTK and BCL2 inhibitors. Blood Adv. 2022;6(2):503–508. doi: 10.1182/bloodadvances.2021006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rahal R, Frick M, Romero R, et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med. 2014;20(1):87–92. doi: 10.1038/nm.3435. [DOI] [PubMed] [Google Scholar]
- 124.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106(1):296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chesi M, Stein CK, Garbitt VM, et al. Monosomic loss of MIR15A/MIR16-1 is a driver of multiple myeloma proliferation and disease progression. Blood Cancer Discov. 2020;1(1):68–81. doi: 10.1158/0008-5472.BCD-19-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Broyl A, Hose D, Lokhorst H, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116(14):2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 128.Walker BA, Mavrommatis K, Wardell CP, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132(6):587–597. doi: 10.1182/blood-2018-03-840132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chng WJ, Dispenzieri A, Chim CS, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28(2):269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 130.Corre J, Munshi NC, Avet-Loiseau H. Risk factors in multiple myeloma: is it time for a revision? Blood. 2021;137(1):16–19. doi: 10.1182/blood.2019004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sonneveld P, Goldschmidt H, Rosiñol L, et al. Bortezomib-based versus nonbortezomib-based induction treatment before autologous stem-cell transplantation in patients with previously untreated multiple myeloma: a meta-analysis of phase III randomized, controlled trials. J Clin Oncol. 2013;31(26):3279–3287. doi: 10.1200/JCO.2012.48.4626. [DOI] [PubMed] [Google Scholar]
- 132.Giri S, Grimshaw A, Bal S, et al. Evaluation of daratumumab for the treatment of multiple myeloma in patients with high-risk cytogenetic factors: a systematic review and meta-analysis. JAMA Oncol. 2020;6(11):1759–1765. doi: 10.1001/jamaoncol.2020.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cavo M, Gay F, Beksac M, et al. Autologous haematopoietic stem-cell transplantation versus bortezomib-melphalan-prednisone, with or without bortezomib-lenalidomide-dexamethasone consolidation therapy, and lenalidomide maintenance for newly diagnosed multiple myeloma (EMN02/HO95): a multicentre, randomised, open-label, phase 3 study. Lancet Haematol. 2020;7(6):e456–e468. doi: 10.1016/S2352-3026(20)30099-5. [DOI] [PubMed] [Google Scholar]
- 134.Kumar SK, Harrison SJ, Cavo M, et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2020;21(12):1630–1642. doi: 10.1016/S1470-2045(20)30525-8. [DOI] [PubMed] [Google Scholar]
- 135.Shaughnessy JD, Jr., Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2006;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 136.Kuiper R, Broyl A, de Knegt Y, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012;26(11):2406–2413. doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- 137.Egan JB, Shi C-X, Tembe W, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120(5):1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Samur MK, Samur AA, Fulciniti M, et al. Genome-wide somatic alterations in multiple myeloma reveal a superior outcome group. J Clin Oncol. 2020;38(27):3107–3118. doi: 10.1200/JCO.20.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Misund K, Keane N, Stein CK, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia. 2020;34(1):322–326. doi: 10.1038/s41375-019-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Oben B, Froyen G, Maclachlan KH, et al. Whole-genome sequencing reveals progressive versus stable myeloma precursor conditions as two distinct entities. Nat Commun. 2021;12(1):1861. doi: 10.1038/s41467-021-22140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maclachlan KH, Rustad EH, Derkach A, et al. Copy number signatures predict chromothripsis and clinical outcomes in newly diagnosed multiple myeloma. Nat Commun. 2021;12(1):5172. doi: 10.1038/s41467-021-25469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rustad EH, Yellapantula VD, Glodzik D, et al. Revealing the impact of structural variants in multiple myeloma. Blood Cancer Discov. 2020;1(3):258–273. doi: 10.1158/2643-3230.BCD-20-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bustoros M, Sklavenitis-Pistofidis R, Park J, et al. Genomic profiling of smoldering multiple myeloma identifies patients at a high risk of disease progression. J Clin Oncol. 2020;38(21):2380–2389. doi: 10.1200/JCO.20.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boyle EM, Deshpande S, Tytarenko R, et al. The molecular make up of smoldering myeloma highlights the evolutionary pathways leading to multiple myeloma. Nat Commun. 2021;12(1):293. doi: 10.1038/s41467-020-20524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mateos M-V, Kumar S, Dimopoulos MA, et al. International Myeloma Working Group risk stratification model for smoldering multiple myeloma (SMM) Blood Cancer J. 2020;10(10):102. doi: 10.1038/s41408-020-00366-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Merz M, Hielscher T, Schult D, et al. Cytogenetic subclone formation and evolution in progressive smoldering multiple myeloma. Leukemia. 2020;34(4):1192–1196. doi: 10.1038/s41375-019-0634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bolli N, Maura F, Minvielle S, et al. Genomic patterns of progression in smoldering multiple myeloma. Nat Commun. 2018;9(1):3363. doi: 10.1038/s41467-018-05058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Treon SP, Xu L, Guerrera ML, et al. Genomic landscape of Waldenstrom macroglobulinemia and its impact on treatment strategies. J Clin Oncol. 2020;38(11):1198–1208. doi: 10.1200/JCO.19.02314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu L, Hunter ZR, Tsakmaklis N, et al. Clonal architecture of CXCR4 WHIM-like mutations in Waldenström macroglobulinaemia. Br J Haematol. 2016;172(5):735–744. doi: 10.1111/bjh.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Varettoni M, Zibellini S, Defrancesco I, et al. Pattern of somatic mutations in patients with Waldenström macroglobulinemia or IgM monoclonal gammopathy of undetermined significance. Haematologica. 2017;102(12):2077–2085. doi: 10.3324/haematol.2017.172718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Varettoni M, Arcaini L, Zibellini S, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenström’s macroglobulinemia and related lymphoid neoplasms. Blood. 2013;121(13):2522–2528. doi: 10.1182/blood-2012-09-457101. [DOI] [PubMed] [Google Scholar]
- 152.Castillo JJ, Itchaki G, Gustine JN, et al. A matched case-control study comparing features, treatment and outcomes between patients with non-IgM lymphoplasmacytic lymphoma and Waldenström macroglobulinemia. Leuk Lymphoma. 2020;61(6):1388–1394. doi: 10.1080/10428194.2020.1719100. [DOI] [PubMed] [Google Scholar]
- 153.Hunter ZR, Xu L, Tsakmaklis N, et al. Insights into the genomic landscape of MYD88 wild-type Waldenström macroglobulinemia. Blood Adv. 2018;2(21):2937–2946. doi: 10.1182/bloodadvances.2018022962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Treon SP, Cao Y, Xu L, Yang G, Liu X, Hunter ZR. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenström macroglobulinemia. Blood. 2014;123(18):2791–2796. doi: 10.1182/blood-2014-01-550905. [DOI] [PubMed] [Google Scholar]
- 155.Treon SP, Gustine J, Xu L, et al. MYD88 wild-type Waldenstrom macroglobulinaemia: differential diagnosis, risk of histological transformation, and overall survival. Br J Haematol. 2018;180(3):374–380. doi: 10.1111/bjh.15049. [DOI] [PubMed] [Google Scholar]
- 156.Abeykoon JP, Paludo J, King RL, et al. MYD88 mutation status does not impact overall survival in Waldenström macroglobulinemia. Am J Hematol. 2018;93(2):187–194. doi: 10.1002/ajh.24955. [DOI] [PubMed] [Google Scholar]
- 157.Laribi K, Poulain S, Willems L, et al. Bendamustine plus rituximab in newly-diagnosed Waldenström macroglobulinaemia patients. A study on behalf of the French Innovative Leukaemia Organization (FILO) Br J Haematol. 2019;186(1):146–149. doi: 10.1111/bjh.15718. [DOI] [PubMed] [Google Scholar]
- 158.Dimopoulos M, Sanz RG, Lee H-P, et al. Zanubrutinib for the treatment of MYD88 wild-type Waldenström macroglobulinemia: a substudy of the phase 3 ASPEN trial. Blood Adv. 2020;4(23):6009–6018. doi: 10.1182/bloodadvances.2020003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kofides A, Hunter ZR, Xu L, et al. Diagnostic next-generation sequencing frequently fails to detect MYD88L265P in Waldenström macroglobulinemia. HemaSphere. 2021;5(8):e624. doi: 10.1097/HS9.0000000000000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Treon SP, Meid K, Gustine J, et al. Long-term follow-up of ibrutinib monotherapy in symptomatic, previously treated patients with Waldenström macroglobulinemia. J Clin Oncol. 2021;39(6):565–575. doi: 10.1200/JCO.20.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Castillo JJ, Meid K, Gustine JN, et al. Long-term follow-up of ibrutinib monotherapy in treatment-naive patients with Waldenstrom macroglobulinemia. Leukemia. 2022;36(2):532–539. doi: 10.1038/s41375-021-01417-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tam CS, Opat S, D'Sa S, et al. A randomized phase 3 trial of zanubrutinib vs ibrutinib in symptomatic Waldenström macroglobulinemia: the ASPEN study. Blood. 2020;136(18):2038–2050. doi: 10.1182/blood.2020006844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Poulain S, Roumier C, Venet-Caillault A, et al. Genomic landscape of CXCR4 mutations in Waldenström macroglobulinemia. Clin Cancer Res. 2016;22(6):1480–1488. doi: 10.1158/1078-0432.CCR-15-0646. [DOI] [PubMed] [Google Scholar]
- 164.Castillo JJ, Xu L, Gustine JN, et al. CXCR4 mutation subtypes impact response and survival outcomes in patients with Waldenström macroglobulinaemia treated with ibrutinib. Br J Haematol. 2019;187(3):356–363. doi: 10.1111/bjh.16088. [DOI] [PubMed] [Google Scholar]
- 165.Wang Y, Gali VL, Xu-Monette ZY, et al. Molecular and genetic biomarkers implemented from next-generation sequencing provide treatment insights in clinical practice for Waldenström macroglobulinemia. Neoplasia. 2021;23(4):361–374. doi: 10.1016/j.neo.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gustine JN, Xu L, Yang G, et al. Bone marrow involvement and subclonal diversity impairs detection of mutated CXCR4 by diagnostic next-generation sequencing in Waldenström macroglobulinaemia. Br J Haematol. 2021;194(4):730–733. doi: 10.1111/bjh.17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Guerrera ML, Tsakmaklis N, Xu L, et al. MYD88 mutated and wild-type Waldenström’s macroglobulinemia: characterization of chromosome 6q gene losses and their mutual exclusivity with mutations in CXCR4. Haematologica. 2018;103(9):e408–e411. doi: 10.3324/haematol.2018.190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jiménez C, Alonso-Álvarez S, Alcoceba M, et al. From Waldenström's macroglobulinemia to aggressive diffuse large B-cell lymphoma: a whole-exome analysis of abnormalities leading to transformation. Blood Cancer J. 2017;7(8):e591. doi: 10.1038/bcj.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu L, Tsakmaklis N, Yang G, et al. Acquired mutations associated with ibrutinib resistance in Waldenström macroglobulinemia. Blood. 2017;129(18):2519–2525. doi: 10.1182/blood-2017-01-761726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Poulain S, Roumier C, Bertrand E, et al. TP53 mutation and its prognostic significance in Waldenstrom’s macroglobulinemia. Clin Cancer Res. 2017;23(20):6325–6335. doi: 10.1158/1078-0432.CCR-17-0007. [DOI] [PubMed] [Google Scholar]
- 171.Gustine JN, Tsakmaklis N, Demos MG, et al. TP53 mutations are associated with mutated MYD88 and CXCR4, and confer an adverse outcome in Waldenström macroglobulinaemia. Br J Haematol. 2019;184(2):242–245. doi: 10.1111/bjh.15560. [DOI] [PubMed] [Google Scholar]
- 172.Hilton LK, Tang J, Ben-Neriah S, et al. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood. 2019;134(18):1528–1532. doi: 10.1182/blood.2019002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123(8):1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 175.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lionakis MS, Dunleavy K, Roschewski M, et al. Inhibition of B cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell. 2017;31(6):833–843.e835. doi: 10.1016/j.ccell.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Grommes C, Younes A. Ibrutinib in PCNSL: the curious cases of clinical responses and aspergillosis. Cancer Cell. 2017;31(6):731–733. doi: 10.1016/j.ccell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Younes A, Sehn LH, Johnson P, et al. Randomized phase III trial of ibrutinib and rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in non-germinal center B-cell diffuse large B-cell lymphoma. J Clin Oncol. 2019;37(15):1285–1295. doi: 10.1200/JCO.18.02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell. 2021;39(12):1643–1653.e1643. doi: 10.1016/j.ccell.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Davies A, Cummin TE, Barrans S, et al. Gene-expression profiling of bortezomib added to standard chemoimmunotherapy for diffuse large B-cell lymphoma (REMoDL-B): an open-label, randomised, phase 3 trial. Lancet Oncol. 2019;20(5):649–662. doi: 10.1016/S1470-2045(18)30935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nowakowski GS, Chiappella A, Gascoyne RD, et al. ROBUST: a phase III study of lenalidomide plus R-CHOP versus placebo plus R-CHOP in previously untreated patients with ABC-type diffuse large B-cell lymphoma. J Clin Oncol. 2021;39(12):1317–1328. doi: 10.1200/JCO.20.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020;135(20):1759–1771. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551–568.e514. doi: 10.1016/j.ccell.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Runge HFP, Lacy S, Barrans S, et al. Application of the LymphGen classification tool to 928 clinically and genetically-characterised cases of diffuse large B cell lymphoma (DLBCL) Br J Haematol. 2021;192(1):216–220. doi: 10.1111/bjh.17132. [DOI] [PubMed] [Google Scholar]
- 189.Kotlov N, Bagaev A, Revuelta MV, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov. 2021;11(6):1468–1489. doi: 10.1158/2159-8290.CD-20-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Copie-Bergman C, Cuilliere-Dartigues P, Baia M, et al. MYC-IG rearrangements are negative predictors of survival in DLBCL patients treated with immunochemotherapy: a GELA/LYSA study. Blood. 2015;126(22):2466–2474. doi: 10.1182/blood-2015-05-647602. [DOI] [PubMed] [Google Scholar]
- 191.Rosenwald A, Bens S, Advani R, et al. Prognostic significance of MYC rearrangement and translocation partner in diffuse large B-cell lymphoma: a study by the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2019;37(35):3359–3368. doi: 10.1200/JCO.19.00743. [DOI] [PubMed] [Google Scholar]
- 192.Chong LC, Ben-Neriah S, Slack GW, et al. High-resolution architecture and partner genes of MYC rearrangements in lymphoma with DLBCL morphology. Blood Adv. 2018;2(20):2755–2765. doi: 10.1182/bloodadvances.2018023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Allahyar A, Pieterse M, Swennenhuis J, et al. Robust detection of translocations in lymphoma FFPE samples using targeted locus capture-based sequencing. Nat Commun. 2021;12(1):3361. doi: 10.1038/s41467-021-23695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cucco F, Barrans S, Sha C, et al. Distinct genetic changes reveal evolutionary history and heterogeneous molecular grade of DLBCL with MYC/BCL2 double-hit. Leukemia. 2020;34(5):1329–1341. doi: 10.1038/s41375-019-0691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Evrard SM, Pericart S, Grand D, et al. Targeted next generation sequencing reveals high mutation frequency of CREBBP, BCL2 and KMT2D in high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements. Haematologica. 2019;104(4):e154–e157. doi: 10.3324/haematol.2018.198572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Collinge BJ, Hilton LK, Wong J, et al. The mutational landscape of double/triple hit high-grade B-cell lymphoma with BCL2 rearrangement (DH/TH-BCL2) - an LLMPP project. Hematol Oncol. 2021;39(S2) [Google Scholar]
- 197.Johnson SM, Umakanthan JM, Yuan J, et al. Lymphomas with pseudo-double-hit BCL6-MYC translocations due to t(3;8)(q27;q24) are associated with a germinal center immunophenotype, extranodal involvement, and frequent BCL2 translocations. Hum Pathol. 2018;80:192–200. doi: 10.1016/j.humpath.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 198.Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022;36(7):1720–1748. doi: 10.1038/s41375-022-01620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sha C, Barrans S, Cucco F, et al. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J Clin Oncol. 2019;37(3):202–212. doi: 10.1200/JCO.18.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Collinge BJ, Hilton LK, Wong J, et al. Characterization of the genetic landscape of high-grade B-cell lymphoma,–NOS - an LLMPP project. Hematol Oncol. 2021;39(S2) [Google Scholar]
- 201.Richter J, John K, Staiger AM, et al. Epstein-Barr virus status of sporadic Burkitt lymphoma is associated with patient age and mutational features. Br J Haematol. 2022;196(3):681–689. doi: 10.1111/bjh.17874. [DOI] [PubMed] [Google Scholar]
- 202.Crombie J, LaCasce A. The treatment of Burkitt lymphoma in adults. Blood. 2021;137(6):743–750. doi: 10.1182/blood.2019004099. [DOI] [PubMed] [Google Scholar]
- 203.Abate F, Ambrosio MR, Mundo L, et al. Distinct viral and mutational spectrum of endemic Burkitt lymphoma. PLoS Pathog. 2015;11(10):e1005158. doi: 10.1371/journal.ppat.1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Grande BM, Gerhard DS, Jiang A, et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood. 2019;133(12):1313–1324. doi: 10.1182/blood-2018-09-871418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hutcheson RL, Chakravorty A, Sugden B. Burkitt lymphomas evolve to escape dependencies on Epstein-Barr virus. Front Cell Infect Microbiol. 2021;10 doi: 10.3389/fcimb.2020.606412. 606412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Newman AM, Zaka M, Zhou P, et al. Genomic abnormalities of TP53 define distinct risk groups of paediatric B-cell non-Hodgkin lymphoma. Leukemia. 2022;36(3):781–789. doi: 10.1038/s41375-021-01444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gong C, Krupka JA, Gao J, et al. Sequential inverse dysregulation of the RNA helicases DDX3X and DDX3Y facilitates MYC-driven lymphomagenesis. Mol Cell. 2021;81(19):4059–4075.e4011. doi: 10.1016/j.molcel.2021.07.041. [DOI] [PubMed] [Google Scholar]
- 209.Schmidt J, Gong S, Marafioti T, et al. Genome-wide analysis of pediatric-type follicular lymphoma reveals low genetic complexity and recurrent alterations of TNFRSF14 gene. Blood. 2016;128(8):1101–1111. doi: 10.1182/blood-2016-03-703819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Martin-Guerrero I, Salaverria I, Burkhardt B, et al. Recurrent loss of heterozygosity in 1p36 associated with TNFRSF14 mutations in IRF4 translocation negative pediatric follicular lymphomas. Haematologica. 2013;98(8):1237–1241. doi: 10.3324/haematol.2012.073916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Louissaint A, Jr., Schafernak KT, Geyer JT, et al. Pediatric-type nodal follicular lymphoma: a biologically distinct lymphoma with frequent MAPK pathway mutations. Blood. 2016;128(8):1093–1100. doi: 10.1182/blood-2015-12-682591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ozawa MG, Bhaduri A, Chisholm KM, et al. A study of the mutational landscape of pediatric-type follicular lymphoma and pediatric nodal marginal zone lymphoma. Mod Pathol. 2016;29(10):1212–1220. doi: 10.1038/modpathol.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schmidt J, Ramis-Zaldivar JE, Nadeu F, et al. Mutations of MAP2K1 are frequent in pediatric-type follicular lymphoma and result in ERK pathway activation. Blood. 2017;130(3):323–327. doi: 10.1182/blood-2017-03-776278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Salmeron-Villalobos J, Egan C, Borgmann V, et al. PNMZL and PTFL: morphological variants with a common molecular profile - a unifying hypothesis. Blood Adv. 2022;6(16):4661–4674. doi: 10.1182/bloodadvances.2022007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lim S, Lim KY, Koh J, et al. Pediatric-type indolent B-cell lymphomas with overlapping clinical, pathologic, and genetic features. Am J Surg Pathol. 2022 doi: 10.1097/PAS.0000000000001932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Salaverria I, Philipp C, Oschlies I, et al. Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood. 2011;118(1):139–147. doi: 10.1182/blood-2011-01-330795. [DOI] [PubMed] [Google Scholar]
- 217.Klapper W, Kreuz M, Kohler CW, et al. Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood. 2012;119(8):1882–1887. doi: 10.1182/blood-2011-10-388470. [DOI] [PubMed] [Google Scholar]
- 218.Ramis-Zaldivar JE, Gonzalez-Farré B, Balagué O, et al. Distinct molecular profile of IRF4-rearranged large B-cell lymphoma. Blood. 2020;135(4):274–286. doi: 10.1182/blood.2019002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Salaverria I, Martin-Guerrero I, Burkhardt B, et al. High resolution copy number analysis of IRF4 translocation-positive diffuse large B-cell and follicular lymphomas. Genes Chromosom Cancer. 2013;52(2):150–155. doi: 10.1002/gcc.22014. [DOI] [PubMed] [Google Scholar]
- 220.Liu Q, Salaverria I, Pittaluga S, et al. Follicular lymphomas in children and young adults: a comparison of the pediatric variant with usual follicular lymphoma. Am J Surg Pathol. 2013;37(3):333–343. doi: 10.1097/PAS.0b013e31826b9b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Frauenfeld L, Castrejon-de-Anta N, Ramis-Zaldivar JE, et al. Diffuse large B-cell lymphomas in adults with aberrant coexpression of CD10, BCL6, and MUM1 are enriched in IRF4 rearrangements. Blood Adv. 2022;6(7):2361–2372. doi: 10.1182/bloodadvances.2021006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Salaverria I, Martin-Guerrero I, Wagener R, et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014;123(8):1187–1198. doi: 10.1182/blood-2013-06-507996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Grygalewicz B, Woroniecka R, Rymkiewicz G, et al. The 11q-gain/loss aberration occurs recurrently in MYC-negative Burkitt-like lymphoma with 11q aberration, as well as MYC-positive Burkitt lymphoma and MYC-positive high-grade B-cell lymphoma, NOS. Am J Clin Pathol. 2017;149(1):17–28. doi: 10.1093/ajcp/aqx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wagener R, Seufert J, Raimondi F, et al. The mutational landscape of Burkitt-like lymphoma with 11q aberration is distinct from that of Burkitt lymphoma. Blood. 2019;133(9):962–966. doi: 10.1182/blood-2018-07-864025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gonzalez-Farre B, Ramis-Zaldivar JE, Salmeron-Villalobos J, et al. Burkitt-like lymphoma with 11q aberration: a germinal center-derived lymphoma genetically unrelated to Burkitt lymphoma. Haematologica. 2019;104(9):1822–1829. doi: 10.3324/haematol.2018.207928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pienkowska-Grela B, Rymkiewicz G, Grygalewicz B, et al. Partial trisomy 11, dup(11)(q23q13), as a defect characterizing lymphomas with Burkitt pathomorphology without MYC gene rearrangement. Med Oncol. 2011;28(4):1589–1595. doi: 10.1007/s12032-010-9614-0. [DOI] [PubMed] [Google Scholar]
- 227.Bonetti P, Testoni M, Scandurra M, et al. Deregulation of ETS1 and FLI1 contributes to the pathogenesis of diffuse large B-cell lymphoma. Blood. 2013;122(13):2233–2241. doi: 10.1182/blood-2013-01-475772. [DOI] [PubMed] [Google Scholar]
- 228.Rinaldi A, Capello D, Scandurra M, et al. Single nucleotide polymorphism-arrays provide new insights in the pathogenesis of post-transplant diffuse large B-cell lymphoma. Br J Haematol. 2010;149(4):569–577. doi: 10.1111/j.1365-2141.2010.08125.x. [DOI] [PubMed] [Google Scholar]
- 229.Mottok A, Wright G, Rosenwald A, et al. Molecular classification of primary mediastinal large B-cell lymphoma using routinely available tissue specimens. Blood. 2018;132(22):2401–2405. doi: 10.1182/blood-2018-05-851154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Duns G, Viganò E, Ennishi D, et al. Characterization of DLBCL with a PMBL gene expression signature. Blood. 2021;138(2):136–148. doi: 10.1182/blood.2020007683. [DOI] [PubMed] [Google Scholar]
- 231.Bobee V, Ruminy P, Marchand V, et al. Determination of molecular subtypes of diffuse large B-cell lymphoma using a reverse transcriptase multiplex ligation-dependent probe amplification classifier: a CALYM study. J Mol Diagn. 2017;19(6):892–904. doi: 10.1016/j.jmoldx.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 232.Mottok A, Hung SS, Chavez EA, et al. Integrative genomic analysis identifies key pathogenic mechanisms in primary mediastinal large B-cell lymphoma. Blood. 2019;134(10):802–813. doi: 10.1182/blood.2019001126. [DOI] [PubMed] [Google Scholar]
- 233.Chapuy B, Stewart C, Dunford AJ, et al. Genomic analyses of PMBL reveal new drivers and mechanisms of sensitivity to PD-1 blockade. Blood. 2019;134(26):2369–2382. doi: 10.1182/blood.2019002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pittaluga S, Nicolae A, Wright GW, et al. Gene expression profiling of mediastinal gray zone lymphoma and its relationship to primary mediastinal B-cell lymphoma and classical Hodgkin lymphoma. Blood Cancer Discov. 2020;1(2):155–161. doi: 10.1158/2643-3230.BCD-20-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Dunleavy K, Grant C, Eberle FC, Pittaluga S, Jaffe ES, Wilson WH. Gray zone lymphoma: better treated like Hodgkin lymphoma or mediastinal large B-cell lymphoma? Curr Hematol Malig Rep. 2012;7(3):241–247. doi: 10.1007/s11899-012-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sarkozy C, Hung SS, Chavez EA, et al. Mutational landscape of gray zone lymphoma. Blood. 2021;137(13):1765–1776. doi: 10.1182/blood.2020007507. [DOI] [PubMed] [Google Scholar]
- 237.Sánchez-Espiridión B, Sánchez-Aguilera A, Montalbán C, et al. A TaqMan low-density array to predict outcome in advanced Hodgkin's lymphoma using paraffin-embedded samples. Clin Cancer Res. 2009;15(4):1367–1375. doi: 10.1158/1078-0432.CCR-08-1119. [DOI] [PubMed] [Google Scholar]
- 238.Scott DW, Chan FC, Hong F, et al. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J Clin Oncol. 2013;31(6):692–700. doi: 10.1200/JCO.2012.43.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jachimowicz RD, Klapper W, Glehr G, et al. Gene expression-based outcome prediction in advanced stage classical Hodgkin lymphoma treated with BEACOPP. Leukemia. 2021;35(12):3589–3593. doi: 10.1038/s41375-021-01314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Burton CH, Scott D, Kirkwood AA, et al. Application of a gene expression-based model in combination with FDG-PET imaging to predict treatment response in advanced Hodgkin lymphoma in the RATHL study (CRUK/07/03) Hematol Oncol. 2017;35(S2) [Google Scholar]
- 241.Scott DW, Li H, Harvey Y, et al. The 23-gene gene expression-based assay does not predict interim PET scan results after ABVD in advanced stage classical Hodgkin lymphoma in the US Intergroup S0816 trial. Hematol Oncol. 2017;35(S2) [Google Scholar]
- 242.Chan FC, Mottok A, Gerrie AS, et al. Prognostic model to predict post-autologous stem-cell transplantation outcomes in classical Hodgkin lymphoma. J Clin Oncol. 2017;35(32):3722–3733. doi: 10.1200/JCO.2017.72.7925. [DOI] [PubMed] [Google Scholar]
- 243.Johnston RL, Mottok A, Chan FC, et al. A gene expression–based model predicts outcome in children with intermediate-risk classical Hodgkin lymphoma. Blood. 2022;139(6):889–893. doi: 10.1182/blood.2021011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Calvente L, Tremblay-LeMay R, Xu W, et al. Validation of the RHL30 digital gene expression assay as a prognostic biomarker for relapsed Hodgkin lymphoma. Br J Haematol. 2020;190(6):864–868. doi: 10.1111/bjh.16777. [DOI] [PubMed] [Google Scholar]
- 245.Tiacci E, Ladewig E, Schiavoni G, et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood. 2018;131(22):2454–2465. doi: 10.1182/blood-2017-11-814913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131(22):2413–2425. doi: 10.1182/blood-2017-11-812073. [DOI] [PubMed] [Google Scholar]
- 247.Wienand K, Chapuy B, Stewart C, et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv. 2019;3(23):4065–4080. doi: 10.1182/bloodadvances.2019001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Roemer MGM, Redd RA, Cader FZ, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol. 2018;36(10):942–950. doi: 10.1200/JCO.2017.77.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sobesky S, Mammadova L, Cirillo M, et al. In-depth cell-free DNA sequencing reveals genomic landscape of Hodgkin’s lymphoma and facilitates ultrasensitive residual disease detection. Med. 2021;2(10):1171–1193.e1111. doi: 10.1016/j.medj.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 250.Foss H, Anagnostopoulos I, Araujo I, et al. Anaplastic large-cell lymphomas of T-cell and null-cell phenotype express cytotoxic molecules. Blood. 1996;88(10):4005–4011. [PubMed] [Google Scholar]
- 251.Benharroch D, Meguerian-Bedoyan Z, Lamant L, et al. ALK-positive lymphoma: a single disease with a broad spectrum of morphology. Blood. 1998;91(6):2076–2084. [PubMed] [Google Scholar]
- 252.Melchers RC, Willemze R, Bekkenk MW, et al. Frequency and prognosis of associated malignancies in 504 patients with lymphomatoid papulosis. J Eur Acad Dermatol Venereol. 2020;34(2):260–266. doi: 10.1111/jdv.16065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Melchers RC, Willemze R, van de Loo M, et al. Clinical, histologic, and molecular characteristics of anaplastic lymphoma kinase-positive primary cutaneous anaplastic large cell lymphoma. Am J Surg Pathol. 2020;44(6):776–781. doi: 10.1097/PAS.0000000000001449. [DOI] [PubMed] [Google Scholar]
- 254.Prokoph N, Larose H, Lim MS, Burke GAA, Turner SD. Treatment options for paediatric anaplastic large cell lymphoma (ALCL): current standard and beyond. Cancers (Basel) 2018;10(4):99. doi: 10.3390/cancers10040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Larose H, Prokoph N, Matthews JD, et al. Whole exome sequencing reveals NOTCH1 mutations in anaplastic large cell lymphoma and points to Notch both as a key pathway and a potential therapeutic target. Haematologica. 2021;106(6):1693–1704. doi: 10.3324/haematol.2019.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124(9):1473–1480. doi: 10.1182/blood-2014-04-571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pedersen MB, Hamilton-Dutoit SJ, Bendix K, et al. DUSP22 and TP63 rearrangements predict outcome of ALK-negative anaplastic large cell lymphoma: a Danish cohort study. Blood. 2017;130(4):554–557. doi: 10.1182/blood-2016-12-755496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Luchtel RA, Zimmermann MT, Hu G, et al. Recurrent MSCE116K mutations in ALK-negative anaplastic large cell lymphoma. Blood. 2019;133(26):2776–2789. doi: 10.1182/blood.2019000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hapgood G, Ben-Neriah S, Mottok A, et al. Identification of high-risk DUSP22-rearranged ALK-negative anaplastic large cell lymphoma. Br J Haematol. 2019;186(3):e28–e31. doi: 10.1111/bjh.15860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Feldman AL, Dogan A, Smith DI, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. 2011;117(3):915–919. doi: 10.1182/blood-2010-08-303305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Luchtel RA, Dasari S, Oishi N, et al. Molecular profiling reveals immunogenic cues in anaplastic large cell lymphomas with DUSP22 rearrangements. Blood. 2018;132(13):1386–1398. doi: 10.1182/blood-2018-03-838524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Karai LJ, Kadin ME, Hsi ED, et al. Chromosomal rearrangements of 6p25.3 define a new subtype of lymphomatoid papulosis. Am J Surg Pathol. 2013;37(8):1173–1181. doi: 10.1097/PAS.0b013e318282d01e. [DOI] [PubMed] [Google Scholar]
- 263.Wada DA, Law ME, Hsi ED, et al. Specificity of IRF4 translocations for primary cutaneous anaplastic large cell lymphoma: a multicenter study of 204 skin biopsies. Mod Pathol. 2011;24(4):596–605. doi: 10.1038/modpathol.2010.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fauconneau A, Pham-Ledard A, Cappellen D, et al. Assessment of diagnostic criteria between primary cutaneous anaplastic large-cell lymphoma and CD30-rich transformed mycosis fungoides; a study of 66 cases. Br J Dermatol. 2015;172(6):1547–1554. doi: 10.1111/bjd.13690. [DOI] [PubMed] [Google Scholar]
- 265.Vasmatzis G, Johnson SH, Knudson RA, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. 2012;120(11):2280–2289. doi: 10.1182/blood-2012-03-419937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Boi M, Rinaldi A, Kwee I, et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. 2013;122(15):2683–2693. doi: 10.1182/blood-2013-04-497933. [DOI] [PubMed] [Google Scholar]
- 267.Klairmont MM, Ward N. Co-occurring rearrangements of DUSP22 and TP63 define a rare genetic subset of ALK-negative anaplastic large cell lymphoma with inferior survival outcomes. Leuk Lymphoma. 2022;63(2):506–508. doi: 10.1080/10428194.2021.1984457. [DOI] [PubMed] [Google Scholar]
- 268.Scarfò I, Pellegrino E, Mereu E, et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood. 2016;127(2):221–232. doi: 10.1182/blood-2014-12-614503. [DOI] [PubMed] [Google Scholar]
- 269.Chiarle R, Simmons WJ, Cai H, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11(6):623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 270.Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516–532. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Fitzpatrick MJ, Massoth LR, Marcus C, et al. JAK2 rearrangements are a recurrent alteration in CD30+ systemic T-cell lymphomas with anaplastic morphology. Am J Surg Pathol. 2021;45(7):895–904. doi: 10.1097/PAS.0000000000001708. [DOI] [PubMed] [Google Scholar]
- 272.Hu G, Dasari S, Asmann YW, et al. Targetable fusions of the FRK tyrosine kinase in ALK-negative anaplastic large cell lymphoma. Leukemia. 2018;32(2):565–569. doi: 10.1038/leu.2017.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lobello C, Tichy B, Bystry V, et al. STAT3 and TP53 mutations associate with poor prognosis in anaplastic large cell lymphoma. Leukemia. 2021;35(5):1500–1505. doi: 10.1038/s41375-020-01093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Velusamy T, Kiel MJ, Sahasrabuddhe AA, et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood. 2014;124(25):3768–3771. doi: 10.1182/blood-2014-07-588434. [DOI] [PubMed] [Google Scholar]
- 275.Quesada AE, Zhang Y, Ptashkin R, et al. Next generation sequencing of breast implant-associated anaplastic large cell lymphomas reveals a novel STAT3-JAK2 fusion among other activating genetic alterations within the JAK-STAT pathway. Breast J. 2021;27(4):314–321. doi: 10.1111/tbj.14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Laurent C, Nicolae A, Laurent C, et al. Gene alterations in epigenetic modifiers and JAK-STAT signaling are frequent in breast implant–associated ALCL. Blood. 2020;135(5):360–370. doi: 10.1182/blood.2019001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Los-de Vries GT, de Boer M, van Dijk E, et al. Chromosome 20 loss is characteristic of breast implant–associated anaplastic large cell lymphoma. Blood. 2020;136(25):2927–2932. doi: 10.1182/blood.2020005372. [DOI] [PubMed] [Google Scholar]
- 278.Moskowitz AJ, Ghione P, Jacobsen E, et al. A phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood. 2021;138(26):2828–2837. doi: 10.1182/blood.2021013379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Vallois D, Dobay MPD, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell–derived lymphomas. Blood. 2016;128(11):1490–1502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 280.Heavican TB, Bouska A, Yu J, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133(15):1664–1676. doi: 10.1182/blood-2018-09-872549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Maura F, Dodero A, Carniti C, et al. CDKN2A deletion is a frequent event associated with poor outcome in patients with peripheral T-cell lymphoma not otherwise specified (PTCL-NOS) Haematologica. 2020;106(11):2918–2926. doi: 10.3324/haematol.2020.262659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Drieux F, Ruminy P, Sater V, et al. Detection of gene fusion transcripts in peripheral T-cell lymphoma using a multiplexed targeted sequencing assay. J Mol Diagn. 2021;23(8):929–940. doi: 10.1016/j.jmoldx.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 283.Debackere K, Marcelis L, Demeyer S, et al. Fusion transcripts FYN-TRAF3IP2 and KHDRBS1-LCK hijack T cell receptor signaling in peripheral T-cell lymphoma, not otherwise specified. Nat Commun. 2021;12(1):3705. doi: 10.1038/s41467-021-24037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Watatani Y, Sato Y, Miyoshi H, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33(12):2867–2883. doi: 10.1038/s41375-019-0473-1. [DOI] [PubMed] [Google Scholar]
- 285.Syrykh C, Gorez P, Péricart S, et al. Molecular diagnosis of T-cell lymphoma: a correlative study of PCR-based T-cell clonality assessment and targeted NGS. Blood Adv. 2021;5(22):4590–4593. doi: 10.1182/bloodadvances.2021005249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Dobson R, Du PY, Rásó-Barnett L, et al. Early detection of T-cell lymphoma with T follicular helper phenotype by RHOA mutation analysis. Haematologica. 2021;107(2):489–499. doi: 10.3324/haematol.2020.265991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Nicolae A, Bouilly J, Lara D, et al. Nodal cytotoxic peripheral T-cell lymphoma occurs frequently in the clinical setting of immunodysregulation and is associated with recurrent epigenetic alterations. Mod Pathol. 2022;35(8):1126–1136. doi: 10.1038/s41379-022-01022-w. [DOI] [PubMed] [Google Scholar]
- 288.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Tiacci E, Venanzi A, Ascani S, et al. High-risk clonal hematopoiesis as the origin of AITL and NPM1-mutated AML. N Engl J Med. 2018;379(10):981–984. doi: 10.1056/NEJMc1806413. [DOI] [PubMed] [Google Scholar]
- 290.Lewis NE, Petrova-Drus K, Huet S, et al. Clonal hematopoiesis in angioimmunoblastic T-cell lymphoma with divergent evolution to myeloid neoplasms. Blood Adv. 2020;4(10):2261–2271. doi: 10.1182/bloodadvances.2020001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lemonnier F, Dupuis J, Sujobert P, et al. Treatment with 5-azacytidine induces a sustained response in patients with angioimmunoblastic T-cell lymphoma. Blood. 2018;132(21):2305–2309. doi: 10.1182/blood-2018-04-840538. [DOI] [PubMed] [Google Scholar]
- 292.Ghione P, Faruque P, Mehta-Shah N, et al. T follicular helper phenotype predicts response to histone deacetylase inhibitors in relapsed/refractory peripheral T-cell lymphoma. Blood Adv. 2020;4(19):4640–4647. doi: 10.1182/bloodadvances.2020002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Falchi L, Ma H, Klein S, et al. Combined oral 5-azacytidine and romidepsin are highly effective in patients with PTCL: a multicenter phase 2 study. Blood. 2021;137(16):2161–2170. doi: 10.1182/blood.2020009004. [DOI] [PubMed] [Google Scholar]
- 294.Küçük C, Jiang B, Hu X, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK cells. Nat Commun. 2015;6(1):6025. doi: 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Hu G, Phillips JL, Dasari S, et al. Targetability of STAT3-JAK2 fusions: implications for T-cell lymphoproliferative disorders of the gastrointestinal tract. Leukemia. 2020;34(5):1467–1471. doi: 10.1038/s41375-019-0678-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Xiao W, Gupta GK, Yao J, et al. Recurrent somatic JAK3 mutations in NK-cell enteropathy. Blood. 2019;134(12):986–991. doi: 10.1182/blood.2019001443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sharma A, Oishi N, Boddicker RL, et al. Recurrent STAT3-JAK2 fusions in indolent T-cell lymphoproliferative disorder of the gastrointestinal tract. Blood. 2018;131(20):2262–2266. doi: 10.1182/blood-2018-01-830968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Craig RS, Nupam P, Vundavalli VM, et al. Genetic and phenotypic characterization of indolent T-cell lymphoproliferative disorders of the gastrointestinal tract. Haematologica. 2020;105(7):1895–1906. doi: 10.3324/haematol.2019.230961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Montes-Moreno S, King RL, Oschlies I, et al. Update on lymphoproliferative disorders of the gastrointestinal tract: disease spectrum from indolent lymphoproliferations to aggressive lymphomas. Virchows Arch. 2020;476(5):667–681. doi: 10.1007/s00428-019-02704-8. [DOI] [PubMed] [Google Scholar]
- 300.Roberti A, Dobay MP, Bisig B, et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat Commun. 2016;7:12602. doi: 10.1038/ncomms12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Moffitt AB, Ondrejka SL, McKinney M, et al. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J Exp Med. 2017;214(5):1371–1386. doi: 10.1084/jem.20160894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Tomita S, Kikuti YY, Carreras J, et al. Monomorphic epitheliotropic intestinal T-cell lymphoma in Asia frequently shows SETD2 alterations. Cancers (Basel) 2020;12(12):3539. doi: 10.3390/cancers12123539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Veloza L, Cavalieri D, Missiaglia E, et al. Monomorphic epitheliotropic intestinal T-cell lymphoma comprises morphologic and genomic heterogeneity impacting outcome. Haematologica. 2022 doi: 10.3324/haematol.2022.281226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Cording S, Lhermitte L, Malamut G, et al. Oncogenetic landscape of lymphomagenesis in coeliac disease. Gut. 2022;71(3):497–508. doi: 10.1136/gutjnl-2020-322935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Verkarre V, Romana SP, Cellier C, et al. Recurrent partial trisomy 1q22-q44 in clonal intraepithelial lymphocytes in refractory celiac sprue. Gastroenterology. 2003;125(1):40–46. doi: 10.1016/s0016-5085(03)00692-9. [DOI] [PubMed] [Google Scholar]
- 306.Soderquist CR, Lewis SK, Gru AA, et al. Immunophenotypic spectrum and genomic landscape of refractory celiac disease type II. Am J Surg Pathol. 2021;45(7):905–916. doi: 10.1097/PAS.0000000000001658. [DOI] [PubMed] [Google Scholar]
- 307.Tometten I, Felgentreff K, Hönig M, et al. Increased proportions of γδ T lymphocytes in atypical SCID associate with disease manifestations. Clin Immunol. 2019;201:30–34. doi: 10.1016/j.clim.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 308.Roden AC, Morice WG, Hanson CA. Immunophenotypic attributes of benign peripheral blood gammadelta T cells and conditions associated with their increase. Arch Pathol Lab Med. 2008;132(11):1774–1780. doi: 10.5858/132.11.1774. [DOI] [PubMed] [Google Scholar]
- 309.Zhang S, Bayerl MG. Florid splenic γ/δ T-cell proliferation in patients with splenomegaly and cytopenias: a "high stakes" diagnostic challenge. Hum Pathol. 2017;66:216–221. doi: 10.1016/j.humpath.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 310.Wlodarska I, Martin-Garcia N, Achten R, et al. Fluorescence in situ hybridization study of chromosome 7 aberrations in hepatosplenic T-cell lymphoma: isochromosome 7q as a common abnormality accumulating in forms with features of cytologic progression. Genes Chromosom Cancer. 2002;33(3):243–251. doi: 10.1002/gcc.10021. [DOI] [PubMed] [Google Scholar]
- 311.Weidmann E. Hepatosplenic T cell lymphoma. A review on 45 cases since the first report describing the disease as a distinct lymphoma entity in 1990. Leukemia. 2000;14(6):991–997. doi: 10.1038/sj.leu.2401784. [DOI] [PubMed] [Google Scholar]
- 312.Pro B, Allen P, Behdad A. Hepatosplenic T-cell lymphoma: a rare but challenging entity. Blood. 2020;136(18):2018–2026. doi: 10.1182/blood.2019004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Nicolae A, Xi L, Pittaluga S, et al. Frequent STAT5B mutations in γδ hepatosplenic T-cell lymphomas. Leukemia. 2014;28(11):2244–2248. doi: 10.1038/leu.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.McKinney M, Moffitt AB, Gaulard P, et al. The genetic basis of hepatosplenic T-cell lymphoma. Cancer Discov. 2017;7(4):369–379. doi: 10.1158/2159-8290.CD-16-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hong M, Lee T, Young Kang S, Kim S-J, Kim W, Ko Y-H. Nasal-type NK/T-cell lymphomas are more frequently T rather than NK lineage based on T-cell receptor gene, RNA, and protein studies: lineage does not predict clinical behavior. Mod Pathol. 2016;29(5):430–443. doi: 10.1038/modpathol.2016.47. [DOI] [PubMed] [Google Scholar]
- 316.Li Z, Xia Y, Feng LN, et al. Genetic risk of extranodal natural killer T-cell lymphoma: a genome-wide association study. Lancet Oncol. 2016;17(9):1240–1247. doi: 10.1016/S1470-2045(16)30148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Lin GW, Xu C, Chen K, et al. Genetic risk of extranodal natural killer T-cell lymphoma: a genome-wide association study in multiple populations. Lancet Oncol. 2020;21(2):306–316. doi: 10.1016/S1470-2045(19)30799-5. [DOI] [PubMed] [Google Scholar]
- 318.Tian X-P, Ma S-Y, Young KH, et al. A composite single-nucleotide polymorphism prediction signature for extranodal natural killer/T-cell lymphoma. Blood. 2021;138(6):452–463. doi: 10.1182/blood.2020010637. [DOI] [PubMed] [Google Scholar]
- 319.Jiang L, Gu Z-H, Yan Z-X, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47(9):1061–1066. doi: 10.1038/ng.3358. [DOI] [PubMed] [Google Scholar]
- 320.Koo GC, Tan SY, Tang T, et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. 2012;2(7):591–597. doi: 10.1158/2159-8290.CD-12-0028. [DOI] [PubMed] [Google Scholar]
- 321.Lee S, Park HY, Kang SY, et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget. 2015;6(19):17764–17776. doi: 10.18632/oncotarget.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Song TL, Nairismägi M-L, Laurensia Y, et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood. 2018;132(11):1146–1158. doi: 10.1182/blood-2018-01-829424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Montes-Mojarro IA, Chen BJ, Ramirez-Ibarguen AF, et al. Mutational profile and EBV strains of extranodal NK/T-cell lymphoma, nasal type in Latin America. Mod Pathol. 2020;33(5):781–791. doi: 10.1038/s41379-019-0415-5. [DOI] [PubMed] [Google Scholar]
- 324.Dong G, Liu X, Wang L, et al. Genomic profiling identifies distinct genetic subtypes in extra-nodal natural killer/T-cell lymphoma. Leukemia. 2022;36(8):2064–2075. doi: 10.1038/s41375-022-01623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Gao Y, Li Y, Ma G, Zhao G, Liu H. KMT2D and TP53 mutation status improve the prognostic value of the International Prognostic Index (IPI) stratification in ENKTL patients. Neoplasma. 2020;67(3):636–643. doi: 10.4149/neo_2020_190307N194. [DOI] [PubMed] [Google Scholar]
- 326.Xiong J, Cui BW, Wang N, et al. Genomic and transcriptomic characterization of natural killer T cell lymphoma. Cancer Cell. 2020;37(3):403–419.e406. doi: 10.1016/j.ccell.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 327.Arai A. Chronic active Epstein-Barr virus infection: the elucidation of the pathophysiology and the development of therapeutic methods. Microorganisms. 2021;9(1):180. doi: 10.3390/microorganisms9010180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.El Hussein S, Medeiros LJ, Khoury JD. Aggressive NK cell leukemia: current state of the art. Cancers (Basel) 2020;12(10):2900. doi: 10.3390/cancers12102900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kataoka K, Miyoshi H, Sakata S, et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia. 2019;33(7):1687–1699. doi: 10.1038/s41375-019-0380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lim JQ, Huang D, Tang T, et al. Whole-genome sequencing identifies responders to pembrolizumab in relapse/refractory natural-killer/T cell lymphoma. Leukemia. 2020;34(12):3413–3419. doi: 10.1038/s41375-020-1000-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kwong Y-L, Chan TSY, Tan D, et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood. 2017;129(17):2437–2442. doi: 10.1182/blood-2016-12-756841. [DOI] [PubMed] [Google Scholar]
- 332.Li X, Cheng Y, Zhang M, et al. Activity of pembrolizumab in relapsed/refractory NK/T-cell lymphoma. J Hematol Oncol. 2018;11(1):15. doi: 10.1186/s13045-018-0559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Cho J, Kim SJ, Park WY, et al. Immune subtyping of extranodal NK/T-cell lymphoma: a new biomarker and an immune shift during disease progression. Mod Pathol. 2020;33(4):603–615. doi: 10.1038/s41379-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 334.Gayden T, Sepulveda FE, Khuong-Quang DA, et al. Germline HAVCR2 mutations altering TIM-3 characterize subcutaneous panniculitis-like T cell lymphomas with hemophagocytic lymphohistiocytic syndrome. Nat Genet. 2018;50(12):1650–1657. doi: 10.1038/s41588-018-0251-4. [DOI] [PubMed] [Google Scholar]
- 335.Sonigo G, Battistella M, Beylot-Barry M, et al. HAVCR2 mutations are associated with severe hemophagocytic syndrome in subcutaneous panniculitis-like T-cell lymphoma. Blood. 2020;135(13):1058–1061. doi: 10.1182/blood.2019003811. [DOI] [PubMed] [Google Scholar]
- 336.Koh J, Jang I, Mun S, et al. Genetic profiles of subcutaneous panniculitis-like T-cell lymphoma and clinicopathological impact of HAVCR2 mutations. Blood Adv. 2021;5(20):3919–3930. doi: 10.1182/bloodadvances.2021004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Cook LB, Melamed A, Niederer H, et al. The role of HTLV-1 clonality, proviral structure, and genomic integration site in adult T-cell leukemia/lymphoma. Blood. 2014;123(25):3925–3931. doi: 10.1182/blood-2014-02-553602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 339.Kogure Y, Kameda T, Koya J, et al. Whole-genome landscape of adult T-cell leukemia/lymphoma. Blood. 2022;139(7):967–982. doi: 10.1182/blood.2021013568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Marçais A, Lhermitte L, Artesi M, et al. Targeted deep sequencing reveals clonal and subclonal mutational signatures in adult T-cell leukemia/lymphoma and defines an unfavorable indolent subtype. Leukemia. 2021;35(3):764–776. doi: 10.1038/s41375-020-0900-3. [DOI] [PubMed] [Google Scholar]
- 341.Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534(7607):402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 342.Nakagawa M, Schmitz R, Xiao W, et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J Exp Med. 2014;211(13):2497–2505. doi: 10.1084/jem.20140987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Kataoka K, Iwanaga M, Iasunaga J-i, et al. Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood. 2018;131(2):215–225. doi: 10.1182/blood-2017-01-761874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Sakamoto Y, Ishida T, Masaki A, et al. CCR4 mutations associated with superior outcome of adult T-cell leukemia/lymphoma under mogamulizumab treatment. Blood. 2018;132(7):758–761. doi: 10.1182/blood-2018-02-835991. [DOI] [PubMed] [Google Scholar]
- 345.Tanaka N, Mori S, Kiyotani K, et al. Genomic determinants impacting the clinical outcome of mogamulizumab treatment for adult T-cell leukemia/lymphoma. Haematologica. 2022 doi: 10.3324/haematol.2021.280352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sakamoto Y, Ishida T, Masaki A, et al. Clinical significance of TP53 mutations in adult T-cell leukemia/lymphoma. Br J Haematol. 2021;195(4):571–584. doi: 10.1111/bjh.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Koskela HLM, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Rajala HLM, Eldfors S, Kuusanmäki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–4550. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Teramo A, Barilà G, Calabretto G, et al. STAT3 mutation impacts biological and clinical features of T-LGL leukemia. Oncotarget. 2017;8(37):61876–61889. doi: 10.18632/oncotarget.18711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Bhattacharya D, Teramo A, Gasparini VR, et al. Identification of novel STAT5B mutations and characterization of TCRβ signatures in CD4+ T-cell large granular lymphocyte leukemia. Blood Cancer J. 2022;12(2):31. doi: 10.1038/s41408-022-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Andersson EI, Tanahashi T, Sekiguchi N, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood. 2016;128(20):2465–2468. doi: 10.1182/blood-2016-06-724856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Barilà G, Teramo A, Calabretto G, et al. Stat3 mutations impact on overall survival in large granular lymphocyte leukemia: a single-center experience of 205 patients. Leukemia. 2020;34(4):1116–1124. doi: 10.1038/s41375-019-0644-0. [DOI] [PubMed] [Google Scholar]
- 353.Teramo A, Barilà G, Calabretto G, et al. Insights into genetic landscape of large granular lymphocyte leukemia. Front Oncol. 2020;10:152–159. doi: 10.3389/fonc.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Shi M, He R, Feldman AL, et al. STAT3 mutation and its clinical and histopathologic correlation in T-cell large granular lymphocytic leukemia. Hum Pathol. 2018;73:74–81. doi: 10.1016/j.humpath.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 355.Muñoz-García N, Jara-Acevedo M, Caldas C, et al. STAT3 and STAT5B mutations in T/NK-cell chronic lymphoproliferative disorders of large granular lymphocytes (LGL): association with disease features. Cancers (Basel) 2020;12(12):3508–3530. doi: 10.3390/cancers12123508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–3057. doi: 10.1182/blood-2012-06-435297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Baer C, Kimura S, Rana MS, et al. CCL22 mutations drive natural killer cell lymphoproliferative disease by deregulating microenvironmental crosstalk. Nat Genet. 2022;54(5):637–648. doi: 10.1038/s41588-022-01059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Gasparini VR, Binatti A, Coppe A, et al. A high definition picture of somatic mutations in chronic lymphoproliferative disorder of natural killer cells. Blood Cancer J. 2020;10(4):42–52. doi: 10.1038/s41408-020-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Pastoret C, Desmots F, Drillet G, et al. Linking the KIR phenotype with STAT3 and TET2 mutations to identify chronic lymphoproliferative disorders of NK cells. Blood. 2021;137(23):3237–3250. doi: 10.1182/blood.2020006721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Olson TL, Cheon H, Xing JC, et al. Frequent somatic TET2 mutations in chronic NK-LGL leukemia with distinct patterns of cytopenias. Blood. 2021;138(8):662–673. doi: 10.1182/blood.2020005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kawakami T, Sekiguchi N, Kobayashi J, et al. STAT3 mutations in natural killer cells are associated with cytopenia in patients with chronic lymphoproliferative disorder of natural killer cells. Int J Hematol. 2019;109(5):563–571. doi: 10.1007/s12185-019-02625-x. [DOI] [PubMed] [Google Scholar]
- 362.Sun Y, Tang G, Hu Z, et al. Comparison of karyotyping, TCL1 fluorescence in situ hybridisation and TCL1 immunohistochemistry in T cell prolymphocytic leukaemia. J Clin Pathol. 2018;71(4):309–315. doi: 10.1136/jclinpath-2017-204616. [DOI] [PubMed] [Google Scholar]
- 363.Virgilio L, Lazzeri C, Bichi R, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci U S A. 1998;95(7):3885–3889. doi: 10.1073/pnas.95.7.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Gritti C, Dastot H, Soulier J, et al. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood. 1998;92(2):368–373. [PubMed] [Google Scholar]
- 365.Herling M, Patel KA, Teitell MA, et al. High TCL1 expression and intact T-cell receptor signaling define a hyperproliferative subset of T-cell prolymphocytic leukemia. Blood. 2008;111(1):328–337. doi: 10.1182/blood-2007-07-101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Hu Z, Medeiros LJ, Fang L, et al. Prognostic significance of cytogenetic abnormalities in T-cell prolymphocytic leukemia. Am J Hematol. 2017;92(5):441–447. doi: 10.1002/ajh.24679. [DOI] [PubMed] [Google Scholar]
- 367.Stilgenbauer S, Schaffner C, Litterst A, et al. Biallelic mutations in the ATM gene in T-prolymphocytic leukemia. Nat Med. 1997;3(10):1155–1159. doi: 10.1038/nm1097-1155. [DOI] [PubMed] [Google Scholar]
- 368.Stoppa-Lyonnet D, Soulier J, Laugé A, et al. Inactivation of the ATM gene in T-cell prolymphocytic leukemias. Blood. 1998;91(10):3920–3926. [PubMed] [Google Scholar]
- 369.Schrader A, Crispatzu G, Oberbeck S, et al. Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL. Nat Commun. 2018;9(1):697. doi: 10.1038/s41467-017-02688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Andersson EI, Pützer S, Yadav B, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2018;32(3):774–787. doi: 10.1038/leu.2017.252. [DOI] [PubMed] [Google Scholar]
- 371.Wahnschaffe L, Braun T, Timonen S, et al. JAK/STAT-activating genomic alterations are a hallmark of T-PLL. Cancers (Basel) 2019;11(12):1833. doi: 10.3390/cancers11121833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Berres M-L, Lim KPH, Peters T, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med. 2014;211(4):669–683. doi: 10.1084/jem.20130977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Milne P, Bigley V, Bacon CM, et al. Hematopoietic origin of Langerhans cell histiocytosis and Erdheim-Chester disease in adults. Blood. 2017;130(2):167–175. doi: 10.1182/blood-2016-12-757823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Allen CE, Merad M, McClain KL. Langerhans-cell histiocytosis. N Engl J Med. 2018;379(9):856–868. doi: 10.1056/NEJMra1607548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.McClain KL, Bigenwald C, Collin M, et al. Histiocytic disorders. Nat Rev Dis Primers. 2021;7(1):73. doi: 10.1038/s41572-021-00307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Badalian-Very G, Vergilio J-A, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Haroche J, Cohen-Aubart F, Emile J-F, et al. Dramatic efficacy of vemurafenib in both multisystemic and refractory Erdheim-Chester disease and Langerhans cell histiocytosis harboring the BRAF V600E mutation. Blood. 2013;121(9):1495–1500. doi: 10.1182/blood-2012-07-446286. [DOI] [PubMed] [Google Scholar]
- 378.Brown NA, Furtado LV, Betz BL, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E–negative Langerhans cell histiocytosis. Blood. 2014;124(10):1655–1658. doi: 10.1182/blood-2014-05-577361. [DOI] [PubMed] [Google Scholar]
- 379.Chakraborty R, Burke TM, Hampton OA, et al. Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood. 2016;128(21):2533–2537. doi: 10.1182/blood-2016-08-733790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Facchetti F, Pileri SA, Lorenzi L, et al. Histiocytic and dendritic cell neoplasms: what have we learnt by studying 67 cases. Virchows Arch. 2017;471(4):467–489. doi: 10.1007/s00428-017-2176-1. [DOI] [PubMed] [Google Scholar]
- 381.Garces S, Medeiros LJ, Patel KP, et al. Mutually exclusive recurrent KRAS and MAP2K1 mutations in Rosai-Dorfman disease. Mod Pathol. 2017;30(10):1367–1377. doi: 10.1038/modpathol.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Durham BH, Lopez Rodrigo E, Picarsic J, et al. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat Med. 2019;25(12):1839–1842. doi: 10.1038/s41591-019-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Shanmugam V, Griffin GK, Jacobsen ED, Fletcher CDM, Sholl LM, Hornick JL. Identification of diverse activating mutations of the RAS-MAPK pathway in histiocytic sarcoma. Mod Pathol. 2019;32(6):830–843. doi: 10.1038/s41379-018-0200-x. [DOI] [PubMed] [Google Scholar]
- 384.Egan C, Nicolae A, Lack J, et al. Genomic profiling of primary histiocytic sarcoma reveals two molecular subgroups. Haematologica. 2020;105(4):951–960. doi: 10.3324/haematol.2019.230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Haroche J, Cohen-Aubart F, Amoura Z. Erdheim-Chester disease. Blood. 2020;135(16):1311–1318. doi: 10.1182/blood.2019002766. [DOI] [PubMed] [Google Scholar]
- 386.Sengal A, Velazquez J, Hahne M, et al. Overcoming T-cell exhaustion in LCH: PD-1 blockade and targeted MAPK inhibition are synergistic in a mouse model of LCH. Blood. 2021;137(13):1777–1791. doi: 10.1182/blood.2020005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Gatalica Z, Bilalovic N, Palazzo JP, et al. Disseminated histiocytoses biomarkers beyond BRAFV600E: frequent expression of PD-L1. Oncotarget. 2015;6(23):19819–19825. doi: 10.18632/oncotarget.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Kemps PG, Picarsic J, Durham BH, et al. ALK-positive histiocytosis: a new clinicopathologic spectrum highlighting neurologic involvement and responses to ALK inhibition. Blood. 2022;139(2):256–280. doi: 10.1182/blood.2021013338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124(19):3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Nelson DS, Quispel W, Badalian-Very G, et al. Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood. 2014;123(20):3152–3155. doi: 10.1182/blood-2013-06-511139. [DOI] [PubMed] [Google Scholar]
- 391.Chakraborty R, Abdel-Wahab O, Durham BH. MAP-kinase-driven hematopoietic neoplasms: a decade of progress in the molecular age. Cold Spring Harb Perspect Med. 2021;11(5):a034892. doi: 10.1101/cshperspect.a034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Milne P, Abhyankar HA, Scull BP, et al. Cellular distribution of mutations and association with disease risk in Langerhans cell histiocytosis without BRAFV600E. Blood Adv. 2022;6(16):4901–4904. doi: 10.1182/bloodadvances.2022007519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Feldman AL, Arber DA, Pittaluga S, et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood. 2008;111(12):5433–5439. doi: 10.1182/blood-2007-11-124792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Shao H, Xi L, Raffeld M, et al. Clonally related histiocytic/dendritic cell sarcoma and chronic lymphocytic leukemia/small lymphocytic lymphoma: a study of seven cases. Mod Pathol. 2011;24(11):1421–1432. doi: 10.1038/modpathol.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Cohen Aubart F, Roos-Weil D, Armand M, et al. High frequency of clonal hematopoiesis in Erdheim-Chester disease. Blood. 2021;137(4):485–492. doi: 10.1182/blood.2020005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.McClain KL, Picarsic J, Chakraborty R, et al. CNS Langerhans cell histiocytosis: common hematopoietic origin for LCH-associated neurodegeneration and mass lesions. Cancer. 2018;124(12):2607–2620. doi: 10.1002/cncr.31348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Massoth LR, Hung YP, Ferry JA, et al. Histiocytic and dendritic cell sarcomas of hematopoietic origin share targetable genomic alterations distinct from follicular dendritic cell sarcoma. Oncol. 2021;26(7):e1263–e1272. doi: 10.1002/onco.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Griffin GK, Sholl LM, Lindeman NI, Fletcher CD, Hornick JL. Targeted genomic sequencing of follicular dendritic cell sarcoma reveals recurrent alterations in NF-κB regulatory genes. Mod Pathol. 2016;29(1):67–74. doi: 10.1038/modpathol.2015.130. [DOI] [PubMed] [Google Scholar]
- 399.Laginestra MA, Tripodo C, Agostinelli C, et al. Distinctive histogenesis and immunological microenvironment based on transcriptional profiles of follicular dendritic Cell sarcomas. Mol Cancer Res. 2017;15(5):541–552. doi: 10.1158/1541-7786.MCR-16-0301. [DOI] [PubMed] [Google Scholar]
- 400.Diamond EL, Subbiah V, Lockhart AC, et al. Vemurafenib for BRAF V600-mutant Erdheim-Chester disease and Langerhans cell histiocytosis: analysis of data from the histology-independent, phase 2, open-label VE-BASKET study. JAMA Oncol. 2018;4(3):384–388. doi: 10.1001/jamaoncol.2017.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Diamond EL, Durham BH, Ulaner GA, et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature. 2019;567(7749):521–524. doi: 10.1038/s41586-019-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Donadieu J, Larabi IA, Tardieu M, et al. Vemurafenib for refractory multisystem Langerhans cell histiocytosis in children: an international observational study. J Clin Oncol. 2019;37(31):2857–2865. doi: 10.1200/JCO.19.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Eckstein OS, Visser J, Rodriguez-Galindo C, Allen CE, Group tN-LS Clinical responses and persistent BRAF V600E+ blood cells in children with LCH treated with MAPK pathway inhibition. Blood. 2019;133(15):1691–1694. doi: 10.1182/blood-2018-10-878363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Schwentner R, Kolenová A, Jug G, et al. Longitudinal assessment of peripheral blood BRAFV600E levels in patients with Langerhans cell histiocytosis. Pediatr Res. 2019;85(6):856–864. doi: 10.1038/s41390-018-0238-y. [DOI] [PubMed] [Google Scholar]
- 405.Hyman DM, Diamond EL, Vibat CR, et al. Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov. 2015;5(1):64–71. doi: 10.1158/2159-8290.CD-14-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Cohen Aubart F, Emile JF, Carrat F, et al. Targeted therapies in 54 patients with Erdheim-Chester disease, including follow-up after interruption (the LOVE study) Blood. 2017;130(11):1377–1380. doi: 10.1182/blood-2017-03-771873. [DOI] [PubMed] [Google Scholar]
- 407.Arthur SE, Jiang A, Grande BM, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun. 2018;9(1):4001. doi: 10.1038/s41467-018-06354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Hübschmann D, Kleinheinz K, Wagener R, et al. Mutational mechanisms shaping the coding and noncoding genome of germinal center derived B-cell lymphomas. Leukemia. 2021;35(7):2002–2016. doi: 10.1038/s41375-021-01251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Morin RD, Arthur SE, Hodson DJ. Molecular profiling in diffuse large B-cell lymphoma: why so many types of subtypes? Br J Haematol. 2022;196(4):814–829. doi: 10.1111/bjh.17811. [DOI] [PubMed] [Google Scholar]
- 410.Hilton LK, Dreval K, Soudi S, et al. Constrained FL: a genetically distinct subgroup of follicular lymphoma with low rates of somatic hypermutation and a reduced propensity for histologic transformation. Blood. 2021;138(S1) Abstract 807. [Google Scholar]
- 411.Thomas N, Dreval K, Gerhard DS, et al. Genetic subgroups inform on pathobiology in adult and pediatric Burkitt lymphoma. medRxiv. Preprint posted online 8 December 2021 doi: 10.1101/2021.12.05.21267216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Fuster C, Martín-Garcia D, Balagué O, et al. Cryptic insertions of the immunoglobulin light chain enhancer region near CCND1 in t(11;14)-negative mantle cell lymphoma. Haematologica. 2020;105(8):e408–e411. doi: 10.3324/haematol.2019.237073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Polonis K, Schultz MJ, Olteanu H, et al. Detection of cryptic CCND1 rearrangements in mantle cell lymphoma by next generation sequencing. Ann Diagn Pathol. 2020;46:151533. doi: 10.1016/j.anndiagpath.2020.151533. [DOI] [PubMed] [Google Scholar]
- 414.Barwick BG, Neri P, Bahlis NJ, et al. Multiple myeloma immunoglobulin lambda translocations portend poor prognosis. Nat Commun. 2019;10(1):1911. doi: 10.1038/s41467-019-09555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Shuai S, Suzuki H, Diaz-Navarro A, et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature. 2019;574(7780):712–716. doi: 10.1038/s41586-019-1651-z. [DOI] [PubMed] [Google Scholar]
- 416.Wiestner A, Tehrani M, Chiorazzi M, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109(11):4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Diaz LA, Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 419.Kurtz DM, Soo J, Co Ting Keh L, et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol. 2021;39(12):1537–1547. doi: 10.1038/s41587-021-00981-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Deveson IW, Gong B, Lai K, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol. 2021;39(9):1115–1128. doi: 10.1038/s41587-021-00857-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Alig S, Macaulay CW, Kurtz DM, et al. Short diagnosis-to-treatment interval is associated with higher circulating tumor DNA levels in diffuse large B-cell lymphoma. J Clin Oncol. 2021;39(23):2605–2616. doi: 10.1200/JCO.20.02573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Kurtz DM, Scherer F, Jin MC, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol. 2018;36(28):2845–2853. doi: 10.1200/JCO.2018.78.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Huet S, Salles G. Potential of circulating tumor DNA for the management of patients with lymphoma. JCO Oncol Pract. 2020;16(9):561–568. doi: 10.1200/JOP.19.00691. [DOI] [PubMed] [Google Scholar]
- 424.Roschewski M, Rossi D, Kurtz DM, Alizadeh AA, Wilson WH. Circulating tumor DNA in lymphoma: principles and future directions. Blood Cancer Discov. 2022;3(1):5–15. doi: 10.1158/2643-3230.BCD-21-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Gohil SH, Iorgulescu JB, Braun DA, Keskin DB, Livak KJ. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol. 2021;18(4):244–256. doi: 10.1038/s41571-020-00449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Zheng L, Qin S, Si W, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374(6574) doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 427.Haebe S, Shree T, Sathe A, et al. Single-cell analysis can define distinct evolution of tumor sites in follicular lymphoma. Blood. 2021;137(21):2869–2880. doi: 10.1182/blood.2020009855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Hanna BS, Llaó-Cid L, Iskar M, et al. Interleukin-10 receptor signaling promotes the maintenance of a PD-1(int) TCF-1(+) CD8(+) T cell population that sustains anti-tumor immunity. Immunity. 2021;54(12):2825–2841.e2810. doi: 10.1016/j.immuni.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 429.Gut’errez C, Al'Khafaji AM, Brenner E, et al. Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat Cancer. 2021;2(7):758–772. doi: 10.1038/s43018-021-00222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Gaiti F, Chaligne R, Gu H, et al. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature. 2019;569(7757):576–580. doi: 10.1038/s41586-019-1198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Milpied P, Cervera-Marzal I, Mollichella ML, et al. Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat Immunol. 2018;19(9):1013–1024. doi: 10.1038/s41590-018-0181-4. [DOI] [PubMed] [Google Scholar]
- 432.Holmes AB, Corinaldesi C, Shen Q, et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J Exp Med. 2020;217(10):e20200483. doi: 10.1084/jem.20200483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Cohen YC, Zada M, Wang S-Y, et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat Med. 2021;27(3):491–503. doi: 10.1038/s41591-021-01232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Gohil SH, Wu CJ. Dissecting CLL through high-dimensional single-cell technologies. Blood. 2019;133(13):1446–1456. doi: 10.1182/blood-2018-09-835389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Duran-Ferrer M, Clot G, Nadeu F, et al. The proliferative history shapes the DNA methylome of B-cell tumors and predicts clinical outcome. Nat Cancer. 2020;1(11):1066–1081. doi: 10.1038/s43018-020-00131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Roos-Weil D, Giacopelli B, Armand M, et al. Identification of 2 DNA methylation subtypes of Waldenström macroglobulinemia with plasma and memory B-cell features. Blood. 2020;136(5):585–595. doi: 10.1182/blood.2020005081. [DOI] [PubMed] [Google Scholar]
- 437.Queiros AC, Villamor N, Clot G, et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia. 2015;29(3):598–605. doi: 10.1038/leu.2014.252. [DOI] [PubMed] [Google Scholar]
- 438.Giacopelli B, Zhao Q, Ruppert AS, et al. Developmental subtypes assessed by DNA methylation-iPLEX forecast the natural history of chronic lymphocytic leukemia. Blood. 2019;134(8):688–698. doi: 10.1182/blood.2019000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Duy C, Béguelin W, Melnick A. Epigenetic mechanisms in leukemias and lymphomas. Cold Spring Harb Perspect Med. 2020;10(12):a034959. doi: 10.1101/cshperspect.a034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Oakes CC, Martin-Subero JI. Insight into origins, mechanisms, and utility of DNA methylation in B-cell malignancies. Blood. 2018;132(10):999–1006. doi: 10.1182/blood-2018-02-692970. [DOI] [PubMed] [Google Scholar]
- 441.Lemonnier F, Couronné L, Parrens M, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 442.Zang S, Li J, Yang H, et al. Mutations in 5-methylcytosine oxidase TET2 and RhoA cooperatively disrupt T cell homeostasis. J Clin Invest. 2017;127(8):2998–3012. doi: 10.1172/JCI92026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98(12):1912–1920. doi: 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Liu P, Jiang W, Zhao J, Zhang H. Integrated analysis of genome-wide gene expression and DNA methylation microarray of diffuse large B-cell lymphoma with TET mutations. Mol Med Rep. 2017;16(4):3777–3782. doi: 10.3892/mmr.2017.7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Rosikiewicz W, Chen X, Dominguez PM, et al. TET2 deficiency reprograms the germinal center B cell epigenome and silences genes linked to lymphomagenesis. Sci Adv. 2020;6(25):eaay5872. doi: 10.1126/sciadv.aay5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Dominguez PM, Ghamlouch H, Rosikiewicz W, et al. TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation, and promotes B-cell lymphomagenesis. Cancer Discov. 2018;8(12):1632–1653. doi: 10.1158/2159-8290.CD-18-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Dominguez PM, Teater M, Chambwe N, et al. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep. 2015;12(12):2086–2098. doi: 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.De S, Shaknovich R, Riester M, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013;9(1) doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Chambwe N, Kormaksson M, Geng H, et al. Variability in DNA methylation defines novel epigenetic subgroups of DLBCL associated with different clinical outcomes. Blood. 2014;123(11):1699–1708. doi: 10.1182/blood-2013-07-509885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Landau DA, Clement K, Ziller MJ, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26(6):813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Pan H, Jiang Y, Boi M, et al. Epigenomic evolution in diffuse large B-cell lymphomas. Nat Commun. 2015;6:6921. doi: 10.1038/ncomms7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Alhejaily A, Day AG, Feilotter HE, Baetz T, Lebrun DP. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin Cancer Res. 2014;20(6):1676–1686. doi: 10.1158/1078-0432.CCR-13-2175. [DOI] [PubMed] [Google Scholar]
- 453.Clozel T, Yang S, Elstrom RL, et al. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov. 2013;3(9):1002–1019. doi: 10.1158/2159-8290.CD-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Stelling A, Wu CT, Bertram K, et al. Pharmacological DNA demethylation restores SMAD1 expression and tumor suppressive signaling in diffuse large B-cell lymphoma. Blood Adv. 2019;3(20):3020–3032. doi: 10.1182/bloodadvances.2019000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Beekman R, Chapaprieta V, Russiñol N, et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018;24(6):868–880. doi: 10.1038/s41591-018-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Chapuy B, McKeown MR, Lin CY, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Ott CJ, Federation AJ, Schwartz LS, et al. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell. 2018;34(6):982–995.e987. doi: 10.1016/j.ccell.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Ordoñez R, Kulis M, Russiñol N, et al. Chromatin activation as a unifying principle underlying pathogenic mechanisms in multiple myeloma. Genome Res. 2020;30(9):1217–1227. doi: 10.1101/gr.265520.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Béguelin W, Popovic R, Teater M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Béguelin W, Teater M, Meydan C, et al. Mutant EZH2 induces a pre-malignant lymphoma niche by reprogramming the immune response. Cancer Cell. 2020;37(5):655–673.e611. doi: 10.1016/j.ccell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Caganova M, Carrisi C, Varano G, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123(12):5009–5022. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Leung W, Teater M, Durmaz C, et al. SETD2 haploinsufficiency enhances germinal center-associated AICDA somatic hypermutation to drive B cell lymphomagenesis. Cancer Discov. 2022;12(7):1782–1803. doi: 10.1158/2159-8290.CD-21-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Liu H, Wang W, Tang G, et al. Lymphomatous variant of hairy cell leukaemia: a distinctive presentation mimicking low-grade B-cell lymphoma. Histopathology. 2015;67(5):740–745. doi: 10.1111/his.12694. [DOI] [PubMed] [Google Scholar]
- 464.Davies FE, Pawlyn C, Usmani SZ, et al. Perspectives on the risk-stratified treatment of multiple myeloma. Blood Cancer Discov. 2022;3(4):273–284. doi: 10.1158/2643-3230.BCD-21-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Kurtz DM, Esfahani MS, Scherer F, et al. Dynamic risk profiling using serial tumor biomarkers for personalized outcome prediction. Cell. 2019;178(3):699–713.e619. doi: 10.1016/j.cell.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Read JA, Koff JL, Nastoupil LJ, Williams JN, Cohen JB, Flowers CR. Evaluating cell-of-origin subtype methods for predicting diffuse large B-cell lymphoma survival: a meta-analysis of gene expression profiling and immunohistochemistry algorithms. Clin Lymphoma Myeloma Leuk. 2014;14(6):460–467.e462. doi: 10.1016/j.clml.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Petrich AM, Gandhi M, Jovanovic B, et al. Impact of induction regimen and stem cell transplantation on outcomes in double-hit lymphoma: a multicenter retrospective analysis. Blood. 2014;124(15):2354–2361. doi: 10.1182/blood-2014-05-578963. [DOI] [PubMed] [Google Scholar]
- 468.Rodrigues JM, Hassan M, Freiburghaus C, et al. p53 is associated with high-risk and pinpoints TP53 missense mutations in mantle cell lymphoma. Br J Haematol. 2020;191(5):796–805. doi: 10.1111/bjh.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Amador C, Bouska A, Wright G, et al. Gene expression signatures for the accurate diagnosis of peripheral T-cell lymphoma entities in the routine clinical practice. J Clin Oncol. 2022 doi: 10.1200/JCO.21.02707. Jco2102707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Hare L, Burke GAA, Turner SD. Resistance to targeted agents used to treat paediatric ALK-positive ALCL. Cancers (Basel) 2021;13(23):6003. doi: 10.3390/cancers13236003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Lemonnier F, Safar V, Beldi-Ferchiou A, et al. Integrative analysis of a phase 2 trial combining lenalidomide with CHOP in angioimmunoblastic T-cell lymphoma. Blood Adv. 2021;5(2):539–548. doi: 10.1182/bloodadvances.2020003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Amador C, Greiner TC, Heavican TB, et al. Reproducing the molecular subclassification of peripheral T-cell lymphoma-NOS by immunohistochemistry. Blood. 2019;134(24):2159–2170. doi: 10.1182/blood.2019000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Artesi M, Marcais A, Durkin K, et al. Monitoring molecular response in adult T-cell leukemia by high-throughput sequencing analysis of HTLV-1 clonality. Leukemia. 2017;31(11):2532–2535. doi: 10.1038/leu.2017.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Kataoka K, Koya J. Clinical application of genomic aberrations in adult T-cell leukemia/lymphoma. J Clin Exp Hematop. 2020;60(3):66–72. doi: 10.3960/jslrt.20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Gambacorti Passerini C, Farina F, Stasia A, et al. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J Natl Cancer Inst. 2014;106(2):djt378. doi: 10.1093/jnci/djt378. [DOI] [PubMed] [Google Scholar]
- 476.Fukano R, Mori T, Sekimizu M, et al. Alectinib for relapsed or refractory anaplastic lymphoma kinase-positive anaplastic large cell lymphoma: an open-label phase II trial. Cancer Sci. 2020;111(12):4540–4547. doi: 10.1111/cas.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Yoshida N, Shigemori K, Donaldson N, et al. Genomic landscape of young ATLL patients identifies frequent targetable CD28 fusions. Blood. 2020;135(17):1467–1471. doi: 10.1182/blood.2019001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Sekulic A, Liang WS, Tembe W, et al. Personalized treatment of Sézary syndrome by targeting a novel CTLA4:CD28 fusion. Mol Genet Genomic Med. 2015;3(2):130–136. doi: 10.1002/mgg3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Dierks C, Adrian F, Fisch P, et al. The ITK-SYK fusion oncogene induces a T-cell lymphoproliferative disease in mice mimicking human disease. Cancer Res. 2010;70(15):6193–6204. doi: 10.1158/0008-5472.CAN-08-3719. [DOI] [PubMed] [Google Scholar]
- 480.Pechloff K, Holch J, Ferch U, et al. The fusion kinase ITK-SYK mimics a T cell receptor signal and drives oncogenesis in conditional mouse models of peripheral T cell lymphoma. J Exp Med. 2010;207(5):1031–1044. doi: 10.1084/jem.20092042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Fathi NN, Mohammad DK, Görgens A, et al. Translocation-generated ITK-FER and ITK-SYK fusions induce STAT3 phosphorylation and CD69 expression. Biochem Biophys Res Commun. 2018;504(4):749–752. doi: 10.1016/j.bbrc.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 482.Zhang LL, Pan HX, Wang YX, Guo T, Liu L. Genome profiling revealed the activation of IL2RG/JAK3/STAT5 in peripheral T-cell lymphoma expressing the ITK-SYK fusion gene. Int J Oncol. 2019;55(5):1077–1089. doi: 10.3892/ijo.2019.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Debackere K, van der Krogt JA, Tousseyn T, et al. FER and FES tyrosine kinase fusions in follicular T-cell lymphoma. Blood. 2020;135(8):584–588. doi: 10.1182/blood.2019002401. [DOI] [PubMed] [Google Scholar]
- 484.Boddicker RL, Razidlo GL, Dasari S, et al. Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma. Blood. 2016;128(9):1234–1245. doi: 10.1182/blood-2016-03-707141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Bachy E, Camus V, Thieblemont C, et al. Romidepsin plus CHOP versus CHOP in patients with previously untreated peripheral T-cell lymphoma: results of the Ro-CHOP phase III study (conducted by LYSA) J Clin Oncol. 2022;40(3):242–251. doi: 10.1200/JCO.21.01815. [DOI] [PubMed] [Google Scholar]
- 486.Horwitz SM, Koch R, Porcu P, et al. Activity of the PI3K-δ,γ inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood. 2018;131(8):888–898. doi: 10.1182/blood-2017-08-802470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Khodadoust MS, Feldman TA, Yoon DH, et al. Cpi-818, an oral interleukin-2-inducible T-cell kinase inhibitor, is well-tolerated and active in patients with T-cell lymphoma. Blood. 2020;136(Supplement 1):19–20. [Google Scholar]
- 488.Rossi D, Kurtz DM, Roschewski M, Cavalli F, Zucca E, Wilson WH. The development of liquid biopsy for research and clinical practice in lymphomas: report of the 15-ICML workshop on ctDNA. Hematol Oncol. 2020;38(1):34–37. doi: 10.1002/hon.2704. [DOI] [PubMed] [Google Scholar]
- 489.Esfahani MS, Hamilton EG, Mehrmohamadi M, et al. Inferring gene expression from cell-free DNA fragmentation profiles. Nat Biotechnol. 2022;40(4):585–597. doi: 10.1038/s41587-022-01222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Meriranta L, Alkodsi A, Pasanen A, et al. Molecular features encoded in the ctDNA reveal heterogeneity and predict outcome in high-risk aggressive B-cell lymphoma. Blood. 2022;139(12):1863–1877. doi: 10.1182/blood.2021012852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.