Abstract
The binary Clostridium botulinum C2 toxin consists of two separate proteins, the binding component C2II (80.5 kDa) and the actin-ADP-ribosylating enzyme component C2I (49.4 kDa). For its cytotoxic action, C2II binds to a cell membrane receptor and induces cell entry of C2I via receptor-mediated endocytosis. Here we studied the structure-function relationship of C2II by constructing truncated C2II proteins and producing polyclonal antisera against selective regions of C2II. An antibody raised against the C terminus (amino acids 592 to 721) of C2II inhibited binding of C2II to cells. The antibody prevented pore formation by C2II oligomers in artificial membranes but did not influence the properties of existing channels. To further define the region responsible for receptor binding, we constructed proteins with deletions in C2II; specifically, they lacked amino acid residues 592 to 721 and the 7 C-terminal amino acid residues. The truncated proteins still formed sodium dodecyl sulfate-stable oligomers but were unable to bind to cells. Our data indicate that the C terminus of C2II mediates binding of the protein to cells and that the 7 C-terminal amino acids are structurally important for receptor binding.
The actin-ADP-ribosylating C2 toxin from Clostridium botulinum types C and D belongs to the family of toxins which consist of two separate proteins, an enzyme component, C2I, and a binding component, C2II (2, 7, 32). Further members of this toxin family are iota toxin from Clostridium perfringens (27), Clostridium difficile ADP-ribosyltransferase (24), Clostridium spiroforme toxin (23), and the vegetative insecticidal proteins produced by Bacillus cereus (13). The C2I enzyme component of C2 toxin ADP-ribosylates G-actin at Arg-177 (1). ADP-ribosylation inhibits actin polymerization (1) and actin ATPase activity (12) and turns actin into a capping protein that binds to the barbed ends of actin filaments, inhibiting fast polymerization (30). Moreover, ADP-ribosylation of actin complexed with gelsolin alters the nucleation of the gelsolin-actin complex (32). In intact cells, C2 toxin causes redistribution of the actin cytoskeleton, depolymerization of actin filaments, and rounding up (25, 31, 33).
Cellular uptake of C2I depends on the binding and translocation component C2II. C2II binds specifically to asparagine-linked complex carbohydrates, which act as toxin receptors on the surfaces of target cells (9). For efficient binding and translocation, C2II has to be activated by trypsin cleavage; thereby, an N-terminal 20-kDa fragment of C2II is released (20). Trypsin-activated C2II (59.8 kDa) oligomerizes to heptamers and forms channels in artificial membranes (3). After endocytosis of the C2II-C2I complex, translocation of the enzyme component into the cytosol occurs most likely from an acidic endosomal compartment (3).
Recent cloning and sequencing of the gene encoding the binding component of C2 toxin revealed significant sequence similarities with the genes of binding components of the other actin-ADP-ribosylating toxins but also with the gene of the protective antigen (PA) of Bacillus anthracis (15). PA is the binding component of the tripartite anthrax toxin (18) and translocates the edema factor, an adenylyl cyclase (17), and/or the lethal factor, a mitogen-activated protein kinase-cleaving metalloprotease (8), into the cytosol. Basing our work mainly on the crystal structure of PA (22), which is characterized by a four-domain structure, and its sequence similarity with the binding component of C2 toxin, we performed a structure-function analysis of C2II. Deduced from the primary sequence, C2II could be divided into four domains like those of PA. Whereas domains 1 to 3 (D1 to D3) of C2II show sequence similarities with those domains in PA, D4 is dissimilar to D4 in PA. Here we report that C-terminal D4 of C2II (C2II-D4), which covers amino acid residues 592 to 721, mediates cell surface binding of C2 toxin. Deletion analysis suggested that the 7 C-terminal amino acid residues of this domain are essential for cell binding.
MATERIALS AND METHODS
Materials.
Oligonucleotides were obtained from MWG Biotech (Ebersberg, Germany). The pGEX-2T vector was included in the glutathione S-transferase (GST) gene fusion system from Pharmacia Biotech (Uppsala, Sweden). PCRs were performed with the GeneAmp PCR system 2400 from Perkin-Elmer (Langen, Germany), and DNA sequencing was done by G. Igloi, University of Freiburg, Freiburg, Germany. Taq polymerase was purchased from Roche Molecular Diagnostics.
Donkey anti-rabbit antibody coupled to horseradish peroxidase and an enhanced chemiluminescence detection kit were from Amersham (Braunschweig, Germany). The nitrocellulose-blotting membrane was from Schleicher and Schuell (Dassel, Germany). Glutathione-Sepharose 4B and protein A–Sepharose-CL 4B were obtained from Pharmacia Biotech. Cell culture medium was purchased from Biochrom (Berlin, Germany), and fetal calf serum was obtained from PAN Systems (Aidenbach, Germany). Thrombin was obtained from Sigma (Deisenhofen, Germany). Trypsin and trypsin inhibitor were from Boehringer. Hanks' balanced salt solution (HBSS) contained (concentrations in grams per liter in parentheses) CaCl2 (0.185), MgSO4 (0.089), KCl (0.4), KH2PO4 (0.06), NaCl (8.0), Na2HPO4 (0.048), and glucose (1.0), to which 10 mM HEPES (pH 7.4) was added. C2II of C. botulinum was purified as described previously (10). The N-terminal sequencing of trypsin-activated C2II was carried out by C. C. Shone Centre for Applied Microbiology and Research, Salisbury, United Kingdom).
Cloning of the C2II gene.
The C2II gene was amplified by PCR with 30 ng of partially SmaI-digested chromosomal DNA from C. botulinum KZZ1577(92-13) in a total volume of 100 μl with 1 U of Taq DNA polymerase in a reaction mixture that included deoxynucleoside triphosphates (100 μM each) and 50 pmol of the primers C2II-pos (5′-GATGGACCATGGCGGTTTCAAAATTTGAGAAC-3′), which contains an NcoI site (underlined), and C2II-neg (5′-TCGATCGGATCCGATATTATTAATTTATCTAATTC-3′), which contains a BamHI site (underlined). Amplification was done by using 3 cycles of denaturing at 94°C for 1 min, primer annealing at 35°C for 1 min, and extension at 72°C for 3 min followed by 27 cycles of denaturing at 94°C for 1 min, primer annealing at 50°C for 1 min, and extension at 72°C for 3 min. The PCR product was digested with NcoI and BamHI and cloned in an NcoI- and BamHI-cut pET22b vector, which resulted in the plasmid pET22b-C2II. For subcloning of the C2II gene in the vector pGEX-2T, we amplified C2II from pET22b by PCR with 100 ng of plasmid DNA in a total volume of 50 μl with 2 U of Taq DNA polymerase in a reaction mixture that included deoxynucleoside triphosphates (100 μM each) and 50 pmol of the primers C2II-5′ (5′-GCTTCGGGATCCATGTTAGTTTCAAAATTTGAG-3′), which contains a BamHI site, and C2II-3′ (5′-TCGATCGAATTCTATATTATTAATTTATCTAATTC-3′), which contains an EcoRI site. Amplification was done by performing 30 cycles of denaturing at 94°C for 1 min, primer annealing at 50°C for 1 min, and extension at 70°C for 3 min. The resulting PCR product was digested with EcoRI and BamHI and cloned into an EcoRI- and BamHI-cut pGEX-2T vector, resulting in the plasmid pGEX-2T-C2II. The sequence of the construct was confirmed by DNA sequencing.
Construction of proteins with deletions in C2II.
For structure-function analysis, several C2II fragments were constructed (see Fig. 3B). The C2II fragments were obtained by PCR with pGEX-2T-C2II as the template by using the following primers: for C2II-D1, which contains amino acids 1 to 263, we used C2II-5′ and C2II-D1-3′ (5′-TATGAATTCAGCAGATATCATTGGATC-3′); for C2II-D2, which contains amino acids 264 to 486, we used C2II-D2-5′ (5′-CGCGGATCCTATCCTATAGTTGGAGTCCAA-3′) and C2II-D2-3′ (5′-TGAGAATTCTGTACTTTTTATAGTACC-3′); for C2II-D3, which contains amino acids 487 to 591, we used C2II-D3-5′ (5′-AAAGGATCCACAGCTTCATTAAC-3′) and C2II-D3-3′ (5′-TTCGAATTCAGTAATTACTTTTACTAA-3′); for C2II-D4, which contains amino acids 592 to 721, we used C2II-D4-5′ (5′-GTAGGATCCTTCAAAGAAAATATATC-3′) and C2II-3′; for C2II with D4 deleted (C2II-ΔD4), we used C2II-5′ and C2II-D3-3′; for C2II with 7 C-terminal amino acids deleted (C2II-d7), we used C2II-5′ and C2II-d7 (5′-TCGGGGGTTTTCTTAATATAGGAATTCAAA-3′), which contains a stop codon (shown in bold); for C2II with 16 C-terminal amino acids deleted (C2II-d16), we used C2II-5′ and C2II-d16 (5′-GATATTATAAATTCTATTAATTAGGAATTCGGG-3′), which contains a stop codon (shown in bold); and for C2II with 181 N-terminal amino acids deleted (C2IIreca), we used C2IIa-5′ (5′-AAAGGATCCGCTAATGCAAATAGA-3′) and C2II-3′. The 5′ primer always contained a BamHI site, and the 3′ primer always contained an EcoRI site (restriction sites are underlined). Amplification was done as described for the C2II gene. The PCR products were digested with EcoRI and BamHI and cloned into an EcoRI- and BamHI-cut pGEX-2T vector, resulting in the plasmids pGEX-C2II-D1, pGEX-C2II-D2, pGEX-C2II-D3, pGEX-C2II-D4, pGEX-C2IIreca, pGex-C2II-ΔD4, pGEX-C2II-d7, and pGEX-C2II-d16. The sequences of the constructs were confirmed by DNA sequencing.
FIG. 3.
(A) Sequence similarities between the binding components of C2 toxin (C2II) and anthrax toxin (PA). Based on sequence homologies to PA, C2II was divided into four domains. Sequence identities and homologies (in parentheses) between C2II and PA are shown below the diagrams. Numbers above the diagrams indicate amino acid residues. Arrows indicate the protease cleavage sites for activation. (B) Generation of C2II fragments and antisera. D1, D3, and D4 of C2II were amplified from pGEX-C2II by PCR. The resulting PCR products were cloned in the pGEX-2T vector, resulting in plasmids pGEX-C2II-D1, pGEX-C2II-D3, and pGEX-C2II-D4, respectively. The respective antisera against the resulting C2II domains were anti-C2II-D1, anti-C2II-D3, and anti-C2II-D4 antisera. Anti-C2II182–196, antiserum against an N-terminal peptide of C2IIa (amino acids 182 to 197); anti-C2II707–721, antiserum against a C-terminal peptide of C2II (amino acids 706 to 721).
Expression and purification of recombinant proteins.
Various recombinant proteins were expressed as GST fusion proteins in Escherichia coli BL21 cells harboring the separate DNA fragments in plasmid pGEX-2T. Proteins were purified as described previously (4) and eluted with 10 mM glutathione–100 mM NaCl–50 mM Tris (pH 8.0) or incubated with thrombin (3.25 National Institutes of Health units/ml of bead suspension) for cleavage of the fusion proteins from GST. Thereafter, the suspension was centrifuged at 500 × g (10 min, room temperature) and an aliquot of the resulting supernatant was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). C. botulinum C2II, recombinant C2II, C2II-d7, and C2II-ΔD4 were activated with 0.2 μg of trypsin per μg of protein for 30 min at 37°C.
Generation of C2II antisera.
An N-terminal peptide (amino acids 182 to 196, ANANRDTDRDGIPDE) and a C-terminal peptide (amino acids 707 to 721, RLSGVFLIELDKLII) of C2II were synthesized and coupled to keyhole lympet hemocyanin by H. R. Rackwitz (German Cancer Research Center, Heidelberg, Germany). Antibodies against these peptides and the whole protein of C2II were raised in rabbits and designated anti-C2II182–196, anti-C2II707–721, and anti-C2II antibodies (see Fig. 3B). D1, D3, and D4 of C2II were expressed and purified as described above. Antibodies against the domain peptides (C2II-D1, C2II-D3, and C2II-D4) were raised in rabbits and designated anti-C2II-D1, anti-C2II-D3, and anti-C2II-D4 antibodies, respectively (Fig. 3B). The immunoglobulin G fraction of the C2II-D4 antiserum was affinity purified with protein A-Sepharose.
SDS-PAGE.
SDS-PAGE was performed according to the methods of Laemmli (16). The gels were stained with Coomassie brilliant blue R-250.
Immunoblot analysis.
The proteins were transferred by electroblotting from the gel onto a nitrocellulose membrane using a semidry system. The membranes were blocked for 30 min with 5% nonfat dry milk in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-T), followed by a 1-h incubation with the appropriate antiserum (rabbit antiserum diluted 1:5,000 in PBS-T). After being washed with PBS-T, the blots were probed for 1 h with donkey anti-rabbit antibody coupled to horseradish peroxidase (1:3,000 dilution in PBS-T) and washed and proteins were detected using the enhanced chemiluminescence system according to the manufacturer's instructions.
Cell culture and cytotoxicity assay.
All cells were cultivated in tissue culture flasks at 37°C and 5% CO2. CHO-K1 cells were maintained in Dulbecco's minimal essential medium–Ham's F-12 medium (1:1) containing 5% fetal calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. HeLa and NIH 3T3 cells were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum and antibiotics as described above. Cells were routinely trypsinized and reseeded three times a week. For cytotoxicity assays, cells were grown as subconfluent monolayers and treated with different concentrations of several C2II proteins and 100 ng of C2I per ml. For microscopy, cells were washed with PBS, fixed in 4% paraformaldehyde in PBS for 30 min, and washed and the coverslips were embedded in Kaiser's gelatin on glass.
In vitro ADP-ribosylation of actin.
Cells were lysed by sonication in a solution containing 25 mM HEPES and 2 mM MgCl2. Protein (50 μg) of each lysate was subjected to the ADP-ribosylation assay as described previously (4). Briefly, samples were incubated with 300 ng of C2I in 35 mM HEPES (pH 7.5)–0.2 mM MgCl2–0.1 mM dithiothreitol–0.5 μM [adenylate-32P]NAD (about 25 nCi) for 20 min at 37°C. Radiolabeled proteins were precipitated with chloroform-methanol and detected by SDS-PAGE and subsequent phosphorimaging.
Binding of C2II to cells.
Cells were grown as subconfluent monolayers, prechilled to 4°C, and then incubated with proteins in HBSS at 4°C for 1.5 h. Thereafter, cells were washed five times with ice-cold PBS and lysed by sonication in 25 mM HEPES (pH 7.5)–2 mM MgCl2. Protein (50 μg) of each lysate was subjected to SDS-PAGE, and Western blot analysis was done as described above.
Competition experiments.
Cells were grown as subconfluent monolayers, prechilled to 4°C, and incubated with the separate truncated C2II proteins (5 μg/ml) in HBSS at 4°C for 1.5 h. Thereafter, the trypsin-cleaved activated binding component of C2 toxin (C2IIa) was added (200 ng/ml) and the cells were incubated on ice for an additional 1.5 h. Next the cells were washed five times with ice-cold PBS. Competition of C2IIa activity by C2II truncations was analyzed either by immunoblot analysis or by cytotoxicity assay. For immunoblot analysis, cells were lysed and binding of C2IIa and truncated proteins was detected with the appropriate antiserum. For the cytotoxicity assay, 100 ng of C2I per ml was added and cells were shifted to 37°C. Pictures of the cells were taken after 3 h of incubation. In a second approach, 5-μg quantities of the separate truncated C2II proteins per ml in HBSS were added together with C2IIa (200 ng/ml) and C2I (100 ng/ml) to cells and incubated for 3 h at 37°C.
Artificial-membrane experiments.
Black lipid bilayer membranes were formed as described previously (5). The instrument consisted of a Teflon chamber with two aqueous compartments connected by a small circular hole. The hole had a surface area of about 0.5 mm2. Membranes were formed across the hole by painting onto a 1% solution of diphytanoyl phosphatidylcholine (Avanti Polar Lipids, Alabaster, Ala.) in n-decane. The single-channel recordings were performed using calomel electrodes (with salt bridges) connected in series to a voltage source and a current amplifier. The amplified signal was monitored on a storage oscilloscope (Tektronix 7633) and recorded on a strip chart or tape recorder.
All experiments were performed at least three times. Data from representative experiments are shown.
RESULTS
Cloning, expression, and characterization of C2II.
In order to perform a structure-function analysis of the binding component of C. botulinum C2 toxin (C2II), C2II was amplified from chromosomal DNA of C. botulinum KZZ1577(92-13) and expressed as a GST fusion protein in E. coli. The C2II protein was purified as described in Materials and Methods and analyzed by SDS-PAGE (Fig. 1A) and immunoblot analysis with anti-C2II182–196 antibody (Fig. 1B). Like C. botulinum C2II, recombinant C2II formed SDS-stable oligomers after tryptic activation. These oligomers could be detected when C2IIa was subjected to SDS–3 to 12.5% PAGE without prior heating of the proteins (Fig. 1C, lane 2). When C2IIa was applied together with C2I to NIH 3T3 cells, cell rounding was observed (Fig. 2A). The C2I-catalyzed ADP-ribosylation of actin in intact cells was confirmed by subsequent in vitro [32P]ADP-ribosylation in cell lysates (Fig. 2B). In the absence of C2I, C2IIa did not induce any cytotoxic effect. C2II which had not been activated by trypsin was biologically inactive. Comparing C. botulinum C2IIa and recombinant C2IIa in cell experiments, we could not detect any differences in the cytotoxicities of these proteins and in their channel-forming activities in artificial membranes (not shown).
FIG. 1.
Analysis of recombinant C2II protein. C2II was expressed as a GST fusion protein in E. coli and cleaved with thrombin from glutathione-Sepharose beads. Proteins were subjected to SDS–12.5% PAGE and either stained with Coomassie blue (A) or detected by Western blot analysis with anti-C2II182–196 antiserum (B). Lane 1, GST-C2II fusion protein; lane 2, C2II; lane 3, C2II after activation with 0.2 μg of trypsin per μg of protein. (C) C2IIa was subjected to SDS–3 to 12.5% PAGE with and without prior heating (lanes 1 and 2, respectively) and stained with Coomassie blue. Lane 1, C2IIa monomer; lane 2, C2IIa oligomer. Relative molecular weights (Mr) (in thousands) are noted at the left.
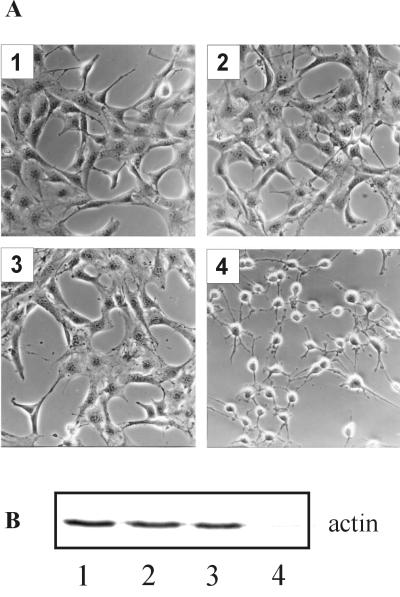
FIG. 2.
Cytotoxic effect of recombinant C2 toxin on NIH 3T3 cells. NIH 3T3 cells were incubated at 37°C in HBSS with C2 toxin components. Three hours after toxin addition cells were photographed (A), and cell lysates were analyzed by subsequent in vitro ADP-ribosylation of actin (B). Cell lysates were incubated with C2I and [32P]NAD. Actin that had not been modified by the toxin pretreatment was [32P]ADP-ribosylated in the in vitro assay and detected by SDS-PAGE and phosphorimaging. (A) Image 1, control cells; image 2, cells treated with 200 ng of C2II per ml plus 100 ng of C2I per ml; image 3, cells treated with 200 ng of C2IIa per ml; image 4, cells treated with 200 ng of C2IIa per ml plus 100 ng of C2I per ml.
Analysis of C2II truncations.
Sequence comparisons revealed high sequence similarities of C2II to the PA of anthrax toxin and, as was shown earlier (15), to the binding components of C. perfringens iota toxin, C. spiroforme toxin, and C. difficile ADP-ribosyltransferase. On the basis of sequence homologies to PA, C2II could be divided into four domains (Fig. 3A). D1 of PA contains the activating protease cleavage site, D2 contains a putative membrane insertion loop, and D4 is responsible for receptor binding (22, 28). The function of D3 is still unknown. To obtain more insight into the functions of the four separate C2II domains, we analyzed truncated C2II proteins and raised several C2II antisera against these domains.
Sequencing of trypsin-cleaved C2II revealed the N-terminal sequence ANANRDTDRDGIPDE of activated C2IIa, indicating trypsin cleavage after lysine 181. We constructed and expressed the corresponding C2II deletion protein lacking the 181 N-terminal amino acids of the full-length toxin. However, genetically activated C2II (C2IIreca) did not form oligomers and was not toxic when it was applied to cells together with C2I (data not shown). Binding experiments revealed that C2IIreca was not able to bind to cells. We therefore propose that the 181 N-terminal amino acids of C2II are important for the correct folding of the protein. Next, we constructed toxin fragments covering D1, D2, D3, and D4 of C2II (Fig. 3B). Whereas D1, D3, and D4 were stably expressed, D2 was not stable. In testing the biological activities of D1, D3, and D4 on NIH 3T3 cells, we did not observe any cytotoxicity or competitive effects with the C2II truncations.
Influence of various C2II antisera on cytotoxic effects of C2 toxin.
Antisera were raised against the three truncated C2II proteins (C2II-D1, -D3, and -D4), against an N-terminal and a C-terminal peptide covering amino acids 182 to 196 and 707 to 772, respectively, and against the full-length C2II protein (Fig. 3B). Western blot analysis of these antisera exhibited specific recognition of C2II, C2IIa, and the parts of C2II against which they were raised (Table 1). As an exception, anti-C2II antiserum did not recognize C2II-D3.
TABLE 1.
Analysis of various C2II antiseraa
| Protein | Test result with indicated antiserum
|
|||||
|---|---|---|---|---|---|---|
| Anti- C2II | Anti- C2II-D1 | Anti- C2II182–196 | Anti- C2II-D3 | Anti- C2II-D4 | Anti- C2II707–721 | |
| C2II | + | + | + | + | + | + |
| C2IIa | + | + | + | + | + | + |
| C2II-D1 | − | + | + | − | − | − |
| C2II-D3 | + | − | − | + | − | − |
| C2II-D4 | + | − | − | − | + | + |
C2II, C2IIa, C2II-D1, C2II-D3, and C2II-D4 (200 ng each) were subjected to SDS–12.5% PAGE and blotted to a nitrocellulose membrane. Proteins were detected with anti-C2II, anti-C2II182–196, anti-C2II-D1, anti-C2II-D3, anti-C2II-D4, and anti-C2II707–721 antisera. +, protein was recognized by antiserum; −, protein was not recognized by antiserum.
To test the effects of the various antisera on the biological activity of C2IIa, the activated binding component C2IIa (200 ng) was preincubated with 20 μl of the separate antisera for 1 h on ice and applied together with 100 ng of C2I in 1 ml of HBSS to NIH 3T3 cells. After 3 h of incubation at 37°C, cells were washed and analyzed. Anti-C2II and anti-C2II-D4 antisera completely inhibited the cytotoxic C2 effect, even after an extended incubation of cells for up to 12 h (Table 2). The same inhibitory effect was observed when the protein A-Sepharose-purified immunoglobulin G antibody was used. Incubation of cells with the various antisera did not lead to any morphological alterations. The activity of C2I was not influenced when C2I was preincubated with the different antisera and subsequently applied to NIH 3T3 cells together with C2IIa.
TABLE 2.
Influence of various C2II antisera and cytotoxic C2 effects on NIH 3T3 cellsa
| Antiserum | Inhibition |
|---|---|
| Anti-C2II | + |
| Anti-C2II-D1 | − |
| Anti-C2II182–196 | − |
| Anti-C2II-D3 | − |
| Anti-C2II-D4 | + |
| Anti-C2II707–721 | − |
C2IIa was preincubated with different C2II antisera (or PBS as a control) for 1 h on ice. Together with 100 ng of C2I, 200 ng of pretreated C2IIa was added to cells. After 3 h of incubation at 37°C, the cells were photographed and cell rounding was determined. +, inhibition of cytotoxic C2 effect by antiserum; −, no inhibition of cytotoxic C2 effect by antiserum.
Antibody against D4 inhibits binding of C2IIa to cells.
To test whether the loss of toxicity after preincubation of C2IIa with anti-C2II and anti-C2II-D4 antisera was due to a reduced ability of C2IIa to bind the cell receptor, we performed binding experiments with antibody-pretreated C2IIa. Western blot analysis revealed that anti-C2II-D4 antiserum inhibited and anti-C2II antiserum significantly decreased binding of C2IIa to cells (Fig. 4). Based on the observation that antiserum against D4 prevented C2IIa from binding to cells, we propose that D4 of C2II (amino acids 592 to 721) is involved in receptor recognition and binding. In order to confirm this result, we deleted D4 from C2II. The resulting protein (C2II-ΔD4) was still able to form SDS-stable oligomers after tryptic activation (Fig. 5A) and channels in artificial membranes (not shown). However, the migration behavior of oligomers of the C2II-ΔD4 protein activated by trypsin treatment (C2II-ΔD4a) differed from that of C2IIa oligomers. C2II-ΔD4 was nontoxic when it was applied to cells together with C2I and was not able to bind the cellular receptor (Fig. 5B). This result confirmed that D4 of C2II mediates receptor binding. Since C2II-ΔD4 still oligomerized, we concluded that D4 is not necessary for oligomer formation of the protein.
FIG. 4.
Pretreatment of C2IIa with anti-C2II-D4 antiserum inhibits binding of C2IIa to NIH 3T3 cells. Two hundred nanograms of C2IIa was preincubated with 20 μl of anti-C2II-D4 or anti-C2II antiserum (or PBS as a control) for 1 h on ice. Pretreated C2IIa was added to prechilled cells in 1 ml of HBSS. After incubation for 1.5 h on ice, cells were washed five times with ice-cold PBS and lysed in 25 mM HEPES–2 mM MgCl2. Equal amounts of protein were subjected to SDS-PAGE and Western blot analysis with anti-C2II-D4 antiserum. Lane 1, control; lane 2, cells incubated with C2IIa which had been pretreated with anti-C2II antiserum; lane 3, cells incubated with C2IIa which had been pretreated with anti-C2II-D4 antiserum; lane 4, cells incubated with C2IIa.
FIG. 5.
Characterization of C2II-ΔD4. C2II-ΔD4 was expressed as a GST fusion protein in E. coli and cleaved with thrombin from glutathione-Sepharose beads. (A) C2II-ΔD4 and C2II were activated with trypsin and subjected to SDS–3 to 12.5% PAGE without prior heating and stained with Coomassie blue. Lane 1, C2IIa oligomer; lane 2, C2II-ΔD4a oligomer. Relative molecular weights (in thousands) are noted. (B) Binding of truncated C2IIa to NIH 3T3 cells. Two hundred nanograms of C2IIa and 200 ng of C2II-ΔD4a were separately added to prechilled NIH 3T3 cells in 1 ml of ice-cold HBSS. Cells were incubated for 1.5 h on ice, washed five times with ice-cold PBS, and lysed in 25 mM HEPES–2 mM MgCl2. Proteins were subjected to SDS–12.5% PAGE and analyzed by Western blotting with anti-C2II182–196 antiserum. Lane 1, C2IIa; lane 2, C2II-ΔD4a; lane 3, control cells; lane 4, cells incubated with C2IIa; lane 5, cells incubated with C2II-ΔD4a.
Effect of anti-C2II-D4 antiserum on reconstitution of C2II oligomers in lipid bilayer membranes.
In a previous study we showed that activated C2II is able to form ion-permeable channels in lipid bilayer membranes (26). Similar effects were observed with the recombinant C2IIa (not shown). Addition of anti-C2II-D4 antiserum to the cis side of the membrane blocked the channel-forming activity of C2IIa (Fig. 6). Interestingly, the addition of the antibody inhibited the incorporation of channels into the membranes but had no effect on channels already existing. No influence on the single-channel characteristic or the voltage dependence was observed. Moreover, addition to the trans side of the membrane had no influence on the reconstitution rate. These results indicated that the antibody bound to free C2IIa oligomers but did not influence the channel properties of inserted C2IIa oligomers.
FIG. 6.
Influence of anti-C2II-D4 antiserum on channel formation in artificial membranes by C2IIa. Shown are results of increasing the current as a function of time after the addition of 500 ng of C2IIa per ml to one side (the cis side) of a black diphytanoyl phosphatidylcholine–n-decane membrane bathed in 0.1 M KCl (left-side arrow) (squares). The circles represent the results of another experiment in which 10 min after the addition of 500 ng of C2IIa per ml the anti-C2II-D4 polyclonal antibody was added in excess to the same side of the membrane as that containing C2IIa (right-side arrow). The diamonds show the results of a third experiment in which 500 ng of C2IIa per ml was preincubated with an excess of anti-C2II-D4 antibody and then added to one side of the membrane (left-side arrow). The applied voltage was 50 mV, and the temperature was 20°C. C2IIa and C2IIa preincubated with the antibodies were added in all three cases 10 min (corresponding to the start of the record in the figure) after the membrane was in the black state.
Deletion of 7 C-terminal amino acids prevents binding of C2II to cells.
To define the region responsible for receptor binding of C2II more closely, we deleted the 16 and 7 C-terminal amino acids. Attempts to purify the protein lacking 16 amino acids (C2II-d16) were unsuccessful because of extensive degradation. The protein lacking only 7 amino acids (C2II-d7) was stable. It formed SDS-stable oligomers after activation with trypsin, which could be detected when trypsin-activated C2II-d7 was subjected to SDS-PAGE without prior heating (C2II-d7a) (Fig. 7A). However, the migration behavior was clearly different from that of oligomers formed by complete C2IIa. We tested whether C2II-d7a was functional in membrane experiments. Single-channel measurements were taken in the presence of C2IIa and C2II-d7a. Figure 7B shows that C2II-d7a was able to form channels in the artificial membranes which did not differ from C2IIa channels. To analyze the biological activity, we incubated NIH 3T3 cells with C2II-d7a and C2I (100 ng/μl). Even after incubation for up to 24 h and at concentrations of C2II-d7a up to 1 μg of HBSS per ml, no cytotoxic effects occurred (Fig. 7C). Therefore, the binding of C2II-d7a to NIH 3T3 cells was analyzed. The results of Western blot analysis in Fig. 7D show that the binding activity was completely lost upon deletion of the 7 C-terminal amino acids. However, this result was not in line with the finding that an antiserum against the 15 C-terminal amino acids of C2II (anti-C2II707–721) did not inhibit binding of C2IIa to cells. We therefore tested by immunoblot analysis whether anti-C2II707–721 antiserum was able to bind C2IIa monomers and oligomers. With and without prior heating, full-length C2II and C2IIa were subjected to SDS-PAGE, blotted to nitrocellulose membranes, and probed with anti-C2II707–721 antiserum. The immunoblot revealed interaction of anti-C2II707–721 antiserum with full-length C2II and monomeric trypsin-cleaved C2IIa but not with C2IIa oligomers (Fig. 8). By contrast, all other C2II antisera used in this study did recognize C2IIa monomers as well as C2IIa oligomers (results shown in Fig. 8 are for anti-C2II-D4 antiserum).
FIG. 7.
Characterization of C-terminally truncated C2II. C2II-d7 was expressed as GST fusion protein in E. coli and cleaved with thrombin from glutathione-Sepharose beads. (A) Following tryptic activation, C2II-d7a and C2IIa were subjected to SDS–3 to 12.5% PAGE without prior heating and stained with Coomassie blue. Lane 1, C2IIa oligomer; lane 2, C2II-d7a oligomer. Relative molecular weights (in thousands) are noted at the left. (B) Single-channel recording of a diphytanoyl phosphatidylcholine–n-decane membrane in the presence of recombinant C2IIa oligomers (a) and C2II-d7a oligomers (b). Ten minutes after the formation of the membrane, 100 ng of oligomers per ml was added to the aqueous phase on one side of the membrane. The aqueous phase contained 1 M KCl (pH 6). The applied membrane potential was 20 mV, and the temperature was 20°C. (C) Cytotoxic effect of C2II-d7 on NIH 3T3 cells. Cells were incubated with 100 ng of C2I per ml together with either 200 ng of C2IIa per ml or 200 ng of C2II-d7a per ml in HBSS at 37°C, respectively. After 3 h, cells were fixed and photographed. Image 1, control; image 2, C2I plus C2IIa; image 3, C2I plus C2II-d7a. (D) Binding of truncated C2IIa to NIH 3T3 cells. Two hundred nanograms of C2IIa and 200 ng of C2II-d7a were separately added to prechilled NIH 3T3 cells in 1 ml of ice-cold HBSS. After incubation for 1.5 h on ice, cells were washed five times with ice-cold PBS and lysed in 25 mM HEPES–2 mM MgCl2. Proteins were subjected to SDS-PAGE and analyzed by Western blotting with anti-C2II182–196 antiserum. Lane 1, C2IIa; lane 2, C2II-d7a; lane 3, control cells; lane 4, cells incubated with C2IIa; lane 5, cells incubated with C2II-d7a.
FIG. 8.
Recognition of monomeric and oligomeric C2IIa by anti-C2II707–721 antiserum. Both C2II and C2IIa oligomers (200 ng of each protein) were subjected to SDS–3 to 12.5% PAGE without (lanes 1 and 2) and with (lanes 3 and 4) prior heating at 95°C. Proteins were blotted onto a nitrocellulose membrane and detected with anti-C2II707–721 antiserum. As a control, the same blot was probed with anti-C2II-D4 antiserum.
DISCUSSION
Recent sequencing (15) of the binding component of C. botulinum, C2II, revealed high sequence similarity with the PA of anthrax toxin (22), suggesting a similar four-domain structure for C2II. Here we studied the role of D4 of C2II in receptor binding, heptamerization, and pore formation. An antibody (anti-C2II-D4 antibody) raised against D4 of the toxin blocked cell binding and inhibited channel formation of C2IIa in a lipid bilayer. Only the formation of new channels was inhibited, whereas properties of existing channels were not changed, suggesting a role of D4 in receptor binding and membrane insertion. The C2II-related PA also forms channels in artificial lipid bilayers which are similar to the channels formed by C2IIa (6, 19). Membrane insertion of PA is suggested to be mediated by a flexible loop (2β2-2β3 loop) located in D2 (22) which contains a conserved pattern of alternating hydrophobic and hydrophilic residues. Kimura et al. (15) proposed a transmembrane segment in D2 of C2II, covering residues 260 to 277. We believe that a different sequence of alternating hydrophobic and hydrophilic residues (303-TVGAEVSGSLQLAGGIFPVFSMSASANYS-331, with the hydrophilic residues underlined), which is similar to the proposed transmembrane sequence of PA, is involved in membrane insertion. However, the inhibitory effect of anti-C2II-D4 antiserum on channel formation is most likely not caused by direct interaction with the inserting sequence but rather due to sterical hindrance of membrane insertion by a bulky group at the putative receptor-binding domain D4.
Further evidence for the location of the receptor-binding domain at the C terminus of C2II was obtained by C-terminal deletions. Truncation of D4 (residues 592 to 721) or of the 7 C-terminal amino acids of C2II blocked the binding and cytotoxicities of trypsin-activated proteins. At least two explanations are possible. First, the C-terminal part of C2II is important for the whole structure of the toxin, and second, the C terminus (and the most C-terminal part in the case of C2II-d7) of C2II is directly involved in receptor binding. The results of our experiments argue against the first possibility, because the C-terminally truncated proteins (C2II-ΔD4 and C2II-d7) were still activated by trypsin and formed SDS-stable oligomers as shown by SDS-PAGE. Furthermore, both truncated proteins induced pores in artificial membranes very similar to those of C2IIa, suggesting that at least major parts of the proteins are structurally preserved and are able to oligomerize and to insert into membranes. Although PA differs in its receptor-binding properties from C2II and shows no sequence similarity in D4 with C2II, it is noteworthy that 3 to 5 amino acid residues at the most C-terminal part of D4 of PA are crucial for receptor binding and cytotoxicity (29). For PA, it was suggested that this region plays a role in stabilizing a conformation needed for receptor-binding activity. It remains to be tested whether the most C-terminal residues of C2II are necessary for the stabilization of the receptor-binding domain of C2II. Notably, the truncation of the 7 C-terminal amino acids caused a significant shift in the migration of the oligomer on SDS-PAGE, suggesting major changes in the structure of the protein. This view is in line with the findings that a peptide antibody (anti-C2II707–721 antibody) produced against the 15 C-terminal amino acids of C2II did not inhibit cell binding of C2IIa and recognized only C2IIa monomers and not oligomers. Therefore, we propose that after oligomerization, amino acids 706 to 721 of C2IIa are stabilized in a specific conformation or are located inside the oligomer not accessible by the antibody. Since only C2IIa oligomers are able to bind to cells (3), the site responsible for high-affinity receptor binding presumably arises by oligomerization. Deletion of 7 C-terminal amino acids may alter this structure, thereby rendering the C2IIa oligomer inactive.
Recently, it has been reported that C2IIa binds specifically to asparagine-linked complex carbohydrates (9). Although binding to carbohydrates may explain why C2IIa binds to essentially all cell types tested, it does not exclude the possibility of the presence of a specific protein receptor which is essential for uptake. The receptor of PA clearly shows properties of a protein (10). Because the putative receptor-binding domains of C2II and of PA have no homology to each other, it is suggested that the two proteins bind to different receptors. D4 of C2II also differs from the C termini of the binding components of C. perfringens iota toxin, C. spiroforme toxin, and C. difficile toxin, whereas D4 of these three toxins are closely related. For C2 toxin and iota toxin, binding to different cell surface receptors has been shown (11), which is in line with the model that the C terminus mediates receptor recognition and binding.
ACKNOWLEDGMENTS
We thank Otilia Wunderlich, Brigitte Neufang, and Ulrike Müller for expert technical assistance. The anti-C2II antiserum was produced by Ingo Just. The N-terminal amino acid sequencing of C2IIa by C. C. Shone (CAMR, Salisbury, United Kingdom) is gratefully acknowledged. The peptides were kindly synthesized by Hans-Richard Rackwitz (German Cancer Research Center, Heidelberg, Germany).
This work was financially supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 388).
REFERENCES
- 1.Aktories K, Bärmann M, Ohishi I, Tsuyama S, Jakobs K H, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 2.Aktories K, Wegner A. Mechanisms of the cytopathic action of actin-ADP-ribosylating toxins. Mol Microbiol. 1992;6:2905–2908. doi: 10.1111/j.1365-2958.1992.tb01749.x. [DOI] [PubMed] [Google Scholar]
- 3.Barth, H., D. Blöcker, J. Behlke, W. Bergsma-Schutter, A. Brisson, R. Benz, and K. Aktories. Cellular uptake of Clostridium botulinum C2 toxin requires oligomerization and acidification. J. Biol. Chem., in press. [DOI] [PubMed]
- 4.Barth H, Preiss J C, Hofmann F, Aktories K. Characterization of the catalytic site of the ADP-ribosyltransferase Clostridium botulinum C2 toxin by site-directed mutagenesis. J Biol Chem. 1998;273:29506–29511. doi: 10.1074/jbc.273.45.29506. [DOI] [PubMed] [Google Scholar]
- 5.Benz R, Janko K, Lauger K. Ionic selectivity of pores formed by the matrix protein (porin) of Escherichia coli. Biochim Biophys Acta. 1979;551:238–247. doi: 10.1016/0005-2736(89)90002-3. [DOI] [PubMed] [Google Scholar]
- 6.Blaustein R O, Koehler T M, Collier R J, Finkelstein A. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc Natl Acad Sci USA. 1989;86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Considine R V, Simpson L L. Cellular and molecular actions of binary toxins possessing ADP-ribosyltransferase activity. Toxicon. 1991;29:913–936. doi: 10.1016/0041-0101(91)90076-4. [DOI] [PubMed] [Google Scholar]
- 8.Duesbery N S, Webb C P, Leppla S H, Gordon V M, Klimpel K R, Copeland T D, Ahn N G, Oskarsson M K, Fukasawa K, Paull K D, Woude G F V. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 9.Eckhardt M, Barth H, Blöcker D, Aktories K. Binding of Clostridium botulinum C2 toxin to asparagine-linked complex and hybrid carbohydrates. J Biol Chem. 2000;275:2328–2334. doi: 10.1074/jbc.275.4.2328. [DOI] [PubMed] [Google Scholar]
- 10.Escuyer V, Collier R J. Anthrax protective antigen interacts with a specific receptor on the surface of CHO-K1 cells. Infect Immun. 1991;59:3381–3386. doi: 10.1128/iai.59.10.3381-3386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritz G, Schroeder P, Aktories K. Isolation and characterization of a Clostridium botulinum C2 toxin-resistant cell line: evidence for possible involvement of the cellular C2II receptor in growth regulation. Infect Immun. 1995;63:2334–2340. doi: 10.1128/iai.63.6.2334-2340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geipel U, Just I, Schering B, Haas D, Aktories K. ADP-ribosylation of actin causes increase in the rate of ATP exchange and inhibition of ATP hydrolysis. Eur J Biochem. 1989;179:229–232. doi: 10.1111/j.1432-1033.1989.tb14545.x. [DOI] [PubMed] [Google Scholar]
- 13.Han S, Craig J A, Putnam C D, Carozzi N B, Tainer J A. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 14.Kimura K, Fujii N, Tsuzuki K, Murakami T, Indoh T, Yokosawa N, Oguma K. Cloning of the structural gene for Clostridium botulinum type C1 toxin and whole nucleotide sequence of its light-chain component. Appl Environ Microbiol. 1991;57:1168–1172. doi: 10.1128/aem.57.4.1168-1172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura K, Kubota T, Ohishi I, Isogai E, Isogai H, Fujii N. The gene for component-II of botulinum C2 toxin. Vet Microbiol. 1998;62:27–34. doi: 10.1016/s0378-1135(98)00195-3. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Leppla S. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations in eukaryotic cells. Proc Natl Acad Sci USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leppla S H. Anthrax toxins. In: Moss J, Iglewski B, Vaughan M, Tu A T, editors. Bacterial toxins and virulence factors in disease. New York, N.Y: Marcel Dekker; 1995. pp. 543–572. [Google Scholar]
- 19.Milne J C, Collier R J. pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol Microbiol. 1993;10:647–653. doi: 10.1111/j.1365-2958.1993.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 20.Ohishi I. Activation of botulinum C2 toxin by trypsin. Infect Immun. 1987;55:1461–1465. doi: 10.1128/iai.55.6.1461-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohishi I, Iwasaki M, Sakaguchi G. Purification and characterization of two components of botulinum C2 toxin. Infect Immun. 1980;30:668–673. doi: 10.1128/iai.30.3.668-673.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddingtom R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 23.Popoff M R, Boquet P. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem Biophys Res Commun. 1988;152:1361–1368. doi: 10.1016/s0006-291x(88)80435-2. [DOI] [PubMed] [Google Scholar]
- 24.Popoff M R, Rubin E J, Gill D M, Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988;56:2299–2306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuner K H, Presek P, Boschek C B, Aktories K. Botulinum C2 toxin ADP-ribosylates actin and disorganizes the microfilament network in intact cells. Eur J Cell Biol. 1987;43:134–140. [PubMed] [Google Scholar]
- 26.Schmid A, Benz R, Just I, Aktories K. Interaction of Clostridium botulinum C2 toxin with lipid bilayer membranes: formation of cation-selective channels and inhibition of channel function by chloroquine and peptides. J Biol Chem. 1994;269:16706–16711. [PubMed] [Google Scholar]
- 27.Simpson L L, Stiles B G, Zapeda H H, Wilkins T D. Molecular basis for the pathological actions of Clostridium perfringens iota toxin. Infect Immun. 1987;55:118–122. doi: 10.1128/iai.55.1.118-122.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh Y, Klimpel K R, Quinn C P, Chaudhary V K, Leppla S H. The Carboxyl-terminal end of protective antigen is required for receptor binding and anthrax toxin activity. J Biol Chem. 1991;266:15493–15497. [PubMed] [Google Scholar]
- 29.Varughese M, Teixeira A V, Liu S H, Leppla S H. Identification of a receptor-binding region within domain 4 of the protective antigen component of anthrax toxin. Infect Immun. 1999;67:1860–1865. doi: 10.1128/iai.67.4.1860-1865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegner A, Aktories K. ADP-ribosylated actin caps the barbed ends of actin filaments. J Biol Chem. 1988;263:13739–13742. [PubMed] [Google Scholar]
- 31.Wiegers W, Just I, Müller H, Hellwig A, Traub P, Aktories K. Alteration of the cytoskeleton of mammalian cells cultured in vitro by Clostridium botulinum C2 toxin and C3 ADP-ribosyltransferase. Eur J Cell Biol. 1991;54:237–245. [PubMed] [Google Scholar]
- 32.Wille M, Just I, Wegner A, Aktories K. ADP-ribosylation of the gelsolin-actin complex by clostridial toxins. J Biol Chem. 1992;267:50–55. [PubMed] [Google Scholar]
- 33.Zepeda H, Considine R V, Smith H L, Sherwin J, Ohishi I, Simpson L L. Actions of the Clostridium botulinum binary toxin on the structure and function of Y-1 adrenal cells. J Pharmacol Exp Ther. 1988;246:1183–1189. [PubMed] [Google Scholar]