Abstract
The French National Reference Centre for Escherichia coli, Shigella and Salmonella (FNRC-ESS) detected two human clusters of 33 cases (median age: 10 years; 17 females) infected by Salmonella enterica serotype Bovismorbificans, ST142, HC5_243255 (EnteroBase HierCC‑cgMLST scheme) in September–November 2020 and of 11 cases (median age: 11 years; seven males) infected by S. enterica serotype 4,12:i:-, ST34, HC5_198125 in October–December 2020. Epidemiological investigations conducted by Santé publique France linked these outbreaks to the consumption of dried pork sausages from the same manufacturer. S. Bovismorbificans and S. 4,12:i:- were isolated by the National Reference Laboratory from different food samples, but both strains were identified in a single food sample only by qPCR. Three recalls and withdrawals of dried pork products were issued by the French general directorate of food of the French ministry for agriculture and food in November 2020, affecting eight supermarket chains. A notification on the European Rapid Alert System for Food and Feed and a European urgent enquiry on the Epidemic Intelligence Information System for Food and Waterborne Diseases and Zoonoses (EPIS-FWD) were launched. No cases were reported outside France. Outbreaks caused by multiple serotypes of Salmonella may go undetected by protocols in standard procedures in microbiology laboratories.
Keywords: Salmonella, multi-serotype, outbreak, Bovismorbificans, Typhimurium, cgMLST
Key public health message.
What did you want to address in this study?
Salmonella enterica subsp. enterica is the main cause of food-borne diarrhoeal infections in Europe. Present diagnostic protocols target single-agent infections, but not multi-serotype infections. We wanted to describe a recent multiple-serotype Salmonella outbreak in humans in France in 2020, which led to an international investigation.
What have we learnt from this study?
The outbreak affected 44 people in association with the consumption of dried pork sausages contaminated by two different serotypes of Salmonella. The investigations identifying multiple serotypes of Salmonella in the food were complicated and required the collaboration of seven different institutions. The identification of the two serotypes was only possible in a single food sample.
What are the implications of your findings for public health?
This study highlights the need to improve the procedures to better detect mixed contaminations of food products by different serotypes of Salmonella, that we believe may go undetected with the present standard laboratory procedures.
Background
Salmonella enterica is a major cause of gastroenteritis, with 180 million cases globally per year (9% of all infectious gastroenteritis cases) and is responsible for almost half (41%) of the deaths associated to the diarrhoeal disease. Salmonella shows the highest rates of demonstrated association to food-borne infection, i.e. 52% for non-typhoidal salmonellosis [1]. In 2019, 87,923 confirmed cases of salmonellosis in humans were reported in Europe, with a European Union (EU) notification rate of 20.0 cases per 100,000 population; Salmonella caused 26.6% of all food-borne outbreaks [2]. In France, Salmonella remains the main cause of food-borne illness–associated hospitalisation and death [3,4].
Three serotypes are responsible for the majority of Salmonella infections in Europe: Enteritidis, Typhimurium and its monophasic variant (1,4,[5],12:i:-), together representing 70.3% of the 79,300 confirmed human cases with a known serotype in 2019. After poultry, pork is the most frequent source for salmonellosis in Europe (31%), and it has become the most frequent source for Salmonella enterica serotype Typhimurium and its monophasic variant 1,4,[5],12:i:-. In France, pork is suspected to be responsible for half of the salmonellosis cases reported every year [2,5,6].
S. enterica serotype Bovismorbificans is a relatively frequent food-borne pathogen (57 cases/year in France from 2012–20, and it was the 13th most frequently isolated serotype among human-identified Salmonella infections in Europe in 2019 [2]. Serotype Bovismorbificans is often identified in association with consumption of contaminated vegetables [7-11]. However, it has also been recently involved in outbreaks linked to horse and pork meat in Australia and France [12,13].
Outbreak detection
Between 27 October and 6 November 2020, the French National Reference Centre for E. coli, Shigella and Salmonella (FNRC-ESS), at the Institut Pasteur in Paris, France, detected through the routine sequencing and clustering of Salmonella spp. a cluster of 14 human isolates of S. Bovismorbificans ST142, with HC5_243255. Cases resided in four different regions in France, with ages ranging from 1 month to 48 years. On 24 December 2020, the FNRC-ESS identified another cluster of nine cases of S. 4,12:i:-, ST34, HC5_198125. Patients lived in seven regions in France, with ages ranging from 4 to 42 years old.
The French National Institute for Public Health (Santé publique France, SpF), initiated the outbreak investigations, in collaboration with the French general directorate of food (DGAL) and the National Reference Laboratory at the French National Agency for Food Security (LNR-ANSES) to identify the source of the outbreak and implement control measures. Epidemiological investigations showed the consumption of the same food product was associated to most of cases.
We present here our investigation of two associated food-borne outbreak clusters of salmonellosis in France, one caused by S. Bovismorbificans ST142 and the other caused by S. 4,12:i:-, ST34. We also aimed to draw attention to the possibility of multi-serotype Salmonella outbreaks occurring and the challenges associated with their identification.
Methods
Case definition
A confirmed case was defined as a person with a laboratory-confirmed isolate of S. Bovismorbificans, ST142 or S. 4,12:i:-, ST34, belonging to the outbreak phylogenetic clusters: HierCC HC5_243255 or HC5_198125, respectively, or at five or less allelic distance (AD) to another isolate in the cluster, by the EnteroBase HierCC scheme on core genome multilocus sequence typing (cgMLST) [14].
All cases were notified by the FNRC-ESS to SpF upon identification; name, date of birth, sex and postal code were included in the notification. There was no timeframe in the case definition.
Epidemiological investigation
Following the first notifications of the two clusters, and then after each new case detection by the FNRC-ESS, SpF interviewed patients using a standardised Salmonella trawling questionnaire, to collect information on dates of symptoms onset, hospitalisation, food consumption, other exposures in the 7 days before illness, places of food purchase and numbers of loyalty cards, if available. A descriptive analysis of the cases was conducted, and relevant information were shared with the DGAL for an investigation of food products.
No further analytical studies (case control or cohort) were carried out for this outbreak and no follow-up was performed for the identified cases, because the outbreak ceased following the food recalls and withdrawals.
Human microbiological investigation
Clinical sample collection
Bacterial isolates were sent to the FNRC-ESS by laboratories of clinical microbiology. As part of the routine procedure, all laboratories in France are encouraged to send all Salmonella isolates for surveillance on a voluntarily basis. In 2020, 7,181 isolates were received and analysed at the FNRC-ESS, of which 80 belonged to serotype Bovismorbificans and 1,820 to the monophasic variant of serotype Typhimurium (4,[5]12:i:-).
All 44 isolates in this outbreak were obtained from stool samples, except for two isolates of the S. Bovismorbificans ST142 HC5_243255 cluster that were obtained from blood cultures.
Whole genome sequencing
Whole genome sequencing (WGS) was performed as part of routine procedures at the FNRC-ESS. WGS was carried out at the Plateforme de microbiologie mutualisée (P2M) from the Pasteur International Bioresources network (PIBnet, Institut Pasteur, Paris, France). The MagNAPure 96 system (Roche Diagnostics) was used for DNA extraction, libraries were prepared using the Nextera XT kit (Illumina) and sequencing was done with the NextSeq 500 system (Illumina). Serotype prediction was done by in-house scripts based on MLST [15], fliC and fljB gene databases (FNRC-ESS internal flagellin database, unpublished).
Genomic sequences from all S. Bovismorbificans ST142 and S. 4,12:i:- ST34 isolates received at the FNRC-ESS were deposited, per routine, into EnteroBase (https://enterobase.warwick.ac.uk) [16]. Phylogenetic analysis was performed by two different approaches integrated into EnteroBase: single nucleotide polymorphism (SNP) analysis (maximum-likelihood trees) and cgMLST (HierCC and minimum spanning trees based on AD.
Antimicrobial resistance profiles were inferred using the Resfinder 4.1 tool (https://cge.food.dtu.dk/services/ResFinder) [17].
Classical sero-agglutination was performed on three selected isolates of the ST34 cluster.
Food product investigation
The epidemiological investigations carried out by SpF pointed towards dried pork sausages produced by Manufacturer X. Trace-back food investigations on those sausages were directed by the DGAL in November 2020. Biological food sampling was performed at different levels: (i) self-controls by Manufacturer X (1 sample per batch and environmental), (ii) reinforced self-controls by Manufacturer X (8 samples from Batch I), (iii) self-controls by Supermarket Chain 1 and (iv) official controls by the local food agri-food laboratories (Laboratoire Départemental d’Analyses (LDA)) (1–5 samples per batch for 12 batches, including Batch I, with no samples available from Batch II).
Additionally, a leftover food sample provided by one patient was sent to the LNR-ANSES for analysis. All analyses followed the NF EN ISO 6579–1 standards [18]. Salmonella isolates were serotyped according to the White-Kauffmann-Le Minor scheme [19]. Non-routine in-house quantitative real-time PCR (qPCR) assays targeting S. Bovismorbificans and S. Typhimurium/S. 1,4,[5],12:i:- were performed on the leftover food samples. Briefly, 1 ml of the pre-enrichment broths were DNA extracted using the InstaGene matrix following the recommendation of the manufacturer (Bio-Rad) and 5 µl of the DNA were tested by qPCR in a CFX apparatus (BioRad) (see Supplementary Table S1 for the primer and probe sequences used in this study).
The Salmonella food isolates were eventually sequenced by the LNR-ANSES using Illumina technology as described elsewhere [20]. Three tools available at the Center for Genomic Epidemiology (CGE) (http://genomicepidemiology.org) were used to analyse the genomes: SeqSero 1.2, MLST and ResFinder 2.1. These isolates were compared with the human isolates on EnteroBase (both SNP and cgMLST approaches).
Results
Epidemiological investigation
To determine the source of the outbreak, epidemiological investigations were conducted during the 3 months following the identification of the first genomic cluster, starting on 6 November 2020, and until the outbreak ended. Of the 33 cases of S. Bovismorbificans, 19 were children less than 13 years of age, median age was 10 years (range: 2 months–69 years), and 17 were female. SpF successfully interviewed 23 cases (or their parents); the remaining nine cases were not reachable (no contact information available or not answering the phone call). Symptom onset ranged from 22 September to 10 November 2020 (Figure 1) and seven patients required hospitalisation. Twenty-three cases reported grocery shopping at one specific supermarket chain; 22 cases indicated the consumption of dried pork sausages and 17 allegedly purchased the same Brand A. For 19 cases, the loyalty card number for Supermarket Chain 1 was provided, which confirmed the purchase for 15 cases and helped to identify unreported purchases by four additional cases.
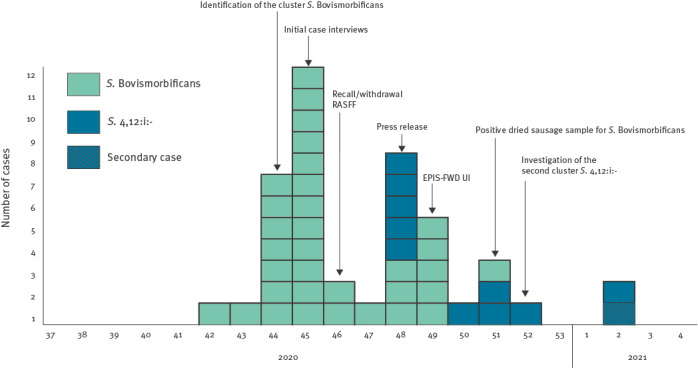
Figure 1.
Epidemic curve of S. Bovismorbificans ST142 HC5_243255 (n = 33) and S. 4,12:i:- ST34 HC5_198125 (n = 11) cases by week of identification at the FNRC-ESS, France, September 2020–January 2021
EPIS-FWD: Epidemic Intelligence Information System or Food and Waterborne Diseases; RASFF: Rapid Alert System for Food and Feed; S.: Salmonella, UI: urgent enquiry.
Secondary case refers to a 2-month-old infant who may have acquired the infection through cross contamination from another case in the household.
The second cluster, caused by S. 4,12:i:- ST34, had 11 cases with ages ranging from 2 months to 49 years (median: 11 years), seven of whom were male. Ten cases were interviewed; Case 11 was an infant aged 2 months, thus a secondary case. Three cases were hospitalised, and symptom onset ranged from 27 October to 30 November. One case reported a later date of onset, on 12 December (Figure 2). Nine cases reported the recent consumption of dried pork sausages and eight of them indicated Brand A, purchased in Supermarket Chain 1 stores. All nine patients provided the number of their loyalty card, and the purchase was thus confirmed for seven of them. There were no deaths associated to either outbreak.
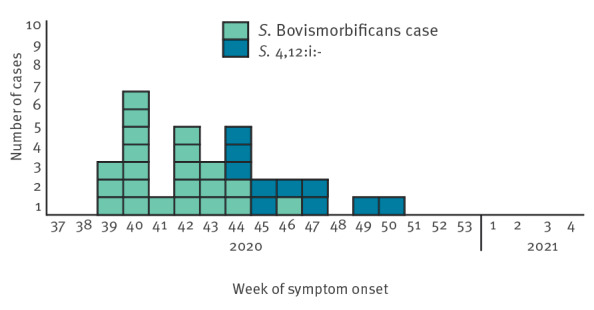
Figure 2.
Epidemic curve of S. Bovismorbificans ST142 HC5_243255 (n = 23a) and S. 4,12:i:- ST34 HC5_198125 (n = 10) interviewed cases by week of symptom onset, France, September–December 2020
S.: Salmonella.
a Date of symptom onset unknown for one case.
Human microbiological investigation
On 27 October 2020, the FNRC-ESS notified to SpF a cluster of eight S. Bovismorbificans ST142 isolates sharing a new HC5 profile (HC5_243255) (Figure 3), all sampled within a period of 12 days (22 September–5 October). By the end of 2020, a total of 33 isolates were identified, with isolation dates ranging between 22 September and 16 November. The isolates were inferred to be susceptible to all antibiotics.
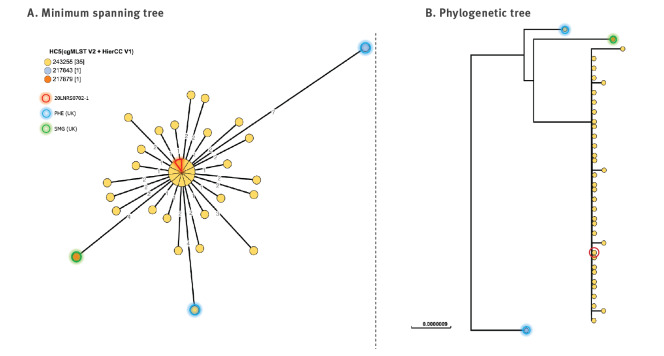
Figure 3.
Phylogenetic visualisation of the S. Bovismorbificans HC5_243255 cluster (n = 33) in relation to other ST142 genomes (n = 4) up to 17 February 2021, France
AD: allelic distance; cgMLST: core genome multilocus sequence typing; SNP: single nucleotide polymorphism.
The trees were constructed on EnteroBase (http://enterobase.warwick.ac.uk), selecting ST142 (n = 37) genomes clustering at less than 10 AD from the central node of the HC5_243255 cluster. Two genomes were uploaded by the United Kingdom (UK) Health Security Agency, appearing on the EnteroBase site as Public Health England (PHE; blue circles) and one genome was uploaded by the Scottish National Health System (NHS, SMG on EnteroBase), from the UK (green circle). The isolate from the food sample is 20LNRS0702–1 (red circle).
A. Minimum spanning tree on cgMLST by the EnteroBase ‘MSTree V2’ algorithm. Dots in the tree represent genomes. When genomes are at 0 AD, they form a bigger dot, where each genome is a sector in the dot. All HC5_243255 genomes are coloured in yellow, and that includes the 33 outbreak genomes – all at 0 to 3 alleles from one another – n = 1 genome from the food sample (outlined in red, in the centre of the main node), n = 1 genome from the UK (at 4 AD, circled in blue).
B. Phylogenetic tree constructed with the ‘create SNP project’ tool in EnteroBase, based on the 196 non-repetitive SNPs present in 95% or more of all the queried genomes. Each dot in the tree represents one genome. Colours correspond to different HC5, and HC5_243255 (to which the outbreak cluster belongs) is here represented in yellow.
Between 24 December 2020 and 8 January 2021, the FNRC-ESS identified another cluster of 11 cases of S. 4,12:i:- ST34 HC5_198125 (isolation dates ranging from 31 October–14 December) (Figure 4). They were genomically assigned to S. Typhimurium because of the presence of both fliC:i and fljB:1,2 flagellin genes in all the isolates. However, classical sero-agglutination performed on three selected isolates revealed that they were monophasic (i.e. fljB:1,2 was not expressed). Further analyses revealed that this was due to an insertion sequence that prevented fragment H (including hin and the fljB promoter) from inverting its genetic configuration and therefore, expressing fljB. The outbreak genetic configuration of invertible Fragment H scheme is shown in Supplementary Figure S1.
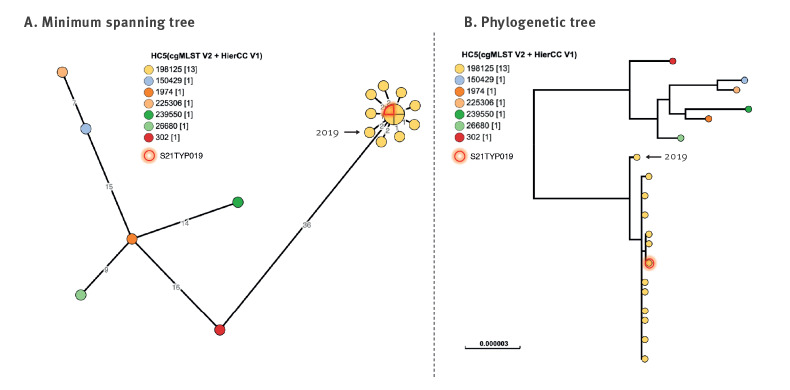
Figure 4.
Phylogenetic visualisation of the S. 4,12:i:- ST34 HC5_198125 cluster (n = 11) in relation to other ST34 HC50_2 genomes (n = 7) up to 18 March 2021, France
AD: allelic distance; cgMLST: core genome multilocus sequence typing; SNP: single nucleotide polymorphism.
The trees were constructed on EnteroBase (http://enterobase.warwick.ac.uk), selecting ST34 genomes (n = 19) clustering at less than 10 AD from the node of the HC5_198125 cluster. The isolate from food origin is S21TYP019 (circled in red).
A. Minimum spanning tree on cgMLST by the EnteroBase ‘MSTree V2’ algorithm. Dots in the tree represent genomes. When genomes are at 0 AD; they form a bigger dot, where each genome is a sector in the dot. All cluster members (yellow circles) are at 0 to 2 alleles from one another, which corresponds to HC5_198125. One genome clustering together with the outbreak genomes dated from 2019 (arrows) and was therefore not considered as part of the outbreak.
B. Phylogenetic tree constructed with the ‘create SNP project’ tool in EnteroBase, based on the 125 non-repetitive SNPs present in 95% or more of all the queried genomes. Each dot in the tree represents one genome. Colours correspond to different HC5, and the outbreak cluster (HC5_198125) is here represented as yellow. The 2019 isolate was well separated from the 2020 outbreak cluster, while the food genome S21TYP019 clustered with the genomes of human origin in 2020.
Food product trace-back investigation
Trace-back investigations on Batch I (suspected batch for cases belonging to the S. Bovismorbificans outbreak cluster) performed in November 2020 by the manufacturer (8 samples) and by the LDA (3 samples) were negative for Salmonella spp.
However, dried pork sausage leftover samples belonging to Batch I provided by one of the cases were sent to the LNR-ANSES. S. Bovismorbificans was isolated and identified by the standard methods, including sero-agglutination, and detected directly in the food sample by an in-house qPCR targeting serotype Bovismorbificans (Ct values: ca 20). Supplementary Figure S2 shows the qPCR analysis of the food sample.
In January 2021, following the identification of the second human outbreak cluster caused by S. 4,12:i:-, the epidemiological and food investigations uncovered a new batch of incriminated products (Batch II) from the same manufacturer. Reinforced self-control analyses were then performed by Manufacturer X (n = 5 samples) and S. 4,12:i:- was isolated from one sample and identified by classical sero-agglutination. No other Salmonella serotypes were found.
A new qPCR assay targeting serotype Typhimurium/1,4,[5],12:i:- was then performed on the remaining DNA extracts from the leftover samples from Batch I that had been previously positive to S. Bovismorbificans, and the assay identified S. Typhimurium/1,4,[5],12:i:- (Ct value: ca 30) (Supplementary Figure S2). However, by this time, there were no samples available for culture to determine whether S. 4,12:i:- ST34 HC5_198125 could be also isolated in the same samples from Batch I in which S. Bovismorbificans was previously isolated.
Supplementary Figure S2 shows the qPCR analysis of the food sample, where four genomic targets were aimed: A. S. enterica, the species to which both outbreak serotypes belong, B. serotype Bovismorbificans and C. and D. two different targets for serotype Typhimurium/4,[5],12:i:-. While qPCR for S. enterica and serotype Bovismorbificans had Ct values of ca 20 cycles, the qPCR for S. Typhimurium/1,4,[5],12:i:- had Ct values of ca 30 cycles, which means that there was less amount of S. Typhimurium/1,4,[5],12:i:-DNA than of S. Bovismorbificans in the sample.
WGS allowed the confirmation of the identity between human and food Salmonella isolates. The isolate first obtained from the patient’s leftover samples (Batch I) was S. Bovismorbificans, ST142, HC5_243255, clustering together with the human isolates (Figure 3). The second food isolate, obtained during the Manufacturer X’s analyses in January (Batch II), was characterised as S. 4,12:i:- ST34 and HC5_198125, clustering together with the second epidemic outbreak isolates detected in humans (Figure 4).
Outbreak control measures
A first recall and withdrawal were issued on 13 November 2020 by Supermarket Chain 1, 2 weeks after the initial alert by the FNRC-ESS, and 1 week after the first SpF questionnaires [21]. The recall implied two types of Brand A dried pork sausages. On 16 November 2020, as a complement and preventive measure, two additional batches of sausages from Brand B and Brand C produced with the same raw pork meat as Brand A were recalled and withdrawn. They were sold by Supermarket Chains 2 and 3. A third recall and withdrawal were issued in November 2020 for sandwiches of Brand D made with sausages from Manufacturer X, and sold in Supermarket Chains 4, 5, 6, 7 and 8. All suspected batches of dried pork sausages from Manufacturer X with production dates up to 8 December 2020 were removed from the market.
A Rapid Alert System for Food and Feed [22] (RASFF: 2020.5038) alert was posted by the French authorities on 16 November 2020, at the same time as the second recall and withdrawal were issued in France. The contaminated dried pork sausages had been distributed in other five countries: Belgium, Luxemburg, Poland, Portugal and Slovenia.
On 26 November 2020, the French Ministry for agriculture and food delivered a press release reporting these recalls and withdrawals.
An urgent inquiry (UI-688, later renamed as 2020-FWD-00065) on the Epidemic Intelligence Information System for Food and Waterborne Diseases and Zoonoses (EPIS-FWD, now called EpiPulse) platform operated by the European Centre for Disease Prevention and Control (ECDC) was issued by SpF on 1 December 2020 concerning the S. Bovismorbificans outbreak. Germany, Ireland, Norway, and the UK responded with one case each, at 4 to 5 AD from the representative genome provided by the FNRC-ESS. SNP analysis and epidemiological data excluded them from being part of the cluster.
No additional recalls and withdrawals were initiated following the investigations of the second cluster in end of December 2020 and January 2021, because the products were no longer on the market. No related cases were identified outside of France afterwards. Three S. Bovismorbificans genomes genetically close to the epidemiological outbreak were identified in EnteroBase from the UK, but no epidemiological link was established with the French outbreak. The last bacterial isolate of human origin for the S. Bovismorbificans cluster dates to 16 November, the day of the second and largest product withdrawal and recall. The last bacterial isolate of human origin for the S. 4,12:i:- cluster dates to 14 December. Given the lack of new cases in January 2021, the investigations were closed by the end of the month.
As no other manufacturing plants supplied ingredients from common origin were affected, we speculate with a local contamination at Manufacturer X. However, no investigations upstream were performed, as they are not required, and the staff resources were scarce at that time.
Discussion
We describe a nationwide outbreak of salmonellosis involving 44 cases in two microbiological clusters by two serotypes: 33 cases with S. Bovismorbificans ST142 and 11 cases with the monophasic variant of S. Typhimurium ST34. Both clusters were associated with the consumption of dried pork sausages produced by the same manufacturer between September and November 2020. Epidemiological investigations pointed to the consumption of Brand A sausages marketed by Supermarket Chain 1. Moreover, the investigation of the supermarket loyalty cards reinforced this association by recording the purchase of the suspected product a few days before the date of symptom onset in most records.
Control measures were implemented following the epidemiological investigations of the S. Bovismorbificans cluster with three recalls/withdrawals in November 2020. The S. 4,12:i:- cluster was identified in late December, but symptom onset of most cases occurred by 30 November (except for one case in December). Following the recalls and withdrawals of the suspected products, detection in human cases declined rapidly. International health agencies were informed of the first outbreak cluster (S. Bovismorbificans) through RASFF and EpiPulse messages because of the exportation of the product in other European countries. No cases belonging to the first cluster were reported outside France, and no genomes belonging to the second cluster were identified on EnteroBase for any other country.
S. Bovismorbificans was only isolated from a food sample provided by a patient. Although post-purchase contamination by the patient might be possible, the epidemiological data and the absence of new cases following the recalls and withdrawals strongly supported the hypothesis that the product was contaminated before the sale.
S. Bovismorbificans is not an unusual serotype infecting humans in France, with a mean of 57 cases per year (median: 51 cases) for the past 10 years. The outbreak here presented reflects an increase of 40% over the average in 2020. This serotype has been often involved in human outbreaks in recent years, mostly associated to the consumption of vegetables like alfalfa sprouts, salads, hummus or tahini in Australia, Finland, Switzerland, Germany and the United States [7-11]. However, the association of S. Bovismorbificans with pork products has been described previously, in the Netherlands in 2017 [13]. In that outbreak, trace-back investigations led to a Belgian meat processor. Two more outbreaks caused by S. Bovismorbificans have occurred in France in 2019 and 2020: an outbreak of 11 cases in which the source could not be identified, at nine AD from the present outbreak (data not shown), and a cluster of 14 cases associated to the consumption of uncooked horse meat, at more than 50 AD from the present cluster [12]. The ANSES Salmonella network, as part of the French Laboratory for Food Safety, has collected 147 S. Bovismorbificans isolates in France since 2010, principally obtained from pork meat (n = 32 strains), turkey (n = 23 strains), chicken (n = 16 strains) and horse meat (n = 6) (https://www.ANSES.fr/fr/content/inventaire-des-salmonella-dorigine-non-humaine). S. Bovismorbificans should be considered as an important hazard in meat, and measures should be reinforced to control its presence, as is done with other serotypes more frequently identified in association with meat consumption.
Outbreaks of salmonellosis in humans caused by S. 1,4,[5],12:i:- ST34 are frequent (over 6,400 cases in 2018 in Europe) and in France they are often linked to the consumption of contaminated raw dried pork meat (39.6%) [2]. The Commission Regulation (EC) No 218/2014 provides information on the mandatory monitoring of Salmonella in pork at the production chain [23]. Thereby, S. Typhimurium and its monophasic variant 1,4,[5],12:i:- are found as the third and second most frequent serotype of Salmonella isolated from the swine sector (animals and food), respectively [24]. However, the isolation of the pathogen in food is not always possible, presumably because of the meat curing (salting and drying) process. Our findings suggest that perhaps microbiological standard procedures should be enhanced by implementing checks at earlier steps of the food processing [25,26].
Outbreaks caused by food simultaneously contaminated by several serotypes of Salmonella have been reported, but mostly linked to vegetables. In 2008, Finland reported a nationwide outbreak with up to 106 patients infected by either serotypes Newport, Reading or both, in association to the consumption of contaminated iceberg lettuce [27]. In 2009, an outbreak of nine cases infected by serotypes Schwarzengrund and Typhimurium, was linked to a contaminated potato salad served at a reception in the United States [28]. In 2012, a contaminated tahini paste imported from Turkey caused 27 cases of salmonellosis in New Zealand, with three serotypes of Salmonella involved: Montevideo, Maastricht and Mbandaka [29]. A large international outbreak with 94 cases infected in the United States and Canada between 2013 and 2014, implied four different serotypes of Salmonella (Newport, Hartford, Oranienburg and Saintpaul) contaminating sprouted chia seed powder distributed by several brands in both countries [30]. More recently, in 2019, an international outbreak affecting more than 100 cases in five countries was associated to the consumption of sesame-based products imported from Syria, contaminated by six different serotypes of Salmonella [31].
In this outbreak, although it was not possible to isolate both Salmonella serotypes from one single food or human sample, a qPCR identified both serotypes in the same food sample. This suggested that the food products could have been contaminated at different rates by both serotypes, simultaneously, escaping the standard procedures of microbiology laboratories for the identification of multi-strain contaminations. At this end, the detection of a positive Salmonella signal by qPCR in a pre-enrichment sample may be a powerful approach for fast detection and investigation [32-34]. Nevertheless, the method would need to be standardised and a confirmation should always be performed by strain isolation, identification, and genomic characterisation.
Our study has some limitations. The standard laboratory methods for identification of Salmonella in clinical microbiology laboratories do not include the testing of several colonies per sample [28], and that could explain the low numbers of multiple serotype infections reported in the literature. However, it seems logical to expect that processed food may be contaminated by a mixture of Salmonella strains more frequently than it is observed. In the outbreak presented here, two different serotypes of Salmonella were obtained from two different batches of dried pork sausages produced by the same manufacturer during a short period of time. Date of symptom onset and date of purchase of the product in the two clusters overlapped, and both serotypes were detected in the sausage leftovers kept by one case. Unfortunately, it was not possible to isolate the two serotypes from any human or food samples, which is a clear limitation in our study. Similarly, nine cases could not be reached for interviews during the epidemiological investigation, although the 24 successful interviews gave sufficient evidence to initiate the trace-back food investigations.
Conclusions
We present here two concurrent Salmonella enterica outbreaks linked to the consumption of dried pork sausages produced during a short period of time by the same manufacturer: the former caused by S. Bovismorbificans and the latter by the monophasic variant of S. Typhimurium, 4,12:i:-. However, molecular methods revealed a dual contamination of a single food sample analysed. Multi-strain outbreaks may go undetected by the current bacteriological detection approaches. We believe that standard laboratory protocols, both at the clinical and the food control levels, should include procedures to enhance the detection of mixed Salmonella serotypes contaminations. Moreover, our report highlights the utility of serotype-specific qPCR assays for investigating food-borne outbreaks.
Ethical statement
The data used for the outbreak investigation were collected as part of the infectious disease surveillance and control, defined by national legislation and ethical approval was therefore not necessary.
Data availability
Raw reads of the 34 genomes of S. Bovismorbificans and 12 genomes of S. Typhimurium from human and food origin in this study were submitted to the European Nucleotide Archive under project numbers PRJEB41645 and PRJEB49565.
Acknowledgements
The authors would like to thank the FNRC-ESS team, particularly Laetitia Fabre, Véronique Guibert, Magali Ravel and Estelle Serre, for technical assistance in the verification of genomic and phenotypic typing of the ST34 HC5_198125 cluster strains.
We also thank Maï-Lan Tran from the IdentyPath Genomics Platform of ANSES for technical assistance in the implementation of the PCR tests used in that study.
We also thank the United Kingdom Health Security Agency (UKSHA), former Public Health England (PHE) and the National Health System (NHS) Greater Glasgow and Clyde, from the United Kingdom, for placing on EnteroBase the three genomes that were used in Figure 3.
Finally, we would like to thank Karine Gratia from the Mission des Urgences Sanitaires at the DGAL for her help in completing food trace-back information.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: MPG: conceived and drafted the manuscript and performed the microbial analyses on the human bacterial isolates. NF: performed the epidemiological investigations and contributed to the drafting of the manuscript, and critically revised the manuscript. LB: performed the microbial analyses on the food bacterial isolates and contributed to the drafting of the manuscript. SL: performed microbiological analyses and critically revised the manuscript. MC: critically revised the manuscript. CG: contributed to the trace back investigations and critically revised the manuscript. SCS: performed genetic analyses of the food bacterial isolates by qPCR and participated in the drafting of the manuscript. PF: performed genetic analyses of the food bacterial isolates by qPCR and revised the manuscript. AP: coordinated the trace back investigations and critically revised the manuscript. AB: critically revised the manuscript. NJS: coordinated the epidemiological investigations and critically revised the manuscript. FXW: oversaw the genomic analyses and critically revised the manuscript.
References
- 1.World Health Organization (WHO). WHO Estimates of the Global Burden of Foodborne Diseases: Food-borne Disease Burden Epidemiology Reference Group 2007-2015. Geneva: WHO; 2015. Available from: https://www.who.int/publications/i/item/9789241565165
- 2. European Food Safety Authority. European Centre for Disease Prevention and Control . The European Union One Health 2019 Zoonoses Report. EFSA J. 2021;19(2):e06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Cauteren D, Le Strat Y, Sommen C, Bruyand M, Tourdjman M, Da Silva NJ, et al. Estimated annual numbers of foodborne pathogen-associated illnesses, hospitalizations, and deaths, France, 2008-2013. Emerg Infect Dis. 2017;23(9):1486-92. 10.3201/eid2309.170081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Cauteren D, Le Strat Y, Sommen C, Bruyand M, Tourdjman M, Jourdan-Da Silva N, et al. Estimation de la morbidité et de la mortalité liées aux infections d’origine alimentaire en France métropolitaine, 2008-2013. [Estimates of food-related morbidity and mortality in metropolitan France, 2008-2013]. Bull Epidemiol Hebd (Paris). 2018;(1):2-10. French. Available from: http://beh.santepubliquefrance.fr/beh/2018/1/2018_1_1.html [Google Scholar]
- 5. Bonardi S. Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol Infect. 2017;145(8):1513-26. 10.1017/S095026881700036X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DE Knegt LV, Pires SM, Hald T. Attributing foodborne salmonellosis in humans to animal reservoirs in the European Union using a multi-country stochastic model. Epidemiol Infect. 2015;143(6):1175-86. 10.1017/S0950268814001903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention (CDC) . Multistate outbreak of Salmonella serotype Bovismorbificans infections associated with hummus and tahini--United States, 2011. MMWR Morb Mortal Wkly Rep. 2012;61(46):944-7. [PubMed] [Google Scholar]
- 8. Knoblauch AM, Bratschi MW, Zuske MK, Althaus D, Stephan R, Hächler H, et al. Cross-border outbreak of Salmonella enterica ssp. enterica serovar Bovismorbificans: multiple approaches for an outbreak investigation in Germany and Switzerland. Swiss Med Wkly. 2015;145:w14182. 10.4414/smw.2015.14182 [DOI] [PubMed] [Google Scholar]
- 9. Oberreuter H, Rau J. Artificial neural network-assisted Fourier transform infrared spectroscopy for differentiation of Salmonella serogroups and its application on epidemiological tracing of Salmonella Bovismorbificans outbreak isolates from fresh sprouts. FEMS Microbiol Lett. 2019;366(15):fnz193. 10.1093/femsle/fnz193 [DOI] [PubMed] [Google Scholar]
- 10. Rimhanen-Finne R, Niskanen T, Lienemann T, Johansson T, Sjöman M, Korhonen T, et al. A nationwide outbreak of Salmonella Bovismorbificans associated with sprouted alfalfa seeds in Finland, 2009. Zoonoses Public Health. 2011;58(8):589-96. 10.1111/j.1863-2378.2011.01408.x [DOI] [PubMed] [Google Scholar]
- 11. Stafford RJ, McCall BJ, Neill AS, Leon DS, Dorricott GJ, Towner CD, et al. A statewide outbreak of Salmonella Bovismorbificans phage type 32 infection in Queensland. Commun Dis Intell Q Rep. 2002;26(4):568-73. [PubMed] [Google Scholar]
- 12.Santé publique France. Épidémie de salmonellose à Salmonella sérotype Bovismorbificans liée à la consommation de viande chevaline, Hauts‐de‐France. [Outbreak of Salmonella serotype Bovismorbificans linked to horse meat consumption]. Saint‐Maurice: Santé publique France; 2021. French. Available from: https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-infectieuses-d-origine-alimentaire/salmonellose/documents/rapport-synthese/epidemie-de-salmonelloses-a-salmonella-serotype-bovismorbificans-liee-a-la-consommation-de-viande-chevaline
- 13. Brandwagt D, van den Wijngaard C, Tulen AD, Mulder AC, Hofhuis A, Jacobs R, et al. Outbreak of Salmonella Bovismorbificans associated with the consumption of uncooked ham products, the Netherlands, 2016 to 2017. Euro Surveill. 2018;23(1):17-00335. 10.2807/1560-7917.ES.2018.23.1.17-00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Z, Charlesworth J, Achtman M. HierCC: A multi-level clustering scheme for population assignments based on core genome MLST. Bioinformatics. 2021;37(20): 3645–6. Epub ahead of print. 10.1093/bioinformatics/btab234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8(6):e1002776. 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14(4):e1007261. 10.1371/journal.pgen.1007261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491-500. 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Organization for Standardization (ISO). NF EN ISO 6579-1/A1 Mars 2020. Microbiologie de la chaîne alimentaire - Méthode horizontale pour la recherche, le dénombrement et le sérotypage des Salmonella - Partie 1: recherche des Salmonella spp. Available from: https://www.iso.org/standard/56712.html
- 19.Grimont PA, Weill FX. Antigenic formulae of the Salmonella serovars. Paris: WHO collaborating centre for reference and research on Salmonella; 2007. Available from https://www.pasteur.fr/sites/default/files/veng_0.pdf
- 20. Sévellec Y, Granier SA, Radomski N, Felten A, Le Hello S, Feurer C, et al. Complete genome sequence of Salmonella enterica subsp. enterica serotype derby, associated with the pork sector in France. Microbiol Resour Announc. 2018;7(12):e01027-18. 10.1128/MRA.01027-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ministère de l’Agriculture et de l’Alimentation. Communiqué de presse. Retrait et rappel de saucisses sèches et de rosettes tranchées contaminées par des salmonelles. [Removal and recall of salmonella-contaminated dry sausages and sliced rosettes]. Paris: Ministère de l’Agriculture et de l’Alimentation; 26 Nov 2020. French. Available from: https://agriculture.gouv.fr/alerte-sanitaire-retrait-et-rappel-de-saucisses-seches-et-de-rosettes-tranchees-contaminees-par-des
- 22.European Commission. Rapid Alert System for Food and Feed (RASFF) Window: Notification 2020.5038. Dry sausage -Salmonella Bovismorbivicans. Brussels: European Commission; 8 Dec 2020. Available from: https://webgate.ec.europa.eu/rasff-window/screen/notification/450578
- 23.European Commission. Commission Regulation (EU) No 218/2014 of 7 March 2014 amending Annexes to Regulations (EC) No 853/2004 and (EC) No 854/2004 of the European Parliament and of the Council and Commission Regulation (EC) No 2074/2005. Text with EEA relevance. Official Journal of the European Union. Luxembourg: Publications Office of the European Union. 8.3.2014: L 69, p. 95–98. Available from: http://data.europa.eu/eli/reg/2014/218/oj
- 24. Delannoy S, Cadel-Six S, Bonifait L, Tran ML, Cherchame E, Baugé L, et al. Closed genome sequence of a Salmonella enterica serovar Bovismorbificans strain isolated from dried pork sausage associated with an outbreak in France. Microbiol Resour Announc. 2021;10(40):e0066221. 10.1128/MRA.00662-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bone A, Noel H, Le Hello S, Pihier N, Danan C, Raguenaud ME, et al. Nationwide outbreak of Salmonella enterica serotype 4,12:i:- infections in France, linked to dried pork sausage, March-May 2010. Euro Surveill. 2010;15(24):19592. 10.2807/ese.15.24.19592-en [DOI] [PubMed] [Google Scholar]
- 26. Gossner CM, van Cauteren D, Le Hello S, Weill FX, Terrien E, Tessier S, et al. Nationwide outbreak of Salmonella enterica serotype 4,[5],12:i:- infection associated with consumption of dried pork sausage, France, November to December 2011. Euro Surveill. 2012;17(5):20071. 10.2807/ese.17.05.20071-en [DOI] [PubMed] [Google Scholar]
- 27. Lienemann T, Niskanen T, Guedes S, Siitonen A, Kuusi M, Rimhanen-Finne R. Iceberg lettuce as suggested source of a nationwide outbreak caused by two Salmonella serotypes, Newport and Reading, in Finland in 2008. J Food Prot. 2011;74(6):1035-40. 10.4315/0362-028X.JFP-10-455 [DOI] [PubMed] [Google Scholar]
- 28. Centers for Disease Control and Prevention (CDC) . Multiple-serotype Salmonella gastroenteritis outbreak after a reception --- Connecticut, 2009. MMWR Morb Mortal Wkly Rep. 2010;59(34):1093-7. [PubMed] [Google Scholar]
- 29. Paine S, Thornley C, Wilson M, Dufour M, Sexton K, Miller J, et al. An outbreak of multiple serotypes of Salmonella in New Zealand linked to consumption of contaminated tahini imported from Turkey. Foodborne Pathog Dis. 2014;11(11):887-92. 10.1089/fpd.2014.1773 [DOI] [PubMed] [Google Scholar]
- 30. Harvey RR, Heiman Marshall KE, Burnworth L, Hamel M, Tataryn J, Cutler J, et al. International outbreak of multiple Salmonella serotype infections linked to sprouted chia seed powder - USA and Canada, 2013-2014. Epidemiol Infect. 2017;145(8):1535-44. 10.1017/S0950268817000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. European Food Safety Authority . Multi-country outbreak of multiple Salmonella enterica serotypes linked to imported sesame-based products. EFSA J. 2021;18(10):6922E. 10.2903/sp.efsa.2021.EN-6922 [DOI] [Google Scholar]
- 32.Blackstone GM, Nordstrom JL, DePaola A. 2005. A same day detection method for Salmonella species in shrimp using real time PCR, poster P-079. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 33. Bugarel M, Tudor A, Loneragan GH, Nightingale KK. Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J Microbiol Methods. 2017;134:14-20. 10.1016/j.mimet.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 34.International Organization for Standardization (ISO). Microbiology of the food chain – Horizontal method for the detection, enumeration and serotyping of Salmonella. Part 4: Identification of monophasic Salmonella Typhimurium (1,4,[5],12,i:-) by polymerase chain reaction (PCR). Draft ISO/TS CD 6579-4. Geneva: ISO; 2021. Available from: https://www.iso.org/standard/56712.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.